
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 October 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1006375
This article is part of the Research Topic Worldwide Emergence of Drug-Resistant fungi: from Basic to Clinic, Volume II View all 13 articles
 Qiqi Wang1†
Qiqi Wang1† Xuan Cai2†
Xuan Cai2† Yun Li3
Yun Li3 Jianhong Zhao4
Jianhong Zhao4 Zhiyong Liu5
Zhiyong Liu5 Yan Jiang6
Yan Jiang6 Ling Meng7
Ling Meng7 Yanming Li8
Yanming Li8 Shiyang Pan9
Shiyang Pan9 Xiaoman Ai10
Xiaoman Ai10 Fang Zhang11
Fang Zhang11 Ruoyu Li1
Ruoyu Li1 Bo Zheng3
Bo Zheng3 Zhe Wan1
Zhe Wan1 Wei Liu1* on behalf of the China Antifungal Resistance Surveillance Trial (CARST-fungi) Study Group
Wei Liu1* on behalf of the China Antifungal Resistance Surveillance Trial (CARST-fungi) Study GroupTo have a comprehensive understanding of epidemiology and antifungal susceptibilities in pathogenic yeasts, the China Antifungal Resistance Surveillance Trial (CARST-fungi) study was conducted. All yeast isolates were identified by ribosomal DNA sequencing. Antifungal susceptibilities were performed using CLSI M27-A4 broth microdilution method. Sequence and expression level of resistant-related genes in resistant/non-wide-type (NWT) Candida isolates were analyzed. Totally 269 nonduplicate yeast isolates from 261 patients were collected. About half of the yeast isolates (127, 47.2%) were recovered from blood, followed by ascetic fluid (46, 17.1%). C. albicans remained the most prevalent (120, 44.6%), followed by C. parapsilosis complex (50, 18.6%), C. tropicalis (40, 14.9%), and C. glabrata (36, 13.4%). Fourteen (11.7%) C. albicans isolates and 1 (2.0%) C. parapsilosis isolate were resistant/NWT to triazoles. Only 42.5% (17/40) C. tropicalis were susceptible/WT to all the triazoles, with 19 (47.5%) isolates NWT to posaconazole and 8 (20%) cross-resistant to triazoles. Among C. glabrata, 20 (55.6%) and 8 (22.2%) isolates were resistant/NWT to voriconazole and posaconazole, respectively, and 4 (10.3%) isolates were cross-resistant to triazoles. Isavuconazole was the most active triazole against common Candida isolates. Except for 2 isolates of C. glabrata cross-resistant to echinocandins which were also NWT to POS and defined as multidrug-resistant, echinocandins exhibit good activity against common Candida species. All isolates were WT to AMB. For less common species, Rhodotorula mucilaginosa exhibited high MICs to echinocandins and FLC, and 1 isolate of Trichosporon asahii showed high MICs to all the antifungals except AMB. Among triazole-resistant Candida isolates, ERG11 mutations were detected in 10/14 C. albicans and 6/23 C. tropicalis, while 21/23 C. tropicalis showed MDR1 overexpression. Overexpression of CDR1, CDR2, and SNQ2 exhibited in 14, 13, and 8 of 25 triazole-resistant C. glabrata isolates, with 5 isolates harboring PDR1 mutations and 2 echinocandins-resistant isolates harboring S663P mutation in FKS2. Overall, the CARST-fungi study demonstrated that although C. albicans remain the most predominant species, non-C. albicans species accounted for a high proportion. Triazole-resistance is notable among C. tropicalis and C. glabrata. Multidrug-resistant isolates of C. glabrata and less common yeast have been emerging.
Invasive fungal diseases (IFDs) are life-threatening diseases with considerable morbidity and mortality, primarily occurring in immunocompromised and critically ill hosts (Bassetti et al., 2017). About 80% of IFDs were caused by Candida species, the third most frequently isolated microorganism of all infections in a worldwide ICU prevalence study (Kett et al., 2011). Although C. albicans continues to be the most prevalent Candida spp. causing IFDs, the past decades have witnessed changing epidemiology of IFDs shift to non-albicans spp., which may be more resistant to antifungal therapy (Bassetti et al., 2017). Other uncommon yeasts, such as Cryptococcus spp., Saccharomyces spp., Trichosporon spp., and Rhodotorula spp., although remained relatively rare in IFDs, were emerging as opportunistic pathogens and made diagnosis challenging and treatment suboptimal (Miceli et al., 2011). The management of IFDs was hindered owing to the paucity of rapid diagnostic assays (such as molecular identification methods). However, numerous clinicians still rely on traditional culture-based methods which are not rapid and sensitive. Therefore, empirical antifungal therapy often needs to be initiated and may contribute to increasing and inappropriate use of antifungals, which will not only alter Candida spp. distribution but also decrease antifungal susceptibility, making antifungal resistance an emerging problem worldwide (Lamoth et al., 2018).
There are 4 major classes of antifungals available to treat IFDs, including (1) polyenes, such as amphotericin B (AMB), destabilize membrane by binding to ergosterol; (2) triazoles, including fluconazole (FLC) itraconazole (ITC), voriconazole (VRC), posaconazole (POS), isavuconazole (ISA), target the enzyme 14-α-demethylase (Erg11p), a key step in the biosynthesis of ergosterol; (3) echinocandins, including caspofungin (CAS), anidulafungin (ANF), micafungin (MCF), block the catalytic subunit of the β-1,3 glucan synthase and thus inhibit cell wall biosynthesis; (4) pyrimidine analogs, such as 5-fluorocytosine (5-FC), are metabolized by fungal cells into fluorinated pyrimidines, which destabilize nucleic acids and result in growth arrest. Among these antifungals, triazoles represent the most widely prescribed antifungal class for IFDs. However, triazole-resistance has become a growing problem worldwide. The major mechanisms of triazole-resistance in Candida spp. include alteration in the ERG11 gene encoding Erg11p, upregulation of the ERG11 gene due to mutations in transcriptional factors such as UPC2, and overexpression of drug efflux pumps such as Mdr1p and Cdr1p/Cdr2p occurring mainly as a result of gain-of-function (GOF) mutations in transcription factors genes such as MRR1, TAC1 and PDR1 in C. glabrata. Echinocandins have now been recommended as first-line drugs for candidiasis (Pappas et al., 2016). Although the resistance rate remains generally low, echinocandin-resistance is rising as usage broadens among Candida spp., most notably C. glabrata, which was reported as high as >13% in some centers (Alexander et al., 2013). The mechanism of echinocandin resistance involves mutations in FKS genes encoding subunits of glucan synthase (Arastehfar et al., 2020). Different resistance mechanisms vary by species and geographic distribution, and the underlying mechanisms in some species have not been well defined. A better understanding of antifungal resistance mechanisms will provide insights to reclaim those antifungal classes as an option for empiric treatment of IFDs.
Under these circumstances, surveillance of IFDs plays a significant role in understanding epidemiology and antifungal susceptibility data to guide empirical therapy and aid antifungal stewardship efforts (Pfaller et al., 2019). The China Antifungal Resistance Surveillance Trial (CARST-fungi) study was a prospective national surveillance program for IFDs in mainland China. The study described the epidemiology and antifungal susceptibilities of clinical yeast isolates, including Candida spp., Cryptococcus neoformans, Trichosporon asahii, and other less common yeast species recovered from patients across major cities in China during 2019 and 2020. Additionally, to preliminarily elucidate the mechanisms underlying the resistant phenotypes, sequences of resistance-related genes ERG11, PDR1, FKS, and expression profiles of ERG11 and efflux pump genes including CDR1, CDR2, MDR1, and SNQ2, were analyzed.
