
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 November 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1003244
This article is part of the Research Topic Community Series in Plants and Microbial Communities: Diversity, Pathogens and Biological Control, volume II View all 25 articles
 Lei Shao1
Lei Shao1 Yumei Tan2,3*
Yumei Tan2,3* Shiying Song2,3
Shiying Song2,3 Yuchen Wang2,3
Yuchen Wang2,3 Yongxiang Liu2,3
Yongxiang Liu2,3 Yonghui Huang2,3
Yonghui Huang2,3 Xiyi Ren2,3
Xiyi Ren2,3 Zuoyi Liu2,3,4*
Zuoyi Liu2,3,4*Aspergillus cristatus is the dominant fungus during the fermentation of Fuzhuan brick tea; hypotonic conditions only induce its sexual development to produce ascospores, while hypertonic conditions only induce its asexual development to produce conidia, indicating that osmotic stress can regulate spore production in A. cristatus. However, the underlying regulatory mechanism is unclear. In this study, the role of Achog1, which is homologous to hog1 from Saccharomyces cerevisiae, in sporulation, different kinds of stress responses and pigment production was investigated. Deletion mutants of Achog1 were obtained by homologous recombination. Phenotypic observations showed that the time required to produce conidia was delayed, and the number of conidia produced was significantly reduced in the deletion mutants of Achog1 in hypertonic media, indicating that Achog1 plays a positive role in asexual development. Stress sensitivity tests showed that ΔAchog1 strains were sensitive to hyperosmolarity, and the order of the sensitivity of ΔAchog1 to different osmotic regulators was 3 M sucrose >3 M NaCl >3 M sorbitol. Moreover, the deletion mutants were sensitive to high oxidative stress. pH sensitivity tests indicated that Achog1 inhibited the growth of A. cristatus under alkaline stress. Additionally, pigmentation was decreased in the Achog1 deletion mutants compared with the WT. All the above developmental defects were reversed by the reintroduction of the Achog1 gene in ΔAchog1. Pull-down and LC–MS/MS analysis showed that the expression levels of proteins interacting with Achog1 were significantly different under low and high osmotic stress, and proteins related to conidial development were present only in the cultures treated with hyperosmotic stress. Transcription profiling data showed that Achog1 suppressed the expression of several genes related to asexual development, osmotic and oxidative stress resistance. On the basis of gene knockout, pull-down mass spectrometry and RNA-seq analyses, a regulatory pathway for Achog1 was roughly identified in A. cristatus.
Aspergillus cristatus is the dominant fungus during the fermentation of Fuzhuan brick tea and is known as the “Golden Flower Fungus” because of its golden yellow cleistothecia (Mo et al., 2008; Tan et al., 2017). Studies have shown that A. cristatus has antitumor activity (Deng et al., 2007), bacteriostatic effects (Li et al., 2011), antioxidant activity (Ouyang et al., 2011) and digestive enzyme activity (Yang, 2005; Ding, 2012). A. cristatus is a homothallic filamentous fungus and performs both sexual and asexual reproduction. Unlike Aspergillus nidulans, light does not significantly affect the development of A. cristatus (Tan et al., 2018). Interestingly, hypotonic conditions can only induce sexual development, while hypertonic conditions can only induce asexual development. Thus, A. cristatus provides an excellent genetic system for studying the mechanism of sporulation of filamentous fungi.
The sexual development of other fungi in Aspergillus is mainly affected by light, temperature, nutrition, and pH. For example, a complex protein, VeA/VelB/LaeA, called “Velvet,” plays a central role in the regulation of sexual development under light and dark conditions (Bayram et al., 2008). The regulation of asexual development is mainly regulated by the core regulatory network composed of brlA, abaA, and wetA, and other genes are directly or indirectly regulated by this network (Han and Adams, 2001; Tao and Yu, 2011; Alkhayyat et al., 2015). brlA was mainly found in vesicle and pedicel but not in mycelia and conidia. brlA is located upstream of the central regulatory pathway, and the expression of several genes related to asexual development, such as rodA, is regulated by brlA (Twumasi et al., 2009). We have been trying to explore the functions of genes related to sporulation in A. cristatus since 2013. Many genes associated with sexual and asexual spore production have been reported, such as flbA, preA, and AcndtA (Wang et al., 2014; Xiang et al., 2020; Wang et al., 2021). Nevertheless, it is not clear that why A. cristatus can produce ascospores under hypotonic conditions and conidia under hypertonic conditions. We speculate that there is a certain association between osmotic stress and spore production in A. cristatus.
It is well known that the high osmolarity glycerol mitogen-activated protein kinase signal transduction pathway (HOG-MAPK, HOG) is the main way to adapt to changes in osmolarity in the environment. Hog1 (MAPK) is the core protein kinase in this pathway (Krantz et al., 2006). It was first reported in S. cerevisiae. Cells respond to increases in osmolarity in the extracellular environment by activating hog1 (Wurgler-Murphy et al., 1997). hog1 was also reported to be involved in osmotic stress regulation in other fungi. osm1 is a functional homolog of hog1, the hyphal growth of Δosm1 is very sensitive to osmotic stress, and the hyphal morphology changes under hypertonic conditions in Magnaporthe grisea (Dixon et al., 1999; Xu, 2000). Thhog1 regulates the hypertonic stress response in Trichoderma harzianum (Delgado-Jarana et al., 2006). In A. fumigatus, sakA, the homolog of hog1, could regulate conidial germination (Du et al., 2006). A similar phenomenon was found in Botrytis. cinerea (Segmüller et al., 2007; Heller et al., 2012). hog1 also plays a role in the oxidative stress response (Calmes et al., 2015). Cpmk1, a homolog of hog1, can regulate the production of pigment in Cryphonectria parasitica (Park et al., 2004). The effect of hog1 on PH stress has also been reported. The deletion mutant of hog1 slowed down significantly in the medium with alkaline pH in A. fumigatus (Ma et al., 2012). In summary, hog1 plays an important role in the stress response, asexual sporulation and pigment production in fungi.
Sequence alignment revealed a gene homologous to hog1, SI65_07698, in the A. cristatus genome database. In this study, SI65_07698 was named Achog1 based on the results of a phylogenetic analysis. Achog1 deletion and complementation strains were generated to verify the role of Achog1. Under different osmotic stress conditions, pull-down mass spectrometry was carried out, and RNA sequencing (RNA-seq) was performed on the ΔAchog1 and WT strains to explore the connection between the HOG pathway and spore production of A. cristatus.
The WT strains of A. cristatus (CGMCC 7.193) used in this study were isolated from Fuzhuan brick tea produced by the Yiyang Tea Factory in Yiyang, China. The strains were grown on low-concentration sodium chloride solid MYA media (20 g of malt extract, 5 g of yeast extract, 30 g of sucrose, 50 g of sodium chloride, and 1,000 mL of water) at 28°C to induce sexual development and high-concentration sodium chloride solid media (20 g of malt extract, 5 g of yeast extract, 30 g of sucrose, 170 g of sodium chloride, and 1,000 mL of water) at 37°C to induce asexual development. SD/−Trp, SD/−Leu, SD/−Trp/−Leu, SD/−Trp/− Leu/-His and SD/−Trp/−Leu/-His/−Ade (purchased from Beijing Cool Lab Technology, China). The colonies were imaged using a Canon EOS 7D Mark II camera (Canon, Tokyo, Japan).
The Achog1 gene fragment was ligated into the expression vector pGEX-6p-1, and GST fusion protein expression and transformation were performed. Then, the total proteins of A. cristatus cultivated under normal osmotic stress and hypertonic stress were extracted, and pull-down assays were carried out. The proteins were subsequently detected via LC–MS/MS. Proteins were verified by yeast two-hybrid assays. To facilitate subsequent analysis, the samples of A. cristatus cultivated under normal stress and hypertonic stress were named WT-D and WT-H, respectively.
Phylogenetic analysis was performed using MEGA 6.06 software with the conserved domains of homologous AcHog1 proteins (Tamura et al., 2013). ClustalW was employed for multiple sequence alignment with the default values. A maximum likelihood with a bootstrap value of 1,000 was used to generate a phylogenetic tree.
Homologous recombination was employed to delete the whole open reading frame (ORF) of Achog1. An Achog1 deletion cassette containing hph as a selective marker was constructed via the fusion of the 5′-untranslated region (5’-UTR) and the 3′-untranslated region (3’-UTR). Briefly, the 5’-BamHI-XhoI UTR and 3’-SpeI-XbaI UTR were amplified using specific primer pairs (Supplementary Table S1) from genomic DNA of the WT strain and cloned into the pDHt/sk-hyg cloning sites to generate the final Achog1-L-pDHt/sk-hyg-Achog1-R knockout vector. To complement the Achog1-null mutant, the Achog1 gene with its own promoter was amplified from the genomic DNA with the primer pair qc-hog1-F/R (Supplementary Table S1) and then inserted into pDHt/sknt at the enzyme sites HindIII/KpnI.
The plasmids were confirmed via PCR, restriction enzyme digestion and sequencing. The knockout plasmids were transformed into the WT strains, and complementary plasmids were transformed into the ΔAchog1 knockout strains by Agrobacterium tumefaciens-mediated transformation (ATMT), as previously described (Tan et al., 2018). The transformants were selected on MYA media that included 50 ug/mL hygromycin B or 80 ug/mL geneticin (G418) and were confirmed via PCR (the primers used are listed in Supplementary Table S1).
The transgene copy number in ΔAchog1 was estimated by using RT-PCR based on the methods of Song et al. (2002). Using the genomic DNA of ΔveA, a standard curve was established, in which the copy number was determined by Southern blot analysis (Tan et al., 2018). The genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and HYG were selected from the A. cristatus genome as candidate reference genes (the primers used are listed in Supplementary Table S1).
RNA-seq was performed on the ΔAchog1 and WT strains. A conidial suspension (concentration 1 × 106 conidia/mL) was used for inoculation. Three biological replicates were grown on MYA media comprising 17% NaCl at 37°C. The WT strain produced a large number of conidial heads, but the ΔAchog1 knockout strain did not after 39 h of culture. Based on these observations, all the samples were collected from the cellulose membrane, flash frozen in liquid nitrogen and stored at-80°C.
Total RNA was extracted as described previously using TRIzol (Gilbert et al., 2016). An Agilent Bioanalyzer 2,100 was subsequently used to assess the concentration and quality of the total RNA. mRNA libraries were prepared as described previously (Parkhomchuk et al., 2009). The quality of the final cDNA libraries was verified using an Agilent Bioanalyzer 2,100. The libraries were sequenced using an Illumina NovaSeq 6,000 (Illumina, United States). After filtering the raw data and checking the sequencing error rate and GC content distribution, the clean reads used for the subsequent analysis were obtained. HISAT2 software was used to quickly and accurately align the clean reads with the reference genome to obtain the positioning information. Differentially expressed genes were analysed using DESeq2 (Love et al., 2014); gene ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the differentially expressed genes were performed by using ClusterProfiler software (Kanehisa and Goto, 2000; Young et al., 2010).
Total RNA was extracted at the tested time point. Then, 2 mg of RNA was utilized to synthesize cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, catalogue#K1622). RT-qPCR was performed using a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules CA, United States) in a total volume of 10 μL, which consisted of 5 μL of SsoFast EvaGreen SuperMix (Bio-Rad, catalogue# 172–5,201), 1 μL of each primer (10 pmol/ml) and 1 μL of template. GAPDH was selected as a candidate reference gene. The primers employed in the RT-qPCR experiments were designed using the Primer 3 online program,1 and the resulting RT-qPCR products were tested via agarose gel electrophoresis. Each primer pair was tested with serial dilutions of cDNA to determine the linear range of the RT-qPCR assays. Three biological replicates were analysed. All the RT-qPCR primers used are listed in Supplementary Table S1.
