
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 December 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.789550
Pseudomonas aeruginosa chronically infects in the lungs of people with cystic fibrosis and other forms of lung disease. Infections are treated with antibiotics, but over time, the bacteria acquire mutations that reduce their antibiotic susceptibility. The effects of inhibitory amounts of antibiotics in selecting for antibiotic-resistant mutants have been well studied. However, the concentrations of antibiotics that reach infecting bacteria can be sub-inhibitory and but may nonetheless promote emergence of antibiotic-resistant bacteria. Therefore, the aim of this research was to investigate the effects of sub-inhibitory concentrations of antibiotics on the antibiotic susceptibility of P. aeruginosa. Two P. aeruginosa reference strains, PAO1 and PA14, and six isolates from individuals with cystic fibrosis were studied. The bacteria were passaged in the presence of antibiotics (ceftazidime, ciprofloxacin, meropenem or tobramycin) at sub-inhibitory amounts. Fifteen populations of bacteria (up to five per strain) were exposed to each of the four antibiotics. Antibiotic susceptibility was determined following 10 passages on agar supplemented with antibiotic and compared with susceptibility prior to antibiotic exposure. Antibiotic exposure resulted in susceptibility being significantly (>2-fold) reduced for 13 of the 60 populations. Seven samples had reduced susceptibility to ciprofloxacin, three to tobramycin, two to ceftazidime and one to meropenem. Whole-genome sequencing revealed the mutations arising following antibiotic exposure. Mutants with reduced antibiotic susceptibility had mutations in genes known to affect antibiotic resistance, including regulators of efflux pumps (mexR, mexS, mexZ and nalC) and the fusA1 gene that is associated with aminoglycoside resistance. Genes not previously associated with resistance, including gacS, sigX and crfX and two genes with no known function, were also mutated in some isolates with reduced antibiotic susceptibility. Our results show that exposure to sub-inhibitory amounts of antibiotics can select for mutations that reduce the susceptibility of P. aeruginosa to antibiotics and that the profile of mutations is different from that arising during selection with inhibitory antibiotic concentrations. It is likely that exposure to sub-inhibitory amounts of antibiotics during infection contributes to P. aeruginosa becoming antibiotic-resistant.
Pseudomonas aeruginosa is a widespread environmental bacterium that commonly infects the lungs of individuals with cystic fibrosis (CF) and other forms of lung disease (Garcia-Clemente et al., 2020). By adulthood, the majority of individuals with CF will have chronic P. aeruginosa respiratory infections (Smith et al., 2017; Malhotra et al., 2019). Acquisition of P. aeruginosa is associated with increased occurrence of sudden worsening of pulmonary symptoms (exacerbations), accelerated rate of lung function decline and decreased quality of life and reduced life expectancy (Elborn, 2016; Malhotra et al., 2019; Garcia-Clemente et al., 2020). Ongoing treatment with a range of antibiotics is used to improve lung function and limit the number of pulmonary exacerbations related to P. aeruginosa infections (Doring et al., 2012; Langan et al., 2015; Smith et al., 2018). The most commonly used antibiotics include aminoglycosides, such as tobramycin that inhibit protein synthesis, fluoroquinolones, such as ciprofloxacin that inhibit DNA synthesis, and β-lactams, such as ceftazidime and meropenem that inhibit cell wall synthesis (Smith et al., 2017). Although antibiotics are administered in amounts sufficient to inhibit bacterial growth in vitro, antibiotic effectiveness can be reduced within the infected airways due to a range of pharmacokinetic and pharmacodynamic factors including lung damage, blockage of airways and consequent poor oxygenation, co-morbidities and additional treatments that alter absorption and bioavailability of antibiotics (Akkerman-Nijland et al., 2021). These factors, combined with the tendency of P. aeruginosa to form biofilms in which the bacterial cells, are encased in a polysaccharide matrix, reduce antibiotic access to the target bacteria and potentially result in exposure of infecting P. aeruginosa to sub-lethal doses of antibiotics (Rogers et al., 2011; Maurice et al., 2018; Malhotra et al., 2019; Akkerman-Nijland et al., 2021). Exposure of bacteria to sub-lethal amounts of antibiotic has potential to result in the emergence of antibiotic resistance (Wistrand-Yuen et al., 2018).
Once long-term infections are established, antibiotic treatment is rarely sufficient to eradicate P. aeruginosa from the airways of individuals with CF. During long-term infections, P. aeruginosa becomes increasingly antibiotic-resistant, enhancing its survival (Sherrard et al., 2014; Smith et al., 2017). To better understand the genetic changes that lead to resistance, several research groups have exposed antibiotic-sensitive reference strains of P. aeruginosa to lethal concentrations of antibiotics and selected resistant mutants. Whole-genome sequencing (WGS) of the mutants identified the underlying resistance-causing mutations [Feng et al., 2016, Barbosa et al., 2017; Jorth et al., 2017; Melnyk et al., 2017; Yen and Papin, 2017; Sanz-Garcia et al., 2018a,b; Wardell et al., 2019; reviewed in (Lopez-Causape et al., 2018a)]. Mutations typically arose in genes that had previously been associated with resistance in clinical settings, such as oprD (carbapenem resistance) and gyrAB (fluoroquinolone resistance), and also in previously unrecognised resistance-associated genes, such as ftsI (carbapenem resistance) and fusA1 (aminoglycoside resistance).
