
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 04 March 2022
Sec. Virology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.805472
This article is part of the Research Topic The Viral Evasion of Antiviral Innate Immunity View all 71 articles
In the past few decades, newly evolved coronaviruses have posed a global threat to public health and animal breeding. To control and prevent the coronavirus-related diseases, understanding the interaction of the coronavirus and the host immune system is the top priority. Coronaviruses have evolved multiple mechanisms to evade or antagonize the host immune response to ensure their replication. As the first line and main component of innate immune response, type I IFN response is able to restrict virus in the initial infection stage; it is thus not surprising that the primary aim of the virus is to evade or antagonize the IFN response. Gaining a profound understanding of the interaction between coronaviruses and type I IFN response will shed light on vaccine development and therapeutics. In this review, we provide an update on the current knowledge on strategies employed by coronaviruses to evade type I IFN response.
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been spreading worldwide since December 2019 and has led to the ongoing pandemic, which has infected 271 million individuals and has resulted in 5.32 million deaths until December 14, 2021. Coronaviruses also pose a fatal threat to animal health, including livestock, poultry, and pets, causing enormous economic losses. Given the high similarity between SARS-CoV-2 and bat SARS-CoV-like coronaviruses, it is not surprising that a few mutations in the S protein may confer animal coronaviruses the ability to jump from animals to humans (Andersen et al., 2020; Malaiyan et al., 2021). The innate immune responses are the host’s first defense line and are essential for controlling and eliminating virus infection. It is essential to understand the mechanisms employed by coronaviruses to evade or antagonize the innate immune system. To prevent and control coronaviruses-related diseases, we summarize the recent research on human and animal coronaviruses’ innate immune evasion strategies.
Coronaviruses belong to the family Coronaviridae and are divided into four genera. α-coronavirus and β-coronavirus only infect mammals, while γ-coronavirus and δ-coronavirus (but not all) are avian coronaviruses (Gorbalenya et al., 2004; Woo et al., 2010, 2012). The three highly pathogenic human coronaviruses SARS-CoV, SARS-CoV-2, and Middle East respiratory syndrome (MERS-CoV) belong to β-coronavirus and cause severe symptoms in the lung, with high mortality. The low or mild pathogenic human coronaviruses such as HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63 continually circulate in the human population worldwide and produce mild symptoms of the common cold (Fouchier et al., 2004; Van der Hoek et al., 2004, 2006; Woo et al., 2005).
Coronaviruses also impose great threat to economic animals, resulting in tremendous economic losses (Cui et al., 2019; Adachi et al., 2020). For example, the infectious bronchitis virus (IBV) has been circulating in poultry farms for many years since the 1930s, with high mortality, and is highly contagious. It mainly causes a decline in laying eggs and meat production, resulting in a substantial economic loss in the poultry industry (Cavanagh, 2007; Jordan, 2017). Coronaviruses also affect porcine breeding and cattle raising. Porcine epidemic diarrhea virus (PEDV) has emerged recently and causes diarrhea in pigs, with 100% mortality in piglets (Wood, 1977; Jung et al., 2020). Transmissible gastroenteritis virus (TGEV) is another coronavirus which has circulated in porcine farm for many years and causes diarrhea in young pigs (Katsuda et al., 2006; Xia et al., 2018). In the cattle farm, bovine coronavirus (BCoV) is responsible for severe enteritis in young calves (Saif, 2010). Coronaviruses are also present in domestic pets such as cats, dogs, and ferrets, among which, feline infectious peritonitis coronavirus (FIPV) causes high mortality in felines (Tekes and Thiel, 2016).
Coronaviruses have the largest genomes of all known RNA viruses, ranging from 25 to 32 kb (Mihindukulasuriya et al., 2008). This virus family contains a positive-sense, single-stranded, linear genome that possesses a conserved gene order with 5′-replicase-Spike(S)-Envelope(E)-Membrane(M)-Nucleocapsid(N)-3′. Viral replicase gene covers about two-thirds of 5′ genome, with open reading frames (ORFs) 1a and 1b. Replicase gene produces polyprotein 1a and 1ab, which are involved in coronavirus replication after cleaving into 15–16 nonstructural proteins (nsps) (Wang et al., 2020). Papain-like protease (PLpro, nsp3) and 3C-like protease (3CLpro, nsp5) are responsible for cleaving the polyprotein 1a and 1ab. Nsps are conserved across coronaviruses, except for nsp1, encoded only in α-coronavirus and β-coronavirus, which is associated with suppressing host protein translation (Nakagawa and Makino, 2021). IBV nsp2 is a weak protein kinase R (PKR) inhibitor, involving in evading host innate immune system (Wang et al., 2009). Double-membrane vesicles (DMVs) are the “replication factory” of coronaviruses, closely associated with nsp3 (PLpro), nsp4, and nsp6 (Thiel et al., 2003; Neuman et al., 2014; Neuman, 2016; Oudshoorn et al., 2017). Moreover, coronavirus replicase gene encodes RNA-dependent RNA polymerase (RdRp, nsp12), helicase (nsp13), and three RNA-processing enzymes 3′-5′ exonuclease (nsp14), poly(U)-specific endonuclease (nsp15) and 2′-O-methyltransferase (nsp16) (Decroly et al., 2008; Chen et al., 2009; Zust et al., 2011; Kindler and Thiel, 2014; Fehr and Perlman, 2015).
The downstream of the replicase gene is the last one-third genome encoding structural and accessory proteins. These proteins are not directly translated from genomic RNA (gRNA), but from 3′ nested sub-genomic mRNAs (sgRNAs), which are generated by dis-continuous transcription (Masters, 2006; Hidalgo et al., 2021). Genes encoding accessory protein are interspersed within genes encoding structural protein. Different genera of coronaviruses harbor different numbers of accessory protein genes, and the accessory proteins are not necessary for coronavirus replication (Wang et al., 2020). Structural proteins (S, M, E, and N) are mainly involved in the initial (attachment and entry) and last (assembly and release) steps of the coronavirus life cycle. The start signal of the coronavirus’ life cycle is the binding to specific cell surface receptors by the S protein. It is the most diverse protein among coronavirus structural proteins and is responsible for cross-species transmission when mutation happens. This is, a type I membrane glycoprotein with two functional subunits, S1 and S2, which are in charge of receptor binding and membrane fusion, respectively (Yurkovetskiy et al., 2020). For SARS-CoV and SARS-CoV-2, the cell surface receptor is angiotensin converting enzyme 2 (ACE2) (Mathewson et al., 2008; Benton et al., 2020; Coate et al., 2020), whereas, for MERS-CoV, the receptor is dipeptidyl peptidase 4 (DPP4) (Van Doremalen et al., 2014; Lim et al., 2016; Letko et al., 2020). During coronavirus replication, a replication–transcription complex (RTC) is formed by gRNA, N protein, and replicases, to support gRNA replication and mRNA transcription, associated with DMVs (Gosert et al., 2002; Snijder et al., 2006; Knoops et al., 2008; Ulasli et al., 2010; Lundin et al., 2014; Avila-Perez et al., 2016). After all materials are prepared, the M protein triggers virion assembly in the ER-Golgi intermediate compartment (ERGIC) (Klumperman et al., 1994). During the entire assembly process, the M protein acts as a scaffold protein to interact with the S protein and N protein to recruit structural components, gRNA is brought into the virion by N protein; while the M–E protein interaction is responsible for membrane bending (Lim and Liu, 2001; Fung and Liu, 2019). Finally, mature virions are budded from ERGIC are released by exocytosis (Neuman et al., 2011; McBride et al., 2014; Ujike and Taguchi, 2015; Figure 1).
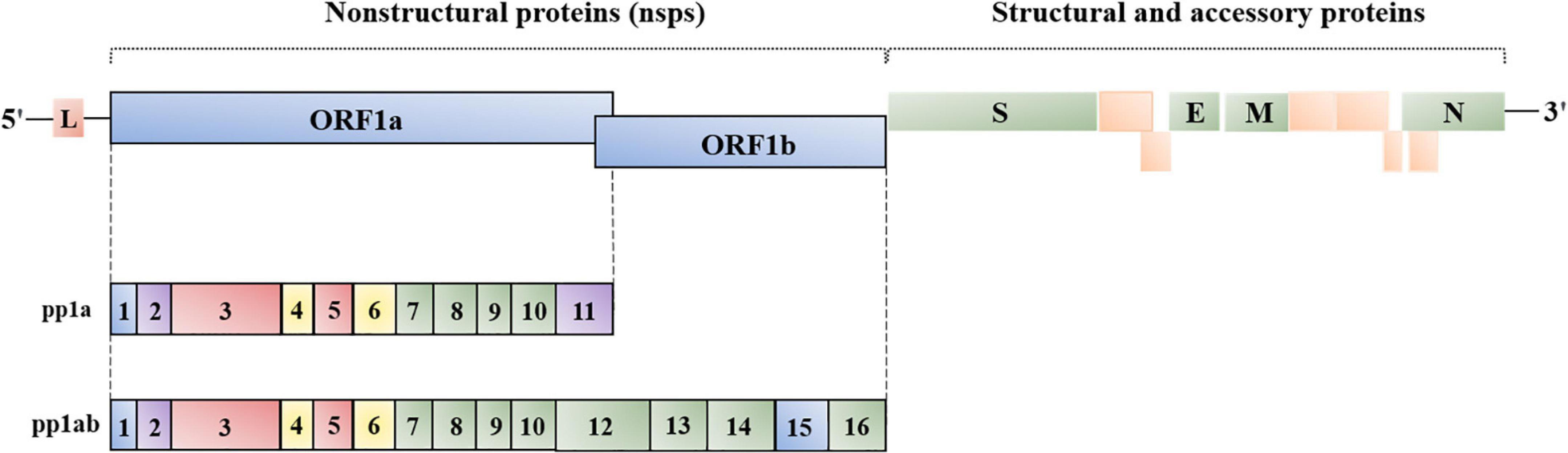
Figure 1. Genome organization of coronavirus. L, leader; ORF1a/1b, replicase, encoding nsp1-16 (γ- and δ-coronavirus lack nsp1); S, spike; E, envelope; M, membrane; N, nucleocapsid. The location and number of accessory proteins vary in different coronaviruses.
When SARS-CoV and MERS-CoV infect mice, rapid virus replication and delayed type I IFN expression are observed in the lungs. The pretreatment of IFNα/β protects mice from lethal clinical diseases, implying that coronaviruses are sensitive to IFN treatment (Channappanavar et al., 2016, 2019). In addition, the delayed IFN response is also observed during IBV, PEDV, MHV, and PDCoV infection (Roth-Cross et al., 2007; Cao et al., 2015; Kint et al., 2015b; Luo et al., 2016). Thus, inhibiting type I IFN response is conserved among coronaviruses and is essential for the successful infection of coronavirus.
As a pioneer in eradicating pathogens, type I IFN response plays an important role in the innate immune system. The type I IFN signaling pathway is mainly composed of the IFN induction pathway and the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway; the latter is responsible for the expression of IFN-stimulated genes (ISGs). Activating the type I IFN response is a delicate and complicated process that involves subtle molecular interactions, the formation of functional complexes, the post-translational modifications, and the assistance of the cytoplasmic–nuclear transport system.
