
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 January 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.789280
This article is part of the Research Topic From Genomics to Antibiotic Resistance in Emerging Pathogens View all 10 articles
Antimicrobial resistance (AMR) is a growing concern in public health, particularly for the clinically relevant extended-spectrum beta-lactamase (ESBL) and AmpC-producing Enterobacteriaceae. Studies describing ESBL-producing Escherichia coli clinical samples from Finland to the genomic level and investigation of possible zoonotic transmission routes are scarce. This study characterizes ESBL-producing E. coli from clinical samples in Finland using whole genome sequencing (WGS). Comparison is made between animal, food, and environmental sources in Finland to gain insight into potential zoonotic transmission routes and to recognize successful AMR genes, bacterial sequence types (STs), and plasmids. ESBL-producing E. coli isolates (n = 30) obtained from the Eastern Finland healthcare district between 2018 and 2020 underwent WGS and were compared to sequences from non-human and healthy human sources (n = 67) isolated in Finland between 2012 and 2018. A majority of the clinical isolates belonged to ST131 (n = 21; 70%), of which 19 represented O25:H4 and fimH30 allele, and 2 O16:H5 and fimH41 allele. Multidrug resistance was common, and the most common bla gene identified was blaCTX–M–27 (n = 14; 47%) followed by blaCTX–M–15 (n = 10; 33%). blaCTX–M–27 was identified in 13 out of 21 isolates representing ST131, with 12 isolates belonging to a recently discovered international E. coli ST131 C1-M27 subclade. Isolates were found to be genetically distinct from non-human sources with core genome multilocus sequence typing based analysis. Most isolates (n = 26; 87%) possessed multiple replicons, with IncF family plasmids appearing in 27 (90%) and IncI1 in 5 (17%) isolates. IncF[F1:A2:B20] replicon was identified in 11, and IncF[F-:A2:B20] in 4 isolates. The results indicate the ST131-C1-M27 clade gaining prevalence in Europe and provide further evidence of the concerning spread of this globally successful pathogenic clonal group. This study is the first to describe ESBL-producing E. coli in human infections with WGS in Finland and provides important information on global level of the spread of ESBL-producing E. coli belonging to the C1-M27 subclade. The results will help guide public health actions and guide future research.
Antimicrobial resistance (AMR) is an increasing public health concern worldwide. Especially extended-spectrum beta-lactamase (ESBL) and AmpC-producing Enterobacteriaceae have spread globally, and infections caused by resistant bacteria are associated with prolonged hospital stays, increased mortality, and healthcare costs (Ray et al., 2018). Clinically relevant Escherichia coli and Klebsiella pneumoniae have also become common in community-acquired infections in recent years (Devi et al., 2020). The success of these pathogens is highly attributable to epidemic plasmids, which enable AMR spread via horizontal gene transfer (Mathers et al., 2015; Rozwandowicz et al., 2018). Transmission of ESBL-producing E. coli from animal, food, and environmental sources have previously been found to account for a limited amount of human ESBL-carriage in selected countries (Börjesson et al., 2016; Mughini-Gras et al., 2019), but gaps in knowledge regarding the wider epidemiology of these bacteria and the role in human carriage and infections still exist. Whole genome sequencing (WGS) allows for in-depth analysis of possible genetic links between different sources, and the role of plasmids in the spread of AMR.
Extraintestinal pathogenic E. coli (ExPEC) of sequence type (ST) 131 is a globally spread clonal lineage often associated with multidrug resistance (MDR), conferring resistance to ESBLs and fluoroquinolones (Nicolas-Chanoine et al., 2014). A recently recognized subclade within the dominant ST131 clade C, termed C1-M27, has emerged as a common cause of infection globally (Matsumura et al., 2016; Decano and Downing, 2019) but the occurrence of this subclade in Finland remains unknown.
Carbapenemase-producing E. coli isolates of human origin have been described using WGS in Finland previously (Räisänen et al., 2020), but to the best of our knowledge, publications covering ESBL-producing E. coli isolates from Finland of human clinical origin to a genomic level are scarce. WGS studies on healthy individuals have been conducted in Finland (Verkola et al., 2019; Gröndahl-Yli-Hannuksela et al., 2020) and a national surveillance program routinely implements only phenotypic characterization for ESBL-producers (Räisänen et al., 2019). The proportion of ESBL-producing E. coli in blood and urine specimens in patients in Finland has been relatively low, but steadily increasing during recent years (Räisänen et al., 2019). The national surveillance has found 7.3% of all blood specimens and 3.1 and 7.2% of urine specimens in women and men, respectively, positive for ESBL-producing E. coli in 2019. A study investigating fecal samples from healthy, adult volunteers in Finland found the prevalence of ESBL-producing E. coli or K. pneumoniae to be 6.3% (Gröndahl-Yli-Hannuksela et al., 2020). A gap in knowledge regarding bla genes and further genomic characterization of ESBL-producing E. coli isolates involved in clinical cases in Finland exists.
This study aimed to characterize ESBL-producing E. coli isolates obtained from clinical specimens in Finland. This involves genetic comparisons between previously sequenced isolates from healthy human, animal, food, and environmental sources in Finland to evaluate possible genetic overlap and provide genome-level information of the ESBL-producing E. coli found in Finland.
Altogether 30 ESBL-producing E. coli strains were obtained retrospectively from the Eastern Finland Laboratory Centre Joint Authority Enterprise (ISLAB), covering the healthcare district in Eastern Finland. The samples were part of routine practice collected during 2018–2020 from clinical samples, each originating from a different patient. Stored samples were recultivated in March 2020 and transported to the University of Helsinki for further studies with Copan M40 Transystem sterile transport swabs (Copan Transystem, Copan Diagnostics, Italy).
Briefly, antimicrobial susceptibility testing for specimens other than blood and urine was performed using disk diffusion method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) standards with third-generation cephalosporins (cefpodoxime, ceftazidime, and ceftriaxone), together with amoxicillin-clavulanic acid (Oxoid, Basingstoke, Hampshire, United Kingdom). Specimens other than blood and urine consisted of samples originating from various body sites and tissue types, including joint, scrotum, maxillary sinus, eye conjunctiva, wound, bile, abscess, bronchoalveolar lavage, and abdominal cavity. Additionally, combination disk method (cefotaxime 30 μg and cefotaxime 30 μg + clavulanic acid 10 μg; ceftazidime 30 μg and ceftazidime 30 μg + clavulanic acid 10 μg) and AmpC disk test (Mast Group Ltd., Bootle, United Kingdom) were used for presumptive ESBL-producing E. coli isolates. Susceptibility testing for urine samples was performed with a Vitek 2 AST-N385 card (bioMeriéux, Marcy-L’Etoile, France), and for blood samples with both Vitek 2 and disk diffusion method according to EUCAST standards, to ensure rapid and accurate diagnosis in possible septicaemia cases.
Bacterial DNA was extracted and purified with a PureLink Genomic DNA Mini Kit (Invitrogen by Thermo Fischer Scientific, Carlsbad, CA, United States) according to manufacturer’s instructions. The assessment of DNA quality was carried out using a NanoDrop ND-1000 spectrophotometer (Thermo Fischer Scientific, Wilmington, DE, United States) and DNA quantity was measured using a Qubit 2.0 fluorometer (Invitrogen, Life Technologies, Carlsbad, CA, United States). An optical density of 1.8–2.0 at 260/280 nm and a concentration of ≥10 ng/μl with a minimum amount of 0.2 μg were set as thresholds. Library preparation was performed with NEBNext Ultra DNA Library Prep Kit for Illumina (Cat No. E7370L). Sequencing was performed with an Illumina NovaSeq 6000 (Novogene, Cambridge, United Kingdom) with 100 × coverage and 2 × 150 bp read length and a Phred score of Q30 ≥ 80%.
Raw reads have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB47797. Accession numbers are provided in Supplementary Table 1.