The homolog of hog1, SI65-07698, was identified based on the genome of A. cristatus (accession number [JXNT01000000]) (Ge et al., 2016). SI65_07698 is 1,689 bp in length, includes six introns and encodes a predicted protein of 366 amino acids showing 98% identity to S. cerevisiae Hog1 and the stress-activating kinase SakA of A. nidulans. The results of multisequence alignment revealed that Hog1 was very conserved across Aspergillus species (Supplementary Figure S1A). The predicted SI65_07698 protein was found to contain a Pkc-like superfamily domain (Supplementary Figure S1C), which was consistent with the conserved domain of hogA in A. nidulans. SI65_07698 was named Achog1 based on these analysises. It is worth noting that halophilic Aspergillus contains 3 Hog1 homologs, which are differentially regulated during different salinity conditions (Rodríguez-Pupo et al., 2021). A. cristatus does not contain a second Hog1 homolog, as it occurs in most aspergilli (Kawasaki et al., 2002; Garrido-Bazán et al., 2018). Achog1 deletion mutants were obtained by gene replacement with the hygromycin B phosphotransferase gene (hph) as a selective marker to verify the function of Achog1 in A. cristatus. Six ΔAchog1 transformants were ultimately obtained (Supplementary Figure S2B shows five of the transformants). The expression of Achog1 in ΔAchog1 strain was detected and the results showed that Achog1 was not expressed in ΔAchog1 strain, which further indicated that Achog1 had been successfully knocked out (Supplementary Figure S2D, the last column). Real-time fluorescent PCR showed that the numbers of hph in ΔAchog1 were single copies (Supplementary Figure S3 and Supplementary Table S2). Eighteen ΔAchog1-C strains with the WT-like phenotype were verified by PCR. The results are shown in Supplementary Figure S2C.
Aspergillus cristatus could produce pure sexual/asexual spores under hypotonic and hypertonic stress. It was speculated that there was an association between osmotic stress and sporulation. Therefore, the sexual and asexual development of WT and ΔAchog1 was observed under different osmotic stresses by inoculating an equal amount of conidia (concentration: 1 × 106 conidia/mL). When hyphae of the WT strain formed conidiophores, most hyphae of ΔAchog1 only formed vesicles at 39 h on MYA media containing 3 M NaCl (Figures 1A,B, indicating that deleting Achog1 delays the germination of conidia. A large number of grey-green conidia were produced by WT, while only a small number of conidia was produced by ΔAchog1 in the center of the colony as the incubation time increased (Figure 1C). The conidial number produced by WT was nearly 2 times that produced by the ΔAchog1 strain (Figure 1D). No significant difference was found in sexual development between WT and ΔAchog1 on hypotonic media (Figure 1C: the first column). However, hyphal curling appeared in ΔAchog1 after culture with 3 M NaCl for 7 days, and a small amount of golden yellow cleistothecia appeared in the ΔAchog1 strain after culturing for 14 days (Figure 2), which was an interesting phenomenon. These results indicated that Achog1 positively regulated asexual sprouting and might inhibit sexual development under hypertonic stress in A. cristatus.
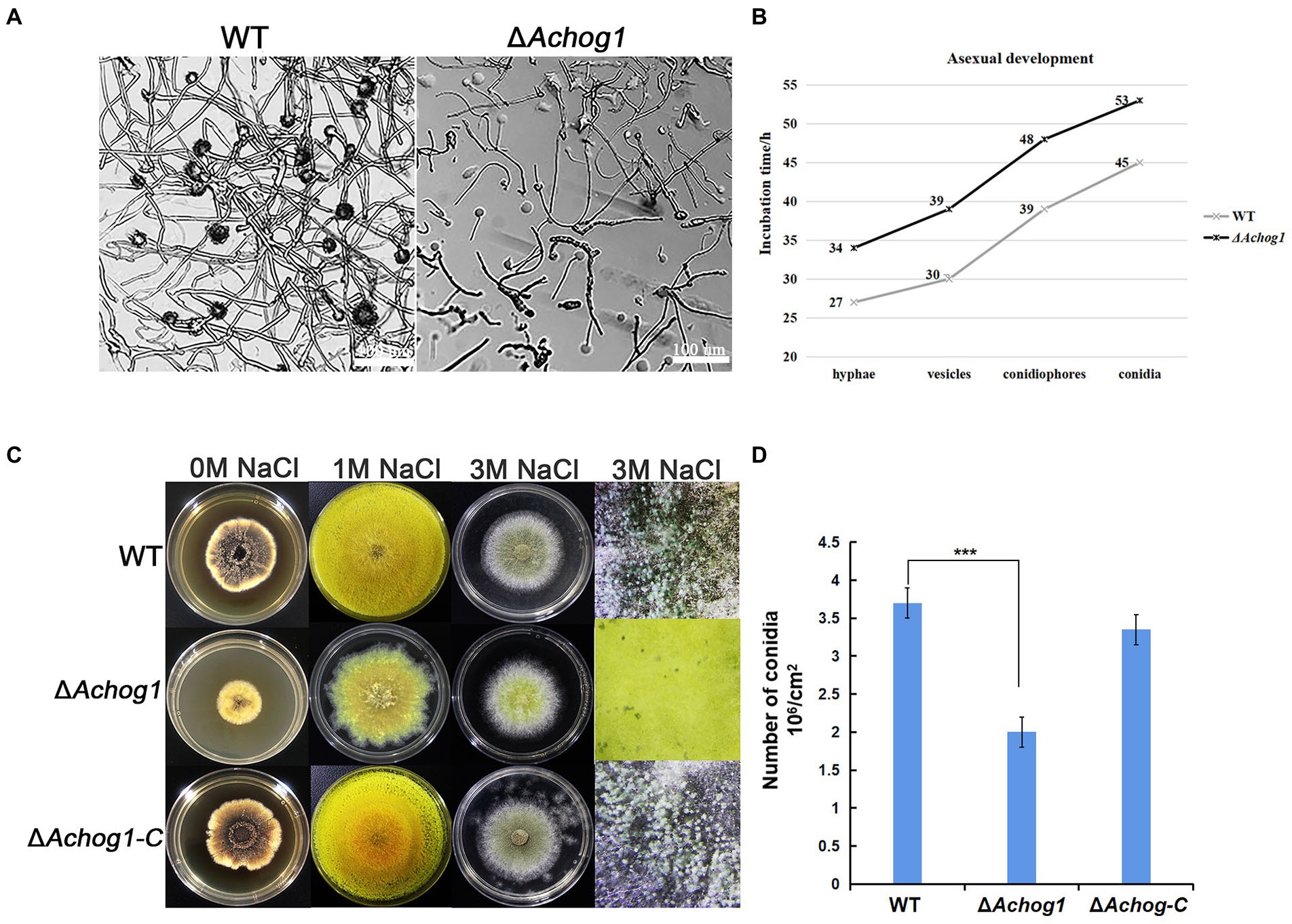
Figure 1. The effect of Achog1 on asexual sporulation. (A) The morphology of WT and ΔAchog1. All strains were cultivated on MYA medium containing 3 M NaCl at 37°C for 39 h. (B) Incubation time of asexual development. (C) The morphology of WT and ΔAchog1. All strains were cultivated on MYA medium containing 1 M and 3 M NaCl at 37°C for 7 days. (D) Conidia of WT and ΔAchog1 under stereoscopic microscopy. Statistics of the number of conidia produced by the WT, ΔAchog1 and ΔAchog1-C strains cultivated on MYA medium containing 3 M NaCl at 37°C for 7 days. The total number of conidia per fixed area was counted with a haematocytometer under a microscope, and the data were used to calculate the number of conidia per cm2. Error bars represent the standard deviation of at least three replicates. Values that differed significantly from the value for the WT strain according to a t test are indicated with asterisks (***p < 0.001).
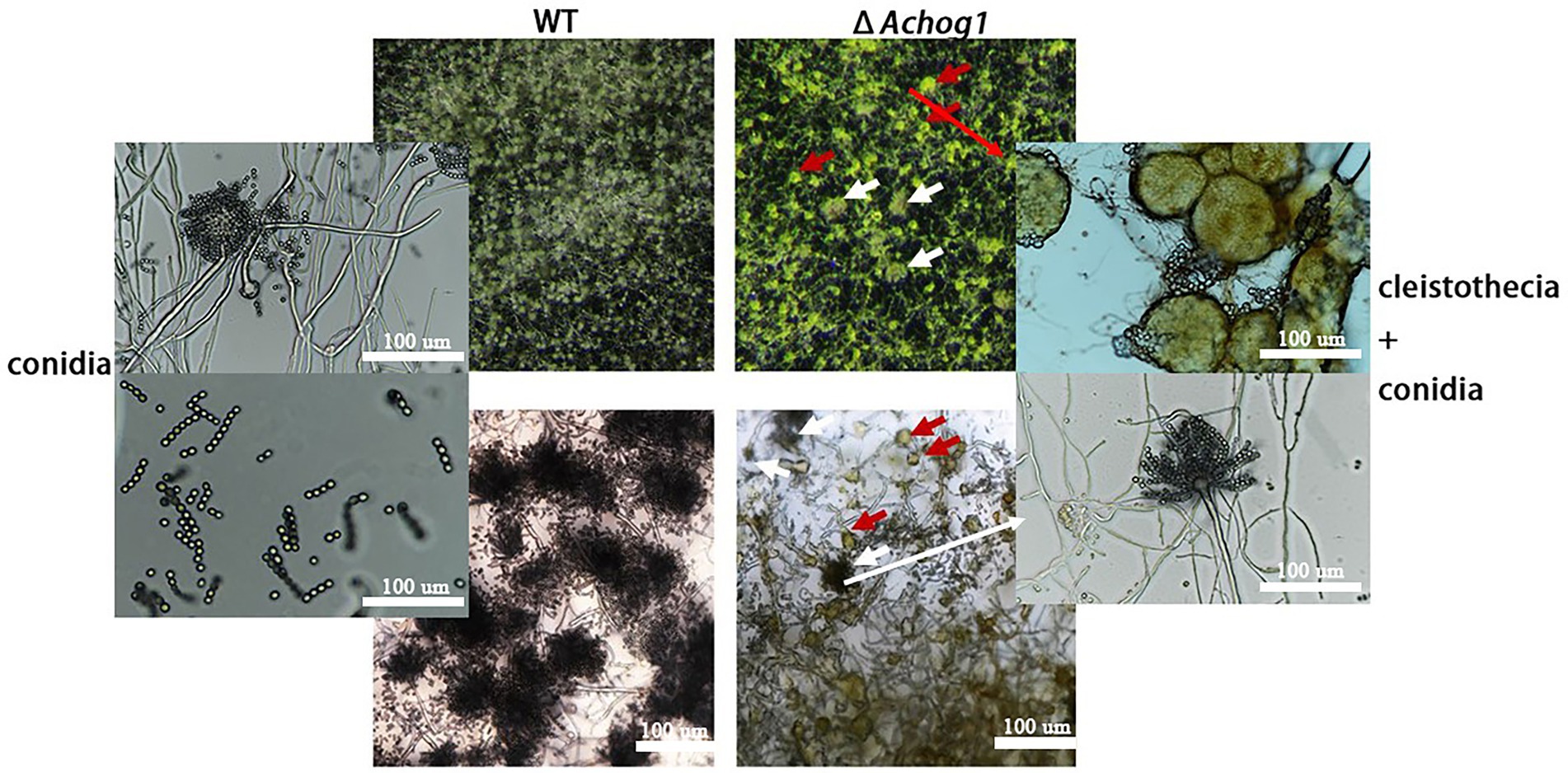
Figure 2. Microscopic observation of WT and ΔAchog1. WT and ΔAchog1 strains cultivated on MYA medium containing 3 M NaCl at 37°C for 14 days. The red arrows refer to cleistothecia, and the white arrows refer to conidia.
