- Department of Horticulture and Crop Science, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH, United States
High fertilizer rates are often applied to horticulture crop production systems to produce high quality crops with minimal time in production. Much of the nutrients applied in fertilizers are not taken up by the plant and are leached out of the containers during regular irrigation. The application of plant growth promoting rhizobacteria (PGPR) can increase the availability and uptake of essential nutrients by plants, thereby reducing nutrient leaching and environmental contamination. Identification of PGPR can contribute to the formulation of biostimulant products for use in commercial greenhouse production. Here, we have identified Serratia plymuthica MBSA-MJ1 as a PGPR that can promote the growth of containerized horticulture crops grown with low fertilizer inputs. MBSA-MJ1 was applied weekly as a media drench to Petunia×hybrida (petunia), Impatiens walleriana (impatiens), and Viola×wittrockiana (pansy). Plant growth, quality, and tissue nutrient concentration were evaluated 8weeks after transplant. Application of MBSA-MJ1 increased the shoot biomass of all three species and increased the flower number of impatiens. Bacteria application also increased the concentration of certain essential nutrients in the shoots of different plant species. In vitro and genomic characterization identified multiple putative mechanisms that are likely contributing to the strain’s ability to increase the availability and uptake of these nutrients by plants. This work provides insight into the interconnectedness of beneficial PGPR mechanisms and how these bacteria can be utilized as potential biostimulants for sustainable crop production with reduced chemical fertilizer inputs.
Introduction
The plant rhizosphere, the microscopic environment surrounding plant roots, hosts a diversity of microorganisms. Plants secrete a variety of root exudates into the rhizosphere including sugars, vitamins, and amino acids, which assist in recruiting specific microbial populations (Lugtenberg and Kamilova, 2009; Martín-Sánchez et al., 2020). In return, many of these microbes play essential roles in the rhizosphere ecosystem by influencing both plant and soil health (Vessey, 2003). The study of these beneficial plant growth promoting rhizobacteria (PGPR) and their contribution to agriculture has gained significant interest in recent years (Bhardwaj et al., 2014). PGPR can stimulate plant growth through a variety of mechanisms including modulating phytohormone levels, inhibiting growth of plant pathogens, producing secondary metabolites, and increasing the availability and uptake of nutrients for their plant host (Raklami et al., 2019; Martín-Sánchez et al., 2020).
Plants require 14 essential mineral nutrients to support proper growth and development, and agricultural systems often rely on the addition of chemical fertilizers to provide adequate nutrients to support crop production (Marschner, 2011). However, high rates of fertilizer are frequently attributed to surface and groundwater pollution due to nutrient leaching and runoff (Adesemoye and Kloepper, 2009). The bioavailability of nutrients for plant uptake plays a significant role in plant nutrient use efficiency (NUE), and therefore, the lack of bioavailability increases the susceptibility of nutrients to leaching and runoff (Adesemoye and Kloepper, 2009; Bindraban et al., 2015). This is of particular concern for horticulture crops grown in peat-based soilless substrates, which have lower ion exchange capacities than field soil, increasing the possibility that nutrients will be removed from the substrate via leaching (Bachman and Metzger, 2008). Therefore, it is important that crop producers find sustainable ways to produce crops with high yield and quality while minimizing chemical fertilizer inputs.
Studies have shown that the application of PGPR can increase the NUE of plants, thereby reducing the amount of nutrients that are leached into the environment (Khan, 2005; Adesemoye et al., 2008, 2010; Shaharoona et al., 2008; Arif et al., 2017; Pereira et al., 2020). A meta-analysis of studies evaluating PGPR application on plant NUE showed an average increase in NUE of 5.8kg yield per kg nitrogen fertilizer applied (Schütz et al., 2018). Direct mechanisms that PGPR can utilize to facilitate increased nutrient bioavailability and uptake by plants include nitrogen fixation, nutrient solubilization, sulfur oxidation, and chelation of metals (Lugtenberg and Kamilova, 2009).
The availability of essential plant nutrients including phosphorus, potassium, and zinc is dependent on their solubility in the rhizosphere (Ruzzi and Aroca, 2015). When these nutrients are bound by other salts and metals, plants are unable to access them for growth and development. However, bacterial solubilization can make them readily accessible for plant utilization and less likely to be removed from the growing substrate via leaching (Adesemoye and Kloepper, 2009). Many bacterial species that are plant growth promoters have been identified as efficient solubilizers of phosphorus, potassium, and zinc (Sharma et al., 2013, 2016; Kamran et al., 2017; Borgi et al., 2020). The application of phosphorus-solubilizing Bacillus and Aspergillus strains reduces the level of fertilizer required for optimal yield and tissue nutrient content in strawberry (Güneş et al., 2014). Although many different bacterial enzymes can solubilize these nutrients, bacterial-produced organic acids are thought to be the primary mechanism involved in increasing the availability of insoluble phosphorus, potassium, and zinc (Rodríguez et al., 2006; Hussain et al., 2015; Parmar and Sindhu, 2019).
Iron is an essential nutrient for both plants and bacteria; however, it is not readily available in the required concentrations in its predominant form as a ferric ion (Gupta et al., 2015). Bacteria can produce siderophores that act as chelating agents to make the element available for uptake by plants (Crowley et al., 1988; Sharma et al., 2003; Lurthy et al., 2020). Additionally, siderophores can act similarly to chelate other essential metals, such as copper (Yoon et al., 2010). Application of the siderophore-producing PGPR Chryseobacterium C138 increased the iron content of tomato plants supplied solely with ferric iron (Radzki et al., 2013). Siderophore production by PGPR allows for reduced chemical inputs to meet the demand of available iron to plants (Gupta et al., 2015).
It is advantageous to crop producers and the environment that PGPR increase the availability or uptake of multiple essential plant nutrients, allowing for a reduction in total fertilizer inputs. Formulation of these PGPR into biostimulant products would then make them widely available for commercial greenhouse operations. Therefore, in this work, we have evaluated the model PGPR strain Serratia plymuthica MBSA-MJ1 for its ability to increase plant growth and tissue nutrient concentration of three economically important horticulture crop species produced with low fertilizer inputs. MBSA-MJ1 was previously evaluated for its ability to increase plant growth during recovery from severe water stress (Nordstedt and Jones, 2021). In addition, we have characterized the in vitro and genomic characteristics of this PGPR to begin elucidating different putative mechanisms used to promote plant growth with an emphasis on nutrient availability. This work demonstrates the potential of this PGPR as a commercial biostimulant, allowing growers to reduce chemical fertilizer inputs without sacrificing crop health or quality.
Materials and Methods
Bacterial Strain
Serratia plymuthica MBSA-MJ1 originated from a bacteria collection in the laboratory of Dr. Christopher Taylor (Aly et al., 2007), although the environmental source of this strain is unknown. MBSA-MJ1 was recently evaluated for its ability to reduce Botrytis cinerea infection in Petunia×hybrida (South et al., 2020) and to stimulate the growth of plants recovering from severe water stress (Nordstedt and Jones, 2021).
Low-Nutrient Greenhouse Trial
A previously established greenhouse trialing protocol was utilized to evaluate the ability of S. plymuthica MBSA-MJ1 to increase the growth of three economically important ornamental plant species produced with low fertilizer inputs (Nordstedt et al., 2020). Petunia×hybrida ‘Picobella Blue’ (petunia; Syngenta Flowers Gilroy, CA), Impatiens walleriana ‘Super Elfin Ruby’ (impatiens; PanAmerican Seed, West Chicago, IL), and Viola×wittrockiana ‘Delta Pure Red’ (pansy; Syngenta Flowers) seeds were sown in Pro-Mix PGX soilless media (Premier Tech Horticulture, Quakertown, PA) and grown for 3weeks. Seedlings were then transplanted to 11.4cm diameter pots containing Pro-Mix PGX. All plants were fertilized with 25mgL−1 N from 15N–2.2P–12.5K–2.9Ca–1.2Mg water soluble fertilizer (JR Peters Inc., Allentown, PA) at each irrigation to provide low-nutrient conditions. Plants were arranged in a Randomized Complete Block Design by species. Each block contained one bacterial-treated and one untreated control plant with n=13 for petunia, n=14 for pansy, and n=18 for impatiens. Greenhouse temperatures were set at 24/18°C (day/night) and supplemental lighting was provided by high-pressure sodium and metal halide lights (GLX/GLS e-systems GROW lights, PARSource, Petaluma, CA, United States) to maintain light levels above 250mmolm−2 s−1 and provide a 16h photoperiod.
Each plant was treated weekly with 120ml working inoculum of S. plymuthica MBSA-MJ1. Working inoculum was prepared by diluting an overnight culture grown in LB media (OD595=0.8) 1:100 in reverse osmosis (RO) water as described previously (Nordstedt et al., 2020). Diluted uninoculated LB media was used as the control. Plants were grown under low-nutrient conditions with weekly bacterial treatments for 8weeks, at which point all plants had open flowers. Plant performance was then evaluated by counting flower number (open flowers and flower buds showing color) and collecting shoots (including stems and leaves) and flowers for biomass measurements. Shoot and flower biomass was combined to get the total shoot biomass. Potting media was removed from the roots by rinsing in water, and root tissue was collected separately from shoot tissue. Roots, shoots, and flowers were dried at 49°C for at least 96h and then weighed to determine dry weights. Root dry weights and shoot dry weights were used to calculate the root:shoot ratio.
