
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 17 December 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.771170
This article is part of the Research TopicGlobal Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and TreatmentView all 11 articles
 Yingying Hao1,2
Yingying Hao1,2 Xuguang Zhao3
Xuguang Zhao3 Cui Zhang4
Cui Zhang4 Yuanyuan Bai2
Yuanyuan Bai2 Zhen Song2
Zhen Song2 Xinglun Lu2
Xinglun Lu2 Ran Chen1
Ran Chen1 Yaoyao Zhu2
Yaoyao Zhu2 Yueling Wang2*
Yueling Wang2*Treatment strategies of infection by carbapenem-resistant Klebsiella pneumoniae (CRKP) are limited. Fosfomycin, a broad-spectrum antibiotic, has attracted renewed interest in combination therapy to fight K. pneumoniae infections. However, reports on fosfomycin-resistant K. pneumoniae are increasing. Among the 57 CRKP strains, 40 (70.2%) were resistant to fosfomycin. Thus, whole-genome sequencing and bioinformatics analysis were conducted to reveal molecular characteristics of fosfomycin-resistant K. pneumoniae. Twenty-three isolates coharbored fosAkp and fosA3, with K. pneumoniae carbapenemase (KPC)-producing ST11-KL64-wzi64-O2 (n = 13) and ST11-KL47-wzi209-OL101 (n = 8), the predominating clonal groups, while fosA3 was not detected in isolates carrying class B carbapenemase genes. Twenty-two (out of 26) ST11-KL64 strains were positive for rmpA2, of which 12 carried fosA3. Four of the 23 fosA3-positive isolates could successfully transfer their fosfomycin-resistant determinants to Escherichia coli J53AziR. All four strains belonged to ST11-KL47 with the same pulsed-field gel electrophoresis profile, and their transconjugants acquired fosfomycin, carbapenem, and aminoglycoside resistance. A 127-kb conjugative pCT-KPC-like hybrid plasmid (pJNKPN52_KPC_fosA) coharboring fosA3, blaKPC–2, blaCTX–M–65, blaSHV–12, rmtB, and blaTEM–1 was identified. ST11-KL64 and ST11-KL47 K. pneumoniae, with higher resistance and virulence, should be critically monitored to prevent the future dissemination of resistance.
Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a serious challenge in clinical medicine (Zhou et al., 2020). CRKP can cause bloodstream infections that are difficult to treat, with dramatically high hospital mortality rates of 55.8% (Xiao et al., 2020). In 2017, the WHO published a list of critical priority pathogens, where CRKP was placed in the Priority 1 group (Ekwanzala et al., 2019). Drug resistance is spreading rapidly across microbial species; however, new antibiotics are not discovered as frequently. Therefore, reassessing the utility of and the mechanism of drug resistance among microbes against long-established antibiotics is necessary (Vardakas et al., 2016).
Fosfomycin, discovered in 1969, is a broad-spectrum antimicrobial agent targeting peptidoglycan synthesis (Liu et al., 2020). It is effective against carbapenem-resistant Enterobacterales (CRE) and extended-spectrum β-lactamase-producing (ESBL) Enterobacterales and CRE, in vitro, and is approved for the management of systemic infections in Spain, Germany, and France (Mączyńska et al., 2021). In China, intravenous fosfomycin has been employed in the treatment of systemic infectious diseases since the 1990s. However, resistance has gradually increased over the past decade; in China, 80% of K. pneumoniae carbapenemase-producing K. pneumoniae (KPC-KP) strains are resistant to fosfomycin, which is much higher than in other countries (Huang et al., 2021).
Routine susceptibility test for fosfomycin in non-Escherichia coli Enterobacterales is not feasible (Elliott et al., 2019; Mączyńska et al., 2021). Clinical and Laboratory Standards Institute (CLSI) recommends glucose-6-phosphate (G6P)-supplemented broth microdilution and agar dilution for accurate susceptibility testing. Even tests like Kirby–Bauer Disk Diffusion (DD) and E-tests cannot yield reliable results and screen the fosfomycin resistance phenotype. Moreover, the epidemiology of fosfomycin resistance in clinical CRKP isolates from Shandong, China, is unclear.
Three mechanisms of fosfomycin resistance have been reported (Huang et al., 2021). Mutations in glpT and uhpT genes affect L-a-glycerophosphate and hexose-6-phosphate uptake, respectively. Mutations in MurA binding site, most notably Asp369Asn and Leu370lle, can also confer fosfomycin resistance. However, the most potent mechanism of resistance is drug hydrolysis by various chromosomal or plasmid-borne fosfomycin hydrolases, including FosA (fosA2, fosA3, fosA4, foskp96, fosAkp, and fosA7), fosB, fosC, and fosX (Liu et al., 2020).
Although the high fosfomycin resistance rate among KPC-KP was considered predominantly caused by clonal dissemination, horizontal transfer of fosA3-encoding plasmids among KPC-KP was also documented; in particular, the emergence of conjugative plasmids carrying a combination of the fosA3 and blaKPC–2 genes could accelerate the spread of antibiotic resistance (Jiang et al., 2015; Chen et al., 2019; Zhang et al., 2019). However, little is known regarding the prevalence of plasmid-mediated fosA3 and blaKPC–2 co-dissemination among KPC-KP.
In this study, we aimed to elucidate the molecular epidemiology of fosfomycin resistance among clinical CRKP isolates in China and to determine their genetic lineages. One of the self-transmissible plasmid pJNKPN52_KPC_fosA harboring fosA3, blaKPC–2, blaCTX–M–65, blaSHV–12, rmtB, and blaTEM–1 was fully sequenced and characterized. To the best of our knowledge, this is the first report of conjugative pCT-KPC-like plasmid co-carrying fosA3 and blaKPC–2.
Fifty seven non-duplicate CRKP clinical isolates from urine (n = 6), sputum or bronchoalveolar lavage fluid (n = 35), abscess (n = 2), blood (n = 5), pus (n = 2), abdominal fluid (n = 3), and other patient samples (n = 6) were collected from Shandong Provincial Hospital of China, between January 2017 and June 2020. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (BioMérieux, Marcy-l’Étoile, France) was used to identify the isolates. Carbapenemases were detected using carbapenem inactivation method (CIM) and EDTA-modified CIM (eCIM).
Antibiotic susceptibility was analyzed with a VITEK-2 compact system (BioMérieux, France) for aztreonam (ATM), cefepime (FEP), ceftriaxone (CRO), ceftazidime (CAZ), ertapenem (ETP), imipenem (IMP), piperacillin-tazobactam (TZP), trimethoprim-sulfamethoxazole (SXT), ciprofloxacin (CIP), levofloxacin (LVX), gentamicin (GEN), and amikacin (AMK); broth microdilution for polymyxin B (POL), tigecycline (TGC); and agar dilution for fosfomycin (FOS), using Mueller–Hinton agar supplemented with 25 μg/ml of G6P (CLSI, 2020). Susceptibility assay results were interpreted by CLSI breakpoints (CLSI, 2020), except for TGC, which were defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2020) guidelines (EUCAST, 2020). Fosfomycin susceptibility was interpreted according to CLSI breakpoints for E. coli urinary isolates. Phenotypic detection of carbapenemases was performed using the CIM and eCIM tests (CLSI, 2020).
Genomic DNA from clinical strains embedded in gel plugs was digested with QuickCut XbaI (Takara, Shiga, Japan), and restriction fragments, ranging from 50 to 500 kb, were separated using CHEF Mapper apparatus (Bio-Rad, Hercules, CA, United States) for 19 h with the pulse time switched from 6 to 36 s. Pulsed-field gel electrophoresis (PFGE) patterns were compared using Gel-J software, version 2.0 (Heras et al., 2015). Pulsotypes were assigned to the clusters with 80% similarity (Chen et al., 2019).
Conjugation experiments were carried out with sodium azide-resistant E. coli J53AziR being used as the recipient. Transconjugants harboring fosfomycin resistance genes were selected on Mueller–Hinton agar plates containing 64 mg/ml of fosfomycin, 100 mg/ml of sodium azide, and 25 μg/ml of G6P (Xiang et al., 2015). Transconjugants harboring carbapenemase resistance genes were selected on Mueller–Hinton agar plates containing 6 μg/ml of CAZ and 100 mg/ml of sodium azide. Antibiotic susceptibility test and PCR analysis were performed to confirm the fosA3 and/or carbapenemase gene transfer (Poirel et al., 2011; Xiang et al., 2015). Furthermore, PCR-based replicon typing (PBRT) was used to characterize the plasmid harbored by the transconjugants (Carattoli et al., 2005).
