- 1Carolinas Medical Center—Atrium Health, Charlotte, NC, United States
- 2Section of Oral Medicine–University of Connecticut Health, Farmington, CT, United States
Objectives: Human papillomavirus (HPV) is a known etiological factor of oropharyngeal head and neck cancer (HNC). HPV positivity and periodontal disease have been associated with higher HNC risk, suggesting a role for oral bacterial species. Our objective was to determine oral microbiome profiles in HNC patients (HPV-positive and HPV-negative) and in healthy controls (HC).
Methods: Saliva samples and swabs of buccal mucosa, supragingival plaque, and tongue were collected from HNC patients (N = 23 patients, n = 92 samples) before cancer therapy. Next-generation sequencing (16S-rRNA gene V3–V4 region) was used to determine bacterial taxa relative abundance (RA). β-Diversities of HNC HPV+ (N = 16 patients, n = 64 samples) and HNC HPV– (N = 7 patients, n = 28 samples) groups were compared using PERMANOVA (pMonte Carlo < 0.05). LEfSe discriminant analysis was performed to identify differentiating taxa (Log LDA > 2.0). RA differences were analyzed by Mann–Whitney U-test (α = 0.05). CombiROC program was used to determine multi-marker bacterial signatures. The Microbial Interaction Network Database (MIND) and LitSuggest online tools were used for complementary analyses.
Results: HNC vs. HC and HNC HPV+ vs. HNC HPV– β-diversities differed significantly (pMonte Carlo < 0.05). Streptococcus was the most abundant genus for HNC and HC groups, while Rothia mucilaginosa and Haemophilus parainfluenzae were the most abundant species in HNC and HC patients, respectively, regardless of antibiotics treatment. LEfSe analysis identified 43 and 44 distinctive species for HNC HPV+ and HNC HPV– groups, respectively. In HNC HPV+ group, 26 periodontal disease-associated species identified by LefSe had a higher average RA compared to HNC HPV– group. The significant species included Alloprevotella tannerae, Fusobacterium periodonticum, Haemophilus pittmaniae, Lachnoanaerobaulum orale, and Leptotrichia spp. (Mann–Whitney U-test, p < 0.05). Of 43 LEfSe-identified species in HPV+ group, 31 had a higher RA compared to HPV– group (Mann–Whitney U-test, p < 0.05). MIND analysis confirmed interactions between Haemophilus and Leptotrichia spp., representing a multi-marker signature per CombiROC analysis [area under the curve (AUC) > 0.9]. LitSuggest correctly classified 15 articles relevant to oral microbiome and HPV status.
Conclusion: Oral microbiome profiles of HNC HPV+ and HNC HPV– patients differed significantly regarding periodontal-associated species. Our results suggest that oral bacterial species (e.g., Leptotrichia spp.), possessing unique niches and invasive properties, coexist with HPV within HPV-induced oral lesions in HNC patients. Further investigation into host–microbe interactions in HPV-positive HNC patients may shed light into cancer development.
Introduction
Head and neck cancer (HNC) is the sixth most common cancer worldwide with over 95% comprising squamous cell carcinomas (SCCs) (Jemal et al., 2008; Kumarasamy et al., 2019). Head and neck SCCs are characterized by a locoregional development mainly diagnosed at an advanced stage of the disease, resulting in difficult treatment and eradication of both pre-neoplastic and neoplastic tissue (Carvalho et al., 2005; Ganci et al., 2015). Despite advancements in chemoradiation, ionizing radiation, and surgical resection techniques, HNC has an overall mortality rate of approximately 50% and is characterized by high recurrence rates (Carvalho et al., 2005). While triggers of HNC development have not been fully elucidated, two primary risk factors have been identified, namely, alcohol and tobacco consumptions (Božinović et al., 2019). Most recent studies have identified infection with human papillomavirus (HPV) as a third and more prominent cause of tumor formation (Božinović et al., 2019). HPV-associated SCCs represent the most common HPV-related cancer in the US and are classified by a new staging system for oropharyngeal cancers (National Cancer Institute1 : van Gysen et al., 2019).
HPV-positive (HPV+) HNC patients are often younger and present with a more advanced cancer stage than HPV-negative (HPV–) HNC patients (Blitzer et al., 2014). It has been reported that the majority of HPV-associated HNCs are caused by HPV16, though more than 220 HPV serotypes have been identified (Tumban, 2019). Two HPV genes, E6 and E7, have been the matter of extensive research due to their role as oncogenes (Yim and Park, 2005). These genes are involved in multiple pathways such as transmembrane signaling, cell cycle regulation, and cell transformation (Yim and Park, 2005). E6 has been shown to promote degradation of tumor suppressor TP53 (Crook et al., 1991), while E7 is able to inhibit retinoblastoma protein (Pal and Kundu, 2020). Aside from E6 and E7, the E5 HPV gene promotes malignancy, has anti-apoptotic effects and plays a role in epidermal growth factor (EGF) receptor-regulated cell proliferation (Venuti et al., 2011).
There is a mounting body of evidence that a synergistic interaction between periodontal disease-associated pathogens and HPV exists. Indeed, a case–control study by Tezal et al. (2009) found that HPV+ tumors in 21 patients had a significantly higher alveolar bone loss mean and a fourfold increased risk for HPV+ tumor status for every millimeter of alveolar bone loss caused by periodontal disease.
Overall, without implying a causal effect, a link between oral microbiome dysbiosis and cancer has been suggested by several studies (Kudo et al., 2016; Hayes et al., 2018; Mohammed et al., 2018; Wu et al., 2018; Mougeot et al., 2020). For instance, Fusobacterium nucleatum was found overabundant in the oral cavity of patients with colon cancer and lymph node metastasis (Kudo et al., 2016). F. nucleatum might initiate oncogenic and proinflammatory responses that stimulate the growth of colon cancer cells (Kudo et al., 2016). Increased levels of blood serum antibodies against the oral bacterial species Porphyromonas gingivalis was associated with a twofold higher risk of pancreatic cancer when compared to healthy individuals (Mohammed et al., 2018).
Furthermore, the increased prevalence of P. gingivalis and Aggregatibacter actinomycetemcomitans was shown to initiate a Toll-like receptor signaling pathway predictive of pancreatic cancer in animal models (Mohammed et al., 2018). Higher levels of firmicutes and bacteroidetes also constitute a potential risk for gastric cancer (Wu et al., 2018). A study by Hayes et al. (2018) suggested an association between the oral microbiome and HNC.
HNC has also been associated with oral cavity diseases such as periodontal disease and dental caries (Michaud et al., 2017; Gasparoni et al., 2021). Periodontitis disrupts the normal oral microbial environment, thereby leading to dysbiosis. Dysbiosis can translate into an abundance shift of opportunistic species, like P. gingivalis, which produce several virulence factors resulting in the destruction of periodontal tissues (Rafiei et al., 2017). Chronic periodontitis can result in the release of proinflammatory cytokines from squamous cells, causing inflammation and possible decreased apoptosis (Gholizadeh et al., 2016). In a meta-analysis by Zeng et al. (2013) HNC cancer risk was found to be increased by 2.63-fold in patients with periodontitis.
Using 16S rRNA gene next-generation sequencing and computational approaches, the purpose of this study was to compare microbiome profiles of a limited cohort of HNC patients to those of healthy control subjects, and profiles of HNC HPV+ patients to those of HNC HPV– patients, within the HNC cohort. We also aimed to determine whether bacterial species differentiating HPV+ from HPV– HNC patients are associated with periodontal disease-associated species.
Materials and Methods
Patient Recruitment
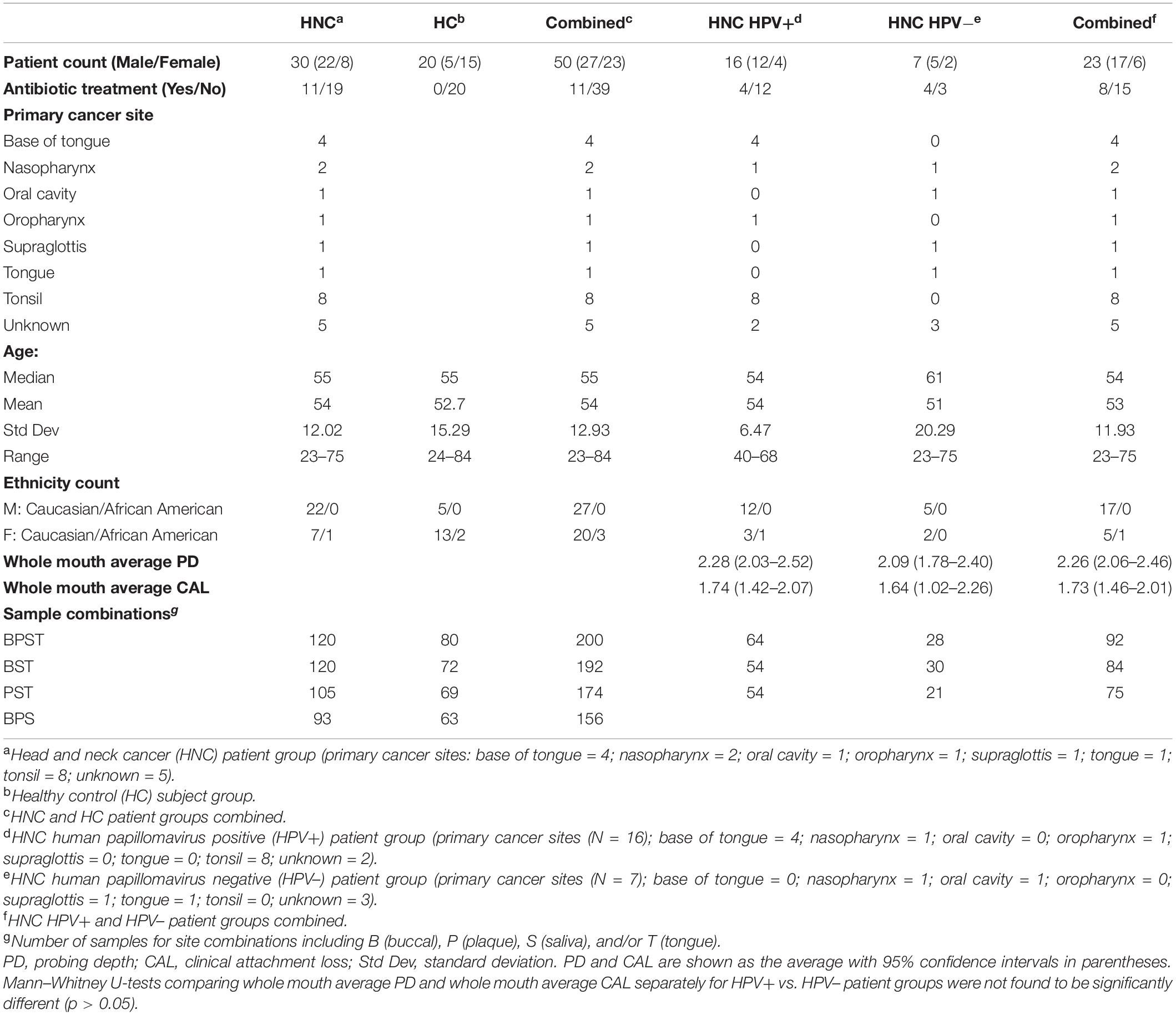
HNC patients with SCC [N = 30; 8 females, 22 males, age range = 23–75 years (SD = ± 12.02)] were recruited from the OraRad study (U01DE022939) (Brennan et al., 2017; Lalla et al., 2017). OraRad was a multicenter cohort study that collected longitudinal data on radiation-treated HNC patients at 6-month intervals for 2 years. Primary cancer site origin included the base of the tongue, tonsil, neck, tongue, and oral cavity.
