
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 December 2021
Sec. Systems Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.782621
This article is part of the Research Topic Community Assembly Mechanisms Shaping Microbiome Spatial or Temporal Dynamics View all 13 articles
The bacterial communities of the root-zone soil are capable of regulating vital biogeochemical cycles and the succession of plant growth. Stipa as grassland constructive species is restricted by the difference features of east–west humidity and north–south heat, which shows the population substituting distribution. The distribution, turnover, and potential driving factors and ecological significance of the root-zone bacterial community along broad spatial gradients of Stipa taxa transition remain unclear. This paper investigated seven Stipa species root-zone soils based on high-throughput sequencing combined with the measurements of multiple environmental parameters in arid and semi-arid steppe. The communities of soil bacteria in root zone had considerable turnover, and some regular variations in structure along the Stipa taxa transition are largely determined by climatic factors, vegetation coverage, and pH at a regional scale. Bacterial communities had a clear Stipa population specificity, but they were more strongly affected by the main annual precipitation, which resulted in a biogeographical distribution pattern along precipitation gradient, among which Actinobacteria, Acidobacteria, Proteobacteria, and Chloroflexi were the phyla that were most abundant. During the transformation of Stipa taxa from east to west, the trend of diversity shown by bacterial community in the root zone decreased first, and then increased sharply at S. breviflora, which was followed by continuous decreasing toward northwest afterwards. However, the richness and evenness showed an opposite trend, and α diversity had close association with altitude and pH. There would be specific and different bacterial taxa interactions in different Stipa species, in which S. krylovii had the simplest and most stable interaction network with the strongest resistance to the environment and S. breviflora had most complex and erratic. Moreover, the bacterial community was mainly affected by dispersal limitation at a certain period. These results are conducive to the prediction of sustainable ecosystem services and protection of microbial resources in a semi-arid grassland ecosystem.
Soil microorganisms play an important role in regulating and maintaining biogeochemical cycles in a terrestrial ecosystem, and are sensitive to environmental changes (Chen et al., 2020). Arid and semi-arid steppe ecosystem, as one of the most important components of the terrestrial system (Kang et al., 2007; Wang et al., 2021), is becoming fragmented and degraded due to climate changes and anthropogenic activity (Gao et al., 2018; Zhang Y. et al., 2018). Climates, vegetation, and soil properties in such steppes vary greatly (Lv et al., 2016), and these changes will strongly affect the structure and function of the soil microbial community (Tu et al., 2017). Numerous studies have focused on soil microbial diversity and assembly processes in arid and semi-arid ecosystems, as well as on the variations and distribution of microorganisms along spatio-temporal, altitude, longitude, plant diversity, and drought gradients (Tu et al., 2017; Yao et al., 2017; Richter-Heitmann et al., 2020; Wang et al., 2021). However, plant root-zone soils have been relatively neglected in complex natural communities. To be specific, the rhizosphere acts as the region of interactions between soil microorganisms and the plant root system's own biological activities (Verma et al., 2019). Moreover, rhizosphere soil microbes are involved in the slow ecological process of vegetation succession and evolution to adapt to the regional environments (Song et al., 2019). This study systematically investigates the turnover and distribution of root-zone soil microbial communities and the correlation with plants along broad spatial gradients of the plant community succession distribution, which is critical to indicating and predicting ecosystem functions.
Abundant rhizosphere microbial resources create a highly evolved external functional environment for plants, thereby allowing organic matters to degrade, nutrients to be released from minerals, nitrogen to be fixed, and elemental forms to be transformed (Lau and Lennon, 2011; Lakshmanan et al., 2014; Na et al., 2018; Krishna et al., 2020). In turn, plants affect microorganisms by the decomposition of litter, turnover of roots, and the release of exudates (Philippot et al., 2013). The interactions help maintain the stability of ecosystem structures and functions, while positively affecting plant fitness and resistance, especially in extreme environments (Marasco et al., 2018). Bacteria comprise a large part of rhizosphere soil biodiversity and participate in most of the material transformation processes (Chu et al., 2020), hence greatly affecting plant growth and establishment. Studies have shown that the size, structure, and activity of the soil bacterial communities could be greatly affected by individual plant species (Liu et al., 2021). Thus, gaining insights into the variation and interactions of plant root-zone soil bacterial communities will enhance our understanding of the ecology and function of soil bacteria. Over the past few decades, spatial and temporal variation in bacterial communities and linkages with plant communities have been extensively investigated in a wide variety of habitats (Tian et al., 2017; Na et al., 2018; Zeng et al., 2019; Chu et al., 2020), whereas such studies have been rare for the root zone of natural forages.
As indicated from existing studies, the soil rhizosphere bacterial composition and relative abundance are highly affected by the biotic and abiotic environments (Fan et al., 2017; Zhang B. et al., 2018). Edaphic factors, particularly soil pH and nutrient availability, vegetation composition and species, regional climate, and altitude, have been shown to shape soil bacterial communities (Na et al., 2018; Zhalnina et al., 2018; Clairmont et al., 2019; Mohanram and Kumar, 2019). Moreover, bacterial evolutionary responses may be driven by edaphic and non-edaphic variables that function as selective pressures; for example, the relationship between the adaptive range of pH and biogeographical patterns (Na et al., 2018), or the greater sensitivity of bacteria in drier areas to environmental stimuli (Maestre et al., 2015). Recently, chemical element indices in soil (e.g., Fe, Cu, Ca, and Mg) have also been proposed to directly impact or predict bacterial diversity and composition (Corneo et al., 2013; Wang et al., 2020). Microbial community assembly (both deterministic and stochastic processes) is critical to maintaining the spatial distribution and composition of microbial communities from a local to a global scale (Stegen et al., 2013; Zhang et al., 2016). Microbial taxa interactions also describe the underlying ecological processes, which can affect the response of communities to environmental variations and may be more important than environmental variables in determining community structures (Zhang B. et al., 2018). For example, it is predicted that ecological networks that consist of weak interactions are more stable than those with strong interactions (Coyte et al., 2015). Yet little is known about how the mentioned ecological processes govern rhizosphere soil bacterial community turnover during the obviously geographic substitution of the plants.
The genus Stipa refers to a grazing tolerant, drought-resistant perennial bunchgrass species (Gao et al., 2018; Chen et al., 2020) that is an important ecological barrier and basically in the highly specialized stage of the grassland community succession and evolution (Lv et al., 2016; Liu et al., 2019). From eastern to western Inner Mongolia, across various combinations of hydro-thermal characteristic, different moisture ecotypes of Stipa occupy zonal habitats and form meadow steppe, typical steppe, and desert steppe. Accordingly, the S. grandis community is generally replaced by the mesophytic S. baicalensis community that connects the forest grassland subzone in the east under the progressively humid habitat conditions. As opposed to the mentioned, the S. grandis community is largely distributed in the eastern part of the typical steppe, and is often replaced by the S. krylovii community when the habitat conditions tend to be dry or frequently grazed. Also, the S. krylovii community in the western region of the typical steppe exhibits more xerophytic characteristics and is replaced by the warm-loving S. breviflora when the habitat is transmitted to the desert steppe to the west and southwest. Furthermore, S. breviflora crosses the southeastern edge of the desert steppe zone from east to west, and then the drier communities of S. gobica, S. klemenzii, and S. glareosa appear to the northwest. Moreover, S. gobica extends westward to the desert, and redundantly is combined with S. klemenzii and S. glareosa communities (Lv et al., 2016; Li et al., 2018; Nan et al., 2020). Stipa taxa show divergences in morphological and physiological parameters (Han and Tian, 2016). The mentioned typical divergent evolution processes provide a unique opportunity and ideal study area to investigate the root-zone bacterial communities in natural ecosystems. The following hypotheses were verified here: (1) There is considerable turnover of root-zone soil bacterial communities along the Stipa taxa transition and largely determined by climatic factors, vegetation coverage, and pH at a regional scale. (2) Bacterial communities in root zone have a clear Stipa population specificity and the biogeographic distribution pattern would not be affected solely by climatic factors. (3) There would be specific and different bacterial taxa interactions and different resistance to environment in different Stipa species, and that the bacterial community assembly is dominated by dispersal limitation. This paper aimed to investigate the geographic distribution, turnover, and interaction of root-zone bacterial community across a transect of zonal Stipa species across broad spatial gradients. The identification of potential drivers is critical to elucidating between soil bacterial and plant populations, which can more effectively predict grassland ecosystem responses and functions under a changing climate, and lay a theoretical basis for gaining insights into the community structure and influencing factors of rhizosphere soil bacteria.
This study was conducted along a 1,700-km east–west transect across Inner Mongolia (41.52–50.12 N, 108.46–120.21 E) in northern China. A total of 32 sampling sites that had nearly natural plant communities with only light animal grazing were selected along the zonal distribution of Stipa spp.; six for S. baicalensis (S1–S6), 11 for S. grandis (S7–S17), eight for S. krylovii (S18–S25), three for S. breviflora (S28–S30), two for S. gobica (S31–S32), and one each for S. glareosa (S26) and S. klemenzii (S27). Samples were gradually collected from west to east from early August 2019 through the entire transect, to ensure that Stipa were in the same mature phenological stage (Figure 1A). Identification of Stipa species was according to FLORA INTRAMONGOLICA (EDITIO TERTIA Tomus 6: 205–217). Spatial geographic coordinates, climatic information, and the plant indexes (Plant coverage and Biomass of Stipa) for each sampling site are listed in Supplementary Table S2. The region has a temperate continental arid and semi-arid monsoonal climate with a mean annual average precipitation of 172.19–472.06 mm and a mean annual temperature of −0.12–6.56°C (https://disc.gsfc.nasa.gov/). The soil types in the study area include forest grassland black soil and chernozem, chestnut soil in the dry grassland, and brown calcareous soil and lime-calcium soil in the desert steppe.