The CARST-fungi study was a multi-center, prospective, observational, and laboratory-based study of IFDs with its inception in July 2019 and finished in June 2020. Nine “rank-A tertiary” hospitals distributed throughout 9 major cities in China took part in the study. All Candida, Cryptococcus, and other yeast isolates recovered from sterile sites including blood, other sterile body fluids (ascitic fluid, pleural fluid, cerebrospinal fluid [CSF]), pus, tissue from patients with invasive yeast diseases, bronchoalveolar lavage fluid (BALF), central venous catheter (CVC) tips, biliary tract fluid were collected. Additionally, yeast strains from considered colonizers such as urine, feces, sputum, and the genital tract were also included. For each episode of yeast isolation, the information including the patient’s age, gender, the ward location (e.g., emergency department, surgical, medical, and ICU), the time of sample collection, the specimen type, and the initial species identification made by the referring laboratory were collected. All isolates were sent to the Research Center for Medical Mycology at Peking University First Hospital, Beijing, China, for further study.
To ensure the accuracy of identification, all clinical yeast isolates were identified to the species level in the central laboratory. Six colonies from primary culture plates were subcultured and identified using sequence-based methods for the internal transcribed spacer (ITS) region, 28S ribosomal subunit (D1/D2), and the intergenic spacer (IGS, for Trichosporon spp. and Cryptococcus spp.), primers used for identification were listed in Supplementary Table S1. Those sequences were aligned using CBS database1 (Pfaller et al., 2012).
Antifungal susceptibility testing was performed according to the Clinical and Laboratory Standards Institute (CLSI) M27-A4 microbroth dilution method (CLSI, 2017), and the tested drugs including AMB (from North China Pharmaceutical Co. Ltd., Shijiazhuang, China), FLC, ITC, VRC, POS, ISA, CAS, ANF, MCF and 5-FC, (all from Harveybio Gene Technology Co. Ltd., Beijing, China) were prepared according to CLSI methods.
Minimum inhibitory concentrations (MICs) were determined after 24 h incubation at 35°C for Candida spp. and Trichosporon spp. and after 72 h incubation for Cryptococcus spp. MICs were read as the lowest drug concentration producing a prominent decrease in turbidity translating to 50% (triazoles, echinocandins, and 5-FC) or 100% (AMB) growth reduction compared with the drug-free control. Quality control was performed with each test run using C. parapsilosis ATCC22019 and C. krusei ATCC6258.
The interpretation of susceptibility was performed by applying the updated species-specific clinical breakpoints (CBPs) according to CLSI M60 document (CLSI, 2020a). In the absence of CBPs, isolates were defined as having a wild-type (WT) or a non-WT (NWT) drug susceptibility phenotype according to the epidemiological cutoff values (ECVs) as determined by the CLSI M59 document (CLSI, 2020b). Cross-resistance was defined as resistance to at least two antifungals of the same drug class. Multidrug-resistance was defined as resistance to at least two classes of antifungal drugs (Orasch et al., 2014).
Genomic DNA of resistant or non-WT yeast strains was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Primers to amplify the full open reading frame of ERG11, PDR1 (for C. glabrata), and FKS1, FKS2 for (C. glabrata) were designed (Supplementary Table S1) according to the genome of C. albicans SC5314, C. glabrata CBS138, C. tropicalis MYA-3404, and C. parapsilosis CDC317 from the Candida Genome Database2 as reference. The amplified products were sent to the BGI Company (Beijing, China) for sequencing and the sequences were aligned with the reference strain using the Clustal Omega.3
Suspensions of Candida isolates cells (OD600, 0.1) freshly prepared in YPD medium were grown at 35°C to reach the mid-exponential phase (OD600, 0.6–0.8). The cells were washed twice with sterile water. Total RNA was extracted using the RNeasy Mini kit (QIAGEN Science, Maryland, USA) following the manufacturer’s instructions. The RNA was then treated with RNase-free DNase (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendations. cDNA was synthesized using an Advantage RT-for-PCR kit (Clontech) according to the manufacturer’s instructions. RT-qPCR was performed on an Applied Biosystems ViiA7 Real-Time PCR system using SYBR green reagent (Applied Biosystems). Optimal thermal cycling conditions consisted of a 10-min initial denaturation at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s, and annealing/extension at 60°C for 10 s. The experiments were carried out in triplicate for each data point. The cycle threshold (CT) value of the gene was normalized to that of internal control ACT1 gene for C. albicans, C. tropicalis, C. tropicalis, and RND5.8 gene for C. glabrata as ΔCT value. Relative gene expression (2ΔΔCt) was calculated as the fold change in expression of the isolates compared to the mean expression values in drug-susceptible control strains including C. albicans SC5314, C. glabrata CBS138, C. tropicalis ATCC01463, and C. parapsilosis ATCC22019. The primers used were listed in Supplementary Table S1.
Comparisons were performed using SPSS software version 22 (SPSS, Chicago, IL, USA). Continuous variables were compared using the Mann–Whitney test, and categorical variables were analyzed using the χ2 test or Fisher’s exact test. A value of p <0.05 was considered statistically significant.
A total of 269 nonduplicate yeast isolates from 261 patients were collected, among which more than 1 isolate was recovered from 8 patients. Of the yeast isolates investigated, 161 (59.9%) were cultured from male patients, and 108 (40.1%) were from female patients. The patients’ ages ranged from 0 to 98 years (median, 57 years; interquartile range, 36 to 72 years). Most isolates (252/269, 93.7%) were from hospital inpatients (including those in ICUs [23.4%], medical wards [28.3%], surgical wards [33.8%], obstetrics and gynecology departments [5.2%], paediatrics departments [3.0%]), and remaining 6.3% were from patients in emergency settings.
Of the various specimen types, about half of the yeast isolates (127, 47.2%) were recovered from blood, followed by ascitic fluid (46, 17.1%), pus (17, 6.3%), CVC (13, 4.8%), CSF (11, 4.1%), bile (9, 3.3%), tissue (7, 2.6%), and other sterile sites (3, 1.1%, including bone marrow, BALF; Figure 1). Thirty-six yeast isolates (13.4%) were from non-sterile samples and considered colonizers (such as urine, feces, sputum, and genital tract secretion). The proportion of non-albicans Candida isolates recovered from blood (78/127, 61.4%) was significantly higher than that recovered from other specimen types (58/142, 40.8%) (p < 0.01).
