
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 October 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.769012
This article is part of the Research Topic Assembly and Functions of Gut Microbiota in Aquatic Animals View all 28 articles
 Shijie Bai1
Shijie Bai1 Peijun Zhang2*
Peijun Zhang2* Changhao Zhang3
Changhao Zhang3 Jiang Du4
Jiang Du4 Xinyi Du3
Xinyi Du3 Chengwei Zhu1
Chengwei Zhu1 Jun Liu2,5
Jun Liu2,5 Peiyu Xie2,5
Peiyu Xie2,5 Songhai Li2*
Songhai Li2*Despite an increasing appreciation in the importance of host–microbe interactions in ecological and evolutionary processes, information on the gut microbial communities of some marine mammals is still lacking. Moreover, whether diet, environment, or host phylogeny has the greatest impact on microbial community structure is still unknown. To fill part of this knowledge gap, we exploited a natural experiment provided by an aquarium with belugas (Delphinapterus leucas) affiliated with family Monodontidae, Pacific white-sided dolphins (Lagenorhynchus obliquidens) and common bottlenose dolphin (Tursiops truncatus) affiliated with family Delphinidae, and Cape fur seals (Arctocephalus pusillus pusillus) affiliated with family Otariidae. Results show significant differences in microbial community composition of whales, dolphins, and fur seals and indicate that host phylogeny (family level) plays the most important role in shaping the microbial communities, rather than food and environment. In general, the gut microbial communities of dolphins had significantly lower diversity compared to that of whales and fur seals. Overall, the gut microbial communities were mainly composed of Firmicutes and Gammaproteobacteria, together with some from Bacteroidetes, Fusobacteria, and Epsilonbacteraeota. However, specific bacterial lineages were differentially distributed among the marine mammal groups. For instance, Lachnospiraceae, Ruminococcaceae, and Peptostreptococcaceae were the dominant bacterial lineages in the gut of belugas, while for Cape fur seals, Moraxellaceae and Bacteroidaceae were the main bacterial lineages. Moreover, gut microbial communities in both Pacific white-sided dolphins and common bottlenose dolphins were dominated by a number of pathogenic bacteria, including Clostridium perfringens, Vibrio fluvialis, and Morganella morganii, reflecting the poor health condition of these animals. Although there is a growing recognition of the role microorganisms play in the gut of marine mammals, current knowledge about these microbial communities is still severely lacking. Large-scale research studies should be undertaken to reveal the roles played by the gut microbiota of different marine mammal species.
Microorganisms, as a resident group that has colonized the mammalian body, especially the gut, outnumber mammalian cells by as many as ten to one, and therefore encode 100-fold more unique genes than the genome of their hosts (Ley et al., 2006). The gut microbiota and their hosts always evolve together, forming a symbiotic immune system, where the microbiota plays an essential role in the development stage of the immune system, functioning both locally and systemically (Dang and Marsland, 2019). For instance, host immune systems have complex mechanisms to exclude invading pathogens that interact with the hosts. Furthermore, many physiological processes, such as obesity, energy metabolism, blood pressure, glucose homeostasis, clotting risks, and different behaviors of hosts, are influenced by gut microbiota. The metabolites generated by gut microbes and host’s receptors work together through a crucial cross talk between the gut microbiota and different organs to response their different phenotypes (Schroeder and Bäckhed, 2016). The majority of microbes residing in the gut have a profound influence on host physiology and nutrition, both of which are crucial for host health (Bäckhed et al., 2005; Flint et al., 2012). Therefore, revealing the gut microbiota of mammals is vital to fully understand the physiological and health status of mammals themselves. To date, the majority of research has focused on the gut microbiota of humans with very limited information on the composition of gut microbiota from other mammals, especially marine mammals, often due to sampling constraints.
Belugas (Delphinapterus leucas), affiliated with family Monodontidae, are omnivorous toothed cetaceans, and the most abundant odontoceti in Arctic waters. The Arctic cod (Boreogadus saida) is the most important food source of some beluga populations, such as the Beaufort Sea beluga population (Loseto et al., 2009). However, redfish (Sebastes marinus), halibut (Reinhardtius hippoglossoides), shrimp (Pandalus borealis), saffron cod (Eleginus gracilis), rainbow smelt (Osmerus mordax) and Pacific salmon (Oncorhynchus spp.), are also considered as the potential food of belugas (Frost and Lowry, 1981; Heide-Jorgensen and Teilmann, 1994; Quakenbush et al., 2015). The Pacific white-sided dolphin (Lagenorhynchus obliquidens) and the common bottlenose dolphin (Tursiops truncatus) are delphinids. The Pacific white-sided dolphins are one of the most abundant, widely distributed small delphinids in cold temperate waters of the North Pacific Ocean, and their scope of activities includes the eastern North Pacific, ranging from the Gulf of California to the Gulf of Alaska, and the western North Pacific, including the East China Sea, Sea of Japan, and Sea of Okhotsk (Hayano et al., 2004). Pacific white-sided dolphins primarily consume high-energy fishes, including anchovy, sardine, herring, and salmon, and other prey such as squid (Rechsteiner et al., 2013). The common bottlenose dolphins are globally distributed in warm temperate waters as one of the top predators in the marine food chain (Wells and Scott, 1999). Their diet composition includes different fish, such as the herring and mackerel (Scomber scombrus), and other organisms like squid (Kastelein et al., 2002). The Cape fur seals (Arctocephalus pusillus pusillus), affiliated with familly Otariidae (Kirkman and Arnould, 2018), are the only pinniped endemic to the African continent and widely distribute in the range from the southeast coast of South Africa to southern Angola (Kirkman et al., 2013). Their food source includes teleost fish, cephalopods, elasmobranchs, and seabirds, specifically including hake (Merluccius spp.), sardine (Sardinops sagax), chokka squid (Loligo vulgaris reynaudii), horse mackerel (Trachurus trachurus capensis), anchovy (Engraulis encrasicolus), and West Coast rock lobster (Jasuslalandii) (David, 1987; Makhado et al., 2006; Mecenero et al., 2006). Phylogeny of these three marine mammal lineages is shown in Supplementary Figure S1 (modified from Yuan et al., 2021).
Food is not only essential for the survival and growth of mammals but also impacts the symbiotic microbial communities in the gut. Food can shape the gut microbiota, modulating microbial composition and function, impact host–microbe interactions, and lead to changes in health status (McKenzie et al., 2017). Environment is another important factor, which impacts gut microbiota diversity, as underlying environmental changes will either increase or decrease the diversity of gut microbiota (McKenzie et al., 2017; Makki et al., 2018). A network-based analysis of microbial communities from the fecal microbiota of humans and 59 other mammalian species indicated that both host diet and phylogeny impact microbial diversity and composition, the intensity of influence increasing from carnivory to omnivory to herbivory. However, no marine mammals were included in that study (Ley et al., 2008). A subsequent study found that the gut microbiomes of different mammalian lineages have diverged at equally rate over the past 75 million years, but the gut microbiomes of Cetartiodactyla (ruminants, whales, and hippopotami) have evolved much faster than that of Chiropterans, and the gut microbiota are more likely to be associated with a strict mammalian lineage than with a particular dietary category, resulting in a strong phylogenetic influence on gut microbiomes (Nishida and Ochman, 2018). So far, research focused on the gut microbiota of marine mammals has primarily been based on next generation sequencing technologies, rather than the culture-dependent techniques, the latter of which just can reveal only a small fraction of total microbial communities of target samples (Bai et al., 2021; Glaeser et al., 2021). To the best of our knowledge, there have been no studies regarding the gut microbiota of Belugas, Pacific white-sided dolphins, and Cape fur seals. However, few studies have explored the gut microbial communities of common bottlenose dolphins, including captive and wild individuals, based on culture-independent approaches, which have shown that the Proteobacteria (Gamma), Firmicutes, and Fusobacteria dominate (Suzuki et al., 2019; Robles-Malagamba et al., 2020).
The first step toward developing health indices for the rapid assessment of marine mammal health is to explore their gut microbial communities, especially for those housed in aquariums, which are sampled on a regular basis. Characterizing the microbial communities of different marine mammals in aquariums can provide insight into their health status. The aim of this study was to explore the gut microbiota of four different marine mammal species in a shared environment with the same food sources, including herring (Clupea harengus) and capelin (Mallotus villosus). Thirty-five fecal samples from two whales, four dolphins, and nine fur seals were used in this study to address two questions: (i) what are the gut microbial compositions and diversity of belugas, Pacific white-sided dolphins, and Cape fur seals; moreover, what are the similarities and differences of gut microbial community structure and composition of common bottlenose dolphins collected from the aquarium in Sanya, China, with previous studies? and (ii) with the same living environment and food sources, could we find the similar gut microbial communities between different marine mammal species? Of food sources, environment, and phylogeny, which is the most influential factor that shapes the gut microbiota?
Two belugas, three Pacific white-sided dolphins, one common bottlenose dolphin, and nine Cape fur seals kept in the Marine and Waterpark in Atlantis hotel, Sanya, China, were surveyed in this study (Supplementary Table S1). All animals were maintained for public display in indoor pools with natural seawater filtered by the same filtration system based on mechanical filtration with percolator filters, pressurized sand filter, and protein skimmers. All seawater was treated with chlorination and ozone sterilization. The diet of all surveyed animals consisted of whole frozen fish including herring (Clupea harengus) and capelin (Mallotus villosus).
For each individual animal, at least one fecal sample was collected during June to July, 2020. All 35 fecal samples were collected from animals under operant conditioning (behavioral training) during routine medical examinations. Fecal samples were harvested by veterinarians between 8:00 and 9:00 in the morning. Whales and dolphins were asked for a “belly-up” position and feces were collected directly from the rectum with a Levin’s tube (Vygon, France), with a diameter of 4mm, inserted 10–20cm into the rectum. Fecal samples of fur seals were collected using anal swabs, with a diameter of 12mm, and inserted 10–15cm into the rectum. All fecal samples were frozen at −20°C until analysis.
DNA was extracted using MoBio PowerSoil extraction kits (Mo Bio Laboratories, Carlsbad, CA, United States), according to the manufacturer’s instructions. Three extraction blank control samples were also included. The extracted DNA was quantified with a Qubit fluorometer (Invitrogen Inc. Manufacturer: Life Technologies Holdings Pte Ltd., Singapore) and used for amplification of the V4 region of the 16S rRNA gene with the primer pair 515f Modified and 806r Modified (Walters et al., 2015). The PCR cycling conditions were as follows: denaturation at 95°C for 3min, followed by 27cycles at 95°C for 30s, 55°C for 30s, and 72°C for 45s, and a final extension at 72°C for 10min. Triplicate PCR amplifications were combined after purification with a TaKaRa purification kit (TaKaRa, Japan). The PCR products were prepared for library construction with the TruSeq DNA sample preparation kit (Illumina, San Diego, CA, United States), according to the manufacturer’s instructions. The libraries were sequenced at MajorBio Co. Ltd. (Shanghai, China) using the HiSeq platform (Illumina) with a paired-end 250bp sequence read run.
After sequencing, the raw reads were assigned to their respective samples according to their barcodes and forward and reverse primers (one mismatch of each was allowed). Paired-end reads of sufficient length with at least a 30-bp overlap were combined into full-length sequences by using FLASH program version 1.2.8 (Magoč and Salzberg, 2011). The average fragment length was 253bp. Btrim program (version 0.2.0) was used to filter out low-quality sequences. The quality score was set to >20 with a 5-base window size as the standard and any sequences containing Ns or<200bp were discarded. Sequences with lengths of 245–260bp were retained (Kong, 2011). We used UNOISE3 to correct sequencing errors to determine the real biological sequences at single-nucleotide resolution by generating amplicon sequence variants (ASVs) with default settings (Edgar, 2016). A representative sequence from each ASV was selected for taxonomic annotation by comparison with the SILVA 132 database (Quast et al., 2013), which includes bacterial, archaeal, and eukaryotic sequences. To account for varying sequencing depths, the ASVs were randomly resampled to normalize the reads of each sample. The raw sequencing reads of all samples were deposited to the NCBI database1 under BioProject accession number: PRJNA743584.