It is well known that one of the main functions of hog1 is to respond to hyperosmolarity in other fungi. The conidia of WT and ΔAchog1 (concentration: 1 × 106 conidia/mL) were cultivated on MYA media with 3 M sorbitol and sucrose to confirm whether Achog1 is involved in hypertonic stress. The growth of ΔAchog1 was significantly slower than that of WT on MAY media with 3 M sucrose, and ΔAchog1 also grew slowly on media with 3 M NaCl and sorbitol (Figures 3A,B). Furthermore, the expression of genes related to hypertonic stress was detected. The results showed that the expression of genes related to hypertonic stress in ΔAchog1 was lower than that of in WT under hypertonic stress (Figure 3D). Fewer conidia were produced by ΔAchog1 (Figure 3C). The results of sensitivity tests showed that Achog1 could positively regulate the response to hypertonic stress, and the sensitivity of ΔAchog1 to different osmotic stress regulators was in descending order: 3 M sucrose >3 M NaCl >3 M sorbitol.
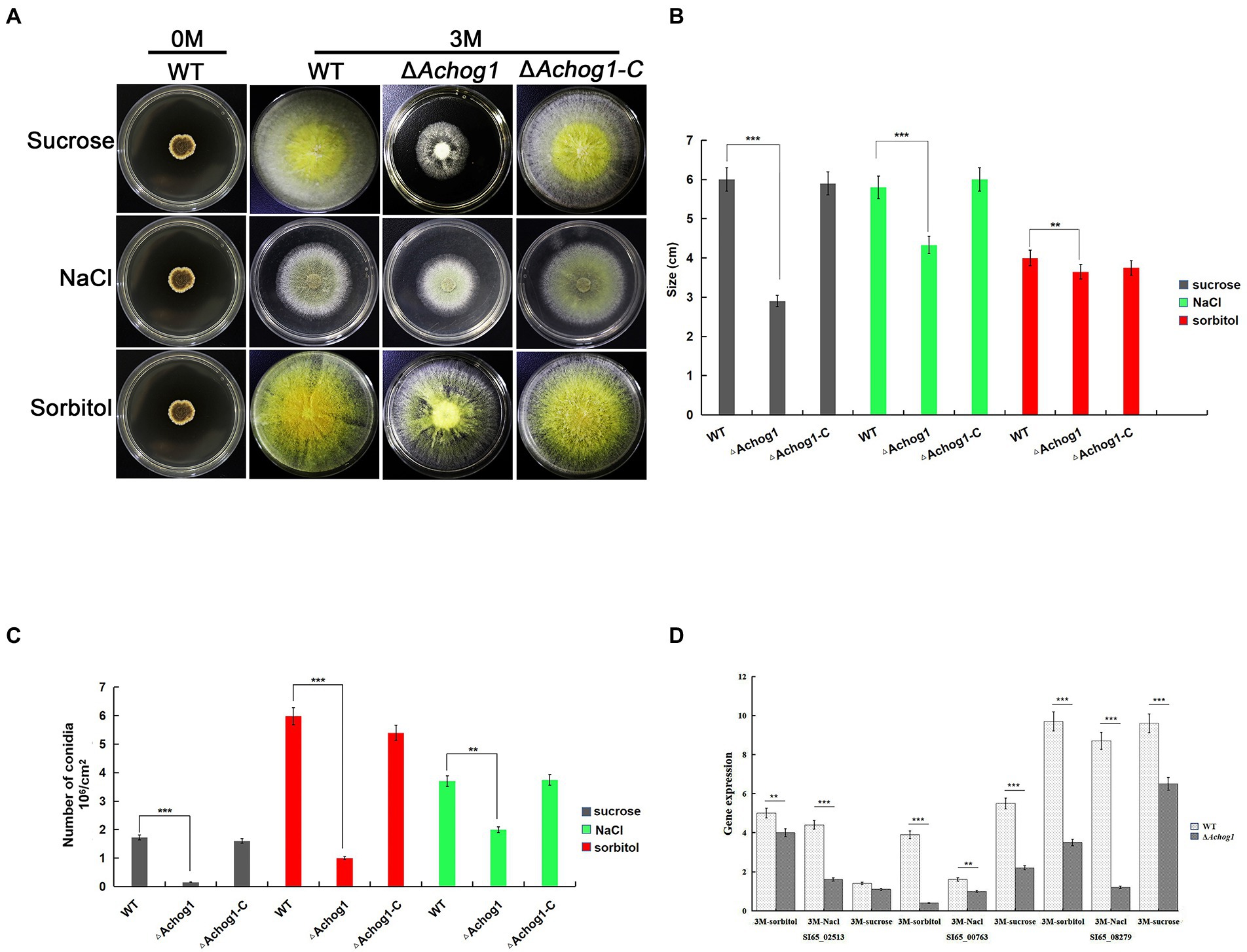
Figure 3. Effect of Achog1 on the osmotic stress response. (A) Colony morphology of WT, ΔAchog1 and ΔAchog1-C strains grown in medium with 1 M,3 M sucrose,sorbitol and NaCl at 37°C for 7 days; (B) Statistics of the number of conidia produced by WT, ΔAchog1 and ΔAchog1-C strains grown in medium with 1 M,3 M sucrose, sorbitol and NaCl at 37°C for 7 days; (C) Statistics of the colony diameters in medium with 1 M, 3 M sucrose, sorbitol and NaCl at 37°C for 7 days; (D) Expression of genes related to the response of high osmotic stress. Error bars indicate the standard deviation of at least three replicates. Values that differed significantly from the value for the WT strain according to a t test are indicated with asterisks (**p < 0.01, ***p < 0.001).
A study showed that hog1 mediated the oxidative stress pathway in C. albicans (Thomas et al., 2013). The conidia of WT and ΔAchog1 (concentration: 1 × 106/mL) were cultivated in MYA with different concentrations of H2O2 (0 mM, 10 mM, 30 mM, and 50 mM) to verify whether Achog1 was involved in oxidative stress. The colony diameter of ΔAchog1 decreased, and its edges were irregular (Figures 4A,B), suggesting that the tolerance of ΔAchog1 to oxidative stress was significantly reduced. We detected the expression of genes related to oxidative stress. The results showed that the expression of genes related to oxidative stress in ΔAchog1 was lower than that of in WT under oxidative stress (Figure 4C). When the Achog1 gene was complemented, the sensitivity to H2O2 was essentially restored. These results indicated that Achog1 positively regulated the response to oxidative stress.
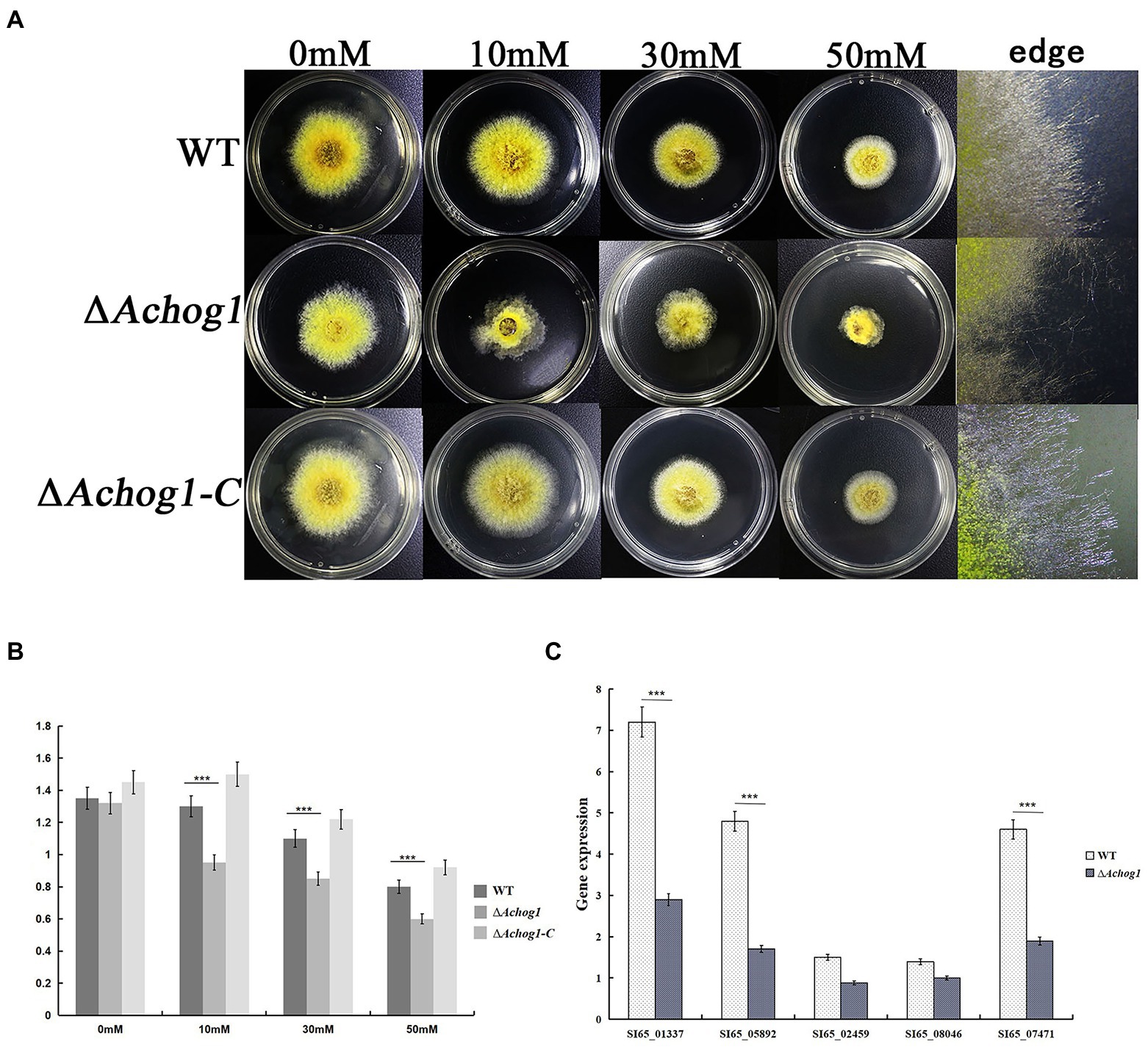
Figure 4. Effect of Achog1 on the oxidative stress response. (A) Colony morphology of WT and ΔAchog1 under MYA medium with different concentrations of H2O2 at 28°C for 3 days. (B) Colony diameters of WT and ΔAchog1. (C) Expression of genes related to the response of oxidative stress. Error bars indicate the standard deviation of at least three replicates. Values that differed significantly from the value for the WT strain according to a t test are indicated with asterisks (***p < 0.001).
It was shown that the ΔAfhog1 deletion mutant strain grew slower than the wild type strain in medium containing high PH in A. fumigatus (Ma et al., 2012). To test whether Achog1 was involved in PH stress, equal amounts of conidia (1.0 × 106 conidia/mL) of WT, ΔAchog1, and ΔAchog1-C were inoculated on MYA media containing 5% NaCl with different pH values (pH 3, pH 4, pH 5, pH 8, and pH 11). ΔAchog1 grew normally in media at pH 3–5, its growth was just slower than that of the WT and ΔAchog1-C strains after culturing for 6 days at 28°C. However, the growth of ΔAchog1 significantly slowed down in the media at pH 8, and ΔAchog1 could not grow at pH 11 (Figures 5A,B). Hyphae samples cultured in the medium with pH 8 were collected to detect the expression of genes related to pH stress. The results showed that the expression of genes related to pH stress in ΔAchog1 was lower than that of in WT (Figure 5C); indicating that Achog1 is mainly involved in alkali stress.
