- 1Nanshan Center for Disease Control and Prevention, Shenzhen, China
- 2State Key Laboratory of Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
Arcobacter spp. is a globally emerging zoonotic and foodborne pathogen. However, little is known about its prevalence and antimicrobial resistance in China. To investigate the prevalence of Arcobacter spp. isolated from various sources, 396 samples were collected from human feces, chicken cecum, and food specimens including chicken meat, beef, pork, lettuce, and seafood. Arcobacter spp. was isolated by the membrane filtration method. For 92 strains, the agar dilution method and next-generation sequencing were used to investigate their antimicrobial resistance and to obtain whole genome data, respectively. The virulence factor database (VFDB) was queried to identify virulence genes. ResFinder and the Comprehensive Antibiotic Resistance Database (CARD) were used to predict resistance genes. A phylogenetic tree was constructed using the maximum likelihood (ML) method with core single-nucleotide polymorphisms (SNPs). We found that 27.5% of the samples (n = 109) were positive for Arcobacter spp., comprising Arcobacter butzleri (53.0%), Arcobacter cryaerophilus (39.6%), and Arcobacter skirrowii (7.4%). Chicken meat had the highest prevalence (81.2%), followed by seafood (51.9%), pork (43.3%), beef (36.7%), lettuce (35.5%), chicken cecum (8%), and human fecal samples (0%, 0/159). Antimicrobial susceptibility tests revealed that 51 A. butzleri and 40 A. cryaerophilus strains were resistant to streptomycin (98.1, 70%), clindamycin (94.1, 90%), tetracycline (64.7, 52.5%), azithromycin (43.1%, 15%), nalidixic acid (33.4, 35%), and ciprofloxacin (31.3, 35%) but were susceptible to erythromycin, gentamicin, chloramphenicol, telithromycin, and clindamycin (≤10%). A. skirrowii was sensitive to all experimental antibiotics. The virulence factors tlyA, mviN, cj1349, ciaB, and pldA were carried by all Arcobacter spp. strains at 100%, and the following percentages were cadF (95.7%), iroE (23.9%), hecB (2.2%), hecA, and irgA (1.1%). Only one A. butzleri strain (F061-2G) carried a macrolide resistance gene (ereA). One A. butzleri and one A. cryaerophilus harbored resistance island gene clusters, which were isolated from pork and chicken. Phylogenetic tree analysis revealed that A. butzleri, A. cryaerophilus, and A. skirrowii were separated from each other. To our knowledge, this is the first report of the isolation of Arcobacter spp. from vegetables and seafood in China. The resistance island gene cluster found in pork and chicken meat and the presence of virulence factors could be a potential risk to human health.
Introduction
Arcobacter is a globally emerging foodborne pathogen causing diarrhea, enteritis, and bacteremia in humans and diarrhea, mastitis, and abortion in animals (Ramees et al., 2017; Zambri et al., 2019; Chieffi et al., 2020; Khodamoradi and Abiri, 2020). Humans are mainly infected with Arcobacter via the consumption of contaminated food and water (Collado and Figueras, 2011; Šilha et al., 2019). The main strains causing diseases in humans are A. butzleri, A. cryaerophilus, and A. skirrowii (Van den Abeele et al., 2014; Ferreira et al., 2019).
Arcobacter-infected poultry is considered the major source of infection (Jribi et al., 2020; Khodamoradi and Abiri, 2020). The prevalence of Arcobacter in broiler chickens was reported as 26.0% (26/100) in Iran (Khodamoradi and Abiri, 2020), 55.7% (54/97) in the south of Chile (Vidal-Veuthey et al., 2021), and 73.33% (44/60) in Beijing, China (Wang et al., 2016). Moreover, Arcobacter is also found in beef, pork, vegetables, and seafood (Mottola et al., 2016, 2021; Córdoba-Calderón et al., 2017; Kim et al., 2019; Zhang et al., 2019; Jasim et al., 2021), which represent possible transmission sources to humans. However, there have been a few reports regarding Arcobacter isolated from various sources in China.
Although most Arcobacter infections are self-limited, antibiotic treatment is required for severe clinical infections (Ferreira et al., 2019). This treatment usually includes quinolones, tetracyclines, macrolides, and β-lactamase (Figueras et al., 2014). However, high resistance rates of Arcobacter isolates to quinolones and macrolides have been reported (Ferreira et al., 2016; Dekker et al., 2019; Jribi et al., 2020).
Currently, our knowledge is limited concerning the pathogenic mechanisms and virulence features of Arcobacter strains (Oliveira et al., 2018; Parisi et al., 2019; Šilha et al., 2019). It was found that 10 potential virulence genes (iroE, irgA, tlyA, pldA, mviN, hecB, hecA, ciaB, cj1349, and cadF) were considered important for the virulence of this pathogen (Miller et al., 2007; Rathlavath et al., 2017; Kietsiri et al., 2021). Different virulence genes have different effects on disease (Kietsiri et al., 2021). The existence of virulence factors in Arcobacter spp. isolated from food could threaten human health.
This study aimed to determine the prevalence of Arcobacter spp. in various sources in Shenzhen, China, and to identify the virulence and antibiotic resistance profiles of Arcobacter spp. using whole genome sequencing (WGS). Furthermore, we aimed to determine the minimum inhibitory concentrations (MICs) of 11 common antibiotics to identify the most appropriate and effective treatment for Arcobacter infections.
Methods
Sample collection
Between June and September 2019, 159 fecal samples were collected from adult patients with diarrhea at the top three local hospitals. In this study, informed consent was obtained from each adult patient with diarrhea. Patients >16 years of age and who experienced acute diarrhea three times or more in the previous 24 h were included. Meanwhile, a collection of 237 samples from chicken meat (n = 69), beef (n = 30), pork (n = 30), lettuce (n = 31), and seafood (n = 27) were purchased from two retail markets in the Nanshan center; chicken cecum samples (n = 50) were collected from a poultry wholesale market.
Fecal samples (~0.5 g each) were collected into the Cary-Blair medium, and food samples (~250 g each) were placed in a sterile plastic bag, and all samples were transported to the laboratory at 4°C within 4 h for bacterial isolation.
Bacterial culture, isolation, and identification
Arcobacter was isolated by an Arcobacter isolation kit using the enrichment and membrane filter method (ZC-ARCO-001, Qingdao Sinova Biotechnology Co., Ltd., Qingdao, China) for stool samples and a direct filtration method for food samples. Briefly, stool specimens were transferred into a 4-ml enrichment buffer, which was provided in the kit. The principal component of the enrichment buffer was the modified Preston broth containing vancomycin, trimethoprim, and amphotericin B, as described in the manual book. The enriched suspension from stool samples was incubated for 24–48 h at 37°C in a microaerophilic atmosphere consisting of 5% O2, 10% CO2, and 85% N2. Subsequently, 300 μl of cultured enrichment suspension was spotted on Karmali and Columbia agar with a 0.45-μm cellulose membrane filter. At the same time, food samples were fully washed with buffered peptone water (100 ml per 250-g sample), which was concentrated by centrifugation at a low speed of 1,500 g for 15 min. Then, 300–500 ml of concentrated suspension was spotted on Karmali and Columbia agar with a 0.45-μm cellulose membrane filter. After air-drying for 40 min in a biological safety cabinet, the filter membrane was removed and the plates were incubated in an aerobic atmosphere at 30°C for 48 h.
