- Beijing Key Laboratory of Pediatric Respiratory Infection Diseases, Key Laboratory of Major Diseases in Children, Ministry of Education, National Key Discipline of Pediatrics (Capital Medical University), National Clinical Research Center for Respiratory Diseases, National Center for Children’s Health, Beijing Pediatric Research Institute, Beijing Children’s Hospital, Capital Medical University, Beijing, China
Background: The isolation rate of serogroup 15 Streptococcus pneumoniae has been increasing since developing countries began administering the 13-valent pneumococcal conjugate vaccine.
Methods: We detected the antibiotic resistance and molecular characteristics of 126 serogroup 15 S. pneumoniae strains isolated from children in China. Serotypes were determined via the Quellung reaction. Antibiotic resistance was tested using the E-test or disc diffusion method. Sequence types were assigned via multilocus sequence typing. Data were analyzed using WHONET 5.6 software.
Results: The frequencies of S. pneumoniae serotypes 15A, 15B, 15C, and 15F were 29.37, 40.48, 28.57, and 1.59%, respectively. Continuous-monitoring data from Beijing showed that the annual isolation rates of serogroup 15 S. pneumoniae were 7.64, 7.17, 2.58, 4.35, 3.85, 7.41, and 10.53%, respectively, from 2013 to 2019. All 126 serogroup 15 strains were susceptible to vancomycin and ceftriaxone. The non-susceptibility rate to penicillin was 78.57%. All strains were resistant to erythromycin with high minimum inhibitory concentrations (MICs). The multidrug resistance rate was 78.57%. The most common clonal complexes were CC3397, CC6011, CC10088, CC9785, and ST8589.
Conclusion: Serogroup 15 S. pneumoniae is common among children in China, and these strains should be continuously monitored.
Introduction
Streptococcus pneumoniae (S. pneumoniae) is among the most important pathogenic bacteria in children. More than 100 serotypes within 46 serogroups have been discovered according to the biochemical structure of the capsular polysaccharide (Ganaie et al., 2020). As of 12 October 2020, the 13-valent pneumococcal conjugate vaccine (PCV13), which protects against 13 serotypes (1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, and 23F) has been incorporated into national immunization programs in 160 countries (World Health Organization, 2020). Research on the efficacy of PCV13 showed that PCV13 can reduce the incidence of pneumococcal disease and cause changes in the serotype distributions of pathogenic S. pneumoniae strains (Janoir et al., 2016; Schroeder et al., 2017; Ubukata et al., 2018; Kim et al., 2020). Many regions worldwide have reported increased isolation rates of serogroup 15 S. pneumoniae strains after PCV13 use (Linden et al., 2015; Liyanapathirana et al., 2015; Sheppard et al., 2016; Lo et al., 2019; Nakano et al., 2020).
PCV13 was approved in China in November 2016 and was available in the country in May 2017. Because PCV13 is expensive and has not been included in China’s national immunization program, the PCV13 vaccination rate among Chinese children is low. Another 23-valent pneumococcal polysaccharide vaccine, PPV23, which protects against serotypes 1, 2, 3, 4, 5, 6B, 7F, 8, 9N, 9V, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19A, 19F, 20, 22F, 23F, and 33F, is available in China for children aged > 2 years. The vaccination rates for both vaccines are low. Recipients are concentrated in cities and are children whose parents can afford to pay for the vaccines. Reports on the vaccination data are scarce, and the vaccine coverage is unclear.
We sought to determine the prevalence of serogroup 15 S. pneumoniae among Chinese children as well as the characteristics of these strains. Here, we report the serotype distribution, antibiotic-resistance patterns and molecular biological characteristics of 126 serogroup 15 S. pneumoniae strains collected from children in China.
Materials and Methods
Bacterial Strains
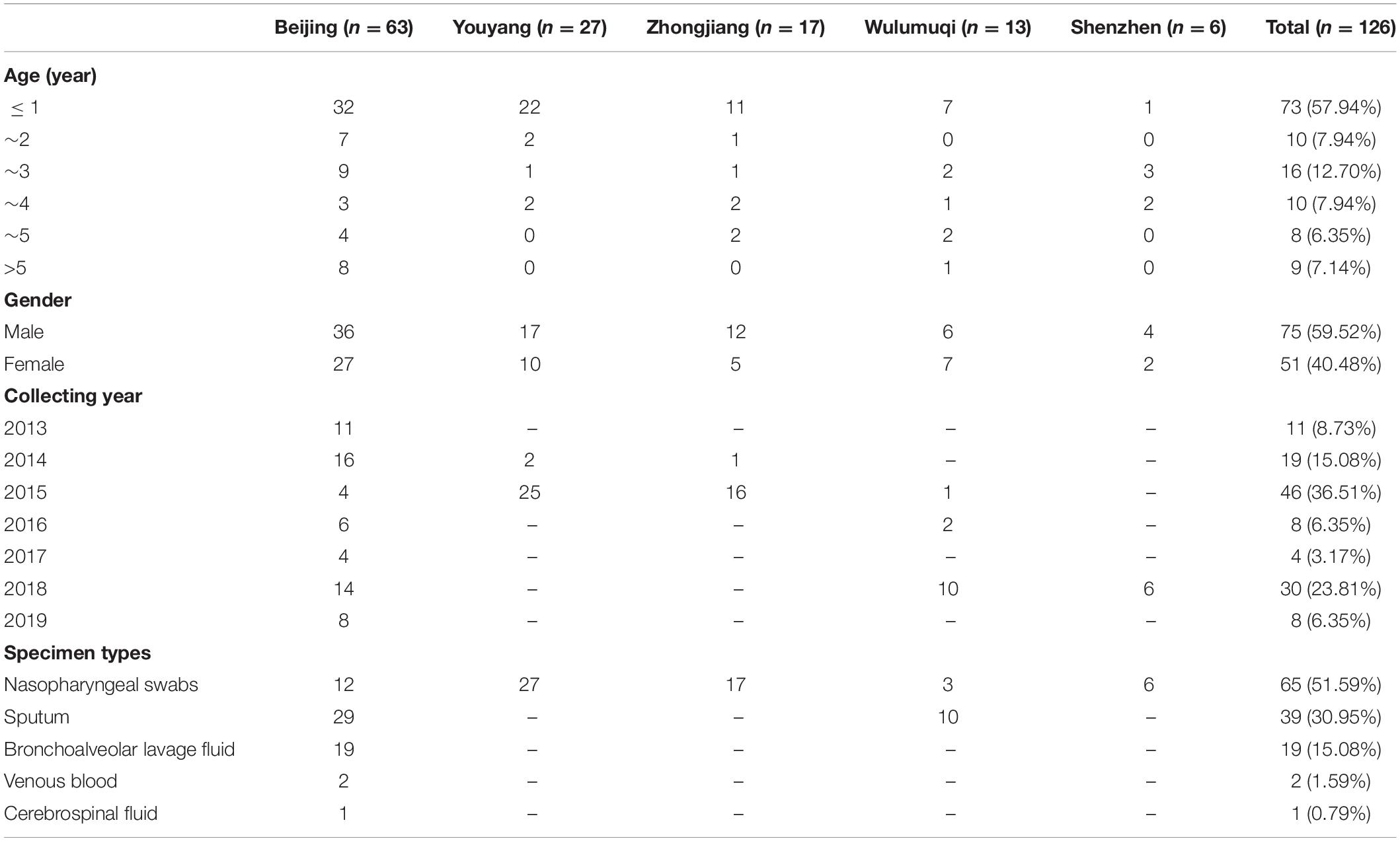
We collected 126 unduplicated strains of serogroup 15 S. pneumoniae from Beijing (Beijing Children’s Hospital affiliated to Capital Medical University; hereinafter referred to as Beijing Children’s Hospital), Zhongjiang (People’s Hospital of Zhongjiang County), Youyang (People’s Hospital of Chongqing Youyang County), Wulumuqi (Wulumuqi Children’s Hospital) and Shenzhen (Shenzhen Children’s Hospital) from 2013 to 2019. Table 1 lists the detailed clinical information on these children, including age, sex, culture time and specimen type isolated.
