
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 February 2022
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.793345
This article is part of the Research Topic Population and Comparative Genomics of Plant Pathogenic Bacteria View all 16 articles
Clavibacter michiganensis, a Gram-positive, plant-pathogenic bacterium belonging to Actinobacteria, is a causal agent of bacterial canker in tomatoes. Although LMG7333T is the type strain of C. michiganensis, it has not been used in many studies, probably because of a lack of the complete genome sequence being available. Therefore, in this study, the complete genome sequence of this type strain was obtained, and comparative genome analysis was conducted with the genome sequences of two other C. michiganensis strains and type strains of Clavibacter species, of which their complete genome sequences are available. C. michiganensis LMG7333T carries one chromosome and two plasmids, pCM1 and pCM2, like two other C. michiganensis strains. All three chromosomal DNA sequences were almost identical. However, the DNA sequences of two plasmids of LMG7333T are similar to those of UF1, but different from those of NCPPB382, indicating that both plasmids carry distinct gene content among C. michiganensis strains. Moreover, 216 protein-coding sequences (CDSs) were only present in the LMG7333T genome compared with type strains of other Clavibacter species. Among these 216 CDSs, approximately 83% were in the chromosome, whereas others were in both plasmids (more than 6% in pCM1 and 11% in pCM2). However, the ratio of unique CDSs of the total CDSs in both plasmids were approximately 38% in pCM1 and 30% in pCM2, indicating that the high gene content percentage in both plasmids of C. michiganensis are different from those of other Clavibacter species, and plasmid DNAs might be derived from different origins. A virulence assay with C. michiganensis LMG7333T using three different inoculation methods, root-dipping, leaf-clipping, and stem injection, resulted in typical disease symptoms, including wilting and canker in tomato. Altogether, our results indicate that two plasmids of C. michiganensis carry distinct gene content, and the genome information of the type strain LMG7333T will help to understand the genetic diversity of the two plasmids of Clavibacter species, including C. michiganensis.
Plant pathogenic bacteria belonging to the genus Clavibacter are Gram-positive and often cause severe damage to crops (Eichenlaub and Gartemann, 2011). This genus was reclassified from Corynebacterium and newly identified as a new genus, including several phytopathogenic bacteria (Davis et al., 1984). The genus Clavibacter contained the single species, C. michiganensis with nine subspecies, according to its host specificity (Eichenlaub et al., 2007; Gonzalez and Trapiello, 2014; Yasuhara-Bell and Alvarez, 2015; Oh et al., 2016). Recently, seven out of eight subspecies except C. michiganensis subsp. chilensis (non-pathogenic bacterium) were reclassified as separate species based on the genomic analysis in addition to its host specificity (Li et al., 2018; Méndez et al., 2020): C. michiganensis (bacterial canker and wilting in tomato) (Smith, 1910), C. sepedonicus (ring rot in potato) (Manzer and Genereux, 1981), C. nebraskensis (Goss’s leaf blight and wilting in maize) (Vidaver and Mandel, 1974), C. insidiosus (bacterial wilt in alfalfa) (McCulloch, 1925), C. tessellarius (leaf freckles and leaf spots in wheat) (Carlson and Vidaver, 1982), C. phaseoli (bacterial leaf yellowing in beans) (Gonzalez and Trapiello, 2014), and C. capsici (bacterial canker in pepper) (Oh et al., 2016) and a non-pathogenic C. californiensis (Yasuhara-Bell and Alvarez, 2015). Although Méndez et al. (2020) also propose reclassifying C. michiganensis subsp. chilensis to C. phaseoli, we kept the original subspecies name to distinguish this strain from the original C. phaseoli in this study.
Disease through Clavibacter bacterial infection resulted in economic damage due to the significant yield loss. For example, C. michiganensis was first reported in 1910 in North America (Smith, 1910), and since then, this pathogen has caused significant economic loss (Xu et al., 2012; Peritore-Galve et al., 2020). Additionally, C. sepedonicus was designated a quarantine pathogen in Europe owing to its severe economic damage annually (Van der Wolf et al., 2005). C. michiganensis has been studied relatively well among Clavibacter species. Although virulence mechanisms of this pathogen are not well known yet, previous research shows that two plasmid-borne genes, celA and pat-1, play essential roles in C. michiganensis pathogenicity (Dreier et al., 1997; Jahr et al., 2000; Burger et al., 2005; Hwang et al., 2019). Moreover, a pathogenicity island (PAI) region, which contains putative virulence genes such as the ppa, chp, and pel gene families, was found in the chromosome (Gartemann et al., 2008; Stork et al., 2008). In tomato seeds, C. michiganensis ssp. chilensis and C. californiensis were isolated and exhibited non-pathogenic effects to tomato plants (Yasuhara-Bell and Alvarez, 2015). Notably, these two non-pathogenic bacteria missed most virulence genes and/or putative virulence genes possessed by pathogenic C. michiganensis (Méndez et al., 2020).
So far, the complete genome sequences of type strains of only three Clavibacter species have been reported: C. sepedonicus ATCC33113T (Bentley et al., 2008), C. nebraskensis NCPPB2581T (GenBank accession #: HE614873), and C. capsici PF008T (Bae et al., 2015). In the case of C. insidiosus species, the complete genome sequence of strain R1-1, which is not a type strain, is available in the public database (Lu et al., 2015). Based on these whole genome sequences of Clavibacter species in the public database, studies on Clavibacter species at the genomic level have recently increased. Moreover, newly sequenced genome data of Clavibacter strains have allowed us to conduct comparative genome analysis at the species or strain level (Tambong, 2017; Osdaghi et al., 2020). Indeed, several comparative genomic approaches have been conducted to target Clavibacter species isolated from specific geographic regions using genome database (Thapa et al., 2017; Hwang et al., 2020a; Méndez et al., 2020), and the genomic analysis is becoming more important to the genus Clavibacter research.
In the case of a tomato pathogen C. michiganensis species, only four strains, NCPPB382 (Gartemann et al., 2008), UF1, and two Chile isolates, VL527 and MSF322 (Méndez et al., 2020), were completely sequenced. Most of C. michiganensis genome sequences have not been completely assembled (Thapa et al., 2017). Although the draft sequence of C. michiganensis type strain LMG7333T (= NCPPB 2979T, CFBP 2352T, DSM 46364T) was already reported, it has not yet been completed (Li and Yuan, 2017; Li et al., 2018). The draft genome sequencing of C. michiganensis LMG7333T was conducted previously by PacBio-SMRT sequencing technology (Li and Yuan, 2017), resulting in five contigs. Because this genome sequence was not complete, there was a limitation to use it as a reference genome of C. michiganensis species. For these reasons, NCPPB382 rather than a type strain of C. michiganensis was mainly used for many studies that have focused on molecular mechanisms to understand virulence mechanism(s) even though this type strain can induce typical disease as severe as C. michiganensis NCPPB382 on tomato plants (Hwang et al., 2019).
In this study, we report the completed genome sequence of C. michiganensis type strain LMG7333T and use this sequence for comparison with other previously published genomes of C. michiganensis to obtain more valid insights onto genetic diversity among pathogenic Clavibacter species. Additionally, we conducted a virulence assay with this type strain using three inoculation methods. Our results suggest that comparative genome analysis with the complete genome sequence of C. michiganensis type strains LMG7333T can provide a genomic resource to understand the genetic diversity among Clavibacter species better.
