
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 January 2022
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.818228
Pogostemon cablin (patchouli), an important medicinal and aromatic plant, is widely used in traditional Chinese medicine as well as in perfume industry. Patchouli plants are susceptible to bacterial wilt disease, which causes significant economic losses by reduction in yield and quality of the plant products. However, few studies focus on the pathogens causing bacterial wilt on patchouli. In this study, strain Pa82 was isolated from diseased patchouli plants with typical bacterial wilt symptoms in Guangdong province, China, and was confirmed to be a highly virulent pathogen of patchouli bacterial wilt. Comparative sequence analysis of 16S rRNA gene showed that the strain was closely related to Kosakonia sp. CCTCC M2018092 (99.9% similarity) and Kosakonia cowanii Esp_Z (99.8% similarity). Moreover, phylogenetic tree based on 16S rRNA gene sequences showed that the strain was affiliated with genus Kosakonia. Further, the whole genome of strain Pa82 was sequenced, and the sequences were assembled and annotated. The complete genome of the strain consists of one chromosome and three plasmids. Average nucleotide identity (ANI) and phylogenetic analysis revealed that the strain belongs to Kosakonia cowanii (designated Kosakonia cowanii Pa82). Virulence-related genes of the strain involved in adherence, biofilm formation, endotoxin and other virulence factors were predicted. Among them, vgrG gene that encodes one of the type VI secretion system components was functionally validated as a virulence factor in Kosakonia cowanii Pa82 through construction of Tn5 insertion mutants and identification of mutant defective in virulence.
Pogostemon cablin (Blanco) Benth. (patchouli), a herbaceous plant species belonging to family Lamiaceae, is well known for its medicinal and aromatic properties. Patchouli is native to tropical Southeast Asia and now widely cultivated in many tropical and subtropical countries, especially China, India, Philippines, Indonesia, Vietnam, Malaysia, and Thailand (Swamy and Sinniah, 2016). In traditional Chinese medicine, the dried aerial parts of patchouli are frequently used for the treatment of headache, common cold, fever, nausea, vomiting, diarrhea, and so forth (Chen et al., 2021). Recently, extensive studies have indicated that patchouli possesses diverse pharmacological activities, including antimicrobial, antioxidative, antitumor, sedative, and gastrointestinal protective activities (Hu et al., 2017; Kim et al., 2019; Xie et al., 2020). Moreover, patchouli is also an economically important aromatic plant producing essential oil, which is widely used in fragrance and cosmetic industries (Chen et al., 2013).
Patchouli plants are susceptible to bacterial wilt disease, which causes significant economic losses by reduction in yield and quality of the plant products. Bacterial wilt is a devastating soil-borne disease that is widely distributed in tropical, subtropical, and some temperate regions, affecting hundreds of plant species, particularly the crops of family Solanaceae. Typical symptoms of the disease are leaf wilt, browning of vascular tissues, and even collapsing of the plant. Ralstonia solanacearum is known to be the most prominent causal agent of bacterial wilt worldwide, infecting a wide range of hosts, including solanaceous and non-solanaceous plants (Peeters et al., 2013; Jiang et al., 2017). In recent years, other causal organisms such as members of genus Enterobacter, genus Erwinia and genus Kosakonia have been reported to cause similar symptoms of bacterial wilt (Wang et al., 2008; Zhu et al., 2010; Sanogo et al., 2011; Sarkar and Chaudhuri, 2015).
In this study, strain Pa82 was isolated from diseased patchouli plants with typical bacterial wilt symptoms in Guangdong province, China, and was confirmed to be a highly virulent pathogen of patchouli bacterial wilt. And then, the complete genome of strain Pa82 was sequenced, and the sequences were assembled and annotated. Moreover, the approach for functional validation of potential virulence-related genes in strain Pa82 was established through construction of Tn5 insertion mutants and identification of mutants defective in virulence. In this paper, we presented a novel pathogen causing bacterial wilt on patchouli and unraveled its phylogeny and genetic basis.
Naturally infected patchouli plants with typical symptoms of bacterial wilt were collected from Guangdong province, China. Vascular tissues at the base of the stems from these diseased plants were rinsed with sterile water, and then cut into several fragments. The fragments were soaked in sterile water, and the suspension was diluted and streaked on the medium containing 0.005% 2, 3, 5-triphenyltetrazolium chloride (TTC) (Swanson et al., 2005). From the plates, bacterial colonies were picked based on their morphology and were subcultured on fresh TTC medium until pure isolates were obtained. The pure isolates were stored at −80°C in nutrient agar (NA) liquid medium containing 7% dimethyl sulfoxide (DMSO).
The bacterial isolates were streaked on TTC medium, and were cultured at 30°C for 36 h. Colony morphology of the bacterial isolates was then observed. Bacterial cells from each isolate were suspended in 0.9% NaCl solution. Following the Gram stain, cell morphology was observed under fluorescence microscopy using 100 × oil-immersion objective lens (1,000 × total magnification) (Azabou et al., 2007).
The pathogenicity test of the bacterial isolates was conducted by inoculating the bacteria to patchouli plants. Surface sterilized leaf and stem explants of patchouli were cultured on Murashige and Skoog (MS) medium to achieve shoot regeneration, seedling propagation and regenerated shoot rooting. Plantlets with well-developed shoots and roots were transplanted into pots containing sterile soil and grown in greenhouse for about 40 days. A single colony of each bacterial isolate on NA solid medium was picked and transferred to NA liquid medium. After 24 h incubation at 28°C with continuous shaking (200 rpm), the bacterial suspension was diluted to approximately 5 × 108 cfu mL–1. The adjusted bacterial suspension was inoculated into roots of patchouli plants using the soaking method while control plants were mock-inoculated sterile water (Vailleau et al., 2007). The plants were grown under greenhouse conditions maintained at 28 ± 2°C with 85% relative humidity.