Of the 126 strains isolated, 63 were collected via continuous surveillance in Beijing Children’s Hospital. Beijing Children’s Hospital is a national children’s medical center, with more than 3 million outpatients and 70,000 inpatients annually. Since 2013, Beijing Children’s Hospital has been monitoring the serotype and antibiotic susceptibility characteristics of S. pneumoniae strains isolated from children in the hospital. In this study, the isolation rate of serogroup 15 S. pneumoniae strains from Beijing Children’s Hospital was used to reflect the isolation of serogroup 15 S. pneumoniae in children in China.
A parent and/or legal guardian of each participant signed a written informed consent document before enrollment and before the study procedures were performed. The Ethics Committees of Beijing Children’s Hospital affiliated to Capital Medical University, People’s Hospital of Zhongjiang County, People’s Hospital of Chongqing Youyang County, Wulumuqi Children’s Hospital, and Shenzhen Children’s Hospital approved the study. No ethical problems were encountered.
Serotyping
Serotypes were determined using the Quellung reaction with Pneumotest kits (Statens Serum Institute, Copenhagen, Denmark). Serotyping was interpreted based on capsular swelling under phase-contrast microscopy with an oil-immersion lens (magnification, 100×), as described previously (Sørensen, 1993).
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of penicillin, amoxicillin, ceftriaxone, imipenem, vancomycin and erythromycin for each strain were determined using E-test strips (AB Biodisk, Solna, Sweden). Disc diffusion tests (Oxoid Ltd., Basingstoke, United Kingdom) were performed to ascertain antimicrobial susceptibilities to tetracycline, sulfamethoxazole-trimethoprim, and chloramphenicol. The results were interpreted in accordance with the Clinical and Laboratory Standards Institute 2019 guidelines, and oral breakpoints (susceptible, ≤ 0.06 mg/L; intermediate, 0.12–1.0 mg/L; resistant, ≥ 2.0 mg/L) were used for penicillin (Clinical Laboratory Standards Institute [CLSI], 2019). S. pneumoniae American Type Culture Collection 49619 was used as the quality-control strain and was included in each test set to ensure accuracy of the results. Multidrug-resistant S. pneumoniae (MDRSP) was defined as S. pneumoniae isolates that were resistant to three or more kinds of antibiotics tested in this study.
Multilocus Sequence Typing
Strains were characterized using multilocus sequence typing (MLST). Chromosomal DNA was extracted from overnight cultures of S. pneumoniae strains grown on 5% trypticase soy agar (Oxoid Ltd.) using the SiMax™ Genomic DNA Extraction Kit (SBS Genetech Co., Ltd., Beijing, China) per the manufacturer’s instructions. Seven housekeeping genes (aroE, gdh, gki, recP, spi, xpt, and ddl) were amplified via polymerase chain reaction from the chromosomal DNA as described previously (Enrigh and Spratt, 1998). The products were sent to BGI Company (Beijing, China) for sequencing of both strands. The resulting sequences were compared with those of all known alleles at each locus, as well as with the sequence types (STs) in the pneumococcal MLST database.1 New alleles and allelic profiles were submitted to the MLST database for name assignment. goeBURST v1.2.12 was used to investigate relationships among the strains and assign strains to a clonal complex (CC) based on the stringent group definition of six of the seven shared alleles.
Statistical Analysis
Antimicrobial susceptibility and MLST data were analyzed using WHONET 5.6 as recommended by the World Health Organization.
