- 1Department of Comparative Biomedical Sciences, College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
- 2Department of Clinical Sciences, College of Veterinary Medicine, Mississippi State University, Mississippi State, MS, United States
- 3Department of Poultry Science, Mississippi State University, Mississippi State, MS, United States
Listeria monocytogenes, the causative agent of listeriosis, displays a lifestyle ranging from saprophytes in the soil to pathogenic as a facultative intracellular parasite in host cells. In the current study, a random transposon (Tn) insertion library was constructed in L. monocytogenes strain F2365 and screened to identify genes and pathways affecting in vitro growth and fitness in minimal medium (MM) containing different single carbohydrate as the sole carbon source. About 2,000 Tn-mutants were screened for impaired growth in MM with one of the following carbon sources: glucose, fructose, mannose, mannitol, sucrose, glycerol, and glucose 6-phosphate (G6P). Impaired or abolished growth of L. monocytogenes was observed for twenty-one Tn-mutants with disruptions in genes encoding purine biosynthesis enzymes (purL, purC, purA, and purM), pyrimidine biosynthesis proteins (pyrE and pyrC), ATP synthase (atpI and atpD2), branched-chain fatty acids (BCFA) synthesis enzyme (bkdA1), a putative lipoprotein (LMOF2365_2387 described as LP2387), dUTPase family protein (dUTPase), and two hypothetical proteins. All Tn-mutants, except the atpD2 mutant, grew as efficiently as wild-type strain in a nutrient rich media. The virulence of twenty-one Tn-mutants was assessed in mice at 72 h following intravenous (IV) infection. The most attenuated mutants had Tn insertions in purA, hypothetical protein (LMOf2365_0064 described as HP64), bkdA1, dUTPase, LP2387, and atpD2, confirming the important role of these genes in pathogenesis. Six Tn-mutants were then tested for ability to replicate intracellularly in murine macrophage J774.1 cells. Significant intracellular growth defects were observed in two Tn-mutants with insertions in purA and HP64 genes, suggesting that an intact purine biosynthesis pathway is important for intracellular growth of L. monocytogens. These findings may not be fully generalized to all of L. monocytogenes strains due to their genetic diversity. In conclusion, Tn-mutagenesis identified that biosynthesis of purines, pyrimidines, ATP, and BCFA are important for L. monocytogens pathogenesis. Purine and pyrimidine auxotrophs play an important role in the pathogenicity in other bacterial pathogens, but our study also revealed new proteins essential for both growth in MM and L. monocytogenes strain F2365 virulence.
Introduction
Listeria monocytogenes is a facultative intracellular pathogen that is responsible for listeriosis, a severe food-borne illness with a high mortality rate in immunocompromised people, children, older adults, and pregnant women (in whom it causes miscarriages, stillbirths or infection of newborns) (Vázquez-Boland et al., 2001; Lecuit, 2020). The route of infection in humans is usually through consumption of L. monocytogenes-contaminated food products, both processed and unprocessed, and the bacterium is frequently found and isolated from processed and ready-to-eat (RTE) food products (Radoshevich and Cossart, 2018). An increasing number of food product recalls have been issued recently due to adulteration with L. monocytogenes including ice cream, lettuce and/or packaged salads, fully cooked chicken, and deli meat (CDC, 2022). According to the Centers for Disease Control, about 1,600 people get sick from L. monocytogenes each year in the United States alone, with a mortality rate of 15–20%, making it one of the deadliest foodborne pathogens (CDC, 2011). For example, L. monocytogenes was responsible for the deaths of 10 people in Texas in 2010 due to consumption of chopped celery (Gaul et al., 2013) and 33 people in 28 different states in the US due to consumption of contaminated melons (CDC, 2016).
L. monocytogenes is regarded as a model species capable of growth in the environment, soil, food processing environments, and as a facultative intracellular pathogen of mammals (Anaya-Sanchez et al., 2021). A successful transition of L. monocytogenes from the extracellular to the intracellular environment requires sensing the surrounding environment and activating relevant metabolic pathways in a timely manner, especially with respect to carbon metabolism and utilization of various nutrients.
Upon encountering a host, L. monocytogenes expresses virulence factors responsible for host cell entry, escape from the entry vacuole, intracellular replication in the cytosol, and spread to systemic sites (Cahoon and Freitag, 2014). In the entry phagosomal vacuoles, intracellular L. monocytogenes have limited access to the host cells’ nutrients, while L. monocytogenes replicating in the cytosolic compartment have free access to the host cell nutrients (Chen and Pensinger, 2017). Thus, metabolic adaptations and utilization of varying externally available carbon sources and host-derived metabolites are essential for survival of L. monocytogenes and establishment of infection.
Much is known about virulence factors that are primarily regulated by transcriptional regulator PrfA (positive regulatory factor A) (Radoshevich and Cossart, 2018). However, little is known about L. monocytogenes metabolic adaptations and utilization of nutrients during listerial pathogenesis. The genome of L. monocytogenes reflects its ability to acquire and metabolize a range of different carbon sources, but previous isotopic labeling studies demonstrated that its intracellular metabolism relies mostly on glycerol and glucose 6-phosphate (G6P), which are generated in host cells (Eylert et al., 2008; Gillmaier et al., 2012). Glycerol is the primary carbon substrate for generation of energy and synthesis of some amino acids and fatty acids. G6P is an important carbon source for synthesis of sugar components essential for biosynthesis of the cell envelope and nucleotides (Grubmüller et al., 2014). A previous study showed that L. monocytogenes can utilize glucose-1-phosphate, G6P, fructose-6-phosphate, and mannose-6-phosphate as carbon sources and the uptake of these sugars is mediate by hexose phosphate transporter (Hpt), which is tightly controlled by PrfA (Ripio et al., 1997; Chico-Calero et al., 2002).
Carbon utilization and metabolism have been linked to pathogenicity in L. monocytogenes (Joseph et al., 2008; Muchaamba et al., 2019). Indeed, the availability of specific metabolites and sugars control the transcription, translation, and activation of PrfA and the expression of PrfA-dependent virulence genes. The objective of this study was to identify bacterial genes required for efficient growth and proliferation under limited nutrient conditions by screening for transposon (Tn) mutants that have a defect in growth in defined MM with different carbohydrates (glucose, fructose, mannose, mannitol, sucrose, and G6P, and glycerol, a non-carbohydrate carbon source. Tn-mutagenesis is a high-throughput technique used to identify determinants in a bacterial genome that are required for bacterial fitness (Fu et al., 2013). Using this method, we identified 21 Tn-mutants that have defective growth in MM with a sole carbon source. Further, some of these genes were evaluated for L. monocytogenes virulence and intracellular replication.
Materials and methods
Bacterial strains, media, and growth conditions
Escherichia coli (NF-E1130) harboring the pMC38 plasmid (Dr. Daniel Portnoy, UCLA) was propagated at 37°C in Luria-Bertani (LB) broth (Cao et al., 2007). L. monocytogenes wild-type F2365 strain (serotype 4b) was used as the parent strain for Tn mutagenesis (Nelson et al., 2004). The virulent F2365 strain was initially identified during a listeriosis outbreak in California in 1985, this outbreak was one of the deadliest foodborne outbreaks in the United States and serovar 4b represents the majority of the outbreak isolates (Melo et al., 2015). L. monocytogenes strains were grown at 37°C under aerobic conditions with shaking in brain heart infusion (BHI) broth or in chemically-defined minimal medium (MM) (Premaratne et al., 1991). For Tn-mutant screening, MM containing L-glutamine (4 mM) as nitrogen source and one of the following carbon sources was used: glucose (55.5 mM); fructose (25 mM); sucrose (125 mM); G6P (30 mM); mannose (20 mM); glycerol (41 mM); and maltose (30 mM). When appropriate, erythromycin (10 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml) was supplemented to the MM.
Cell culture and media
Mouse fibroblast L cells (American Type Culture Collection, Manassas, Virginia) were propagated in ATCC-formulated Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The murine macrophage cell line J774.1 was grown in DMEM supplemented with 10% FBS. Both the cell lines were maintained at 37°C and 5% CO2.
Transposon mutagenesis
Transposon mutagenesis was performed with a mariner-based transposon pMC38 delivery vector following previously published methods (Cao et al., 2007; Azizoglu et al., 2014). Briefly, the electro-competent L. monocytogenes F2365 strain was transformed with pMC38 and selected on BHI agar supplemented with erythromycin and incubated at 30°C. Transformant positive colonies were grown overnight in BHI broth supplemented with kanamycin and erythromycin. Then the culture was diluted into fresh BHI medium supplemented with erythromycin and grown at 30°C for 1 hour. The temperature was increased to 42°C for ∼6 h until an OD600 of ∼0.5 was reached. These cultures were then diluted and spread on BHI agar with erythromycin and incubated at 42°C for transposon integration. Individual colonies from the Tn library were separately examined for kanamycin sensitivity to test for plasmid loss. Colonies resistant to only erythromycin were chosen for library construction and screening. For long-term storage before any phenotypic screening, approximately 2,000 Tn-insertion mutants were picked and inoculated in 96-well plates containing BHI broth (180 μl) and grown overnight at 37°C to an OD600 of approximately 1.0. Then glycerol (20 μl) was added to each well, and plates were stored at −80°C. L. monocytogenes wild-type was included in each plate as a positive control.
Screening of transposon-mutants for growth-deficiency in defined minimal medium
For growth analysis in MM containing a single carbon source, 10 μl of Tn-insertion mutants (OD600 ≈1.0) from frozen libraries were transferred in duplicate into 96-well plates containing 90 μl of MM supplemented with one of the following carbon sources: glucose, mannose, fructose, sucrose, maltose, glycerol, or G6P. Following inoculation, 96-well plates were covered with Breathe-Easy film (Diversified Biotech) and incubated at 37°C in a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek, Winooski, VT, United States). The OD600 for each Tn-mutant was recorded every hour for 48 h and compared to the OD600 of L. monocytogenes wild-type. A total of 21 Tn-mutants with reduced growth in 96-well plates containing MM were considered candidates and tested individually using culture tubes (four replicates) for further validation. Briefly, L. monocytogenes wild-type and Tn-mutants from overnight cultures (OD600 ≈1.0) were harvested by centrifugation and resuspended in PBS. After washing with PBS, bacterial suspension was used to inoculate 5 ml of MM with the respective carbon source at 1:100 dilution. The culture tubes were incubated at 37°C and the growth of each strain was determined by measuring OD600 after 48–72 h of incubation. Tn-mutants that grew on BHI but showed reduced or no growth in MM containing a single carbohydrate were selected for further analysis. For growth kinetics in BHI, overnight cultures of wild-type and Tn-mutants were normalized based on optical density at 600 nm (OD600) and used to inoculate fresh BHI broth at 1:100 dilution in a 24-well plate. Cytation 5 was used to monitor the growth by measuring OD600 every hour for 24-h. The growth assays were conducted in three independent experiments, and each experiment was run with four replicates.
