
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 February 2022
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.809431
 Olga I. Nedashkovkaya1*
Olga I. Nedashkovkaya1* Song-Gun Kim2
Song-Gun Kim2 Larissa A. Balabanova1*
Larissa A. Balabanova1* Natalia V. Zhukova3
Natalia V. Zhukova3 Oksana M. Son4
Oksana M. Son4 Liudmila A. Tekutyeva4
Liudmila A. Tekutyeva4 Valery V. Mikhailov1
Valery V. Mikhailov1A strictly aerobic, Gram-stain-negative, rod-shaped, and motile bacterium, designated strain 16-SW-7, isolated from a seawater sample, was investigated in detail due to its ability to produce a unique α-galactosidase converting B red blood cells into the universal type blood cells. The phylogenetic analysis based on 16S rRNA gene sequences revealed that the strain 16-SW-7 is a member of the Gammaproteobacteria genus Pseudoalteromonas. The closest relatives of the environmental isolate were Pseudoalteromonas distincta KMM 638T and Pseudoalteromonas paragorgicola KMM 3548T, with the plural paralogous 16S rRNA genes of 99.87–100% similarity. The strain 16-SW-7 grew with 1–10% NaCl and at 4–34°C, and hydrolyzed casein, gelatin, tyrosine, and DNA. The genomic DNA G+C content was 39.3 mol%. The prevalent fatty acids were C16:1 ω7c, C16:0, C17:1 ω8c, C18:1 ω7c, C17:0, and C12:0 3-OH. The polar lipid profile was characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, two unidentified amino lipids, and three unidentified lipids. The major respiratory quinone was Q-8. The finished genome of the strain 16-SW-7 (GenBank assembly accession number: GCA_005877035.1) has a size of 4,531,445 bp and comprises two circular chromosomes L1 and S1, deposited in the GenBank under the accession numbers CP040558 and CP040559, respectively. The strain 16-SW-7 has the ANI values of 98.2% with KMM 638T and KMM 3548T and the DDH values of 84.4 and 83.5%, respectively, indicating clearly that the three strains belonged to a single species. According to phylogenetic evidence and similarity for the chemotaxonomic and genotypic properties, the strain 16-SW-7 (= KCTC 52772 = KMM 701) represents a novel member of the species Pseudoalteromonas distincta. Also, we have proposed to reclassify Pseudoalteromonas paragorgicola as a later heterotypic synonym of P. distincta based on the rules of priority with the emendation of the species.
The genus Pseudoalteromonas was proposed by Gauthier et al. (1995) by splitting the genus Alteromonas into two genera due to its high level of heterogeneity. At the time of writing, the genus Pseudoalteromonas comprises 49 validly published species, including Pseudoalteromonas haloplanktis as the type species. Cells of the genus were described as Gram-stain-negative, aerobic, chemoorganotrophic, non-spore-forming, straight and curved rods or ovoid, those are motile by means of a single polar flagellum. All species of the genus were oxidase-positive, required a seawater base for growth, and did not accumulate poly-p-hydroxybutyrate. Subsequently, the genus was emended due to the newly obtained data, including the presence of polar, bipolar, or lateral flagella, gelatin and Tween 80 hydrolysis, glucose fermentation, and the ability of some strains to produce buds and prosthecae (Ivanova et al., 2002a; Hwang et al., 2016; Beurmann et al., 2017). In addition, the G+C content of DNA was extended up to 37–55 mol% (Park et al., 2016). Currently, the G+C content of the genomic DNA ranges from 34.8 mol% for Pseudoalteromonas denitrificans DSM 6059T (NCBI RefSeq: NZ_FOLO00000000.1) to 54.9 mol% for Pseudoalteromonas aestuariivivens DB-2T (Park et al., 2016). Members of the genus Pseudoalteromonas are often isolated from different marine environments, including surface and deep seawater, sediments and sea ice samples, ascidians, coral, a surface slime of a puffer fish, mussels, brown and green algae, diatoms, and the halophyte plants (Bowman, 1998; Sawabe et al., 2000; Egan et al., 2001; Ivanova et al., 2002b,c, 2004; Romanenko et al., 2003a,b; Park et al., 2005; Matsuyama et al., 2013; Wu et al., 2017; Navarro-Torre et al., 2020). In this study, we characterized a non-pigmented strain 16-SW-7, isolated from seawater of the Okhotsk Sea. This strain is attracting the attention of researchers for its ability to convert B red blood cells into the universal type of blood cells (Bakunina et al., 1998; Balabanova et al., 2010). A preliminary study of the taxonomic position of the strain by Sanger sequencing of the 16S rRNA gene indicated its closest relationship with the type strains of the recognized species of the genus Pseudoalteromonas, P. distincta, and P. paragorgicola, with a similarity of 99.9% by the taxonomic EzBioCloud 16S rRNA database (Yoon et al., 2017). The type and single strain of P. distincta KMM 638T (formerly Alteromonas distincta) was originally isolated from a marine sponge collected at a depth of 350 m near the Komandorskie Islands, Russia (Romanenko et al., 1995). This strain formed non-pigmented colonies and was shown to produce the dark-gray diffusible melanin-like pigments. The type of strain of P. paragorgicola KMM 3548T was originally isolated from a gorgonian, Paragorgia arborea, collected from the Pacific Ocean, and formed lightly orange-pigmented colonies (Ivanova et al., 2002a). Further investigation presented here on the whole-genome sequences has shown the affiliation of the strain 16-SW-7 and the above-mentioned species with validly published names to the single species. In the present study, we clarify the taxonomic position of the strain 16-SW-7, reclassify the species P. paragorgicola as a later heterotypic synonym of P. distincta, and specify the description of the species P. distincta based on the results of phylogenetic analysis, and genotypic and phenotypic characterization.
The strain 16-SW-7 was isolated from a seawater sample collected near Island Paramushir (Kuril Islands), the Okhotsk Sea, during the 16th cruise of the Research Vessel Academician Oparin by plating 0.1 ml of seawater directly onto nutrient medium as described previously (Nedashkovskaya et al., 2007). After primary isolation and purification, the bacterium was cultivated at 28°C on the same medium or marine agar 2216 (Difco, bioMérieux, Pacific, Biosciences) and stored at −80°C in artificial seawater or marine broth (Difco, bioMérieux, Pacific, Biosciences) supplemented with 20% (v/v) glycerol. The strain 16-SW-7 was deposited in the collection of marine microorganisms (KMM) at the G.B. Elyakov Pacific Institute of Bioorganic Chemistry FEB RAS (Vladivostok, Russia), Korean Collection for the type cultures (KCTC) and VKM under deposit numbers KMM 701, KCTC 52772, and VKM B-2135 D, respectively. The type strains P. distincta KMM 638T (=ATCC 700518T) and P. paragorgicola KMM 3548T (=DSM 26439T) were obtained from the collection of marine microorganisms (KMM) and used as the reference strains for comparative taxonomic analysis.
The physiological, morphological, and biochemical properties of the strain 16-SW-7 were studied using the standard methods. The novel isolate was also examined in the API 20E, API 20NE, API 50 CH, API 32 ID GN, and API ZYM galleries (bioMérieux, France) according to the manufacturer’s instructions, except that the inoculum was prepared using ASW (Bruns et al., 2001) and the galleries were incubated at 28°C. Gram-staining was performed as recommended by Gerhardt et al. (1994). Oxidative or fermentative utilization of glucose was determined on Hugh and Leifson’s medium modified for marine bacteria (Lemos et al., 1985). Catalase activity was tested by the addition of 3% (v/v) H2O2 solution to a bacterial colony and observation for the appearance of gas. Oxidase activity was determined by using tetramethyl-p-phenylenediamine. Degradation of agar, starch, casein, gelatin, chitin, DNA and urea and production of acid from carbohydrates, hydrolysis of Tween 80, nitrate reduction, production of hydrogen sulfide, acetoin (Voges–Proskauer reaction), and indole were tested according to standard methods (Gerhardt et al., 1994). The temperature range for growth was assessed on MA. Tolerance to NaCl was assessed in medium containing 5 g Bacto Peptone (Difco), 2 g Bacto Yeast Extract (Difco), 1 g glucose, 0.02 g KH2PO4, and 0.05 g MgSO4.7H2O per liter of distilled water with 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3, 4, 5, 6, 8, 10, 12. 15, 17, 19, and 20% (w/v) of NaCl. Susceptibility to antibiotics was examined on MA plates at 28°C by the routine disk diffusion plate method. Disks were impregnated with the following antibiotics: ampicillin (10 μg), benzylpenicillin (10U), carbenicillin (100 μg), cefalexin (30 μg), cefazolin (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), lincomycin (15 μg), nalidixic acid (30 μg), neomycin (30 μg), ofloxacin (5 μg), oleandomycin (15 μg), oxacillin (10 μg), polymyxin B (300 U), rifampicin (5 μg), streptomycin (30 μg), tetracycline (5 μg), and vancomycin (30 μg).
