- 1Department of Laboratory Medicine, Linkou Chang Gung Memorial Hospital, Taoyuan, Taiwan
- 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 3Department of Medicine, College of Medicine, Chang Gung University, Taoyuan, Taiwan
Oxacillin-resistant Staphylococcus lugdunensis (ORSL) is considered a life-threatening isolate in healthcare settings. Among ORSL clones, ST6-SCCmec II strains are associated with an endemic spread in hospitals. We analyzed the complete genome of ORSL CGMH-SL118, a representative strain. Results revealed that this strain contained three MGEs (two prophages and one plasmid) other than the SCCmec II element, which showed remarkable differences in genome organization compared to the reference strains from NCBI. Eight multidrug-resistant genes were identified. All but blaZ were carried by MGEs, such as the SCCmec II element [mecA, ant (9)-Ia, and ermA] and the prophage φSPbeta [aac (6')-aph (2'), aph (3')-III, and ant (6)-Ia], indicating that MGEs carrying multidrug-resistant genes may be important for ST6 strains. The prophage φSPbeta contains sasX gene, which was responsible for the pathogenesis of Staphylococcus aureus. A phage-mediated resistant island containing fusB (SlRIfusB-118) was found near φSPbeta, which was highly homologous to type III SeRIfusB-5907 of Staphylococcus epidermidis. In contrast to previous studies, over 20% of ST6 isolates showed a fusidic acid-resistant phenotype, suggesting that phage-mediated intraspecies transmission of resistant islands may become an important issue for ST6 strains. Sixty-eight clinical isolates of ST6 Staphylococcus lugdunensis (50 OSSL, oxacillin-sensitive S. lugdunensis, and 18 ORSL, including CGMH-SL118) collected from various types of specimens in the hospital were studied. Among these isolates in this study, ORSL showed similar drug-resistant genes and phenotypes as CGMH-SL118. The comparative genomic analyses highlight the contribution of MGEs in the development and dissemination of antimicrobial resistance in ST6 strains, suggesting that resistance determinants and virulence factors encoded by MGEs provide a survival advantage for successful colonization and spread in healthcare settings.
Introduction
Staphylococcus lugdunensis was considered a commensal coagulase-negative staphylococcal species (CoNS) until the emergence of nosocomial infections, after which it became an important pathogen (Yeh et al., 2015). Unlike other CoNS, S. lugdunensis showed similar pathogenicity to Staphylococcus aureus (Argemi et al., 2015; Douiri et al., 2016; Heilbronner and Foster, 2021) and caused various infections, such as skin and soft tissue infections, bone and joint infections, bacteraemia, and infective endocarditis (Wu et al., 2011; Yeh et al., 2015, 2016; Douiri et al., 2016). The mortality rate of endocarditis caused by S. lugdunensis infection is nearly 40% (Liu et al., 2010), while that caused by other CoNS is only 14.3% (Fernandez-Rufete et al., 2012; Molina et al., 2013), suggesting that S. lugdunensis is more virulent than other CoNS. Previous epidemiological surveillance showed that most isolates remained susceptible to oxacillin (Lin et al., 2015; Yeh et al., 2015, 2016); however, a significantly high proportion of oxacillin-resistant S. lugdunensis (ORSL) strains were found in nosocomial infections (Lin et al., 2015), indicating that transfer of SCCmec (staphylococcal cassette chromosome mec) should be monitored in hospitals. In fact, our previous investigation identified an endemic spreading in northern Taiwan caused by a group of SCCmec II, ST6 strains (Cheng et al., 2015). Further characterization of their SCCmec cassette structure revealed similarities with S. aureus, which suggested that ORSL may act as an interspecies for SCCmec transfer to S. aureus in hospitals (Chang et al., 2019).
It is interesting to note that the endemic spread was caused by SCCmec II, ST6 strains, since most ORSL isolates belong to SCCmec V strains (Yeh et al., 2015, 2016). According to our previous studies regarding ORSL antimicrobial susceptibilities, SCCmec II strains showed more multidrug-resistant phenotypes than SCCmec V strains (Yeh et al., 2015), suggesting that drug resistance may enhance SCCmec II endemic spread. In addition to the drug resistance, virulence factors may also enhance the endemic spread. Although few studies have revealed virulence factors associated with their pathogenicity (Giormezis et al., 2015; Argemi et al., 2017b; Heilbronner and Foster, 2021), the above evidence originated from OSSL, while studies involving ORSL infections are still limited.
Whole-genome sequencing analysis can provide more comprehensive information to investigate virulence factors, drug resistance, pathogenesis, and other factors, which have been widely used in S. aureus studies (Kuroda et al., 2001; Laabei et al., 2014). A prospective study of these reports found that most virulence factors were encoded by mobile genetic elements (MGEs), such as plasmids, prophages, or pathogenicity islands (Malachowa and DeLeo, 2010). Staphylococcus lugdunensis N920143 was one of the earlier strains that provided complete genome sequence and information about virulence factors, although few MGEs were found in this strain compared to S. aureus (Heilbronner et al., 2011). Recent comparative genomic analyses have identified several novel MGEs in S. lugdunensis, and few of the pathogenicity islands have been reported thus far (Argemi et al., 2017a, 2018; Lebeurre et al., 2019).
Recently, the first complete genome sequence of S. lugdunensis ORSL JICS135 revealed the presence of several virulence features compared to other OSSLs (Shibuya et al., 2020). The SCCmec element of JICS135 contained two genes encoding microbial surface components recognizing adhesive matrix molecules (MSCRAMM)-like proteins, which are considered responsible for strain pathogenesis (Patti et al., 1994). However, structural comparisons indicated that partial regions of SCCmecJICS135 were similar to those of two previously reported ORSL SCCmec V strains: CMUH 22 and CMUH25 (Chang et al., 2017). To elucidate the undiscovered virulence factors in SCCmec II, ST6 ORSL, whole-genome sequence analysis was adopted to decipher its genome structure. The present study demonstrated that SCCmec II and ST6 strains contained unique MGEs encoding a putative virulence factor and antimicrobial resistance genes, which may be responsible for its endemic spread.
Materials and Methods
Bacterial Isolates
All S. lugdunensis isolates were collected from 2009 to 2014 at Taiwan Linkou Chang Gung Memorial Hospital. Strain CGMH-SL118 was isolated from a blood sample and selected for whole-genome sequencing analysis. Sixty-eight clinical isolates of ST6 S. lugdunensis (50 OSSL, oxacillin-sensitive S. lugdunensis, and 18 ORSL, oxacillin-resistant S. lugdunensis, including CGMH-SL118) collected from various types of specimens in the hospital were studied, which had been published (Yeh et al., 2015, 2016); detailed information is included in Supplementary Table 1.