Strain Pa82, a highly virulent strain isolated from diseased patchouli plants, was used for further strain analysis. After incubation at 28°C for 24 h, bacterial cells of strain Pa82 in NA liquid medium were harvested by centrifugation at 4,000 rpm for 10 min. Genomic DNA was extracted from the bacterial cells using Biospin Bacteria Genomic DNA Extraction Kit. The 16S rRNA gene sequence of strain Pa82 was amplified by polymerase chain reaction (PCR) using the forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′). PCR reaction was conducted in a total volume of 25 μL containing 1 μL of DNA template, 1 μL of each primer, 12.5 μL of Premix Ex Taq and 9.5 μL of double-distilled water. PCR protocol was as follows: 94°C for 2 min; followed by 35 cycles at 94°C for 30 s, 57°C for 30 s, 72°C for 2 min; and a final extension at 72°C for 10 min. The PCR product was sequenced, and the sequence was compared with 16S rRNA gene sequences in the GenBank database. A phylogenetic tree based on the 16S rRNA gene sequences from strain Pa82 and its relative species was constructed using neighbor-joining method in MEGA X software and bootstrap analysis was performed with 1,000 replicates (Russo and Selvatti, 2018).
Genomic DNA of strain Pa82 was extracted using the Wizard Genomic DNA Purification Kit according to the manufacturer’s instructions. The quality and quantity of the genomic DNA was determined using TBS-380 fluorometer. Short-read libraries (2 × 150 bp) were sequenced on the Illumina HiSeq X Ten platform and long-read libraries (about 10 kb) were sequenced on the Pacific Bioscience (PacBio) Sequel platform. The high-quality PacBio clean reads were then used for genome assembly by using Canu (Koren et al., 2017). Meanwhile the Illumina clean reads were used to correct errors for further improve the accuracy of the PacBio assembly results. The final genome assembly comprised seamless chromosomes and plasmids.
Coding sequences (CDS), ribosomal RNA (rRNA) genes, and transfer RNA (tRNA) genes were predicted using Glimmer 3.02, Barrnap 0.8, and tRNAscan-SE 2.0, respectively (Delcher et al., 2007; Seemann, 2013; Lowe and Chan, 2016). The circular genome map was drawn using CGView 2 (Stothard and Wishart, 2005). All predicted CDS were searched and annotated by BLAST against the publicly available protein databases including NCBI non-redundant protein (NR), Swiss-Prot, Pfam, and Clusters of Orthologous Groups (COG). Average nucleotide identity (ANI) values between genomes of strain Pa82 and its relative strains were calculated using JSpeciesWS (Richter et al., 2016). Potential virulence-related genes were predicted by the BLAST search against the virulence factor database (VFDB) (Chen et al., 2016).
Functional validation of predicted virulence-related genes in strain Pa82 requires the generation of gene disruption mutants. Mutants of strain Pa82 were constructed by using an EZ-Tn5™ < KAN-2 > Tnp Transposome™ Kit. Competent cells of the strain were prepared and mixed with Tn5 transposomes, then the mixture was electrically transformed at 2.5 kV, 25 μF and 400 Ω. Transformed cells were recovered in SOC liquid medium at 30°C for 1 h with 200 rpm shaking and then plated on NA solid medium supplemented with 10 μg mL–1 kanamycin. Positive clones were identified by PCR with the specific primer pairs Kan-F and Kan-R, which were designed according to the sequence of the kanamycin resistance gene (Kanr) of Tn5. From these mutants, the flanking sequences adjacent to Tn5 transposon insertion-site were amplified by inverse PCR using transposon-specific primer pairs KAN-2 FP-1 and KAN-2 RP-1, and the PCR products were sequenced (Malamud et al., 2012). The sequences were then compared with corresponding sequences available in GenBank database using BLAST.
Bacteria were isolated from the patchouli plants infected with bacterial wilt by the streak plate method. The pathogenic isolates were evaluated by molecular, physiological and pathological characterization. Among them, strain Pa82 was found to be a highly virulent pathogen to cause patchouli bacterial wilt. The morphological characteristics of strain Pa82 are shown in Figure 1. The colonies are smooth, moist, circular disc with white edge and red pigmentation in the center when incubated on TTC medium at 30°C for 36 h (Figure 1A). The bacterial cells are Gram-negative and rod-shaped, ranging in length from 0.7 to 2.1 μm under the oil-immersion lens (Figure 1B). The virulence characteristics of strain Pa82 were investigated by conventional pathogenicity test. Patchouli plants inoculated with strain Pa82 developed initial symptoms as drooping and shriveling leaves, followed by complete wilting or collapsing of the whole plants, showing typical symptoms of bacterial wilt. Whereas control plants treated with sterile water did not exhibit disease-like symptoms and remained healthy. The plant phenotypes of pathogenicity test at 7 days post-inoculation are shown in Figure 2.