The genomic DNA (gDNA) of C. michiganensis LMG7333T was extracted using the solution-based HiGene Genomic DNA Prep kit (BIOFACT, Daejeon, Korea), according to the manufacturer’s instructions. Using extracted gDNA, whole genome sequencing of LMG7333T was conducted using combined methods of two sequencing platforms, PacBio RSII and Illumina HiSeqXten (Macrogen, Seoul, Korea), for genome sequence completion. The reads obtained from PacBio RSII were assembled by Hierarchical Genome Assembly Process (Chin et al., 2013). HiseqXten reads were also used to correct assembly by Pilon v1.21 (Walker et al., 2014). Based on the assembled sequences, gene prediction and annotation were conducted by Prokka v1.13 (Seemann, 2014). Then, InterProScan v5.30–69.0 (Jones et al., 2014) and psiblast v2.4.0 (Camacho et al., 2009) with EggNOG DB v4.5 (Huerta-Cepas et al., 2016) and the Clustering Orthologous Groups (COG) database v2014 (Tatusov et al., 2000) were used to confirm the predicted proteins. Circos program v0.69.3 (Krzywinski et al., 2009) was used for drawing circular maps of the genome.
The completed whole genome sequence of C. michiganensis LMG7333T type strain was first compared by aligning the EasyFig program (Sullivan et al., 2011) and NCBI blast program with those of C. michiganensis NCPPB382 (GenBank accession #: AM711865, AM711866, and AM711867) (Gartemann et al., 2008) and UF1 (GenBank accession #: CP033724, CP033725, and CP033726). Then, it was also compared with completed genome sequences of nine other Clavibacter species, including several type strains such as C. sepedonicus ATCC33113T (GenBank accession #: AM849034) (Bentley et al., 2008), C. capsici PF008T (GenBank accession #: CP012573, CP012574, and CP012575) (Bae et al., 2015), C. insidiosus R1-1 (GenBank accession #: CP011043, CP011044, CP011045, and CP011046) (Lu et al., 2015), C. nebraskensis NCPPB2581T (GenBank accession #: HE614873), C. insidiosus LMG3663T (GenBank accession #: MZMO01000001, MZMO01000002, and MZMO01000003), C. tessellarius ATCC33566T (GenBank accession #: MZMQ01000001 and MZMQ01000002) (Li and Yuan, 2017; Li et al., 2018), C. phaseoli CFBP8627T (GenBank accession #: QWGV00000000), C. michiganensis subsp. chilensis CFBP8217T (GenBank accession #: QWGS01000000), and C. californiensis CFBP8216T (GenBank accession #: QWEE01000000) (Méndez et al., 2020). The genome sequence of Leifsonia xyli subsp. cydontis DSM46306T (GenBank accession #: CP006734) (Monteiro-Vitorello et al., 2013), which belongs to the same family as genus Clavibacter, was used as an out group. To understand the phylogenetic relationships among the analyzed strains, OrthoANI values were calculated, and UPGMA dendrograms were generated using the Orthologous ANI Tool of ChunLab (Lee et al., 2016). Moreover, the pan-genome analysis was conducted, and results were shown through a Venn diagram (Bardou et al., 2014).
C. michiganensis strain LMG7333T was grown at 26°C in KB medium (20 g proteose peptone No. 3, 1.5 g K2HPO4, 6 mL 1 M MgSO4, 16 mL 50% glycerol, and 15 g agar in the total volume of 1 L) for 3 days and inoculated into tomato plants using three different inoculation methods, which are leaf-clipping inoculation, stem injection, and root-dipping inoculation. Ten-day-old and 3- and 4-week-old tomato plants (Solanum lycopersicum L. cv. “Betatini”) were used for the root-dipping inoculation, leaf-clipping inoculation, and stem injection, respectively. For the leaf-clipping method, the bacterial inoculum was prepared and adjusted to a concentration of 2 × 108 CFU/mL (OD600 = 0.4). Sterilized scissors were dipped into this inoculum, and then half of the leaves were cut using scissors (Hwang et al., 2020b). Three leaves located in different directions of each plant and at least five plants were used for each treatment. For stem injection, a bacterial concentration was adjusted to 109 CFU/mL (OD600 = 2.0), and the 20 μL bacterial suspension was injected into tomato stems above 2 cm from the cotyledon after making a hole with a needle. At least four plants were used for each treatment. For the root-dipping inoculation method, 10-day-old seedlings were pulled out from soil, and their roots were dipped in 1 mL of bacterial suspension (109 CFU/mL) for 30 min. The seedlings were then replanted into pots. Five plants were used for each treatment. C. michiganensis LMG7333T was resuspended in sterilized 10 mM MgCl2 buffer, and this buffer was used as a negative control in all inoculation methods.
After inoculation with C. michiganensis LMG7333T by stem injection, tomato stems were collected at 0, 7, 14, 21, and 28 days after inoculation (dai). For sample collection, 3 cm lengths of inoculated stems were harvested from four different sites, which are the inoculated site and 10, 15, and 20 cm above from the inoculated site. Then, 2 mL of 10 mM MgCl2 was added to ground samples and the supernatant was dotted onto KB media after 10-fold serial dilution. Three plants were used for each repetition, and experiments were repeated at least three times.
For statistical analysis of bacterial growth of Clavibacter michiganensis LMG7333T in planta, Duncan’s multiple range test was conducted (p < 0.05).
The complete genome sequence of C. michiganensis LMG7333T was obtained through the combination of PacBio RSII and Illumina HiSeqXten sequencing methods, and the genome coverage was 320X. From HiSeqXten, we got 9,154,590 reads, and after assembly, the N50 value was 3,271,838. Finally, we obtained three contigs, one circular chromosome (GenBank accession #: CP080437), and two plasmids, pCM1 and pCM2 (GenBank accession #: CP080438 and CP080439). Their genome sizes and G + C contents are 3.27 Mb with 72.74% G + C content, 32 kb with 67.75% G + C content, and 73 kb with 66.99% G + C content, respectively (Figures 1A,B). This genomic composition of C. michiganensis LMG7333T is similar to those of NCPPB382 and UF1 strains, of which the complete genome sequences are available. However, their sizes are different from one another, particularly, plasmid sizes (Figure 1B). Genome annotation revealed 3,033 protein-coding sequences (CDSs) as well as 45 tRNAs and 6 rRNAs in the chromosome and 32 and 67 CDSs in pCM1 and pCM2, respectively (Figure 1B). The annotated proteins were categorized into 21 COG. Approximately 40% of CDSs were categorized into two groups for the function unknown (S; 891 CDSs) and the general function only (R; 349 CDSs), which are poorly characterized, in the COG database (Figure 2). COG groups for carbohydrate transport and metabolism (G; 255 CDSs); transcription (K; 205 CDSs); amino acid transport and metabolism (E; 198 CDSs); inorganic ion transport and metabolism (P; 179 CDSs); translation, ribosomal structure, and biogenesis (J; 146 CDSs); and cell wall/membrane/envelope biogenesis (M; 137 CDSs) were found to be dominant groups. In contrast, only two CDSs were categorized into cell motility groups (N) (Figure 2). Some CDSs were not grouped into specific COG or grouped to two or more COG groups. There are no CDSs for flagella formation, which is consistent with a previous study reporting that C. michiganensis LMG7333T did not have flagella (Oh et al., 2016).