Whole-Genome Sequencing and Annotation
Staphylococcus lugdunensis strain CGMH-SL118 was grown on TSB medium overnight for genomic DNA extraction. The extracted genomic DNA was sequenced using the PacBio™ method (Pacific Biosciences, Menlo Park, CA, United States). A single total length 2,818,231 base pair contig was generated using three software; a de novo assembler Flye (Kolmogorov et al., 2019) was used for contig assembly, contigs scaffolding was applied using SSPACE (Boetzer and Pirovano, 2014), and scaffolds were finally polished using arrow algorithm.1 Gene annotation was generated using “Prokka v1.12”,2 which is designed for bacterial or viral genome annotation. The quality of the assembled genome was evaluated using “Quast v4.5” (Gurevich et al., 2013). The annotated data were further verified using the RAST web annotation service3 to determine the function of each gene. CGview web service4 was used for visualization of the circular genome and comparative genomic analysis of three individual strains: CGMH-SL118, N920143 (Heilbronner et al., 2011), and JICS135 (Shibuya et al., 2020; Figure 1). The prophage search was performed using “PHAST” (PHAge Search Tool)5 and “PHASTER” analyses.6 The Cas-Crispr system was verified using the CRISPR web server.7 Virulence factors were identified using the “Virulence Factors of Pathogenic Bacteria Database” (VFDB)8 web service, and antimicrobial resistance genes were analyzed using “The Comprehensive Antibiotic Resistance Database” (CARD).9
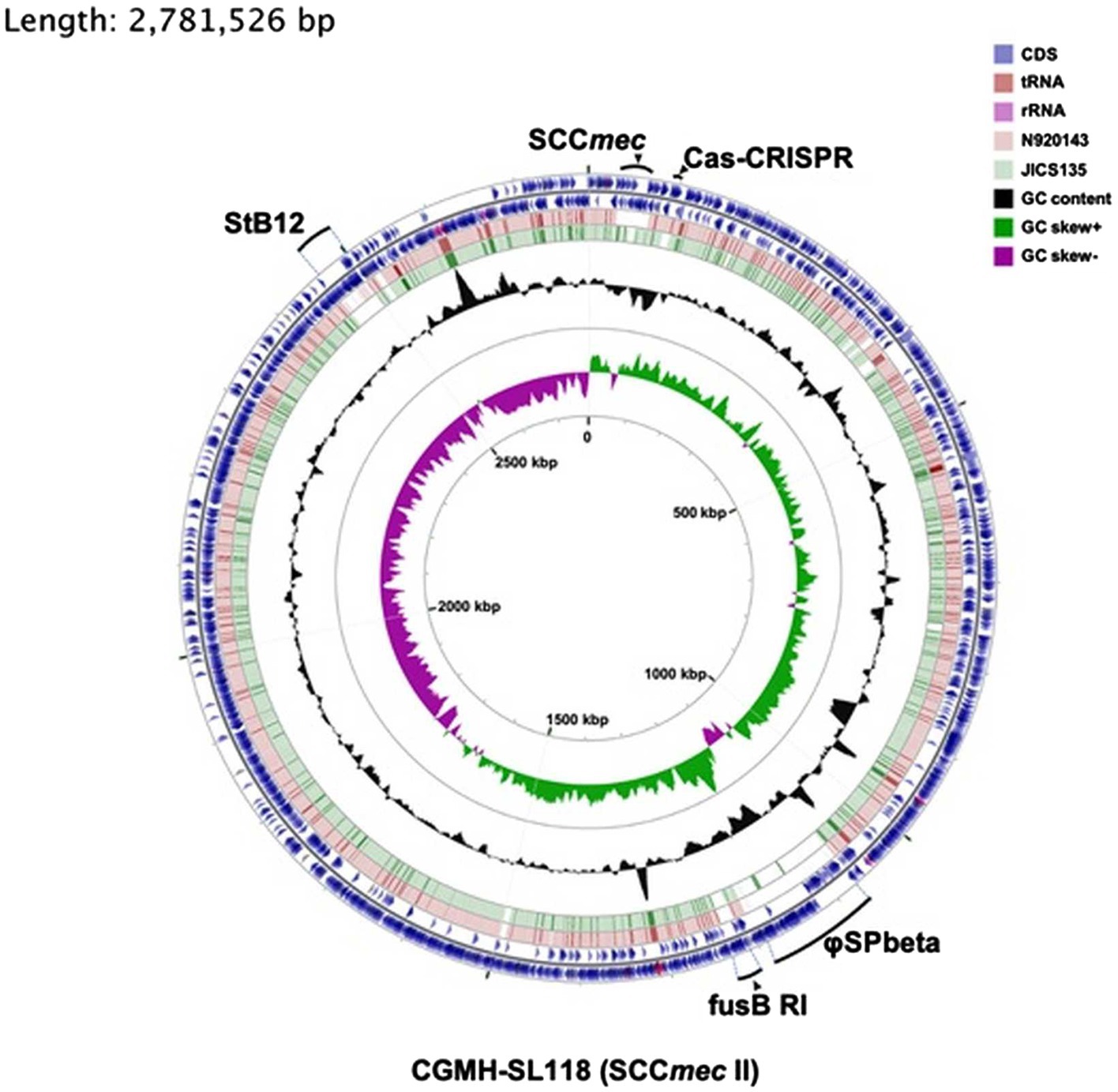
Figure 1. Comparative genomic analysis of three Staphylococcus lugdunensis strains. Six rings for the circular diagram (inner to outer): GC skew of the CGMH-SL118, GC content of the CGMH-SL118, green ring representing the genome of JICS135, pink ring representing the genome of N920143, and two of the outer blue rings representing two coding strands (forward and reverse) of CGMH-SL118. On these two outer blue circles, three colors represent different gene types: blue represents coding sequence, shallow pink represents tRNA, and deep pink represent rRNA. mobile genetic elements (MGEs; prophage; StB12 and phi-SPbeta, SCCmec, fusB-resistant island) and CRISPR-Cas regions were labeled with bold dark.
Antimicrobial Phenotype and Relative Drug-Resistant Genes Characterization
The antimicrobial phenotype was characterized using the disk diffusion method. Gentamicin, clindamycin, erythromycin, and fusidic acid disks were placed on the surface of the bacterium-grown medium, and their susceptibilities were evaluated via the inhibition zone. All procedures and evaluations were performed according to the CLSI guideline (Wayne, 2018). The prevalence of drug-resistant genes among the collected isolates was examined using PCR. Primers used in this study are listed in Table 1. PCR conditions for the detection of drug-resistant genes [including aac (6′)-aph (2′), aph (3′)-III, ant (6)-Ia, ant (9)-Ia, fusB, and ermA] were performed as previously described (Sutcliffe et al., 1996; Kao et al., 2000; Sepulveda et al., 2007; Castanheira et al., 2010; Fessler et al., 2010; Emaneini et al., 2013).