DNA from clinical isolates was extracted and sequenced using an Illumina Hiseq platform at Novogene Co., Ltd. (Beijing, China). Illumina sequences were assembled de novo using the SPAdes v3.10 (Nurk et al., 2013).
For the JNKPN52 and JNKPN30 isolates, genome sequencing was also performed on a PacBio RSII sequencer at Biozeron Biological Technology Co., Ltd. (Shanghai, China). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread, and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo.1 Sequence annotation was conducted using RAST 2.02 combined with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot and RefSeq databases. Annotation of resistance genes and mobile elements was carried out using the online databases, including CARD3 and ISfinder.4
Antimicrobial resistance genes and multilocus sequence typing (MLST) were analyzed in silico by using Abricate software5 (Sherry et al., 2019). Virulence scores, capsular (K) serotypes, and lipopolysaccharide (LPS) O antigen serotype were predicted using Kleborate v0.3.06 (Wyres et al., 2020). Single-nucleotide polymorphism (SNP) calling was performed using Snippy 3.1,7 and recombinant variants were excluded using ClonalFrameML 1.0 (Lu et al., 2019). Maximum likelihood phylogenetic trees were constructed with RAxML,8 from the recombination-free SNPs.
For our dataset, core-genome MLST (cgMLST) analysis was performed using SeqSphere+ software (8.0.2 version; Ridom, Münster, Germany) according to the ‘‘K. pneumoniae sensu lato cgMLST’’ version 1.0 scheme9 (Weber et al., 2019). A total of 2,358 target genes were used to characterize the gene-by-gene allelic profile of the K. pneumoniae strains. The resulting set of target genes was then used for interpreting the clonal relationship displayed in a minimum spanning tree using the “pairwise ignoring missing values” parameter during distance calculations.
The fosfomycin resistance-related proteins MurA, GlpT, and UhpT of the genomes were aligned with K. pneumoniae reference strain ATCC 700721 using local BLAST software. Multiple sequence alignments were performed by MAFFT, with the 4-kb fosAkp gene-related fragment of JNKPN10 as a reference.
The complete sequence of pJNKPN52_KPC_fosA has been deposited in GenBank under accession number MZ709016. Eleven fully sequenced pCT-KPC-like plasmids harboring blaKPC–2 were compared with pJNKPN52_KPC_fosA by BLAST Ring Image Generator,10 including pCT-KPC (GenBank accession no. KT185451), p69-2 (GenBank accession no. CP025458), p1068-KPC (GenBank accession no. MF168402), p20049-KPC (GenBank accession no. MF168404), p675920-1 (GenBank accession no. MF133495), pC2414-2-KPC (GenBank accession no. CP039820), pKP1034 (GenBank accession no. NZ_KP893385), pKSH203-KPC (GenBank accession no. CP034324), p16HN-263_KPC (GenBank accession no. CP045264), pEBSI036-2-KPC (GenBank accession no. MT648513), and pKP19-2029-KPC2 (GenBank accession no. CP047161). More detailed genome alignment between closely related plasmids was conducted by local BLAST and visualized with Easyfig.11
The complete sequence of pJNKPN30_NDM has been deposited in GenBank under accession number OL389795. Four fully sequenced IncA/C type plasmids harboring blaNDM–1 were compared with pJNKPN30_NDM by BLAST Ring Image Generator (see text footnote 10), including pNDM_KN (GenBank accession no. JN157804), pMS6198A (GenBank accession no. CP015835), p1605752AC2 (GenBank accession no. CP022126), and pT1 (GenBank accession no. KX147633).
Among the 57 tested CRKP strains, 40 were resistant to fosfomycin. High resistance (> 70%) was observed against β-lactam antibiotics, fosfomycin, and quinolones. The highest resistance (> 98%) was observed against TZP, CRO, FEP, and ETP. All the strains remained 100% susceptible to TGC, and only one isolate showed resistance to POL [minimal inhibitory concentration (MIC) ≥ 64]. The antibiotic susceptibility results are shown in Table 1 and Supplementary Table 4.
The 57 K. pneumoniae strains had 18 sequence types (STs), with ST11 being the most common (n = 36, 63%) (Figure 1 and Supplementary Table 2). The others were ST101 (n = 2), ST15 (n = 2), ST24 (n = 2), ST37 (n = 2), ST1031, ST133, ST152, ST1537, ST2246, ST25, ST258, ST29, ST323, ST392, ST3924, ST485, and ST528.
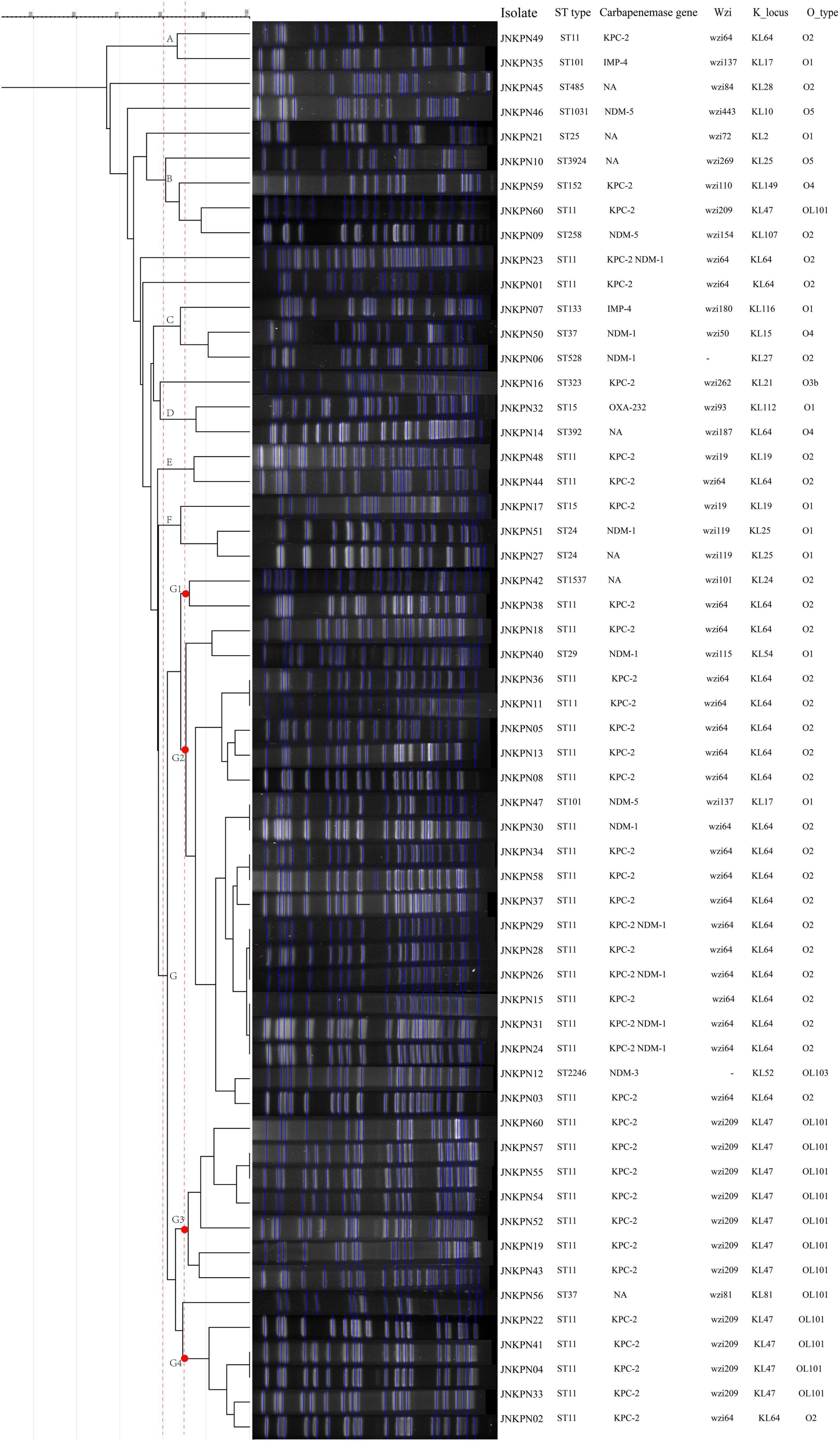
Figure 1. The dendrogram is based on the similarity of pulsed-field gel electrophoresis (PFGE) patterns in the 57 clinical carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates. The right panel shows results from isolate name, sequence type, carbapenemase gene, K_locus, and O_type. NA, not available.