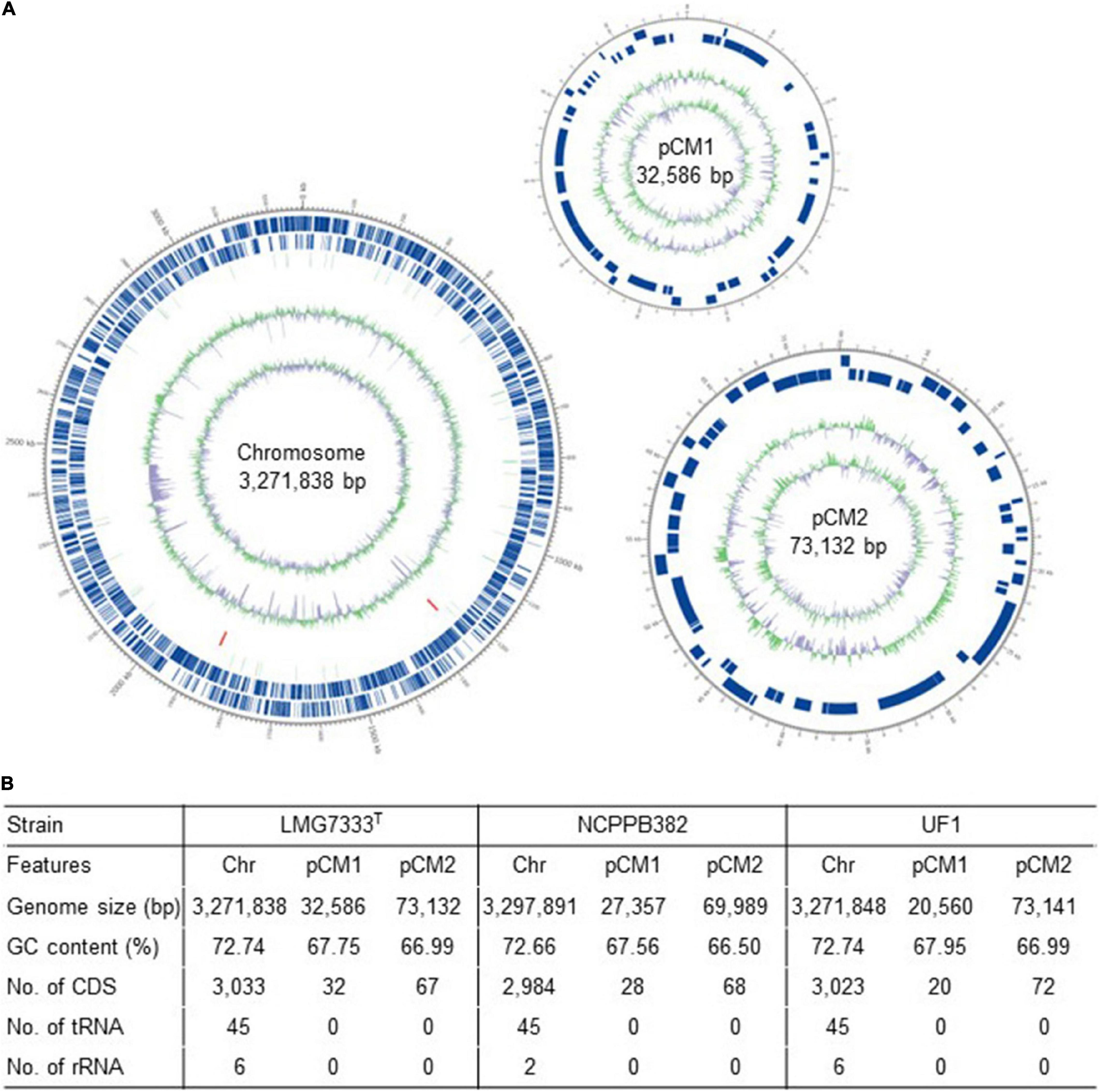
Figure 1. Genome information of C. michiganensis type strain LMG7333T. (A) A circular map drawn by Circos program. Circles outside to the center are represented as CDS on the forward strand, CDS on the reverse strand, tRNA, rRNA, GC content, and GC skew. (B) Genome summary of three C. michiganensis strains, LMG7333T, NCPPB382, and UF1. Chr, chromosome; pCM1 and pCM2, plasmids; CDS, coding sequences.
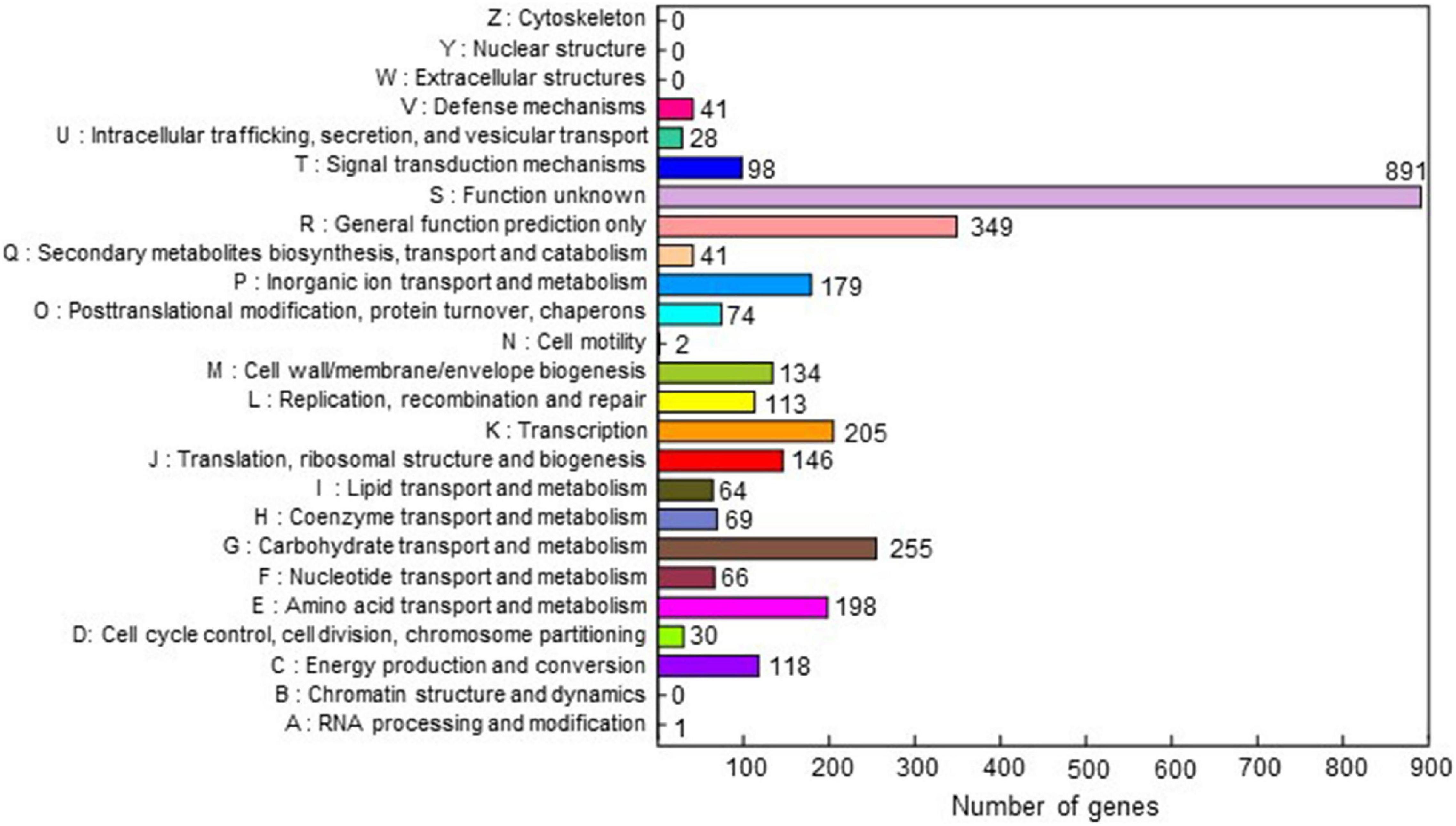
Figure 2. Clusters of orthologous groups (COG) of the annotated CDSs of C. michiganensis type strain LMG7333T. COG database v2014 was used for analysis. Annotated genes were classified into 21 categories of 25. The X-axis represents the number of genes, and the Y-axis indicates each COG cluster. The numbers next to each bar indicate the number of genes in each COG cluster.
