- 1Department of Microbiology and Infectious Diseases, Nara Medical University, Kashihara, Japan
- 2Division of Central Clinical Laboratory, Iwate Medical University Hospital, Yahaba, Japan
- 3Department of Bacteriology, BML Inc., Kawagoe, Japan
- 4Hamamatsu Pharmaceutical Association, Hamamatsu, Japan
- 5Department of Laboratory Medicine, Iwate Medical University School of Medicine, Yahaba, Japan
Various carbapenemases have been identified in the Enterobacteriaceae. However, the induction and corresponding regulator genes of carbapenemase NmcA has rarely been detected in the Enterobacter cloacae complex (ECC). The NmcA-positive isolate ECC NR1491 was first detected in Japan in 2013. It was characterized and its induction system elucidated by evaluating its associated regulator genes nmcR, ampD, and ampR. The isolate was highly resistant to all β-lactams except for third generation cephalosporins (3GC). Whole-genome analysis revealed that blaNmcA was located on a novel 29-kb putatively mobile element called EludIMEX-1 inserted into the chromosome. The inducibility of β-lactamase activity by various agents was evaluated. Cefoxitin was confirmed as a strong concentration-independent β-lactamase inducer. In contrast, carbapenems induced β-lactamase in a concentration-dependent manner. All selected 3GC-mutants harboring substitutions on ampD (as ampR and nmcR were unchanged) were highly resistant to 3GC. The ampD mutant strain NR3901 presented with a 700 × increase in β-lactamase activity with or without induction. Similar upregulation was also observed for ampC and nmcA. NR1491 (pKU412) was obtained by transforming the ampR mutant (135Asn) clone plasmid whose expression increased by ∼100×. Like NR3901, it was highly resistant to 3GC. Overexpression of ampC, rather than nmcA, may have accounted for the higher MIC in NR1491. The ampR mutant repressed nmcA despite induction and it remains unclear how it stimulates nmcA transcription via induction. Future experiments should analyze the roles of nmcR mutant strains.
Introduction
The Enterobacter cloacae complex (ECC) have become clinically significant opportunistic bacteria and are now common nosocomial pathogens causing pneumonia, urinary tract infections, and septicemia (Davin-Regli and Pages, 2015). Six Enterobacter species are assigned to the ECC: E. cloacae, E. asburiae, E. hormaechei, E. kobei, E. ludwigii, and E. nimipressuralis (Mezzatesta et al., 2012).
Multidrug resistance (MDR) has been observed for the last-resort carbapenems and has led to an increased global interest in Enterobacteriaceae in general and carbapenem-resistant ECC, in particular (Annavajhala et al., 2019). ECC are innately resistant to penicillins, first- and second-generation cephalosporins, and cephamycin due to the chromosomally encoded AmpC β-lactamase genes (serine β-lactamase, Ambler class C). AmpC β-lactamase expression is low but inducible in response to β-lactam exposure and is closely linked to a peptidoglycan recycling system, with the β-lactams imipenem, cefoxitin, and clavulanic acid strong ampC inducers (Jacoby, 2009). Regulation of AmpC β-lactamase expression is complex and involves AmpR (a transcriptional regulator of the LysR family), AmpD (a cytosolic amidase), and AmpG (a transmembrane permease) (Guerin et al., 2015). AmpR usually represses ampC in the absence of β-lactam inducers, whereas mutations at specific sites in AmpR derepresses AmpC synthesis and results in constitutive AmpC β-lactamase overexpression. Asp135Asn AmpR substitution is correlated with substantial increases in β-lactamase activity in several Gram-negative organisms including ECC, Citrobacter freundii, and Pseudomonas aeruginosa (Kuga et al., 2000; Caille et al., 2014; Nakano et al., 2017). Mutations that inactivate AmpD permanently induce and increase muropeptide concentrations in the cytoplasm and change the conformation of AmpR so that it becomes a transcriptional activator (Kuga et al., 2000). Specifically, AmpR mutations require site-specific substitution to induce AmpC β-lactamase overexpression whereas AmpD mutations need loss-of-function point mutations (missense mutation) or disruption of the protein carboxy terminus, nonsense mutations, frameshifts, and truncations (Schmidtke and Hanson, 2006). Among the ECC clinical isolates, high-level resistance to third generation cephalosporins (3GC) is caused by constitutive ampC overexpression mainly from ampD mutations and, more rarely, from ampR mutations (Kaneko et al., 2005; Guerin et al., 2015).
Carbapenem resistance in ECC is conferred either through constitutive AmpC β-lactamase overexpression combined with defective outer membrane (porin) permeability or via the acquisition of carbapenemase genes (Annavajhala et al., 2019), with the latter scenario being more common. Carbapenemases hydrolyze most β-lactams including carbapenems and are classified as serine β-lactamases (Ambler class A; KPC type and D; OXA-48 type) or metallo-β-lactamases (Ambler class B; IMP type, VIM type, and NDM type) (Diene and Rolain, 2014; Nakano et al., 2014; Ando et al., 2018). The distributions of these enzymes differ with geographical location: the KPC type occurs in the United States, NDM in the Indian subcontinent, and IMP in Japan (Chavda et al., 2016; Aoki et al., 2018; Peirano et al., 2018). Chromosomally encoded carbapenemase NmcA (Ambler class A) has been sporadically detected in ECC (Walther-Rasmussen and Hoiby, 2007).
NmcA was originally detected in the carbapenem-resistant E. cloacae strain NOR-1 isolated in France in 1990 (Nordmann et al., 1993). NmcA has occasionally been detected in E. cloacae, E. asburiae, and E. ludwigii from Europe, North America, and South America (Pottumarthy et al., 2003; Radice et al., 2004; Antonelli et al., 2015; Boyd et al., 2017). A recent study revealed that blaNmcA is associated with a novel 29-kb putative Xer-dependent integrative mobile element (EludIMEX-1) inserted into the ECC chromosome (Antonelli et al., 2015). This enzyme hydrolyses different β-lactam agents except for 3GC and has particularly high hydrolytic activity against carbapenems (Nordmann et al., 1993; Mariotte-Boyer et al., 1996). The inducibility of NmcA is similar to AmpC β-lactamase (Pottumarthy et al., 2003), where the LysR family transcriptional regulator gene nmcR upstream of blaNmcA regulates nmcA in the same manner as the ampR–ampC regulatory system does for AmpC. Additionally, AmpD co-regulates nmcA (Naas et al., 2001).
