- 1Vetsuisse Faculty, Institute for Food Safety and Hygiene, University of Zurich, Zurich, Switzerland
- 2Kantonales Labor Basel-Stadt, Basel, Switzerland
- 3Department of Pathobiology, Functional Microbiology, Institute of Microbiology, University of Veterinary Medicine Vienna, Vienna, Austria
Bacillus thuringiensis is a microbial insecticide widely used to control agricultural pests. Although generally regarded as safe, B. thuringiensis is phylogenetically intermingled with the foodborne pathogen B. cereus sensu stricto and has been linked to foodborne outbreaks. Limited data on the pathogenicity potential of B. thuringiensis and the occurrence of biopesticide residues in food compromise a robust consumer risk assessment. In this study, we analyzed whole-genome sequences of 33 B. thuringiensis isolates from biopesticides, food, and human fecal samples linked to outbreaks. All food and outbreak-associated isolates genomically matched (≤ 6 wgSNPs; ≤ 2 cgSNPs) with one of six biopesticide strains, suggesting biopesticide products as their source. Long-read sequencing revealed a more diverse virulence gene profile than previously assumed, including a transposase-mediated disruption of the promoter region of the non-hemolytic enterotoxin gene nhe and a bacteriophage-mediated disruption of the sphingomyelinase gene sph in some biopesticide strains. Furthermore, we provide high-quality genome assemblies of seven widely used B. thuringiensis biopesticide strains, which will facilitate improved microbial source tracking and risk assessment of B. thuringiensis-based biopesticides in the future.
Introduction
Bacillus thuringiensis is one of the most popular plant protection agents worldwide, accounting for 90% of the bioinsecticide market (Chattopadhyay et al., 2004). The organism synthesizes parasporal crystals as well as other entomopathogenic factors with insecticidal activity, leading to its widespread application in agriculture and vector control (Chattopadhyay et al., 2004; Jouzani et al., 2017). B. thuringiensis-based biopesticides represent an important alternative to broad-spectrum chemical pesticides, as they exhibit a high degree of host specificity, with only limited harmful effects to non-target species (Lacey et al., 2015). In the coming decade, the use of these already highly popular biopesticides will likely increase further. The agrosystems transformation and major political agendas are committed to promoting healthy foods and preserving biodiversity. Major drivers are the United Nations’ Sustainable Development Goals and the Farm to Fork Strategy, which is a vital part of the European Green Deal: By the year 2030, a decrease of chemical pesticides by 50% is envisaged, while at the same time increasing organic farming to 25% of total farmland. These ambitious agendas will inevitably generate additional demand for biopesticides and may substantially boost the use of B. thuringiensis-based formulations.
However, B. thuringiensis lacks Qualified Presumption of Safety (QPS) status and the role of B. thuringiensis as a causative agent of diarrheal disease is controversially discussed (McIntyre et al., 2008; EFSA BIOHAZ Panel, 2016; Raymond and Federici, 2017; Bonis et al., 2021). B. thuringiensis belongs to the B. cereus sensu lato group and is genetically intermingled with the widely recognized foodborne pathogen B. cereus sensu stricto (Ehling-Schulz et al., 2019; Carroll et al., 2020b). In addition, B. thuringiensis strains from biopesticides can express medium to high levels of enterotoxins and were linked to outbreaks of diarrheal disease (Johler et al., 2018; Schwenk et al., 2020; Bonis et al., 2021).
B. thuringiensis is ubiquitous in the environment, which may result in the contamination of agricultural products (Ankolekar et al., 2009; Kim et al., 2017; Bağcıoğlu et al., 2019). Thus, it has been suggested that B. cereus-like spores detected on foods largely stem from contamination with naturally occurring wild-type strains (Raymond and Federici, 2017). While there are indications that residues from biopesticides could also be detected (Frederiksen et al., 2006; Hendriksen and Hansen, 2006; Zhou et al., 2008; Johler et al., 2018; Frentzel et al., 2020; Bonis et al., 2021), the extent to which these are present in foods is unclear. In this study, we aimed to utilize genomic analysis to assess a potential link of biopesticides to B. thuringiensis strains detected on foods.
Materials and Methods
Isolation of B. cereus sensu lato From Food Samples
A total of 100 samples of tomatoes, bell peppers, and salads from a central retail distribution facility in Switzerland were screened for B. cereus sensu lato (Table 1 and Supplementary Table 1). 10 g of each sample was added to 90 g peptone salt solution (0.1% enzymatic casein digest) and homogenized in a stomacher@ 400 circulator (Seward) for 60 s. Subsequently, B. cereus s.l. counts were determined by plating in serial dilutions on selective media (Mannitol-Egg Yolk-Polymyxin [MYP] agar) and incubated at 30°C overnight. Up to five presumptive B. cereus s.l. colonies (mannitol negative, phospholipase C positive phenotype) per sample were selected and whenever different colony morphologies were observed, these were included in the selection. Isolates were purified and preserved in glycerol stocks at −80°C until further characterization.
