- 1Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 2Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Disease and Zoonoses, Yangzhou University, Yangzhou, China
The recombinant bivalent live vectored vaccine rPRRSV-E2 has been proved to be a favorable genetic engineering vaccine against classical swine fever (CSF) and highly pathogenic porcine reproductive and respiratory syndrome (HP-PRRS). NADC30-like strains have recently emerged in China and caused severe disease, and it is necessary to evaluate the vaccine candidate for the currently circulating viruses. This study established a good challenge model to evaluate the candidate rPRRSV-E2 vaccine in preventing infection with a representative NADC30-like strain (ZJqz21). It was shown that the challenge control piglets displayed clinical signs typical of PRRSV, including a persistent fever, dyspnea, moderate interstitial pneumonia, lymph node congestion, and viremia. In contrast, the rPRRSV-E2 vaccination significantly alleviated the clinical signs, yielded a high level of antibodies, provided adequate protection against challenge with ZJqz21, and inhibited viral shedding and the viral load in target tissues. Our results demonstrated that the recombinant bivalent live vectored vaccine strain rPRRSV-E2 can provide efficient protection against the challenge of heterologous circulating NADC30-like strain and could be a promising vaccine candidate for the swine industry.
Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is the causal agent of the porcine reproductive and respiratory syndrome (PRRS) and is one of the most economically infectious illnesses of the swine industry worldwide. PRRSV belongs to the order Nidovirales, family Arteriviridae and can be classified into PRRSV-1 and PRRSV-2 with Lelystad and VR-2332 as representative strains (Cavanagh, 1997; Lunney et al., 2016). These two genotypes share approximately 60% nucleotide identity (Darwich et al., 2011).
PRRSV was first reported in 1996 in China. Over the past 20 years, PRRSV has been the cause of major diseases in the swine industry, and PRRSV-2 strains are highly predominant within the field (An et al., 2007). After 2006, highly pathogenic PRRSV (HP-RRRSV) became the predominantly epidemic strain on the farms (Tian et al., 2007, 2009; Zhou et al., 2008). Since 2013, PRRSV infection during routine clinical detection has significantly increased. The recurrent PRRS pandemic was attributed to the emergence of NADC30-like strains, which were likely imported from North America and rapidly spread widely throughout China (Zhao et al., 2015; Zhou et al., 2015; Li et al., 2017). The NADC30-like and HP-PRRSV-like strains currently circulate on pig farms and have high clinical detection rates (Li et al., 2016; Guo et al., 2019).
According to the global PRRSV classification system, the abundance of PRRSV-2 strains in China can be classified into four lineages: NADC30-like (lineage 1), QYYZ-like (lineage 3), VR-2332-like (lineage 5), and JXA1-like/CH-1a-like (lineage 8) lineages (Shi et al., 2010a,b). Extensive genetic variations exist between the strains in each viral subtype, and genetically distinct PRRSV variants display different clinical characteristics and virulence (Guo et al., 2018). rPRRSV-E2 was a recombinant bivalent live vectored vaccine and capable of expressing the classical swine fever virus (CSFV) E2 protein in the backbone of a commercial modified live virus (MLV) vaccine strain, HuN4-F112, which was classified into lineage 8 (Gao et al., 2018). rPRRSV-E2 could provide 100% protection against HP-PRRSV (lineage 8), but it remained to be answered about the protective effect of rPRRSV-E2 for resisting challenge with NADC30-like (lineage 1) strain.
NADC30-like PRRSVs are not as pathogenic as HP-RRRSV but can be distinguished by a greater probability of recombination with other PRRSV strains, leading to different virulence (Zhang et al., 2016). Outbreaks of NADC30-like PRRSV in vaccinated swine herds indicate that the current commercial PRRSV vaccines cannot completely protect against infection with NADC30-like strains (Tian, 2017). This study evaluated the current vaccine candidate rPRRSV-E2 against infection with a heterologous strain and demonstrated that rPRRSV-E2 vaccination exerted superior efficiency at decreasing the clinical signs and viral infection NADC30-like strain in piglets.
Materials and Methods
Virus Isolation and Viral Characteristics Analyses
The disease materials were obtained from a pig farm in Zhejiang province, China, in 2020. Three sick piglets displayed high fever, dyspnea, and tachypnea. Lung samples were ground and subjected to three freeze-thaw cycles in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA). Samples were detected by PRRSV ORF5-specific primers, and the results appeared to be positive (data not shown). (PAMs) were infected with the supernatants for 1 h and subsequently cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco). Daily observations were performed until obvious cytopathic effects (CPEs) appeared. Specific monoclonal antibodies termed SDOW17 (Rural Technologies) were used for an indirect immune fluorescent assay (IFA) to confirm the presence of the PRRSV N protein.
Complete Genome Sequencing and Phylogenetic Analysis of ZJqz21
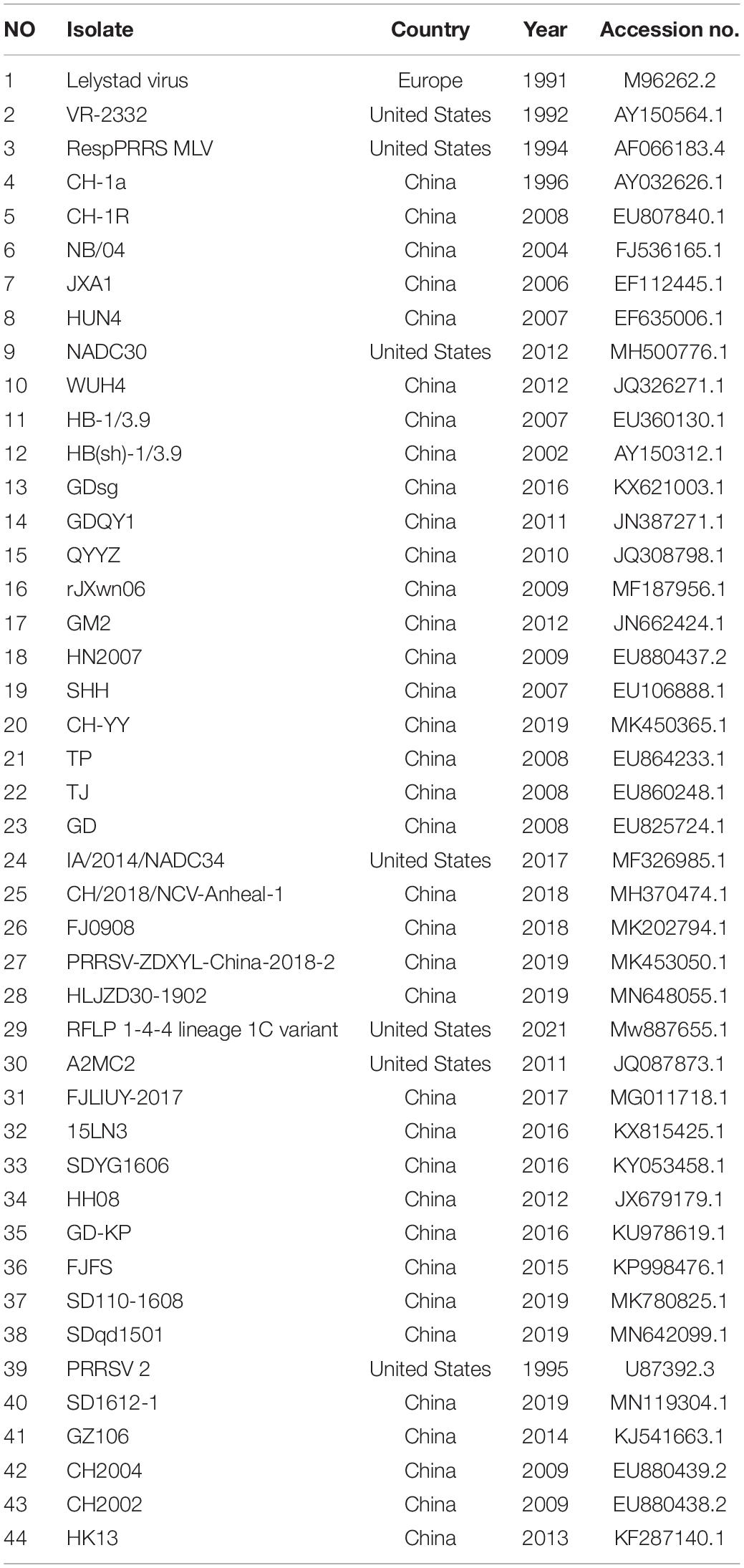
The total RNA was extracted from ZJqz21-infected PAMs using TRIzol (Invitrogen, Thermo Fisher Scientific, Waltham, MA). A PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) was used for reverse transcription. The complete ZJqz21 genome was amplified by seven overlapping primers using the cDNA template. The sequences of all primers used for genome amplification will be made available upon request. Purified PCR products were sequenced by Personalbio (Shanghai, China). A total of 44 representative PRRSV isolates were collected from the GenBank database (Table 1), and multiple sequence alignments were performed using Clustal X. Phylogenetic trees of the complete genome and ORF5 sequences were constructed by MEGA 6.0 using the neighbor-joining method. Bootstrap confidence values from 1,000 replicates were used to analyze the evolutionary relationship of ZJqz21.
