
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 January 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.804935
This article is part of the Research TopicStreptococcus spp. and Corynebacterium spp.: Clinical and Zoonotic Epidemiology, Virulence Potential, Antimicrobial Resistance and Genomic Trends and ApproachesView all 12 articles
 Kadi J. Horn1
Kadi J. Horn1 Alexander C. Jaberi Vivar1,2
Alexander C. Jaberi Vivar1,2 Vera Arenas1,3
Vera Arenas1,3 Sameer Andani1
Sameer Andani1 Edward N. Janoff4,5
Edward N. Janoff4,5 Sarah E. Clark1*
Sarah E. Clark1*
The stability and composition of the airway microbiome is an important determinant of respiratory health. Some airway bacteria are considered to be beneficial due to their potential to impede the acquisition and persistence of opportunistic bacterial pathogens such as Streptococcus pneumoniae. Among such organisms, the presence of Corynebacterium species correlates with reduced S. pneumoniae in both adults and children, in whom Corynebacterium abundance is predictive of S. pneumoniae infection risk. Previously, Corynebacterium accolens was shown to express a lipase which cleaves host lipids, resulting in the production of fatty acids that inhibit growth of S. pneumoniae in vitro. However, it was unclear whether this mechanism contributes to Corynebacterium-S. pneumoniae interactions in vivo. To address this question, we developed a mouse model for Corynebacterium colonization in which colonization with either C. accolens or another species, Corynebacterium amycolatum, significantly reduced S. pneumoniae acquisition in the upper airway and infection in the lung. Moreover, the lungs of co-infected mice had reduced pro-inflammatory cytokines and inflammatory myeloid cells, indicating resolution of infection-associated inflammation. The inhibitory effect of C. accolens on S. pneumoniae in vivo was mediated by lipase-dependent and independent effects, indicating that both this and other bacterial factors contribute to Corynebacterium-mediated protection in the airway. We also identified a previously uncharacterized bacterial lipase in C. amycolatum that is required for inhibition of S. pneumoniae growth in vitro. Together, these findings demonstrate the protective potential of airway Corynebacterium species and establish a new model for investigating the impact of commensal microbiota, such as Corynebacterium, on maintaining respiratory health.
Respiratory tract infections are a prominent a global health problem. Pneumonia, or lung infection, is a major source of mortality as the 4th most common cause of death worldwide (World Health Organization [WHO], 2021). Children are particularly vulnerable, as pneumonia is the leading cause of death in people under 5 years old (Centers for Disease Control and Prevention, 2019). In the United States, over one million emergency room visits are due to pneumonia every year (Centers for Disease Control and Prevention, 2019). The most common bacterial cause of pneumonia is the Gram-positive bacterial pathogen Streptococcus pneumoniae, also known as the pneumococcus (Jose et al., 2015; Torres et al., 2018). S. pneumoniae vaccines are extremely effective against invasive pneumococcal disease but are substantially less protective against pneumococcal pneumonia (Cutts et al., 2005; Pilishvili and Bennett, 2015), highlighting the need for new therapeutic strategies to reduce the burden of disease. S. pneumoniae is an opportunistic pathogen, as colonization in the nasopharynx of the upper airway is asymptomatic and susceptibility to infection differs among individuals. While this variation is not completely understood, the composition and function of the airway microbiome have emerged as important contributing factors (Clark, 2020). The disruption of the airway microbiome following antibiotic therapy correlates with an increased risk of S. pneumoniae acquisition (Biesbroek et al., 2014; Teo et al., 2015). Closely related Streptococcus commensals produce factors such as bacteriocins that kill S. pneumoniae, and serial administration of such commensals has shown some efficacy in reducing the incidence of pneumococcal-associated infections including otitis media in children (Marchisio et al., 2015; La Mantia et al., 2017). However, the contributions of other members of the commensal airway microbiome, particularly non-streptococcal species, to protection against S. pneumoniae are poorly understood.
Next-generation microbiome sequencing studies have identified additional members of the airway microbiome which are proposed to improve protection against pathogens including S. pneumoniae. Carriage of Corynebacterium, frequently together with Dolosigranulum, is associated with fewer upper and lower respiratory tract infections in both children and adults (Chonmaitree et al., 2017; Hasegawa et al., 2017; Copeland et al., 2018; Lappan et al., 2018; Man et al., 2019). Corynebacterium species are Gram-positive bacteria which frequently colonize the skin and upper airway. While Corynebacterium diphtheriae, the causative agent of diphtheria, is an important pathogen (Bernard, 2012), other Corynebacterium species are rarely associated with disease. Case reports have identified some Corynebacterium species associated with pneumonia, most frequently Corynebacterium pseudodiphtheriticum (Yang et al., 2018), indicating opportunistic infections may occur. However, non-C. diphtheriae species are more often associated with respiratory health, as the abundance of Corynebacterium species in nasopharynx correlates with reduced S. pneumoniae colonization and infection in several studies (Laufer et al., 2011; Pettigrew et al., 2012; Bomar et al., 2016; Kelly et al., 2017; Xu et al., 2021). Longitudinal studies tracking upper airway microbiome dynamics in children over time indicate that Corynebacterium abundance is predictive of both S. pneumoniae carriage and risk of respiratory tract infection (Biesbroek et al., 2014; Teo et al., 2015; Bosch et al., 2017; Kelly et al., 2021). Both children and adults with a higher abundance of Corynebacterium also have more stable airway microbiomes, compared to those with a predominance of opportunistic pathogens including S. pneumoniae (Bosch et al., 2017; Piters et al., 2019; Kelly et al., 2021). Together, these studies indicate that Corynebacterium species may play an important role in microbiome-mediated protection against pathogens such as S. pneumoniae, thereby reducing susceptibility to pneumonia.
Interruption of the potential protective benefit of Corynebacterium species in the airway is of particular concern for patients receiving antibiotics, as antibiotic therapy selectively reduces the Corynebacterium population while pathogens including S. pneumoniae are enriched (Teo et al., 2015; Prevaes et al., 2016; Kelly et al., 2021). Mechanistically, several Corynebacterium species can directly inhibit growth of S. pneumoniae in vitro (Bomar et al., 2016; Kelly et al., 2021; Xu et al., 2021). Among these, Corynebacterium accolens inhibits S. pneumoniae growth by expression of a bacterial lipase called LipS1, which hydrolyses host triacylglycerols, releasing free fatty acids that kill S. pneumoniae (Bomar et al., 2016). Fatty acids (FAs) are long-chain hydrocarbons with a terminal carboxyl group which are a major component of lipids. Corynebacterium species use fatty acids for growth, and lipophilic species including C. accolens require exogenous lipids to survive (Bernard, 2012; Tchoupa et al., 2021). Two non-lipophilic Corynebacterium species, which do not require host lipids for growth, were recently shown to inhibit S. pneumoniae in vitro, though the mechanisms were not identified (Xu et al., 2021). Another non-lipophilic species, Corynebacterium amycolatum, is frequently isolated from the human nasopharynx (Kaspar et al., 2016; Kelly et al., 2021) but is unusual in that it lacks mycolic acid, a long chain fatty acid which is a major component of the cell wall in almost all other Corynebacterium (Collins et al., 1988; Marrakchi et al., 2014). It is unclear whether FA production by bacterial lipases are universally required for Corynebacterium-mediated inhibition of S. pneumoniae, or whether diverse Corynebacterium species, including those less dependent on exogenous lipids, utilize similar mechanisms.
Understanding the host and bacterial requirements for Corynebacterium-mediated protection against S. pneumoniae is hampered by the lack of animal models for Corynebacterium colonization. Two related studies employing a five-dose serial inoculation of C. pseudodiphtheriticum in infant mice found that this strategy improves protection against S. pneumoniae (Moyano et al., 2020) as well as secondary pneumococcal infection in Respiratory Syncytial Virus (RSV)-infected mice (Kanmani et al., 2017). However, unknown is whether C. pseudodiphtheriticum actually colonizes the infant mouse airway in this model or if other Corynebacterium species can be used in a similar approach. For this study, we developed a mouse model to investigate the impact of two distinct species, C. accolens and C. amycolatum, on protection against S. pneumoniae in vitro and in vivo. Our findings indicate that both C. accolens and C. amycolatum can colonize the airways of antibiotic treated mice, and that pre-exposure to these Corynebacterium species protects against S. pneumoniae colonization of the upper airway as well as infection and inflammation in the lung. We envision that this co-infection model can be used to further understand how exposure to different Corynebacterium species in the airway influences pneumococcal colonization and disease.
Corynebacterium accolens strain ATCC® 49726™ was obtained from the American Type Culture Collection. Corynebacterium amycolatum strain SK46, HM-109 was obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project. Corynebacterium strains were grown from glycerol stocks on BHI agar plates (BD Difco™ Bacto™ Brain Heart Infusion, Thermo Fisher Scientific) aerobically at 37°C overnight. Growth from fresh plates was used to inoculate BHI broth cultures, which were grown overnight aerobically at 37°C with shaking at 200 rpm in preparation for in vitro assays and in vivo infections. BHI plates and broth were supplemented with 1% Tween® 80 (polysorbate, VWR) to provide an exogenous lipid source, unless otherwise specified. When indicated, 180 mg/mL triolein or oleic acid (VWR) were added to plates or broth cultures.
Streptomycin resistant variants of Streptococcus pneumoniae serotype 2 strain D39 and serotype 4 strain TIGR4 were kind gifts from Dr. Jeffrey N. Weiser (New York University). S. pneumoniae serotype 3 strain ATCC® 6303™ from the American Type Culture Collection was also used. S. pneumoniae was grown in Todd Hewitt Broth supplemented with 5% Yeast Extract (BD Bacto™) and 50 μg/mL streptomycin (MilliporeSigma) for streptomycinR strains. For all experiments, S. pneumoniae cultures were grown from frozen stocks at 37°C with 5% CO2 without shaking to mid-log phase, centrifuged at ≥ 20,000 × g for 10 min, and resuspended in phosphate-buffered saline (PBS). S. pneumoniae CFUs were determined by serial dilution plating on Tryptic Soy Broth agar (TSB, MP Biomedicals) supplemented with neomycin 5 μg/mL, streptomycin 50 μg/mL (streptomycinR strains), and fresh catalase (5,000 Units/plate, Worthington Biomedical Corporation Lakewood, NJ) at 37°C with 5% CO2 overnight.
For supernatant inhibition assays, C. accolens and C. amycolatum were grown on BHI agar supplemented with or without 1% Tween 80 and 180 mg/mL triolein. Triolein was emulsified in 100% ethanol for spreading onto plates, and plates were dried to evaporate ethanol prior to inoculation. Plates were incubated under aerobic conditions at 37°C overnight. Cultures were inoculated from fresh plates in BHI broth with or without 1% Tween 80 and 180 mg/mL triolein and grown aerobically at 37°C overnight with shaking at 200 rpm. OD600 was measured to standardize starting inocula, and cultures were centrifuged at ≥ 20,000 × g for 10 min to pellet bacterial cells. Cell-free supernatants (200 μL/plate) were spread onto TSB agar plates containing neomycin 5 μg/mL, streptomycin 50 μg/mL (streptomycinR strains), and fresh catalase (5,000 Units/plate) and allowed to dry completely prior to S. pneumoniae serial dilution plating. For the dose response curve, supernatants were diluted in BHI broth (100–1.6% supernatant) prior to plating. S. pneumoniae serial dilutions were completed in PBS, with 10 μL spotted onto culture plates per dilution, plated in duplicate. Plates were incubated at 37°C with 5% CO2 overnight for S. pneumoniae CFU enumeration.