The diversity of the microbial communities from the fecal samples of different marine mamamals were determined by statistical analysis of the α-diversity indices. The Shannon and Inverse Simpson indices were calculated using the vegan package in R language version 3.4.3 (R Core Team, 2018). The Chao1 values (Chao, 1984) were generated using the Mothur program (Schloss et al., 2009). The indicator ASVs for microbial communities were classified using IndVal.g analysis with the R package labdsv tool (Roberts, 2007). Only ASVs with highly significant indicator values (IndVal.g index >0.95, p<0.001) were considered as strict habitat specialists (Li et al., 2021). To investigate differences in microbial composition among fecal samples, ß-diversity-based statistical tools and non-metric multidimensional scaling (NMDS) were used for calculating Bray-Curtis and Jaccard distance matrices. We also tested whether there were any dissimilarities among defined groupings, including belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals, by performing permutational multivariate analysis of variance (PERMANOVA), multi response permutation procedure (MRPP), and a one-way ordered analysis of similarity (ANOSIM). Data analyses followed the method previously described in Bai and Hou (Bai and Hou, 2020). Data comparison between different groups was performed by the Wilcoxon rank-sum test using IBM SPSS Statistics 19.
A total of 1,927,942 sequences were obtained from 35 fecal samples of different marine mammals after quality assessment, the marine mammals included two belugas, three Pacific white-sided dolphins, one common bottlenose dolphin, and nine Cape fur seals. An average of 55,084±10,935 sequences was obtained from each sample. In order to obtain a more accurate result of α-diversity, we rarefied to 33,334 sequences per sample, and then used this set for analysis of microbial diversity, composition, and structure. The α-diversities of microbial communities from the gut of different marine mammals were calculated. The Shannon, Inverse Simpson, and Chao1 indices and observed richness indicated that the α-diversity of the gut microbiomes from belugas and Cape fur seals was higher than those from Pacific white-sided dolphins and common bottlenose dolphin (Figure 1).
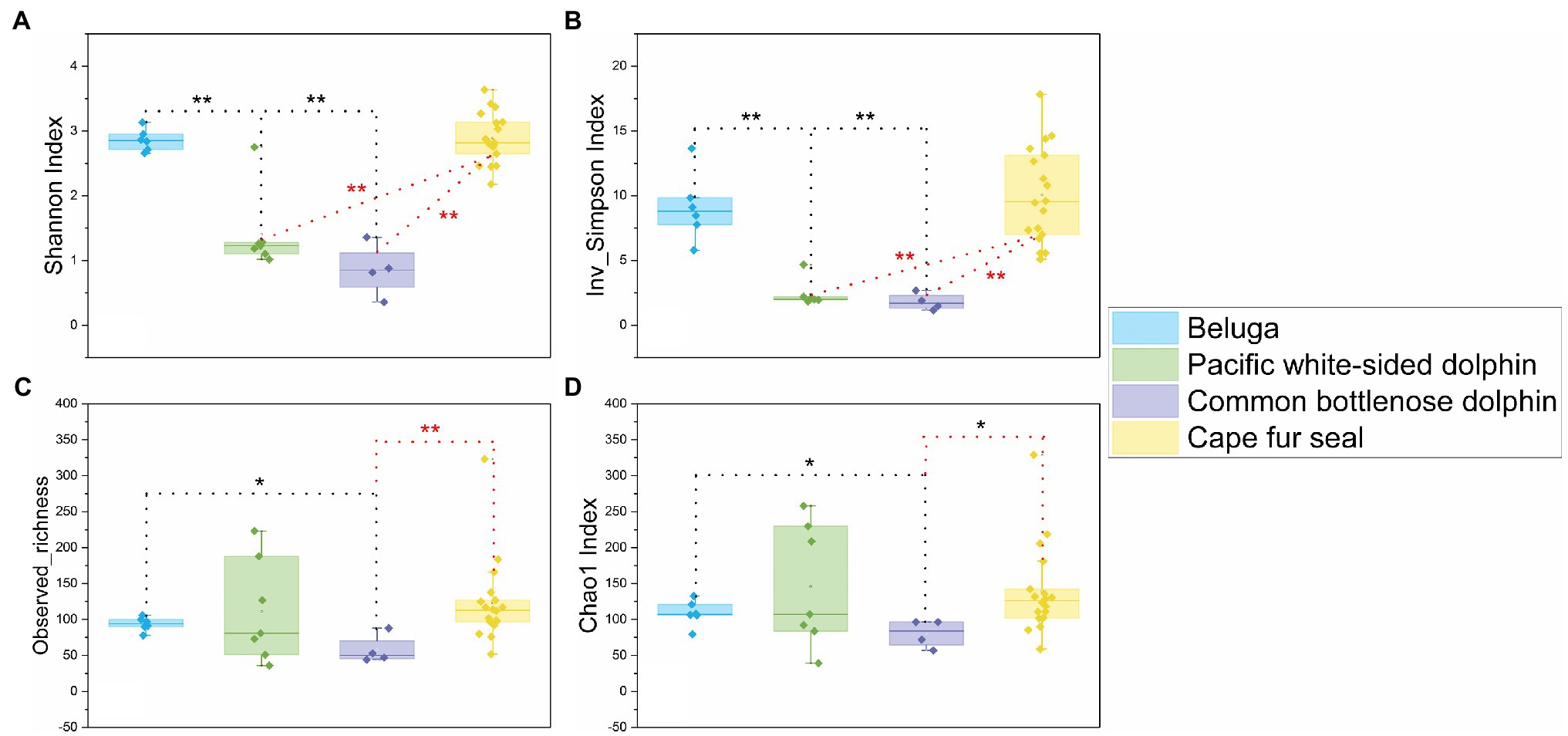
Figure 1. Comparisons of four α-diversity indices, Shannon index (A), Inverse Simpson index (B), observed richness (C), and Chao1 index (D), of the 35 fecal specimens from belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals. Based on Wilcoxon rank-sum test, *difference is significant at 0.05 level, **difference is significant at 0.01 level. The results are based on the ASVs datasets.
NMDS analysis of microbial communities clearly showed three principal groups: belugas, two dolphin species, and Cape fur seals (Figure 2). These results suggest that whales, dolphins, and fur seals possess different gut microbial communities, even in the context of similar food and environment. Furthermore, the MRPP, ANOSIM, and PERMANOVA showed significant differences in the gut microbial communities of whales, dolphins, and fur seals (p<0.01). By contrast, no significant difference was detected between the Pacific white-sided dolphin and the common bottlenose dolphin (p>0.05; Table 1).
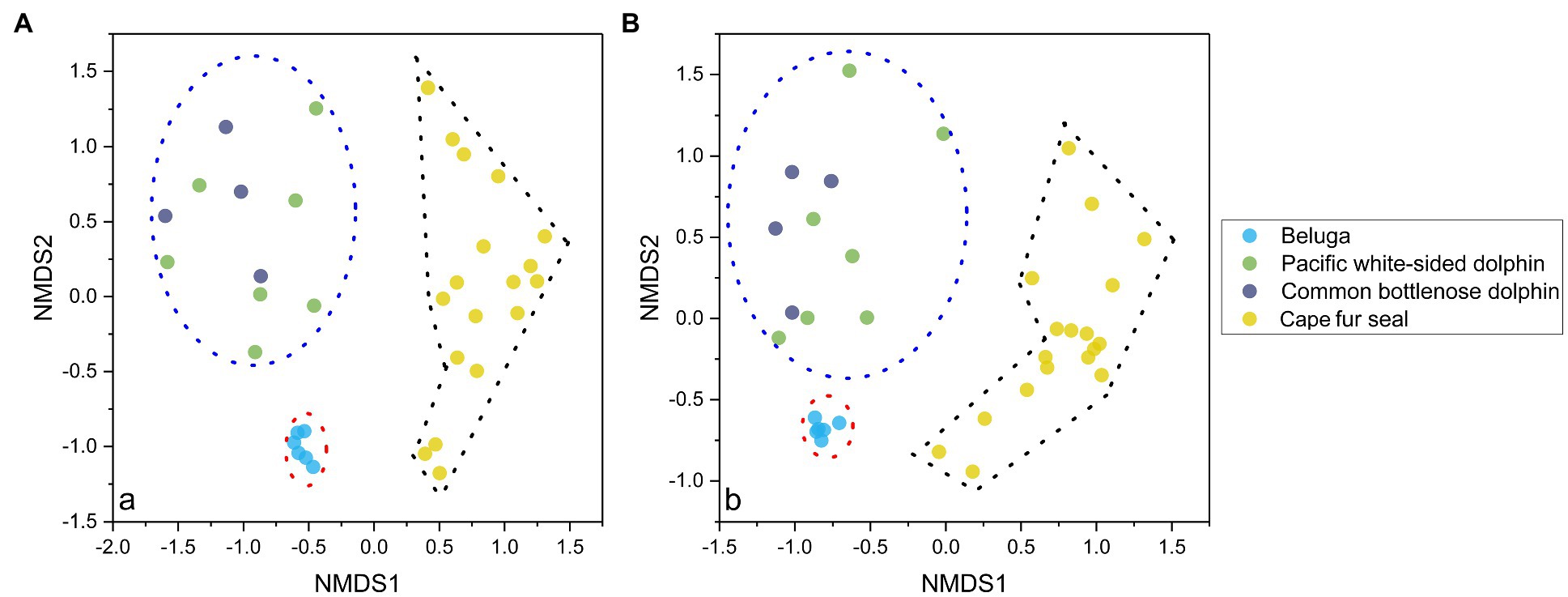
Figure 2. NMDS analysis of the gut microbial communities separated the samples into three principal groups, the first group composed of the fecal samples of belugas, the second group composed of fecal samples from the Pacific white-sided dolphins and common bottlenose dolphin, and the third group composed of fecal samples from Cape fur seals. The results are based on the ASVs datasets, and the left (A) and right (B) plots were calculated based on Bray-Curtis distance and Jaccard distance, respectively.
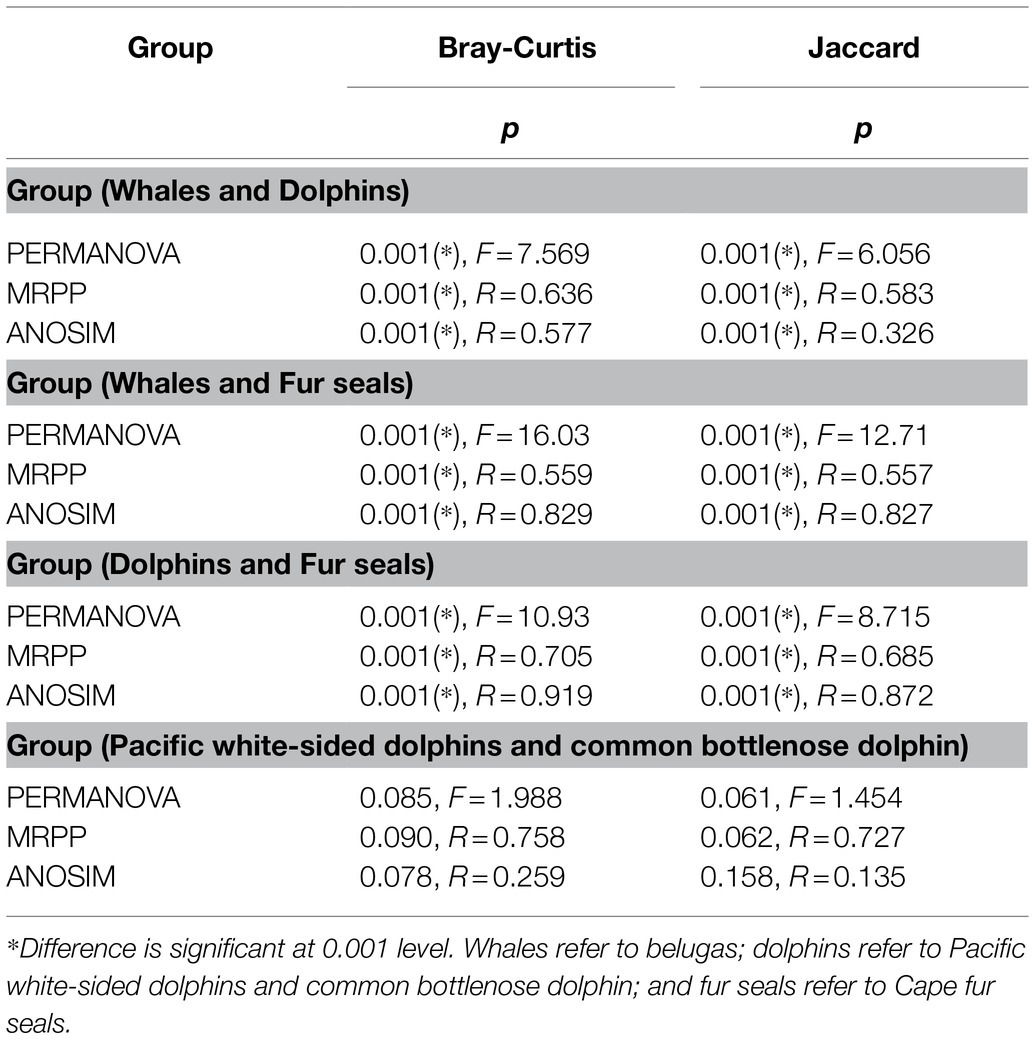
Table 1. Dissimilarity tests of gut microbial communities of whales, dolphins, and fur seals based on Bray-Curtis and Jaccard distance.