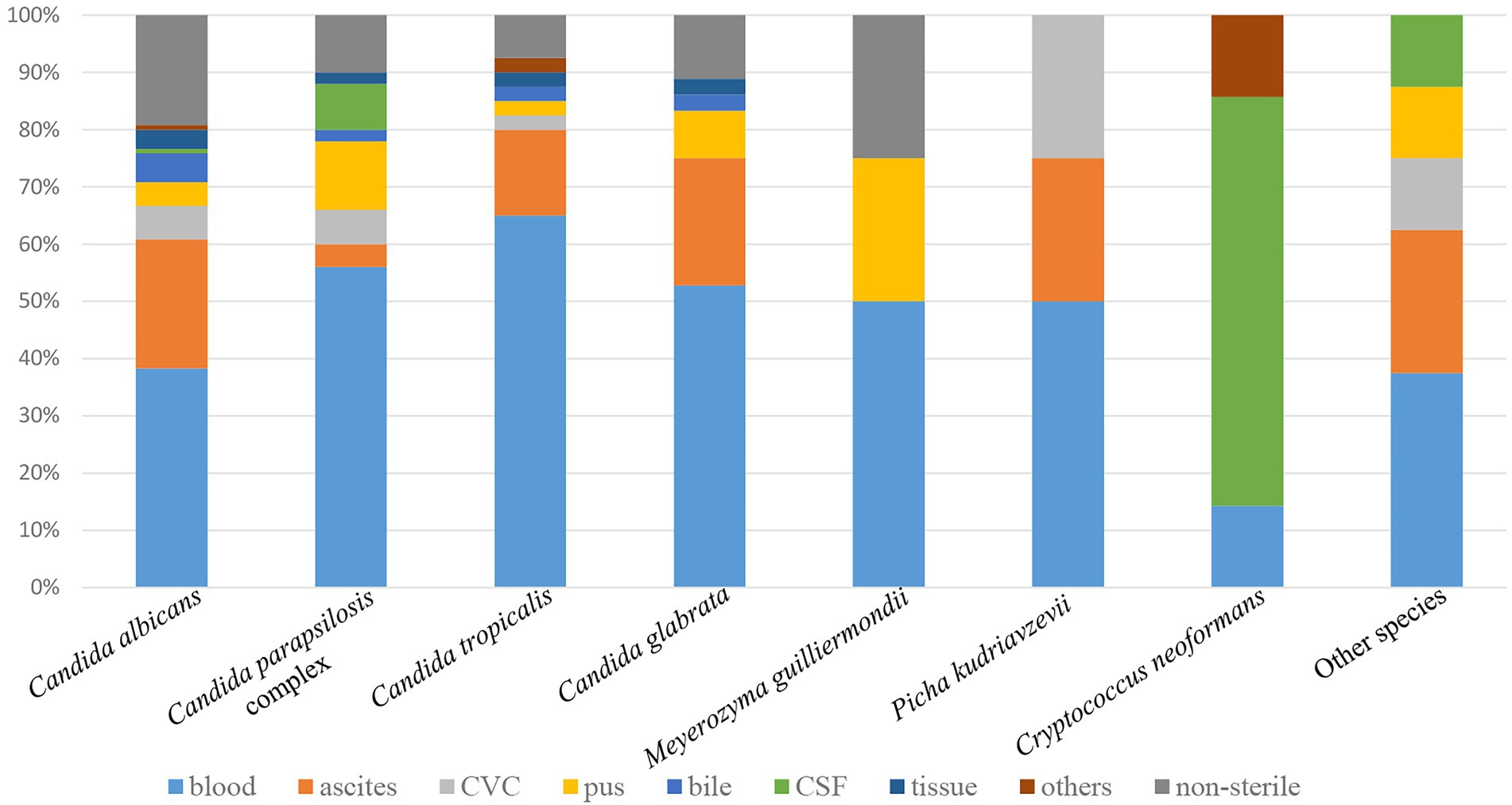
Figure 1. Distribution of yeast species by specimen type. Other species include 2 each of Clavispora lusitaniae, Trichosporon asahii, and Wickerhamomyces anomalus, 1 each of Cyberlindnera fabianii and Rhodotorula mucilaginosa. Other sterile sites include bone marrow and bronchoalveolar lavage fluid (BALF). Non-sterile samples include urine, feces, sputum, and genital tract secretion. CVC, central venous catheter; CSF, cerebrospinal fluid.
Thirteen species were identified among the 269 yeast isolates (Figure 2). C. albicans remained the most prevalent (120, 44.6%), followed by C. parapsilosis complex (including C. parapsilosis sensu stricto [41, 15.2%] and C. metapsilosis [9, 3.3%]), C. tropicalis (40, 14.9%), and C. glabrata (36, 13.4%). Other yeast species including 7 isolates of Cryptococcus neoformans, 4 each of Picha kudriavzevii (Candida krusei) and Meyerozyma guilliermondii (Candida guilliermondii), 2 each of Clavispora lusitaniae (Candida lusitaniae), Trichosporon asahii, and Wickerhamomyces anomalus (Candida pelliculosa), 1 each of Cyberlindnera fabianii (Candida fabianii) and Rhodotorula mucilaginosa, accounted for tiny proportion.
Eight out of 261 samples (3.1%) in our study were co-isolated more than 1 yeast species (Table 1). Of the 8 specimens, 5 were from blood, 1 each from ascites, paracentesis fluid, and urine, among which 4 cases were C. albicans/C. glabrata mixed, 1 each C. albicans/C. tropicalis mixed and C. parapsilosis sensu stricto/C. metapsilosis mixed. Interestingly, 2 isolates of T. asahii in blood specimens were both co-isolated with C. albicans. Of note, most specimens contained yeast isolates that were resistant/NWT to at least one antifungal (Table 1).
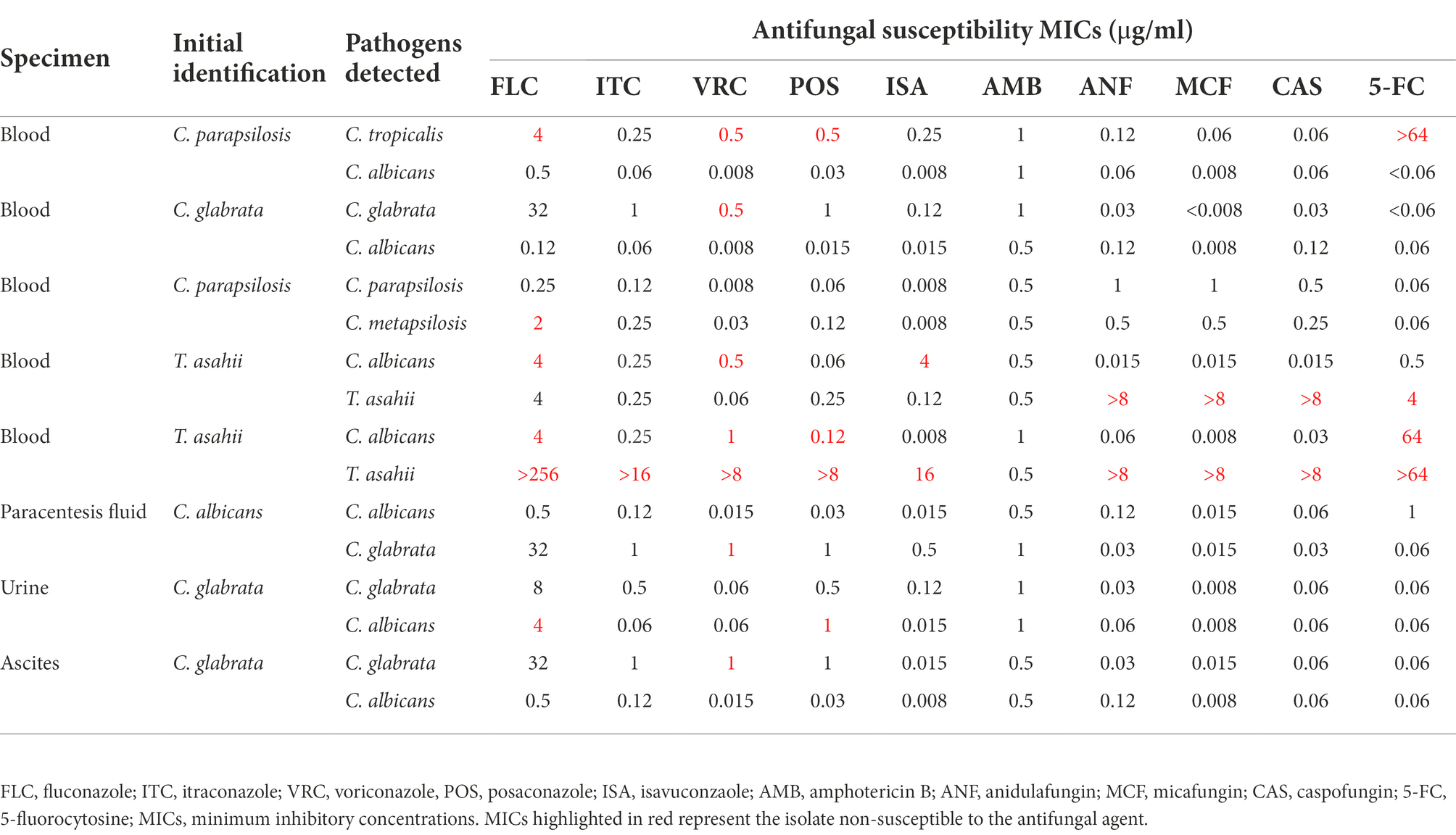
Table 1. Distribution of co-isolated yeast species in specimens and their antifungal susceptibility profiles.
