- 1School of Medical Technology and Nursing, Shenzhen Polytechnic, Shenzhen, China
- 2Department of Gastroenterology, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen, China
- 3Department of Microbiology, School of Public Health, Southern Medical University, Guangzhou, China
- 4Department of Neurosurgery, Peking University Shenzhen Hospital, Shenzhen, China
- 5Key Laboratory of Tropical Disease Research of Guangdong Province, Guangzhou, China
The ppk1 gene encodes polyphosphate kinase (PPK1), which is the major catalytic enzyme that Escherichia coli utilizes to synthesize inorganic polyphosphate (polyP). The aim of this study was to explore the role of PPK1 in the pathogenesis of Enterohemorrhagic E. coli O157:H7 (EHEC O157:H7). An isogenic in-frame ppk1 deletion mutant (Δppk1) and ppk1 complemented mutant (Cppk1) were constructed and characterized in comparison to wild-type (WT) EHEC O157:H7 strain EDL933w by microscope observation and growth curve analysis. Survival rates under heat stress and acid tolerance, both of which the bacteria would face during pathogenesis, were compared among the three strains. LoVo cells and a murine model of intestinal colitis were used as the in vitro and in vivo models, respectively, to evaluate the effect of PPK1 on adhesion and invasion during the process of pathogenesis. Real-time reverse-transcription PCR of regulatory gene rpoS, adhesion gene eae, and toxin genes stx1 and stx2 was carried out to corroborate the results from the in vitro and in vivo models. The ppk1 deletion mutant exhibited disrupted polyP levels, but not morphology and growth characteristics. The survival rate of the Δppk1 strain under stringent environmental conditions was lower as compared with WT and Cppk1. The in vitro assays showed that deletion of the ppk1 gene reduced the adhesion, formation of attaching and effacing (A/E) lesions, and invasive ability of EHEC O157:H7. Moreover, the virulence of the Δppk1 in BALB/c mice was weaker as compared with the other two strains. Additionally, mRNA expression of rpoS, eae, stx1 and stx2 were consistent with the in vitro and in vivo results. In conclusion: EHEC O157:H7 requires PPK1 for both survival under harsh environmental conditions and virulence in vivo.
Introduction
Enterohemorrhagic Escherichia coli O157:H7 (EHEC O157:H7) is a gram-negative bacterium that was identified as a human pathogen in 1982 and has continued to be a worldwide threat to public health. EHEC O157:H7 is known to cause hemorrhagic colitis (HC), thrombotic thrombocytopenic purpura (TTP), and hemolytic uremic syndrome (HUS), which results in high mortality (Riley et al., 1983; Perna et al., 2001; Zhao et al., 2013). The main pathogenic mechanisms of EHEC O157:H7 include the production of Shiga toxin (Stx), which is primarily responsible for the renal complications and neurological sequelae of EHEC infections, as well as a type III secretion system (TTSS) that enables tight adherence of bacteria to host epithelial cells by inducing characteristic actin cytoskeletal rearrangements and loss of microvillus structure (A/E lesions) (Nguyen and Sperandio, 2012; Davis et al., 2014; Melton-Celsa, 2014; Stevens and Frankel, 2014; Pacheco et al., 2018; Menge, 2020). In recent years, progress has been made in understanding the function and mechanism of TTSS effector proteins (Wang et al., 2017; Hua et al., 2020), and factors that have overlapping effects on TTSS and Stx, such as sRNA Esr055, which is involved in the regulation of the preferential colonization in the large intestine and inhibition of the stx2 (Han et al., 2017) as well as targeted loci associated with sphingolipid biosynthesis (Pacheco et al., 2018).
Shiga toxin, including Stx1 and Stx2 produced by bacteria including Shigella dysenteriae serotype 1 and EHEC O157:H7 strain 933 (Lee et al., 2016), act as primary virulence factors. Each ribosome-inactivating holotoxin possess an AB5 molecular configuration consisting of a large monomeric A subunit and small homo-pentameric B subunits (Rangel et al., 2005). Once colonized in intestinal epithelial cells, EHEC induces the delivery of Stx and the production of cytokines and chemokines. Toxins pass through the intestinal mucosa, enter the bloodstream and travel to target organs such as the kidneys and Central Nervous System (CNS). After membrane invasion-mediated endocytosis through the toxin receptor Gb3 on the cell surface, Stxs migrate to the Golgi and endoplasmic reticulum (ER). Stx acts as a multifunctional bacterial protein, promoting ER stress, ribotoxic stress, pro-inflammatory responses, apoptosis, and autophagy in host cells (Lee et al., 2021). In addition to cell death by Stxs, various cells such as neutrophils, induce inflammation in the intestine, which leads to damage.
Adhesion and effacing lesions (A/E lesions) are characterized by intimate bacterial attachment to the surface of intestinal epithelial cells, cytoskeletal rearrangements beneath adherent bacteria, formation of characteristic actin-rich “pedestals” and destruction of proximal microvilli (Stevens and Frankel, 2014).
The EHEC TTSS injects a plethora of effector proteins into host cells that induce alteration or disruption of numerous host cell processes (Pacheco et al., 2018). Tir is the first translocated effector protein inserted into the host cell membrane as a receptor of adhesion intimin. Intimin-Tir interactions are required to cluster Tir, which initiates the process of activating actin assembly and recruiting cytoskeletal proteins, such as clathrin (Hua et al., 2018), at the site of bacterial adherence (Campellone et al., 2004a; Stevens and Frankel, 2014). The main pathway causing actin rearrangement resulting in the formation of actin-rich pedestals eventually in EHEC is the Tir: IRTKS/IRSp53:TccP/EspFu pathway, which triggers Arp2/3-mediated actin polymerization independently of N-WASP (Vingadassalom et al., 2010).
Inorganic polyphosphate (polyP) is abundant in all cells, including bacteria, fungi, parasites, plants, and animals, and plays critical roles (Rao et al., 2009). Polyphosphate kinase 1 (PPK1) reversibly catalases the polymerization of the terminal phosphate of ATP into a polyP chain and is the major catalytic enzyme in E. coli that synthesizes polyP. PPK1, encoded by the ppk1 gene, has been related to stationary phase (SP) survival and pathogenesis in many bacteria species, such as E. coli, Vibrio cholera, Shigella and Salmonella spp., via polyP inducing the expression of DNA repair enzymes (Varas et al., 2017) or inducing the expression of rpoS, which is the selective σS or σ38 factor of the RNA polymerase and regulates the expression of survival and other virulence genes in bacteria, such as E. coli (Shiba et al., 1997). However, RpoS proteins are not the only regulators of survival and virulence genes during the SP in E. coli. A previous study identified a new regulatory gene, ibeR, that mediated stress resistance and pathogenesis SP gene expression in E. coli K1 RS218, which has a loss-of-function mutation in the rpoS gene (Chi et al., 2009). Additionally, Varas et al. (2017) found that the metabolic balance of polyP was necessary for the synthesis of the second messenger (p) ppGpp, which regulated other important cellular and regulatory processes such as the recycling of sigma factors/anti-sigma factors that allowed the bacteria to adapt to a wide range of environmental conditions, including nutritional or stressful conditions (Magnusson et al., 2005; Dalebroux and Swanson, 2012). To investigate the role of PPK1 in the pathogenesis of EHEC O157:H7, an isogenic in-frame ppk1 deletion mutant of EHEC O157:H7 EDL933w and a ppk1-complemented strain, where the ppk1 open reading frame in the plasmid pBAD33 was transferred into the ppk1 deletion mutant, were constructed and characterized. The polyP levels in the ppk1 deletion mutant (Δppk1) indicated that the Δppk1 was deficient in polyP synthesis as compared to the wild-type (WT) and ppk1-complemented (Cppk1) strains. Moreover, the deletion of ppk1 influenced the survival of EHEC O157:H7 under harsh environmental conditions of 55°C and pH 2.0. PPK1 contributed to the adhesion, formation of A/E lesions, and invasion of LoVo cells in vitro. Additionally, deletion of the ppk1 gene reduced the virulence in BALB/c mice. Moreover, deletion of ppk1 resulted in decreased expression of rpoS, as well as the adhesion gene eae and virulence genes stx1 and stx2. Our findings suggest that PPK1 is a determinant in the pathogenesis of EHEC O157:H7.