Results
Serotype Distribution and Antimicrobial Susceptibility
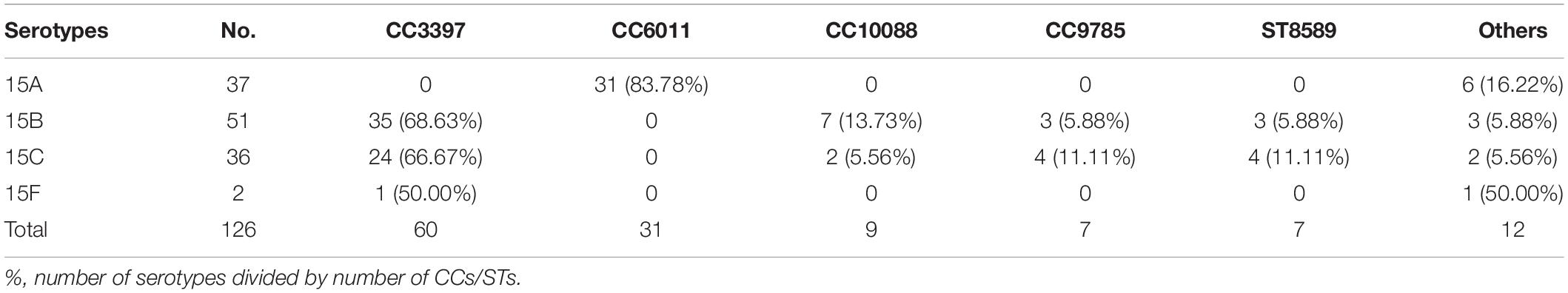
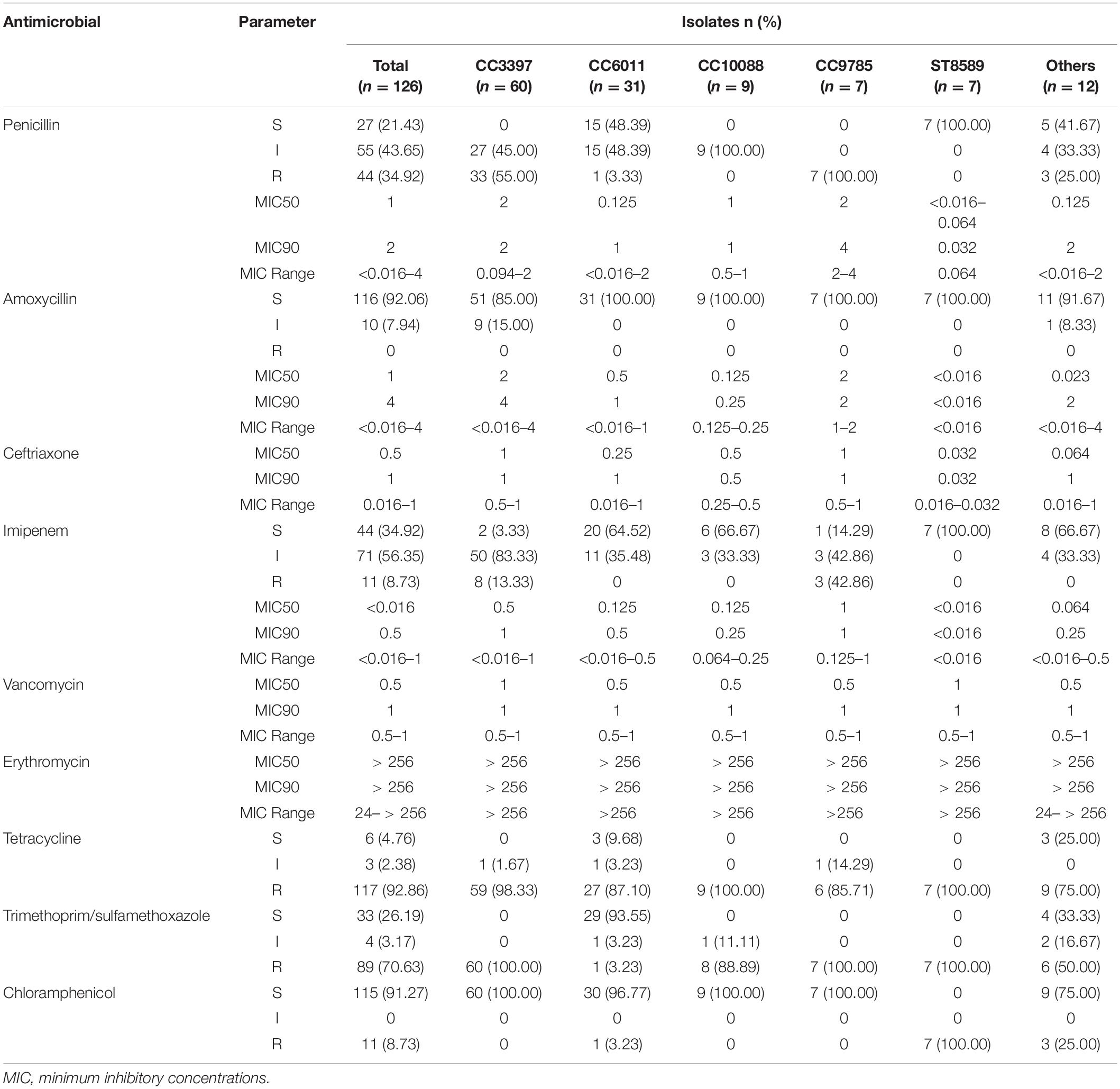
Among the 126 strains, the proportions of serotypes 15A, 15B, 15C, and 15F were 29.37% (37/126), 40.48% (51/126), 28.57% (36/126), and 1.59% (2/126), respectively. Table 2 shows the antimicrobial susceptibility testing results, including MIC values, for the serotypes. All strains were susceptible to vancomycin and ceftriaxone; none were susceptible to amoxycillin. The non-susceptibility rate to penicillin was 78.57%. All 126 strains were resistant to erythromycin, with high MIC values of 24 mg/L for one strain and > 256 mg/L for all other strains. The resistance rate to tetracycline reached 92.86%.
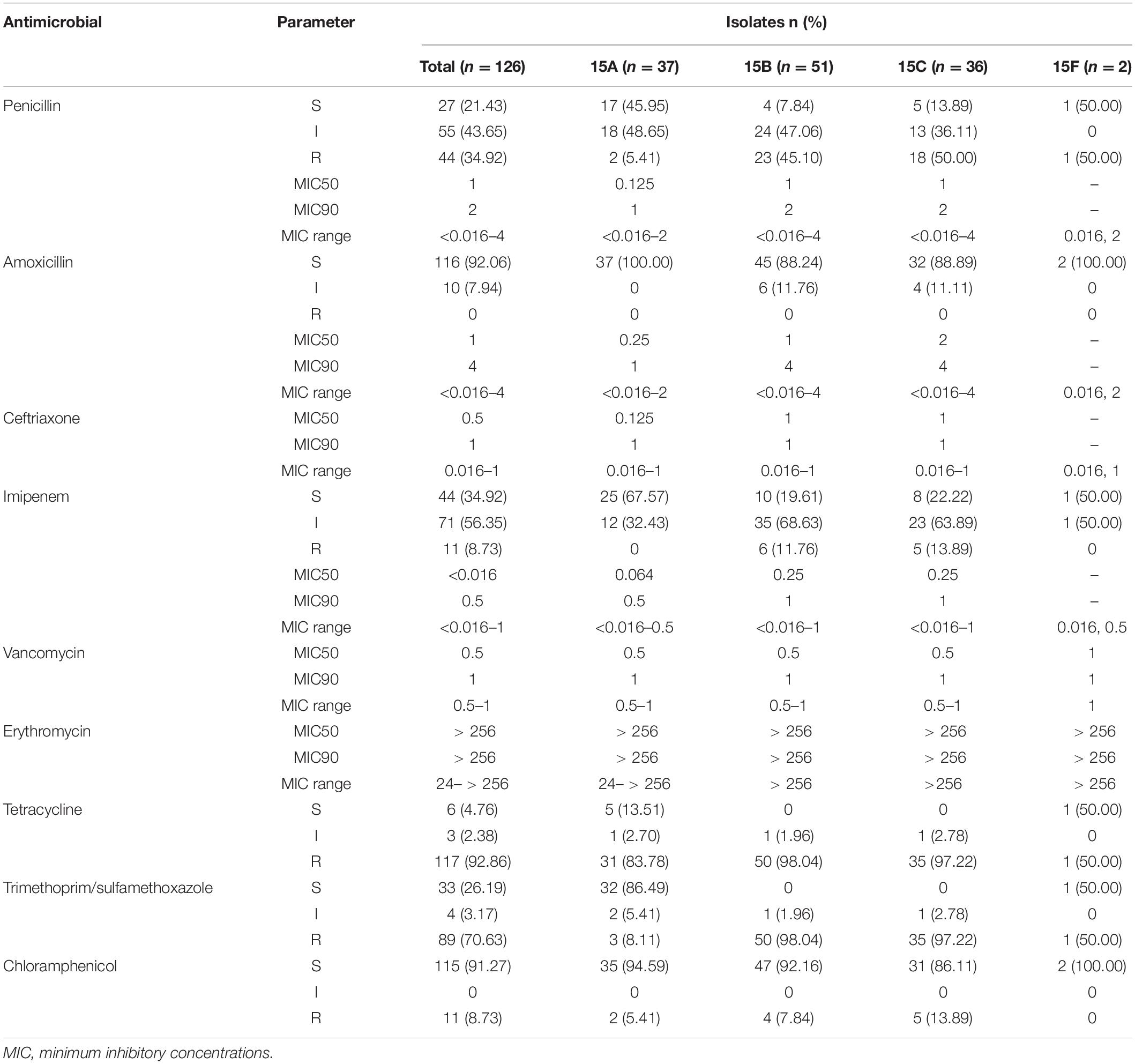
Table 2. Antimicrobial susceptibility pattern of the 126 serogroup 15 Streptococcus pneumoniae strains.