In vitro susceptibility of antifungal drugs against yeast species is shown in Table 2. Among the common Candida species, 5 (4.2%), 6 (5.0%), and 10 (8.4%) isolates of C. albicans were resistant/NWT to FLC, VRC, and POS, respectively. As for the C. parapsilosis complex, only 1 (2.4%) isolate of C. parapsilosis sensu stricto was cross-resistant to FLC and POS, while all the 9 C. metapsilosis isolates were WT to triazoles. However, only 42.5% (17/40) of C. tropicalis were susceptible/WT to all the triazoles, and 12 (30.0%), 3 (7.5%), 8 (20.0%), 19 (47.5%) isolates were resistant/NWT to FLC, ITC, VRC, and POS, respectively. Among C. glabrata, 2 (5.6%) isolates were resistant to FLC while the remaining 34 were susceptible-dose dependent (SDD), 20 (55.6%) and 8 (22.2%) isolates were resistant/NWT to VRC and POS, respectively, and all isolates were WT to ITC.
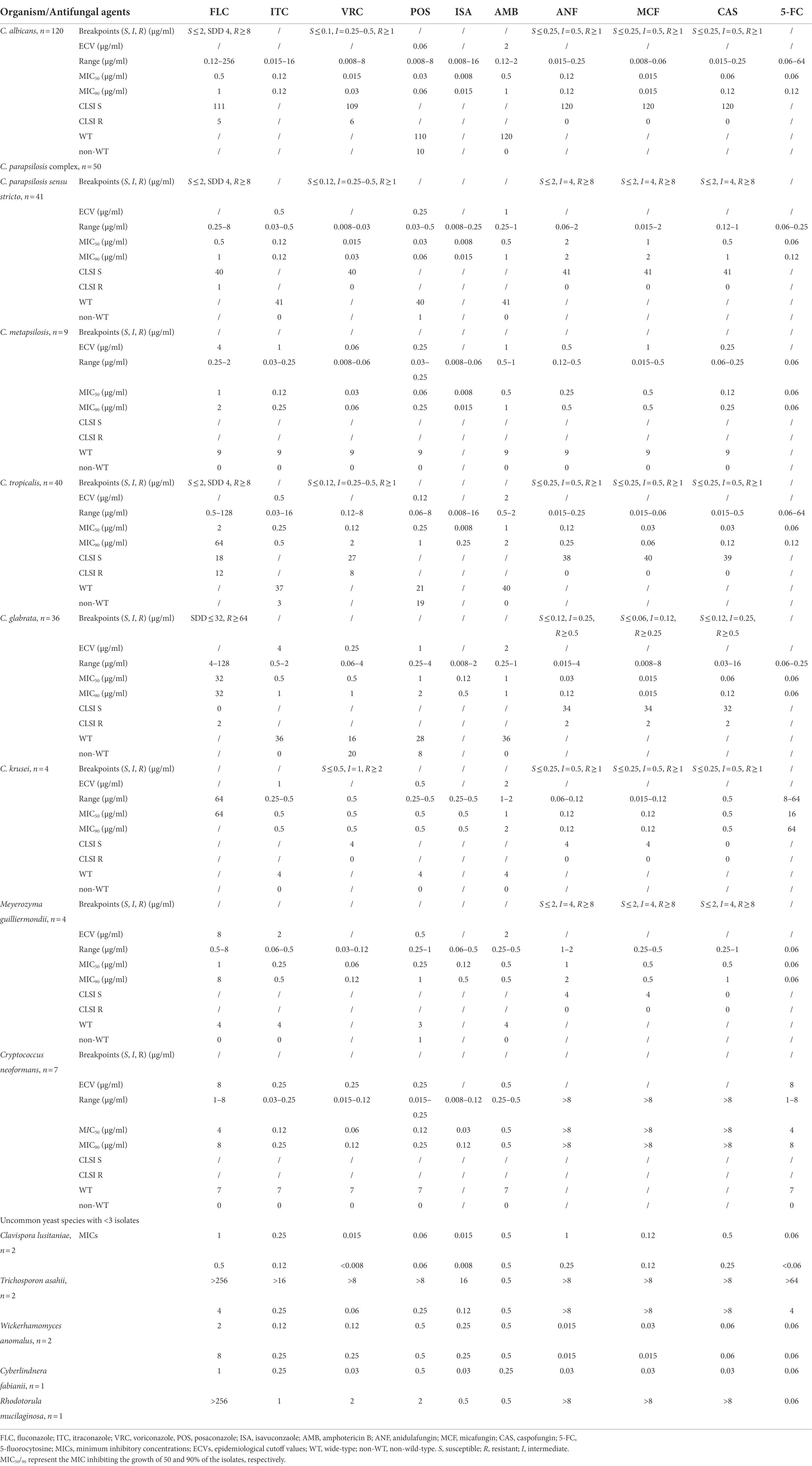
Table 2. Activities of 9 antifungal drugs against yeast species according to CLSI clinical breakpoints or ECVs.
