- 1Department of Infectious Diseases and Public Health, Jockey Club College of Veterinary Medicine and Life Sciences, City University of Hong Kong, Kowloon, Hong Kong SAR, China
- 2State Key Laboratory of Chemical Biology and Drug Discovery, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Kowloon, Hong Kong SAR, China
We recently reported the recovery of a novel IncI1 type conjugative helper plasmid which could target mobile genetic elements (MGE) located in non-conjugative plasmid and form a fusion conjugative plasmid to mediate the horizontal transfer of the non-conjugative plasmid. In this study, interactions between the helper plasmid pSa42-91k and two common MGEs, ISEcp1 and IS15DI, which were cloned into a pBackZero-T vector, were monitored during the conjugation process to depict the molecular mechanisms underlying the plasmid fusion process mediated by insertion sequence (IS) elements. The MinION single-molecule long-read sequencing technology can dynamically reveal the plasmid recombination events and produce valuable information on genetic polymorphism and plasmid heterogeneity in different multidrug resistance (MDR) encoding bacteria. Such data would facilitate the development of new strategies to control evolution and dissemination of MDR plasmids.
Introduction
Bacterial asexual reproduction is known to involve division of its genetic traits equally into daughter cells to maintain the original phenotypic characteristics. However, bacteria may also evolve by elevating genetic plasticity via horizontal gene transfer (HGT), a process in which beneficial genetic elements are acquired from different species and passed onto the offspring of the recipient strain, whereas harmful genes are prevented from being heritable (Thomas and Nielsen, 2005; Pinilla-Redondo et al., 2018). Three processes of HGT, namely transformation, transduction, and conjugation, have been described to date (Frost et al., 2005). During the transformation process, exogenous genes are passively acquired by the recipient cells and mobile genetic elements (MGE) are not involved. Transduction is often accompanied by bacteriophage invasion, yet this infection event is usually detrimental to the recipient strain (Davison, 1999). In contrast, bacterial conjugation is a much more prevalent HGT mechanism which requires an intricate molecular structure to transfer genetic materials from a donor strain to a recipient strain (Garcillán-Barcia et al., 2009; De La Cruz et al., 2010). Conjugation plays a significant role in dissemination of antimicrobial resistance and virulence-encoding elements in bacteria (San Millan, 2018; Xie et al., 2020). Conjugative plasmids normally harbor genes that encode genetic transfer functions and the Type IV secretion system, which are essential elements that drive the conjugation events. A number of non-conjugative plasmids also encode transfer functions via cooperation with co-resident conjugative plasmids that carry the oriT sequence, or via expression of the cognate genes to produce relaxase (Miyano et al., 2018). In previous studies, we reported the discovery of helper plasmids which mediated transmission of non-conjugative plasmids and a chromosomal DNA fragment though fusion mechanism. In these co-integration processes, the insertion sequences (ISs) IS26 and ISPa40 played a critical role in molecular interaction via replicative transposition and homologous recombination, and led to a sharp increase in the incidence of Salmonella resistance to ciprofloxacin (Chen et al., 2018, 2019a,b). In addition, these ISs, particularly IS26, exhibit a vital function in the spreading of antimicrobial resistance elements among gram-negative strains, as it is normally linked with determinants that confer resistance to various categories of antimicrobial agents (Wrighton and Strike, 1987; Kim and Aoki, 1994), or some class 1 integrons (Miriagou et al., 2005). In this work, we investigated the nature of interactions between helper plasmids and MGEs during the conjugation process of Salmonella. Plasmid pSa42-91k is a 91-kbp-conjugative plasmid originated from a strain of Salmonella Meleagridis, which exhibited high homology to pSa44-CRO (MH430883) and was considered a helper plasmid. Two prevalent mobile genetic elements, ISEcp1 and IS15DI, were cloned into the pBackZero-T vector and transferred to a strain known as Sa42, which was then used as the donor strain in conjugation experiments. Recipient strains which have acquired fusion plasmids were subjected to whole genome sequence analysis. Utilizing the long-read sequencing technology, we revealed the functional role of ISEcp1 and IS15DI in mediating recombination events that enhance the genetic plasticity of the resistance-encoding plasmids during the process of conjugative transfer, and hence the dissemination potential of the resistance genes harbored by the plasmids concerned.
Materials and Methods
Cloning of ISEcp1 and IS15DI
DNA segments were amplified using primer pairs targeting the ISEcp1 gene (ISEcp1-F-AGCGTGGTAATGCTGAAAACT and ISEcp1-R-TCCACAGAGCAACACTCAAT) and the IS15DI gene (IS15DI-F-TGTGGTTAATGCAAAGCGGG and IS15DI-R-AAGTCCGCCACATTCGTCTG). The PCR products were ligated to a cloning vector, pBackZero-T, respectively, yielding pBackZero- ISEcp1 and pBackZero-IS15DI, which were then transformed into E. coli DH5α by electroporation. All transformants were selected on LB plates containing 50 μg/mL kanamycin. Transconjugants named DH5α-ISEcp1 and DH5α-IS15DI were obtained, followed by confirmation of genetic identity through PCR screening with the pair of cloning primers described above.
Conjugation Experiments
The transmission potential of the ISEcp1, IS15DI and blaCTX–M–130 genes was assessed by performing a conjugation experiment using the filter mating method as previously described (Chen et al., 2018). Sa42, Sa42-TC1, and Sa42-TC2 were used as donor strains and the recipient strain was the sodium azide-resistant E. coli strain J53. The transconjugant Sa42-TC3 was selected on EMB agar containing cefotaxime (2 μg/ml), and sodium azide (100 μg/ml). For donor strains Sa42-TC1 and Sa42-TC2 carrying different pBackZero-IS vectors, transconjugants Sa42-TC4 and Sa42-TC5 were selected on EMB agar containing kanamycin (50 μg/ml) and sodium azide (100 μg/ml). Antimicrobial susceptibility of both parental strains and their transconjugants was determined (Table 1).