After incubation, small, round, and whitish colonies 2 mm in diameter were plated and confirmed by Gram staining, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and real-time polymerase chain reaction (PCR). Mass spectrometry was performed using Flexcontrol software, and the results were interpreted with IVD MALDI Biotyper 2.3 software (Bruker Daltonik GmbH, Bremen, Germany). The criteria for determining the genus and species of bacteria were as follows: 2.300–3.000 points indicated reliable identification to the species level and 2.000–2.299 points indicated reliable identification to the genus level and possible identification to the species level. In this study, scores≥2.000 were considered credible. For PCR identification, a loop was used to collect suspected pure culture colonies, which were resuspended in 200 μl of ultrapure water, boiled for 10 min, and centrifuged for 10 min at 8,000 × g. Subsequently, the supernatant was removed for PCR species identification by a realtime PCR kit (MABSKY BIO-TECH CO., LTD, Shenzhen, China). PCR amplification conditions were as follows: initial denaturation at 94°C for 5 min, followed by 45 cycles of 94°C for 15 s and 60°C for 1 min.
Antimicrobial susceptibility testing
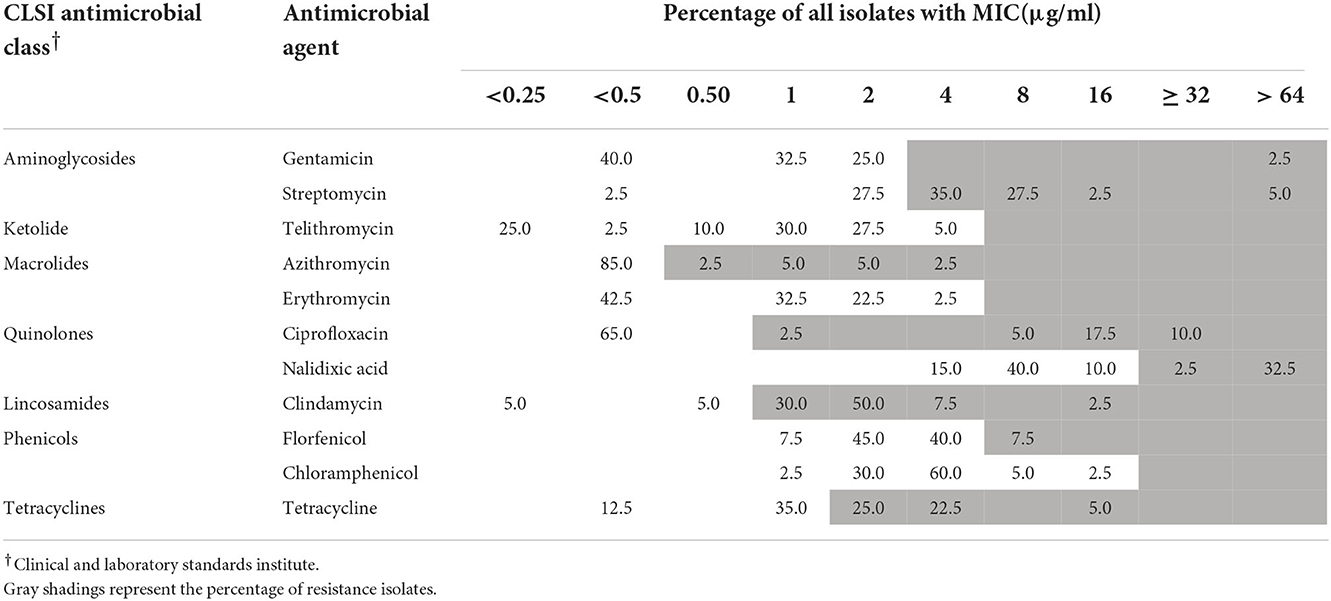
Antimicrobial susceptibility testing was performed using an agar dilution method-based kit (ZC-CAMPY-013, Zhongchuang Biotechnology Ltd. Corp., Qingdao, China). The test was performed two times in parallel. Mueller-Hinton agar containing 11 different antibiotics was coated onto wells in a 96-well-plate to obtain the MIC of 92 Arcobacter strains. The cutoff criteria for each antibiotic were based on the National Antimicrobial Resistance Monitoring System (NARMS-2015: https://www.cdc.gov/narms/pdf/2015-NARMS-Annual-Report-cleared_508.pdf) for Campylobacter jejuni and included clindamycin (≥ 1 μg ml−1), telithromycin (≥ 8 μg ml−1), tetracycline (≥ 2 μg ml−1), florfenicol (≥ 8 μg ml−1), gentamycin (≥ 4 μg ml−1), ciprofloxacin (≥ 1 μg ml−1), nalidixic acid (≥ 32 μg ml−1), azithromycin (≥ 0.5 μg ml−1), and erythromycin (≥ 8 μg ml−1), combined with MIC EUCAST (https://mic.eucast.org/search/?search%5Bmethod%5D=mic&search%5Bantibiotic%5D=-1&search%5Bspecies%5D=100&search%5Bdisk_content%5D=-1&search%5Blimit%5D=50:) streptomycin (≥ 4 μg ml−1) and chloramphenicol (≥ 32 μg ml−1). The quality control bacterial strain was C. jejuni ATCC 33560. In addition, we defined multidrug resistance (MDR) as resistance to ≥ 3 classes of antibiotics.
Extraction of deoxyribonucleic acid and WGS
Deoxyribonucleic acid (DNA) was extracted from Arcobacter isolates. One or two Arcobacter plates from blood agar plates (Huaikai biology, Guangzhou, China) were needed to obtain sufficient material for DNA preparation. Colonies were harvested using fiber swabs and resuspended in 1 ml of phosphate-buffered saline (PBS). The tubes were centrifuged at 16,000 × g for 6 min, and the supernatant was discarded. The resulting pellet was further processed for DNA recovery using the bacterial genomic DNA Extraction kit (T132, Tianlong, Shanxi, China) according to the manufacturer's instructions. The concentration of the double-stranded DNA (dsDNA) was examined using a Microplate spectrophotometer (Epoch, Berten Instruments Co., Ltd., Montigny-le-Bretonneux, France).
Next-generation sequencing was performed using the Illumina NovaSeq PE150 (Illumina, San Diego, CA, USA) by Novo Source Technology Co., Ltd., (Beijing, China) and BGI Genomics Co., Ltd., (Beijing, China). To sequence the genomes, a 270-bp paired-end library was constructed and then 150-bp reads were generated. FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and fastp v0.20.0 (https://github.com/OpenGene/fastp) software tools were applied to evaluate and improve the quality of the raw sequence data, respectively. Low-quality reads were removed in case of the quality scores of ≥3 consecutive bases ≤ Q30. The clean reads were assembled using SOAPdenovo v2.04 (http://soap.genomics.org.cn/soapdenovo.html) and spades v3.13.1 software (Prjibelski et al., 2020). Finally, the assembled sequences were subjected to gene prediction and functional annotation using the Prokka pipeline (Seemann, 2014) and glimmer software (http://ccb.jhu.edu/software/glimmer/index.shtml). The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Clusters of Orthologous Groups (COG) databases were used for functional classification.
