
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 November 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.764105
Staphylococcus aureus (S. aureus) is now regarded as a zoonotic agent. Methicillin-susceptible S. aureus (MSSA) ST398 is a livestock-associated bacterium that is most prevalent in China, but there are currently no data available for Shandong. Therefore, the aim of this study was to investigate the epidemiology and characterization of MSSA ST398 from retail pork and bulk tank milk (BTM) in Shandong. A total of 67 S. aureus isolates were collected from retail pork between November 2017 and June 2018. Among the isolates, high antimicrobial resistance rates were observed for penicillin (97.0%), and 92.5% of the isolates were multi-drug resistant (MDR). Eight sequence types (STs) were identified in the retail pork isolates, and the predominant type was ST15 (n=26), which was followed by ST398 (n=14). Staphylococcal protein A gene (spa) typing identified spa types t034 and t1255 in MSSA ST398 from retail pork. Using whole-genome sequencing analysis, we described the phylogeny of 29 MSSA ST398 isolates that were obtained from retail pork (n=14) and BTM (n=15). The phylogenetic tree showed that the MSSA ST398 isolates from different sources had the same lineage. Among the 29 MSSA ST398 isolates, five resistance genes were detected, and all isolates carried DHA-1. Fifteen toxin genes were detected, and all isolates carried eta, hla, and hlb. In conclusion, this study found that a high risk for MSSA ST398 was present in retail pork and BTM. These findings have major implications for how investigations of MSSA ST398 outbreaks should be conducted in the One-Health context.
Staphylococcus aureus is a well-known commensal pathogen of many animal species, including humans, that causes community and nosocomial infections (Li et al., 2018). It is regarded as one of the world’s leading causes of food consumption-related disease outbreaks (Abdalrahman et al., 2015). Previous studies have revealed that S. aureus has often been isolated from raw milk and retail meat (Johler et al., 2018; Sahibzada et al., 2018).
S. aureus-associated food poisoning in humans and similar intramammary infections in animals are produced by strains with the capacity to produce a wide range of virulence factors, including enterotoxins (se); toxic shock syndrome toxin-1 (tst); exfoliative toxins (eta and etb); leukocidins (lukD/E/FS); and haemolysins (hla, hld, hlg, and hlgv; Le Marechal et al., 2011; Wilson et al., 2011). The role of virulence factors in disease causation is not as definitive as foodborne illness, and a plethora of factors are likely to be involved in disease pathogenesis (Fijalkowski et al., 2013).
Most S. aureus isolates are host-specific, and the potential for animals to act as a source of S. aureus infections for humans has been shown for some clonal lineages, such as ST398 (Mediavilla et al., 2012; Yan et al., 2014). Methicillin-resistant S. aureus (MRSA) ST398 can colonize animals and humans and can be transmitted between them (Li et al., 2015). MRSA ST398 has been isolated in Australia (Mitchell et al., 2014), New Zealand (Deborah et al., 2014), and Spain (Lozano et al., 2012). MSSA ST398 has been recovered from pigs in Germany and northeast China (Kadlec and Schwarz, 2010; Yan et al., 2014), humans in France and Taiwan, China (Rasigade et al., 2010; Huang and Chen, 2020), and retail meat in USA and Shanxi, China (Waters et al., 2011; Li et al., 2015). Recently, infections caused by MSSA ST398 have been described in humans (Valentin-Domelier et al., 2011; Mediavilla et al., 2012; David et al., 2013), and MSSA ST398 has been associated with efficient transmission between people, with greater capacity for adhesion to human skin (Uhlemann et al., 2012).
The food safety challenge we have faced during the current COVID-19 pandemic has increased concerns about the increasing numbers of outbreaks of staphylococcal food-borne intoxication. As a commensal bacterium in livestock, there has been little focus on MSSA ST398 contamination of retail meat and BTM in China. Therefore, the main objective of this study was to investigate the population structure of MSSA ST398 in retail pork and BTM by using whole-genome sequencing.
A total of 15 MSSA ST398 (SA-N1-N15) isolates from BTM in Shandong dairy farms were used in this study. Among the 15 MSSA ST398, 6 isolates were from Weifang, 3 isolates were from Jinan, 2 isolates were from Linyi, 2 isolates were from Dezhou, 1 isolate was from Rizhao, and 1 isolate was from Tai’an. Antimicrobial susceptibility testing and molecular typing were performed, as previously described (Zhao et al., 2021).
From November 2017 to June 2018, a total of 200 retail pork samples were randomly purchased from two supermarkets (100 samples per supermarket) located in Tai’an and Jinan city. Each 25-g retail pork sample was aseptically placed into a sterile Whirl-Pak bag (Nasco, United States) and labelled. All collected samples were transported to the laboratory on ice within 6h after collection and were processed immediately. Isolation of S. aureus was performed as described by the following experimental procedure with some modifications (Sergelidis et al., 2012; Tang et al., 2017). For the isolation and detection of S. aureus, 25g of the retail pork samples were enriched in 100ml of trypticase soy broth (TSB, OXOID, United Kingdom) containing 6.5% NaCl and incubated at 37°C for 24h. Then, a loopful from the incubated tubes was streaked onto Baird-Parker Agar (Hopebiol, Qingdao, China) that was supplemented with 5% egg yolk and tellurite and incubated at 37°C for 24h. The suspected colonies with typical black appearances and surrounded by clear zones were identified as S. aureus. The suspected colonies were confirmed using the Stap identification system (bioMérieux, Marcy-l Étoile, France) and were further identified via amplification of the species-specific nuc gene using a previously described primer set (Louie et al., 2000).