Here, we describe the characteristics of an nmcA-positive ECC isolate first observed in Japan. We also elucidate the nmcA induction system by evaluating nmcA expression in ampD and ampR mutant strains.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
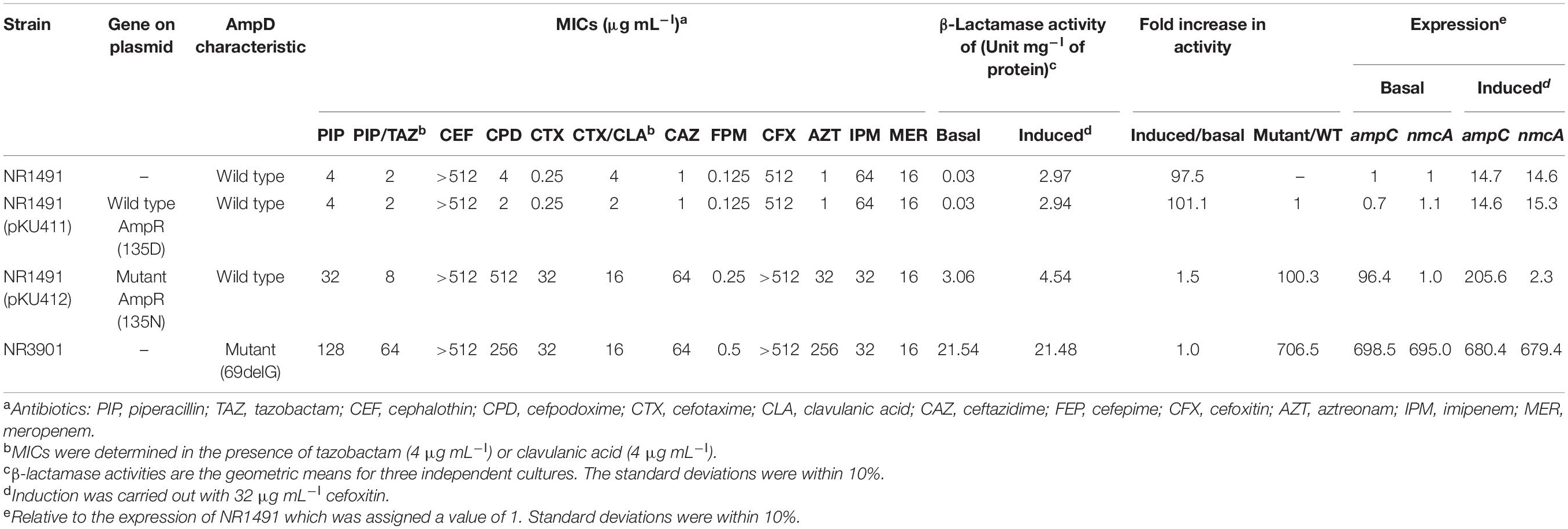
The carbapenem-resistant ECC strain NR1491 was isolated from the urine of a patient in a Japanese hospital in 2013. The species was identified as E. cloacae by MicroScan WalkAway plus (Beckman Coulter, Inc., Brea, CA, United States). To evaluate the effects of ampR mutation on antimicrobial susceptibility and β-lactamase expression, ampR clone plasmids were constructed and used to transform NR1491. The ampR clone plasmids (pKU411 and pKU412) used in this study were already previously constructed (Kuga et al., 2000). An in vitro ceftazidime-resistant mutant strain NR3901 was isolated from NR1491. Characteristics of the bacterial strains and plasmids used in the present study are listed in Table 1.
The minimum inhibitory concentrations (MICs) of the various antimicrobial agents were determined for each strain by the agar dilution method according to CLSI guidelines (CLSI, 2018).
Whole-Genome Sequencing and Analysis
The genomic DNA of NR1491 was prepared with a Qiagen Genomic-tip 500/G kit (Qiagen, Hilden, Germany) and subjected to whole-genome sequencing on the MiSeq X10 platform (Illumina, San Diego, CA, United States). Reads were trimmed in Trimmomatic and assembled to contigs with the SPAdes v. 3.8.1 genome assembler in caution mode (Bankevich et al., 2012).
Species were precisely identified based on their average nucleotide identity (ANI) and in silico DNA–DNA hybridization between strain NR1491 (GenBank accession no. BKZO00000000.1), the E. cloacae type strain ATCC 13047 (GenBank accession no. MTFV00000000.1), the E. ludwigii type strain EN-119 (GenBank accession no. JTLO00000000.1), and the E. ludwigii type strain AOUC-8/14 (GenBank accession no. LGIV00000000.1). Earlier studies recommended ANI of ∼95–96% as a species demarcation cutoff (Goris et al., 2007; Chun and Rainey, 2014).
Antimicrobial resistance genes were identified in the genome sequence with the ResFinder database1 using thresholds of 90% identity and 60% minimum length. β-lactamase genes including carbapenemases and extended-spectrum β-lactamases (ESBLs) were also assessed by PCR. PCR detection of carbapenemases (blaIMP, blaVIM, blaKPC, blaOXA–48–like, blaNDM, blaGES, blaIMI/NmcA, and blaSME) (Poirel et al., 2011; Hong et al., 2012; Nakano et al., 2018) and ESBLs (blaTEM, blaSHV, and blaCTX–M) (Dallenne et al., 2010) were performed as previously described. The sequence surrounding blaNmcA, a carbapenemase-encoding gene, was elucidated by PCR and Sanger sequencing to close the gaps between the contigs. Sequence alignment and analysis were performed with BLAST2 at NCBI (National Centre for Biotechnology Information, Bethesda, MD, United States). Multilocus sequence typing (MLST) of the E. cloacae isolates was performed as previously described (Miyoshi-Akiyama et al., 2013). Sequence types were assigned at the PubMLST database.3 The presence of mobile genetic elements was investigated using the MobileElementFinder (Johansson et al., 2021) and INTEGRALL (Moura et al., 2009). The plasmid content was assessed using PlasmidFinder (Carattoli et al., 2014).
Selection of Third-Generation Cephalosporin-Resistant Mutants and Detection of Sequence Alterations
Third-generation cephalosporin-resistant mutants were obtained by plating ∼109 CFU mL–l late-logarithmic-phase NR1491 grown in Luria-Bertani (LB) broth and on LB agar plates containing ceftazidime or cefotaxime at 2×, 4×, 8×, 16×, and 32× MIC. The mutation frequencies were determined by dividing the colony density in CFU mL–l on LB agar plates containing the antibiotic agents by the total colony density in CFU mL–l.
The DNA sequences of the selected mutants were determined by Sanger sequencing of nmcR, ampR, and ampD amplicons. The primers used are listed in Supplementary Table 1 (Radice et al., 2004). The nucleotides and amino acids of the selected mutants were compared with those of E. ludwigii NR1491 and EN-119.