Viruses Used for Vaccination and Challenge
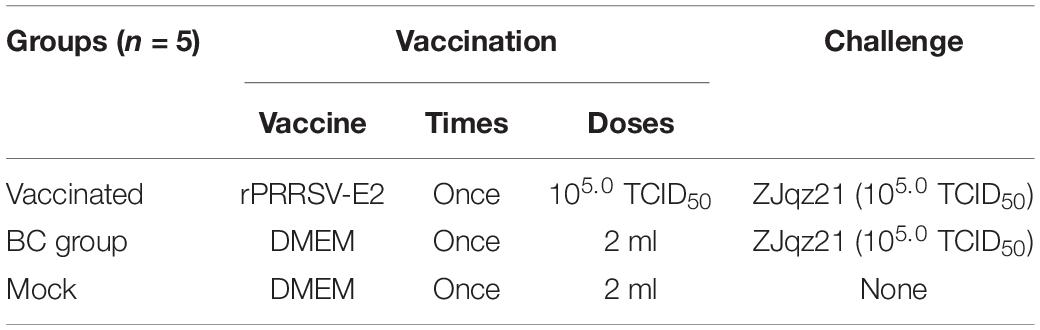
PRRSV-permissive MARC-145 cells were cultured and maintained in Minimum Essential Medium Eagle (MEM; Sigma-Aldrich Corporation, CA, United States) with 10% FBS or 2% FBS at 37°C under humidity of 5% CO2. The vaccination strain, rPRRSV-E2, and challenge virus strain, ZJqz21, were propagated and obtained when approximately 80% CPE of the MARC-145 cells appeared. The two viruses were titrated and used for vaccination and challenge, respectively. The doses used in the experiments are shown in Table 2.
Evaluation of rPRRSV-E2 Immune Efficacy Against ZJqz21 in Piglets
A professional veterinary pathologist was invited to guide and perform professional animal experimental operations. A total of 15, 30-day-old landrace PRRSV- and CSFV-free piglets were selected for immune efficacy experiments to assess the effect of rPRRSV-E2 against ZJqz21 and divided into three groups. Each group contained five piglets fed separately, as shown in Table 2. A 2 ml dose of rPRRSV-E2 (dosage: 105.0 TCID50) was used to vaccinate each piglet in the vaccinated and challenge control piglets (BC groups; PRRSV S/P < 0.4; n = 5) and 2 ml of ZJqz21 (dosage: 105.0 TCID50) was inoculated at 28 days post-vaccination (dpv). A cervical intramuscular injection was used for vaccination and challenge. Each piglet was injected with 2 ml DMEM in the mock group. General health status and clinical symptoms were monitored daily, and the rectal temperatures were measured daily during the experimental period. PRRS clinical symptoms were recorded and scored as described previously (Halbur et al., 1995; Thanawongnuwech et al., 2000; Li et al., 2014). All surviving piglets were euthanized and autopsied 21 days post-infection (dpi). The sera were collected every 7 days after rPRRSV-E2 vaccination. After ZJqz21 inoculation, sera were collected every 2 or 3 days. Real-time RT-qPCR evaluated the level of viral RNA replication in the serum samples containing PRRSV viremia and virus shedding and viral load in the tissues (Jiang et al., 2015; Gao et al., 2018, 2020b). An evaluation of lung lesions in each of the three groups was performed as previously described (Jiang et al., 2015; Gao et al., 2018, 2020a). For pathological observations, a pathological section analysis and detection of the viral load in the tissues of piglets infected with the ZJqz21 strain was performed. The heart, lung, inguinal lymph nodes, mesenteric lymph nodes, mandibular lymph nodes, spleen, kidneys, and tonsils were collected from each piglet.
Detection of Specific Humoral Immunity After Vaccination and Challenge
The PRRSV- and CSFV-specific antibody titers in the serum samples collected at the specified time points were tested with the commercial ELISA kits (IDEXX Laboratories, Westbrook, No. 06-40959-04 and No. 06-43230-06) as previously described (Gao et al., 2018). Based on these values, the level of humoral immunity was displayed by drawing curves with GraphPad Prism 6.0.
Determination of the Viral Load in Tissues
Tissue samples from the heart, lung, inguinal lymph nodes, mesenteric lymph nodes, mandibular lymph nodes, spleen, kidneys, and tonsils were collected by multi-point sample collection. A certain amount of each tissue sample was gathered, 1 ml of sterile PBS was added for tissue homogenates, and grinding treated tissue homogenates were centrifuged at 4°C (9,500 × g for 10 min), and the supernatant was cryopreserved in a 1.5 ml centrifuge tube. The RNA of 200 μL-treated homogenate supernatant was extracted using the QIAcube HT nucleic acid autotractor according to the QIAcube HT Plasticware kit instructions. RT-qPCR was performed, and Shanghai Sunny Biotechnology Co., Ltd., synthesized the primer pairs and probes.
Statistical Analysis of Experimental Data
A Student′s t-test was used for statistical differences analysis in the study. SPSS 14.0 and GraphPad Prism software (version 6.0) was used to analyze and chart the clinical symptom scores, lung lesion scores, viral shedding, and viral load values. A P-value < 0.05 was considered to be statistically significant.
Results
ZJqz21 Was Identified as a Heterologous Strain to rPRRSV-E2
To evaluate the immune efficacy of rPRRSV-E2 against the heterologous strain, we first identified a NADC30 epidemic strain. In 2020, ZJqz21 was successfully isolated from PAMs from a RespPRRS MLV-vaccinated piglet in China. PRRSV-specific CPE was observed in the P2 passage, and the existence of PRRSV was further identified by IFA (Figure 1A). As shown in Figure 1A, ZJqz21 could cause severe CPE. An IFA analysis was carried out on MARC-145 cells, or PAMs infected with ZJqz21 (Figure 1A). The viral growth characteristics of ZJqz21 are presented by multi-step growth curves and a plaque morphology analysis (Figures 1B–D).
