- 1Faculty of Medicine, The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Woolloongabba, QLD, Australia
- 2Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, St Lucia, QLD, Australia
- 3Department of Medicine, University of Cambridge School of Clinical Medicine, Cambridge, United Kingdom
- 4Cellular Genetics, Wellcome Sanger Institute, Hinxton, United Kingdom
- 5The Institute for Molecular Bioscience, The University of Queensland, St Lucia, QLD, Australia
- 6Dermatology Research Centre, The University of Queensland Diamantina Institute, The University of Queensland, Brisbane, QLD, Australia
- 7Dermatology Department, Princess Alexandra Hospital, Brisbane, QLD, Australia
- 8QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 9The University of Queensland Centre for Clinical Research, Faculty of Medicine, The University of Queensland, Brisbane, QLD, Australia
Squamous cell carcinoma (SCC) is a common type of skin cancer that typically arises from premalignant precursor lesions named actinic keratoses (AK). Chronic inflammation is a well-known promoter of skin cancer progression. AK and SCC have been associated with an overabundance of the bacterium Staphylococcus aureus (S. aureus). Certain secreted products from S. aureus are known to promote cutaneous pro-inflammatory responses; however, not all S. aureus strains produce these. As inflammation plays a key role in SCC development, we investigated the pro-inflammatory potential and toxin secretion profiles of skin-cancer associated S. aureus. Sterile culture supernatants (“secretomes”) of S. aureus clinical strains isolated from AK and SCC were applied to human keratinocytes in vitro. Some S. aureus secretomes induced keratinocytes to overexpress inflammatory mediators that have been linked to skin carcinogenesis, including IL-6, IL-8, and TNFα. A large phenotypic variation between the tested clinical strains was observed. Strains that are highly pro-inflammatory in vitro also caused more pronounced skin inflammation in mice. Proteomic characterization of S. aureus secretomes using mass spectrometry established that specific S. aureus enzymes and cytolytic toxins, including hemolysins, phenol-soluble modulins, and serine proteases, as well as currently uncharacterized proteins, correlate with the pro-inflammatory S. aureus phenotype. This study is the first to describe the toxin secretion profiles of AK and SCC-associated S. aureus, and their potential to induce a pro-inflammatory environment in the skin. Further studies are needed to establish whether these S. aureus products promote SCC development by mediating chronic inflammation.
Introduction
Actinic keratoses (AK) are pre-malignant skin lesions that, over time, can progress to intraepidermal carcinoma (IEC) and invasive squamous cell carcinoma (SCC) (Brantsch et al., 2008; Soyer et al., 2012). AK formation on photo-damaged skin is common in fair-skinned individuals and its incidence rises substantially with age (Soyer et al., 2012). Exposure to UV radiation is the primary risk factor driving the progression from AK to SCC. Chronic inflammation is also recognized as a key driver for the malignant transformation of photo-damaged skin (Mueller, 2006; Grivennikov et al., 2010). IEC and SCC more frequently develop in chronically inflamed skin sites, such as burns, wounds, and ulcers, and are common in individuals with inflammatory skin conditions (Mueller, 2006). Inflammation predisposes to genomic instability due to inflammation-linked production of reactive oxygen species which can cause DNA damage (Mantovani et al., 2008). Additionally, inflammation-associated secretion of soluble inflammatory mediators and growth factors, such as IL-6, IL-8, and TNF-α, promote cell proliferation, inhibit apoptosis, and activate survival pathways (Lin and Karin, 2007; Grivennikov et al., 2010). Inflammatory mediators recruit leukocytes to the tumor site where these trigger stroma remodeling and angiogenesis, which, in turn, supports the high demand for nutrients and oxygen of fast proliferating tumor cells (Grivennikov et al., 2010).
Human skin is colonized by bacteria, archaea, fungi, and viruses, and the skin microbiome is known to influence the inflammatory state of the skin (Prescott et al., 2017; Byrd et al., 2018). Hence, it is plausible that cutaneous microbiota impact the malignant transformation of skin epithelial cells by mediating lasting pro-inflammatory signaling. However, relatively little is known about the impact of the local skin microbiota on the development of cutaneous SCC. Several studies have reported skin microbiome changes occurring in AK and SCC, when compared to non-malignant skin, with an overabundance of the Gram-positive bacterium Staphylococcus aureus (S. aureus) in AK and SCC being the most prominent finding (Kullander et al., 2009; Wood et al., 2018; Andersson et al., 2019; Madhusudhan et al., 2020).
S. aureus has the capacity to produce toxins and superantigens that can mediate pronounced cutaneous pro-inflammatory immune responses (Otto, 2014; Liu et al., 2017, 2018). However, this species is also well-known for its wide phenotypic heterogeneity and diverse virulence potential, which is largely mediated via its highly variable exoproteome (Ziebandt et al., 2010; Bonar et al., 2016). Thus, the quality and quantity of secreted bacterial products (the “secretome”) of S. aureus predominately determines the host response and distinct clinical manifestations. To our knowledge, the expression profiles of skin-cancer associated clinical S. aureus strains, and their potential to induce pro-inflammatory responses in the skin, have previously not been investigated. Here we demonstrate that secreted products from AK, IEC, and SCC-associated S. aureus can trigger keratinocytes to produce inflammatory cytokines that are typically upregulated in SCC. The potency of these clinical S. aureus strains to induce pro-tumorigenic inflammatory responses varied widely, due to differences in secretome composition. Our study indicates that certain S. aureus strains and products may mediate inflammation in premalignant skin lesions.
Results
Genetic Characterization of Staphylococcus aureus Isolates From Subjects With Squamous Skin Cancer
S. aureus overabundance is associated with premalignant skin and skin cancers (Kullander et al., 2009; Wood et al., 2018; Andersson et al., 2019; Madhusudhan et al., 2020). To assess the characteristics of S. aureus relevant and specific to the different progression stages of SCC, we established a biobank of 67 S. aureus isolates from non-malignant photo-damaged skin (n = 13), AK (n = 8), IEC (n = 8), or SCC (n = 38) from 16 subjects (Supplementary Table 1). For some subjects, multiple S. aureus cultures from the same skin swab were isolated to determine whether these lesions were typically populated by the same strain or a mix of genetically distinct S. aureus. Draft genome sequences were derived from 34 of our clinical isolates. Multilocus sequence typing and phylogenetic analysis of these isolates showed that S. aureus recovered from the same skin site were closely related, and that S. aureus isolated from the same subject but from different skin sites were also genetically similar (Supplementary Table 1 and Supplementary Figure 1). As strains isolated from the same skin site were genetically and phenotypically alike, we only present data for one representative isolate per skin site in the subsequent functional studies.
Staphylococcus aureus Secretions Trigger Keratinocytes to Produce Cytokines Overexpressed in Squamous Skin Cancer
The pro-inflammatory cytokines IL-6, IL-8, and TNFα possess tumor-promoting functions and increased tissue levels are associated with many types of cancer (Gambichler et al., 2006; Lin and Karin, 2007; Lederle et al., 2011; Moussai et al., 2011; Ghahartars et al., 2021). Consistent with this observation, we found these cytokines were overexpressed in AK, IEC, and SCC biopsies when compared to non-malignant skin (Figure 1A). To determine if S. aureus secreted products can directly contribute to this overexpression of pro-tumorigenic cytokines in epithelial cells, we first measured by qPCR the level of expression of IL-6, IL-8, and TNFα genes in human HaCaT keratinocytes, before and after exposure to filter-sterilized culture supernatants from S. aureus isolated from non-malignant skin (n = 4), AK (n = 4), and SCC (n = 6). IL-6, IL-8, and TNFα transcripts were generally increased in keratinocytes challenged with S. aureus secretomes when compared to keratinocytes exposed to Staphylococcus medium only, with certain secretomes being particularly potent inducers (Figure 1B). The genes encoding each of the three cytokines were induced to a similar extent by any given S. aureus secretome. The gene expression levels by qPCR were mirrored for IL-6 and IL-8 by comparable increases in protein secretion measured using a cytometric bead array (Figure 1C). Further, the levels of secreted IL-6 and IL-8 protein in response to S. aureus secretomes from HaCaT keratinocytes was comparable to those observed for primary human keratinocytes, isolated from normal skin and actinic keratosis (Figure 1C).
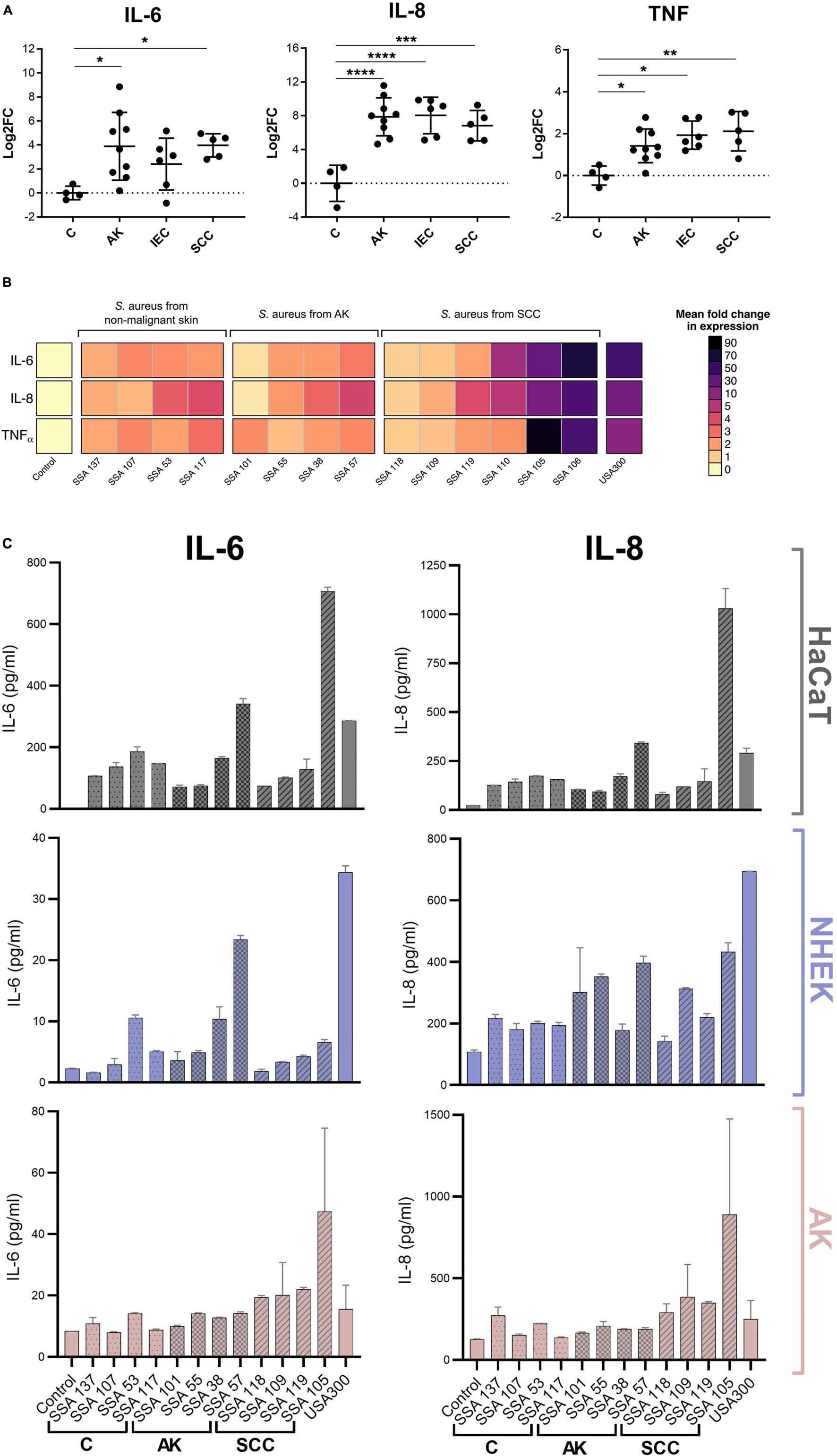
Figure 1. Staphylococcus aureus products induce transcription of cytokines that are commonly overexpressed in AK, IEC, and SCC in human keratinocytes. (A) Biopsies of actinic keratoses (AK; n = 9), intraepithelial cancers (IEC; n = 6), and squamous cell carcinomas (SCC; n = 5) assayed for IL-6, IL-8, and TNFα mRNA. Results (mean ± 1 SD) are normalized to results from healthy skin (C; n = 4). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001; Kruskal-Wallis test. (B) Expression of cytokine mRNAs by HaCaT keratinocytes exposed for 6 h to secretome from S. aureus isolated from control skin, AK or SCC lesions, normalized to mRNA expression induced by the control defined Staphylococcus media (DSM). Mean of three biological replicates ± 1 SD shown. (C) HaCaT keratinocytes and primary human keratinocytes originating from foreskin (NHEK) and actinic keratosis lesions (AK) were exposed to secretome from S. aureus strains isolated from non-malignant skin (C), AK, and SCC, or uninoculated DSM as control. After 24 h, cytokine levels in keratinocyte conditioned media were measured via multiplex bead-based cytometric assay. Displayed is the mean of two biological replicates ± 1 SD.
