- 1Department of Science, Roma Tre University, Rome, Italy
- 2Department of Molecular Medicine, Sapienza University of Rome, Rome, Italy
- 3IRCCS Fondazione Santa Lucia, Rome, Italy
Colistin represents a last-line treatment option for infections caused by multidrug resistant Gram-negative pathogens, including Pseudomonas aeruginosa. Colistin resistance generally involves the modification of the lipid A moiety of lipopolysaccharide (LPS) with positively charged molecules, namely phosphoethanolamine (PEtN) or 4-amino-4-deoxy-L-arabinose (Ara4N), that reduce colistin affinity for its target. Several lines of evidence highlighted lipid A aminoarabinosylation as the primary colistin resistance mechanism in P. aeruginosa, while the contribution of phosphoethanolamination remains elusive. PEtN modification can be due to either endogenous (chromosomally encoded) PEtN transferase(s) (e.g., EptA in P. aeruginosa) or plasmid borne MCR enzymes, commonly found in enterobacteria. By individually cloning eptA and mcr-1 into a plasmid for inducible gene expression, we demonstrated that MCR-1 and EptA have comparable PEtN transferase activity in P. aeruginosa and confer colistin resistance levels similar to those provided by lipid A aminoarabinosylation. Notably, EptA, but not MCR-1, negatively affects P. aeruginosa growth and, to a lesser extent, cell envelope integrity when expressed at high levels. Mutagenesis experiments revealed that PEtN transferase activity does not account for the noxious effects of EptA overexpression, that instead requires a C-terminal tail unique to P. aeruginosa EptA, whose function remains unknown. Overall, this study shows that both endogenous and exogenous PEtN transferases can promote colistin resistance in P. aeruginosa, and that PEtN and MCR-1 mediated resistance has no impact on growth and cell envelope homeostasis, suggesting that there may be no fitness barriers to the spread of mcr-1 in P. aeruginosa.
Introduction
The world is currently facing a worrying threat due to the spread of multidrug resistance in several bacterial pathogens. Emergence of resistance to most available antibiotics, including carbapenems, aminoglycosides, and fluoroquinolones, is increasing worldwide at an alarming rate, especially in Gram-negative bacteria, such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacteriaceae (Theuretzbacher et al., 2020). The lack of new antimicrobials active against critical Gram-negative pathogens has prompted the medical community to re-evaluate the use of the old polymyxin antibiotic colistin (polymyxin E) in clinical practice when no other less toxic or effective antibiotics are available, and colistin is now considered a last-resort treatment option to combat recalcitrant Gram-negative infections (Falagas and Kopterides, 2007; Pogue et al., 2017; Nang et al., 2021).
Structurally, colistin is a decapeptide, consisting of a seven-member cyclic ring and a tripeptide side chain bound to a short fatty acid (Velkov et al., 2010). Its antibacterial activity relies on the interaction with the anionic lipid A moiety of lipopolysaccharide (LPS) in the outer membrane (OM) of Gram-negatives, leading to displacement of Ca2+ and Mg2+ cations that stabilize the LPS layer and, thus, causing derangement of the OM. This results in increased membrane permeability that allows colistin entry, leakage of cell contents and, ultimately, cell death (Poirel et al., 2017). Recent studies showed that the direct interaction of colistin with nascent LPS in the periplasmic leaflet of the inner membrane (IM) is crucial for its bacteriolytic activity (Sabnis et al., 2021).
Most Gram-negative bacteria acquire colistin resistance through genomic mutations in regulatory genes leading to induction of genes responsible for LPS remodeling, mostly through the covalent addition to lipid A of positively charge molecules, such as 4-amino-4-deoxy-L-arabinose (Ara4N) or phosphoethanolamine (PEtN), or by the acquisition of plasmid-borne PEtN transferase genes of the MCR family (Liu et al., 2016; Zhang et al., 2019). The resulting positive charge reduces LPS affinity for colistin, ultimately causing resistance (Olaitan et al., 2014; Jeannot et al., 2017; Moffatt et al., 2019; Sabnis et al., 2021).
Pseudomonas aeruginosa is a Gram-negative pathogen responsible for severe infections in immunocompromised patients and represents a major threat to cystic fibrosis (CF) patients. A hallmark of P. aeruginosa infections is the life-threatening severity and poor responsiveness to antibiotics due to intrinsic and acquired resistance (Poole, 2011). Colistin is currently used for treating multidrug-resistant P. aeruginosa infections, especially in CF (Langan et al., 2015). This, however, has inevitably led to the emergence of colistin-resistant P. aeruginosa strains (Falagas et al., 2010; Giamarellou, 2016), even though colistin resistance rates are overall lower as compared to other anti-pseudomonal agents (Horcajada et al., 2019; Gajdács et al., 2021).
In P. aeruginosa clinical isolates, colistin resistance is always associated with mutations leading to the overexpression of the arn operon, which encodes the enzymes for Ara4N modification of lipid A (Barrow and Kwon, 2009; Schurek et al., 2009; Moskowitz et al., 2012; Jochumsen et al., 2016; Jeannot et al., 2017). Accordingly, different studies have found that a functional Ara4N pathway is required for the acquisition of colistin resistance by P. aeruginosa in in vitro evolution experiments (Jochumsen et al., 2016; Chung et al., 2017; Lo Sciuto and Imperi, 2018), implying that lipid A aminoarabinosylation could represent a possible target for the development of anti-Pseudomonas colistin adjuvants. Indeed, inhibitors of the Ara4N transferase ArnT have been recently demonstrated to potentiate colistin activity against colistin-resistant P. aeruginosa strains (Ghirga et al., 2020; Quaglio et al., 2020).
In contrast, the role of PEtN modification of lipid A in P. aeruginosa colistin resistance is much less clear. The P. aeruginosa genome contains a functional ortholog of the PEtN transferase gene eptA, whose expression is induced by extracellular zinc through the two-component system ColR–ColS (Nowicki et al., 2015). Notably, zinc-induced expression of EptA and the consequent addition of PEtN to lipid A were, however, reported not to affect colistin MIC (Nowicki et al., 2015), leading to hypothesize that EptA-mediated lipid A phosphoethanolamination does not confer colistin resistance in P. aeruginosa. On the other hand, more recently, a couple of studies investigated the effect of the exogenous PEtN transferase MCR-1 on P. aeruginosa colistin resistance, by introducing engineered or native plasmids carrying mcr-1 with its native promoter. PEtN modification of lipid A was invariably observed in MCR-1 expressing strains and conferred colistin resistance, even though the resistance level varied significantly among different isolates and was overall much lower than that observed in other Gram-negative pathogens, such as Escherichia coli, Klebsiella pneumoniae, or A. baumannii (Liu et al., 2016, 2017). Interestingly, whilst until recently mcr plasmids were mainly found in Enterobacteriaceae, in the last couple of years some reports described the sporadic isolation of mcr-positive colistin resistant isolates of P. aeruginosa (Hameed et al., 2019; Abd El-Baky et al., 2020; Tahmasebi et al., 2020a,b; Nitz et al., 2021). Although these studies did not provide genetic and/or molecular evidence that mcr genes were indeed responsible for the colistin resistance phenotype, this body of evidence suggests that lipid A phosphoethanolamination could play a role in colistin resistance also in P. aeruginosa and, thus, that the contribution of endogenous and exogenous PEtN transferases to colistin resistance might be different in this bacterium.
To clarify the role of PEtN modification of lipid A in colistin resistance and to directly compare the in vivo activities of endogenous (EptA) and exogenous (MCR-1) lipid A PEtN transferases in P. aeruginosa, in this work we cloned the eptA and mcr-1 genes in a plasmid for isopropyl-β-D-thiogalactoside (IPTG)-inducible gene expression and analyzed the effect of ectopic EptA or MCR-1 expression on P. aeruginosa colistin resistance and fitness.
Materials and Methods
Bacterial Strains and Culture Media
Strains and plasmids are listed in Supplementary Table 1. Lysogeny broth (LB, Acumedia) was used for genetic manipulation. Mueller–Hinton broth (MH, Difco) was used for Kirby–Bauer assay, while cation-adjusted MH (MH II, Difco) was used for all the other assays. When required, IPTG was added at the indicated concentrations. All chemicals were purchased from Merck.
Generation of Plasmids
To obtain the constructs for IPTG-inducible expression of mcr-1, eptA, or the truncated variant eptAΔC–ter, the coding sequence of each gene was amplified by PCR using the genomic DNA of P. aeruginosa PAO1 (for eptA and eptAΔC–ter) or the plasmid pHNSHP45 (for mcr-1) as the template (Supplementary Table 1). The amplicons were digested with the restriction enzymes EcoRI and XhoI and individually cloned into the sequencing plasmid pBlueScript II KS (pBS; Stratagene), previously digested with the same enzymes. The construct pBSeptA was used as template for PCR-mediated site-specific mutagenesis using the “Q5 Site-Directed Mutagenesis” kit (New England BioLabs), according to manufacturer’s instructions, and a primer pair specifically designed to introduce an A-to-G substitution leading to the mutation T278A, thus obtaining the eptAT278A allele (Supplementary Table 1). After verification of the cloned fragments through restriction analysis and DNA sequencing, the inserts were excised from pBS and sub-cloned, using the same enzymes, into the shuttle vector pME6032, which replicates in P. aeruginosa and allows IPTG-inducible expression of cloned genes (Heeb and Haas, 2001). Primers used for cloning, site-specific mutagenesis and sequencing are listed in Supplementary Table 2. The constructs were transferred into P. aeruginosa strains by transformation.