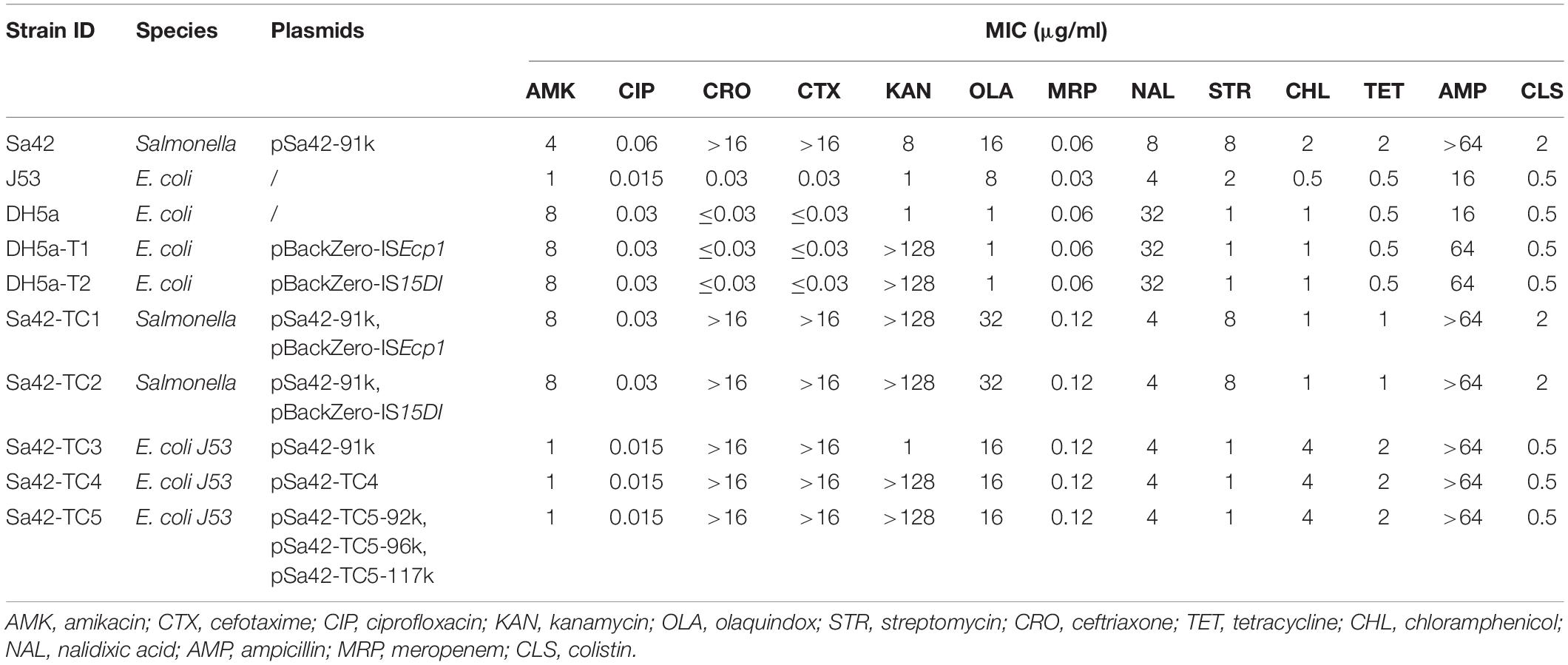
Table 1. Phenotypic characteristics of ISEcp1 and IS15DI–bearing strains and their corresponding transconjugants.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (CHEF -MAP -PER System, Bio-Rad Laboratories, Hercules, CA, United States) was performed to confirm the genetic identity of the parental strains and the corresponding transconjugants as described previously (Ribot et al., 2006). All plasmid-bearing transconjugants were analyzed by S1 nuclease PFGE.
Analysis of Plasmidomes Based on Sequencing Data
Total plasmid samples of strain Sa42 and the corresponding transconjugants were extracted using the Qiagen Midi Plasmid Kit and sequenced with two different platforms, namely the Illumina and Oxford Nanopore Technologies (ONT) MinION platform. Paired-end libraries were constructed and sequenced by using a 300 cycle Illumina NextSeq 500 Kit. Sequencing reads were assembled de novo with the SPAdes 3.5 tool (Bankevich et al., 2012). A Rapid Barcoding Sequencing Kit was used to construct the libraries sequenced in a MinION device as previously reported (Li et al., 2018). To determine the structure of the plasmids, Nanopore raw contigs containing pBackZero-T were selected by sequence homology searches with BLAST + makeblastdb (ver. 2.2.28) (Cock et al., 2015) and relevant sequences were extracted from the initial fasta files by faSomeRecords1. Long reads assembled from Nanopore were used to align and join contigs acquired from Illumina assembly using the CLC Genomics Workbench v10 (CLC bio, Denmark) and then annotated by the Rapid Annotation using Subsystem Technology (RAST) version 2.0 (Aziz et al., 2008). The EasyFig tool was used to compare and visualize the structures of plasmids and long reads (Alikhan et al., 2011).
Results and Discussion
Research Design
To depict the role of ISEcp1 and IS15DI (IS26) in dissemination of antibiotic resistance genes, these IS elements were first cloned into the pBackZero-T vector to obtain pBackZero-ISEcp1 and pBackZero-IS15DI. To differentiate the original ISEcp1 from the one being cloned into pBackZero-T, only a 341 bp fragment (21–361) of ISEcp1 was cloned into pBackZero-T. These two plasmids were then transformed into Sa42, a foodborne Salmonella strain, which carried a IncI1 type plasmid, pSa42-91k, that contains the mobile element ISEcp1-blaCTX–M–130-IS903. The pSa42-91k plasmid was able to form fusion plasmid with other antimicrobial resistance (AMR)-encoding plasmids. We therefore used it as a model plasmid to study the fusion process mediated by IS elements. Upon transformation, two transformants were obtained, namely Sa42-TC1 and Sa42-TC2, which carried pSa42-91k/pBackZero-ISEcp1 and pSa42-91k/pBackZero-IS15DI, respectively. Strains Sa42-TC1 and Sa42-TC2 were then used as donor strains and subjected to conjugation with the recipient E. coli strain J53 to obtain transconjugants Sa42-TC4 and Sa42-TC5, which were then subjected to sequencing by both Illumina and Nanopore MinION to assess the conjugation potential of fusion plasmid pSa42-91k/pBackZero-ISEcp1 and pSa42-91k/pBackZero-IS15DI, as well as to investigate the fusion process (Figure 1).
