- 1Root Biology Center, Fujian Agriculture and Forestry University, Fuzhou, China
- 2National Engineering Research Center of Sugarcane, Fujian Agriculture and Forestry University, Fuzhou, China
Plant microbiota are of great importance for host nutrition and health. As a C4 plant species with a high carbon fixation capacity, sugarcane also associates with beneficial microbes, though mechanisms underlying sugarcane root-associated community development remain unclear. Here, we identify microbes that are specifically enriched around sugarcane roots and report results of functional testing of potentially beneficial microbes propagating with sugarcane plants. First, we analyzed recruitment of microbes through analysis of 16S rDNA enrichment in greenhouse cultured sugarcane seedlings growing in field soil. Then, plant-associated microbes were isolated and assayed for beneficial activity, first in greenhouse experiments, followed by field trials for selected microbial strains. The promising beneficial microbe SRB-109, which quickly colonized both roots and shoots of sugarcane plants, significantly promoted sugarcane growth in field trials, nitrogen and potassium acquisition increasing by 35.68 and 28.35%, respectively. Taken together, this report demonstrates successful identification and utilization of beneficial plant-associated microbes in sugarcane production. Further development might facilitate incorporation of such growth-promoting microbial applications in large-scale sugarcane production, which may not only increase yields but also reduce fertilizer costs and runoff.
Introduction
Plant-associated microbes colonize organs throughout host plants, with distinctive microbial communities forming in the different niches plants present, and the collection of organisms forming a holobiont (Tringe et al., 2005; Hassani et al., 2018). Within this intimate set of associations, plants provide microbes with photosynthates in exchange for a variety of beneficial features from microbes that confer improvements in host fitness across diverse environments (Dong et al., 2018). The composition of host associated microbial communities may act as an important determinant of plant health and yield through impacts on nitrogen fixation, phosphorus solubilization, 3-indoleacetic acid (IAA) production, root growth, and nutrient acquisition efficiency (Beckers et al., 2017; Wang et al., 2021). Recruitment and enrichment of the beneficial members of the plant microbial community are in turn determined by the available pool of soil microbiota, the host plant genotypes, and host plant nutrient status (Mendes et al., 2011; Zhong et al., 2019).
Sugarcane accounts for 90% of the sugar production in China (Li and Yang, 2015), and the byproducts can also be used for biomass energy production and animal husbandry (de Almeida et al., 2018; Vieira et al., 2020). Sugarcane is also a C4 crop with high photosynthetic efficiency and rapid growth, which requires large amounts of mineral nutrients, especially nitrogen, to support growth and coordinate nutrient homeostasis with carbon fixation capacity (Gopalasundaram et al., 2012; Carvalho et al., 2014; de Oliveira et al., 2016). This has led to excessive nitrogen applications in sugarcane production (Boddey et al., 2003). A tradeoff of excessive nitrogen fertilization is serious environment impacts, including soil acidification, water eutrophication, and air pollution (Guo et al., 2010; Le et al., 2010; Liu et al., 2013). Hence, increasing sugarcane production while simultaneously protecting agroecosystems through more efficient nutrient acquisition and decreased fertilization are interconnected practical objectives for improving sugarcane production (Rosenblueth et al., 2018).
Although sugarcane plants need large amounts of nitrogen during vegetative growth, up to 70% of the total nitrogen required for its growth may be acquired through associated nitrogen fixation (Urquiaga et al., 1992). Multiple years of field trials in Brazil sugarcane production systems have demonstrated that associated nitrogen fixation can yield up to 40 kg/ha/y of nitrogen in these fields (Urquiaga et al., 2012). Hence, nitrogen fertilizer applications for sugarcane production in Brazil are significantly lower than in other countries due to the nitrogen fixation associated with Brazilian sugarcane (Boddey et al., 2003; Urquiaga et al., 2012). To date, nitrogen fixing bacteria isolated from sugarcane include Beijerinckia spp., Azospirillum spp., Herbaspirillum spp., Gluconacetobacter spp., Enterobacter spp., Burkholderia spp., Klebsiella spp., and so on (Döbereiner and Day, 1976; Boddey and Döbereiner, 1995; Baldani et al., 1997; Oliveira et al., 2009; Mehnaz, 2013; Carvalho et al., 2014; Lin et al., 2015; Muthukumarasamy et al., 2017; Guo et al., 2020). Beyond nitrogen fixation, plant-associated bacteria may also provide numerous other benefits for host plants (Oliveira et al., 2002; Saravanan et al., 2007; Ahmed and Holmström, 2014; Carvalho et al., 2014).
Over the last two decades, while knowledge of associated nitrogen fixation with sugarcane has greatly improved, most of this work focused on specific nitrogen fixing bacteria and lacked a systematic investigation of sugarcane associated communities (da Silva et al., 2018; Singh et al., 2019). Recently, with the development of high throughput sequencing technology, the structure and activities of sugarcane microbiome components have been gradually revealed (Bertalan et al., 2009; Pedrosa et al., 2011; Sant’Anna et al., 2011; Zhu et al., 2012; de Souza et al., 2016; Yeoh et al., 2016; da Silva et al., 2018; Dong et al., 2018; Schwab et al., 2018). In this study, we decipher the composition of a bacterial community in the root and rhizosphere of sugarcane using 16S rDNA sequencing. Then, potentially beneficial microbes were isolated from sugarcane roots and evaluated for plant growth promotion in both greenhouse pot experiments and a field trial. The results provide significant insights into interactions between sugarcane and root microbiota that may be and further harnessed to utilize beneficial microbes in sugarcane production.
Materials and Methods
Sugarcane Growth and Cultivation
In order to investigate microbes specifically recruited from soil and enriched in the roots of commercial sugarcane variety ROC22, seedlings were propagated to the four-leaf stage in MS medium prior to transferring to rooting medium for another 2 weeks (Supplementary Figure 1). Transplanted sugarcane seedlings were then acclimatized for another 2 weeks to growth in liquid plant nutrition solution (Li et al., 2015) prior to transplanting into soils collected from a sugarcane farm managed by the Fujian Agriculture and Forestry University. Before planting, soils were filtered to remove larger residues and mixed well with equal volumes of sterile vermiculite, which was heated at 121°C for 40 min before use. Sugarcane seedlings were planted 1 seedling per pot (10 * 10 * 15 cm) in the soil mix described above. A total of 16 pots of sugarcane seedlings were reared in a growth chamber (day/night: 14 h/10 h, 26°C/24°C) under 37.5 μE/m2/s of daylight light intensity for 3 months before harvesting samples. During plant growth, liquid plant nutrient solution was supplied according to plant needs.
Rhizosphere and Root-Associated Microorganism Sampling
To sample the rhizosphere and root-associated microorganisms, sugarcane plants were removed from pots, with loosely attached soil being removed from roots prior to collecting rhizosphere soil firmly attached to the roots. Rhizosphere soils were collected through washing in 100 mL sterile phosphate buffer saline (PBS) solution and centrifugation at 12,000 rpm for 15 min (Bulgarelli et al., 2012, 2015; Zhong et al., 2019). After removing rhizosphere soil, roots were further treated with sonication for 20 min to remove microbes from the root surface (Lundberg et al., 2012), which were collected in three rinses of sterile water (Supplementary Figure 2). Each biological replicate of rhizosphere soil and sugarane root sample was collected from four independent sugarcane plants. In total, four biological replicates were collected for each rhizosphere soil and root.
DNA Extraction, Sequencing, and Analysis
Total DNA was extracted from rhizosphere soil and root samples using the PowerSoil DNA extraction kit (Mobio Laboratories, Carlsbad, CA, United States) according to the kit instructions. The DNA concentration was determined using the NanoDrop 1000 (Thermo Scientific, Waltham, EUA). Then, total DNA was subjected to polymerase chain reaction (PCR) (Majorbio Bio-pharm Technology Co., Ltd, Shanghai, China) prior to sequencing PCR fragments using an Illumina Miseq PE300 platform (Illumina, San Diego, CA, United States). Specifically, total DNA was used as PCR templates, and 16S amplicon libraries were generated using the PCR primers 799F (5′-AACMGGATTAGATACCCKG-3′), 1193R (5′-ACGTCATCCCCACCTTCC-3′), and 1392R (5′-ACGGGCGGTGTGTRC-3′), which span approximately 400 bp of the V5–V7 hypervariable regions of the prokaryotic 16S rDNA gene (Bulgarelli et al., 2012, 2015; Lundberg et al., 2012).
The QIIME2 software platform was used for bioinformatics analysis of raw data derived from the Illumina Miseq platform (Bolyen et al., 2019). Specifically, q2-demux and q2-cutadapt trim-pairs were used to remove barcodes and linkers (Martin, 2011) prior to merging paired-end reads in vsearch (Rognes et al., 2016). The quality-filter and deblur plugins in QIIME2 were used to carry out quality control (Quality Score > 25) and denoising, respectively (Amir et al., 2017). OTUs were clustered based on a 97% similarity threshold determined by the q2-feature-classifier using the SILIVA database (Yilmaz et al., 2014). The diversity plugin of QIIME2 was used to calculate the alpha diversity, weight-unifrac distance matrix, and Bray–Curtis distance matrix. The Vegan package (version: 2.5.6) in R (version: 4.0) (R core team) was used to perform ANOSIM (analysis of similarity) analysis. The calculation of R and P-values and the comparison of differences between groups were carried out by permutation testing with permutational anova, with the number of permutations set to 999. Two-sided Welch’s t-tests with Benjaminii–Hochberg FDR corrections were conducted in the STAMP software (version: 2.1.3) to identify significant differences between sugarcane associated microbial groups (Parks et al., 2014). The Kruskal--Wallis rank-sum test, with an alpha value of 0.05 and a threshold value of 4.8, was used to test for significant differences between microbial groups using an online LEfSe (linear discriminant analysis effect size) program1.
