- New England Biolabs, Inc., Ipswich, MA, United States
Strand-specific DNA nicking endonucleases (NEases) typically nick 3–7 bp sites. Our goal is to engineer infrequent NEase with a >8 bp recognition sequence. A BamHI catalytic-deficient mutant D94N/E113K was constructed, purified, and shown to bind and protect the GGATCC site from BamHI restriction. The mutant was fused to a 76-amino acid (aa) DNA nicking domain of phage Gamma HNH (gHNH) NEase. The chimeric enzyme was purified, and it was shown to nick downstream of a composite site 5′ GGATCC-N(4-6)-AC↑CGR 3′ (R, A, or G) or to nick both sides of BamHI site at the composite site 5′ CCG↓GT-N5-GGATCC-N5-AC↑CGG 3′ (the down arrow ↓ indicates the strand shown is nicked; the up arrow↑indicates the bottom strand is nicked). Due to the attenuated activity of the small nicking domain, the fusion nickase is active in the presence of Mn2+ or Ni2+, and it has low activity in Mg2+ buffer. This work provided a proof-of-concept experiment in which a chimeric NEase could be engineered utilizing the binding specificity of a Type II restriction endonucleases (REases) in fusion with a nicking domain to generate infrequent nickase, which bridges the gap between natural REases and homing endonucleases. The engineered chimeric NEase provided a framework for further optimization in molecular diagnostic applications.
Introduction
In nature, DNA nicking endonucleases (NEases) are mainly involved in DNA restriction, rolling circle replication, F plasmid transfer, and DNA damage repair (Zheleznaya et al., 2009; Chan et al., 2011; Xu, 2015). The restriction endonuclease (REase)-derived nicking enzymes typically nick DNA target sites in 3–7 bp, and homing NEases recognition sequences are usually more than 16 bp (Chan et al., 2011). Therefore, there is a significant gap of NEases that nick DNA sites in the range of 8–15 bp. To expand the DNA target sequence range, we propose to utilize the DNA binding specificities of Type II REases (3–8 bp) that are cleavage-deficient with mutations in the catalytic site and fusion of the mutant enzymes to a DNA nicking domain [e.g., phage Gamma HNH (gHNH) nicking endonuclease, gHNH, 3–5 bp recognition (Xu and Gupta, 2013)]. When a REase binding specificity for 7–8 bp target is fused to a gHNH nicking domain of 5 bp (e.g., ACCGR, complementary strand YCGGT, R = A or G, Y = C or T), it is possible to generate an infrequent nicking enzyme with 12–13 bp composite sites. It is shown that the minimal DNA nicking domain of the gHNH endonuclease contains only 76 amino acid (aa) residues and required Mn2+ for stimulated nicking activities (Xu and Gupta, 2013).
For isothermal DNA amplification, thermostable nicking enzymes are needed to be compatible with Bst DNA polymerase active at 50–65°C. Thermostable DNA NEase has been found in thermophilic phage genome isolated from deep sea vent (Xu and Gupta, 2013) and a large number of metagenome sequences from deep sea vent and hot springs can be rich sources for discovery of more thermostable NEases (Zhang et al., 2017; Castelan-Sanchez et al., 2019). Thermostable NEases, such as Nt.BstNBI and Nb.BsrDI, are part of natural occurring restriction systems. When they are combined with the corresponding small subunits, they become double-stranded (ds) DNA cleaving enzymes (Higgins et al., 2001; Xu et al., 2007).
Sequence-specific DNA NEases have been used in DNA optical mapping by nicking, nick translation of the nicked sites with fluorescently labeled dNTP, and religation of the nicked strand (Zhang et al., 2010; Jeffet et al., 2021). Large chromosome rearrangement (insertion, deletion, or translocation) of cancer cells could be “visualized” under high-resolution fluorescent microscope when the abnormal DNA nicking pattern images are compared with the WT (English et al., 2015; Jeffet et al., 2021) (Bionano Genomics, Irys system). NEases are also used in isothermal DNA amplifications, such as strand-displacement DNA amplification (SDA) (Walker et al., 1994), EXPAR (Qian et al., 2012), nicking enzyme assisted amplification (NEAA) (Van Ness et al., 2003; Qian et al., 2019), Nt.CviPII-assisted random whole genome amplification (Chan et al., 2004), and in NicE-seq to study open chromatin profiling (Ponnaluri et al., 2017). NEase-mediated amplification is utilized in FDA-approved clinical diagnostic testing that enabled the first CLIA-waived molecular diagnostic test for rapid identification of influenza A and B. (Nie et al., 2014). Most recently, nicking enzyme-assisted reaction (NEAR) technology has been applied to the detection of COVID-19 viral RNA by reverse transcription, DNA nicking, and isothermal DNA amplification (James and Alawneh, 2020). NEAR (ID NOW™ COVID-19 (Abbott/Alere) was able to detect SARS-CoV2 genomic fragment in a short time (10 min).
In this work, we constructed and purified a catalytic-deficient BamHI mutant D94N/E113K. It was shown previously that D94N or E113K single mutant displayed poor cleavage activity but maintained specific binding to BamHI site in a DNA mobility shift assay (Xu and Schildkraut, 1991). Here, the purified BamHI mutant D94N/E113K was shown to bind and protect against BamHI digestion of a plasmid with a single BamHI site. We then fused the BamHI mutant to a small gHNH nicking domain and purified the fusion enzyme. The fusion nicked the composite site GGATCC-N(4-6)-AC↑CGR in NEB buffer 2 (50 mM NaCl) and buffer 3 (100 mM NaCl) supplement with Mn2+ or Ni2+. The fusion NEase also nicks the composite site CCG↓GT-N5-GGATCC-N5 AC↑CGG both upstream (5′) and downstream (3′), potentially generating a long overhang (20 bases) after melting of the duplex. This work demonstrates a proof of concept for fusion of a catalytic-deficient REase and phage gHNH nicking domain and opened the door for further optimization of infrequent NEases in future research and diagnostic applications.
Materials and Methods
Enzymes, Plasmid DNA, and DNA Sequencing
Restriction endonucleases, Nb.BsrDI, Proteinase K, pBR322, and pUC19 were provided by New England Biolabs, Inc. (NEB). Synthetic gene blocks were purchased from Integrated DNA Technologies (IDT). Big-Dye Sanger sequencing kit was purchased from Thermo-Fisher/ABI. Restriction buffer compositions (1 ×): Buffer 2: 50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, and pH 7.9 at 25°C. Buffer 3: 100 mM NaCl, 50 mM Tris–HCl, 10 mM MgCl2, and pH 7.9 at 25°C. CutSmart buffer (buffer 4 plus BSA): 50 mM Potassium Acetate, 20 mM Tris-acetate, 10 mM Magnesium Acetate, 100 μg/ml BSA, and pH 7.9 at 25°C.
In some nicking reactions, Mn2+ or Ni2+ divalent cations were supplemented in NEB buffer 2 or 3 as indicated. DNA blunting kit and blunt/TA quick ligation kit were provided by NEB. Escherichia coli competent cells T7 Express (C2566) and NEB 10β were also from NEB.
Nicking reactions were usually carried out at 37°C for 30–60 min or as indicated in specific buffers. One forward and one reverse primer were used to sequence (map) the BamHI site and nicking site(s) in pUC19 and the adjacent sequences. The primers were purchased from IDT and have the following sequences:
pUC-F61, 5′ CAGCTTGTCTGTAAGCGGATGCCG 3′.
pUC-R600, 5′ GGGCAGTGAGCGCAACGCAAT 3′.
The DNA strands of the pUC19 sequence near BamHI site is artificially defined as top and bottom strand as following:
top strand: 5′-…AGAGGATCCCCGGGTACCGAGC…-3′.
bottom strand: 3′-…TCTCCTAGGGGCCCATGGCTCG… -5′.
Gene Assembly/Cloning
Synthetic gene blocks were assembled into pTYB1 vector (NdeI and XhoI digested) using Gibson assembly kit (NEB) and the DNA inserts were selected by plasmid DNA transformation and the correct insert was verified by DNA sequencing. Plasmids were prepared by plasmid mini-preparation kits (Qiagen, Sigma, or NEB). DNASTAR Lasergene software was used for sequence editing and visualization of nicking sites (doublet peaks) in DNA run-off sequencing.
