
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 February 2022
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.807512
This article is part of the Research Topic Insights in Microbial Symbioses: 2021 View all 7 articles
 Lulu Han1†
Lulu Han1† Wanchao Xue2†
Wanchao Xue2† Hanwen Cao3
Hanwen Cao3 Xiaoying Chen3
Xiaoying Chen3 Fasheng Qi4
Fasheng Qi4 Tao Ma1
Tao Ma1 Yan Tu1
Yan Tu1 Qiyu Diao1
Qiyu Diao1 Chengfu Zhang3*
Chengfu Zhang3* Kai Cui1,3*
Kai Cui1,3*
Rumen microbiota are closely linked to feed utilization and environmental adaptability of ruminants. At present, little is known about the influence of different extreme environments on the rumen microbiota of yaks. In this study, 30 ruminal fluid samples from 30 healthy female yaks (average 280 kg of BW) in 5–8 years of life were collected from three regions in Tibet, China, and compared by gas chromatography and high-throughput sequencing. Results showed that propionic acid, butyric acid, and total volatile fatty acids were significantly (p < 0.05) higher, while microbial abundance and diversity were significantly (p < 0.05) lower, in the Nagqu (4,500 m altitude) compared with the Xigatse (4,800 m altitude) and Lhasa (3,800 m altitude) regions. Principal coordinate analysis revealed significant (p < 0.05) differences in rumen microbial composition of yaks from different regions. Specifically, Bacteroidetes and Firmicutes were identified by linear discriminant analysis effect size (LDA > 3) as being the signature phyla for Xigatse and Nagqu regions, respectively. In addition, the relative abundance of Rikenellaceae_RC9_gut_group, Quinella, Prevotellaceae_UCG-003, Lachnospiraceae_NK3A20_group, Papillibacter, Ruminococcaceae_UCG-010, Prevotellaceae_NK3B31_group, and Ruminococcaceae_UCG-005 correlated with altitude and rumen fermentation parameters (p < 0.05). Finally, the predicted function of rumen microbiota was found to differ between regions (p < 0.05). In summary, our results reveal that regions located at different altitudes influence microbiota composition and fermentation function of yaks’ rumen. The present findings can provide mechanistic insights on yak adaptation to high altitudes and improve the feeding efficiency of these animals in extreme regions.
Yaks (Bos grunniens) are long-haired ruminants living in high-altitude regions (>3,000 m) such as the Tibetan Plateau in China (Guo et al., 2015). About 90% of the world population of yaks is found in China (Li et al., 2018), where these iconic animals provide meat, milk, wool, and fuel for the local nomadic people (Liu et al., 2008; Long et al., 2008; Qiu et al., 2012; Gao et al., 2013; Zhang J. et al., 2020), who call yaks “boats of the plateau” (Liu et al., 2019).
Owing to the extreme environmental conditions of the Tibetan Plateau and long-term natural selection, yaks have developed a tolerance for cold, low oxygen, rough feeding, and hardship (Long et al., 2008; Qiu et al., 2012). Different environments lead to different metabolic strategies, which allow animals to adapt and minimize the adverse effects of environmental changes (Whiteley and Faulkner, 2005; Oliveira et al., 2021). Recent evidence suggests that such effect is largely due to the microbiota in the digestive tract, which play an important role in nutrient intake, metabolism, and immune response of the host (Furman et al., 2020; O’Hara et al., 2020; Bi et al., 2021). Consequently, the correlation between the adaptability of yaks to environmental changes and their gastrointestinal microbiota composition has attracted increasing attention. Guo et al. (2021) found that the fecal microbiota of yaks reflected seasonal dietary changes, allowing yaks to make better use of low-protein roughage. Whereas fecal microbiota composition of yaks differs across regions, its functional profile is more uniform (Liu et al., 2021). The rumen is one of the most important digestive organs of ruminants and is characterized by an abundance of microbiota (Guan et al., 2008; Khan et al., 2016). Indeed, owing to their microbiota, ruminants can ferment solid feed into volatile fatty acids (VFAs), which provide 70% of the required energy (Anantasook et al., 2013; Khan et al., 2016). At the same time, rumen microbiota and VFAs are the main factors explaining different feed efficiencies among individual cattle (Liang et al., 2017; Zhou et al., 2018; Zhang Y. et al., 2020). Although Wu et al. (2021) found that altitude affected the composition of rumen microbiota in yaks, it remains to be determined how microbiota and VFAs vary under extreme conditions (>3,000 m altitude).
To understand the response of yak rumen microbiota and VFAs to different altitudes, the present study employed microbiome technology to characterize rumen composition in yaks living at different altitudes in three extreme regions of Tibet. We hypothesized that rumen fermentation parameters and microbiota of yaks would change dynamically with increasing altitude.
The experimental protocol was approved by the Chinese Academy of Agricultural Sciences Animal Ethics Committee, and all procedures were performed in accordance with humane animal care and handling (AEC-CAAS-20190905).
Three study regions, including Zhongba County, Xigatse City (84°03′ E, 29°77′ N; about 4,800 m altitude, HAL), Nagqu City (92°07′ E, 31°48′ N; about 4,500 m altitude, MAL), and Dangxiong County, Lhasa City (91°05′ E, 30°51′ N; about 3,800 m altitude, LAL), were selected. The three regions are key breeding grounds for yaks; their average annual air temperature is −4.0°C, −0.5°C, and 2.0°C, while precipitation is 280, 433, and 485 mm, respectively.
In September 2019, ten healthy female yaks (average 280 kg of BW) in 5–8 years of life were randomly selected from every study region. At the study regions, yaks are allowed to live free-range, follow their dams, and are not given any artificial feed, relying instead on typical alpine meadows as the main type of vegetation.
Before morning grazing, ruminal chyme (30 samples in total) was collected through the mouth of yaks using an esophageal tube as described previously (Ramos-Morales et al., 2014). Next, the chyme was filtered through a four-layer woven gauze to collect the ruminal fluid, which was immediately frozen in liquid nitrogen (−80°C) for subsequent microbiota and fermentation characterization. The pH value was detected using a portable pH meter (206-pH1; Testo). For VFA analysis, three tubes containing 1 ml of rumen fluid were taken from each sample after thawing and centrifugation at 20,000 × g and 4°C for 15 min. Then, 0.25 ml metaphosphoric acid (25 g/100 ml) was added per tube and the sample was analyzed by gas chromatography equipping a megabore HP-MOLSIV column (film thickness: 30 m × 0.53 mm × 25 μm) (SP-3420A; Beifenrili Analyzer Associates, Beijing, China) (Yue et al., 2009).
Microbial DNA was extracted from rumen fluid samples (30 samples in total) using PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer’s guidelines. The concentration and purity of the extracted DNA were measured by a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The V3–V4 region of the bacterial 16S rRNA gene was amplified using primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Takahashi et al., 2014). PCR amplification proceeded through an initial denaturing step at 94°C for 5 min; 28 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s; and a final extension at 72°C for 7 min using 25-μl reaction volumes, containing 12.5 μl 2 × Taq PCR MasterMix, 3 μl BSA (2 ng/μl), 1 μl forward primer (5 μM), 1 μl reverse primer (5 μM), 2 μl template DNA, and 5.5 μl ddH2O. The PCR products were detected by 2% agarose gel electrophoresis, purified with the Agencourt AMPure XP kit (Beckman Coulter, La Brea, CA, United States) according to the manufacturer’s instructions, and quantified by QuantiFluor-ST (Promega, Madison, WI, United States). Purified amplicons were sequenced on an Illumina MiSeq-PE300 platform (Illumina Inc., San Diego, CA, United States), generating 2 × 300 bp paired-end reads.