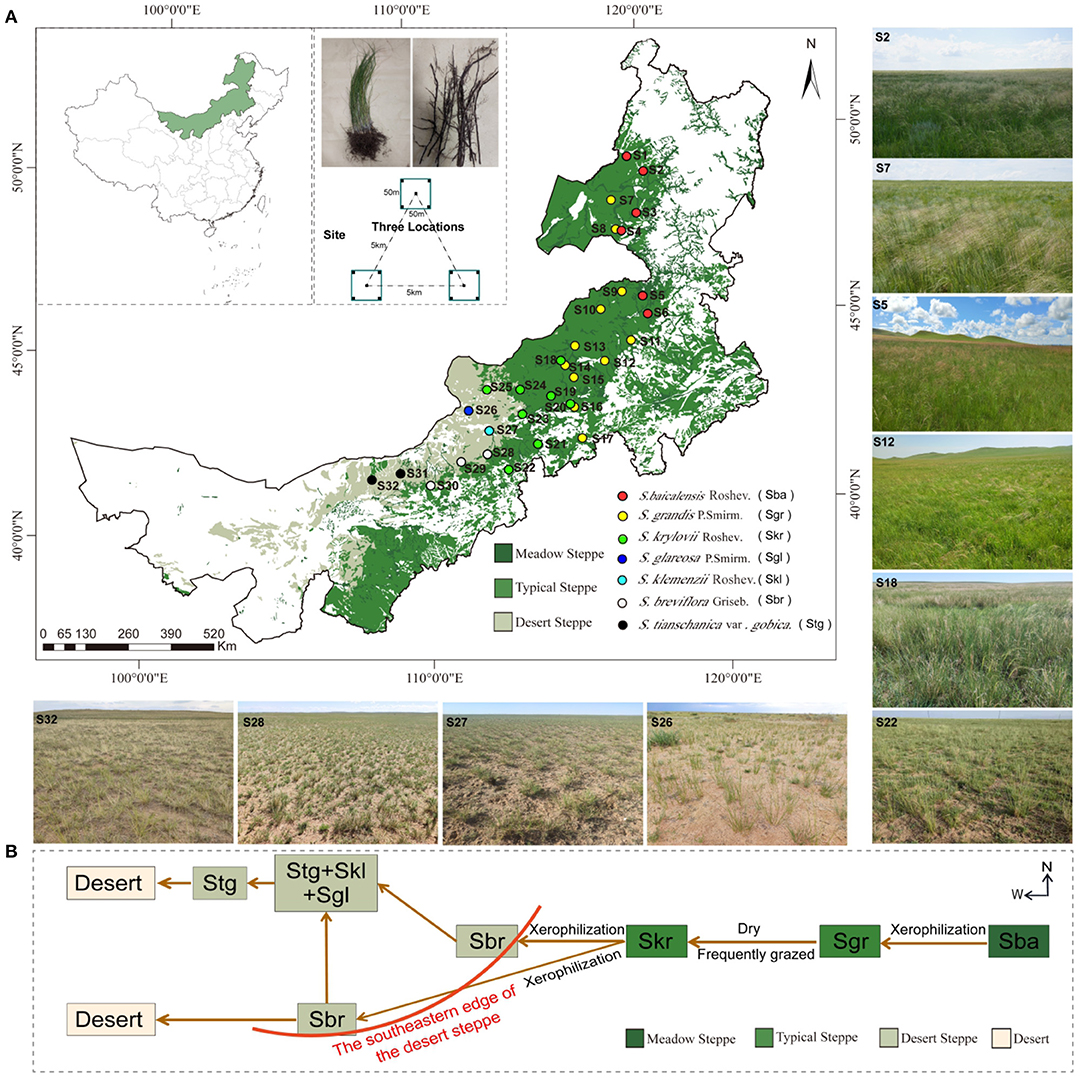
Figure 1. (A) Map of sample locations, landscape, and schematic diagram in arid and semi-arid Stipa steppe. Each circle represents one sampling site, and each color represents a Stipa population; samples collected from different sites and species are referred to as Stipa baicalensis Roshev (Sba: S1–S6), Stipa grandis P. Smirn (Sgr: S7–S17), Stipa krylovii Roshev (Skr: S18–S25), Stipa glareosa P. Smirn (Sgl: S26), Stipa klemenzii Roshev (Skl: S27), Stipa breviflora Griseb (Sbr: S28–S30), and Stipa tianschanica var. gobica (Stg: S31–S32). (B) Schematic diagram of alternative distribution of Stipa population.
At the respective site (n = 32), three independent replicate locations (50 × 50 m, at least 5 km apart) were selected (Figure 1). In accordance with the double diagonal principle, five 1 × 1 m plots were evenly distributed on the center point of the respective independent location and the two diagonals of the four corners for a vegetation survey, which included plant diversity, number, vegetation coverage, and a calculation of the Shannon–Wiener index (, where Pi denotes the ratio of the number of each species to the overall number of all species present). The Stipa species exhibits a caespitose growth form, which contains numerous tillers and culms emerging from a single rootstock. After establishment, soil, litter, roots, and live shoots accumulate to form tussocks (Bai et al., 1999). The active root system of Stipa has rhizosheath (a layer of adhering soil particle to the root surface) that does not easily fall off, creating sufficient water and inorganic salts for the plant and protecting the fibrous roots from mechanical damage (Basirat et al., 2019). The age of perennial bunch grasses is difficult to find through field observation. In the sampling process, plants with tussock fragmentation and self-thinning branches, or too small or excessively large plants should not be selected, as an attempt to collect representative root-zone soil samples and minimize the plant species-age effects on bacterial community diversity and composition (Bai et al., 1999; Na et al., 2018). Given the mentioned information, 25 individual Stipa were collected at the respective sampling site, and the excavation depth should exceed 30 cm (Chen, 1987) to ensure the root system to be as complete as possible. To be specific, 15 individual Stipa were applied for soil samples and 10 individual Stipa for vegetation samples. Several root segments were randomly sampled from each individual Stipa after shaking off the extremely loose root-attached soil. Next, the root segments from 15 individual Stipa were integrated as one site sample. Subsequently, root-zone soil (Edwards et al., 2015; Shi et al., 2019) was collected from the respective site sample with a disposable brush to brush out the soil around the rhizosheath and then divided equally into three parts. On the whole, 32 × 3 root-zone soil samples were collected, and immediately stored in liquid nitrogen until 16S rRNA high-throughput sequencing analysis could be performed. Brushes and gloves were sterile and disposable to avoid contamination during sampling. The mentioned 10 individual Stipa for vegetation samples fell into two equal parts—one part was used for weighing the average aboveground biomass of Stipa with the drying method at 80°C, and the other was mixed, crushed, and sieved through a sieve with 0.154 mm pore size into one sample to determine the nutritive value of the forage. The longitude and latitude of each location were recorded from a portable GPS. The collected soil samples fell into two parts: (1) stored at 4°C for soil physical and chemical analysis, and (2) sieved through pore size of 0.074 mm to analyze of soil chemical elements.
A total of 25 environmental predictors were measured. The pH was determined using an SKW 500 soil monitor (Palintest Co., Ltd, UK). Total nitrogen (TN), ammonium (), nitrate nitrogen (), and plant crude protein (CP) were measured with an automatic Kjeldahl apparatus (K1100 Hanon; Haineng Future Technology Group Co., Ltd, China) after digestion in H2SO4 (ρ = 1.8419 g/L). Soil organic matter (SOM) and soil organic carbon (SOC) were determined by dichromate oxidation (Chen et al., 2015). Alkaline and neutral Olsen-phosphorus (AP) were determined based on the sodium bicarbonate extraction (0.05 mol·L−1) molybdenum antimony anti-colorimetric method (Wang et al., 2020). Available potassium (AK) was determined with a kit according to the instructions (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China). Root-zone soil pretreatment was carried out by microwave digestion system (MARS 6; CEM Co., Ltd, USA) according to a soil and sediment digestion of total metal elements–microwave-assisted acid digestion method (HJ 832−2017, Ministry of Environmental Protection MEP, China), and then soil chemical elements (S, Ca, Mg, Fe, Cu, Zn, and Mn) were determined by ICP-OES (PlasmaQuant PQ9000; Jena Analytical Instrument Co., Ltd, Germany). Plant crude fiber and crude fat were measured by an acid–base heating digestion method (GB/T5009.10-2003; Ministry of Health, P. R. China) and a Soxhlet extraction method (GB/T6433-2006; General Administration of Quality Supervision GAQSIQ, China), respectively. Mean annual temperature and mean annual precipitation was obtained from the GES DISC of NASA (https://disc.gsfc.nasa.gov/).
Triplicate genomic DNA samples were extracted from 0.5 g of the composite soil samples using an E.Z.N.A. soil DNA kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer's instructions. The DNA extract was run on a 1% agarose gel, and the DNA concentration and purity were determined with a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA). To generate bacterial PCR amplicon libraries, universal 16S rRNA gene primers (338 forward: 5′-ACTCCTACGGGAGGCAGCAG-3′ and 806 reverse: 5′-GGACTACHVGGGTWTCTAAT-3′). The PCR amplification of the 16S rRNA gene was performed according to details in Supplementary Materials. Purified amplicons were pooled in equimolar quantities and paired-end sequencing (2 × 300 bp) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). All of the sequence data generated for this study have been deposited in the NCBI Sequence Read Archive (SRR14415681–SRR14415776).
The raw 16S rRNA gene sequences obtained from the MiSeq platform were quality filtered, trimmed, de-noised, and merged using Mothur 1.32.2 (Schloss et al., 2009) where chimeric sequences were identified and removed using the UCHIME de novo algorithm (Perez-Jaramillo et al., 2019). Subsequently, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) using the UPARSE (version 7.0.1090, http://www.drive5.com/uparse/) algorithm, setting a distance limit of 0.03 using the open-reference OTU picking protocol. A representative sequence was aligned using the RDP Classifier (http://rdp.cme.msu.edu/) against the 16S rRNA database (Silva 132; https://www.arb-silva.de/) using a confidence threshold of 0.7. For downstream analysis, OTUs affiliated with chloroplasts and mitochondria were subsequently removed from the bacterial OTU table, and OTUs that were assigned to non-bacteria, including plants and protozoans, were removed from the bacterial OTU table (Perez-Jaramillo et al., 2019). The effects of sampling on diversity were corrected by rarifying the sequence numbers of each sample to that of the sample with the lowest number of reads (17,865 reads). The number of OTUs, community richness (Chao index), community diversity (Simpson index), community evenness (Shannon index), and a sequencing depth index (Good's coverage) were subsequently calculated using MOTHUR (v.1.30.1; www.mothur.org).
All statistical analyses were conducted through a one-way ANOVA with IBM SPSS Statistics 26.0 (p < 0.05 was considered statistically significant). Non-metric multidimensional scaling [NMDS, stress < 0.20 was considered acceptable (Kuczynski et al., 2011)] and hierarchical clustering (unweighted pair group method with arithmetic means, UPGMA) that all used the Bray–Curtis distance to assess the Stipa population-specificity distribution of bacteria, as well as the similarity of bacterial community composition. To demonstrate the effect of Stipa population on the bacterial composition, permutational multivariate analysis of variance (PERMANOVA, p < 0.05) was conducted with the adonis function of the vegan package (Glasl et al., 2019) with 999 permutations. A linear discriminant analysis (LDA score threshold of 4.0) coupled with effect size (LEfSe) was adopted to search for statistically different biomarkers among groups of Stipa taxa root zone. In addition, Circos-0.67-7 was employed to determine the connection between the dominant phylum or genus and the sample group of Stipa species. The variance inflation factor (VIF) was exploited to remove the redundant variables (VIF > 20) (Fan and Xing, 2016) from the explanatory variables and avoid collinearity among a range of factors. The Mantel test and the Pearson correlation analysis were performed to examine the relationship between bacterial communities and environmental factors using the “Vegan” package (Oksanen et al., 2015) and the “ggcor” package (Wang et al., 2021) version 0.9 implemented in R, respectively. A redundancy analysis (RDA) was also performed to determine the most significant environmental variables shaping the microbial community composition.
