
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 January 2022
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.813245
This article is part of the Research Topic Probiotics-Modulated Intestinal Immunity against Infectious Diseases in Animals View all 21 articles
Altered gut microbiota are implicated in inflammatory neonatal calf diarrhea caused by E. coli K99. Beneficial probiotics are used to modulate gut microbiota. However, factors that mediate host-microbe interactions remain unclear. We evaluated the effects of a combination of multispecies probiotics (MSP) on growth, intestinal epithelial development, intestinal immune function and microbiota of neonatal calves infected with E. coli K99. Twelve newborn calves were randomly assigned as follows: C (control, without MSP); D (E. coli O78:K99 + gentamycin); and P (E. coli O78:K99 + supplemental MSP). All groups were studied for 21 d. MSP supplementation significantly (i) changed fungal Chao1 and Shannon indices of the intestine compared with group D; (ii) reduced the relative abundance of Bacteroides and Actinobacteria, while increasing Bifidobacteria, Ascomycetes, and Saccharomyces, compared with groups C and D; (iii) improved duodenal and jejunal mucosal SIgA and total Short Chain Fatty Acids (SCFA) concentrations compared with group D; (iv) increased relative ZO-1 and occludin mRNA expression in jejunal mucosa compared with group D; and (v) enhanced intestinal energy metabolism and defense mechanisms of calves by reducing HSP90 expression in E. coli K99, thereby alleviating the inflammatory response and promoting recovery of mucosal function. Our research may provide direct theoretical support for future applications of MSP in ruminant production.
Newborn calf diarrhea (NCD) can cause huge economic losses due to high morbidity and mortality (Walker et al., 1998; Cho and Yoon, 2014; Brunauer et al., 2021). The risk factors associated with diarrhea include the environmental conditions, nutritional levels, and immune status of calves (Klein-Jöbstl et al., 2014; Al Mawly et al., 2015). The most common enteric pathogens include Escherichia coli, Salmonella, Cryptosporidium and Rotavirus (Moon et al., 1978; Gulliksen et al., 2009; da Silva Medeiros et al., 2015). Enterotoxigenic Escherichia coli (ETEC), which constitute the most common cause of neonatal calf diarrhea globally, have been extensively studied over the past 40 years (Bywater and Logan, 1974; González Pasayo et al., 2019). Neonatal calves are most susceptible E. coli K99 ETEC infections during the first 4 days of life (Krogh, 1983; Acres, 1985).
The E. coli K99 antigen binds to the small intestinal mucosa and gradually decreases from the first 12 h of adhesion. This adhesion ability is related to age (Runnels et al., 1980). However, in ETEC, this ability increases after the 3rd week of age (Bulgin et al., 1982; Izzo et al., 2011). Antibiotics have been used to treat E. coli that cause diarrhea (Sunderland et al., 2003), where these antibiotics not only affect the target pathogens, but also beneficial microorganisms in the intestine, resulting in long-term changes in intestinal microbiota being associated with the disease. Application of antibiotics also exerts many other side effects, such as intestinal barrier dysfunction and the emergence of carcinogenic and drug-resistant bacteria, which greatly affect the usefulness of antibiotics (Raheem et al., 2021). Probiotics, which are considered as sustainable alternatives to antibiotics, can be used to prevent and treat diarrhea in humans and animals (Collinson et al., 2020; Dahlgren et al., 2021; Hrala et al., 2021). Many previous studies have demonstrated that supplementing calves with probiotics early in life effectively prevents diarrhea (Malmuthuge and Guan, 2017; Wu et al., 2021a). Lactobacillus acidophilus (Lépine et al., 2018), Bacillus subtilis (Sambanthamoorthy et al., 2014; Shen et al., 2017; Memon et al., 2021) and Saccharomyces cerevisiae (Chiaro et al., 2017; Bitla et al., 2021) has the ability to resist pathogen adhesion and enhance the intestinal barrier function (Sanchez et al., 2010; Younis et al., 2017).
In this study, we evaluated whether newborn calves were infected with E. coli K99, and whether supplementation with MSP enhanced the integrity of the intestinal barrier and local and systemic immune responses of calves, by regulating intestinal microbiota and ameliorating intestinal dysfunction caused by inflammation. In addition, we investigated the recovery of damaged intestinal function and explored the interaction between the diversity of microbiota and the repair of intestinal mucosa.
The MSP (L. acidophilus 3 × 109 CFU/1 g, B. subtilis 3 × 109 CFU/1 g, and S. cerevisiae 1 × 109 CFU/1 g) was prepared by the Biological Feed Laboratory of the College of Animal Science and Technology, Shihezi University, and the MSP preparation method is described in the previous study (Wu et al., 2021b).
Thirty-six male Holstein calves (body weight 40.1 ± 0.6 kg; age 5 ± 2) were used for this study. Three groups, each containing 12 calves that were randomly assigned using a random number generator (Microsoft Corp., Redmond, WA), were formed as follows: (i) the control group (C) fed a basal diet and not challenged with E. coli K99; (ii) the diarrhea group (D) also fed a basal diet and orally challenged with E. coli K99 (30 mL; 1 × 109 CFU/mL) and antibiotic support therapy (intramuscular gentamicin 20 mL/days, injection lasted for 2 days); and (iii) the MSP group (P): fed MSP every day from the next day (7.0 × 109CFU/g; 2 g/calf) after orally challenged with E. coli K99 (30 mL; 1.0 × 109CFU/mL). This is because the best effect was obtained with 2 g MSP per day according to a previous study of ours (Wu et al., 2021a), MSP which is in the form of a powder, was administered via milk. The basal diet (Supplementary Table 1) was free of antibiotics. The experimental treatments lasted 21 d, during which all the animals had free access to fresh water and starter concentrate. The starter concentrate, provided by Xinjiang Urumqi Zhengda Feed Co., Ltd. (Urumqi, China) was fed to the calves from day 4. The study was conducted between April and May 2020 at the Shu Rui Farm (Shihezi, China). The calves were maintained according to the standards for the professional guidance process for feeding and management of the Shu Rui farm. The health of the calves was monitored and recorded after birth and throughout the experimental period.
Blood samples were obtained from six calves before morning feeding on day 21, 10 mL of blood being collected from the jugular vein of each calf with an anticoagulant vacuum blood collection tube, during the course of 30 min. The serum was centrifuged at 3,000 rpm for 15 min, separated into a centrifuge tube, and stored in the frozen state at –20°C until needed for testing.
Samples of intestinal tissue, mucosa, and contents (mid duodenum, mid jejunum, mid ileum, mid cecum, mid colon, and rectum) were obtained from different intestinal regions of the same six calves on day 21 of the experiment. Next, the 18 calves (6 in each group) were humanely sacrificed by injecting 4% sodium pentobarbital solution. Intestinal tissues were stored in 4% paraformaldehyde fixative (Biosharp, China). Samples of the mucosa and intestinal contents were stored in 50 mL cryotubes, and divided into three 2 mL sterile, enzyme-free cryotubes, which were immediately placed in liquid nitrogen for rapid freezing and stored at –80°C until needed for further analyses. All experimental steps were aseptically conducted and all intestinal tissues and digests were obtained within 20 min after euthanasia (Roth et al., 2009).
Total gut microbial genomic DNA was extracted, wherein the primer sequences as well as PCR conditions used for the amplification of bacterial and fungal DNA were in accordance with those described in a previous study of ours (Wu et al., 2021a). According to the standard protocol of Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China), paired-end sequencing was performed on the Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego, United States). The sequences were submitted to GenBank under accession number SRP329437.
Key steps involved in sequencing data analysis are as follows. First, in order to obtain clean readings by eliminating adapter contamination and low quality data, the data was preprocessed to connect overlapping double-ended (COPE) software (V1.2.1) to obtain clean double-ended readings incorporated into labels (Liu et al., 2012). Bacterial tags were classified as operational taxonomic units (OTUs) based on 97% sequence similarity via Mothur (v1.31.2) softwares (Schloss et al., 2009). Bacterial sequences representative of OTUs was classified using a Mothur software script based on the Ribosomal Database Project (RDP) database (Cole et al., 2009). Fungal tags were clustered into OTUs based on 97% sequence similarity, using USEARCH (v7.0.1090) software (Edgar, 2013). The RDP classifier (v.2.2), based on the UNITE database, was used to classify fungal OTU representative sequences (Abarenkov et al., 2010). Mothur (v1.31.2) was used to calculate the Chao 1 index, the Shannon index, and the Simpson index, and R (v3.0.3) software was used to draw a sparse curve. QIIME (v1.80) Principal coordinate analysis (PCoA) software was used to plot beta diversity, using weighted UniFrac distance.
The double-antigen sandwich method was used to determine immunoglobulin M (IgM), immunoglobulin A (IgA), immunoglobulin G (IgG), tumor necrosis factor-α (TNF-α), and interleukin-2 in calf serum (IL-2). Interleukin-1β (IL-lβ), interleukin-4 (IL-4), and secretory immunoglobulin (SIgA) of small intestine contents were determined using a biochemical kit. ELISA kits and biochemical kits were purchased from Shanghai Enzyme Link Biotechnology Co., Ltd. All tests were performed according to the manufacturer’s instructions.
Tissue preparation for mRNA quantification was recently described by Schäff et al. (2018). The optical density measured at 260:280 using a spectrophotometer (NanoPhotometer, Implen GmbH, Munich, Germany) was used to estimate the quantity and quality of total RNA. For cDNA synthesis, 750 ng RNA used 200 U reverse transcriptase MMLV-RT RNase (H-) point mutant (Promega, Madison, WI) and 250 pmol random hexamer primers (Metabion International AG, Planegg-Steinkirchen, Germany) Dilute the cDNA 1:4 with diethyl pyrocarbonate water and store aliquots at –80°C. Specific primers were used to measure the mRNA expression of interleukin-1β (IL-1β), interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), Toll-like receptors (TLRs), nuclear factor kappa-B (NF-κB), zona occludens-1 (ZO-1), occludin, claudin, and the copy number of 16S rRNA genes of total bacteria, L. acidophilus, B. subtilis, S. cerevisiae, and E. coli K99 in the small intestinal mucosa (duodenum, jejunum and ileum). Primers were designed using Primer 3 version 0.4.0 (Untergasser et al., 2012) or via literature surveys as shown (Supplementary Table 2). β-actin was used as a reference gene to normalize data. Data are presented as the ratio between the copy numbers of relevant genes and the abundance of reference genes. Relative expression of the target genes was determined using the 2-△△ Ct method.
A sample (100 mg) of jejunum contents was weighed, transferred to a 5 mL tube (containing 25% phosphoric acid), and vortexed vigorously until completely dissolved (4:1; v:v). As described in Bi et al. (2021), the concentration of acetate, propionate, butyrate, isobutyrate, isovalerate, and valerate was measured by gas chromatography. The concentration of Short Chain Fatty Acids (SCFA) in the jejunum is expressed as μg/g.
Fecal contents in the jejunum samples were enriched with microbial cells via differential centrifugation, according to the method reported by Tanca et al. (2017). The bicinchoninic acid (Beyotime, Shanghai) method was used to quantitatively extract the protein in the jejunum sample, and SDS polyacrylamide gel electrophoresis was used to evaluate the quality of the protein extract in the sample. The protein sample (100 μg) was digested by the FASP method (Wiśniewski et al., 2009). After enzymolysis, similar amounts from each biological sample were removed and mixed. Of this, 100 μg were classified via high pH RP reversed-phase chromatography (chromatograph: Agilent 1100 (including Chemstation, 214 nm dad detector, and vacuum degasser).
The column was Waters XBridge C18 (5 μm, 4.6 × 250 mm, 120 Å). Protein Discoverer 2.1.0182 (Thermo Fisher Scientific, Rockford, IL, United States) was used for protein retrieval and analysis. Biological samples were collected for data-independent acquisition (DIA) and analyzed quantitatively using Skyline software (Department of Genome Sciences, University of Washington, Seattle, WA) (Egertson et al., 2015).
The sequences were submitted to MassIVE and can be downloaded via the following address: fttp://massive.ucsd.edu/MSV000087975/.
Jejunum content samples were analyzed using an online nanospray Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, MA, United States) with an EASY-nLC system (Thermo Fisher Scientific, MA, United States). The peptides were dissolved in solvent A (0.1% formic acid in water) spiked with 1× iRT standard (iRT Kit; Biognosys, Schlieren, Switzerland). Exactly 1 μg of peptide sample was loaded onto an Acclaim PepMap C18 column (75 μm × 25 cm) and separated using 120-min linear gradient. Column flow rate was maintained at 400 nL/min, while column temperature was maintained at 40°C. An electrospray voltage of 2100 V was applied. A full scan was performed at m/z 350–1,200 with a resolution of 120,000 at m/z = 200, and maximum injection time of 50 ms. The MS/MS scan was performed with higher-energy collision-activated dissociation for m/z 200–2,000 with a resolution of 30,000 at m/z = 200, and maximum injection time of 90 ms. Collision energy was 32%, and the stepped collision energy was 5%. DIA was performed with 25 variable isolation windows with a 1 Da overlap and total cycle time of 3 s.