Of 30 HNC patients, 23 were clinically classified as HPV+/− into 16 HPV+ and 7 HPV– patients. In addition, healthy control subjects (HC group) (N = 20; age range 24–84, SD = –12.93) were recruited through Atrium Health’s Carolinas Medical Center, Charlotte, NC. Of 30 HNC patients, 11 had received antibiotic treatment within 2 weeks of sampling. No HC subject had received antibiotic treatment. The study was approved by the institutional review board, and all participants gave informed consent for the study.
Sample Collection
Saliva (S) samples and swab samples of buccal mucosa (B), supragingival plaque (P), and tongue (T) were collected from HNC patients, pre-cancer treatment at baseline, and from HC subjects. Saliva collection was performed while chewing unsweetened and unflavored gum (The Wrigley Company—Mars, Chicago, IL, United States) for a period of 2 min into a 50-ml conical BD falcon polypropylene centrifuge tube (Corning, Corning, NY, United States).
Buccal mucosal samples were subsequently collected by swabbing both sides of the buccal mucosa for 10 s each. Tongue samples were then obtained by swabbing a 1-cm2 region on both sides of the mid-dorsal region of the tongue for 5 s. Finally, supragingival plaque samples were obtained by swabbing across the lateral surfaces of all maxillary and mandibular teeth at the junction of the tooth and gingiva. All swab collections were performed using OmniSwabs (GE Life Sciences-Buckinghamshire, United Kingdom).
Bacterial DNA Extraction, Processing, and Sequencing
Bacterial genomic DNA was extracted from oral samples using QIAamp DNA Mini Kit procedure (QIAGEN, Valencia, CA, United States) per manufacturers’ instructions. During sample preparation, 50 ng of genomic DNA was used for PCR in which the 16S rRNA gene (V3–V4) region was amplified, followed by purification and processing methods as previously described (Caporaso et al., 2011). Next-generation sequencing was performed using the MiSeq v3 reagent kit and platform (Illumina, Inc., San Diego, CA, United States). To prepare for cluster generation and sequencing, libraries were denatured with NaOH and diluted with a hybridization buffer. Libraries then underwent heat denaturation prior to MiSeq sequencing. Total of 100 ng of each library was pooled together, run on a gel, gel-extracted, and run on a bioanalyzer for quantification. A total concentration of 4 nM of the library was then diluted, and 12 pM of the library was spiked with 20% PhiX. At least 5% PhiX was added as an internal control for low-diversity libraries. Identification of bacterial genera and species was performed using Human Oral Microbe Identification, HOMINGS, which employs a ProbeSeq BLAST program for species/genera identification through recognition of the 16S rRNA gene (V3–V4 region) sequence reads (Caporaso et al., 2011; Mougeot et al., 2016). ProbeSeq loads raw sequence files into a cell array, looping through the array one sequence at a time searching for small sequence strings that 100% match an oligomer (partials are not considered matches). If a match is identified, a counter begins giving counts of the total number of probe-specific “hits.” Hits are then accumulated by species/genera and sample.
The sequence reads were matched to 737 ProbeSeq taxon probes, i.e., to species probes (n = 620) or genus probes (n = 117) if not matched to a species probe, or were otherwise recorded as an unmatched read. Matched and unmatched probe count data were provided per taxon per patient as Excel spreadsheets. Species/genus probes containing zeros for all samples were removed from the dataset. Raw abundance data were then transformed into relative abundance (RA) data for further analysis.
Bioinformatic Analysis
α-Diversity
Shannon and Simpson indices were generated using PRIMERv7 (PRIMER-E Ltd., Ivybridge, United Kingdom) (Clarke and Gorley, 2006), based on microbiome RA data. RA data of HC subjects (HC group: N = 20) were compared to the RA data of HNC patients including those with antibiotic treatment within 2 weeks of sampling (Grp-All: N = 30). RA data of HC group were also compared to RA data of HNC patients excluding those with antibiotic treatment (Grp-NoAB: N = 19). Subsequently, comparisons of HPV+ vs. HPV– HNC sub-cohorts were performed by including or excluding patients who received antibiotic treatment. Mann–Whitney U-tests were then used to determine significant RA comparisons (α = 0.05) using XLSTATv2016.02.29253 (Data Analysis and Statistical Solution for Microsoft Excel, Addinsoft, Paris, France, 2017).
Permutational Multivariate Analysis of Variance
Patient subgroups used for permutational multivariate analysis of variance (PERMANOVA) included Grp-All (HNC: N = 30; HC: N = 20) and Grp-noAB (HNC: N = 19; HC: N = 20). Sub-analyses were performed based on the multiple sample site combinations “BPST,” “BST,” and “PST” which provided sufficient power in PRIMERv7 program (PRIMER-E Ltd., Ivybridge, United Kingdom) (Clarke and Gorley, 2006) for all five relevant comparisons (Figure 1). Species and genera RA data were square root transformed and converted into Bray–Curtis similarity matrices.
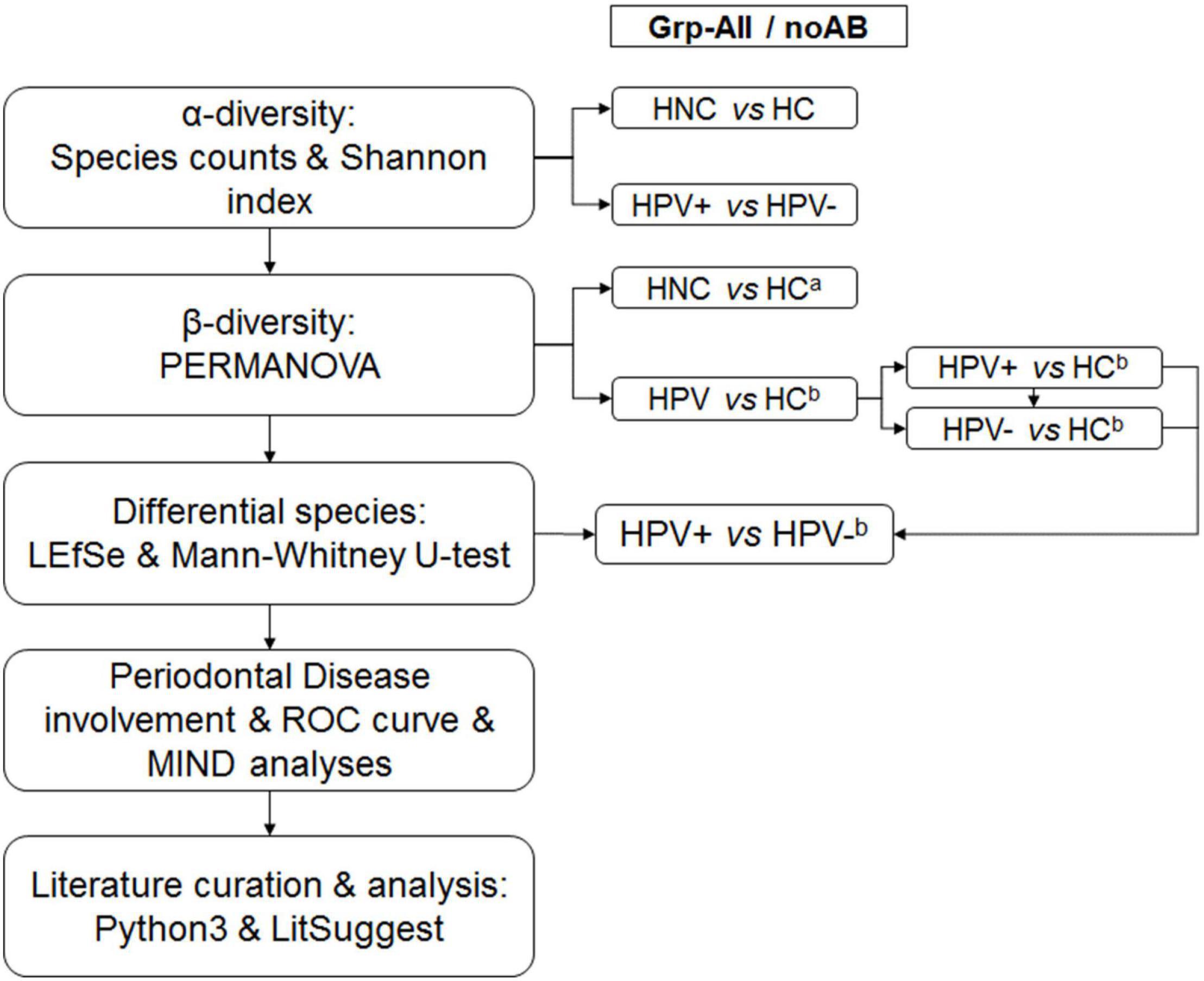
Figure 1. Overall analytical strategy. α-Diversity was calculated for Grp-All HNC vs. HC (N = 30; N = 20) and HPV+ vs. HPV– (N = 16; N = 7) and for Grp-noAB HNC vs. HC (N = 19; N = 20) and HPV+ vs. HPV– (N = 12; N = 3). Sample sites consisted of different combinations of buccal (B), plaque (P), saliva (S), and tongue (T) samples. β-Diversity was calculated using PERMANOVA by two different analytical designs: (1) aAnalytical design with “Diagnosis” and “Sample site” as fixed factors and “Antibiotic” as the nested random factor. Comparisons were Grp-All HNC vs. HC and Grp-noAB HNC vs. HC. (2) bAnalytical design with “HPV status” and “Sample site” as fixed factors and “Antibiotic” as the nested random factor. Grp-All comparisons were as follows: HPV vs. HC (N = 23; N = 20), HPV+ vs. HC (N = 16; N = 20), HPV– vs. HC (N = 7; N = 20), and HPV+ vs. HPV– (N = 16; N = 7). Grp-noAB comparisons were as follows: HPV vs. HC (N = 15; N = 20), HPV+ vs. HC (N = 12; N = 20), HPV– vs. HC (N = 3; N = 20), and HPV+ vs. HPV– (N = 12; N = 3). Distinct species were determined using LEfSe and Mann–Whitney U-tests for Grp-All and Grp-noAB comparisons of HPV+ vs. HPV– for the sample site combination BPST. Significant LEfSe bacterial species for HPV+ group were investigated for their roles in periodontal disease using conventional methods (PubMed, Google Scholar, etc.). Significant LEfSe HPV+ species using the Mann–Whitney U-test were analyzed by using MedCalc ROC curve analysis. Multi-marker combinatorial analysis was completed using CombiROC online tool (http://combiroc.eu/) for the bacterial species identified with MedCalc log(RA + 1) ROC curve AUC of 0.75 or greater (n = 10 species). CombiROC utilized raw RA data and log(RA + 1) transformed data. Microbial Interaction Network Database (MIND) online tool was used to identify possible interactions between most significant bacterial species. Pythonv3.6.2 code was used to extract PubMed abstracts matching key words to determine positive and negative training sets and were validated manually. Using NCBI LitSuggest online tool (https://www.ncbi.nlm.nih.gov/research/litsuggest/), 203 PubMed articles were classified to identify articles involving HPV infection in the oral microbiome.