β-Lactamase Induction Assays
β-lactamase activity was measured in terms of the protein content in the extract and compared among cultures in 50 mM phosphate buffer (pH 7.0) at 30°C by spectrophotometry as previously described (Nakano et al., 2004). The protein concentrations were determined by the Bradford assay (Bradford, 1976). One unit of β-lactamase activity was defined as the amount of β-lactamase hydrolyzing 1 μmol cephalothin in 1 min at 30°C. The β-lactamase induction assays were performed by subjecting mid-logarithmic phase bacteria in Mueller-Hinton broth to β-lactams at 1/16×, 1/8×, 1/4×, 1/2×, and 1× MICs for 2 h (Kuga et al., 2000). The antibiotics cefpodoxime, clavulanic acid, cefoxitin, imipenem, and meropenem were used as inducers. The induction ratios were calculated in terms of the ratio of β-lactamase activity mg–1 protein in induced cells to the β-lactamase activity per mg–1 protein in uninduced cells.
Measurement of ampC and nmcA mRNA Levels by qRT-PCR
The mRNA expression levels of ampC and nmcA with and without induction were determined by qRT-PCR as previously described (Nakano et al., 2017). Total RNA was extracted with the RNeasy protect bacteria mini kit and the RNase-free DNase set (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The qRT-PCR was performed in a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, United States) with a Power SYBR Green RNA-to-CT 1-Step kit (Thermo Fisher Scientific, Waltham, MA, United States) and 100 ng total RNA in a 20-μL reaction, according to the manufacturer’s instructions. The primers used are listed in Supplementary Table 1 (Doumith et al., 2009). The relative gene expression levels were calculated by the 2–△△CT method. The mRNA of the housekeeping gene rpoB was chosen as the endogenous reference for relative quantification. The results are presented as the mRNA expression level compared with that of NR1491. The experiment was performed in triplicate. The final relative expression levels of ampC and nmcA were determined by calculating the averages for their transcripts. The coefficient of variation (SD/mean) among experiments was < 10%.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the genetic regions surrounding blaNmcA and the whole-genome DNA sequences of NR1491 were deposited in the GenBank database under accession numbers LC482123 and BKZO00000000.1, respectively.
Results
Identification of blaNmcA-Harboring Enterobacter ludwigii NR1491
The draft NR1491 genome (GenBank accession no. BKZO00000000.1) was obtained by MiSeq (Illumina, Sa, Diego, CA, United States), and average nucleotide identity (ANI) analysis using E. cloacae strain ATCC13047, E. ludwigii type strain EN-119, and E. ludwigii AOUC-8/14 as reference genomes. Their respective ANI values were 87.82, 98.96, and 98.97%.4 NR1491 was identified as E. ludwigii belonging to ST258.
Antimicrobial Susceptibility and Resistance Genes
Antimicrobial susceptibility assays showed that E. ludwigii NR1491 was highly resistant to cephalothin, cefmetazole, carbapenems, and fosfomycin (> 512 μg mL–l) but susceptible to 3GC, piperacillin–tazobactam, cefepime, aztreonam, levofloxacin (≤0.06 μg mL–l), and gentamicin (0.5 μg mL–l) (Table 2). However, the MIC of cefotaxime increased when the agent was combined with clavulanic acid.
Whole-genome analysis with ResFinder revealed the following resistance-encoding genes: blaNmcA (carbapenemase), ACT-12 (AmpC β-lactamase), and fosA2 (glutathione transferase; fosfomycin resistance). It also disclosed that the regulator genes nmcR and ampR were upstream of blaNmcA and ampC, respectively. The entire nucleotide sequences of blaNmcA and nmcR and the intercistronic region were determined by Sanger sequencing. The sequences were identical to that of E. cloacae NOR-1 (accession no. Z21956). PCR demonstrated that no other acquired β-lactamase gene was harbored. The regulatory gene sequences of blaNmcA (nmcR, ampR, and ampD) were compared with that of the reference strains of ECC (NOR-1, EN-119, and AOUC-8/14); there are no mutations in these genes. Whole-genome analysis indicated that the insertion sequence (IS) elements and an integron were not encoded on the chromosome; the strain did not harbor a plasmid.
Genetic Environment Analysis of blaNmcA
The genetic environment of blaNmcA was determined to be a 48,089-bp nucleotide fragment characterized by whole-genome and Sanger sequencing and deposited into GenBank under accession no. LC482123. A BLASTn analysis showed that the fragment was highly similar to E. ludwigii AOUC-8/14 (accession no. KR919803) (44,766/44,874 nucleotides; 99.76% identity). The blaNmcA was located on a novel putatively mobile 29-kb element designated EludIMEX-1 inserted into the same chromosome location as that in E. ludwigii AOUC-8/14. Two imperfect 29-bp inverted repeat XerC/XerD binding sites associated with EludIMEX were identified at the chromosome–EludIMEX-1 junctions. The genetic regions were compared with the corresponding regions of E. ludwigii P101 (GenBank accession no. CP006580); the schematic representations are depicted in Figure 1. There were highly homologous regions (> 99% nucleotide sequence identity) and the insertion of EludIMEX-1. blaNmcA was putatively integrated into the E. ludwigii chromosome via a Xer-dependent recombination mechanism mediated by EludIMEX-1, as described previously.
