
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 November 2022
Sec. Phage Biology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1001237
Stenotrophomonas maltophilia (S. maltophilia) is widely distributed in nature and frequently causes nosocomial infections. In this work, the biological characteristics and genome of a new S. maltophilia phage BUCT609 isolated from hospital sewage with S. maltophilia strain No. 3015 as host was analyzed and its therapeutic effect in vivo was explored. It was observed by TEM that phage BUCT609 belongs to the Podoviridae with a 10 nm tail structure and a capsid with a diameter of about 50 nm. It has a short latent period (about 10 min) and its burst size is 382 PFU /cell when multiplicity of infection (MOI) is 0.01. Furthermore, it has a high survival rate in the environment with a pH range from 3 to 10 and temperature range from 4°C to 55°C. The complete genome of phage BUCT609 is linear double-stranded DNA of 43,145 bp in length, and the GC content is 58%. The genome sequence of phage BUCT609 shares <45% homology with other phages. No virulence genes and antibiotic resistance genes were found in bacteriophage BUCT609. In vivo animal experiments showed that the survival rate of mice infected with S. maltophilia was significantly improved after the intranasal injection of phage BUCT609. Therefore, our study supports that phage BUCT609 could be used as a promising antimicrobial candidate for treating S. maltophilia infections.
Stenotrophomonas maltophilia is Gram-negative bacteria that widely exists in nature and also resides in the human respiratory and intestinal tract. The isolation rate of S. maltophilia was only less than Acinetobacter and Pseudomonas aeruginosa among non-fermented glycogram negative bacilli (An and Berg, 2018). As a conditioned pathogen, S. maltophilia is a major pathogenic bacterium for iatrogenic infection that can cause many diseases such as infections of the respiratory tract, urinary tract, and wounds. In recent years, with the extensive use of broad-spectrum antibiotics, S. maltophilia has become one of the most important pathogens of nosocomial infection, which brings great difficulties to clinical treatment (Gokhan Gozel et al., 2015). Phages are specific for infection and lysis of host bacteria in the environment and organisms. During the recent years, many countries have made exciting achievements in phage therapy (Bao et al., 2020) which demonstrate the unique advantages of phage therapy and greatly promote further research of phage therapy, indicating that phage therapy has a very bright application prospect (Moelling et al., 2018).
In this study, phage BUCT609 was isolated from hospital sewage. The biological and genomic characteristics of phage BUCT609 were analyzed to evaluate the infectivity and animal experiments were conducted to explore its therapeutic effect on S. maltophilia in vivo. This study will increase the diversity of S. maltophilia phage and provide a potential candidate for phage-based therapy.
S. maltophilia strain No. 3015 in our laboratory bacteria bank was used as the host bacterium of phage BUCT609, which is a clinical strain isolated from Shanghai Public Health Clinical Center. The bacteria were cultured 5 h to reach the exponential phase in Luria-Bertani (LB) medium at 37°C.
Phage BUCT609 was isolated from sewage in the China-Japanese Friendship Hospital of Beijing. Ten milliliters of sewage water were centrifuged at 12,000 × g for 5 min and filtered by membrane filtration with 0.22 μm pore diameter (Ding et al., 2020). The host bacteria were cultured to the exponential phase (OD600 = 0.7) at 37°C for phage infection. Then the filtered phage (100 μl) was mixed evenly with 500 μl of the host bacteria. The mixture was added into 5 ml LB liquid medium and incubated at 37°C for 5 h. Phage was diluted by phosphate-buffered saline (PBS) buffer and verified by the standard double-layer agar method. Eventually, a single plaque was picked up and amplified and purified three times (Chang et al., 2005).
To obtain a high titer of phage solution, 8 ml of 30% sucrose solution was slowly added to phage solution (32 ml). After centrifugation at 30,000 × g for 2 h at 4°C, the supernatant was discarded. The phage was re-suspended with 200 μl of PBS buffer.
The morphological characteristics of phage BUCT609 were observed by transmission electron microscopy (TEM). As reported previously, 10 μl of purified phages were loaded onto a copper grid for 20 min and the phage particles were negatively stained with 10 μl of 2% (w/v) phosphotungstic acid (PTA) for 10 min. The samples were dried for 5 h at room temperature. Morphology of the phage was observed by a transmission electron microscope (JEM-1200EX, Japan) at 80 kV (Li et al., 2019).
MOI refers to the ratio of the number of phages (PFU) to the host bacteria (CFU) and the optimal MOI is the multiplicity of infection when the phage can achieve the optimal growth state (Xing et al., 2017). Five hundred microliters of each purified phage and the host bacteria were mixed (MOI = 100, 10, 1, 0.1, and 0.01) evenly and incubated for 20 min. Then the mixture was added into fresh sterile LB liquid medium for 5 h at 37°C. After incubating, the mixture was centrifugated (12,000 × g, 5 min) and filtered to obtain the phage in the supernatant. The phage titer was determined by the standard double-layer agar method and the MOI with the highest titer is the optimal MOI.
To evaluate the infectivity of phage BUCT609 with the latent period and the burst size, it is necessary to study the one-step growth curve of the phage (Hagens and Loessner, 2010). The latent period is defined as the stage in which the phage proliferates in the bacterial cells. The burst size refers to the ratio of the ultimate titer of phage to the number of infected bacteria in the latent period. Phage BUCT609 was mixed with S. maltophilia strain No. 3015 (OD600 = 0.7) to the optimal MOI and allowed to adsorb at 37°C for 20 min. Then the mixture was centrifugated (12,000 × g, 1 min). The sediment containing phage was resuspended with 20 ml of LB liquid medium. The phage titer was determined by the soft agar overlay method every 10 min during the incubation at 37°C. Three replicates were performed and the results were averaged.
To test the thermal stability of phage BUCT609, the purified phage was incubated at different temperatures (4, 37, 45, 55, 65, or 75°C) for an hour. Then, the phage titer was calculated by the method of soft agar overlay (Fan et al., 2012). Similarly, to explore the effect of pH on phage BUCT609, the phage suspensions were incubated at various pH values ranging from 1 to 12 at 37°C for an hour and the phage titer was calculated. Each experiment was performed three times and the initial phage concentration was about 109 PFU/ml.
Multilocus sequence typing (MLST) is a bacterial typing method based on nucleic acid sequence determination (Larsen et al., 2012). In this experiment, multiple internal fragments of housekeeping genes were amplified by PCR and sequenced to analyze the variation of strains. Using housekeeping gene primers of S. maltophilia (atpD, gapA, guaA, mutM, nuoD, ppsA, and recA) for PCR amplification (Supplementary Table 1) and the amplified products were sequenced and analyzed on the MLST database.
To determine phage BUCT609 host range, 13 strains of S. maltophilia were cultured to the exponential phase (OD600 = 0.7). 500 μl of each sample were added into 5 ml 0.7% LB agar medium. Then, the mixture was immediately poured into the solid medium. After solidification, 2 μl phages were added to a double-layer plate, and PBS was added on the other side as a control. The formation of plaque was observed and recorded after 9 h.