Materials and Methods
Bacterial Strains, Cells, Plasmids and Culture Conditions
Enterohemorrhagic Escherichia coli O157:H7 strain EDL933w was obtained from the Chinese Center for Disease Control and Prevention (CDC) (Beijing, China). The nalidixic acid resistant mutant of EHEC O157:H7 strain EDL933w named EHEC O157:H7 EDL933w (NalR) was selected as described by published paper by Zhao et al. from our lab (Zhao et al., 2013). Plasmids, pKD46 [containing recombinant enzymes including Gam, Bet, and Exo (Exo is an exonuclease that binds to the ends of double-stranded DNA, degrades the DNA from the 5′ end to the 3′ end, and produces 3′ overhangs. Beta binds to the single-stranded DNA and mediates the complementary single-stranded DNA annealing. Gam protein binds with the RecBCD enzyme to inhibit its activity of degrading foreign DNA Jeong et al., 2013)] and pKD4 plasmid (containing the kanamycin resistance gene) were obtained from the Institute of Microbiology and Epidemiology (Beijing, China). LoVo colonic carcinoma cells and DH5α cells with the plasmid pBAD33 (ATCC, 87402) were purchased from ATCC. T4 DNA ligase and restriction endonucleases were purchased from TaKaRa Biotechnology Co., Ltd. (Dalian, China). Standard laboratory reagents and chemicals, such as DAPI, were purchased from Sigma-Aldrich (St. Louis, MO, United States) unless otherwise mentioned. Primers used in this study were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). DNA sequencing was performed by Guangzhou IGE Biotechnology, Ltd. (Guangzhou, China).
All bacterial strains in this study were grown in Luria broth (LB) media (10 g SELECT Peptone 140 (Oxiod, England), 5 g SELECT Yeast Extract (Oxiod, England), and 10 g sodium chloride (Beijing, China) dissolving completely in double-distilled water (ddH2O) with 5mol/L NaOH for pH = 7.0 and a total volume of 1 L by ddH2O, autoclave sterilization by 121°C for 20 min) at 37°C at 200 rpm in a constant-temperature, oscillating shaker with nalidixic acid (50 μg/mL) and appropriate antibiotics (WT: no antibiotics; Δppk1: 100 μg/mL kanamycin; Cppk1: 100 μg/mL kanamycin and 10 μg/mL chloramphenicol) if necessary. LoVo cells were cultured in DMEM (Gibco, Waltham, MA, United States) with 10% fetal bovine serum (FBS, Gibco, Waltham, MA, United States) and 1% penicillin/streptomycin (Gibco, Waltham, MA, United States) in 5% CO2 for routine passage prior to infection.
Female BALB/c (4–5 weeks of age) mice were obtained from the Lab Animal Center of Southern Medical University (Guangzhou, China; Certificate: SCXK (Guangdong Province) 2016-0041, No. 44002100010995]. All mice were housed under specific pathogen-free (SPF) conditions according to the regulations of the animal care committee. This study was conducted in accordance with the recommendations of the Southern Medical University Experimental Animal Ethics Committee (Guangzhou, China).
Construction of an Isogenic In-Frame Deletion Mutant of ppk1 and ppk1-Complemented Strains
A pKD46-mediated λ red homologous recombination system was used to construct EHEC O157:H7 Δppk1. According to the ppk1 gene sequence in GenBank (Accession no. NC_002655) and its upstream and downstream DNA sequences, three pairs of primers were designed and synthesized (Table 1). H1-K1 and H2-K2 primers contained homologous arms, of which the 5′ end was homologous with the ppk1 gene and the 3′ end was homologous with the kanamycin resistant gene. PPK-P1, PPK-P2, N1-F, and N2-R were cross-identifying primers specific for the ppk1 gene in EHEC O157:H7. Kana-F and Kana-R were internal-identifying primers for the kanamycin resistance gene within the pKD4 plasmid. The kanamycin resistance gene was amplified using primers H1-K1 and H2-K2 with the pKD4 plasmid as a template by using overlap extension PCR. The PCR conditions were as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s, and a final extension of 72°C for 10 min. Subsequently, plasmid pKD46 was transformed into EHEC O157:H7 EDL933w and cultured to an OD600 = 0.2–0.3. L-arabinose was then added to a final concentration of 10 mmol/L for 40–60 min to induce the full expression of recombinant enzymes Exo, Bet, and Gam of pKD46. Finally, 10 μL of the targeting fragments (34.67 ng/μL) isolated from the gels and the prepared 100 μL EHEC O157:H7 EDL933w competent cells were mixed in a gene pulser cuvette (Bio-Rad Laboratories, Inc., Hercules, CA, United States) and subjected to an electric shock for 10–20 s (25 μF, 200 Ω, 3 KV). Positive strains were screened using LB plates containing 50 μg/mL kanamycin. The recombinant strain was confirmed by PCR and DNA sequencing.

Table 1. Primer sequences used in the construction of the isogenic in-frame deletion mutant of ppk1 and ppk1-complemented strains.
The complemented strain, Cppk1, was constructed using the prokaryotic expression plasmid pBAD33. A DNA fragment carrying the complete ppk1 open reading frame was amplified using the PPK1-F and PPK1-R primers (Table 1) with the EDL933w genome as the DNA template. After double-enzyme digestion with HindIII and XbaI, the DNA fragment and vector pBAD33 were ligated together with T4 DNA Ligase, and the products were transformed into strain EHEC O157:H7 Δppk1. Positive strains were screened using LB medium containing 100 μg/mL kanamycin and 10 μg/mL chloramphenicol. PCR and DNA sequencing analyses were conducted to identify the complemented strains. Finally, quantitative real-time PCR using Applied Biosystems (ABI) 7500 FAST Real-Time PCR system (Thermo Fisher, Waltham, MA United States) was performed to verify the transcription of ppk1 in the complemented strain. Briefly, the WT, Δppk1 and Cppk1 strains were cultured in liquid LB medium overnight. Total RNA extracted with TRIzol® (Invitrogen, Carlsbad, CA, United States) was reverse transcribed using the ExScript RT reagent kit (Takara, Japan) according to the manufacturer’s instructions. The real-time PCR conditions were: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 32 s. A 16S rRNA gene fragment (68 bp) was used as an internal control, and the amplification of ppk1 (255 bp) in the WT strain was used as a reference. Gene expression levels were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Gram Staining and Growth Curves
Gram staining was performed with the classical steps (Coico, 2005), and reagents needed for this assay were from Nanjing Jiancheng Technology Co., Ltd., China. The growth curves were determined by measuring the optical density at 600 nm (OD600) of the cultures at different times in LB media at 37°C at 200 rpm in a constant-temperature, oscillating shaker. Antibiotics were added if the strain required them as described above.
Measurement of polyP Levels
Quantification of polyP by DAPI was performed in different fields of view as previously published (Aschar-Sobbi et al., 2008; Diaz and Ingall, 2010; Kulakova et al., 2011; Gomes et al., 2013). Based on these researches, we optimized the procedure in E. coli. Briefly, the WT, Δppk1 and Cppk1 strains were grown in LB media for 12-14 h at 37°C with the appropriate antibiotics. The cells were then diluted to an OD600 = 1.0, which equaled approximately 1 × 108 CFU/mL, and centrifuged at 10,000 × g for 15 min at 4°C. After washing with 20 mM HEPES buffer (pH 7.5) twice, the pellets were resuspended in 1 mL of 20 mM HEPES buffer (pH 7.5). In order to release intracellular polyP from bound state for direct quantification without prior extraction, cells were lysed by a freeze-thaw cycle at − 80°C followed by thawing at 24–26°C. Then, 300 μL of the lysed cells were added to new sterilized microcentrifuge tubes containing 600 μL of 20 mM HEPES buffer (pH 7.5). Next, 100 μL of 100 μM DAPI reagent was added, resulting in a final DAPI concentration of 10 μM. The reaction was allowed to incubate in the dark for 15 min to ensure steady fluorescence. Then 200 μL of each reaction was added to a 96-well plate in triplicate. A polyP standard calibration curve using sodium phosphate glass type 45 (short for polyP45) was constructed on the same 96-well plate. PolyP45 dilutions were prepared in 20 mM HEPES buffer (pH 7.5) for a polyP relative unit (ru) of 0–1 ru, and the concentration of polyP ranged from 0–3 μM Pi. The polyP45 dilutions were mixed with 100 μM DAPI by vortexing for 7.5 min, incubated for 15 min at 24–26°C in the dark, vortexed again, and then 200 μL of each mixture was added to the 96-well plate in triplicate. The fluorescence module of a microplate reader (Infinite M200 Pro, Tecan, Switzerland) was used to measure the fluorescence value in the dark. The excitation wavelength was 415 nm, and the emission wavelength was 550 nm. The polyP quantity of the three strains was extrapolated from the standard curve. For bacteria cultured in vitro, we used the polyP concentration in μM Pi/mL, which translates to the amount of polyP in 1 mL of bacteria at an OD600 = 1.0.