Large Heterogeneity of Staphylococcus aureus Clinical Isolates to Promote IL-6 Secretion in Human Keratinocytes
As the induction of pro-tumorigenic cytokines from keratinocytes by S. aureus secretomes appeared highly strain-specific, we next investigated the potency of a larger number of secretomes from different clinical isolates, originating from non-malignant control (C) (n = 5); AK (n = 5); IEC (n = 8); SCC (n = 13), to induce IL-6 secretion from human keratinocytes by using a reporter cell assay. Additionally, keratinocyte viability after exposure to secretomes was assessed, to measure toxicity-dependent effects. S. aureus secretomes varied in their ability to reduce cell viability (Figure 2A) and induce the secretion of IL-6 (Figure 2B) in HaCaT keratinocytes. Cytotoxic S. aureus secretomes tended to induce higher levels of IL-6. However, many secretome samples were non-toxic, but yet triggered IL-6 secretion, indicating that cytokine induction was not directly linked to toxicity (Figure 2C). A significant increase in keratinocyte IL-6 secretion was observed in response to secretomes from S. aureus originating from IEC (p = 0.0001) and SCC (p = 0.0001), when compared to the media control (Figure 2B). Notably, secretomes from other Staphylococcus species isolated from AK, IEC, and SCC swabs, including S. epidermidis and S. capitis, induced lower levels of IL-6 than lesion-associated S. aureus secretomes (Figure 2B). The keratinocyte IL-6 secretion mediated by S. aureus supernatant was not dependent on bacterial culture conditions, or on bacterial cell counts at the time of supernatant collection, and was comparable to IL-6 secretion in response to keratinocyte infection with unfiltered S. aureus supernatant containing live cells (Supplementary Figure 2). To validate the IL-6 results obtained using a cell-based reporter assay, a cytometric bead array was used as an additional method to measure secreted IL-6 levels from keratinocytes after challenge with 12 different S. aureus secretomes. The IL-6 concentration quantified via bead array closely matched the IL-6 concentration measured via cell reporter assay, confirming reliability of the IL-6 results (R2 = 0.97; p < 0.0001) (Supplementary Figure 3).
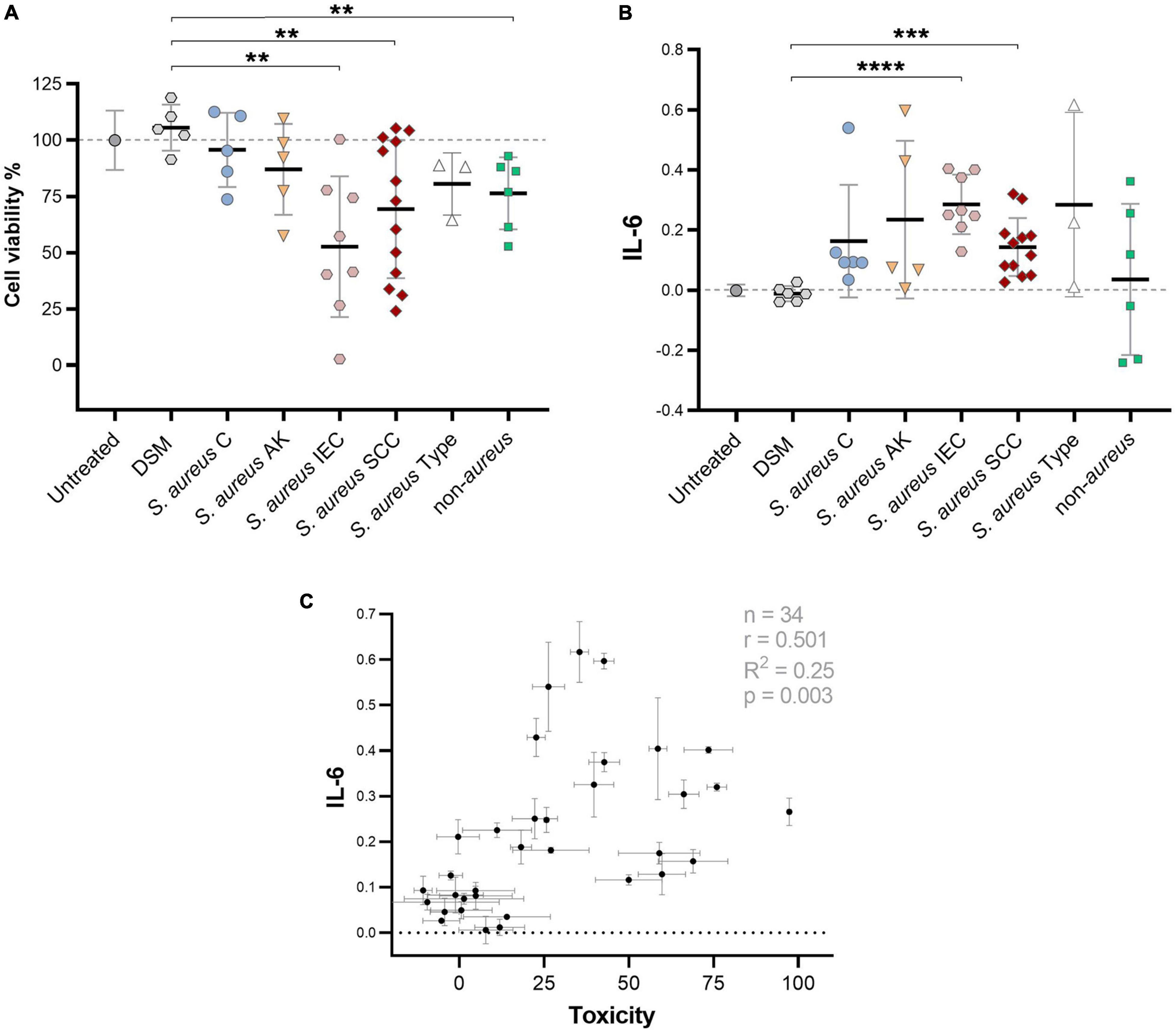
Figure 2. Large variance of clinical S. aureus to induce cytotoxic and IL-6 responses in keratinocytes. (A) Percent viability of HaCaT keratinocytes after challenge with secretome from S. aureus isolates obtained from control skin (C), AK, IEC, and SCC, S. aureus lab type strains USA300, ATCC25923, and ATCC29213, as well as secretome from other Staphylococcus species (S. epidermidis and S. capitis isolated from AK, IEC, or SCC) as measured via MTT assay. The different batches of the bacterial media control (DSM) used to culture the tested staphylococci strains served as negative control. Normalized results are displayed as percentage of viable cells (untreated = 100% viable; cells killed with lysis buffer = 0%). Bars indicate the group mean ± 1 SD; each data point represents the mean of triplicate measurement of a different isolate; DSM n = 3; untreated control n = 12. Statistical significance between DSM and treatment groups determined by unpaired t-test; two-tailed p-value indicated. (B) Relative IL-6 levels as measured by colorimetric cell reporter assay (absorbance at 650 nm) in keratinocyte conditioned media after exposure to secretome from S. aureus or other Staphylococcus species (untreated cells = baseline). S. aureus secretome samples are grouped based on swab origin. One data point represents IL-6 activity of a single isolate secretome (mean of triplicate measurements). Bars indicate mean ± 1 SD. Statistical significance between DSM and treatment groups determined by unpaired t-test; two-tailed p-value indicated. (C) Pearson’s correlation analysis between keratinocyte IL-6 levels and toxicity in response to treatment with 34 S. aureus secretome samples (two-tailed p-value 0.0025). **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
IL-6-Inducing Staphylococcus aureus Stimulate More Pronounced Pro-inflammatory Skin Responses in Mice
IL-6 trans-signaling typically induces local and systemic pro-inflammatory responses, in part by recruiting and activating innate immune cells (Turner et al., 2014). To test whether S. aureus secretomes that promoted a high expression of IL-6 in keratinocytes in vitro were also potent activators of innate immune responses in the skin in vivo, a mouse model of intradermal injection of S. aureus secretome into the ear skin was utilized. Mouse ears were injected either with secretome that had no significant effect on IL-6 production in cultured keratinocytes, or secretome that was highly IL-6-inducing (Figure 3A). At 24 h post-injection, most IL-6-inducing secretomes caused a statistically significant increase in neutrophil, monocyte, and macrophage infiltrates, compared to ears injected with the bacterial media control (Figure 3B). S. aureus secretomes that had no effect on keratinocyte IL-6 production in vitro produced non-significant increases in immune populations in the murine skin (Figure 3B). A substantial positive correlation between the in vitro IL-6 keratinocyte signal and the in vivo infiltrates was observed, though not statistically significant (Figure 3C).