To quickly and effectively detect the sensitivity of pathogenic bacteria to various antibiotics and guide rational drug use in the clinic, the drug sensitivity test of S. maltophilia strain No. 3015 was carried out by disc diffusion method. The host bacteria were cultured to the exponential phase (OD600 = 0.7) at 37°C. 500 μl of bacteria was added into 5 ml 0.7% LB agar medium. The mixture was immediately poured into the solid medium. After solidification, the antibiotic sensitivity testing disc was placed on the plate for incubating at 37°C overnight. If the strain was sensitive to the drug, the formation of the inhibitory zone could be observed. This experiment was repeated three times.
Phage genomic DNA was extracted by the Proteinase K/SDS method (Pickard, 2009). 600 μl of purified phage were incubated with DNase I and RNase A overnight at 37°C after which the enzymes were inactivated at 80°C for 15 min. 24 μl of EDTA (20 mM) with 1.5 μl of proteinase K (50 μg/ml) and 30 μl of SDS (0.5%) were added to the mixture and were incubated at 56°C for 1 h. Phage DNA was extracted with an equal volume of extraction agent (phenol: chloroform: isoamyl alcohol, 25:24:1). 400 μl of isopropanol was added to the upper aqueous layer and then the sample was incubated at −20°C for more than 1 h. 75% ethanol was added to rinse the precipitate. Eventually, the deionized water was used to dissolve the nucleic acids and supposed to be stored at −20°C.
The whole genome was sequenced using Illumina’s MiSeq sequencing platform (Thermo Fisher Scientific, United States; Kozich et al., 2013) and the low-quality sequences were filtered by Trimmomatic (V0.32) program (Bolger et al., 2014). Then, the complete genomic sequence of BUCT609 was assembled by Newbler V3.0 software (Roche, Switzerland; Zhang et al., 2012) and CLC software (QIAGEN, Germany; Liu and Di, 2020). Using the online tools RAST (https://rast.nmpdr.org/; Aziz et al., 2008) to predict the ORFs and ORF Finder (https://www.bioinformatics.org/sms2/orf_find.html; Rombel et al., 2002) to annotate the DNA sequencing result. Sequence similarity analyses and comparisons were performed using the NCBI BLAST algorithm. A phylogenetic tree of phage BUCT609 was conducted by VICTOR (https://ggdc.dsmz.de/victor.php; Meier-Kolthoff and Göker, 2017) and average nucleotide identity (ANI) analysis was analyzed by another online tool VIRIDIC (http://rhea.icbm.uni-oldenburg.de/VIRIDIC/; Moraru et al., 2020).
Specific pathogen-free (SPF) Balb/c mice reared for 6–8 weeks, weighing 17-19 g, were chosen as the animal model in this study. The median lethal dose (LD50) refers to the minimum number of bacteria required to kill half of the animal population through a specified route of infection within a specified period of time (Welkos and Brien, 1994). Pick a single clone of S. maltophilia No.3015 strain on the agar plate and cultivate it in LB liquid medium to exponential phase (37°C, 220 rpm). Then, the bacteria were centrifuged at 10,000 × g for 10 min and resuspended with PBS diluting to 8 × 107 CFU/ml, 6 × 107 CFU/ml, 4 × 107 CFU/ml, and 2 × 107 CFU/ml, respectively, for use. Forty mice were randomly divided into 5 groups (n = 8 in each group), of which 4 groups were used as the experimental group (injected with bacteria) and 1 group was used as the control group (injected with PBS). All mice were immunosuppressed by injection of cyclophosphamide (125 mg/kg) 4 days before infection and injected with cyclophosphamide (125 mg/kg) and dexamethasone (12.5 mg/kg) next day to stabilize the immunosuppression. After above pretreatment, each mouse was anesthetized by intraperitoneal injection of 50 mg/kg of 0.5% sodium pentobarbital. The experimental groups were injected intranasally with 40 μl of No.3015 bacterial solution with titers of 8 × 107 CFU/ml, 6 × 107 CFU/ml, 4 × 107 CFU/ml, and 2 × 107 CFU/ml, respectively, and the control group was injected with 40 μl of sterile PBS solution. The activity status of mice was observed twice a day after infection. The changes in body weight and death situation were recorded.
To explore the therapeutic effect of phage BUCT609 in vivo, 30 mice were equally divided into three groups (n = 10 in each group), two of which were the experimental group (injected with both bacteria and phage) and one was the control group (injected with PBS only). Mice were immunosuppressed and anesthetized in the same manner as described above. 40 μl of No.3015 bacterial solution was injected into the nasal cavity of the experimental group and 40 μl of PBS solution was injected into the control group. After 2 h, 40 μl of phage BUCT609 was injected into one of the experimental groups, 40 μl sterile of PBS solution was injected into other groups. The activity status of mice was observed daily after infection and the changes in body weight and death situation were recorded.
Three replicates were performed for each experiment. Take the mean of the three sets of data and calculate the standard error. The data were subjected to one-way ANOVA test and Tukey test by GraphPad Prism 8.0.2 (GraphPad Software, Inc., La Jolla, United States). p < 0.05 means that the data of repeated groups are statistically significant.
A virulent phage BUCT609 was successfully isolated from untreated sewage in the hospital using S. maltophilia No. 3015 as the host bacterium. Phage BUCT609 can form transparent plaques with a diameter of 1 ~ 2 mm on a double-layer plate (Figure 1A). TEM images showed that the head diameter of phage BUCT609 was about 51.93 ± 1.68 nm, and the tail length was about 12.36 ± 1.37 nm, which can be inferred as a Podoviridae phage (Figure 1B).
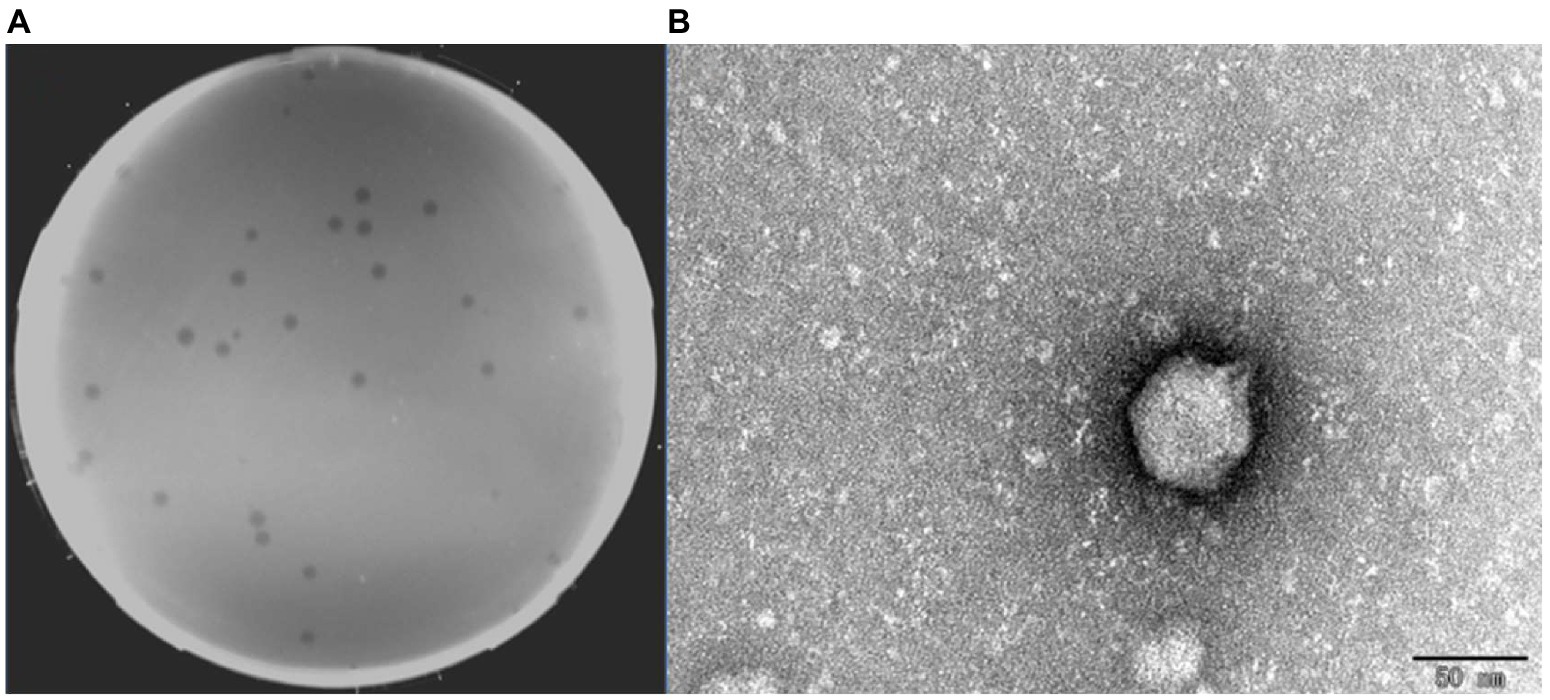
Figure 1. Plaques and transmission electron micrograph of phage BUCT609. (A) Plaques formed by BUCT609. (B) Transmission electron micrograph of phage BUCT609. Scale bar 50 nm.