Phylogenetic Tree Construction
Sequences of 16S rDNA from isolates and similar sequences of known bacteria in GenBank database with NCBI BLAST program were downloaded and used for phylogenetic trees analysis in MEGA 7.0 (Kumar et al., 2016). ClustalX2 was used for multiple alignments, and the neighbor-joining method in MEGA 7.0 was employed for genetic tree construction (Saitou and Nei, 1987) with 1000 replicates of bootstrap testing (Felsenstein, 1985). Finally, phylogenetic trees were visualized in iTol V43 (Letunic and Bork, 2019) or GraPhlAn (Asnicar et al., 2015).
Sugarcane Root-Associated Microbe Isolation and Identification
Roots of sugarcane plants grown in the pot cultures described above were used for root-associated microbe isolation. After harvesting sugarcane plants, root samples were collected after first shaking off loosely attached bulk soil, followed by removal of rhizosphere soil firmly attached to the roots through washing with 50 mL of sterile PBS solution for 15 min and centrifugation at 7000 rpm for 8 min (Bulgarelli et al., 2012, 2015). Then, 5 g of root tissues was homogenized in 15 mL of sterile PBS using a mortar and pestle in a laminar flow hood. Plant debris was removed by filtration, and the homogenized solution was considered as the collection of root-associated microbes. Homogenized solutions were diluted 1000-fold with sterile PBS. Finally, 100 μL of diluted microbial suspension was plated on a panel of four microbial isolation culture media and incubated at 28°C (Bai et al., 2015; Wang et al., 2021). The panel of isolation media consisted of two nitrogen free culture media, SNX (Hu et al., 2017) and JNFb (Baldani et al., 1992), and two other culture media containing nitrogen, TSB and M715 (Bai et al., 2015). Single colonies were picked out and streaked to new plates for further purification. Then, single colonies were picked once again from these solid medium plates. All collected isolates from root-associated compartments were subsequently stored in 40% glycerol at –80°C.
To identify potentially nitrogen fixing isolates, PCR assays were employed to identify the N2 fixing marker gene (NifH) using the primer pair NifH-F: AAAGGYGGWATCGGYAARTCCACCAC and NifH-R: TTGTTSGCSGCRTACATSGCCATC AT (RöSch et al., 2002). For the determination of potential phosphate solubilizing capacity, isolates were cultured on Pikovskaya agar plates containing Ca3(PO4)2 or phytin as the sole Pi source for 3 days at 28°C. The appearance of a transparent zone around a bacterial colony indicated the phosphate solubilizing capacity. The IAA biosynthesis capacity was measured in a color development assay (Littlewood and Bennett, 2003; Qin et al., 2011). Briefly, isolates were propagated in flasks containing liquid growth media for 3 days at 28°C, followed by centrifugation of 1 mL samples. Cell pellets were washed twice with 1 mL PBS and re-suspended to 107 cells/mL prior to mixing 1 mL of cell suspension with 10 mL of liquid medium containing 100 mol/mL of tryptophan. After incubating for 3 days, 50 μL of supernatant was collected and mixed with 50 μL of Salkowski buffer (4.5 g/L FeCl3, 10.8 mol/L H2SO4). When the solution turns red, it has the ability to synthesize IAA. The colony morphology of SRB-109 was recorded using stereoscopic microscopy.
Application of Potential Beneficial Microbes in the Fields
For the field application of potentially beneficial microbes. Strains stored at –80°C were melted on ice, streaked on isolation culture medium, and then incubated at 28°C for 5–7 days. Single colonies were then picked off and inoculated in liquid culture medium at 28°C and shaken at 200 rpm until the concentration of bacteria liquid was 1 × 108 CFU/mL. Microbial cells were collected through centrifugation at 7000 rpm for 10 min. After discarding the supernatant, microbial cells were washed three times with low N (200 μM) nutrient solution and finally suspended in low N nutrient solution for later to the concentration of bacteria solution is 5 × 107 CFU/mL.
For sugarcane plant inoculations, seedlings of ROC22 sugarcane were collected at the six-leaf stage from tissue cultures and then immersed in prepared microbial solutions (5 × 107 CFU/mL) for 12 h prior to transplanting into pots filled with sterile substrate and vermiculite in equal proportions. Seedlings were co-cultured with microbial isolates in a growth chamber for 2 weeks, with pots supplied with 20 mL of low N (200 μM) nutrient solution each day (Li et al., 2018). A total of seven pots, with each containing one seedling, were cultivated for each microbial isolate. Control pots contained seedlings that were not inoculated with any microbial isolates. After 2 weeks in a growth chamber, sugarcane seedlings were transplanted to the field with 50 cm spacing between plants for further growth; soil nutrient content and pH of field experimentation are shown in Supplementary Table 1. After cultivation for 120 days in the field, sugarcane plants were harvested for assessment of plant growth promotion.
Colonization Pattern of SRB-109 on Sugarcane Plants
SRB-109, an isolate from sugarcane roots exhibiting significant growth promotion effects, was selected for further study. To visualize the colonization patterns of SRB-109 on sugarcane plants, SRB-109 was labeled for GUS or red fluorescent protein (RFP) staining with pMG103-NPTII-GUS or pMG103-NPTII-RFP modified from pMG103-NPTII-GFP (Inui et al., 2000) using Sph I and EcoR I restriction endonuclease sites. Bacteria were labeled by electroporation (Liu S. et al., 2020). Positively transformed bacterial colonies were screened through kanamycin resistance and further identified using GUS staining.
Positively identified GUS labeled bacteria were cultured at 28°C in JNFb liquid medium supplied with 50 μg/mL of kanamycin and shaken at 200 rpm until the concentration of bacteria liquid was 1 × 108 CFU/mL. Bacterial inoculants were prepared through centrifugation at 7000 rpm for 8 min, prior to resuspension in low nitrogen liquid solution to 5 × 107 CFU/mL. Then, 5 mL of prepared inoculum (GUS labeled SRB-109) was added to the liquid nutrient solution of five acclimatized sugarcane plants. After 3 days of co-culturing with GUS labeled microbes, the colonization of SRB-109 throughout various organs of sugarcane plants such as leaves, roots, and whole plants was visualized with GUS staining according to the methods of Li et al. (2015), with the GUS signal being recorded using a stereo microscope (Zeiss Axio Zoom. V16).
Plant Growth Promoting Genes Identification With Polymerase Chain Reaction
To further confirm the plant growth promoting genes in SRB-109, the genome DNA of SRB-109 was used as a template, the primers specific target to the nifH (nitrogenase gene) (RöSch et al., 2002), ACC (1-aminocyclopropane-1-carboxylic acid deaminase) (Blaha et al., 2006), and phoD (Alkaline phosphatase D) (Sakurai et al., 2008).
Results
Structure and Composition of Bacterial Communities Colonizing Root-Associated Compartments of Sugarcane Plants
16S rDNA sequencing spanning the V5–V7 regions was employed to investigate the composition of bacterial communities in the rhizospheres and roots of sugarcane plants. A total of 1,149,659 sequences were thusly obtained. After joining paired-end reads, sequences were subjected to quality control and denoising procedures. This yielded 127,810 high-quality 355 bp length reads. Operational taxonomic unit grouping was obtained through clustering of high-quality sequences at the 97% similarity threshold. Rarefaction curves of observed OTU plateaued after 60% sampling (Supplementary Figure 3), suggesting that the sequencing depth for all the samples was enough to cover most of the bacterial in the rhizosphere and root compartments. Bacterial community richness and evenness were significantly higher in the rhizosphere than in the roots, as reflected by the Chao 1 index, the Shannon index, observed numbers of OTU, and the Pielou evenness index (Supplementary Figure 4). This result suggests host involvement in determining root bacterial community members. Further structure analysis based on Bray_Curtis and Weighted unifrac tests all showed that bacterial communities in root compartments can be clearly separated from those in the rhizosphere compartment (Figures 1A,B). An ANOSIM analysis further showed that bacterial communities in the root compartment were significantly different from those in the rhizosphere (Figures 1A,B). All of these results reinforce the notion that roots of sugarcane actively select associated microbial communities.