As shown in Figure 1B, three C. michiganensis strains differed in the sizes of a chromosome and two plasmids. Here, three genome sequences were compared to examine how different they were. First, the sequence identity of chromosomal DNA among the three strains was almost 100% identical although the query cover between LMG7333T and NCPPB382 strains was 97% (Figure 3). As with other C. michiganensis strains, C. michiganensis LMG7333T has about 120 kb of pathogenicity island (PAI) region from 2,334,274 to 2,464,147 bp on its chromosome (Figure 3 and Supplementary Figure 1). In the chp region of PAI, the putative virulence genes, such as the chp, ppa, and pel gene families, were intact like those in the NCPPB382 strain. Moreover, the tomA gene was found in the tomA region of PAI (Supplementary Figure 1). Two other important virulence genes, celA, and pat-1 genes are located from 7,321 to 9,561 bp in pCM1 and from 7,833 to 8,675 bp in pCM2 of C. michiganensis LMG7333T (Figure 3). The presence of an intact PAI region, celA and pat-1 genes, suggests that C. michiganensis LMG7333T can also infect tomato plants to cause typical disease symptoms like the NCPPB382 stain.
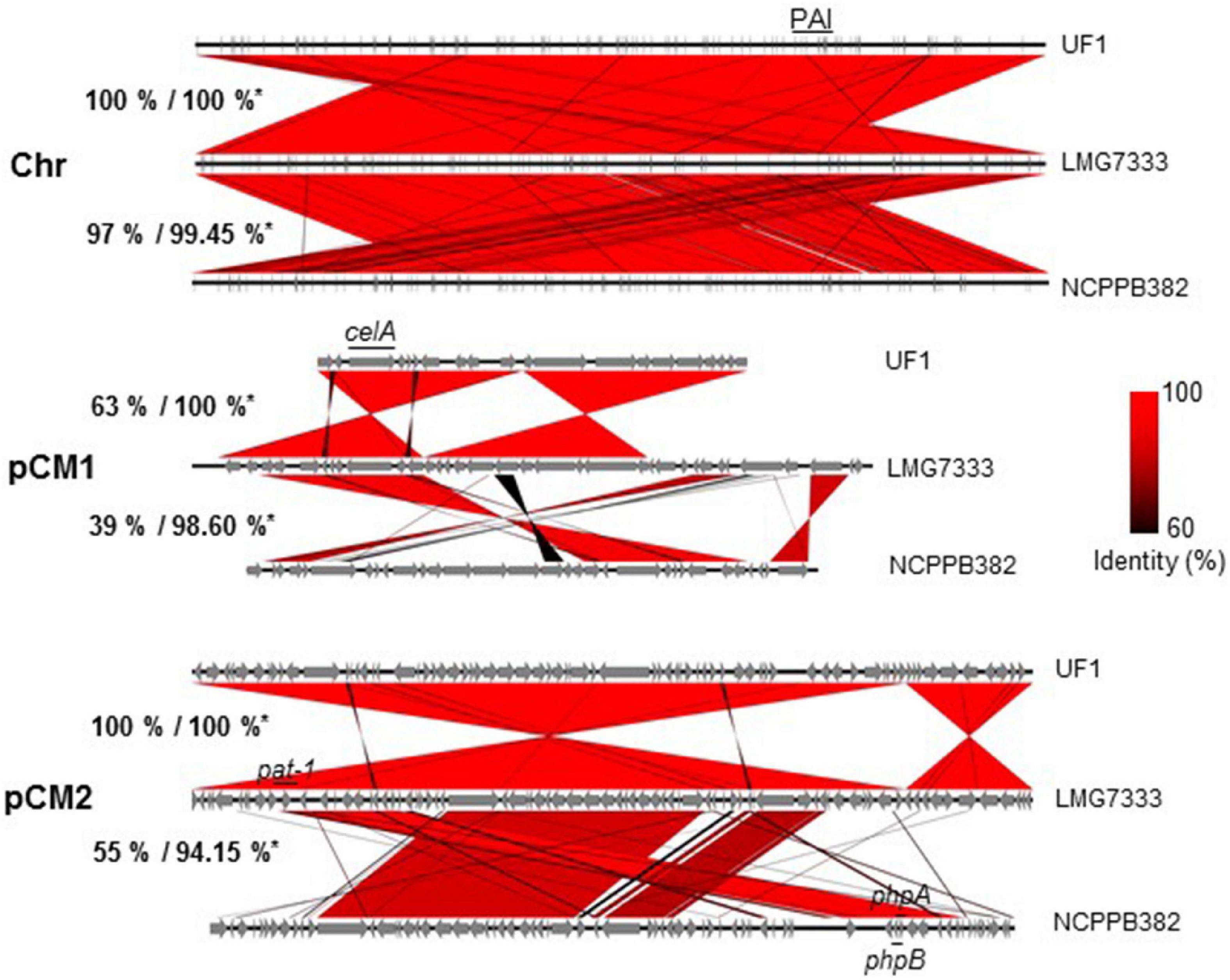
Figure 3. Genome comparison of three C. michiganensis strains. Genome alignment of three C. michiganensis strains. *Percentages indicate represented query cover/identity of a chromosome (Chr) and two plasmids, pCM1 and pCM2, of C. michiganensis NCPPB382 and UF1 based on C. michiganensis LMG7333T. The identical parts were aligned in red, and the key virulence genes such as celA and pat-1 genes or a pathogenicity island (PAI) are shown.
The size and sequence query cover of the pCM1 plasmid of LMG7333T was the most different from those of two other strains although the sequence identity in the matched regions was almost 100% identical. Its size was the biggest among the three strains, and the matched regions were only 63% with UF1 pCM1 and only 39% with NCPPB382 pCM1 (Figure 3). Among unmatched regions in pCM1 of LMG7333T, some regions were unique to this strain, whereas others were only matched with that of NCPPB382. Most of the genes in these unmatched regions are annotated as hypothetical proteins, indicating that the origin and functions of these genes are unknown. In the public database, C. michiganensis NCPPB382 pCM1 has genes for cellulase (celA), portioning protein (parA and parB), and replication origin (repA) and several hypothetical proteins. When the sequence of pCM1 was compared with these annotated genes of pCM1 in C. michiganensis NCPPB382, we found that C. michiganensis LMG7333T pCM1 also contained these annotated genes except the traG gene encoding putative conjugal transfer protein (Załuga et al., 2014). Gene annotation also revealed that the espI gene, which is a component of the type VII ESX-1 secretion system (Zhang et al., 2014), was found in the pCM1 of LMG7333T but not NCPPB382, whereas both pCM2 plasmids have the espI homologous gene.
The size and sequence of LMG7333T pCM2 plasmid were almost 100% identical to that of the UF1 strain, whereas the matched regions with NCPPB382 pCM2 were only approximately 37 kb of 73 kb, which is only 55% of the total plasmid size with 91–99% sequence identity (Figure 3). In the matched region of pCM2 plasmids, all three C. michiganensis strains possess pat-1 genes, encoding a putative serine protease, which was known as a plasmid-borne virulence factor of C. michiganensis (Burger et al., 2005). In the case of C. michiganensis NCPPB382, there are more pat-1 paralogs, phpA and phpB, in pCM2. However, C. michiganensis LMG7333T lacks these two genes. The unmatched region covered by 45% of the total plasmid size was unique to each strain.