The DDA mode was used to construct a spectral library for protein identification and quantification using DIA. One microgram of peptides from each sample was combined, and the mixture was redissolved in 50 μL of buffer C (20 mM ammonium formate in water, pH = 10.0, adjusted by ammonium hydroxide). Next, the combined peptide solution was subjected to high-pH reversed-phase Liquid Chromatography (LC) fractionation via an Ultimate 3000 system (Thermo Fisher Scientific, MA, United States) with a C18 column (4.6 mm × 250 mm, 5 μm). Column flow rate was maintained at 1 mL/min, while column temperature was maintained at 40°C. The fractions were continuously collected. Each fraction was dried in a vacuum-freeze dryer, redissolved in 50 μL of solvent A (0.1% formic acid in water), and subjected to LC-MS/MS analysis; the injection volume was 5 μL. Dynamic exclusion was enabled within a duration of 30 s. An MS/MS scan was performed with 1.6 Da isolation window widths. The other MS parameters, LC gradient conditions, and LC column were similar to those used in the DIA experiments.
All quantifiable microbial protein sequences were annotated using the Clusters of Orthologous Genes (COG) database (version 2014), as previously described (Zhang et al., 2016). KEGG ortholog (KO) annotation of protein sequences was conducted using the GhostKOALA web application (Kanehisa et al., 2016). Taxonomic assignment of the proteins was performed using MEGAN 6 (Huson et al., 2007). The taxonomy of a protein group was assigned using the lowest common ancestor (LCA) of all proteins within that protein group.
Pathway enrichment analysis was performed using STRING (version 10.5) (Szklarczyk et al., 2017). Protein interaction networks were exported from STRING and visualized using Cytoscape software (version 3.4.0).
The Durbin Watson test was used to examine the randomness of initial weight data and the effectiveness of randomization. The GLIMMIX program SAS 9.4 was used to analyze serum immunoglobulin, SCFA concentration, SIgA and fecal microbial data on the basis of repeated measurement and compound symmetrical variance and covariance structure. The fixed effect of treatment, day, the interaction between treatment and day, and the random effect of calf identity were included in the repeated measurement model. A melting curve analysis was generated to verify the specificity of the reaction after each quantitative real-time PCR analysis. Select housekeeping gene β-Actin was used as a reference gene to normalize the mRNA expression of the target gene. The gene expression data of replicated samples were calculated using the CT method (Pfaffl, 2001). The relative expression of target genes in group C was set to 1.0. Each sample was measured in triplicate. The data are expressed as the least squares mean and the standard error of the mean. Tukey’s multi range test was used to evaluate the differences between the treatment groups.
DDA data were analyzed using Proteome Discoverer 2.1.0182 (Thermo Fisher Scientific, Rockford, IL, United States). MS1 tolerance was set to 10 ppm, and MS/MS tolerance was 0.02 Da. All DDA MS/MS spectra were searched against the database of bacterial proteomes downloaded from the UniProt database (23,730,617 protein entries,1 access date July 2019). The Bacteria, Eukaryota, and Archaea databases in the Uniprot database were “searched and compared.” The FDR cut-off for PSM, peptide, and protein group levels was 1%. Raw DIA data were then processed and analyzed via Skyline (Department of Genome Sciences, University of Washington, Seattle, WA, United States) using the default settings. The top three filtered peptides that passed the 1% Q-value cut-off were used to calculate the major group quantities.
Annotation of identified proteins was performed using KOBAS2 in which several databases (i.e., GO, KEGG, and eggNOG) were integrated. Differentially expressed proteins were analyzed using mapDIA, a software package used to preprocess and statistically analyze quantitative proteomics data. The abundance of each phylum was calculated by summing the intensities of all proteins corresponding to that phylum. Data visualization was conducted using R packages, including heatmaps, circles, and ggplot2.
Our data indicated that bacterial amplicon sequencing of the intestinal content microbiota generated 6,978,315 high-quality sequences that were assigned to a total of 33952 OTUs based on a 97% nucleotide sequence similarity, with an average of 32,112 sequences per sample (21,230–35,090 sequences). PCoA (Supplementary Figure 2) and analysis of similarities (ANOSIM) analyses (Supplementary Table 3) revealed that supplementing E. coli infected neonatal calves with MSP induced significant differences in the microbial structure of different gut region digesta-attached microbiota. The diversity of microbiota in the D and P groups was similar to that of control (C) on day 21, whereas the duodenal, ileal and jejunal microbiota of the D and P groups demonstrated a marked shift along principal component 1 compared with that of the C group.
Bacterial diversity on day 21, which was estimated using Chao1, Shannon, and Simpson indices, was lowest in the jejunum, ileum, cecum, and colon of the D group, as compared to those of the H and P groups. In addition, the Chao1 and Shannon indices for the C group and P group were all significantly higher than those for the D group. There were no significant differences between the diversities in the duodenums of the C and D groups (Figure 1 and; Supplementary Table 4).
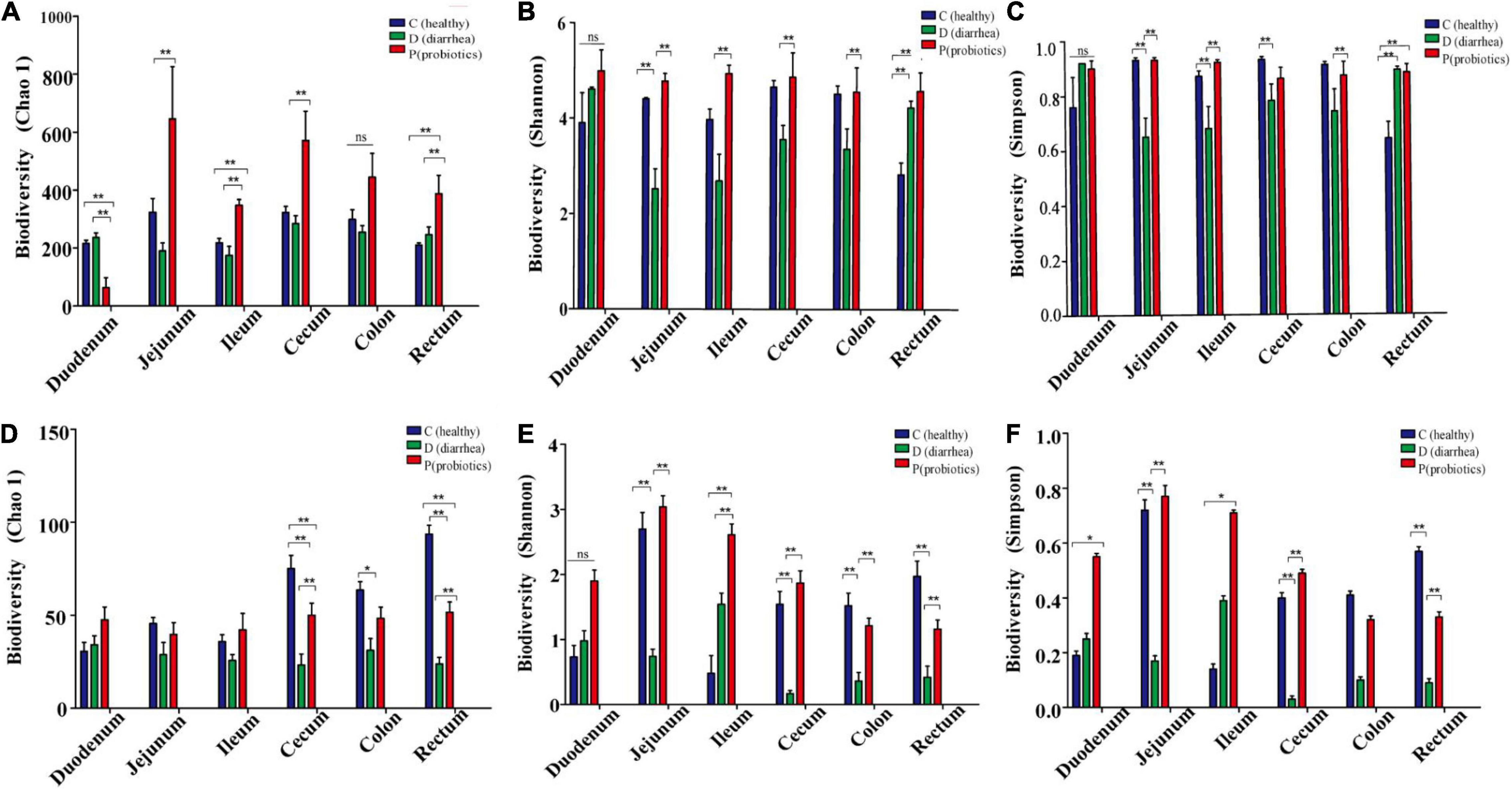
Figure 1. The abundance and diversity index of microorganisms in different intestinal contents of Holstein calves. (A–F) Represent the abundance of Chao1, Shannon and Simpson and the diversity of bacterial and fungal microflora diversity indexes. *P < 0.05, **P < 0.01.
The fungal diversity index, Chao 1, for the cecum, colon, and rectum of the D group was significantly lower than that for the C and P groups (P < 0.01; P = 0.02; and P < 0.01, respectively). The Shannon diversity index for the jejunum, ileum, cecum, and rectum of the D group was significantly lower than that of the C and P groups (P < 0.01; P = 0.01; P < 0.01; and P < 0.01, respectively). There were no significant differences between the C and P groups. The Simpson index of fungal diversity for the jejunum, cecum, and rectum of group D was significantly lower than that of the C and P groups (P < 0.01; P < 0.01; and P < 0.01, respectively) (Figure 1 and Supplementary Table 4). In addition, the results indicated that treatment and gut segment had a significant effect on the α-diversity of bacteria and fungi.
Firmicutes, Actinobacteria, and Bacteroidetes were the dominant bacterial phyla in the duodenum, jejunum, ileum, cecum, colon, and rectum, followed by Proteobacteria and Fusobacteria. The relative abundance of Firmicutes in the duodenum (P = 0.01) and jejunum (P = 0.04) of neonatal calves in the D group was higher than those in the C and P groups. The relative abundance of Actinobacteria in the duodenum (P = 0.01) and jejunum (P = 0.03) of calves in the D group was lower than that in the C and P groups, whereas in the ileum (P = 0.01) it was higher than that in the C and P groups. The relative abundance of Bacteroidetes in the jejunum (P = 0.03) of calves in the D group was lower than that in the C and P groups, while no significant differences were observed between other intestinal segments (Figure 2 and Supplementary Table 5).
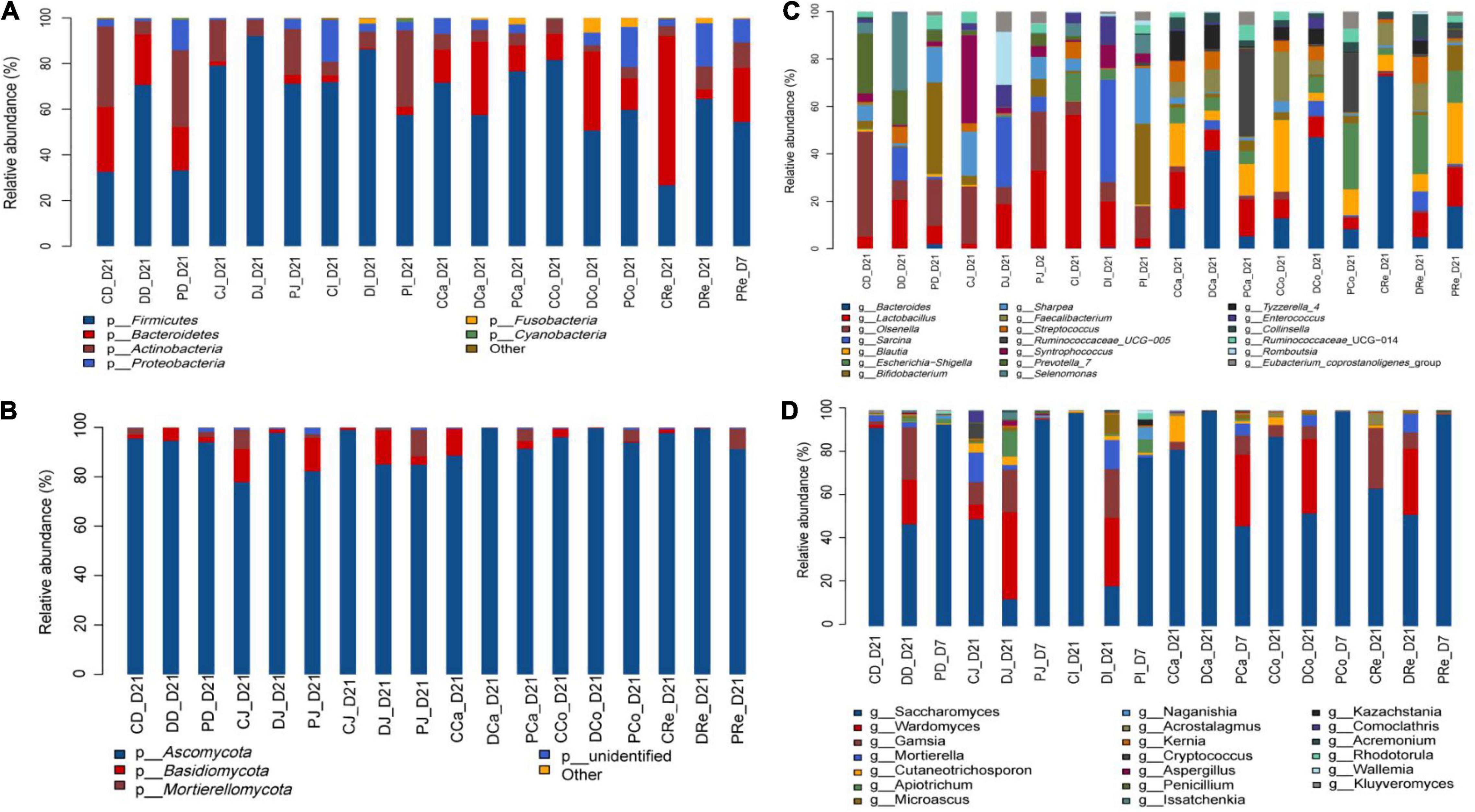
Figure 2. Composition of the different gut region digesta-associated microbial community. (A,C) Show the composition of the microbial community in the three groups (C, D, and P) with respect to bacterial phyla and genera. (B,D) Show the composition of the microbial community in the three groups (C, D, and P) with respect to fungal phyla and genera (CD, DD, and PD represent groups C, D, P dudenum; CJ, DJ, and PJ, represent groups C, D, P jejunum; CI, DI, and PI represent groups C, D, P ileum; CCa, DCa, and PCa, represent groups C, D, P cecum; CCo, DCo, and PCo, represent groups C, D, P colon; CRe, Dre and PRe, represent groups C, D, P rectum).