All S. aureus isolates obtained from retail pork were subjected to antimicrobial susceptibility tests against 18 antimicrobial agents on Muller-Hinton agar with the agar dilution method by following the guidelines of the Clinical and Laboratory Standard Institute (Clinical and Laboratory Standards Institute, 2019). The antimicrobial agents used in this study were penicillin (PEN), amoxicillin/clavulanic acid (AMC), erythromycin (ERY), clindamycin (CLI), enrofloxacin (ENR), ofloxacin (OFL), ceftiofur (EFT), cefoxitin (FOX), sulfisoxazole (SF), vancomycin (VAN), trimethoprim/sulfamethoxazole (SXT), doxycycline (DOX), florfenicol (FFC), tiamulin (TIA), oxacillin (OXA), tilmicosin (TIL), gentamicin (GEN), and linezolid (LEZ). S. aureus ATCC25923 was used as the quality control strain. Antimicrobial resistance was defined as resistance to one or more classes of antimicrobials, whereas MDR was defined as resistance to three or more classes of antimicrobials.
All of the isolates from retail pork were typed by multilocus sequence typing (MLST; Enright et al., 2000). The following seven housekeeping genes were used in the MLST scheme: carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glp), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL). The amplicons were purified and sequenced (Invitrogen, Beijing, China). The alleles and STs were assigned according to the criteria of the MLST database.1 All typing data were imported into the BioNumerics software v6.5 (Applied Maths, Kortrijk, Belgium), clustered using the appropriate settings and the relationships displayed using graphing method called minimum spanning tree as described before (Schouls et al., 2005). All ST398 S. aureus isolates were investigated by spa typing (Harmsen et al., 2003). Typing was performed through the publicly available Ridom SpaServer.2
The genomic DNA of S. aureus isolates from the retail pork samples was extracted with a TIANamp Bacterial DNA extraction kit (Tiangen, Beijing, China). The presence of the mecA and mecC genes was tested by polymerase chain reactions (PCR) using primers and conditions as previously reported (Zdragas et al., 2015).
All identified ST398 S. aureus isolates from retail pork and BTM were subjected to whole genome sequencing (Bankevich et al., 2012). Genomic DNA from each isolate was purified on DNeasy columns (Qiagen), and then sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA, United States), with 100-base paired-end reads and barcodes within the Nextera XT DNA Library Preparation Kit (Illumina, United States). Read sequence quality was assessed with the Fastqc program3 and reads were quality-filtered with fastq-mcf (Ea-utils: https://expressionanalysis.github.io/ea-utils/). Genome assembly was performed with Edena v3 assembler (Hemandez et al., 2014). The assembled genomes were annotated with Prokka v1.10 software (Seemann, 2014). The Illumina sequence reads have been deposited in NCBI’s short read archive and the study accession number is shown in Supplementary Table S1. MUSCLE v3.8.31 software was used to compare the multiple sequences of the core genomes, and the results of these comparisons were used to construct the phylogenetic tree. The phylogenetic tree was constructed by using the maximum likelihood method with phyml v3.0.4 The ST398 isolates were investigated for all resistance and virulence genes in the de novo assembled contigs using ResFinder v2.1 (Zankari et al., 2012) and VirulenceFinder v1.5 (Joensen et al., 2014), respectively. Furthermore, the selected virulence genes (e.g., sea ser, eta, etb, hla, hlb, hld, hlg, hlgv, tst, lukM, lukE, lukD, lukS, and lukF) and resistance genes (e.g., blaZ, DHA-1, msrA, ermA, ermB, ermC, ermT, tetK, tetL, tetM, vanA, fusB, far, dfrA, dfrG, aacA-aphD, aac (6′)-Ie-aph (2″)-Ia, mupA, and mupB) were investigated by mapping the reference genes (Chairat et al., 2015; Ronco et al., 2018). The final phylogenetic tree and data of the carried resistance and virulence genes were input into the interactive Tree Of Life5 for further annotation (Xue et al., 2020).
Among the 200 retail pork samples, a total of 67 S. aureus (67/200, 33.5%) were isolated, which included 42 isolates (42/100, 42.0%) from the Tai’an supermarket and 25 isolates (25/100, 25.0%) from the Jinan supermarket.
The susceptibility of the 67 S. aureus isolates to 18 antimicrobial agents was assessed (Table 1). Resistance to PEN (97.0%) was the most commonly observed in the retail pork isolates. High rates of resistance were also noted for ERY (86.6%) and FFC (83.4%). In contrast, all isolates were susceptible to AMC. The MDR phenotype was observed in 92.5% of the isolates. In addition, the MSSA ST398 isolates from BTM were susceptible to AMC, EFT, FOX, SXT, DOX, TIA, OXA, GEN, and LEZ. But most isolates were resistant to PEN (93.3%), OFL (93.3%), and ERY (86.7%, Table 1).