Bacterial DNA sequences were analyzed with Ridom SeqSphere+ software v7.0.4 (Ridom GmbH, Germany) (Jünemann et al., 2013) and Center for Genomic Epidemiology (CGE) web-based tools (DTU, Denmark) available at http://www.genomicepidemiology.org. Within Ridom SeqSphere+ pipeline, raw reads were assembled with SKESA v2.3.0 (Souvorov et al., 2018) together with quality control with FastQC v0.11.7 (Babraham Institute, 2021) and adapter trimming with Trimmomatic v0.36 (Bolger et al., 2014), AMR genes were identified with NCBI AMRFinderPlus v3.2.3 (Feldgarden et al., 2019), virulence genes with VFDB (Chen et al., 2016), and ST with E. coli MLST Warwick v1.0 (Achtman scheme) based on the PubMLST database (Jolley et al., 2018). Sequences from isolates with novel STs were submitted to the Enterobase database1 (Zhou et al., 2020) to assign new Achtman scheme STs.
Using default values, PlasmidFinder 2.0 (Carattoli et al., 2014) was employed to detect plasmid replicons and pMLST 2.0 (Carattoli et al., 2014) the plasmid multilocus ST for IncF and IncI1 type replicons. FimTyper 1.0 (Roer et al., 2017) was used to identify the fimH allele. ResFinder 4.1 (Camacho et al., 2009; Zankari et al., 2017; Bortolaia et al., 2020), SerotypeFinder 2.0 (Joensen et al., 2015), MLST 2.0 (Larsen et al., 2012), and VirulenceFinder 2.0 (Joensen et al., 2014; Tetzschner et al., 2020) were used to confirm acquired resistance genes, serotype, and ST, respectively.
Isolates were compared with BLASTn to strain KUN5781 (GenBank accession: LC209430) (Matsumura et al., 2016) to determine the presence of M27-C1 clade-specific prophage-like regions M27PP1 and M27PP2. Results were visualized with BRIG v0.95 (Alikhan et al., 2011) for isolates with matching regions.
Core genome multilocus sequence typing (cgMLST) targeting 2520 genes was performed using Ridom SeqSphere+ software v7.0.4 (Ridom GmbH, Germany) (Jünemann et al., 2013) to compare all 30 isolates obtained from ISLAB and results were visualized with a minimum spanning tree (MST).
Isolates were also compared to available previously sequenced ESBL/AmpC-producing E. coli isolates from Finland (total n = 67) collected between 2012 and 2018 from broiler meat (n = 5) (Päivärinta et al., 2020), broiler caecum (n = 5) (Päivärinta et al., 2020), broiler production including broiler parents (n = 8) (Oikarainen et al., 2019), egg surfaces (n = 4) (Oikarainen et al., 2019) and production environment (n = 1) (Oikarainen et al., 2019), imported food products (n = 16) (Kurittu et al., 2021a), barnacle geese (n = 9) (Kurittu et al., 2021b), wastewater (n = 1) (unpublished), cattle (n = 1) (Päivärinta et al., 2016), veterinarians (n = 9) (Verkola et al., 2019), and healthy adults (n = 8) (Gröndahl-Yli-Hannuksela et al., 2020; Supplementary Table 2). cgMLST-based MST and was constructed with Ridom SeqSphere+ software.
Altogether 30 E. coli isolates obtained from human clinical samples confirmed as ESBL-producing with phenotypic screening from the Eastern Finland healthcare district between 2018 and 2020 were subjected to WGS. Most isolates (n = 12; 40%) originated from a urine sample, followed by blood samples (n = 8; 27%). A majority of the isolates (n = 21; 70%) belonged to ST131, with the remaining nine isolates representing a different ST each, including ST38, ST1193, ST162, ST537, ST59, and ST405 (Table 1). A novel ST was identified in three isolates and the sequences were submitted to the Enterobase database (see text footnote 1) to assign new Achtman scheme STs. The newly assigned STs were as follows: ST12704, ST12703, and ST12705 for isolates D2, D4, and D9, respectively. Sequences from two isolates from previous studies, A41.2-1 and C76.1-2 (Kurittu et al., 2021a), were additionally submitted to Enterobase to assign new Achtman scheme STs. All ST131 isolates from the current study were of serotype O25:H4 and possessed the fimH30 allele, except for two isolates, which belonged to serotype O16:H5 and possessed the fimH41 allele.
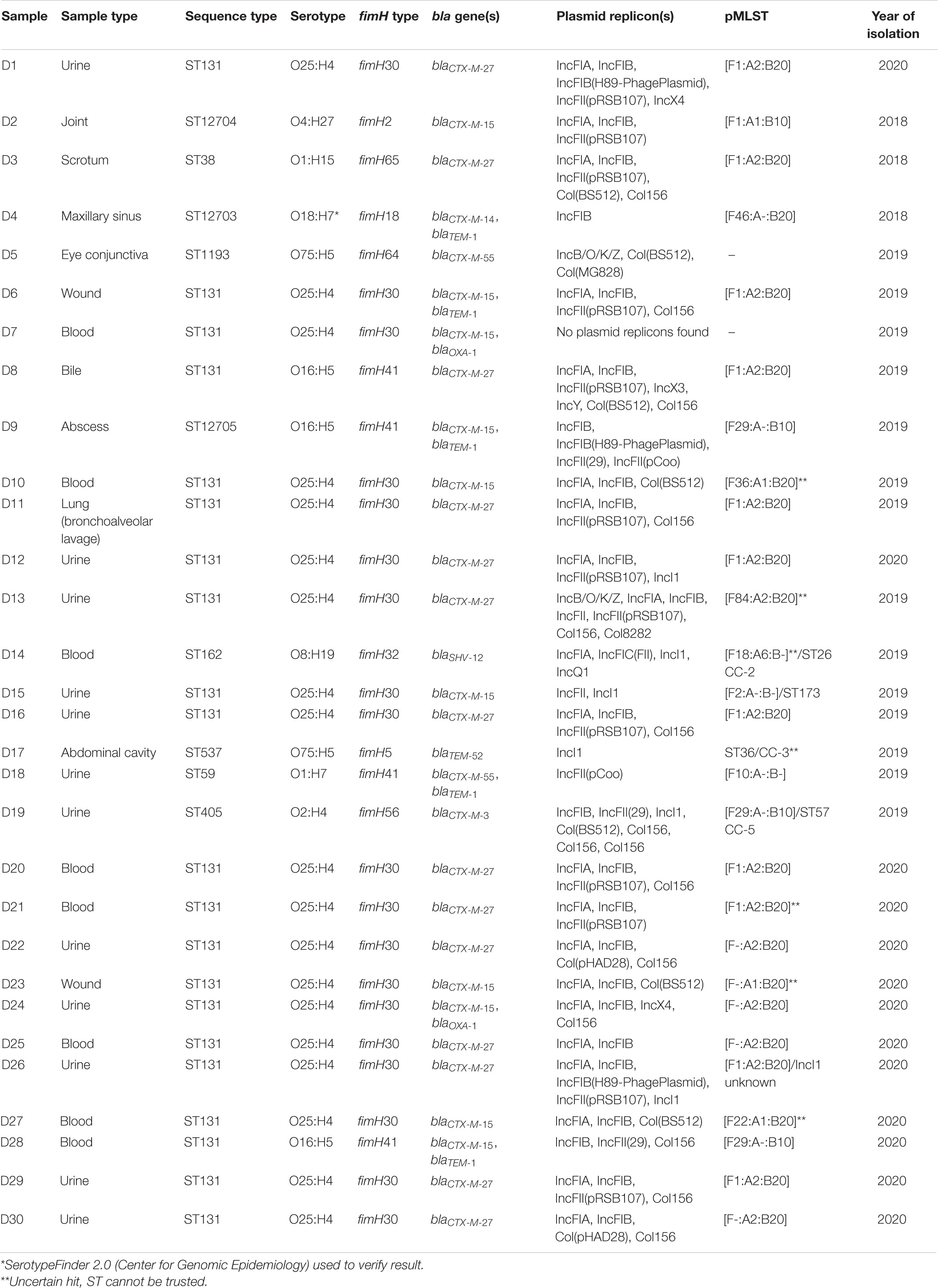
Table 1. Genomic characterization of 30 ESBL-producing Escherichia coli isolates obtained from human clinical samples in the Eastern Finland healthcare district during 2018–2020.