Survival Rates Under Different Stringent Environmental Conditions
The WT, Δppk1 and Cppk1 strains bacteria were grown in LB medium for 12-14 h at 37°C. The bacterial culture was diluted in LB medium without antibiotics until the OD600 was 1.0 (approximately 1 × 108 CFU/mL). For heat stress, the three strains (prepared as stated above) were incubated at 55°C for 0, 1, 2, 3, 4, 5, 8, 10, and 15 min. Samples collected at these points were serially diluted in LB liquid medium until 10–6 and then plated on LB agar plates and cultured for at least for 12 h. The survival rate after heat stress for each culture was calculated by dividing the number of colonies counted after exposure to heat stress by the viable cell number before exposure to stress colonies. For acid tolerance test, the samples (prepared as stated above) were collected by centrifugation. Then they were resuspended in acidified LB (pH 2.0) and incubated at 37°C. Samples were collected at 0, 5, 10, 20, 30, 60, and 120 min. Samples collected at these points were serially diluted in LB liquid medium until 10–6 and then plated on LB agar plates and cultured for at least for 12 h. The survival rate after acid tolerance for each culture was calculated by dividing the number of colonies counted after exposure to heat stress by the viable cell number before exposure to stress colonies (Peng et al., 2012).
Preparation of Bacterial Solution for in vitro Adhesion and Invasion Assays
The WT, Δppk1 and Cppk1 strains were grown overnight in LB medium at 37°C. Then the bacteria solution was diluted with LB medium with no antibiotics until the OD600 was 1.0 (approximately 1 × 108 CFU/mL). The bacteria were collected by centrifugation and resuspended in cell culture medium without antibiotics.
Adherence Quantitation Assay
The adherence assay was carried out according to previous methods (Elsinghorst and Kopecko, 1992; Peng et al., 2012). Briefly, LoVo confluent monolayers in 12-well plates (approximately 1 × 106 cells/well) were incubated with the WT, Δppk1 and Cppk1 strains of EHEC O157:H7 (1 × 108 CFU/well) for 2 h in an incubator with 5% CO2, 95% air [including 78% nitrogen, 21% oxygen, 0.934% rare gases (helium, neon, argon, krypton, xenon, and radon)], 37°C. After incubation, the monolayers were washed at least five times with phosphate buffered saline (PBS). Then the cells were lysed with 0.5% Triton X-100 for 10 min, which had no lethal effect on the bacteria. The lysate was then serially diluted at 1:1000 in PBS, plated on LB plates, and incubated for 12 h. The number of colonies was counted, and the adhesion rate was calculated as follows: ‰ = (number of colonies × 103)/1 × 108. Each sample was assayed in triplicate.
Adherence Assay by Scanning Electron Microscopy
Nine sterile cell slides for 24-well plates were placed in 9 wells of a 24-well plate. Then 200 μl of LoVo cells (approximately 5 × 105/mL) were added into the 9 wells for cultivation in a 5% CO2 incubator at 37°C until the cells formed confluent monolayers (approximately 1.0 × 106 cells/well). The WT, Δppk1 and Cppk1 strains of EHEC O157:H7 with OD600 = 1.0 (approximately 1 × 108 CFU/mL of bacteria) were inoculated to those wells at a 1:100 cell:bacteria ratio. The 24-well plate was placed in a 5% CO2 incubator at 37°C. After washing with PBS five times, the nine wells were placed into wide caliber EP tubers and fixed with 2.5% glutaraldehyde for SEM.
Invasion Assay
LoVo confluent monolayers in 12-well plates (approximately 1 × 106 cells/well) were incubated with bacteria (1 × 108 CFU/well) for 2 h. The monolayers were washed with PBS and incubated with the mixed medium [DMEM with 10% heat-inactivated FBS and gentamicin (100 μg/mL)] for another 2 h at 37°C in order to kill the extracellular bacteria. The monolayers were then washed with PBS and lysed with 0.5% Triton X-100. The released intracellular bacteria were collected, serially diluted, plated on LB plates and cultured for 12 h. The number of bacteria was counted, and the invasion rate is expressed as follows: ‰ = (number of bacteria × 1000)/number of total added bacteria. The assay was performed in triplicate for each strain.
Invasion Assay via Transmission Electron Microscopy
All the procedures were the same as described above (i.e., “Invasion assay” section). After treatment with gentamicin for 2 h at 37°C, the monolayers were washed with warm cell culture medium. The cells were then scraped off the plates and fixed with 2.5% glutaraldehyde for transmission electron microscopy (TEM) imaging.
Intracellular F-Actin Observation in LoVo Cells Infected by Enterohemorrhagic Escherichia coli O157:H7 via Confocal Microscopy
LoVo cells were passaged in confocal culture dishes (35 mm in diameter) and cultured into monolayers. The bacterial culture and invasion processes are described above (i.e., “Invasion assay” section). The culture media was removed, and the cells were gently washed once with phosphate buffered saline (PBS) at 37°C. Then cells were fixed in 4% paraformaldehyde for 10 min at 24–26°C, washed with PBS for 30 s, and then permeabilized with 0.15% Triton X-100 for 5 min. The cells were blocked with 1% bovine serum albumin after washing with PBS at 24–26°C for 5 min, and then 100 nM rhodamine phalloidin was added and incubated with the cells for 30 min in the dark at 37°C. After washing three times in PBS, the DNA was counterstained with 100 nM DAPI in PBS for 30 s. The samples were rinsed with PBS, and then confocal images (LSM710, ZEISS, Germany) of intracellular actin were obtained.
Murine Model of Infectious Enterohemorrhagic Escherichia coli O157:H7 Colitis
The animal experiments were performed strictly according to the guidelines for animal care at Southern Medical University (Guangzhou, China). The murine model of E. coli-induced colitis was performed as described previously (Zhao et al., 2013). In brief, twenty BALB/c mice (4–5 weeks old) were randomly divided into four groups: (1) EHEC WT, (2) Δppk1, (3) Cppk1, and (4) control (uninfected). The mice were provided water containing nalidixic acid (50 μg/mL) for 12 h prior to infection. Mice were then infected twice at an interval of 12 h. Mice were simultaneously administered 300 μL of the bacterial suspension (2 × 1010 CFU/mL total) orally and then intraperitoneally injected with mitomycin C (MMC, 2.5 mg/kg) to improve their infection susceptibility. Mice in the control group were treated with MMC and inoculated equivalently with LB broth under the same conditions. The mice were sacrificed at day 7 post-infection. The distal 5 cm of the colon was harvested. Typical features of the colon, such as the appearance of the colons and solidified feces, were compared among the groups. After cleaning with PBS buffer, the colons were embedded in paraffin and stained with hematoxylin and eosin (H&E) to observe the pathological changes among the groups. Histologic damage scores were performed using previously described methods (Ledwaba et al., 2020) by a pathologist who had no knowledge of this study.
Real-Time Reverse-Transcription PCR
The mRNA expression levels of rpoS (encodes RpoS), eae (encodes intimin), stx1A, stx1B (both encode Stx1), stx2A and stx2B (both encode Stx2) in WT, Δppk1, and Cppk1 strains were determined by quantitative real-time PCR. Primers (Table 2) were synthesized and used for cDNA amplification of 16S rRNA (68 bp), rpoS (225 bp), eae (220 bp), stx1A (240 bp), stx1B (112 bp), stx2A (129 bp), and stx2B (111 bp). A 16S rRNA gene was used as an internal control. All gene expression was normalized against WT of EHEC O157:H7 using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Statistical Analysis
Experimental values are expressed as the mean ± standard deviation. A Student’s t-test was used to analyze values between two groups, while a one-way ANOVA was used to analyze values between three groups. Multiple comparisons were completed by using the Student-Newman-Keuls (S-N-K) and Bonferroni tests. Differences with p < 0.05 were regarded as statistically significant.