The key to turn on the type I IFN system is through “aberrant” nucleic acids, the pathogen-associated molecular patterns (PAMPs), which are recognized by host pattern recognition receptors (PRRs) (Wu and Chen, 2014). The coronavirus PAMP is double-stranded RNA (dsRNA) produced during virus replication, which is usually detected by cytosolic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including RIG-I or melanoma differentiation-associated protein 5 (MDA5) (Zielecki et al., 2013). After using their helicase and C-terminal regulatory domain to sense dsRNA, RIG-I or MDA5 undergoes a conformational change. RIG-I forms the tetramer via N-terminal tandem caspase activation and recruitment domains (2CARD), which are modified by K63-polyubiquitin chains to stabilize the RIG-I 2CARD tetramer. For MDA5, 2CARD forms large oligomers by self-association in a concentration-dependent manner (Zerbe et al., 2020). Subsequently, the similar CARD domain of mitochondrial antiviral signaling protein (MAVS) interacts with 2CARD of RLRs, leading to tripartite motif-containing protein 31 (TRIM31)-mediated K63-linked polyubiquitination of MAVS and then the prion-like aggregation of MAVS CARD, which is essential for the biological function of MAVS (Oshiumi et al., 2010; Hou et al., 2011; Peisley et al., 2014; Liu et al., 2017). Activated MAVS recruits TRAFs, which are pre-associated with TBK1/IKKε; next, TBK1 is activated by MAVS-TRAFs-mediated oligmerization/auto-phosphorylation and NEMO-IKKβ-mediated phosphorylation, resulting in the formation of the TBK1 complex (containing TBK1, IKK i/ε, and NEMO) (Liu S. et al., 2013; Fang et al., 2017). MAVS is phosphorylated by the TBK1 complex, thereby gaining the ability to recruit IRF3. TBK1 promotes IRF3 phosphorylation and homodimerization, and then IRF3 is translocated into the nucleus to induce type I IFN transcription (Liu et al., 2015; Oshiumi, 2020).
IFN-α receptor (IFNAR) consists of two subunits IFNAR1 (low affinity) and IFNAR2 (high affinity) (Piehler et al., 2012; Schreiber and Piehler, 2015). IFNα/β binds to IFNAR1 or IFNAR2 to form a binary complex, bringing the receptor-associated kinases JAK1 and TYK2 into proximity, resulting in kinase transphosphorylation and subsequent phosphorylation of tyrosine on IFNAR1 and IFNAR2 (Deonarain et al., 2002). Phospho-tyrosine residues on IFNAR1 and IFNAR2 provide a docking site for STAT proteins (Hertzog and Williams, 2013). Once recruited, STAT proteins are phosphorylated and form homo- or hetero-dimers. The STAT dimers are released from the IFNAR and then associate with IFN regulatory factor 9 (IRF9) to form a complex, ISGF3. ISGF3 is transported into the nucleus and binds to a specific promoter, IFN-stimulated regulatory elements (ISREs), leading to expression of hundreds of ISGs (Heim et al., 1995; Leung et al., 1995; Darnell, 1997; Li et al., 1997; Hebenstreit et al., 2005; Fink and Grandvaux, 2013; McNab et al., 2015).
ISGs are grouped into three functional classes: type I IFN response reinforcement, antiviral factors, and IFN desensitization. ISGs enhance type I IFN response in different ways. PRRs are one type of ISGs responsible for the detection of PAMPs, and IRFs are the type of ISGs promoting target genes’ transcription. There is also a type of ISGs that reinforce the type I IFN response by assisting signal transduction. For example, the upregulation of E3 ubiquitin ligase TRIM21 through the JAK-STAT pathway promotes K27-linked ubiquitination of MAVS, thereby promoting the ability of MAVS to recruit TBK1 (Xue et al., 2018). Most antiviral ISGs fight against the virus by intervening virus life cycle. For example, myxovirus resistance (Mx), cholesterol-25-hydroxylase (CH25H), and IFN-inducible transmembrane (IFITM) protein inhibit the early stage of the virus life cycle, virus entry (Gao et al., 2010; Park and Scott, 2010; Haller and Kochs, 2011; Anafu et al., 2013; Liu S. Y. et al., 2013; Wilkins et al., 2013; Zang et al., 2020). OAS/RNaseL and PKR detect viral RNA to restrict virus replication at the transcriptional and translational levels, respectively (Chakrabarti et al., 2011; Munir and Berg, 2013; Gal-Ben-Ari et al., 2018). In addition, virus budding and release can be restricted by Viperin and Tetherin (Wang et al., 2007; Perez-Caballero et al., 2009; Szretter et al., 2011; Swiecki et al., 2013). The last ISGs group is responsible for IFN desensitization to avoid over- or persistent activation of the JAK-STAT pathway. SOCS protein is known as a JAK-STAT pathway inhibitor. SOCS1 associates with the IFNAR to prevent STATs from attaching to the receptor, while SOCS3 binds JAKs to inhibit the activity of these kinases (Hong and Carmichael, 2013). Unlike SOCS proteins, USP18 binds IFNAR2 to block the activation of the JAK-STAT pathway induced by IFNα (Francois-Newton et al., 2011). In addition, the protein inhibitor of activated STAT1 (PIAS1) is a broad inhibitor of the type I IFN system. Many studies have shown that PIAS1 inhibits the DNA binding activity of transcription factors, including STAT1, IRF3, and p65 (Liao et al., 2000; Liu et al., 2005; Shuai and Liu, 2005; Shuai, 2006; Li et al., 2013).
To perform the function as a transcription factors, the cytoplasmic STATs and IRF3 must gain access into the nucleus to bind specific DNA promoters. The nuclear translocation process is closely related to the nucleocytoplasmic transport system (Springhower et al., 2020). Transport direction through the nuclear pore complex (NPC) depends on the signal sequence on the cargo proteins: the nuclear localization signal (NLS) guides cargos into the nucleus and the nuclear export signal (NES) guides cargos into the cytoplasm. Karyopherin subunit α (KPNA) acts as an adaptor protein that binds NLS of cargo proteins, and then the KPNA–cargo complex combines with Karyopherin subunit β1 (KPNB1) to form the KPNA:KPNB1:Cargo complex. KPNB1 interacts with NPC and eventually transports cargos into the nucleus (Chook and Blobel, 2001; Cautain et al., 2015). In general, KPNA2, KPNA3, and KPNA4 mediate the import of phospho-IRF3 into the nucleus (Reich, 2002; Cai et al., 2020), whereas the ISGF3 complex enters the nucleus via KPNA1 (Sekimoto et al., 1997; Nardozzi et al., 2010). After transcription factors have completed their mission, they must exit from the nucleus. Most transcription factors are exported into the cytoplasm through exportin1 (CRM1), with the widest range of known substrates (Fornerod et al., 1997; Kutay and Guttinger, 2005). When phospho-STAT1 is dephosphorylated by nuclear phosphatase TC45, the NES of STAT1 binds to CRM1 and goes through the NPC to the cytoplasm (McBride et al., 2000; Ten Hoeve et al., 2002). Another interesting example is the nucleocytoplasmic shuttling of the STAT2-IRF9 complex. IRF9 contains a bipartite basic NLS that directs the unphosphorylated STAT2-IRF9 (U-STAT2-IRF9) complex into the nucleus, while STAT2 harbors a functional and dominant NES that guides the return of the U-STAT2-IRF9 complex to the cytoplasm. As a result, the U-STAT2-IRF9 complex is normally located in the cytoplasm (Lau et al., 2000; Banninger and Reich, 2004).
To guarantee their successful replication in host cells, coronaviruses have evolved rigorous and precise strategies to evade the host’s innate immune system, especially type I IFN response. Viral proteins are the main executors of immune evasion strategies during the infection process. For instance, among the viral proteins encoded by SARS-CoV or SARS-CoV-2, at least 8 proteins have been confirmed harboring the activity of hindering type I IFN response (Totura and Baric, 2012; Lei et al., 2020). Here, we will summarize the strategies exploited by the coronaviruses to evade type I IFN response (Figure 2).
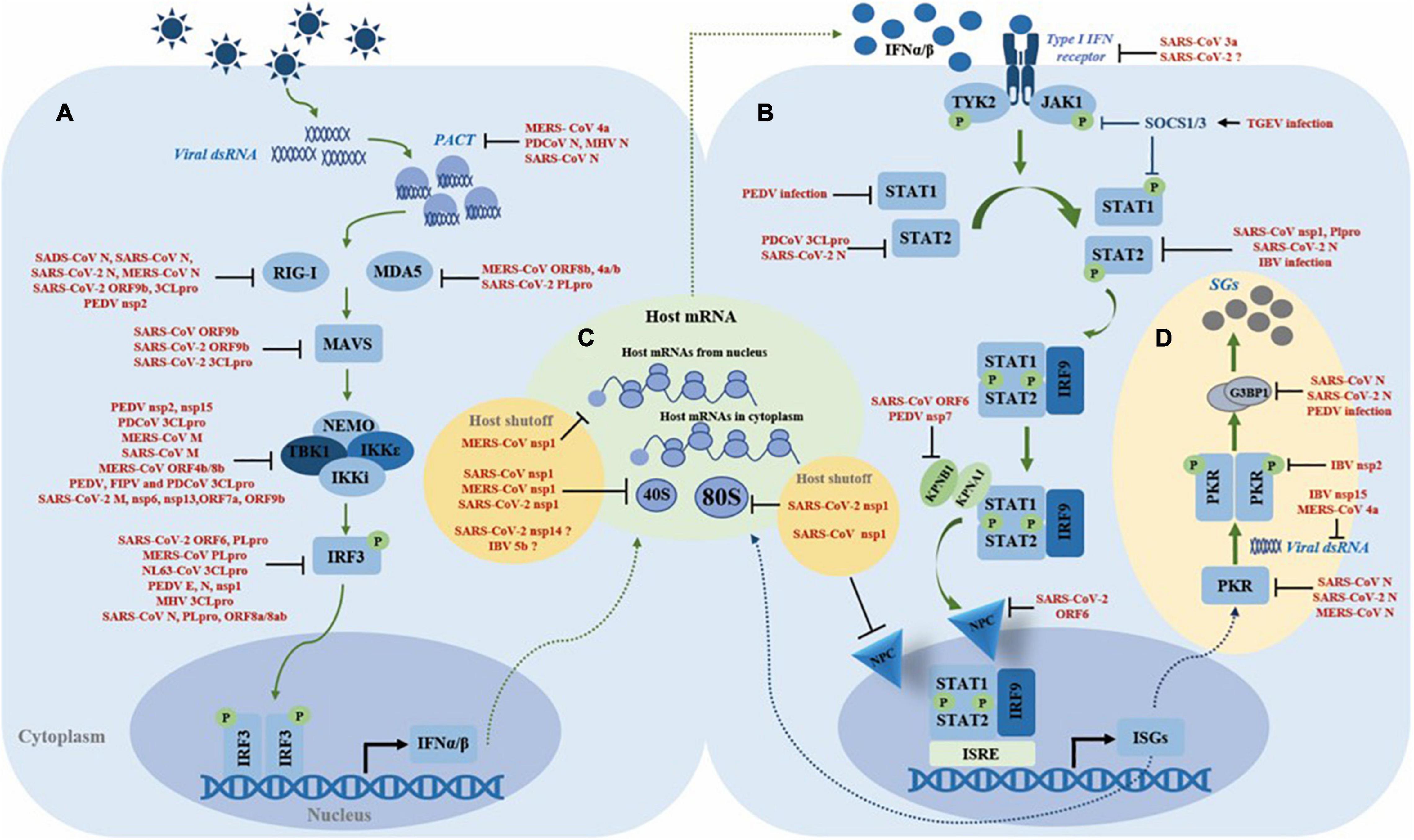
Figure 2. Summary of the mechanisms of coronaviruses antagonizing type I IFN system. (A) Inhibition of IFN induction. (B) Inhibition of the JAK-STAT pathway. (C) Manipulation of the host translation machinery. (D) Regulation of stress granules formation.