Bioinformatic analyses
The presence of virulence genes was assessed by submitting the assembled genomes to the virulence factor database (VFDB) (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer). ResFinder (http://cge.cbs.dtu.dk/services/ResFinder/), and Abricate software (https://github.com/tseemann/abricate), and the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/?q=CARD/ontology/35506) were used to predict resistance genes, with a cutoff comprising an E-value of at least 1e−10. Cutoffs for identification and query coverage values were >80 and >60%, respectively. Individual missense mutations in gyrA conferring ciprofloxacin resistance were detected using BLASTn. Databases including KEGG, COG, SwissProt, and PHI, were used to functionally annotate and classify protein sequences. We extracted the nucleotide sequences of all annotated resistance genes from the genomes, and the genoPlotR package was used to visualize related gene clusters. Based on a previous study (Parisi et al., 2019), 10 common virulence factors (iroE, irgA, tlyA, pldA, mviN, hecB, hecA, ciaB, cj1349, ciaB, and cadF) were analyzed using the BLASTn method.
Phylogenetic tree analysis was performed on 177 Arcobacter strains, including 85 Arcobacter strain sequences (45 A. butzleri, 26 A. cryaerophilus, and 14 A. skirrowii), which were downloaded from the GenBank database and 92 strains obtained in this study. Core single-nucleotide polymorphisms (SNPs) were called using Snippy 4.3.6 software (https://github.com/tseemann/snippy) with default parameters and ICDCAB83 as the reference genome. Gubbins software (Croucher et al., 2015) was used as a recombination-removal tool to gain pure SNPs without recombination. Phylogeny reconstruction was performed using the maximum likelihood (ML) method in MEGA 7 software (Kumar et al., 2016) with 1,000 bootstraps.
Data analysis
SPSS 26.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis, and the chi-squared test (χ2) was used to compare count data between groups. A statistical probability of <0.05 (p < 0.05) indicated a statistically significant difference.
Results
The prevalence of Arcobacter spp. isolated from patients with diarrhea, poultry, beef, pork, vegetables, and seafood
Out of 159 human fecal samples, no Arcobacter strains were isolated. In total, 109 Arcobacter strains were separated from chicken meat, chicken cecum, beef, pork, lettuce, and seafood (Figure 1). A. butzleri was the most prevalent species (33.3%, 79/237), followed by A. cryaerophilus (24.9%, 59/237) and A. skirrowii (4.6%, 11/237). In 37 of the samples, two or three species of Arcobacter were isolated. Moreover, the prevalence percentage of Arcobacters spp. in chicken meat, seafood, pork, beef, lettuce, and chicken cecum was 81.2% (56/69), 51.9% (14/27), 43.3% (13/30), 36.7% (11/30), 35.5% (11/31), and 8% (4/50), respectively (Figure 1). Significantly, Arcobacter spp. isolated from chicken cecum (8%, 4/50) had a lower prevalence than those isolated from poultry meat (81.2%, 56/69) (χ2 = 10.632, p = 0.001).
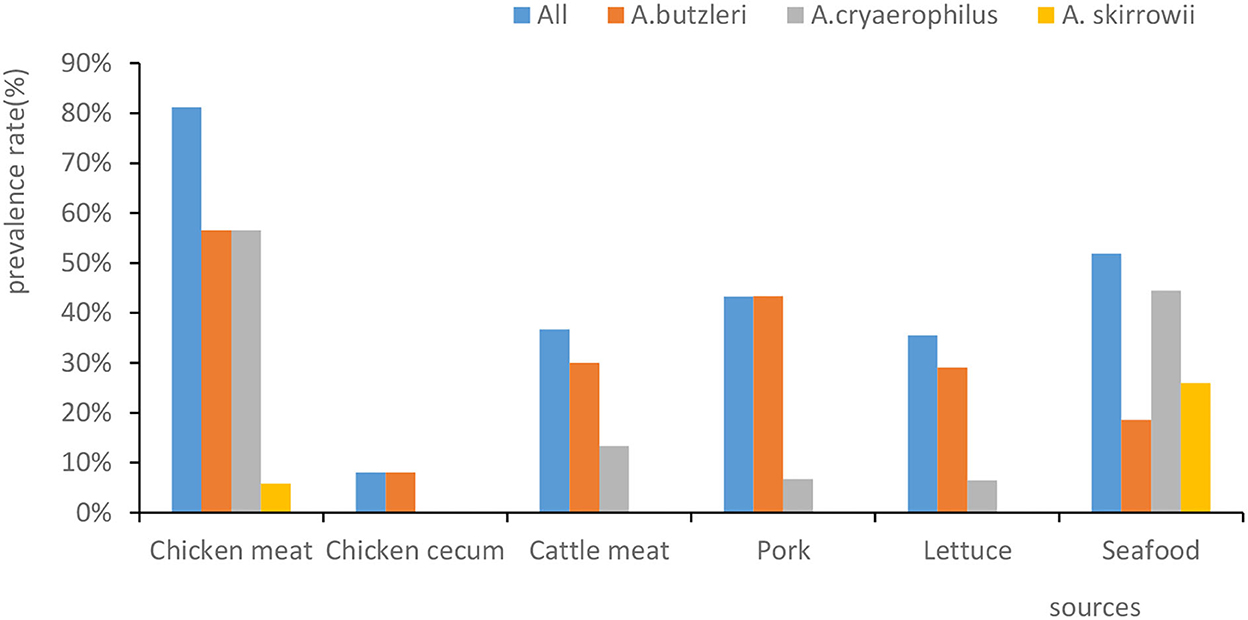
Figure 1. The prevalence rates of Arcobacter spp. isolated from different sources. The x-axis represents the sources. The y-axis represents the prevalence percentage. The color of the columns corresponds to the organisms.
Whole genome sequencing
A total of 92 strains (51 A. butzleri isolated from 26 chicken, eight beef, seven pork, seven lettuce, and three seafood samples; 40 A. cryaerophilus isolated from 29 chicken, three beef, two pork, two lettuce, and four seafood samples; and one A. skirrowii isolated from seafood) were characterized by WGS. WGS of Arcobacter isolates were registered in GenBank with accession numbers SAMN30871879 to SAMN30871970. Assemblies consisted of 10–132 contigs. The sequence length was predicted to be between 1,827,334 and 2,453,640 concordant bases. The estimated sizes of the genomes of the 92 strains varied from 1.82 to 2.34 Mb. The guanine-cytosine (GC) content was determined to be 27.78% for A. skirrowii, 27.15–27.47% for A. cryaerophilus, and 26.65–27.02% for A. butzleri.
Virulence genes and secretion systems
Genome sequencing in VFDB showed that all A. butzleri and A. cryaerophilus isolates had the following virulence factor genes: tlyA, mviN, cj1349, ciaB, and pldA, followed by cadF (95.7%), iroE (23.9%), hecB (2.2%), hecA, and irgA (1.1%) (Figure 2). Moreover, there was no difference in the distribution between A. butzleri and A. cryaerophilus. Almost no A. cryaerophilus strains carried the IrgA, hecA, and hecB genes. None of the strains had a secretion system. A. skirrowii contained the virulence factor genes tlyA, mviN, cj1349, ciaB, and pldA.