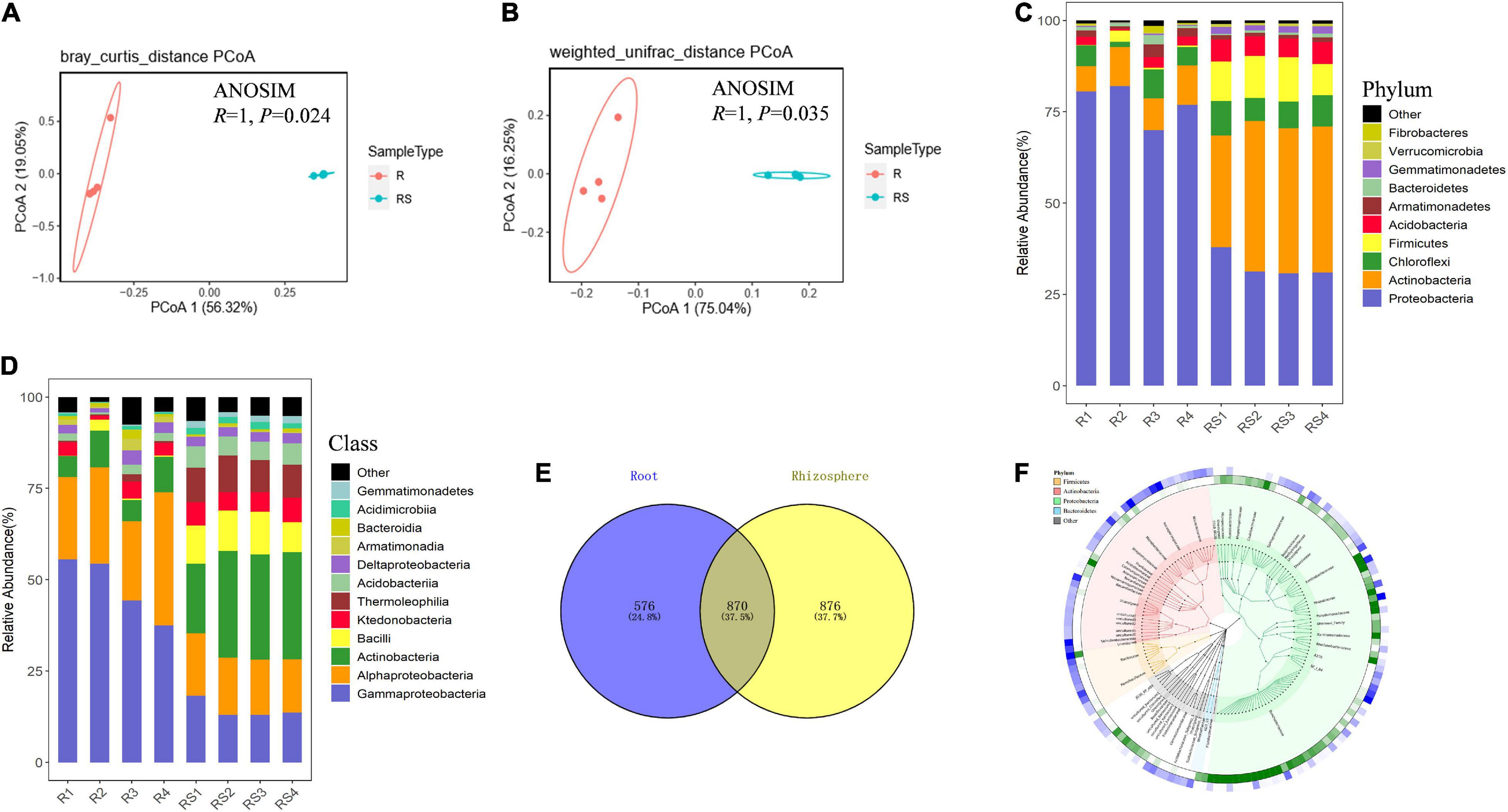
Figure 1. Bacterial communities in the root-associated compartments of sugarcane. Samples were divided into rhizosphere and root samples according to the compartment of origin. Bacterial community composition was determined using 16S rDNA sequencing. Principal coordinates analysis (PCoA) was performed using Bray–Curtis distance (A) and weighted-unifrac distance (B) based on the OTU table. Analysis of similarity (ANOSIM) was performed based on the OTU tables of R and RS samples to calculate differences. In permutation tests, the number of permutations was 999. The composition of microorganisms was analyzed in the rhizosphere at the phylum level (C) and class level (D). R, sugarcane root; RS, sugarcane rhizosphere; OTU, operational taxanomic unit. (E) Venn diagram of microbes in the rhizosphere and root compartments of sugarcane. (F) Cladogram of top 150 microbes (mean relative abundance) based on taxonomy. OTUs relative abundance of two compartments are shown in the outermost rings of the green and red heat map (root samples as green and rhizosphere samples as blue).
Microbes Enriched by Sugarcane
To further investigate which microbes were specifically selected by sugarcane roots. Composition analysis at the phylum taxonomic level showed that Proteobacteria was the dominant bacteria in roots, with members of this group accounting for about 77.31% of the relative abundance of total bacteria in sugarcane roots (Figure 1C). In all, Proteobacteria (32.68% of rhizosphere bacteria; 77.31% of root bacterial), Actinobacteria (37.91%; 9.31%), Chloroflexi (7.88%; 5.02%), and Firmicutes (10.77%; 0.97%) were the dominant bacteria in the rhizosphere and root compartments of sugarcane plants. At the class taxonomic level, Gammaproteobacteria (47.91%; 14.45%) and Alphaproteobacteria (26.79%; 15.63%) were the dominant bacteria in the root and rhizosphere compartments, respectively (Figures 1C,D). In addition, there were 870 OTUs existing in both the rhizosphere and root compartments, while 576 OTU were found exclusively in the root compartment (Figure 1E). The phylogenetic relationship of microbes detected through high throughput sequencing suggests that the sum of the sugarcane associated microbial community originated from a variety of phyla (Figure 1F).
To further investigate which microbes were significantly enriched by sugarcane, LEfSe analysis was employed. Results showed that Gammaproteobacteria, Alphaproteobacteria, Burkholderiaceae, Rhizobials, and Burkholderia were all significantly enriched in the root compartment, while Actinobacteria, Intrasporangiaceae, Firmicutes, and Bacillus were enriched biomarkers in the rhizosphere compartment (Figure 2). Meanwhile, STAMP results were similar to LEfSe results (Supplementary Figure 5). Taken together, the observations herein show that Rhizobials and Burkholderia were significantly enriched in the root compartment and Bacillus taxa were mainly enriched in the rhizosphere.
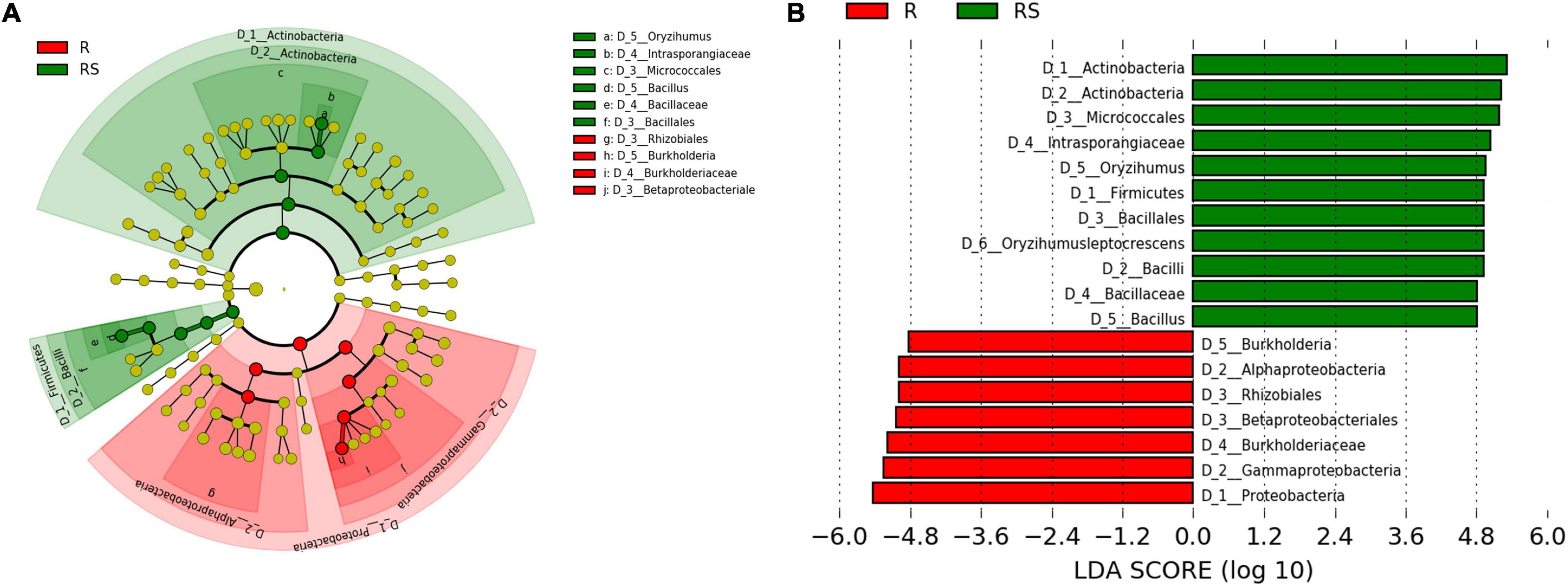
Figure 2. Comparison of bacteria between the rhizosphere and root compartments. (A) Phylogenetic dendrogram of biomarkers in the R and RS sugarcane groups. Circles from inside to outside indicate bacterial taxonomic levels from phylum to genus. Yellow dots represent bacteria not significantly varying in abundance among treatments. Biomarker bacteria are colored according to their corresponding class colors on the right. (B) LDA scores of biomarker bacteria for each combination of sugarcane sites. LDA scores are shown as horizontal bars for the biomarker bacteria with an LDA score > 4.8 as listed on the left, Kruskal–Wallis rank sum test, P < 0.05. LDA, linear discriminant analysis.