Among these strains, we detected 19 different K-loci, the most common being KL64 (n = 26, including 25 ST11) and KL47 (n = 10), together accounting for 63% of all the strains (Figure 1). Seven distinct O antigen encoding loci were detected among the strains, and the most common were O2 (n = 30), OL101 (n = 11), and O1 (n = 9).
According to PFGE profile, seven different clusters as A∼G clone groups and five singletons were identified. The phylogenetic tree revealed that CRKP strains could be broadly clustered into three major clades: clades 1 and clades 2 consisted of ST11 strains alone, while clade 3 consisted of ST11 and the other STs (Figure 2). A minimum spanning tree of the 57 K. pneumoniae isolates was constructed based on cgMLST allelic profiles, showing the presence of five cluster types (≤ 15 allele differences). ST11-KL64-wzi64-O2 isolates mainly belonged to Cluster 1 (n = 17) and Cluster 3 (n = 6), while ST11-KL47-OL101 isolates mainly belonged to Cluster 2 (n = 7) (Figure 3 and Supplementary Table 5).
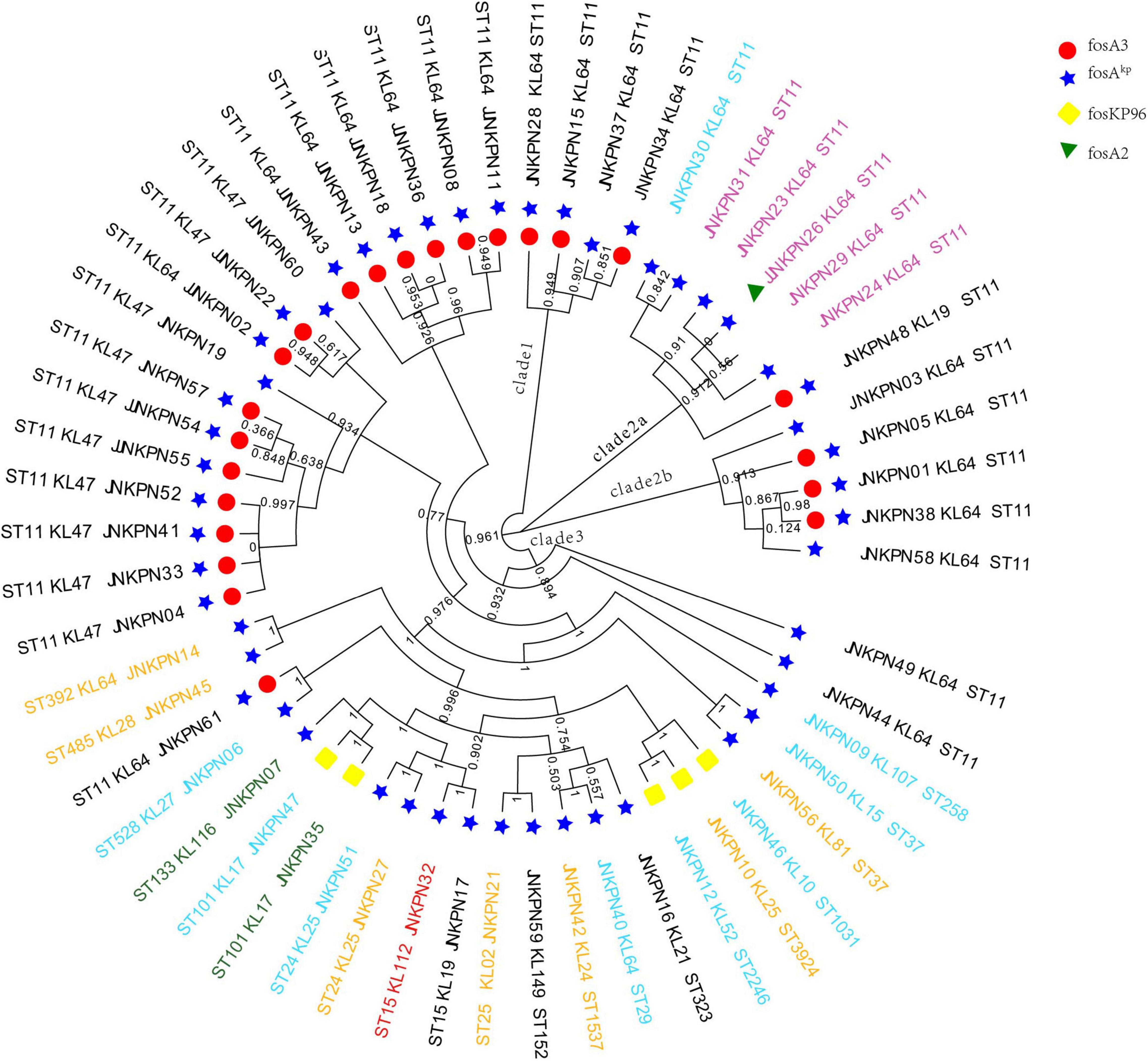
Figure 2. Phylogenetic analysis of carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates. The names of isolates were colored according to the carbapenemase(s) harbored, including blaKPC–2 (black), blaNDM (blue), blaNDM and blaKPC (pink), blaIMP (green), and blaOXA–232 (red).
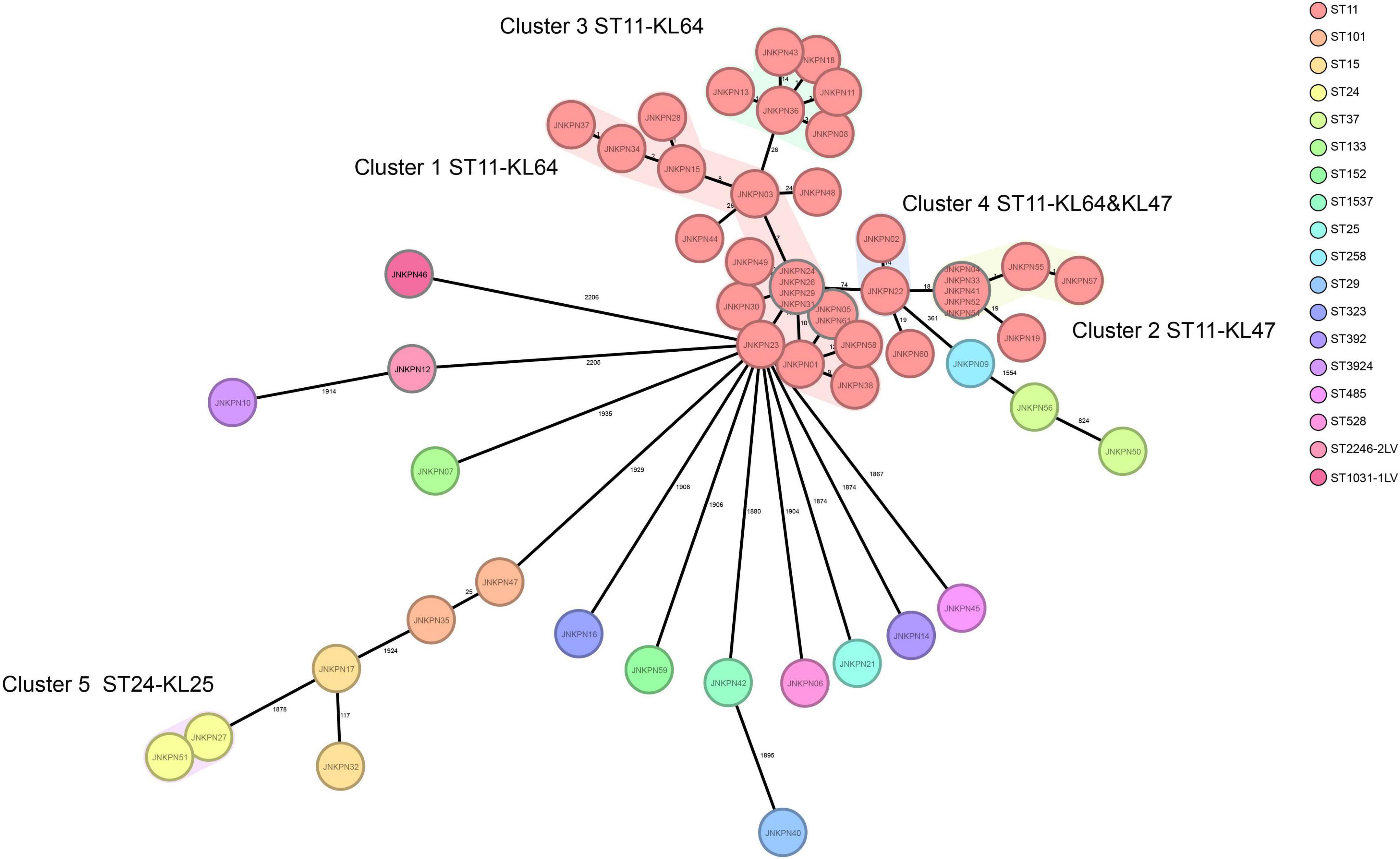
Figure 3. Minimum spanning tree showing core genome multilocus sequence typing (cgMLST) of the 57 carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates, showing five cluster types numbered consecutively. Each circle represents an allelic profile. Colors of circles indicate the different sequence types (STs). Cluster types consist of closely related genotypes (≤15 allele differences). The numbers on the connecting lines illustrate the numbers of target genes with different alleles.