Bacterial co-occurrence network analyses for Stipa species were conducted based on the Molecular Ecological Network Analyses Pipeline (MENAP, http://ieg4.rccc.ou.edu/MENA/main.cgi) at the OTU level (Deng et al., 2012). To simplify the networks for a more effective visualization and unified analysis conditions, only the 300 most abundant OTUs were analyzed and network topology parameters (i.e., network complexity and modularity) were determined to indicate the stability of the network and the resistance from environmental interference. Abundances of OTUs were log transformed. To compare networks under the identical conditions, a threshold of 0.82 (the recommended similarity threshold) was adopted to build the networks for the respective Stipa taxa in the study. Lastly, the networks were visualized with the Gephi 0.9.2 (Zhou et al., 2020). Furthermore, 100 random networks were generated and the properties were compared with the experience network to verify whether the constructed network is reasonable and effective. The different assembly processes of community composition were determined by phylogenetic null model analyses. All OTUs were adopted to build a maximum-likelihood tree in FastTree (Tang et al., 2021) to determine the weighted β-nearest taxon index (βNTI) using the picante package (Dini-Andreote et al., 2015). The βNTI values > +2 (variable selection) or < −2 (homogeneous selection) implies significantly more or less phylogenetic turnover than expected (Tripathi et al., 2018), respectively, thereby demonstrating the predominance of deterministic processes. If the |βNTI| ≤ 2, stochastic processes are predominant. To more specifically differentiate between stochastic scenarios of assemblages of Stipa species root-zone soil bacteria, a Raup–Crick matrix (RCbray) based on the Bray–Curtis matrix was computed with the vegan package (Cheng et al., 2021). |βNTI| < 2 and RCBray < −0.95, |βNTI| < 2 and RCBray > 0.95, and |βNTI| < 2 and |RCBray| < 0.95 denote homogenizing dispersal, dispersal limitation, and undominated processes, respectively (e.g., weak selection, weak dispersal, diversification, and/or drift) (Stegen et al., 2013; Dini-Andreote et al., 2015). Furthermore, unless otherwise indicated, all analyses were conducted in R (version 3.5.1; R Development Core Team).
In accordance with Supplementary Tables S1, S2, the MAP of the semi-arid grassland decreased gradually from east (472.06 mm) to west (152.08 mm). SOM, SOC, TN, -N, VC, and SB tended to increase with precipitation, while pH, altitude, and temperature tended to decrease. The soils were moderately alkaline and the average pH increased significantly from 6.06 to 8.74. Other test indices displayed significant differences among 32 sites, but no obvious rules to follow. There were significant differences in vegetation indexes among seven Stipa species, and the vegetation coverage (66.67 ± 14.38 to 27.50 ± 3.61) and Stipa biomass (3.00 ± 1.31 to 0.85 ± 0.32) decreased significantly under the process of Sbr replacing Sba distribution (Supplementary Table S3). Most soil parameters differed significantly between the various root-zone soils of Stipa, except for TK and , in which macronutrients (SOM, SOC, TN, ) in Sba were significantly higher than for other Stipa taxa (Supplementary Table S3). A Pearson correlation analysis (Figure 6) found that most variables (MAP, MAT, altitude, soil pH, SOC, TN, S, VS, and SB) had strong positive or negative correlations with each other. In addition, there was a significant positive correlation between soil chemical elements.
We obtained a total of 4,665,705 raw reads by Illumina MiSeq high-throughput sequencing. After quality filtering, trimming, and assigning reads to the different samples, 3,150,865 high-quality reads were recovered in the dataset, representing 17,865 bacterial operational taxonomic units (OTUs) based on 97% sequence identity across all samples, with a median of 2,349 OTUs per sample (Supplementary Table S4). The observed OTUs and rarefaction curves of Shannon index, Chao richness, and Good's coverage (values were over 0.94) in all soil samples were plateaued (Supplementary Figure S1), suggesting that the sequencing depth was sufficient.
Circular visualization diagrams were adopted to display the overlapping and differentiating bacterial taxa among the seven Stipa taxa at the phylum and genus levels (Figures 2A,B). The bacterial community consisted of 37 different phyla, and the main bacterial phyla (top 11) collectively accounted for 96.98–98.76% of all taxon sequences (Figure 2A). Moreover, the five most abundant bacterial genera comprised 27.71% of the bacterial communities on average (Figure 2B). Actinobacteria, Acidobacteria, Proteobacteria, and Chloroflexi were the most abundant phyla observed in all of the Stipa taxa analyzed (Figure 2A). Except for Sgl, Actinobacteria (28.34–45.35%) was the dominant phylum in other Stipa species groups, especially in Sbr root zone (45.35%). Acidobacteria had a most significant advantage as dominant phylum in Sgl (32.81%) compared with other six Stipa taxa (10.73–24.10%). When Sba was replaced by Sbr from east to west along the transect, the relative abundance of Chloroflexi (phylum, from 8.17 to 14.80%) and Rubrobacter (genus, from 1.89 to 10.20%) increased significantly, and Rubrobacter was the most dominant genus in Sbr (10.20%); as opposed to the mentioned, the relative abundances of Proteobacteria (from 24.06 to 15.70%) decreased significantly (Figures 2A,B and Supplementary Table S6). Except Sbr, the relative abundances of genus RB41 (7–15.27%) and Subgroup_6 (5.85–13.04%) in the root zone of Stipa taxa were significantly higher than for other genera, especially in Sgl (Figure 2B). Taken together, these results revealed shifts in the bacterial community composition in root-zone soils of different Stipa taxa.
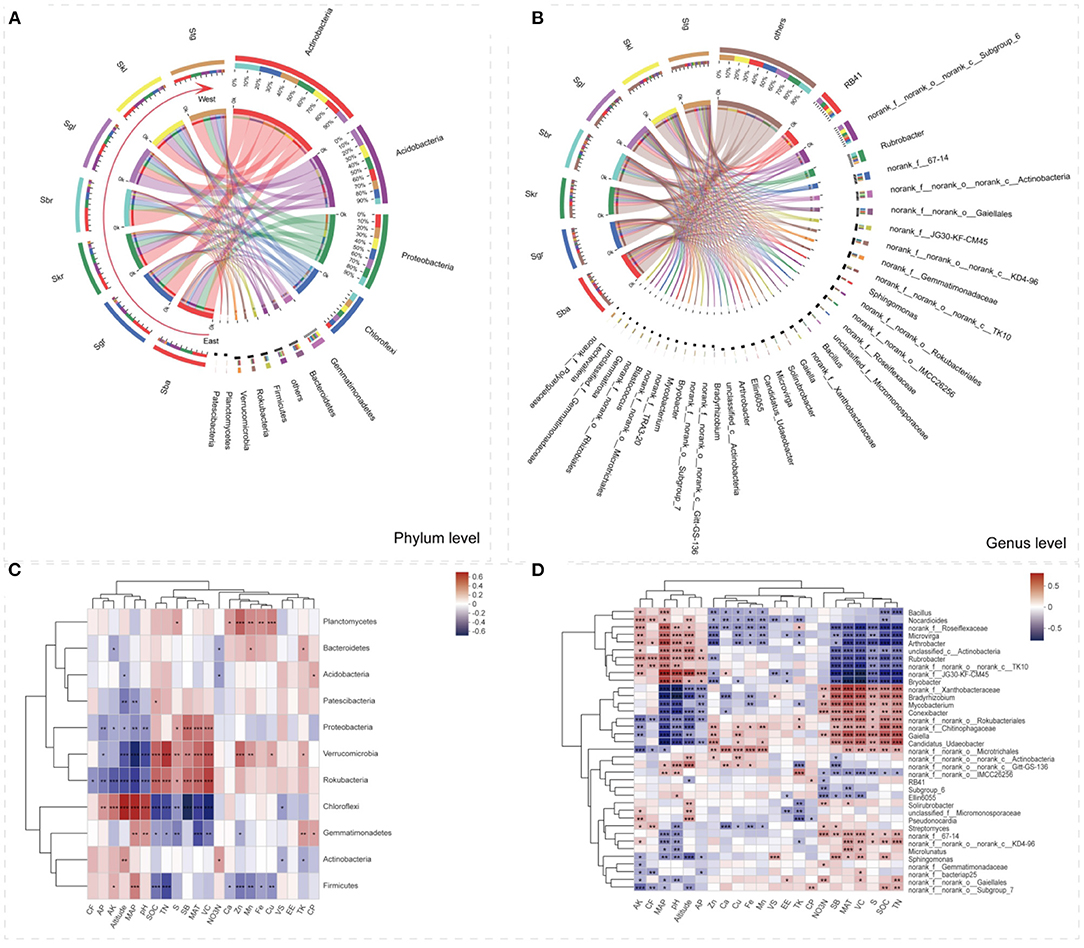
Figure 2. The distributions of root-zone soil bacterial dominant taxa at the different Stipa species. (A) Represents the phylum level; (B) represents the genus level, and Spearman correlation analysis of relative abundance of bacterial keystone taxa with environmental factors: (C) phylum level, (D) genus level. Red represents positive correlation and blue represents negative correlation (significance levels are as follows: *0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001). Arrows refer to the geographic substitution order of Stipa species from east to west. SOC, soil organic carbon; TN, total nitrogen; TK, total kalium; , nitrate nitrogen; AP, alkaline and neutral Olsen-phosphorus; AK, available potassium; Fe, iron element; Cu, copper element; Zn, zinc element; Mn, manganese element; Ca, calcium element; S, sulfur element; VS, Shannon–Wiener index of plot; VC, plant coverage; SB, biomass of Stipa; CP, crude protein; EE, crude fat; CF, crude fiber; MAT, mean annual temperature; MAP, mean annual precipitation. S. baicalensis (Sba), S. grandis (Sgr), S. krylovii (Skr), S. glareosa (Sgl), S. klemenzii (Skl), S. breviflora (Sbr), and S. gobica (Stg).