Gene Expression Analysis
The expression level of eptA was determined by quantitative reverse transcription PCR (qRT-PCR). Bacteria were cultured in MH II in the presence of increasing concentrations of IPTG until mid-log phase, harvested by centrifugation and treated with RNAprotect Bacteria Reagent (Qiagen). RNA was purified using the RNeasy Mini Kit (Qiagen), treated with DNase, and re-purified with the RNeasy MinElute Cleanup Kit (Qiagen). cDNA was reverse transcribed from 0.5 μg of RNA with Prime Script RT Reagent Kit (Takara). Appropriate dilutions of the cDNA were used as template for qRT-PCR in a AriaMx Real-Time PCR System (Agilent) using TB Green Premier EX Taq master mix (Takara) and the primers listed in Supplementary Table 2. Relative gene expression with respect to the housekeeping gene rpoD was calculated using the 2–ΔΔ Ct method (Livak and Schmittgen, 2001).
Lipid A Analysis
Bacteria were cultured in MH II until early stationary phase, harvested by centrifugation and lipid A was extracted using the ammonium hydroxide-isobutyric acid-based procedure (Liu et al., 2017) with previously described modifications (Lo Sciuto et al., 2018). Samples were analyzed by matrix-assisted laser desorption-time of flight (MALDI-TOF) in the negative-ion mode (Lo Sciuto et al., 2018).
Growth Assays
Growth assays were performed in MH II in 96-well microtiter plates at 37°C (200 μL per well). Growth was measured as optical density at 600 nm (OD600) in a Tecan Spark 10M microtiter plate reader.
Pairwise competition experiments were performed by inoculating each strain of the pair of interest at 5 × 105 cells/mL in MH II. After 16 and 20 h at 37°C, cultures were serially diluted in saline and plated onto MH II agar and MH II agar containing 2 μg/mL colistin and 0.5 mM IPTG, to evaluate the number of cells carrying pMEeptA or pMEmcr-1 (that develop colonies both in the presence and absence of colistin) and the number of cells carrying the empty plasmid pME6032 (that develop colonies only in the absence of colistin). Preliminary experiments were performed to select the lowest colistin concentration that inhibits the growth of cells with the empty vector without affecting the plating efficiency of those carrying pMEeptA or pMEmcr-1. The competitive index was calculated as the ratio between the number of cells expressing EptA or MCR-1 and the number of cells carrying pME6032 at 16 or 24 h divided by to same ratio at time 0 (Lo Sciuto et al., 2020).
MIC Assays
Colistin MIC was determined through the broth microdilution method in MH II. Strains were cultured at 37°C for 8 h, and then refreshed at 5 × 105 cells/mL in the presence of increasing concentrations of colistin (up to 128 μg/mL). MIC was recorded after 24 h at 37°C. Each strain was tested in at least six independent experiments.
Kirby–Bauer Disc Diffusion Assay
Bacterial cell suspensions in saline were normalized at 0.5 McFarland standard and swabbed onto MH agar plates. Disks containing ciprofloxacin (5 μg), imipenem (10 μg), gentamicin (10 μg), novobiocin (30 μg), erythromycin (15 μg), rifampicin (5 μg), or tobramycin (10 μg) (Becton Dickinson) were placed on the surface of the inoculated plates, and growth inhibition halo diameters were measured after 24-h incubation at 37°C.
Detergent Sensitivity Assay
Bacteria were cultured in MH II until mid-exponential phase, harvested by centrifugation, resuspended in saline at OD600 of 0.5 and dispensed in microtiter plate wells in the presence of increasing concentrations (0–5%) of sodium dodecyl sulphate (SDS) (200 μL per well). Sensitivity to the SDS lytic effect was assessed by determining the turbidity (OD600) of cell suspensions after 5-min incubation at room temperature (Lo Sciuto et al., 2014), using a Tecan Spark 10M microtiter plate reader.
Fluorescent Probe-Permeability Assays
Permeability assays were performed as previously described (Hancock and Farmer, 1993; Kwon et al., 2019), with few modifications. Bacteria were cultured in MH II until mid-exponential phase, harvested by centrifugation, resuspended in 5 mM HEPES (pH 7.2) at OD600 of 2 and dispensed in microtiter plate wells in the presence of 1-N-phenylnaphthylamine (NPN) or propidium iodide (PI) at 10 μM or 20 μg/mL final concentration, respectively. Fluorescence was measured in a Tecan Spark 10M microtiter plate reader (excitation at 350 nm and emission at 420 nm for NPN; excitation at 580 nm and emission at 620 nm for PI) after 5 min at room temperature, subtracted of the background values of samples without NPN or PI, and normalized to the OD600 of the cell suspension.
In silico Analyses
Sequence alignments were generated with Clustal Omega.1 Residue conservation analysis was performed with the ConSurf server.2 The TMHMM server was used for transmembrane helix prediction.3 Three-dimensional modeling was performed with SWISS-MODEL,4 using the Neisseria meningitidis EptA protein (PDB 5FGN) as the template.
Statistical Analyses
Statistical analysis was performed with GraphPad InStat, using one-way analysis of variance (ANOVA) followed by Tukey–Kramer multiple comparison test.
Results
Effect of EptA and MCR-1 on Lipid A
To investigate the contribution of PEtN-modification of the lipid A moiety of LPS to colistin resistance in P. aeruginosa, and to evaluate the effect of this lipid A modification on bacterial fitness, two plasmids were generated for the IPTG-inducible expression of either the endogenous lipid A PEtN transferase EptA or the exogenous analog MCR-1. These plasmids, named pMEeptA and pMEmcr-1, were individually introduced in two P. aeruginosa reference strains, namely PAO1 and PA14, that are distantly related and are representative of the two major strain groups of the P. aeruginosa population structure (Freschi et al., 2015).
To confirm that phosphoethanolamination of lipid A occurs in the strains expressing MCR-1 or EptA, the lipid A was extracted from PAO1 and PA14 carrying the empty vector pME6032, pMEeptA, or pMEmcr-1 cultured in the presence of 0.5 mM IPTG. Lipid A was then analyzed by MALDI-TOF mass spectrometry (Figure 1). The lipid A spectra were characterized by a major peak corresponding to diphosphorylated penta-acylated lipid A (m/z 1446), that represents the main lipid A form of laboratory P. aeruginosa strains (Lo Sciuto et al., 2018, 2020). Less abundant peaks were also identified, such as tetra-acylated diphosphorylated lipid A (m/z 1276), penta-acylated monophosphorylated lipid A (m/z 1366), penta-acylated triphosphorylated lipid A (m/z 1526), and hexa-acylated lipid A (m/z 1616). Notably, some lipid A forms were found with different hydroxylation states, due to the presence/absence of a hydroxyl group in the two secondary C12 acyl chains (Lo Sciuto et al., 2019).