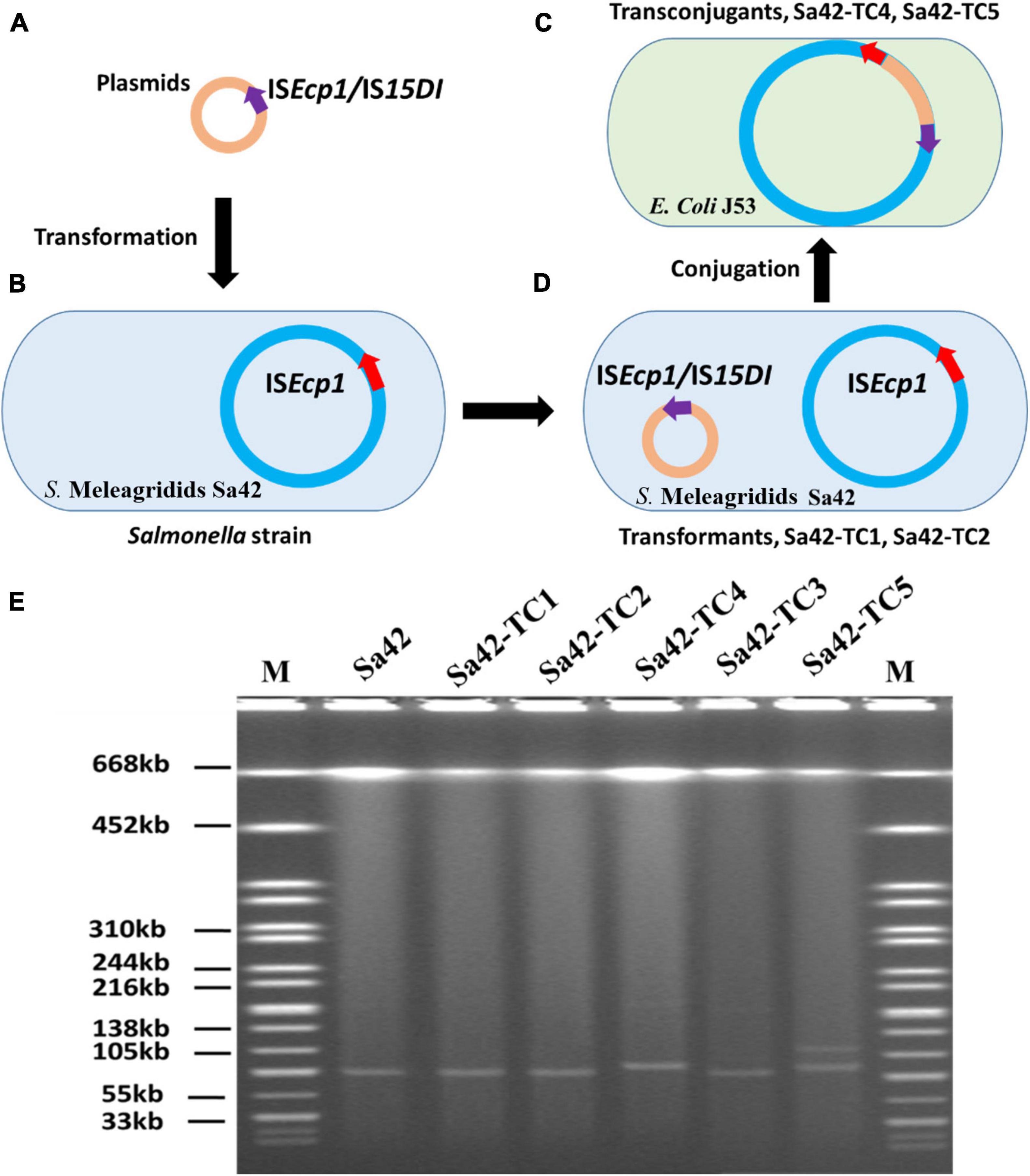
Figure 1. Research design for resolution of the plasmid profile of Salmonella strain Sa42 and the corresponding transconjugants by S1-PFGE. (A) Plasmids pBackZero-ISEcp1 and pBackZero-IS15DI were transferred to (B) strain Sa42 by transformation and generated (C) Sa42-TC1 and Sa42-TC2, respectively, (D) which were then conjugated to E. coli J53 to produce the transconjugants Sa42-TC4 and Sa42-TC5 that carry fusion plasmids. (E) Plasmids in strain Sa42, Sa42-TC1, Sa42-TC2, Sa42-TC3, Sa42-TC4, and Sa42-TC5 by S1-PFGE. Conjugation of Sa42 to E. coli J53 produced Sa42-TC3. The sizes of plasmids in Sa42-TC4 and Sa42-TC5 were different from the one in Sa42-TC3. Red arrow depicts original ISEcp1 in plasmid pSa42-91k, purple arrow denotes insertion sequences ISEcp1 or IS15DI being cloned into pBackZero-T vector.
In vitro Plasmid Fusion and Creation of a Transmission Model
Salmonella Meleagridis Sa42 was resistant to cefotaxime (Table 1). The cefotaxime-resistance phenotype of this strain, which was conferred by blaCTX–M–130, was found to be transferable in a conjugation experiment under the selection of cefotaxime and sodium azide. S1-PFGE revealed that it carried only one plasmid of ∼90 kb, which could be conjugated to E. coli J53 under the selection of ceftazidime and sodium azide to produce transconjugant Sa42-TC3, suggesting that it was a conjugative plasmid (Figure 1E). This plasmid was subjected to whole plasmid sequencing using both the Illumina and Nanopore sequencing platform to obtain the complete plasmid map. Our data showed that this plasmid was 91,229 bp in length, exhibited a GC content of 50.9%, and comprised 98 putative open reading frames (ORFs), most of which were responsible for conjugation, replication, partition and other plasmid maintenance functions. A 530 bp β-lactamase gene blaCTX–M–130 franked by ISEcp1 was also found in plasmid pSa42-91k at position 7434-8039 (Figure 2A).