Phage and host bacteria were cocultured for 5 h at different MOI ratio as shown in Supplementary Table 2 and the average titer of each phage was measured, respectively. As can be seen from Supplementary Table 2, when MOI = 0.01, the phage titer was the highest with 3.1 × 109 PFU/ml. Therefore, the optimal multiplicity of infection was defined as 0.01.
The one-step growth curve showed that the latent period was about 10 min (Figure 2A). In the following 60 min, the phage titer increased rapidly, where was the burst period. About 10 min later, the phage grew into the plateau phase. According to the calculation, phage BUCT609 had a large burst size (382 PFU/cell). Compared with the phage P24 (the latent period was 55 min and the burst size was 147 PFU/cell), phage BUCT609 had a shorter latent period and a larger burst size (Zhang et al., 2020). In conclusion, it was a lytic phage and had the potential to be used for phage therapy.
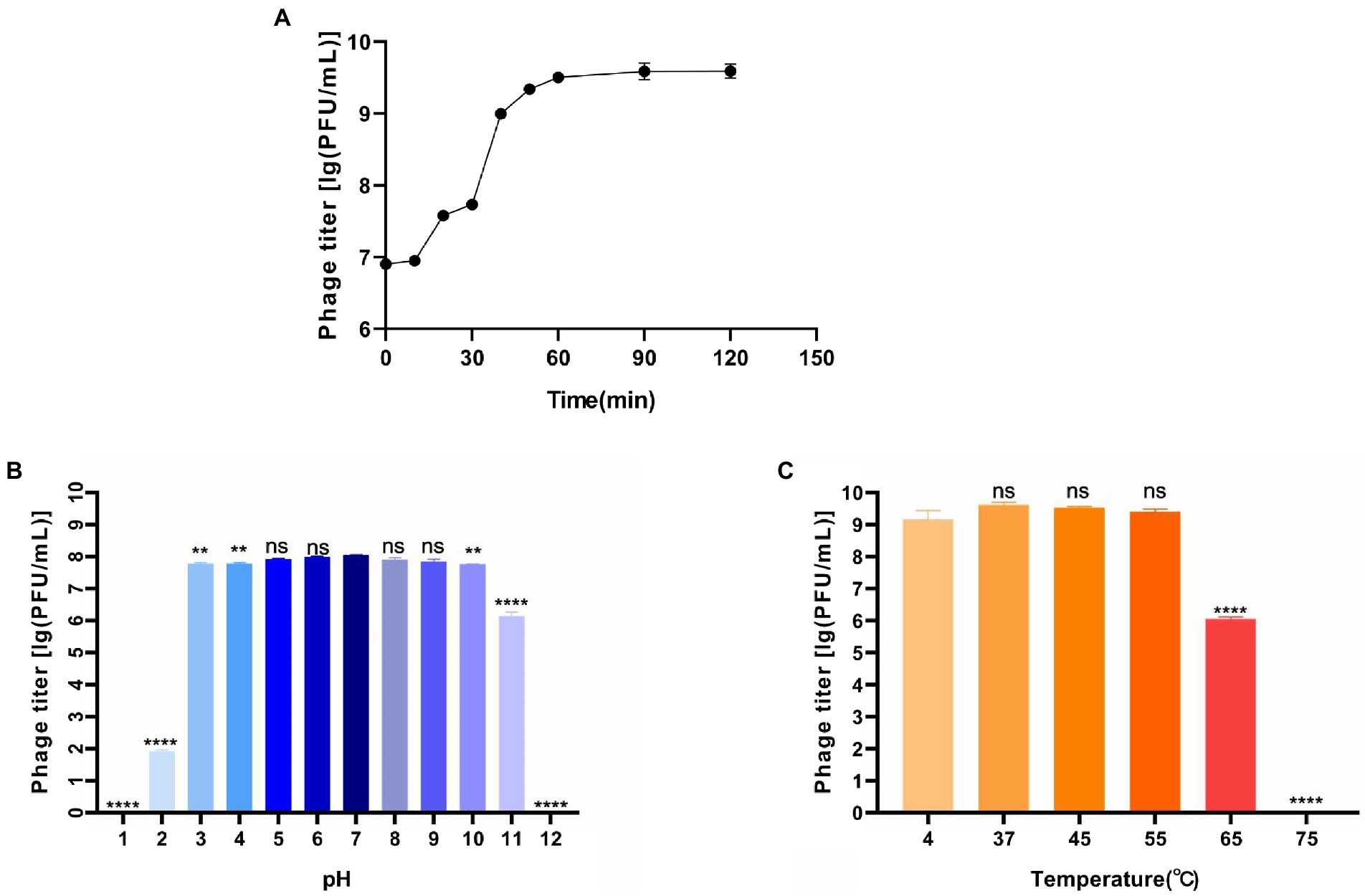
Figure 2. Biological characteristics of phage BUCT609. (A) One-step growth curve. The latent period of the phage was about 10 min, and the burst period of the phage was the following 60 min. And after 70 min, the phage grew into the plateau phase. The data is the average of three parallel experiments. Two bars above and below each point represent the standard deviation of the results of three parallel experiments. (B) Thermal stability. Phage titer of BUCT609 after incubation for 1 h at different temperatures. The data is the average of three replicates. There was no significant difference in the titer between 4 and 55°C. At 65 and 75°C, its titer decreased significantly (p < 0.0001) compared with that at 4°C. (C) pH sensitivity. Phage titer of BUCT609 after incubation at various pH values at 37°C for 1 h. The data is the average of three replicates. There was no significant difference in the titer from pH 5 to 9. Compared with the titer at pH 7, its titer decreased slightly (p < 0.01) at pH 3, 4, and 10 and decreased significantly (p < 0.0001) at pH 1, 2, 11, and 12. There is no significant difference between the two groups of data when p < 0.5, which was showed as ns in the figure. The bar above column represents the standard deviation of the results of three parallel experiments.