Determination of transposon-insertion sites
Transposon insertion sites of the mutants were determined as previously described (Cao et al., 2007; Azizoglu et al., 2014). Briefly, chromosomal DNA was isolated from the Tn-insertion mutants and used to amplify the fragments flanking the transposon through a nested arbitrary PCR reaction. The list of primers used in nested PCR are listed in Table 1. The 1st PCR amplification of the mutant genomic DNA was a touch-up PCR performed using two primer sets Marq207/Marq255 (PCR-1) and Marq207/Marq269 (PCR-2) using reaction conditions: (i) 95°C for 3 min; (ii) 15 cycles of 95°C for 30 s, 35°C for 30 s (increase 1°C every cycle), and 72°C for 3 min; and (iii) 15 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 2 min. The products of the first reaction were used as template for a second PCR reaction with the following primer sets: Marq208/Marq256 for amplification of products from PCR-1 and Marq208/Marq270 for amplification of products from PCR-2. Standard reaction conditions were used for the second PCR: (i) 95°C for 3 min; (ii) 40°C cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 2 min; and (iii) 72°C for 5 min. To remove unincorporated nucleotides and primers prior to sequencing, PCR products were cleaned with ExoSAP-IT (Thermofisher) according to manufacturer instructions. The samples from PCR-1 were sequenced with Marq257, and PCR-2 were sequenced with Marq271. Sequence results were searched with BLAST analysis against the whole genome of L. monocytogenes strain F2365 (AE017262.2) from NBCI (threshold was ≥ 100% identity and E-value of 0).
Southern blot hybridization
Southern blot hybridization for six Tn-mutants was performed by Celplor, a molecular biology CRO company1 to confirm the accuracy of the mapping methodology. Genomic DNA of mutants was digested with EcoRI (New England Biolabs, Beverly, MA, USA) overnight, resolved on agarose gels, and transferred to a nylon membrane. A 400 bp PCR product generated using primers Marq206/254 and pMC38 was used as a probe for the hybridization to locate the inserted DNA in targeted disrupted mutants.
Mutant virulence
All animal experiments were approved by the Institutional Animal Care and Use Committee of Mississippi State University (protocol # IACUC-21-461). Eight-week-old female Swiss Webster mice were obtained from Charles River laboratories (Wilmington, MA, USA) and housed at a rate of 5 mice per cage. Twenty-one Tn-mutants were tested for virulence. Mice were placed in quarantine for 3 days before they were released for the study. Overnight cultures of wild-type and Tn-mutants grown in BHI broth were standardized based on OD600, washed, resuspended in sterile saline. Serial dilutions were spread on agar plates for estimation of colony forming units (CFU). Mice (5 mice per group) were injected intravenously (IV) into a tail vein with 200 μL of the diluted bacterial suspension, achieving infectious dose of 2 x104 CFU/mL. The negative control group received only saline, and the positive control group received L. monocytogenes wild-type. Mice were frequently monitored for signs of clinical deterioration. Mice were euthanized 72-h post infection, and their livers and spleens were harvested, followed by homogenization in sterile PBS. Dilutions of the homogenized suspension were spread on BHI agar and incubated at 37°C. Colonies were counted after 48 h, and CFU/g of tissue was calculated to analyze the virulence of each strain.
Intracellular replication in murine macrophages
The ability of L. monocytogenes mutants to invade and replicate within host cell cytosol was compared to the wild-type strain in the presence and absence of gentamicin as previously described (Alonzo and Freitag, 2010). Confluent 12-well plates containing J774.1 murine macrophage cells were infected with the wild-type and Tn-mutants at a multiplicity of infection (MOI) of 1 bacterium to 1 macrophage. The infected plates were centrifuged and incubated for 1 h at 37°C and 5% CO2. Then the media was removed, and the cells were washed three times with sterile PBS. To measure total counts of bacteria associated with macrophages without gentamicin treatment (zero time points), monolayer cells were lysed with 0.5% Triton X-100 for 10 min followed by spreading on BHI agar to calculate CFU (Abdelhamed et al., 2015). To monitor intracellular survival and replication inside the macrophages, DMEM containing gentamicin (10 μg/ml) was added to each well and incubated at 37°C and 5% CO2 for 2, 4, 6, 24–h. At the end of each time point, media was removed, and the monolayer cells were washed. Cells were dislodged by adding trypsin and scraping. Serial dilutions of the lysed cell suspensions were spread on BHI agar and incubated at 37°C. Bacterial colonies were counted after 48 h, and CFU/ml was calculated for the wild-type and Tn-mutants for each time point. Bacterial intracellular growth and replication were performed in two independent experiments with three replicates.
Cell-to-cell spread
The ability of Tn-mutants to replicate in the host cytosol and spread from cell to cell was evaluated in plaque assays as described (Abdelhamed et al., 2020). Briefly, L2 fibroblast cells were grown in 6-well plates. Overnight cultures of wild-type and Tn-mutants were used to infect the L2 fibroblast cells at an MOI of 1–1. Plates were incubated for 1 h at 37°C and 5% CO2. After 1 h, the infected cells were washed and overlaid with a mixture containing DMEM and 1% Sea Plaque agarose supplemented with gentamicin (10 μg/ml). After solidification of the agarose, the plates were incubated at 37°C and 5% CO2 for 4 days. Staining of the plaques was performed by adding a second overlay of the DMEM and 1% Sea Plaque agarose mixture containing 0.01% Neutral red (Sigma Aldrich, St. Louis, MO). The plates were stained overnight at 37°C. Plaques sizes were measured using Olympus cellSens software (Olympus Corp. Shinjuku City, Tokyo, Japan). Experiments were repeated twice, and a minimum of 10 plaques were measured to determine plaque size.
Statistical analysis
Statistical significance was assessed using the Kruskal-Wallis test with GraphPad Prism 5 software (GraphPad Software San Diego, CA) to compare bacterial concentrations in mice tissues infected with wild-type and Tn-mutants. Statistical significance in intracellular replication was analyzed by two-way ANOVA (multiple groups and different time points) and Dunnett’s multiple comparison test. Statistical significance of mutants’ adhesion/replication to macrophages at zero time point was analyzed by one-way ANOVA and Dunnett’s multiple comparison test. All other data were analyzed with Student’s t-test. Data are presented as the mean ± SEM. A P value of less than 0.05 was considered significant.
Results
Identification of transposon-mutants deficient in growth in minimal medium
A library consisting of ∼2,000 Tn-mutants was screened to identify mutants with impaired growth in MM in the presence of one of the following carbon sources: glucose, mannose, fructose, sucrose, maltose, glycerol, or G6P. The screening identified 21 Tn-mutants that exhibited a significant reduction in growth rate compared to the wild-type in at least one of the media tested. The percentage growth of the identified candidates compared to wild-type in the presence of different carbon sources is presented in Table 2. The average OD600 values of 21 Tn-mutants are listed in Supplementary Table 1. Southern blotting confirmed the presence of a single Tn-insertion in the genome of the selected mutants.
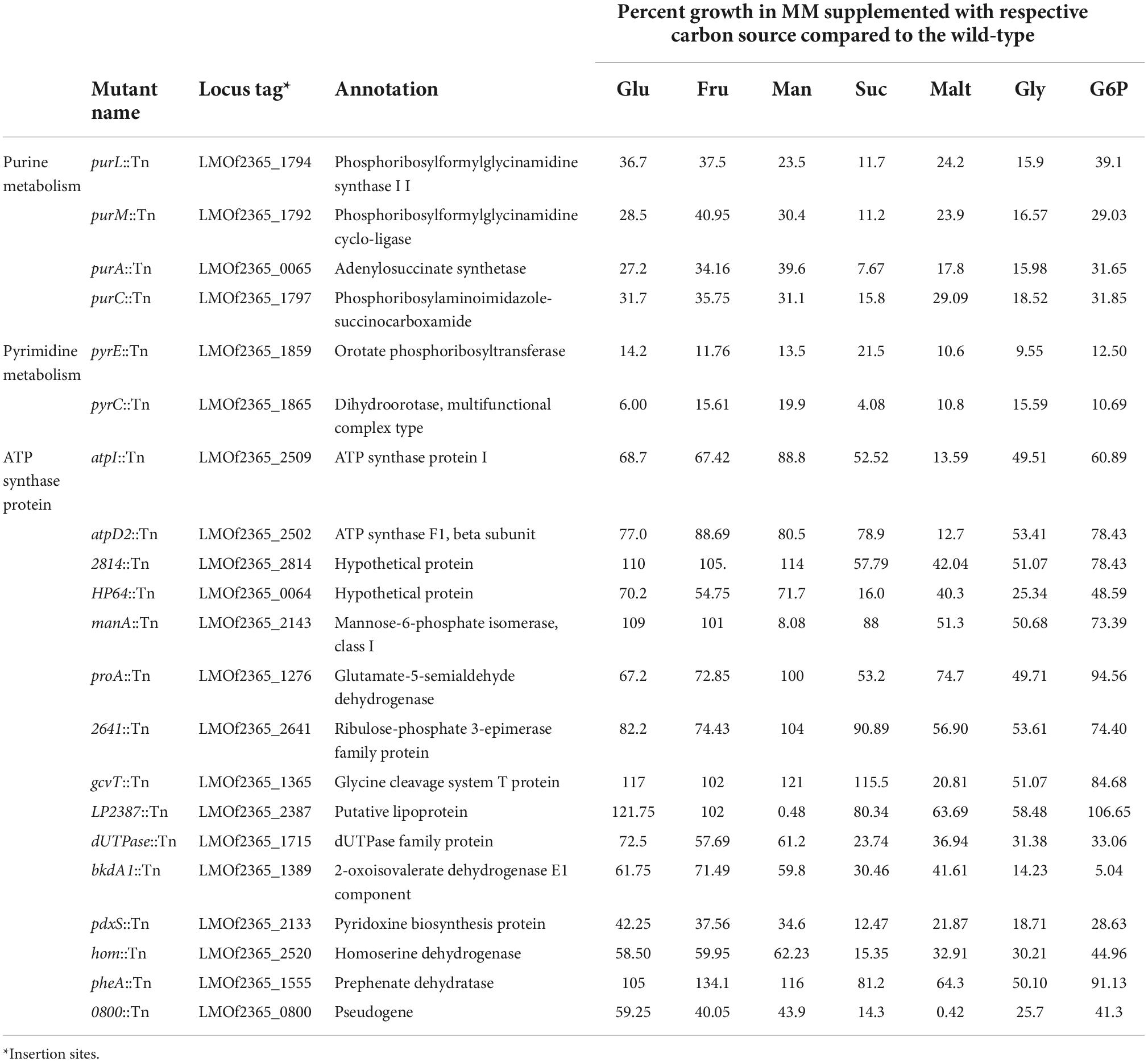
Table 2. Tn-insertion mutants with defective growth in MM containing glucose (Glu), fructose (Fru), mannose (Man), sucrose (Suc), maltose (Malt), glycerol (Gly), or glucose 6-phosphate (G6P) as the sole carbon source.