The relative abundance of microorganisms was evident at the phylum, family, and genus levels with a similarity of 97% for ASV classification and provided detailed information on the composition of the microbial communities (Figures 3–5). Firmicutes were the dominant bacterial lineage in the fecal samples of belugas, accounting for 94%~96%. The dominant microbial phyla for two of the three Pacific white-sided dolphins were also Firmicutes, the remaining animal (Duomi) was dominated by Proteobacteria (Gamma) (76%~83%). The majority of the fecal samples from the lone common bottlenose dolphin in this study were dominated by Proteobacteria (Gamma), with one exception that was dominated by Firmicutes. The gut microbial communities of Cape fur seals were dominated by Firmicutes, Bacteroidetes, Fusobacteria, Epsilonbacteraeota, and Proteobacteria (Gamma). However, the respective compositions of different seal fecal samples were slightly different. At the family taxonomic level, Lachnospiraceae, Ruminococcaceae, and Peptostreptococcaceae, which are affiliated with the Clostridiales, were the dominant bacterial lineages in the beluga fecal samples. Enterobacteriaceae, Enterococcaceae, Clostridiaceae 1, and Peptostreptococcaceae were dominant in the fecal samples of Pacific white-sided dolphins. The gut microbial communities of the common bottlenose dolphin was also dominated by Enterobacteriaceae, Clostridiaceae 1, and Peptostreptococcaceae, but Vibrionaceae was also one of the dominant bacterial lineage in some of the fecal samples. Campylobacteraceae, Fusobacteriaceae, Family XI (affiliated with the Clostridiales), Moraxellaceae, and Bacteroidaceae were the dominant bacterial lineages in the fecal samples of Cape fur seals. Furthermore, at the genus taxonomic level, the gut microbial communities of belugas were mainly composed of Lachnoclostridium, Romboutsia, Fournierella, and Eubacterium fissicatena group. Meanwhile, Escherichia-Shigella, Clostridium sensu stricto 1, and Enterococcus were the dominant bacterial genera in the fecal samples of Pacific white-sided dolphins, while Vibrio, Morganella, and Clostridium sensu stricto 1 were dominant in samples from the common bottlenose dolphin. As for Cape fur seals, Fusobacterium, Campylobacter, Psychrobacter, Bacteroides, Marinifilum, and Ezakiella were the main bacterial genera.
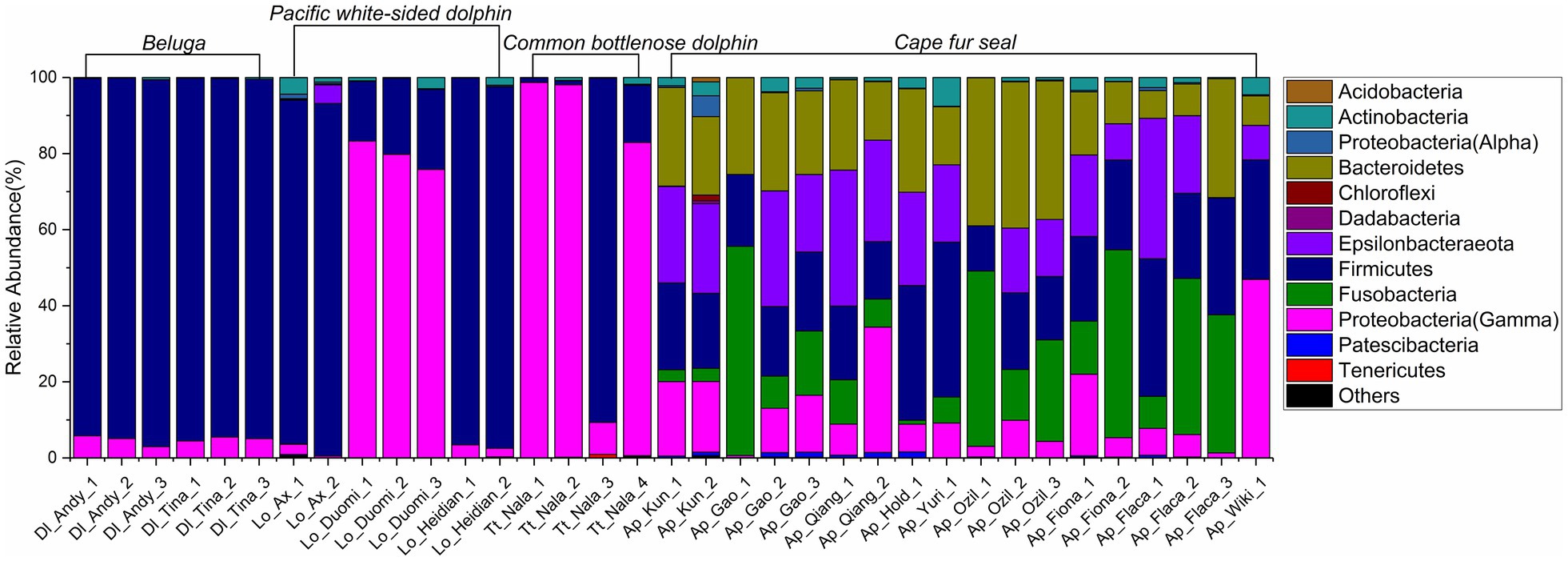
Figure 3. Gut microbial community members of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals at the phylum level.
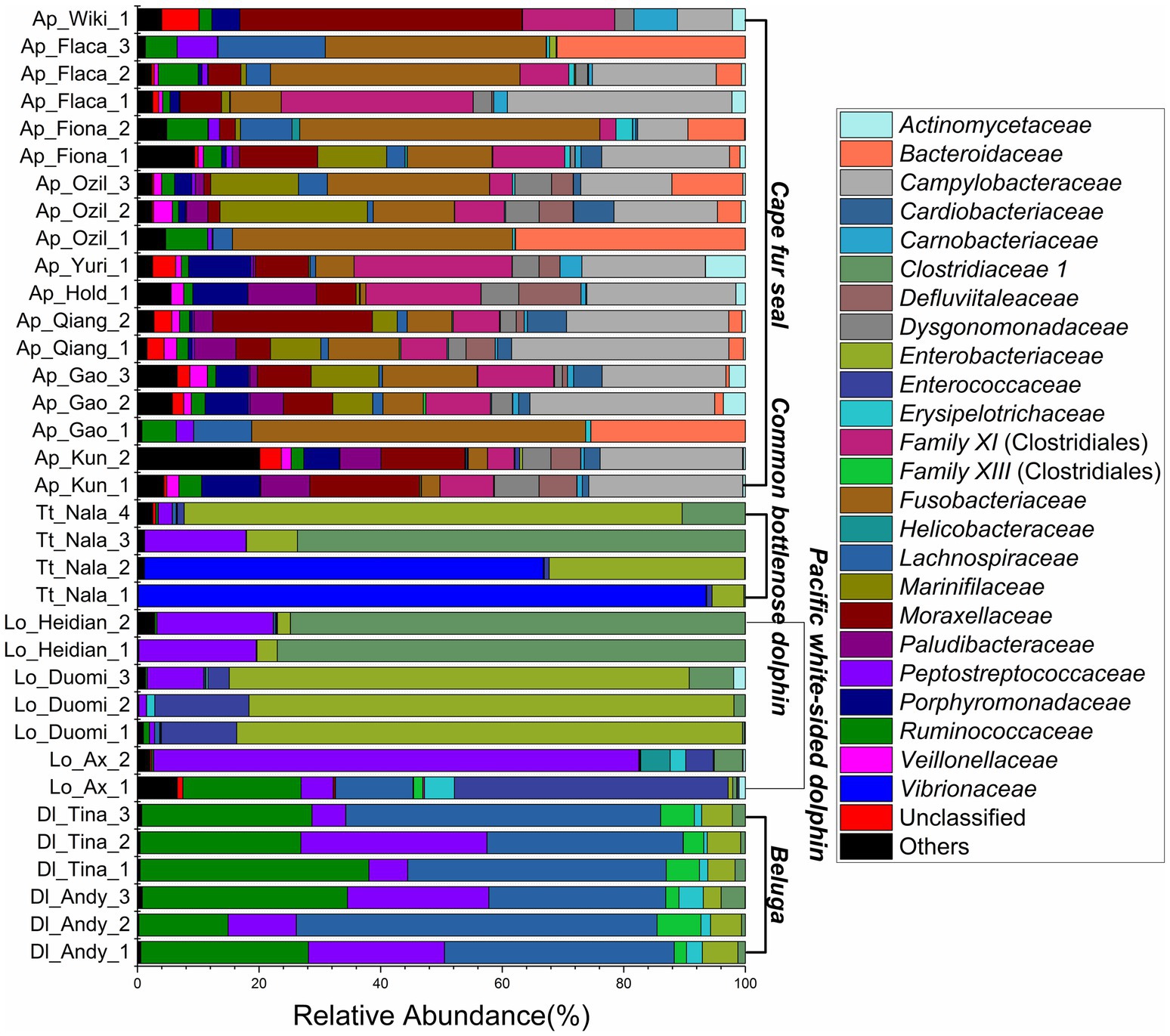
Figure 4. Gut microbial community members of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals at the family level.