PERMANOVA analyses were performed using a mixed model with unrestricted permutation of raw data, 9,999 permutations, and type III partial sum of squares (Clarke and Gorley, 2006), as previously implemented (Mougeot et al., 2019, 2020). Fixed factors were “Diagnosis” (e.g., HNC vs. HC) and “Sample site” (B, P, S, and T). In this design, the “Antibiotic” treatment (yes or no) variable was used as a random factor nested into “Diagnosis” and “Sample site.” Monte Carlo corrected p-values (α = 0.05) were determined, as appropriate for relatively small sample sizes. Principal coordinate analysis (PCoA) was completed for the Grp-All: HNC vs. HC BPST sample site combination.
β Diversity Sub-Analyses
Sub-analyses were completed using subsets of Grp-All and Grp-noAB patients, based on n = 3 or 4 sample sites per patient (BPST, BST, and PST): HPV+ (N = 16) vs. HPV– (N = 7) and HPV+ (N = 12) vs. HPV– (N = 3), respectively. The sample site combinations BPST, BST, and PST and the previously mentioned data transformation were used for PERMANOVAs in PRIMERv7 program. Fixed factors used were “HPV status” (positive and negative) and “Sample site” (B, P, S, and T). “Antibiotic” (yes or no) was used as a random factor and nested into “HPV status” and “Sample site.” Monte Carlo corrected p-values (α = 0.05) were determined. PCoA was completed for the Grp-All: HPV+ vs. HPV– BPST sample site combination.
Linear Discriminant Analysis Effect Size
Taxonomy levels were added manually to ProbeSeq derived datasets for Grp-All (HPV+ vs. HPV–) and Grp-noAB (HPV+ vs. HPV–) subsets. The text files were then formatted for linear discriminant analysis (LDA) effect size (LEfSe) using the Galaxyv1.0 online tool (Jalili et al., 2020). LEfSe data input consisted of “HPV status” as the option “Class” and “Patient ID” as the option “Subject” (Segata et al., 2011). Data were normalized. Using the “one-against-all” strategy for multi-class analysis, the factorial Kruskal–Wallis test and pairwise Wilcoxon signed-rank test were set at a Monte Carlo significance (α = 0.05) to calculate LDA scores. Log LDA scores were set at a threshold > 2.0. Histograms of the differential features (species) were generated, and each species was investigated for its role in periodontal disease.
Receiver-Operating Characteristic Curve Analyses
Conventional Receiver-Operating Characteristic Analysis
Mann–Whitney U-tests were completed for LEfSe differential features for HPV+ species probes from Grp-All and Grp-noAB groups. Significant species probes (α = 0.05) further underwent receiver-operating characteristic (ROC) curve analysis for Grp-All HPV+ and Grp-noAB HPV+ species probes using the BPST sample combination in MedCalc program (MedCalc Software Ltd, Ostend, Belgium).
RA data were log-transformed with the addition of a pseudo-count [i.e., log(RA + 1)]. Analysis was completed for Grp-All (HPV+; n = 64 samples and HPV–; n = 28 samples) and Grp-noAB (HPV+; n = 48 samples and HPV–; n = 12 samples) groups and for each non-zero RA probe in MedCalc program. The area under the curve (AUC) of each probe was calculated, and ROC curves were generated. Significance level was set at α = 0.05, and biomarker accuracy was calculated using methods described by Ray et al. (2010).
CombiROC Analysis
ROC curves from MedCalc that had an AUC greater than 0.75 were subjected to combinatorial analysis using CombiROC online tool2 (Mazzara et al., 2017) based on raw RA data and log(RA + 1) transformed RA data. Using CombiROC, marker profile plots were generated to confirm quality, and the detection threshold was set to 0.001. Using this threshold, combinational analysis was performed which calculated the sensitivity and specificity scores for each marker or combination of markers corresponding to the probability that the microbial data will be positive when HPV is present and the probability that microbial data will be negative when HPV is not present.
A minimum feature filter was set to include at least two markers. Based on a threshold of 10 for sensitivity and 50 for specificity, the best or “gold” combinations of markers were kept, thereby creating optimal multi-marker ROC curves and violin plots. Summary statistics were calculated and recorded for the top two AUC scores of the raw and log(RA + 1) transformed RA data.
Microbial Interaction Network Database Analysis
A microbial interaction network was created to illustrate possible interactions between Haemophilus spp. and Leptotrichia spp. with other bacterial genera or species, by using Microbial Interaction Network Database (MINDv1.0) (Microbial Interaction Network Database, 2019). Default options were selected for human tissue sites, interaction weight, and health or disease conditions.
LitSuggest
An application programming interface was established using the National Center for Biotechnology Information guidelines (National Center for Biotechnology Information, 1988) and Pythonv3.6.2 (van Rossum and Drake, 2009). Pythonv3.6.2 was used to generate classifiers by extracting abstracts from PubMed (1997) through the keywords (i) “oral microbiome” and “HPV” to constitute a positive training set, (ii) “vaginal microbiome” and “HPV” keywords to constitute the negative training set, and (iii) “HPV” and “microbiome” to constitute the test set. Positive and negative training set abstracts were then manually validated. Using the NCBI LitSuggest3 online tool; a total of 19 positively and 104 negatively classified articles were used to train the model (Allot et al., 2021). Test set classification was then completed using LitSuggest, and full articles were manually verified for relevancy.
Results
The overall analytical strategy is presented in Figure 1. Demographics and clinical information of our HNC patient cohort (N = 30 patients, sub-cohort of OraRad study) are presented in Table 1. Clinical information, including caries and periodontal disease status for OraRad HNC patient cohort, associated with HPV status (N = 559 of 572 total patients) has been published elsewhere (Brennan et al., 2021). While no significant differences were noted in age and ethnicity, the male population was over-represented in the HNC patient set in OraRad and this study, as anticipated for oral SCC in general and for HPV-associated oropharyngeal cancers (Fakhry et al., 2018; Mahal et al., 2019). In our sub-cohort, most HPV+ HNC patients had oropharyngeal cancer (e.g., tonsil, base of tongue), whereas most of the HPV– HNC patients had cancer in other sites (Table 1).
Abundance, Species Detection, and α-Diversity
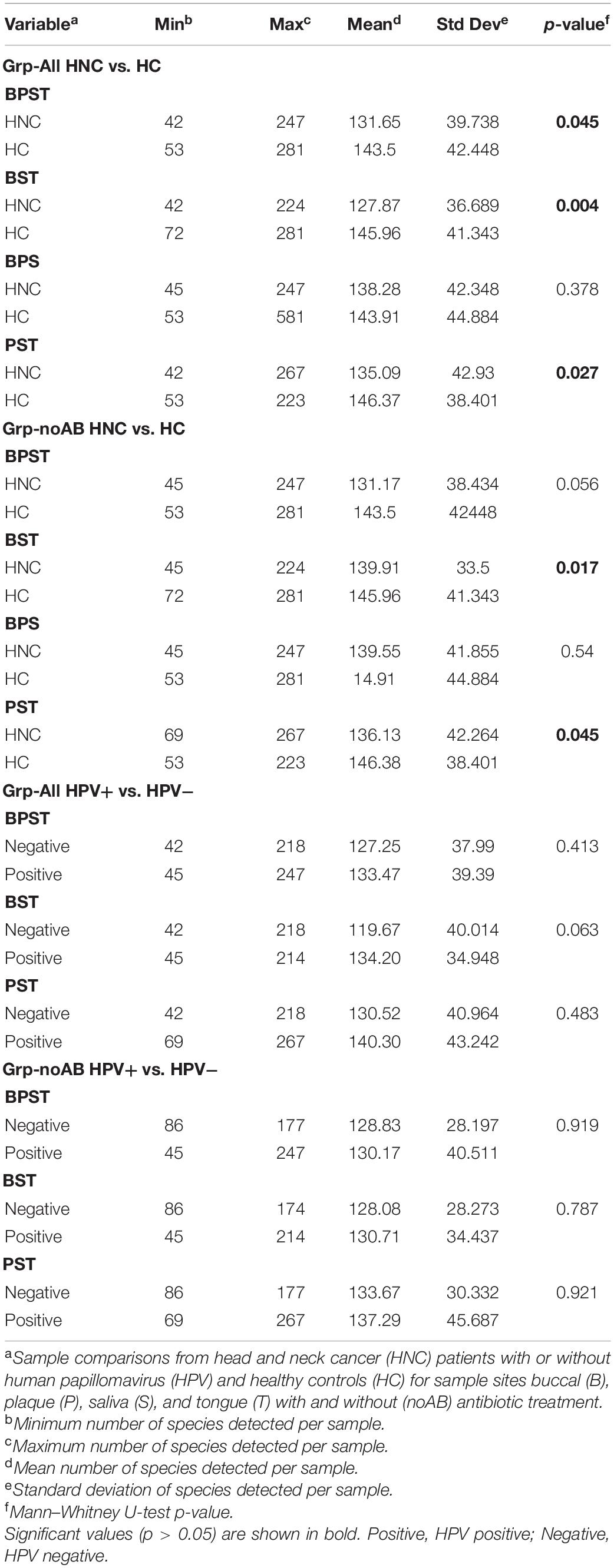
Probe count data are provided as Supplementary Data Files and can be downloaded from our lab’s Github repository4 (Supplementary Data Files 1, 2). Sequencing reads matched 737 total probes (117 genera and 620 species probes) for all samples from HNC and HC groups combined. Comparisons of species and genera detected for HNC vs. HC and HPV+ vs. HPV– are presented in Supplementary Table 1. Unmatched reads were removed from RA determinations. For all samples sequencing data, 442 of 620 species probes and 65 of 117 genus probes had at least one matched read. Significant α-diversity differences were identified for Grp-All and Grp-noAB for HNC vs. HC and Grp-noAB HNC vs. HC for the matched sample site combinations BPST, BST, and PST (Table 2). Streptococcus was the most abundant genus for HNC and HC groups, whereas Rothia mucilaginosa and Haemophilus parainfluenzae were the most abundant species detected in HNC patients and HC, respectively. Excluding HNC patients treated with antibiotics (Grp-noAB) did not affect these results (data not shown). Overall, the highest and lowest average number of taxa detected per sample were 96.08 and 117.66 for species probes and 24.75 and 26.83 for genus probes (Supplementary Table 1).
β-Diversity Analysis
PERMANOVA β-diversity analyses were performed for sample site combinations providing sufficient power based on available oral microbiome data (i.e., BPST, BST, and PST). Significance of β-diversity analyses is presented in Figure 2. For the Grp-All comparisons, including HNC HPV+ vs. HNC HPV– comparisons, all but one (i.e., HPV+ vs. HC, BST, pMonte Carlo = 0.261) were significant, regardless of the sample site combinations analyzed. All Grp-noAB comparisons were found significant for “HPV status” and “Sample site.” Monte Carlo corrected p-values of all comparisons are presented in Supplementary Table 2. PCoA plots describing the variations explaining dissimilarity between groups (i.e., HNC vs. HC and HPV+ vs. HPV–) are presented in Supplementary Figure 1.