Among the list of identified Tn-mutants are four mutants with insertion sites located in genes encoding purine biosynthesis enzymes, including purL::Tn, purC::Tn, purA::Tn, and purM::Tn. In addition, two mutants have Tn-insertion in genes encoding pyrimidine biosynthesis enzymes, including pyrE::Tn and pyrC::Tn. Two of the Tn-mutants have insertions in genes encoding ATP synthase, atpI::Tn and atpD2::Tn. One mutant had a Tn-insertion in a gene encoding 2-oxoisovalerate dehydrogenase E1 component, α subunit of the branched-chain alpha-keto acid dehydrogenase (BKD) complex. Two mutants had Tn-insertions in genes annotated as hypothetical proteins (LMOf2365_2814 and LMOf2365_0064), and one mutant had a Tn-insertion in a gene annotated as a pseudogene (LMOf2365_0800). One transposon mutation mapped to LMOF2365_2387, a putative lipoprotein.
Growth of atpD2::Tn mutant is affected in a nutrient rich media
To understand the growth kinetics of the Tn-mutants, their growth rate in BHI was determined over a 24-h period at 37°C. The atpD2::Tn mutant had marked reduction in the growth with a doubling time of 190.9 min compared to the wild-type that doubled in 84.2 min (Figure 1). The doubling time of dUTPase::Tn and bkdA1::Tn mutants slightly increased by 9.9 and 2.9 min, respectively, compared to wild-type but this difference was not significant. In stationary phase, bkdA1::Tn did not achieve the same peak density as the parental strain. All other Tn-mutants grew as well as wild-type strain in BHI rich media. The growth defect in HP64::Tn and purA::Tn mutants could be restored to levels similar to that obtained for the parent strain by addition of adenine to MM (data not shown).
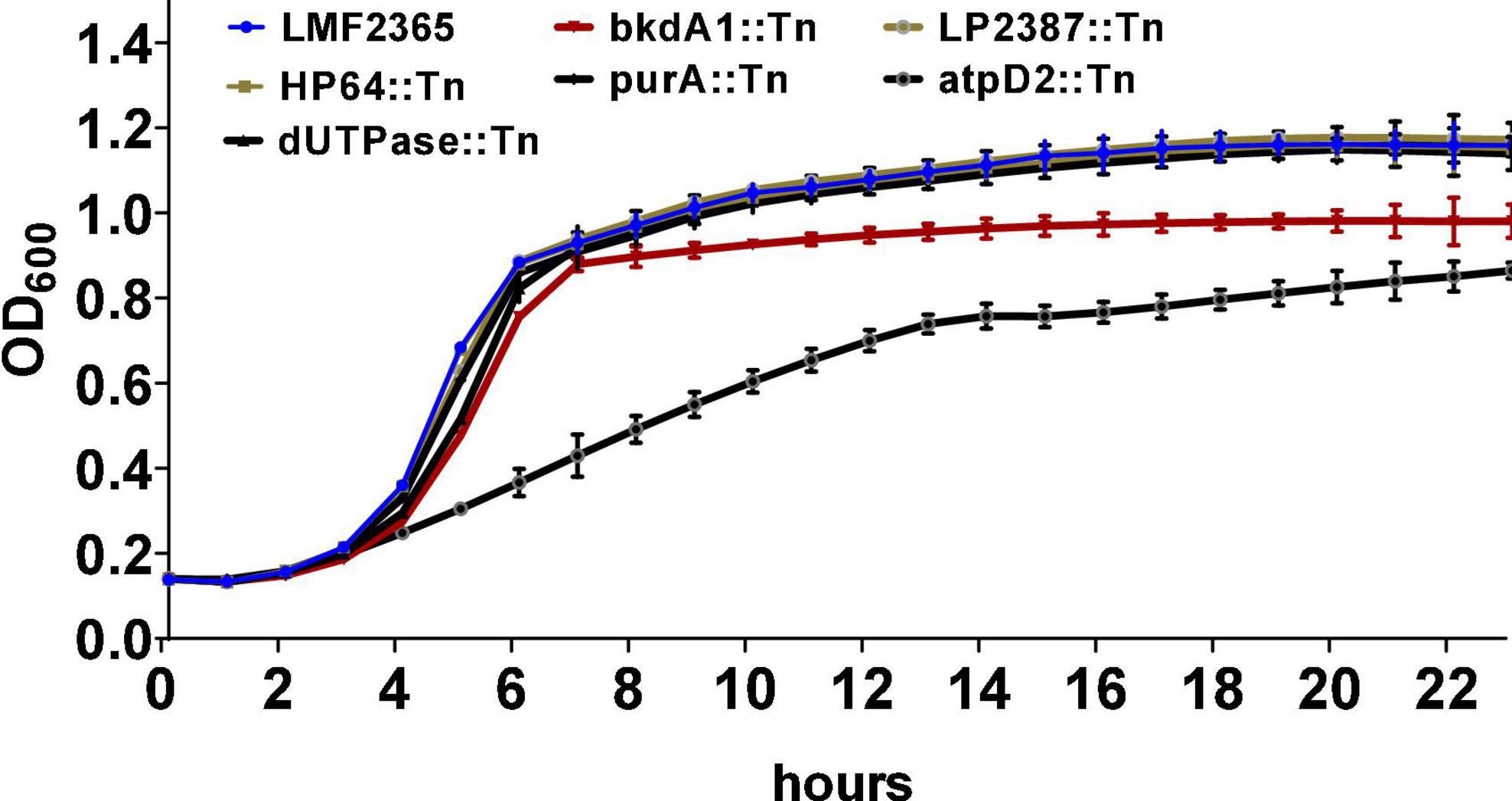
Figure 1. Growth of the Tn-mutants and wild-type L. monocytogenes in BHI medium at 37°C for 24 h. The growth kinetic was monitored using optical density measurements at 600 nm. Data represent he means ± standard error of three independent experiments with four replicates.
Mutants with impaired growth in minimal medium display varied attenuation in mice
Because pathogenesis of L. monocytogenes requires metabolic adaptation and nutrient uptake, we determined whether the MM growth defective Tn-mutants are attenuated in vivo using murine model.
Bacterial concentrations in liver and spleen tissues after 72 h of infection are presented in Figure 2. We detected a reduction in bacteria concentration in several Tn-mutants, but the reduction was only statistically significant in purA::Tn, HP64::Tn, bkdA1::Tn, dUTPase::Tn, LP2387::Tn, and atpD2::Tn. Log10 CFU/g changes in bacterial burden of the tested Tn-mutants are summarized in Table 3.
