- 1World Health Organization Collaborating Centre for Gonorrhoea and Other Sexually Transmitted Infections, National Reference Laboratory for Sexually Transmitted Infections, Department of Laboratory Medicine, Faculty of Medicine and Health, Örebro University, Örebro, Sweden
- 2Public Health Agency of Sweden, Solna, Sweden
- 3Department of Clinical Microbiology, Karolinska University Hospital, Huddinge, Sweden
- 4Department of Laboratory Medicine, Medical Microbiology, Lund University, Skåne Laboratory Medicine, Lund, Sweden
- 5Center for Translational Microbiome Research, Department of Microbiology, Tumor and Cell Biology, Science for Life Laboratory, Karolinska Institutet, Solna, Sweden
The increasing transmission and antimicrobial resistance (AMR) in Neisseria gonorrhoeae is a global health concern with worrying trends of decreasing susceptibility to also the last-line extended-spectrum cephalosporin (ESC) ceftriaxone. A dramatic increase of reported gonorrhea cases has been observed in Sweden from 2016 and onward. The aim of the present study was to comprehensively investigate the genomic epidemiology of all cultured N. gonorrhoeae isolates in Sweden during 2016, in conjunction with phenotypic AMR and clinical and epidemiological data of patients. In total, 1279 isolates were examined. Etest and whole-genome sequencing (WGS) were performed, and epidemiological data obtained from the Public Health Agency of Sweden. Overall, 51.1%, 1.7%, and 1.3% resistance to ciprofloxacin, cefixime, and azithromycin, respectively, was found. No isolates were resistant to ceftriaxone, however, 9.3% of isolates showed a decreased susceptibility to ceftriaxone and 10.5% to cefixime. In total, 44 penA alleles were found of which six were mosaic (n = 92). Using the typing schemes of MLST, NG-MAST, and NG-STAR; 133, 422, and 280 sequence types, respectively, and 93 NG-STAR clonal complexes were found. The phylogenomic analysis revealed two main lineages (A and B) with lineage A divided into two main sublineages (A1 and A2). Resistance and decreased susceptibility to ESCs and azithromycin and associated AMR determinants, such as mosaic penA and mosaic mtrD, were predominantly found in sublineage A2. Resistance to cefixime and azithromycin was more prevalent among heterosexuals and MSM, respectively, and both were predominantly spread through domestic transmission. Continuous surveillance of the spread and evolution of N. gonorrhoeae, including phenotypic AMR testing and WGS, is essential for enhanced knowledge regarding the dynamic evolution of N. gonorrhoeae and gonorrhea epidemiology.
Introduction
Neisseria gonorrhoeae, the causative agent of gonorrhea, is a global public health concern with increasing transmission and development of antimicrobial resistance (AMR) (Rowley et al., 2019; Unemo et al., 2021). The incidence of reported gonorrhea cases in the European Union/European Economic Area (EU/EEA) increased from 6.6 cases per 100 000 inhabitants in 2009 to 26.9 cases per 100 000 inhabitants in 2018 (ECDC). However, the resistance to the extended-spectrum cephalosporins (ESCs) in the EU/EEA has decreased since 2009, as observed in the surveillance performed in the European Gonococcal Antimicrobial Susceptibility Programme, which is funded by the European Centre for Disease Prevention and Control (Cole et al., 2015; Cole et al., 2019; European Centre for Disease Prevention and Control [ECDC], 2021; Jacobsson et al., 2021). This trend may have been partly caused by the changed treatment implemented through the 2012 European gonorrhea guideline, in which cefixime 400 mg was replaced by ceftriaxone 500 mg plus azithromycin 2 g dual therapy as the recommended empirical first-line treatment for uncomplicated gonorrhea (Bignell and Unemo, 2013). In 2020, the recommended first-line treatment was further refined to ceftriaxone 1 g plus azithromycin 2 g or ceftriaxone 1 g monotherapy in well-controlled settings lacking ceftriaxone resistance (Unemo et al., 2020). Nonetheless, several treatment failures have been reported with ceftriaxone monotherapy and occasional failures also with ceftriaxone plus azithromycin/doxycycline dual therapy (Unemo et al., 2019a). Furthermore, the first gonococcal strain with ceftriaxone resistance combined with high-level resistance to azithromycin was detected in the United Kingdom and Australia in 2018 (Jennison et al., 2019) and the ceftriaxone-resistant strain FC428, including its sublineages, has been transmitted internationally since 2015 (Golparian et al., 2018; Lahra et al., 2018; Lee et al., 2019; Chen et al., 2020).
In N. gonorrhoeae, there are many different molecular AMR determinants, for example, target mutations in: penA, encoding penicillin-binding protein 2 (PBP2), associated with resistance and decreased susceptibility to ESCs and penicillins; 23S rRNA associated with macrolide resistance; gyrA and parC, encoding subunits of DNA gyrase and DNA topoisomerase IV, respectively, associated with fluoroquinolone resistance; and 16S rRNA associated with spectinomycin resistance (Dona et al., 2017; Unemo et al., 2019b). Additional mutations in the porin PorB1b (G120/A121 substitutions) and MtrCDE efflux pump (-35 A-deletion in mtrR promoter, MtrR G45D substitution or mosaic mtrRCDE) result in decreased influx and increased efflux of antimicrobials, respectively. These mutations decrease the antimicrobial susceptibility further (Unemo and Shafer, 2014; Shafer, 2018).
The World Health Organization recognizes the challenges of reducing the transmission and AMR development in N. gonorrhoeae and recommends surveillance of both cases and AMR in the population as one of several strategies to manage the infectious burden and control AMR development (World Health Organization [WHO], 2018). Enhanced knowledge concerning N. gonorrhoeae strains circulating nationally and internationally is imperative to make informed decisions regarding prevention, management, and control. Whole-genome sequencing (WGS) is, due to its high resolution and accuracy, an ideal method for molecular epidemiology and enables collection of comprehensive data on gonococcal population characteristics and AMR determinants internationally (De Silva et al., 2016; Grad et al., 2016; Eyre et al., 2017; Harris et al., 2018; Yahara et al., 2018; Gianecini et al., 2019; Lan et al., 2020; Sánchez-Busó et al., 2021). No national WGS-based study for N. gonorrhoeae has previously been performed in Sweden.
In Sweden, an alarming increase in the national gonorrhea incidence has been observed during the recent decade, and the reported incidence increased from 7.8 to 31.4 per 100 000 inhabitants in 2009-2019 (Swedish Society for Dermatology and Venereology [SSDV], 2019; Swedres-Svarm, 2019; European Centre for Disease Prevention and Control [ECDC], 2021). However, the most dramatic increase in the gonorrhea incidence started in 2016-2017, when it increased from 17.8 in 2016 to 25.0 in 2017, and it has subsequently continued to increase. The resistance to ESCs substantially decreased from 2010 to 2019, i.e., the resistance to ceftriaxone decreased from 2% to 0% and resistance to cefixime from 8% to approximately 1%. The resistance to azithromycin initially decreased from 12% in 2010 to 3% in 2016, but then started to increase (12% in 2019). Ciprofloxacin resistance fluctuated between 47 and 62% and no spectinomycin resistance was reported in 2010-2019 (Swedres-Svarm, 2019). In 2019, the main Swedish guideline for treatment of uncomplicated anogenital and pharyngeal gonorrhea was revised to ceftriaxone 1 g single dose Swedish Society for Dermatology and Venereology (SSDV). Previous recommendation of dual therapy, including ceftriaxone 500 mg, plus 2 g azithromycin for pharyngeal infections, (Läkemedelsverket, 2015) has been discontinued, i.e., due to the lack of ceftriaxone resistance in the national AMR surveillance and mandatory test of cure for all gonorrhea patients.
The aim of this study was to perform a genome-based epidemiologic analysis of all N. gonorrhoeae isolates cultured in Sweden during 2016 in conjunction with phenotypic AMR and clinical and epidemiological data of patients, in order to identify more prevalent gonococcal lineages and clones, AMR lineages and patient groups at higher risk of gonorrhea.