The aa sequences of D94N/E113K and phage gHNH nicking domain are shown below. No linker aa residues were inserted between BamHI* and the gHNH nicking domain due to the flexible nature of the BamHI C-terminal region (Newman et al., 1994).
>BamHI D94N/E113K (GGATCC) 213 aa (N and K residues shown in red are the aa substitutions, predicted molecular mass = 24.57 kDa)
MEVEKEFITD EAKELLSKDK LIQQAYNEVK TSICSPI WPA TSKTFTINNT
EKNCNGVVPI KELCYTLLED TYNWYREKPL DILKLEK KKG GPI NVYKEFI
ENSELKRVGM EFKTGNISSA HRSMNKLLLG LKHGE IDLAI ILMPIKQLAY
YLTDRVTNFE ELEPYFELTE GQPFIFIGFN AEAYNSNVPL IPKGSDGMSK
RSIKKWKDKV ENK
>Bacillus phage gHNH small nicking domain (76 aa)
-fyrskawer lreqalmrdk glclhcknnr kikva dmvdh iipikvdpsl
klklenlqsl cnpchnrkta edkkkyg
>WT Bacillus phage Gamma HNH endonuclease 127 aa (GenBank accession number YP_338236)
(the minimal nicking domain sequence is shown in underlined residues).
mkkplrpcce fhcynltrer
yceehrykek etqqdknryy drfkrdkest afyrskawer lreqalmrdk glclhcknnr kikvadmvdh iipikvdpsl klklenlqsl cnpchnrkta edkkkyg.
Protein Expression and Purification
The IMPACT protein expression system, including pTYB1 vector and chitin beads, was provided by NEB. Target protein was cleaved from the intein-CBD fusion by DTT cleavage overnight and eluted from a chitin column (in some cases, a second DTT overnight cleavage step was carried out, and target protein was eluted to increase recovery yield). Enzymes were further purified by chromatography through a heparin column (5 ml HiTrap Heparin HP, GE Healthcare). Proteins were concentrated by low-speed centrifugation in protein concentrators (10 kDa cut-off) and resuspended in enzyme storage buffer (200 mM NaCl, 10 mM Tris–HCl, pH 7.5, 1 mM DTT, and 50% glycerol) and kept in a –20°C freezer. T7 Express cells carrying BamHI mutant gene or BamHI*-gHNH fusion gene in a plasmid was cultured at 37°C to late log phase and enzyme production was initiated by addition of 0.5 mM IPTG final concentration and cells were induced at 18°C overnight in a temperature-controlled shaker. Cells were lysed by sonication in a chitin column buffer (20 mM Tris–HCl, pH 8.5, and 0.5 M NaCl) at 4°C. To remove nucleic acids from enzymes, BamHI*-gHNH fusion protein eluted from the chitin column was diluted in a buffer (0.3 M NaCl, 20 mM Tris–HCl, pH 7.9, 0.1 mM ETDA, and 1 mM DTT) and passed through HiTrap DEAE column (Fast flow, 5 ml). Enzymes were concentrated by low-speed centrifugation in protein concentrators and resuspended in enzyme storage buffer.
Results
Construction of BamHI Mutant D94N/E113K
The structure of BamHI endonuclease dimer in complex with cognate DNA had been solved previously (Figure 1A). The enzyme makes multiple direct hydrogen bonds and water-mediated hydrogen bonds with the G/C and A/T base pairs in the major groove in the recognition sequence GGATCC to form specific binding complex (Newman et al., 1994). In the cocrystal structure, the active site is formed by PD94-E111xE113 residues to bind divalent cations and active a water molecule for nucleophilic attack of the DNA backbone. A typical REase fold consisting of central four-stranded β-sheets flanked by two α-helices on both sides was present in BamHI. The C-terminal BamHI forms a flexible “arm” extension (Newman et al., 1995). The mechanism of REase and DNA interactions include five steps: (1) non-specific binding to dsDNA, (2) one-dimensional diffusion and two-dimensional hopping on DNA to locate a target site, (3) specific binding (hydrogen bonding to GGATCC sequence and DNA backbone), (4) enzyme activation in cleavage, and (5) product release (Aggarwal, 1995).
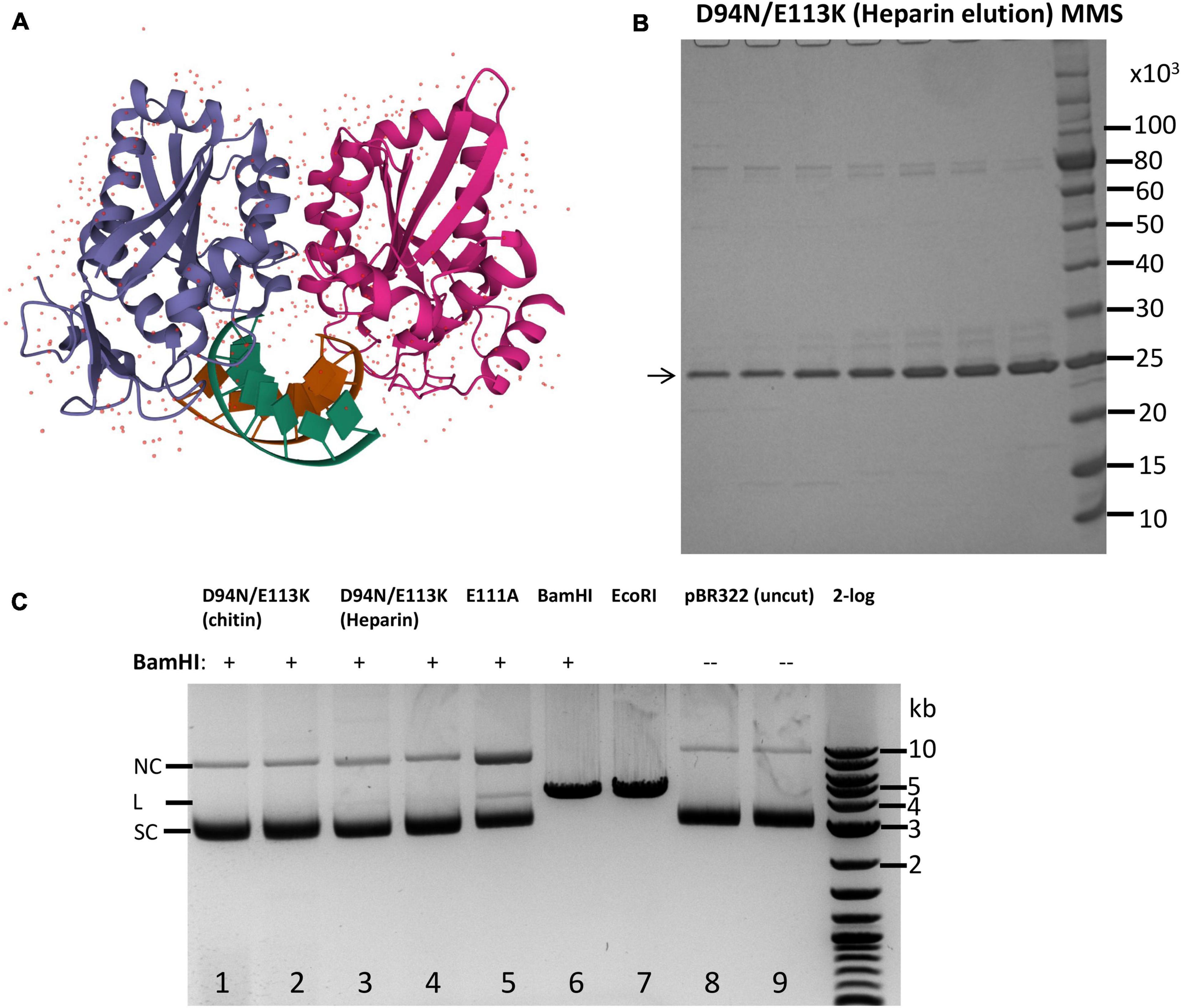
Figure 1. Purification of catalytic-deficient BamHI mutant D94N/E113K and protection of BamHI site by D94N/E113K against BamHI restriction. (A) Crystal structure of a BamHI dimer bound to the cognate site (tatGGATCCata) (PBD ID 1BHM). Two subunits of the BamHI dimer are shown in purple and magenta colors, respectively. The DNA double helix is shown in green and brown colors. The small dots indicate water molecules in the dimer structure. (B) SDS-PAGE analysis of the purified BamHI variant D94N/E113K (∼24.57 kDa). Eluted fractions from a heparin column (HiTrap Heparin 5 ml) were shown. MMS, protein molecular mass standard in Daltons (Da) (NEB). (C) BamHI restriction digestion in the presence of D94N/E113K. Lanes 1–4, pBR322 (1 μg, ∼7.1 nM) with a single BamHI site was first incubated with D94N/E113K (0.5 and 1 μg, ∼0.4 and 0.8 μM) at 25°C for 15 min, and 5 U of BamHI was then added to the bound complex and incubated at 37°C for 30 min. After restriction digestion, Proteinase K (1.6 U) was added and incubated at 37°C for 15 min to remove BamHI and the mutant protein. Lane 5 is the same as in lanes 1–4 except a different BamHI catalytic mutant (E111A, 1 μg, ∼0.8 μM) was used. In lanes 6 and 7, pBR322 was linearized by BamHI (10 U) and EcoRI (10 U), respectively, in the absence of D94N/E113K. Lanes 8, TE (10 mM Tris–HCl, 0.1 mM EDTA) mock digestion; lane 9, uncut pBR322. 2-log, DNA size ladder (100 bp to 10 kb, NEB). SC, supercoiled DNA; L, linear DNA; NC, nicked circular DNA. The size of linear pBR322 is ∼4.4 kb.