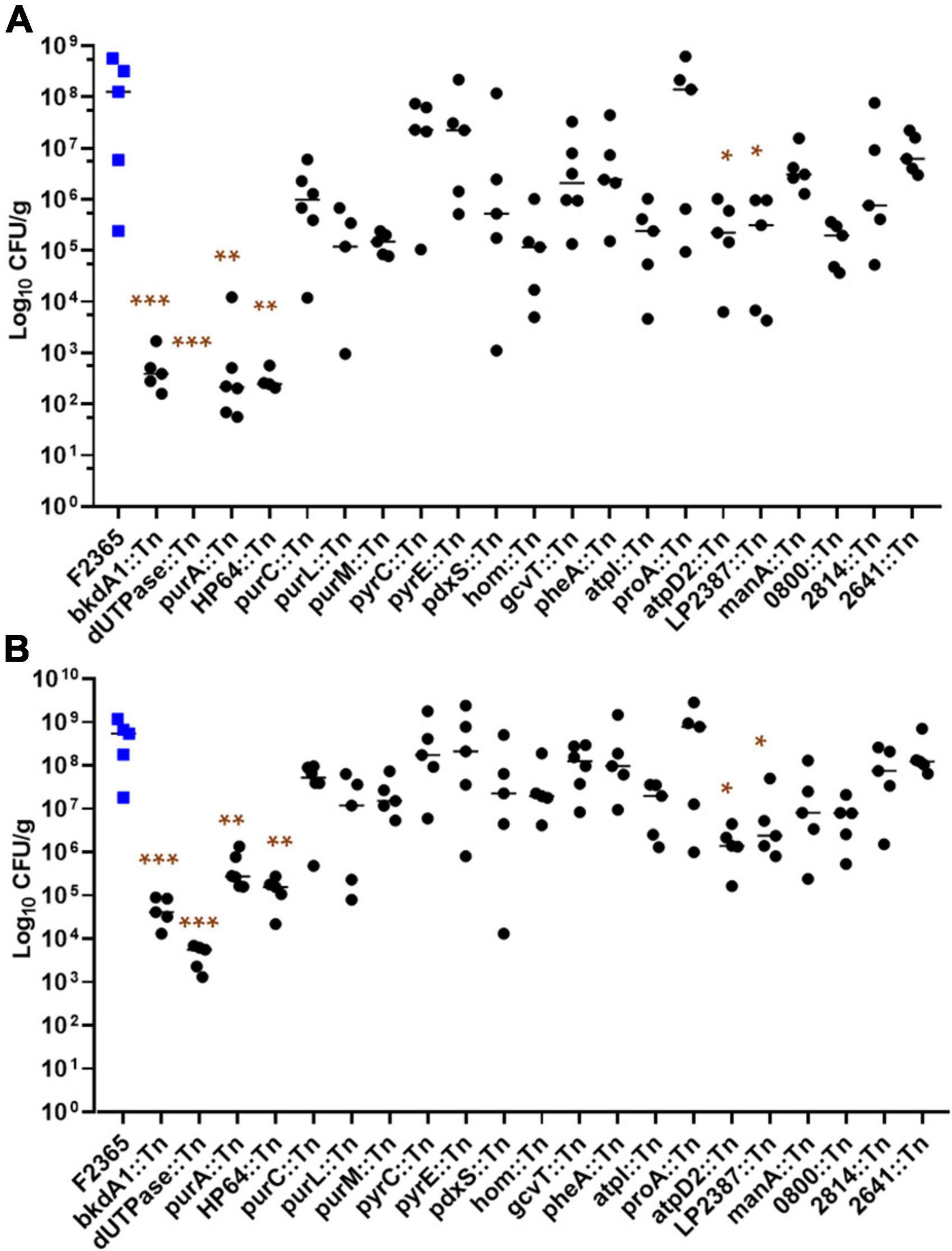
Figure 2. Virulence phenotypes of L. monocytogenes Tn-insertion mutants. Each animal group (5 mice per cage) was intravenously infected with 2 × 104 colony forming units (CFUs) with the indicated strains. At 72 h post-infection, bacterial concentrations were determined for livers (A) and spleens (B). Each point represents one mouse. Statistical analysis was performed with Kruskal-Wallis test in GraphPad Prism. *Indicates a significant difference at P < 0.05, **indicates a significant difference at P < 0.001, and ***indicates a significant difference at P < 0.0001.
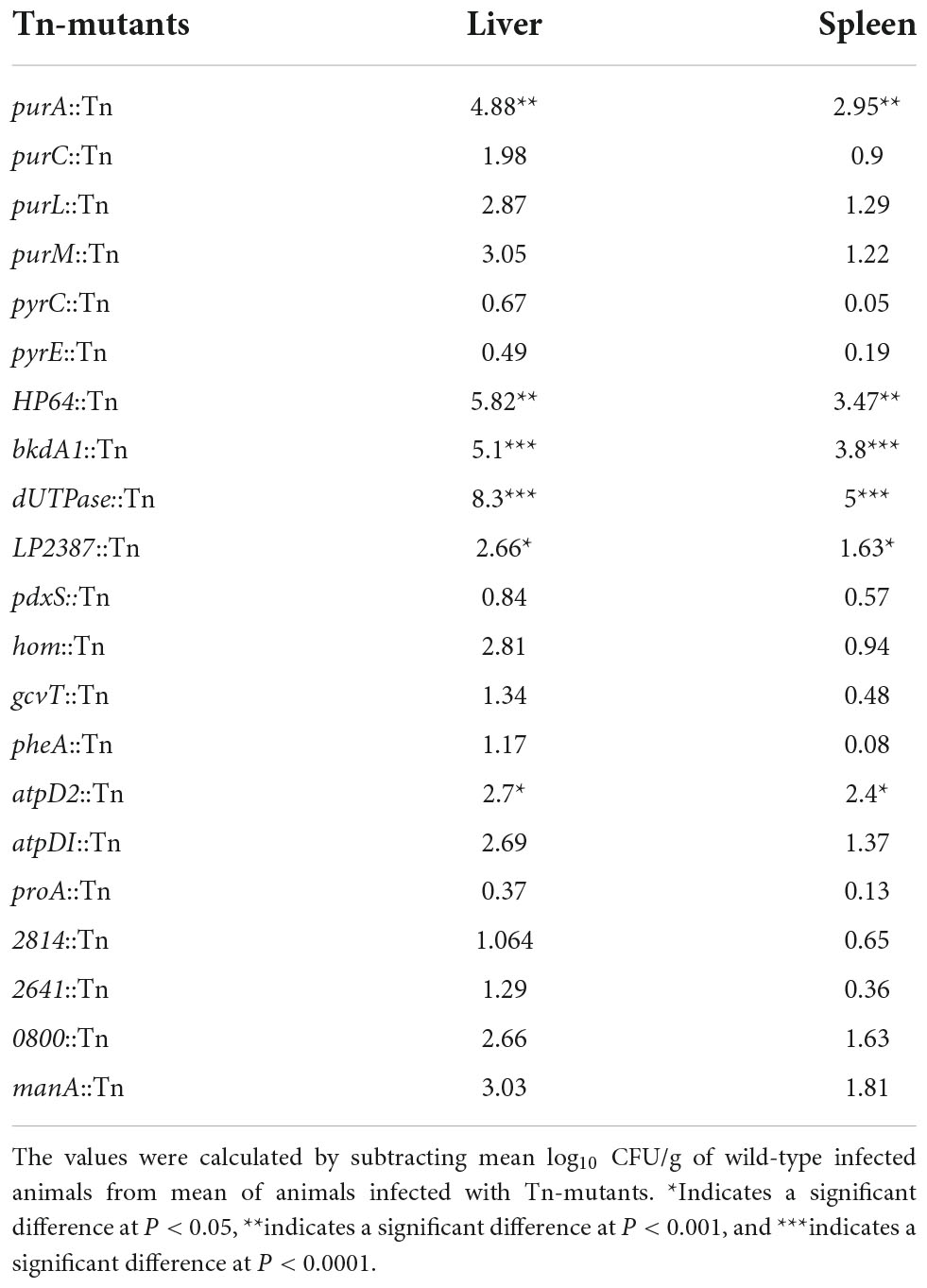
Table 3. Log10 CFU/g changes in concentration of Tn-mutants compared to wild-type in the liver and spleen of infected mice.
The change in the body weight of the mice from the time prior to challenge and at the time of euthanasia reflected the severity of infection caused by each Tn-mutant (Figure 3). The mice infected with purA::Tn, HP64::Tn, bkdA1::Tn, and dUTPase::Tn mutants showed significant weight gain compared to mice infected with wild-type. The mice group challenged with atpD2::Tn neither gained any significant weight nor lost any during their 8 days of incubation with bacteria. However, the mice infected with purC::Tn, purL::Tn, purM::Tn, pyrC::Tn, pyrE::Tn, pdxS::Tn, hom::Tn, gcvT::Tn, pheA::Tn, and atpI::Tn showed reduction in body weight similar to mice infected with wild-type.
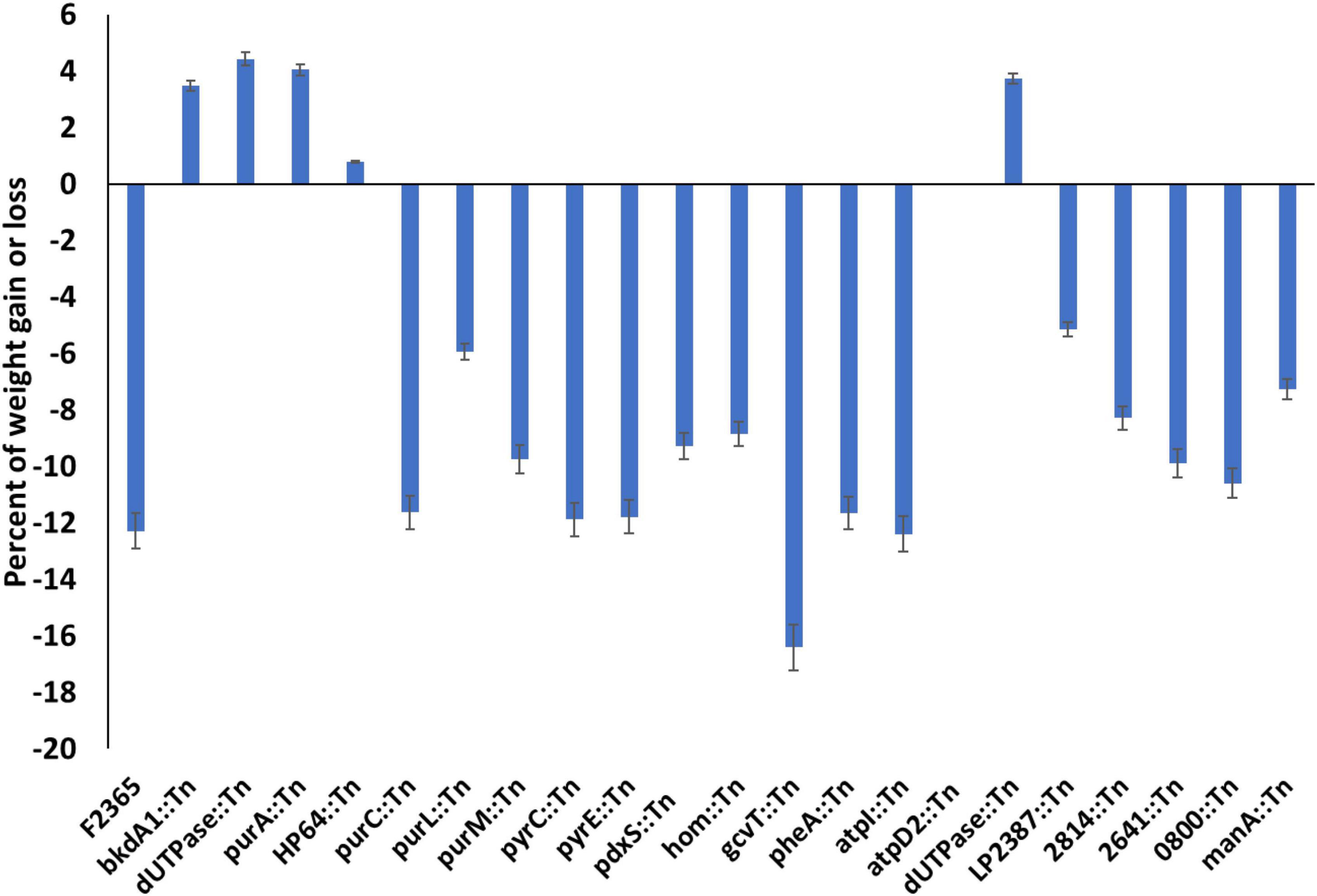
Figure 3. Average changes in body weight in mice infected with Tn-mutants compared to mice infected with wild-type L. monocytogenes. Variation in body weight (gain or loss) was calculated by subtracting body weight of each mouse at end of experiment from the baseline body weight at the time of the injection. Values represent the mean of 5 mice.