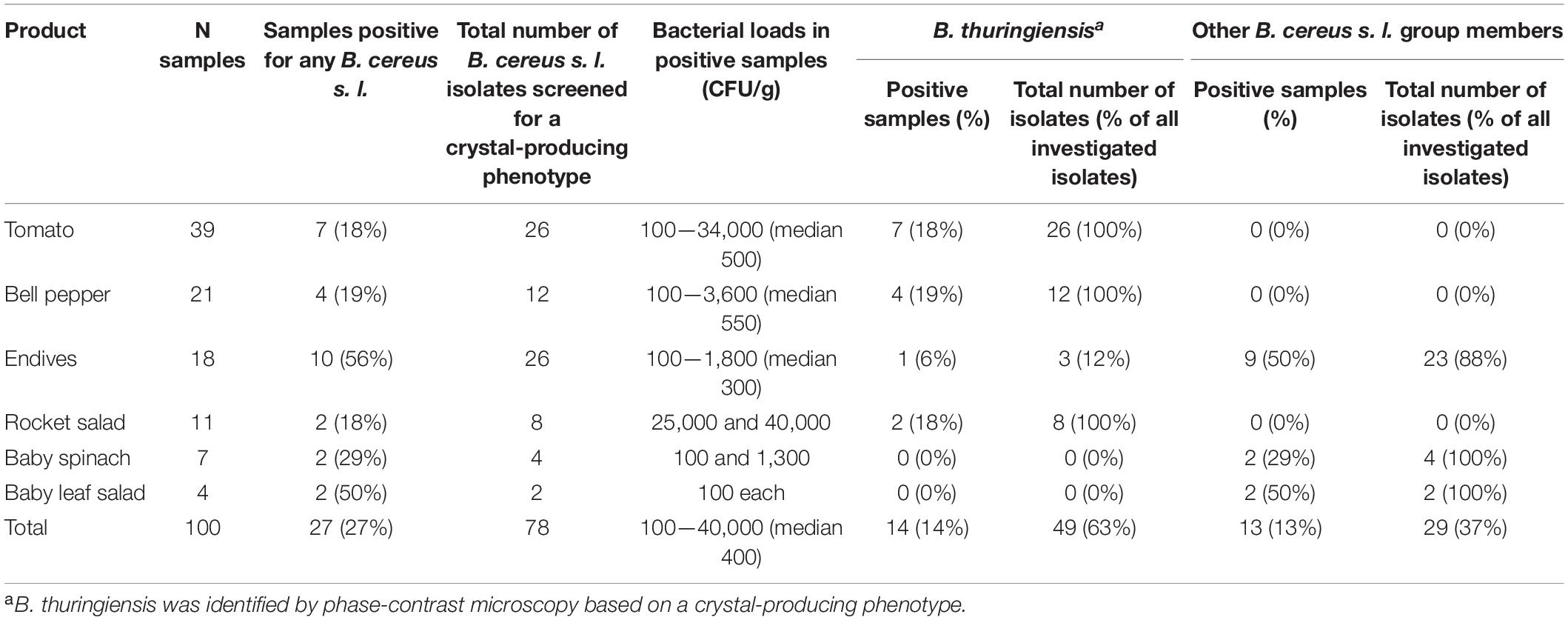
Table 1. Prevalence of B. thuringiensis and other B. cereus group members in 100 food samples collected at the retail level in Switzerland.
Screening for Parasporal Crystals Characteristic for Bacillus thuringiensis
Pure cultures of all isolates were grown on T3 agar (Travers et al., 1987) for 3 days at 30°C. A tiny amount of colony material was resuspended in 10 μL sterile water and screened for crystals of diamond, spherical or bipyramidal shape using phase-contrast microscopy (EFSA BIOHAZ Panel, 2016). Isolates exhibiting characteristic parasporal crystals were defined as B. thuringiensis.
Metabolomic Fingerprinting by Fourier-Transform Infrared Spectroscopic Analysis
All B. thuringiensis were screened by FT-IR spectroscopy to detect clusters of related isolates and select a subset of strains for whole-genome sequencing. To this end, metabolic fingerprints of all isolates were generated as previously described (Ehling-Schulz et al., 2005). Briefly, isolates were cultivated as lawns on tryptone soy agar (TSA) (Oxoid) at 25°C for 24 h. Subsequently, one loop of cell material was suspended in 300 μL sterile deionized water, subjected to ultrasonication for 5 × 1 s at 100% power with a Bandelin Sonopuls HD2200 (Bandelin electronic), and the bacterial suspension was spotted on a zinc selenite (ZnSe) optical plate. After drying the plates, infrared absorption spectra were recorded using an HTS-XT microplate adapter coupled to a Tensor 27 FT-IR spectrometer (Bruker Optics). Spectra in the range of 4,000–500 cm–1 were acquired in transmission mode with the following parameters: 6 cm–1 spectral resolution, zero-filling factor 4, Blackmann-Harris 3-term apodization, and 32 interferograms were averaged with background subtraction for each spectrum. The quality of FTIR spectral data was evaluated using the OPUS software (version 7.5; Bruker Optics). Second derivatives were calculated using the Savitzky-Golay algorithm with 11 smoothing points and the derivative spectra were unit vector normalized for further data processing. The so-called fingerprint region (spectral region of 1,500–800 cm–1) was chosen for chemometric analyses. Hierarchical cluster analysis (HCA) was carried out using Ward’s algorithm.
Whole-Genome Sequencing and Genome Analyses
For short-read sequencing, genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen). Libraries were prepared using the Nextera DNA Flex Library Preparation Kit (Illumina), and sequencing was performed on the Illumina MiniSeq platform with 2 × 150 bp paired-end chemistries. Illumina read adapters and low-quality bases were trimmed with TrimGalore v0.6.61 and quality metrics computed using FastQC v0.11.92. Draft genomes were assembled using SPAdes v3.14.1 (Bankevich et al., 2012) implemented in shovill v1.1.03. Additionally, all seven biopesticide isolates obtained during this study were long-read sequenced. For nanopore sequencing, genomic DNA was extracted using the MasterPure Complete DNA and RNA Purification Kit (Lucigen). Multiplex libraries were prepared using the SQK-LSK109 ligation sequencing kit with the EXP-NBD114 native barcoding expansion kit (Oxford Nanopore Technologies). Libraries were sequenced on a MinION Mk1B device using the FLO-MIN106 (R9) flow cell (Oxford Nanopore Technologies). Adapters were trimmed using Porechop v0.2.44 and quality assessed with LongQC v1.2.0 (Fukasawa et al., 2020). Hybrid assemblies were produced with Unicycler 0.4.8 (Wick et al., 2017). Assembly quality metrics were assessed using QUAST v5.0.3 (Gurevich et al., 2013).
Multi-locus sequence types (MLST), panC types, and the presence of virulence genes were determined using BTyper3 and srst2 0.2.0 (Inouye et al., 2014; Carroll et al., 2020a). The completeness of virulence genes and their promoter regions was additionally investigated using BLAST (Altschul et al., 1990) implemented in abricate 1.0.15 and putative disruptions manually inspected in assemblies annotated with prokka v1.14.6 (Seemann, 2014). Prophages were detected using PHASTER (Arndt et al., 2016). sph genes disrupted by the insertion of a BceA1-like bacteriophage were identified by screening genomes for the sequence of open reading frame BG08_RS31255 (strain HD-1, GCF_000835235.1;>99% alignment coverage,>99% sequence identity in BLAST), which contains elements of sph and of a site-specific integrase from the bacteriophage.
