- 1Academy of Military Medical Sciences, Beijing, China
- 2Center for Disease Control and Prevention of PLA, Beijing, China
- 3School of Life Sciences, Tsinghua University, Beijing, China
- 4Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China
With the rapid emergence of plasmid-mediated colistin resistance gene mcr-1, the increased resistance of Salmonella has attracted extensive attention. This study reports on 11 multidrug-resistant Salmonella enterica serovar Typhimurium strains harboring mcr-1 in China. They all presented resistance to colistin, and additionally, one that was isolated from a child’s stool sample was also resistant to ceftriaxone and azithromycin. We screened 1454 strains of Salmonella for mcr-1 gene through PCR, and these strains are all preserved in our laboratory. Antimicrobial sensitivity analysis was carried out for the screened mcr-1 positive strains. Genetic polymorphism analysis of S. Typhimurium was performed by using the Pulsed-Field Gel Electrophoresis (PFGE). The plasmids harboring mcr-1 were identified by S1-PFGE and southern blotting. Plasmid conjugation assays were used to analyze the transferability of colistin resistance. The plasmids harboring mcr-1 were characterized by sequencing and bioinformatic analysis. Eleven S. Typhimurium strains harboring mcr-1 with colistin resistance (MICs 4μg/ml) were detected, which were isolated from children and pig offal in China. All of them were multidrug-resistant strains. PFGE results revealed that the strains isolated from different samples or locations have identical genotypes. S1-PFGE and southern blotting experiments showed that three plasmids of different sizes (33, 60, and 250 kb) all carried the mcr-1 gene. The plasmid conjugation assays revealed that Salmonella acquired mcr-1 harboring plasmids by horizontal transfer. Sequencing and plasmid type analysis revealed that these plasmids were types IncX4, IncI2, and IncHI2. Among them, IncX4 and IncI2 plasmids had extremely similar backbones and contained one resistant gene mcr-1. IncHI2 plasmid contained multiple resistant genes including blaCTX–M, oqxB, sul, aph, aadA, and blaTEM. We identified 11 mcr-1 harboring S. Typhimurium strains in China and described their characteristics. Our findings indicate that the mcr-1 gene can effectively spread among intestinal bacteria by horizontal transfer of three types of plasmids. Moreover, the IncHI2 plasmid can also mediate the transfer of other drug resistance genes. These results reveal that constant surveillance of mcr-1 harboring S Typhimurium is imperative to prevent the spread of colistin resistance.
Introduction
The rise of multidrug-resistant (MDR, resistance to three or more classes of antimicrobials) bacteria poses a serious threat to public health (Kumar et al., 2014; Jain et al., 2020). Salmonella is one of the common pathogens that can cause bacterial intestinal infections and diarrhea in developed and developing countries (Lokken et al., 2016). Salmonella enterica serovar Typhimurium (S. Typhimurium), one of the most prevalent serovars of Salmonella, is regularly linked to human infections and is frequently reported to be associated with human infections in several industrialized countries (Gomes-Neves et al., 2012), which can result in gastroenteritis and bacteremia. For clinical therapy of Salmonella infection, fluoroquinolones, azithromycin, and cephalosporins have been indicated. However, the extensive use of antibacterial medicines has resulted in the emergence of S. Typhimurium being resistant to antibiotics (Zhu et al., 2017; Wang Y. et al., 2020).
Polymyxin, a colistin antibiotic, acts as the last-line defense against severe infections caused by broad-spectrum active gram-negative bacteria (Lu et al., 2019). Additionally, colistin resistance has developed in S. Typhimurium, involving a variety of mechanisms. The plasmid-mediated colistin resistance gene mcr-1 was first discovered in E. coli in China in 2015 (Liu et al., 2016), and has been the subject of research attention due to the mcr-1 gene’s ability to spread horizontally between bacteria. The colistin resistance gene mcr-1 in the IncI2 plasmid encodes a phosphoethanolamine transferase, which is the modification of the lipid A and provides adequate protection from colistin. Multiple plasmids were used to propagate the colistin resistance gene mcr-1, including IncHI1, IncHI2, IncI2, IncX4, IncF, IncFI, IncFII, and IncP (Zurfluh et al., 2017; Touati and Mairi, 2021). These findings indicate that horizontal transfer of multiple resistance genes in the intestine bacteria may result in bacterial resistance. This study performed a screening analysis for the mcr-1 gene of S. Typhimurium, which was preserved in the laboratory. The present study aimed to characterize the S. Typhimurium harboring mcr-1 plasmids isolated from patients and food.
Materials and Methonds
Bacterial mcr-1 Gene Screening, Serotyping
To clarify the epidemic situation of the colistin resistance gene mcr-1 in critical areas in China, we detected 1454 S. Typhimurium strains stored in our laboratory. All S. Typhimurium strains were isolated from stool samples of patients and food in markets, which were collected from Shanghai City (1046), Guangdong Province (209), and Guangxi Province (199) from 2006 to 2018, respectively. These strains were strictly identified by biochemical tests (API 20E system; bioMérieux Vitek, Marcy-L’Etoile, France) and serotyped on slides by microtiter agglutination tests for O and H antigens (SSI, Copenhagen, Denmark) according to the manufacturer’s instructions. We screened all historical S. Typhimurium strains for mcr-1 gene by PCR using the published primers sequence according to a study by Liu et al. (2016).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was designed using broth microdilution in Sensititre Gram Negative AST Plates for Salmonella strains (Thermo Fisher Scientific, Inc., West Sussex, United Kingdom) including 14 different antimicrobials: ceftriaxone (CRO), tetracycline (TE), ceftiofur (XNL), cefoxitin (FOX), gentamicin (GEN), ampicillin (AMP), chloramphenicol (CHL), ciprofloxacin (CI), trimethoprim/sulfamethoxazole (SXT), sulfisoxazole (SX), nalidixic acid (NAL), streptomycin (SM), azithromycin (AZI), and amoxicillin/clavulanic acid 2:1 ratio (AUG2). The susceptibility to polymyxin is to use the dye WST (Dojindo Molecular Technologies, Inc., Japan) by a microbial viability assay kit. A reference strain of Escherichia coli ATCC 25922 strain was performed in the test as quality control (Wang et al., 2017).