The thermal stability test showed that phage BUCT609 was high-temperature sensitive (Figure 2B). There was no significant difference in the titer between 4 and 55°C for 1 h. At 65°C, phage titer decreased significantly (p < 0.0001) compared with that at 4°C and it was completely inactivated at 75°C. As for pH tolerance (Figure 2C), phage BUCT609 exhibited a strong tolerance from pH 3 to 10. At pH 2 or 11, the phage titer was significantly lower than at pH 7 (p < 0.0001). And at pH 1 or 12, the phage was almost completely inactivated. Compared with phage IME392 (temperature tolerance range: 30–50°C; pH tolerance range: 4–11; Hu et al., 2021), phage BUCT609 had a wider adaptation range of temperature and pH, indicating that it had a good temperature and pH tolerance and could be broadly applied for phage therapy in the future.
The MLST results showed that 13 strains of S. maltophilia had different ST values, suggesting that they were 13 different species. The analysis of the host range results showed that phage BUCT609 could lyse not only S. maltophilia strain No. 3015 but also the other 4 S. maltophilia strains (No. 118, No. 548, No. 992, and No. 1207; Supplementary Table 3).
The main reason S. maltophilia infection is difficult to treat is that the bacteria present low susceptibility to antibiotics (Gil-Gil et al., 2020). Drug susceptibility tests confirmed that the host S. maltophilia No. 3015 of phage BUCT609 was resistant to a variety of antibiotics but sensitive to only a few antibiotics such as ceftriaxone, tetracycline, trimethoprim, minocycline, and levofloxacin (Supplementary Table 4).
The complete sequence of phage BUCT609 was 43,145 bp in length and the CG content of the genome was 58% that had been submitted to the NCBI database with the accession number MW960043. The online comparison tool BlastN showed that it had 42% homology with S. maltophilia phage vB_SmeS_BUCT703 (GenBank: OM735688.1) at nucleotide level. The results of RAST online annotation showed that BUCT609 had 56 open reading frames (ORFs), of which 25 had known functions and the rest were annotated as hypothetical proteins. The majority of ORFs presented an ATG start codon (87.5%) while 2 of them started with TTG and 5 with GTG. There is no tRNA in the whole genome. The 25 functional proteins are shown in different colors on the whole genome map (Figure 3).
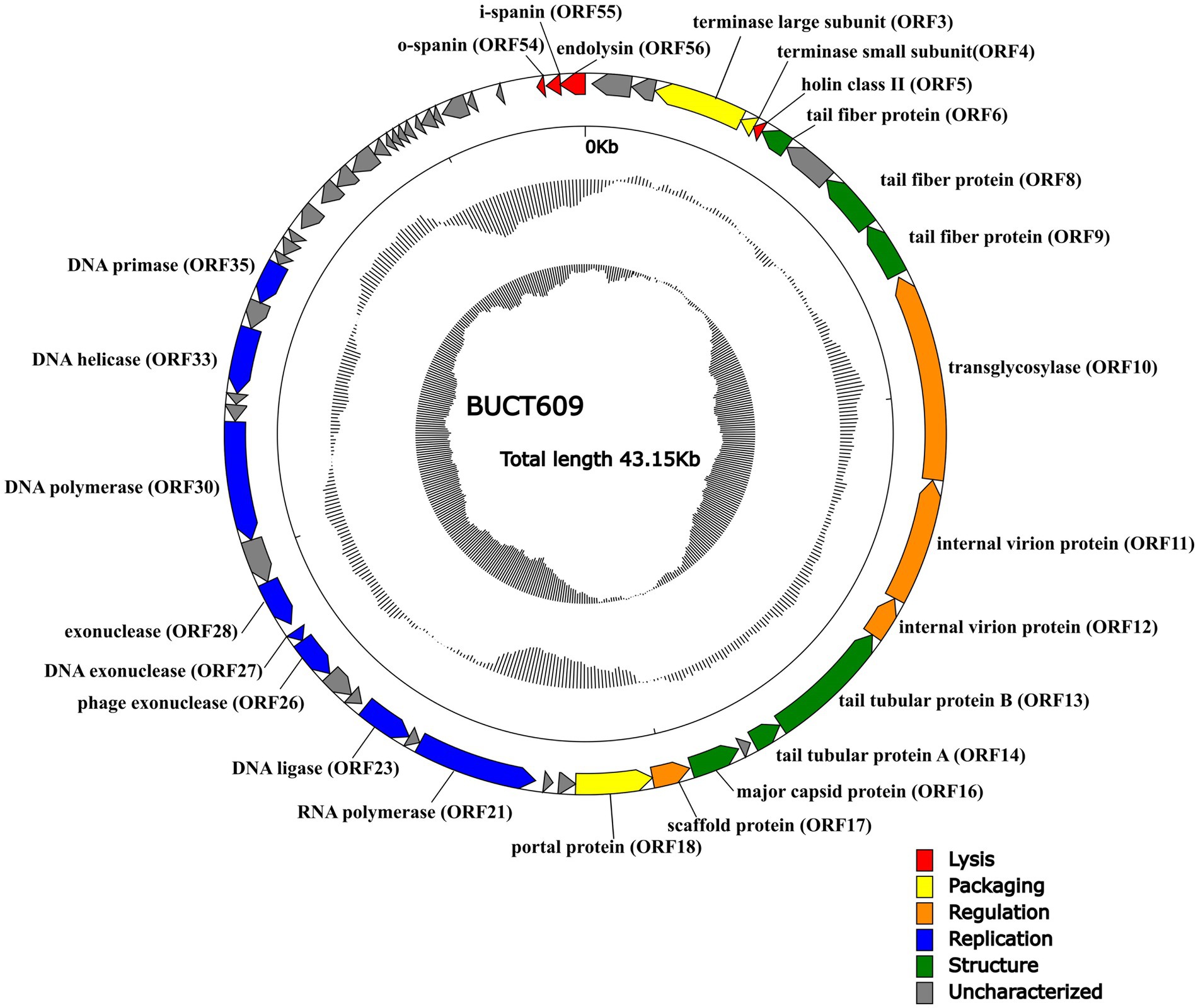
Figure 3. The whole genetic map of phage BUCT609. The outermost circle represents the open reading frame. Colors distinguish different functional genes while arrows represent ORF directions. The middle circle represents (G + C) mol% (outwards larger than the whole-genome average (G + C) mol%, inwards the opposite). The innermost circle represents the G + C tilt of G−C/G + C (outward for greater than 0 and inward for less than 0).
Same as most dsDNA phage, phage BUCT609 has the same modular genomic structure such as DNA replication, regulation, phage packaging, structural, and host lytic proteins (Table 1). The structural proteins of phage BUCT609 are mainly distributed at the front of the genomic sequence. According to previous data analysis, ORF6, ORF8, ORF9, ORF13, and ORF14 were predicted to be tail related proteins. ORF14 encodes tail tubular protein A (TTPA) that is usually responsible for adhering the phage to host cells (Pyra et al., 2017), having 62.98% homology to the protein of S. maltophilia phage Ponderosa at amino acid level (the following homology values are all at amino acid level in this section). Besides, the major capsid protein was encoded by ORF16 and had 75.62% homology with that of S. maltophilia phage Ponderosa.