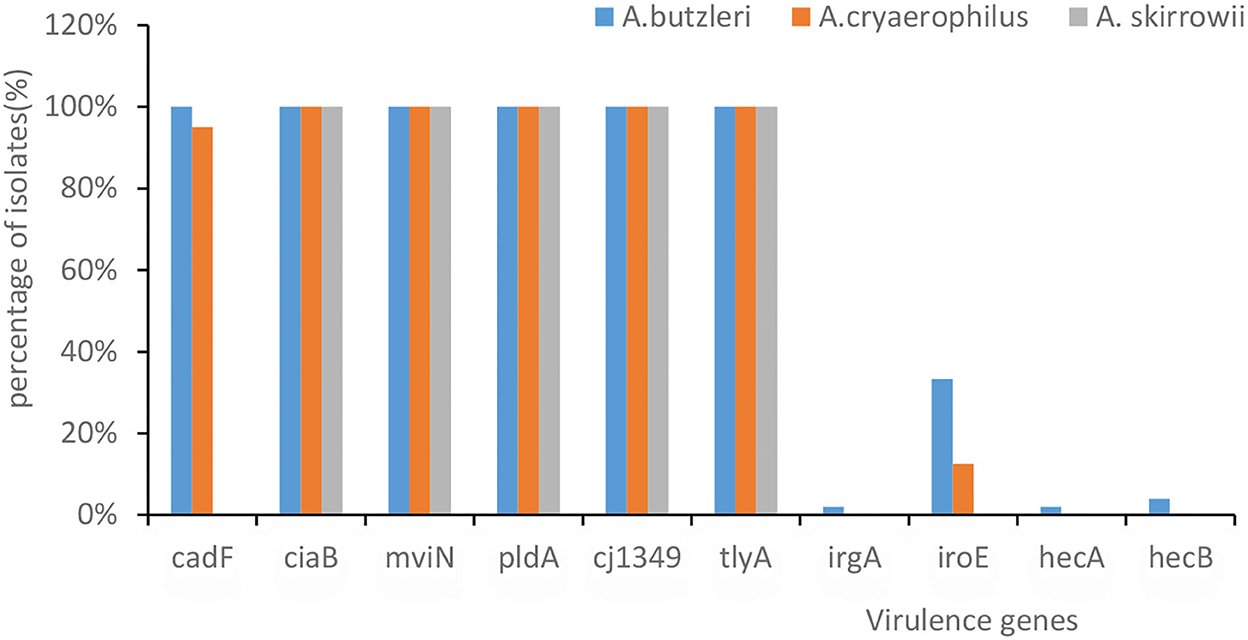
Figure 2. The presence of virulence-associated genes in Arcobacter spp. The x-axis represents the virulence genes. The y-axis represents their percentage of isolates. The color of the columns corresponds to the organisms.
Genetic prediction of antibiotic resistance and concordance with resistance phenotypes
All 51 strains of A. butzleri contained β-lactam antibiotic resistance genes (blaOXA464or blaOXA491), among which 27 strains contained complete genes and the remaining 24 strains contained incomplete genes. Five strains (F034-1G, F050-4G, F061-2G, F101-1G, and F114.2G) contained three tetracycline resistance genes (tet (L), tet (H), and tet (M)). F061-2G contained a macrolide resistance gene (ere (A)). F050-4G, which was isolated from pork, contained aminoglycoside resistance genes (APH (3 ') -IIIa and ant (6) -Ia), streptomycin resistance genes (SAT-4), and tetracycline resistance genes (tet (M)). These genes might exist as gene clusters in bacteria. The gene cluster size was 6,072 bp, with a GC content of 37.17%, and comprised five genes, including four resistance genes (Figure 3).
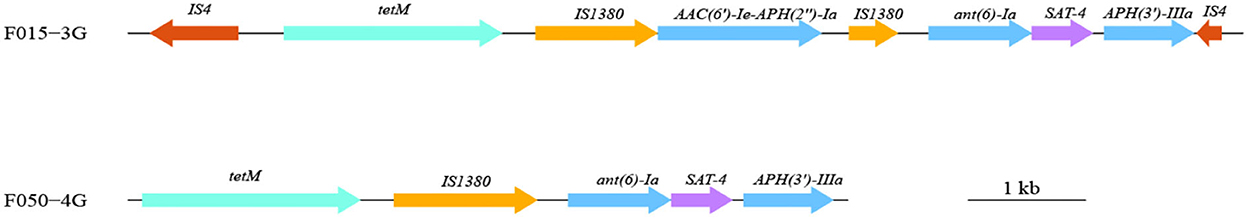
Figure 3. Gene cluster composition in one A. butzleri (F050-4G) and one A. cryaerophilus (F015-3G) isolate. Different colors represent different genes or categories of the resistance gene. The length and direction of the arrows indicate the size and direction of genes.
Seventeen A. cryaerophilus strains contained β-lactam resistance genes, and three of them (F015-3G, F035-7G, and F132-4G) contained tetracycline resistance genes, tet (Y), tet (H), and tet (M). Strain F015-3G isolated from chicken contained one MDR gene island flanked by the insertion sequence IS4 (Figure 3). The size of this MDR gene island was 9,409 bp in length and had a GC content of 33.45%. Seven resistance genes, including three aminoglycosides resistance genes, one tetracycline resistance gene, one streptomycin resistance gene, and two insertion sequences (IS1380), were harbored on this island (Figure 3). This resistance island of F015-3G was similar to that carried by Campylobacter coli SH96 (Sequence ID: MT107516.1).
One strain of A. skirrowii (F198-3G) did not contain resistance genes. Among 51 A. butzleri isolates from five sources, streptomycin had the highest resistance rate (98.1%), followed by clindamycin (94.1%), tetracycline (64.7%), azithromycin (43.1%), nalidixic acid (33.4%), and ciprofloxacin (31.3%); others were <10% (Table 1). Among 40 A. cryaerophilus isolates from five sources, clindamycin resistance was the highest (90%), followed by streptomycin (70%), tetracycline (52.5%), nalidixic acid, and ciprofloxacin (35%); others were <8% (Table 2). In terms of MDR, 33 A. butzleri strains and 23 A. cryaerophilus strains were resistant to three or more classes of antibiotics.
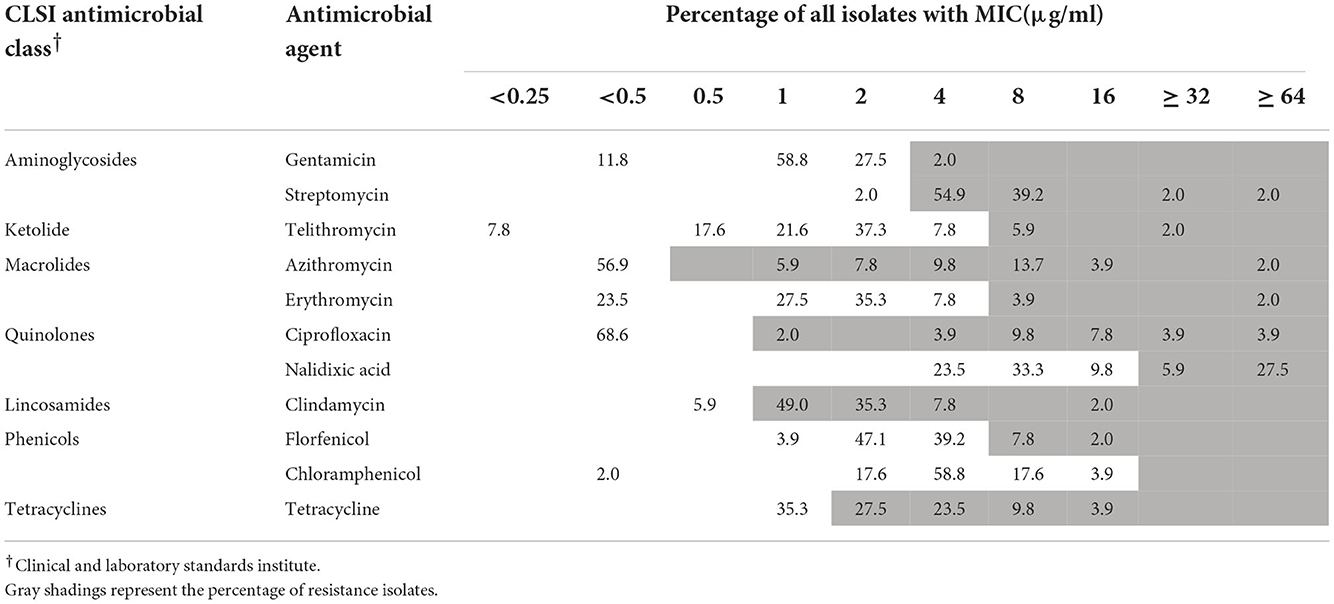
Table 1. Minimum inhibitory concentrations (MICs) of antimicrobial agents toward 51 A. butzleri isolates.
