
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 December 2021
Sec. Systems Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.767895
Advanced physicochemical and chemical absorption methods for chlorinated ethenes are feasible but incur high costs and leave traces of pollutants on the site. Biodegradation of such pollutants by anaerobic or aerobic bacteria is emerging as a potential alternative. Several mycobacteria including Mycolicibacterium aurum L1, Mycolicibacterium chubuense NBB4, Mycolicibacterium rhodesiae JS60, Mycolicibacterium rhodesiae NBB3 and Mycolicibacterium smegmatis JS623 have previously been described as assimilators of vinyl chloride (VC). In this study, we compared nucleotide sequence of VC cluster and performed a taxogenomic evaluation of these mycobacterial species. The results showed that the complete VC cluster was acquired by horizontal gene transfer and not intrinsic to the genus Mycobacterium sensu lato. These results also revealed the presence of an additional xcbF1 gene that seems to be involved in Coenzyme M biosynthesis, which is ultimately used in the VC degradation pathway. Furthermore, we suggest for the first time that S/N-Oxide reductase encoding gene was involved in the dissociation of the SsuABC transporters from the organosulfur, which play a crucial role in the Coenzyme M biosynthesis. Based on genomic data, M. aurum L1, M. chubuense NBB4, M. rhodesiae JS60, M. rhodesiae NBB3 and M. smegmatis JS623 were misclassified and form a novel species within the genus Mycobacterium sensu lato. Mycolicibacterium aurum L1T (CECT 8761T = DSM 6695T) was the subject of polyphasic taxonomic studies and showed ANI and dDDH values of 84.7 and 28.5% with its close phylogenetic neighbour, M. sphagni ATCC 33027T. Phenotypic, chemotaxonomic and genomic data considering strain L1T (CECT 8761T = DSM 6695T) as a type strain of novel species with the proposed name, Mycolicibacterium vinylchloridicum sp. nov.
Chlorinated ethenes (CE) are one of the major contaminants of soil and groundwater due to its excessive use in several industries, such as polyvinylchloride industry and plastic manufactory. The physicochemical properties of CE help for their infiltration from water table to the bottom of aquifer. CE stands in the environment longer than other volatile hydrocarbons and has a carcinogenic effect in animal and human health (Henschler, 1994; Volpe et al., 2007). Therefore, a threshold value of chlorinated ethene compounds in drinking water was set up in several countries. CE are presented in three forms, methanes, ethanes and ethenes with tetrachloride (CT), perchloroethene (PCE), trichloroethene (TCE) and vinyl chloride (VC) as common pollutants (Le and Coleman, 2011). The latter is the results of reductive dechlorination of polychlorinated ethenes by anaerobic bacteria (Le and Coleman, 2011). The reduction of VC to ethenes is not an easy step due to the absence and/or inactive microorganisms in the subsurface of ecosystems which led to an increasing rate of VC in groundwater. In addition, this pollutant can be formed naturally in soil after the oxidative degradation of organic matter. For these reasons, VC is considered as a priority contaminant of the groundwater and regulated by the US Environmental Protection Agency (Sass et al., 2005).
The aerobic degradation pathway of VC is the same as ethene where alkene monooxygenase (AKMO), encoded by etnABCD, is the starting point for degrading VC which is transformed to chlorooxirane after AKMO adds O2 to its double bond (Hartmans and de Bont, 1992; Jin and Mattes, 2010). The epoxide is associated to coenzyme M (CoM) by epoxyalkane, coenzyme M transferase (EaCoMT) which is encoded by etnE, and consequently 2-ketoethyl-CoM is generated (Jin and Mattes, 2010). The latter is transformed to malonate semi aldehyde which is converted to malonate by CoM reductase carboxylase and aldehyde/alcohol dehydrogenase, respectively. CoA-transferase transforms malonate to malonyl-CoA that is converted to acetyl-S-CoA by reductive decarboxylase as the final aerobic VC degradation (de Bont and Harder, 1978). In this regard, two groups of bacteria were identified: (1) VC cometabolizers known for their ability to use ethane as carbon source and found to lack etnE gene. These organisms can only degrade ethene to chlorooxiranes while (2) VC assimilators bacteria are able to use VC as sole carbon source and have etnABCD and etnE genes (Begley et al., 2012). Moreover, the degradation of VC can also be done through a non-specific oxygenase by ammonia, isoprene, methane, propane and toluene-associated bacteria (e.g. Methylosinus trichosponum OB3b and Nitrosomonas europaea).
The actinobacterial VC assimilators include strains, such as: Nocardioides sp. JS614, Mycobacterium rhodesiae JS60 (Coleman et al., 2002), Mycobacterium rhodesiae NBB3 (Coleman et al., 2006), Mycobacterium chubuense NBB4 (Coleman et al., 2006) and Mycobacterium smegmatis JS623 (Coleman and Spain, 2003a). Mycobacterium aurum L1T (CECT 8761T = DSM 6695T) was the first actinobacteria strain described as a remover of VC from waste gases with high rate of degradation (93%; Hartmans et al., 1985). However, little is known about the ethylene gene cluster for these VC assimilator microorganisms.
Mycobacterial strains showed a great adaptability to different contaminated terrestrial and aquatic environments through their ability to produce biosurfactants and to degrade chlorinated pollutants, such as Mycobacterium chubuense DSM 44219T and Mycobacterium obuense DSM 44075T (Tsukamura et al., 1981; Coleman et al., 2006; Satsuma and Masuda, 2012; Das et al., 2015). These latter strains together with Mycobacterium aurum species were transferred to the genus Mycolicibacterium defined after the taxonomic revision of the genus Mycobacterium based on comparative genomic studies by Gupta et al. (2018). Mycolicibacterium genus encompasses fast growing environmental mycobacterial strains of ‘fortuitum-vaccae’ clade which are known for their saprophytic and opportunistic lifestyles and are widely distributed in nature (soil, sediment, water, etc.) including contaminated soils. The genus Mycolicibacterium of the family Mycobacteriaceae (Chester 1897) housed 88 species with validly published name and with Mycolicibacterium fortuitum as the type species.1 Like Mycobacterium sensu lato, members of this taxon showed yellow or orange and white to cream-coloured colonies. They were characterised by the presence of cell wall rich in lipids and waxes; straight-chain saturated, unsaturated and tuberculostearic (10-methyloctadecanoic) fatty acids and mycolic acid with 60–90 carbon atoms (Magee and Ward, 2012). Genome size from 3.95 to 8.0 Mbp and G + C content between 65.4 and 70.3% (Gupta et al., 2018).
In this present study, Mycolicibacterium aurum L1T (CECT 8761T = DSM 6695T), potential degrader of VC, was the subject of a comparative genome mapping of ethylene clusters and polyphasic taxonomic studies.
Mycolicibacterium aurum L1T was isolated from a vinyl chloride polluted soil collected at Arnhem, Netherland (Hartmans and de Bont, 1992). The strain was deposited by Dr. Sybe Hartmans (Wageningen Agricultural University, Netherlands) at the German Collection of Microorganisms and Cell Cultures (DSMZ) and the Spanish Type Culture Collection (CECT) under accession numbers DSM 6695T and CECT 8761T, respectively. The strain included in this study was obtained from CECT and the strain designation L1T was used in the whole manuscript to avoid confusion. Mycolicibacterium sphagni DSM 44076T (obtained from the DSMZ) was found to be the close phylogenetic neighbour of strain L1T. These strains were maintained on proteose peptone-meat extract-glycerol agar (PMG; DSMZ 250 medium) at 28°C and conserved as bacterial suspensions in 30%, v/v glycerol at −80°C.
The cultural properties of strain L1T were evaluated on different agar media: PMG, glucose-yeast extract-malt extract agar (GYM; DSMZ medium 65), International Streptomyces Project (ISP2; Shirling and Gottlieb, 1966), Löwenstein-Jensen medium (LJ; Jensen, 1932), Middlebrook 7H10 agar (MB7H10; Lorian, 1968) and tryptic soy agar (TSA; MacFaddin, 1985) and in presence of at a wide range of temperature 4°C, 10°C, 15°C, 25°C, 28°C, 37°C and 45°C. Anaerobic growth test of strain L1T was examined using an anaerobic bag system (Sigma-Aldrich 68,061).