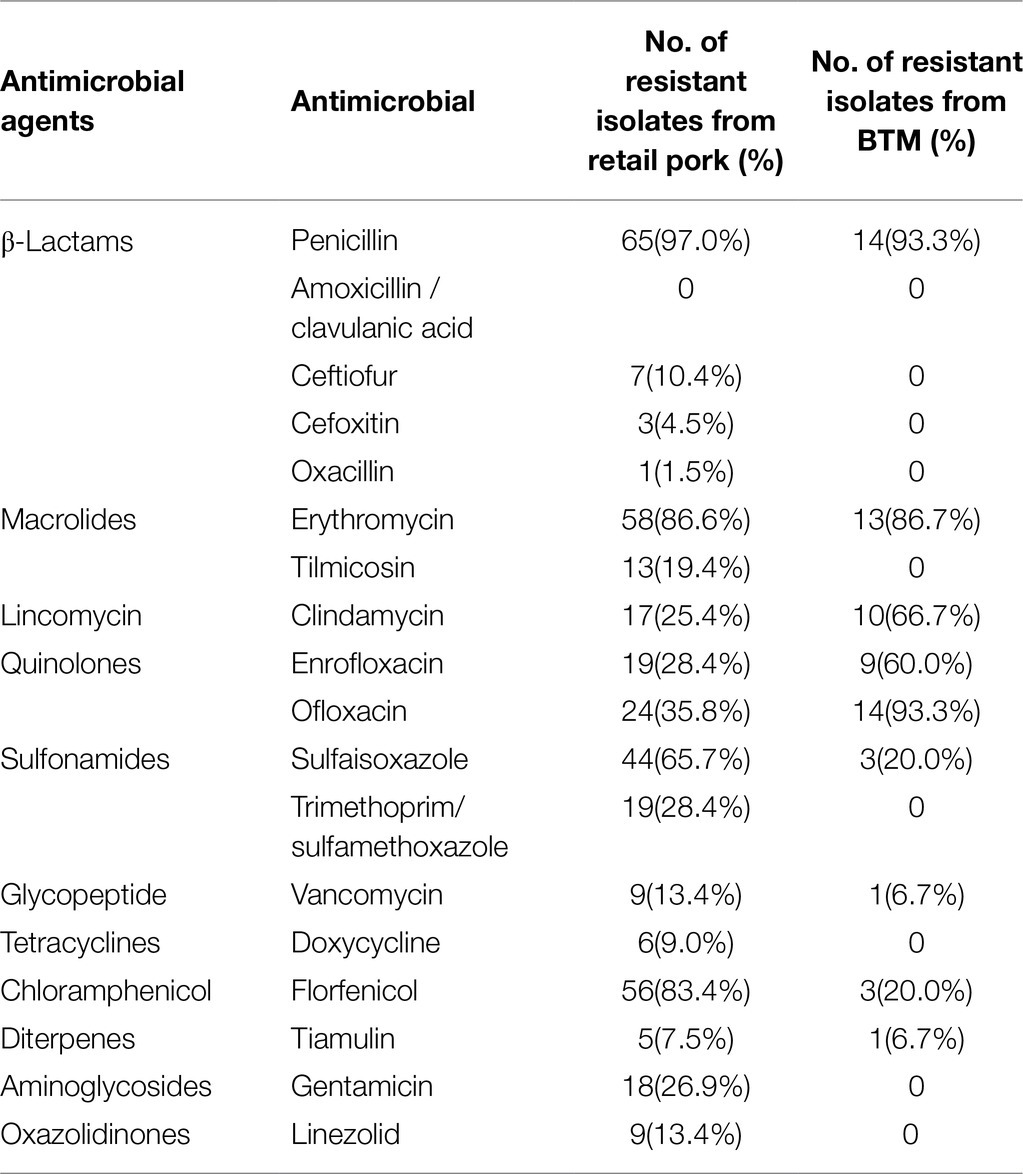
Table 1. Number and percentage of antimicrobial resistance of S. aureus isolated from retail pork and BTM.
The MLST results revealed that a total of eight STs were identified in the retail pork isolates, including ST398, ST1036, ST25, ST15, ST7, ST1, ST9, and ST188 (Figure 1). Among them, ST15 was the most frequent genotype that was recovered from both supermarkets and involved 26 S. aureus isolates, which was followed by ST398 (n=14). The fourteen ST398 isolates from retail pork isolates were assigned to two different spa types. Most isolates belonged to t034 (n=12) and were followed by t1255 (n=2). In addition, the MSSA ST398 isolates from BTM all belonged to t034 (Figure 2).