Five A. butzleri isolates, including tetracycline resistance genes (tet (L), tet (H), and tet (M)), were phenotypically resistant to tetracycline. Thirteen A. butzleri isolates with gyrA (C254T) mutation were phenotypically resistant to ciprofloxacin. One of the A. butzleri isolates carried the ere(A) gene, which is associated with erythromycin resistance (MIC value ≥ 64 μg/ml). One A. butzleri isolate (F050-4G) isolated from pork had gyrA (C254T) mutation, contained a streptomycin resistance gene (Sat-4), a tetracycline resistance gene (tet (M)) and aminoglycoside resistance genes (APH (3 ') -IIIa and ant (6) -Ia), and was phenotypically resistant to ciprofloxacin, streptomycin, and tetracycline.
Three A. cryaerophilus isolates containing tetracycline resistance genes (tet (L), tet (H), and tet (M)) were phenotypically resistant to tetracycline. Fourteen A. cryaerophilus isolates with a gyrA (C254T) mutation were phenotypically resistant to ciprofloxacin. One A. cryaerophilus isolate (F015-3G) isolated from chicken had a gyrA (C254T) mutation, contained a streptomycin resistance gene (Sat-4), a tetracycline resistance gene (tet (M)), and aminoglycoside resistance genes (APH (3 ') -IIIa and ant (6) -Ia), and showed resistance to ciprofloxacin, streptomycin, and tetracycline.
Phylogenetic reconstruction
Phylogenetic tree analysis revealed that the 177 strains were mainly divided into three groups (A. butzleri, A. cryaerophilus, and A. skirrowii). There was no obvious aggregation phenomenon in each group according to the source of the host or the sampling site, indicating that Arcobacter strains showed high genetic diversity (Figure 4).
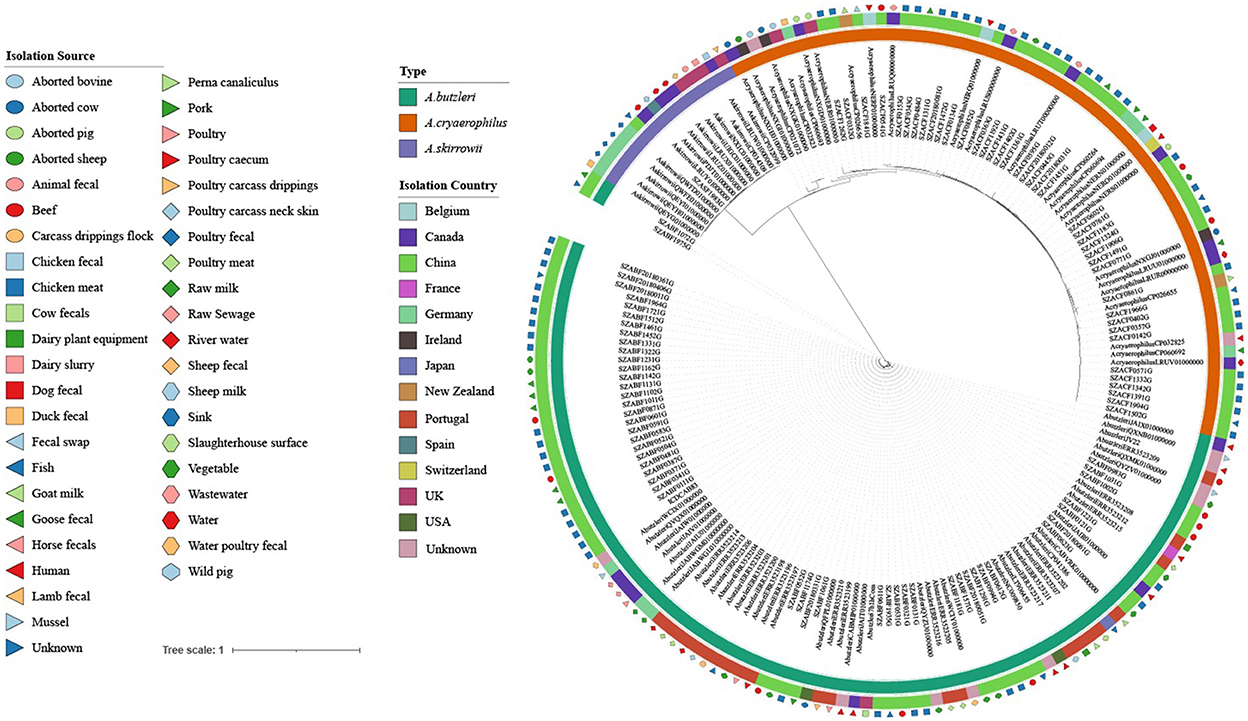
Figure 4. Maximum likelihood (ML) tree of the core genome alignments of strains isolated from various sources and countries with 1,000 bootstraps. Different colors and shapes indicate various sources. From the inner circle to the outer circle, strains, Type, Isolation Country, and Isolation Source are listed.
Discussion
Arcobacter spp. is globally recognized as one of the causes of acute gastroenteritis (Zhang et al., 2019; Brückner et al., 2020). The main transmission source of Arcobacter spp. in humans is via the consumption of contaminated water and food (Collado and Figueras, 2011; Zambri et al., 2019). This study provides updated information on the incidence, genetic diversity, and antimicrobial susceptibility of Arcobacter spp. Samples were collected from various sources in Shenzhen, China.
Our results showed that the prevalence of Arcobacter spp. ranged from high to low in chicken meat (81.2%, 56/69), seafood (51.9%, 14/27), pork (43.3%, 13/30), beef (36.7%, 11/30), lettuce (35.5%,11/31), chicken cecum (8.0%, 4/50), and human feces (0.0%,0/159). The prevalence of Arcobacter spp. in human feces ranged from 0.2 to 3.6% in studies conducted in Germany, Chile, Portugal, India, and Turkey (Patyal et al., 2011; Kayman et al., 2012; Ferreira et al., 2014; Fernandez et al., 2015; Brückner et al., 2020), which was somewhat consistent with our result that Arcobacter spp. was not isolated from any of the 159 fecal samples. In contrast to our findings, the prevalence of Arcobacter isolated from chicken meat was higher than the prevalence found in Iran (26%) (Khodamoradi and Abiri, 2020), Germany (26.8%) (Lehmann et al., 2015), and Tunisia (13.42%) (Jribi et al., 2020), but similar to that found in Beijing (73.33%) (Wang et al., 2016). Jasim et al. (2021) reported that (120/1,293) the prevalence of positive beef samples in Iraq was 9.25%, which was lower than that in the present study. Mottola et al. (2021) reported that its prevalence in ready-to-eat vegetables was 14.5% (16/110), which was lower than that in the present study. Zhang et al. (2019) reported that the contamination rate for seafood was 17.6% (56/318), which was lower than that reported here.