The LEfSe with LDA scores of 4 (Figure 3) revealed that, when compared with the Sba root-zone soil bacterial community (eight biomarkers at finer taxonomic level, one phylum, one class, three orders, two families, and one genera), the other Stipa species root-zone soil communities had less biomarkers (Figure 3A). However, Skr, as an intermediate transitional species, had no biomarkers. Furthermore, Stg and Sgl were distributed over a small area and had no biomarkers. To be specific, root-zone soil of Skl showed two significantly different taxa, Bacteroidia (class) and Bacteroidetes (phylum); Sgr were rich in Rhizobiales (order), Xanthobacteraceae (family), and norank_f__Xanthobacteraceae (genus); Sba were enriched with Sphingomonadales (order), Chthoniobacterales (order), Betaproteobacteriales (order), Sphingomonadaceae (family), Chthoniobacteraceae (family), and Candidatus_Udaeobacter (genus), while the root-zone soil of Sbr was enriched with Thermomicrobiales (order), JG30_KF_CM45 (family and genus) (Figure 3B).
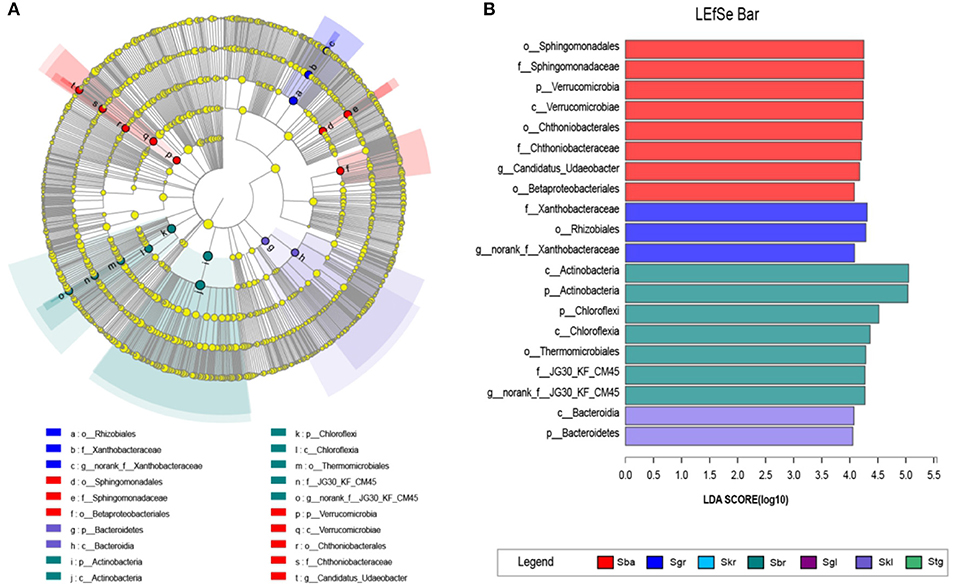
Figure 3. LEfSe analysis of soil bacterial abundance in seven Stipa species root-zone soils. (A) Cladogram of microbial communities. Cladograms indicate the phylogenetic distribution of microbial lineages associated with the study Stipa; circles represent phylogenetic levels from kingdom to genus. (B) Indicator microbial groups with LDA scores > 4 and p-values < 0.05. S. baicalensis (Sba), S. grandis (Sgr), S. krylovii (Skr), S. glareosa (Sgl), S. klemenzii (Skl), S. breviflora (Sbr), and S. gobica (Stg).
The bacterial biodiversity (Simpson index, Figure 4A) of the root-zone soil decreased when Sba was replaced by Skr from southeast to northwest along the transect, increased for Sbr, and then decreased sharply as Sbr transitioned to Stg. The trends of indices richness (Chao index Figure 4C) and evenness (Shannon index Figure 4B) of bacterial were opposite with the variation patterns of the mentioned bacterial diversity. We found that the α diversity of bacterial communities was different across 32 sampling sites with respect to the bacterial richness, diversity, and evenness for 96 soil samples, and a lower bacterial diversity was observed in more arid areas, as indicated by the changes in the Simpson index (Supplementary Table S2). Furthermore, bacterial evenness (Shannon index) and diversity (Simpson index) were more affected by altitude and pH, and bacterial evenness also affected crude protein; bacterial richness, as indicated by the Chao index, was affected mostly by the altitude and was also principally connected with crude fat (Figure 6).
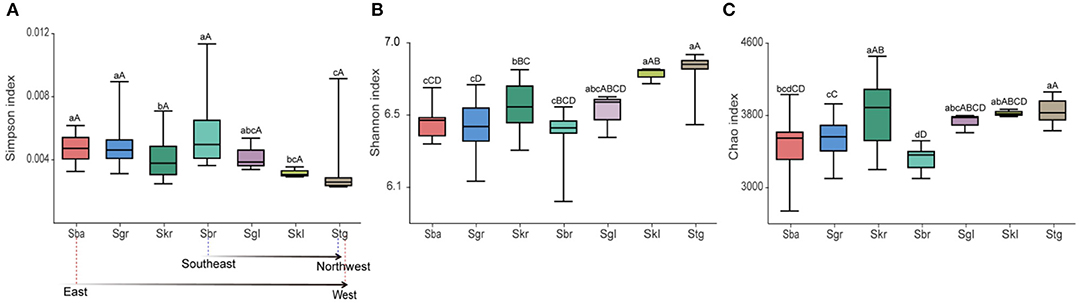
Figure 4. Alpha-diversity indices of bacterial communities in root-zone soils of seven Stipa species along a transect from east to west. Different letters (a–g, p < 0.05/A–D, p < 0.01) above the bars show significant difference among groups based on one-way ANOVA. (A) Simpson index expresses microbial diversity. (B) Shannon index expresses microbial evenness. (C) Chao index expresses microbial richness. S. baicalensis (Sba), S. grandis (Sgr), S. krylovii (Skr), S. glareosa (Sgl), S. klemenzii (Skl), S. breviflora (Sbr), and S. gobica (Stg).
NMDS analysis revealed a clear separation of the bacterial communities from different Stipa taxa (Figure 5B), and Stipa population specificity was further confirmed with a PERMANOVA (p = 0.001, Supplementary Table S5). In addition, the UPGMA clustering analysis also revealed similarity distribution in the bacterial community composition based on sample sites (Figure 5A). The samples formed two clear large clusters; moreover, six clear small clusters (I–VI) were generated, and two of these included only from Sba (VI) and Sgr (IV). The bacterial community was roughly distributed from southeast (VI) to northwest (I) along the reduced precipitation gradient and increased temperature gradient (Figures 5C,D). MAP (R2 = 0.78, p < 0.01) was more dominant than the Stipa taxa (R2 = 0.71, p < 0.01) for the distribution of bacterial community, which was indicated by the NMDS plot ordination analysis (Supplementary Table S7). In addition, soil pH (R2 = 0.63, p < 0.01), vegetation coverage (R2 = 0.64, p < 0.01), and MAT (R2 = 0.68, p < 0.01) also had a strong effect on the distribution of the bacterial communities across the geographical replacement gradient of the Stipa taxa (Supplementary Table S7). The mentioned results indicate that the composition of root-zone bacterial community not only had the Stipa population specificity, but also was strongly regulated by climate, and had a change pattern along the hydrothermal gradient. Especially in the process of transition from Sba to Sgr and then to Skr, root-zone bacteria distribution has a certain phenomenon of cross overlap.
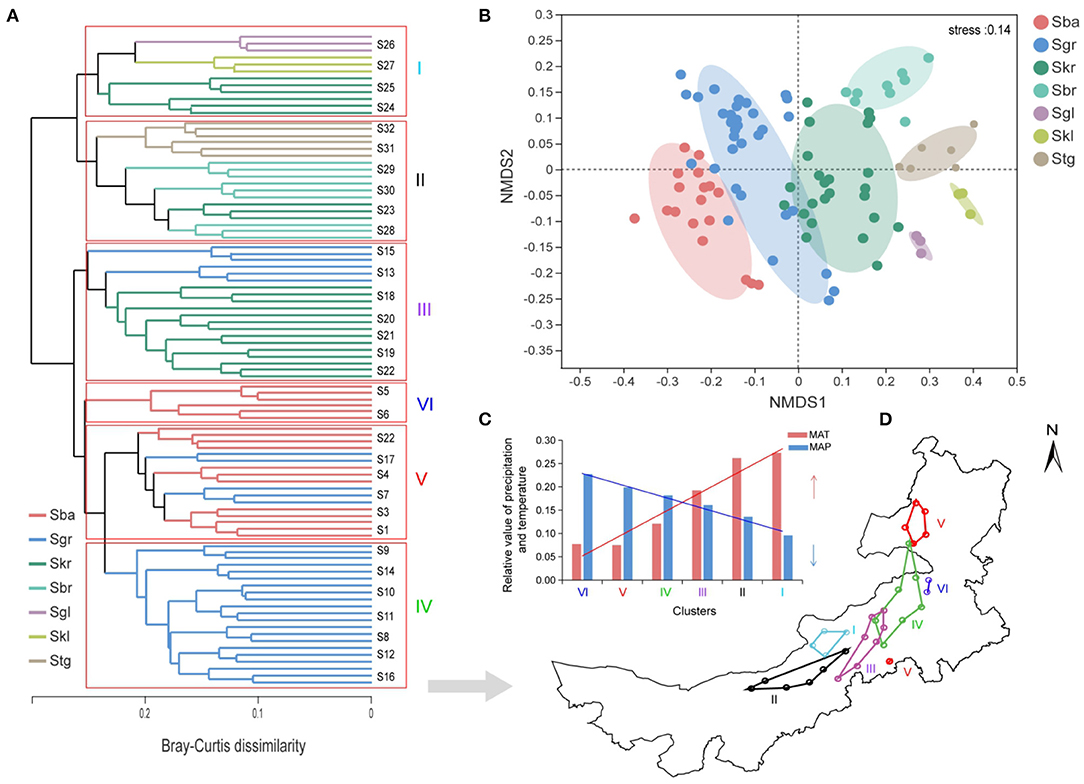
Figure 5. Bacterial community differentiation in the root-zone soils from seven Stipa species sampled from different sites of arid and semi-arid steppe. (A) Similarity analysis of bacterial community in Stipa taxa root zone based on UPGMA clustering using Bray–Curtis distances, clustered into six clusters (I–VI), roman numeral color corresponds to (C). (B) Population specificity of Stipa root-zone soil bacterial communities. (C) Columnar trend graphs of relative mean annual precipitation (MAP) and temperature (MAT) in different clusters; roman numerals correspond to (A,D). (D) Biogeographical patterns of bacterial communities, with same color clustering. S. baicalensis (Sba), S. grandis (Sgr), S. krylovii (Skr), S. glareosa (Sgl), S. klemenzii (Skl), S. breviflora (Sbr), and S. gobica (Stg).