However, much less is known about the effects of low-level sub-inhibitory amounts of antibiotics on the genome of P. aeruginosa even though the bacteria likely encounter this situation during chronic infection. There is evidence that sub-lethal doses of antibiotics can increase the antimicrobial resistance of P. aeruginosa (Jorgensen et al., 2013; Wright et al., 2013; Ahmed et al., 2018, 2020; Moore et al., 2021). However, these studies have largely been limited to use of a single antibiotic (ciprofloxacin or in one case tobramycin) in conjunction with reference strains of P. aeruginosa rather than isolates from chronically infected patients. There has also been only limited investigation of the mutations arising following exposure of bacteria to sub-inhibitory amounts of antibiotics.
Fully understanding the effects of sub-lethal amounts of antibiotics on P. aeruginosa is of high clinical importance as incorrect treatment may facilitate emergence of antibiotic-resistant P. aeruginosa within the airways. Consequently, the aims of this research were to determine how often exposure to antibiotics at sub-lethal concentrations increases the amount of antibiotic that can be tolerated by these bacteria and to identify genetic changes associated with any changes in resistance phenotype.
Two well-characterised reference strains (PAO1 from our laboratory collection and PA14, kindly provided by Dr. Andrea Battistoni [University of Rome Tor Vergata, Rome, Italy]; Stover et al., 2000; Lee et al., 2006) and isolates of P. aeruginosa from sputum samples of six adults with CF (Table 1), collected under the approval of the New Zealand Health and Disability Ethics Committees (NTY/10/12/106), have been described previously (Martin et al., 2018; Freschi et al., 2019; Wardell et al., 2021). The selected isolates were sensitive to all of the antibiotics used in this study.
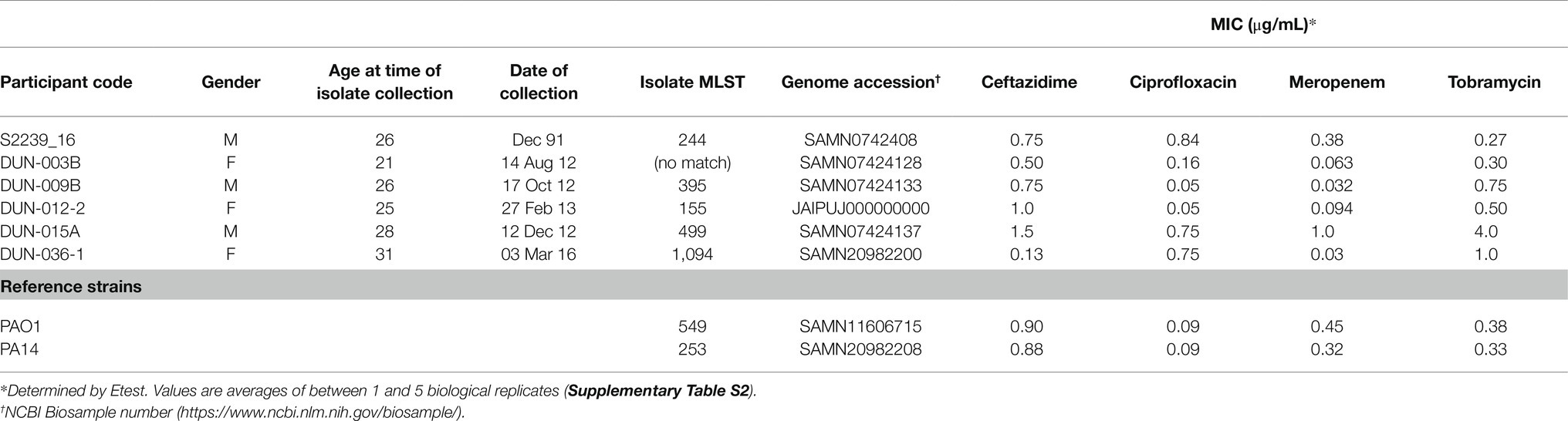
Table 1. Demographic details of Pseudomonas aeruginosa isolates obtained from people with cystic fibrosis.
Ciprofloxacin (Ciplox), ceftazidime and tobramycin (Mylan New Zealand Ltd) and meropenem (Penembact, Venus Remedies Limited) were used. Overnight cultures grown in Luria Bertani broth (LB broth; Miller, 1972) were adjusted to OD600nm 0.01 and spot inoculated onto Mueller-Hinton (MH) agar plates (Difco) supplemented with doubling concentrations of antibiotic. The SIC was defined as being ¼ of the lowest antibiotic concentration that inhibited visible growth after overnight incubation at 37°C (Supplementary Table S1).
Antibiotic sensitivity testing was carried out using ETEST strips (bioMérieux, North Ryde, Australia) in accordance with the manufacturer’s instructions. Briefly, bacteria from an overnight culture grown in LB broth were diluted to OD600nm 0.1 in 0.9% normal saline (NaCl) and inoculated onto an LB agar plate. ETEST strips were added and plates incubated at 37°C for 18 h under aerobic conditions. The minimum inhibitory concentration (MIC) was determined at the point where the inhibition ellipse intersected the side of the antibiotic strip. In some cases, replicate tests were undertaken to confirm reproducibility.