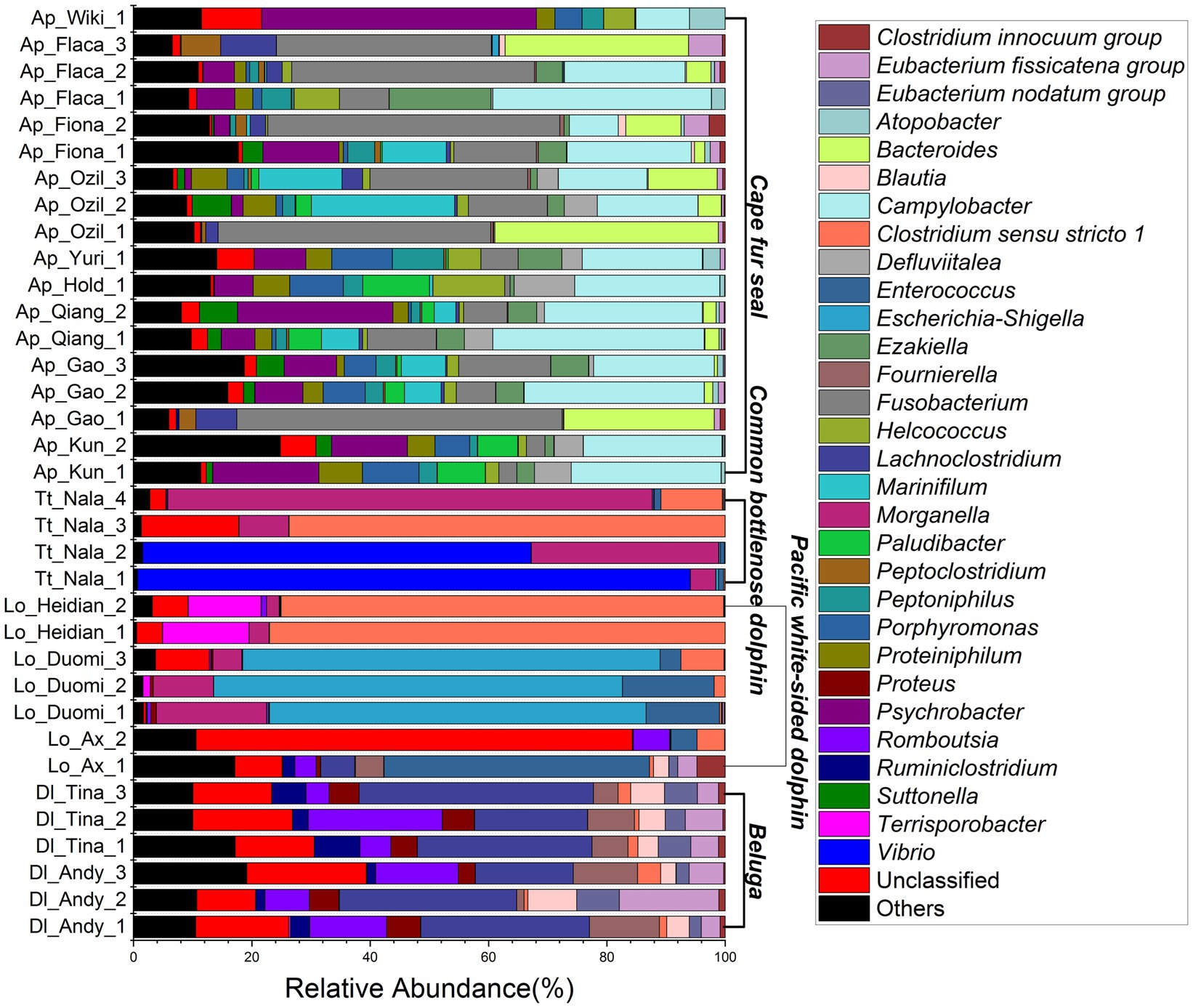
Figure 5. Gut microbial community members of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals at the genus level.
To better understand the core microbial communities of different marine mammals, we explored the detailed differences at the ASV level between different marine mammal fecal samples. Quality control and random resampling of the 35 samples were conducted, and the sequence reads were clustered into 572 ASVs at the 97% similarity level. As shown in Figure 6, in the fecal samples of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals, contained 13, 44, 7, and 206 unique ASVs, respectively. Cape fur seals shared 132, 20, and 15 ASVs with Pacific white-sided dolphins, belugas, and common bottlenose dolphin, respectively, while Pacific white-sided dolphins shared 16 and 13 ASVs with common bottlenose dolphin and belugas, respectively. By contrast, only two ASVs were shared between belugas and common bottlenose dolphin. Furthermore, 44 ASVs were detected in all four different marine mammal species.
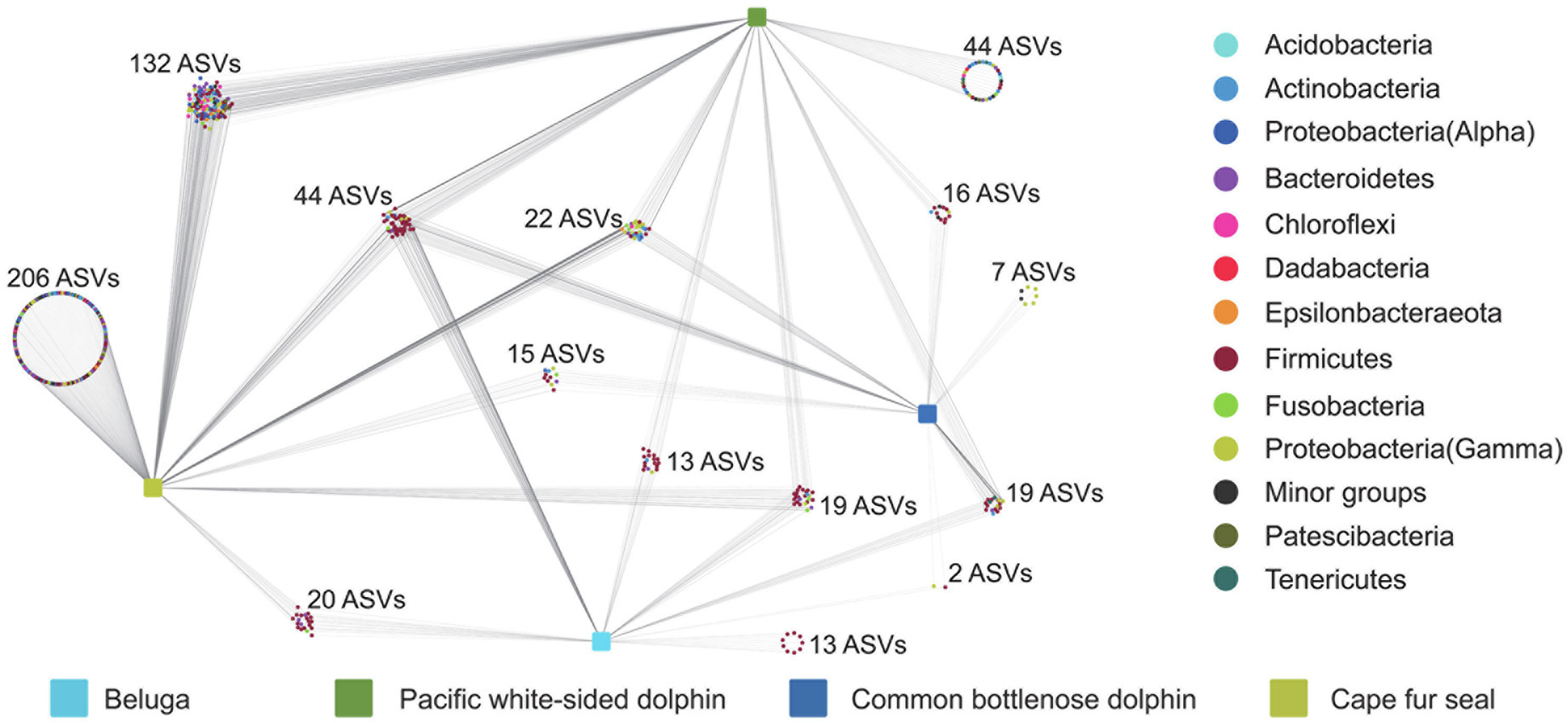
Figure 6. Distribution of ASVs in the gut of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals.
Indicator organisms analysis showed that Lachnoclostridium, Proteus, Eubacterium nodatum group, Tyzzerella, Ruminiclostridium, Oscillospira, Eubacterium fissicatena group, Romboutsia, Hydrogenoanaerobacterium, GCA-900066225 (Ruminococcaceae), Ruminococcus gauvreauii group, Negativibacillus, Mycobacterium, Fournierella, Faecalibacterium, Butyricicoccus, and some other unclassified genera were highly associated with the gut microbial communities of belugas (Figure 7). Enterococcus was the only indicator genus in the gut microbial communities of Pacific white-sided dolphins. In the gut of the common bottlenose dolphin, Ureaplasma and one unclassified genus were the indicator organisms. Three indicator genera, Fusobacterium, Bacteroides, and Peptoclostridium, were observed in the fecal samples of Cape fur seals. The heatmap based on these indicator ASVs reveals that belugas and Cape fur seals harbor distinct gut microbial communities (Figure 8). Meanwhile, the gut microbial communities of Pacific white-sided dolphins and common bottlenose dolphins could not be separated (Figure 8), consistent with the NMDS results.
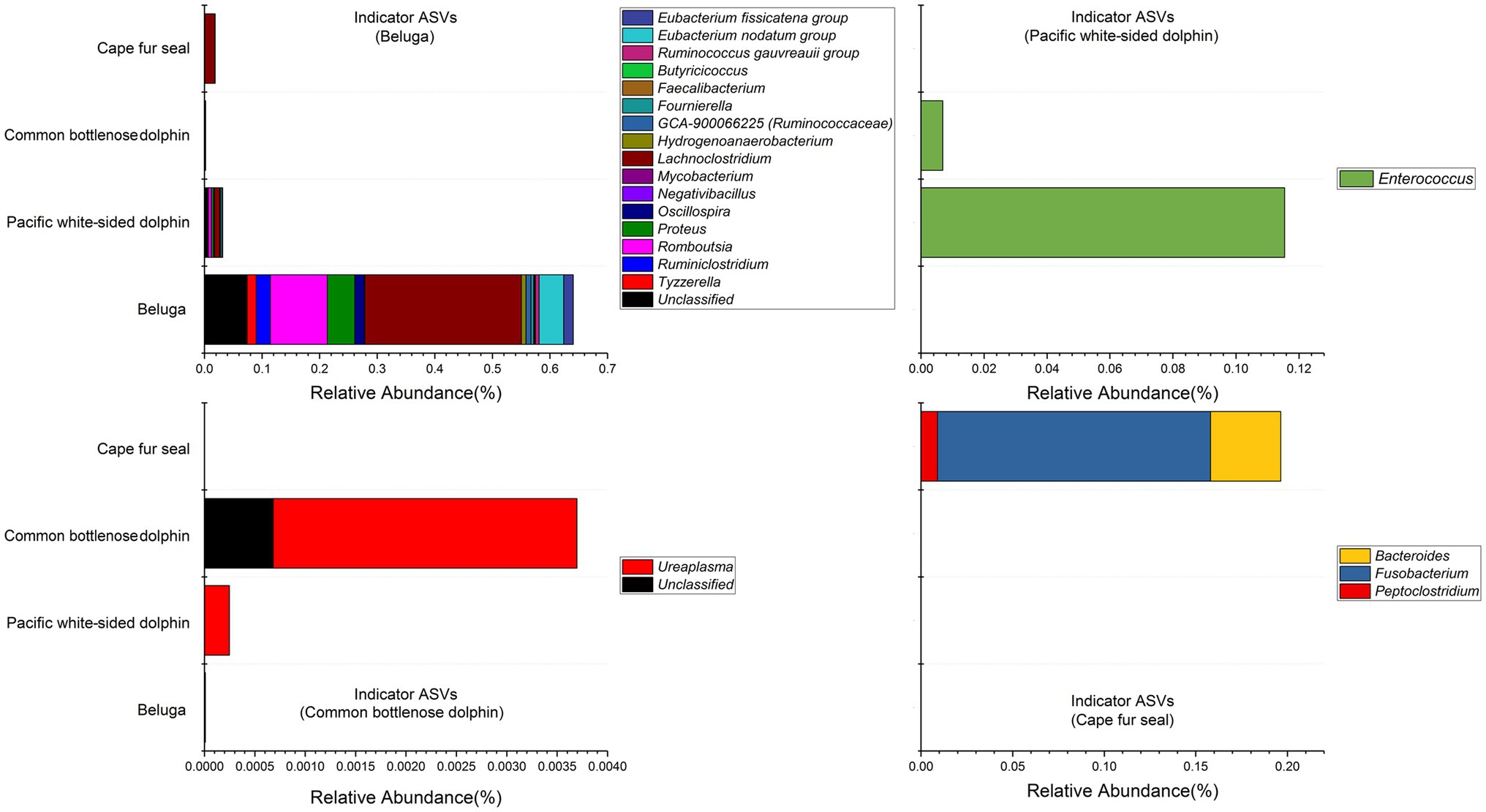
Figure 7. Stacked bar chart showing relative abundance of indicator ASVs in the gut of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals. The indicator organisms being shown was at the genus level.