Three redundant variables (-N, SOM, and Mg) were eliminated based on VIF analysis to reveal the major environmental variables shaping bacterial community composition and turnover. RDA and Mantel test were conducted to distinguish the impact of soil properties, climate, and vegetation factors on bacterial communities. RDA1 and RDA2 axis, respectively, explained 22.54 and 9.97% of the variance in soil bacterial community structures (Supplementary Figure S2A). The Mantel tests (Figure 6) denoted that the compositions of soil bacterial communities were significantly constructed by MAP (r = 0.58), MAT (r = 0.65), pH (r = 0.50), and VC (r = 0.53). These results were also supported by the RDA (Supplementary Figure S2A). The soil chemical elements (i.e., S, Fe, and Cu) also had an effect on the bacterial community structure, but not on the bacterial community distribution (Figure 6, Supplementary Figure S2A, and Supplementary Table S7). SOC, TN, and SB were significantly related to the spatial turnover and distribution of the root-zone soil bacterial community (Figure 6, Supplementary Figure S2A, and Supplementary Table S7). In accordance with the RDA examined, the bacteria phyla at seven Stipa root zones were mainly effected by VC (conditional effect = R2 = 0.26, p = 0.001), SB (R2 = 0.22, p = 0.001), and MAT (R2 = 0.19, p = 0.001; Supplementary Figure S2B). At genus level, MAP (conditional effect = R2 = 0.60, p =0.001), MAT (R2 = 0.52, p = 0.001), MAT (R2 = 0.51, p = 0.001), pH (R2 = 0.41, p = 0.001), SB (R2 = 0.39, p = 0.001), SOC (R2 = 0.38, p = 0.001), and TN (R2 = 0.38, p = 0.001) affected bacteria, and all bacteria with relative abundance >1% could be explained by one soil property at least (Supplementary Figure S2C). The correlation heatmap denoted the relationship between relative abundance of bacterial keystone taxa with environmental factor (Figures 2C,D). It was found that the relative abundances of most dominant bacterial phyla and genus were significantly related to most soil physicochemical properties (i.e., soil pH, TN, AP, AK, S, Zn, Mn, Fe, Cu, and SOC), plant factors (i.e., SB and VC), and other indicators (i.e., MAT, MAP, and altitude). Furthermore, Firmicutes was significantly positively correlated with soil chemical elements (except S); on the contrary, Planctomycetes was significantly negatively correlated.
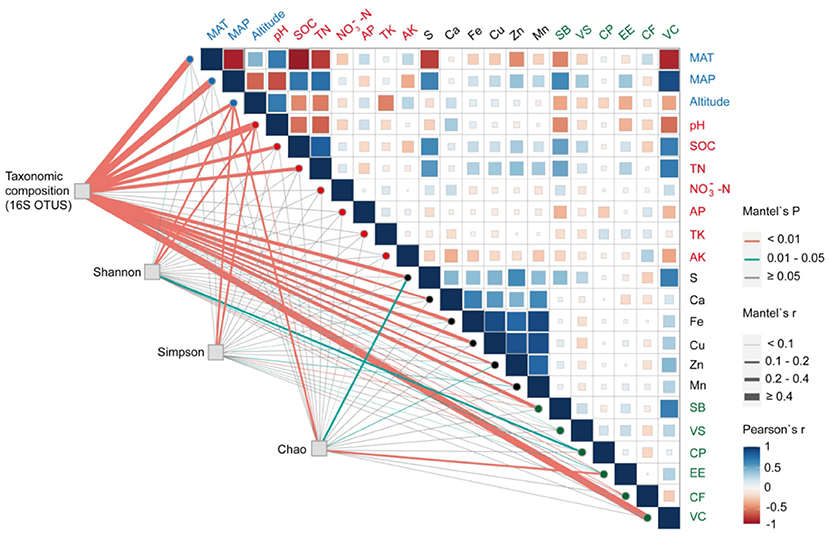
Figure 6. Mantel correlations among bacterial community composition (OTU level), bacterial richness (Chao index), bacterial diversity (Simpson index), bacterial evenness (Shannon index), and environmental factors (climate, plant, and soil variables); Pearson correlation analysis was used among environmental factors. The width of edges represents the size of the correlation coefficient (Mantel's r), while edge color represents the statistical significance based on Mantel's p. Abbreviations are consistent with Figure 2.
To explore the complex bacterial taxa interactions in different Stipa taxa root-zone soils and to identify the response to environmental change, a co-occurrence network analysis for abundant OTUs (top 300 OTUs) was performed. The Sgl, Stg, and Skl root-zone soils were excluded from the analyses because of limited samples. Based on the number of links (Sba = 648, Sgr = 527), avgK (Sba = 7.16, Sgr = 6.47), and the avgCC (Sba = 0.38, Sgr = 0.33), the network complexity was comparable between the Sba and Sgr root-zone soils, with nodes in the control network, respectively, grouped into 21 and 25 modules with respective quality values of 0.59 and 0.38 (Figures 7A,B and Supplementary Table S8). Moreover, the complexity (links = 226, avgK = 2.83, avgCC = 0.32) was less in Skr root-zone soils (Figure 7C), and the network grouped into 22 (quality value of 0.81) modules. However, the complexity (links = 2593, avgK = 18.13, avgCC = 0.49) greatly increased in Sbr root-zone soils (Figure 7D), which contained six modules, with a quality value of 0.49. There were absolutely more positive correlations in each network than negative correlations. R square of power-law values of Sba, Sgr, and Skr networks were obtained for 0.87, 0.78, and 0.76, respectively, indicating the occurrence of scale-free network characteristics (Supplementary Table S8).
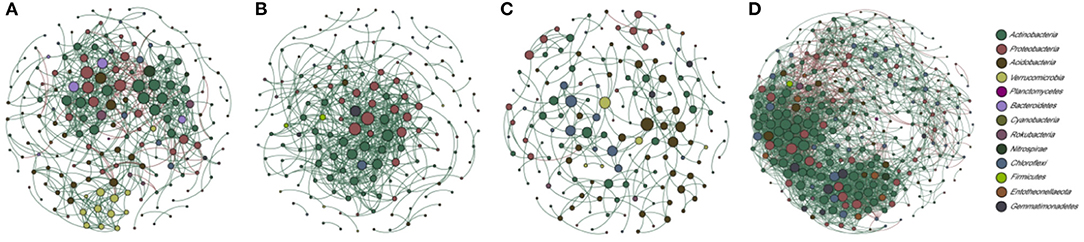
Figure 7. Co-occurrence networks of the high-abundant bacteria from four Stipa species root-zone soil; in turn is S. baicalensis (Sba: A), S. grandis (Sgr: B), S. krylovii (Skr: C), and S. breviflora (Sbr: D). A connection stands for a robust (Spearman's r > |0.82|) and significant (p < 0.05) correlation. Green and red lines represent positive and negative associations between two nodes, respectively. Nodes are colored according to microbial phylum and the size of each node is proportional to the degree.
The βNTI values were determined, to report that the stochastic process was dominant in the bacterial communities of seven Stipa species root-zone soils (Supplementary Figure S3A; values >-2 and < 2). We calculated the taxonomic-diversity metric RCBray to further quantify the relative effects of various ecological processes, including homogenizing dispersal, dispersal limitation and undominated processes, on the stochastic community assembly of every group (Supplementary Figure S3B). Consequently, the relative effects of dispersal limitation (24–49%) and undominated processes (27–41%) increased significantly across the transition from Sba to Skr, while the relative role of homogenizing dispersal (10–48%) was significantly reduced. In arid areas, the relative role of homogenizing dispersal (33–100%) was dominant in the bacterial community assembly in the root zone of Stipa species, except for Stg. Overall, the relative effects of dispersal limitation (73%) of Stipa genus root-zone soil bacteria were the most important in the grassland system.
Diverse bacterial composition and structure were detected in root-zone soils when the Stipa taxa replace each other in space. According to the dominant bacteria taxon, Actinobacteria achieved the maximal relative abundance in most Stipa species root-zone soils (except for Sgl), which might be attributed to the climate and the soil characteristics of the research area. The members of the bacterial phylum Actinobacteria have been proven as an indicator of drought sensitivity and drought tolerance, and exhibit a strong metabolic capacity at low temperatures (Cheng et al., 2021). Given the Circos analysis, the top two Stipa taxa in Acidobacteria group were Sgl and Skl, which were located in the desert steppe that was characterized by low soil nutrition; in addition, Acidobacteria was the most dominant phylum in the Sgl root-zone soil; Acidobacteria was composed of various oligotrophs (Zeng et al., 2019), capable of tolerating harsh conditions (Yao et al., 2017). The phylum Proteobacteria comprises diverse groups of copiotrophic organisms in soil normally identified under nutrient-rich conditions (Janssen, 2006). The nutrient richness of root-zone soil of Sba is much higher than for other Stipa taxa (Supplementary Table S3), and the relative abundance of the phylum Proteobacteria decreased significantly in the process of Sbr replacing Sba. Moreover, the phylum Proteobacteria was negatively correlated with MAT, pH, and AP. The relative abundance of Proteobacteria in Sbr root zone was lower than that of several Stipa taxa (Sgl, Skl, and Stg) with higher content of AP. Root-zone bacterial phyla and genera of the Stipa varied significantly following the vegetation replacement distribution gradient (Supplementary Table S6). For instance, when Sba was replaced by Sbr from east to west following the transect, the relative abundance of the phylum Chloroflexi and the genus Rubrobacter was improved significantly, and Rubrobacter was reported as the most dominant genus in Sbr root-zone soil (Supplementary Table S6 and Figure 2). The phylum Chloroflexi pertains to the group of heterotrophic oligotrophs in soil (Hug et al., 2013), and the genus Rubrobacter is significantly radiation resistant, halotolerant, thermotolerant, or even thermophilic (Kourilova et al., 2021). Most bacteria exhibited a relatively narrow growth tolerance, thereby causing individual taxa to be unable to adapt to soil pH variations (Fierer and Jackson, 2006), and then probably inducing variations of soil bacterial communities (Shen et al., 2013). pH (near neutral range) is suggested to exert a mediating effect in grassland system (Tripathi et al., 2018). This may explain why the relative abundance of Actinobacteria and Acidobacteria in this paper was considerable, whereas this has no significant correlation with neutral soil pH, while the areas exhibiting an acidic soil pH showed a significant correlation [e.g., the bacterial community in Changbai Mountain (Han et al., 2018)]. Together with the significant correlations between soil pH and SOC, TN and -N of root-zone process of grassland might explain why soil pH affected the root-zone bacterial community by affecting the availability of soil carbon and nitrogen or the number of biogeochemical processes involving carbon and nitrogen cycles (Jiao and Lu, 2020), thereby complying consistent with the research on Caragana spp. rhizosphere soil (Na et al., 2018). All the mentioned results collectively suggested that the variations of whole bacterial communities are strongly ecological-variable dependent and various phyla and genus may show different variation patterns.