The obtained paired-end reads in the original DNA fragments were merged using Flash version 1.20 (Mago and Salzberg, 2011), and then each sample was separated according to a unique barcode. After removing barcodes, primers, and splice variants, raw reads were obtained. To generate high-quality reads, a specific sliding window strategy was adopted for Trimmomatic version 0.36, the window size was set to 50 bp, the average quality value was 20, and the minimum reserved sequence length was 120 bp; in addition, sequences containing N were removed by Pear version 0.9.6 (Bolger et al., 2014). Afterward, Flash and Pear were used to merge the sequences at both ends according to PE overlap correlation, the minimum overlap was set to 10 bp, and the mismatch rate was 0.1, allowing for fasta sequences to be obtained. Finally, the chimera containing fasta sequences was eliminated by comparison with the Gold database using the UCHIME algorithm, whereas the unknown database was removed by de novo means (Knight, 2011). Simultaneously, the short sequences, failing to meet the requirements, were removed, while high-quality sequences of clean reads were obtained. Subsequently, these clean tags were clustered into operational taxonomic units (OTUs) based on a 97% sequence similarity threshold using the UPARSE algorithm in Vsearch version 2.7.1. In each OTU, the richest sequence was filtered as the representative sequence (Edgar, 2013). To derive species classification information corresponding to each OTU, the representative sequences were compared and analyzed by the RDP Classifier algorithm version 2.2 (Wang, 2007) and Silva database1 (Quast et al., 2012), allowing community annotation at kingdom, phylum, class, order, family, and genus level. After the sample number of the lowest sequence was flattened, alpha diversity (Chao1, Shannon, Simpson, and observed species) was calculated by QIIME 2, while intergroup alpha index variability was demonstrated by the Kruskal–Wallis test in R version 4.0.2 (Miller et al., 2016). Principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity matrices and QIIME 2 was used to calculate differences in bacterial communities among groups. The linear discriminant analysis effect size (LEfSe, LDA > 3) was used to identify significant bacteria among the three groups (Miller et al., 2016). PICRUSt2 software was used to predict microbiota function and explore differences among the three groups (Douglas et al., 2019).
Rumen fermentation parameters were analyzed using one-way ANOVA and Duncan’s multiple comparison in SPSS 22.0 software (IBM, Chicago, IL, United States). Differences in alpha diversity, relative abundance at phylum, family, and genus level, as well as microbiota function among the three groups were tested using the Kruskal–Wallis method in R version 4.0.3. PCoA, Venn, column, and LEfSe results were visualized using the “ape” and “ggplot2,” “limma,” “ggplot2,” and “ggtree” packages in R, respectively. The “corrplot” package in R was used to analyze the correlation between the top 20 genera of all samples, rumen fermentation parameters, and altitude (based on Spearman’s coefficient). The network containing the top 20 genera was visualized using the “igraph” package in R version 4.0.3. All data are reported as means, and intergroup differences of p < 0.05 were considered significant.
As reported in Table 1, rumen fermentation parameters differed significantly between the three groups (p < 0.05), whereas the pH value had no difference between the three groups (p > 0.05). Also, the MAL group exhibited the highest (p < 0.05) total VFA content and molar proportions of propionic and butyric acids, whereas the molar proportion of acetic acid and A/P (acetic acid/propionic acid) were lower (p < 0.05) than in other groups. Interestingly, rumen fermentation parameters were similar between HAL and LAL groups (p > 0.05).
Gradual stabilization of the OTU rank curve chart indicates high coverage of test samples (Supplementary Figure 1). The rumen microbiota consisted of 5,491,884 high-quality reads, with an average of 183,063 reads per sample. A total of 7,977 OTUs were obtained based on 97% nucleotide sequence identification: 6,895 OTUs in HAL, 5,544 in MAL, and 6,109 in LAL groups (Figure 1A). Of these, 4,219 OTUs (52.89% of the total) were shared among samples from different groups. Alpha diversity calculations (Table 2) revealed significantly higher indices of Shannon and community richness (Chao1 and observed species) in HAL and LAL groups compared with the MAL group (p < 0.05), whereas no significant difference was detected between HAL and LAL groups (p > 0.05). Interestingly, we found Simpson index in HAL significantly higher (p < 0.05) than MAL group, whereas no significant difference was detected between LAL and other groups (p > 0.05). PCoA plots of bacterial structure profiles (Figure 1B) revealed distinct clustering of the three regions. The segregation and dissimilarities observed at OTU level were revealed using Bray–Curtis matrices (PERMANOVA, p < 0.001).
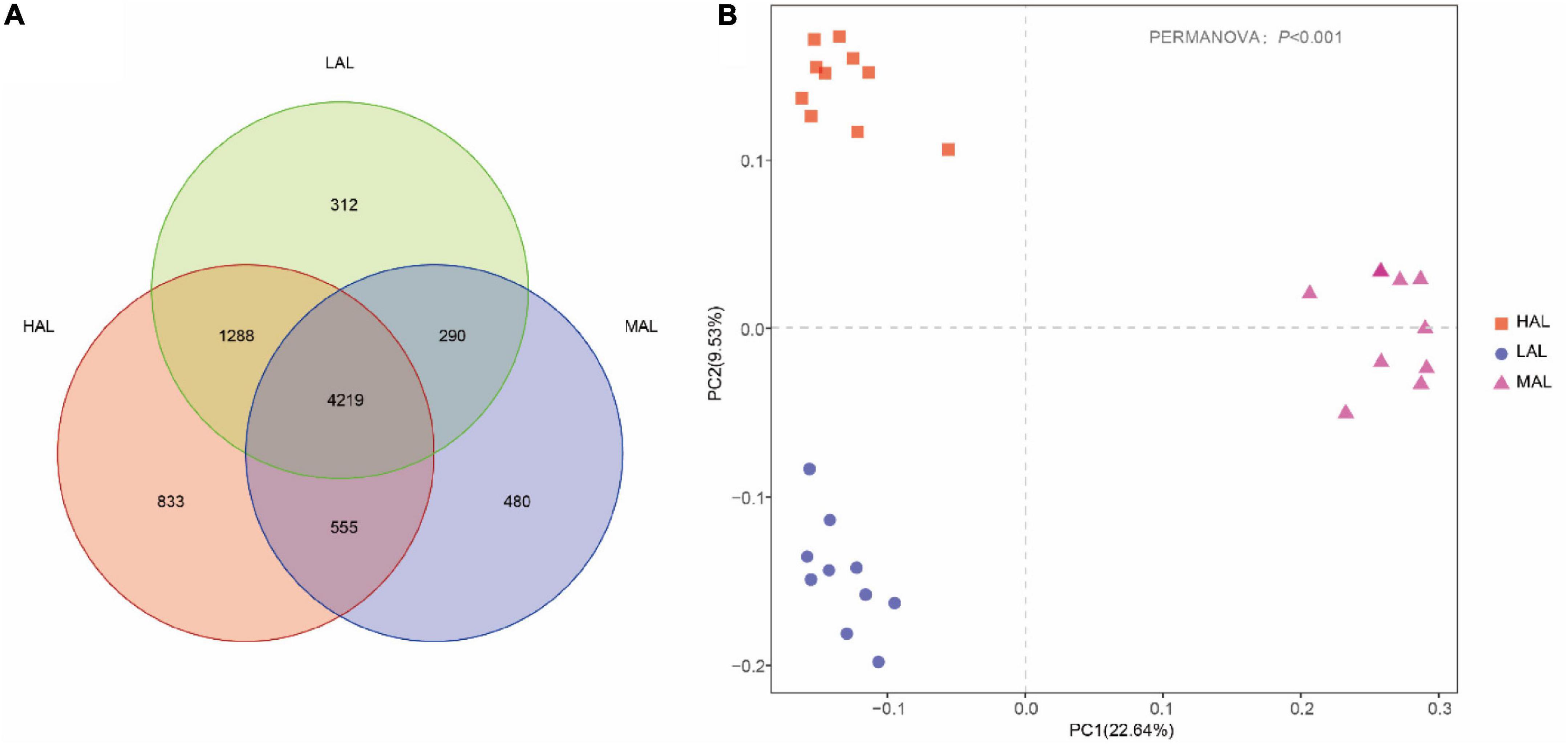
Figure 1. Response of rumen microbiota from yaks among the three regions. (A) Composition of rumen microbiota from yaks inhabiting regions located at distinct altitudes (OTU-level analysis). (B) PCoA analysis of rumen microbiota from yaks inhabiting regions located at distinct altitudes. HAL, high-altitude region (Zhongba County, Xigatse City; 4,800 m); MAL, medium-altitude region (Nagqu City; 4,500 m); LAL, low-altitude region (Dangxiong County, Lhasa City; 3,800 m).