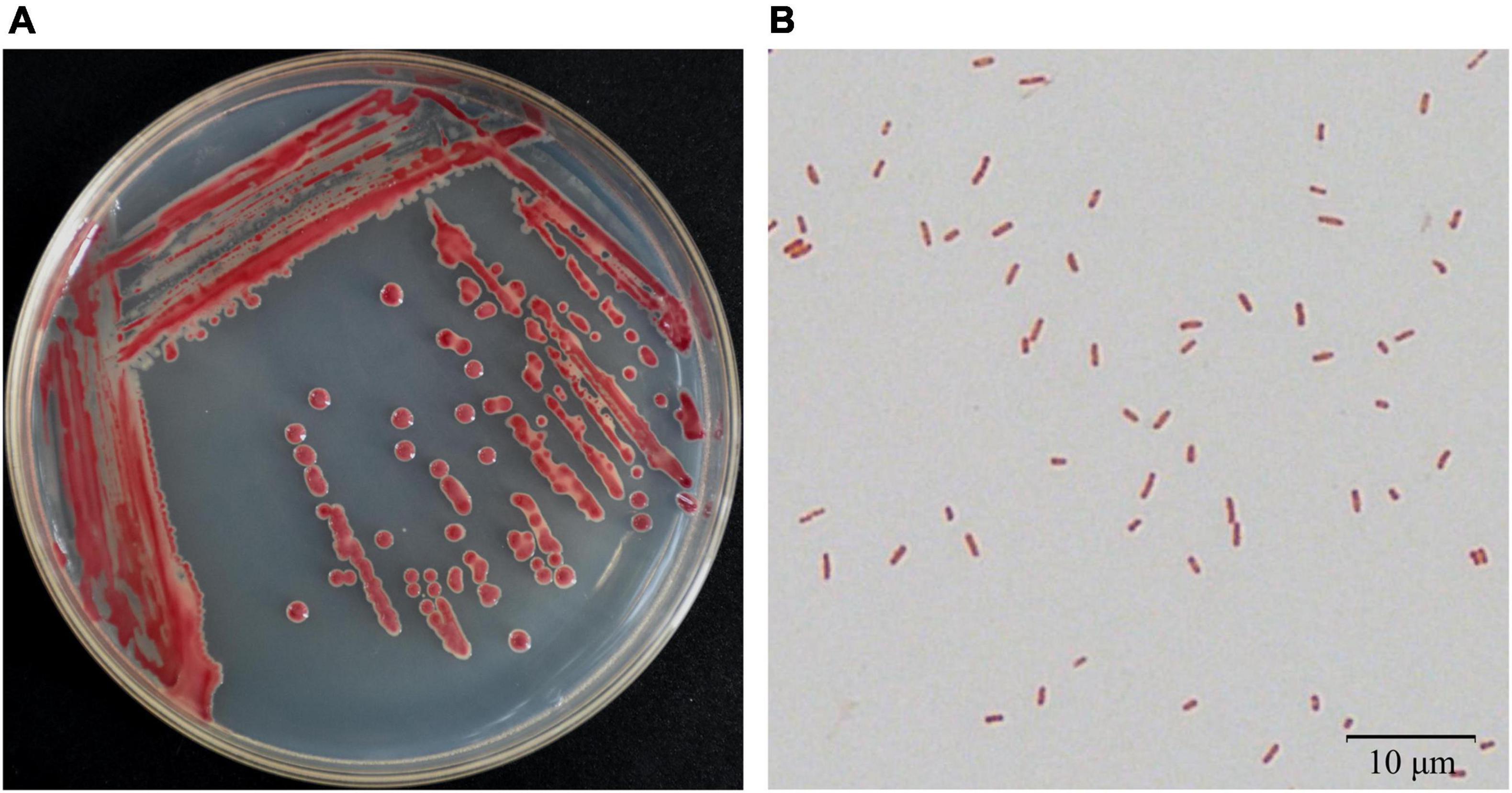
Figure 1. Morphological characteristics of strain Pa82. (A) Colony appearance on TTC medium; (B) Cell morphology under 100 × oil-immersion objective lens (1,000 × total magnification).
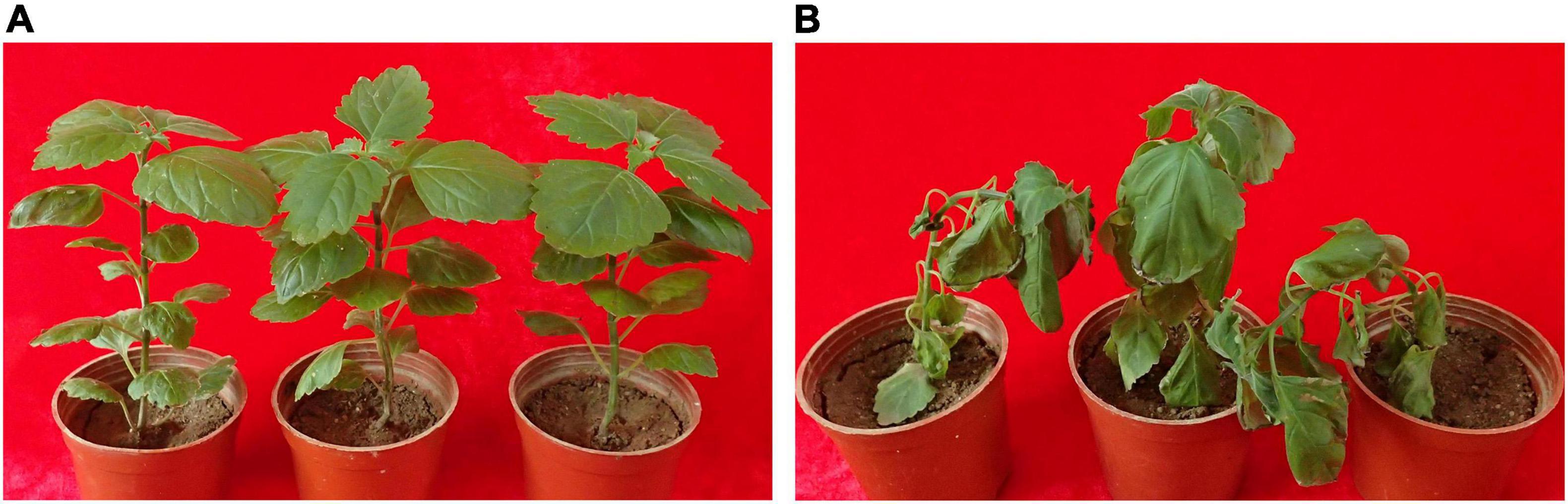
Figure 2. The plant phenotypes of pathogenicity test at 7 days post-inoculation. (A) Plants mock-inoculated with sterile water; (B) Plants inoculated with strain Pa82.