The multidrug-resistance rate was 78.57% (99/126). Figure 1 shows the antibiotic-resistance pattern distributions of the MDRSP strains. The most common multidrug-resistance pattern was macrolides-β-lactams-tetracyclines, accounting for 64.65% (64/99) of the MDRSP.
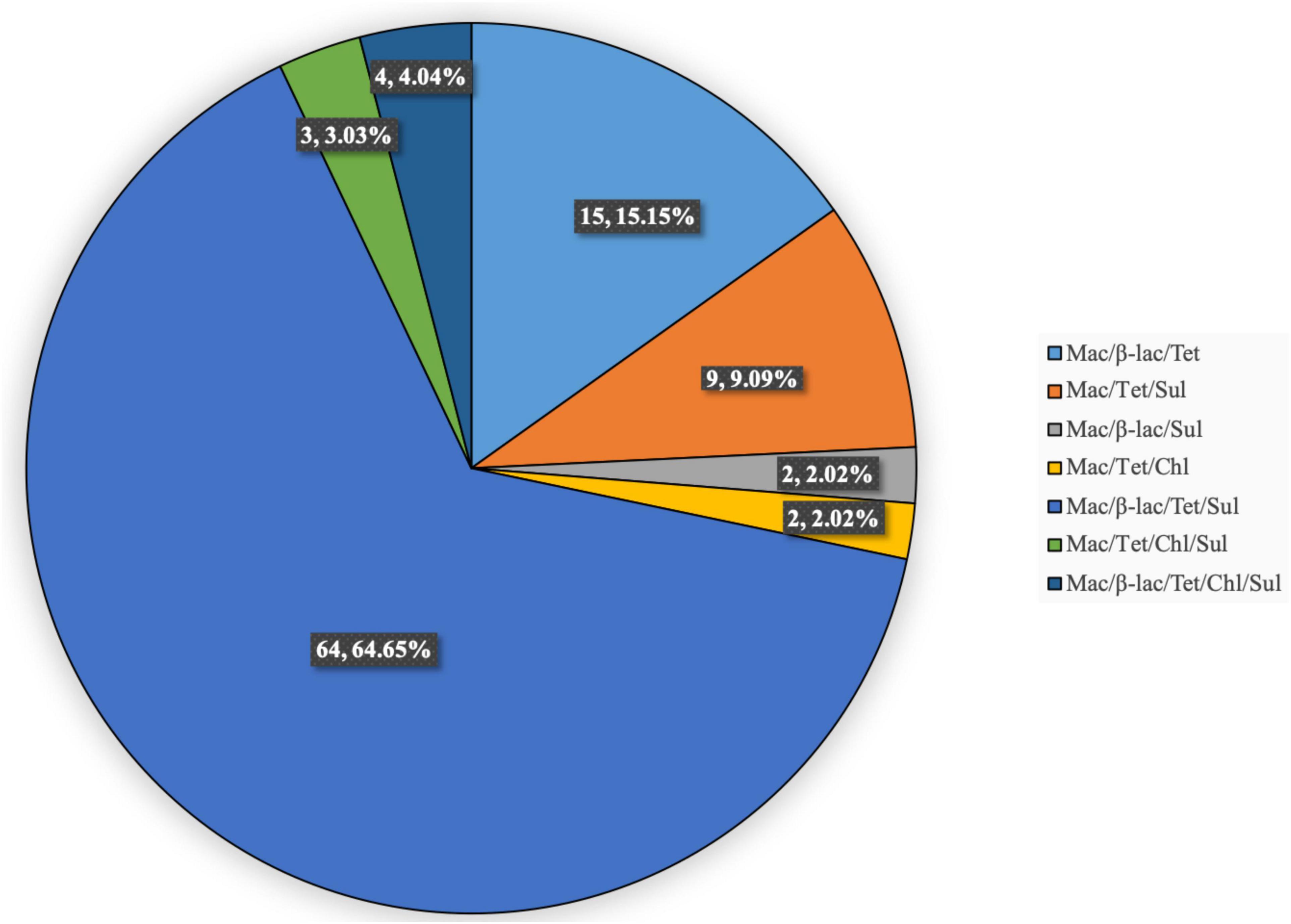
Figure 1. Distribution of antibiotic-resistance patterns of MDRSP strains. Mac, macrolides; β-lac, β-lactams; Tet, tetracyclines; Sul, sulfonamides; Chl, chloramphenicols.