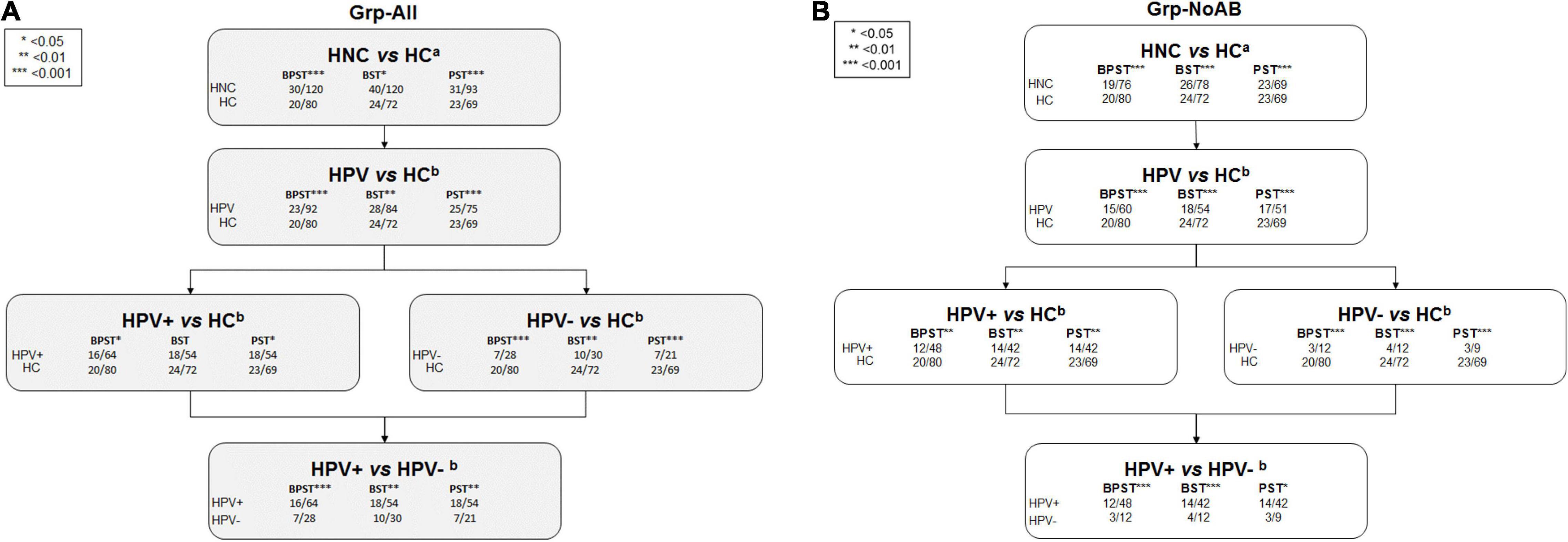
Figure 2. PERMANOVA results comparisons flowchart. (A) Grp-All. (B) Grp-noAB. β-diversity analyses were performed using PERMANOVA in PRIMERv7 software (PRIMER-E Ltd., IvyBridge, United Kingdom) to compare microbial profiles of head and neck cancer (HNC) patients to healthy controls (HC) and to compare microbial profiles of HNC HPV-positive (HPV+) patients to HNC HPV– patients. Sample sites consisted of three to four site combinations of buccal (B), plaque (P), saliva (S), and tongue (T). PERMANOVA analysis was completed using two different analytical designs based on Bray–Curtis similarity matrices determined from square root transformed relative abundance data of 737 probes (620 species and 117 genus probes). Sample site combinations consisted of BPST, BST, and PST for (A) Grp-All HNC vs. HC (N = 30; N = 20) and (B) Grp-no Antibiotics (Grp-noAB) HNC vs. HC (N = 19; N = 20). aFor the Grp-All HNC vs. HC comparison, “Diagnosis” was the main fixed factor, and “Sample Site” was the secondary fixed factor. “Antibiotics” was nested into “Diagnosis” and “Sample site” factors as a random variable. Grp-noAB analytical design did not include antibiotics as a factor. bFor the analytical design considering HPV, “HPV status” and “Sample site” were set as fixed factors and “Antibiotics” as nested as random factor. Grp-noAB analytical design did not include antibiotics as a factor. Level of significance is denoted using an asterisk (*): * < 0.05 = p-value less than 0.05; ** < 0.01 = p-value less than 0.01; *** < 0.001 = p-value less than 0.001.
LEfSe Analysis
A total of 44 and 43 species were identified for Grp-All HPV– and HPV+, respectively. A histogram of the differential features is presented in Figure 3A. Species of the genera Actinomyces and Leptotrichia were the most representative of HPV– and HPV+ patient groups, respectively. A total of 52 and 38 species were identified for Grp-noAB HPV– and HPV+, respectively (Figure 3B). Leptotrichia spp. were the most represented taxa for Grp-noAB HPV+ patients, and Prevotella spp. were the most represented ones for Grp-noAB HPV– patients. A total of 26 of 43 species in Grp-All HPV+ group (60.5%) and 24 of 38 species in Grp-noAB HPV+ group (63.2%) were recognized for their involvement in periodontal disease by performing manual searches in PubMed (Supplementary Table 3).
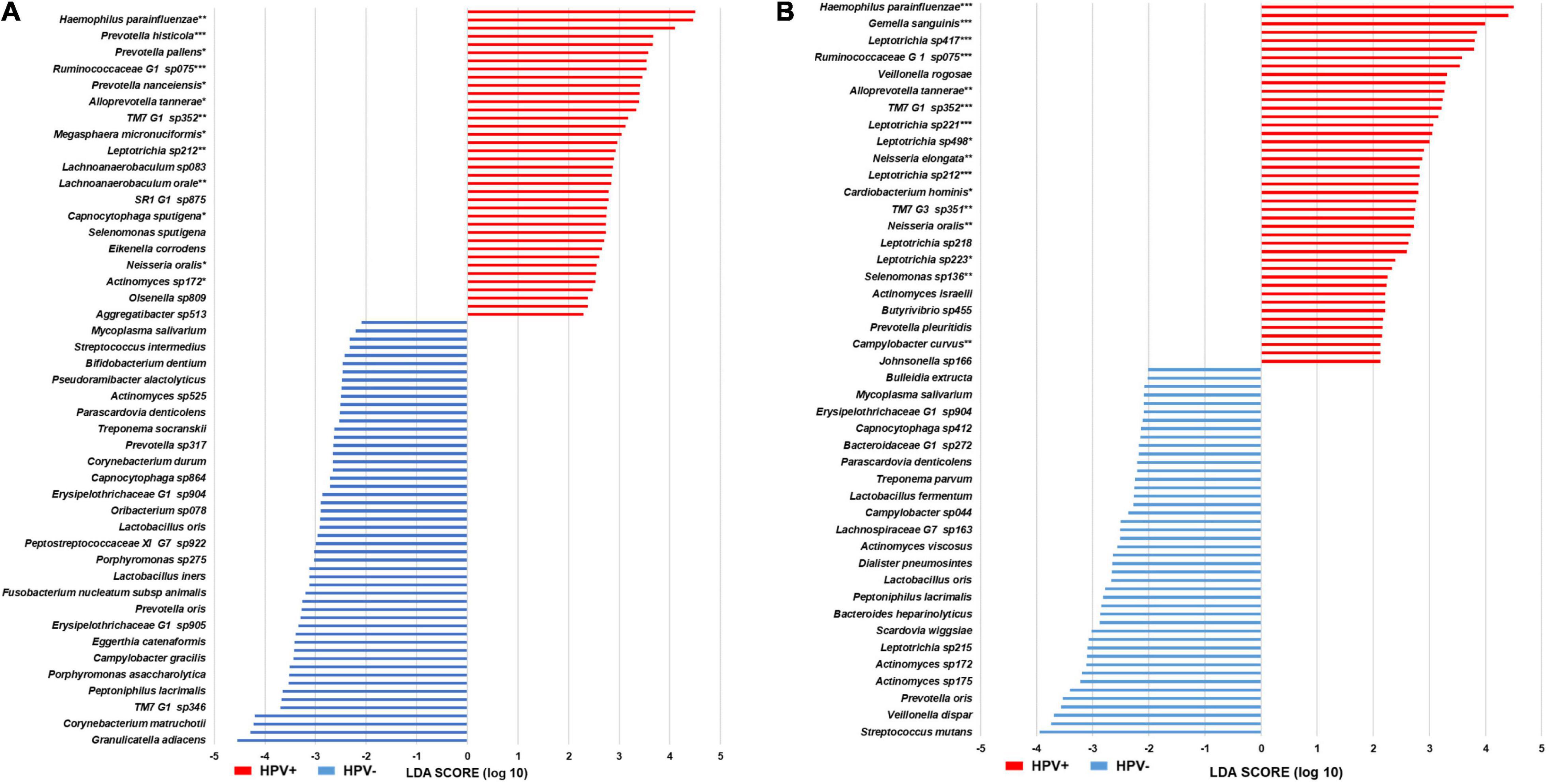
Figure 3. LEfSe histograms of differential features in head and neck cancer patients. (A) Grp-All BPST. (B) Grp-noAB BPST. Linear discriminant analysis Effect Size (LEfSe) was performed to determine distinct microbiome features in oral samples [stimulated saliva (S) samples and swabs of buccal mucosa (B), supragingival plaque (P), and tongue (T)] of the following: (A) Grp-All patient cohort (N = 23) for the head and neck cancer (HNC) HPV-positive (HPV+; N = 16) vs. HPV-negative (HPV–; N = 7) comparisons. (B) Grp-noAB patient cohort (N = 15) for HNC HPV+ (N = 12) vs. HPV– (N = 3) that did not receive antibiotics within 2 weeks of sampling. Horizontal histograms depict the discriminant features, i.e., bacterial species, for Grp-All and Grp-noAB potential biomarkers for HPV+ (red) and HPV– (blue). Mann–Whitney U-tests were used to determine significance of HPV+ distinctive species. Level of significance is depicted by an asterisk (*): *p < 0.05; **p < 0.01; ***p < 0.001.
Receiver-Operating Characteristic Determination
Using the Mann–Whitney U-test, 31 of 43 bacterial species in Grp-All HPV+ and 29 of 38 bacterial species in Grp-noAB HPV+ LEfSe were significant (p < 0.05) (Figures 3A,B). Using MedCalc ROC curve analysis, one species (Lachnoanaerobaculum sp083) in Grp-All HPV+ was found not significant. All species in Grp-noAB HPV+ were, however, significant (p < 0.01). By minimizing zero inflation, we found 17 of the 31 Grp-All HPV+ species and 16 of the 29 Grp-noAB HPV+ species to be significant (MedCalc Software Ltd, Ostend, Belgium). Minimization of zero inflation is required to optimize ROC analysis for the bacterial species which are more consistently detected across subjects and to increase the “signal-to-noise” ratio for a panel of select candidate bacterial taxa biomarkers. Indeed, Haemophilus pittmaniae, Rumonococcaceae G1 sp. HOT 075, and three Leptotrichia spp. were determined to be “Excellent” biomarkers in terms of sensitivity, specificity, and accuracy in the Grp-noAB using the log(RA + 1) transformed data with minimized zero inflation (Supplementary Table 4). Descriptive statistics of all the species with significant ROC curves are presented in Supplementary Table 4. Notably, Leptotrichia was the most represented significant genus for both Grp-All and Grp-noAB groups HPV+ vs. HPV– comparisons.