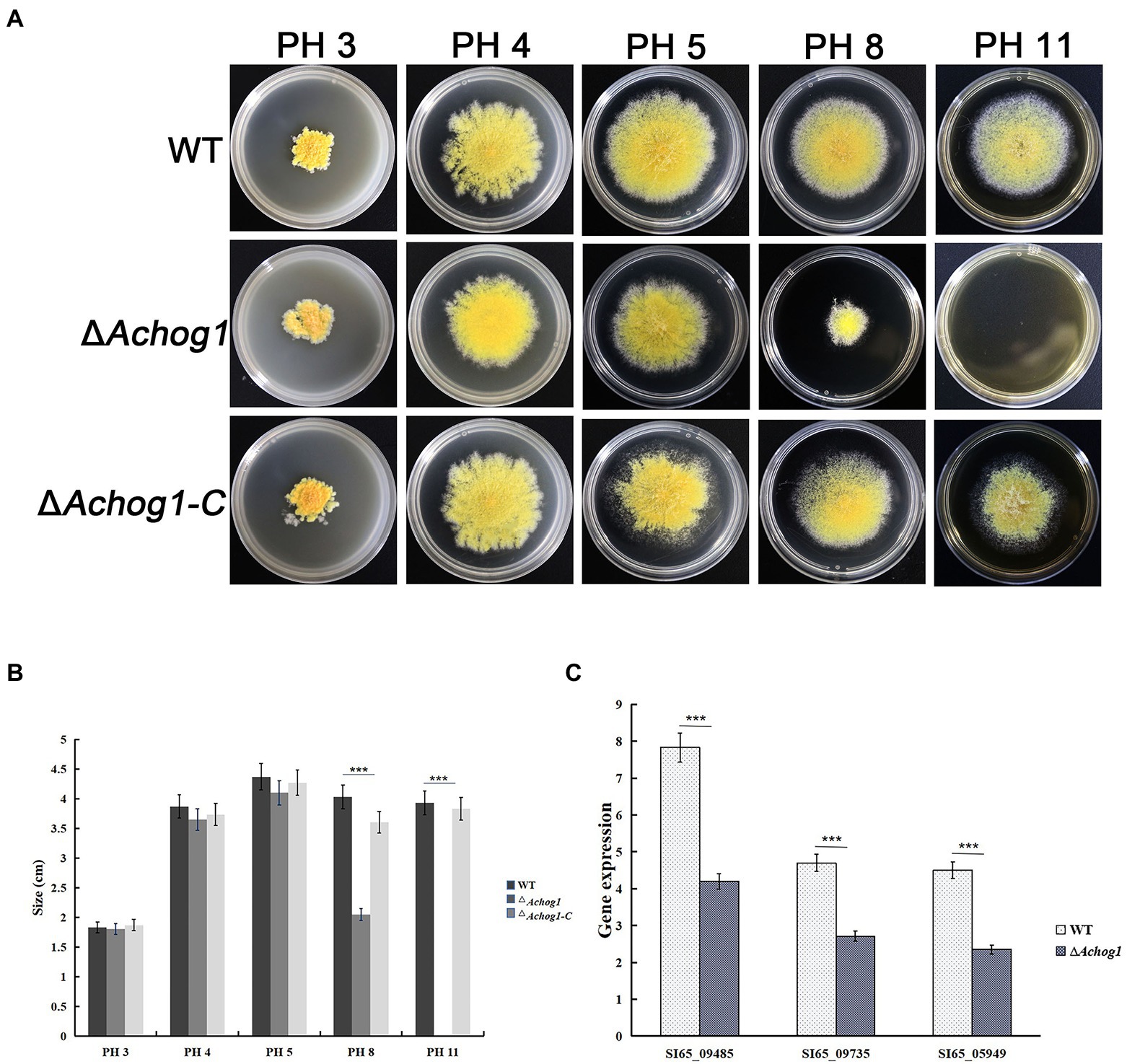
Figure 5. Effect of AChog1 on the pH stress response. (A) Colony morphology of the WT, ΔAchog1 and ΔAchog1-C strains on MYA containing 5% NaCl with different pH values at 28°C for 6 days. (B) Colony diameter. (C) Expression of genes related to the response of pH stress. Error bars indicate the standard deviation of at least three replicates. Values that differed significantly from the value for the WT strain according to a t test are indicated with asterisks (***p < 0.001).
The colonies of the WT strain were brown-black in the center and yellow at the edge on MYA media. However, the colonies of the ΔAchog1 strain were brown, and their edges were pale yellow. The pigments produced by the ΔAchog1-C strains were similar to those produced by the WT (Figure 6A). Genes involved in pigment synthesis, such as SI65_05588 (ayg1), SI65_05589 (arp2), SI65_05591 (abr1) and SI65_05592 (abr2), were downregulated according to RNA-seq data (Figure 6B). Furthermore, we detected the expression of four genes related to pigment synthesis. The results showed that the expression of these genes in ΔAchog1 strain was lower than that of in WT (Figure 6C). These results suggested that the Achog1 gene played a role in regulating pigment synthesis in A. cristatus.
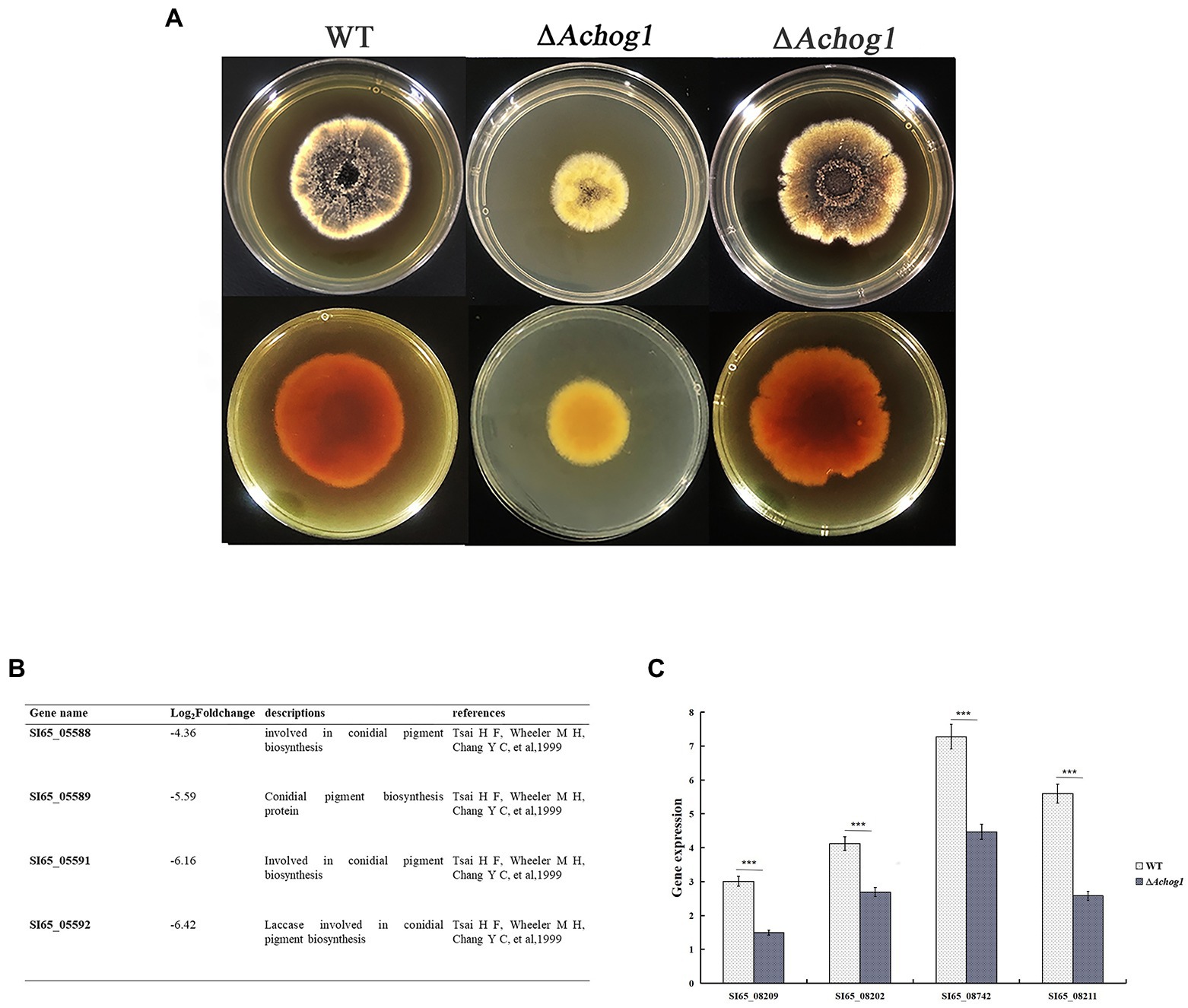
Figure 6. Pigmentation of the WT, ΔAchog1, ΔAchog1-C strains. (A) Pigmentation of the WT, ΔAchog1, and ΔAchog1-C strains on MYA medium at 28°C for 7 days. (B) Genes related to pigmentation according to the results of RNA-seq. (C) Expression of genes related to pigment synthesis.
A pull-down mass spectrometry experiment was performed to determine the proteins in WT-D and WT-H (Figure 7A). The total ion chromatogram and evaluation chart of the identified proteins showed that the mass spectrometry data were normal, indicating that the identified proteins were accurate (Supplementary Figure S4). A total of 940 proteins and 2,980 peptides were identified (Supplementary Tables S8, S9). A total of 530 proteins were upregulated, and 291 proteins were downregulated (proteins with a fold change (FC) ≥ 2 were upregulated, and those with an FC ≤ 1/2 were downregulated; Figure 7B). Comparing WT-H with WT-D, 483 were present only in WT-H, and 251 were present only in WT-D, demonstrating that there were significant differences in the proteins interacting with AcHog1 between the normal stress and hypertonic stress conditions.
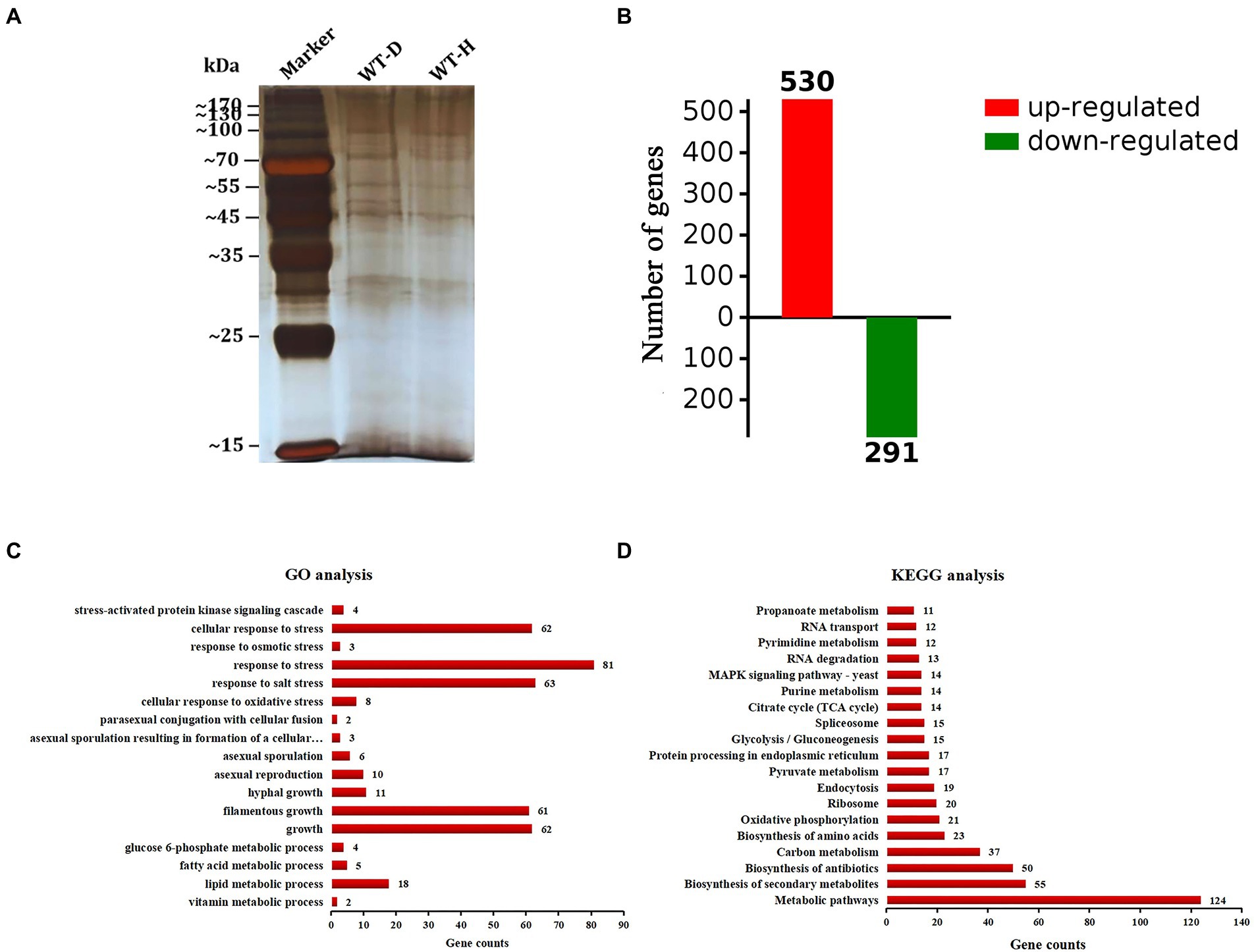
Figure 7. Analysis of differentially expressed proteins. (A) Proteins were pulled down in WT-D and WT-H. (B) Differential proteins (FC ≥ 2 were considered up-regulatregulated, and FC ≤ 0.5 proteins were considered down-regulreglated). (C) GO enrichment analysis. (D) KEGG enrichment analysis. The X-axis shows the number of proteins. The Y-axis shows the pathway names.