Results
Construction of Enterohemorrhagic Escherichia coli O157:H7 Δppk1 and Cppk1 Strains
The ppk1 isogenic in-frame deletion mutant strain of EHEC O157:H7 EDL933w was successfully constructed. As shown in Figure 1A, ppk1 was replaced by a kanamycin resistance cassette, both of which contained the same homologous arm H1 and H2. Deletion of the ppk1 gene was initially confirmed by colony PCR (Figure 1B). All three pairs of primers verified the successful deletion of ppk1 from the Δppk1 strain, and DNA sequencing further confirmed this deletion.
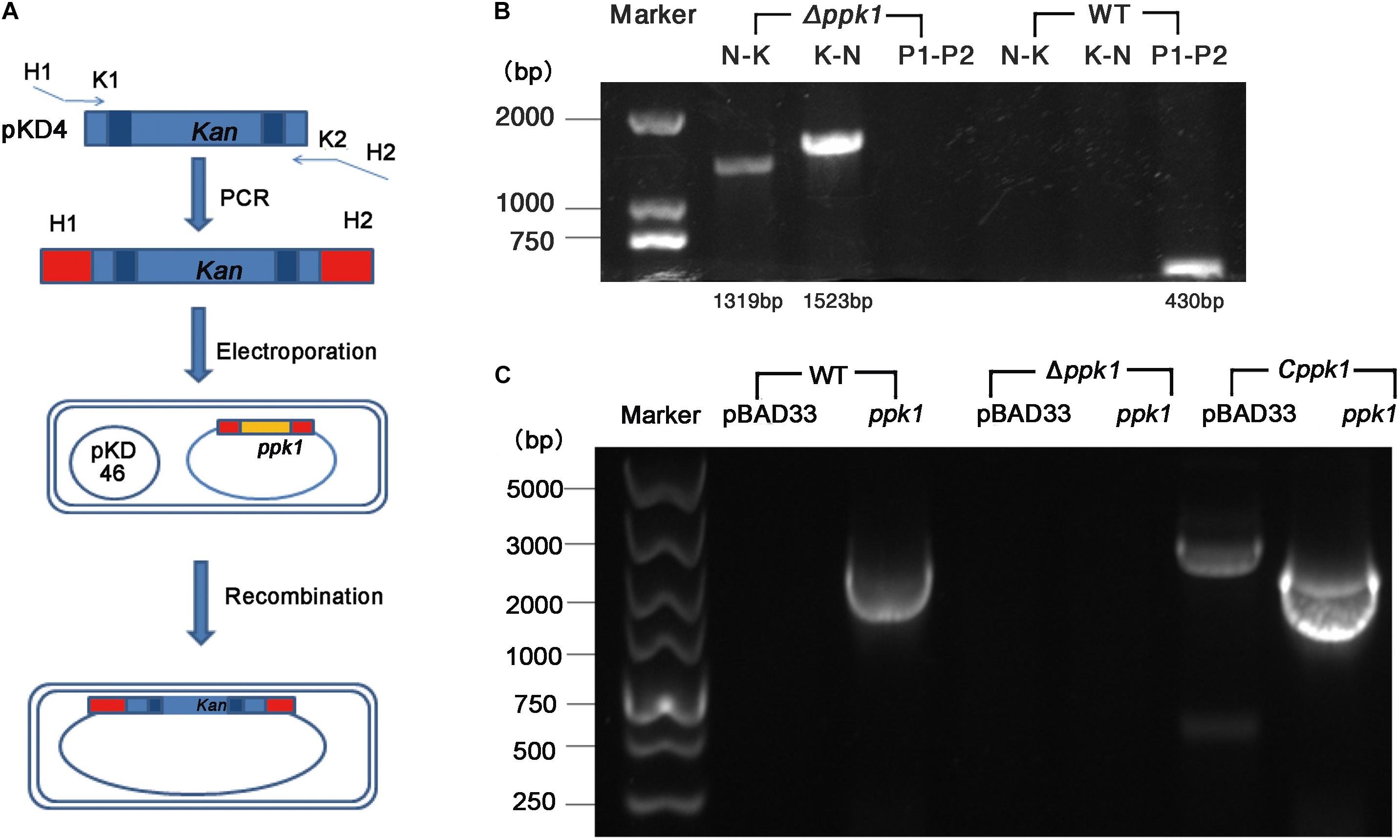
Figure 1. Construction of the ppk1 deletion mutant (Δppk1) and complemented strain (Cppk1) of EHEC O157:H7. (A) Schematic diagram of the λ red system homologous recombination. (B) The primers of lane 2 and 5 (N-K) were N1-F and Kana-R. The primers of lane 3 and 6 (K-N) were Kana-F and N2-R. The primers of lane 4 and 7 (P1-P2) were PPK-P1 and PPK-P2. (C) Two pairs of primers (pBAD33-F/R and ppk-F/R) were used to verify the complemented strain, which produced products of 2092 bp and 2268 bp, respectively. The Cppk1 strain contained both fragments, while WT strain contained only the 2092 bp, and the Δppk1 strain contained neither.
Similarly, the Cppk1 strain was constructed based on the Δppk1 and verified by colony PCR (Figure 1C) and DNA sequencing. The results of real-time reverse-transcription PCR displayed that the ppk1 gene in the Δppk1 was minimally expressed as compared with WT (t = 18.227, P < 0.001), which indicated that the ppk1 gene was successfully knocked out in Δppk1. The ppk1 gene was increased 58-fold in the Cppk1 strain as compared to WT (t = −16.869, P < 0.001), demonstrating that 0.2% L-arabinose could induce a high expression of recombinant plasmid in the Cppk1 strain.
The Influence of the ppk1 Deletion Mutant in polyP Levels, Morphology and Growth Characteristics
The growth curve of the EHEC O157:H7 strain Δppk1 displayed an identical trend of growth as that of the WT and Cppk1 strains in LB medium (Figure 2A). All three strains entered the exponential phase at 3 h post-inoculation and stationary phase at 12 h post-inoculation. In addition, there were no significant differences in the morphology of the bacteria as revealed by optical microscopy and scanning electron microscopy (data not shown). Yet, based on the results Δppk1 was defective in polyP synthesis. DAPI staining was used to quantify the polyP levels of the three strains. The results demonstrated that polyP levels were significantly different (F = 38.90, P < 0.001) among the strains. The fluorescence of the Δppk1 strain [304.19 relative fluorescence units (rfu)] was significantly lower than that in the WT (835.38 rfu) and Cppk1 (899.36 rfu) strains (Figure 2B), with no significant differences between WT and Cppk1. This indicated that complementation of the Δppk1 with the ppk1 gene enabled the Cppk1 strain to produce polyP. All factors above indicated that the deletion of ppk1 did not change the morphology and growth rate of the Δppk1 in nutritive medium but significantly reduced the polyP level.