For whole-cell fatty acid and polar lipid analysis, the strains 16-SW-7, P. distincta KMM 638T and P. paragorgicola KMM 3548T were grown under optimal physiological conditions for all strains (at 28°C for 24 h on MA). Cellular fatty acid methyl esters (FAMEs) were prepared according to the methods described by Sasser (1990), using the standard protocol of Sherlock Microbial Identification System (version 6.0, MIDI), and analyzed with the use of a GC-21A chromatograph (Shimadzu) equipped with a fused-silica capillary column (30 m × 0.25 mm) coated with Supelcowax-10 and SPB-5 phases (Supelco) at 210°C. FAMEs were identified by using equivalent chain-length measurements and comparing the retention times to those of authentic standards. The polar lipids of the strains studied were extracted using the chloroform/methanol extraction method of Bligh and Dyer (1959). Two-dimensional TLC of polar lipids was carried out on silica gel 60 F254 (10 cm × 10 cm; Merck) using chloroform/methanol/water (65: 25: 4, by vol.) in the first dimension, and chloroform/methanol/acetic acid/water (80: 12: 15: 4, by vol.) in the second dimension (Collins and Shah, 1984). For detection of the lipids, 10% sulfuric acid in methanol, molybdenum blue, ninhydrin, and a-naphthol were applied. Isoprenoid quinones were extracted with chloroform/methanol (2:1, v/v) and purified by TLC, using a mixture of n-hexane and diethyl ether (85:15, v/v) as the solvent. Isoprenoid quinone composition of the strain 16-SW-7 was characterized by HPLC (Shimadzu LC-10A) using a reversed-phase type Supelcosil LC-18 column (15 cm × 4.6 mm) and acetonitrile/2-propanol (65:35, v/v) as a mobile phase at a flow rate of 0.5 ml min–1 as described previously (Komagata and Suzuki, 1988).
The genomic DNA of the strain Pseudoalteromonas sp. 16-SW-7 (=KMM 701 = KCTC 52772) was extracted from the cells grown on marine agar (25°C, 72 h), using a NucleoSpin microbial DNA kit (Macherey-Nagel, 54 Germany), and sequenced at Macrogen, Inc. (Seoul, South Korea). To construct libraries, the high-molecular-weight DNA (15 μg) was fragmented to generate 20-kb SMRTbell™ templates, and then, the fragments were annealed using a PacBio DNA polymerase binding kit and sequenced by the PacBio RS II platform (Pacific Biosciences, United States), with the use of PacBio version 4.0 sequencing kit with single-molecule real-time cells. Hierarchical Genome Assembly Process 3 (HGAP3) was used to perform de novo assembly of the PacBio reads for Pseudoalteromonas sp. 16-SW-7. The circular shape of the contigs was formed by testing the overlap ability of the contig ends. Because of the mapping reads against the assembled contigs and error correction using Quiver, the final sequence with the highest quality was generated.
The 16S rRNA gene and genome phylogenetic analyses were performed by the Type (Strain) Genome Server (TYGS), an automated high-throughput platform for state-of-the-art genome-based taxonomy (Meier-Kolthoff and Göker, 2019). The genome of the strain 16-SW-7 (GenBank assembly accession: GCA_005877035.1) was compared against all types of strain genomes available in the TYGS database via the MASH algorithm, a fast approximation of intergenomic relatedness (Ondov et al., 2016), and the 10 types of strains with the smallest MASH distances were chosen. In addition, the set of 10 closely related type strains was determined via the 16S rRNA gene sequences, extracted from the genomes using RNAmmer (Lagesen and Hallin, 2007), and BLASTed (Camacho et al., 2009) against the 16S rRNA gene of each of 14,309 type strains. This was used as a proxy to find the best 50 matching type strains, according to the bit score for the 16-SW-7 genome and to subsequently calculate precise distances using the Genome BLAST Distance Phylogeny approach (GBDP) under the algorithm “coverage” and distance formula d5 (Meier-Kolthoff et al., 2013). These distances were finally used to determine the top 10 closest genomes of the type strains. For the phylogenomic inference, all pairwise comparisons among the set of genomes were conducted using GBDP and accurate intergenomic distances inferred under the algorithm “trimming” and distance formula d5. One hundred distance replicates were calculated each. Digital DNA-DNA hybridization (DDH) values and confidence intervals were calculated using the recommended settings of the GGDC 2.1 (Meier-Kolthoff et al., 2013). The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FastME 2.1.6.1 including SPR post-processing (Lefort et al., 2015). Branch support was inferred from 100 pseudo-bootstrap replicates each. The trees were rooted at the midpoint (Farris, 1972) and visualized with PhyD3 (Kreft et al., 2017). The type-based species clustering, using a 70% dDDH radius around each of the 13 type strains, was done as previously described (Meier-Kolthoff and Göker, 2019). Subspecies clustering was done using a 79% dDDH threshold as previously introduced (Meier-Kolthoff et al., 2014).
The whole-genome average nucleotide identity (ANI) values were calculated with the use of ChunLab’s ANI calculator (Yoon et al., 2017). The average amino acid identity (AAI) values were calculated by an AAI-profiler available at http://ekhidna2.biocenter.helsinki.fi/AAI (Medlar et al., 2018). Biosynthetic gene clusters (BGCs) were identified with antiSMASH version 5.1.1 (Medema et al., 2011). The whole-genome sequence analyses and comparative genomics of Pseudoalteromonas sp. 16-SW-7, P. paragorgicola KMM 3548T (GenBank assembly accession: GCA_014918315.1), and P. distincta ATCC 700518T (GenBank assembly accession: GCA_000814675.1) were additionally carried out using the high-performance computing servers Rapid Annotation of microbial genomes using Subsystems Technology (RAST; Overbeek et al., 2014), EzBioCloud (Yoon et al., 2017), and Integrated Microbial Genomes and Microbiomes (IMG/M) system (Chen et al., 2021).
The strain 16-SW-7 was shown to be a strictly aerobic, heterotrophic, Gram-stain-negative, and motile bacterium, which formed non-pigmented colonies on marine agar and required NaCl or seawater for growth. It was positive for cytochrome oxidase and catalase and hydrolyzed aesculin, casein, gelatin, Tweens 20, 40, and 80, DNA, and tyrosine (Table 1).
