
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 November 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.767769
Ruminants may suffer from rumen acidosis when fed with high-concentrate diets due to the higher proliferation and overproduction of lactate by Streptococcus bovis. The catabolite control protein A (CcpA) regulates the transcription of lactate dehydrogenase (ldh) and pyruvate formate-lyase (pfl) in S. bovis, but its role in response to different carbon concentrations remains unclear. To characterize the regulatory mechanisms of CcpA in S. bovis S1 at different levels of carbon, herein, we analyzed the transcriptomic and physiological characteristics of S. bovis S1 and its ccpA mutant strain grown in glucose-excess and glucose-limited conditions. A reduced growth rate and a shift in fermentation pattern from homofermentation to heterofermentation were observed under glucose-limited condition as compared to glucose-excess condition, in S. bovis S1. Additionally, the inactivation of ccpA significantly affected the growth and end metabolites in both conditions. For the glycolytic intermediate, fructose 1,6-bisphosphate (FBP), the concentration significantly reduced at lower glucose conditions; its concentration decreased significantly in the ccpA mutant strain. Transcriptomic results showed that about 46% of the total genes were differentially transcribed between the wild-type strain and ccpA mutant strain grown in glucose-excess conditions; while only 12% genes were differentially transcribed in glucose-limited conditions. Different glucose concentrations led to the differential expression of 38% genes in the wild-type strain, while only half of these were differentially expressed in the ccpA-knockout strain. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses showed that the substrate glucose concentration significantly affected the gene expression in histidine metabolism, nitrogen metabolism, and some carbohydrate metabolism pathways. The deletion of ccpA affected several genes involved in carbohydrate metabolism, such as glycolysis, pyruvate metabolism, fructose and mannose metabolism, as well as in fatty acid biosynthesis pathways in bacteria grown in glucose-excess conditions; this effect was attenuated under glucose-limited conditions. Overall, these findings provide new information on gene transcription and metabolic mechanisms associated with substrate glucose concentration and validate the important role of CcpA in the regulation of carbon metabolism in S. bovis S1 at differential glucose availability.
In ruminants, undesirable lactate accumulation in the rumen due to high-concentrate diets can cause rumen acidosis. Previous studies show that Streptococcus bovis, in presence of sufficient highly digestible carbohydrates, proliferates rapidly, and predominates over a short period in the rumen with the accumulation of lactate as the major fermentation product (Marounek and Bartos, 1987; Nocek, 1997). This causes a large drop in ruminal pH (Kenney et al., 2015). Thus, S. bovis may play an important role in the progress of rumen acidosis. A better understanding of the factors affecting lactate production and overgrowth of S. bovis will be beneficial for preventing rumen acidosis.
Streptococcus bovis, an amylolytic and lactate-producing bacterium in the rumen, can rapidly break down starch into glucose and produce lactate, formate, acetate, and ethanol as the end metabolites (McAllister et al., 1990; Asanuma and Hino, 2002b). Following the Embden–Meyerhof–Parnas (EMP) pathway, pyruvate in S. bovis is either metabolized to lactate, by lactate dehydrogenase (LDH) or to formate and acetyl-CoA by pyruvate formate-lyase (PFL) (Russell and Hino, 1985); acetyl-CoA is converted subsequently to acetate or ethanol. Thus, the proportion of organic acids produced by S. bovis is dependent on the activity ratio of LDH to PFL (Asanuma et al., 1999; Asanuma and Hino, 2000). The activities of LDH and PFL are not only dependent on some glycolytic intermediates, such as fructose 1,6-bisphosphate (FBP), glyceraldehyde 3-phosphate (GAP), and dihydroxyacetone phosphate (DHAP) (Asanuma and Hino, 1997, 2002a), but also on the amount of enzyme protein, which are affected by energy supply and the intracellular pH at the transcription level (Asanuma et al., 1997, 1999; Asanuma and Hino, 2000). When the pH is low and the glucose is sufficiently available, the specific activity and amount of LDH increase, while the PFL synthesis and activity decrease (Asanuma et al., 1999), causing higher lactate production and lesser production of acetate, ethanol, and formate.
Additionally, the synthesis of LDH and PFL in S. bovis is also controlled at the transcriptional level by the catabolite control protein A (CcpA), which give a potential to control rumen acidosis due to overproduction of lactate by S. bovis at high-concentrate diets (Asanuma et al., 2004a). CcpA is a pleiotropic regulatory protein in low-GC Gram-positive bacteria and has a key role in the regulation of carbon and nitrogen metabolism, biofilm formation, toxic gene expression, and other physiological processes (Li et al., 2016). In S. bovis 12U1, the ldh mRNA level and LDH specific activity of the ccpA-knockout strain were significantly lower than those of the wild-type strain when the bacteria were grown on glucose; however, there were no significant difference when lactose was used as the substrate (Asanuma et al., 2004a). These findings show that the target gene regulation by CcpA depends on the source of carbon. However, whether the regulation of CcpA on the production of organic acids and expression of target genes is dependent on energy availability, remains unknown.
In the present study, to better understand the global regulation of CcpA in the carbohydrate metabolism of S. bovis S1 in response to carbon availability, the transcriptomic and physiological characteristics were analyzed for S. bovis S1 and its ccpA mutant strain grown under glucose-excess or glucose-limited conditions. Based on these results, we identified a large number of genes controlled by CcpA at different carbon substrate concentrations, thereby revealing further details of CcpA-mediated regulatory networks in S. bovis S1.
Streptococcus bovis S1 used in this study was previously isolated from the rumen fluid of Saanen goats in our laboratory (Chen et al., 2016b). The ccpA mutant of S. bovis S1 was constructed by homologous recombination in this experiment. Before inoculation, both the S. bovis S1 and its ccpA mutant were revived in a modified de Man, Rogosa and Sharpe (MRS) medium (Chen et al., 2016b) in an anaerobic workstation (DG250, Don Whitley Scientific, England) at 37°C. These cultures (at exponential phase) were transferred using 1% (v/v) inoculum into 200 mL anaerobic serum bottles containing 100 mL basal medium, respectively. The basal medium was prepared according to the methods described in Chen et al. (2016b); and it contained: 0.45 g/L KH2PO4, 0.9 g/L NaCl, 0.9 g/L (NH4)2SO4, 0.12 g/L CaCl2⋅2H2O, 0.19 g/L MgSO4⋅7H2O, 1.0 g/L tryptone, 1.0 g/L yeast extract, and 0.6 g/L cysteine hydrochloride. Glucose solutions were filter-sterilized and added to the sterile basal medium at a final concentration of 5 or 50 mM. After inoculation, the culture bottles were sealed and transferred to a thermostat shaker (TS-1102C, Bosheng Scientific Instrument Co., Ltd., Yangzhou, China) and grown at 37°C and 160 rpm. The pH of the medium was constantly maintained at 6.5 by continuous titration with 10% NaOH. Three replicates were set for each treatment.
The ccpA gene was disrupted by homologous recombination as follows. First, DNA fragments corresponding to the upstream (1053 bp fragment; primer pairs ccpA Up F/ccpA Up R, Supplementary Table 2) and downstream (1101 bp fragment; primer pairs ccpA Down F/ccpA Down R, Supplementary Table 2) sequences of ccpA were amplified by PCR using S. bovis S1 genomic DNA as a template. The erythromycin resistance gene erm was amplified with the primers erm F and erm R (Supplementary Table 2). The PCR product was purified using a PCR purification kit (Qiagen, Beijing, China) according to the manufacturer’s instructions. The amplified fragments were, respectively, cloned into EcoRI, BamHI, and SacI restriction sites of pUC19 vector to generate pUC19-ccpA up-erm-ccpA down (pCE). The recombinant vector pCE was electroporated into S. bovis S1 cells using an electroporation system at 2.5 kV, 200 Ω, and 25 μF. Finally, knockout mutants were selected on MRS plates containing 1 μg/mL erythromycin at 37°C for 3–4 days. The results of knockout were validated by qRT-PCR and DNA sequencing as previously described (Chen et al., 2015; Supplementary Figure 1).
Cell growth was monitored by measuring OD values at 600 nm using SpectraMax M5 plate reader (Molecular Devices Corporation, United States) at 1-h intervals, and the maximal growth rate (μmax) was estimated according to the logistic model (Perni et al., 2005; Dai et al., 2020). The cultures of each sample were harvested by centrifugation (12,000 rpm, 2 min, 4°C) when they reached exponential phase (at OD600 of 0.6 for the wild-type and ccpA mutant strains grown in glucose-excess condition, at OD600 of 0.2 for both strains grown in glucose-limited condition) and stationary phase (at OD600 of 0.9 for both strains grown in glucose-excess condition, at OD600 of 0.3 for both strains grown in glucose-limited condition). The cell pellets obtained at exponential phase were quickly frozen in liquid nitrogen for 15 min, and stored at −80°C for further RNA isolation; the supernatants at both growth phases were filtered using a 0.22 μm filter membrane and stored at −80°C for the determination of metabolites.