All isolates were found to match their respective phenotype genotypically, as all were genotypically confirmed to carry at least one bla gene representing an ESBL phenotype. The most common bla gene identified was blaCTX–M–27 (n = 14; 47%) followed by blaCTX–M–15 (n = 10; 33%). blaCTX–M–27 was identified in 13 out of the 21 isolates representing ST131 and additionally from one isolate of ST 38. Eight out of the 10 blaCTX–M–15 were harbored by ST131, with 7 representing the serotype O25:H4 together with fimH allele 30, while 1 was of serotype O16:H5 with fimH41 allele. Two isolates of novel STs, ST12704 (isolate D2) and ST12705 (isolate D9), were found to possess blaCTX–M–15. Regarding other blaCTX–M genes, blaCTX–M–55 occurred in two isolates (D5 of ST1193 and D18 of ST59), and blaCTX–M–14 and blaCTX–M–3 each in one isolate each (D4 of ST12703 and D19 of ST405, respectively). blaSHV–12 was found from one isolate (D14) of ST 162 and blaTEM–52 from one isolate (D17) of ST 537.
All isolates were found to harbor at least one plasmid replicon, except for D7, from which no replicons were identified. Altogether 17 different replicons were detected with IncFIB (n = 24), IncFIA (n = 21), IncFII (n = 13), and Col156 (n = 13) type replicons were most prevalent. The majority of the isolates (n = 26; 87%) possessed multiple replicons, with IncF family plasmids appearing in 27 (90%) isolates. IncI1 plasmids were recovered from five isolates (D14, D15, D17, D19, and D26), all with varying pMLST profiles. Plasmids of pMLST IncF[F1:A2:B20] type were identified in 11 isolates, and pMLST IncF[F-:A2:B20] in 4 isolates.
Multidrug resistance, resistance to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012), was common among the isolates with multiple acquired resistance genes identified in 21 (70%) isolates, including genes against aminoglycosides, tetracycline, sulfonamides, macrolides, and trimethoprim (Table 2). Acquired sulfonamide resistance genes, sul1 and sul2, were found either alone or together in 18 (60%) of the isolates.
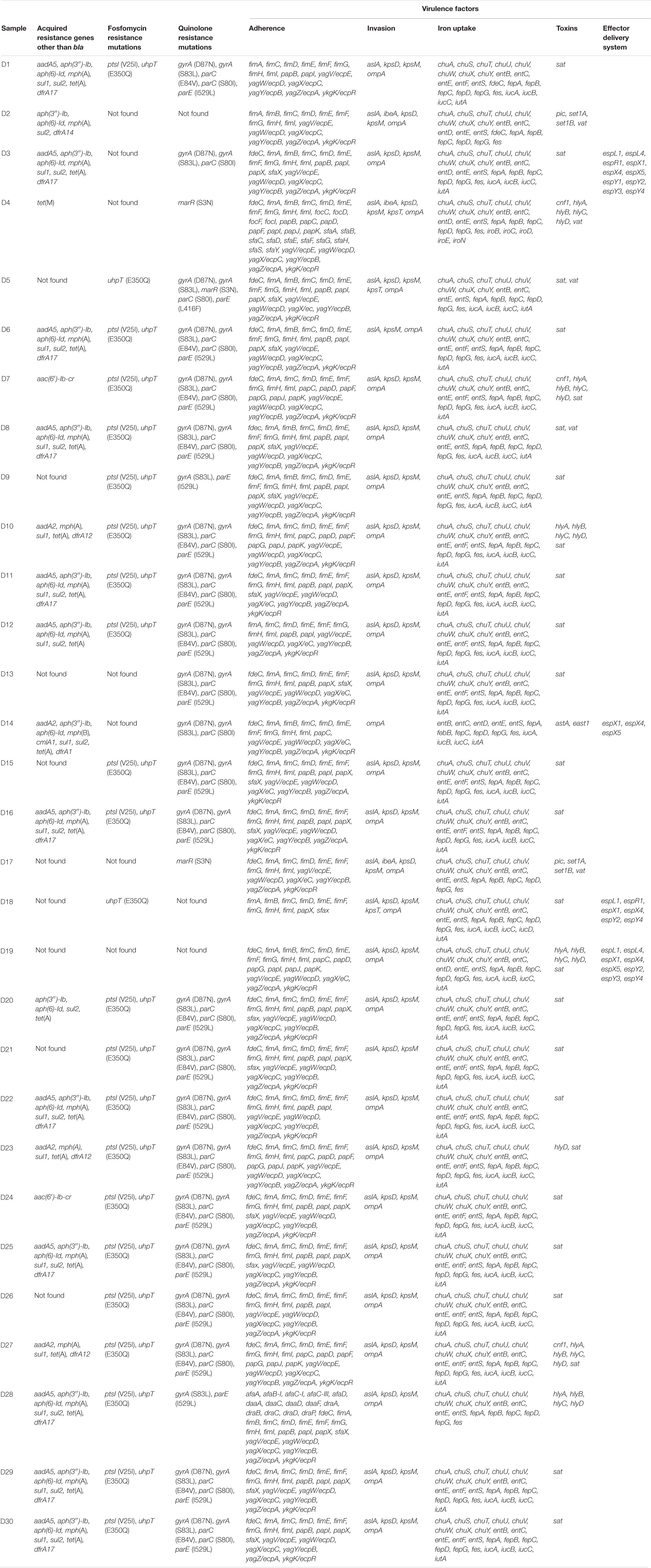
Table 2. Virulence and antimicrobial resistance genes other than bla identified in 30 ESBL-producing Escherichia coli isolates obtained from Finnish patients collected in the Eastern Finland healthcare district during 2018–2020.
Genes conferring trimethoprim resistance, either dfrA17, dfrA12, dfrA1, or dfrA14, were detected in 16 (53%) isolates. Of these genes, dfrA17 was the most prevalent (n = 11; 37%), followed by dfrA12 (n = 3; 10%). dfrA1 and dfrA14 appeared in one isolate each.
Aminoglycoside resistance [aadA5, aadA2, aph(3″)-Ib, aph(6)-Id, and/or aac(6′)-Ib-cr] was detected in 20 (67%) isolates and tetracycline resistance in 18 (60%) isolates [tet(A) in 17 and tet(M) in one isolate]. No carbapenemase genes were detected.
Chromosomal quinolone resistance mutations in gyrA, parC, parE, or marR were recovered from 27 of the 30 isolates, whereas plasmid-mediated quinolone resistance (PMQR) gene aac(6′)-Ib-cr was additionally identified in two isolates (D7 and D24). Chromosomal mutations in ptsI and uhpt associated with fosfomycin resistance were discovered in 23 (77%) isolates.
All the isolates harbored multiple virulence factors, with extraintestinal pathogenic E. coli (ExPEC) associated virulence genes (Johnson et al., 2003) pap (P fimbrial adhesin), kpsMII (polysialic acid transport protein; group 2 capsule), iutA (ferric aerobactin receptor), and sfa (S and F1C fimbriae) recovered from 29, 28, 26, and 18 isolates, respectively. Only isolate D2 lacked the previously described threshold of two or more of the five virulence genes (pap, kps, iutA, sfa/foc, afa/dra) defined as discriminatory for ExPEC classification (Johnson et al., 2003; Kanamori et al., 2017). No Shiga toxin (stx) genes were found. Virulence factors are presented according to their pathogenicity factor groups (Nesta et al., 2012; Pitout, 2012; Chen et al., 2016; Sarowska et al., 2019; Duan et al., 2020; Dekker et al., 2021) in Table 2 together with resistance genes other than bla.