CombiROC and Microbial Interaction Network Database Investigation
From the ROC MedCalc analyses, 10 bacterial species from Grp-All HPV+ group had an AUC of at least 0.75, distinguishing HNC HPV+ from HNC HPV– group (Supplementary Table 4). Using RA data in CombiROC program (Mazzara et al., 2017), 24 “gold” combinations were generated out of 2,036 possible combinations containing at least two markers. The best two combinations (greatest AUC) were “Combo XXII” consisting of the microbial species probes Ruminococcaceae sp075, H. parainfluenzae, H. pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 and “Combo XV” consisting of Ruminococcaceae sp075, H. pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 (Figure 4A). “Combo XXII” and “Combo XV” had AUCs of 0.941 and 0.928, accuracies of 0.88 and 0.85, and positive predictive values of 0.69 and 0.65, respectively (Figure 4A). Using log(RA + 1) data, 46 “gold” combinations were created out of 1,013 possible combinations with the best two combinations of microbial probes being “Combo XLII” and “Combo XXXVI.” “Combo XLII” contained a combination of Gemella sanguinis, H. pittmaniae, Leptotrichia sp212, and Ruminococcaceae sp075, while “Combo XXXVI” contained TM7 G1 sp352, H. pittmaniae, Leptotrichia sp221, Leptotrichia sp417, and Ruminococcaceae sp075 (Figure 4B). “Combo XLII” and “Combo XXXVI” had AUCs of 0.943 and 0.938 and positive predictive values of 0.68 and 0.66, respectively. Both of these combinations from log(RA + 1) transformed data had an accuracy of 0.86 (Figure 4B). ROC curves, violin plots, and descriptive statistics of each data type are presented in Figure 4. Using MIND, Haemophilus and Leptotrichia were found to have many interactions in common (Figure 5).
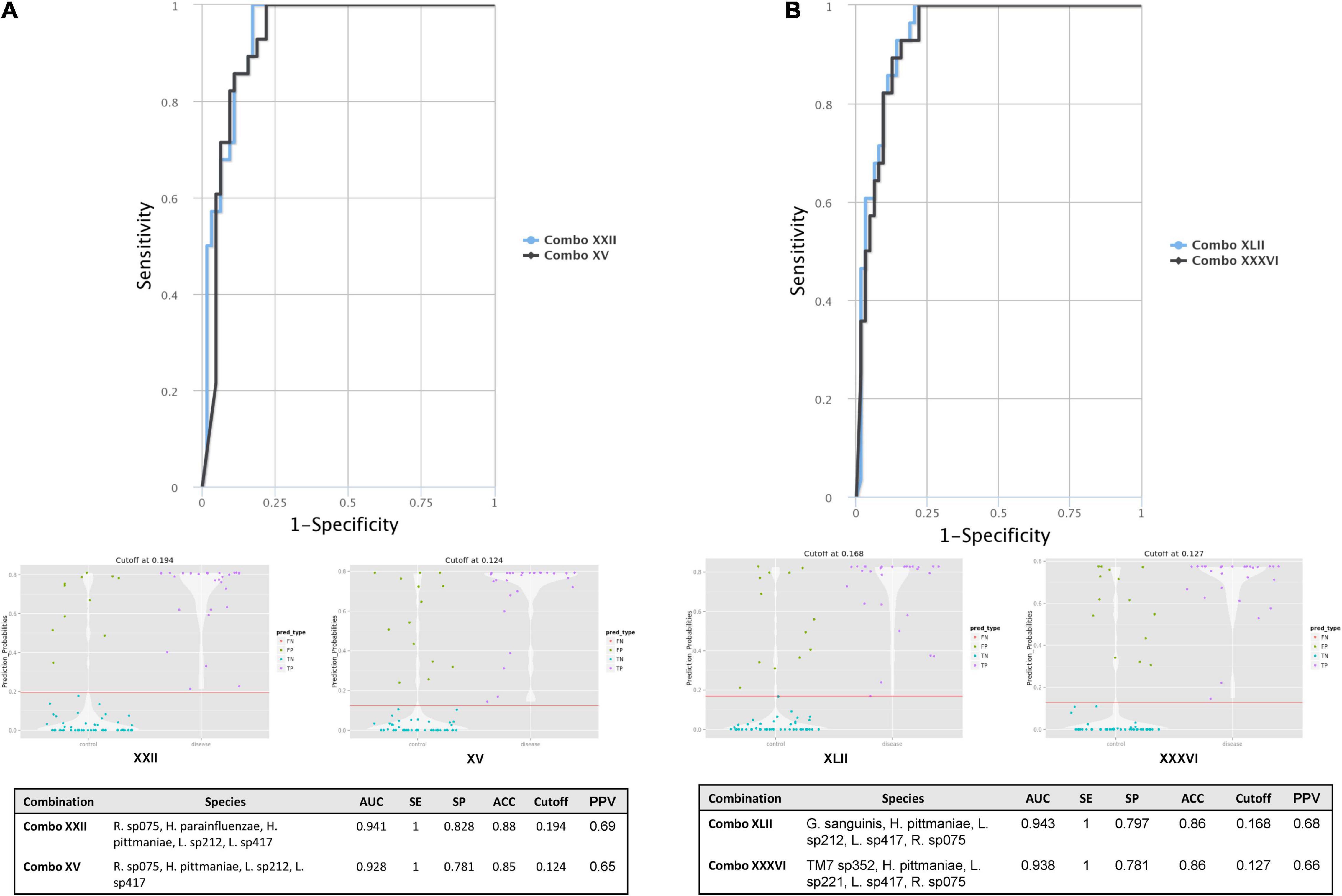
Figure 4. HNC HPV+ vs. HNC HPV– CombiROC analysis using RA and log(RA + 1) transformed abundance data. (A) CombiROC analysis for biomarkers with AUC > 0.75 on RA data. (B) CombiROC analysis for biomarkers with AUC > 0.75 on log(RA + 1) data. Significant (α = 0.05) CombiROC program-generated receiver operator characteristic (ROC) curves for (A) RA data and (B) log(RA + 1) transformed abundance data of candidate microbial biomarkers (AUC > 0.75; n = 10 bacterial species) identified via MedCalc Software for Grp-All HPV+ vs. HPV– group are shown. (A) ROC curves, violin plots, and descriptive statistic summary table for the best two combinations from 10 potential markers using the RA are shown. “Combo XXII” consists of a combination of the markers Ruminococcaceae sp075, Haemophilus parainfluenzae, Haemophilis pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 (ROC curve blue line). “Combo XV” contains markers Ruminococcaceae sp075, H. pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 (ROC curve black line). (B) ROC curves, violin plots, and descriptive statistic summary table for the best two combinations from 10 potential markers using the log(RA + 1) transformed data are shown. “Combo XLII” contains potential markers Gemella sanguinis, H. pittmaniae, Leptotrichia sp212, and Ruminococcaceae sp075 (ROC curve blue line). “Combo XXXVI” consists of species probes TM7 G1 sp352, H. pittmaniae, Leptotrichia sp221, Leptotrichia sp417, and Ruminococcaceae sp075 (ROC curve black line). Cut-off refers to the optimal value or the highest true positive rate that has the lowest false positive rate. FN, false negative; FP, false positive; TN, true negative; TP, true positive; AUC, area under the curve; SE, sensitivity; SP, specificity; ACC, accuracy; PPV, positive predictive value.
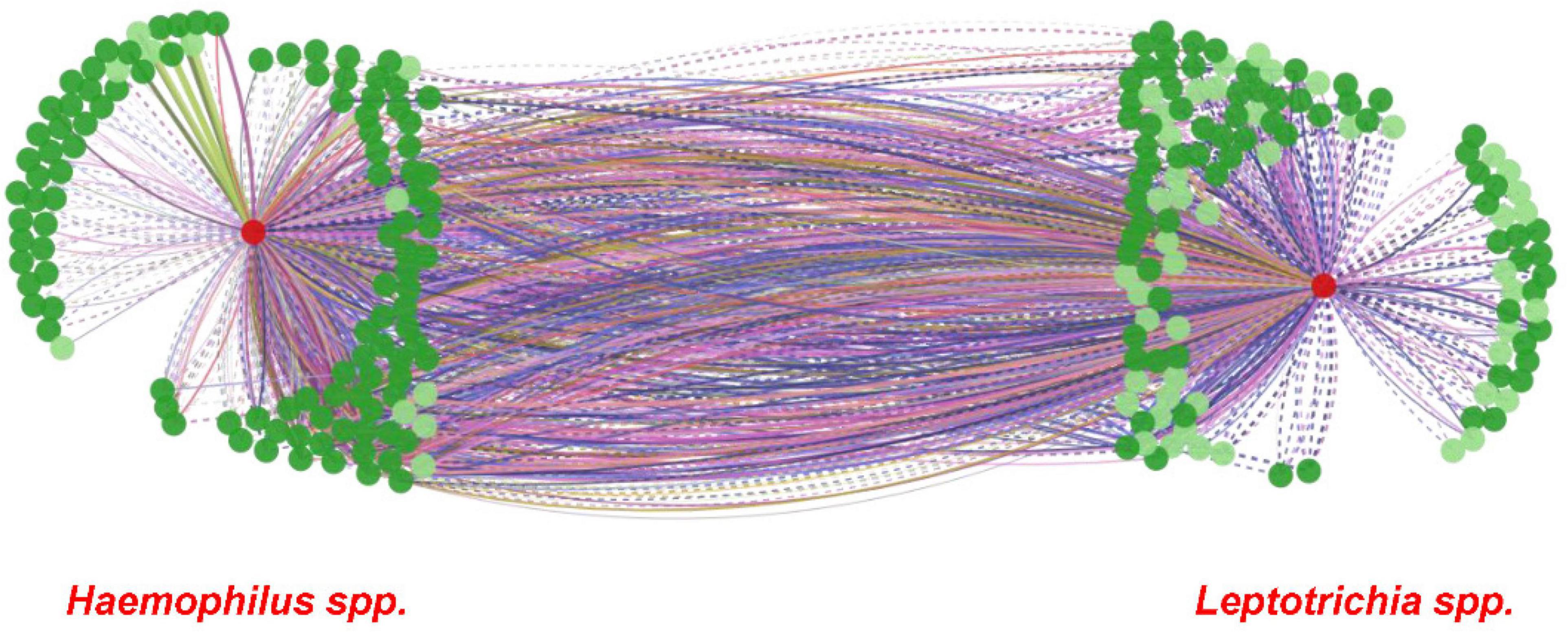
Figure 5. MIND interaction network of Haemophilus and Leptotrichia spp. Microbial Interaction Network Database (MIND; http://www.microbialnet.org/mind_home.html) results, illustrating a network between Haemophilus spp. (left red circle) and Leptotrichia spp. (right red circle) by using MIND default options, are presented. Dark green circles depict genera that interact with Haemophilus spp./Leptotrichia spp., and light green circles depict species that interact with Haemophilus spp./Leptotrichia spp. The lines between Haemophilus spp. and Leptotrichia spp. show interactions in common with other genera and species. The different color lines depict the human tissue sites where interactions have been demonstrated in previous studies (MIND; http://www.microbialnet.org/mind_home.html).