Results
Information of CGMH-SL118 Whole-Genome Sequence
To understand the difference in genome structure composition with that of S. lugdunensis, the whole-genome sequence of CGMH-SL118 was compared with those of S. lugdunensis strains N920143 and JICS135 (Table 2 and Figure 1). The genome size of CGMH-SL118 was 2,818,231bp, which is larger than that of the other two strains (JICS135, 2,687,768bp and N920143, 2,595,888bp, Table 2). The CGMH-SL118 genome also contained more coding sequences (2,659 encoded proteins) than the other two strains (N920143, 2,406 and JICS135, 2,498, Table 2). The overall genome coverage of CGMH-SL118 was 93 and 97% for JICS135 and N920143 strains, respectively. Both coverage regions showed >99% identity, indicating that most genome regions were conserved among the three strains. The total numbers of tmRNA, tRNA, and rRNA, and GC content percentages were identical between CGMH-SL118 and JICS135, which were also similar to N920143 (Table 2). In addition to these conserved regions, structural genome differences can still be found among these three strains, and most belonged to MGEs (Figure 1). Both the CGMH-SL118 and JICS135 strains were ORSL with the SCCmec element, but N920143 was OSSL without the SCCmec element (Table 2). Among these three strains, only CGMH-SL118 contained a plasmid, which was similar to the plasmid SAP107A in Staphylococcus epidermidis. In addition to the SCCmec element and plasmid, PHASTER analysis showed that both CGMH-SL118 and N920143 contained prophages, with CGMH-SL118 carrying φSPbeta and StB12 and N920143 carrying φSL1 (Heilbronner et al., 2011). Figure 1 shows the genomic sequence comparison of CGMH-SL118 with JICS135 and N920143. Two spaces on JICS135 and N920143 represented the prophage regions (StB12 and φSPbeta-like), which only existed on CGMH-SL118. A region near the prophage φSPbeta, which appeared as a blank space in the corresponding regions of N920143 and JICS135, was further investigated. This region contained a novel fusidic acid-resistant island (named SlRIfusB-118) of S. lugdunensis (fusB RI, Figure 1), which was almost identical to type III SeRIfusB-5907 (JF777506) of S. epidermidis, but partially similar to type I SaRI (AM292600) of S. aureus (Chen et al., 2011; Figure 2). Sequence blast results showed that all three fusidic acid-resistant islands were inserted near the groEL gene. The SaRI was only partially aligned with SlRIfusB-118 in two regions: the left insertion region (83–90% identity) and fusB encoding region (>99% identity). In contrast, SeRIfusB-5907 showed high similarity with SlRIfusB-118 across all islands.
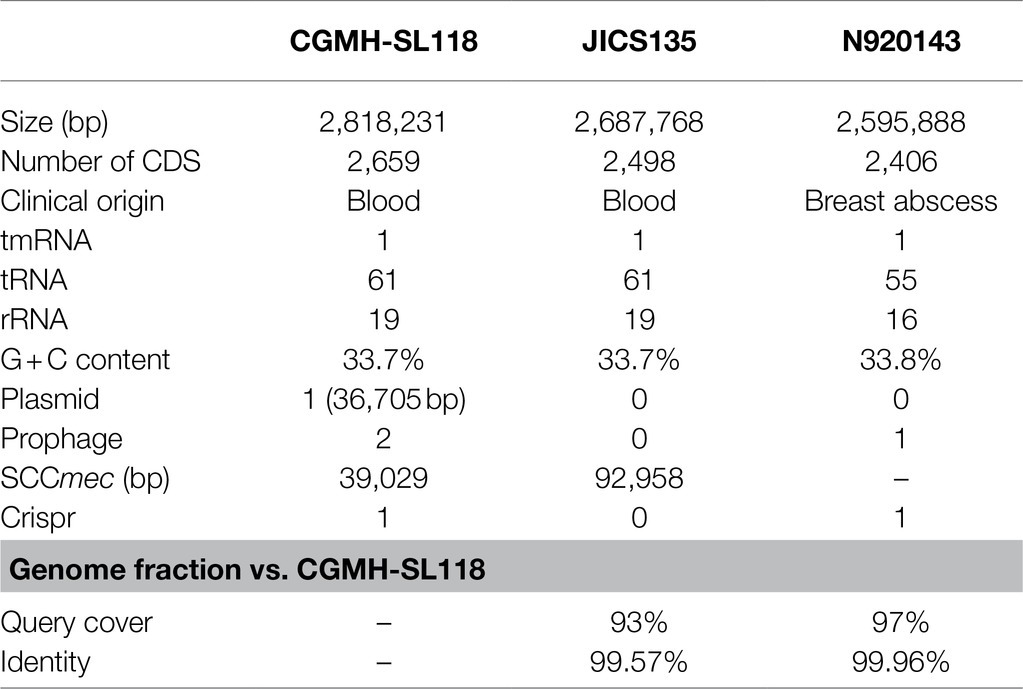
Table 2. Basic information of CGMH-SL118 whole-genome sequence analysis and comparison with other reference strains.
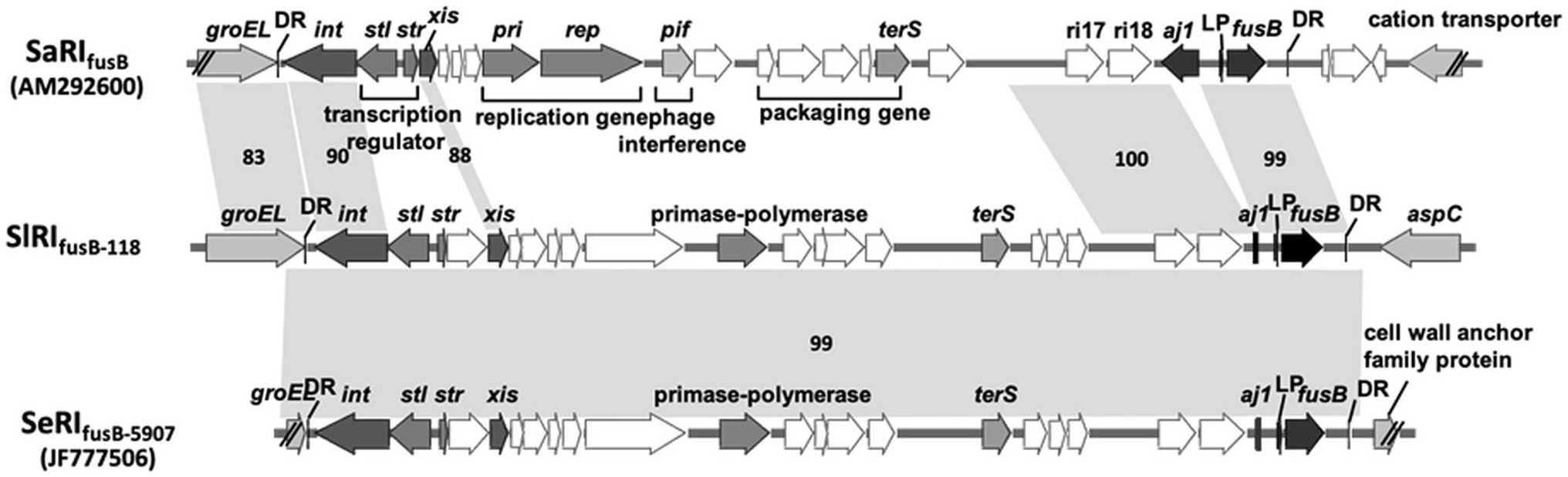
Figure 2. Comparison of fusB-resistant islands between CGMH-SL118 and the other species. Three of the fusB resistance island from Staphylococcus lugdunensis CGMH-SL118 (SlRIfusb-118), Staphylococcus epidermidis JF777506 (SeRIfusb-5907), and Staphylococcus aureus AM292600 (SlRIfusb-118) were comparatively analyzed the structure similarities. The gray color and double slash were representing the inserted location on each genome; the right side of these three islands was inserted below the groEL: the left side was varied by each strain. The dark color represents integrase (int), excisionase (xis), fusB core region genes (aj1 and fusB), and two sequences (LP; aj1-leader peptide, and DR; direct repeat sequence). The other genes were labeled with white color. Critical genes are additionally labeled with the abbreviation; stl and str are transcription regulators; pri and rep are responsible for replication; pif is response for phage interference; terS is phage terminase small subunit, which is responsible for phage packaging. The number accompanied with shadow connected to each fusB resistance island represents the sequence similarities.