Pulsed-Field Gel Electrophoresis (PFGE), S1-PFGE, and Southern Blotting
Genomic polymorphism analysis of Salmonella strains was performed using the pulsed field gel electrophoresis (PFGE) after a slight modification of the pulseNet standardized PFGE protocol for Salmonella (Ribot et al., 2006). To study the relationship between strains, mcr-1-negative S. Typhimurium Bacteria at different times in the same location and at the same time in other regions were used as the reference bacteria in the laboratory. These isolates were digested with XbaI (Takara, Dalian, China) at 37°C, and the Salmonella enterica var. Braendrup H9812 strain was used as the reference. Electrophoresis performed on a CHEF MAPPER variable angle system (Bio-Rad, California, America) with the parameters set at 2.16–63.8 s for 19 h performed following previously described methods (Liu et al., 2018). The plasmid profiles were characterized by S1-PFGE. The endonuclease S1 nuclease (Takara, Dalian, China) was used to digest at 37°C, and electrophoresis running set at 0.22–26.29 s for 15 h. The images were captured by a Gel Doc 2000 system (Bio-Rad), and imported into the BioNumerics software (v6.0) database for further processing and analysis. The southern blotting with digoxigenin-labeled mcr-1 probe using published primer sequences (Liu et al., 2016) was performed to membrane transfer, molecular hybridization, and probe detection following a previously reported method (Zou et al., 2015).
Plasmid Conjugation Assays
To verify the mcr-1 positive plasmid’s transfer capacity, plasmid conjugation experiments were performed by utilizing a standard E. coli J53 as the recipient, and the mcr-1 positive S. Typhimurium strains as donors. The donor bacteria cultured overnight were mixed with the recipient bacteria in a ratio of 1:3 and harvested, re-suspended in 80μL. The mixture was incubated for mating at 37°C for 12–18 h in 5 ml LB liquid broth. Then a Muller-Hinton agar (BD Biosciences, San Jose, CA) plate containing 100 mg/L sodium azide and 2 mg/L polymyxin B was to a selective medium for E. coli J53 transconjugants. Putative transconjugants were confirmed by antimicrobial susceptibility testing and detection of mcr-1 with PCR.
Whole Genome Sequencing and Bioinformatic Analysis
Using Next-Generation Sequencing (NGS), we sequenced plasmids of S. Typhimurium harboring the mcr-1 gene. DNA was extracted from the overnight cultured strains using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). The mate-pair library was constructed by nucleic acid protein analyzer Qsep100 to obtain DNA fragments (not less than 500 bp, not more than 800 bp) and sequenced by MiSeq sequencer. The raw reads were assembled into draft continuous sequences (contigs) by Newbler (Arredondo-Alonso et al., 2018) and NxTrim (O’Connell et al., 2015), and then spliced with Cytoscape’s GapFiller. Complete plasmid genomes were annotated using the online annotation server RAST. Identification of insertion sequence (IS), plasmid replicons, and resistance genes were performed by ISfinder,1 PlasmidFinder,2 and ResFinder (Bortolaia et al., 2020), respectively. Multiple plasmids were compared by Mauve, Brig, and CLC Genomics Workbench. The circled figure of multiple plasmids for comparison was drawn by DNAploter (Roberts et al., 2008).
Results
Antimicrobial Susceptibility Testing of S. Typhimurium Harboring mcr-1
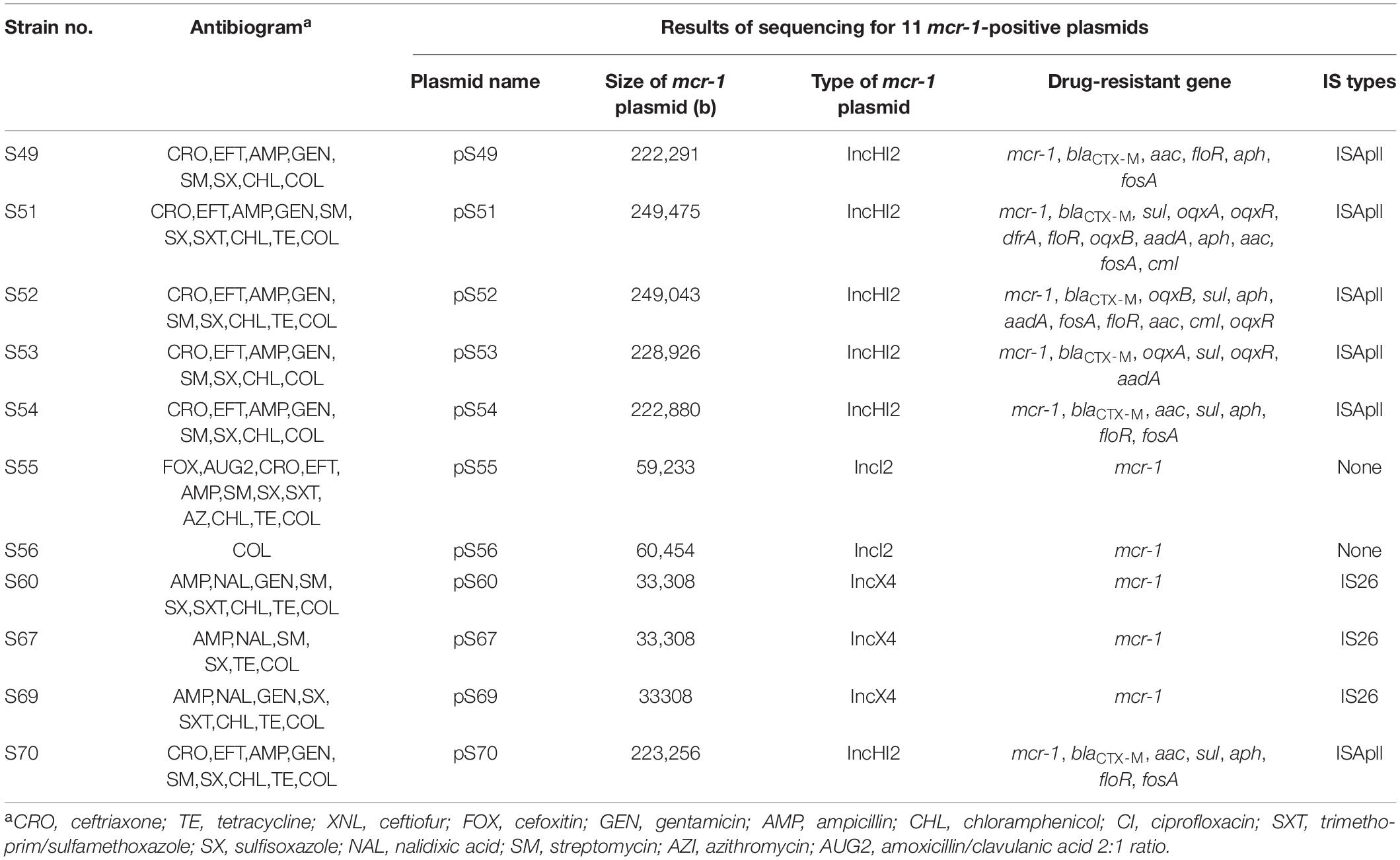
Among the 1454 strains of S. Typhimurium maintained in our laboratory, 11 strains harboring colistin resistance gene mcr-1 were identified. Eight strains were isolated from the feces of children under the age of five, while the remaining strains were isolated from pork offal. These strains exhibited multidrug resistance, including polymyxin (MICs 4μg/ml). Additionally, the majority (63.6%) of S. Typhimurium harboring mcr-1 were resistant to the third-generation cephalosporins. Notably, one of them exhibited co-resistant to azithromycin and third-generation cephalosporins (Table 1).