The 16S rRNA gene of strain Pa82 was amplified by PCR, and the sequencing data (1,460 bp in length) was submitted to NCBI database (accession number: OL795994.1). Comparative sequence analysis of 16S rRNA gene showed that the strain was closely related to Kosakonia sp. CCTCC M2018092 (99.9% similarity) and Kosakonia cowanii Esp_Z (99.8% similarity). As genus Kosakonia belongs to family Enterobacteriaceae, a phylogenetic tree based on the 16S rRNA gene sequences from strain Pa82 and its relative species belonging to family Enterobacteriaceae was constructed. The 16S rRNA gene sequence from Dickeya chrysanthemi SD17-11 was used as the outgroup. Horizontal branch lengths represent relative evolutionary distances. Numbers above branch nodes are bootstrap values. The phylogenetic tree showed that all the species were divided into three clades, and one of which included strain Pa82 and species from genus Enterobacter or genus Kosakonia. The results also showed that strain Pa82 was most closely related to Kosakonia sp. CCTCC M2018092, indicating that the strain was affiliated with genus Kosakonia (Figure 3).

Figure 3. Phylogenetic tree based on 16S rRNA gene sequences from strain Pa82 and its relative strains of family Enterobacteriaceae. The 16S rRNA gene sequence from Dickeya chrysanthemi SD17-11 was used as the outgroup.
To better understand the genetic basis of the virulence properties of strain Pa82, the complete genome sequencing was performed, and the sequences were assembled and annotated. The complete genome of the strain was found to contain one chromosome of 4,895,354 bp and three plasmids of 141,409, 6,175, and 5,538 bp, respectively (Figure 4). The general genome features of strain Pa82 are summarized in Table 1. In total, 4,669 coding sequences (CDS) were predicted with 4,505 in the chromosome and 164 in the three plasmids. Among them, 4,284 CDS were assigned into the functional categories in Clusters of Orthologous Groups (COG) database (Table 2). The main categories are G (carbohydrate transport and metabolism; 8.15%), K (transcription; 8.15%), E (amino acid transport and metabolism; 7.82%), P (inorganic ion transport and metabolism; 6.63%), M (cell wall/membrane/envelope biogenesis; 5.86%) and C (energy production and conversion; 5.30%). However, the results also showed that a large number of CDS were assigned into COG category S (function unknown, 29.48%), which may be revealed by further functional studies.
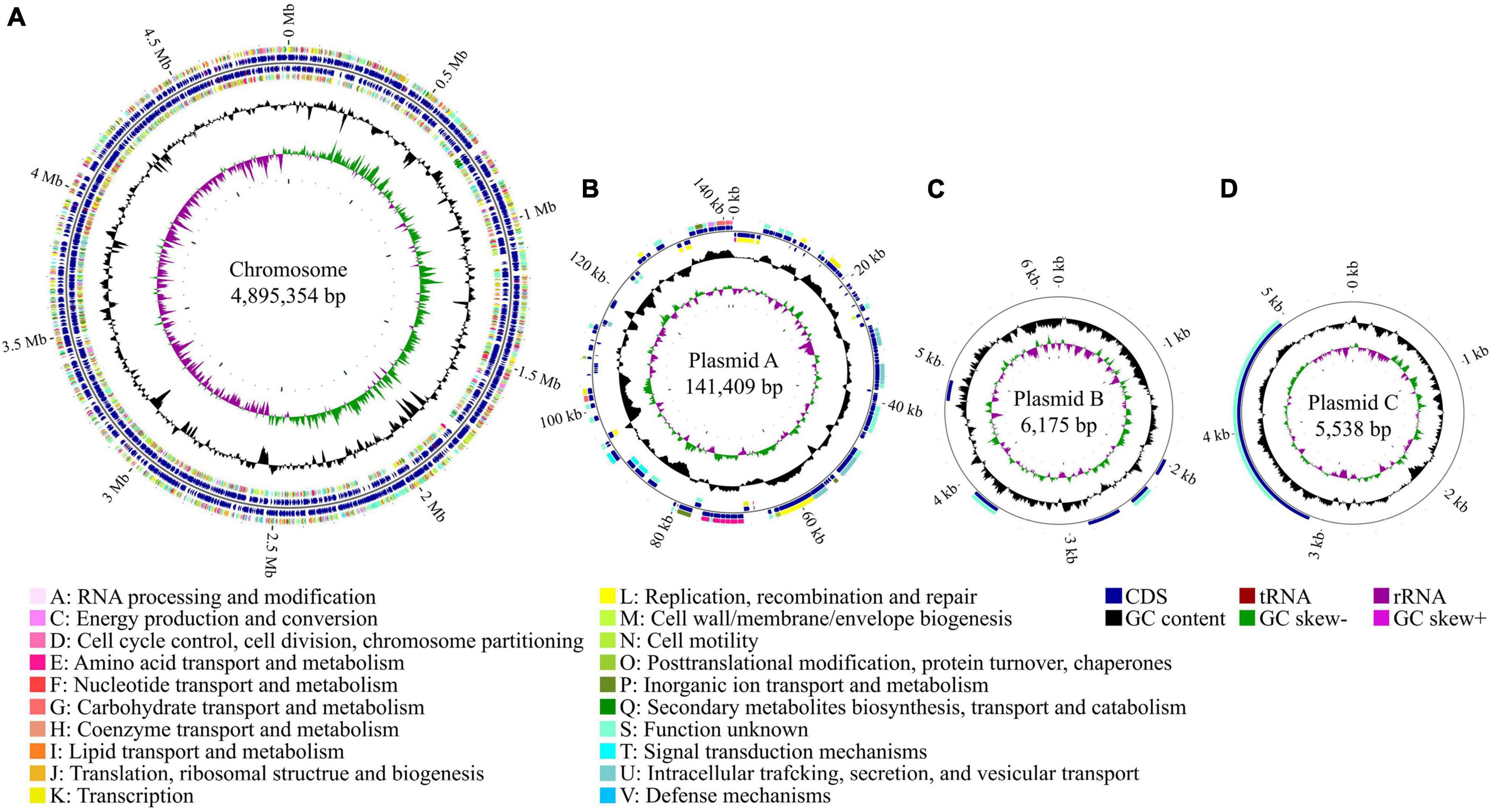
Figure 4. Circular maps of strain Pa82 genome. (A) Chromosome; (B) Plasmid A; (C) Plasmid B; (D) Plasmid C. The distribution of circles from inside to outside, circle 1, GC skew (green indicates a region with G content greater than C content; purple indicates a region with G content less than C content); circle 2, GC content (outward means above the average GC content of the whole genome; inward means below the average GC content of the whole genome); circles 3 and 6, CDS on the reverse and forward strands were annotated by Clusters of Orthologous Groups (COG) database (different colors represent different COG functional categories); circles 4 and 5, CDS, tRNA and rRNA on the reverse and forward strands; circle 7, scale marks of the genome size.