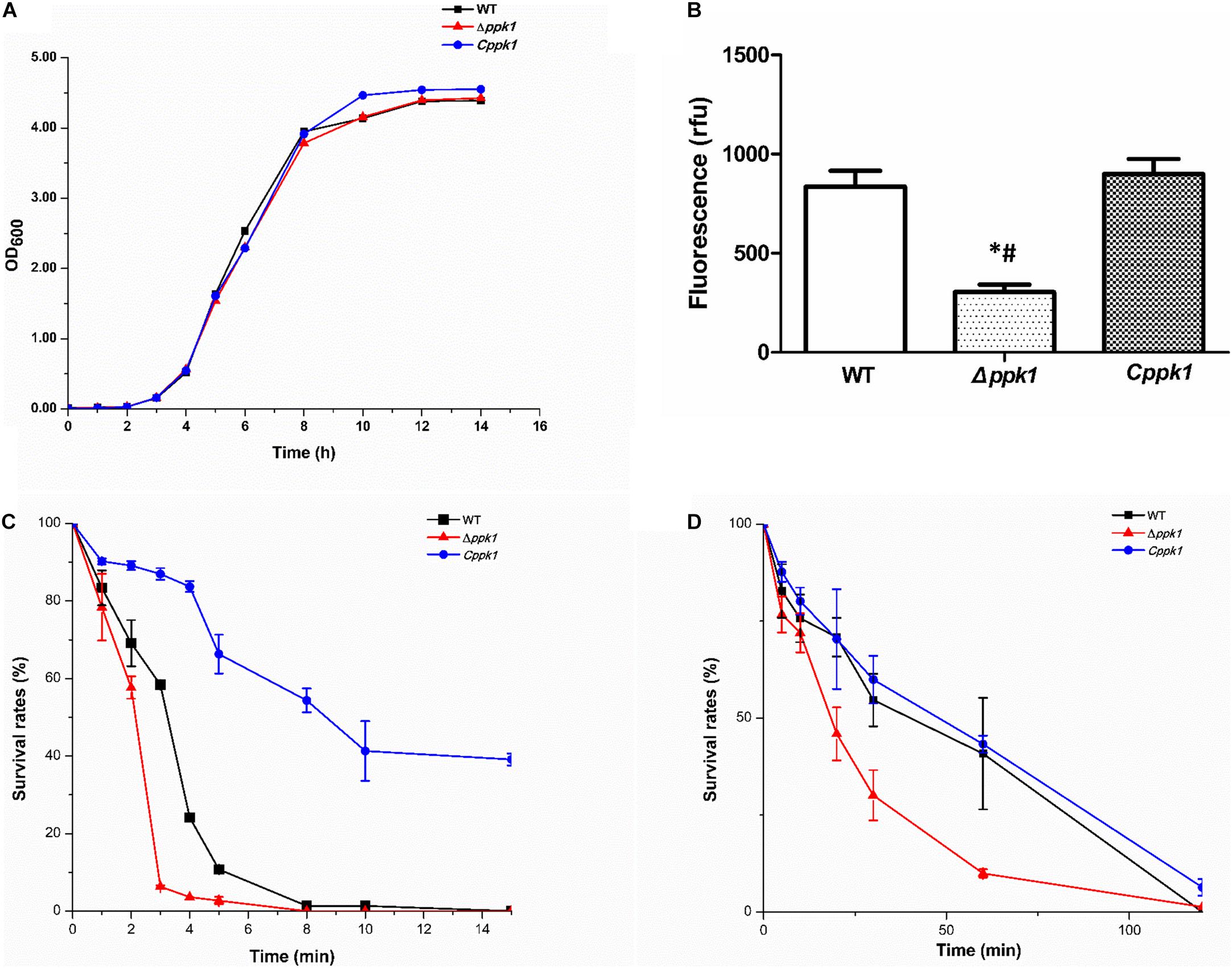
Figure 2. Comparison of the biological characteristics and stringent stress survival assays of the three EHEC O157:H7 strains. (A) Growth curve analysis of the WT, Δppk1, and Cppk1 strains. OD: Optical density. (B) The polyP levels of WT, Δppk1, and Cppk1 strains. Error bars indicate standard deviations. *, p < 0.05 as compared with the WT strain; #, p < 0.05 as compared with the Cppk1 strain. (C) The survival rates of WT, Δppk1, and Cppk1 strains after exposure to 55°C at different time points. (D) The survival rates of WT, Δppk1, and Cppk1 strains under acid stress at pH 2.0 in liquid LB at different time points.
Deletion of the ppk1 Gene Affected the Ability of Enterohemorrhagic Escherichia coli O157:H7 to Survive Under Stringent Environmental Conditions
When the Δppk1 strain was incubated at 55°C (EHEC O157:H7 can withstand this temperature) for 3 min, the survival rate was only 6.31% as compared with 58.39% for the WT strain. Cppk1 (86.96%) was more tolerant to heat than that of WT and Δppk1 strains (Figure 2C). When the bacterial strains were suspended and cultured in liquid LB at pH 2 (close to the acidic environment of the human stomach) for 60 min, similar results to the heat shock experiment were obtained. The survival rate of the Δppk1 was only 9.97% as compared with 40.84% of the WT and 43.27% of Cppk1 (Figure 2D). Both the heat shock and strong acid resistance results suggested that ppk1 played an important role in the stringent response in EHEC O157:H7.
Deletion of the ppk1 Gene Changed the Adhesion Capacity to Cells, F-Actin Rearrangement in Cells and Adhesion and Effacing Lesion Formation of Enterohemorrhagic Escherichia coli O157:H7
Scanning electron microscopy (SEM) results of the adhesion assay demonstrated that the three bacterial strains adhered to LoVo cells, but Δppk1 accumulated on and adhered to LoVo cells at a lower amount as compared to the WT and Cppk1 strains (Figure 3A) under each magnification. These results were verified through an adhesion quantitative assay (F = 24.680, P < 0.001). The adhesion rate of the Δppk1 was only 4.09‰ as compared with WT (26.02‰) and Cppk1 (33.88‰) strains (Figure 3B).
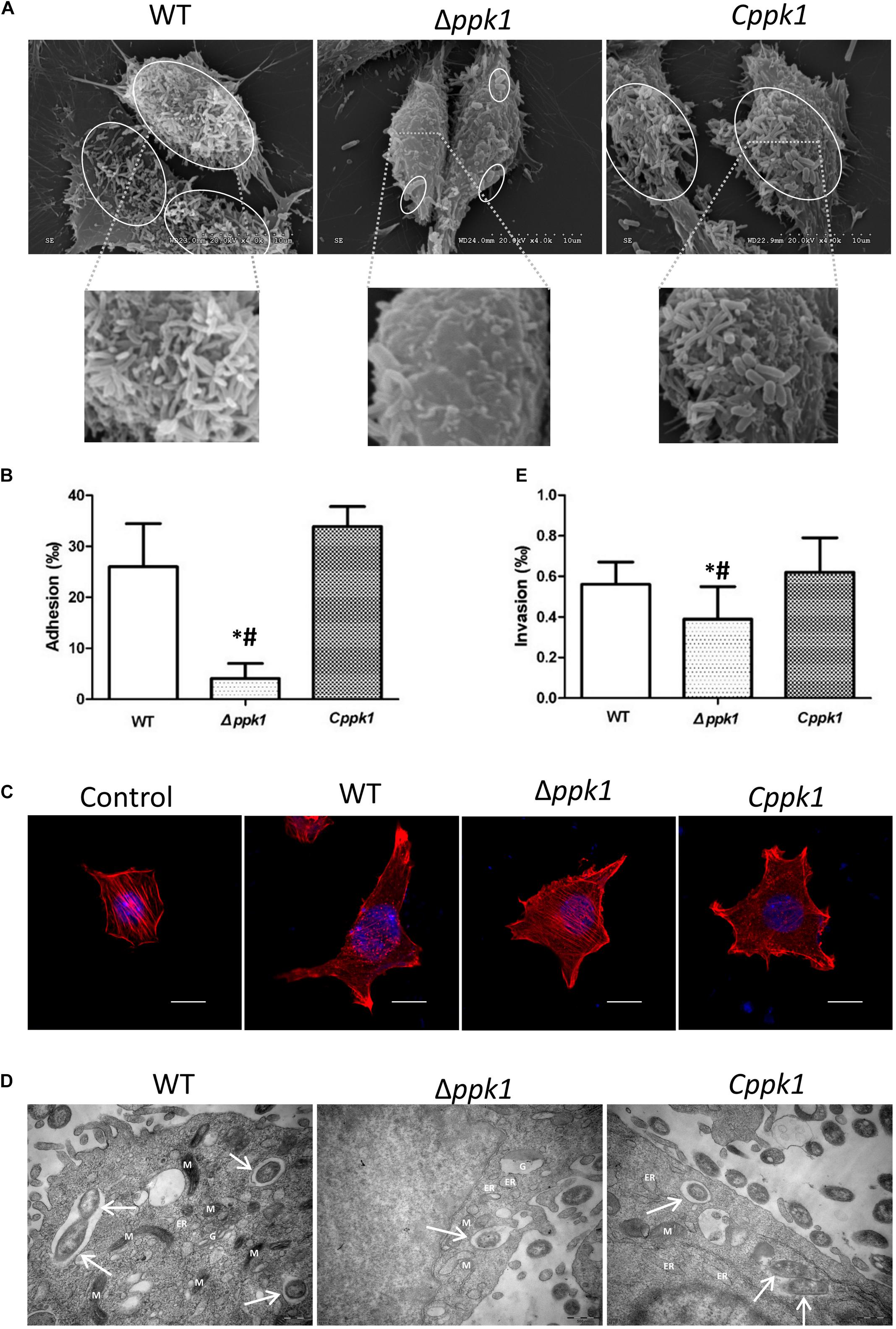
Figure 3. Deletion of the ppk1 gene reduced the adhesion, A/E lesions, and invasive ability. (A) High-resolution SEM images of LoVo cells infected with WT, Δppk1, or Cppk1 strains. Scale bar: 10 μm. (B) Comparison of adhesion rates between the WT, Δppk1, or Cppk1 strains. (C) LoVo cells were stained with rhodamine phalloidin (actin) and DAPI (nuclei) after infection. Scale bar: 10 μm. (D) High-resolution TEM images of LoVo cells infected with WT, Δppk1, or Cppk1 strains. White arrows point to the bacteria. ER, endoplasmic reticulum; M, mitochondria; G, Golgi apparatus. Scale bar: 500 nm. (E) Comparison of invasion rates between the WT, Δppk1, and Cppk1 strains. Error bars in both (B) and (E) indicate standard deviations. *, p < 0.05 as compared with the WT strain; #, p < 0.05 as compared with the Cppk1 strain. (E).