SNP Analysis and Phylogenies
Core genome SNPs (cgSNPs) among CC8 and CC23 isolates were detected from Illumina read data using the CFSAN SNP pipeline v2.2.1 (Davis et al., 2015) with chromosomes of strains HD-1 (CC8, GCF_000835235.1), and BGSC 4Q7rifR (CC23, GCF_013113775.1) as references, respectively. Phages, IS elements, and repeat regions in reference chromosomes were identified using phastaf v0.1.06, ISEScan v1.7.2.3 (Xie and Tang, 2017), and NUCmer v3.1 (Marçais et al., 2018) and masked prior to read-mapping. cgSNP, i.e., SNP sites present in all genomes, were extracted from the SNP alignment by removing missing sites using SNP-sites v2.5.1 (Page et al., 2016), and core-genome SNP (cgSNP) distances were determined with snp-dists v0.7.07. Maximum-likelihood phylogenetic trees were constructed from the SNP alignment using IQ-TREE v2.0.3 (Nguyen et al., 2015) with the generalized time-reversible (GTR) model and gamma distribution with 100 bootstraps. The number of invariant sites was estimated from core genome alignments generated with parsnp v1.5.3 (Treangen et al., 2014) and passed to IQ-TREE. A phylogenetic tree of all 33 isolates was created using IQ-TREE directly from a core genome alignment generated with parsnp.
Whole-genome SNPs (wgSNPs) were detected by mapping reads of food/human isolates to the assembly of the phylogenetically closest biopesticide isolate (without any masking) using the CFSAN SNP pipeline. Read data of the respective reference biopesticide isolate were included in the pipeline as a reference to calculate wgSNP distances. The percentage of reference assembly bases covered by sequencing reads was determined with BBMap8.
Results
Occurrence of Bacillus thuringiensis in Foods at the Retail Level in Switzerland
A total of 100 food samples (tomatoes, bell pepper, endives, rocket salad, baby spinach, and baby leaf salad) were collected at the retail level. Presumptive B. cereus s. l. isolates were identified in 27 samples. From these 27 samples, a total of 78 isolates (up to 5 isolates per sample) were screened by phase-contrast microscopy. A crystal-producing phenotype, characteristic for B. thuringiensis, was identified for 49 isolates (63%) from 14 food samples (14%). Positive samples included tomatoes (7/39, 18%), bell pepper (4/21, 19%), rocket salad (2/11, 18%), and endives (1/18, 6%) (Table 1). No B. thuringiensis was detected in samples from baby spinach (n = 7) and baby leaf salad (n = 4). Remarkably, all investigated B. cereus s. l. isolates from tomatoes (n = 26), bell pepper (n = 12), and rocket salad (n = 8) were identified as B. thuringiensis. In the 14 samples positive for B. thuringiensis, presumptive B. cereus counts ranged from 100 to 40,000 CFU/g and were thus below the alert threshold of 105 CFU/g (Table 1 and Supplementary Table 1). In 13 food samples, only non-crystal-producing B. cereus s. l. isolates were detected. These were particularly common in endives (9/18, 50%) and baby leaf salad (2/4, 50%) and reached bacterial loads of up to 1,800 CFU/g.
Genetic Similarity of Bacillus thuringiensis Isolates From Biopesticide Products, Food, and Outbreaks
B. cereus s. l. isolates from all food samples were subjected to FT-IR spectroscopy for dereplication and selection of representative isolates to be included in the WGS analysis. B. thuringiensis isolates from the biopesticide products Agree, B401, Delfin, DiPel, Novodor, Solbac, and XenTari were included as references. Closely related isolates were grouped into clusters using Ward’s hierarchical clustering method. Out of each cluster containing B. thuringiensis, either all (for small clusters and singletons) or a minimum of three isolates from distinct samples were subjected to WGS analysis for further characterization. Two samples were represented by two isolates each which fell into different FT-IR clusters (Supplementary Table 2).
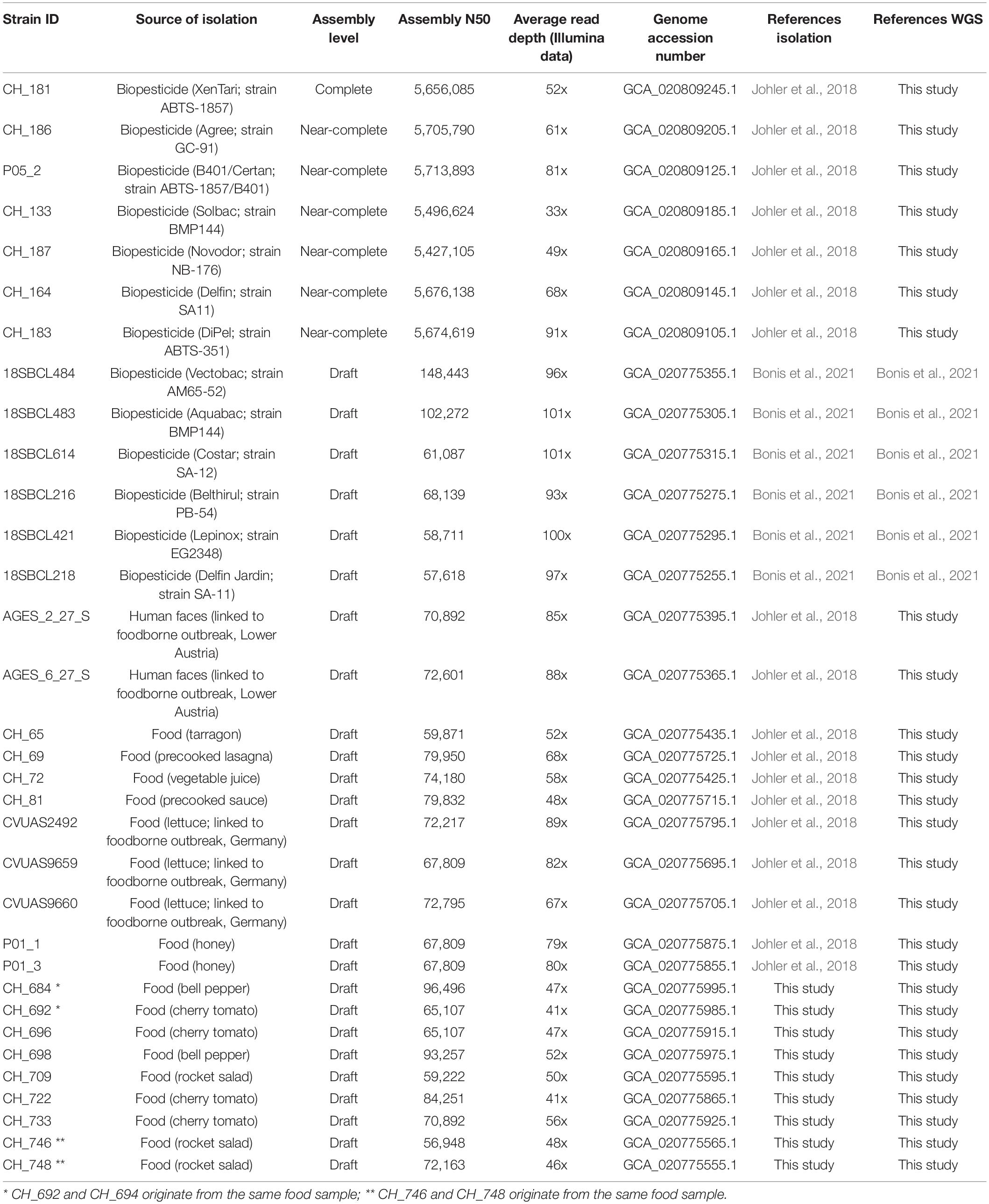
Overall, whole-genome sequences of 13 biopesticide product isolates, 15 food isolates, and 5 isolates linked to two foodborne outbreaks were investigated to identify potential matches of food/outbreak isolates with biopesticide strains. Of these 33 B. thuringiensis isolates, 27 were sequenced as part of this study (Table 2). These included 9 food isolates obtained from 7 samples as part of this study. To obtain a more comprehensive picture of possible links between biopesticide strains, food isolates, and outbreaks, the collection was supplemented with 18 previously described isolates originating from biopesticide products (n = 7), food (n = 9), or human fecal samples (n = 2) (Johler et al., 2018). Three of these food isolates and the two fecal isolates were linked to foodborne outbreaks of diarrheal disease (Johler et al., 2018); three food isolates were recovered during controls by a cantonal laboratory (CH_69, CH_72, CH_81) from lasagna, vegetable juice, and sauce; two isolates were obtained from honey during the production process; and one isolate was obtained from tarragon at the retail level. Furthermore, publicly available WGS data of 6 previously sequenced isolates from additional biopesticide products (Bonis et al., 2021) were included for genomic analyses.