Prior to the crystal structure, random mutagenesis of the bamHIR gene identified three catalytic residues: E77, D94, and E113 because aa substitutions at these positions generated mutants (E77K, D94N, and E113K) with diminished catalytic activity (Xu and Schildkraut, 1991). Later, E111 was also identified as a catalytic residue by a genetic selection because E111A could bind to the BamHI site and served as a “repressor” (Dorner and Schildkraut, 1994). However, E77K and E111A still showed a low residual cleavage activity at high enzyme concentration in overdigestion. To construct a BamHI mutant with absolutely zero cleavage activity, we combined the two mutations to generate a double mutant D94N/E113K. The mutant enzyme was purified by chromatography through chitin and Heparin columns (Figure 1B). The partially purified protein was used to protect a single BamHI site in pBR322 against BamHI digestion. Figure 1C shows that the purified D94N/E113K protein protected pBR322 against BamHI restriction (lanes 1–4). Purified E111A binding also protects the BamHI site against BamHI digestion although a small fraction of DNA was linearized, and some nicked circular DNA was also generated (lane 5). BamHI alone digested pBR322 to linear DNA (lane 6). The mutant enzyme also blocked SmaI digestion of pUC19 because the BamHI site overlaps with a SmaI site in GGATCCCGGG sequence (data not shown). No BamHI cleavage activity was detected using 4 μg of the double mutant protein to digest pBR322 (data not shown) (4 μg of the WT BamHI should have ∼2,000 U at ∼5 × 105 U/mg specific activity) (Jack et al., 1991). We concluded from this preliminary experiment that BamHI double mutant D94N/E113K did not show any cleavage activity, but it could bind to BamHI site and protect the site against BamHI restriction. The mutant protein can also protect SmaI site against SmaI digestion when BamHI and SmaI sites overlap.
Construction of BamHI Catalytic Mutant and Gamma HNH Fusion
Our goal is to construct a chimeric NEase with infrequent nicking sites. The minimal phage gHNH nicking domain consists of 76-aa residues and has attenuated activity (the WT enzyme has 127-aa residues). The truncated nicking domain is more active in the presence of Mn2+ and it has poor activity in Mg2+ buffers (Xu and Gupta, 2013). A schematic diagram of the fusion construct is shown in Figure 2A. A synthetic gene block encoding BamHI*-gHNH fusion (BamHI* denotes D94N/E113K double mutant) was cloned into pTYB1 and the fusion enzyme was partially purified by chitin column chromatography and analyzed by SDS-PAGE (Figure 2B). The fusion enzyme yield is rather low, producing approximately 1 mg/L of IPTG-induced cells, which might be caused by the toxicity of the fusion enzyme as compared with the yield of ∼4 mg per L of IPTG-induced cells for the BamHI double mutant (D94N/E113K) alone. The purified enzyme is active in nicking of pUC19 in a time course (5–60 min digestion) in NEB buffer 2 supplemented with 1 mM MnCl2 (Figure 2C). It is estimated that more than 80% of supercoiled DNA was converted to nicked circular form after 1 h digestion (under limited digestion condition). The fusion enzyme has relatively low nicking activity in standard restriction buffers with 10 mM MgCl2 (Supplementary Figure 1A). It is also active in Ni2+ buffer and the substrate DNA was partially nicked after 1 h digestion (Supplementary Figure 1B). In an enzyme titration experiment vs. fixed amount of pUC19 DNA, it was estimated that 1 μg of the fusion enzyme was required to completely nick 1 μg of the substrate into nicked circular DNA (Supplementary Figure 2). Therefore, the specific activity was estimated at ∼1–2 × 103 U/mg protein (1 U is defined as the amount of enzyme required to completely nick 1 μg pUC19 into nicked circular form at 37°C for 1 h in NEB buffer 3 supplemented with 1 mM MnCl2). The full-length phage gHNH endonuclease is fully active in Mg2+ buffer and nicks the sites AC↑CGR (or AC↑CG in over-digestion) (Xu and Gupta, 2013). The specific nicking activity of gHNH endonuclease on pUC19 was estimated at 4–8 × 103 U/mg in NEB buffer 3 (Supplementary Figure 2, bottom panel). The specific nicking activity of the short 76-aa nicking domain could not be determined due to its attenuated activity (estimated at less than 500 U/mg, Supplementary Figure 2, top panel). We conclude from this experiment that the activity of BamHI*-gHNH fusion nickase was low in Mg2+ buffer, but its activity was stimulated in Mn2+ and Ni2+ buffers. Short incubation time (30–60 min) and medium to high salt are required for the chimeric enzyme to minimize non-specific (star) nicking activity (see below).
