- 1Biotechnology Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 2Nanping Institute of Agricultural Sciences, Sanming Agricultural Bureau, Fujian, China
- 3Jiangsu Collaborative Innovation Center of Regional Modern Agriculture and Environmental Protection, Jiangsu Key Laboratory for Eco-Agricultural Biotechnology Around Hongze Lake, Huaiyin Normal University, Huai’an, China
- 4College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
Utilization of rice blast-resistance (R) genes is the most economical and environmentally friendly method to control blast disease. However, rice varieties with R genes influence the outcome of genetic architectures of Magnaporthe oryzae (M. oryzae), and mutations in avirulence (AVR) genes of M. oryzae may cause dysfunction of the corresponding R genes in rice varieties. Although monitoring and characterizing rice R genes and pathogen AVR genes in field populations may facilitate the implementation of effective R genes, little is known about the changes of R genes over time and their ultimate impact on pathogen AVR genes. In this study, 117 main cultivated rice varieties over the past five decades and 35 M. oryzae isolates collected from those diseased plants were analyzed by PCR using gene-specific markers of the nine R genes and six primer pairs targeting the coding sequence or promoter of AVR genes, respectively. The R genes Pigm, Pi9, Pi2, Piz-t, Pi-ta, Pik, Pi1, Pikp, and Pikm were identified in 5, 0, 1, 4, 18, 0, 2, 1, and 0 cultivars, respectively. Significantly, none of these R genes had significant changes that correlated to their application periods of time. Among the four identified AVR genes, AVR-Pik had the highest amplification frequency (97.14%) followed by AVR-Pita (51.43%) and AVR-Pi9 (48.57%); AVR-Piz-t had the lowest frequency (28.57%). All these AVR genes except AVR-Pi9 had 1–2 variants. Inoculation mono-genic lines contained functional genes of Pi2/9 and Pik loci with 14 representative isolates from those 35 ones revealed that the presence of certain AVR-Piz-t, AVR-Pita variants, and AVR-Pik-E + AVR-Pik-D in M. oryzae populations, and these variants negated the ability of the corresponding R genes to confer resistance. Importantly, Pi2, Pi9, and Pigm conferred broad-spectrum resistance to these local isolates. These findings reveal that the complex genetic basis of M. oryzae and some effective blast R genes should be considered in future rice blast-resistance breeding programs.
Introduction
Rice is the main staple food crop for more than 50% of the world population. Rice blast disease, caused by Magnaporthe oryzae B.C. Couch, is a major threat to rice production in most rice-planting regions. The most efficient tool for rice blast disease control is the implementation of host R genes (Velent and Chumley, 1991). In principle, each rice R gene has a cognate AVR gene in M. oryzae (Flor, 1971), the utilization of R genes in agriculture has a selective effect on the corresponding AVR genes, leading to the rapid evolution of fitness of AVR genes toward virulence to their respective R genes. Thus, the identification and utilization of R genes that confer broad-spectrum resistance against a large number of pathogen races is the most effective approach to manage the disease.
To date, more than 100 blast R genes have been identified, and at least 28 have been cloned and characterized (Deng et al., 2017; Meng et al., 2021). Pigm, Pi9, Pi2, Piz-t in the Pi2/9 locus, and Pik, Pi1, Pikp, and Pikm in the Pik locus, which are well-known broad-spectrum resistance NBS-LRR (nucleotide-binding site-leucine-rich repeat) genes with good resistance against M. oryzae in rice-planting region of southern China (Tian et al., 2020, 2021). With respect to the cognate AVR genes, a total of 12 have been cloned: PWL1 (Kang et al., 1995), AVR-Pita (Orbach et al., 2000), AVR-Pib (Zhang et al., 2015), AVR1-Co39 (Farman et al., 2002), PWL2, ACE1 (Böhnert et al., 2004), AVR-Piz-t (Li et al., 2019), AVR-Pii, AVR-Pia, AVR-Pik/km/kp (Yoshida et al., 2009), AVR-Pi54 (Ray et al., 2016), and AVR-Pi9 (Wu et al., 2015). In general, there are three modes by which R genes recognize their respective AVR partners. In the first mode, a single R gene recognizes a single AVR gene, e.g., Pi-ta recognizes AVR-Pita (Jia et al., 2000). In the second mode, a single R gene recognizes two AVR genes, such as Pia/AVR1-Co39 and Pia/AVR-Pia (Farman et al., 2002; Yoshida et al., 2009; Okuyama et al., 2011; Cesari et al., 2013). In the third mode, multiple R alleles recognize multiple AVR alleles, e.g., the six haplotypes (A–F) of the AVR-Pik allele in different isolates can be recognized by Pik/kp/km/ks/kh, Pik/kp/km/kh, and Pik/km/kh, etc (Yoshida et al., 2009; Kanzaki et al., 2012; Wu et al., 2014; Hu et al., 2022). In fact, a rice variety with a particular R gene may lose its resistance if a mutation(s) occurs in the corresponding AVR gene (Lee and Cho, 1990). Particularly, three AVR genes, namely AVR-Pita, AVR-Pik, and AVR-Piz-t, have a relatively large number of polymorphisms, including point mutations, transposon insertions, and partial or total gene deletions, and hence usually escape recognition by the corresponding R genes in rice (Dean et al., 2005; Zhou et al., 2007; Chuma et al., 2011; Huang et al., 2014; Zhang et al., 2015). For example, Wang et al. (2020) identified 11 haplotypes encoding three new AVR-Piz-t variants in 100 isolates among 312 isolates of M. oryzae in Yunnan, China, including a 39-bp deletion and a 192-bp solo-LTR (long terminal repeat) homolog of the retrotransposon inago2 that had inserted into the promoter region and created a frameshift in the coding sequence. Each of these genetic alterations pushed AVR-Piz-t to a virulent form. Additionally, Meng et al. (2020) identified 5 virulent isolates of AVR-Pita from 202 isolates of M. oryzae in Heilongjiang, China. They also found 7 AVR-Pik alleles and 4 AVR-Piz-t variants among another 335 isolates of M. oryzae in Heilongjiang, China, leading to the loss of function of the corresponding R genes (Meng et al., 2019). In contrast, variants of AVR-Pia and AVR-Co39, which recognize the same R gene Pia, mostly are deletion mutants and thus can be easily lost among isolates of M. oryzae (Yoshida et al., 2009; Zheng et al., 2011).
The complex rice-growing environment in southern of China has resulted in a rich genetic diversity and complexity of rice blast fungus, and thus, the disease is prone to recurrence. Therefore, the local varieties as well as those that have come from breeding programs must contain cognate R genes to ensure their survival (Tian et al., 2020). To date, no study has correlated the evolution of AVR genes in M. oryzae with the application of R genes in rice. Therefore, to identify effective R genes in the specific rice-growing region, we utilized 117 representative cultivars from the main rice-planting regions in South China over the past 50 years to carry out a field evaluation of resistance to rice blast disease and characterize the genetic basis of R genes in these rice cultivars and AVR genes. The results provide a reference for rational distribution of rice blast R genes.
Materials and methods
Rice cultivars and Magnaporthe oryzae isolates
The cultivars comprised 117 Oryza sativa accessions and positive controls including IRBLzt-t (Piz-t), C101A51 (Pi2), IRBL9-W/75–1-127 (Pi9), Gumei 4 (Pigm), IRBLta-K1 (Pi-ta), IRBLZK-Ka (Pik), IRBLZKp-K60 (Pikp), IRBL1-CL (Pi1), and IRBLKM-Ts (Pikm), IRBLkh-K3 (Pikh), and IRBLks-F5 (Piks) collected from the Fujian Provincial Key Laboratory of Genetic Engineering for Agriculture, Fujian Academy of Agricultural Sciences, Fuzhou, China. The 117 cultivars comprised 27 three-line male sterility lines, 11 two-lined male sterility lines, and 79 restorer lines or conventional varieties. Forty-six accessions were bred in the 2000s, 19 before the 1980s, 16 in the 1980s, 28 in the 1990s, and 8 in the 2010s. These selected cultivars were from large acreages planted in the past several decades. Supplementary Table 1 lists detailed information regarding the accessions.