A high-performance liquid chromatographer (HPLC, Shimadzu, Japan) equipped with an acclaim OA column (Sepax Carbomix H-NP) and a UV detector was used to detect concentrations of organic acids (lactate, formate, and acetate) in the supernatant. The column temperature was maintained at 55°C; the mobile phase was 2.5 mM H2SO4, and its flow rate was set at 0.5 mL/min. Organic acids were then measured with a UV detector set at 210 nm. The concentration of FBP was determined using a commercial kit (Comin Biotechnology Co., Ltd., Suzhou, China), according to the manufacturer’s instructions.
Total RNA from S. bovis S1 wild-type and mutant strains, grown in the different conditions were extracted using TRIzol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. The quality and integrity of total RNA were determined using NanoDrop spectrophotometer (Thermo Scientific, United States) and Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, United States). Total RNA was purified using Zymo-Seq RiboFree Total RNA Library Kit. The first-strand cDNA was synthesized using random oligonucleotides and SuperScript III and the second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends by exonuclease/polymerase activities and the enzymes were removed. After adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated for subsequent hybridization. To select cDNA fragments of the preferred length (400–500 bp), the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA, United States). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using Illumina PCR Primer Cocktail in a 15-cycle PCR reaction. Products were purified (AMPure XP system) and quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, United States). The library obtained was sequenced on NovaSeq 6000 platform (Illumina) by Shanghai Personal Biotechnology Co., Ltd.
The raw data was filtered using Cutadapt (v1.15) software to obtain high-quality data, free of adapter sequences, primers, poly-A tails and unwanted artifacts from the high-throughput sequencing reads. The filtered reads were mapped onto the reference genome of Streptococcus equinus S1, using Bowtie2.1 The gene read count value was calculated using HTSeq (v0.9.1) as the original expression level of the gene. To normalize the gene expression levels of different genes and different samples, fragments per kilobase of exon per million fragments mapped (FPKM) was used. DESeq (v1.30.0) was used to analyze the differentially expressed mRNA transcripts with | log2FoldChange | > 1. P-value < 0.05 was considered as a statistically significant differential expression.
Primers for qRT-PCR were designed using Beacon Designer 7.0 software. Total RNA was isolated using the method described above, and reverse-transcribed to cDNA using Quant reverse transcriptase (Tiangen, Biotech Co., Ltd., China), as per the manufacturer’s instructions. The qPCR was performed on the ABI Step-One-Plus RT-PCR system (ABI 7500, Applied Biosystems, Foster City, CA) using the TB Green Premix Ex Taq™ II Kit (TaKaRa Biotechnology Co., Ltd., Dalian, China). A 20 μL reaction was set; reaction solution contained 10 μL 2 × TB Green Premix Ex Taq II, 1.6 μL primer, 1 μL cDNA, 0.4 μL 50 × ROX, and 7.0 μL Rnase-free water. The RT-qPCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of the amplification at 95°C for 5 s and 60°C for 34 s. All samples were evaluated in triplicates. Relative gene expression was normalized with the expression of the 16S rRNA gene and calculated using the 2–ΔΔCT method.
Results were expressed as “mean ± SD (standard deviation).” All data were analyzed with SPSS 25.0 (IBM, United States) and plotted using GraphPad Prism 8. Statistical significances were evaluated with the Student’s t-test (unpaired Student’s t-test, P < 0.05).
The growth curves for S. bovis S1 wild strain and the ccpA deletion strain under glucose-excess or -limited conditions are shown in Figure 1. In the stationary phase, as expected, the optical density (OD600) values of both strains in glucose-excess were significantly higher than those in glucose-limited conditions. Moreover, the growth yield of the two strains in same glucose levels exhibited no differences in the stationary phase. However, the inactivation of ccpA significantly reduced the maximal growth rate (μmax) of bacteria in the exponential phase (P < 0.05) in both glucose-excess (0.37 ± 0.002 h–1 versus 0.23 ± 0.007 h–1, respectively) and glucose-limited conditions (0.29 ± 0.008 h–1 versus 0.18 ± 0.008 h–1, respectively); μmax of both the strains grown in glucose-excess were significantly higher than those in glucose-limited conditions (P < 0.05). These data suggested that the growth of S. bovis S1 was dependent on both CcpA and glucose availability.
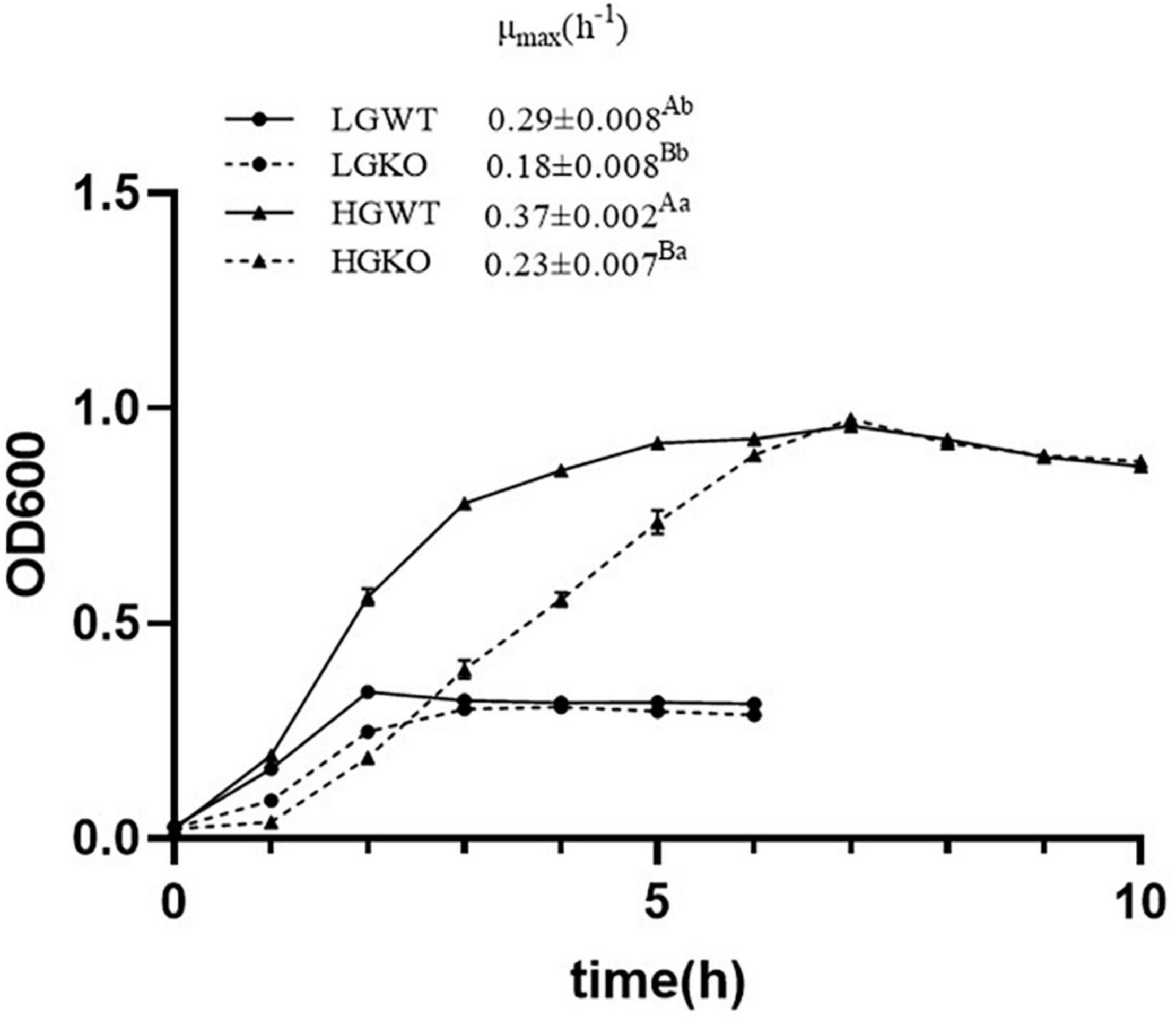
Figure 1. Growth curve of wild-type strain (WT) and ccpA-knockout strain (KO) of S. bovis S1 in glucose-excess or -limited conditions measured as optical density at 600 nm (OD600). Error bars indicate SD. The maximal growth rate (μmax) was estimated and shown in the figure. Values marked with different superscript uppercase letters (AB) indicate those are statistically significant differences (P < 0.05) between wild-type strain (WT) and ccpA-knockout strain (KO); values marked with different superscript lowercase letters (ab) indicate those are statistically significant differences (P < 0.05) between strains grown in glucose-excess or -limited conditions. LGWT, the wild-type strain grown in the media with 5 mM glucose; LGKO, the ccpA-knockout strain grown in the media with 5 mM glucose; HGWT, the wild-type strain grown in the media with 50 mM glucose; HGKO, the ccpA-knockout strain grown in the media with 50 mM glucose.