Inhibiting IFNα/β production is the first and essential step for coronavirus to evade the host immune defense. Due to the delayed type I IFN response, the host misses the best timing to clear the virus. In turn, the virus achieves successful replication (Rose et al., 2010; Menachery et al., 2014; Kint et al., 2015b). Escaping viral RNA recognition, inhibiting innate immune sensors, targeting the TBK1 complex, and interfering with IRF3 are the main strategies used by coronaviruses to inhibit IFNα/β expression.
During replication, viral RNA or intermediate products like dsRNA are recognized by host immune sensors, such as RIG-I, MDA5, or TLR. Coronaviruses have evolved three tricks to circumvent host recognition to evade immune sensors’ detection (Ulasli et al., 2010). The first way is “hiding.” Coronavirus creates “a replication factory” called DMVs derived from ER membranes, to hide the viral RNA and to enwrap the replication complex. Nsp3 and nsp4 are involved in the formation of DMVs (Gosert et al., 2002; Knoops et al., 2008; Oudshoorn et al., 2017). DMVs keep viral replication machinery, intermediates, and products away from host sensors, creating an isolated and safe environment for viral replication (Snijder et al., 2006). The second trick is “disguising”. 2′-O-methylation of mRNA is an important modification that provides a molecular marker for host cells to distinguish self mRNA from non-self mRNA, and coronaviruses cleverly evade this discrimination. Coronaviruses have evolved nsp16 with 2′-O-methyltransferase activity, which autonomously modifies viral mRNAs with a 5′ cap structure, mimicking host mRNA. Relevant studies showed that human and mouse coronaviruses without nsp16 2′-O-methyltransferase activity trigger higher expression of type I IFN. Therefore, nsp16 helps the virus to evade the IFN response by modifying viral RNA with a 5′ cap (Zust et al., 2011). The third strategy is “removing”. It has been reported that the highly conserved endoribonuclease nsp15 is responsible for reducing the accumulation of negative-stranded RNA and dsRNA. Recombinant coronavirus lacking the nsp15 endoribonuclease activity produces more dsRNA during infection (Kindler et al., 2017; Gao et al., 2021). It is speculated that coronavirus utilizes its endoribonuclease to cleave its negative-stranded RNA to maintain the ratio of negative-positive stranded RNA to facilitate virus genome replication and transcription, thereby reducing the accumulation of dsRNA and avoiding the activation of host RNA sensors (Deng et al., 2017). This hypothesis was confirmed by Hackbart et al. (2020) who demonstrates that nsp15 cleaves viral 5′-poly(U) negative-stranded RNA at uridine residues to limit the abundance and length of the negative-sense RNA, thereby escaping host immune sensor detection. In conclusion, the DMV, nsp16 and nsp15 play an important role in subverting the recognition of viral RNA by host sensors.
In addition, coronaviruses also have evolved a series of strategies to directly antagonize the host sensors. For MERS-CoV, co-expression of ORF8b with ORF4a/b significantly downregulates the protein level of MDA5 (Lee et al., 2019), whereas for SARS-CoV-2, current studies show that papain-like protease (PLpro) antagonizes ISG15-dependent MDA5 activation through de-ISGylation (Liu et al., 2021a). Several reports show that the N protein from SARS-CoV, SARS-CoV-2, and MERS-CoV interferes with K63-linked ubiquitination of RIG-I by interacting with TRIM25 E3 ubiquitin ligase, thereby preventing the activation of RIG-I; as for SADS-CoV, the N protein directly associates with RIG-I and leads to RIG-I degradation by ubiquitination (Hu et al., 2017; Chang et al., 2020; Chen et al., 2020; Liu et al., 2021b; Oh and Shin, 2021). SARS-CoV-2 main protease 3CLpro not only cleaves the N-terminal amino acids from RIG-I and deprives its ability to activate MAVS, but also promotes the ubiquitination and proteasome-mediated degradation of MAVS; in addition, the SARS-CoV-2 unique accessory protein 9b interferes with RIG-I and MAVS interaction (Kouwaki et al., 2021; Liu et al., 2021c). PACT is a dsRNA-binding protein that binds dsRNA to activate RIG-I or MDA5, thereby initiating the type I IFN induction pathway. The N protein from PDCoV, MHV, and SARS-CoV directly interacts with PACT, thus impeding the association of RIG-I or MDA5 with dsRNA or/and PACT (Ding et al., 2017; Chen J. et al., 2019), whereas the MERS-CoV 4a protein contains a dsRNA-binding domain that competes with PACT for dsRNA binding (Siu et al., 2014b). The E3 ubiquitin ligase FBXW7 is verified as an innate antiviral factor capable of enhancing the expression of RIG-I and TBK1. A recent study reports that PEDV nsp2 interacts with and targets FBXW7 for degradation through the K48-linked ubiquitination, thereby reducing the levels of RIG-I and TBK1, which are confirmed in vivo (Li et al., 2021; Figure 2A). Above reports show that the N protein’s antagonism to the dsRNA sensors is conserved in several coronaviruses. As the N protein is a structural and functional conserved protein, we speculate that the N protein plays an important role in innate immune evasion during different genera coronaviruses infection, which needs further investigation. The virus proteases, nsps, and unique accessory proteins play a certain role in the antagonism of dsRNA sensors.
Once immune sensors have been activated, the next step is the formation of the TBK1 complex (containing TBK1, IKK i/ε, and NEMO). Coronaviruses also use their proteins as weapons to prevent TBK1 complex formation. For example, SARS-CoV-2 M, nsp6, and nsp13 directly interact with TBK1 to suppress IRF3 phosphorylation (Xia et al., 2020; Zheng et al., 2020); SARS-CoV-2 unique accessory protein ORF7a destabilizes TBK1; while PEDV endoribonuclease nsp15 targets TBK1 mRNA for degradation (Wu et al., 2020; Kouwaki et al., 2021). SARS-CoV and SARS-CoV-2 ORF9b not only inhibit IFN induction pathway by antagonizing MAVS, but also restrain K63-linked ubiquitination of NEMO (Shi et al., 2014; Kouwaki et al., 2021; Wu et al., 2021). MERS-CoV M protein destroys the interaction of TRAF3, TBK1, and IKKε, and so do SARS-CoV M and PDCoV 3CLpro (Siu et al., 2009, 2014a; Lui et al., 2016). Unlike MERS-CoV ORF4b, which binds TBK1 and IKKε directly (Yang et al., 2015), MERS-CoV ORF8b competes with IKKε for interacting with HSP70 that is vital for activation of IKKε, thus blocking the downstream signal transduction (Wong et al., 2020). PEDV, FIPV, and PDCoV 3CLpro cleaves NEMO at glutamine 231 (Q231), thereby inhibiting TBK1 complex formation (Wang et al., 2016; Zhu et al., 2017a; Chen S. et al., 2019; Figure 2A). Altoghther, the M protein, main protease 3CLpro, accessory proteins, and several nsps play the major role in antagonizing the TBK1-NEMO-IKKε complex, by impeding protein–protein interaction, preventing the post-translation modification, such as ubiquitination, or directly cleaving the signaling molecules.
Inhibiting nuclear translocation of IRF3 is a common trick for the coronaviruses to compromise type I IFN expression. PLpro from MERS-CoV, SARS-CoV, and HCoV-NL63 not only harbor the proteolytic processing but also process deubiquitinating activity, which have been confirmed to affect K48- and K63-linked ubiquitination and ISG15-linked ISGylation of host proteins, thereby inhibiting type I IFN signaling. Furthermore, these PLpro impair IRF3 activation and its nuclear translocation, independently of its protease activity (Barretto et al., 2005; Chen et al., 2007; Yang et al., 2014). SARS-CoV-2 PLpro directly cleaves IRF3 to prevent the transcription of type I IFN (Moustaqil et al., 2021). SARS-CoV ORF8a/8ab protein mediate ubiquitin-dependent rapid degradation of IRF3, whereas PEDV and SARS-CoV-2 N protein curb IRF3 phosphorylation and nuclear translocation by sequestering the interaction between TBK1 and IRF3 interaction (Ding et al., 2014; Wong et al., 2018; Zhao et al., 2021). PEDV E protein is also associated with IRF3 directly to inhibit IRF3 nuclear translocation (Zheng L. et al., 2021). The MHV PLpro targets TBK1 by deubiquitination, thereby reducing its kinase activity and forming an inactive complex with TBK1 and IRF3 in the hypo-phosphorylated form in the cytoplasm (Wang et al., 2011). SARS-CoV-2 ORF6 chooses another way to inhibit nuclear translocation of IRF3, by binding importin Karyopherin α2 (KPNA2), a nuclear transport importer, to antagonize the expression of type I IFN (Xia et al., 2020). Even if phospho-IRF3 successfully enters the nucleus, PEDV nsp1 induces degradation of CREB-binding protein (CBP) through ubiquitination, thereby disrupting the binding of IRF3 to CPB and ultimately inhibiting type I IFN expression (Zhang et al., 2016; Figure 2A). It seems that the PLpro and N protein play the major role in impeding the IRF3 activation or nuclear translocation.
In summary, coronaviruses employ several conserved strategies to evade host sensor’s detection of viral RNA or dsRNA, by forming DMVs (nsp3 and nsp4 are involved), by disguising viral RNA (nsp16 is involved), or by removing dsRNA/negative intermediates (nsp15 is involved). Several conserved viral proteins are involved in antagonism of innate immune responses: the N protein employs similar strategies to interfere with the activation of sensors; protease PLpro and 3CLpro are involved in many steps of type I IFN signaling, by directly the signaling proteins or by promoting the degradation of signaling proteins via manipulation of the ubiquination modification. These conserved viral proteins are excellent targets for therapeutics and vaccine design. The compounds targeting nsp15 (Canal et al., 2021; Kumar et al., 2021; Sixto-Lopez and Martinez-Archundia, 2021), nsp16 (Krafcikova et al., 2020; Lin et al., 2020; Rosas-Lemus et al., 2020), PLpro (Baez-Santos et al., 2015; Jo et al., 2020; Paul et al., 2021), and 3CLpro (Hakami et al., 2021; Milligan et al., 2021) are under screening and investigation.
