- 1Department of Microbiology, School of Basic Medical Science, Xinxiang Medical University, Xinxiang, China
- 2Department of Clinical Laboratory, The Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People’s Hospital, Qingyuan, China
- 3Department of Microbiology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
- 4Key Laboratory of Tropical Diseases Control (Sun Yat-sen University), Ministry of Education, Guangzhou, China
- 5School of Laboratory Medicine, Chengdu Medical College, Chengdu, China
- 6Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University, Guangzhou, China
Type I and type II CRISPR-Cas systems are employed to evade host immunity by targeting interference of bacteria’s own genes. Although Mycobacterium tuberculosis (M. tuberculosis), the causative agent of tuberculosis, possesses integrated type III-A CRISPR-Cas system, its role in mycobacteria remains obscure. Here, we observed that seven cas genes (csm2∼5, cas10, cas6) were upregulated in Mycobacterium bovis BCG under oxidative stress treatment, indicating the role of type III-A CRISPR-Cas system in oxidative stress. To explore the functional role of type III-A CRISPR-Cas system, TCC (Type III-A CRISPR-Cas system, including cas6, cas10, and csm2-6) mutant was generated. Deletion of TCC results in increased sensitivity in response to hydrogen peroxide and reduced cell envelope integrity. Analysis of RNA-seq dataset revealed that TCC impacted on the oxidation-reduction process and the composition of cell wall which is essential for mycobacterial envelop integrity. Moreover, disrupting TCC led to poor intracellular survival in vivo and in vitro. Finally, we showed for the first time that TCC contributed to the regulation of regulatory T cell population, supporting a role of TCC in modulating host immunity. Our finding reveals the important role of TCC in cell envelop homeostasis. Our work also highlights type III-A CRISPR-Cas system as an important factor for intracellular survival and host immunoregulation in mycobacteria, thus may be a potential target for therapy.
Introduction
CRISPR-Cas (Clustered regularly interspaced short palindromic repeats-CRISPR associated) are found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea (Hille et al., 2018). Based on the architectures of effector modules, the CRISPR-Cas systems can be divided into two major classes. The class 1 CRISPR-Cas systems consist of large, multisubunit Cas protein complexes, whereas class 2 CRISPR-Cas systems perform the function by a single large protein. These systems provide adaptive immunity against foreign nucleic acids. In addition to their role in bacterial immunity, increasing evidence reveals that the CRISPR-Cas systems impact bacterial physiology and virulence. For example, Francisella novicida employ type II CRISPR/Cas to dampen a proinflammatory innate immune response by repressing transcription of a bacterial lipoprotein (Sampson et al., 2013), which suppresses TLR2-dependent proinflammatory responses in macrophages (Jones et al., 2012). Pseudomonas aeruginosa type I CRISPR-Cas system dampen the recognition by toll-like receptor 4 via targeting the mRNA of the bacterial quorum-sensing regulator lasR (Li et al., 2016). These findings support the importance of CRISPR-Cas system in bacterial pathogenesis.
It has been shown that type III CRISPR-Cas system belongs to class 1 CRISPR-Cas systems and contains diverse and polymorphic CRISPR-Cas loci with high divergence between the subtypes (Makarova et al., 2015), such as Csm (Type III-A) complexes and Cmr (Type III-B) complexes. Well-established type III CRISPR-Cas system have a unique targeting mechanism that is able to degrade both the DNA and RNA of invader (Pyenson and Marraffini, 2017). For example, mature crRNA was generated by the Cas6 endonuclease, the complementary target RNA was recognized by crRNA-guided Csm/Cmr complexes and then Csm3 or Cmr4 subunit cleave target RNA into 6 nt nucleotide intervals, while the DNA cleavage was catalyzed by the single-stranded DNase activity of Cas10 (Samai et al., 2015; Park et al., 2017; Tamulaitis et al., 2017). Once Cas10 degraded DNA, cleavage of the protospacer RNA by Csm6/1 resulted in inactivation of Cas10. In addition, the Palm domain of Cas10 synthesized cyclic oligoadenylate (cOA), which was required for modulating the endoribonuclease activity of Csm6 (Rostol and Marraffini, 2019). These systems can broadly eliminate foreign targets with multiple mutations but circumvent damage to the host genome. Although recent studies have shown that the molecular mechanism of type III CRISPR-Cas systems against exogenous DNA, it is unclear whether type III CRISPR-Cas system is associated with bacterial physiology to date.
Mycobacterium tuberculosis is one of the leading causes of death from an infectious disease. Integrated type III-A CRISPR-Cas system is predominantly presented in the slow-growing mycobacteria, such as M. tuberculosis and M. bovis (Singh et al., 2021), rather than non-pathogenic mycobacteria (He et al., 2012). In the virulent M. tuberculosis strain H37Rv, the type III-A system is comprised of Csm2-6, Cas10, Cas6, Cas1, Cas2, and two CRISPR loci. In addition to its role in M. tuberculosis strain genotyping, recent studies have focused on the activity of M. tuberculosis CRISPR interference system in invader defense and potential for an endogenous genome editing tool.
A recent study demonstrated that deletion of cpnB, which encodes a multifunctional protein that hydrolyzes c-di-AMP, c-di-GMP, and nanoRNAs, upregulated all the cas (CRISPR-associated) genes compared to the WT (Zhang et al., 2018). Of note, the expression of cas gene varies in response to environmental stresses, such as oxidative stress and antibiotic treatment (Miryala et al., 2019), suggesting a link between the activity of type III-A CRISPR-Cas and stresses. Indeed, it has been reported that overexpression of cas genes impacts mycobacterial physiology (Huang et al., 2016; Zhai et al., 2018; Wei et al., 2019). For example, overexpression of cas2 decreased the survival ability of Mycobacterium smegmatis under oxidative stress (Huang et al., 2016). Cas1 impaired the repair of DNA damage and reduced the ability of M. smegmatis to resist oxidative stress. Although these studies indicated that a role for type III-A CRISPR-Cas in adapting to stressful environments in fast-growing mycobacteria, the function of type III-A CRISPR-Cas system in slowly growing mycobacteria is unclear.
The current study aimed to explore the role of type III-A CRISPR-Cas system in slowly growing mycobacteria. By studying Mycobacterium bovis BCG, we found that type III-A CRISPR-Cas system is required for adaptation to oxidative stress and cell envelope integrity. We also proved the involvement of type III-A CRISPR in regulating gene expression by RNA-seq. As expected, intracellular fitness significantly declines due to loss of type III-A CRISPR system. Moreover, mice infected with ΔTCC (type III-A CRISPR system mutant) increased the regulatory T cell population compared to WT control. This study could shed light on the molecular mechanisms of type III-A CRISPR system in mycobacteria.
Materials and Methods
Strains and Media
Mycobacterium bovis BCG and M. smegmatis MC2 155 were grown at 37°C in Middlebrook 7H9 broth (BD Biosciences), 7H10 agar plates (BD Biosciences) supplemented with 0.5% glycerol, 0.05% Tween-80, 10% oleic acid ADC and appropriate antibiotics. When indicated, antibiotics and inducers were used at the following concentrations: kanamycin (20 μg/ml, Sangon Biotech), hygromycin (50 μg/ml, Sangon Biotech), and anhydrotetracycline (ATc; 50 ng/ml, Abcam). The Escherichia coli strain DH5α was used for plasmid cloning and was cultivated at 37°C in Luria-Bertani (LB) broth or agar plates.
Ethics Statement
All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental procedures were approved by the Ethics Committee of Zhongshan School of Medicine on Laboratory Animal Care (reference number: 2016-159), Sun Yat-sen University.
Strain Construction
To generate ΔTCC, or cas6 knock-out in M. bovis BCG strain, deletion strain was generated by phage specialized transduction as described previously (Jain et al., 2014). Briefly, PCR was performed to synthesize fragments bearing the ∼1,000 and ∼1,000 bps of flanking regions of endogenous gene of M. bovis BCG, resulting in a deletion of the gene. The primers used in this study are listed in Supplementary Table 1. Amplicons corresponding to upstream and downstream flanking regions were digested with Van91I and cloned into the Van91I (Thermo Fisher Scientific) digested p0004s plasmid that contains a hygromycin resistance cassette and the sacB gene to be able to select for sucrose sensitivity. This allelic exchange substrate was introduced into the PacI (Thermo Fisher Scientific) site of phasmid phAE159 and electroporated into M. smegmatis mc2155 to obtain high titers of phage phAE159. Subsequently, the M. bovis BCG wild-type strain was incubated with high titers of the corresponding phage to create TCC, or cas6 knockouts, respectively. Colonies that had deleted endogenous TCC, or cas6 were selected on hygromycin-containing plates and verified for sucrose sensitivity, respectively. The deletion was confirmed by PCR analysis and sequencing (Supplementary Figure 2).
For construction of the complementation strain, TCC with its native promoter was first cloned into a pMV306 (mycobacterial integration vector; integrates into the attB site; KanR), and the recombinant plasmid pMV306-TCC was integrated into the chromosomes of the deletion strains, resulting in comp strain. The comp strain was selected in Middlebrook 7H10 agar medium (complemented with 10% OADC) containing 20 μg/ml kanamycin.
Construction of CRISPR Interference (CRISPRi) Targeting Constructs in M. bovis BCG
CRISPRi targeting constructs in M. bovis BCG was performed as previously described (Rock et al., 2017). Briefly, we designed sgRNA to target the non-target strand of whiB6 and bcg_1807 gene. For each individual sgRNA, two complementary oligonucleotides with appropriate sticky end overhangs were annealed to generate a double-stranded insertion which was phosphorylated by T4 polynucleotide kinase (Thermo Fisher Scientific). PCR conditions were: 37°C for 30 min, 95°C for 5 min, ramp down to 25°C at 5°C min–1. And then PCR products was ligated (T4 DNA ligase, Thermo Fisher Scientific) into the BsmBI-v2 (NEB) digested pLJR965 to obtain pLJR965-sgRNA. Successful cloning was confirmed by Sanger sequencing. pLJR965 containing sgRNA-targeting the genes of interest was transformed into M. bovis BCG competent cells. For each transformation, 100 ng plasmid and 100 μl pf electrocompetent mycobacteria were used. Transformants were selected by plating cells on 7H10-OADC plates containing 50 μg/ml hygromycin and 20 μg/ml kanamycin. A single-clone was picked and cultivated to log phase in the absence of ATc and then diluted back to OD600 = 0.015 with or without 100 ng/ml ATc. After 10 days of outgrowth, 0.4 OD600 units of cells was harvested by centrifugation and processed for RNA extraction.