Table 1 shows the organic acid production characteristics of S. bovis S1 wild-type and ccpA deletion strains in glucose-excess or -limited conditions. In the exponential growth phase, the lactate yield and percentage of S. bovis S1 wild-type strain were significantly higher than those of ccpA deletion strain in glucose-excess condition (P < 0.05); corresponding values for formate and acetate of wild-type strain were lower than those of the mutant strain (P < 0.05). Similar results were observed for organic acid production in the low-glucose conditions. Indeed, the yields of organic acids for both strains were greater in the high-glucose condition, however, the percentages of formate and acetate were much higher in glucose-limited culture. The observations in the stationary phase were similar to those in the exponential growth phase, except for the difference in acetate yield and percentage between both strains grown in low-glucose condition, which were statistically non-significant. As shown in Figure 2, the FBP concentration in S. bovis S1 wild-type strain was significantly higher than that of ccpA deletion strain in both the glucose-excess and glucose-limited conditions (P < 0.05); the concentrations of FBP for both strains were significantly higher in high-glucose conditions (P < 0.05).
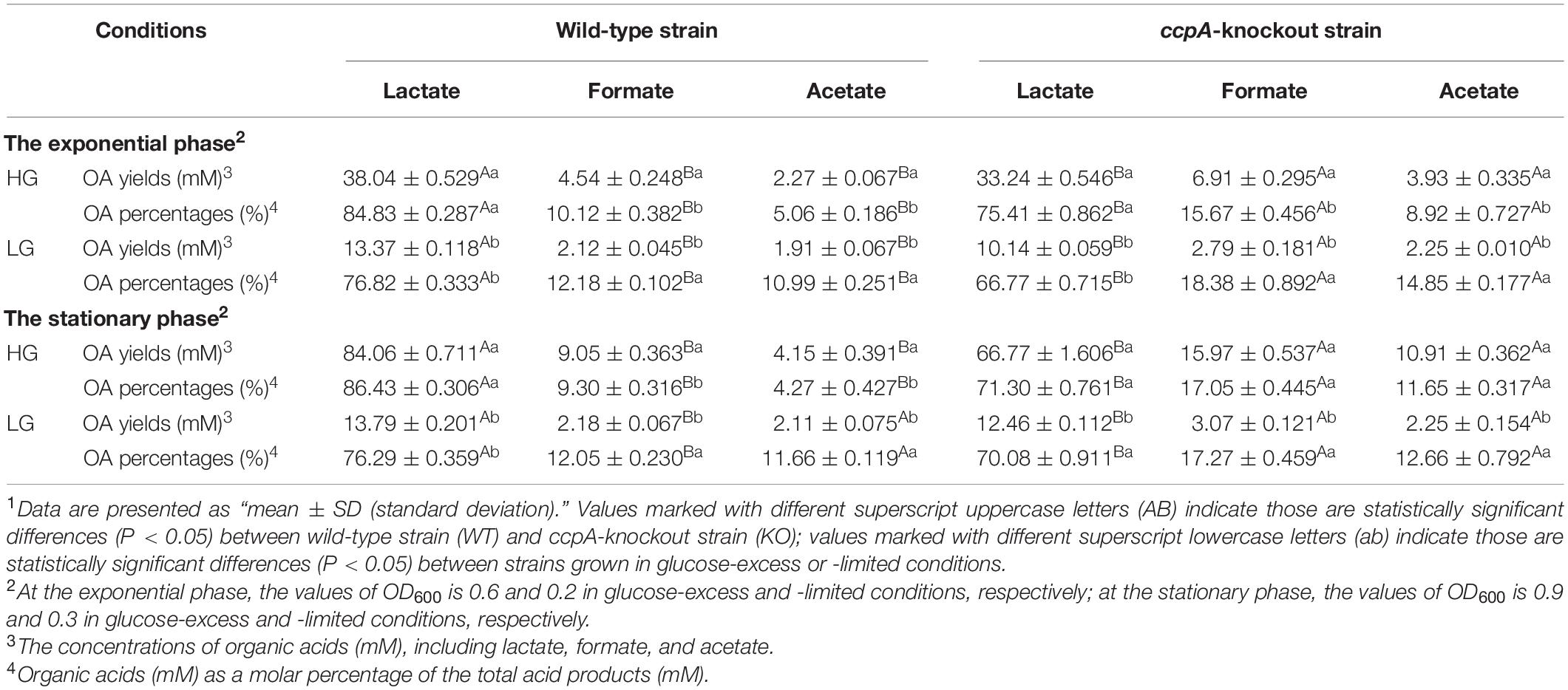
Table 1. The organic acid production characteristics of S. bovis S1 wild-type strain and ccpA-knockout strain at the exponential and stationary phases in glucose-excess or -limited conditions.1
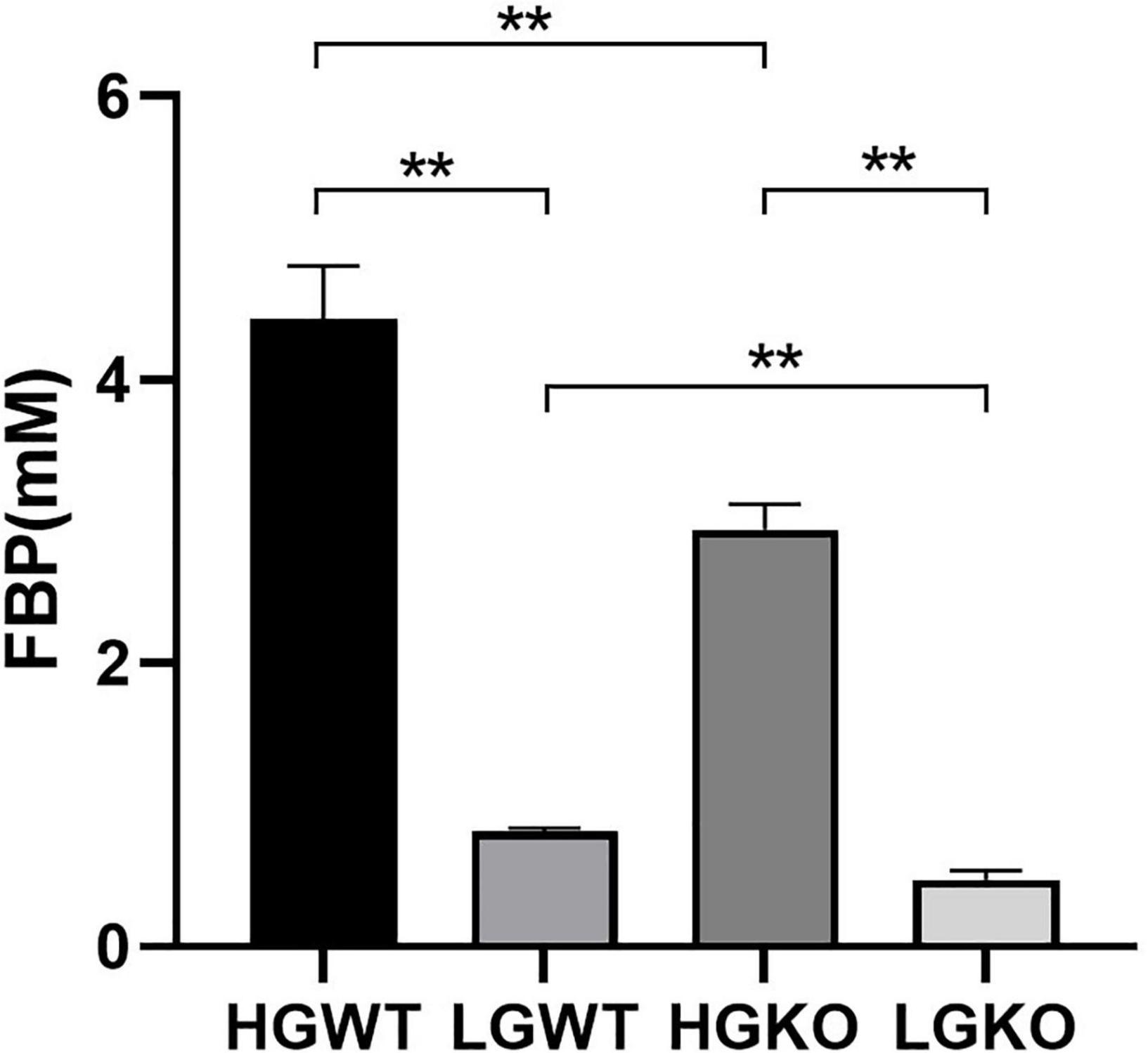
Figure 2. Intracellular concentration of FBP in wild-type strain (WT) and ccpA-knockout strain (KO) of S. bovis S1 in glucose-excess or -limited conditions. HGWT, the wild-type strain grown in the media with 50 mM glucose; HGKO, the ccpA-knockout strain grown in the media with 50 mM glucose; LGWT, the wild-type strain grown in the media with 5 mM glucose; LGKO, the ccpA-knockout strain grown in the media with 5 mM glucose. **Means P < 0.01.