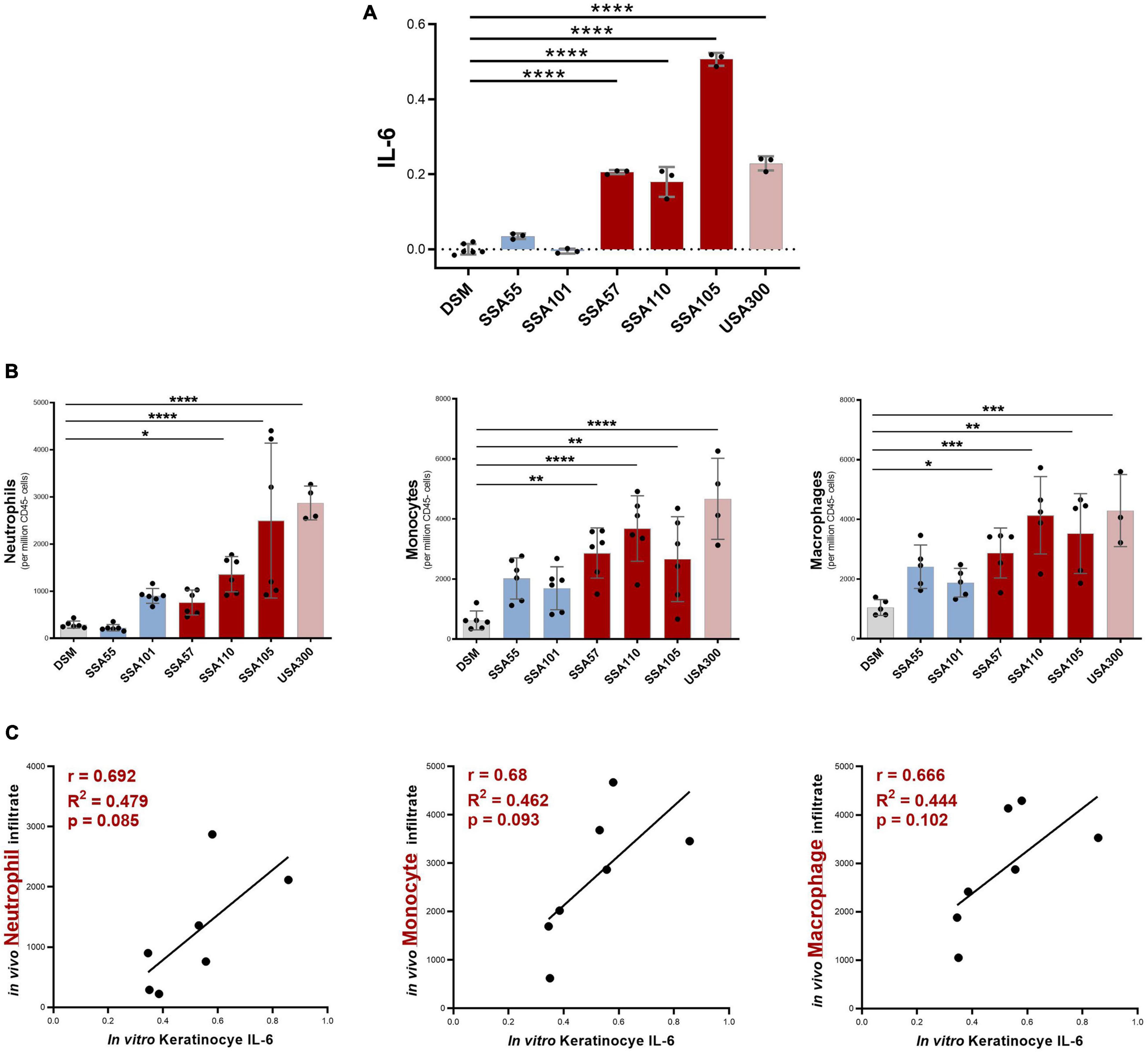
Figure 3. Staphylococcus aureus secretome-induced IL-6 levels from keratinocyte in vitro predict neutrophil, monocyte, and macrophage recruitment in response to secretome in vivo. (A) Six S. aureus secretomes were selected for in vivo studies which had no significant effect (blue), or a strong positive effect (red), on keratinocyte IL-6 secretion in vitro (relative IL-6 concentration based on OD value from cell reporter assay; values normalized to baseline = DSM; mean ± 1 SD: n = 3 per secretome; Dunnett’s multiple comparisons test ****P < 0.0001). (B) Immune infiltrate in mouse ear skin 24 h post injection with S. aureus secretomes, determined by flow cytometry (mean ± 1 SD: n = 6 per secretome); Statistical significance determined by Dunnett’s multiple comparisons test. (C) Correlation between in vitro keratinocyte IL-6 induction and in vivo neutrophil, monocyte, and macrophage infiltrate in response to S. aureus secretome exposure. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001.
A Specific Secretome Profile Is Linked to the Pro-inflammatory Staphylococcus aureus Phenotype
To identify bacterial proteins contributing to the cytokine inducing S. aureus phenotype, the protein content of the secretome from 28 clinical isolates derived from non-malignant photo-damaged skin (n = 10); AK (n = 8); SCC (n = 10), and three S. aureus type strains, was characterized by shotgun mass spectrometry (MS) following trypsin digest. The IL-6 levels secreted by keratinocytes in response to these MS-characterized secretomes was used as representative readouts for inflammatory potential, and ranged from non-inflammatory to highly pro-inflammatory secretomes (Supplementary Table 2). A total of 348 S. aureus proteins were identified across the 31 secretomes and the relative quantity of these proteins in each secretome sample was determined by MaxQuant label-free quantification. Using Spearman’s correlation with an adjusted p < 0.05 as cut off, we identified 37 S. aureus proteins positively correlated, and three negatively correlated, with IL-6 induction (Table 1). IL-6-inducing potential of S. aureus secretomes was correlated with levels of potent cytolytic toxins including leucocidins, hemolysins, and phenol-soluble modulins (PSMs), as well as the SarA protein, which is involved in the regulation of these toxins. Additionally, multiple serine proteases and cell wall modifying enzymes such as lipoteichoic acid synthase were associated with the ability of S. aureus to cause pro-inflammatory skin responses. Interestingly, we also identified six as-yet uncharacterized proteins associated with the pro-inflammatory S. aureus phenotype, including a probable CtpA-like serine protease, a putative phosphoesterase, and an uncharacterized peptidase and lipoprotein.
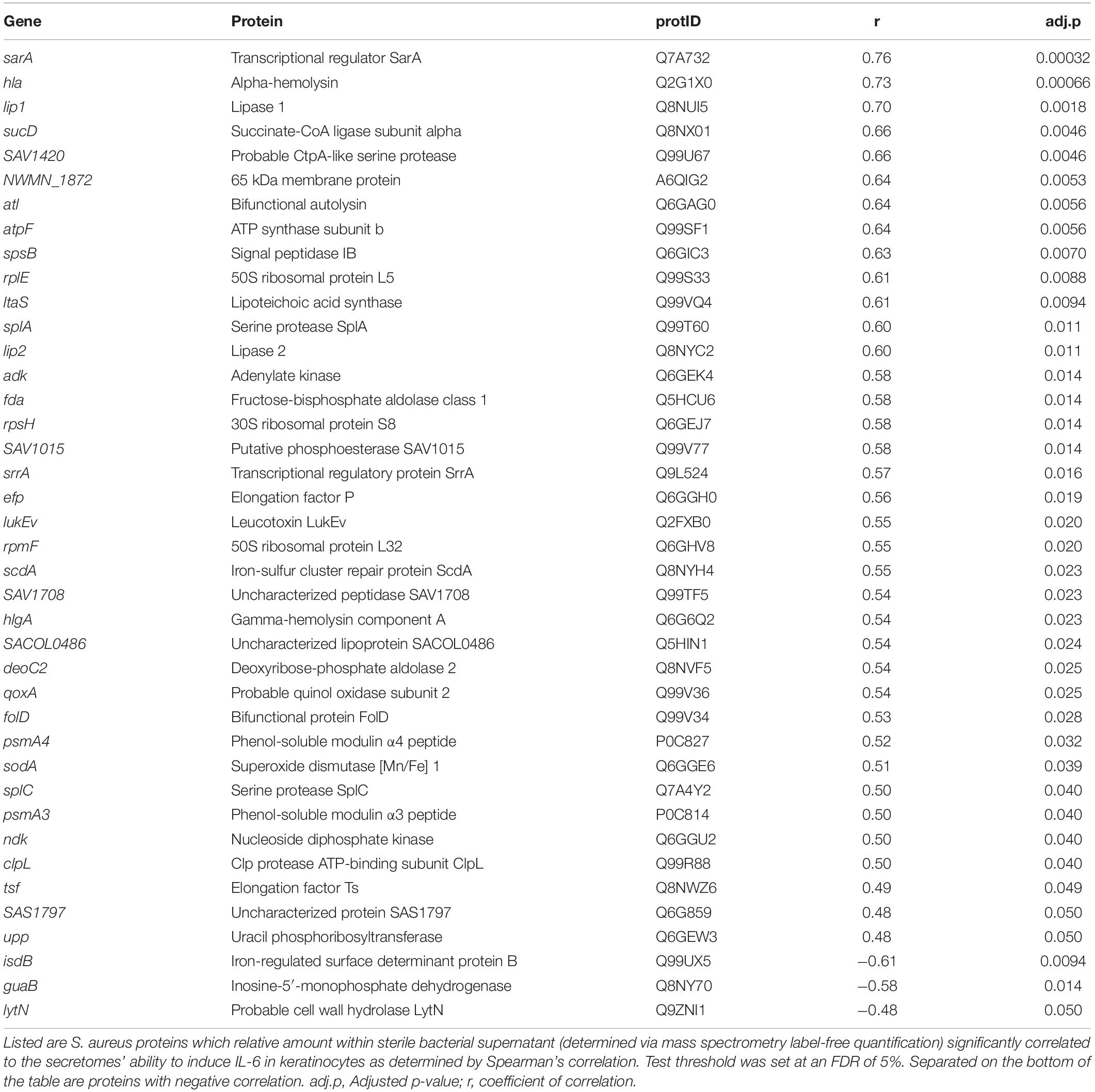
Table 1. S. aureus peptides and proteins that are significantly correlated with S. aureus-mediated IL-6 induction in human keratinocytes.
Neutralization of one of the most positively correlated proteins, α-toxin, caused a significant reduction in the ability of secretomes to induce IL-6 in cultured keratinocytes (Figures 4A,B). Conversely, the addition of recombinant α-toxin to S. aureus secretomes that typically had no impact on keratinocyte IL-6 levels caused these secretomes to become potent activators of IL-6 secretion (Figure 4C).
