
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 October 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1004733
This article is part of the Research Topic Rising Stars in Microbial Physiology and Metabolism: 2022 View all 5 articles
 Wichanan Wannasrichan1
Wichanan Wannasrichan1 Htut Htut Htoo2
Htut Htut Htoo2 Rubsadej Suwansaeng2
Rubsadej Suwansaeng2 Joe Pogliano3
Joe Pogliano3 Poochit Nonejuie2
Poochit Nonejuie2 Vorrapon Chaikeeratisak1*
Vorrapon Chaikeeratisak1*Pseudomonas aeruginosa, a major cause of nosocomial infections, has been categorized by World Health Organization as a critical pathogen urgently in need of effective therapies. Bacteriophages or phages, which are viruses that specifically kill bacteria, have been considered as alternative agents for the treatment of bacterial infections. Here, we discovered a lytic phage targeting P. aeruginosa, designated as JJ01, which was classified as a member of the Myoviridae family due to the presence of an icosahedral capsid and a contractile tail under TEM. Phage JJ01 requires at least 10 min for 90% of its particles to be adsorbed to the host cells and has a latent period of 30 min inside the host cell for its replication. JJ01 has a relatively large burst size, which releases approximately 109 particles/cell at the end of its lytic life cycle. The phage can withstand a wide range of pH values (3–10) and temperatures (4–60°C). Genome analysis showed that JJ01 possesses a complete genome of 66,346 base pairs with 55.7% of GC content, phylogenetically belonging to the genus Pbunavirus. Genome annotation further revealed that the genome encodes 92 open reading frames (ORFs) with 38 functionally predictable genes, and it contains neither tRNA nor toxin genes, such as drug-resistant or lysogenic-associated genes. Phage JJ01 is highly effective in suppressing bacterial cell growth for 12 h and eradicating biofilms established by the bacteria. Even though JJ01-resistant bacteria have emerged, the ability of phage resistance comes with the expense of the bacterial fitness cost. Some resistant strains were found to produce less biofilm and grow slower than the wild-type strain. Among the resistant isolates, the resistant strain W10 which notably loses its physiological fitness becomes eight times more susceptible to colistin and has its cell membrane compromised, compared to the wild type. Altogether, our data revealed the potential of phage JJ01 as a candidate for phage therapy against P. aeruginosa and further supports that even though the use of phages would subsequently lead to the emergence of phage-resistant bacteria, an evolutionary trade-off would make them more sensitive to antibiotics.
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen responsible for nosocomial infections that represent a leading cause of Healthcare-Associated Infections (HAIs) (Magill et al., 2014; Weiner et al., 2016) and death in cystic fibrosis (CF) patients (Moradali et al., 2017). It also occupies the top position among Gram-negative bacteria as they are responsible for healthcare-associated pneumonia, the most common type of nosocomial infection in the United States (Magill et al., 2014). P. aeruginosa can naturally resist various classes of antibiotics due to its both intrinsic and extrinsic mechanisms for resistance, and thus, the treatment of P. aeruginosa infection tends to be less successful than other bacterial infections (Lister et al., 2009; Pang et al., 2019). Not only does it show intrinsic and acquired resistant mechanisms, P. aeruginosa also possesses an adaptive resistance in which the gene expression pattern is altered in response to stresses, growth stages, and environments (Breidenstein et al., 2011; Coleman et al., 2020). The ability of biofilm formation is one of the adaptive resistance mechanisms of P. aeruginosa whose biofilms serve as a virulence factor for its attachment to the surfaces and each other, assembling as a microenvironment. The biofilm also serves as a barrier that limits the diffusion of antibiotics, making it hard for them to reach the bacterial cells inside, further empowering its antibiotic resistance (Drenkard, 2003; Das et al., 2013). The drug-resistant P. aeruginosa has been announced by WHO as being “first priority pathogens” for global human health, for which new antibiotics or approaches are urgently needed (Duval et al., 2019). Unfortunately, the lack of discovery of novel drugs in comparison to the swift rate of the emergence of antibiotic resistance has made the fight against P. aeruginosa harder to handle (Gordillo Altamirano and Barr, 2019).
Also known as phage therapy, bacteriophages (phages), prokaryotic viruses that specifically kill bacteria, have been considered as an alternative way to combat drug-resistant bacteria (Kortright et al., 2019). The phages were claimed as the most abundant entities in the biosphere with a rough estimation of 1031 particles (Hatfull, 2015). Therefore, they can be easily found and used to fight against bacteria. Auto-dosing and specificity to the hosts are among the advantages of using phages in therapies. Phages have the ability to penetrate through biofilms, which are the drug-resistant mechanisms of P. aeruginosa, and kill persistent cells (Loc-Carrillo and Abedon, 2011). Phage therapies using Pseudomonas phages against P. aeruginosa infections have been proven to be successful in many cases worldwide, such as in the treatment of mouse models that were infected with the pathogen in multi-infection sites including lungs (Debarbieux et al., 2010; Morello et al., 2011; Chen et al., 2021; Yang et al., 2021), burn wounds (McVay et al., 2007), cornea (Fukuda et al., 2012), gut (Watanabe et al., 2007), and blood (Watanabe et al., 2007; Yang et al., 2021). Phage therapies for P. aeruginosa infection in humans have also been reported (Wright et al., 2009; Khawaldeh et al., 2011; Chan et al., 2018; Maddocks et al., 2019). For example, in England, a single dose of the six-phage cocktail was exploited to treat 24 patients with chronic otitis that was caused by antibiotic-resistant P. aeruginosa. After the therapy, the majority of the patients have significantly improved symptoms and also show a significant reduction in bacteria in patient samples (Wright et al., 2009). Due to recent breakthroughs in systematic phage therapy in clinical setup, many researchers have been focusing on the isolation of novel phages in order to create a strong foundation for phage therapy against this pathogen. As a result, many virulent P. aeruginosa phages have been discovered and characterized, giving rise to great potential for therapeutic purposes (Garbe et al., 2010, 2011; Pires et al., 2011; Ahiwale et al., 2012; Kim et al., 2012; Danis-Wlodarczyk et al., 2015; Yu et al., 2017; de Melo et al., 2019; Ong et al., 2020).
Albeit numerous successful cases of phage therapy against antibiotic-resistant bacterial infection, the development of phage-resistant P. aeruginosa after phage treatment is inevitable. A phage therapy study in endocarditis rats with a phage cocktail containing 12 phages revealed that the frequency of phage-resistant P. aeruginosa subpopulation occurred around 10–4 at 6 h and 10–2 at 24 h after phage treatment (Oechslin et al., 2016). P. aeruginosa is able to rapidly evolve to resist the phage infection through the modification of its outer membrane structure, such as glycosylation of its pilus and loss of O-antigen, where the phage first attaches (Johnson and Lory, 1987; Le et al., 2015; Yang et al., 2020). Hall et al. (2012) performed both single- and multi-phage therapy in wax moth larvae against P. aeruginosa infection, and they demonstrated that the single-phage use was not as effective as the multi-phage therapy to suppress bacterial growth. However, phage-resistant bacteria with impaired fitness were able to eventually emerge in both therapies (Hall et al., 2012). This raises concerns regarding the ability of P. aeruginosa which trades off its physiological fitness to rapidly evolve to resist phages (Hosseinidoust et al., 2013; Markwitz et al., 2021).
Since P. aeruginosa is highly capable of developing resistance to phages, it is very crucial to expand the variety of phages that effectively target this pathogen. This will allow scientists to have more choices to formulate efficient phage cocktails, which are highly preferred in the treatment of bacterial infections to suppress possible resistance (Chan et al., 2013). Here, we discovered a novel virulent Pseudomonas bacteriophage, named JJ01, which is classified as a member of Pbunavirus genus, Myoviridae family. This phage not only exhibits a strong lytic activity but also has the ability to eradicate biofilm formation of P. aeruginosa. Both conventional and molecular characterizations of phage JJ01 were conducted to explore the potential of the phage for therapeutic purposes, including adsorption assay, one-step growth experiment, burst size determination, viral particle stability, host range analysis, and whole-genome analysis. Moreover, JJ01-resistant P. aeruginosa strains were isolated and characterized for the physiological trade-off for phage resistance. Our finding revealed that this novel virulent Pseudomonas phage JJ01 could be a candidate for phage therapy in the future.
Pseudomonas aeruginosa strain PAO1 was used as the host for phage isolation. Before experiments, bacterial cells were grown overnight in Luria-Bertani (LB) broth (Tryptone; Himedia™, Cat. No. RM027, and Yeast extract; Himedia™, Cat. No. RM014) at 37°C with shaking at 250 rpm. Environmental samples were enriched by adding LB medium, 0.5 mM CaCl2, and P. aeruginosa culture, and incubated at 37°C for 48 h with shaking at 200 rpm. After the incubation, the mixture was centrifuged at 8,500 rpm and 4°C for 10 min and filtered through a 0.45 μm filter. A spot test was performed to indicate the existence of P. aeruginosa phage. For the phage purification, the filtrates were diluted in SM buffer and mixed with P. aeruginosa before preparing the double-layer agar (DLA) plates. After that, the single plaques were selected and kept in SM buffer. Plaques with the same morphology were selected and purified at least 3–5 times to ensure that identical phages were derived. To propagate high-titer bacteriophage, phage-containing SM buffer was diluted into a suitable dilution. Ten microliters of the phage suspension was mixed with the overnight culture of P. aeruginosa and 5 mL of 0.35% LB agar (top agar). Then, the mixtures were transferred onto LB plates (bottom agar). The plates that displayed web lysis of plaques were then selected and soaked with 5 mL of SM buffer for 5 h (Chaikeeratisak et al., 2017). After that, the phage lysate was collected and centrifuged at 8,500 rpm and 4°C for 10 min before filtration with a 0.45 μm filter. The phage lysate was kept at 4°C until use, and the phage titer was counted by spot test.
This work has been reviewed and approved by Chulalongkorn University-Institutional Biosafety Committee (CU-IBC) in accordance with the levels of risk in pathogens and animal toxins, listed in the Risk Group of Pathogen and Animal Toxin (2017) published by the Department of Medical Sciences (Ministry of Public Health), the Pathogen and Animal Toxin Act (2015) and Biosafety Guidelines for Modern Biotechnology BIOTEC (2016), with approval number: SC CU-IBC-028/2020 Ex1.
Ten microliters of early-log phase PAO1 (OD600 ≈ 0.2) was mixed with 100 μL of 1010 PFU/mL of phage JJ01. The mixture was incubated for 5 min and diluted before spreading on LB plates (Le et al., 2015). After 24 h of incubation, the colonies were selected and purified at least three times. The phage resistance of the candidates was confirmed by drawing a straight line of each isolate on the LB agar plate and dropping 5 μL of phage lysate onto the bacterial stripe. After incubation, if a clear zone does not appear as observed in the wild-type PAO1, the bacterial isolates are considered to be phage-resistant. These isolates were then confirmed to be P. aeruginosa by MALDI Biotyper and were cryo-frozen for later experiments.
Prophage induction by mitomycin C and prophage detection in bacterial host genomes were conducted to determine whether phage JJ01 was a temperate phage. In prophage induction by mitomycin C, mid-log phase cultures (OD600 ≈ 0.4) of JJ01-resistant isolates and wild-type PAO1 (internal control) were mixed with 10 μg/mL (final concentration) of mitomycin C. The mixtures were incubated at 37°C for 2 h before centrifugation at 12,000 × g for 5 min. The supernatants were filtered with 0.45 μm filters. The filtrates were serially diluted, followed by a spot test on the PAO1 lawn (Webb et al., 2003). The plates were incubated at 37°C for 24 h. Prophage detection in bacterial host genomes, colony PCR of genes gp44 and gp56 that are solely found in bacteriophages (particularly phage JJ01), was performed in phage-resistant P. aeruginosa isolates using specific primer sets. PCR products of gp44 and gp56 were then run on agarose gel electrophoresis and checked for the amplicon size. If a prophage integrates into the bacterial genome, its gene would be amplified and detected.