The top 20 proteins with the highest scores are listed in Supplementary Table S3. Among them, 5 proteins are uncharacterized, and the remaining 15 proteins are known functions. For example, the protein A0A1E3B957 is 6-phosphogluconate dehydrogenase. It has been reported that 6-phosphogluconate dehydrogenase is mainly involved in the salt stress response in plants (Nemoto and Sasakuma, 2000; Huang et al., 2003). AcHog1 interacted with 6-phosphogluconate dehydrogenase in samples incubated in MYA containing 3 M NaCl, suggesting that AcHog1 could regulate 6-phosphogluconate dehydrogenase to participate in the salt stress response in A. cristatus. RodA/RodB-related conidia (Pedersen et al., 2011), Ste20 and Cla4 located upstream of the HOG1 pathway were also found in WT-H (Dan et al., 2001; Joshua and Höfken, 2019). The data proved AcHog1 could interact with 6-phosphogluconate dehydrogenase and might be involved in the salt stress response. In addition, Achog1 could interact with RodA/RodB, Ste20 and Cla4 and Achog1 might be also involved in the production of conidia and the response of HOG pathway.
GO enrichment analysis showed that the differentially expressed proteins were mainly involved in various metabolic processes, asexual development processes and stress responses (Figure 7C). Nine proteins (Supplementary Table S4) related to asexual sporulation and stress responses were randomly selected for yeast two-hybrid validation. PGBKT7-AcHog1 and PGADT7 plasmids containing 9 proteins were co-transformed into yeast AH109 receptive cells. After 5 days of culturing in SD/−Leu/−Trp, a single colony was cultured on SD/−Trp/−Leu/-His and SD/−Trp/−Leu/-His/−Ade medium containing X-α-Gal and AbA for 5 days. PGBKT7-53 + PGADT7-T was chosen as a positive control (Figure 8A), and PGBKT7-lam + PGADT7-T was a negative control (Figure 8G). Both the colonies in the positive control and in the experimental groups (Figures 8B–F,H–K) were blue, indicating that all the selected proteins could interact with AcHog1 (Figure 8). These results showed that AcHog1 might regulate asexual sporulation and the response to osmotic stress and oxidative stress in A. cristatus. KEGG enrichment analysis showed that 14 differentially expressed proteins were enriched in the MAPK pathway and were significantly upregulated (Figure 7D). The AA1E3BER4 (AcSko1) protein is located downstream of AcHog1 in the HOG pathway in WT-H, and its interaction with AcHog1 was verified by using yeast two-hybrid assays (Figures 8B,K). Therefore, we speculated that Sko1 was the target protein of AcHog1, which will be a focus of our future studies.
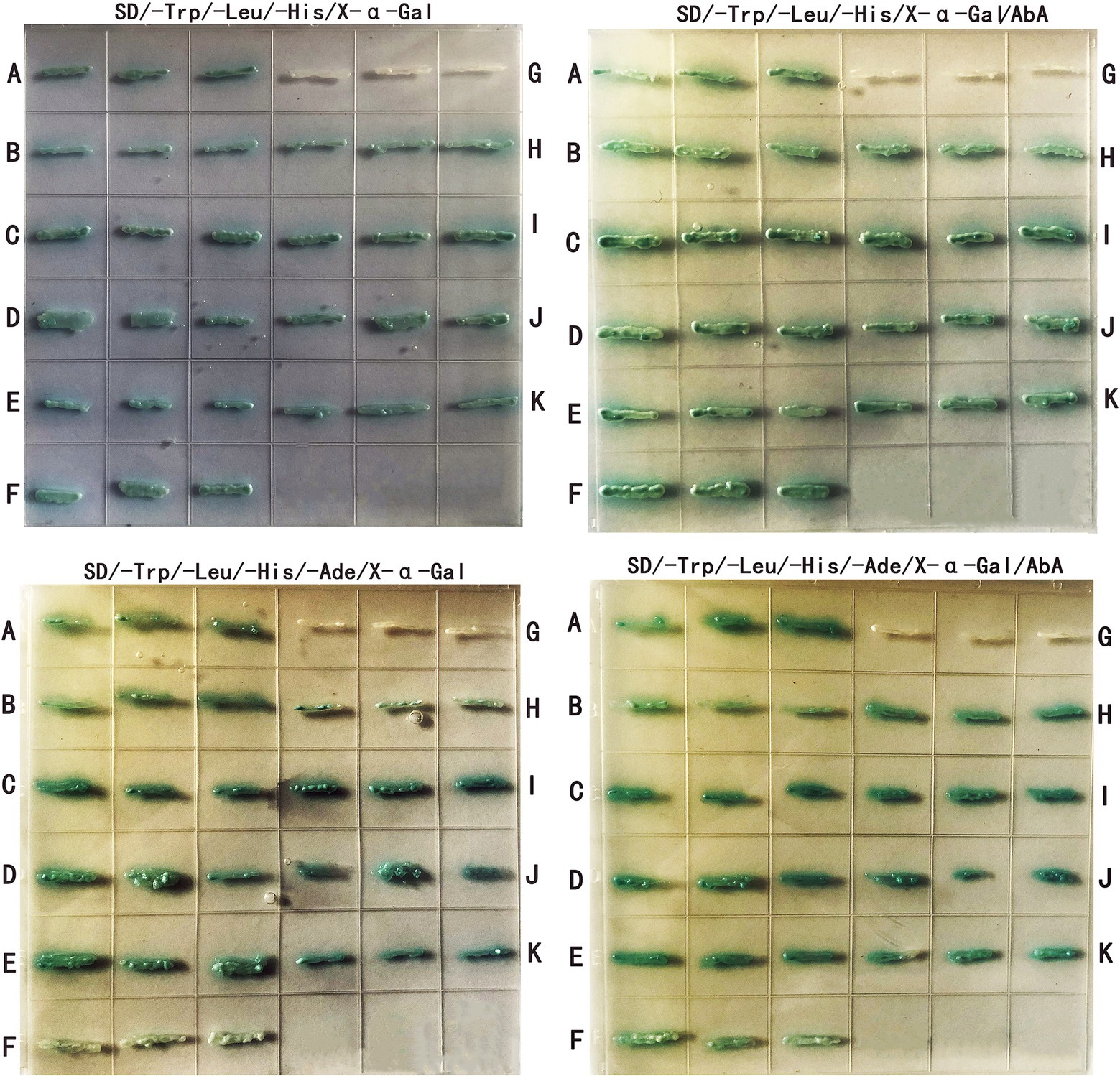
Figure 8. Differential protein yeast two-hybrid validation. (A–K) correspond to positive control (PGBKT7-53 + PGADT7-T), SI65_07477 (AcRodB), SI65_06242 (AcSte7), SI65_05510 (AcBmh1), SI65_09934 (AcCsp), SI65-06799 (AcPhnA), negative control (PGBKT7-lam + PGADT7-T), SI65_10255 (AcRodA), SI65_03085 (AcCla4), SI65_07597 (AcSte20), SI65_05898 (AcSko1), respectively.
To investigate the roles of Achog1 in A. cristatus, RNA-seq of ΔAchog1 and WT was performed at selected time points. Conidiophores and chain-like conidia appeared on the WT strain; however, only some vesicles were formed in ΔAchog1, when the ΔAchog1 and WT strains were cultivated on MYA media containing 3 M NaCl for 39 h (Figure 1A). Therefore, mycelial samples of the cultures at 39 h were collected for RNA-seq. All sequences obtained by sequencing were uploaded to the NCBI database: Sequence Read Archive,2 and get the accession number: PRJNA877470. A summary of the RNA-seq data is reported in Supplementary Table S5. In this study, a total of 1,074 differentially expressed genes (Supplementary Table S10) were obtained from the Achog1 transcriptome data (|log2(FC)| > = 1 and padj < = 0.05). Among these differentially expressed genes, 480 were upregulated, and 594 were downregulated. The top 20 genes upregulated and downregulated in ΔAchog1 are listed in Supplementary Tables S6, S7, respectively. Among them, SI65_00200 (phiA) is a cell wall protein (Schachtschabel et al., 2012); SI65_01919 encodes a low-affinity glucose transporter, and a study showed that low-affinity glucose required the coordinated activities of the HOG and glucose signaling pathways in S. cerevisiae (Tomás-Cobos et al., 2004). The protein kinases Dsk1 (SI65_03917) contributed to cell cycle and pre-mRNA splicing (Tang et al., 2012). In addition, genes responding to the HOG pathway and related to sporulation were screened from the RNA-seq data by referring to the relevant literature reports and the genome database of Aspergillus. The results were shown in Table 1. It was noted that genes related to asexual development were downregulated, such as brlA (SI65_02778) and wetA (SI65_00383) (Tsai et al., 1999), were involved in asexual sporulation and downregulated in the ΔAchog1 mutant. We speculated that Achog1 could affect the conidia production of A. cristatus by down-regulating the expression of these genes.
GO enrichment analysis showed that the differentially expressed genes were mainly enriched in carbohydrate metabolism, transmembrane transport, hydrolase activity, binding and other processes. Carbohydrate metabolic process (GO: 0005975), hydrolase activity (GO: 0016798, GO: 0016798) and oxidoreductase activity (GO: 0016614) were significantly upregulated. Transmembrane transport (GO: 0055085) was significantly downregulated (Figure 9A). The differentially expressed genes were mainly enriched in various metabolic pathways and MAPK pathways by KEGG enrichment analysis. Galactose metabolism (ani00052) and amino sugar and nucleotide sugar metabolism (ani00520) were significantly upregulated, glycerophospholipid metabolism (ani00564) and glyoxylate and dicarboxylate metabolism (ani00564) were significantly downregulated (Figure 9B). In addition, all the genes enriched in the MAPK pathway were downregulated. We speculated that Achog1 also played an important role in hydrolase activity, various metabolic processes and MAPK pathways in A. cristatus.