Mutation of the lipase encoding lipS1 in C. accolens was performed as described previously (Bomar et al., 2016). Briefly, ∼1 kb regions flanking the coding sequence region for WP_005285206.1 in C. accolens ATCC 49726 (nucleotides 2,348,522–2,349,946) were amplified using Q5 high fidelity DNA polymerase (NEB) with primers designed to add a 21 bp scar sequence on the end of the 5′ fragment and beginning of the 3′ fragment, respectively, in addition to a stop codon at the end of the 3′ fragment (see Supplementary Table 1 for primer sequences). PCR products were purified using the Monarch PCR and DNA cleanup kit (NEB). We used the plasmid pKO (KAN) as a suicide vector for lips1 cloning. pKO (KAN) was a gift from Markus Seeger (Addgene #110086; RRID:Addgene_110086).1 pKO was digested with PstI and treated with calf intestinal phosphatase according to manufacturer’s instructions (NEB) and purified using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific). Purified plasmid and PCR products were assembled by Gibson assembly using the HiFi DNA Assembly Cloning Kit (NEB), and the assembled sequence was transformed into high efficiency NEB 5-alpha competent E. coli (NEB). Clones containing the assembly sequence inserted into pKO were identified by sequencing (Quintara Biosciences, Hayward CA). Competent cells of C. accolens were prepared and pKO containing the lipS1 flanking regions was introduced by electroporation as previously described for Corynebacterium glutamicum (Eggeling et al., 2005). Electroporated cells were resuspended in BHI Tween 80 broth containing sorbitol (9.1 g/100 mL) and heat shocked at 46°C for 6 min followed by incubation at 37°C for 1 h prior to plating on selective media. Transformants were grown on BHI agar supplemented with Tween 80, sorbitol, and kanamycin 20 μg/mL to select for clones which integrated the lipS1 flanking regions and kanamycin resistance gene from the modified pKO plasmid into the C. accolens genome by homologous recombination (kanamycinR) followed by 5% sucrose to select for clones which lost the plasmid vector backbone (sucrose selection against sacB in pKO). Deletions in lipS1 were confirmed by PCR amplification and sequencing (Quintara Biosciences, Hayward CA). The same process was used to construct an in-frame deletion in the coding sequence region for WP_005510233.1 in C. amycolatum (nucleotides 16,770–17,957) using primers specific for the flanking sequence regions (Supplementary Table 1).
Male and female 6–12 week old C57BL/6J (WT) mice, purchased from The Jackson Laboratory (RRID:IMSR_JAX:000664), were bred in-house. Mouse colonies were maintained in the University of Colorado Office of Laboratory Animal Resources.
All mice were treated with ingested broad-spectrum antibiotics (ampicillin 1 g/L, neomycin 1 g/L, metronidazole 1 g/L, vancomycin 0.5 g/L, MilliporeSigma and McKesson) ad libitum for 7 days until 48 h prior to infection. Bacterial loads in the stool pre- vs. post-antibiotic treatment were detected by qPCR as described previously (Ricchi et al., 2017). Briefly, stools were weighed and genomic DNA was extracted using the PureLink™ Genomic DNA Mini Kit (Thermo Fisher Scientific). Quantitative PCR was performed using iTaq™ Universal SYBR® Green Supermix (BioRad) together with 200 nM forward and reverse primers (Supplementary Table 1; Jimeno et al., 2018) and 2.5 μL template DNA. Reactions were performed using a CFX Connect™ Real-Time System (BioRad) with cycling conditions of: (1) 94°C for 4 min; (2) 40 cycles of 15 s at 95°C, 30 s at 60°C, and (3) 72°C for 10 min. A standard curve was generated using a known concentration of S. pneumoniae D39 gDNA, which together with the input ng/μL DNA and Ct values was used to calculate total 16S rRNA gene copy number as described previously (Gomes-Neto et al., 2017). qPCR data was analyzed using CFX Manager Software (version 2.1, BioRad). Nasal lavages, which can only be collected upon sacrifice, were obtained from separate groups of mice with and without antibiotic treatment for quantification of bacterial load by qPCR as above. Lavages were performed by the instillation of 200 μL PBS into cannulated trachea through the nasal cavity and collected from the nares.
Mice were infected with C. accolens, C. amycolatum, and/or S. pneumoniae, as indicated, at 107 CFU/mouse i.n. under inhaled isoflurane anesthesia. Nasal lavages were collected as above. Lungs were homogenized using a Bullet Blender tissue homogenizer (Stellar Scientific, Baltimore, MD). S. pneumoniae nasal lavage and lung CFUs were calculated following serial dilution and growth on Tryptic Soy agar plates containing neomycin (5 μg/mL, MilliporeSigma) and streptomycin (50 μg/mL, only streptomycinR strains), supplemented with fresh catalase (5,000 Units/plate, Worthington Biochemical Corporation, Lakewood, NJ) grown overnight at 37°C with 5% CO2. Corynebacterium CFUs were calculated following serial dilution and growth on BHI plates containing 1% Tween 80 grown overnight aerobically at 37°C. In co-infected mice, Corynebacterium CFUs were determined by subtracting S. pneumoniae CFUs (on streptomycin plates) from total growth (on non-selective BHI plates).
Single cells were prepared from lungs following transcardial perfusion with PBS as described previously (Clark et al., 2020). Briefly, lungs were minced and digested with DNAseI (30 μg/mL, MilliporeSigma) and type 4 collagenase (1 mg/mL, Worthington Biochemical Corporation). Lung preparations were filtered with a 70 μM strainer prior to lysis of red blood cells by brief (1 min) incubation in RBC lysis buffer (0.15M NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA, pH 7.4). Cell Fc receptors were blocked with anti-CD16/32 2.4G2 hybridoma supernatant prior to staining in FACS buffer (1% BSA, 0.01% NaN3 in PBS). Cells were fixed in 1% paraformaldehyde. Antibodies used included: Siglec F (clone E50-2440), MHCII (clone M5/114.15.2), Ly6G (clone 1A8), Ly6C (clone HK1.4), CD45.2 (clone 104), CD11c (clone N418), CD11b (clone M1/70), CD64 (clone X54-5/7.1), and CD103 (clone 2E7). Antibodies were purchased from eBioscience, BD, BioLegend, or Thermo Fisher Scientific. Flow cytometry was performed using an LSRII (BD) in the Rocky Mountain Regional VAMC Research Core, Aurora CO and data were analyzed using FlowJo™ v10.8 Software (BD Life Sciences).
BAL cytokines and chemokines were measured using a LEGENDplex™ Mouse Inflammation Panel, and data were analyzed using the LEGENDplex™ Data Analysis Software Suite (BioLegend). Analytes were detected on the LSR Fortessa X-20 in the ImmunoMicro Flow Cytometry Shared Resource Laboratory at the University of Colorado Anschutz Medical Campus (RRID:SCR_021321). Serum cytokines were measured using mouse TNFα and IFNγ ELISA kits (BD), with analytes detected on a Synergy™ HT Microplate Reader (BioTek).
Samples were analyzed via ultra-high pressure liquid chromatography coupled to mass spectrometry (UHPLC-MS) at the University of Colorado School of Medicine Metabolomics Core. Nasal lavages were vigorously vortexed to homogenize, then diluted 1:1 in Optima LC-MS (Fisher) grade ice-cold MeOH and vortexed briefly again. Next, samples were incubated at –20°C for 30 min, then centrifuged at 4°C, 30,130 × g for 10 min to remove remaining solids as previously described (Nemkov et al., 2017; Reisz et al., 2019). Supernatants were transferred to pre-cooled autosampler vials, randomized, and analyzed on a Vanquish UHPLC coupled to a Q Exactive MS (Thermo Fisher Scientific, San Jose, CA, United States). Extracts (15 μL) were resolved with flow rate of 300–400 μL/min over an Acuity HSS T3 column (150 × 2.1 mm, 1.8 μm, Waters, Milford, MA, United States) with a 17-min gradient in negative ion polarity mode as previously described (Reisz et al., 2019). Samples were introduced to the MS via electrospray ionization with the MS scanning in full MS mode over the range of 150–1,500 m/z. As a quality control measure, technical replicates were injected every six samples to verify instrument stability (Nemkov et al., 2019). Fatty acids were annotated and integrated with Maven (Princeton University) in reference to the KEGG database. Peak quality was validated using blanks, technical mixes, and 13C natural isotope abundance (Nemkov et al., 2015).
These studies were approved by the Animal Care and Use Committee (Protocol #00927) and the Institutional Biosafety Committee (Protocol #1418) of the University of Colorado School of Medicine.
Graphing and statistical analysis were completed using GraphPad Prism (Version 8.4.3, GraphPad Software, LLC, San Diego, CA). Statistical tests included t-tests for single comparisons, ANOVA with multiple group comparisons of normally distributed data and Kruskal-Wallis tests with multiple group comparisons of non-normal (non-Gaussian) distributions of infectious burden data (due to the limit of detection cut-off). For all statistical tests p < 0.05 was considered significant. All in vivo infections were conducted with 3–6 mice per group and repeated 3 times. For in vitro assays, experiments were completed with 2–3 technical replicates per condition and repeated 3 times.
A recent survey of morphologically distinct Corynebacterium strains collected from infants showed that cell-free supernatants from four different lipophilic species, including C. accolens, inhibited S. pneumoniae growth in vitro (Kelly et al., 2021). However, supernatants from only 20% of the Corynebacterium isolates tested in this study were inhibitory, indicating that not all Corynebacterium strains mediate S. pneumoniae growth restriction in vitro. We considered whether C. amycolatum, which unlike C. accolens does not require exogenous FAs for either growth or mycolic acid production, inhibits S. pneumoniae growth in vitro. Cell-free supernatants from C. accolens or C. amycolatum were spread onto culture plates prior to S. pneumoniae inoculation, and S. pneumoniae colony forming units (CFUs) were determined following overnight incubation (Figure 1A). In contrast to robust growth of type 2 S. pneumoniae (strain D39) on untreated culture plates, no growth of S. pneumoniae was detected on plates pre-treated with supernatants from either C. accolens or C. amycolatum (Figure 1B and Supplementary Figure 1A). S. pneumoniae growth was also not detected on plates treated with oleic acid, a FA that kills S. pneumoniae (Clementi et al., 2013), included as a positive control. Supernatants from both C. accolens and C. amycolatum demonstrated similar inhibition of type 4 and type 3 S. pneumoniae strains (Figures 1C,D), indicating that multiple serotypes of S. pneumoniae are vulnerable to C. accolens and C. amycolatum-mediated growth restriction. Thus, both C. accolens and the non-lipophilic C. amycolatum inhibit S. pneumoniae growth in vitro.