To determine the time required for JJ01 attachment to P. aeruginosa, the bacterial culture with OD600 ≈ 0.4 (≈4 × 108 CFU/mL) (Bucior et al., 2014) was infected with phage JJ01 at a multiplicity of infection (MOI) of 0.01 and incubated at 37°C. At each time point of incubation (0, 1, 2, 5, 7.5, 10, 15, 20, 25, and 30 min), the JJ01-PAO1 mixture was aliquoted and pelleted by centrifugation at 15,000 × g for 2 min. The supernatant was serially diluted in SM buffer, and a spot titer test was performed to determine the number of free phage particles in the supernatant. The experiment was performed in triplicates (Thammatinna et al., 2020).
Pseudomonas aeruginosa PAO1 in exponential phase (OD600 ≈ 0.4) was infected with phage JJ01 at MOI of 0.01 for 10 min at 37°C. Then, the cells were centrifuged at 12,000 × g for 5 min. The pellet was resuspended in LB broth and incubated in a shaker at 250 rpm for 2 h. Every 10 min including time point at 0 min, the suspension was collected, serially diluted, and the phage titer was counted by plaque assay. The PFU/mL was calculated and plotted against time in minutes post-infection (mpi). The phage burst size was calculated from the plotted curve. Briefly, the average value of the highest phage titer after the burst was divided by the average value of baseline phage titer during the latent period. The result was described as particles per cell (Kropinski, 2018).
For thermal stability, 100 μL of phage JJ01 lysate was incubated at different temperatures: 4, 20, 30, 37, 40, 50, 60, 70, 80, and 100°C. For pH stability, 10 μL of phage JJ01 was mixed with 90 μL of different pH values of SM buffer ranging from pH 2 to 10. After 1 h of incubation, phage JJ01 from both experiments was serially diluted and enumerated by spot titer test. The experiments were performed in triplicates.
To determine the host of the phage JJ01, 5 μL of lysate was spotted on the lawn of P. aeruginosa strains derived from the Department of Medical Sciences (DMST), Ministry of Public Health, Thailand, and American Type Culture Collection (ATCC). Other hosts including Vibrio cholerae, Escherichia coli, Burkholderia thailandensis, and Acinetobacter baumannii were also used as indicated in Table 1. The efficiency of plating (EOP) was also performed in JJ01-susceptible bacteria by the spot test of different concentrations of phage JJ01 onto each tested bacterial lawn. The total number of plaques produced in each bacterial strain was then compared. The EOP was calculated (average PFU on target bacteria/average PFU on host bacteria) from triplicate along with the standard deviation. The average EOP value for a particular combination of phage and bacterium was classified according to the efficiency of killing as “High production” (the ratio was 0.5 or more), “Medium production” (the ratio was between 0.1 and 0.5), “Low production” (the ratio was between 0.001 and 0.1), and “inefficient” (the ratio was below 0.001) (Khan Mirzaei and Nilsson, 2015).
Phage JJ01 lysate (∼1011 PFU/mL) was precipitated overnight with 10% w/v polyethylene glycol and 1 M NaCl. The precipitated phage was pelleted at 8,500 rpm for 10 min and resuspended in SM buffer. The phage was negatively stained with 2% uranyl acetate on a carbon-coated grid, and the morphology of the phages was visualized by transmission electron microscopy (TEM) (HITACHI model HT7700). Then, the phages were classified based on their structure observed under TEM.
To determine the lytic activity of phage JJ01 against the host, P. aeruginosa PAO1 at the early exponential phase was infected with phage JJ01 in 96-well plates at different MOIs (0.01, 0.1, 1, 10, and 100) (Imam et al., 2019; Sharma et al., 2021). The mixtures were incubated at 37°C and were collected every 10 min for 12 h. During this, bacterial growth was monitored through the measurement of optical density (OD) using a Microplate reader (Synergy™ H1, Biotek) at 600 nm. The bacterial culture without phage infection was used as a control. The experiment was performed in triplicates.
To conduct the biofilm eradication assay, 5 μL of overnight cultured P. aeruginosa PAO1 was seeded into each well of 96-well plates that contained 145 μL of LB broth. After incubation at 37°C for 48 h, planktonic cells were removed and the plate was washed two times with 300 μL of PBS buffer. Next, 200 μL of LB broth containing phage JJ01 (106, 107, and 108 PFU/mL) was added to the wells, and the plate was further incubated at 37°C for 48 h. The phage suspension was then aspirated followed by washing with 300 μL of PBS buffer three times. To measure the biofilm amount remaining after eradication by the phage, 200 μL of absolute ethanol was added to the wells and the plate was then incubated for 15 min to fix the biofilm. After the ethanol was removed and dried, 200 μL of a 1% solution of crystal violet was added to the wells. The plate was incubated for 15 min at room temperature before rinsing with water three times in a water tub, followed by tapping on a paper towel to remove the excess water. After the plate was dried, 150 μL of 30% acetic acid solution was added to the wells, and the plate was further incubated for 1 h at room temperature to solubilize the crystal violet-staining biofilm. Finally, the absorbance of the biofilm content was measured at 580 nm using a Microplate reader (Synergy™ H1, Biotek). To measure biofilm formation of wild-type P. aeruginosa and JJ01-resistant isolates, 5 μL of bacterial overnight culture was added to 145 μL of LB broth. The cell suspension was cultured in the well plate at 37°C for 48 h, followed by the steps for biofilm measurement described above (Chaudhry et al., 2017; Bisht et al., 2021).
The DNA of phage JJ01 was extracted using phenol–chloroform–isoamyl alcohol method. Briefly, the phage lysate with at least 109 PFU/mL was mixed with 10% w/v polyethylene glycol and 1M NaCl overnight to precipitate the phage particles. Phage precipitant was centrifuged at 8,500 rpm for 10 min (Sambrook et al., 1989). After that, it was re-suspended with SM buffer and treated with 10 U of DNase I and 0.1 mg/mL of RNase A to eliminate host DNA and RNA at 37°C. The DNase and RNase activities were inhibited by 20 mM EDTA, and the phage capsid was digested by 0.5 mg/mL of proteinase K and 0.5% sodium dodecyl sulfate (SDS). After the incubation at 55°C for 1 h, phenol–chloroform–isoamyl alcohol (25:24:1) (Sigma Aldrich, Switzerland) was added to the mixture at equal volumes. The mixture was gently mixed before centrifugation at 15,000 × g for 10 min, and the top aqueous layer was collected. The steps of phenol–chloroform–isoamyl alcohol and centrifugation were repeated before being treated with 1/10 volume of 3 M sodium acetate and 2 volumes of cold absolute ethanol. The genomic DNA was allowed to precipitate at –20°C for 3 h followed by centrifugation at 21,000 × g for 20 min. The DNA pellet was washed with 70% ethanol, centrifuged at 15,000 × g for 5 min, and the pellet was air-dried until the remaining ethanol evaporated. The phage DNA was dissolved in TE buffer. The DNA quality and quantity were analyzed using a NanoDrop 2000 spectrophotometer (Thermo Scientific) (Nale et al., 2016).
Whole-genome sequencing (WGS) was carried out using the MiSeq (Illumina). FASTQ files derived from the sequencing process were assembled with SPAdes. The single contig of the phage genome was annotated with DNA master version 5.23.6 (Glimmer 3.02 and GeneMark HMM). The functional prediction was manually performed using viral genomes available in the NCBI database. PHASTER and NCBI conserved domains were additionally used for the annotation. The phage genome map was constructed and visualized using Artemis: DNAPlotter version 18.1.0. Genome phylogenetic tree was constructed by aligning phage JJ01 genome sequence against all Pseudomonas phage genomes designated as genus Pbunavirus approved by International Committee on Taxonomy of Viruses (ICTV), including NC_048676.1 (SL1), NC_011756.1 (SN), NC_048745.1 (vB_PaeM_SCUT-S1), NC_048663.1 (R26), NC_048662.1 (R12), NC_028939.1 (vB_Pae_PS44), NC_011810.1 (PB1), NC_048806.1 (PA8P1), NC_048626.1 (PA01), NC_011166.1 (LMA2), NC_041865.1 (phiKTN6), NC_007810.1 (F8), NC_048744.1 (EPa61), NC_042079.1 (vB_PaeM_E217), NC_042080.1 (vB_PaeM_E215), NC_041870.1 (vB_PaeM_CEB_DP1), NC_028971.1 (DL68), NC_028745.1 (DL60), NC_048675.1 (BrSP1), NC_026600.1 (vB_PaeM_C1-14_Ab28), NC_041902.1 (PA5), FM897211.1 (14-1), AB560486.1 (KPP12), JN254800.1 (NH-4), NC_048699.1 (vB_PaeM_LS1), NC_011165.1 (LBL3), and NC_017674.1 (JG024). The sequences were aligned with the iterative refinement method, and the tree was constructed using MAFFT version 7 with Neighbor-Joining algorithm (NJ) with Bootstrap 1,000x. The T4 phage genome was used as an outgroup to construct the phylogenetic tree. To elucidate the genetic relation between phage JJ01 and other phages in Pbunavirus, genome comparison was performed by Easyfig software version 2.1 (Imam et al., 2019). Pairwise intergenomic similarities between related phage genomes were performed using the Virus Intergenomic Distance Calculator (VIRIDIC1) (Moraru et al., 2020). The VIRIDIC algorithm relies on ICTV criteria to calculate intergenomic similarities among phages for classification.
Minimal inhibitory concentrations (MICs) were determined for the following antibiotics using the microdilution method: ciprofloxacin (CIP), piperacillin (CIP), tobramycin (TOB), colistin (COL), meropenem (MER), and gentamicin (GEN). The antibiotics were serially diluted in a 96-well plate. Overnight cultures of wild-type PAO1 and phage-resistant isolates were diluted 100-fold in LB and allowed to grow until they reached exponential growth (OD600 ≈ 0.2). The bacterial culture was further diluted in LB medium, in order to obtain 200,000 cells per well. The cells were dispensed to each well of a 96-well plate that contained different concentrations of antibiotics in LB. A positive growth control that contained bacteria without antibiotics and a negative media control of LB medium only were included for each antibiotic that was tested. The 96-well plate was then incubated at 30°C for 24 h. The MIC for each antibiotic was determined, in comparison with both the negative and positive controls, as the lowest concentration of antibiotic that could visibly inhibit the growth of bacteria. All MIC determinations were carried out at least in triplicates (Htoo et al., 2019).
As previously described (Htoo et al., 2019), well-separated, single colonies of bacteria were inoculated in LB medium and grown overnight at 30°C on a roller at 50 rpm. The following day, the overnight culture was diluted 100-fold in fresh LB medium and further cultured until the early log phase of growth (OD600 of 0.2) was obtained. Colistin was added to the bacterial culture, at a concentration of 1.25 μg/mL, for both the wild-type PAO1 and W10 strains. Each colistin treatment was accompanied by an untreated control that consisted of the bacterial culture only. Both the control and the treated cultures were further incubated at 30°C on the roller at 50 rpm for 1 h. The cells were harvested by centrifugation at 8,000 x g for 30 s and concentrated by resuspending in 1:10 of the original volume. A few microliters of the concentrated culture was loaded onto a concave glass slide that was previously prepared with a 1.2% agarose pad on it, made with 10% LB, and containing fluorescent dyes: 1 μg/mL FM 4-64, 2 μg/mL DAPI, and 0.125 μM SYTOX Green. A cover slip was applied, and fluorescent microscopy was carried out on DeltaVision™ Ultra, with consistent imaging parameters throughout. Three independent experiments were performed for fluorescence microscopy. For image analysis, images from fluorescent microscopy were first pre-processed on the Fiji application (Schindelin et al., 2012), and then analyzed on CellProfiler software version 4.2.1 (Carpenter et al., 2006). From the parameters generated, a nucleoid outline was used to define the area from which SYTOX Green intensity is to be measured. The background intensity of each corresponding cell was then subtracted in order to obtain the intensity of SYTOX Green for each individual cell. Data for SYTOX Green intensity were obtained from the unadjusted, pre-deconvolved image from fluorescent microscopy. Data for the percentage of cells stained with SYTOX Green was collected from three independent experiments. Three to five images were randomly chosen from each condition (colistin-treated and untreated control) for every individual experiment. The percentage of cells stained with SYTOX Green was determined as the cells having SYTOX Green intensities more than twofold above (3x) the mean intensity of the untreated control for that strain and presented as mean ± standard deviation (STDEV).