Bacteria underwent 10 passages in the presence of antibiotic at SIC concentrations. In a subset of experiments, half or twice the SIC concentration was used. Briefly, a single colony inoculum in antibiotic-free LB broth was cultured overnight and adjusted to OD600nm 0.1. An MH plate was surface flooded with a 750 μl aliquot of the prepared inoculum and excess culture removed. Following aerobic incubation (37°C, overnight), bacteria were harvested from the plate and used to inoculate antibiotic-free LB broth which was incubated overnight. This process was repeated for 10 passages. Four strains were also passaged 10 times under antibiotic-free conditions to assess any effect of passaging itself on changes to MIC or occurrence of genetic mutations.
Genome assemblies for isolates S2239, DUN003B, DUN009B, DUN012-2 and DUN015A (Freschi et al., 2015) were generously provided by Prof Roger Levesque and co-workers. WGS of DUN036-1, the reference strains and passaged bacteria and identification of mutations was carried out as described previously (Wardell et al., 2019). Briefly, extracted DNA (MoBio UltraClean® Microbial DNA isolation kit) underwent sequencing on the Illumina MiSeq platform with a minimum of 40-fold coverage across each genome. Raw sequence reads were trimmed using Trimmomatic version 0.36 (Bolger et al., 2014). Draft genome assemblies were created using SPAdes version 3.12.0 (Prjibelski et al., 2020) with the careful flag enabled. Draft genome assemblies were ordered relative to P. aeruginosa PAO1 using mauve version 2.4.0 (Darling et al., 2010) and then annotated using Prokka version 1.13 (Seemann, 2014). The resulting annotated draft genomes were used as a reference for mapping the sequence reads of passaged bacteria using Breseq version 0.35.0 (Deatherage and Barrick, 2014), allowing for contig reference (−c flag) and polymorphism mode enabled (−p flag). MLST types were determined from the genome sequences using pubMLST.1 For strain PA14, the reference genome was derived from the refseq genome sequence (NC 008463) using GDtools version 0.30 (Deatherage and Barrick, 2014) to incorporate sequence differences in the isolate used in this study. The genome of the reference strain PAO1 used in this study was previously derived from the refseq genome sequence (NC 002516) in the same way (Wardell et al., 2019).
All raw sequencing reads from mutants generated in this study are available under SRA BioProject PRJNA757894.
Five biological replicates of the laboratory reference strain PAO1 were passaged in the presence of sub-inhibitory concentrations (SIC) of each of four antibiotics. There were no significant changes in the MIC following passaging with tobramycin, ceftazidime or meropenem, where a significant change was defined as being a difference of more than one doubling dilution (> 2-fold; Brennan-Krohn et al., 2017; Table 2). In contrast, the MIC increased 4-fold or greater for four of five replicates passaged in the presence of SIC of ciprofloxacin.
For comparison, we passaged biological duplicates of reference strain PA14 in the presence of SIC antibiotics. As with strain PAO1, there was no significant change in the MIC for ceftazidime, meropenem or tobramycin. However, there was an increase of 10.6-fold in the MIC in one of the duplicates passaged with ciprofloxacin.
To determine whether these findings were applicable to clinical isolates of P. aeruginosa, six susceptible isolates from respiratory infections of people with CF were passaged under SIC antibiotic conditions. The initial isolates studied, S2339_16 and DUN-003B, were tested in duplicate. In contrast to the reference strains, neither S2239_16 replicate showed a significant increase in MIC when passaged in the presence of ciprofloxacin, whereas there was a 3.5-fold increase for one of the DUN-003B replicates. In addition, there were slight increases in the MIC for tobramycin for each S2239_16 replicate (2.4- and 2.8-fold) and one replicate of strain DUN-003B (2.5-fold). One DUN-003B replicate had a 3.5-fold increase in the MIC following passaging in the presence of ceftazidime.
Passaging of an additional four isolates (DUN-009B, DUN-012-2, DUN-015A and DUN-036-1) was undertaken to further assess the impact of antibiotics at SIC levels on clinical isolates. Similar to the results for the initial two clinical isolates, exposure to antibiotics at SIC generally had no significant impact on the MIC. As exceptions, exposure to ceftazidime, ciprofloxacin and meropenem resulted in a 2.7–16- and 6.3-fold increase in the MIC for one replicate of DUN-015A, DUN-009B and DUN-036-1, respectively.
In total, 13 of the 60 passaged cultures had a significant (>2-fold) increase in MIC. Most of these were for ciprofloxacin (n = 7), followed by tobramycin (n = 3), ceftazidime (n = 2) and meropenem (n = 1). None of the increases in MIC were sufficiently high to cause the bacteria to be classified as clinically resistant (CLSI, 2018).
Passaging under antibiotic-free conditions was performed on a subset of strains and isolates (PAO1, PA14, DUN-003B and S2239_16), to determine whether passaging alone resulted in a change to MIC. None of the passaged bacteria had a significant change in MIC (Supplementary Table S3).