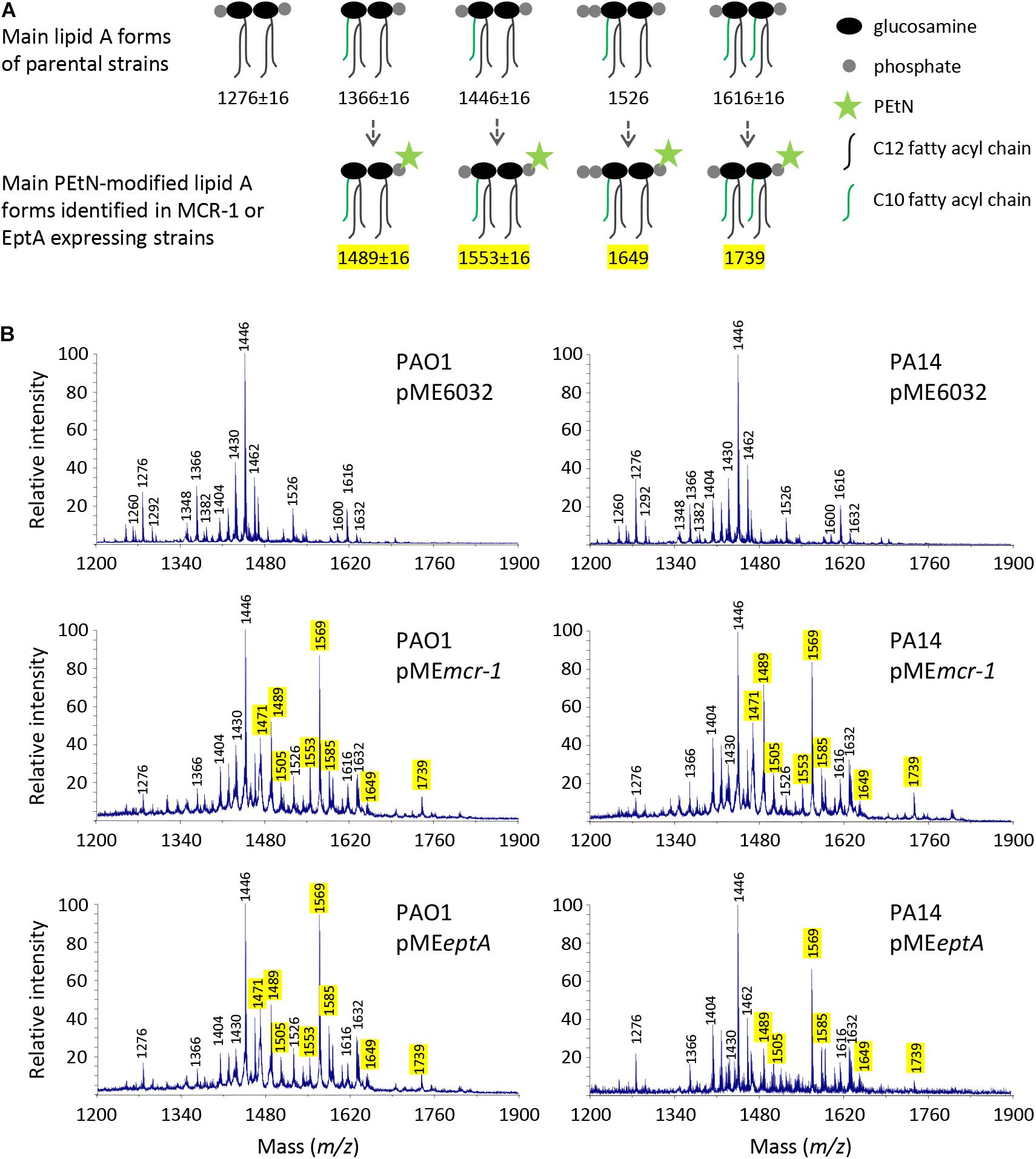
Figure 1. Effect of MCR-1 or EptA expression on lipid A in P. aeruginosa PAO1 and PA14. (A) Cartoons showing the relevant lipid A forms identified by mass spectrometry. (B) MALDI-TOF spectra of lipid A extracted from cells cultured in the presence of 0.5 mM IPTG. Spectra were obtained in the ion negative mode, so m/z values correspond to (molecular mass – 1)/1. m/z = 1276, diphosphorylated tetra-acylated lipid A; m/z = 1366, monophosphorylated penta-acylated lipid A; m/z = 1446, diphosphorylated penta-acylated lipid A; m/z = 1526, triphosphorylated penta-acylated lipid A; and m/z = 1616, diphosphorylated hexa-acylated lipid A. m/z + 123 indicates the addition of a PEtN group, while m/z ± 16 indicates lipid A forms that vary for the presence/absence of a hydroxyl group in the secondary C12 acyl chains (Nowicki et al., 2015; Liu et al., 2017; Lo Sciuto et al., 2019). The peak at m/z = 1471 likely corresponds to the phosphoethanolaminated form of a still non-identified lipid A species at m/z = 1348 (Ghirga et al., 2020; Lo Sciuto et al., 2020). The m/z values of peaks corresponding to the phosphoethanolaminated lipid A forms here identified are highlighted in yellow.
As expected, additional peaks were exclusively detected in the lipid A spectra of cells carrying pMEeptA or pMEmcr-1. The addition of a PEtN group results in a + 123 shift in the m/z value of lipid A peaks (Nowicki et al., 2015; Liu et al., 2017). The peaks at m/z 1489 and 1569 correspond to the phosphoethanolaminated penta-acylated mono- or diphosphorylated form, respectively, while the peak at m/z 1739 corresponds to the phosphoethanolaminated hexa-acylated diphosphorylated form. Overall, lipid A analysis confirmed that both MCR-1 and EptA expressed from the pMEmcr-1/eptA constructs are functional and that they efficiently modify lipid A with PEtN. Notably, PEtN-modified lipid A was not observed in both PAO1 and PA14 carrying the empty plasmid (Figure 1), confirming that the endogenous copy of eptA, that is present in the genome of both strains,5 is not or only poorly expressed in standard laboratory media (Nowicki et al., 2015; Liu et al., 2017; Lo Sciuto et al., 2018).
Effect of Lipid A Phosphoethanolamination on Colistin Resistance
To investigate the effect of MCR-1 or EptA expression on colistin resistance, MIC assays were performed in the presence of different IPTG concentrations (Table 1). As expected, strains carrying the empty plasmid were susceptible to colistin, as inferred from MIC values lower than the clinical breakpoint for P. aeruginosa (2 μg/mL; EUCAST). P. aeruginosa expressing MCR-1 showed an IPTG-dose dependent increase in colistin MIC, confirming the capability of MCR-1 mediated lipid A phosphoethanolamination to confer colistin resistance in P. aeruginosa (Liu et al., 2017). Notably, colistin MIC was fourfold higher for PA14 pMEmcr-1 than for PAO1 pMEmcr-1 (Table 1). This is in line with the results of a recent study about the contribution of Ara4N-modified lipid A to colistin resistance, that was found to be significantly greater in PA14 with respect to PAO1 (Lo Sciuto et al., 2020). These results confirm previous evidence that PA14 is more responsive than PAO1 to lipid A modifications that confer colistin resistance (Gutu et al., 2015). Notably, when colistin MIC was directly compared between MCR-1 expressing strains, cultured in the presence of 0.5 mM IPTG, and recombinant strains that constitutively express the arn operon responsible for Ara4N modification of lipid A (PAO1/PA14 PrpsA:arn), in which the arn promoter was replaced with the promoter of the housekeeping gene rpsA (Lo Sciuto et al., 2020; Supplementary Table 1), it appeared that lipid A phosphoethanolamination is as efficient as aminoarabinosylation in conferring colistin resistance to P. aeruginosa (Table 1).
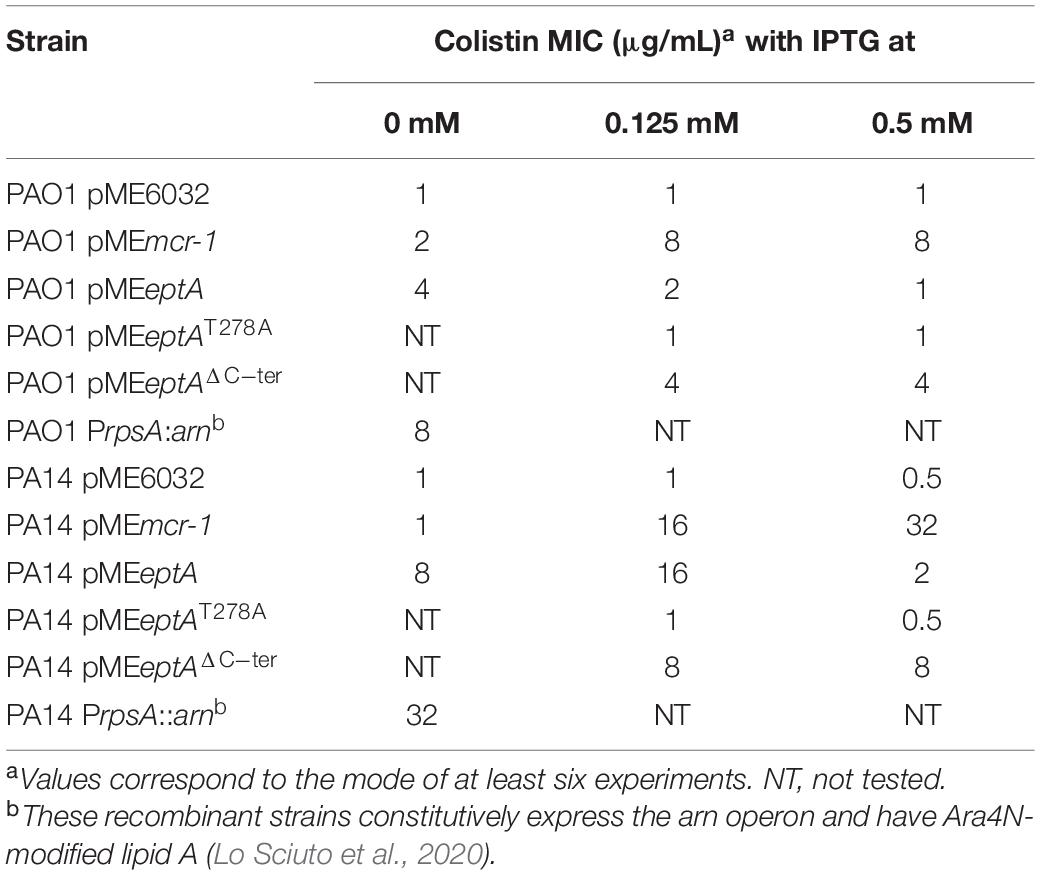
Table 1. MIC of colistin for P. aeruginosa PAO1 and PA14 expressing or not MCR-1, EptA or EptA variants.