Strain L1T and its phylogenetic neighbour, M. sphagni DSM 44076T, were examined for a broad range of biochemical tests known to be of value in mycobacterial systematics: arylsulfatase after 3 and 14 days (Tomioka et al., 1990), catalase (de Waard and Robledo, 2007), heat stable catalase (Sequeira de Latini and Barrera, 2008), nitrate reduction (Vincent et al., 2003) and potassium tellurite tolerance (Kilburn et al., 1969; Kent and Kubica, 1985). Moreover, the growth of these strains was also evaluated in the presence of a wide range of carbon, nitrogen substrates and inhibitory compounds using GENIII microplates. The latter were inoculated with a bacterial suspension as described by Nouioui et al. (2017) and then incubated at 28°C for 5 days in an Omnilog device (Biolog Inc., Hayward, United States). The resultants data were analysed using opm package version 1.3 (Vaas et al., 2012, 2013). Furthermore, the enzymatic activities of strains L1T and M. sphagni DSM 44076 T were determined using API coryne kit and following the manufacturer’s instruction (Biomérieux, France). Each test was performed in duplicate.
The chemotaxonomic markers relevant to the genus Mycolicibacterium were examined for strain L1T and its neighbour M. sphagni DSM 44076T. Biomass was harvested from cultures, prepared on medium DSMZ 250 and shaked at 250 rpm for 5 days at 28°C. The pellets were washed twice with sterile distilled water and then freeze-dried. Diaminopimelic acid (Schleifer and Schleifer and Kandler, 1972), whole-organism sugars (Lechevalier and Lechevalier, 1970; Staneck and Roberts, 1974) and polar lipids (Minnikin et al., 1984; Kroppenstedt and Goodfellow, 2006) were performed. Cellular fatty acids were extracted following the protocol of Miller (1982) modified by Kuykendall et al. (1988). Gas chromatography (Agilent 6,890 N instrument) was used to analyse the fatty acid methyl esters which were identified using microbial identification (MIDI) system version 4.5 and the MYCO 6 database (Sasser, 1990). Thin-layer chromatographic analyses of mycolic acid extracts of strains L1T and M. sphagni DSM 44076T were performed following the protocol of Goodfellow et al. (1976).
The genomic DNA extraction was performed according to Amaro et al. (2008) and the 16S rRNA gene sequence was generated using the Sanger method (Sanger and Coulson, 1975; Sanger et al., 1977) as a quality control step for the identity of the strain. The genome sequencing was performed on a MiSeq instrument (Illumina) as previously described by Sangal et al. (2015). 300 bp paired-end reads were assembled into contigs using SPAdes 3.9.0 with a k-mer length of 127 (Bankevich et al., 2012). The draft genome sequence was annotated through RAST server (Aziz et al., 2012) and deposited in GenBank database under accession number JACBJQ000000000. The pairwise comparison of average nucleotide identity (ANI) values was performed using the OrthoANIu algorithm and ANI Calculator web tool (Yoon et al., 2017a), at the EzBioCloud portal. Digital DNA–DNA hybridization (dDDH) between the draft genome sequence of strain L1T and their close phylogenetic neighbours were estimated according to the methods described by Meier-Kolthoff et al. (2013).
An almost complete 16S rRNA gene sequence (1531 bp, accession number MT478173) was extracted from the draft genome of strain L1T and found to be identical to the sequence obtained by Sanger method. However, the 16S rRNA gene sequence of the nearest neighbours was retrieved from the EzTaxon database (Yoon et al., 2017b). BLAST of the full 16S rRNA gene sequence of isolate L1T was performed against those of validly named species available in EzBioCloud portal (Yoon et al., 2017b). A multiple sequence alignment of all 16S rRNA gene sequences was performed using MUSCLE (Multiple Sequence Comparison by Log- Expectation) algorithm (Edgar, 2004). Phylogenetic trees were constructed using MEGA X software (Kumar et al., 2018) and including Neighbour-Joining (NJ; Saitou and Nei, 1987) and Maximum-Likelihood (ML; Nei and Kumar, 2000) methods with 1,000 bootstrap iterations. The evolutionary distances were calculated using Kimura’s two parameter (Kimura, 1980) and General time reversible (Nei and Kumar, 2000) models.
Phylogenomic tree was inferred from the genome distances calculated with the BLAST distance phylogeny approach (GBDP) using the Type Strain Genome Server pipeline (Meier-Kolthoff and Göker, 2019). The type-based species and subspecies affiliation of strain L1T and the 17 type strains included in the analysis were performed based on the pairwise comparisons mentioned above and according to the thresholds previously reported (Meier-Kolthoff et al., 2013; Yoon et al., 2017a). Tree annotations and visualisations were carried out using the Interactive Tree Of Life (iTOL) webtool (Letunic and Bork, 2021).
The coding sequences comprising the putative gene cluster of ethene (ETH) and vinyl chloride (VC) assimilation pathway were manually mapped and annotated on the draft genome sequence of strain L1T considering criteria of GC reading-frame content (Bibb et al., 1984) and protein domain similarity using ARTEMIS (Berriman and Rutherford, 2003). The ORFs were screened based on their similarity with protein domains of the previously described etnEABCD gene cluster of ‘Mycobacterium smegmatis JS623’ (accession number: FJ602754.1) and ‘Mycobacterium chubuense NBB4’ genome (accession number: NC_018027), which were evidenced after comparison with the Conserved Domains Database (CDD) of NCBI (Marchler-Bauer et al., 2012). The genomic sub-regions of the clusters were compared using BLASTN (Johnson et al., 2008) and visualised with EasyFig 2.2 software (Sullivan et al., 2011).
A sequence similarity network (SSN) of the epoxyalkane coenzyme M transferase (EaCoMT) encoded by the etnE gene (Coleman and Spain, 2003b) was constructed to evaluate the taxonomic distribution of the Mycolicibacterium etnEABCD gene cluster based on the functional-sequence space of EaCoMT in homologous protein families and its genome context (Copp et al., 2018, 2019). The SSN was generated using EFI-EST (Zallot et al., 2019),2 with 1000 homologous proteins from UniProtKB database3 and an e-value clustering threshold of 1E-3. The final network was processed and visualised using the organic layout within Cytoscape v. 3.2.0 (Shannon et al., 2003). The genome context of the closest proteins of validly named species was clustered along with the EaCoMT of strain L1T. The EaCoMT clusters were preliminary visualised through EFI-GNT4 (Zallot et al., 2019) and fully mapped as described above. Only strains with publicly available genome sequences were analysed in this present report. The sequence of EaCoMT of the five mutants, Mycolicibacterium smegmatis JS623 (M1-M5) and the partial one of Mycolicibacterium rhodesiae JS60 (Coleman and Spain, 2003a,b), was included for the SSN analysis but not considered for further studies.
Smooth colonies of strain L1T acquired yellow-orange colour, after 5 days of incubation on DSMZ 65 and 250, LJ and MB7H10 media at 28°C and 37°C. Optimal growth was observed on DSMZ 250 medium, pH 7 after 5 days of incubation at 28°C. No colonies were developed under anaerobic condition and neither at 4°C, 15°C, 25°C nor 45°C.
Strain L1T and M. sphagni DSM 44076T were unable to reduce nitrate but were able to produce arylsulfatase (after 3 and 14 days) and catalase and reduce potassium tellurite. However, only strain L1T produced a heat stable catalase at 68°C and could be distinguished from its close neighbour by a wide range of metabolic features as shown in Table 1. Strain L1T metabolised D-trehalose and methyl pyruvate (carbon source); butyric acid and citric acid (organic acids); and D-serine and L-arginine (amino acids). It was found to be resistant to nalidixic acid, vancomycin and was able to grow in the presence of guanidine hydrochloride, lithium chloride, up to 4% NaCl and 1% sodium lactate (Table 1).