In a small number of experiments, passaging was carried out with half or double the SIC. In one of these experiments, half of the SIC resulted in an increased MIC for tobramycin in isolate DUN-003B. The MIC did not change in other experiments where passaging was carried out with different amounts of antibiotic (Table 2).
Bacteria that had a > 2-fold increase in MIC following passaging, and a selection of bacteria that did not, underwent WGS to identify any changes to the genome (Table 2, Supplementary Table S2).
All five of the reference strain replicates that had an increased MIC for ciprofloxacin had mutations in genes that control efflux pump activity (mexZ, mexR, mexS and nalC [Table 2]), consistent with the known role of efflux pumps in contributing to ciprofloxacin resistance (Rehman et al., 2019). No changes were identified in the only clinical isolate (DUN-009B) with an increased ciprofloxacin MIC after passaging. Failure to detect a mutation may have been because the passaged bacteria comprised a mixture of strain DUN-009B and a mutant with increased MIC. In a mixture, the mutant able to tolerate a higher concentration of antibiotic would be detected in MIC testing but a genetic mutation may go undetected, due to the presence of a high proportion of wild-type allele in WGS.
Passaged bacteria with an increased MIC for ceftazidime had mutations in the sigX gene or the crfX gene. So far as we are aware, these genes have not previously been associated with antibiotic resistance. A mutation in the cmpX gene, that is operonic with crfX and likely to have a related function, reduces expression of sigX (Bhagirath et al., 2018). Conversely, SigX upregulates the operon containing cmpX and crfX (Bouffartigues et al., 2020). These observations suggest a common pathway for increased ceftazidime MIC in the crfX and sigX mutants. SigX affects expression of genes encoding a variety of cellular functions including expression of the oprF gene that encodes an outer membrane porin (Bouffartigues et al., 2012) and genes involved in carbon metabolism, membrane fluidity, stress response and c-di-GMP signalling (Gicquel et al., 2013; Blanka et al., 2014; Flechard et al., 2018). How changes in SigX/CrfX alter susceptibility to ceftazidime is not yet clear.
Increased MIC following tobramycin exposure resulted in a range of mutations. Two of the three mutants with an increased MIC for tobramycin had mutations in fusA1, a known contributor to tobramycin resistance (Bolard et al., 2018; Lopez-Causape et al., 2018b; Scribner et al., 2020). One of these mutants also had a mutation in ptsP that has also been associated with tobramycin resistance (Feng et al., 2016; Sanz-Garcia et al., 2018b; Scribner et al., 2020). The third mutant had a mutation in the gacS global regulator gene, that controls a wide variety of cellular functions and intriguingly is influenced by the cmpX gene (Bhagirath et al., 2018) although its relationship with tobramycin resistance is not clear.
Only one mutant with an increased MIC was obtained following passaging of a clinical isolate in the presence of meropenem. This mutant had mutations in two genes, both of unknown function and neither of which have previously been associated with meropenem resistance.
Derivatives of clinical isolates S2239_16 and DUN-003B had mutations in genes (mucA, mucD and algU) associated with alginate biosynthesis following passaging in the presence of antibiotics. Clinical isolates frequently undergo changes in mucoidy that are associated with altered alginate production during subculture (Govan and Deretic, 1996). Indeed, two replicates of isolate S2239_16 passaged in the presence of ceftazidime had mutations in mucA, with no change in MIC (Table 2) and two replicates of the same isolate passaged in the absence of antibiotic also had mutations in mucA (data not shown). It is therefore likely that the mucA mutations are associated with adaptation of the bacteria to growth under laboratory conditions.
In chronic lung infection, the physiology of the airways, changes resulting from disease progression, and the biofilm growth of P. aeruginosa all inhibit delivery of antibiotics to the bacterial cells (Maurice et al., 2018; Akkerman-Nijland et al., 2021). It is therefore difficult to determine the dosage of antibiotics experienced by P. aeruginosa during infection in the airways of people with lung disease, but as antibiotics usually fail to clear established chronic infection, it is likely that the bacteria are exposed to SIC amounts. Furthermore, exposure to SIC amounts of antibiotic can increase bacterial ability to resist antibiotics (Wistrand-Yuen et al., 2018) and this likely causes the increased antibiotic resistance associated with prolonged infection (Sherrard et al., 2014; Smith et al., 2017). We therefore investigated the effects of prolonged antibiotic exposure at SIC. A total of 60 passaging experiments were carried out across four antibiotics and eight isolates of P. aeruginosa, and there was a significant increase in MIC in 13 of the passaged lines, showing that SIC exposure can increase ability of the bacteria to tolerate antibiotics. This finding is consistent with previous studies that indicated that exposure to antibiotics at SIC can impact the amounts of antibiotic that can be tolerated by P. aeruginosa (Jorgensen et al., 2013; Wright et al., 2013; Ahmed et al., 2018, 2020; Moore et al., 2021).