Prevalence of Antibiotic-Resistant Genes Between CGMH-SL118 and Other Reference Strains
An antibiotic resistance gene survey identified several drug resistance genes involved in various resistance mechanisms in CGMH-SL118, which differed from strains N920143 and JICS135 (Table 3). A previous study has shown that OSSL N920143 has no antibiotic resistance gene (Heilbronner et al., 2011). In contrast, ORSL JICS135 has only three antibiotic resistance genes [blaZ, mecA, and aac (6')-aph (2'); Shibuya et al., 2020], all of which were also identified in CGMH-SL118. Interestingly, except for mecA carried on the SCCmec element, the other two genes were located in the genomic regions of CGMH-SL118, which differed from JICS135. In CGMH-SL118, blaZ was located close to the SCCmec element, and aac (6')-aph (2') was found in the prophage φSPbeta region (Figure 3A). In addition to these three genes, other antibiotic resistance genes were also identified in CGMH-SL118, including three aminoglycoside [aph (3′)-III, ant (6)-Ia, ant (9)-Ia], one fusidic acid (fusB), and one macrolide (ermA)-resistant genes. Among these drug-resistant genes, three aminoglycoside-resistant genes were located on the prophage φSPbeta-like, which was located close to SlRIfusB-118. CGMH-SL118 harbored multiple copies of ermA and ant (9)-Ia; one was located on the SCCmec cassette, and the other two were located aside to two prophage regions. Moreover, we observed ant (9)-Ia co-localized with ermA on the transposon Tn554 (Figure 3A).
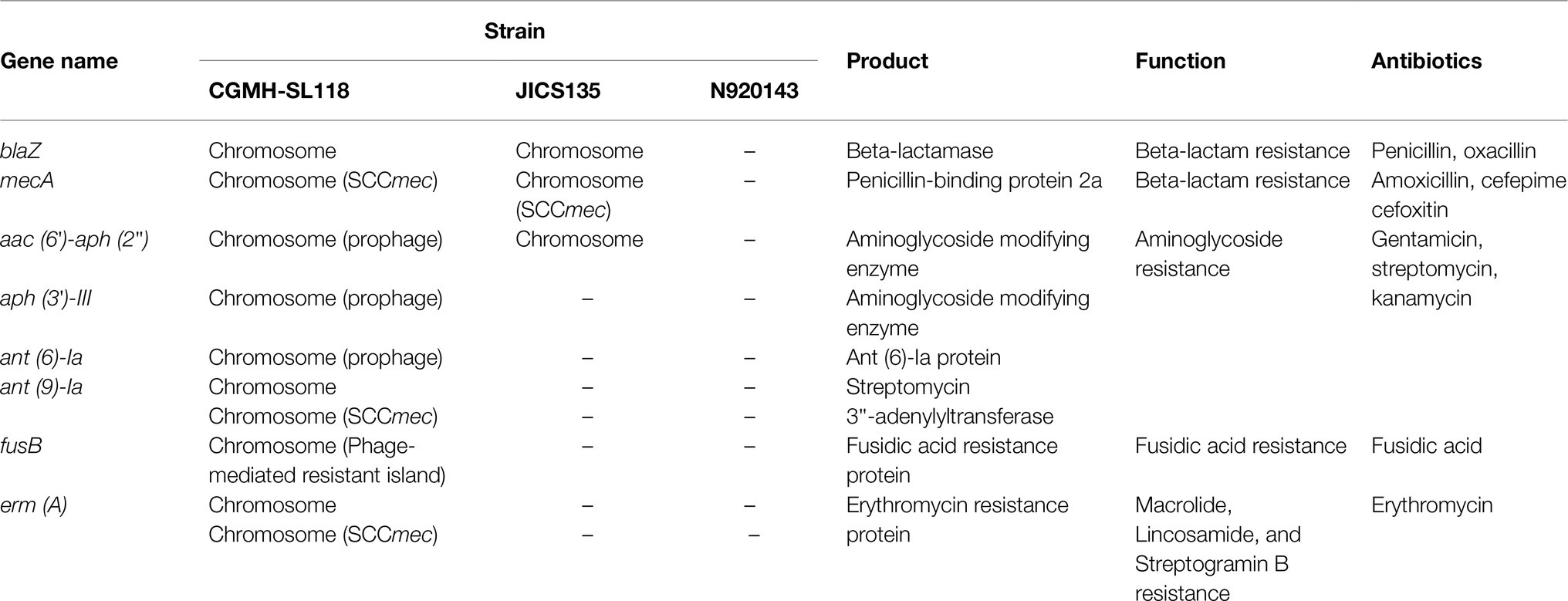
Table 3. Comparison of antibiotic-resistant gene distribution between CGMH-SL118 and other reference strains.
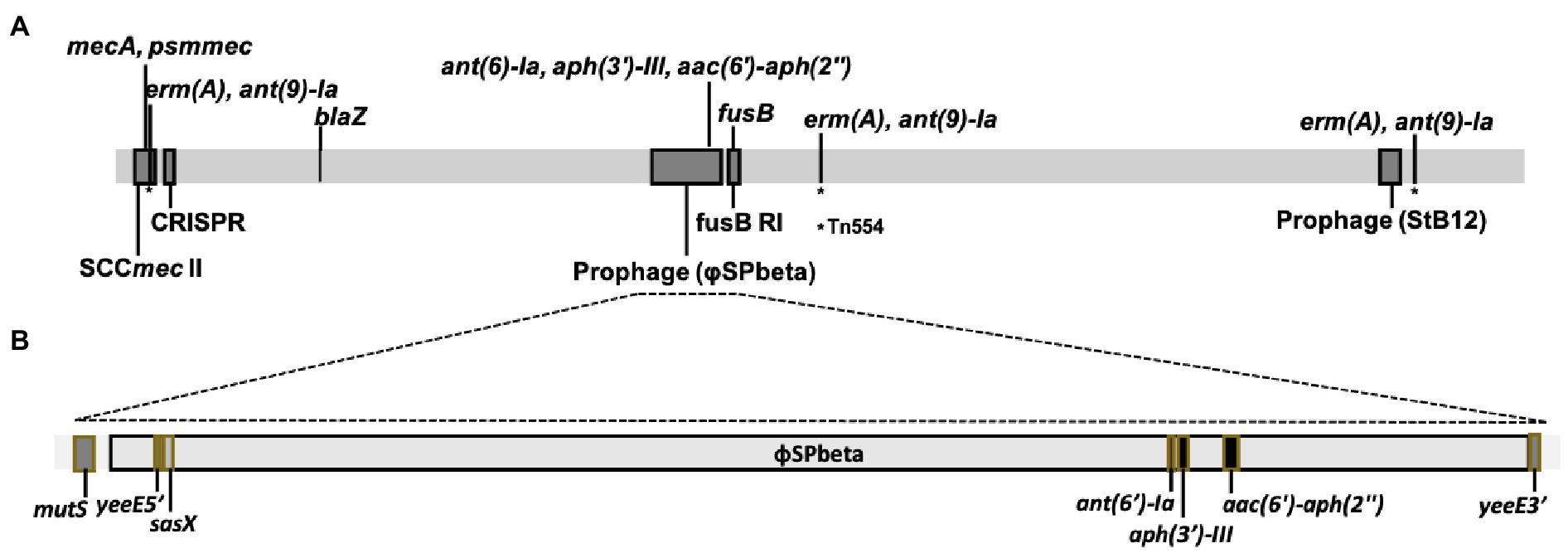
Figure 3. Schematic presentation of genome structure of CGMH-SL118. (A) The linear schematic presentation of the CGMH-SL118 genome; which containing all of the antibiotic resistance genes, MGEs (SCCmec and prophage), and CRISPR. Detailed structure of prophage Spbeta-like was shown in (B), containing each antibiotic resistance genes and a putative virulence gene. This prophage was inserted in the yeeE (nearby the mutS), which caused the yeeE separated into two parts on this structure. The asterisk (*) represents the position of Tn554.