Multilocus Sequence Typing
Thirty-one STs were identified among the 126 strains; the most common were ST3397 (n = 16, 12.70%), ST6011 (n = 16, 12.70%), ST11972 (n = 15, 11.90%), ST7768 (n = 14, 11.11%), ST6555 (n = 11, 8.73%), ST10088 (n = 8, 6.35%), ST8589 (n = 7, 5.56%), ST11950 (n = 7, 5.56%), and ST9785 (n = 6, 4.76%). The 31 STs were assigned to five CCs and nine singletons using goeBURST analysis (Figure 2). The most predominant CCs were CC3397 (n = 60, 47.62%) and CC6011 (n = 31, 24.60%), constituting 72.22% of all strains.
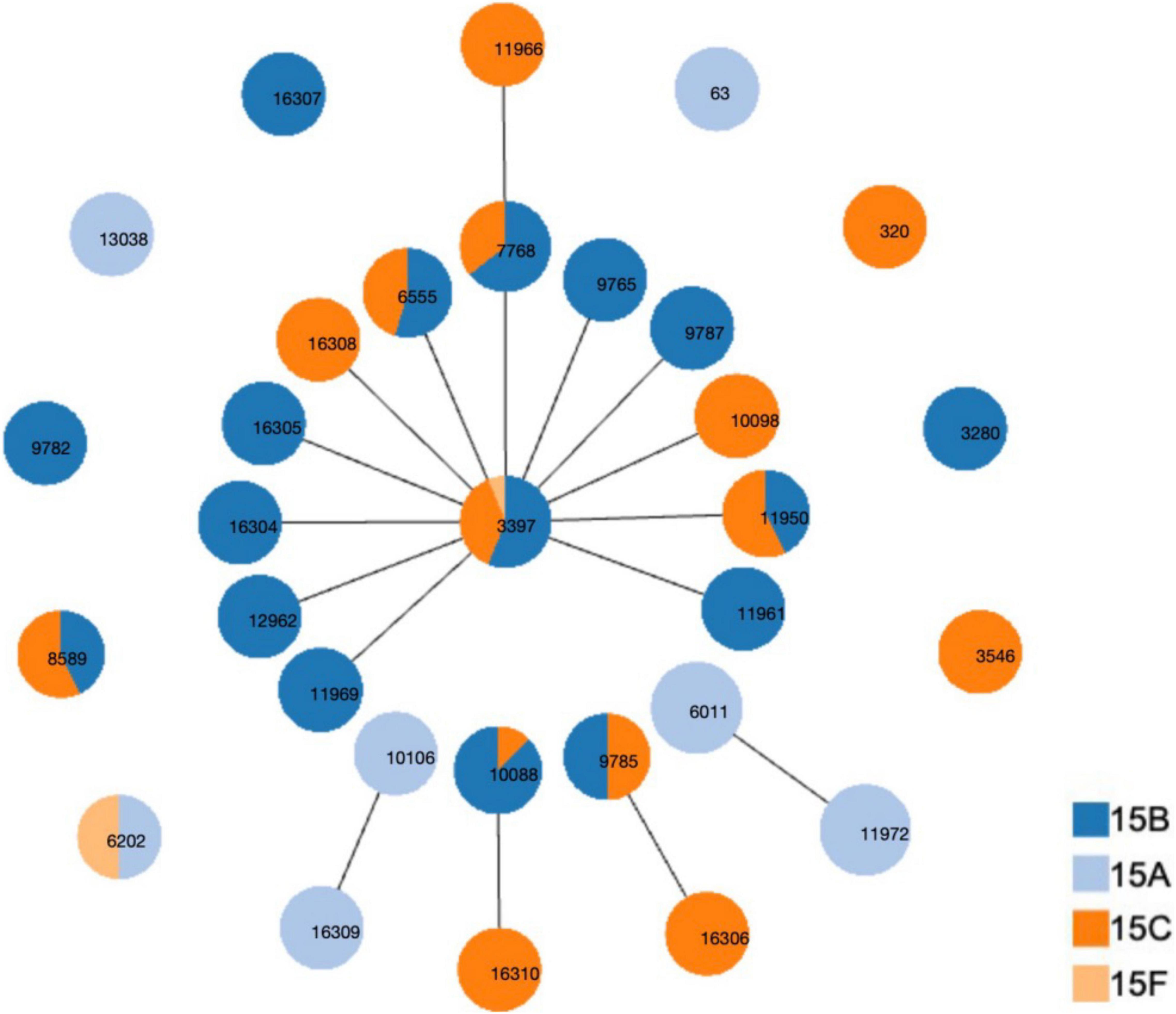
Figure 2. Distribution of sequence types (STs) amongst the clonal complexes of serogroup 15 Streptococcus pneumoniae strains. goeBURST analysis was performed on the multilocus sequence typing data generated from all 126 strains analyzed. STs linked by a line belong to the same cluster.
Table 3 shows the CCs/ST distribution by serotype. Each serotype had a predominant CC that included most of the isolates. CC6011 was the main CC for the 15A strains (83.78%, 31/37). CC3397 was the main CC for 15B and 15C, accounting for 68.63% (35/51) and 66.67% (24/36) of these strains, respectively.
Relationship Between Antimicrobial Susceptibility and Clonal Complexes
Table 4 summarizes the antibiotic-resistance profiles according to CCs. Different CCs showed different antimicrobial-resistance characteristics. No CC3397 strains were susceptible to penicillin or tetracycline, and all were 100% resistant to sulfamethoxazole-trimethoprim and 100% sensitive to chloramphenicol. All CC6011 and CC10088 strains were susceptible to amoxicillin and resistant to imipenem. All CC10088 strains were intermediate to penicillin, resistant to tetracycline and susceptible to chloramphenicol. All CC9785 strains were resistant to penicillin. All ST8589 strains were susceptible to penicillin, amoxicillin, and imipenem and resistant to tetracycline, sulfamethoxazole-trimethoprim, and chloramphenicol.
Fluctuation of Serotypes by Years
We isolated 1029 S. pneumoniae strains from Beijing Children’s Hospital during the study period, and 63 were determined to be serogroup 15. The proportion of serogroup 15 S. pneumoniae strains was 6.12%. Figure 3 shows the number of S. pneumoniae strains collected yearly and the serogroup 15 isolation rates.