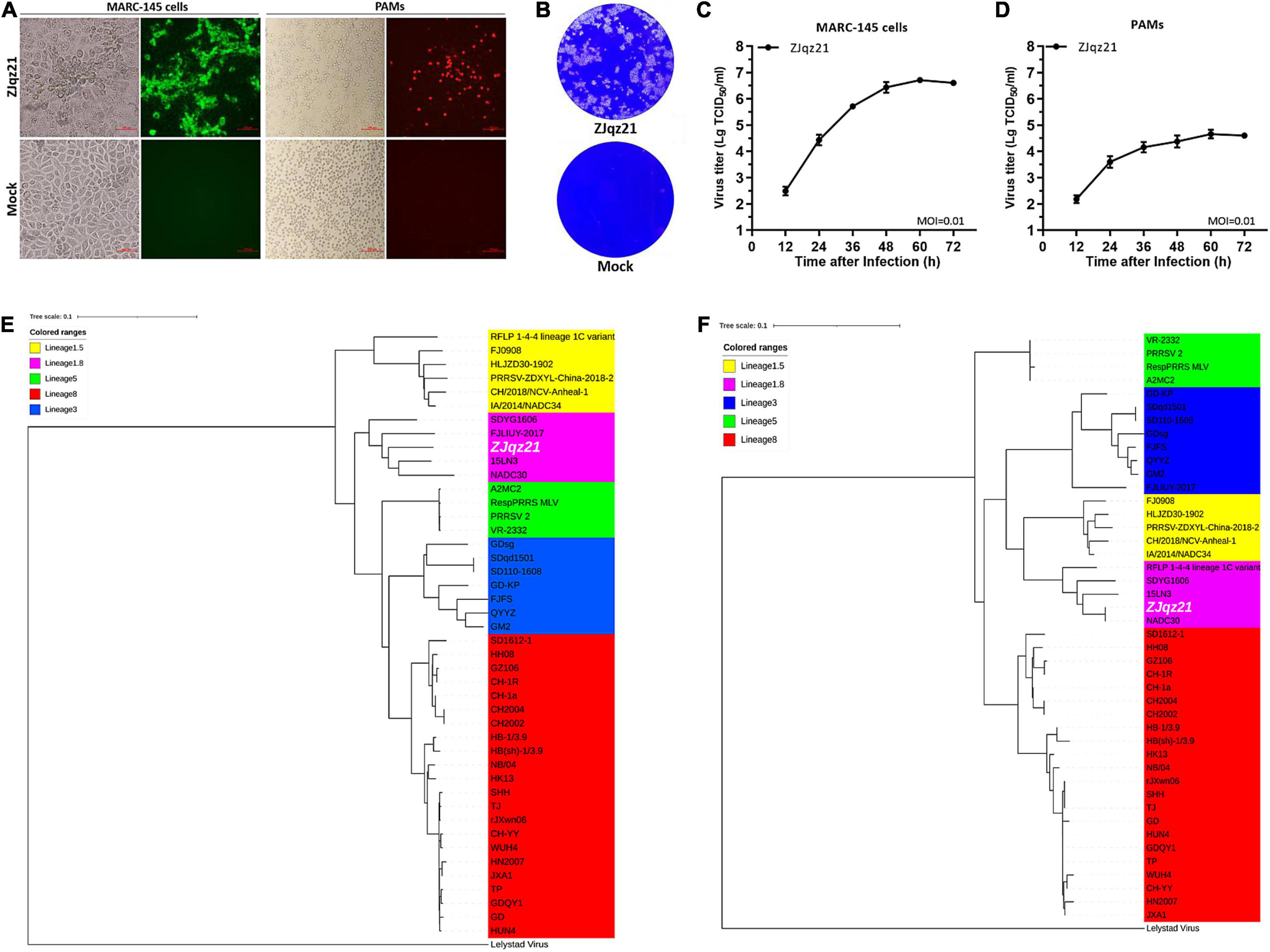
Figure 1. (A) Cytopathic effects (CPE) and immunofluorescence staining against the N protein in Primary porcine alveolar macrophages (PAMs) and MARC-145 cells at 36 hpi with ZJqz21 (MOI = 0.1). Scale bar = 100 μm. (B) Plaque morphology. MARC-145 cells were infected with ZJqz21 (MOI = 0.001). The mock control represented non-infected MARC-145 cells. The MARC-145 cells were stained with crystal violet at 5 days post-infection. Virus growth kinetics in MARC-145 cells (C) and PAMs (D). Data represent the mean ± standard deviation (SD) of three independent experiments. Phylogenetic trees of ZJqz21 based on the complete genome (E) and GP5 (F) nucleotide sequences. A white color highlighted the ZJqz21 strain. The scale bar indicated the number of nucleotide substitutions per site.
The complete sequences of the ZJqz21 genome have been uploaded into GenBank (accession no. OK274266). The complete ZJqz21 genome is 15,042 nt in length, including poly(A) tails. The ZJqz21 genome shared 86.9, 87.0, 84.4, 86.8, and 90.1% homology in the nucleotide sequence with CH-1a, JXA1, GM2, VR2332, and NADC30, respectively (Table 3). Each region of the ZJqz21 genome was further compared with those representative strains, and the results showed that ZJqz21 shared the highest identity in each region with the NADC30 strain. Amino acid sequence alignment results revealed that Nsp2 of ZJqz21 has a discontinuous 131-aa (111 + 1 + 19-aa) deletion compared with VR-2332 (data not shown), which are the typical characteristics of the NADC30-like PRRSV. The ORF5 of ZJqz21 displayed higher nucleotide and amino acid identity (94.7%, 93.0%) with NADC30 (Table 3).
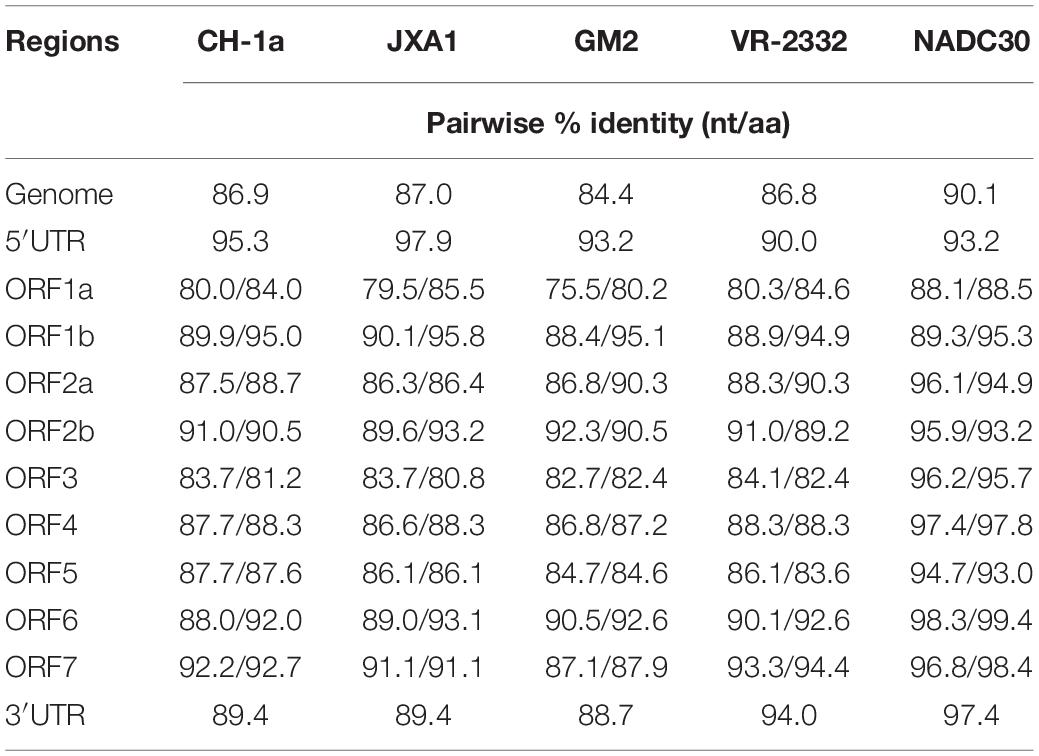
Table 3. Nucleotide and amino acid sequence identity (%) of ZJqz21 as compared to five representative PRRSV strains.
