
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 February 2022
Sec. Extreme Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.822229
This article is part of the Research Topic Microbial Life Under Stress: Biochemical, Genomic, Transcriptomic, Proteomic, Bioinformatics, Evolutionary Aspects and Biotechnological Applications of Poly-Extremophilic Bacteria, Volume II View all 15 articles
Extreme acidophiles thrive in environments rich in protons (pH values <3) and often high levels of dissolved heavy metals. They are distributed across the three domains of the Tree of Life including members of the Proteobacteria. The Acidithiobacillia class is formed by the neutrophilic genus Thermithiobacillus along with the extremely acidophilic genera Fervidacidithiobacillus, Igneacidithiobacillus, Ambacidithiobacillus, and Acidithiobacillus. Phylogenomic reconstruction revealed a division in the Acidithiobacillia class correlating with the different pH optima that suggested that the acidophilic genera evolved from an ancestral neutrophile within the Acidithiobacillia. Genes and mechanisms denominated as “first line of defense” were key to explaining the Acidithiobacillia acidophilic lifestyle including preventing proton influx that allows the cell to maintain a near-neutral cytoplasmic pH and differ from the neutrophilic Acidithiobacillia ancestors that lacked these systems. Additional differences between the neutrophilic and acidophilic Acidithiobacillia included the higher number of gene copies in the acidophilic genera coding for “second line of defense” systems that neutralize and/or expel protons from cell. Gain of genes such as hopanoid biosynthesis involved in membrane stabilization at low pH and the functional redundancy for generating an internal positive membrane potential revealed the transition from neutrophilic properties to a new acidophilic lifestyle by shaping the Acidithiobacillaceae genomic structure. The presence of a pool of accessory genes with functional redundancy provides the opportunity to “hedge bet” in rapidly changing acidic environments. Although a core of mechanisms for acid resistance was inherited vertically from an inferred neutrophilic ancestor, the majority of mechanisms, especially those potentially involved in resistance to extremely low pH, were obtained from other extreme acidophiles by horizontal gene transfer (HGT) events.
Microbial cells present a variety of genetic mechanisms allowing them to manage stressful situations such as changes in temperature, pH values, and oxidative stress (Li et al., 2011). Organisms with an acidic optimal growth pH are termed “acidophiles,” and this classification has been further divided into moderate acidophiles with a pH optimum ≤5 and extreme acidophiles that grow optimally at pH ≤3. To survive and grow in low pH environments, acidophiles must maintain a near-neutral cytoplasm when faced with an external to internal proton gradient across the cytoplasmic membrane up to 105-fold (Slonczewski et al., 2009; Zammit and Watkin, 2016; Quatrini and Johnson, 2018). Different pH homeostasis mechanisms have been proposed to explain the acidophilic resistance including some that are shared with neutrophiles. These are (i) proton export pumps and antiporters (Michels and Bakker, 1985); (ii) cytoplasmic buffering via an overproduction of alkaline amino acids (Parro et al., 2007); (iii) proton consuming reactions such as glutamate decarboxylase (Mangold et al., 2013); (iv) alterations in the membrane structure to reduce fluidity by the inclusion of hopanoids (Mykytczuk et al., 2010); (v) reduction in the outer membrane permeability via porins using polyamines such as spermidine (Samartzidou et al., 2003); and (vi) pH homeostatic systems such as an internal positive membrane potential thought to be generated by potassium ions (Buetti-Dinh et al., 2016). However, as pH homeostatic mechanisms are often shared between neutrophiles and acidophiles, those most important to mediating homeostasis in highly acidic conditions are not always apparent.
Low pH mitigation mechanisms have been investigated in acidophiles (Baker-Austin and Dopson, 2007; Li et al., 2011; Ullrich et al., 2016a; Colman et al., 2018; Vergara et al., 2020; Chen et al., 2021; Mayer et al., 2021; Sriaporn et al., 2021). These studies show that horizontal gene transfer (HGT), gene mutation, and gene loss trajectories in evolution allow adaptation and survival of prokaryotes in extreme conditions (Foster, 2004; Ullrich et al., 2016a; Zhang et al., 2017; Vergara et al., 2020). Extensive HGT events are also documented in acidophiles such as Sulfolobus islandicus (Whitaker et al., 2005), Leptospirillum (Simmons et al., 2008; Vergara et al., 2020), and the extremophilic red alga Galdieria sulphuraria (Schonknecht et al., 2013; Colman et al., 2018) that acquired genes by HGT from bacteria and archaea to be able to thrive in thermophilic, acidic, and metal-rich environments. To cope with the high acidity, G. sulphuraria lowers the proton permeability of its plasma membrane by having a single copy of a voltage-gated ion channel gene compared to three copies in the neutrophilic Cyanidioschyzon merolae from which it diverged one billion years ago (Schonknecht et al., 2013). However, the events that led to the evolution of acidophiles and their role in the generation of acidic habitats are underexplored.
The Acidithiobacillia is a newly recognized class of Proteobacteria at the root of the Betaproteobacteria/Gammaproteobacteria division (Williams and Kelly, 2013). Two families make up this class, family I comprising the acidophilic Acidithiobacillaceae and the neutrophilic family II Thermithiobacillaceae. Due to their acidophilic nature, the Acidithiobacillaceae are largely recalcitrant to standard genetic manipulation (Inaba et al., 2018; Jung et al., 2021), and consequently, the use of bioinformatics approaches advances our understanding of the biology of these extremophiles. The research of Moya-Beltrán et al. (2021) describes four Acidithiobacillaceae genera (Nuñez et al., 2016), namely, (i) sulfur oxidizing Ambacidithiobacillus that includes the recently described species Am. sulfuriphilus (ex Acidithiobacillus sulfuriphilus) (Falagán et al., 2019); (ii) Igneacidithiobacillus including species I. copahuensis (Moya-Beltrán et al., 2021), Candidatus I. taiwanensis (Moya-Beltrán et al., 2021), and Candidatus I. yellowstonensis (Moya-Beltrán et al., 2021); (iii) Fervidacidithiobacillus with F. caldus (ex-Acidithiobacillus caldus) (Hallberg and Lindström, 1994; Valdes et al., 2009; You et al., 2011; Zhang et al., 2016b); and (iv) the Acidithiobacillus genus formed by A. thiooxidans (Waksman and Joffe, 1922; Valdes et al., 2011; Travisany et al., 2014; Yin et al., 2014; Zhang et al., 2016a,2018; Quatrini et al., 2017; Camacho et al., 2020), A. albertensis (Bryant et al., 1983; Castro et al., 2017), Acidithiobacillus sp. SH (Kamimura et al., 2018) plus the iron/sulfur oxidizers A. ferrooxidans (Temple and Colmer, 1951; Valdés et al., 2008; Chen P. et al., 2015; Yan et al., 2015; Kucera et al., 2016; Ulloa et al., 2019), A. ferridurans (Hedrich and Johnson, 2013; Miyauchi et al., 2018), A. ferrivorans (Hallberg et al., 2010; Liljeqvist et al., 2011; Talla et al., 2014; Tran et al., 2017), and A. ferriphilus (Falagán and Johnson, 2016) represented by A. ferrooxidans BY0502 for which phylogenetic and nucleotide identity analysis is suggested to be reclassified as an A. ferriphilus-like species (González et al., 2016; Fariq et al., 2019).
Genome sequences are available for many of the acidophilic species (Neira et al., 2020) including A. ferrooxidans (Valdés et al., 2008; Chen P. et al., 2015; Yan et al., 2015; Kucera et al., 2016; Ulloa et al., 2019), A. ferrivorans (Liljeqvist et al., 2011; Talla et al., 2014; Tran et al., 2017), A. ferridurans (Hedrich and Johnson, 2013; Miyauchi et al., 2018), A. ferrianus (Norris et al., 2020), A. thiooxidans (Valdes et al., 2011; Travisany et al., 2014; Yin et al., 2014; Zhang et al., 2016a,2018; Quatrini et al., 2017; Camacho et al., 2020), F. caldus (Valdes et al., 2009; You et al., 2011; Zhang et al., 2016b), and Am. sulfuriphilus (Falagán et al., 2019) along with the neutrophile Thermithiobacillus tepidarius (Boden et al., 2016).
Three populations of uncultivated hot spring Acidithiobacillus strains and ten publicly available genomes were analyzed for the presence or absence of predicted genes for acid resistance, identifying a similar number of gene copies encoding for K+ transporters, deiminases/deaminase group and adenosine deaminase shared between genomes, and a difference in the number of amino acid decarboxylases, Na+/H+ antiporters, and plasma membrane proton-efflux P-type ATPases (Sriaporn et al., 2021). In addition, the mechanisms of resistance to low temperature and pH from an extreme microbial community where A. ferrivorans was the dominant member revealed an adaptation to low temperature by the presence trehalose synthase pathways, oxidative stress pathways, cold shock proteins, and genes encoding for biofilm formation (González et al., 2014; Liljeqvist et al., 2015). This study investigates the genes present in the Acidithiobacillia class by phylogenomic and comparative genomic analyses deepening our understanding of how the acidophilic genera Fervidacidithiobacillus, Igneacidithiobacillus, Ambacidithiobacillus, and Acidithiobacillus obtained genes coding for pH homeostasis that allows them to survive at extremely low pH values of ≤3.
Thirty-seven permanent draft or complete genomes from the Acidithiobacillia class were downloaded from NCBI with those genomes passing CheckM (Parks et al., 2015) quality control for completeness (>90%) and lack of contamination (<10%) retained. The Acidithiobacillia class conserved core genome was identified using the GET_Homologues (Contreras-Moreira and Vinuesa, 2013) software suite, selecting conserved proteins with at least 50% identity and alignment coverage. Core protein families were aligned using MAFFT (Katoh, 2002; Katoh and Standley, 2013; Katoh et al., 2019) with L-INS-i iterative refinement. Alignments were masked to remove unreliable aligned regions with GBLOCK (Castresana, 2000) followed by concatenation of families. A maximum likelihood tree was constructed based on concatenated alignment of core genes using IQ-TREE (Nguyen et al., 2015; Hoang et al., 2018) with 1,000 replicates and the best-fit model predicted by IQ-TREE (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017) according to Bayesian information criterion and the T. tepidarius DSM 3134 genome as an outgroup for the Acidithiobacillaceae (Boden et al., 2016). The phylogenomic tree was visualized with iTOL6 (Letunic and Bork, 2021).
A representative genome was selected from each clade of core genomes according to the following criteria (Supplementary Figure 1): (i) using the type strain genome if available and complete; (ii) if this was unavailable or a draft, a strain with a complete genome was selected; and (iii) a strain genome sequence was selected according to the quality as calculated by CheckM.
Conserved orthologous proteins were selected based on the classification of all protein-coding genes in protein families. The selected proteins were sorted using the GET_Homologues (Contreras-Moreira and Vinuesa, 2013) software suite, with BLAST (Altschul, 1997) and orthoMCL (Fischer et al., 2011) programs. Protein families were constructed using a 50/50 rule of 50% of identity and 50% of alignment coverage (Altschul et al., 1990; Altschul, 1997; Snipen and Ussery, 2010) and each protein was then assigned to one protein family. The protein families were classified in core (proteins shared by all strains), dispensable (proteins assigned to some strains), and unique (proteins assigned to only a single strain) genomes according to their distribution (Supplementary Data Sheet 1). The sum of all three groups form the pangenome, i.e., the union of the genomes under consideration (Tettelin et al., 2005; Snipen and Ussery, 2010).
Genes and protein sequences that were either predicted or experimentally validated to be involved in low pH responses in extreme acidophiles were extracted from the literature (Gassel et al., 1998; Foster, 2004; Zhou et al., 2010; Krulwich et al., 2011; Mangold et al., 2013; De Biase and Lund, 2015; Mühling et al., 2016; Vergara et al., 2020). The list of potential genes was extended by keyword literature searches in Google Scholar and species description journals (e.g., International Journal of Systematic and Evolutionary Microbiology). Additionally, the JGI pH metadata and NCBI’s BIOSAMPLE database were used to collect gene and protein information linked to low pH environments. The collected assemblage of potential acid resistance gene/protein sequences was used to formulate Blast searches against Acidithiobacillia genomes using a minimal E-value cutoff of 1e–5 (Supplementary Figure 1).
Prediction of transmembrane regions in protein sequences was carried out using TMHMM (Sonnhammer et al., 1998; Krogh et al., 2001) and TMPRED (Chen et al., 2002). Signal peptide and subcellular localization predictions were made with SignalP 5.0 (Almagro Armenteros et al., 2019), PSORTb (Yu et al., 2010), and CELLO (Yu et al., 2006). Prediction of lipoprotein signals, search of lipobox, plus identification of conserved sequences, motifs, and domains were made with LipoP Server (Juncker et al., 2003) and WebLogo (Schneider and Stephens, 1990; Crooks et al., 2004) using AliView (Larsson, 2014) and MAFFT (Katoh, 2002; Katoh and Standley, 2013; Katoh et al., 2019) as alignment tools.
Mobile genetic elements as insertion sequences in Acidithiobacillia genomes were predicted with ISsaga (Varani et al., 2011) of ISFINDER (Kichenaradja et al., 2010; Varani et al., 2011) and TnpPred (Riadi et al., 2012). Potential HGT events were predicted by HGTector (Zhu et al., 2014). Genes and pathways related to acid resistance were analyzed to identify mobile genetic elements as transposases, integrases, or phage elements in close genomic context using IslandViewer (Bertelli et al., 2017), Artemis (Carver et al., 2012), MAUVE (Darling et al., 2004, 2010), and STRING (von Mering et al., 2005; Szklarczyk et al., 2019).
Trees of acid resistance proteins were constructed for proteins identified in the Acidithiobacillia class and orthologs from other microorganisms to propose a common or different evolutionary origin. Orthologous proteins from acid resistance genes were obtained by selection of best hits from BlastP comparison of Acidithiobacillia acid resistance genes versus the NCBI database. Phylogenies were constructed using acid resistance genes from Acidithiobacillia plus orthologous proteins from NCBI, which were aligned with MAFFT using L-INS-i iterative refinement (Katoh, 2002; Katoh and Standley, 2013; Katoh et al., 2019) and IQ-TREE (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017; Hoang et al., 2018) for prediction of best suited model and phylogenetic construction. Visualization of the trees was with Figtree1 and iTOL6 (Letunic and Bork, 2021).
Thirty-seven Acidithiobacillia class genomes met the CheckM quality criteria while the pangenome analysis showed a core genome of 440 protein families (Supplementary Data Sheet 1). A maximum likelihood phylogenetic tree of Acidithiobacillia core concatenated proteins showed ten clades, separating the neutrophilic T. tepidarius from the acidophilic genera Ambacidithiobacillus, Fervidacidithiobacillus, and Acidithiobacillus (Figure 1). The acidophiles were further grouped into nine clades, of which two represented different genera (clades 2 and 3); meanwhile, clades 4–10 grouped species from the Acidithiobacillus genus, which is divided into clades according to their capacity to obtain energy such as sulfur oxidizers (clades 4 and 5) and iron/sulfur oxidizers (clades 6–10). While the association between the Acidithiobacillaceae and Thermithiobacillaceae families was consistent with previous analyses (Williams et al., 2010; Williams and Kelly, 2013; Boden et al., 2016; Moya-Beltrán et al., 2021), the tree also suggested a need for deeper phylogenomic analysis such as for the Acidithiobacillus clade, questioning if this clade includes more than a single genus according to their energy properties. Representative genomes from the ten clades of the core tree were selected for this study with their accession numbers and isolation data summarized in Table 1.