The effects of antibiotic exposure varied between isolates and between antibiotics. With the reference strains PAO1 and PA14, only ciprofloxacin exposure resulted in higher MIC, whereas across the 6 clinical isolates, all four antibiotics resulted in at least one derivative with a higher MIC. This difference may be the result of genetic or biological differences between isolates from people with CF and the reference strains PAO1 and PA14 that were initially derived from acute infections. Sub-MIC antibiotics can induce stress responses that increase the frequency of mutations (Blazquez et al., 2018), and stress responses may vary between isolates, and with different antibiotics, influencing the frequency of mutations. MIC has been reported to increase following exposure of strain PAO1 to SIC ciprofloxacin (Ahmed et al., 2018, 2020), of isolates from CF to SIC tobramycin (Moore et al., 2021), and of epidemic strain LESB58 to a range of antibiotics (Wright et al., 2013). More extensive analysis is required to determine whether there is a genuine difference between the clinical isolates and the PAO1 and PA14 reference strains. The strains and isolates used in this study were sensitive to all of the antibiotics, and none of the mutants that arose during passaging had MICs that would be classified as being resistant (CLSI, 2018). More extensive passaging, including passaging of the mutant bacteria generated here, may well lead to clinical resistance (Jorgensen et al., 2013; Moore et al., 2021).
Whole-genome sequencing identified genetic changes underlying increases in MIC. Mutations had occurred in a number of genes associated with antibiotic resistance. For example, mutations in the fusA1 gene are frequently associated with tobramycin resistance (Bolard et al., 2018; Wardell et al., 2019). Mutations also occurred in genes mexR and nalC that encode direct and indirect regulators of the MexAB-OprM efflux pump (Poole et al., 1996; Cao et al., 2004), mexZ that encodes a repressor of the MexXY-OprM efflux pump (Morita et al., 2012) and mexS that encodes a putative oxidoreductase that influences production of the MexEF-OprN efflux pump (Morita et al., 2015). Overexpression of MexAB-OprM reduces susceptibility to ciprofloxacin and β-lactam antibiotics, overexpression of MexXY-OprM to aminoglycosides and fluoroquinolones and overexpression of MexEF-OprN to fluoroquinolones (Lopez-Causape et al., 2018b).
A number of passaged bacteria had mutations in mucA, mucD or algU, genes that are associated with the production of alginate and the development of mucoidy, a hallmark of chronic infections (Govan and Deretic, 1996). Others have also reported that mutations in algU are associated with altered alginate production and biofilm architecture and a decrease in susceptibility of P. aeruginosa to aminoglycosides (Hentzer et al., 2001). However, it should be noted that mutations arose in mucA in isolate S2239_16 that did not result in an increased MIC and were likely selected as part of the passaging process, independent of the presence of antibiotic.
A number of mutants with increased MICs had mutations in genes not so clearly associated with resistance. One mutant with reduced susceptibility to ciprofloxacin had a pilB gene mutation. Ciprofloxacin-resistant P. aeruginosa can have mutations in pilin-encoding genes (Wardell et al., 2019; Ahmed et al., 2020), although how pilin gene mutations reduce susceptibility to ciprofloxacin is not known. Mutations in the sigX gene and in the cfrX gene, a potential regulator of sigX expression, were present in mutants arising from ceftazidime exposure. SigX contributes to a wide range of cellular properties (Bouffartigues et al., 2012; Gicquel et al., 2013; Blanka et al., 2014; Flechard et al., 2018) and how it might influence ceftazidime susceptibility is not known. Intriguingly, the SigX and AlgU sigma factors are both involved in expression of the operon containing the crfX gene (Bouffartigues et al., 2020) but it remains to be determined whether this connection extends to mechanisms of antibiotic susceptibility. Lastly, a mutant with increased MIC for tobramycin had a mutation in the gacS gene. This gene is associated with regulation of biofilm formation and control of virulence factor production (Valentini et al., 2018) but has not previously been associated with antibiotic resistance. How these genes contribute to antibiotic susceptibility phenotype will require further investigation.
Interestingly, several genes that are frequently mutated during experimental evolution in the presence of inhibitory amounts of antibiotics were not mutated following the SIC selection procedure. These include mutations associated with ciprofloxacin resistance, such as gyr and par gene mutations (Melnyk et al., 2017; Rehman et al., 2019). Similarly, resistance to meropenem and ceftazidime is often associated with mutations in the oprD and dacB genes, respectively (Lister et al., 2009; Chalhoub et al., 2016; Lopez-Causape et al., 2018b; Sanz-Garcia et al., 2018a; Wardell et al., 2019), mutations that did not arise in this study. Although relatively small numbers of mutants with increased MIC were analysed, these findings suggest that the selection that occurs during passaging with SIC amounts of antibiotic is different from that which occurs during passaging in lethal amounts of antibiotic. They also demonstrate that development of antibiotic resistance is complex and multifactorial.
While this study furthers the understanding of the antibiotic resistance of P. aeruginosa, there are some limitations. Firstly, inclusion of a larger panel of isolates would more strongly test the effects of SIC amounts of antibiotics. Secondly, isolates were passaged 10 times in the presence of SIC amounts of antibiotics and increasing the number of passages would likely have increased the proportion of bacteria with increased MIC. Lastly, sequencing of populations of passaged bacteria may have masked mutations that were present in a subset within a population. Growth of bacterial populations in the absence of antibiotic, during the passaging process, may also have reduced the proportion of bacteria containing any mutations that negatively affect fitness. Greater sequencing depth, or characterisation of multiple individual clones from each passaged population, may have identified more genetic changes.