ANI was used to assess the relationship between strain Pa82 and its relative species from genus Kosakonia, and a threshold of 95–96% was set as a boundary for species delineation (Richter and Rosselló-Móra, 2009). The genome comparison between strain Pa82 and its relative species from genus Kosakonia showed ANI values ranging from 81.18 to 97.48% (Figure 5). Notably, the ANI value (97.48%) between the genome sequences of strain Pa82 and its closest related strain Kosakonia cowanii Esp_Z was above the accepted 95–96% cut-off threshold for species delineation. Therefore, combined with the ANI and phylogenetic analysis, strain Pa82 was identified as Kosakonia cowanii and designated as Kosakonia cowanii Pa82.
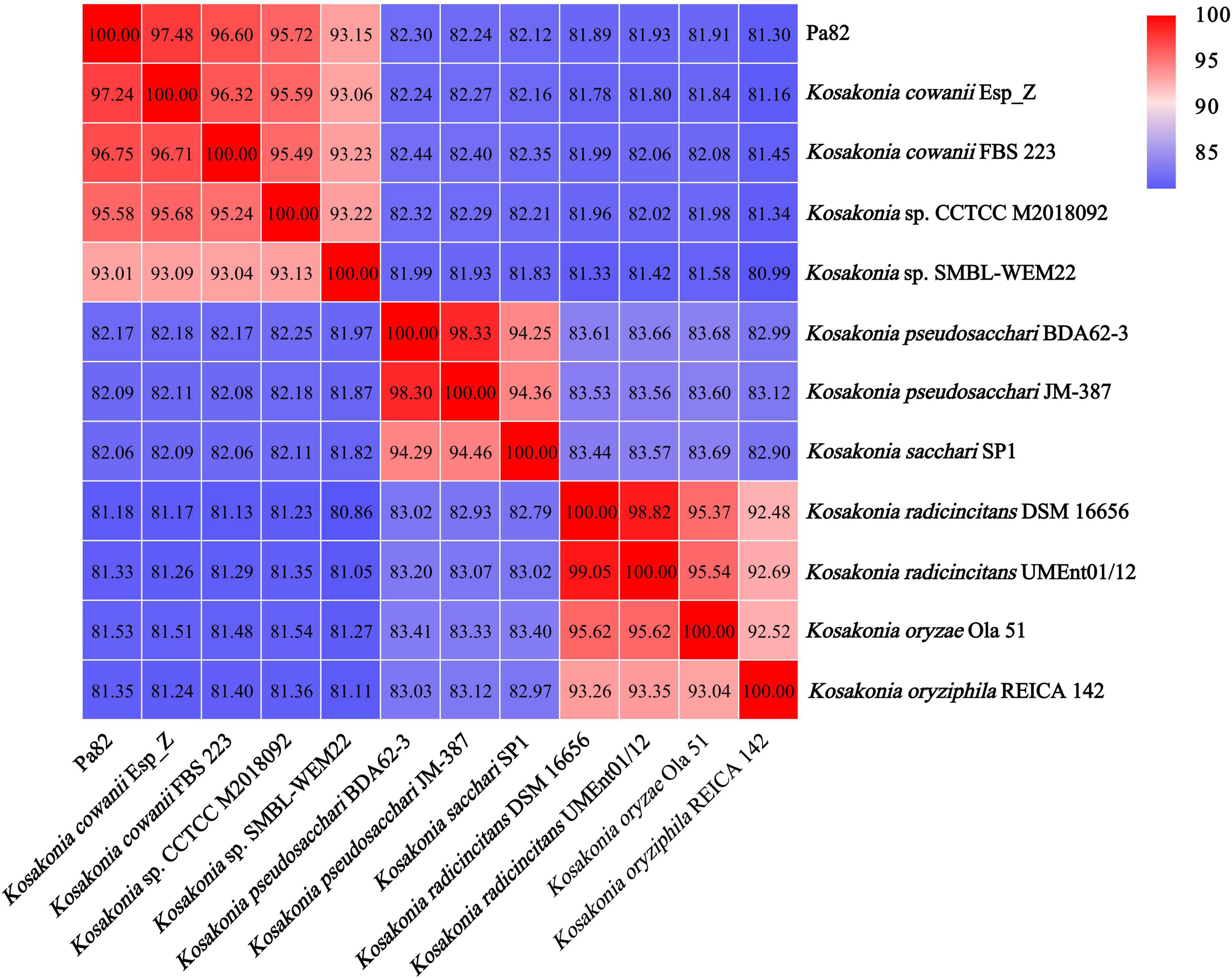
Figure 5. Heat map based on average nucleotide identity (ANI) values between each pair of genome sequences from strain Pa82 and strains within genus Kosakonia.
Genes potentially involved in virulence of strain Pa82 were predicated using the VFDB. A total of 610 potential virulence-related genes were found in strain Pa82 genome with 601 in the chromosome and 9 in the plasmid A. Among them, genes involved in known and putative virulence factors which are important for bacterial infections are summarized in Table 3. These genes are classified into 6 categories, including adherence (9 genes), biofilm formation (3 genes), endotoxin (4 genes), invasion (23 genes), secretion system (6 genes), and toxin (2 genes).