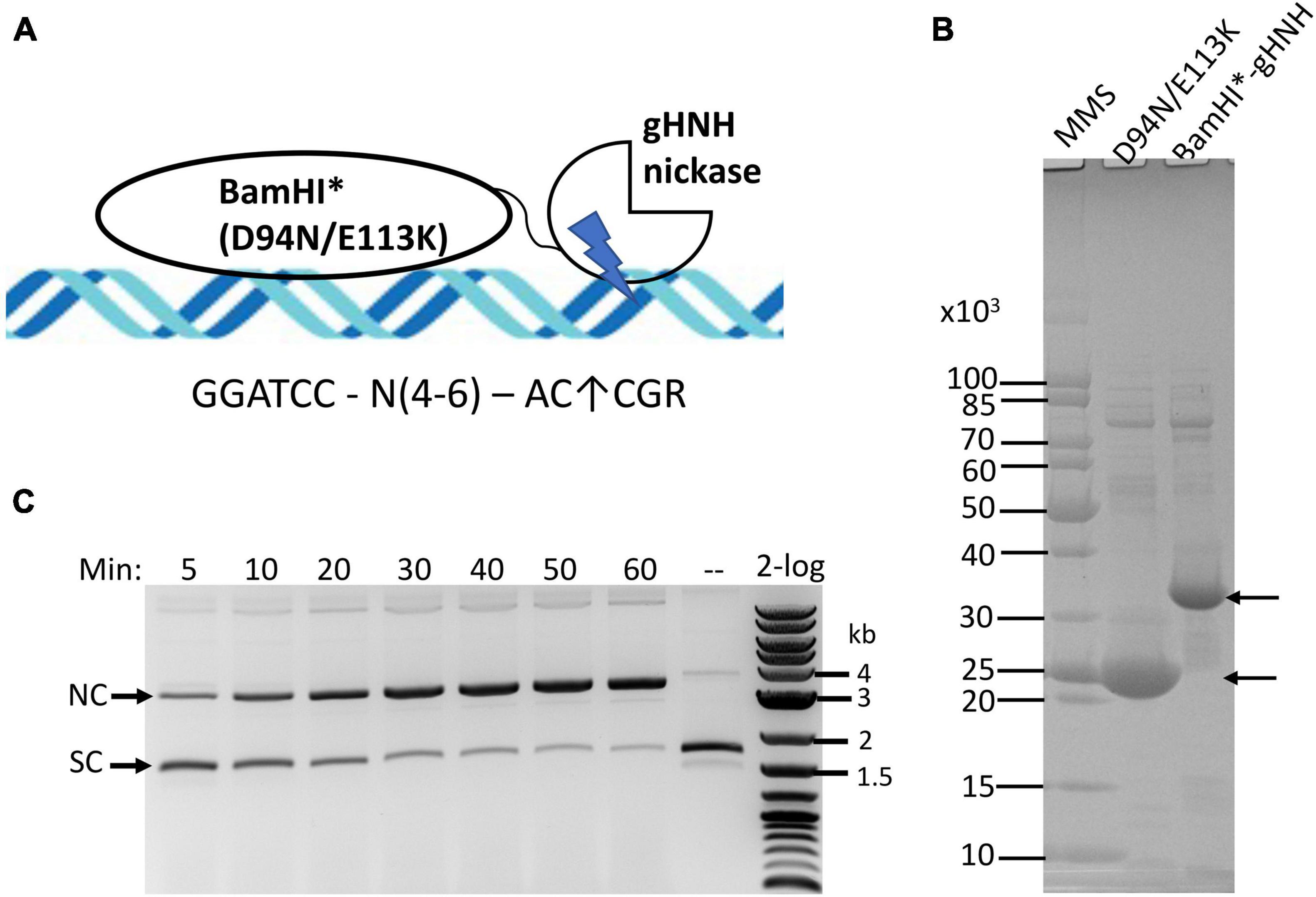
Figure 2. Purification of BamHI catalytic mutant (BamHI*)-gHNH fusion enzyme and nicking activity assay in the presence of divalent cations. (A) Schematic diagram of BamHI catalytic mutant (BamHI*) in fusion with phage gHNH nicking domain (the size of DNA and protein domains are not shown in scale). The diagram is simplified to show the monomeric fusion only. (B) SDS-PAGE analysis of partially purified BamHI*-gHNH fusion protein. Lane 3, chitin column purified fusion protein (predicted molecular mass in ∼33.53 kDa). Following DTT cleavage overnight, eluants 1 and 2 from the chitin column were concentrated by low-speed centrifugation in centrifugal filter unit (Millipore, 10 kDa cut-off) and resuspended in an enzyme storage buffer. Approximately 20 and 5 μg of the BamHI mutant and fusion proteins were loaded in the gel. MMS, protein molecular mass standard in Daltons (Da) (NEB). (C) A time course (5–60 min) of pUC19 nicking digestion (partial) by the fusion enzyme in NEB buffer 2 supplemented with MnCl2. Approximately 80% of supercoiled DNA was converted to nicked circular form after 1 h nicking reaction. A weak linear band also appeared. SC and NC, supercoiled and nicked circular DNA, respectively. 2-log, DNA size ladder (100 bp to 10 kb, NEB).
Mapping of Nicking Sites in pUC19
We used DNA run-off sequencing to map the nicking site near the BamHI binding sequence. It is based on the appearance of doublets in sequencing base calling. When the template strand is broken (nicked or cleaved), the sequencing reads with doublet A/C, A/G, A/T, or extra high A peak are generated by the template-independent terminal nucleotide transferase activity of the Taq DNA polymerase [or extra T peak in doublet (T/C, T/G, T/A) in reverse strand sequencing] (Too et al., 2010). One example of BspQI cleavage site mapping is shown in Supplementary Figure 3. BspQI recognition sequence is 5′ GCTCTTC 3′; but it cuts downstream at N1 (top strand)/N4 (bottom strand). When the bottom strand template was cleaved four nucleotides downstream of the site, Taq DNA polymerase “sensed” the broken template and added an extra “A” peak in the sequencing read (see a schematic diagram in panel A, and the actual sequencing read in panel B). If the cleavage/nicking reaction was partial (a mixture of uncut and cut templates), the run-off position would be indicated as a doublet (A/T). The top-strand cleavage position is marked by an extra “T” peak and sudden drop-off of sequencing peaks at the cleavage site. The bottom panel in Supplementary Figure 3C shows the control pBR322 (uncut) sequencing reactions in which the base calls are normally continuous and no apparent doublets were detected. Supplementary Figure 4 shows the run-off sequencing of BamHI partially digested pUC19 and the appearance of T/G doublet in the cleaved BamHI site 5′-G↓GATCC-3′.
We used the same run-off sequencing to determine the nicking site of the chimeric NEase as described above. The schematic diagram of the sequencing reaction is shown on the top of Figure 3A, in which the bottom strand serves as the template (nicked strand) and the top strand is the sequencing read. A doublet A/C is read as “N” by the sequence editing software, indicating bottom-strand nicking at GGATCC-N6-AC↑CGA (Figure 3B). In the uncut control DNA, the sequence reads as continuous bases and no doublet was detected (Figure 3B, top). It is noted that no cleavage or nicking occurred within the BamHI site as one expects. In the bottom-strand sequencing, a nick (T/A doublet) is detected near the BamHI site (GGATCC N5 AG↑TCG) (Figure 3C). The same sequence is shown as a reverse complement in Figure 3D (CGN↓CT N5 GGATCC). The nicking site AGTCG can be considered as a star nicking site of gHNH in Mn2+ buffer, which is somewhat analogous to ACCGG with the combined sequence of AS↑YSG (S = C or G, Y = C or T). Nicking at this star site was diminished in NEB buffer 3 supplemented with 1 mM MnCl2 or when this site is mutated to CTATA sequence (data not shown). DNA sequences ACCGA or ACCGC further downstream of the BamHI site were not nicked. Some intervening sequences were manually deleted (as marked by Del1 and Del2) to show the downstream nicking sites. This mapping experiment showed that the fusion nicking enzyme recognizes GGATCC as an anchor site and nicks DNA 5–6 bp downstream (3′) or upstream (5′) in the cognate site ACCGR or a nearby site AGTCG. The stand-alone nicking sites (ACCGA, ACCGC, or other star sites) farther away from the BamHI site are poor substrates for the fusion enzyme.