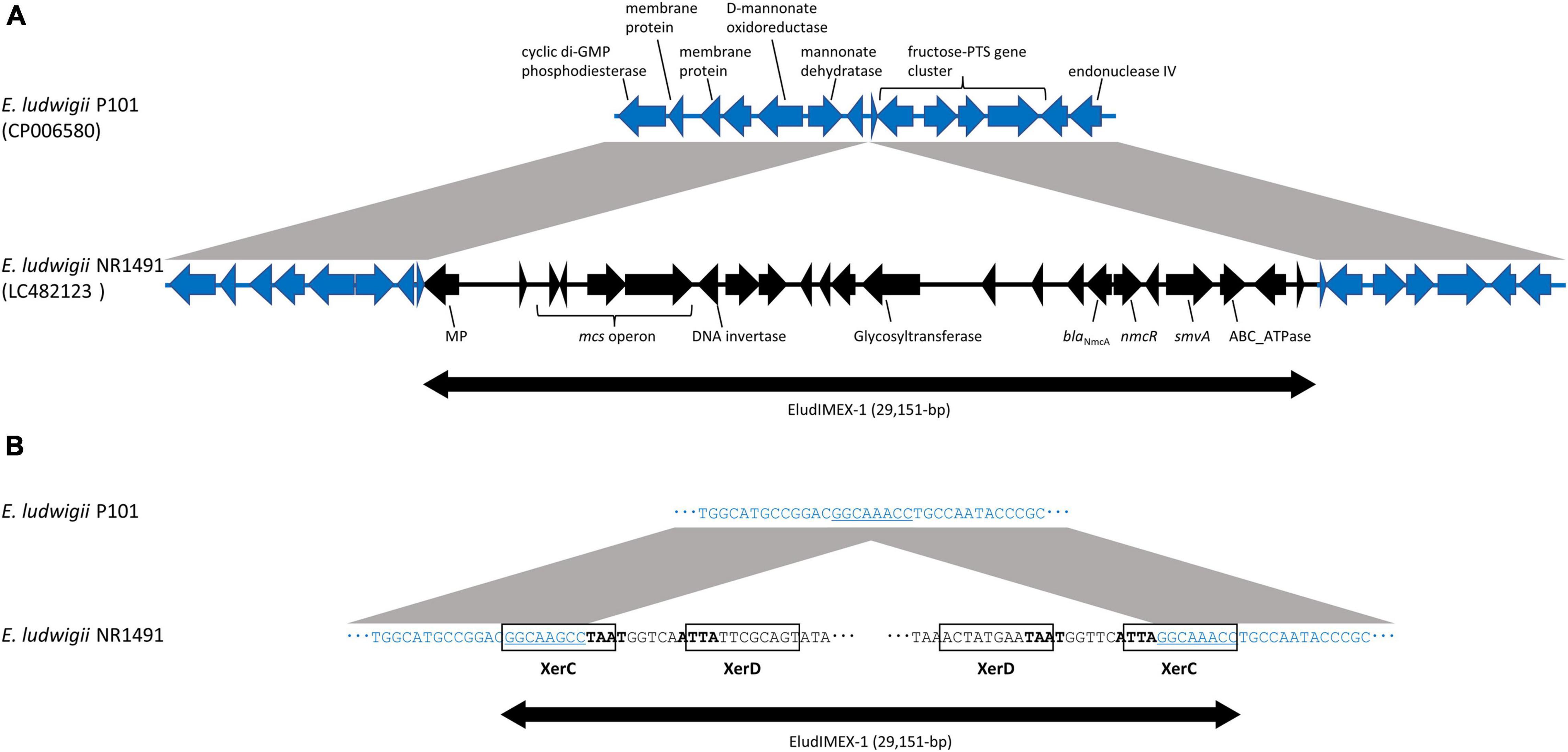
Figure 1. Schematic representations and nucleotide sequences of the genetic elements surrounding EludIMEX-1 of E. ludwigii NR1491 (GenBank accession no. LC482123) and the corresponding region of E. ludwigii P101 (GenBank accession no. CP006580). (A) Schematic representation of the genetic region of E. ludwigii NR1491 (black arrow) and E. ludwigii P101 (blue arrow). The gray regions between the NR1491 and P101 indicate > 99% nucleotide sequence identity. Insertion of the EludIMEX-1 element (double-headed arrow) in E. ludwigii NR1491 was observed. (B) Nucleotide sequences at the junctions of EludIMEX-1 (a part of left and right of the junctions) of E. ludwigii NR1491 and the corresponding region of E. ludwigii P101. The XerC/XerD binding sites are boxed, the conserved regions are boldfaced, and the consensus repeat sequences in XerC binding site of E. ludwigii NR1491 and corresponding sequences of E. ludwigii P101 are indicated using underlined letters.
BLASTn analysis of NR1491 indicated that there were 10 strains of NmcA-positive ECC with > 99% nucleotide sequence identities (Supplementary Table 2). The query coverage includes the gene regions of the EludIMEX-1 element and the surrounding regions with consensus repeat sequences and a binding site. The EludIMEX-1 was integrated into the chromosome at the same site, in these strains. The genotypes of these strains were including ST258 (n = 2) same with NR1491, ST282 (n = 2), ST257, ST260, ST374, ST714 (AOUC-8/14), ST748, and ST1724.
Antibiotic Inducibility of β-Lactamase
The antibiotic inducibility of β-lactamase was analyzed in NR1491 (Figure 2). Cefpodoxime and clavulanic acid were slightly inducer in a concentration-dependent manner. They yielded only a maximum 3.3 × induction of the MIC. Conversely, cefoxitin, imipenem, and meropenem were strong β-lactamase inducers. The carbapenems imipenem and meropenem induced β-lactamase in a concentration-dependent manner to 159× and 202×, respectively, at half their MIC. In contrast, the cefoxitin induction rate was concentration-independent and remained virtually unchanged (98–113 ×) across the tested concentrations (1/16–1 × MIC).
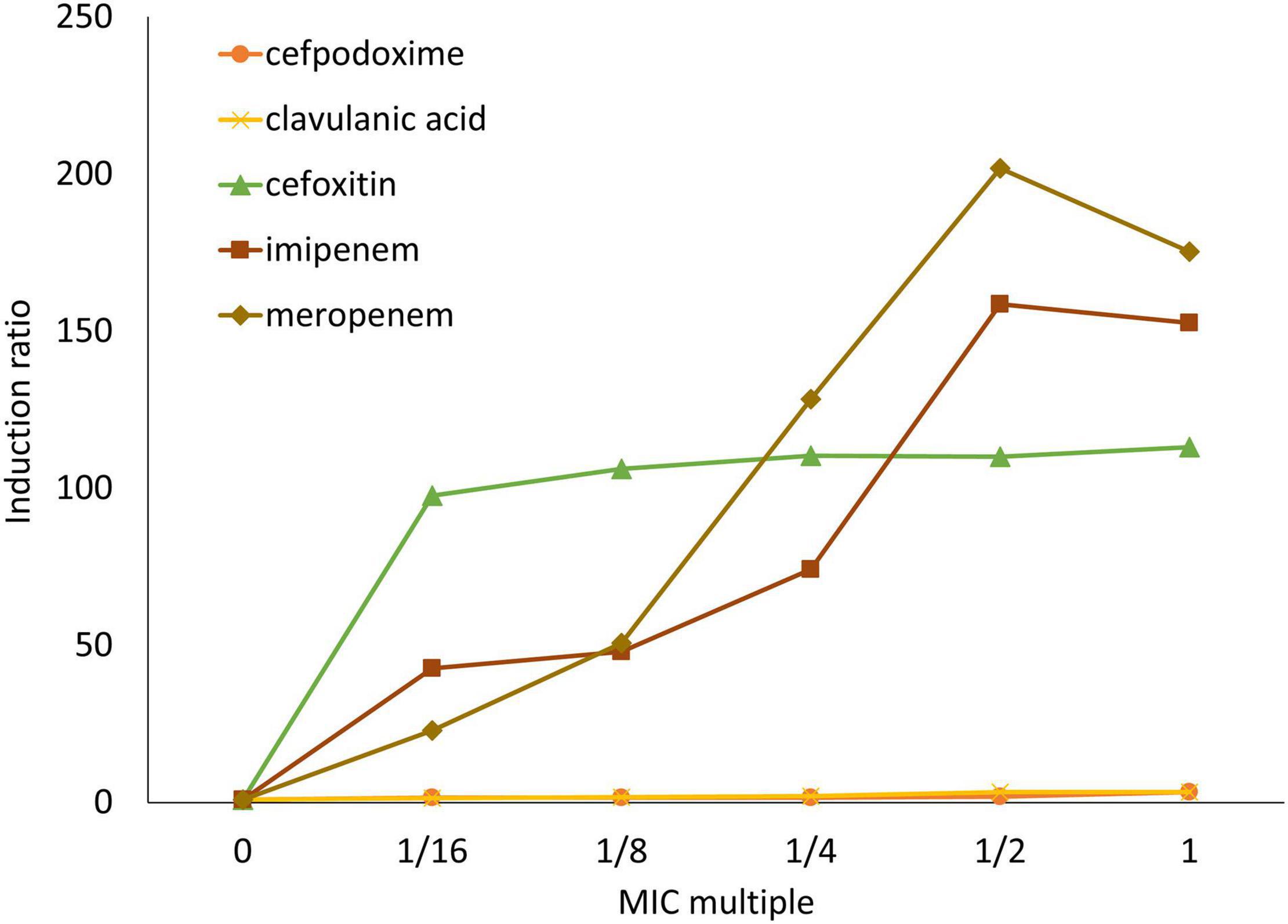
Figure 2. Induction ratios of β-lactamase activity in response to antibiotics on E. ludwigii NR1491. β-lactamase inducers were cefpodoxime (○), clavulanic acid (×), cefoxitin (△), imipenem (□), and meropenem (◇).