Isolation and Identification of Beneficial Microbes in Sugarcane
To isolate microbes enriched within the root compartment of sugarcane plants, four bacterial culture media were employed, including two nitrogen-free and two of nitrogen-rich media. Isolation and purification procedures yielded a total of 519 isolates from sugarcane roots. These strains were tested for potential nitrogen fixation through PCR amplification of nifH genes. Microbes with positive nifH gene PCR results were further screened for phosphate-solubilizing capacity and phytohormone IAA production as described in the methods. In these tests, 92 isolates were identified with potential nitrogen fixing capabilities. Among these potential nitrogen fixing members of the sugarcane root flora, 52 returned positive results in one or more of the three other functional assays. Of these, 17 isolates were able to produce IAA, 43 exhibited the capacity of solubilizing inorganic phosphate, and 43 could solubilize organic phosphate. Forty-one potential nitrogen fixing isolates returned positive results in at least two of the IAA production and phosphate solubilizing assays. Ten isolates yielded positive results in all four functional assays (Figure 3A and Table 1). All 92 of the potentially beneficial isolates were subjected to taxonomic assignments and phylogenetic analysis based on 16S rDNA sequences. These results showed that the 92 potentially beneficial strains were Actinobacteria (14.13%), Bacilli (28.26%), Alphaproteobacteria (10.86%), Betaproteobacteria (6.52%), Gammaproteobacteria (38.06%), and Sphingobacteriia (2.17%) (Figure 3B and Supplementary Table 2). The taxonomic grouping of these isolates was mostly consistent with the distribution of all dominant bacterial taxa in the root associated compartments of sugarcane plants (Figure 1F).
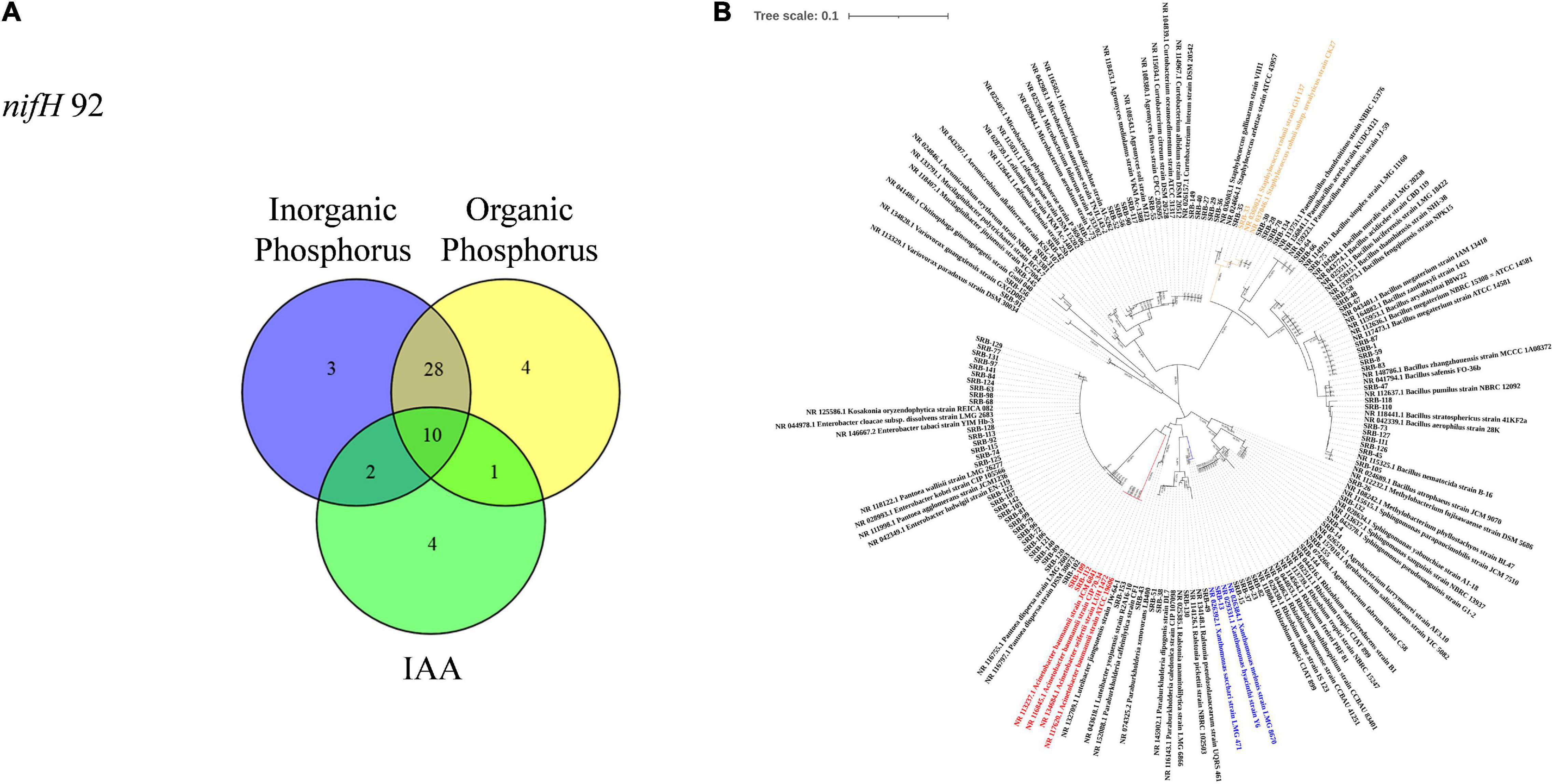
Figure 3. Isolation and identification of microbes from roots of sugarcane. (A) Distribution of potentially beneficial functions among isolated root-associated microbes represented in a Venn diagram. (B) Phylogenetic tree was constructed using known sequences exhibited similar sequence similarity in GenBank database with NCBI BLAST program. Multiple sequence alignment was done using ClustalX 1.8 software package (http://wwwigbmc.ustrasbg.fr/BioInfo/clustalx), tree was constructed by neighbor-joining method using MEGA 7.0 (Kumar et al., 2016).
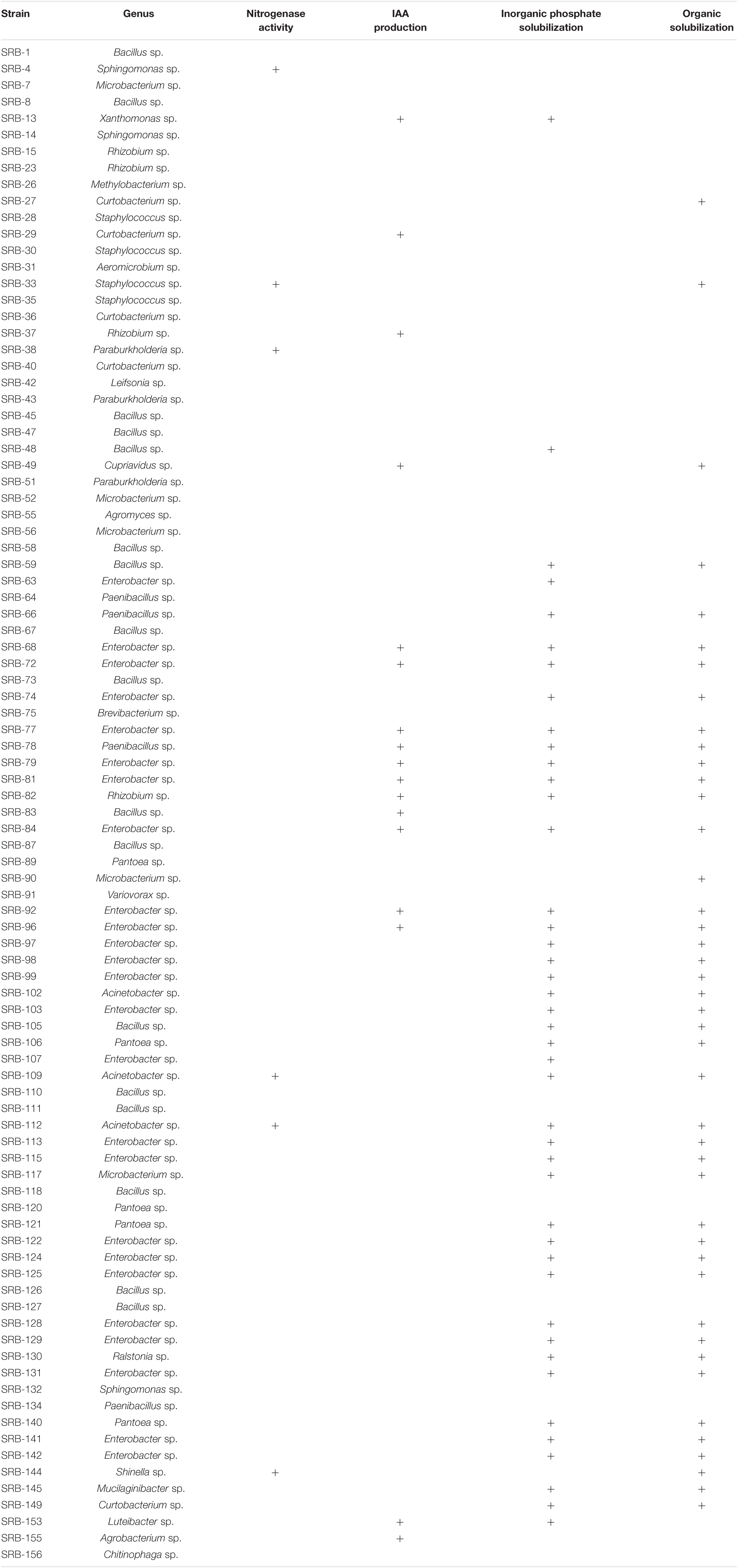
Table 1. 16S rDNA sequence and functional characteristics of nitrogen-fixing bacteria in sugarcane roots.