Surveillance of Antibiotic-Resistant Phenotypes Among ST6 Clinical Isolates
The presence of multidrug-resistant genes in CGMH-SL118 made us consider whether this phenotype is a general distribution scheme among other clinical isolates of ST6 S. lugdunensis. Sixty-eight clinical isolates of ST6 were screened for the presence of aminoglycoside, macrolide, and fusidic acid resistance genes (Table 4). All 18 ORSL isolates carried SCCmec II. The prevalence of four aminoglycoside-resistant genes was over 90% among ORSL clinical isolates, and the prevalence was consistent with the results of the antibiotic susceptibility test, indicating that nearly 90% of ORSL isolates were resistant to gentamycin (Table 4). Similar trends were also observed for the macrolide-resistant phenotype. All 18 ORSL isolates with the ermA gene were resistant to clindamycin and erythromycin. However, only five isolates harbored the fusB gene, and four also showed a fusidic acid-resistant phenotype.
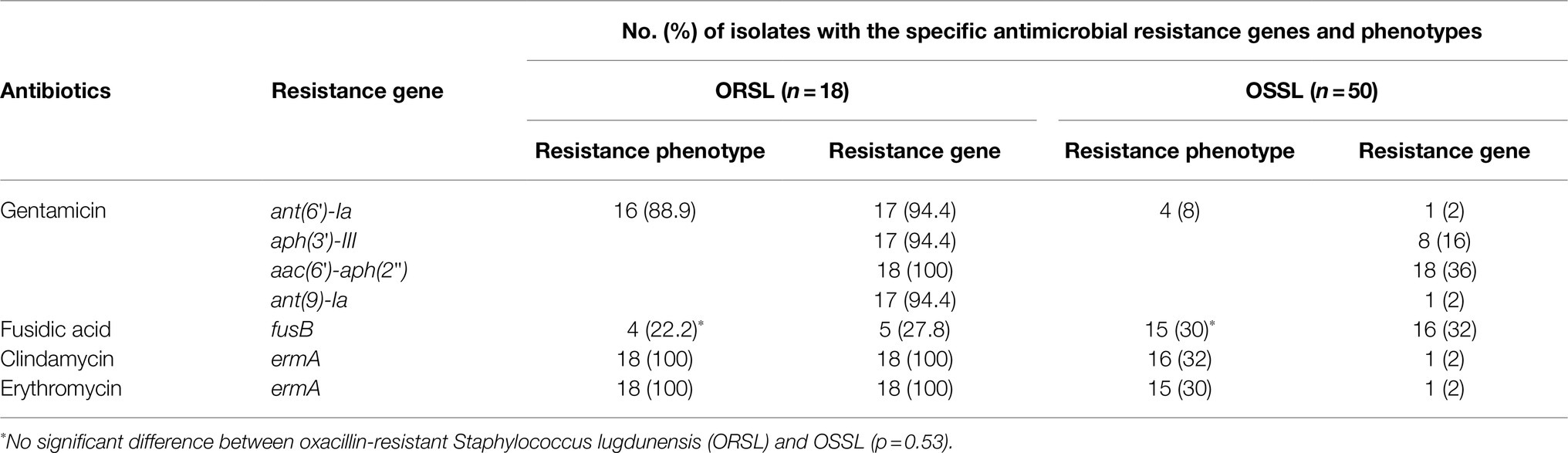
Table 4. Prevalence of antibiotic resistance genes and phenotype among clinically collected ST6 strains.
Previous studies have shown that the ST6-SCCmec II strains are the major persistent clones in hospitals (Cheng et al., 2015). Therefore, we were also interested in the antibiotic susceptibilities of ST6 OSSL isolates (Table 4). Compared with the ORSL, most antibiotic resistance genes were not present in ST6 OSSL isolates, which was consistent with their antibiotic-resistant phenotypes. The fusB was the only exception; both the antibiotic-resistant phenotype and prevalence of fusB were higher than ORSL, and statistical analysis showed no significant difference between them.
Prevalence of Prophage φSPbeta and Its Encoded Virulent and Antibiotic-Resistant Genes Among ST6 Clinical Isolates
The presence of prophage was the major difference between CGMH-SL118 and JICS135. Although two prophage regions were identified in CGMH-SL118, only the φSPbeta region contained antibiotic-resistant genes (Figure 3B). Three aminoglycoside resistance genes [aph (3′)-III, ant (6)-Ia, and aac (6′)-aph (2′)] within the prophage φSPbeta indicated its contribution to the drug-resistant phenotype. A previous study has shown that this prophage carries the sasX gene, which is an important virulence factor for S. aureus pathogenesis (Li et al., 2012). To understand the prevalence of this prophage among ST6 OSSL and ORSL clinical isolates, gene-specific PCRs were used to examine the existence of φSPbeta and its encoding virulence factor sasX and three antibiotic resistance genes (Table 5). A high prevalence of φSPbeta was found in ST6 ORSL (16/18), and these 16 isolates also had sasX and three antibiotic-resistant genes. Only two ST6 ORSL isolates did not have this prophage, which is quite different from the distribution in ST6 OSSL isolates. Although some of the above genes could still be found in isolates lacking this prophage, most of these four genes were found simultaneously with the φSPbeta in the isolates.
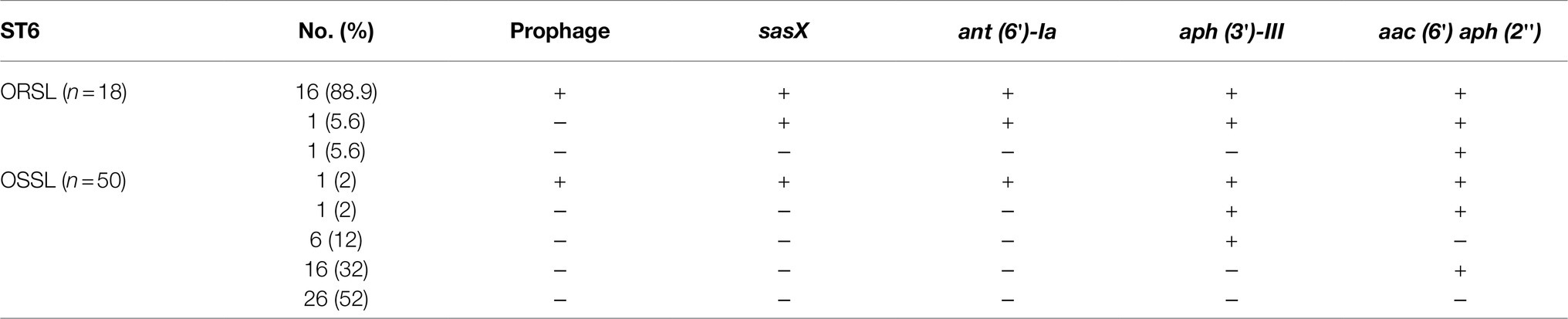
Table 5. Distribution of prophage and its relative genes in ST6 Staphylococcus lugdunensis clinical isolates.