Tissue Nutrient Concentration
Dried leaf and stem tissue (shoots minus the flowers) collected from the low-nutrient greenhouse trial were pooled for tissue nutrient analysis. Each sample consisted of tissue pooled separately for each plant species and treatment (MBSA-MJ1 and control) and contained tissue from three blocks located spatially together in the greenhouse (n=3). Tissue nutrient analyses were conducted at the Service Testing and Research Laboratory (The Ohio State University/OARDC, Wooster, OH). Total tissue nitrogen (N) concentration was determined from a 100mg sample using the Dumas combustion method (Vario Max combustion analyzer, Elementar America, Inc., Germany; Sweeney, 1989). The phosphorus (P), potassium (K), magnesium (Mg), calcium (Ca), sulfur (S), boron (B), copper (Cu), iron (Fe), sodium (Na), molybdenum (Mo), manganese (Mn), and zinc (Zn) concentration of the tissue was then determined by inductively coupled plasma spectrometry (model PS3000, Leeman Labs Inc., Hudson, NH) of a 250mg tissue sample following nitric acid microwave digestion (Discover SP-D, CEM Corporation; Isaac and Johnson, 1985).
In vitro Characterization
Sole Carbon Source Utilization
Growth of S. plymuthica MBSA-MJ1was evaluated on media containing different sole sources of carbon, including cellobiose, fructose, galactose, glucose, glycerol, mannitol, sucrose, and ribose. Basal media was prepared according to Pridham and Gottlieb (1948) supplemented with 1% w/v (1% w/w for glycerol) of each carbon source. Cells from an overnight culture of MBSA-MJ1 were resuspended in PBS buffer, and 5μl of inoculum was struck out into each carbon source plate in triplicate (n=3). Plates were incubated at 28°C for 96h and growth on each carbon source was recorded as yes/no.
Phosphate Solubilization
Phosphate solubilization capabilities of S. plymuthica MBSA-MJ1 were evaluated according to Mehta and Nautiyal (2001). An overnight culture grown in LB media was diluted to OD595=0.2 and 80μl was used to inoculate 8ml NBRIP media in triplicate (n=3). Uninoculated LB in NBRIP media was used for the negative control. NBRIP media consisted of (per liter): 10.0g glucose, 5.0g Ca3(PO4)2, 5.0g MgCl2 • 6H2O, 0.25g MgSO4 • 7H2O, 0.2g KCl, and 0.1g (NH4)2SO4. The pH of the media was adjusted to 7.0 before autoclaving. Following inoculation, samples were incubated at 30°C for 72h with 200rpm shaking. After incubation, samples were centrifuged at 4°C and 5,000rpm for 10min to collect bacterial cells and remaining insoluble phosphate. The supernatant was collected, and the pH was measured as an indicator of solubilized phosphate.
Potassium Solubilization
The potassium solubilization ability of S. plymuthica MBSA-MJ1 was evaluated according to a modified protocol adapted from Rajawat et al. (2016). Cultures were prepared similar to those described above for the phosphate solubilization assay, and 80μl was used to inoculate 8ml Aleksandrov (AKV) liquid media. Uninoculated LB in AKV media served as the negative control. AKV media consisted of (per liter): 5.0g glucose, 2.0g Ca3(PO4)2, 0.5g MgSO4 • 7H2O, 0.1g CaCO3, 0.005g FeCl3, and 2.0g montmorillonite as the insoluble form of potassium. The pH of the media was adjusted to 7.2 before autoclaving. After inoculation, samples were incubated at 30°C for 72h with 200rpm shaking. Following incubation, bacterial cells and insoluble montmorillonite were collected by centrifugation at 4°C and 5,000rpm for 10min, and the supernatant was collected. The pH of the supernatant was measured as an indicator of solubilized potassium.
Quantification of Solubilized Phosphate and Potassium
Total solubilized orthophosphate and potassium in defined media were measured as validation of the nutrient solubilization capabilities of S. plymuthica MBSA-MJ1. Cultures were grown and inoculated in NBRIP and AKV media to evaluate phosphate and potassium solubilization, respectively. Samples were prepared similar to the solubilization assays, and after incubation and centrifugation, the supernatant was filtered through a 0.45μm nylon filter and total orthophosphate (PO4) or potassium in the filtrate was determined using Dionex ICS-6000 Ion Chromatograph (Thermo Fisher Scientific Inc., Waltham, MA).
Iron and Copper Chelation
The iron and copper chelation capabilities of S. plymuthica MBSA-MJ1 were evaluated according to Schwyn and Neilands (1987). All labware for media preparation and culturing the samples was washed in a 6M HCl acid bath for at least 24h prior to use. Succinic acid (SA) media consisted of (per liter): 6.0g K2HPO4, 3.0g KH2PO4, 1.0g (NH4)2SO4, 0.2g MgSO4 • 7H2O, and 4.0g succinic acid disodium salt, and the pH of the media was adjusted to 7.0 before autoclaving. The chrome azurol S (CAS) assay solution was prepared in two solutions. Solution one contained 20ml H2O, 6.0ml 10mm hexadecyltrimethylammonium bromide, 7.5ml 2mm CAS, and 1.5ml 1mm FeCl3 or CuCl2 to test for iron and copper chelation, respectively. Solution two contained 4.307g anhydrous piperazine in 30ml H2O, pH adjusted to 5.6 with 12M HCl. Once prepared, solution one was added to solution two and the final volume was brought to 100ml with H2O to make up the working CAS assay solution. The CAS assay solution was stored in a dark polyethylene bottle.
For the assay, 5ml SA media was inoculated with a single colony of MBSA-MJ1 and incubated for 24h. After incubation, cultures were diluted to OD595=0.5, and 80μl diluted culture was inoculated in 8ml SA media in triplicate (n=3). Uninoculated SA media was used as the negative control. Samples were incubated at 28°C for 24h with 200rpm shaking. After incubation, bacterial cells were collected by centrifugation at 4°C and 5,000rpm for 10min. After centrifugation, 750μl supernatant was mixed with 750μl CAS assay solution and incubated in the dark for 15min with 100rpm shaking to allow for color development. Absorbance of each sample was then measured at 630nm, with a decrease in absorbance corresponding to chelation of the respective metal and a color change from blue to orange. The chelation percentage of the MBSA-MJ1 sample was calculated using [(Ac-Ab)/Ac]×100%, where Ac is the absorbance at 630nm of the control sample, and Ab the absorbance at 630nm of the MBSA-MJ1 sample (Dimkpa, 2016).
Statistical Analysis
Statistical analyses for the greenhouse trial and in vitro characterization experiments were conducted in R Studio version 3.5.2 using an ANOVA with the model: Y=μ+treatment+block. The Tukey’s HSD test was used to determine statistical significance between MBSA-MJ1 treatment and the negative control. For the greenhouse trial, each plant species was analyzed independently of each other.
Genome Analyses
Genome sequence data for S. plymuthica MBSA-MJ1 (Nordstedt and Jones, 2021) were used to search for genes putatively involved in increasing nutrient availability, carbon and amino acid metabolism, and heavy metal resistance. Genome sequence data can be found in the National Center for Biotechnology Information data base accession # PRJNA669647. AntiSMASH (v 5.0) was used to identify secondary metabolite biosynthetic gene clusters within the genome of MBSA-MJ1 (Blin et al., 2019). BlastKOALA was used for annotation of KEGG pathways involved in nutrient transport, amino acid synthesis, and carbon metabolism and transport (Kanehisa et al., 2016).
Results
Low-Nutrient Greenhouse Trial
Application of S. plymuthica MBSA-MJ1 increased the visual quality of all three plant species. Overall, plants treated with MBSA-MJ1 were larger and had greener leaves than the control plants that were not treated with bacteria (Figure 1). In addition to increasing the visual quality of plants, impatiens treated with MBSA-MJ1 had significantly more flowers than the uninoculated control, an average increase of six flowers per plant (Figure 2A). Application of MBSA-MJ1 significantly increased the total shoot biomass of all three plant species. Petunia, impatiens, and pansy grown under low-nutrient conditions and treated with MBSA-MJ1 had 24, 41, and 51% greater biomass, respectively (Figure 2B). Petunia and pansy plants treated with MBSA-MJ1 also had significantly lower root:shoot biomass ratio, whereas there was no statistically significant difference in impatiens (Figure 2C).

Figure 1. Petunia×hybrida (petunia), Impatiens walleriana (impatiens), and Viola×wittrockiana (pansy) plants grown under low-nutrient conditions had increased visual quality when treated with Serratia plymuthica MBSA-MJ1. Plants were treated weekly for 8weeks with S. plymuthica MBSA-MJ1 or uninoculated LB (control) as a media drench and fertilized at each irrigation with 25mgL−1 N from 15N–2.2P–12.5K–2.9Ca–1.2Mg water soluble fertilizer to induce low-nutrient conditions.