The top 20 genera at the genus level, were detected and further analyzed: Olsenella, Prevotella_7, Selenomonas, Bifidobacterium, Lactobacillus, Blautia, Faecalibacterium, Syntrophococcus, and Ruminococcaceae_UCG-005 were the predominant bacterial genera in the duodenum and jejunum. The relative abundances of Sarcina (P < 0.01), Streptococcus (P < 0.01), Selenomonas (P < 0.01), and Lactobacillus (P < 0.01) in the duodenums of the D group were significantly higher than those of the C and P groups.
However, the relative abundance of Bifidobacterium in the duodenum (P = 0.02), jejunum (P = 0.02), and ileum (P = 0.01) of the P group was significantly higher than that of the C and D groups. The relative abundance of Sharpea in the jejunum (P = 0.04) of the D group was significantly lower than that of the C and P groups (P = 0.02), while the relative abundance of Prevotella_7 in the jejunum of the P group was significantly higher than that of the C and D groups. No significant differences were observed between other gut regions. In addition, no significant difference was observed between the hindguts of the three groups (Figure 2 and Supplementary Table 6).
The microbial composition of the calf gut indicated that Ascomycota was the dominant phylum followed by Basidiomycota and Mucoromycota (Figure 2 and Supplementary Table 6). A trend toward greater relative abundance of Ascomycota was observed in MSP supplemented neonatal calves compared to that in the C and D groups (P < 0.10). However, the relative abundance of Mucoromycota in the colon of group D neonatal calves was higher than that in the group C and P neonatal calves (P = 0.03). In addition, the relative abundance of Basidiomycota in the C group was significantly higher than that in the other two groups (P = 0.01).
Calf gut microbial composition indicated that Saccharomyces, Aspergillus, Wardomyces, and Acaulium were the predominant genera, followed by Cladosporium, Filobasidium, Geotrichum, Mortierella, Kazachstania, Kernia, Cutaneotrichosporon, and Scopulariopsis (Figure 2 and Supplementary Table 6). The relative abundances of Acaulium in the duodenum, jejunum, ileum, and cecum of the D group tended to be higher than those of the C and P groups (P = 0.01; P < 0.01; P < 0.01; and P = 0.01, respectively). The relative abundance of Saccharomyces in the duodenum, jejunum, and ileum of the P group was significantly higher than that of the C and D groups (P < 0.01; P < 0.01; and P = 0.03, respectively). The relative abundance of Wardomyces in the jejunum, ileum, cecum, colon, and rectum of group D was significantly higher than that of the C and P groups (P < 0.01; P = 0.02; P = 0.03; P = 0.02; and P = 0.02, respectively). The relative abundances of Kazachstania and Acremonium in group P were significantly higher than those in the C and D groups (P < 0.01). The relative abundance of Cladosporium and Pichia in the jejunum of group D was significantly higher than that of the C and P groups (P < 0.01). In addition, the relative abundance of fungi at the phylum and genus levels was significantly different in the different treatment groups of neonatal calves infected with E. coli K99.
MSP supplementation caused significant differences in the levels of IgM and TNF-α in the sera of neonatal calves infected with E. coli K99 (P < 0.05), wherein the C group differed significantly from the P group (P < 0.01), while the calf sera of the C, D, and P groups showed significant differences. The concentrations of IgG, IgA, IL-1, IL-2, and IL-4 were not significantly affected (P > 0.05) (Table 1).
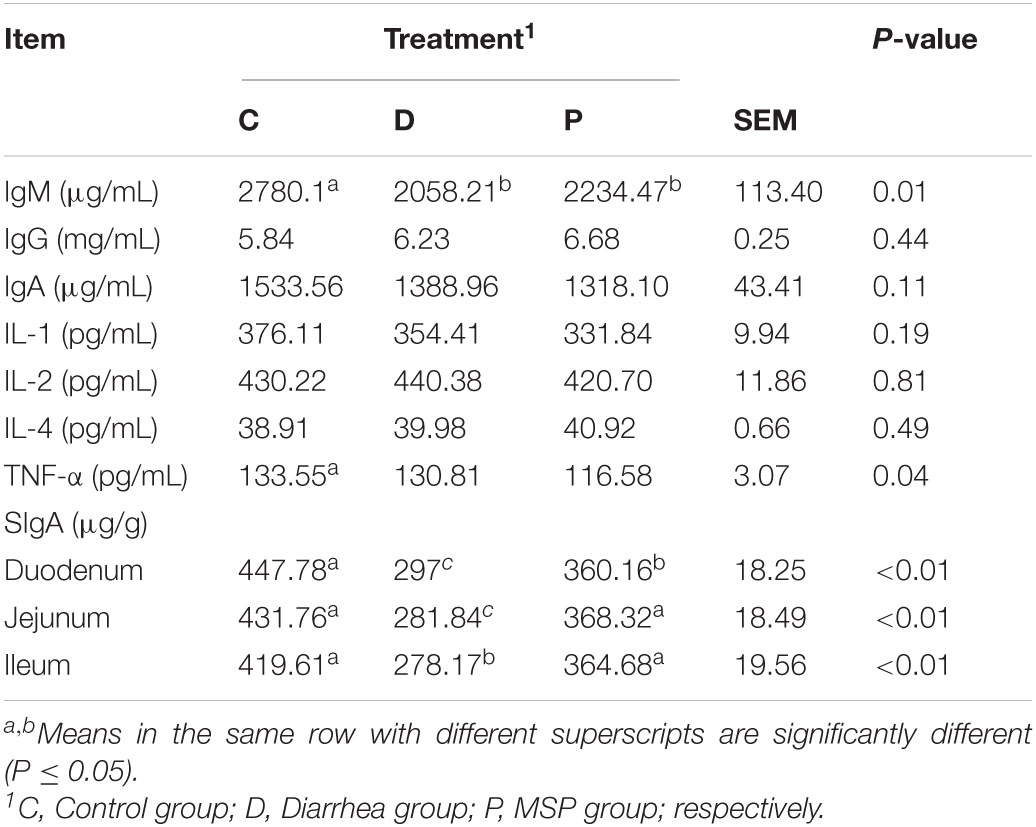
Table 1. Effect of MSP supplementation on the serum immune function and the content of SIgA in neonatal calves intestinal mucosa of neonatal calves infected by E. coli K99.
The effect of MSP supplementation on SIgA in the duodenum, jejunum, and ileum mucosa of neonatal calves infected with E. coli K99 was significantly different from that of the D and P groups (P < 0.01). In addition, SIgA concentrations in the duodenum, jejunum, and ileum mucosa of calves in group P were significantly higher than those in group D (P < 0.01). The SIgA concentrations in the duodenum and jejunum mucosa of calves in the C group were significantly higher than those in the D and P groups (P < 0.01) (Table 1).
The relative expression of IL-1β, NF-κB, and TNF-α mRNA in the duodenal and jejunal mucosa of the D group was significantly higher than those of the C and P groups (P < 0.01). The relative expression of IL-2 mRNA in the jejunum of calves in the P group was significantly higher than that in the D group (P < 0.01). The relative expression of ZO-1 mRNA in the jejunum mucosa of calves in group P was significantly higher than that in groups C and D (P < 0.01). The relative expression of claudin mRNA in the mucosa of calves in the duodenum, jejunum, and ileum of the P group was significantly higher. The expression of occludin mRNA was significantly higher in the P group than in the C and D groups (P < 0.01), while the relative expression of occludin mRNA in the jejunum and ileum mucosa of calves in group P was significantly lower than that in groups C and D (P < 0.01) (Figure 3).
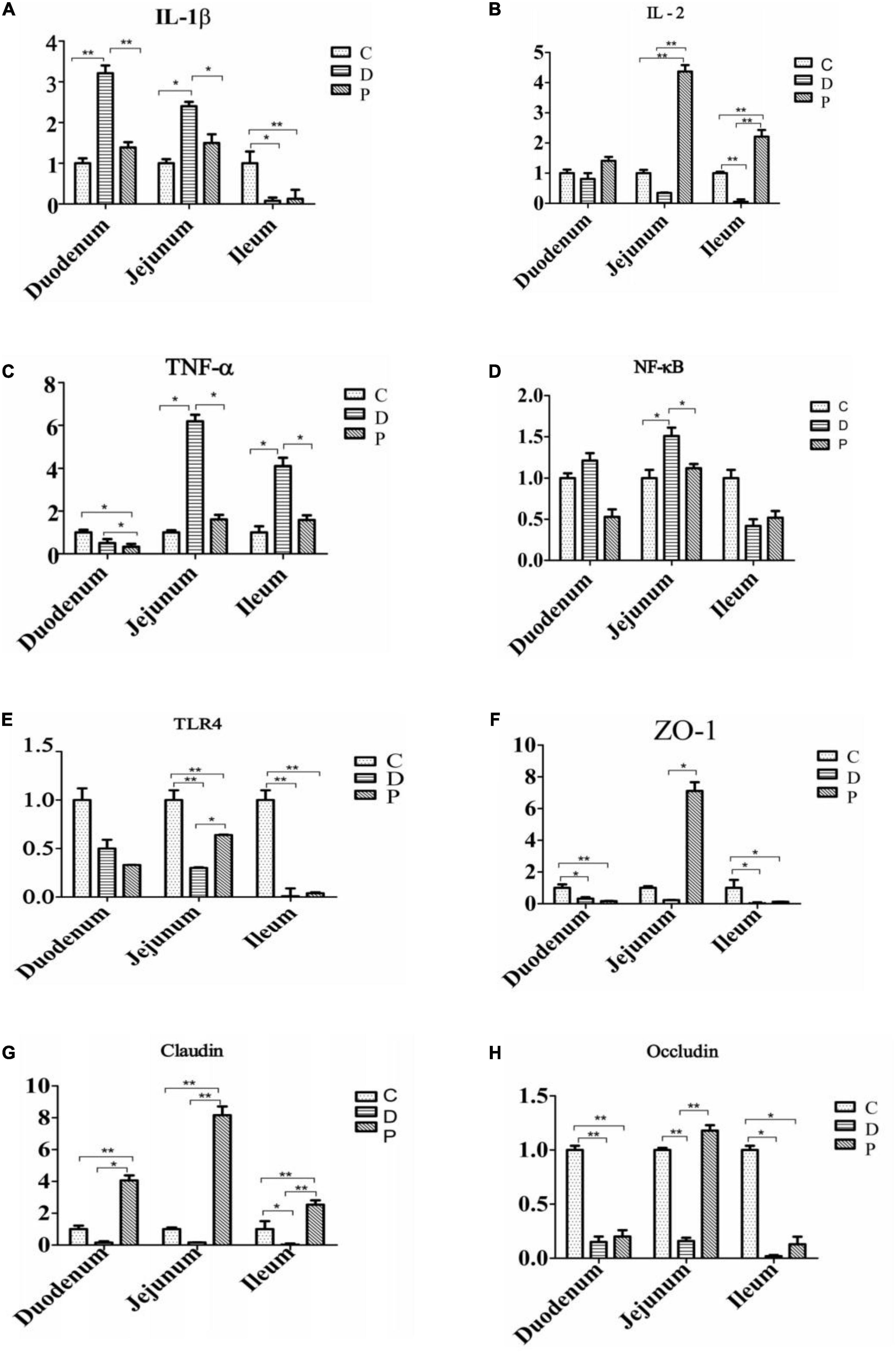
Figure 3. Effect of supplementation with MSP on the relative expression of small intestinal mucosal immune factors and tight junction protein mRNA in neonatal calves infected by E. coli K99 (C, Control group; D, Diarrhea group; P, MSP group; respectively). (A) Interleukin-1β, IL-1β. (B) Interleukin-2, IL-2. (C) Tumor necrosis factor-α, TNF-α. (D) Nuclear factor kappa-B, NF-κB. (E) Toll-like receptor 4,TLR4. (F) Zonula occludens 1, ZO-1. (G) Claudin. (H) Occludin. *P < 0.05, **P < 0.01.