Statistical analysis was performed, and statistical significance was determined by student’s t-test and one-way ANOVA with Tukey’s HSD post hoc test. The analyses for each experiment are mentioned in the figure legends.
The nucleotide sequence of the Pseudomonas phage JJ01 genome was deposited in the GenBank database with the accession number: ON324181.1. All data needed to evaluate the conclusions in the article are present in the article and/or the Supplementary material.
The newly isolated phage named “JJ01” was isolated from soil using P. aeruginosa PAO1 as a host. The individual plaque of phage JJ01 appears as either a clear or a turbid area at the center surrounded by a narrow opaque zone with a diameter of around 1.5 mm (Figure 1A and Supplementary Figure 1). As observed by TEM, phage JJ01 belongs to the Myoviridae family according to the structure that consists of an icosahedral capsid and a contractile tail, following the classification according to the International Committee on Taxonomy of Viruses (ICTV) guidelines. Phage JJ01 possesses an icosahedral head with an estimated height and width of 69.6 ± 3 nm and 65 ± 0.8 nm, respectively, and a tail that is approximately 118.9 ± 2.4 nm in length (Figure 1B; n = 5).
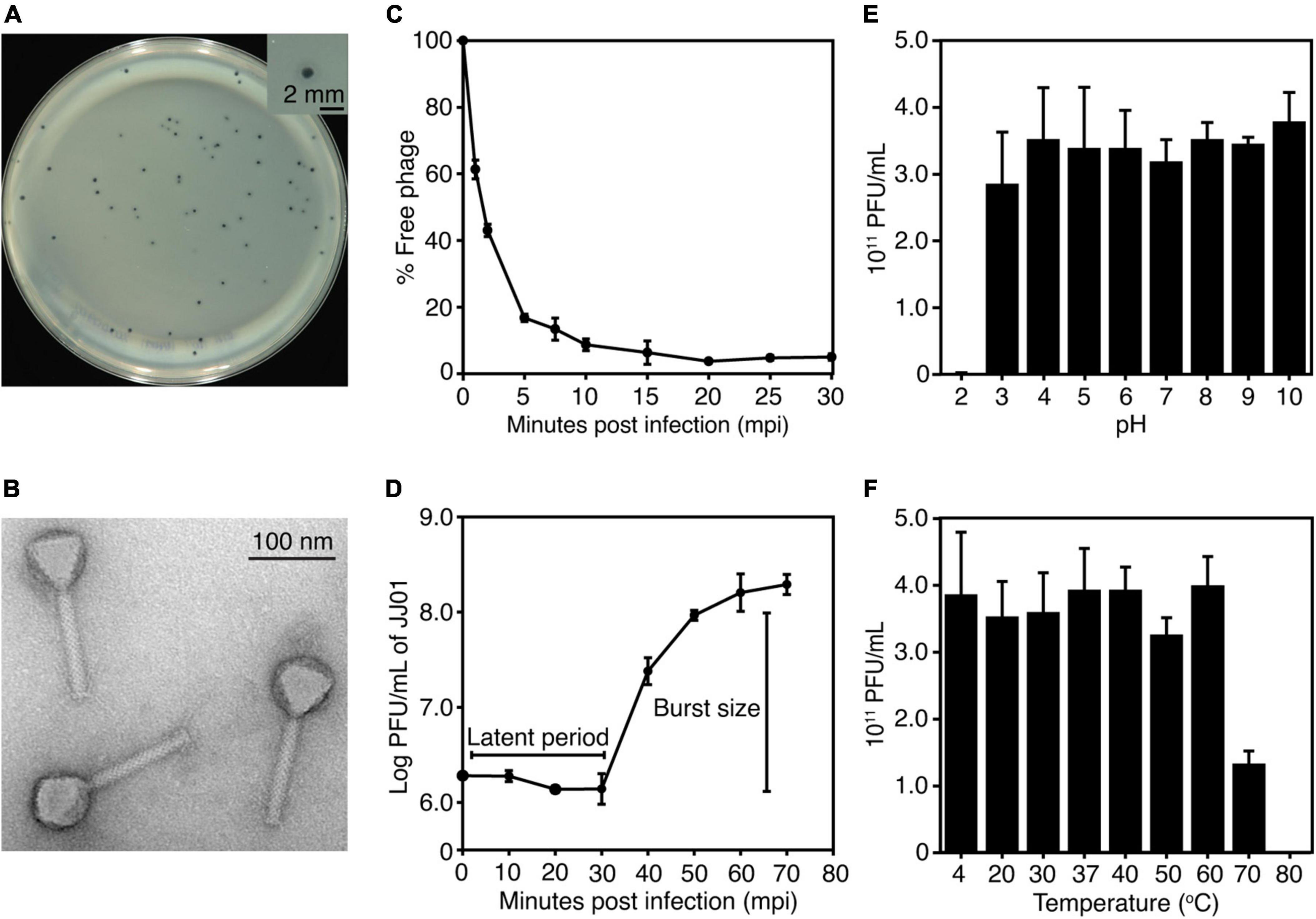
Figure 1. (A) Plaque morphology of phage JJ01. The right panel shows an individual plaque of JJ01. Scale bar equals 2 mm. (B) A transmission electron microscopy (TEM) image of the purified phage JJ01. Scale bar equals to 100 nm. (C–F) Biological properties of phage JJ01. (C) Adsorption assay, (D) One-step growth curve, (E,F) pH and thermal stability. (C–F) Experiments were performed in at least three replicates and data were represented as the mean ± standard deviation.
To better understand the biological properties of phage JJ01, we performed an adsorption assay, a one-step growth experiment, and a pH and thermal stability assay. The results of the adsorption assay demonstrated that phage JJ01 requires at least 10 min for around 90% of phage particles to get adsorbed onto the host cells (Figure 1C). One-step growth curve showed that phage JJ01 undergoes subcellular activities and propagates inside the host cells during the latent period, for 30 min, before releasing its progenies with a burst size of 109 particles per cell (Figure 1D). Furthermore, pH and thermal stability tests revealed that phage JJ01 withstands a wide range of pH from 3 to 10 and temperatures from 4°C to 60°C, where the numbers of viable phages are all comparable. Viable phages are completely inactivated at pH 2 and at 80°C; however, viable phages remain but decrease around 0.3 log at 70°C (Figures 1E,F). In addition, the host range of phage JJ01 was examined with 16 strains of various bacteria, including Pseudomonas spp. and other genera of bacteria. The result revealed that the phage is highly specific to P. aeruginosa, as it is capable of producing plaques only with P. aeruginosa PAO1, ATCC 15442, and ATCC 27853. No plaques were observed in other Pseudomonas strains and bacteria of different genera (Table 1).
Whole-genome sequencing by Illumina platform resulted in 1,287,392 reads of FastQ sequence after host DNA subtraction. After assembly by SPAdes with coverage over 2000x, a complete genome of phage JJ01 was obtained. Phage JJ01 has a double-stranded circularly permuted genome. Its genome is 66,346 base pairs long with a GC content of 55.7% and encodes 92 open reading frames (ORFs), as annotated by DNA master software (Figure 2 and Supplementary Table 2). For functional annotation, 38 ORFs with significant hits (E-values less than 10–5 according to Blastx search) were designated as functionally predicted genes (Table 2), while the rest (54 ORFs) were identified as hypothetical proteins. The functionally predicted ORFs were found to be composed of 9 genes related to DNA replication, transcription, and translation; 4 genes related to DNA metabolism and modification; 13 genes related to virion structure and assembly; 5 genes related to host cell lysis proteins; and 7 genes for other phage-related proteins. tRNA-encoding genes were not found in the genome (Figure 2 and Table 2). Phage JJ01 encodes a set of necessary enzymes involved in DNA replication (e.g., DNA primase; ORF13, DNA helicase; ORF15, DNA ligase; ORF81, and DNA polymerase III alpha and epsilon subunits; ORF88 and ORF89), DNA metabolism and modification (e.g., DNA adenine methyltransferase, ORF30, and thymidylate synthase, ORF92). Proteins involved in the DNA packaging machinery, including terminase large (ORF8) and small (ORF10) subunits, and the cell lysis module, including holin (ORF42), transglycosylase SLT domain-containing protein (ORF71), endolysin (ORF79), and U-spanin (ORF85), were also identified in the genome. In addition, toxin, drug-resistant, or lysogenic-associated genes were not found in the JJ01 genome. Since our genome annotation suggested that phage JJ01 is potentially a lytic phage due to the absence of lysogenic-associated genes in the genome, we therefore validated our annotation by the lysogeny test. We first isolated 10 JJ01-resistant P. aeruginosa strains and then confirmed the phage resistance and bacterial species by MALDI Biotyper (Supplementary Figure 2). After the activation of lysogens by mitomycin C, no clear zone was found around the spot of all JJ01-resistant P. aeruginosa isolates, indicating that no phage is released from the bacterial cells after stimulation (Supplementary Figure 3). To further confirm that phage JJ01 strictly enters the lytic cycle and is not capable of integrating its genome into the host cells, we screened for JJ01 prophage in the genomes of the resistant isolates using primer sets specific to ORF44 and ORF56 (Supplementary Figure 3). The result revealed that none of the resistant strains contain an integrated genome of phage JJ01, suggesting that phage JJ01 is indeed a lytic bacteriophage.