Analysis of in vitro Response to Oxidative Stress
Exponentially growing bacterial cultures (OD600 0.4–0.6) were pelleted and washed three times with 7H9 containing 0.05% Tween-80, then diluted to OD600 = 0.5. The cells were treated only in 7H9 broth medium with different amounts of H2O2 (0, 5, 10, and 50 mM) and were incubated for 24 h. Then OD600 were recorded by using Nano-photometer (IMPLEN) and normalized to the corresponding control without oxidant treatment. When indicated, bacteria strains were serially diluted ten-fold with PBS immediately and spotted on 7H10-OADC agar. CFUs of M. bovis were counted after 4 weeks of incubation at 37°C.
Synthesis of cDNA and RT-PCR
Exponentially growing bacterial cultures (OD600 0.4–0.6) were pelleted and washed once with 7H9 containing 0.05% Tween-80, then diluted to OD600 = 0.5. The cells were treated with different stresses, including 5 mM H2O2, 5 mM NO, acidic medium (pH4.5), 1%SDS and were incubated for 2 h. Equal amounts of bacteria of M. bovis strains were collected. Total RNA and cDNA from M. bovis BCG were extracted as previously described (Rustad et al., 2009). Briefly, the collected cultures were harvested by centrifugation. After removal of the supernatant, the cell pellet was resuspended in RNA-easy Isolation Reagent (Vazyme) and disrupted by bead beating with a TissueLyser-II (QIAGEN). Total RNA was precipitated by the addition of isopropanol and collected by centrifugation, the supernatant was discarded, and the mRNA pellet was washed with 75% ethanol. After drying of the pellet, mRNA was dissolved in RNase-free H2O. The contaminating genomic DNA was digested with gDNA wiper Mix (Vazyme). The cDNA was prepared from purified mRNA with HiScriptII qRT SuperMix II (Vazyme) through reverse transcription. The cDNA levels of target genes were then quantified by quantitative real-time PCR (qRT-PCR) on a CFX96 cycler (BIO RAD) by using AceQ Universal SYBR qPCR Master Mix (Vazyme) as a dye for fluorescence signal detection. All qPCR primers were determined to be >95% efficient, and the cDNA masses tested were experimentally validated to be within the linear dynamic range of the assay. Signals were normalized to those of the housekeeping sigA transcript and quantified with the ΔΔCt method. Error bars are 95% confidence intervals of the three technical replicates.
Growth Curves
Mycobacterium bovis strains were grown in 7H9 medium containing OADC-Tween 80 to mid-log phase (OD600, 0.5–0.8). The precultures were pelleted at 3,000 g for 10 min and washed once in PBS, then diluted to OD 0.1 in 7H9 medium containing OADC-Tween 80. A Nano-photometer (IMPLEN) was used to measure the optical density at indicated time. The growth curve was generated based on the OD600. This experiment was performed in three replicates.
In vitro Permeability
Mycobacteria strains were grown to an OD600 0.5–1.0. bacteria were collected by centrifugation at 8,000 g for 10 min at 4°C. The collected cells were further washed three times using 1× PBS buffer then suspended in 1× PBST containing 50 μg/ml EtBr (Sangon Biotech) or 20 μM Nile Red (Sangon Biotech). Fluorescence was measured immediately in a Spectra Max M5 (Molecular Devices) using an excitation of 540 nm and emission of 590 nm, correcting with samples lacking bacteria. All experiments were repeated in triplicate.
RNA Preparation and RNA-Seq Analysis
RNA preparation and RNA-seq analysis were performed as previously described (Feng et al., 2020). Briefly, wild-type M. bovis BCG as well as the TCC deletion strain were grown to the log-phase (OD600, 0.5) in Middlebrook 7H9-OADC. Total RNA was prepared from wild-type and mutant strains using TRIzol reagent (Invitrogen), followed by DNase I treatment. Approximately 1-μg total RNA samples were treated by the Ribo-Zero rRNA removal procedure (Illumina) to enrichment for mRNA. Approximately 1 μg of RNA was used for library preparation using a ScriptSeq (v2) RNA-seq kit and high-throughput sequencing on an Illumina NextSeq platform. All raw sequence reads by RNA-Seq were initially pre-processed by Trimmomatic (v0.36) to trim the adaptor sequences and remove low-quality sequences. The remaining clean reads were mapped to the M. bovis BCG genomes using Tophat2. The alignment results were input into Cufflinks (v2.2.1). Unless otherwise stated, the unit of expression level in our analyses is FPKM. Cuffdiff (v2.2.1) was used to test for differential expression. The genome sequences and the annotations of M. bovis BCG were obtained from the GenBank1. The genes with a p-value ≤ 0.05 and |log2 fold change (ΔTCC/WT)| ≥ 1 were identified as differentially expression genes. The data for visualization was generated by R (R Development Core Team, Vienna, Austria). The functional annotations of differentially expression genes were carried out by gene ontology (GO) from an internationally established system (GO2). This system comprehensively describes the genes’ properties and provides their products in any organism. The corrected p-value of GO terms (p < 0.05) was considered significantly enriched by differentially expression genes. Differentially expression genes analyses were performed on the basis of the biological process, molecular function, and cellular components. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to determine the molecular pathways for the differentially expression genes.
Biofilm Growth Conditions
Mycobacteria strains were cultured in Sauton’s medium without Tween 80 in 12-well polystyrene tissue culture plates (Corning). Plates were incubated at 37°C under 5% CO2 for 5 weeks.
Cell Culture
Murine macrophage-like RAW264.7 cells (ATCC; TIB-71) were cultured on Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO, Invitrogen) with 5% CO2 at 37°C.
Macrophage Infection Study
RAW264.7 cells were seeded at 3 × 105 cells/well in a 24-well plate and incubated for 18 h. Bacterial cells were expanded to OD600 = 1, pooled and passed through a 10 μm cell strainer to obtain a single cell suspension. Adherent RAW264.7 cells were infected with homogenized bacterial suspension at a multiplicity of infection (MOI) of 10. The extracellular bacteria were removed by washing three times with phosphate buffer saline (PBS) after incubation for 2 h. Cells were further incubated for 48 h in a CO2 incubator at 37°C. Then cell lysates were serially diluted 10-fold with PBS immediately and spotted on 7H10-OADC agar. CFUs of M. bovis were counted after 4 weeks of incubation at 37°C.
Animal Experimentation
Female C57BL/6 mice were purchased from Animal Supply Center, Sun Yat-sen University. Mice were bred and maintained under specific pathogen-free conditions at the animal facility of the Sun Yat-sen University. Adult mice between 6 and 8 weeks of age were used, and infected mice were maintained under biosafety 2 conditions. To induce acute BCG infection, mice were infected with 1 × 107 colony-forming units (CFUs) of WT, ΔTCC and comp, through the intraperitoneal injection. Four mice from each group were sacrificed after 21-day post-infection, and the spleen homogenates [prepared by homogenizing aseptically removed organs in 1 ml of sterile saline with Tissue-Tearor (Biospec Products) at a maximum of 2,500 g] were plated on 7H10 agar supplemented with 10% OADC, in triplicates. CFUs of M. bovis were counted after 4 weeks of incubation at 37°C.
Flow Cytometry
Spleens of infected C57BL/6 mice were collected at day 21. Single-cell suspensions were generated from spleens in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal calf serum (FCS) and Penicillin-Streptomycin, red blood cells were lysed with red blood cell lysis buffer (BioLegend) followed by washing step. Cells were resuspended in FACS buffer and incubated with the Live/Dead marker DRAQ7 (BioLegend) for 5 min at 4°C in the dark. Cells were resuspended in FACS buffer and flow cytometry was performed to quantify cell populations. Details of antibodies used are as follows: CD4 phycoerythrin (PE)-Cy7 (BioLegend; clone RM4-5), CD8 V500 (BD Horizon, clone 53-6.7), CD62 allophycocyanin (APC) (BD Pharmingen; clone MEL-14), CD44 Pacific Blue (BioLegend, clone IM7), CD25-APC (eBioscience, clone PC61.5), Foxp3-eFluor450 (eBioscience, clone FJK-16S). Central memory CD4+ T cells were CD3+ CD4+ CD44high CD62high, effector memory CD4+ T cells were CD3+ CD4+ CD44high CD62Llow, regulatory T cells were CD4+ CD25+ FoxP3+. To analyze CD4, CD25 and FoxP3 expression, cells were stained with anti-CD4, anti-CD25); then followed by intracellular staining with anti-FoxP3. Isotype-matched antibodies (eBioscience) were used as controls. After staining, the cells were analyzed using a BD FACS Aria II (BD Biosciences) and analyzed using FlowJo software (version 8.6.3). The gating strategy has been described in Li et al. (2021). The CD4+ CD25+ lymphocyte population was gated for FoxP3 expression analysis. The proportion of FoxP3+ cells in CD4+ CD25+ T cells were compared among the different groups.
Immunohistochemical Analysis
Spleen specimens were fixed in 4% PFA (paraformaldehyde) and embedded in paraffin. Tissue blocks were sliced into 5 μm sections, air dried and fixed with cold acetone. Tissue slides were stained with primary antibodies against Foxp3 (CST, clone D6O8R, 1:800) for overnight incubation at 4°C. After incubation with primary antibody, staining was followed by ABC detection kit (Abcam) using biotinylated anti-rabbit IgG. AEC was used as chromogen. After counterstaining with hematoxylin, sections were dehydrated, mounted and examined under the Olympus BX63 microscope (Olympus). Specimens were scored according to the intensity of the dye color and the number of positive cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3 (brown), and the number of positive cells was graded as 0 (<5%), 1 (5–25%), 2 (25–50%), 3 (51–75%), or 4 (>75%). The two grades were added together and specimens were assigned to one of 4 levels: 0–1 score (−), 2 scores (+), 3–4 scores (++), more than 5 scores (+++). The positive expression rate was expressed as the percent of the addition of (++) and (+++) to the total number.