The chemotaxonomic properties of strain L1T were consistent with its affiliation to the genus Mycolicibacterium. Strain L1T showed quantitative and qualitative variations in polar lipid pattern comparing to its close phylogenetic neighbour M. sphagni DSM 44076T. The major polar lipids for strains L1T and DSM 44076T were diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylinositol (Supplementary Figure S1). The whole-cell hydrolysates of both strains were rich in meso-diaminopimelic acid (Supplementary Figure S2), galactose, glucose, mannose and ribose as whole-cell sugars (Supplementary Figure S3). The mycolic acid profiles of strain L1T and M. sphagni DSM 44076T contained α-mycolate, methoxymycolate and ketomycolate (Supplementary Figure S4).
The fatty acids patterns of the strain L1T and M. sphagni DSM 44076T consisted of C16:0, C17:1 ω7c/18 alcohol, C18:1ω9c, 10Me-C18:0 (tuberculostearic) and 20:0 ALC 18.838/ 20:0 ALC as shown in Supplementary Table S1.
Strain L1T showed 16S rRNA gene sequence (1531pb) similarity values of 98.5% with M. sphagni DSM 44076T and 98.8% with Mycolicibacterium houstonense ATCC 49403T, Mycolicibacterium senagalense CIP 104941T and Mycolicibacterium setense DSM 45070T. These results were not in line with their phylogenetic positions based on the ML and NJ phylogenetic trees (Figures 1A,B). Strain L1T formed a poorly supported distinct branch that is loosely associated to a clade housed the type strains of Mycolicibacterium alvei, Mycolicibacterium fortuitum subsp. fortuitum, M. houstonense, Mycolicibacterium lutetiense, Mycolicibacterium peregrinum, Mycolicibacterium setense and M. senagalense (Figures 1A,B). Hartmans et al. (1985) proposed strain L1T as Mycobacterium aurum which was emended as Mycolicibacterium aurum (Gupta et al., 2018) as stated above. However, the 16S rRNA gene sequence similarity between strain L1T and the type strain Mycolicibacterium aurum DSM 43999T was 97.3% which well below the cut-off point of 98.65% for prokaryotic species demarcation (Kim et al., 2014). Based on this data, strain L1T potentially needs to be defined as a different species.
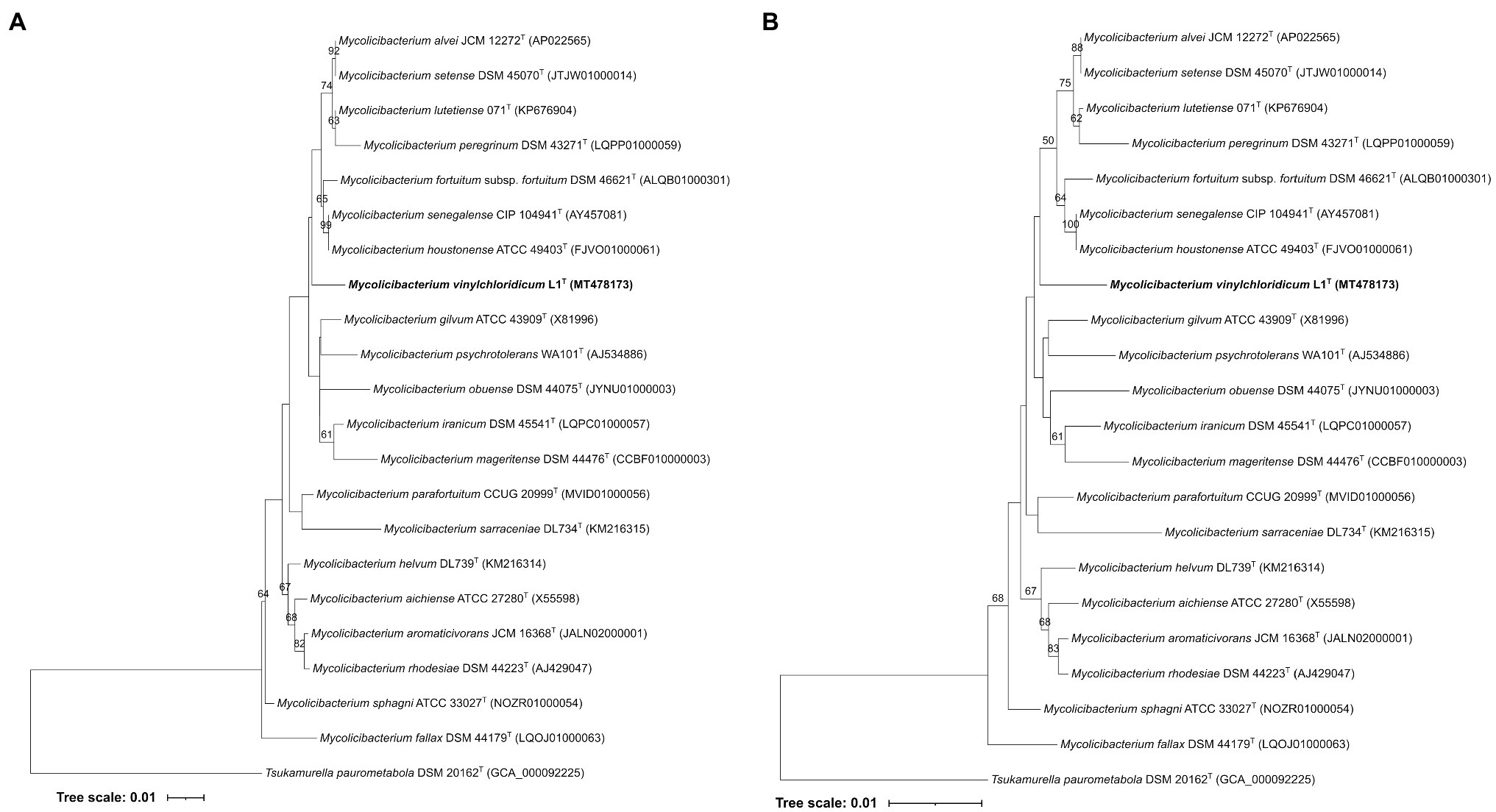
Figure 1. (A) Maximum-Likelihood and Neighbour-Joining (B) phylogenetic tree based on 16S rDNA gene sequences, showing the taxonomic position of strain L1T within the evolutionary radiation of the genus Mycolicibacterium. The numbers above branches are bootstrap support values.
In the genome-based phylogeny, strain L1T occupied a well-supported distinct branch closely related to M. sphagni ATCC 33027T (GenBank accession number: GCA_002250655) which was next to a subclade housing Mycolicibacterium sarraceniae JCM 30395T and Mycolicibacterium helvum JCM 30396T (Figure 2). However, M. senegalense DSM 43656T, M. houstonenese ATCC 49403T and M. setense DSM 45070T were in distant clade (Figure 2). More confidence can be attributed to the topology of the phylogenomic tree since it is generated from millions of unit characters (Nouioui et al., 2018) and therefore, M. sphagni ATCC 33027T is considered as the close neighbour to strain L1T.