For less common yeast species, all 4 isolates of P. kudriavzevii were susceptible/WT to other triazoles which were assumed to be intrinsically resistant to FLC; M. guilliermondii and C. lusitaniae were WT to triazoles except 1 isolate of M. guilliermondii was NWT to POS. Based on MIC90 since neither CBPs nor ECV values have been established for ISA and Candida spp., ISA was the most active triazole against C. albicans (MIC90, 0.015 μg/ml), C. parapsilosis (MIC90, 0.015 μg/ml), C. tropicalis (MIC90, 0.25 μg/ml), and C. glabrata (MIC90, 0.5 μg/ml), while comparable to other triazoles against P. kudriavzevii, M. guilliermondii, and other non-Candida spp. Two isolates of W. anomalus exhibited relatively less susceptible to FLC (MICs 2–8 μg/ml) and POS (MICs 0.5–1 μg/ml) than ITC, VRC, and ISA (MICs 0.12–0.25 μg/ml).
All isolates of C. albicans and C. parapsilosis were susceptible to echinocandins. Among C. glabrata, 2 isolates were cross-resistant to CAS, MCF, ANF (MICs 32 μg/ml, 8 μg/ml, 4 μg/ml, respectively) and 2 isolates were intermediate to CAS (MICs 0.25 μg/ml). There were 1 and 2 isolates of C. tropicalis exhibiting intermediate to CAS and ANF, respectively. Among the non-Candida yeasts, MICs of the echinocandins were consistently high for all isolates of C. neoformans, R. mucilaginosa, and T. asahii (all MICs >16 μg/ml) (Table 2).
All yeast isolates tested in this study were WT to AMB (MICs ≤2 μg/ml). No ECVs were defined for 5-FC against Candida spp., the MICs of 5-FC against 7 isolates of C. albicans and 2 isolates of C. tropicalis were > 64 μg/ml, while the MICs against the remaining Candida isolates were all ≤1 μg/ml. All isolates of C. neoformans were WT to 5-FC (ECV 8 μg/ml).
For yeast species with established CBPs or ECVs, 18 isolates were cross-resistant to at least 2 triazoles, including 5 (4.2%) isolates of C. albicans, 8 (20.0%) isolates of C. tropicalis, 4 (10.3%) isolates of C. glabrata, and 1 (2.4%) isolate of C. parapsilosis sensu stricto. Among triazole-resistant Candida isolates, 2 isolates of C. albicans and 1 isolate of C. tropicalis were also non-susceptible to 5-FC (MICs >64 μg/ml), and 2 isolates of C. glabrata were resistant to 3 echinocandins, which were defined as multidrug-resistance.
For less common species or those without CBPs or ECVs, 1 isolate of R. mucilaginosa exhibited high MICs to echinocandins and triazoles, and 1 isolate of T. asahii showed high MICs to all the antifungals tested except AMB (Table 2).
From the sequencing results of ERG11 in the 14 triazole-resistant/NWT C. albicans isolates, missense mutations were detected in 10 isolates, among which the most common substitution is Y132H exhibiting in 6 isolates. Four isolates without ERG11 mutations exhibited overexpression of ERG11, CDR1, CDR2, or MDR1 (Table 3). One isolate of C. parapsilosis sensu stricto cross-resistant to FLC and POS harbored R398I substitution in Erg11p as well as CDR1 overexpression. Among the 23 triazole-resistant/NWT C. tropicalis isolates, only 6 isolates exhibited amino acid substitutions in Erg11p, among which S154F was detected in 1 isolate, Y132F with S154F was present in 5 isolates. Overexpression of MDR1 was most frequent in triazole-resistant isolates (21/23), followed by ERG11, CDR1, and CDR2. In 25 triazole-resistant/NWT C. glabrata isolates, no mutations in ERG11 despite of 5 isolates showed ERG11 overexpression. Five isolates had PDR1 modifications, among which P76S, P143T, D243N were detected in 3 isolates, 1 each harboring R250K and Y682C, respectively. Overall, there were 14, 13, and 8 isolates showed overexpression of CDR1, CDR2, and SNQ2, respectively. The entire sequences of FKS1 and FKS2 were determined in 2 echinocandin-resistant C. glabrata isolates, and S663P substitutions in FKS2 were detected in both isolates (Table 3).
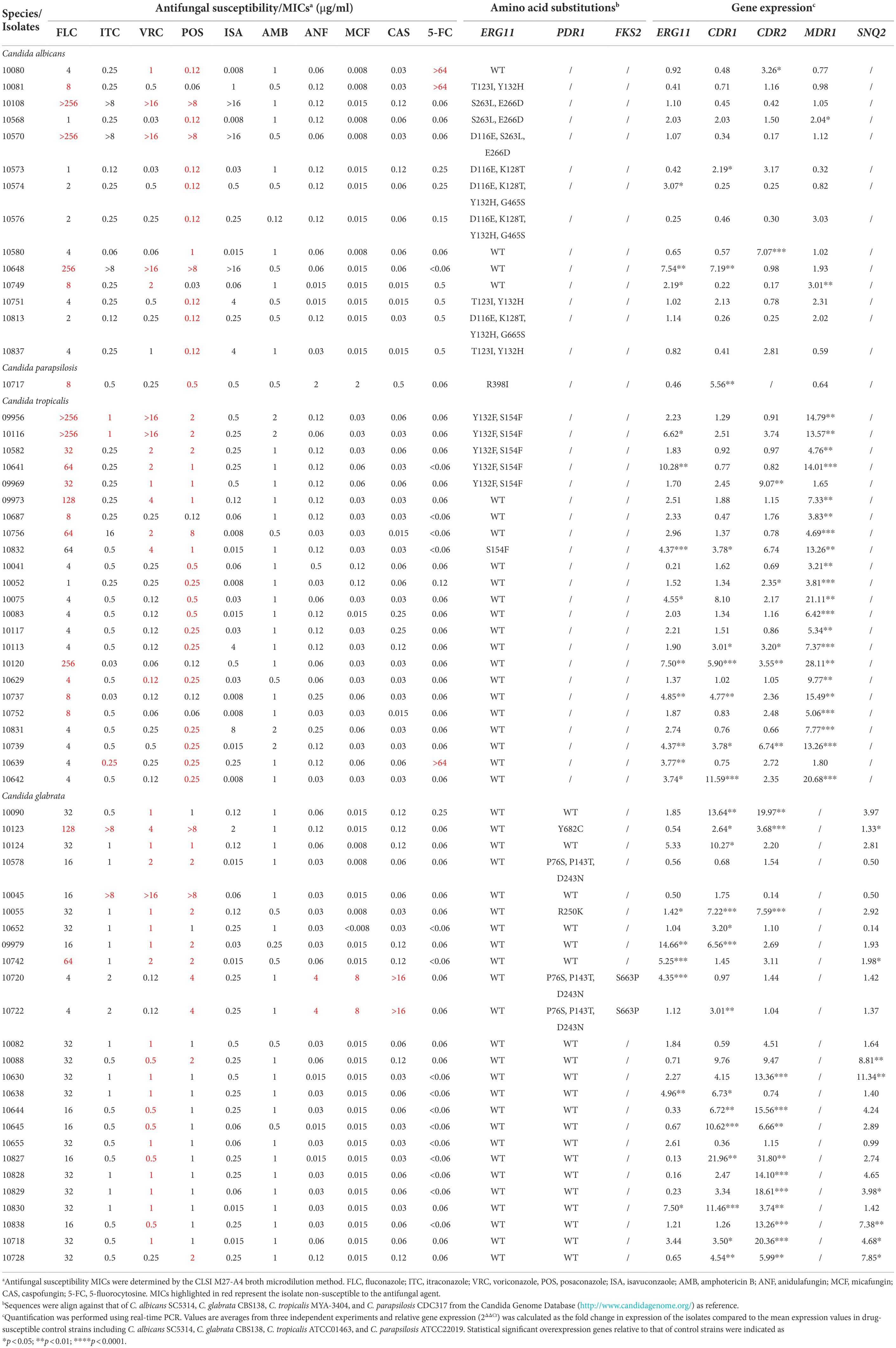
Table 3. Mutations and expression level of resistance-related genes in resistant/NWT Candida isolates.
