
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 14 October 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.768283
This article is part of the Research Topic Advances in Bioleaching and Bioremediation View all 21 articles
 Li-Jun Liu1,2
Li-Jun Liu1,2 Zhen Jiang3,4
Zhen Jiang3,4 Pei Wang3,4
Pei Wang3,4 Ya-Ling Qin3,4
Ya-Ling Qin3,4 Wen Xu1,2
Wen Xu1,2 Yang Wang1,2
Yang Wang1,2 Shuang-Jiang Liu3,4*
Shuang-Jiang Liu3,4* Cheng-Ying Jiang3,4*
Cheng-Ying Jiang3,4*The order Sulfolobales (phylum Crenarchaeota) is a group of thermoacidophilic archaea. The first member of the Sulfolobales was discovered in 1972, and current 23 species are validly named under the International Code of Nomenclature of Prokaryotes. The majority of members of the Sulfolobales is obligately or facultatively chemolithoautotrophic. When they grow autotrophically, elemental sulfur or reduced inorganic sulfur compounds are their energy sources. Therefore, sulfur metabolism is the most important physiological characteristic of the Sulfolobales. The functions of some enzymes and proteins involved in sulfur reduction, sulfur oxidation, sulfide oxidation, thiosulfate oxidation, sulfite oxidation, tetrathionate hydrolysis, and sulfur trafficking have been determined. In this review, we describe current knowledge about the physiology, taxonomy, and sulfur metabolism of the Sulfolobales, and note future challenges in this field.
On the basis of analysis of 16S (18S) rRNA gene sequences, Woese proposed in 1977 that archaebacteria are a different group from eubacteria and eukaryotes (Woese and Fox, 1977). In 1990, life on Earth was then divided into three domains: Bacteria, Archaea, and Eukarya. Crenarchaeota, one of the original phyla of the Archaea, mainly comprise sulfur-dependent thermoacidophiles (Woese et al., 1990).
Sulfolobales are an order within the class Thermoprotei, phylum Crenarchaeota, superphylum TACK (Stetter, 1989; Guy and Ettema, 2011). Since the first member of the Sulfolobales was isolated and identified in 1972, only one family, the Sulfolobaceae was constructed, which included nine validly described genera: Acidianus, Metallosphaera, Saccharolobus, Stygiolobus, Sulfodiicoccus, Sulfolobus, Sulfuracidifex, Sulfurisphaera, and Sulfurococcus (Brock et al., 1972; Segerer et al., 1986, 1991; Huber et al., 1989; Karavaĭko et al., 1994; Kurosawa et al., 1998; Sakai and Kurosawa, 2017, 2018; Itoh et al., 2020). Several species of the Sulfolobales have been reassigned to new phylogenetic position according to phylogenetic data and physiological characters during these years, which are detailed below. Members of the Sulfolobales grow autotrophically by oxidizing elemental sulfur (S0), hydrogen (H2), sulfidic ores, and reduced inorganic sulfur compounds (RISCs), such as thiosulfate. Heterotrophic growth occurs by aerobic respiration, anaerobic sulfur respiration, or by fermentation of organic substrates (Huber and Prangishvili, 2006).
The element sulfur exists in various chemical valence ranging from −2 to +6, and RISCs include sulfides (S2−, HS−, and H2S), polysulfide (−S-Sn-S−), elemental sulfur (S0), sulfite (SO32−), thiosulfate (S2O32−), and tetrathionate (S4O62−). Because of the diversity of the available forms of sulfur, many enzymes and proteins exist in Sulfolobales for sulfur metabolism, including sulfur-reducing enzymes, sulfur-oxidizing enzymes, sulfur carrier proteins, and sulfur transferases, which cooperate with each other as shown in Figure 1. The sulfur metabolism summarized here contains the reduction of S0, the oxidation of RISCs (including sulfide, S0, S2O32−, and SO32−), hydrolysis of S4O62−, and sulfur trafficking. The investigation of functions of these enzymes and proteins in sulfur metabolism is one of the main research aspects regarding the Sulfolobales. Significant research progress has been made over the past decades.
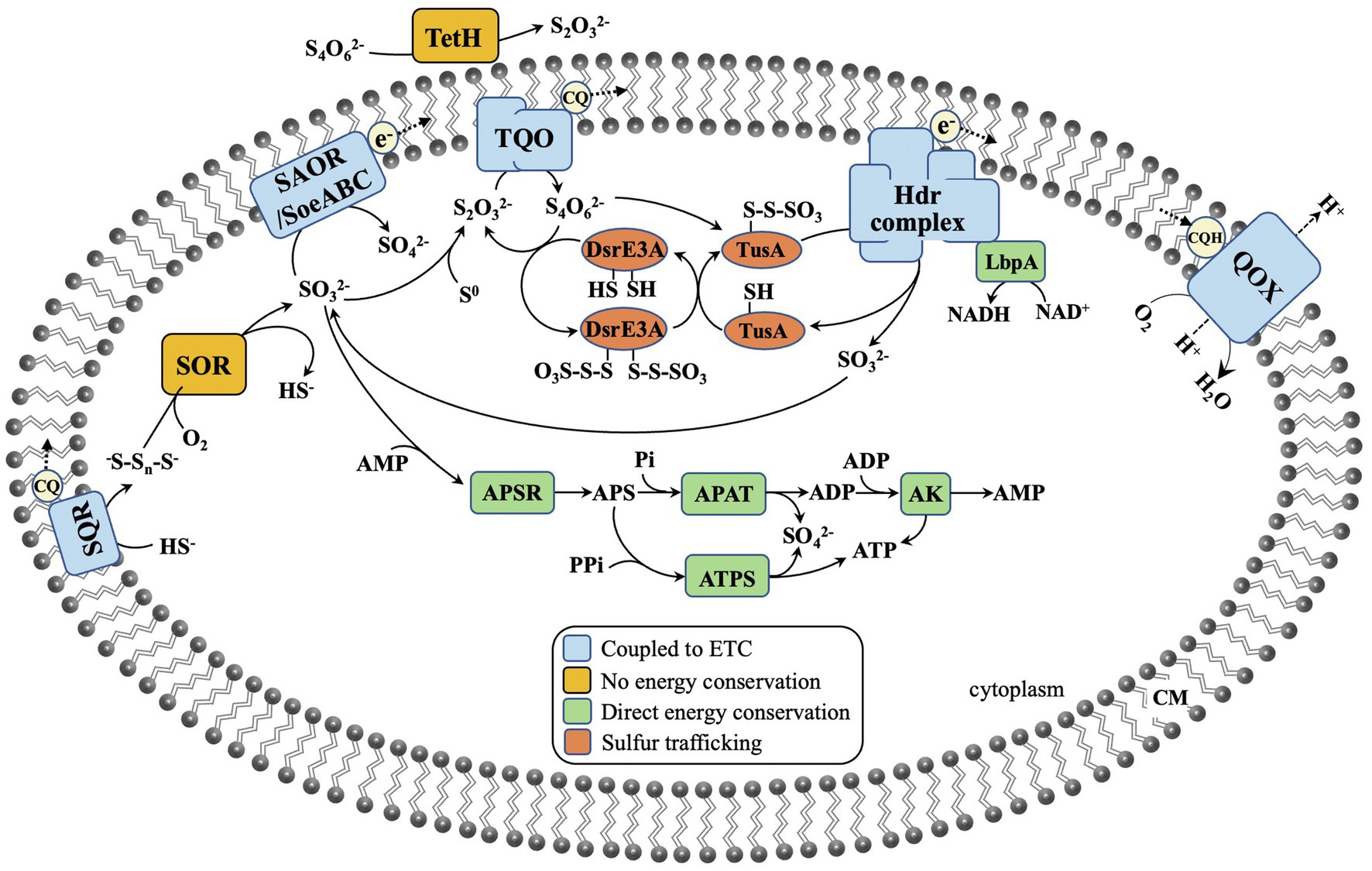
Figure 1. The proposed sulfur oxidation pathway in Sulfolobales. SQR, sulfide:quinone oxidoreductase; SOR, sulfur oxygenase reductase; SAOR, sulfite: acceptor oxidoreductase; SoeABC, sulfite oxidizing enzyme; TetH, tetrathionate hydrolase; TQO, thiosulfate: quinone oxidoreductase; Hdr, heterodisulfide reductase; LbpA, lipoate-binding protein A; Dsr, disulfide reductase; Tus, tRNA 2-thiouridine synthesizing protein; APSR, APS reductase; APAT, adenylylsulfate: phosphate adenylyltransferase; ATPS, ATP sulfurylase; AK, adenylate kinase; CQ, caldariellaquinones; and QOX, quinone oxidoreductase.
The first complete genome of a member of the Sulfolobales, Saccharolobus solfataricus P2, was published in 2001 (She et al., 2001). Now, complete genomic data are available for 69 strains within eight genera (except Sulfurococcus) in the order Sulfolobales. From genomic information and enzyme activity analysis, we can better understand the characteristics of sulfur metabolism in these organisms. Here, we summarize the key points to provide a clearer understanding of the taxonomy of Sulfolobales and sulfur oxidation in these organisms.
Nine genera have been identified in the order Sulfolobales. The first, Sulfolobus, was described in 1972 (Brock et al., 1972). In recent years, some species were reclassified to new genera based on morphology, physiology, and phylogenetic evidence. Herein, we summarize the latest progress in taxonomy within the Sulfolobales. The main features of the nine genera and the main species within each genus are described below and in Tables 1 and 2.