Functional validation of potential virulence-related genes in strain Pa82 requires construction of gene disruption mutants and identification of mutants defective in virulence. Mutants of strain Pa82 were created by random Tn5 transposon mutagenesis and confirmed by PCR method. The flanking fragments adjacent to Tn5 transposon insertion site in each mutant was individually amplified by inverse PCR, and amplified product was sequenced. The obtained sequences were compared for similarity to those available in GenBank database, and the potential virulence-related genes were selected. One such mutant (Pa82-87-1) was obtained, and the Tn5 insertion junction was mapped to vgrG, a gene encoding one of type VI secretion system (T6SS) components (as shown in Table 3). Gene vgrG (2,613 bp in length) of mutant Pa82-87-1 was inserted by a 1,221 bp Tn5 transposon (Figure 6). Pathogenicity test showed that mutant Pa82-87-1 was significantly reduced in virulence on patchouli plants compared to the wild type strain Pa82 (Figure 7). The results showed that vgrG gene in strain Pa82 was functionally validated as a virulence factor. In this way, more virulence-related genes of strain Pa82 may be further validated.
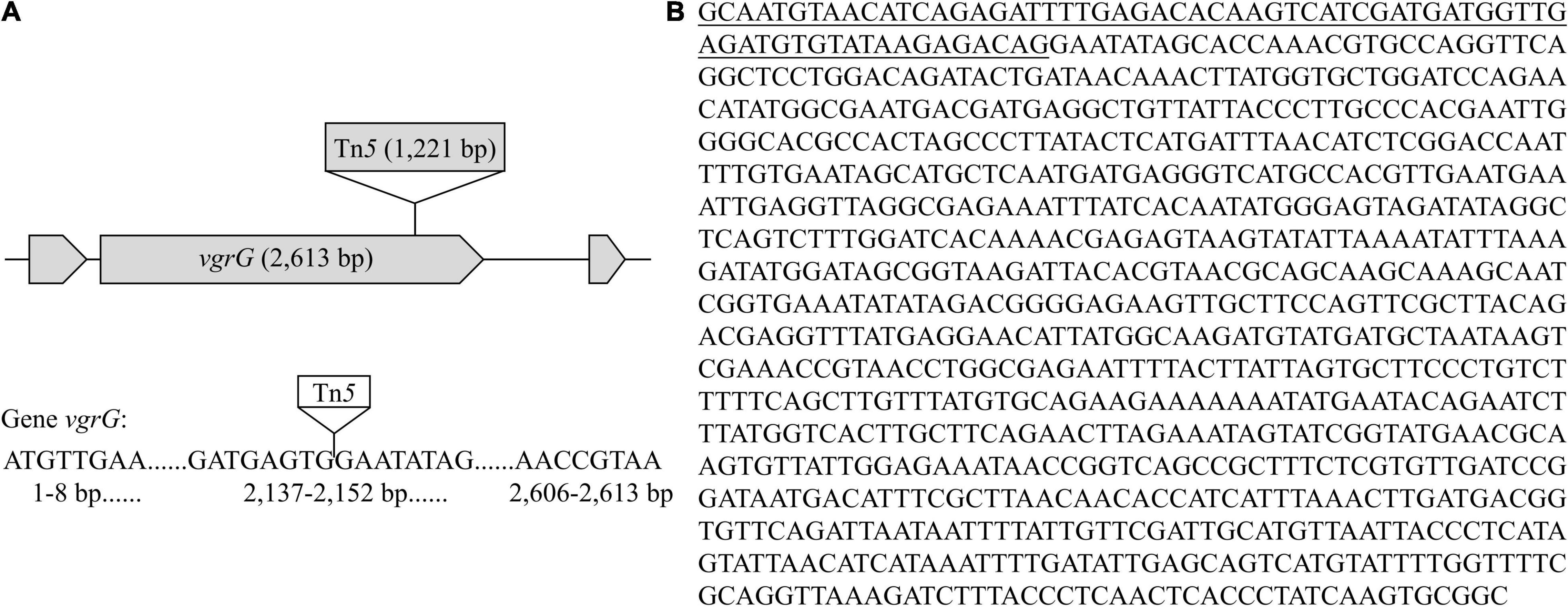
Figure 6. Characterization and sequence analysis of mutant Pa82-87-1. (A) Schematic diagram of Tn5 transposon insertion in the vgrG gene; (B) Flanking sequence adjacent to Tn5 transposon insertion site in mutant Pa82-87-1. The underlined part is the residual sequence of Tn5 transposon.

Figure 7. Pathogenicity test of mutant Pa82-87-1 on patchouli plants (at 5 days post-inoculation). (A) Plants mock-inoculated with sterile water (negative control); (B) Plants inoculated with wild type strain Pa82 (positive control); (C) Plants inoculated with mutant Pa82-87-1.
The genus Kosakonia has been recently derived from reclassification of genus Enterobacter, and several species, including Enterobacter arachidis, Enterobacter cowanii, Enterobacter oryzae, Enterobacter radicincitans, previously included in genus Enterobacter have been transferred to the novel genus Kosakonia (Brady et al., 2013). The genus Kosakonia has been reported to associate with plant growth-promoting bacteria (Kämpfer et al., 2005; Peng et al., 2009), and nowadays, some species from genus Kosakonia were found to be phytopathogen causing a variety of plant diseases (Furtado et al., 2012; Wu et al., 2016; Krawczyk and Borodynko-Filas, 2020). Bacterial wilt is one of the most serious threats to hundreds of plant species worldwide. For decades, the disease has been reported to be caused by Ralstonia solanacearum. However, several studies in recent years have indicated that some species from genus Kosakonia are capable of causing similar symptoms of bacterial wilt. For example, Kosakonia cowanii was identified as the causal agent of bacterial wilt on tomatoes (Sarkar and Chaudhuri, 2015). In this study, strain Pa82 was isolated from diseased patchouli plants with typical bacterial wilt symptoms and identified as Kosakonia cowanii. To our knowledge, this is the first report of Kosakonia cowanii Pa82 causing patchouli bacterial wilt in China.