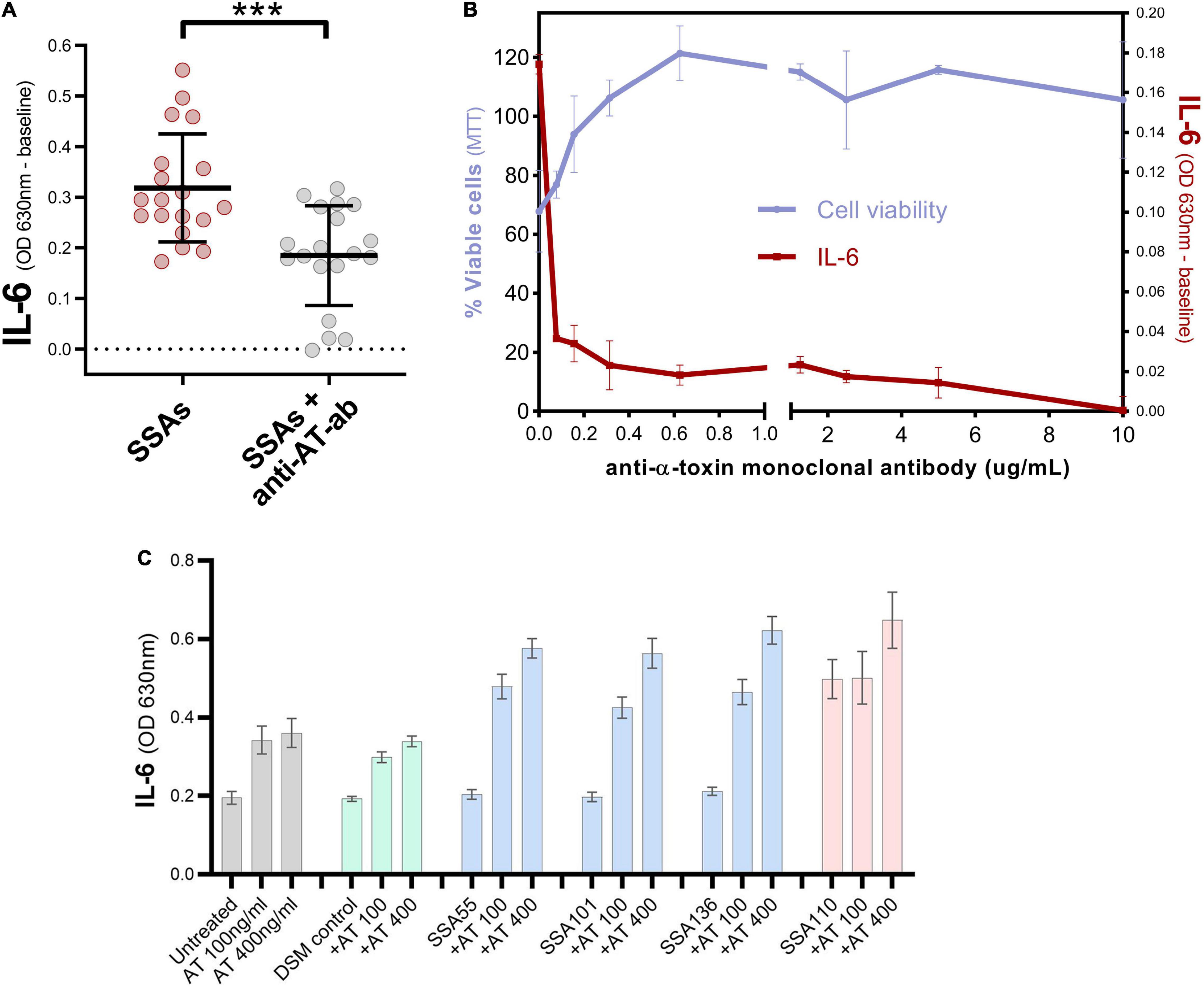
Figure 4. α-toxin contributes to the induction of IL-6 by S. aureus secretomes. (A) Keratinocyte secreted IL-6 levels after exposure to 19 different S. aureus secretomes with or without addition of anti-α-toxin polyclonal antibody (mean ± 1 SD: n = 3 per secretome). IL-6 absorbance values were normalized to baseline i.e., IL-6 value for untreated cells. Unpaired t-test; two-tailed ***p = 0.0003. (B) Validation of polyclonal antibody data, by testing different concentrations of anti-α-toxin monoclonal antibody in combination with secretome from a SCC-derived S. aureus isolate SSA105 known to contain high levels of α-toxin. Percentage of viable keratinocytes (left axis) and keratinocyte IL-6 response (IL-6 absorbance values baseline-corrected to untreated cells, right axis) after exposure to secretome from SSA105 ± anti-α-toxin monoclonal antibody. Neutralizing α-toxin results in loss of cytotoxic action of the tested S. aureus secretome and diminishes its ability to stimulate IL-6 secretion from HaCaT cells. Error bars indicate mean and standard deviation (n = 2). (C) Keratinocyte secreted IL-6 levels after exposure to S. aureus secretomes with or without addition of 100 or 400 ng/ml recombinant α-toxin (mean ± 1 SD: n = 3 per secretome), compared to untreated cells (gray) and control cells treated with uninoculated bacterial media DSM (green). Secretome from S. aureus isolates SSA55, SSA55, and SSA55 naturally contain very low levels of α-toxin (blue), whereas SSA110 naturally produces high levels of α-toxin (red).
Some S. aureus strains may generally secrete higher levels of bacterial proteins than others, which could affect keratinocyte responses. Indeed, the keratinocyte IL-6 readout also showed a significantly positive correlation to the total protein content of S. aureus secretomes (Supplementary Figure 4A). However, this was not a culturing-dependent artifact caused by differences in growth since the overall protein content of S. aureus secretome did not correlate with bacterial cell density at time of harvest (Supplementary Figure 4B).
Pro-inflammatory Staphylococcus aureus Secretome Regulation Is Independent of Genotype
To address whether the observed phenotypic differences in pro-inflammatory potential of S. aureus clinical isolates was associated with defined differences in the bacterial genome, an overlay matrix (Figure 5) was generated to compare the presence or absence of specific genes with the relative abundance of the corresponding protein for the S. aureus candidates presented in Table 1 that were significantly associated with keratinocyte IL-6 induction. If S. aureus isolates with no or minimal effect on IL-6 lack the relevant gene for proteins positively correlated with IL-6 induction, this would indicate that the observed phenotypic differences are due to genetic variance. However, non-IL-6 inducing S. aureus supernatants showed some expression of bacterial proteins correlated with IL-6 expression (Figure 5), and even when bacterial protein expression was not detected, a functional gene copy was often present. Therefore, the observed heterogeneity in induction of IL-6 by S. aureus supernatants is more readily explained by a variation in bacterial protein expression, post-translational modifications, and/or targeted protein degradation or stabilization, rather than the presence or absence of a specific bacterial gene.
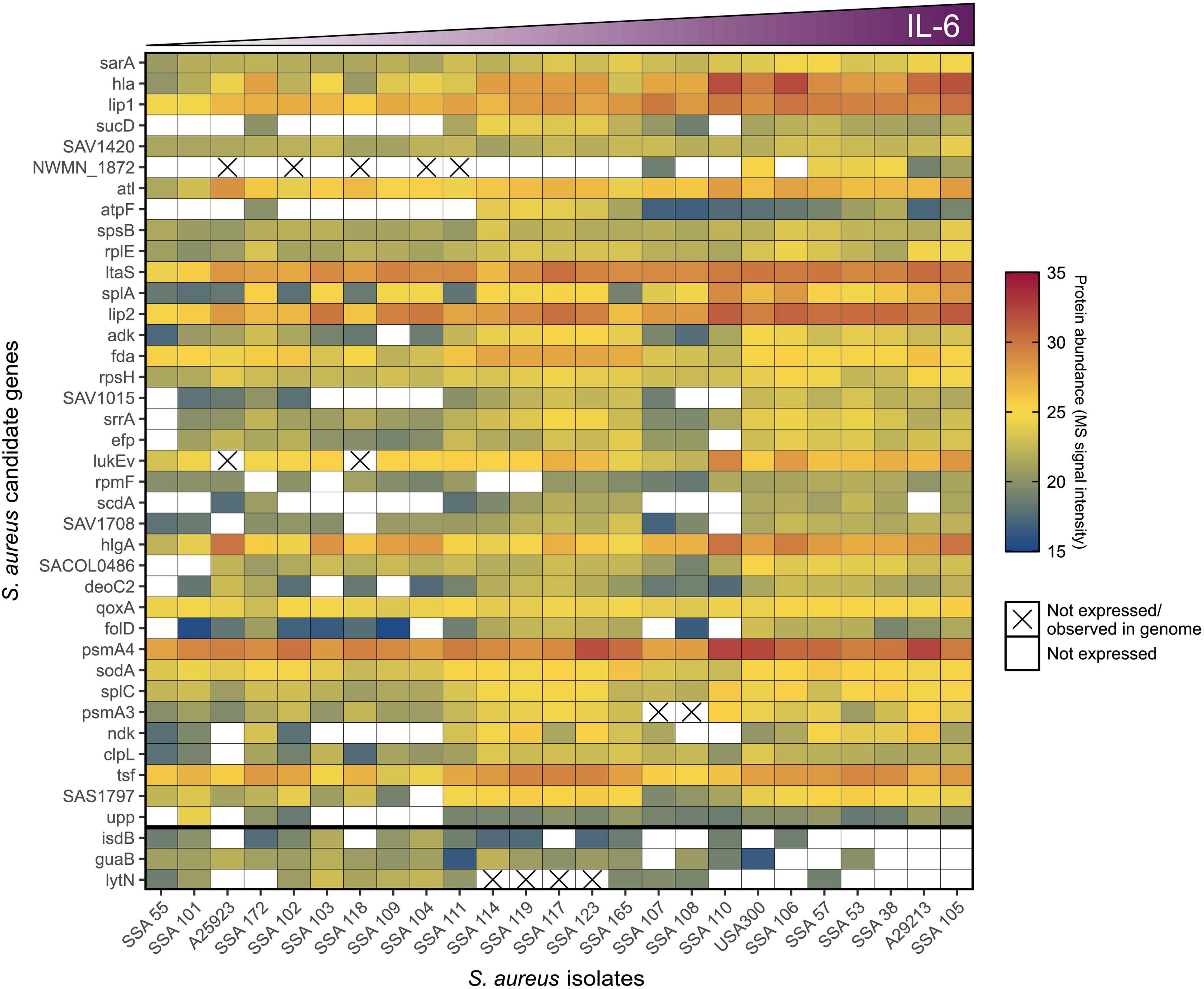
Figure 5. The pro-inflammatory potential of S. aureus is determined by differences in gene regulation. Genetic and proteomic characteristics of statistically significant candidates (Table 1) that positively (n = 37, ordered by significance top to bottom) or negatively (n = 3, separated to bottom of heat map) correlated to S. aureus secretomes’ ability to induce IL-6 in keratinocytes (ordered by level of IL-6 induction left to right). Protein abundance, i.e., MS signal intensity of candidates within the different S. aureus secretome samples, is indicated by color scale, where white is no recorded signal. The cross signifies the absence of a functional gene.
Discussion
Keratinocyte cancer and its precursor stages are associated with an increased abundance of Staphylococcus aureus (Kullander et al., 2009; Wood et al., 2018; Andersson et al., 2019; Madhusudhan et al., 2020). S. aureus can secrete products which induce pro-inflammatory signaling in the skin (Otto, 2014; Liu et al., 2017, 2018), and chronic inflammation is linked to skin cancer development and progression (Mueller, 2006; Grivennikov et al., 2010). It therefore seems plausible that S. aureus colonizing pre-malignant lesions may chronically activate pro-inflammatory signaling pathways that could aid cancer progression. However, S. aureus has considerable strain-level heterogeneity (Ziebandt et al., 2010; Bonar et al., 2016), and not all strains may adversely impact the inflammatory state of the skin. The phenotypes of skin lesion-associated S. aureus in relation to cutaneous inflammation have previously not been reported. Here, we demonstrate that secretome components of S. aureus isolated from AK, IEC, and SCC lesions can induce cancer-linked pro-inflammatory responses in cultured human keratinocytes and murine skin.