In contrast, colistin resistance in EptA overexpressing strains did not correlate with the IPTG concentration in the medium. Indeed, colistin MIC was higher at 0 (for PAO1) or 0.125 mM IPTG (for PA14) and reverted at levels comparable to the empty plasmid controls at 0.5 mM IPTG (Table 1). The significant increase in colistin MIC observed in strains carrying pMEeptA under non-inducing conditions prompted us to verify lipid A phosphoethanolamination and eptA expression levels at different IPTG concentrations. Mass spectrometry analysis revealed that lipid A was efficiently phosphoethanolaminated even in the absence of IPTG (Supplementary Figure 1). This is in line with the results of the qRT-PCR analysis, which showed that eptA expression was hundreds-fold higher in pMEeptA carrying strains under non-inducing conditions as compared to empty plasmid controls (Figure 2). Overall, these experiments revealed that P. aeruginosa EptA can promote colistin resistance at levels almost comparable to those conferred by the exogenous PEtN transferase MCR-1, even if proper EptA levels seem to be critical for optimal induction of resistance.
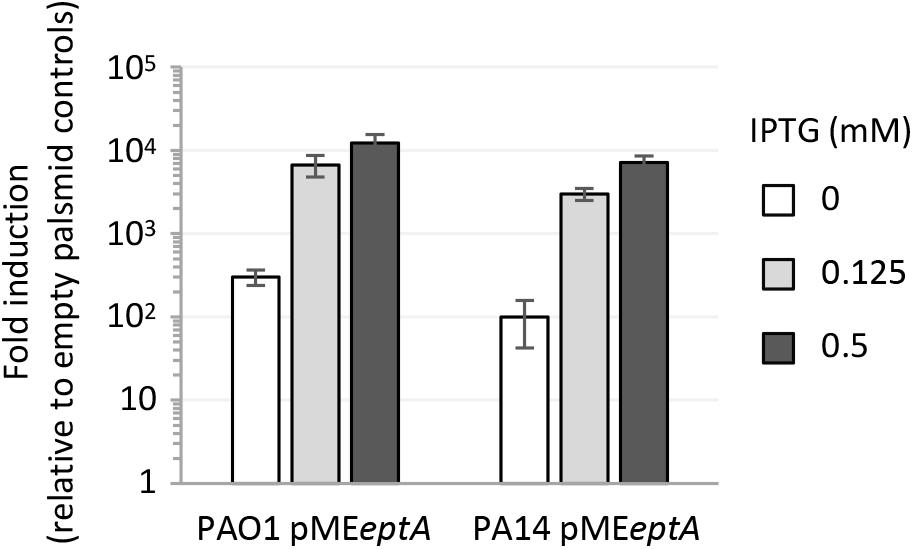
Figure 2. Expression level of eptA, determined by qRT-PCR, in P. aeruginosa PAO1 or PA14 carrying pMEeptA cultured in the presence of increasing IPTG concentrations. Relative eptA gene expression is shown as fold induction with respect to the cognate strain carrying the empty plasmid pME6032. Data represent the mean (±SD) of two independent experiments performed in triplicate.
Effect of EptA and MCR-1 on Bacterial Fitness
The relevant decrease in colistin MIC observed in EptA expressing cells cultured in the presence of 0.5 mM IPTG (Table 1) led us to hypothesize that EptA could exert some toxicity when expressed at very high levels (about 10,000-fold higher than the expression level of the empty plasmid controls; Figure 2), that could somehow counterbalance the positive effect of PEtN-modified lipid A on colistin resistance. To evaluate whether EptA overexpression causes any fitness costs, we compared planktonic and colony growth between EptA/MCR-1 expressing cells and the empty plasmid controls. As shown in Figure 3A and Supplementary Figure 2, MCR-1 expression did not affect growth of any strain at any IPTG concentration. In contrast, the expression of EptA caused a reduction in the growth kinetics of both PAO1 and PA14 in an IPTG-dose dependent manner, with maximum growth inhibition observed at IPTG concentrations ≥0.5 mM (Figure 3A and Supplementary Figure 2). Colony growth over time was also slightly delayed for EptA overexpressing strains as compared to the other strains (Figure 3B), corroborating the negative effect of EptA on P. aeruginosa growth kinetics. The growth defects of EptA overexpressing cells were further investigated through competition experiments, by co-culturing cells carrying pMEeptA (or pMEmcr-1 as the control) with those with the empty vector in the presence of 0.5 mM IPTG. The results are shown as competitive index, representing the relative growth of EptA (or MCR-1) overexpressing strains with respect to the empty plasmid controls (Figure 3C). In agreement with the results of mono-culture assays, cells expressing MCR-1 showed a competitive index close to 1, indicating that their growth kinetic was overall comparable to that of cells lacking mcr-1. On the other hand, both PAO1 pMEeptA and PA14 pMEeptA showed a much lower competitive index (0.1 and 0.2, respectively), implying that EptA overexpressing cells were outcompeted by cells harboring the empty plasmid.
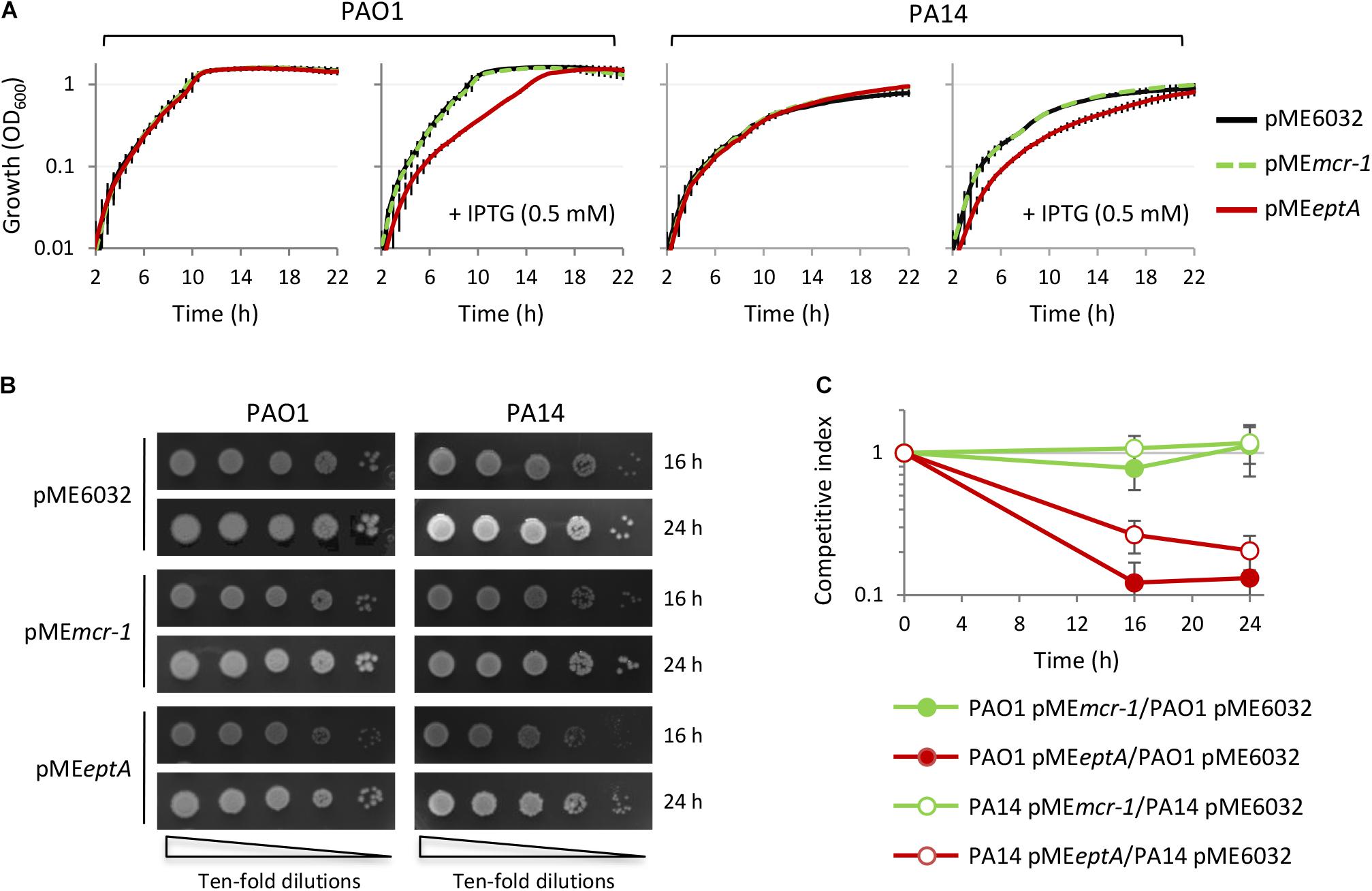
Figure 3. Effect of MCR-1 or EptA overexpression on P. aeruginosa growth. (A) Growth curves of PAO1 or PA14 containing the empty vector pME6032 (black lines), pMEmcr-1 (green lines), or pMEeptA (red lines), cultured in the absence or in the presence of 0.5 mM IPTG. (B) Colony growth of the same strains described in (A) on agar plates containing 0.5 mM IPTG. (C) In vitro competition between PAO1 or PA14 cells with the empty vector and cells overexpressing MCR-1 or EptA in the presence of 0.5 mM IPTG. Data represent the mean (±SD) or are representative of at least three independent experiments.