To our knowledge, there are several excellent surveillance studies of IFDs worldwide, such as the SENTRY Antimicrobial Surveillance Program (Pfaller et al., 2019), the ARTEMIS DISK study (Pfaller et al., 2010), the SCOPE Program (Wisplinghoff et al., 2014), and so on. In China, the national-wide surveillance studies include the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study, which has provided useful data on the epidemiology of IFDs in mainland China (Xiao et al., 2018b), and China-SCAN study determining species distribution and antifungal susceptibility of invasive Candida infection (ICI) in ICU across China (Liu et al., 2014). The CARST-fungi study is another multi-center surveillance study of IFDs in mainland China with its inception in July 2019. Totally 9 hospitals participated during the first year with the collection of 269 yeast isolates. As expected, the four major Candia species, C. albicans, C. parapsilosis complex, C. tropicalis, and C. glabrata, accounted for predominate proportion. Although C. albicans remained the most prevalent (120, 44.6%), the proportion of non-albicans Candida species was over 50%, especially have been the primary causative pathogen of candidemia, in accordance with previous studies from China (Xiao et al., 2018a; Song et al., 2020), Asia-Pacific regions, and European countries (Pappas et al., 2018).
Mixed species detection in the same specimen was another significant result of our study. Totally, 8 out of 261 samples (3.1%) in our study were co-isolated with more than 1 yeast species, among which 4 cases were C. albicans/C. glabrata mixed, 1 each C. albicans/C. tropicalis mixed and C. parapsilosis sensu stricto/C. metapsilosis mixed, and 2 cases of C. albicans /T. asahii co-isolated. Indeed, mixed yeast infections have been detected in 8.78% of the culture-positive samples in an extensive study of 6,192 clinical yeast isolates (Cassagne et al., 2016). Similarly, an 18-year report from a tertiary-care university hospital revealed the incidence of mixed fungaemia was 3.7% (33/883) (Gulmez et al., 2020). Most mixed species in our study were C. albicans/C. glabrata, in accordance with the above-mentioned studies. Several studies revealed that C. albicans and C. glabrata are frequently co-isolated and co-adhesion in vitro (Tati et al., 2016), demonstrating enhanced invasion and increased tissue damage (Silva et al., 2011; Alves et al., 2014), which probably can be explained for the high proportion of the co-culture of these two species. Of note, most specimens contained yeast isolates that were non-susceptible to one or more antifungals. Mixed infections may further bring additional issues for treatment, especially infected by more than one fungal species with different drug susceptibilities, which highlights that mycological analysis of clinical samples should also reliably detect mixed fungal species, especially those involving yeasts species with a particular antifungal resistance profile. A previous study suggested that detection of mixed infection increased significantly after subculture from yeast-positive blood-culture bottles and routine use of chromogenic agar (Gulmez et al., 2020).
First-line drugs treating IFDs, including echinocandins and triazoles, are relatively effective, but the emergence of antifungal resistance is a matter of concern and poses a global threat (Pappas et al., 2018). In our study, most C. albicans and C. parapsilosis complex are susceptible to all the antifungals tested. Only 1 isolate (2.0%) of C. parapsilosis sensu stricto exhibited triazole-resistance, similar to the global (Pfaller et al., 2019) and domestic data (Wang et al., 2016; Xiao et al., 2018a; Song et al., 2020). The overall triazole-resistant rate of C. albicans was 11.6% (14/120), higher than the global data (<1%) (Pfaller et al., 2019). However, triazole susceptibility of C. tropicalis and C. glabrata was low, with only 42.5% (17/40) C. tropicalis and 30.5% (11/36) C. glabrata susceptible/WT to all tested triazoles. Of particular note, the POS-resistant rate of C. tropicalis isolates was 47.5% and the VRC-resistant rate of C. glabrata isolates was 55.6%, consistent with previous reports (Xiao et al., 2018a; Song et al., 2020) and much higher than that of past decade (Liu et al., 2014).
Another noteworthy finding of our study was the emergence of echinocandin resistance, although exhibiting excellent activity to most common Candida isolates. Two isolates of C. glabrata were cross-resistant to all the echinocandins tested, which also NWT to POS and defined as multidrug-resistant. Echinocandin-resistance among C. glabrata isolates ranges from 3–5% in population-based studies, and some centers even report as high as 10–15% (Farmakiotis et al., 2014), but it is less than 1% in China (Hou et al., 2017). Alarmingly, C. glabrata often presents as multidrug-resistance, with nearly one-third of echinocandin-resistant isolates also being non-susceptible to triazoles, leaving extremely few options to treat patients infected with multidrug-resistant isolates (Healey and Perlin, 2018). Emerging of multidrug-resistance is a significant threat to the treatment of IFD and urges our research group to explore the resistance mechanism of the C. glabrata isolates, demonstrating mutations in FKS2 explaining echinocandin-resistance and overexpression of CDR1 and ERG11 contributing to triazole-resistance (Wang et al., 2021).
Except for the above-mentioned common Candida species, less common and cryptic yeast species were also identified in this study, among which C. metapsilosis accounted for a high proportion of C. parapsilosis complex (9/50), indicating the proportion was underestimated. Interestingly, C. metapsilosis and C. parapsilosis were co-cultured, highlight the significance of subculture and accurate identification. One isolate of C. fabianii was identified in our study. C. fabianii infections have rarely been reported (Park et al., 2019) and only one case has been reported in China which causes blood infection in a premature infant (Wu et al., 2013). Additionally, W. anomalus (previously named C. pelliculosa) accounted for a low rate (<1%) and was similar to previous national surveillance in China (Liu et al., 2014), and to our knowledge, one of which in our study is the first isolate of W. anomalus reported to be cultured from CSF in China. W. anomalus isolates in our study were less susceptible to FLC (MICs 2–8 μg/ml) and POS (MIC, 0.5 μg/ml), while AMB, echinocandins, and 5-FC showed good in vitro activity, but recent research revealed that W. anomalus isolates showed high MICs against all triazoles tested and 5-FC (Zhang et al., 2021). As for other yeast genera, C. neoformans, R. mucilaginosa, and T. asahii were included in our study. As expected, most C. neoformans isolates were isolated from CSF, which is believed to be the prevalent pathogen of fungal meningitis. R. mucilaginosa is a multidrug-resistant pathogen with the ability to cause nosocomial infection (Huang et al., 2022), which exhibited high MICs to all the triazoles and echinocandins. Recently, Trichosporon spp. which is usually associated with superficial mycosis has recently been recognized as an emergent fungal pathogen capable of causing invasive infections (Padovan et al., 2019). Besides intrinsic echinocandins-resistance, one isolate of T. asahii in our study showed high MICs to all triazoles and 5-FC. Several studies have estimated antifungal profiles of rare yeast species (Stavrou et al., 2020), demonstrated that high MICs against azole drugs and echinocandins were common and resistance rates in yeast species are dynamic and variable between medical institutions and countries (Desnos-Ollivier et al., 2021), highlighting the need for an accurate species identification of yeast isolates, which is essential for proper management of patients and prevention of emergence of drug resistance.