Materials and Methods
Neisseria gonorrhoeae Isolates, Antimicrobial Resistance Testing, and Patient Data
All viable cultured and species-verified clinical isolates of N. gonorrhoeae (n = 1279; one per patient or infection episode if the same patient had multiple gonorrhea episodes during the year) in Sweden in 2016, representing 72.0% of all reported gonorrhea cases (n = 1777) in the year, were included. When multiple isolates from the same gonorrhea episode were available, isolates from extragenital sites were prioritized. Accordingly, the examined gonococcal isolates were cultured from the urethra (n = 521), rectum (n = 291), pharynx (n = 150), or other/not reported (n = 65) in men and from cervix (n = 119), pharynx (n = 44), urethra (n = 34), rectum (n = 21), vaginal swabs (n = 14), or other/not reported (n = 20) in women. Multiple gonococcal infections of the same patient were considered separate episodes if the infections were > 3 weeks apart. AMR testing was conducted using Etest, according to the instructions of the manufacturer (bioMérieux, Marcy-l′Étoile, France). Minimum inhibitory concentrations (MICs; mg/L) of ceftriaxone, cefixime, ciprofloxacin and spectinomycin were interpreted according to the clinical breakpoints recommended by the European Committee on Antimicrobial Susceptibility Testing. For azithromycin, no clinical breakpoints are recommended and the epidemiological cut-off value of 1 mg/L (1 v11.0) was applied (isolates with azithromycin MIC > 1 mg/L are referred to as resistant hereafter). Additionally, isolates with MIC values of 0.064 to 0.125 mg/L for ceftriaxone and cefixime were interpreted to have a decreased susceptibility (Gianecini et al., 2019; Unemo et al., 2019a; Golparian et al., 2020a). Only whole MIC doubling dilutions are reported in the present study. All isolates were subcultured on non-selective GCAGP agar plates (Foerster et al., 2019) overnight at 37°C in a humid 5% CO2-enriched atmosphere prior to DNA extraction.
Epidemiological data for the gonorrhea patients were obtained from the infectious diseases register SmiNet at the Public Health Agency of Sweden. Data on age, sex, anatomical site of infection, sexual orientation, geographical region, type of clinic the patient visited, reason for testing (such as symptomatic infection or contact tracing), and country of infection were extracted.
Statistical analysis was performed using logistic regression in IBM SPSS Statistics (version 25) with calculation of a 95% confidence interval and odds ratio (OR). Pearson’s χ2 was used to calculate significance and significance was set at p < 0.05.
Ethical Approval
Ethical approval for the study was obtained from the Swedish Ethical Review Authority (Approval number 2020-05008).
Whole Genome Sequencing and Analysis
Genomic DNA was extracted using the QIASymphony DSP virus/pathogen kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions, including RNase treatment and elution with Tris-HCl (pH 8), on the QIASymphony instrument (Qiagen). All DNA extractions were paired-end sequenced using Illumina HiSeq X platform (Illumina, Inc., San Diego, CA, United States). DNA extractions and sequencing libraries were quality controlled using Qubit BR/HS assays (Thermo Fischer Scientific, Waltham, MA, United States) and appropriate assays on the TapeStation according to manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, United States).
All N. gonorrhoeae genomes (n = 1279) were sequenced with 50-100-fold coverage and initially quality controlled for contamination using Kraken (v1.1.1) (Wood and Salzberg, 2014). All reads were subsequently analyzed using a customized workflow in CLC Genomic Workbench (v20.0.4, Qiagen), which includes additional quality controls, assembly, mapping to reference sequence [WHO F (Unemo et al., 2016)], genetic characterization of AMR determinants, and characterization of alleles for genotyping, i.e., Multi-locus sequence typing (MLST) (Maiden et al., 1998), N. gonorrhoeae sequence typing for antimicrobial resistance (NG-STAR) (Demczuk et al., 2017) and N. gonorrhoeae multi-antigen sequence typing (NG-MAST) (Martin et al., 2004) sequence types (STs) (Golparian et al., 2020b). The NG-STAR STs were assigned to clonal complexes (CCs) as previously described (Golparian et al., 2021) based on the NG-STAR database2; obtained 2021-05-18. An additional assembly was generated using SPAdes (v3.14.1) (Bankevich et al., 2012) with the --careful option to resolve or confirm any partially extracted or novel alleles, respectively.
All reads were mapped to N. gonorrhoeae reference strain FA1090 (accession number LT591897) to obtain a multiple sequence alignment using multiple_mappings_to_bam pipeline3 with BWA-MEM (Li, 2013) as the aligner. Furthermore, 8095 recombinant blocks were removed using Gubbins (v 1.4.10) (Croucher et al., 2015) with an average size of 5876 bp per recombinant region. Phylogenomic tree was created using IQ-TREE v1.6.1041 (Nguyen et al., 2015) based on 47,583 informative sites.
The phylogenomic tree was midpoint rooted using Figtree (v1.4.4) visualized using Microreact (Argimón et al., 2016).
All WGS sequence reads are available from the ENA (PRJEB47922).
Results
Patient Population Characteristics
The study population included 252 women and 1027 men with median (mean) age of 24 (27.4) years and 30 (32.1) years, respectively (Table 1). In total, 50.3% of patients were reported as men who have sex with men (MSM). MSM and heterosexual men had a median (mean) age of 30 (31.6) years and 30 (33.2) years, respectively. The majority of patients (81.1%) were diagnosed in regions with major metropolitan areas, i.e., Stockholm (55.4%), Västra Götaland (15.6%), and Skåne (10.2%). However, all Swedish regional councils (n = 21) except one (Västerbotten) were represented. Regarding origin of infection, 68.4% and 29.6% were reported as domestic (n = 875) and foreign (n = 379), respectively (Table 1). The most common foreign countries of infection were Thailand (n = 70), Germany (n = 32), Spain (n = 31), Denmark (n = 25), and Turkey (n = 18) (Supplementary Figure S1). A higher proportion of MSM was infected in Sweden (76.2%) compared to heterosexual men (52.6%) (domestic infection in women was 76.3%).
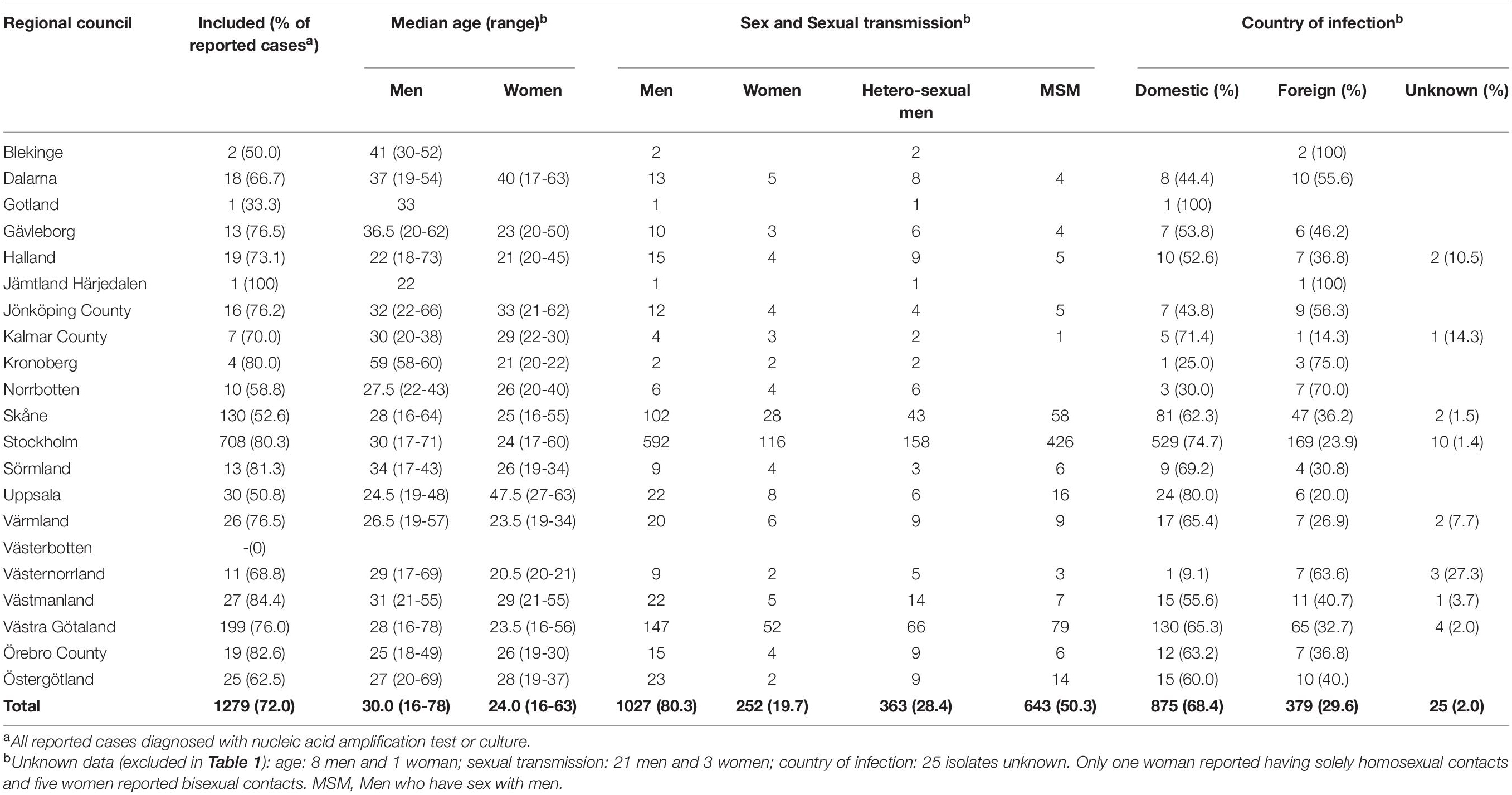
Table 1. Demographics and epidemiological characteristics of included gonorrhea patients in Sweden in 2016 by regional council.