The genus Sulfolobus was established in 1972 and is the type genus of the order Sulfolobales. Members of the Sulfolobus were first isolated from acid thermal soils and acid hot springs in Yellowstone National Park (United States), El Salvador, Dominica, and Italy (Brock et al., 1972). Eight species of Sulfolobus have been characterized, described, and validly named under the International Code of Nomenclature of Prokaryotes (ICNP)1; however, six of them were later reassigned to other genera. The type species is Sulfol. acidocaldarius, which was isolated from Locomotive Spring in Yellowstone National Park. Sulfol. yangmingensis and Sulfol. tengchongensis (which is not validly named) were isolated from a geothermal vent in Yang-Ming National Park in northern Taiwan, and an acidic hot spring in Tengchong, Yunnan, China, respectively (Jan et al., 1999; Xiang et al., 2003). Two species, Sulfol. islandicus isolated from Icelandic solfataras (Zillig et al., 1993; Reno et al., 2009) and Sulfol. sp. A20 isolated from a hot spring in Costa Rica (Dai et al., 2016), were also described and sequenced, although they were not validly named under the ICNP.
Cells of Sulfolobus are irregular cocci with frequent lobes, with diameter 0.8–1.5μm. Cells grow in the temperature range 55–95°C (optimal 65–85°C) and pH range 1.0–6.5 (optimal 2.0–4.0). Aerobic and facultatively chemolithoautotrophic growth occurs on S0 or a variety of complex organic compounds and sugars. Anaerobic growth of this genus has not been detected (Jan et al., 1999). The type strain Sulfol. acidocaldarius cannot oxidize elemental sulfur autotrophically in aerobic conditions (Brock et al., 1972; Huber et al., 1989; Huber and Prangishvili, 2006; Sakai and Kurosawa, 2018). Corresponding to this, genes encoding sulfur oxygenase for sulfur oxidation were not found in its genome (Sakai and Kurosawa, 2018).
Metallosphaera contains five members with valid name. The type species M. sedula was isolated from a continental solfataric field in Italy (Huber et al., 1989). Metallosphaera prunae was isolated from a smoldering slag heap of a uranium mine in Thuringen (Germany; Fuchs et al., 1995), and M. hakonensis was isolated from an acidic hot spring at a geothermal area in Hakone (Japan; Takayanagi et al., 1996; Kurosawa, 2003). Metallosphaera cuprina (Liu et al., 2011a) and M. tengchongensis (Peng et al., 2015) were both isolated from sulfuric hot springs in Tengchong (Yunnan, China). Metallosphaera hakonensis originally belonged to Sulfolobus, but it was reclassified to Metallosphaera in 2003 by Kurosawa (2003) based on phylogenetic evidence, DNA G+C content, and phenotypic properties (Table 2; Takayanagi et al., 1996; Kurosawa, 2003). Metallosphaera yellowstonensis was isolated from Yellowstone National Park and introduced in 2011 (Kozubal et al., 2011), which has not been validly named under the ICNP.
Cells of Metallosphaera are cocci or irregular cocci with diameter 0.8–1.2μm. Growth happens at 50–80°C (optimal 65–75°C), and pH 1.0–6.5 (optimal 2.5–3.5). Metallosphaera are aerobic and facultatively chemolithoautotrophic (Table 1). They can extract metal ions from several kinds of sulfidic ore, such as pyrite, chalcopyrite, and sphalerite. They can also oxidize S0 to sulfate but cannot reduce S0 (with or without the presence of H2). Heterotrophic growth occurs on complex organic compounds, such as beef extract, casamino acids, peptone, tryptone, and yeast extract. Metallosphaera cannot use sugars and amino acids (with the exception of M. cuprina, which can use a few types of sugar and amino acid, such as D-glucose, D-xylose, L-arabinose, and L-tryptophan; Table 2; Huber et al., 1989; Fuchs et al., 1995; Takayanagi et al., 1996; Liu et al., 2011a; Peng et al., 2015).
The genus Sulfuracidifex was proposed by Itoh in 2020. The type species is Sulfura. tepidarius, which was isolated from a solfataric field at Hakone, Japan (Itoh et al., 2020). Another member is Sulfura. metallicus, isolated from continental solfataric fields in Iceland (Huber and Stetter, 1991). Sulfura. metallicus was reclassified from Sulfolobus because its phenotypic properties and 16S rRNA gene sequences are closer to those of Sulfura. tepidarius than to other members of the order Sulfolobales (Itoh et al., 2020).
Cells of Sulfuracidifex are irregular cocci with diameter 0.8–1.2μm. Growth occurs at 45–75°C (optimal around 65°C), and at pH 0.4–5.5 (optimal 2.5–3.5). Sulfuracidifex is obligate aerobes. Cells grow autotrophically on S0, reduced sulfur compounds, or sulfide ores. When grown mixotrophically, Sulfura. tepidarius uses several complex organics and sugars, whereas Sulfura. metallicus uses only yeast extract or glycogen as carbon sources (Tables 1 and 2; Huber and Stetter, 1991; Itoh et al., 2020).
The type species of genus Sulfodiicoccus is Sulfod. acidiphilus, isolated from the Hakone Ohwaku-dani hot spring in Japan in 2017 (Sakai and Kurosawa, 2017). At present, it is the only member of the Sulfodiicoccus. Growth of the species is significantly inhibited in the presence of S0. The DNA G+C content is 52.0mol%, which is remarkably higher than that of the other known species of the Sulfolobales (30.6–46.2mol%; Sakai and Kurosawa, 2017).
Cells of Sulfodiicoccus are cocci to irregular cocci with diameter 0.8–1.5μm. Cells grow at 50–70°C (optimal 65–70°C), pH 1.4–5.5 (optimal 3.0–3.5), and 0–2.5% (w/v) NaCl. Sulfod. acidiphilus is strictly aerobic and heterotrophic. Growth occurs on various complex substrates or sugars as carbon sources. Chemolithoautotrophic growth does not occur by oxidation of S0, pyrite, K2S4O6, Na2S2O3, or FeSO4·7H2O, or on H2 (Sakai and Kurosawa, 2017).
There are six species described in the genus Acidianus: A. ambivalens (Zillig et al., 1986; Fuchs et al., 1996), A. brierleyi (Segerer et al., 1986), A. infernus (Segerer et al., 1986), A. manzaensis (Yoshida et al., 2006), A. sulfidivorans (Plumb et al., 2007), and A. tengchongenses, which is not validly named (He et al., 2004). Among them, A. infernus is the type species of Acidianus (Segerer et al., 1986). Acidianus ambivalens was previously named Desulfurolobus ambivalens (Zillig et al., 1986), but, as it is very similar to A. infernus in physiological and biochemical features, it was reassigned to the genus Acidianus (Fuchs et al., 1996). Members of the Acidianus occur in acidic solfataras and marine hydrothermal systems. Cells of Acidianus are irregular cocci with diameter 0.5–2.0μm. Cells grow at 45–96°C (optimal 70–90°C), pH 1.0–6.0 (optimal 0.8–2.5), and 0.1–4% (w/v) NaCl. Chemolithoautotrophic growth occurs aerobically by means of S0 oxidation or anaerobically via S0 reduction with H2 as electron donor (Tables 1 and 2). Acidianus infernus (Segerer et al., 1986), A. ambivalens (Zillig et al., 1986), and A. tengchongenses (He et al., 2004) are obligately chemolithotrophic. A. brierleyi (Segerer et al., 1986), A. manzaensis (Yoshida et al., 2006), and A. sulfidivorans (Plumb et al., 2007) are facultatively autotrophic and can grow heterotrophically on yeast extract in the absence of S0 in aerobic conditions.
The genus Sulfurisphaera contains three species at present, Sulfuri. javensis (Tsuboi et al., 2018), Sulfuri. ohwakuensis (Kurosawa et al., 1998), and Sulfuri. tokodaii (Suzuki et al., 2002), which were all isolated from acidic hot springs (Kurosawa et al., 1998; Suzuki et al., 2002; Tsuboi et al., 2018). Sulfuri. tokodaii formerly belonged to Sulfolobus but was reclassified to Sulfurisphaera by Tsuboi et al. (2018) based on the latest phylogenetic data (Suzuki et al., 2002; Tsuboi et al., 2018). The type species of this genus is Sulfuri. ohwakuensis (Kurosawa et al., 1998).
Cells of Sulfurisphaera are irregular cocci with diameter approximately 1μm and grow at 60–96°C (optimal 80–84°C), pH 1.5–6.0 (optimal 2.0–4.0), and 0–1.5% (w/v) NaCl. Cells are facultatively anaerobic. Anaerobic growth occurs on FeCl3 in the presence of yeast extract. Chemolithoautotrophic growth occurs on S0, S4O62−, and pyrite in aerobic conditions. The G+C content is in the range 30.6–33.7mol% (Tsuboi et al., 2018).
The type species of the genus is Sa. solfataricus, which was first described by Zillig et al. (1980). The other two species are Sa. shibatae and Sa. caldissimus (Sakai and Kurosawa, 2018). Sa. solfataricus and Sa. shibatae were originally classified into the genus Sulfolobus (Zillig et al., 1980; Grogan et al., 1990). However, later study demonstrated that their abilities to use various sugars were quite different from that of Sulfol. acidocaldarius, the type species of Sulfolobus. The growth temperature and pH, and facultatively anaerobic characteristics, of Sa. solfataricus and Sa. shibatae are almost identical to those of Sa. caldissimus. Phylogenetic evidence based on 16S rRNA and 23S rRNA gene sequences also helped distinguish Sa. solfataricus, Sa. shibatae, and Sa. caldissimus from Sulfol. acidocaldarius. Therefore, Sa. solfataricus and Sa. shibatae were reclassified as Saccharolobus (Sakai and Kurosawa, 2018).
Cells of Saccharolobus are irregular cocci. The temperature and pH ranges for growth are 50–93°C and pH 1.5–6.0 (optima 80–85°C and 3.0–4.5), respectively. Cells are facultatively anaerobic, using FeCl3 as an electron acceptor and yeast extract as an electron donor. Heterotrophic growth occurs on complex substrates, such as yeast extract and various kinds of sugar. Chemolithoautotrophic growth occurs on pyrite or, poorly, by oxidation of H2. S0 and K2S4O6 cannot be used as electron donors. The G+C content of this genus is in the range 31.7–35.8mol% (Sakai and Kurosawa, 2018).