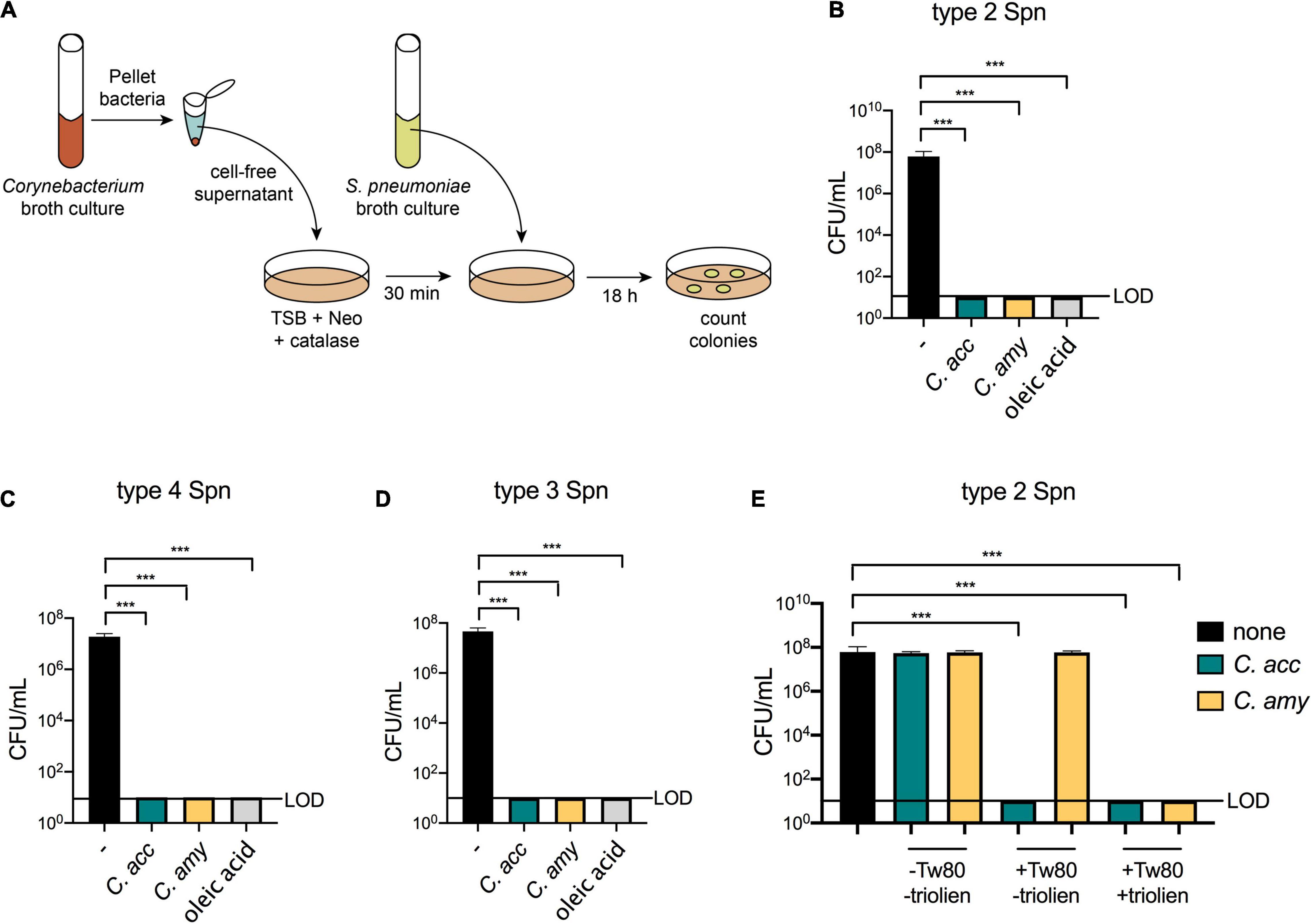
Figure 1. Inhibition of S. pneumoniae growth in vitro by lipophilic and non-lipophilic Corynebacterium species. Supernatant inhibition assay schematic using tryptic soy broth (TSB), neomycin (Neo), and catalase plates (A). Growth of type 2 S. pneumoniae (Spn), (B), type 4 Spn (C), and type 3 Spn (D) on untreated plates (–) or on plates pre-treated with supernatants from C. accolens (C. acc) or C. amycolatum (C. amy) or with 180 mg/mL oleic acid. Growth of type 2 Spn on untreated plates (–) or on plates pre-treated with supernatants from C. acc or C. amy grown in BHI with or without 1% Tween 80 and 180 mg/mL triolein (E). Limit of detection (LOD) is indicated. Data are pooled from three independent experiments in duplicate. ***p < 0.001, ANOVA.
We next considered whether the source or amount of FAs determined Corynebacterium-mediated inhibition of S. pneumoniae growth. In the supernatant assays described above, Corynebacterium cultures were supplemented with the synthetic surfactant Tween 80 and triolein, a naturally occurring triacylglycerol. Both Tween 80 and triolein are hydrolyzed by bacterial enzymes, resulting in the release of oleic acid, which is used by Corynebacterium as a carbon source for growth (Plou et al., 1998; Taoka et al., 2011). Triolein is hydrolyzed by bacterial lipases, while Tween 80 can be hydrolyzed by both lipases and esterases (Plou et al., 1998). As a result, the bacterial lipase LipS1 is required for growth of lipophilic C. accolens in triolein as the sole source of FAs, whereas other enzymes suffice for growth in Tween 80 (Bomar et al., 2016). To compare the relative importance of Tween 80 vs. triolein for Corynebacterium-mediated inhibition of S. pneumoniae growth, we investigated the inhibitory capacity of supernatants from Corynebacterium grown in BHI supplemented with none, one, or both of these lipid sources. For C. accolens, the addition of Tween 80 to cultures was sufficient for supernatant inhibition of S. pneumoniae. In contrast, C. amycolatum inhibition required the addition of both Tween 80 and triolein (Figure 1E). Titration of the amount of Corynebacterium supernatants added to culture plates revealed that at lower concentrations, C. accolens supernatants from cultures with triolein were more inhibitory than those without triolein (Supplementary Figure 1B). For C. amycolatum, triolein was required for inhibition, which only occurred when supernatants were added above a threshold concentration (Supplementary Figure 1B). We also confirmed that BHI alone, supplemented with Tween 80 or Tween 80 and triolein, was not inhibitory (Supplementary Figure 1B). These results indicate a dose-dependence for C. accolens and C. amycolatum-mediated inhibition of S. pneumoniae. In both cases, the inclusion of triolein increased the inhibitory effect, although the efficiency of C. amycolatum-mediated inhibition was lower than that of C. accolens, which was inhibitory in the presence of Tween 80 alone. This difference in substrate usage likely relates to the composition and expression of bacterial lipases and esterases responsible for triolein vs. Tween 80 cleavage in C. amycolatum compared with C. accolens. Therefore, despite the ability of C. amycolatum to grow without an external source of FAs, these are required for the inhibitory effect on S. pneumoniae. In aggregate, these results confirm and extend the importance of FA release following lipid hydrolysis for Corynebacterium-mediated inhibition of S. pneumoniae in vitro and identify differential requirements in distinct Corynebacterium species.
In order to investigate whether Corynebacterium species mediate inhibition of S. pneumoniae in vivo, we first established a model of short-term Corynebacterium colonization. Mice were treated for 1 week with water containing an antibiotic cocktail to reduce the endogenous flora. One week of antibiotic treatment significantly decreased the total microbial burden in both the upper airway and gut (Supplementary Figure 2). Mice were taken off antibiotics 2 days prior to intranasal (i.n.) infection with C. accolens, C. amycolatum, or PBS (control), and Corynebacterium burdens were determined 2 days following infection (Figure 2A). Both C. accolens and C. amycolatum were detected in the upper airway (nasal lavage) and lungs of mice 48 h post-infection as measured by live CFU counts from nasal lavage fluid and homogenized lung tissue of mice infected with Corynebacterium species, but not PBS-treated controls (Figure 2B). Both sites were successfully colonized, albeit with generally higher numbers in the lungs than in the upper airway. These data indicate that both lipophilic and non-lipophilic Corynebacterium species can successfully colonize the airway of antibiotic-treated mice. In order to determine the availability of FAs in the murine upper airway, as well as whether Corynebacterium colonization influenced FA content, nasal lavage fluids were analyzed by mass spectrometry (MS). Several FAs were detected in the nasal lavage fluid, including oleic acid, myristic acid, and palmitoleic acid (Figure 2C), all of which are also present in human nasal lavage fluid (Do et al., 2008). These data are consistent with the presence of FAs in the upper airway of mice which are capable of supporting Corynebacterium growth. However, Corynebacterium colonization did not significantly alter the relative abundances of any of the FAs detected (Figure 2C). These data suggest that either short-term colonization is not sufficient to alter the FA landscape, or that FA turnover by host or bacterial metabolic processes obscured any temporal changes. This model for short-term Corynebacterium colonization of the murine airway provided the opportunity to characterize the physiologic and pathologic impact of these commensals on pneumococcal infection.
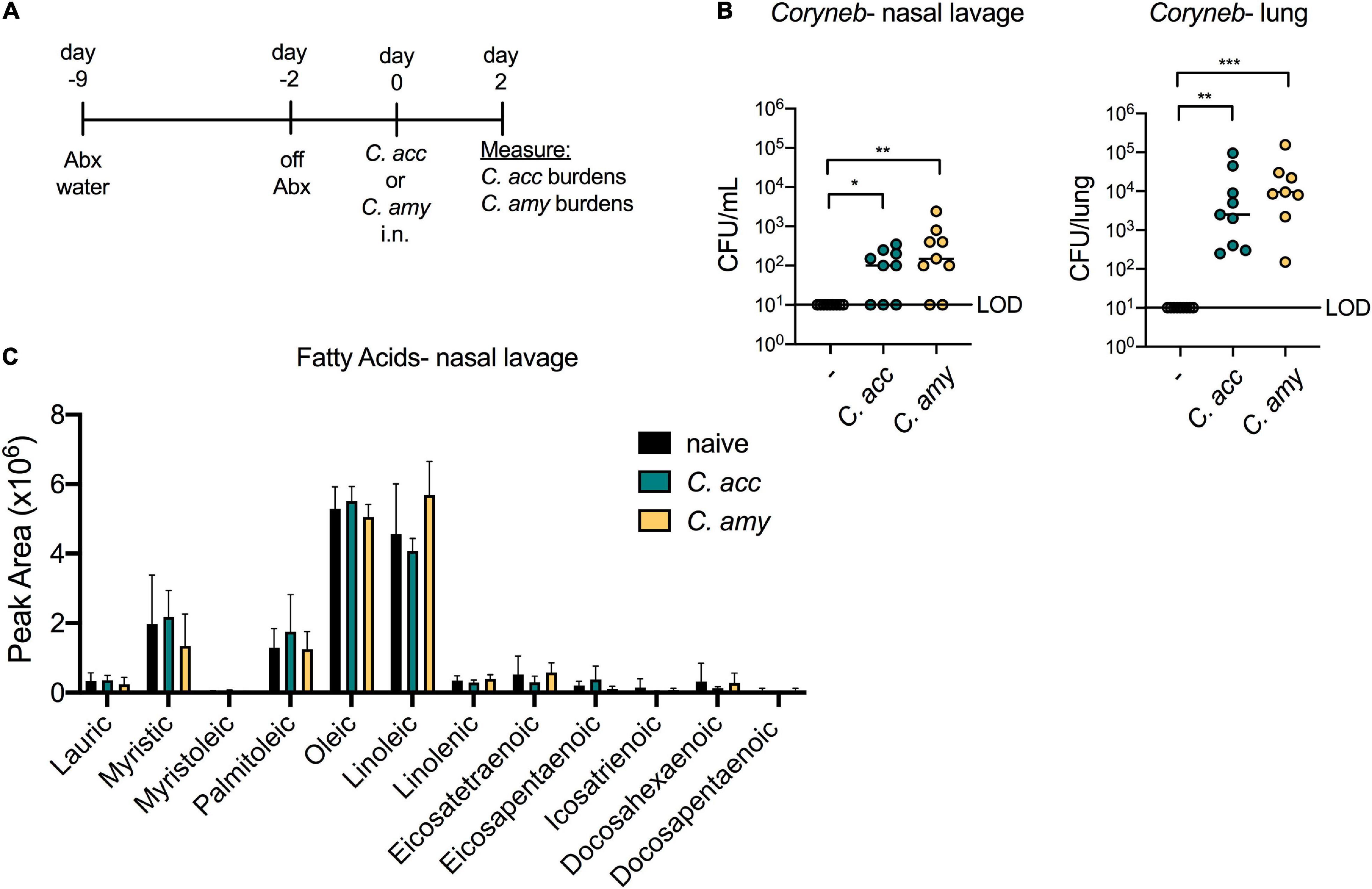
Figure 2. Corynebacterium species colonize the upper and lower airway of antibiotic treated mice. Timeline for antibiotic treatment followed by infection with C. acc or C. amy (A). Burden of Corynebacterium (Coryneb) in the nasal lavage fluid and lungs of mice 48 h post-infection with C. acc, C. amy, or PBS (–) detected by live growth (B). Relative abundance of fatty acids detected in the nasal lavage fluid of naïve mice and mice 48 h post-infection with C. acc or C. amy (C). Data are pooled from three independent experiments with 3–6 mice per group. No significant differences detected in relative FA abundances between groups, ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, Kruskal-Wallis test.