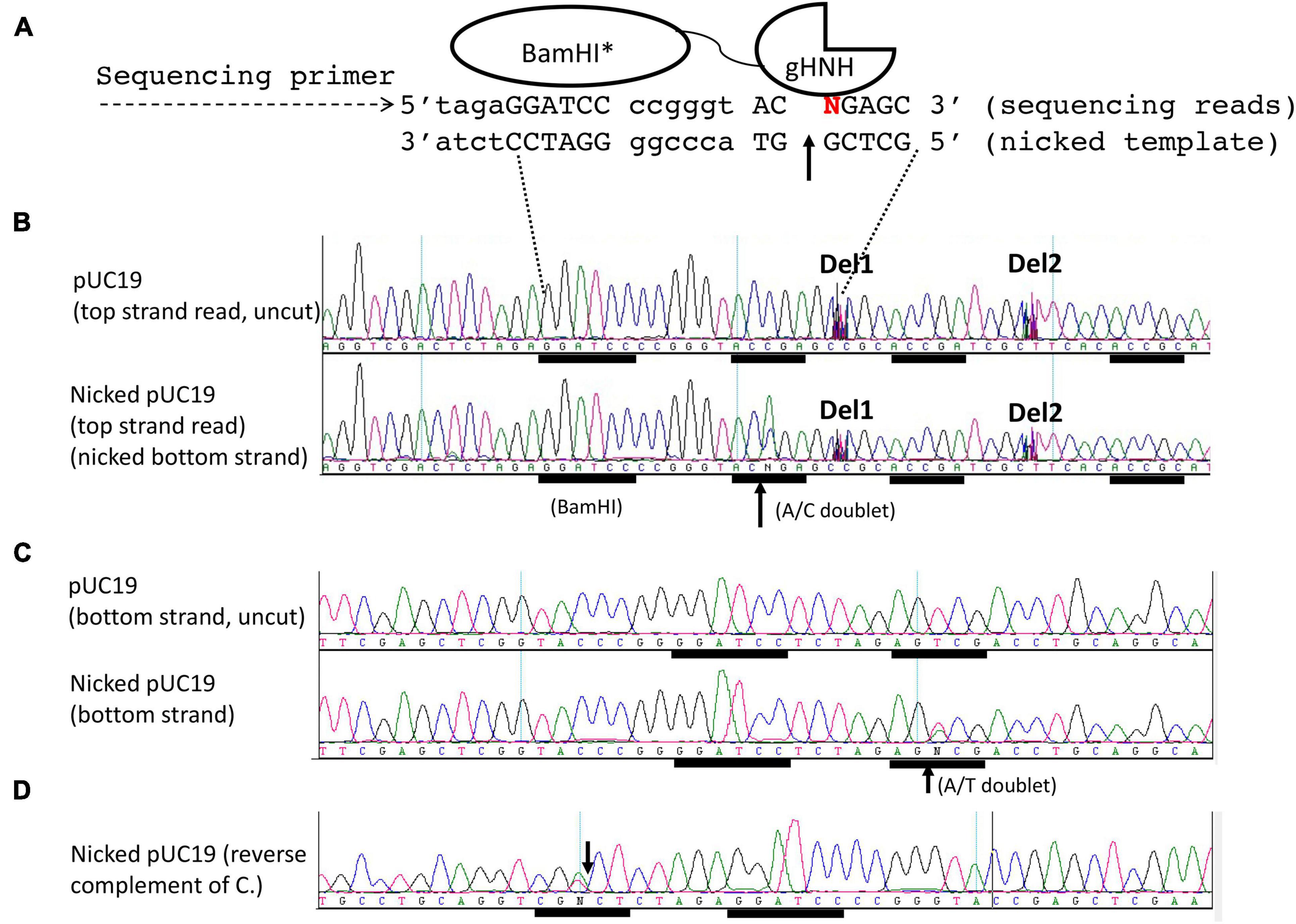
Figure 3. DNA run-off sequencing to map the nicking site(s) and nicking distance from a BamHI site in pUC19. The plasmid substrate was digested by the fusion nickase at 37°C for 30 min in NEB buffer 2 supplemented with 1 mM MnCl2. (A) The composite nicking site GGATCC-N6-AC↑CGA is shown in a schematic diagram. (B) Top-strand sequencing read from the nicked bottom strand. The up arrow indicates the bottom strand of AC↑CGA was nicked and the appearance of an extra adenine (A) peak in the doublet as “A/C.” The addition of an extra peak “A” was catalyzed by the Taq DNA polymerase (template-independent terminal nucleotide transferase) when the template DNA is broken (nicked). Del1 and Del2 indicate some intervening sequences were manually deleted to show the downstream sites (ACCGA and ACCGC). (C) Bottom-strand sequencing read form nicked top-strand DNA. A star nicking site 5′ GGATCC N5 AGTCG 3′ was detected by the presence of an “A/T” doublet. The same sequence was shown as a reverse-complement in (D). The down arrow indicates the nicked position.
To analyze the nicking activity dependence on a BamHI site, we deleted the BamHI site in pUC19 by treatment of BamHI-linearized plasmid with a blunting kit and religated the DNA and transferred the DNA into NEB 10β by transformation. Individual plasmids were prepared and confirmed the absence of BamHI site by BamHI digestion (data not shown). The pUC19-ΔBamHI was used as a substrate for BamHI*-gHNH nicking digestion. Only a small fraction of the DNA was nicked in 30 min digestion, and a majority of the DNA remained intact as supercoiled form (Supplementary Figure 5, lanes 1–4), suggesting weak non-specific nicking of star sites. In control digestions, pUC19-ΔBamHI was resistant to BamHI restriction, but it was linearized by EcoRI, and nicked completely by nicking enzyme Nb.BsrDI. Nicked circular form of pUC19-ΔBamHI was accumulated after long digestion (4 h to overnight) (data not shown), indicating possible non-specific nicking activity directed by the nicking domain or by some contaminating E. coli nucleases. In a control digestion, the fusion nickase completely nicked pUC19 in Mn2+ buffer although a small amount of linear DNA was also generated. We concluded from this experiment that efficient nicking of a plasmid required the presence of BamHI site and gHNH nicking site. Some non-specific nicking may occur by the gHNH nicking domain during prolonged digestion.
Mapping of Nicking Sites 4–5 bp Downstream of BamHI Site
To further map composite nicking sites, we constructed two pUC19 derivatives with GGATCC N(4–5) ACCGG and replace the star site AGTCG (complement CGACT) with CTATA (complement TATAG) upstream. The plasmids with one composite nicking site GGATCC N4 ACCGG or GGATCC N5 ACCGG were digested by the fusion enzyme and the nicked DNA was subjected to run-off sequencing. Figure 4 shows that only the downstream site AC↑CGG was nicked with a spacer N4 (top panel) or N5 (bottom panel) as a strong doublet (A/C) was detected. No apparent nicking was detected on the site ACCGA farther downstream or at the sequence TATAG (CTATA) upstream (data not shown). We concluded from this experiment that efficient DNA nicking required both a BamHI site and a gHNH nicking site. No nicking was detected upstream at TATAG (complement CTATA) after replacing the star nicking site. However, we have not analyzed all possible star sites for the gHNH nicking domain in Mn2+ buffer.
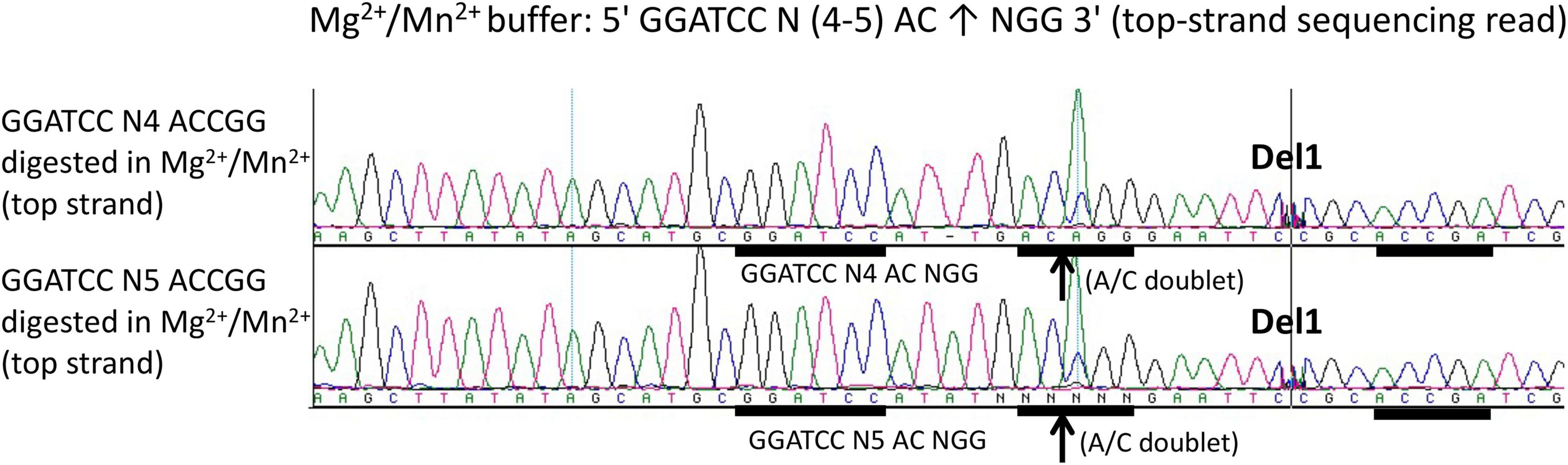
Figure 4. DNA run-off sequencing to map the nicking site and nicking distance from BamHI in the engineered composite site GGATCC-N(4-5)-AC↑CGG in pUC19 (1xACCGG). Top-strand sequencing read using nicked bottom-strand template. DNA nicking reactions were carried out in NEB buffer 2 supplemented with Mn2+ (1 mM) at 37°C for 30 min. The up arrows indicate the nicked position where a strong A/C doublet was present. Del1 indicates the intervening sequence had been manually deleted to show the downstream site ACCGA (where no nicking had occurred). The composite sites GGATCC N4 ACCGG and GGATCC N5 ACCGG were inserted into pUC19 multiple cloning sites and flanked by HindIII and EcoRI sites. In addition, a star site (AGTCG) upstream of the BamHI site was changed to the sequence TATAG to eliminate the star nicking activity.