Phylogenetic trees were constructed according to the complete genome and ZJqz21 ORF5 sequences. The results demonstrated that PRRSV-2 strains could be divided into the following five lineages: RFLP 1-4-4 (lineage 1.5), NADC30-like (lineage 1.8), VR-2332-like (lineage 5), GM2-like (lineage 3), and JXA1-like/CH-1a-like (lineage 8) based on the complete genome sequence. ZJqz21 was classified into lineage 1.8, represented by NADC30 based on full nucleotide genome and ORF5 sequences (Figures 1E,F). These findings revealed that ZJqz21 belonged to lineage 1.8 and had characteristics of the NADC30 strain, which can be used as the heterologous challenge strain to rPRRSV-E2.
rPRRSV-E2 Protected the Piglets From the Challenge of the Heterologous NADC30-Like Strain ZJqz21
Next, the efficacy of rPRRSV-E2 against the heterologous strain ZJqz21 in piglets was tested. The BC group and vaccinated piglets (vaccinated group) were challenged with the ZJqz21 strain. In the BC group, the body temperature rose at 3 days post-challenge (dpc) and reached the peak temperature at about 11 dpc. Among them, three of the pigs reached 41.2°C. The rectal temperatures of the piglets in the mock group were below 40.5°C throughout the experimental period. In the vaccinated group, one piglet developed symptoms of fever, which lasted from 8 to 12 dpc, before returning to normal. The rectal temperature of the other pigs in this group was regular (Figure 2A). Piglets in the BC group showed typical PRRS symptoms (e.g., high fever, cough, loss of appetite, lethargy, and frequent paralysis) from 4 dpc (Figure 2B). The clinical sign scores in the BC group from 8 to 21 dpc were significantly higher than those in the vaccinated and mock groups (P < 0.001, Figure 2B). Of note, all ZJqz21-infected piglets survived throughout the study. The ZJqz21 strain was found to be a moderately virulent isolate. Compared with piglets in the BC group, the piglets vaccinated with rPRRSV-E2 were challenged with the ZJqz21 strain, and there were almost no signs of a high fever or other abnormal clinical symptoms. Only one pig in the vaccinated group developed symptoms of clinical fever, which lasted for 5 days. The rectal temperature was approximately 41°C; however, the mental state was normal and did not show serious clinical symptoms (Figure 2A).
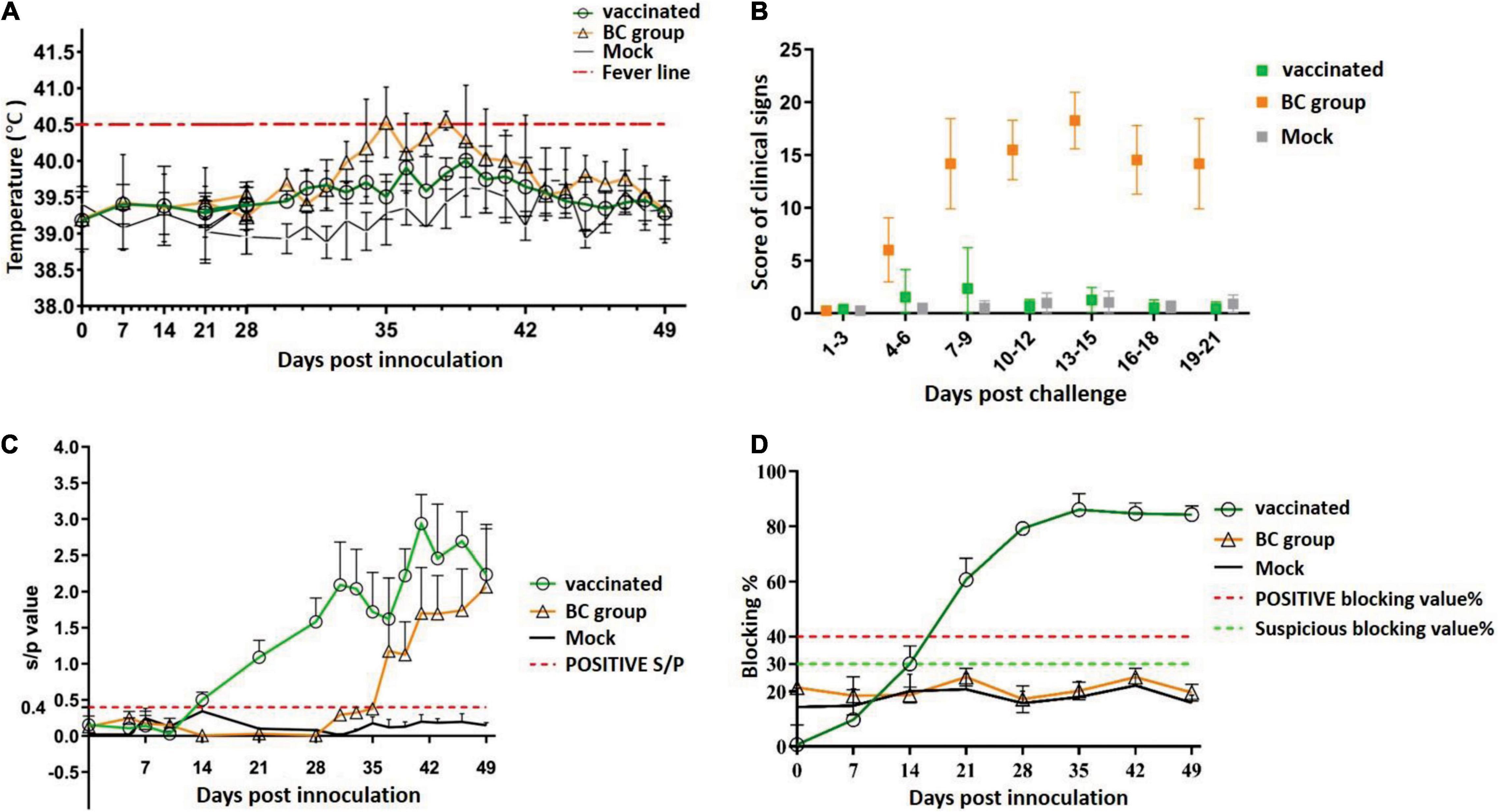
Figure 2. Serological response after rPRRSV-E2 vaccination and scoring of clinical signs following challenge with the novel NADC30-like strain, ZJqz21. (A) Daily rectal temperature record and analysis following rPRRSV-E2 vaccination and ZJqz21 challenge. (B) Scoring and charting for clinical symptoms following rPRRSV-E2 vaccination and ZJqz21 challenge. Scoring data were indicated as the means ± standard deviations. (C) The S/P value assessed the PRRSV-specific humoral immune response (IDEXX PRRS X3) identified from serum samples collected at the indicated time points from piglets in the vaccinated, BC, and mock groups. (D) CSFV-specific antibodies in the three groups were tested in the serum samples collected in the experiments using an IDEXX CSFV Ab ELISA kit.
The antibody reaction induced by rPRRSV-E2 and the subsequent challenge by ZJqz21 were examined to determine the protective efficacy of rPRRSV-E2 against the heterologous strain ZJqz21. Based on the serum collected at 0, 7, 14, 21, 28, 35, 42, and 49 dpi, the immune response to PRRSV and CSFV was analyzed. The group inoculated with rPRRSV-E2 produced PRRSV-specific immune effects. All of the piglets vaccinated with rPRRSV-E2 showed seroconversion by 14 dpv, and the average S/P showed a peak of 1.75 ± 0.39 at 28 dpv (Figure 2C), demonstrating that rPRRSV-E2 vaccination-induced high-level PRRSV-specific antibodies. At the same time, the level of CSFV-specific antibodies in the piglets inoculated with rPRRSV-E2 began to increase at 7 dpv and rapidly climbed to the average peak titer at 35 dpv. At about 2 weeks after vaccination with the rPRRSV-E2 vaccine, CSFV-specific antibodies in piglets were seroconverted in the vaccination group and reached 82.6 ± 2.1% after 28 dpv (Figure 2D). Following the viral challenge, the S/P values indicated that the PRRSV-specific antibodies in the BC group increased from 10 dpc and were higher than those of the mock group (Figure 2C). During the study period, PRRSV-specific and CSFV-specific antibodies in the piglets of the mock group remained negative (Figures 2C,D).