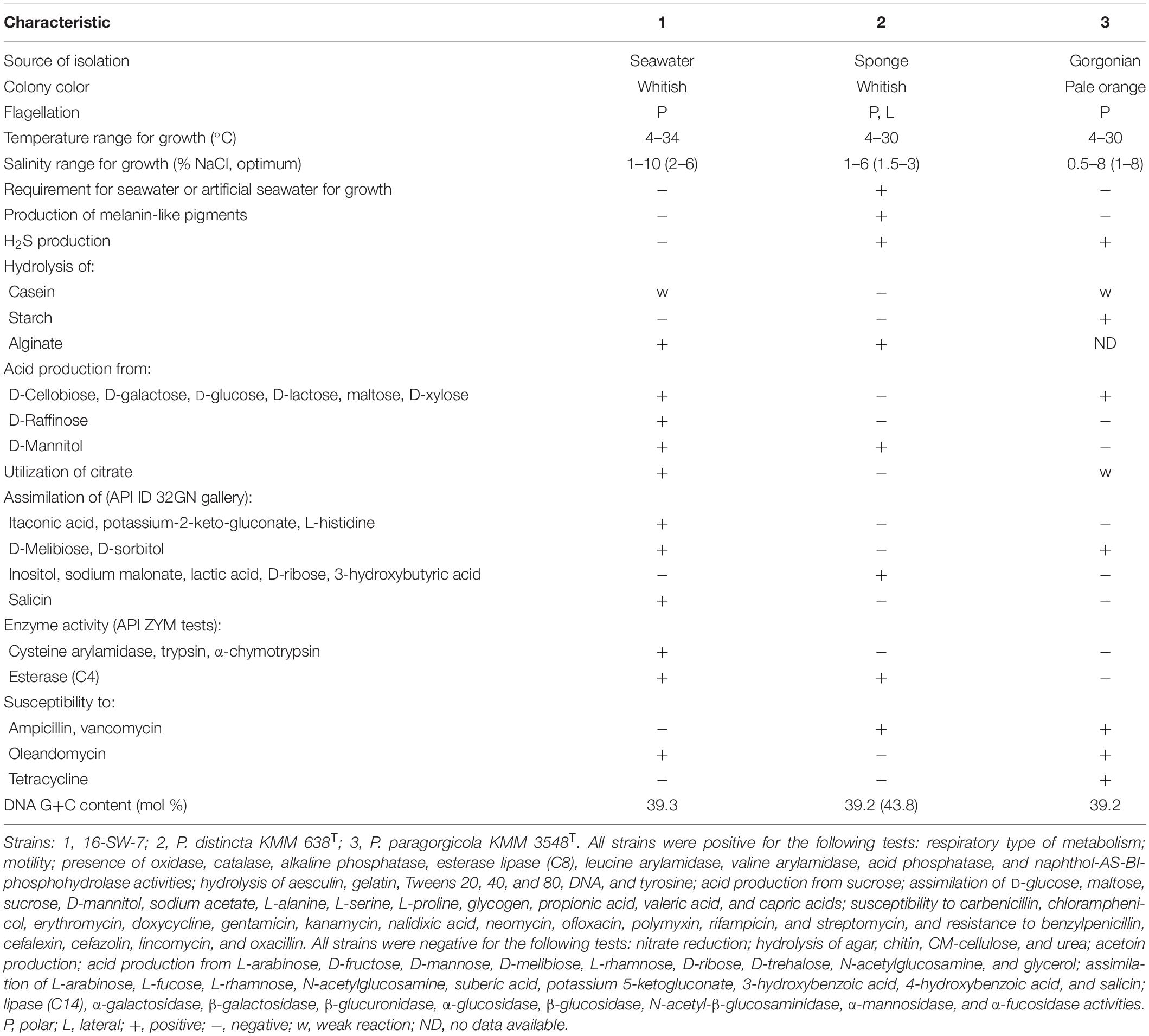
Table 1. Phenotypic characteristics of the strain 16-SW-7 and the closest relatives of the genus Pseudoalteromonas.
The strains 16-SW-7, P. distincta KMM 638T, and P. paragorgicola KMM 3548T shared many common phenotypic features, such as respiratory type of metabolism, motility by means of flagella, the presence of catalase, alkaline phosphatase, esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase activities (Table 1). They could not synthesize lipase (C14), N-acetyl-β-glucosaminidase, β-glucosidase, α-galactosidase, β-glucuronidase, α-mannosidase and α-fucosidase, hydrolyse agar, chitin, and urea and reduce nitrate to nitrite. However, the strain 16-SW-7 can be distinguished from its closest phylogenetic relatives by the several phenotypic traits, including the ability to form acid from D-raffinose, to produce hydrogen sulfide and cysteine arylamidase, trypsin and α-chymotrypsin, and to be resistant to ampicillin and vancomycin (Table 1). The above findings can extend the phenotypic characteristics those were reported for the species P. distincta (Romanenko et al., 1995; Ivanova et al., 2000, 2004) after justification of the placement of the strains 16-SW-7 and P. paragorgicola KMM 3548T in the species P. distincta.
The fatty acid profiles of the strains 16-SW-7, P. distincta KMM 638T, and P. paragorgicola KMM 3548T were similar (Table 2). The predominant fatty acids (>5% of the total fatty acids) of the strain 16-SW-7 and its closest relatives were C16:1 ω7c (29–32.1%), C16:0 (15.4–18.2%), C17:1 ω8c (11.7–17.9%), C18:1 ω7c (5.2–11%), C17:0 (5.8–10.3%), and C12:0 3-OH (4.8–7.5%). The composition of other fatty acids presented in Table 2 was also similar except that the strain 16-SW-7 contained higher proportions of C14:0, C12:0, and iso-C16:0, and lower proportions of C15:1 ω8c, C12:0 3-OH, and C13:0 3-OH. These values were consistent with the results of phylogenetic analysis and confirmed the affiliation of the strains studied to the same species. The polar lipid profile of the strain 16-SW-7 was characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, two unidentified amino lipids, and three unidentified lipids (Table 2 and Supplementary Figure 1). It was similar to that of P. paragorgicola KMM 3548T and it can be distinguished from another relative, P. distincta KMM 638T, by the presence of unknown lipids L1 and L2. The nearest neighbors of the strains under study, Pseudoalteromonas aliena LMG 22059T and Pseudoalteromonas fuliginea KMM 216T, distinguished from them by the presence of unknown phospholipids and unknown aminophospholipid and two unknown glycolipids, respectively (Machado et al., 2016; Zhang et al., 2016). The main respiratory quinone of the strains under study was ubiquinone Q-8 that is consistent with those reported for the members of the family Pseudoalteromonadaceae (Ivanova et al., 2004).
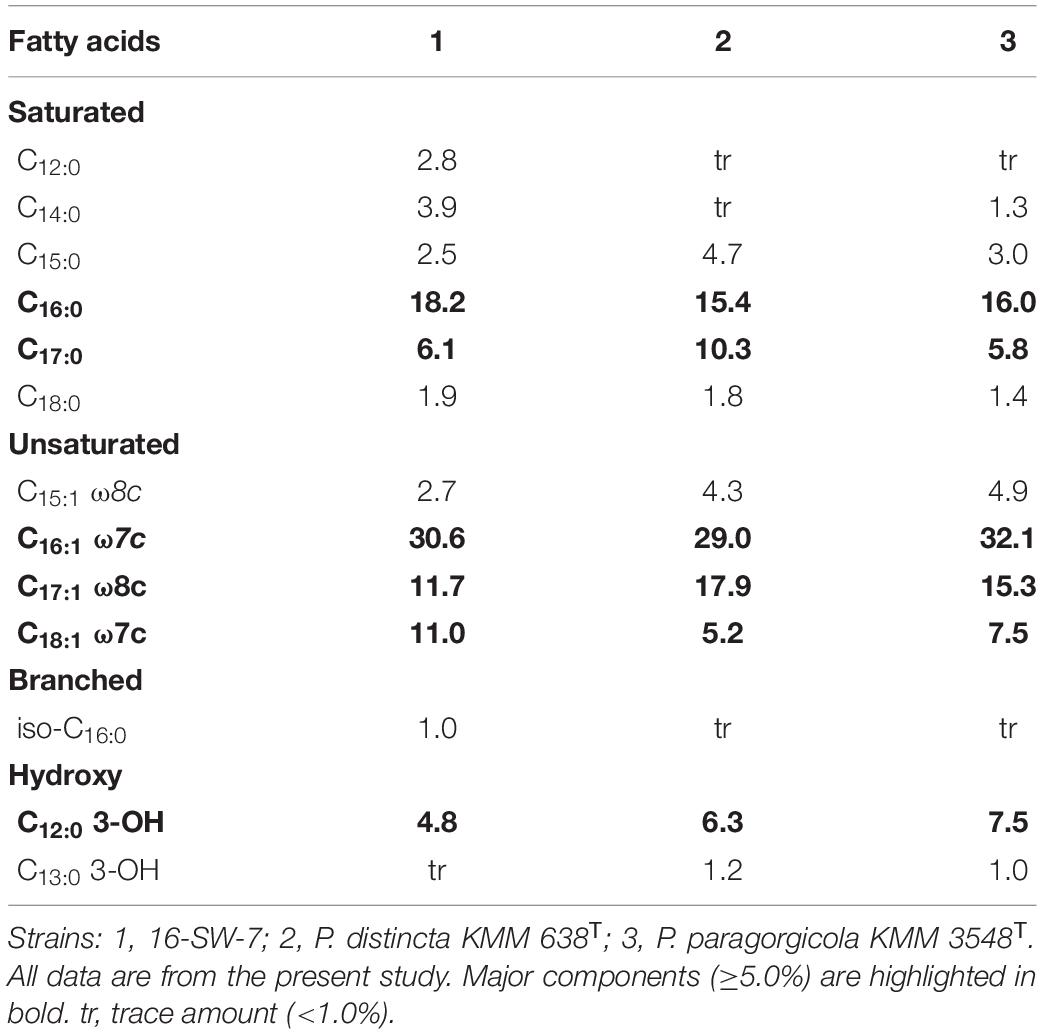
Table 2. Fatty acid composition (%) of the strain 16-SW-7 and closely related strains of the genus Pseudoalteromonas.