To further analyze microbial activity in jejunum contents, we measured the concentrations of short-chain fatty acids (SCFAs), using them as a proxy reflecting microbial metabolic activity in our gut samples. The concentrations of acetate, propionate, butyrate, and isovaleric acid were significantly higher than those in the C and P groups (Table 2). The concentration of total SCFA in the P group was increased compared to that in the D group, while being significantly higher in the C group than that in the D group (Table 2). In addition, there were significant differences between the concentrations of acetic acid, propionic acid, butyric, isobutyric, isovaleric, and isocaproic acid contents among the three groups.
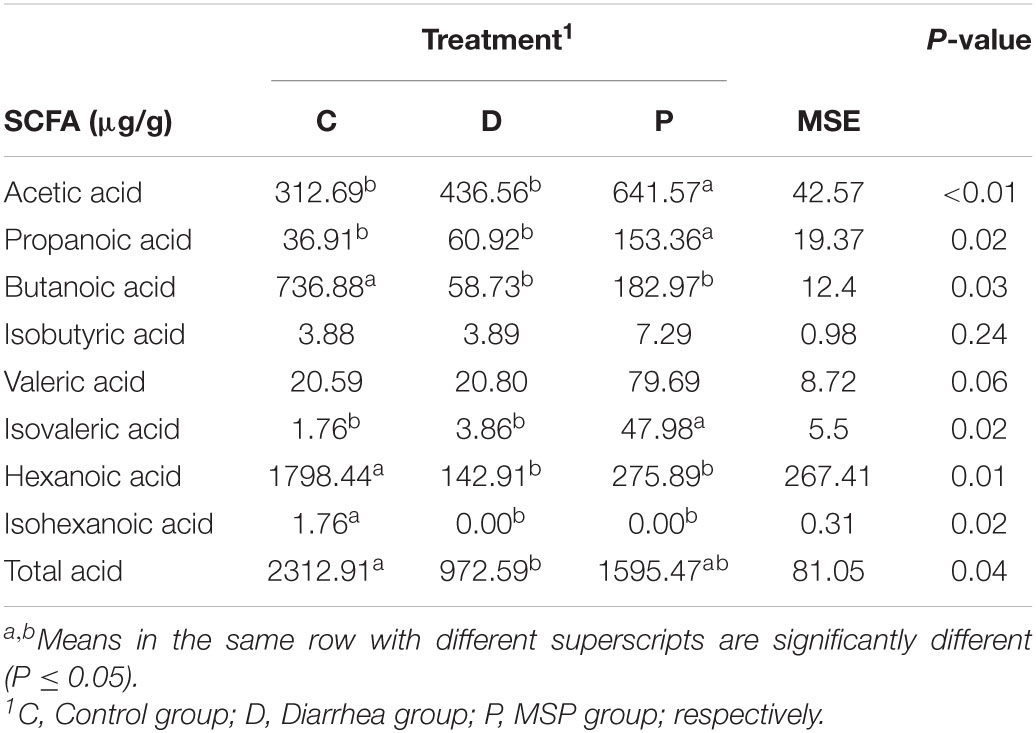
Table 2. Short chain fatty acid (SCFA) concentration and molar proportion in the jejunum contents of neonatal calves.
An advantage associated with metaproteomics is that the microbial population as well as host proteins can be measured in a single analysis. Therefore, we further analyzed microbial function in the calf jejunum, via a metaproteomics analysis aimed at detecting microbial and host proteins in our samples.
We quantified 22,080 unique peptides and 4,414 proteomes in nine samples, with an average recognition rate of 39 ± 8% (mean ± standard deviation). On average, 2,453 ± 89 unique peptides and 735 ± 33 proteomes were identified for each sample. Among the 4,414 proteomes, 3,679 proteins (83%) were from host calves, and only 735 (17%) were from calf jejunum-content microorganisms. However, host proteins accounted for 5.54 times the intensity of the total proteins. We used macroproteomics to characterize the microbiomes of 9 jejunum samples from groups C, D, and P (three samples in each group). The results indicated that 226 showed expressions specific for each group.
Data obtained via further analyses of the gut microbiome function in the jejunum indicated that probiotics caused shifts in intestinal microbiome function, as evidenced by the scatterplot generated from the principal component analysis (PCA) and the Cluster Dendrogram (Figure 4). PCA was performed using calf-derived proteins or non-calf proteins (i.e., microbiota-derived) quantified in ≥ 50% of the samples. The PCA score plot for calf proteins showed an obvious separation among the three groups (Figure 4A). Furthermore, PCA analysis of microbiota-derived proteins also showed obvious separation among the three groups (Figure 4C).
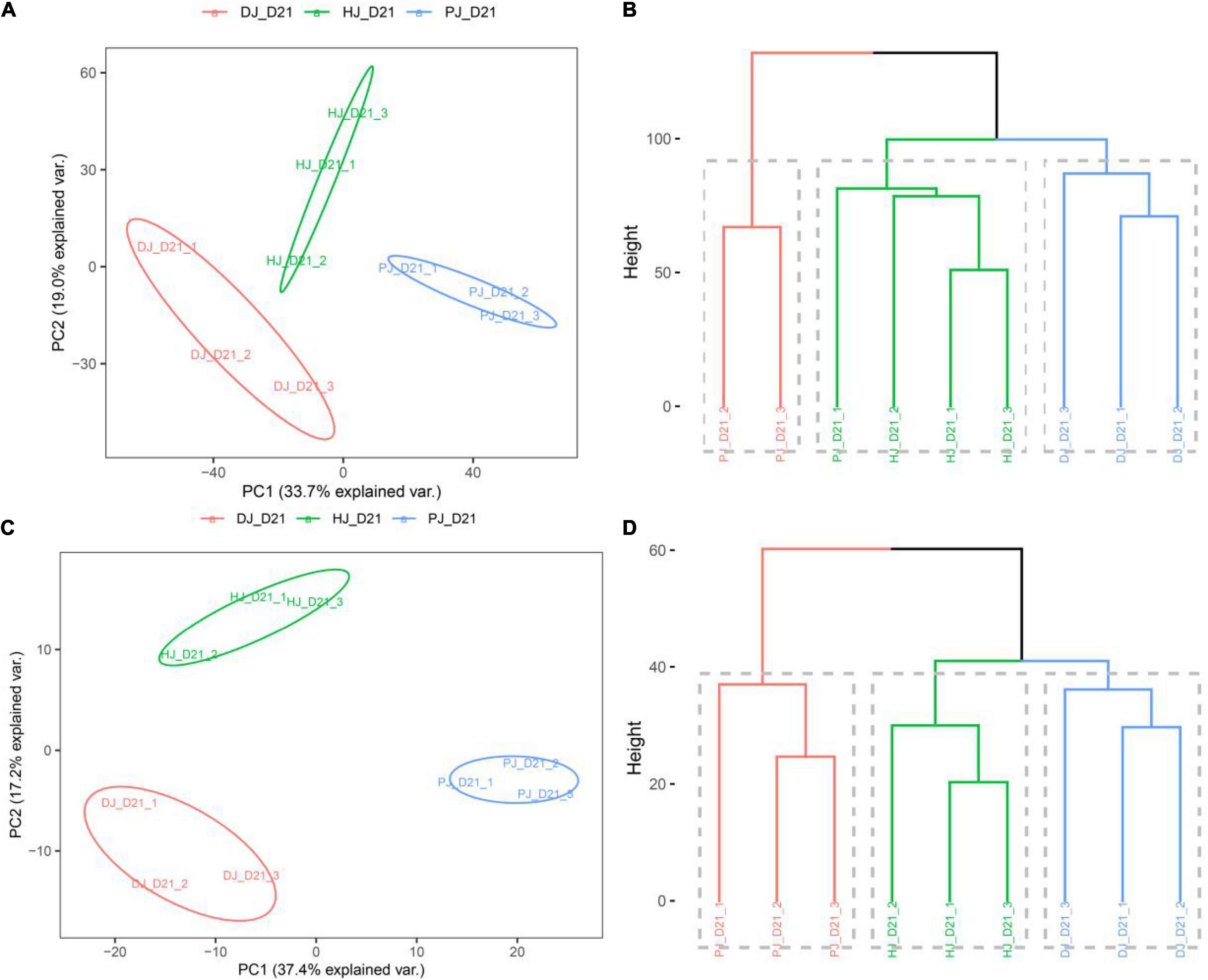
Figure 4. Host and microbiota proteome landscape alterations in neonatal calves infected by E. coli K99. (A,C) PCA score plot of calf-derived and microbiome-derived proteins quantified in ≥ 50% of the samples; (B,D) group mean clustering based on Mahalanobis distances calculated with MANOVA test using the first 10 PCs of PCA on calf-derives proteins, respectively.
Multivariate analysis of variance (MANOVA) of calf-derived proteins yielded greater differences, revealing significant differences (P < 0.01) between groups C and D (Figure 4B). By contrast, MANOVA of microbiota-derived proteins revealed significant differences between C, D, and P groups (Figure 4D). To identify treatment-related alterations in host and microbiota, we combined samples from C, D, and P in jejunal function recovery for further analysis.
To assess microbial function, we annotated all quantified microbial proteins using the COG database and obtained 330 COGs from 24 COG categories. A total of 154 COGs were identified in all three groups (Figures 5, 6).

Figure 5. Functional compositions of microbiome in neonatal calves infected by E. coli K99. Heatmap of differentially abundant COGs in neonatal calves. Representative COG categories are shown, and the colors indicate the average label free quantitation (LFQ) intensity for each subgroup of samples. Each row corresponds to a COG with the COG ID and the name indicated.
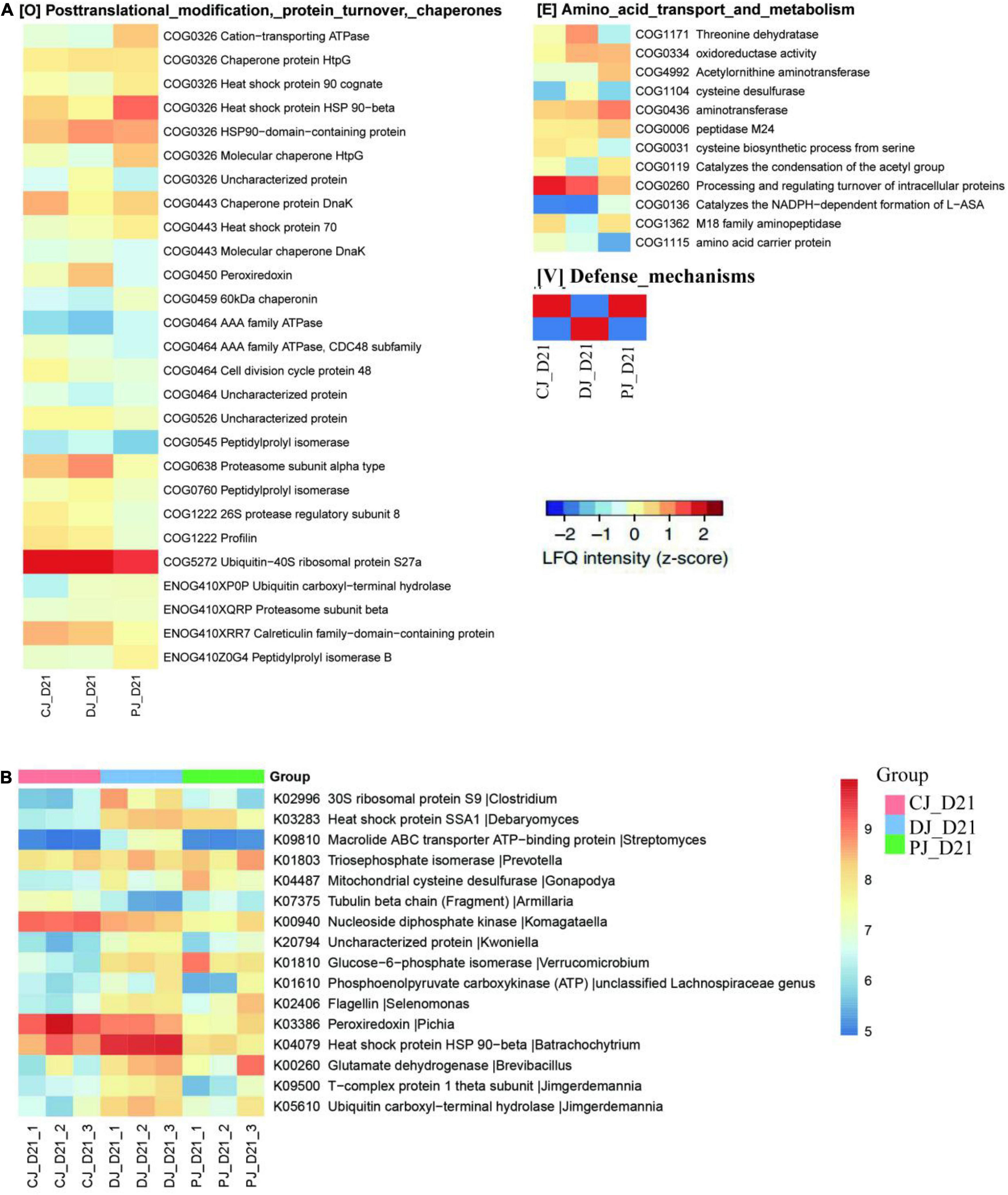
Figure 6. Functional compositions of the microbiome in neonatal calves infected by E. coli K99. (A) Heatmap of differentially abundant COGs in neonatal calves. Representative COG categories are shown, and the colors indicate the average LFQ intensity for each subgroup of samples. Each row corresponds to a COG with the COG ID and name indicated. (B) Heatmap shows the average LFQ intensity for each group of samples. Each row represents one protein group, and the corresponding KO ID, KO name and taxonomic assignment are shown in the right panel.