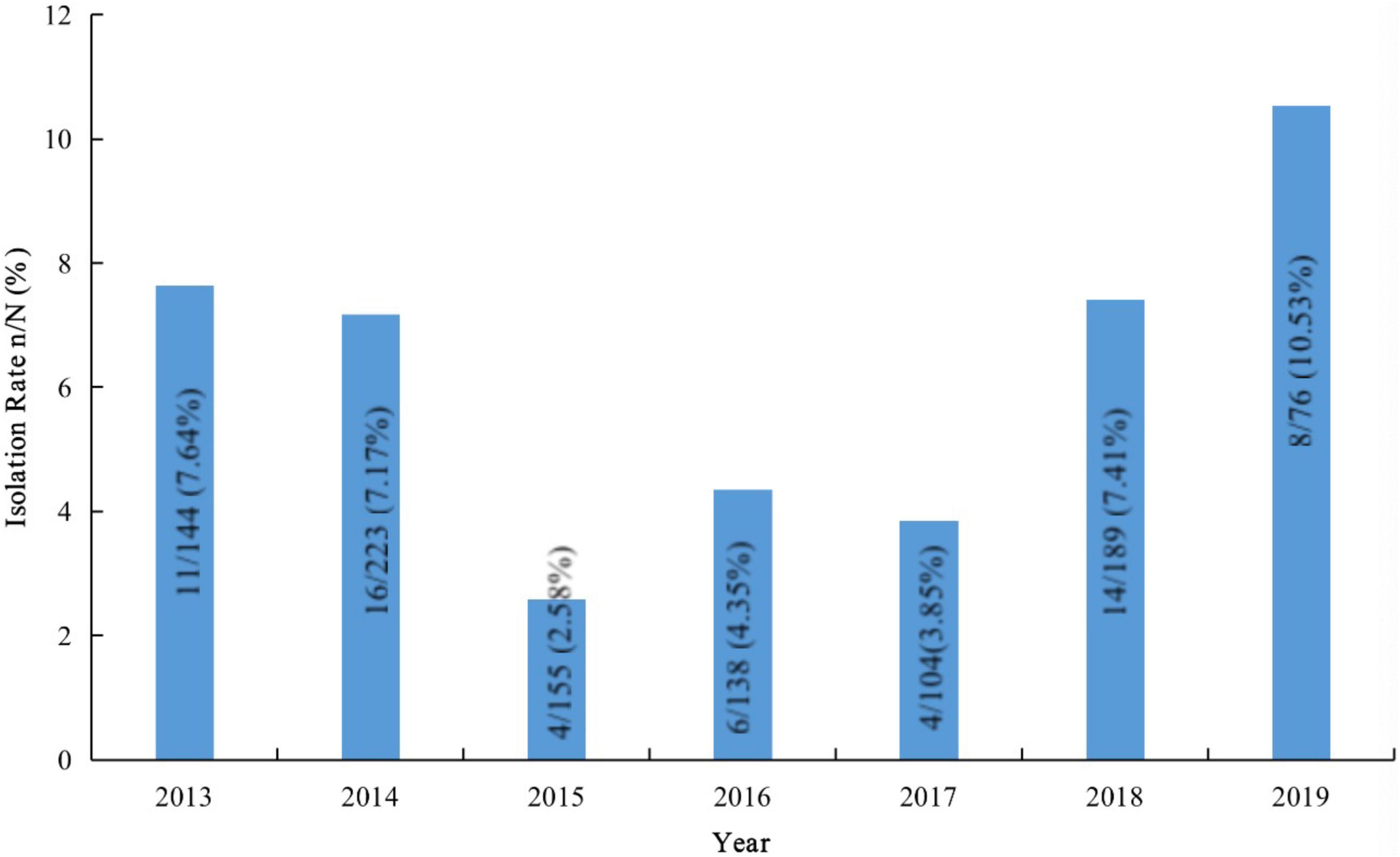
Figure 3. Isolation rates of serogroup 15 Streptococcus pneumoniae strains in samples from Beijing Children’s Hospital.
In 2014 and 2015, the proportions of serogroup 15 S. pneumoniae were 6.67% (2/30) and 12.20% (25/205), respectively, in Youyang and 6.25% (1/16) and 9.09% (16/176), respectively, in Zhongjiang. In 2015, 2016, and 2018, the proportions of serogroup 15 were 6.25% (1/16), 12.50% (2/16), and 3.88% (10/258), respectively, in Wulumuqi. Six of 66 (9.09%) strains were isolated in Shenzhen in 2018.
Discussion
Since the introduction of PCV13, many countries and regions that included PCV13 in their national immunization programs have reported increased infections from serogroup 15 S. pneumoniae, which is not covered by the PCV13 vaccine (Linden et al., 2015; Sheppard et al., 2016; Nakano et al., 2020). Serogroup 15 S. pneumoniae strains have caused outbreaks (Katherine et al., 2012) and deaths (Arushothy et al., 2020) among children. China has not yet included PCV13 in the national immunization program, and the PCV13 vaccination rate among Chinese children is low. Additionally, the epidemiological monitoring data for S. pneumoniae in Chinese children are inexact; thus, the prevalence of serogroup 15 S. pneumoniae and the characteristics of these strains are unclear. Here, we reported the serotype distribution, antibiotic-resistance patterns, and ST characteristics of 126 serogroup 15 S. pneumoniae strains isolated from children in China. Half of these strains came from the continuous monitoring of Beijing Children’s Hospital, which is one of the few continuous-monitoring data programs in mainland China and can partially reflect the prevalence of S. pneumoniae in Chinese children. During the study period, the overall isolation rate of serogroup 15 S. pneumoniae in the child population of Beijing Children’s Hospital was 6.12%. After PCV13 was launched in China in May 2017, the isolation rates of serogroup 15 S. pneumoniae in 2018 and 2019 were 7.41 and 10.53%, respectively, showing an increasing trend.
The Pneumococcal Molecular Epidemiology Network (PMEN)3 was established in 1997 for global surveillance of antibiotic-resistant S. pneumoniae and to standardize the nomenclature and classification of resistant clones. One 15A serotype was included among the 43 drug-resistant clones spread worldwide and published by the PMEN: Sweden15A-25 (ST63). Over the past few years, several studies have reported increasing numbers of serotype 15A-ST63 strains (Ho et al., 2015; Sheppard et al., 2016; Nakano et al., 2018, 2019, 2020). In this study, ST63 accounted for 5.41% (2/37) of the serotype 15A strains, which was far lower than the 88.24% reported in Hong Kong (Ho et al., 2015) and the 98.28% reported in Japan (Nakano et al., 2020). Some studies reported that ST63 showed high multidrug-resistance rates (Sheppard et al., 2016; Nakano et al., 2018). Both ST63 strains in this study were resistant to erythromycin; one of these was also resistant to tetracycline, and the other was non-susceptible to imipenem, but both strains were sensitive to other antibiotics. The significant differences in isolation rates and antibiotic susceptibility characteristics of ST63 between our study and the other studies may be due to regional differences.