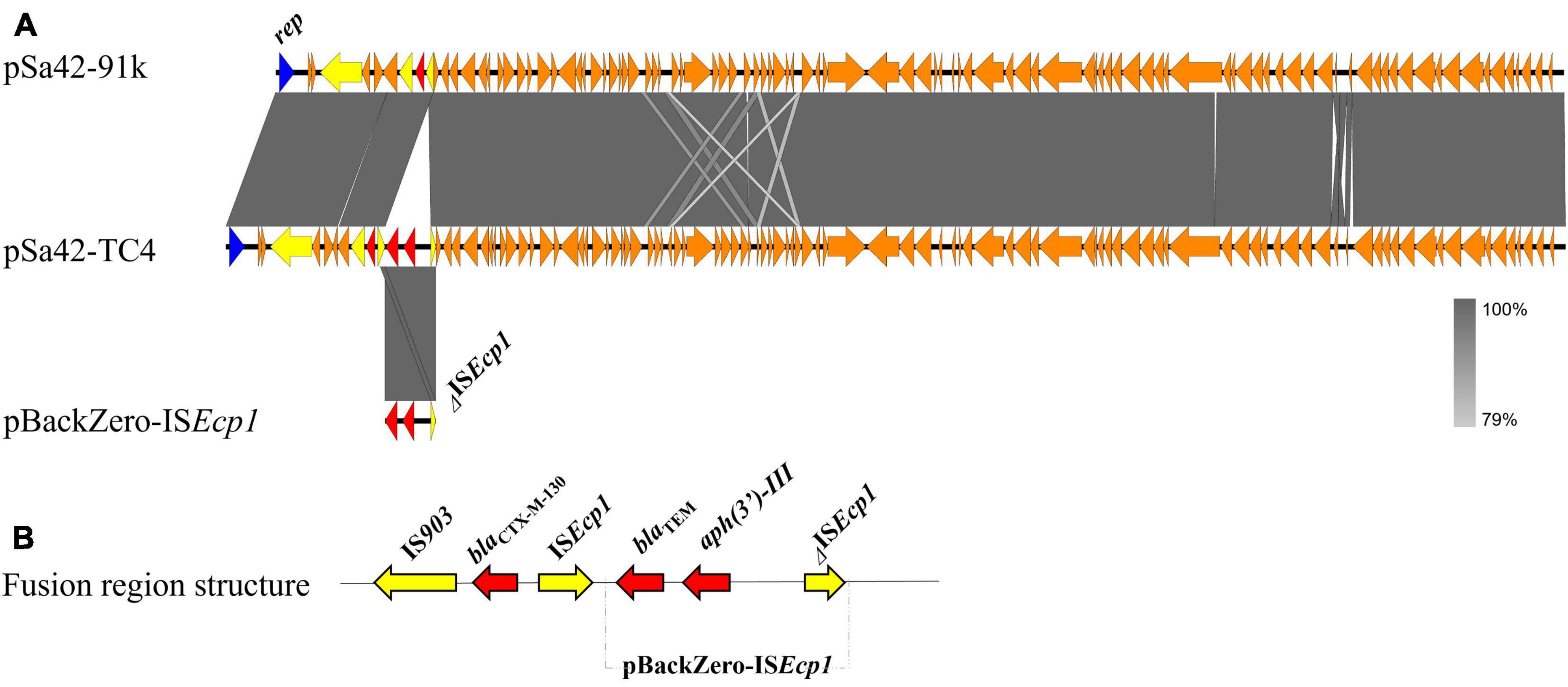
Figure 2. Alignment and structure of the relevant plasmids in the donor strain and transconjugant. (A) Linear alignment of plasmids pSa42-91k, pSa42-TC4, and pBackZero-ISEcp1 assembled de novo by Nanopore long reads. IS elements were highlighted in yellow and drug-resistance genes were depicted in red arrows. (B) Structure of pBackZero-ISEcp1 upon being merged with the plasmid pSa42-91k. Red denoted the resistance genes and yellow denoted the ISs gene.
We next cloned ISEcp1 and IS15DI into the pBackZero-T vector to produce pBackZero-ISEcp1 and pBackZero-IS15DI, respectively, followed by conjugation to the pSa42-91k-bearing strain S. Meleagridis Sa42 to produce Sa42-TC1 and Sa42-TC2. The conjugation experiment was further performed to obtain Sa42-TC4 and Sa42-TC5, which were E. coli J53 which have acquired a variety of fusion plasmids formed by interaction between pSa42-91k and pBackZero-ISEcp1 or pBackZero-IS15DI. It was expected that Sa42-TC4 and Sa42-TC5 contained the fusion plasmid of pSa42-91k and pBackZero-ISEcp1 or pBackZero-IS15DI. S1-PFGE was performed on these five transconjugants, with results showing that Sa42, Sa42-TC1, and Sa42-TC2 carried only one plasmid with a size similar to that of pSa42-91k. However, Sa42-TC4 carried only one plasmid but the size was slightly larger than that of plasmid pSa42-91k, suggesting that this plasmid should be the fusion product of pSa42-91k and pBackZero-ISEcp1. Interestingly, Sa42-TC5 carried two plasmids of different sizes and both were larger than pSa42-91k. These two plasmids should be the fusion product of pSa42-91k and pBackZero-IS15DI, but exhibited different structural arrangement (Figure 1E). Antimicrobial susceptibilities were determined for Sa42, Sa42-TC1, Sa42-TC2, Sa42-TC3, Sa42-TC4, Sa42-TC5 to confirm that these transconjugants exhibited the right antimicrobial resistance phenotypes encoded by the plasmids they harbored, namely ceftazidime resistance for pSa42-91k and kanamycin resistance for pBackZero-ISEcp1 and pBackZero-IS15DI. Antimicrobial susceptibility results showed that the resistance phenotypes of these transconjugants matched well with those encoded by the plasmids that they harbored (Table 1).
Genetic Features of Conjugative Fusion Plasmids in Sa42-TC4 and Sa42-TC5
The plasmids harbored by E. coli DH5α-T1 and DH5α-T2 were sequenced by Illumina, with results showing that pBackZero-ISEcp1 from DH5α-T1 was 3,596 bp in length and contained the pBackZero-T vector and a 341 bp DNA fragment containing ISEcp1; pBackZero-IS15DI from DH5α-T2 was 4,157 bp in length, containing the pBackZero-T vector and a 820 bp DNA fragment harboring IS15DI. Both plasmids had a GC content of 47.8%, comprised 3 coding sequences (CDSs) and harbored the kanamycin resistance gene.
In a previous study, ISEcp1 was found to mediate genetic transposition events that involve homologous recombination (Zong et al., 2010). Using the Illumina and Nanopore sequencing platforms, complete sequence of the plasmid in Sa42-TC4 was obtained. The plasmid in Sa42-TC4, designated as pSa42-TC4, was a fusion plasmid of pBackZero-ISEcp1 and pSa42-91k (Figure 2). The fusion site was found to be at ISEcp1, in which a structure of ISEcp1-pBackZero-ISEcp1 was replaced by the original ISEcp1 in plasmid pSa42-91k. To differentiate between the ISEcp1 element harbored by plasmid pSa42-91k from the one we cloned to pBackZero-T, the latter was designed to be shorter than the one in pSa42-91k, which included 21–362 bp of the full length of ISEcp1 (Figure 3A). This design will lead to production of ISEcp1 with a size of 341 bp in pBackZero-T and 530 bp in pSa42-91k, respectively (Figure 3A).