All 33 genomes contained the B. thuringiensis-defining cry genes and were assigned to panC group IV. The 5 outbreak-associated isolates and 15 food isolates belonged to one of two closely related lineages ST8 (subspecies kurstaki) or ST15 (subspecies aizawai), which together form CC8. Most (11 out of 15) biopesticide isolates investigated in our study also fell into these two lineages. SNP analyses suggested that all food/fecal isolates originated from one of six distinct biopesticide strains: 19 of the 20 isolates differed by ≤ 4 wgSNPs (≤ 2 cgSNPs) from their phylogenetically closest biopesticide isolate (Figure 1) and food isolate CH_65 differed by 6 wgSNPs (2 cgSNPs) from its closest biopesticide isolate. The food isolates matched with biopesticide strains ABTS-351, ABTS-1857, ABTS-1857/B401, GC-91, SA-11, or SA-12. The five outbreak-associated isolates clustered in a sublineage of ST15 and differed by 0–3 wgSNPs (0–2 cgSNPs) from the biopesticide strain ABTS-1857, suggesting this biopesticide as a highly likely origin. Whereas most biopesticides are applied on a wide spectrum of crops, the biopesticide B401 is specifically recommended for the control of wax moths on honeycombs. Accordingly, the two isolates recovered from honey products corresponded to the B401 isolate (0 and 2 wgSNP/cgSNP difference, respectively).
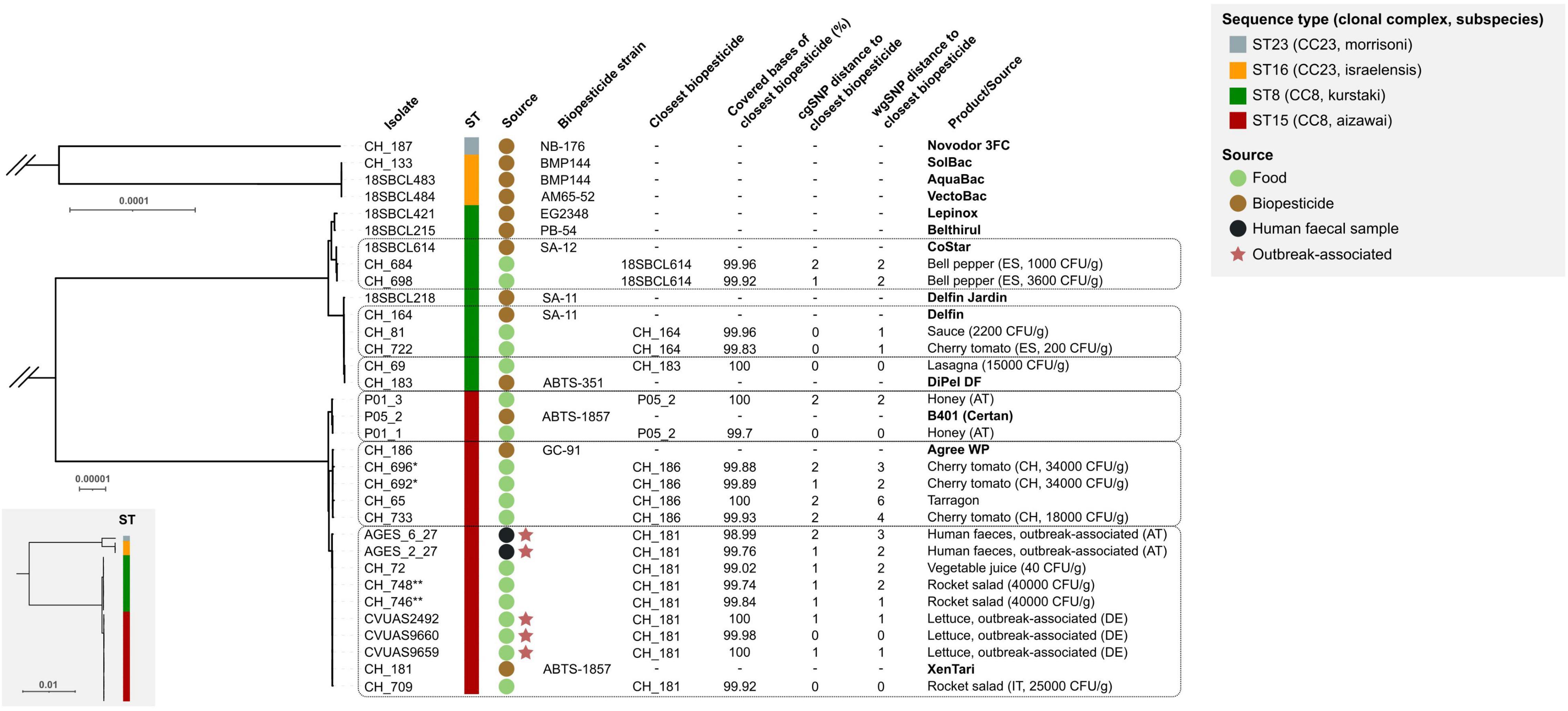
Figure 1. Genetic similarity of B. thuringiensis isolates from food, biopesticide products, and outbreaks. Maximum-likelihood phylogenetic trees for CC8 (ST8 and ST15) and CC23 (ST16 and ST23) were constructed from two separate core genome SNP (cgSNP) matrices. Branch lengths between CC8 and CC23 are shown in the full tree (bottom left box). Scale bars indicate the number of variant sites in core genomes of 3.8 Mbp (full tree), 4.6 Mbp (CC8), and 4.1 Mbp (CC23) lengths. The isolates’ sequence type (ST), clonal complex (CC), subspecies, and source of each isolate are labeled according to the legend. Dashed boxes group food/fecal isolates with their putative biopesticide source strain. Whole-genome SNPs (wgSNPs) were identified by mapping sequencing reads from each food/clinical isolate to the genome of the closest biopesticide isolate. The genome coverage refers to the number of bases in the reference biopesticide genome covered by reads of the food/clinical isolate, excluding repeat regions. The phylogeny was visualized in iTOL (Letunic and Bork, 2019). CFU: colony-forming units; AT, Austria; DE, Germany; ES, Spain; IT, Italy. * CH_692 and CH_694 originate from the same food sample ** CH_746 and CH_748 originate from the same food sample.