Stygiolobus isolates were obtained from solfataric fields in the Azores and described by Segerer et al. (1991). The type species is S. azoricus, the only member of Stygiolobus, which is an obligate anaerobe. Cells are irregular cocci or lobed and are approximately 0.5–1.8μm wide in exponential growth phase, and frequently surrounded by pilus- or fimbria-like appendages. The growth temperature and pH ranges are 57–89°C and 1.0–5.5 (optimum around 80°C and 2.5–3.0), respectively. Stygiolobus azoricus is obligately chemolithotroph and grow by means of H2–S0 lithotrophy. Growth was stimulated by a trace amount of yeast extract (0.005–0.02%; Segerer et al., 1991).
The genus Sulfurococcus contains two species: Sulfuro. mirabilis and Sulfuro. yellowsonensis. The original characterization manuscripts are both in Russian. However, the abstract (written in English) states that Sulfuro. yellowsonensis was isolated from the hydrotherm of Yellowstone National Park (United States) and is a spherical, sulfur-oxidizing thermoacidophile. It grows at 40–80°C. Sulfuro. yellowsonensis is a facultative autotroph that grows autotrophically by oxidizing elemental sulfur, ferrous sulfate, and sulfide minerals, and heterotrophically on organic compounds. The DNA G+C content is 44.6mol% (Karavaĭko et al., 1994).
All members of the order Sulfolobales are acidothermophiles. Most of them were isolated from terrestrial or aquatic solfatara aeras, which are hot and acidic. They have many phenotypic characteristics in common, but also numerous differences. The phylogenetic relationships of some species were revised in recent years. The main characteristics of the nine genera in the order Sulfolobales are listed in Tables 1 and 2.
As shown in Table 2, the cells of all Sulfolobales are cocci or irregular cocci, 0.5–2.2μm in diameter. However, they have diverse O2 requirements and nutrition types. In general, Sulfolobus, Metallosphaera, Sulfuracidifex, and Sulfodiicoccus are obligate aerobes. Acidianus, Sulfurisphaera, and Saccharolobus are facultative anaerobes. Stygiolobus is the only obligate anaerobic genus within the Sulfolobales. Most members within Sulfolobales are facultatively chemolithoautotrophic, but Sulfodiicoccus species are heterotrophs (Sakai and Kurosawa, 2017). Stygiolobus and some species of Acidianus are obligately chemolithoautotrophic.
Compared with members of Sulfolobus, most Metallosphaera have greater ability to oxidize RISCs, such as S0, S4O62−, and sulfidic ores, but lesser ability to use sugars. The members of Sulfolobus can use different types of sugar. However, compared with Sa. solfataricus, which shows high metabolic versatility and is able to use a broad spectrum of substrates, including mono-, di-, oligo-, and polysaccharides, Sulfol. acidocaldarius has a much narrower substrate spectrum. This could be attributed to its relatively small genome, which lacks numerous transport systems for substrate uptake (Lewis et al., 2021).
Both Sulfurisphaera and Saccharolobus are facultatively anaerobic and facultatively chemolithoautotrophic. They have similar abilities in using FeCl3 as an electron acceptor in anaerobic conditions, while their abilities to use RISCs vary. In the case of Sulfurisphaera, chemolithoautotrophic growth occurs on various kinds of RISC in aerobic conditions. However, Saccharolobus can only oxidize pyrite poorly (Kurosawa et al., 1998; Sakai and Kurosawa, 2018). Although Zillig et al. (1980) described that Sa. solfataricus can use S0 as an energy source, the data of Sakai and Kurosawa (2018) indicate that this species cannot use pyrite or S0 at all (Zillig et al., 1980; Sakai and Kurosawa, 2018).
With the increasing number of isolates and phylogenetic data, several species of Sulfolobales have been taxonomically reclassified. Sulfolobus was the first described genus of the Sulfolobales. Six species that were originally classified as Sulfolobus species – A. brierleyi (Zillig et al., 1980), M. hakonensis (Takayanagi et al., 1996), Sa. solfataricus (Zillig et al., 1980), Sa. shibatae (Grogan et al., 1990), Sulfura. metallicus (Huber and Stetter, 1991), and Sulfuri. tokodaii (Suzuki et al., 2002) – have been reclassified into new genera according to later physiological and phylogenetic evidences (Segerer et al., 1986; Kurosawa, 2003; Sakai and Kurosawa, 2018; Tsuboi et al., 2018; Itoh et al., 2020; Table 2; Figures 2, 3).
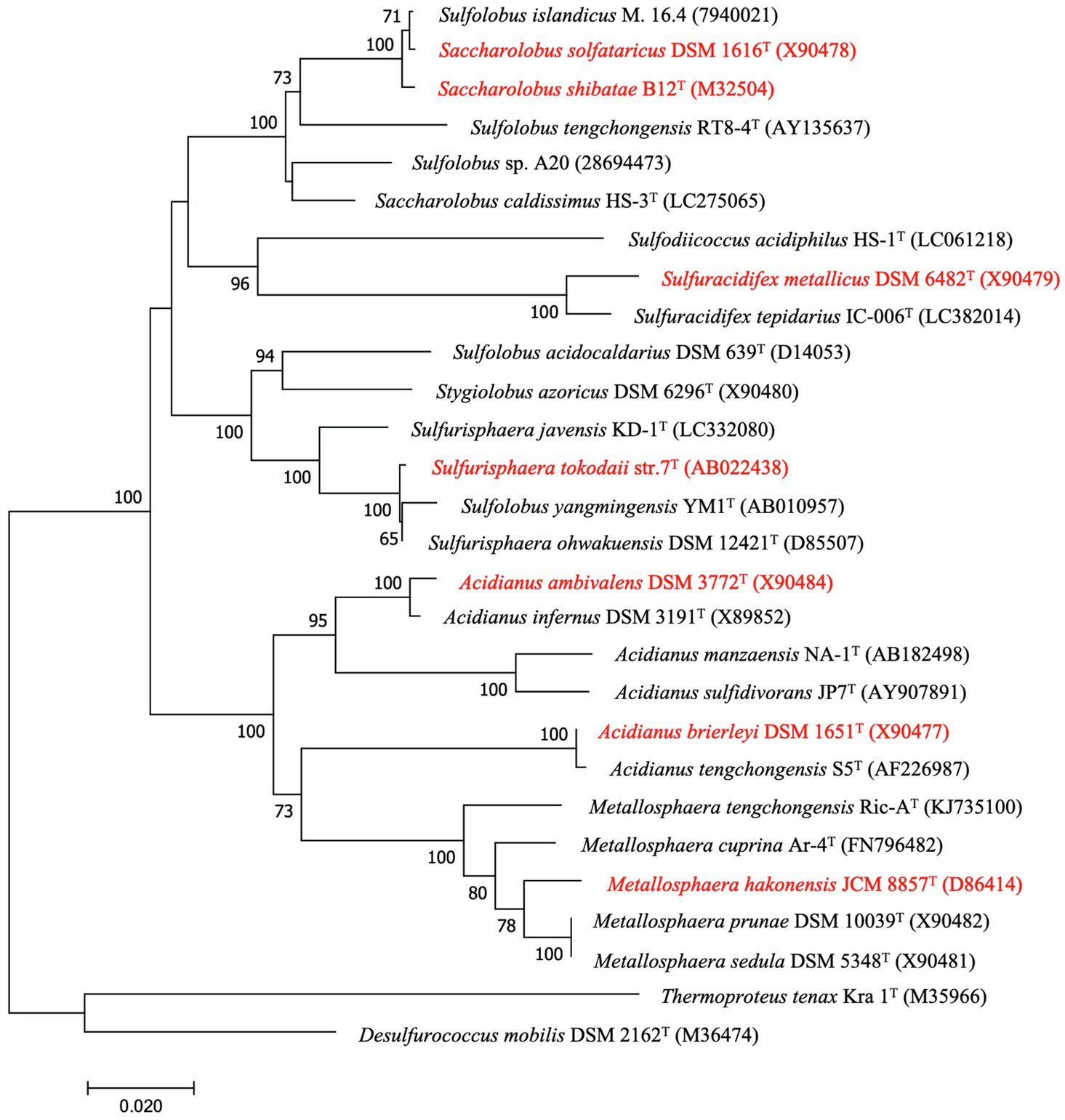
Figure 2. Neighbor-joining phylogenetic tree of the members of Sulfolobales described in this manuscript based on 16S rRNA gene sequences by Mega 7. Numbers at branch nodes represent confidence levels based on 1,000 replicates bootstrap samplings (values greater than 50% are shown), Bar, 0.02 substitutions per nucleotide position. GenBank accession numbers are given in parentheses. The reclassified species are highlighted in red.