LitSuggest Performance
From the Pythonv3.6.2 data extraction code, 203 PubMed articles were identified for classification from the model matching the search terms “HPV” and “microbiome.” From the classification set of these articles, 36 were determined as positively associated with the search terms “oral microbiome” and “HPV.” LitSuggest program found 171 articles negatively classified. Manual validation of the 36 positively associated articles resulted in 21 articles being discarded. A total of 15 remaining articles were correctly determined as positively associated with HPV and the oral microbiome. Of these articles, three were reviews and 12 were research articles evaluating the HPV status in the context of oral tumor and microbiome relationship in SCC patients with oropharynx, including tonsil specifically, as the primary tumor site (Table 3).
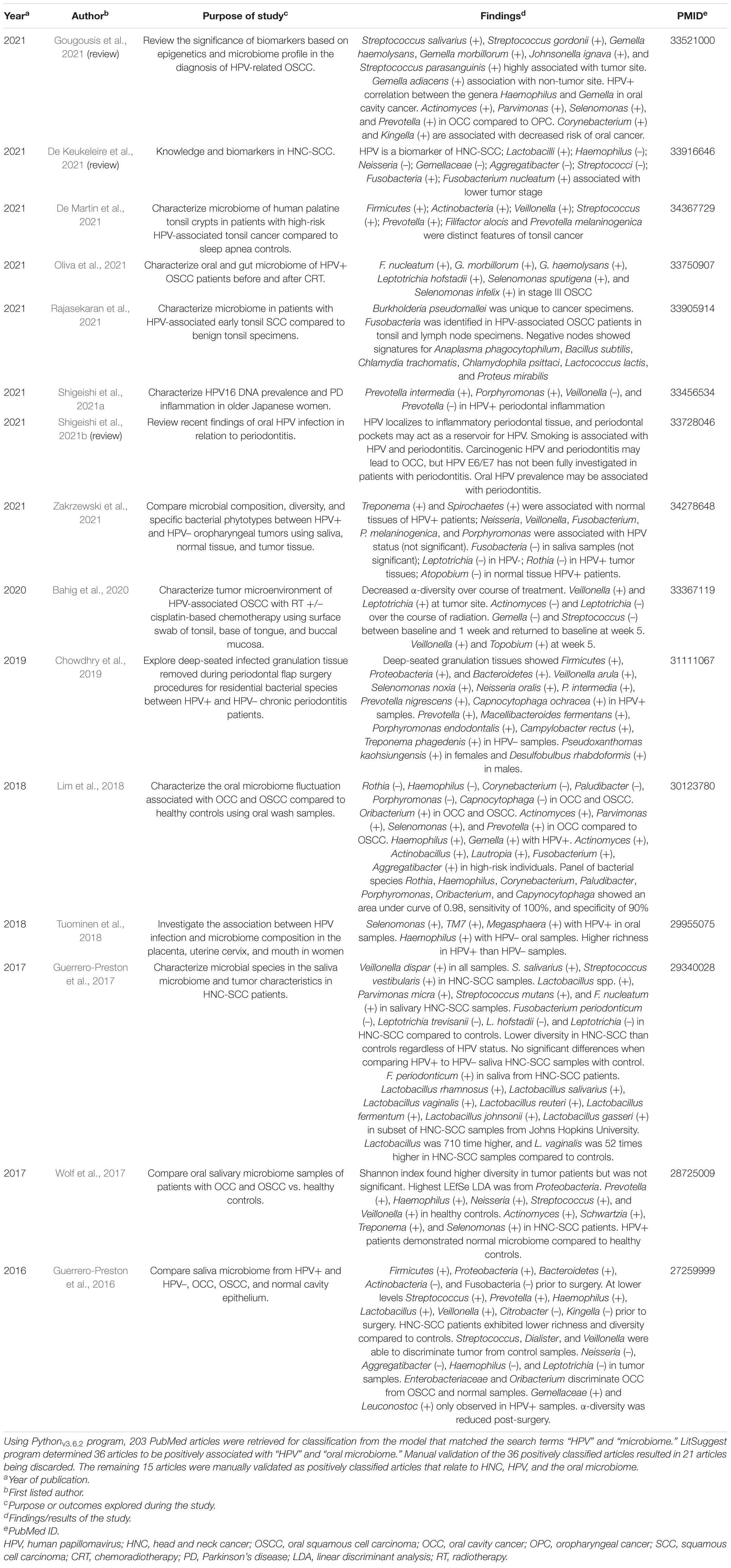
Table 3. LitSuggest positively classified articles (n = 15) involving HPV and the oral microbiome in HNC patients.
Discussion
This is the first study to evaluate the microbial differences in HNC HPV+ patients compared to those of healthy individuals and HNC HPV– patients by means of oral samples including saliva, buccal mucosa, supragingival plaque, and tongue swabs using multivariate analysis. We were able to identify 442 species and 65 genera detected based on HOMINGS sequencing data. We confirmed findings from multiple studies indicating that shifts in microbiome profiles which may be defined as “dysbiosis” occur in HNC patients compared to HC subjects (Guerrero-Preston et al., 2016, 2017; Tuominen et al., 2018).
Furthermore, we were able to establish that HNC HPV+ patients have significantly different microbiome than that of HNC HPV– patients (Supplementary Table 2). While α-diversity was not significantly different between HNC HPV+ and HNC HPV– patients, α-diversity differed between HNC patients and HC subjects (Table 2). Additionally, β-diversity differences were significant for all comparisons in this study except for one out of 30 comparisons (Supplementary Table 2). There was a clear separation between the Grp-All HNC patients and HC subjects (Supplementary Figure 1A). We were also able to determine that although antibiotic treatment within 2 weeks of sampling is a confounding variable, excluding antibiotic-treated HNC patients did not affect the main results, by comparing the microbiome data of HNC HPV+ patients with the data of HNC HPV– patients (i.e., GrpAll and GrpNoAB) (Supplementary Table 1). A visualization of the division of the Grp-All HNC HPV+ vs. HNC HPV– can be seen in Supplementary Figure 1B.
Regarding periodontal disease and dental caries status, our sub-cohort is similar to that of the larger OraRad cohort (Table 1 and Brennan et al., 2021). A study identified by LitSuggest, reviewing findings pertaining to oral HPV infection in relation to periodontitis, suggests periodontal pockets may act as a reservoir for HPV and that oral HPV prevalence may be associated with periodontitis (Shigeishi et al., 2021a). Another recent study characterizing HPV16 DNA prevalence and periodontal disease inflammation in a population of older Japanese women identified an increase of Prevotella intermedia and Porphyromonas and a decrease of Veillonella and Prevotella to be associated with periodontal disease inflammation (Shigeishi et al., 2021b).
Furthermore, a study by Chowdhry et al. (2019) exploring deep-seated infected tissues removed during periodontal flap surgery in chronic periodontitis patients, observed an increased abundance of Veillonella arula, Selenomonas noxia, Neisseria oralis, P. intermedia, Prevotella nigrescens, and Capnocytophaga ochracea in HPV+ samples. Interestingly, in our study we found species of Capnocytophaga, Neisseria, Prevotella, Selenomonas, and Veillonella spp. to represent distinct taxa for HNC HPV+ patients through LEfSe analysis (Figure 3). Species from these genera were also found to be significant using Mann–Whitney U-test (p < 0.05) (Figure 3). While ROC curves were also significant (p < 0.01), we determined none of these species to be excellent biomarkers (Supplementary Table 4). We agree with Shigeishi et al. (2021b) suggesting that sampling methods of the oral microbiome should be carefully selected for periodontal tissue to ensure detection of HPV DNA directly along with the associated periodontal microbiome.
LEfSe analysis showed 43 bacterial species differentiating Grp-All HNC HPV+ from Grp-All HNC HPV– patients (Figure 3). Grp-noAB group analysis confirmed these findings since 24 bacterial species characterizing the HNC HPV+ patients were in common with the species distinctive of Grp-All HNC HPV+ patients (Figure 3). Leptotrichia spp. were the most prominent and significant species in comparisons performed for both Grp-All and Grp-noAB groups, precluding the possibility that antibiotics alone account for differences between HNC HPV+ and HPV– groups. In addition to LEfSe, Mann–Whitney U-tests comparing the RA of HNC HPV+ to HNC HPV– found 5/6 (83%) and 6/6 (100%) Leptotrichia spp. for Grp-All and Grp-noAB, respectively, to be significant (p < 0.05) (Figure 3). For these Leptotrichia spp., the RA was found to be greater in samples of HNC HPV+ compared to HNC HPV– patients for Grp-All and Grp-noAB (data not shown). ROC analysis on log(RA + 1) data further confirmed these findings with five Grp-All Leptotrichia spp. and six Grp-noAB Leptotrichia spp. to have an AUC significantly different from that of 0.5 (p < 0.01) (Supplementary Table 4). By minimizing zero inflation on log(RA + 1) transformed abundance data for the Grp-noAB group, we determined three Leptotrichia spp. (Leptotrichia sp215, sp392, and sp417) to be excellent biomarkers distinguishing HNC HPV+ from HNC HPV– patients, with a sensitivity >95%, a specificity = 100%, and an accuracy >95% (Supplementary Table 4D).
A previous study by Bahig et al. (2020) investigating the tumor microenvironment of HPV-associated SCC patients, determined an increased abundance of Leptotrichia genus in oral samples at baseline which declined over the course of radiation. This study was positively classified in our LitSuggest analysis (Table 3). A study by Oliva et al. (2021) found that Leptotrichia hofstadii was abundant in stage III oropharynx cancer, while Zakrzewski et al. (2021) determined Leptotrichia genus to be decreased in oropharynx HPV- tumor samples.
Surprisingly, other studies have found Leptotrichia spp. to be absent or less abundant at SCC primary tumor sites (Schmidt et al., 2014; Guerrero-Preston et al., 2016, 2017). Leptotrichia spp. have also been investigated for their role in periodontal disease (i.e., gingivitis and periodontitis) (Supplementary Table 3; Eribe and Olsen, 2017). A systematic review by Pérez-Chaparro et al. (2014) described a study by Griffen et al. (2012) that correlated an increased abundance of Leptotrichia genera, Leptotrichia oral taxon 210, Leptotrichia EX103, and Leptotrichia IK040 to be associated with deep pockets of patients with periodontal disease. Accordingly, Leptotrichia species consist of non-motile facultative anaerobic and anaerobic species mostly present in the oral cavity (Eribe and Olsen, 2017).