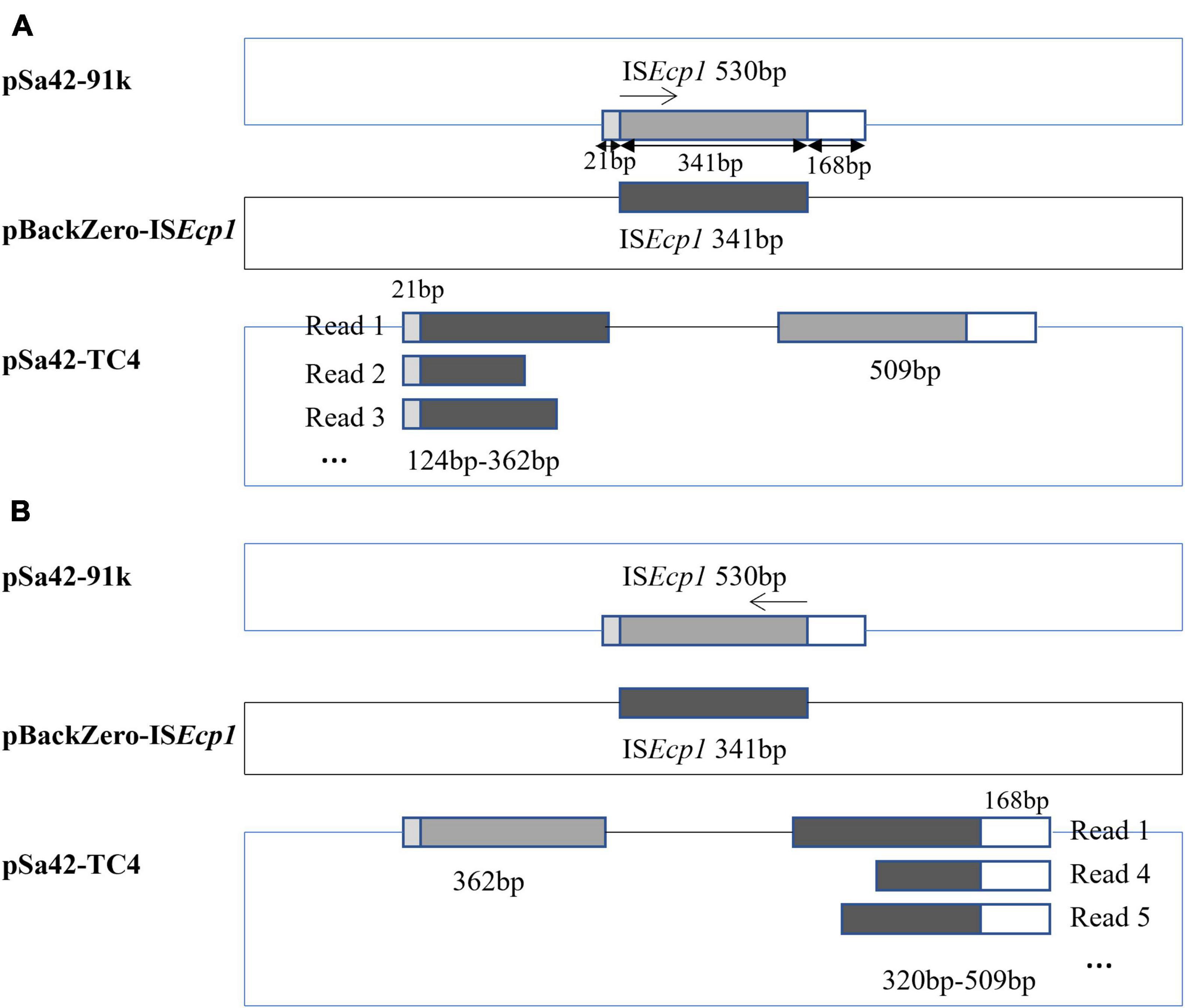
Figure 3. Mechanism of ISEcp1–mediated homologous recombination depicted by analysis of Nanopore long-reads. ISEcp1 was found to exhibit a size of 530 bp in pSa42-91k. A fragment of ISEcp1 was cloned into pBackZero-T vector (depicted in black). Such fragment exhibited sequence homology with ISEcp1 from pSa42-91k (depicted in gray). A 21 bp and 168 bp region were located at each terminal. (A) When the homologous recombination process started from the left, various sizes of ISEcp1 linked with the 21 bp sequence were observed in Nanopore long reads; oppositely oriented terminal side of a 509 bp constant size could be detected in another ISEcp1. (B) Homologous recombination began in the opposite direction, Nanopore long reads were also found to contain various sizes of ISEcp1 at 341 bp, which were connected to a 168 bp fragment in the original ISEcp1, whereas another part of 362 bp in the original ISEcp1 whose size was stable and not variable was located in the other end.
Unlike ISEcp1, IS15DI was found to belong to the IS6 family and exhibited genetic characteristics similar to those of IS26, which can undergo replicative transposition during the co-integration process. In this study, two different sizes of plasmids from strain Sa42-TC5 were observed by S1-PFGE, indicating that IS15DI may target different insertion sites during conjugation. Nanopore long reads sequencing was performed to solve this problem, with sequencing reads being assembled de novo by Canu (Koren et al., 2017). Complete sequencing of the large plasmid was obtained, for which a size of 117,221 bp was consistent with the S1-PFGE result. The plasmid contained 147 CDSs with a GC content of 50.8% and was designated as plasmid pSa42-TC5-117k. Compared with the original plasmid pSa42-91k, an IS15DI element and an IS15DI-pBackZero-IS15DI element were found to be inserted into the traJ and traF genes in plasmid pSa42-TC5-117k, respectively. Another fusion plasmid in strain Sa42-TC5, designated as pSa42-TC5-96k, was formed by insertion of the IS15DI-pBackZero-IS15DI into the traF gene of pSa42-91k, which was 95,624 bp in length, contained 126 CDSs and exhibited a GC content of 50.8%. However, an extra ∼92 kb plasmid could be identified from our sequencing data and was designated pSa42-TC5-92k. This plasmid could not be observed in S1-PFGE as its size was similar to that of plasmid pSa42-TC5-96k. Plasmid pSa42-TC5-92k was 91,784 bp in length, contained only one IS15DI inserted into the traJ gene, and exhibited a GC content of 50.9%. For the fusion plasmid pSa42-TC5-117k, in addition to an insertion event in the traJ gene, another insertion site was also observed in traF in which a nearly 20 kb DNA sequence upstream of traJ was found to be repeatedly inserted beyond the traJ gene, which was responsible for causing an increase in the plasmid size to around ∼117 kb.