F-actin was stained with rhodamine phalloidin and observed by confocal microscopy. Results showed that the actin filaments of LoVo cells in the control group were typically present, distributed evenly, and arranged neatly in the cells (Figure 3C), whereas the F-actin in the WT and Cppk1 groups were significantly reduced and rearranged with significant aggregating at the two ends of cells. When the ppk1 gene was knocked out, the ability to induce actin rearrangement was significantly weaker than that of WT and Cppk1 strains. These results demonstrated that PPK1 plays an important role in EHEC O157:H7 in the development of A/E lesions.
Deletion of the ppk1 Gene Affected the Invasive Ability of Enterohemorrhagic Escherichia coli O157:H7
Transmission electron microscopy results demonstrated that the three strains all invaded LoVo cells and were wrapped in vacuoles (Figure 3D). However, the Δppk1 was less invasive as compared to the WT and Cppk1 strains. We thus performed gentamicin protection experiments to quantify the invasion rates. The WT reached an invasion rate of 0.56‰, which was not significantly different than the Cppk1 strain (0.62‰). However, the invasion rate of the Δppk1 was 0.39‰, which confirmed that the ppk1 gene was also required for bacteria invasion (F = 5.224, P = 0.014; Figure 3E).
The Δppk1 Strain Was Less Pathogenic in BALB/c Mice
Twenty BALB/c mice were divided into four groups and infected with EHEC O157:H7 WT, Δppk1, Cppk1, or LB (control). Mice infected with the WT strain for 2 days showed some symptoms of illness, such as listlessness, anorexia, slow movements, and diarrhea. These symptoms, including a disheveled coat, became more severe the days following the infection until euthanasia on day 7 post-infection. The symptoms of mice in the Cppk1 group were similar to the WT group, with one mouse dying 5 days post-infection and others exhibiting bloody diarrhea. Mice infected with the Δppk1, however, exhibited listlessness, rage, and slow movements without obvious diarrhea 3 days post-infection. All mice began to recover gradually in the following days. The uninfected control group treated only with LB were healthy.
After 7 days of infection, the distal 5-cm of the colon was collected, and the pathological features were compared among the groups (Figure 4A). The colon of the control group was normal with solidified stool (Figure 4A), while the WT group displayed edematous and congested intestinal colitis with very minimal solidified stool (Figure 4A). The colon of the mice infected with the Cppk1 strain exhibited typical intestinal colitis with severe edema and congestion, without any solidified feces (Figure 4A). However, the colon of the mice infected with the Δppk1 group appeared healthier with a small amount of solid stool, although some portions appeared slightly swollen (Figure 4A).
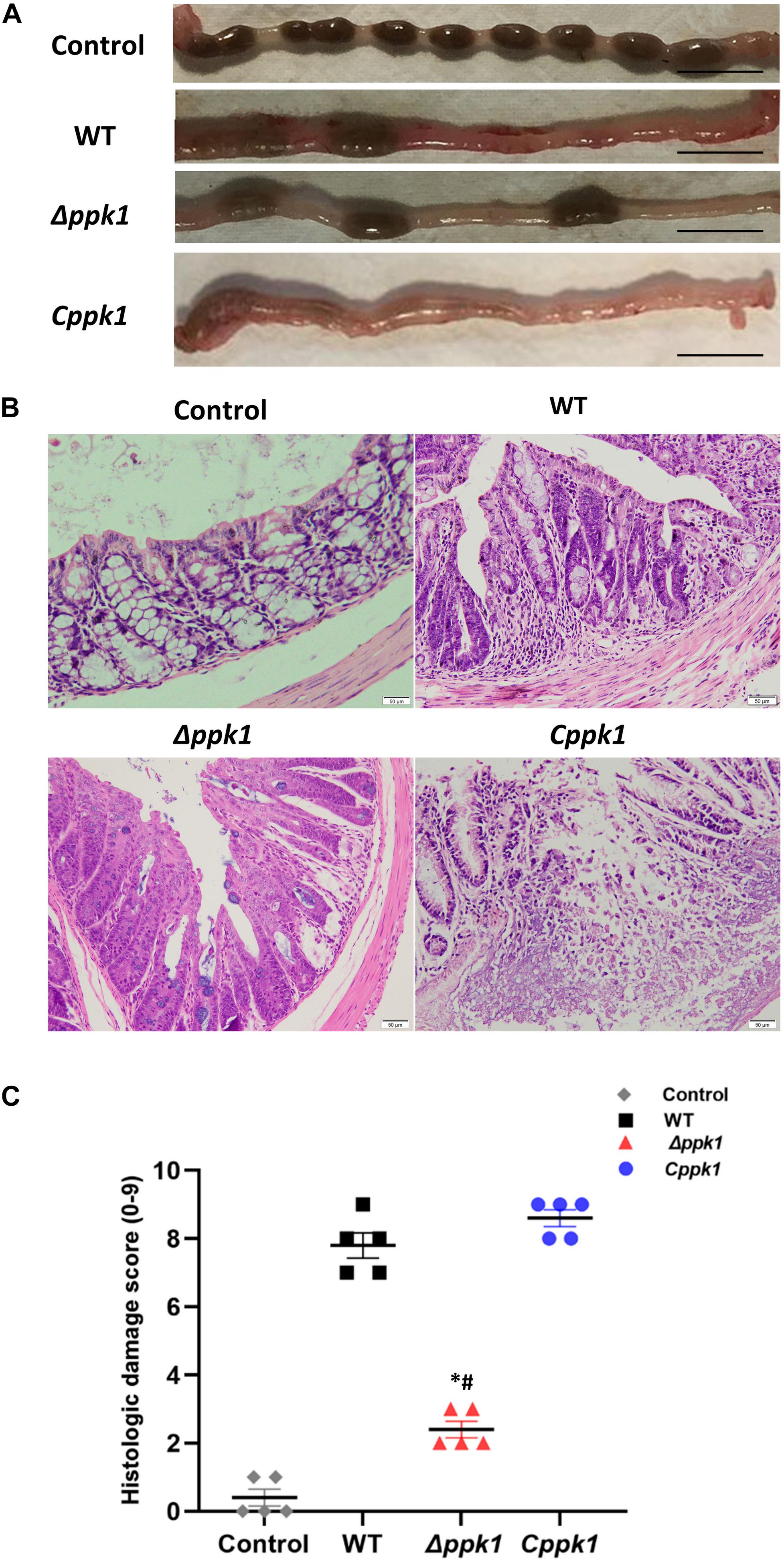
Figure 4. The Δppk1 strain was less pathogenic in BALB/c mice. (A) The colonic morphology of BALB/c mice after the infection. Scale bar: 1 cm. (B) Images of colons after infection. Scale bar: 50 μm. (C) Histologic damage scores of uninfected (control group), WT, Δppk1, and Cppk1 strains. *, p < 0.05 as compared with the WT strain; #, p < 0.05 as compared with the Cppk1 strain.