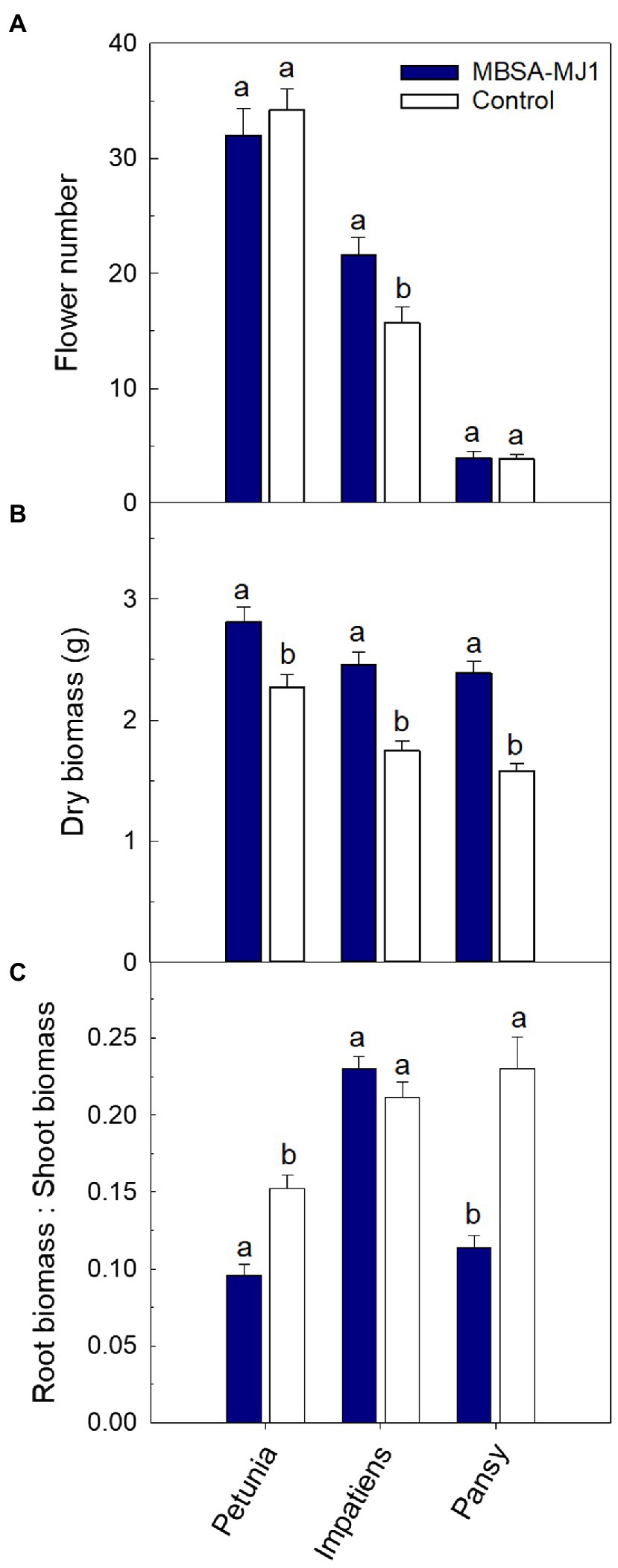
Figure 2. Treatment with S. plymuthica MBSA-MJ1 influenced plant growth parameters for Petunia×hybrida (petunia), Impatiens walleriana (impatiens), and Viola×wittrockiana (pansy) plants grown under low-nutrient conditions. Plants were treated weekly with a media drench of S. plymuthica MBSA-MJ1 (blue bars) or uninoculated LB (control; white bars). Total number of flowers (A) and total shoot biomass (dry weight) (B) was measured 8weeks post-transplant. The root:shoot ratio (C) was calculated with root and total shoot dry weights. Bars represent mean (±SE) with different letters representing significant differences (p<0.05), and n=13 for petunia, n=14 for pansy, and n=18 for impatiens.
Tissue Nutrient Concentration
Plants grown under low-nutrient conditions and treated with S. plymuthica MBSA-MJ1 had significantly higher concentrations of certain nutrients in the shoot tissue; however, differences varied depending on plant species (Figures 3, 4). Petunia, impatiens, and pansy plants had an increase of 32, 74, and 82% in tissue nitrogen concentration when treated with MBSA-MJ1, respectively. No other nutrients were significantly greater in petunia plants treated with MBSA-MJ1 when compared to the control. The concentrations of potassium, calcium, and sulfur were significantly greater in bacteria-treated impatiens and pansy plants. Only impatiens showed a significant increase in magnesium, boron, copper, molybdenum, and zinc when treated with MBSA-MJ1. Phosphorus was the only element where an increase was only observed in pansy. No significant differences in iron concentration were observed in any of the three plant species, and manganese concentration was found to be significantly greater in the control plants of petunia and pansy compared to those treated with MBSA-MJ1.
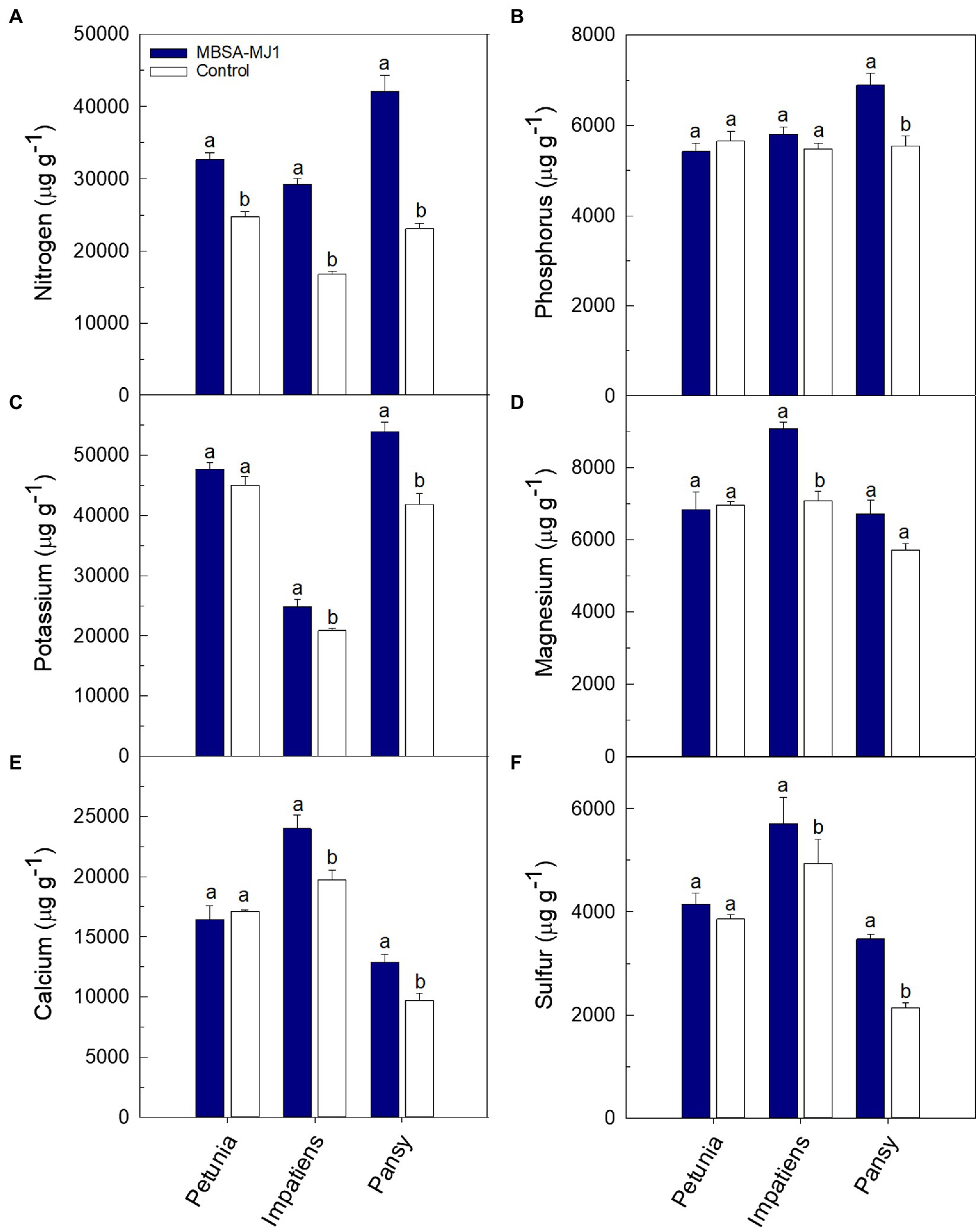
Figure 3. Tissue macronutrient concentration of Petunia×hybrida (petunia), Impatiens walleriana (impatiens), and Viola×wittrockiana (pansy) plants grown under low-nutrient conditions: nitrogen (A), phosphorus (B), potassium (C), magnesium (D), calcium (E), and sulfur (F). Plants were treated weekly with a media drench of S. plymuthica MBSA-MJ1 (blue bars) or uninoculated LB (control; white bars). Tissue nutrient concentration was evaluated 8weeks post-transplant. Bars represent the mean (±SE) with different letters representing significant differences (p<0.05) and n=3.