Mapping of Nicking Sites Near GGATCC 5 Nucleotides Upstream and Downstream
We next constructed a symmetric composite site CCGGT N5 GGATCC N5 ACCGG in a pUC19 derivative and examined the fusion enzyme nicking activity in NEB buffer 2 supplemented with Mn2+. Figure 5 shows that the enzyme can nick the symmetric substrate both upstream and downstream (CCG↓GT N5 GGATCC N5 AC↑CGG), generating a long 5′ overhang of 20 nucleotides (nt) if the duplex is melted at high temperature. Such a long overhang might be useful for DNA cloning and DNA fragment end labeling. Figure 5A (sequencing panels 1 and 2) shows the nicking activity generated in the Mg2+/Mn2+ and Mg2+/Ni2+ buffers (i.e., top-strand sequencing read from nicked bottom-strand template). The bottom strand also contains a nick near the BamHI site (Figure 5B) as evidenced by the presence of an A/C doublet, indicating both strands were nicked by the fusion nickase. The same sequence is shown in Figure 5C as a reverse complement (T/G doublet at the nicked position). Due to limited digestions to minimize star activity, the plasmid substrate was only partially nicked and did not reach complete nicking.
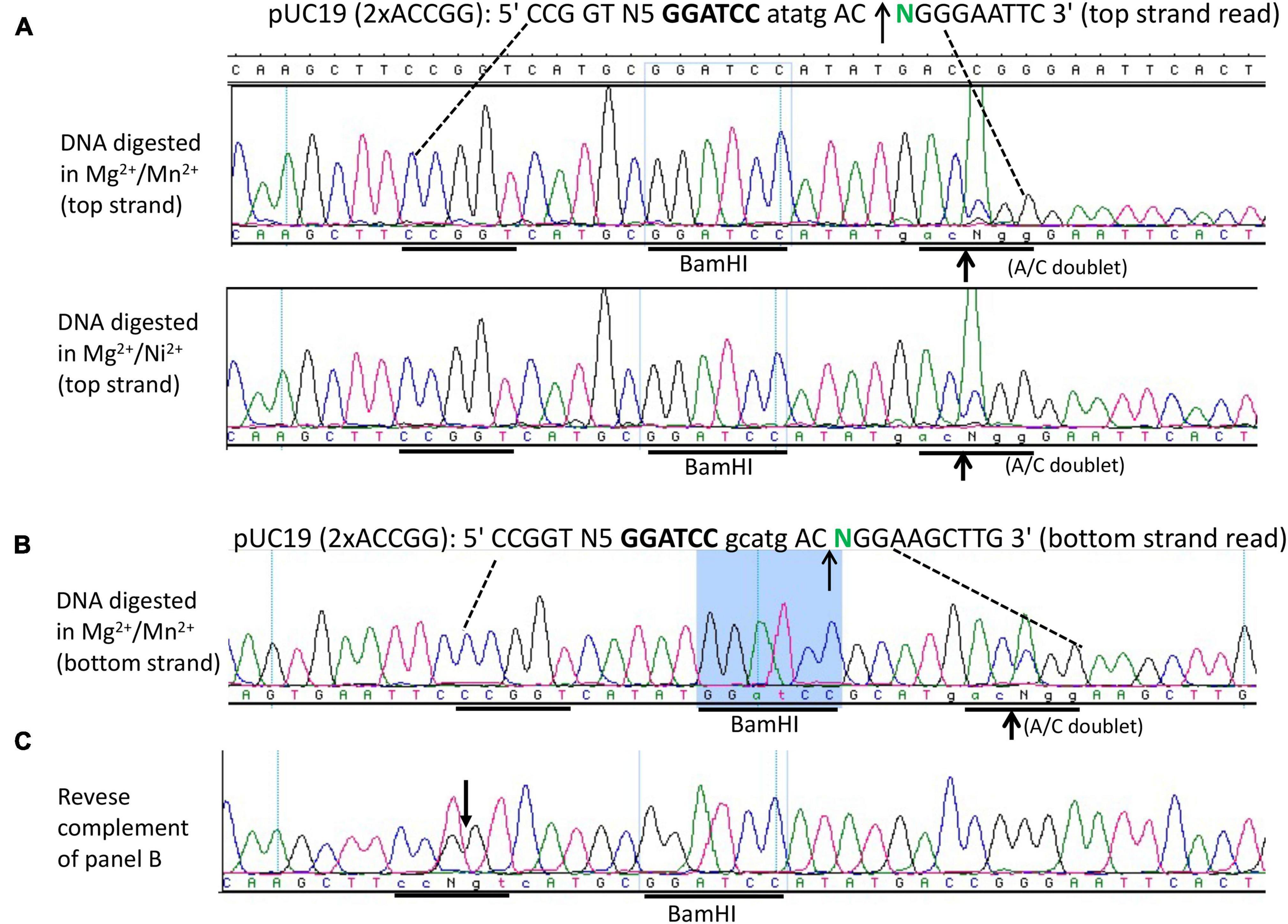
Figure 5. DNA run-off sequencing of the nicking sites and nicking distance from the BamHI site. (A) Top-strand sequencing read from nicked bottom strand. Nicking reactions were carried out in NEB buffer 2 supplemented with MnCl2 (1 mM) or NiCl2 (1 mM) using pUC19 (2xACCGG) containing the composite site 5′ CCGGT-N5-GGATCC-N5 ACCGG 3′. The duplex oligos containing the composite site was inserted into pUC19 and flanked by HindIII (AAGCTT) and EcoRI (GAATTC) sites. (B) Bottom-strand sequencing read (nicked top-strand template) near the BamHI site. The composite site contains a nick as indicated by a strong A/C doublet. If the bottom strand is shown as a reverse-complement, a T/G doublet would be present (C) in the nicking site.
We next tested the fusion enzyme nicking activity in high salt buffers (100 mM to 200 mM NaCl + KCl) supplemented with 1 mM MnCl2 under limited digestion. The enzyme is partially active in the high salt buffers as shown in Supplementary Figure 6 although small inhibition was observed in 200 mM salt. The nicking site in pUC19 was mapped to the sequence 5′ GGATCC N6 AC↑CGA 3′.
Discussion
In this work, we constructed an infrequent NEase by fusion of cleavage-deficient BamHI mutant D94N/E113K and a phage gHNH nicking domain (∼76 aa). The fusion enzyme nicks near the BamHI site N4-6 when a nicking site ACCGR/YCGGT is present either downstream or upstream or on both sides. The activity on the gHNH nicking site ACCGR or its variants alone is rather low, and no doublets were detected in run-off sequencing of ACCGR sites further downstream or upstream under limited digestion, which is likely due to the attenuated activity of the small nicking domain that requires a strong DNA binding partner to anchor onto DNA. The nicking site recognition could be relaxed to nick star sites (ASYSR) in Mn2+ buffer, which is consistent with the star activity of the full-length gHNH endonuclease in Mn2+ buffer (1–2 base off from ACCGR sites) (Xu and Gupta, 2013). To minimize the star activity of the fusion enzyme, high salt buffer (100–200 mM NaCl or KCl) may be required in digestion in Mg2+/Mn2+ buffer or Ni2+ buffer. The fusion enzyme is active in nicking reactions with 50–150 mM NaCl (200 mM NaCl slightly inhibits activity). The fusion NEases is active in Mn2+ or Ni2+ buffer, and it has low nicking activity in the presence of 10 mM Mg2+. To study the nicking site profile of gHNH minimal nicking domain in Mn2+ and Ni2+ buffers, it would be necessary to construct NGS libraries from nicked DNA and sequencing the inserts and map the sites back to the undigested DNA by examination of thousands of sites. The divalent cation Mg2+ is probably not required for specific binding to the BamHI site for the D94N/E113K mutant as the single mutant D94N or E113K can bind to DNA in the absence of divalent cations (Xu and Schildkraut, 1991). For most of REases, they do not require divalent cations for binding, but for EcoRV-specific binding to the cognate site (GATATC), a divalent cation is required (Vipond et al., 1995). For example, Ca2+ supports binding but not cleavage for EcoRV on cognate DNA substrate.