Even though IFNα/β are successfully expressed, the coronavirus still employs strategies to impede the JAK-STAT pathway (Figure 2B). SARS-CoV accessory protein 3a induces serine phosphorylation of IFNAR1, resulting in ubiquitination and lysosomal degradation of IFNAR1 (Minakshi et al., 2009). IFNAR1 ubiquitination and degradation can also be induced by SARS-CoV-2 infection, but the precise mechanism is unknown yet (Chen et al., 2021). During TGEV infection, the level of miR-30a-5p is downregulated and then the negative regulators SOCS1/3 are increased to block the type I IFN response (Ma et al., 2018). Coronaviruses also employ multiple ways to inhibit STAT1 and STAT2 nuclear translocation, including downregulating the molecules involved, interfering with post-translational modifications of signaling molecules, and disrupting the function of nuclear transport receptors. PEDV infection causes degradation of STAT1 (Guo et al., 2016), while PDCoV 3CLpro cleaves STAT2 via its protease activity (Zhu et al., 2017b). SARS-CoV-2 N interacts with and downregulates STAT2 protein through an unknown mechanism (Chen et al., 2021). SARS-CoV nsp1 inhibits STAT1 phosphorylation and nuclear translocation, and IBV infection STAT1 phosphorylation and nuclear translocation by unclear mechanisms (Wathelet et al., 2007; Kint et al., 2015a). Since γ-coronavirus lacks nsp1, IBV definitely encodes an alternative protein to play a similar role to nsp1 (Kint et al., 2015a). STAT1 phosphorylation is also impeded by SARS-CoV PLpro, which disrupts the interaction between extracellular signal-regulated kinase (ERK) 1 and STAT1 (Li et al., 2011). Both SARS-CoV and SARS-CoV-2 ORF6 impact the nuclear transport system in diverse ways: SARS-CoV ORF6 interacts with KPNA2 and promotes KPNA2 to hijack KPNB1 to the ER/Golgi membrane, resulting in the shortage of KPNB1 that is responsible for carrying the ISGF3 complex to the nucleus (Frieman et al., 2007); while SARS-CoV-2 ORF6 inhibits STAT1 and STAT2 nuclear translocation by preventing the ISGF3/KPNA1/KPNB1 complex from docking with the Nup98-Rae1 nuclear pore complex (Miorin et al., 2020). Another study showed that PEDV nsp7 disrupts the interaction between KPNA1 and STAT1/2, hence impeding ISGF3 nuclear translocation (Li et al., 2020). Interestingly, recent studies showed that in contrast to the decrease of STAT1/2 phosphorylation, high concentration of SARS-CoV-2 N increases phosphorylation by low concentration of SRAS-CoV-2 N protein and nuclear translocation of STAT1/2 (Mu et al., 2020; Zhao et al., 2021).
Although coronaviruses use a series of strategies to inhibit ISGs expression, the JAK-STAT pathway is activated during cytokine release syndrome (CRS) caused by SARS-CoV and SARS-CoV-2 (Schwartz et al., 2017; Huang et al., 2020; Zhou et al., 2020). An animal test showed that lethal doses of SARS-CoV-infected mice lacking IFNα/β receptor displayed a high survival rate and mild clinical disease (Channappanavar et al., 2016). Thus, the activation of the JAK-STAT pathway might be associated with the pathogenicity of coronaviruses. Compared with the number of studies about evasion or antagonism of type I IFN induction by coronaviruses, reports on the antagonism of the JAK-STAT pathway are limited. One reason for this phenomenon is the lower cost-effectiveness, since the JAK-STAT pathway is generally silent during coronavirus infection due to the failure of IFNα/β induction. Another reason is the research on the JAK-STAT pathway is a contradictory and tough task. Anyway, the interplay between coronaviruses and the JAK-STAT pathway remains to be further explored.
Shutting down host translation kills two birds with one stone for the coronavirus, by hijacking the host translation system for their own use and inhibiting host antiviral genes expression (Lokugamage et al., 2012, 2015; Tanaka et al., 2012; Kint et al., 2016; Finkel et al., 2021). Inhibiting host antiviral genes expression is an important strategy to antagonize the IFN response. Nsp1 encoded by α- and β-coronavirus have similar biological functions to trigger host translation shutoff (Shen et al., 2019). SARS-CoV nsp1 inactivates translation by tightly binding with the 40S ribosomal subunit, while SARS-CoV-2 nsp1 associates with both 40S and 80S ribosomal subunits to achieve host translation shutoff (Kamitani et al., 2009; Schubert et al., 2020; Thoms et al., 2020). In addition, when combining with 40S ribosomes, SARS-CoV nsp1 is also adept at inducing endo-nucleolytic cleavage of 5′ capped non-viral mRNA and triggers the degradation of the cleaved host mRNA by exonuclease Xrn I (Kamitani et al., 2009; Huang et al., 2011; Gaglia et al., 2012). Similar to SARS-CoV nsp1, MERS-CoV nsp1 exerts a parallel mechanism to induce host mRNA degradation (Lokugamage et al., 2015). For SARS-CoV-2 nsp1, which has a high amino acid sequence identity with SARS-CoV nsp1, whether to employ the same way to degrade non-viral RNA remains to be further investigated (Lei et al., 2020; Tidu et al., 2020; Shen et al., 2021). In addition to these common strategies, SARS-CoV nsp1 plays other tricks to prevent RNA nuclear export, by interfering with nucleocytoplasmic transport of RNA binding protein, nucleolin, or by altering the localization of Nup93 (Gomez et al., 2019). SARS-CoV-2 nsp1 directly interacts with mRNA export receptor heterodimer NXF1-NXT1, leading to nuclear retention of host mRNA, further supporting host translation shutdown (Zhang et al., 2021). MERS-CoV nsp1 also inhibits host translation in its unique way, that is, by halting translation of host mRNA through targeting mRNA derived from the nucleus, thereby hijacking the translation machine for the translation of viral RNA that is synthesized in the cytoplasm; however, the detailed mechanism on how MERS-CoV nsp1 specifically recognizes host mRNA transported from the nucleus is unknown (Lokugamage et al., 2015; Figure 2C). As a vital pathogenic determinant, nsp1 is also an excellent drug and vaccine design target. A study demonstrated that SARS-CoV nsp1 with two amino acid residues (KS) deletion possesses a decreased antagonism ability of type I IFN response (Jauregui et al., 2013). Such property may exist in both α- and β-coronavirus (Benedetti et al., 2020). Recently, it has been confirmed that several SARS-CoV-2 proteins function as translation inhibitors. The translation inhibition activity of SARS-CoV-2 nsp14 is more significant than nsp1 (Hsu et al., 2021), and nsp16 suppresses global mRNA splicing and prevents the production of mature host mRNA (Banerjee et al., 2020). Since nsp14 and nsp16 are conserved viral proteins among coronaviruses, the mechanisms of these two proteins as translation inhibitors may be a breakthrough in finding common strategies employed by coronaviruses. IBV, a γ-coronavirus, which lacks nsp1, also inhibits host translation. It has been shown that IBV accessory protein 5b is responsible for inducing host translation shutoff (Kint et al., 2016). Further research is needed on how coronaviruses block the host translation systems, especially for γ- and δ-coronavirus.
Coronavirus infection is usually accompanied by a host stress response (Kikkert, 2020). When facing infection stress, host cells instinctively limit protein synthesis to restrict virus replication and minimize the cell damage (McCormick and Khaperskyy, 2017). When the protein translation is attenuated, the cell untranslated host mRNA trigger the formation of stress granules (SGs). SGs are large cytoplasmic foci containing stalled 48S preinitiation complexes (PICs) that consist of untranslated mRNAs, translation initiation factors, small ribosomal subunits, and the RNA-binding proteins (RBPs), such as Ras-GAP SH3 domain-binding protein (G3BP) and poly A-binding protein (PABP) (Kedersha et al., 1999; Moser and Fritzler, 2010; Cao et al., 2020). SGs serve as an exhibition of robust translation inhibition and act as a platform for innate immune sensors like PKR, MDA5, and RIG-I, to transmit signaling downstream (Kim et al., 2019; Hofmann et al., 2021). Thus, SGs are also regarded as an antiviral platform involved in PAMP recognition, signaling transduction, and IFN induction (Onomoto et al., 2014; Yoneyama et al., 2016; McCormick and Khaperskyy, 2017; Eiermann et al., 2020; Gao et al., 2021). Since host translation shutoff is the main cause of SG formation, and translation machinery is essential for viral protein synthesis, the coronavirus employs multiple strategies to fine-tune the protein translation efficiency and to manipulate the formation of SGs (Figure 2D).
Most studies have focused on how coronaviruses regulate the PKR-eIF2α-G3BP1 axis, the main pathway responsible for triggering SG formation. Blocking PKR recognition of viral dsRNA and disrupting the assembly of SGs are common techniques used by coronaviruses to manipulate SG formation. MERS-CoV 4a accessory protein binds viral dsRNA to inhibit the activation of PKR, thereby impeding SG formation. MERS-CoV’s inefficient replication was observed while MERS-CoV 4a lost its RNA binding motif. The reason for this phenomenon is that 4a RNA binding defect results in plenty of RNA binding to and in trans activation of PKR, and high level of PKR-mediated eIF2α phosphorylation lead to the protein shutoff and formation of SGs, resulting in inefficient viral protein translation (Rabouw et al., 2016; Nakagawa et al., 2018). Hackbart et al. (2020) showed that coronavirus nsp15 cleaves its 5′-poly(U) negative-sense RNA to evade innate immune responses. This mechanism is also utilized by IBV nsp15 to impede the accumulation of dsRNA, thereby decreasing its dsRNA to evade PKR sensor activation (Gao et al., 2021). Gao et al. (2021) also showed that nsp15 of PEDV, TGEV, SARS-CoV, SARS-CoV-2, and MERS-CoV interferes with the formation of SGs induced by chemical stimulation, probably by targeting to host RNA or other host factors. IBV manipulates the protein translation and probably also impedes SG formation by upregulating the expression of phosphatase subunit GADD34 to restrict eIF2α phosphorylation (Wang et al., 2009). The assembly of SGs can be compromised when G3BP1 is impaired. SARS-CoV and SARS-CoV-2 N protein interact with PKR and G3BP1 directly, while MERS-CoV N only associates with PKR to impede SGs assembly (Cai et al., 2021; Luo et al., 2021; Zheng Z. Q. et al., 2021). PEDV infection triggers the cleavage of G3BP1 by caspase-8, which subverts SG formation and promotes virus replication (Sun et al., 2021). Although some studies have reported that SGs induced by TGEV and MHV may be positively correlated with its replication, these studies do not provide sufficient evidence to prove this conclusion, and further studies are needed (Raaben et al., 2007; Sola et al., 2011). The relationship between coronaviruses and SGs appears complicated and needs further study.
As the first line of defense against pathogens, the innate immune system inevitably becomes the coronaviruses’ primary target, to ensure virus replication and produce infectious progeny. Most coronavirus infection delay type I IFN response, making the host miss the best time to clear the virus. Because of this, the mechanisms of coronaviruses antagonizing the type I IFN response have been a research hotspot. Coronaviruses have been prevalent in livestock and poultry for many years, since the finding of the first coronavirus, IBV, in the 1930s. This virus family mainly causes economic loss in domestic animal farming and is important in the veterinary field. Human coronaviruses only infect the upper respiratory tract and cause mild respiratory symptoms, with low risk to public health, until the outbreak of SARS-CoV in 2003 and MERS-CoV in 2012. With the disappearance of SARS-CoV and MERS-CoV, people seem to have forgotten the suffering of these coronaviruses brought us. The worldwide epidemic of SARS-CoV-2 brought people’s attention back to this virus family: we have never really defeated the coronavirus. Although the vaccine targeting SARS-CoV-2 has been successfully developed and has a protective effect on existing strains, we still face the risk of immune failure, with the emergence of SARS-CoV-2 mutants. To fight the coronavirus, it is important to find common conservative mechanisms for the coronavirus to antagonize the innate immune system. Furthermore, SARS-CoV, MERS-CoV, and SARS-CoV-2 are typical examples of interspecies transmission of viruses from animal to human. Therefore, besides paying attention to human coronaviruses, we should also pay effort to prevent and control these pathogens in animal. Understanding various or common mechanisms employed by coronaviruses to evade or antagonize the host innate immune system could offer therapeutic targets and strategies for controlling coronavirus-associated diseases. This review offers a panoramic view of the mechanisms of fighting against the host innate immune system by human and animal coronaviruses, especially type I IFN response.