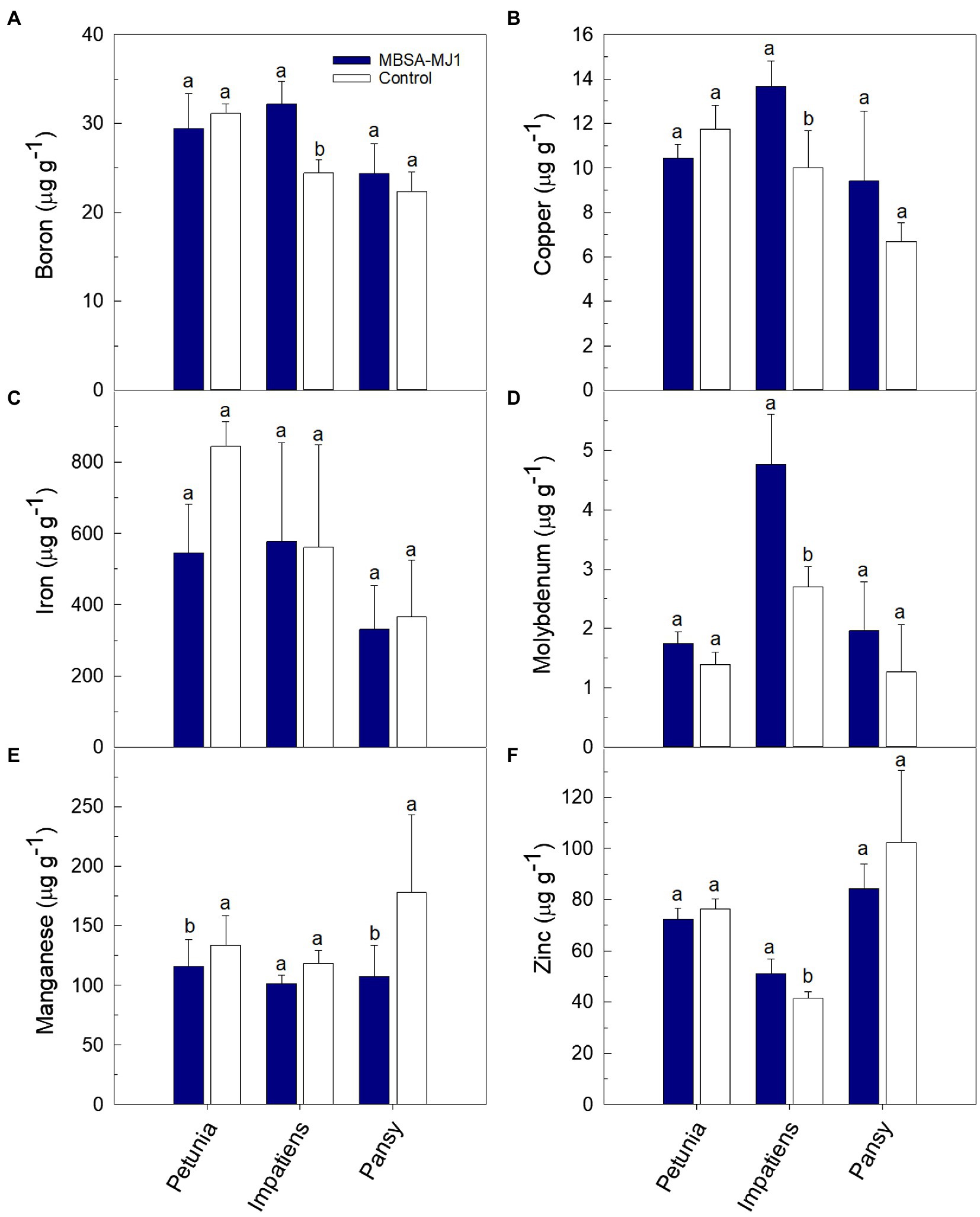
Figure 4. Tissue micronutrient concentration of Petunia×hybrida (petunia), Impatiens walleriana (impatiens), and Viola×wittrockiana (pansy) plants grown under low-nutrient conditions: boron (A), copper (B), iron (C), molybdenum (D), manganese (E), and zinc (F). Plants were treated weekly with a media drench of S. plymuthica MBSA-MJ1 (blue bars) or uninoculated LB (control; white bars). Tissue nutrient concentration was evaluated 8weeks post-transplant. Bars represent the mean (±SE) with different letters representing significant differences (p<0.05) and n=3.
In vitro Characterization
S. plymuthica MBSA-MJ1 was able to grow on basal media containing either cellobiose, fructose, galactose, glucose, glycerol, mannitol, sucrose, or ribose as the sole carbon source, indicating its ability to utilize each as an energy source.
In vitro characterization of MBSA-MJ1’s ability to increase the availability of nutrients showed that the strain was able to solubilize phosphate and potassium. Media containing insoluble forms of phosphorus and potassium had a significant decrease in pH when inoculated with MBSA-MJ1 compared to the uninoculated control (Table 1). The pH of the media decreased by 29 and 32% for the phosphate and potassium solubilization assays when inoculated with MBSA-MJ1, respectively. In addition, defined media with insoluble forms of phosphorus or potassium had a significant increase in available orthophosphate and potassium after inoculation with MBSA-MJ1 (Table 1). MBSA-MJ1 was also able to chelate copper and iron in vitro as indicated by sample color change when evaluated with the CAS assay. Absorbance values of the bacteria and control samples converted to chelation percentages show that MBSA-MJ1 was able to chelate over 62 and 63% of the iron and copper in the media, respectively (Table 1).
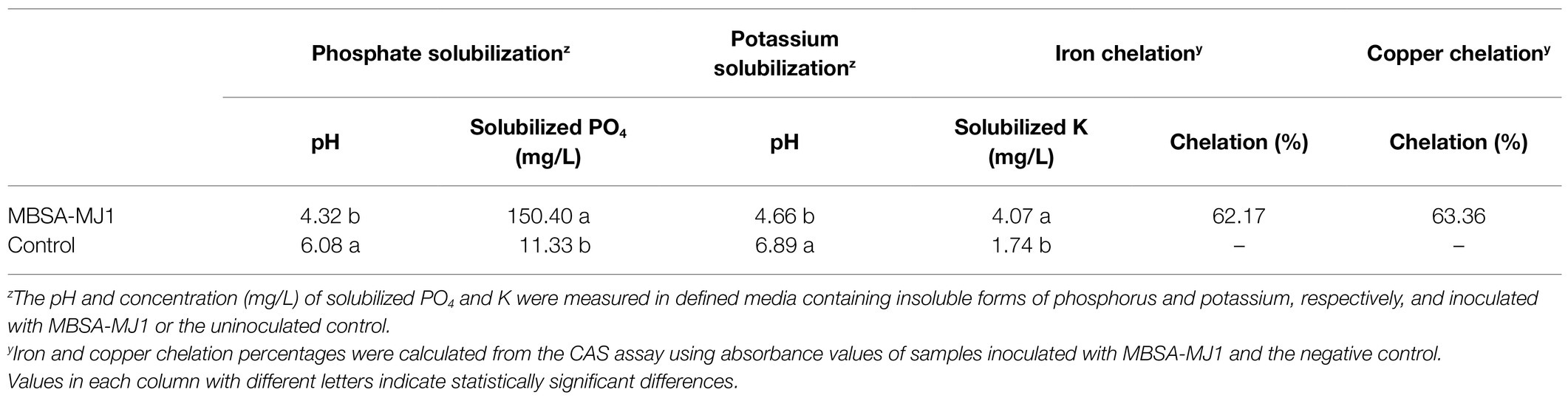
Table 1. In vitro characterization of nutrient solubilization and chelation abilities of S. plymuthica MBSA-MJ1.
Genomic Analyses to Identify Genes Putatively Involved in Growth Promotion
Nitrogen Availability and Transport
Annotation files for S. plymuthica MBSA-MJ1 were used to identify genes within the genome that are putatively involved in nutrient metabolism and increasing availability to plants (Supplementary Table S1). Genes encoding for the nitrogen starvation transcription regulation system that are also involved in ammonium transport (glnD, glnK, and amtB) were identified in the genome of MBSA-MJ1(Supplementary Table S1). The genome of MBSA-MJ1 encodes for genes involved in nitrate transport (nrtA), nitrite uptake and reduction to ammonium (nirC), nitrate reduction (nasA), nitrite reduction (nirD) and its cofactor (cysG), and the operons responsible for nitrate and nitrite reductase (narLXKGHJI) and fumarate reductase (frdABCD; Supplementary Table S1). Genes involved in nitrogen assimilation (gdhA, glnA, glnB, glnD, glnL, gltB, and gltD) and periplasmic nitrate reductase (napA) were also identified (Supplementary Table S1).
Phosphate Availability and Transport
Bacteria can convert phosphorus to inorganic bioavailable forms via nonspecific phosphatases, phytases, and phosphonatases (Liu et al., 2018). The genome of MBSA-MJ1 encodes for the appA enzyme, which has both acid phosphatase and phytase activity, a polyphosphate kinase (ppk), exopolyphosphatases (ppx and gppA), a pyrophosphatase (ppa), alkaline phosphatase (phoA), and a nonspecific acid phosphatase (phoC; Supplementary Table S1). Genes encoding for both low-affinity (pitA) and high-affinity phosphate transporters (phnCDE1E2 and pstSCAB) were identified within the genome (Supplementary Tables S1 and S3). We identified components of the bacterial P signaling pathway including the phosphate starvation two-component system (phoBR) and the negative regulator of phosphate transport (phoU; Supplementary Table S1). Genes involved in the catabolism of phosphonates and phosphites (phnGHIJKLMOP) and the negative regulator of PhnCDE (phnF) were also identified (Supplementary Table S1).
Zinc Availability and Transport
The MBSA-MJ1 genome encodes for the high-affinity ABC zinc transporter (znuABC) and its regulator (zur), which are responsible for zinc transport under low-zinc conditions (Supplementary Table S1). Genes encoding for both constitutive (zitB) and regulated (zntABR) zinc export were also identified (Supplementary Tables S1 and S3).
Organic Acid Synthesis
Mineral nutrient solubilization is most often attributed to the production of organic acids (Rodríguez and Fraga, 1999; Rodríguez et al., 2006). Genes involved in the metabolism of gluconic, ketogluconic, acetic, glyoxylic, lactic, and glycolic acid were identified (Supplementary Table S1). In particular, the genome included multiple genes putatively involved in the synthesis of gluconic acid (pqqB, pqqC, pqqD, gcd, gdh, gnl, kdgK, gnd, gntR, and ylil; Supplementary Table S1).