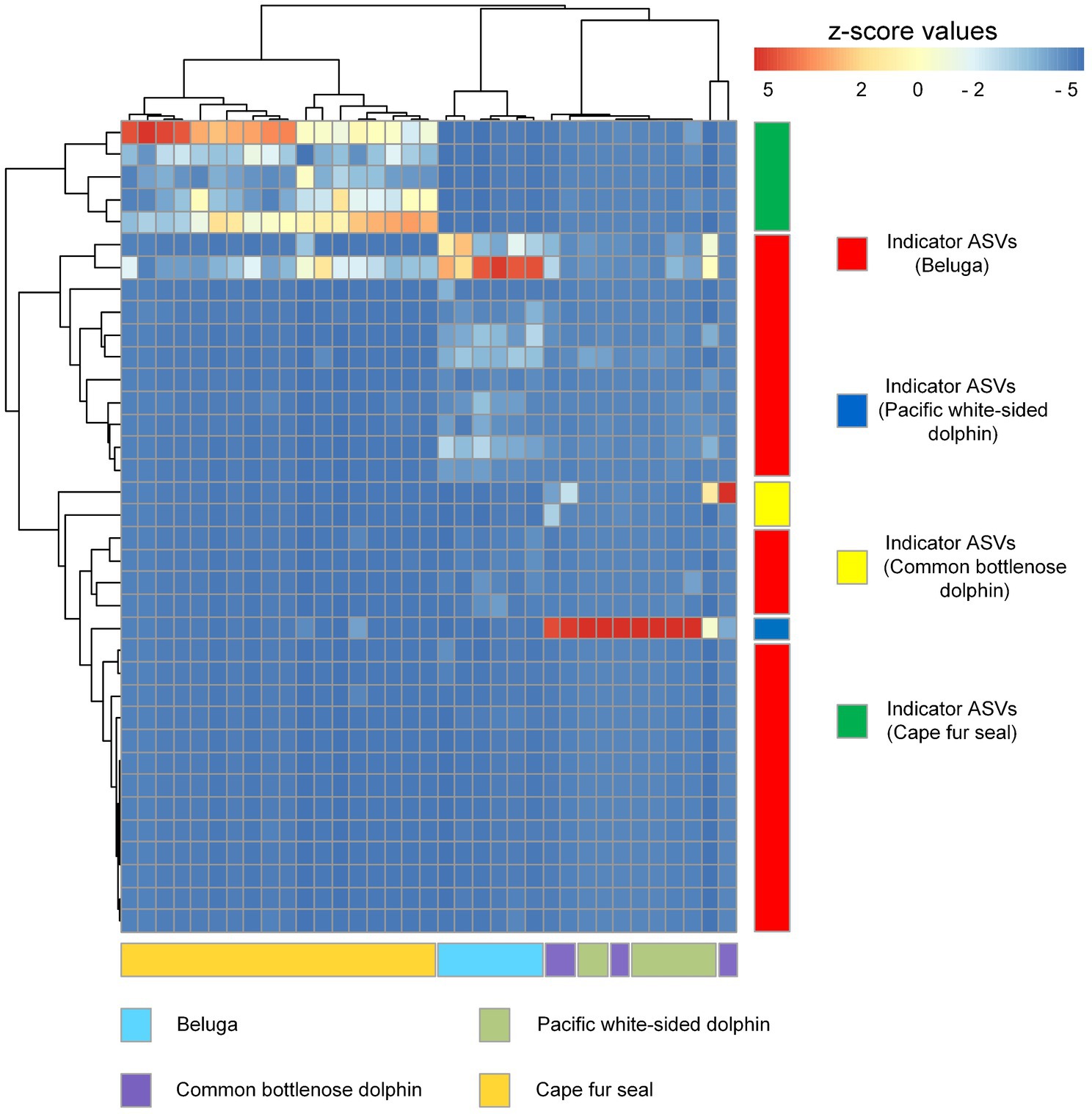
Figure 8. Heatmap diagram showing the distribution of indicator ASVs of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals. Each row and column of the heatmap diagram corresponds to a single indicator and samples, respectively. The row data for each indicator were z-score transformed. Dendrograms were constructed based on Pearson correlation clustering.
In this study, we reported the gut microbial communities in two belugas, three Pacific white-sided dolphins, one common bottlenose dolphin, and nine Cape fur seals. These marine mammals resided at the Marine and Waterpark in Atlantis hotel, Sanya, China. Moreover, these creatures also shared the same water filtration system and diet including herring (Clupea harengus) and capelin (Mallotus villosus). The α-diversity indices showed that the gut microbial diversity of belugas and Cape fur seals were higher than that of Pacific white-sided dolphins and common bottlenose dolphin, with the gut microbial diversity of the two dolphin species being similar. Given the same food sources and water quality, we speculate that the low gut microbial diversity of Pacific white-sided dolphins and common bottlenose dolphin may attribute to the poor health status of the animals themselves.
In their pioneering work, Ley et al. (2008) explored the fecal microbiota of humans and 59 other mammalian species living in captivity and in the wild. Results indicated that host diet and phylogeny both influence microbial diversity, and the influence increases from carnivory to omnivory, and to herbivory (Ley et al., 2008). The gut microbiota makes an important contribution to host metabolism by contributing enzymes that are not encoded by the host genome such as the decomposition of polysaccharides and polyphenols and the synthesis of vitamins (Rowland et al., 2018). Different diets can shape gut microbial communities in both the short and long term (Flint et al., 2012). For instance, low intake of accessible carbohydrates and dietary fibers may reduce the microbial diversity and change the microbial communities, contributing to the depletion of specific microbial taxa (Sonnenburg and Sonnenburg, 2014; Sonnenburg et al., 2016). In our study, four different marine mammals are carnivores, living in the same environment, and eating the same food. Therefore, if diet and environment affects gut microbial community composition and structure more than the phylogeny, the gut microbial β-diversity of belugas, Pacific white-sided dolphins, common bottlenose dolphin, and Cape fur seals should tend to be similar. However, according to the results of NMDS and different statistical analyses, belugas, dolphins, and Cape fur seals possess significantly different gut microbial communities. Whereas, the gut microbial communities of Pacific white-sided dolphins and common bottlenose dolphin were statistically undifferentiated, we propose the reason for the similarities in β-diversity may attributed to the animals’ health status and medical treatments prior to sampling; however, further study is required.
Members of Firmicutes, Bacteroidetes, Fusobacteria, and Epsilonbacteraeota constituted the vast majority of the microorganisms in the gut of Cape fur seals. The most common bacterial genera were Fusobacterium, Campylobacter, Psychrobacter, Bacteroides, Marinifilum, and Ezakiella. However, to date, no Cape fur seal gut microbial communities have been reported. The skin microbial communities of Antarctic fur seal (Arctocephalus gazella) showed that Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria were the dominant bacterial lineages (Grosser et al., 2019). Although there are definite similarities, the Fusobacteria, rather than the Actinobacteria were the dominant bacterial lineage in the gut of Cape fur seals. Nevertheless, at the genus level, only Psychrobacter was detected as a dominant bacterial lineage both in the gut and the skin microbial communities of fur seals, and Chryseobacterium and Jeotgalibaca, which were dominant in the skin microbial communities, were either not detected (Chryseobacterium) in the gut microbial dataset or identified as a single ASV (Jeotgalibaca) within in the gut of AX (0.003–0.05%, relative abundance, Pacific white-sided dolphin), and Kun, Flaca (0.003–0.02%, relative abundance, Cape fur seals). The Fusobacterium, as an animal-associated genus affiliated with the Fusobacteriaceae, are widely found in the gut of humans and other animals (Brennan and Garrett, 2019) including rats (Zhang et al., 2020), macaques (Nugeyre et al., 2019), pigs (Wylensek et al., 2020), penguins (Tian et al., 2021), striped dolphin (Godoy-Vitorino et al., 2017), and whales (Li et al., 2019), and some birds and fishes (Michl et al., 2017; Rasheeda et al., 2017; Chen et al., 2018; Michel et al., 2018). These organisms may play a symbiotic or pathogenic role (Brennan and Garrett, 2019).
The genus Campylobacter are very common bacteria in animal digestive tracts, whose powerful flagellar motility is very important in nutrient acquisition, whereby bacterial cells attach to surfaces and rotate their flagella to increase nutrient flux for metabolism (Beeby, 2015). Different species of Campylobacter can have variable characteristics of either pathogen or commensal, depending on their host immune status and the pathotype of the bacteria themselves (Wigley, 2015). For example members of the Campylobacter, such as Campylobacter jejuni and Campylobacter coli, are among some of the most common human gastrointestinal pathogens, causing bacterial diarrheal illness (Fitzgerald, 2015). In addition, the ASVs classified into Campylobacter in this study were further identified by phylogenetic analysis. The results showed that these ASVs clustered together far from the common species of Campylobacter (Supplementary Figure S2). Therefore, the role of Campylobacter in the gut of Cape fur seals remains unclear. The members of the Psychrobacter, another common genus, can be found in different marine environments and marine creatures, such as Arctic fjord (Zeng et al., 2015), Arctic glacier (Zeng et al., 2016), deep sea (Evans-Yamamoto et al., 2019), penguins (Kämpfer et al., 2020), corals (Zachariah et al., 2016), and marine crustaceans (Romanenko et al., 2009). In this and previous studies, the genus Psychrobacter was dominant in the gut of Cape fur seals and the skin of Antarctic fur seal (Arctocephalus gazella). At the same time, we barely detected this bacterium in the gut of belugas, Pacific white-sided dolphins, and common bottlenose dolphin. The members of Psychrobacter, as one of the commensal bacteria, could improve the microbial diversity of gastrointestinal tract, and upregulate the expression of immune-related genes and is therefore considered to be probiotic (Sun et al., 2011; Yang et al., 2011; Makled et al., 2017). According to previous study, it is interesting that the decrease in the relative abundance of Psychrobacter may associate with depressive conditions of hosts. Meanwhile, the results of metabolomics showed that the lower levels of amino acids, and fatty acids, and higher amounts of bile acids, hypoxanthine, and stercobilins were detected in the depression groups. However, the roles of these bacteria in the different stages of depression are not clear (Yu et al., 2017). Moreover, the Psychrobacter, one of the key genera of young-like gut microbial community of African turquoise killifish (Nothobranchius furzeri), was considered to play an important role in extending host life span (Smith et al., 2017). Additionally, after infection by porcine epidemic diarrhea virus, the relative abundance of Psychrobacter, which is a symbiotic group of bacteria in the gut of pigs, was decreased.
Members of the Bacteroidetes, one of the major lineages of friendly bacterial commensuals, possess large numbers of genes encoding carbohydrate active enzymes, which allows them to switch readily between different energy sources in the gut, depending on availability, using sophisticated regulatory mechanisms to control gene expression (Thomas et al., 2011). Polysaccharides comprise the most abundant type of biological polymer and, as such, also the most abundant biological food source. Carbohydrate fermentation by Bacteroides and other intestinal bacteria results in the production of a pool of volatile fatty acids that are absorbed through the large intestine and utilized by the host as an energy source, providing a significant proportion of the host’s daily energy requirement (Hooper et al., 2002). Although most of the Bacteroides are commensals in the gut, some species can also be responsible for infection, including Bacteroides fragilis, Bacteroides distasonis, Bacteroides ovatus, Bacteroides thetaiotaomicron, Bacteroides vulgatus, and Bacteroides uniformis, with significant morbidity and mortality (Wexler, 2007). However, in this study, the genus Bacteroides was only found to be dominant in the fecal samples of Cape fur seals and was rarely detected in the other three marine mammals. Moreover, ASV 12, ASV 24, and ASV 35 were the most predominant ASVs and were classified to Bacteroides at the genus level. The phylogenetic analysis, based on the neighbor-joining method, showed that the strain Bacteroides plebeius DSM 17135 was most closely related to ASV 12, ASV 24, and ASV 35 (Supplementary Figure S3). Bacteroides plebeius is a common bacterium in human faces, with numerous strains (Kitahara et al., 2005). Furthermore, compared to healthy cats, the relative abundance of B. plebeius was significantly decreased in the fecal microbiome of animals with Feline chronic enteropathy (CE; Marsilio et al., 2019). The genus of Marinifilum is often isolated or detected in coastal sea water and sediments (Na et al., 2009; Ruvira et al., 2013; Xu et al., 2016) and is most likely involved in the hydrolysis and fermentation of proteins, carbohydrates, and lipids (Müller et al., 2018). The members of Ezakiella, a genus of Gram-stain positive, coccus-shaped, anaerobic bacteria are often found in the intestinal and vaginal tracts of healthy people (Patel et al., 2015; Diop et al., 2018). However, based on the very limited reports currently available, it is not clear what role(s) this genus may play in the gut and vagina.