Pulsed Field Gel Electrophoresis, Plasmid Profiling, and Southern Blotting
We studied the PFGE results of 11 mcr-1-positive S. Typhimurium strains and 12 mcr-1-negative S. Typhimurium strains. The majority of these (10/11) belong to ST34, while one (S55) belongs to ST19. In total, 13 distinct PFGE genotypes were discovered among 23 strains using the 85% cutoff (Supplementary Figure 1). The mcr-1 positive strains were distributed in 10 different genotypes. Furthermore, numerous isolates obtained from diverse samples and provinces were identified as belonging to the same genotype. For instance, S. Typhimurium harboring mcr-1 strains isolated from patient samples and food in Shanghai and the strains isolated from patients in Zhejiang and Henan were in the same cluster. S1-PFGE analysis showed that two of 11 mcr-1 positive S. Typhimurium carried two plasmids, while the remaining eight carried single plasmid (Supplementary Figure 2). Southern blotting revealed that all of them carried one plasmid harboring mcr-1 (Supplementary Figure 2).
Plasmid Conjugation Assays
To determine the transferability of the plasmid harboring mcr-1, plasmid conjugation assays were performed using 11 mcr-1 positive S. Typhimurium as donors and E. coli J53 as the recipient. Five of eleven recipients tested positive for the resistance gene mcr-1 via PCR amplification and sequencing analysis. Five plasmids harboring mcr-1 were identified (pS49, pS51, pS52, pS55, pS56). Three were IncHI2 and two were IncI2 plasmids (Supplementary Figure 1). The MIC of colistin for the transconjugants (the E. coli J53 harboring mcr-1 gene) was increased to 4 μg/ml, which was significantly more than the colistin resistance levels of the original J53 strains (which have MIC values of 0.125 μg/ml). It can be speculated that the transconjugants acquire the donor strains’ colistin resistance gene.
The Complete Sequence of Plasmid Harboring mcr-1
We sequenced the plasmid of 11 S. Typhimurium strains. As a control, we downloaded five highly comparable plasmids harboring mcr-1 from Salmonella strains for comparison (at above 95% coverage and above 99% identity) from NCBI (full name and cited reference). Three IncX4, two IncI2, and six IncHI2 plasmids harboring the mcr-1 gene were identified by analyzing the plasmid sequences. Plasmid sequence comparison revealed that the lengths of the three IncX4 plasmids were approximately 33 kb and the sequences were completely identical; the lengths of the two IncI2 plasmids were 59,233 and 60,454 b, respectively, and the sequence differences were within 1.3 kb, and the 6 IncHI2 plasmid sequences were around 220-250 kb in length and the length variance was less than 2.8 kb. The three IncX4 plasmids with the identical sequence had a typical IncX4 backbone and were extremely similar to pNG14043 from Salmonella in Taiwan (at above 99% homology). The IS26 was upstream of resistance gene mcr-1 in our isolates. The IncX4 plasmids had only the resistance gene mcr-1 and no other identifiable resistance genes (Figures 1, 2). In contrast to pHNSHP45, three IncX4 plasmids lacked an ISApl1 insertion element upstream of mcr-1 but had an IS26 insertion element. The IncI2 plasmids were similar to pHNSHP45 by E. coli strains from Shanghai in July 2013. The sequence of mcr-1 on these plasmids was identical to pHNSHP45. However, two IncI2 plasmids lacked an ISApl1 insertion element upstream of mcr-1, and an IS683 region was found to be missing in all two IncI2-type plasmids isolated in this study (Figures 2, 3). Unlike the IncX4 and IncI2 plasmids, IncHI2 plasmids exhibited the most genetic diversity (Figures 2, 4). Compared with the pHNSHP45-2 plasmid, these IncHI2 plasmids with the common backbone were 250 kb in length. All the IncHI2 plasmids contained a single copy of mcr-1, and the sequence surrounding mcr-1 shared 100% sequence identity. However, these plasmids contain numerous variable resistance genes, integrons, and ISs. The reference plasmid contained a variety of resistance genes, including blaCTX–M, oqxA, oqxB, oqxR, sul, aph, aadA, dfrA, floR, aac, fosA, hph. The IncHI2 plasmids we investigated had different resistance genes and insert sequences with the reference plasmid (Table 1). Compared to the reference plasmid, several drug-resistant genes were missing, including oqxA, oqxB, oqxR, sul, aadA in three IncHI2 plasmids (pS49, pS54, pS70) and dfrA12 in the five IncHI2 plasmids (pS49, pS52, pS53, pS54, pS70) (Figure 4). Five of six IncHI2 plasmids contained the insertion sequence ISApl1 on the upstream of mcr-1, but another plasmid lacked ISApl1 around mcr-1 (Figure 2).