Besides the differences in the relative abundance of the identical bacterial taxa, soils in different Stipa root zones also harbor different potential biomarkers (Figure 3), which have been characteristically isolated from soil samples or plant root systems. The Skl only had two significant taxa, Bacteroidia (class) and Bacteroidetes (phylum), that were involved in C and N metabolism and occurring extensively across various ecological niches (Wang et al., 2020). Several species of the family Sphingomonadaceae (Sphingomonadales) utilize a wide range of organic compounds for growth and survival under low-nutrient conditions (Glaeser and Kämpfer, 2014). The family Chthoniobacteraceae (Chthoniobacterales), represented by the genera Chthoniobacter and Candidatus Xiphinematobacter, can metabolize organic carbon (Victoria et al., 2012) and prevent nutrient leaching and erosion (Kant et al., 2011). The order Betaproteobacteriales exhibited a large relative abundance in these bacterial community studies. However, it is significantly enriched in the Sba than other Stipa taxa. Given the vulnerability of several species pertaining to Betaproteobacteriales to salinity, acidity, and grazing pressure, Sba achieves a better growth and living environment than other Stipa taxa. Moreover, Sba itself is easily replaced in heavily grazing and salinized areas. Several species of the order Thermomicrobiales (the family and genus of JG30_KF_CM45) are characterized as thermophilic, neutral, and basophilic (Bergey's Manual of Systematics of Archaea and Bacteria). Norank_f__JG30_KF_CM45 acts as a potential genus biomarker for the Sbr, with a significant positive correlation with MAT, pH, and AP, but a negative correlation with MAP, SOC, TN, and S (Figure 2B), while Candidatus_Udaeobacter (Sba) and norank_f__Xanthobacteraceae (Sgr) exhibit opposite behaviors (Figure 2B). The mentioned explains why the warm-loving, drought-resistant, alkali-resistant, and barren-resistant norank_f__JG30_KF_CM45 is enriched, which may indicate available phosphorus transformation and utilization. The critical thing is that Sbr also likes warmth (Chen et al., 2020). Candidatus_Udaeobacter and norank_f__Xanthobacteraceae act as potential biomarkers for the Sba and Sgr, respectively, which might indicate vital ecosystem multifunctionality involved in SOC accumulation, S cycle, and N stock across the chronosequence (Li et al., 2021). The family Xanthobacteraceae (Rhizobiales and norank_f__Xanthobacteraceae) in the Sgr grows with aerobic chemoheterotrophs and fix nitrogen (Oren, 2014; Yu et al., 2019). The mentioned functional bacteria in root zone can directly indicate the soil nutrient transformation and the response of bacterial to environmental variations.
Climate has been recognized as a vital driver of soil bacterial community structure in regional and large-scale studies (Zeng et al., 2019). Yao proved climate drives the differentiation of bulk soil bacterial communities in the eastern Inner Mongolia steppe (Yao et al., 2017). Likewise, temperature and precipitation are two major climatic factors affecting the Stipa species root-zone soil bacterial community structure and distribution in this paper. This is related to the influence of precipitation on plant community composition and soil nutrient availability. For instance, higher precipitation may upregulate soil organic matter decomposition rate, thereby reducing soil organic matter availability (Tian et al., 2017; Na et al., 2019), and increasing microbial metabolic rates and biochemical processes due to temperature (Zhou et al., 2016), which subsequently affects the soil bacterial community structure. Furthermore, the spatial pattern of root-zone soil bacterial communities was distributed along the hydrothermal combination gradient. The mentioned result was attributed to the long-term influence of the southeast ocean monsoon and the northeast–southwest arc mountain barriers (Greater Khingan Mountains and the Yin Mountains), thereby causing the climatic factors to form an arc belt distribution. Such a peculiar water–heat combination causes the zonation of soil and vegetation, which makes the differentiation of vegetation zones in this area roughly coincide with the distribution of the climatic zones (Han and Tian, 2016).
Plant and soil parameters were other crucial factors of the bacterial community structure and distribution. As indicated from existing studies, soil pH exerts a dominant effect on various ecosystems in shaping bacterial structures and large-scale spatial distributions (Fierer and Jackson, 2006; Fan et al., 2017), mainly in acidic and neutral soil environments (Yang et al., 2018). Different soil properties play a leading role in an alkaline environment (Wang et al., 2017); for example, the soil carbon content was the dominant factor of bacteria distribution in the Ali area (Chu et al., 2016). The result of this paper partially complies with the existing studies indicating that soil pH was the most crucial soil parameter, whereas SOC, SOM, TN, and -N significantly affected the structure and distribution of Stipa taxa root-zone soil bacterial communities. In this paper, chemical element (S, Fe, and Cu) also impacted the bacterial community structure, whereas it did not impact the bacterial community distribution except for S (Figure 6 and Supplementary Table S7). Thus, the soil sulfur cycle plays a partial role in the ecological distribution of bacteria, whereas microbes in rhizosphere are critical to allowing plants to access soil organosulfur (Kertesz et al., 2007). This paper also reported that the plant coverage and biomass of Stipa had a significant impact on the distribution and structure of the bacterial community at the phylum and genus levels were particularly prominent (Supplementary Figures S2, S6); complying with Han's view (Han et al., 2018), while the species of Stipa significantly impacted the distribution of bacteria. This is because a higher plant biomass and differences in the plant cover are generally directly affecting the quality and quantity of organic matter input, with subsequent effects on bacterial communities (Fierer and Jackson, 2006). In arid systems, the changes in plant coverage are also especially important for nutrient and moisture maintenance (Yao et al., 2017). Furthermore, different vegetation types can determine litter composition, soil conditions, and resource acquisition strategies, thereby indirectly regulating the structure of the bacterial community through a superposition effect with soil factors (Yao et al., 2018). In contrast, the soil microbial community structure will also affect the natural selection pattern of plant traits, while regulating the response of plants to abiotic environmental pressure, thereby affecting the evolutionary process of the whole ecosystem (Berg and Smalla, 2009). Besides, high interspecific clustering of the bacterial root-zone communities was identified. Such an interspecific difference could be partially determined by the genetic diversity among host species (Yu and Hochholdinger, 2018).
On the whole, the effect of precipitation and Stipa species on the structure and distribution of the root-zone soil bacterial community exceeded that of soil pH on a large scale. It is noteworthy that the bacterial community composition in the root zone of Stipa taxa complied with the previous reports of bulk or rhizosphere soil in a semi-arid system (Wang et al., 2015; Nan et al., 2020), while the bacteria community structure was different. Many studies have confirmed that the rhizosphere microbiome consists of a subset of the bulk soil microbiome; besides, microbial community diversity decreased rapidly from bulk soil to rhizosphere (Reinhold-Hurek et al., 2015; Trivedi et al., 2020). During the assembly of microbial communities from bulk soil to rhizosphere, the bulk soil provides a microbial seed bank, the physical–chemical properties, the biogeography, and climate conditions (Zhang et al., 2017). Based on reasons discussed previously, the differences observed in the root zone of Stipa also might be mediated by the bulk soil microbiota in their respective regions.
The diversity, evenness, and richness of rhizosphere bacteria were inconsistent in the seven plant species. There was a sudden change in the root-zone soil bacterial diversity at Sbr in desert steppe. According to Dijkstra et al. (2012), bacterial diversity changed suddenly in arid areas, and soil microbes living in zones with scarce water availability might be easily activated by even small rainfall events, which did not reach a level that would satisfy the plants' needs. As reported from numerous studies, soil pH acts as a vital factor driving the diversity of rhizosphere soil bacterial community (Na et al., 2018; Wang et al., 2020). This paper further examined the previously found strong correlation between soil pH and bacterial root-zone community diversity (Figure 6), as the pH conditions affect the adaptation and selection of particular phylogenic groups (Wang et al., 2020). In turn, bacterial α diversity, as indicated by the Shannon and Chao indexes, significantly affects the nutrient composition of forage grasses, especially the accumulation of crude fat and crude protein, whereas the correlation coefficient is relatively small. In addition, the elevation is correlated with the variables (SOC, TN, TK, and pH) affecting the ecosystem and plays a key role in influencing root-zone bacterial diversity, evenness, and richness. Wang reported that microbial richness in the Qinghai–Tibet Plateau showed a significant negative correlation with altitude. The pattern of microbial diversity and altitude was inconsistent due to the differences in the primary factors regulating the bacterial diversity in soils from different regions (Wang et al., 2020). Furthermore, this paper reported that root-zone soil bacterial α diversity was positively correlated with altitude.
Both deterministic and stochastic processes were governing the spatial distribution of microbial communities concurrently, and different processes were found to be dominant in various cases (Zhang et al., 2016). The root-zone bacterial community assembly was dominated by stochastic processes at Stipa taxa, which also complied with the results of several studies, i.e., the effects of stochastic processes were commonly dominant at larger geographic scales (Li et al., 2021). Previous research has shown that the spatio-temporal distribution of soil bacteria in a temperate grassland was dominated by stochastic dispersal (Richter-Heitmann et al., 2020). According to other studies, the variation of microbial community structure will be dominated by different assembly factors in different periods (Zhou et al., 2014; Liu et al., 2020). The results of this paper may only represent the assembly of Stipa rhizosphere microorganisms in such a period, which can be exploited to determine the stability of different Stipa rhizosphere bacterial communities of the period (Zhou and Ning, 2017). The significant variations of drought span and soil structure in the study area may impose the natural limitation on the undirected dispersal of microbes like others have reported (Li and Hu, 2021), which also explains why the relative effects of dispersal limitation of Stipa genus root-zone soil bacteria showed the absolute advantage in the study. As expected, the relative effects of dispersal limitation and undominated processes increased significantly across the transition from Sba to Skr, while the relative role of homogenizing dispersal decreased significantly (Supplementary Figure S3B), thereby demonstrating that the bacterial community structure tended to be increasingly stable, and environmental variations exerted fewer effects (Zhou and Ning, 2017). Homogeneous selection was the main process driving the assembly of Sbr, Sgl, and Skl bacterial communities, showing that community structure would change largely with environmental conditions (Cheng et al., 2021).