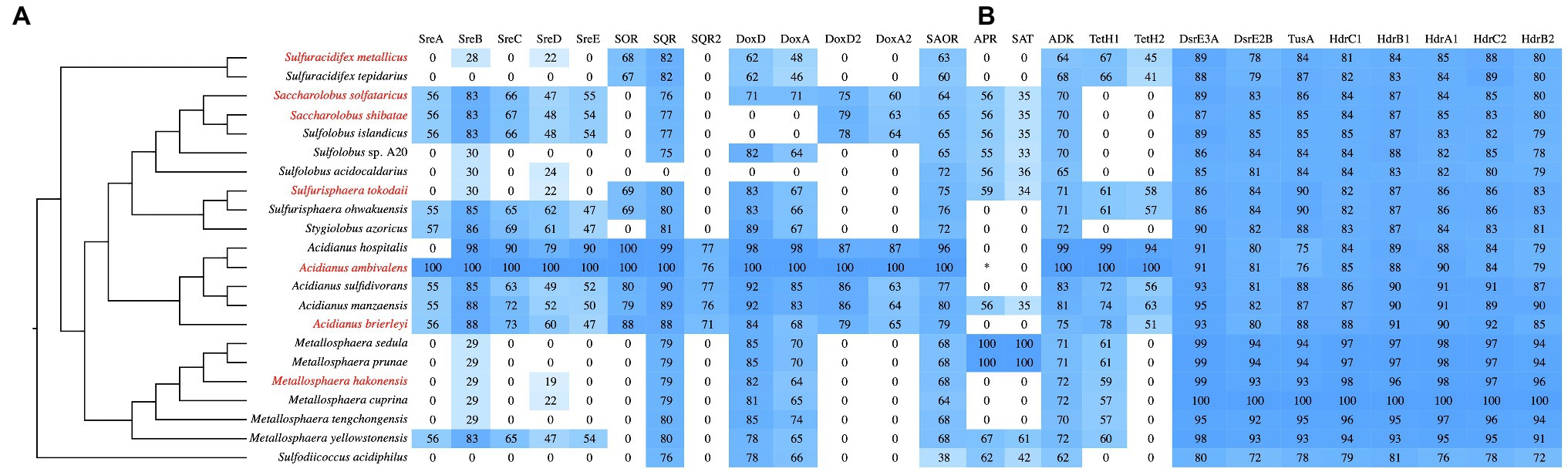
Figure 3. Phylogenetic relationship of the proteins related to sulfur metabolism in the Sulfolobales. (A) Phylogenetic tree based on whole-genome sequences constructed by using a Composition Vector (CV) approach (Qi et al., 2004; Zuo and Hao, 2015). K-tuple length: 5. The reclassified species are highlighted in red. (B) Protein related to sulfur metabolism within the Sulfolobales and their percentage identity at the amino acid level (blastp, coverage >80%). Acidianus ambivalens, Metallosphaera sedula, and Metallosphaera cuprina proteins are used as queries. Sre, sulfur reductase; SOR, sulfur oxygenase reductase; SQR, sulfide:quinone oxidoreductase; DoxD, thiosulfate:quinone oxidoreductase (TQO) small subunit; DoxA, TQO large subunit; SAOR, sulfite:acceptor oxidoreductase; APR, APS reductase; SAT, ATP sulfurylase; ADK, adenylate kinase; TetH, tetrathionate dehydrogenase; Dsr, disulfide reductase; Tus, tRNA 2-thiouridine synthesizing protein; and Hdr, heterodisulfide reductase. *, the APR activity was detected in Acidianus ambivalens (Zimmermann et al., 1999), while no homologous sequence was found when use APR sequence of Metallosphaera sedula as query.
Although features, such as morphology, temperature and pH for growth, O2 requirements, and nutrition types of Sulfurisphaera, resemble those of the Acidianus, phylogenetic analyses including 16S rRNA gene similarities and DNA–DNA hybridization data distinguish it from the other genera of the Sulfolobales (Segerer et al., 1986; Kurosawa et al., 1998; Tsuboi et al., 2018).
As the phylogenetic tree in Figures 2, 3 show, members of the other genera of Sulfolobales each cluster together or closely, with the exception of the members of Sulfolobus, which are dispersed in different clusters. Notably, the 16S rRNA gene sequence of Sulfol. yangmingensis is more similar to that of Sulfuri. tokodaii and Sulfuri. ohwakuensis, and these three species form a clade in the phylogenetic tree (Figure 2). Furthermore, the use of organic compounds and RISCs by Sulfol. yangmingensis is also similar to that by Sulfurisphaera, although the G+C content of Sulfol. yangmingensis (42%) is much higher than that of Sulfurisphaera spp. (30–33%; Kurosawa et al., 1998; Jan et al., 1999; Tsuboi et al., 2018). Sulfol. islandicus, Sulfol. tengchongensis, and Sulfol. sp. A20, have been charactered or sequenced but not validly named, are all far from the type strain Sulfol. acidocaldarius but related to the clade containing Saccharolobus, according to phylogenetic analysis based on 16S rRNA gene sequences and whole genome sequences (Figures 2, 3). Average amino acid identity (AAI) and conserved multi-locus sequence alignment (MLSA) also indicate that Sulfol. acidocaldarius is distinct from the other species of Sulfolobus (Counts et al., 2021).
In addition, AAI and MLSA data for Sulfolobales suggest that Sulfod. acidophilus should be classified into a new family (Counts et al., 2021). All these observations indicate that the phylogenetic positions of the members of Sulfolobales need to be reconsidered.
Sulfur metabolism is an important physiological process of many members of Sulfolobales. From genomic information and enzyme activity analysis, several enzymes and proteins related with the metabolism of different RISC have been recognized.
Reduction of elemental sulfur is universal among hyperthermophilic archaea. Three genera of Sulfolobales – Acidianus, Sulfurisphaera, and Stygiolobus – reduce S0 to H2S with H2 as the electron donor (Table 2). Two membrane-bound, multisubunit enzymes are involved in S0 reduction in Acidianus: sulfur reductase (SR) and NiFe hydrogenase. SR is composed of five subunits encoded by the sreABCDE gene cluster: a large subunit (SreA), an Fe–S-cluster-containing subunit (SreB), a membrane-anchor subunit (SreC), and SreD and SreE, whose functions are unknown. Both SreA and SreB share sequence similarity with molybdopterin oxidoreductases belonging to the dimethylsulfoxide reductase family. sreABC gene clusters are found in Sulfol. islandicus, A. ambivalens, A. brierleyi, A. manzaensis, A. sulfidivorans, and Sa. solfataricus. The NiFe hydrogenase is encoded by an operon with 12 open reading frames, hynS–isp1–isp2–hynL–hynYZ–hypDCE–hypYZ–hoxM. HynS, HynL, and Isp1 are the small subunit and large subunit of the hydrogenase and the membrane-anchor protein, respectively. HynS and HynL contain [NiFe] and Fe–S clusters, respectively. HypDCE and HoxM are proteins required for hydrogenase maturation. Isp2, HynYZ, and HypYZ are proteins with unknown functions. Electron transfer between NiFe hydrogenase and SR is probably mediated by quinones in Acidianus (Laska, 2003).
Sulfides (S2−, HS−, and H2S) are widely distributed in soils, ore, wastewater, and marine environments. They are produced partly from mineral deposits, and partly by biological metabolism, including as products of eukaryotic and prokaryotic endogenous catabolism of cysteine and iron–sulfur proteins and dissimilatory metabolism of sulfur-containing inorganic compounds (Kabil and Banerjee, 2010; Lencina et al., 2013; Gao et al., 2017; Bełtowski, 2019). H2S is an important electron donor in prokaryotes, such as phototrophic or chemotrophic microorganisms (Reinartz et al., 1998; Sakurai et al., 2010; Klatt et al., 2015). The enzymes involved in maintaining sulfide homeostasis and providing bioenergy in Sulfolobales are sulfide:quinone oxidoreductase (SQR), which are found distributed widely in all domains (they are found in archaea, bacteria, and mitochondria; Hell et al., 2008; Sousa et al., 2018).
SQR homologs are present in all the members of the Sulfolobales listed in Figure 3 except Sulfol. acidocaldarius. Only one SQR-encoding gene is present in the genomes of most members of the Sulfolobales. However, Acidianus species harbor a second SQR (SQR2), which share 71–77% identities (100% coverage) with SQR from A. ambivalens (AaSQR; Figure 3). SQRs are classified into six types based on their structures and sequences (Marcia et al., 2010; Sousa et al., 2018). AaSQR belongs to Type V SQRs (Sousa et al., 2018), which is the first X-ray crystal structure of an archaeal SQR (PDB ID: 3H8L; Brito et al., 2009). AaSQR has one extended capping loop and a cysteine–flavin adenine dinucleotide (FAD) linkage, and contains two monomers in the asymmetric unit (Sousa et al., 2018). It has two redox centers: the covalently bound FAD and a pair of cysteine residues (C178 and C350) bridged by a chain of three sulfur atoms. A channel on the surface of SQR, at the re-side of the FAD, is for substrate entry or product exit. The oxidation reaction product, a polysulfide chain (comprising four or five sulfur atoms) or sulfane, is the substrate for SOR. The reduction part of the reaction occurs on the si-side of FAD, where the primary electron acceptor, a quinone, is reduced by electrons from sulfide. This process feeds electrons into the respiratory chain and is coupled to energy conservation (Brito et al., 2009). AaSQR is a membrane-anchored protein, most likely facing the cytoplasm (Brito et al., 2009). McSQR (Mcup_0231) from M. cuprina Ar-4 was upregulated when cells grew autotrophically on S0 compared with growth heterotrophically on yeast extract (Jiang et al., 2014). The ability of McSQR to oxidize sulfide to polysulfide has been shown (data not published by our group).
SQRs are involved in sulfide-dependent energy conservation and in sulfide detoxification to maintain sulfide homeostasis. Microbial oxidation of sulfide is a hot topic in wastewater bioremediation technology and for sulfide removal from soil. Nevertheless, the catalytic mechanism and the function of most SQRs in cells remain to be uncovered.
Elemental sulfur (S0), existing mainly in the most stable form, cyclo-S8, is insoluble in water (Boulegue, 1978; Suzuki, 1999; Sosa Torres et al., 2020). In Sulfolobales, S0 oxidation is catalyzed by SOR, which was first characterized in A. ambivalens. SOR catalyzes oxygen-dependent S0 disproportionation, with hydrogen sulfide, sulfite, and thiosulfate as the products. Thiosulfate is produced mainly due to the chemical reaction between sulfite and S0 (Kletzin, 1989; Kletzin et al., 2004). S0 serves both as electron donor and acceptor, and no external cofactors are required by SOR. The reaction is not coupled with energy conservation (Kletzin et al., 2004; Urich et al., 2004).