Overall, these results indicate that, although three C. michiganensis strains have similar genome size and major virulence factors, like PAI, celA, and pat-1, in their genomes, the plasmid sequences are varied, and both pCM1 and pCM2 of LMG7333T contain unique DNA sequences mostly annotated as hypothetical proteins. Moreover, genome comparison among three C. michiganensis strains using complete genome sequences indicates that C. michiganensis LMG7333T genome sequence can be used as another reference genome of C. michiganensis.
Based on genomic information, average nucleotide identity (ANI) values were calculated among C. michiganensis LMG7333T and nine other Clavibacter bacteria, including type strains of other Clavibacter species and Leifsonia xyli subsp. cydontis as an outgroup to measure the genetic distance among genomes. Based on ANI data, three C. michiganensis strains were clustered into the same clade with more than 99% ANI value, and non-pathogenic Clavibacter californiensis was the closest to C. michiganensis with almost 95% ANI value (Figure 4 and Supplementary Table 1). There was the second clade, including C. insidiosus, C. nebraskensis, C. phaseoli, and C. michiganensis subsp. chilensis, which is close to the clade of C. michiganensis. Type strains of C. capsici PF008T, C. sepedonicus ATCC33113T, and C. tessellarius ATCC33566T are phylogenetically distinct from the C. michiganensis clade.

Figure 4. Phylogenetic tree of C. michiganensis type strain LMG7333T with other Clavibacter species and subspecies based on ANI. This analysis was conducted using OrthoANI software. Leifsonia xyli subsp. cydontis DSM46306T was used for outgroup. Numbers indicate genetic similarity (GS) coefficient value. T, type strain.
Using genome sequences of pathogenic Clavibacter strains, pan-genome analysis was also conducted. Three C. michiganensis strains were first compared, and they have 2,959 common genes. C. michiganensis LMG7333T only has seven unique genes in its genome. Moreover, it has 8 and 84 genes overlapped with C. michiganensis NCPPB382 and UF1, respectively (Figure 5A). Among the seven unique genes in C. michiganensis LMG7333T, only one gene was annotated as RNA helicase, and the other six genes were annotated as hypothetical proteins. The specific genes in other two C. michiganensis strains were also annotated as hypothetical proteins. C. michiganensis LMG7333T has 91 unique genes compared with C. michiganensis NCPPB382. Among these genes, 50 genes are located in the chromosome of the type strain, and the other 41 genes exist in two plasmids, 17 and 24 genes in pCM1 and pCM2, respectively. Among these 91 genes, only 17 genes were annotated (Supplementary Table 2).
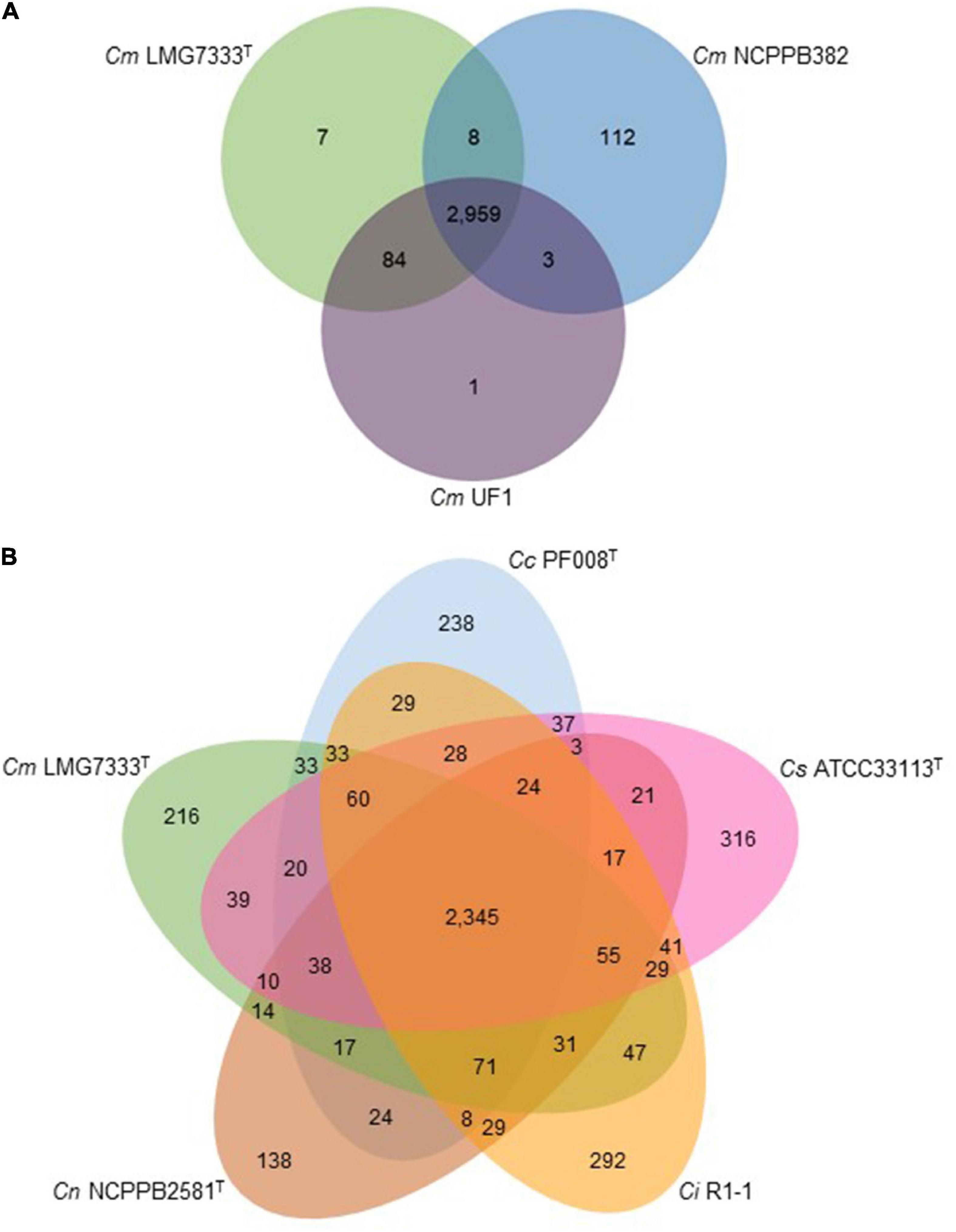
Figure 5. Comparative genome analysis of C. michiganensis LMG7333T and other Clavibacter strains. (A) Venn diagram of three C. michiganensis strains. (B) Venn diagram of C. michiganensis LMG7333T and four other type strains of Clavibacter species. Numbers represent gene numbers, which are common or unique to each strain. Cm, C. michiganensis; Cc, C. capsici; Cs, C. sepedonicus; Ci, C. insidiosus; Cn, C. nebraskensis.