Toxin Gene Profiles of Biopesticide Isolates
The toxin gene profile varied between biopesticide isolates (Table 3). All biopesticide isolates except CH_187 (NB-176) carried intact genes and associated 5’ intergenic regions encoding cytotoxin CytK2 (cytK2), hemolysin BL (hblA, hblB, hblC, hblD), and non-hemolytic enterotoxin (nheA, nheB, nheC). In CH_187 (NB-176), cytK was absent and the nheA promoter disrupted by a transposase inserted 13 bp upstream of the translation initiation site, likely preventing effective transcription. As expected for B. thuringiensis, none of the isolates possessed the ces locus (encoding the emetic toxin cereulide).
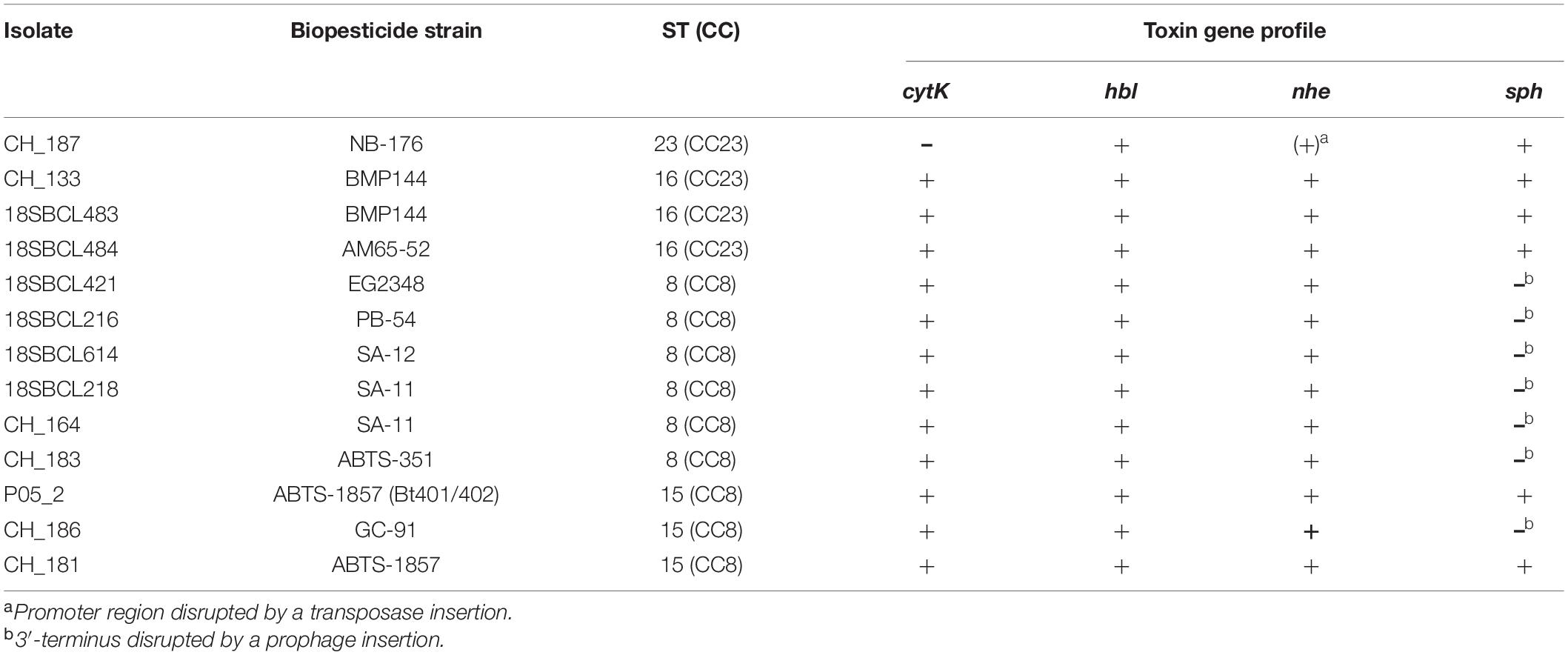
Table 3. Presence of intact toxin genes and their 5′ intergenic regions in genomes from biopesticide isolates.
In multiple B. thuringiensis genomes, the sph gene (sphingomyelinase) was disrupted by the insertion of prophages (Figure 2 and Table 3). Hybrid assemblies from biopesticide isolates CH_183 (ABTS-351, ST8) and CH_164 (SA-11, ST8) revealed the presence of the intact 42 kb Bacillus prophage BtCS33 (NC_018085.1, >99% alignment coverage and identity) inserted into the 3′-terminus of sph. Long-read assemblies from isolate CH_186 (GC-91, ST15) generated using Canu or Raven contained the intact 42 kb Bacillus prophage BceA1 (GCF_003047795.1, > 99% alignment coverage and identity) inserted into the 3′-terminus of sph. By contrast, sph was located at the end of a split contig in a hybrid assembly of CH_186 generated with Unicycler. Verification by PCR suggested that both intact and phage-disrupted sph loci are detectable in CH_186, possibly resulting from subpopulations of lysogenic, pseudo-lysogenic, lytic, and/or naïve cells. In sequencing read mapping analyses, the average base coverage in prophage regions of CH_186 was > 4-fold higher than in flanking regions, further indicating the presence of extrachromosomal DNA of active bacteriophages. Almost half of the BtCS33 and BceA1 genomes are homologous, including the identical site-specific integrases GP25 and P30, respectively (Figure 2). A 19 bp identical region in sph and downstream of the integrase genes was identified as the insertion site. In short-read assemblies, phage insertions could be identified by an incomplete sph sequence (96.85% alignment coverage). Accordingly, disrupted sph was also detected in all other biopesticide isolates from ST8, whereas biopesticide isolates from CC23 and two out of three biopesticide isolates from ST15 harbored intact sph genes. The toxin gene profile (sph completeness and nhe, hbl, cytK, ces) of food and fecal isolates always corresponded to their respective closest biopesticide isolate (shown in Figure 1).