The analysis of the 16S rRNA gene sequence of the strain 16-SW-7 (GenBank accession number: OL587468) in the EzTaxon database application (Yoon et al., 2017) revealed 100% similarity with Pseudoalteromonas arctica A 37-1-2T (CP011026) and Pseudoalteromonas elyakovii KMM 162T (AF082562), and 99.9% similarity with P. distincta KMM 638T (JWIG01000030) and P. paragorgicola KMM 3548T (AY040229). However, the 16S rRNA gene sequences obtained by the Sanger method are recommended to be compared with the genome sequences, as well as the use of overall genome data for the taxonomy of prokaryotes, such as average nucleotide identity (ANI) and digital DDH (dDDH) and relatedness between the strains and type of strain of a species (Chun et al., 2018). The closed genome of 16-SW-7 was found to contain nine full-length sequences of 16S rRNA genes with 99.87–100% similarity between each other (Table 3). The multiple 16S rRNA genes seem to be a characteristic of the type species of the family Pseudoalteromonadaceae, including P. distincta KMM 638T and P. paragorgicola KMM 3548T (Table 3). Among the nine 16S rRNA genes found in the genome of P. distincta KMM 638T (=ATCC 700518T), only one—the length-comparable gene in a contig 30—was extracted for the analysis, probably due to an incomplete genome sequencing (GenBank WGS accession: JWIG00000000.1). In the P. paragorgicola KMM 3548T genome, two 16S rRNA genes were completely sequenced among three found (GenBank WGS accession: AQHE00000000.1).
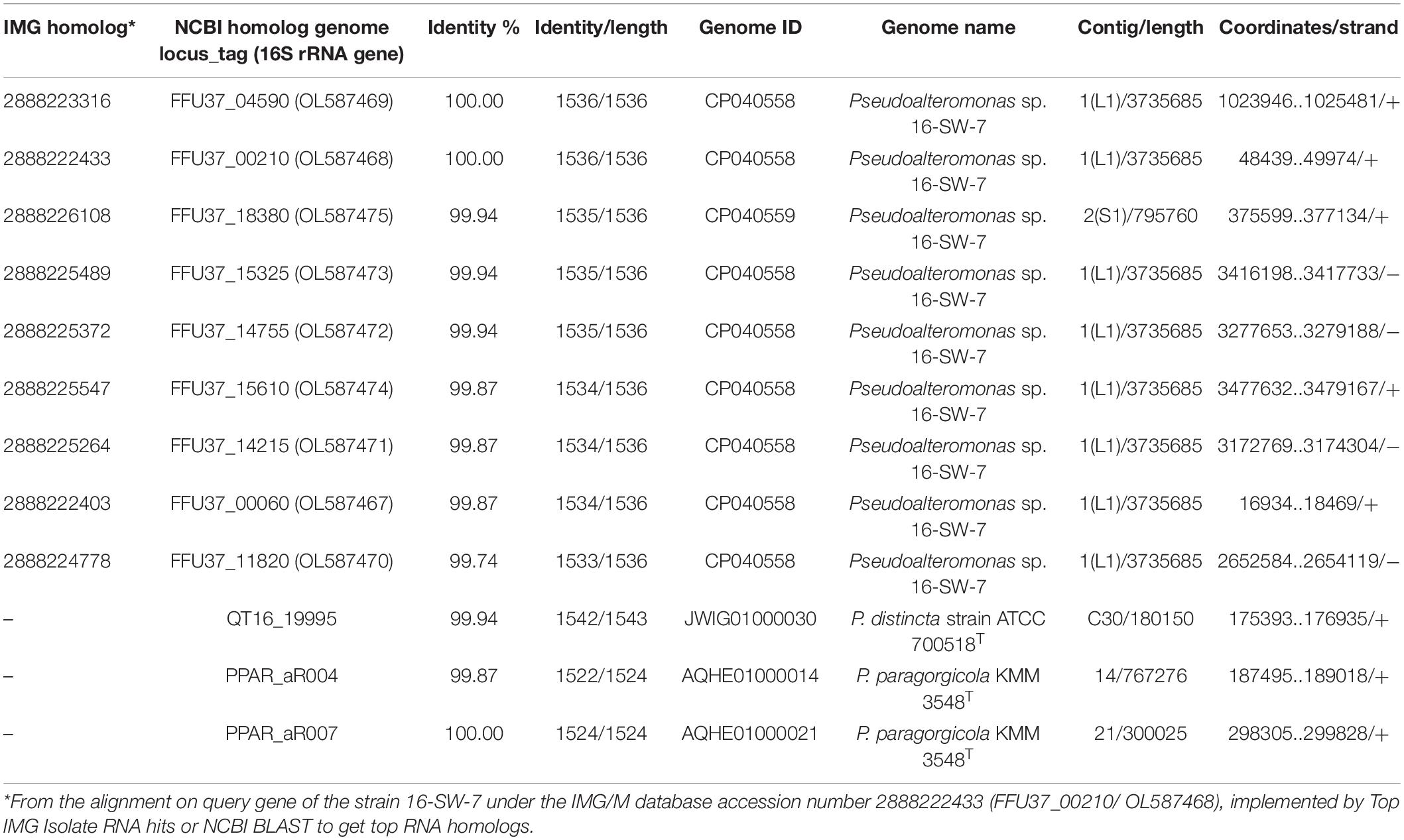
Table 3. The 16S rRNA gene sequences content and similarity for the strains Pseudoalteromonas sp. 16-SW-7, P. distincta ATCC 700518T, and P. paragorgicola KMM 3548T.
The whole-genome sequence of the strain 16-SW-7 was 4,531,445 bp, with a G+C content of 39.3 mol% and comprised of two circular chromosomes L1 and S1, deposited in the GenBank under the accession numbers CP040558 and CP040559, respectively (assembly accession: GCA_005877035.1). The genome size of P. paragorgicola KMM 3548T (GenBank assembly accession: GCA_014918315.1) was 4,322,351 bp, with the G+C content 39.2 mol%. In comparison, the genome size of P. distincta KMM 638T (GenBank assembly accession: GCA_000814675.1) was 4,532,748 bp and the G+C content of 39.2 mol% (Supplementary Table 1). The GBDP phylogenomic tree is consistent with the branching patterns, observed for only the strains 16-SW-7 and P. paragorgicola KMM 3548T (=DSM 26439T) in the 16S rRNA gene sequence-based tree, generated by TYGS, because of the use of the P. distincta KMM 638T (=ATCC 700518T) gene with lower identity (Table 3, Figure 1, and Supplementary Figure 2).