Colon paraffin sections were created and stained with H&E to observe the architecture of the intestinal epithelium (Figure 4B). In the control group, the colonic epithelium was complete and continuous, with regular, clear, and structural glands without inflammatory cell infiltration. Infection with WT of EHEC resulted in mucosal reactionary hyperplasia, disordered cell arrangement, decreased goblet cells, and inflammatory cell infiltration. The Cppk1 strain caused mucosal necrosis, and the muscle layer was involved and destructed. The colonic mucosa in mice infected with the Δppk1 strain appeared to have reactive hyperplasia, but the glands were regular with a clear structure. The histopathological scores were consistent with the morphological results observed by H&E staining (Figure 4C). The above results demonstrated that the Δppk1 was less pathogenic and induced milder A/E lesions than the WT and Cppk1 strains. The mild microvilli effacement in the Δppk1 indicated an important role of PPK1 in colonization of the intestinal epithelium.
ppk1 Was Required in Modulating the Expression of rpoS, eae, stx1, and stx2
To determine whether ppk1 regulated the expression of regulatory gene rpoS, adhesion gene eae, and virulence genes stx1 and stx2, real-time PCR was conducted. A 16S rRNA gene was used as the internal reference, and WT EHEC was used as the control. The relative level of mRNA in the Δppk1 and Cppk1 strains was also calculated (Figure 5). The relative copies of rpoS in the Δppk1 (0.36 ± 0.03) were less than that in the WT strain (1.00 ± 0.13), while there were no differences between Cppk1 (1.14 ± 0.05) and WT. These results were consistent with the amount of polyP among the three strains. Moreover, the ppk1 deletion significantly reduced the relative copy numbers of eae (0.32 ± 0.02), stx1A (0.60 ± 0.02), stx1B (0.46 ± 0.06), stx2A (0.42 ± 0.02), and stx2B (0.86 ± 0.01) as compared to that of WT (1.00 ± 0.02, 1.00 ± 0.08, 1.00 ± 0.04, 1.00 ± 0.06 and 1.00 ± 0.01, respectively). Similarly, these relative expression levels increased to 1.06 ± 0.03, 1.87 ± 0.04, 1.69 ± 0.09, 1.53 ± 0.01, and 1.36 ± 0.03, respectively, in the Cppk1 strain as compared with WT. These results indicated that ppk1 plays an important role in regulating the expression of rpoS, eae, stx1, and stx2.
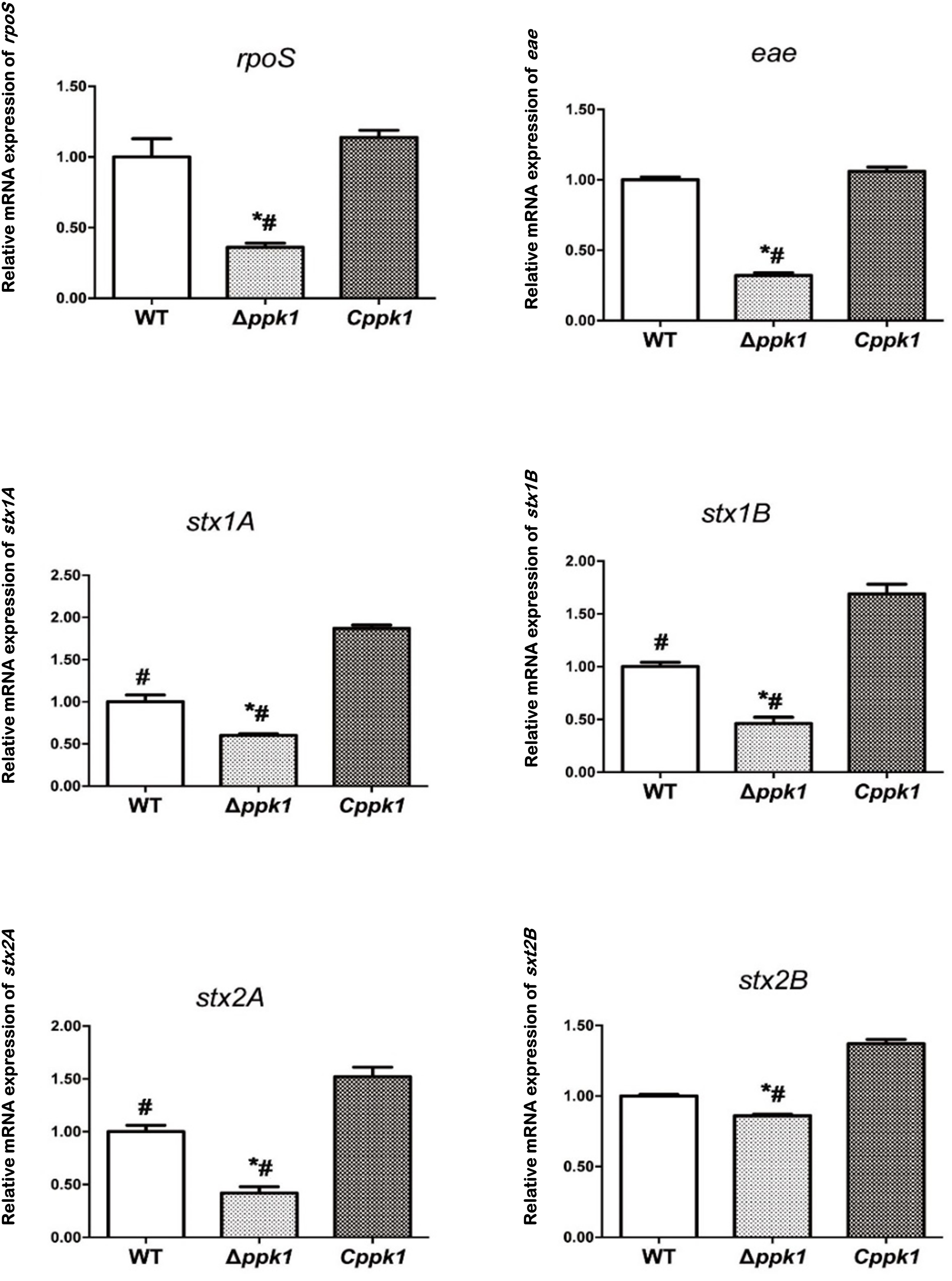
Figure 5. The ppk1 gene regulated the mRNA expression of rpoS, eae, stx1A, stx1B, stx2A, and stx2B. The relative mRNA expression of rpoS, eae, stx1A, stx1B, stx2A, and stx2B of WT, ppk1 and Cppk1 were measured via real-time PCR. Data were normalized to the relative RNA expression of the 16S rRNA. *, p < 0.05 as compared with the WT strain; #, p < 0.05 as compared with the Cppk1 strain.
Discussion
Polyphosphate is available in living cells and plays an important role in the survival, stress response, and pathogenicity of prokaryotic cells (Rao et al., 2009), which is regulated mainly through the regulation of rpoS, a key transcriptional factor. PPK1, encoded by the ppk1 gene, catalyzes the synthesis of polyP in E. coli. At present, the role of PPK1 in the pathogenesis of EHEC O157:H7 has not yet been reported. To determine the role of PPK1 in the pathogenesis of EHEC O157:H7, we successfully constructed a Δppk1 (Figure 1B) and a complemented strain Cppk1 (Figure 1C) to test the biological functions of PPK1 via in vitro and in vivo models.
Evaluation of the growth curve is an important but indirect method to compare the genomic expression of each strain, as the expression of many genes are affected by growth rate (Dong and Schellhorn, 2009). Our results showed that deletion of ppk1 had little effect on the growth rate or morphology of EHEC in nutritive medium (LB) (Figure 2A). The Δppk1 entered the exponential phase and stationary phase (SP) at the same time as the WT and Cppk1 strains. However, the ppk1 deletion mutant had reduced polyP levels as compared to the WT and Cppk1 strains (Figure 2B). These results confirmed that at the genetic and protein level, we successfully constructed the Δppk1 and Cppk1 strains.