We also observed that H. pittmaniae had a higher RA in HNC HPV+ than HNC HPV– patients in Grp-All and Grp-noAB (data not shown). This species was identified as a differential feature of HPV+ by LEfSe and was found to have a significant ROC curve using log(RA + 1) data with an AUC of 0.824 (Figure 3 and Supplementary Table 4). Additionally, this species was determined to be an excellent biomarker in the Grp-All and Grp-noAB log(RA + 1) ROC curve analysis with zero inflation minimized as well as a good biomarker when zero inflation was not minimized (Supplementary Table 3). H. pittmaniae was also included in all four multi-marker ROC combinations including Leptotrichia species, suggesting it is a contributor to HPV+ SCC progression (Figure 4). H. pittmaniae has been suggested as a pathogen possibly responsible for respiratory tract infections in patients with lung diseases (Boucher et al., 2012) but has also been identified at significantly higher levels in male children with active caries (Ortiz et al., 2019). While little is known about H. pittmaniae and its role in periodontal disease, the Haemophilus genus was identified in many positively classified studies per our LitSuggest text mining analysis. In a recent study by De Keukeleire et al. (2021) a decrease in Haemophilus was associated with HNC-SCC, confirming findings by Wolf et al. (2017) and Lim et al. (2018). However, studies by Guerrero-Preston et al. (2016) and Gougousis et al. (2021) found the opposite to be true. In our study, we were able to verify findings by Lim et al. (2018) and Tuominen et al. (2018) that an increase in abundance of Haemophilus in the oral cavity is associated with HNC-SCC HPV+ samples. Altogether, a microbiome metagenome/metatranscriptome survey focused on Haemophilus and Leptotrichia at the species and strain levels could provide reliable biomarker signatures with clinical implications for HNC HPV+ patients in the future.
MIND analysis found many microbial interactions between the genera Leptotrichia and Haemophilus connected through many species and genera (Figure 5). These genera have also been shown as two of the nine taxa that facilitate structures in oral plaque and intermingle at the micron scale (Mark Welch et al., 2016). The study by Mark Welch et al. (2016) also suggests that Corynebacterium forms long structures with Streptococcus and Porphyromonas in direct contact. Streptococcus creates an environment rich in CO2, lactate, and acetate, facilitating the contact with Haemophilus, Aggregatibacter, and Neisseriaceae (Mark Welch et al., 2016). With the exception of Neisseriaceae, these species are likely essential to aerobic metabolism, allowing Fusobacterium and Leptotrichia to thrive as key participants in the metabolism of sugars, producing lactic acid (Mark Welch et al., 2016). It is suggested that this metabolism is involved in degradation of oral tissues, possibly leading to dental caries and/or periodontal disease (Eribe and Olsen, 2017).
Text mining used in this study suggests a lack of information involving HPV and the oral microbiome of HNC patients. Of 203 HPV microbiome articles, only 15 were verified as relevant to HPV and the oral microbiome in SCC. Only two studies identified by LitSuggest related oral SCC with HPV and periodontitis (Chowdhry et al., 2019; Shigeishi et al., 2021a). Furthermore, few studies investigated multiple primary tumor sites in SCC patients. Most identified studies in our analysis focused on investigating oropharyngeal SCC (Guerrero-Preston et al., 2016; Wolf et al., 2017; Lim et al., 2018; Bahig et al., 2020; Gougousis et al., 2021; Oliva et al., 2021; Zakrzewski et al., 2021). Only two positively classified articles were found to characterize the microbiome in the tonsil of SCC patients (Table 3; De Martin et al., 2021; Rajasekaran et al., 2021). In the future, more studies on HNC-SCC HPV+ patients using larger patient cohorts will be required to determine HNC risk in relation to the oral microbiome and HPV status.
Limitations
While this study was able to show the significance of microbial composition in HNC-SCC HPV+ patients compared to HNC-SCC HPV– patients, we were unable to account for the immune status of the patients. Furthermore, our patient cohort was relatively small, and our design was not optimally balanced due to the various primary cancer sites in our patient cohort. In addition, many factors not addressed in this study may affect HNC progression, such as genetics, oral hygiene practices, and periodontal treatment. However, our main conclusion remains pertinent, in that the species identified as multi-marker combinations, i.e., H. pittmaniae and Leptotrichia spp., increase in HNC-SCC HPV+ patients regardless of the primary cancer site.
Data Availability Statement
The data presented in the study are provided as Supplementary Material and have been deposited in a public GitHub repository. The data can be found here: https://github.com/mbeckm01/HPV_HNC.git.
Ethics Statement
The studies involving human participants were reviewed and approved by the Atrium Health Institutional Review Board and University of Connecticut Health Institutional Review Board. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
J-LM and FB conceived this microbiome study. MTB and RL had previously established the cited clinical outcomes study “OraRad” and provided clinical insights for this study. J-LM directed the statistical analyses implemented and verified by MFB and HL. MFB, HL, J-LM, and FB contributed to the writing of the manuscript, the overall analysis, and biological interpretation. All authors participated in the revisions of the manuscript and interpretation of the results, gave their final approval, and agreed to be accountable for all aspects of the work.
Funding
Funding was obtained from the Carolinas HealthCare Foundation Research Fund-Atrium Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to James Davis, Cynthia Rybczyk, and Jennifer Plascencia for collection of patient samples and the clinical data; Becca Mitchell for providing periodontal status data; Darla Morton for initial steps of sample processing; and Alexis Kokaras for conducting next-generation sequencing and providing the HOMINGS ProbeSeq data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.794546/full#supplementary-material
Footnotes
- ^ https://www.cancer.gov/about-cancer/causes-prevention/risk/infectious-agents/hpv-and-cancer
- ^ http://combiroc.eu
- ^ https://www.ncbi.nlm.nih.gov/research/litsuggest
- ^ https://github.com/mbeckm01/HPV_HNC.git
References
Allot, A., Lee, K., Chen, Q., Luo, L., and Lu, Z. (2021). LitSuggest: a web-based system for literature recommendation and curation using machine learning. Nucleic Acids Res. 49, W352–W358. doi: 10.1093/nar/gkab326
Bahig, H., Fuller, C. D., Mitra, A., Yoshida-Court, K., Solley, T., Ping Ng, S., et al. (2020). Longitudinal characterization of the tumoral microbiome during radiotherapy in HPV-associated oropharynx cancer. Clin. Transl. Radiat. Oncol. 26, 98–103. doi: 10.1016/j.ctro.2020.11.007
Blitzer, G. C., Smith, M. A., Harris, S. L., and Kimple, R. J. (2014). Review of the clinical and biologic aspects of human papillomavirus-positive squamous cell carcinomas of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 88, 761–770. doi: 10.1016/j.ijrobp.2013.08.029
Boucher, M. B., Bedotto, M., Couderc, C., Gomez, C., Reynaud-Gaubert, M., and Drancourt, M. (2012). Haemophilus pittmaniae respiratory infection in a patient with siderosis: a case report. J. Med. Case Rep. 6:120. doi: 10.1186/1752-1947-6-120
Božinović, K., Sabol, I., Dediol, E., Milutin Gašperov, N., Manojlović, S., Vojtechova, Z., et al. (2019). Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci. Rep. 9:2306. doi: 10.1038/s41598-019-38797-z
Brennan, M. T., Treister, N. S., Sollecito, T. P., Schmidt, B. L., Patton, L. L., Mohammadi, K., et al. (2017). Dental disease before radiotherapy in patients with head and neck cancer: clinical registry of dental outcomes in head and neck cancer patients. J. Am. Dent. Assoc. 148, 868–877. doi: 10.1016/j.adaj.2017.09.011
Brennan, M. T., Treister, N. S., Sollecito, T. P., Schmidt, B. L., Patton, L. L., Yang, Y., et al. (2021). Epidemiologic factors in patients with advanced head and neck cancer treated with radiation therapy. Head Neck. 43, 164–172. doi: 10.1002/hed.26468
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Carvalho, A. L., Nishimoto, I. N., Califano, J. A., and Kowalski, L. P. (2005). Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int. J. Cancer 114, 806–816. doi: 10.1002/ijc.20740
Chowdhry, R., Singh, N., Sahu, D. K., Tripathi, R. K., Mishra, A., Singh, A., et al. (2019). Dysbiosis and variation in predicted functions of the granulation tissue microbiome in HPV positive and negative severe chronic periodontitis. Biomed. Res. Int. 2019:8163591. doi: 10.1155/2019/8163591
Clarke, K. R., and Gorley, R. N. (2006). PRIMER v7: User Manual/Tutorial (PRIMER v7). Plymouth: PRIMER-E, 192.