Phenotypic Antimicrobial Susceptibility/Resistance and Molecular Antimicrobial Resistance Determinants Derived From the Whole-Genome Sequencing Sequences
The phenotypic antimicrobial susceptibility of all gonococcal isolates by regional council is summarized in Supplementary Table S1. The prevalence of the main AMR determinants derived from the WGS sequences among all isolates by regional council of the cases is presented in Figure 14 and Table 2.
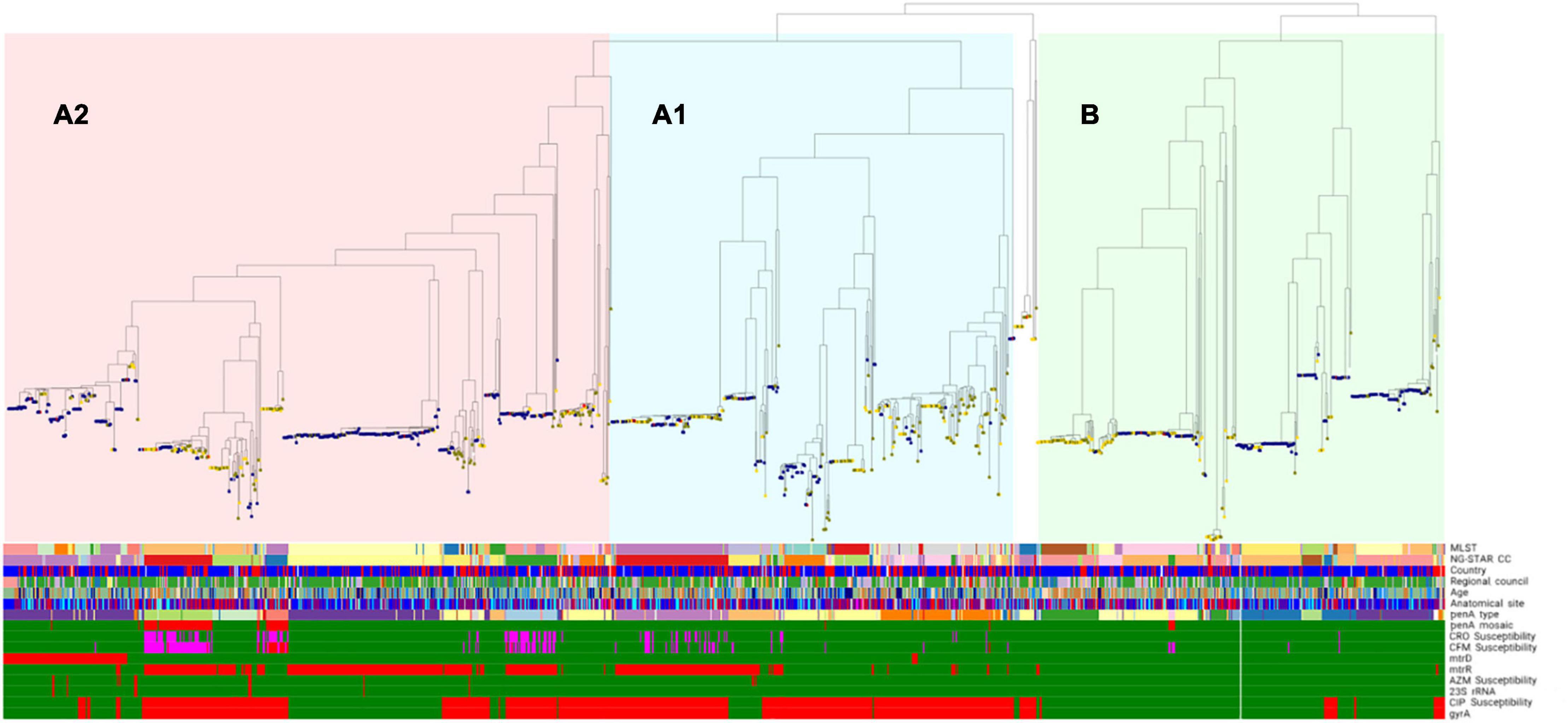
Figure 1. Phylogenomic tree including genomic lineages, molecular typing and phenotypic antimicrobial resistance (AMR) of cultured Neisseria gonorrhoeae isolates (n = 1278) in Sweden 2016. One isolate was excluded from the phylogeny due to low coverage (<70%). Nodes denote sexual orientation (blue, homosexual males; yellow, females; green, heterosexual males; red, bisexual males; white, not reported). Bars for phenotypic antimicrobial susceptibility (or presence/absence of AMR determinants): green, susceptible (or AMR determinant absent); red, resistant (or AMR determinant present); and pink, decreased susceptibility. See line-listed details about the isolates and each determinant, including their color coding, at: https://microreact.org/project/Hadad_et_al_Swe2016. MLST, multi-locus sequence typing; NG-STAR CC, Neisseria gonorrhoeae sequence typing for antimicrobial resistance clonal complex; CFM, cefixime; CRO, ceftriaxone; AZM, azithromycin; CIP, ciprofloxacin.

Table 2. Main antimicrobial resistance determinants in Neisseria gonorrhoeae isolates (n = 1279) cultured in Sweden in 2016, by regional council.
No resistance to ceftriaxone was found, however, 9.3% (n = 119) of all isolates (n = 1279) showed a decreased susceptibility. Cefixime resistance was found in 1.7% of isolates (n = 22, MIC = 0.25-0.5 mg/L), and the highest resistance levels were in Östergötland (4.0%), Stockholm (2.4%), and Skåne (2.3%) (Supplementary Table S1). Furthermore, 10.5% (n = 134) of isolates had a decreased susceptibility to cefixime. Eighty (6.3%) isolates had a decreased susceptibility to both ceftriaxone and cefixime. One of these 80 isolates was also resistant to azithromycin, i.e., a high-level azithromycin-resistant (MIC > 256 mg/L) strain cultured from a heterosexual male with a sexual contact in Thailand. Based on the WGS sequences, 20 (90.9%) of the cefixime-resistant isolates (n = 22) had mosaic penA alleles [penA-10.001 (n = 18) and penA-34.001 (n = 2)] (Figure 1; see text footnote 4), the main determinant for decreased ESC susceptibility, and 81.8% (18/22) and 86.4% (19/22) had mtrR and porB1b mutations, respectively. Two (9.1%) cefixime-resistant isolates did not contain any mosaic penA, however, these isolates harbored PBP2 A501V and P551S alterations (penA-13.001) as well as mtrR and porB1b mutations. Overall, 44 penA alleles were found among the 1279 isolates, of which the most common were penA-2.001 (n = 325), penA-2.002 (n = 232), penA-19.001 (n = 101), penA-44.001 (n = 98), and penA-14.001 (n = 94). Ninety-two isolates (7.2%) had mosaic penA alleles: penA-34.001 (n = 60), penA-10.001 (n = 23), penA-34.008 (n = 6), penA-72.001, penA-105.001, and penA-180.001 (n = 1 each) resulting in ceftriaxone MICs < 0.002-0.125 mg/L and cefixime MICs < 0.016-0.5 mg/L. The combination of mosaic penA with mtrR and/or porB1b mutations, which further decrease the ESC susceptibility, was found in 81 isolates, of which 49 (60.5%) isolates had decreased susceptibility to ceftriaxone and 17 (21.0%) and 60 (74.1%) isolates had resistance and decreased susceptibility to cefixime, respectively. PBP2 A501V/T, P551S and G542S alterations were found in 162 (12.7%), 56 (4.4%), and 45 (3.5%) isolates, respectively. The ceftriaxone and cefixime MIC range (mean) was 0.004-0.125 (0.035) mg/L and < 0.016-0.125 (0.040) mg/L for A501V/T, 0.004-0.064 (0.034) mg/L and < 0.016-0.064 (0.029) mg/L for P551S, and 0.004-0.032 (0.016) mg/L and < 0.016-0.032 (0.020) mg/L for G542S, respectively. The combination of PBP2 A501V/T and P551S alterations was present in 48 isolates (3.8%) with ceftriaxone and cefixime MIC range (mean) of 0.004-0.125 (0.07) mg/L and < 0.016-0.25 (0.07) mg/L, respectively. The combination of PBP2 A501T and G542S alterations was present in six isolates (0.5%) with ceftriaxone and cefixime MIC range (mean) of 0.016-0.064 (0.051) mg/L and 0.016-0.064 (0.045) mg/L, respectively.