All the 57 strains harbored at least one fosfomycin-modifying enzymes, including fosA2, fosA3, foskp96, and fosAkp (Figure 2). fosAkp was the most prevalent chromosomal-encoded enzyme, detected in 52 isolates, followed by foskp96, in five isolates. fosAkp and fosA3 were detected in 23 isolates. foskp96 was detected among 5 isolates, though only one isolate showed fosfomycin-resistant phenotype in vitro.
Among the isolates, 3 isolates had Ser148Asn and Ser209Thr substitutions in MurA, and two isolates had MurA deletion. One variation, Val434Ile, in uhpT was detected in two isolates. Three substitutions in glpT, Ile260Val, Val337Ile, and Ile429Val, were detected in one isolate.
As illustrated in Figure 1 and Supplementary Table 1, 87.7% of the isolates produced carbapenemase in accordance with the results of CIM test. blaKPC–2 was the main type of carbapenemase (n = 33). Five strains coharboring blaKPC–2 and blaNDM–1 were detected. Meanwhile, blaNDM–1 (n = 5), blaNDM–5 (n = 3), blaNDM–3 (n = 1), blaIMP–4 (n = 2), and blaOXA–232 (n = 1) also contributed to the carbapenem resistance. Twenty transconjugants harboring carbapenemase genes were acquired. One transconjugant harbored blaIMP–4, ten transconjugants harbored blaKPC–2, and nine transconjugants harbored blaNDM. ESBL resistance genes, such as blaTEM, blaCTX–M, blaSHV, blaCMY, and blaSHV, were also detected, with blaTEM, blaCTX–M, and blaSHV being the most prevalent. The strains coharboring blaTEM, blaCTX–M, and blaSHV made 52.6% of all the CRKP strains (Figure 4 and Supplementary Table 1).
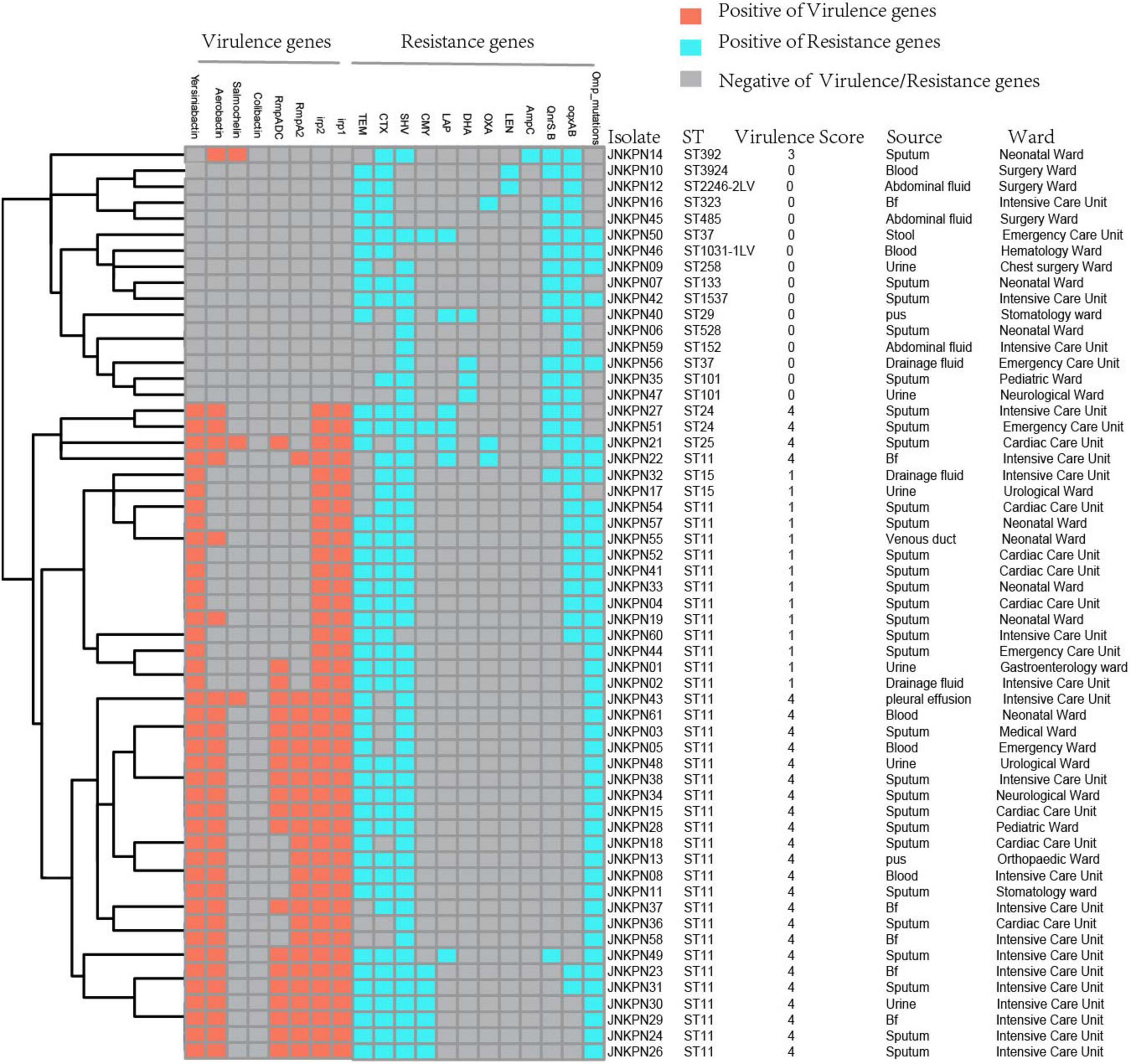
Figure 4. Detection of resistance genes and virulence genes in carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates. The sequence type, virulence score, source, and ward of each isolate were marked on the right of the squares. Bf, bronchoalveolar lavage fluids.
Plasmid-encoded fluoroquinolone resistance genes (QnrS and QnrB) were detected in 18 isolates, while chromosomal oqxA and oqxB were detected in 33 isolates (Figure 4 and Supplementary Table 1). A high prevalence of specific porin defects was detected, and only 14 isolates were without any mutation in ompK5 or ompK36. It was observed that ompK35 truncations and ompK36GD mutations coexisted in 59.6% of the strains (Figure 4 and Supplementary Table 2).
According to the Katholt criterion, 27 isolates were assigned a virulence score of 4, which were closely related to ST11 (n = 24) (Figure 4). Analysis of virulence genes showed that 41 isolates possessed yersiniabactin genes located on ICEKp3 (n = 38), ICEKp1 (n = 1), ICEKp5 (n = 1), and ICEKp12 (n = 1), with ICEKp3 being the most prevalent carrier. Of 25 ST11-KL64 isolates, 22 (88%) carried rmpA2 gene, but only three were positive for the string test, suggesting that rmpA2 was inactive in most isolates. Among 10 ST11-KL47 isolates, only one was positive for rmpA2 gene (Supplementary Table 2).
Four types of genetic environments existed in the 52 fosAkp-positive CRKP isolates. The upstream genes of fosAkp among the strains are identical, but those downstream are variable. The intergenic regions between fosAkp and the downstream MocR gene could be DNA helicase-related genes, a Type I restriction–modification system, or a hypothetical Protein. As shown in Supplementary Figure 1, the genetic environment of foskp96 was similar to that of fosAkp, with a backbone of YrkL-LysR-FosA-MocR-YjiS. The genetic environment of fosA3 was consistent, where fosA3 is flanked by IS26 at both ends, in a transposon-like structure.