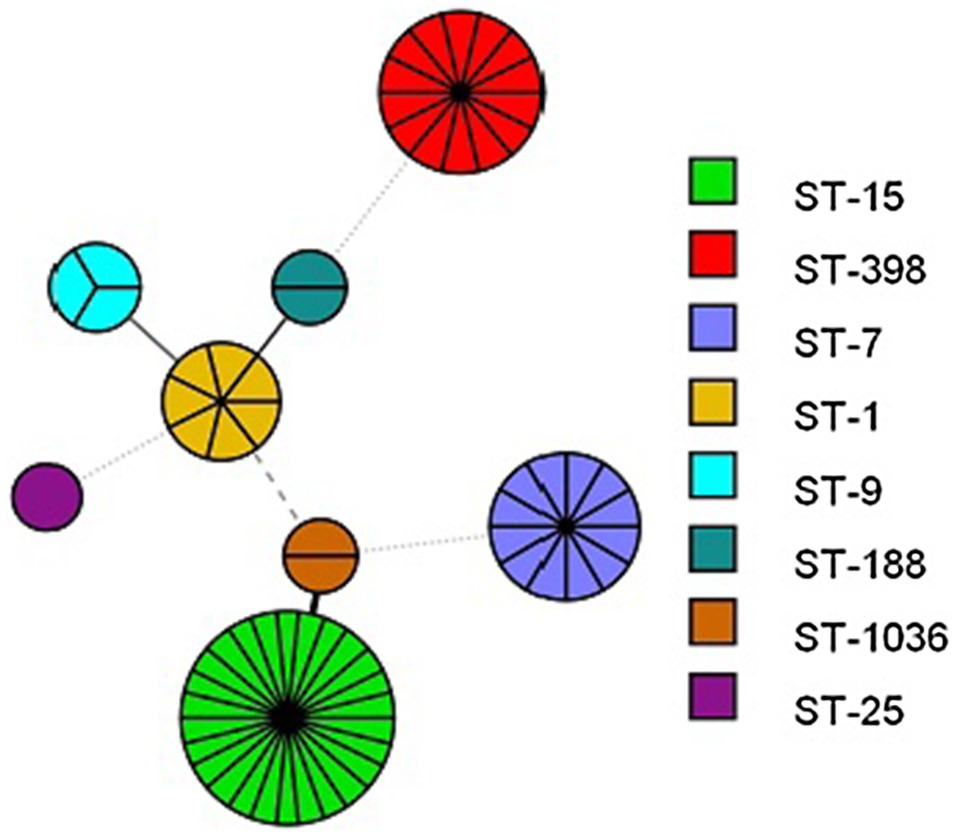
Figure 1. Minimum Spanning Tree based on the MLST data for each isolate. Numbers indicate ST of each node.
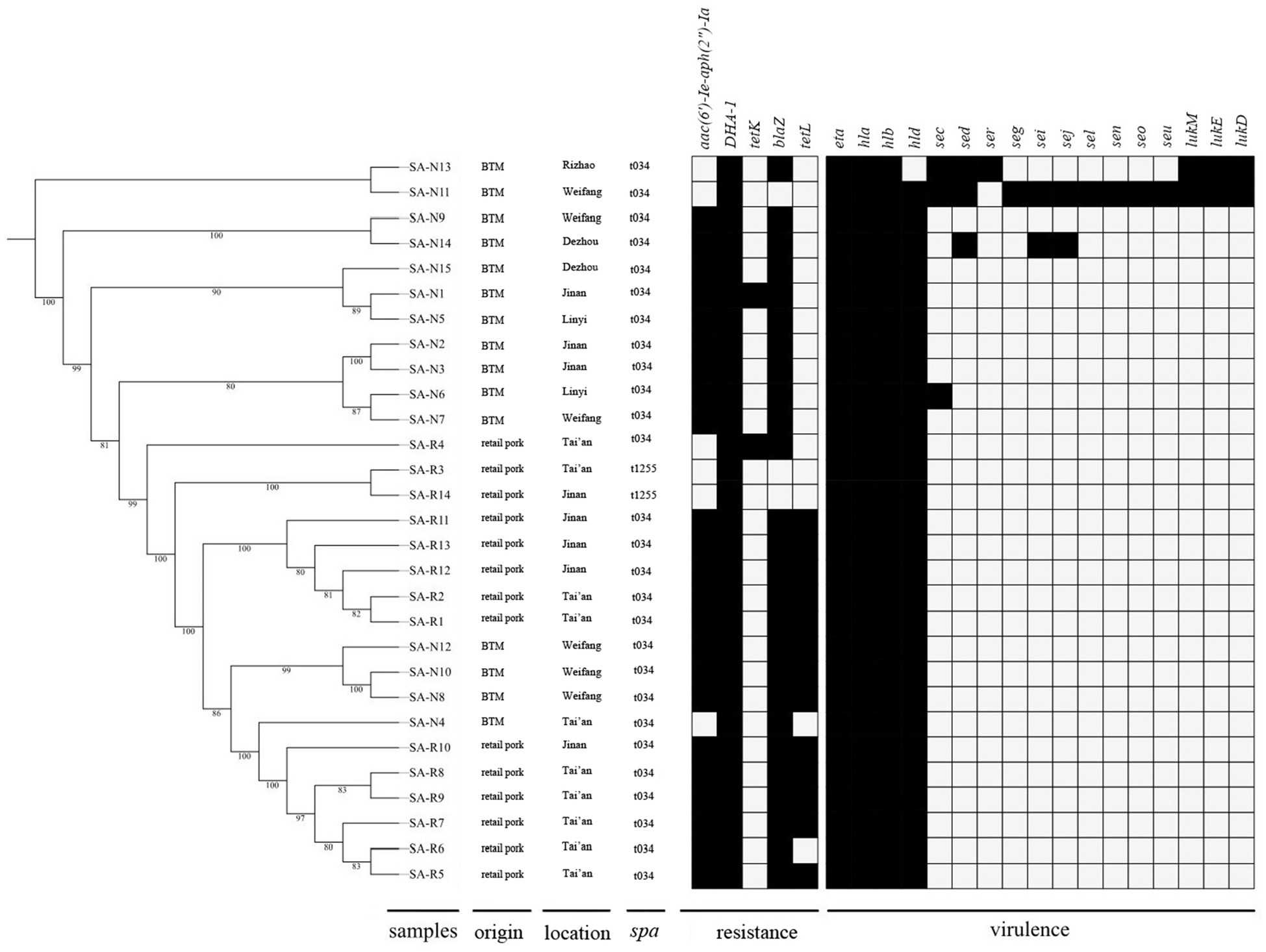
Figure 2. The distributions of the virulence and resistance genes identified in the 29 MSSA ST398 isolates from retail pork and BTM.