Azithromycin resistance was found in 16 (1.3%) isolates, and the highest resistance levels were found in Dalarna (11.1%) and Stockholm (1.8%) (Supplementary Table S1). High-level azithromycin resistance (MIC > 256 mg/L) was found in only one isolate, which based on the WGS sequences had the A2059G mutation in all four alleles of the 23S rRNA gene. Ten resistant isolates with azithromycin MICs of 2-16 mg/L had the C2611T mutation in all four 23S rRNA alleles. The remaining five isolates lacked 23S rRNA mutations, however mtrR -35 A-deletion was found in all of them, two isolates had additionally a mosaic mtrD and the remaining three isolates had porB1b mutations. Mosaic mtrR was found in 1.1% (n = 14) of isolates, mosaic mtrC in 1.1% (n = 14), mosaic mtrD in 8.4% (n = 107), and mosaic mtrE in 1.0% (n = 13). The combination of mosaic alleles in all genes of the mtrRCDE operon was found in 1.0% (n = 13) of isolates with azithromycin MICs of 0.032-1.0 mg/L. One isolate harbored mosaic alleles in all genes except mtrE (azithromycin MIC = 1.0 mg/L). Notably, one isolate, which lacked known azithromycin resistance determinants, contained the previously described GC deletion in mtrC (Ma et al., 2020) (azithromycin MIC = 0.5 mg/L).
Ciprofloxacin resistance was found in 653 (51.1%) isolates (Supplementary Table S1). All isolates with ciprofloxacin resistance harbored GyrA S91F and D95A/G/N mutations, and no ciprofloxacin-susceptible isolates had the GyrA S91F mutation (Figure 1 and Table 2; see text footnote 4). One isolate harbored only a GyrA D95N mutation, and this isolate was susceptible to ciprofloxacin (MIC = 0.032 mg/L). Mutations in parC was found in 451 (35.3%) isolates, all except one had the GyrA S91F mutation, consisting of the amino acid substitutions D86N (n = 137), S87I/N/R (n = 311), S88P (n = 27), and E91K (n = 10).
No resistance was found to spectinomycin, including no spectinomycin resistance determinants in the 16S rRNA or rpsE genes.
In total, mtrR (-35 deletion, G45D and/or mosaic mtrR) and porB1b mutations (G120 and/or A121 alterations) were found in 540 (42.2%) and 356 (27.8%) isolates, respectively.
Molecular Epidemiology and Genome-Based Phylogeny
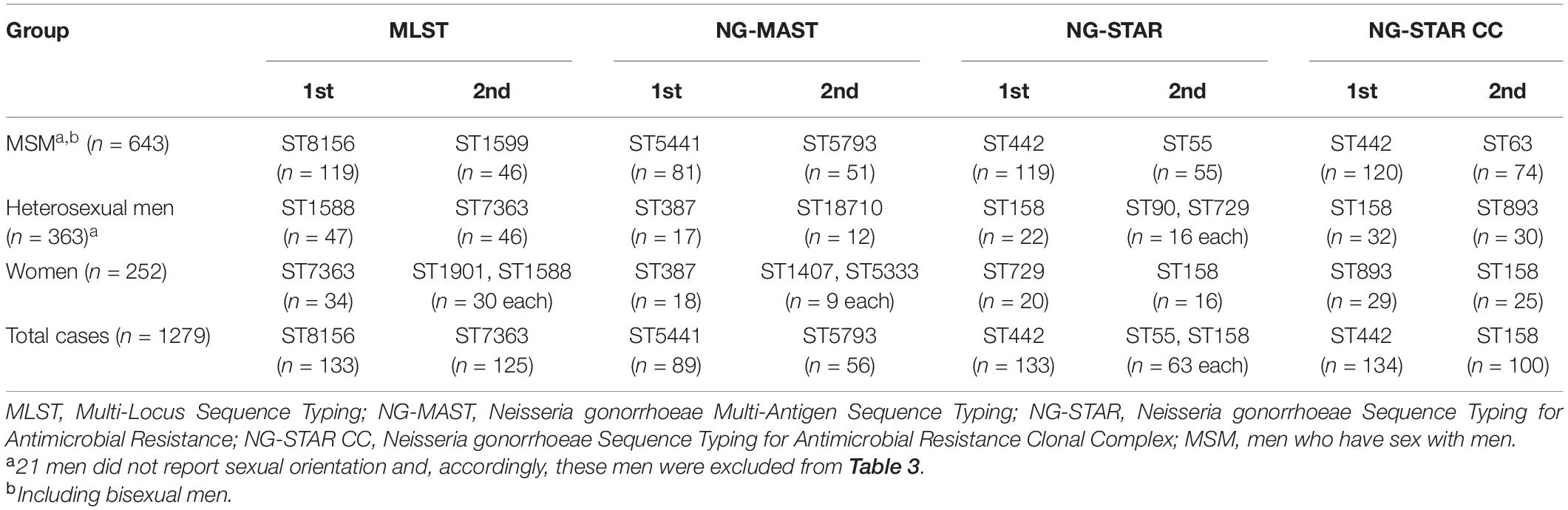
Based on the WGS sequences, the most common MLST, NG-MAST, NG-STAR STs and NG-STAR CCs by sex and sexual orientation is summarized in Table 3. Overall, 133 MLST STs were found and the five most common STs were ST8156 (n = 133), ST7363 (n = 125), ST1901 (n = 93), ST1588 (n = 89), and ST7359 (n = 67). Fifty-five MLST STs represented by single isolates were found. Using NG-MAST, 422 STs were identified and the five most prevalent STs were ST5441 (n = 89), ST5793 (n = 56), ST2992 (n = 41), ST11461 (n = 39), and ST387 (n = 35). In total, 260 NG-MAST STs were found in only one isolate. With NG-STAR, 280 STs were observed and the five most prevalent ones were ST442 (n = 133), ST55 (n = 63), ST158 (n = 63), ST231 (n = 61), and ST520 (n = 52). One hundred and forty-eight NG-STAR STs were represented by single isolates. Lastly, 92 NG-STAR CCs were found and 19 isolates were ungroupable. The five most common CCs were CC442 (n = 134), CC158 (n = 100), CC63 (n = 91), CC42 (n = 70), and CC390 (n = 69). Twenty-six NG-STAR CCs were represented by single isolates.
Phylogenomic analysis revealed two main lineages (A and B), of which one (A) was subdivided into two main sublineages (A1 and A2) (Figure 1; see text footnote 4). The distributions of patient age, sexual orientation, anatomical site of infection, MLST STs, NG-STAR CCs, and penA types in sublineages A1 and A2 and linage B are summarized in Figure 2. In A2 (539 isolates), the most prevalent molecular types were MLST ST8156 (n = 133) and ST1901 (n = 90); NG-MAST ST5441 (n = 89) and ST2992 (n = 41); NG-STAR ST442 (n = 133) and ST90 (n = 51); and NG-STAR CCs 442 (n = 134) and 63 (n = 91). The sexual orientation of the patients in A2 was homosexual male (57.9%), bisexual male (2.6%), heterosexual male (23.4%), women (14.5%), and not reported (1.7%) (Figure 2). Isolates with resistance and decreased susceptibility to ceftriaxone and cefixime, including isolates containing mosaic penA (49/81) predominantly belonged to A2. The isolates with decreased susceptibility to ceftriaxone (n = 89) were cultured from MSM (51.7%, n = 46), heterosexual men (24.7%, n = 22), women (20.2%, n = 18) and not reported (3.4%, n = 3). The majority (71.9%, n = 64) of these infections were domestic and 73.0% (n = 65) were diagnosed in Stockholm. The most common molecular types among the isolates with decreased susceptibility to ceftriaxone were MLST ST1901 (n = 34), NG-MAST ST1407 (n = 25), NG-STAR ST90 (n = 33), and NG-STAR CC90 (n = 35). The 21 (95.5%) cefixime-resistant isolates in A2 were cultured from infections acquired heterosexually [18/21; men (n = 13) and women (n = 5)] or MSM (3/21). Furthermore, 14 isolates were from domestic and seven from foreign infections, in six different countries. The majority (81.0%, n = 17) were diagnosed in Stockholm, however, all cefixime-resistant isolates were from patients in larger metropolitan areas. The most common molecular types among the cefixime-resistant isolates were MLST ST7363 (n = 16), NG-MAST ST13876 (n = 9), NG-STAR ST232 (n = 10), and NG-STAR CC348 (n = 15).