The plasmids harboring fosA3 from strains JNKPN52, JNKPN54, JNKPN55, and JNKPN57 were successfully transferred into E. coli J53AziR by conjugation. All the four transconjugants were resistant to CRO, FEP, CAZ, ATM, TZP, ETP, IMP, AMK, GEN, and FOS but were susceptible to SXT, CIP, LVX, TGC, and POL (Supplementary Table 3). JNKPN52 and JNKPN54 were isolated from 2-month-old pediatric patients after cardiac surgery enrolled in the cardiac care unit in December 2019. JNKPN55 and JNKPN57 were isolated from premature babies enrolled in the neonatal intensive care unit in December 2019. A nosocomial outbreak caused by a clone of ST-KL47 KPC-KP strains was considered according to the high similarities of PFGE patterns of group G3 (> 85%) (Figure 1) and the high level of correlation within cgMLST Cluster 2 (up to 1 allele difference) (Figure 3).
The complete sequence of plasmid pJNKPN52_KPC_fosA from clinical strain JNKPN52 was determined to better characterize the self-transmissible plasmid coharboring fosA3 and blaKPC–2. pJNKPN52_KPC_fosA is a 127,668-base pair (bp) multireplicon plasmid that belongs to the IncR:IncFII-type and shares a similar structure with pHN7A8/pKPC-LK30 hybrid plasmid pCT-KPC (Figure 5).
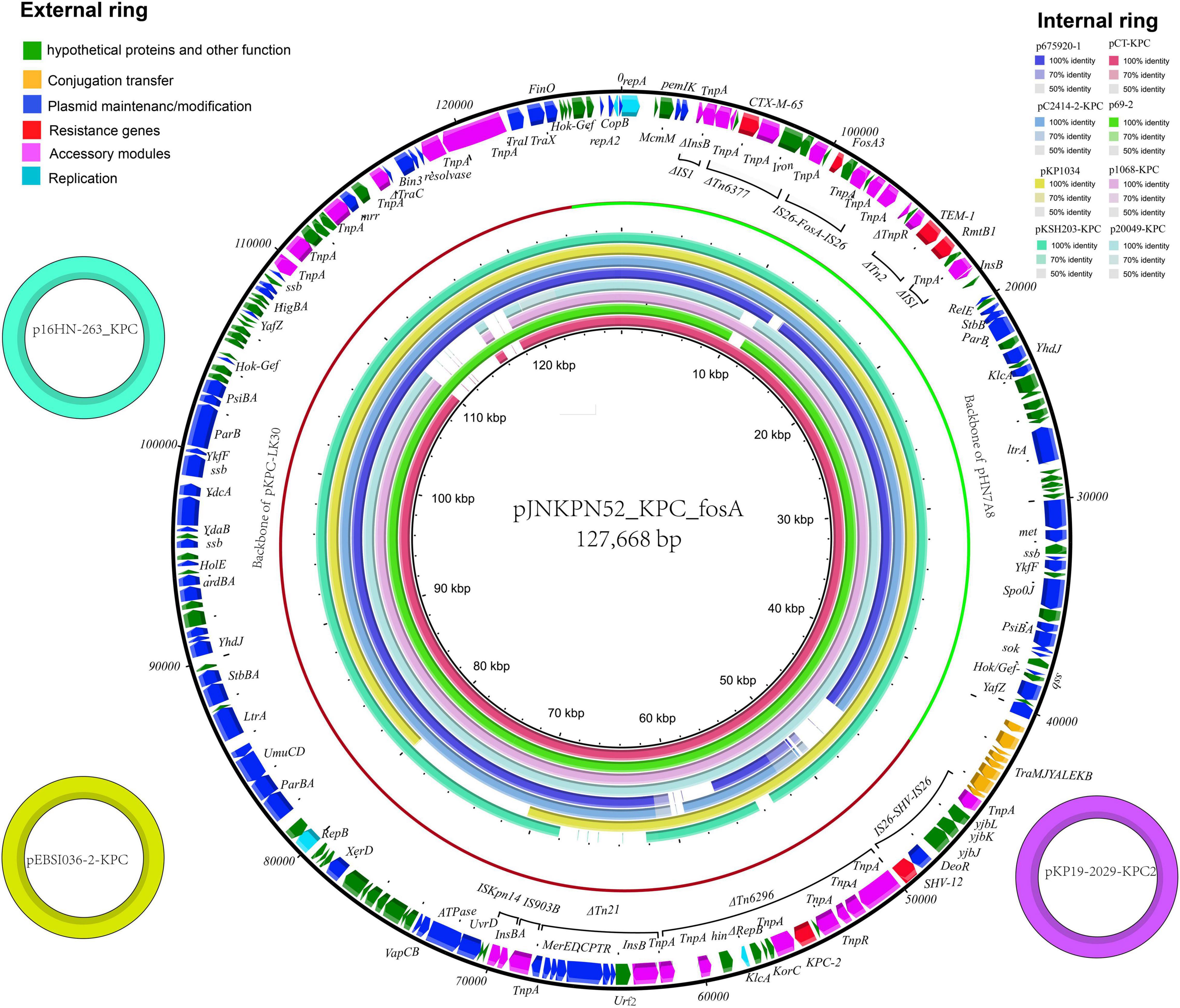
Figure 5. The external ring is the main structural feature of plasmid pJNKPN52_KPC_fosA (GenBank accession no. MZ709016). The three small external rings display plasmids coharboring blaKPC–2 and fosA with identity > 99%. The internal eight rings showed the comparative analysis of blaKPC-harboring plasmids with pJNKPN52_KPC_fosA (constructed by BRIG).
pJNKPN52_KPC_fosA contains two major accessory resistance regions, including the blaKPC–2 region harboring blaKPC–2 and blaSHV–12, and the multidrug-resistant (MDR) region carrying rmtB (aminoglycoside resistance), fosA3, blaTEM–1, and blaCTX–M–65. The MDR region was generated from the insertion of ΔTn6377–blaCTX–M–65, IS26–fosA3–IS26 unit, ΔTn2-rmtB element within IS1. The blaKPC–2 region was organized in order of a truncated IS26–blaSHV–12–IS26 unit, ΔTn6296, ΔTn21 with insertion of IS5075, an IS903B remnant, and ISKpn14 (Figure 5).
According to sequence alignment by BRIG, pJNKPN52_KPC_fosA showed 99% nucleotide identity with the previously reported plasmids p16HN-263_KPC and pKP19-2029-KPC2 isolated from China and pEBSI036-2-KPC isolated from Egypt (Ahmed et al., 2021) (Figure 5).
The MDR regions of the eleven plasmids were similar, with pJNKPN52_KPC_fosA, p675920-1, p69-2, and p20049-KPC slightly differing from one another. To determine the detailed structural differences between these plasmids, additional linear comparative genomics analysis was performed by BLAST. Compared with pJNKPN52_KPC_fosA, p675920-1 lacked a ΔIS1294 region, possibly because of recombination of IS26–fosA3–IS26 region. In p20049-KPC, the deletion of IS26–fosA3–IS26 region and insertion of partial plasmid backbone genes were observed within the MDR region (Figure 6).