This paper reported that the potential correlation patterns of different Stipa taxa root-zone bacterial groups varied substantially for network complexity and modularity. Moreover, the M, GD, and avgCC of the empirical networks were clearly higher than those of the respective random networks (Supplementary Table S8), suggesting that empirical networks had a noticeable hierarchy and modularity in their topological properties. Some studies have reported that the interactions of root-associated microbes are more complex than those of microbes in bulk soil, which may be related to rhizosphere microbial diversity, and the main interactions are positive (>80%), which also indicates that rhizosphere may have greater potential for cooperative or mutualistic associations (Coyte et al., 2015; Fan et al., 2018), and so are the root zones of Stipa, because four networks had a large proportion of members that are connected through positive links (Supplementary Table S8). However, such bacterial community is considered unstable since bacterial taxa may make a coincident response to environmental fluctuations, thereby inducing positive feedback and co-oscillation (Coyte et al., 2015). According to the bacterial co-occurrence pattern characterized by a high average degree, an average clustering coefficient, and fewer modules and modularity at extremely arid areas indicated that the networks of Sbr root-zone soil exhibit a more complex bacterial community structure with closer and better-connected nodes, whereas such topological features are of low stability (De Vries et al., 2018). Nevertheless, the Skr network was simplest but most stable based on topological characteristics. This network exhibited stronger resistance to environmental variations. Skr is easy to replace other Stipa taxa when the growth environment is stressed, thereby also indicating that the population can strongly adapt to the environment. This also reveals that there may be potential co-evolution relationship between vegetation and their root-zone bacteria. In summary, network analysis and RCbray results exert a good indication and prediction effect on environmental interference. More network complexity and fewer modules were reported in the severe environment than in the mild environment, thereby demonstrating that network complexity of bacteria was facilitated by various environmental factors, whereas the clustering of modules was not.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
XM: investigation, data curation, methodology, visualization, writing—original draft, writing—review, and editing. LC, SW, and JL: investigation. ZD and JL: data curation. FL: field investigation. YB: funding acquisition, supervision, resources, review, and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (31760005) and Science and Technology Major Project of Inner Mongolia Autonomous Region of China (Grant No. zdzx2018065).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the reviewers who helped improve the article with constructive comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.782621/full#supplementary-material
Bai, Y. F., Xu, Z. X., Li, D. X., and Zhao, G. (1999). Study on age and bunch structure of four stipa species in inner mongolia plateau. Acta Bot. Sin. 41, 1125–1131.
Basirat, M., Mousavi, S. M., Abbaszadeh, S., Ebrahimi, M., and Zarebanadkouki, M. (2019). The rhizosheath: a potential root trait helping plants to tolerate drought stress. Plant Soil 445, 565–575. doi: 10.1007/s11104-019-04334-0
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Chen, L., Saixi, Y., Yi, R., and Baoyin, T. (2020). Characterization of soil microbes associated with a grazing-tolerant grass species, Stipa breviflora, in the Inner Mongolian desert steppe. Ecol. Evol. 10, 10607–10618. doi: 10.1002/ece3.6715
Chen, S. H. (1987). Root System Types of Steppe Plants in Inner Mongolia. Inner Mongolia: Inner Mongolia People's Publishing House. p. 42–50.
Chen, Z., Wu, W., Shao, X., Li, L., Guo, Y., and Ding, G. (2015). Shifts in abundance and diversity of soil ammonia-oxidizing bacteria and archaea associated with land restoration in a semi-arid ecosystem. PLoS ONE 10:e0132879. doi: 10.1371/journal.pone.0132879
Cheng, X., Yun, Y., Wang, H., Ma, L., Tian, W., Man, B., et al. (2021). Contrasting bacterial communities and their assembly processes in karst soils under different land use. Sci. Total Environ. 751:142263. doi: 10.1016/j.scitotenv.2020.142263
Chu, H., Gao, G. F., Ma, Y., Fan, K., and Delgado-Baquerizo, M. (2020). Soil microbial biogeography in a changing world: recent advances and future perspectives. mSystems 5:e00803–19. doi: 10.1128/mSystems.00803-19
Chu, H., Sun, H., Tripathi, B. M., Adams, J. M., Huang, R., Zhang, Y., et al. (2016). Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environ. Microbiol. 18, 1523–1533. doi: 10.1111/1462-2920.13236
Clairmont, L. K., Stevens, K. J., and Slawson, M. R. (2019). Site-specific differences in microbial community structure and function within the rhizosphere and rhizoplane of wetland plants is plant species dependent. Rhizosphere 9, 56–68. doi: 10.1016/j.rhisph.2018.11.006
Corneo, P. E., Pellegrini, A., Cappellin, L., Roncador, M., Chierici, M., Gessler, C., et al. (2013). Microbial community structure in vineyard soils across altitudinal gradients and in different seasons. FEMS Microbiol. Ecol. 84, 588–602. doi: 10.1111/1574-6941.12087
Coyte, K. Z., Schluter, J., and Foster, R. K. (2015). The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
De Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. doi: 10.1038/s41467-018-05516-7
Deng, Y., Jiang, Y. H., Yang, Y. F., He, Z. L., Luo, F., and Zhou, Z. J. (2012). Molecular ecological network analyses. BioMed. Central 13:113. doi: 10.1186/1471-2105-13-113
Dijkstra, F. A., Augustine, D. J., Brewer, P., and von Fischer, C. J. (2012). Nitrogen cycling and water pulses in semiarid grasslands: are microbial and plant processes temporally asynchronous? Oecologia 170, 799–808. doi: 10.1007/s00442-012-2336-6
Dini-Andreote, F., Stegen, J. C., van Elsas, J. D., and Salles, F. J. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U.S.A. 112, E1326–E1332. doi: 10.1073/pnas.1414261112
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Fan, K., Cardona, C., Li, Y., Shi, Y., Xiang, X., Shen, C., et al. (2017). Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol. Biochem. 113, 275–284. doi: 10.1016/j.soilbio.2017.06.020
Fan, K., Weisenhorn, P., Gilbert, J. A., and Chu, H. (2018). Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 125, 251–260. doi: 10.1016/j.soilbio.2018.07.022
Fan, X., and Xing, P. (2016). The vertical distribution of sediment archaeal community in the “black bloom” disturbing Zhushan Bay of Lake Taihu. Archaea 2016:8232135. doi: 10.1155/2016/8232135
Fierer, N., and Jackson, B. R. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Gao, S. B., Mo, L. D., Zhang, L. H., Zhang, J. L., Wu, J. B., Wang, J. L., et al. (2018). Phenotypic plasticity vs. local adaptation in quantitative traits differences of Stipa grandis in semi-arid steppe, China. Sci. Rep. 8:3148. doi: 10.1038/s41598-018-21557-w
Glaeser, S. P., and Kämpfer, P. (2014). “The family sphingomonadaceae,” in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer Berlin Heidelberg), 641–707.
Glasl, B., Bourne, D. G., Frade, P. R., Thomas, T., Schaffelke, B., and Webster, S. N. (2019). Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 7:94. doi: 10.1186/s40168-019-0705-7
Han, B., and Tian, S. Q. (2016). Ecological Adaptability of Main Stipa Plants in Inner Mongolia. Beijing: China Agricultural Science and Technology Press.
Han, D., Wang, N., Sun, X., Hu, Y., and Feng, F. (2018). Biogeographical distribution of bacterial communities in Changbai Mountain, Northeast China. Microbiol. Open 7:e00529. doi: 10.1002/mbo3.529
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. doi: 10.1186/2049-2618-1-22
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006
Jiao, S., and Lu, Y. (2020). Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 22, 1052–1065. doi: 10.1111/1462-2920.14815
Kang, L., Han, X., Zhang, Z., and Sun, J. O. (2007). Grassland ecosystems in China: review of current knowledge and research advancement. Philos. Trans. R. Soc. Lond,. B,. Biol. Sci. 362, 997–1008. doi: 10.1098/rstb.2007.2029
Kant, R., van Passel, M. W., Palva, A., Lucas, S., Lapidus, A., Glavina del Rio, T., et al. (2011). Genome sequence of Chthoniobacter flavus Ellin428, an aerobic heterotrophic soil bacterium. J. Bacteriol. 193, 2902–2903. doi: 10.1128/JB.00295-11
Kertesz, M. A., Fellows, E., and Schmalenberger, A. (2007). Rhizobacteria and plant sulfur supply. Adv. Appl. Microbiol. 62, 235–268. doi: 10.1016/S0065-2164(07)62008-5
Kourilova, X., Schwarzerova, J., Pernicova, I., Sedlar, K., Mrazova, K., Krzyzanek, V., et al. (2021). The first insight into polyhydroxyalkanoates accumulation in multi-extremophilic Rubrobacter xylanophilus and Rubrobacter spartanus. Microorganisms 9:909. doi: 10.3390/microorganisms9050909
Krishna, M., Gupta, S., Delgado-Baquerizo, M., Morrien, E., Garkoti, S. C., Chaturvedi, R., et al. (2020). Successional trajectory of bacterial communities in soil are shaped by plant-driven changes during secondary succession. Sci. Rep. 10:9864. doi: 10.1038/s41598-020-66638-x
Kuczynski, J., Stombaugh, J., Walters, W. A., Gonzalez, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics Chapter 10:Unit 107. doi: 10.1002/0471250953.bi1007s36
Lakshmanan, V., Selvaraj, G., and Bais, P. H. (2014). Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 166, 689–700. doi: 10.1104/pp.114.245811
Lau, J. A., and Lennon, T. J. (2011). Evolutionary ecology of plant-microbe interactions: soil microbial structure alters selection on plant traits. New Phytol. 192, 215–224. doi: 10.1111/j.1469-8137.2011.03790.x
Li, M., Mi, T., He, H., Chen, Y., Zhen, Y., and Yu, Z. (2021). Active bacterial and archaeal communities in coastal sediments: biogeography pattern, assembly process and co-occurrence relationship. Sci. Total Environ. 750:142252. doi: 10.1016/j.scitotenv.2020.142252
Li, X., Wang, J., Zhang, S., Wang, H., Li, X., Li, X., et al. (2018). Distribution of fungal endophytes in roots of Stipa krylovii across six vegetation types in grassland of northern China. Fungal Ecol. 31, 47–53. doi: 10.1016/j.funeco.2017.11.001
Li, Y., and Hu, C. (2021). Biogeographical patterns and mechanisms of microbial community assembly that underlie successional biocrusts across northern China. NPJ Biofilms Microbiomes 7:15. doi: 10.1038/s41522-021-00188-6
Liu, C., Wentworth, T. R., Qiao, X., Guo, K., and Hou, D. (2019). Vegetation classification at the association level under the China Vegetation Classification System: an example of six Stipa steppe formations in China. J. Plant Ecol. 12, 1009–1024. doi: 10.1093/jpe/rtz028
Liu, M., Li, X., Zhu, R., Chen, N., Ding, L., and Chen, C. (2021). Vegetation richness, species identity and soil nutrients drive the shifts in soil bacterial communities during restoration process. Environ. Microbiol. Rep. 13, 411–424. doi: 10.1111/1758-2229.12913
Liu, W., Graham, E. B., Zhong, L., Zhang, J., Li, S., Lin, X., et al. (2020). Long-term stochasticity combines with short-term variability in assembly processes to underlie rice paddy sustainability. Front. Microbiol. 11:873. doi: 10.3389/fmicb.2020.00873
Lv, X., Zhou, G., Wang, Y., and Song, X. (2016). Sensitive indicators of zonal stipa species to changing temperature and precipitation in Inner Mongolia Grassland, China. Front. Plant Sci. 7:73. doi: 10.3389/fpls.2016.00073
Maestre, F. T., Delgado-Baquerizo, M., Jeffries, T. C., Eldridge, D. J., Ochoa, V., Gozalo, B., et al. (2015). Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. U.S.A. 112, 15684–15689. doi: 10.1073/pnas.1516684112
Marasco, R., Mosqueira, M. J., Fusi, M., Ramond, J. B., Merlino, G., Booth, J. M., et al. (2018). Rhizosheath microbial community assembly of sympatric desert speargrasses is independent of the plant host. Microbiome 6:215. doi: 10.1186/s40168-018-0597-y
Mohanram, S., and Kumar, P. (2019). Rhizosphere microbiome: revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 69, 307–320. doi: 10.1007/s13213-019-01448-9
Na, X., Xu, T., Li, M., Zhou, Z., Ma, S., Wang, J., et al. (2018). Variations of bacterial community diversity within the rhizosphere of three phylogenetically related perennial shrub plant species across environmental gradients. Front. Microbiol. 9:709. doi: 10.3389/fmicb.2018.00709
Na, X., Yu, H., Wang, P., Zhu, W., Niu, Y., and Huang, J. (2019). Vegetation biomass and soil moisture coregulate bacterial community succession under altered precipitation regimes in a desert steppe in northwestern China. Soil Biol. Biochem. 136:107520. doi: 10.1016/j.soilbio.2019.107520
Nan, J., Chao, L., Ma, X., Xu, D., Mo, L., Zhang, X., et al. (2020). Microbial diversity in the rhizosphere soils of three Stipa species from the eastern Inner Mongolian grasslands. Global Ecol. Conserv. 22:e00992. doi: 10.1016/j.gecco.2020.e00992
Oksanen, J., Blanchet, F. G., Kindt, R., and Minchin, R. P. (2015). Vegan: Community Ecology Package. R Package Version 22–1.