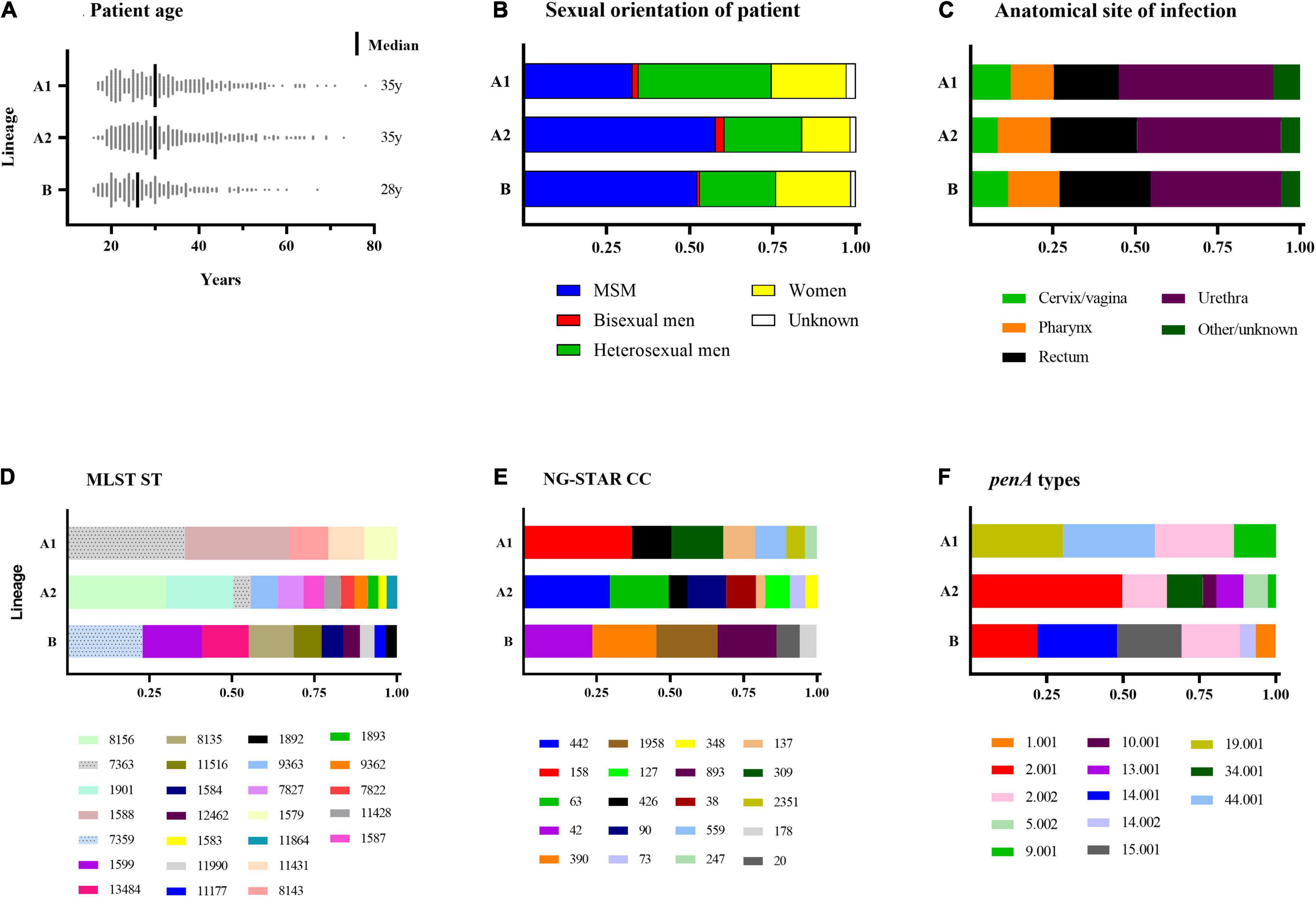
Figure 2. Summary of patient characteristics (A–C) and Neisseria gonorrhoeae sequence types (D–F) of sublineages A1 and A2 and lineage B. Figures (D–F) Include sequence types of ≥10 isolates. MSM, men who have sex with men; MLST, multi-locus sequence typing; ST, sequence type; NG-STAR CC, Neisseria gonorrhoeae sequence typing for antimicrobial resistance clonal complex.
The majority of azithromycin-resistant isolates (81.3%, 13/16) also belonged to A2, and contained 23S rRNA gene mutations (n = 11), or mtrR -35 A-deletion plus mosaic mtrD (n = 2). Sexual orientation of these patients was homosexual male (46.2%), heterosexual male (15.4%) and women (38.5%). These azithromycin-resistant isolates did not belong to any predominant ST nor did they cluster together in the phylogenomic tree (Figure 1; see text footnote 4). Notably, 95.3% (102/107) of the isolates with mosaic mtrD belonged to A2.
Resistance to ciprofloxacin was also prevalent (53.2%) in A2. The most common molecular types among the ciprofloxacin-resistant isolates in A2 were MLST ST1901 (n = 90), NG-MAST ST1407 (n = 33), NG-STAR ST90 (n = 51), and NG-STAR CC90 (n = 61).
In lineage A1 (360 isolates), the most common molecular types were MLST ST7363 (n = 101) and ST1588 (n = 89); NG-MAST ST12001 (n = 21), ST9184 (n = 19), and ST18710 (n = 19); NG-STAR ST158 (n = 63) and ST426 (n = 22); and NG-STAR CC158 (n = 100) and CC309 (n = 48). The sexual orientation of the patients in A1 was homosexual male (32.8%), bisexual male (1.9%), heterosexual male (40.0%), women (22.5%), and not reported (2.8%) (Figure 2). A1 included the remaining cefixime-resistant isolate (with mosaic penA). The remaining three (18.8%, 3/16) azithromycin-resistant isolates also belonged to A1 and appeared to represent the same gonococcal strain (MLST ST1579, NG-MAST 21, NG-STAR CC137). All these isolates were also lacking 23S rRNA mutations and mosaic mtrD, but had mtrR -35 A-deletion plus porB1b mutations (azithromycin MICs of 2 mg/L). Furthermore, ciprofloxacin resistance was very common in A1 (90%). The most common molecular types among the ciprofloxacin-resistant isolates in A1 were MLST ST7363 (n = 101), NG-MAST ST12001 (n = 21), NG-STAR ST158 (n = 63), and NG-STAR CC158 (n = 100) (Figure 2).
In lineage B (359 isolates), the most frequent molecular types were MLST ST7359 (n = 67) and ST1599 (n = 53), NG-MAST ST5793 (n = 56) and ST11461 (n = 39), NG-STAR ST55 (n = 63) and ST231 (n = 61), and NG-STAR CC42 (n = 69) and CC390 (n = 64). The sexual orientation of the patients was homosexual male (52.4%), bisexual male (0.8%), heterosexual male (22.8%), women (22.9%), and not reported (1.4%) (Figure 2). Lineage B did not include any isolates expressing resistance to cefixime or azithromycin. Six isolates contained mosaic penA but no isolates comprised mosaic mtrD. The majority (93.3%) of isolates were ciprofloxacin susceptible.
Statistical analysis was performed on the most common NG-STAR CCs (n = 5) and epidemiological data (Supplementary Table S2) and phenotypic AMR (Supplementary Table S3), respectively, as well as phenotypic AMR and epidemiological data AMR (Supplementary Table S4). NG-STAR CC442, CC63, CC42, and CC390 were significantly associated with MSM. CC442, CC158 and CC42 were significantly more prevalent in domestic infections, and CC442 and CC390 were significantly more frequent in age groups of 16-24 years and 25-44 years, respectively. CC158 was significantly associated with decreased susceptibility to ceftriaxone and cefixime. Decreased susceptibility to ceftriaxone was significantly associated with age groups between 16 and 44 years and resistance and decreased susceptibility to cefixime was significantly associated with age groups of 16-24 and 35-44 years. Notably, CC348 was strongly associated with decreased susceptibility to ceftriaxone (OR 16.15) and resistance and decreased susceptibility to cefixime (all isolates). Ciprofloxacin susceptibility was significantly associated with age groups of 16-35 years, MSM and domestic infections. Ciprofloxacin resistance was significantly associated with heterosexual transmissions.