Terminase is a key protein in phage DNA packaging that includes large subunit and small subunit. Generally, terminase large subunit and terminase small subunit are adjacent and both involved in the splicing and packaging process of phage DNA. The large subunit is responsible for ATP-driven DNA translocations while the small subunit interacts with terminase large subunit and initiate packaging through binding and cleaving specifically near the initial package site (Weiditch et al., 2019 Gao et al., 2020). Terminase small subunit specifically recognizes viral DNA, while the terminase large subunit plays an important role in ATP recognition and hydrolysis (Loredo-Varela et al., 2013). As is shown from the chart, ORF3 from phage BUCT609 encodes the terminase large subunit that has 69.26% homology with that of S. maltophilia phage Ponderosa. The terminase small subunit is the product of ORF14 and exhibited 39% identity to that of Xanthomonas phage Xaa_vb_phi31. The portal control protein is encoded by ORF18 and forms a channel at the phage tail attachment site through where the phage can inject its genome into the host cell (Prevelige Jr and Cortines, 2018).
Replication is a complex process involving a variety of proteins and enzymes. In the initiation phase, the DNA helicase encoded by OFR33 can unlock double-stranded DNA by hydrolyzing ATP for energy, with 63.53% homology to that of S. maltophilia phage Ponderosa. DNA primer enzyme encoded by ORF35 has 63.44% homology with that of Xanthomonas phage Xaa_vb_phi31, which can catalyze the synthesis of RNA primer. Different DNA polymerases play different roles in DNA replication (Hoitsma et al., 2020). Exhibiting 69.95% identity to DNA polymerase of S. maltophilia phage Ponderosa, the DNA polymerase encoded by ORF30 is mainly used for DNA replication and repair. Furthermore, exonuclease is a class of enzymes that degrade nucleotides one by one from the end of a polynucleotide chain (Dean et al., 2001). In phage BUCT609, ORF26, ORF27, and ORF28 encode three different exonuclease enzymes, with 44.4% homology to that of Xylella phage Cota, 65% homology to that of Xanthomonas phage Xaj24 and 65.12% homology to that of Xanthomonas phage XAA VB phi31, respectively. The DNA ligase encoded by ORF23 plays an important role in the process of DNA replication and repair, catalyzing the reaction that two adjacent bases connect consuming ATP and has 42.42% homology to the protein of S. maltophilia phage Ponderosa by comparison (Shi et al., 2018). ORF21 encodes RNA polymerase whose main function is to synthesize RNA using DNA or RNA as templates and triphosphate ribonucleoside as substrates. Due to that RNA is involved in the transcription of genetic information about genes within a cell, it is also called a transcriptase (Griesenbeck et al., 2017). The RNA polymerase ORF21 has 100% coverage but only 72% identity with that of S. maltophilia phage Ponderosa.
For the lytic mechanism, the phage lyses the host bacteria under the combined action of holin, lyase, and spanin. Holin encoded by OFR5 plays a crucial role in the penetration of phages after attaching to the host. Holin forms pores in the cell membrane and begins the process of osmosis and dissolution (Palmer et al., 2021). By the BlastN comparison, the holin of phage BUCT609 has 59.68% homology with S. maltophilia phage Ponderosa. Most phages produce a two-component protein complex consisting of outer membrane lipoprotein (o-spanin) and inner membrane protein (i-spanin) which are necessary for the host outer membrane destruction (Cahill and Young, 2019). Phage BUCT609 has an o-spanin ORF54 with 57.14% homology to the protein of S. maltophilia phage Ponderosa and i-spanin ORF55 with 30.85% homology to the protein of Xylella phage Paz. The endolysin transcribed by ORF56, which showed 65.06% identity to that of S. maltophilia phage Ponderosa, was synthesized by host bacteria and can result in bacteria death through inducing the lysis of bacteria cell walls specifically and effectively (Ghose and Euler, 2020).
A total of 21 phages in NCBI had homology (>0%) with BUCT609. To study the evolution of phage BUCT609 and its relationship with other phages, a phylogenetic tree of complete genome sequences of above 22 phages including BUCT609 was constructed. According to VICTOR’s classification of family, genus, and species, 21 strains of phages homologous to phage BUCT609 belonged to the same genus. From NCBI records, all of these strains were presumed to be Autographiviridae family and Pradovirus genus. However, at the species level, there were 16 different clusters, among which BUCT609 independently represented a species (Figure 4A). To determine whether phage BUCT609 could form an independent clade as a new genus, average nucleotide identity (ANI) analysis with other 21 phages was analyzed. The results showed that BUCT609 had a maximum similarity with Stenotrophomonas phage vB_SmaS_P15 (57.3%), which was sufficient for classification at the level of a new genus (Figure 4B).
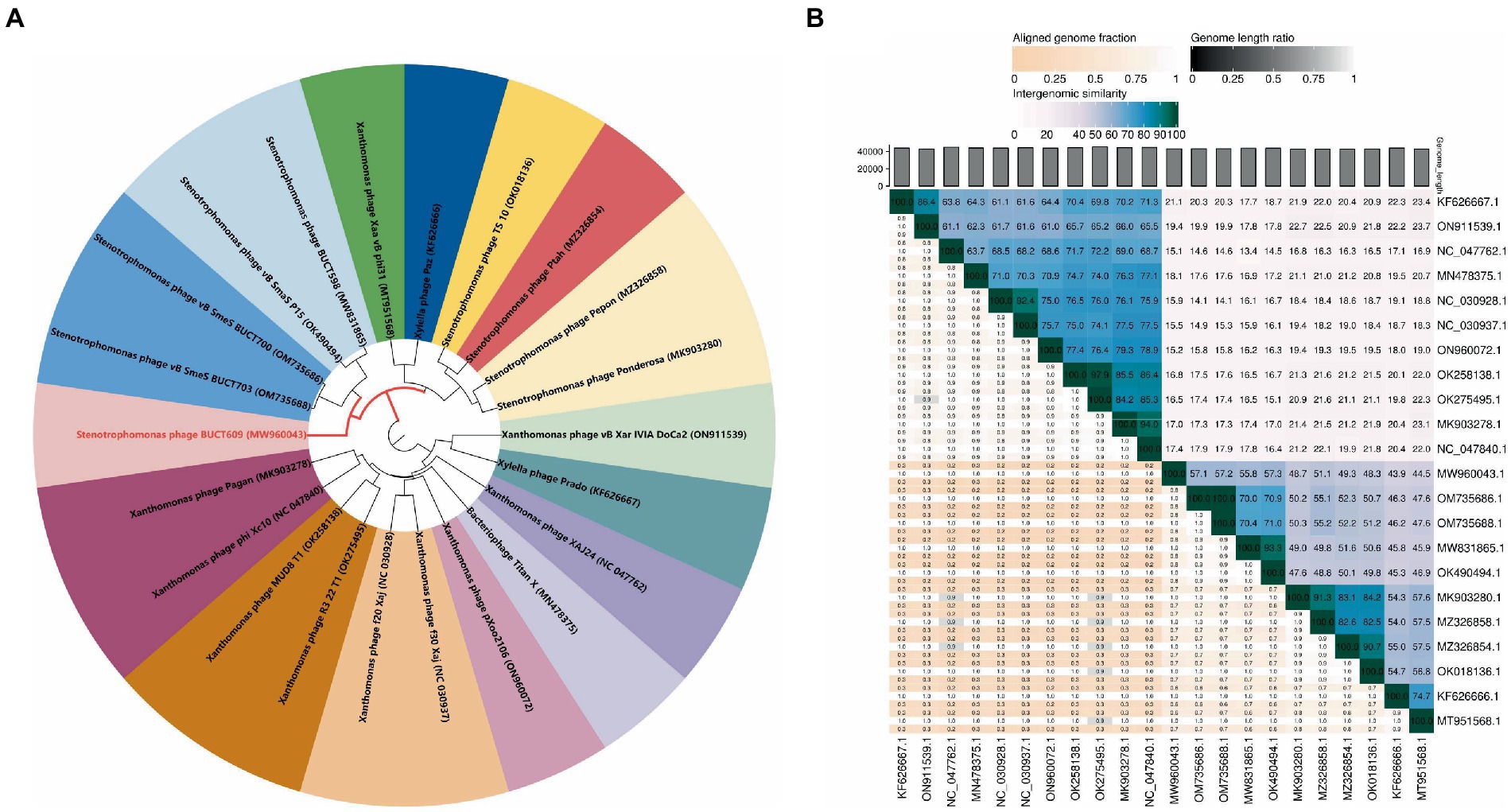
Figure 4. Phylogenetic analysis of phage BUCT609. (A) A phylogenetic tree was constructed based on the whole genome sequence by VICTOR which indicated phage BUCT609 could be a new species. (B) Percentage sequence similarity between phages calculated using VIDIRIC which shows phage BUCT609 could be a new clade. The horizontal and vertical coordinates indicate the corresponding phage Genebank number.