Statistical Analysis
Statistical analysis was performed using Prism (version 7.0c; GraphPad Software). Statistical significance was assessed using Welch’s t-tests, unpaired t-tests, one-way ANOVA followed by Dunnett’s multiple comparison test, or two-way ANOVA followed by Turkey’s multiple comparisons test wherever appropriate. The statistical tests used, exact p-values, and sample sizes are indicated in each figure legend. A p-value of 0.05 or less was considered statistically significant.
Results
Type III-A CRISPR-Cas System Is Required for Adaptation to Oxidative Stress
Mycobacterium bovis BCG CRISPR-Cas locus possesses two CRISPR regions and nine cas genes (Figure 1A and Supplementary Figure 1), similar to M. tuberculosis type III-A CRISPR-Cas system. Since M. tuberculosis encounters different degrees of adverse niche, such as the acidic, nitro-oxidative stresses, when growing in the intracellular environment, we first measured transcription level of nine cas genes in M. bovis BCG under such stresses. The mRNA levels of cas1, csm5, csm4, csm3, csm2, and cas10 were significantly increased in response to oxidative stress (Figure 1B), and the expression of csm6 was increased but without significance. In addition, only cas1 and csm5 was significantly upregulated upon acidic medium treatment. CRISPR-associated genes were not induced after NO or SDS exposure. These results indicated that potential role of type III-A CRISPR-Cas in resistance to oxidative stress. To further explore the function of type III-A CRISPR-Cas system in mycobacteria, two mutant strains, Δcas6 and ΔTCC (total III-A CRISPR-Cas, including cas6, cas10, and csm2-6), were generated in M. bovis BCG by specialized transducing mycobacteriophages (Supplementary Figure 2). To confirm the phenotype caused by deletion of TCC, the complement strain (comp) was constructed by expression of TCC on the chromosome. The strains of WT, ΔTCC, comp, and Δcas6 were treated with hydrogen peroxide. Consistently, deletion of TCC conferred reduced survivability after exposure to hydrogen peroxide compared to parental bacteria and complement strain (Figure 1C). In addition, the percentage growth of Δcas6 mutant strain was significantly reduced compared to WT strain upon the treatment with 50 mM. These results indicated that type III-A CRISPR-Cas system contributed to oxidative stress resistance.
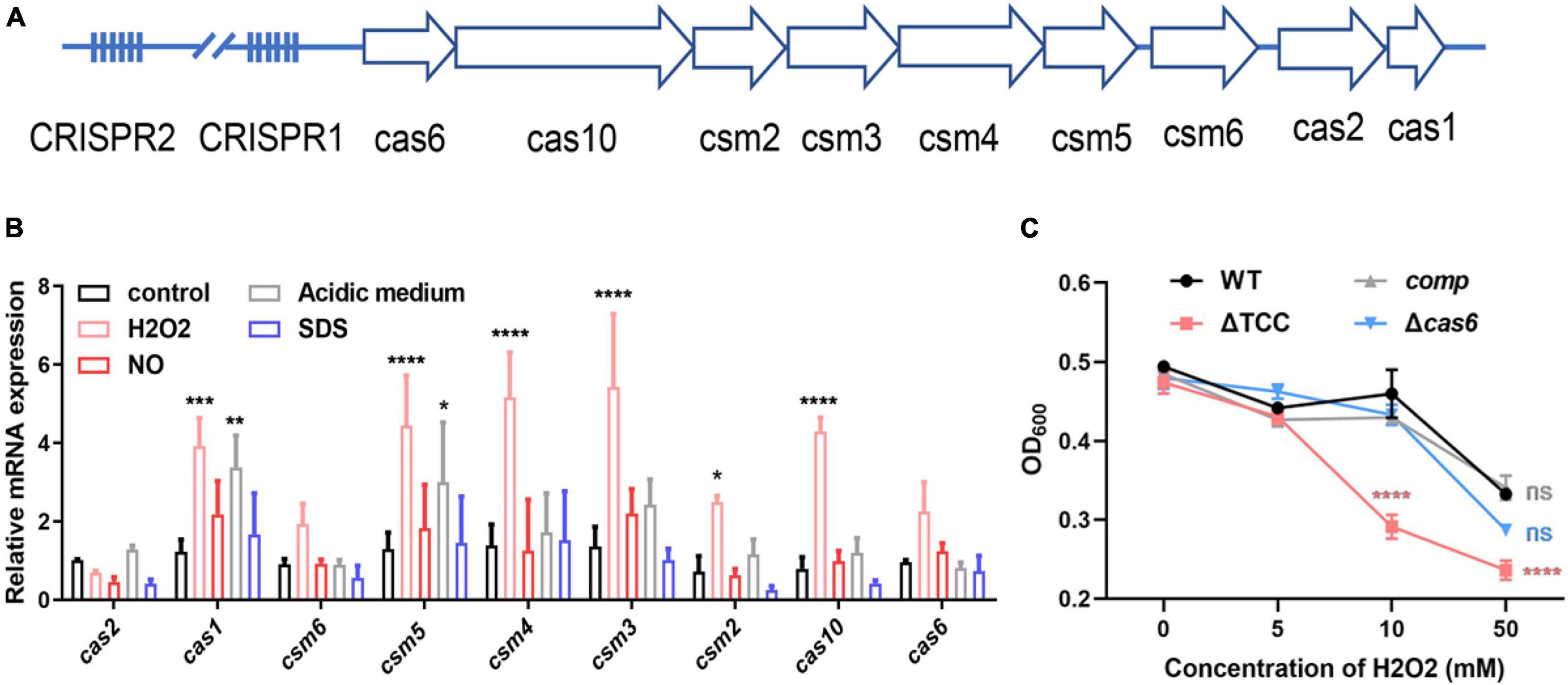
Figure 1. Inactivation of type III-A CRISPR-Cas system decreased bacterial survivability under oxidative stress. (A) Schematic of the Mycobacterium bovis BCG type III-A CRISPR-Cas operon. The type III-A CRISPR-Cas loci comprise csm2-6, cas10, cas6, cas1, cas2, and two CRISPR locus. (B) Fold changes in mRNA level of nine cas genes during different stresses. BCG was grown in 7H9 medium containing OADC and Tween 80 to mid-log phase. These strains were treated with different stresses, including 5mM NO, acidic medium (pH4.5), 1% SDS, 5mM H2O2. The expression of cas genes was analyzed by real time PCR (n = 3 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with control group; *p < 0.05, **p < 0.01, ****p < 0.0001). (C) Differential susceptibility of WT BCG, ΔTCC, comp, or Δcas6 strains to H2O2 compared to control was determined by OD600 measurements. M. bovis BCG were grown in 7H9 medium containing OADC and Tween 80 to mid-log phase. The cultures were treated with different amounts of H2O2. OD600 were recorded (n = 3 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with WT group; ns, not significant, ***p < 0.001, ****p < 0.0001).
TCC Enhances Mycobacterial Envelope Integrity
Next, we sought to explore the phenotype caused by TCC mutant. It has been previously demonstrated that cas genes deletion impacts bacterial biofilm formation (Cui et al., 2020). To examine whether type III-A CRISPR-Cas system can regulate biofilm formation, WT, ΔTCC, and comp strains were cultured in Sauton’s medium. Compared to WT and comp strains, no significant biofilm change was observed in ΔTCC (Supplementary Figure 3). Interestingly, the colony of ΔTCC was rougher than WT and complement strain. In liquid media, ΔTCC grew slightly faster with similar growth curves (Figure 2A). In light of the observation of abnormal morphology due to loss of TCC, we speculated whether TCC enhanced mycobacterial envelope integrity. To explore this, the permeability of ΔTCC was analyzed by measuring its uptake of Nile red. TCC deletion mutant exhibited a significant increase in fluorescence intensity compared with WT bacteria, indicating that it is more permeable to Nile red (Figure 2B). Furthermore, similar experiments were performed with the nucleic acid-staining dye ethidium bromide (Figure 2C) and a significant increase in fluorescence intensity was observed in the TCC mutant. Thus, our results demonstrated that TCC contributed to mycobacterial envelope integrity.
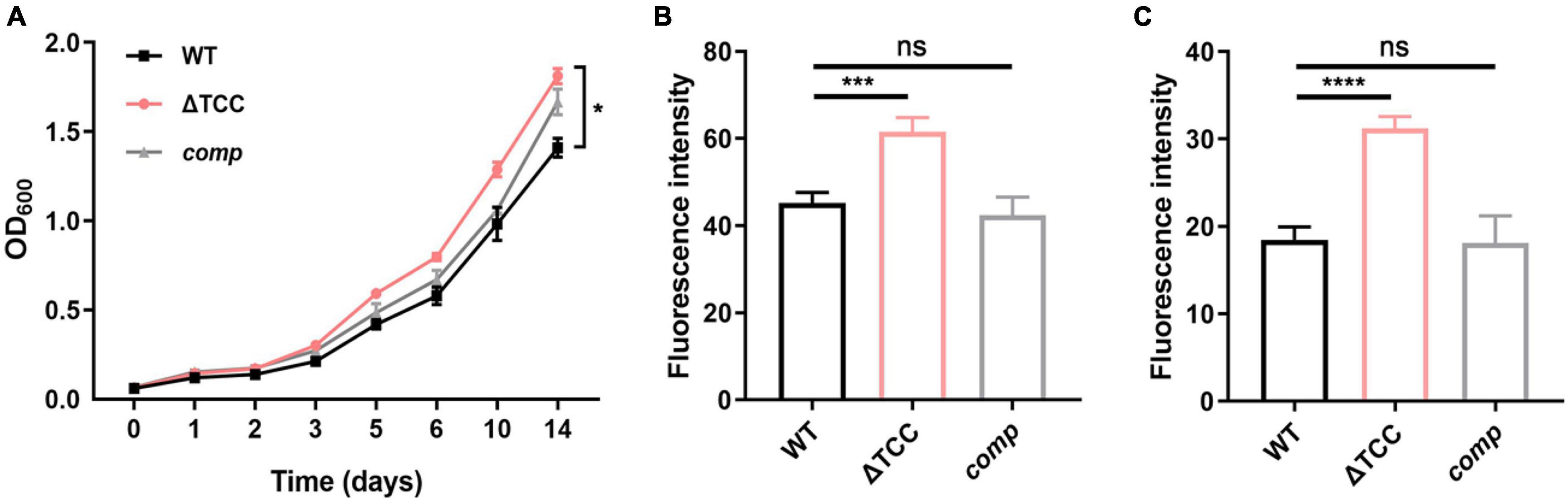
Figure 2. The effect of TCC deletion on mycobacterial cell wall permeability. (A) WT, TCC deletion mutant, comp strains were grown to mid-log phase, washed. Bacteria were diluted to an initial OD600 of 0.05 and the OD600 was monitored at the designated time points. Cell growth was monitored by OD600 measurements (n = 3 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with WT group; *p < 0.05). (B,C) WT, TCC deletion mutant, comp strains were grown to mid-log phase, washed, and stained with (B) Nile red or (C) ethidium bromide, and fluorescence was measured (n = 3 biological replicates per group, data are shown as the mean ± SEM, one-way ANOVA with Dunnett’s multiple comparison test, compared with WT group; ns, not significant; ***p < 0.001, ****p < 0.0001).