In conclusion, our findings demonstrate that exposure to SIC levels of antibiotics can result in increased MIC of a range of P. aeruginosa strains and also identifies the underlying mutations. Many of the mutated genes have known roles in antibiotic resistance, although some of those in clinical isolates have not been previously associated with resistance. Prolonged exposure to SIC amounts of antibiotic during infection are likely to result in the emergence of antibiotic-resistant P. aeruginosa. Our findings emphasise the importance of achieving high antibiotic concentrations in the lungs of patients.
The datasets presented in this study can be found in online repositories (see Table 1). The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, JAIPUJ000000000 https://www.ncbi.nlm.nih.gov/, PRJNA757894.
KR: data curation, formal analysis, funding acquisition, investigation, supervision, validation, writing – original draft, and writing – review and editing. SM: data curation, investigation, and writing – review and editing. SW: formal analysis and writing – review and editing. IL: conceptualisation, data curation, formal analysis, project administration, supervision, writing – original draft, and writing – review and editing. All authors contributed to the article and approved the submitted version.
This work was supported by the Otago Medical Research Foundation (AG 374) and the Health Research Council of New Zealand (17/372). SW was supported by a Postgraduate Scholarship from the University of Otago.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We gratefully acknowledge Professor Roger Levesque and co-workers for providing raw sequence reads and genome assemblies for isolates.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articless/10.3389/fmicb.2021.789550/full#supplementary-material
Ahmed, M. N., Abdelsamad, A., Wassermann, T., Porse, A., Becker, J., Sommer, M. O. A., et al. (2020). The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance. NPJ Biofilms Microbiomes 6:28. doi: 10.1038/s41522-020-00138-8
Ahmed, M. N., Porse, A., Sommer, M. O. A., Hoiby, N., and Ciofu, O. (2018). Evolution of antibiotic resistance in biofilm and planktonic Pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 62, e00320–e00318. doi: 10.1128/CMR.00138-18
Akkerman-Nijland, A. M., Akkerman, O. W., Grasmeijer, F., Hagedoorn, P., Frijlink, H. W., Rottier, B. L., et al. (2021). The pharmacokinetics of antibiotics in cystic fibrosis. Expert Opin. Drug Metab. Toxicol. 17, 53–68. doi: 10.1080/17425255.2021.1836157
Barbosa, C., Trebosc, V., Kemmer, C., Rosenstiel, P., Beardmore, R., Schulenburg, H., et al. (2017). Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244. doi: 10.1093/molbev/msx158
Bhagirath, A. Y., Somayajula, D., Li, Y., and Duan, K. (2018). CmpX affects virulence in Pseudomonas aeruginosa through the Gac/Rsm signaling pathway and by modulating c-di-GMP levels. J. Membr. Biol. 251, 35–49. doi: 10.1007/s00232-017-9994-6
Blanka, A., Schulz, S., Eckweiler, D., Franke, R., Bielecka, A., Nicolai, T., et al. (2014). Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J. Bacteriol. 196, 345–356. doi: 10.1128/JB.01034-13
Blazquez, J., Rodriguez-Beltran, J., and Matic, I. (2018). Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu. Rev. Microbiol. 72, 209–230. doi: 10.1146/annurev-micro-090817-062139
Bolard, A., Plesiat, P., and Jeannot, K. (2018). Mutations in gene fusA1 as a novel mechanism of aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62, e01835–e01817. doi: 10.1128/AAC.01835-17
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouffartigues, E., Gicquel, G., Bazire, A., Bains, M., Maillot, O., Vieillard, J., et al. (2012). Transcription of the oprF gene of Pseudomonas aeruginosa is dependent mainly on the SigX sigma factor and is sucrose induced. J. Bacteriol. 194, 4301–4311. doi: 10.1128/JB.00509-12
Bouffartigues, E., Si Hadj Mohand, I., Maillot, O., Tortuel, D., Omnes, J., David, A., et al. (2020). The temperature-regulation of Pseudomonas aeruginosa cmaX-cfrX-cmpX operon reveals an intriguing molecular network involving the sigma factors AlgU and SigX. Front. Microbiol. 11:579495. doi: 10.3389/fmicb.2020.579495
Brennan-Krohn, T., Smith, K. P., and Kirby, J. E. (2017). The poisoned well: enhancing the predictive value of antimicrobial susceptibility testing in the era of multidrug resistance. J. Clin. Microbiol. 55, 2304–2308. doi: 10.1128/JCM.00511-17
Cao, L., Srikumar, R., and Poole, K. (2004). MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53, 1423–1436. doi: 10.1111/j.1365-2958.2004.04210.x
Chalhoub, H., Saenz, Y., Rodriguez-Villalobos, H., Denis, O., Kahl, B. C., Tulkens, P. M., et al. (2016). High-level resistance to meropenem in clinical isolates of Pseudomonas aeruginosa in the absence of carbapenemases: role of active efflux and porin alterations. Int. J. Antimicrob. Agents 48, 740–743. doi: 10.1016/j.ijantimicag.2016.09.012
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing Wayne, PA, USA, Clinical and Laboratory Standards Institute.