Mechanism That Gives Rise to ISEcp1 Polymorphisms and Enhances Dissemination Potential
To depict the detailed mechanism of interaction between the ISEcp1 elements located in two plasmids, MinION nanopore long reads were generated from Sa42-TC4 and reads baring the fusion region were extracted for further analysis (Table 2). Nanopore raw fasta reads (>50 kb) were selected and the chromosomal contamination reads were removed by matching against the pSa42-91k reference plasmid via BLASTN. To check the fusion structure in each read, two genes harbored by the plasmid pBackZero-T including blaTEM and aph(3′)-III were used as the marker genes to filter out reads that did not span the fusion regions of ISEcp1-pBackZero-ISEcp1. Among the Nanopore reads from sample Sa42-TC4, 256 out of 376 reads were found to harbor complete structure of ISEcp1-pBackZero-ISEcp1 in various sizes. Different sizes of reads were selected for BLASTN analysis against relevant sequences in donor strains (Figure 3). ISEcp1 in ISEcp1-pBackZero-ISEcp1 structures of various sizes was observed among different reads and shown to be polymorphic by Rast tools (Overbeek et al., 2013). These reads could be grouped into two categories based on alignment to two different sizes of ISEcp1, which originated from pBackZero-T and pSa42-91k, respectively. One category of reads contained a 509 bp ISEcp1, with the first 21 bp of DNA located on the right of the reads, whereas various sizes of ISEcp1 sequences ranging from 124 to 362 bp were found located at the left side of the reads (Figure 3A). ISEcp1 elements located at the left are often truncated at different sites. These data suggested that (1) fusion of pBackZero-ISEcp1 to pSA42-91k was mediated by homologous recombination; (2) the truncated ISEcp1 located in pBackZero aligned to the full length of ISEcp1 at 21 bp site, which initiated the homologous recombination; and (3) the length of the homologous region varied from 103 bp to the full length of the truncated ISEcp1 (341 bp); and upon insertion of pBackZero-ISEcp1 into the fusion plasmid, the ISEcp1 was duplicated (Figure 3B).
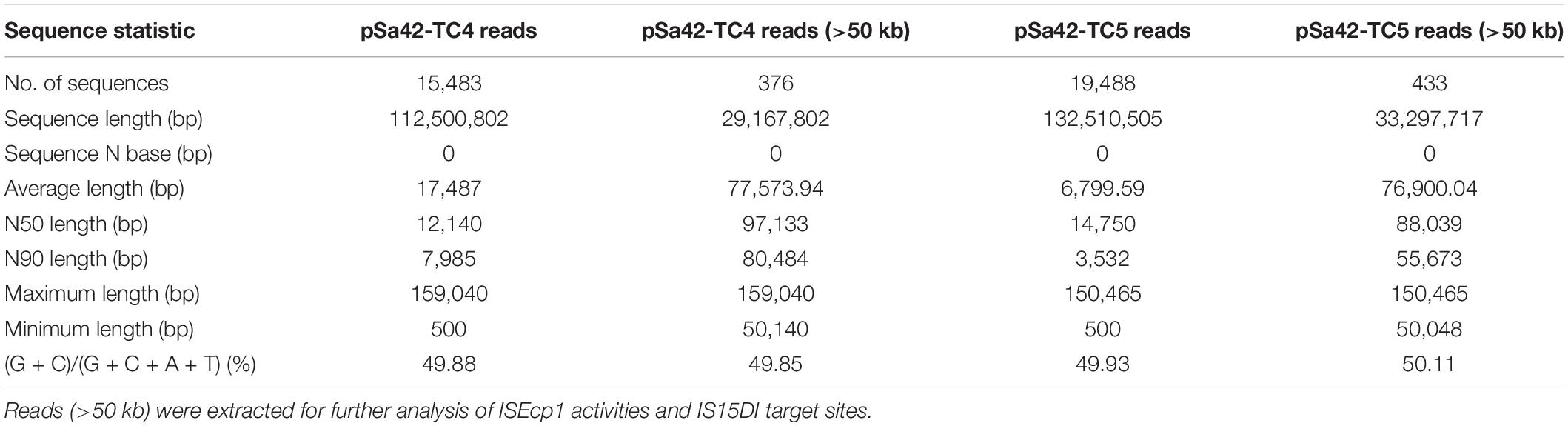
Table 2. Statistical summary of MinION Nanopore long reads generated from plasmid samples of strains Sa42-TC4 and Sa42-TC5.
Another category of reads had a stable 362 bp region of ISEcp1 located on the left, whereas the right side of ISEcp1 exhibited polymorphism, with sizes ranging from 320∼509 bp. Sequence analysis showed that the truncated region on the left covered base 1∼362 of ISEcp1, yet the terminal region of the truncated ISEcp1 was located in pBackZero-ISEcp1. The right hand side of ISEcp1 existed in various sizes and always ended at position 530, even though the start sites in ISEcp1 were variable. These findings suggest that the 3′-terminal of the truncated ISEcp1 could also align with the ISEcp1 element located in a site at 362 bp in plasmid pSa42-91k, as well as the full length of ISEcp1. Homologous recombination could occur at short sequence at any site within a 152 bp fragment (from position 210 to 362) and regions up to the start codon of ISEcp1. Upon homologous recombination, the rest of ISEcp1 (from 1 to 362) will remain at the left side of the fusion plasmid (Figure 3B). Among the reads from nanopore, the most dominant reads were 362 bp of the truncated ISEcp1 at the left side and 590 bp at the right side, which could be generated through molecular interaction by targeting either the 5′- or 3′-terminal of the truncated ISEcp1 located in pBackZero-ISEcp1 or the ISEcp1 located in pSa42-91k. Based on our understanding of the homologous recombination process, we believe that both copies of ISEcp1, located in pBackZero-ISEcp1, and pSA42-91k were identical. In this case, upon homologous recombination, the ISEcp1 located at the left could become variable in size, whereas the ISEcp1 element located at the right remains full length.
Mechanism of Generating IS15DI Polymorphisms and Enhancing Dissemination Potential
Similarly, nanopore long reads containing IS15DI were extracted and analyzed by the aforementioned method (Table 2). Nanopore raw fasta reads (>50 kb) were also selected and chromosomal contamination reads were screened out by mapping against the pSa42-91k reference plasmid via BLASTN. Unlike ISEcp1, IS15DI was known to undergo replicative transposition during the co-integration process like IS26, but IS15DI did not exist in the original plasmid pSa42-91k, so it can be regarded as a marker for direct selection from raw reads. Among the nanopore reads from Sa42-TC5, 222 out of 433 reads were found to harbor complete IS15DI sequences. Different forms of reads of various sizes were observed in the case of ISEcp1, among which three type of reads could be identified. The first type was a structure in which IS15DI-pBackZero-IS15DI was inserted into the traF gene by replicative transposition (Figure 4C; Arthur and Sherratt, 1979). During the process, the IS15DI element cleaved both terminal inverted repeats (TIRs), resulting in the formation of nicks in both strands of DNA and generating 3-Diols groups (3 = -OH) that attack the hot spot of the traF gene, leading to formation of a Shapiro intermediate. The DNA complex then underwent intermolecular replicative transposition, in which the target site was duplicated. The second type involved insertion of one copy of IS15DI into the traJ gene. The process probably involved the insertion of IS15DI-pBackZero-IS15DI into the traJ gene and then the element was excised out, leaving a single scar copy of IS15DI in the traJ gene (Figure 4B). Interestingly, a 8 bp target site duplication sequence (ATACAGAT) was found to flank the IS15DI elements, indicating that this process may occur in two steps. The first step was mediated by the attack at the hot spot of the traJ gene by the insertion element IS15DI, leading to insection of IS15DI-pBackZero-IS15DI into the traJ gene, followed by the excision of pBackZero-IS15DI from the fusion plasmid through homologous recombination, leaving one single copy of the IS15DI scar in traJ gene (Harmer et al., 2020). This type of plasmid contained only one copy IS15DI, which existed in a circular form observable in Nanopore long reads, but the number of such plasmid was too low to be seen in S1-PFGE. The third type is more complicated. Insertion events occurred in both the traF and traJ genes and duplication of a DNA fragment from the pSa42-91k led to the formation of a plasmid of a much larger size (∼117 kb) designated as pSa42-TC5-117k (Figure 4A). The mechanism of DNA duplication in this type of plasmid is not clear and entails further investigation.