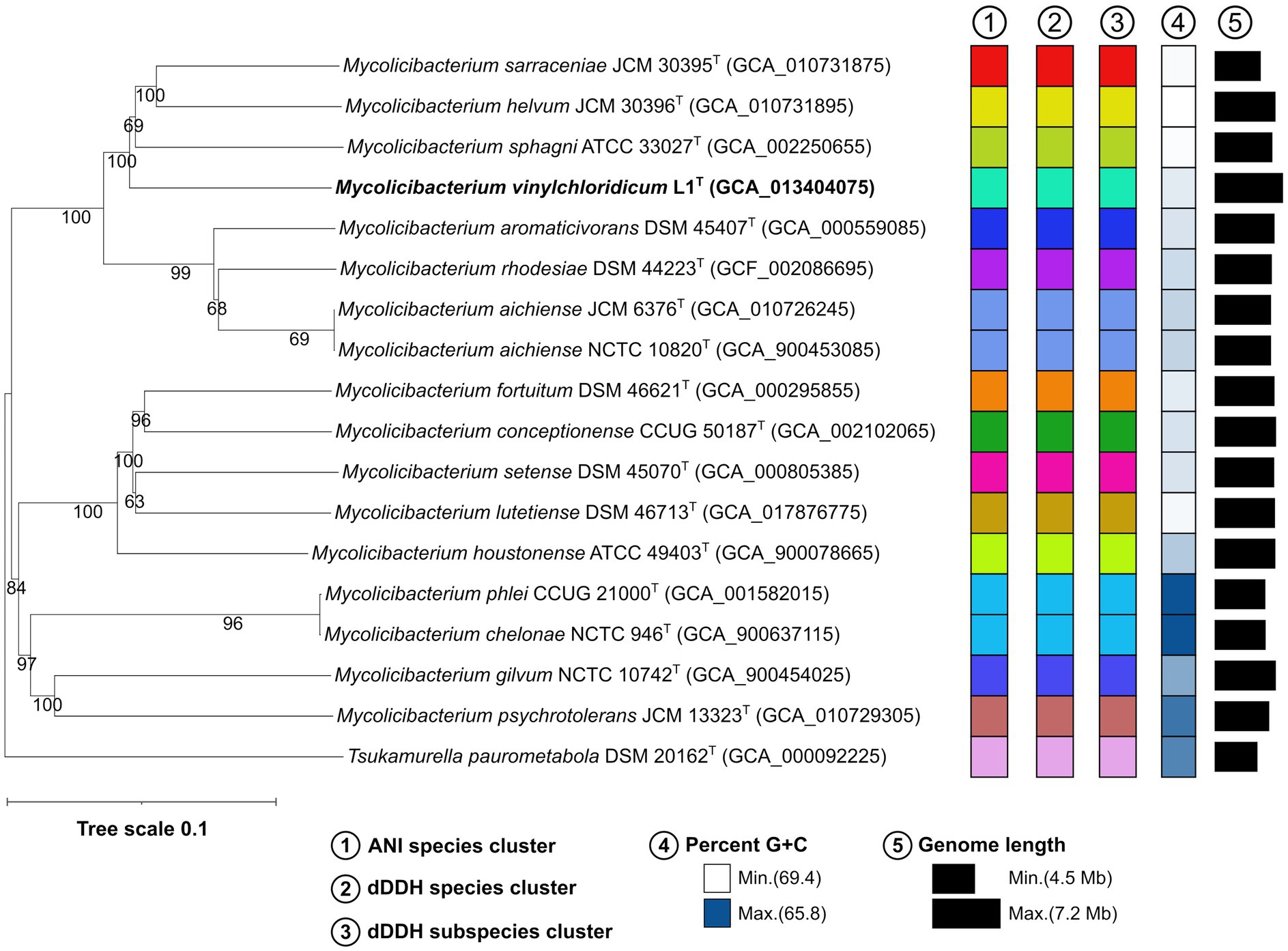
Figure 2. Phylogenomic tree based on GBDP distances calculated from genome sequences, showing the phylogenetic relationship of strain L1T with its close phylogenetic relatives. The numbers above branches are GBDP pseudo-bootstrap support values >60% from 100 iterations, with an average branch support of 89.4%.
Strain L1T and M. sphagni DSM 44076 T have genome sizes of 7.1 Mb and 6.0 Mb with 66.6 and 65.9% G + C content, 6,914 and 5,690 coding sequences and 52 and 56 RNAs, respectively. The ANI and dDDH values between the draft genome sequences of strain L1T and its close relative, M. sphagni ATCC 33027T, were 84.7% and 28.5%, values well below the threshold of 95–96% and 70% used for prokaryotic species delineation, respectively (Wayne et al., 1987; Goris et al., 2007; Richter and Rosselló-Móra, 2009; Lee et al., 2016; Jain et al., 2018).
Since VC degradation trait has been associated with strains belong to M. aurum, M. chubuense, M. rhodesiae and M. smegmatis species, the taxonomic affiliation of the reported mycobacterial strains as assimilators of VC was evaluated based on genomic approaches. The dDDH and ANI values between the genome sequence of M. rhodesiae NBB3, M. rhodesiae JS60, M. smegmatis JS623 and M. chubuense NBB4 and those of the type strains of their corresponding species were below the defined threshold cited above and confirm that these strains were misclassified and form novel species within the genus Mycobacterium sensu lato (Supplementary Table S2). The misclassification of strain JS623 to M. smegmatis species was already reported by Garcia and Gola (2016). Therefore, these strains should be referred as Mycolicibacterium sp. to avoid confusion and misleading conclusion. In addition, genome mining for VC gene cluster of the reference strains showed that the type strains of M. aurum, M. chubuense, M. rhodesiae and M. smegmatis species devoid from VC gene cluster.
The resulting SSN for the EaCoMT (etnE) of strain L1T was filtered to include edges with a minimum edge alignment score of 170 (identity >73.5%) and proteins within a minimum and maximum length of 330 and 500 residues (Figure 3). The full SSN170 was integrated by 995 protein sequences, segregated into 45 isofunctional clusters and all of them correspond to the catalytic domain of the cobalamin-independent synthase II family (PF01717; Figure 3A). The cluster 5 contained the EaCoMT aminoacidic sequence of strain L1T along with those of Mycobacteriaceae and Nocardioidaceae (Figure 3B) species, with exception of Mycolicibacterium moriokaense GAS496 whose EaCoMT protein sequence showed low similarity with all the other sequences included in the analysis and was subsequently not grouped into any cluster. The sequences incorporated into the cluster 5 belong to strains from the genus Mycolicibacterium (6 sequences), Amycolatopsis (2 sequences) and Carbonactinospora, Nocardioides, Rhodococcus, Kribella and Marmoricola (1 sequence for each); all these taxa belong to the class of Actinobacteria.
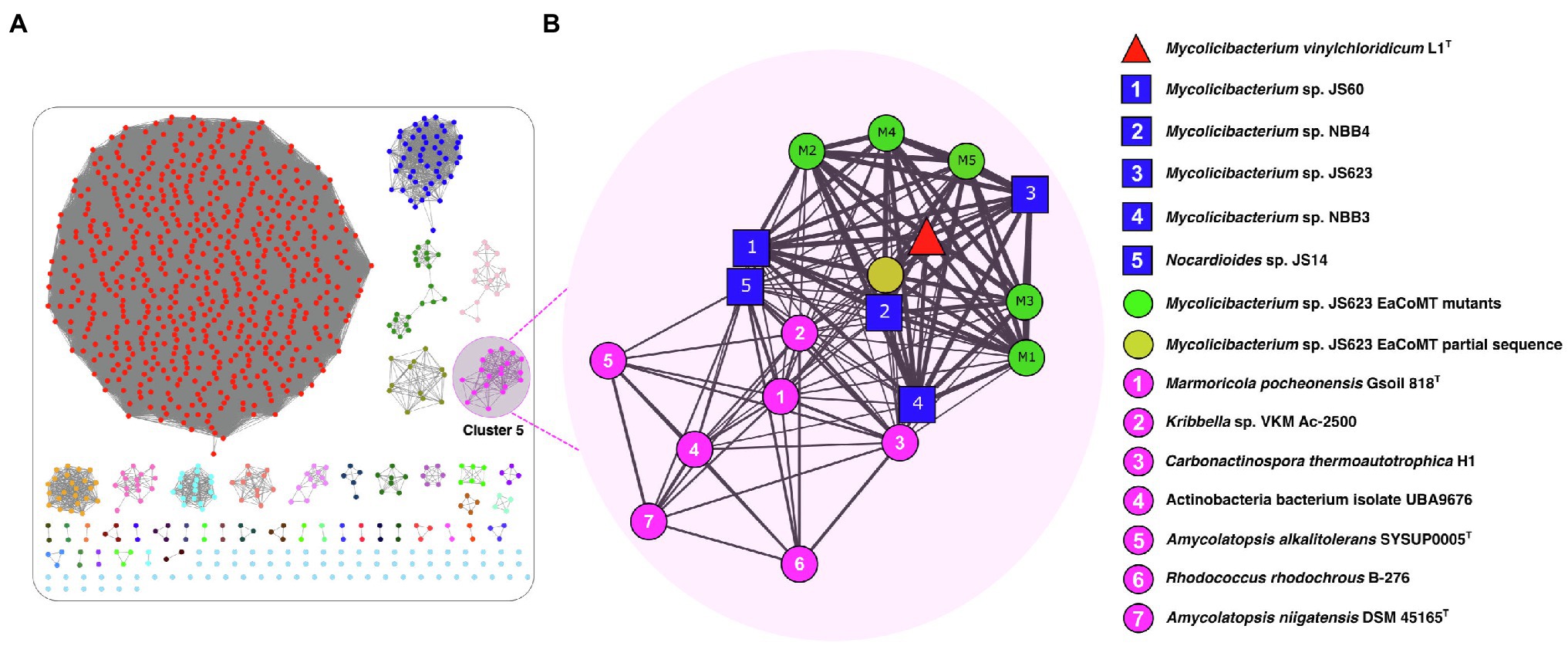
Figure 3. UniProt SSN for the epoxyalkane coenzyme M transferase (EaCoMT) of strain L1T. (A) Full SSN170 with an edge cut-off value of 10−3, showing the pairwise sequence similarity relationships among EaCoMT from the strain L1T and their 1,000 closest homologues proteins from UniProtKB database. (B) The Cluster 5 (Pink) containing the EaCoMT from the strain L1T (red triangle) and 18 EaCoMTs closest homologues including members of the families Mycobacteriaceae and Nocardioidaceae (blue squares). Each protein is represented by a circle (node), connected by a line (edge) according to their alignment score and identity reflected by the edge thickness.