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
Deatherage, D. E., and Barrick, J. E. (2014). “Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq,” in Engineering and Analyzing Multicellular Systems: Methods and Protocols. eds. L. Sun and W. Shou (New York, NY: Springer New York), 165–188.
Doring, G., Flume, P., Heijerman, H., Elborn, J. S., and Consensus Study, G (Eds.) (2012). Treatment of lung infection in patients with cystic fibrosis: current and future strategies. J. Cyst. Fibros. 11, 461–479. doi: 10.1128/AAC.01835-17
Feng, Y., Jonker, M. J., Moustakas, I., Brul, S., and Ter Kuile, B. H. (2016). Dynamics of mutations during development of resistance by Pseudomonas aeruginosa against five antibiotics. Antimicrob. Agents Chemother. 60, 4229–4236. doi: 10.1128/AAC.00434-16
Flechard, M., Duchesne, R., Tahrioui, A., Bouffartigues, E., Depayras, S., Hardouin, J., et al. (2018). The absence of SigX results in impaired carbon metabolism and membrane fluidity in Pseudomonas aeruginosa. Sci. Rep. 8:17212. doi: 10.1038/s41598-018-35503-3
Freschi, L., Jeukens, J., Kukavica-Ibrulj, I., Boyle, B., Dupont, M. J., Laroche, J., et al. (2015). Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 6:1036. doi: 10.3389/fmicb.2015.01036
Freschi, L., Vincent, A. T., Jeukens, J., Emond-Rheault, J. G., Kukavica-Ibrulj, I., Dupont, M. J., et al. (2019). The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol. Evol. 11, 109–120. doi: 10.1093/gbe/evy259
Garcia-Clemente, M., de la Rosa, D., Maiz, L., Giron, R., Blanco, M., Olveira, C., et al. (2020). Impact of Pseudomonas aeruginosa infection on patients with chronic inflammatory airway diseases. J. Clin. Med. 9:3800. doi: 10.3390/jcm9123800
Gicquel, G., Bouffartigues, E., Bains, M., Oxaran, V., Rosay, T., Lesouhaitier, O., et al. (2013). The extra-cytoplasmic function sigma factor SigX modulates biofilm and virulence-related properties in Pseudomonas aeruginosa. PLoS One 8:e80407. doi: 10.1371/journal.pone.0080407
Govan, J. R. W., and Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539, 574. doi: 10.1128/mr.60.3.539-574.1996
Hentzer, M., Teitzel, G. M., Balzer, G. J., Heydorn, A., Molin, S., Givskov, M., et al. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001
Jorgensen, K. M., Wassermann, T., Jensen, P. O., Hengzuang, W., Molin, S., Hoiby, N., et al. (2013). Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 4215–4221. doi: 10.1128/AAC.00493-13
Jorth, P., McLean, K., Ratjen, A., Secor, P. R., Bautista, G. E., Ravishankar, S., et al. (2017). Evolved Aztreonam resistance is multifactorial and can produce Hypervirulence in Pseudomonas aeruginosa. MBio 8:e00517-17. doi: 10.1128/mBio.00517-17
Langan, K. M., Kotsimbos, T., and Peleg, A. Y. (2015). Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr. Opin. Infect. Dis. 28, 547–556. doi: 10.1097/QCO.0000000000000217
Lee, D. G., Urbach, J. M., Wu, G., Liberati, N. T., Feinbaum, R. L., Miyata, S., et al. (2006). Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. doi: 10.1186/gb-2006-7-10-r90
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. doi: 10.1128/CMR.00040-09
Lopez-Causape, C., Cabot, G., Del Barrio-Tofino, E., and Oliver, A. (2018a). The versatile mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 9:685. doi: 10.3389/fmicb.2018.00685
Lopez-Causape, C., Rubio, R., Cabot, G., and Oliver, A. (2018b). Evolution of the Pseudomonas aeruginosa aminoglycoside mutational Resistome In vitro and in the cystic fibrosis setting. Antimicrob. Agents Chemother. 62:e02583-17. doi: 10.1128/AAC.02583-17
Malhotra, S., Hayes, D. Jr., and Wozniak, D. J. (2019). Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe Interface. Clin. Microbiol. Rev. 32, 1–46. doi: 10.1128/CMR.00138-18
Martin, L. W., Robson, C. L., Watts, A. M., Gray, A. R., Wainwright, C. E., Bell, S. C., et al. (2018). Expression of Pseudomonas aeruginosa antibiotic resistance genes varies greatly during infections in cystic fibrosis patients. Antimicrob. Agents Chemother. 62, e01789–e01718. doi: 10.1128/AAC.01789-18
Maurice, N. M., Bedi, B., and Sadikot, R. T. (2018). Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 58, 428–439. doi: 10.1165/rcmb.2017-0321TR
Melnyk, A. H., McCloskey, N., Hinz, A. J., Dettman, J., and Kassen, R. (2017). Evolution of cost-free resistance under fluctuating drug selection in Pseudomonas aeruginosa. mSphere 2:e00158-17. doi: 10.1128/mSphere.00158-17
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, New York, Cold Spring Harbor Laboratory.