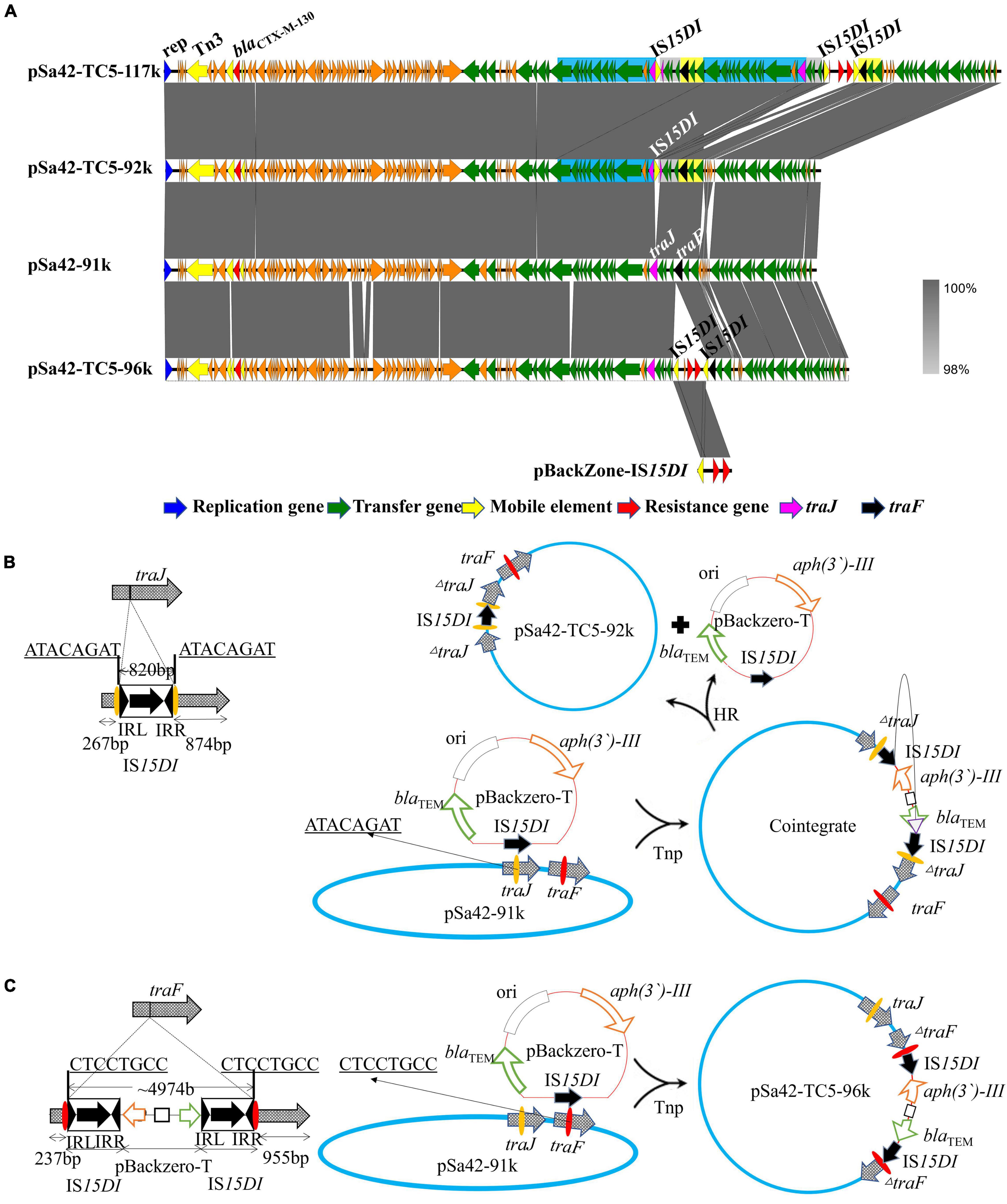
Figure 4. Alignment of three plasmids in transconjugant Sa42-TC5 and schematic representation of the two insertion events. (A) Alignment of various plasmids harbored by strain Sa42-TC5. Plasmids pSa42-TC5-117k, pSa42-TC5-92k, and pSa42-TC5-96k were obtained from transconjugant Sa42-TC5; pSa42-TC5-117k had two target site duplications (TSD) and was assembled de novo with Canu, the slight blue, gray, and yellow color depicted various repeated regions. Plasmids pSa42-TC5-92k and pSa42-TC5-96k with one TSD were selected directly from nanopore long reads and calibrated by matching with known plasmid sequences. (B) Genetic mechanisms of insertion of IS15DI into traJ. IS15DI led to an attack on the hot spot in the traJ gene. Resolution of traJ via transposase (Tnp) mediated cointegration, which was followed by homologous recombination (HR). (C) Proposed IS element-mediated fusion in traF only via replicative transposition event at the hot spot (CTCCGTCC). Target site duplications from different genes were underlined with bold letters. The left and right inverted repeats 14 bp (IRL and IRR) of IS15DI were shown in black triangles. IS15DI was denoted in black with an arrow indicating the orientation and length of the transposase gene.