Analysis of Transcriptome Profile Alterations in ΔTCC
To understand the described phenotypes of the ΔTCC mutant, we next sought to identify the genes regulated by TCC. Although it has been reported that RNA targeting in type III-A CRISPR-Cas systems in Thermus thermophilus (Staals et al., 2014), no apparent homology was found between the genome of M. bovis BCG and spacer sequences in type III-A CRISPR locus. To shed light on the functional roles of TCC, we performed whole-transcriptome RNA-seq analyses to identify differentially expression genes in ΔTCC strains. Wild-type M. bovis BCG and ΔTCC were collected at log-phase (OD600, 0.5) for RNA-seq (see section “Materials and Methods”). With our significant criteria (|log2 fold change (ΔTCC/WT)| ≥ 1 and p-value < 0.05), 590 genes were identified as differentially expressed genes, 220 of which were upregulated and 370 were downregulated in ΔTCC. Differentially expressed genes were shown in the volcano plot (Figure 3A, and Supplementary Table 2). Of note, the positive control in our experiments, cas genes, were the most down-regulated genes in the entire dataset and 84 cell wall proteins were differential expressed (Supplementary Table 3). Consistently, mRNA expression of whiB6, whiB3, bcg_1807, ctpV, and otsB that downregulated in ΔTCC, significantly decreased as shown in Figure 3B. Also, mce4B belonging to cholesterol transport system mce4, was increased in ΔTCC. Moreover, differentially expressed genes were mapped to the KEGG database and enriched KEGG pathways were analyzed. The 62 enriched pathways (FDR > 0.05) are shown in Supplementary Table 4. In addition, to investigate the potential biological functions in which differentially expressed genes might be involved, the differentially expressed genes were enriched in GO (Gene Ontology). The top three categories were oxidation-reduction process (BP, 36 genes), response to stimulus (BP, 70 genes), and oxidoreductase activity (MF, 30 genes) (Figure 3C). Of note, pathways related to fatty acid metabolism were significantly enriched in CC, such as cholesterol catabolic process and lipid catabolic process, indicating the impact of TCC on lipid metabolism. All the enriched GO terms are presented in Supplementary Table 5. These outcomes demonstrated that inactivation of TCC has a profound effect on cellular gene regulation, especially on fatty acid metabolism.
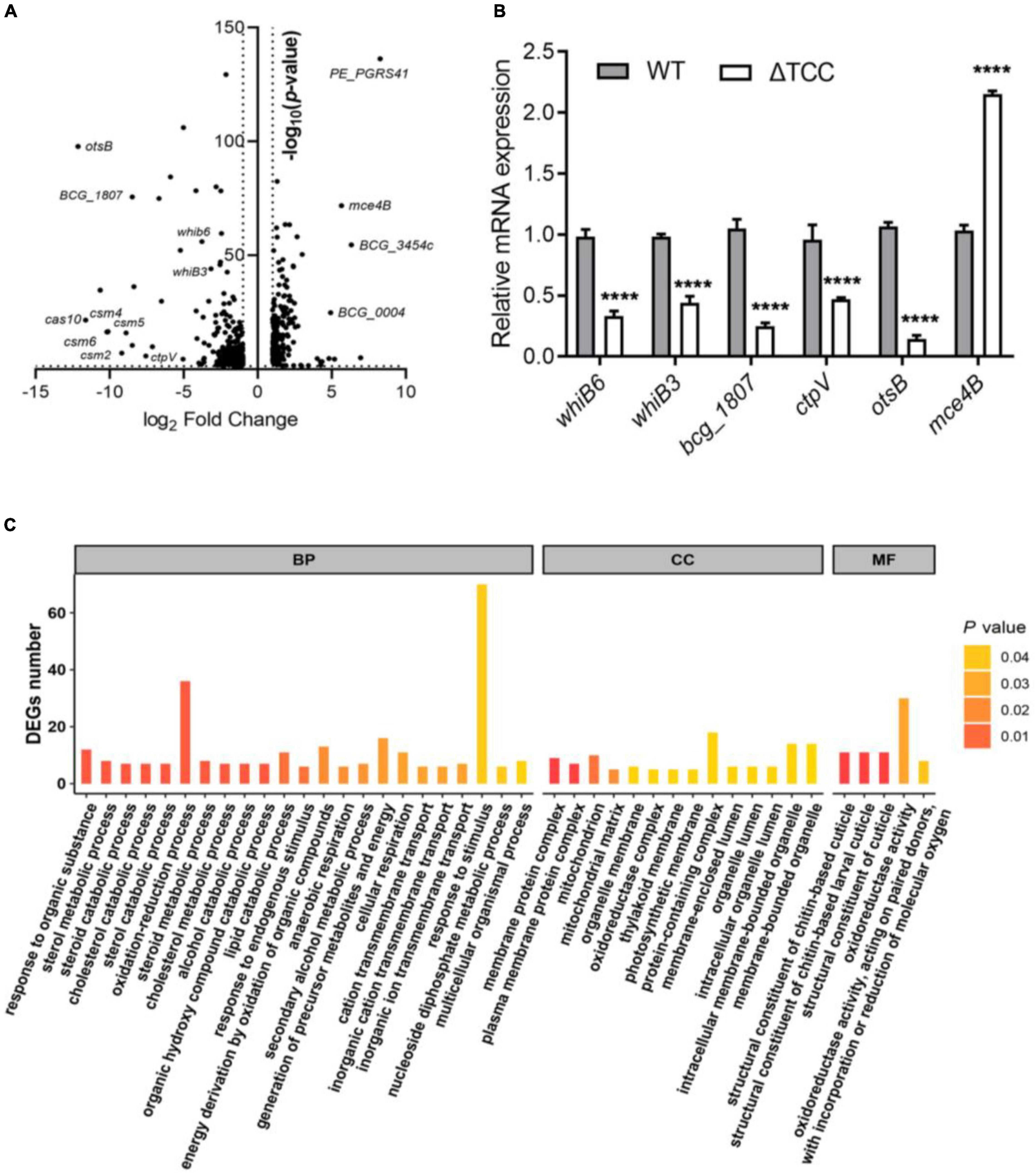
Figure 3. Gene-expression differences between WT and ΔTCC. (A) The volcanic diagram of gene expression difference between WT and ΔTCC strains determined by RNA-seq. The significantly upregulated and downregulated genes [p ≤ 0.05 and | log2 fold change (ΔTCC/WT) | ≥ 1] were shown. (B) The mRNA level of differential genes was tested by qRT-PCR. WT and ΔTCC strain were grown to log phase, the bacterial pellet was harvest for mRNA detection (n = 3 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with control group; ****p < 0.0001). (C) Gene ontology (GO) annotations of differentially expressed genes and classification graph of GO terms for differentially expressed genes. GO includes the molecular function (MF), cellular component (CC), and biological process (BP).
Knockdown of TCC-Regulated Genes Conferred Reduced Survivability Upon Oxidative Stress
To determine whether disruption of TCC resulted in decreased survivability upon oxidative stress by regulation of gene expression, whiB6 and bcg_1807 were silenced by CRISPR interference (Rock et al., 2017), a robust tool for gene knockdown. WhiB6, a transcriptional regulator is important for M. tuberculosis virulence by modulating ESX-1 and the Dos regulon (Chen et al., 2016). BCG_1807 is an oxidoreductase which dysregulated response to stress (Leversen et al., 2012). We designed sgRNA to target whiB6 and bcg_1807 in BCG, respectively. After induction of CRISPRi, the knockdown efficiency was assessed by real-time PCR. The mRNA level of whiB6 and bcg_1807 was 10- and 20-fold lower than ctl-sgRNA, suggesting successful knockdown of target genes (Figures 4A,B). Next, knockdown strains were treated with H2O2. Consistent with our previous results, the survivability of ΔTCC was significantly decreased when compared to WT. Also, knockdown of whiB6 showed slightly decreased survival, and the bcg_1807 knockdown was significantly compromised for oxidative stress (Figure 4C), suggesting that downregulated whiB6 and bcg_1807 to some extent participated in ΔTCC-conferred reduced survivability in oxidative stress conditions.