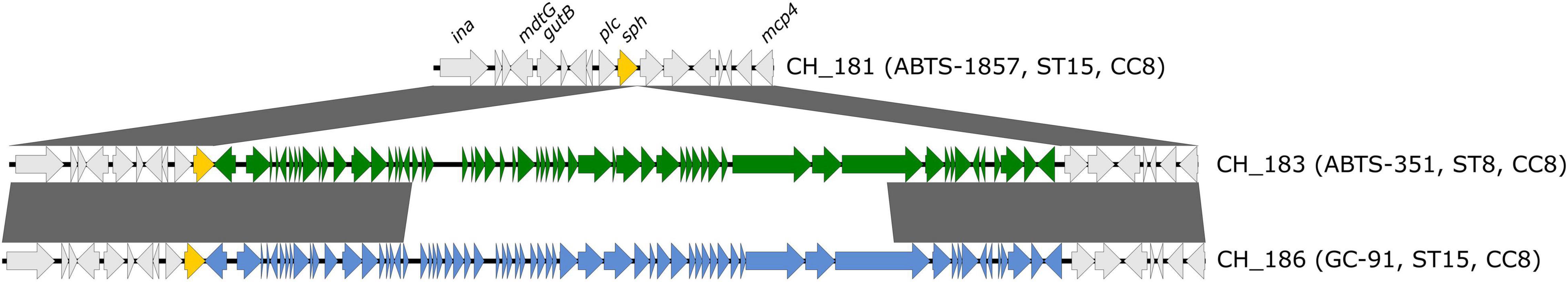
Figure 2. Disruption of the sphingomyelinase-encoding gene sph by prophage insertions. Genetic context of sph in B. thuringiensis biopesticide isolates CH_181 (ABTS-1857, ST15), CH_183 (ABTS-351, ST8), and CH_186 (GC-91, ST15). Shaded boxes between sequences indicate homologous regions (>90% sequence identity). CH_183 and CH_186 harbor the prophages BtCS33 (42 kb, green) and BceA1 (42 kb, blue), respectively, inserted into the 3′-terminus of sph. The figure was generated with Easyfig 2.1 (Sullivan et al., 2011).
Discussion
B. thuringiensis is frequently found on fresh vegetables, particularly on tomatoes, and has also been detected in processed foods such as lasagna, ratatouille, or strawberry cake (Johler et al., 2018; Bonis et al., 2021). Reliable source attribution is essential to assess a potential consumer risk posed by B. thuringiensis-based biopesticides. In this study, we investigated a potential link between 13 biopesticide isolates and 20 B. thuringiensis isolates from food and human fecal samples. SNP analyses revealed that all food/fecal isolates genomically matched (≤ 6 wgSNPs, median 2 wgSNPs; ≤ 2 cgSNPs, median 1 cgSNPs) with one of six widely used biopesticide strains, strongly suggesting biopesticide products as their source. While there is no specific SNP cutoff to define closely related isolates, pairwise distances among outbreak-linked emetic B. cereus isolates, for comparison, ranged from 0 to 8 cgSNPs when applying the CFSAN pipeline (Carroll et al., 2019). In some cases, the food/fecal isolates investigated here shared the same wgSNPs: for instance, food isolates CH_65, CH_692, CH_696, and CH_733 had two wgSNPs (CH_186 C3822937T and pCH_186-a T177153C; confirmed by Sanger sequencing) in common that distinguished them from their closest biopesticide isolate CH_186 (GC-91), suggesting that these isolates originate from a GC-91-like aizawai biopesticide product not included in this study. Alternatively, the investigated biopesticide isolate CH_186 may have acquired SNPs at these positions, for instance during storage and subculturing in the sequencing laboratory. A single SNP difference was found between the same-sample isolates CH_692 and CH_696, demonstrating that the identification of a limited number of SNPs among same-source isolates is plausible.
Consistent with our finding, the biopesticide residues were detected on food samples that corresponded to the recommended field of application of the respective products: B401, authorized for moth control in beekeeping, was re-isolated from honey; biopesticide strains authorized for pest control on various crops were re-isolated from fruits, vegetables, spices, and processed food; B. thuringiensis strains used for mosquito control (B. thuringiensis ssp. israelensis) or on potato foliage (B. thuringiensis ssp. morrisoni var. tenebrionis) were not found. B. thuringiensis isolates from lettuce linked to a foodborne outbreak in 2012 in Germany (EFSA BIOHAZ Panel, 2016) and two B. thuringiensis isolates from feces associated with a foodborne outbreak in Austria (Schmid et al., 2016) genomically matched ABTS-1857 (≤ 3 wgSNP; ≤ 2 cgSNP).
Our findings are consistent with other recent genomic analyses of B. thuringiensis food isolates. Among 83 B. thuringiensis isolates from tomatoes and bell pepper from retailers in Germany, 82 (99%) belonged to the ST8 or ST15 lineages (Frentzel et al., 2020), which also comprise the biopesticide strains commonly used for agricultural pest management. Of 42 isolates that were further characterized by WGS, 25 matched (4 cgSNPs) with one of the two included biopesticide strains, with the remaining 17 isolates possibly originating from biopesticide strains that were not covered by this study (Frentzel et al., 2020). In a comprehensive analysis of outbreak-associated B. thuringiensis isolates from food, Bonis et al. (2021) reported that most isolates phylogenetically clustered (0 – 10 cgSNPs) with biopesticide strains (Bonis et al., 2021). A plausible link between food and biopesticide strains was also established using FT-IR spectroscopy or low-resolution genotyping in earlier studies (Frederiksen et al., 2006; Hendriksen and Hansen, 2006; Zhou et al., 2008; Johler et al., 2018).