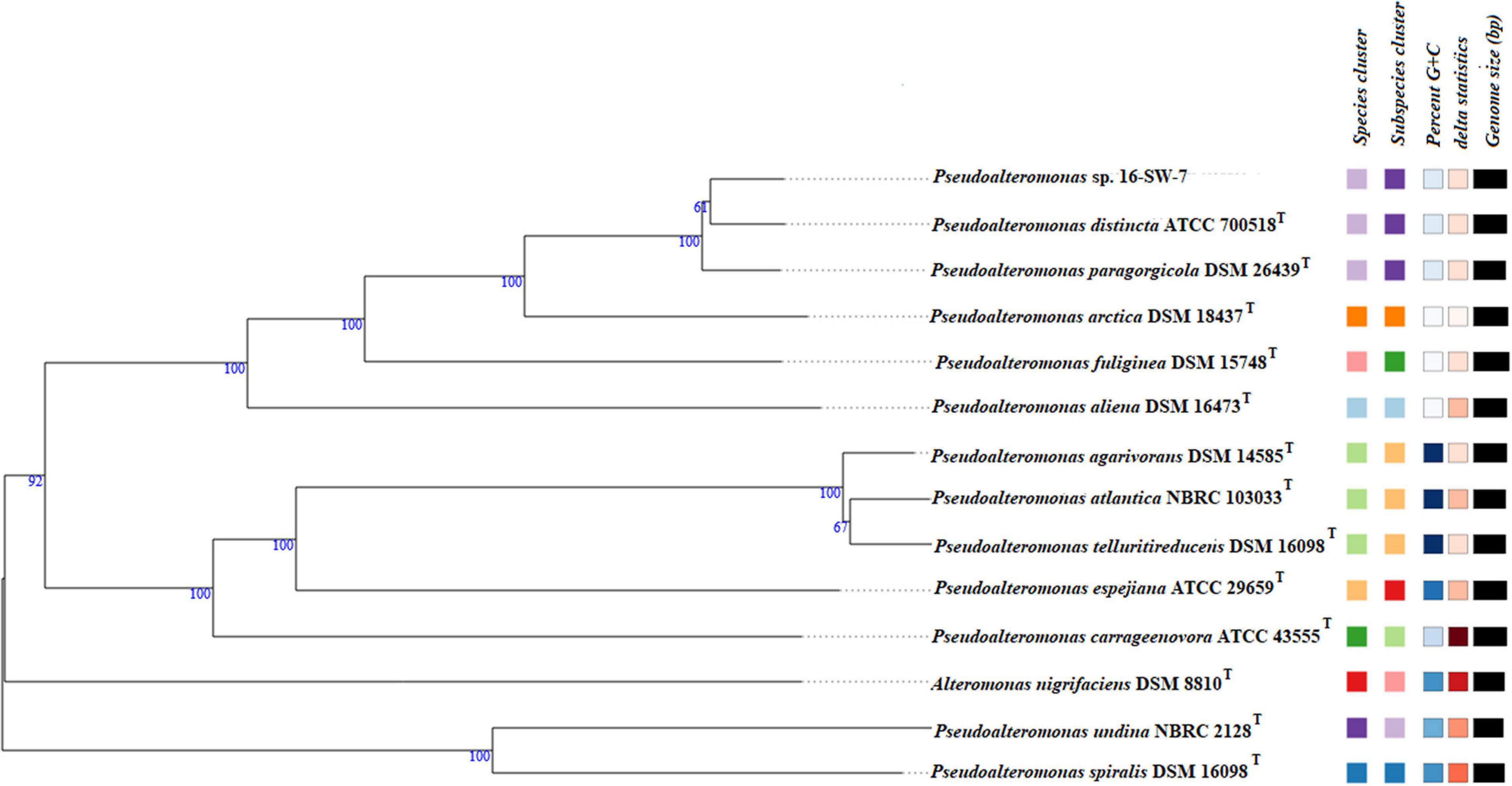
Figure 1. Tree inferred with FastME 2.1.6.1 from GBDP distances calculated from genome sequences by the TYGS server (Lefort et al., 2015). The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above the branches are GBDP pseudo-bootstrap support values >60% from 100 replications, with an average branch support of 92.7%. The tree was rooted at the midpoint (Farris, 1972). The square labels colored by the same color indicate the genomes with the same species (light lilac) and subspecies (dark lilac) gene clusters, G+C content (white), and delta statistics: Pseudoalteromonas_GCF_005877035.1 (the strain 16-SW-7), P. distincta ATCC 700518T, and P. paragorgicola DSM 26439T form one phylogenomic clade separated from other species of the genus.
However, the species-specific gene clusters for the strains 16-SW-7, P. paragorgicola KMM 3548T (=DSM 26439T), and P. distincta KMM 638T (=ATCC 700518T) were identical, indicating that the three strains belonged to a single species (Figure 1 and Supplementary Figure 2). The ANI calculator in EzBioCloud, based on the use of OrthoANIu algorithm (Lee et al., 2016), showed the similar high ANI values (98.04–98.2%) for the strain 16-SW-7 and the reference strains P. distincta KMM 638T and P. paragorgicola KMM 3548T (Supplementary Table 2), which are higher than the species-level cutoff value of 95–96% (Richter and Rosselló-Móra, 2009). According to the TYGS results, the dDDH (d4) values between the 16-SW-7 genome and genomes of P. distincta and P. paragorgicola were 84.4 and 83.5%, respectively (Supplementary Table 3), that are sufficiently higher than the suggested species boundary of 70% (Wayne et al., 1987). The proteome-wide sequence search results, implemented by a web server AAI-profiler (Medlar et al., 2018), showed 98.6 (87% matched proteins) and 98.8% (85.9% matches) average amino acid identity (AAI) of the strain 16-SW-7 with P. distincta and P. paragorgicola, which are higher than 85% proposed as a threshold for delimitation of a species (Goris et al., 2007).
The target count for P. paragorgicola proteins showed 98.8 and 98.5% AAI (83.5 and 83.1% matches) against the proteins of Pseudoalteromonas sp. 16-SW-7 and P. distincta, respectively. The calculated genetic similarities are presented in Supplementary Tables 1–3. These values support the proposed affiliation of the three strains to a single species of the genus Pseudoalteromonas.
The core-gene content (about 3200 genes according to EzBioCloud) and coding sequences (CDSs) similarity confirmed the affiliation of the strains 16-SW-7 and P. paragorgicola KMM 3548T to the species P. distincta (Supplementary Figures 3, 4 and Supplementary Tables 4–6). They include genes for use of some common mechanisms to respond to high salt stress, such as a high-affinity choline and betaine uptake system (Supplementary Table 4, Column 1: lines 769, 898, 1257, 2699, 3969–3974), the doubled genes for the glutamate synthase small and large subunits (Supplementary Table 4, Column 1: lines 1761, 3562, 2003, 2033), K+ transporters Trk H/A (Supplementary Table 4, Column 1: lines 3064, 3069), other ABC transporters/permeases, and transcription factors (Fu et al., 2014). However, analysis of the subsystem features, annotated by RAST, indicated the presence of some individual functions for the strains 16-SW-7, P. distincta KMM 638T (=ATCC 700518T), and P. paragorgicola KMM 3548T (Table 4).
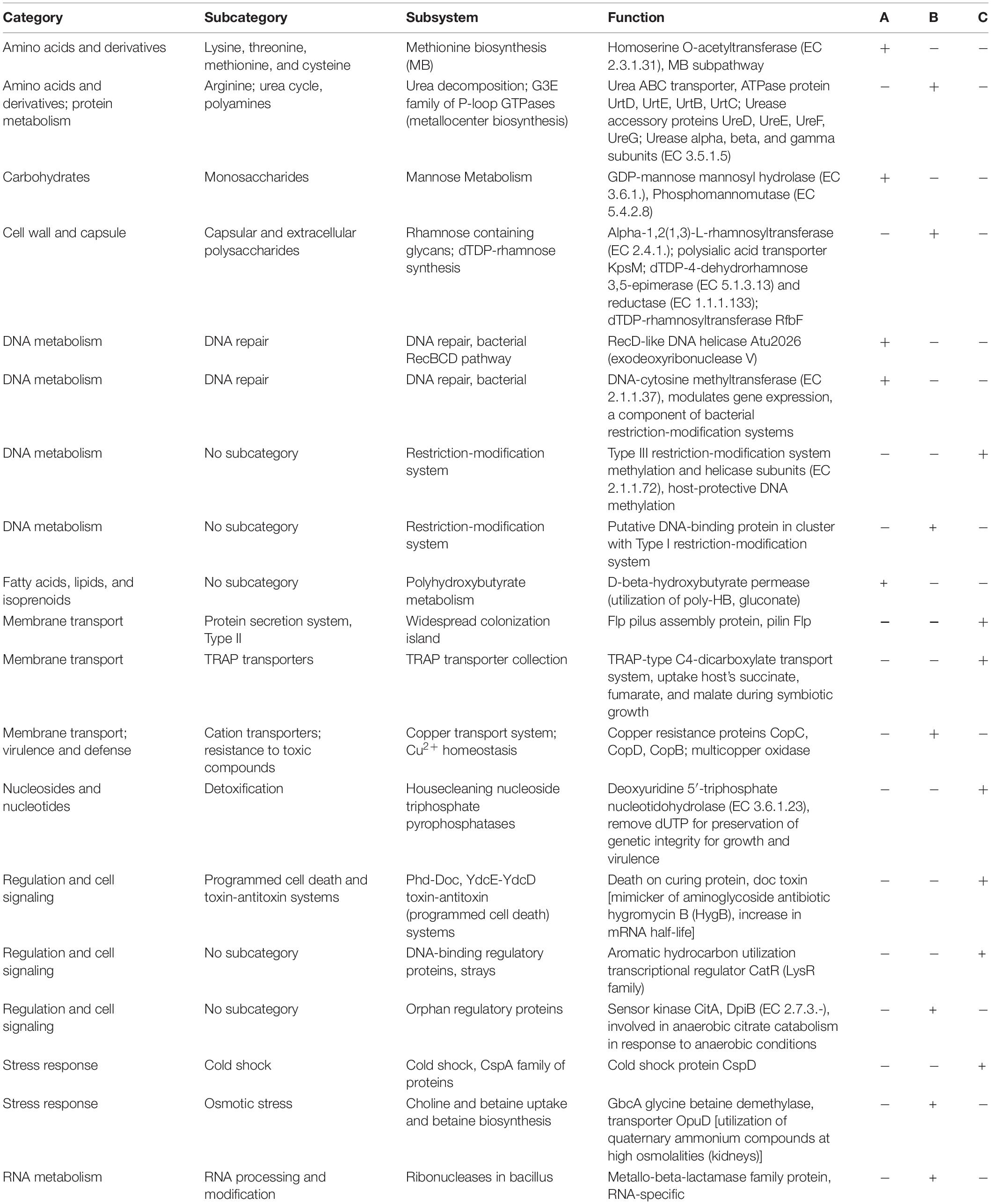
Table 4. Genome-based comparison for the presence/absence of metabolic subsystem functions in Pseudoalteromonas sp. 16-SW-7 (A), P. distincta ATCC 700518T (B), and P. paragorgicola KMM 3548T (C).