Attenuated transposon-mutants showed a defect in cellular invasion, intracellular replication and/or cell-to-cell spread
The first steps of L. monocytogenes pathogenesis rely on the ability of the bacteria to invade the host cells, escape the vacuole, replicate within the nutrient-rich host cell cytosol, and move from one cell to another without exposing itself to the extracellular environment. We determined the ability of six Tn-mutants (purA::Tn, HP64::Tn, dUTPase::Tn, bkdA1::Tn, LP2387::Tn, and atpD2::Tn) for cellular invasion, intracellular replication, and cell-cell spread in comparison to the wild-type. These six mutants were selected because they showed significant attenuation in mice. By 1-h post-infection and without gentamicin treatment, there was no significant difference (P > 0.05) in the total number of CFU between the six mutants compared to the wild-type (Figure 4).
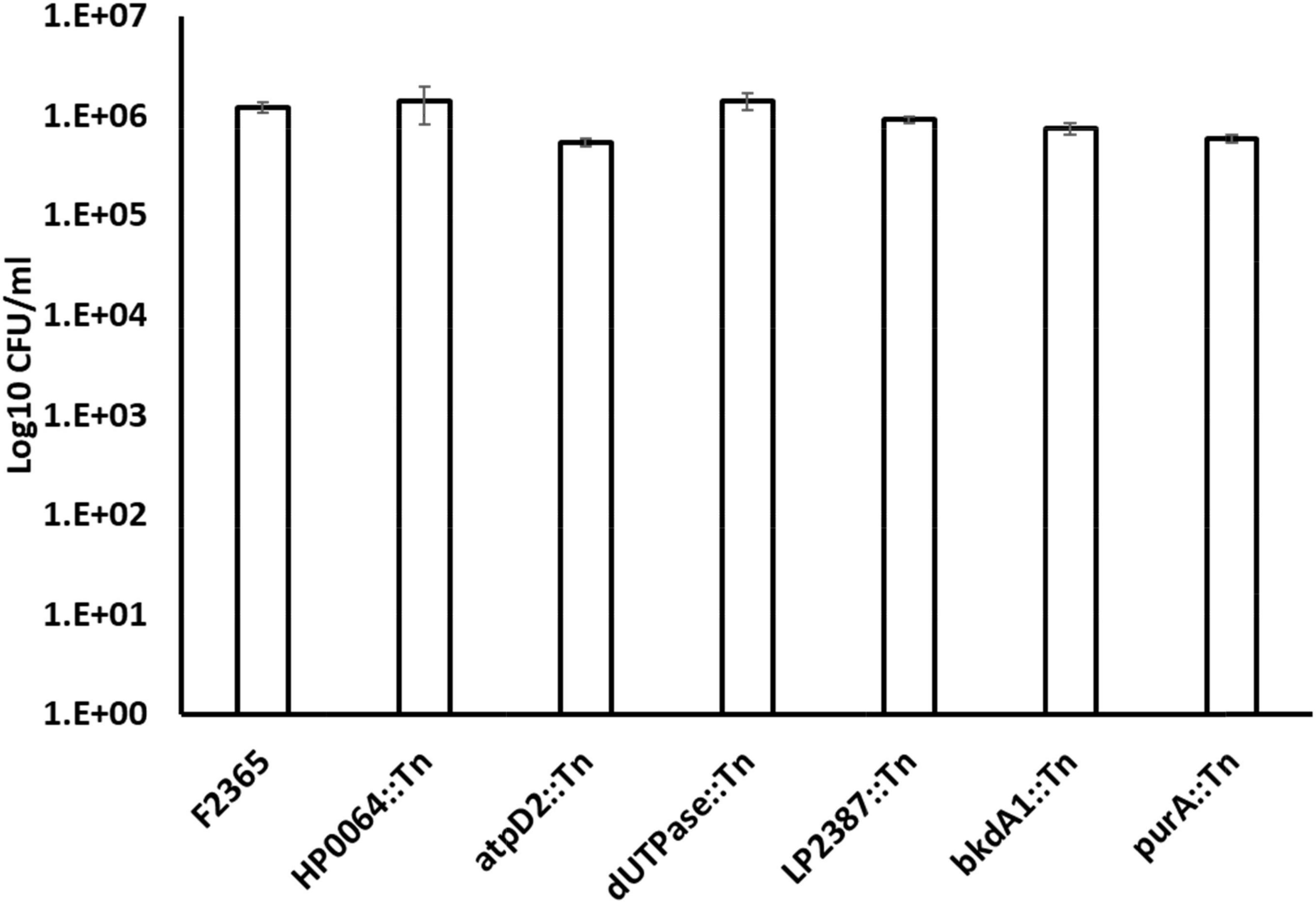
Figure 4. The total number of CFU/ml of wild-type L. monocytogenes strain F2365 and Tn-mutant strains obtained without gentamicin treatment to J774.1 murine macrophage cells at 1 h post-infection. Numbers on the Y axis indicate bacterial quantities (CFU/ml). Statistical analysis did not show any significant differences (P > 0.05). The experiment was repeated twice with three replicates each.
We next determined intracellular replication of the Tn-mutants by gentamicin protection assays in J774.1 murine macrophages at 2-, 4-, 6-, and 24-h post-infection (HPI). The purA::Tn and HP64::Tn mutants exhibited significant deficiency to replicate within J774.1 macrophages compared to the wild-type strain (Figure 5). The reduction in the intracellular replication of purA::Tn and HP64::Tn strains was 1.13 and 1.32-log10 at 2 h, 2.21 and 1.68-log10 at 4 h, 2.65 and 2.56-log10 at 6 h, and 1,83 and 1.68-log10 at 24 h compared to the wild-type strain, respectively. At 2-, 4-, 6-, and 24-h after infection, the intracellular bacterial numbers of the atpD2::Tn mutant were lower than those of the wild-type strain with log10 reduction of 0.45, 0.59, 0.64, and 0.12.
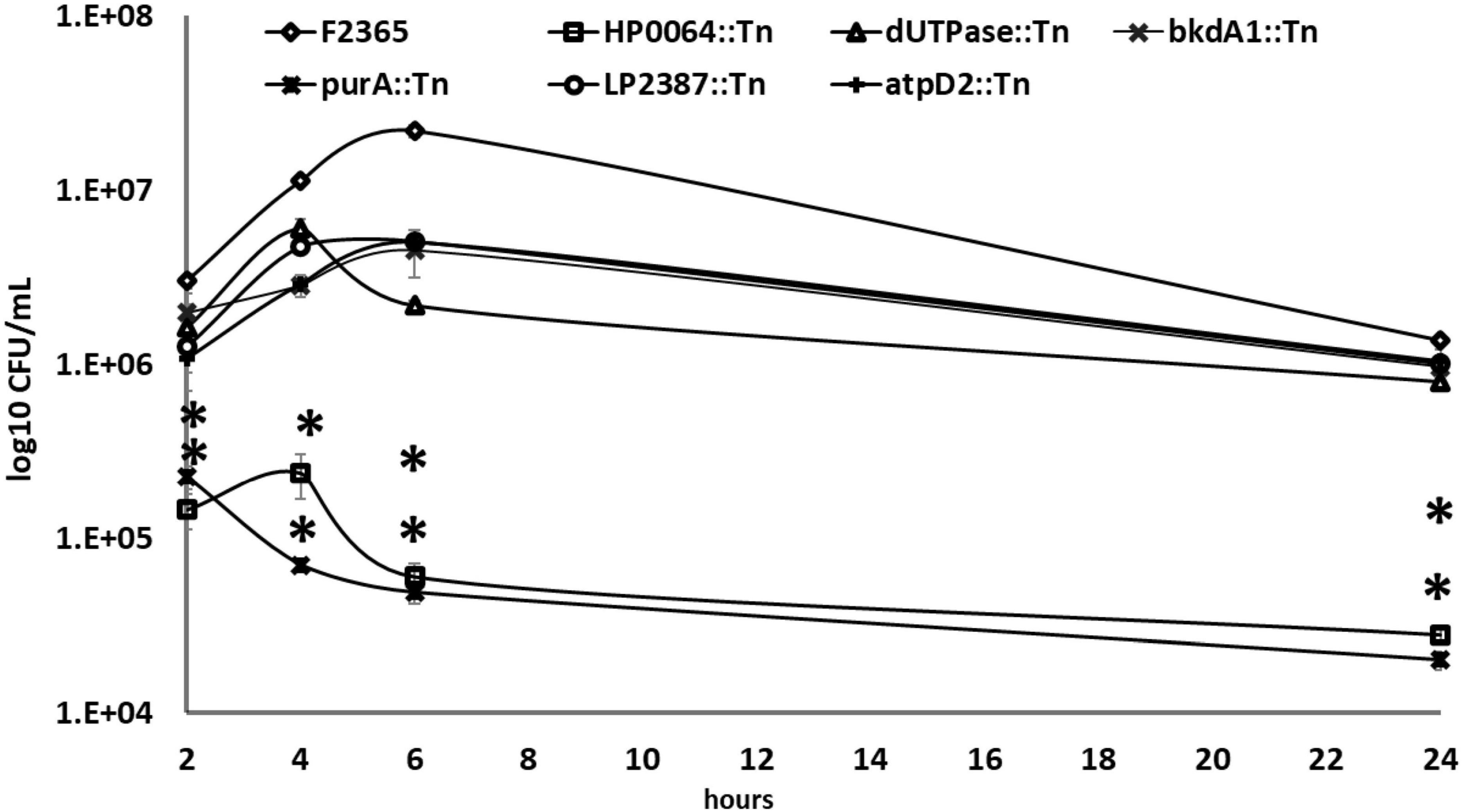
Figure 5. Intracellular growth of Tn-mutants compared to the wild-type L. monocytogenes strain in J774 macrophage-cells using an MOI of 10:1. At 2, 4, 6, and 24 h pot-infection, cells were lysed, and CFUs were enumerated. The experiment was repeated twice with three replicates each. Error bars are the standard error of mean. Two-way ANOVA and Dunnett’s multiple comparison test were used to measure the statistical significance between Tn-mutants and wild-type at different time points. Asterisks (*) indicate statistically significant differences (P < 0.05).