WX and YL: conceptualization and writing—original draft. CD, KQ, and YL: funding acquisition. YL: project administration and writing—review and editing. CD and YL: supervision. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key Research and Development Program (No. 2021YFD1801104), the National Natural Science Foundation of China (32172834, 31772724, and 31530074), and the Open Project Program of Jiangsu Key Laboratory of Zoonosis (R2007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adachi, S., Koma, T., Doi, N., Nomaguchi, M., and Adachi, A. (2020). Commentary: origin and evolution of pathogenic coronaviruses. Front. Immunol. 11:811. doi: 10.3389/fimmu.2020.00811
Anafu, A. A., Bowen, C. H., Chin, C. R., Brass, A. L., and Holm, G. H. (2013). Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 288, 17261–17271. doi: 10.1074/jbc.M112.438515
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., and Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi: 10.1038/s41591-020-0820-9
Avila-Perez, G., Rejas, M. T., and Rodriguez, D. (2016). Ultrastructural characterization of membranous torovirus replication factories. Cell. Microbiol. 18, 1691–1708. doi: 10.1111/cmi.12620
Baez-Santos, Y. M., St John, S. E., and Mesecar, A. D. (2015). The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 115, 21–38. doi: 10.1016/j.antiviral.2014.12.015
Banerjee, A. K., Blanco, M. R., Bruce, E. A., Honson, D. D., Chen, L. M., Chow, A., et al. (2020). SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 183, 1325–1339.e21. doi: 10.1016/j.cell.2020.10.004
Banninger, G., and Reich, N. C. (2004). STAT2 nuclear trafficking. J. Biol. Chem. 279, 39199–39206. doi: 10.1074/jbc.M400815200
Barretto, N., Jukneliene, D., Ratia, K., Chen, Z., Mesecar, A. D., and Baker, S. C. (2005). The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79, 15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005
Benedetti, F., Snyder, G. A., Giovanetti, M., Angeletti, S., Gallo, R. C., Ciccozzi, M., et al. (2020). Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J. Transl. Med. 18:329. doi: 10.1186/s12967-020-02507-5
Benton, D. J., Wrobel, A. G., Xu, P., Roustan, C., Martin, S. R., Rosenthal, P. B., et al. (2020). Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588, 327–330. doi: 10.1038/s41586-020-2772-0
Cai, T., Yu, Z., Wang, Z., Liang, C., and Richard, S. (2021). Arginine methylation of SARS-Cov-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication. J. Biol. Chem. 297:100821. doi: 10.1016/j.jbc.2021.100821
Cai, Z., Zhang, M. X., Tang, Z., Zhang, Q., Ye, J., Xiong, T. C., et al. (2020). USP22 promotes IRF3 nuclear translocation and antiviral responses by deubiquitinating the importin protein KPNA2. J. Exp. Med. 217:e20191174. doi: 10.1084/jem.20191174
Canal, B., Fujisawa, R., McClure, A. W., Deegan, T. D., Wu, M., Ulferts, R., et al. (2021). Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem. J. 478, 2465–2479. doi: 10.1042/BCJ20210199
Cao, L., Ge, X., Gao, Y., Herrler, G., Ren, Y., Ren, X., et al. (2015). Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 12:127. doi: 10.1186/s12985-015-0345-x
Cao, X., Jin, X., and Liu, B. (2020). The involvement of stress granules in aging and aging-associated diseases. Aging Cell 19:e13136. doi: 10.1111/acel.13136
Cautain, B., Hill, R., De Pedro, N., and Link, W. (2015). Components and regulation of nuclear transport processes. FEBS J. 282, 445–462. doi: 10.1111/febs.13163
Cavanagh, D. (2007). Coronavirus avian infectious bronchitis virus. Vet. Res. 38, 281–297. doi: 10.1051/vetres:2006055
Chakrabarti, A., Jha, B. K., and Silverman, R. H. (2011). New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 31, 49–57. doi: 10.1089/jir.2010.0120
Chang, C. Y., Liu, H. M., Chang, M. F., and Chang, S. C. (2020). Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J. Virol. 94:e00099-20. doi: 10.1128/JVI.00099-20
Channappanavar, R., Fehr, A. R., Vijay, R., Mack, M., Zhao, J., Meyerholz, D. K., et al. (2016). Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193. doi: 10.1016/j.chom.2016.01.007
Channappanavar, R., Fehr, A. R., Zheng, J., Wohlford-Lenane, C., Abrahante, J. E., Mack, M., et al. (2019). IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 129, 3625–3639. doi: 10.1172/JCI126363
Chen, D. Y., Khan, N., Close, B. J., Goel, R. K., Blum, B., Tavares, A. H., et al. (2021). SARS-CoV-2 disrupts proximal elements in the JAK-STAT pathway. J. Virol. 95:e0086221. doi: 10.1128/JVI.00862-21
Chen, J., Fang, P., Wang, M., Peng, Q., Ren, J., Wang, D., et al. (2019). Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus Genes. 55, 520–531. doi: 10.1007/s11262-019-01673-z
Chen, K., Xiao, F., Hu, D., Ge, W., Tian, M., Wang, W., et al. (2020). SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-beta production. Viruses 13:47. doi: 10.3390/v13010047
Chen, S., Tian, J., Li, Z., Kang, H., Zhang, J., Huang, J., et al. (2019). Feline infectious peritonitis virus Nsp5 inhibits type I interferon production by cleaving NEMO at multiple sites. Viruses 12:43. doi: 10.3390/v12010043
Chen, Y., Cai, H., Pan, J., Xiang, N., Tien, P., Ahola, T., et al. (2009). Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 106, 3484–3489. doi: 10.1073/pnas.0808790106
Chen, Z., Wang, Y., Ratia, K., Mesecar, A. D., Wilkinson, K. D., and Baker, S. C. (2007). Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81, 6007–6018. doi: 10.1128/JVI.02747-06
Chook, Y. M., and Blobel, G. (2001). Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715. doi: 10.1016/s0959-440x(01)00264-0
Coate, K. C., Cha, J., Shrestha, S., Wang, W., Goncalves, L. M., Almaca, J., et al. (2020). SARS-CoV-2 cell entry factors ACE2 and TMPRSS2 are expressed in the microvasculature and ducts of human pancreas but are not enriched in beta cells. Cell Metab. 32, 1028–1040.e4. doi: 10.1016/j.cmet.2020.11.006
Cui, J., Li, F., and Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 17, 181–192. doi: 10.1038/s41579-018-0118-9
Darnell, J. E. Jr. (1997). STATs and gene regulation. Science 277, 1630–1635. doi: 10.1126/science.277.5332.1630
Decroly, E., Imbert, I., Coutard, B., Bouvet, M., Selisko, B., Alvarez, K., et al. (2008). Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 82, 8071–8084. doi: 10.1128/JVI.00407-08
Deng, X., Hackbart, M., Mettelman, R. C., O’Brien, A., Mielech, A. M., Yi, G., et al. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 114, E4251–E4260. doi: 10.1073/pnas.1618310114
Deonarain, R., Chan, D. C., Platanias, L. C., and Fish, E. N. (2002). Interferon-alpha/beta-receptor interactions: a complex story unfolding. Curr. Pharm. Des. 8, 2131–2137. doi: 10.2174/1381612023393288
Ding, Z., Fang, L., Jing, H., Zeng, S., Wang, D., Liu, L., et al. (2014). Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 88, 8936–8945. doi: 10.1128/JVI.00700-14
Ding, Z., Fang, L., Yuan, S., Zhao, L., Wang, X., Long, S., et al. (2017). The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-beta antagonizing mechanism: attenuation of PACT-mediated RIG-I/MDA5 activation. Oncotarget 8, 49655–49670. doi: 10.18632/oncotarget.17912
Eiermann, N., Haneke, K., Sun, Z., Stoecklin, G., and Ruggieri, A. (2020). Dance with the devil: stress granules and signaling in antiviral responses. Viruses 12:984. doi: 10.3390/v12090984
Fang, R., Jiang, Q., Zhou, X., Wang, C., Guan, Y., Tao, J., et al. (2017). MAVS activates TBK1 and IKKepsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 13:e1006720. doi: 10.1371/journal.ppat.1006720
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi: 10.1007/978-1-4939-2438-7_1
Fink, K., and Grandvaux, N. (2013). STAT2 and IRF9: Beyond ISGF3. JAKSTAT. 2, e27521. doi: 10.4161/jkst.27521
Finkel, Y., Gluck, A., Nachshon, A., Winkler, R., Fisher, T., Rozman, B., et al. (2021). SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594, 240–245. doi: 10.1038/s41586-021-03610-3
Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060. doi: 10.1016/s0092-8674(00)80371-2
Fouchier, R. A., Hartwig, N. G., Bestebroer, T. M., Niemeyer, B., De Jong, J. C., Simon, J. H., et al. (2004). A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101, 6212–6216. doi: 10.1073/pnas.0400762101
Francois-Newton, V., Magno de Freitas Almeida, G., Payelle-Brogard, B., Monneron, D., Pichard-Garcia, L., Piehler, J., et al. (2011). USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One 6:e22200. doi: 10.1371/journal.pone.0022200
Frieman, M., Yount, B., Heise, M., Kopecky-Bromberg, S. A., Palese, P., and Baric, R. S. (2007). Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81, 9812–9824. doi: 10.1128/JVI.01012-07
Fung, T. S., and Liu, D. X. (2019). Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 73, 529–557. doi: 10.1146/annurev-micro-020518-115759