Properties of the Selected Third Generation Cephalosporins-Resistant Mutants
The 3GC-resistant mutants were selected with cefotaxime and ceftazidime at 2×, 4×, 8×, 16×, and 32× MIC. The antibiotic-resistant mutants were consistently obtained at a mutation frequency of ∼10–6–10–7 both for cefotaxime and ceftazidime (Table 3).
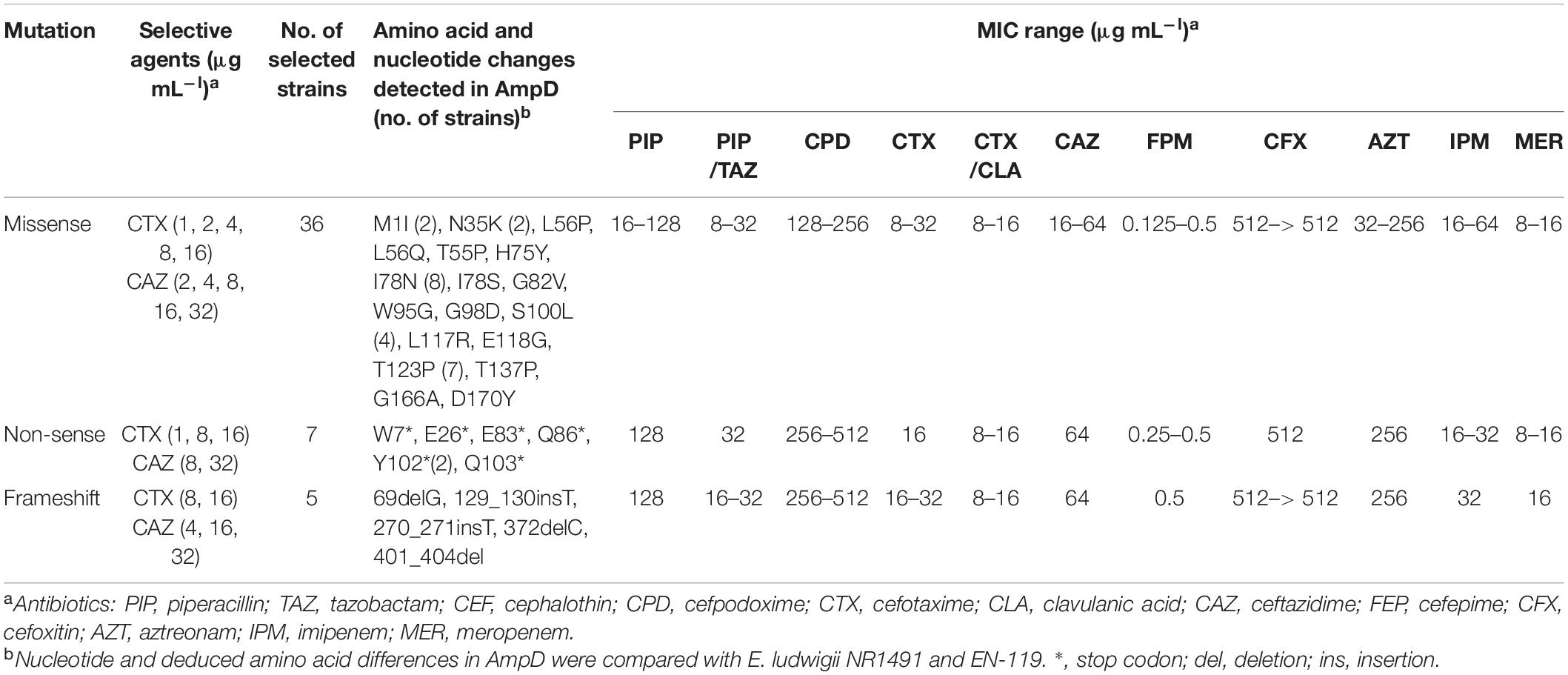
Table 3. Amino acid and nucleotide changes in AmpD of ceftazidime- or cefotaxime-resistant mutants of E. ludwigii NR1491 and their antimicrobial susceptibilities.
To investigate the molecular mechanism of 3GC resistance in these mutants, 48 clones were randomly selected from each condition. The regulator genes ampR, nmcR, and ampD were sequenced and compared to those of the parent E. ludwigii NR1491 and wild type EN-119 strains. Sequence data revealed that only ampD was altered in all cases whereas neither ampR nor nmcR was changed. Of the 48 3GC-resistant mutants, 34 had possible loss-of-function caused by missense mutations including 18 amino acid substitutions at 16 positions in ampD (Table 3). Premature termination of the AmpD protein was found in 14 mutants. Seven had nonsense mutation at six positions, five had frameshift mutations (three deletions and two insertions), and two had missense mutations in which the start codon (ATG) was changed to Ile (ATA) at position 1. Its effect was transcriptional decay. These mutations were scattered throughout the entire ampD sequence. Moreover, the nucleotide substitutions and mutation types and locations did not differ among selective agents and concentrations. However, certain mutants had the same missense mutation positions. Eight were I78N, seven were T123P, and four were S100L. These mutation positions may have been influential to AmpD activity. These mutants were resistant to 3GC presumably as a consequence of loss of AmpD function via the introduction of substitution mutations or decay of the transcript containing the premature stop codon.
For the mutants, the MICs were determined for the selected β-lactams (Table 3). Compared with the MICs for the parent strain NR1491, the MICs of 3GC (cefotaxime and ceftazidime) and aztreonam for the mutants had increased by ≥ 16 ×. Thus, these strains were reclassified from susceptible to resistant. The MICs of cefepime increased by only 1–4 × relative to NR1491 and were beyond the clinical resistance breakpoint. In contrast, the MICs of the carbapenems were almost always the same as those for NR1491.