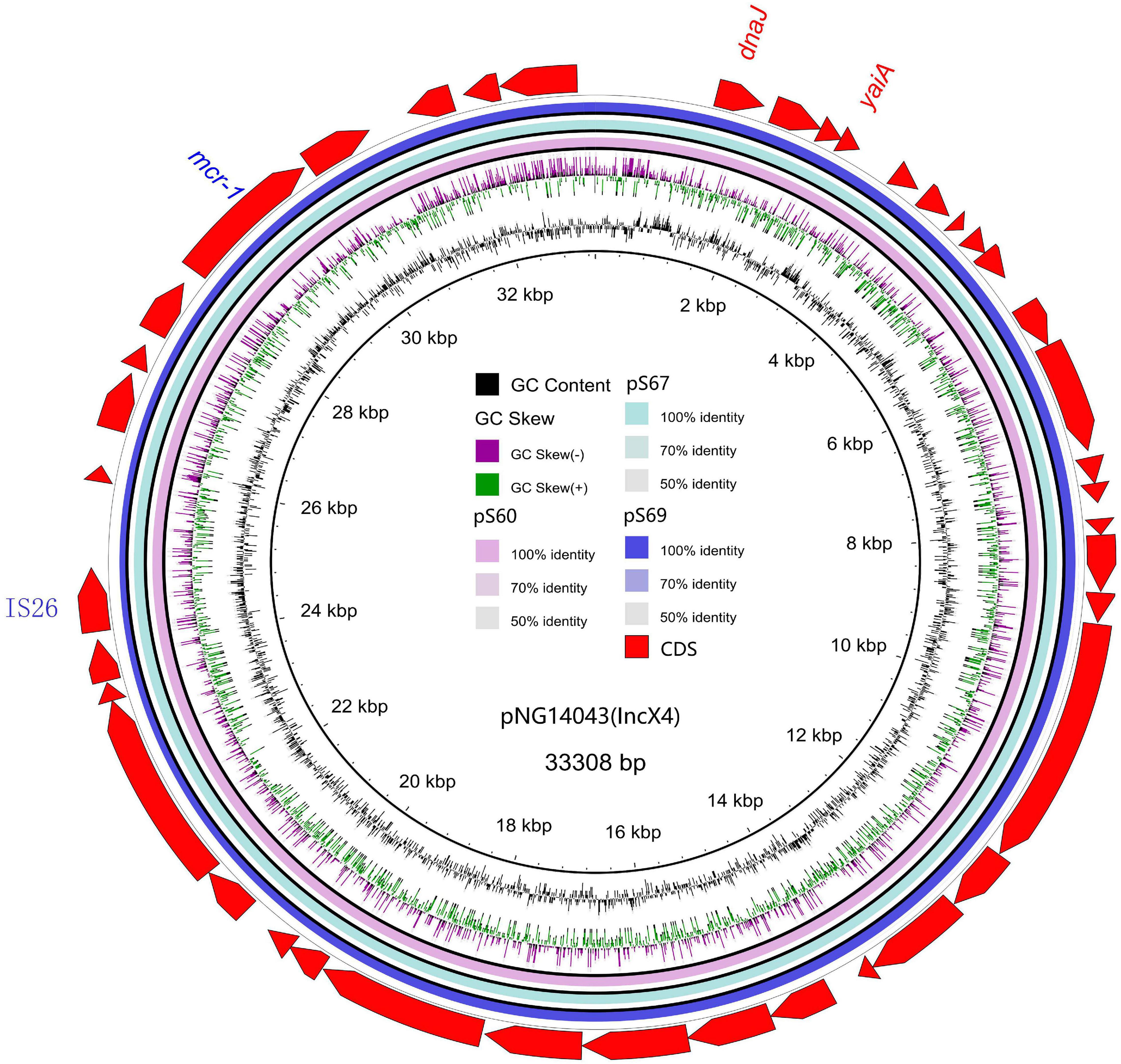
Figure 1. Sequencing alignment of mcr-1 harboring IncX4 plasmids. The mcr-1 harboring plasmid pNG14043 with GenBank no. KY120364 which was isolated from S. Typhimurium in Taiwan was used as refrence plasmid (black circle). The outmost circle in red arrows denots the annotations of refrence plasmid. The figure shows the extremly high degree of homology of the four mcr-1 harboring IncX4 plasmids. Detailed information of mcr-1 location of plasmids is provided in Figure 2.
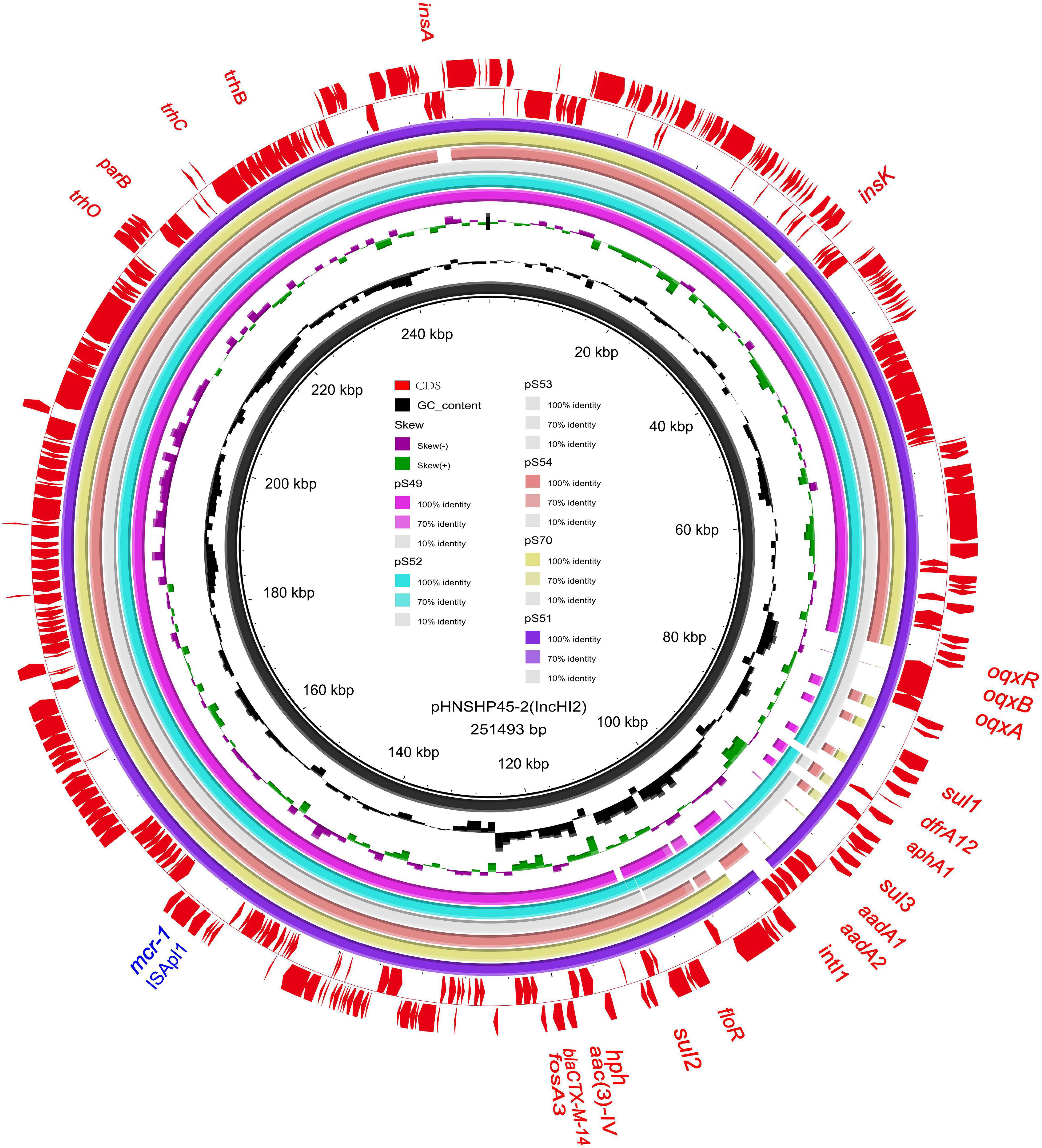
Figure 2. Genetic context of 11 plasmids surrounding the mcr-1 gene. In CDS, red and blue arrows represent mcr-1 and IS, respectively, black arrows present plasmid bone. The light blue and orange shaded regions indicate genetic regions that show the direct and reverse nucleotide identity homology between different segments (> 99%).