We next interrogated the impact of short-term Corynebacterium colonization on local and systemic cytokine production as well as the recruitment of inflammatory myeloid cells to the lung. Both C. accolens and C. amycolatum induced a significant increase in systemic levels of the pro-inflammatory cytokines TNFα and IFNγ, compared with PBS-treated mice (Figure 3A). In the lung, however, we found that Corynebacterium colonization had little impact on cytokine expression in the bronchoalveolar lavage fluid (BAL) at 48 h post-infection, as measured by a 13-plex inflammatory cytokine/chemokine array. Of the 13 cytokines and chemokines tested, only MCP-1 (CCL2), an important chemokine for monocyte and macrophage recruitment (Gschwandtner et al., 2019), was significantly upregulated in the BAL of C. accolens and C. amycolatum-colonized mice (Figure 3B and not shown). These findings indicate that Corynebacterium colonization is associated with increased circulating pro-inflammatory cytokines, but a minimal such response in the lung.
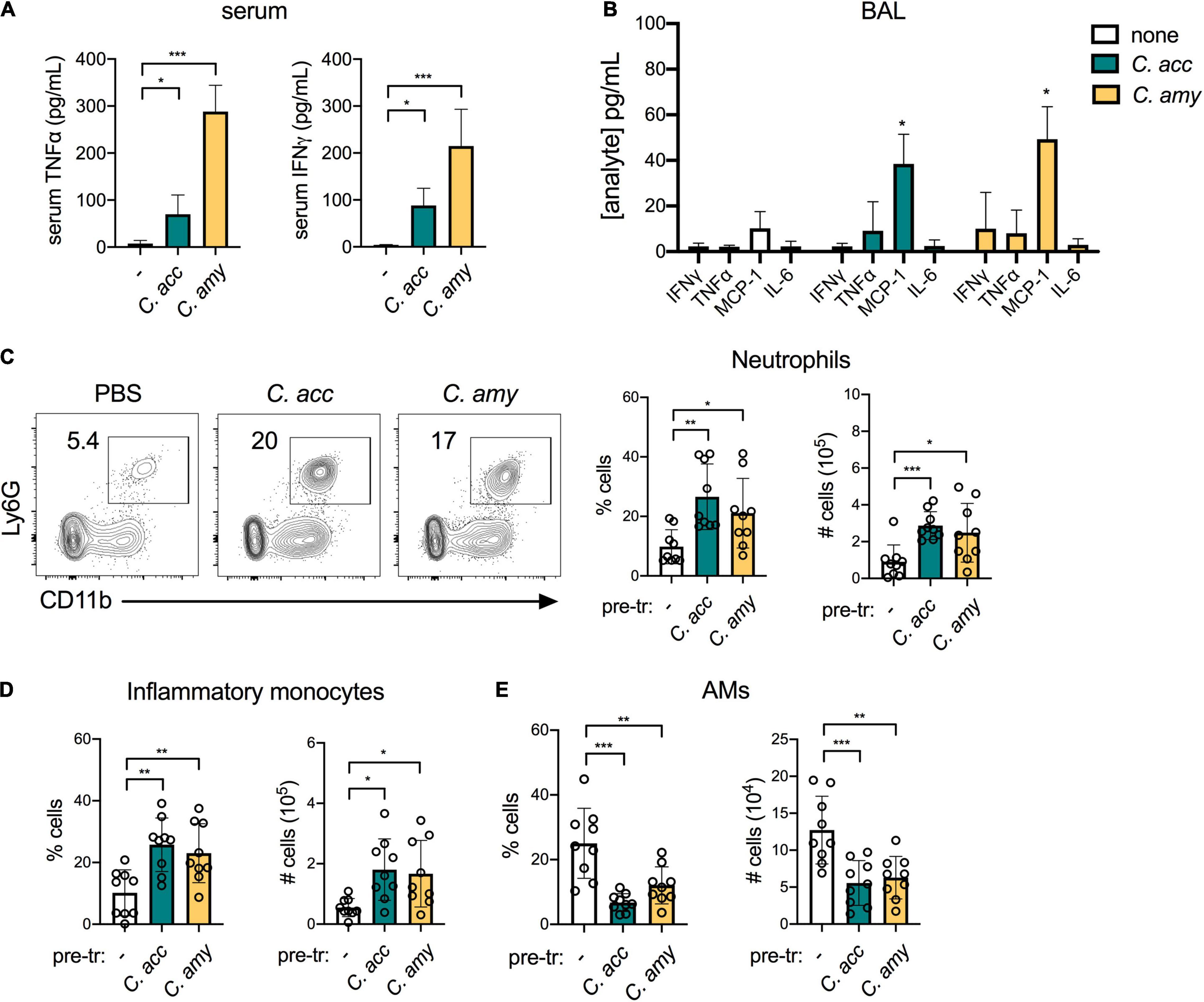
Figure 3. Inflammatory myeloid cells are recruited to the lungs of Corynebacterium-colonized mice. Systemic TNFα and IFNγ detected in mice 48 h post-infection with C. acc, C. amy, or PBS (–) (A). Cytokines and chemokines detected in lung BAL in mice 48 h post-infection with C. acc or C. amy (B), with significance indicated relative to mice pre-exposed to PBS (none). Percentage and total number of neutrophils (CD45+SiglecF–Ly6G+CD11b+ cells) (C), inflammatory monocytes (CD45+ Ly6G–Ly6C+CD11b+ cells) (D), and alveolar macrophages (AMs, CD45+SiglecF+CD11blowCD11c+ cells) (E) detected by flow cytometry in the lungs of mice 48 h post-infection with C. acc, C. amy, or PBS (–). Data are pooled from three independent experiments with 3–6 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, ANOVA.
We determined the impact of Corynebacterium colonization on lung innate immune cell populations by flow cytometry (see gating strategy, Supplementary Figure 3). Corynebacterium colonization was associated with an increase of both neutrophils (CD45+Ly6G+CD11b+ cells) and inflammatory monocytes (CD45+Ly6G–Ly6C+CD11b+ cells) in the lung (Figures 3C,D). These increases parallel the elevated MCP-1, which mobilizes the recruitment of inflammatory monocytes from the bone marrow (Gschwandtner et al., 2019), detected in the BAL of Corynebacterium-infected mice. Neutrophils are also recruited to the lung from the bone marrow, though responses to other chemokines such as MIP-1 (CXCL2) (Lin and Fessler, 2021) were not measured here. In contrast, the lungs of mice infected with C. accolens or C. amycolatum had reduced alveolar macrophages (AMs, CD45+SiglecF+CD11blowCD11c+ cells), compared with PBS treated controls (Figure 3E). Unlike neutrophils and inflammatory monocytes, AMs are largely self-renewing locally in the lung. As the first immune cell to encounter inhaled particles, AMs frequently undergo cell death following bacterial uptake (FitzGerald et al., 2020), consistent with our observation of reduced AMs in the lungs of Corynebacterium infected mice. We detected no changes in CD103+ dendritic cells (DCs, CD45+MHCII+SiglecF–CD11c+CD64– cells) or CD11bhi DCs, or the expression of MHCII in either population as an indicator of activation, in Corynebacterium-infected mice compared with PBS-treated controls (Supplementary Figure 4). Collectively, these data indicate that Corynebacterium colonization is associated with selective mucosal responses, including increased chemokine-associated myeloid cell recruitment to the lung but minimal changes to the local pro-inflammatory cytokine landscape.
Our goal in establishing a model for Corynebacterium colonization was to evaluate whether Corynebacterium species inhibit the acquisition and growth of S. pneumoniae in vivo, similar to Corynebacterium-mediated inhibition of S. pneumoniae observed by our group and others in vitro. We exposed antibiotic-treated mice to C. accolens or C. amycolatum 1 day prior to intranasal infection with streptomycin-resistant type 2 S. pneumoniae and plated nasal lavage contents and lung homogenates on selective media containing streptomycin 1 day later (Figure 4A). Compared with mice pre-exposed to PBS, colonization with either C. accolens or C. amycolatum significantly reduced the burden of S. pneumoniae in both the upper airway (nasal lavage) and lung by 24 h post-infection (Figure 4B). In mice pre-exposed to Corynebacterium, S. pneumoniae burdens were reduced by ∼1 log in the upper airway, and S. pneumoniae was not detected in the lungs of 50–60% of the animals in each Corynebacterium exposure group. C. accolens and C. amycolatum burdens remained detectable following growth in non-selective media in the majority of co-infected mice at this timepoint (Figure 4C). Further, mice with the highest Corynebacterium burdens had the lowest, frequently undetectable, S. pneumoniae burdens in the lung (Supplementary Figures 5A,B). Our data are consistent with longitudinal microbiome sequencing studies which suggest that Corynebacterium abundance predicts S. pneumoniae acquisition and infection risk in children (Biesbroek et al., 2014; Teo et al., 2015; Bosch et al., 2017; Kelly et al., 2021). Of particular note, these studies demonstrate the potential for live animal modeling of Corynebacterium-S. pneumoniae dynamics in vivo in order to elucidate the bacterial and host requirements for Corynebacterium-mediated protection against respiratory tract infection.