Discussion
This study is the first comprehensive national description of N. gonorrhoeae isolates in Sweden using WGS in conjunction with phenotypic AMR and patient epidemiological data. The majority of cases and gonococcal AMR (except for ciprofloxacin) were diagnosed in metropolitan areas, especially in the capital city Stockholm. In 2016, resistance to ceftriaxone and cefixime remained low nationally; i.e., no resistance to ceftriaxone and 1.7% to cefixime. Resistance to azithromycin was also low (1.3%). The cefixime resistance was caused by penA-10.001 (n = 18), penA-34.001 (n = 2), or PBP2 A501V and P551S alterations in combination with mtrR and porB1b mutations (n = 2). The azithromycin resistance was caused by 23S rRNA A2059G (n = 1), 23S rRNA C2611T (n = 10) or mtrR -35 A-deletion plus mosaic mtrD (n = 2) or mtrR -35 A-deletion plus porB1b mutations (n = 3). None of the cefixime-resistant isolates was resistant to azithromycin. Prior to 2019, the Swedish gonorrhea guideline recommended ceftriaxone 500 mg, plus azithromycin 2 g if pharyngeal infection, for treatment of uncomplicated gonorrhea. However, from 2019, ceftriaxone 1 g monotherapy is the recommended treatment (Läkemedelsverket, 2015; Swedish Society for Dermatology and Venereology [SSDV], 2019), i.e., due to the lack of ceftriaxone resistance in the AMR surveillance, mandatory test of cure, and use of the first-line doxycycline if Chlamydia trachomatis infection has not been excluded. A regular, systematic and quality-assured surveillance of the N. gonorrhoeae AMR, nationally and internationally, remains imperative for evidence-based refinements of management guidelines.
Phylogenomic analysis showed that the isolates grouped into two main lineages as previously described (Harris et al., 2018; Sánchez-Busó et al., 2019; Golparian et al., 2020a), of which one (A) was divided into two main sublineages (A1 and A2) (Golparian et al., 2020a). The vast majority of isolates with resistance to cefixime and azithromycin and decreased susceptibility to ESCs, including associated AMR determinants, was found within the lineage A2. Although a higher proportion of MSM (60.5%) were present in A2, the cefixime-resistant isolates were predominantly (86.4%, 19/22) cultured from heterosexual patients, which is in line with a previous European paper (Jacobsson et al., 2021). However, isolates with decreased susceptibility to ceftriaxone and cefixime were more prevalent among MSM (57.1%) compared to heterosexual patients (38.1%). Sublineage A2 also harbored the majority of mtrR mutations and mtrD mosaic alleles. In sublineage A1, only a few cefixime- and azithromycin-resistant isolates as well as isolates containing mosaic penA alleles and mosaic mtrD alleles were present. Notably, the vast majority of isolates in sublineage A1 were resistant to ciprofloxacin (90%) and A1 contained the lowest proportion of MSM. Lineage B harbored most of the antimicrobial-susceptible isolates.
The most prevalent MLST STs found in the present study have been described in previous studies. A national genomic Norwegian study described 21 STs with ≥ 10 isolates (Alfsnes et al., 2020), all of which were also present in Sweden. Accordingly, both the prevalence and distribution of MLST STs were similar in the two Scandinavian countries. For example, MLST ST8156 and ST7359, common STs in the Norwegian population (Alfsnes et al., 2020), were also among the most frequent STs in Sweden. However, these STs have been less frequently observed in other EU/EEA countries. Limited observations have been made in the United Kingdom and United States (ST8156) and Japan (ST7359) but the majority of isolates has been found in Australia (ST8156 and ST7359) (Buckley et al., 2018; Hanao et al., 2021; Pathogenwatch, 2021). In 2013, MLST ST7363, ST1901 and ST1588 were the most prevalent STs in isolates from 20 different EU/EEA countries (Harris et al., 2018). Notably, a small cluster of 7 isolates belonging to MLST ST7363 in the European study, phylogenetically unrelated from the main cluster of ST7363, were resistant to cefixime. Similarly, the majority of cefixime-resistant isolates in Sweden clustered together, belonged to ST7363, and were phylogenomically unrelated to the main cluster of ST7363 isolates. The emergence of mosaic penA alleles in MLST ST1901 and ST7363 was originally documented in Japan (Shimuta et al., 2015) and these strains have since then spread internationally and have accounted for most resistance and decreased susceptibility to ESCs in Europe and North America (present study; Harris et al., 2018; Thomas et al., 2019, 2021). It has been shown that mosaic penA-34 and mosaic penA-10 have been the dominating penA alleles in MLST ST1901 and ST7363, respectively (Harris et al., 2018; Yahara et al., 2018; Town et al., 2020). The majority of MLST ST1901 in Sweden harbored mosaic penA-34 and all also belonged to NG-MAST ST1407, a clone with resistance or decreased susceptibility to ESCs that has been internationally disseminated. Cefixime-resistant MLST ST7363 strains frequently harbor mosaic penA-10. Both mosaic penA-10 and mosaic penA-34 have been described in ESC-resistant N. gonorrhoeae strains worldwide, e.g., in Japan, Vietnam, Argentina, United States, Canada and Europe (Demczuk et al., 2015; Grad et al., 2016; Harris et al., 2018; Gianecini et al., 2019; Lan et al., 2020; Hanao et al., 2021; Pathogenwatch, 2021). In Sweden, 85% of all isolates with mosaic penA alleles belonged to MLST ST1901 and ST7363 and these isolates were mostly associated with domestic heterosexual transmission. These gonococcal strains were found in 13 (61.9%) of the 21 Swedish regions, making preventions targeted on single sexual networks or risk group and in general gonorrhea control very challenging.
The newly proposed classification method for N. gonorrhoeae, NG-STAR CC (Golparian et al., 2021), assigned most (72.7%) cefixime-resistant isolates as the novel CC348. NG-STAR CC90 and CC38 accounted for two cefixime-resistant isolates each and, together with CC158, most of the isolates with decreased susceptibility to ESCs. CC90 has been described previously in Brazil (Golparian et al., 2020a), where it was a common type with elevated MICs of ESCs. Similar to other Brazilian isolates, CC63 in Sweden harbored the highest proportion of mosaic mtrD alleles. Azithromycin-resistant isolates did not belong to any specific NG-STAR CC(s) due to the lack of phylogenomic similarity of the isolates. NG-STAR with CC designation, which clusters based on AMR determinants and the closest alleles, appeared to correspond relatively well with the genome-based phylogeny and can also inform regarding resistance or decreased susceptibility to cefixime and ceftriaxone and presence of associated AMR determinants.
Conclusion
In conclusion, the Swedish N. gonorrhoeae population in 2016 had a low prevalence of resistance to cefixime and no resistance to ceftriaxone. However, the limited cefixime resistance and especially the high level of decreased susceptibility to ESCs, predominantly domestically transmitted, remain worrisome and is a risk for development of resistance to ceftriaxone. The prevalence of azithromycin resistance was also low. Resistance and decreased susceptibility to ESCs and azithromycin and associated AMR determinants, such as mosaic penA and mosaic mtrD, were mainly found in the phylogenomic sublineage A2. Resistance to cefixime and azithromycin was more prevalent among heterosexuals and MSM, respectively, and both were predominantly spread through domestic transmission. Continuous surveillance of the spread and evolution of N. gonorrhoeae, including phenotypic AMR testing and WGS, is essential for enhanced knowledge regarding the dynamic evolution of N. gonorrhoeae and gonorrhea epidemiology.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB47922.
Ethics Statement
The studies involving human participants were reviewed and approved by the Swedish Ethical Review Authority (Approval number 2020-05008). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
MU and RH designed, initiated, and coordinated the study. RH, DG, and LE were involved in the laboratory work, WGS, and/or bioinformatic analysis. A-KO, E-LE, and YL provided gonococcal isolates. IV and HF contributed with epidemiological data. RH, DG, and MU analyzed and interpreted all the data. RH and MU wrote the first draft of the manuscript. All authors read, commented, and approved the submitted manuscript.