In the present study, most of the serotype 15A strains belonged to CC6011, which was composed of ST6011 and ST11972. ST6011 was popular in both serotype 3 (Ho et al., 2019) and serogroup 15 strains. There were 28 ST6011 strains recorded in the MLST database,4 among which 22 were serotype 3, 5 were serotype 15A, and 1 strain was serotype 15B/15C. Both serotype 3 and serogroup 15 are non-PCV13 vaccine serotypes, and have an increasing trend in recent years (Linden et al., 2015; Liyanapathirana et al., 2015; Sheppard et al., 2016; LeBlanc et al., 2019; Lo et al., 2019; Goettler et al., 2020; Nakano et al., 2020). Therefore, close attention should be paid to ST6011 prevalent in these two serotypes.
We found that serogroup 15 S. pneumoniae strains showed good sensitivity to common antibiotics. However, the most common CC, CC3397, was 100% non-susceptible to penicillin. We wondered whether the prevalence of CC3397 in serogroup 15 S. pneumoniae results from antibiotic pressure. Many studies have reported clonal shift phenomena in other serotypes, as CC271 replaced CC983 among serotype 19F strains (Li et al., 2013), ST81 replaced ST342 amongst serotype 23F pneumococcal isolates (Ma et al., 2013), CC320 replaced CC230 in 19A strains (Beall et al., 2011; Golden et al., 2015; Mayanskiy et al., 2017), and CC876 replaced CC875 in serotype 14 strains (He et al., 2015). These examples of clonal shift phenomena in the same serotype may be caused by antibiotic pressure, as they followed the principle that CCs/STs expressing high antibiotic resistance will replace CCs/STs with lower antibiotic resistance. Presently, the epidemiological characteristics of serogroup 15 S. pneumoniae are not being continuously monitored to access the changes in CC/ST prevalence in China. Whether CC3397 is the most drug-resistant of the serogroup 15 S. pneumoniae clones or whether CC3397 will be replaced by more drug-resistant clones in the future is unclear. Therefore, clinicians must strengthen surveillance of the antibiotic resistance and other epidemiological characteristics of serogroup 15 S. pneumoniae.
Conclusion
Serogroup 15 S. pneumoniae is common among children in China. Because CC3397, the main CC of serogroup 15 S. pneumoniae, showed high non-susceptibility to penicillin, long-term monitoring of antibiotic susceptibility and other epidemiological characteristics of serogroup 15 S. pneumoniae is essential.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
WS, QD, LY, and WG conducted the experiments. WS and QW were responsible for the laboratory analysis. WS and KY designed the study, collected the data, analyzed the data, interpreted the results, and drafted the manuscript. All authors reviewed and revised the manuscript and approved the final version.
Funding
This study was supported by the Beijing Natural Science Foundation (L202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank the staff of the following esteemed institutions for providing isolates during the several collection programs: Beijing Children’s Hospital affiliated to Capital Medical University (Fang Dong), People’s Hospital of Zhongjiang County (Denian Wen), People’s Hospital of Chongqing Youyang County (Changhui Chen), Wulumuqi Children’s Hospital (Juling Tian), and Shenzhen Children’s Hospital (Jikui Deng).
Footnotes
- ^ https://pubmlst.org/spneumoniae/
- ^ http://www.phyloviz.net/goeburst/
- ^ https://www.pneumogen.net/pmen
- ^ https://pubmlst.org
References
Arushothy, R., Ramasamy, H., Hashim, R., Raj, A. S. S., Amran, F., Samsuddin, N., et al. (2020). Multidrug-resistant Streptococcus pneumoniae causing invasive pneumococcal disease isolated from a paediatric patient. Int. J. Infect. Dis. 90, 219–222. doi: 10.1016/j.ijid.2019.10.037
Beall, B. W., Gertz, R. E., Hulkower, R. L., Whitney, C. G., Moore, M. R., and Brueggemann, A. B. (2011). Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J. Infect. Dis. 203, 1360–1368. doi: 10.1093/infdis/jir052
Clinical Laboratory Standards Institute [CLSI] (2019). Performance Standards for Antimicrobial Susceptibility Testing. In: Twenty-Ninth Informational Supplement. M100-S29. Wayne, PA: Clinical and Laboratory Standards Institute.
Enrigh, M. C. T., and Spratt, B. G. (1998). A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clone associated with serious invasive disease. Microbiology 144(Pt. 11) 3049–3060. doi: 10.1099/00221287-144-11-3049
Ganaie, F., Saad, J. S., McGee, L., van Tonder, A. J., Bentley, S. D., Lo, S. W., et al. (2020). A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral streptococcus. mBio 11:e00937–20. doi: 10.1128/mBio.00937-20
Goettler, D., Streng, A., Kemmling, D., Schoen, C., von Kries, R., Rose, M. A., et al. (2020). Increase in Streptococcus pneumoniae serotype 3 associated parapneumonic pleural effusion/empyema after the introduction of PCV13 in Germany. Vaccine 38, 570–577. doi: 10.1016/j.vaccine.2019.10.056
Golden, A. R., Rosenthal, M., Fultz, B., Nichol, K. A., Adam, H. J., Gilmour, M. W., et al. (2015). Characterization of MDR and XDR Streptococcus pneumoniae in Canada, 2007–13. J. Antimicrob. Chemother. 70, 2199–2202.