After conducting greenhouse assays of plant-microbe interactions, four isolates (SRB-13, SRB-33, SRB-109, SRB-112) displaying obvious growth promotion capacities were selected as candidate strains for further functional validation in a field experiment (Supplementary Figure 6). Phylogenetic analysis showed that the four selected strains were a Xanthomonas sp. (SRB-13), a Staphylococcus sp. (SRB-33), and 2 Acinetobacter sp. (SRB-109 and SRB-112) (Figure 3B). Relative to control treated plots, isolate SRB-109 significantly increased plant height by 27.6%, the leaf SPAD value by 11.7%, and the number of tillers by 6 ± 1. The only other isolate to produce significant improvements in sugarcane traits in these trials was SRB-33, which significantly increased plant height by 27.6% and the number of tillers by 7 ± 1 (Supplementary Figures 8A–C). No isolate significantly impacted dry weight of individual tillers relative to control tillers (Figures 4A,B). Nutrient acquisition relative to control plants was significantly enhanced with SRB-109 treatment by 35.7% for nitrogen and by 28.4% for potassium, but not for phosphorus with this isolate, nor for any nutrients with any of the other three tested isolates (Figures 4C–E). Taken together, these results suggest that SRB-109 may be the most promising isolate identified for promoting sugarcane growth and nutrient acquisition.
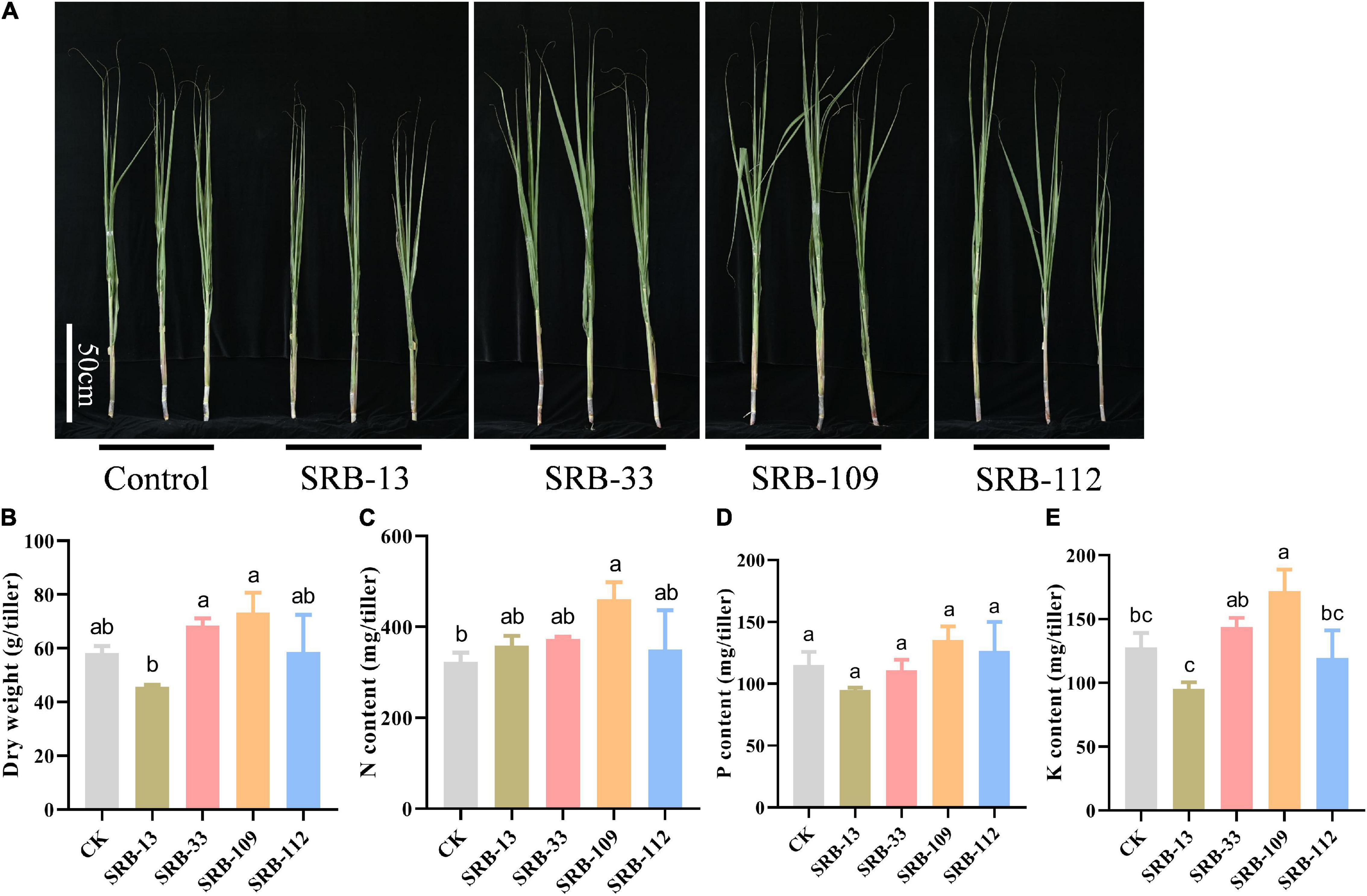
Figure 4. Results of applying potentially beneficial microbes in a field experiment. (A) Growth performance of single stem of sugarcane plants in the field trial 120 days after inoculation with microbial isolates, bar = 50 cm. (B) Biomass of single stem of sugarcane under differnet treatment conditions. Total nitrogen content (C), phosphate content (D), and potassium content (E) of sugarcane plants under different microbial applications under field conditions. Different letters indicate significant differences among different treatments in Duncan’s multiple range comparison test.
Colonization Patterns of SRB-109 on Sugarcane Plants
Since SRB-109 exhibited the most promising impacts on sugarcane growth-promotion capacity among tested isolates, this isolate was, therefore, selected for further study of interactions with sugarcane plants. Colony morphology showed SRB-109 with light yellow color, shiny smooth surface, and clear colony edge (Supplementary Figure 7A). And the plant growth promoting genes such as phoD, ACC deaminase and nifH (Supplementary Figure 7B). Taxonomic identification using 16S rDNA showed that SRB-109 is an Acinetobacter sp. (Figure 3B). This isolate not only possesses a NifH gene but also exhibits the capacity to solubilize both inorganic and organic forms of normally insoluble phosphate (Table 1). Further investigation of SRB-109 colonization patterns using the labeled GUS and RFP reporter gene (Supplementary Figure 9) showed that 3 days after SRB-109/pMG103-NPTII-GUS inoculation, an obvious blue signal was detectable on the different tissues of roots, including root tips and root hairs, suggesting that SRB-109 could quickly colonize roots of sugarcane (Figures 5A–C), which was further confirmed by RFP labeled SRB-109 and visualized with confocal microscopy (Figures 5K,L). At the same time, the blue signal of GUS staining was also observed in sugarcane leaves, which suggests that SRB-109 may also colonize the aboveground parts of sugarcane plants (Figures 5D–J).