Moore, J. E., Millar, B. C., Ollman-Selinger, M., and Cambridge, L. (2021). The role of suboptimal concentrations of nebulized tobramycin in driving antimicrobial resistance in Pseudomonas aeruginosa isolates in cystic fibrosis. Respir. Care 66, 1446–1457. doi: 10.4187/respcare.08671
Morita, Y., Tomida, J., and Kawamura, Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408. doi: 10.3389/fmicb.2012.00408
Morita, Y., Tomida, J., and Kawamura, Y. (2015). Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front. Microbiol. 6:8. doi: 10.3389/fmicb.2015.00008
Poole, K., Tetro, K., Zhao, Q., Neshat, S., Heinrichs, D. E., and Bianco, N. (1996). Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40, 2021–2028. doi: 10.1128/AAC.40.9.2021
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., and Korobeynikov, A. (2020). Using SPAdes De Novo assembler. Curr. Protoc. Bioinformatics 70:e102. doi: 10.1002/cpbi.102
Rehman, A., Patrick, W. M., and Lamont, I. L. (2019). Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J. Med. Microbiol. 68, 1–10. doi: 10.1099/jmm.0.000873
Rogers, G. B., Hoffman, L. R., and Doring, G. (2011). Novel concepts in evaluating antimicrobial therapy for bacterial lung infections in patients with cystic fibrosis. J. Cyst. Fibros. 10, 387–400. doi: 10.1016/j.jcf.2011.06.014
Sanz-Garcia, F., Hernando-Amado, S., and Martinez, J. L. (2018a). Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either Ceftazidime or Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 62:e01379-18. doi: 10.1128/AAC.01379-18
Sanz-Garcia, F., Hernando-Amado, S., and Martinez, J. L. (2018b). Mutational evolution of Pseudomonas aeruginosa resistance to ribosome-targeting antibiotics. Front. Genet. 9:451. doi: 10.3389/fgene.2018.00451
Scribner, M. R., Santos-Lopez, A., Marshall, C. W., Deitrick, C., and Cooper, V. S. (2020). Parallel evolution of tobramycin resistance across species and environments. MBio 11:e00932-20. doi: 10.1128/mBio.00932-20
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sherrard, L. J., Tunney, M. M., and Elborn, J. S. (2014). Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384, 703–713. doi: 10.1016/S0140-6736(14)61137-5
Smith, W. D., Bardin, E., Cameron, L., Edmondson, C. L., Farrant, K. V., Martin, I., et al. (2017). Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol. Lett. 364:fnx121. doi: 10.1093/femsle/fnx121
Smith, S., Rowbotham, N. J., and Regan, K. H. (2018). Inhaled anti-pseudomonal antibiotics for long-term therapy in cystic fibrosis. Cochrane Database Syst. Rev. 3:CD001021. doi: 10.1002/14651858.CD001021.pub3
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Valentini, M., Gonzalez, D., Mavridou, D. A., and Filloux, A. (2018). Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 41, 15–20. doi: 10.1016/j.mib.2017.11.006
Wardell, S. J. T., Gauthier, L. W., Martin, J., Potvin, M., Brockway, B., Levesque, R. C., et al. (2021). Integration of genomic, transcriptomic and phenotypic analyses reveals how Pseudomonas aeruginosa adapts to the cystic fibrosis lung during a 20-year infection. Microb Genom. 7. doi: 10.1099/mgen.0.000681
Wardell, S. J. T., Rehman, A., Martin, L. W., Winstanley, C., Patrick, W. M., and Lamont, I. L. (2019). A large-scale whole-genome comparison shows that experimental evolution in response to antibiotics predicts changes in naturally evolved clinical Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63:e01619. doi: 10.1128/AAC.01619-19
Wistrand-Yuen, E., Knopp, M., Hjort, K., Koskiniemi, S., Berg, O. G., and Andersson, D. I. (2018). Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 9, 1599. doi: 10.1038/s41467-018-04059-1
Wright, E. A., Fothergill, J. L., Paterson, S., Brockhurst, M. A., and Winstanley, C. (2013). Sub-inhibitory concentrations of some antibiotics can drive diversification of Pseudomonas aeruginosa populations in artificial sputum medium. BMC Microbiol. 13:170. doi: 10.1186/1471-2180-13-170
Keywords: Pseudomonas aeruginosa, antibiotic resistance, sub-inhibitory concentration, genetic mutations, resistance mechanisms, cystic fibrosis, sub-lethal concentration
Citation: Ramsay KA, McTavish SM, Wardell SJT and Lamont IL (2021) The Effects of Sub-inhibitory Antibiotic Concentrations on Pseudomonas aeruginosa: Reduced Susceptibility Due to Mutations. Front. Microbiol. 12:789550. doi: 10.3389/fmicb.2021.789550
Received: 05 October 2021; Accepted: 05 November 2021;
Published: 20 December 2021.
Edited by:
Yuji Morita, Meiji Pharmaceutical University, JapanReviewed by:
Benno H. Ter Kuile, University of Amsterdam, NetherlandsCopyright © 2021 Ramsay, McTavish, Wardell and Lamont. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iain L. Lamont, aWFpbi5sYW1vbnRAb3RhZ28uYWMubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.