To further investigate the global transcriptional regulation of CcpA in S. bovis S1 at different glucose concentrations, the transcriptome of S. bovis S1 wild-type and ccpA mutant were sequenced in glucose-excess or -limited conditions. RNA-seq was performed in the exponential growth phase of the bacterium. The statistical data for the transcriptome analysis is presented in Supplementary Table 1. A total of 364 million raw reads were generated from all samples. After the removal of low-quality reads, about 334 million clean reads with an average read length of 150 bp were obtained and used for subsequent analysis. Of these, 316 million were mapped onto the annotated S. equinus S1 genome with an average mapping ratio of at least 94.9%; the sequence reads matched with all the 1802 coding genes in the S. equinus S1 genome, which indicated that the sequencing depth was sufficient to cover all the transcripts in the cells (Supplementary Figure 2).
Based on the annotation of the S. equinus S1 genome, the read counts for each gene were calculated using HTSeq 0.6.1p2. The gene expression levels were normalized for different samples using FPKM. The gene expression profile of all samples was evaluated using principal component analysis (PCA). As expected, the three biological replicates cluster together closely for each treatment condition (Supplementary Figure 3). Differentially expressed genes (DEGs) were identified using the DESeq software with an absolute value of log2FoldChange > 1 and the false discovery rate (FDR) <0.05. DEGs between S. bovis S1 wild-type and its ccpA mutant in glucose-excess or -limited conditions were identified by four pair-wise comparisons of the overall transcriptome profiles as follows: between wild-type strains grown in glucose-excess and -limited conditions; between ccpA mutants grown in glucose-excess and -limited conditions; between wild-type and its ccpA mutant in glucose-excess conditions; between wild-type and its ccpA mutant in glucose-limited conditions (Supplementary Figure 4). Global gene expression patterns were visualized by volcano plots (Supplementary Figure 5). To better understand the molecular mechanisms of CcpA-regulated phenotypic differences of S. bovis S1 at different glucose concentrations, an enrichment analysis for the differential expressed genes was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Figure 3 and Supplementary Table 3).
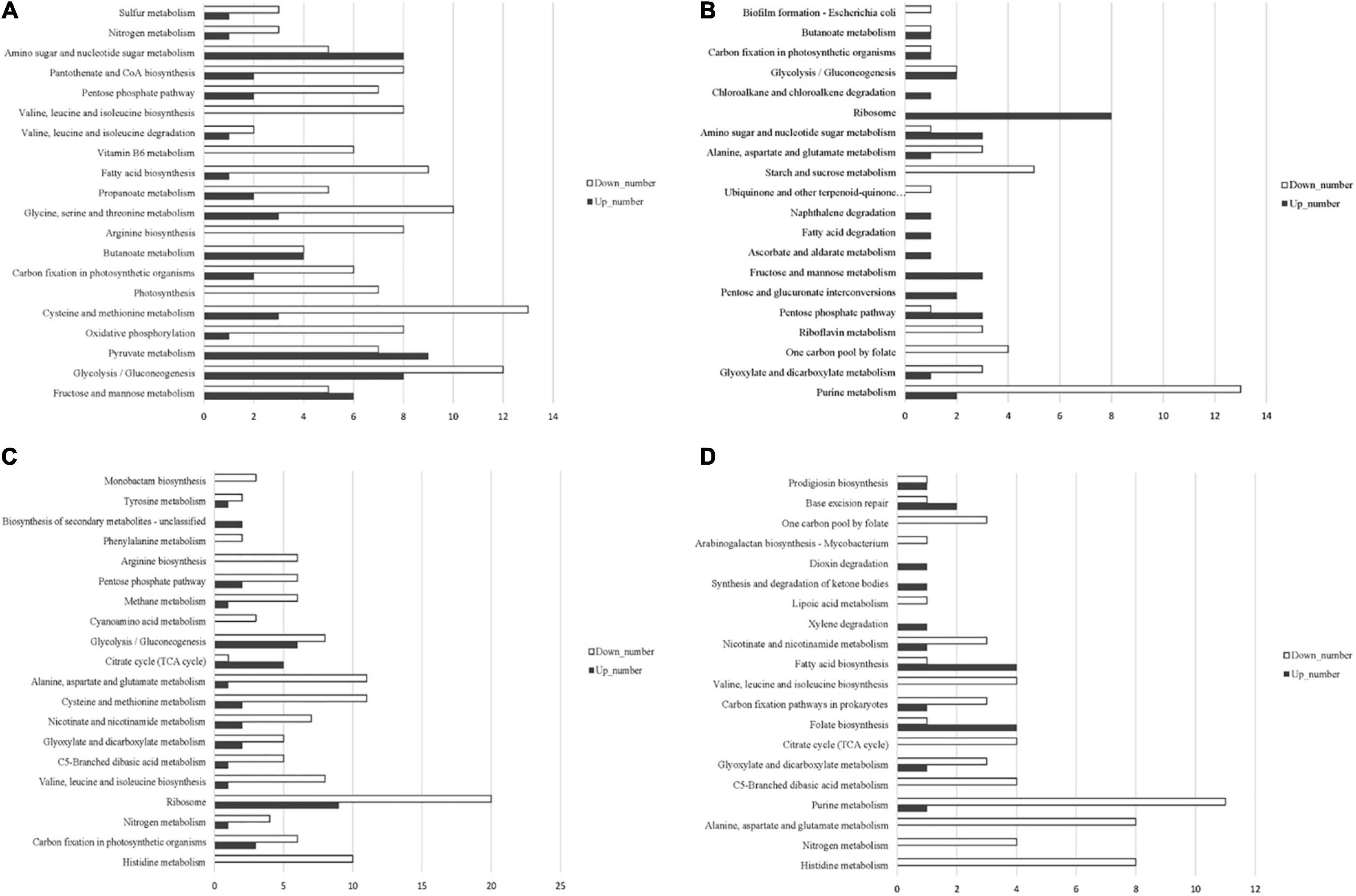
Figure 3. Distribution of upregulated and downregulated genes in the four pair-wise comparisons based on KEGG pathway categories. (A) The comparison between wild-type and its ccpA mutant grown in glucose-excess condition; (B) the comparison between wild-type and its ccpA mutant grown in glucose-limited condition; (C) the comparison between wild-type strain grown in glucose-excess and -limited conditions; (D) the comparison between ccpA mutant grown in glucose-excess and -limited conditions.
To validate the RNA-seq results, we randomly selected 10 genes and examined their transcript levels by qPCR (Supplementary Table 2). Although the magnitude of the genetic variation was different between the two analyses, the qPCR results showed similar trends in upregulations or downregulations as in the transcriptomic analysis. This confirmed the reliability of the transcriptome data.
A total of 822 differential genes were expressed in the wild-type and ccpA mutant strains grown in glucose-excess conditions. Among them, 401 genes were downregulated while 421 were upregulated. This comparison showed the largest number of genetic changes among the four comparisons studied. Sixty-one percent of these DEGs (502 genes) corresponded to 98 specific expression pathways, including metabolism pathways for carbohydrates, energy, and amino acids. Significantly altered carbohydrate metabolism pathways in KEGG following ccpA knockout were fructose and mannose metabolism, glycolysis/gluconeogenesis, pyruvate metabolism, and butanoate metabolism. In the fructose and mannose metabolism pathway (ko00051), four genes involved in fructose and mannose transport and gene encoding fructose-1-phosphate kinase were upregulated, while the genes encoding five enzymes involved in the metabolism of fructose and mannose were downregulated. These findings showed that CcpA regulated the transport and metabolism of fructose and mannose (Figure 4). The expression levels of 20 genes in the glycolysis/gluconeogenesis pathway (ko00010) were altered in absence of ccpA (Figure 4). In comparison with the wild-type strain, genes encoding the pyruvate dehydrogenase (PDH) complex, galactose mutarotase, phosphoenolpyruvate carboxykinase, and alcohol-acetaldehyde dehydrogenase were significantly upregulated in the ccpA mutant. The genes encoding L-LDH, phosphopyruvate hydratase, phosphoglycerate kinase, 6-phosphofructokinase, pyruvate kinase, two glyceraldehyde-3-phosphate dehydrogenases, 6-phospho-beta-glucosidases, triose-phosphate isomerase, phosphoglycerate mutase, and fructose-1,6-bisphosphate aldolase were significantly downregulated. We also identified DEGs involved in the pyruvate metabolism pathway (ko00620) (Figure 4). Among these DEGs, all the upregulated genes were also involved in the glycolysis/gluconeogenesis pathway except for those encoding for NAD-dependent malic enzyme, D-3-phosphoglycerate dehydrogenase, and formate acetyltransferase; genes encoding acetyl-CoA carboxylase biotin and acyl phosphatase were downregulated in the ccpA mutant. In addition, the inactivation of ccpA also led to the downregulation of the gene encoding α-amylase. Taken together, these results showed that deletion of ccpA affected the pathways in glycolysis and pyruvate metabolism of S. bovis S1. The downregulation of ldh and upregulation of pfl led to higher formate and lower lactate levels, which suggested that the absence of ccpA could switch the fermentation pattern from homolactic to mixed fermentation.