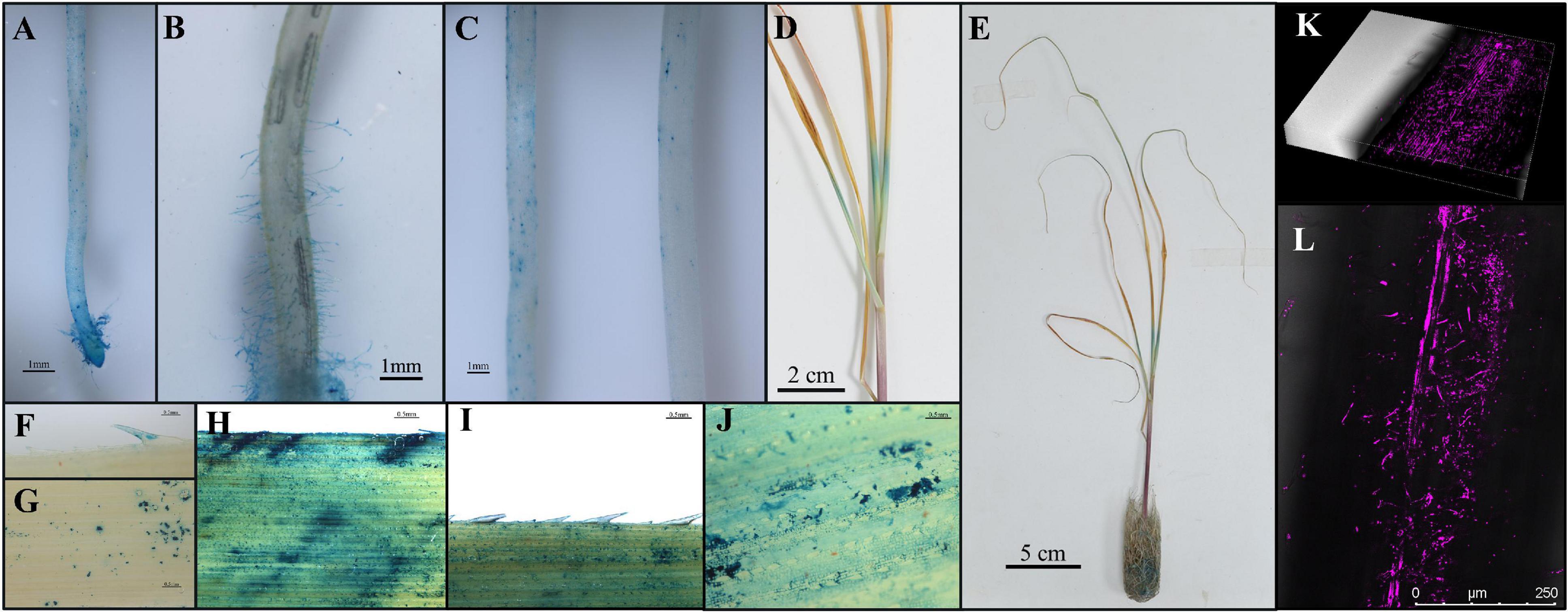
Figure 5. Colonization pattern analysis of SRB-109 on sugarcane. GUS labeled SRB-109 was inoculated and co-cultured with sugarcane for 3 days in the growth chamber. Then, different organs of sugarcane plants were harvested for GUS staining. The blue color indicates the colonization of SRB-109 on different organs, including roots (A–C) and leaves (D–G), along with GUS signals detected near inoculation wounds on leaves (H–J). Results of colonization patterns of SRB-109 (RFP labeled) on the roots of sugarcane visualized with confocal microscopy (K,L).
Discussion
Plant-associated microbiota are essential for proper host plant growth and health (Niu et al., 2017). During the long history of co-evolution between host plants and their particular suites of microbiota, host plants have evolved several strategies to recruit specific microbes from surrounding soil environments (Mendes et al., 2011; Prashar et al., 2014). These strategies include changing rhizosphere soil pH and texture, production of antibiotic and Quorum-sensing mimicry substances, and specific signaling based on the composition of individual root exudates and cell debris (Marschner et al., 1986; Dennis et al., 2010). Overall, plant associated microbial community composition and activities result from the complex interactions between soil types, geographic conditions, nutrient status, and genotypes of host plants and the available microbial pool (Zhang et al., 2019).
In this study, Proteobacteria, Chloroflexi, Actinobacteria, and Firmicutes were the dominant bacteria in the rhizosphere of sugarcane plants at the phylum taxonomic level (Figure 1C). Microbes in these phyla have previously been reported as the core microbiota of sugarcane plants along with Bacteroidetes, Spirochaetae, and Verrucomicrobia (Yeoh et al., 2016). The latter three listed taxa from the previous study were only found in low abundance in this study (Figure 1C), which may due to variation in the soil types and sugarcane genotypes between that published work and this report (Yeoh et al., 2016; Zhang et al., 2019; Liu W. et al., 2020). Consistent with previous studies, Actinobacteria was much more abundant than other taxa detected in the root compartments of sugarcane (Figure 2; Dong et al., 2018; Gao et al., 2019). At the genus level, Leptothrix sp. were the most abundant microbe in sugarcane roots, though its functions there remain mysterious due to a lack of reports on its functions or interactions with plants. In addition, Burkholderia sp. and Bradyrhizobium sp. also exhibited higher relative abundances in the sugarcane roots observed herein, with LEfSe analysis showing that these bacteria were significantly enriched in the roots relatvive to the rhizosphere. Burkholderia sp. and Bradyrhizobium sp. have also been detected from sugarcane plants cultivated in Yunnan Province, China (Dong et al., 2018), though they have been rare or absent in sugarcane grown in Brazil and Australia (de Souza et al., 2016; Yeoh et al., 2016; Gao et al., 2019; Liu W. et al., 2020). Previous work has also concluded that Burkholderia sp. and Bradyrhizobium sp. may be common plant growth promoting bacteria (Bernabeu et al., 2015; Cagide et al., 2018). However, these two genera were rarely isoalted in this study (Figure 3B and Supplementary Figure 2), which might be due to the composition of the culture medium used to isolate microbes (Bonnet et al., 2019). These results suggest that geographic location, culture conditions, and genotypic variation among of sugarcane plants coordinately regulate the composition of microbiota associated with the sugarcane hosts.
Pure culturing of microbes is important for accurate determination of specific microbial functions (Singh et al., 2019). Over the course of recent decades, with the development of high throughput next-generation sequencing, amplicon sequencing and metagenomics have been widely applied in microbial ecology investigations, with large numbers of microbes being discovered and assigned predicted functions (Giovannoni et al., 1990). To fully assess these predictions requires studying these microbes in controlled settings, typically in isolated cultures. Soil microbes have been historically perceived as largely unculturable (Woese et al., 1990). However, Bai et al. (2015) have demonstrated that 52–65% of Arabidopsis associated microbes may be cultured using proper culture media. More recent investigation suggests that up to 97.3% of microbes can be cultured, though most of them have not been investigated (Yang and Jia, 2021). Finally, in sugarcane, 56.1–64.5% of associated microbes may be isolated using broad spectrum microbial media (Armanhi et al., 2017). Therefore, culturing might not be as limiting to functionally characterizing microbial communities as previously perceived.
In this study, considering the importance of associated nitrogen fixation for sugarcane growth (Bai et al., 2015; Hu et al., 2017), two nitrogen-free and two nitrogen-rich media were used to culture and isolate microbes from root-associated compartments. In addition, nitrogen-fixing bacteria associated with sugarcane might also promote sugarcane growth even after the loss of nitrogen fixing capacity, which suggests that associated microbes could also promote plant growth independent of nitrogen fixation (Sevilla et al., 2001). Therefore, after the identification of the nitrogen fixation marker gene NifH (Gaby et al., 2018), microbes isolated in this study were also subjected to assays for other potentially beneficial functions relevant to the low phosphorus bioavailability acid soil regions of sugarcane production in South China (Kochian, 2012; Figure 3A). Since not all potentially beneficial microbes actually promote host plant growth, and given the complex interactions between microbes and host plants (Finkel et al., 2017; Wang et al., 2021), isolated microbe–plant interaction assays were necessary in this study to ultimately verify the potentially beneficial functions of isolated microbes. After co-culturing sugarcane seedlings with different selected microbes in greenhouse testing, four isolates were further observed under field conditions. The results of this field trial suggest that SRB-109 exhibited better plant growth-promotion than the other isolates identified in this study (Figure 4). Differences in the performance of the other isolates between greenhouse testing and the field trial might be due to the complex and dynamic field conditions (Finkel et al., 2017). Nevertheless, SRB-109, an apparent Acinetobacter sp. (Supplementary Figure 8), significantly promoted sugarcane growth in both greenhouse and field conditions. Laboratory assays suggested that SRB-109 might assist host plants in nutrient acquisition through solubilizing phosphate, as well as through fixing nitrogen (Table 1). However, whether the nitrogen fixing capacity of SRB-109 higher or lower than the previously identified sugarcane associative nitrogen fixing bacteria, such as Azospirillum spp. Herbaspirillum spp., Gluconacetobacter spp., Enterobacter spp., and Burkholderia spp., it will need further invesstigation and comparision of their nitrogen fixing capacity under the same treament conditions. Previous studies also suggest that Acinetobacter sp. may promote host plant growth through phytohormone production along with solubilizing phosphate (Kang et al., 2009; Rokhbakhsh-Zamin et al., 2011; Ishizawa et al., 2020). Consistently, in our study, the application of SRB-109 significanly enhanced the acquisition of nitrogen and potassium (Figure 4), which might be due to the functions of nitrogen fixation (Table 1). In addition, although phosphate solubilizing capacity in SRB-109 was significant in the lab, application of this isolate in the field did not significantly enhance phosphorus acquisition (Figure 4D). Further observation with GUS staining to determine which tissues SRB-109 colonizes suggests that SRB-109 can colonize sugarcane roots and root hairs, as well as, aboveground compartments. These observations indicate that SRB-109 establishes an intimate relationship with sugarcane that benefits the plants through gains in nutirent acquisition capabilities (Figure 5).
Conclusion
In this study, we used tissue culture of sugarcane seedlings and 16S rDNA sequencing of associated microbiota to systematically decipher the structure and composition of bacterial communities recruited and enriched from soils by sugarcane roots. Isolation of root-associated microbes and screening of the potentially beneficial members allowed for further evaluation of these isolates for the promotion of sugarcane growth in both greenhouse and field experiments. This led to the identification of SRB-109 as a rapid colonizer of both sugarcane roots and shoots that may significantly increase tillering and nitrogen acquisition by sugarcane plants. In this study, we outlined a strategy for functionally studying potentially beneficial plant-associated microbes, and demonstrated effects under field conditions. Building on these results might lead to applications of beneficial plant-associated microbes to decrease fertilizer use and promote the development of sustainable agriculture.
Data Availability Statement
The 16s rDNA sequencing files for all samples used in this study have been deposited in the public database of the National Center for Biotechnology Information (NCBI) under project number BioProject ID: PRJNA748846.