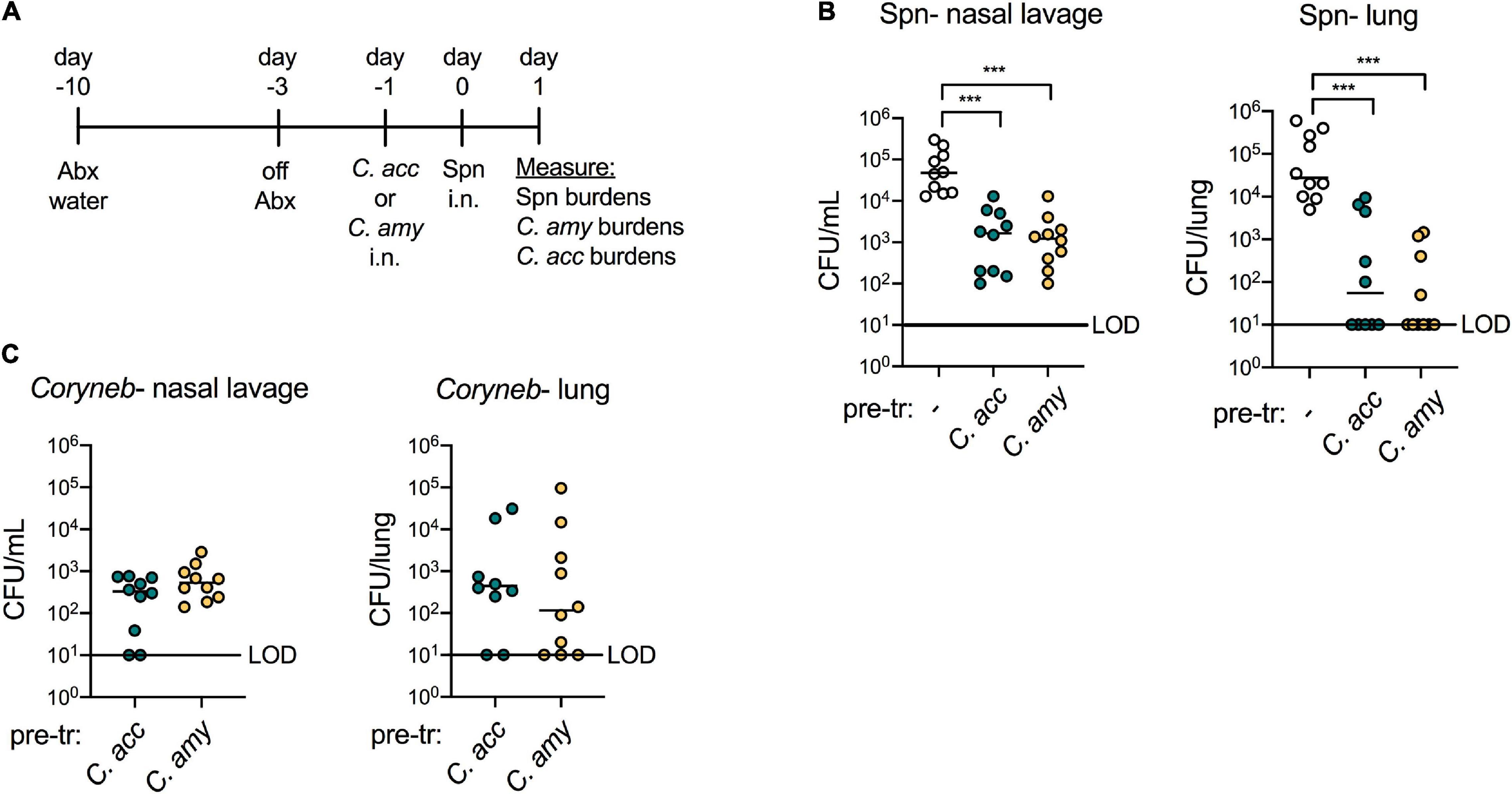
Figure 4. Colonization with Corynebacterium inhibits colonization and infection with S. pneumoniae in vivo. Timeline for antibiotic treatment followed by pre-exposure to PBS (–), C. acc or C. amy prior to infection with S. pneumoniae (Spn) (A). Burden of Spn (B) or Corynebacterium (Coryneb) (C) in nasal lavage fluid and lungs of mice pre-exposed to PBS (–), C. acc or C. amy at 24 h post-Spn infection detected by live growth. Limit of detection (LOD) is indicated. Data are pooled from three independent experiments with 3–5 mice per group. ***p < 0.001, Kruskal-Wallis test.
Corynebacterium co-infected mice also had a reduced inflammatory response in the lung compared to mice infected with S. pneumoniae alone. In contrast to S. pneumoniae challenged mice pre-exposed to PBS, systemic TNFα and IFNγ were reduced in mice pre-exposed to C. accolens or C. amycolatum (Figure 5A). Similarly, in the BAL of co-infected mice, levels of several pro-inflammatory cytokines including TNFα, IFNγ, and IL-6 as well as the chemokine MCP-1 were reduced in mice pre-exposed to Corynebacterium compared to mice infected with S. pneumoniae alone (Figure 5B), while 9 other cytokines were not affected (not shown). Total neutrophils and inflammatory monocytes in the lungs of co-infected mice were also reduced compared with mice infected with S. pneumoniae alone (Figures 5C,D). In contrast, AMs were increased in co-infected mice (Figure 5E). Overall populations of lung DCs, which are important for mobilizing the adaptive immune response but may also facilitate pneumococcal dissemination (Rosendahl et al., 2013), remained the same, although CD103+ DC expression of the activation marker MHCII was significantly reduced in Corynebacterium co-infected mice compared to mice infected with S. pneumoniae alone (Supplementary Figure 6). The reduction in inflammatory myeloid cells together with the recovery of AMs, which are important for the resolution of inflammation in the lung, is consistent with a pro-resolving immune response in the lungs of co-infected mice. We conclude that colonization with either C. accolens or C. amycolatum improves early clearance of S. pneumoniae from the upper and lower airways of mice, in association with reduced, rather than enhanced, inflammation in the lung, likely limiting lung injury.
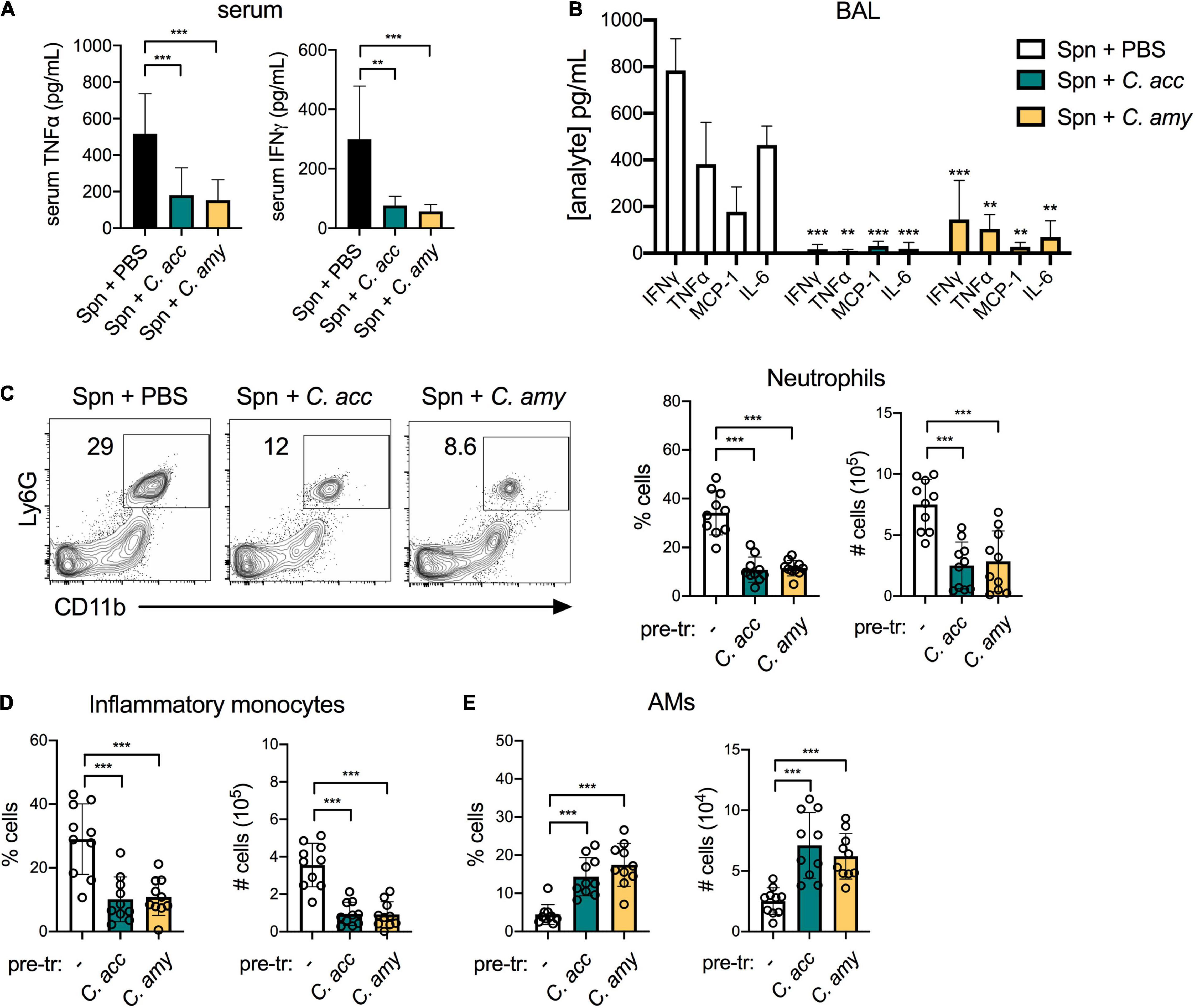
Figure 5. Reduced inflammation and inflammatory myeloid cell recruitment in the lungs of Corynebacterium-S. pneumoniae co-infected mice. Systemic TNFα and IFNγ detected in mice pre-exposed to PBS (–), C. acc or C. amy at 24 h post-S. pneumoniae (Spn) infection (A). Cytokines and chemokines detected in lung BAL in mice pre-exposed to PBS (–), C. acc or C. amy at 24 h post-Spn infection (B), significance indicated relative to mice pre-exposed to PBS (none). Percentage and total number of neutrophils (C), inflammatory monocytes (D), and alveolar macrophages (AMs) (E) detected by flow cytometry in the lungs of mice pre-exposed to PBS (–), C. acc or C. amy at 24 h post-Spn infection. Data are pooled from three independent experiments with 3–5 mice per group. **p < 0.01, ***p < 0.001, ANOVA.
We next sought to confirm the importance of the lipase LipS1 for C. accolens-mediated inhibition of S. pneumoniae growth in vitro and to determine whether C. amycolatum expresses a lipase with similar activity. We first constructed a lipS1 mutant in our strain of C. accolens by in-frame gene deletion. As described for the original ΔlipS1 C. accolens, our ΔlipS1 C. accolens grew poorly in triolein as the sole source of FAs (Bomar et al., 2016), indicating LipS1 as critical for lipase-mediated hydrolysis and growth in C. accolens (Supplementary Figure 7A). In contrast, growth of ΔlipS1 C. accolens in Tween 80 was not impaired relative to wild-type (WT) C. accolens, presumably due to the hydrolysis of Tween 80 by bacterial esterases.
Bacterial lipases in both C. accolens and C. amycolatum are essential for Corynebacterium-mediated inhibition of S. pneumoniae growth in vitro but have distinct characteristics. Supernatants from ΔlipS1 C. accolens did not inhibit S. pneumoniae growth in vitro, whereas those from WT C. accolens did (Figure 6A), confirming the importance of LipS1 for S. pneumoniae inhibition in vitro. A search for similar bacterial lipases in C. amycolatum in the NCBI database identified the protein WP_005510233.1 as sharing the greatest homology with that of C. accolens LipS1 (WP_005285206.1). Though overall protein sequence homology to C. accolens LipS1 was low (36%), the NCBI Conserved Domain Database (CDD, version cdd.v.3.19) (Lu et al., 2019) classified WP_005510233.1 as a member of the LIP family of secretory lipases with an E-value of 2.94e–33, similar to C. accolens LipS1. C. amycolatum WP_005510233.1 also contains the lipase motif G-X-S-X-G with a predicted active site serine residue (Jaeger et al., 1994), a shared feature among bacterial lipases. Analysis using the SignalP program (version 5.0) (Petersen et al., 2011) indicated that while both C. accolens LipS1 and C. amycolatum WP_005510233.1 are predicted to be secreted by the general secretion pathway (Sec), LipS1 contains an SP1 cleavage site, whereas C. amycolatum WP_005510233.1 contains an SPII cleavage site. Based on this analysis, we generated an in-frame deletion of WP_005510233.1 in C. amycolatum, referred to as Δlip C. amycolatum, to determine whether this putative lipase contributes to C. amycolatum-mediated inhibition of S. pneumoniae growth. As C. amycolatum is non-lipophilic, Δlip C. amycolatum grew similarly to WT in either Tween 80 or triolein (Supplementary Figure 7B). However, supernatant from Δlip C. amycolatum no longer inhibited S. pneumoniae growth in vitro (Figure 6A), suggesting that this lipase is critical for C. amycolatum-mediated inhibition in vitro.