To investigate whether the detrimental effect EptA overexpression on P. aeruginosa growth correlated with defects in the integrity and/or permeability barrier of the cell envelope, we compared the sensitivity profile to different antibiotics belonging to different classes of P. aeruginosa PAO1 and PA14 carrying the empty plasmid pME6032 or the pMEmcr-1/pMEeptA derivatives cultured in the presence of 0.5 mM IPTG, by using the Kirby–Bauer disc diffusion test. MCR-1 expressing strains did not show any difference in the resistance profile with respect to the empty plasmid controls, while EptA overexpressing strains showed a slight increase in susceptibility to some antibiotics, i.e., ciprofloxacin, gentamicin, tobramycin, and imipenem (Figure 4A). All strains remained completely insensitive to high molecular weight antibiotics, such as novobiocin, erythromycin, and rifampicin, that are poorly active against P. aeruginosa because of poor diffusion across the OM and that are generally used to highlight defects in the OM integrity (Nikaido, 2005; Lo Sciuto et al., 2014, 2018). This suggests that EptA overexpressing cells have minor defects (if any) in cell envelope homeostasis. This was confirmed by the analysis of the sensitivity profile to the lytic effect of SDS, that was overall comparable between MCR-1/EptA overexpressing cells and the empty plasmid controls (Figure 4B). To further investigate the possible effect of PEtN and/or PEtN transferases on the P. aeruginosa cell envelope, the OM and IM permeability was assessed through the fluorescent probes NPN and PI, respectively (Hancock and Farmer, 1993; Kwon et al., 2019; Figure 4C). No differences were observed in NPN and PI fluorescence between MCR-1 expressing cells and empty plasmid controls. In contrast, EptA overexpression slightly increased NPN and PI uptake in PA14 but not in PAO1, suggesting that EptA might have a strain-specific effect on OM and IM integrity. Notably, such increase in membrane permeability can hardly explain the observed growth defects of EptA expressing cells, as both mono- and co-culture assays revealed that the negative impact of EptA overexpression on growth is more severe in PAO1 than in PA14 (Figure 3).
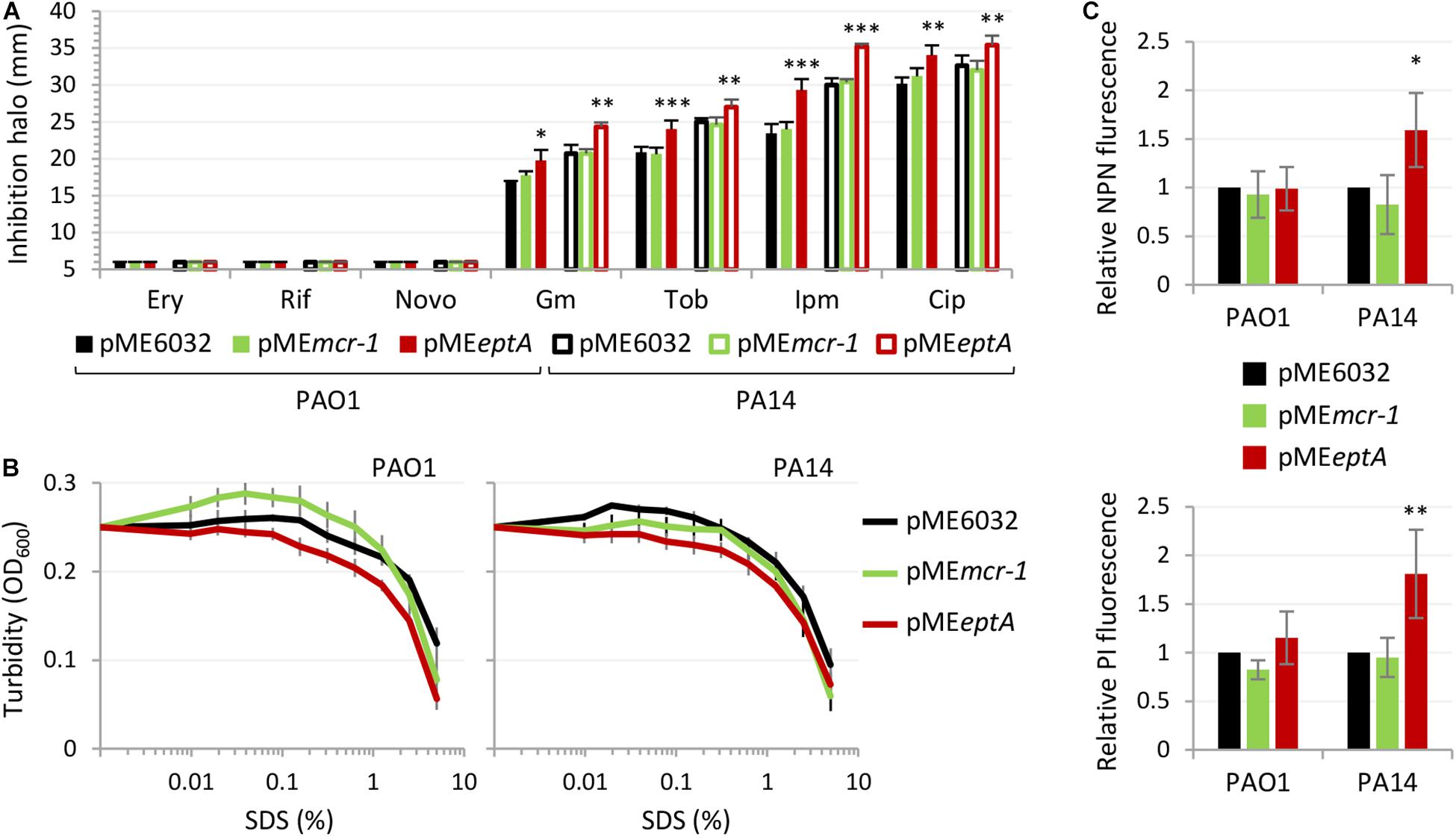
Figure 4. Effect of MCR-1 or EptA overexpression on antibiotic sensitivity and cell envelope integrity. (A) Inhibition halos in the Kirby–Bauer disc diffusion assay of erythromycin (Ery), rifampicin (Rif) novobiocin (Novo), gentamicin (Gm), tobramycin (Tob), imipenem (Ipm), or ciprofloxacin (Cip) for PAO1 or PA14 containing pME6032, pMEmcr-1, or pMEeptA. (B) Lytic effect of SDS, measured as decrease in cell suspension turbidity (OD600), on PAO1 or PA14 cells containing pME6032, pMEmcr-1, or pMEeptA. (C) Uptake of NPN (upper panel) and PI (lower panel) by PAO1 or PA14 cells containing pME6032, pMEmcr-1, or pMEeptA, reported as fluorescence emission relative to the empty plasmid controls. In all the assays, strains were cultured in the presence of 0.5 mM IPTG. Data represent the mean (±SD) of three (A,B) or five (C) independent experiments. Asterisks indicate a statistically significant difference with respect to the corresponding empty plasmid control (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001; ANOVA).
Overall, these assays revealed that MCR-1 and lipid A phosphoethanolamination do not influence cell envelope homeostasis in P. aeruginosa, while EptA overexpression has a marginal effect on the cell envelope permeability barrier, which is, however, unrelated to the growth defects of EptA expressing cells.