A total of 35 M. oryzae isolates were purified from leaf-diseased of 13 diseased rice cultivars at the mature stage in Shanghang, China, in 2017. Isolate purification was achieved with a single-spore isolation method (Jia et al., 2000). Subsequently, the purified mycelia derived from a single spore were grown on the surface of oatmeal agar medium. The mycelia and spores were allowed to colonize the entire filter paper. Each filter paper with the fungus was then transferred to a distilled envelope and stored at −20°C.
Field and greenhouse evaluation of blast resistance
The 117 cultivars were planted in the disease nursery beds with 20 plants per plot; the highly susceptible variety Guangluai 4 was sown on the plot borders. Disease severity was scored using leaves of seedlings and at the maximum tillering stage using a 1–9 scale: 0, no symptoms, i.e., a highly resistant (HR) phenotype; 1 and 2, only very small spots on leaves, indicating a resistant (R) phenotype; 3, small elliptical lesions, indicating a moderately resistant (MR) phenotype; 4, an elliptical lesion area of <2% of the entire leaf, indicating a moderately susceptible (MS) phenotype; 5, lesion area < 10%, indicating a susceptible (S) phenotype; 6 and 7, lesion area 10–25% or 26–50%, respectively, indicating susceptible phenotype; 8 and 9, lesion area 51–75% or > 75%, respectively, indicating highly susceptible (HS) phenotype. Scale values of 0, 1, 3, 5, 7, and 9 were used to score disease phenotype on spikes at maturity: 0, no evidence of infection; 1, diseased spike rate of <5%; 3, rate 5%–10%; 5, rate 10%–25%; 7, rate 25%–50%; and 9, rate 50%–100%. The comprehensive resistance evaluation was scored as 25%, 25%, and 50% of the diseased scoring at the seedling stage, maximum tillering stage, and maturity, respectively.
The greenhouse evaluation of blast resistance is shown by Tian et al. (2016). Briefly, the M. oryzae isolates were prepared in the dark for approximately 5 to 7 days at 28°C, and then under light for 5 to 7 days at room temperature for sporulation. Twenty rice seedlings of each variety were grown in a greenhouse for about 2 weeks under natural conditions and then were spray inoculated with spores at a concentration of 5 × 105 spores mL−1 until run-off. After inoculation, seedlings were maintained in the dark for 24 h and then kept under high humidity for 1 week to evaluate their symptoms. The disease score standard scale is above described. The recurrent parent variety Co39 was susceptibility control.
DNA extraction, marker evaluation, and genotyping
DNA from rice plant leaves were isolated using the cetyltrimethylammonium bromide (CTAB) method with some modifications (Tai and Tanksley, 1990). DNA from M. oryzae was extracted using the DNeasy Plant mini kit (QIAGEN, Germantown, MD). Six major AVR genes and nine R genes were validated with the markers listed in Supplementary Table 2. PCR amplification was carried out in a 25 μl reaction volume containing 20–200 ng template DNA, 1 μl of each forward and reverse primers (10 mM), 12.5 μl 2 × Mix buffer (Mg2+plus), 0.2 μl Taqase (5 U/ml), and 10.0 ul ddH2O. The PCR program was set up as follows: initial denaturation of 5 min at 94°C; followed by 32 cycles of denaturation for 30 s at 94°C, primers annealing for 30 s at varied temperatures according to the specific marker, and extension for 30 s at 72°C, followed by a final extension for 10 min at 72°C. For scoring the marker loci, the amplified PCR products of markers Pi9-Pro, Pikp-Del, Pi1-In, and Sepigm-4 were separated by 8% polyacrylamide gel electrophoresis along with a DL500 DNA ladder stained with silver staining. After electrophoresis, the gels were documented under UV using gel documentation system (AlphaImager, United States). PCR amplicons from the CAPS markers 2-LRR, PikFNP, and PikmFNP were digested with the restriction enzyme PstI, KpnI, and MboI, respectively. The digested products were separated by electrophoresis with 8% polyacrylamide gels with subsequent visualization by silver staining. PCR products from M. oryzae were sequenced using an ABI3730 XL automatic DNA sequencer. DNA sequences were aligned using Clustal Omega (Sievers et al., 2011). Those PCR products with double-peak sequences were cloned into pMD18-T Vector (Takara, Japan) for further analysis. Figure 1 and Supplementary Table 2 list detailed information regarding the markers.
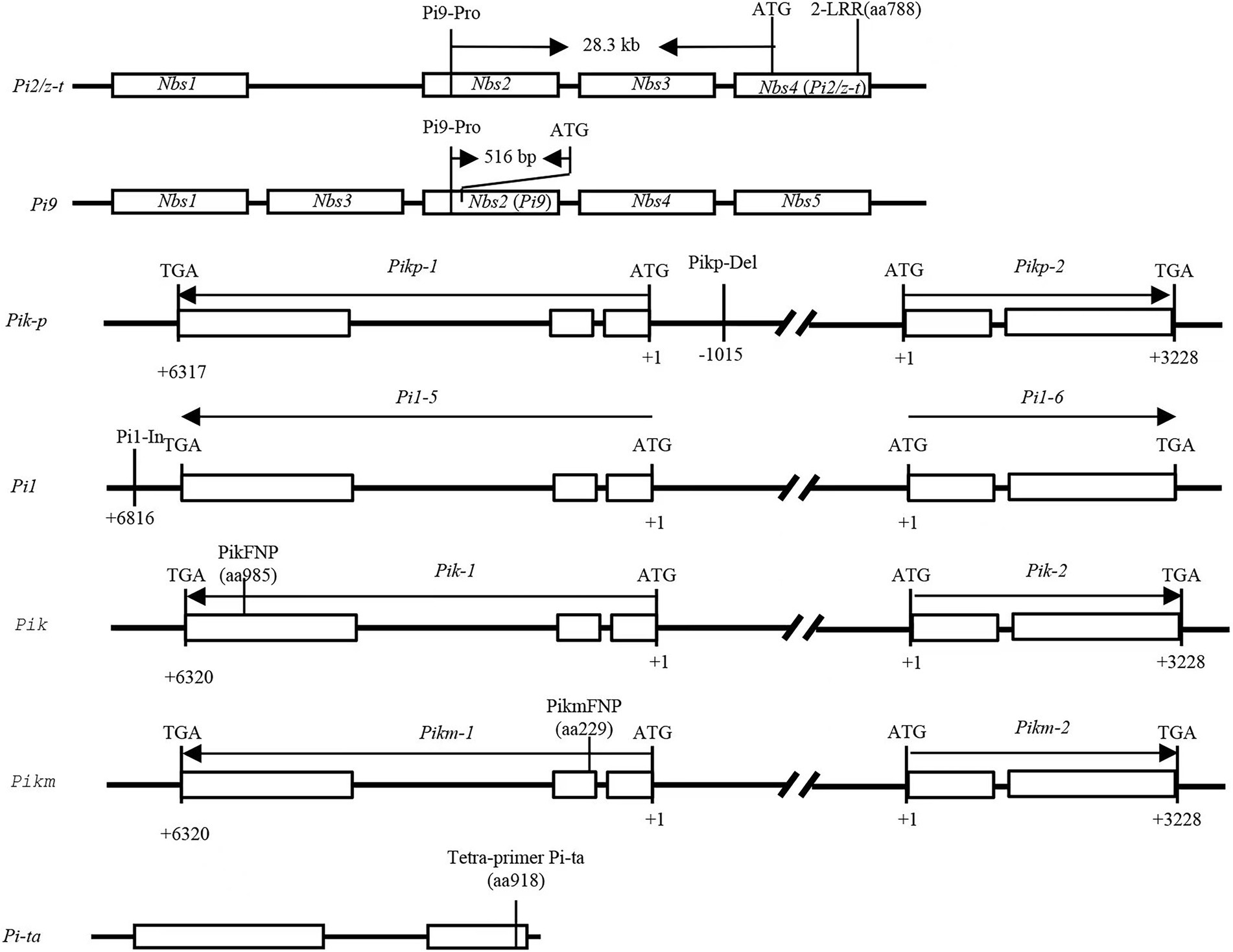
Figure 1. The physical maps of nine genes and their molecular markers in the study. The position of the nine molecular markers on the corresponding genes.