Discussion
Epidemiological surveillance showed that most clinical isolates of S. lugdunensis belonged to OSSL, and only a few isolates belonged to ORSL (McHardy et al., 2017). Among the ORSL clinical isolates, SCCmec V strains were the most frequently isolated strains, yet our previous studies showed that some isolates belonging to SCCmec II were the strains possibly involved in the endemic transmission, which makes them important in the hospital environment (Cheng et al., 2015; Yeh et al., 2015). To further understand the factors contributing to transmission of SCCmec II strains, one of these isolates, CGMH-SL118, was selected for whole-genome sequencing and compared with other OSSLs and SCCmec V ORSL deposited in the NCBI database. The complete genome sequence of S. lugdunensis CGMH-SL118 showed a different genome composition compared to that of OSSL or SCCmec V ORSL. Comparative genome analysis showed that CGMH-SL118 contained two prophages, which may cause an increase in genome size (Table 2). In addition to the prophage, only CGMH-SL118 contained one plasmid, and the above information indicated that MGEs played key roles in differences observed in SCCmec II ORSL compared to other ORSLs and OSSLs.
Mobile genetic elements are known for their horizontal gene transfer abilities and are involved in the dissemination of drug-resistant genes and virulence factors among various strains. Our results showed that multiple drug-resistant genes were present in the CGMH-SL118 genome, and most were located inside the MGEs. The SCCmec II element of CGMH-SL118 contained not only the mecA gene, but also two additional drug-resistant genes, ermA and ant (9)-Ia. Since these two genes confer erythromycin and clindamycin resistance, SCCmec II strains are probably more difficult to treat than SCCmec V strains. According to epidemiological surveillance, few resistant isolates of the above three antibiotics have been reported in previous studies (Taha et al., 2019). Therefore, these two resistant genes were not commonly found in S. lugdunensis. Among ST6 clinical isolates, almost all ORSLs showed resistance to erythromycin and clindamycin, but only a few OSSLs showed similar phenotypes (Table 4), indicating that erythromycin and clindamycin resistances were highly correlated with the SCCmec cassette.
Most of these antibiotic-resistant genes were located on MGEs, which suggested that antibiotic resistance genes horizontally transferred between bacterial strain and indeed species. This possibility is supported by our previous studies showing that the SCCmec cassette of CGMH-SL118 was structurally similar to that of S. aureus N315, and both belonged to SCCmec II (Chang et al., 2019), but structurally different from SCCmec V carried by JICS135 (SCCmecMRSL-JICS135) (Shibuya et al., 2020). In this study, a transposon Tn554 containing ermA and adjacent ant(9)-Ia was identified in the SCCmec II element of CGMH-SL118 (Chang et al., 2019), and three copies of Tn554 were found in the CGMH-SL118 genome. This phenomenon was also found in the S. aureus N315 genome, suggesting that multiple copies of Tn554 were commonly observed in SCCmec II containing staphylococci.
In addition to the SCCmec cassette, our genome sequencing data revealed the presence of two prophage regions, which was the major difference between SCCmec II and SCCmec V ORSL. The φSPbeta region contained three antibiotic resistance genes and a putative virulence factor sasX gene, all of which might able to enhance the competitiveness of this strain in the healthcare environment. Ant (6′)-Ia, aph (3′)-III, and aac (6′)-aph (2′) all belong to the aminoglycoside-resistant genes, which also contributed to the gentamicin-resistant phenotype. The varying prevalence of these three genes among ST6 ORSL and OSSL indicated that their distribution was highly correlated with the MGEs, since all three genes were located in the φSPbeta region (Table 3). This result suggesting that the dissemination of drug-resistant determinants may carried out by the prophage φSPbeta through a horizontal transfer process.
In addition to aminoglycoside resistance genes, a putative virulence factor SasX reportedly plays critical roles in strain pathogenesis (Li et al., 2012). The SasX found in S. aureus is a homolog of SesI (S. epidermidis surface protein) and contains the LPXTG motif, which can be recognized by sortase A (SrtA) and anchored on the cell wall (Soderquist et al., 2009). Virulence studies of S. epidermidis showed that the sesI gene was only present in clinical isolates but not in the flora of healthy individuals, which indicates a virulence factor associated with strain pathogenesis (Soderquist et al., 2009). Further epidemiological surveillance of S. aureus showed that sasX was transferred by prophage φSPbeta from S. epidermidis and broadly existed in ST239 S. aureus, which was continuously transferred into ST59 S. aureus and considered to be a critical factor for increased clonal spreading (Li et al., 2012). In coagulase-negative staphylococci, sasX has been observed mainly in S. haemolyticus and S. epidermidis isolates, but not in S. lugdunensis isolates (Tekeli et al., 2020). To the best of our knowledge, this is the first report of sasX found in S. lugdunensis. Moreover, further epidemiological surveillance of ST6 clinical isolates showed that both sasX and prophage φSPbeta were present in most ORSLs, but only in one OSSL clinical isolate, suggesting that this putative virulence factor may enhance the ST6 ORSL during the endemic infection.
Among the various drug resistances identified in S. lugdunensis, fusidic acid resistance is one of the phenotypes that should be noted. Surveillance of CoNS found that less than 10% of CoNS showed fusidic acid-resistant phenotypes (Farrell et al., 2016). Similar results have been reported in previous studies of S. lugdunensis and indicated that fusidic acid resistance was low in S. lugdunensis (Hellbacher et al., 2006; Zaaroura et al., 2018). The whole-genome sequence revealed that CGMH-SL118 contained the fusB gene, which is responsible for the fusidic acid-resistant phenotype. Further investigation showed that both OSSL and ORSL ST6 isolates were more than 20% resistant (Table 4), suggesting that fusidic acid resistance is becoming a threatening issue in S. lugdunensis. Previous studies of fusidic acid resistance in S. epidermidis showed that fusB-mediated fusidic acid resistance was carried by various types of phage-related resistant islands (Chen et al., 2011, 2013), which were responsible for the high fusidic acid resistance phenotype in S. epidermidis (Chen et al., 2011). It has been suggested that fusB RI may be easily transferred and spread among different species (Chen et al., 2011). The structure of SlRIfusB-118 was almost identical to that of SeRIfusB-5907 and contained partial portions of SaRI, suggesting that the fusB-resistant island may be transferred between two CoNS species and further transferred to S. aureus. Although this phenomenon was rarely mentioned in S. lugdunensis (Bocher et al., 2009; Taha et al., 2019), it appears consistent with our previous studies that CGMH-SL118 may act as a reservoir for interspecies transfer of MGEs, such as SCCmec elements, to S. aureus in hospitals. This drug resistance gene was only found in SCCmec II ORSL in our analysis, suggesting that the antibiotic resistance properties of SCCmec II strains may differ from those of other SCCmec type ORSLs. In addition, our ST6 clinical isolates showed an increased fusidic acid-resistant phenotype, which may be considered as an alternative factor causing ST6 endemic transmission in hospital environments.