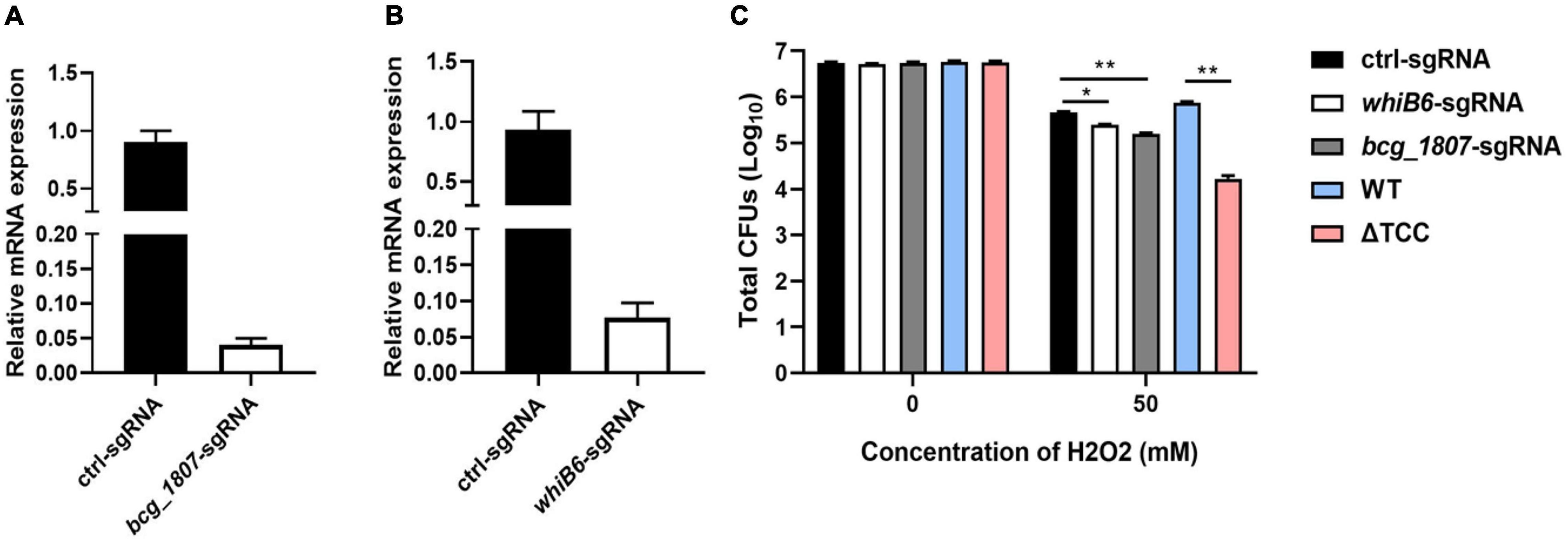
Figure 4. Knockdown of TCC-regulated genes decreased the resistance of BCG to hydrogen peroxide. (A,B) Using CRISPRi to target gene silence. Bacteria was collected at logarithmic phase in the presence of ATc. Gene knockdown was quantified by qRT-PCR (n = 3 biological replicates per each group, data are shown as the mean ± SEM). (C) Testing the survivability of knockdown strain. After induction, these knockdown strains were treated with 50 mM hydrogen peroxide. Bacteria strains were serially diluted 10-fold and spotted on 7H10-OADC agar. CFUs of M. bovis BCG were counted after 4 weeks days of incubation at 37°C (n = 3 biological replicates per group, data are shown as the mean ± SEM by unpaired t-test with two tailed p-value; *p < 0.05, **p < 0.01).
Intracellular Fitness of ΔTCC Mutant Strain Was Evaluated in vivo and in vitro
Mycobacterium tuberculosis use macrophages to traverse host epithelial barriers to enter deeper tissues, where they recruit additional macrophages to form granulomas. M. tuberculosis-infected macrophages generate reactive oxygen species (ROS) such as hydrogen peroxides and nitric oxide to control mycobacterial growth in the hyperinflammatory environment of the granuloma (Dallenga et al., 2017). Given that type III-A CRISPR-Cas system is required for resistance to oxidative stress which mimicks intracellular niche, we postulated that TCC mutant might exhibit reduced bacterial survivability in vitro and in vivo. To test this, RAW264.7 were infected with WT, ΔTCC, or comp strains and the numbers of surviving intracellular bacteria were assessed at 0, 24, and 48 h after infection. We found deletion of TCC significantly reduced the number of surviving intracellular mycobacteria at 48 h (Figure 5A). In addition, the expression of cytokines was examined following macrophage infection and we found that IL-10 mRNA expression was slightly increased in the macrophage cell line RAW264.7 infected with ΔTCC compared to WT or comp strains (Supplementary Figure 4).
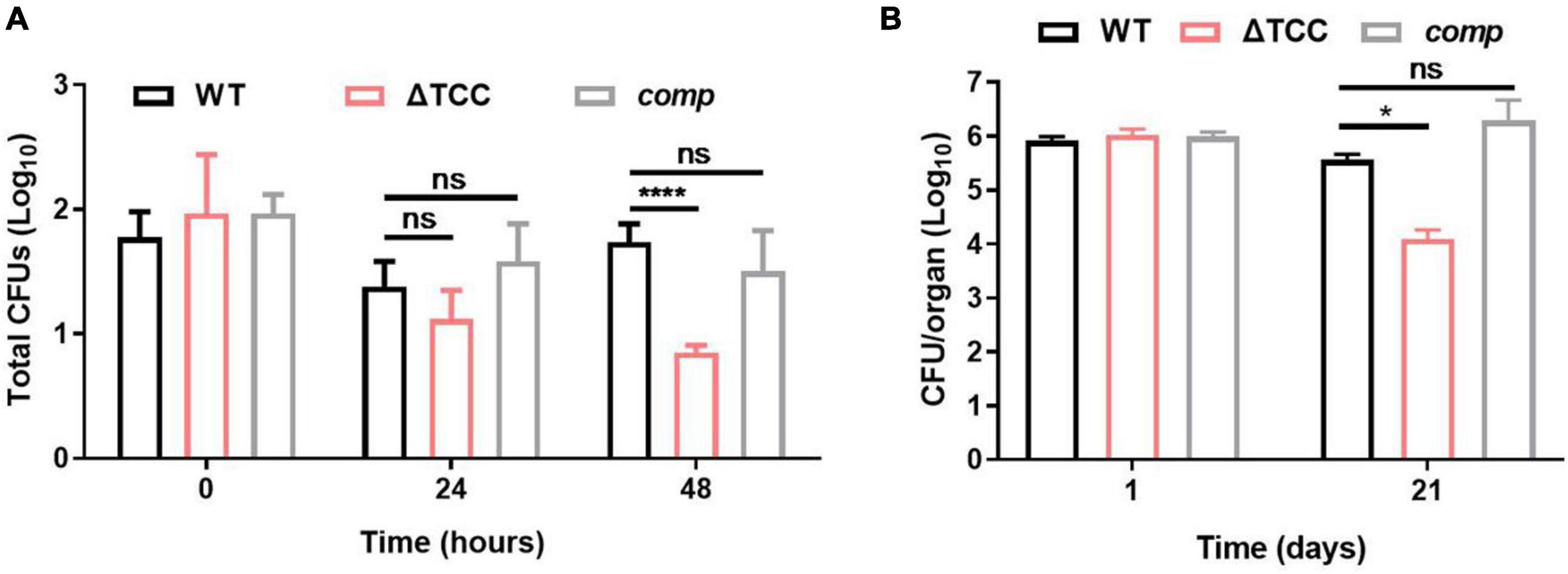
Figure 5. Evaluation intracellular fitness of ΔTCC mutant strain in vivo and in vitro. (A) RAW264.7 cells were seeded at 3 × 105 cells per well in 24-well culture plates. After adhesion, cells were infected with bacteria at MOI of 10. The extracellular bacteria were removed 2 h post infection by washing with PBS. The cells of M. bovis BCG were further incubated for 24 and 48h at 37°C. Then serially diluted (10-fold) cells lysates were spotted on a 7H10-OADC agar and cultured in 37°C. CFUs were counted after incubation for the indicated time (n = 3 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with WT group; ns, not significant, ****p < 0.0001). (B) Mice were infected i.p. (1 × 107) with WT, ΔTCC, or comp strains of M. bovis BCG. The number of surviving bacilli by CFU counting at the indicated time (n = 4 biological replicates per group, data are shown as the mean ± SEM, two-way ANOVA with Tukey’s multiple comparison test, compared with WT group; ns, not significant, *p < 0.05).
Next, survival of WT, ΔTCC, and comp were assessed by CFU in mice. At 1-day postinfection there were no differences in the number of bacteria from the spleen. After 21-day of infection, we observed a decrease of bacterial burden in mice infected with the TCC mutant strain (Figure 5B). Together, these data suggested the essential role of type III-A CRISPR-Cas system in intracellular mycobacterial survival.
Deletion of TCC in M. bovis BCG Is Associated With Increased Regulatory T Cell Population in Animal Model of Infection
It is known that the type I and type II CRISPR-Cas systems contribute to dampen the host immune response (Sampson et al., 2013; Li et al., 2016), but the role of type III CRISPR-Cas system in host immune response is unclear. M. tuberculosis possesses an unusual cell wall dominated by lipids and carbohydrates that is crucial for its survival and virulence. For example, mannose-capped lipoarabinomannan, a mycobacterial wall component, enabled expansion of regulatory T cell. Given that altered colony morphology and changing in transcript of cell wall component caused by TCC mutant, we hypothesized that type III CRISPR-Cas system may be involved in regulating host immune response. To verify our hypothesis, mice were infected with M. bovis BCG, ΔTCC, or comp strains. At day 21, no significant effect was observed in frequencies of central memory and effector memory CD8+ T cells, central memory CD4+ T, and effector memory CD4+ T cells between WT and ΔTCC (Supplementary Figure 5). Interestingly, frequencies were significantly increased for CD4+ CD25+ Foxp3+ T cells in the spleen of mice infected with ΔTCC (Figure 6A). Moreover, increasing Foxp3+ cells in the spleen of mice infected with ΔTCC was verified by immunohistochemical analysis (Figure 6B), which is in line with our previous observation. Collectively, these observations support the conclusion that TCC contributed to reduced CD4+ CD25+ Foxp3+ regulatory T cell population in BCG infected mice.