Results
Correlation between the field blast resistance and the bred periods of the 117 rice cultivars
A total of 117 rice cultivars were collected from the major rice-producing areas in China: 17 were from Guangdong, 23 from Fujian, 22 from Hunan, 6 from Jiangsu, 4 from Guangxi, 25 from Sichuan, 12 from Zhejiang, and 8 from other areas (Supplementary Table 1). These cultivars were evaluated for resistance to rice blast disease under field conditions in Shanghang, Fujian.
Figure 2 displays the distribution of blast disease severity scores from the field evaluation. The Pearson’s correlation values between the seedling stage and maximum tillering stage, seedling stage and fully ripe stage, and maximum tillering stage and fully ripe stage were 0.907, 0.596, and 0.604, respectively (p-value < 0.01; Supplementary Table 1), indicating a close relationship among the data acquired for the seedling stage and maximum tillering stage. The S, MS, and HS accessions were evenly distributed in every period of time, and the number of R, MR, MS, S, and HS cultivars changed from 6 to 62 with a rising tendency. Combining the results of blast disease severity and bred periods of time, there was no significant correlation between them (Figure 2). The percentage of MR, R, and HR cultivars changed from 5.26%, 12.5%, 3.57, 21.74%, and 0 across the past five decades with a random tendency (Supplementary Table 1), indicating that blast resistance among these rice cultivars has not obviously improved over time.
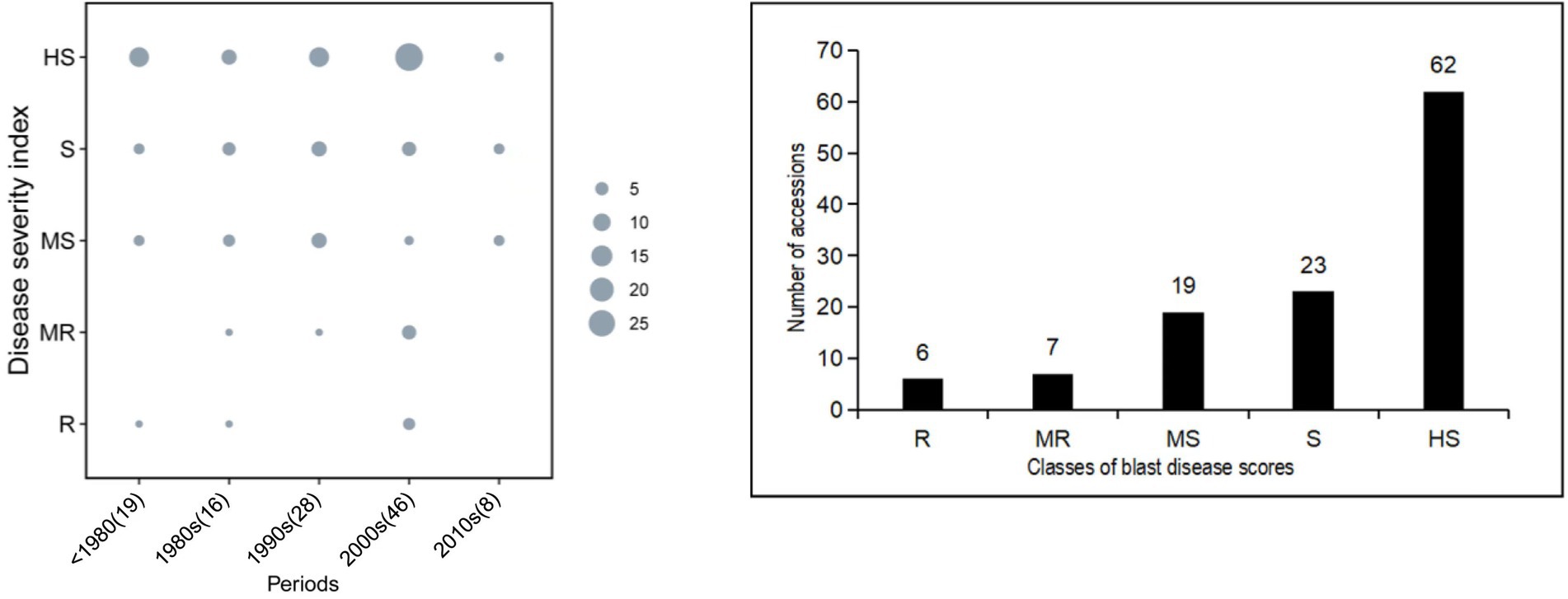
Figure 2. Distribution of blast-resistance scores for the 117 cultivars in the field test. Period: A, >1980; B, 1980s; C, 1990s, D, 2000s; E, 2010s. HR, highly resistant; R, resistant; MR, moderately resistant; MS, moderately susceptible; S, susceptible; HS, highly susceptible.
The identification and distribution of nine rice blast R genes across the periods of time
Specific markers for R genes Piz-t/Pi2, Pi9, Pi1, Pigm, Pik-p, Pik, Pik-m, and Pit-a are shown in Figure 1. The frequency of the 9 major rice blast R genes ranged from 0 to 18 among the 117 cultivars (Figure 3). The presence of Pi9 and Piz-t/Pi2 was determined based on amplicons of 128 bp and 111 bp, respectively, using the allele-specific marker Pi9-Pro along with the positive control IRBL9-W/75–1-127 as well as IRBLzt-t/C101A51. Pi9 was absent in all 117 cultivars. The PCR results for Piz-t and Pi2 revealed amplicons of 439 and 399 bp, respectively, corresponding to the respective positive controls IRBLZT-t and C101A51. The results revealed the presence of Pi2 in one variety and Piz-t in three cultivars (Figure 3). Additionally, five cultivars Minbeiwanxian, Guangkang 13A, Gufeng A, Fuyi A, and Guanghui 128 were identified to contain Pigm (Zeng et al., 2018). For the Pik locus, the presence of Pik, Pikp, and Pikm was determined using the dCAPS markers T1-2944G and T1-786A/G plus the markers A2-1879G and C1-685A/G corresponding to the positive control IRBLZK-Ka as well as IRBLZKp-k60 and IRBLKm-Ts, respectively (Zhai et al., 2011). Furthermore, two cultivars RGD-7S and Jinkang1A contained Pi1, and only one variety of Huifeng A had Pikp (Figure 3), and all other cultivars were negative for both Pik and Pikm. The presence of Pi-ta was determined based on amplicons of 286 and 406 bp (Zhang et al., 2013), which correspond to the positive control IRBLta-K1. Pi-ta was found in 18 cultivars with 286/406 bp (Figure 3). Taken together, the two cultivars Huazhan and R2115 possessed 2 of the positive alleles out of 6 R genes, and 30 cultivars had positive bands for a single R gene.