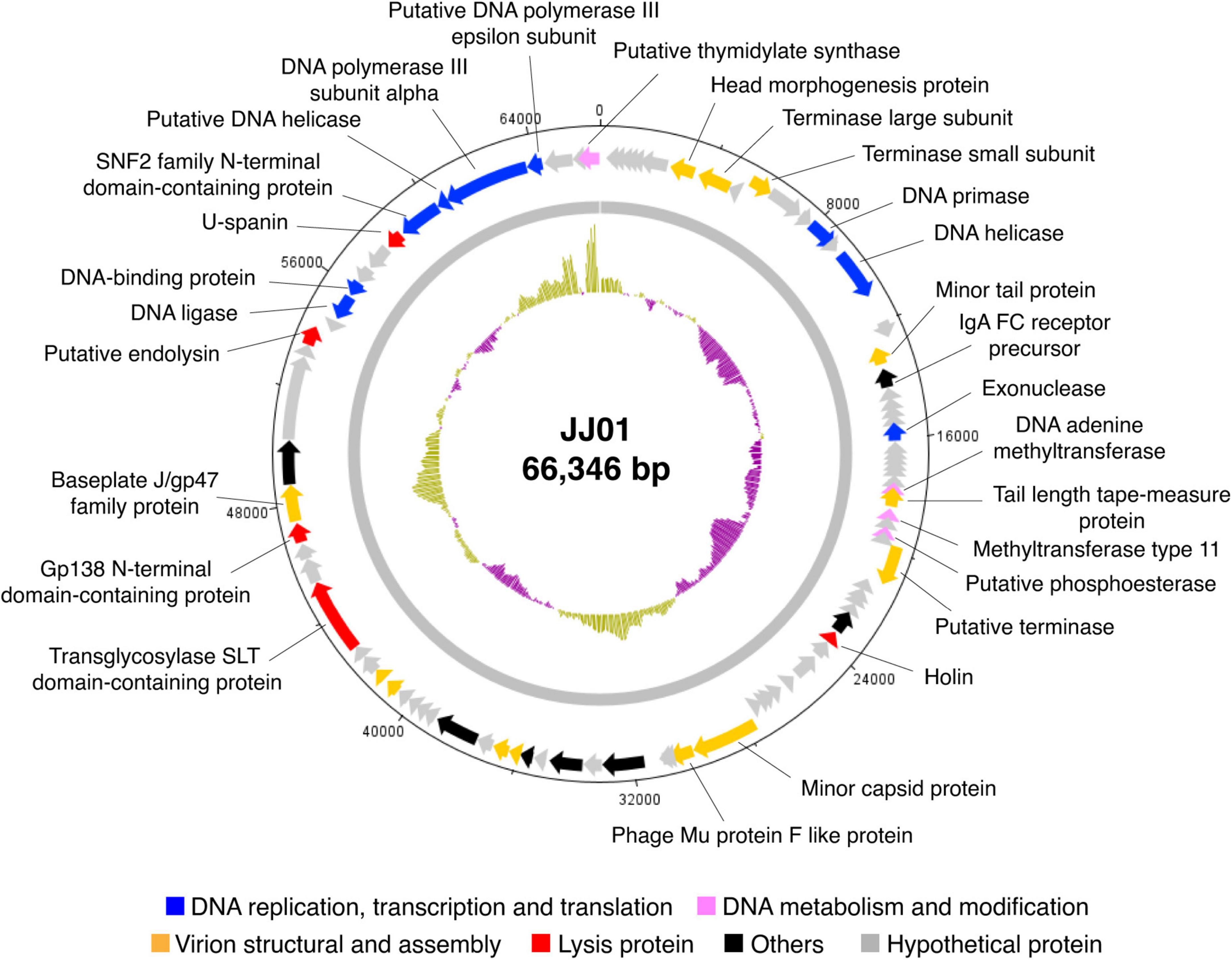
Figure 2. Circular genomic map of phage JJ01. The genome size is 66,346 base pairs, composed of 92 ORFs which are functionally annotated and are divided into different groups as indicated by colors as follows: blue, DNA replication and transcription, and translation; pink, DNA metabolism and modification; orange, virion structural and assembly; red, lysis protein; black, others; and gray, hypothetical proteins. An inner plot presents the GC content across the genome (yellow above average and purple below average). The direction of arrows indicates gene arrangement in the JJ01 genome. The map was generated using Artemis: DNAPlotter version 18.1.0.
According to Blastn search, the JJ01 genome sequence is highly similar to those of the members of the genus Pbunavirus (Order Caudovirales; Family Myoviridae). Genome to genome phylogenetic tree of phage JJ01 and other phages in the Pbunavirus genus was therefore constructed. The results showed that JJ01 has a very close relationship with phages LS1, E217, phiKTN6, and KPP12 (Figure 3A). VIRIDIC program was used to calculate pairwise intergenomic similarities among the phages, and it revealed that the genome similarity of phage JJ01 against KPP12, phiKTN6, E217, and LS1 is 93.89, 95.46, 96.04, and 96.44%, respectively (Supplementary Table 1). Even though all were clustered into the same genus, phage JJ01 was classified into a different species from KPP12 and phiKTN6. However, JJ01, E217, and LS1 were clustered into the same species due to the very high genome similarity (more than 96%) (Supplementary Table 1). To further elucidate whether their genomes are similarly organized, complete genomes of phages JJ01, E217, and LS1 were compared. The result revealed that, although JJ01, E217, and LS1 are likely the same species, based on genome similarity, their genomes are differently organized (Figure 3B). Specifically, the gene encoding for a terminase large subunit (Figure 3B, yellow arrow) of JJ01 is located at a different locus, compared to that of the other phages. Moreover, the large subunit of the terminase gene of LS1 is found to be located on the complementary strand of DNA, which is opposite to the direction of the JJ01 terminase gene. Altogether, these results suggested that phage JJ01 is a novel Pseudomonas phage that belongs to the genus Pbunavirus, whose genome sequence is highly similar to phages E217 and LS1.
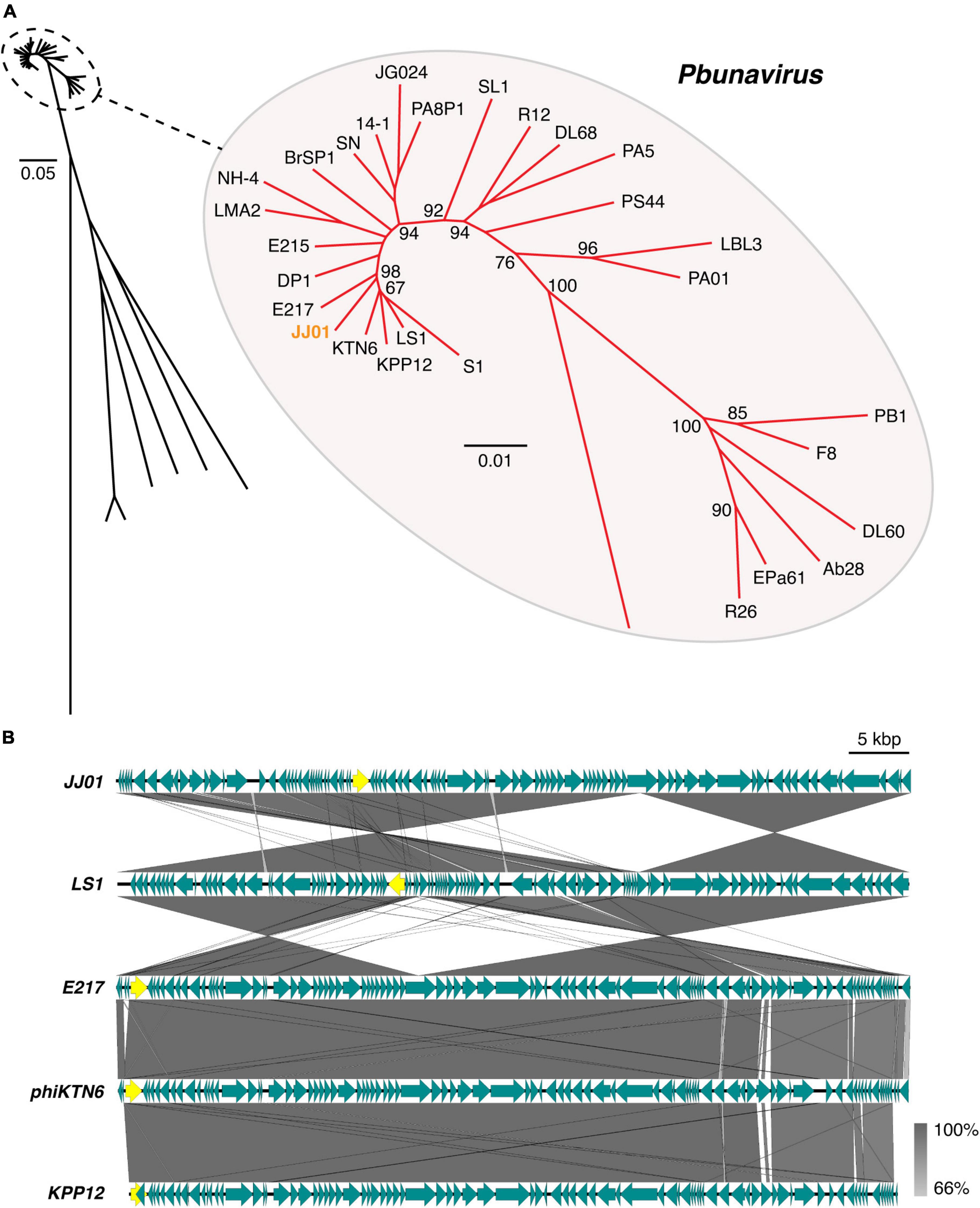
Figure 3. (A) Genome phylogenetic tree between species in Pbunavirus according to ICTV approval constructed using MAFFT version 7 with 1,000x Bootstrap. Numbers at branches represent Bootstrap values (%). Scale bars represent the branch length of taxa. (B) Comparative genomic analysis shows the gene organization of phage JJ01 compared to phages LS1, E217, phiKTN6, and KPP12 whose genomes are closely related to phage JJ01. Yellow labeled arrows represent the location and direction of the large subunit terminase gene.
To determine the killing activity of phage JJ01 against P. aeruginosa PAO1, we measured the bacterial growth during phage infection for 12 h at different MOIs (Figure 4A). The results demonstrated that MOI at 100 appears to be the best ratio to inhibit and efficiently suppress the growth of P. aeruginosa throughout the period of 12 h. As expected, the lower the MOIs, the lower the lytic activity. The graph showed descending lytic activities of the phage at MOIs from 10 to 0.01, with a pattern of bacterial growth quite similar to an MOI of 100. OD values of the cultures at MOI from 10 to 0.01 only increase at the beginning of incubation until 6 h, followed by a gradual decline until the end of the experiment (Figure 4A). In addition, during the experiment of 12 h, phage-resistant P. aeruginosa does not seem to emerge. To further investigate whether phage JJ01 plays any role in eliminating biofilm production of the bacteria, we first allowed P. aeruginosa to produce biofilm followed by the phage addition at various titers (106-108 PFU/mL) to test the ability of phage to eradicate the biofilm. The result revealed that phage JJ01 is able to eliminate the established biofilms with an almost 3-fold reduction when compared to the control (Figure 4B). Our findings that Pseudomonas phage JJ01 is highly efficient not only in killing the bacteria but also in eradicating the biofilm formation of the bacteria. These demonstrate the potential use of phage JJ01 in the clinical setting.
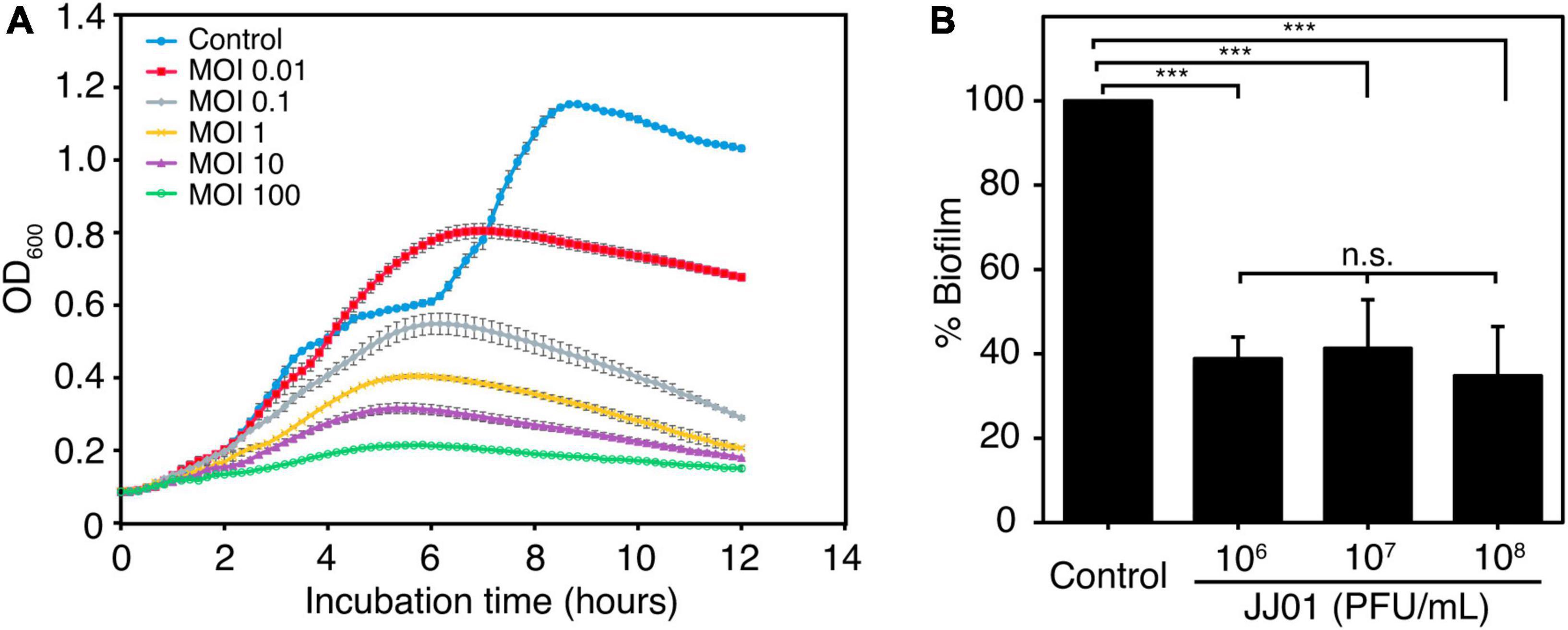
Figure 4. (A) Killing assay shows the ability of phage JJ01 to lyse P. aeruginosa cells at different multiplicity of infections (MOIs) at 0.01, 0.1, 1, 10, and 100 in LB medium at 37oC for 12 h. Each experiment was performed in triplicate. Data and error bars were represented as the mean ± standard deviation. (B) Biofilm eradication assay. Crystal violet staining reveals the grown biofilm level of P. aeruginosa PAO1 after incubation with phage JJ01 at 106, 107, and 108 PFU/mL for 48 h compared with the control (absence of phage JJ01). Asterisks represent significant differences in data (p ≤ 0.001) according to one-way ANOVA followed by Tukey’s HSD post hoc tests. n.s. represents statistical non-significance of data.