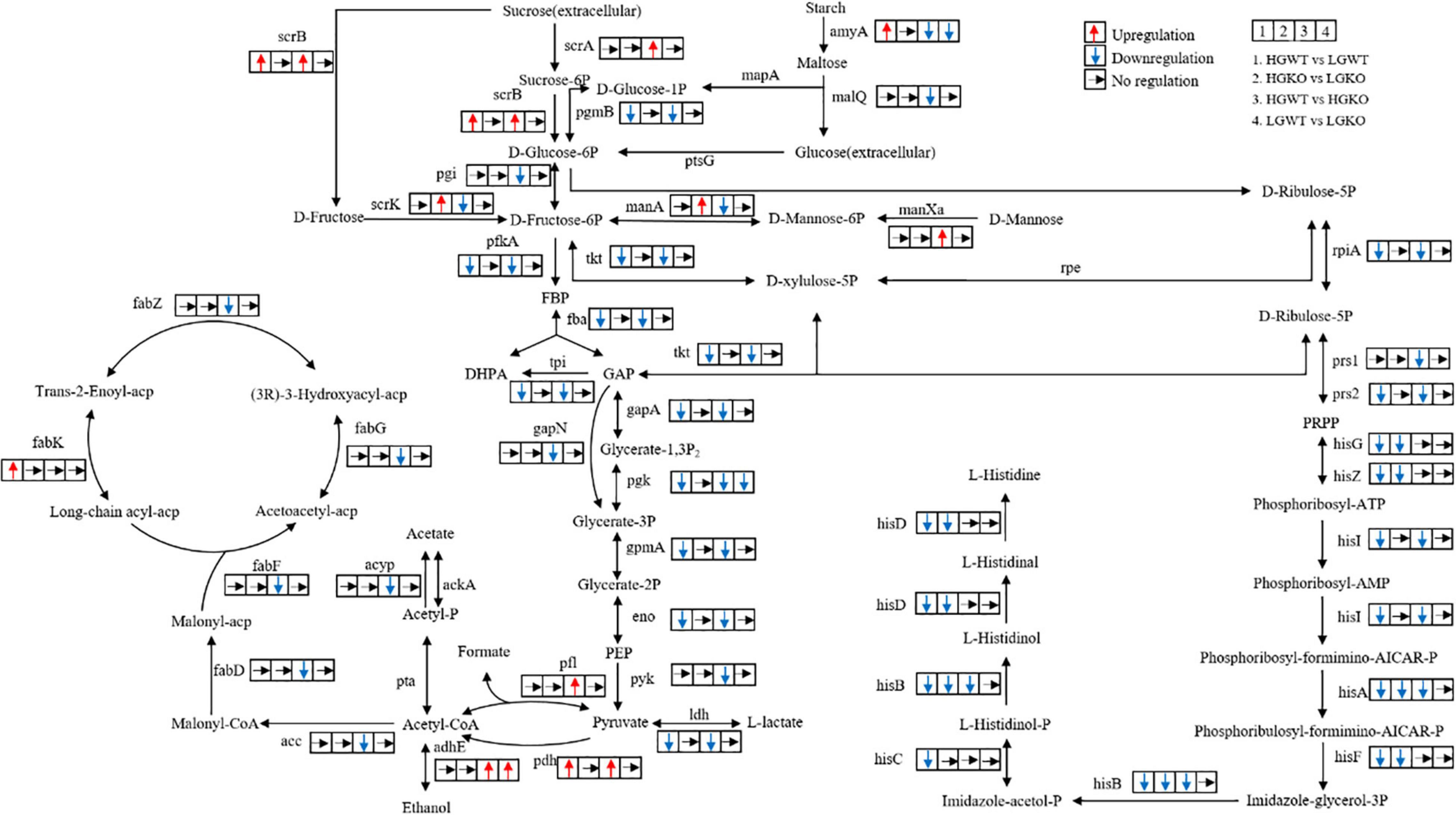
Figure 4. Overview of the key genes and related pathways changed in the transcriptomic analysis. 1, the comparison between wild-type strain grown in glucose-excess and -limited conditions; 2, the comparison between ccpA mutant grown in glucose-excess and -limited conditions. 3, the comparison between wild-type and its ccpA mutant grown in glucose-excess condition; 4, the comparison between wild-type and its ccpA mutant grown in glucose-limited condition. Gene annotation: scrA, sucrose PTS system EIIBCA or EIIBC component; scrB, beta-fructofuranosidase; scrK, fructokinase; amyA, alpha-amylase; malQ, 4-alpha-glucanotransferase; mapA, maltose phosphorylase; pgmB, beta-phosphoglucomutase; ptsG, glucose PTS system EIICB or EIICBA component; pgi, glucose-6-phosphate isomerase; manXa, mannose PTS system EIIA component; manA, mannose-6-phosphate isomerase; pfkA, 6-phosphofructokinase; fba, fructose-bisphosphate aldolase; tpi, triosephosphate isomerase; gapA, glyceraldehyde 3-phosphate dehydrogenase; gapN, glyceraldehyde-3-phosphate dehydrogenase (NADP+); pgk, phosphoglycerate kinase; gpmA, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase; eno, enolase; pyk, pyruvate kinase; ldh, L-lactate dehydrogenase; pfl, formate C-acetyltransferase; pdh, pyruvate dehydrogenase complex (pyruvate dehydrogenase E1 component, dihydrolipoamide dehydrogenase, pyruvate dehydrogenase E2 component); adhE, acetaldehyde dehydrogenase/alcohol dehydrogenase; pta, phosphate acetyltransferase; acyP, acylphosphatase; ackA, acetate kinase; acc, acetyl-CoA carboxylase; fabD, [acyl-carrier-protein] S-malonyltransferase; fabF, 3-oxoacyl-[acyl-carrier-protein] synthase II; fabG, 3-oxoacyl-[acyl-carrier protein] reductase; fabZ, 3-hydroxyacyl-[acyl-carrier protein] dehydratase; fabK, enoyl-[acyl-carrier protein] reductase II; tkt, transketolase; rpe, ribulose-phosphate 3-epimerase; rpiA, ribose 5-phosphate isomerase A; prs, ribose-phosphate pyrophosphokinase; hisG, ATP phosphoribosyltransferase; hisZ, ATP phosphoribosyltransferase regulatory subunit; hisI, phosphoribosyl-AMP cyclohydrolase; hisA, phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase; hisF, imidazole glycerol-phosphate synthase subunit; hisB, imidazoleglycerol-phosphate dehydratase; hisC, histidinol-phosphate aminotransferase; hisD, histidinol dehydrogenase.
We compared the transcriptomic profiles of wild-type and ccpA mutant strains in glucose-limited conditions. In this comparison, only 210 genes were significantly differentially expressed; 129 genes were upregulated while 81 genes were downregulated in the ccpA mutant. Compared with the transcriptomic comparison in glucose-excess condition, the DEGs reduced by approximately three quarters in glucose-limited condition. This indicated that the regulation of some genes by CcpA depended on glucose availability. Among these 210 DEGs, 118 could be grouped into different metabolic pathways in the KEGG database. In this comparison, only 4 significantly altered pathways were identified (P < 0.05), which included purine metabolism (ko00230), glyoxylate and dicarboxylate metabolism (ko00630), one carbon pool by folate (ko00670), and riboflavin metabolism (ko00740); only glyoxylate and dicarboxylate metabolism pathway was related to carbohydrate metabolism. Interestingly, there were almost no DEGs associated with pyruvate metabolism. Despite this, there were a significant number of DEGs involved in carbohydrate metabolism. The genes encoding 1-phosphofructokinase, glycosyl hydrolase family 32, PTS fructose transporter subunit IIC, galactose mutarotase, and alcohol-acetaldehyde dehydrogenase were upregulated in the absence of ccpA, while the genes encoding PTS cellobiose transporter subunit IIB, two alpha-amylases, glucose-6-phosphate isomerase, starch phosphorylase, phosphoglycerate kinase, and glucose-6-phosphate isomerase were significantly downregulated. These DEGs were mainly associated with fructose and mannose metabolism (ko00051), starch and sucrose metabolism (ko00500), and glycolysis/gluconeogenesis (ko00010), which was consistent with the results of comparison between the wild-type strain and ccpA mutant grown in glucose-excess condition. Notably, the expression of ldh and pfl was not significantly different in this comparison, which implied that the inactivation of ccpA might have little effect on the fermentation products in glucose-limited conditions.