Sulfur Availability and Transport
The genome of MBSA-MJ1 encodes for the master regulator and transcriptional activator under sulfur starvation (cysB) and the sulfate ABC-type transporter complex (sbp and cysPWAT; Supplementary Table S1). The operon responsible for alkanesulfonate transport under sulfur limiting conditions (ssuACBDE), which includes an ABC-like transport system, was also identified (Supplementary Table S1). Additionally, the genome encodes for multiple genes involved in sulfur metabolism (cysND, cysC, and cysHIJ; Supplementary Tables S1 and S3).
Iron Chelation and Transport
The antiSMASH analysis predicted two biosynthetic gene clusters that encode for the siderophores malleobactin and amonabactin (Supplementary Table S2). The genome of MBSA-MJ1 also encodes for other genes involved in iron transport, such as the two-component system to transport ferric iron (basSR), the operon responsible for ferric hydroxamate uptake (fhuADCB), the ferrous iron transporters (feoABC and efeUOB), and two copies of the ferrous ion efflux pump (fieF; Supplementary Table S1). Additionally, the KEGG pathway analysis identified ABC transporters for iron (II; sitACDB), iron (III; afuABC), iron-enterobactin complex (fepBDGC), and the iron (III) hydroxamate complex (fhuDBC; Supplementary Table S3).
Amino Acid and Carbon Metabolism
KEGG pathway analysis identified genes responsible for the synthesis of several amino acids within the genome of MBSA-MJ1, including isoleucine, valine, leucine, lysine, arginine, proline, phenylalanine, tryptophan, and tyrosine. Further, amino acid uptake systems were identified for tryptophan, tyrosine, phenylalanine (aroP), isoleucine (brnQ), asparagine (gltP), glutamate (gltP and gltS), and serine and threonine (sstT; Supplementary Table S1). ABC transporters were also identified for lysine/arginine/ornithine (argT and hisMQP), histidine (hisJMQP), glutamine (glnHPQ), arginine (artJMIQP), glutamate/aspartate (gltIKJL), cystine (tcyABC), methionine (metQIN), proline (proVWX), and branched-chain amino acids (livKHMGF; Supplementary Table S3). In addition to amino acids, genes were identified for both the transport and metabolism of a variety of sugars (Table 2).
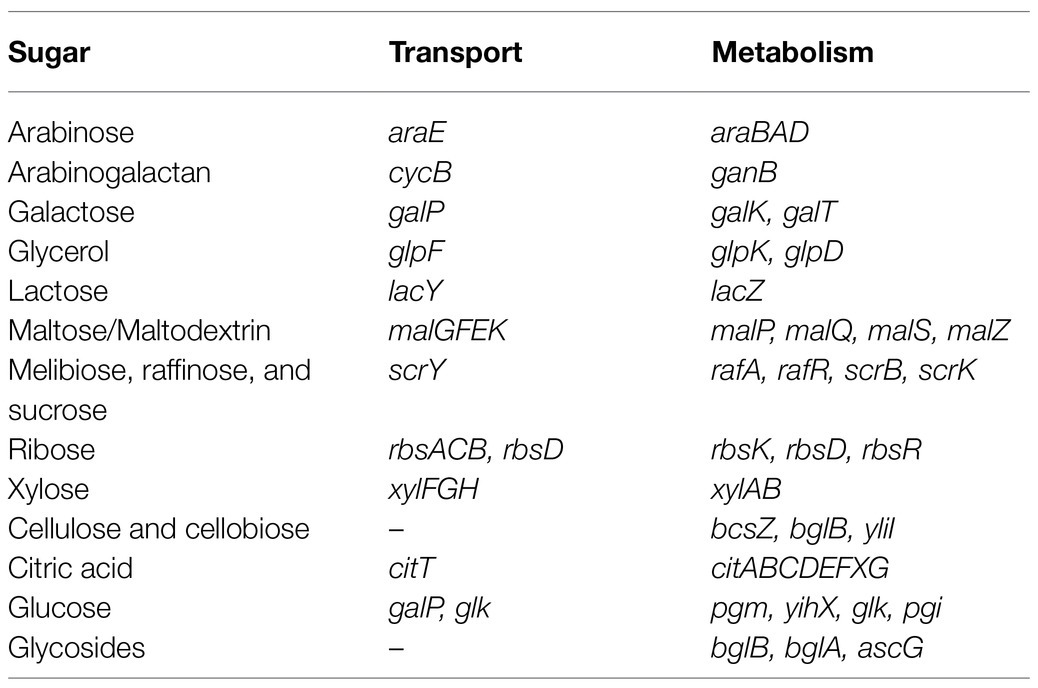
Table 2. Genes identified in the genome of S. plymuthica MBSA-MJ1 involved in sugar transport and metabolism.
Heavy Metal Resistance
Multiple copper resistance proteins were identified (copA, cueO, cueR, and pcoC; Supplementary Table S1). Additionally, the genome of MBSA-MJ1 encodes for two lead/cadmium/zinc/mercury resistance proteins (zntA and zntR), two copies of the chromate resistance protein (chrR), two nickel transporters (hypA and hypB), and the two-component system involved in response to heavy metals (basRS; Supplementary Table S1).
Discussion
Although the benefits of fertilizer application are easily recognizable in agricultural production systems, excessive application of these chemicals has negative effects on the environment. Negative environmental impacts of fertilization can often be attributed to low bioavailability and uptake by plants, leaving excess nutrients more prone to leaching (Yang et al., 2009). Therefore, it is important for crop producers to have sustainable options to ensure plants have an adequate supply of nutrients for proper growth and development, without relying on increasing applications of chemical fertilizers. We used a rate of 25mgL−1 N from 15N–2.2P–12.5K–2.9Ca–1.2Mg fertilizer in our greenhouse experiments because it is less than 20% of the average recommended fertilizer rate for ornamental crop species (150–200mgL−1 N; Beytes and Hamrick, 2003). The application of biostimulant products containing plant growth promoting bacteria (PGPR) with the ability to increase the bioavailability of nutrients provides a sustainable approach for growers to ensure proper nutrient supply while reducing chemical inputs (Saravanan et al., 2007; Park et al., 2009; Kraiser et al., 2011; Gupta et al., 2015; Ruzzi and Aroca, 2015; Drobek et al., 2019; Kramer et al., 2020). In this work, we determined that S. plymuthica MBSA-MJ1 can increase the growth and quality of containerized horticulture crops produced with low fertilizer inputs. Our work has utilized the biological and genomic characteristics of this beneficial bacteria to identify different putative mechanisms that MBSA-MJ1 may utilize to increase the availability of nutrients to plants and promote growth under otherwise limiting conditions.
The results obtained from the low-nutrient greenhouse trial provide evidence that MBSA-MJ1 can significantly improve the growth and quality of different plant species grown under low-nutrient conditions (Figures 1, 2). Multiple factors influence host specificity of both endophytic and rhizospheric bacteria, including attraction to plant root exudates, root colonization, and functioning of plant growth promoting mechanisms, contributing to varied host specificity between strains (Drogue et al., 2012; Tabassum et al., 2017; Afzal et al., 2019). Broad host specificity is particularly important for application in greenhouse production, which is characterized by the production of many different plant species. Although the application of MBSA-MJ1 only increased the flower number of impatiens compared to the uninoculated control, application did significantly increase the shoot biomass of all three plant species. Similarly, previous work with the same plant species showed application of Pseudomonas poae 29G9 or Pseudomonas fluorescens 90F12-2 increased the flower number of impatiens, while also increasing the total biomass of petunia, impatiens, and pansy plants grown under similar low-nutrient conditions (Nordstedt et al., 2020).
The growth differences observed between plants treated with MBSA-MJ1 and the uninoculated control are likely a result of MBSA-MJ1’s ability to increase plant nutrient levels. This is particularly apparent with the difference in the tissue nitrogen concentration, where all three plant species had a very large increase in foliar nitrogen when treated with MBSA-MJ1 (Figure 3A), with up to 82% more nitrogen in pansy. This is similar to other reports that show Serratia spp. increase tissue nitrogen and leaf chlorophyll content compared to uninoculated control plants (Martínez et al., 2018). Due to high concentrations of carbon but low concentrations of other nutrients in soilless media, microorganisms have the potential to deplete available nitrogen sources leading to plant nutrient deficiency (Jackson et al., 2009). Nitrogen deficiency in plants often leads to a decrease in leaf chlorophyll content, and therefore a yellowing of the leaves (Boussadia et al., 2010). In our study, leaf yellowing was observed in the uninoculated control plants, but not in the three plant species treated with MBSA-MJ1, even though all plants were grown under similar low-nutrient conditions (Figure 1). Although bacterial nitrogen fixation can be very complex, our genomic analyses identified a variety of genes involved in ammonium, nitrate, and nitrite transport and reduction that likely play a role in MBSA-MJ1’s ability to increase bioavailable nitrogen to the plant when grown under low-nutrient conditions, contributing to the growth promotion observed in the greenhouse trial.