The pro-inflammatory cytokines IL-6, IL-8, and TNFα possess cancer-promoting functions and are commonly overexpressed in many malignancies including keratinocyte cancers (Gambichler et al., 2006; Lin and Karin, 2007; Lederle et al., 2011; Moussai et al., 2011; Ghahartars et al., 2021). In line with this, we show these cytokines are overexpressed in skin biopsies of AK, IEC, and SCC. We demonstrate that secretomes from S. aureus isolated from photo-damaged skin, AK, and SCC can cause enhanced gene expression of IL-6, IL-8, and TNFα in human keratinocytes. Some secretomes were particularly potent at inducing these pro-tumorigenic inflammatory mediators, and hence, colonization with certain S. aureus strains may more readily contribute to a tumor-promoting microenvironment in the skin than others. Further, we show that keratinocytes challenged with IEC or SCC-associated S. aureus secretome secrete significantly increased levels of IL-6 protein. Interestingly, the overexpression of IL-6 in cultured human keratinocytes has previously been found to result in enhanced angiogenesis, proliferation and migration, and resistance to apoptosis (Jee et al., 2001; Lederle et al., 2011). Type 1 cytokine signaling through IL-6 typically propagates systemic pro-inflammatory responses (Turner et al., 2014). Consistent with this, S. aureus secretomes identified to be strong IL-6 inducers in vitro also triggered pronounced innate immune responses in murine skin. Thus, skin colonization with IL-6-inducing S. aureus may cause a persistent accumulation of lingering immune cells, which has been linked to cancer progression (Grivennikov et al., 2010).
In both our in vitro and in vivo experiments, there were clear strain-level differences in the potency of S. aureus to induce pro-inflammatory skin responses. Proteomic and genetic analyses revealed that the observed difference in S. aureus phenotypes was due to variances in regulation and secretion of potent cytolytic toxins, such as leucocidins, hemolysins, and PSMs, as well as multiple serine proteases. The pro-inflammatory properties of these factors have been well-documented in the past (Zdzalik et al., 2012; Otto, 2014; Liu et al., 2017; Stentzel et al., 2017). PSM α3 and α4 were significantly correlated with IL-6 induction in keratinocytes, consistent with a recent study showing α-type PSMs to be potent inducers of a range of pro-inflammatory cytokines, including IL-6, in human primary keratinocytes and skin explants (Damour et al., 2021). PSMα peptides are known to mediate the release of membrane-embedded S. aureus lipoproteins (Hanzelmann et al., 2016), and lipoproteins trigger TLR2-dependent immune activation (Stoll et al., 2005; Nguyen and Götz, 2016; Mohammad et al., 2021). Interestingly, we identified a to-date uncharacterized lipoprotein (SACOL0486) associated with pro-inflammatory skin responses, alongside a number of other uncharacterized proteins that could play as-yet-unknown roles in the virulence of this important pathogen. We also found that lipoteichoic acid synthase was positively correlated with the pro-inflammatory S. aureus phenotype. Lipoteichoic acid is a separable cell wall-anchored amphiphilic glycolipid with pro-inflammatory and pathogenic properties that, of note, has been shown to encourage enhanced proliferation of adeno- and squamous cell carcinoma cells (Hattar et al., 2017). Lastly, further experimental evidence supports the results of our correlation analysis as the antibody neutralization of α-toxin, one of our identified candidates most strongly associated with IL-6 activity, in the S. aureus supernatants caused a significant reduction in IL-6-inducing potency. This is consistent with findings from previous studies that support α-toxins’ direct, and indirect, contributions to IL-6 induction in keratinocytes and a range of other cell types in vitro as well as in vivo mouse models (Onogawa, 2002, 2005; Hruz et al., 2009; Hong et al., 2014).
This study provides valuable insights into the secreted proteome of S. aureus isolates recovered from photo-damaged skin and different SCC developmental stages, and shows that strain-specific pro-inflammatory effects are most likely explained by differential regulation of toxins. A caveat of our study is that filter-sterilized culture supernatant from S. aureus not only contains secreted proteins but also intracellular proteins released by the lysis and turnover of bacterial cells, as well as other S. aureus-derived metabolites and molecules, such as DNA and RNA, that could promote immune activation. Moreover, the growth of S. aureus in liquid culture may produce different secreted products and/or at different quantities than during skin colonization due to higher nutrient availability, less environmental stress, and lacking interactions with other microbial species and host cells. Therefore, in vivo studies are required to establish whether S. aureus skin colonization indeed mediates lasting pro-inflammatory effects that can aid SCC development and progression. This could have important implications in the treatment of AK and prevention of IEC and SCC.
Materials and Methods
RNA Sequencing of Skin Cancer Biopsies
RNA sequencing was performed on four healthy skin samples and nine AK, six IEC, and five SCC lesion biopsies. Part of the biopsy was sectioned to confirm diagnosis via histology. The 24 skin samples were collected from 15 individuals at the Dermatology department in Princess Alexandra Hospital. Ethical approval was obtained by Metro South Human Research Ethics Committee and the University of Queensland Human Research Ethics Committee (HREC-11-QPAH-236, HREC-11-QPAH-477, HREC-12-QPAH-217, and HREC-12-QPAH-25). Written, informed consent was obtained from all patients prior to participation. Fresh biopsy tissues were immersed in RNA later (Life Technologies, Carlsbad, CA) and stored at −80°C until processing. RNA isolation was performed using the QIAGEN RNeasy Plus Mini kit. RNA concentration was measured on the Qubit fluorometer (Life Technologies, Carlsbad, CA) and RNA integrity determined using the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) on RNA Pico chips. RIN > 6 was set as minimum acceptable quality to proceed. RNA-Seq libraries of poly (A) RNA from 500 ng total RNA were generated using the TruSeq unstranded mRNA library prep KIT for AK, IEC, and SCC samples. TruSeq stranded mRNA library preparation kit was used to generate poly (A) RNA libraries from RNA extracted from normal skin. Libraries were sequenced (100 bp, paired-end) on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, United States). Quality of raw data was assessed using FastQC (version 0.11.2) (Andrews, 2010). Adapters and poor-quality sequences (∼6% of reads) were removed using Trim Galore v0.3.7 (Krueger, 2015). Trimmed reads were then aligned using the unstranded protocol in TopHat (Trapnell et al., 2012) against the Human Genome (hg19 build) guided with a human transcriptome generated from the GENCODE Gene annotation v17 (Harrow et al., 2012). Raw counts were normalized using TMM method using Bioconductor package edgeR version 3.3. Quantification of gene expression was based on counting the overlaps of mapped reads with genes annotated in the GENCODE gene annotation v17 and was conducted via HTSeq (Anders et al., 2015).
Clinical Sampling of Skin Cancer and Precancerous Lesions
Individuals with AK, IEC, and/or SCC lesions were recruited at the Princess Alexandra hospital (Brisbane, Australia) under approved protocols HREC/16/QPAH/364 and HREC/11/QPAH/477. Exclusion criteria were the presence of chronic skin disorders, as well as current or recent use of antibiotics and/or topical agents to treat AK. Skin swabs from AK were collected from the forearm only, whereas those from IEC and invasive SCC were collected from any UV-exposed body site. Non-malignant control swabs were collected adjacent to each sampled lesion on an anatomically similar skin site. Sterile swabs were first dipped into 0.15 M sodium chloride solution, firmly rotated over the skin sampling area for 30 s, and then collected in sterile glycerol-saline solution and stored immediately at −80°C. Suspected IEC and SCC lesions were excised after swabbing and the diagnosis confirmed via histopathology.
Isolation of Staphylococcus Clinical Strains
Swab samples preserved in glycerol were plated onto Staphylococcus genus-selective mannitol salt agar (MSA). After 48 h at 36°C, a representative sample of colonies from each MSA-positive plate were picked and restreaked onto new MSA plates. Golden colonies that formed a yellow halo on the red MSA agar were selected as indicative of the S. aureus species. After 24 h of growth, a single colony was transferred into tryptic soy broth and grown overnight. The next morning, a subsample of the live bacterial suspension was stored in 100% glycerol (1:1) and the remaining biomass was collected by centrifugation and resuspended in lysis buffer (500 mM NaCl, 50 mM TRIS-HCl [pH 8.0], 50 mM EDTA, 4% Sodium Dodecyl Sulfate) for DNA extraction and sequencing. Both the live glycerol stock and lysed bacteria were stored at −80°C.
Species Identification
The MSA-positive isolates were first subjected to Gram staining, and Gram positive cocci were chosen for coagulase and catalase test following a standard protocol (Moyes et al., 2009; Reiner, 2010). For coagulase and catalase-positive isolates, a PCR was carried out using S. aureus-specific primers targeting the nuc gene as previously published (Kullander et al., 2009; Reuschenbach et al., 2011). A thermal extraction method was used to collect DNA from liquid cultures of the chosen isolates. Overnight cultures were pelleted by centrifugation and resuspended in 200 μL of ultrapure water. The cell suspension was then incubated in a heating block at 100°C for 10 min. Lysates were immediately transferred onto ice and centrifuged at 10,000 × g for 3 min. In addition, 2 μL of the supernatant were used in a 15 μL PCR assay using Taq DNA polymerase (0.1 μL per sample) with standard Taq buffer and 20 μm of each nuc primer (nuc forward 5′-GCGATTGATGGTGATACGGTT-3′; nuc reverse 5′-AGCCAAGCCTTGACGAACTAAAGC-3′). PCR cycle setting were as follows: 94°C for 2 min, followed by 37 cycles of 94°C for 1 min, 55°C for 30 s and 72°C for 1.5 min, and lastly 72°C for 3.5 min followed by a 4°C hold. The 263-bp PCR product was confirmed via visualization on a 1.5% agarose gel. Lastly, nuc-positive isolates were confirmed as S. aureus via 16S rRNA gene sequencing. To purify DNA for sequencing, the frozen vial of bacteria resuspended in lysis buffer was thawed and DNA extracted using a repeated bead beating step for cell lysis, followed by DNA purification utilizing the Maxwell® 16 MDx instrument (Promega). The 16S rRNA gene was amplified using 27F and 1492R primers (27F 5′-AGAGTTTGATCCTGGCTCAG-3′; 1492R 5′-GGTTACCTTGTTACGACTT-3′), and subsequently the reaction mixture was purified and cleaned from unincorporated dyes, nucleotides, salts, and contaminants using the Agencourt® CleanSEQ® kit according to standard protocol. The PCR product was subjected to Sanger sequencing and the resulting amplicon sequence was used to query the GenBank database. Isolates that were Gram-positive cocci, catalase and coagulase-positive, were nuc PCR-positive, and confirmed affiliation with S. aureus via 16S RNA gene sequencing were included in this study and selected for secretome production.