Assembly statistics including the number of bases and contigs, the N50 value and average coverage for each isolate are available in Supplementary Table 3.
All 12 ST131 E. coli isolates with blaCTX–M–27 and fimH30 allele were found to possess the C1-M27 clade-specific prophage-like 11,894-bp region M27PP1 together with the 7 bp direct repeats (Matsumura et al., 2016; Figure 1). Four of these 12 samples additionally possessed the 19,352-bp prophage-like region M27PP2.
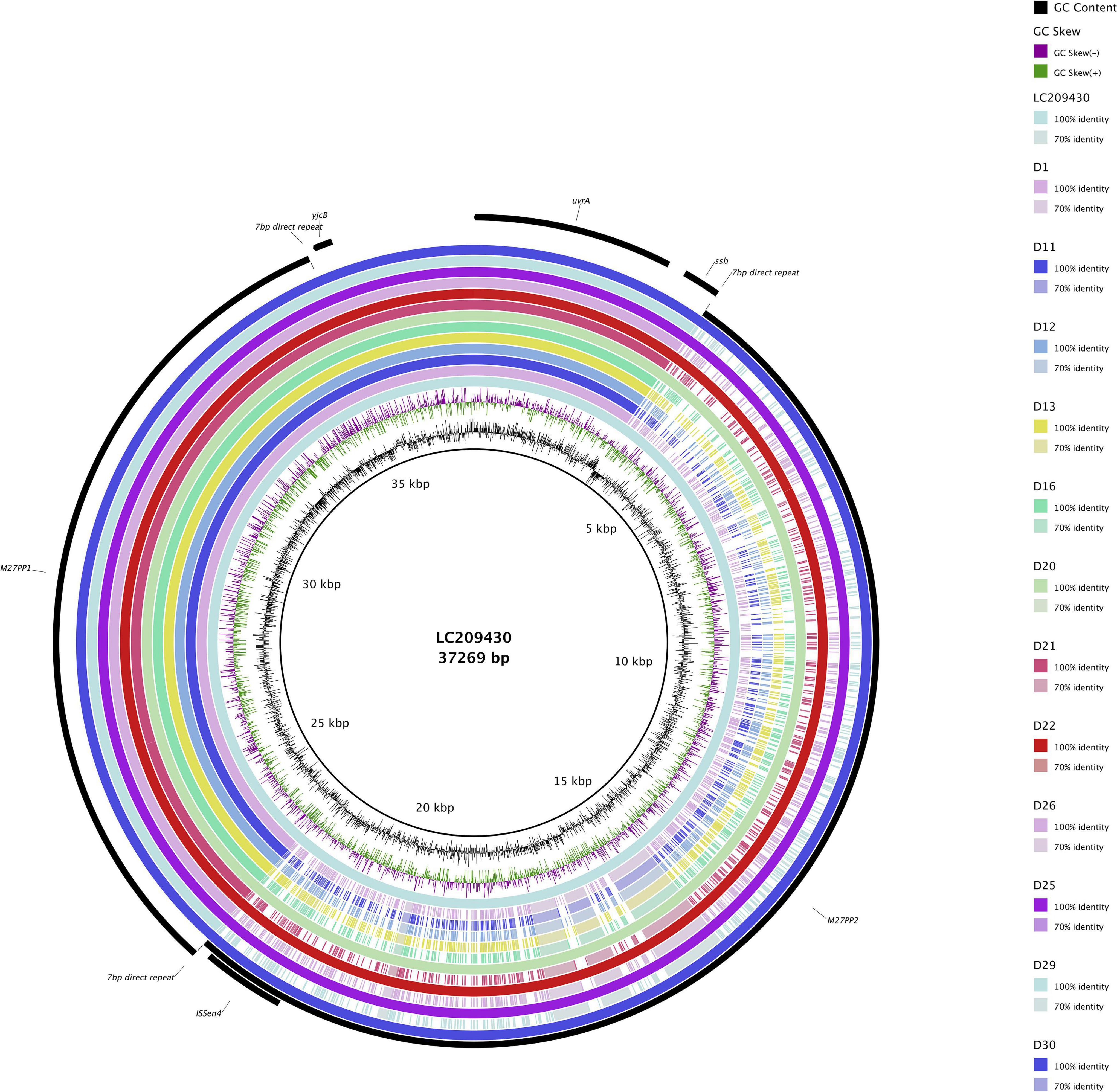
Figure 1. ST131 Escherichia coli C1-M27 clade-specific regions, prophage-like genomic islands M27PP1 and M27PP2, of KUN5781 (GenBank accession: LC209430) compared to CTX-M-27-producing ST131 E. coli with fimH30 allele from the current study. GC content and GC skew are depicted on the inner map with distance scale and predicted coding sequences depicted on the outer ring.
All of the 30 human ESBL-producing E. coli isolates obtained from the Eastern Finland healthcare district were compared with a cgMLST-based MST (Figure 2). Results indicate isolates within ST131 form clusters, meaning allelic differences were ≤10, whereas isolates of different STs are genetically distant with allelic differences ranging from 640 to over 2000 between two isolates. Three different clusters were observed within isolates belonging to the ST131 C1-M27 clade: D11 and D16, D26 and D12, and D25 together with D30 and D22. Additionally, one cluster between three isolates (D23, D27, and D10) was identified, comprising isolates with the same ST (ST131), serotype (O25:H4), fimH type (fimH30), and AMR gene resistance profile [aadA2, blaCTX–M–15, mph(A), sul1, tet(A), dfrA12].
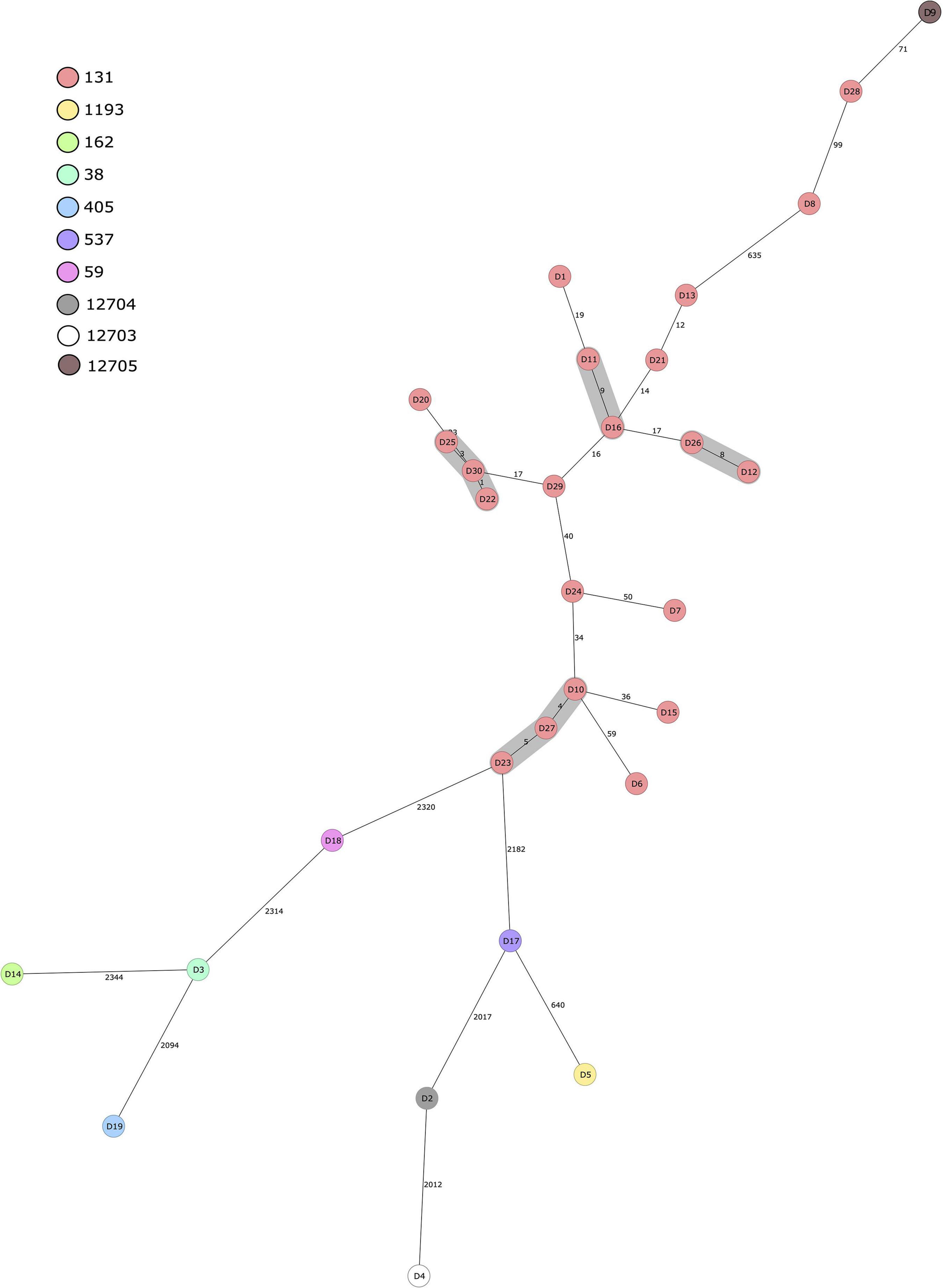
Figure 2. Minimum spanning tree of 30 human ESBL-producing Escherichia coli isolates obtained from patients in Eastern Finland during 2018–2020. Tree was calculated in Ridom SeqSphere+ with 2513 core genome multilocus sequence typing (cgMLST) targets and 7 E. coli MLST Warwick targets (pairwise ignoring missing values, logarithmic scale). Nodes are colored according to sequence type. Number of allelic differences between isolates are indicated on the connecting lines. Clusters are defined as ≤10 allelic difference and shaded in gray.