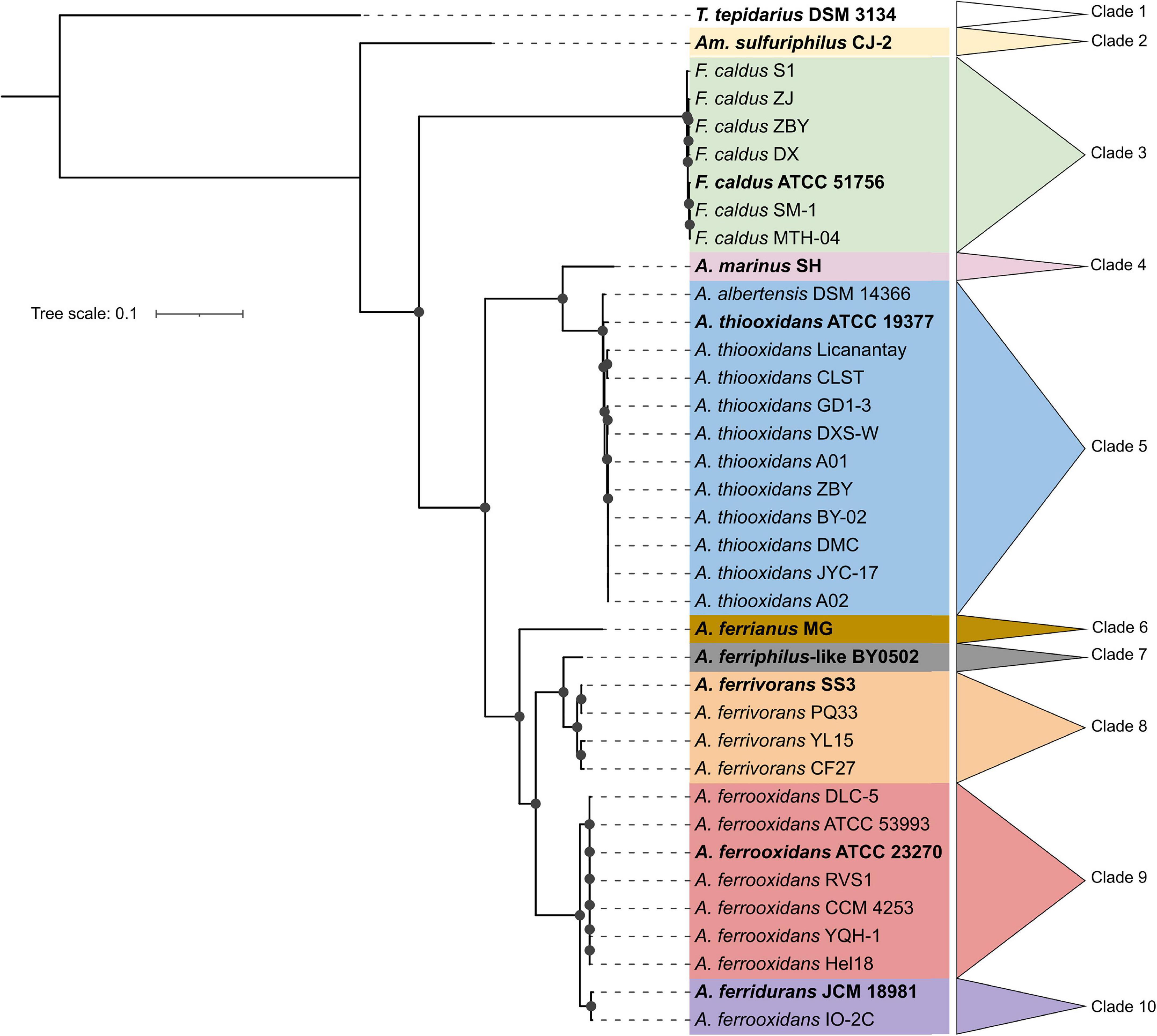
Figure 1. Maximum likelihood phylogenetic tree showing the neutrophilic ancestral origins of the Acidithiobacillia class. The bold genomes were selected to represent the ten clades from Acidithiobacillia class core tree, including T. tepidarius DSM 3134 for the neutrophilic Thermithiobacillus genus and nine genomes for the acidophilic genera Ambacidithiobacillus (Am.), Fervidacidithiobacillus (F.), and Acidithiobacillus (A.). The tree was constructed using 440 conserved proteins using 1,000 replicates. Bootstraps values ≥60% are represented by black dots on the nodes. The scale bar represents 0.1 amino acid substitution per position.
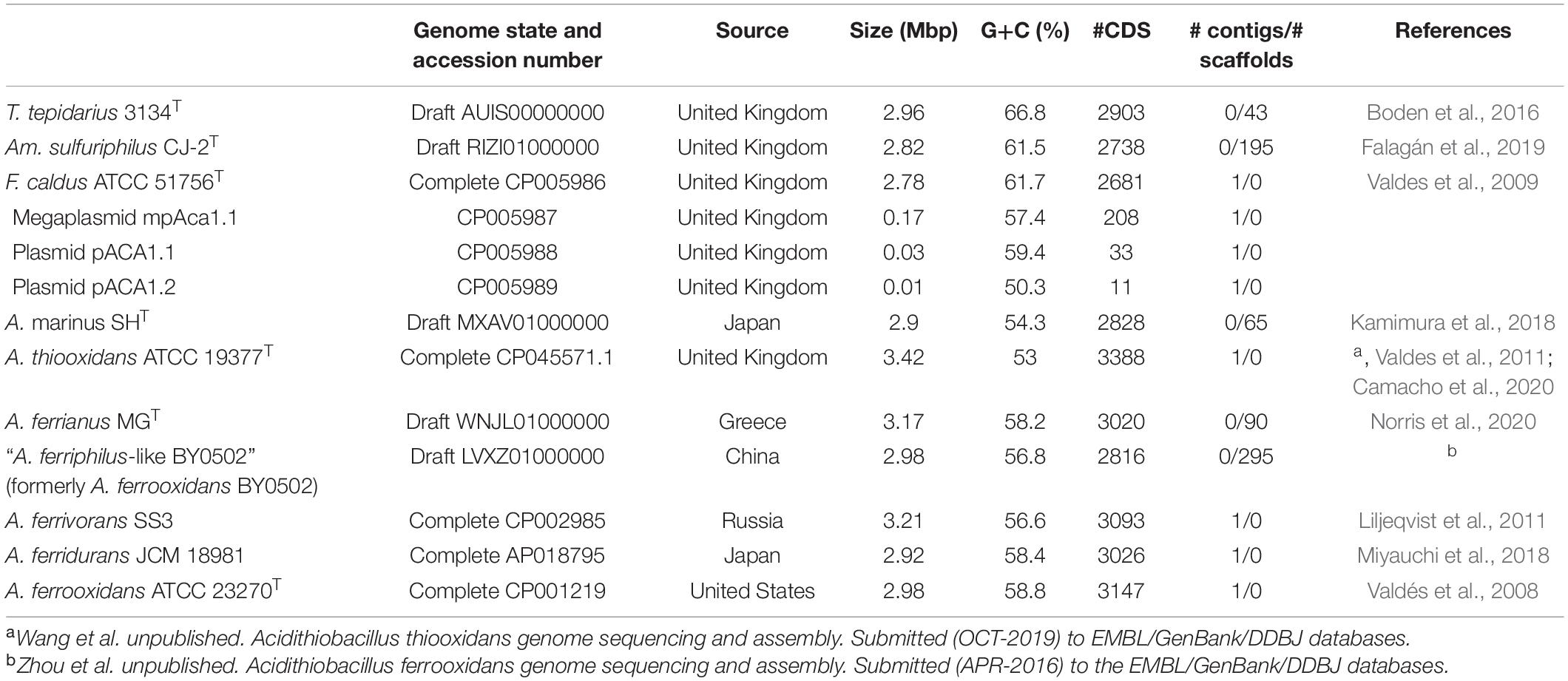
Table 1. Strains, genome accession, and features of the selected representatives of the Acidithiobacillia class.
Genes with predicted or experimental evidence for acid resistance mechanisms were searched in the Acidithiobacillia class using a bioinformatic pipeline (Supplementary Figure 1) that generated the list in Supplementary Data Sheet 2. The acid resistance genes were further labeled into first and second line of defense, according to the classification system proposed by the research of Vergara et al. (2020), which proposes that genes involved in the prevention of the entry of proton into the cell correspond to the “first line of defense,” and those related to neutralization or expulsion of protons inside the cell belong to the “second line of defense.”
Genes related to the first line of defense involved in the prevention of entry of protons into the cell, membrane rigidity, reduced membrane permeability to protons, and maintenance of cellular integrity are summarized in Figure 2.
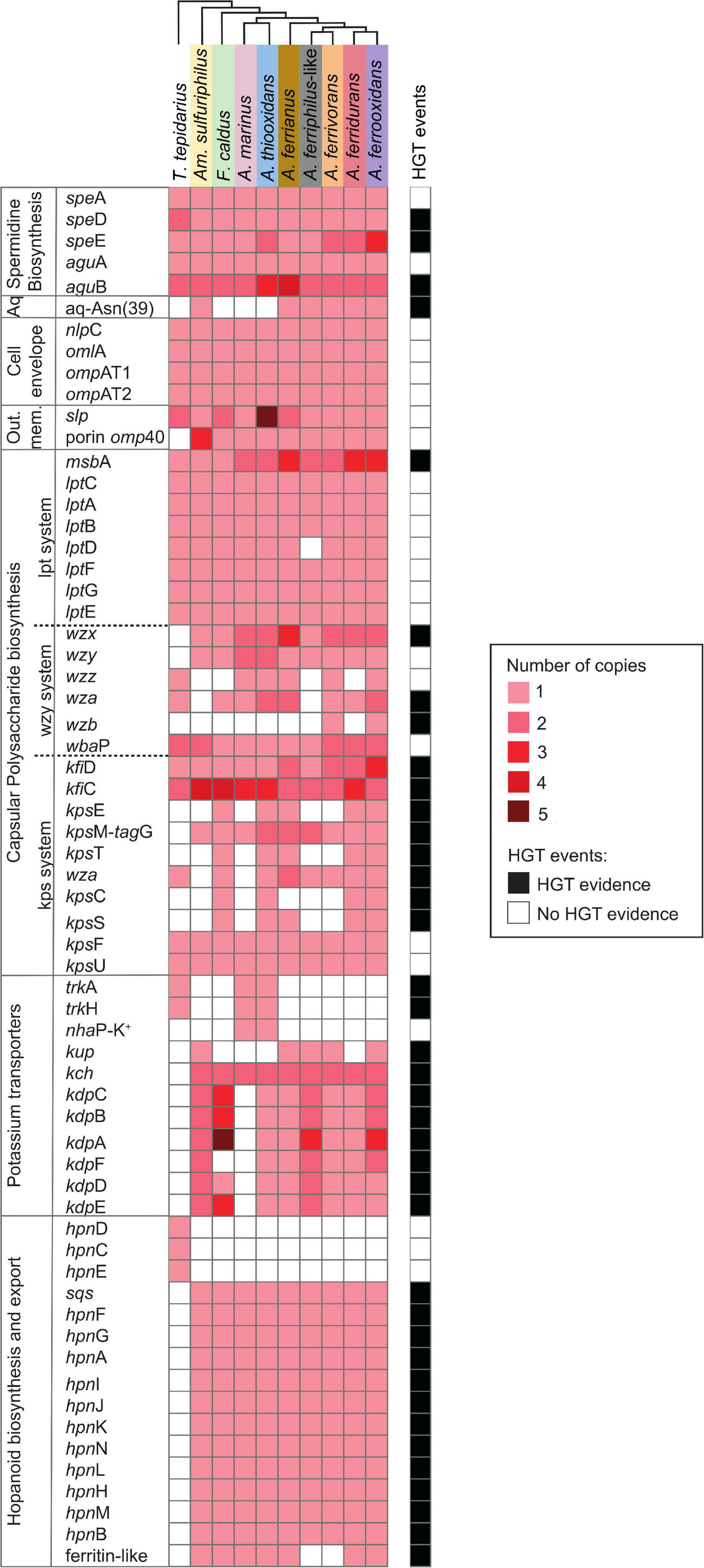
Figure 2. Heatmap of presence (red) and absence (white) of genes involved in the first line of Acidithiobacillia class acid resistance including spermidine biosynthesis; aquaporine (Aq) with an asparagine residue (Asn39); cell envelope; outer membrane proteins (Out. mem.); capsular polysaccharide biosynthesis with pathways Lpt-, Wzy-, and Kps-systems; potassium transporters; and hopanoid biosynthesis plus export. The color intensity represents higher gene copy number per genome. The presence of genes (black; right-hand side) denotes HGT events by genome context with mobile genetic elements, phylogenetic trees, or if it was signaled as part of an HGT event by HGTector (Zhu et al., 2014). T. tepidarius, Thermithiobacillus tepidarius DSM 3134; Am. Sulfuriphilus, Ambacidithiobacillus sulfuriphilus CJ-2; F. caldus, Fervidacidithiobacillus caldus ATCC 51756; A. marinus, Acidithiobacillus marinus SH; A. thiooxidans, A. thiooxidans ATCC 19377; A. ferrianus, A. ferrianus MG; A. ferriphilus-like, A. ferriphilus-like BY0502; A. ferrivorans, A. ferrivorans SS3; A. ferridurans, A. ferridurans JCM 18981; and A. ferrooxidans, A. ferrooxidans ATCC 23270 (shaded colors correspond to those used in Figure 1).
The putrescine biosynthesis pathway was identified in the Acidithiobacillia class including genes speA, aguA, and one or more copies of aguB (Figure 3A). Acidophilic genomes from the Acidithiobacillaceae family present several copies of aguB clustered in four clades aguB1–aguB4 (Figure 3B). The aguB1 and aguB3 clades associated aguB from acidophiles with the neutrophile T. tepidarius although the aguB3 genome context in the acidophiles displayed a cupin domain gene with high similarity to copies in the acidophiles Acidocella and Acidiphilium. In addition, aguB3 genes were predicted to result from HGT in the Acidithiobacillus genus. The clade of acidophile aguB2 genes were downstream from an feoABC operon related to iron management (Osorio et al., 2008).