Gaglia, M. M., Covarrubias, S., Wong, W., and Glaunsinger, B. A. (2012). A common strategy for host RNA degradation by divergent viruses. J. Virol. 86, 9527–9530. doi: 10.1128/JVI.01230-12
Gal-Ben-Ari, S., Barrera, I., Ehrlich, M., and Rosenblum, K. P. K. R. (2018). A kinase to remember. Front. Mol. Neurosci. 11:480. doi: 10.3389/fnmol.2018.00480
Gao, B., Gong, X., Fang, S., Weng, W., Wang, H., Chu, H., et al. (2021). Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathog. 17:e1008690. doi: 10.1371/journal.ppat.1008690
Gao, S., Von der Malsburg, A., Paeschke, S., Behlke, J., Haller, O., Kochs, G., et al. (2010). Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506. doi: 10.1038/nature08972
Gomez, G. N., Abrar, F., Dodhia, M. P., Gonzalez, F. G., and Nag, A. (2019). SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochem. Cell Biol. 97, 758–766. doi: 10.1139/bcb-2018-0394
Gorbalenya, A. E., Snijder, E. J., and Spaan, W. J. (2004). Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78, 7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004
Gosert, R., Kanjanahaluethai, A., Egger, D., Bienz, K., and Baker, S. C. (2002). RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76, 3697–3708. doi: 10.1128/jvi.76.8.3697-3708.2002
Guo, L., Luo, X., Li, R., Xu, Y., Zhang, J., Ge, J., et al. (2016). Porcine epidemic diarrhea virus infection inhibits interferon signaling by targeted degradation of STAT1. J. Virol. 90, 8281–8292. doi: 10.1128/JVI.01091-16
Hackbart, M., Deng, X., and Baker, S. C. (2020). Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U.S.A. 117, 8094–8103. doi: 10.1073/pnas.1921485117
Hakami, A. R., Bakheit, A. H., Almehizia, A. A., and Ghazwani, M. Y. (2021). Selection of SARS-CoV-2 main protease inhibitor using structure-based virtual screening. Future Med. Chem. 14, 61–79. doi: 10.4155/fmc-2020-0380
Haller, O., and Kochs, G. (2011). Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. 31, 79–87. doi: 10.1089/jir.2010.0076
Hebenstreit, D., Horejs-Hoeck, J., and Duschl, A. J. A. K. (2005). /STAT-dependent gene regulation by cytokines. Drug News Perspect. 18, 243–249. doi: 10.1358/dnp.2005.18.4.908658
Heim, M. H., Kerr, I. M., Stark, G. R., and Darnell, J. E. Jr. (1995). Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science 267, 1347–1349. doi: 10.1126/science.7871432
Hertzog, P. J., and Williams, B. R. (2013). Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 24, 217–225. doi: 10.1016/j.cytogfr.2013.04.002
Hidalgo, P., Valdes, M., and Gonzalez, R. A. (2021). Molecular biology of coronaviruses: an overview of virus-host interactions and pathogenesis. Bol. Med. Hosp. Infant. Mex. 78, 41–58. doi: 10.24875/BMHIM.20000249
Hofmann, S., Kedersha, N., Anderson, P., and Ivanov, P. (2021). Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 1868:118876. doi: 10.1016/j.bbamcr.2020.118876
Hong, X. X., and Carmichael, G. G. (2013). Innate immunity in pluripotent human cells: attenuated response to interferon-beta. J. Biol. Chem. 288, 16196–16205. doi: 10.1074/jbc.M112.435461
Hou, F., Sun, L., Zheng, H., Skaug, B., Jiang, Q. X., and Chen, Z. J. (2011). MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461. doi: 10.1016/j.cell.2011.06.041
Hsu, J. C., Laurent-Rolle, M., Pawlak, J. B., Wilen, C. B., and Cresswell, P. (2021). Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U.S.A. 118:e2101161118. doi: 10.1073/pnas.2101161118
Hu, Y., Li, W., Gao, T., Cui, Y., Jin, Y., Li, P., et al. (2017). The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 91:e02143-16. doi: 10.1128/JVI.02143-16
Huang, C., Lokugamage, K. G., Rozovics, J. M., Narayanan, K., Semler, B. L., and Makino, S. (2011). SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 7:e1002433. doi: 10.1371/journal.ppat.1002433
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5
Jauregui, A. R., Savalia, D., Lowry, V. K., Farrell, C. M., and Wathelet, M. G. (2013). Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One 8:e62416. doi: 10.1371/journal.pone.0062416
Jo, S., Kim, S., Shin, D. H., and Kim, M. S. (2020). Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 35, 145–151. doi: 10.1080/14756366.2019.1690480
Jordan, B. (2017). Vaccination against infectious bronchitis virus: a continuous challenge. Vet. Microbiol. 206, 137–143. doi: 10.1016/j.vetmic.2017.01.002
Jung, K., Saif, L. J., and Wang, Q. (2020). Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 286:198045. doi: 10.1016/j.virusres.2020.198045
Kamitani, W., Huang, C., Narayanan, K., Lokugamage, K. G., and Makino, S. (2009). A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 16, 1134–1140. doi: 10.1038/nsmb.1680
Katsuda, K., Kohmoto, M., Kawashima, K., and Tsunemitsu, H. (2006). Frequency of enteropathogen detection in suckling and weaned pigs with diarrhea in Japan. J. Vet. Diagn. Invest. 18, 350–354. doi: 10.1177/104063870601800405
Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442. doi: 10.1083/jcb.147.7.1431
Kikkert, M. (2020). Innate immune evasion by human respiratory RNA viruses. J. Innate Immun. 12, 4–20. doi: 10.1159/000503030
Kim, S. S., Sze, L., Liu, C., and Lam, K. P. (2019). The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-beta response. J. Biol. Chem. 294, 6430–6438. doi: 10.1074/jbc.RA118.005868
Kindler, E., Gil-Cruz, C., Spanier, J., Li, Y., Wilhelm, J., Rabouw, H. H., et al. (2017). Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13:e1006195. doi: 10.1371/journal.ppat.1006195
Kindler, E., and Thiel, V. (2014). To sense or not to sense viral RNA–essentials of coronavirus innate immune evasion. Curr. Opin. Microbiol. 20, 69–75. doi: 10.1016/j.mib.2014.05.005
Kint, J., Fernandez-Gutierrez, M., Maier, H. J., Britton, P., Langereis, M. A., Koumans, J., et al. (2015b). Activation of the chicken type I interferon response by infectious bronchitis coronavirus. J. Virol. 89, 1156–1167. doi: 10.1128/JVI.02671-14
Kint, J., Dickhout, A., Kutter, J., Maier, H. J., Britton, P., Koumans, J., et al. (2015a). Infectious bronchitis coronavirus inhibits STAT1 signaling and requires accessory proteins for resistance to type I interferon activity. J. Virol. 89, 12047–12057. doi: 10.1128/JVI.01057-15
Kint, J., Langereis, M. A., Maier, H. J., Britton, P., Van Kuppeveld, F. J., Koumans, J., et al. (2016). Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J. Virol. 90, 7519–7528. doi: 10.1128/JVI.00627-16
Klumperman, J., Locker, J. K., Meijer, A., Horzinek, M. C., Geuze, H. J., and Rottier, P. J. (1994). Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68, 6523–6534. doi: 10.1128/JVI.68.10.6523-6534.1994
Knoops, K., Kikkert, M., Worm, S. H., Zevenhoven-Dobbe, J. C., Van der Meer, Y., Koster, A. J., et al. (2008). SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. doi: 10.1371/journal.pbio.0060226
Kouwaki, T., Nishimura, T., Wang, G., and Oshiumi, H. (2021). Like receptor-mediated recognition of viral genomic RNA of severe acute respiratory syndrome coronavirus-2 and viral escape from the host innate immune responses. Front. Immunol. 12:700926. doi: 10.3389/fimmu.2021.700926
Krafcikova, P., Silhan, J., Nencka, R., and Boura, E. (2020). Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 11:3717. doi: 10.1038/s41467-020-17495-9
Kumar, S., Kashyap, P., Chowdhury, S., Kumar, S., Panwar, A., and Kumar, A. (2021). Identification of phytochemicals as potential therapeutic agents that binds to Nsp15 protein target of coronavirus (SARS-CoV-2) that are capable of inhibiting virus replication. Phytomedicine 85:153317. doi: 10.1016/j.phymed.2020.153317
Kutay, U., and Guttinger, S. (2005). Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15, 121–124. doi: 10.1016/j.tcb.2005.01.005
Lau, J. F., Parisien, J. P., and Horvath, C. M. (2000). Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. U.S.A. 97, 7278–7283. doi: 10.1073/pnas.97.13.7278
Lee, J. Y., Bae, S., and Myoung, J. (2019). Middle East respiratory syndrome coronavirus-encoded ORF8b strongly antagonizes IFN-beta promoter activation: its implication for vaccine design. J. Microbiol. 57, 803–811. doi: 10.1007/s12275-019-9272-7
Lei, X., Dong, X., Ma, R., Wang, W., Xiao, X., Tian, Z., et al. (2020). Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11:3810. doi: 10.1038/s41467-020-17665-9
Letko, M., Marzi, A., and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569. doi: 10.1038/s41564-020-0688-y
Leung, S., Qureshi, S. A., Kerr, I. M., Darnell, J. E. Jr., and Stark, G. R. (1995). Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 15, 1312–1317. doi: 10.1128/MCB.15.3.1312
Li, M., Wu, Y., Chen, J., Shi, H., Ji, Z., Zhang, X., et al. (2021). Innate immune evasion of porcine epidemic diarrhea virus through degradation of F-box and WD repeat domain-containing 7 protein via Ubiquitin-proteasome Pathway. J. Virol. JVI0088921. doi: 10.1128/JVI.00889-21 [Epub ahead of print].