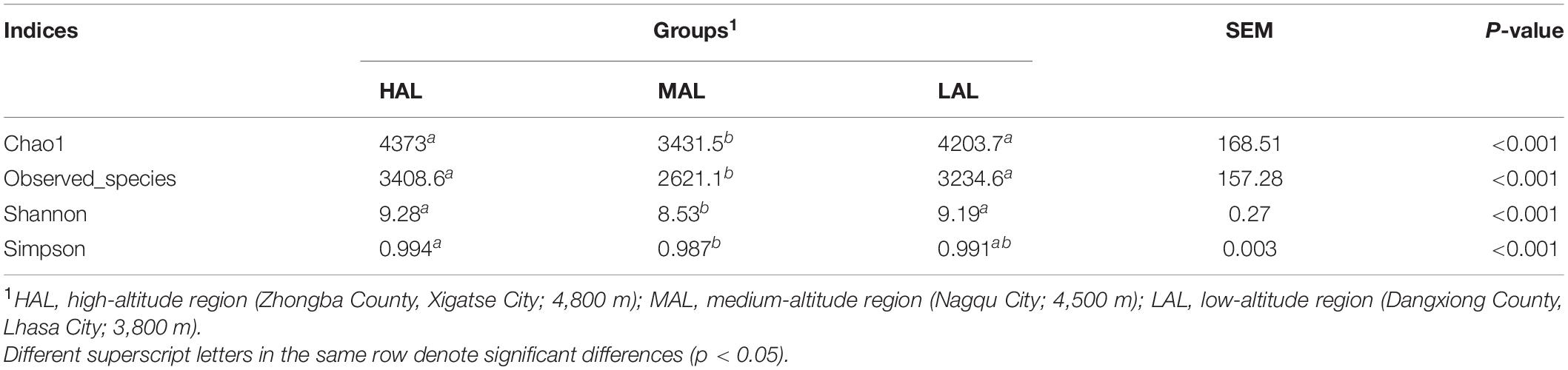
Table 2. Effect of regions located at distinct altitudes on alpha diversity of yak rumen microbiota.
Next, we proceeded with bacterial identification of the 30 samples at different levels. At the phylum level, the top five ruminal bacteria of yaks across all samples were Bacteroidetes (50.69%), Firmicutes (41.11%), Proteobacteria (1.45%), Tenericutes (1.40%), and Actinobacteria (1.05%) (Figure 2A and Supplementary Table 1). The MAL group presented the lowest (p < 0.05) relative abundances of Bacteroidetes and Verrucomicrobia, but the highest (p < 0.05) relative abundance of Firmicutes compared with the HAL and LAL groups. At the family level, Prevotellaceae (24.67%) was the most abundant taxon, followed by Ruminococcaceae (14.76%) and Rikenellaceae (11.67%) across all samples; other families included Lachnospiraceae (9.15%), Christensenellaceae (8.42%), Bacteroidales_BS11_gut_group (7.20%), Veillonellaceae (4.05%), and Bacteroidales_S24-7_group (3.44%) (Figure 2B and Supplementary Table 2). Notably, the relative abundances of Lachnospiraceae, Christensenellaceae, and Veillonellaceae were higher (p < 0.05) in the MAL compared with other groups, whereas the relative abundance of Rikenellaceae increased significantly (p < 0.05) according to the following order: MAL < LAL < HAL. At the genus level, the dominant genera across all groups were Prevotella_1 (16.82%), Rikenellaceae_RC9_gut_group (11.26%), and Christensenellaceae_R-7_group (8.06%) (Figure 2C and Supplementary Table 3). Specifically, the MAL group displayed the highest (p < 0.05) relative abundances of Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214 _group, Quinella, Ruminococcaceae_UCG-005, Olsenella, Butyrivibrio_2, Lachnospiraceae_NK3A20_group, and Acetitomaculum, but the lowest (p < 0.05) relative abundances of Prevotellaceae_UCG-003, Rikenellaceae_RC9_gut_group, Eubacterium_coprostanoligenes_group, Saccharofermentans, and Prevotellaceae_NK3B31_group compared with the LAL and HAL groups. The relative abundance of Ruminococcaceae_UCG-010 was significantly (p < 0.05) higher in the HAL group compared with the MAL and LAL groups, whereas the relative abundance of Papillibacter was higher (p < 0.05) in the LAL group than in the other groups.