To examine the effects of glucose concentration at the transcriptional level in the S. bovis S1, the transcriptomes of wild-type strains grown in glucose-excess and -limited conditions were compared. A total of 680 genes were differentially expressed (P < 0.05): 316 genes were upregulated while 364 were downregulated in the wild-type strain grown in glucose-limited conditions. To assess their biological functions, 418 of the 680 DEGs were clustered in 92 special pathways in KEGG; 12 significant pathways were primarily involved in the metabolism of amino acids, energy, carbohydrates, and cofactors and vitamins. Here, we focused on the KEGG pathways associated with carbohydrate metabolism. Compared to the wild-type strain grown at high-glucose concentrations, the DEGs involved in the citrate cycle including genes encoding phosphoenolpyruvate carboxykinase and the PDH complex were significantly upregulated in the low-glucose condition, while the gene encoding citrate synthase was downregulated. The glycolysis/gluconeogenesis pathway in the S. bovis S1 wild-type strain was dependent on glucose concentrations in the culture. In this pathway, the upregulated genes were consistent with those in the citrate cycle pathway except for the gene encoding galactose mutarotase. The genes encoding enzymes including L-LDH, type I glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, 6-phosphofructokinase, phosphopyruvate hydratase, triose-phosphate isomerase, phosphoglycerate mutase, and fructose-1,6-bisphosphate aldolase were downregulated in low-glucose conditions. Besides, the low-glucose condition also upregulated the expression of genes encoding alpha-amylase, pyruvate formate lyase-activating protein, as well as CcpA. All of these results indicated that, although to a lesser extent, glucose availability could affect the fermentation pattern by transcriptional regulation in S. bovis S1 wild-type strain. This was also confirmed by the fermentation profiles.
Finally, the transcriptome profiles of ccpA mutant strains grown in glucose-excess and -limited conditions were compared. Here, a total of 340 genes were differentially expressed; 144 genes were upregulated while 196 were downregulated; 210 genes were annotated to different KEGG pathways. Unlike in the wild-type strain, no gene in the citrate cycle pathway was significantly upregulated; the gene encoding phosphoenolpyruvate carboxykinase and a cluster of three genes that encoded aconitate hydratase, citrate synthase and NADP-dependent isocitrate dehydrogenase were significantly downregulated in this comparison. Besides, the number of DEGs in pyruvate metabolism, starch and sucrose metabolism, fructose and mannose metabolism, and glycolysis was considerably lesser than the comparison of wild-type strains grown in glucose-excess and -limited conditions. These findings suggested that CcpA mediated the differential expression of these genes. The gene encoding L-LDH was downregulated in this comparison, which was consistent with the results of the comparison in wild-type strain. This result indicated that in low-glucose conditions, the yield of lactate of S. bovis S1 could reduce even in the absence of ccpA.
In the ruminants, inhibition of the overgrowth of S. bovis in the rumen is critical for the prevention of rumen acidosis (Wang et al., 2015). In the present study, deletion of ccpA resulted in a decreased growth rate of S. bovis S1, which implied that the overgrowth of S. bovis could be inhibited by controlling CcpA synthesis, consequently relieving rumen acidosis. The results are consistent with the results obtained in other lactic acid-producing bacteria, such as Lactobacillus bulgaricus (Li et al., 2016) and Lactobacillus casei (Esteban et al., 2004), although the growth differences between S. bovis 12U1 wild strain and ccpA-disrupted mutants have not been previously observed (Asanuma et al., 2004a). Previous study reported that CcpA regulated the growth rate of bacteria depending on the extracellular glucose concentration in Streptococcus intermedius (Imaki et al., 2014). However, in the present study, we observed a reduced growth due to the inactivation of ccpA in both glucose-excess and glucose-limited conditions.
In addition, suppressing the overproduction of lactate by S. bovis is also important to prevent rumen acidosis. Bacteria can shift fermentation patterns to produce lactate instead of mixed acids depending on the growth condition (McLeod et al., 2017). In the present study, we observed an inconsistent fermentation pattern for organic acids in S. bovis S1 grown in glucose-excess and glucose-limited conditions; fermentation products, including lower acetate and formate and higher lactate percentages, were obtained in excess-glucose conditions, which was consistent with the results of Chen et al. (2016a). This finding suggested that increasing glucose concentrations could induce a shift in fermentation patterns from heterofermentation to homofermentation in S. bovis S1. This is also observed in other lactic acid bacteria (Thomas et al., 1979; Bitoun et al., 2012; McLeod et al., 2017). CcpA has been reported to mediate the transcription of the gene encoding LDH (ldh) (Luesink et al., 1998; van den Bogaard et al., 2000; Asanuma et al., 2004a), PFL (pfl) (Asanuma et al., 2004a; Reed et al., 2018), and acetate kinase (ack) (Wunsche et al., 2012; Kim and Burne, 2017; Reed et al., 2018), thereby altering the production of organic acids. Here, we showed that the deletion of ccpA led to an increase in the percentage of acetate and formate and a significant reduction in lactate percentage. Notably, there was over a 10% reduction of lactate production in the ccpA-mutant in both exponential and stationary phases in glucose-excess conditions; while in glucose-limited conditions, only a 7% decrease was observed. The results of fermentation end-products indicated that CcpA could be a key factor for changes in fermentation pattern of S. bovis S1, and this was dependent on the glucose concentration.
In the EMP pathway, glucose-6-phosphate is converted to FBP by glucose-6-phosphate isomerase and phosphofructokinase, and FBP is then split into GAP and DHAP by FBP aldolase (FBA) (Bond and James, 1998). The intracellular FBP concentration fluctuates depending on the rate of glucose influx (Asanuma and Hino, 2000, 2002a). When S. bovis is growing in the presence of excess glucose, the intracellular FBP concentration is high, and as the availability of glucose decreases, the intracellular FBP concentration also decreases (Bond and James, 1998). Correspondingly, we observed a decrease in FBP in S. bovis S1 growing in glucose-limited media as compared to glucose-rich media. LDH activity can be induced by FBP in S. bovis (Wolin, 1964), and FBP concentration is positively correlated with the levels of ldh-mRNA and inversely correlated with the pfl-mRNA levels (Asanuma et al., 2004b). In this study, the increased lactate and decreased formate production in glucose-rich media might be associated with the increased FBP concentration. In addition, the concentration of FBP was reduced in ccpA deletion strains, which might be attributed to the negative effect of ccpA inactivation on the glycolysis process.
Regulation of CcpA on the target genes is dependent on glucose level in Clostridium difficile (Antunes et al., 2012) and Streptococcus mutans (Kim et al., 2019). To better understand the transcriptional regulation of CcpA with glucose concentration in S. bovis S1, we performed a transcriptomic analysis of S. bovis S1 wild-type and ccpA mutant strains at different glucose levels. Among the four pair-wise comparisons, the DEGs were the highest between S. bovis S1 wild-type and ccpA mutant strains grown at high glucose concentrations and were the lowest between these two strains grown in low glucose concentrations. The findings showed that CcpA could indeed regulate the gene transcription of S. bovis S1 depending on the extracellular glucose concentration. This was consistent with our metabolite analyses. Also, these results implied that controlling the growth and metabolism of S. bovis by CcpA may have great potential for prevention of rumen acidosis in ruminants fed a high-concentrate diet. Moreover, the DEGs in S. bovis S1 wild-type grown in glucose-excess and glucose-limited conditions were twice as high as those of ccpA mutants. This indicated that the gene expression of S. bovis S1 was dependent on extracellular glucose concentration, and the observed effect was attenuated by the inactivation of ccpA.
Glucose is metabolized to pyruvate via glycolysis after uptake, then pyruvate is further converted to lactate, formate, acetate, and ethanol in S. bovis (Asanuma and Hino, 2000). Interestingly, the transcriptomic analyses in this study focused on glycolysis and pyruvate metabolism, especially in the comparison between S. bovis S1 wild-type strain grown in glucose-excess and glucose-limited conditions and the comparison between S. bovis S1 wild-type and ccpA mutant strains grown at high glucose concentrations. For S. bovis S1 wild-type strain, most PDH complex genes were significantly upregulated in low glucose conditions, and genes related to the glycolysis and lactate production pathways were downregulated. This was consistent with the result for metabolites, where low glucose concentrations altered the metabolism of S. bovis S1 wild-type strain to produce less lactate. In lactic acid bacteria, the metabolic shifts during growth are associated with multiple factors, among which the intracellular redox potential reflected by NADH/NAD+ ratio is a key sensor (Goel et al., 2012; van Hoek and Merks, 2012). It is reported that ldh appears to be regulated by the global regulators Rex and CcpA in Enterococcus faecalis (Opsata et al., 2010; Mehmeti et al., 2011), and both of which are sensitive to NADH/NAD+ levels. In this study, though we observed a reduced transcription level of rex and elevated ccpA in response to glucose limitation, the repression or activation of these two transcriptional regulators on target genes responding to glucose concentration need to be further investigated. In addition, herein, the ldh gene showed reduced expression, which probably was a consequence of low FBP concentration at low glucose concentration (Abbe et al., 1982). Interestingly, the differential expressions of those genes were eliminated after the ccpA gene deletion, which indicated that CcpA played an important regulatory role at different levels of glucose. Notably, despite glucose being the main carbon source for S. bovis S1 in the present study, the genes involved in starch degradation including starch phosphorylase and alpha-amylase were significantly upregulated in low glucose conditions. This phenomenon might be a potential response of the bacteria to insufficient energy resources.