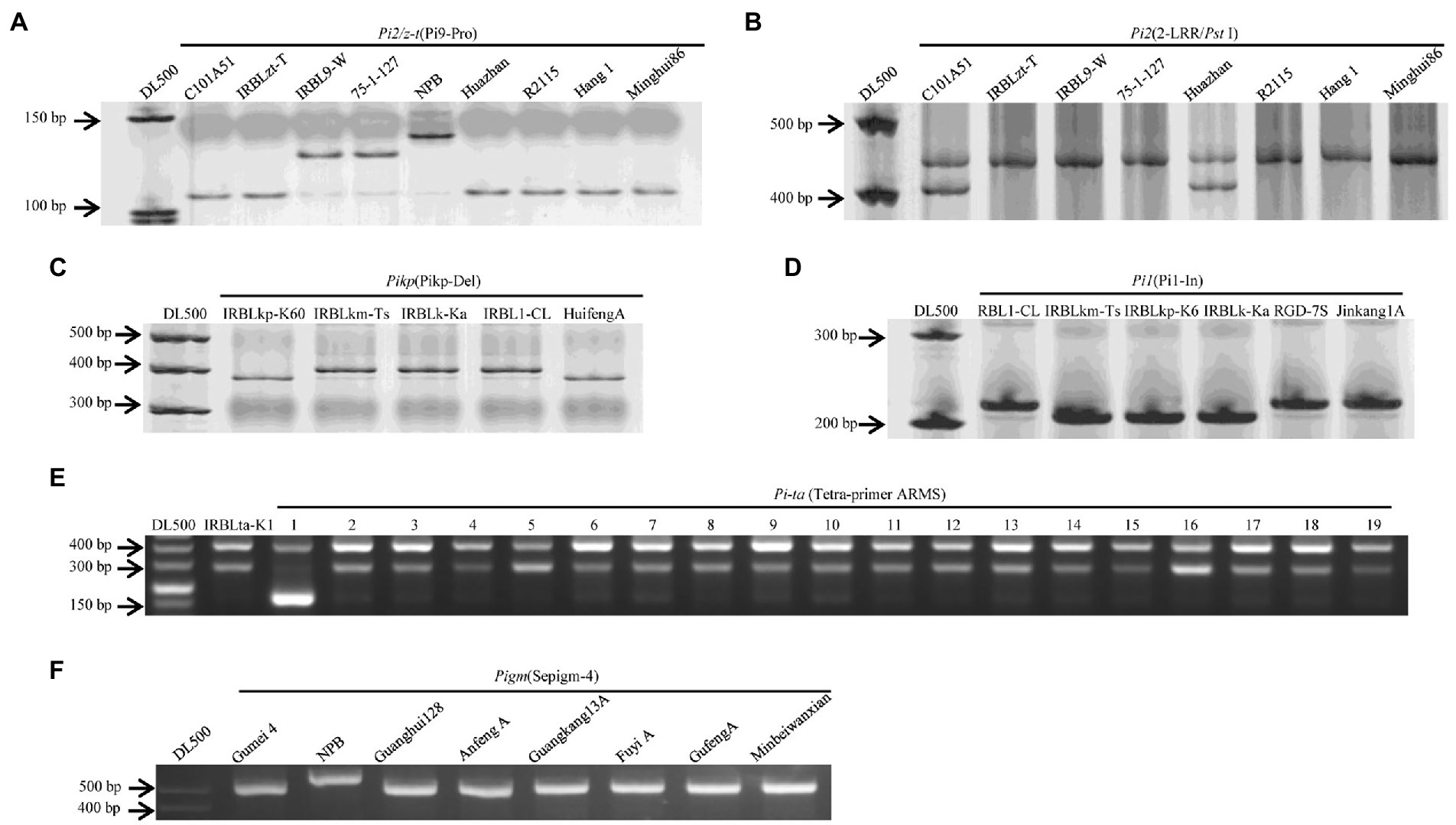
Figure 3. Electrophoretic gel of nine genes containing in the 117 cultivars. E, Number in Pi-ta: Positive control, IRBLta-K1; 1, NPB: Nipponbare; 2, Wanlixian; 3, Shuhui527; 4, Huazhan; 5, Minhui 3301; 6, Jiayu293; 7, RGD-7S; 8, Nanhui 511; 9, Shuhui 316; 10, Shen 08S; 11, Zhefu 802; 12, Duoxi1; 13,Ezao11; 14, Minghui82; 15, Shuhui 881; 16, Zhongxiang 1; 17, Chenghui 3203; 18, Lehui 188; 19, R2155. The other gene-containing cultivars in A–D and F are showen in gel.
Through identifying the 117 cultivars based on the nine R genes specific primer sets, Pi-ta, Pigm, Piz-t, Pik-p, Pi2, and Pi1 were revealed to present in 18, 6, 3, 1, 1, and 2 cultivars, respectively (Figure 3). Pi-ta and Pigm were distributed at a comparable level in these periods among the identified genes, further to find the cultivars in the 2000s had the most number of R genes among the five periods. However, the irregular application of R genes was present in modern cultivars at low frequency, suggesting that they just could be randomly utilized in breeding program.
Haplotypes variance of six AVR genes in the Magnaporthe oryzae isolates corresponding to R genes
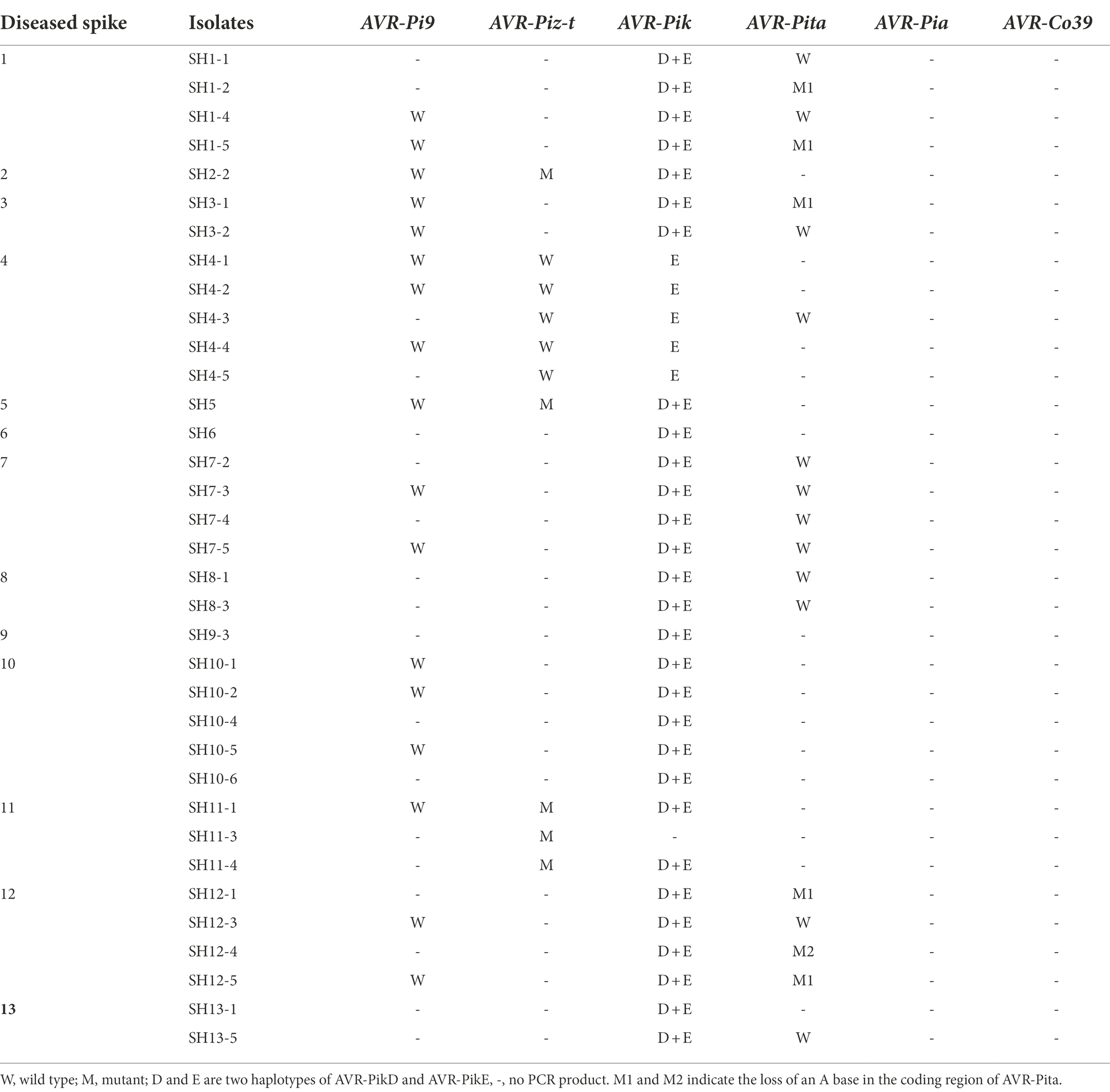
The haplotypes of six AVR genes were determined using PCR and amplicon sequencing. PCR amplification utilized primer pairs (Supplementary Table 2) targeting the coding and promoter regions of the AVR genes, revealing three patterns: amplification yielded a product with the expected size, the unexpected size, or no amplification. AVR-Pik was amplified in 34 of these isolates, and the AVR-Pita, AVR-Piz-t, and AVR-Pi9 fragments were present in 18, 10, and 17 isolates, respectively. Neither AVR-Pia nor AVR1-Co39 was amplified in these isolates. Amplicon sequencing revealed that the AVR-Pi9 sequence was identical to that of the avirulent strain KM004023, as described previously (Wu et al., 2015). The two haplotypes of AVR-Pik, namely AVR-Pik-E (AB498879) and its combination with AVR-Pik-D (AB498875), were identified in 4 and 30 isolates, respectively (Figure 4). Notably, AVR-Pik-E was identified in 4 isolates of the same diseased plant (Table 1). The AVR-Pita sequences in 18 isolates were compared with that of AVR-Pita in an avirulent strain (AF207841), revealing a loss of two nucleotides in the coding region (301A, and 333A) in two isolates (Figure 4; Table 1) that resulted in the loss of function of the avirulence effect of AVR-Pita. The inclusion of the promoter region in the PCR validation assay enabled the identification of a solo-LTR (Inagos homolog) insertion in the promoter of five AVR-Piz-t genes (Figure 4; Table 1).