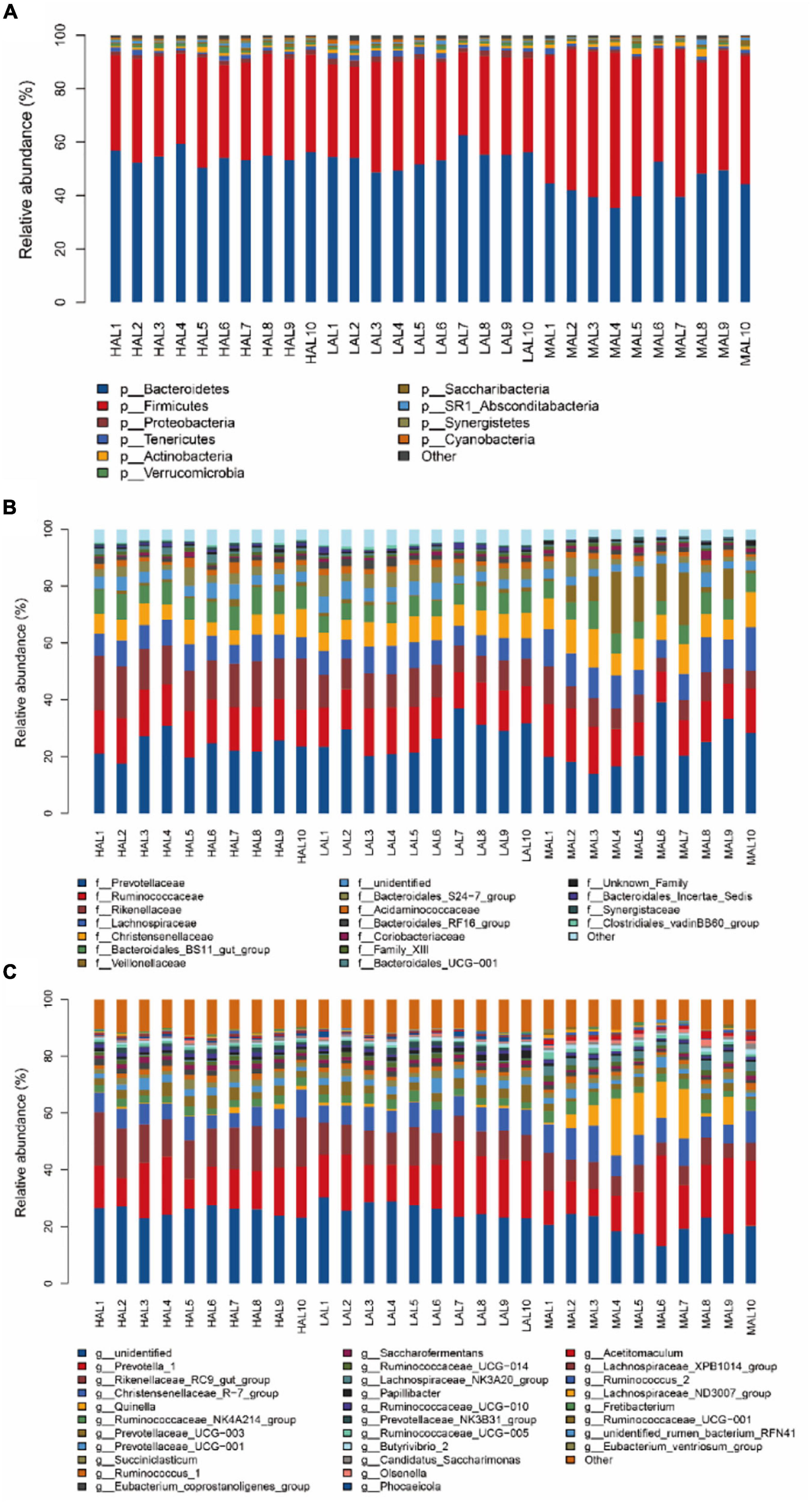
Figure 2. Compositional profiles of rumen microbiota from yaks inhabiting regions located at distinct altitudes. (A) Phylum level, (B) family level, and (C) genus level. HAL, high-altitude region (Zhongba County, Xigatse City; 4,800 m); MAL, medium-altitude region (Nagqu City; 4,500 m); LAL, low-altitude region (Dangxiong County, Lhasa City; 3,800 m).
To better understand the dominance of specific bacteria within the three groups, we used the LEfSe method (Figure 3). The Firmicutes and Saccharibacteria phyla, including genera Quinella, Christensenellaceae_R_7_group, Lachnospiraceae _NK3A20_group, Acetitomaculum, Olsenella, Ruminococcaceae _UCG_005, Butyrivibrio_2, and Ruminococcus_2, were abundant in the MAL group. The Bacteroidetes, Verrucomicrobia, and Synergistetes phyla, including Rikenellaceae_RC9_gut _group, Saccharofermentans, Lachnospiraceae_AC2044_group, Ruminococcaceae_UCG_010, Anaerovorax, and Family_XIII, were enriched in the HAL group. Finally, the Papillibacter, Tenericutes, Cyanobacteria, and Bacteroidales_S24_7_group phyla, including Prevotellaceae_UCG_003, Prevotellaceae_NK3 B31_group, Papillibacter, Anaeroplasma, and Lachnospiraceae _UCG_006, were over-represented in the LAL group.
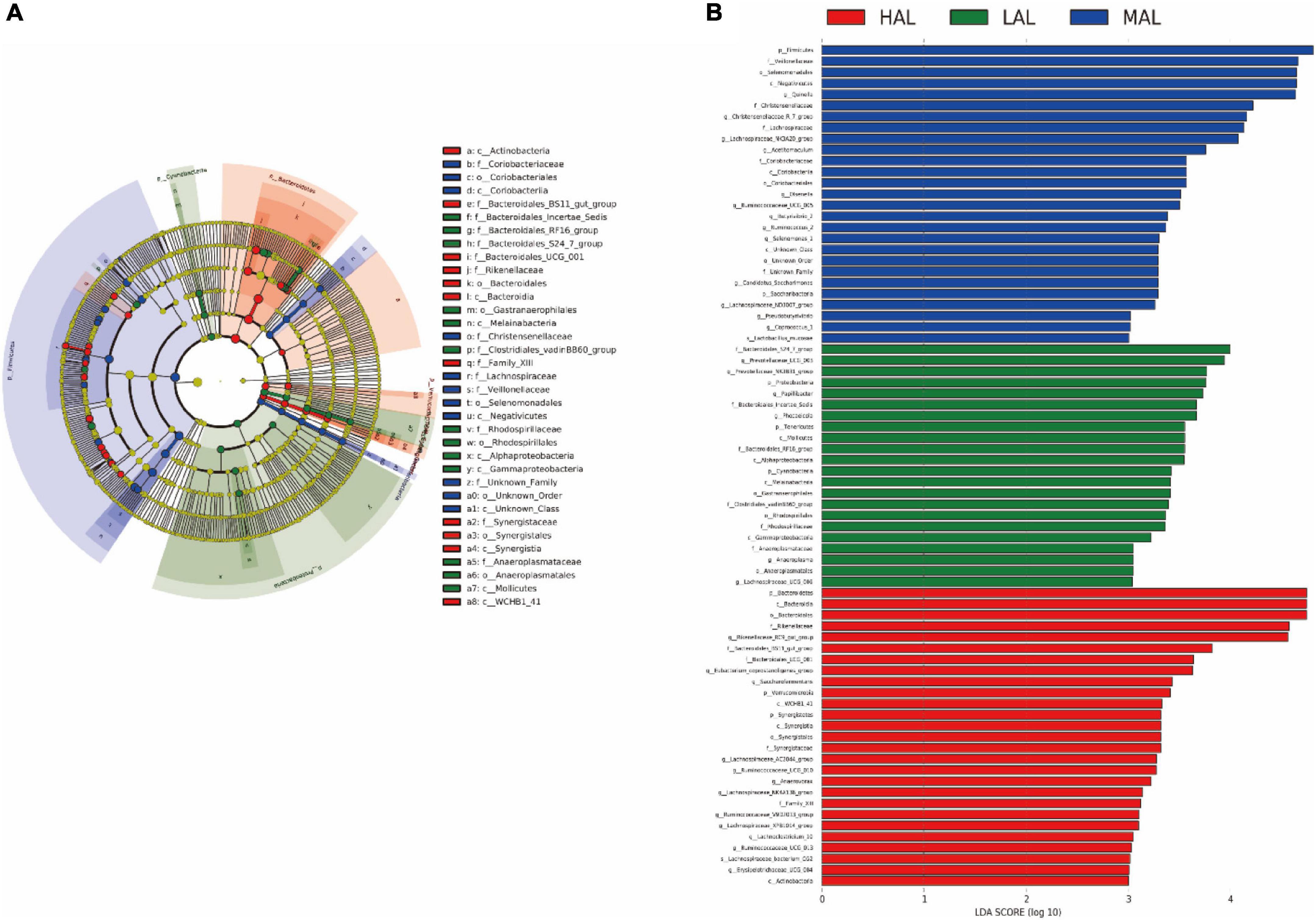
Figure 3. LEfSe analysis of rumen microbiota from yaks inhabiting regions located at distinct altitudes. (A) Histogram of linear discriminant analysis scores based on Classification Information. (B) Linear discriminant analysis effect size cladogram based on Classification Information. HAL, high-altitude region (Zhongba County, Xigatse City; 4,800 m); MAL, medium-altitude region (Nagqu City; 4,500 m); LAL, low-altitude region (Dangxiong County, Lhasa City; 3,800 m).