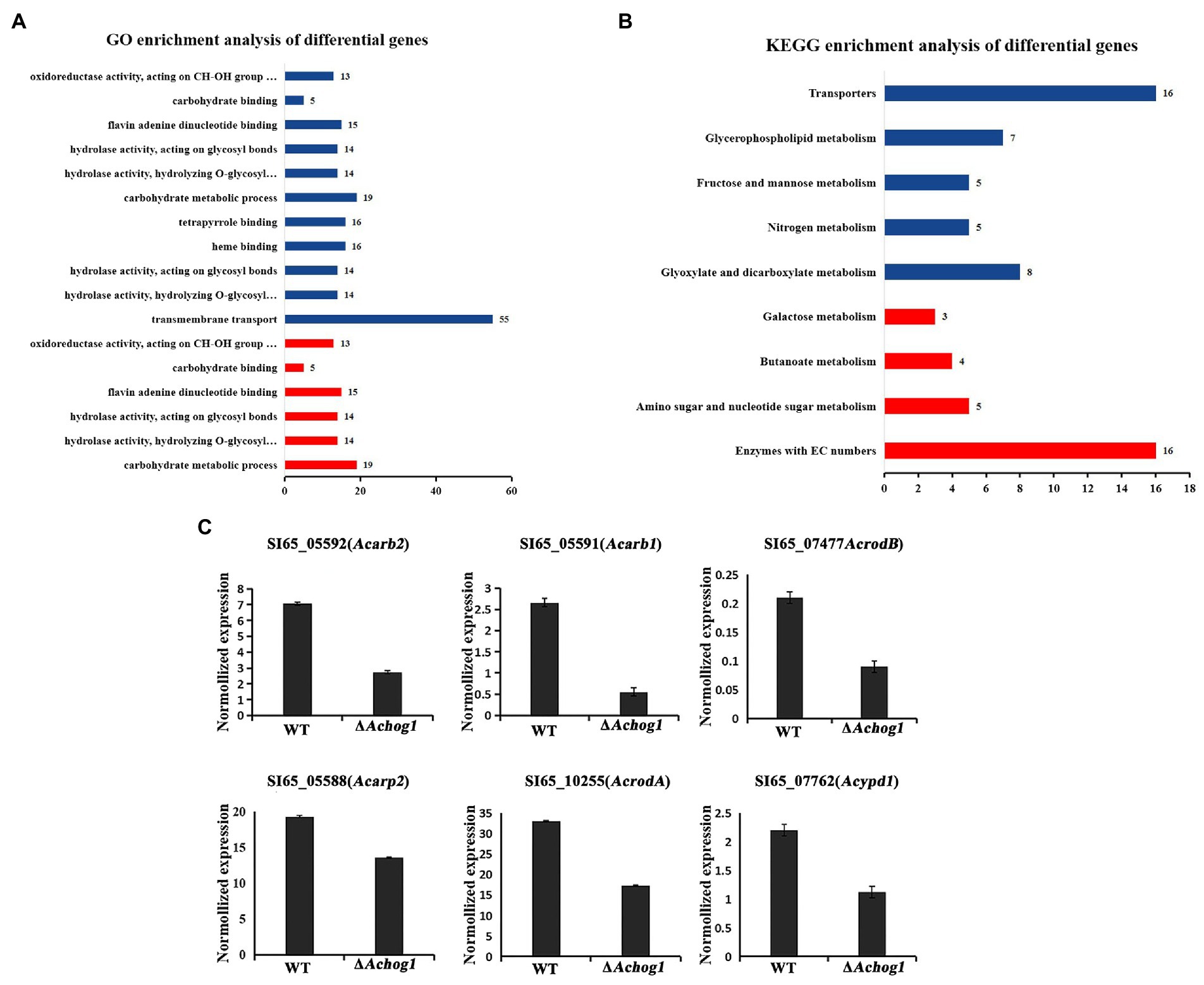
Figure 9. Transcriptional analysis of ΔAchog1 and WT strains. (A) GO analysis of differentially significant enrichment. (B) KEGG analysis of differentially significant enrichment. The X-axis shows the number of differentially expressed genes. The Y-axis shows the pathway names. Red indicates significant upregulation; blue indicates significant downregulation. (C) RT-qPCR verification of selected genes.
To verify the reliability of the expression changes in the transcriptome, 6 genes were selected for RT-qPCR detection (Figure 9C). The gene expression patterns of the gene analysis were similar in the RT-qPCR and RNA-seq, indicating that the transcriptome sequencing results obtained in this study were reliable.
Aspergillus cristatus is a homothallic filamentous fungus that produces sexual or asexual spores under different osmotic conditions. However, the mechanism that controls reproduction in spores is currently unknown. We speculated that osmotic stress was involved in regulating this process based on the fact that A. cristatus can only produce ascospores in medium containing 1 M NaCl, while it can only produce conidia in medium containing 3 M NaCl. It is well known that the HOG pathway is one of the main pathways in response to osmotic stress (Gustin et al., 1998; Estruch, 2000; Welsh, 2000; Sean et al., 2004). The key gene of the HOG pathway is hog1, which belongs to PKc kinase. A PKc-like gene, SI65_07698, was found in the A. cristatus genome database by sequence alignment. The conserved domain of the gene was consistent with that of hog1 in S. cerevisiae, so it was named Achog1. In this study, we constructed the ΔAchog1 and ΔAchog1-C strains to further investigate the function of Achog1. The results indicate that Achog1 plays important roles in asexual development, osmotic stress, oxidative stress, PH stress and pigmentation in A. cristatus. Moreover, we roughly describe the regulatory pathway (Figure 10) of Achog1 in A. cristatus combined with Pull-down mass spectrometry and RNA-Seq.
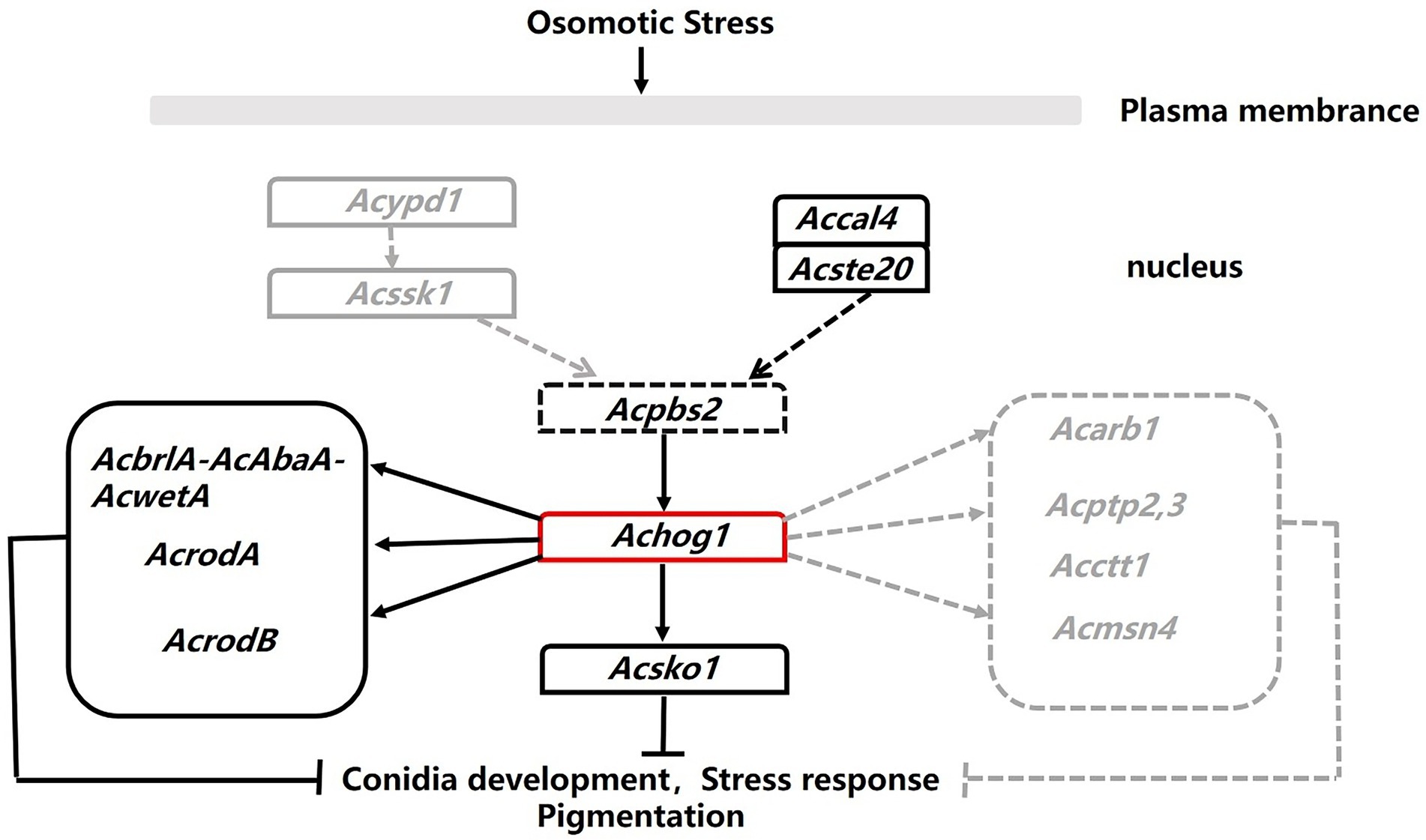
Figure 10. Regulation map of Achog1. The black solid lines represented genes from pull-down assays. The gray dotted lines represented genes from the RNA-seq.
PKc kinase plays an important role in the growth, development and conidial formation of filamentous fungi (Dickman and Yarden, 1999). When osm1, a functional homolog of the yeast hog1, was deleted, the number of conidia increased exponentially in mutants under hypertonic conditions (Xu, 2000); In A. nidulans, the sakA mutation showed no apparent effect on asexual development (Kawasaki et al., 2002). In contrast to these studies, ΔAchog1 produced 2 times fewer conidia under hypertonic stress compared with the WT in our study. The deletion of hog1 homologs affects the germination of spores in A. fumigatus (Xue et al., 2004; Du et al., 2006). And ΔsakA conidiospores lost viability more rapidly than those from the wild-type strain in A. nidulans (Kawasaki et al., 2002; Garrido-Bazán et al., 2018). Similarly, a large number of conidiophores were produced in WT strains in media with 3 M NaCl at 39 h, while ΔAchog1 could produce only vesicles, indicating that conidial germination was delayed. In addition, it was found that a total of 16 genes related to sporulation were significant difference in the expression according to the RNA-seq data. Among them, SI65_10255, SI65_02778, SI65_07477 and SI65_00383 were genes involved in the development of phialides and conidiophores, and their expression levels were significantly down-regulated in ΔAchog1 strains. Four proteins (A0A1E3B4I1, A0A1E3B9P1, A0A1E3BK58 and A0A1E3BGW2) enriched in the glucose 6-phosphate metabolic process (GO: 0051156) were identified in LC–MS/MS differential protein analysis. The glucose 6-phosphate metabolic process is known to be a key branch of the HMP pathway, which is important in conidial formation of A. nidulans (Broek, 1997). The HMP pathway is also very active during the conidial formation of Aspergillus niger (Ruijter and Visser, 1999). Because the samples selected for Pull-down experiment were A. cristatus cultured for 7 days under hypertonic stress. Under this culture condition, A. crsitatus could only produce a large number of conidia. We speculated that the HMP pathway existed in A. cristatus according to the LC/MS–MS data. And Achog1 might negatively regulate the four proteins to affect the formation of conidia. hogA plays an important role in the regulation of sexual development in A. nidulans. The progression of cleistothecia formation in ΔhogA strains was somewhat precocious (Kawasaki et al., 2002). However, the sexual development between WT and ΔAchog1 was not significant on hypotonic medium. Interestingly, hyphal curling appeared in ΔAchog1 at 7 days. ΔAchog1 strains produced cleistothecia on MYA medium containing 3 M NaCl after 14 days. This phenomenon was rarely observed in the Δhog1 strains of other fungi. These results suggested that Achog1 played a positive role in the asexual development of A. cristatus and that Achog1 may regulate the transition between sexual and asexual development, but the specific regulatory mechanism still needs to be further studied.