The Growth Inhibitory Effect of EptA Is Not Related to Phosphoethanolamine Transferase Activity
The finding that EptA but not MCR-1 overexpression is deleterious to P. aeruginosa strongly suggests that the growth defects might not be related to PEtN-modified lipid A. This, however, does not rule out that the PEtN transferase activity might be involved in the noxious effects of EptA overexpression. In fact, while most PEtN transferases have high substrate specificity (Zhang et al., 2019), a PEtN transferase of Campylobacter jejuni is more promiscuous, being able to add PEtN to (1) lipid A, (2) heptose I of the LPS inner core, (3) a Thr residue of the flagellar protein FlgG, and (4) the N-terminally linked glycans of some periplasmic proteins (Cullen et al., 2013). Considering that P. aeruginosa EptA has been poorly characterized so far (Nowicki et al., 2015), we attempted to verify the relevance of PEtN transferase activity for EptA-dependent toxicity. To this aim, we generated a construct (pMEeptAT278A) to express a catalytically inactive EptA variant (EptAT278A), mutagenized in a conserved catalytic residue essential for N. meningitidis EptA activity (Xu et al., 2018a; Supplementary Figure 3). As expected, lipid A was not modified with PEtN in EptAT278A overexpressing cells (Figure 5A and Supplementary Figure 4) and, accordingly, colistin MIC for these cells was identical to that for the empty plasmid controls (Table 1). Interestingly, overexpression of EptAT278A caused growth defects comparable to those caused by wild type EptA (Figure 5B), indicating that PEtN transferase activity is not responsible for P. aeruginosa EptA deleterious effects. We therefore decided to search for possible unique feature(s) of P. aeruginosa EptA, by evaluating the evolutionary conservation among 150 representative EptA orthologs through the ConSurf server (Ashkenazy et al., 2016). This analysis highlighted a 25-aa C-terminal tail in the periplasmic domain of P. aeruginosa EptA that is absent from all other EptA orthologs (Figure 6). The IPTG-inducible expression of a truncated EptA variant lacking the last 22 amino acids (EptAΔC–ter) in P. aeruginosa cells, followed by mass spectrometry analysis of lipid A, revealed that the C-terminal extension is not required for lipid A phosphoethanolamination (Figure 5A). In contrast, cells expressing the EptAΔC–ter variant showed less severe growth defects as compared to cells expressing wild type EptA (Figure 5B), implying that the C-terminal tail is partly responsible for the growth inhibitory effect of EptA overexpression. Moreover, EptAΔC–ter expressing strains showed high colistin MIC even in the presence of high IPTG concentrations (Table 1), supporting the hypothesis that the inverse proportion between wild type EptA expression and colistin resistance is likely due to the negative impact of high EptA levels on P. aeruginosa growth. Although previous studies have shown that also other PEtN transferases can negatively impact bacterial physiology (Xu et al., 2018b; Yang et al., 2020), this is the first report that associates PEtN transferase toxicity to a specific and evolutionary divergent protein domain. Further studies are required to elucidate the mechanistic basis of EptA-mediated toxicity in P. aeruginosa.
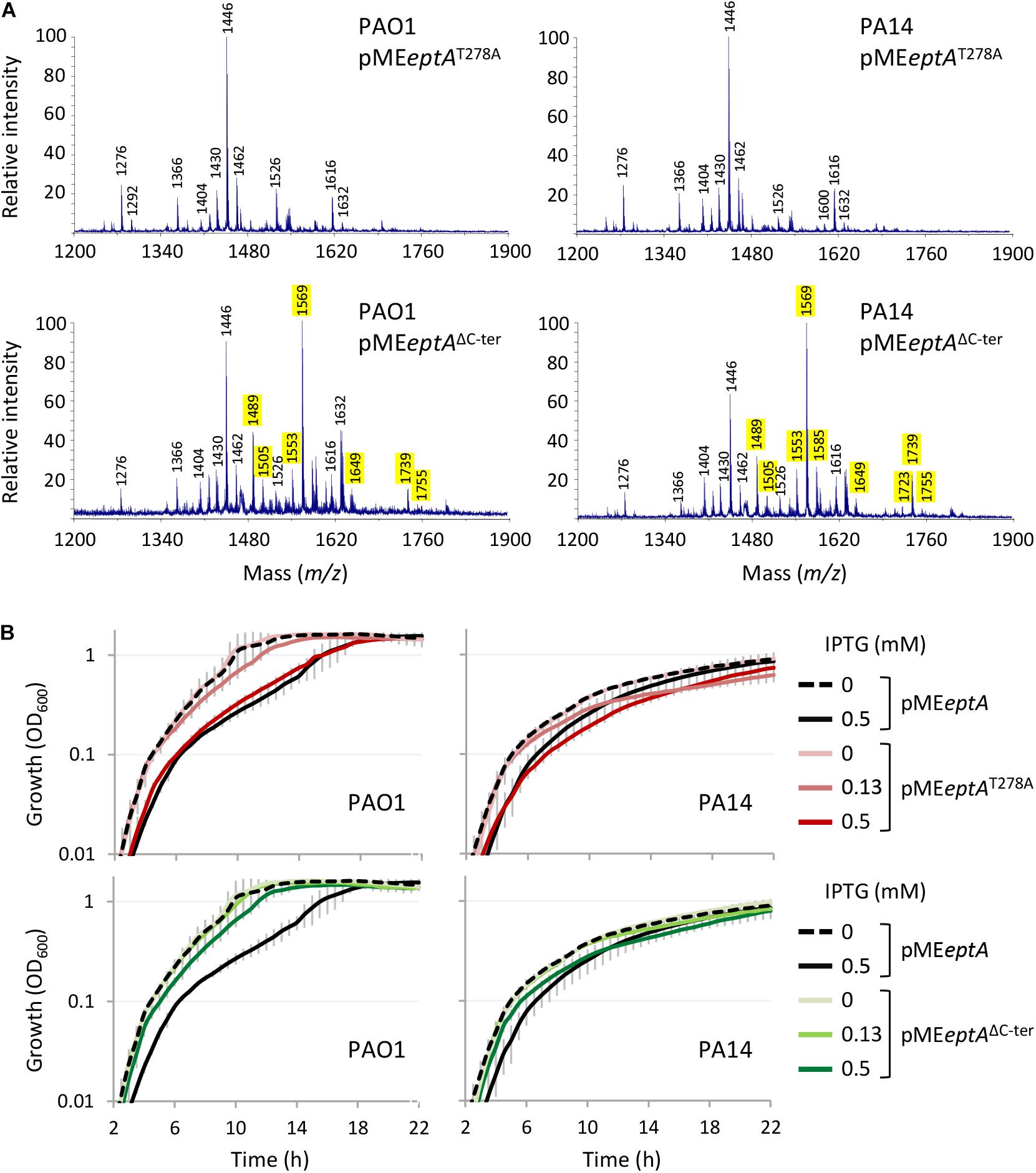
Figure 5. Effect of the expression of EptA variants mutated in a catalytic residue (EptAT278A) or deleted of 22 amino acids at the C-terminus (EptAΔC–ter) in P. aeruginosa. (A) MALDI-TOF spectra of lipid A extracted from PAO1 or PA14 containing pMEeptA, pMEeptAT278A, or pMEeptAΔC–ter cultured in the presence of 0.125 mM IPTG. The m/z values of peaks corresponding to phosphoethanolaminated lipid A forms are highlighted in yellow. See the legend to Figure 1 for further details. (B) Growth curves of PAO1 or PA14 containing pMEeptA, pMEeptAT278A, or pMEeptAΔC–ter cultured in the presence of the indicated IPTG concentrations. Data represent the mean (±SD) or are representative of at least three independent experiments.
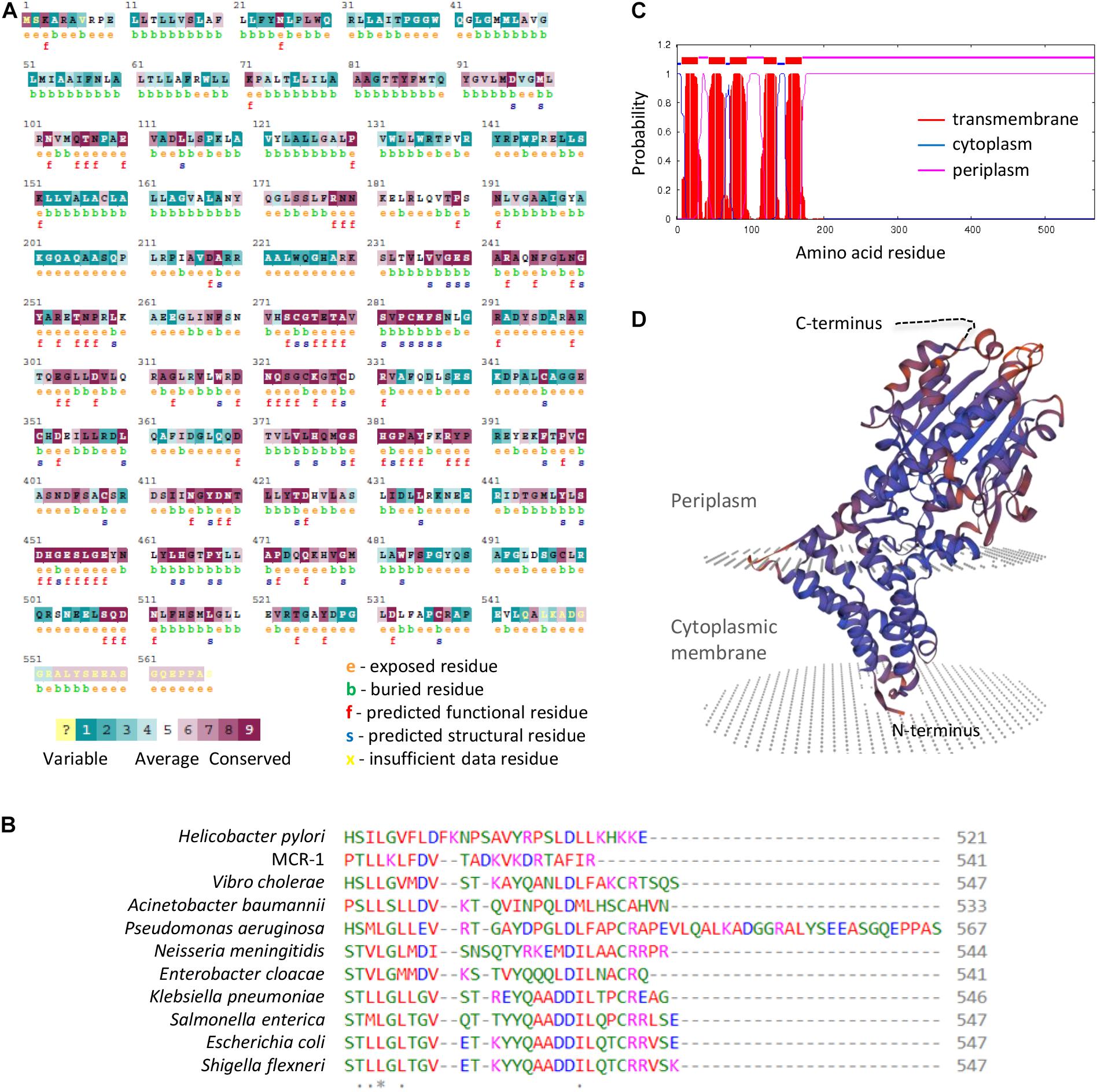
Figure 6. (A) ConSurf analysis of evolutionary residue conservation in P. aeruginosa EptA. (B) Alignment of the C-terminus of selected EptA orthologs from the indicated bacteria. The MCR-1 protein has been included as control. The corresponding full-length alignment is shown in Supplementary Figure 3. (C) Prediction of transmembrane helices in P. aeruginosa EptA by TMHMM. (D) Modeling of the P. aeruginosa EptA structure by SWISS-MODEL using the N. meningitidis EptA protein (PDB 5FGN) as the template. The C-terminal tail of P. aeruginosa EptA cannot be modeled and is represented as a dashed black line.