The RAST comparative genomics revealed 242 and 426 strain-specific CDSs (singletons) in the strain 16-SW-7 against the strains KMM 638T (=ATCC 700518T) and KMM 3548T, respectively, most of which encoded hypothetical proteins, and only 40 and 50 genes, respectively, had a function (Supplementary Table 4). Meanwhile, 1378 and 1229 CDSs of KMM 638T (=ATCC 700518T) and KMM 3548T, respectively, had 100% identity with the CDSs of 16-SW-7 (Supplementary Table 5). In the genome of the strain KMM 3548T, 1259 identical CDS and 481 singletons were found vs. the strain KMM 638T (ATCC 700518T), and 420 singletons vs. the strain 16-SW-7 (Supplementary Table 6). All three strains putatively have xylan and xylose degradation specialization, but differ from each other by exopolysaccharide and lipopolysaccharide biosynthesis pathways (Table 4 and Supplementary Tables 4, 6), which are known to be responsible for serotypes in clinical bacterial strains (Balabanova et al., 2020). Thus, the strain 16-SW-7 exclusively contains the doubled genes encoding for GDP-mannose mannosyl hydrolase, phosphomannomutase, and several sugar and glycosyltransferases, as well as the low-identical capsular polysaccharide synthases (up to 78–79%) of the type strain KMM 638T (ATCC 700518T): A, B, C, D, UDP-glucose 4-epimerase, lipid carrier: UDP-N-acetylgalactosaminyltransferase, O-antigen acetylase, N-acetylneuraminate cytidylyltransferase (sialic acid synthesis), polysaccharide deacetylase, dTDP-4-dehydrorhamnose reductase, and specific lipoproteins which are absent in KMM 3548T (Supplementary Tables 4, 6). Meanwhile, KMM 3548T includes many genes for rhamnosyltransferases, some of which are similar only to KMM 638T (ATCC 700518T), and related hydrolases, lipoproteins, and capsular polysaccharide synthesis and export systems (Supplementary Table 6). In addition, 16-SW-7 and KMM 638T (ATCC 700518T) differ from KMM 3548T by some genes responsible for xanthine and gluconate metabolism, carbon starvation, catechol pathway, and phosphate metabolism (Supplementary Table 4, Column 1: lines 3684–3691). Contrarily, the strain KMM 3548T has almost all genes found in the strains 16-SW-7 and ATCC 700518T, but mostly distinguishes from them by the large number of mobile elements, transposases, integrases, chaperone proteins of heat shock (HtpG), components of fatty acid synthases, enlarged pectin degradation system, and hypothetical proteins of unknown function (Supplementary Table 6). In general, the different level of identity and numbers of the genes for motility functions, metal resistance, TonB-related receptors, transporters, beta-lactamases, bactericins, signal transduction (sensors, receptors, transporters, enzymes), mobile elements, and DNA repair systems reflect eco-physiological diversity and different adaptive lifestyles of the free-living 16-SW-7, and host-associated KMM 3548T and KMM 638T (ATCC 700518T) (Table 4 and Supplementary Tables 4–6). Indeed, KMM 638T (ATCC 700518T) has acquired the additional pathogenic-like systems for osmotic stress response, urea decomposition, and sensor kinase CitA (Table 4), indicating an ability to live in the host tissues with a high osmolality and use citrate fermentation under anaerobic conditions (Wetzel et al., 2011; Scheu et al., 2012). KMM 3548T rather contains rhizobia-like symbiotic traits (Table 4), such as TRAP-type C4-dicarboxylate transport system for facultatively anaerobically fumarate and nitrate respiration (Janausch et al., 2002), aromatic hydrocarbon utilization regulator CatR, Type III restriction-modification system that confers host DNA protection via methylation (Murray et al., 2021). Meanwhile, the free-living strain 16-SW-7 is enforced by the modification and repair systems of its own DNA and methylation-dependent biosynthetic pathways, such as two bacteriocin BGCs (found by antiSMASH), defending it from UV excess, environmental competitors, and facilitating the rapid genetic rearrangement (Table 4).
In addition, the strains KMM 638T (ATCC 700518T) and KMM 3548T produce brown and lightly orange pigments, respectively, in comparison with the colorless free-living 16-SW-7 colonies, probably due to its active dye-decolorization peroxidase (Supplementary Table 4, Column 1: 2637) or inactive pigment-producing genes. P. distincta KMM 638T (ATCC 700518T) was reported to produce diffusible dark-gray-colored melanin-like pigment (pyomelanin) (Romanenko et al., 1995), the responsible genes of which are found in each strain (Supplementary Table 4, column 1: 2123, 3887). For many pathogens, melanin production is associated with virulence and completely suppressed at 30–35°C (Turick et al., 2009). According to the gene content (Supplementary Tables 4–6), P. distincta can produce different colored polyketide secondary metabolites from simple fatty acids (pigments, antioxidants, toxins), which increase virulence for many pathogens by improving its intracellular survival (Woo et al., 2012; Hug et al., 2020). Probably, KMM 638T (ATCC 700518T) and KMM 3548T developed and fixed their pigment-producing phenotypic traits due to colonizing psychrophilic marine organisms (Romanenko et al., 1995; Ivanova et al., 2002a). Thus, the slightly orange pigments in KMM 3548T could belong to antioxidant xanthomonadin-like pigments (carotenoids, anthraquinones, zeaxanthin, flexirubins), which protect against oxidative stress and UV exposure, establishing or maintaining commensal relationships between bacteria and their hosts (He et al., 2020). At least, the resorcinol and arylpolyene biosynthetic gene clusters (66209 nucleotide region), which are functionally related to antioxidative carotenoids, were found in the KMM 3548T genome by antiSMASH (Medema et al., 2011; Schöner et al., 2016).
In any case, all three genomes of P. distincta possess many polyketide synthases and concomitant proteins with a high level of identity (97–100%) (Supplementary Tables 4–6). The dark shade of pigments in KMM 638T (ATCC 700518T) may be due to the degree of expansion of the pyomelanin production and weakening xanthine metabolism by some genes lost, compared to the KMM 3548T pathway (Supplementary Table 6).
In conclusion, based on the results of the above phylogenetic, phenotypic, and chemotaxonomic study, we suggest that the strain 16-SW-7 is affiliated to the species P. distincta. Moreover, the high similarities in the genomic sequences and phenotypic characteristics found between the species P. distincta and P. paragorgicola places them in the same species. Therefore, it is proposed to reclassify the species P. paragorgicola as a later heterotrophic synonym of P. distincta in accordance with the rules of priority of prokaryotic names, governed by the International Code of Nomenclature of Bacteria (Parker et al., 2019), and to emend the description of the species P. distincta.