The EaCoMT sequence of strain L1T showed 98.9, 98.6, 92.6 and 92.3% similarities to those of M. rhodesiae JS60, M. chubuense NBB4, M. smegmatis JS623 and M. rhodesiae NBB3, respectively (Figure 3B). However, the sequence identity value decreased to 76.7% between EaCoMT sequence of strain L1T and Nocardioides sp. JS614. The latter is known as ETH and VC assimilators (Coleman et al., 2002, 2006; Coleman and Spain, 2003b; Figure 3B).
The genome neighbourhood analysis of the etnE gene (EaCoMT) for strain L1T revealed the presence of etnABCD genes, encoding for the protein components of the AkMO (Hartmans et al., 1991), consisting on the monooxygenase β-subunit (EtnA), the monooxygenase coupling-effector protein (EtnB), the monooxygenase α-subunit (EtnC) and the alkene reductase (EtnD; Figure 4). The AkMO complex is responsible for the oxidations of ETH and VC to epoxyethane and chlorooxirane, which are conjugated to CoM yielding 2-hydroxyethyl-CoM and the putative 2-chloro-hydroxyethyl-CoM, respectively. The latter involves in the first steps of the catabolic pathway (Coleman and Spain, 2003a; Mattes et al., 2005). All mycobacterial strains of cluster 5 (Figure 3B) presented the AkMO genes (etnABCD) in their genomes (Figure 4) while the type strains of M. aurum, M. chubuense, M. rhodesiae, M. smegmatis and M. sphagni lacked etnACBD cluster.
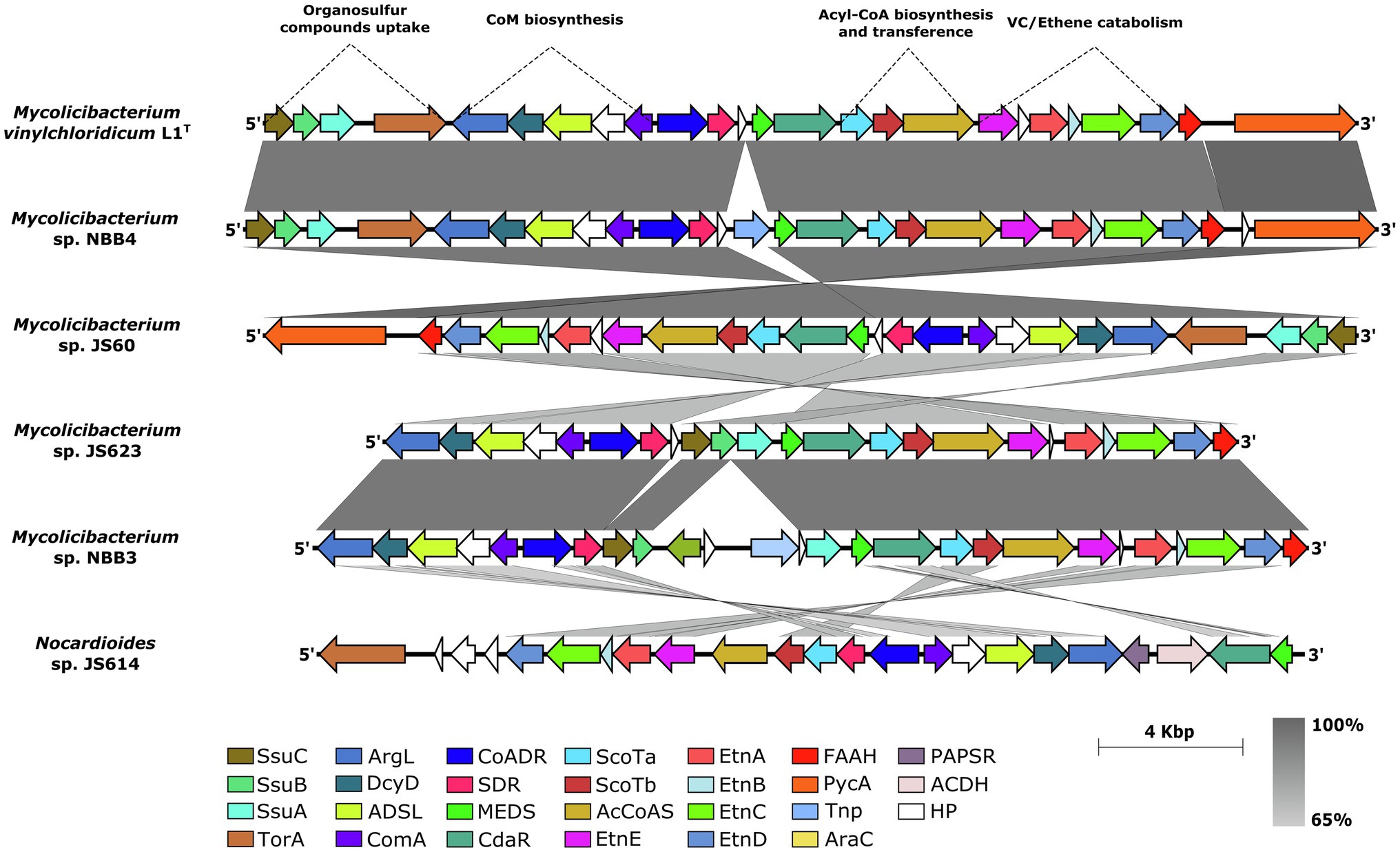
Figure 4. Synteny of the etnEABCD cluster sub-region in the genome sequence of strain L1T and five vinyl chloride assimilator strains. SsuB and SsuC: ABC transporter subunits; SsuA: periplasmic aliphatic sulfonate-binding protein; ArgL, argininosuccinate lyase; DcyD, D-cysteine desulfhydrase; AdsL, adenylosuccinate lyase; ComA, phosphosulfolactate synthase; CoADR, CoA-disulphide reductase; SDR, alcohol dehydrogenase; MEDS, methanogen/methylotroph DcmR Sensory domain; CdaR, transcription factor; CoATa and CoATb, two component CoA-transferase; ACoAS, acyl-CoA synthetase; EtnA, monoxygenase β-subunit; EtnB, monoxygenase coupling-effector protein; EtnC, monoxygenase α-subunit; EtnD, alkene reductase; FAAH, fumarylacetoacetate hydrolase; Tnp, transposase; PAPSR, Phosphoadenylyl-sulfate reductase; ACDH, NAD-dependent aldehyde dehydrogenases; and HP, hypothetical proteins.