The dUTPase::Tn mutant showed a substantial reduction of 0.27-log10, 027-log10, 1-log10, 0.23-log10 in the CFU recovered from infected macrophages compared to those infected with the wild-type after 2, 4, 6, and 24 h of infection. The intracellular replications of bkdA1::Tn and LP2387::Tn were almost indistinguishable from those of the wild-type strain with a log10 reduction of 0.18 and 0.38 at 2 HPI, 0.60 and 0.38 at 4 HPI, and 0.68 and 0.63 at 6 HPI, and 0.15, 0.13, and 0.12 at 24 HPI compared to the wild-type, respectively.
To assess the ability of the Tn-mutants to spread from cell to cell, a plaque assay in L2 fibroblast cells was conducted for six Tn-mutants, and plaque diameters were compared to wild-type. The plaque area formed by HP64::Tn, purA::Tn, dUTPase::Tn, bkdA1::Tn, atpD2::Tn, and LP2387::Tn mutants was significantly smaller compared to the wild-type, measuring 35.73, 29.3, 64.1, 85.58, 77.55, and 82.64% of wild-type plaque diameter, respectively (Figure 6). The small plaque-phenotype was the most prominent in the purA::Tn and HP64::Tn mutants where the plaque sizes were approximately 30% of the wild-type F2365, reflecting that purA and HP64 are required not only for cell-to-cell spread, but also for intracellular survival and growth.
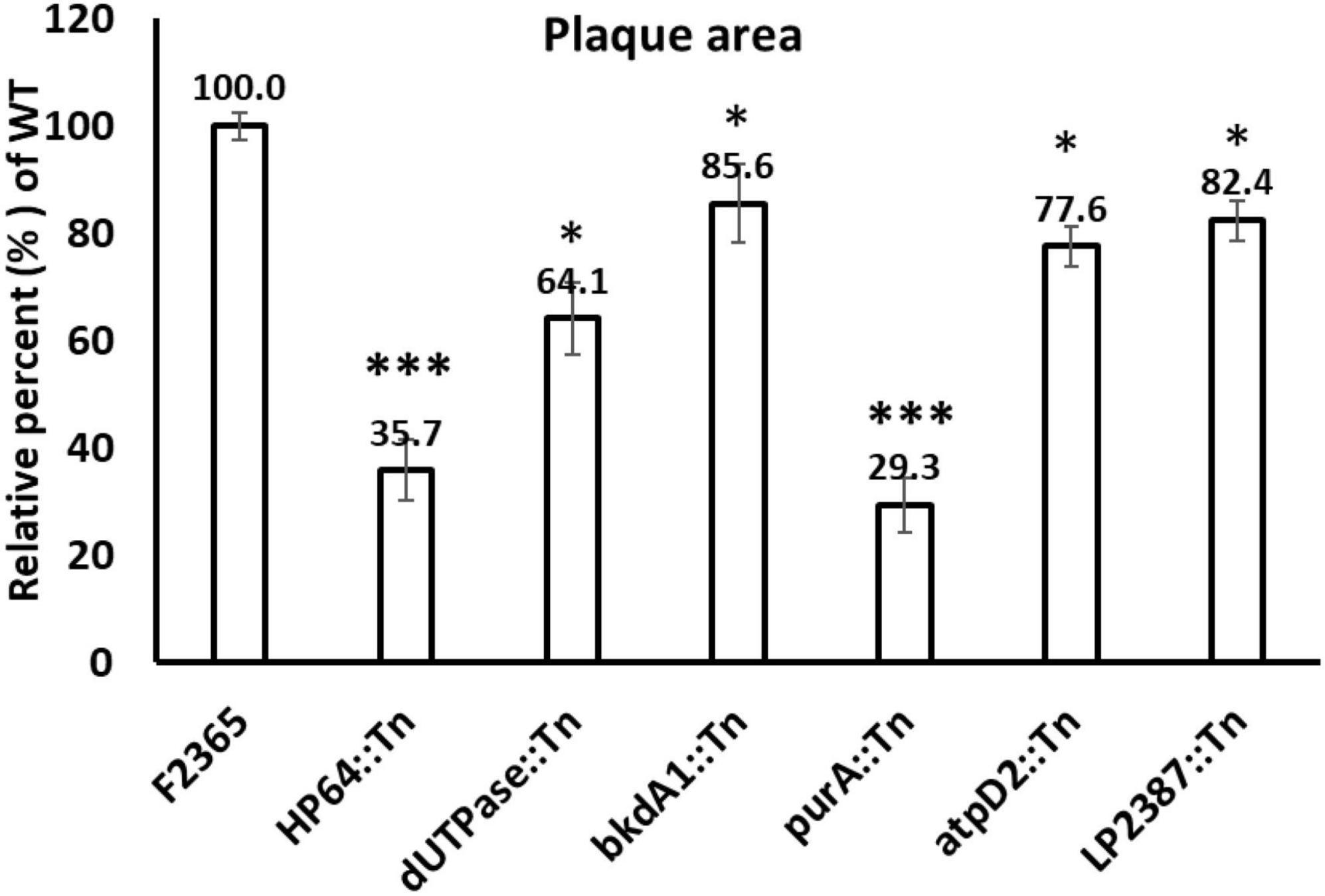
Figure 6. Plaque formation of wild-type L. monocytogenes and Tn-mutants in monolayers of murine L2 fibroblast cells. The mean plaque size of each strain is shown as a percentage relative to the wild-type plaque size. Error bars represent standard deviations of the mean plaque size from two independent experiments. Statistical analysis was performed using Student’s t-test. Asterisks indicate statistically significant differences (*P < 0.05, ***P < 0.001).
Discussion
Listeria monocytogenes fitness during pathogenesis requires efficient utilization of available nutrients. Furthermore, the function of PrfA, the master regulator of virulence genes in L. monocytogenes, is dependent on the metabolic state of the bacterium. Carbon metabolism is among several metabolic pathways that has been associated with functional state of PrfA; glucose and other sugars are known to inhibit PrfA activity, but metabolism of non-carbohydrate sources such glycerol activates PrfA (Mertins et al., 2007; Chen et al., 2017). However, our understanding of nutritional signaling, metabolism, and virulence in L. monocytogenes is still incomplete.
To further bridge this knowledge gap, we conducted a genetic screening and isolated insertional mutants that are deficient in growth in MM supplemented with different carbohydrate substrates but can grow in BHI (nutrient rich media). Growth in MM requires the biosynthesis of several metabolites that can be scavenged from a complex growth media. We identified 21 Tn-mutants that show growth deficiency when grown in MM supplemented with various carbon sources. We have characterized the mutants and their role in establishing infection.
By Tn-mutagenesis, we defined four genes encoding purine biosynthesis enzymes which are essential for L. monocytogenes in growth in MM. The products of these genes are phosphoribosylformyl glycinamidine synthase subunit (purL), phosphoribosylformylglycinamidine cyclo-ligase (purM), phosphoribosylaminoimidazole-succinocarboxamide synthase (purC), and adenylosuccinate synthetase (purA). In addition, HP64::Tn has Tn-insertion in a hypothetical protein (LMOf2365_0064) that is 68 bp upstream of purA (LMOf2365_0065). The growth defect in HP64::Tn and purA::Tn mutants could be restored by addition of adenine to MM. Thus, the phenotypic effect shown by HP64::Tn is apparently due to a defect in the downstream purA gene or may be HP64 has its own function in purine biosynthesis.
Interestingly, Tn-mutants for purine and pyrimidine biosynthesis showed severely impaired growth in MM; however, the mutants grew as well as wild-type bacteria in BHI broth. These findings are typical for purine and pyrimidine auxotrophs. Because they are unable to synthesize purines or pyrimidines, the respective auxotrophic strains must rely on salvage of nutrients, which are not readily available in MM, but they are available in nutrient-rich BHI medium.
PurA catalyzes the conversion of inosine mono phosphate to adenylosuccinate in the purine metabolic pathway for the synthesis of adenine. Purine auxotrophs of several bacterial pathogens are attenuated. For example, the following mutants are all attenuated: purA and purB mutants of L. monocytogens (Faith et al., 2012); purF, purG, purC, and purA mutants in Salmonella (McFarland and Stocker, 1987), purL mutant in Brucella abortus (Alcantara et al., 2004), and purE mutant in B. melitensis (Crawford et al., 1996). Purine biosynthesis is indispensable for Staphylococcus aureus growth in human blood and in vivo pathogenicity (Connolly et al., 2017).
In the present study, we observed a critical role of purA in intracellular multiplication and cell to cell spread of L. monocytogenes within infected host cells. The reduced plaque sizes and intracellular replication in the purA::Tn mutant was partially restored when the media was supplemented with adenine (data not shown). This finding suggests that adenine is not available for listerial salvage in the intracellular environment. Several research groups have reported a reduced ability of a purA Tn-mutant to multiply intracellularly and within intestinal epithelial cells (Sauer et al., 2011; Faith et al., 2012).