At the COG category level, we found that category G (carbohydrate transport and metabolism), category O (posttranslational modification, protein turnover, chaperones) category V (defense mechanisms), and category S (function unknown) were significantly increased in the MSP group compared to those of the C and D groups (Figure 7). Defense mechanisms in the P group, in particular, were significantly increased compared to those in the C and D groups. Two COGs belonging to defense mechanisms were identified as differentially abundant in the P group microbiome compared to that in the C and D groups (Figure 7). KO annotation identified 516 KOs corresponding to 735 proteins. The Wilcoxon rank-sum test identified 116 proteins that were significantly different among the three groups, among which six were upregulated in the D group and downregulated in the C and P groups, where ATP-binding cassette (ABC) transporters (KO:K09810), ubiquitin carboxyl-terminal hydrolases (UCHs)(KO:K05610), Glucose-6-phosphate isomerase (KO:K01810), 30S ribosomal protein S9 (A0A099S8L6), and heat shock protein 90 OS (KO:K04079) were significantly upregulated in group D compared with groups C and P (Figure 6B). Interestingly, dietary supplementation with MSP only decreased the expression of both HSPs, where the difference was significant in the case of HSP90.
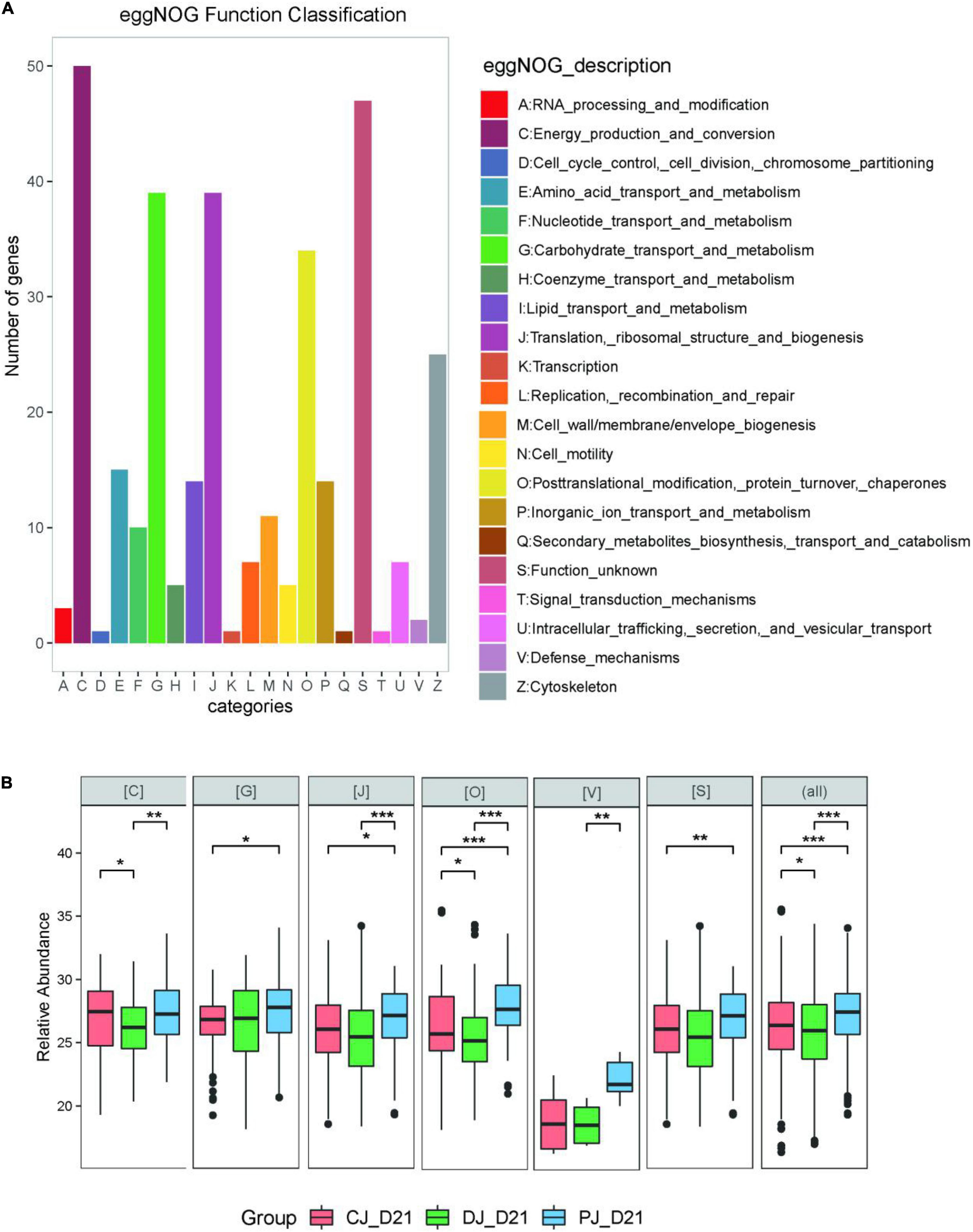
Figure 7. (A) eggNOG classifications of the neonatal calves gut Metaproteomics. (B) LFQ intensity of COG category C (Energy production and conversion), category G (Carbohydrate transport and metabolism), category J (Translation, ribosomal structure and biogenesis), category O (Posttranslational modification, protein turnover, chaperones), and category V (Defense mechanisms) in microbiome of neonatal calves infected by E. coli K99. For the box plot, the bottom and top of the box represent the first and third quartiles, respectively. The middle line represents the sample median. Whiskers are drawn from the ends of the IQR to the furthest observations within 1.5 times the IQR range. Outliers > 1.5 times the IQR are indicated with circle. *p < 0.05, **p < 0.01, ***p < 0.001.
We identified 666 microbial species (666 ± 27 species per sample) with a minimum of one distinctive peptide from three different kingdoms (bacteria, fungi, and archaea). Linear discriminant analysis effect size (LEfSe) identified seven phyla and 71 species as differentially abundant among the three groups. All seven differentially abundant phyla, namely Proteobacteria, Firmicutes, Chytridiomycota, Ascomycota, and Basidiomycota, were decreased in group P, compared to those in D, while Blastocladiomycota, Zoopagomycota, and Crenarchaeota, were increased compared to the D and C groups. Of the four most significantly changed strains in the D group, Prevotella and Streptomyces, were decreased and Pichia and Selenomonas were increased, respectively, compared to C and P groups. This finding agreed with those of a 16S rRNA and ITS sequencing study using a similar type of calves. However, in contrast to the 16s rRNA and internal transcribed spacer (ITS) sequencing study using a similar type of calves, Batrachochytrium in the D group was higher than that in the C and P groups. Previous studies have indicated that genera identified via metagenomics may differ from those identified via the 16S rRNA and ITS sequencing (Walker and Lue, 2015; Hubert et al., 2017; Hu et al., 2018).
To identify calf proteins that were differentially abundant in group D compared to groups C or P, or in groups D or P compared to C, a Venn diagram was conducted for each pair of groups. We identified 55, 57 and 52 differentially abundant calf proteins for C vs. D, P vs. D, and P vs. C, respectively (Supplementary Figure 2 and Supplementary Table 7). Among the differentially abundant calf proteins, 11 were upregulated and 12 were downregulated in the other groups compared with group D. The proteins, Q3ZBF3, P62248, Q9XSC9, and Q3ZC02, in group D were significantly downregulated compared to those in C, and the proteins, Q3ZBF3, Q3ZC02, and Q9XSC9, in group P were significantly upregulated compared to those in group D (Supplementary Table 7).
Gene ontology enrichment analysis of differentially abundant calf proteins in P and D groups demonstrated that the most highly enriched biological functions involved metabolic and cellular processes (Supplementary Figure 4 and Supplementary Table 8).
To gain insights into host-microbiome interactions, we performed co-occurrence analysis for all identified differentially abundant calf proteins and microbial functions and found that both calf proteins and microbial functions were substantially associated with each other (Supplementary Figure 4). In the protein interaction networks (Supplementary Figure 4 and Supplementary Table 9), the microbial heat stress protein, HSPCOG 0326, which displayed the highest association (n = 5) with calf proteins, acted as an important microbial hub connecting the host and microbiome nodes, thereby suggesting a “hub” role for these proteins. As mentioned above, 54 of the 154 significantly increased microbial COGs belonged to categories G, O, and V, which were related to energy production and conversion and DNA damage/mismatch responses, indicating that in calves, microbes are utilizing a considerable amount of energy production and conversion as well as defensive mechanisms against stressors derived by the host.
HSP90 is associated with five host proteins, and generates an inflammatory response via the necroptosis pathway, thereby facilitating the upregulation of IL-1β expression (KO4079). The microbial host-protein interaction map indicated that HSP90 was associated with the host protein, Q3MHN2, which participates in the immune system process and responds to stimuli affecting the host. Q3MHN2 contains a membrane attack complex (MAC)/perforin domain, and the MAC of the complement system is a well-characterized innate immune effector (Merselis et al., 2021). These findings proved that the relationship between neonatal calves infected by E. coli K99 and Batrachochytrium salamandrivorans may be at the strain level instead of at the species level, and that its sub-species level dysbiosis may be related to differences between immune functions in response to oxidative stress.
The diversity of intestinal microbiota showed a gradually increasing trend during the growth of newborn calves. Shannon diversity differs between animals of different ages (Alipour et al., 2018; Dias et al., 2018). The establishment of intestinal microflora in calves within 7 weeks of birth is closely related to calf health and growth (Oikonomou et al., 2013). Beta diversity decreased with age, while alpha diversity increasing (Dill-McFarland et al., 2019; Li et al., 2020). In addition, the Shannon and Chao1 indices showed significant changes in diversity from birth until the 8th week. Previous studies on dairy cows have shown that the Chao1 index gradually increases with body weight over time from the 1st to the 7th week before weaning. Older calves have a higher Chao1 index (Oikonomou et al., 2013). Diarrhea or an infection may cause a decrease in microbial diversity (Kim et al., 2021; Xin et al., 2021). While most previous studies focused on investigating and interpreting calf manure samples, only a relatively few studies, such as ours, have focused on investigating the effects of feeding calves with compound probiotics on the intestinal microflora of these animals. The results of our current study on the diversity of microbiota in calf fecal samples were consistent with those of previous studies (Li et al., 2018, Dill-McFarland et al., 2019). Our results showed that the Chao1 and Shannon indices of the fecal microbiota gradually changed over time. However, the diversity of microbiota in the small intestine of D calves was higher than that of the fungi, while Chao1 and Shannon indices were lower than those of the C and P groups, in addition to which α-diversity was significantly reduced. Thus, intestinal microbiota diversity of calves infected with E. coli can be significantly improved by adding compound probiotics.
However, calves have a large amount of mucosal-associated E. coli in their intestines in the first week after birth, which causes them to be more likely infected with pathogens. Therefore, reducing pathogen colonization and intestinal infections has become an effective way to improve intestinal health (Song et al., 2018). This study shows that Streptococcus and Enterococcus can use the available oxygen in the intestine and create an anaerobic environment, which is beneficial to the colonization of strict anaerobic intestinal residents such as Bifidobacterium and Bacteroides (Conroy et al., 2009). However, recent studies have indicated that gut microbiota possibly begin colonizing during the birth process (Malmuthuge et al., 2015; Song et al., 2018). The transmission process of maternal microbiota to newborn calves may originate at the amniotic fluid stage, in a manner similar to that reported for NB human infant gut microbiota (DiGiulio, 2012) and meconium (Moles et al., 2013). Significant individual variations observed among the NB calves may be due to variability in the transmission process from the mother cow to the birth environment (uterus, vaginal canal, and fetal membranes) (Klein-Jöbstl et al., 2019). The study found that the relative abundance of Lactobacillu and Shigella was higher in newborn calves than in D21 calves (Song et al., 2018).