The Coenzyme M (2-mercaptoethanesulfonic acid), which is necessary for hydroxyalkyl-CoM derivatives generation, is produced by the xcbB1,C1,D1,E1 biosynthetic gene cluster (Mattes et al., 2010; Partovi et al., 2018) and was also present in all mycobacterial genomes of cluster 5, and Nocardioides sp. JS614, with a slightly different synteny (Figure 4). The gene xcbB1 encodes for the phosphosulfolactate synthase (ComA) that catalyses a nucleophilic addition of a sulphite to phosphoenolpyruvate yielding (R)-phosphosulfolactate while argininosuccinate lyase encoded by gene xcbC1 is responsible for releasing the phosphate group by mean of β-elimination and generating sulphoacrylic acid. However, the adenylosuccinate lyase encoded by xcbD1 was found to presumably catalyse an undetermined co-substrate addition across the sulphoacrylic acid double bond. The xcbE1 gene product, D-cysteine desulfhydrase, should be the responsible for the thiolation of the unknown intermediary substrate generated after the XcbD1 reaction, in order to produce the final CoM (Partovi et al., 2018). A new xcbF1 gene associated to hypothetical protein of 308 amino acid residues is placed between genes xcbB1 and xcbE1, showing a strong synteny in all the analysed genomes (Figure 4). This protein found to have a high degree of conservation between the studied strains and an amino acid sequence identity value over 62% (Supplementary Figure S5). Further studies are necessary to determine the function of this protein in the Coenzyme M biosynthesis. Its high conservation within the cluster could be a starting point to decipher the missing intermediaries in the CoM biosynthesis (Partovi et al., 2018).
Two genes encoding for acyl-CoA synthetase (ACoAS) and two component CoA-transferase (CoATa and CoATb; Figure 4) were found in the upstream of gene etnE for strain L1T. These genes are presumably involved in the generation of Acyl-CoA and the subsequent transference of a CoA group to the malonate in the last steps of Ethene/Vinyl chloride pathway (Mattes et al., 2010).
Following upstream in the genome sequences, strain L1T together with all mycobacterial strains of cluster 5, showed the putative CdaR family transcription factor and the methanogen/methylotroph DcmR Sensory domain (MEDS), which found to be involved in the negative regulation of dichloromethane degradation on methylotrophic bacteria (La Roche and Leisinger, 1991). Both genes are responsible for the transcription of all the catabolic genes downstream including the etnEABCD genes (Figure 4).
Furthermore, in the upstream of MEDS regulator, strain L1T housed an alcohol dehydrogenase-associated gene (SDR; Figure 4) which is involved in the dehydrogenation of 2-hydroxyethyl-CoM to 2-ketoethyl-CoM and a CoA-disulphide reductase which could act as a reductive decarboxylase needed to complete the malonyl-CoA assimilation reactions (Mattes et al., 2010). All the analysed strains showed sequence identities above 65% for the conserved etnEABCD gene cluster, accessory epoxyalkane catabolic genes (ScoTa and ScoTb), regulators and the genes involved in the coenzyme M biosynthesis.
The ssuABC genes responsible for the uptake of organosulfur compounds, such as sulphate esters, sulfamates, sulfonates and alkanesulfonates during sulphur limited conditions (Beale et al., 2010), were detected approximately 2.7 Kb downstream of CoM biosynthesis genes in the genome of L1T. The product of ssuA gene corresponds to a periplasmic aliphatic sulfonate-binding protein which binds to the extracellular organosulfur compounds in order to be incorporated by an ABC transporter (proteins SsuB and SsuC) to the cell (Figure 4). An additional gene encoding for S/N-Oxide reductase (T or A; Cheng and Weiner, 2007) was also found next to the ssuABC genes suggesting its participation in the reduction of the uptaked extracellular organosulfur compounds (Figure 4). The sulphate ABC transporter encoding genes (SsuABC) were present in all the analysed strains except Nocardioides sp. JS614.
The boundaries of the described gene cluster showed the presence of genes encoding for transposases, integrases, mobile elements and also flanking direct repeats in most of the studied genomes with exception of M. smegmatis JS623 (Figure 4). These genetic elements are known to be associated with genomic islands and lead to further mobilisation, deletion or/and insertion of a complete genomic region (Schmidt and Hensel, 2004; Juhas et al., 2009; da Silva Filho et al., 2018). These findings indicate that these mycobacterial studied strains have acquired the VC degrader feature via horizontal gene transfer in order to adapt and survive in the environment.
M. vinylchloridicum (vi.nyl.chlo.ri’di.cum. N.L. neut. n. vinylchloridicum, vinyl chloride; N.L. neut. Adj. vinylchloridicum, related to vinyl chloride).
Aerobic, fast growing actinobacterium that develops colonies with yellow-orange colour, after 5 days of incubation on DSMZ 65 and 250, LJ and MB7H10 media. Optimal growth is observed after 3 days of incubation at 28°C on DSMZ 250 medium, pH 7. It is able to metabolise D-fructose, D-glucose, D-mannitol, glycerol and myo- inositol, D-trehalose (carbon source); acetic acid, butyric acid, β-hydroxy-butyric acid, citric acid, bromo-succinic acid, l-malic acid, propionic acid, sodium formate and methyl pyruvate (organic acids); and D-serine and L-arginine (amino acids). It is resistant to nalidixic acid, rifamycin sv, vancomycin and able to grow in the presence of aztreonam, guanidine hydrochloride, lithium chloride, 1% sodium lactate and tetrazolium blue, tetrazolium violet, and up to 4% (w/v) NaCl. It produces alkaline phosphatase, arylsulfatase after 3 and 14 days, catalase and heat stable catalase and reduce potassium tellurite. Whole-cell hydrolysates are rich in meso-diaminopimelic acid and galactose, glucose, mannose and ribose as cell sugars. The polar lipid pattern of strain L1T contains diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, phosphoglycolipid, unidentified glycolipids and unknown phosphoaminolipid. The mycolic acid profile of strain L1T contains α-mycolate, methoxymycolate and ketomycolate. The major fatty acids (>10%) consist of C16:0, C18:1 ω9c, C17:1 ω 7c/18 alcohol and 10Me-C18:0. The G + C content is 66.6 mol% and the genome size is 7.1 Mbp.
The type strain L1T (DSM 6695T = CECT 8761T) was isolated from vinyl chloride polluted soil, collected at Arnhem, Netherland. The GenBank accession number of the 16S rRNA gene is MT478173. The Whole-Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JACBJQ000000000. The version described in this paper is version JACBJQ010000000.
Improvements of the systematics of these mycobacterial VC assimilator strains and their assignment to the corresponding species rank are crucial for their prospective roles in bioremediation. M. aurum L1T, an actinobacteria degrader of VC, could be distinguished from its close neighbour, M. sphagni DSM 44076T, by its phenotypic and genomic features and therefore, it merits to be affiliated to a novel species with the proposed name Mycolicibacterium vinylchloridicum sp. nov. Genome comparison based on dDDH and ANI showed that M. rhodesiae NBB3, M. rhodesiae JS60, M. smegmatis JS623 and M. chubuense NBB4 were misclassified and are new candidate species within the genus Mycolicibacterium (Supplementary Table S2). The genome sequence of the type strains of M. aurum NCTC 10437T, M. chubuense DSM 44219T, M. rhodesiae DSM 44223T, M. smegmatis NCTC 8159T and M. sphagni ATCC 33027T devoid from VC gene cluster. This present report clarifies which mycobacterial species have the VC degrader feature and highlights the importance in attaching a species name to a strain that has potential application in bioremediation as example.