Genes encoding SOR homologs in Sulfolobales are widespread in all sequenced Acidianus species and are also found in Sulfuri. tokodaii and Sulfura. metallicus. Three SORs within the Sulfolobales have been structurally characterized: AaSOR from A. ambivalens (PDB ID: 2CB2; Urich et al., 2004, 2006), AtSOR from A. tengchongensis (PDB ID: 3BXV; He et al., 2000; Li et al., 2008), and StSOR from Sulfuri. tokodaii (PDB ID: 6M3X, 6M35; Sato et al., 2020).
The SORs are homomultimers, each composed of 24 identical subunits, which form a large hollow sphere enclosing a positively charged nanocompartment, where the disproportionation reaction takes place. Six chimney-like protrusions, each composed of four helices that belong to individual monomers, referred to as tetramer channels, are the entry routes of the substrate S0; S0 enters the tetramer reaction pocket via the apolar tetramer channels as a linear polysulfone, rather than as an S8 ring. Each monomer possesses an active site pocket comprising a mononuclear non-heme iron site and three conserved cysteine residues (C31, C101, and C104; Figure 4; Urich et al., 2006; Li et al., 2008; Sato et al., 2020). In AaSOR, C31 is involved in binding the substrate S0 via a cysteine persulfide, and only this residue (among the three cysteines) was essential for the catalytic activity of AaSOR (Urich et al., 2006; Veith et al., 2011). However, the cysteine residues are present as free thiols in AtSOR and StSOR structures. Mutation of any of the three cysteine residues completely abolished the catalytic activity of AtSOR (Li et al., 2008). Mutation of C101 or C104 in StSOR significantly decreased the activity of the enzyme (Sato et al., 2020). The polar reaction products hydrogen sulfide, sulfite, and thiosulfate were proposed to exit the sphere via channels located at threefold symmetry axes (Urich et al., 2006; Li et al., 2008; Veith et al., 2011; Sato et al., 2020). SOR activity was detected only in the cytoplasm of A. ambivalens, while it is partially located in the cytoplasmic membrane of A. tengchongensis (Chen et al., 2005).
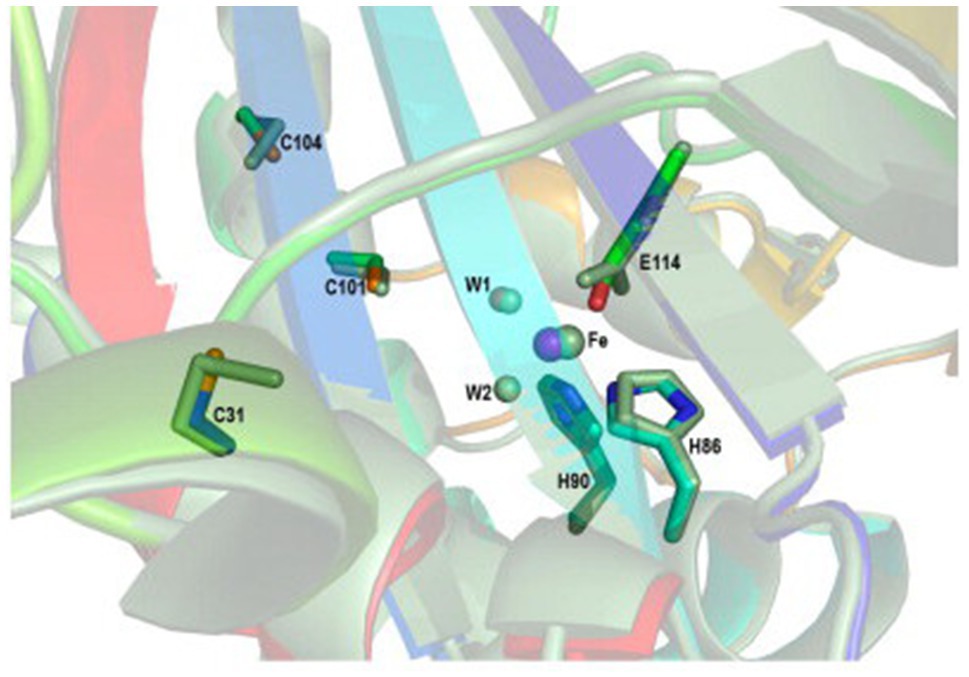
Figure 4. Superposition of the active site between the AtSOR (I432 form) and AaSOR structures. The iron atom is represented as a large sphere and water molecules as small spheres. The residues constituting the active site are shown as sticks. The AaSOR structure is shown in green, while the color of the AtSOR structure is represented by the atom type: yellow, carbon atom; red, oxygen atom (water molecule is included); blue, nitrogen atom; orange, sulfur atom; and magenta, iron atom (Li et al., 2008).
SOR is found in all Acidianus species and a few other members of the Sulfolobales, such as Saccharolobus and Sulfurisphaera (Kletzin, 1989; Chen et al., 2007). All archaea harboring SORs have the ability to oxidize S0. Notably, although Metallosphaera can oxidize S0 for growth, no SOR-coding genes are found in their genomes (Figure 3; Auernik et al., 2008; Liu et al., 2011b; Wang et al., 2020). SOR is indicated to be a supplementary but not necessary enzyme for cytoplasmic elemental sulfur oxidation in the sulfur-oxidizing bacteria Acidithiobacillus spp. (Wang et al., 2019). Other enzymes may exist that perform S0 oxidation in aerobic sulfur-oxidizing Sulfolobales; this requires further research.
Thiosulfate is further oxidized to tetrathionate by the membrane-bound protein TQO in A. ambivalens. A. ambivalens TQO consists of two 28-kD DoxD and two 16-kD DoxA subunits, forming an α2β2 tetramer. TQO oxidizes thiosulfate to tetrathionate with caldariellaquinone (CQ) as the electron acceptor. TQO and the terminal quinol:oxygen oxidoreductase, comprised of two major subunits (DoxB and DoxC) and one minor subunit (DoxE), may form a loose aggregation in the membrane and transfer electrons via CQ to reduce O2, producing a transmembrane proton gradient for coupled ATP synthesis (Müller et al., 2004).
doxDA homologs are found in several genera of Sulfolobales, including Acidianus, Metallosphaera, Saccharolobus, Sulfodiicoccus, Sulfolobus, Sulfuracidifex, and Sulfurisphaera. DoxD (Mcup_1713) and DoxA (Mcup_1712) in M. cuprina Ar-4 were upregulated when cells grew in autotrophic conditions compared with heterotrophic conditions, as determined by quantitative proteomics (Jiang et al., 2014). Genes encoding DoxD2 and DoxA2, which have low similarity (around 40%) of amino acid sequences to DoxD and DoxA, are present in Acidianus, Saccharolobus, and Sulfol. islandicus (Figure 3). DoxD2 and DoxA2 are separated from DoxDA phylogenetically, and their functions are still unclear (Müller et al., 2004).
There are two pathways of oxidation of sulfite to sulfate: direct and indirect oxidation. The direct oxidation of sulfite to sulfate in A. ambivalens is catalyzed by sulfite:acceptor oxidoreductase (SAOR), a membrane-bound molybdenum protein. The electrons from sulfite oxidation are probably transferred to CQ, feeding into the respiratory chain. Genes encoding SAOR homologs are found in all the sequenced Sulfolobales (Figure 3). The sulfite produced during sulfur metabolism is in the cytoplasm, while sulfate produced by the membrane-bound SAOR is released to the outside of the cell. However, it is still unknown whether SAOR transports sulfate across the membrane, or if a sulfate transporter is present (Zimmermann et al., 1999). It was reported that sulfite was readily oxidized to sulfate through the direct pathway in the purple sulfur bacterium Allochromatium vinosum, catalyzed by the heterotrimeric membrane-bound sulfite-oxidizing enzyme complex SoeABC (Dahl et al., 2013). The sequences of SoeABC subunits were detected in all Metallosphaera species. Whether this direct sulfite oxidation pathway works in sulfur-oxidizing archaea remains to be established (Wang et al., 2020).
The indirect sulfite oxidation pathway is catalyzed by adenylylsulfate or adenosine 5'-phosphosulfate (APS) reductase and ATP sulfurylase (also named ATP:sulfate adenylyltransferase, encoded by the sat gene) or adenylylsulfate:phosphate adenylyltransferase (APAT, formerly named ADP sulfurylase). APS is an intermediate, involved in substrate-level phosphorylation. In the first reaction, APS reductase catalyzes APS formation from sulfite and AMP, and releases two electrons. The APS can be used in two ways: One is reacting with pyrophosphate (Ppi) catalyzed by ATP sulfurylase, forming ATP and sulfate; the other is in production of ADP and sulfate catalyzed by APAT in the presence of phosphate (Pi). ADP is then converted to ATP by adenylate kinase (Kappler and Dahl, 2001). The activities of APS reductase, APAT, and adenylate kinase were detected in the cytoplasm in Acidianus ambivalens, revealing indirect oxidation of sulfite via the APS and ADP pathway (Zimmermann et al., 1999). According to our BLAST search results, genes encoding APS reductase and ATP sulfurylase are also present in A. manzaensis, M. sedula, M. yellowstonensis, Sulfuri. tokodaii, Sulfod. acidophilus, Saccharolobus, and Sulfolobus species, indicating indirect sulfite oxidation occurs in these organisms, probably via APS to form ATP and sulfate (Figure 3), although biochemical evidence for this is still lacking. Neither APS reductase- nor ATP sulfurylase-encoding genes are found in Acidianus species, M. cuprina, M. hakonensis, Sulfuri. ohwakuensis, and Sulfuracidifex species (Figure 3). The indirect sulfite oxidation pathways in these organisms are still unclear.