We conducted animal experiments to further explore the possibility of phage BUCT609 for clinical treatment. In mouse models, there was no significant change before and after No.3015 infection (fluctuation within 1 gram). Remarkably, the hair of the mice before bacterial infection was white and shiny while sparse and coarse after infection. Besides, they appeared weaker and clustered together, suggesting that S. maltophilia infections could have adverse effects on the organism. The results of infection in mice with different doses were shown in Figure 5A. No mice died in the control group. The survival rate of mice gradually decreased with the increase of injected bacterial titer. When the titer of No.3015 was 4 × 107 CFU/ml, the survival rate of mice was 50%, which was defined as the value of LD50.
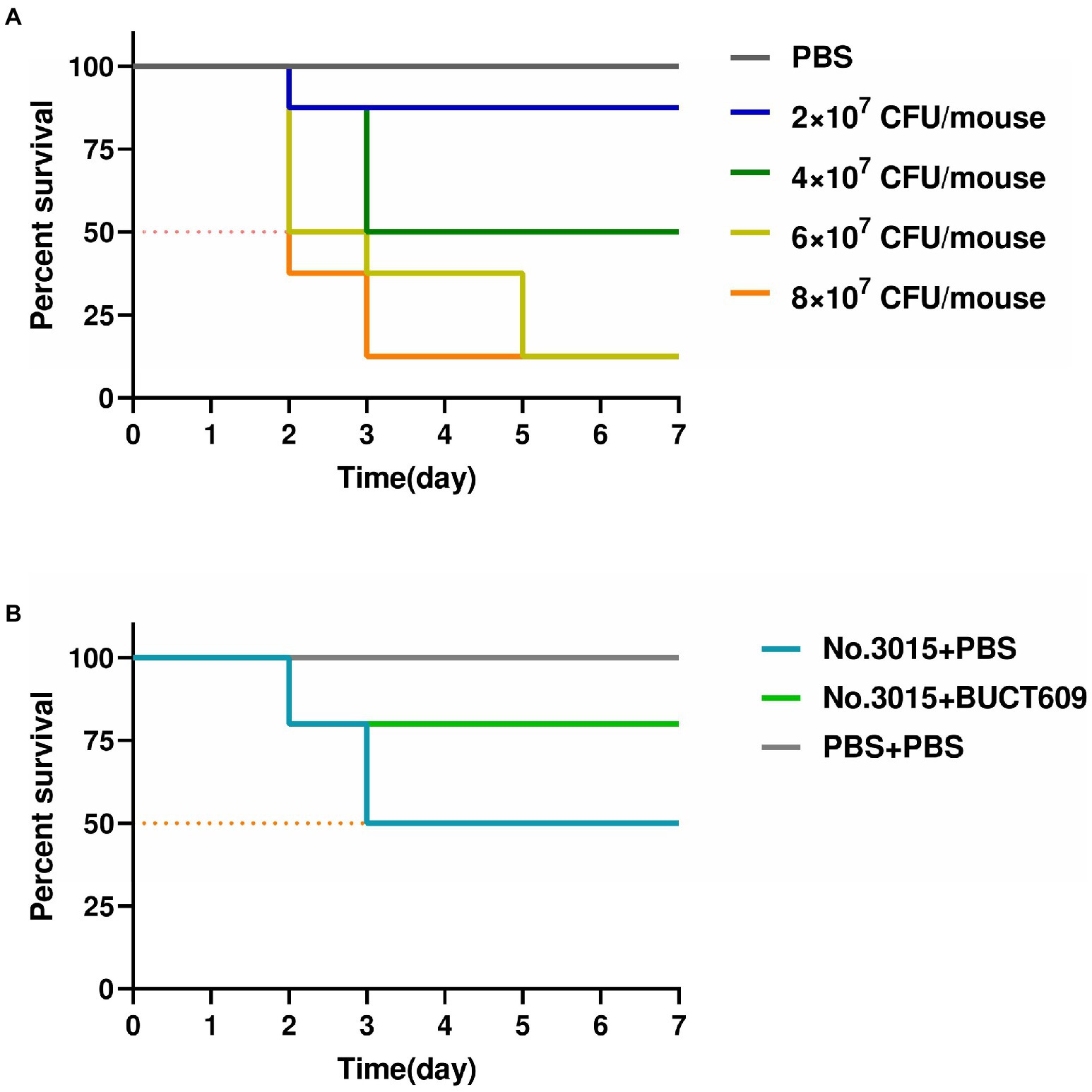
Figure 5. Antibacterial effect of BUCT609 in vivo. (A) Determination of median lethal dose. The daily percent survival of mice at different titers of bacterial doses. The LD50 was determined as 4 × 107 CFU/ml. When the titer of No.3015 was 4 × 107 CFU/ml, the survival rate of mice was 50%. (B) Antibacterial effect of BUCT609 in vivo. The daily percent survival of mice before and after treatment. After the treatment, the mice had a survival rate of up to 80%, which was 30% higher than no treatment.