Catabolite control protein A is a pleiotropic regulator involved in controlling in carbon metabolism in many bacteria species in response to changes in overall energy levels and amount of carbohydrate (Abranches et al., 2008; Willenborg et al., 2014; Hofmann et al., 2021). For example, >80% of the genes are controlled by CcpA in response to glucose in Bacillus subtilis (Moreno et al., 2001). The results of the present study showed that the genes involved in the pyruvate metabolism were upregulated for the production of formate, while downregulated for lactate production in absence of ccpA, which was consistent with the alteration in metabolite levels in ccpA mutants grown in both glucose-excess or glucose-limited conditions. These results implied that the fermentation patterns of S. bovis S1 could be regulated by CcpA via modifying the enzyme transcription, similar to the previous findings in other lactic acid bacteria (Lu et al., 2018; Chen et al., 2019). Transcription of acetate kinase is enhanced in ccpA mutants of Lactobacillus plantarum (Lu et al., 2018) and S. mutans (Kim and Burne, 2017). Interestingly, though the transcriptomic showed non-significant differences in acetate kinase levels between S. bovis S1 wild-type and ccpA mutant strains, the qPCR results showed an elevated expression of acetate kinase after ccpA inactivation at the high glucose concentrations, which validated the metabolic shifts. However, this elevation was attenuated when cells were grown at low glucose concentrations, which suggested that the acetate kinase regulation by CcpA depended on glucose availability. Furthermore, the knockout of ccpA also largely influenced fructose and mannose metabolism pathway of S. bovis S1 grown under glucose-excess condition; genes involved in the transportation of fructose and mannose were upregulated, and those related to the metabolism of fructose and mannose were downregulated. However, this effect was also attenuated when cells were grown in the low-glucose conditions. As found in C. difficile, the attenuated regulation of target genes by CcpA in the low-glucose conditions may point toward inactive CcpA regulatory systems of S. bovis S1 at low glucose concentrations (Antunes et al., 2011, 2012).
Apart from carbon metabolism, the glucose concentrations also influenced histidine metabolism and nitrogen metabolism in both S. bovis S1 wild-type and ccpA mutant strains. Almost all the genes in these pathways were downregulated at the high glucose concentrations, which suggested that the amino acid metabolism and nitrogen metabolism of S. bovis S1 were also altered in response to extracellular glucose concentration. Fatty acids are essential components of membranes in all organisms, and their biosynthesis and degradation are important for maintaining membrane lipid homeostasis in response to environmental changes (Fujita et al., 2007; Tojo et al., 2011). It has been reported that CcpA regulates the fatty acid metabolism in B. subtilis and S. mutans in response to environmental changes (Tojo et al., 2011; Faustoferri et al., 2015). In our present study, nine DEGs involved in fatty acid biosynthesis were downregulated in ccpA mutant as compared to the wild-type strains at high glucose concentrations, similar to the previous findings in L. plantarum (Lu et al., 2018). Interestingly, this phenomenon was abolished when cells were grown in glucose-limited conditions. These results indicated that the absence of ccpA attenuated the fatty acid synthesis of S. bovis S1 only when the energy was sufficient.
In conclusion, high glucose concentration in the media led to the rapid growth of S. bovis S1 and a shift to produce more lactate. The inactivation of ccpA slowed down the proliferation of S. bovis S1 and shifted the fermentation pattern toward the production of less lactate and more formate and acetate. The whole-transcriptome analyses showed that the pathways for histidine metabolism, nitrogen metabolism, and carbohydrate metabolism were significantly altered in response to glucose concentration. CcpA was involved in the regulation of metabolic processes, including glycolysis, pyruvate metabolism, fructose- and mannose-metabolism, and fatty acid biosynthesis in S. bovis S1. The transcriptional regulation by CcpA was more potent for S. bovis S1 grown in glucose-excess conditions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA746134.
YJ designed and conducted the experiments, analyses, and wrote the manuscript. YF, HS, and YZ helped to collect the samples. HW directed in experiments design and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (NSFC Nos. 31872988 and 31572429) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PADA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.767769/full#supplementary-material
Abbe, K., Takahashi, S., and Yamada, T. (1982). Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J. Bacteriol. 152, 175–182.
Abranches, J., Nascimento, M. M., Zeng, L., Browngardt, C. M., Wen, Z. T., Rivera, M. F., et al. (2008). CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 190, 2340–2349. doi: 10.1128/Jb.01237-07
Antunes, A., Camiade, E., Monot, M., Courtois, E., Barbut, F., Sernova, N. V., et al. (2012). Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 40, 10701–10718. doi: 10.1093/nar/gks864
Antunes, A., Martin-Verstraete, I., and Dupuy, B. (2011). CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79, 882–899. doi: 10.1111/j.1365-2958.2010.07495.x
Asanuma, N., and Hino, T. (1997). Tolerance to low pH and lactate production in rumen bacteria. Animal Sci. Technol. 68, 367–376.
Asanuma, N., and Hino, T. (2000). Effects of pH and energy supply on activity and amount of pyruvate formate-lyase in Streptococcus bovis. Appl. Environ. Microbiol. 66, 3773–3777. doi: 10.1128/aem.66.9.3773-3777.2000
Asanuma, N., and Hino, T. (2002b). Regulation of fermentation in a ruminal bacterium, Streptococcus bovis, with special reference to rumen acidosis. Anim. Sci. J. 73, 313–325.
Asanuma, N., and Hino, T. (2002a). Fructose bisphosphate aldolase activity and glycolytic intermediate concentrations in relation to lactate production in Streptococcus bovis. Anaerobe 8, 1–8. doi: 10.1006/anae.2001.0398
Asanuma, N., Iwamoto, M., and Hino, T. (1997). Regulation of lactate dehydrogenase synthesis in a ruminal bacterium, Streptococcus bovis. J. General Appl. Microbiol. 43, 325–331. doi: 10.2323/jgam.43.325
Asanuma, N., Iwamoto, M., and Hino, T. (1999). Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiol. UK 145, 151–157. doi: 10.1099/13500872-145-1-151
Asanuma, N., Yoshii, T., and Hino, T. (2004a). Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 70, 5244–5251. doi: 10.1128/aem.70.9.5244-5251.2004
Asanuma, N., Yoshii, T., Kikuchi, M., and Hino, T. (2004b). Effects of the overexpression of fructose-1,6-bisphosphate aldolase on fermentation pattern and transcription of the genes encoding lactate dehydrogenase and pyruvate formate-lyase in a ruminal bacterium, Streptococcus bovis. J. General Appl. Microbiol. 50, 71–78. doi: 10.2323/jgam.50.71
Bitoun, J. P., Liao, S., Yao, X., Xie, G. G., and Wen, Z. T. (2012). The redox-sensing regulator Rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS One 7:e44766. doi: 10.1371/journal.pone.0044766
Bond, D. R. R., and James, B. (1998). Relationship between intracellular phosphate, proton motive force, and rate of nongrowth energy dissipation (Energy Spilling) in Streptococcus bovis JB1. Appl. Environ. Microbiol. 64, 976–981.
Chen, C., Wang, L., Lu, Y., Yu, H., and Tian, H. (2019). Comparative transcriptional analysis of lactobacillus plantarum and its ccpA-knockout mutant under galactooligosaccharides and glucose conditions. Front. Microbiol. 10:1584. doi: 10.3389/fmicb.2019.01584
Chen, C., Zhao, G., Chen, W., and Guo, B. (2015). Metabolism of fructooligosaccharides in lactobacillus plantarum ST-III via differential gene transcription and alteration of cell membrane fluidity. Appl. Environ. Microbiol. 81, 7697–7707. doi: 10.1128/AEM.02426-15
Chen, L., Luo, Y., Wang, H., Liu, S., Shen, Y., and Wang, M. (2016b). Effects of glucose and starch on lactate production by newly isolated streptococcus bovis s1 from saanen goats. Appl. Environ. Microbiol. 82, 5982–5989. doi: 10.1128/AEM.01994-16
Chen, L., Liu, S., Wang, H., Wang, M., and Yu, L. (2016a). Relative significances of pH and substrate starch level to roles of Streptococcus bovis S1 in rumen acidosis. AMB Express 6:80. doi: 10.1186/s13568-016-0248-2
Dai, X., Hackmann, T. J., Lobo, R. R., and Faciola, A. P. (2020). Lipopolysaccharide stimulates the growth of bacteria that contribute to ruminal acidosis. Appl. Environ. Microbiol. 86, 19. doi: 10.1128/AEM.02193-19
Esteban, Carlos, D., Mahr, Kerstin, Monedero, et al. (2004). Complementation of ΔccpA mutant of Lactobacillus casei with CcpA mutants affected in the DNA- and cofactor-binding domains. Microbiology 150, 613–620.