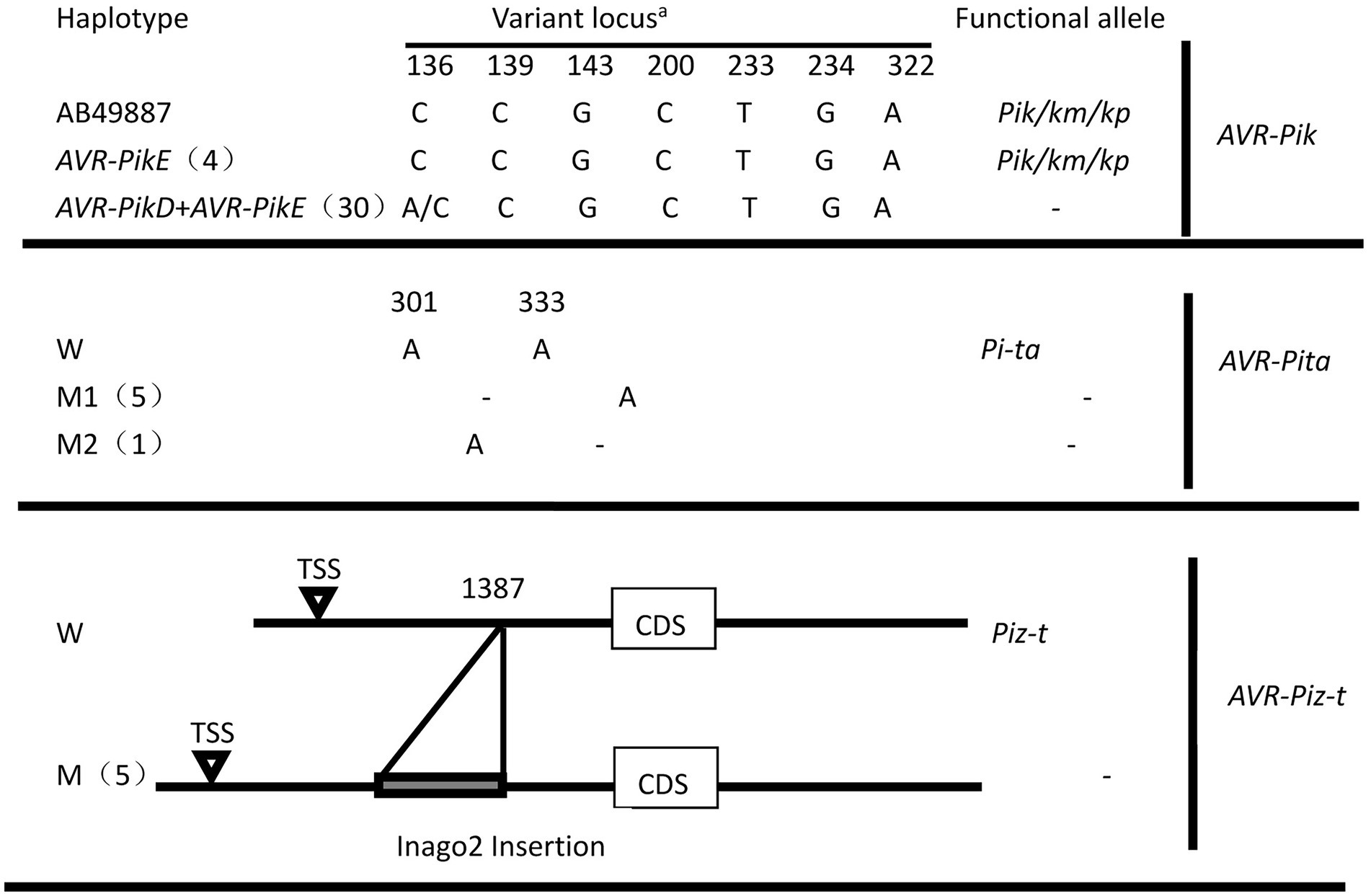
Figure 4. Characterization of allelic variations at AVR-Pik, AVR-Pita, and AVR-Piz-t. aThe positions were based on the reference sequence of AVR-PikD (AB498875), AVR-PikE (AB498879), AVR-Pita (AF207841), and AVR-Piz-t(EU837058). -, avirulent genes without resistance genes; W, wide type; M, M1, and M2, mutant type.
In total, 2, 1, and 1 novel variants were found in AVR-Pita, AVR-Pik, and AVR-Piz-t haplotypes, respectively (Figure 4), among which the 2 new haplotypes of AVR-Pita were distributed in 5 and 1 isolates, respectively. The new haplotypes of AVR-Piz-t were comprised 5 isolates of 3 diseased rice plants. Whereas the combination of AVR-Pik-D + AVR-Pik-E of AVR-Pik was comprised 85.7% isolates and widely distributed in 12 diseased rice plants (Table 1).
Association analysis between the AVR genotypes in Magnaporthe oryzae and the field resistance of cultivars containing cognate R genes in rice
As the pathogenicity of the isolates depends on the haplotypes of both the AVR genes and corresponding R genes, field blast resistance of these 117 cultivars was identified in the disease nursery bed in Shanghang. All of these cultivars were scored for resistance in 2017. The correlation between the haplotypes of AVR genes and the field resistance of those cultivars containing cognate R genes was analyzed. Nearly all of the rice cultivars containing Piz-t or Pi-ta were susceptible to the field isolates in the test, it was possible that these variants of AVR-Piz-t and AVR-Pita led to loss of function of Piz-t and Pi-ta, respectively. Whereas 3 Pik cultivars that contain Pi1 and Pik-p were HS to MR. In contrast, those cultivars possessing Pi2 and Pigm exhibited good resistance to the M. oryzae isolates from the Fujian fields (Supplementary Table 2). Additionally, a number of cultivars such as Chenghui 178 and Neihui 99-14 that lacked a identified R gene expressed resistance, which suggests that other blast R genes in these cultivars may play important roles in resisting blast (Supplementary Table 1).