Author Contributions
YZ and HL designed the experiments and managed the projects. ML, RL, YL, CW, WM, LZ, KZ, and XF performed the experiments. XL, YS, and GH provided the tissue culture seedling and suggestion in the experiment and manuscript revision. YZ and ML performed the data analysis. YZ, HL, and ML wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
ML and KZ were supported by the K+S Group scholarship from the International Magnesium Institute. This work was supported by the Natural Science Foundation of Fujian Province (2017J01603).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Z. X. Chen and M. J. Zeng for help during the outbreak of Coronavirus-2019 and help from other members in the Root Biology Center. We thank J. X. Xiang at the Yangzhong field experimental station of Fujian Agriculture and Forestry University for assistance with field trials. We also thank Thomas Walk for the valuable suggestions and comments and F. Tang at the Inner Mongolia Agricultural University for kindly providing the pMG103-NPTII-GFP construct.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.783925/full#supplementary-material
Footnotes
- ^ http://huttenhower.sph.harvard.edu/galaxy/root/index
- ^ http://www.clustal.org/
- ^ https://itol.embl.de/
References
Ahmed, E., and Holmström, S. J. M. (2014). Siderophores in environmental research: roles and applications. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Amir, A., Mcdonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191–16. doi: 10.1128/msystems.00191-16
Armanhi, J. S. L., de Souza, R. S. C., Damasceno, N. B., de Araújo, L. M., Imperial, J., and Arruda, P. A. (2017). A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front. Plant Sci. 8:2191. doi: 10.3389/fpls.2017.02191
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C., and Segata, N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Baldani, J., Caruso, L., Baldani, V. L. D., Goi, S. R., and Döbereiner, J. (1997). Recent advances in BNF with non-legume plants. Soil Biol. Biochem. 29, 911–922.
Baldani, V. L. D., Baldani, J. I., and Olivares, F. L. (1992). Identification and ecology of Herbaspirillum seropedicae and the closely related Pseudomonas rubrisubalbicans. Symbiosis 13, 65–73.
Beckers, B., Op De Beeck, M., Weyens, N., Boerjan, W., and Vangronsveld, J. (2017). Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:25. doi: 10.1186/s40168-017-0241-2
Bernabeu, P. R., Pistorio, M., Torres-Tejerizo, G., Paulina, E. D. L. S., Galar, M. L., Boiardi, J. L., et al. (2015). Colonization and plant growth-promotion of tomato by Burkholderia tropica. Sci. Hortic. 191, 113–120. doi: 10.1016/j.scienta.2015.05.014
Bertalan, M., Albano, R., De Pádua, V., Rouws, L., Rojas, C., Hemerly, A., et al. (2009). Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450. doi: 10.1186/1471-2164-10-450
Blaha, D., Prigent-Combaret, C., Mirza, M. S., and Moënne-Loccoz, Y. (2006). Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 56, 455–470.
Boddey, R. M., and Döbereiner, J. (1995). Nitrogen fixation associated with grasses and cereals: recent progress and perspectives for the future. Fertilizer Research 42, 241–250.
Boddey, R. M., Urquiaga, S., Alves, B. J. R., and Reis, V. (2003). Endophytic nitrogen fixation in sugarcane: present knowledge and future applications. Plant Soil 252, 139–149. doi: 10.1023/a:1024152126541
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bonnet, M., Lagier, J. C., Raoult, D., and Khelaifia, S. (2019). Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes. New Infect. 34:100622. doi: 10.1016/j.nmni.2019.100622
Bulgarelli, D., Garrido-Oter, R., Münch, P. C., Weiman, A., Dröge, J., Pan, Y., et al. (2015). Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403. doi: 10.1016/j.chom.2015.01.011
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren Van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Cagide, C., Riviezzi, B., Minteguiaga, M., Morel, M. A., and Castro-Sowinski, S. (2018). Identification of plant compounds involved in the microbe-plant communication during the coinoculation of soybean with Bradyrhizobium elkanii and Delftia sp. strain JD2. Mol. Plant Microbe Interact. 31, 1192–1199. doi: 10.1094/mpmi-04-18-0080-cr
Carvalho, T. L. G., Balsemao-Pires, E., Saraiva, R. M., Ferreira, P. C. G., and Hemerly, A. S. (2014). Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 65, 5631–5642. doi: 10.1093/jxb/eru319
da Silva, P. R. A., Simões-Araújo, J. L., Vidal, M. S., Cruz, L. M., Souza, E. M., and Baldani, J. I. (2018). Draft genome sequence of Paraburkholderia tropica Ppe8 strain, a sugarcane endophytic diazotrophic bacterium. Braz. J. Microbiol. 49, 210–211. doi: 10.1016/j.bjm.2017.07.005
de Almeida, G. A. P., Ferreira, M. D. A., Silva, J. D. L., Chagas, J. C. C., Véras, A. S. C., De Barros, L. J. A., et al. (2018). Sugarcane bagasse as exclusive roughage for dairy cows in smallholder livestock system. Asian Australas. J. Anim. Sci. 31, 379–385. doi: 10.5713/ajas.17.0205
de Oliveira, R. I., de Medeiros, M. R. F. A., Freire, C. S., Freire, F. J., Neto, D. E. S., and de Oliveira, E. C. A. (2016). Nutrient partitioning and nutritional requirement in sugarcane. Australian Aust. J. Crop. Science 10, 69–75. doi: 10.3316/informit.936795392927333
de Souza, R. S. C., Okura, V. K., Armanhi, J. S. L., Jorrín, B., Lozano, N., da Silva, M. J., et al. (2016). Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 6:28774. doi: 10.1038/srep28774
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. doi: 10.1111/j.1574-6941.2010.00860.x
Döbereiner, J., and Day, J. M. (1976). “Associative symbiosis in tropical grasses: characterization of microorganisms and dinitrogen fixing sites,” in Proceedings of the 1st International Symposium on Nitrogen Fixation. (Pullman: Washington State University Press), 518–538.
Dong, M., Yang, Z., Cheng, G., Peng, L., Xu, Q., and Xu, J. (2018). Diversity of the bacterial microbiome in the roots of four Saccharum species: S. spontaneum, S. robustum, S. barberi, and S. officinarum. Front. Microbiol. 9:267. doi: 10.3389/fmicb.2018.00267
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I., and Dangl, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant. Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Gaby, J. C., Rishishwar, L., Valderrama-Aguirre, L. C., Green, S. J., Valderrama-Aguirre, A., Jordan, I. K., et al. (2018). Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl. Environ. Microbiol. 84, e01512–e01517. doi: 10.1128/aem.01512-17
Gao, X., Wu, Z., Liu, R., Wu, J., Zeng, Q., and Qi, Y. (2019). Rhizosphere bacterial community characteristics over different years of sugarcane ratooning in consecutive monoculture. Biomed. Res. Int. 2019, 1–10. doi: 10.1155/2019/4943150
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L., and Field, K. G. (1990). Genetic diversity in sargasso sea bacterioplankton. Nature 345, 60–63. doi: 10.1038/345060a0
Gopalasundaram, P., Bhaskaran, A., and Rakkiyappan, P. (2012). Integrated nutrient management in sugarcane. Sugar. Tech. 14, 3–20. doi: 10.1007/s12355-011-0097-x
Guo, D. J., Singh, R. K., Singh, P., Li, D. P., Sharma, A., Xing, Y. X., et al. (2020). Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 22:580081. doi: 10.3389/fmicb.2020.580081
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327, 1008–1010. doi: 10.1126/science.1182570
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Hu, H. N., Ao, J. H., Huang, X. C., Li, X. X., and Liao, H. (2017). Evaluation on associative nitrogen fixation capability in different tissues of sugarcane. Zhiwu Shengli Xuebao Plant Physiol. J. 53, 437–444. doi: 10.13592/j.cnki.ppj.2016.0407
Inui, M., Roh, J. H., Zahn, K., and Yukawa, H. (2000). Sequence analysis of the cryptic plasmid pMG101 from Rhodopseudomonas palustris and construction of stable cloning vectors. Appl. Environ. Microbiol. 66, 54–63. doi: 10.1128/aem.66.1.54-63.2000
Ishizawa, H., Ogata, Y., Hachiya, Y., Tokura, K. I., Kuroda, M., Inoue, D., et al. (2020). Enhanced biomass production and nutrient removal capacity of duckweed via two-step cultivation process with a plant growth-promoting bacterium, Acinetobacter calcoaceticus P23. Chemosphere 238:124682. doi: 10.1016/j.chemosphere.2019.124682
Kang, S. M., Joo, G. J., Hamayun, M., Na, C. I., Shin, D. H., Kim, H. Y., et al. (2009). Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol. Lett. 31, 277–281. doi: 10.1007/s10529-008-9867-2
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, C., Zha, Y., Li, Y., Sun, D., Lu, H., and Yin, B. (2010). Eutrophication of lake waters in China: cost, causes, and control. Environ. Manage. 45, 662–668. doi: 10.1007/s00267-010-9440-3
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259. doi: 10.1093/nar/gkz239
Li, X., Zhao, J., Tan, Z., Zeng, R., and Liao, H. (2015). GmEXPB2, a cell wall beta-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 169, 2640–2653. doi: 10.1104/pp.15.01029
Li, X., Zheng, J., Yang, Y., and Liao, H. (2018). Increasing NODULE SIZE1 expression is required for normal rhizobial symbiosis and nodule development. Plant Physiol. 178, 1233–1248. doi: 10.1104/pp.18.01018
Li, Y. R., and Yang, L. T. (2015). Sugarcane agriculture and sugar industry in China. Sugar Tech. 17, 1–8. doi: 10.1007/s12355-014-0342-1
Lin, L., Wei, C., Chen, M., Wang, H., Li, Y., Li, Y., et al. (2015). Complete genome sequence of endophytic nitrogen-fixing Klebsiella variicola strain DX120E. Stand. Genomic Sci. 10, 22. doi: 10.1186/s40793-015-0004-2
Littlewood, T. D., and Bennett, M. R. (2003). Apoptotic cell death in atherosclerosis. Curr. Opin. Lipidol. 14, 469–475. doi: 10.1097/00041433-200310000-00007
Liu, S., Liao, L. L., Nie, M. M., Peng, W. T., Zhang, M. S., Lei, J. N., et al. (2020). A VIT-like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. New Phytol. 226, 1413–1428. doi: 10.1111/nph.16506
Liu, W., Jiang, L., Yang, S., Wang, Z., Tian, R., Peng, Z., et al. (2020). Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment. Ecology 101:e03053. doi: 10.1002/ecy.3053
Liu, X., Zhang, Y., Han, W., Tang, A., Shen, J., Cui, Z., et al. (2013). Enhanced nitrogen deposition over China. Nature 494, 459–462. doi: 10.1038/nature11917
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Marschner, H., Römheld, V., Horst, W. J., and Martin, P. (1986). Root-induced changes in the rhizosphere: importance for the mineral nutrition of plants. Z. Pflanzenernährung Bodenkunde 149, 441–456. doi: 10.1002/jpln.19861490408
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Mehnaz, S. (2013). Microbes-friends and foes of sugarcane. J. Basic Microbiol. 53, 954–971. doi: 10.1002/jobm.201200299
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Muthukumarasamy, R., Revathi, E., Vadivelu, M., and Arun, K. (2017). Isolation of bacterial strains possessing nitrogen-fixation, phosphate and potassium-solubilization and their inoculation effects on sugarcane. Indian J. Exp. Biol. 55, 161–170.