The 30 human ESBL-producing E. coli isolates obtained from the Eastern Finland healthcare district were additionally compared to available, previously sequenced ESBL/AmpC-producing E. coli isolates obtained from different sources in Finland (Figure 3). A cgMLST-based MST included 2520 gene targets.
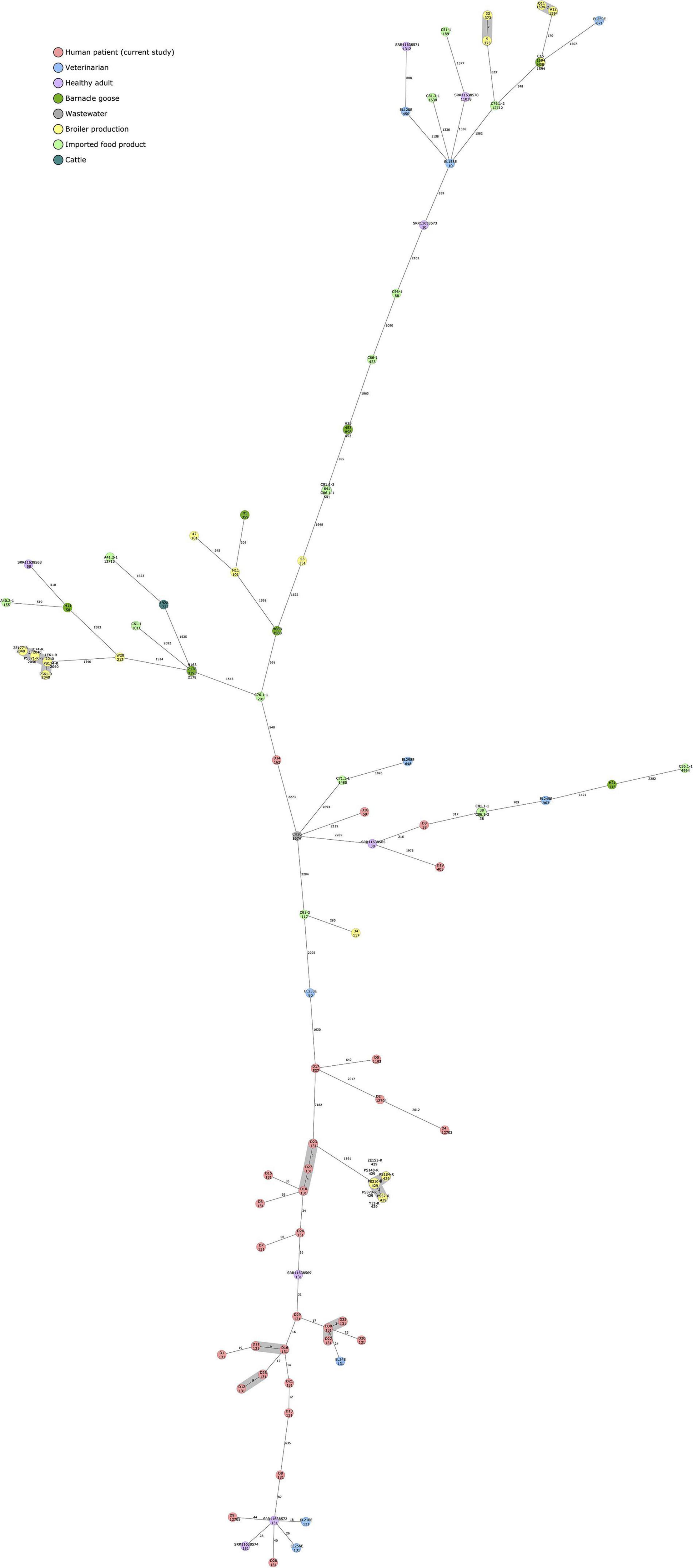
Figure 3. Minimum spanning tree of 97 ESBL/AmpC-producing Escherichia coli isolates calculated in Ridom SeqSphere+ with 2513 core genome multilocus sequence typing (cgMLST) targets and 7 E. coli MLST Warwick targets (pairwise ignoring missing values, logarithmic scale). Nodes are colored according to isolation source. Sequence type is indicated under the isolate name. Number of allelic differences between isolates are indicated on the connecting lines. Clusters are defined as ≤10 allelic difference and shaded in gray.
Isolates obtained in the current study failed to form close clusters with isolates recovered from earlier studies, although the least distance (24 allelic difference) was observed between isolate D30 and EL24E, an isolate from a healthy veterinarian volunteer. Both isolates were of ST 131 and harbored blaCTX–M–27.
Other relatively close connections were also observed among isolates originating from human samples (18–44 allelic differences), all of ST 131, and positive for blaCTX–M–15. Two isolates from the current study, D9 and D28, differed by 44 and 40 alleles, respectively, from SRR11638572, an isolate recovered from a healthy Finnish volunteer. Two veterinarian isolates from a previously published study, EL216E and EL256E, differed by 18 and 26 alleles, respectively, from the same Finnish volunteer sample.
In addition to clusters observed within the isolates from the current study, isolates originating from poultry production (Oikarainen et al., 2019) formed two clusters with samples from the same study, and broiler meat (isolates 5 and 33) and broiler caecum (Q11 and A12) (Päivärinta et al., 2020) formed a cluster each. C15, a blaCMY–2-carrying ST1594 E. coli isolate from broiler caecum (Päivärinta et al., 2020), did not differ at all with cgMLST-based MST analysis from H58, an isolate originating from barnacle goose (Kurittu et al., 2021b).