Finally, sequence of ERG11 genes (and PDR1 in C. glabrata) and expression level of ERG11 and efflux pump genes in triazole-resistant Candida isolates and sequence of FKS genes in echinocandin-resistant isolates were determined. ERG11 mutations were found in 10 of 14 triazole-resistant C. albicans isolates and 6 of 23 C. tropicalis isolates, the most common substitution is Y132H in C. albicans, and Y132F with S154F in C. tropicalis, similar to Y132F and S154F were the most common substitutions in Erg11p of C. tropicalis and proven to mediate triazole resistance (Jiang et al., 2012; Castanheira et al., 2020). Except for mutations of drug target, overexpression of MDR1 seems as the predominant mechanism of C. tropicalis triazole-resistance in our study. Despite of no ERG11 mutation was detected in triazole-resistant C. glabrata isolates, 5 isolates exhibited ERG11 overexpression, which was considered to be less common in C. glabrata. PDR1 modifications were detected in 5 isolates, with 3 isolates harboring P76S, P143T, D243N and 1 each harboring R250K and Y682C, respectively. The latter two isolates showed high expression level of ABC transporter genes including CDR1, CDR2, and SNQ2, indicating those PDR1 modifications maybe GOF mutations resulting in up-regulation of downstream ABC transporter genes, which need further verification. The entire sequences of FKS1 and FKS2 were determined in 2 echinocandin-resistant C. glabrata isolates and S663P substitutions in FKS2 were detected in both isolates. Notably, there are numerous isolates without modifications in well-known drug targets or up-regulation of transporter genes, the resistant mechanisms remain to be elucidated.
There are some limitations in our study. First, this paper concluded the first-year data of the CARST-fungi study, which encompass only nine hospitals cannot reveal the dynamic change year by year. The study will continue and more centers will join in to enhance the comprehension of epidemiology and antifungal susceptibilities in pathogenic yeasts across China. Second, the information of the patients is incomplete including mycoses, antibiotics used, basal conditions and other more detailed clinical characteristics. These data need to be included in future study. Nevertheless, this study provides important epidemiological findings of species distribution, antifungal susceptibility profile, and preliminary resistant mechanism exploration, which are instrumental in designing strategies for better management of yeast infections in China.
In conclusion, the CARST-fungi study demonstrated that although C. albicans remain the most predominant species, non-C. albicans species accounted for a high proportion, especially as the causative pathogen of fungemia. Triazole-resistance is notable among C. tropicalis and C. glabrata and multidrug-resistant isolates of C. glabrata and less common yeast have been emerging.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
WL designed the experiments, supervised the data analysis, and contributed to funding acquisition. WL, YL, BZ, and RL administered the project. XC, JZ, ZL, YJ, LM, YML, SP, XA, and FZ collected all the isolates and provided clinical information. QW contributed to the species identification, antifungal susceptibility testing, molecular biology experiments, and original draft preparation. YL and ZW supervised the transformation, collection, and storage of the isolates. QW, XC, and WL contributed to writing and reviewing the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key Research and Development Program of China (2021YFC2302005) and National Natural Science Foundation of China (81971912).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1006375/full#supplementary-material
Alexander, B. D., Johnson, M. D., Pfeiffer, C. D., Jiménez-Ortigosa, C., Catania, J., Booker, R., et al. (2013). Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56, 1724–1732. doi: 10.1093/cid/cit136
Alves, C. T., Wei, X. Q., Silva, S., Azeredo, J., Henriques, M., and Williams, D. W. (2014). Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J. Infect. 69, 396–407. doi: 10.1016/j.jinf.2014.06.002
Arastehfar, A., Lass-Flörl, C., Garcia-Rubio, R., Daneshnia, F., Ilkit, M., Boekhout, T., et al. (2020). The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. J. Fungi (Basel) 6:138. doi: 10.3390/jof6030138
Bassetti, M., Garnacho-Montero, J., Calandra, T., Kullberg, B., Dimopoulos, G., Azoulay, E., et al. (2017). Intensive care medicine research agenda on invasive fungal infection in critically ill patients. Intensiv. Care Med. 43, 1225–1238. doi: 10.1007/s00134-017-4731-2
Cassagne, C., Normand, A.-C., Bonzon, L., L'Ollivier, C., Gautier, M., Jeddi, F., et al. (2016). Routine identification and mixed species detection in 6, 192 clinical yeast isolates. Med. Mycol. 54, 256–265. doi: 10.1093/mmy/myv095
Castanheira, M., Deshpande, L. M., Messer, S. A., Rhomberg, P. R., and Pfaller, M. A. (2020). Analysis of global antifungal surveillance results reveals predominance of erg 11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 55:105799. doi: 10.1016/j.ijantimicag.2019.09.003
CLSI (2017). Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. Approved Standard-Fourth Edition. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2020a). Clinical and Laboratory Standards Institute. Performance Standards for Antifungal Susceptibility Testing of Yeasts, CLSI Supplement M60. 2nd edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2020b). Epidemiological Cutoff Values for Antifungal Susceptibility Testing. CLSI Supplement M59. 3rd edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Desnos-Ollivier, M., Lortholary, O., Bretagne, S., and Dromer, F. (2021). Azole susceptibility profiles of more than 9,000 clinical yeast isolates belonging to 40 common and rare species. Antimicrob. Agents Chemother. 65, e02615–e02620. doi: 10.1128/aac.02615-20
Farmakiotis, D., Tarrand, J. J., and Kontoyiannis, D. P. (2014). Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 20, 1833–1840. doi: 10.3201/eid2011.140685
Gulmez, D., Alp, S., Gursoy, G., Ayaz, C. M., Dogan, O., Arikan-Akdagli, S., et al. (2020). Mixed fungaemia: an 18-year report from a tertiary-care university hospital and a systematic review. Clin. Microbiol. Infect. 26, 833–841. doi: 10.1016/j.cmi.2020.03.030
Healey, K. R., and Perlin, D. S. (2018). Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J. Fungi (Basel) 4:105. doi: 10.3390/jof4030105
Hou, X., Xiao, M., Chen, S. C., Kong, F., Wang, H., Chu, Y. Z., et al. (2017). Molecular epidemiology and antifungal susceptibility of Candida glabrata in China (August 2009 to July 2014): a multi-center study. Front. Microbiol. 8:880. doi: 10.3389/fmicb.2017.00880