Comparative genomic mapping and analyses showed that the complete VC gene cluster of strain L1T, M. chubuense NBB4 and M. rhodesiae JS60 was acquired by lateral gene transfer and it is not intrinsic to the mycobacterial taxa. These findings are in line with its absence in the genome of other Mycolicibacterium strains, such as its close neighbour. The comparative analyses of the coenzyme M biosynthetic gene cluster of the studied strains highlighted the presence of a well conserved hypothetical protein-associated gene between xcbB1 and xcbE1 genes. The detected gene could be a starting point for further molecular studies to determine its function and decipher the remaining intermediary substrates for the CoM biosynthesis. Moreover, the conserved genomic position of the S/N-Oxide reductase encoding gene (torA) next to the ssuABC gene cluster in all the Mycobacterium genomes questions the role of torA related to the uptaken organosulfur by ssuABC proteins. Therefore, we propose that torA gene product involves in the dissociation of the organosulfur from the ssuABC transporters and consequently, it can be used by cells for the Coenzyme M biosynthesis. These data can be used for further molecular and biochemical research studies to decipher all the remaining steps in the VC degradation pathways.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
IN: conceptualization, writing—original draft preparation, supervision, and project administration. IN, CC-A, VS, and H-PK: methodology, validation, and writing—review and editing. IN, CC-A, and VS: software and data curation. IN, CC-A, VS and H-PK: formal analysis and visualisation. IN, CC-A, and H-PK: investigation. IN and H-PK: resources. All authors have read and agreed to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are indebted to Dr. Meina Neumann-Schaal, Ms. Gabriele Pötter and Ms. Marlen Jando (Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) for their help with fatty acid analysis and growth cultures.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.767895/full#supplementary-material
1. ^https://lpsn.dsmz.de/genus/mycolicibacterium
2. ^http://efi.igb.illinois.edu/efi-est/
Amaro, A., Duarte, E., Amado, A., Ferronha, H., and Botelho, A. (2008). Comparison of three DNA extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and Mycobacterium avium subsp. avium. Lett. Appl. Microbiol. 47, 8–11. doi: 10.1111/j.1472-765X.2008.02372.x
Aziz, R. K., Devoid, S., Disz, T., Edwards, R. A., Henry, C. S., Olsen, G. J., et al. (2012). SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS. One. 7:e48053. doi: 10.1371/journal.pone.0048053
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beale, J., Lee, S. Y., Iwata, S., and Beis, K. (2010). Structure of the aliphatic sulfonate-binding protein SsuA from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 391–396. doi: 10.1107/S1744309110006226
Begley, J. F., Czarnecki, M., Kemen, S., Verardo, A., Robb, A. K., Fogel, S., et al. (2012). Oxygen and ethene biostimulation for a persistent dilute vinyl chloride plume. Ground. Water. Monit. Remediat. 32, 99–105. doi: 10.1111/j.1745-6592.2011.01371.x
Berriman, M., and Rutherford, K. M. (2003). Viewing and annotating sequence data with Artemis. Brief. Bioinform. 4, 124–132. doi: 10.1093/bib/4.2.124
Bibb, M. J., Findlay, P. R., and Johnson, M. W. (1984). The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30, 157–166. doi: 10.1016/0378-1119(84)90116-1
Cheng, V. W., and Weiner, J. H. (2007). S- and N-oxide reductases. EcoSal Plus 2:2. doi: 10.1128/ecosalplus.3.2.8
Coleman, N. V., Bui, N. B., and Holmes, A. J. (2006). Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ. Microbiol. 8, 1228–1239. doi: 10.1111/j.1462-2920.2006.01015.x
Coleman, N. V., Mattes, T. E., Gossett, J. M., and Spain, J. C. (2002). Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68, 6162–6171. doi: 10.1128/AEM.68.12.6162-6171.2002
Coleman, N. V., and Spain, J. C. (2003a). Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 69, 6041–6046. doi: 10.1128/aem.69.10.6041-6046
Coleman, N. V., and Spain, J. C. (2003b). Epoxyalkane: coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185, 5536–5545. doi: 10.1128/jb.185.18.5536-5545.2003
Copp, J. N., Akiva, E., Babbitt, P. C., and Tokuriki, N. (2018). Revealing unexplored sequence-function space using sequence similarity networks. Biochemistry 57, 4651–4662. doi: 10.1021/acs.biochem.8b00473
Copp, J. N., Anderson, D. W., Akiva, E., Babbitt, P. C., and Tokuriki, N. (2019). Exploring the sequence, function, and evolutionary space of protein superfamilies using sequence similarity networks and phylogenetic reconstructions. Meth Enzymol. 620, 315–347. doi: 10.1016/bs.mie.2019.03.015
da Silva Filho, A. C., Raittz, R. T., Guizelini, D., De Pierri, C. R., Augusto, D. W., Dos Santos-Weiss, I., et al. (2018). Comparative analysis of genomic island prediction tools. Front. Genet. 9:619. doi: 10.3389/fgene.2018.00619
de Waard, J. H., and Robledo, J. (2007). “Conventional Diagnostic Methods” in Tuberculosis 2007. From Basic Science to patient care 1st Edn. J. C. Palomino, S. C. Leao, and V. Ritacco (New York: Springer), 401–424.
Das, S., Pettersson, B. M. F., Krishna Behra, P. R., Ramesh, M., Dasgupta, S., Bhattacharya, A., et al. (2015). Characterization of three Mycobacterium spp. with potential use in bioremediation by genome sequencing and comparative genomics. Genome Biol. Evol. 7, 1871–1886. doi: 10.1093/gbe/evv111
de Bont, J. A. M., and Harder, W. (1978). Metabolism of ethylene by Mycobacterium E20. FEMS Microbiol. Letts. 3, 89–93. doi: 10.1111/j.1574-6968.1978.tb01890.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Garcia, M. J., and Gola, S. (2016). Gene and whole genome analyses reveal that the mycobacterial strain JS623 is not a member of the species Mycobacterium smegmatis. Microbial. Biotech. 9, 269–274. doi: 10.1111/1751-7915.12336
Goodfellow, M., Collins, M. D., and Minnikin, D. E. (1976). Thin-layer chromatographic analysis of mycolic acid and other long-chain components in whole-organism methanolysates of coryneform and related taxa. J. Gen. Microbiol. 96, 351–358. doi: 10.1099/00221287-96-2-351
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Hartmans, S., and de Bont, J. A. M. (1992). Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ. Microb. 58, 1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992
Hartmans, S., de Bont, J. A. M., Tramper, J., and Luyben, K. C. A. M. (1985). Bacterial degradation of vinyl chloride. Biotechnol. Le. 7, 383–388. doi: 10.1007/BF01166208
Hartmans, S., Weber, F. J., Somhorst, D. P., and de Bont, J. A. (1991). Alkene monooxygenase from Mycobacterium: a multicomponent enzyme. J. Gen. Microbiol. 137, 2555–2560. doi: 10.1099/00221287-137-11-2555
Henschler, D. (1994). Toxicity of chlorinated organic compounds: effects of the introduction of chlorine in organic molecules. Angew. Chem. Int. Ed. Engl. 33, 1920–1935. doi: 10.1002/anie.199419201
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jensen, K. A. (1932). Reinzuch und typen bestimmung von tuberkelbazillenstamen. Zentralbl. Bakteriol. 125, 222–239.
Jin, Y. O., and Mattes, T. E. (2010). A quantitative PCR assay for aerobic, vinyl chloride- and ethene-assimilating microorganisms in groundwater. Environ. Sci. Technol. 44, 9036–9041. doi: 10.1021/es102232m
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic. Acids. Res. 1, W5–W9. doi: 10.1093/nar/gkn201
Juhas, M., van der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kent, P. T., and Kubica, G. P. (1985). Public Health Mycobacteriology: A Guide for the Level III Laboratory. Atlanta, GA: Centers for Disease control and Prevention.