Our screening of Tn-mutants identified the gene coding for 2-oxoisovalerate dehydrogenase E1 component, α subunit of the branched-chain alpha-keto acid dehydrogenase (BKD) complex, which is a necessary enzyme for branched-chain fatty acids (BCFA) synthesis (Giotis et al., 2007; Sun and O’Riordan, 2010; Sun et al., 2012). BCFA represent more than 90% of the membrane composition in L. monocytogenes, and modulating BCFA membrane composition is a critical mechanism for regulating membrane fluidity to allow adaptation to low temperatures (Annous et al., 1997; Zhu et al., 2005; Giotis et al., 2007). The importance of BKD enzymes in bacterial virulence has been well characterized in several pathogens (Annous et al., 1997; Zhu et al., 2005; Giotis et al., 2007). In L. monocytogenes, Tn-mutants in BKD encoding genes ipd and bkdB (cld-1 and cld-2 mutants) exhibited defects in BCFA composition, cold sensitivity, and a significantly less-fluid membrane than the parent strain (Jones et al., 2002). In addition, L. monocytogenes cld-1 and cld-2 mutants exhibit lower tolerance to acidic and alkaline pH than parent strain (Giotis et al., 2007).
In our study, we demonstrated that inactivation of bkdA1 resulted in complete attenuation of virulence in mice. The mice infected with bkdA1::Tn exhibited no signs of morbidity and had low concentrations of bacteria in spleen and liver. This phenotypic effect may be due to a reduction in anteiso-BCFAs, which provide fluidity to the listerial membrane. We showed that a bkdA1::Tn mutant was defective for growth in MM supplemented with glucose, sucrose, maltose, glycerol, or G6P. In BHI broth, the bkdA1::Tn mutant showed no significant difference in doubling time compared to the wild-type at 37°C. Our findings indicate that MM does not support the scavenging of an exogenous nutrient necessary for growth of a BCFA auxotrophic strain.
Deoxyuridine triphosphatase (dUTPase) hydrolyzes dUTP to dUMP (deoxyuridine monophosphate) and pyrophosphate (Bessman et al., 1958). It is an important enzyme that protects DNA against uracil incorporation (Dubois et al., 2011). Through catalytic conversion of dUTP to dUMP, dUTPase also provides precursors for synthesis of thymidine nucleotides essential in DNA replication. It also maintains a low cellular concentration of dUTP, thus preventing its incorporation into DNA (Vértessy and Tóth, 2009). In E. coli and yeast, mutation of dUTPase is lethal (el-Hajj et al., 1988; Gadsden et al., 1993). In the current study, a L. monocytogenes dUTPase::Tn mutant displayed a 30–40% reduction in growth compared to the wild-type in MM with glucose, fructose, and mannose. Its growth was more affected in MM with sucrose, maltose, glycerol, and glucose 6-phosphate (65–80% reduction compared to the wild-type). When J774.1 macrophage cells were infected with the dUTPase::Tn mutant, the number of bacteria reached a peak at 4 h followed by a decline in intracellular replication. These results were substantiated by plaque assay that indicated a 60% reduction in plaque number and size compared to wild-type, indicating a defect in intracellular replication and/or cell-to-cell spread. Mice infected with dUTPase::Tn cleared the bacteria completely from liver after 72 h of infection with a 5-log reduction in viable bacterial cells observed in spleen. Further investigation will focus on the mechanisms that allow the dUTPase::Tn mutant to grow in nutrient rich media but prevent their proliferation in mice with an intact immune system.
Adenosine triphosphate synthase beta 2 subunit (atpD2) is a part of the plasma membrane ATP synthase complex catalytic core (F1), which catalyzes the phosphorylation of ADP using inorganic phosphate to form ATP at the expense of a proton gradient across the membrane (Revington et al., 2002). L. monocytogenes also generates ATP through substrate level phosphorylation that occurs during the glycolytic pathway (Sauer et al., 2019). In the present study, inactivation of atpD2 gene significantly reduced the doubling time of L. monocytogenes F2365 strain in nutrient rich BHI media, indicating the requirement of atpD2 gene for efficient replication of L. monocytogens. This is likely due to an impairment in ATP synthesis and resulting insufficient ATP generation for aerobic growth (Müller-Herbst et al., 2014). In our study, the atpD2::Tn mutant was grown under aerobic conditions, and its anaerobic growth dynamics remains to be determined. The atpD2::Tn mutant was also defective for growth in mice liver and spleen, suggesting that generation of ATP via substrate level phosphorylation alone is not sufficient for L. monocytogenes replication in the host.
The LP2387::Tn mutant has a mutation mapped to LMOF2365_2387, which encodes a putative lipoprotein. Lipoproteins are a major category of bacterial surface proteins that have diverse functions and often have important effects on pathogen/host interactions (García-del Portillo and Cossart, 2007; Machata et al., 2008). Up to 68 putative lipoproteins have been predicted in the genome of L. monocytogenes include ABC transporter lipoproteins for acquisition of micronutrients (Baumgärtner et al., 2007). In the present study, the LP2387::Tn mutant showed a decreased capacity to colonize in mice and displayed growth defect in MM. Mutation of individual lipoproteins affects Streptococcus pneumoniae sensitivity to environmental stress, adhesion to host tissues, and interactions with host phagocytes (Tseng et al., 2002). Deletion of the S. pneumoniae lgt results in reduced growth in physiological media with glucose, sucrose, raffinose, or maltotriose as the only carbon source (Chimalapati et al., 2012). Our result suggested that LP2387 may be part of ABC transporter lipoproteins that play a role in uptake of micronutrients for the growth and survival of L. monocytogenes F2365 strain in the host.
In summary, we report the construction and screening of a Tn-mutant library in L. monocytogenes to identify genes essential for growth under limited nutrient conditions. These include genes necessary for purine biosynthesis, pyrimidine biosynthesis, and BCFA metabolism, and ATP synthase protein. Purine biosynthesis and BCFA synthesis mutants were also defective for L. monocytogens virulence. This work indicates that L. monocytogenes metabolism affects its ability to cause disease and replicate in the host, and it provides evidence for the pathogen’s need to salvage specific nutrients during infection. Finally, our findings may not be fully generalized to all L. monocytogenes strains due to genetic and phenotypic diversity between strains. Further work will focus on the construction of clean deletion mutants and complementation strains corresponding to 21 genes identified in the present study and testing their ability to grow in MM and virulence in a mouse model of L. monocytogenes infection. Although intravenous infection is commonly used as a murine model to characterize the pathogenicity of L. monocytogenes strains it completely bypasses the gastrointestinal phase of the infection and bloodstream invasion. While sharing similarities, mouse and human macrophage cells exhibit numerous differences in microbial responses (Murray and Wynn, 2011). These differences must be considered when extrapolating mouse results to humans. Thus, evaluating the survival and replication of the Tn-mutants in human macrophage are warranted.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
All animal experiments were done in accordance with a protocol (18-508) approved by the Institutional Animal Care and Use Committee at Mississippi State University.
Author contributions
LN: library construction, data analyses, and writing first draft. OO: mutants screening and mice experiment. NA: cell cultures. RR: mice experiment. ML: experimental design, review, and edit. HA: experimental design, library construction, data analysis, review, and edit. All authors wrote and approved the manuscript.
Funding
This project was supported by the Center for Biomedical Research Excellence in Pathogen–Host Interactions, National Institute of General Medical Sciences, and National Institutes of Health awarded (grant no. P20GM103646-08).
Acknowledgments
We thank the Laboratory Animal Resources and Care Unit at Mississippi State University for animal and veterinary care. We thank Daniel Portnoy’ laboratory for sharing plasmid and for providing assistance with library construction. We also thank Stephen Pruett for proof reading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1007657/full#supplementary-material
Footnotes
References
Abdelhamed, H., Lawrence, M. L., and Karsi, A. (2015). A novel suicide plasmid for efficient gene mutation in Listeria monocytogenes. Plasmid 81, 1–8.
Abdelhamed, H., Ramachandran, R., Narayanan, L., Ozdemir, O., Cooper, A., Olivier, A. K., et al. (2020). Contributions of a LysR transcriptional regulator to Listeria monocytogenes virulence and identification of its regulons. J. Bacteriol. 202:e00087-20. doi: 10.1128/JB.00087-20
Alcantara, R. B., Read, R. D. A., Valderas, M. W., Brown, T. D., and Roop, R. M. II (2004). Intact purine biosynthesis pathways are required for wild-type virulence of Brucella abortus 2308 in the BALB/c mouse model. Infect. Immun. 72, 4911–4917. doi: 10.1128/IAI.72.8.4911-4917.2004
Alonzo, F., and Freitag, N. E. (2010). Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect. Immun. 78, 4944–4957.
Anaya-Sanchez, A., Feng, Y., Berude, J. C., and Portnoy, D. A. (2021). Detoxification of methylglyoxal by the glyoxalase system is required for glutathione availability and virulence activation in Listeria monocytogenes. PLoS Pathogens 17:e1009819. doi: 10.1371/journal.ppat.1009819
Annous, B. A., Becker, L. A., Bayles, D. O., Labeda, D. P., and Wilkinson, B. J. (1997). Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63, 3887–3894. doi: 10.1128/aem.63.10.3887-3894.1997
Azizoglu, R. O., Elhanafi, D., and Kathariou, S. (2014). Mutant construction and integration vector-mediated gene complementation in Listeria monocytogenes. Methods Mol. Biol. 1157, 201–211. doi: 10.1007/978-1-4939-0703-8_17
Baumgärtner, M., Kärst, U., Gerstel, B., Loessner, M., Wehland, J., and Jänsch, L. (2007). Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189, 313–324. doi: 10.1128/JB.00976-06
Bessman, M. J., Lehman, I. R., Adler, J., Zimmerman, S. B., Simms, E. S., and Kornberg, A. (1958). Enzymatic synthesis of deoxyribonucleic acid. Iii. the incorporation of pyrimidine and purine analogues into deoxyribonucleic acid. Proc. Natl. Acad. Sci. U S A. 44, 633–640.