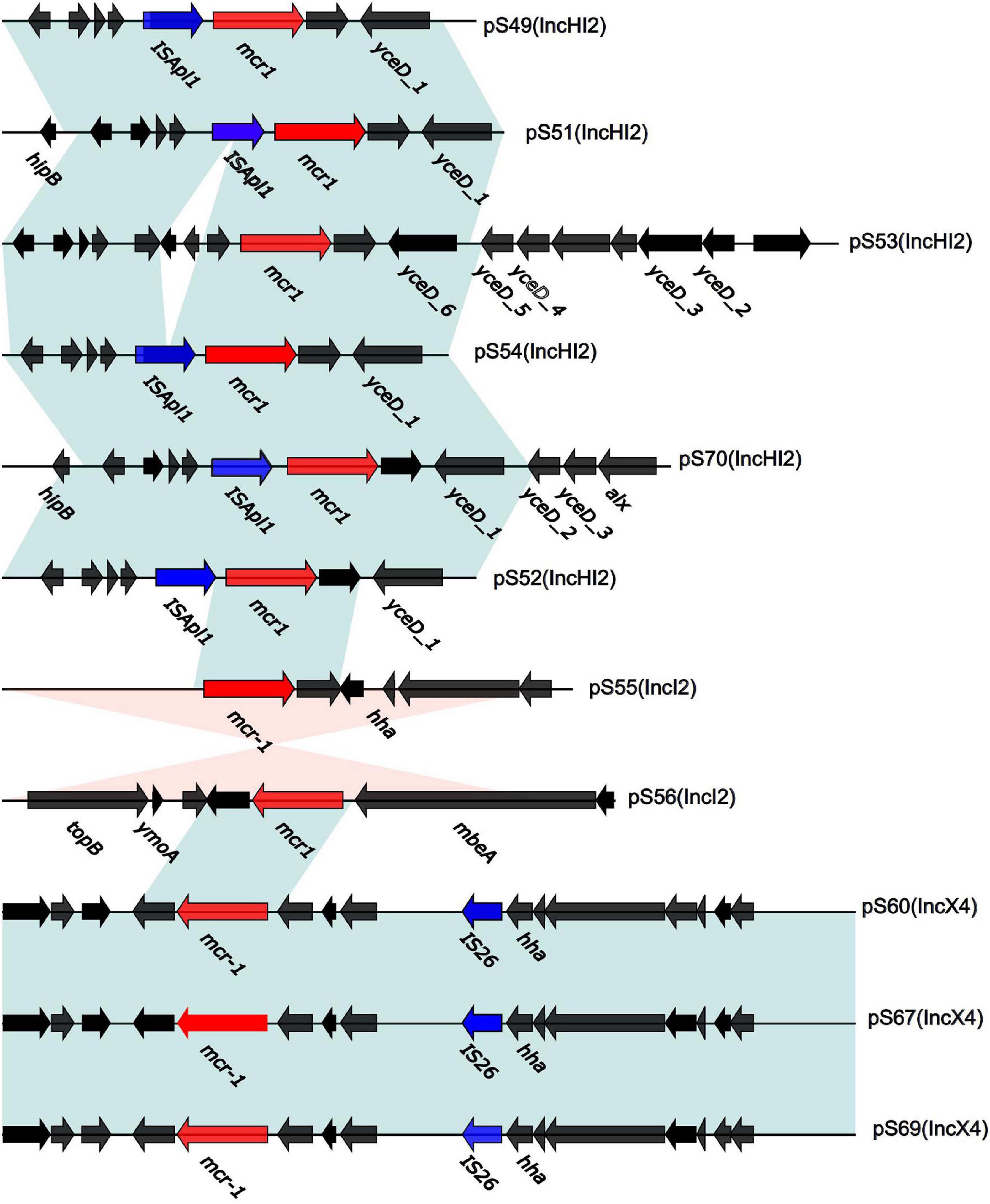
Figure 3. Sequencing alignment of mcr-1 harboring IncI2 plasmids. The first mcr-1 harboring plasmid, pHNSHP45 with GenBank no. KP347127 which was isolated from E. coli strains from Shanghai in July, 2013 was used as refrence plasmid (black circle). The outmost circle in red arrows denots the annotations of refrence plasmid. The IS683 and ISApl1 are absent in two IncI2 plasmids in this study. Detailed information of mcr-1 location of plasmids is provided in Figure 2.