BLAST analysis with 91 genes unique to C. michiganensis LMG7333T compared with C. michiganensis NCPPB382 was conducted to identify the possible origin of these genes. Based on the analysis results, these 91 genes exhibited significant homology with diverse organisms, including other Clavibacter species (Supplementary Table 2). One gene exists in only C. michiganensis LMG7333T, and nine genes are found in only two C. michiganensis strains, LMG7333T and UF1. Twenty-seven genes were shared with other C. michiganensis strains, MSF322 and VL527 (Méndez et al., 2020). Twenty-two genes are shared with other Clavibacter bacteria, such as C. insidiosus (10), C. capsici (7), C. nebraskensis (4), and C. sepedonicus (1). The remaining 32 genes are shared with diverse organisms, 11 families, and 18 genera. Most of these genes are shared with bacteria, including four families, Actinomycetaceae (1), Cellulomonadaceae (1), Microbacteriaceae (18), and Micrococcaceae (2), which belong to the phylum Actinobacteria. In particular, the 18 genes shared with the Microbacteriaceae family, including Clavibacter genus, were shared with six genera, Microbacterium (5), Cryobacterium (1), Curtobacterium (8), Leifsonia (2), Leucobacter (1), and Rathayibacter (1). Finally, among 91 C. michiganensis LMG7333T genes, 77 genes are found only in the Microbacteriaceae family. These results indicate that the C. michiganensis LMG7333T might obtain most foreign genes from the same bacterial family.
The C. michiganensis LMG7333T genome was also compared with those of four other Clavibacter type strains. Type strains of five Clavibacter species have 2,345 core genes. Interestingly, each Clavibacter has 138–316 specific genes in their genomes (Figure 5B). Most of these genes were annotated as hypothetical proteins and categorized mostly into unknown function genes, carbohydrate transport, and metabolism as well as transcription proteins in the COG database (Supplementary Table 3). When pan-genome analysis was conducted, duplicated genes were regarded as one gene. Therefore, the total CDS number (Figure 1B) and CDS number of pan-genome analysis (Figure 5B) were slightly different.
In our previous study, C. michiganensis LMG7333T and its mutant strain were used to investigate the gene function involved in virulence (Hwang et al., 2019). For the virulence assay or mutant screening, the root-dipping method was the best way to observe typical wilting symptoms within a short period. Here, we examined the virulence of C. michiganensis LMG7333T on tomato plants in various ways. Tomato leaves infected with C. michiganensis LMG7333T using the leaf-clipping method exhibited wilting symptoms from the cut part of leaves (Figure 6A). As bacteria are directly inoculated into the plants, it was successfully spread along the leaf veins and induced wilting symptoms on the inoculated leaves. Eventually, this bacterium spread throughout the plant and caused typical wilting symptoms even on non-inoculated leaves and branches (Figure 6A). In contrast, no disease symptoms and no physiological disorders were found in buffer-treated tomato plants.
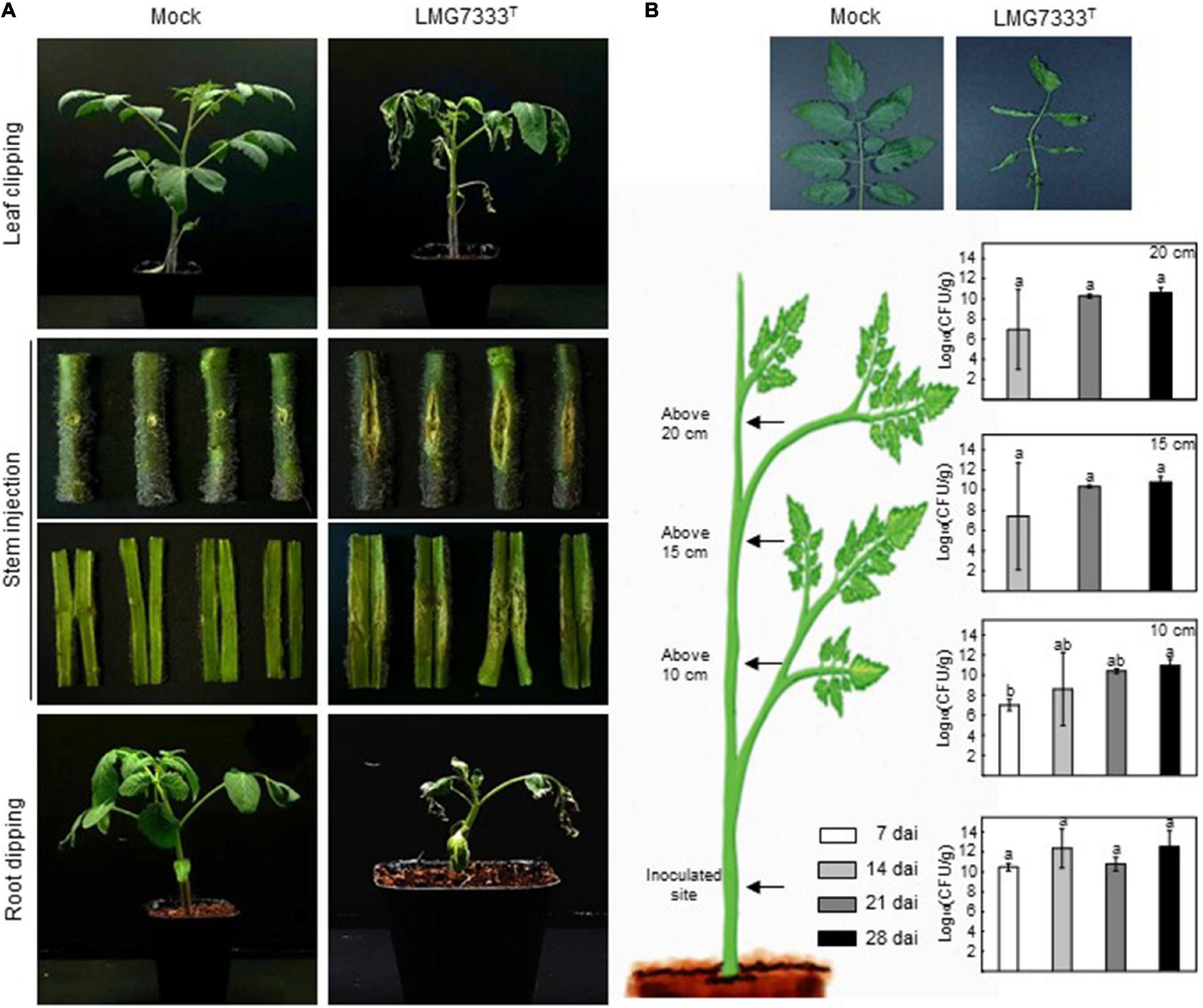
Figure 6. Virulence of C. michiganensis LMG7333T on tomato plants. (A) Typical disease symptoms of C. michiganensis LMG7333T, according to inoculation methods. Photos were taken at 30 days after inoculation (dai) for leaf clipping and stem inoculation and for root-dipping inoculation at 10 dai. (B) Bacterial numbers after stem inoculation at the indicated time points. The graphs from bottom to top indicate cfu/g at the inoculation site, 10, 15, and 20 cm above site from the inoculation point. Error bars indicate standard deviation (n = 3). The different letters indicate statistically significant differences as determined by Duncan’s multiple range test (p < 0.05).
Stem inoculation after artificial wounding, which is a conventional method for studying C. michiganensis, induced large canker lesions at the site of inoculation (Figure 6A). The inoculated bacteria were gradually spread along the xylem toward the tip, and approximately from 21 dai, leaves on the tomato tip exhibited wilting symptoms (Figure 6B). Movement of C. michiganensis LMG7333T within the stem was confirmed by bacterial growth in planta. From 7 dai, bacteria were detected at 10 cm above the inoculation site, and from 14 dai, it was isolated at the topper site of the stem, indicating that C. michiganensis LMG7333T could move and colonize in areas higher than the inoculated site (Figure 6B). Altogether, typical bacterial cankers at the site of injection as well as wilting symptoms at the branch and leaves were observed in stem-inoculated tomato plants.