Huang, J. J., Chen, X. F., Tsui, C. K. M., Pang, C. J., Hu, Z. D., Shi, Y., et al. (2022). Persistence of an epidemic cluster of Rhodotorula mucilaginosa in multiple geographic regions in China and the emergence of a 5-flucytosine resistant clone. Emerg. Microbes. Infect. 11, 1079–1089. doi: 10.1080/22221751.2022.2059402
Jiang, C., Dong, D., Yu, B., Cai, G., Wang, X., Ji, Y., et al. (2012). Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 68, 778–785. doi: 10.1093/jac/dks481
Kett, D. H., Azoulay, E., Echeverria, P. M., and Vincent, J.-L. (2011). Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 39, 665–670. doi: 10.1097/CCM.0b013e318206c1ca
Lamoth, F., Lockhart, S. R., Berkow, E. L., and Calandra, T. (2018). Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 73, i4–i13. doi: 10.1093/jac/dkx444
Liu, W., Tan, J., Sun, J., Xu, Z., Li, M., Yang, Q., et al. (2014). Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. J. Antimicrob. Chemother. 69, 162–167. doi: 10.1093/jac/dkt330
Miceli, M. H., Díaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Orasch, C., Marchetti, O., Garbino, J., Schrenzel, J., Zimmerli, S., Muhlethaler, K., et al. (2014). Candida species distribution and antifungal susceptibility testing according to European committee on antimicrobial susceptibility testing and new vs. old clinical and laboratory standards institute clinical breakpoints: a 6-year prospective candidaemia survey from the fungal infection network of Switzerland. Clin. Microbiol. Infect. 20, 698–705. doi: 10.1111/1469-0691.12440
Padovan, A. C. B., Rocha, W., Toti, A. C. M., Freitas de Jesus, D. F., Chaves, G. M., and Colombo, A. L. (2019). Exploring the resistance mechanisms in Trichosporon asahii: triazoles as the last defense for invasive trichosporonosis. Fungal Genet. Biol. 133:103267. doi: 10.1016/j.fgb.2019.103267
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 62, e1–e50. doi: 10.1093/cid/civ933
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L., and Kullberg, B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers. 4:18026. doi: 10.1038/nrdp.2018.26
Park, J.-H., Oh, J., Sang, H., Shrestha, B., Lee, H., Koo, J., et al. (2019). Identification and antifungal susceptibility profiles of Cyberlindnera fabianii in Korea. Mycobiology 47, 449–456. doi: 10.1080/12298093.2019.1651592
Pfaller, M. A., Diekema, D. J., Gibbs, D. L., Newell, V. A., Ellis, D., Tullio, V., et al. (2010). Results from the ARTEMIS DISK global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48, 1366–1377. doi: 10.1128/jcm.02117-09
Pfaller, M. A., Diekema, D. J., Turnidge, J. D., Castanheira, M., and Jones, R. N. (2019). Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997–2016. Open Forum Infect. Dis. 6, S79–S94. doi: 10.1093/ofid/ofy358
Pfaller, M. A., Woosley, L. N., Messer, S. A., Jones, R. N., and Castanheira, M. (2012). Significance of molecular identification and antifungal susceptibility of clinically significant yeasts and moulds in a global antifungal surveillance programme. Mycopathologia 174, 259–271. doi: 10.1007/s11046-012-9551-x
Silva, S., Henriques, M., Hayes, A., Oliveira, R., Azeredo, J., and Williams, D. W. (2011). Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J. Oral Pathol. Med. 40, 421–427. doi: 10.1111/j.1600-0714.2010.00981.x
Song, Y., Chen, X., Yan, Y., Wan, Z., Liu, W., and Li, R. (2020). Prevalence and antifungal susceptibility of pathogenic yeasts in China: a 10-year retrospective study in a teaching hospital. Front. Microbiol. 11:1401. doi: 10.3389/fmicb.2020.01401
Stavrou, A. A., Perez-Hansen, A., Lackner, M., Lass-Florl, C., and Boekhout, T. (2020). Elevated minimum inhibitory concentrations to antifungal drugs prevail in 14 rare species of candidemia-causing Saccharomycotina yeasts. Med. Mycol. 58, 987–995. doi: 10.1093/mmy/myaa005
Tati, S., Davidow, P., McCall, A., Hwang-Wong, E., Rojas, I. G., Cormack, B., et al. (2016). Candida glabrata binding to Candida albicans hyphae enables its development in oropharyngeal candidiasis. PLoS Pathog. 12:e1005522. doi: 10.1371/journal.ppat.1005522
Wang, H., Zhang, L., Kudinha, T., Kong, F., Ma, X. J., Chu, Y. Z., et al. (2016). Investigation of an unrecognized large-scale outbreak of Candida parapsilosis sensu stricto fungaemia in a tertiary-care hospital in China. Sci. Rep. 6:27099. doi: 10.1038/srep27099
Wang, Q., Li, Y., Cai, X., Li, R., Zheng, B., Yang, E., et al. (2021). Two sequential clinical isolates of Candida glabrata with multidrug-resistance to posaconazole and echinocandins. Antibiotics (Basel) 10:1217. doi: 10.3390/antibiotics10101217
Wisplinghoff, H., Ebbers, J., Geurtz, L., Stefanik, D., Major, Y., Edmond, M. B., et al. (2014). Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features and antifungal susceptibilities. Int. J. Antimicrob. Agents 43, 78–81. doi: 10.1016/j.ijantimicag.2013.09.005
Wu, Y., Wang, J., Li, W., Jia, H., Che, J., Lu, J., et al. (2013). Pichia fabianii blood infection in a premature infant in China: case report. BMC. Res. Notes 6:77. doi: 10.1186/1756-0500-6-77
Xiao, M., Chen, S. C. A., Kong, F., Fan, X., Cheng, J.-W., Hou, X., et al. (2018a). Five-year China Hospital invasive fungal surveillance net (CHIF-NET) study of invasive fungal infections caused by noncandidal yeasts: species distribution and azole susceptibility. Infect. Drug Resist. 11, 1659–1667. doi: 10.2147/IDR.S173805
Xiao, M., Sun, Z. Y., Kang, M., Guo, D. W., Liao, K., Chen, S. C., et al. (2018b). Five-year national surveillance of invasive candidiasis: species distribution and azole susceptibility from the China Hospital invasive fungal surveillance net (CHIF-NET) study. J. Clin. Microbiol. 56, e00577–e00518. doi: 10.1128/jcm.00577-18
Zhang, L., Xiao, M., Arastehfar, A., Ilkit, M., Zou, J., Deng, Y., et al. (2021). Investigation of the emerging nosocomial Wickerhamomyces anomalus infections at a Chinese tertiary teaching hospital and a systemic review: clinical manifestations, risk factors, treatment, outcomes, and anti-fungal susceptibility. Front. Microbiol. 12:744502. doi: 10.3389/fmicb.2021.744502
Keywords: pathogenic yeasts, invasive fungal diseases, Candida spp., antifungal susceptibility, triazoles, echinocandins, multidrug resistance
Citation: Wang Q, Cai X, Li Y, Zhao J, Liu Z, Jiang Y, Meng L, Li Y, Pan S, Ai X, Zhang F, Li R, Zheng B, Wan Z and Liu W (2022) Molecular identification, antifungal susceptibility, and resistance mechanisms of pathogenic yeasts from the China antifungal resistance surveillance trial (CARST-fungi) study. Front. Microbiol. 13:1006375. doi: 10.3389/fmicb.2022.1006375
Received: 29 July 2022; Accepted: 20 September 2022;
Published: 06 October 2022.
Edited by:
Weihua Pan, Shanghai Changzheng Hospital, ChinaCopyright © 2022 Wang, Cai, Li, Zhao, Liu, Jiang, Meng, Li, Pan, Ai, Zhang, Li, Zheng, Wan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Liu, bGl1d2VpQGJqbXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.