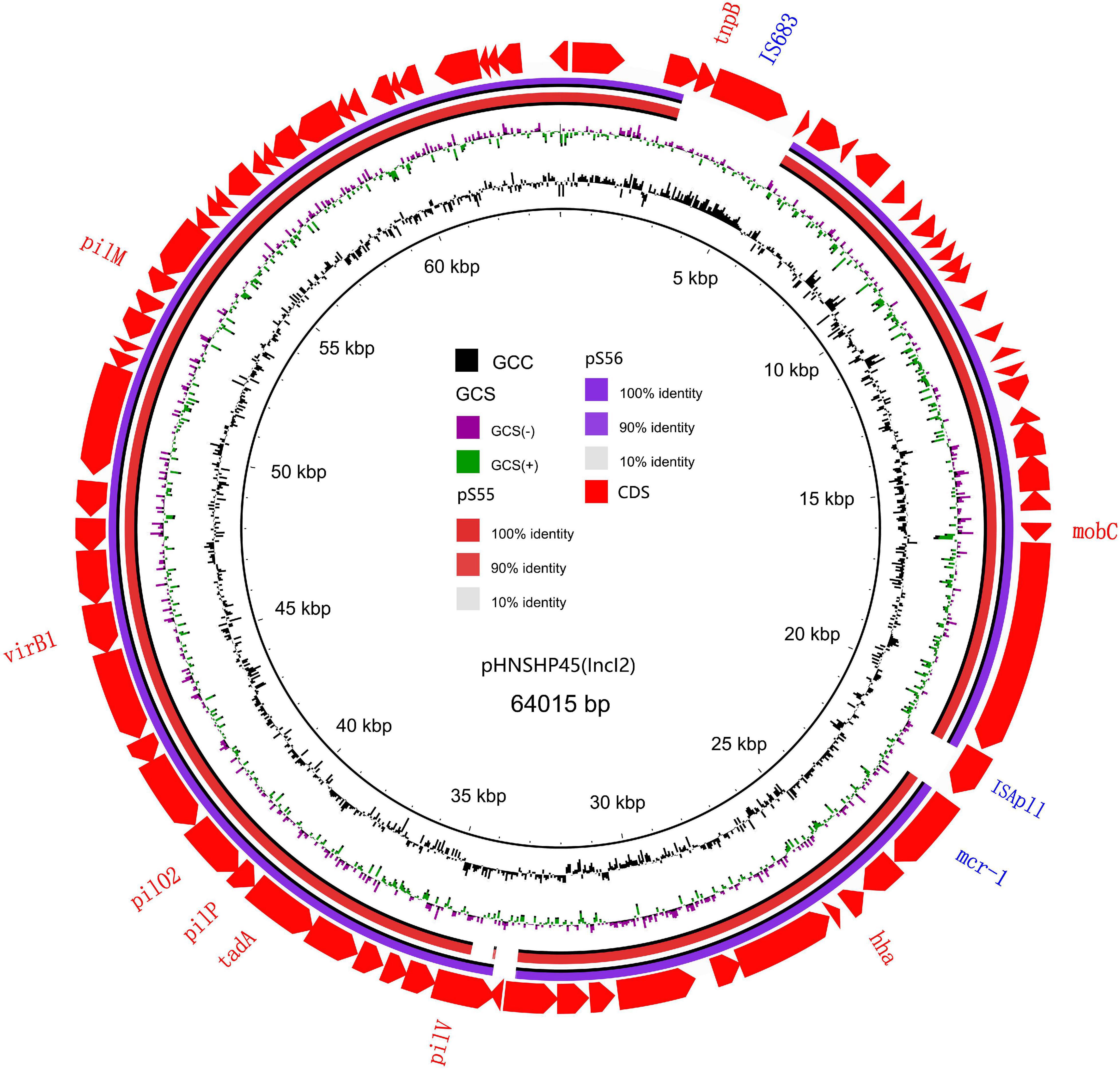
Figure 4. Sequencing alignment of mcr-1 harboring IncHI2 plasmids. pHNSHP45-2 with GenBank no. KU341381 which was isolated from E. coli strains was used as refrence plasmid (black circle). The outmost circle in red arrows denots the annotations of refrence plasmid. Among the six IncHI2 plasmids, pSH51, pS52, and pS53 exhibit sequence with the refrence sequence, others are low sequence homology. An MDR region is exhibieted in the five IncHI2 plasmids. Detailed information of mcr-1 location of plasmids is provided in Figure 2.
Discussion
Salmonella is a widespread zoonotic pathogen that can cause human food poisoning and diarrhea (Ling et al., 2020). In general, food poisoning and diarrhea caused by non-typhoidal Salmonella (NTS) are self-limited. However, if the patient is young children, older people, and people with weak immune systems, antibiotic therapy will be preferred with multi-drug resistant Salmonella infection if the patient is a young child, elderly, or has a weak immune system. Studies have demonstrated that the multidrug resistance rate of Salmonella increased to 40% in the last decade of the twentieth century (Elbediwi et al., 2020). Resistance to fluoroquinolones, azithromycin, and third-generation cephalosporins in NTS species has been reported from numerous countries in the world (Tack et al., 2020; Appiah et al., 2021). Among a large number of Salmonella serotypes, S. Typhimurium and the rapid growth of multidrug-resistant has been a subject of concern globally (Huang et al., 2020). Colistin is considered as a last-line therapy for multidrug-resistant S. Typhimurium infection based on its prevalence and has been listed as a significant antibiotic by WHO since 2015 (Li et al., 2017). As colistin is widely used, bacteria have developed resistance to colistin. 37 Salmonella strains were identified harboring the mcr-1 gene among 12,053 Salmonella strains collected from diarrhea outpatients under surveillance (Lu et al., 2019), and our finding appeared consistent with previous studies in Shanghai. We identified 11 mcr-1-positive strains among 1454 strains of S. Typhimurium (0.76%). Notably, eight strains of S. Typhimurium harboring mcr-1 were isolated from the feces of children under the age of five (Supplementary Table 1). This observation is consistent with the finding of Luo et al. (2020), in which the majority of the Salmonella infection occurs in children under the age of five and patients with inadequate immunity. All mcr-1 harboring S. Typhimurium strains from various sources were resistant to multiple antibiotics. In total, 63.6% of them were resistant to colistin and third-generation cephalosporin. Moreover, one of these strains was isolated from children under 5 years old was resistant to colistin, azithromycin, and third-generation cephalosporins. Considering the important role of azithromycin and third-generation cephalosporins in clinical treatment, this causes concern. ST19 and ST34 were common genotypes in S. Typhimurium (Zhang et al., 2021). The MDR ST34 S. Typhimurium has become a threat to public health due to its carriage of mcr-1 and mcr-3 (Biswas et al., 2019), and has been frequently detected in human clinical samples and food samples in China (Sun et al., 2014). The PFGE results indicated that S. Typhimurium that were isolated from various samples and provinces had identical genotypes. For example, S. Typhimurium harboring mcr-1 strains isolated from children and food in Shanghai were clustered with the S. Typhimurium in Zhejiang and Henan strains. This result indicated that S. Typhimurium was prevalent in a number of regions in China. Thus, the monitoring of multidrug-resistant S. Typhimurium strains should significantly prevent their spread.