Disease reaction of mono-genic lines against the local field blast isolates
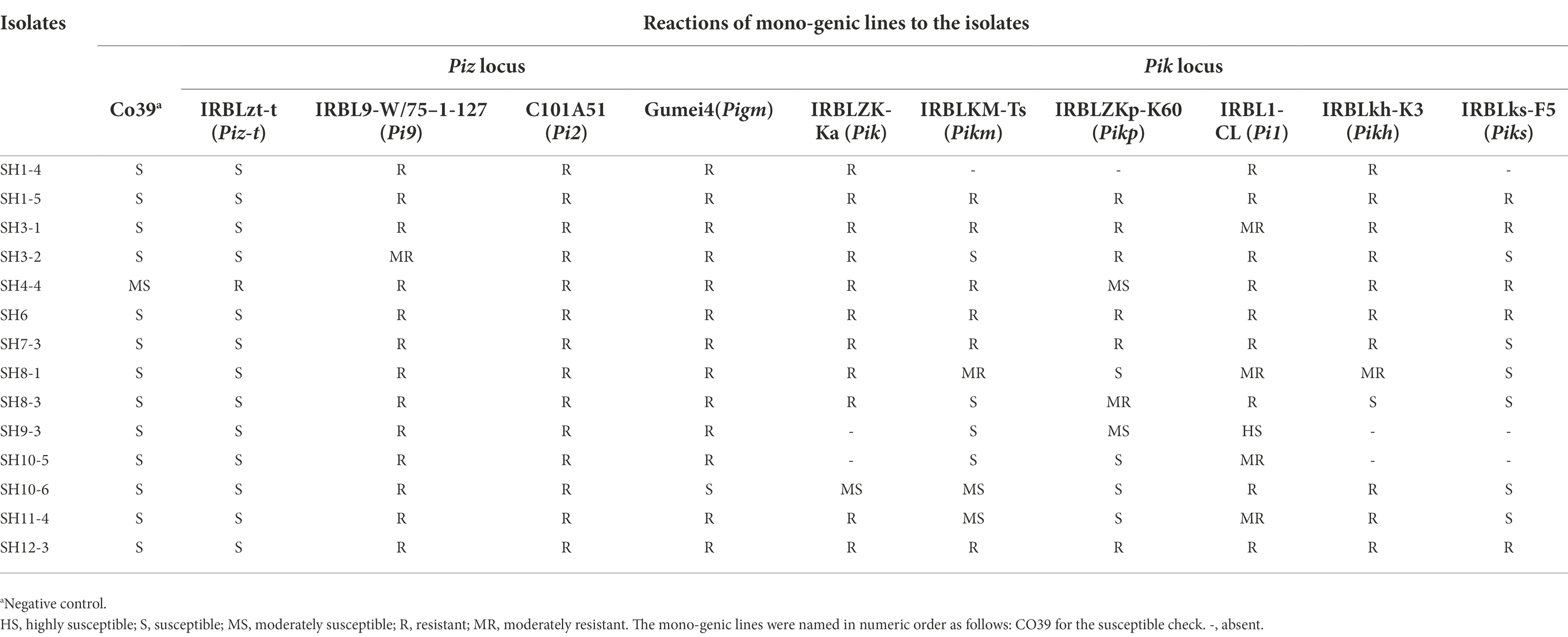
The above data show that those cultivars containing Pi2, Pi9, and Pigm express broad-spectrum resistance in the field test, and few of functional genes in Pik locus were existed in these materials. Thus, 10 mono-genic lines containing these R genes and Co39 were inoculated with 14 local blast isolates, including isolates with AVR-Piz-t, AVR-Pik-E, AVR-Pik-D + AVR-Pik-E, and AVR-Pi9, etc. The isolate of SH4-4 containing AVR-Piz-t and AVR-Pik-E showed avirulence to Piz-t and Pik/km/kh in the pathotype test (Table 2), reiterating a perfect correlation between the existence of AVR and their cognate R genes. However, those mono-genic lines containing Pi2, Pi9, and Pigm were resistant to all of these isolates except that Gumei 4 was susceptible to SH10-6, suggesting that these three R genes express broad-spectrum/high resistance (Table 2). Additionally, as at least 1 isolate containing AVR-Pik-D + AVR-Pik-E exhibited MS, S, or HS to those mono-genic lines containing functional alleles of the Pik locus, suggesting that AVR-Pik-D + AVR-Pik-E was virulence to these genes (Table 2).
Discussion
Rice is one of the most important food crops in Fujian, accounting for 78% of the planting area and 80% of the total grain output (Huang et al., 2012). The diverse geography and variable climate of the Fujian region have stimulated the adaptive variation of M. oryzae in rice fields, and R genes in rice varieties have influenced the formation of the variation of M. oryzae genotypes (Selisana et al., 2017). Thus, monitoring the R genes in rice cultivars and AVR genotypes of M. oryzae is critical for effective utilization of R genes to improve rice yields. However, little effort has been directed toward the simultaneous characterization of both R genes of rice and AVR genes of M. oryzae (Kolmer and Hughes, 2018).
Conventional pathogenicity analyses of M. oryzae populations and the evaluation of resistance levels of rice varieties have been carried out in blast nurseries or by pathotyping in a greenhouse, and differential varieties have been derived from cultivars LTH and Co39-IRTL (Telebanco-Yanoria et al., 2008), but such analyses are both time-consuming and laborious (Vasudevan et al., 2014). Recently, PCR-based gene diagnosis has been applied to infer the effectiveness of R genes, and this approach has been shown to be suitable for large-scale pathogen identification because the PCR method does not require live M. oryzae strains (Imam et al., 2015). The present study categorized 117 cultivars into five group at 10-year level of planting period. Our results showed that the genotype distributions for nine R genes were randomly distributed in these periods based on their corresponding markers (Figures 1, 3). As a matter of fact, most of the rice cultivars in the early decades possessed few R genes, and although more recent cultivars such as in 2000s possess more R genes, all express similar resistance to the field isolates as shown in Figure 2. The smaller class of blast disease scores HR consists of six cultivars, whereas the major group HS included 62 ones (Figure 2-right). Accordingly, avirulence genes of blast fungal were differentiated into wild and mutant genotypes by using sequencing analysis (Table 1). The corresponding disease reaction in mono-genic lines was found concurrent with the avirulent and virulent genes, respectively (Table 2). The consistent results in the study showed by the individual markers for selected blast-resistance genes and AVR genes make them a suitable marker for genotyping of rice blast resistant genes in the rice germplasm and avirulence genes in the blast fungal, respectively.
Some previous studies have revealed that the rice blast pathogen population, which are related to the divergence of AVR genes that have been shaped by changing environmental conditions, can have a clear impact on R gene-mediated resistance to blast disease (Li et al., 2019). Our data demonstrate that several mutational events were brought about at the AVR-Pita and AVR-Piz-t loci, leading to those cultivars containing Pi-ta and Piz-t exhibited susceptible to the field isolates. Additionally, previous studies revealed that AVR-Pita alleles are highly diversified, and transposon insertions, point mutations, frame-shift mutations, partial or complete deletions, and sequence variations in the coding or promoter region caused a change from avirulence to virulence of AVR genes (Xing et al., 2013; Chen et al., 2014). Similarly, the loss of function of AVR-Piz-t was also observed in 312 isolates collected from the rice-producing areas of Yunnan, and a 198-bp insertion homologous to solo-LTR of the retrotransposon inago2 in the promoter region of AVR-Piz-t in one isolate was found to have evolved from avirulence to virulence (Wang et al., 2020). Meantime, the changes of virulence frequencies of M. oryzae for Pi-ta and Piz-t happened in different years, suggesting that the disease reaction of rice cultivars were changing overtime (Shen, 2018).
Even though Pikm, Pikh, Pikp, Pik, and Pi1 exhibited a high level of resistance to blast fungus from Fujian, Guangdong, Sichuan provinces and Japanese, etc. (Yang et al., 2008; Wang et al., 2017; Zhang et al., 2017; Tian et al., 2021), the stepwise evolution with the AVR-Pik allele found in the United States, Philippines, and Japanese isolates populations and attempt to overcome them (Raffaele et al., 2010; Kanzaki et al., 2012; Selisana et al., 2017; Wang et al., 2017). To date, six AVR-Pik variants (A–F) have been identified, and they differ in just five amino acid positions. The generation and diversity of mutational events may be influenced by introducing an R gene in new rice varieties (Zhou et al., 2007; Daverdin et al., 2012). M. oryzae isolates carrying AVR-PikD trigger immune responses in rice lines containing Pikp/km/kh/Pik, isolates carrying AVR-PikE only elicit a response in rice lines with Pikm/kh. AVR-Pik has evolved via gene duplication and substitution mutations in the coding regions of AVR-Pik/km/kp in M. oryzae populations (Wu et al., 2014; Li et al., 2019; Longya et al., 2019). However, no known Pik alleles respond to the combination effectors of AVR-PikD + AVR-PikE identified in the present study. Subsequently, the corresponding rice R genes may have become blind to the new genotypes (Kanzaki et al., 2012). Consistent with these results, most of those mono-genic lines carrying Pikm/kp/ks/1 were partly susceptible to those isolates containing AVR-PikD + AVR-PikE; however, whether the combination of AVR-PikD + AVR-PikE would affect gene function needs to be further validated by transcript or translation level.