Disease symptoms through this bacterial infection were also caused by the root-dipping methods (Figure 6A). Although this method has not been widely used, disease symptoms were shown within 10 days by the root-dipping method in 8–10-day-old tomato plants. Because this pathogen can enter the tomato plant through its roots during natural infection, the virulence or pathogenicity assay in tomato seedlings by root infection may be more significant than the stem inoculation (Figure 6A). The observation that C. michiganensis LMG7333T infection in tomato plants resulted in typical disease symptoms by three different inoculation methods indicates that this pathogen with the completed whole genome sequence might be valuable as a standard strain for C. michiganensis study and its interaction with the plants.
So far, genome sequences of many Clavibacter bacteria have been reported and used for understanding these bacteria at the genomic level. For C. michiganensis, the complete whole genome sequence of C. michiganensis strain NCPPB382 was first reported (Gartemann et al., 2008), and this genomic information has provided the opportunity to reveal many putative virulence genes in a PAI region, such as the chp, ppa, and pel gene families, and also the presence of two plasmids, pCM1 and pCM2. Since the NCPPB382 genome sequence became available, many draft genome sequences of C. michiganensis strains have been reported (Thapa et al., 2017; Méndez et al., 2020). Genome sequencing of C. michiganensis type strain C. michiganensis LMG7333T was previously conducted with PacBio-SMRT sequencing technology, and its draft genome sequence was reported (Li and Yuan, 2017). However, this report showed five contigs. Because this genome sequence was not complete, there was a limitation in using it as a reference genome of C. michiganensis species to understand the genetic diversity in either chromosome or plasmids among strains of this species. In this study, for completion of genome sequencing of type strain LMG7333T, we combined two sequencing platforms, PacBio RSII and Illumina HiSeqXten, and could successfully obtain the complete genome sequence with three contigs, one chromosome and two plasmids (Figure 1). This could make it possible to perform comparative genomic analyses among strains within C. michiganensis species and among other Clavibacter species.
Comparative genomic analyses revealed that type strain LMG7333T has a genome composition similar to NCPPB382, one chromosome and two plasmids. Overall G + C content and the presence of a PAI region and major virulence genes such as celA and pat-1 were also similar. However, the plasmid size and gene contents were different between the two strains: only 39 and 55% of plasmid DNA of pCM1 and pCM2, respectively, were identical. These findings indicate that high genetic diversity is more present in plasmids than in a chromosome (almost 100% identical between two strains). Moreover, some genes, such as phpA and phpB genes in pCM2 of NCPPB382, which are pat-1 paralogs, were missing in pCM2 of type strains LMG7333T (Figure 3), indicating that these two strains might use different virulence factors interaction with host plants.
Among C. michiganensis, of which their genome sequences are available in the public genome database, the UF1 strain was most similar to C. michiganensis LMG7333T (Figures 1, 3). Only 4 and 15 genes within the whole genome were specific to the UF1 strain and a type strain LMG7333T, respectively (Figure 5). Based on the genome comparison, the UF1 strain might be pathogenic although its pathogenicity has not been reported. Moreover, the genetic diversity between type strain LMG7333T and NCPPB382 was bigger because 91 and 115 genes were specific to each strain, respectively. More analysis of 91 type strain LMG7333T-specific genes showed that most of them encode hypothetical proteins, and their homologous are primarily present in bacteria belonging to Microbacteriaceae family, including the genus Clavibacter. This might indicate that gene exchange or transfer among bacteria in this family occurs.
As expected, the genetic difference among Clavibacter species was bigger than that of strains within the single species, C. michiganensis (Figure 5). The number of core genes among five Clavibacter species is 2,345, whereas its number among strains of C. michiganensis is 2,959. Moreover, in each species, the number of unique genes ranged from 138 to 316. Notably, among C. michiganensis-specific genes, tomA encodes tomatinase. Although the function of these genes is shown to be critical to C. michiganensis virulence (Kaup et al., 2005), this gene has been defined as a virulence factor in C. michiganensis. Seipke and Loria (2008) show that Streptomyces scabies have tomatinase, and it is related to plant-microbe interactions rather than the pathogenicity of bacterial pathogen, implying that tomA in C. michiganensis might affect the interaction with host plants. There are many genes specific to each Clavibacter species (Figure 5B and Supplementary Table 3). However, many of them encode hypothetical proteins with unknown functions. The function of these genes remains determined.
Infection with C. michiganensis LMG7333T induced typical disease symptoms, canker, and wilting on tomato plants (Figure 6) as observed on tomato plants infected with other pathogenic C. michiganensis. This means that type strain LMG7333T is also fully pathogenic, and it can be used to understand C. michiganensis–host plant interactions. Furthermore, the availability of the complete genome information of type strain LMG7333T would make this strain more useful for studies to reveal virulence genes and understand virulence mechanisms against host plants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
E-JO, IP, IH, and C-SO: investigation, writing—review and editing. E-JO, IP, and IH: methodology and formal analysis. C-SO: validation, project administration, and funding acquisition. All authors have read and agreed to the submitted version of the manuscript.
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (2019R1A2C2004568).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.793345/full#supplementary-material
Bae, C., Oh, E.-J., Lee, H.-B., Kim, B.-Y., and Oh, C.-S. (2015). Complete genome sequence of the cellulase-producing bacterium Clavibacter michiganensis PF008. J. Biotechnol. 214, 103–104. doi: 10.1016/j.jbiotec.2015.09.021
Bardou, P., Mariette, J., Escudié, F., Djemiel, C., and Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. doi: 10.1186/1471-2105-15-293
Bentley, S. D., Corton, C., Brown, S. E., Barron, A., Clark, L., Doggett, J., et al. (2008). Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J. Bacteriol. 190, 2150–2160. doi: 10.1128/JB.01598-07
Burger, A., Gräfen, I., Engemann, J., Niermann, E., Pieper, M., Kirchner, O., et al. (2005). Identification of homologues to the pathogenicity factor Pat-1, a putative serine protease of Clavibacter michiganensis subsp. michiganensis. Microbiol. Res. 160, 417–427. doi: 10.1016/j.micres.2005.03.006
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carlson, R. R., and Vidaver, A. K. (1982). Taxonomy of Corynebacterium plant pathogens, including a new pathogen of wheat, based on polyacrylamide gel electrophoresis of cellular proteins. Int. J. Syst. Evol. Microbiol. 32, 315–326. doi: 10.1099/00207713-32-3-315
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Davis, M. J., Gillaspie, A. G. Jr., Vidaver, A. K., and Harris, R. W. (1984). Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Evol. Microbiol. 34, 107–117. doi: 10.1099/00207713-34-2-107