rPRRSV-E2 Vaccination Alleviated the Clinical Signs Typical of PRRSV Caused by ZJqz21
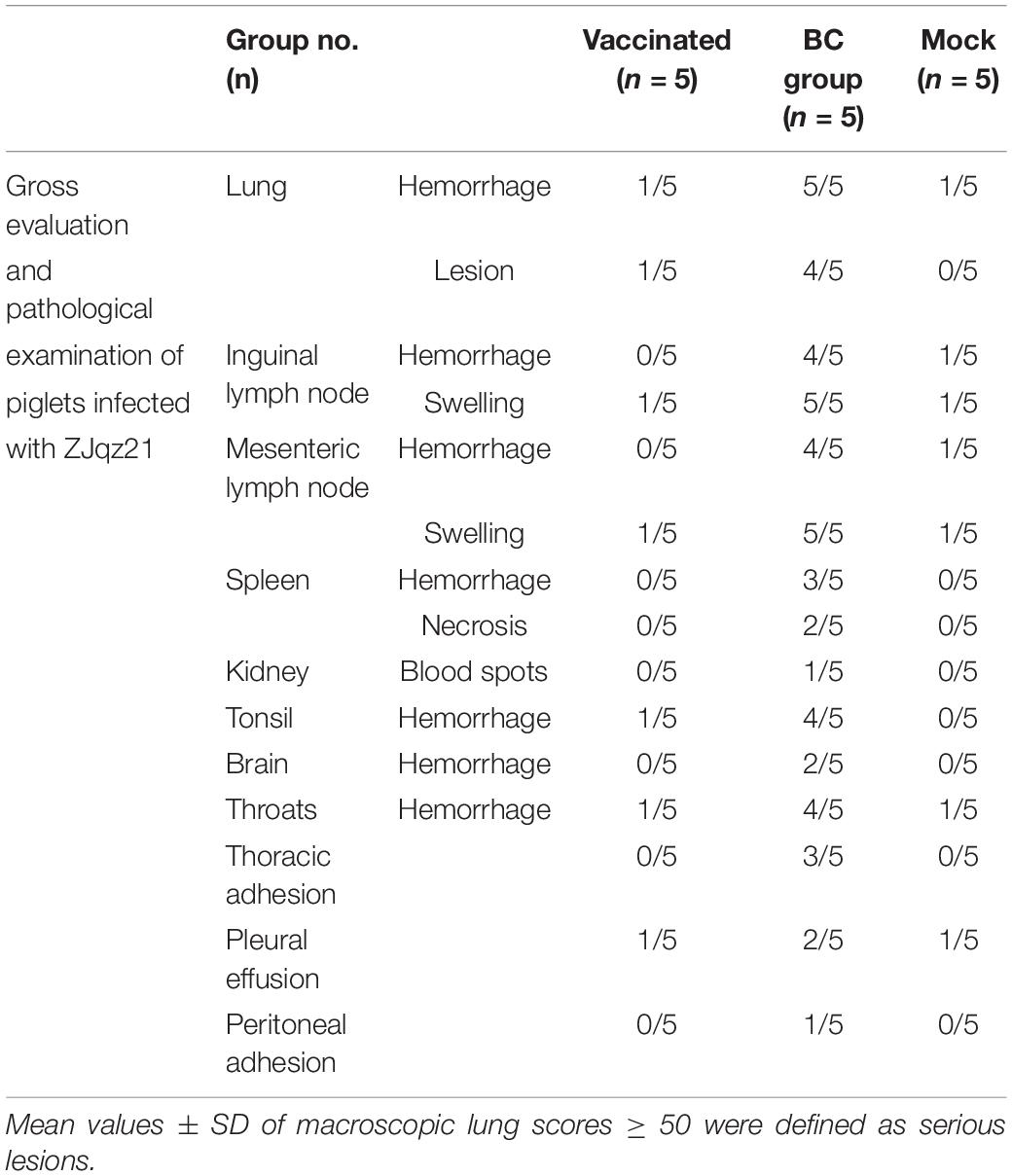
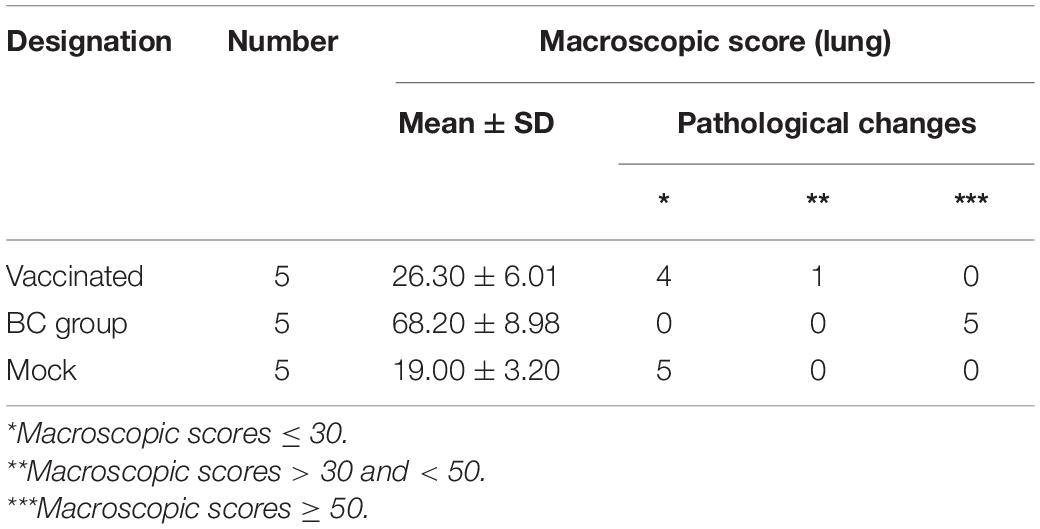
All piglets in rPRRSV-E2 vaccinated, and mock groups were generally normal and did not show obvious pathological damage typical of PRRS disease. Piglets inoculated with the ZJqz21 strain (BC group) showed severe pathological changes, including tonsil necrosis and hemorrhage, congestion, and swelling of the lymph nodes, bladder, and petechial kidney hemorrhage. Table 4 describes the overall pathological changes in the three groups. Typical pathological signs of PRRSV were observed in the BC group, such as lung lesions, enlarged bleeding of the mesenteric and inguinal lymph nodes, hemorrhage and necrosis of the spleen, and hemorrhaging in the throat and kidney, and tonsil (Table 4). An analysis of the macroscopic score in the lungs showed that infected piglets in the BC group showed severe pathological changes in the lung, significantly higher than those in the inoculation and mock groups (Table 5). The BC group displayed typical interstitial pneumonia and obvious microscopic lung lesions, including exfoliated epithelial cells in the bronchioles and collapsed alveoli infiltrating numerous inflammatory cells in alveolar spaces (Figure 3). The lymph nodes of piglets inoculated with ZJqz21 showed lower proliferation of cortical lymphocytes and enlargement and hyperplasia of the lymphatic nodules (Figure 3). During the entire study period, neither macroscopic nor histological changes in the lung and lymph node lesions were observed in the vaccinated piglets and negative control piglets (Figure 3), indicating that rPRRSV-E2 vaccination significantly alleviated the clinical signs caused by ZJqz21.