Osmotic stress is an important factor regulating the growth and development of filamentous fungi (Bahn, 2008; Duran et al., 2010). In this study, the Achog1 deletion mutant grew significantly slower than the wild type in medium containing a high concentration of NaCl (2 M/3 M), indicating that the deletion of Achog1 reduces the resistance of A. cristatus to hyperosmolarity. The same results were obtained when we replaced the osmotic pressure regulators with sucrose and sorbitol. We demonstrate that Achog1 plays a positive role in osmotic stress according to these results. However, the role of Achog1 in the osmotic response of A. cristatus contrasts sharply with that of sakA in A. nidulans and hog1 in other fungi. In A. nidulans, sakA is even dispensable for osmotic stress resistance (Kawasaki et al., 2002; Garrido-Bazán et al., 2018). In other fungi, the resistance of hog1 to hyperosmolarity is much lower than that in A. cristatus. For example, 0.4 M NaCl can greatly inhibit colony growth, although it has been shown to be not completely lethal to deletion mutants of hog1 in M. oryzae (Montemurro, 1992). After hog1 is deleted from F. graminearum, the mutants cannot grow in media with 0.7 M NaCl (Morawetz et al., 1996). In Botrytis cinerea, hog1 mutant strains cannot grow on media containing 1.5 M NaCl (Morawetz et al., 1996). Although the functions of hog1-type MAPKs are relatively conserved, they might play diverse roles in different fungi. In addition, the Achog1 deletion mutant exhibited a significant decrease in conidiation compared with the WT strain in the presence of 3 M NaCl, indicating that Achog1 can promote conidia production under hyperosmotic stress.
hog1 responds not only to osmotic stress but also to oxidative stress (Alonso-Monge et al., 2003). An oxidative stress sensitivity test was performed on Achog1 deletion mutants. It was found that ΔAchog1 grew slowly in media containing different concentrations of H2O2, especially at high concentrations of H2O2. Our experiments indicate that Achog1 plays a positive role in the regulation of the oxidative stress response. This finding was similar to that observed in A. fumigatus and F. graminearum. In A. fumigatus, H2O2, as a mimic of oxidative stress, significantly inhibited the growth of hog1 mutant strains (Du et al., 2006). The growth of deletion mutants of each individual gene involved in the HOG pathway was suppressed under oxidative stress in F. graminearum (Montemurro, 1992). Moreover, the hyphae of ΔAchog1 were extremely irregular at the edge, suggesting that the polar growth of hyphae was affected under oxidative stress. In the hypertonic sample (WT-H), many proteins interacting with AcHog1 were enriched in the stress response pathway. Moreover, some differentially expressed genes related to the stress response were also identified among the ΔAchog1 transcriptome data. Sixty-five genes were enriched in oxidoreductase activity molecular function, and 30 genes were downregulated. We speculate that Achog1 can maintain the balance of intracellular and extracellular stress by downregulating the expression of these proteins and genes so that ΔAchog1 can grow normally under hypertonic stress and oxidative stress when high-concentration stress stimulates A. cristatus.
Achog1 is also involved in pH stress. In our study, both acid and alkali stress tests were carried out and found that ΔAchog1 grew slower in the media at pH 8 than WT. ΔAchog1 could not even grow in media at pH 11, which was similar to the results of a study in A. fumigatus (Ma et al., 2012). However, acid stress has not been studied in A. fumigatus. Studies showed that low pH-induced structural changes were dependent on hog1 when yeast cells were cultivated in media in which the pH was shifted from 5.5 to 3.5 in yeast (Bilsland et al., 2004; Lawrence et al., 2004; Sotelo and Rodriguez-Gabriel, 2006; Kapteyn et al., 2010). However, our acid stress test showed that ΔAchog1 could grow normally. These results showed that Achog1 mainly participates in the response to alkaline stress.
Pigment is a type of secondary metabolite produced by fungi during their growth and development. On MYA media, compared with the WT strain, the Achog1 deletion mutant produced less pigment. Fifty-seven genes were annotated to secondary metabolite synthesis according to the RNA-seq data. Among these genes, SI65_05588 (ayg1), SI65_05589 (arp2), SI65_05591 (abr1) and SI65_05592 (abr2) were involved in the regulation of pigment synthesis, and they all were downregulated in ΔAchog1, indicating that Achog1 participates in the regulation of secondary metabolism and may be involved in the regulation of pigment synthesis. hog1 plays an important role in regulating fungicide resistance. Os-2 is a gene with high homology to hog1 in N. ceassa. The deletion mutant of Os-2 was found to be resistant to N-moncootyl dicycloheptene dimethylphthalide and phenylpyrroliac fungicides (Kojima et al., 2004). However, A. cristatus is a beneficial fungus, we did not performed the fungicide sensitivity test.
The pull-down mass spectrometry and RNA-seq data were consistent with the phenotypic changes observed in the Achog1 deletion mutant. In this study, pull-down combined mass spectrometry under normal and hypertonic conditions was performed. The proteins interacting with AcHog1 were quite different under normal stress and hypertonic stress conditions. Proteins associated with conidial development could interact with AcHog1 under hypertonic stress. Among them, AcRodB and AcRodA were verified by yeast two-hybrid assays. We speculate that AcHog1 can regulate them to promote conidia production. We also found proteins Ste20 and Cla4 located upstream of Hog1 and protein Sko1 located downstream of Hog1, and yeast two-hybrid assays showed that all three proteins could interact with AcHog1. The interactions between AcHog1 and them are represented by using solid lines in Figure 10. RNA-seq results showed that the expression of genes related to conidial formation, such as SI65_02778 (AcbrlA), SI65_07477 (AcrodB), SI65_10255 (AcrodA) and other genes, was significantly reduced, indicating that Achog1 could regulate these genes to control the conidial development of A. cristatus. Consequently, Achog1 could respond to H2O2 stress by regulating the gene SI65_02778 (Acmsn4), which regulates the H2O2 stress response (Hasan et al., 2010). ypd1 and ssk1 were located upstream of hog1, and ptp2 and ptp3 were located downstream of hog1. When the Achog1 gene was deleted, the expression of ypd1, ssk1, and ptp2,3 was downregulated. We speculate that Achog1 can regulate the expression of these genes to affect conidia production and stress responses in A. cristatus, which requires further study. Therefore, it is represented by gray dotted lines in Figure 10. The results of GO enrichment analysis and KEGG analysis showed that both differentially expressed genes and differentially expressed proteins were significantly enriched in secondary metabolic pathways, and four genes were related to pigment synthesis, indicating that Achog1 also participates in secondary metabolism.
In conclusion, this report is the first to present a functional analysis of Achog1 encoding a Pkc-like superfamily in A. cristatus. Our data suggest that Achog1 plays a key role in positively regulating asexual sporulation, stress response, pH response and pigmentation. Pull-down combined mass spectrometry and RNA-seq data provide a foundation for future genetic and biochemical analyses, which will be required for regulatory mechanisms and links between the control of asexual development, stress sensing and secondary metabolism in A. cristatus.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
LS, YT, and ZL conceived and designed the experiments. LS and SS performed the experiments. YW and YL took part in the data analysis. LS drafted the manuscript. YT and ZL revised the manuscript. All authors read, corrected and approved the final manuscript.
This study was supported by the National Natural Science Funds of China (Nos. 31960018 and 31660021) and the Subsidy from NSFC of Guizhou Academy of Agricultural Sciences (Nos. 202104 and 202138).
We especially thank Chengshu Wang (Institute of Plant Physiology and Ecology, Shanghai Institute for Biology Science, Chinese Academy of Sciences) for providing plasmid pDHt/sk-hyg.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1003244/full#supplementary-material
Alkhayyat, F., Chang, K. S., and Yu, J. H. (2015). Genetic control of asexual development in Aspergillus fumigatus. Adv. Appl. Microbiol. 90, 93–107. doi: 10.1016/bs.aambs.2014.09.004
Alonso-Monge, R., Navarro-García, F., Román, E., Negredo, A. I., Eisman, B., Nombela, C., et al. (2003). The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2, 351–361. doi: 10.1128/EC.2.2.351-361.2003
Bahn, Y. S. (2008). Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot. Cell 7, 2017–2036. doi: 10.1128/EC.00323-08
Bayram, Ö., Min, N., Jin, W. B., Kerstinm, H., Oliver, V., Susanna, B. S., et al. (2008). velB/veA/laeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Bilsland, E., Molin, C., Swaminathan, S., Ramne, A., and Sunnerhagen, P. (2004). Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53, 1743–1756. doi: 10.1111/j.1365-2958.2004.04238.x
Broek, P. (1997). The Glucose-6-phosph.Ate dehydrogenase encoding genes from Aspergillus niger and Aspergillus nidulans. [Doctoral dissertation]. ed V. den Broker (SL: Van den Broker).
Calmes, B., N’Guyen, G., Dumur, J., Brisach, C. A., Campion, C., Iacomi, B., et al. (2015). Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery. Front. Plant Sci. 06:414. doi: 10.3389/fpls.2015.00414
Dan, I., Watanabe, N. M., and Kusumi, A. (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230. doi: 10.1016/S0962-8924(01)01980-8
Delgado-Jarana, J., Sousa, S., Gonzalez, F., Rey, M., and Llobell, A. (2006). Thhog1 controls the hyperosmotic stress response in Trichoderma harzianum. Microbiology 152, 1687–1700. doi: 10.1099/mic.0.28729-0
Deng, F. M., Gong, S. L., and Yang, W. L. (2007). High-throughput screening test for extracellular polysaccharide bioactivity of Xanthacoccus coronary. Food Machin. 23, 48–51. doi: 10.3969/j.issn.1003-5788.2007.06.013
Dickman, M. B., and Yarden, O. (1999). Serine/threonine protein kinases and phosphatases in filamentious fungi. Fungal Genet. Biol. 26, 99–117. doi: 10.1006/fgbi.1999.1118
Ding, T. (2012). Biological characteristics and digestive enzyme-producing activity of "Golden flower fungus" in Fuzhuan tea. [Doctoral dissertation]. Shanxi: Shanxi University of Science and Technology.