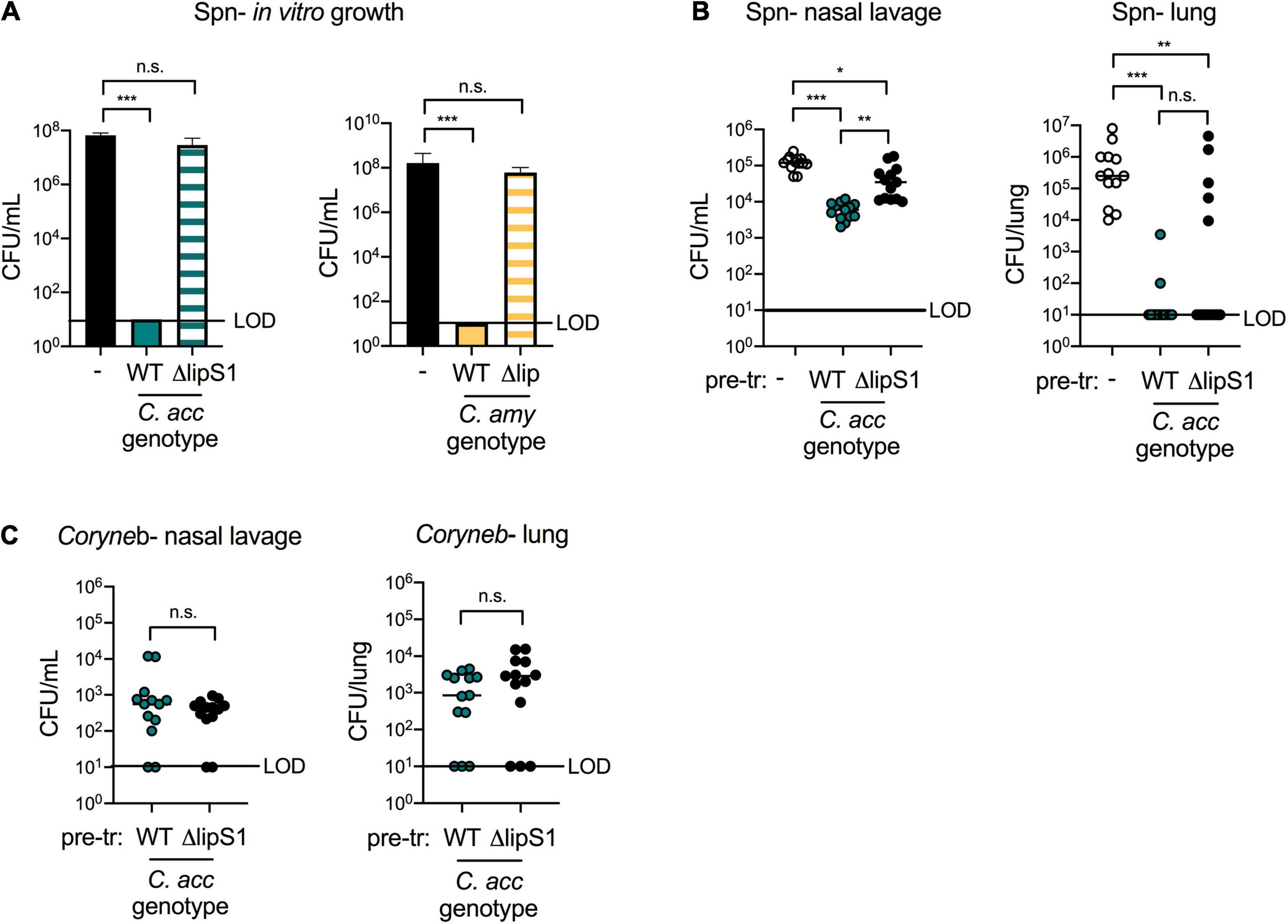
Figure 6. Corynebacterium lipases contribute to, but are not required for, protection against S. pneumoniae in vivo. Growth of type 2 S. pneumoniae (Spn) on untreated plates (–) or on plates pre-treated with supernatants from WT vs. ΔlipS1 C. acc or WT vs. Δlip C. amy grown in BHI with 1% Tween 80 and 180 mg/mL triolein (A). Burden of Spn (B) or Corynebacterium (Coryneb) (C) in nasal lavage fluid and lungs of mice pre-exposed to PBS (–), WT C. acc or ΔlipS1 C. acc at 24 h post-Spn infection detected by live growth. Limit of detection (LOD) is indicated. Data are pooled from three independent experiments with 3–5 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, ANOVA (A) or Kruskal-Wallis test (B,C).
Lipases such as LipS1 and/or other bacterial factors may contribute to Corynebacterium-mediated protection against S. pneumoniae in vivo. Using our co-infection animal model, we compared the impact of pre-exposure to WT vs. ΔlipS1 C. accolens on S. pneumoniae colonization and infection. S. pneumoniae burdens in the upper airway (nasal lavage) were higher in mice pre-exposed to ΔlipS1 C. accolens compared to mice pre-exposed to WT C. accolens (Figure 6B). However, there was still a significant reduction in upper airway S. pneumoniae burdens in mice pre-exposed to ΔlipS1 C. accolens compared to mice infected with S. pneumoniae alone. These findings indicate that C. accolens expression of LipS1 contributes to, but is not solely required for, inhibition of S. pneumoniae colonization in the upper airway. In the lungs, mice pre-exposed to either ΔlipS1 or WT C. accolens had significantly reduced burdens of S. pneumoniae compared with mice infected with S. pneumoniae alone (Figure 6B). There was a slight increase in the percentage of mice with detectable S. pneumoniae burdens in the lung following pre-exposure to ΔlipS1 C. accolens (38%) vs. WT C. accolens (15%), similar to the upper airway, however, this difference was not statistically significant. Burdens of WT and ΔlipS1 C. accolens in the nasal lavage and lungs of co-infected mice were similar (Figure 6C), indicating no colonization defect for ΔlipS1 C. accolens. Also, mice with the highest burdens of either WT or ΔlipS1 C. accolens correlated with lower and frequently undetectable burdens of S. pneumoniae in the lung (Supplementary Figure 5C), confirming a relationship between C. accolens abundance and reduced S. pneumoniae infection of the lung. These data indicate that multiple mechanisms, both LipS1-dependent and independent, contribute to Corynebacterium-mediated protection against S. pneumoniae in vivo.
Our findings provide novel evidence in support of the potential benefit of Corynebacterium abundance in the airway. Both C. accolens and C. amycolatum inhibited S. pneumoniae growth in vitro, observations corroborated by protection in vivo using a new co-infection model. The only mechanism for Corynebacterium-mediated protection against S. pneumoniae described thus far is the liberation of host FAs by LipS1 in C. accolens (Bomar et al., 2016), as confirmed herein both in vitro and in vivo. Beyond direct killing of S. pneumoniae, FAs induce expression of the antimicrobial peptide β-defensin-2 by human sebocytes (Nakatsuji et al., 2010), which may further enhance protection against S. pneumoniae in the airway. We demonstrate that FAs are present in the upper airway of mice, as in humans (Do et al., 2008), consistent with their availability to serve as a nutrient source for lipophilic Corynebacterium such as C. accolens. The finding that Corynebacterium colonization did not influence FA abundance may result from a lack of accessible triacylglycerols for FA production, as has been suggested (Bomar et al., 2016), metabolic turnover, or temporal fluctuations which were missed. Alternatively, we did not determine whether the expression of LipS1 is upregulated in co-infected mice. Regardless, the observation that LipS1-deficient C. accolens is less protective against S. pneumoniae suggests that lipase activity contributes to S. pneumoniae killing in vivo. Others have confirmed that the mouse airway can be colonized with endogenous Corynebacterium, which becomes the dominant genus following treatment with the antibiotic neomycin (Ichinohe et al., 2011). This finding indicates that competition with the resident microbiota limits Corynebacterium colonization, consistent with our inability to establish Corynebacterium colonization in non-antibiotic-treated mice (not shown). Thus, while host lipids, including FAs, are present in the mouse airway, microbial depletion is likely a requirement for modeling Corynebacterium colonization.
The partial protective effect of LipS1-deficient C. accolens indicates a role for additional mechanisms of Corynebacterium-mediated protection in vivo. S. pneumoniae and other opportunistic pathogens are enriched following antibiotic therapy in children, whereas Corynebacterium are reduced (Teo et al., 2015; Kelly et al., 2021), suggesting that S. pneumoniae is better equipped for expansion into the microbiome-depleted airway. We found that colonizing mice with Corynebacterium prior to S. pneumoniae exposure is sufficient to reverse this expansion. In addition to competition with pathogens through the expression of bacterial lipases, some Corynebacterium species produce siderophores, which reduce growth of the pathogen Staphylococcus aureus in iron-limiting environments (Stubbendieck et al., 2019), although the potential impact on S. pneumoniae is not known. Beyond competition for niche space and nutrients, interactions between Corynebacterium and airway epithelial cells may contribute to enhanced protection against S. pneumoniae in vivo. For example, some species of Corynebacterium express the molecule phosphorylcholine (ChoP) (Gillespie et al., 1996), which binds to platelet activating factor receptor (PAFR), a critical adherence receptor for S. pneumoniae (Rijneveld et al., 2004). ChoP expression by selected Corynebacterium species may therefore interfere with S. pneumoniae access to PAFR, reducing S. pneumoniae acquisition and infection. Moreover, Corynebacterium activation of airway epithelial cells may enhance their production of antimicrobial peptides in a FA-independent manner, as with other airway commensals (Liu et al., 2019). Finally, Corynebacterium-mediated recruitment of inflammatory myeloid cells, as we observed in the lung, may serve as an immune priming signal to enhance clearance of S. pneumoniae, as neutrophils and inflammatory monocytes are critical for early S. pneumoniae killing (Calbo and Garau, 2010). The development of an animal model for Corynebacterium colonization, which mirrors the relationship between Corynebacterium abundance and risk of S. pneumoniae acquisition observed in humans, should facilitate investigation of these and other mechanisms in future studies. However, translation of such findings in mice into novel therapeutic approaches will require verification that similar effects are active during human colonization with Corynebacterium.