Even though phage resistance was not detected during infection at 12 h (Figure 4A), the emergence of JJ01-resistant P. aeruginosa was observed after incubation with phage JJ01 overnight. Previous studies showed that, due to an evolutionary trade-off, bacterial resistance to certain stressors required drastic physiological changes that commonly led to some degree of vulnerabilities, such as reduced bacterial overall fitness and susceptibility toward antibiotics (Le et al., 2015; Pires et al., 2017; Markwitz et al., 2021). Thus, we first explored if resistance to the phage affects any aspects of bacterial growth and physiology. Among the 10 JJ01-resistant strains (Supplementary Figures 2, 3), we selected JJ01-resistant strains W10, W11, and W12 as representatives of the study. We sequenced the whole genomes of these isolates, and the genomic data showed no sequences of JJ01 prophage in the genomes, assuring that W10, W11, and W12 are not lysogens and truly phage-resistant P. aeruginosa (data not shown). By measuring a bacterial growth curve in the presence of phage JJ01 at MOI of 100, the growth of the wild-type isolate is completely suppressed by phage JJ01 (Figures 5A,B; blue line). However, the growth of W10, W11, and W12 is not affected by the phage, indicating that the phage is not capable of killing the bacteria (Figures 5A,B; red, gray, and yellow lines). The trade-off of resistance to the phage seems to affect the growth and physiological properties of the isolates. Among the three representatives of resistant isolates, even though W11 and W12 appear to grow faster than the wild type, W10 grows much slower (Figure 5B; red line). The growth of W10 was also confirmed by smaller colony size when compared to the others (Figure 5C; upper panel). The defect in the bacterial growth was also observed in the other JJ01-resistant isolates, which accounted for approximately 85% of the isolates (six out of the additional seven tested isolates) (Supplementary Figure 4A). Moreover, some resistant isolates are impaired in their ability to produce biofilm, as W10, W132, W138, W142, and W143 produce up to 3-fold less biofilm than the wild-type PAO1 (Figure 5C, lower panel and Supplementary Figure 4B), and none is capable of producing higher amount of biofilm than the wild type statistically. Altogether, these results suggested that, to gain resistance toward the phage JJ01, some bacteria undergo a physiological trade-off that would result in the defect in bacterial growth and biofilm production.
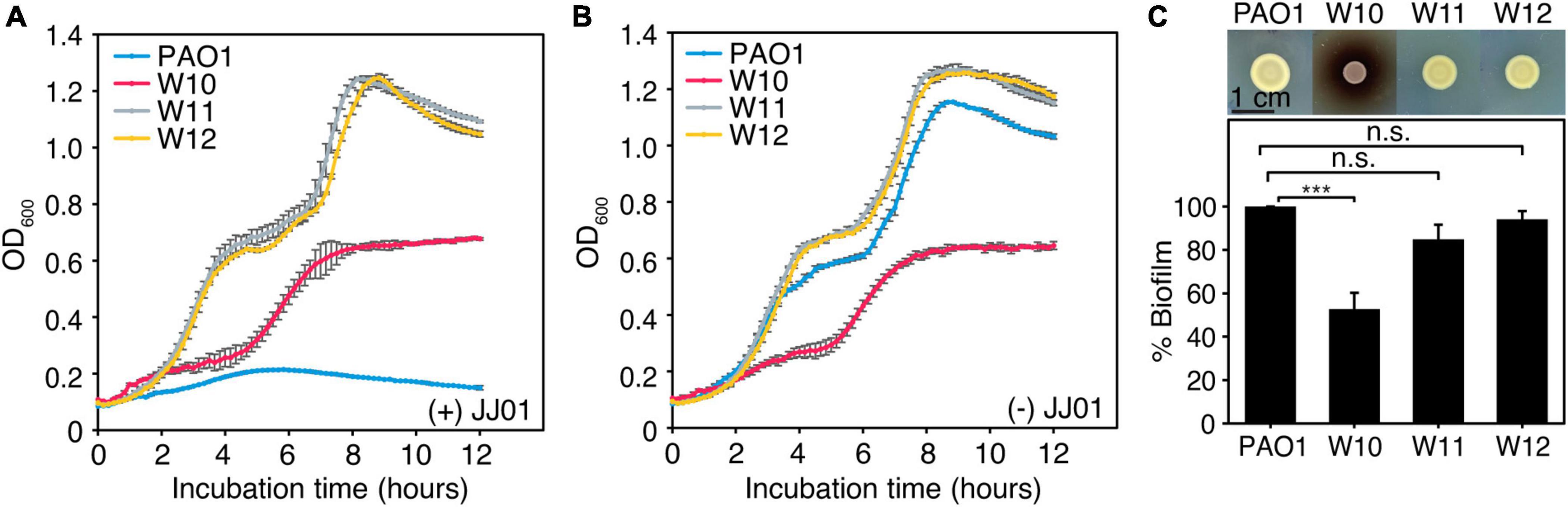
Figure 5. (A,B) Growth curve of wild-type PAO1 and representatives of phage-resistant isolates (W10, W11, and W12) (A) in the presence of phage JJ01 at MOI 100 and (B) in the absence of phage JJ01. (C) Colony morphology of JJ01-resistant P. aeruginosa isolates compared to wild-type PAO1 (upper panel). Scale bar equals 1 cm. The biofilm production assay shows the amount of biofilm production in JJ01-resistant P. aeruginosa isolates (W10, W11, and W12) compared to wild-type PAO1 (lower panel). Asterisks represent significant differences in data (p-value ≤ 0.001) according to one-way ANOVA followed by Tukey’s HSD post hoc tests. n.s. represents statistical non-significance of data.
Because biofilm serves as a barrier that limits the diffusion of antibiotics to access the bacterial cells (Pang et al., 2019), it was tempting to explore if the phage-resistant isolates that have lost their capability to produce biofilms become more sensitive to antibiotics. We then performed a minimum inhibitory concentration (MIC) assay of JJ01-resistant isolates and wild-type PAO1 against six antibiotics, which are commonly used to treat P. aeruginosa infections (Table 3 and Figure 6). The result showed that JJ01-resistant strains display various drug susceptibility profiles in the range of 2x increase (PIP, TOB, and MER) and 2x decrease (CIP, COL, MER, and GEN) in MIC, compared to those of the wild-type PAO1. Nevertheless, by the definition of Clinical and Laboratory Standard Institute (CLSI) standard, none of the JJ01-resistant isolates is defined as resistant to antibiotics according to their MIC level. Interestingly, 6 out of 10 resistant strains become 4–8 times more susceptible to COL as indicated by their lower MIC levels (Table 3). Among these strains, W10 and W132, which were observed to grow slower and produce brownish pigments, were noticeably more sensitive to COL with an 8-fold decrease in MIC, compared to wild-type PAO1 (Figure 5C, Supplementary Figures 2A, 4A, and Table 3). The hypersensitivity of W10 to COL was further confirmed by single-cell death analysis via impermeable SYTOX Green staining. In principle, when the membrane is compromised by COL, SYTOX Green can enter the cells and stain the nucleoid, thus emitting green fluorescence signals (Htoo et al., 2022). In other words, the more the cell membranes are damaged, the higher the number of cells that emit green fluorescence can be observed. This sensitivity of W10 toward COL, as tested in MIC assay, is in agreement with our microscopy data. In the untreated wild-type and W10 strains (Figures 6A,B; left panels), SYTOX Green did not well stain the bacterial nucleoids of both PAO1 and W10, as indicated by the very low percentage of stained nucleoids at 1.605 ± 1.522 and 2.369 ± 2.856, respectively (Figure 6C), indicating the uncompromised bacterial cell barrier. During the treatment of COL in the wild-type PAO1, the percentage of positive staining slightly increased, when compared to the untreated condition (Figures 6A,C). However, treatment of W10 with COL resulted in a substantial increase (above 14 times higher) in the percentage of stained nucleoids than in the treated wild-type PAO1 (Figures 6A–C), suggesting that the cell membrane of the majority of the W10 population was compromised and thus is more fragile than the wild-type cells when exposed to the same concentration of COL. Altogether, the results demonstrated that the evolution of phage resistance of W10 against phage JJ01 affects bacterial physiological properties, some of which, in turn, lead to antibiotic hypersensitivity of the bacteria.