Whole Genome Sequencing of Staphylococcus aureus Isolates
The genomic DNA prepared from 34 of our S. aureus isolates was used to produce draft genome assemblies. DNA libraries were prepared with a Nextera Flex library preparation kit (Illumina, #20018705). Library preparation was run on the Mantis Liquid Handler (Formulatrix). Resulting amplified libraries were cleaned-up as per the “Clean Up Libraries” section in the manufacturer’s protocol. Each library was then quantified using the Quant-iT™ dsDNA HS Assay Kit (Invitrogen) and quality assessed via Agilent D1000 HS tapes (#5067-5582) on the TapeStation 4200 (Agilent # G2991AA) as per the manufacturer’s protocol. Nextera DNA Flex libraries were pooled at equimolar amounts of 0.5 nM per library to create a sequencing pool. The library pool was then again quantified in triplicates using the Qubit™ dsDNA HS Assay Kit (Invitrogen). Library quality control was performed using the Agilent D1000 HS tapes (#5067-5582) on the TapeStation 4200 (Agilent # G2991AA). The library was prepared for sequencing on the NextSeq500 (Illumina) using NextSeq 500/550 High Output v2 2 × 150bp paired end chemistry according to manufacturer’s protocol. Approximately 0.5 GB of sequence data was produced per sample. Raw reads for isolated strains were processed with Trimmomatic (ver. 0.39; LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:50) for quality filtering (Bolger et al., 2014). Quality filtered reads were then assembled using SPAdes (ver. 3.14.0) (Bankevich et al., 2012) as part of the Shovill assembly pipeline (ver. 1.1.0, T. Seeman, unpublished, https://github.com/tseemann/shovill “–isolate,” –minlen 1000), with the completeness and contamination of each assembly evaluated using CheckM (ver. 1.1.2) (Parks et al., 2015) and taxonomy assigned using GTDB-Tk (ver. 1.3) (Chaumeil et al., 2019).
A phylogenetic tree was constructed for the assembled 34 S. aureus draft genomes based on a core-alignment of single nucleotide polymorphisms (SNPs) following the removal of recombinant regions. Draft genomes were aligned against the S. aureus USA300_FPR3757 reference genome (RefSeq accession GCF_000013465.1) using Parsnp (ver. 1.5.6) (Treangen et al., 2014), with recombinant regions identified and removed using Gubbins (ver. 3.0.0) (Croucher et al., 2015). A maximum-likelihood phylogenetic tree was constructed using RAxML (ver. 8.2.12) using a general time-reversible nucleotide substitution model with gamma correction for site variation (GTRGAMMA) and 1,000 bootstraps. The phylogenetic tree was rooted on the reference USA300_FPR3757 genome and visualized using the ggtree package (ver. 2.2.4) (Yu et al., 2017) in R (ver. 4.0.2). Multi-locus sequence typing (MLST) was performed with MLST ver. 2.17.6, https://github.com/tseemann/mlst, which scans against PubMLST typing schemes1 (Jolley et al., 2018).
Production of Staphylococcus aureus Sterile Culture Supernatant
For secretome production, S. aureus strains were cultured in chemically defined staphylococcal medium (DSM) (adapted from Hussain et al., 1991; composition in Supplementary Table 3) as this medium without added proteins or peptides is compatible with mass spectrometry. Clinical isolates were plated fresh from glycerol stock onto MSA and left to grow for 24 h at 37°C and 5% CO2. A single colony was then used to inoculate 6 ml DSM within 15 mL round bottom tubes and grown with shaking (200 rpm) at 37°C. After overnight growth, the optical density at 600 nm (OD 600) of the cultures was measured, and a volume of the culture was used to inoculate 100 mL DSM so as to provide an initial OD 600 of 0.15. The cultures were shaken at 200 rpm and incubated at 37°C for 24 h, with growth monitored via OD readings. After 24 h, the final OD 600 value was measured. Then, the cultures were centrifuged at 800 × g for 20 min at 4°C to pellet the cell biomass. The supernatant was carefully transferred into a fresh tube and centrifuged at 3,000 x g for 20 min at 4°C. The resulting supernatant was filter-sterilized through 0.2 μm regenerated cellulose (Corning; CLS431222) and partitioned into single-use aliquots, which were stored immediately at −80°C. Uninoculated DSM was treated precisely the same way to use in experiments as a negative control. The sterility of the secretome samples was routinely confirmed by plating aliquots of the filtered bacterial supernatant onto agar and monitored for growth at 37°C. For experiments, aliquots were freshly thawed, used immediately, and discarded after use.
Initially, sterile culture supernatant was produced from four of our S. aureus isolates in three biological replicates, i.e., three different colonies were inoculated, grown for 24 h, and sterile supernatant collected as described above. As the effect of the matching replicate supernatants on keratinocyte cytokine induction was highly consistent, there was only one biological replicate of supernatant produced and tested from the remaining isolates in our functional assays (methods below).
Keratinocyte Cell Culture and Experiments
HaCaT cells were cultured in DMEM medium supplemented with 10% fetal calf serum (FCS) and 100 U/mL penicillin/streptomycin at 37°C and 5% CO2. HaCaT experiments were performed up to passage 30 in serum-free culture conditions using DermaLife K keratinocyte basal medium (Lifeline; SKU: LL-0007). Primary human keratinocytes were isolated from abdominal skin, foreskin, or actinic keratosis lesions collected at the Princess Alexandra Hospital Dermatology Centre (Brisbane, Australia) with patient consent and institutional approval (HREC/11/QPAH/477). Primary keratinocytes were cultured in DermaLife K keratinocyte basal medium supplemented with 10 μM Rho Kinase inhibitor Y-27632 (Sigma Aldrich; SCM075). Experiments on primary keratinocytes were performed up to passage eight. For S. aureus secretome stimulation assays, keratinocytes were seeded in 96-well plates and, unless otherwise stated, experiments were initiated at approximately 70% cell confluence. The keratinocytes were then incubated with sterile culture supernatant of S. aureus and bacterial medium control, diluted 1:40 in DermaLife medium, for 24 h. By end point, keratinocyte cultures reached confluence and cell supernatant was collected for cytokine quantification and the cell viability assessed as described below in 4.8 and 4.9, respectively.
Assessment of Cytokine Protein Levels
To measure IL-6 in keratinocyte conditioned media after challenge with S. aureus secretome, the HEK-Blue™ IL-6 reporter cell line from InvivoGen was used according to the provided standard protocol. This cell system employs HEK293 cells stably transfected with the human IL-6R gene and STAT3-inducible reporter gene of secreted embryonic alkaline phosphatase (SEAP). The presence of IL-6 in keratinocyte supernatant triggers STAT3 activation in transfected HEK293 cells and subsequent production of SEAP. SEAP levels were monitored by QUANTI-Blue™ colorimetric assay at an absorbance of 650 nm. Statistical analyses were performed with GraphPad Prism version 8.3.1 for Windows (GraphPad Software, San Diego, CA), with p ≤ 0.05 considered statistically significant. To validate cytokine results, a multiplex bead array kit by BioLegend was used according to the provided manual (LEGENDplex™ Human Inflammation Panel 1 [13-plex] with V-bottom Plate; Cat#740809). The assay was performed on 25 μL of undiluted keratinocyte culture supernatant and results were acquired on the CytoFLEX flow cytometer platform (Beckman Coulter; Cat#B53000). Data was analyzed with BioLegend’s LEGENDplexTM Data Analysis Software. Standard was prepared in technical duplicates and samples in biological triplicates. The standard curves for each monitored marker had a CV ≤ 1.7%.
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) Assay
To assess keratinocyte cell activity, the MTT assay was employed as developed by Mossmann (1983) with MTT purchased by Invitrogen™ (M6494). Keratinocyte culture medium was replaced with phenol-free medium containing 0.5 mg/ml MTT solution. After 4-h incubation at 37°C, the purple formazan crystals were dissolved in 60 μL dimethyl sulfoxide. The plates were incubated at 37°C for 10 min, then shaken for 1 min on a plate shaker, and finally absorbance measured spectrophotometrically at 540 nm. Healthy, untreated cells served as control to normalize to (=100% activity) and cells that were killed with lysis solution (Promega, G1821) served as 0% activity control.
RNA Extraction and Real-Time Reverse Transcription Quantitative PCR
Keratinocytes were cultured in a 12-well dish until 60–70% confluent followed by treatment with S. aureus secretome diluted 1:40 in keratinocyte medium. Cells were washed with PBS and collected after 6 h by dislodging cells directly in RLT lysis buffer (Qiagen, 79216). RNA extraction was performed using the RNeasy Mini Kit (Qiagen, 74106) as per given protocol. Samples were eluted in 50 μL of nuclease-free water (Ambion, AM9937). To remove potential DNA contamination, the TURBO DNA-free kit by Ambion (AM1907) was used according to the manufacturers user guide. RNA concentration was measured using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, 0716). cDNA was synthesized from 1 μg of RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, 18080051). Real-Time Reverse Transcription Quantitative PCRs (RT-qPCRs) were run on Quantstudio7 Flex real-time PCR system, 384-well plate reader using Sybr Green assay (Applied biosystems, 4309849). Primers were synthesized by Integrated DNA Technologies and sequences were as follows: IL-6 forward GGCACTGGCAGAAAACAACC and reverse CACCAGGCAAGTCTCCTCAT; IL-8 forward ACCACCGGAAGGAACCATCT and reverse GGCAAAACTGCACCTTCACAC; TNF-α forward GTTCCC CAGGGACCTCTCTC and reverse GAGGGTTTGCT ACAACATGGG. To account for variations in RNA yield and cDNA synthesis efficiency, TATA box-binding protein (TBP) was chosen as the housekeeping gene to normalize against (forward CCGGCTGTTTAACTTCGCTT and reverse CCAAGAAACAGTGATGCTGGGT). Fold change in gene expression was calculated via the ΔΔCt method. Keratinocytes treated with bacterial media DSM served as control. Additionally, a reverse transcriptase negative cDNA sample and a water control were included in each assay.
Keratinocyte Infection Experiment
Single colonies of four S. aureus isolates were inoculated in DSM and growth curves monitored for 24 h. The cultures were then clarified by centrifugation at 800 × g for 20 min and the supernatant of each culture was collected and divided into two tubes. One aliquot was filter-sterilized through 0.2 μm regenerated cellulose (Corning; CLS431222) to remove bacteria but retain S. aureus secretome, and the other was left untreated to retain live bacterial cells. Both unfiltered and filtered supernatants were spread on agar plates for bacterial live cell counts and to confirm sterility, respectively. HaCaT keratinocytes, which were seeded in 96-well plates the day prior, were then exposed to either the sterile S. aureus supernatant diluted 1 in 10 in keratinocyte media, or the supernatant containing live S. aureus diluted 1 in 10 in keratinocyte media, corresponding to an MOI of 1. After 24-h co-incubation, the keratinocyte cell viability and IL-6 secretion was assessed via MTT assay and HEK-Blue™ IL-6 assay as described above, respectively. This experiment was carried out in three biological replicates, i.e., three independent cultures of S. aureus, each in at least two technical replicates of HaCaT cells.