Plasmids play a vital role in the acquisition of colistin resistance caused by drug resistant genes (McGann et al., 2016). In 2016, China reported the first case of Plasmid-mediated colistin resistance in the form of mcr-1 (Liu et al., 2016). Colistin resistant gene mcr-1 was widely spread in animals, the environment, and food in a number of nations and areas throughout the world by plasmid horizontal transfer (Zurfluh et al., 2017; Huang et al., 2020; Wang Z. et al., 2020; Touati and Mairi, 2021). During plasmid transfer, the plasmid harboring mcr-1 exhibited significant diversity in terms of antibiotic resistance patterns, incompatibility groups, and genetic content (Touati and Mairi, 2021). In our study, three types of plasmids (IncI2, IncX4, and IncHI2) harboring mcr-1 were identified from 11 S. Typhimurium strains. The first reported mcr-1 gene was identified in an IncI2 Plasmid. IncI2 and IncX4 plasmids, which promote Salmonella resistance, are the two major types of plasmids spreading globally (Arredondo-Alonso et al., 2018). IncHI2 plasmids are well-known for their role in clinically significant antibiotic resistant genes (Hammad et al., 2019). According to a previous study on Enterobacteriaceae (Zingali et al., 2020), IncHI2 plasmid (216–280 kb) is the fifth most common plasmid family containing a multidrug resistance region. it is also one of the major plasmid groups harboring mcr-1 gene variants. The present study observed a coexistence of plasmids harboring mcr-1 and multiple drug resistance genes, including oqxB, blaTEM, and blaCTX resistance genes. These genes in drug-resistant plasmids were one of the significant factors for the decreased sensitivity of colistin, quinolones, and third-generation cephalosporins. The rapid spread of antibiotic resistance in a particular area was caused by IncHI2 plasmids transfer carrying multiple drug resistance genes between bacteria. MDR IncHI2 plasmids containing mcr-1 are widely distributed in human pathogens and are the efficient vector for the transmission of mcr-1 and other drug-resistant genes (Hammad et al., 2019). Whether MDR IncHI2 plasmids can alter the Salmonella resistance phenotype and disseminate rapidly is concerning.
The insertion sequence (IS) family in the bacterial genome with a widely variable DNA sequence in nature is also one of the significant modes for resistance gene transmission between bacterial pathogens. The 11 plasmids are classified into distinct patterns based on the existence of IS elements and the connection with mcr-1 sites. In our study, IS26 elements are closely related to the IncX4 type plasmid, while the ISApl1 element is closely related to the IncHI2 type plasmid. ISApl1 is consistently associated with the mcr-1 gene and the mcr-1 gene cassette can be inserted into a variety of genetic loci in different plasmids. Five of the six IncHI2 plasmids contained the ISApl1 insertion sequence located upstream of mcr-1, and only one plasmid lacked ISApl1 around mcr-1. The IncHI2 backbone structure is considered to be stable (Garcia-Fernandez and Carattoli, 2010), and the presence of different resistance genes in these plasmids is probably due to the acquisition of different mobile genetic elements (Cain and Hall, 2012). As such, the existence of ISApl1 in our study offers a potential hotspot for involving novel antibiotic resistant genes.
Conclusion
The present study describes the genetic characterization of mcr-1-positive multidrug-resistant S. Typhimurium isolated from intestinal infection in children and pork offal in China. Our results indicated that mcr-1-positive S. Typhimurium strains were multidrug-resistant, and one strain was additionally resistant to ceftriaxone and azithromycin. Three types of plasmids harboring mcr-1 have respective characteristics regarding IS and resistance genes. Plasmids harboring mcr-1 and other resistance genes confer resistance to colistin and other multiple antibiotics. Therefore, the findings of this study are critical to estimating the transmission of mcr-1 and monitoring the international epidemic.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JL, SQ, and HS designed the study. HZ wrote the main manuscript. YX, BL, and XX participated in the specimen collection and revised the manuscript. YH and LW contributed to the bioinformatics data analysis. JX, XD, XH, and LJ participated in data collection. XD, CY, HL (9th author), HL (10th author), HW, and MY performed the experiments. YJ critically revised important knowledge content. YJ, SQ, and HS gave final approval of the version to be submitted. All authors made substantial contributions to preparation and submission of the manuscript.
Funding
The present study was funded by grants from the National Key R&D Program of China (nos. 2018YFC1603801 and 2017YFC1600105), the National Science and Technology Major Project (nos. 2018ZX10101003 and 2018ZX10714002), and the National Nature Science Foundation of China (nos. 81872678 and 81673237).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.774797/full#supplementary-material
Footnotes
References
Appiah, G. D., Mpimbaza, A., Lamorde, M., Freeman, M., Kajumbula, H., Salah, Z., et al. (2021). Salmonella bloodstream infections in hospitalized children with acute febrile illness-Uganda, 2016-2019. Am. J. Trop. Med. Hyg. 105, 37–46. doi: 10.4269/ajtmh.20-1453
Arredondo-Alonso, S., Rogers, M., Braat, J. C., Verschuuren, T. D., Top, J., Corander, J., et al. (2018). Mlplasmids: a user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 4:e000224. doi: 10.1099/mgen.0.000224
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella typhimurium monophasic clone ST34. Microorganisms 7:298. doi: 10.3390/microorganisms7090298
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). Resfinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Cain, A. K., and Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Elbediwi, M., Pan, H., Biswas, S., Li, Y., and Yue, M. (2020). Emerging colistin resistance in Salmonella enterica serovar newport isolates from human infections. Emerg. Microbes Infect. 9, 535–538. doi: 10.1080/22221751.2020.1733439
Garcia-Fernandez, A., and Carattoli, A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. doi: 10.1093/jac/dkq101
Gomes-Neves, E., Antunes, P., Tavares, A., Themudo, P., Cardoso, M. F., Gartner, F., et al. (2012). Salmonella cross-contamination in swine abattoirs in Portugal: carcasses, meat and meat handlers. Int. J. Food Microbiol. 157, 82–87. doi: 10.1016/j.ijfoodmicro.2012.04.015
Hammad, A. M., Hoffmann, M., Gonzalez-Escalona, N., Abbas, N. H., Yao, K., Koenig, S., et al. (2019). Genomic features of colistin resistant Escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect. Genet. Evol. 73, 126–131. doi: 10.1016/j.meegid.2019.04.021
Huang, H., Dong, N., Shu, L., Lu, J., Sun, Q., Chan, E. W., et al. (2020). Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014-2019. Emerg. Microbes Infect. 9, 237–245. doi: 10.1080/22221751.2020.1717380
Jain, P., Chowdhury, G., Samajpati, S., Basak, S., Ganai, A., Samanta, S., et al. (2020). Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000-2016. Braz. J. Microbiol. 51, 613–627. doi: 10.1007/s42770-019-00213-z
Kumar, A., Browne, C., Scotland, S., Krishnamurthy, K., and Nielsen, A. L. (2014). Selected enteropathogens and clinical course in children hospitalized with severe acute gastroenteritis in Barbados. Int. J. Health Sci. 8, 409–417.