None of the S. aureus isolates from the retail pork carried mecA and mecC, and they were all MSSA.
The core genome-based phylogeny of the 29 MSSA ST398 isolates from retail pork (SA-R1-R14) and BTM is shown in Figure 2. In general, the phylogenetic tree showed that the MSSA ST398 isolates from different sources had the same lineage. In most cases, isolates from the same source clustered together. In general, the virulence and resistance genes were identified with thresholds of 90% nucleotide identity and 90% coverage of the query sequence length. The distributions of the virulence and resistance genes that were carried by these strains are shown in Figure 2. The resistance profiles were distinct between the MSSA ST398 isolates from retail pork and BTM, while the toxin profiles were relatively similar. Among the 29 MSSA ST398 isolates, five resistance genes were detected, and all strains carried DHA-1, which was followed by blaZ (n=26), aac (6′)-Ie-aph (2″)-Ia (n=23), tetL (n=13), and tetK (n=2). Fifteen toxin genes were detected, and all isolates carried eta, hla, and hlb, which were followed by hld (n=28), sec (n=3), sed (n=3), sei (n=2), lukE (n=2), and lukD (n=2), while seg, sej, seo, seu, and ser were represented only once.
In this study, we carried out whole-genome sequencing to investigate the phylogeny and characterization of MSSA ST398 from retail pork and BTM in China. To our knowledge, this is the first time that such type of MSSA ST398 isolates have been whole-genome sequenced and made publicly available. The prevalence of S. aureus in retail pork products from other studies has ranged from 12.0 to 59.7% (Hanson et al., 2011; Kelman et al., 2011; Waters et al., 2011). The results from this study were similar to those in other published reports, since S. aureus was found in 33.5% of retail pork, and they also coincide with other reports on poultry products that the prevalence of S. aureus in poultry products that has been reported worldwide has ranged from 23.5 to 50.5% (Ou et al., 2017). The isolates from the retail pork samples may originate from S. aureus-positive animals, the surrounding environment, humans, and other sources during processing and commercialization in meat counters at supermarkets and retail stores. Our results indicated that these meat items may serve as reservoirs of S. aureus, as with other retail meat samples.
Due to the excessive use of antibiotics, the prevalence of drug-resistant bacteria is increasing. In this study, we observed high resistance rates. The resistance rates to PEN (97.0%), ERY (86.6%), and FFC (83.4%) that were detected in this study have commonly been reported among S. aureus isolates from meat samples (Buyukcangaz et al., 2013; Ge et al., 2017). These high resistance rates may be related to the use of antimicrobials for treating disease and as growth promoters or feed additives in livestock. In this study, the MDR isolate rate of S. aureus (92.5%) from retail pork was consistent with the report that indicated that 90.0% of the strains were isolated from ready-to-eat meat sandwiches in Egypt (Mahmoud et al., 2021). However, this rate was higher than those reported for BTM (55.4%) and retail ready-to-eat foods (75.8%) in China (Zhao et al., 2021; Yang et al., 2016). Retail pork that is contaminated by MDR S. aureus is potentially hazardous, and the food chain may be the key site where resistance is transmitted between the environment and humans. In this regard, it was estimated that 2.8 million patients in the USA will be treated each year for resistant bacteria, and more than 35,000 will die as a result (CDC, 2019).
Several molecular typing methods were used to characterize the isolates, including MLST and spa typing. MLST is a DNA sequencing technology that uses sequence analyses of housekeeping genes to discriminate between isolates. MLST also offers the advantage that it is highly reproducible, which makes it an excellent tool for global comparisons of population structures (Enright et al., 2000). spa typing is specific to staphylococci and analyses the polymorphisms in the protein A gene (Frenay et al., 1996). In this study, eight STs were identified, and ST15 was the predominant type, which was different from the results of other studies. Previous studies have reported that ST5 has been found in retail pork in the United States, while ST9 was predominantly obtained from Asian countries, including China (Bhargava et al., 2011; Li et al., 2017). Importantly, it has been reported that MSSA ST398 isolates collected from retail pork are primarily associated with community- and hospital-acquired MSSA infections in humans (Chen and Huang, 2014). Two spa types were identified among the MSSA ST398 isolates from retail pork, and the predominant type was t034, which is a common and dominant type in Europe and North America (Pantosti, 2012).