In vivo Experiments: Mouse Tissue Collection, Cell Isolation, Antibody Staining, and Flow Cytometry
Six- to eight-week-old female C57BL/6J mice were purchased from Animal Resources Centre (Perth, Australia). Mice were housed at the Translational Research Institute Biological Research Facility which is operated as Specific Pathogen Free. Animal experiments were approved by The University of Queensland Animal Ethics Committee in accordance with the National Health and Medical Research Council of Australia guidelines for care and use of laboratory animals (UQDI/TRI/153/17). After anesthetizing with isoflurane, mice ear pinnae were injected intradermally with 20 μL of sterile S. aureus secretome or the controls PBS or DSM. To ensure precise delivery of treatment into the dermal layer of ear pinnae, a 30-gauge needle (BRAUN Omnican 50 Insulin Syringes 30G × 5/16) was used. Intradermal injection was considered successful when the formation of a wheal was observed and a sample was excluded if the injected secretome leaked excessively.
Then, 24 h after intradermal injection with S. aureus secretome or control, mice were euthanized by CO2 asphyxiation and the ears were harvested. Mice ears were split into dorsal and ventral halves with fine curved forceps and placed with the dermis side down in 2.5 mg/mL Dispase II (Roche) in PBS for 45 min at 37°C in 5% CO2. The dermis and epidermis were then separated and placed in PBS containing 1 mg/mL collagenase D (Roche) and 0.2 mg/mL DNase I (Thermo Fisher Scientific). The tissue was further disrupted into small pieces using surgical scissors to aid with isolation of single cell suspension. Samples were incubated for 45 min at 37°C and filtered through a 70 μm nylon cell strainer and rinsed through with 10 mL PBS buffer. The cell suspension was centrifuged at 350 × g for 5 min at 4°C. The supernatant was discarded and cell pellets disrupted and transferred into fluorescence-activated cell sorting (FACS) tubes.
The cells were stained with 1:100 Fcγ receptor blocking anti-CD16/32 (2.4 G2; eBioscience) and 1:250 LIVE/DEAD Fixable Aqua Dead Cell Stain (Thermo Fisher Scientific) in PBS. After 20-min incubation at 4°C in the dark, cells were washed with FACS buffer (2% fetal bovine serum (FBS), 2 mM EDTA, PBS) and centrifuged at 350 × g for 5 min at 4°C. After discarding the supernatant, cell pellets were resuspended in 50 μL of FACS buffer containing the following fluorescently conjugated antibodies: APC-anti-F4/80 (clone BM8; AbNo 570; eBioscience 1:200); Percp Cy5.5-anti-CD45 (clone 30-F11; AbNo 719; Biolegend 1:200) 1; PE/Cy7-anti-CD11c (clone HL3; AbNo 262; BD; 1:200); APC/Cy7-anti-MHC Class II (clone M5/114.15.2; AbNo 482; Biolegend 1:400); PE-anti-EPCAM PE (clone G8.8; AbNo 328; Biolegend 1:200); PB-anti-CD11b (clone M1/70; AbNo 564; Biolegend; 1:200); AF594-anti-TCRβ (clone H57-597; AbNo 808; Biolegend 1:200); AF700-anti-Ly6G (clone 1A8; AbNo 717; Biolegend; 1:200); BV711-anti-Ly6C (clone HK1.4; AbNo 718; Biolegend 1:400). After a 30-min incubation at 4°C in the dark, the cells were washed twice with 500 μL FACS Buffer and then again centrifuged at 350 × g for 5 min at 4°C. The supernatant was discarded and samples gently vortexed to dissociate the cell pellet which was then resuspended in 250 μL FACS buffer before analysis on the BD LSRFortessa™ X-20 flow cytometer.
Mass Spectrometry Analysis of Staphylococcus aureus Supernatant
Protein concentration of S. aureus secretome was determined via DirectDetect® infrared spectrometer (Merck) as well as BCA assay according to manufacturer’s protocol. S. aureus secretomes were diluted to 1 μg protein per μL in 10 mM tris(2-carboxyethyl)phosphine (TCEP), 40 mM chloroacetamide, and 1% sodium deoxycholate in 100 mM Tris pH 8.5. An aliquot of 10 μg of protein was heated for 5 min at 95°C and diluted 1 in 10 in water, then 0.2 μg of sequencing grade trypsin (Promega) was added and incubated overnight at 37°C. The next day, trifluoroacetic acid was added to 0.5%, and the samples were centrifuged at 13,000 × g for 10 min. The supernatant collected and the peptides were cleaned-up using 20 μg capacity C-18 tips (Glygen). After rehydrating the membrane twice with buffer B (80% acetonitrile, 0.1% formic acid), and equilibrating in buffer A (0.1% formic acid) 3 times, the peptides was bound using 10 pipetting cycles. The tip was washed once with buffer A and then peptides were eluted in buffer B.
Peptides were analyzed on a Thermo U3000 nano-HPLC system coupled to a Thermo Q Exactive Plus Orbitrap mass spectrometer. The HPLC setup used a C-18 trap column (DX160454) and a 50 cm EasySpray C-18 analytical column (ES803A) from Thermo Fisher Scientific. Mobile phases were A: 0.1% formic acid, and B: 80% acetonitrile with 0.1% formic acid. A total of 4 μL of sample equivalent to 1 μg of peptides were loaded in 2% B, and peptides eluted over a gradient from 2 to 50% B over 45 min at 250 nL/min. An EasySpray source was ran in positive ion data-dependent analysis mode with settings typical of peptide analyses. Briefly, full MS scans were acquired at 70,000 resolution, automatic gain control target was 3e6, with a maximum injection time of 50 ms. MS2 fragmentation was carried out on the top 10 precursors, excluding 1+ precursors. Precursor isolation width was 1.4 m/z and normalized collision energy was 28. MS2 resolution was 17,500 with an automatic gain control target of 5e5. Maximum injection time was 50 ms and the exclusion window was 10 s.
MaxQuant (Tyanova et al., 2016a) was used for peptide identification against the reviewed UniProtKB database (SwissProt; S. aureus; 11.10.2018; 11,075 proteins) using the “match between runs” function, normalization by label-free quantification and refining to a minimum of one unique peptide. The MaxQuant outputs were imported into Perseus (Tyanova et al., 2016b) and after the removal of proteins only identified by site, reverse sequences, and contaminants, the remaining 348 proteins were subjected to further analyses. The LFQ normalized MS intensity values were log2 transformed and missing values were imputed from normal distribution.
To establish whether there was a relationship between S. aureus protein abundance and the previously established keratinocyte IL-6 response, a Spearman’s rank correlation analysis for each individual protein was performed using the rcorr() function in the Hmisc package for R v3.6.1. Raw p-values were corrected for multiple testing using the “fdr” method and considered significantly correlated if the FDR was below 5% (FDR < 0.05).
To determine the presence/absence of significantly IL-6 correlated genes within individual isolates, translated coding sequences for the 34 S. aureus genomes were predicted by Prodigal (ver. 2.6.4) (Hyatt et al., 2010) as part of Prokka (ver. 1.14.6) (Seemann, 2014). Translated proteins were then aligned with blast + (ver. 2.9.0; e-value = 0.00001) (Camacho et al., 2009) against the IL-6 correlated protein sequences, with a gene considered present when at least 70% of its sequence was aligned with 70% sequence identity.
Alpha Toxin Experiments
Recombinant S. aureus α-toxin and mouse monoclonal antibody [8B7] against α-toxin N-terminal were purchased from abcam (ab233724 and ab190467). Polyclonal anti-Staphylococcal α-toxin antibody produced in rabbit, and normal rabbit serum as control, were obtained from Sigma-Aldrich (S7531 and R9133). For the α-toxin neutralization experiments, the monoclonal antibody was serial diluted in serum-free keratinocyte media and added to diluted S. aureus secretome. The polyclonal antiserum and rabbit serum control were added to diluted S. aureus secretome at a 1:200 dilution. Antibody was allowed to bind for 30 min at 37°C with occasional shaking. The mixture was then added to keratinocytes grown in 96-well plate and after 24 h, IL-6 measurement via HEKblue cell reporter assay and viability assessment via MTT was carried out as described previously.
In a separate experiment, recombinant α-toxin (100 or 400 ng/ml) was added to diluted S. aureus secretome that typically does not induce an IL-6 response in keratinocytes, as well as one IL-6-inducing secretome. The S. aureus secretomes ± recombinant α-toxin were added to HaCaT cells in a 96-well plate and after 24 h of coincubation, the secreted IL-6 levels were measured by HEKblue cell reporter assay.
Data Availability Statement
The genome sequencing data of our S. aureus clinical isolates have been deposited in the NCBI Sequence Read Archive under the Bioproject ID PRJNA754839. The raw MS data, the database search results, as well as the filtered and normalized protein data have been deposited to the publicly accessible platform ProteomeXchange Consortium via the PRIDE partner repository Perez-Riverol et al. (2019) under the dataset identifier PXD028604.
Ethics Statement
The studies involving human participants were reviewed and approved by the Metro South Human Research Ethics Committee. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the University of Queensland Animal Ethics Committee.
Author Contributions
IF, MM, HS, PH, MH, and AK conceived and designed the experiments. AK, SC, and ST performed the experiments. AK and JZ analyzed the data. JZ, RL, NL, ZT, and SL contributed materials and/or analysis tools. AK wrote the manuscript. IF, MM, HS, PH, MH, and JZ helped editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by NHMRC project grants APP1071822 and APP1059012, the Merchant Foundation and an Australian Research Council Linkage Project LP160100731. HS holds an NHMRC MRFF Next Generation Clinical Researchers Program Practitioner Fellowship (APP1137127).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the Translational Research Institute for providing the excellent research environment and core facilities that enabled this research. We particularly thank Dorothy Loo-Oey and Janelle Hanock from the Proteomics Core Facility. We would like to acknowledge the study participants and the staff at the Princess Alexandra Hospital for their involvement. We also thank Chenhao Zhou, Lynn Tolley, Katharina Devitt, Megha Budhwani, and Burhan Khan for their assistance and Kiarash Khosrotehrani for insightful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.789042/full#supplementary-material
Footnotes
References
Anders, S., Pyl, P. T., and Huber, W. (2015). Htseq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Andersson, T., Bergdahl, G. E., Saleh, K., Magnúsdóttir, H., Stødkilde, K., Andersen, C. B. F., et al. (2019). Common skin bacteria protect their host from oxidative stress through secreted antioxidant RoxP. Sci. Rep. 9: 3596. doi: 10.1038/s41598-019-40471-3
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed December 1, 2016).