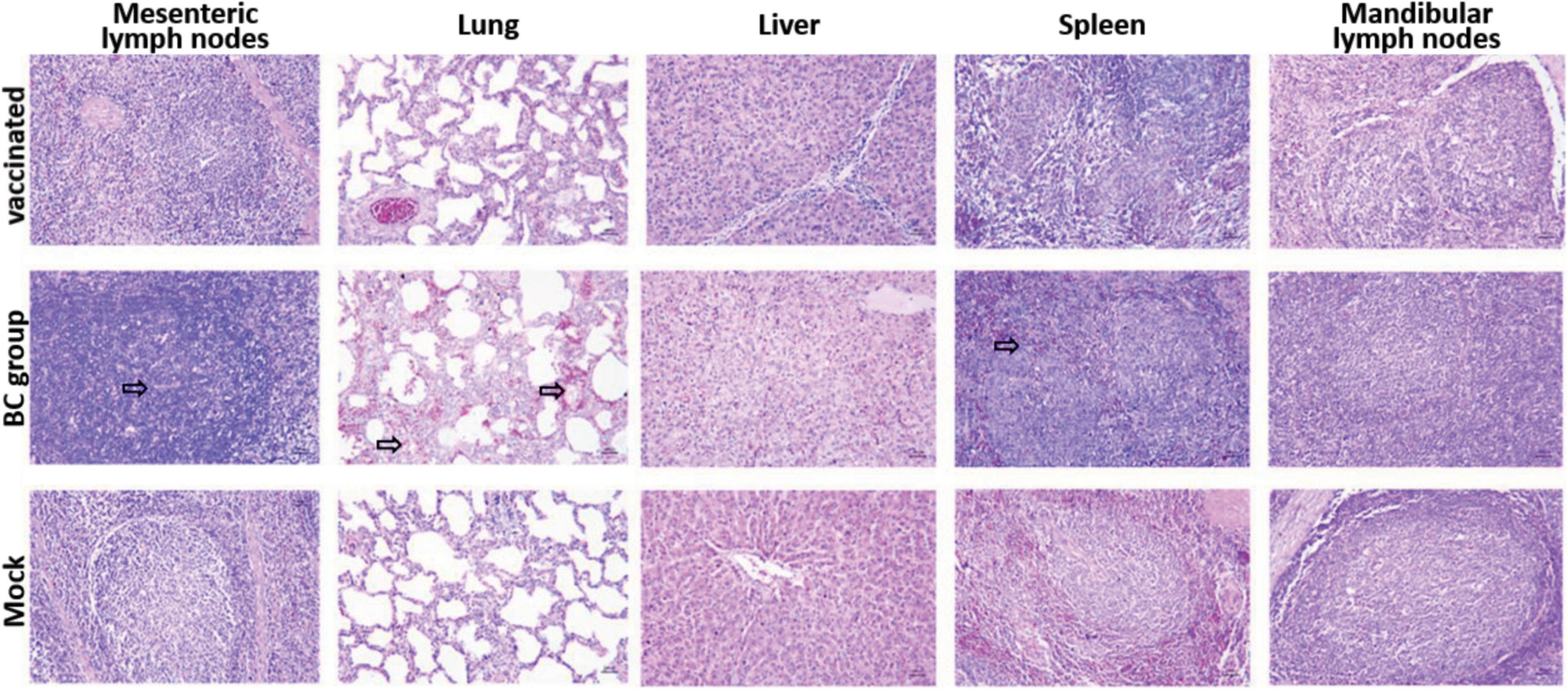
Figure 3. Histopathology analysis by hematoxylin-eosin staining in three groups. Each group’s microscopic figures of the lung, liver, spleen, mesenteric lymph nodes, and mandibular lymph nodes are shown and observed at a magnification of × 200. The infiltration of inflammatory cells around the alveolar septa and alveolar spaces was shown clearly by black arrows.
rPRRSV-E2 Vaccination Decreased Viral Replication and Virus Shedding of ZJqz21 in Piglets
RT-qPCR was used to monitor the viral titer in the sera and excreta (including nasal and anal swab samples) and to monitor and evaluate the viral load in tissues and organs. Viremia was detected in all three groups. In the BC group, all piglets inoculated with the ZJqz21 strain showed viremia, which started from 3 dpc and continued to 21 dpc. One of five vaccinated piglets in the vaccinated group exhibited PRRSV-viremia, which started from 3 dpc and lasted for 5 days. Viremia was lower than the negative line at 10 dpc and decreased to an undetectable level until the end of the experiment (Figure 4A), indicating that rPRRSV-E2 vaccination mitigated the viremia caused by ZJqz21. During the entire experiment, it was determined that the Ct value related to virus shedding was close to the detection limit for the nasal and anal samples from the vaccinated group. The level of virus shedding identified in both the nasal and anal swabs of the BC group was high (Figures 4B,C). The numerical values in the viral load of the tissues and organs exhibited similar trends with the detection of virus shedding (Figure 4D). The viral load was extremely high in the mesenteric, inguinal, mandibular lymph nodes, and tonsils of the BC group, indicating that rPRRSV-E2 vaccination also inhibited virus shedding of ZJqz21.
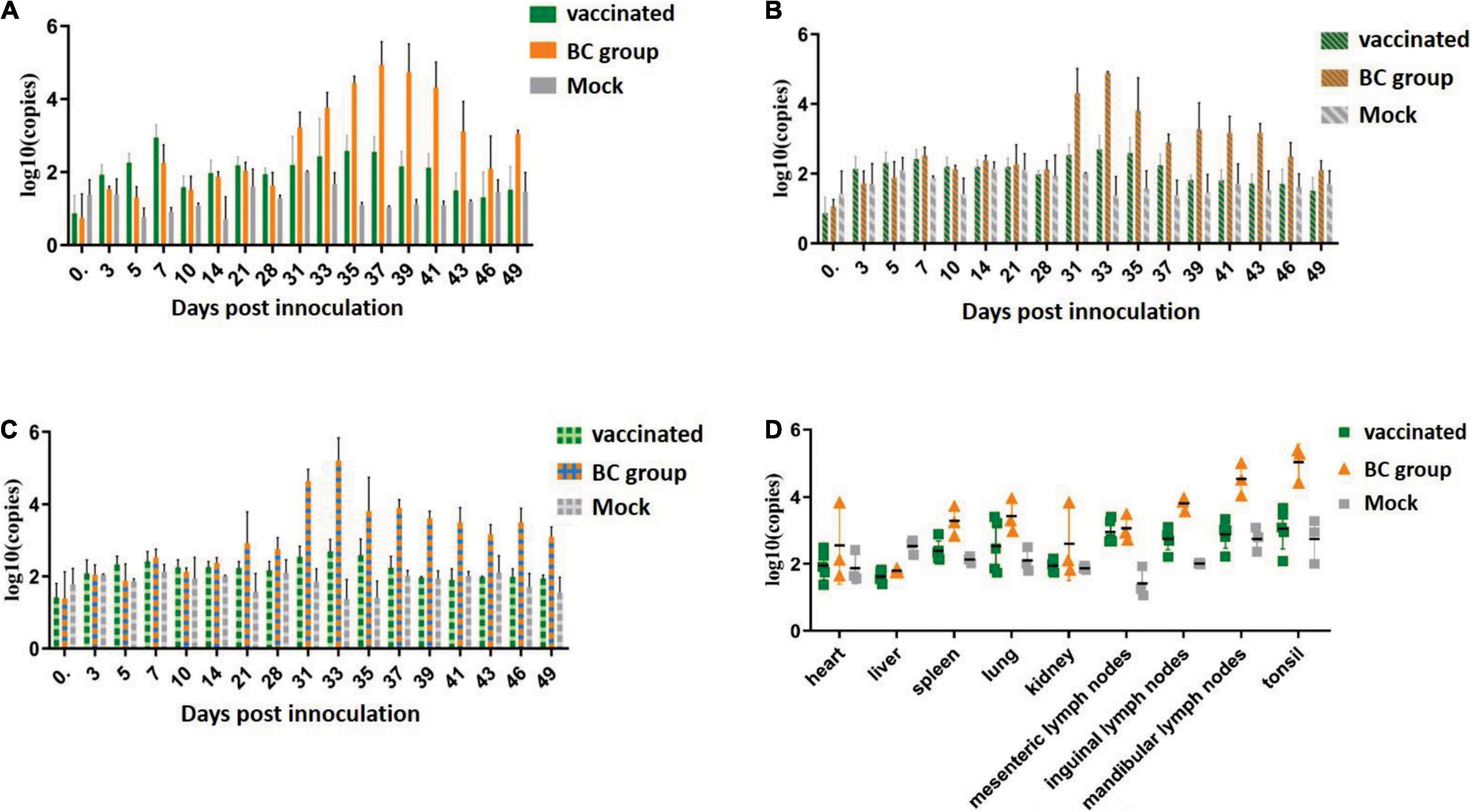
Figure 4. Detection of viremia, virus shedding, and viral load in the tissues by RT-qPCR. (A) Viremia detection in experiments assessing the immune efficacy of rPRRSV-E2 against ZJqz21. (B) Identification of viral RNA in nasal swabs to assess viral shedding. (C) Viral RNA detection in anal swabs to assess viral shedding. (D) Detection of the viral load in the lungs, heart, spleen, liver, kidneys, mesenteric lymph nodes, inguinal lymph nodes, mandibular lymph nodes, and tonsils.