We used WGS to characterize 30 ESBL-producing E. coli isolates obtained from clinical samples in Eastern Finland and performed cgMLST-based genomic comparisons to ESBL/AmpC-producing E. coli isolates of human, animal, food, and environmental origins isolated in Finland previously. ESBL-producing E. coli isolates from human sources were found to be genetically distinct from non-human sources in Finland. However, most ST131 blaCTX–M–27-positive E. coli isolates from human clinical samples were found to belong to a recently discovered international E. coli ST131 C1-M27 subclade, providing important insight to the epidemiology and increasing spread of this globally successful pathogenic clonal group. Strains within the C1-M27 clade carry blaCTX–M–27, possess fimH allele 30 and a prophage-like genomic island termed M27PP1, sometimes together with another prophage-like region, M27PP2 (Matsumura et al., 2016). blaCTX–M–27 has been noted to rival the globally dominant human-associated blaCTX–M–15 in many parts of the world, having been isolated from human, animal, food, or environmental sources in multiple countries in Europe, North America, and Asia (Bevan et al., 2017).
ST131 has become the dominant ExPEC lineage causing infections in humans worldwide (Banerjee and Johnson, 2014; Nicolas-Chanoine et al., 2014; Mathers et al., 2015). Subclones of ST131 E. coli, mainly H30 and H30Rx, are associated with fluoroquinolone resistance, and blaCTX–M–15 in the case of H30Rx (Banerjee et al., 2013). Previously blaCTX–M–15 has been the most prevalent bla gene identified from human isolates, but recent studies have noted a rise in the prevalence of blaCTX–M–27, starting from Japan in the late 2000s (Matsumura et al., 2016), and more recently a rapid increase in fecal carriage was observed in children in France (Birgy et al., 2017), along with human isolates from Germany (Ghosh et al., 2017), as well as from samples from hospitalized patients from four European cities (Berlin, Geneva, Madrid, and Utrecht) (Merino et al., 2018). Worldwide distribution is further demonstrated with the recent finding of ST131 E. coli belonging to C1-M27 clade in Brazil from a marine sample (Fernandes et al., 2020). Worryingly, ST131-blaCTX–M–27-E. coli has been noted to have a higher transmission rate compared to ST131-blaCTX–M–15-E. coli in an Israeli hospital setting (Adler et al., 2012). Our findings support the notion of a shift in the most dominant blaCTX–M observed in human samples, and the emergence of blaCTX–M–27 as a challenger for blaCTX–M–15. Our findings regarding isolates within C1-M27 clade also are in line with previous studies where a majority of isolates were found to possess only the M27PP1 prophage-like region, instead of possessing both M27PP1 and M27PP2 (Matsumura et al., 2016; Decano and Downing, 2019). Interestingly, M27PP1 was also found with 100% coverage and 99.95% identity with BLASTn from sample D18, which carries blaCTX–M–55 and blaTEM–1 and is of ST 59 and of fimH type H41.
blaCTX–M–55 was identified in two of our isolates, D5 and D18, with different STs (ST1193 and ST59, respectively). This bla gene was additionally identified from two previously sequenced ESBL-producing E. coli isolates from Finland, one from an imported food sample (coriander from Malaysia) representing ST155 (Kurittu et al., 2021a) and one from a healthy, human adult fecal sample, representing ST58 (Gröndahl-Yli-Hannuksela et al., 2020). blaCTX–M–55-harboring E. coli has been reported especially in samples from meat and food-producing animals, as well as in humans in Asian countries (Zheng et al., 2012; Zhang et al., 2014; Zeng et al., 2021). Studies conducted in China have noted an increase in the proportion of blaCTX–M–55 compared to other prevalent blaCTX–M genes, such as blaCTX–M–15 and blaCTX–M–14, in human patient material (Zhang et al., 2014; Zeng et al., 2021), depicting the rapidly evolving epidemiology of these enzymes. In addition to blaCTX–M–55, ST1193 E. coli was notably recognized as the most prevalent ST among uropathogenic E. coli (UPEC) isolates in female patients in China (Zeng et al., 2021). A rapid increase in ST1193 has also been detected in the United States from clinical fluoroquinolone-resistant E. coli isolates from urine samples (Tchesnokova et al., 2019). The only ST1193 isolate recovered in our study from an eye conjunctive sample (D5) harbored chromosomal quinolone resistance genes (gyrA, marR, parC, parE) in addition to blaCTX–M–55. Isolates from the previously mentioned study were, however, rarely resistant to beta-lactams, which was not the case in our sample. An Australian study has described fluoroquinolone-resistant ST131 and ST1193 E. coli as being less prevalent in animals compared to humans, and considers humans most likely as the source for possible findings of these bacteria in animals (Kidsley et al., 2020). Although not found in the comparative analysis of our study, ST131 E. coli isolates belonging to C1-M27 subclade have also been isolated previously from animal sources, more specifically from two pig isolates in the United Kingdom (Duggett et al., 2021) and companion animals in France (Melo et al., 2019).
A recent study in the United States found blaCTX–M–27 to be the second most common bla gene after blaCTX–M–15 in clinical human isolates, and notably blaCTX–M–27 was associated with ST38 E. coli (Mostafa et al., 2020). The blaCTX–M–27 gene on ST38 E. coli was found to be mostly plasmid-borne, residing in an IncF[F2:A-:B10] or IncF[F1:A2:B20] plasmid (Mostafa et al., 2020). One isolate (D3) in our study was positive for blaCTX–M–27-carrying ST38 E. coli, and this isolate harbored several IncF type replicons [IncFIA, IncFIB, IncFII(pRSB107)] together with Col plasmids [Col(BS512), Col156] and represented the replicon ST [F1:A2:B20].
The majority of our human clinical isolates harbored plasmid replicons belonging to the IncF family, which have been identified as important carriers of globally successful AMR genes, especially those encoding for ESBLs (Villa et al., 2010; Rozwandowicz et al., 2018). Thirteen of our isolates harbored an IncFII replicon together with FIA and FIB replicons, which together form a typical IncF multireplicon (Villa et al., 2010). The most common IncF replicon type identified in our isolates was [F1:A2:B20], which was found from 10 ST131 E. coli isolates carrying blaCTX–M–27 (n = 9) and blaCTX–M–15 (n = 1), and from one ST38 carrying blaCTX–M–27, similar to the findings of a study conducted in the United States (Mostafa et al., 2020). This supports the observation that plasmids belonging to pMLST [F1:A2:B20] are associated with the C1-M27 subclade (Ghosh et al., 2017; Mostafa et al., 2020). IncI1 plasmid replicons belonging to different pMLST profiles were identified in five isolates (D14, D15, D17, D19, and D26), together with blaSHV–12, blaCTX–M–15, blaTEM–52, blaCTX–M–3, and blaCTX–M–27, respectively. IncI type plasmids, especially with blaCTX–M–1, are frequently found from E. coli from poultry sources (Rozwandowicz et al., 2018).
The pMLST results should, however, be interpreted with care, since long-read sequencing would allow for more robust and accurate identification of plasmid structures and gene locations. As plasmids are important mediators of AMR worldwide (Carattoli, 2013; Rozwandowicz et al., 2018), further plasmid characterization through hybrid sequencing methods is warranted to investigate the epidemiological events in more detail in future studies. Another limitation of our study is the limited number of human clinical isolates analyzed, and the confined geographical origin of the samples. Our results do, however, represent a period of several years and multiple different specimen types, which provide an initial overview of the situation of ESBL-producing E. coli in clinical samples in Finland. Furthermore, our results strengthen the finding of blaCTX–M–27-harboring E. coli belonging to C1-M27 subclade gaining prevalence in Europe and describe the first published finding of C1-M27-clade isolates in Finland.
Multidrug resistance was common among the human clinical isolates analyzed in our study. Trimethoprim resistance is often associated with UPEC isolates, and our frequent finding of dfrA17 and dfrA12 genes is in line with a previous study conducted in Korea, which observed dfrA17 and dfrA12 as the most prevalent trimethoprim resistance genes in urinary tract isolates (Lee et al., 2001). Chromosomal quinolone resistance, as well as acquired tetracycline, aminoglycoside, and sulfonamide resistance genes, was also common in our human isolates.