Effects of Regulator Genes on β-Lactamase Expression
To evaluate the effects of ampR and ampD on NR1491 drug susceptibility, β-lactamase activity and mRNA expression of AmpC and NmcA with and without induction were analyzed in the ampR mutant NR1491 (pKU412) and the ampD mutant NR3901 (Table 2). Induction was carried out with 32 μg mL–l cefoxitin. The β-lactamase activity of NR1491 increased by ∼100 × and AmpC and NmcA were upregulated by ∼14.7× and 14.6×, respectively, in response to induction. ampC and nmcA expression levels were equally influenced by the induction.
NR3901 bore an ampD mutant (69delG) and was isolated by selection with 32 μg mL–l ceftazidime. The MICs of 3GC and aztreonam increased by ≥ 64 × and they were reclassified as highly resistant. The β-lactamase activity of NR3901 increased by ∼706.5 and ∼704.5 × with and without induction, respectively, compared with the NR1491 basal condition. The ampC and nmcA expression levels in NR3901 both increased by ∼700 × in the presence and absence of induction. The ampC and nmcA expression levels in NR1491 both increased by ∼15 × in response to induction. Hence, ampD equally induced ampC and nmcA expression. NR3901 was highly drug-resistant because it acquired the ampD mutation which derepressed ampC and nmcA expression.
NR1491 (pKU412) was obtained by transfecting the ampR mutant (135Asn) clone plasmid pKU412 into NR1491. The MICs of NR1491 (pKU412) were elevated as they were for NR3901. The β-lactamase activity had increased by ∼100 × at basal condition. ampC expression also increased by ∼100 × whereas that of nmcA did not change. AmpR may induce AmpC β-lactamase but does not affect nmcA expression. NR1491 (pKU412) induction resulted in a 1.5 × increase which suggests partial derepression. ampC expression increased by ∼200 × after induction. However, nmcA expression only doubled despite NR1491 expression increasing by ∼15 ×. Plasmid pKU411 comprising the wild type ampR (135Asp)-harboring strain NR1491 (pKU411) was compared with the ampR mutant strain and used to verify it. The MICs and β-lactamase activity of NR1491 (pKU411) were nearly the same as those for NR1491.
Discussion
The incidence of CPE is increasing globally. However, it has seldom (0.34%) been detected in Japan (Ohno et al., 2017). The most common carbapenemase genotype detected in Japan is IMP. Here, we isolated NmcA carbapenemase-producing ECC. NmcA carbapenemase has occasionally been reported for ECC in Europe, North America, and South America (Radice et al., 2004; Antonelli et al., 2015; Boyd et al., 2017). To the best of our knowledge, this is the first reported clinical isolation of an NmcA carbapenemase producer in Japan.
ANI revealed that NR1491 was, in fact, E. ludwigii belonging to ST258. A previous study reported that blaNmcA was highly associated with E. ludwigii. ST258 is a genotype of the NmcA carbapenemase producer (Boyd et al., 2017). The genetic environment of blaNmcA was nearly identical to that of E. ludwigii AOUC-8/14. Thus, blaNmcA was putatively integrated into the chromosome by EludIMEX-1 via a Xer-dependent recombination mechanism as previously described for E. ludwigii AOUC-8/14 (Antonelli et al., 2015). Interestingly, E. ludwigii AOUC-8/14 was isolated from a Japanese tourist in Italy. These strains may have been concealed in Japan and unintentionally isolated in the present study. Comparative genome analysis revealed that there were 10 strains including AOUC-8/14, which have high homology regions with NR1491. These EludIMEX-1 was integrated in the chromosome at the same site as in NR1491. The genotypes of these strains were different; the EludIMEX-1 insertion event has possibility occurred in these STs strains.
As with blaNmcA, NR1491 coexists with the regulator gene nmcR. A β-lactamase induction assay on NR1491 showed that it was weakly induced by clavulanic acid which was already known to be an inhibitor of Class A β-lactamases. Thus, it is inhibitory against NmcA β-lactamase (Mariotte-Boyer et al., 1996). On the other hand, clavulanic acid also induces chromosomally mediated AmpC β-lactamases in several Enterobacteriaceae (Drawz and Bonomo, 2010). Here, clavulanic acid induced β-lactamases via transcriptional regulator genes and not by inhibiting NmcA. The MIC of cefotaxime was increased in combination with clavulanic acid while imipenem and meropenem induced β-lactamase in a concentration-dependent manner. Previous study of its kinetic parameters show that NmcA demonstrated unusually strong hydrolytic activity toward imipenem and meropenem (Mariotte-Boyer et al., 1996). Therefore, NmcA producers are highly resistant to carbapenems as their inducers are upregulated and they are potently hydrolytic to carbapenems. Cefoxitin is a strong, stable, dose-independent β-lactamase inducer (100 × induction). The catalytic efficiency (kcat/Km) of cefoxitin is lower than those of the carbapenems but its MIC is comparatively higher (Mariotte-Boyer et al., 1996) possibly because of its high and stable inducibility.
To elucidate the mechanism of β-lactamase induction in NR1491, 3GC-resistant mutants were selected with cefotaxime and ceftazidime. The mutation frequencies were 10–6–10–7 as previously described (Naas et al., 2001). Forty-eight randomly selected clones had variable susceptibilities to 3GC, piperacillin–tazobactam, and aztreonam (Table 3). A DNA sequence analysis revealed that all mutants presented with nucleotide substitutions (frameshift, missense, or nonsense) in ampD alone. In contrast, ampR and nmcR were unchanged. The mutants were resistant to 3GC and probably had loss of AmpD function. Premature AmpD termination with a stop codon or frameshift induced strong 3GC resistance. The MICs of the ampD mutant strains with missense mutation had different 3GC resistance levels. The degree of 3GC resistance depended on the position of the substitution at the core residues of the active site of AmpD.
In the present study, no mutants of the transcriptional regulator genes ampR and nmcR were obtained. We investigated the effects of ampR in the presence of a mutant or wild type ampR clone plasmid. AmpR is a member of the lysR family and regulates the expression of chromosomal AmpC β-lactamase. Nevertheless, ampR mutants cause constitutive AmpC overproduction (75–470× increase) irrespective of induction (Kuga et al., 2000). In the enzyme activity assay, NR1491 (pKU412) increased β-lactamase activity by 100 × at the basal level compared with NR1491, also upregulating ampC. β-lactamase activity in the NR1491 strain with wild type ampR was increased by 100 × by induction and ampC and nmcA were each upregulated 15×. Whereas NR1491 with wild type ampR upregulated nmcA 15×, nmcA expression in NR1491 (pKU412) only doubled. Mutant ampR may negatively regulate nmcA expression. Putative ampR binding sequences in the E. cloacae ampR–ampC intergenic region were highly conserved with nmcR–blaNmcA and cross-reaction may have occurred (Naas and Nordmann, 1994). Earlier studies suggested that ampR is a global transcriptional regulator affecting the expression of several genes as well as ampC (Balasubramanian et al., 2012). NmcR was described as a positive regulator both in the absence and especially in the presence of a β-lactam inducer. In the absence of inducer, ampR is a negative regulator of ampC expression. In its presence, it positively regulates ampC expression (Naas and Nordmann, 1994). These findings suggest that even with available induction, mutant ampR represses the expression of nmcA. We believe this is the first study to describe the association between ampR and nmcA expression.