Tetrathionate, the product of TQO, is further hydrolyzed by tetrathionate hydrolase (TetH), a pseudoperiplasmic protein attached to the S-layer, with an overall β-propeller structure. In A. ambivalens, TetH was found only in cells grown on tetrathionate; the gene is poorly expressed in cells grown on sulfur (Protze et al., 2011). TetH secreted by A. hospitalis YS8 forms zipper-like particles (ZLPs). The amounts of ZLPs that increased after cells were treated by mitomycin C, UV light, or by freezing in liquid nitrogen and rapid thawing, and they decreased to nondetectable levels after cells adapted to their growth conditions. TetH from A. hospitalis YS8 has 99% identity with that from A. ambivalens; both are stimulated by general stress (Krupovic et al., 2012).
TetH-coding genes exist in strictly or facultatively chemolithoautotrophic members of the Sulfolobales, which can grow in tetrathionate (Figure 3). Two copies of TetH-encoding genes (tetH1 and tetH2) are found in Acidianus, Sulfuracidifex, and Sulfurisphaera species. TetH1 and TetH2, the function of which is unknown, cluster in distinct clades in a dendrogram (Protze et al., 2011).
Heterodisulfide reductase (Hdr) is an iron–sulfur protein first discovered in methanogenic archaea that catalyzes reversible reduction of the heterodisulfide (CoM–S–S–CoB) of the thiol-coenzymes M (CoM–SH) and B (CoB–SH), coupled with energy conservation. Hdr is composed of three subunits, HdrA, HdrB, and HdrC. HdrA contains a typical FAD-binding motif and four [4Fe–4S] cluster-binding motifs. HdrB harbors two similar non-cubane [4Fe–4S] clusters and each cluster consist of fused [3Fe–4S]-[2Fe–2S] subcluster sharing one iron and one sulfur. The ferredoxin-like HdrC contains two [4Fe–4S] cluster-binding motifs (Hedderich et al., 2005; Wagner et al., 2017). Hdr complex-like proteins in sulfur-oxidizing bacteria and archaea are encoded by the gene cluster hdrC1B1A-hyp-hdrC2B2 (Liu et al., 2014). The Hdr complex in the thermophilic bacterium Aquifex aeolicus is a membrane-bound protein composed of at least five subunits: HdrA, HdrB1, HdrB2, HdrC1, and HdrC2 (Boughanemi et al., 2016). The Hdr complex is supposed to oxidize disulfide intermediates to sulfite and deliver the collected electrons to the membrane quinol pool. Furthermore, sulfur trafficking proteins, such as TusA and DsrE, are involved in transferring the sulfur groups to Hdr (Quatrini et al., 2009). Recent evidence showed that the Hdr complex oxidized thiosulfate to sulfite in Hyphomicrobium denitrificans, and the electrons produced may be transferred via a lipoate-binding protein (LbpA) to generate NADH (Cao et al., 2018; Koch and Dahl, 2018). The expression of Hdr subunits in M. cuprina Ar-4 increases when cells are grown in autotrophic conditions compared with heterotrophic conditions, indicating the participation of the Hdr-like complex in sulfur oxidation in M. cuprina (Jiang et al., 2014).
Sulfur trafficking is normally required for delivery of sulfur-containing groups as protein-bound forms to the sulfur-catalyzing enzymes. During this process, the unstable sulfur groups can be protected. The active site of TQO is suggested to face the cytoplasm; the tetrathionate produced by thiosulfate oxidation is thus released to the cytoplasm. However, tetrathionate is unstable at the near-neutral pH in the cytoplasm (Protze et al., 2011). Whereas the (rhd–)tusA–dsrE2 gene cluster is widely distributed in phototrophic and chemotrophic sulfur-oxidizing bacteria for transfer of sulfane sulfur, the dsrE3A–tusA–hdr gene cluster is ubiquitous in Sulfolobales (Figure 3). TusA appears to be a central and common protein for sulfur trafficking in sulfur-oxidizing pathways (Dahl, 2015). It has been proven in M. cuprina that the dsrE3A–tusA–hdr gene cluster is important in trafficking the sulfane sulfur of tetrathionate to prevent its biological toxicity. As shown in Figure 1, DsrE3A and TusA can both react with tetrathionate to form protein–Cys–S–thiosulfonate, which is stable in the cytoplasm. Then, DsrE3A–Cys–S–thiosulfonate transfers one thiosulfonate to TusA, forming TusA–Cys–S–thiosulfonate, and releases another thiosulfonate to TQO. The reverse transfer reaction does not happen. Next, the thiosulfonate combined with TusA serves as the substrate of the Hdr-like complex to produce sulfite for SAOR/SoeABC. The sulfane group remaining on TusA is then oxidized and released (Liu et al., 2014; Dahl, 2015).
The order Sulfolobales, phylum Crenarchaeota, is distributed in acidic and hot terrestrial or aquatic solfatara aeras and includes nine validly named genera. On the basis of new physiological data and phylogenetic analysis, several species have been reassigned to new taxa over the years. Furthermore, Sulfol. yangmingensis should be reclassified in genus Sulfurisphaera. Sulfol. islandicus, Sulfol. tengchongensis, Sulfol. sp. A20, and some other Sulfol. sp. strains might be placed in the genus Saccharolobus according to phylogenetic analysis. Moreover, it is proposed that Sulfod. acidiphilus should be used as the type strain of a new family. More newly isolates and their physiological and phylogenetic data are needed to support the reclassification.
Sulfolobales possess a broad array of physiological traits, such as a pH range for growth of 0.4–6.5, a temperature range for growth from 45 to 96°C, different O2 requirements (including obligate aerobes, facultative aerobes, and obligate anaerobes), different nutrition types (including heterotrophs, mixotrophs, and chemolithoautotrophs), and DNA G+C content from 30 to 52mol% (Tables 1 and 2). Most Sulfolobales are sulfur or RISC oxidizers or reducers, and they are considered to play important roles in the sulfur cycle of Earth. Some proteins and enzymes involved in sulfur metabolism have been characterized (Figure 1). It seems no universal pathway exists, and the proteins involved in sulfur metabolism vary in different species (Figure 3). Gaps remain in the sulfur metabolism pathways of Sulfolobales: (i) How does element sulfur access to the cytoplasm or do cytomembrane proteins exist to directly oxidize element sulfur? (ii) Which enzyme catalyzes S0 oxidation in the species without SOR? (iii) What are the functions of DoxD2 and DoxA2? (iv) How does sulfate transport across the membrane? Further research is required in the above area for understanding these questions.
S-JL and C-YJ modified and edited the manuscript. ZJ constructed the phylogenetic tree. PW and Y-LQ provided the information of SQR, SAOR, and TetH. WX and YW contributed to the final version of the manuscript. L-JL wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (grants no. 91851206, 31600040, and 31670124), the fellowship of China Postdoctoral Science Foundation (2021M692614), the Key Research Program of Chinese Academy of Sciences (ZDRW-ZS-2018-1), the Joint Funds of Innovation Academy for Green Manufacture, Chinese Academy of Sciences (IAGM2020C24), the CAS Engineering Laboratory for Advanced Microbial Technology of Agriculture, Chinese Academy of Sciences (KFJ-PTXM-016), and the Supporting Foundation of Xi’an Medical University (grants nos. 2017PT29 and 2017PT40).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Auernik, K. S., and Kelly, R. M. (2008). Identification of components of electron transport chains in the extremely thermoacidophilic crenarchaeon Metallosphaera sedula through iron and sulfur compound oxidation transcriptomes. Appl. Environ. Microbiol. 74, 7723–7732. doi: 10.1128/AEM.01545-08
Auernik, K. S., Maezato, Y., Blum, P. H., and Kelly, R. M. (2008). The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl. Environ. Microbiol. 74, 682–692. doi: 10.1128/AEM.02019-07
Bełtowski, J. (2019). “Synthesis, metabolism, and signaling mechanisms of hydrogen sulfide: an overview,” in Vascular Effects of Hydrogen Sulfide Methods in Molecular Biology. ed. J. Bełtowski (New York, NY: Springer New York), 1–8.