A number of cleavage-deficient REases have been isolated before as reported in the literature (e.g., EcoO109I, EcoRI, EcoRV, BsoBI, BspQI/SapI, HincII, NotI, PacI, PvuII, and PpuMI) (reviewed in Pingoud et al., 2016). The cleavage-deficient and binding-proficient variants (4–8 bp) can be potentially coupled to the 76-aa nicking domain to generate NEases with 9–14 bp nicking specificities. Restriction enzyme catalytic residues can also be predicted among the PD-D/ExK and HNH enzyme superfamilies and catalytic mutants can be constructed with relative ease by site-directed mutagenesis or cloning of mutant gene blocks (Roberts et al., 2003; Pingoud et al., 2016).
Natural NEases have been found as one subunit of restriction endonucleases (e.g., Nb.BsrDI and Nt.BstNBI) (Higgins et al., 2001; Xu et al., 2007), in bacteriophages (phages) and pathogenicity island involved in cos/pac site cleavage/nicking and DNA packaging (e.g., gp74 of HK97, gp37 of ΦSLT, and Φ12 HNH endonucleases) (Kala et al., 2014), in mobile genetic elements involved in DNA nicking and F element transfer (Luo et al., 1995), in Chlorella viruses that catalyzed the frequent nicking of host genomic DNA (e.g., Nt.CviPII) (Chan et al., 2004, 2006), and in introns encoded homing NEases that catalyze intron mobility [e.g., I-BasI (Landthaler and Shub, 2003), I-TslI (Bonocora and Shub, 2009), I-PfoP3I (Kong et al., 2012), reviewed in (Chan et al., 2011; Xu, 2015)]. NEases can also be engineered from REases and homing endonucleases (HEases) by alteration of catalytic sites (e.g., Nt.BbvCI, Nt.BsrDI) (Heiter et al., 2005; Xu et al., 2007; McConnell Smith et al., 2009) or by nicking domain swapping (Nt.AlwI) (Xu et al., 2001).
It is not surprising that the fusion NEase nicks a few bases (N4–6) downstream of the BamHI site. In FokI footprint analysis using methylation protection experiment with dimethyl sulfate (DMS) modification and hydroxyl radical Fe(II)-EDTA foot-printing study, FokI protected ∼3 bp 5′ upstream of its recognition sequence GGATG and some additional bases at the 3′ end (the cleavage domain reaches out and cleaves DNA downstream) (Yonezawa and Sugiura, 1994). It is very likely that most REases occupy and protect a few more bp in addition to their recognition sequences. We found D94N/E113K could partially block SmaI digestion of pUC19 plasmid with the overlapping sites in GGATCCCGGG (SmaI site = CCCGGG), suggesting BamHI occupies more than 6 bp.
The infrequent NEase reported in this work might be useful for end labeling and DNA optical mapping when its non-specific nicking activity could be minimized. To enhance specific nicking of the composite site, it may be necessary to incubate DNA substrate with the fusion enzyme for 10–20 min to form enzyme-DNA complex and then add divalent cations to activate nicking for a short period of time (5–10 min). Another possibility is to utilize mutants of the 76-aa nicking domain with further weakened DNA binding affinity. Future study and optimization of the infrequent NEase is required prior to any biotechnology applications.
The cleavage-deficient BamHI double mutant may be used in in vivo study of DNA binding proteins overlapping a BamHI site or it can serve as a repressor (Dorner and Schildkraut, 1994). As reported previously that BamHI E113K mutant could also block exonuclease progression on linear DNA (Xu and Schildkraut, 1993). Similarly, we expect D94N/E113K binding to BamHI site can also physically block exonuclease degradation of linear DNA. By using similar protein fusion strategy, it is now possible to construct catalytic-deficient PvuRts1I mutant (Type IV REase) and gHNH nicking domain fusion which binds to 5-hydroxylmethylcytocine (5hmC)-modified DNA and introduces nicks near 5hmC modified sites for labeling and detection on mammalian DNA (Janosi et al., 1994).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
S-YX designed and performed the experiments, interpreted the data, and wrote the manuscript.
Funding
This work was supported by New England Biolabs, Inc. Open access publication fee is paid for by New England Biolabs, Inc.
Conflict of Interest
S-YX is an employee of New England Biolabs, Inc. a company developing restriction enzyme and nicking enzyme reagents for the research and diagnostic community.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Andy Gardner, Ira Schildkraut, and Nathan Tanner for critical reading of the manuscript; Don Comb, James Ellard, Richard Roberts, Bill Jack, and Tom Evans for support; Siu-Hong Chan, Dan Heiter, Geoff Wilson, and Andy Gardner for helpful discussions; NEB DNA core lab for sequencing. We are grateful to Lydia Dorner, Rebecca Kucera, and Ira Schildkraut for providing the BamHI E111A mutant. We appreciate two reviewers’ suggestions to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.787073/full#supplementary-material
References
Aggarwal, A. K. (1995). Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 5, 11–19. doi: 10.1016/0959-440x(95)80004-k
Bonocora, R. P., and Shub, D. A. (2009). A likely pathway for formation of mobile group I introns. Curr. Biol. 19, 223–228. doi: 10.1016/j.cub.2009.01.033
Castelan-Sanchez, H. G., Lopez-Rosas, I., Garcia-Suastegui, W. A., Peralta, R., Dobson, A. D. W., Batista-Garcia, R. A., et al. (2019). Extremophile deep-sea viral communities from hydrothermal vents: structural and functional analysis. Mar. Genom. 46, 16–28. doi: 10.1016/j.margen.2019.03.001
Chan, S. H., Stoddard, B. L., and Xu, S. Y. (2011). Natural and engineered nicking endonucleases–from cleavage mechanism to engineering of strand-specificity. Nucleic Acids Res. 39, 1–18. doi: 10.1093/nar/gkq742
Chan, S. H., Zhu, Z., Dunigan, D. D., Van Etten, J. L., and Xu, S. Y. (2006). Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: m.CviQII methylates AG sequences. Protein Expr. Purif. 49, 138–150. doi: 10.1016/j.pep.2006.04.002
Chan, S. H., Zhu, Z., Van Etten, J. L., and Xu, S. Y. (2004). Cloning of CviPII nicking and modification system from chlorella virus NYs-1 and application of Nt.CviPII in random DNA amplification. Nucleic Acids Res. 32, 6187–6199. doi: 10.1093/nar/gkh958
Dorner, L. F., and Schildkraut, I. (1994). Direct selection of binding proficient/catalytic deficient variants of BamHI endonuclease. Nucleic Acids Res. 22, 1068–1074. doi: 10.1093/nar/22.6.1068
English, A. C., Salerno, W. J., Hampton, O. A., Gonzaga-Jauregui, C., Ambreth, S., Ritter, D. I., et al. (2015). Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics 16:286. doi: 10.1186/s12864-015-1479-3
Heiter, D. F., Lunnen, K. D., and Wilson, G. G. (2005). Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 348, 631–640. doi: 10.1016/j.jmb.2005.02.034