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U.S.A. 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Oliveira, A. L. M., Stoffels, M., Schmid, M., Reis, V. M., Baldani, J. I., and Hartmann, A. (2009). Colonization of sugarcane plantlets by mixed inoculations with diazotrophic bacteria. Eur. J. Soil Biol. 45, 106–113. doi: 10.1016/j.ejsobi.2008.09.004
Oliveira, A. L. M., Urquiaga, S., Döbereiner, J., and Baldani, J. I. (2002). The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242, 205–215. doi: 10.1023/a:1016249704336
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Pedrosa, F. O., Monteiro, R. A., Wassem, R., Cruz, L. M., Ayub, R. A., Colauto, N. B., et al. (2011). Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 7:e1002064. doi: 10.1371/journal.pgen.1002064
Prashar, P., Kapoor, N., and Sachdeva, S. (2014). Rhizosphere: its structure, bacterial diversity and significance. Rev. Environ. Sci. Biol. 13, 63–77. doi: 10.1007/s11157-013-9317-z
Qin, L., Jiang, H., Tian, J., Zhao, J., and Liao, H. (2011). Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant Soil 349, 25–36. doi: 10.1007/s11104-011-0947-z
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rokhbakhsh-Zamin, F., Sachdev, D., Kazemi-Pour, N., Engineer, A., Pardesi, K. R., Zinjarde, S., et al. (2011). Characterization of plant-growth-promoting traits of Acinetobacter Species isolated from rhizosphere of Pennisetum glaucum. J. Microbiol. Biotechnol. 21, 556–566. doi: 10.1111/j.1439-0523.2012.01954.x
RöSch, C., Mergel, A., and Bothe, H. (2002). Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68, 3818–3829. doi: 10.1128/aem.68.8.3818-3829.2002
Rosenblueth, M., Ormeño-Orrillo, E., López-López, A., Rogel, M. A., Reyes-Hernández, B. J., Martínez-Romero, J. C., et al. (2018). Nitrogen fixation in cereals. Front. Microbiol. 9:1794. doi: 10.3389/fmicb.2018.01794
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sakurai, M., Wasaki, J., Tomizawa, Y., Shinano, T., and Osaki, M. (2008). Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci. Plant Nut. 54, 62–71.
Sant’Anna, F. H., Almeida, L. G., Cecagno, R., Reolon, L. A., Siqueira, F. M., Machado, M. R., et al. (2011). Genomic insights into the versatility of the plant growth-promoting bacterium Azospirillum amazonense. BMC Genomics 12:409. doi: 10.1186/1471-2164-12-409
Saravanan, V. S., Madhaiyan, M., and Thangaraju, M. (2007). Solubilization of zinc compounds by the diazotrophic, plant growth promoting bacterium Gluconacetobacter diazotrophicus. Chemosphere 66, 1794–1798. doi: 10.1016/j.chemosphere.2006.07.067
Schwab, S., Terra, L. A., and Baldani, J. I. (2018). Genomic characterization of Nitrospirillum amazonense strain CBAmC, a nitrogen-fixing bacterium isolated from surface-sterilized sugarcane stems. Mol. Genet. Genomics 293, 997–1016. doi: 10.1007/s00438-018-1439-0
Sevilla, M., Burris, R. H., Gunapala, N., and Kennedy, C. (2001). Comparison of benefit to sugarcane plant growth and 15N2 incorporation following inoculation of sterile plants with Acetobacter diazotrophicus wild-type and Nif- mutants strains. Mol. Plant Microbe Interact. 14, 358–366. doi: 10.1094/mpmi.2001.14.3.358
Singh, R., Ryu, J., and Kim, S. W. (2019). Microbial consortia including methanotrophs: some benefits of living together. J. Microbiol. 57, 939–952. doi: 10.1007/s12275-019-9328-8
Tringe, S. G., von Mering, C., Kobayashi, A., Salamov, A. A., Chen, K., Chang, H. W., et al. (2005). Comparative metagenomics of microbial communities. Science 308, 554–557. doi: 10.1126/science.1107851
Urquiaga, S., Cruz, K. H. S., and Boddey, R. M. (1992). Contribution of nitrogen fixation to sugar cane: Nitrogen-15 and nitrogen-balance estimates. Soil Sci. Soc. Am. J. 56, 105–114. doi: 10.2136/sssaj1992.03615995005600010017x
Urquiaga, S., Xavier, R. P., De Morais, R. F., Batista, R. B., Schultz, N., Leite, J. M., et al. (2012). Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil 356, 5–21. doi: 10.1007/s11104-011-1016-3
Vieira, S., Barros, M. V., Sydney, A., Piekarski, C. M., de Francisco, A. C., Vandenberghe, L., et al. (2020). Sustainability of sugarcane lignocellulosic biomass pretreatment for the production of bioethanol. Bioresour. Technol. 299:122635. doi: 10.1016/j.biortech.2019.122635
Wang, C., Li, Y., Li, M., Zhang, K., Ma, W., Zheng, L., et al. (2021). Functional assembly of root-associated microbial consortia improves nutrient efficiency and yield in soybean. J. Integr. Plant Biol. 63, 1021–1035. doi: 10.1111/jipb.13073
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Yang, L. J., and Jia, Z. J. (2021). The history and definition of 99% unculturability paradigm and its assessment using nitrogen-fixing bacteria. Acta Microbiol. Sin. 61, 903–922. doi: 10.13343/j.cnki.wsxb.20200258
Yeoh, Y. K., Paungfoo-Lonhienne, C., Dennis, P. G., Robinson, N., Ragan, M. A., Schmidt, S., et al. (2016). The core root microbiome of sugarcanes cultivated under varying nitrogen fertilizer application. Environ. Microbiol. 18, 1338–1351. doi: 10.1111/1462-2920.12925
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zhang, J., Liu, Y.-X., Zhang, N., Hu, B., Jin, T., Xu, H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684. doi: 10.1038/s41587-019-0104-4
Zhong, Y., Yang, Y., Liu, P., Xu, R., Rensing, C., Fu, X., et al. (2019). Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 42, 2028–2044. doi: 10.1111/pce.13519
Keywords: sugarcane, root-associated microbes, beneficial function, nitrogen, growth promotion
Citation: Li M, Liu R, Li Y, Wang C, Ma W, Zheng L, Zhang K, Fu X, Li X, Su Y, Huang G, Zhong Y and Liao H (2022) Functional Investigation of Plant Growth Promoting Rhizobacterial Communities in Sugarcane. Front. Microbiol. 12:783925. doi: 10.3389/fmicb.2021.783925
Received: 27 September 2021; Accepted: 30 November 2021;
Published: 04 January 2022.
Edited by:
Ying Ma, University of Coimbra, PortugalReviewed by:
José David Flores Félix, Universidade da Beira Interior, PortugalMonali C. Rahalkar, Agharkar Research Institute, India
Copyright © 2022 Li, Liu, Li, Wang, Ma, Zheng, Zhang, Fu, Li, Su, Huang, Zhong and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjia Zhong, WW9uZ2ppYXpob25nQGZhZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Mingjia Li
Mingjia Li Ran Liu1†
Ran Liu1† Xinxin Li
Xinxin Li