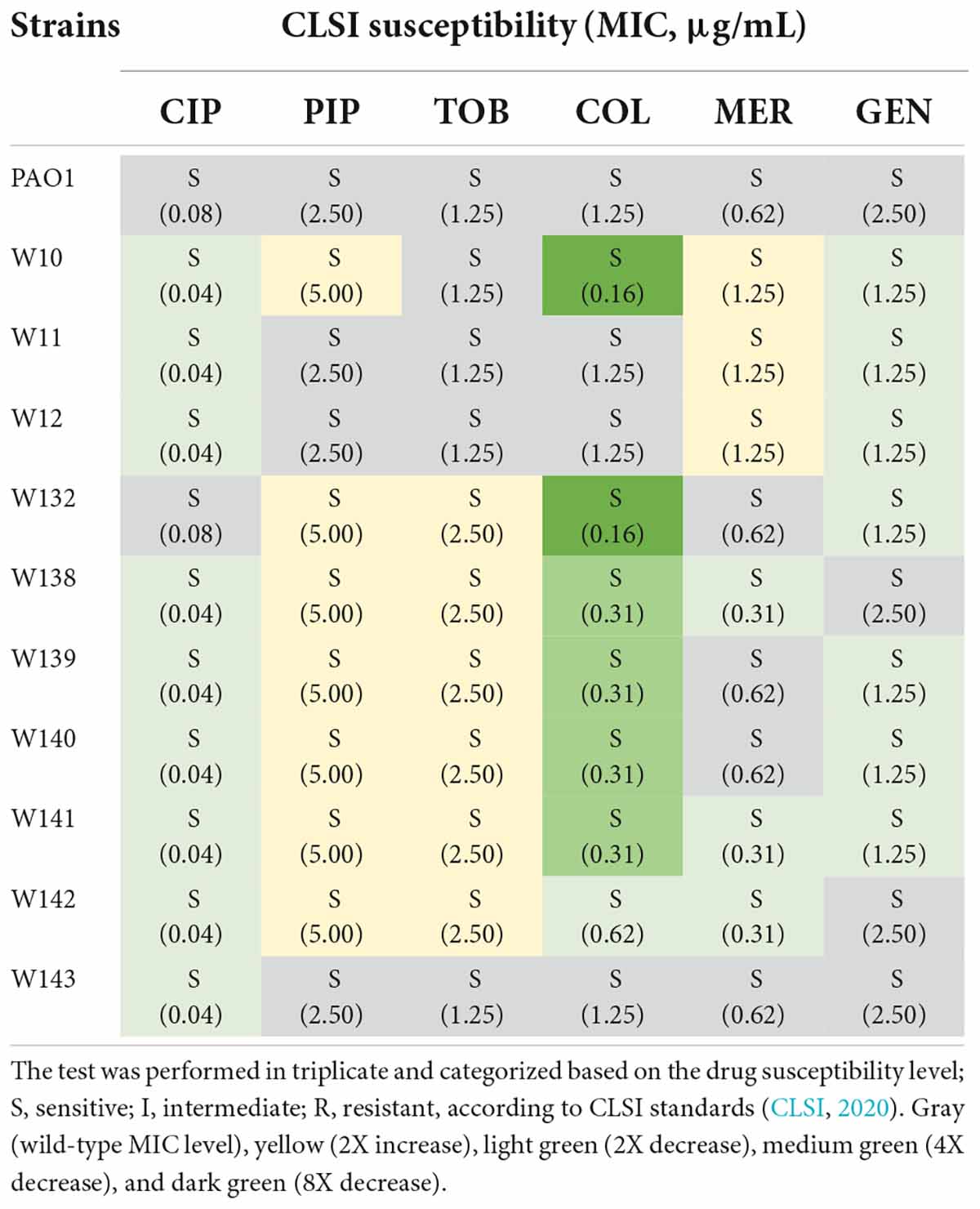
Table 3. Minimum inhibitory concentration (MIC) determination of six main antibiotics used for the treatment of P. aeruginosa infection, ciprofloxacin (CIP), piperacillin (PIP), tobramycin (TOB), colistin (COL), meropenem (MER), and gentamicin (GEN), against wild-type PAO1 and 10 strains of phage-resistant isolates.
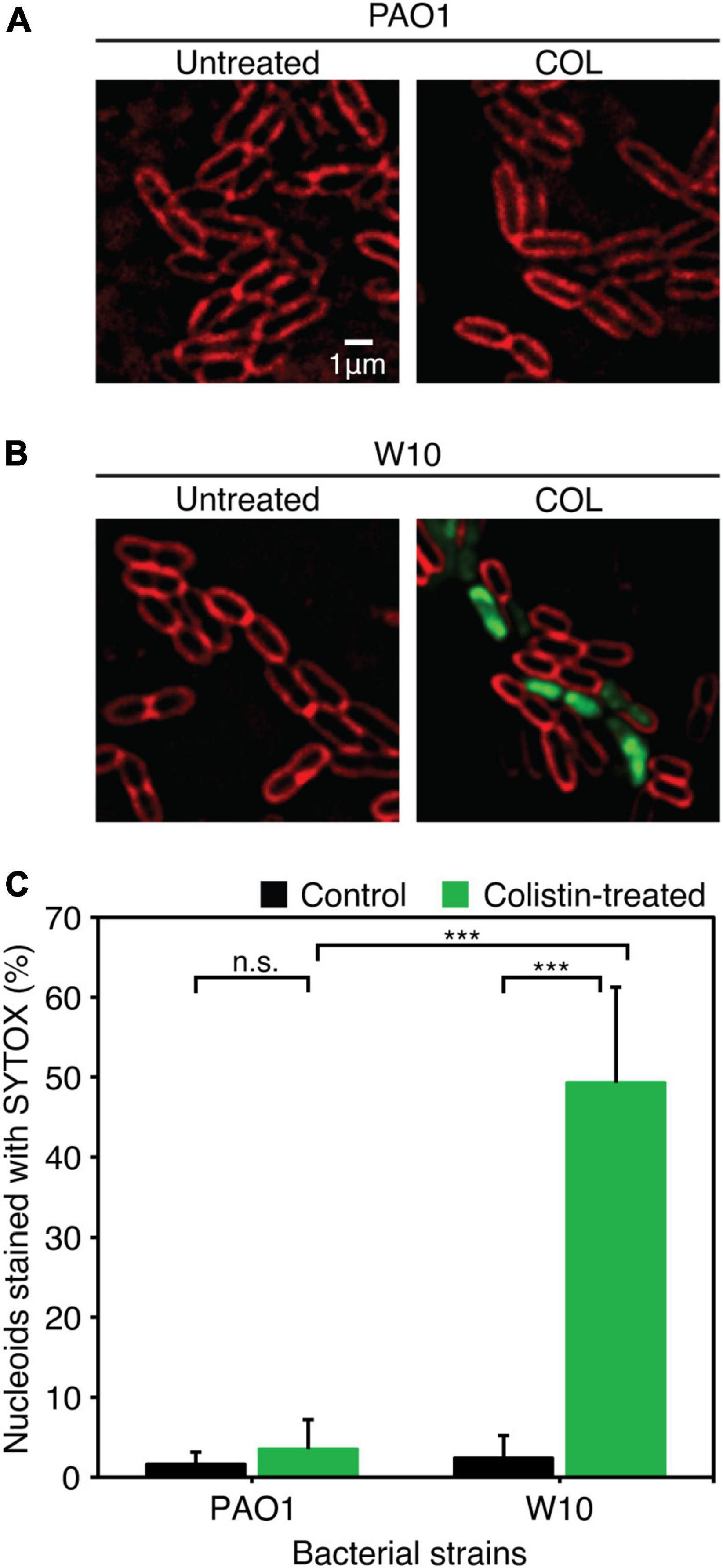
Figure 6. (A,B) Fluorescence images of wild-type PAO1 (A) and phage-resistant isolates (W10) (B) upon treatment with COL. The cell membrane was stained with FM4-64 (red) and nucleoid of cells with impaired membrane integrity was stained with impermeable SYTOX (Green). (C) Percentage of nucleoids strained with SYTOX Green after treatment with COL in wild-type PAO1 and phage-resistant isolation (W10) compared to untreated cells. SYTOX Green was used to stain DNA as an indicator for the cells with permeabilized membranes. Asterisks represent significant differences in data at p ≤ 0.001 according to the student’s t-test and n.s. represents statistical non-significance of data. The test was performed in triplicate. Scale bar equals 1 μm.
The emergence of antibiotic-resistant P. aeruginosa together with the shortage of novel antibiotic discoveries urges scientists around the world to find other alternative ways to deal with this worsening situation. Using bacteriophages for therapeutic purposes is one of the alternatives that have been proven to be a promising tool to cope with drug-resistant bacteria (Schooley et al., 2017; Dedrick et al., 2019). Here, we successfully isolated a novel virulent P. aeruginosa phage, designated as JJ01, from soil and showed its characteristic feature, which can be classified as a myovirus in the Myoviridae family. The myoviruses constitute about 25% of all tailed phages in the Caudovirales order (Ackermann, 2003), and are frequently found in Gamma-proteobacteria, the phylum of Pseudomonas genus. According to VIRIDIC analysis among phages in the Pbunavirus genus, JJ01 is very closely related to the phage E217 (96.04%) and with the highest similarity toward LS1 (96.44%). As the ICTV species demarcation criteria that classify the genome sequences whose sequence similarity is above 95% as organisms in the same species (Adriaenssens and Brister, 2017; Moraru et al., 2020), this raises some concerns on whether or not the phage JJ01 is a novel phage. Phylogenetic tree analysis revealed that JJ01 and E217 are classified as different species because they are in separate branches of the tree with a strong-supporting bootstrap value of over 95% (Soltis and Soltis, 2003). Even though the tree weakly supports the claim of JJ01 and LS1 as different species since the bootstrap number on their nodes is lower than 95 but higher than 50 (Soltis and Soltis, 2003), genome comparison between these phages indeed assured that they are different due to the distinct genome organizations. Our finding is not the only case. It is possible that different Pseudomonas phages isolated at different periods and from distant geographical areas contain high genome sequence similarities. For example, distinct giant phages that were classified in different families (siphophage SD1-M and nucleus-forming myophage phiKZ) shared a very high genome identity up to 99% with only two DNA segments different between their genomes (Kwan et al., 2006; Krylov et al., 2007; Chaikeeratisak et al., 2021). Specifically, in the Pbunavirus genus, PB1-like phages, which are the very common virulent P. aeruginosa phages, shared a remarkable genome similarity to one another (over 95%). Despite the very high similarity of their genomes, they are organized in different patterns and contain unequal gene clusters (Ceyssens and Lavigne, 2010). Therefore, our finding further supports this statement and revealed that the genome organization of phage JJ01, as one of the Pbunavirus members, has its gene orientation and genome organized differently from the nearest-related phage, LS1.
In the attempt to employ phages for therapeutic purposes for P. aeruginosa infection, a variety of Pseudomonas phages and phage cocktails have been isolated and investigated for their roles in the treatment of the infections (Wright et al., 2009; Khawaldeh et al., 2011; Chan et al., 2018; Maddocks et al., 2019). Many Pseudomonas phages in Pbunavirus genus have been proven to be successful in phage therapy. For example, phage KPP12, whose genome is highly similar to JJ01 (93.89%), displayed a broad host range over the clinical isolates of P. aeruginosa, and the phage use by single-dose administration was proven to improve the symptom outcome of ophthalmic infections in mice (Fukuda et al., 2012). Phages E215 and E217 which are genetically close to JJ01 were used as ingredients of a six-phage cocktail to treat acute respiratory infection in mice and cure bacteremia in wax moth larvae infected with P. aeruginosa. The phage cocktail was able to rescue and prolong the survival of the treated animals, suggesting the high efficiency of the cocktail (Forti et al., 2018). Our novel P. aeruginosa phage JJ01 does also display suitable characteristics and would be considered as a decent candidate for therapy. The phage JJ01 is a lytic bacteriophage. The phage is highly specific to P. aeruginosa and is capable of killing 75% of the tested strains of P. aeruginosa. With the fast adsorption to the bacterial host cells, it spends relatively short periods of replication and produces many phage progenies. Genome annotation further demonstrated that lysogenic-associated genes, as well as other unwanted genes such as toxin, virulence, or antibiotic-resistant genes, are not present in the JJ01 genome. The lysogeny test experimentally confirmed that JJ01 is not capable of lysogenizing the host and strictly enters the lytic cycle. Together with the stability of the phage over broad ranges of pH and temperatures, we have assured that phage JJ01 is potentially a great candidate for therapy against P. aeruginosa. Further investigation of phage JJ01 in targeting Pseudomonas clinical isolates will be needed to explore its potential in therapy.
However, the genome databases of prokaryotic viruses are not fully annotated and many phage-encoding genes in the database remain unknown, rendering it improper to use therapeutically. Rather than using the whole phage, the phage genome is also considered as an untapped resource for antimicrobial discovery (Liu et al., 2004; Van den Bossche et al., 2014; De Smet et al., 2021). Even though phage JJ01 harbors 54 out of 92 genes that were predicted to encode hypothetical proteins, the phage encodes 38 proteins with known functions. We found that the phage encodes putative DNA polymerase III alpha (ORF88) and epsilon subunits (ORF89), suggesting that its DNA metabolism is possibly independent of the host DNA replication system. This also implies that JJ01 might use these subunits to modulate or even interfere with host DNA polymerase during infection which would subsequently impair the host DNA synthesis. Phage JJ01 also harbors five genes that are involved in phage invasion and bacterial cell lysis: U-spanin (ORF85) disrupts bacterial outer membrane (Young, 2014), putative endolysin (ORF79) degrades peptidoglycan, gp138 N-terminal-domain containing protein (ORF74) pierces bacterial cell membrane during phage invasion (Browning et al., 2012), transglycosylase SLT domain-containing protein (ORF71) cleaves glycosidic bonds between N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) (Thunnissen et al., 1995), and holin (ORF42) creates holes on the bacterial cell membrane (Young, 2014). Indeed, these antimicrobials potentially support the use of alternative proteins instead of whole phage particles to combat pathogenic bacteria. Further investigation on whether these phage proteins are bactericidal would be needed to explore their antibacterial function.