Discussion
PRRS is an important swine disease rapidly becoming prevalent worldwide and has caused huge economic losses to the pig industry since the 1990s (Lunney et al., 2010). In 2006, there was an HP-PRRSV outbreak in China, posing a serious threat to the Chinese pig industry (Tian et al., 2007, 2009; Zhou et al., 2008). Subsequently, HP-PRRSV with a 30-aa discontinuous deletion in the Nsp2-coding region gradually emerged as the major epidemic strain in China (Zhou et al., 2009, 2014). The emergence of the new lineage 3 (QYYZ-like) and lineage 1 (NADC30-like) viruses in mainland China in 2010 and 2013, respectively, complicated the epidemic situation of PRRS (Lu et al., 2015; Zhao et al., 2015; Zhou et al., 2015). The diversity of PRRSV strains poses a substantial challenge to protecting existing live vaccines. The NADC30-like strain, which is easy to recombine with other PRRSV strains, has become more complex and challenging on pig farms since it became popular in China but shows moderate or relatively low clinical virulence. Since its first outbreak, researchers in China have paid close attention to NADC30-like PRRSV.
Seven commercial MLV vaccines are currently used to control and prevent PRRSV infection in China with their corresponding virus strains, including Ingelvac PRRS MLV/RespPRRS MLV (VR-2332), CH-1R (parental strain CH-1a), HuN4-F112 (HuN4), JXA1-P80 (JXA1), R98 (R98), TJM-F92 (TJ), and GDr180 (GD). These PRRS MLV vaccines are obtained from the classical or HP-PRRSV strains, which can provide adequate protection against genetically homologous strains (Wang et al., 2021); however, the protective effect of MLV vaccines for resisting challenge with heterologous strains is far from conclusive. Zhou et al. (2017) found that the cross-protective efficacy of three different attenuated vaccine strains (including an HP-PRRSV attenuated vaccine strain) against the NADC30-like strain was extremely limited against infection with the NADC30-like virus. Among these strains, Ingelvac PRRS MLV was a commercial vaccine based on the classical PRRSV-2 strain that appeared to exert superior efficiency at reducing clinical fever (Zhou et al., 2017). As a recombinant bivalent live vectored vaccine, rPRRSV-E2 was constructed from the PRRS vaccine virus HuN4-F112, providing 100% protection against HP-PRRSV and CSFV (Tian et al., 2009; Gao et al., 2018). This study demonstrated that rPRRSV-E2 could resist the heterologous NADC30-like strain ZJqz21 infection. Vaccination with rPRRSV-E2 provided good piglet protection and decreased viral replication and virus shedding against heterologous infection with PRRSV strain ZJqz21 (Figures 2–4).
NADC30-like PRRSVs are epidemic in numerous provinces in China. The current commercial vaccines show limited protection in infected piglets. The extensive recombination phenomenon among NADC30-like PRRSVs is a distinctive characteristic of PRRSV (Wang et al., 2018; Zhou et al., 2018; Han et al., 2020). In dealing with complex epidemic situations, there is an urgent need to establish a good challenge model to evaluate the current vaccine candidates in preventing infection with a heterologous strain. In this study, we isolated a novel PRRSV strain, ZJqz21, and identified its genomic characterization, pathogenicity, and application as a challenge model for the heterologous protection of rPRRSV-E2. ZJqz21 was identified and clustered into the NADC30-like lineage (Figure 1). High genetic diversity is a significant characteristic of PPRSV. A previous study constructed the global classification system of PRRSV based on a comprehensive analysis of the complete ORF5 gene sequence. PRRSV-1 was divided into three subtypes (subtypes 1-3) according to the classification system, and PRRSV-2 was classified into 9 lineages with several sublineages in each lineage (Shi et al., 2010b; Zhou et al., 2014). ZJqz21 had a discontinuous 131-aa (111 + 1 + 19-aa) deletion in Nsp2 compared to VR-2332, which was identical to NADC30. A phylogenetic analysis of the full genome and ORF5 sequences showed that ZJqz21 was classified as lineage 1.8 with genetic features from NADC30 (Figure 1). ZJqz21 and NADC30 also belonged to lineage 1.8. The structural proteins only presented 5–8% differences between each other in the amino acid sequence (Table 3). NADC30-like PRRSVs have been recognized as moderately pathogenic isolate strains (Tian, 2017). The pathogenicity of ZJqz21 was evaluated to explore whether it could be applied as the challenge model for the heterologous protection of PRRSV candidate vaccines. Our results showed that none of the piglets died upon challenge with ZJqz21; however, all piglets exhibited typical PRRS clinical symptoms, including fever, cough, inappetence, dyspnea, and tachypnea (Tables 4, 5). Microscopic lung lesions and interstitial pneumonia were observed in ZJqz21-infected piglets (Figure 3). The above results demonstrated that the novel ZJqz21 strain exhibited moderate virulence in piglets and could be used as a challenge model to evaluate the current vaccine candidates in preventing infection with NADC30-like strain.
Nevertheless, in this experiment, rPRRSV-E2 could not perfectly prevent the symptoms of clinical fever after a challenge with the ZJqz21 strain. One out of five pigs in the vaccinated group exhibited clinical fever from day 5 dpc, which lasted for 5 days, and then returned to normal. However, the rectal temperature was not particularly high at around 41°C. Although one piglet in the rPRRSV-E2 vaccinated group had fever symptoms, there were no other more serious clinical symptoms, and there were no significant differences in the clinical symptoms and lung lesions between the vaccinated and mock group (Table 4). There were no typical pathological PRRSV-specific changes in histopathologic slides’ results (Figure 3).
Moreover, there were no significant differences in viremia, viral shedding, or viral load between this pig and other pigs in the same group and those in the mock group (Figure 4). This finding may be related to the infection of piglets with other pathogens. Future studies should confirm that our vaccine candidate strain rPRRSV-E2 displays a good cross-protection effect against heterologous strains and is a good choice for clinical prevention and control of NADC30-like strains.
In conclusion, the current vaccine candidate rPRRSV-E2 can provide good protection against the attack of the moderately pathogenic heterologous PRRSV strain ZJqz21 in piglets. rPRRSV-E2 vaccination alleviated the clinical signs, viral replication, and virus shedding of ZJqz21 in piglets. Our research emphasizes the importance of developing a broad-spectrum vaccine to prevent and control further complicated PRRS epidemics in China.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
All experimental programs involving pigs were carried out in accordance with the Guidelines for the Nursing and Use of Experimental Animals, and approved by the Ethics Committee of Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (number SV-20201021-Y02).
Author Contributions
FG and GT conceived and designed the study. LL and JC wrote the main manuscript text and prepared the figures. ZC, YC, ZG, SQ, and JL performed the experiments. WT, YZ, GL, YJ, CL, and LY prepared the manuscript. All authors reviewed the manuscript and participated in the experiments.
Funding
We are very grateful for the support from the Key-Area Research and Development Program of Guangdong Province (2019B020211003), the Natural Science Foundation of Shanghai (21ZR1476900), the Central Public-Interest Scientific Institution Basal Research Fund (2021JB01 and Y2020YJ15), and the National Natural Science Foundation of China (31941017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, T. Q., Zhou, Y. J., Liu, G. Q., Tian, Z. J., Li, J., Qiu, H. J., et al. (2007). Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: coexistence of two NA-subgenotypes with great diversity. Vet. Microbiol. 123, 43–52. doi: 10.1016/j.vetmic.2007.02.025
Cavanagh, D. (1997). Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142, 629–633.