Boughanemi, S., Lyonnet, J., Infossi, P., Bauzan, M., Kosta, A., Lignon, S., et al. (2016). Microbial oxidative sulfur metabolism: biochemical evidence of the membrane-bound heterodisulfide reductase-like complex of the bacterium Aquifex aeolicus. FEMS Microbiol. Lett. 363:fnw156. doi: 10.1093/femsle/fnw156
Boulegue, J. (1978). Solubility of elemental sulfur in water at 298 K. Phosphorus Sulfur Silicon Relat. Elem. 5, 127–128. doi: 10.1080/03086647808069875
Brito, J. A., Sousa, F. L., Stelter, M., Bandeiras, T. M., Vonrhein, C., Teixeira, M., et al. (2009). Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48, 5613–5622. doi: 10.1021/bi9003827
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68. doi: 10.1007/BF00408082
Cao, X., Koch, T., Steffens, L., Finkensieper, J., Zigann, R., Cronan, J. E., et al. (2018). Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. elife 7:e37439. doi: 10.7554/eLife.37439
Chen, Z. W., Jiang, C. Y., She, Q. X., Liu, S. J., and Zhou, P. J. (2005). Key role of cysteine residues in catalysis and subcellular localization of sulfur oxygenase-reductase of Acidianus tengchongensis. Appl. Environ. Microbiol. 71, 621–628. doi: 10.1128/AEM.71.2.621-628.2005
Chen, Z. W., Liu, Y. Y., Wu, J. F., She, Q. X., Jiang, C. Y., and Liu, S. J. (2007). Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl. Microbiol. Biotechnol. 74, 688–698. doi: 10.1007/s00253-006-0691-0
Counts, J. A., Willard, D. J., and Kelly, R. M. (2021). Life in hot acid: a genome-based reassessment of the archaeal order Sulfolobales. Environ. Microbiol. 23, 3568–3584. doi: 10.1111/1462-2920.15189
Dahl, C. (2015). Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes: sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67, 268–274. doi: 10.1002/iub.1371
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Dai, X., Wang, H., Zhang, Z., Li, K., Zhang, X., Mora-López, M., et al. (2016). Genome sequencing of Sulfolobus sp. A20 from Costa Rica and comparative analyses of the putative pathways of carbon, nitrogen, and sulfur metabolism in various Sulfolobus strains. Front. Microbiol. 7:1902. doi: 10.3389/fmicb.2016.01902
Fuchs, T., Huber, H., Burggraf, S., and Stetter, K. O. (1996). 16S rDNA-based phylogeny of the archaeal order sulfolobales and reclassification of Desulfurolobus ambivalens as Acidianus ambivalens comb. nov. Syst. Appl. Microbiol. 19, 56–60. doi: 10.1016/S0723-2020(96)80009-9
Fuchs, T., Huber, H., Teiner, K., Burggraf, S., and Stetter, K. O. (1995). Metallosphaera prunae, sp. nov., a novel metal-mobilizing, thermoacidophilic archaeum, isolated from a uranium mine in Germany. Syst. Appl. Microbiol. 18, 560–566. doi: 10.1016/S0723-2020(11)80416-9
Gao, R., Liu, H., and Xun, L. (2017). Cytoplasmic localization of sulfide:quinone oxidoreductase and persulfide dioxygenase of Cupriavidus pinatubonensis JMP134. Appl. Environ. Microbiol. 83, e01820–e01917. doi: 10.1128/AEM.01820-17
Grogan, D., Palm, P., and Zillig, W. (1990). Isolate B12, which harbours a virus-like element, represents a new species of the archaebacterial genus Sulfolobus, Sulfolobus shibatae, sp. nov. Arch. Microbiol. 154, 594–599. doi: 10.1007/BF00248842
Guy, L., and Ettema, T. J. G. (2011). The archaeal “TACK” superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587. doi: 10.1016/j.tim.2011.09.002
He, Z., Li, Y., Zhou, P., and Liu, S. (2000). Cloning and heterologous expression of a sulfur oxygenase/reductase gene from the thermoacidophilic archaeon Acidianus sp. S5 in Escherichia coli. FEMS Microbiol. Lett. 193, 217–221. doi: 10.1111/j.1574-6968.2000.tb09427.x
He, Z. G., Zhong, H., and Li, Y. (2004). Acidianus tengchongensis sp. nov., a new species of acidothermophilic archaeon isolated from an acidothermal spring. Curr. Microbiol. 48, 159–163. doi: 10.1007/s00284-003-4155-9
Hedderich, R., Hamann, N., and Bennati, M. (2005). Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol. Chem. 386, 961–970. doi: 10.1515/BC.2005.112
Hell, R., Dahl, C., Knaff, D., and Leustek, T. (2008). Sulfur Metabolism in Phototrophic Organisms. Dordrecht: Springer Netherlands.
Huber, H., and Prangishvili, D. (2006). “Sulfolobales,” in The Prokaryotes. eds. M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 23–51.
Huber, G., Spinnler, C., Gambacorta, A., and Stetter, K. O. (1989). Metallosphaera sedula gen, and sp. nov. represents a new genus of aerobic, metal-mobilizing, thermoacidophilic archaebacteria. Syst. Appl. Microbiol. 12, 38–47. doi: 10.1016/S0723-2020(89)80038-4
Huber, G., and Stetter, K. O. (1991). Sulfolobus metallicus, sp. nov., a novel strictly chemolithoautotrophic thermophilic archaeal species of metal-mobilizers. Syst. Appl. Microbiol. 14, 372–378. doi: 10.1016/S0723-2020(11)80312-7
Itoh, T., Miura, T., Sakai, H. D., Kato, S., Ohkuma, M., and Takashina, T. (2020). Sulfuracidifex tepidarius gen. Nov., sp. nov. and transfer of Sulfolobus metallicus Huber and Stetter 1992 to the genus Sulfuracidifex as Sulfuracidifex metallicus comb. nov. Int. J. Syst. Evol. Microbiol. 70, 1837–1842. doi: 10.1099/ijsem.0.003981
Jan, R. L., Wu, J., Chaw, S. M., Tsai, C. W., and Tsen, S. D. (1999). A novel species of thermoacidophilic archaeon, Sulfolobus yangmingensis sp. nov. Int. J. Syst. Bacteriol. 49, 1809–1816. doi: 10.1099/00207713-49-4-1809
Jiang, C. Y., Liu, L. J., Guo, X., You, X. Y., Liu, S. J., and Poetsch, A. (2014). Resolution of carbon metabolism and sulfur-oxidation pathways of Metallosphaera cuprina Ar-4 via comparative proteomics. J. Proteome 109, 276–289. doi: 10.1016/j.jprot.2014.07.004
Kabil, O., and Banerjee, R. (2010). Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 285, 21903–21907. doi: 10.1074/jbc.R110.128363
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9. doi: 10.1111/j.1574-6968.2001.tb10813.x
Karavaĭko, G. I., Golyshina, O. V., Troitskiĭ, A. V., Val'ekho-Roman, K. M., Golovacheva, R. S., and Pivovarova, T. A. (1994). Sulfurococcus yellowstonii sp. nov.: a new species of iron- and sulfur-oxidizing thermoacidophilic archaeobacterium. Mikrobiologiia 63, 668–682.
Klatt, J. M., Haas, S., Yilmaz, P., de Beer, D., and Polerecky, L. (2015). Hydrogen sulfide can inhibit and enhance oxygenic photosynthesis in a cyanobacterium from sulfidic springs. Environ. Microbiol. 17, 3301–3313. doi: 10.1111/1462-2920.12791
Kletzin, A. (1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 171, 1638–1643. doi: 10.1128/jb.171.3.1638-1643.1989
Kletzin, A., Urich, T., Müller, F., Bandeiras, T. M., and Gomes, C. M. (2004). Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36, 77–91. doi: 10.1023/B:JOBB.0000019600.36757.8c
Koch, T., and Dahl, C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12:2479. doi: 10.1038/s41396-018-0209-7
Kozubal, M. A., Dlakic, M., Macur, R. E., and Inskeep, W. P. (2011). Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the Sulfolobales. Appl. Environ. Microbiol. 77, 1844–1853. doi: 10.1128/AEM.01646-10
Krupovic, M., Peixeiro, N., Bettstetter, M., Rachel, R., and Prangishvili, D. (2012). Archaeal tetrathionate hydrolase goes viral: secretion of a sulfur metabolism enzyme in the form of virus-like particles. Appl. Environ. Microbiol. 78, 5463–5465. doi: 10.1128/AEM.01186-12
Kurosawa, N. (2003). Reclassification of Sulfolobus hakonensis Takayanagi et al. 1996 as Metallosphaera hakonensis comb. nov. based on phylogenetic evidence and DNA G+C content. Int. J. Syst. Evol. Microbiol. 53, 1607–1608. doi: 10.1099/ijs.0.02716-0
Kurosawa, N., Itoh, Y. H., Iwai, T., Sugai, A., Uda, I., Kimura, N., et al. (1998). Sulfurisphaera ohwakuensis gen. Nov., sp. nov., a novel extremely thermophilic acidophile of the order Sulfolobales. Int. J. Syst. Bacteriol. 48, 451–456. doi: 10.1099/00207713-48-2-451
Laska, S. (2003). Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149, 2357–2371. doi: 10.1099/mic.0.26455-0
Lencina, A. M., Ding, Z., Schurig-Briccio, L. A., and Gennis, R. B. (2013). Characterization of the type III sulfide:quinone oxidoreductase from Caldivirga maquilingensis and its membrane binding. Biochim. Biophys. Acta 1827, 266–275. doi: 10.1016/j.bbabio.2012.10.010
Lewis, A. M., Recalde, A., Bräsen, C., Counts, J. A., Nussbaum, P., Bost, J., et al. (2021). The biology of thermoacidophilic archaea from the order Sulfolobales. FEMS Microbiol. Rev. 45:fuaa063. doi: 10.1093/femsre/fuaa063
Li, M., Chen, Z., Zhang, P., Pan, X., Jiang, C., An, X., et al. (2008). Crystal structure studies on sulfur oxygenase reductase from Acidianus tengchongensis. Biochem. Biophys. Res. Commun. 369, 919–923. doi: 10.1016/j.bbrc.2008.02.131
Liu, L. J., Stockdreher, Y., Koch, T., Sun, S. T., Fan, Z., Josten, M., et al. (2014). Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J. Biol. Chem. 289, 26949–26959. doi: 10.1074/jbc.M114.591669
Liu, L. J., You, X. Y., Guo, X., Liu, S. J., and Jiang, C. Y. (2011a). Metallosphaera cuprina sp. nov., an acidothermophilic, metal-mobilizing archaeon. Int. J. Syst. Evol. Microbiol. 61, 2395–2400. doi: 10.1099/ijs.0.026591-0