Following nitrogen, phosphorus is the second most important element in plant nutrition. Although phosphorus is often present in abundant amounts, the total phosphorus that is available to plants is typically at very low concentrations due to its poor solubility (Gupta et al., 2015; Wang et al., 2018; Borgi et al., 2020). Phosphorus is usually applied in excess in greenhouse production and is more easily leached from the soilless substrates, leading to increasing environmental pollution (Kim and Li, 2016). Bacterial phosphate solubilization has been widely accepted as a mechanism for PGPR to increase phosphorus bioavailability to plants (Rodríguez et al., 2006; Park et al., 2009; Chen et al., 2014; Goswami et al., 2016), and Serratia spp. have been reported to be phosphate solubilizers (Ben Farhat et al., 2009). Our in vitro characterization of MBSA-MJ1 showed that the strain is an efficient phosphate solubilizer, as indicated by its ability to convert insoluble phosphorus to bioavailable orthophosphate in defined media (Table 1). The tissue nutrient analyses confirmed that this increased availability resulted in increased total phosphorus in the shoots of pansy plants (Figure 3B). Similarly, S. plymuthica BMA1 is an efficient phosphate solubilizer that increased plant growth of Vicia faba (Borgi et al., 2020). The pH reduction observed in the in vitro phosphate solubilization assay (Table 1), coupled with the identification of multiple genes involved in gluconic acid synthesis (Silva et al., 2021), indicates that MBSA-MJ1 could be producing organic acids as a mechanism to solubilize phosphate (Rodríguez et al., 2006). This mechanism for phosphate solubilization has been previously reported in Serratia marcescens strains (Chen et al., 2006; Selvakumar et al., 2008; Ben Farhat et al., 2009). Impatiens and pansy plants treated with MBSA-MJ1 also had significantly greater tissue potassium concentration than the uninoculated control (Figure 3C). Application of Serratia marcescens strains NBRI1213 and MTCC 8708 similarly show increases of phosphorus and potassium tissue nutrient content in maize and wheat plants (Selvakumar et al., 2008; Lavania and Nautiyal, 2013). Our in vitro characterization showed a similar decrease in media pH and increase in soluble potassium in defined media when inoculated with MBSA-MJ1 (Table 1); therefore, it is probable that MBSA-MJ1 is able to utilize organic acid production as a mechanism to solubilize and increase the bioavailability of potassium similar to phosphate (Sharma et al., 2016).
Zinc is essential to many plant physiological processes including chlorophyll synthesis, and zinc deficiency can negatively impact plant growth and development (Hussain et al., 2015). In our work, we showed that impatiens grown under low-nutrient conditions had significantly higher levels of zinc in shoot tissues when treated with MBSA-MJ1 as compared to the uninoculated control (Figure 4F). Bacterial zinc solubilization is one mechanism that can increase the bioavailability of this valuable nutrient to plants, and research has shown that zinc solubilizing bacteria can increase plant growth (Ramesh et al., 2014; Kamran et al., 2017). Similar to phosphate and potassium, zinc becomes more soluble and available to plants at a lower pH (Hussain et al., 2015). Research has shown that phosphate solubilizing bacteria can also increase plant zinc uptake (Salimpour et al., 2012). Considering MBSA-MJ1’s ability to solubilize phosphate and potassium through a reduction in pH, the production of organic acids by MBSA-MJ1 is a potential mechanism allowing this strain to solubilize phosphorus, potassium, and zinc, making them more bioavailable and increasing tissue nutrient levels.
Iron is usually present in abundant amounts in soil, but its availability to plants is often dependent on the production of iron chelators, such as bacterial-produced siderophores (Powell et al., 1980). Microbial-produced siderophores have been shown to increase iron acquisition and reduce iron deficiency symptoms in multiple crops grown under iron-deficient conditions (Sharma et al., 2003; Jin et al., 2006; Zhou et al., 2018; Lurthy et al., 2020). Our in vitro characterization showed that MBSA-MJ1 acts as an efficient iron chelator (Table 1). Additionally, genomic analyses identified biosynthetic gene clusters for two siderophores and multiple genes involved in bacterial iron transport and metabolism. However, our tissue nutrient analysis did not show any significant differences between bacterial-treated plants and the control (Figure 4C). Lurthy et al. (2020) suggests that the impact of siderophores on plant iron acquisition is highly dependent on the type of siderophore and the plant host. Therefore, it is probable that MBSA-MJ1 could assist in iron acquisition when colonizing plant hosts not evaluated in this study.
Copper is another essential metal element; however, it can be toxic to cells above certain levels (Vita et al., 2016). Our tissue nutrient analysis showed that impatiens plants treated with MBSA-MJ1 had significantly higher copper levels than the untreated control. However, the plants did not exhibit any signs of copper toxicity, such as stunted growth or the inhibition of photosynthesis, as these plants were larger and greener than untreated plants (Figures 1, 2). The optimal tissue copper concentration in impatiens is 10–15μgg−1 (Dole and Wilkins, 1999). The control plants had only 10μgg−1, whereas plants treated with MBSA-MJ1 had 13.7μgg−1 (Figure 4B). This indicates that MBSA-MJ1 can increase levels of this essential plant nutrient without reaching toxic levels. Our in vitro characterization of MBSA-MJ1 showed that like iron, the strain could efficiently chelate copper (Table 1). This likely provides a mechanism for MBSA-MJ1 to provide safe levels of copper to its plant host and augment growth under low-nutrient conditions. Notably, our genome analyses also identified four copper resistance proteins that may be involved in protecting the bacteria from potentially toxic exposure to copper (Supplementary Table S1; Lee et al., 2002).
Recent work has shown that application of sulfur oxidizing bacteria increases soil sulfate concentration and increases plant growth and sulfur uptake in maize and garlic (Youssif et al., 2015; Pourbabaee et al., 2020). In our study, we observed that impatiens and pansy plants grown under low-nutrient conditions and treated with MBSA-MJ1 had significantly greater levels of sulfur when compared to the uninoculated control (Figure 3F). Additionally, our genomic analyses identified both the master transcriptional regulator under sulfur starvation (cysB) and the sulfate transporter complex (sbp-cysPWAT) encoded in the genome of MBSA-MJ1 (Supplementary Table S1). These genes could potentially play a role in MBSA-MJ1’s ability to sense limited sulfur in the rhizosphere and supply bioavailable sulfur to the plant host, corroborating the increase in plant tissue concentration that was observed.
In addition to the nutrients already discussed in detail, it should be noted that treatment with MBSA-MJ1 significantly increased the tissue concentration of other essential nutrients, such as boron, calcium, magnesium, and molybdenum (Figures 3, 4). Impatiens and pansy treated with MBSA-MJ1 also had significantly greater tissue sodium concentration (data not shown). Interestingly, the untreated plants had higher manganese content than bacterial-treated plants (Figure 4E). When plants are grown under phosphorus-deficient conditions, as were induced by our low fertilizer treatment, many plant species increase root exudation of carboxylates, which increase the availability and uptake of manganese (Lambers et al., 2014). Bacteria that increase the availability of phosphorus in the rhizosphere may prevent the release of carboxylates from the roots and subsequently these plants will have reduced Mn uptake and Mn tissue concentration. Although the emphasis on understanding bacterial nutrition-related plant growth promoting mechanisms have focused on nutrients, such as nitrogen and phosphorus, our work has shown the bacterial application can increase the tissue levels of these other important macro and micronutrients. Therefore, these results provide justification for future work to begin understanding bacterial mechanisms involved in increasing the availability or uptake of nutrients to plants grown under low-nutrient conditions.
Amino acids and carbon secreted by plant roots serve as an important method to recruit and sustain beneficial rhizospheric bacteria. Therefore, bacteria with robust amino acid and carbon source metabolism are more likely to persist in the rhizosphere and be able to positively influence the growth of their host (Lugtenberg and Kamilova, 2009). Genomic analyses conducted in this work identified a series of genes related to amino acid and carbon metabolism and transport. In addition to the genomic analyses, our in vitro characterization assays provided evidence that MBSA-MJ1 could utilize a variety of carbon sources for growth. Not only is this robust carbon metabolism useful for persistence in the rhizosphere, but it also has the potential to increase MBSA-MJ1’s host range, a valuable trait for potential commercial biostimulant products.
Conclusion
Serratia plymuthica MBSA-MJ1 significantly increased the shoot biomass of all three plant species, the flower number of impatiens plants, and the tissue concentrations of certain nutrients in different plant species grown under low-nutrient conditions. In addition, the comprehensive genomic analyses shed light on different genes encoded within MBSA-MJ1’s genome that are putatively involved in mechanisms conferring plant growth promotion under low-nutrient conditions. Plant growth promotion by rhizospheric bacteria is likely the result of multiple coordinated mechanisms, and this work begins to highlight how interconnected mechanisms can increase overall plant health and quality. Increasing plant health under low-nutrient conditions without increasing chemical inputs provides an exciting option for crop producers to improve environmental sustainability. Formulation of biostimulant products containing well-characterized PGPR is a viable solution to accomplish this goal. Using MBSA-MJ1 as a model strain, future work should be invested in characterizing how PGPR increase the bioavailability or uptake of different essential nutrients by plants. Our work has defined the biology and genomic characteristics of this agriculturally important PGPR, contributing to a comprehensive understanding of this strain’s ability to increase plant growth under low-nutrient stress and the mechanisms that might be allowing this interaction to take place.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA669647.