Faustoferri, R. C., Hubbard, C. J., Santiago, B., Buckley, A. A., Seifert, T. B., and Quivey, R. G. (2015). Regulation of fatty acid biosynthesis by the global regulator CcpA and the local regulator FabT in Streptococcus mutans. Mol. Oral Microbiol. 30, 128–146. doi: 10.1111/omi.12076
Fujita, Y., Matsuoka, H., and Hirooka, K. (2007). Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66, 829–839. doi: 10.1111/j.1365-2958.2007.05947.x
Goel, A., Wortel, M. T., Molenaar, D., and Teusink, B. (2012). Metabolic shifts: a fitness perspective for microbial cell factories. Biotechnol. Lett. 34, 2147–2160. doi: 10.1007/s10529-012-1038-9
Hofmann, J. D., Biedendieck, R., Michel, A. M., Schomburg, D., Jahn, D., and Neumann-Schaal, M. (2021). Influence of L-lactate and low glucose concentrations on the metabolism and the toxin formation of Clostridioides difficile. PLoS One 16:e0244988. doi: 10.1371/journal.pone.0244988
Imaki, H., Tomoyasu, T., Yamamoto, N., Taue, C., Masuda, S., Takao, A., et al. (2014). Identification and characterization of a novel secreted glycosidase with multiple glycosidase activities in streptococcus intermedius. J. Bacteriol. 196, 2817–2826. doi: 10.1128/Jb.01727-14
Kenney, N. M., Vanzant, E. S., Harmon, D. L., and McLeod, K. R. (2015). Direct-fed microbials containing lactate-producing bacteria influence ruminal fermentation but not lactate utilization in steers fed a high-concentrate diet. J. Anim. Sci. 93, 2336–2348. doi: 10.2527/jas.2014-8570
Kim, H. M., Waters, A., Turner, M. E., Rice, K. C., and Ahn, S. J. (2019). Regulation of cid and lrg expression by CcpA in Streptococcus mutans. Microbiol 165, 113–123. doi: 10.1099/mic.0.000744
Kim, J. N., and Burne, R. A. (2017). CcpA and CodY coordinate acetate metabolism in streptococcus mutans. Appl. Environ. Microbiol. 83:16. doi: 10.1128/AEM.03274-16
Li, C., Sun, J. W., Zhang, G. F., and Liu, L. B. (2016). Effect of the absence of the CcpA gene on growth, metabolic production, and stress tolerance in Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 99, 104–111. doi: 10.3168/jds.2015-10321
Lu, Y., Song, S., Tian, H., Yu, H., Zhao, J., and Chen, C. (2018). Functional analysis of the role of CcpA in Lactobacillus plantarum grown on fructooligosaccharides or glucose: a transcriptomic perspective. Microb. Cell Fact. 17:201. doi: 10.1186/s12934-018-1050-4
Luesink, E. J., van Herpen, R. E., Grossiord, B. P., Kuipers, O. P., and de Vos, W. M. (1998). Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30, 789–798. doi: 10.1046/j.1365-2958.1998.01111.x
Marounek, M., and Bartos, S. (1987). Interactions between rumen amylolytic and lactate-utilizing bacteria in growth on starch. J. Appl. Bacteriol. 63, 233–238. doi: 10.1111/j.1365-2672.1987.tb04941.x
McAllister, T. A., Cheng, K. J., Rode, L. M., and Forsberg, C. W. (1990). Digestion of barley, maize, and wheat by selected species of ruminal bacteria. Appl. Environ. Microbiol. 56, 3146–3153. doi: 10.1128/aem.56.10.3146-3153.1990
McLeod, A., Mosleth, E. F., Rud, I., Branco Dos Santos, F., Snipen, L., Liland, K. H., et al. (2017). Effects of glucose availability in Lactobacillus sakei; metabolic change and regulation of the proteome and transcriptome. PLoS One 12:e0187542. doi: 10.1371/journal.pone.0187542
Mehmeti, I., Jonsson, M., Fergestad, E. M., Mathiesen, G., Nes, I. F., and Holo, H. (2011). Transcriptome, proteome, and metabolite analyses of a lactate dehydrogenase-negative mutant of Enterococcus faecalis V583. Appl. Environ. Microbiol. 77, 2406–2413. doi: 10.1128/AEM.02485-10
Wolin, M. J. (1964). Fructose-1,6-diphosphate requirement of streptococcal lactic dehydrogenases. Science 146, 775–777.
Moreno, M. S., Schneider, B. L., Maile, R. R., Weyler, W., and Saier, M. H. Jr. (2001). Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39, 1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x
Nocek, J. E. (1997). Bovine acidosis: implications on laminitis. J. Dairy Sci. 80, 1005–1028. doi: 10.3168/jds.S0022-0302(97)76026-0
Opsata, M., Nes, I. F., and Holo, H. (2010). Class IIa bacteriocin resistance in Enterococcus faecalis V583: the mannose PTS operon mediates global transcriptional responses. BMC Microbiol. 10:224. doi: 10.1186/1471-2180-10-224
Perni, S., Andrew, P. W., and Shama, G. (2005). Estimating the maximum growth rate from microbial growth curves: definition is everything. Food Microbiol. 22, 491–495. doi: 10.1016/j.fm.2004.11.014
Reed, J. M., Olson, S., Brees, D. F., Griffin, C. E., Grove, R. A., Davis, P. J., et al. (2018). Coordinated regulation of transcription by CcpA and the Staphylococcus aureus two-component system HptRS. PLoS One 13:e0207161. doi: 10.1371/journal.pone.0207161
Russell, J. R., and Hino, T. (1985). Regulation of lactate production in Streptococcus bovis: A spiraling effect that contributes to rumen acidosis. J. Dairy Sci. 68, 1712–1721. doi: 10.3168/jds.S0022-0302(85)81017-1
Thomas, T. D., Ellwood, D. C., and Longyear, V. M. (1979). Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J. Bacteriol. 138, 109–117. doi: 10.1128/jb.138.1.109-117.1979
Tojo, S., Satomura, T., Matsuoka, H., Hirooka, K., and Fujita, Y. (2011). Catabolite repression of the bacillus subtilis FadR regulon, which is involved in fatty acid catabolism. J. Bacteriol. 193, 2388–2395. doi: 10.1128/Jb.00016-11
van den Bogaard, P. T., Kleerebezem, M., Kuipers, O. P., and de Vos, W. M. (2000). Control of lactose transport, beta-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J. Bacteriol. 182, 5982–5989. doi: 10.1128/JB.182.21.5982-5989.2000
van Hoek, M. J. A., and Merks, R. M. H. (2012). Redox balance is key to explaining full vs. partial switching to low-yield metabolism. BMC Syst. Biol. 6:22.
Wang, H. R., Pan, X. H., Wang, C., Wang, M. Z., and Yu, L. H. (2015). Effects of different dietary concentrate to forage ratio and thiamine supplementation on the rumen fermentation and ruminal bacterial community in dairy cows. Animal Prod. Sci. 55, 189–193. doi: 10.1071/An14523
Willenborg, J., de Greeff, A., Jarek, M., Valentin-Weigand, P., and Goethe, R. (2014). The CcpA regulon of Streptococcus suis reveals novel insights into the regulation of the streptococcal central carbon metabolism by binding of CcpA to two distinct binding motifs. Mol. Microbiol. 92, 61–83. doi: 10.1111/mmi.12537
Keywords: Streptococcus bovis S1, catabolite control protein A, transcriptome, glucose concentration, metabolism regulation
Citation: Jin Y, Fan Y, Sun H, Zhang Y and Wang H (2021) Transcriptome Analysis Reveals Catabolite Control Protein A Regulatory Mechanisms Underlying Glucose-Excess or -Limited Conditions in a Ruminal Bacterium, Streptococcus bovis. Front. Microbiol. 12:767769. doi: 10.3389/fmicb.2021.767769
Received: 31 August 2021; Accepted: 20 October 2021;
Published: 18 November 2021.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Greg Somerville, University of Nebraska–Lincoln, United StatesCopyright © 2021 Jin, Fan, Sun, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongrong Wang, aHJ3YW5nQHl6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.