E. coli infection can destroy the integrity of the intestinal tract and severely damage its intestinal function, thereby disrupting the balance of intestinal microbiota and leading to a decline in immune function (Dubreuil, 2017; Braz et al., 2020). Antibiotics play a vital role in many bacterial infections (Liu et al., 2017). However, antibiotics are known to cause intestinal microbiota imbalance, affect host metabolism, and lead to chronic inflammation (Scott et al., 2018). Relevant studies have shown that probiotics have a positive effect in reducing the susceptibility of calves to intestinal infections before weaning (Markowiak and Śliżewska, 2018; Mazloom et al., 2019; Plaza-Diaz et al., 2019). The anti-diarrheal function of MSP may be associated with its role in immunity (Inatomi et al., 2017; Villot et al., 2020). Concentrations of circulating immunoglobulins, especially those of IgG, IgM, and IgA, are important indicators of immune function (Crassini et al., 2018). Some studies have indicated that supplementation with probiotics promotes the immune responsiveness of calves by improving serum IgG concentration (Song et al., 2019). Other studies have shown that supplementing with probiotics can reduce the rate of calf diarrhea and increase the serum IgM concentration on the 14th day, while increasing the serum IgA, IgG and IgM concentrations on the 28 day, respectively (Wang et al., 2019).
Intestinal SIgA has become the main regulator of intestinal microbiota through agglutination and enhancement of the opportunity to eliminate pathogens (Moor et al., 2017; Guo et al., 2020, 2021). Furthermore, Karamzadeh-Dehaghani et al. (2021) demonstrated that supplementation with probiotics improved the growth performance and immune response of calves. Consistent with these findings, we found that the gut SIgA concentration in the jejunum of calves in the MSP supplemented P group was increased by 86.48 μg/g compared to that in the D group. Studies have indicated that the regulation of potential intestinal pathogens and fungal colonization may enhance immunity (Nakawesi et al., 2020; Guo et al., 2021), whereas antibiotic exposure disturbs early GM, leading to intestinal dysbiosis and reduced SIgA (Guo et al., 2021), and that supplementation with additional probiotics may help enhance normal intestinal functioning (Tanaka et al., 2017; Plaza-Diaz et al., 2019; Roggero et al., 2020). The results of our study indicated that MSP supplementation improved the diversity and structure of the small intestinal microbiota in neonatal calves infected with E. coli K99, and simultaneously increased the secretion of SIgA compared to that in the antibiotics group.
The gut tract, which is one of the most microbiologically active ecosystems, plays a crucial role in the functioning of the mucosal immune system (MIS). Probiotics are known to exert positive effects on intestinal diseases. Among other effects, probiotics exert barrier-modulating effects and alter the inflammatory response toward pathogens in in vitro and in vivo intestinal infection models (Lodemann et al., 2017; Wang et al., 2019; Maldonado Galdeano et al., 2019), prevent or ameliorate damage to epithelial integrity caused by pathogenic challenge (Sherman et al., 2005; Johnson-Henry et al., 2008), and modulate the ease of cytokine secretion (Bahrami et al., 2011; Paszti-Gere et al., 2012). There is a close connection between the gut microbial community and the host immune system (Duparc et al., 2017; Cani and Jordan, 2018; Cani, 2019). Several reports have validated the fact that manipulating gut microbiota using probiotics, such as Bacillus subtilis (Jia et al., 2021) and Lactobacillus acidophilus (Lépine et al., 2018) may affect host gut barrier function and gut microbiota composition (Régnier et al., 2021). A growing number of studies have shown that certain probiotics (such as Lactobacillus acidophilus) can regulate the obstruction of intestinal epithelial fluid absorption and secretion caused by diarrhea (Borthakur et al., 2008; Singh et al., 2014). The results of the present study showed that MSP supplementation significantly upregulated relative mRNA expression of ZO-1, claudin-1, and occludin in the jejunum of E. coli-infected neonatal calves. Among them, a downward trend was observed in group D, indicating that in group P, claudin mRNA levels were increased to reduce intestinal E. coli adhesion, and to protect the mechanical barrier and paracellular permeability of the small intestine.
SCFA has important physiological effects on the host (Perego et al., 2018; Chen and Vitetta, 2019). Acetic acid is a short-chain fatty acid produced by intestinal bacteria. It inhibits the growth of pathogens and regulates the metabolism and gene expression of B cells, and promotes the secretion of SIgA, thereby maintaining and repairing the intestinal morphology of animals (Garcia et al., 2007; Kim et al., 2016; Wu et al., 2017; Bourassa et al., 2018). The results of this study indicate that MSP maintains intestinal health by increasing the acetic acid content in the jejunum of newborn calves infected with E. coli K99. Previous studies have shown that most Bacteroides, Bifidobacterium, Prevotella, and Rumenococci produce acetic acid through “pyruvate via the acetyl-CoA pathway” (Louis et al., 2014; Koh et al., 2016). In our study, the relative abundances of Bacteroides, Bifidobacterium, Prevotella, and Rumenococcus in the calf jejunum content of the MSP group was significantly higher than those of the antibiotic group. The results of this study indicated that MSP promotes the recovery of the intestinal function of calves following E. coli infection. Further, the results of this study are consistent with those of previous studies (Louis et al., 2014; Koh et al., 2016; Heeney et al., 2019).
Various functions of HSPs that target microbial stimuli include maintaining protein stability and functionality, activating, potent immune responses, immunomodulation of host-microbe relationships, and acting as biomarkers (diagnosis of infection) (Bolhassani and Agi, 2019). Various functions of HSPs that may be used to control bacterial infections have been studied (Bolhassani and Agi, 2019). Studies have shown that high expression of HSP90 may lead to the secretion of inflammatory cytokines in the body (Abaza, 2014). Studies have shown that some HSPs are effective inducers of innate and adaptive immunity. In addition, HSPs exert cytoprotection against inflammatory reactions by modulating inflammatory cascades which activate pro-inflammatory cytokines, such as TNF-α, thus attenuating chronic inflammation (Zugel and Kaufmann, 1999; Ikwegbue et al., 2019). In our study, HSP expression levels which were further decreased in the C and P groups, were upregulated in group D, thus alleviating diarrhea in neonatal calves. In agreement with the results of our study, Giri et al. (2018) and Kong et al. (2020) demonstrated that dietary supplementation with a probiotic mixture consisting of B. subtilis, L. acidophilus, and S. cerevisiae decreased the expression of HSP90 in in probiotic treated groups resulting in better growth and immunity.
Overexpression of HSP90S impairs the function of T cells, leading to a decrease in IL-2 expression (Bae et al., 2013). Enteric pathogens that invade a host are recognized by pattern recognition receptors (Kan et al., 2021). Probiotic supplementation can promote toll like receptors (TLR) to recognize various pathogens and activate NF-κB. Then NF-κB is transferred to the nucleus to induce the expression of target genes, thereby regulating immune and inflammatory responses (Huebener and Schwabe, 2013; Elghandour et al., 2020). This study shows that probiotic supplementation can effectively down regulate IL-1 in jejunal mucosa β, TNF-α, and NF-κ. The expression of B mRNA and inflammatory factors further indicates that probiotic supply can effectively resist E. coli infection by enhancing the intestinal innate immune function of newborn calves. This is consistent with the results of previous studies (Dong et al., 2019; Boranbayeva et al., 2020; Kan et al., 2021). However, some studies believe that the physiological functions of different probiotic strains are different (Kang and Im, 2015). Although the addition of probiotics can activate the TLR-NF-κB signaling pathway, the gene expression of IL-1β and TNF-α cytokines was not significantly affected (Wang et al., 2017). In conclusion, the gene expression of the TLR-NF-κB signaling pathway, pro-inflammatory cytokines, and heat shock protein in the jejunum of healthy newborn calves in group D in this article is down-regulated, indicating that antibiotic treatment of calves with diarrhea may reduce their immune levels and cause infections in the large intestine. The probiotic supplementation of bacillus new calves may focus on promoting the recovery of their intestinal function.
We demonstrated that supplementation with MSP promotes immune function and microbiota function in neonatal dairy calves. MSP supplementation significantly changed the microbial structure and diversity of the small intestine of calves infected with E. coli K99 MSP supplementation improved intestinal immune function, especially the production of SIgA and SCFA concentrations in the jejunum of infected calves, decreased the relative mRNA expression of IL-1β and TLR4 and increased the relative mRNA expression of ZO-1 and occludin in the jejunal mucosa. Supplementation with MSP significantly enhanced intestinal energy metabolism and defense mechanisms of calves infected with E. coli K99 by reducing the expression of HSP90, which reduced inflammation and promoted the recovery of intestinal mucosal function. In view of these results, we recommend adding 2 g/day of MSP supplementation to the diets of dairy calves infected with E. coli K99. Our results are expected to provide a theoretical basis for the rational use of MSP supplements in calf production and provide substitutes for reducing the use of antibiotics.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
This study was approved by the Ethics Committee of the College of Animal Science and Technology, Shihezi University (Shihezi, China) (No. A2020-171-01).
YW and WZ: conceptualization, writing—original draft preparation. YW, CN, RL, HC, JN, CC, and XB: investigation, writing—review and editing. YW and CN: formal analysis. All authors have read and approved the final version of the manuscript.
This work was supported by the Key Scientific and Technological Research Project in Key Fields of Xinjiang Production and Construction Corps (No. 2018AB041), the Modern Agricultural Science and Technology Research and Achievement Transformation Project of the Eighth Division (No. 2018NY05), the Program for Science and Technology Innovation Talents in Xinjiang Production and Construction Corps (No. 2020CB023), and the Major Science and Technology Project of Xinjiang Production and Construction Corps (No. 2021AA004).
RL and HC were employed by Xinjiang Tianshan Junken Animal Husbandry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.813245/full#supplementary-material
Abarenkov, K., Tedersoo, L., Nilsson, R. H., Vellak, K., Saar, I., Veldre, V., et al. (2010). PlutoF-a web based workbench for ecological and taxonomic research, with an online implementation for fungal ITS sequences. Evol. Bioinform. Online 6, 189–196. doi: 10.4137/EBO.S6271
Abaza, M. (2014). Heat shock proteins and parasitic diseases: part 1: helminths. Parasitol. United J. 7, 93–103.
Acres, S. D. (1985). Enterotoxigenic Escherichia coli infections in newborn calves: a review. J. Dairy Sci. 68, 229–256. doi: 10.3168/jds.S0022-0302(85)80814-6
Al Mawly, J., Grinberg, A., Prattley, D., Moffat, J., Marshall, J., and French, N. (2015). Risk factors for neonatal calf diarrhoea and enteropathogen shedding in New Zealand dairy farms. Vet. J. 203, 155–160. doi: 10.1016/j.tvjl.2015.01.010
Alipour, M. J., Jalanka, J., Pessa-Morikawa, T., Kokkonen, T., Satokari, R., Hynönen, U., et al. (2018). The compo-sition of the perinatal intestinal microbiota in cattle. Sci. Rep. 8:10437. doi: 10.1038/s41598-018-28733-y
Bae, J., Munshi, A., Li, C., Samur, M., Prabhala, R., Mitsiades, C., et al. (2013). Heat shock protein 90 is critical for regulation of phenotype and functional activity of human t lymphocytes and nk cells. J. Immunol. 190:3009. doi: 10.4049/jimmunol.1200593
Bahrami, B., Macfarlane, S., and Macfarlane, G. T. (2011). Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J. Appl. Microbiol. 110, 353–363. doi: 10.1111/j.1365-2672.2010.04889.x
Bi, Y., Tu, Y., Zhang, N., Wang, S., Zhang, F., Suen, G., et al. (2021). Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut 70, 853–864. doi: 10.1136/gutjnl-2020-320951
Bitla, S., Gayatri, A. A., Puchakayala, M. R., Kumar Bhukya, V., Vannada, J., Dhanavath, R., et al. (2021). Design and synthesis, biological evaluation of bis-(1,2,3- and 1,2,4)-triazole derivatives as potential antimicrobial and antifungal agents. Bioorg. Med. Chem. Lett. 41:128004. doi: 10.1016/j.bmcl.2021.128004
Bolhassani, A., and Agi, E. (2019). Heat shock proteins in infection. Clin. Chim. Acta 498, 90–100. doi: 10.1016/j.cca.2019.08.015
Boranbayeva, T., Karahan, A. G., Tulemissova, Z., Myktybayeva, R., and Özkaya, S. (2020). Properties of a New probiotic candidate and lactobacterin-TK2 against diarrhea in calves. Probiotics Antimicrob. Proteins 12, 918–928. doi: 10.1007/s12602-020-09649-4
Borthakur, A., Gill, R. K., Tyagi, S., Koutsouris, A., Alrefai, W. A., Hecht, G. A., et al. (2008). The probiotic Lactobacillus acidophilus stimulates chloride/hydroxyl exchange activity in human intestinal epithelial cells. J. Nutr. 138, 1355–1359. doi: 10.1093/jn/138.7.1355
Bourassa, D. V., Wilson, K. M., Ritz, C. R., Kiepper, B. K., and Buhr, R. J. (2018). Evaluation of the addition of organic acids in the feed and/or water for broilers and the subsequent recovery of Salmonella typhimurium from litter and ceca. Poult. Sci. 97, 64–73. doi: 10.3382/ps/pex289
Braz, V. S., Melchior, K., and Moreira, C. G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Front. Cell Infect. Microbiol. 10:548492. doi: 10.3389/fcimb.2020.548492
Brunauer, M., Roch, F. F., and Conrady, B. (2021). Prevalence of worldwide neonatal calf diarrhoea caused by bovine rotavirus in combination with bovine coronavirus, Escherichia coli K99 and Cryptosporidium spp.: a meta-analysis. Animals (Basel) 11:1014. doi: 10.3390/ani11041014
Bulgin, M. S., Anderson, B. C., Ward, A. C. S., and Evermann, J. F. (1982). Infectious agents associated with neonatal calf disease in southwestern Idaho and eastern Oregon. J. Am. Vet. Med. Assoc. 180, 1222–1226.