At least six functional genes, including Pigm, Pi2, Pi9, and Piz-t, reside within the Pi2/9 locus. In contrast to the fact that Piz-t exhibited relatively lower resistance to M. oryzae in Fujian, China, Pigm, Pi9, and Pi2 have been shown to confer higher and broad-spectrum resistance against a wide range of blast isolates present in this planting region (Jiang et al., 2012; Tian et al., 2016, 2020). Previous studies showed that Piz-t was the first widely used in the 1970s (Fjellstrom et al., 2006), but with the evolution of AVR-Piz-t, the gene has lost its resistance (Marchetti et al., 1987; Conaway-Bormans et al., 2003; Tian et al., 2016); whereas Pi2 and Pigm were distributed in recent rice varieties from Fujian and Guangdong as they exhibited good resistance to rice blast from the two provinces (Tian et al., 2020); Additionally, Vasudevan et al. (2014) identified 289 accessions containing Pi2 from a total collection of over 120,000 ones originating from 13 major rice-growing countries showed broad-spectrum resistance against all five single rice blast isolates. In contrast, Pi9 is not widely utilized in Chinese rice varieties. In our study, the rice cultivars with Pi2, Pi9, and Pigm can be classified R or MR, whereas those cultivars containing Piz-t were relatively more susceptible to disease. Further to find that those mono-genic lines carrying Pi9, Pi2, or Pigm exhibited good resistance to the local isolates. Intriguingly, the association between the AVR gene(s) and the disease reaction was not completely understood in our study, such as some isolates containing AVR-Pi9 show negative band by gene amplification, which could be explained by addition of more number of markers needs to be tested for new R genes or Avr genes. All these aforementioned findings provide valuable insights pertaining to the selection of desirable R genes-particularly those with durable resistance.
Conclusion
In the study of nine R genes in 117 representative cultivars over past 5 decade years and six cognate AVR genes in 35 M. oryzae isolates. Nine R genes have not been conscious utilization in these periods of time. The three observed variants of AVR-Pita, AVR-Piz-t, and AVR-PikD + AVR-PikE confer virulence to their cognate R genes, which may be explained by the rapid evolution of local M. oryzae isolate in respond to these genes. Our results also indicated that effective control of blast disease will require the functional allele of Pi2/9 locus.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
DT conceived the initial screening and research plans. DT, LY, and HZ designed the experiments. YD, XY, GL, QL, ZC, XG, and YS performed most of the experiments and analyzed the data. DT wrote the manuscript with the contributions of all the authors. All authors contributed to the article and approved the submitted version.
Funding
The work was funded by the Key Program of the National Natural Science of Fujian province (2022J02010), the Youth Program of Fujian Academy of Agricultural Sciences (YC2019004), the ‘5511’ Collaborative Innovation project for High-quality Development and Surpasses of Agriculture between Government of Fujian Province and Chinese Academy of Agricultural Sciences (Grant no. XTCXGC2021002), the Opening Foundation of Jiangsu Key Laboratory for Eco-Agricultural Biotechnology around Hongze Lake (HZHLAB2101), and the Natural Science Foundation of Jiangsu Provincial Department of Education (17KJA180002 and 19KJB180011).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YD declared a shared affiliation with several of the authors, DT, XY, ZC, and XG, to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1007492/full#supplementary-material
Supplementary Table 1 | Primers used for PCR diagnosis of avirulence and resistance genes.
Supplementary Table 2 | List of 117 cultivars bred over five decades periods of time that were used for field evaluation of M. oryzae.
References
Böhnert, H. U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J. L., and Lebrun, M. H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513. doi: 10.1105/tpc.104.022715
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. The Plant cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Chen, C., Chen, M., Hu, J., Zhang, W., Zhong, Z., Jia, Y., et al. (2014). Sequence variation and recognition specificity of the avirulence gene AvrPiz-t in Magnaporthe oryzae field populations. Fungal Genom. Biol. 4:113. doi: 10.4172/2165-8056.1000113
Chuma, I., Isobe, C., Hotta, Y., Ibaragi, K., Futamata, N., Kusaba, M., et al. (2011). Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7:e1002147. doi: 10.1371/journal.ppat.1002147
Conaway-Bormans, C. A., Marchetti, M. A., Johnson, C. W., McClung, A. M., and Park, W. D. (2003). Molecular markers linked to the blast resistance gene pi-z in rice for use in marker-assisted selection. Theoret. Appl. Genet. 107, 1014–1020. doi: 10.1007/s00122-003-1338-5
Daverdin, G., Rouxel, T., Gout, L., Aubertot, J. N., Fudal, I., Meyer, M., et al. (2012). Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathog. 8:e1003020. doi: 10.1371/journal.ppat.1003020
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Farman, M. L., Eto, Y., Nakao, T., Tosa, Y., Nakayashiki, H., Mayama, S., et al. (2002). Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant Microbe Interact. 15, 6–16. doi: 10.1094/MPMI.2002.15.1.6
Fjellstrom, R., McClung, A. M., and Shank, A. R. (2006). SSR markers closely linked to the pi-z locus are useful for selection of blast resistance in a broad Array of Rice germplasm. Mol. Breed. 17, 149–157. doi: 10.1007/s11032-005-4735-4
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Hu, Z. J., Huang, Y. Y., Lin, X. Y., Feng, H., Zhou, S. X., Xie, Y., et al. (2022). Loss and natural variations of blast fungal Avirulence genes breakdown Rice resistance genes in the Sichuan Basin of China. Front. Plant Sci. 13:788876. doi: 10.3389/fpls.2022.788876
Huang, J., Si, W., Deng, Q., Li, P., and Yang, S. (2014). Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. BMC Genet. 15:45. doi: 10.1186/1471-2156-15-45
Huang, T. X., Zheng, J. T., You, Q. R., Yang, D., Zhang, S. J., Dong, R. X., et al. (2012). Status and prospect of the research on the breeding of hybrid rice in Fujian province. Fujian J. Agric. Sci. 27, 312–318. doi: 10.3969/j.issn.1008-0384.2012.03.020
Imam, J., Alam, S., Mandal, N. P., Shukla, P., Sharma, T. R., and Variar, M. (2015). Molecular identification and virulence analysis of AVR genes in rice blast pathogen, Magnaporthe oryzae from eastern India. Euphytica 206, 21–31. doi: 10.1007/s10681-015-1465-5
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Jiang, N., Li, Z., Wu, J., Wang, Y., Wu, L., Wang, S., et al. (2012). Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice 5:29. doi: 10.1186/1939-8433-5-29
Kang, S., Sweigard, J. A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8, 939–948. doi: 10.1094/mpmi-8-0939
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Kolmer, J. A., and Hughes, M. E. (2018). Physiologic specialization of Puccinia triticina on wheat in the United States in 2016. Plant Dis. 102, 1066–1071. doi: 10.1094/PDIS-11-17-1701-SR
Lee, E., and Cho, S. Y. (1990). Variation in races of rice blast pathogen and varietal resistance in Korea. In: Int. Rice Res. Conf. International Rice Research Institute, Los Baños, Philippines.