This study investigated the whole-genome sequence of SCCmec II, ST6 ORSL, and identified several MGEs containing multidrug-resistant genes and virulence factor, which could account for the differences observed when compared to SCCmec V ORSL and OSSL. The unique genome composition of CGMH-SL118 suggests that it may take advantage of these multidrug-resistant genes and virulence factor to compete with other OSSLs and ORSLs in hospital environments. After further surveillance of clinical isolates, we found that drug-resistant phenotypes similar to that of CGMH-SL118 were commonly found among ST6 ORSLs. Taken together, our results demonstrate that MGEs containing multidrug-resistant genes and virulence factor may play critical roles in ST6-SCCmec II ORSL infections and may be one of the reasons for endemic transmission in hospitals.
Data Availability Statement
Raw data of the CGMH-SL118 complete genome sequence were submitted to the NCBI BioProject and approved with accession number CP048008.
Ethics Statement
This study was approved by the Ethics Committee of Linkou Chang Gung Memorial Hospital. Because this study only experimented on bacteria and did not affect the patients adversely, the Review Board agreed the usage of the requested bacterial materials from Linkou Chang Gung Memorial Hospital bacterial storage.
Author Contributions
S-CC, L-CL, and J-JL conceived the study, analyzed the data, prepared the tables and figures, and contributed to the writing of the manuscript. All authors read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 110-2320-B-182A-006-MY3, 108-2320-B-182A-013, and 110-2811-B-182A-505).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for the bacterial isolates supported by Chang Gung Memorial Hospital bacterial storage bank program (CLRPG3E0025).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.765437/full#supplementary-material
Footnotes
1. ^https://github.com/PacificBiosciences/GenomicConsensus
2. ^https://github.com/tseemann/prokka/
5. ^http://phast.wishartlab.com/index.html
7. ^https://crispr.i2bc.paris-saclay.fr/
References
Argemi, X., Martin, V., Loux, V., Dahyot, S., Lebeurre, J., Guffroy, A., et al. (2017a). Whole-genome sequencing of seven strains of Staphylococcus lugdunensis allows identification of mobile genetic elements. Genome Biol. Evol. 9, 1183–1189. doi: 10.1093/gbe/evx077
Argemi, X., Matelska, D., Ginalski, K., Riegel, P., Hansmann, Y., Bloom, J., et al. (2018). Comparative genomic analysis of Staphylococcus lugdunensis shows a closed pan-genome and multiple barriers to horizontal gene transfer. BMC Genomics 19:621. doi: 10.1186/s12864-018-4978-1
Argemi, X., Prevost, G., Riegel, P., Keller, D., Meyer, N., Baldeyrou, M., et al. (2017b). VISLISI trial, a prospective clinical study allowing identification of a new metalloprotease and putative virulence factor from Staphylococcus lugdunensis. Clin. Microbiol. Infect. 23, 334.E1–334.E8. doi: 10.1016/j.cmi.2016.12.018
Argemi, X., Riegel, P., Lavigne, T., Lefebvre, N., Grandpre, N., Hansmann, Y., et al. (2015). Implementation of matrix-assisted laser desorption ionization-time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J. Clin. Microbiol. 53, 2030–2036. doi: 10.1128/JCM.00177-15
Bocher, S., Tonning, B., Skov, R. L., and Prag, J. (2009). Staphylococcus lugdunensis, a common cause of skin and soft tissue infections in the community. J. Clin. Microbiol. 47, 946–950. doi: 10.1128/JCM.01024-08
Boetzer, M., and Pirovano, W. (2014). SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211. doi: 10.1186/1471-2105-15-211
Castanheira, M., Watters, A. A., Bell, J. M., Turnidge, J. D., and Jones, R. N. (2010). Fusidic acid resistance rates and prevalence of resistance mechanisms among Staphylococcus spp. isolated in North America and Australia, 2007–2008. Antimicrob. Agents Chemother. 54, 3614–3617. doi: 10.1128/AAC.01390-09
Chang, S. C., Lee, M. H., Yeh, C. F., Liu, T. P., Lin, J. F., Ho, C. M., et al. (2017). Characterization of two novel variants of staphylococcal cassette chromosome mec elements in oxacillin-resistant Staphylococcus lugdunensis. J. Antimicrob. Chemother. 72, 3258–3262. doi: 10.1093/jac/dkx291
Chang, S. C., Lin, L. C., Ge, M. C., Liu, T. P., and Lu, J. J. (2019). Characterization of a novel, type II staphylococcal cassette chromosome mec element from an endemic oxacillin-resistant Staphylococcus lugdunensis clone in a hospital setting. J. Antimicrob. Chemother. 74, 2162–2165. doi: 10.1093/jac/dkz189
Chen, H. J., Chang, Y. C., Tsai, J. C., Hung, W. C., Lin, Y. T., You, S. J., et al. (2013). New structure of phage-related islands carrying fusB and a virulence gene in fusidic acid-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 57, 5737–5739. doi: 10.1128/AAC.01433-13
Chen, H. J., Tsai, J. C., Hung, W. C., Tseng, S. P., Hsueh, P. R., and Teng, L. J. (2011). Identification of fusB-mediated fusidic acid resistance islands in Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 55, 5842–5849. doi: 10.1128/AAC.00592-11
Cheng, C. W., Liu, T. P., Yeh, C. F., Lee, M. H., Chang, S. C., and Lu, J. J. (2015). Persistence of a major endemic clone of oxacillin-resistant Staphylococcus lugdunensis sequence type 6 at a tertiary medical Centre in northern Taiwan. Int. J. Infect. Dis. 36, 72–77. doi: 10.1016/j.ijid.2015.05.022
Douiri, N., Hansmann, Y., Lefebvre, N., Riegel, P., Martin, M., Baldeyrou, M., et al. (2016). Staphylococcus lugdunensis: a virulent pathogen causing bone and joint infections. Clin. Microbiol. Infect. 22, 747–748. doi: 10.1016/j.cmi.2016.05.031
Emaneini, M., Bigverdi, R., Kalantar, D., Soroush, S., Jabalameli, F., Noorazar Khoshgnab, B., et al. (2013). Distribution of genes encoding tetracycline resistance and aminoglycoside modifying enzymes in Staphylococcus aureus strains isolated from a burn center. Ann. Burns Fire Disasters 26, 76–80.
Farrell, D. J., Mendes, R. E., Castanheira, M., and Jones, R. N. (2016). Activity of fusidic acid tested against staphylococci isolated from patients in U.S. medical centers in 2014. Antimicrob. Agents Chemother. 60, 3827–3831. doi: 10.1128/AAC.00238-16
Fernandez-Rufete, A., Garcia-Vazquez, E., Hernandez-Torres, A., Canteras, M., Ruiz, J., and Gomez, J. (2012). Coagulase-negative Staphylococcus bacteraemia: prognosis factors and influence of antibiotic treatment. Rev. Esp. Quimioter. 25, 199–205.