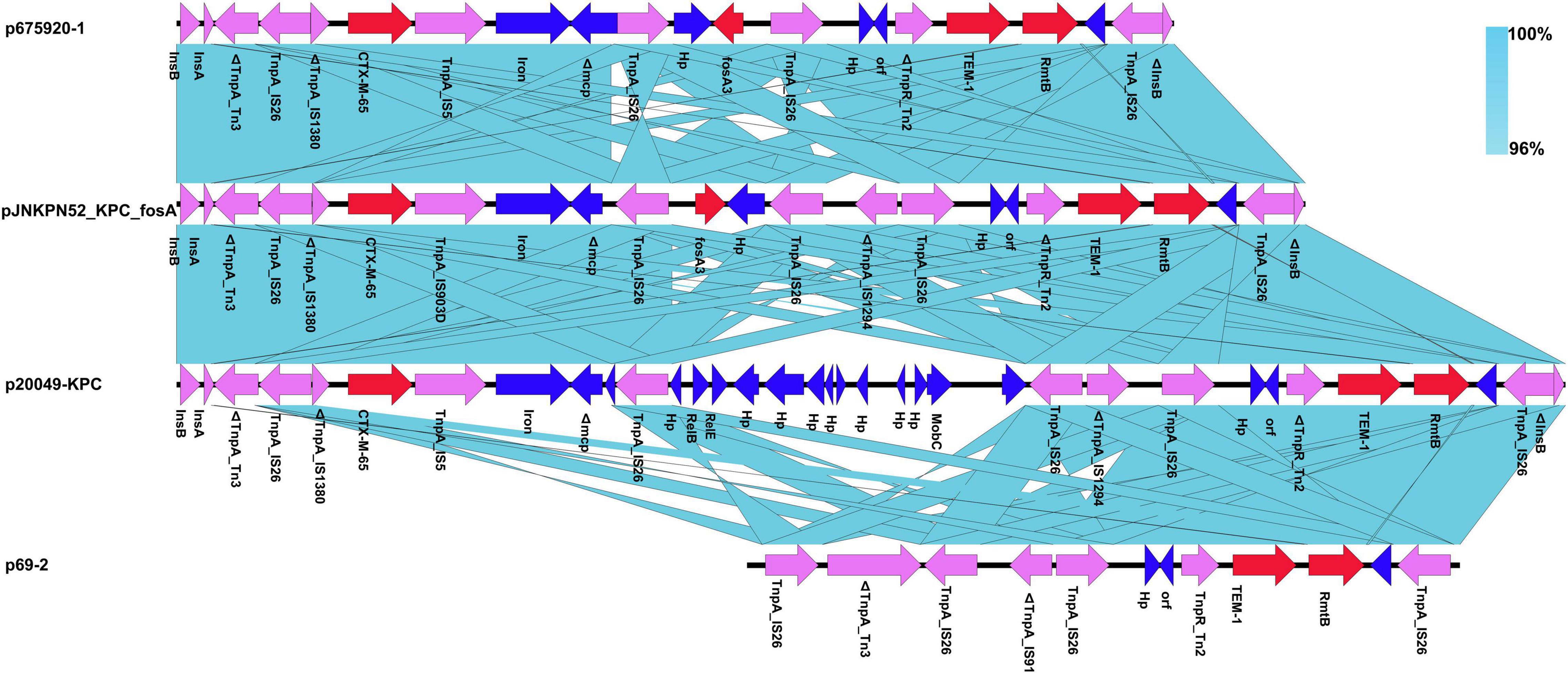
Figure 6. Comparison of genetic context of the multidrug-resistant (MDR) region with related regions.
The blaKPC–2 region of the pJNKPN52_KPC_fosA was similar to that of p20049-KPC and pKP1034, with the inversion of ΔTn6296. Compared with pJNKPN52_KPC_fosA, the deletion of the truncated IS26–blaSHV–12–IS26 unit was observed in p675920-1, probably due to IS26-mediated deletion (Figure 7).
The plasmids harboring blaNDM from strains JNKPN23, JNKPN24, JNKPN26, JNKPN29, JNKPN30, JNKPN31, JNKPN46, JNKPN47, and JNKPN51 were successfully transferred into E. coli J53AziR by conjugation. All the nine transconjugants were resistant to CRO, CAZ, TZP, ETP, and IMP, but were susceptible to FOS, CIP, and LVX (Supplementary Table 6). All the plasmids harboring blaNDM–1 were shown to belong to IncA/C2 type through plasmid typing (Supplementary Table 6).
The complete sequence of plasmid pJNKPN30_NDM from clinical strain JNKPN30 was determined to better characterize the self-transmissible IncA/C2 type plasmid harboring blaNDM–1. pJNKPN30_NDM contains two main accessory resistance regions, including the IS1380–blaCMY–6 region and the MDR region carrying Tn6196, class 1 integron structure bearing arr-3, dfrA1, AadA16, ErmE, and sul1, and partial of Tn125-bearing blaNDM–1 interrupted by the insertion of ΔISKpn14, followed by the 16S rRNA methylase rmtC gene (Figure 8).
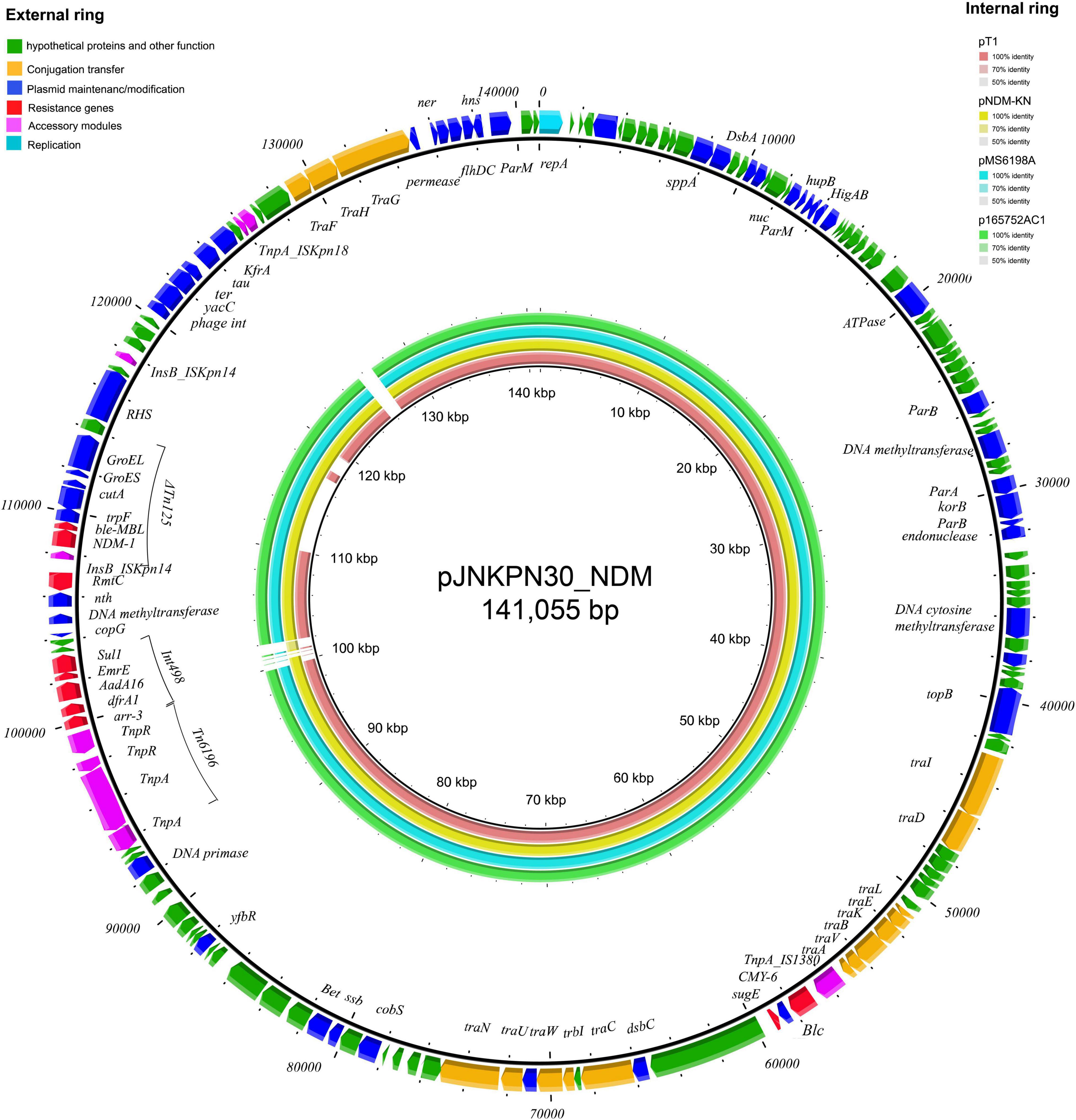
Figure 8. The external ring is the main structural feature of plasmid pJNKPN30_NDM (GenBank accession no. OL389795). The internal four rings showed the comparative analysis of blaNDM-harboring plasmids with pJNKPN30_NDM (constructed by BRIG).
According to sequence alignment by BRIG, pJNKPN30_NDM was with a backbone similar to that of other IncA/C2 plasmids (84–99% query coverage, > 99% nucleotide sequence identity). The main regions of discontinuity were within the Int498 region and the ISKpn18 region (Figure 8).
Raw reads of all 57 isolates have been deposited in GenBank (BioProject PRJNA769451). The complete sequence of pJNKPN52_KPC_fosA and pJNKPN30_NDM has been deposited in GenBank under accession numbers MZ709016 and OL389795, respectively.
The emergence of CRKP has become a crucial public health problem, as it limits treatment options and requires novel active agents or combination therapies (Ekwanzala et al., 2019). K. pneumoniae isolates have shown susceptibility to fosfomycin; hence, the “old” antibiotic agent is being re-considered as a possible auxiliary drug (Vardakas et al., 2016). Pontikis et al. (2014) reported that intravenous fosfomycin for nosocomial CRKP infections has a good clinical outcome. Intravenous fosfomycin is used in many countries and has completed phase 3 clinical trials for the treatment of urinary tract infection and acute pyelonephritis (Ito et al., 2017). However, information on resistance to fosfomycin among CRKP in China is inadequate. The occurrence of fosfomycin-resistant CRKP in China ranges from 18.7 to 80% (Tseng et al., 2017; Huang et al., 2021). These inconsistent data indicate that resistance of K. pneumoniae isolates to fosfomycin, especially to CRKP, requires further evaluation. Our results indicated that 70.2% of CRKP strains were resistant to fosfomycin in Shandong, which is much higher than that in most regions in China.