Bywater, R. J., and Logan, E. F. (1974). The site and characteristics of intestinal water and electrolyte loss in Escherichia coli—induced diarrhoea in calves. J. Comp. Pathol. 84, 599–610. doi: 10.1016/0021-9975(74)90051-6
Cani, P. D. (2019). Severe obesity and gut microbiota: does bariatric surgery really reset the system? Gut 68, 5–6. doi: 10.1136/gutjnl-2018-316815
Cani, P. D., and Jordan, B. F. (2018). Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 15, 671–682. doi: 10.1038/s41575-018-0025-6
Chen, J., and Vitetta, L. (2019). Activation of t-regulatory cells by a synbiotic may be important for its anti-inflammatory effect. Eur. J. Nutr. 58, 3379–3380. doi: 10.1007/s00394-019-02080-8
Chiaro, T. R., Soto, R., Zac Stephens, W., Kubinak, J. L., Petersen, C., Gogokhia, L., et al. (2017). A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044
Cho, Y.-I. I., and Yoon, K.-J. J. (2014). An overview of calf diarrhea—Infectious etiology, diagnosis, and intervention. J. Vet. Sci. 15, 1–17. doi: 10.4142/jvs.2014.15.1.1
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Collinson, S., Deans, A., Padua-Zamora, A., Gregorio, G. V., Li, C., Dans, L. F., et al. (2020). Probiotics for treating acute infectious diarrhoea. Cochrane Datab. Syst. Rev. 12:CD003048. doi: 10.1002/14651858.CD003048.pub4
Conroy, M. E., Shi, H. N., and Walker, W. A. (2009). Long-term health effects of the neonatal microbialmicrobiota. Curr. Opin. Allergy Clin. Immunol. 9, 197–201. doi: 10.1097/ACI.0b013e32832b3f1d
Crassini, K. R., Zhang, E., Balendran, S., Freeman, J. A., Best, O. G., Forsyth, C. J., et al. (2018). Humoral immune failure defined by immunoglobulin class and immunoglobulin G subclass deficiency is associated with shorter treatment-free and overall survival in chronic lymphocytic leukaemia. Br. J. Haematol. 181, 97–101. doi: 10.1111/bjh.15146
da Silva Medeiros, T. N., Lorenzetti, E., Alfieri, A. F., and Alfieri, A. A. (2015). Phylogenetic analysis of a G6P[5] bovine rotavirus strain isolated in a neonatal diarrhea outbreak in a beef cattle herd vaccinated with G6P[1] and G10P[11] genotypes. Arch. Virol. 160, 447–451. doi: 10.1007/s00705-014-2271-4
Dahlgren, D., Sjöblom, M., Hellström, P. M., and Lennernäs, H. (2021). Chemotherapeutics-induced intestinal mucositis: pathophysiology and potential treatment strategies. Front. Pharmacol. 12:681417. doi: 10.3389/fphar.2021.681417
Dias, J., Marcondes, M. I., De Souza, S. M., Silva, B. C. D. M. E., Noronha, M. F., Resende, R. T., et al. (2018). Bacterial community dynamics across the gastrointestinal tracts of dairy calves during preweaning development. Appl. Environ. Microbiol. 84:e02675-17. doi: 10.1128/AEM.02675-17
DiGiulio, D. B. (2012). Diversity of microbes in amniotic fluid. Semin. Fetal Neonatal Med. 17, 2–11. doi: 10.1016/j.siny.2011.10.001
Dill-McFarland, K. A., Weimer, P. J., Breaker, J. D., and Suen, G. (2019). Diet inflfluences early microbiota development in dairy calves without long-term impacts on milk production. Appl. Environ. Microbiol. 85:e02141-18. doi: 10.1128/AEM.02141-18
Dong, Y., Zhang, F., Wang, B., Gao, J., Zhang, J., and Shao, Y. (2019). Laboratory evolution assays and whole-genome sequencing for the development and safety evaluation of lactobacillus plantarum with stable resistance to gentamicin. Front Microbiol. 10:1235. doi: 10.3389/fmicb.2019.01235
Dubreuil, J. D. (2017). Enterotoxigenic Escherichia coli targeting intestinal epithelial tight junctions: An effective way to alter the barrier integrity. Microb. Pathogen. 113, 129–134. doi: 10.1016/j.micpath.2017.10.037
Duparc, T., Plovier, H., Marrachelli, V. G., Van Hul, M., Essaghir, A., Ståhlman, M., et al. (2017). Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut 66, 620–632. doi: 10.1136/gutjnl-2015-310904
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Egertson, J. D., MacLean, B., Johnson, R., Xuan, Y., and MacCoss, M. J. (2015). Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat. Protoc. 10, 887–903. doi: 10.1038/nprot.2015.055
Elghandour, M., Tan, Z. L., Abu Hafsa, S. H., Adegbeye, M. J., Greiner, R., Ugbogu, E. A., et al. (2020). Saccharomyces cerevisiae as a probiotic feed additive to non and pseudo-ruminant feeding: a review. J. Appl. Microbiol. 128, 658–674. doi: 10.1111/jam.14416
Garcia, V., Catala-Gregori, P., Hernandez, F., Megias, M. D., and Madrid, J. (2007). Effffect of formic acid and plant extracts on growth, nutrient digestibility, intestine mucosa morphology, and meat yield of broilers. J. Appl. Poultry Res. 16, 555–562. doi: 10.3382/japr.2006-00116
Giri, S. S., Yun, S., Jun, J. W., Kim, H. J., Kim, S. G., Kang, J. W., et al. (2018). Therapeutic effect of intestinal autochthonous Lactobacillus reuteri P16 against waterborne lead toxicity in Cyprinus carpio. Front. Immunol. 9:1824.
González Pasayo, R. A., Sanz, M. E., Padola, N. L., and Moreira, A. R. (2019). Phenotypic and genotypic characterization of enterotoxigenic Escherichia coli isolated from diarrheic calves in Argentina. Open Vet. J. 9, 65–73. doi: 10.4314/ovj.v9i1.12
Gulliksen, S. M., Jor, E., Lie, K. I. I, Hamnes, S., Loken, T., Akerstedt, J., et al. (2009). Enteropathogens and risk factors for diarrhea in Norwegian dairy calves. J. Dairy Sci. 92, 5057–5066. doi: 10.3168/jds.2009-2080
Guo, J., Huang, W., Han, X., You, Y., and Jicheng, Z. (2020). Interaction between IgA and gut microbiota and its role in control ling metabolic syndrome. Obesity Rev. 22, 1–15. doi: 10.1111/obr.13155
Guo, J., Ren, C., Han, X., Huang, W., You, Y., and Zhan, J. (2021). Role of IgA in the early-life establishment of the gut microbiota and immunity: Implications for constructing a healthy start. Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1908101
Heeney, D. D., Zhai, Z., Bendiks, Z., Barouei, J., Martinic, A., Slupsky, C., et al. (2019). Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 10, 382–397. doi: 10.1080/19490976.2018.1534513
Hrala, M., Bosák, J., Micenková, L., Křenová, J., Lexa, M., Pirková, V., et al. (2021). Escherichia coli strains producing selected Bacteriocins inhibit porcine enterotoxigenic Escherichia coli (ETEC) under both in vitro and in vivo conditions. Appl. Environ. Microbiol. 87:e0312120. doi: 10.1128/AEM.03121-20
Hu, J., Chen, L., Zheng, W., Shi, M., Liu, L., Xie, C., et al. (2018). Lactobacillus frumenti facilitates intestinal epithelial barrier function maintenance in early-weaned piglets. Front. Microbiol. 9:897. doi: 10.3389/fmicb.2018.00897
Hubert, J., Erban, T., Kopecky, J., Sopko, B., Nesvorna, M., Lichovnikova, M., et al. (2017). Comparison of microbiomes between red poultry mite populations (Dermanyssus gallinae): predominance of Bartonella-like bacteria. Microb. Ecol. 74, 947–960. doi: 10.1007/s00248-017-0993-z
Huebener, P., and Schwabe, R. F. (2013). Regulation of wound healing and organ fibrosis by toll-like receptors. Biochim. Biophys. Acta 1832, 1005–1017. doi: 10.1016/j.bbadis.2012.11.017
Huson, D. H., Auch, A. F., Qi, J., and Schuster, S. C. (2007). MEGAN analysis of metagenomic data. Genome Res. 17, 377–386. doi: 10.1101/gr.5969107
Ikwegbue, P. C., Masamba, P., Mbatha, L. S., Oyinloye, B. E., and Kappo, A. P. (2019). Interplay between heat shock proteins, inflammation and cancer: a potential cancer therapeutic target. Am. J. Cancer Res. 9, 242–249.
Inatomi, T., Amatatsu, M., Romero-Pérez, G. A., Inoue, R., and Tsukahara, T. (2017). Dietary probiotic compound improves reproductive performance of porcine epidemic diarrhea virus-infected sows reared in a Japanese commercial swine farm under vaccine control condition. Front. Immunol. 8:1877. doi: 10.3389/fimmu.2017.01877
Izzo, M. M., Kirkland, P. D., Mohler, V. L., Perkins, N. R., Gunn, A. A., and House, J. K. (2011). Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 89, 167–173. doi: 10.1111/j.1751-0813.2011.00692.x
Jia, R., Sadiq, F. A., Liu, W., Cao, L., and Shen, Z. (2021). Protective effects of Bacillus subtilis ASAG 216 on growth performance, antioxidant capacity, gut microbiota and tissues residues of weaned piglets fed deoxynivalenol contaminated diets. Food Chem. Toxicol. 148:111962. doi: 10.1016/j.fct.2020.111962
Johnson-Henry, K. C., Donato, K. A., Shen-Tu, G., Gordanpour, M., and Sherman, P. M. (2008). Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect. Immun. 76, 1340–1348. doi: 10.1128/IAI.00778-07
Kan, L., Guo, F., Liu, Y., Pham, V. H., Guo, Y., and Wang, Z. (2021). Probiotics Bacillus licheniformis improves intestinal health of subclinical necrotic enteritis-challenged broilers. Front. Microbiol. 12:623739. doi: 10.3389/fmicb.2021.623739
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kang, H. J., and Im, S. H. (2015). Probiotics as an immune modulator. J. Nutr. Sci. Vitaminol. (Tokyo) 61, S103–S105. doi: 10.3177/jnsv.61.S103
Karamzadeh-Dehaghani, A., Towhidi, A., Zhandi, M., Mojgani, N., and Fouladi-Nashta, A. (2021). Combined effect of probiotics and specific immunoglobulin Y directed against Escherichia coli on growth performance, diarrhea incidence, and immune system in calves. Animal 15:100124. doi: 10.1016/j.animal.2020.100124
Kim, E. T., Lee, S. J., Kim, T. Y., Lee, H. G., Atikur, R. M., Gu, B. H., et al. (2021). Dynamic changes in fecal microbial communities of neonatal dairy calves by aging and diarrhea. Animals (Basel) 11:1113. doi: 10.3390/ani11041113
Kim, M., Qie, Y., Park, J., and Kim, C. H. (2016). Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214. doi: 10.1016/j.chom.2016.07.001
Klein-Jöbstl, D., Quijada, N. M., Dzieciol, M., Feldbacher, B., Wagner, M., Drillich, M., et al. (2019). Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves’ gastrointestinal microbiota. PLoS One 14:e0220554. doi: 10.1371/journal.pone.0220554
Klein-Jöbstl, D., Schornsteiner, E., Mann, E., Wagner, M., Drillich, M., and Schmitz-Esser, S. (2014). Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 5:622. doi: 10.3389/fmicb.2014.00622
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kong, Y., Gao, C., Du, X., Zhao, J., Li, M., Shan, X., et al. (2020). Effects of single or conjoint administration of lactic acid bacteria as potential probiotics on growth, immune response and disease resistance of snakehead fish (Channa argus). Fish Shellfish Immunol. 102, 412–421. doi: 10.1016/j.fsi.2020.05.003
Krogh, H. V. (1983). Occurrence of enterotoxigenic Escherichia coli in calves with acute neonatal diarrhoea. Nord. Vet. Med. 35, 346–352.