The phylogenetic analysis showed that MSSA ST398 from retail pork clustered together with MSSA ST398 from BTM, which suggested that ST398 has a wide range of hosts and is considered to be adapted to the colonization of nonhuman mammals (Golding et al., 2010). In this study, we found that all MSSA ST398 isolates carried DHA-1, which belongs to class C of the Ambler classification and to group 1 of the functional classification of Bush, Jacoby, and Medeiros. These results were different from those of another study in which, among the 56 S. aureus isolates, only one in Iran carried DHA-1 (Shahnaz et al., 2016). In our study, most of the MSSA ST398 isolates harboured blaZ (26/29), which was consistent with the result that most isolates were resistant to PEN. This observation is in line with a previous report that the blaZ gene confers PEN resistance (Johler et al., 2018). According to the results of this study, we suggested that MSSA ST398 could be a reservoir for the resistance genes. With regard to the risk of pathogenicity, the presence of virulence genes among the MSSA ST398 isolates was assessed in this study. All isolates carried eta, hla, and hlb, which coincided with the results of another study (Sun et al., 2019). SEs in particular are involved in human food poisoning (Hennekinne et al., 2012). In this study, we only detected SEs in BTM but none in retail pork, which can be attributed to the differences in geographical regions, sample sources and environments and needs to be further monitored. The detection of virulence genes in MSSA ST398 isolates reveals the lurking threat of retail pork and BTM, suggesting the need for implementing surveillance programs and prevention strategies.
Our findings suggested that the retail pork examined was highly contaminated with S. aureus. In addition, the MSSA ST398 harboured multiple virulence and exhibited multiple antimicrobial resistance. To our knowledge, this is the first report of the detection of MSSA ST398 in retail pork and BTM in China. The whole genome sequencing studies are crucial to survey the global epidemiology of infectious agents, including MSSA ST398, and provided a deeper knowledge of the epidemiology of this bacterium and may help in understanding how to prevent and treat infections without boosting antibiotic resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical review and approval was not required for the animal study because The Ethical Statement is not applicable because sample collection from animals has been gathered.
XZ and YLi designed the work. XZ, MH, YZ, QZ, LL, and YLu collected samples. XZ and YLi analyzed and interpreted data. XZ drafted the article. XZ and YLi critically reviewed the article. XZ and CZ revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Project (2019YFA0904004); Shandong Agricultural Major Applied Technology Innovation Program (SD2019XM009); Major Scientific and Technological Innovation Project in Shandong Province (2019JZZY010719); and The High-Level Talents and Innovative Team Recruitment Program of the Shandong Academy of Agricultural Sciences, China (CXGC2018E10).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.764105/full#supplementary-material
3. ^http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Abdalrahman, L. S., Stanley, A., Wells, H., and Fakhr, M. K. (2015). Isolation, virulence, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin sensitive Staphylococcus aureus (MSSA) strains from Oklahoma retail poultry meats. Int. J. Environ. Res. Public Health 12, 6148–6161. doi: 10.3390/ijerph120606148
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bhargava, K., Wang, X., Donabedian, S., Zervos, M., de Rocha, L., and Zhang, Y. (2011). Methicillin-resistant Staphylococcus aureus in retail meats, Detroit, Michigan, USA. Emerg. Infect. Dis. 17, 1135–1137. doi: 10.3201/eid1706.101905
Buyukcangaz, E., Velasco, V., Sherwood, J. S., Stepan, R. M., Koslofsky, R. J., and Logue, C. M. (2013). Molecular typing of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) isolated from animals and retail meat in North Dakota, United States. Foodborne Pathog. Dis. 10, 608–617. doi: 10.1089/fpd.2012.1427
CDC. (2019). Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services, CDC, Atlanta, GA. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
Chairat, S., Gharsa, H., Lozano, C., Gomez-Sanz, E., Gonez, P., Zarazaga, M., et al. (2015). Characterization of Staphylococcus aureus from raw meat samples in Tunisia: detection of clonal lineage ST398 from the African continent. Foodborne Pathog. Dis. 12, 686–692. doi: 10.1089/fpd.2015.1958
Chen, C. J., and Huang, Y. C. (2014). New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 20, 605–623. doi: 10.1111/1469-0691.12705
Clinical and Laboratory Standards Institute. (2019). Performance Standards for Antimicrobial Susceptibility Testing. 29th informational supplement, M100-S29. CLSI, Pennsylvania, USA.
David, M. Z., Siegel, J., Lowy, F. D., Zychowski, D., Taylor, A., and Lee, C. J.. (2013). Asymptomatic carriage of sequence type 398, spa type t571 methicillin-susceptible Staphylococcus aureus in an urban jail: a newly emerging transmissible pathogenic strain. J. Clin. Microbiol. 51, 2443–2447.