The correlation between relative abundance of the top 20 bacterial genera, fermentation parameters, and altitude (Figure 4A) revealed that the relative abundances of Rikenellaceae_RC9_gut_group, Quinella, Prevotellaceae_UCG-003, Lachnospiraceae_NK3A20_group, Papillibacter, Ruminococcaceae_UCG-010, Prevotellaceae_NK3B31_group, and Ruminococcaceae_UCG-005 correlated with altitude and rumen fermentation (p < 0.05). For example, the relative abundance of Rikenellaceae_RC9_gut_group correlated positively with altitude (r = 0.84) and A/P (r = 0.47), whereas the relative abundance of Quinella correlated negatively with altitude (r = −0.49) and butyric acid (r = −0.43). The relative abundance of Papillibacter correlated positively with altitude (r = 0.41), total VFA (r = 0.62), propionic acid (r = 0.45), and butyric acid (r = 0.62), but negatively with acetic acid (r = −0.52) and A/P (r = −0.48). Interestingly, we found a significant negative correlation between Papillibacter and Ruminococcaceae_UCG-005 (|r|> 0.6, p < 0.05; Figure 4B).
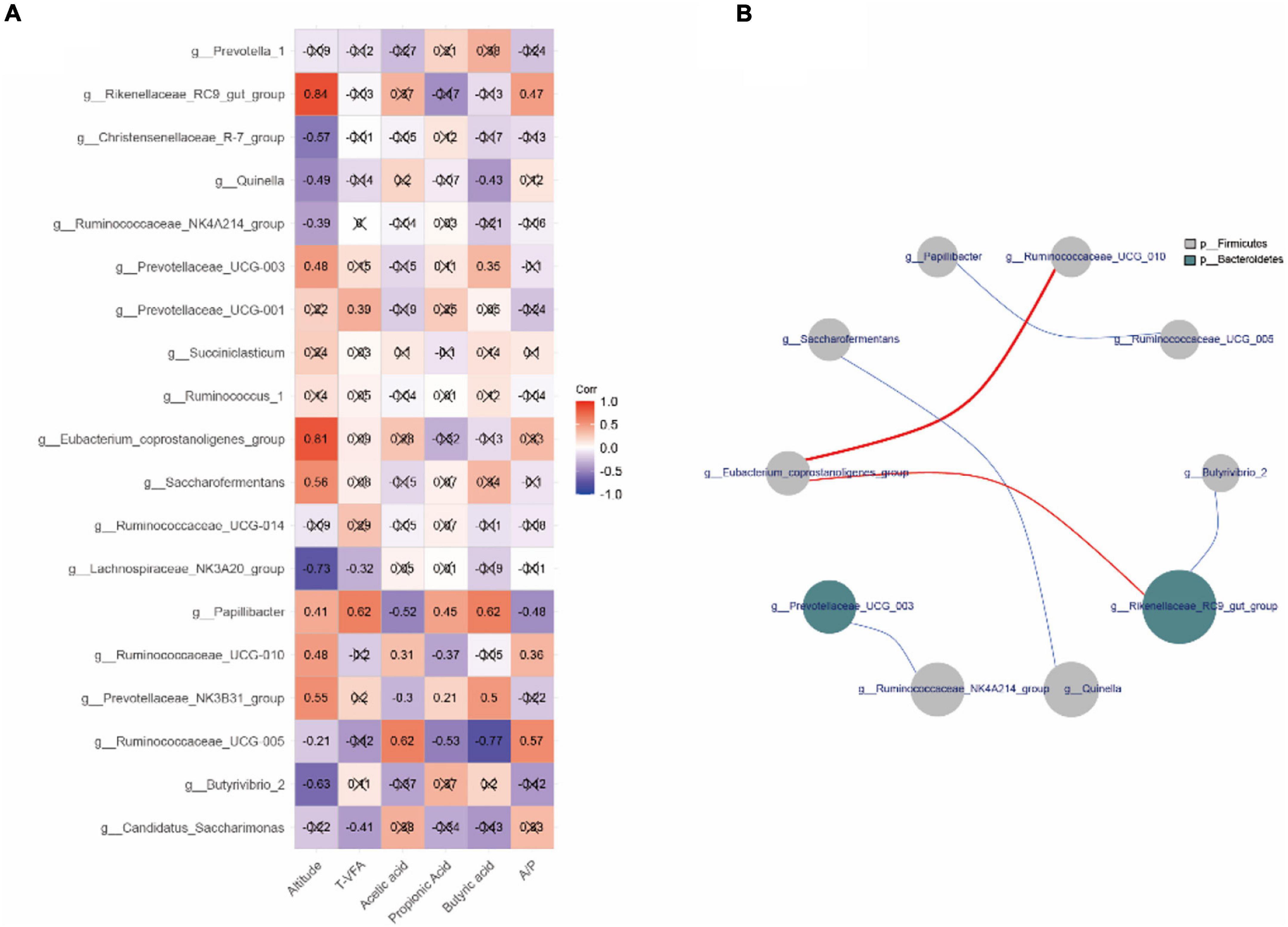
Figure 4. Correlation analysis of rumen genera in yaks inhabiting regions located at distinct altitudes. (A) Correlation between the top 20 relative abundances of genera, fermentation parameters, and altitude. (B) Interaction among the top 20 relative abundances at genus level. Red represents a positive correlation and blue a negative correlation. The number represents the correlation coefficient. The × sign represents p > 0.05. The size and color of the circle represent the relative abundance of the respective genus and the phylum.
The function of rumen microbial communities in yaks from three different regions was predicted by PICRUSt2 software and related to KEGG pathways (Table 3). Notably, “Biosynthesis of ansamycins” (4.47%) was the most abundant pathway in all three groups, followed by “Biosynthesis of vancomycin group antibiotics” (2.47%), “Valine, leucine and isoleucine biosynthesis” (2.25%), and “D-Glutamine and D-glutamate metabolism” (2.19%). The Kruskal–Wallis test confirmed that a total of 25 pathways showed significant (p < 0.05) differences among the three groups (Table 3). Specifically, “Amino acid metabolism,” “D-Alanine metabolism,” and “Thiamine metabolism” pathways, as well as “Carbohydrate metabolism,” “Cell motility,” and “Folding, sorting and degradation” gene categories were significantly (p < 0.05) more abundant in the MAL than in the other two groups.