Dixon, K. P., Xu, J. R., Smirnoff, N., and Talbot, N. J. (1999). Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058. doi: 10.1105/tpc.11.10.2045
Du, C., Sarfati, J., Latge, J. P., and Calderone, R. (2006). The role of the sakA (hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med. Mycol. 44, 211–218. doi: 10.1080/13693780500338886
Duran, R., Cary, J. W., and Calvo, A. M. (2010). Role of the osmotic stress regulatory pathway in morphogenesis and secondary metabolism in filamentous fungi. Toxins 2, 367–381. doi: 10.3390/toxins2040367
Estruch, F. (2000). Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24, 469–486. doi: 10.1016/S0168-6445(00)00035-8
Garrido-Bazán, V., Jaimes-Arroyo, R., Sánchez, O., Lara-Rojas, F., and Aguirre, J. (2018). sakA and mpkC stress MAPKs show opposite and common functions during stress responses and development in Aspergillus nidulans. Front. Microbiol. 9:2518. doi: 10.3389/fmicb.2018.02518
Ge, Y. Y., Wang, Y. C., Liu, Y. X., Tan, Y. M., Ren, X. X., Zhang, X. Y., et al. (2016). Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus Aspergillus cristatus. BMC Genomics 17:428. doi: 10.1186/s12864-016-2637-y
Gilbert, M. K., Mack, B. M., Wei, Q., Bland, J. M., Bhatnagar, D., and Cary, J. W. (2016). RNA sequencing of an nsdC mutant reveals global regulation of secondary metabolic gene clusters in Aspergillus flavus. Microbiol. Res. 182, 150–161. doi: 10.1016/j.micres.2015.08.007
Gustin, M. C., Alberty, N. J., Alexander, M., and Davenport, K. (1998). MAP kinase pathway in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300. doi: 10.1128/MMBR.62.4.1264-1300.1998
Han, S., and Adams, T. H. (2001). Complex control of the developmental regulatory locus brlA in aspergillus nidulans. Mol. Gen. Genomics. 266, 260–270. doi: 10.1007/s004380100552
Hasan, R., Leroy, C., Isnard, A. D., La Ba Rre, J., and Toledano, M. B. (2010). The control of the yeast H2O2 response by the msn2/4 transcription factors. Mol. Microbiol. 45, 233–241. doi: 10.1046/j.1365-2958.2002.03011.x
Heller, J., Ruhnke, N., Espino, J. J., Massaroli, M., Collado, I. G., and Tudzynski, P. (2012). The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant-Microbe Interact. 25, 802–816. doi: 10.1094/MPMI-11-11-0299
Huang, J., Zhang, H., Wang, J., and Yang, J. (2003). Molecular cloning and characterization of rice 6-phosphogluconate dehydrogenase gene that is up-regulated by salt stress. Mol. Biol. Rep. 30, 223–227. doi: 10.1023/a:1026392422995
Joshua, I. M., and Höfken, T. (2019). Ste20 and Cla4 modulate the expression of the glycerol biosynthesis enzyme gpd1 by a novel MAPK-independent pathway. Biochem. Biophys. Res. Commun. 517, 611–616. doi: 10.1016/j.bbrc.2019.07.072
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kapteyn, J. C., Riet, B. T., Vink, E., Blad, S., and Klis, F. M. (2010). Low external pH induces hog1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39, 469–479. doi: 10.1046/j.1365-2958.2001.02242.x
Kawasaki, L., Sanchez, O., Shiozaki, K., and Aguirre, J. (2002). SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45, 1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x
Kojima, K., Takano, Y., Yoshimi, A., Tanaka, C., Kikuchi, T., and Tetsuro, O. (2004). Fungicide activity through activation of a fungal signialing pathway. Mol. Microbiol. 53, 1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x
Krantz, M., Becit, E., and Hohmann, S. (2006). Comparative genomic of the HOG-signaling system in fungi. Curr. Genet. 49, 137–151. doi: 10.1007/s00294-005-0038-x
Lawrence, C. L., Botting, C. H., Antrobus, R., and Coote, P. J. (2004). Evidence of a new role for the high-osmolarity glycerol mitogen-activated protein kinase pathway in yeast: regulating adaptation to citric acid stress. Mol. Cell. Biol. 24, 3307–3323. doi: 10.1128/MCB.24.8.3307-3323.2004
Li, G. L., Hu, B. H., Zhao, Y. B., Liu, S. C., Jiang, Y. J., and Liu, Z. H. (2011). Bacteriostatic effect of fermentation broth of fungus coronus. Food Sci. 11, 165–168. doi: 10.CNKI:SUN:SPKX.0.2011-11-035
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–521. doi: 10.1186/s13059-014-0550-8
Ma, D. M., Ji, Y. J., and Yang, F. (2012). Effects of extra copies of Aspergillus fumigatus sho1 and pbs2 genes on several stress capacities. Chin. J. Microbiol. 7, 192–198. doi: 10.3969/j.issn.1673-3827.2012.04.001
Mo, H. Z., Zhu, Y., and Chen, Z. M. (2008). Microbial fermented tea-a potential source of natural food preservatives. Trends Food Sci. Technol. 19, 124–130. doi: 10.1016/j.tifs.2007.10.001
Morawetz, R., Lendenfeld, T., Mischak, H. M., Mühlbauer, F., Gruber, G., and Goodnight, J. (1996). Cloning and characterization of genes (pkc1 and pkcA) encoding protein kinase C homologs from Trichoderma reesei and Aspergillus niger. Mol. Gen. Genet. 250, 17–28. doi: 10.1007/BF02191821
Nemoto, Y., and Sasakuma, T. (2000). Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L). Plant Sci. 158, 53–60. doi: 10.1016/S0168-9452(00)00305-8
Ouyang, M., Xiong, C. Y., Tu, Y. Y., Cheng, L., Shu, H., and Tang, W. (2011). Effects of fungus coronus on tea quality composition and antioxidant activity. Mycosystema 30, 343–348.
Park, S. M., Choi, E. S., Kim, M. J., Cha, B. J., Yang, M. S., and Kim, D. H. (2004). Characterization of hog1 homolog, Cpmk1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277. doi: 10.1111/j.1365-2958.2004.03919.x
Parkhomchuk, D., Borodina, T., Amstislavskiy, V., Banaru, M., Hallen, L., Krobitsch, S., et al. (2009). Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37:e123. doi: 10.1093/nar/gkp596
Pedersen, M. H., Borodina, I., Frisvad, J. C., and Søndergaard, I. (2011). Fed-batch production of hydrophobin RodB from Aspergillus fumigatus in host Pichia pastoris. Abstract from Biofilms in Nosocomial Fungal Infections: ESCMID Postgraduate Education Course, Paris, France.
Rodríguez-Pupo, E. C., Pérez-Llano, Y., Tinoco-Valencia, J. R., Sánchez, N. S., Padilla-Garfias, F., Calahorra, M., et al. (2021). Osmolyte signatures for the protection of Aspergillus sydowii cells under halophilic conditions and osmotic shock. J Fungi. 7:414. doi: 10.3390/jof7060414
Ruijter, G., and Visser, J. (1999). Characterization of Aspergillus niger phosphoglucose isomerase use for quantitative determination of erythrose 4-phosphate. Biochimie 81, 267–272. doi: 10.1016/S0300-9084(99)80061-3
Schachtschabel, D., Arentshorst, M., Lagendijk, E. L., and Ram, A. (2012). Vacuolar H+-ATPase plays a key role in cell wall biosynthesis of Aspergillus niger. Fungal Genet. Biol. 49, 284–293. doi: 10.1016/j.fgb.2011.12.008
Sean, M., Rourke, O., and Ira, H. (2004). Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15, 532–542. doi: 10.1091/mbc.E03-07-0521
Segmüller, N., Ellendorf, U., Tudzynski, B., and Tudzynski, P. (2007). Bcsak1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 6, 211–221. doi: 10.1128/EC.00153-06
Song, P., Cai, C., Skokut, M., Kosegi, B., and Petolino, J. (2002). Quantitative real-time PCR as a screening tool for estimating transgene copy number in WHISKERS™-derived transgenic maize. Plant Cell Rep. 20, 948–954. doi: 10.1007/s00299-001-0432-x
Sotelo, J., and Rodriguez-Gabriel, M. A. (2006). Mitogenactivated protein kinase hog1 is essential for the response to arsenite in Saccharomyces cerevisiae. Eukaryot. Cell 5, 1826–1830. doi: 10.1128/EC.00225-06
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). Mega6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, Y., Wang, Y., Ge, Y., Ren, X., Wang, Y., and Liu, Z. Y. (2017). Isolation and molecular identification of Aspergillus cristatus in fermented “fuzhuan” brick tea from Guizhou Province. Mycosystema 36, 154–163. doi: 10.13346/j.mycosystema.160125
Tan, Y., Wang, H., Wang, Y., Ge, Y., Ren, X., Ren, C., et al. (2018). The role of the veA gene in adjusting developmental balance and environmental stress response in Aspergillus cristatus. Fungal Biol. 122, 952–964. doi: 10.1016/j.funbio.2018.05.010
Tang, Z., Luca, M., Taggart-Murphy, L., Portillio, J., Chang, C., Guven, A., et al. (2012). Interacting factors and cellular localization of SR protein-specific kinase Dsk1. Exp. Cell Res. 318, 2071–2084. doi: 10.1016/j.yexcr.2012.05.020
Tao, L., and Yu, J. H. (2011). abaA and wetA govern distinct stages of Aspergillus fumigatus development. Microbiology 157, 313–326. doi: 10.1099/mic.0.044271-0
Thomas, E., Roman, E., Claypool, S., Manzoor, N., Pla, J., and Panwar, S. L. (2013). Mitochondria influence cdr1 efflux pump activity, hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob. Agents Chemother. 57, 5580–5599. doi: 10.1128/AAC.00889-13
Tomás-Cobos, L., Casadomé, L., Mas, G., Sanz, P., and Posas, F. (2004). Expression of the hxt1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signaling pathways. J. Biol. Chem. 279, 22010–22019. doi: 10.1074/jbc.M400609200
Tsai, H. F., Wheeler, M. H., and Chang, Y. C. (1999). A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181, 6469–6477. doi: 10.1128/JB.181.20.6469-6477.1999
Twumasi, B. K., Yu, Y. D., Gravelat, F. N., and Nierman, W. C. (2009). Transcriptional profiling identifies a role for brlA in the response to nitrogen depletion and for stuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8, 104–115. doi: 10.1016/j.vetpar.2004.10.004
Wang, Y. C., Tan, Y. M., Liu, Z. Y., and Liu, Y. X. (2014). Bioinformatics analysis of genes related to sexual sporulation in Eurotium cristatum. Guizhou Agric. Sci. 32–36. doi: 10.3969/j.issn.1001-3601.2014.10.009
Wang, Y., Tan, Y., Wang, Y., Ge, Y., Liu, Y., Liu, H., et al. (2021). Role of AcndtA in cleistothecium formation, osmotic stress response, pigmentation and carbon metabolism of Aspergillus cristatus. Fungal Biol. 125, 749–763. doi: 10.1016/j.funbio.2021.04.009
Welsh, D. T. (2000). Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol. Rev. 24, 263–290. doi: 10.1016/S0168-6445(99)00038-8
Wurgler-Murphy, S. M., Maeda, T., Witten, E. A., and Saito, H. (1997). Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the Ptp2 and Ptp3 protein tyrosine phosphatases. Mol. Cell. Biol. 17, 1289–1297. doi: 10.1128/MCB.17.3.1289
Xiang, T., Yu, F. M., and Yang, Z. J. (2020). Functional analysis of Aspergillus cristatus preA gene. Genomics Appl. Biol. 39:9. doi: 10.13417/j.gab.039.003070
Xu, J. R. (2000). MAP Kinase in Fungal Pathogens Fungal. Genomics Appl. Biol. 31, 137–152. doi: 10.1006/fgbi.2000.1237
Xue, T., Nguyen, C. K., Romans, A., and May, G. S. (2004). Amitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryot. Cell 3, 557–560. doi: 10.1128/EC.3.2.557-560.2004
Yang, F. L. (2005). Study on the liquid fermentation process of Eurotium cristatum and its effect on the activity of digestive enzymes. [Doctoral dissertation]. Guangzhou: Hunan Agricultural University.
Keywords: pull down, LC–MS/MS, asexual sporulation, stress response, RNA-seq
Citation: Shao L, Tan Y, Song S, Wang Y, Liu Y, Huang Y, Ren X and Liu Z (2022) Achog1 is required for the asexual sporulation, stress responses and pigmentation of Aspergillus cristatus. Front. Microbiol. 13:1003244. doi: 10.3389/fmicb.2022.1003244
Received: 26 July 2022; Accepted: 03 November 2022;
Published: 25 November 2022.
Edited by:
Amin Uddin Mridha, University of Chittagong, BangladeshReviewed by:
Kang Zhang, Hebei Agricultural University, ChinaCopyright © 2022 Shao, Tan, Song, Wang, Liu, Huang, Ren and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumei Tan, NTQwNzY0MDM5QHFxLmNvbQ==; Zuoyi Liu, Z3psaXV6dW95aUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.