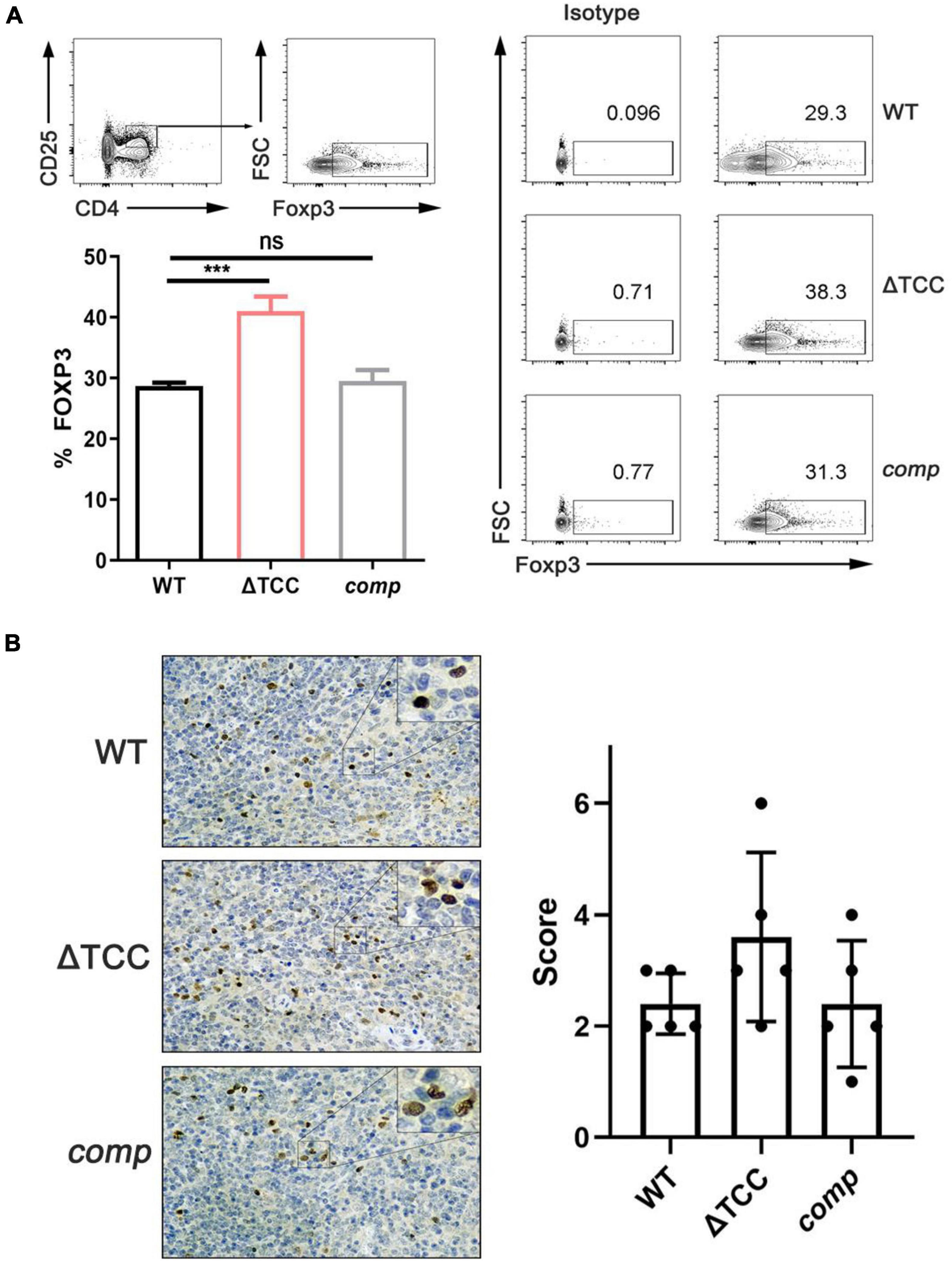
Figure 6. TCC is required for inhibition of regulatory T cells. (A) Flow cytometry evaluation of CD4+ CD25+ FOXP3+ cells in spleens from mycobacteria-infected mice. FOXP3+ cells were determined by intracellular staining with anti-FOXP3 after surface staining with anti-CD4 and anti-CD25+. Cells were stained with FOXP3 as a marker for regulatory T cells. Isotypic antibodies were used as controls. The proportion of FoxP3+ T cells within total CD4+ CD25+ T cells was significantly higher in ΔTCC compared to WT (n = 3 biological replicates per each group, data are shown as the mean ± SEM, one-way ANOVA with Dunnett’s multiple comparison test performed for ΔTCC, compared with WT group; ns, not significant, ***p < 0.001). (B) Immunohistochemical evaluation of FOXP3+ cells in mice spleens. 400× magnifications, respectively. Similar results were observed in all spleens evaluated (n = 5). Bar = 200 μm. Specimens were scored according to the intensity of the dye color and the number of positive cells.
Discussion
As a remarkably successful pathogen, M. tuberculosis is capable of persisting in host tissues for decades. Accumulating evidence described that M. tuberculosis employed several virulence factors to evade host immunity (Conrad et al., 2017; Wang et al., 2017; Sanchez et al., 2020). Type III-A CRISPR-Cas system was exclusively found in pathogenic mycobacteria, suggesting its unique role of type III-A CRISPR-Cas system in mycobacterial pathogenicity. In this study, we preliminarily described the important role of type III-A CRISPR-Cas system in mycobacteria. We found that the type III-A CRISPR-Cas system of M. bovis BCG is required for adaptation to oxidative stress and bacterial envelope integrity. Moreover, we found that TCC deletion increases the sensitivity of BCG to H2O2 through modulation of cell wall component. Consistent with the above findings, TCC deficiency attenuates the intracellular survivability, suggesting that the type III-A CRISPR-Cas system may be a novel target for therapy. Furthermore, we demonstrated in vivo that TCC contributes to regulate the regulatory T cell population during infection in vivo.
It has been described that the expression of CRISPR-Cas system was controlled by quorum sensing signaling molecules in P. aeruginosa and Chromobacterium violaceum (Patterson et al., 2016). Unlike P. aeruginosa, the LuxS synthase that is responsible for synthesis the quorum sensing signaling molecules autoinducer-2 is absent in M. tuberculosis. Therefore, autoinducer-2 failed to act as an autoinducer in M. avium (Geier et al., 2008). A recent study demonstrated that control of the CRISPR-Cas system by CnpB may be mediated through nanoRNAs (2–5 nucleotides), instead of hydrolysis of c-di-AMP or c-di-GMP, indicating that the accumulation of nanoRNAs may initiate the mRNA transcript of cas genes. Indeed, various stresses, such as low pH and hydron peroxide, to some extent, contributes to nanoRNAs accumulation (Nickels, 2012). Intriguingly, we found that cas genes were upregulated after H2O2 treatment, but not after NO treatment. It appears M. tuberculosis exhibited different transcriptional response after H2O2 and NO, respectively. For instance, M. tuberculosis exposed to 5 mM H2O2 resulted in massive transcriptional changes without an effect on growth or survival. Unlike H2O2 exposure, 5 mM NO exposure caused bacteriostatic activity. Moreover, after exposure to the same concentration of H2O2 and NO, a similar but different transcriptional response of M. tuberculosis was observed. After H2O2 exposure, DNA damage response and several PE and PPE genes were differentially expression that was not observed upon NO treatment (Voskuil et al., 2011).
Our data also demonstrated that inactivation of TCC resulted in reduced resistance to hydrogen peroxide. There are two possible explanations of increased sensitivity of ΔTCC to hydrogen peroxide. First, deletion of TCC significantly increased the envelope permeability. Increased cell-envelope permeability may lead to the enhanced uptake of compounds (Arts et al., 2015; Dulberger et al., 2020), such as hydron peroxide. Second, TCC influenced oxidation-reduction process. Indeed, in comparison to WT strain, several known transcriptional regulators that linked to oxidative stress, were significantly downregulated in ΔTCC (Figure 3A and Supplementary Table 2). For example, WhiB3 (BCG_3486) as an intracellular redox sensor maintained redox homeostasis (Saini et al., 2016). Deletion of whiB3 resulted in reduced viability in response to H2O2 and NO (Mehta and Singh, 2019). A RNA polymerase sigma factor SigE was induced upon oxidative stress (Jensen-Cain and Quinn, 2001) and sigE mutant was more sensitive to oxidative stress than its parent strain (Manganelli et al., 2001).
Accumulating evidence demonstrated that CRISPR-mediate transcriptional interference of the endogenous target could facilitate bacterial pathogenesis (Sampson et al., 2013; Li et al., 2016; Ratner et al., 2019). Although no apparent homology was found between the genome of M. bovis BCG and spacer sequences in CRISPR loci, whole-transcriptome RNA-seq identified 590 differentially expressed genes in TCC deletion strains, of which the majority were associated with metabolism, suggesting the profound effect of type III-A CRISPR-Cas system on mycobacteria viability. Of note, several virulence associated genes were significantly downregulated in ΔTCC. For example, CtpV is a metal cation-transporting ATPase and mutant for ctpV attenuated bacterial survival in vivo and in vitro (Ward et al., 2010). otsB is a gene involved in trehalose biosynthesis and the otsB deficiency in M. tuberculosis led to intracellular accumulation of trehalose-6-phosphate and therefore resulted in reduced bacterial survival in mice (Vanaporn and Titball, 2020). In addition, transcriptional regulator WhiB6 (BCG_3925c), was significantly down-regulated in TCC mutant strain, it raised the possibility that type III-A CRISPR-Cas system may involve in the regulation of bacterial virulence due to WhiB6-mediated control of type VII secretion system ESX-1 modulated mycobacteria virulence (Chen et al., 2016; Bosserman et al., 2017). This suggested that type III-A CRISPR-Cas system of BCG may be involved in bacterial virulence through WhiB6-mediated regulation of type VII secretion system ESX-1, indicating importance of type III-A CRISPR-Cas system in mycobacterial survival in vivo. Because type VII secretion system ESX-1 is absent in the avirulent strain M. bovis BCG, it would be interesting in future studies to examine the role of type III-A CRISPR-Cas system in the virulence of pathogenic M. tuberculosis.