Conclusion
Insertion of MDR-encoding mobile elements into the chromosome and plasmids has been widely reported following the wide usage of the whole genome sequencing (WGS) technology. With the increasing use of MinION nanopore sequencing technology and Illumina sequencing, IS element-mediated plasmid fusion events have also been increasingly reported. However, most of these previous studies only reported the final plasmid sequences based on the assembly of the most dominant reads and contigs generated from the sequencing platforms. The underlying mechanisms of the fusion and integration processes remain unknown. In this study, we have designed a set of experiments to investigate the molecular mechanisms underlying two of the most common fusion processes mediated by ISEcp1 and IS26-like elements, which involve homologous recombination and replicative transposition, respectively. Our data showed that both the 5′- or 3′-terminal of ISEcp1 could attack another copy of ISEcp1 through interaction with homologous sequences as short as 100 bp, resulting in the formation of a truncated ISEcp1. This finding helps explain why IS elements are commonly found in MDR-encoding genetic fragments. The IS15DI element mainly attacks the hot spots located in different target genes and mediates the conjugation process of plasmids that contain the traF and traJ genes. The hot spot site seems to be conservative for specific IS element, whereas the type of the hot spot site determines the mode of the integration. Findings in this study showed that detailed analysis of Nanopore reads would enable us to depict the dynamic DNA fusion processes mediated by different IS. This approach can be applied to study the mechanism of DNA fusion process mediated by other important IS elements.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW567497, https://www.ncbi.nlm.nih.gov/genbank/, MW567498, https://www.ncbi.nlm.nih.gov/genbank/, MW881232, https://www.ncbi.nlm.nih.gov/genbank/, MW881 230, and https://www.ncbi.nlm.nih.gov/genbank/, MW881231.
Author Contributions
KC performed sequencing and bioinformatic analysis. MX helped with conjugation and other experiments. EC, KC, and SC participated in research design and manuscript writing. SC supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key R&D Program of China (2018YFD0500300), Basic Research Fund of Shenzhen (20170410160041091), Collaborative Research Fund from the Research Grant Council of the Government of Hong Kong SAR (C5026-16G), and Research Impact Fund (R5011-18F).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Alikhan, N.-F., Petty, N. K., Zakour, N. L. B., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:1. doi: 10.1186/1471-2164-12-402
Arthur, A., and Sherratt, D. (1979). Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mole. Gen. Genet. MGG 175, 267–274. doi: 10.1007/BF00397226
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 1–15. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Chen, K., Dong, N., Chan, E. W.-C., and Chen, S. (2019a). Transmission of ciprofloxacin resistance in Salmonella mediated by a novel type of conjugative helper plasmids. Emerg. Microb. Infect. 8, 857–865. doi: 10.1080/22221751.2019.1626197
Chen, K., Dong, N., Zhao, S., Liu, L., Li, R., Xie, M., et al. (2018). Identification and characterization of conjugative plasmids that encode ciprofloxacin resistance in Salmonella. Antimicrob. Agents Chemother. 62:8. doi: 10.1128/AAC.00575-18
Chen, K., Wai Chi, Chan, E., and Chen, S. (2019b). Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg. Microb. Infect. 8, 396–403. doi: 10.1080/22221751.2019.1585965
Cock, P. J., Chilton, J. M., Grüning, B., Johnson, J. E., and Soranzo, N. (2015). NCBI BLAST+ integrated into Galaxy. Gigascience 4:3747. doi: 10.1186/s13742-015-0080-7
Davison, J. (1999). Genetic exchange between bacteria in the environment. Plasmid 42, 73–91. doi: 10.1006/plas.1999.1421
De La Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Garcillán-Barcia, M. P., Francia, M. V., and De La Cruz, F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microb. Rev. 33, 657–687. doi: 10.1111/j.1574-6976.2009.00168.x
Harmer, C. J., Pong, C. H., and Hall, R. M. (2020). Structures bounded by directly-oriented members of the IS26 family are pseudo-compound transposons. Plasmid 2020:102530. doi: 10.1016/j.plasmid.2020.102530
Kim, E.-H., and Aoki, T. (1994). The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microb. Immunol. 38, 31–38. doi: 10.1111/j.1348-0421.1994.tb01741.x
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017:215116. doi: 10.1101/gr.215087.116
Li, R., Xie, M., Dong, N., Lin, D., Yang, X., Wong, M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. GigaScience 7:gix132.
Miriagou, V., Carattoli, A., Tzelepi, E., Villa, L., and Tzouvelekis, L. S. (2005). IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob. Agents Chemother. 49, 3541–3543. doi: 10.1128/AAC.49.8.3541-3543.2005
Miyano, M., Tanaka, K., Ishikawa, S., Takenaka, S., Miguel-Arribas, A., Meijer, W. J., et al. (2018). Rapid conjugative mobilization of a 100 kb segment of Bacillus subtilis chromosomal DNA is mediated by a helper plasmid with no ability for self-transfer. Microb. Cell Fact. 17, 1–10. doi: 10.1186/s12934-017-0855-x
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Pinilla-Redondo, R., Cyriaque, V., Jacquiod, S., Sørensen, S. J., and Riber, L. (2018). Monitoring plasmid-mediated horizontal gene transfer in microbiomes: recent advances and future perspectives. Plasmid 99, 56–67. doi: 10.1016/j.plasmid.2018.08.002
Ribot, E. M., Fair, M., Gautom, R., Cameron, D., Hunter, S., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodbourne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
San Millan, A. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microb. 26, 978–985. doi: 10.1016/j.tim.2018.06.007
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microb. 3, 711–721. doi: 10.1038/nrmicro1234
Wrighton, C. J., and Strike, P. (1987). A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17, 37–45. doi: 10.1016/0147-619x(87)90006-0
Xie, M., Chen, K., Ye, L., Yang, X., Xu, Q., Yang, C., et al. (2020). Conjugation of virulence plasmid in Clinical Klebsiella pneumoniae strains through formation of a fusion plasmid. Adv. Biosyst. 4:1900239. doi: 10.1002/adbi.201900239
Keywords: helper plasmid, ISEcp1, IS15DI, polymorphism, plasmid fusion
Citation: Chen K, Xie M, Chan EW-C and Chen S (2022) Delineation of ISEcp1 and IS26-Mediated Plasmid Fusion Processes by MinION Single-Molecule Long-Read Sequencing. Front. Microbiol. 12:796715. doi: 10.3389/fmicb.2021.796715
Received: 17 October 2021; Accepted: 17 December 2021;
Published: 07 February 2022.
Edited by:
Jesus L. Romalde, University of Santiago de Compostela, SpainReviewed by:
Joseph Nesme, University of Copenhagen, DenmarkSabela Balboa, University of Santiago de Compostela, Spain
Copyright © 2022 Chen, Xie, Chan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Chen, c2hlY2hlbkBjaXR5dS5lZHUuaGs=
†These authors have contributed equally to this work
 Kaichao Chen
Kaichao Chen Miaomiao Xie
Miaomiao Xie Edward Wai-Chi Chan
Edward Wai-Chi Chan Sheng Chen
Sheng Chen