NmcR mutant strain has not been identified yet; therefore, the effect of nmcR mutations on the expression of nmcA could not be assessed. Point mutations in nmcR may be required to enhance its efficacy as an activator of nmcA in the same way as mutant ampR (such as that with a change in Asp135Asn). Alternatively, it may repress nmcA expression in the same way as wild type ampR. In a previous study, ampR mutants were obtained from the ampD mutant strain at a very low frequency (Kuga et al., 2000). The ampD mutant strain NR3901 selected nmcR mutants using ceftazidime at double and quadruple the MIC (128 and 256 μg mL–l). Nevertheless, no nmcR mutants were obtained (data not shown). nmcR mutants may be selected using ampD-mutant E. coli strains carrying blaNmcA and nmcR cloning plasmids. NR1491 co-harbors ampC on the chromosome. The observed differences in β-lactamase substrate specificity may influence selection conditions, and further investigation is needed to clarify whether nmcR mutation increases resistance by upregulating nmcA. This study has certain limitations. The analysis included only one strain and the conclusions were based on the results from this strain. Therefore, further studies are required to clarify the mechanisms of nmcR by selecting the nmcR mutant strains using other NmcA-producing ECC or IMI (closely related carbapenemase NmcA) producing E. coli. Moreover, the ampR mutation in NR1491 resulted in strong 3GC resistance via ampC overexpression. However, it remains unclear as to how mutant ampR stimulates nmcA transcription through induction. Future research should aim to elucidate the function of ampR.
NR3901 with an ampD mutation presented with a 700 × increase in β-lactamase activity as well as upregulated ampC and nmcA. Consequently, the MICs of 3GC were elevated. It indicates that ampD mutation has a similar influence on the expression of both ampC and nmcA, suggesting that structurally unrelated genes could be under the control of an identical regulatory system (Naas et al., 2001). The MICs for NR3901 and NR1491 (pKU412) bearing the ampR mutant plasmid were nearly equal. The upregulated nmcA in NR3901 had no effect on MIC compared with NR1491 (pKU412). Moreover, the MICs of the carbapenems for NR3901 were almost always the same as those of the parent strain despite the ampD mutant constitutively upregulating ampC and nmcA. High-level nmcA expression may elevate the MICs of carbapenems; in fact, the MICs were same as those of the parent strain. NmcA metabolism and its associated physiology may be connected with MICs. However, further experimentation is required to clarify this mechanism.
Here, we detected the NmcA-producing strain NR1491 in a hospital patient. Examination of MIC patterns showed high resistance to carbapenems but susceptibility to 3GC. The ampD mutant strains were identified among clinical isolates of ceftazidime-resistant E. cloacae as previously reported (Kaneko et al., 2005). Therefore, ampD-mutant NmcA producers may occur and interfere with the clinical detection of their wild type counterparts. We characterized NmcA producers that were highly resistant to carbapenems and yet susceptible to cefepime, whether they acquired the ampD mutation. In future works, it would be informative to compare these strains with the Big Five carbapenemases (KPC, IMP, NDM, VIM, and OXA).
Conclusion
In the present study, we identified the E. ludwigii isolate NR1491 in Japan that produces NmcA. The blaNmcA was located on a novel 29-kb putatively mobile element designated EludIMEX-1 identical in structure to that previously described in Europe. Induction studies revealed that the ampD mutants equally upregulated β-lactamases nmcA and ampC and were highly resistant to 3GC. However, the observed increase in the MIC value of 3GC was caused mainly by ampC overexpression. The ampR mutants also upregulated ampC, however, that of nmcA seemed to be repressed. Further research is necessary to elucidate the functions of ampR and nmcR.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
RN conceived and designed the experiments, performed the experiments, analyzed and interpreted the data, and wrote the manuscript. YY contributed reagents and materials and performed the experiments. AN, YS, KS, RS, MO, and KN performed the experiments, validation, and interpreted the data. AK contributed reagents and materials, conceived, and designed the experiments. AS and HY supervision and project administration. All authors contributed to manuscript, read, and approved the submitted version.
Funding
This work was supported by JSPS KAKENHI (Grants Nos. 17K10027 and 21K10403).
Conflict of Interest
RS and MO were employed by the company BML Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.794134/full#supplementary-material
Footnotes
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://pubmlst.org/ecloacae/
- ^ https://www.ezbiocloud.net/tools/ani
References
Ando, S., Nakano, R., Kuchibiro, T., Yamasaki, K., Suzuki, Y., Nakano, A., et al. (2018). Emergence of VIM-2-producing Citrobacter freundii in Japan. Infect. Dis. (Lond.). 50, 862–863. doi: 10.1080/23744235.2018.1498592
Annavajhala, M. K., Gomez-Simmonds, A., and Uhlemann, A. C. (2019). Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 10:44. doi: 10.3389/fmicb.2019.00044
Antonelli, A., D’andrea, M. M., Di Pilato, V., Viaggi, B., Torricelli, F., and Rossolini, G. M. (2015). Characterization of a novel putative xer-dependent integrative mobile element carrying the blaNMC-A carbapenemase gene, inserted into the chromosome of members of the Enterobacter cloacae complex. Antimicrob. Agents Chemother. 59, 6620–6624. doi: 10.1128/AAC.01452-15
Aoki, K., Harada, S., Yahara, K., Ishii, Y., Motooka, D., Nakamura, S., et al. (2018). Molecular characterization of IMP-1-producing Enterobacter cloacae complex isolates in Tokyo. Antimicrob. Agents Chemother. 62, e2091–e2017. e02091-17, doi: 10.1128/AAC.02091-17
Balasubramanian, D., Schneper, L., Merighi, M., Smith, R., Narasimhan, G., Lory, S., et al. (2012). The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes. PLoS One 7:e34067. doi: 10.1371/journal.pone.0034067
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Boyd, D. A., Mataseje, L. F., Davidson, R., Delport, J. A., Fuller, J., Hoang, L., et al. (2017). Enterobacter cloacae complex isolates harboring blaNMC-A or blaIMI-type class A carbapenemase genes on novel chromosomal integrative elements and plasmids. Antimicrob. Agents Chemother. 61:e02578-16. doi: 10.1128/AAC.02578-16
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Caille, O., Zincke, D., Merighi, M., Balasubramanian, D., Kumari, H., Kong, K. F., et al. (2014). Structural and functional characterization of Pseudomonas aeruginosa global regulator AmpR. J. Bacteriol. 196, 3890–3902. doi: 10.1128/JB.01997-14
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob. Agents Chemother. 58, 3895–3903.