Complete genome sequencing could provide novel insight into the genetic basis of virulence in strain Pa82. In this study, a total of 610 potential virulence-related genes were predicted in the genome of strain Pa82 through VFDB analysis. The roles of some genes in bacterial attachment, motility and virulence have been investigated in several animal and plant pathogenic bacteria. Gene fimA and gene fimH encoding adhesin mediating attachment of type 1 fimbriae are commonly associated with bacterial adhesion to host cells (Spaulding et al., 2017; Hasegawa and Nagano, 2021). Gene flhC and gene flgK, the regulating and structural flagellar genes, participate in the regulation of bacterial motility (Xu et al., 2014; Weller-Stuart et al., 2017). Gene tssB and gene vgrG related to the assembly of type VI secretion system (T6SS) have been reported to be important in the virulence of many animal and plant pathogens (Zhang et al., 2014; Wang et al., 2018). Detection of genes associated with virulence of strain Pa82 facilitates a better understanding how the bacterial pathogen has evolved virulence strategies to invade patchouli.
The genome of strain Pa82 has been completely sequenced and virulence-related genes have been predicted, the next step is often to validate roles of those genes in pathogenicity of the strain. The construction of gene disruption mutants generated by Tn5 transposon insertion and their individual phenotype analysis is a common approach for the functional studies on the role of genes (Prelich, 2012). If Tn5 transposon inserts into the virulence-related genes, the pathogenicity of the mutants may decrease or even disappear, which may identify these genes (Sakata et al., 2019). Our results showed that one such mutant with an insertion in vgrG gene (a gene encoding one of T6SS components) was obtained, it was therefore predicted to be affected in virulence phenotype. Pathogenicity test showed that the mutant was significantly reduced in virulence on patchouli plants compared to the wild type strain Pa82. In this study, an efficient method for functional validation of potential virulence-related genes in strain Pa82 was established.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP069319, CP069320, CP069321, and CP069322.
HH was the principal investigator, conceived, and designed the experiments, and contributed to the writing and revision of the manuscript. YZ, BW, and QL performed the experiments, analyzed the data, and wrote the manuscript. DH wrote parts of the manuscript. YYZ and GL performed parts of the experiments. All authors approved the final version of the manuscript.
This study was financially supported by the National Natural Science Foundation of China (81373901) and the Natural Science Foundation of Guangdong province, China (2019A1515012151).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Azabou, S., Mechichi, T., Patel, B. K. C., and Sayadi, S. (2007). Isolation and characterization of a mesophilic heavy-metals-tolerant sulfate-reducing bacterium Desulfomicrobium sp. from an enrichment culture using phosphogypsum as a sulfate source. J. Hazard. Mater. 140, 264–270. doi: 10.1016/j.jhazmat.2006.07.073
Brady, C., Cleenwerck, I., Venter, S., Coutinho, T., and Vos, P. D. (2013). Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 36, 309–319. doi: 10.1016/j.syapm.2013.03.005
Chen, J. R., Xie, X. F., Li, M. T., Qiong, Q. Y., Li, G. M., Zhang, H. Q., et al. (2021). Pharmacological activities and mechanisms of action of Pogostemon cablin Benth: a review. Chin. Med. 16:5. doi: 10.1186/s13020-020-00413-y
Chen, L. H., Zheng, D. D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chen, M. W., Zhang, J. M., Lai, Y. F., Wang, S. P., Li, P., Xiao, J., et al. (2013). Analysis of Pogostemon cablin from pharmaceutical research to market performances. Expert Opin. Investig. Drugs 22, 245–257. doi: 10.1517/13543784.2013.754882
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Furtado, G. Q., Guimarães, L. M. S., Lisboa, D. O., Cavalcante, G. P., Arriel, D. A. A., Alfenas, A. C., et al. (2012). First report of Enterobacter cowanii causing bacterial spot on Mabea fistulifera, a native forest species in Brazil. Plant Dis. 96:1576. doi: 10.1094/PDIS-02-12-0160-PDN
Hasegawa, Y., and Nagano, K. (2021). Porphyromonas gingivalis FimA and Mfa1 fimbriae: current insights on localization, function, biogenesis, and genotype. Jpn. Dent. Sci. Rev. 57, 190–200. doi: 10.1016/j.jdsr.2021.09.003
Hu, G. Y., Peng, C., Xie, X. F., Zhang, S. Y., and Cao, X. Y. (2017). Availability, pharmaceutics, security, pharmacokinetics, and pharmacological activities of patchouli alcohol. Evid. Based Compl. Alternat. Med. 2017:4850612. doi: 10.1155/2017/4850612
Jiang, G. F., Wei, Z., Xu, J., Chen, H. L., Zhang, Y., and She, X. M. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8:1549. doi: 10.3389/fpls.2017.01549
Kämpfer, P., Ruppel, S., and Remus, R. (2005). Enterobacter radicincitans sp. nov., a plant growth promoting species of the family Enterobacteriaceae. Syst. Appl. Microbiol. 28, 213–221. doi: 10.1016/j.syapm.2004.12.007
Kim, E. K., Kim, J. H., Jeong, S., Choi, Y. W., Choi, H. J., Kim, C. Y., et al. (2019). Pachypodol, a methoxyflavonoid isolated from Pogostemon cablin Bentham exerts antioxidant and cytoprotective effects in HepG2 cells: possible role of ERK-dependent Nrf2 activation. Int. J. Mol. Sci. 20:4082. doi: 10.3390/ijms20174082
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Krawczyk, K., and Borodynko-Filas, N. (2020). Kosakonia cowanii as the new bacterial pathogen affecting soybean (Glycine max Willd.). Eur. J. Plant Pathol. 157, 173–183.