Since biofilm provides a niche environment for bacteria to embed and serves as a barrier that limits the diffusion of antibiotics to bacterial cells (Drenkard, 2003), the bacteria growing in biofilms are more resistant to antibiotics and the host immune response than the cells that grow freely in suspension (Stewart and William Costerton, 2001). P. aeruginosa is one of those that alter its microenvironment by producing biofilms in which, with this protection, it becomes highly resistant to antibiotics up to 1,000 times more than the planktonic cells (Drenkard, 2003; Rumbaugh and Sauer, 2020). In our in vitro killing assay, phage JJ01 efficiently suppressed the bacterial growth at various doses even at very low MOI. Even though the emergence of phage resistance can be observed, half of the JJ01-resistant strains isolated in this study clearly exhibit defects in biofilm formation. Some of these strains also lose their ability to grow normally, when compared to the wild type. These characteristics observed in the phage JJ01-resistant strains would be beneficial for therapy. As the bacteria develop resistance to phage and lose the ability to establish biofilms, they would be easier to be eliminated by antibiotics.
A previous study revealed that most phage-resistant bacteria with a reduction of biofilm formation tend to be more susceptible to antibiotics. Moreover, some isolates also had less virulence and were unable to kill G. mellonella larvae (Markwitz et al., 2021). This implies that the susceptibility of JJ01-resistant strains to antibiotics might also be altered due to the impaired biofilm-producing ability. According to our antibiotic susceptibility test, W10 and W132, the phage-resistant strains that produce brown pigments (so-called brown phenotype), exhibited eight times higher sensitivity to colistin when compared to the wild type. The brown phenotype as observed in these strains is similar to the previously studied phage-resistant P. aeruginosa strains that were isolated during phage PaP1 (Le et al., 2015) and phage PaoP5 (Shen et al., 2018) infections. These mutants had been proven to contain an approximately 200 – 600 kb long genomic fragment deletion that was driven by MutL activity and non-homologous end joining (NHEJ) of the bacteria to avoid phage infection (Shen et al., 2018). The two genes that are always deleted among the brown phenotype mutants include hmgA, which is related to the brown pigment accumulation (Rodríguez-Rojas et al., 2009; Hosseinidoust et al., 2013), and galU, which is involved in lipopolysaccharide production, bacterial virulence, and synthesis of a component by which the phage recognizes (Le et al., 2015; Pires et al., 2017; Yang et al., 2020). Furthermore, the brown phenotype-producing P. aeruginosa strains that are resistant to phages AM.P2, Mat, and Kat have been recently shown to also exhibit hypersusceptibility toward human antimicrobial peptide LL-37 (Menon et al., 2022). Even though the deletion of this particular genomic fragment has been well conserved among the phage-resistant P. aeruginosa strains, it is still elusive how the deletion of this genomic region would contribute to the antibiotic hypersensitivity of phage-resistant P. aeruginosa. Whole-genome sequencing and analysis of these mutants will be further performed to investigate how the bacteria would trade its physiological fitness, which consequently weakens it, for bacteriophage resistance and results in hypersensitivity to antibiotics.
Phage JJ01 is a novel lytic bacteriophage that belongs to the Pbunavirus genus under the Myoviridae family. Phage JJ01 requires about 10 min for 90% adsorption to host cells and 30 min for replication during the latent period, and it produces a relatively large burst size at around 109 particles/cell. It tolerates a wide range of pH and temperature. Although phage-resistant P. aeruginosa emerges, it comes at a cost to bacterial survival, by altering some physiological fitness of the bacteria. Some JJ01-resistant isolates produce much less amount of biofilm than the wild type, and some of them become much more susceptible to colistin, which is the last resort antibiotic for Gram-negative bacterial infections. Therefore, phage JJ01 would be considered as a promising candidate for therapeutic purposes in the treatment of P. aeruginosa infection.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
WW, PN, and VC: conceptualization. WW, HH, PN, and VC: methodology. WW, HH, RS, PN, and VC: investigation, formal analysis, and visualization. WW and HH: validation. WW, HH, PN, and VC: writing of the original draft. All authors: reviewing and editing. PN and VC: project administration, resource, and funding acquisition. JP, PN, and VC: supervision.
This project was funded by the National Research Council of Thailand (NRCT) (Grant no. N41A640136) (VC), Sci-Super VII Grant, Faculty of Science Chulalongkorn University (VC), and Second Century Fund (C2F) of Chulalongkorn University (WW). This research project was also supported by Mahidol University under the New Discovery and Frontier Research Grant and by the National Research Council of Thailand (NRCT) and Mahidol University: N42A650368 (PN).
We acknowledge the support for scientific instruments by the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA), Science and Technology Research Partnership for Sustainable Development, SATREPS JPMJSA1806 (VC). We thank Mahidol University Frontier Research Facility (MU-FRF) for Instrumentation Support for DeltaVision™ Ultra and the Advanced Cell Imaging Center, Institute of Molecular Biosciences, Mahidol University, for the fluorescent microscopy imaging. We acknowledge Watcharapong Hongjamrassilp at the Marine Science Department, Faculty of Science, Chulalongkorn University, for intellectual advice on the phylogenetic analysis of the phage.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1004733/full#supplementary-material
Ackermann, H.-W. (2003). Bacteriophage observations and evolution. Res. Microbiol. 154, 245–251. doi: 10.1016/S0923-2508(03)00067-6
Adriaenssens, E., and Brister, J. R. (2017). How to name and classify your phage: An informal guide. Viruses 9:70. doi: 10.3390/v9040070
Ahiwale, S., Prakash, D., Gajbhiye, M., Jagdale, S., Patil, N., and Kapadnis, B. (2012). BVPaP-3, a T7-like lytic phage of Pseudomonas aeruginosa: Its isolation and characterisation. Curr. Microbiol. 64, 305–311. doi: 10.1007/s00284-011-0071-6
Bisht, K., Moore, J. L., Caprioli, R. M., Skaar, E. P., and Wakeman, C. A. (2021). Impact of temperature-dependent phage expression on Pseudomonas aeruginosa biofilm formation. NPJ Biofilms Microbiomes 7:22. doi: 10.1038/s41522-021-00194-8
Breidenstein, E. B. M., de la Fuente-Núñez, C., and Hancock, R. E. W. (2011). Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 19, 419–426. doi: 10.1016/j.tim.2011.04.005
Browning, C., Shneider, M. M., Bowman, V. D., Schwarzer, D., and Leiman, P. G. (2012). Phage pierces the host cell membrane with the iron-loaded spike. Structure 20, 326–339. doi: 10.1016/j.str.2011.12.009
Bucior, I., Tran, C., and Engel, J. (2014). “Assessing Pseudomonas virulence using host cells,” in Pseudomonas methods and protocols. Methods in molecular biology, eds A. Filloux and J.-L. Ramos (New York, NY: Springer New York), 741–755. doi: 10.1007/978-1-4939-0473-0_57
Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., et al. (2006). CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. doi: 10.1186/gb-2006-7-10-r100
Ceyssens, P.-J., and Lavigne, R. (2010). Bacteriophages of Pseudomonas. Future Microbiol. 5, 1041–1055. doi: 10.2217/fmb.10.66
Chaikeeratisak, V., Birkholz, E. A., and Pogliano, J. (2021). The phage nucleus and PhuZ spindle: Defining features of the subcellular organization and speciation of nucleus-forming jumbo phages. Front. Microbiol. 12:641317. doi: 10.3389/fmicb.2021.641317
Chaikeeratisak, V., Nguyen, K., Egan, M. E., Erb, M. L., Vavilina, A., and Pogliano, J. (2017). The phage nucleus and tubulin spindle are conserved among large Pseudomonas phages. Cell Rep. 20, 1563–1571. doi: 10.1016/j.celrep.2017.07.064
Chan, B. K., Abedon, S. T., and Loc-Carrillo, C. (2013). Phage cocktails and the future of phage therapy. Future Microbiol. 8, 769–783. doi: 10.2217/fmb.13.47
Chan, B. K., Turner, P. E., Kim, S., Mojibian, H. R., Elefteriades, J. A., and Narayan, D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66. doi: 10.1093/emph/eoy005
Chaudhry, W. N., Concepción-Acevedo, J., Park, T., Andleeb, S., Bull, J. J., and Levin, B. R. (2017). Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615. doi: 10.1371/journal.pone.0168615
Chen, F., Cheng, X., Li, J., Yuan, X., Huang, X., Lian, M., et al. (2021). Novel lytic phages protect cells and mice against Pseudomonas aeruginosa infection. J. Virol. 95:e01832-20. doi: 10.1128/JVI.01832-20
CLSI (2020). Performance standards for antimicrobial susceptibility testing, 30th Edn. Wayen, PA: Clinical and laboratory standards institude.