The effect of phage BUCT609 in lysing host bacteria in vivo was studied by injecting phage BUCT609 to mice infused with LD50. The vital signs of all mice stabilized after 3dpi after the treatment (Figure 5B). After injection of phage BUCT609, the survival rate of mice increased to 80%, which was 30% higher than that of the other group of mice that did not receive treatment. There was no significant change in the weight of the mice before and after phage treatment (fluctuation within 1 gram). Even though the survival rate of the mice improved after the treatment, the coarseness of the hair was not cured. However, the data were sufficient to demonstrate that phage BUCT609 was bacteriostatic in vivo and could be used as a potential clinical antimicrobial agent.
Phage BUCT609 was successfully isolated from hospital sewage using S. maltophilia No. 3015 as the host bacterium. The host range test showed that 5 of 13 strains of S. maltophilia could be lysed by phage BUCT609. Though it does not have a broad host range as phage DLP3, one of the most widespread S. maltophilia phages infecting 22 out of 29 S. maltophilia strains (Peters et al., 2020), BUCT609 still have a relatively wide host range compared with phage AXL3 that can lyse 5 out of 29 S. maltophilia strains (McCutcheon et al., 2020). In previous studies, phage SM1 was found to be effective in animal S.maltophilia infection model with 184 PFU/cell burst size (Zhang and Li, 2013). However, the burst size of phage BUCT609 is 382 PFU/cell, which was much higher than that of SM1. Meanwhile, given its temperature stability and wide pH tolerance, BUCT609 shows promising potential to use against multi-drug resistant S. maltophila infections. In addition, phylogenetic tree and ANI results showed that phage BUCT609 is an independent new clade that can be classified in subsequent phage discoveries.
For Autographiviridae phages that can lyse Gram-negative bacteria, there are two systems for lysing bacteria: the holin-endolysin and pinholin-SAR endolysin pathways (Cahill and Young, 2019). In the former, holin first forms micron-scale pores in the inner membrane, releasing active endolysin into the periplasm to degrade peptidoglycan, thus completing the first step of lysing bacteria. According to genome analysis, the cleavage mechanism of phage BUCT609 was mainly mediated by holin protein encoded by ORF5. Endolysin ORF56 could promote the lysis of host bacteria as well. At present, there are many studies on holin protein, showing that holin protein can be used as a new antibacterial agent. HolGH15, the holin protein in Staphylococcus aureus phage GH15, can cause changes in the structural properties of Listeria monocytogenes leading to shrinkage, resulting in the release and removal of cellular contents and ultimately leading to the host death. HolGH15 (the final concentration: 240 μg/ml) can reduce L. monocytogenes (the initial concentration: 106 CFU/ml) to undetectable levels at 4°C (Song et al., 2021), which shows that the holin protein of phage can potentially become a new type of antibacterial drug by spraying or soaking. BUCT609 can provide a new antibacterial agent in the field of antibacterial as well.
However, phage therapy has many challenges such as safety, ethics, intellectual property rights, and stability (Skurnik et al., 2007). Although this study initially explored the therapeutic effect of phage BUCT609 in mice, the stability of its treatment was not explored. In vivo experiments demonstrated that phages can reduce the bacterial load and weight of abscesses (Capparelli et al., 2007). At the same time, increasing evidence suggests that phages may have a major impact on the immune system by interacting with macrophages, neutrophils, and T-cell polarization (Chechushkov et al., 2021). Phage therapy as a potential regimen against drug-resistant bacteria still requires adequate clinical trials.
Phage therapy has become a new possibility to treat clinically drug-resistant bacteria. The diversity of phages may contribute to the development of phage-based therapies. In vivo animal studies demonstrated that phage BUCT609 as a new member to the S. maltophilia bacteriophage provided empirical data for phage cocktail therapy in combination with antibiotics against multi-drug resistant bacterial infections.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by the Seventh Medical Center of the PLA General Hospital.
KH: data curation, writing – original draft, investigation, and validation. YD: data curation, writing – original draf, investigation and validation. LS: supervision and writing – review and editing. ML: supervision, validation, writing – review and editing. HF: conceptualization, methodology, supervision, validation, and writing – review and editing. YT: conceptualization, supervision, validation, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This research was supported by Funds for First-class Discipline Construction (nos. XK1805 and XK1803-06), National Key Research and Development Program of China (nos. 2018YFA0903000, 2020YFC2005405, 2020YFA0712100, 2020YFC0840805, 19SWAQ06, 20SWAQX27, and 20SWAQK22), Inner Mongolia Key Research and Development Program (nos. 2019ZD006), NSFC-MFST project (China-Mongolia; no. 31961143024), Fundamental Research Funds for Central Universities (no. BUCTRC201917 and BUCTZY2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1001237/full#supplementary-material
An, S. Q., and Berg, G. (2018). Stenotrophomonas maltophilia. Trends Microbiol. 26, 637–638. doi: 10.1016/j.tim.2018.04.006
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bao, J., Wu, N., Zeng, Y., Chen, L., Li, L., Yang, L., et al. (2020). Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg Microbes Infect 9, 771–774. doi: 10.1080/22221751.2020.1747950
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cahill, J., and Young, R. (2019). Phage Lysis: multiple genes for multiple barriers. Adv. Virus Res. 103, 33–70. doi: 10.1016/bs.aivir.2018.09.003
Capparelli, R., Parlato, M., Borriello, G., Salvatore, P., and Iannelli, D. (2007). Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob. Agents Chemother. 51, 2765–2773. doi: 10.1128/AAC.01513-06
Chang, H. C., Chen, C. R., Lin, J. W., Shen, G. H., Chang, K. M., Tseng, Y. H., et al. (2005). Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl. Environ. Microbiol. 71, 1387–1393. doi: 10.1128/AEM.71.3.1387-1393.2005
Chechushkov, A., Kozlova, Y., Baykov, I., Morozova, V., Kravchuk, B., Ushakova, T., et al. (2021). Influence of Caudovirales phages on humoral immunity in mice. Viruses 13:1241. doi: 10.3390/v13071241
Dean, F. B., Nelson, J. R., Giesler, T. L., and Lasken, R. S. (2001). Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11, 1095–1099. doi: 10.1101/gr.180501
Ding, T., Sun, H., Pan, Q., Zhao, F., Zhang, Z., and Ren, H. (2020). Isolation and characterization of Vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res. 286:198080. doi: 10.1016/j.virusres.2020.198080