Deborah, A. W., Sarah, B., Geoffrey, W. C., Hui, T., Stefan, M., and Helen, H. (2014). Emergence and molecular characterization of clonal complex 398 (CC398) methicillin-resistant Staphylococcus aureus (MRSA) in New Zealand. J. Antimicrob. Chemother. 499, 1428–1430. doi: 10.1093/jac/dkt499
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000
Fijalkowski, K., Masiuk, H., Czernomysy-Furowicz, D., Karakulska, J., Nawrotek, P., Paszkowska, A., et al. (2013). Superantigen gene profiles, genetic relatedness and biological activity of exosecretions of Staphylococcus aureus isolates obtained from milk of cows with clinical mastitis. Microbiol. Immunol. 57, 674–683. doi: 10.1111/1348-0421.12088
Frenay, H. M., Bunschoten, A. E., Schouls, L. M., van Leeuwen, W., Vandenbroucke-Grauls, C. M., Verhoef, J., et al. (1996). Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15, 60–64. doi: 10.1007/BF01586186
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Golding, G. R., Bryden, L., Levett, P. N., McDonald, R. R., Wong, A., Wylie, J., et al. (2010). Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg. Infect. Dis. 16, 587–594. doi: 10.3201/eid1604.091435
Hanson, B. M., Dressler, A. E., Harper, A. L., Scheibel, R. P., Wardyn, S. E., Roberts, L. K., et al. (2011). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) on retail meat in Iowa. J. Infect. Public Health 4, 169–174. doi: 10.1016/j.jiph.2011.06.001
Harmsen, D., Claus, H., Witte, W., Rothgänger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
Hemandez, D., Tewhey, R., Veyrieras, J. B., Farinelli, L., Osteras, M., Francois, P., et al. (2014). De novo finished 2.8 Mbp Staphylococcus aureus genome assembly from 100 bp short and long range paired-end reads. Bioinformatics 30, 40–49. doi: 10.1093/bioinformatics/btt590
Hennekinne, J. A., De Buyser, M. L., and Dragacci, S. (2012). Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36, 815–836. doi: 10.1111/j.1574-6976.2011.00311.x
Huang, Y. C., and Chen, C. J. (2020). Detection and phylogeny of Staphylococcus aureus sequence type 398 in Taiwan. J. Biomed. Sci. 27:15. doi: 10.1186/s12929-019-0608-8
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Johler, S., Macori, G., Bellio, A., Acutis, P. L., Gallina, S., and Decastelli, L. (2018). Characterization of Staphylococcus aureus isolated along the raw milk cheese production process in artisan dairies in Italy. J. Dairy Sci. 101, 2915–2920. doi: 10.3168/jds.2017-13815
Kadlec, K., and Schwarz, S. (2010). Identification of the novel dfrK-carrying transposon Tn559 in a porcine methicillin-susceptible Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54, 3475–3477. doi: 10.1128/AAC.00464-10
Kelman, A., Soong, Y. A., Dupuy, N., Shafer, D., Richbourg, W., Johnson, K., et al. (2011). Antimicrobial susceptibility of Staphylococcus aureus from retail ground meats. J. Food Prot. 74, 1625–1629. doi: 10.4315/0362-028X.JFP-10-571
Le Marechal, C., Seyffert, N., Jardin, J., Hernandez, D., Jan, G., Rault, L., et al. (2011). Molecular basis of virulence in Staphylococcus aureus mastitis. PLoS One 6:e27354. doi: 10.1371/journal.pone.0027354
Li, W., Liu, J. H., Zhang, X. F., Wang, J., Ma, Z. B., Chen, L., et al. (2018). Emergence of methicillin-resistant Staphylococcus aureus ST398 in pigs in China. Int. J. Antimicrob. Ag. 51, 275–276.
Li, G., Wu, C., Wang, X., and Meng, J. (2015). Prevalence and characterization of methicillin susceptible Staphylococcus aureus ST398 isolates from retail foods. Int. J. Food Microbiol. 196, 94–97. doi: 10.1016/j.ijfoodmicro.2014.12.002
Louie, L., Matsumura, S. O., Choi, E., Louie, M., and Simor, A. E. (2000). Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 38, 2170–2173. doi: 10.1128/JCM.38.6.2170-2173.2000
Lozano, C., Rezusta, A., Gomez, P., Gomez-Sanz, E., Baez, N., Martin-Saco, G., et al. (2012). High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J. Antimicrob. Chemother. 67, 330–334. doi: 10.1093/jac/dkr497
Mahmoud, A. M., Samir, M. A., and Khalid, I. S. (2021). Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health concern. Int. J. Food Microbiol. 346:109165. doi: 10.1016/j.ijfoodmicro.2021.109165
Mediavilla, J. R., Chen, L., Uhlemann, A. C., Hanson, B. M., Rosenthal, M., Stanak, K., et al. (2012). Methicillin-susceptible Staphylococcus aureus ST398, New York and New Jersey, USA. Emerg. Infect. Dis. 18, 700–702. doi: 10.3201/eid1804.111419
Mitchell, D. G., Matthew, V. N. O., Huub, J. M. B., Toni, A. C., Sam, A., Darren, J. T., et al. (2014). Staphylococcus aureus ST398 detected in pigs in Australia. J. Antimicrob. Chemother. 529, 1426–1428. doi: 10.1093/jac/dkt526
Ou, Q., Peng, Y., Lin, D., Bai, C., Zhang, T., Lin, J., et al. (2017). A meta-analysis of the global prevalence rates of Staphylococcus aureus and methicillin-resistant S. aureus contamination of different raw meat products. J. Food Prot. 80, 763–774. doi: 10.4315/0362-028X.JFP-16-355