Chavda, K. D., Chen, L., Fouts, D. E., Sutton, G., Brinkac, L., Jenkins, S. G., et al. (2016). Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio 7:e02093-16. doi: 10.1128/mBio.02093-16
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the bacteria and archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Davin-Regli, A., and Pages, J. M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392. doi: 10.3389/fmicb.2015.00392
Diene, S. M., and Rolain, J. M. (2014). Carbapenemase genes and genetic platforms in gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 20, 831–838. doi: 10.1111/1469-0691.12655
Doumith, M., Ellington, M. J., Livermore, D. M., and Woodford, N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63, 659–667. doi: 10.1093/jac/dkp029
Drawz, S. M., and Bonomo, R. A. (2010). Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201.
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Guerin, F., Isnard, C., Cattoir, V., and Giard, J. C. (2015). Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob. Agents Chemother. 59, 7753–7761. doi: 10.1128/AAC.01729-15
Hong, S. S., Kim, K., Huh, J. Y., Jung, B., Kang, M. S., and Hong, S. G. (2012). Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann. Lab. Med. 32, 359–361. doi: 10.3343/alm.2012.32.5.359
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: mobile element finder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Kaneko, K., Okamoto, R., Nakano, R., Kawakami, S., and Inoue, M. (2005). Gene mutations responsible for overexpression of AmpC β-lactamase in some clinical isolates of Enterobacter cloacae. J. Clin. Microbiol. 43, 2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005
Kuga, A., Okamoto, R., and Inoue, M. (2000). ampR gene mutations that greatly increase class C β-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 44, 561–567. doi: 10.1128/AAC.44.3.561-567.2000
Mariotte-Boyer, S., Nicolas-Chanoine, M. H., and Labia, R. (1996). A kinetic study of NMC-A β-lactamase, an Ambler class A carbapenemase also hydrolyzing cephamycins. FEMS Microbiol Lett 143, 29–33. doi: 10.1111/j.1574-6968.1996.tb08457.x
Mezzatesta, M. L., Gona, F., and Stefani, S. (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7, 887–902. doi: 10.2217/fmb.12.61
Miyoshi-Akiyama, T., Hayakawa, K., Ohmagari, N., Shimojima, M., and Kirikae, T. (2013). Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Naas, T., Massuard, S., Garnier, F., and Nordmann, P. (2001). AmpD is required for regulation of expression of NmcA, a carbapenem-hydrolyzing β-lactamase of Enterobacter cloacae. Antimicrob. Agents Chemother. 45, 2908–2915. doi: 10.1128/AAC.45.10.2908-2915.2001
Naas, T., and Nordmann, P. (1994). Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 91, 7693–7697. doi: 10.1073/pnas.91.16.7693
Nakano, A., Nakano, R., Suzuki, Y., Saito, K., Kasahara, K., Endo, S., et al. (2018). Rapid Identification of blaIMP-1 and blaIMP-6 by multiplex amplification refractory mutation system PCR. Ann. Lab. Med. 38, 378–380.
Nakano, R., Nakano, A., Hikosaka, K., Kawakami, S., Matsunaga, N., Asahara, M., et al. (2014). First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob. Agents Chemother. 58, 7611–7612.
Nakano, R., Nakano, A., Yano, H., and Okamoto, R. (2017). Role of AmpR in the high expression of the plasmid-encoded AmpC β-lactamase CFE-1. mSphere 2:e00192-17. doi: 10.1128/mSphere.00192-17
Nakano, R., Okamoto, R., Nakano, Y., Kaneko, K., Okitsu, N., Hosaka, Y., et al. (2004). CFE-1, a novel plasmid-encoded AmpC β-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob. Agents Chemother. 48, 1151–1158. doi: 10.1128/AAC.48.4.1151-1158.2004
Nordmann, P., Mariotte, S., Naas, T., Labia, R., and Nicolas, M. H. (1993). Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 37, 939–946. doi: 10.1128/AAC.37.5.939
Ohno, Y., Nakamura, A., Hashimoto, E., Matsutani, H., Abe, N., Fukuda, S., et al. (2017). Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J. Infect. Chemother. 23, 224–229.
Peirano, G., Matsumura, Y., Adams, M. D., Bradford, P., Motyl, M., Chen, L., et al. (2018). Genomic epidemiology of global carbapenemase-producing Enterobacter spp., 2008-2014. Emerg. Infect. Dis. 24, 1010–1019. doi: 10.3201/eid2406.171648
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pottumarthy, S., Moland, E. S., Juretschko, S., Swanzy, S. R., Thomson, K. S., and Fritsche, T. R. (2003). NmcA carbapenem-hydrolyzing enzyme in Enterobacter cloacae in North America. Emerg. Infect. Dis. 9, 999–1002. doi: 10.3201/eid0908.030096
Radice, M., Power, P., Gutkind, G., Fernandez, K., Vay, C., Famiglietti, A., et al. (2004). First class a carbapenemase isolated from enterobacteriaceae in Argentina. Antimicrob. Agents Chemother. 48, 1068–1069. doi: 10.1128/AAC.48.3.1068-1069.2004
Schmidtke, A. J., and Hanson, N. D. (2006). Model system to evaluate the effect of ampD mutations on AmpC-mediated β-lactam resistance. Antimicrob. Agents Chemother. 50, 2030–2037. doi: 10.1128/AAC.01458-05
Keywords: carbapenemase, NmcA, AmpC β-lactamase, Enterobacter cloacae complex, induction, regulator genes
Citation: Nakano R, Yamada Y, Nakano A, Suzuki Y, Saito K, Sakata R, Ogawa M, Narita K, Kuga A, Suwabe A and Yano H (2022) The Role of nmcR, ampR, and ampD in the Regulation of the Class A Carbapenemase NmcA in Enterobacter ludwigii. Front. Microbiol. 12:794134. doi: 10.3389/fmicb.2021.794134
Received: 13 October 2021; Accepted: 23 December 2021;
Published: 12 January 2022.
Edited by:
Mullika Traidej Chomnawang, Mahidol University, ThailandReviewed by:
Valentine Usongo, Health Canada, CanadaSubhasree Roy, National Institute of Cholera and Enteric Diseases (ICMR), India
Copyright © 2022 Nakano, Yamada, Nakano, Suzuki, Saito, Sakata, Ogawa, Narita, Kuga, Suwabe and Yano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryuichi Nakano, cm5ha2Fub0BuYXJhbWVkLXUuYWMuanA=
 Ryuichi Nakano
Ryuichi Nakano Yuki Yamada2
Yuki Yamada2 Akiyo Nakano
Akiyo Nakano Yuki Suzuki
Yuki Suzuki