The bacterial genus of Lachnoclostridium is commonly found in the healthy human gut (Tidjani Alou et al., 2016a,b; Traore et al., 2017). But, Lachnoclostridium was also isolated from the gut microbiota of an obese patient (Amadou et al., 2016). A previous study reported that the genus of Romboutsia was predominant in the samples of mammalian gut microbiota (Gerritsen et al., 2018). However, this genus can be treated as one of the gut microbiotal markers for obesity in patients (Zeng et al., 2019). The Fournierella can be found in the fecal samples of humans, and it has also been reported to be significantly associated with intrahepatic fat accumulation (Yaskolka Meir et al., 2021). The Eubacterium fissicatena group is closely associated with obesity and obesity-related metabolic disorders of the host. Previous studies found that medium-, long-, and medium-chain (MLM) structured lipids have anti-obesity effects (Song et al., 2021). After supplementation with MLM structured lipids, the relative abundance of E. fissicatena group were decreased (Yue et al., 2020). The dominant gut bacterial lineages of belugas in this study indicated that they were obese or overweight, which we propose to be an essential environmental adaption strategy for the belugas to defense extremely cold in the arctic waters.
Escherichia-Shigella, Clostridium sensu stricto 1, and Enterococcus were the dominant bacterial lineages in the fecal samples of Pacific white-sided dolphins. However, only one ASV (ASV3) was assigned to Escherichia-Shigella. The results of phylogenetic analysis indicate that ASV3 is closely related to either Cronobacter turicensis or Erwinia iniecta (Supplementary Figure S4). Cronobacter turicensis is an opportunistic foodborne pathogen, which can lead to severe disease manifestations, such as brain abscesses, meningitis, necrotizing enterocolitis, and systemic sepsis (Lehner and Stephan, 2004; Stephan et al., 2011). Only one paper has reported that E. iniecta possess virulence to the Russian wheat aphid (Campillo et al., 2015). The most abundant ASV, ASV2, was assigned to Clostridium sensu stricto 1 at the genus level and was closely related to Clostridium perfringens based on the phylogenetic analysis (Supplementary Figure S5). ASV11 was present in the fecal samples of Pacific white-sided dolphins and was closely related to Enterococcus lactis (Supplementary Figure S6), which can produce an antimicrobial substance that is stable over a wide range of pH (2–10) and after heating at 100°C for 15min (Ben Braïek et al., 2018a). Therefore, E. lactis is classified as a probiotic (Sharma et al., 2012; Ben Braïek et al., 2018b). Clostridium perfringens is a typical anaerobic, Gram-positive, spore-forming bacillus that can secrete more than 20 virulence toxins and has been associated with intestinal diseases ranging in severity from diarrhea to necrotizing enterocolitis and myonecrosis in both animals and humans (Ohtani and Shimizu, 2016; Kiu and Hall, 2018). Clostridium perfringens was found to be dominant in the fecal samples of the common bottlenose dolphin of this study. The other two dominant bacterial lineages were Vibrio and Morganella. Phylogenetic analysis showed that ASV40 was closely related to Vibrio fluvialis (Supplementary Figure S7), and ASV 5 was closely related to Morganella morganii (Supplementary Figure S8). Vibrio fluvialis and M. morganii are both pathogenic bacteria. Vibrio fluvialis is a halophilic, Gram-negative pathogen commonly found in coastal environments including sea water and seafood. Several outbreaks and sporadic cases of acute diarrhea caused by V. fluvialis have been reported extensively (Igbinosa and Okoh, 2010; Chowdhury et al., 2012, 2013; Liang et al., 2013). Morganella morganii is a gram-negative bacterium found in the gut of humans and is associated with many diseases including cellulitis, abscessation, sepsis, bacteremia, and diarrhea (Falagas et al., 2006; Kim et al., 2007; Liu et al., 2016). These results indicated that the Pacific white-sided dolphins and common bottlenose dolphin are in poor health condition.
Indicator species refers to a species constrained to one or a few habitat types, and as “specialists” have a greater susceptibility to local or regional extinction. These organisms can also potentially represent a better ecological indicator of environmental change than habitat generalists (Carignan and Villard, 2002). For example, in the vegetation studies, the species of plant that are absolutely inclined to present in a single or, at the most, a few vegetation types are identified as indicator species, and this method is beneficial to the identification of vegetation types in research surveys (Chytrý et al., 2002). To find out the association between single species and one or several groups of sites representing habitat types, community types, or other categories, the indicator value index (IndVal) is used to assess these relationships (De Cáceres and Legendre, 2009). Because there were an uneven number of fecal samples from the different groups of marine mammals in this study, we used the index of IndVal.g to eliminate the effect of different numbers in each group. According to the set threshold (IndVal.g>0.95, p<0.001), our results show that the gut microbial communities of belugas harbor 28 indicator species representing by ASVs. While five and two ASVs were identified in the gut microbial communities of Cape fur seals and common bottlenose dolphin, respectively. However, only a single ASV was detected in the gut microbiota of Pacific white-sided dolphins. Heatmap results based on these indicator species show that different species of marine mammals can harbor distinct microbial communities with the exception of Pacific white-sided dolphins and common bottlenose dolphin, which cannot be separated. Interestingly, in our study, the microbial phyla with the highest relative abundance in the fecal samples of common bottlenose dolphin were Proteobacteria (Gamma) and Firmicutes at the phylum level, and Vibrionaceae, Enterobacteriaceae, Clostridiaceae 1, and Peptostreptococcaceae at the family level. These findings were consistent with previous studies (Soverini et al., 2016;Suzuki et al., 2019 ; Robles-Malagamba et al., 2020). The Fusobacteriaceae of Fusobacteria was also documented in the previous studies as a dominant bacterial lineage (Suzuki et al., 2019; Robles-Malagamba et al., 2020), however, we barely detected Fusobacteria in the gut microbial communities of common bottlenose dolphin but found that it dominated the fecal samples of Cape fur seals. Moreover, at the genus level, previous work has shown that the genera Cetobacterium (70.9%), Clostridium XI (15.3%), and Clostridium sensu stricto (4.2%) had the highest relative abundance in the fecal samples of common bottlenose dolphins, while in our microbial datasets, Vibrio (Vibrio fluvialis), Morganella (Morganella morganii), and Clostridium sensu stricto 1 (Clostridium perfringens) were the dominant bacterial lineages, all of which are pathogenic bacteria, especially the C. perfringens, which have been reported to be the causative agent of common bottlenose dolphins (Buck et al., 1987; Jaing et al., 2015). Furthermore, we also detected lager number of C. perfringens in the fecal samples of Pacific white-sided dolphins, because the C. perfringens can enter other dolphins via skin wounds, which serve as the focus for these bacteria to penetrate and spread (Buck et al., 1987). Meanwhile, the spread of same bacteria between Pacific white-sided dolphins and common bottlenose dolphin may contribute the similarity of microbial community structure between them. Unfortunately, we did not find any published reports on the gut microbiota of Pacific white-sided dolphins. For a comparative and comprehensive understanding, further studies of the microbial communities of Pacific white-sided dolphins from different places should be conducted in the future.
In this study, we systematically compared gut microbiota of four different marine mammals in an aquarium; they shared the same food, and depended on the same water filtration system. We found the belugas and Cape fur seals harbored remarkably greater gut microbial diversity than Pacific white-sided dolphins and common bottlenose dolphin. Diet, environment, and host phylogeny, all of these have been reported to have impact on microbial community structure. Our results demonstrated that phylogeny (>family level) has the greatest influence on the shaping of microbial communities. Although diet and water quality were the same for all subjects, we still observed significantly different gut microbial community structure among whales, dolphins, and fur seals. Nevertheless, the microbial communities of Pacific white-sided dolphins and common bottlenose dolphin showed similarities, but most of the dominant bacterial lineages were pathogenic bacteria revealed by phylogenetic analysis. Moreover, some potential pathogens were found in the guts of both Pacific white-sided dolphins and common bottlenose dolphin, and this could be one of the reasons for the similarity in gut microbial community structure of these two dolphins. Our results also indicated that the Pacific white-sided dolphins and common bottlenose dolphin are in poor health condition, and that the gut microbiota of belugas could aid in defense against cold conditions. Our study provides foundamental information on the composition of gut microbial communities in marine mammals; however, knowledge about this subject is still scarce. For better understanding of the gut microbial community structure and composition of marine mammals, further studies should be focused on the spatial and temporal of gut microbiota in different marine mammal species in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA743584.
The animal study was performed under an Ethical Statement from Institute of Deep-sea Science and Engineering, CAS, with the number of IDSSE-SYLL-MMMBL-01.
SB, PZ, and SL conceived and designed this study. PZ, CZ, XD, JL, and PX collected the samples. SB did the experiments, prepared the figures, and wrote the manuscript. SB, JD, and CZ analyzed the data. PZ and SL revised the manuscript. All authors contributed to the article and approved the submitted version.
This project was supported by grants of the Youth Innovation Promotion Association of Chinese Academy of Sciences (2020363).
CZ and XD are employed by Atlantis Sanya.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.769012/full#supplementary-material
Amadou, T., Hosny, M., La Scola, B., and Cassir, N. (2016). Lachnoclostridium bouchesdurhonense, a new bacterial species isolated from human gut microbiota. New Microbes New Infect. 13, 69–70. doi: 10.1016/j.nmni.2016.06.015
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Bai, S., and Hou, G. (2020). Microbial communities on fish eggs from Acanthopagrus schlegelii and Halichoeres nigrescens at the XuWen coral reef in the Gulf of Tonkin. PeerJ. 8:e8517. doi: 10.7717/peerj.8517
Bai, S., Zhang, P., Lin, M., Lin, W., Yang, Z., and Li, S. (2021). Microbial diversity and structure in the gastrointestinal tracts of two stranded short-finned pilot whales (Globicephala macrorhynchus) and a pygmy sperm whale (Kogia breviceps). Integr. Zool. 16, 324–335. doi: 10.1111/1749-4877.12502
Beeby, M. (2015). Motility in the epsilon-proteobacteria. Curr. Opin. Microbiol. 28, 115–121. doi: 10.1016/j.mib.2015.09.005
Ben Braïek, O., Cremonesi, P., Morandi, S., Smaoui, S., Hani, K., and Ghrairi, T. (2018a). Safety characterisation and inhibition of fungi and bacteria by a novel multiple enterocin-producing enterococcus lactis 4CP3 strain. Microb. Pathog. 118, 32–38. doi: 10.1016/j.micpath.2018.03.005
Ben Braïek, O., Morandi, S., Cremonesi, P., Smaoui, S., Hani, K., and Ghrairi, T. (2018b). Safety, potential biotechnological and probiotic properties of bacteriocinogenic enterococcus lactis strains isolated from raw shrimps. Microb. Pathog. 117, 109–117. doi: 10.1016/j.micpath.2018.02.021
Brennan, C. A., and Garrett, W. S. (2019). Fusobacterium nucleatum – symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Buck, J. D., Shepard, L. L., and Spotte, S. (1987). Clostridium perfringens as the cause of death of a captive Atlantic bottlenosed dolphin (Tursiops truncatus). J. Wildl. Dis. 23, 488–491. doi: 10.7589/0090-3558-23.3.488
Campillo, T., Luna, E., Portier, P., Fischer-Le Saux, M., Lapitan, N., Tisserat, N. A., et al. (2015). Erwinia iniecta sp. nov., isolated from Russian wheat aphid (Diuraphis noxia). Int. J. Syst. Evol. Microbiol. 65, 3625–3633. doi: 10.1099/ijsem.0.000466
Carignan, V., and Villard, M. A. (2002). Selecting indicator species to monitor ecological integrity: a review. Environ. Monit. Assess. 78, 45–61. doi: 10.1023/A:1016136723584
Chao, A. (1984). Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 11, 265–270.