Funding
The present work was supported by grants from the Örebro County Council Research Committee, Örebro, Sweden; and the Foundation for Medical Research at the Örebro University Hospital, Örebro, Sweden.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.820998/full#supplementary-material
Footnotes
- ^ www.eucast.org/clinical_breakpoints
- ^ https://ngstar.canada.ca/
- ^ https://github.com/sanger-pathogens/bact-gen-scripts
- ^ https://microreact.org/project/Hadad_et_al_Swe2016
References
Alfsnes, K., Eldholm, V., Olsen, A. O., Brynildsrud, O. B., Bohlin, J., Steinbakk, M., et al. (2020). Genomic epidemiology and population structure of Neisseria gonorrhoeae in Norway, 2016-2017. Microb. Genom 6:e000359. doi: 10.1099/mgen.0.000359
Argimón, S., Abudahab, K., Goater, R. J. E., Fedosejev, A., Bhai, J., Glasner, C., et al. (2016). Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom 2:e000093. doi: 10.1099/mgen.0.000093
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bignell, C., and Unemo, M. (2013). 2012 European guideline on the diagnosis and treatment of gonorrhoea in adults. Int. J. STD AIDS 24, 85–92. doi: 10.1177/0956462412472837
Buckley, C., Forde, B. M., Trembizki, E., Lahra, M. M., Beatson, S. A., and Whiley, D. M. (2018). Use of whole genome sequencing to investigate an increase in Neisseria gonorrhoeae infection among women in urban areas of Australia. Sci. Rep. 8:1503. doi: 10.1038/s41598-018-20015-x
Chen, S. C., Yuan, L. F., Zhu, X. Y., Van Der Veen, S., and Yin, Y. P. (2020). Sustained transmission of the ceftriaxone-resistant Neisseria gonorrhoeae FC428 clone in China. J. Antimicrob. Chemother. 75, 2499–2502. doi: 10.1093/jac/dkaa196
Cole, M. J., Quinten, C., Jacobsson, S., Day, M., Amato-Gauci, A. J., Woodford, N., et al. (2019). The European gonococcal antimicrobial surveillance programme (Euro-GASP) appropriately reflects the antimicrobial resistance situation for Neisseria gonorrhoeae in the European Union/European Economic Area. BMC Infect. Dis. 19:1040. doi: 10.1186/s12879-019-4631-x
Cole, M. J., Spiteri, G., Jacobsson, S., Pitt, R., Grigorjev, V., and Unemo, M. (2015). Is the tide turning again for cephalosporin resistance in Neisseria gonorrhoeae in Europe? Results from the 2013 European surveillance. BMC Infect. Dis. 15:321. doi: 10.1186/s12879-015-1013-x
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
De Silva, D., Peters, J., Cole, K., Cole, M. J., Cresswell, F., Dean, G., et al. (2016). Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect. Dis. 16, 1295–1303. doi: 10.1016/s1473-3099(16)30157-8
Demczuk, W., Lynch, T., Martin, I., Van Domselaar, G., Graham, M., Bharat, A., et al. (2015). Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J. Clin. Microbiol. 53, 191–200. doi: 10.1128/JCM.02589-14
Demczuk, W., Sidhu, S., Unemo, M., Whiley, D. M., Allen, V. G., Dillon, J. R., et al. (2017). Neisseria gonorrhoeae sequence typing for antimicrobial resistance (NG-STAR): a novel antimicrobial resistance multilocus typing scheme for tracking the global dissemination of N. gonorrhoeae strains. J. Clin. Microbiol. 55, 1454–1468. doi: 10.1128/JCM.00100-17
Dona, V., Low, N., Golparian, D., and Unemo, M. (2017). Recent advances in the development and use of molecular tests to predict antimicrobial resistance in Neisseria gonorrhoeae. Expert Rev. Mol. Diagn. 17, 845–859. doi: 10.1080/14737159.2017.1360137
European Centre for Disease Prevention and Control [ECDC] (2021). Data from the ECDC Surveillance Atlas of Infectious Diseases. Available online at: https://atlas.ecdc.europa.eu/public/index.aspx?Dataset=27&HealthTopic=4 (accessed August 7, 2021).
Eyre, D. W., De Silva, D., Cole, K., Peters, J., Cole, M. J., Grad, Y. H., et al. (2017). WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J. Antimicrob. Chemother. 72, 1937–1947. doi: 10.1093/jac/dkx067
Foerster, S., Drusano, G., Golparian, D., Neely, M., Piddock, L. J. V., Alirol, E., et al. (2019). In vitro antimicrobial combination testing of and evolution of resistance to the first-in-class spiropyrimidinetrione zoliflodacin combined with six therapeutically relevant antimicrobials for Neisseria gonorrhoeae. J. Antimicrob. Chemother. 74, 3521–3529. doi: 10.1093/jac/dkz376
Gianecini, R. A., Golparian, D., Zittermann, S., Litvik, A., Gonzalez, S., Oviedo, C., et al. (2019). Genome-based epidemiology and antimicrobial resistance determinants of Neisseria gonorrhoeae isolates with decreased susceptibility and resistance to extended-spectrum cephalosporins in Argentina in 2011-16. J. Antimicrob. Chemother. 74, 1551–1559. doi: 10.1093/jac/dkz054
Golparian, D., Bazzo, M. L., Golfetto, L., Gaspar, P. C., Schörner, M. A., Schwartz Benzaken, A., et al. (2020a). Genomic epidemiology of Neisseria gonorrhoeae elucidating the gonococcal antimicrobial resistance and lineages/sublineages across Brazil, 2015-16. J. Antimicrob. Chemother. 75, 3163–3172. doi: 10.1093/jac/dkaa318
Golparian, D., Harris, S. R., Sánchez-Busó, L., Hoffmann, S., Shafer, W. M., Bentley, S. D., et al. (2020b). Genomic evolution of Neisseria gonorrhoeae since the preantibiotic era (1928-2013): antimicrobial use/misuse selects for resistance and drives evolution. BMC Genomics 21:116. doi: 10.1186/s12864-020-6511-6
Golparian, D., Rose, L., Lynam, A., Mohamed, A., Bercot, B., Ohnishi, M., et al. (2018). Multidrug-resistant Neisseria gonorrhoeae isolate, belonging to the internationally spreading Japanese FC428 clone, with ceftriaxone resistance and intermediate resistance to azithromycin, Ireland, August 2018. Euro Surveill. 23:1800617. doi: 10.2807/1560-7917.ES.2018.23.47.1800617
Golparian, D., Sánchez-Busó, L., Cole, M., and Unemo, M. (2021). Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance (NG-STAR) clonal complexes are consistent with genomic phylogeny and provide simple nomenclature, rapid visualization and antimicrobial resistance (AMR) lineage predictions. J. Antimicrob. Chemother. 76, 940–944. doi: 10.1093/jac/dkaa552
Grad, Y. H., Harris, S. R., Kirkcaldy, R. D., Green, A. G., Marks, D. S., Bentley, S. D., et al. (2016). Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000-2013. J. Infect. Dis. 214, 1579–1587. doi: 10.1093/infdis/jiw420
Hanao, M., Aoki, K., Ishii, Y., Shimuta, K., Ohnishi, M., and Tateda, K. (2021). Molecular characterization of Neisseria gonorrhoeae isolates collected through a national surveillance programme in Japan, 2013: evidence of the emergence of a ceftriaxone-resistant strain from a ceftriaxone-susceptible lineage. J. Antimicrob. Chemother. 76, 1769–1775. doi: 10.1093/jac/dkab104
Harris, S. R., Cole, M. J., Spiteri, G., Sanchez-Buso, L., Golparian, D., Jacobsson, S., et al. (2018). Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect. Dis. 18, 758–768. doi: 10.1016/S1473-3099(18)30225-1
Jacobsson, S., Cole, M. J., Spiteri, G., Day, M., and Unemo, M. (2021). Associations between antimicrobial susceptibility/resistance of Neisseria gonorrhoeae isolates in European Union/European Economic Area and patients’ gender, sexual orientation and anatomical site of infection, 2009-2016. BMC Infect. Dis. 21:273. doi: 10.1186/s12879-021-05931-0
Jennison, A. V., Whiley, D., Lahra, M. M., Graham, R. M., Cole, M. J., Hughes, G., et al. (2019). Genetic relatedness of ceftriaxone-resistant and high-level azithromycin resistant Neisseria gonorrhoeae cases, United Kingdom and Australia, February to April 2018. Euro Surveill. 24:1900118. doi: 10.2807/1560-7917.ES.2019.24.8.1900118
Lahra, M. M., Martin, I., Demczuk, W., Jennison, A. V., Lee, K. I., Nakayama, S. I., et al. (2018). Cooperative recognition of internationally disseminated ceftriaxone-resistant Neisseria gonorrhoeae strain. Emerg. Infect. Dis. 24, 735–740. doi: 10.3201/eid2404.171873
Läkemedelsverket (2015). Sexuellt Överförbara Bakteriella Infektioner – Behandlingsrekommendation: Information Från Läkemedelsverket. 26. Available online at: https://www.lakemedelsverket.se/sv/behandling-och-forskrivning/behandlingsrekommendationer/sok-behandlingsrekommendationer/antibiotika-vid-sexuellt-overforbara-bakteriella-infektioner–behandlingsrekommendation (accessed December 23, 2021).