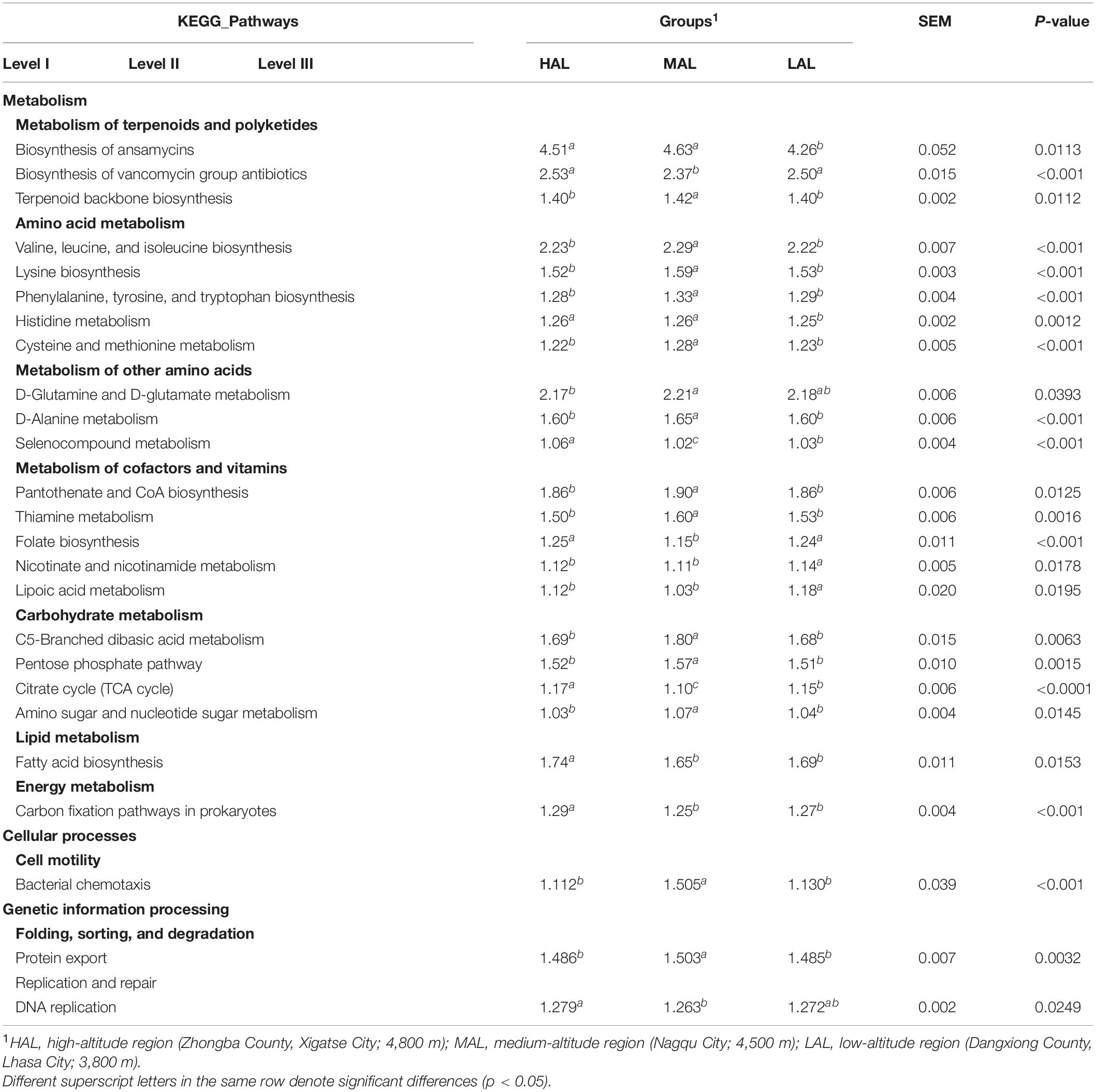
Table 3. Functional predictions for rumen microbiota in yaks inhabiting regions located at distinct altitudes; only significantly different KEGG pathways are shown.
This is the first study to evaluate the influence of extreme environments, corresponding to different altitudes of the Tibetan Plateau, on rumen microbiota in yaks. The study highlights how rumen fermentation, bacterial composition, and function are related to the regions in which the yaks live.
Ruminants rely on VFAs produced in the rumen during fermentation as a source of energy. Indeed, their feed efficiency is related to such VFAs (Lam et al., 2018; Zhou et al., 2018). While propionic acid can supply additional energy to ruminants, acetic and butyric acids are converted to methane and carbon dioxide during the consumption process, lowering energy efficiency (Gunun et al., 2018). Propionic acid and butyric acid compete with methane for hydrogen during fermentation, reducing methane energy consumption while improving energy supply (Pragna et al., 2018). Acetic acid and butyric acid originate mainly from the fermentation of fiber, whereas propionic acid derives from the fermentation of sugar and starch (Astawa et al., 2011). In this study, VFAs and propionic acid were highest in the rumen of MAL animals, suggesting that feed efficiency might be better in this than in the other groups. Contrary to the present results, Zhang et al. (2016) found that ruminants living at elevated altitude had higher VFA content and significantly upregulated genes responsible for VFA absorption in the rumen. Diet is the most important factor regulating development and colonization of rumen microbiota (Zhou et al., 2018; Lin et al., 2019; Myer, 2019). Ram (2005) reported that meadows found at different elevation in the Uttaranchal Himalaya exhibited distinct species richness. Similarly, Bao et al. (2021) linked the alpine meadows between Dangxiong and Nagqu regions of Tibet with a varied nutritional value. Regional variation could be related to temperature differences between day and night, photosynthesis, and topography (Han et al., 1997; Skapetas et al., 2004). Accordingly, further experiments are required to ascertain whether the observed differences are due to the extreme climatic conditions and altitude tested in this study or to the quality of grass in the different regions. Furthermore, the alpha and beta diversity indices of rumen microbiota differed significantly between the three groups, indicating a close relationship between the diversity of rumen microbiota in yaks and geographic location. Interestingly, unlike VFAs, the diversity of ruminal microbiota first decreased and then increased with elevation. Shabat et al. (2016) found that microbiota diversity correlated negatively with feed efficiency, with efficient dairy cattle exhibiting lower rumen microbiota diversity, methane emission, and acetic acid concentration, but higher propionic acid content. Taking into account those and the present results, it appears that yaks in the MAL group are more efficient at utilizing their pasture and emit less methane.
As observed previously (Zhang et al., 2016; Guo et al., 2021; Liu et al., 2021), Bacteroidetes and Firmicutes were the dominant taxa identified in this study. Guo et al. (2020) reported that Bacteroidetes were the most important bacteria in yak rumen from birth to adulthood. Here, we found that Firmicutes were the signature bacteria of the MAL group as they were significantly more abundant than in the other two groups. Because Firmicutes play an important role in energy conversion (Turnbaugh et al., 2006), our results indicated that they might be the key enablers of high feed utilization by yaks in the MAL group. Furthermore, the relative abundance of Papillibacter was found to correlate positively with altitude, whereas Ruminococcaceae_UCG_005 showed a negative correlation. Mao et al. (2013) found that dairy cattle had lower levels of Papillibacter when in a state of subacute ruminal acidosis, which indirectly suggests that yaks may be better suited to living at higher altitudes. Ruminococcaceae_UCG_005 enterotype has been associated with yak diets rich in proteins but low in fibers (Guo et al., 2021). Combined with the correlation results of VFAs, we hypothesize that Papillibacter and Ruminococcaceae_UCG_005 may be the hub genera allowing for adaptation to high altitude. By antagonizing each other, they may allow yaks to adjust to different altitudes through modulation of the ruminal VFA content. The genera Rikenellaceae_RC9_gut_group, Quinella, Prevotellaceae_UCG-003, Lachnospiraceae_NK3A20_group, Prevotellaceae_NK3B31_group, and Ruminococcaceae_UCG-005 exhibited a correlation with different altitudes and rumen fermentation parameters, suggesting that they, too, may play a key part in the adaptation to extreme environments. As there are other reports pointing to the involvement of altitude-related genera in VFA production, rumen development, immunity, and methane output (Petri et al., 2013; Zhai et al., 2017; Pragna et al., 2018; Lin et al., 2019; Sha et al., 2021), we believe that altitude affects the microbial composition of yaks’ rumen and enables adaptation to extreme environments by modulating rumen fermentation products.