Author Contributions
NN and MJ conceived the project and experimental design. NN performed the greenhouse production trial, in vitro assays, genomic analyses, and led the writing of the manuscript. MJ acquired funding and resources, served as project administrator, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Salaries and research support were provided in part by the State and Federal funds appropriated to the OARDC, the Ohio State University. Journal Article Number HCS 21–01. This work was financially supported, in part, by the American Floral Endowment, the Floriculture and Nursery Research Initiative, USDA-ARS cooperative agreement #58–5082–0-006 as part of the Application Technology Research Unit project #5082–21000-001-00D, and the OSU D.C. Kiplinger Floriculture Endowment. Support was also provided to NN by the Ohio State University Distinguished Fellowship, the OARDC Director’s Graduate Associateship, and the Altman Family Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Laura Chapin for assistance with the greenhouse experiments, Dr. Chris Taylor for his advice and for sharing his bacteria collections, and Dr. James Altland and Leslie Morris from the USDA-ARS Application Technology Research Unit (Wooster, OH) and Juan Quijia Pillajo for assistance with the in vitro nutrient measurements. We also thank Ball Horticultural Company and Syngenta Flowers for providing seeds for the greenhouse experiment.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmicb.2021.788198/full#supplementary-material
References
Adesemoye, A. O., and Kloepper, J. W. (2009). Plant-microbes interactions in enhanced fertilizer-use efficiency. Appl. Microbiol. Biotechnol. 85, 1–12. doi: 10.1007/s00253-009-2196-0
Adesemoye, A. O., Torbert, H. A., and Kloepper, J. W. (2008). Enhanced plant nutrient use efficiency with PGPR and AMF in an integrated nutrient management system. Can. J. Microbiol. 54, 876–886. doi: 10.1139/W08-081
Adesemoye, A. O., Torbert, H. A., and Kloepper, J. W. (2010). Increased plant uptake of nitrogen from 15N-depleted fertilizer using plant growth-promoting rhizobacteria. Appl. Soil Ecol. 46, 54–58. doi: 10.1016/j.apsoil.2010.06.010
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Aly, H., Kamalay, J., Walter, N., Okubara, P., and Taylor, C. (2007). “Characterization of the pseudomonas genus of bacteria for plant-parasitic nematode control.” in ASM Conferences “Pseudomonas 2007”; August 26, 2007 (Seattle, WA, USA).
Arif, M. S., Shahzad, S. M., Riaz, M., Yasmeen, T., Shahzad, T., Akhtar, M. J., et al. (2017). Nitrogen-enriched compost application combined with plant growth-promoting rhizobacteria (PGPR) improves seed quality and nutrient use efficiency of sunflower. J. Plant Nutr. Soil Sci. 180, 464–473. doi: 10.1002/jpln.201600615
Bachman, G. R., and Metzger, J. D. (2008). Growth of bedding plants in commercial potting substrate amended with vermicompost. Bioresour. Technol. 99, 3155–3161. doi: 10.1016/j.biortech.2007.05.069
Ben Farhat, M., Farhat, A., Bejar, W., Kammoun, R., Bouchaala, K., Fourati, A., et al. (2009). Characterization of the mineral phosphate solubilizing activity of Serratia marcescens CTM 50650 isolated from the phosphate mine of Gafsa. Arch. Microbiol. 191, 815–824. doi: 10.1007/s00203-009-0513-8
Beytes, C., and Hamrick, D. (2003). in Ball Redbook. 17th Edn. eds. C. Beytes and D. Hamrick Batavia (IL: Ball Publishing).
Bhardwaj, D., Ansari, M. W., Sahoo, R. K., and Tuteja, N. (2014). Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Factories 13, 1–10. doi: 10.1186/1475-2859-13-66
Bindraban, P. S., Dimkpa, C., Nagarajan, L., Roy, A., and Rabbinge, R. (2015). Revisiting fertilisers and fertilisation strategies for improved nutrient uptake by plants. Biol. Fertil. Soils 51, 897–911. doi: 10.1007/s00374-015-1039-7
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Borgi, M. A., Saidi, I., Moula, A., Rhimi, S., and Rhimi, M. (2020). The attractive Serratia plymuthica BMA1 strain with high rock phosphate-solubilizing activity and its effect on the growth and phosphorus uptake by Vicia faba L. plants. Geomicrobiol J. 37, 437–445. doi: 10.1080/01490451.2020.1716892
Boussadia, O., Steppe, K., Zgallai, H., Ben El Hadj, S., Braham, M., Lemeur, R., et al. (2010). Effects of nitrogen deficiency on leaf photosynthesis, carbohydrate status and biomass production in two olive cultivars “Meski” and “Koroneiki”. Sci. Hortic. 123, 336–342. doi: 10.1016/j.scienta.2009.09.023
Chen, Y., Fan, J. B., Du, L., Xu, H., Zhang, Q. H., and He, Y. Q. (2014). The application of phosphate solubilizing endophyte Pantoea dispersa triggers the microbial community in red acidic soil. Appl. Soil Ecol. 84, 235–244. doi: 10.1016/j.apsoil.2014.05.014
Chen, Y. P., Rekha, P. D., Arun, A. B., Shen, F. T., Lai, W. A., and Young, C. C. (2006). Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 34, 33–41. doi: 10.1016/j.apsoil.2005.12.002
Crowley, D. E., Reid, C. P. P., and Szaniszlo, P. J. (1988). Utilization of microbial siderophores in iron acquisition by oat. Plant Physiol. 87, 680–685. doi: 10.1104/pp.87.3.680
Dimkpa, C. (2016). Microbial siderophores: production, detection and application in agriculture and environment. Endocytobiosis Cell Res. 27, 7–16.
Dole, J. M., and Wilkins, H. F. (1999). Floriculture Priciples and Species. New Jersey: Prentice-Hall, Inc.
Drobek, M., Frąc, M., and Cybulska, J. (2019). Plant biostimulants: importance of the quality and yield of horticultural crops and the improvement of plant tolerance to abiotic stress-A review. Agronomy 9:9060335. doi: 10.3390/agronomy9060335
Drogue, B., Doré, H., Borland, S., Wisniewski-Dyé, F., and Prigent-Combaret, C. (2012). Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 163, 500–510. doi: 10.1016/j.resmic.2012.08.006
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2, 1–19. doi: 10.1080/23311932.2015.1127500
Güneş, A., Turan, M., Güllüce, M., and Şahin, F. (2014). Nutritional content analysis of plant growth-promoting rhizobacteria species. Eur. J. Soil Biol. 60, 88–97. doi: 10.1016/j.ejsobi.2013.10.010
Gupta, G., Parihar, S. S., Ahirwar, N. K., Snehi, S. K., and Singh, V. (2015). Plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 07, 96–102. doi: 10.4172/1948-5948.1000188
Hussain, A., Arshad, M., Zahir, Z. A., and Asghar, M. (2015). Prospects of zinc solubilizing bacteria for enhancing growth of maize. Pakistan J. Agric. Sci. 52, 915–922.
Isaac, R. A., and Johnson, W. C. (1985). Elemental analysis of plant tissue by plasma emission spectroscopy: collaborative study. J. Assoc. Off. Anal. Chem. 68, 499–505. doi: 10.1093/jaoac/68.3.499
Jackson, B. E., Wright, R. D., and Alley, M. M. (2009). Comparison of fertilizer nitrogen availability, nitrogen immobilization, substrate carbon dioxide efflux, and leaching in peat-lite, pine bark, and pine tree substrates. HortScience 44, 781–790. doi: 10.21273/hortsci.44.3.781
Jin, C. W., He, Y. F., Tang, C. X., Wu, P., and Zheng, S. J. (2006). Mechanisms of microbially enhanced Fe acquisition in red clover (Trifolium pratense L.). Plant Cell Environ. 29, 888–897. doi: 10.1111/j.1365-3040.2005.01468.x
Kamran, S., Shahid, I., Baig, D. N., Rizwan, M., Malik, K. A., and Mehnaz, S. (2017). Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 8:2593. doi: 10.3389/fmicb.2017.02593
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Khan, A. G. (2005). Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace Elem. Med. Biol. 18, 355–364. doi: 10.1016/j.jtemb.2005.02.006
Kim, H. J., and Li, X. (2016). Effects of phosphorus on shoot and root growth, partitioning, and phosphorus utilization efficiency in lantana. HortScience 51, 1001–1009. doi: 10.21273/hortsci.51.8.1001
Kraiser, T., Gras, D. E., Gutiérrez, A. G., González, B., and Gutiérrez, R. A. (2011). A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 62, 1455–1466. doi: 10.1093/jxb/erq425
Kramer, J., Özkaya, Ö., and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Lambers, H., Hayes, P. E., Lailiberte, E., Oliveira, R. S., and Turner, B. L. (2014). Leaf manganese accumulation and phosphorus-acquisition efficiency. Trends Plant Sci. 20, 83–90. doi: 10.1016/j.tplants.2014.10.007
Lavania, M., and Nautiyal, C. S. (2013). Solubilization of tricalcium phosphate by temperature and salt tolerant Serratia marcescens NBRI1213 isolated from alkaline soils. African J. Microbiol. Res. 7, 4403–4413. doi: 10.5897/AJMR2013.5773
Lee, S. M., Grass, G., Rensing, C., Barrett, S. R., Yates, C. J. D., Stoyanov, J. V., et al. (2002). The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem. Biophys. Res. Commun. 295, 616–620. doi: 10.1016/S0006-291X(02)00726-X
Liu, J., Cade-Menun, B. J., Yang, J., Hu, Y., Liu, C. W., Tremblay, J., et al. (2018). Long-term land use affects phosphorus speciation and the composition of phosphorus cycling genes in agricultural soils. Front. Microbiol. 9, 1–14. doi: 10.3389/fmicb.2018.01643