There are a few caveats of our study that are worth discussing. First, we observed that TCC genes were significantly induced upon oxidative stress, and disruption of TCC resulted in reduced viability after hydrogen peroxide treatment. Our data may give rise to three independent lines of evidence in support of our hypothesis that TCC maintained the envelope integrity. (i) ΔTCC exhibited a distinct colony morphology compared to WT and complement strain (Supplementary Figure 3). Actually, abnormal morphology is associated with the alteration of cell wall components (Bhatt et al., 2007; Reddy et al., 2013; Singh et al., 2016); (ii) TCC deletion mutant exhibited a significant increase in fluorescence intensity compared with WT bacteria (Figures 2B,C); (iii) Analysis of GO terms revealed that pathways related to fatty acid metabolism were significantly enriched in CC (Figure 3C), such as cholesterol catabolic process and lipid catabolic process. Moreover, in KEGG pathway, fatty acid degradation (Down_P-value = 0.0019, and Down_FDR = 0.058) and steroid degradation (Down_P-value = 0.048, and Down_FDR = 0.57) were enriched (Supplementary Table 4), indicating the impact of TCC on lipid metabolism. It is well known that M. tuberculosis possesses complex lipids on the cell wall surface that are critical for the integrity of mycobacterial cell envelope (Dulberger et al., 2020). Intriguingly, 84 cell wall proteins were differentialy expressed between ΔTCC and WT (Supplementary Table 3). Among these genes, mce4B, mce4C, and mce4F that belong to mce4 operon were upregulated due to loss of TCC. It is generally accepted that host lipids are the primary carbon source for M. tuberculosis in vivo. Cholesterol transport system mce4 was essential for growth with macrophage (Pandey and Sassetti, 2008). The mce4 mutants unable to metabolize cholesterol also showed important defects in intracellular growth or survival (Griffin et al., 2012). Together, these results are consistent with the possibility that type III-A CRISPR-Cas system is involved in the biosynthesis of cell wall lipids. Exploring these interactions would be an interesting future research direction. Second, ΔTCC mutant exhibited a reduced intracellular fitness in vivo and we only tested the inflammatory response in BCG-infected macrophage. It is unknown whether other immune cells, such as Th1 and Th17 cells, may involve in reduced survivability caused by TCC deletion. It is well-established that Th1 and Th17 cells are needed in order to control infection by M. tuberculosis (Cardona and Cardona, 2019). Mycobacterial cell envelope components, such as lipoarabinomannan (Sarmiento et al., 2019) and phosphatidyl-inositol mannosides (Toyonaga et al., 2016), were able to promote a Th1 response during infection. Investigating the impact of TCC on Th1 and Th17 response would be an interesting future research direction. Third, type III-A CRISPR-Cas system of BCG decreased the CD4+ CD25+ Foxp3+ T regulatory cell population. Although ΔTCC upregulated the expression of anti-inflammatory cytokine IL-10, a key cytokine involved in the immunosuppressive function of Tregs, the relationship between TCC and Treg cells remained unknown. Garg et al. demonstrated that treatment with mannose-capped lipoarabinomannan, a mycobacterial wall component, enabled expansion of regulatory T cell (Garg et al., 2008). It appeared that the expansion of regulatory T cell may link to ΔTCC-mediated cell wall modeling. In addition, there is a recurring debate as to whether Tregs are beneficial or detrimental in M. tuberculosis infection. For example, when CD25+ cells were depleted in DBA/2 mice prior to M. tuberculosis infection, a reduction in the bacterial load of lungs and spleen was observed (Ozeki et al., 2010). Depletion of CD25+ cells 3 days prior to infection in M. bovis BCG had no effect on bacterial load (Quinn et al., 2006). However, in another study, the adoptive transfer of M. tuberculosis-specific Treg before T cell expression results in higher bacterial load (Shafiani et al., 2010). Also, it is not yet clear whether high level of Tregs are a consequence of inflammation for development of tuberculosis (Cardona and Cardona, 2019). Thus, the detailed interesting mechanism by which type III-A CRISPR-Cas of M. tuberculosis impacts Treg expansion requires further investigation.
Conclusion
Our study revealed the important role of type III-A CRISPR-Cas system in mycobacterial biology. The deletion of TCC resulted in decreased resistance to oxidative stress, damage of cell envelope, reduction of intracellular fitness, and increased T regulatory cells. The unappreciated role of type III-A CRISPR-Cas systems would help us to understand the pathogenesis of M. tuberculosis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE166137.
Ethics Statement
All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental procedures were approved by the Ethics Committee of Zhongshan School of Medicine on Laboratory Animal Care (reference number: 2016-159), Sun Yat-sen University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
SF: conceptualization. YL and MD: formal analysis. WL, LX, and DZ: investigation. FY, LL, and JL: methodology. G-BT: resources. SF, YC, and HZ: writing-original draft. DL: writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work has been supported by grants from National Natural Science Foundation of China (Grant Numbers 81722030, 81830103, 8201101256, and 82002173), the Qingyuan People’s Hospital Medical Scientific Research Fund Project (Grant Number 20190209), the Guangdong Provincial Bureau of Traditional Chinese Medicine research fund (Grant Number 20201407), National Key Research and Development Program (Grant Number 2017ZX10302301), Guangdong Natural Science Foundation (Grant Number 2017A030306012), project of high-level health teams of Zhuhai at 2018 (The Innovation Team for Antimicrobial Resistance and Clinical Infection), 111 Project (Grant Number B12003), and Open Project of Key Laboratory of Tropical Disease Control (Sun Yat-sen University), Ministry of Education (Grant Numbers 2020kfkt04 and 2020kfkt07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.774492/full#supplementary-material
Footnotes
References
Arts, I. S., Gennaris, A., and Collet, J. F. (2015). Reducing systems protecting the bacterial cell envelope from oxidative damage. FEBS Lett. 589, 1559–1568. doi: 10.1016/j.febslet.2015.04.057
Bhatt, A., Fujiwara, N., Bhatt, K., Gurcha, S. S., Kremer, L., Chen, B., et al. (2007). Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 104, 5157–5162. doi: 10.1073/pnas.0608654104
Bosserman, R. E., Nguyen, T. T., Sanchez, K. G., Chirakos, A. E., Ferrell, M. J., Thompson, C. R., et al. (2017). WhiB6 regulation of ESX-1 gene expression is controlled by a negative feedback loop in Mycobacterium marinum. Proc. Natl. Acad. Sci. U.S.A. 114, E10772–E10781. doi: 10.1073/pnas.1710167114
Cardona, P., and Cardona, P. J. (2019). Regulatory T cells in Mycobacterium tuberculosis infection. Front. Immunol. 10:2139.
Chen, Z. K., Hu, Y. B., Cumming, B. M., Lu, P., Feng, L. P., Deng, J. Y., et al. (2016). Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos Regulon to modulate granuloma formation and virulence in Zebrafish. Cell Rep. 16, 2512–2524. doi: 10.1016/j.celrep.2016.07.080
Conrad, W. H., Osman, M. M., Shanahan, J. K., Chu, F., Takaki, K. K., Cameron, J., et al. (2017). Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. U.S.A. 114, 1371–1376. doi: 10.1073/pnas.1620133114
Cui, L., Wang, X., Huang, D., Zhao, Y., Feng, J., Lu, Q., et al. (2020). CRISPR-cas3 of Salmonella upregulates bacterial biofilm formation and virulence to host cells by targeting quorum-sensing systems. Pathogens 9:53. doi: 10.3390/pathogens9010053
Dallenga, T., Repnik, U., Corleis, B., Eich, J., Reimer, R., Griffiths, G. W., et al. (2017). M. tuberculosis-induced necrosis of infected neutrophils promotes bacterial growth following phagocytosis by macrophages. Cell Host Microbe 22, 519–530.e3. doi: 10.1016/j.chom.2017.09.003
Dulberger, C. L., Rubin, E. J., and Boutte, C. C. (2020). The mycobacterial cell envelope - a moving target. Nat. Rev. Microbiol. 18, 47–59.
Feng, S. Y., Liu, Y., Liang, W. F., Ahmed, M. A. G. E., Zhao, Z. H., Shen, C., et al. (2020). Involvement of transcription elongation Factor GreA in Mycobacterium viability, antibiotic susceptibility, and intracellular fitness. Front. Microbiol. 11:413. doi: 10.3389/fmicb.2020.00413
Garg, A., Barnes, P. F., Roy, S., Quiroga, M. F., Wu, S., et al. (2008). Mannose-capped lipoarabinomannan- and prostaglandin E2-dependent expansion of regulatory T cells in human Mycobacterium tuberculosis infection. Eur. J. Immunol. 38, 459–469. doi: 10.1002/eji.200737268
Geier, H., Mostowy, S., Cangelosi, G. A., Behr, M. A., and Ford, T. E. (2008). Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 74, 1798–1804. doi: 10.1128/Aem.02066-07
Griffin, J. E., Pandey, A. K., Gilmore, S. A., Mizrahi, V., McKinney, J. D., Bertozzi, C. R., et al. (2012). Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227. doi: 10.1016/j.chembiol.2011.12.016
He, L., Fan, X., and Xie, J. (2012). Comparative genomic structures of Mycobacterium CRISPR-Cas. J. Cell. Biochem. 113, 2464–2473. doi: 10.1002/jcb.24121
Hille, F., Richter, H., Wong, S. P., Bratovic, M., Ressel, S., and Charpentier, E. (2018). The Biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259.