Lépine, A., de Wit, N., Oosterink, E., Wichers, H., Mes, J., and de Vos, P. (2018). Lactobacillus acidophilus attenuates Salmonella-induced stress of epithelial cells by modulating tight-junction genes and cytokine responses. Front. Microbiol. 9:1439. doi: 10.3389/fmicb.2018.01439
Li, H., Limenitakis, J. P., Greiff, V., Yilmaz, B., Schären, O., Urbaniak, C., et al. (2020). Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature 584, 274–278. doi: 10.1038/s41586-020-2564-6
Li, M., Chen, Z., Wu, T., and Lu, J. (2018). Li2S- or S-based lithium-ion batteries. Adv. Mater. 30:e1801190. doi: 10.1002/adma.201801190
Liu, B., Yuan, J., Yiu, S. M., Li, Z., Xie, Y., Chen, Y., et al. (2012). COPE: an accurate k-mer-based pair-end reads connection tool to facilitate genome assembly. Bioinformatics 28, 2870–2874. doi: 10.1093/bioinformatics/bts563
Liu, Y., Ji, P., Lv, H., Qin, Y., and Deng, L. (2017). Gentamicin modified chitosan film with improved antibacterial property and cell biocompatibility. Int. J. Biol. Macromol. 98, 550–556. doi: 10.1016/j.ijbiomac.2017.01.121
Lodemann, U., Amasheh, S., Radloff, J., Kern, M., Bethe, A., Wieler, L. H., et al. (2017). Effects of ex vivo infection with ETEC on jejunal barrier properties and cytokine expression in probiotic-supplemented pigs. Dig. Dis. Sci. 62, 922–933. doi: 10.1007/s10620-016-4413-x
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Maldonado Galdeano, C., Cazorla, S. I., Lemme Dumit, J. M., Vélez, E., and Perdigón, G. (2019). Beneficial effects of probiotic consumption on the immunesystem. Ann. Nutr. Metab. 74, 115–124. doi: 10.1159/000496426
Malmuthuge, N., Griebel, P. J., and Guan, L. (2015). The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front. Vet. Sci. 2:36. doi: 10.3389/fvets.2015.00036
Malmuthuge, N., and Guan, L. L. (2017). Understanding the gut microbiome of dairy calves: opportunities to improve early-life gut health. J. Dairy Sci. 100, 5996–6005. doi: 10.3168/jds.2016-12239
Markowiak, P., and Śliżewska, K. (2018). The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 10:21. doi: 10.1186/s13099-018-0250-0
Mazloom, K., Siddiqi, I., and Covasa, M. (2019). Probiotics: how effffective are they in the fight against obesity? Nutrients 11:258. doi: 10.3390/nu11020258
Memon, F. U., Yang, Y., Leghari, I. H., Lv, F., Soliman, A. M., Zhang, W., et al. (2021). Transcriptome analysis revealed ameliorative effects of bacillus based probiotic on immunity, gut barrier system, and metabolism of chicken under an experimentally induced Eimeria tenella Infection. Genes 12:536. doi: 10.3390/genes12040536
Merselis, L. C., Rivas, Z. P., and Munson, G. P. (2021). Breaching the bacterial envelope: the pivotal role of perforin-2 (MPEG1) within phagocytes. Front. Immunol. 12:597951. doi: 10.3389/fimmu.2021.597951
Moles, L., Gómez, M., Heilig, H., Bustos, G., Fuentes, S., de Vos, W., et al. (2013). Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 8:e66986. doi: 10.1371/journal.pone.0066986
Moon, H. W., McClurkin, A. W., Isaacson, R. E., Pohlenz, J., Skartvedt, S. M., Gillette, K. G., et al. (1978). Pathogenic relationships of rotavirus, Escherichia coli, and other agents in mixed infections in calves. J. Am. Vet. Med. Assoc. 173, 577–583.
Moor, K., Diard, M., Sellin, M. E., Felmy, B., Wotzka, S. Y., Toska, A., et al. (2017). High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502. doi: 10.1038/nature22058
Nakawesi, J., This, S., Hütter, J., Boucard-Jourdin, M., Barateau, V., Muleta, K. G., et al. (2020). αvβ8 integrin-expression by BATF3-dependent dendritic cells facilitates early IgA responses to Rotavirus. Mucosal Immunol. 14, 53–67. doi: 10.1038/s41385-020-0276-8
Oikonomou, G., Teixeira, A. G., Foditsch, C., Bichalho, M. L., Machado, V. S., and Bicalho, R. C. (2013). Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS One 8:e63157. doi: 10.1371/journal.pone.0063157
Paszti-Gere, E., Szeker, K., Csibrik-Nemeth, E., Csizinszky, R., Marosi, A., Palocz, O., et al. (2012). Metabolites of Lactobacillus plantarum 2142 prevent oxidative stress-induced overexpression of proinflammatory cytokines in IPEC-J2 cell line. Inflammation 35, 1487–1499. doi: 10.1007/s10753-012-9462-5
Perego, S., Sansoni, V., Banfi, G., and Lombardi, G. (2018). Sodium butyrate has anti-proliferative, pro-differentiating, and immunomodulatory effects in osteosarcoma cells and counteracts the TNFα-induced low-grade inflammation. Int. J. Immunopathol. Pharmacol. 32:394632017752240. doi: 10.1177/0394632017752240
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RTPCR. Nucleic Acids Res. 29, 900–905. doi: 10.1093/nar/29.9.e45
Plaza-Diaz, J., Ruiz-Ojeda, F. J., Gil-Campos, M., and Gil, A. (2019). Mechanisms of action of probiotics. Adv Nutr. 10, S49–S66. doi: 10.1093/advances/nmy063
Raheem, A., Liang, L., Zhang, G., and Cui, S. (2021). Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front. Immunol. 12:616713. doi: 10.3389/fimmu.2021.616713
Régnier, M., Van Hul, M., Knauf, C., and Cani, P. D. (2021). Gut microbiome, endocrine control of gut barrier function and metabolic diseases. J. Endocrinol. 248, R67–R82. doi: 10.1530/JOE-20-0473
Roggero, P., Liotto, N., Pozzi, C., Braga, D., Troisi, J., Menis, C., et al. (2020). Analysis of immune, microbiota and metabolome maturation in infants in a clinical trial of Lactobacillus paracasei CBA L74-fermented formula. Nat. Commun. 11:2703.
Roth, B. A., Keil, N. M., Gygax, L., and Hillmann, E. (2009). Influence of weaning method on health status and rumen development in dairy calves. J. Dairy Sci. 92, 645–656. doi: 10.3168/jds.2008-1153
Runnels, P. L., Moon, H. W., and Schneider, R. A. (1980). Development of resistance with host age to adhesion of K99+ Escherichia coli to isolated intestinal epithelial cells. Infect. Immun. 28, 298–300.
Sambanthamoorthy, K., Feng, X., Patel, R., Patel, S., and Paranavitana, C. (2014). Antimicrobial and antibiofifilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 14:197. doi: 10.1186/1471-2180-14-197
Sanchez, B., Urdaci, M. C., and Margolles, A. (2010). Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosabacteria interactions. Microbiology 156, 3232–3242. doi: 10.1099/mic.0.044057-0
Schäff, C. T., Gruse, J., Maciej, J., Pfuhl, R., Zitnan, R., Rajsky, M., et al. (2018). Effects of feeding unlimited amounts of milk replacer for the first 5 weeks of age on rumen and small intestinal growth and development in dairy calves. J. Dairy Sci. 101, 783–793. doi: 10.3168/jds.2017-13247
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Scott, N. A., Andrusaite, A., Andersen, P., Lawson, M., Alcon-Giner, C., Leclaire, C., et al. (2018). Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci. Transl. Med. 10:eaao4755. doi: 10.1126/scitranslmed.aao4755
Shen, Z., Mustapha, A., Lin, M., and Zheng, G. (2017). Biocontrol of the internalization of Salmonella enterica and Enterohaemorrhagic Escherichia coli in mung bean sprouts with an endophytic Bacillus subtilis. Int. J. Food Microbiol. 250, 37–44. doi: 10.1016/j.ijfoodmicro.2017.03.016
Sherman, P. M., Johnson-Henry, K. C., Yeung, H. P., Ngo, P. S., Goulet, J., and Tompkins, T. A. (2005). Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 73, 5183–5188.
Singh, V., Kumar, A., Raheja, G., Anbazhagan, A. N., Priyamvada, S., Saksena, S., et al. (2014). Lactobacillus acidophilus attenuates downregulation of DRA function and expression in inflammatory models. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G623–G631. doi: 10.1152/ajpgi.00104.2014
Song, L., Qiao, X., Zhao, D., Xie, W., Bukhari, S. M., Meng, Q., et al. (2019). Effects of Lactococcus lactis MG1363 producing fusion proteins of bovine lactoferricin-lactoferrampin on growth, intestinal morphology and immune function in weaned piglet. J. Appl. Microbiol. 127, 856–866. doi: 10.1111/jam.14339
Song, Y., Malmuthuge, N., Steele, M. A., and Guan, L. L. (2018). Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol Ecol. 94:fix179. doi: 10.1093/femsec/fix179
Sunderland, S. J., Sarasola, P., Rowan, T. G., Giles, C. J., and Smith, D. G. (2003). Efficacy of danofloxacin 18% injectable solution in the treatment of Escherichia coli diarrhoea in young calves in Europe. Res. Vet. Sci. 74, 171–178. doi: 10.1016/s0034-5288(02)00186-8
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Tanaka, K., Tsukahara, T., Yanagi, T., Nakahara, S., Furukawa, O., Tsutsui, H., et al. (2017). Bifidobacterium bifidum OLB6378 simultaneously enhances systemic and mucosal humoral immunity in low birth weight infants: a non-randomized study. Nutrients 9:195. doi: 10.3390/nu9030195
Tanca, A., Abbondio, M., Palomba, A., Fraumene, C., Manghina, V., Cucca, F., et al. (2017). Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome 5:79. doi: 10.1186/s40168-017-0293-3
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—New capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Villot, C., Chen, Y., Pedgerachny, K., Chaucheyras-Durand, F., Chevaux, E., Skidmore, A., et al. (2020). Early supplementation of Saccharomyces cerevisiae boulardii CNCM I-1079 in newborn dairy calves increases IgA production in the intestine at 1 week of age. J. Dairy Sci. 103, 8615–8628. doi: 10.3168/jds.2020-18274
Walker, D. G., and Lue, L. F. (2015). Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimers Res. Ther. 7:56. doi: 10.1186/s13195-015-0139-9
Walker, P. G., Constable, P. D., Morin, D. E., Drackley, J. K., Foreman, J. H., and Thurmon, J. C. (1998). A reliable, practical, and economical protocol for inducing diarrhea and severe dehydration in the neonatal calf. Can. J. Vet. Res. 62, 205–213.
Wang, K., Cao, G., Zhang, H., Li, Q., and Yang, C. (2019). Effects of Clostridium butyricum and Enterococcus faecalis on growth performance, immune function, intestinal morphology, volatile fatty acids, and intestinal microbiota in a piglet model. Food Funct. 10, 7844–7854. doi: 10.1039/c9fo01650c
Wang, S., Peng, Q., Jia, H. M., Zeng, X. F., Zhu, J. L., Hou, C. L., et al. (2017). Prevention of Escherichia coli infection in broiler chickens with Lactobacillus plantarum B1. Poult. Sci. 96, 2576–2586. doi: 10.3382/ps/pex061
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Wu, W., Sun, M., Chen, F., Cao, A. T., Liu, H., Zhao, Y., et al. (2017). Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 10, 946–956. doi: 10.1038/mi.2016.114
Wu, Y., Wang, L., Luo, R., Chen, H., Nie, C., Niu, J., et al. (2021a). Effect of a multispecies probiotic mixture on the growth and incidence of diarrhea, immune function, and fecal microbiota of pre-weaning dairy calves. Front. Microbiol. 12:681014. doi: 10.3389/fmicb.2021.681014
Wu, Y., Wenju, Z., Hongli, C., and Qicheng, L. (2021b). Optimization of 3 strains of the compound microecological preparation for preventing calf diarrhea. J. Livestock Ecol. 42, 115–120.
Xin, H., Ma, T., Xu, Y., Chen, G., Chen, Y., Villot, C., et al. (2021). Characterization of fecal branched-chain fatty acid profiles and their associations with fecal microbiota in diarrheic and healthy dairy calves. J. Dairy Sci. 104, 2290–2301. doi: 10.3168/jds.2020-18825
Younis, G., Awad, A., Dawod, R. E., and Yousef, N. E. (2017). Antimicrobial activity of yeasts against some pathogenic bacteria. Vet. World 10, 979–983. doi: 10.14202/vetworld.2017.979-983
Zhang, X., Ning, Z., Mayne, J., Moore, J. I., Li, J., Butcher, J., et al. (2016). MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 4:31. doi: 10.1186/s40168-016-0176-z
Keywords: multispecies probiotic, immune function, microbiota function, neonatal calves, E. coli K99
Citation: Wu Y, Nie C, Luo R, Qi F, Bai X, Chen H, Niu J, Chen C and Zhang W (2022) Effects of Multispecies Probiotic on Intestinal Microbiota and Mucosal Barrier Function of Neonatal Calves Infected With E. coli K99. Front. Microbiol. 12:813245. doi: 10.3389/fmicb.2021.813245
Received: 11 November 2021; Accepted: 25 November 2021;
Published: 26 January 2022.
Edited by:
Yigang Xu, Zhejiang A&F University, ChinaReviewed by:
Gui-Lian Yang, Jilin Agricultural University, ChinaCopyright © 2022 Wu, Nie, Luo, Qi, Bai, Chen, Niu, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenju Zhang, emhhbmd3ajEwMjJAc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.