Fessler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S., and Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65, 619–625. doi: 10.1093/jac/dkq021
Giormezis, N., Kolonitsiou, F., Makri, A., Vogiatzi, A., Christofidou, M., Anastassiou, E. D., et al. (2015). Virulence factors among Staphylococcus lugdunensis are associated with infection sites and clonal spread. Eur. J. Clin. Microbiol. Infect. Dis. 34, 773–778. doi: 10.1007/s10096-014-2291-8
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Heilbronner, S., and Foster, T. J. (2021). Staphylococcus lugdunensis: a skin commensal with invasive pathogenic potential. Clin. Microbiol. Rev. 34:e00205-20. doi: 10.1128/CMR.00205-20
Heilbronner, S., Holden, M. T., van Tonder, A., Geoghegan, J. A., Foster, T. J., Parkhill, J., et al. (2011). Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 322, 60–67. doi: 10.1111/j.1574-6968.2011.02339.x
Hellbacher, C., Tornqvist, E., and Soderquist, B. (2006). Staphylococcus lugdunensis: clinical spectrum, antibiotic susceptibility, and phenotypic and genotypic patterns of 39 isolates. Clin. Microbiol. Infect. 12, 43–49. doi: 10.1111/j.1469-0691.2005.01296.x
Kao, S. J., You, I., Clewell, D. B., Donabedian, S. M., Zervos, M. J., Petrin, J., et al. (2000). Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in enterococcus faecium. Antimicrob. Agents Chemother. 44, 2876–2879. doi: 10.1128/AAC.44.10.2876-2879.2000
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. doi: 10.1016/S0140-6736(00)04403-2
Laabei, M., Recker, M., Rudkin, J. K., Aldeljawi, M., Gulay, Z., Sloan, T. J., et al. (2014). Predicting the virulence of MRSA from its genome sequence. Genome Res. 24, 839–849. doi: 10.1101/gr.165415.113
Lebeurre, J., Dahyot, S., Diene, S., Paulay, A., Aubourg, M., Argemi, X., et al. (2019). Comparative genome analysis of Staphylococcus lugdunensis shows clonal complex-dependent diversity of the putative virulence factor, ess/type VII locus. Front. Microbiol. 10:2479. doi: 10.3389/fmicb.2019.02479
Li, M., Du, X., Villaruz, A. E., Diep, B. A., Wang, D., Song, Y., et al. (2012). MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18, 816–819. doi: 10.1038/nm.2692
Lin, J. F., Cheng, C. W., Kuo, A. J., Liu, T. P., Yang, C. C., Huang, C. T., et al. (2015). Clinical experience and microbiologic characteristics of invasive Staphylococcus lugdunensis infection in a tertiary center in northern Taiwan. J. Microbiol. Immunol. Infect. 48, 406–412. doi: 10.1016/j.jmii.2013.12.010
Liu, P. Y., Huang, Y. F., Tang, C. W., Chen, Y. Y., Hsieh, K. S., Ger, L. P., et al. (2010). Staphylococcus lugdunensis infective endocarditis: a literature review and analysis of risk factors. J. Microbiol. Immunol. Infect. 43, 478–484. doi: 10.1016/S1684-1182(10)60074-6
Malachowa, N., and DeLeo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071. doi: 10.1007/s00018-010-0389-4
McHardy, I. H., Veltman, J., Hindler, J., Bruxvoort, K., Carvalho, M. M., and Humphries, R. M. (2017). Clinical and microbiological aspects of beta-lactam resistance in Staphylococcus lugdunensis. J. Clin. Microbiol. 55, 585–595. doi: 10.1128/JCM.02092-16
Molina, J., Penuela, I., Lepe, J. A., Gutierrez-Pizarraya, A., Gomez, M. J., Garcia-Cabrera, E., et al. (2013). Mortality and hospital stay related to coagulase-negative staphylococci bacteremia in non-critical patients. J. Infect. 66, 155–162. doi: 10.1016/j.jinf.2012.10.021
Patti, J. M., Allen, B. L., McGavin, M. J., and Hook, M. (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617. doi: 10.1146/annurev.mi.48.100194.003101
Sepulveda, M. A., Bello, H. T., Dominguez, M. Y., Mella, S. M., Zemelman, R. Z., and Gonzalez, G. R. (2007). Molecular identification of aminoglycoside-modifying enzymes among strains of enterococcus spp. isolated in hospitals of the VIII region of Chile. Rev. Med. Chil. 135, 566–572. doi: 10.4067/s0034-98872007000500003
Shibuya, R., Uehara, Y., Baba, T., Teruya, K., Satou, K., Hirano, T., et al. (2020). Complete genome sequence of a methicillin-resistant Staphylococcus lugdunensis strain and characteristics of its staphylococcal cassette chromosome mec. Sci. Rep. 10:8682. doi: 10.1038/s41598-020-65632-7
Soderquist, B., Andersson, M., Nilsson, M., Nilsdotter-Augustinsson, A., Persson, L., Friberg, O., et al. (2009). Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J. Med. Microbiol. 58, 1395–1397. doi: 10.1099/jmm.0.008771-0
Sutcliffe, J., Grebe, T., Tait-Kamradt, A., and Wondrack, L. (1996). Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40, 2562–2566. doi: 10.1128/AAC.40.11.2562
Taha, L., Stegger, M., and Soderquist, B. (2019). Staphylococcus lugdunensis: antimicrobial susceptibility and optimal treatment options. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1449–1455. doi: 10.1007/s10096-019-03571-6
Tekeli, A., Ocal, D. N., and Dolapci, I. (2020). Detection of sasX gene and distribution of SCCmec types in invasive and non-invasive coagulase-negative staphylococci. Balkan Med. J. 37, 215–221. doi: 10.4274/balkanmedj.galenos.2020.2019.8.21
Wayne, P. (2018). Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement.
Wu, A. B., Wang, M. C., Tseng, C. C., Lin, W. H., Teng, C. H., Huang, A. H., et al. (2011). Clinical and microbiological characteristics of community-acquired Staphylococcus lugdunensis infections in southern Taiwan. J. Clin. Microbiol. 49, 3015–3018. doi: 10.1128/JCM.01138-11
Yeh, C. F., Chang, S. C., Cheng, C. W., Lin, J. F., Liu, T. P., and Lu, J. J. (2016). Clinical features, outcomes, and molecular characteristics of community- and health care-associated Staphylococcus lugdunensis infections. J. Clin. Microbiol. 54, 2051–2057. doi: 10.1128/JCM.00847-16
Yeh, C. F., Liu, T. P., Cheng, C. W., Chang, S. C., Lee, M. H., and Lu, J. J. (2015). Molecular characteristics of disease-causing and commensal Staphylococcus lugdunensis isolates from 2003 to 2013 at a tertiary Hospital in Taiwan. PLoS One 10:e0134859. doi: 10.1371/journal.pone.0134859
Keywords: oxacillin-resistant Staphylococcus lugdunensis, mobile genetic elements, prophage, multidrug-resistant genes, resistant islands
Citation: Chang S-C, Lin L-C and Lu J-J (2021) Comparative Genomic Analyses Reveal Potential Factors Responsible for the ST6 Oxacillin-Resistant Staphylococcus lugdunensis Endemic in a Hospital. Front. Microbiol. 12:765437. doi: 10.3389/fmicb.2021.765437
Edited by:
Daniel Yero, Universidad Autónoma de Barcelona, SpainReviewed by:
Maria De Lourdes Ribeiro De Souza Da Cunha, São Paulo State University, BrazilTimothy J. Foster, Trinity College Dublin, Ireland
Sandrine Dahyot, Centre Hospitalier Universitaire (CHU) de Rouen, France
Copyright © 2021 Chang, Lin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jang-Jih Lu, amFuZ2x1NDVAZ21haWwuY29t
†These authors have contributed equally to this work
 Shih-Cheng Chang
Shih-Cheng Chang Lee-Chung Lin
Lee-Chung Lin Jang-Jih Lu
Jang-Jih Lu