Darwich, L., Gimeno, M., Sibila, M., Diaz, I., de la Torre, E., Dotti, S., et al. (2011). Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet. Microbiol. 150, 49–62. doi: 10.1016/j.vetmic.2011.01.008
Gao, F., Jiang, Y., Li, G., Li, L., Zhang, Y., Yu, L., et al. (2020a). Evaluation of immune efficacy of recombinant PRRSV vectored vaccine rPRRSV-E2 in piglets with maternal derived antibodies. Vet. Microbiol. 248:108833. doi: 10.1016/j.vetmic.2020.108833
Gao, F., Jiang, Y., Li, G., Zhang, Y., Zhao, K., Zhu, H., et al. (2020b). Immune duration of a recombinant PRRSV vaccine expressing E2 of CSFV. Vaccine 38, 7956–7962. doi: 10.1016/j.vaccine.2020.10.057
Gao, F., Jiang, Y., Li, G., Zhou, Y., Yu, L., Li, L., et al. (2018). Porcine reproductive and respiratory syndrome virus expressing E2 of classical swine fever virus protects pigs from a lethal challenge of highly-pathogenic PRRSV and CSFV. Vaccine 36, 3269–3277. doi: 10.1016/j.vaccine.2018.04.079
Guo, Z., Chen, X. X., Li, R., Qiao, S., and Zhang, G. (2018). The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol. J. 15:2. doi: 10.1186/s12985-017-0910-6
Guo, Z., Chen, X. X., Li, X., Qiao, S., Deng, R., and Zhang, G. (2019). Prevalence and genetic characteristics of porcine reproductive and respiratory syndrome virus in central China during 2016-2017: nADC30-like PRRSVs are predominant. Microb Pathog. 135:103657. doi: 10.1016/j.micpath.2019.103657
Halbur, P. G., Paul, P. S., Frey, M. L., Landgraf, J., Eernisse, K., Meng, X. J., et al. (1995). Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet. Pathol. 32, 648–660. doi: 10.1177/030098589503200606
Han, G., Lei, K., Xu, H., and He, F. (2020). Genetic characterization of a novel recombinant PRRSV2 from lineage 8, 1 and 3 in China with significant variation in replication efficiency and cytopathic effects. Transbound Emerg. Dis. 67, 1574–1584. doi: 10.1111/tbed.13491
Jiang, Y. F., Xia, T. Q., Zhou, Y. J., Yu, L. X., Yang, S., Huang, Q. F., et al. (2015). Characterization of three porcine reproductive and respiratory syndrome virus isolates from a single swine farm bearing strong homology to a vaccine strain. Vet. Microbiol. 179, 242–249. doi: 10.1016/j.vetmic.2015.06.015
Li, C., Zhuang, J., Wang, J., Han, L., Sun, Z., Xiao, Y., et al. (2016). Outbreak Investigation of NADC30-Like PRRSV in South-East China. Transbound. Emerg. Dis. 63, 474–479. doi: 10.1111/tbed.12530
Li, X., Bao, H., Wang, Y., and Tian, K. (2017). Widespread of NADC30-like PRRSV in China: another Pandora’s box for Chinese pig industry as the outbreak of highly pathogenic PRRSV in 2006? Infect Genet. Evol. 49, 12–13. doi: 10.1016/j.meegid.2016.12.021
Li, Y., Zhou, L., Zhang, J., Ge, X., Zhou, R., Zheng, H., et al. (2014). Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog. 10:e1004216. doi: 10.1371/journal.ppat.1004216
Lu, W. H., Tun, H. M., Sun, B. L., Mo, J., Zhou, Q. F., Deng, Y. X., et al. (2015). Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet. Microbiol. 175, 332–340. doi: 10.1016/j.vetmic.2014.11.016
Lunney, J. K., Benfield, D. A., and Rowland, R. R. (2010). Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 154, 1–6. doi: 10.1016/j.virusres.2010.10.009
Lunney, J. K., Fang, Y., Ladinig, A., Chen, N., Li, Y., Rowland, B., et al. (2016). Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 4, 129–154. doi: 10.1146/annurev-animal-022114-111025
Shi, M., Lam, T. T., Hon, C. C., Hui, R. K., Faaberg, K. S., Wennblom, T., et al. (2010a). Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. 154, 7–17. doi: 10.1016/j.virusres.2010.08.014
Shi, M., Lam, T. T., Hon, C. C., Murtaugh, M. P., Davies, P. R., Hui, R. K., et al. (2010b). Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 84, 8700–8711. doi: 10.1128/JVI.02551-09
Thanawongnuwech, R., Brown, G. B., Halbur, P. G., Roth, J. A., Royer, R. L., and Thacker, B. J. (2000). Pathogenesis of porcine reproductive and respiratory syndrome virus-induced increase in susceptibility to Streptococcus suis infection. Vet. Pathol. 37, 143–152. doi: 10.1354/vp.37-2-143
Tian, K. (2017). NADC30-Like Porcine Reproductive and Respiratory Syndrome in China. Open Virol. J. 11, 59–65. doi: 10.2174/1874357901711010059
Tian, K., Yu, X., Zhao, T., Feng, Y., Cao, Z., Wang, C., et al. (2007). Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526
Tian, Z. J., An, T. Q., Zhou, Y. J., Peng, J. M., Hu, S. P., Wei, T. C., et al. (2009). An attenuated live vaccine based on highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) protects piglets against HP-PRRS. Vet. Microbiol. 138, 34–40. doi: 10.1016/j.vetmic.2009.03.003
Wang, H. M., Liu, Y. G., Tang, Y. D., Liu, T. X., Zheng, L. L., Wang, T. Y., et al. (2018). A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains. Transbound Emerg. Dis. 65, 1078–1086. doi: 10.1111/tbed.12852
Wang, H., Xu, Y., and Feng, W. (2021). Porcine Reproductive and Respiratory Syndrome Virus: immune Escape and Application of Reverse Genetics in Attenuated Live Vaccine Development. Vaccines 9:480. doi: 10.3390/vaccines9050480
Zhang, Q., Jiang, P., Song, Z., Lv, L., Li, L., and Bai, J. (2016). Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Vet. Microbiol. 197, 93–101. doi: 10.1016/j.vetmic.2016.11.010
Zhao, K., Ye, C., Chang, X. B., Jiang, C. G., Wang, S. J., Cai, X. H., et al. (2015). Importation and Recombination Are Responsible for the Latest Emergence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus in China. J. Virol. 89, 10712–10716. doi: 10.1128/JVI.01446-15
Zhou, L., Kang, R., Yu, J., Xie, B., Chen, C., Li, X., et al. (2018). Genetic Characterization and Pathogenicity of a Novel Recombined Porcine Reproductive and Respiratory Syndrome Virus 2 among Nadc30-Like, Jxa1-Like, and Mlv-Like Strains. Viruses 10:551. doi: 10.3390/v10100551
Zhou, L., Wang, Z., Ding, Y., Ge, X., Guo, X., and Yang, H. (2015). NADC30-like Strain of Porcine Reproductive and Respiratory Syndrome Virus, China. Emerg. Infect Dis. 21, 2256–2257. doi: 10.3201/eid2112.150360
Zhou, L., Yang, B., Xu, L., Jin, H., Ge, X., Guo, X., et al. (2017). Efficacy evaluation of three modified-live virus vaccines against a strain of porcine reproductive and respiratory syndrome virus NADC30-like. Vet. Microbiol. 207, 108–116. doi: 10.1016/j.vetmic.2017.05.031
Zhou, L., Yang, X., Tian, Y., Yin, S., Geng, G., Ge, X., et al. (2014). Genetic diversity analysis of genotype 2 porcine reproductive and respiratory syndrome viruses emerging in recent years in China. Biomed. Res. Int. 2014:748068. doi: 10.1155/2014/748068
Zhou, L., Zhang, J., Zeng, J., Yin, S., Li, Y., Zheng, L., et al. (2009). The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 83, 5156–5167. doi: 10.1128/JVI.02678-08
Keywords: rPRRSV-E2, live vectored vaccine, NADC30-like strain, heterologous protection, challenge model
Citation: Li L, Chen J, Cao Z, Cao Y, Guo Z, Tong W, Zhou Y, Li G, Jiang Y, Liu C, Yu L, Qiao S, Liu J, Tong G and Gao F (2022) Recombinant Bivalent Live Vectored Vaccine Against Classical Swine Fever and HP-PRRS Revealed Adequate Heterogeneous Protection Against NADC30-Like Strain. Front. Microbiol. 12:822749. doi: 10.3389/fmicb.2021.822749
Received: 26 November 2021; Accepted: 13 December 2021;
Published: 05 January 2022.
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Bin Zhou, Nanjing Agricultural University, ChinaQinfeng Huang, University of Minnesota Twin Cities, United States
Copyright © 2022 Li, Chen, Cao, Cao, Guo, Tong, Zhou, Li, Jiang, Liu, Yu, Qiao, Liu, Tong and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangzhi Tong, Z3p0b25nQHNodnJpLmFjLmNu; Fei Gao, ZmVpZ2FvQHNodnJpLmFjLmNu
†These authors have contributed equally to this work
 Liwei Li
Liwei Li Jinxia Chen1†
Jinxia Chen1† Fei Gao
Fei Gao