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Malamud, F., Conforte, V. P., Rigano, L. A., Castagnaro, A. P., Marano, M. R., Alexandre, M. D. A., et al. (2012). HrpM is involved in glucan biosynthesis, biofilm formation and pathogenicity in Xanthomonas citri ssp. citri. Mol. Plant Pathol. 13, 1010–1018. doi: 10.1111/j.1364-3703.2012.00809.x
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
Peng, G. X., Zhang, W., Luo, H. F., Xie, H. W., Lai, W. H., and Tan, Z. Y. (2009). Enterobacter oryzae sp. nov., a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia. Int. J. Syst. Evol. Microbiol. 59, 1650–1655. doi: 10.1099/ijs.0.65484-0
Prelich, G. (2012). Gene overexpression: uses, mechanisms, and interpretation. Genetics 190, 841–854. doi: 10.1534/genetics.111.136911
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Russo, C. A. D. M., and Selvatti, A. P. (2018). Bootstrap and rogue identification tests for phylogenetic analyses. Mol. Biol. Evol. 35, 2327–2333. doi: 10.1093/molbev/msy118
Sakata, N., Ishiga, T., Saito, H., Nguyen, V. T., and Ishiga, Y. (2019). Transposon mutagenesis reveals Pseudomonas cannabina pv. alisalensis optimizes its virulence factors for pathogenicity on different hosts. PeerJ 7:e7698. doi: 10.7717/peerj.7698
Sanogo, S., Etarock, B. F., and Clary, M. (2011). First report of bacterial wilt caused by Erwinia tracheiphila on pumpkin and watermelon in New Mexico. Plant Dis. 95:1583. doi: 10.1094/PDIS-06-11-0507
Sarkar, S., and Chaudhuri, S. (2015). New report of additional enterobacterial species causing wilt in West Bengal. India Can. J. Microbiol. 61, 477–486. doi: 10.1139/cjm-2015-0017
Spaulding, C. N., Klein, R. D., Ruer, S., Kau, A. L., Schreiber, H. L., and Cusumano, Z. T. (2017). Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546, 528–532. doi: 10.1038/nature22972
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Swamy, M. K., and Sinniah, U. R. (2016). Patchouli (Pogostemon cablin Benth.): botany, agrotechnology and biotechnological aspects. Ind. Crop Prod. 87, 161–176.
Swanson, J. K., Yao, J., Tans-Kersten, J., and Allen, C. (2005). Behavior of Ralstonia solanacearum race 3 niovar 2 during latent and active infection of geranium. Phytopathology 95, 136–143. doi: 10.1094/PHYTO-95-0136
Vailleau, F., Sartorel, E., Jardinaud, M. F., Chardon, F., Genin, S., Huguet, T., et al. (2007). Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula. Mol. Plant Microbe Interact. 20, 159–167. doi: 10.1094/MPMI-20-2-0159
Wang, G. F., Xie, G. L., Zhu, B., Huang, J. S., Liu, B., Praphat, K., et al. (2008). Identification and characterization of the Enterobacter complex causing mulberry (Morus alba) wilt disease in China. Eur. J. Plant Pathol. 126, 465–478.
Wang, J. F., Zhou, Z. H., He, F., Ruan, Z., Jiang, Y., Hua, X. T., et al. (2018). The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One 13:e0192288. doi: 10.1371/journal.pone.0192288
Weller-Stuart, T., Toth, I., Maayer, P. D., and Coutinho, T. (2017). Swimming and twitching motility are essential for attachment and virulence of Pantoea ananatis in onion seedlings. Mol. Plant Pathol. 18, 734–745. doi: 10.1111/mpp.12432
Wu, H. M., Wu, L. K., Wang, J. Y., Zhu, Q., Lin, S., Xu, J. H., et al. (2016). Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front. Microbiol. 7:335. doi: 10.3389/fmicb.2016.00335
Xie, L., Guo, Y. L., Chen, Y. R., Zhang, L. Y., Wang, Z. C., Zhang, T., et al. (2020). A potential drug combination of omeprazole and patchouli alcohol significantly normalizes oxidative stress and inflammatory responses against gastric ulcer in ethanol-induced rat model. Int. Immunophar. 85:106660. doi: 10.1016/j.intimp.2020.106660
Xu, T., Su, Y., Xu, Y., He, Y., Wang, B., Dong, X., et al. (2014). Mutations of flagellar genes fliC12, fliA and flhDC of Edwardsiella tarda attenuated bacterial motility, biofilm formation and virulence to fish. J. Appl. Microbiol. 116, 236–244. doi: 10.1111/jam.12357
Zhang, L. Q., Xu, J. S., Xu, J., Zhang, H., He, L. Y., and Feng, J. (2014). TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb. Pathog. 74, 1–7. doi: 10.1016/j.micpath.2014.06.006
Keywords: Pogostemon cablin (patchouli), bacterial wilt, pathogen isolation, complete genome sequencing, Kosakonia cowanii, virulence-related genes, functional validation
Citation: Zhang Y, Wang B, Li Q, Huang D, Zhang Y, Li G and He H (2022) Isolation and Complete Genome Sequence Analysis of Kosakonia cowanii Pa82, a Novel Pathogen Causing Bacterial Wilt on Patchouli. Front. Microbiol. 12:818228. doi: 10.3389/fmicb.2021.818228
Received: 19 November 2021; Accepted: 23 December 2021;
Published: 14 January 2022.
Edited by:
Bernardo González, Adolfo Ibáñez University, ChileReviewed by:
Krzysztof Krawczyk, Institute of Natural Fibres and Medicinal Plants, PolandCopyright © 2022 Zhang, Wang, Li, Huang, Zhang, Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong He, aGVob25nNjdAaG90bWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.