Our findings are in line with earlier studies investigating the possible origins of ESBL-producing E. coli in humans. ESBL-producing E. coli isolates from human sources were found to be genetically distant from isolates obtained from food, animal, and environmental sources with a cgMLST-based MST approach. The total sample size, however, was relatively small, and closer genetic connections could possibly have been observed with a larger dataset. The only relatively close connections between human clinical isolates sequenced in this study and previously sequenced ESBL-producing E. coli isolates from Finland were observed among human-derived samples from veterinarians and a healthy, adult volunteer. The isolates represented ST131 and carried either blaCTX–M–27 or blaCTX–M–15, representing typical results for a human-derived sample. The only close connections among the other previously sequenced isolates from non-human sources in Finland were observed between isolates originating from poultry sources. Interestingly, ST1594 with AmpC type beta-lactamase, blaCMY–2, was identified from a broiler caecal sample and a barnacle goose fecal sample and showed no allelic variation in the cgMLST-based MST analysis.
A population-based modeling study with a larger dataset conducted in the Netherlands investigating the community-acquired ESBL-carriage and its attributable sources concluded that human-to-human transmission is the main route for acquiring ESBL E. coli, even though food, animals, and environmental sources were found to account for transmission to a lesser extent (Mughini-Gras et al., 2019). Another study conducted in Sweden found no evidence for clonal transmission events between ESBL/AmpC-producing E. coli in humans, animals, and the environment, but similarities were discovered in resistance genes and plasmids, indicating possible limited transmission potential (Börjesson et al., 2016).
In conclusion, blaCTX–M–27 was found to be the most prevalent ESBL gene in human clinical samples, and no clear evidence for animal, food, or environmental genetic overlap was observed in our dataset. Our results prove the spread of E. coli belonging to the C1-M27 clade has been successful, and WGS-based methods for surveillance of AMR trends and is effective and warranted for future studies. Surveillance studies are needed to detect the rapid evolution and epidemiology of ESBL genes, and future studies focusing on plasmid-mediated AMR spread are needed to assess the epidemiological links between different bacterial sources further.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PK, AH, BK, and JJ contributed to the concept and design of the study. JK performed the sample collection. PK and BK analyzed the whole genome sequence data and performed the subsequent analysis and constructed images of the sequence analysis. PK drafted the manuscript. BK, AH, and JK revised the manuscript. All authors have read and approved the final draft of the manuscript.
This study received funding from the Finnish Foundation for Veterinary Research and the Doctoral School of Environmental, Food and Biological Sciences in University of Helsinki.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MB declared a past collaboration with one of the authors, PK, to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank senior laboratory technician Kirsi Ristkari for valuable help with laboratory work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.789280/full#supplementary-material
Adler, A., Gniadkowski, M., Baraniak, A., Izdebski, R., Fiett, J., Hryniewicz, W., et al. (2012). Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin. Microbiol. Infect. 18, E497–E505. doi: 10.1111/j.1469-0691.2012.03999.x
Alikhan, N. F., Petty, N. K., ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Babraham Institute (2021). Babraham Bioinformatics–FastQC A Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed August 18, 2021)
Banerjee, R., and Johnson, J. R. (2014). A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 58, 4997–5004. doi: 10.1128/AAC.02824-14
Banerjee, R., Robicsek, A., Kuskowski, M. A., Porter, S., Johnston, B. D., Sokurenko, E., et al. (2013). Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob. Agents Chemother. 57, 6385–6388. doi: 10.1128/AAC.01604-13
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Birgy, A., Bidet, P., Levy, C., Sobral, E., Cohen, R., and Bonacorsi, S. (2017). CTX-M-27–producing Escherichia coli of sequence type 131 and clade C1-M27, France. Emerg. Infect. Dis. 23:885. doi: 10.3201/eid2305.161865
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Börjesson, S., Ny, S., Egervärn, M., Bergström, J., Rosengren, Å, Englund, S., et al. (2016). Limited dissemination of extended-spectrum β-lactamase- and plasmid-encoded AmpC-producing Escherichia coli from food and farm animals, Sweden. Emerg. Infect. Dis. 22, 634–640. doi: 10.3201/eid2204.151142
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Decano, A. G., and Downing, T. (2019). An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-54004-5
Dekker, D., Pankok, F., Thye, T., Taudien, S., Oppong, K., Akenten, C. W., et al. (2021). Clonal clusters, molecular resistance mechanisms and virulence factors of Gram-negative bacteria isolated from chronic wounds in Ghana. Antibiotics 10:339. doi: 10.3390/antibiotics10030339
Devi, L. S., Broor, S., Rautela, R. S., Grover, S. S., Chakravarti, A., and Chattopadhya, D. (2020). Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-Type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: a 3-year study. Int. J. Appl. Basic Med. Res. 10, 156–163. doi: 10.4103/ijabmr.IJABMR_360_19
Duan, Y., Gao, H., Zheng, L., Liu, S., Cao, Y., Zhu, S., et al. (2020). Antibiotic resistance and virulence of extraintestinal pathogenic Escherichia coli (ExPEC) vary according to molecular types. Front. Microbiol. 11:2960. doi: 10.3389/fmicb.2020.598305
Duggett, N., Ellington, M. J., Hopkins, K. L., Ellaby, N., Randall, L., Lemma, F., et al. (2021). Detection in livestock of the human pandemic Escherichia coli ST131 fimH 30(R) clone carrying bla CTX-M-27. J. Antimicrob. Chemother. 76, 263–265. doi: 10.1093/jac/dkaa407
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19. doi: 10.1128/AAC.00483-19
Fernandes, M. R., Sellera, F. P., Cunha, M. P. V., Lopes, R., Cerdeira, L., and Lincopan, N. (2020). Emergence of CTX-M-27-producing Escherichia coli of ST131 and clade C1-M27 in an impacted ecosystem with international maritime traffic in South America. J. Antimicrob. Chemother. 75, 1647–1649. doi: 10.1093/jac/dkaa069
Ghosh, H., Doijad, S., Falgenhauer, L., Fritzenwanker, M., Imirzalioglu, C., and Chakraborty, T. (2017). BlaCTX-M-27–encoding escherichia coli sequence type 131 lineage c1-m27 clone in clinical Isolates, Germany. Emerg. Infect. Dis. 23, 1754–1756. doi: 10.3201/eid2310.170938
Gröndahl-Yli-Hannuksela, K., Lönnqvist, E., Marttila, H., Rintala, E., Rantakokko-Jalava, K., and Vuopio, J. (2020). Performance of the check-direct ESBL screen for BD MAXTM for detection of asymptomatic faecal carriage of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 22, 408–413. doi: 10.1016/j.jgar.2020.04.015
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johnson, J. R., Murray, A. C., Gajewski, A., Sullivan, M., Snippes, P., Kuskowski, M. A., et al. (2003). Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47, 2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; referees: 2 approved]. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jünemann, S., Sedlazeck, F. J., Prior, K., Albersmeier, A., John, U., Kalinowski, J., et al. (2013). Updating benchtop sequencing performance comparison. Nat. Biotechnol. 31, 294–296. doi: 10.1038/nbt.2522