In these studies, two morphologically and metabolically distinct species of Corynebacterium protected against S. pneumoniae growth. The non-lipophilic C. amycolatum was only inhibitory in vitro upon the addition of multiple FA sources, indicating that screens which do not incorporate enough exogenous lipids may miss the inhibitory potential of additional Corynebacterium clinical isolates. Indeed, while airway microbiome sequencing studies indicate a broadly protective role for Corynebacterium species against respiratory tract infections and S. pneumoniae, mechanistic studies have revealed heterogeneity among different Corynebacterium-pathogen pairs. For example, both C. pseudodiphtheriticum and Corynebacterium striatum inhibit growth of the pathogen S. aureus, whereas C. accolens instead promotes S. aureus growth in vitro (Yan et al., 2013; Ramsey et al., 2016). C. pseudodiphtheriticum mediates a broadly protective effect against primary and secondary S. pneumoniae infections (Kanmani et al., 2017; Moyano et al., 2020) and also inhibits growth of another pathogen, Moraxella catarrhalis, in vitro (Lappan and Peacock, 2019). The potential therapeutic effect of C. pseudodiphtheriticum has been evaluated in humans, where delivery by nasal spray was shown to reduce colonization with S. aureus in healthy adults (Kiryukhina et al., 2013). While this was only tested in four subjects, another Corynebacterium designated as strain Co304 had a similar effect in another 17 subjects (Uehara et al., 2000). The impact of other Corynebacterium species, including those such as C. accolens which may inhibit some pathogens but promote others, on a broader infection profile will be important to determine in future studies.
Colonization of mice by both species of Corynebacterium (C. accolens and C. amycolatum) elicited comparably robust myeloid cell responses in the lung in parallel with elevations in the chemotactic protein MCP-1. However, neither species induced appreciable expression of pro-inflammatory cytokines in lung BAL. As reported following colonization with C. pseudodiphtheriticum in infant mice (Kanmani et al., 2017), both TNFα and IFNγ were increased in the serum, as in our study, but, unlike our results, several pro-inflammatory cytokines were also increased in the BAL, highlighting the importance of investigating species-specific outcomes. Colonization at different host sites may also induce distinct immune activation profiles. We did not detect an IL-17A response in the lungs of Corynebacterium-exposed mice, whereas skin colonization with C. accolens, but not C. amycolatum, was associated with the induction of a γδT cell IL-17A response (Ridaura et al., 2018). The lung is particularly sensitive to the detrimental effects of inflammation, which causes damage to the integrity of the barrier, reducing oxygen exchange (Muller-Redetzky et al., 2014). S. pneumoniae co-infections, particularly with viral pathogens, are associated with an enhanced inflammatory response, promoting inflammation-mediated damage in the co-infected lung and leading to increased morbidity and mortality (Kash et al., 2011; Aguilera and Lenz, 2020). In contrast, we find that, unlike with respiratory viral and other pathogens, co-infection with Corynebacterium tempers the inflammatory response to S. pneumoniae, as described for C. pseudodiphtheriticum in the context of sterile lung injury (Kanmani et al., 2017; Moyano et al., 2020), along with S. pneumoniae clearance, indicating the potential to improve lung recovery from infection and injury.
A limitation of this study is that we did not investigate co-colonization with Dolosigranulum, which frequently associates with Corynebacterium in the human upper airway of humans (Bogaert et al., 2011; Laufer et al., 2011; Copeland et al., 2018). Abundances of several different Corynebacterium species are increased in both child and adult carriers of Dolosigranulum (Brugger et al., 2020). In vitro, these authors demonstrated that some Corynebacterium species support growth of Dolosigranulum pigrum, although not consistently (Brugger et al., 2020). Further, S. pneumoniae growth was inhibited in vitro by D. pigrum and C. pseudodiphtheriticum, neither of which were inhibitory on their own under the assay conditions used (Brugger et al., 2020). This study alludes to greater complexity in the human airway, where Corynebacterium-pathogen interactions are likely influenced by the presence of other commensal species including Dolosigranulum. In addition, Corynebacterium and Dolosigranulum must themselves compete with other airway commensals. For example, Staphylococcus epidermidis can inhibit both D. pilgrim and C. pseudodiphtheriticum growth in vitro (Janek et al., 2016). These findings highlight the importance of considering multiple commensal-pathogen dynamics in the airway, beyond interactions between specific commensal-pathogen pairs.
In summary, these studies identify C. accolens and C. amycolatum as Corynebacterium species which are capable of short-term colonization in antibiotic treated mice. Further, pre-exposure to either of these Corynebacterium species significantly reduces S. pneumoniae infection while mitigating lung inflammation. Although the host and bacterial requirements for Corynebacterium-mediated protection against S. pneumoniae are not fully resolved, both C. accolens and C. amycolatum express bacterial lipases which contribute to growth inhibition in vitro, a mechanism responsible, in part, for C. accolens-mediated protection in vivo. These findings indicate that therapeutic strategies to enhance Corynebacterium colonization may reduce the risk of S. pneumoniae acquisition and infection. We envision that such approaches would be particularly beneficial following antibiotic therapy, when the endogenous airway microbiome is disrupted. However, it is also important to consider potential adverse effects associated with enhanced Corynebacterium colonization, as some species may serve as a source of opportunistic infections. Improved understanding of the mechanisms of Corynebacterium-mediated protection will therefore facilitate harnessing the potential therapeutic benefit of these endogenous “protector” bacteria.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Animal studies were reviewed and approved by the Animal Care and Use Committee of the University of Colorado School of Medicine.
SC conceived the study and designed the experiments together with VA, EJ, and SA. SC, KH, VA, and SA performed the experiments. SC wrote the manuscript. SC and EJ edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by a Career Transition Award K22AI143922 (SC) and an R21 award AI146295 (EJ) from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, an American Lung Association Innovation Award (SC), the Boettcher Foundation Webb-Waring Biomedical Research Award (SC), and the Veterans Affairs Research Service (Merit Review I01BX004320) (EJ). AJ was supported by the MARC U-STAR T34 Program grant GM141639.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We wish to acknowledge members of the University of Colorado School of Medicine Metabolomics Core, including Micaela Roy and Julie Haines, for their contributions to this manuscript. We also acknowledge the National Summer Undergraduate Research Project (NSURP) and the Colorado Undergraduate Summer Research Program (CUSP) for programmatic support of VA and AJ, respectively. Finally, we thank their colleagues in the Department of Otolaryngology and Department of Immunology and Microbiology for contributing to critical analysis and discussions of these data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.804935/full#supplementary-material
Aguilera, E. R., and Lenz, L. L. (2020). Inflammation as a modulator of host susceptibility to pulmonary influenza, pneumococcal, and co-infections. Front. Immunol. 11:105. doi: 10.3389/fimmu.2020.00105
Bernard, K. (2012). The genus Corynebacterium and other medically relevant coryneform-like bacteria. J. Clin. Microbiol. 50, 3152–3158. doi: 10.1128/JCM.00796-12
Biesbroek, G., Tsivtsivadze, E., Sanders, E. A. M., Montijn, R., Veenhoven, R. H., Keijser, B. J. F., et al. (2014). Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190, 1283–1292. doi: 10.1164/rccm.201407-1240OC
Bogaert, D., Keijser, B., Huse, S., Rossen, J., Veenhoven, R., van Gils, E., et al. (2011). Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6:e17035. doi: 10.1371/journal.pone.0017035
Bomar, L., Brugger, S. D., Yost, B. H., Davies, S. S., and Lemon, K. P. (2016). Corynebacterium accolens Releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7:e01725-15.
Bosch, A. A. T. M., Piters, W. A. A., de, S., van Houten, M. A., Chu, M. L. J. N., Biesbroek, G., et al. (2017). Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. American J. Respir. Crit. Care Med. 196, 1582–1590.
Brugger, S. D., Eslami, S. M., Pettigrew, M. M., Escapa, I. F., Henke, M. T., Kong, Y., et al. (2020). Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere 5:e00852-20. doi: 10.1128/mSphere.00852-20
Calbo, E., and Garau, J. (2010). Of mice and men: innate immunity in pneumococcal pneumonia. Int. J. Antimicrob. Agents 35, 107–113. doi: 10.1016/j.ijantimicag.2009.10.002
Centers for Disease Control and Prevention (2019). Deaths: final data for 2017. Natl. Vital Stat. Rep. 68, 1–80.
Chonmaitree, T., Jennings, K., Colovko, G., Khanipov, K., Pimenova, M., Patel, J. A., et al. (2017). Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One 12:e0180630. doi: 10.1371/journal.pone.0180630
Clark, S. E. (2020). Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr. Opin. Immunol. 66, 42–49. doi: 10.1016/j.coi.2020.03.010
Clark, S. E., Schmidt, R. L., Aguilera, E. R., and Lenz, L. L. (2020). IL-10-producing NK cells exacerbate sublethal Streptococcus pneumoniae infection in the lung. Transl. Res. 226, 70–82. doi: 10.1016/j.trsl.2020.07.001
Clementi, E. A., Wilhelm, K. R., Schleucher, J., Morozova-Roche, L. A., and Hakansson, A. P. (2013). A complex of equine lysozyme and oleic acid with bactericidal activity against Streptococcus pneumoniae. PLoS One 8:e80649. doi: 10.1371/journal.pone.0080649
Collins, M. D., Burton, R. A., and Jones, D. (1988). Corynebacterium amycolatum sp. nov. a new mycolic acid-less Corynebacterium species from human skin. FEBS Microbiol. Lett. 49, 349–352. doi: 10.1111/j.1574-6968.1988.tb02755.x
Copeland, E., Leonard, K., Carney, R., Kong, J., Forer, M., Naidoo, Y., et al. (2018). Chronic rhinosinusitis: potential role of microbial dysbiosis and recommendations for sampling sites. Front. Cell. Infect. Microbiol. 8:57. doi: 10.3389/fcimb.2018.00057
Cutts, F. T., Zaman, S. M. A., Enwere, G., Jaffar, S., Levine, O. S., Okoko, J. B., et al. (2005). Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet 365, 1139–1146. doi: 10.1016/S0140-6736(05)71876-6
Do, T. Q., Moshkani, S., Castillo, P., Anunta, S., Pogosyan, A., Cheung, A., et al. (2008). Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J. Immunol. 181, 4177–4187. doi: 10.4049/jimmunol.181.6.4177
Eggeling, L., Bott, M., and Daffe, M. (2005). Handbook of Corynebacterium glutamicum. Boca Raton, FL: Taylor & Francis Group, LLC.
FitzGerald, E. S., Luz, N. F., and Jamieson, A. M. (2020). Competitive cell death interactions in pulmonary infection: host modulation versus pathogen manipulation. Front. Immunol. 11:814. doi: 10.3389/fimmu.2020.00814
Gillespie, S. H., Ainscough, S., Dickens, A., and Lewin, J. (1996). Phosphorylcholine-containing antigens in bacteria from the mouth and respiratory tract. J. Med. Microbiol. 44, 35–40. doi: 10.1099/00222615-44-1-35
Gomes-Neto, J. C., Mantz, S., Held, K., Sinha, R., Munoz, R. R. S., Schmaltz, R., et al. (2017). A real-time PCR assay for accurate quantification of the individual members of the Altered Shaedler Flora microbiota in gnotobiotic mice. J. Microbiol. Methods 135, 52–62. doi: 10.1016/j.mimet.2017.02.003
Gschwandtner, M., Derler, R., and Midwood, K. S. (2019). More than just attractive: how CCL2 influences myeloid cell behavior beyond chemotaxis. Front. Immunol. 10:2759. doi: 10.3389/fimmu.2019.02759
Hasegawa, K., Linnemann, R. W., Mansbach, J. M., Ajami, N. J., Espinola, J., Petrosino, J., et al. (2017). Nasal airway microbiota profile and severe bronchiolitis in infants a case-control study. Pediatr. Infect. Dis. J. 36, 1044–1051. doi: 10.1097/INF.0000000000001500
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108/-/DCSupplemental/pnas.201019378SI.pdf
Jaeger, K.-E., Ransac, S., Dijkstra, B. W., Colson, C., van Heuvel, M., and Misset, O. (1994). Bacterial lipases. FEMS Microbiol. Rev. 15, 29–63.