Dreier, J., Meletzus, D., and Eichenlaub, R. (1997). Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol. Plant Microbe Interact. 10, 195–206. doi: 10.1094/MPMI.1997.10.2.195
Eichenlaub, R., and Gartemann, K.-H. (2011). The Clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 49, 445–464. doi: 10.1146/annurev-phyto-072910-095258
Eichenlaub, R., Gartemann, K.-H., and Burger, A. (2007). “Clavibacter michiganensis, a group of Gram-positive phytopathogenic bacteria,” in Plant-associated bacteria, ed. S. S. Gnanamanickam (Dordrecht: Springer), 385–421. doi: 10.1007/978-1-4020-4538-7_12
Gartemann, K.-H., Abt, B., Bekel, T., Burger, A., Engemann, J., Flügel, M., et al. (2008). The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190, 2138–2149. doi: 10.1128/JB.01595-07
Gonzalez, A. J., and Trapiello, E. (2014). Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int. J. Syst. Evol. Microbiol. 64, 1752–1755. doi: 10.1099/ijs.0.058099-0
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hwang, I. S., Lee, H. M., Oh, E. J., Lee, S., Heu, S., and Oh, C. S. (2020a). Plasmid composition and the chpG gene determine the virulence level of Clavibacter capsici natural isolates in pepper. Mol. Plant Pathol. 21, 808–819. doi: 10.1111/mpp.12932
Hwang, I. S., Oh, E.-J., and Oh, C.-S. (2020b). Transcriptional Changes of Plant Defense-Related Genes in Response to Clavibacter Infection in Pepper and Tomato. Plant Pathol. J. 36, 450–458. doi: 10.5423/PPJ.OA.07.2020.0124
Hwang, I. S., Oh, E.-J., Lee, H. B., and Oh, C.-S. (2019). Functional characterization of two cellulase genes in the Gram-positive pathogenic bacterium Clavibacter michiganensis for wilting in tomato. Mol. Plant Microbe Interact. 32, 491–501. doi: 10.1094/MPMI-08-18-0227-R
Jahr, H., Dreier, J., Meletzus, D., Bahro, R., and Eichenlaub, R. (2000). The endo-β-1, 4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant Microbe Interact. 13, 703–714. doi: 10.1094/MPMI.2000.13.7.703
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kaup, O., Gräfen, I., Zellermann, E.-M., Eichenlaub, R., and Gartemann, K.-H. (2005). Identification of a tomatinase in the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382. Mol. Plant Microbe Interact. 18, 1090–1098. doi: 10.1094/MPMI-18-1090
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lee, I., Kim, Y. O., Park, S.-C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, X., Tambong, J., Yuan, K. X., Chen, W., Xu, H., Lévesque, C. A., et al. (2018). Re-classification of Clavibacter michiganensis subspecies on the basis of whole-genome and multi-locus sequence analyses. Int. J. Syst. Evol. Microbiol. 68, 234–240. doi: 10.1099/ijsem.0.002492
Li, X. S., and Yuan, X. K. (2017). Genome sequences for multiple Clavibacter strains from different subspecies. Genome Announc. 5, e00721–17. doi: 10.1128/genomeA.00721-17
Lu, Y., Samac, D. A., Glazebrook, J., and Ishimaru, C. A. (2015). Complete genome sequence of Clavibacter michiganensis subsp. insidiosus R1-1 using PacBio single-molecule real-time technology. Genome Announc. 3, e00396–15. doi: 10.1128/genomeA.00396-15
Manzer, F., and Genereux, H. (1981). “Ring rot,” in Compendium of potato disease, ed. W. J. Hooker (Saint Paul, MN: American Phytopathological Society Press), 31–32.
McCulloch, L. (1925). Aplanobacter insidiosum n. sp., the cause of an Alfalfa disease. Phytopathology 15, 496–497.
Méndez, V., Valenzuela, M., Salvà-Serra, F., Jaén-Luchoro, D., Besoain, X., Moore, E. R., et al. (2020). Comparative genomics of pathogenic Clavibacter michiganensis subsp. michiganensis strains from Chile reveals potential virulence features for tomato plants. Microorganisms 8:1679. doi: 10.3390/microorganisms8111679
Monteiro-Vitorello, C. B., Zerillo, M. M., Van Sluys, M.-A., Camargo, L. E. A., and Kitajima, J. P. (2013). Complete genome sequence of Leifsonia xyli subsp. cynodontis strain DSM46306, a gram-positive bacterial pathogen of grasses. Genome Announc. 1, e00915–13. doi: 10.1128/genomeA.00915-13
Oh, E.-J., Bae, C., Lee, H.-B., Hwang, I. S., Lee, H.-I., Yea, M. C., et al. (2016). Clavibacter michiganensis subsp. capsici subsp. nov., causing bacterial canker disease in pepper. Int. J. Syst. Evol. Microbiol. 66, 4065–4070. doi: 10.1099/ijsem.0.001311
Osdaghi, E., Rahimi, T., Taghavi, S. M., Ansari, M., Zarei, S., Portier, P., et al. (2020). Comparative genomics and phylogenetic analyses suggest several novel species within the genus Clavibacter, including nonpathogenic tomato-associated strains. Appl. Environ. Microbiol. 86, e02873–19. doi: 10.1128/AEM.02873-19
Peritore-Galve, F. C., Tancos, M. A., and Smart, C. D. (2020). Bacterial canker of tomato: revisiting a global and economically damaging seedborne pathogen. Plant Dis. 105, 1581–1595. doi: 10.1094/PDIS-08-20-1732-FE
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seipke, R. F., and Loria, R. (2008). Streptomyces scabies 87-22 possesses a functional tomatinase. J. Bacteriol. 190, 7684–7692. doi: 10.1128/JB.01010-08
Stork, I., Gartemann, K. H., Burger, A., and Eichenlaub, R. (2008). A family of serine proteases of Clavibacter michiganensis subsp. michiganensis: chpC plays a role in colonization of the host plant tomato. Mol. Plant Pathol. 9, 599–608. doi: 10.1111/j.1364-3703.2008.00484.x
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tambong, J. T. (2017). Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS One 12:e0172295. doi: 10.1371/journal.pone.0172295
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Thapa, S. P., Pattathil, S., Hahn, M. G., Jacques, M.-A., Gilbertson, R. L., and Coaker, G. (2017). Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a Gram-positive bacterial pathogen. Mol. Plant Microbe Interact. 30, 786–802. doi: 10.1094/MPMI-06-17-0146-R
Van der Wolf, J., Elphinstone, J., Stead, D., Metzler, M., Müller, P., Hukkanen, A., et al. (2005). Epidemiology of Clavibacter michiganensis subsp. sepedonicus in relation to control of bacterial ring rot. Report no. 95. Wageningen: Plant Research International,
Vidaver, A. K., and Mandel, M. (1974). Corynebacterium nebraskense, a New, Orange-Pigmented Phytopathogenic Species1. Int. J. Syst. Evol. Microbiol. 24, 482–485. doi: 10.1099/00207713-24-4-482
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Xu, X., Rajashekara, G., Paul, P. A., and Miller, S. A. (2012). Colonization of tomato seedlings by bioluminescent Clavibacter michiganensis subsp. michiganensis under different humidity regimes. Phytopathology 102, 177–184. doi: 10.1094/PHYTO-03-11-0090
Yasuhara-Bell, J., and Alvarez, A. M. (2015). Seed-associated subspecies of the genus Clavibacter are clearly distinguishable from Clavibacter michiganensis subsp. michiganensis. Int. J. Syst. Evol. Microbiol. 65, 811–826. doi: 10.1099/ijs.0.000022
Załuga, J., Stragier, P., Baeyen, S., Haegeman, A., Van Vaerenbergh, J., Maes, M., et al. (2014). Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics 15:392. doi: 10.1186/1471-2164-15-392
Keywords: Clavibacter michiganensis, comparative genomics, complete genome sequence, Gram-positive bacteria, type strain
Citation: Oh E-J, Hwang IS, Park IW and Oh C-S (2022) Comparative Genome Analyses of Clavibacter michiganensis Type Strain LMG7333T Reveal Distinct Gene Contents in Plasmids From Other Clavibacter Species. Front. Microbiol. 12:793345. doi: 10.3389/fmicb.2021.793345
Received: 12 October 2021; Accepted: 16 December 2021;
Published: 01 February 2022.
Edited by:
Ralf Koebnik, Plant Health Institute, FranceReviewed by:
Mohammad Arif, University of Hawai’i at MānoaCopyright © 2022 Oh, Hwang, Park and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Sik Oh, Y28zNUBraHUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.