The functional properties of microbiota dictate the host–microbiome interaction (Lin et al., 2019; Myer, 2019). Here, the PICRUSt2 result revealed that the function of yak rumen microbiota differed between the three regions. In contrast, Liu et al. (2021) found a similar functional gene composition of yak intestinal microbes across the three regions, suggesting that the yak rumen may be more sensitive than the intestinal tract in terms of environmental adaptability. Due to limited experimental conditions, this study did not further explore how diet and extreme environment affected microbiota composition beyond the correlation with altitude. The mechanism regulating adaptation between yak genome and extreme environment has been described previously (Qiu et al., 2012), and the microbiome is closely related to animal activities (Wu et al., 2011). Therefore, future metagenomics studies may help untangle the contribution of gut microorganisms to mammalian adaptation to extreme environments.
This study has analyzed the rumen microbiota and fermentation products of yaks living at different altitudes in Tibet, China. Indeed, rumen fermentation, composition, and function were found to vary across regions, with some genera correlating strongly with altitude. Based on the results, we found the middle-altitude region was suitable for rumen fermentation end-products and microbiota composition compared with the low- or high-altitude regions in Tibet. These results provide an insight on the mechanism enabling yaks to adapt to high altitude and maximize feed efficiency in extreme environments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://submit.ncbi.nlm.nih.gov/subs/, PRJNA776716.
The animal study was reviewed and approved by the Chinese Academy of Agricultural Sciences Animal Ethics Committee approved the experimental protocol, and all the methods conducted in this experiment were in accordance with humane animal care and handling procedures (AEC-CAAS-20190905). Written informed consent was obtained from the owners for the participation of their animals in this study.
WX: conceptualization. KC: methodology, supervision, and funding acquisition. LH: software, resources, data curation, and writing—original draft preparation. YT, QD, and XC: validation. HC: formal analysis and visualization. FQ: investigation. LH, WX, TM, KC, and CZ: writing—review and editing. KC and CZ: project administration. All authors contributed to the article and approved the submitted version.
This research was funded by the State Key Laboratory of Hulless Barley and Yak Germplasm Resources and Genetic Improvement (XZNKY-2020-C-007K02). The Key R&D Program of Tibet Autonomous Region (XZ201901NB01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Shuo Wang from the College of Animal Science and Technology, China Agricultural University, for his guidance on the article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.807512/full#supplementary-material
Anantasook, N., Wanapat, M., Cherdthong, A., and Gunun, P. (2013). Changes of microbial population in the rumen of dairy steers as influenced by plant containing tannins and saponins and roughage to concentrate ratio. Asian Australas J. Anim. Sci. 26, 1583–1591. doi: 10.5713/ajas.2013.13182
Astawa, P. A., Partama, I., Suyadnya, P., and Sutarpa, I. (2011). Effect of vitamin - mineral supplementation in commercial feed on the digestibility coefficient and rumen fermentation of bali cattle. J. Indones Trop Anim. 36, 69–74. doi: 10.14710/jitaa.36.1.69-74
Bao, Y. H., Wang, W. B., Yuan, X. J., Pubu, Z. M., Luo, Z., and Can, M. Y. (2021). Comparative study on nutritive value of Kobresia tibetica of alpine meadow in dangxiong and naqu regions of qinghai-tibet plateau. Chin. J. Anim. Nutr. 33, 6233–6244. doi: 10.3969/j.issn.1006-267x.2021.11.024
Bi, Y. L., Tu, Y., Zhang, N. F., Wang, S. Q., Zhang, F., Suen, G., et al. (2021). Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut 70, 853–864. doi: 10.1136/gutjnl-2020-320951
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Douglas, G. M., Maffei, V. J., Zaneveld, J., Yurgel, S. N., and Langille, M. (2019). PICRUSt2: an improved and extensible approach for metagenome inference. BioRxiv [preprint]. *BioRxiv. 672295, doi: 10.1101/672295
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Furman, O., Shenhav, L., Sasson, G., Kokou, F., Honig, H., Jacoby, S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11:1904. doi: 10.1038/s41467-020-15652-8
Gao, J., Liu, M., and Meng, X. (2013). Seroprevalence of bovine viral diarrhea infection in yaks (bos grunniens) on the qinghai-tibetan plateau of China. Trop Anim. Health Pro. 45, 791–793. doi: 10.1007/s11250-012-0290-2
Guan, L. L., Nkrumah, J. D., Basarab, J. A., and Moore, S. S. (2008). Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 288, 85–91. doi: 10.1111/j.1574-6968.2008.01343.x
Gunun, P., Gunun, N., Cherdthong, A., Wanapat, M., Polyorach, S., Sirilaophaisan, S., et al. (2018). In vitro rumen fermentation and methane production as affected by rambutan peel powder. J. Appl. Anim. Res. 46, 626–631. doi: 10.1080/09712119.2017.1371608
Guo, N., Wu, Q., Shi, F., Niu, J., Zhang, T., Degen, A. A., et al. (2021). Seasonal dynamics of diet-gut microbiota interaction in adaptation of yaks to life at high altitude. NPJ Biofilms Microbiomes. 7:38. doi: 10.1038/s41522-021-00207-6
Guo, W., Zhou, M., Ma, T., Bi, S., Wang, W., Zhang, Y., et al. (2020). Survey of rumen microbiota of domestic grazing yak during different growth stages revealed novel maturation patterns of four key microbial groups and their dynamic interactions. Anim Microbiome. 2, 23. doi: 10.1186/s42523-020-00042-8
Guo, X., Pei, J., Bao, P. J., Chu, M., Wu, X. Y., Ding, X. Z., et al. (2015). The complete mitochondrial genome of the qinghai plateau yak bos grunniens (cetartiodactyla: bovidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 2889–2890. doi: 10.3109/19401736.2015.1060423
Han, F., Chen, G. Y., and Shi, S. B. (1997). Contents of protein, fat and starch of kobresia humilis plants grown at different altitude in qinghai-xizang plateau. Acta Ecol. Sin. 21, 97–104.
Khan, M. A., Bach, A., Weary, D. M., and von Keyserlingk, M. A. G. (2016). Invited review: transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 99, 885–902. doi: 10.3168/jds.2015-9975
Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Lam, S., Munro, J. C., Zhou, M., Guan, L. L., Schenkel, F. S., Steele, M. A., et al. (2018). Associations of rumen parameters with feed efficiency and sampling routine in beef cattle. Animal 12, 1442–1450. doi: 10.1017/S1751731117002750
Li, K., Mehmood, K., Zhang, H., Jiang, X., Shahzad, M., Dong, X., et al. (2018). Characterization of fungus microbial diversity in healthy and diarrheal yaks in gannan region of tibet autonomous prefecture. Acta Tropica 182, 14–26. doi: 10.1016/j.actatropica.2018.02.017
Liang, Y. S., Li, G. Z., Li, X. Y., Lv, J. Y., Li, F. D., Tang, D. F., et al. (2017). Growth performance, rumen fermentation, bacteria composition, and gene expressions involved in intracellular pH regulation of rumen epithelium in finishing hu lambs differing in residual feed intake phenotype. J. Anim. Sci. 95, 1727–2738. doi: 10.2527/jas.2016.1134
Lin, L., Xie, F., Sun, D. M., Liu, J. H., Zhu, W. Y., and Mao, S. Y. (2019). Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 7:83. doi: 10.1186/s40168-019-0701-y
Liu, C., Wu, H., Liu, S., Chai, S., Meng, Q., and Zhou, Z. (2019). Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 10:1116. doi: 10.3389/fmicb.2019.01116
Liu, J., Cai, J. Z., Zhang, W., Chen, D., and Liu, Q. R. (2008). Seroepidemiology of neospora caninum and toxoplasma gondii infection in yaks (bos grunniens) in qinghai, china. Vet. Parasitol. 152, 330–332. doi: 10.1016/j.vetpar.2007.12.010
Liu, W. W., Wang, Q., Song, J. J., Xin, J. W., Zhang, S. S., Lei, Y. H., et al. (2021). Comparison of gut microbiota of yaks from different geographical regions. Front. Microbiol. 12:666940. doi: 10.3389/fmicb.2021.666940
Long, R. J., Ding, L. M., Shang, Z. H., and Guo, X. H. (2008). The yak grazing system on the qinghai-tibetan plateau and its status. Rangeland J. 30, 241–246.