He, M., Yao, K., Shi, W., Gao, W., Yuan, L., Yu, S., et al. (2015). Dynamic of serotype 14 Streptococcus pneumoniae population causing acute respiratory infections among children in China (1997-2012). BMC Infect. Dis. 15:266. doi: 10.1186/s12879-015-1008-7
Ho, P. L., Chiu, S. S., Law, P. Y., Chan, E. L., Lai, E. L., and Chow, K. H. (2015). Increase in the nasopharyngeal carriage of non-vaccine serogroup 15 Streptococcus pneumoniae after introduction of children pneumococcal conjugate vaccination in Hong Kong. Diagn. Microbiol. Infect. Dis. 81, 145–148. doi: 10.1016/j.diagmicrobio.2014.11.006
Ho, P. L., Law, P. Y., and Chiu, S. S. (2019). Increase in incidence of invasive pneumococcal disease caused by serotype 3 in children eight years after the introduction of the pneumococcal conjugate vaccine in Hong Kong. Hum. Vaccin. Immunother. 15, 455–458. doi: 10.1080/21645515.2018.1526555
Janoir, C., Lepoutre, A., Gutmann, L., and Varon, E. (2016). Insight into resistance phenotypes of emergent non 13-valent pneumococcal conjugate vaccine type pneumococci isolated from invasive disease after 13-valent pneumococcal conjugate vaccine implementation in France. Open Forum Infect. Dis. 3:ofw020. doi: 10.1093/ofid/ofw020
Katherine, F. D., Chukwuma, M., Ruth, L. G., Alexander, N., Guh, A., Forbes, E., et al. (2012). Streptococcus pneumoniae serotype 15A in psychiatric unit, Rhode Island, USA, 2010–2011. Emerg. Infect. Dis. 18, 1889–1893. doi: 10.3201/eid1811.120454
Kim, S. H., Chung, D. R., Song, J.-H., Baek, J. Y., Thamlikitkul, V., Wang, H., et al. (2020). Changes in serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates from adult patients in Asia: emergence of drug-resistant non-vaccine serotypes. Vaccine 38, 6065–6073. doi: 10.1016/j.vaccine.2019.09.065
LeBlanc, J. J., ElSherif, M., Ye, L., MacKinnon-Cameron, D., Ambrose, A., Hatchette, T. F., et al. (2019). Streptococcus pneumoniae serotype 3 is masking PCV13-mediated herd immunity in Canadian adults hospitalized with community acquired pneumonia: a study from the Serious Outcomes Surveillance (SOS) Network of the Canadian immunization research Network (CIRN). Vaccine 37, 5466–5473. doi: 10.1016/j.vaccine.2019.05.003
Li, Q. H., Yao, K. H., Yu, S. J., Ma, X., He, M. M., Shi, W., et al. (2013). Spread of multidrug-resistant clonal complex 271 of serotype 19F Streptococcus pneumoniae in Beijing, China: characterization of serotype 19F. Epidemiol. Infect. 141, 2492–2496. doi: 10.1017/S0950268813000514
Linden, M., Perniciaro, S., and Imöhl, M. (2015). Increase of serotypes 15A and 23B in IPD in Germany in the PCV13 vaccination era. BMC Infect. Dis. 15:207. doi: 10.1186/s12879-015-0941-9
Liyanapathirana, V., Nelson, E. A., Ang, I., Subramanian, R., Ma, H., and Ip, M. (2015). Emergence of serogroup 15 Streptococcus pneumoniae of diverse genetic backgrounds following the introduction of pneumococcal conjugate vaccines in Hong Kong. Diagn. Microbiol. Infect. Dis. 81, 66–70. doi: 10.1016/j.diagmicrobio.2014.09.028
Lo, S. W., Gladstone, R. A., van Tonder, A. J., Lees, J. A., du Plessis, M., Benisty, R., et al. (2019). Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect. Dis. 19, 759–769. doi: 10.1016/S1473-3099(19)30297-X
Ma, X., Yao, K. H., Yu, S. J., Zhou, L., Li, Q. H., Shi, W., et al. (2013). Genotype replacement within serotype 23F Streptococcus pneumoniae in Beijing, China: characterization of serotype 23F. Epidemiol. Infect. 141, 1690–1696. doi: 10.1017/S0950268812002269
Mayanskiy, N., Savinova, T., Alyabieva, N., Ponomarenko, O., Brzhozovskaya, E., Lazareva, A., et al. (2017). Antimicrobial resistance, penicillin-binding protein sequences, and pilus islet carriage in relation to clonal evolution of Streptococcus pneumoniae serotype 19A in Russia, 2002–2013. Epidemiol. Infect. 145, 1708–1719. doi: 10.1017/S0950268817000541
Nakano, S., Fujisawa, T., Ito, Y., Chang, B., Matsumura, Y., Yamamoto, M., et al. (2020). Nationwide surveillance of paediatric invasive and non-invasive pneumococcal disease in Japan after the introduction of the 13-valent conjugated vaccine, 2015–2017. Vaccine 38, 1818–1824. doi: 10.1016/j.vaccine.2019.12.022
Nakano, S., Fujisawa, T., Ito, Y., Chang, B., Matsumura, Y., Yamamoto, M., et al. (2019). Whole-genome sequencing analysis of lultidrug-resistant serotype 15A Streptococcus pneumoniae in Japan and the emergence of a highly resistant serotype 15A-ST9084 clone. Antimicrob. Agents Chemother. 63:e02579–18. doi: 10.1128/AAC.02579-18
Nakano, S., Fujisawa, T., Ito, Y., Chang, B., Matsumura, Y., Yamamoto, M., et al. (2018). Spread of meropenem-resistant Streptococcus pneumoniae serotype 15A-ST63 clone in Japan, 2012–2014. Emerg. Infect. Dis. 24, 275–283. doi: 10.3201/eid2402.171268
Schroeder, M. R., Chancey, S. T., Thomas, S., Kuo, W. H., Satola, S. W., Farley, M. M., et al. (2017). A population-based assessment of the impact of 7- and 13-valent pneumococcal conjugate vaccines on macrolide-resistant invasive pneumococcal disease: emergence and decline of Streptococcus pneumoniae serotype 19A (CC320) with dual macrolide resistance mechanisms. Clin. Infect. Dis. 65, 990–998. doi: 10.1093/cid/cix446
Sheppard, C., Fry, N. K., Mushtaq, S., Woodford, N., Reynolds, R., Janes, R., et al. (2016). Rise of multidrug-resistant non-vaccine serotype 15A Streptococcus pneumoniae in the United Kingdom, 2001 to 2014. Euro Surveill. 21:30423.
Sørensen, U. B. (1993). Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31, 2097–2100. doi: 10.1128/jcm.31.8.2097-2100.1993
Ubukata, K., Takata, M., Morozumi, M., Chiba, N., Wajima, T., Hanada, S., et al. (2018). Effects of pneumococcal conjugate vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010–2017. Emerg. Infect. Dis. 24, 2010–2020. doi: 10.3201/eid2411.180326
Keywords: Streptococcus pneumoniae, serotype 15, PCV13, vaccine, antibiotic resistance, children
Citation: Shi W, Du Q, Yuan L, Gao W, Wang Q and Yao K (2022) Antibiotic Resistance and Molecular Biological Characteristics of Non-13-Valent-Pneumococcal Conjugate Vaccine Serogroup 15 Streptococcus pneumoniae Isolated From Children in China. Front. Microbiol. 12:778985. doi: 10.3389/fmicb.2021.778985
Received: 17 September 2021; Accepted: 10 December 2021;
Published: 05 January 2022.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Anusak Kerdsin, Kasetsart University, ThailandLesley McGee, Centers for Disease Control and Prevention (CDC), United States
Copyright © 2022 Shi, Du, Yuan, Gao, Wang and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaihu Yao, aml1aHUyNjU1QHNpbmEuY29t
 Wei Shi
Wei Shi Qianqian Du
Qianqian Du Kaihu Yao
Kaihu Yao