Huang, Q. Q., Luo, H. P., Liu, M. Q., Zeng, J., Abdalla, A. E., Duan, X. K., et al. (2016). The effect of Mycobacterium tuberculosis CRISPR-associated Cas2 (Rv2816c) on stress response genes expression, morphology and macrophage survival of Mycobacterium smegmatis. Infect. Genet. Evol. 40, 295–301. doi: 10.1016/j.meegid.2015.10.019
Jain, P., Hsu, T., Arai, M., Biermann, K., Thaler, D. S., Nguyen, A., et al. (2014). Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio 5:e01245-14. doi: 10.1128/mBio.01245-14
Jensen-Cain, D. M., and Quinn, F. D. (2001). Differential expression of sigE by Mycobacterium tuberculosis during intracellular growth. Microb. Pathog. 30, 271–278. doi: 10.1006/mpat.2001.0431
Jones, C. L., Sampson, T. R., Nakaya, H. I., Pulendran, B., and Weiss, D. S. (2012). Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell. Microbiol. 14, 1531–1543. doi: 10.1111/j.1462-5822.2012.01816.x
Leversen, N. A., Sviland, L., Wiker, H. G., and Mustafa, T. (2012). Long-term persistence of BCG Pasteur in lungs of C57BL/6 mice following intranasal infection. Scand. J. Immunol. 75, 489–499. doi: 10.1111/j.1365-3083.2012.02683.x
Li, R., Fang, L., Tan, S., Yu, M., Li, X., He, S., et al. (2016). Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res. 26, 1273–1287. doi: 10.1038/cr.2016.135
Li, W., Hang, S., Fang, Y., Bae, S., Zhang, Y., Zhang, M., et al. (2021). A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377.e9. doi: 10.1016/j.chom.2021.07.013
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Manganelli, R., Voskuil, M. I., Schoolnik, G. K., and Smith, I. (2001). The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437. doi: 10.1046/j.1365-2958.2001.02525.x
Mehta, M., and Singh, A. (2019). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis and survival in response to reactive oxygen and nitrogen species. Free Radic. Biol. Med. 131, 50–58. doi: 10.1016/j.freeradbiomed.2018.11.032
Miryala, S. K., Anbarasu, A., and Ramaiah, S. (2019). Impact of bedaquiline and capreomycin on the gene expression patterns of multidrug-resistant Mycobacterium tuberculosis H37Rv strain and understanding the molecular mechanism of antibiotic resistance. J. Cell. Biochem. 120, 14499–14509. doi: 10.1002/jcb.28711
Nickels, B. E. (2012). A new way to start: nanoRNA-mediated priming of transcription initiation. Transcription 3, 300–304.
Ozeki, Y., Sugawara, I., Udagawa, T., Aoki, T., Osada-Oka, M., Tateishi, Y., et al. (2010). Transient role of CD4(+)CD25(+) regulatory T cells in mycobacterial infection in mice. Int. Immunol. 22, 179–189. doi: 10.1093/intimm/dxp126
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Park, K. H., An, Y., Jung, T. Y., Baek, I. Y., Noh, H., Ahn, W. C., et al. (2017). RNA activation-independent DNA targeting of the Type III CRISPR-Cas system by a Csm complex. EMBO Rep. 18, 826–840. doi: 10.15252/embr.201643700
Patterson, A. G., Jackson, S. A., Taylor, C., Evans, G. B., Salmond, G. P. C., Przybilski, R., et al. (2016). Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol. Cell 64, 1102–1108. doi: 10.1016/j.molcel.2016.11.012
Pyenson, N. C., and Marraffini, L. A. (2017). Type III CRISPR-Cas systems: when DNA cleavage just isn’t enough. Curr. Opin. Microbiol. 37, 150–154. doi: 10.1016/j.mib.2017.08.003
Quinn, K. M., McHugh, R. S., Rich, F. J., Goldsack, L. M., de Lisle, G. W., Buddle, B. M., et al. (2006). Inactivation of CD4(+)CD25(+) regulatory T cells during early mycobacterial infection increases cytokine production but does not affect pathogen load. Immunol. Cell Biol. 84, 467–474.
Ratner, H. K., Escalera-Maurer, A., Le Rhun, A., Jaggavarapu, S., Wozniak, J. E., Crispell, E. K., et al. (2019). Catalytically Active Cas9 mediates transcriptional interference to facilitate bacterial virulence. Mol. Cell 75, 498–510.e5. doi: 10.1016/j.molcel.2019.05.029
Reddy, P. V., Puri, R. V., Chauhan, P., Kar, R., Rohilla, A., Khera, A., et al. (2013). Disruption of mycobactin biosynthesis leads to attenuation of Mycobacterium tuberculosis for growth and virulence. J. Infect. Dis. 208, 1255–1265. doi: 10.1093/infdis/jit250
Rock, J. M., Hopkins, F. F., Chavez, A., Diallo, M., Chase, M. R., Gerrick, E. R., et al. (2017). Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2:16274. doi: 10.1038/nmicrobiol.2016.274
Rostol, J. T., and Marraffini, L. A. (2019). Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat. Microbiol. 4, 656–662. doi: 10.1038/s41564-018-0353-x
Rustad, T. R., Roberts, D. M., Liao, R. P., and Sherman, D. R. (2009). Isolation of mycobacterial RNA. Methods Mol. Biol. 465, 13–21. doi: 10.1007/978-1-59745-207-6_2
Saini, V., Cumming, B. M., Guidry, L., Lamprecht, D. A., Adamson, J. H., Reddy, V. P., et al. (2016). Ergothioneine maintains redox and bioenergetic homeostasis essential for drug susceptibility and virulence of Mycobacterium tuberculosis. Cell Rep. 14, 572–585. doi: 10.1016/j.celrep.2015.12.056
Samai, P., Pyenson, N., Jiang, W., Goldberg, G. W., Hatoum-Aslan, A., and Marraffini, L. A. (2015). Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174.
Sampson, T. R., Saroj, S. D., Llewellyn, A. C., Tzeng, Y. L., and Weiss, D. S. (2013). A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257. doi: 10.1038/nature12048
Sanchez, K. G., Ferrell, M. J., Chirakos, A. E., Nicholson, K. R., Abramovitch, R. B., Champion, M. M., et al. (2020). EspM is a conserved transcription factor that regulates gene expression in response to the ESX-1 System. mBio 11:e02807-19. doi: 10.1128/mBio.02807-19
Sarmiento, M. E., Alvarez, N., Chin, K. L., Bigi, F., Tirado, Y., Garcia, M. A., et al. (2019). Tuberculosis vaccine candidates based on mycobacterial cell envelope components. Tuberculosis 115, 26–41. doi: 10.1016/j.tube.2019.01.003
Shafiani, S., Tucker-Heard, G., Kariyone, A., Takatsu, K., and Urdahl, K. B. (2010). Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J. Exp. Med. 207, 1409–1420. doi: 10.1084/jem.20091885
Singh, A., Gaur, M., Sharma, V., Khanna, P., Bothra, A., Bhaduri, A., et al. (2021). Comparative genomic analysis of Mycobacteriaceae reveals horizontal gene transfer-mediated evolution of the CRISPR-Cas System in the Mycobacterium tuberculosis complex. mSystems 6:e00934-20. doi: 10.1128/mSystems.00934-20
Singh, P., Rao, R. N., Reddy, J. R. C., Prasad, R. B. N., Kotturu, S. K., Ghosh, S., et al. (2016). PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci. Rep. 6:21624. doi: 10.1038/srep21624
Staals, R. H., Zhu, Y., Taylor, D. W., Kornfeld, J. E., Sharma, K., Barendregt, A., et al. (2014). RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 56, 518–530. doi: 10.1016/j.molcel.2014.10.005
Tamulaitis, G., Venclovas, C., and Siksnys, V. (2017). Type III CRISPR-Cas immunity: major differences brushed aside. Trends Microbiol. 25, 49–61. doi: 10.1016/j.tim.2016.09.012
Toyonaga, K., Torigoe, S., Motomura, Y., Kamichi, T., Hayashi, J. M., Morita, Y. S., et al. (2016). C-Type Lectin Receptor DCAR recognizes Mycobacterial Phosphatidyl-Inositol Mannosides to Promote a Th1 response during infection. Immunity 45, 1245–1257. doi: 10.1016/j.immuni.2016.10.012
Vanaporn, M., and Titball, R. W. (2020). Trehalose and bacterial virulence. Virulence 11, 1192–1202. doi: 10.1080/21505594.2020.1809326
Voskuil, M. I., Bartek, I. L., Visconti, K., and Schoolnik, G. K. (2011). The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2:105. doi: 10.3389/fmicb.2011.00105
Wang, J., Ge, P. P., Qiang, L. H., Tian, F., Zhao, D. D., Chai, Q. Y., et al. (2017). The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation. Nat. Commun. 8:244.
Ward, S. K., Abomoelak, B., Hoye, E. A., Steinberg, H., and Talaat, A. M. (2010). CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77, 1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x
Wei, J., Lu, N., Li, Z., Wu, X., Jiang, T., Xu, L., et al. (2019). The Mycobacterium tuberculosis CRISPR-associated Cas1 involves persistence and tolerance to anti-tubercular drugs. Biomed Res. Int. 2019:7861695. doi: 10.1155/2019/7861695
Zhai, X., Luo, T., Peng, X., Ma, P., Wang, C., Zhang, C., et al. (2018). The truncated Rv2820c of Mycobacterium tuberculosis Beijing family augments intracellular survival of M. smegmatis by altering cytokine profile and inhibiting NO generation. Infect. Genet. Evol. 59, 75–83. doi: 10.1016/j.meegid.2018.01.027
Keywords: type III-A CRISPR-Cas, Mycobacterium, envelope integrity, oxidative stress, immunoregulation
Citation: Yang F, Xu L, Liang L, Liang W, Li J, Lin D, Dai M, Zhou D, Li Y, Chen Y, Zhao H, Tian G-b and Feng S (2021) The Involvement of Mycobacterium Type III-A CRISPR-Cas System in Oxidative Stress. Front. Microbiol. 12:774492. doi: 10.3389/fmicb.2021.774492
Received: 12 September 2021; Accepted: 15 November 2021;
Published: 09 December 2021.
Edited by:
Nelson da Cruz Soares, University of Sharjah, United Arab EmiratesReviewed by:
Rifat Hamoudi, University of Sharjah, United Arab EmiratesEvgeniya V. Nazarova, Genentech Inc., United States
Clemens Hermann, University of Basel, Switzerland
Copyright © 2021 Yang, Xu, Liang, Liang, Li, Lin, Dai, Zhou, Li, Chen, Zhao, Tian and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siyuan Feng, ZnN5MDU5M0AxNjMuY29t; ZmVuZ3N5OEBtYWlsLnN5c3UuZWR1LmNu
†These authors have contributed equally to this work
 Fan Yang
Fan Yang Lingqing Xu2†
Lingqing Xu2† Lujie Liang
Lujie Liang Wanfei Liang
Wanfei Liang Jiachen Li
Jiachen Li Min Dai
Min Dai Guo-bao Tian
Guo-bao Tian Siyuan Feng
Siyuan Feng