Fosfomycin resistance may be due to chromosome-encoded murA, glpT, uhpT, uhpA, ptsI, and cyaA mutations, or plasmid-encoded or chromosomal inactivation by fosfomycin-modifying enzymes (Liu et al., 2020). We found that fosA homologs were widely distributed among the CRKP strains, and all the strains harbored fosAkp or foskp96. Moreover, 40.3% of the strains had both fosAkp and fosA3. It was difficult to discriminate fosA variants located on plasmids from those on chromosomes because the analysis was based on draft sequences of genomes. fosAkp or foskp96 is intrinsically distributed on K. pneumoniae chromosomes (Ito et al., 2017). According to the previous report, MIC50/90 values of fosfomycin for K. pneumoniae clinical strains producing KPC-type carbapenemase were 16/64 μg/ml (Ito et al., 2017). In this study, 34 CRKP isolates without fosA3 had a MIC range of ≤ 32 to ≥ 1,024 μg/ml and MIC50/90 values of 256/1,024 μg/ml. Not all the isolates showed fosfomycin resistance. We speculated that the dissimilarity between fosfomycin-resistant genotype and phenotype was caused by the expression level of fosA gene.
fosA3 is the most common acquired fosA, encoded by plasmids (Ito et al., 2017). Apart from fosA production, fosfomycin resistance is also related to MurA, glpT, and uhpT mutations, which were rare in our isolates, and only 2, 3, and 1 isolates, respectively, were detected. Therefore, our results strongly suggest that fosA3 and fosA contribute mainly to fosfomycin resistance.
Further, pLVPK-like-positive ST11-KL64 isolates show better survival in the environment (Zhou et al., 2020). Fourteen fosA3-positive strains were screened from the ST11-KL64-wzi64-O2 subgroup, and eight fosA3-positive strains were screened from the ST11-KL47-wzi209-OL101 subgroup. Twenty-two out of 25 ST11-KL64 strains and only one ST11-KL47 strain contained rmpA2. All the seven isolates belonging to cgMLST Cluster 2 and Cluster 6 isolates belonging to cgMLST Cluster 3 coharbored blaKPC–2 and fosA3. The isolates belonging to cgMLST Cluster 1 and Cluster 3 except JNKPN01 and JNKPN30 were positive for blaKPC–2 and rmpA2. PFGE profiles and cgMLST confirmed that the clonal relation may predominantly be due to clonal dissemination.
It was noteworthy that blaNDM-producing CRKP isolates were increasingly reported (Qamar et al., 2021). In this study, 24.6% (14/57) of CRKP isolates were positive for blaNDM (including the isolates co-producing blaKPC–2 and blaNDM), which were higher than that in the previous research in China (11.5%) (Wang et al., 2018). Plasmids harboring blaNDM from 64.3% (9/14) of blaNDM-carrying isolates could be transferred to the recipients. The conjugative IncA/C2 type plasmids played an important role in the rapid and efficient dissemination of the blaNDM–1 gene among CRKP isolates in this study. According to the previous report, IncA/C type plasmids were known to be of broad host range and had been detected in numerous MDR Gram-negative species (Carattoli et al., 2012). Moreover, an outbreak caused by a clone of Citrobacter freundii strains bearing blaNDM–1 located on IncA/C plasmids and secondary in vivo spread of an IncA/C2 plasmid with blaNDM–1 to E. coli, K. pneumoniae, and Klebsiella oxytoca from the individual patients was reported in Denmark (Hammerum et al., 2016). Thus, more attention should be paid to monitoring and controlling the horizontal transmission of blaNDM mediated by IncA/C type plasmids among K. pneumoniae isolates.
Interestingly, fosA3 was not detected in any CRKP strains due to class B carbapenemase, including blaIMP or blaNDM. According to the MLST results, KPC-2-producing ST11 was the only clone in our study closely related to fosA3. Analysis of the genetic environment confirmed that the mobile element IS26–fosA3–IS26 played an important role in the dissemination of fosfomycin resistance. Moreover, most of the fosA3-positive KPC-2 producing strains (22/23) carried at least two kinds of ESBLs, indicating that ST11 type K. pneumoniae might be a good reservoir of resistance genes.
According to previous research, the high prevalence of fosfomycin resistance in KPC-producing isolates from China is associated with plasmids coharboring fosA3 and blaKPC (Singkham-In et al., 2020). Recently, fosA3 and blaKPC–2 genes located on non-conjugative pCT-KPC-like plasmids have been sporadically reported in China (Xiang et al., 2015; Liu et al., 2018; Shi et al., 2018; Zhai et al., 2021). Furthermore, Shi et al. (2018) proved that blaKPC–2- and rmtB-carrying pCT-KPC-like plasmids were prevalent among clonal K. pneumoniae CG258 strains collected from five different hospitals and were associated with the dissemination of blaKPC–2 and rmtB. It seemed that the spread of the pCT-KPC-like plasmids was mainly due to the clonal dissemination of ST11 KPC-producing K. pneumoniae, as the conjugation tests failed to recover transconjugants in all reports (Zhai et al., 2021).
Recently, a plasmid pEBSI036-2-KPC from a high-risk clone ST11 KL47 serotype of a CR-HvKP strain isolated from an Egyptian hospital was reported. pJNKPN52_KPC_fosA showed 99% identity with pEBSI036-2-KPC. But the transferability of plasmid pEBSI036-2-KPC was not determined (Ahmed et al., 2021). In our study, four ST11-KL47 type K. pneumoniae strains, JNKPN52, JNKPN54, JNKPN55, and JNKPN57, could co-transfer fosA3 and blaKPC–2 genes into recipient E. coli J53AziR by conjugation tests, indicating that the fosA3 gene could be co-disseminated with blaKPC–2. The four strains isolated from different patients were identified from the same clone according to the PFGE profiles and cgMLST cluster. Notably, blaKPC–2 and fosA3 genes were confirmed to be located on a plasmid pJNKPN52_KPC_fosA, which shared similar backbones with the previously reported pCT-KPC-like plasmids, including pKP1034, pCT-KPC, pKPC-LK30, p69-2, and p675920-1. For the first time, we confirmed the pCT-KPC-like plasmid-mediated horizontal transmission of blaKPC–2 and fosA3 resistance. We noticed that ten copies of IS26 were detected in plasmid pJNKPN52_KPC_fosA and would mediate homologous recombination and mobilization of accessory resistance regions within and among different plasmids (Zhai et al., 2021). Therefore, IS26 may have played a vital role in the generation process of pJNKPN52_KPC_fosA.
Our findings indicate that fosA is intrinsically distributed in the genome of clinically isolated K. pneumoniae and might contribute to fosfomycin resistance. The coexistence of plasmid-mediated fosA3 and chromosomal-encoded fosAkp was observed commonly among ST11 CRKP strains. The emerging conjugative pCT-KPC-like plasmids coharboring blaKPC–2 and fosA3 would exacerbate the fosfomycin resistance among CRKP strains. ST11-KL64 and ST11-KL47 K. pneumoniae, the so-called “super-bug,” with higher resistance and virulence, should be monitored by more effective strategies to prevent the future dissemination of resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YW contributed to experiment conception and design. YH, YZ, and RC conducted bioinformatics analysis and wrote the manuscript. XZ and YB performed the data analysis. ZS carried out the bacteria identification. XL and CZ prepared the tables and figures. YW is responsible for submitting a competing interest statement on behalf of all authors of the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the National Natural Science Foundation of China (81902119) and the Shandong Province Natural Science Foundation (ZR2020MH306).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Mingju Hao of The First Affiliated Hospital of Shandong, First Medical University, and Shandong Provincial Qianfoshan Hospital, for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.771170/full#supplementary-material
Supplementary Figure 1 | Comparison of genetic environments of the fosA. (A) The genetic environment of fosAkp and foskp96; (B) The genetic environment of fosA2; (C) The genetic environment of fosA3.