Furthermore, our results showed a lower prevalence of Arcobacter spp. in the chicken cecum (8%, 4/50) than in chicken meat (81.2%, 56/69) (χ2 = 62.073, p = 0.000), which was possibly due to cross-contamination of meat in both the slaughter and retail market environments.
Barboza et al. (2017) reported that the prevalence of chicken cecal content was 5.26% (8/152). Schönknecht et al. (2020) reported that the prevalence of Arcobacter spp. in the chicken cecum was 3% (1/29), which was lower than other intestinal contents. Cecal contents might not be the main Arcobacter reservoir inside chickens. The prevalence of Arcobacter spp. isolated from various sources in this study might change with seasons and climate. We will continue to conduct relevant pathogen monitoring research in the future.
In the current study, almost all strains of Arcobacter spp. have β-lactam antibiotic resistance genes. It has been speculated that Arcobacter spp. may be resistant to β-lactamase. Several studies reported that β-lactam resistance may be caused by the presence of three putative β-lactamases (AB0578, AB1306, and AB1486) identified in the RM4018 genome, which are enhanced by the occurrence of the lrgAB operon (ab0179 and ab0180) and may regulate tolerance to penicillin in Staphylococcus (Bayles, 2000; Groicher et al., 2000; Miller et al., 2007). However, we did not perform an experiment showing the phenotypic resistance of Arcobacter spp. to β-lactam antibiotics. Therefore, further investigation on the resistance phenotype and mechanism of β-lactam antibiotics is needed. For severe clinical diseases caused by Arcobacter spp., fluoroquinolones, tetracyclines, macrolides, and aminoglycosides are recommended for treatment (Ferreira et al., 2016). A meta-analysis of Arcobacter spp. antibiotic resistance in 2019 (Ferreira et al., 2019) reported that fluoroquinolone resistance ranged from 4.3 to 14.0%, whereas it was 0.8–7.1% for tetracyclines, 10.7–39.8% for macrolides, and 1.8–12.9% for aminoglycosides. In this study, the resistance rates of A. butzleri and A. cryaerophilus to nalidixic acid (33.4%, 35%), ciprofloxacin (31.3%, 35%), and tetracycline (64.7%, 52.5%) were much higher than previously reported (Ferreira et al., 2019). However, Jribi et al. (2020) reported a high level of resistance to tetracycline (100%) in Arcobacter spp. isolated from poultry in Tunisia.
The resistance rates of 51 A. butzleri isolates to erythromycin, azithromycin, and telithromycin were 5.9, 43.1, and 7.9%, respectively. All A. cryaerophilus isolates were completely sensitive to erythromycin. The resistance rate of Arcobacter spp. to erythromycin is consistent with a previous study (3.6%, 3/84) (Kietsiri et al., 2021). Moreover, streptomycin resistance rates for A. butzleri and A. cryaerophilus were 41.2 and 32.5%, respectively, which were higher than those reported in a previous study (Ferreira et al., 2016, 2019). Therefore, further attention should be paid to streptomycin resistance.
One strain each of A. butzleri (F050-4G) and A. cryaerophilus (F015-3G) contained resistance island gene clusters, which contained multiple antibiotic resistance genes and were located near the transposon. The resistance island in A. cryaerophilus (F015-3G) was inserted into IS4. The GC content of the two resistance islands was significantly higher than that of the genome of Arcobacter spp. We suspected that the resistance island might have been obtained by the horizontal gene transfer. Several studies reported that the use of antibiotics in animals might cause MDR and then transfer to humans (Chang et al., 2015; Dekker et al., 2019).
Several studies (Webb et al., 2018; Hodges et al., 2021) showed that the base mutation in the gyrA gene was associated with a higher level of resistance to ciprofloxacin. In our study, 13 A. butzleri and 14 A. cryaerophilus isolates with gyrA (C254T) mutation were 100% phenotypically resistant to ciprofloxacin. Moreover, the genes aph (3 ') -IIIa and ant (6) –Ia were reported to correlate with resistance to kanamycin and streptomycin, respectively (Ntilde et al., 2018; Cho et al., 2020), and in our study, two isolates carried the ant (6) –Ia gene and were phenotypically resistant to streptomycin. In addition, isolates that carried tetracycline resistance genes were phenotypically resistant to tetracycline, whereas the occurrence of ere(A) gene was associated with erythromycin resistance, which was consistent with previous studies (Gao et al., 2015; Zhao et al., 2016; Webb et al., 2018).
The number of antibiotic resistance genes varied greatly between the isolates. Some isolates harbored multiple antibiotic resistance genes, and specific resistance genes were detected in the corresponding antibiotic resistance isolates. Strains containing resistance genes were resistant to the corresponding antibiotics or had higher MIC values. A C254T mutation was found in some strains, resulting in a Thr to Ile substitution at position 85 of the deduced protein sequence. This substitution in A. butzleri and A. cryaerophilus isolates could be responsible for the observed fluoroquinolone resistance. A C254T mutation in gyrA, which resulted in a Thr to Ile substitution in gyrA were found in all ciprofloxacin resistance strains. This substitution in A. butzleri and A. cryaerophilus isolates could be responsible for the observed fluoroquinolone resistance.
Arcobacter skirrowii, A. cryaerophilus, and A. butzleri isolated from food and originating from animals commonly carry tlyA, pldA, mviN, ciaB, cj1349, and cadF (Douidah et al., 2012; Khoshbakht et al., 2014; Parisi et al., 2019; Khodamoradi and Abiri, 2020). Rathlavath et al. (2017) reported that the majority of A. butzleri isolated from seafood and the coastal environment contained six common virulence genes [cadF (89.7%), cj1349 (97.2%), ciaB (95.9%), mviN (100%), pldA (91.1%), and tlyA (91.8%)] but relatively lower amounts of hecA (10.8%), hecB (19%), iroE (12.9%), and irgA (17.6%). Similarly, our study found that more than 90% Arcobacter contained these six common virulence factors. It was found that different virulence genes had different functions, e.g., tlyA encoding hemolysin and pldA encoding the outer membrane phospholipase A are involved in erythrocyte lysis; mviN is required for the biosynthesis of peptidoglycan; ciaB is required for the biosynthesis of peptidoglycan; and both cadF and cj1349 encode fibronectin-binding protein (Parisi et al., 2019). A. butzleri and A. cryaerophilus showed no differences in the distribution of these virulence factors, which was in contrast to the results of a previous study (Sekhar et al., 2017) in which A. butzleri was observed to carry more of these virulence factors than A. cryaerophilus.
In addition to poultry, vegetables and seafood are also important transmission routes for Arcobacter infection in humans. The resistance island gene cluster found in pork and chicken meat and the carriage of virulence factors could be a potential health risk to human health.
Author's note
The first author, YM MD, female, was born in 1991, whose specialty is pathogenic microbiology; CJ, female, was born in 1982, majoring in analysis of genetic characteristics of Campylobacter and Arcobacter.
Data availability statement
The data presented in the study are deposited in the GenBank with accession numbers SAMN30871879 to SAMN30871970.
Author contributions
YD and MZ designed the experiments. YM, CJ, MY, HC, and JH participated in the sample collection and performed the experiments. GZ performed the genome bioinformatic analysis. YM and CJ wrote this paper. All authors read and approved the submitted manuscript.