Higgins, L. S., Besnier, C., and Kong, H. (2001). The nicking endonuclease N.BstNBI is closely related to type IIs restriction endonucleases MlyI and PleI. Nucleic Acids Res. 29, 2492–2501. doi: 10.1093/nar/29.12.2492
Jack, W. E., Greenough, L., Dorner, L. F., Xu, S.-Y., Strzelecka, T., Aggarwal, A. K., et al. (1991). Overexpression, purification and crystallization of BamHI endonuclease. Nucleic Acids Res. 19, 1825–1829. doi: 10.1093/nar/19.8.1825
James, A. S., and Alawneh, J. I. (2020). COVID-19 Infection Diagnosis: potential Impact of Isothermal Amplification Technology to Reduce Community Transmission of SARS-CoV-2. Diagnostics 10:399. doi: 10.3390/diagnostics10060399
Janosi, L., Yonemitsu, H., Hong, H., and Kaji, A. (1994). Molecular cloning and expression of a novel hydroxymethylcytosine-specific restriction enzyme (PvuRts1I) modulated by glucosylation of DNA. J. Mol. Biol. 242, 45–61. doi: 10.1006/jmbi.1994.1556
Jeffet, J., Margalit, S., Michaeli, Y., and Ebenstein, Y. (2021). Single-molecule optical genome mapping in nanochannels: multidisciplinarity at the nanoscale. Essays Biochem. 65, 51–66. doi: 10.1042/EBC20200021
Kala, S., Cumby, N., Sadowski, P. D., Hyder, B. Z., Kanelis, V., Davidson, A. R., et al. (2014). HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. U S A. 111, 6022–6027. doi: 10.1073/pnas.1320952111
Kong, S., Liu, X., Fu, L., Yu, X., and An, C. (2012). I-PfoP3I: a novel nicking HNH homing endonuclease encoded in the group I intron of the DNA polymerase gene in Phormidium foveolarum phage Pf-WMP3. PLoS One 7:e43738. doi: 10.1371/journal.pone.0043738
Landthaler, M., and Shub, D. A. (2003). The nicking homing endonuclease I-BasI is encoded by a group I intron in the DNA polymerase gene of the Bacillus thuringiensis phage Bastille. Nucleic Acids Res. 31, 3071–3077. doi: 10.1093/nar/gkg433
Luo, Y., Gao, Q., and Deonier, R. C. (1995). Boundaries of the nicking region for the F plasmid transfer origin, oriT. Mol. Microbiol. 15, 829–837. doi: 10.1111/j.1365-2958.1995.tb02353.x
McConnell Smith, A., Takeuchi, R., Pellenz, S., Davis, L., Maizels, N., Monnat, R. J. Jr. et al. (2009). Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc. Natl. Acad. Sci. U S A. 106, 5099–5104. doi: 10.1073/pnas.0810588106
Newman, M., Strzelecka, T., Dorner, L. F., Schildkraut, I., and Aggarwal, A. K. (1994). Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature 368, 660–664. doi: 10.1038/368660a0
Newman, M., Strzelecka, T., Dorner, L. F., Schildkraut, I., and Aggarwal, A. K. (1995). Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science 269, 656–663. doi: 10.1126/science.7624794
Nie, S., Roth, R. B., Stiles, J., Mikhlina, A., Lu, X., Tang, Y. W., et al. (2014). Evaluation of Alere i Influenza A&B for rapid detection of influenza viruses A and B. J. Clin. Microbiol. 52, 3339–3344.
Pingoud, A., Wilson, G. G., and Wende, W. (2016). Type II restriction endonucleases - a historical perspective and more. Nucleic Acids Res. 44:8011.
Ponnaluri, V. K. C., Zhang, G., Esteve, P. O., Spracklin, G., Sian, S., Xu, S. Y., et al. (2017). NicE-seq: high resolution open chromatin profiling. Genome. Biol. 18:122. doi: 10.1186/s13059-017-1247-6
Qian, C., Wang, R., Wu, H., Ji, F., and Wu, J. (2019). Nicking enzyme-assisted amplification (NEAA) technology and its applications: a review. Anal. Chim. Acta 1050, 1–15. doi: 10.1016/j.aca.2018.10.054
Qian, J., Ferguson, T. M., Shinde, D. N., Ramirez-Borrero, A. J., Hintze, A., Adami, C., et al. (2012). Sequence dependence of isothermal DNA amplification via EXPAR. Nucleic Acids Res. 40:e87. doi: 10.1093/nar/gks230
Roberts, R. J., Belfort, M., Bestor, T., Bhagwat, A. S., Bickle, T. A., Bitinaite, J., et al. (2003). A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31, 1805–1812. doi: 10.1093/nar/gkg274
Too, P. H., Zhu, Z., Chan, S. H., and Xu, S. Y. (2010). Engineering Nt.BtsCI and Nb.BtsCI nicking enzymes and applications in generating long overhangs. Nucleic Acids Res. 38, 1294–1303. doi: 10.1093/nar/gkp1092
Van Ness, J., Van Ness, L. K., and Galas, D. J. (2003). Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. U S A. 100, 4504–4509. doi: 10.1073/pnas.0730811100
Vipond, I. B., Baldwin, G. S., and Halford, S. E. (1995). Divalent metal ions at the active sites of the EcoRV and EcoRI restriction endonucleases. Biochemistry 34, 697–704. doi: 10.1021/bi00002a037
Walker, G. T., Nadeau, J. G., Spears, P. A., Schram, J. L., Nycz, C. M., and Shank, D. D. (1994). Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res. 22, 2670–2677. doi: 10.1093/nar/22.13.2670
Xu, S., and Schildkraut, I. (1993). Protecting recognition sequences on DNA by a cleavage-deficient restriction endonuclease. BioTechniques 15, 310–315.
Xu, S. Y. (2015). Sequence-specific DNA nicking endonucleases. Biomol. Concepts 6, 253–267. doi: 10.1515/bmc-2015-0016
Xu, S. Y., and Gupta, Y. K. (2013). Natural zinc ribbon HNH endonucleases and engineered zinc finger nicking endonuclease. Nucleic Acids Res. 41, 378–390. doi: 10.1093/nar/gks1043
Xu, S. Y., Zhu, Z., Zhang, P., Chan, S. H., Samuelson, J. C., Xiao, J., et al. (2007). Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res. 35, 4608–4618. doi: 10.1093/nar/gkm481
Xu, S.-y, and Schildkraut, I. (1991). Isolation of BamHI variants with reduced cleavage activities. J. Biol. Chem. 266, 4425–4429.
Xu, Y., Lunnen, K. D., and Kong, H. (2001). Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl. Acad. Sci. U.S.A. 98, 12990–12995. doi: 10.1073/pnas.241215698
Yonezawa, A., and Sugiura, Y. (1994). DNA binding mode of class-IIS restriction endonuclease FokI revealed by DNA footprinting analysis. Biochim. Biophys. Acta 1219, 369–379. doi: 10.1016/0167-4781(94)90061-2
Zhang, L., Xu, D., Huang, Y., Zhu, X., Rui, M., Wan, T., et al. (2017). Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci. Rep. 7:42542. doi: 10.1038/srep42542
Zhang, P., Too, P. H., Samuelson, J. C., Chan, S. H., Vincze, T., Doucette, S., et al. (2010). Engineering BspQI nicking enzymes and application of N.BspQI in DNA labeling and production of single-strand DNA. Protein Expr. Purif. 69, 226–234. doi: 10.1016/j.pep.2009.09.003
Keywords: BamHI catalytic mutant, phage Gamma HNH nicking domain, infrequent nicking enzyme, protein engineering, DNA nicking assay
Citation: Xu S-y (2022) Engineering Infrequent DNA Nicking Endonuclease by Fusion of a BamHI Cleavage-Deficient Mutant and a DNA Nicking Domain. Front. Microbiol. 12:787073. doi: 10.3389/fmicb.2021.787073
Received: 30 September 2021; Accepted: 17 December 2021;
Published: 01 February 2022.
Edited by:
Howard Junca, RG Microbial Ecology: Metabolism, Genomics & Evolution, Microbiomas Foundation, ColombiaCopyright © 2022 Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang-yong Xu, eHVzQG5lYi5jb20=
 Shuang-yong Xu
Shuang-yong Xu