Pantosti, A. (2012). Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. 3:127. doi: 10.3389/fmicb.2012.00127
Rasigade, J. P., Laurent, F., Lina, G., Meugnier, H., Bes, M., Vandenesch, F., et al. (2010). Global distribution and evolution of panton-valentine leucocidin-positive methicillin-susceptible Staphylococcus aureus, 1981-2007. J. Infect. Dis. 201, 1589–1597. doi: 10.1086/652008
Ronco, T., Klaas, I. C., Stegger, M., Svennesen, L., Astrup, L. B., Farre, M., et al. (2018). Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis. Vet. Microbiol. 215, 35–42. doi: 10.1016/j.vetmic.2018.01.003
Sahibzada, S., Hernández-Jover, M., Jordan, D., Thomson, P. C., and Heller, J. (2018). Emergence of highly prevalent CA-MRSA ST93 as an occupational risk in people working on a pig farm in Australia. PLoS One 13:e0195510. doi: 10.1371/journal.pone.0195510
Schouls, L. M., van der Ende, A., van de Pol, I., Schot, C., Spanjaard, L., Vauterin, P., et al. (2005). Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43, 2741–2749. doi: 10.1128/JCM.43.6.2741-2749.2005
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sergelidis, D., Abrahim, A., Anagnostou, V., Govaris, A., Papadopoulos, T., and Papa, A. (2012). Prevalence, distribution, and antimicrobial susceptibility of Staphylococcus aureus in ready-to-eat salads and in the environment of a salad manufacturing plant in northern Greece. Czech J. Food Sci. 30, 285–291. doi: 10.17221/37/2011-CJFS
Shahnaz, A., Fatemeh, F., Masoumeh, N., and Sahar, V. (2016). Prevalence of blaOXA-1 and blaDHA-1 AmpC β-lactamase-producing and methicillin-resistant Staphylococcus aureus in Iran. Archives Pediatric Infect. Dis. 5:36778. doi: 10.5812/pedinfect.36778
Sun, C., Chen, B., Hulth, A., Schwarz, S., Ji, X., Nilsson, L., et al. (2019). Genomic analysis of Staphylococcus aureus along a pork production chain and in the community, Shandong province, China. Int. J. Antimicrob. Agents 54, 8–15. doi: 10.1016/j.ijantimicag.2019.03.022
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
Uhlemann, A. C., Porcella, S. F., Trivedi, S., Sullivan, S. B., Hafer, C., Kennedy, A. D., et al. (2012). Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. MBio 3:e00027-12. doi: 10.1128/mBio.00027-12
Valentin-Domelier, A., Girard, M., Bertrand, X., Violette, J., Francois, P., Donnio, P., et al. (2011). Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: An emerging human-adapted subclone? PLoS One 6:e28369. doi: 10.1371/journal.pone.0028369
Waters, A. E., Contente-Cuomo, T., Buchhagen, J., Liu, C. M., Watson, L., Pearce, K., et al. (2011). Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 52, 1227–1230. doi: 10.1093/cid/cir181
Wilson, G. J., Seo, K. S., Cartwright, R. A., Connelley, T., Chuang-Smith, O. N., Merriman, J. A., et al. (2011). A novel core genome encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7:e1002271. doi: 10.1371/journal.ppat.1002271
Xue, J. Y., Zhao, T., Liu, Y., Liu, Y., Zhang, Y. X., Zhang, G. Q., et al. (2020). Genome-wide analysis of the nucleotide binding site leucine-rich repeat genes of four orchids revealed extremely low number of disease resistance genes. Front. Genet. 10:1286. doi: 10.3389/fgene.2019.01286
Yan, X., Yu, X., Tao, X., Zhang, J., Zhang, B., Dong, R., et al. (2014). Staphylococcus aureus ST398 from slaughter pigs in Northeast China. Int. J. Med. Microbiol. 304, 379–383. doi: 10.1016/j.ijmm.2013.12.003
Yang, X. J., Zhang, J. M., Yu, S. B., Wu, Q. P., Guo, W. P., Huang, J. H., et al. (2016). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail ready to-eat foods in China. Front. Microbiol. 7:816. doi: 10.3389/fmicb.2016.00816
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zdragas, A., Papadopoulos, T., Mitsopoulos, I., Samouris, G., Vafeas, G., Boukouvala, E., et al. (2015). Prevalence, genetic diversity, and antimicrobial susceptibility profiles of Staphylococcus aureus isolated from bulk tank milk from Greek traditional ovine farms. Small Rumin. Res. 125, 120–126. doi: 10.1016/j.smallrumres.2015.02.009
Keywords: methicillin-susceptible Staphylococcus aureus, sequence type 398, antimicrobial susceptibility testing, whole-genome epidemiology, spa
Citation: Zhao X, Hu M, Zhao C, Zhang Q, Li L, Zhang Y, Luo Y and Liu Y (2021) Whole-Genome Epidemiology and Characterization of Methicillin-Susceptible Staphylococcus aureus ST398 From Retail Pork and Bulk Tank Milk in Shandong, China. Front. Microbiol. 12:764105. doi: 10.3389/fmicb.2021.764105
Received: 25 August 2021; Accepted: 03 November 2021;
Published: 30 November 2021.
Edited by:
Spiros Paramithiotis, Agricultural University of Athens, GreeceReviewed by:
Chunlei Shi, Shanghai Jiao Tong University, ChinaCopyright © 2021 Zhao, Hu, Zhao, Zhang, Li, Zhang, Luo and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaonan Zhao, emhhb3hpYW9uYW4xMjE0QDE2My5jb20=; Yuqing Liu, bGl1aXVxaW5nQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.