Li, J., Wang, Q., Li, C., Bi, Y., Fu, X., and Wang, R. (2019). Novel haplotypes and networks of AVR-Pik alleles in Magnaporthe oryzae. BMC Plant Biol. 19:204. doi: 10.1186/s12870-019-1817-8
Longya, A., Chaipanya, C., Franceschetti, M., Maidment, J., Banfield, M. J., and Jantasuriyarat, C. (2019). Gene duplication and mutation in the emergence of a novel aggressive allele of the AVR-Pik effector in the Rice blast fungus. Mol. Plant Microbe Interact. 32, 740–749. doi: 10.1094/MPMI-09-18-0245-R
Marchetti, M. A., Lai, X., and Bollich, C. N. (1987). Inheritance of resistance to Pyricularia oryzae in rice cultivars grown in the United States. Phytopathology 77, 799–804. doi: 10.1094/Phyto-77-799
Meng, F., He, Y., Chen, J., Long, X., Wang, H., Zhu, M., et al. (2021). Analysis of natural variation of the rice blast resistance gene Pike and identification of a novel allele Pikg. Mol. Genet. Genomics 296, 939–952. doi: 10.1007/s00438-021-01795-w
Meng, F., Zhang, J. L., and Jin, X. H. (2020). Detection and analysis of Magnaporthe oryzae avirulent gene AVR-Pita and its homologous genes in Heilongjiang province. Chin. J. Rice Sci. 34, 143–149. doi: 10.16819/j.1001-7216.2020.9085, (In Chinese with English abstract)
Meng, F., Zhang, Y. L., Jin, X. H., Zhang, X. Y., and Jiang, J. (2019). Detection and analysis of Magnaporthe oryzae avirulence genes AVR-Pib, AVR-Pik and AvrPiz-t in Heilongjiang province. Sci. Agric. Sin. 52, 4262–4273. doi: 10.3864/j.issn.0578-1752.2019.23.007
Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, M., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. Cell Mol. Biol. 66, 467–479. doi: 10.1111/j.1365-313X.2011.04502.x
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., and Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene pi-ta. Plant Cell 12, 2019–2032. doi: 10.1105/tpc.12.11.2019
Raffaele, S., Farrer, R., Cano, L., Studholme, D., Maclean, D., Thines, M., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330, 1540–1543. doi: 10.1126/science.1193070
Ray, S., Singh, P. K., Gupta, D. K., Mahato, A. K., Sarkar, C., Rathour, R., et al. (2016). Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic Rice line containing resistance gene, Pi54. Front. Plant Sci. 7:1140. doi: 10.3389/fpls.2016.01140
Selisana, S. M., Yanoriam, M. J., Quime, B., Chaipanya, C., Lu, G., Opulencia, R., et al. (2017). Avirulence (AVR) gene-based diagnosis complements existing pathogen surveillance tools for effective deployment of resistance (R) genes against Rice blast disease. Phytopathology 107, 711–720. doi: 10.1094/PHYTO-12-16-0451-R
Shen, L. R. (2018). Analysis on population structure of Magnaporthe oryzae in Jiangsu province and functional analysis of melatonin synthase Moasmt2 in Magnaporthe oryzae (D). Jiangsu, Nanjing Agriculture University.
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Tai, T., and Tanksley, S. D. (1990). A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol. Biol. Report. 8, 297–303. doi: 10.1007/BF02668766
Telebanco-Yanoria, M. J., Ohsawa, R., Senoo, S., Kobayashi, N., and Fukuta, Y. (2008). Diversity analysis for resistance of rice (Oryza sativa L.) to blast disease Magnaporthe grisea (Hebert) Barr. using differential isolates from the Philippines. Plant Breeding 127, 355–363. doi: 10.1111/j.1439-0523.2008.01497.x
Tian, D., Chen, Z., Chen, Z., Zhou, Y., Wang, Z., Wang, F., et al. (2016). Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice 9:19. doi: 10.1186/s12284-016-0091-8
Tian, D., Chen, Z., Lin, Y., Chen, Z., Luo, J., Ji, P., et al. (2021). Two novel gene-specific markers at the Pik locus facilitate the application of rice blast resistant alleles in breeding. J. Integr. Agric. 20, 1554–1562. doi: 10.1016/s2095-3119(20)63272-5
Tian, D., Lin, Y., Chen, Z., Chen, Z., Yang, F., Wang, F., et al. (2020). Exploring the distribution of blast resistance alleles at the Pi2/9 locus in major Rice-producing areas of China by a novel Indel marker. Plant Dis. 104, 1932–1938. doi: 10.1094/PDIS-10-19-2187-RE
Vasudevan, K., Vera Cruz, C. M., Gruissem, W., and Bhullar, N. K. (2014). Large scale germplasm screening for identification of novel rice blast resistance sources. Front. Plant Sci. 5:505. doi: 10.3389/fpls.2014.00505
Velent, B., and Chumley, F. G. (1991). Molecular genetic analysis of the rice blast fungus Magnaporthe grisea. Annu. Rev. Phytopathol. 29, 443–467. doi: 10.1146/annurev.py.29.090191.002303
Wang, X., Jia, Y., Wamishe, Y., Jia, M. H., and Valent, B. (2017). Dynamic changes in the Rice blast population in the United States over six decades. Mol. Plant Microbe Interact. 30, 803–812. doi: 10.1094/MPMI-04-17-0101-R
Wang, Q., Li, J., Lu, L., He, C., and Li, C. (2020). Novel variation and evolution of AvrPiz-t of Magnaporthe oryzae in field isolates. Front. Genet. 11:746. doi: 10.3389/fgene.2020.00746
Wu, J., Kou, Y., Bao, J., Li, Y., Tang, M., Zhu, X., et al. (2015). Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 206, 1463–1475. doi: 10.1111/nph.13310
Wu, W., Wang, L., Zhang, S., Li, Z., Zhang, Y., Lin, F., et al. (2014). Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant Microbe Interact. 27, 759–769. doi: 10.1094/MPMI-02-14-0046-R
Xing, J., Jia, Y., Correll, J. C., Lee, F. N., Cartwright, R., Cao, M., et al. (2013). Analysis of genetic and molecular identity among field isolates of the Rice blast fungus with an international differential system, rep-PCR, and DNA sequencing. Plant Dis. 97, 491–495. doi: 10.1094/PDIS-04-12-0344-RE
Yang, J., Chen, S., Zeng, L., Li, Y., Chen, Z., and Zhu, X. (2008). Evaluation on resistance of major rice blast resistance genes to Magnaporthe grisea isolates collected from indica rice in Guangdong Province, China. Chin. J. Rice Sci. 22, 190–196. Available at: http://en.cnki.com.cn/Article_en/CJFDTOTALZGSK200802013.htm
Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., et al. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591. doi: 10.1105/tpc.109.066324
Zeng, S., Li, C., Du, C., Sun, L., Jing, D., Lin, T., et al. (2018). Development of specific markers for Pigm in marker-assisted breeding of panicle blast resistant japonica rice. Chin. J. Rice Sci. 32, 453–461. doi: 10.16819/j.1001-7216.2018.7135
Zhai, C., Lin, F., Dong, Z., He, X., Yuan, B., Zeng, X., et al. (2011). The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. The New phytologist 189, 321–334. doi: 10.1111/j.1469-8137.2010.03462.x
Zhang, Y., Wang, S., Liu, X., and Deng, E. (2013). Allele specific amplification detection of rice blast resistance pi-ta. J. Northwest Agric. 22, 44–49.
Zhang, S., Wang, L., Wu, W., He, L., Yang, X., and Pan, Q. (2015). Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 5:11642. doi: 10.1038/srep11642
Zhang, S., Zhong, X., Qiao, G., Shen, L., Zhou, T., and Peng, Y. (2017). Difference in virulence of Magnaporthe oryzae from Sichuan, Chongqing and Guizhou. Southwest China J. Agric. Sci. 30, 359–365. Available at: http://en.cnki.com.cn/Article_en/CJFDTotal-XNYX201702020.htm
Zheng, Y., Zheng, W., Lin, F., Zhang, Y., Yi, Y., Wang, B., et al. (2011). AVR1-CO39 is a predominant locus governing the broad avirulence of Magnaporthe oryzae 2539 on cultivated rice (Oryza sativa L.). molecular plant-microbe interactions. MPMI 24, 13–17. doi: 10.1094/MPMI-10-09-0240
Keywords: rice blast, Magnaporthe oryzae, avirulence gene, resistance gene, resistance breeding
Citation: Tian D, Deng Y, Yang X, Li G, Li Q, Zhou H, Chen Z, Guo X, Su Y, Luo Y and Yang L (2022) Association analysis of rice resistance genes and blast fungal avirulence genes for effective breeding resistance cultivars. Front. Microbiol. 13:1007492. doi: 10.3389/fmicb.2022.1007492
Edited by:
Tika Adhikari, North Carolina State University, United StatesReviewed by:
Yixin Du, Fujian Academy of Agricultural Sciences, ChinaTao Guo, South China Agricultural University, China
Chatchawan Jantasuriyarat, Kasetsart University, Thailand
Copyright © 2022 Tian, Deng, Yang, Li, Li, Zhou, Chen, Guo, Su, Luo and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dagang Tian, dGRnQGZqYWdlLm9yZw==; Liming Yang, eWFuZ2xpbWluZ0BuamZ1LmVkdS5jbg==
 Dagang Tian
Dagang Tian Yun Deng2
Yun Deng2 Ziqiang Chen
Ziqiang Chen Xinrui Guo
Xinrui Guo Liming Yang
Liming Yang