Kilburn, J. O., Silcox, V. A., and Kubica, G. P. (1969). Differential identification of mycobacteria. V. The tellurite reduction test. Am. Rev. Respir. Dis. 99, 94–100. doi: 10.1164/arrd.1969.99.1.94
Kim, M., Oh H-S., Park, S.-C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kroppenstedt, R. M., and Goodfellow, M. (2006). “The family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillispora and Thermomonospora. Archaea, bacteria, Firmicutes, Actinomycetes,” in The Prokaryotes: A Handbook on the Biology of Bacteria. M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (Eds.) (New York, United States: Springer), 682–724.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuykendall, L. D., Roy, M. A., O’Neill, J. J., and Devine, T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Evol. Microbiol. 38, 358–361. doi: 10.1099/00207713-38-4-358
La Roche, S. D., and Leisinger, T. (1991). Identification of dcmR, the regulatory gene governing expression of dichloromethane dehalogenase in Methylobacterium sp. strain DM4. J. Bacteriol. 173, 6714–6721. doi: 10.1128/jb.173.21.6714-6721.1991
Le, N. B., and Coleman, N. V. (2011). Biodegradation of vinyl chloride, cis-dichloroethene and 1,2-dichloroethane in the alkene/alkane-oxidising mycobacterium strain NBB4. Biodegradation 22, 1095–1108. doi: 10.1007/s10532-011-9466-0
Lechevalier, M. P., and Lechevalier, H. A. (1970). Chemical composition asa criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 20, 435–443. doi: 10.1099/00207713-20-4-435
Lee, I., Kim, Y. O., and Y, Park., S.C., Chun, J., (2016). OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Letunic, I., and Bork, P. (2021). Interactive tree Of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lorian, V. (1968). Differentiation of Mycobacterium tuberculosis and Runyon group 3 “V” strains on direct cord-reading agar. Am. Rev. Respir. Dis. 97, 1133–1135. doi: 10.1164/arrd.1968.97.6P1.1133
MacFaddin, J. F. (1985). Media for Isolation–Cultivation–Identification–Maintenance of Medical Bacteria. Baltimore: Williams and Wilkins.
Magee, J. G., and Ward, A. C. (2012). “Genus I. Mycobacterium Lehmann and Neumann 1896, 363AL,” in Bergey’s Manual of Systematics Bacteriology. The Actinobacteria part A and B. 2nd Edn, Vol. 5. M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, K.-I. Suzuki, and W. Ludwig, et al. (New York: Springer), 312–375.
Marchler-Bauer, A., Zheng, C., Chitsaz, F., Derbyshire, M. K., Geer, L. Y., Geer, R. C., et al. (2012). CDD: conserved domains and protein three-dimensional structure. Nucleic. Acids. Res. 41, D348–D352. doi: 10.1093/nar/gks1243
Mattes, T. E., Alexander, A. K., and Coleman, N. V. (2010). Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS. Microbiol. Rev. 34, 445–475. doi: 10.1111/j.1574-6976.2010.00210.x
Mattes, T. E., Coleman, N. V., Spain, J. C., and Gossett, J. M. (2005). Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch. Microbiol. 183, 95–106. doi: 10.1007/s00203-004-0749-2
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC. Bioinform. 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Miller, L. T. (1982). Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J. Clin. Microbiol. 16, 584–586. doi: 10.1128/jcm.16.3.584-586.1982
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods. 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. New York: Oxford University Press.
Nouioui, I., Carro, L., Teramoto, K., Igual, J. M., Jando, M., Montero-Calasanz, M. D. C., et al. (2017). Mycobacterium eburneum sp. nov., a non-chromogenic, fast-growing strain isolated from sputum. Int. J. Syst. Evol. Microbiol. 67, 3174–3181. doi: 10.1099/ijsem.0.002033
Nouioui, I., Carro, L., Garcia-Lopez, M., Meier-Kolthoff, J. P., Woyke, T., and Kyrpides, N. C.,, et al. (2018). Genome-based taxonomic classification of the Phylum, Actinobacteria. Front. Microbiol. 9, 2007.
Palomino, J. C., Leao, S. C., and Ritacco, V. (2007). Tuberculosis 2007 – From basic science to patient care. Available at: www.Tuberculosistextbook.com.
Partovi, S. E., Mus, F., Gutknecht, A. E., Martinez, H. A., Tripet, B. P., Lange, B. M., et al. (2018). Coenzyme M biosynthesis in bacteria involves phosphate elimination by a functionally distinct member of the aspartase/fumarase superfamily. J. Biol. Chem. 293, 5236–5246. doi: 10.1074/jbc.RA117.001234
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sangal, V., Jones, A. L., Goodfellow, M., Hoskisson, P., Kämpfer, P., and Sutcliffe, I. C. (2015). Genomic analyses confirm close relatedness between Rhodococcus defluvii and Rhodococcus equi (Rhodococcus hoagii). Arch. Microbiol. 197, 113–116. doi: 10.1007/s00203-014-1060-5
Sanger, F., and Coulson, A. R. (1975). A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94, 441–448. doi: 10.1016/0022-2836(75)90213-2
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Sass, J. B., Castleman, B., and Wallinga, D. (2005). Vinyl chloride: a case study of data suppression and misrepresentation. Environ. Health Perspect. 113, 809–812. doi: 10.1289/ehp.7716
Sasser, M. J. (1990). Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, Technical Note 101. United States: Microbial ID.
Satsuma, K., and Masuda, M. (2012). Reductive dechlorination of methoxychlor by bacterial species of environmental origin: evidence for primary biodegradation of methoxychlor in submerged environments. J. Agric. Food Chem. 60, 2018–2023. doi: 10.1021/jf2048614
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Schmidt, H., and Hensel, M. (2004). Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17, 14–56. doi: 10.1128/CMR.17.1.14-56.2004
Sequeira de Latini, M. D., and Barrera, L. (2008). Manual para el Diagnóstico Bacteriológico de la Tuberculosis: Normas y Guía Tecnica. Parte I Baciloscopía. Organización Panamericana de la Salud. Uruguay: Universidad de la República Montevideo.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Staneck, J. L., and Roberts, G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin layer chromatography. J. Appl. Microbiol. 28, 226–231. doi: 10.1128/am.28.2.226-231.1974
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tomioka, H., Saito, H., Sato, K., and Dawson, D. J. (1990). Arylsulfatase activity for differentiating Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 28, 2104–2106. doi: 10.1128/jcm.28.9.2104-2106.1990
Tsukamura, M., Mizuno, S., and Tsukamura, S. (1981). Numerical analysis of rapidly growing, scotochromogenic mycobacteria, including Mycobacterium obuense sp. nov., nom. rev., Mycobacterium rhodesiae sp. nov., nom. rev., Mycobacterium aichiense sp. nov., nom. rev., Mycobacterium chubuense sp. nov., nom. rev., and Mycobacterium tokaiense sp. nov., nom. rev. Int. J. Syst. Bacteriol. 31, 263–275.
Vaas, L. A., Sikorski, J., Hofner, B., Fiebig, A., and Buddruhs, N., Klenk, H-P.,, et al. (2013). OPM: an R package for analysing OmniLog(R) phenotype microarray data. Bioinformatics 29, 1823–1824. doi: 10.1093/bioinformatics/btt291
Vincent, V., Brown-Elliot, B., Jost, K. C., and Wallace, R. J. (2003). “Mycobacterium: phenotypic and genotypic identification,” in Manual of Clinical Microbiology. 8th edn. P. R. Murray, E. Jorgensen, M. A. Pfaller, and R. H. Yolken. (Washington, DC: ASM Press), 560–584.
Vaas, L. A., Sikorski, J., Michael, V., Göker, M., and Klenk, H.-P. (2012). Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS. One. 7:e34846. doi: 10.1371/journal.pone.0034846
Volpe, A., Moro, G., Rossetti, S., Tandoi, V., and López, A. (2007). Remediation of PCE-contaminated groundwater from an industrial site in southern Italy: A laboratory-scale study. Process. Biochem. 42, 1498–1505. doi: 10.1016/j.procbio.2007.07.017
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017b). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017a). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie. van. Leeuwenhoek. 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Keywords: bioremediation, polyphasic taxonomy, actinobacteria, bioprospecting, nontuberculous mycobacteria
Citation: Cortés-Albayay C, Sangal V, Klenk H-P and Nouioui I (2021) Comparative Genomic Study of Vinyl Chloride Cluster and Description of Novel Species, Mycolicibacterium vinylchloridicum sp. nov. Front. Microbiol. 12:767895. doi: 10.3389/fmicb.2021.767895
Received: 31 August 2021; Accepted: 16 November 2021;
Published: 22 December 2021.
Edited by:
George Tsiamis, University of Patras, GreeceCopyright © 2021 Cortés-Albayay, Sangal, Klenk and Nouioui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Imen Nouioui, aW1lbi5ub3Vpb3VpQGRzbXouZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.