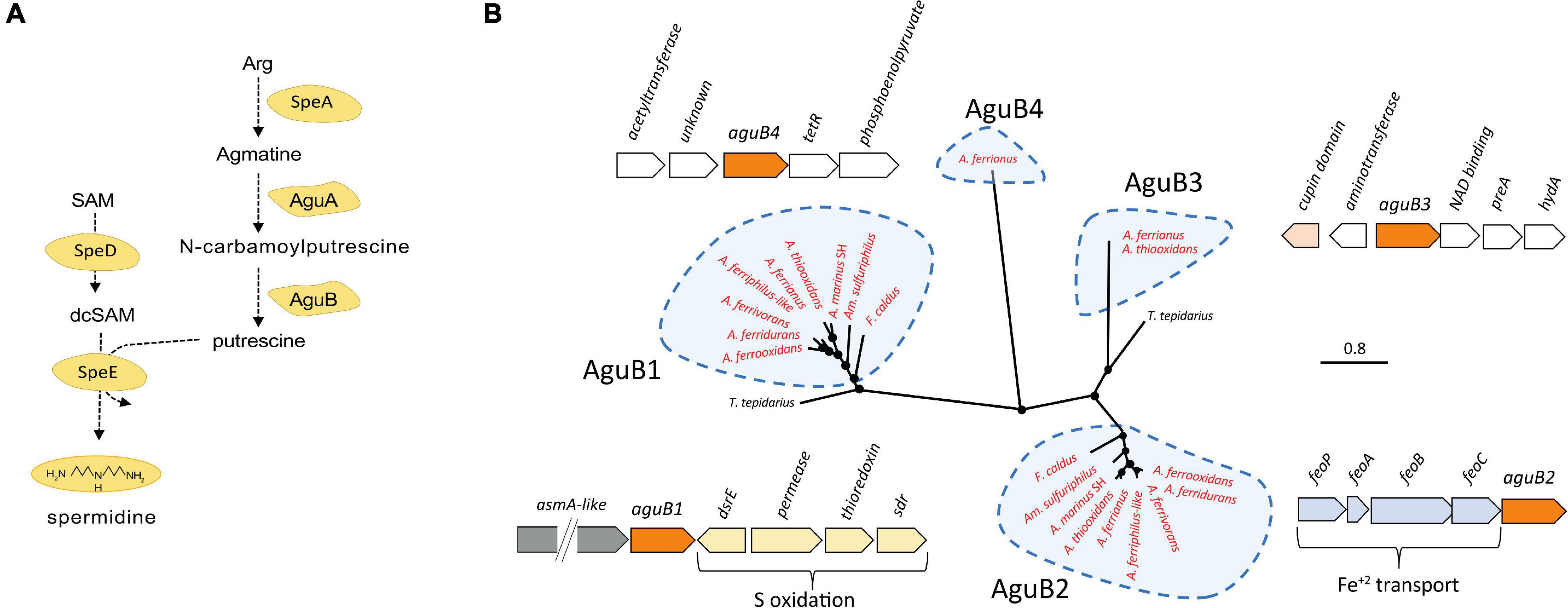
Figure 3. (A) Putrescine and spermidine biosynthesis pathway. (B) Unrooted phylogenetic tree of Acidithiobacillia class aguB amino acid sequences and their best hits in the NCBI databases plus their genomic context in the Acidithiobacillaceae (aguB in orange color). Sequences of aguB group in four clades denominated aguB1–aguB4. Acidophilic sequences are highlighted in red. Filled circles at the nodes indicate bootstrap support ≥60%. The scale bar represents 0.8 amino acid substitution per site.
Putrescine is required for spermidine synthesis (Figure 3A), which protects the cell against acid stress by cytoplasmic buffering (Rhee et al., 2007; Kanjee and Houry, 2013; Vergara et al., 2020) such as in F. caldus (Mangold et al., 2013). S-adenosylmethionine decarboxylase (speD) and spermidine synthase (speE) are essential for the biosynthesis of spermidine (Xie et al., 1989), which are significantly upregulated in acidic environments (Li et al., 2012). This gene cluster was conserved in the Acidithiobacillia, separating into two clades between the neutrophile T. tepidarius and acidophiles of Acidithiobacillaceae with different genome contexts. The acidophile spermidine synthesis genes were downstream of mlaDCEF (Figure 4); the latter acts as an ABC transporter driving translocation of phospholipids between the inner and outer membrane of Gram-negative bacteria (Hughes et al., 2019; Chi et al., 2020). The mlaD outer membrane lipid and mlaC phospholipid ABC transporter maintain outer membrane integrity in osmotic stress conditions in the halotolerant acidophile Acidihalobacter prosperus (Malinverni and Silhavy, 2009; Dopson et al., 2017; Khaleque et al., 2019). Phylogenetic analysis of concatenated speD-speE genes showed similarity to the extreme acidophile Acidiferrobacter (Figure 4). The close genomic context between speDE and the mlaDCEF gene complex observed in acidophiles highlights the relevance of these genes for membrane maintenance as a stress response.
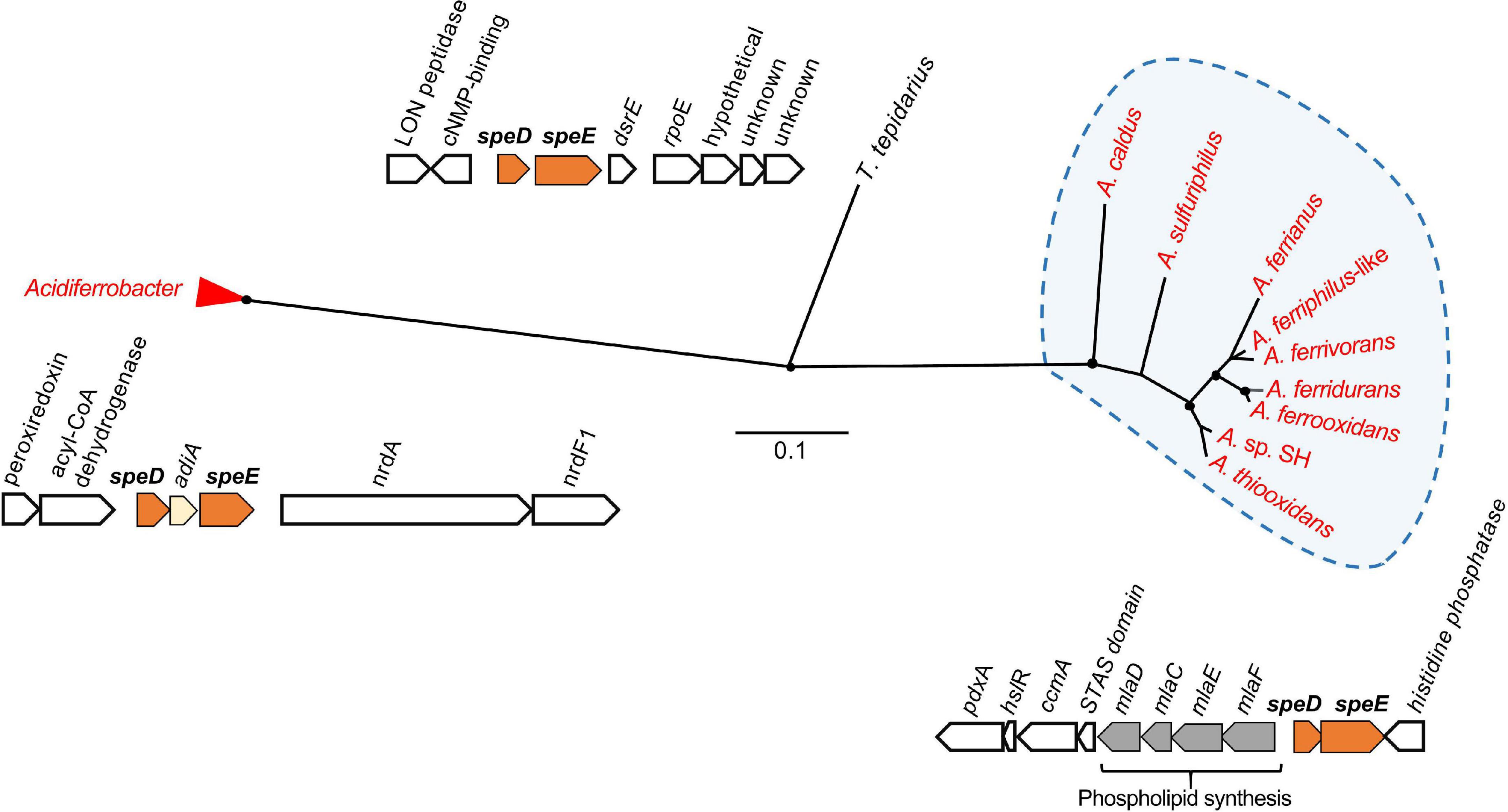
Figure 4. Unrooted phylogenetic tree of concatenated amino acid sequences of speD and speE in the Acidithiobacillia and their best hits in the NCBI databases. Acidithiobacillaceae sequences group in a clade from neutrophilic T. tepidarius, and had highest similarity to homologs in extreme acidophiles of the Acidiferrobacter group. Acidophilic sequences are highlighted in red. Filled circles at the nodes indicate bootstrap support ≥60%. The scale bar represents 0.1 amino acid substitution per site.
An aquaporin AqpF was conserved in the Ambacidithiobacillus and Acidithiobacillus genera (Supplementary Figure 2), including three copies in A. ferrivorans. The aquaporin AqpF with an asparagine residue (Asn39) is proposed to be involved in enhancing the capability for proton-blocking in extreme acidophiles (Duarte et al., 2009). This difference was identified in the Acidithiobacillia where the AqpF in the neutrophiles T. tepidarius lacked this residue in contrast to AqpF in acidophiles Am. sulfuriphilus and Acidithiobacillus iron oxidizers. Aquaporin genes identified in A. ferrianus and A. ferriphilus-like presented signals of HGT events with the extreme acidophile Acidihalobacter-like as the potential donor.
Cell envelope nlpC, omlA, ompAT1, and ompAT2 genes were conserved in the Acidithiobacillia class with a single copy per genome that is upregulated in response to low pH conditions in A. ferrooxidans (Chao et al., 2008). The gene omp40 was exclusively identified in acidophilic members of the Acidithiobacillia including three copies in Am. sulfuriphilus CJ-2. The Omp40 is a major outer membrane ion channel protein that increases in expression in response to low pH and phosphate starvation in F. caldus and A. ferrooxidans (Amaro et al., 1991; Mangold et al., 2013; Hu et al., 2020), suggesting that it controls proton influx across the outer membrane (Guiliani and Jerez, 2000; Baker-Austin and Dopson, 2007). A further cell envelope gene slp prevents the flux of organic acids across the outer membrane counteracting their toxic effect in acidophiles (Alexander et al., 1987; Mates et al., 2007; Vergara et al., 2020) and ameliorating low pH stress conditions in Escherichia coli (Mates et al., 2007). The slp genes contain a characteristic lipobox motif with an Asn amino acid residue in the +2 position, indicative that the protein is located at the inner membrane, and a different residue if Slp protein is exported (Zückert, 2014; Vergara et al., 2020). The slp gene was present in all Acidithiobacillia genomes (including several copies in some species; Supplementary Figure 3A) and was predicted to be exported. Phylogenetic analysis grouped slp into three clades (Supplementary Figure 3B) with the neutrophilic slp genes separated from those in the acidophile genomes.
Capsule polysaccharides constitute a mechanical defense layer (Feng et al., 2015) that acts as a protective barrier around the cell (Li et al., 2011), preventing influx of protons (Baker-Austin and Dopson, 2007; Mykytczuk et al., 2010) and other harmful external factors (Plante, 2000; Feng et al., 2015; Hu et al., 2020). The A. thiooxidans capsule became significantly thicker at the sub-optimal acid pH 0.8, and at super acid pH 0.4 began to fade, owing to that extreme acid stress became an unbearable threat to the cell survival (Feng et al., 2015, 2019). Three Acidithiobacillia capsule polysaccharide pathways were identified. These were the Lpt (Ma et al., 2008; Renzi et al., 2016), Wzy-dependent (Yuan et al., 2013; Barahona et al., 2014; Wen and Zhang, 2015; Renzi et al., 2016; Oetiker et al., 2018), and Kps (Barton, 2005; Vera et al., 2013; Nzakizwanayo et al., 2015) systems (Figure 2). The Lpt system was conserved in the Acidithiobacillia, with additional copies of the ABC transporter msbA in the Acidithiobacillaceae and absence of lptD in the A. ferriphilus-like genome (probably due to its draft state). The Acidithiobacillaceae family encode genes for the Wzy system such as wzx for flippases that transport capsule polysaccharides across the cytoplasmic membrane along with the accessory genes wzz, wza, wzb, and wbaP. The accessory genes wzx, wzz, and wza were predicted to result from HGT events from potential Proteobacteria, Acidihalobacter-like, and Bacteroidetes donors. The Kps system includes KfiC and KfiD required for polysaccharide biosynthesis and the export pathway consisting of the Wza integral outer membrane protein that acts in conjunction with KpsE to move polysaccharide between the KpsMT cytoplasmic membrane transporter and the outer membrane (Dong et al., 2006). The genes encoding polysaccharide biosynthesis and transport by the Kps system were identified in F. caldus, A. thiooxidans, A. ferrianus [absence of kpsC encoding Kdo linker (Willis, 2013)], A. ferridurans, and A. ferrooxidans. Analysis of HGT events suggested that the Kps system was from donor acidophiles, such as kpsE, wza, and kpsS genes from Acidihalobacter-like (Khaleque et al., 2019, 2020); kfiC and kpsT from Acidiferrobacter-like (Issotta et al., 2018); plus kfiD, kpsM, and kpsC from Proteobacteria. Finally, the capsule formation systems Wzy and Kps were exclusively identified in Acidithiobacillaceae acidophiles and presented signals of HGT events from other extreme acidophiles.