Conclusion
While several studies highlighted the importance of lipid A aminoarabinosylation for colistin resistance in P. aeruginosa, the role of lipid A phosphoethanolamination in this bacterium remained elusive. Here, we have demonstrated that (1) exogenous and endogenous PEtN transferases (MCR-1 and EptA) have overall comparable lipid A PEtN transferase activity in P. aeruginosa, (2) both are effective in promoting colistin resistance, and (3) modification of lipid A with PEtN confers colistin resistance levels comparable to Ara4N. EptA overexpression was found to negatively impact P. aeruginosa fitness, in a manner that is independent of the lipid A phosphotransferase activity but dependent on the presence of a C-terminal extension specific to P. aeruginosa EptA. The mechanism by which this C-terminal tail affects growth has not been investigated in this study, but we are tempted to speculate that it could not be physiologically relevant, as growth defects were only observed at very high EptA expression levels, thousands-fold higher than the basal levels. For comparison, the only cue that has been proven to trigger P. aeruginosa eptA transcription to date (i.e., extracellular zinc) only caused a 20-fold increase in eptA gene expression (Nowicki et al., 2015).
Given the efficacy of EptA in conferring colistin resistance, the question arises as to why lipid A aminoarabinosylation is preferred over lipid A phosphoethanolamination as colistin resistance mechanism in P. aeruginosa during in vitro evolution experiments as well as in the clinical setting (Barrow and Kwon, 2009; Schurek et al., 2009; Moskowitz et al., 2012; Jochumsen et al., 2016; Chung et al., 2017; Lo Sciuto and Imperi, 2018). Even if we cannot exclude that the fitness cost associated with the EptA C-terminal domain could somehow favor Ara4N producing cells over PEtN producing ones, an alternative hypothesis is that the evolutionary pathways to Ara4N-mediated resistance are more likely to occur due to the high number of genes and, thus, of possible mutations directly or indirectly triggering arn gene expression (Barrow and Kwon, 2009; Schurek et al., 2009; Moskowitz et al., 2012; Jochumsen et al., 2016; Jeannot et al., 2017). Since very few is known about eptA regulation in P. aeruginosa (Nowicki et al., 2015), further studies are needed to decipher the regulatory circuit(s) controlling eptA expression and the likelihood of eptA-inducing mutations in P. aeruginosa and, therefore, to support or deny such hypothesis.
On the other hand, the present study has also shown that MCR-1 mediated colistin resistance in P. aeruginosa has no impact on relevant physiological aspects, such as growth, drug sensitivity and cell envelope integrity, suggesting that there would be no fitness barriers to the spread of mcr genes in the P. aeruginosa population. This is an issue that deserves further investigation, especially in view of the recent identification of mcr harboring P. aeruginosa clinical isolates in different geographic areas (Hameed et al., 2019; Abd El-Baky et al., 2020; Tahmasebi et al., 2020a,b; Nitz et al., 2021).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AL and FI conceived and designed the experiments. MC, AL, and CB performed the experiments. MC, AL, CM, and FI analyzed the data. CM and FI contributed reagents, materials, and analysis tools. FI wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Pasteur Institute-Cenci Bolognetti Foundation (call 2018), the PRIN 2017 grant protocol 20177J5Y3P, the Italian Cystic Fibrosis Foundation (grant FFC#15/2019), the Excellence Departments grant from the Italian Ministry of Education, University and Research (MIUR, Italy) (Art. 1, commi 314-337 Legge 232/2016) to the Department of Science, Roma Tre University, and the Regione Lazio (“Gruppi di Ricerca 2020,” POR A0375E0026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to thank Robert Ernst (University of Maryland, Baltimore) for sharing the protocol for lipid A extraction and analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.778968/full#supplementary-material
Footnotes
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://consurf.tau.ac.il/index_proteins.php
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ https://swissmodel.expasy.org/interactive
- ^ www.pseudomonas.com
References
Abd El-Baky, R. M., Masoud, S. M., Mohamed, D. S., Waly, N. G., Shafik, E. A., Mohareb, D. A., et al. (2020). Prevalence and some possible mechanisms of colistin resistance among multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 13, 323–332. doi: 10.2147/IDR.S238811
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi: 10.1093/nar/gkw408
Barrow, K., and Kwon, D. H. (2009). Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 5150–5154. doi: 10.1128/AAC.00893-09
Chung, E. S., Lee, J. Y., Rhee, J. Y., and Ko, K. S. (2017). Colistin resistance in Pseudomonas aeruginosa that is not linked to arnB. J. Med. Microbiol. 66, 833–841. doi: 10.1099/jmm.0.000456
Cullen, T. W., O’Brien, J. P., Hendrixson, D. R., Giles, D. K., Hobb, R. I., Thompson, S. A., et al. (2013). EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infect. Immun. 81, 430–440. doi: 10.1128/IAI.01046-12
Falagas, M. E., and Kopterides, P. (2007). Old antibiotics for infections in critically ill patients. Curr. Opin. Crit. Care 13, 592–597. doi: 10.1097/MCC.0b013e32827851d7
Falagas, M. E., Rafailidis, P. I., and Matthaiou, D. K. (2010). Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13, 132–138. doi: 10.1016/j.drup.2010.05.002
Freschi, L., Jeukens, J., Kukavica-Ibrulj, I., Boyle, B., Dupont, M. J., Laroche, J., et al. (2015). Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 6:1036. doi: 10.3389/fmicb.2015.01036
Gajdács, M., Baráth, Z., Kárpáti, K., Szabó, D., Usai, D., Zanetti, S., et al. (2021). No Correlation between biofilm formation, virulence factors, and antibiotic resistance in Pseudomonas aeruginosa: results from a laboratory-based in vitro study. Antibiotics 10:1134. doi: 10.3390/antibiotics10091134
Ghirga, F., Stefanelli, R., Cavinato, L., Lo Sciuto, A., Corradi, S., Quaglio, D., et al. (2020). A novel colistin adjuvant identified by virtual screening for ArnT inhibitors. J. Antimicrob. Chemother. 75, 2564–2572. doi: 10.1093/jac/dkaa200
Giamarellou, H. (2016). Epidemiology of infections caused by polymyxin-resistant pathogens. Int. J. Antimicrob. Agents 48, 614–621. doi: 10.1016/j.ijantimicag.2016.09.025
Gutu, A. D., Rodgers, N. S., Park, J., and Moskowitz, S. M. (2015). Pseudomonas aeruginosa high-level resistance to polymyxins and other antimicrobial peptides requires cprA, a gene that is disrupted in the PAO1 strain. Antimicrob. Agents Chemother. 59, 5377–5387. doi: 10.1128/AAC.00904-15
Hameed, F., Khan, M. A., Muhammad, H., Sarwar, T., Bilal, H., and Rehman, T. U. (2019). Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev. Soc. Bras. Med. Trop. 52:e20190237. doi: 10.1590/0037-8682-0237-2019
Hancock, R. E., and Farmer, S. W. (1993). Mechanism of uptake of deglucoteicoplanin amide derivatives across outer membranes of Escherichia coli and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 3, 453–456. doi: 10.1128/AAC.37.3.453
Heeb, S., and Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant Microbe Interact. 14, 1351–1363. doi: 10.1094/MPMI.2001.14.12.1351
Horcajada, J. P., Montero, M., Oliver, A., Sorlí, L., Luque, S., Gómez-Zorrilla, S., et al. (2019). Epidemiology and treatment of multidrug-Resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32, e00031–19. doi: 10.1128/CMR.00031-19
Jeannot, K., Bolard, A., and Plésiat, P. (2017). Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 49, 526–535. doi: 10.1016/j.ijantimicag.2016.11.029
Jochumsen, N., Marvig, R. L., Damkiær, S., Jensen, R. L., Paulander, W., Molin, S., et al. (2016). The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 7:13002. doi: 10.1038/ncomms13002
Kwon, J. Y., Kim, M. K., Mereuta, L., Seo, C. H., Luchian, T., and Park, Y. (2019). Mechanism of action of antimicrobial peptide P5 truncations against Pseudomonas aeruginosa and Staphylococcus aureus. AMB. Express. 9:122. doi: 10.1186/s13568-019-0843-0
Langan, K. M., Kotsimbos, T., and Peleg, A. Y. (2015). Managing Pseudomonas aeruginosa respiratory infections in cystic fibrosis. Curr. Opin. Infect. Dis. 28, 547–556. doi: 10.1097/QCO.0000000000000217
Liu, Y. Y., Chandler, C. E., Leung, L. M., McElheny, C. L., Mettus, R. T., Shanks, R. M. Q., et al. (2017). Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-Negative ESKAPE pathogens. Antimicrob. Agents Chemother. 61, e00580–17. doi: 10.1128/AAC.00580-17
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408.