Mago, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mao, S. Y., Zhang, R. Y., Wang, D. S., and Zhu, W. Y. (2013). Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe 24, 12–19. doi: 10.1016/j.anaerobe.2013.08.003
Miller, G. E., Engen, P. A., Gillevet, P. M., Maliha, S., Masoumeh, S., Forsyth, C. B., et al. (2016). Lower neighborhood socioeconomic status associated with reduced diversity of the colonic microbiota in healthy adults. PLoS One 11:e148952. doi: 10.1371/journal.pone.0148952
Myer, P. R. (2019). Bovine genome-microbiome interactions: metagenomic frontier for the selection of efficient productivity in cattle systems. mSystems 4, e103–e119. doi: 10.1128/mSystems.00103-19
O’Hara, E., Neves, A. L. A., Song, Y., and Guan, L. L. (2020). The role of the gut microbiome in cattle production and health: driver or passenger? Annu Rev. Anim Biosci. 8, 199–220. doi: 10.1146/annurev-animal-021419-083952
Oliveira, A. C., Rebelo, A. R., and Homem, C. (2021). Integrating animal development: how hormones and metabolism regulate developmental transitions and brain formation. Dev. Biol. 475, 256–264. doi: 10.1016/j.ydbio.2021.01.016
Petri, R. M., Schwaiger, T., Penner, G. B., Beauchemin, K. A., Forster, R. J., Mckinnon, J. J., et al. (2013). Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microb. 79, 3744–3755. doi: 10.1128/AEM.03983-12
Pragna, P., Chauhan, S. S., Sejian, V., Leury, B. J., and Dunshea, F. R. (2018). Climate change and goat production: enteric methane emission and its mitigation. Animals 8:235. doi: 10.3390/ani8120235
Qiu, Q., Zhang, G., Auvil, L., Capitanu, B., Ma, J., Lewin, H. A., et al. (2012). The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949. doi: 10.1038/ng.2343
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., and Glckner, F. O. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ram, J. (2005). Biodiversity and conservation of high altitude meadows of uttaranchal himalaya. Bull. Natl. Inst. Ecol. 15, 11–17.
Ramos-Morales, E., Arco-Pérez, A., Martín-García, A. I., Ez-Ruiz, D. Y., Frutos, P., and Hervás, G. (2014). Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim Feed Sci. Tech. 198, 57–66. doi: 10.1016/j.anifeedsci.2014.09.016
Sha, Y. Z., Hu, J., Shi, B. G., Dingkao, R. Q., Wang, J. Q., Li, S. B., et al. (2021). Supplementary feeding of cattle-yak in the cold season alters rumen microbes, volatile fatty acids, and expression of SGLT1 in the rumen epithelium. PeerJ. 9:e11048. doi: 10.7717/peerj.11048
Shabat, S., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Miller, M. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Skapetas, B., Nitas, D., Karalazos, A., and Hatziminaoglou, I. (2004). A study on the herbage mass production and quality for organic grazing sheep in a mountain pasture of northern Greece. Livest Prod. Sci. 87, 277–281. doi: 10.1016/j.livprodsci.2003.08.002
Takahashi, S., Tomita, J., Nishioka, K., Hisada, T., Nishijima, M., and Bourtzis, K. (2014). Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One 9:e105592. doi: 10.1371/journal.pone.0105592
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Wang, Q. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Whiteley, N. M., and Faulkner, L. S. (2005). Temperature influences whole-animal rates of metabolism but not protein synthesis in a temperate intertidal isopod. Physiol. Biochem Zool. 78, 227–238. doi: 10.1086/427054
Wu, D. W., Vinitchaikul, P., Deng, M. Y., Zhang, G. R., Sun, L. Y., Wang, H. X., et al. (2021). Exploration of the effects of altitude change on bacteria and fungi in the rumen of yak (bos grunniens). Arch. Microbiol. 203, 1–12. doi: 10.1007/s00203-020-02072-x
Wu, G. D., Chen, J., Hoffmann, C., Chen, Y. Y., Keilbaugh, S. A., Bewtra, M., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Yue, Q., Yang, H. J., Cao, Y. C., Zhang, D. F., Jiang, Y. H., and Wang, J. Q. (2009). Feruloyl and acetyl esterase production of an anaerobic rumen fungus Neocallimastix sp. YQ2 effected by glucose and soluble nitrogen supplementations and its potential in the hydrolysis of fibrous feedstuffs. Anim. Feed Sci. Tech. 153, 263–277. doi: 10.1016/j.anifeedsci.2009.06.019
Zhai, Q. X., Li, T. Q., Yu, L. L., Xiao, Y., Feng, S. S., Wu, J. P., et al. (2017). Effects of subchronic oral toxic metal exposure on the intestinal microbiota of mice. Sci. Bull. 62, 831–840. doi: 10.1016/j.scib.2017.01.031
Zhang, J., Yang, M., Cai, D. Y. C., Hao, Y. J., Zhao, X., Zhu, Y. H., et al. (2020). Composition, coagulation characteristics, and cheese making capacity of yak milk. J. Dairy Sci. 103, 1276–1288. doi: 10.3168/jds.2019-17231
Zhang, Y., Li, F., Chen, Y., Wu, H., Meng, Q., and Guan, L. L. (2020). Metatranscriptomic profiling reveals the effect of breed on active rumen eukaryotic composition in beef cattle with varied feed efficiency. Front. Microbiol. 11:367. doi: 10.3389/fmicb.2020.00367
Zhang, Z. G., Xu, D. M., Wang, L., Hao, J. J., Wang, J. F., Zhou, X., et al. (2016). Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 26, 1873–1879. doi: 10.1016/j.cub.2016.05.012
Keywords: yak, altitude, region, rumen, microbiota
Citation: Han L, Xue W, Cao H, Chen X, Qi F, Ma T, Tu Y, Diao Q, Zhang C and Cui K (2022) Comparison of Rumen Fermentation Parameters and Microbiota of Yaks From Different Altitude Regions in Tibet, China. Front. Microbiol. 12:807512. doi: 10.3389/fmicb.2021.807512
Received: 02 November 2021; Accepted: 20 December 2021;
Published: 10 February 2022.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
Anusorn Cherdthong, Khon Kaen University, ThailandCopyright © 2022 Han, Xue, Cao, Chen, Qi, Ma, Tu, Diao, Zhang and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengfu Zhang, dGliZXR6Y2ZAMTI2LmNu; Kai Cui, Y3Vpa2FpQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.