Lan, P. T., Golparian, D., Ringlander, J., Van Hung, L., Van Thuong, N., and Unemo, M. (2020). Genomic analysis and antimicrobial resistance of Neisseria gonorrhoeae isolates from Vietnam in 2011 and 2015-16. J. Antimicrob. Chemother. 75, 1432–1438. doi: 10.1093/jac/dkaa040
Lee, K., Nakayama, S. I., Osawa, K., Yoshida, H., Arakawa, S., Furubayashi, K. I., et al. (2019). Clonal expansion and spread of the ceftriaxone-resistant Neisseria gonorrhoeae strain FC428, identified in Japan in 2015, and closely related isolates. J. Antimicrob. Chemother. 74, 1812–1819. doi: 10.1093/jac/dkz129
Li, H. (2013). Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv. 1303.3997v1. Available online at: https://arxiv.org/abs/1303.3997 (accessed December 23, 2021).
Ma, K. C., Mortimer, T. D., Hicks, A. L., Wheeler, N. E., Sánchez-Busó, L., Golparian, D., et al. (2020). Adaptation to the cervical environment is associated with increased antibiotic susceptibility in Neisseria gonorrhoeae. Nat. Commun. 11:4126. doi: 10.1038/s41467-020-17980-1
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Martin, I. M., Ison, C. A., Aanensen, D. M., Fenton, K. A., and Spratt, B. G. (2004). Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 189, 1497–1505. doi: 10.1086/383047
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pathogenwatch (2021). A Global Platform for Genomic Surveillance. Available: https://pathogen.watch/ (accessed August 7, 2021).
Rowley, J., Vander Hoorn, S., Korenromp, E., Low, N., Unemo, M., Abu-Raddad, L. J., et al. (2019). Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull. World Health Organ. 97, 548–562. doi: 10.2471/BLT.18.228486
Sánchez-Busó, L., Golparian, D., Corander, J., Grad, Y. H., Ohnishi, M., Flemming, R., et al. (2019). The impact of antimicrobials on gonococcal evolution. Nat. Microbiol. 4, 1941–1950. doi: 10.1038/s41564-019-0501-y
Sánchez-Busó, L., Yeats, C. A., Taylor, B., Goater, R. J., Underwood, A., Abudahab, K., et al. (2021). A community-driven resource for genomic epidemiology and antimicrobial resistance prediction of Neisseria gonorrhoeae at Pathogenwatch. Genome Med. 13:61. doi: 10.1186/s13073-021-00858-2
Shafer, W. M. (2018). Mosaic drug efflux gene sequences from commensal neisseria can lead to low-level azithromycin resistance expressed by Neisseria gonorrhoeae clinical isolates. mBio 9, e11747–e11718. doi: 10.1128/mBio.01747-18
Shimuta, K., Watanabe, Y., Nakayama, S., Morita-Ishihara, T., Kuroki, T., Unemo, M., et al. (2015). Emergence and evolution of internationally disseminated cephalosporin-resistant Neisseria gonorrhoeae clones from 1995 to 2005 in Japan. BMC Infect. Dis. 15:378. doi: 10.1186/s12879-015-1110-x
Swedish Society for Dermatology and Venereology [SSDV] (2019). Gonorré. Available online at: https://www.ssdv.se/images/Riktlinjer_Gonorre_2019.pdf (accessed December 23, 2021).
Swedres-Svarm (2019). Sales of Antibiotics and Occurrence of Resistance in Sweden. Solna/Uppsala ISSN1650-6332. Uppsala: Public HEALTH AGENCY OF Sweden and National Veterinary Institute.
Thomas, J. C., Seby, S., Abrams, A. J., Cartee, J., Lucking, S., Vidyaprakash, E., et al. (2019). Evidence of recent genomic evolution in gonococcal strains with decreased susceptibility to cephalosporins or azithromycin in the United States, 2014-2016. J. Infect. Dis. 220, 294–305. doi: 10.1093/infdis/jiz079
Thomas, J. C. T., Joseph, S. J., Cartee, J. C., Pham, C. D., Schmerer, M. W., Schlanger, K., et al. (2021). Phylogenomic analysis reveals persistence of gonococcal strains with reduced-susceptibility to extended-spectrum cephalosporins and mosaic penA-34. Nat. Commun. 12:3801. doi: 10.1038/s41467-021-24072-1
Town, K., Harris, S., Sánchez-Busó, L., Cole, M. J., Pitt, R., Fifer, H., et al. (2020). Genomic and phenotypic variability in Neisseria gonorrhoeae antimicrobial susceptibility, England. Emerg. Infect. Dis. 26, 505–515. doi: 10.3201/eid2603.190732
Unemo, M., Golparian, D., Sanchez-Buso, L., Grad, Y., Jacobsson, S., Ohnishi, M., et al. (2016). The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J. Antimicrob. Chemother. 71, 3096–3108. doi: 10.1093/jac/dkw288
Unemo, M., Lahra, M. M., Cole, M., Galarza, P., Ndowa, F., Martin, I., et al. (2019a). World health organization global gonococcal antimicrobial surveillance program (WHO GASP): review of new data and evidence to inform international collaborative actions and research efforts. Sex Health 16, 412–425. doi: 10.1071/SH19023
Unemo, M., Seifert, H. S., Hook, E. W. 3rd., Hawkes, S., Ndowa, F., and Dillon, J. R. (2019b). Gonorrhoea. Nat. Rev. Dis. Primers 5:79. doi: 10.1038/s41572-019-0128-6
Unemo, M., Lahra, M. M., Escher, M., Eremin, S., Cole, M. J., Galarza, P., et al. (2021). WHO global antimicrobial resistance surveillance for Neisseria gonorrhoeae 2017-18: a retrospective observational study. Lancet Microbe 2:e627–36. doi: 10.1016/S2666-5247(21)00171-3
Unemo, M., Ross, J., Serwin, A. B., Gomberg, M., Cusini, M., and Jensen, J. S. (2020). 2020 European guideline for the diagnosis and treatment of gonorrhoea in adults. Int. J. STD AIDS [Online ahead of print] 956462420949126. doi: 10.1177/0956462420949126
Unemo, M., and Shafer, W. M. (2014). Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin. Microbiol. Rev. 27, 587–613. doi: 10.1128/CMR.00010-14
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46. doi: 10.1186/gb-2014-15-3-r46
World Health Organization [WHO] (2018). Report on Global Sexually Transmitted Infection Surveillance 2018 [Online]. Available online at: https://www.who.int/reproductivehealth/publications/stis-surveillance-2018/en/ (accessed August 7, 2021).
Keywords: Neisseria gonorrhoeae, molecular epidemiology, antimicrobial resistance, whole-genome sequencing, Sweden
Citation: Hadad R, Golparian D, Velicko I, Ohlsson A-K, Lindroth Y, Ericson E-L, Fredlund H, Engstrand L and Unemo M (2022) First National Genomic Epidemiological Study of Neisseria gonorrhoeae Strains Spreading Across Sweden in 2016. Front. Microbiol. 12:820998. doi: 10.3389/fmicb.2021.820998
Received: 23 November 2021; Accepted: 14 December 2021;
Published: 13 January 2022.
Edited by:
Karsten Becker, University Medicine Greifswald, GermanyReviewed by:
Phil Giffard, Charles Darwin University, AustraliaChristophe Van Dijck, Institute of Tropical Medicine Antwerp, Belgium
Copyright © 2022 Hadad, Golparian, Velicko, Ohlsson, Lindroth, Ericson, Fredlund, Engstrand and Unemo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magnus Unemo, bWFnbnVzLnVuZW1vQHJlZ2lvbm9yZWJyb2xhbi5zZQ==
 Ronza Hadad
Ronza Hadad Daniel Golparian
Daniel Golparian Inga Velicko2
Inga Velicko2 Ylva Lindroth
Ylva Lindroth Magnus Unemo
Magnus Unemo