An inside positive membrane potential (Baker-Austin and Dopson, 2007; Vergara et al., 2020) suppresses the inflow of protons by creation of a chemical permeation barrier (Hu et al., 2020) and is suggested to be generated by potassium ions accumulated by Trk, Kch, Kup, and Kdo potassium channel proteins (Cholo et al., 2015; Buetti-Dinh et al., 2016; Christel et al., 2018; Wang et al., 2019). While the neutrophile T. tepidarius contained trkAH genes for TrK activity (Supplementary Figure 4), one or more of these genes were disrupted in the acidophiles A. marinus SH and A. thiooxidans. The disrupted A. thiooxidans trkAH genes were preceded by a NhaP antiporter and surrounded by mobile genetic elements, suggesting that they were gained by HGT events. The gene encoding NhaP antiporter, denominated nhaP-K+, was classified within the first line of defense as it was in a cluster with a TrK system, suggesting a relation to potassium transport. The TrK system is a rapid-transport system at neutral or alkaline pH (Bossemeyer et al., 1989; Trchounian and Kobayashi, 2000; Epstein, 2003; Su et al., 2009) corresponding with its presence in T. tepidarius. Despite the disruption of the TrK system in acidophiles, the Acidithiobacillaceae contain three potassium transport systems that were not identified in the closest neutrophile. These were Kup (Trchounian and Kobayashi, 1999, 2000; Zakharyan and Trchounian, 2001; You et al., 2011), Kch (Voges and Jap, 1998; Munsey et al., 2002), and Kdp (Gassel et al., 1998; Yan et al., 2011; Cholo et al., 2015). The presence of three potassium transport systems in extreme acidophiles can be explained by the need to induce K+ uptake genes at different pH. An example is E. coli where the K+ transport TrK system is the most powerful system for K+ accumulation upon hyper-osmotic stress at neutral pH (Bossemeyer et al., 1989; Trchounian and Kobayashi, 2000; Epstein, 2003; Su et al., 2009; Yan et al., 2011) that is downregulated at low pH and Kup acts to compensate (Trchounian and Kobayashi, 1999; Yan et al., 2011), highlighting the role of different K+ uptake according to pH environment (Trchounian and Kobayashi, 1999; Yan et al., 2011). Phylogenetic analysis confirmed the similarity of Kup and Kch (Figure 5) to other extreme acidophiles such as Acidiferrobacter (Issotta et al., 2018), Methylacidimicrobium (van Teeseling et al., 2014), Verrucomicrobium (Schmitz et al., 2021), Leptospirillum (Vergara et al., 2020), and Acidiphilium (Li et al., 2020). This similarity was supported by HGTector prediction that suggested that Kup and Kch were transferred from Gammaproteobacteria and the acidophile Acidihalobacter-like. The Kup system was identified in Am. sulfuriphilus CJ-2 and the Acidithiobacillus iron oxidizers with the exception of A. ferridurans (Figure 5A). The Kch voltage-gated potassium channel was coded by two gene copies, kch1 and kch2, which were conserved in the Acidithiobacillaceae (Supplementary Figure 5). These gene copies had the highest similarity to homologs in the extreme acidophiles Leptospirillum, Acidiphilium, Acidihalobacter, and Acidiferrobacter (Figure 5B). The Kdp is a complex of four inner membrane subunits KdpF, KdpA, KdpB, and kdpC, with a KdpD sensor kinase and KdpE response regulator (Gassel et al., 1998) that are upregulated at low extracellular pH as a survival strategy for Mycobacterium tuberculosis (Cholo et al., 2015). The genes coding for the Kdp complex and regulatory proteins were identified exclusively in acidophiles from the class with three forms according to the gene contexts (Supplementary Figure 6). These were (i) the kdpCBAFED cluster conserved in Acidithiobacillaceae genomes except A. marinus SH that lacked a complete system and Am. sulfuriphilus with kdpDE genes disrupted by transposases; (ii) a second copy of the kdpCBAFED cluster in Am. sulfuriphilus and A. ferriphilus-like, a kdpABC cluster in F. caldus, and kdpFABC in A. ferrooxidans; and (iii) a kdpABCE gene cluster in the F. caldus plasmid. Mobile genetic elements were identified in the genome context of three forms of Kdp systems and BLAST analysis of Kdp encoding genes showed similarity to acidophile Acidiferrobacter, suggesting that the Kdp complex could be the result of an HGT event from Acidiferrobacter-like to Acidithiobacillaceae acidophiles as a mechanism to uptake potassium ions and thus improve acid resistance.
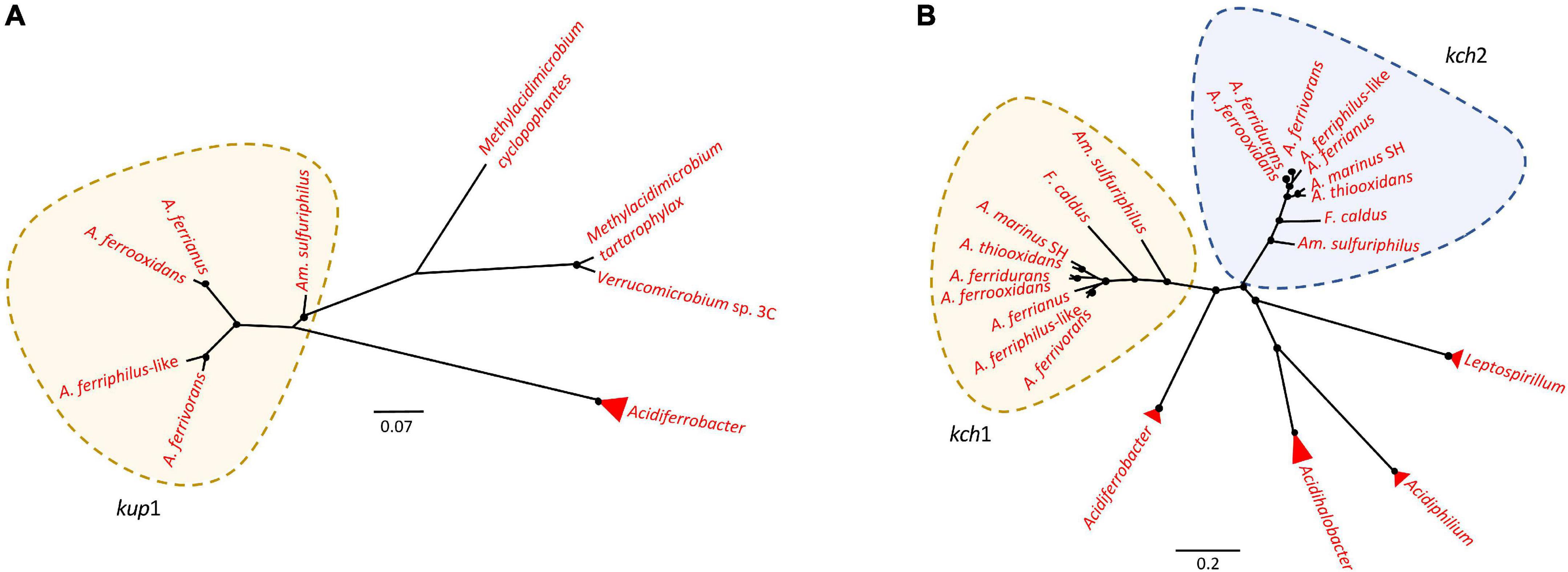
Figure 5. Unrooted phylogenetic tree of potassium transporters in Acidithiobacillia and their best hits in NCBI. (A) kup gene coding for Kup system had the highest similarity to homologs in extreme acidophiles Acidiferrobacter, Methylacidimicrobium, and Verrucomicrobium. (B) kch coding for Kch system had the highest similarity to Kch homologs in acidophiles Leptospirillum, Acidiphilium, Acidihalobacter, and Acidiferrobacter. Acidophilic sequences are highlighted in red. Filled circles at nodes represent bootstrap support of ≥60%. The scale bar represents amino acid substitution per site.
Comparative genomic analysis also revealed major differences between the hopanoid biosynthesis pathways of Acidithiobacillia and neutrophiles. Hopanoid biosynthesis regulates membrane fluidity, maintaining membrane integrity and permeability of cells and promoting resistance to antibiotics, detergents, extreme pH, high osmolarity (Welander et al., 2009; Wu et al., 2015), and surviving general environmental stressors such as in the cyanobacterium genus Nostoc (Ricci et al., 2017; Mayer et al., 2021). The neutrophile T. tepidarius synthesizes squalene via the hpnCDE gene products compared to the one step through sqs gene product in the acidophiles (Figure 6). A further hopanoid cluster was conserved in the Acidithiobacillaceae, including the hpnABFGHIJKLMN genes conferring the ability to produce several hopanoids such as bacteriohopanetetrol (BHT), cyclitol, and hopan-22-ol. This was consistent with A. thiooxidans as a source of bacteriohopanepolyols (BHPs) including BHT, aminotriol, and BHT cyclitol ether (Jones et al., 2012) and the Acidithiobacillales for BHT cyclitol ether, aminotriol, BHT, and adenosylhopane at geothermal vents (Gibson et al., 2014). RNA transcripts confirmed the expression and functionality of sqs gene in A. ferrivorans (Christel et al., 2016). Phylogenomic analysis of the hpn cluster showed similarity with hpn from the acidophile Acidiferrobacter sp. SPIII (Supplementary Figure 7), highlighting the role of this pathway for extreme acidophiles.
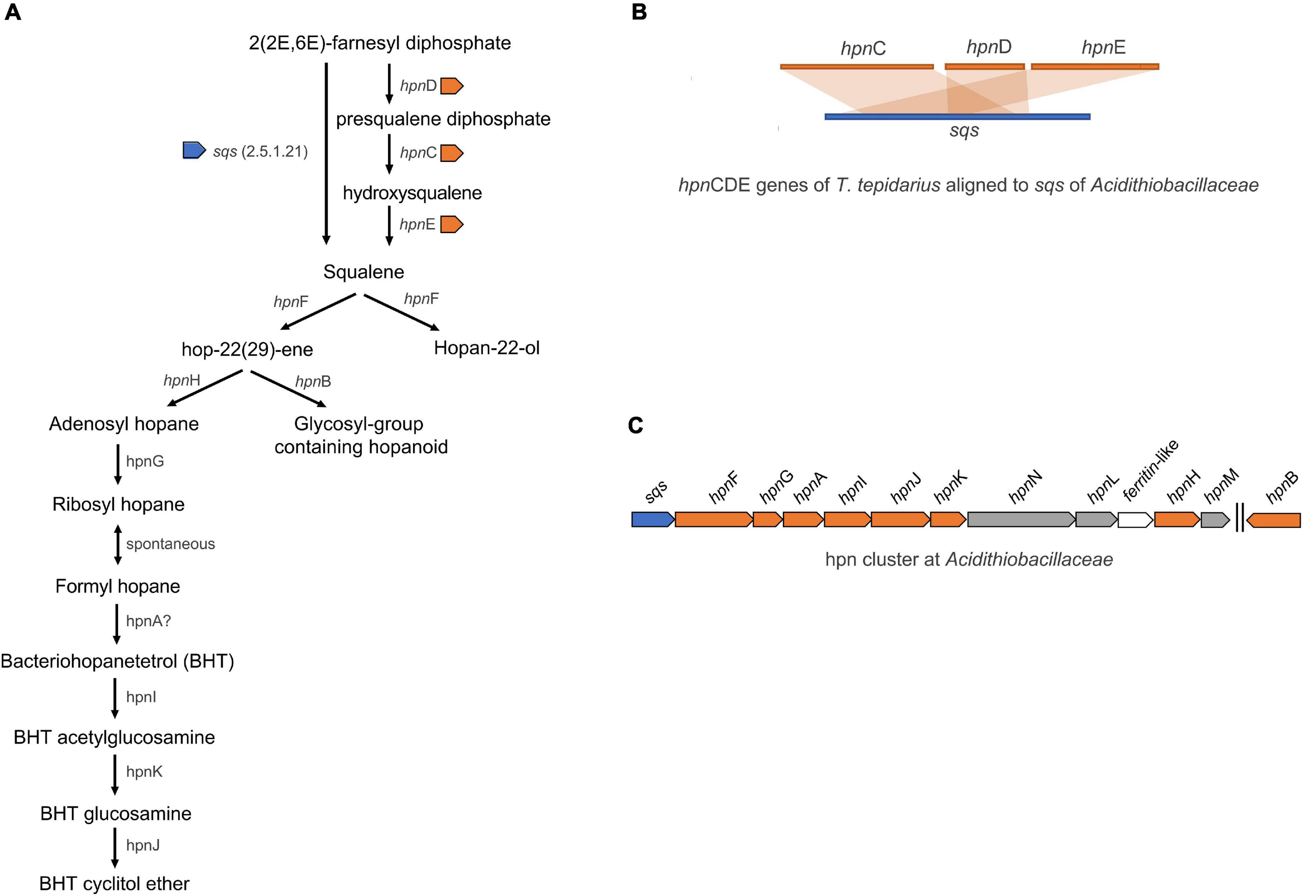
Figure 6. (A) Hopanoid biosynthesis in Acidithiobacillaceae. (B) Biosynthesis of squalene, that is required for hopanoid, is synthetized by sqs in acidophiles in contrast to hpnCDE in the neutrophile T. tepidarius. (C) The hpn cluster was identified in Acidithiobacillaceae including sqs-hpnFGAIJKNL-ferritin-hpnHM genes.
The second line of defense includes genes coding for proton export from the cell (Figure 7). Phosphorus is of utmost importance for living organisms (Li et al., 2011) and may also ameliorate low pH stress via cytoplasmic buffering of protons (Baker-Austin and Dopson, 2007; Chen L. et al., 2015). The phosphate transport system genes pstSCAB are abundant in acidophilic Acidithiobacillus, Leptospirillum, and Acidiphilium taxa in acid mine drainage (Chen L. et al., 2015). A complete repertoire of phosphate uptake genes was identified in the Acidithiobacillia class including the two-component regulatory proteins PhoB/PhoR, Pst-transport system PstSCAB (Valdés et al., 2008), and auxiliary PhoU (Vuppada et al., 2018). The Acidithiobacillaceae genomes maintained two copies of the pstSCA gene cluster except A. thiooxidans and A. ferriphilus-like BY0502. HGT event signals were identified for pstSCA genes in A. ferriphilus-like BY0502 from a Bacteria donor.
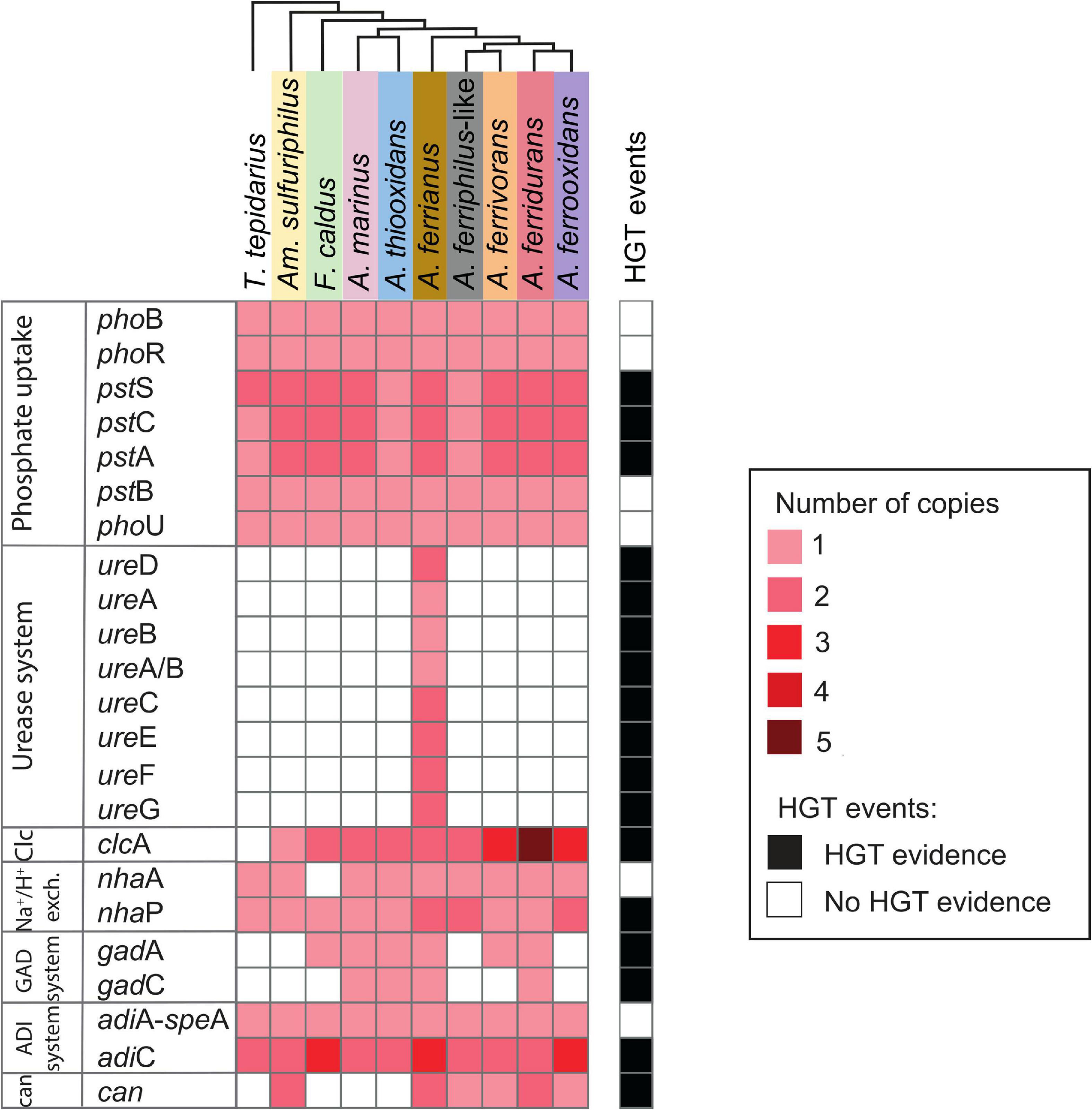
Figure 7. Heatmap of presence (red) and absence (white) of genes involved in second line of defense acid resistance in the Acidithiobacillia class. The red color intensity represents gene copy number per genome. The presence of genes (black; right-hand side) denotes HGT events by genome context with mobile genetic elements, phylogenetic trees, or if it was signaled as part of HGT event by HGTector (Zhu et al., 2014). Species abbreviations are as described in the Figure 3 legend. Second line of acid resistance genes include phosphate uptake, urease system, ClcA antiporter (Clc), proton antiporter (Na+/H+ exch.), amino acid decarboxylation (GAD and ADI systems), and carbonic anhydrase (can) (shaded colors correspond to those used in Figure 1).