B. thuringiensis cytotoxicity can vary widely. While many strains exhibit low or medium cytotoxicity, a wild-type strain has been demonstrated to surpass the cytotoxicity of a B. cereus outbreak strain by a factor of 1.5 (Johler et al., 2018). Genes encoding the heat-labile enterotoxins Nhe, Hbl, and CtyK were identified in all biopesticide strains except NB-176 (ST23), which lacks the cytK gene. In addition, long-read sequencing revealed that the nhe promoter region of NB-176 is disrupted by a transposase insertion, likely preventing Nhe expression. In a previous study, NB-176 showed notably lower cytotoxicity in a Vero cell assay, indicating lower enterotoxin levels compared to other tested biopesticide strains (Johler et al., 2018).
Many B. cereus s.l. strains produce sphingomyelinase, a virulence factor contributing to B. cereus cytotoxicity that was shown to interact synergistically with Nhe and Hbl (Beecher and Wong, 2000; Doll et al., 2013). In biopesticide strains from ST8 and one (out of three) biopesticide strains from ST15, the sphingomyelinase-encoding sph gene was here found to be disrupted by bacteriophage insertions. In agreement with this disruption, no sphingomyelinase activity was detected in selected biopesticide isolates from ST8 [CH_164 (SA-11) and CH_183 (ABTS-351)] and in CH_186 (GC-91, ST15), whereas ST15 biopesticide isolates with an intact sph gene [CH_181 (ABTS-1857), P05_2 (Bt401)] showed low but detectable sphingomyelinase activities in a previous study (Johler et al., 2018). The presence of prophages integrated into sph may result from stochastic effects rather than being a strain-specific characteristic.
This study has notable limitations. No information was available on the prior use of B. thuringiensis-based biopesticides on the food samples, and there is no generally accepted SNP cut-off value to define a common origin of B. cereus isolates. Likewise, complete epidemiological investigations are needed for outbreak-associated isolates to substantiate our findings. The prevalence of B. thuringiensis was assessed on a limited number of samples from few food categories. Larger screenings across a wide range of food categories from various geographical origins are required to determine how frequently consumers are exposed to B. thuringiensis. Moreover, in our study, B. thuringiensis isolates were identified by phase-contrast microscopy. While this method is recognized as a tool for the identification of B. thuringiensis (EFSA BIOHAZ Panel, 2016), it cannot be excluded that additional isolates carried cry or cyt genes.
In conclusion, this study suggests that B. thuringiensis isolated from foods typically stem from biopesticide residues and we were able to genomically match outbreak-associated strains to Bacillus thuringiensis biopesticides. In addition, we present high-quality genome assemblies of seven widely used biopesticide strains, which provided novel insights into genetic mechanisms underlying differences in their cytotoxicity phenotype and will facilitate improved source-tracking in future studies.
Data Availability Statement
Sequencing data and genome assemblies generated as part of this study are available under BioProject PRJNA757250 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA757250). Genome accession numbers of all investigated isolates are listed in Supplementary Table 2.
Author Contributions
MB, ME-S, RS, and SJ designed the study. DE, PB, SC, and RS isolated strains and conducted laboratory experiments aimed at further characterization of the strains. MB performed bioinformatic analyses. MB and SJ drafted the manuscript that was revised and approved by all authors.
Funding
This research was supported by funding from the Swiss Federal Food Safety and Veterinary Office (project grant 0714001582).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Janine Carmen Steiner, Nicole Cernela, and Tatjana Svoboda for their excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.775669/full#supplementary-material
Supplementary Table 1 | Food samples screened for contamination with B. cereus sensu lato.
Supplementary Table 2 | B. thuringiensis isolates included in the genomic analyses.
Footnotes
- ^ https://github.com/FelixKrueger/TrimGalore
- ^ www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://github.com/tseemann/shovill
- ^ https://github.com/rrwick/Porechop/
- ^ https://github.com/tseemann/abricate
- ^ https://github.com/tseemann/phastaf
- ^ https://github.com/tseemann/snp-dists
- ^ sourceforge.net/projects/bbmap/
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ankolekar, C., Rahmati, T., and Labbé, R. G. (2009). Detection of toxigenic Bacillus cereus and Bacillus thuringiensis spores in U.S. rice. Int. J. Food Microbiol. 128, 460–466. doi: 10.1016/j.ijfoodmicro.2008.10.006
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bağcıoğlu, M., Fricker, M., Johler, S., and Ehling-Schulz, M. (2019). Detection and Identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via Machine Learning Based FTIR Spectroscopy. Front. Microbiol. 10:902. doi: 10.3389/fmicb.2019.00902
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beecher, D. J., and Wong, A. C. L. (2000). Cooperative, synergistic and antagonistic haemolytic interactions between haemolysin BL, phosphatidylcholine phospholipase C and sphingomyelinase from Bacillus cereus. Microbiology 146, 3033–3039. doi: 10.1099/00221287-146-12-3033
Bonis, M., Felten, A., Pairaud, S., Dijoux, A., Maladen, V., Mallet, L., et al. (2021). Comparative phenotypic, genotypic and genomic analyses of Bacillus thuringiensis associated with foodborne outbreaks in France. PLoS One 16:e0246885. doi: 10.1371/journal.pone.0246885
Carroll, L. M., Wiedmann, M., and Kovac, J. (2020b). Proposal of a Taxonomic Nomenclature for the Bacillus cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. mBio 11, e00034–20. doi: 10.1128/mBio.00034-20
Carroll, L. M., Cheng, R. A., and Kovac, J. (2020a). No Assembly Required: using BTyper3 to Assess the Congruency of a Proposed Taxonomic Framework for the Bacillus cereus Group With Historical Typing Methods. Front. Microbiol. 11:580691. doi: 10.3389/fmicb.2020.580691