Funding
This work was supported by the Sanming Project of Medicine in Shenzhen, China (SZSM201803081) and the Key Discipline Construction Subsidy of Nanshan District, Shenzhen, China.
Acknowledgments
We thank the colleagues in the top three local hospitals for collecting the patient stool samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barboza, K., Angulo, I., Zumbado, L., Redondo-Solano, M., Castro, E., and Arias, M. L. (2017). Isolation and identification of Arcobacter species from costa rican poultry production and retail sources. J. Food Prot. 80, 779–782. doi: 10.4315/0362-028X.JFP-16-394
Bayles, K. W. (2000). The bactericidal action of penicillin: New clues to an unsolved mystery. Trends Microbiol. 8, 274–278. doi: 10.1016/S0966-842X(00)01762-5
Brückner, V., Fiebiger, U., Ignatius, R., Friesen, J., Eisenblätter, M., Höck, M., et al. (2020). Prevalence and antimicrobial susceptibility of Arcobacter species in human stool samples derived from out- and inpatients: the prospective German Arcobacter prevalence study Arcopath. Gut Pathog. 12, 21. doi: 10.1186/s13099-020-00360-x
Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M., and Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Chieffi, D., Fanelli, F., and Fusco, V. (2020). Arcobacter butzleri: up-to-date taxonomy, ecology, and pathogenicity of an emerging pathogen. Compr. Rev. Food Sci. Food Saf. 19, 2071–2109. doi: 10.1111/1541-4337.12577
Cho, S., Barrett, J. B., Frye, J. G., and Jackson, C. R. (2020). Antimicrobial resistance gene detection and plasmid typing among multidrug resistant enterococci isolated from freshwater environment. Microorganisms 8, E1338. doi: 10.3390/microorganisms8091338
Collado, L., and Figueras, M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24, 174–192. doi: 10.1128/CMR.00034-10
Córdoba-Calderón, O., Redondo-Solano, M., Castro-Arias, E., and Arias-Echandi, M. L. (2017). Arcobacter isolation from minced beef samples in costa rica. J. Food Prot. 80, 775–778. doi: 10.4315/0362-028X.JFP-16-493
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15. doi: 10.1093/nar/gku1196
Dekker, D., Eibach, D., Boahen, K. G., Akenten, C. W., Pfeifer, Y., Zautner, A. E., et al. (2019). Fluoroquinolone-resistant Salmonella enterica, Campylobacter spp., and Arcobacter butzleri from local and imported poultry meat in Kumasi, Ghana. Foodborne Pathog. Dis. 16, 352–358. doi: 10.1089/fpd.2018.2562
Douidah, L., de Zutter, L., Bare, J., De Vos, P., Vandamme, P., Vandenberg, O., et al. (2012). Occurrence of putative virulence genes in arcobacter species isolated from humans and animals. J. Clin. Microbiol. 50, 735–741. doi: 10.1128/JCM.05872-11
Fernandez, H., Villanueva, M. P., Mansilla, I., Gonzalez, M., and Latif, F. (2015). Arcobacter butzleri and A. Cryaerophilus in human, animals and food sources, in southern Chile. Braz. J. Microbiol. 46, 145–147. doi: 10.1590/S1517-838246120140095
Ferreira, S., Julio, C., Queiroz, J. A., Domingues, F. C., and Oleastro, M. (2014). Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among Portuguese patients. Diagn. Microbiol. Infect. Dis. 78, 220–225. doi: 10.1016/j.diagmicrobio.2013.11.021
Ferreira, S., Luís, Â., Oleastro, M., Pereira, L., and Domingues, F. C. (2019). A meta-analytic perspective on Arcobacter spp. Antibiotic resistance. J. Glob. Antimicrob. Resist. 16, 130–139. doi: 10.1016/j.jgar.2018.12.018
Ferreira, S., Queiroz, J. A., Oleastro, M., and Domingues, F. C. (2016). Insights in the pathogenesis and resistance of Arcobacter: a review. Crit. Rev. Microbiol. 42, 364–383. doi: 10.3109/1040841X.2014.954523
Figueras, M. J., Levican, A., Pujol, I., Ballester, F., Rabada, Q. M., and Gomez-Bertomeu, F. (2014). A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2, 31–37. doi: 10.1002/2052-2975.35
Gao, P., He, S., Huang, S., Li, K., Liu, Z., Xue, G., et al. (2015). Impacts of coexisting antibiotics, antibacterial residues, and heavy metals on the occurrence of erythromycin resistance genes in urban wastewater. Appl. Microbiol. Biotechnol. 99, 3971–3980. doi: 10.1007/s00253-015-6404-9
Groicher, K. H., Firek, B. A., Fujimoto, D. F., and Bayles, K. W. (2000). The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182, 1794–1801. doi: 10.1128/JB.182.7.1794-1801.2000
Hodges, L. M., Taboada, E. N., Koziol, A., Mutschall, S., Blais, B. W., Inglis, G. D., et al. (2021). Systematic evaluation of whole-genome sequencing based prediction of antimicrobial resistance in Campylobacter jejuni and C. coli. Front. Microbiol. 12, 776967. doi: 10.3389/fmicb.2021.776967
Jasim, S. A., Al-Abodi, H. R., and Ali, W. S. (2021). Resistance rate and novel virulence factor determinants of Arcobacter spp., from cattle fresh meat products from Iraq. Microb. Pathog. 152, 104649. doi: 10.1016/j.micpath.2020.104649
Jribi, H., Sellami, H., Amor, S. B., Ducournau, A., SifrÉ, E., Benejat, L., et al. (2020). Occurrence and antibiotic resistance of arcobacter species isolates from poultry in tunisia. J. Food Prot. 83, 2080–2086. doi: 10.4315/JFP-20-056
Kayman, T., Abay, S., Hizlisoy, H., Atabay, H. I., Diker, K. S., and Aydin, F. (2012). Emerging pathogen Arcobacter spp. in acute gastroenteritis: molecular identification, antibiotic susceptibilities and genotyping of the isolated arcobacters. J. Med. Microbiol. 61, 1439–1444. doi: 10.1099/jmm.0.044594-0
Khodamoradi, S., and Abiri, R. (2020). The incidence and antimicrobial resistance of Arcobacter species in animal and poultry meat samples at slaughterhouses in Iran. Iran. J. Microbiol. 12, 531–536. doi: 10.18502/ijm.v12i6.5027
Khoshbakht, R., Tabatabaei, M., Shirzad, A. H., and Seifi, S. (2014). Occurrence of Arcobacter in Iranian poultry and slaughterhouse samples implicates contamination by processing equipment and procedures. Br. Poult. Sci. 55, 732–736. doi: 10.1080/00071668.2014.971223
Kietsiri, P., Muangnapoh, C., Lurchachaiwong, W., Lertsethtakarn, P., Bodhidatta, L., Suthienkul, O., et al. (2021). Characterization of Arcobacter spp. isolated from human diarrheal, non-diarrheal and food samples in Thailand. PLoS ONE. 16, e246598. doi: 10.1371/journal.pone.0246598