The urease complex can act as buffering capacity of the intracellular pH using ammonia derived from urea hydrolysis such in Ferrovum group 2 strains (Ullrich et al., 2016a). The role of urease in pH homeostasis has been shown for Helicobacter pylori during gastric colonization (Scott et al., 1998; Stingl et al., 2002; Schoep et al., 2010) and suggested for “Ferrovum” strain JA12 (Ullrich et al., 2016b) and Thiomonas sp. CB2 (Farasin et al., 2015). The three sub-unit cytoplasmic apoenzyme urease (UreABC) synthesizes CO2 and NH3 from urea, interacting with UreDFG and UreE (Carter et al., 2009). Urease complexes were found in A. ferrianus MG (Figure 8) encoded by ureDABCEFG (urease 1) and ureEF(A/B)CGD (urease 2) where ureA and ureB genes were fused in a single gene denominated ure(A/B) as identified in H. pylori (Voland et al., 2003). A manual curation of genomic context of urease 2 displayed closeness with transposase elements that suggested an origin by HGT event, which was confirmed by HGTector displaying transfer of ureF(A/B)CG genes from Proteobacteria donors. HGT signal events were also identified for all urease 1 complex genes that were associated with Bacteria and Cyanobacteria donors. Other urease clusters, with the same gene distribution as urease 1, were identified in other Acidithiobacillaceae species including A. ferrooxidans IO-2C (new proposed annotation A. ferridurans), F. caldus BC13, and A. ferriphilus DSM 100412 (Supplementary Figure 8).
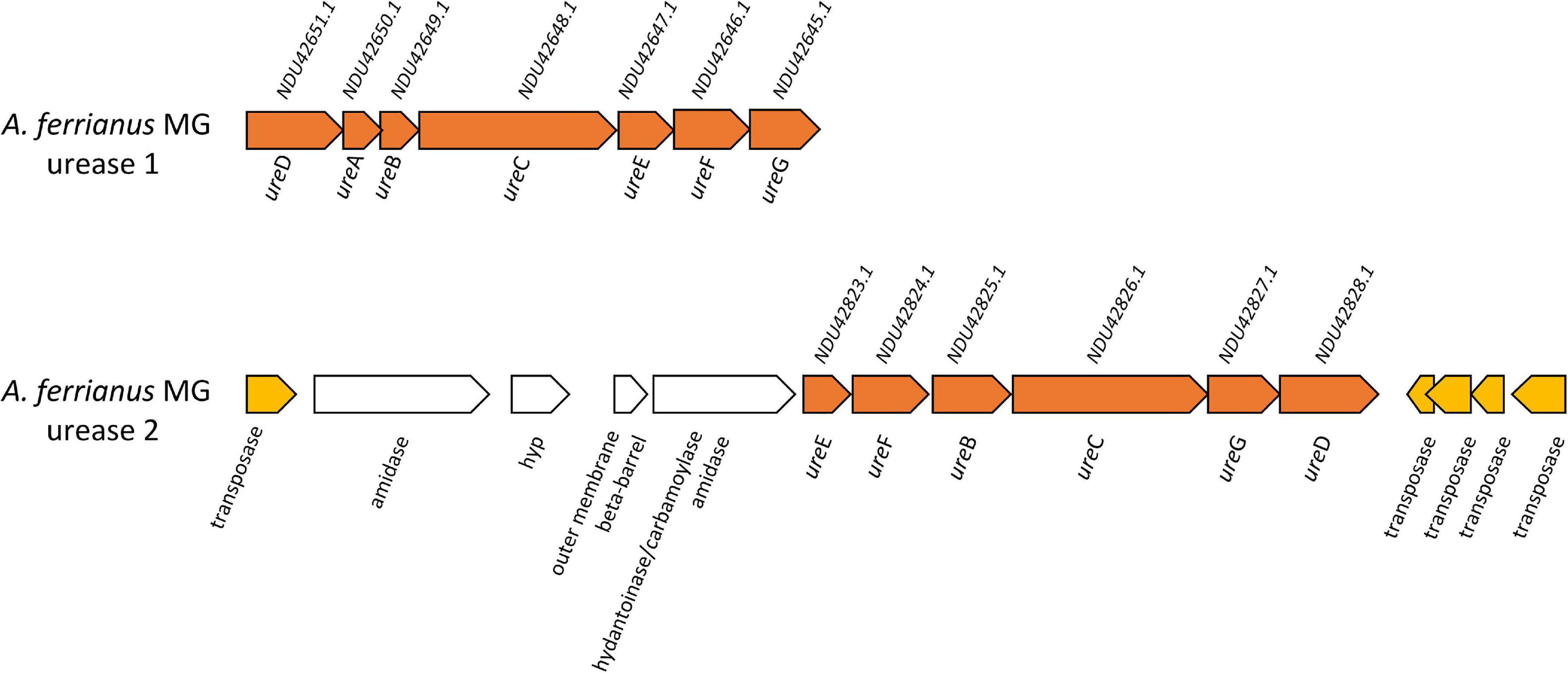
Figure 8. Genetic organization of urease 1 and urease 2 system in A. ferrianus MG. Genome context of urease 1 and extended urease 2 in A. ferrianus MG formed by ureABCDEFG (orange) with their corresponding accession numbers. Mobile genetic elements are highlighted by yellow arrows.
The Cl–/H+ antiporter ClcA prevents inner-membrane hyperpolarization at extreme acidic pH in E. coli (Richard and Foster, 2004) and Bacillus coagulans (McLaggan et al., 1990). This antiporter was gained by the extreme acidophile Leptospirillum as demonstrated by remaining mobile elements (Vergara et al., 2020). The acidophilic Acidithiobacillaceae contained two contiguous clcA genes with the exception of A. ferrianus where the genes were present in separate genomic regions (different contigs) and Am. sulfuriphilus that only had a single copy. In addition, the iron oxidizing species A. ferrivorans, A. ferrooxidans, and A. ferridurans contained more than two copies in their genomes. Phylogenetic trees indicated an association of Acidithiobacillaceae ClcA with other extreme acidophiles (Supplementary Figure 9) such as Acidihalobacter prosperus, Ac. ferrooxydans (Khaleque et al., 2019), Ac. yilgarnensis (Khaleque et al., 2020), Thermodesulfobium acidiphilum (Frolov et al., 2017), and Sulfolobus acidocaldarius (Chen et al., 2005). In addition, HGTector predicted an HGT signal from Bacteria, Proteobacteria, and acidophilic Ferrovum-like donors. The neutrophilic T. tepidarius lacked the clcA antiporter, which agreed with Vergara et al. (2020) who suggested that clcA could be gained by HGT events as a mechanism to resist extreme acid environments. A proton antiporter NhaA was identified in T. tepidarius and the Acidithiobacillaceae with the exception of F. caldus, which was suggested to have lost this gene according to MAUVE synteny analysis. A second proton antiporter, NhaP, was identified in the Acidithiobacillia class with additional copies in the acidophiles A. ferrianus, A. ferriphilus-like, and A. ferrooxidans. Finally, both nhaP genes from A. ferriphilus-like were from HGT events with possible Bacteria and Ferrovum-like donors.
Amino acid decarboxylation systems such as glutamic acid-dependent acid resistance (GDAR) catalyze proton consumption. These systems consist of glutamic acid decarboxylases GadA/GadB enzymes and the glutamate/γ-aminobutyric acid (GABA) antiporter GadC while the arginine-dependent acid resistance (ADAR) system involves arginine decarboxylase AdiA and the arginine/agmatine antiporter AdiC (Foster, 2004; Kanjee and Houry, 2013). The GAD system encoded by gadA and gadC was identified in A. marinus SH, A. thiooxidans, A. ferrianus, and A. ferridurans. Phylogenetic analysis of GadA showed a clade of Acidithiobacillaceae genes sharing a common ancestor with Acidihalobacter acidophiles (Supplementary Figure 10), which was supported by the prediction of an HGT event from Gammaproteobacteria to Acidithiobacillaceae for gadC in A. marinus and A. ferridurans, and the presence of a mobile genetic elements (MGE) upstream of gadC in A. ferrianus. These results suggested that the GAD system was gained by acidophiles by obtention of decarboxylase GadA and amino acid permease GadC. Analysis revealed that the F. caldus and A. ferrivorans genomes only encoded gadA and they lacked the gadC antiporter (Mangold et al., 2011). However, it is proposed that the decarboxylated GABA product from glutamate decarboxylation might be retained in the cell where it can be incorporated into the TCA cycle (Karatzas et al., 2010; Feehily and Karatzas, 2013; Sriaporn et al., 2021). Even though a complete glutamate decarboxylase exporting system was not identified, previous studies have shown that amino acid decarboxylation is highly expressed in F. caldus under acid stress conditions (Mangold et al., 2013; Sriaporn et al., 2021). The adiA gene contained the domain speA that also participates in putrescine synthesis and explains why this gene is associated with both pathways (adiA-speA; Figure 7). The ADAR system was identified in both Acidithiobacillia neutrophiles and acidophiles with a single gene encoding arginine-decarboxylate AdiA and amino acid permease AdiC. However, F. caldus, A. ferrianus, and A. ferrooxidans had three copies of adiC. Finally, adiC genes were suggested to be acquired by HGT transfer from Proteobacteria and Acidiferrobacter-like donors that suggested an evolutionary gain event for acidophiles.
Carbonic anhydrase (can) aids pH homeostasis by catalyzing the interconversion of CO2 to HCO3– (Bury-Moné et al., 2008; Valdés et al., 2008; Frost and Mckenna, 2014; Ansari and Yamaoka, 2017; Hu et al., 2020) and was identified in acidophilic microorganisms such as Leptospirillum, Ferrovum spp., S. thermosulfidooxidans, and Sulfobacillus sp. (Ullrich et al., 2016a,b; Zhang et al., 2017; Hu et al., 2020). Depending on the direction of reaction, the β-carbonic anhydrase may prevent cytoplasmic acidification by breakdown of HCO3– (consuming H+) or conversion of CO2 to HCO3– for carbon fixation (Lehtovirta-Morley et al., 2016). A cytoplasmic carbonic anhydrase of the β-class clade B (Valdés et al., 2008; Esparza et al., 2019) was identified in Am. sulfuriphilus and iron oxidizer genomes with two copies in Am. sulfuriphilus, A. ferrianus, and A. ferridurans. The additional copy of can in the A. ferridurans genome was proposed to result from a transfer event from a Bacteria donor according to HGTector. This concept was reinforced by interpretation of phylogenetic trees (clade can1–can2, Supplementary Figure 11) displaying similarity of Can from A. ferrianus and Am. sulfuriphilus with acidophiles Acidihalobacter and Sulfobacillus. Finally, a truncated can was found in the neutrophile T. tepidarius. However, its activity needs to be experimentally investigated.
The model proposes the transition of a neutrophilic Acidithiobacillia ancestor to the extremely acidophilic Acidithiobacillaceae family by reinforcing the outer membrane and generating a positive membrane potential to restrict the influx of protons into the cytoplasm (Figure 9). Three mechanisms were exclusively identified in extreme Acidithiobacillaceae acidophiles, namely, (i) capsular biosynthesis pathways, (ii) influx of potassium ions, and (iii) hopanoid biosynthesis. NhaP, Kup, Kch, and Kdp were identified in acidophiles generating a transmembrane electrical potential and redundancy of genes for potassium influx. The presence of sqs gene in Acidithiobacillaceae coding for squalene biosynthesis in a single reaction (as compared with the three reactions in T. tepidarius) may also represent an important evolutionary energetic advantage for hopanoid biosynthesis, which, along with the hpnFGAIJKNLHM gene cluster, allows to synthetize and modify hopanoids. The second line in Acidithiobacillaceae presented a pool of accessory genes for buffering of intracellular pH, including decarboxylation of glutamate, urea hydrolysis, and hydration of CO2, and antiporters export excess protons by coupling the uptake of Na+ (Chen et al., 2021). This functional redundancy may represent a key strategy for acidophiles to live across different pH ranges and “hedge bet” in rapidly changing acidic environments, where it is hypothesized that the cost of maintaining genetic redundancy is offset by the ability to expeditiously adjust to environmental fluxes.