Oren, A. (2014). “The family xanthobacteraceae,” in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer Berlin Heidelberg), 709–726.
Perez-Jaramillo, J. E., de Hollander, M., Ramirez, C. A., Mendes, R., Raaijmakers, J. M., and Carrion, J. V. (2019). Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 7:114. doi: 10.1186/s40168-019-0727-1
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, H. W. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Reinhold-Hurek, B., Bunger, W., Burbano, C. S., Sabale, M., and Hurek, T. (2015). Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Richter-Heitmann, T., Hofner, B., Krah, F.-S., Sikorski, J., Wüst, P. K., Bunk, B., et al. (2020). Stochastic dispersal rather than deterministic selection explains the spatio-temporal distribution of soil bacteria in a temperate grassland. Front. Microbiol. 11:1391. doi: 10.3389/fmicb.2020.01391
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shen, C., Xiong, J., Zhang, H., Feng, Y., Lin, X., Li, X., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211. doi: 10.1016/j.soilbio.2012.07.013
Shi, W., Li, M., Wei, G., Tian, R., Li, C., Wang, B., et al. (2019). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 7:14. doi: 10.1186/s40168-019-0629-2
Song, Z., Liu, G., and Zhang, C. (2019). Response of rhizosphere microbial communities to plant succession along a grassland chronosequence in a semiarid area. J. Soils Sediments 19, 2496–2508. doi: 10.1007/s11368-019-02241-6
Stegen, J. C., Freestone, A. L., Crist, T. O., Anderson, M. J., Chase, J. M., Comita, L. S., et al. (2013). Stochastic and deterministic drivers of spatial and temporal turnover in breeding bird communities. Global Ecol. Biogeogr. 22, 202–212. doi: 10.1111/j.1466-8238.2012.00780.x
Tang, H., Zhang, N., Ni, H., Xu, X., Wang, X., Sui, Y., et al. (2021). Increasing environmental filtering of diazotrophic communities with a decade of latitudinal soil transplantation. Soil Biol. Biochem. 154:108119. doi: 10.1016/j.soilbio.2020.108119
Tian, J., He, N., Hale, L., Niu, S., Yu, G., Liu, Y., et al. (2017). Soil organic matter availability and climate drive latitudinal patterns in bacterial diversity from tropical to cold temperate forests. Funct. Ecol. 32, 61–70. doi: 10.1111/1365-2435.12952
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, K. Y. (2018). Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 12, 1072–1083. doi: 10.1038/s41396-018-0082-4
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, K. B. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Tu, B., Domene, X., Yao, M., Li, C., Zhang, S., Kou, Y., et al. (2017). Microbial diversity in Chinese temperate steppe: unveiling the most influential environmental drivers. FEMS Microbiol. Ecol. 93. doi: 10.1093/femsec/fix031
Verma, M., Mishra, J., and Arora, N. K. (2019). “Plant growth-promoting rhizobacteria: diversity and applications,” in Environmental Biotechnology: For Sustainable Future, eds R. A. B. Khalid, R. Hakeem, and m. A. Dervash (Cham: Springer), 129–173.
Victoria, R., Banwart, S. A., Black, H., Ingram, J., Joosten, H., Milne, E., et al. (2012). Benefits of Soil Carbon: Foresight Chapter in UNEP Yearbook 2012. (Philadelphia, PA: United Nations Environment Programme), 19–33.
Wang, S. K., Zuo, X. A., Awada, T., Medima-Roldán, E., Feng, K. T., Yue, P., et al. (2021). Changes of soil bacterial and fungal community structure along a natural aridity gradient in desert grassland ecosystems, Inner Mongolia. Catena 205:105470. doi: 10.1016/j.catena.2021.105470
Wang, X., Van Nostrand, J. D., Deng, Y., Lu, X., Wang, C., Zhou, J., et al. (2015). Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China's grasslands. FEMS Microbiol. Ecol. 91:fiv133. doi: 10.1093/femsec/fiv133
Wang, X., Zhang, Z., Yu, Z., Shen, G., Cheng, H., and Tao, S. (2020). Composition and diversity of soil microbial communities in the alpine wetland and alpine forest ecosystems on the Tibetan Plateau. Sci. Total Environ. 747:141358. doi: 10.1016/j.scitotenv.2020.141358
Wang, X. B., Lu, X. T., Yao, J., Wang, Z. W., Deng, Y., Cheng, W. X., et al. (2017). Habitat-specific patterns and drivers of bacterial beta-diversity in China's drylands. ISME J. 11, 1345–1358. doi: 10.1038/ismej.2017.11
Yang, F., Wu, J., Zhang, D., Chen, Q., Zhang, Q., and Cheng, X. (2018). Soil bacterial community composition and diversity in relation to edaphic properties and plant traits in grasslands of southern China. Appl. Soil Ecol. 128, 43–53. doi: 10.1016/j.apsoil.2018.04.001
Yao, M., Rui, J., Niu, H., Heděnec, P., Li, J., He, Z., et al. (2017). The differentiation of soil bacterial communities along a precipitation and temperature gradient in the eastern Inner Mongolia steppe. Catena 152, 47–56. doi: 10.1016/j.catena.2017.01.007
Yao, X., Zhang, N., Zeng, H., and Wang, W. (2018). Effects of soil depth and plant-soil interaction on microbial community in temperate grasslands of northern China. Sci. Total Environ. 630, 96–102. doi: 10.1016/j.scitotenv.2018.02.155
Yu, P., and Hochholdinger, F. (2018). The role of host genetic signatures on root-microbe interactions in the rhizosphere and endosphere. Front. Plant Sci. 9:1896. doi: 10.3389/fpls.2018.01896
Yu, Z., Jin, J., Li, Y., Yang, Y., Zhao, Y., Liu, C., et al. (2019). Distinct effects of short-term reconstructed topsoil on soya bean and corn rhizosphere bacterial abundance and communities in Chinese Mollisol. R. Soc. Open Sci. 6:181054. doi: 10.1098/rsos.181054
Zeng, Q., An, S., Liu, Y., Wang, H., and Wang, Y. (2019). Biogeography and the driving factors affecting forest soil bacteria in an arid area. Sci. Total Environ. 680, 124–131. doi: 10.1016/j.scitotenv.2019.04.184
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, B., Zhang, J., Liu, Y., Shi, P., and Wei, G. (2018). Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 118, 178–186. doi: 10.1016/j.soilbio.2017.12.011
Zhang, R., Vivanco, J. M., and Shen, Q. (2017). The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Zhang, X., Johnston, E. R., Liu, W., Li, L., and Han, X. (2016). Environmental changes affect the assembly of soil bacterial community primarily by mediating stochastic processes. Glob. Chang. Biol. 22, 198–207. doi: 10.1111/gcb.13080
Zhang, Y., Loreau, M., He, N., Wang, J., Pan, Q., Bai, Y., et al. (2018). Climate variability decreases species richness and community stability in a temperate grassland. Oecologia 188, 183–192. doi: 10.1007/s00442-018-4208-1
Zhou, H., Gao, Y., Jia, X., Wang, M., Ding, J., Cheng, L., et al. (2020). Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, Northwestern China. Soil Biol. Biochem. 144:107782. doi: 10.1016/j.soilbio.2020.107782
Zhou, J., Deng, Y., Shen, L., Wen, C., Yan, Q., Ning, D., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 7:12083. doi: 10.1038/ncomms12083
Zhou, J., Deng, Y., Zhang, P., Xue, K., Liang, Y., Van Nostrand, J. D., et al. (2014). Stochasticity, succession, and environmental perturbations in a fluidic ecosystem. Proc. Natl. Acad. Sci. U.S.A. 111, E836–E845. doi: 10.1073/pnas.1324044111
Keywords: Stipa taxa, root-zone, bacterial communities turnover, assembly processes, environmental factor
Citation: Ma X, Chao L, Li J, Ding Z, Wang S, Li F and Bao Y (2021) The Distribution and Turnover of Bacterial Communities in the Root Zone of Seven Stipa Species Across an Arid and Semi-arid Steppe. Front. Microbiol. 12:782621. doi: 10.3389/fmicb.2021.782621
Received: 24 September 2021; Accepted: 26 November 2021;
Published: 24 December 2021.
Edited by:
Stilianos Fodelianakis, Swiss Federal Institute of Technology Lausanne, SwitzerlandReviewed by:
Hannes Peter, Swiss Federal Institute of Technology Lausanne, SwitzerlandCopyright © 2021 Ma, Chao, Li, Ding, Wang, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuying Bao, bmRieXlAaW11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.