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, H., Zhu, B., Liang, B., Xu, X., Qiu, S., Jia, L., et al. (2018). A novel mcr-1 variant carried by an IncI2-type plasmid identified from a multidrug resistant enterotoxigenic Escherichia coli. Front. Microbiol. 9:815. doi: 10.3389/fmicb.2018.00815
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lokken, K. L., Walker, G. T., and Tsolis, R. M. (2016). Disseminated infections with antibiotic-resistant non-typhoidal Salmonella strains: contributions of host and pathogen factors. Pathog. Dis. 74:ftw103. doi: 10.1093/femspd/ftw103
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Luo, Q., Wan, F., Yu, X., Zheng, B., Chen, Y., Gong, C., et al. (2020). Mdr Salmonella enterica serovar Typhimurium ST34 carrying mcr-1 isolated from cases of bloodstream and intestinal infection in children in China. J. Antimicrob. Chemother. 75, 92–95. doi: 10.1093/jac/dkz415
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX–M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
O’Connell, J., Schulz-Trieglaff, O., Carlson, E., Hims, M. M., Gormley, N. A., and Cox, A. J. (2015). Nxtrim: optimized trimming of illumina mate pair reads. Bioinformatics 31, 2035–2037. doi: 10.1093/bioinformatics/btv057
Ribot, E. M., Fair, M. A., Gautom, R., Cameron, D. N., Hunter, S. B., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Sun, J., Ke, B., Huang, Y., He, D., Li, X., Liang, Z., et al. (2014). The molecular epidemiological characteristics and genetic diversity of Salmonella typhimurium in Guangdong, China, 2007-2011. PLoS One 9:e113145. doi: 10.1371/journal.pone.0113145
Tack, B., Phoba, M. F., Barbe, B., Kalonji, L. M., Hardy, L., Van Puyvelde, S., et al. (2020). Non-typhoidal Salmonella bloodstream infections in Kisantu, DR Congo: emergence of o5-negative Salmonella typhimurium and extensive drug resistance. PLoS Negl. Trop. Dis. 14:e8121. doi: 10.1371/journal.pntd.0008121
Touati, A., and Mairi, A. (2021). Plasmid-determined colistin resistance in the north African countries: a systematic review. Microb. Drug Resist. 27, 121–133. doi: 10.1089/mdr.2019.0471
Wang, J., Li, Y., Xu, X., Liang, B., Wu, F., Yang, X., et al. (2017). Antimicrobial resistance of Salmonella enterica serovar Typhimurium in Shanghai, China. Front. Microbiol. 8:510. doi: 10.3389/fmicb.2017.00510
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, Z., Xu, H., Tang, Y., Li, Q., and Jiao, X. (2020). A multidrug-resistant monophasic Salmonella typhimurium co-harboring mcr-1, fosA3, blaCTX–M–14 in a transferable IncHI2 plasmid from a healthy catering worker in China. Infect. Drug Resist. 13, 3569–3574. doi: 10.2147/IDR.S272272
Zhang, K., Ge, H., He, J., Hu, M., Xu, Z., Jiao, X., et al. (2021). Salmonella typhimurium ST34 isolate was more resistant than the ST19 isolate in China, 2007 – 2019. Foodborne Pathog. Dis. doi: 10.1089/fpd.2021.0047 [Epub ahead of print].
Zhu, Y., Lai, H., Zou, L., Yin, S., Wang, C., Han, X., et al. (2017). Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 259, 43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023
Zingali, T., Chapman, T. A., Webster, J., Roy, C. P., and Djordjevic, S. P. (2020). Genomic characterisation of a multiple drug resistant IncHI2 ST4 plasmid in Escherichia coli ST744 in Australia. Microorganisms 8:896. doi: 10.3390/microorganisms8060896
Zou, D., Huang, Y., Zhao, X., Liu, W., Dong, D., Li, H., et al. (2015). A novel New Delhi metallo-beta-lactamase variant, NDM-14, isolated in a Chinese hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 59, 2450–2453. doi: 10.1128/AAC.05168-14
Keywords: plasmid, colistin, Salmonella typhimurium, multidrug-resistant (MDR), bioinformatic analysis
Citation: Zhang H, Xiang Y, Huang Y, Liang B, Xu X, Xie J, Du X, Yang C, Liu H, Liu H, Wang H, Yang M, Wang L, Hu X, Jin L, Li J, Jiang Y, Qiu S and Song H (2022) Genetic Characterization of mcr-1-Positive Multidrug-Resistant Salmonella enterica Serotype Typhimurium Isolated From Intestinal Infection in Children and Pork Offal in China. Front. Microbiol. 12:774797. doi: 10.3389/fmicb.2021.774797
Received: 13 September 2021; Accepted: 16 November 2021;
Published: 10 January 2022.
Edited by:
Guojie Cao, Center for Food Safety and Applied Nutrition, United States Food and Drug Administration, United StatesReviewed by:
Elaa Maamar, Tunis El Manar University, TunisiaHongxia Jiang, South China Agricultural University, China
Copyright © 2022 Zhang, Xiang, Huang, Liang, Xu, Xie, Du, Yang, Liu, Liu, Wang, Yang, Wang, Hu, Jin, Li, Jiang, Qiu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Jiang, jiangyq710327@sina.com; Shaofu Qiu, qiushf0613@hotmail.com; Hongbin Song, hongbinsong@263.net
†These authors have contributed equally to this work
 Haoran Zhang1,2†
Haoran Zhang1,2† Yong Huang
Yong Huang Beibei Liang
Beibei Liang Xuebin Xu
Xuebin Xu Chaojie Yang
Chaojie Yang Hongbo Liu
Hongbo Liu Shaofu Qiu
Shaofu Qiu Hongbin Song
Hongbin Song