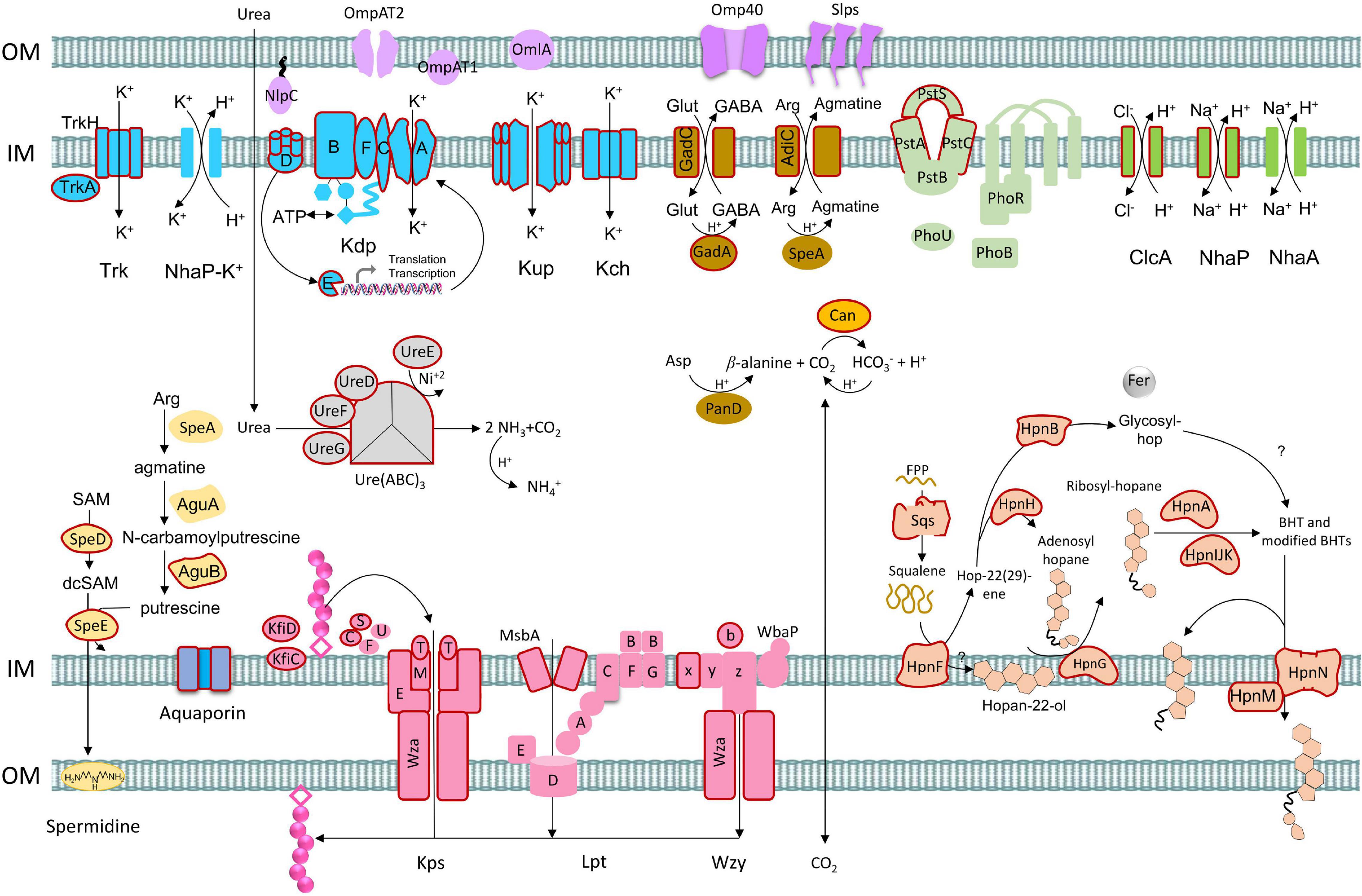
Figure 9. Model of acid resistance genes and mechanisms in acidophiles from the Acidithiobacillaceae including first and second lines of defense. Gene gain events proposed by HGT are represented with outline colors in red according to phylogenetic/phylogenomic and prediction (Zhu et al., 2014) analysis.
The core genome phylogenomic and acid resistance protein phylogenetic trees confirmed the transition across the Acidithiobacillia class from an ancestral neutrophile to an extreme acidophile (Williams et al., 2010; Williams and Kelly, 2013; González et al., 2016). Acidithiobacillia acid resistance genes were classified into first and second lines of defense, where most were identified in the extremely acidophilic Acidithiobacillaceae family. The analysis suggested that whereas a major fraction of genes involved in acid resistance were inherited vertically, genome reprograming including duplication, gain by HGT from extreme acidophiles, and mutation of genes played a role in the evolution of the acidophilic lifestyle. Especially prominent in our analysis was the prediction of a large number of HGT events from other extreme acidophiles, suggesting that this mode of gene acquisition played a major role in the evolution of an inferred neutrophilic ancestor into a clade of extreme acidophiles.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
DH and JV conceived and designed the research. CG-R performed the research. CG-R, EV, MD, JV, and DH analyzed the data. All authors participated in the writing and approval of the final manuscript.
This work was supported by the ANID FONDECYT 3190792 (CG-R), FONDECYT 1181717 (DH), Centro Ciencia & Vida, and FB210008 Financiamiento Basal para Centros Científicos y Tecnológicos de Excelencia de ANID.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.822229/full#supplementary-material
Supplementary Data Sheet 1 | List of conserved core genes in Acidithiobacillia class.
Supplementary Data Sheet 2 | List of genes involved into first and second line of defense acid resistance in the Acidithiobacillia class.
Supplementary Image 1 | Supplementary figures.
Alexander, B., Leach, S., and Ingledew, W. J. (1987). The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. Microbiology 133, 1171–1179. doi: 10.1099/00221287-133-5-1171
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Altschul, S. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Amaro, A. M., Chamorro, D., Seeger, M., Arredondo, R., Peirano, I., and Jerez, C. A. (1991). Effect of external pH perturbations on in vivo protein synthesis by the acidophilic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 173, 910–915. doi: 10.1128/jb.173.2.910-915.1991
Ansari, S., and Yamaoka, Y. (2017). Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22:12386. doi: 10.1111/hel.12386
Baker-Austin, C., and Dopson, M. (2007). Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15, 165–171. doi: 10.1016/j.tim.2007.02.005
Barahona, S., Dorador, C., Zhang, R., Aguilar, P., Sand, W., Vera, M., et al. (2014). Isolation and characterization of a novel Acidithiobacillus ferrivorans strain from the Chilean Altiplano: attachment and biofilm formation on pyrite at low temperature. Res. Microbiol. 165, 782–793. doi: 10.1016/j.resmic.2014.07.015
Barton, L. L. (2005). “Capsules, pili, and internal structures,” in Structural and Functional Relationships in Prokaryotes, (New York, NY: Springer), 190–233. doi: 10.1007/0-387-27125-2_5
Bertelli, C., Laird, M. R., Williams, K. P. Fraser University Research Computing Group Lau, B. Y., Hoad, G., et al. (2017). Island viewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Boden, R., Hutt, L. P., Huntemann, M., Clum, A., Pillay, M., Palaniappan, K., et al. (2016). Permanent draft genome of Thermithiobacillus tepidarius DSM 3134T, a moderately thermophilic, obligately chemolithoautotrophic member of the acidithiobacillia. Stand. Genomic Sci. 11:74. doi: 10.1186/s40793-016-0188-0
Bossemeyer, D., Borchard, A., Dosch, D. C., Helmer, G. C., Epstein, W., Booth, I. R., et al. (1989). K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 264, 16403–16410.
Bryant, R. D., McGroarty, K. M., Costerton, J. W., and Laishley, E. J. (1983). Isolation and characterization of a new acidophilic Thiobacillus species (T. albertis). Can. J. Microbiol. 29, 1159–1170. doi: 10.1139/m83-178
Buetti-Dinh, A., Dethlefsen, O., Friedman, R., and Dopson, M. (2016). Transcriptomic analysis reveals how a lack of potassium ions increases Sulfolobus acidocaldarius sensitivity to pH changes. Microbiology 162, 1422–1434. doi: 10.1099/mic.0.000314
Bury-Moné, S., Mendz, G. L., Ball, G. E., Thibonnier, M., Stingl, K., Ecobichon, C., et al. (2008). Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect. Immun. 76, 497–509. doi: 10.1128/IAI.00993-07
Camacho, D., Frazao, R., Fouillen, A., Nanci, A., Lang, B. F., Apte, S. C., et al. (2020). New insights into Acidithiobacillus thiooxidans sulfur metabolism through coupled gene expression, solution chemistry, microscopy, and spectroscopy analyses. Front. Microbiol. 11:411. doi: 10.3389/fmicb.2020.00411
Carter, E. L., Flugga, N., Boer, J. L., Mulrooney, S. B., and Hausinger, R. P. (2009). Interplay of metal ions and urease. Metallomics 1, 207–221. doi: 10.1039/B903311D
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., and McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Castro, M., Moya-Beltrán, A., Covarrubias, P. C., Gonzalez, M., Cardenas, J. P., Issotta, F., et al. (2017). Draft genome sequence of the type strain of the sulfur-oxidizing acidophile, Acidithiobacillus albertensis (DSM 14366). Stand. Genom. Sci. 12:77. doi: 10.1186/s40793-017-0282-y
Chao, J., Wang, W., Xiao, S., and Liu, X. (2008). Response of Acidithiobacillus ferrooxidans ATCC 23270 gene expression to acid stress. World J. Microbiol. Biotechnol. 24, 2103–2109. doi: 10.1007/s11274-008-9715-5
Chen, C. P., Kernytsky, A., and Rost, B. (2002). Transmembrane helix predictions revisited. Protein Sci. 11, 2774–2791. doi: 10.1110/ps.0214502
Chen, J., Liu, Y., Diep, P., and Mahadevan, R. (2021). Genomic analysis of a newly isolated Acidithiobacillus ferridurans JAGS strain reveals its adaptation to acid mine drainage. Minerals 11:74. doi: 10.3390/min11010074
Chen, L., Brügger, K., Skovgaard, M., Redder, P., She, Q., Torarinsson, E., et al. (2005). The genome of Sulfolobus acidocaldarius, a model organism of the crenarchaeota. J. Bacteriol. 187, 4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005
Chen, L., Hu, M., Huang, L., Hua, Z., Kuang, J., Li, S., et al. (2015). Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 9, 1579–1592. doi: 10.1038/ismej.2014.245
Chen, P., Yan, L., Wu, Z., Xu, R., Li, S., Wang, N., et al. (2015). Draft genome sequence of extremely acidophilic bacterium Acidithiobacillus ferrooxidans DLC-5 isolated from acid mine drainage in northeast China. Genom. Data 6, 267–268. doi: 10.1016/j.gdata.2015.10.018
Chi, X., Fan, Q., Zhang, Y., Liang, K., Wan, L., Zhou, Q., et al. (2020). Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Res. 30, 1127–1135. doi: 10.1038/s41422-020-00404-6
Cholo, M. C., van Rensburg, E. J., Osman, A. G., and Anderson, R. (2015). Expression of the genes encoding the Trk and Kdp potassium transport systems of Mycobacterium tuberculosis during growth in vitro. BioMed Res. Int. 2015:e608682. doi: 10.1155/2015/608682
Christel, S., Fridlund, J., Watkin, E. L., and Dopson, M. (2016). Acidithiobacillus ferrivorans SS3 presents little RNA transcript response related to cold stress during growth at 8 °C suggesting it is a eurypsychrophile. Extremophiles 20, 903–913. doi: 10.1007/s00792-016-0882-2
Christel, S., Herold, M., Bellenberg, S., Hajjami, M. E., Buetti-Dinh, A., Pivkin, I. V., et al. (2018). Multi-omics reveals the lifestyle of the acidophilic, mineral-oxidizing model species Leptospirillum ferriphilumT. Appl. Environ. Microbiol. 84, e02091–17. doi: 10.1128/AEM.02091-17
Colman, D. R., Poudel, S., Hamilton, T. L., Havig, J. R., Selensky, M. J., Shock, E. L., et al. (2018). Geobiological feedbacks and the evolution of thermoacidophiles. ISME J. 12, 225–236. doi: 10.1038/ismej.2017.162
Contreras-Moreira, B., and Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
Crooks, G. E., Hon, G., Chandonia, J.-M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Darling, A. E., Mau, B., and Perna, N. T. (2010). Progressive mauve: multiple genome alignment with gene gain. Loss and Rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
De Biase, D., and Lund, P. A. (2015). “Chapter two - the Escherichia coli acid stress response and its significance for pathogenesis,” in Advances in Applied Microbiology, eds S. Sariaslani and G. M. Gadd (Academic Press), 49–88. doi: 10.1016/bs.aambs.2015.03.002
Dong, C., Beis, K., Nesper, J., Brunkan, A. L., Clarke, B. R., Whitfield, C., et al. (2006). The structure of Wza, the translocon for group 1 capsular polysaccharides in Escherichia coli, identifies a new class of outer membrane protein. Nature 444, 226–229. doi: 10.1038/nature05267
Dopson, M., Holmes, D. S., Lazcano, M., McCredden, T. J., Bryan, C. G., Mulroney, K. T., et al. (2017). Multiple osmotic stress responses in Acidihalobacter prosperus result in tolerance to chloride ions. Front. Microbiol. 7:2132. doi: 10.3389/fmicb.2016.02132
Duarte, F., Araya-Secchi, R., González, W., Perez-Acle, T., González-Nilo, D., and Holmes, D. S. (2009). Protein function in extremely acidic conditions: molecular simulations of a predicted aquaporin and a potassium channel in Acidithiobacillus ferrooxidans. Adv. Mat. Res. 7, 211–214.