Kim, N. H., Park, S. M., Kim, H. W., Cho, T. J., Kim, S. H., Choi, C., et al. (2019). Prevalence of pathogenic Arcobacter species in South Korea: comparison of two protocols for isolating the bacteria from foods and examination of nine putative virulence genes. Food Microbiol. 78, 18–24. doi: 10.1016/j.fm.2018.09.008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lehmann, D., Alter, T., Lehmann, L., Uherkova, S., Seidler, T., and Golz, G. (2015). Prevalence, virulence gene distribution and genetic diversity of Arcobacter in food samples in Germany. Berl. Munch. Tierarztl. Wochenschr. 128, 163–168. doi: 10.2376/0005-9366-128-163
Miller, W. G., Parker, C. T., Rubenfield, M., Mendz, G. L., Wosten, M. M., Ussery, D. W., et al. (2007). The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS ONE. 2, e1358. doi: 10.1371/journal.pone.0001358
Mottola, A., Bonerba, E., Bozzo, G., Marchetti, P., Celano, G. V., Colao, V., et al. (2016). Occurrence of emerging food-borne pathogenic Arcobacter spp. isolated from pre-cut (ready-to-eat) vegetables. Int. J. Food Microbiol. 236, 33–37. doi: 10.1016/j.ijfoodmicro.2016.07.012
Mottola, A., Ciccarese, G., Sinisi, C., Savarino, A. E., Marchetti, P., Terio, V., et al. (2021). Occurrence and characterization of Arcobacter spp. from ready-to-eat vegetables produced in Southern Italy. Ital. J. Food Saf. 10, 8585. doi: 10.4081/ijfs.2021.8585
Ntilde, L. H. O., Iacute, M., Ugarte-Ruiz, A., Palomo, G., Borge, C., Florez-Cuadrado, D., et al. (2018). ant(6)-I genes encoding aminoglycoside O-nucleotidyltransferases are widely spread among streptomycin resistant strains of Campylobacter jejuni and Campylobacter coli. Front. Microbiol. 9, 2515. doi: 10.3389/fmicb.2018.02515
Oliveira, M., Gomes, V., Cunha, M., Moreno, L. Z., Moreno, A. M., and Knöbl, T. (2018). Genotypic characterization of Arcobacter spp. isolated from chicken meat in Brazil. Foodborne Pathog. Dis. 15, 293–299. doi: 10.1089/fpd.2017.2368
Parisi, A., Capozzi, L., Bianco, A., Caruso, M., Latorre, L., Costa, A., et al. (2019). Identification of virulence and antibiotic resistance factors in Arcobacter butzleri isolated from bovine milk by Whole Genome Sequencing. Ital. J. Food Saf. 8, 7840. doi: 10.4081/ijfs.2019.7840
Patyal, A., Rathore, R. S., Mohan, H. V., Dhama, K., and Kumar, A. (2011). Prevalence of Arcobacter spp. in humans, animals and foods of animal origin including sea food from India. Transbound. Emerg. Dis. 58, 402–410. doi: 10.1111/j.1865-1682.2011.01221.x
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., and Korobeynikov, A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinformatics. 70, e102. doi: 10.1002/cpbi.102
Ramees, T. P., Dhama, K., Karthik, K., Rathore, R. S., Kumar, A., Saminathan, M., et al. (2017). Arcobacter: an emerging food-borne zoonotic pathogen, its public health concerns and advances in diagnosis and control - a comprehensive review. Vet. Q. 37, 136–161. doi: 10.1080/01652176.2017.1323355
Rathlavath, S., Kohli, V., Singh, A. S., Lekshmi, M., Tripathi, G., Kumar, S., et al. (2017). Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. Int. J. Food Microbiol. 263, 32–37. doi: 10.1016/j.ijfoodmicro.2017.10.005
Schönknecht, A., Alter, T., and Gölz, G. (2020). Detection of Arcobacter species in different intestinal compartments of broiler chicken during slaughter and processing. Microbiologyopen 9, e1106. doi: 10.1002/mbo3.1106
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sekhar, M. S., Tumati, S. R., Chinnam, B. K., Kothapalli, V. S., and Sharif, N. M. (2017). Virulence gene profiles of Arcobacter species isolated from animals, foods of animal origin, and humans in Andhra Pradesh, India. Vet World. 10, 716–720. doi: 10.14202/vetworld.2017.716-720
Šilha, D., Vacková, B., and Šilhová, L. (2019). Occurrence of virulence-associated genes in Arcobacter butzleri and Arcobacter cryaerophilus isolates from foodstuff, water, and clinical samples within the Czech Republic. Folia Microbiol. 64, 25–31. doi: 10.1007/s12223-018-0628-x
Van den Abeele, A. M., Vogelaers, D., Van Hende, J., and Houf, K. (2014). Prevalence of Arcobacter species among humans, Belgium, 2008-2013. Emerging Infect. Dis. 20, 1731–1734. doi: 10.3201/eid2010.140433
Vidal-Veuthey, B., Jara, R., Santander, K., Mella, A., Ruiz, S., and Collado, L. (2021). Antimicrobial resistance and virulence genes profiles of Arcobacter butzleri strains isolated from back yard chickens and retail poultry meat in Chile. Lett. Appl. Microbiol. 72, 126–132. doi: 10.1111/lam.13404
Wang, M., Gu, Y. X., Liang, H., Fu, Y. Y., He, L. H., Zhang, J. Z., et al. (2016). Isolation and identification of Arcobacter from retail chicken meat in Beijing. Dis. Surveil. 31, 1050–1054. doi: 10.3784/j.issn.1003-9961.2016.12.017 (In Chinese)
Webb, A. L., Selinger, L. B., Taboada, E. N., and Inglis, G. D. (2018). Subtype-specific selection for resistance to fluoroquinolones but not to tetracyclines is evident in Campylobacter jejuni isolates from beef cattle in confined feeding operations in Southern Alberta, Canada. Appl. Environ. Microb. 84, e02713-17. doi: 10.1128/AEM.02713-17
Zambri, M., Cloutier, M., Adam, Z., Lapen, D. R., Wilkes, G., Sunohara, M., et al. (2019). Novel virulence, antibiotic resistance and toxin gene-specific PCR-based assays for rapid pathogenicity assessment of Arcobacter faecis and Arcobacter lanthieri. BMC Microbiol. 19, 11. doi: 10.1186/s12866-018-1357-7
Zhang, X., Alter, T., and Gölz, G. (2019). Characterization of Arcobacter spp. isolated from retail seafood in Germany. Food Microbiol. 82, 254–258. doi: 10.1016/j.fm.2019.02.010
Keywords: Arcobacter, whole genome sequencing, antibiotic resistance, phylogenomic analysis, zoonotic pathogen
Citation: Ma Y, Ju C, Zhou G, Yu M, Chen H, He J, Zhang M and Duan Y (2022) Genetic characteristics, antimicrobial resistance, and prevalence of Arcobacter spp. isolated from various sources in Shenzhen, China. Front. Microbiol. 13:1004224. doi: 10.3389/fmicb.2022.1004224
Received: 27 July 2022; Accepted: 07 November 2022;
Published: 01 December 2022.
Edited by:
Xiangru Wang, Huazhong Agricultural University, ChinaReviewed by:
Seçil Abay, Erciyes University, TurkeyValentine Usongo, Health Canada, Canada
Haiquan Kang, Affiliated Hospital of Xuzhou Medical University, China
Susana Ferreira, University of Beira Interior, Portugal
Copyright © 2022 Ma, Ju, Zhou, Yu, Chen, He, Zhang and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongxiang Duan, c3pkdWFueXhAMTYzLmNvbQ==; Maojun Zhang, emhhbmdtYW9qdW5AaWNkYy5jbg==
†These authors share first authorship
 Yanping Ma
Yanping Ma Changyan Ju1†
Changyan Ju1† Guilan Zhou
Guilan Zhou Maojun Zhang
Maojun Zhang Yongxiang Duan
Yongxiang Duan