Li, R., Pan, Y., Shi, D. D., Zhang, Y., and Zhang, J. (2013). PIAS1 negatively modulates virus triggered type I IFN signaling by blocking the DNA binding activity of IRF3. Antiviral Res. 100, 546–554. doi: 10.1016/j.antiviral.2013.09.001
Li, S., Yang, J., Zhu, Z., and Zheng, H. (2020). Porcine epidemic diarrhea virus and the host innate immune response. Pathogens 9:367. doi: 10.3390/pathogens9050367
Li, S. W., Lai, C. C., Ping, J. F., Tsai, F. J., Wan, L., Lin, Y. J., et al. (2011). Severe acute respiratory syndrome coronavirus papain-like protease suppressed alpha interferon-induced responses through downregulation of extracellular signal-regulated kinase 1-mediated signalling pathways. J. Gen. Virol. 92, 1127–1140. doi: 10.1099/vir.0.028936-0
Li, X., Leung, S., Kerr, I. M., and Stark, G. R. (1997). Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol. Cell. Biol. 17, 2048–2056. doi: 10.1128/MCB.17.4.2048
Liao, J., Fu, Y., and Shuai, K. (2000). Distinct roles of the NH2- and COOH-terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine-induced PIAS1-Stat1 interaction. Proc. Natl. Acad. Sci. U.S.A. 97, 5267–5272. doi: 10.1073/pnas.97.10.5267
Lim, K. P., and Liu, D. X. (2001). The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276, 17515–17523. doi: 10.1074/jbc.M009731200
Lim, Y. X., Ng, Y. L., Tam, J. P., and Liu, D. X. (2016). Human coronaviruses: a review of virus-host interactions. Diseases 4:26. doi: 10.3390/diseases4030026
Lin, S., Chen, H., Ye, F., Chen, Z., Yang, F., Zheng, Y., et al. (2020). Crystal structure of SARS-CoV-2 nsp10/nsp16 2′-O-methylase and its implication on antiviral drug design. Signal Transduct. Target Ther. 5:131. doi: 10.1038/s41392-020-00241-4
Liu, B., Yang, R., Wong, K. A., Getman, C., Stein, N., Teitell, M. A., et al. (2005). Negative regulation of NF-kappaB signaling by PIAS1. Mol. Cell. Biol. 25, 1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005
Liu, B., Zhang, M., Chu, H., Zhang, H., Wu, H., Song, G., et al. (2017). The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 18, 214–224. doi: 10.1038/ni.3641
Liu, G., Lee, J. H., Parker, Z. M., Acharya, D., Chiang, J. J., Van Gent, M., et al. (2021a). ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 6, 467–478. doi: 10.1038/s41564-021-00884-1
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630
Liu, S., Chen, J., Cai, X., Wu, J., Chen, X., Wu, Y. T., et al. (2013). MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. doi: 10.7554/eLife.00785
Liu, S. Y., Aliyari, R., Chikere, K., Li, G., Marsden, M. D., Smith, J. K., et al. (2013). Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38, 92–105. doi: 10.1016/j.immuni.2012.11.005
Liu, Y., Liang, Q. Z., Lu, W., Yang, Y. L., Chen, R., Huang, Y. W., et al. (2021b). A comparative analysis of coronavirus nucleocapsid (N) proteins reveals the SADS-CoV N protein antagonizes IFN-beta production by inducing ubiquitination of RIG-I. Front. Immunol. 12:688758. doi: 10.3389/fimmu.2021.688758
Liu, Y., Qin, C., Rao, Y., Ngo, C., Feng, J. J., Zhao, J., et al. (2021c). SARS-CoV-2 Nsp5 demonstrates two distinct mechanisms targeting RIG-I and MAVS to evade the innate immune response. mBio 12:e0233521. doi: 10.1128/mBio.02335-21
Lokugamage, K. G., Narayanan, K., Huang, C., and Makino, S. (2012). Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 86, 13598–13608. doi: 10.1128/JVI.01958-12
Lokugamage, K. G., Narayanan, K., Nakagawa, K., Terasaki, K., Ramirez, S. I., Tseng, C. T., et al. (2015). Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 89, 10970–10981. doi: 10.1128/JVI.01352-15
Lui, P. Y., Wong, L. Y., Fung, C. L., Siu, K. L., Yeung, M. L., Yuen, K. S., et al. (2016). Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microbes Infect. 5:e39. doi: 10.1038/emi.2016.33
Lundin, A., Dijkman, R., Bergstrom, T., Kann, N., Adamiak, B., Hannoun, C., et al. (2014). Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the Middle East respiratory syndrome virus. PLoS Pathog. 10:e1004166. doi: 10.1371/journal.ppat.1004166
Luo, J., Fang, L., Dong, N., Fang, P., Ding, Z., Wang, D., et al. (2016). Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology 495, 10–17. doi: 10.1016/j.virol.2016.04.025
Luo, L., Li, Z., Zhao, T., Ju, X., Ma, P., Jin, B., et al. (2021). SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci. Bull. (Beijing) 66, 1194–1204. doi: 10.1016/j.scib.2021.01.013
Ma, Y., Wang, C., Xue, M., Fu, F., Zhang, X., Li, L., et al. (2018). The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1alpha-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J Virol. 92:e00728-18. doi: 10.1128/JVI.00728-18
Malaiyan, J., Arumugam, S., Mohan, K., and Gomathi Radhakrishnan, G. (2021). An update on the origin of SARS-CoV-2: despite closest identity, bat (RaTG13) and pangolin derived coronaviruses varied in the critical binding site and O-linked glycan residues. J. Med. Virol. 93, 499–505. doi: 10.1002/jmv.26261
Masters, P. S. (2006). The molecular biology of coronaviruses. Adv. Virus Res. 66, 193–292. doi: 10.1016/S0065-3527(06)66005-3
Mathewson, A. C., Bishop, A., Yao, Y., Kemp, F., Ren, J., Chen, H., et al. (2008). Interaction of severe acute respiratory syndrome-coronavirus and NL63 coronavirus spike proteins with angiotensin converting enzyme-2. J. Gen. Virol. 89, 2741–2745. doi: 10.1099/vir.0.2008/003962-0
McBride, K. M., McDonald, C., and Reich, N. C. (2000). Nuclear export signal located within theDNA-binding domain of the STAT1transcription factor. EMBO J. 19, 6196–6206. doi: 10.1093/emboj/19.22.6196
McBride, R., Van Zyl, M., and Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses 6, 2991–3018. doi: 10.3390/v6082991
McCormick, C., and Khaperskyy, D. A. (2017). Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 17, 647–660. doi: 10.1038/nri.2017.63
McNab, F., Mayer-Barber, K., Sher, A., Wack, A., and O’Garra, A. (2015). Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103. doi: 10.1038/nri3787
Menachery, V. D., Eisfeld, A. J., Schafer, A., Josset, L., Sims, A. C., Proll, S., et al. (2014). Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5:e01174-14. doi: 10.1128/mBio.01174-14
Mihindukulasuriya, K. A., Wu, G., St Leger, J., Nordhausen, R. W., and Wang, D. (2008). Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 82, 5084–5088. doi: 10.1128/JVI.02722-07
Milligan, J. C., Zeisner, T. U., Papageorgiou, G., Joshi, D., Soudy, C., Ulferts, R., et al. (2021). Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp5 main protease. Biochem. J. 478, 2499–2515. doi: 10.1042/BCJ20210197
Minakshi, R., Padhan, K., Rani, M., Khan, N., Ahmad, F., and Jameel, S. (2009). The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. doi: 10.1371/journal.pone.0008342
Miorin, L., Kehrer, T., Sanchez-Aparicio, M. T., Zhang, K., Cohen, P., Patel, R. S., et al. (2020). SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 28344–28354. doi: 10.1073/pnas.2016650117
Moser, J. J., and Fritzler, M. J. (2010). Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 42, 828–843. doi: 10.1016/j.biocel.2009.11.018
Moustaqil, M., Ollivier, E., Chiu, H. P., Van Tol, S., Rudolffi-Soto, P., Stevens, C., et al. (2021). SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infect. 10, 178–195. doi: 10.1080/22221751.2020.1870414
Mu, J., Fang, Y., Yang, Q., Shu, T., Wang, A., Huang, M., et al. (2020). SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 6:65. doi: 10.1038/s41421-020-00208-3
Munir, M., and Berg, M. (2013). The multiple faces of proteinkinase R in antiviral defense. Virulence 4, 85–89. doi: 10.4161/viru.23134
Nakagawa, K., and Makino, S. (2021). Mechanisms of coronavirus Nsp1-mediated control of host and viral gene expression. Cells 10:300. doi: 10.3390/cells10020300
Nakagawa, K., Narayanan, K., Wada, M., and Makino, S. (2018). Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol. 92:e00902-18. doi: 10.1128/JVI.00902-18
Nardozzi, J., Wenta, N., Yasuhara, N., Vinkemeier, U., and Cingolani, G. (2010). Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. J. Mol. Biol. 402, 83–100. doi: 10.1016/j.jmb.2010.07.013
Neuman, B. W. (2016). Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 135, 97–107. doi: 10.1016/j.antiviral.2016.10.005
Neuman, B. W., Chamberlain, P., Bowden, F., and Joseph, J. (2014). Atlas of coronavirus replicase structure. Virus Res. 194, 49–66. doi: 10.1016/j.virusres.2013.12.004
Neuman, B. W., Kiss, G., Kunding, A. H., Bhella, D., Baksh, M. F., Connelly, S., et al. (2011). A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174, 11–22. doi: 10.1016/j.jsb.2010.11.021
Oh, S. J., and Shin, O. S. (2021). SARS-CoV-2 nucleocapsid protein targets RIG-I-like receptor pathways to inhibit the induction of interferon response. Cells 10:530. doi: 10.3390/cells10030530
Onomoto, K., Yoneyama, M., Fung, G., Kato, H., and Fujita, T. (2014). Antiviral innate immunity and stress granule responses. Trends Immunol. 35, 420–428. doi: 10.1016/j.it.2014.07.006
Oshiumi, H. (2020). Recent advances and contradictions in the study of the individual roles of ubiquitin ligases that regulate RIG-I-like receptor-mediated antiviral innate immune responses. Front. Immunol. 11:1296. doi: 10.3389/fimmu.2020.01296
Oshiumi, H., Miyashita, M., Inoue, N., Okabe, M., Matsumoto, M., and Seya, T. (2010). The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe 8, 496–509. doi: 10.1016/j.chom.2010.11.008
Oudshoorn, D., Rijs, K., Limpens, R., Groen, K., Koster, A. J., Snijder, E. J., et al. (2017). Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio 8:e01658-17. doi: 10.1128/mBio.01658-17
Park, K., and Scott, A. L. (2010). Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 88, 1081–1087. doi: 10.1189/jlb.0610318
Paul, A., Sarkar, A., Saha, S., Maji, A., Janah, P., and Kumar Maity, T. (2021). Synthetic and computational efforts towards the development of peptidomimetics and small-molecule SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. 46:116301. doi: 10.1016/j.bmc.2021.116301
Peisley, A., Wu, B., Xu, H., Chen, Z. J., and Hur, S. (2014). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. doi: 10.1038/nature13140
Perez-Caballero, D., Zang, T., Ebrahimi, A., McNatt, M. W., Gregory, D. A., Johnson, M. C., et al. (2009). Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511. doi: 10.1016/j.cell.2009.08.039
Piehler, J., Thomas, C., Garcia, K. C., and Schreiber, G. (2012). Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250, 317–334. doi: 10.1111/imr.12001
Raaben, M., Groot Koerkamp, M. J., Rottier, P. J., and De Haan, C. A. (2007). Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 9, 2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x
Rabouw, H. H., Langereis, M. A., Knaap, R. C., Dalebout, T. J., Canton, J., Sola, I., et al. (2016). Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog. 12:e1005982. doi: 10.1371/journal.ppat.1005982
Reich, N. C. (2002). Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J. Interferon Cytokine Res. 22, 103–109. doi: 10.1089/107999002753452719
Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N. L., Kiryukhina, O., Brunzelle, J., et al. (2020).. High-resolution structures of the SARS-CoV-2 2′-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 13:eabe1202. doi: 10.1126/scisignal.abe1202
Rose, K. M., Elliott, R., Martinez-Sobrido, L., Garcia-Sastre, A., and Weiss, S. R. (2010). Murine coronavirus delays expression of a subset of interferon-stimulated genes. J. Virol. 84, 5656–5669. doi: 10.1128/JVI.00211-10
Roth-Cross, J. K., Martinez-Sobrido, L., Scott, E. P., Garcia-Sastre, A., and Weiss, S. R. (2007). Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81, 7189–7199. doi: 10.1128/JVI.00013-07
Saif, L. J. (2010). Bovine respiratory coronavirus. Vet. Clin. North Am. Food Anim. Pract. 26, 349–364. doi: 10.1016/j.cvfa.2010.04.005
Schreiber, G., and Piehler, J. (2015). The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 36, 139–149. doi: 10.1016/j.it.2015.01.002