Epstein, W. (2003). The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75, 293–320. doi: 10.1016/S0079-6603(03)75008-9
Esparza, M., Jedlicki, E., González, C., Dopson, M., and Holmes, D. S. (2019). Effect of CO2 concentration on uptake and assimilation of inorganic carbon in the extreme acidophile Acidithiobacillus ferrooxidans. Front. Microbiol. 10:603. doi: 10.3389/fmicb.2019.00603
Falagán, C., and Johnson, D. B. (2016). Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 66, 206–211. doi: 10.1099/ijsem.0.000698
Falagán, C., Moya-Beltrán, A., Castro, M., Quatrini, R., and Johnson, D. B. (2019). Acidithiobacillus sulfuriphilus sp. nov.: an extremely acidophilic sulfur-oxidizing chemolithotroph isolated from a neutral pH environment. Int. J. Syst. Evol. Microbiol. 69, 2907–2913. doi: 10.1099/ijsem.0.003576
Farasin, J., Andres, J., Casiot, C., Barbe, V., Faerber, J., Halter, D., et al. (2015). Thiomonas sp. CB2 is able to degrade urea and promote toxic metal precipitation in acid mine drainage waters supplemented with urea. Front. Microbiol. 6:993. doi: 10.3389/fmicb.2015.00993
Fariq, A., Blazier, J. C., Yasmin, A., Gentry, T. J., and Deng, Y. (2019). Whole genome sequence analysis reveals high genetic variation of newly isolated Acidithiobacillus ferrooxidans IO-2C. Sci. Rep. 9:13049. doi: 10.1038/s41598-019-49213-x
Feehily, C., and Karatzas, K. A. G. (2013). Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 114, 11–24. doi: 10.1111/j.1365-2672.2012.05434.x
Feng, S., Li, K., Huang, Z., Tong, Y., and Yang, H. (2019). Specific mechanism of Acidithiobacillus caldus extracellular polymeric substances in the bioleaching of copper-bearing sulfide ore. PLoS One 14:e0213945. doi: 10.1371/journal.pone.0213945
Feng, S., Yang, H., and Wang, W. (2015). System-level understanding of the potential acid-tolerance components of Acidithiobacillus thiooxidans ZJJN-3 under extreme acid stress. Extremophiles 19, 1029–1039. doi: 10.1007/s00792-015-0780-z
Fischer, S., Brunk, B. P., Chen, F., Gao, X., Harb, O. S., Iodice, J. B., et al. (2011). Using OrthoMCL to assign proteins to OrthoMCL-db groups or to cluster proteomes into new ortholog groups. Curr. Prot. Bioinform. 35, 6.12.1–6.12.19. doi: 10.1002/0471250953.bi0612s35
Foster, J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. doi: 10.1038/nrmicro1021
Frolov, E. N., Kublanov, I. V., Toshchakov, S. V., Samarov, N. I., Novikov, A. A., Lebedinsky, A. V., et al. (2017). Thermodesulfobium acidiphilum sp. nov., a thermoacidophilic, sulfate-reducing, chemoautotrophic bacterium from a thermal site. Int. J. Syst. Evol. Microbiol. 67, 1482–1485. doi: 10.1099/ijsem.0.001745
Frost, S., and Mckenna, R. (2014). Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications. Dordrecht: Springer. doi: 10.1007/978-94-007-7359-2
Gassel, M., Siebers, A., Epstein, W., and Altendorf, K. (1998). Assembly of the Kdp complex, the multi-subunit K+-transport ATPase of Escherichia coli. Biochimica et Biophysica Acta 1415, 77–84. doi: 10.1016/S0005-2736(98)00179-5
Gibson, R. A., Sherry, A., Kaur, G., Pancost, R. D., and Talbot, H. M. (2014). Bacteriohopanepolyols preserved in silica sinters from champagne pool (new zealand) indicate a declining temperature gradient over the lifetime of the vent. Org. Geochem. 69, 61–69. doi: 10.1016/j.orggeochem.2014.02.004
González, C., Lazcano, M., Valdés, J., and Holmes, D. S. (2016). Bioinformatic analyses of unique (orphan) core genes of the genus Acidithiobacillus: functional inferences and use as molecular probes for genomic and metagenomic/transcriptomic interrogation. Front. Microbiol. 7:e02035. doi: 10.3389/fmicb.2016.02035
González, C., Yanquepe, M., Cardenas, J. P., Valdes, J., Quatrini, R., Holmes, D. S., et al. (2014). Genetic variability of psychrotolerant Acidithiobacillus ferrivorans revealed by (meta)genomic analysis. Res. Microbiol. 165, 726–734. doi: 10.1016/j.resmic.2014.08.005
Guiliani, N., and Jerez, C. A. (2000). Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 66, 2318–2324. doi: 10.1128/AEM.66.6.2318-2324.2000
Hallberg, K. B., González-Toril, E., and Johnson, D. B. (2010). Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14, 9–19. doi: 10.1007/s00792-009-0282-y
Hallberg, K. B., and Lindström, E. B. (1994). Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology (Reading) 140, 3451–3456. doi: 10.1099/13500872-140-12-3451
Hedrich, S., and Johnson, D. B. (2013). Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 63, 4018–4025. doi: 10.1099/ijs.0.049759-0
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hu, W., Feng, S., Tong, Y., Zhang, H., and Yang, H. (2020). Adaptive defensive mechanism of bioleaching microorganisms under extremely environmental acid stress: advances and perspectives. Biotechnol. Adv. 42:107580. doi: 10.1016/j.biotechadv.2020.107580
Hughes, G. W., Hall, S. C. L., Laxton, C. S., Sridhar, P., Mahadi, A. H., Hatton, C., et al. (2019). Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 4, 1692–1705. doi: 10.1038/s41564-019-0481-y
Inaba, Y., Banerjee, I., Kernan, T., and Banta, S. (2018). Transposase-mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 84, e01381–18. doi: 10.1128/AEM.01381-18
Issotta, F., Moya-Beltrán, A., Mena, C., Covarrubias, P. C., Thyssen, C., Bellenberg, S., et al. (2018). Insights into the biology of acidophilic members of the Acidiferrobacteraceae family derived from comparative genomic analyses. Res. Microbiol. 169, 608–617. doi: 10.1016/j.resmic.2018.08.001
Jones, D. S., Albrecht, H. L., Dawson, K. S., Schaperdoth, I., Freeman, K. H., Pi, Y., et al. (2012). Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J. 6, 158–170. doi: 10.1038/ismej.2011.75
Juncker, A. S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H., and Krogh, A. (2003). Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12, 1652–1662. doi: 10.1110/ps.0303703
Jung, H., Inaba, Y., and Banta, S. (2021). Genetic engineering of the acidophilic chemolithoautotroph Acidithiobacillus ferrooxidans. Trends Biotechnol. [Epub ahead of print]. doi: 10.1016/j.tibtech.2021.10.004
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). Model finder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kamimura, K., Sharmin, S., Yoshino, E., Tokuhisa, M., and Kanao, T. (2018). Draft genome sequence of Acidithiobacillus sp. strain SH, a marine acidophilic sulfur-oxidizing bacterium. Genome Ann. 6:e01603-17. doi: 10.1128/genomeA.01603-17
Kanjee, U., and Houry, W. A. (2013). Mechanisms of acid resistance in Escherichia coli. Annu Rev. Microbiol. 67, 65–81. doi: 10.1146/annurev-micro-092412-155708
Karatzas, K.-A. G., Brennan, O., Heavin, S., Morrissey, J., and O’Byrne, C. P. (2010). Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 76, 3529–3537. doi: 10.1128/AEM.03063-09
Katoh, K. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Br. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khaleque, H. N., González, C., Johnson, D. B., Kaksonen, A. H., Holmes, D. S., and Watkin, E. L. J. (2020). Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov. Int. J. Syst. Evol. Microbiol. 70, 6226–6234. doi: 10.1099/ijsem.0.004519
Khaleque, H. N., González, C., Kaksonen, A. H., Boxall, N. J., Holmes, D. S., and Watkin, E. L. J. (2019). Genome-based classification of two halotolerant extreme acidophiles, Acidihalobacter prosperus V6 (=DSM 14174 =JCM 32253) and “Acidihalobacter ferrooxidans” V8 (=DSM 14175 =JCM 32254) as two new species, Acidihalobacter aeolianus sp. nov. and Acidihalobacter ferrooxydans sp. nov., respectively. Int. J. Syst. Evol. Microbiol. 69, 1557–1565. doi: 10.1099/ijsem.0.003313
Kichenaradja, P., Siguier, P., Pérochon, J., and Chandler, M. (2010). ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 38, D62–D68. doi: 10.1093/nar/gkp947
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kucera, J., Sedo, O., Potesil, D., Janiczek, O., Zdrahal, Z., and Mandl, M. (2016). Comparative proteomic analysis of sulfur-oxidizing Acidithiobacillus ferrooxidans CCM 4253 cultures having lost the ability to couple anaerobic elemental sulfur oxidation with ferric iron reduction. Res. Microbiol. 167, 587–594. doi: 10.1016/j.resmic.2016.06.009
Larsson, A. (2014). AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. doi: 10.1093/bioinformatics/btu531
Lehtovirta-Morley, L. E., Sayavedra-Soto, L. A., Gallois, N., Schouten, S., Stein, L. Y., Prosser, J. I., et al. (2016). Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra”. Appl. Environ. Microbiol. 82, 2608–2619. doi: 10.1128/AEM.04031-15
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, L., Liu, Z., Zhang, M., Meng, D., Liu, X., Wang, P., et al. (2020). Insights into the metabolism and evolution of the genus acidiphilium, a typical acidophile in acid mine drainage. mSystems 5, e00867–20. doi: 10.1128/mSystems.00867-20
Li, Q., Li, N., Liu, X., Zhou, Z., Fang, Y., Fan, X., et al. (2012). Characterization of the acid stress response of Acidithiobacillus ferrooxidans ATCC 23270 based on the method of microarray. J. Biol. Res. 17, 3–15.
Li, Q., Ren, Y., Qiu, G., Li, N., Liu, H., Dai, Z., et al. (2011). Insights into the pH up-shift responsive mechanism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Folia Microbiol. (Praha) 56, 439–451. doi: 10.1007/s12223-011-0067-4
Liljeqvist, M., Ossandon, F. J., González, C., Rajan, S., Stell, A., Valdes, J., et al. (2015). Metagenomic analysis reveals adaptations to a cold-adapted lifestyle in a low-temperature acid mine drainage stream. FEMS Microbiol. Ecol. 91:fiv011. doi: 10.1093/femsec/fiv011
Liljeqvist, M., Valdes, J., Holmes, D. S., and Dopson, M. (2011). Draft genome of the psychrotolerant acidophile Acidithiobacillus ferrivorans SS3. J. Bacteriol. 193, 4304–4305. doi: 10.1128/JB.05373-11
Ma, B., Reynolds, C. M., and Raetz, C. R. H. (2008). Periplasmic orientation of nascent lipid A in the inner membrane of an Escherichia coli LptA mutant. Proc. Natl. Acad. Sci. U.S.A. 105, 13823–13828. doi: 10.1073/pnas.0807028105
Malinverni, J. C., and Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014. doi: 10.1073/pnas.0903229106
Mangold, S., Rao Jonna, V., and Dopson, M. (2013). Response of Acidithiobacillus caldus toward suboptimal pH conditions. Extremophiles 17, 689–696. doi: 10.1007/s00792-013-0553-5
Mangold, S., Valdés, J., Holmes, D. S., and Dopson, M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. doi: 10.3389/fmicb.2011.00017
Mates, A. K., Sayed, A. K., and Foster, J. W. (2007). Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 189, 2759–2768. doi: 10.1128/JB.01490-06
Mayer, M. H., Parenteau, M. N., Kempher, M. L., Madigan, M. T., Jahnke, L. L., and Welander, P. V. (2021). Anaerobic 3-methylhopanoid production by an acidophilic photosynthetic purple bacterium. Arch. Microbiol. 203, 6041–6052. doi: 10.1007/s00203-021-02561-7
McLaggan, D., Keyhan, M., and Matin, A. (1990). Chloride transport pathways and their bioenergetic implications in the obligate acidophile Bacillus coagulans. J. Bacteriol. 172, 1485–1490. doi: 10.1128/jb.172.3.1485-1490.1990
Michels, M., and Bakker, E. P. (1985). Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J. Bacteriol. 161, 231–237. doi: 10.1128/jb.161.1.231-237.1985
Miyauchi, T., Kouzuma, A., Abe, T., and Watanabe, K. (2018). Complete genome sequence of Acidithiobacillus ferridurans JCM 18981. Microbiol. Res. Ann. 7:e01028-18. doi: 10.1128/MRA.01028-18
Moya-Beltrán, A., Beard, S., Rojas-Villalobos, C., Issotta, F., Gallardo, Y., Ulloa, R., et al. (2021). Genomic evolution of the class Acidithiobacillia: deep-branching Proteobacteria living in extreme acidic conditions. ISME J. 15, 3221–3238. doi: 10.1038/s41396-021-00995-x
Mühling, M., Poehlein, A., Stuhr, A., Voitel, M., Daniel, R., and Schlömann, M. (2016). Reconstruction of the metabolic potential of acidophilic Sideroxydans strains from the metagenome of an microaerophilic enrichment culture of acidophilic iron-oxidizing bacteria from a pilot plant for the treatment of acid mine drainage reveals metabolic versatility and adaptation to life at low pH. Front. Microbiol. 7:e02082. doi: 10.3389/fmicb.2016.02082
Munsey, T. S., Mohindra, A., Yusaf, S. P., Grainge, A., Wang, M.-H., Wray, D., et al. (2002). Functional properties of Kch, a prokaryotic homologue of eukaryotic potassium channels. Biochem. Biophys. Res. Commun. 297, 10–16. doi: 10.1016/S0006-291X(02)02095-8
Mykytczuk, N. C. S., Trevors, J. T., Ferroni, G. D., and Leduc, L. G. (2010). Cytoplasmic membrane fluidity and fatty acid composition of Acidithiobacillus ferrooxidans in response to pH stress. Extremophiles 14, 427–441. doi: 10.1007/s00792-010-0319-2
Neira, G., Cortez, D., Jil, J., and Holmes, D. S. (2020). AciDB 1.0: a database of acidophilic organisms, their genomic information and associated metadata. Bioinformatics 36, 4970–4971. doi: 10.1093/bioinformatics/btaa638
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Norris, P. R., Falagán, C., Moya-Beltrán, A., Castro, M., Quatrini, R., and Johnson, D. B. (2020). Acidithiobacillus ferrianus sp. nov.: an ancestral extremely acidophilic and facultatively anaerobic chemolithoautotroph. Extremophiles 24, 329–337. doi: 10.1007/s00792-020-01157-1
Nuñez, H., Covarrubias, P. C., Moya-Beltrán, A., Issotta, F., Atavales, J., Acuña, L. G., et al. (2016). Detection, identification and typing of Acidithiobacillus species and strains: a review. Res. Microbiol. 167, 555–567. doi: 10.1016/j.resmic.2016.05.006
Nzakizwanayo, J., Kumar, S., Ogilvie, L. A., Patel, B. A., Dedi, C., Macfarlane, W. M., et al. (2015). Disruption of Escherichia coli Nissle 1917 K5 capsule biosynthesis, through loss of distinct kfi genes, modulates interaction with intestinal epithelial cells and impact on cell health. PLoS One 10:e0120430. doi: 10.1371/journal.pone.0120430
Oetiker, N., Norambuena, R., Martínez-Bussenius, C., Navarro, C. A., Amaya, F., Álvarez, S. A., et al. (2018). Possible role of envelope components in the extreme copper resistance of the biomining Acidithiobacillus ferrooxidans. Genes 9:347. doi: 10.3390/genes9070347
Osorio, H., Martínez, V., Nieto, P. A., Holmes, D. S., and Quatrini, R. (2008). Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol. 8:203. doi: 10.1186/1471-2180-8-203
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Parro, V., Moreno-Paz, M., and González-Toril, E. (2007). Analysis of environmental transcriptomes by DNA microarrays. Environ. Microbiol. 9, 453–464. doi: 10.1111/j.1462-2920.2006.01162.x
Plante, C. (2000). Role of bacterial exopolymeric capsules in protection from deposit-feeder digestion. Aquat. Microb. Ecol. 21, 211–219. doi: 10.3354/ame021211
Quatrini, R., Escudero, L. V., Moya-Beltrán, A., Galleguillos, P. A., Issotta, F., Acosta, M., et al. (2017). Draft genome sequence of Acidithiobacillus thiooxidans CLST isolated from the acidic hypersaline gorbea salt flat in northern Chile. Stand. Genom. Sci. 12:84. doi: 10.1186/s40793-017-0305-8
Quatrini, R., and Johnson, D. B. (2018). Microbiomes in extremely acidic environments: functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr. Opin. Microbiol. 43, 139–147. doi: 10.1016/j.mib.2018.01.011
Renzi, F., Ittig, S. J., Sadovskaya, I., Hess, E., Lauber, F., Dol, M., et al. (2016). Evidence for a LOS and a capsular polysaccharide in Capnocytophaga canimorsus. Sci. Rep. 6:38914. doi: 10.1038/srep38914
Rhee, H. J., Kim, E.-J., and Lee, J. K. (2007). Physiological polyamines: simple primordial stress molecules. J. Cell. Mol. Med. 11, 685–703. doi: 10.1111/j.1582-4934.2007.00077.x
Riadi, G., Medina-Moenne, C., and Holmes, D. S. (2012). TnpPred: a web service for the robust prediction of prokaryotic transposases. Comparat. Funct. Genom. 2012:e678761. doi: 10.1155/2012/678761
Ricci, J. N., Morton, R., Kulkarni, G., Summers, M. L., and Newman, D. K. (2017). Hopanoids play a role in stress tolerance and nutrient storage in the cyanobacterium nostoc punctiforme. Geobiology 15, 173–183. doi: 10.1111/gbi.12204
Richard, H., and Foster, J. W. (2004). Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal ph and reverse transmembrane potential. J. Bacteriol. 186, 6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004
Samartzidou, H., Mehrazin, M., Xu, Z., Benedik, M. J., and Delcour, A. H. (2003). Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J. Bacteriol. 185, 13–19. doi: 10.1128/JB.185.1.13-19.2003
Schmitz, R. A., Peeters, S. H., Versantvoort, W., Picone, N., Pol, A., Jetten, M. S. M., et al. (2021). Verrucomicrobial methanotrophs: ecophysiology of metabolically versatile acidophiles. FEMS Microbiol. Rev. 45:fuab007. doi: 10.1093/femsre/fuab007
Schneider, T. D., and Stephens, R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. doi: 10.1093/nar/18.20.6097
Schoep, T. D., Fulurija, A., Good, F., Lu, W., Himbeck, R. P., Schwan, C., et al. (2010). Surface properties of Helicobacter pylori urease complex are essential for persistence. PLoS One 5:e15042. doi: 10.1371/journal.pone.0015042
Schonknecht, G., Chen, W.-H., Ternes, C. M., Barbier, G. G., Shrestha, R. P., Stanke, M., et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210. doi: 10.1126/science.1231707
Scott, D. R., Weeks, D., Hong, C., Postius, S., Melchers, K., and Sachs, G. (1998). The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114, 58–70. doi: 10.1016/s0016-5085(98)70633-x
Simmons, S. L., DiBartolo, G., Denef, V. J., Goltsman, D. S. A., Thelen, M. P., and Banfield, J. F. (2008). Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 6:e177. doi: 10.1371/journal.pbio.0060177
Slonczewski, J. L., Fujisawa, M., Dopson, M., and Krulwich, T. A. (2009). “Cytoplasmic pH measurement and homeostasis in bacteria and archaea,” in Advances in Microbial Physiology, ed. R. K. Poole (Academic Press), 1–317. doi: 10.1016/S0065-2911(09)05501-5
Snipen, L., and Ussery, D. W. (2010). Standard operating procedure for computing pangenome trees. Stand. Genom. Sci. 2, 135–141. doi: 10.4056/sigs.38923
Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1998). A hidden markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Int. Syst. Mol. Biol. 6, 175–182.