Kanamori, H., Parobek, C. M., Juliano, J. J., Johnson, J. R., Johnston, B. D., Johnson, T. J., et al. (2017). Genomic analysis of multidrug-resistant Escherichia coli from North Carolina community hospitals: ongoing circulation of CTX-M-producing ST131-H30Rx and ST131-H30R1 Strains. Antimicrob. Agents Chemother. 61:e00912-17. doi: 10.1128/AAC.00912-17
Kidsley, A. K., White, R. T., Beatson, S. A., Saputra, S., Schembri, M. A., Gordon, D., et al. (2020). Companion animals are spillover hosts of the multidrug-resistant human extraintestinal Escherichia coli pandemic clones ST131 and ST1193. Front. Microbiol. 11:1968. doi: 10.3389/fmicb.2020.01968
Kurittu, P., Khakipoor, B., Aarnio, M., Nykäsenoja, S., Brouwer, M., Myllyniemi, A.-L., et al. (2021a). Plasmid-borne and chromosomal ESBL/AmpC genes in Escherichia coli and Klebsiella pneumoniae in global food products. Front. Microbiol. 12:125. doi: 10.3389/fmicb.2021.592291
Kurittu, P., Khakipoor, B., Brouwer, M. S. M., and Heikinheimo, A. (2021b). Plasmids conferring resistance to extended-spectrum beta-lactamases including a rare IncN+IncR multireplicon carrying blaCTX-M-1 in Escherichia coli recovered from migrating barnacle geese (Branta leucopsis). Open Res. Eur. 1:46. doi: 10.12688/openreseurope.13529.1
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lee, J. C., Oh, J. Y., Cho, J. W., Park, J. C., Kim, J. M., Seol, S. Y., et al. (2001). The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 47, 599–604. doi: 10.1093/jac/47.5.599
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mathers, A. J., Peirano, G., and Pitout, J. D. D. (2015). The role of epidemic resistance plasmids and international high- risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Matsumura, Y., Pitout, J. D. D., Gomi, R., Matsuda, T., Noguchi, T., Yamamoto, M., et al. (2016). Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg. Infect. Dis. 22:1900. doi: 10.3201/EID2211.160519
Melo, L. C., Haenni, M., Saras, E., Duprilot, M., Nicolas-Chanoine, M.-H., and Madec, J.-Y. (2019). Emergence of the C1-M27 cluster in ST131 Escherichia coli from companion animals in France. J. Antimicrob. Chemother. 74, 3111–3113. doi: 10.1093/jac/dkz304
Merino, I., Hernández-García, M., Turrientes, M.-C., Pérez-Viso, B., López-Fresneña, N., Diaz-Agero, C., et al. (2018). Emergence of ESBL-producing Escherichia coli ST131-C1-M27 clade colonizing patients in Europe. J. Antimicrob. Chemother. 73, 2973–2980. doi: 10.1093/jac/dky296
Mostafa, H. H., Cameron, A., Taffner, S. M., Wang, J., Malek, A., Dumyati, G., et al. (2020). Genomic surveillance of ceftriaxone-resistant Escherichia coli in Western New York suggests the extended-spectrum β-Lactamase blaCTX-M-27 is emerging on distinct plasmids in ST38. Front. Microbiol. 11:1747. doi: 10.3389/fmicb.2020.01747
Mughini-Gras, L., Dorado-García, A., van Duijkeren, E., van den Bunt, G., Dierikx, C. M., Bonten, M. J. M., et al. (2019). Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet. Health 3, e357–e369. doi: 10.1016/S2542-5196(19)30130-5
Nesta, B., Spraggon, G., Alteri, C., Moriel, D. G., Rosini, R., Veggi, D., et al. (2012). FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. doi: 10.1128/mBio.00010-12
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli st131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Oikarainen, P. E., Pohjola, L. K., Pietola, E. S., and Heikinheimo, A. (2019). Direct vertical transmission of ESBL/pAmpC-producing Escherichia coli limited in poultry production pyramid. Vet. Microbiol. 231, 100–106. doi: 10.1016/j.vetmic.2019.03.001
Päivärinta, M., Latvio, S., Fredriksson-Ahomaa, M., and Heikinheimo, A. (2020). Whole genome sequence analysis of antimicrobial resistance genes, multilocus sequence types and plasmid sequences in ESBL/AmpC Escherichia coli isolated from broiler caecum and meat. Int. J. Food Microbiol. 315:108361. doi: 10.1016/J.IJFOODMICRO.2019.108361
Päivärinta, M., Pohjola, L., Fredriksson-Ahomaa, M., and Heikinheimo, A. (2016). Low occurrence of extended-spectrum β-lactamase-producing Escherichia coli in finnish food-producing animals. Zoonoses Public Health 63, 624–631. doi: 10.1111/zph.12277
Pitout, J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front. Microbiol. 3:9. doi: 10.3389/fmicb.2012.00009
Räisänen, K., Ilmavirta, H., and Bakteerien Mikrobilääkeresistenssi Suomessa (2019). Bakteerien mikrobilääkeresistenssi Suomessa - Finres 2019. Available online at: http://urn.fi/URN:ISBN:978-952-343-588-9 (accessed August 10, 2021).
Räisänen, K., Lyytikäinen, O., Kauranen, J., Tarkka, E., Forsblom-Helander, B., Grönroos, J. O., et al. (2020). Molecular epidemiology of carbapenemase-producing Enterobacterales in Finland, 2012–2018. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1651–1656. doi: 10.1007/s10096-020-03885-w
Ray, S., Anand, D., Purwar, S., Samanta, A., Upadhye, K. V., Gupta, P., et al. (2018). Association of high mortality with extended–spectrum β-lactamase (ESBL) positive cultures in community acquired infections. J. Crit. Care 44, 255–260. doi: 10.1016/j.jcrc.2017.10.036
Roer, L., Tchesnokova, V., Allesoe, R., Muradova, M., Chattopadhyay, S., Ahrenfeldt, J., et al. (2017). Development of a web tool for Escherichia coli subtyping based on fimh alleles. J. Clin. Microbiol. 55, 2538–2543. doi: 10.1128/JCM.00737-17
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 11:10. doi: 10.1186/s13099-019-0290-0
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Tchesnokova, V. L., Rechkina, E., Larson, L., Ferrier, K., Weaver, J. L., Schroeder, D. W., et al. (2019). Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin. Infect. Dis. 68, 334–337. doi: 10.1093/cid/ciy525
Tetzschner, A. M. M., Johnson, J. R., Johnston, B. D., Lund, O., and Scheutz, F. (2020). In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58:e01269-20. doi: 10.1128/JCM.01269-20
Verkola, M., Pietola, E., Järvinen, A., Lindqvist, K., Kinnunen, P. M., and Heikinheimo, A. (2019). Low prevalence of zoonotic multidrug-resistant bacteria in veterinarians in a country with prudent use of antimicrobials in animals. Zoonoses Public Health 66, 667–678. doi: 10.1111/zph.12619
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zeng, Q., Xiao, S., Gu, F., He, W., Xie, Q., Yu, F., et al. (2021). Antimicrobial resistance and molecular epidemiology of uropathogenic Escherichia coli isolated from female patients in Shanghai, China. Front. Cell. Infect. Microbiol. 11:751. doi: 10.3389/fcimb.2021.653983
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Zheng, H., Zeng, Z., Chen, S., Liu, Y., Yao, Q., Deng, Y., et al. (2012). Prevalence and characterisation of CTX-M β-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 39, 305–310. doi: 10.1016/j.ijantimicag.2011.12.001
Keywords: antimicrobial resistance, whole genome sequencing, extended-spectrum beta-lactamases, multidrug resistance, one health
Citation: Kurittu P, Khakipoor B, Jalava J, Karhukorpi J and Heikinheimo A (2022) Whole-Genome Sequencing of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli From Human Infections in Finland Revealed Isolates Belonging to Internationally Successful ST131-C1-M27 Subclade but Distinct From Non-human Sources. Front. Microbiol. 12:789280. doi: 10.3389/fmicb.2021.789280
Received: 04 October 2021; Accepted: 10 December 2021;
Published: 04 January 2022.
Edited by:
Jørgen J. Leisner, University of Copenhagen, DenmarkReviewed by:
Michael Brouwer, Wageningen University and Research, NetherlandsCopyright © 2022 Kurittu, Khakipoor, Jalava, Karhukorpi and Heikinheimo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula Kurittu, cGF1bGEua3VyaXR0dUBoZWxzaW5raS5maQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.