Chen, L., Hu, C., Lok-Shun Lai, N., Zhang, W., Hua, J., Lam, P. K. S., et al. (2018). Acute exposure to PBDEs at an environmentally realistic concentration causes abrupt changes in the gut microbiota and host health of zebrafish. Environ. Pollut. 240, 17–26. doi: 10.1016/j.envpol.2018.04.062
Chowdhury, G., Pazhani, G. P., Dutta, D., Guin, S., Dutta, S., Ghosh, S., et al. (2012). Ramamurthy T. Vibrio fluvialis in patients with diarrhea, Kolkata. India. Emerg. Infect. Dis. 18, 1868–1871. doi: 10.3201/eid1811.120520
Chowdhury, G., Sarkar, A., Pazhani, G. P., Mukhopadhyay, A. K., Bhattacharya, M. K., and Ramamurthy, T. (2013). An outbreak of foodborne gastroenteritis caused by dual pathogens, salmonella enterica serovar Weltevreden and Vibrio fluvialis in Kolkata. India. Foodborne Pathog. Dis. 10, 904–906. doi: 10.1089/fpd.2013.1491
Chytrý, M., Tichý, L., Holt, J., and Botta-Dukát, Z. (2002). Determination of diagnostic species with statistical fidelity measures. J. Veg. Sci. 13, 79–90. doi: 10.1111/j.1654-1103.2002.tb02025.x
Dang, A. T., and Marsland, B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
David, J. H. M. (1987). Diet of south African fur seal (1974–1985) and an assessment of competition with fisheries in southern Africa. Afr. J. Mar. Sci. 5, 693–713. doi: 10.2989/025776187784522568
De Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Diop, K., Andrieu, C., Michelle, C., Armstrong, N., Bittar, F., Bretelle, F., et al. (2018). Characterization of a new Ezakiella isolated from the human vagina: genome sequence and description of Ezakiella massiliensis sp. nov. Curr. Microbiol. 75, 456–463. doi: 10.1007/s00284-017-1402-z
Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv dontidae. doi: 10.1101/081257
Evans-Yamamoto, D., Takeuchi, N., Masuda, T., Murai, Y., Onuma, Y., Mori, H., et al. (2019). Complete genome sequence of Psychrobacter sp. strain KH172YL61, isolated from deep-sea sediments in the Nankai trough, Japan. Microbiol. Resour. Announc. 8, e00326–e00319. doi: 10.1128/MRA.00326-19
Falagas, M. E., Kavvadia, P. K., Mantadakis, E., Kofteridis, D. P., Bliziotis, I. A., Saloustros, E., et al. (2006). Morganella morganii infections in a general tertiary hospital. Infection 34, 315–321. doi: 10.1007/s15010-006-6682-3
Flint, H. J., Scott, K. P., Louis, P., and Duncan, S. H. (2012). The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589. doi: 10.1038/nrgastro.2012.156
Frost, K. J., and Lowry, L. F. (1981). Trophic importance of some marine gadids in northern Alaska and their body-otolith size relationships. Fish. Bull. 79, 187–192.
Gerritsen, J., Umanets, A., Staneva, I., Hornung, B., Ritari, J., Paulin, L., et al. (2018). Romboutsia hominis sp. nov., the first human gut-derived representative of the genus Romboutsia, isolated from ileostoma effluent. Int. J. Syst. Evol. Microbiol. 68, 3479–3486. doi: 10.1099/ijsem.0.003012
Glaeser, S. P., Silva, L. M. R., Prieto, R., Silva, M. A., Franco, A., Kämpfer, P., et al. (2021). A preliminary comparison on faecal microbiomes of free-ranging large baleen (Balaenoptera musculus, B. physalus, B. borealis) and toothed (Physeter macrocephalus) whales. Microb. Ecol. doi: 10.1007/s00248-021-01729-4 [Epub ahead of print]
Godoy-Vitorino, F., Rodriguez-Hilario, A., Alves, A. L., Gonçalves, F., Cabrera-Colon, B., Mesquita, C. S., et al. (2017). The microbiome of a striped dolphin (Stenella coeruleoalba) stranded in Portugal. Res. Microbiol. 168, 85–93. doi: 10.1016/j.resmic.2016.08.004
Grosser, S., Sauer, J., Paijmans, A. J., Caspers, B. A., Forcada, J., Wolf, J. B. W., et al. (2019). Fur seal microbiota are shaped by the social and physical environment, show mother-offspring similarities and are associated with host genetic quality. Mol. Ecol. 28, 2406–2422. doi: 10.1111/mec.15070
Hayano, A., Yoshioka, M., Tanaka, M., and Amano, M. (2004). Population differentiation in the Pacific white-sided dolphin Lagenorhynchus obliquidens inferred from mitochondrial DNA and microsatellite analyses. Zool. Sci. 21, 989–999. doi: 10.2108/zsj.21.989
Heide-Jorgensen, M. P., and Teilmann, J. (1994). Growth, reproduction, age structure and feeding habits of white whales (Delphinapterus leucas) in West Greenland waters. Meddr Gronland, Biosci. 39, 195–212. Copenhagen 1994-04-22.
Hooper, L. V., Midtvedt, T., and Gordon, J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307. doi: 10.1146/annurev.nutr.22.011602.092259
Igbinosa, E. O., and Okoh, A. I. (2010). Vibrio fluvialis: an unusual enteric pathogen of increasing public health concern. Int. J. Environ. Res. Public Health 7, 3628–3643. doi: 10.3390/ijerph7103628
Jaing, C., Thissen, J. B., Gardner, S., McLoughlin, K., Slezak, T., Bossart, G. D., et al. (2015). Pathogen surveillance in wild bottlenose dolphins Tursiops truncatus. Dis. Aquat. Org. 116, 83–91. doi: 10.3354/dao02917
Kämpfer, P., Glaeser, S. P., Irgang, R., Fernández-Negrete, G., Poblete-Morales, M., Fuentes-Messina, D., et al. (2020). Psychrobacter pygoscelis sp. nov. isolated from the penguin Pygoscelis papua. Int. J. Syst. Evol. Microbiol. 70, 211–219. doi: 10.1099/ijsem.0.003739
Kastelein, R. A., Vaughan, N., Walton, S., and Wiepkema, P. R. (2002). Food intake and body measurements of Atlantic bottlenose dolphins (Tursiops truncates) in captivity. Mar. Environ. Res. 53, 199–218. doi: 10.1016/S0141-1136(01)00123-4
Kim, J. H., Cho, C. R., Um, T. H., Rhu, J. Y., Kim, E. S., Jeong, J. W., et al. (2007). Morganella morganii sepsis with massive hemolysis. J. Korean Med. Sci. 22, 1082–1084. doi: 10.3346/jkms.2007.22.6.1082
Kirkman, S. P., and Arnould, J. P. Y. (2018). Cape and Australian Fur Seals: Arctocephalus Pusillus Pusillus and A. P. Doriferus. Encyclopedia of Marine Mammals. 3rd Edn. London: Academic Press.
Kirkman, S. P., Yemane, D., Oosthuizen, W. H., Meÿer, M. A., Kotze, P. G. H., Skrypzeck, H., et al. (2013). Spatio-temporal shifts of the dynamic cape fur seal population in southern Africa, based on aerial censuses (1972–2009). Mar. Mamm. Sci. 29, 497–524. doi: 10.1111/j.1748-7692.2012.00584.x
Kitahara, M., Sakamoto, M., Ike, M., Sakata, S., and Benno, Y. (2005). Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 55, 2143–2147. doi: 10.1099/ijs.0.63788-0
Kiu, R., and Hall, L. J. (2018). An update on the human and animal enteric pathogen Clostridium perfringens. Emerg. Microbes Infect. 7, 1–15. doi: 10.1038/s41426-018-0144-8
Kong, Y. (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153. doi: 10.1016/j.ygeno.2011.05.009
Lehner, A., and Stephan, R. (2004). Microbiological, epidemiological, and food safety aspects of Enterobacter sakazakii. J. Food Prot. 67, 2850–2857. doi: 10.4315/0362-028X-67.12.2850
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848. doi: 10.1016/j.cell.2006.02.017
Li, J., Hu, A., Bai, S., Yang, X., Sun, Q., Liao, X., et al. (2021). Characterization and performance of lactate-feeding consortia for reductive dechlorination of trichloroethene. Microorganisms. 9, 751. doi: 10.3390/microorganisms9040751
Li, C., Tan, X., Bai, J., Xu, Q., Liu, S., Guo, W., et al. (2019). A survey of the sperm whale (Physeter catodon) commensal microbiome. PeerJ. 7:e7257. doi: 10.7717/peerj.7257
Liang, P., Cui, X., Du, X., Kan, B., and Liang, W. (2013). The virulence phenotypes and molecular epidemiological characteristics of Vibrio fluvialis in China. Gut Pathog. 5, 6. doi: 10.1186/1757-4749-5-6
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Loseto, L. L., Stern, G. A., Connelly, T. L., Deibel, D., Gemmill, B., Prokopowicz, A., et al. (2009). Summer diet of beluga whales inferred by fatty acid analysis of the eastern Beaufort Sea food web. J. Exp. Mar. Biol. Ecol. 374, 12–18. doi: 10.1016/j.jembe.2009.03.015
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Makhado, A. B., Crawford, R. J. M., and Underhill, L. J. (2006). Impact of predation by cape fur seals Arctocephulus pusillus pusillus on the cape gannets Morus capensis at Malgas Island, Western cape, South Africa. Afr. J. Mar. Sci. 28, 681–687. doi: 10.2989/18142320609504216
Makki, K., Deehan, E. C., Walter, J., and Bäckhed, F. (2018). The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715. doi: 10.1016/j.chom.2018.05.012
Makled, S. O., Hamdan, A. M., El-Sayed, A. M., and Hafez, E. E. (2017). Evaluation of marine psychrophile, Psychrobacter namhaensis SO89, as a probiotic in Nile tilapia (Oreochromis niloticus) diets. Fish Shellfish Immunol. 61, 194–200. doi: 10.1016/j.fsi.2017.01.001
Marsilio, S., Pilla, R., Sarawichitr, B., Chow, B., Hill, S. L., Ackermann, M. R., et al. (2019). Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma. Sci. Rep. 9, 19208. doi: 10.1038/s41598-019-55691-w
McKenzie, V. J., Song, S. J., Delsuc, F., Prest, T. L., Oliverio, A. M., Korpita,, et al.(2017). The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690–704. doi: 10.1093/icb/icx090
Mecenero, S., Roux, J.-P., Underhill, L. G., and Bester, M. N. (2006). Diet of cape fur seals Arctocephalus pusillus pusillus at three mainland breeding colonies in Namibia. 1. Spatial variation. Afr. J. Mar. Sci. 28, 57–71. doi: 10.2989/18142320609504134
Michel, A. J., Ward, L. M., Goffredi, S. K., Dawson, K. S., Baldassarre, D. T., Brenner, A., et al. (2018). The gut of the finch: uniqueness of the gut microbiome of the Galápagos vampire finch. Microbiome. 6, 167. doi: 10.1186/s40168-018-0555-8
Michl, S. C., Ratten, J. M., Beyer, M., Hasler, M., LaRoche, J., and Schulz, C. (2017). The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): diet-dependent shifts of bacterial community structures. PLoS One 12:e0177735. doi: 10.1371/journal.pone.0177735
Müller, A. L., Pelikan, C., de Rezende, J. R., Wasmund, K., Putz, M., Glombitza, C., et al. (2018). Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ. Microbiol. 20, 2927–2940. doi: 10.1111/1462-2920.14297
Na, H., Kim, S., Moon, E. Y., and Chun, J. (2009). Marinifilum fragile gen. nov., sp. nov., isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 59, 2241–2246. doi: 10.1099/ijs.0.009027-0
Nishida, A. H., and Ochman, H. (2018). Rates of gut microbiome divergence in mammals. Mol. Ecol. 27, 1884–1897. doi: 10.1111/mec.14473
Nugeyre, M. T., Tchitchek, N., Adapen, C., Cannou, C., Contreras, V., Benjelloun, F., et al. (2019). Dynamics of vaginal and rectal microbiota over several menstrual cycles in female cynomolgus macaques. Front. Cell. Infect. Microbiol. 9:188. doi: 10.3389/fcimb.2019.00188
Ohtani, K., and Shimizu, T. (2016). Regulation of toxin production in Clostridium perfringens. Toxins 8:207. doi: 10.3390/toxins8070207
Patel, N. B., Tito, R. Y., Obregón-Tito, A. J., O'Neal, L., Trujillo-Villaroel, O., Marin-Reyes, L., et al. (2015). Ezakiella peruensis gen. nov., sp. nov. isolated from human fecal sample from a coastal traditional community in Peru. Anaerobe 32, 43–48. doi: 10.1016/j.anaerobe.2014.12.002
Quakenbush, L. T., Suydam, R. S., Bryan, A. L., Lowry, L. F., Frost, K. J., and Mahoney, B. A. (2015). Diet of beluga whales, Delphinapterus leucas, in Alaska from stomach contents. Marine Fisheries Review. 77, 70–84. doi: 10.7755/MFR.77.1.7
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team. (2018). R: A Language and Environment for Statistical Computing. Vienna: The R Foundation for Statistical Computing.