The description of the species Pseudoalteromonas distincta is as given by Romanenko et al. (1995) and Ivanova et al. (2000, 2002a) with the following modifications and amendments. Cells are Gram-stain-negative, non-spore-forming, strictly aerobic rods, motile by means of a single polar or four to seven lateral flagella. On marine agar, colonies are non-pigmented or slightly orange colored. They can produce diffusible melanin-like pigments. Cells are catalase- and oxidase-positive. They require Na+ ions or sea water for growth. Growth occurs in media with 0.5–10% NaCl. Temperature for growth ranges from 4 to 34°C. Aesculin, gelatin, Tweens 20, 40, and 80, DNA, alginate, and tyrosine are hydrolyzed but agar, chitin, CM-cellulose, and urea are not hydrolyzed. Hydrolysis of casein and starch is strain dependent. Acid is formed from sucrose but not from L-arabinose, D-fructose, D-mannose, D-melibiose, L-rhamnose, D-ribose, D-trehalose, N-acetylglucosamine, and glycerol. Some strains can produce acid from D-cellobiose, D-galactose, D-glucose, D-lactose, maltose, D-raffinose, D-xylose, and D-mannitol and utilize citrate. In API ID 32GN gallery, they are positive for D-glucose, maltose, sucrose, D-mannitol, sodium acetate, sodium citrate, L-alanine, L-serine, L-proline, glycogen, propionic acid, valeric acid, and capric acids. Assimilation of inositol, sodium malonate, lactic acid, D-ribose, 3-hydroxybutyric acid, itaconic acid, potassium-2-keto-gluconate, L-histidine, and salicin is variable. In API ZYM gallery, alkaline phosphatase, esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase activities are present but lipase (C14), α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities are absent. Esterase (C4), cysteine arylamidase, trypsin, and α-chymotrypsin can be produced. Nitrate is not reduced to nitrite. Acetoin and indole are not produced. Production of hydrogen sulfide is strain dependent. The predominant fatty acids (>5% of the total fatty acids) were C16:1 ω7c, C16:0, C17:1 ω8c, C18:1 ω7c, C17:0, and C12:0 3-OH. The polar lipid profile was characterized by the presence of phosphatidylethanolamine, phosphatidylglycerol, two unidentified amino lipids, and three unidentified lipids. The major respiratory quinone is ubiquinone Q-8. The genomic DNA G+C content is 39.2–39.3 mol%. The genome size is 4.3–4.5 Mb. The type of strain is KMM 638T (=ATCC 700518T), isolated from a marine sponge collected at a depth of 350 m near the Komandorskie Islands, Russia. The GenBank/EMBL/DDBJ assembly accession number for the genome of the type of strain is GCA_000814675.1.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ON and LB contributed to conception and designed of the study. ON, S-GK, LB, and NZ performed the experimental works. ON, S-GK, LB, NZ, OS, LT, and VM wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This research was funded by a grant from the Ministry of Science and Higher Education, Russian Federation 15.BRK.21.0004 (Contract No. 075-15-2021-1052).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.809431/full#supplementary-material
Bakunina, I. Y., Sova, V. V., Nedashkovskaya, O. I., Kuhlmann, R. A., Likhosherstov, L. M., Martynova, M. D., et al. (1998). α-Galactosidase of the marine bacterium Pseudoalteromonas sp. KMM 701. Biochemistry 63, 1209–1215.
Balabanova, L. A., Bakunina, I. Y., Nedashkovskaya, O. I., Makarenkova, I. D., Zaporozhets, T. S., Besednova, N. N., et al. (2010). Molecular characterization and therapeutic potential of a marine bacterium Pseudoalteromonas sp. KMM 701 α-galactosidase. Mar. Biotechnol. 12, 111–120. doi: 10.1007/s10126-009-9205-2
Balabanova, L., Shkryl, Y., Slepchenko, L., Cheraneva, D., Podvolotskaya, A., Bakunina, I., et al. (2020). Genomic features of a food-derived Pseudomonas aeruginosa strain PAEM and biofilm-asociated gene expression under a marine bacterial α-galactosidase. Int. J. Mol. Sci. 21:7666. doi: 10.3390/ijms21207666
Beurmann, S., Ushijima, B., Svoboda, C. M., Videau, P., Smith, A. M., Donachie, S. P., et al. (2017). Pseudoalteromonas piratica sp. nov., a budding, prosthecate bacterium from diseased Montipora capitata, and emended description of the genus Pseudoalteromonas. Int. J. Syst. Evol. Microbiol. 67, 2683–2688. doi: 10.1099/ijsem.0.001995
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bowman, J. P. (1998). Pseudoalteromonas prydzensis sp. nov., a psychrotrophic, halotolerant bacterium form Antarctic sea ice. Int. J. Syst. Bacteriol. 48, 1037–1041. doi: 10.1099/00207713-48-3-1037
Bruns, A., Rohde, M., and Berthe-Corti, L. (2001). Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int. J. Syst. Evol. Microbiol. 51, 1997–2006. doi: 10.1099/00207713-51-6-1997
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chen, I.-M. A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., et al. (2021). The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763. doi: 10.1093/nar/gkaa939
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Collins, M. D., and Shah, H. M. (1984). Fatty acid, menaquinone and polar lipid composition of Rothia dentocariosa. Arch. Microbiol. 137, 247–249. doi: 10.1007/BF00414552
Egan, S., Holmstrom, C., and Kjelleberg, S. (2001). Pseudoalteromonas ulvae sp. nov., a bacterium with antifouling activities isolated from the surface of a marine alga. Int. J. Syst. Evol. Microbiol. 51, 1499–1504. doi: 10.1099/00207713-51-4-1499
Farris, J. S. (1972). Estimating phylogenetic trees from distance matrices. Am. Nat. 106, 645–667. doi: 10.1086/282802
Fu, X., Liang, W., Du, P., Yan, M., and Kan, B. (2014). Transcript changes in Vibrio cholerae in response to salt stress. Gut Pathog. 6:47. doi: 10.1186/s13099-014-0047-8
Gauthier, G., Gauthier, M., and Christen, R. (1995). Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45, 755–761. doi: 10.1099/00207713-45-4-755
Gerhardt, P., Murray, R. G. E., Wood, W. A., and Krieg, N. R. (eds) (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
He, Y.-W., Cao, X.-Q., and Poplawsky, A. R. (2020). Chemical structure, biological roles, biosynthesis and regulation of the yellow xanthomonadin pigments in the phytopathogenic genus Xanthomonas. Mol. Plant Microbe Interact. 33, 705–714. doi: 10.1094/MPMI-11-19-0326-CR
Hug, J. J., Krug, D., and Müller, R. (2020). Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 4, 172–193. doi: 10.1038/s41570-020-0176-1
Hwang, C. Y., Lee, I., Hwang, Y. J., Yoon, S. J., Lee, W. S., and Cho, B. C. (2016). Pseudoalteromonas neustonica sp. nov., isolated from the sea surface microlayer of the Ross Sea (Antarctica), and emended description of the genus Pseudoalteromonas. Int. J. Syst. Evol. Microbiol. 66, 3377–3382. doi: 10.1099/ijsem.0.001202
Ivanova, E. P., Chun, J., Romanenko, L. A., Matte, M. E., Mikhailov, V. V., Frolova, G. M., et al. (2000). Reclassification of Alteromonas distincta Romanenko et al., 1995 as Pseudoalteromonas distincta comb. nov. Int. J. Syst. Evol. Microbiol. 50, 141–144. doi: 10.1099/00207713-50-1-141
Ivanova, E. P., Flavier, S., and Christen, R. (2004). Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54, 1773–1788. doi: 10.1099/ijs.0.02997-0
Ivanova, E. P., Sawabe, T., Lysenko, A. M., Gorshkova, N. M., Hayashi, K., Zhukova, N. V., et al. (2002a). Pseudoalteromonas translucida sp. nov. and Pseudoalteromonas paragorgicola sp. nov., and emended description of the genus. Int. J. Syst. Evol. Microbiol. 52, 1759–1766. doi: 10.1099/00207713-52-5-1759
Ivanova, E. P., Sawabe, T., Lysenko, A. M., Gorshkova, N. M., Svetashev, V. I., Nicolau, D. V., et al. (2002b). Pseudoalteromonas ruthenica sp. nov., isolated from marine invertebrates. Int. J. Syst. Evol. Microbiol. 52, 235–240. doi: 10.1099/00207713-52-1-235
Ivanova, E. P., Shevchenko, L. S., Sawabe, T., Lysenko, A. M., Svetashev, V. I., Gorshkova, N. M., et al. (2002c). Pseudoalteromonas maricaloris sp. nov., isolated from an Australian sponge, and reclassification of [Pseudoalteromonas aurantia] NCIMB 2033 as Pseudoalteromonas flavipulchra sp. nov. Int. J. Syst. Evol. Microbiol. 52, 263–271. doi: 10.1099/00207713-52-1-263