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Lurthy, T., Cantat, C., Jeudy, C., Declerck, P., Gallardo, K., Barraud, C., et al. (2020). Impact of bacterial siderophores on iron status and ionome in pea. Front. Plant Sci. 11, 1–12. doi: 10.3389/fpls.2020.00730
Martínez, O. A., Encina, C., Tomckowiack, C., Droppelmann, F., Jara, R., Maldonado, C., et al. (2018). Serratia strains isolated from the rhizosphere of raulí (Nothofagus alpina) in volcanic soils harbour PGPR mechanisms and promote raulí plantlet growth. J. Soil Sci. Plant Nutr. 18, 804–819. doi: 10.4067/S0718-95162018005002302
Martín-Sánchez, L., Ariotti, C., Garbeva, P., and Vigani, G. (2020). Investigating the effect of belowground microbial volatiles on plant nutrient status: perspective and limitations. J. Plant Interact. 15, 188–195. doi: 10.1080/17429145.2020.1776408
Mehta, S., and Nautiyal, C. S. (2001). An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 43, 51–56. doi: 10.1007/s002840010259
Nordstedt, N. P., Chapin, L. J., Taylor, C. G., and Jones, M. L. (2020). Identification of pseudomonas spp. that increase ornamental crop quality during abiotic stress. Front. Plant Sci. 10, 1–12. doi: 10.3389/fpls.2019.01754
Nordstedt, N. P., and Jones, M. L. (2021). Genomic analysis of Serratia plymuthica MBSA-MJ1, a plant growth promoting rhizobacteria that improves water-stress tolerance in greenhouse ornamentals. Front. Microbiol. 12:653556. doi: 10.3389/fmicb.2021.653556
Park, K. H., Lee, C. Y., and Son, H. J. (2009). Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Lett. Appl. Microbiol. 49, 222–228. doi: 10.1111/j.1472-765X.2009.02642.x
Parmar, P., and Sindhu, S. S. (2019). The novel and efficient method for isolating potassium solubilizing bacteria from rhizosphere soil. Geomicrobiol J. 36, 130–136. doi: 10.1080/01490451.2018.1514442
Pereira, S. I. A., Abreu, D., Moreira, H., Vega, A., and Castro, P. M. L. (2020). Plant growth-promoting rhizobacteria (PGPR) improve the growth and nutrient use efficiency in maize (Zea mays L.) under water deficit conditions. Heliyon 6:e05106. doi: 10.1016/j.heliyon.2020.e05106
Pourbabaee, A. A., Koohbori Dinekaboodi, S., Seyed Hosseini, H. M., Alikhani, H. A., and Emami, S. (2020). Potential application of selected sulfur-oxidizing bacteria and different sources of sulfur in plant growth promotion under different moisture conditions. Commun. Soil Sci. Plant Anal. 51, 735–745. doi: 10.1080/00103624.2020.1729377
Powell, P. E., Cline, G. R., Reid, C. P. P., and Szaniszlo, P. J. (1980). Occurrence of hydroxamate siderophore iron chelators in soils. Nature 287, 833–834. doi: 10.1038/287833a0
Pridham, T. G., and Gottlieb, D. (1948). The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–114. doi: 10.1128/jb.56.1.107-114.1948
Radzki, W., Gutierrez Mañero, F. J., Algar, E., Lucas García, J. A., García-Villaraco, A., and Ramos Solano, B. (2013). Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 104, 321–330. doi: 10.1007/s10482-013-9954-9
Rajawat, M. V. S., Singh, S., Tyagi, S. P., and Saxena, A. K. (2016). A modified plate assay for rapid screening of potassium-solubilizing bacteria. Pedosphere 26, 768–773. doi: 10.1016/S1002-0160(15)60080-7
Raklami, A., Bechtaoui, N., Tahiri, A. I., Anli, M., Meddich, A., and Oufdou, K. (2019). Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front. Microbiol. 10, 1–11. doi: 10.3389/fmicb.2019.01106
Ramesh, A., Sharma, S. K., Sharma, M. P., Yadav, N., and Joshi, O. P. (2014). Inoculation of zinc solubilizing bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in Vertisols of Central India. Appl. Soil Ecol. 73, 87–96. doi: 10.1016/j.apsoil.2013.08.009
Rodríguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rodríguez, H., Fraga, R., Gonzalez, T., and Bashan, Y. (2006). Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287, 15–21. doi: 10.1007/s11104-006-9056-9
Ruzzi, M., and Aroca, R. (2015). Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 196, 124–134. doi: 10.1016/j.scienta.2015.08.042
Salimpour, S., Khavazi, K., Nadian, H., Besharati, H., and Miransari, M. (2012). Canola oil production and nutrient uptake as affected by phosphate solubilizing and sulfur oxidizing bacteria. J. Plant Nutr. 35, 1997–2008. doi: 10.1080/01904167.2012.716892
Saravanan, V. S., Kalaiarasan, P., Madhaiyan, M., and Thangaraju, M. (2007). Solubilization of insoluble zinc compounds by Gluconacetobacter diazotrophicus and the detrimental action of zinc ion (Zn2+) and zinc chelates on root knot nematode Meloidogyne incognita. Lett. Appl. Microbiol. 44, 235–241. doi: 10.1111/j.1472-765X.2006.02079.x
Schütz, L., Gattinger, A., Meier, M., Müller, A., Boller, T., Mäder, P., et al. (2018). Improving crop yield and nutrient use efficiency via biofertilization—A global meta-analysis. Front. Plant Sci. 8, 1–13. doi: 10.3389/fpls.2017.02204
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophore. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Selvakumar, G., Mohan, M., Kundu, S., Gupta, A. D., Joshi, P., Nazim, S., et al. (2008). Cold tolerance and plant growth promotion potential of Serratia marcescens strain SRM (MTCC 8708) isolated from flowers of summer squash (Cucurbita pepo). Lett. Appl. Microbiol. 46, 171–175. doi: 10.1111/j.1472-765X.2007.02282.x
Shaharoona, B., Naveed, M., Arshad, M., and Zahir, Z. A. (2008). Fertilizer-dependent efficiency of pseudomonads for improving growth, yield, and nutrient use efficiency of wheat (Triticum aestivum L.). Appl. Microbiol. Biotechnol. 79, 147–155. doi: 10.1007/s00253-008-1419-0
Sharma, A., Johri, B. N., Sharma, A. K., and Glick, B. R. (2003). Plant growth-promoting bacterium pseudomonas sp. strain GRP3 influences iron acquisition in mung bean (Vigna radiata L. Wilzeck). Soil Biol. Biochem. 35, 887–894. doi: 10.1016/S0038-0717(03)00119-6
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2, 1–14. doi: 10.1186/2193-1801-2-587
Sharma, A., Shankdhar, D., and Shankhdhar, S. C. (2016). “Potassium-solubilizing microorganisms: Mechanism and their role in potassium solubilization and uptake,” in Potassium Solubilizing Microorganisms for Sustainable Agriculture. eds. V. Meena, B. Maurya, and J. Verma (New Delhi: Springer), 1–17.
Silva, U. C., Cuadros-Orellana, S., Silva, D. R. C., Freitas-Junior, L. F., Fernandes, A. C., Leite, L. R., et al. (2021). Genomic and phenotypic insights into the potential of rock phosphate solubilizing bacteria to promote millet growht in vivo. Front. Microbiol. 11:574550. doi: 10.3389/fmicb.2020.574550
South, K. A., Hand, F. P., and Jones, M. L. (2020). Beneficial bacteria identified for the control of Botrytis cinerea in petunia greenhouse production. Plant Dis. 104, 1801–1810. doi: 10.1094/PDIS-10-19-2276-RE
Sweeney, R. A. (1989). Generic combustion method for determination of crude protein in feeds: collaborative study. J. Assoc. Off. Anal. Chem. 72, 770–774. doi: 10.1093/jaoac/72.5.770
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Iqbal Khan, M. S., Shahid, N., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Vita, N., Landolfi, G., Baslé, A., Platsaki, S., Lee, J., Waldron, K. J., et al. (2016). Bacterial cytosolic proteins with a high capacity for cu(I) that protect against copper toxicity. Sci. Rep. 6, 1–11. doi: 10.1038/srep39065
Wang, F., Deng, M., Xu, J., Zhu, X., and Mao, C. (2018). Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 74, 114–122. doi: 10.1016/j.semcdb.2017.06.013
Yang, J., Kloepper, J. W., and Ryu, C. M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14, 1–4. doi: 10.1016/j.tplants.2008.10.004
Yoon, S., Kraemer, S. M., DiSpirito, A. A., and Semrau, J. D. (2010). An assay for screening microbial cultures for chalkophore production. Environ. Microbiol. Rep. 2, 295–303. doi: 10.1111/j.1758-2229.2009.00125.x
Youssif, B. D., Hosna, M. A. F., and Mervat, A. A. T. (2015). Effect of Sulphur and Sulphur oxidizing bacteria on growth and production of garlic (Allium sativum, L.) under saline conditions. Middle East J. Agric. Res. 4, 446–459.
Keywords: biofertilizer, biostimulant, floriculture, genome analysis, horticulture, nutrient stress, nutrient solubilization, PGPR
Citation: Nordstedt NP and Jones ML (2021) Serratia plymuthica MBSA-MJ1 Increases Shoot Growth and Tissue Nutrient Concentration in Containerized Ornamentals Grown Under Low-Nutrient Conditions. Front. Microbiol. 12:788198. doi: 10.3389/fmicb.2021.788198
Edited by:
José David Flores Félix, Universidade da Beira Interior, PortugalReviewed by:
Gayathri Ilangumaran, McGill University, CanadaSowmyalakshmi Subramanian, McGill University, Canada
Naeem Khan, University of Florida, United States
Copyright © 2021 Nordstedt and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle L. Jones, am9uZXMuMTk2OEBvc3UuZWR1
 Nathan P. Nordstedt
Nathan P. Nordstedt Michelle L. Jones
Michelle L. Jones