Liu, L. J., You, X. Y., Zheng, H., Wang, S., Jiang, C. Y., and Liu, S. J. (2011b). Complete genome sequence of Metallosphaera cuprina, a metal sulfide-oxidizing archaeon from a hot spring. J. Bacteriol. 193, 3387–3388. doi: 10.1128/JB.05038-11
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083. doi: 10.1002/prot.22665
Müller, F. H., Bandeiras, T. M., Urich, T., Teixeira, M., Gomes, C. M., and Kletzin, A. (2004). Coupling of the pathway of Sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160. doi: 10.1111/j.1365-2958.2004.04193.x
Peng, T. J., Liu, L. J., Liu, C., Yang, Z. F., Liu, S. J., and Jiang, C. Y. (2015). Metallosphaera tengchongensis sp. nov., an acidothermophilic archaeon isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 65, 537–542. doi: 10.1099/ijs.0.070870-0
Plumb, J. J., Haddad, C. M., Gibson, J. A. E., and Franzmann, P. D. (2007). Acidianus sulfidivorans sp. nov., an extremely acidophilic, thermophilic archaeon isolated from a solfatara on Lihir Island, Papua New Guinea, and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 57, 1418–1423. doi: 10.1099/ijs.0.64846-0
Protze, J., Müller, F., Lauber, K., Naß, B., Mentele, R., Lottspeich, F., et al. (2011). An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front. Microbiol. 2:68. doi: 10.3389/fmicb.2011.00068
Qi, J., Wang, B., and Hao, B. I. (2004). Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J. Mol. Evol. 58, 1–11. doi: 10.1007/s00239-003-2493-7
Quatrini, R., Appia-Ayme, C., Denis, Y., Jedlicki, E., Holmes, D. S., and Bonnefoy, V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394
Reinartz, M., Tschäpe, J., Brüser, T., Trüper, H. G., and Dahl, C. (1998). Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170, 59–68. doi: 10.1007/s002030050615
Reno, M. L., Held, N. L., Fields, C. J., Burke, P. V., and Whitaker, R. J. (2009). Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106, 8605–8610. doi: 10.1073/pnas.0808945106
Sakai, H. D., and Kurosawa, N. (2017). Sulfodiicoccus acidiphilus gen. Nov., sp. nov., a sulfur-inhibited thermoacidophilic archaeon belonging to the order Sulfolobales isolated from a terrestrial acidic hot spring. Int. J. Syst. Evol. Microbiol. 67, 1880–1886. doi: 10.1099/ijsem.0.001881
Sakai, H. D., and Kurosawa, N. (2018). Saccharolobus caldissimus gen. Nov., sp. nov., a facultatively anaerobic iron-reducing hyperthermophilic archaeon isolated from an acidic terrestrial hot spring, and reclassification of Sulfolobus solfataricus as Saccharolobus solfataricus comb. nov. and Sulfolobus shibatae as Saccharolobus shibatae comb. nov. Int. J. Syst. Evol. Microbiol. 68, 1271–1278. doi: 10.1099/ijsem.0.002665
Sakurai, H., Ogawa, T., Shiga, M., and Inoue, K. (2010). Inorganic sulfur oxidizing system in green sulfur bacteria. Photosynth. Res. 104, 163–176. doi: 10.1007/s11120-010-9531-2
Sato, Y., Yabuki, T., Adachi, N., Moriya, T., Arakawa, T., Kawasaki, M., et al. (2020). Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase from Sulfurisphaera tokodaii. J. Struct. Biol. 4:100030. doi: 10.1016/j.yjsbx.2020.100030
Segerer, A., Neuner, A., Kristjansson, J. K., and Stetter, K. O. (1986). Acidianus infernus gen. Nov., sp. nov., and Acidianus brierleyi comb. nov.: facultatively aerobic, extremely acidophilic thermophilic sulfur-metabolizing archaebacteria. Int. J. Syst. Bacteriol. 36, 559–564. doi: 10.1099/00207713-36-4-559
Segerer, A. H., Trincone, A., Gahrtz, M., and Stetter, K. O. (1991). Stygiolobus azoricus gen. Nov., sp. nov. represents a novel genus of anaerobic, extremely thermoacidophilic archaebacteria of the order Sulfolobales. Int. J. Syst. Bacteriol. 41, 495–501. doi: 10.1099/00207713-41-4-495
She, Q., Singh, R. K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M. J., et al. (2001). The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98, 7835–7840. doi: 10.1073/pnas.141222098
Sosa Torres, M. E., Rito Morales, A., Solano Peralta, A., and Kroneck, P. M. H. (2020). “Transition metals and sulfur–a strong relationship for life: 2-sulfur, the versatile non-metal,” in Metal Ions in Life Sciences. eds. M. S. Torres and P. Kroneck (Berlin: De Gruyter), 19–50.
Sousa, F. M., Pereira, J. G., Marreiros, B. C., and Pereira, M. M. (2018). Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. BBA-Bioenergetics 1859, 742–753. doi: 10.1016/j.bbabio.2018.04.004
Stetter, K. O. (1989). “Order III: Sulfolobales Ord. Nov. family Sulfolobaceae fam. Nov.,” in Bergey’s Manual of Systematic Bacteriology. eds. J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt, vol. 3 (Baltimore, MD: The Williams and Wilkins Co.), 2250–2251.
Suzuki, I. (1999). Oxidation of inorganic sulfur compounds: chemical and enzymatic reactions. Can. J. Microbiol. 45, 97–105. doi: 10.1139/w98-223
Suzuki, T., Iwasaki, T., Uzawa, T., Hara, K., Nemoto, N., Kon, T., et al. (2002). Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6, 39–44. doi: 10.1007/s007920100221
Takayanagi, S., Kawasaki, H., Sugimori, K., Yamada, T., Sugai, A., Ito, T., et al. (1996). Sulfolobus hakonensis sp. nov., a novel species of acidothermophilic archaeon. Int. J. Syst. Bacteriol. 46, 377–382. doi: 10.1099/00207713-46-2-377
Tsuboi, K., Sakai, H. D., Nur, N., Stedman, K. M., Kurosawa, N., and Suwanto, A. (2018). Sulfurisphaera javensis sp. nov., a hyperthermophilic and acidophilic archaeon isolated from Indonesian hot spring, and reclassification of Sulfolobus tokodaii Suzuki et al. 2002 as Sulfurisphaera tokodaii comb. nov. Int. J. Syst. Evol. Microbiol. 68, 1907–1913. doi: 10.1099/ijsem.0.002765
Urich, T., Bandeiras, T. M., Leal, S. S., Rachel, R., Albrecht, T., Zimmermann, P., et al. (2004). The Sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron Centre. Biochem. J. 381, 137–146. doi: 10.1042/BJ20040003
Urich, T., Gomes, C. M., Kletzin, A., and Frazão, C. (2006). X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science 311, 996–1000. doi: 10.1126/science.1120306
Veith, A., Urich, T., Seyfarth, K., Protze, J., Frazão, C., and Kletzin, A. (2011). Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens. Front. Microbiol. 2:37. doi: 10.3389/fmicb.2011.00037
Wagner, T., Koch, J., Ermler, U., and Shima, S. (2017). Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357, 699–703. doi: 10.1126/science.aan0425
Wang, P., Li, L. Z., Qin, Y. L., Liang, Z. L., Li, X. T., Yin, H. Q., et al. (2020). Comparative genomic analysis reveals the metabolism and evolution of the thermophilic archaeal genus Metallosphaera. Front. Microbiol. 11:1192. doi: 10.3389/fmicb.2020.01192
Wang, R., Lin, J.-Q., Liu, X.-M., Pang, X., Zhang, C.-J., Yang, C.-L., et al. (2019). Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 9:3290. doi: 10.3389/fmicb.2018.03290
Woese, C. R., and Fox, G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74, 5088–5090. doi: 10.1073/pnas.74.11.5088
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Xiang, X., Dong, X., and Huang, L. (2003). Sulfolobus tengchongensis sp. nov., a novel thermoacidophilic archaeon isolated from a hot spring in Tengchong, China. Extremophiles 7, 493–498. doi: 10.1007/s00792-003-0355-2
Yoshida, N., Nakasato, M., Ohmura, N., Ando, A., Saiki, H., Ishii, M., et al. (2006). Acidianus manzaensis sp. nov., a novel thermoacidophilic archaeon growing autotrophically by the oxidation of H2 with the reduction of Fe3+. Curr. Microbiol. 53, 406–411. doi: 10.1007/s00284-006-0151-1
Zillig, W., Kletzin, A., Schleper, C., Holz, I., Janekovic, D., Hain, J., et al. (1993). Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 16, 609–628. doi: 10.1016/S0723-2020(11)80333-4
Zillig, W., Stetter, K. O., Wunderl, S., Schulz, W., Priess, H., and Scholz, I. (1980). The Sulfolobus-“Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch. Microbiol. 125, 259–269. doi: 10.1007/BF00446886
Zillig, W., Yeats, S., Holz, I., Böck, A., Rettenberger, M., Gropp, F., et al. (1986). Desulfurolobus ambivalens, gen. Nov., sp. nov., an autotrophic archaebacterium facultatively oxidizing or reducing sulfur. Syst. Appl. Microbiol. 8, 197–203. doi: 10.1016/S0723-2020(86)80077-7
Zimmermann, P., Laska, S., and Kletzin, A. (1999). Two modes of sulfite oxidation in the extremely thermophilic and acidophilic archaeon Acidianus ambivalens. Arch. Microbiol. 172, 76–82. doi: 10.1007/s002030050743
Keywords: Crenarchaeota, Sulfolobales, taxonomy, sulfur metabolism, sulfur trafficking
Citation: Liu L-J, Jiang Z, Wang P, Qin Y-L, Xu W, Wang Y, Liu S-J and Jiang C-Y (2021) Physiology, Taxonomy, and Sulfur Metabolism of the Sulfolobales, an Order of Thermoacidophilic Archaea. Front. Microbiol. 12:768283. doi: 10.3389/fmicb.2021.768283
Received: 31 August 2021; Accepted: 22 September 2021;
Published: 14 October 2021.
Edited by:
Qian Li, Guangzhou University, ChinaReviewed by:
Nan Peng, Huazhong Agricultural University, ChinaCopyright © 2021 Liu, Jiang, Wang, Qin, Xu, Wang, Liu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang-Jiang Liu, bGl1c2pAaW0uYWMuY24=; Cheng-Ying Jiang, amlhbmdjeUBpbS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.