Ahmed, M., Yang, Y., Yang, Y., Yan, B., Chen, G., Hassan, R. M., et al. (2021). Emergence of hypervirulent carbapenem-resistant Klebsiella pneumoniae coharboring a bla NDM-1-carrying virulent plasmid and a bla KPC-2-carrying plasmid in an egyptian hospital. mSphere 6:e88-21. doi: 10.1128/mSphere.00088-21
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Villa, L., Poirel, L., Bonnin, R. A., and Nordmann, P. (2012). Evolution of IncA/C blaCMY-(2)-carrying plasmids by acquisition of the blaNDM-(1) carbapenemase gene. Antimicrob. Agents Chemother. 56, 783–786. doi: 10.1128/AAC.05116-11
Chen, J., Wang, D., Ding, Y., Zhang, L., and Li, X. (2019). Molecular epidemiology of plasmid-mediated fosfomycin resistance gene determinants in Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae isolates in China. Microb. Drug Resist. 25, 251–257. doi: 10.1089/mdr.2018.0137
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing document M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Ekwanzala, M. D., Dewar, J. B., Kamika, I., and Momba, M. N. B. (2019). Tracking the environmental dissemination of carbapenem-resistant Klebsiella pneumoniae using whole genome sequencing. Sci. Total Environ. 691, 80–92. doi: 10.1016/j.scitotenv.2019.06.533
Elliott, Z. S., Barry, K. E., Cox, H. L., Stoesser, N., Carroll, J., Vegesana, K., et al. (2019). The role of fosA in challenges with fosfomycin susceptibility testing of multispecies Klebsiella pneumoniae carbapenemase-producing clinical isolates. J. Clin. Microbiol. 57:e634-19. doi: 10.1128/JCM.00634-19
EUCAST (2020). Breakpoints Tables for Interpretation of MICs and Zone Diameters. in Version 10.0. Växjö: EUCAST.
Hammerum, A. M., Hansen, F., Nielsen, H. L., Jakobsen, L., Stegger, M., Andersen, P. S., et al. (2016). Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 71, 3117–3124. doi: 10.1093/jac/dkw289
Heras, J., Dominguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16:270. doi: 10.1186/s12859-015-0703-0
Huang, L., Cao, M., Hu, Y., Zhang, R., Xiao, Y., and Chen, G. (2021). Prevalence and mechanisms of fosfomycin resistance among KPC-producing Klebsiella pneumoniae clinical isolates in China. Int. J. Antimicrob. Agents 57:106226. doi: 10.1016/j.ijantimicag.2020.106226
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., McElheny, C. L., Mettus, R. T., et al. (2017). Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e749-17. doi: 10.1128/mBio.00749-17
Jiang, Y., Shen, P., Wei, Z., Liu, L., He, F., Shi, K., et al. (2015). Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int. J. Antimicrob. Agents 45, 66–70. doi: 10.1016/j.ijantimicag.2014.08.010
Liu, J., Xie, J., Yang, L., Chen, D., Peters, B. M., Xu, Z., et al. (2018). Identification of the KPC plasmid pCT-KPC334: new insights on the evolutionary pathway of epidemic plasmids harboring fosA3-blaKPC-2 genes. Int. J. Antimicrob. Agents 52, 510–511. doi: 10.1016/j.ijantimicag.2018.04.013
Liu, P., Chen, S., Wu, Z. Y., Qi, M., Li, X. Y., and Liu, C. X. (2020). Mechanisms of fosfomycin resistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 22, 238–243. doi: 10.1016/j.jgar.2019.12.019
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006–2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Mączyńska, B., Paleczny, J., Oleksy-Wawrzyniak, M., Choroszy-Król, I., and Bartoszewicz, M. (2021). In vitro susceptibility of multi-drug resistant klebsiellapneumoniae strains causing nosocomial infections to fosfomycin. A comparison of determination methods. Pathogens 10:512. doi: 10.3390/pathogens10050512
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Pontikis, K., Karaiskos, I., Bastani, S., Dimopoulos, G., Kalogirou, M., Katsiari, M., et al. (2014). Outcomes of critically ill intensive care unit patients treated with fosfomycin for infections due to pandrug-resistant and extensively drug-resistant carbapenemase-producing Gram-negative bacteria. Int. J. Antimicrob. Agents 43, 52–59. doi: 10.1016/j.ijantimicag.2013.09.010
Qamar, M. U., Ejaz, H., Walsh, T. R., Shah, A. A., Al, F. D., Alkufeidy, R. M., et al. (2021). Clonal relatedness and plasmid profiling of extensively drug-resistant New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae clinical isolates. Future Microbiol. 16, 229–239. doi: 10.2217/fmb-2020-0315
Sherry, N. L., Lane, C. R., Kwong, J. C., Schultz, M., Sait, M., Stevens, K., et al. (2019). Genomics for Molecular Epidemiology and Detecting Transmission of Carbapenemase-Producing Enterobacterales in Victoria, Australia, 2012 to 2016. J. Clin. Microbiol. 57:e573-19. doi: 10.1128/JCM.00573-19
Shi, L., Feng, J., Zhan, Z., Zhao, Y., Zhou, H., Mao, H., et al. (2018). Comparative analysis of bla KPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple Chinese hospitals. Infect. Drug. Resist 11, 1783–1793. doi: 10.2147/IDR.S171953
Singkham-In, U., Muhummudaree, N., and Chatsuwan, T. (2020). fosA3 overexpression with transporter mutations mediates high-level of fosfomycin resistance and silence of fosA3 in fosfomycin-susceptible Klebsiella pneumoniae producing carbapenemase clinical isolates. PLoS One 15:e0237474. doi: 10.1371/journal.pone.0237474
Tseng, S., Wang, S., Ma, L., Wang, T., Yang, T., Siu, L. K., et al. (2017). The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. J. Microbiol. Immunol. Infect. 50, 653–661. doi: 10.1016/j.jmii.2017.03.003
Vardakas, K. Z., Legakis, N. J., Triarides, N., and Falagas, M. E. (2016). Susceptibility of contemporary isolates to fosfomycin: a systematic review of the literature. Int. J. Antimicrob. Agents 47, 269–285. doi: 10.1016/j.ijantimicag.2016.02.001
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE STUDY in China (2012-2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Weber, R. E., Pietsch, M., Fruhauf, A., Pfeifer, Y., Martin, M., Luft, D., et al. (2019). IS26-mediated transfer of bla NDM-1 as the main route of resistance transmission during a polyclonal, multispecies outbreak in a german hospital. Front. Microbiol. 10:2817. doi: 10.3389/fmicb.2019.02817
Wyres, K. L., Nguyen, T. N. T., Lam, M. M. C., Judd, L. M., van Vinh Chau, N., Dance, D. A. B., et al. (2020). Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 12:11. doi: 10.1186/s13073-019-0706-y
Xiang, D. R., Li, J. J., Sheng, Z. K., Yu, H. Y., Deng, M., Bi, S., et al. (2015). Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob. Agents Chemother. 60, 1343–1348. doi: 10.1128/AAC.01488-15
Xiao, T., Zhu, Y., Zhang, S., Wang, Y., Shen, P., Zhou, Y., et al. (2020). A retrospective analysis of risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae bacteremia in nontransplant patients. J. Infect. Dis. 221, S174–S183. doi: 10.1093/infdis/jiz559
Zhai, Y., Li, D., Du, P., Zhang, Z., He, Z., Guo, Y., et al. (2021). Complete sequences of two new KPC-harbouring plasmids in Klebsiella pneumoniae ST11 strains in China. J. Glob. Antimicrob. Resist. 24, 114–120. doi: 10.1016/j.jgar.2020.11.023
Zhang, W., Zhu, Y., Wang, C., Liu, W., Li, R., Chen, F., et al. (2019). Characterization of a multidrug-resistant porcine Klebsiella pneumoniae sequence type 11 strain coharboring bla KPC-2 and fosA3 on two novel hybrid plasmids. mSphere 4:e590-19. doi: 10.1128/mSphere.00590-19
Keywords: Klebsiella pneumoniae, fosfomycin, carbapenem resistant, fosA3, blaKPC–2
Citation: Hao Y, Zhao X, Zhang C, Bai Y, Song Z, Lu X, Chen R, Zhu Y and Wang Y (2021) Clonal Dissemination of Clinical Carbapenem-Resistant Klebsiella pneumoniae Isolates Carrying fosA3 and blaKPC–2 Coharboring Plasmids in Shandong, China. Front. Microbiol. 12:771170. doi: 10.3389/fmicb.2021.771170
Received: 06 September 2021; Accepted: 17 November 2021;
Published: 17 December 2021.
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Valentine Usongo, Health Canada, CanadaCopyright © 2021 Hao, Zhao, Zhang, Bai, Song, Lu, Chen, Zhu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueling Wang, d2FuZ3l1ZWxpbmc4OEAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.