Rasheeda, M. K., Rangamaran, V. R., Srinivasan, S., Ramaiah, S. K., Gunasekaran, R., Jaypal, S., et al. (2017). Comparative profiling of microbial community of three economically important fishes reared in sea cages under tropical offshore environment. Mar. Genomics 34, 57–65. doi: 10.1016/j.margen.2017.04.003
Rechsteiner, E. U., Rosen, D. A. S., and Trites, A. W. (2013). Energy requirements of Pacific white-sided dolphins (Lagenorhynchus obliquidens) as predicted by a bioenergetic model. J. Mammal. 94, 820–832. doi: 10.1644/12-MAMM-A-206.1
Roberts, D. W. (2007). labdsv: Ordination and multivariate analysis for ecology. R Package Version 2007, 1. Available at: https://CRAN.R-project.org/package=labdsv ().
Robles-Malagamba, M. J., Walsh, M. T., Ahasan, M. S., Thompson, P., Wells, R. S., Jobin, C., et al. (2020). Characterization of the bacterial microbiome among free-ranging bottlenose dolphins (Tursiops truncatus). Heliyon. 6:e03944. doi: 10.1016/j.heliyon.2020.e03944
Romanenko, L. A., Tanaka, N., Frolova, G. M., and Mikhailov, V. V. (2009). Psychrobacter fulvigenes sp. nov., isolated from a marine crustacean from the sea of Japan. Int. J. Syst. Evol. Microbiol. 59, 1480–1486. doi: 10.1099/ijs.0.007195-0
Rowland, I., Gibson, G., Heinken, A., Scott, K., Swann, J., Thiele, I., et al. (2018). Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24. doi: 10.1007/s00394-017-1445-8
Ruvira, M. A., Lucena, T., Pujalte, M. J., Arahal, D. R., and Macián, M. C. (2013). Marinifilum flexuosum sp. nov., a new Bacteroidetes isolated from coastal Mediterranean Sea water and emended description of the genus Marinifilum Na et al., 2009. Syst. Appl. Microbiol. 36, 155–159. doi: 10.1016/j.syapm.2012.12.003
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schroeder, B., and Bäckhed, F. (2016). Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22, 1079–1089. doi: 10.1038/nm.4185
Sharma, S., Chaturvedi, J., Chaudhari, B. P., Singh, R. L., and Kakkar, P. (2012). Probiotic enterococcus lactis IITRHR1 protects against acetaminophen-induced hepatotoxicity. Nutrition 28, 173–181. doi: 10.1016/j.nut.2011.02.012
Smith, P., Willemsen, D., Popkes, M., Metge, F., Gandiwa, E., Reichard, M., et al. (2017). Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. elife 6:e27014. doi: 10.7554/eLife.27014
Song, Y., Shen, H., Liu, T., Pan, B., De Alwis, S., Zhang, W., et al. (2021). Effects of three different mannans on obesity and gut microbiota in high-fat diet-fed C57BL/6J mice. Food Funct. 12, 4606–4620. doi: 10.1039/D0FO03331F
Sonnenburg, E. D., Smits, S. A., Tikhonov, M., Higginbottom, S. K., Wingreen, N. S., and Sonnenburg, J. L. (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. doi: 10.1038/nature16504
Sonnenburg, E. D., and Sonnenburg, J. L. (2014). Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 20, 779–786. doi: 10.1016/j.cmet.2014.07.003
Soverini, M., Quercia, S., Biancani, B., Furlati, S., Turroni, S., Biagi, E., et al. (2016). The bottlenose dolphin (Tursiops truncatus) faecal microbiota. FEMS Microbiol. Ecol. 92:fiw055. doi: 10.1093/femsec/fiw055
Stephan, R., Lehner, A., Tischler, P., and Rattei, T. (2011). Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 193, 309–310. doi: 10.1128/JB.01162-10
Sun, Y. Z., Yang, H. L., Ma, R. L., Zhang, C. X., and Lin, W. Y. (2011). Effect of dietary administration of Psychrobacter sp. on the growth, feed utilization, digestive enzymes and immune responses of grouper Epinephelus coioides. Aquac. Nutr. 17, e733–e740. doi: 10.1111/j.1365-2095.2010.00837.x
Suzuki, A., Segawa, T., Sawa, S., Nishitani, C., Ueda, K., Itou, T., et al. (2019). Comparison of the gut microbiota of captive common bottlenose dolphins Tursiops truncatus in three aquaria. J. Appl. Microbiol. 126, 31–39. doi: 10.1111/jam.14109
Thomas, F., Hehemann, J. H., Rebuffet, E., Czjzek, M., and Michel, G. (2011). Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2:93. doi: 10.3389/fmicb.2011.00093
Tian, J., Du, J., Zhang, S., Li, Y., Gao, X., Han, J., et al. (2021). Age-associated variation in the gut microbiota of chinstrap penguins (Pygoscelis antarctica) reveals differences in food metabolism. Microbiology 10:e1190. doi: 10.1002/mbo3.1190
Tidjani Alou, M., Khelaifia, S., La Scola, B., and Cassir, N. (2016b). Lachnoclostridium touaregense, a new bacterial species isolated from the human gut microbiota. New Microbes New Infect. 14, 51–52. doi: 10.1016/j.nmni.2016.07.007
Tidjani Alou, M., Lagier, J. C., La Scola, B., and Cassir, N. (2016a). Lachnoclostridium massiliosenegalense, a new bacterial species isolated from the human gut microbiota. New Microbes New Infect. 14, 4–5. doi: 10.1016/j.nmni.2016.07.014
Traore, S. I., Azhar, E. I., Yasir, M., Bibi, F., Fournier, P. E., Jiman-Fatani, A. A., et al. (2017). Description of Blautia phocaeensis sp. nov. and Lachnoclostridium edouardi sp. nov., isolated from healthy fresh stools of Saudi Arabia Bedouins by culturomics. New Microbes New Infect. 19, 129–131. doi: 10.1016/j.nmni.2017.05.017
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2015). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15
Wells, R. S., and Scott, M. D. (1999). “Bottlenose dolphin Tursiops truncatus (Montagu, 1821),” in Handbook of Marine Mammals, vol. 6, the Second Book of Dolphins and Porpoises. eds. S. H. Ridgway and R. Harrison (San Diego, California: Academic Press), 137–183.
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Wigley, P. (2015). Blurred lines: pathogens, commensals, and the healthy gut. Front. Vet. Sci. 2:40. doi: 10.3389/fvets.2015.00040
Wylensek, D., Hitch, T. C. A., Riedel, T., Afrizal, A., Kumar, N., Wortmann, E., et al. (2020). A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat. Commun. 11:6389. doi: 10.1038/s41467-020-19929-w
Xu, Z. X., Mu, X., Zhang, H. X., Chen, G. J., and Du, Z. J. (2016). Marinifilum albidiflavum sp. nov., isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 66, 4589–4593. doi: 10.1099/ijsem.0.001395
Yang, H. L., Sun, Y. Z., Ma, R. L., Li, J. S., and Huang, K. P. (2011). Probiotic Psychrobacter sp. improved the autochthonous microbial diversity along the gastrointestinal tract of grouper Epinephelus coioides. J. Aquac. Res. Dev. S1:001. doi: 10.4172/2155-9546.S1-001
Yaskolka Meir, A., Rinott, E., Tsaban, G., Zelicha, H., Kaplan, A., Rosen, P., et al. (2021). Effect of green-Mediterranean diet on intrahepatic fat: the DIRECT PLUS randomised controlled trial. Gut. doi: 10.1136/gutjnl-2020-323106 [Epub ahead of print]
Yu, M., Jia, H., Zhou, C., Yang, Y., Zhao, Y., Yang, M., et al. (2017). Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J. Pharm. Biomed. Anal. 138, 231–239. doi: 10.1016/j.jpba.2017.02.008
Yuan, Y., Zhang, Y. L., Zhang, P. J., Liu, C., Wang, J. H., Gao, H. Y., et al. (2021). Comparative genomics provides insights into the aquatic adaptations of mammals. Proc. Natl. Acad. Sci. U S A 118:e2106080118. doi: 10.1073/pnas.2106080118
Yue, C., Li, M., Li, J., Han, X., Zhu, H., Yu, G., et al. (2020). Medium-, long- and medium-chain-type structured lipids ameliorate high-fat diet-induced atherosclerosis by regulating inflammation, adipogenesis, and gut microbiota in ApoE−/− mice. Food Funct. 11, 5142–5155. doi: 10.1039/D0FO01006E
Zachariah, S., Kumari, P., and Das, S. K. (2016). Psychrobacter pocilloporae sp. nov., isolated from a coral, Pocillopora eydouxi. Int. J. Syst. Evol. Microbiol. 66, 5091–5098. doi: 10.1099/ijsem.0.001476
Zeng, Q., Li, D., He, Y., Li, Y., Yang, Z., Zhao, X., et al. (2019). Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 9:13424. doi: 10.1038/s41598-019-49462-w
Zeng, Y. X., Yu, Y., Li, H. R., and Luo, W. (2015). Psychrobacter fjordensis sp. nov., a psychrotolerant bacterium isolated from an Arctic fjord in Svalbard. Antonie Van Leeuwenhoek 108, 1283–1292. doi: 10.1007/s10482-015-0580-6
Zeng, Y. X., Yu, Y., Liu, Y., and Li, H. R. (2016). Psychrobacter glaciei sp. nov., isolated from the ice core of an Arctic glacier. Int. J. Syst. Evol. Microbiol. 66, 1792–1798. doi: 10.1099/ijsem.0.000939
Keywords: gut microbial communities, beluga, Pacific white-sided dolphin, common bottlenose dolphin, Cape fur seal
Citation: Bai S, Zhang P, Zhang C, Du J, Du X, Zhu C, Liu J, Xie P and Li S (2021) Comparative Study of the Gut Microbiota Among Four Different Marine Mammals in an Aquarium. Front. Microbiol. 12:769012. doi: 10.3389/fmicb.2021.769012
Received: 01 September 2021; Accepted: 23 September 2021;
Published: 21 October 2021.
Edited by:
Jinbo Xiong, Ningbo University, ChinaReviewed by:
Tongtong Li, Zhejiang University of Technology, ChinaCopyright © 2021 Bai, Zhang, Zhang, Du, Du, Zhu, Liu, Xie and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peijun Zhang, cGp6aGFuZ0BpZHNzZS5hYy5jbg== Songhai Li, bGlzaEBpZHNzZS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.