Crook, T., Tidy, J. A., and Vousden, K. H. (1991). Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67, 547–556. doi: 10.1016/0092-8674(91)90529-8
De Keukeleire, S. J., Vermassen, T., Hilgert, E., Creytens, D., Ferdinande, L., and Rottey, S. (2021). Immuno-oncological biomarkers for squamous cell cancer of the head and neck: current state of the art and future perspectives. Cancers (Basel) 13:1714. doi: 10.3390/cancers13071714
De Martin, A., Lütge, M., Stanossek, Y., Engetschwiler, C., Cupovic, J., Brown, K., et al. (2021). Distinct microbial communities colonize tonsillar squamous cell carcinoma. Oncoimmunology 10:1945202. doi: 10.1080/2162402X.2021.1945202
Eribe, E. R. K., and Olsen, I. (2017). Leptotrichia species in human infections II. J. Oral. Microbiol. 9:1368848. doi: 10.1080/20002297.2017.1368848
Fakhry, C., Krapcho, M., Eisele, D. W., and D’Souza, G. (2018). Head and neck squamous cell cancers in the United States are rare and the risk now is higher among white individuals compared with black individuals. Cancer 124, 2125–2133. doi: 10.1002/cncr.31322
Ganci, F., Sacconi, A., Manciocco, V., Spriano, G., Fontemaggi, G., Carlini, P., et al. (2015). “Radioresistance in head and neck squamous cell carcinoma: possible molecular markers for local recurrence and new putative therapeutic strategies,” in Contemporary Issues in Head and Neck Cancer Management, ed. L. G. Marcu (London: IntechOpen), 3–34. doi: 10.5772/60081
Gasparoni, L. M., Alves, F. A., Holzhausen, M., Pannuti, C. M., and Serpa, M. S. (2021). Periodontitis as a risk factor for head and neck cancer. Med. Oral. Patol. Oral. Cir. Bucal. 26, e430–e436. doi: 10.4317/medoral.24270
Gholizadeh, P., Eslami, H., Yousefi, M., Asgharzadeh, M., Aghazadeh, M., and Kafil, H. S. (2016). Role of oral microbiome on oral cancers, a review. Biomed. Pharmacother. 84, 552–558. doi: 10.1016/j.biopha.2016.09.082
Gougousis, S., Mouchtaropoulou, E., Besli, I., Vrochidis, P., Skoumpas, I., and Constantinidis, I. (2021). HPV-related oropharyngeal cancer and biomarkers based on epigenetics and microbiome profile. Front. Cell Dev. Biol. 8:625330. doi: 10.3389/fcell.2020.625330
Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185. doi: 10.1038/ismej.2011.191
Guerrero-Preston, R., Godoy-Vitorino, F., Jedlicka, A., Rodríguez-Hilario, A., González, H., Bondy, J., et al. (2016). 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget 7, 51320–51334. doi: 10.18632/oncotarget.9710
Guerrero-Preston, R., White, J. R., Godoy-Vitorino, F., Rodríguez-Hilario, A., Navarro, K., González, H., et al. (2017). High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget 8, 110931–110948. doi: 10.18632/oncotarget.20677
Hayes, R. B., Ahn, J., Fan, X., Peters, B. A., Ma, Y., Yang, L., et al. (2018). Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 4, 358–365. doi: 10.1001/jamaoncol.2017.4777
Jalili, V., Afgan, E., Gu, Q., Clements, D., Blankenberg, D., Goecks, J., et al. (2020). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 48, W395–W402. doi: 10.1093/nar/gkaa554
Jemal, A., Siegel, R., Ward, E., Hao, Y., Xu, J., Murray, T., et al. (2008). Cancer statistics, 2008. Cancer J. Clin. 58, 71–96. doi: 10.3322/CA.2007.0010
Kudo, Y., Tada, H., Fujiwara, N., Tada, Y., Tsunematsu, T., Miyake, Y., et al. (2016). Oral environment and cancer. Genes Environ. 38:13. doi: 10.1186/s41021-016-0042-z
Kumarasamy, C., Madhav, M. R., Sabarimurugan, S., Krishnan, S., Baxi, S., Gupta, A., et al. (2019). Prognostic value of miRNAs in head and neck cancers: a comprehensive systematic and meta-analysis. Cells 8:772. doi: 10.3390/cells8080772
Lalla, R. V., Treister, N., Sollecito, T., Schmidt, B., Patton, L. L., Mohammadi, K., et al. (2017). Oral complications at 6 months after radiation therapy for head and neck cancer. Oral Dis. 23, 1134–1143. doi: 10.1111/odi.12710
Lim, Y., Fukuma, N., Totsika, M., Kenny, L., Morrison, M., and Punyadeera, C. (2018). The performance of an oral microbiome biomarker panel in predicting oral cavity and oropharyngeal cancers. Front. Cell Infect. Microbiol. 8:267. doi: 10.3389/fcimb.2018.00267
Mahal, B. A., Catalano, P. J., Haddad, R. I., Hanna, G. J., Kass, J. I., Schoenfeld, J. D., et al. (2019). Incidence and demographic burden of HPV-associated oropharyngeal head and neck cancers in the United States. Cancer Epidemiol. Biomark. Prev. 28, 1660–1667. doi: 10.1158/1055-9965.EPI-19-0038
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800. doi: 10.1073/pnas.1522149113
Mazzara, S., Rossi, R. L., Grifantini, R., Donizetti, S., Abrignani, S., and Bombaci, M. (2017). CombiROC: an interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 7:45477. doi: 10.1038/srep45477
Michaud, D. S., Fu, Z., Shi, J., and Chung, M. (2017). Periodontal disease, tooth loss, and cancer risk. Epidemiol. Rev. 39, 49–58. doi: 10.1093/epirev/mxx006
Microbial Interaction Network Database (2019). THAMBAN G. Available online at: http://www.microbialnet.org/mind_home.html (accessed September 9, 2021).
Mohammed, H., Varoni, E. M., Cochis, A., Cordaro, M., Gallenzi, P., Patini, R., et al. (2018). Oral dysbiosis in pancreatic cancer and liver cirrhosis: a review of the literature. Biomedicines 6:115. doi: 10.3390/biomedicines6040115
Mougeot, J. C., Beckman, M. F., Langdon, H. C., Brennan, M. T., and Bahrani Mougeot, F. (2020). Oral microbiome signatures in hematological cancers reveal predominance of actinomyces and rothia species. J. Clin. Med. 9:4068. doi: 10.3390/jcm9124068
Mougeot, J. C., Stevens, C. B., Almon, K. G., Paster, B. J., Lalla, R. V., Brennan, M. T., et al. (2019). Caries-associated oral microbiome in head and neck cancer radiation patients: a longitudinal study. J. Oral. Microbiol. 11:1586421. doi: 10.1080/20002297.2019.1586421
Mougeot, J. L., Stevens, C. B., Cotton, S. L., Morton, D. S., Krishnan, K., Brennan, M. T., et al. (2016). Concordance of HOMIM and HOMINGS technologies in the microbiome analysis of clinical samples. J. Oral. Microbiol. 8:30379. doi: 10.3402/jom.v8.30379
National Center for Biotechnology Information (1988). THAMBAN G. Available online at: https://www.ncbi.nlm.nih.gov/home/develop/api/ (accessed August 13, 2021).
Oliva, M., Schneeberger, P. H. H., Rey, V., Cho, M., Taylor, R., Hansen, A. R., et al. (2021). Transitions in oral and gut microbiome of HPV+ oropharyngeal squamous cell carcinoma following definitive chemoradiotherapy (ROMA LA-OPSCC study). Br. J. Cancer 124, 1543–1551. doi: 10.1038/s41416-020-01253-1
Ortiz, S., Herrman, E., Lyashenko, C., Purcell, A., Raslan, K., Khor, B., et al. (2019). Sex-specific differences in the salivary microbiome of caries-active children. J. Oral. Microbiol. 11:1653124. doi: 10.1080/20002297.2019.1653124
Pal, A., and Kundu, R. (2020). Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front. Microbiol. 10:3116. doi: 10.3389/fmicb.2019.03116I
Pérez-Chaparro, P. J., Gonçalves, C., Figueiredo, L. C., Faveri, M., Lobão, E., Tamashiro, N., et al. (2014). Newly identified pathogens associated with periodontitis: a systematic review. J. Dent. Res. 93, 846–858. doi: 10.1177/0022034514542468
PubMed (1997). THAMBAN G. Available online at: https://pubmed.ncbi.nlm.nih.gov/ (accessed August 13, 2021).
Rafiei, M., Kiani, F., Sayehmiri, F., Sayehmiri, K., Sheikhi, A., and Zamanian Azodi, M. (2017). Study of Porphyromonas gingivalis in periodontal diseases: a systematic review and meta-analysis. Med. J. Islam. Repub. Iran. 31:62. doi: 10.14196/mjiri.31.62
Rajasekaran, K., Carey, R. M., Lin, X., Seckar, T. D., Wei, Z., Chorath, K., et al. (2021). The microbiome of HPV-positive tonsil squamous cell carcinoma and neck metastasis. Oral. Oncol. 117:105305. doi: 10.1016/j.oraloncology.2021.105305
Ray, P., Le Manach, Y., Riou, B., and Houle, T. T. (2010). Statistical evaluation of a biomarker. Anesthesiology 112, 1023–1040. doi: 10.1097/ALN.0b013e3181d47604
Schmidt, B. L., Kuczynski, J., Bhattacharya, A., Huey, B., Corby, P. M., Queiroz, E. L., et al. (2014). Changes in abundance of oral microbiota associated with oral cancer. PLoS One 9:e98741. doi: 10.1371/journal.pone.0098741
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shigeishi, H., Su, C. Y., Kaneyasu, Y., Matsumura, M., Nakamura, M., Ishikawa, M., et al. (2021a). Association of oral HPV16 infection with periodontal inflammation and the oral microbiome in older women. Exp. Ther. Med. 21:167. doi: 10.3892/etm.2020.9598
Shigeishi, H., Sugiyama, M., and Ohta, K. (2021b). Relationship between the prevalence of oral human papillomavirus DNA and periodontal disease (Review). Biomed. Rep. 14:40.
Tezal, M., Sullivan Nasca, M., Stoler, D. L., Melendy, T., Hyland, A., Smaldino, P. J., et al. (2009). Chronic periodontitis-human papillomavirus synergy in base of tongue cancers. Arch. Otolaryngol. Head. Neck Surg. 135, 391–396. doi: 10.1001/archoto.2009.6
Tumban, E. (2019). A current update on human papillomavirus-associated head and neck cancers. Viruses 11:922. doi: 10.3390/v11100922
Tuominen, H., Rautava, S., Syrjänen, S., Collado, M. C., and Rautava, J. (2018). HPV infection and bacterial microbiota in the placenta, uterine cervix and oral mucosa. Sci. Rep. 8:9787. doi: 10.1038/s41598-018-27980-3
van Gysen, K., Stevens, M., Guo, L., Jayamanne, D., Veivers, D., Wignall, A., et al. (2019). Validation of the 8th edition UICC/AJCC TNM staging system for HPV associated oropharyngeal cancer patients managed with contemporary chemo-radiotherapy. BMC Cancer 19:674. doi: 10.1186/s12885-019-5894-8
Venuti, A., Paolini, F., Nasir, L., Corteggio, A., Roperto, S., Campo, M. S., et al. (2011). Papillomavirus E5: the smallest oncoprotein with many functions. Mol. Cancer 10:140. doi: 10.1186/1476-4598-10-140
Wolf, A., Moissl-Eichinger, C., Perras, A., Koskinen, K., Tomazic, P. V., and Thurnher, D. (2017). The salivary microbiome as an indicator of carcinogenesis in patients with oropharyngeal squamous cell carcinoma: a pilot study. Sci. Rep. 7:5867. doi: 10.1038/s41598-017-06361-2
Wu, J., Xu, S., Xiang, C., Cao, Q., Li, Q., Huang, J., et al. (2018). Tongue coating microbiota community and risk effect on gastric cancer. J. Cancer 9, 4039–4048. doi: 10.7150/jca.25280
Yim, E. K., and Park, J. S. (2005). The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res. Treat. 37, 319–324. doi: 10.4143/crt.2005.37.6.319
Zakrzewski, M., Gannon, O. M., Panizza, B. J., Saunders, N. A., and Antonsson, A. (2021). Human papillomavirus infection and tumor microenvironment are associated with the microbiota in patients with oropharyngeal cancers-pilot study. Head Neck 43, 3324–3330. doi: 10.1002/hed.26821
Keywords: head and neck cancer, HPV, oral microbiome, next generation sequencing, Leptotrichia spp.
Citation: Mougeot J-LC, Beckman MF, Langdon HC, Lalla RV, Brennan MT and Bahrani Mougeot FK (2022) Haemophilus pittmaniae and Leptotrichia spp. Constitute a Multi-Marker Signature in a Cohort of Human Papillomavirus-Positive Head and Neck Cancer Patients. Front. Microbiol. 12:794546. doi: 10.3389/fmicb.2021.794546
Received: 13 October 2021; Accepted: 10 November 2021;
Published: 18 January 2022.
Edited by:
Florence Carrouel, Université Claude Bernard Lyon 1, FranceReviewed by:
Divyashri Baraniya, Temple University, United StatesStéphane Viennot, Université Claude Bernard Lyon 1, France
Copyright © 2022 Mougeot, Beckman, Langdon, Lalla, Brennan and Bahrani Mougeot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Luc C. Mougeot, amVhbi1sdWMubW91Z2VvdEBhdHJpdW1oZWFsdGgub3Jn; Farah K. Bahrani Mougeot, ZmFyYWgubW91Z2VvdEBhdHJpdW1oZWFsdGgub3Jn
 Jean-Luc C. Mougeot
Jean-Luc C. Mougeot Micaela F. Beckman1
Micaela F. Beckman1 Michael T. Brennan
Michael T. Brennan Farah K. Bahrani Mougeot
Farah K. Bahrani Mougeot