Carroll, L. M., Wiedmann, M., Mukherjee, M., Nicholas, D. C., Mingle, L. A., Dumas, N. B., et al. (2019). Characterization of Emetic and Diarrheal Bacillus cereus Strains From a 2016 Foodborne Outbreak Using Whole-Genome Sequencing: addressing the Microbiological, Epidemiological, and Bioinformatic Challenges. Front. Microbiol. 10:144. doi: 10.3389/fmicb.2019.00144
Chattopadhyay, A., Bhatnagar, N. B., and Bhatnagar, R. (2004). Bacterial Insecticidal Toxins. Crit. Rev. Microbiol. 30, 33–54. doi: 10.1080/10408410490270712
Davis, S., Pettengill, J. B., Luo, Y., Payne, J., Shpuntoff, A., Rand, H., et al. (2015). CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput. Sci. 1:e20. doi: 10.7717/peerj-cs.20
Doll, V. M., Ehling-Schulz, M., and Vogelmann, R. (2013). Concerted Action of Sphingomyelinase and Non-Hemolytic Enterotoxin in Pathogenic Bacillus cereus. PLoS One 8:e61404. doi: 10.1371/journal.pone.0061404
EFSA BIOHAZ Panel (2016). Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 14:e04524. doi: 10.2903/j.efsa.2016.4524
Ehling-Schulz, M., Lereclus, D., and Koehler, T. M. (2019). “The Bacillus cereus Group: Bacillus Species with Pathogenic Potential,” in Gram-Positive Pathogens, (Washington: ASM Press), 875–902. doi: 10.1128/9781683670131.ch55
Ehling-Schulz, M., Svensson, B., Guinebretiere, M.-H., Lindbäck, T., Andersson, M., Schulz, A., et al. (2005). Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151, 183–197. doi: 10.1099/mic.0.27607-0
Frederiksen, K., Rosenquist, H., Jørgensen, K., and Wilcks, A. (2006). Occurrence of Natural Bacillus thuringiensis Contaminants and Residues of Bacillus thuringiensis -Based Insecticides on Fresh Fruits and Vegetables. Appl. Environ. Microbiol. 72, 3435–3440. doi: 10.1128/AEM.72.5.3435-3440.2006
Frentzel, H., Juraschek, K., Pauly, N., Kelner-Burgos, Y., and Wichmann-Schauer, H. (2020). Indications of biopesticidal Bacillus thuringiensis strains in bell pepper and tomato. Int. J. Food Microbiol. 321:108542. doi: 10.1016/j.ijfoodmicro.2020.108542
Fukasawa, Y., Ermini, L., Wang, H., Carty, K., and Cheung, M.-S. (2020). LongQC: a Quality Control Tool for Third Generation Sequencing Long Read Data. G3 10, 1193–1196. doi: 10.1534/g3.119.400864
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hendriksen, N. B., and Hansen, B. M. (2006). Detection of Bacillus thuringiensis kurstaki HD1 on cabbage for human consumption. FEMS Microbiol. Lett. 257, 106–111. doi: 10.1111/j.1574-6968.2006.00159.x
Inouye, M., Dashnow, H., Raven, L.-A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6:90. doi: 10.1186/s13073-014-0090-6
Johler, S., Kalbhenn, E. M., Heini, N., Brodmann, P., Gautsch, S., Bağcioğlu, M., et al. (2018). Enterotoxin Production of Bacillus thuringiensis Isolates From Biopesticides, Foods, and Outbreaks. Front. Microbiol. 9:1915. doi: 10.3389/fmicb.2018.01915
Jouzani, G. S., Valijanian, E., and Sharafi, R. (2017). Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl. Microbiol. Biotechnol. 101, 2691–2711. doi: 10.1007/s00253-017-8175-y
Kim, J.-B., Choi, O.-K., Kwon, S.-M., Cho, S.-H., Park, B.-J., Jin, N. Y., et al. (2017). Prevalence and Toxin Characteristics of Bacillus thuringiensis Isolated from Organic Vegetables. J. Microbiol. Biotechnol. 27, 1449–1456. doi: 10.4014/jmb.1703.03063
Lacey, L. A., Grzywacz, D., Shapiro-Ilan, D. I., Frutos, R., Brownbridge, M., and Goettel, M. S. (2015). Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132, 1–41. doi: 10.1016/j.jip.2015.07.009
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Marçais, G., Delcher, A. L., Phillippy, A. M., Coston, R., Salzberg, S. L., and Zimin, A. (2018). MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14:e1005944. doi: 10.1371/journal.pcbi.1005944
McIntyre, L., Bernard, K., Beniac, D., Isaac-Renton, J. L., and Naseby, D. C. (2008). Identification of Bacillus cereus Group Species Associated with Food Poisoning Outbreaks in British Columbia, Canada. Appl. Environ. Microbiol. 74, 7451–7453. doi: 10.1128/AEM.01284-08
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genomics 2:e000056. doi: 10.1099/mgen.0.000056
Raymond, B., and Federici, B. A. (2017). In defence of Bacillus thuringiensis, the safest and most successful microbial insecticide available to humanity—a response to EFSA. FEMS Microbiol. Ecol. 93:fix084. doi: 10.1093/femsec/fix084
Schmid, D., Rademacher, C., Kanitz, E. E., Frenzel, E., Simons, E., Allerberger, F., et al. (2016). Elucidation of enterotoxigenic Bacillus cereus outbreaks in Austria by complementary epidemiological and microbiological investigations, 2013. Int. J. Food Microbiol. 232, 80–86. doi: 10.1016/j.ijfoodmicro.2016.05.011
Schwenk, V., Riegg, J., Lacroix, M., Märtlbauer, E., and Jessberger, N. (2020). Enteropathogenic Potential of Bacillus thuringiensis Isolates from Soil, Animals, Food and Biopesticides. Foods 9:1484. doi: 10.3390/foods9101484
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Travers, R. S., Martin, P. A. W., and Reichelderfer, C. F. (1987). Selective Process for Efficient Isolation of Soil Bacillus spp. Appl. Environ. Microbiol. 53, 1263–1266. doi: 10.1128/aem.53.6.1263-1266.1987
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xie, Z., and Tang, H. (2017). ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33, 3340–3347. doi: 10.1093/bioinformatics/btx433
Keywords: Bacillus thuringiensis, B. cereus group, foodborne outbreaks, biopesticide, enterotoxin
Citation: Biggel M, Etter D, Corti S, Brodmann P, Stephan R, Ehling-Schulz M and Johler S (2022) Whole Genome Sequencing Reveals Biopesticidal Origin of Bacillus thuringiensis in Foods. Front. Microbiol. 12:775669. doi: 10.3389/fmicb.2021.775669
Received: 14 September 2021; Accepted: 13 December 2021;
Published: 12 January 2022.
Edited by:
Mostafa Ghanem, University of Maryland, College Park, United StatesReviewed by:
Christophe Nguyen-The, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceMieke Uyttendaele, Ghent University, Belgium
Copyright © 2022 Biggel, Etter, Corti, Brodmann, Stephan, Ehling-Schulz and Johler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophia Johler, c29waGlhLmpvaGxlckB1emguY2g=
 Michael Biggel
Michael Biggel Danai Etter
Danai Etter Sabrina Corti1
Sabrina Corti1 Peter Brodmann
Peter Brodmann Roger Stephan
Roger Stephan Monika Ehling-Schulz
Monika Ehling-Schulz Sophia Johler
Sophia Johler