Fan, H., Huang, Y., Mi, Z., Yin, X., Wang, L., Fan, H., et al. (2012). Complete genome sequence of IME13, a Stenotrophomonas maltophilia bacteriophage with large burst size and unique plaque polymorphism. J. Virol. 86, 11392–11393. doi: 10.1128/JVI.01908-12
Gao, M., Wang, C., Qiang, X., Liu, H., Li, P., Pei, G., et al. (2020). Isolation and characterization of a novel bacteriophage infecting carbapenem-resistant Klebsiella pneumoniae. Curr. Microbiol. 77, 722–729. doi: 10.1007/s00284-019-01849-8
Ghose, C., and Euler, C. W. (2020). Gram-negative bacterial Lysins. Antibiotics (Basel, Switzerland) 9, 2–74. doi: 10.3390/antibiotics9020074
Gil-Gil, T., Martínez, J. L., and Blanco, P. (2020). Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: a review of current knowledge. Expert Rev. Anti-Infect. Ther. 18, 335–347. doi: 10.1080/14787210.2020.1730178
Gokhan Gozel, M., Celik, C., and Elaldi, N. (2015). Stenotrophomonas maltophilia infections in adults: primary bacteremia and pneumonia. Jundishapur J Microbiol 8:e23569. doi: 10.5812/jjm.23569
Griesenbeck, J., Tschochner, H., and Grohmann, D. (2017). Structure and function of RNA polymerases and the transcription machineries. Subcell. Biochem. 83, 225–270. doi: 10.1007/978-3-319-46503-6_9
Hagens, S., and Loessner, M. J. (2010). Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr. Pharm. Biotechnol. 11, 58–68. doi: 10.2174/138920110790725429
Hoitsma, N. M., Whitaker, A. M., Schaich, M. A., Smith, M. R., Fairlamb, M. S., and Freudenthal, B. D. (2020). Structure and function relationships in mammalian DNA polymerases. Cell. Mol. Life Sci. 77, 35–59. doi: 10.1007/s00018-019-03368-y
Hu, Y., Tong, S., Li, P., An, X., Song, L., Fan, H., et al. (2021). Characterization and genome sequence of the genetically unique Escherichia bacteriophage vB_EcoM_IME392. Arch. Virol. 166, 2505–2520. doi: 10.1007/s00705-021-05160-5
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, F., Xing, S., Fu, K., Zhao, S., Liu, J., Tong, Y., et al. (2019). Genomic and biological characterization of the vibrio alginolyticus-infecting "Podoviridae" bacteriophage, vB_ValP_IME271. Virus Genes 55, 218–226. doi: 10.1007/s11262-018-1622-8
Liu, C. H., and Di, Y. P. (2020). Analysis of RNA sequencing data using CLC genomics workbench. Methods Mol. Biol. 2102, 61–113. doi: 10.1007/978-1-0716-0223-2_4
Loredo-Varela, J., Chechik, M., Levdikov, V. M., Abd-El-Aziz, A., Minakhin, L., Severinov, K., et al. (2013). The putative small terminase from the thermophilic dsDNA bacteriophage G20C is a nine-subunit oligomer. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 876–879. doi: 10.1107/S1744309113017016
McCutcheon, J. G., Lin, A., and Dennis, J. J. (2020). Isolation and characterization of the novel bacteriophage AXL3 against Stenotrophomonas maltophilia. Int. J. Mol. Sci. 21, 17–6338. doi: 10.3390/ijms21176338
Meier-Kolthoff, J. P., and Göker, M. (2017). VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics (Oxford, England) 33, 3396–3404. doi: 10.1093/bioinformatics/btx440
Moelling, K., Broecker, F., and Willy, C. (2018). A wake-up call: we need phage therapy now. Viruses 10, 12–688. doi: 10.3390/v10120688
Moraru, C., Varsani, A., and Kropinski, A. M. (2020). VIRIDIC-A novel tool to calculate the Intergenomic similarities of prokaryote-infecting viruses. Viruses 12, 11–1268. doi: 10.3390/v12111268
Palmer, T., Finney, A. J., Saha, C. K., Atkinson, G. C., and Sargent, F. (2021). A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol. Microbiol. 115, 345–355. doi: 10.1111/mmi.14599
Peters, D. L., McCutcheon, J. G., and Dennis, J. J. (2020). Characterization of novel broad-host-range bacteriophage DLP3 specific to Stenotrophomonas maltophilia as a potential therapeutic agent. Front. Microbiol. 11:1358. doi: 10.3389/fmicb.2020.01358
Pickard, D. J. (2009). Preparation of bacteriophage lysates and pure DNA. Methods Mol. Biol. 502, 3–9. doi: 10.1007/978-1-60327-565-1_1
Prevelige, P. E. Jr., and Cortines, J. R. (2018). Phage assembly and the special role of the portal protein. Curr. Opin. Virol. 31, 66–73. doi: 10.1016/j.coviro.2018.09.004
Pyra, A., Brzozowska, E., Pawlik, K., Gamian, A., Dauter, M., and Dauter, Z. (2017). Tail tubular protein a: a dual-function tail protein of Klebsiella pneumoniae bacteriophage KP32. Sci. Rep. 7:2223. doi: 10.1038/s41598-017-02451-3
Rombel, I. T., Sykes, K. F., Rayner, S., and Johnston, S. A. (2002). ORF-FINDER: a vector for high-throughput gene identification. Gene 282, 33–41. doi: 10.1016/s0378-1119(01)00819-8
Shi, K., Bohl, T. E., Park, J., Zasada, A., Malik, S., Banerjee, S., et al. (2018). T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction. Nucleic Acids Res. 46, 10474–10488. doi: 10.1093/nar/gky776
Skurnik, M., Pajunen, M., and Kiljunen, S. (2007). Biotechnological challenges of phage therapy. Biotechnol. Lett. 29, 995–1003. doi: 10.1007/s10529-007-9346-1
Song, J., Niu, W., Wu, R., Wang, J., Lei, L., Han, W., et al. (2021). The phage Holin HolGH15 exhibits potential as an antibacterial agent to control listeria monocytogenes. Foodborne Pathog. Dis. 18, 574–581. doi: 10.1089/fpd.2020.2833
Weiditch, S. A., Seraphim, T. V., Houry, W. A., and Kanelis, V. (2019). Strategies for purification of the bacteriophage HK97 small and large terminase subunits that yield pure and homogeneous samples that are functional. Protein Expr. Purif. 160, 45–55. doi: 10.1016/j.pep.2019.03.017
Welkos, S., and Brien, A. (1994). Determination of median lethal and infectious doses in animal model systems. Meth. Enzymol. 235, 29–39. doi: 10.1016/0076-6879(94)35128-7
Xing, S., Zhang, X., Sun, Q., Wang, J., Mi, Z., Pei, G., et al. (2017). Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 162, 3843–3847. doi: 10.1007/s00705-017-3503-1
Zhang, J., and Li, X. (2013). Biological characteristics of phage SM1 for Stenotrophomonas maltophilia and its effect in animal infection model. Zhejiang Da Xue Xue Bao Yi Xue Ban 42, 331–336. (in Chinese)
Zhang, X., Liu, Y., Wang, M., Wang, M., Jiang, T., Sun, J., et al. (2020). Characterization and genome analysis of a novel marine Alteromonas phage P24. Curr. Microbiol. 77, 2813–2820. doi: 10.1007/s00284-020-02077-1
Keywords: Stenotrophomonas maltophilia, bacteriophage, podoviridae, genomic analysis, phage thearpy
Citation: Han K, Dong Y, An X, Song L, Li M, Fan H and Tong Y (2022) Potential application of a newly isolated phage BUCT609 infecting Stenotrophomonas maltophilia. Front. Microbiol. 13:1001237. doi: 10.3389/fmicb.2022.1001237
Received: 23 July 2022; Accepted: 31 October 2022;
Published: 21 November 2022.
Edited by:
Chengmin Wang, Guangdong Academy of Sciences, ChinaReviewed by:
Steven Batinovic, Yokohama National University, JapanCopyright © 2022 Han, Dong, An, Song, Li, Fan and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yigang Tong, dG9uZy55aWdhbmdAZ21haWwuY29t; Huahao Fan, ZmFuaHVhaGFvQG1haWwuYnVjdC5lZHUuY24=; Mengzhe Li ZnV0dXJlbG16MTIzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.