Schubert, K., Karousis, E. D., Jomaa, A., Scaiola, A., Echeverria, B., Gurzeler, L. A., et al. (2020). SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 27, 959–966. doi: 10.1038/s41594-020-0511-8
Schwartz, D. M., Kanno, Y., Villarino, A., Ward, M., Gadina, M., and O’Shea, J. J. (2017). JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 17:78. doi: 10.1038/nrd.2017.267
Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T., and Yoneda, Y. (1997). Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16, 7067–7077. doi: 10.1093/emboj/16.23.7067
Shen, Z., Wang, G., Yang, Y., Shi, J., Fang, L., Li, F., et al. (2019). A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 294, 13606–13618. doi: 10.1074/jbc.RA119.009713
Shen, Z., Zhang, G., Yang, Y., Li, M., Yang, S., and Peng, G. (2021). Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J. Gen. Virol. 102:jgv001513. doi: 10.1099/jgv.0.001513
Shi, C. S., Qi, H. Y., Boularan, C., Huang, N. N., Abu-Asab, M., Shelhamer, J. H., et al. (2014). SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 193, 3080–3089. doi: 10.4049/jimmunol.1303196
Shuai, K. (2006). Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 16, 196–202. doi: 10.1038/sj.cr.7310027
Shuai, K., and Liu, B. (2005). Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 5, 593–605. doi: 10.1038/nri1667
Siu, K. L., Kok, K. H., Ng, M. J., Poon, V. K. M., Yuen, K. Y., Zheng, B. J., et al. (2009). Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 284, 16202–16209. doi: 10.1074/jbc.M109.008227
Siu, K. L., Yeung, M. L., Kok, K. H., Yuen, K. S., Kew, C., Lui, P. Y., et al. (2014b). Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 88, 4866–4876. doi: 10.1128/JVI.03649-13
Siu, K. L., Chan, C. P., Kok, K. H., Chiu-Yat Woo, P., and Jin, D. Y. (2014a). Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 11, 141–149. doi: 10.1038/cmi.2013.61
Sixto-Lopez, Y., and Martinez-Archundia, M. (2021). Drug repositioning to target NSP15 protein on SARS-CoV-2 as possible COVID-19 treatment. J. Comput. Chem. 42, 897–907. doi: 10.1002/jcc.26512
Snijder, E. J., Van der Meer, Y., Zevenhoven-Dobbe, J., Onderwater, J. J., Van der Meulen, J., Koerten, H. K., et al. (2006). Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80, 5927–5940. doi: 10.1128/JVI.02501-05
Sola, I., Galan, C., Mateos-Gomez, P. A., Palacio, L., Zuniga, S., Cruz, J. L., et al. (2011). The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J. Virol. 85, 5136–5149. doi: 10.1128/JVI.00195-11
Springhower, C. E., Rosen, M. K., and Chook, Y. M. (2020). Karyopherins and condensates. Curr. Opin. Cell Biol. 64, 112–123. doi: 10.1016/j.ceb.2020.04.003
Sun, L., Chen, H., Ming, X., Bo, Z., Shin, H. J., Jung, Y. S., et al. (2021). Porcine epidemic diarrhea virus infection induces caspase-8-mediated G3BP1 cleavage and subverts stress granules to promote viral replication. J. Virol. 95:e02344-20. doi: 10.1128/JVI.02344-20
Swiecki, M., Omattage, N. S., and Brett, T. J. (2013). BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol. Immunol. 54, 132–139. doi: 10.1016/j.molimm.2012.11.008
Szretter, K. J., Brien, J. D., Thackray, L. B., Virgin, H. W., Cresswell, P., and Diamond, M. S. (2011). The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol. 85, 11557–11566. doi: 10.1128/JVI.05519-11
Tanaka, T., Kamitani, W., DeDiego, M. L., Enjuanes, L., and Matsuura, Y. (2012). Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 86, 11128–11137. doi: 10.1128/JVI.01700-12
Tekes, G., and Thiel, H. J. (2016). Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv. Virus Res. 96, 193–218. doi: 10.1016/bs.aivir.2016.08.002
Ten Hoeve, J., De Jesus Ibarra-Sanchez, M., Fu, Y., Zhu, W., Tremblay, M., David, M., et al. (2002). Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22, 5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002
Thiel, V., Ivanov, K. A., Putics, A., Hertzig, T., Schelle, B., Bayer, S., et al. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84, 2305–2315. doi: 10.1099/vir.0.19424-0
Thoms, M., Buschauer, R., Ameismeier, M., Koepke, L., Denk, T., Hirschenberger, M., et al. (2020). Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369, 1249–1255. doi: 10.1126/science.abc8665
Tidu, A., Janvier, A., Schaeffer, L., Sosnowski, P., Kuhn, L., Hammann, P., et al. (2020). The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27, 253–264. doi: 10.1261/rna.078121.120
Totura, A. L., and Baric, R. S. (2012). SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2, 264–275. doi: 10.1016/j.coviro.2012.04.004
Ujike, M., and Taguchi, F. (2015). Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses 7, 1700–1725. doi: 10.3390/v7041700
Ulasli, M., Verheije, M. H., De Haan, C. A., and Reggiori, F. (2010). Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 12, 844–861. doi: 10.1111/j.1462-5822.2010.01437.x
Van der Hoek, L., Pyrc, K., and Berkhout, B. (2006). Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 30, 760–773. doi: 10.1111/j.1574-6976.2006.00032.x
Van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J., Wolthers, K. C., et al. (2004). Identification of a new human coronavirus. Nat. Med. 10, 368–373. doi: 10.1038/nm1024
Van Doremalen, N., Miazgowicz, K. L., Milne-Price, S., Bushmaker, T., Robertson, S., Scott, D., et al. (2014). Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 88, 9220–9232. doi: 10.1128/JVI.00676-14
Wang, D., Fang, L., Shi, Y., Zhang, H., Gao, L., Peng, G., et al. (2016). Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 90, 2090–2101. doi: 10.1128/JVI.02514-15
Wang, G., Chen, G., Zheng, D., Cheng, G., and Tang, H. (2011). PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS One 6:e17192. doi: 10.1371/journal.pone.0017192
Wang, X., Hinson, E. R., and Cresswell, P. (2007). The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105. doi: 10.1016/j.chom.2007.06.009
Wang, X., Liao, Y., Yap, P. L., Png, K. J., Tam, J. P., and Liu, D. X. (2009). Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 83, 12462–12472. doi: 10.1128/JVI.01546-09
Wang, Y., Grunewald, M., and Perlman, S. (2020). Coronaviruses: an updated overview of their replication and pathogenesis. Methods Mol. Biol. 2203, 1–29. doi: 10.1007/978-1-0716-0900-2_1
Wathelet, M. G., Orr, M., Frieman, M. B., and Baric, R. S. (2007). Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 81, 11620–11633. doi: 10.1128/JVI.00702-07
Wilkins, C., Woodward, J., Lau, D. T., Barnes, A., Joyce, M., McFarlane, N., et al. (2013). IFITM1 is a tight junction protein that inhibits hepatitis C virus entry. Hepatology 57, 461–469. doi: 10.1002/hep.26066
Wong, H. H., Fung, T. S., Fang, S., Huang, M., Le, M. T., and Liu, D. X. (2018). Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 515, 165–175. doi: 10.1016/j.virol.2017.12.028
Wong, L. R., Ye, Z. W., Lui, P. Y., Zheng, X., Yuan, S., Zhu, L., et al. (2020). Middle East respiratory syndrome coronavirus ORF8b accessory protein suppresses type I IFN expression by impeding HSP70-dependent activation of IRF3 kinase IKKepsilon. J. Immunol. 205, 1564–1579. doi: 10.4049/jimmunol.1901489
Woo, P. C., Huang, Y., Lau, S. K., and Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses 2, 1804–1820. doi: 10.3390/v2081803
Woo, P. C., Lau, S. K., Chu, C. M., Chan, K. H., Tsoi, H. W., Huang, Y., et al. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895. doi: 10.1128/JVI.79.2.884-895.2005
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wood, E. N. (1977). An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 100, 243–244. doi: 10.1136/vr.100.12.243
Wu, J., and Chen, Z. J. (2014). Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488. doi: 10.1146/annurev-immunol-032713-120156
Wu, J., Shi, Y., Pan, X., Wu, S., Hou, R., Zhang, Y., et al. (2021). SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 34:108761. doi: 10.1016/j.celrep.2021.108761
Wu, Y., Zhang, H., Shi, Z., Chen, J., Li, M., Shi, H., et al. (2020). Porcine epidemic diarrhea virus nsp15 antagonizes interferon signaling by RNA degradation of TBK1 and IRF3. Viruses 12:599. doi: 10.3390/v12060599
Xia, H., Cao, Z., Xie, X., Zhang, X., Chen, J. Y., Wang, H., et al. (2020). Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 33:108234. doi: 10.1016/j.celrep.2020.108234
Xia, L., Yang, Y., Wang, J., Jing, Y., and Yang, Q. (2018). Impact of TGEV infection on the pig small intestine. Virol. J. 15:102. doi: 10.1186/s12985-018-1012-9
Xue, B., Li, H., Guo, M., Wang, J., Xu, Y., Zou, X., et al. (2018). TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J. Virol. 92:e00321-18. doi: 10.1128/JVI.00321-18
Yang, X., Chen, X., Bian, G., Tu, J., Xing, Y., Wang, Y., et al. (2014). Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 95, 614–626. doi: 10.1099/vir.0.059014-0
Yang, Y., Ye, F., Zhu, N., Wang, W., Deng, Y., Zhao, Z., et al. (2015). Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 5:17554. doi: 10.1038/srep17554
Yoneyama, M., Jogi, M., and Onomoto, K. (2016). Regulation of antiviral innate immune signaling by stress-induced RNA granules. J. Biochem. 159, 279–286. doi: 10.1093/jb/mvv122
Yurkovetskiy, L., Wang, X., Pascal, K. E., Tomkins-Tinch, C., Nyalile, T. P., Wang, Y., et al. (2020). Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183:e8. doi: 10.1016/j.cell.2020.09.032
Zang, R., Case, J. B., Yutuc, E., Ma, X., Shen, S., Gomez Castro, M. F., et al. (2020). Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 117, 32105–32113. doi: 10.1073/pnas.2012197117
Zerbe, C. M., Mouser, D. J., and Cole, J. L. (2020). Oligomerization of RIG-I and MDA5 2CARD domains. Protein Sci. 29, 521–526. doi: 10.1002/pro.3776
Zhang, K., Miorin, L., Makio, T., Dehghan, I., Gao, S., Xie, Y., et al. (2021). Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 7:eabe7386. doi: 10.1126/sciadv.abe7386
Zhang, Q., Shi, K., and Yoo, D. (2016). Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology 489, 252–268. doi: 10.1016/j.virol.2015.12.010
Zhao, Y., Sui, L., Wu, P., Wang, W., Wang, Z., Yu, Y., et al. (2021). A dual-role of SARS-CoV-2 nucleocapsid protein in regulating innate immune response. Signal Transduct. Target Ther. 6:331. doi: 10.1038/s41392-021-00742-w
Zheng, L., Wang, X., Guo, D., Cao, J., Cheng, L., Li, X., et al. (2021). Porcine epidemic diarrhea virus E protein suppresses RIG-I signaling-mediated interferon-beta production. Vet. Microbiol. 254:108994. doi: 10.1016/j.vetmic.2021.108994
Zheng, Y., Zhuang, M. W., Han, L., Zhang, J., Nan, M. L., Zhan, P., et al. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target Ther. 5:299. doi: 10.1038/s41392-020-00438-7
Zheng, Z. Q., Wang, S. Y., Xu, Z. S., Fu, Y. Z., and Wang, Y. Y. (2021). CoV-2 nucleocapsid protein impairs stress granule formation to promote viral replication. Cell Discov. 7:38. doi: 10.1038/s41421-021-00275-0
Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062. doi: 10.1016/S0140-6736(20)30566-3
Zhu, X., Fang, L., Wang, D., Yang, Y., Chen, J., Ye, X., et al. (2017a). Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology. 502, 33–38. doi: 10.1016/j.virol.2016.12.005
Zhu, X., Wang, D., Zhou, J., Pan, T., Chen, J., Yang, Y., et al. (2017b). Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 91:e00003-17. doi: 10.1128/JVI.00003-17
Zielecki, F., Weber, M., Eickmann, M., Spiegelberg, L., Zaki, A. M., Matrosovich, M., et al. (2013). Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 87, 5300–5304. doi: 10.1128/JVI.03496-12
Keywords: innate immunity, coronavirus, type I IFN response, immune evasion, viral replication
Citation: Xue W, Ding C, Qian K and Liao Y (2022) The Interplay Between Coronavirus and Type I IFN Response. Front. Microbiol. 12:805472. doi: 10.3389/fmicb.2021.805472
Received: 30 October 2021; Accepted: 24 December 2021;
Published: 04 March 2022.
Edited by:
Zhi-Ming Zheng, National Cancer Institute (NCI), Frederick, United StatesReviewed by:
Juan C. De La Torre, The Scripps Research Institute, United StatesCopyright © 2022 Xue, Ding, Qian and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Liao, bGlhb3lpbmdAc2h2cmkuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.