Janausch, I. G., Zientz, E., Tran, Q. H., Kröger, A., and Unden, G. (2002). C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta Bioenerg. 1553, 39–56. doi: 10.1016/s0005-2728(01)00233-x
Komagata, K., and Suzuki, K.-I. (1988). Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207. doi: 10.1016/S0580-9517(08)70410-0
Kreft, L., Botzki, A., Coppens, F., Vandepoele, K., and Van Bel, M. (2017). PhyD3: a phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 33, 2946–2947. doi: 10.1093/bioinformatics/btx324
Lagesen, K., and Hallin, P. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Lemos, M. L., Toranzo, A. E., and Barja, J. L. (1985). Modified medium for oxidation-fermentation test in the identification of marine bacteria. Appl. Environ. Microbiol. 40, 1541–1543. doi: 10.1128/aem.49.6.1541-1543.1985
Machado, H., Vynne, N. G., Christiansen, G., and Gram, L. (2016). Reclassification of Alteromonas fuliginea (Romanenko et al., 1995) as Pseudoalteromonas fuliginea comb. nov. and an emended description. Int. J. Syst. Evol. Microbiol. 66, 3737–3742. doi: 10.1099/ijsem.0.001259
Matsuyama, H., Minami, H., Kasahara, H., Kato, Y., Murayama, M., and Yumoto, I. (2013). Pseudoalteromonas arabiensis sp. nov., a marine polysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 63, 1805–1809. doi: 10.1099/ijs.0.043604-0
Medema, M. H., Blin, K., Cimermancic, P., De Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucl. Acids Res. 39(Suppl. 2), 339–346. doi: 10.1093/nar/gkr466
Medlar, A. J., Törönen, P., and Holm, L. (2018). AAI-profiler: fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification and contamination. Nucleic Acids Res. 46, W479–W485. doi: 10.1093/nar/gky359
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Hahnke, R. L., Petersen, J., Scheuner, C., Michael, V., Fiebig, A., et al. (2014). Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic Sci. 9:2. doi: 10.1186/1944-3277-9-2
Murray, I. A., Luyten, Y. A., Fomenkov, A., Dai, N., Corrêa, I. R. Jr., Farmerie, W. G., et al. (2021). Structural and functional diversity among Type III restriction-modification systems that confer host DNA protection via methylation of the N4 atom of cytosine. PLoS One 16:e0253267. doi: 10.1371/journal.pone.0253267
Navarro-Torre, S., Carro, L., Rodriguez-Llorente, I. D., Pajuelo, E., Caviedes, M. A., Igual, J. M., et al. (2020). Pseudoalteromonas rhizosphaerae sp. nov., a novel plant growth-promoting bacterium with potential use in phytoremediation. Int. J. Syst. Evol. Microbiol. 70, 3287–3294. doi: 10.1099/ijsem.0.004167
Nedashkovskaya, O. I., Kim, S. B., Shin, D. S., Beleneva, I. A., Vancanneyt, M., and Mikhailov, V. V. (2007). Echinicola vietnamensis sp. nov., a new bacterium of the phylum Bacteroidetes isolated from sea water. Int. J. Syst. Evol. Microbiol. 57, 761–763. doi: 10.1099/ijs.0.64546-0
Ondov, B. D., Treangen, T. J., Melsted, P., Mallonee, A. B., Bergman, N. H., Koren, S., et al. (2016). Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 17:132. doi: 10.1186/s13059-016-0997-x
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Park, S., Jung, Y. T., Park, D. S., and Yoon, J. H. (2016). Pseudoalteromonas aestuariivivens sp. nov., isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 66, 2078–2083. doi: 10.1099/ijsem.0.000995
Park, Y. D., Baik, K. S., Yi, H., Bae, K. S., and Chun, J. (2005). Pseudoalteromonas byunsanensis sp. nov., isolated from tidal flat sediment in Korea. Int. J. Syst. Evol. Microbiol. 55, 2519–2523. doi: 10.1099/ijs.0.63750-0
Parker, C. T., Tindall, B. J., and Garrity, G. M. (2019). International code of nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 69, S1–S111. doi: 10.1099/ijsem.0.000778
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Romanenko, L. A., Mikhailov, V. V., Lysenko, A. M., and Stepanenko, V. I. (1995). A new species of melanin-producing bacteria of the genus Alteromonas. Mikrobiologiya 64, 74–77.
Romanenko, L. A., Zhukova, N. V., Lysenko, A. M., Mikhailov, V. V., and Stackebrandt, E. (2003b). Assignment of ‘Alteromonas marinoglutinosa’ NCIMB 1770 to Pseudoalteromonas mariniglutinosa sp. nov., nom. rev., comb. nov. Int. J. Syst. Evol. Microbiol. 53, 1105–1109. doi: 10.1099/ijs.0.02564-0
Romanenko, L. A., Zhukova, N. V., Rohde, M., Lysenko, A. M., Mikhailov, V. V., and Stackebrandt, E. (2003a). Pseudoalteromonas agarivorans sp. nov., a novel marine agarolytic bacterium. Int. J. Syst. Evol. Microbiol. 53, 125–131. doi: 10.1099/ijs.0.02234-0
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20, 1–6.
Sawabe, T., Tanaka, R., Iqbal, M. M., Tajima, K., Ezura, Y., Ivanova, E. P., et al. (2000). Assignment of Alteromonas elyakovii KMM 162T and five strains isolated from spot-wounded fronds of Laminaria japonica to Pseudoalteromonas elyakovii comb. nov. and the extended description of the species. Int. J. Syst. Evol. Microbiol. 50, 265–271. doi: 10.1099/00207713-50-1-265
Scheu, P. D., Witan, J., Rauschmeier, M., Graf, S., Liao, Y. F., Ebert-Jung, A., et al. (2012). CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo. J. Bacteriol. 194, 636–645. doi: 10.1128/JB.06345-11
Schöner, T. A., Gassel, S., Osawa, A., Tobias, N. J., Okuno, Y., Sakakibara, Y., et al. (2016). Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. Chembiochem. 17, 247–253. doi: 10.1002/cbic.201500474
Turick, C. E., Beliaev, A. S., Zakrajsek, B. A., Reardon, C. L., Lowy, D. A., Poppy, T. E., et al. (2009). The role of 4-hydroxyphenylpyruvate dioxygenase in enhancement of solid-phase electron transfer by Shewanella oneidensis MR-1. FEMS Microbiol. Ecol. 68, 223–235. doi: 10.1111/j.1574-6941.2009.00670.x
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., and Krichevsky, M. I, et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464. doi: 10.1099/00207713-37-4-463
Wetzel, K. J., Bjorge, D., and Schwan, W. R. (2011). Mutational and transcriptional analyses of the Staphylococcus aureus low-affinity proline transporter OpuD during in vitro growth and infection of murine tissues. FEMS Immunol. Med. Microbiol. 61, 346–355. doi: 10.1111/j.1574-695X.2011.00781.x
Woo, P. C. Y., Lam, C.-W., Tam, E. W. T., Leung, C. K. F., Wong, S. S. Y., Lau, S. K. P., et al. (2012). First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl. Trop. Dis. 6:e1871. doi: 10.1371/journal.pntd.0001871
Wu, Y. H., Cheng, H., Xu, L., Jin, X. B., Wang, C. S., and Xu, X. W. (2017). Physiological and genomic features of a novel violacein-producing bacterium isolated from surface seawater. PLoS One 12:e0179997. doi: 10.1371/journal.pone.0179997
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: marine bacteria, taxonogenomics, phenotype, emended description of Pseudoalteromonas distincta, Pseudoalteromonas paragorgicola
Citation: Nedashkovkaya OI, Kim S-G, Balabanova LA, Zhukova NV, Son OM, Tekutyeva LA and Mikhailov VV (2022) Genome-Based Classification of Strain 16-SW-7, a Marine Bacterium Capable of Converting B Red Blood Cells, as Pseudoalteromonas distincta and Proposal to Reclassify Pseudoalteromonas paragorgicola as a Later Heterotypic Synonym of Pseudoalteromonas distincta. Front. Microbiol. 12:809431. doi: 10.3389/fmicb.2021.809431
Received: 08 November 2021; Accepted: 23 December 2021;
Published: 08 February 2022.
Edited by:
Damien Paul Devos, Andalusian Center for Development Biology, Spanish National Research Council (CSIC), SpainReviewed by:
Stefanie P. Glaeser, University of Giessen, GermanyCopyright © 2022 Nedashkovkaya, Kim, Balabanova, Zhukova, Son, Tekutyeva and Mikhailov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga I. Nedashkovkaya, b2xnYW5lZGFzaGtvdnNrYUBwaWJvYy5kdm8ucnU=; b25lZDIwMDRAbWFpbC5ydQ==; Larissa A. Balabanova, YmFsYWJhbkBwaWJvYy5kdm8ucnU=; bGJhbGFiYW5vdmExQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.