Sriaporn, C., Campbell, K. A., Van Kranendonk, M. J., and Handley, K. M. (2021). Genomic adaptations enabling Acidithiobacillus distribution across wide-ranging hot spring temperatures and pHs. Microbiome 9:135. doi: 10.1186/s40168-021-01090-1
Stingl, K., Altendorf, K., and Bakker, E. P. (2002). Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10, 70–74. doi: 10.1016/s0966-842x(01)02287-9
Su, J., Gong, H., Lai, J., Main, A., and Lu, S. (2009). The potassium transporter Trk and external potassium modulate Salmonella enterica protein secretion and virulence. Infect. Immun. 77, 667–675. doi: 10.1128/IAI.01027-08
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Talla, E., Hedrich, S., Mangenot, S., Ji, B., Johnson, D. B., Barbe, V., et al. (2014). Insights into the pathways of iron- and sulfur-oxidation, and biofilm formation from the chemolithotrophic acidophile Acidithiobacillus ferrivorans CF27. Res. Microbiol. 165, 753–760. doi: 10.1016/j.resmic.2014.08.002
Temple, K. L., and Colmer, A. R. (1951). The autotrophic oxidation of iron by a new bacterium, Thiobacillus ferrooxidans. J. Bacteriol. 62, 605–611. doi: 10.1128/jb.62.5.605-611.1951
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Tran, T. T. T., Mangenot, S., Magdelenat, G., Payen, E., Rouy, Z., Belahbib, H., et al. (2017). Comparative genome analysis provides insights into both the lifestyle of Acidithiobacillus ferrivorans strain CF27 and the chimeric nature of the iron-oxidizing acidithiobacilli genomes. Front. Microbiol. 8:e01009. doi: 10.3389/fmicb.2017.01009
Travisany, D., Cortés, M. P., Latorre, M., Di Genova, A., Budinich, M., Bobadilla-Fazzini, R. A., et al. (2014). A new genome of Acidithiobacillus thiooxidans provides insights into adaptation to a bioleaching environment. Res. Microbiol. 165, 743–752. doi: 10.1016/j.resmic.2014.08.004
Trchounian, A., and Kobayashi, H. (1999). Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett. 447, 144–148. doi: 10.1016/s0014-5793(99)00288-4
Trchounian, A., and Kobayashi, H. (2000). K+ uptake by fermenting Escherichia coli cells: pH dependent mode of the TrkA system operating. Biosci. Rep. 20, 277–288. doi: 10.1023/A:1026493024066
Ulloa, R., Moya-Beltrán, A., Rojas-Villalobos, C., Nuñez, H., Chiacchiarini, P., Donati, E., et al. (2019). Domestication of local microbial consortia for efficient recovery of gold through top-down selection in airlift bioreactors. Front. Microbiol. 10:60. doi: 10.3389/fmicb.2019.00060
Ullrich, S. R., González, C., Poehlein, A., Tischler, J. S., Daniel, R., Schlömann, M., et al. (2016a). Gene Loss and horizontal gene transfer contributed to the genome evolution of the extreme acidophile “Ferrovum”. Front. Microbiol. 7:797. doi: 10.3389/fmicb.2016.00797
Ullrich, S. R., Poehlein, A., Tischler, J. S., González, C., Ossandon, F. J., Daniel, R., et al. (2016b). Genome analysis of the biotechnologically relevant acidophilic iron oxidising strain JA12 indicates phylogenetic and metabolic diversity within the novel genus “Ferrovum.”. PLoS One 11:e0146832. doi: 10.1371/journal.pone.0146832
Valdes, J., Ossandon, F., Quatrini, R., Dopson, M., and Holmes, D. S. (2011). Draft genome sequence of the extremely acidophilic biomining bacterium Acidithiobacillus thiooxidans ATCC 19377 provides insights into the evolution of the Acidithiobacillus genus. J. Bacteriol. 193, 7003–7004. doi: 10.1128/JB.06281-11
Valdés, J., Pedroso, I., Quatrini, R., Dodson, R. J., Tettelin, H., Blake, R., et al. (2008). Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genom. 9:597. doi: 10.1186/1471-2164-9-597
Valdes, J., Quatrini, R., Hallberg, K., Dopson, M., Valenzuela, P. D. T., and Holmes, D. S. (2009). Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 191, 5877–5878. doi: 10.1128/JB.00843-09
van Teeseling, M. C. F., Pol, A., Harhangi, H. R., van der Zwart, S., Jetten, M. S. M., and Op, etal. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 80, 6782–6791. doi: 10.1128/AEM.01838-14
Varani, A. M., Siguier, P., Gourbeyre, E., Charneau, V., and Chandler, M. (2011). ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12:R30. doi: 10.1186/gb-2011-12-3-r30
Vera, M., Krok, B., Bellenberg, S., Sand, W., and Poetsch, A. (2013). Shotgun proteomics study of early biofilm formation process of Acidithiobacillus ferrooxidans ATCC 23270 on pyrite. Proteomics 13, 1133–1144. doi: 10.1002/pmic.201200386
Vergara, E., Neira, G., González, C., Cortez, D., Dopson, M., and Holmes, D. S. (2020). Evolution of predicted acid resistance mechanisms in the extremely acidophilic leptospirillum genus. Genes (Basel) 11:389. doi: 10.3390/genes11040389
Voges, D., and Jap, B. K. (1998). Recombinant expression, purification and characterization of Kch, a putative Escherichia coli potassium channel protein. FEBS Lett. 429, 104–108. doi: 10.1016/S0014-5793(98)00509-2
Voland, P., Weeks, D. L., Marcus, E. A., Prinz, C., Sachs, G., and Scott, D. (2003). Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastr. Liver Physiol. 284, G96–G106. doi: 10.1152/ajpgi.00160.2002
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–D437. doi: 10.1093/nar/gki005
Vuppada, R. K., Hansen, C. R., Strickland, K. A. P., Kelly, K. M., and McCleary, W. R. (2018). Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol. 18:8. doi: 10.1186/s12866-017-1126-z
Waksman, S. A., and Joffe, J. S. (1922). Microörganisms concerned in the oxidation of sulfur in the soil: II. Thiobacillus thiooxidans, a new sulfur-oxidizing organism isolated from the soil. J. Bacteriol. 7, 239–256. doi: 10.1128/jb.7.2.239-256.1922
Wang, X., Cai, X., Ma, H., Yin, W., Zhu, L., Li, X., et al. (2019). A c-di-AMP riboswitch controlling kdpFABC operon transcription regulates the potassium transporter system in Bacillus thuringiensis. Commun. Biol. 2, 1–10. doi: 10.1038/s42003-019-0414-6
Welander, P. V., Hunter, R. C., Zhang, L., Sessions, A. L., Summons, R. E., and Newman, D. K. (2009). Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 191, 6145–6156. doi: 10.1128/JB.00460-09
Wen, Z., and Zhang, J.-R. (2015). “Chapter 3 - bacterial capsules,” in Molecular Medical Microbiology (Second Edition), eds Y.-W. Tang, M. Sussman, D. Liu, I. Poxton, and J. Schwartzman (Boston: Academic Press), 33–53. doi: 10.1016/B978-0-12-397169-2.00003-2
Whitaker, R. J., Grogan, D. W., and Taylor, J. W. (2005). Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol. Biol. Evol. 22, 2354–2361. doi: 10.1093/molbev/msi233
Williams, K. P., Gillespie, J. J., Sobral, B. W. S., Nordberg, E. K., Snyder, E. E., Shallom, J. M., et al. (2010). Phylogeny of gamma Proteobacteria. J. Bacteriol. 192, 2305–2314. doi: 10.1128/JB.01480-09
Williams, K. P., and Kelly, D. P. (2013). Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 2901–2906. doi: 10.1099/ijs.0.049270-0
Willis, L. M. (2013). KpsC and KpsS are retaining 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) transferases involved in synthesis of bacterial capsules. Proc. Natl. Acad. Sci. U.S.A. 110, 20753–20758. doi: 10.1073/pnas.1312637110
Wu, C.-H., Bialecka-Fornal, M., and Newman, D. K. (2015). Methylation at the C-2 position of hopanoids increases rigidity in native bacterial membranes. eLife 4:e05663. doi: 10.7554/eLife.05663
Xie, Q. W., Tabor, C. W., and Tabor, H. (1989). Spermidine biosynthesis in Escherichia coli: promoter and termination regions of the speED operon. J. Bacteriol. 171, 4457–4465. doi: 10.1128/jb.171.8.4457-4465.1989
Yan, H., Fukamachi, T., Saito, H., and Kobayashi, H. (2011). Expression and activity of Kdp under acidic conditions in Escherichia coli. Biol. Pharmaceut. Bull. 34, 426–429. doi: 10.1248/bpb.34.426
Yan, L., Zhang, S., Wang, W., Hu, H., Wang, Y., Yu, G., et al. (2015). Draft genome sequence of Acidithiobacillus ferrooxidans YQH-1. Genom. Data 6, 269–270. doi: 10.1016/j.gdata.2015.10.009
Yin, H., Zhang, X., Li, X., He, Z., Liang, Y., Guo, X., et al. (2014). Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 14:179. doi: 10.1186/1471-2180-14-179
You, X.-Y., Guo, X., Zheng, H.-J., Zhang, M.-J., Liu, L.-J., Zhu, Y.-Q., et al. (2011). Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J. Genet. Genom. 38, 243–252. doi: 10.1016/j.jgg.2011.04.006
Yu, C.-S., Chen, Y.-C., Lu, C.-H., and Hwang, J.-K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Yuan, B., Cheng, A., and Wang, M. (2013). Polysaccharide export outer membrane proteins in Gram-negative bacteria. Future Microbiol. 8, 525–535. doi: 10.2217/fmb.13.13
Zakharyan, E., and Trchounian, A. (2001). K+ influx by kup in Escherichia coli is accompanied by a decrease in H+ efflux. FEMS Microbiol. Lett. 204, 61–64. doi: 10.1111/j.1574-6968.2001.tb10863.x
Zammit, C. M., and Watkin, E. L. J. (2016). “Adaptation to extreme acidity and osmotic stress,” in Acidophiles: Life in Extremely Acidic Environments, eds R. Quatrini and D. B. Johnson (Caister Academic Press), 49–62. doi: 10.21775/9781910190333.03
Zhang, X., Liu, X., He, Q., Dong, W., Zhang, X., Fan, F., et al. (2016b). Gene turnover contributes to the evolutionary adaptation of Acidithiobacillus caldus: insights from comparative genomics. Front. Microbiol. 7:e01960. doi: 10.3389/fmicb.2016.01960
Zhang, X., Feng, X., Tao, J., Ma, L., Xiao, Y., Liang, Y., et al. (2016a). Comparative genomics of the extreme acidophile Acidithiobacillus thiooxidans reveals intraspecific divergence and niche adaptation. Int. J. Mol. Sci. 17:1355. doi: 10.3390/ijms17081355
Zhang, X., Liu, X., Liang, Y., Guo, X., Xiao, Y., Ma, L., et al. (2017). Adaptive evolution of extreme acidophile Sulfobacillus thermosulfidooxidans potentially driven by horizontal gene transfer and gene loss. Appl. Environ. Microbiol. 83, e03098–16. doi: 10.1128/AEM.03098-16
Zhang, X., Liu, Z., Wei, G., Yang, F., and Liu, X. (2018). In silico genome-wide analysis reveals the potential links between core genome of Acidithiobacillus thiooxidans and its autotrophic lifestyle. Front. Microbiol. 9:e01255. doi: 10.3389/fmicb.2018.01255
Zhou, X., Chua, T. K., Tkaczuk, K. L., Bujnicki, J. M., and Sivaraman, J. (2010). The crystal structure of Escherichia coli spermidine synthase SpeE reveals a unique substrate-binding pocket. J. Struct. Biol. 169, 277–285. doi: 10.1016/j.jsb.2009.12.024
Zhu, Q., Kosoy, M., and Dittmar, K. (2014). HGTector: an automated method facilitating genome-wide discovery of putative horizontal gene transfers. BMC Genom. 15:717. doi: 10.1186/1471-2164-15-717
Keywords: acidophiles, pH homeostasis, extremophiles, acid mine drainage (AMD), evolution, comparative genomics
Citation: González-Rosales C, Vergara E, Dopson M, Valdés JH and Holmes DS (2022) Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle. Front. Microbiol. 12:822229. doi: 10.3389/fmicb.2021.822229
Received: 25 November 2021; Accepted: 30 December 2021;
Published: 15 February 2022.
Edited by:
Martina Cappelletti, University of Bologna, ItalyReviewed by:
Aleksandr G. Bulaev, Federal Center Research Fundamentals of Biotechnology, Russian Academy of Sciences (RAS), RussiaCopyright © 2022 González-Rosales, Vergara, Dopson, Valdés and Holmes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David S. Holmes, ZHNob2xtZXMyMDAwQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.