Janek, D., Zipperer, A., Kulik, A., Krismer, B., and Peschel, A. (2016). High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog. 12:e1005812. doi: 10.1371/journal.ppat.1005812
Jimeno, R., Brailey, P. M., and Barral, P. (2018). Quantitative polymerase chain reaction-based analyses of murine intestinal microbiota after oral antibiotic treatment. J. Vis. Exp. 141:e58481. doi: 10.3791/58481
Jose, R. J., Periselneris, J. N., and Brown, J. S. (2015). Community-acquired pneumonia. Curr. Opin. Pulm. Med. 21, 212–218. doi: 10.1097/MCP.0000000000000150
Kanmani, P., Clua, P., Vizoso-Pinto, M. G., Rodriguez, C., Alvarez, S., Melnikov, V., et al. (2017). Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front. Microbiol. 8:1613.
Kash, J. C., Walters, K.-A., Davis, A. S., Sandouk, A., Schwartzman, L. M., Jagger, B. W., et al. (2011). Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. mBio 2:e00172-11. doi: 10.1128/mBio.00172-11
Kaspar, U., Kriegeskorte, A., Schubert, T., Peters, G., Rudack, C., Pieper, D. H., et al. (2016). The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ. Microbiol. 18, 2130–2142. doi: 10.1111/1462-2920.12891
Kelly, M. S., Plunkett, C., Yu, Y., Aquino, J. N., Patel, S. M., Hurst, J. H., et al. (2021). Non-diphtheriae Corynebacterium species are associated with decreased risk of pneumococcal colonization during infancy. ISME J. 1–11. doi: 10.1038/s41396-021-01108-4 [Epub ahead of print].
Kelly, M. S., Surette, M. G., Smieja, M., Pernica, J. M., Rossi, L., Luinstra, K., et al. (2017). The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr. Infect. Dis. J. 36, e211–e218.
Kiryukhina, N. V., Melnikov, V. G., Suvorov, A. V., Morozova, Y. A., and Ilyin, V. K. (2013). Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiot. Antimicrob. Proteins 5, 233–238. doi: 10.1007/s12602-013-9147-x
La Mantia, I., Varricchio, A., and Ciprandi, G. (2017). Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for preventing recurrent acute otitis media in children: a real-life clinical experience. Int. J. Gen. Med. 10, 171–175. doi: 10.2147/IJGM.S137614
Lappan, R., and Peacock, C. S. (2019). Corynebacterium and Dolosigranulum: future probiotic candidates for upper respiratory tract infections. Microbiol. Austr. 40, 172–177. doi: 10.1111/1469-0691.12373
Lappan, R., Imbrogno, K., Sikazwe, C., Anderson, D., Mok, D., Coates, H., et al. (2018). A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 18:13. doi: 10.1186/s12866-018-1154-3
Laufer, A. S., Metlay, J. P., Gent, J. F., Fennie, K. P., Kong, Y., and Pettigrew, M. M. (2011). Microbial communities of the upper respiratory tract and otitis media in children. mBio 2:e00245-10. doi: 10.1128/mBio.00245-10
Lin, W.-C., and Fessler, M. B. (2021). Regulatory mechanisms of neutrophil migration from the circulation to the airspace. Cell. Mol. Life Sci. 78, 4095–4124. doi: 10.1007/s00018-021-03768-z
Liu, Q., Liu, Q., Meng, H., Lv, H., Liu, Y., Liu, J., et al. (2019). Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27, 68–78.e5.
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2019). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Man, W. H., van Houten, M. A., Mérelle, M. E., Vlieger, A. M., Chu, M. L. J. N., Jansen, N. J. G., et al. (2019). Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir. Med. 7, 417–426.
Marchisio, P., Santagati, M., Scillato, M., Baggi, E., Fattizzo, M., Rosazza, C., et al. (2015). Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2377–2383. doi: 10.1007/s10096-015-2491-x
Marrakchi, H., Lanéelle, M.-A., and Daffé, M. (2014). Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85.
Moyano, R. O., Tonetti, F. R., Tomokiyo, M., Kanmani, P., Vizoso-Pinto, M. G., Kim, H., et al. (2020). The ability of respiratory commensal bacteria to beneficial modulate the lung innate immune response is a strain dependent characteristic. Microorganisms 8:727. doi: 10.3390/microorganisms8050727
Muller-Redetzky, H. C., Suttorp, N., and Witzenrath, M. (2014). Dynamics of pulmonary endothelial barrier function in acute inflammation: mechanisms and therapeutic perspectives. Cell Tissue Res. 355, 657–673. doi: 10.1007/s00441-014-1821-0
Nakatsuji, T., Kao, M. C., Zhang, L., Zouboulis, C. C., Gallo, R. L., and Huang, C.-M. (2010). Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating β-defensin-2 expression. J. Investig. Dermatol. 130, 985–994. doi: 10.1038/jid.2009.384
Nemkov, T., D’Alessandro, A., and Hansen, K. C. (2015). Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids 47, 2345–2357. doi: 10.1007/s00726-015-2019-9
Nemkov, T., Hansen, K. C., and D’Alessandro, A. (2017). A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun. Mass. Spectrom. 31, 663–673. doi: 10.1002/rcm.7834
Nemkov, T., Reisz, J. A., Gehrke, S., Hansen, K. C., and D’Alessandro, A. (2019). High-throughput metabolomics: isocratic and gradient mass spectrometry-based methods. Methods Mol. Biol. 1978, 13–26.
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pettigrew, M. M., Laufer, A. S., Gent, J. F., Kong, Y., Fennie, K. P., and Metlay, J. P. (2012). Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78, 6262–6270. doi: 10.1128/AEM.01051-12
Pilishvili, T., and Bennett, N. M. (2015). Pneumococcal disease prevention among adults: strategies for the use of pneumococcal vaccines. Vaccine 33, D60–D65.
Piters, W. A. A. S., Jochems, S. P., Mitsi, E., Rylance, J., Pojar, S., Nikolaou, E., et al. (2019). Interaction between the nasal microbiota and S. pneumoniae in the context of live-attenuated influenza vaccine. Nat. Commun. 10:2981. doi: 10.1038/s41467-019-10814-9
Plou, F. J., Ferrer, M., Nuero, O. M., Calvo, M. V., Alcalde, M., Reyes, F., et al. (1998). Analysis of tween 80 as an esterase/lipase substrate for lipolytic activity assay. Biotechnol. Techniq. 12, 183–186. doi: 10.1023/A:1008809105270
Prevaes, S. M. P. J., de Winter-de Groot, K. M., Janssens, H. M., de Steenhuijsen Piters, W. A. A., Tramper-Stranders, G. A., Wyllie, A. L., et al. (2016). Development of the nasopharyngeal microbiota in infants with cystic fibrosis. American J. Respir. Crit. Care Med. 193, 504–515.
Ramsey, M. M., Freire, M. O., Gabrilska, R. A., Rumbaugh, K. P., and Lemon, K. P. (2016). Staphylococcus aureus shifts toward commensalism in response to Corynebacterium Species. Front. Microbiol. 7:1230.
Reisz, J. A., Zheng, C., D’Alessandro, A., and Nemkov, T. (2019). “Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics,” in High-Throughput Metabolomics: Methods and Protocols, ed. A. D’Alessandro (New York, NY: Springer), 121–135.
Ricchi, M., Bertasio, C., Boniotti, M. B., Vicari, N., Russo, S., Tilola, M., et al. (2017). Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front. Microbiol. 8:1174. doi: 10.3389/fmicb.2017.01174
Ridaura, V. K., Bouladoux, N., Claesen, J., Chen, Y. E., Byrd, A. L., Constantinides, M. G., et al. (2018). Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 215, 785–799.
Rijneveld, A. W., Weijer, S., Florquin, S., Speelman, P., Shimizu, T., Ishii, S., et al. (2004). Improved host defense against pneumococcal pneumonia in platelet-activating factor receptor–deficient mice. J. Infect. Dis. 189, 711–716. doi: 10.1086/381392
Rosendahl, A., Bergmann, S., Hammerschmidt, S., Goldmann, O., and Medina, E. (2013). Lung dendritic cells facilitate extrapulmonary bacterial dissemination during pneumococcal pneumonia. Front. Cell. Infect. Microbiol. 3:21. doi: 10.3389/fcimb.2013.00021/abstract
Stubbendieck, R. M., May, D. S., Chevrette, M. G., Temkin, M. I., Wendt-Pienkowski, E., Cagnazzo, J., et al. (2019). Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl. Environ. Microbiol. 85:e02406-18. doi: 10.1128/AEM.02406-18
Taoka, Y., Nagano, N., Okita, Y., Izumida, H., Sugimoto, S., and Hayashi, M. (2011). Effect of Tween 80 on the growth, lipid accumulation and fatty acid composition of Thraustochytrium aureum ATCC 34304. J. Biosci. Bioeng. 111, 420–424. doi: 10.1016/j.jbiosc.2010.12.010
Tchoupa, A. K., Eijkelkamp, B. A., and Peschel, A. (2021). Bacterial adaptation strategies to host-derived fatty acids. Trends Microbiol. 1–13. doi: 10.1016/j.tim.2021.06.002 [Epub ahead of print].
Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715. doi: 10.1016/j.chom.2015.03.008
Torres, A., Cillóniz, C., Blasi, F., Chalmers, J. D., Gaillat, J., Dartois, N., et al. (2018). Burden of pneumococcal community-acquired pneumonia in adults across Europe: a literature review. Respir. Med. 137, 6–13. doi: 10.1016/j.rmed.2018.02.007
Uehara, Y. E. A., Nakama, H., Agematsu, K., Uchida, M., Kawakami, Y., Fattah, A. S. M. A., et al. (2000). Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureusfrom nasal cavities by artificial implantation of Corynebacterium sp. J. Hosp. Infect. 44, 127–133. doi: 10.1053/jhin.1999.0680
World Health Organization [WHO] (2021). World Health Statistics 2021: Monitoring Health for the SDGs, Sustainable Development Goals. Geneva: World Health Organization, 1–136.
Xu, L., Earl, J., and Pichichero, M. E. (2021). Nasopharyngeal microbiome composition associated with Streptococcus pneumoniae colonization suggests a protective role of Corynebacterium in young children. PLoS One 16:e0257207. doi: 10.1371/journal.pone.0257207
Yan, M., Pamp, S. J., Fukuyama, J., Hwang, P. H., Cho, D.-Y., Holmes, S., et al. (2013). Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 14, 631–640. doi: 10.1016/j.chom.2013.11.005
Keywords: Corynebacterium, Streptococcus pneumoniae, nasopharyngeal colonization, lung infection, pneumonia, pneumococcus, bacterial lipases
Citation: Horn KJ, Jaberi Vivar AC, Arenas V, Andani S, Janoff EN and Clark SE (2022) Corynebacterium Species Inhibit Streptococcus pneumoniae Colonization and Infection of the Mouse Airway. Front. Microbiol. 12:804935. doi: 10.3389/fmicb.2021.804935
Received: 29 October 2021; Accepted: 03 December 2021;
Published: 10 January 2022.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Anders P. Hakansson, Lund University, SwedenCopyright © 2022 Horn, Jaberi Vivar, Arenas, Andani, Janoff and Clark. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah E. Clark, c2FyYWguZS5jbGFya0BjdWFuc2NodXR6LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.