Cahoon, L., and Freitag, N. (2014). Listeria monocytogenes virulence factor secretion: don’t leave the cell without a chaperone. Front. Cell. Infect. Microbiol. 4:13. doi: 10.3389/fcimb.2014.00013
Cao, M., Bitar, A. P., and Marquis, H. (2007). A mariner-based transposition system for Listeria monocytogenes. Appl. Environ. Microbiol. 73, 2758–2761. doi: 10.1128/AEM.02844-06
CDC (2011). Multistate outbreak of listeriosis associated with Jensen Farms cantaloupe–United States, August-September 2011. Atlanta, GA: CDC.
CDC (2016). Multistate Outbreak of Listeriosis Linked to Whole Cantaloupes from Jensen Farms, Colorado. Atlanta, GA: CDC.
Chen, G. Y., and Pensinger, D. A. (2017). Listeria monocytogenes cytosolic metabolism promotes replication, survival, and evasion of innate immunity. Cell Microbiol. 19:10.1111/cmi.12762. doi: 10.1111/cmi.12762
Chen, G. Y., Pensinger, D. A., and Sauer, J.-D. (2017). Listeria monocytogenes cytosolic metabolism promotes replication, survival, and evasion of innate immunity. Cell Microbiol 19:10.1111/cmi.12762. doi: 10.1111/cmi.12762
Chico-Calero, I., Suárez, M., González-Zorn, B., Scortti, M., Slaghuis, J., Goebel, W., et al. (2002). Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U S A. 99, 431–436. doi: 10.1073/pnas.012363899
Chimalapati, S., Cohen, J. M., Camberlein, E., MacDonald, N., Durmort, C., Vernet, T., et al. (2012). Effects of deletion of the Streptococcus pneumoniae lipoprotein diacylglyceryl transferase gene lgt on ABC transporter function and on growth in vivo. PLoS One 7:e41393. doi: 10.1371/journal.pone.0041393
Connolly, J., Boldock, E., Prince, L. R., Renshaw, S. A., Whyte, M. K., Foster, S. J., et al. (2017). Identification of Staphylococcus aureus factors required for pathogenicity and growth in human blood. Infect. Immun. 85, e337–e317. doi: 10.1128/IAI.00337-17
Crawford, R. M., Van De Verg, L., Yuan, L., Hadfield, T. L., Warren, R. L., Drazek, E. S., et al. (1996). Deletion of purE attenuates Brucella melitensis infection in mice. Infect. Immun. 64, 2188–2192.
Dubois, E., Córdoba-Cañero, D., Massot, S., Siaud, N., Gakière, B., Domenichini, S., et al. (2011). Homologous recombination is stimulated by a decrease in dUTPase in Arabidopsis. PLoS One 6:e18658. doi: 10.1371/journal.pone.0018658
el-Hajj, H. H., Zhang, H., and Weiss, B. (1988). Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170, 1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988
Eylert, E., Schar, J., Mertins, S., Stoll, R., Bacher, A., Goebel, W., et al. (2008). Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol. Microbiol. 69, 1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x
Faith, N. G., Kim, J. W., Azizoglu, R., Kathariou, S., and Czuprynski, C. (2012). Purine biosynthesis mutants (purA and purB) of serotype 4b Listeria monocytogenes are severely attenuated for systemic infection in intragastrically inoculated A/J Mice. Foodborne Pathogens Dis. 9, 480–486. doi: 10.1089/fpd.2011.1013
Fu, Y., Waldor, M. K., and Mekalanos, J. J. (2013). Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663. doi: 10.1016/j.chom.2013.11.001
Gadsden, M. H., McIntosh, E. M., Game, J. C., Wilson, P. J., and Haynes, R. H. (1993). dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12, 4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x
García-del Portillo, F., and Cossart, P. (2007). An important step in listeria lipoprotein research. J. Bacteriol. 189, 294–297.
Gaul, L. K., Farag, N. H., Shim, T., Kingsley, M. A., Silk, B. J., and Hyytia-Trees, E. (2013). Hospital-acquired listeriosis outbreak caused by contaminated diced celery–Texas, 2010. Clin. Infect. Dis. 56, 20–26. doi: 10.1093/cid/cis817
Gillmaier, N., Gotz, A., Schulz, A., Eisenreich, W., and Goebel, W. (2012). Metabolic responses of primary and transformed cells to intracellular Listeria monocytogenes. PLoS One 7:e52378. doi: 10.1371/journal.pone.0052378
Giotis, E. S., McDowell, D. A., Blair, I. S., and Wilkinson, B. J. (2007). Role of branched-chain fatty acids in pH stress tolerance in Listeria monocytogenes. Appl. Environ. Microbiol. 73, 997–1001. doi: 10.1128/AEM.00865-06
Grubmüller, S., Schauer, K., Goebel, W., Fuchs, T. M., and Eisenreich, W. (2014). Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front. Cell. Infect. Microbiol. 4:156. doi: 10.3389/fcimb.2014.00156
Jones, S. L., Drouin, P., Wilkinson, B. J., and Pd, I. I. M. (2002). Correlation of long-range membrane order with temperature-dependent growth characteristics of parent and a cold-sensitive, branched-chain-fatty-acid-deficient mutant of Listeria monocytogenes. Arch. Microbiol. 177, 217–222. doi: 10.1007/s00203-001-0380-4
Joseph, B., Mertins, S., Stoll, R., Schär, J., Umesha, K. R., Luo, Q., et al. (2008). Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190, 5412–5430. doi: 10.1128/JB.00259-08
Machata, S., Tchatalbachev, S., Mohamed, W., Jänsch, L., Hain, T., and Chakraborty, T. (2008). Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-Mediated immune activation. J. Immunol. 181, 2028–2035. doi: 10.4049/jimmunol.181.3.2028
McFarland, W. C., and Stocker, B. A. (1987). Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 3, 129–141. doi: 10.1016/0882-4010(87)90071-4
Melo, J., Andrew, P. W., and Faleiro, M. L. (2015). Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: the role of stress responses. Food Res. Int. 67, 75–90.
Mertins, S., Joseph, B., Goetz, M., Ecke, R., Seidel, G., Sprehe, M., et al. (2007). Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189, 473–490. doi: 10.1128/JB.00972-06
Muchaamba, F., Eshwar, A. K., Stevens, M. J. A., von, Ah, U., and Tasara, T. (2019). Variable carbon source utilization, stress resistance, and virulence profiles among Listeria monocytogenes strains responsible for listeriosis outbreaks in Switzerland. Front. Microbiol. 10:957. doi: 10.3389/fmicb.2019.00957
Müller-Herbst, S., Wüstner, S., Mühlig, A., Eder, D. M., Fuchs, T., Held, C., et al. (2014). Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology 160, 752–765. doi: 10.1099/mic.0.075242-0
Murray, P. J., and Wynn, T. A. (2011). Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737.
Nelson, K. E., Fouts, D. E., Mongodin, E. F., Ravel, J., DeBoy, R. T., Kolonay, J. F., et al. (2004). Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32, 2386–2395. doi: 10.1093/nar/gkh562
Premaratne, R. J., Lin, W. J., and Johnson, E. A. (1991). Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57, 3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Revington, M., Dunn, S. D., and Shaw, G. S. (2002). Folding and stability of the b subunit of the F(1)F(0) ATP synthase. Protein Sci. 11, 1227–1238. doi: 10.1110/ps.3200102
Ripio, M. T., Brehm, K., Lara, M., Suárez, M., and Vázquez-Boland, J. A. (1997). Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179, 7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997
Sauer, J. D., Sotelo-Troha, K., von Moltke, J., Monroe, K. M., Rae, C. S., Brubaker, S. W., et al. (2011). The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694. doi: 10.1128/IAI.00999-10
Sauer, J.-D., Herskovits, A. A., and O’Riordan, M. X. D. (2019). Metabolism of the gram-positive bacterial pathogen Listeria monocytogenes. Microbiol. Spectr. 7:10.1128/microbiolspec.GPP3-0066-2019. doi: 10.1128/microbiolspec.GPP3-0066-2019
Sun, Y., and O’Riordan, M. X. (2010). Branched-chain fatty acids promote Listeria monocytogenes intracellular infection and virulence. Infect. Immun. 78, 4667–4673. doi: 10.1128/IAI.00546-10
Sun, Y., Wilkinson, B. J., Standiford, T. J., Akinbi, H. T., and O’Riordan, M. X. (2012). Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J. Bacteriol. 194, 5274–5284. doi: 10.1128/JB.00045-12
Tseng, H. J., McEwan, A. G., Paton, J. C., and Jennings, M. P. (2002). Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect. Immun. 70, 1635–1639. doi: 10.1128/IAI.70.3.1635-1639.2002
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domıìnguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640.
Vértessy, B. G., and Tóth, J. (2009). Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 42, 97–106. doi: 10.1021/ar800114w
Zhu, K., Ding, X., Julotok, M., and Wilkinson, B. J. (2005). Exogenous isoleucine and fatty acid shortening ensure the high content of anteiso-C15:0 fatty acid required for low-temperature growth of Listeria monocytogenes. Appl. Environ. Microbiol. 71, 8002–8007. doi: 10.1128/AEM.71.12.8002-8007.2005
Keywords: Listeria monocytogenes, transposon-insertion, virulence, auxotroph, fitness
Citation: Narayanan L, Ozdemir O, Alugubelly N, Ramachandran R, Banes M, Lawrence M and Abdelhamed H (2022) Identification of genetic elements required for Listeria monocytogenes growth under limited nutrient conditions and virulence by a screening of transposon insertion library. Front. Microbiol. 13:1007657. doi: 10.3389/fmicb.2022.1007657
Received: 30 July 2022; Accepted: 22 September 2022;
Published: 13 October 2022.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Ruixi Chen, Massachusetts Institute of Technology, United StatesFrancis Muchaamba, University of Zurich, Switzerland
Copyright © 2022 Narayanan, Ozdemir, Alugubelly, Ramachandran, Banes, Lawrence and Abdelhamed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossam Abdelhamed, YWJkZWxoYW1lZEBjdm0ubXNzdGF0ZS5lZHU=
 Lakshmi Narayanan1,2
Lakshmi Narayanan1,2 Ozan Ozdemir
Ozan Ozdemir Reshma Ramachandran
Reshma Ramachandran Michelle Banes
Michelle Banes Mark Lawrence
Mark Lawrence Hossam Abdelhamed
Hossam Abdelhamed