Lo Sciuto, A., Cervoni, M., Stefanelli, R., Mancone, C., and Imperi, F. (2020). Effect of lipid A aminoarabinosylation on Pseudomonas aeruginosa colistin resistance and fitness. Int. J. Antimicrob. Agents 55:105957. doi: 10.1016/j.ijantimicag.2020.105957
Lo Sciuto, A., Cervoni, M., Stefanelli, R., Spinnato, M. C., Di Giamberardino, A., Mancone, C., et al. (2019). Genetic Basis and Physiological Effects of Lipid A Hydroxylation in Pseudomonas aeruginosa PAO1. Pathogens 8:291. doi: 10.3390/pathogens8040291
Lo Sciuto, A., Fernández-Piñar, R., Bertuccini, L., Iosi, F., Superti, F., and Imperi, F. (2014). The periplasmic protein TolB as a potential drug target in Pseudomonas aeruginosa. PLoS One 9:e103784. doi: 10.1371/journal.pone.0103784
Lo Sciuto, A., and Imperi, F. (2018). Aminoarabinosylation of Lipid A Is Critical for the Development of Colistin Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62, e01820–17. doi: 10.1128/AAC.01820-17
Lo Sciuto, A., Martorana, A. M., Fernández-Piñar, R., Mancone, C., Polissi, A., and Imperi, F. (2018). Pseudomonas aeruginosa LptE is crucial for LptD assembly, cell envelope integrity, antibiotic resistance and virulence. Virulence 9, 1718–1733. doi: 10.1080/21505594.2018.1537730
Moffatt, J. H., Harper, M., and Boyce, J. D. (2019). Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 1145, 55–71. doi: 10.1007/978-3-030-16373-0_5
Moskowitz, S. M., Brannon, M. K., Dasgupta, N., Pier, M., Sgambati, N., Miller, A. K., et al. (2012). PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents. Chemother. 56, 1019–1030. doi: 10.1128/AAC.05829-11
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q. T., and Li, J. (2021). Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 73, 679–728. doi: 10.1124/pharmrev.120.000020
Nikaido, H. (2005). Restoring permeability barrier function to outer membrane. Chem. Biol. 12, 507–509. doi: 10.1016/j.chembiol.2005.05.001
Nitz, F., de Melo, B. O., da Silva, L. C. N., de Souza Monteiro, A., Marques, S. G., Monteiro-Neto, V., et al. (2021). Molecular detection of drug-resistance genes of blaOXA-23-blaOXA-51 and mcr-1 in clinical isolates of Pseudomonas aeruginosa. Microorganisms 9:786. doi: 10.3390/microorganisms9040786
Nowicki, E. M., O’Brien, J. P., Brodbelt, J. S., and Trent, M. S. (2015). Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol. Microbiol. 97, 166–178. doi: 10.1111/mmi.13018
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Pogue, J. M., Ortwine, J. K., and Kaye, K. S. (2017). Clinical considerations for optimal use of the polymyxins: a focus on agent selection and dosing. Clin. Microbiol. Infect. 23, 229–233. doi: 10.1016/j.cmi.2017.02.023
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Quaglio, D., Mangoni, M. L., Stefanelli, R., Corradi, S., Casciaro, B., Vergine, V., et al. (2020). ent-Beyerane Diterpenes as a Key Platform for the Development of ArnT-Mediated Colistin Resistance Inhibitors. J. Org. Chem. 85, 10891–10901. doi: 10.1021/acs.joc.0c01459
Sabnis, A., Hagart, K. L., Klöckner, A., Becce, M., Evans, L. E., Furniss, R. C. D., et al. (2021). Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife. 10:e65836. doi: 10.7554/eLife.65836
Schurek, K. N., Sampaio, J. L., Kiffer, C. R., Sinto, S., Mendes, C. M., and Hancock, R. E. (2009). Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4345–4351. doi: 10.1128/AAC.01267-08
Tahmasebi, H., Dehbashi, S., and Arabestani, M. R. (2020a). Co-harboring of mcr-1 and β-lactamase genes in Pseudomonas aeruginosa by high-resolution melting curve analysis (HRMA): molecular typing of superbug strains in bloodstream infections (BSI). Infect. Genet. Evol. 85:104518. doi: 10.1016/j.meegid.2020.104518
Tahmasebi, H., Dehbashi, S., and Arabestani, M. R. (2020b). Prevalence and Molecular Typing of colistin-resistant Pseudomonas aeruginosa (CRPA) Among β-Lactamase-Producing isolates: a study based on high-resolution melting curve analysis method. Infect. Drug Resist. 13, 2943–2955. doi: 10.2147/IDR.S264796
Theuretzbacher, U., Bush, K., Harbarth, S., Paul, M., Rex, J. H., Tacconelli, E., et al. (2020). Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 18, 286–298. doi: 10.1038/s41579-020-0340-0
Velkov, T., Thompson, P. E., Nation, R. L., and Li, J. (2010). Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916. doi: 10.1021/jm900999h
Xu, Y., Wei, W., Lei, S., Lin, J., Srinivas, S., and Feng, Y. (2018a). An Evolutionarily Conserved Mechanism for Intrinsic and Transferable Polymyxin Resistance. mBio 9, e02317–17. doi: 10.1128/mBio.02317-17
Xu, Y., Zhong, L. L., Srinivas, S., Sun, J., Huang, M., Paterson, D. L., et al. (2018b). Spread of MCR-3 colistin resistance in china: an epidemiological, genomic and mechanistic study. EBioMedicine 34, 139–157. doi: 10.1016/j.ebiom.2018.07.027
Yang, Q. E., MacLean, C., Papkou, A., Pritchard, M., Powell, L., Thomas, D., et al. (2020). Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J. 14, 861–865. doi: 10.1038/s41396-019-0578-6
Keywords: cell envelope, EptA, lipid A, MCR, phosphoethanolamine (PEtN), polymyxin, 4-amino-4-deoxy-L-arabinose
Citation: Cervoni M, Lo Sciuto A, Bianchini C, Mancone C and Imperi F (2021) Exogenous and Endogenous Phosphoethanolamine Transferases Differently Affect Colistin Resistance and Fitness in Pseudomonas aeruginosa. Front. Microbiol. 12:778968. doi: 10.3389/fmicb.2021.778968
Received: 17 September 2021; Accepted: 07 October 2021;
Published: 27 October 2021.
Edited by:
Márió Gajdács, University of Szeged, HungaryReviewed by:
Payam Behzadi, Islamic Azad University, IranMatthew Gavino Donadu, University of Sassari, Italy
Copyright © 2021 Cervoni, Lo Sciuto, Bianchini, Mancone and Imperi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Imperi, ZnJhbmNlc2NvLmltcGVyaUB1bmlyb21hMy5pdA==
†These authors have contributed equally to this work
 Matteo Cervoni
Matteo Cervoni Alessandra Lo Sciuto
Alessandra Lo Sciuto Chiara Bianchini
Chiara Bianchini Carmine Mancone2
Carmine Mancone2 Francesco Imperi
Francesco Imperi