Coleman, S. R., Blimkie, T., Falsafi, R., and Hancock, R. E. W. (2020). Multidrug adaptive resistance of Pseudomonas aeruginosa swarming cells. Antimicrob. Agents Chemother. 64:e01999-19. doi: 10.1128/AAC.01999-19
Danis-Wlodarczyk, K., Olszak, T., Arabski, M., Wasik, S., Majkowska-Skrobek, G., Augustyniak, D., et al. (2015). Characterization of the newly isolated lytic bacteriophages KTN6 and KT28 and their efficacy against Pseudomonas aeruginosa biofilm. PLoS One 10:e0127603. doi: 10.1371/journal.pone.0127603
Das, T., Sehar, S., and Manefield, M. (2013). The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development: The roles of eDNA in the bacterial biofilm development. Environ. Microbiol. Rep. 5, 778–786. doi: 10.1111/1758-2229.12085
de Melo, A. C. C., da Mata Gomes, A., Melo, F. L., Ardisson-Araújo, D. M. P., de Vargas, A. P. C., Ely, V. L., et al. (2019). Characterization of a bacteriophage with broad host range against strains of Pseudomonas aeruginosa isolated from domestic animals. BMC Microbiol. 19:134. doi: 10.1186/s12866-019-1481-z
De Smet, J., Wagemans, J., Boon, M., Ceyssens, P.-J., Voet, M., Noben, J.-P., et al. (2021). The bacteriophage LUZ24 “Igy” peptide inhibits the Pseudomonas DNA gyrase. Cell Rep. 36:109567. doi: 10.1016/j.celrep.2021.109567
Debarbieux, L., Leduc, D., Maura, D., Morello, E., Criscuolo, A., Grossi, O., et al. (2010). Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J. Infect. Dis. 201, 1096–1104. doi: 10.1086/651135
Dedrick, R. M., Guerrero-Bustamante, C. A., Garlena, R. A., Russell, D. A., Ford, K., Harris, K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z
Drenkard, E. (2003). Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 5, 1213–1219. doi: 10.1016/j.micinf.2003.08.009
Duval, R. E., Grare, M., and Demoré, B. (2019). Fight against antimicrobial resistance: We always need new antibacterials but for right bacteria. Molecules 24:3152. doi: 10.3390/molecules24173152
Forti, F., Roach, D. R., Cafora, M., Pasini, M. E., Horner, D. S., Fiscarelli, E. V., et al. (2018). Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob. Agents Chemother. 62:e02573-17. doi: 10.1128/AAC.02573-17
Fukuda, K., Ishida, W., Uchiyama, J., Rashel, M., Kato, S., Morita, T., et al. (2012). Pseudomonas aeruginosa keratitis in mice: Effects of topical bacteriophage KPP12 administration. PLoS One 7:e47742. doi: 10.1371/journal.pone.0047742
Garbe, J., Bunk, B., Rohde, M., and Schobert, M. (2011). Sequencing and characterization of Pseudomonas aeruginosa phage JG004. BMC Microbiol. 11:102. doi: 10.1186/1471-2180-11-102
Garbe, J., Wesche, A., Bunk, B., Kazmierczak, M., Selezska, K., Rohde, C., et al. (2010). Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 10:301. doi: 10.1186/1471-2180-10-301
Gordillo Altamirano, F. L., and Barr, J. J. (2019). Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32:e00066-18. doi: 10.1128/CMR.00066-18
Hall, A. R., De Vos, D., Friman, V.-P., Pirnay, J.-P., and Buckling, A. (2012). Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl. Environ. Microbiol. 78, 5646–5652. doi: 10.1128/AEM.00757-12
Hatfull, G. F. (2015). Dark matter of the biosphere: The amazing world of bacteriophage diversity. J. Virol. 89, 8107–8110. doi: 10.1128/JVI.01340-15
Hosseinidoust, Z., Tufenkji, N., and van de Ven, T. G. M. (2013). Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 79, 2862–2871. doi: 10.1128/AEM.03817-12
Htoo, H. H., Brumage, L., Chaikeeratisak, V., Tsunemoto, H., Sugie, J., Tribuddharat, C., et al. (2019). Bacterial cytological profiling as a tool to study mechanisms of action of antibiotics that are active against Acinetobacter baumannii. Antimicrob. Agents Chemother. 63, e2310–e2318. doi: 10.1128/AAC.02310-18
Htoo, H. H., Tuyet, N. N. T., Nakprasit, K., Aonbangkhen, C., Chaikeeratisak, V., Chavasiri, W., et al. (2022). Mansonone G and its derivatives exhibit membrane permeabilizing activities against bacteria. PLoS One 17:e0273614. doi: 10.1371/journal.pone.0273614
Imam, M., Alrashid, B., Patel, F., Dowah, A. S. A., Brown, N., Millard, A., et al. (2019). vB_PaeM_MIJ3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front. Microbiol. 10:2772. doi: 10.3389/fmicb.2019.02772
Johnson, K., and Lory, S. (1987). Characterization of Pseudomonas aeruginosa mutants with altered piliation. J. Bacteriol. 169, 5663–5667. doi: 10.1128/jb.169.12.5663-5667.1987
Khan Mirzaei, M., and Nilsson, A. S. (2015). Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 10:e0118557. doi: 10.1371/journal.pone.0118557
Khawaldeh, A., Morales, S., Dillon, B., Alavidze, Z., Ginn, A. N., Thomas, L., et al. (2011). Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 60, 1697–1700. doi: 10.1099/jmm.0.029744-0
Kim, S., Rahman, M., Seol, S. Y., Yoon, S. S., and Kim, J. (2012). Pseudomonas aeruginosa bacteriophage PA1Ø requires type IV pili for infection and shows broad bactericidal and biofilm removal activities. Appl. Environ. Microbiol. 78, 6380–6385. doi: 10.1128/AEM.00648-12
Klockgether, J., Munder, A., Neugebauer, J., Davenport, C. F., Stanke, F., Larbig, K. D., et al. (2010). Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192, 1113–1121. doi: 10.1128/JB.01515-09
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Kropinski, A. M. (2018). “Practical advice on the one-step growth curve,” in Bacteriophages methods in molecular biology, eds M. R. J. Clokie, A. M. Kropinski, and R. Lavigne (New York, NY: Springer New York), 41–47. doi: 10.1007/978-1-4939-7343-9_3
Krylov, V. N., Dela Cruz, D. M., Hertveldt, K., and Ackermann, H.-W. (2007). “φKZ-like viruses”, a proposed new genus of myovirus bacteriophages. Arch. Virol. 152, 1955–1959. doi: 10.1007/s00705-007-1037-7
Kwan, T., Liu, J., DuBow, M., Gros, P., and Pelletier, J. (2006). Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188, 1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006
Le, S., Yao, X., Lu, S., Tan, Y., Rao, X., Li, M., et al. (2015). Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci. Rep. 4:4738. doi: 10.1038/srep04738
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa?: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Liu, J., Dehbi, M., Moeck, G., Arhin, F., Bauda, P., Bergeron, D., et al. (2004). Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22, 185–191. doi: 10.1038/nbt932
Loc-Carrillo, C., and Abedon, S. T. (2011). Pros and cons of phage therapy. Bacteriophage 1, 111–114. doi: 10.4161/bact.1.2.14590
Maddocks, S., Fabijan, A. P., Ho, J., Lin, R. C. Y., Ben Zakour, N. L., Dugan, C., et al. (2019). Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 200, 1179–1181. doi: 10.1164/rccm.201904-0839LE
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Markwitz, P., Olszak, T., Gula, G., Kowalska, M., Arabski, M., and Drulis-Kawa, Z. (2021). Emerging phage resistance in Pseudomonas aeruginosa PAO1 is accompanied by an enhanced heterogeneity and reduced virulence. Viruses 13:1332. doi: 10.3390/v13071332
McVay, C. S., Velásquez, M., and Fralick, J. A. (2007). Phage therapy of Pseudomonas aeruginosa infection in a mouse burn wound model. Antimicrob. Agents Chemother. 51, 1934–1938. doi: 10.1128/AAC.01028-06
Menon, N. D., Penziner, S., Montaño, E. T., Zurich, R., Pride, D. T., Nair, B. G., et al. (2022). Increased innate immune susceptibility in hyperpigmented bacteriophage-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 66:e0023922. doi: 10.1128/aac.00239-22
Moradali, M. F., Ghods, S., and Rehm, B. H. A. (2017). Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
Moraru, C., Varsani, A., and Kropinski, A. M. (2020). VIRIDIC—a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. doi: 10.3390/v12111268
Morello, E., Saussereau, E., Maura, D., Huerre, M., Touqui, L., and Debarbieux, L. (2011). Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: First steps towards treatment and prevention. PLoS One 6:e16963. doi: 10.1371/journal.pone.0016963
Nale, J. Y., Spencer, J., Hargreaves, K. R., Buckley, A. M., Trzepiñski, P., Douce, G. R., et al. (2016). Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob. Agents Chemother. 60, 968–981. doi: 10.1128/AAC.01774-15
Oechslin, F., Piccardi, P., Mancini, S., Gabard, J., Moreillon, P., Entenza, J. M., et al. (2016). Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 215, 703–712. doi: 10.1093/infdis/jiw632
Ong, S. P., Azam, A. H., Sasahara, T., Miyanaga, K., and Tanji, Y. (2020). Characterization of Pseudomonas lytic phages and their application as a cocktail with antibiotics in controlling Pseudomonas aeruginosa. J. Biosci. Bioeng. 129, 693–699. doi: 10.1016/j.jbiosc.2020.02.001
Pang, Z., Raudonis, R., Glick, B. R., Lin, T.-J., and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Pires, D., Sillankorva, S., Faustino, A., and Azeredo, J. (2011). Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 162, 798–806. doi: 10.1016/j.resmic.2011.06.010
Pires, D. P., Dötsch, A., Anderson, E. M., Hao, Y., Khursigara, C. M., Lam, J. S., et al. (2017). A genotypic analysis of five P. aeruginosa strains after biofilm infection by phages targeting different cell surface receptors. Front. Microbiol. 8:1229. doi: 10.3389/fmicb.2017.01229
Rodríguez-Rojas, A., Mena, A., Martín, S., Borrell, N., Oliver, A., and Blázquez, J. (2009). Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology 155, 1050–1057. doi: 10.1099/mic.0.024745-0
Rumbaugh, K. P., and Sauer, K. (2020). Biofilm dispersion. Nat. Rev. Microbiol. 18, 571–586. doi: 10.1038/s41579-020-0385-0
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold spring harbor laboratory press.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61:e00954-17. doi: 10.1128/AAC.00954-17
Serwer, P., Hayes, S. J., Zaman, S., Lieman, K., Rolando, M., and Hardies, S. C. (2004). Improved isolation of undersampled bacteriophages: Finding of distant terminase genes. Virology 329, 412–424. doi: 10.1016/j.virol.2004.08.021
Sharma, S., Datta, S., Chatterjee, S., Dutta, M., Samanta, J., Vairale, M. G., et al. (2021). Isolation and characterization of a lytic bacteriophage against Pseudomonas aeruginosa. Sci. Rep. 11:19393. doi: 10.1038/s41598-021-98457-z
Shen, M., Zhang, H., Shen, W., Zou, Z., Lu, S., Li, G., et al. (2018). Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 46, 4505–4514. doi: 10.1093/nar/gky160
Soltis, P. S., and Soltis, D. E. (2003). Applying the bootstrap in phylogeny reconstruction. Stat. Sci. 18, 256–267.
Stewart, P. S., and William Costerton, J. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Thammatinna, K., Egan, M. E., Htoo, H. H., Khanna, K., Sugie, J., Nideffer, J. F., et al. (2020). A novel vibriophage exhibits inhibitory activity against host protein synthesis machinery. Sci. Rep. 10:2347. doi: 10.1038/s41598-020-59396-3
Thunnissen, A.-M. W. H., Isaacs, N. W., and Dijkstral, B. W. (1995). The catalytic domain of a bacterial lytic transglycosylase defines a novel class of lysozymes. Proteins Struct. Funct. Genet. 22, 245–258. doi: 10.1002/prot.340220305
Van den Bossche, A., Ceyssens, P.-J., De Smet, J., Hendrix, H., Bellon, H., Leimer, N., et al. (2014). Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of Pseudomonas aeruginosa. J. Proteome Res. 13, 4446–4456. doi: 10.1021/pr500796n
Watanabe, R., Matsumoto, T., Sano, G., Ishii, Y., Tateda, K., Sumiyama, Y., et al. (2007). Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 51, 446–452. doi: 10.1128/AAC.00635-06
Webb, J. S., Thompson, L. S., James, S., Charlton, T., Tolker-Nielsen, T., Koch, B., et al. (2003). Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003
Weiner, L. M., Webb, A. K., Limbago, B., Dudeck, M. A., Patel, J., Kallen, A. J., et al. (2016). Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174
Wright, A., Hawkins, C. H., Änggård, E. E., and Harper, D. R. (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357. doi: 10.1111/j.1749-4486.2009.01973.x
Yang, X., Haque, A., Matsuzaki, S., Matsumoto, T., and Nakamura, S. (2021). The efficacy of phage therapy in a murine model of Pseudomonas aeruginosa pneumonia and sepsis. Front. Microbiol. 12:682255. doi: 10.3389/fmicb.2021.682255
Yang, Y., Shen, W., Zhong, Q., Chen, Q., He, X., Baker, J. L., et al. (2020). Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 11:327. doi: 10.3389/fmicb.2020.00327
Young, R. (2014). Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 52, 243–258. doi: 10.1007/s12275-014-4087-z
Keywords: P. aeruginosa, bacteriophage, biofilm reduction, colistin susceptibility, phage resistance
Citation: Wannasrichan W, Htoo HH, Suwansaeng R, Pogliano J, Nonejuie P and Chaikeeratisak V (2022) Phage-resistant Pseudomonas aeruginosa against a novel lytic phage JJ01 exhibits hypersensitivity to colistin and reduces biofilm production. Front. Microbiol. 13:1004733. doi: 10.3389/fmicb.2022.1004733
Received: 27 July 2022; Accepted: 12 September 2022;
Published: 06 October 2022.
Edited by:
Nicole Buan, University of Nebraska–Lincoln, United StatesReviewed by:
Boo Shan Tseng, University of Nevada, Las Vegas, United StatesCopyright © 2022 Wannasrichan, Htoo, Suwansaeng, Pogliano, Nonejuie and Chaikeeratisak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vorrapon Chaikeeratisak, dm9ycmFwb24uY0BjaHVsYS5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.