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonar, E., Wojcik, I., Jankowska, U., Kedracka-Krok, S., Bukowski, M., Polakowska, K., et al. (2016). Identification of secreted exoproteome fingerprints of highly-virulent and non-virulent Staphylococcus aureus strains. Front. Cell. Infect. Microbiol. 6:51. doi: 10.3389/fcimb.2016.00051
Brantsch, K. D., Meisner, C., Schönfisch, B., Trilling, B., Wehner-Caroli, J., Röcken, M., et al. (2008). Analysis of risk factors determining prognosis of cutaneous squamous-cell carcinoma: a prospective study. Lancet Oncol. 9, 713–720. doi: 10.1016/S1470-2045(08)70178-5
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Damour, A., Robin, B., Deroche, L., Broutin, L., Bellin, N., Verdon, J., et al. (2021). Phenol-soluble modulins α are major virulence factors of Staphylococcus aureus secretome promoting inflammatory response in human epidermis. Virulence 12, 2474–2492. doi: 10.1080/21505594.2021.1975909
Gambichler, T., Skrygan, M., Hyun, J., Bechara, F., Tomi, N., Altmeyer, P., et al. (2006). Cytokine mRNA expression in basal cell carcinoma. Arch. Dermatol. Res. 298, 139–141. doi: 10.1007/s00403-006-0673-1
Ghahartars, M., Abtahi, S., Zeinali, Z., Fattahi, M. J., and Ghaderi, A. (2021). Investigation of TNF-α and IL-6 levels in the sera of non-melanoma skin cancer patients. Iran. Biomed. J. 25, 88–92. doi: 10.29252/ibj.25.2.88
Grivennikov, S. I., Greten, F. R., and Karin, M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899. doi: 10.1016/j.cell.2010.01.025
Hanzelmann, D., Joo, H.-S., Franz-Wachtel, M., Hertlein, T., Stevanovic, S., Macek, B., et al. (2016). Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat. Commun. 7:12304. doi: 10.1038/ncomms12304
Harrow, J., Frankish, A., Gonzalez, J. M., Tapanari, E., Diekhans, M., Kokocinski, F., et al. (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774. doi: 10.1101/gr.135350.111
Hattar, K., Reinert, C. P., Sibelius, U., Gökyildirim, M. Y., Subtil, F. S., Wilhelm, J., et al. (2017). Lipoteichoic acids from Staphylococcus aureus stimulate proliferation of human non-small-cell lung cancer cells in vitro. Cancer Immunol. Immunother. 66, 799–809. doi: 10.1007/s00262-017-1980-4
Hong, S.-W., Choi, E.-B., Min, T.-K., Kim, J.-H., Kim, M.-H., Jeon, S. G., et al. (2014). An important role of α-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One 9:e100499. doi: 10.1371/journal.pone.0100499
Hruz, P., Zinkernagel, A. S., Jenikova, G., Botwin, G. J., Hugot, J.-P., Karin, M., et al. (2009). NOD2 contributes to cutaneous defense against Staphylococcus aureus through α-toxin-dependent innate immune activation. Proc. Natl. Acad. Sci. U.S.A. 106, 12873–12878. doi: 10.1073/pnas.0904958106
Hussain, M., Hastings, J., and White, P. (1991). A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 34, 143–147. doi: 10.1099/00222615-34-3-143
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jee, S.-H., Shen, S.-C., Chiu, H.-C., Tsai, W.-L., and Kuo, M.-L. (2001). Overexpression of interleukin-6 in human basal cell carcinoma cell lines increases anti-apoptotic activity and tumorigenic potency. Oncogene 20, 198–208. doi: 10.1038/sj.onc.1204076
Jolley, K. A., Bray, J. E., and Maiden, M. C. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST. org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Krueger, F. (2015). Trim Galore: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed April 28, 2016).
Kullander, J., Forslund, O., and Dillner, J. (2009). Staphylococcus aureus and squamous cell carcinoma of the skin. Cancer Epidemiol. Biomarkers Prev. 18, 472–478. doi: 10.1158/1055-9965.EPI-08-0905
Lederle, W., Depner, S., Schnur, S., Obermueller, E., Catone, N., Just, A., et al. (2011). IL-6 promotes malignant growth of skin SCCs by regulating a network of autocrine and paracrine cytokines. Int. J. Cancer 128, 2803–2814. doi: 10.1002/ijc.25621
Lin, W.-W., and Karin, M. (2007). A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 117, 1175–1183. doi: 10.1172/JCI31537
Liu, H., Archer, N. K., Dillen, C. A., Wang, Y., Ashbaugh, A. G., Ortines, R. V., et al. (2017). Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 22, 653–666.e5. doi: 10.1016/j.chom.2017.10.006
Liu, Q., Mazhar, M., and Miller, L. S. (2018). Immune and inflammatory reponses to Staphylococcus aureus skin infections. Curr. Dermatol. Rep. 7, 338–349. doi: 10.1007/s13671-018-0235-8
Madhusudhan, N., Pausan, M. R., Halwachs, B., Durdević, M., Windisch, M., Kehrmann, J., et al. (2020). Molecular profiling of keratinocyte skin tumors links Staphylococcus aureus overabundance and increased human β-Defensin-2 expression to growth promotion of squamous cell carcinoma. Cancers 12:541. doi: 10.3390/cancers12030541
Mantovani, A., Allavena, P., Sica, A., and Balkwill, F. (2008). Cancer-related inflammation. Nature 454, 436–444. doi: 10.1038/nature07205
Mohammad, M., Na, M., Hu, Z., Nguyen, M.-T., Kopparapu, P. K., Jarneborn, A., et al. (2021). Staphylococcus aureus lipoproteins promote abscess formation in mice, shielding bacteria from immune killing. Commun. Biol. 4:432. doi: 10.1038/s42003-021-01947-z
Moussai, D., Mitsui, H., Pettersen, J. S., Pierson, K. C., Shah, K. R., Suárez-Fariñas, M., et al. (2011). The human cutaneous squamous cell carcinoma microenvironment is characterized by increased lymphatic density and enhanced expression of macrophage-derived VEGF-C. J. Invest. Dermatol. 131, 229–236. doi: 10.1038/jid.2010.266
Mossmann, T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immun. Method. 65, 55–63. doi: 10.1016/0022-1759(83)90303-4
Moyes, R. B., Reynolds, J., and Breakwell, D. P. (2009). Differential staining of bacteria: gram stain. Curr. Protoc. Microbiol. 15, A.3C.1–A.3C.8. doi: 10.1002/9780471729259.mca03cs15
Mueller, M. M. (2006). Inflammation in epithelial skin tumours: old stories and new ideas. Eur. J. Cancer 42, 735–744. doi: 10.1016/j.ejca.2006.01.014
Nguyen, M. T., and Götz, F. (2016). Lipoproteins of Gram-positive bacteria: key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 80, 891–903. doi: 10.1128/MMBR.00028-16
Onogawa, T. (2002). Staphylococcal α-toxin synergistically enhances inflammation caused by bacterial components. FEMS Immunol. Med. Microbiol. 33, 15–21. doi: 10.1016/S0928-8244(01)00308-X
Onogawa, T. (2005). Local delivery of soluble interleukin-6 receptors to improve the outcome of alpha-toxin producing Staphylococcus aureus infection in mice. Immunobiology 209, 651–660. doi: 10.1016/j.imbio.2004.09.006
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. doi: 10.1093/nar/gky1106
Prescott, S. L., Larcombe, D.-L., Logan, A. C., West, C., Burks, W., Caraballo, L., et al. (2017). The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 10:29. doi: 10.1186/s40413-017-0160-5
Reiner, K. (2010). Catalase Test Protocol. Available online at: https://asm.org/getattachment/72a871fc-ba92-4128-a194-6f1bab5c3ab7/Catalase-Test-Protocol.pdf (accessed December 1, 2016).
Reuschenbach, M., Tran, T., Faulstich, F., Hartschuh, W., Vinokurova, S., Kloor, M., et al. (2011). High-risk human papillomavirus in non-melanoma skin lesions from renal allograft recipients and immunocompetent patients. Br. J. Cancer 104, 1334–1341. doi: 10.1038/bjc.2011.95
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Soyer, H. P., Rigel, D., and Wurm, E. M. (2012). “Actinic keratosis, basal cell carcinoma and squamous cell carcinoma,” in Dermatology, eds J. L. Bolognia J. L. Jorizzo J. V. Schaffer (Edinburgh: Elsevier Saunders), 1773–1794.
Stentzel, S., Teufelberger, A., Nordengrün, M., Kolata, J., Schmidt, F., Van Crombruggen, K., et al. (2017). Staphylococcal serine protease–like proteins are pacemakers of allergic airway reactions to Staphylococcus aureus. J Allergy Clin Immunol. 139, 492–500.e8. doi: 10.1016/j.jaci.2016.03.045
Stoll, H., Dengjel, J., Nerz, C., and Götz, F. (2005). Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73, 2411–2423. doi: 10.1128/IAI.73.4.2411-2423.2005
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Turner, M. D., Nedjai, B., Hurst, T., and Pennington, D. J. (2014). Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 1843, 2563–2582. doi: 10.1016/j.bbamcr.2014.05.014
Tyanova, S., Temu, T., and Cox, J. (2016a). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319. doi: 10.1038/nprot.2016.136
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016b). The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Wood, D. L., Lachner, N., Tan, J.-M., Tang, S., Angel, N., Laino, A., et al. (2018). A natural history of actinic keratosis and cutaneous squamous cell carcinoma microbiomes. mBio 9:e01432-18. doi: 10.1128/mBio.01432-18
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zdzalik, M., Karim, A. Y., Wolski, K., Buda, P., Wojcik, K., Brueggemann, S., et al. (2012). Prevalence of genes encoding extracellular proteases in Staphylococcus aureus—important targets triggering immune response in vivo. FEMS Immunol. Med. Microbiol. 66, 220–229. doi: 10.1111/j.1574-695X.2012.01005.x
Keywords: skin microbiota, microbiome, squamous cell carcinoma, actinic keratosis, S. aureus, proteomics, multi-omics, inflammation
Citation: Krueger A, Zaugg J, Chisholm S, Linedale R, Lachner N, Teoh SM, Tuong ZK, Lukowski SW, Morrison M, Soyer HP, Hugenholtz P, Hill MM and Frazer IH (2022) Secreted Toxins From Staphylococcus aureus Strains Isolated From Keratinocyte Skin Cancers Mediate Pro-tumorigenic Inflammatory Responses in the Skin. Front. Microbiol. 12:789042. doi: 10.3389/fmicb.2021.789042
Received: 04 October 2021; Accepted: 08 December 2021;
Published: 25 January 2022.
Edited by:
Joseph Alex Duncan, University of North Carolina at Chapel Hill, United StatesReviewed by:
Silke Niemann, University Hospital Münster, GermanyMinh-Thu Nguyen, University Hospital Münster, Germany
Copyright © 2022 Krueger, Zaugg, Chisholm, Linedale, Lachner, Teoh, Tuong, Lukowski, Morrison, Soyer, Hugenholtz, Hill and Frazer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian H. Frazer, aS5mcmF6ZXJAdXEuZWR1LmF1
 Annika Krueger
Annika Krueger Julian Zaugg
Julian Zaugg Sarah Chisholm
Sarah Chisholm Richard Linedale1
Richard Linedale1 Siok Min Teoh
Siok Min Teoh Zewen K. Tuong
Zewen K. Tuong Samuel W. Lukowski
Samuel W. Lukowski Mark Morrison
Mark Morrison H. Peter Soyer
H. Peter Soyer Philip Hugenholtz
Philip Hugenholtz Michelle M. Hill
Michelle M. Hill Ian H. Frazer
Ian H. Frazer