According to previous studies, SP is a critical period for bacteria to resist external stress and invade the host during which the function of the bacteria change significantly (Siegele and Kolter, 1992). For E. coli, SP is an important stage for the expression of pathogenic invasion-associated virulence genes (Wang and Kim, 2000; Peng et al., 2012). We next verified the role of PPK1 in the survival and pathogenesis of EHEC O157:H7 during SP. EHEC O157:H7 is a fecal-mouth transmitted intestinal pathogen that enters the human stomach from the external environment, passes through the intestine, and finally colonizes the colon to cause diarrhea and hemorrhagic colitis (HC), even hemolytic uremic syndrome (HUS). The premise that bacteria, such as EHEC O157:H7, causes disease was based on the thought that they survive all types of environmental stress, such as high environmental temperatures and low pH values found in the human stomach (Rao and Kornberg, 1996; Ault-Riche et al., 1998; Rao et al., 1998; Kim et al., 2002). The ability to quickly adapt to changing environments is critical for bacteria to successfully transmit and infect hosts (Dong and Schellhorn, 2009). Studies have shown that polyP is involved in the regulation of many microbial stress responses (Magnusson et al., 2005; Rao et al., 2009; Dalebroux and Swanson, 2012; Varas et al., 2017). To determine the effect of ppk1 deletion on the bacterial stress response of EHEC O157:H7, WT, Δppk1, and Cppk1 strains were exposed to 55°C and pH 2.0 during SP. The survival rates were compared at corresponding time points. Our results showed that the survival rates of the Δppk1 strain were significantly lower than that of WT and Cppk1 (Figures 2C,D), which indicated that PPK1 played a key role in stress response during the infection process of EHEC O157:H7.
In certain external environmental stresses, some genes of E. coli are induced during SP (Lange and Hengge-Aronis, 1991; Siegele and Kolter, 1992; Hengge-Aronis, 1993), such as adAXW, gadB, and yhbO, but are not induced during the exponential growth phase. A previous study reported that polyP was associated with the expression of RpoS, which plays a central role in SP adaptations (Abramov et al., 2007; Hoac et al., 2013). RpoS is a conserved stress regulator of virulence and pressure adaptation during SP of E. coli, Salmonella typhimurium, Shigella, Yersinia colitis, Vibrio cholerae, and Borrelia burgdorferi (Tzeng and Kornberg, 1998; Hengge-Aronis, 2002; Dong and Schellhorn, 2009). Our results show that loss of ppk1 decreased the ability of E. coli to endure the stress conditions. Furthermore, real-time PCR showed decreased levels of rpoS in the Δppk1 as compared to the WT and Cppk1 strains (Figure 5A). This suggested that RpoS might be the stress regulator of EHEC O157:H7 and its expression was induced by polyP.
When E. coli is under environmental stress, RpoS initiates a complex regulatory network, inducing and regulating the expression of nearly 600 genes (Dong and Schellhorn, 2009). The expression of RpoS-regulated genes was shown to differ according to the environment. For example, the genes gadAXW, gadB, and gadE were found important for acid resistance (Ma et al., 2003), and yhbO for heat and UV resistance (Weber et al., 2005; Abdallah et al., 2007). The mechanism underlying the influence of polyP on RpoS in EHEC O157:H7 is a current focus of our research. The (p) ppGpp pathway is known to regulate other important cellular and regulatory processes that allow bacteria to adapt to a wide range of environmental conditions, including nutritional or stressful conditions (Magnusson et al., 2005; Dalebroux and Swanson, 2012). Proteomic studies conducted with E. coli K12 strains have identified 151 significantly differentially expressed proteins, among which RelA, SpoT, and ArcA protein levels, which influence the metabolic pathways of (p)ppGpp, were significantly decreased in the Δppk1 (Varas et al., 2017). Therefore, the relationships among PPK1/polyP, RpoS and (p)ppGpp are our interest.
PPK1 was shown to be essential not only for the synthesis of polyP and metabolism of ATP, but also for adhesion/invasion and virulence factor expression in bacterial pathogens, such as E. coli K1 (RS218) (Peng et al., 2012), Campylobacter jejuni (Candon et al., 2007), Burkholderia pseudomallei (Srisanga et al., 2019), and Mycobacterium tuberculosis (Tiwari et al., 2019). The pathogenesis of EHEC O157:H7 is mainly reflected in the adhesion and effacing lesions (A/E lesions) as well as the secretion of Shiga-like toxin (Stx) (Nguyen and Sperandio, 2012).
Adhesion and effacing lesions were characterized by EHEC O157:H7 adhesion to host epithelium and then induction of extensive F-actin cytoskeletal rearrangements within cells, formation of pedestal-like structures underneath the bacteria and loss of microvillus structure (Caprioli et al., 2005). Successful adhesion and colonization of EHEC O157:H7 in the large intestine is dependent upon the type TTSS (Pacheco et al., 2018; Liu et al., 2020) by which Tir was the first translocated effector protein to the epithelial cells. Tir is inserted into the membrane of cells, where it then binds to the bacterial adhesion receptor intimin (encoded by eae) to mediate the adhesion between bacteria and host cells (Yi et al., 2010). Interactions between intimin and Tir were also required for recruitment and rearrangement of actin and other cytoskeletal proteins underneath adherent bacteria, which results in characteristic actin-rich “pedestals” through triggering host signal events (Ritchie et al., 2003; Golan et al., 2011).
To gain insight into the effect of ppk1 deficiency in EHEC O157:H7 on the interaction with host cells, we elucidated whether the Δppk1 mutant possessed a defect in adhesion and A/E lesion formation into host cells. Our results showed that the deletion of the ppk1 gene reduced the adhesion as compared with WT (4.09‰ vs. 26.02‰, respectively) (Figures 3A,B). Moreover, the F-actin filaments of cells infected with the WT or Cppk1 strains were significantly reduced and aggregated to the edge, showing a typical rearrangement phenomenon, whereas the Δppk1 strain was weaker in actin filament rearrangement (Figure 3C). Accordingly, real-time PCR was carried out to test expression levels of the intimin gene eae in the three strains. The ppk1 deletion significantly reduced the relative copy number of eae (0.32), as compared with WT (1.00) and Cppk1 (1.06) (Figure 5), which was consistent with these results at the cellular level. In animal models, deletions of eae (the intimin locus) and mutations that render the TTSS inactive markedly reduced the pathogen’s capacity to colonize the intestine (Ritchie et al., 2003), which was similar to our results above.
When colonizing the human intestine, EHEC O157:H7 forms A/E lesions on the colonic epithelial cells. The genes required for A/E lesions were encoded within the chromosomal pathogenicity island known as the locus for enterocyte effacement (LEE) (Elliott et al., 1998; Muller et al., 2009; Nguyen and Sperandio, 2012). The secreted effector proteins by the type III secretion system within LEE were directly injected into the cells through a “molecular injector”. One of the important secreted proteins injected into the host was the translocated intimin receptor (Tir) (Nguyen and Sperandio, 2012). Once released into the host cytoplasm, Tir localizes to the host cytoplasmic membrane and binds to the LEE-encoded surface protein intimin (encoded by eae) to intimately attach the bacteria to the cell (Kenny et al., 1997; Deibel et al., 1998; Nguyen and Sperandio, 2012). Together, with other effector proteins secreted into the cell, F-actin was aggregated, leading to rearrangement of the cytoskeleton of the colonic epithelial cell (Campellone et al., 2004b; Weiss et al., 2009). Actin rearrangement was followed by the appearance of characteristic pathological manifestation of A/E lesions, including intestinal epithelial cytoskeletal (F-actin) rearrangement, destruction of tight junctions, intestinal microvilli actin dimerization, microvilli loss (Rao and Kornberg, 1996), and a pedestal-like structure formation, which cup the individual bacteria (Nataro and Kaper, 1998; Nguyen and Sperandio, 2012). Our results indicated that ppk1 deletion mutant downregulated the eae gene and then reduced the expression of intimin. As a result, Tir could not bind to sufficient intimin, leading to the decrease in adhesion of Δppk1 to LoVo cells. Adhesion is an important step of A/E lesions, which directly affects the rearrangement of F-actin and formation of the pedestal. Therefore, the rearrangement of F-actin in Δppk1 was not obvious, resulting in weak A/E lesions. Thus, PPK1 played an important role in the adhesion and formation of A/E lesions which was verified in other bacteria (Candon et al., 2007; Peng et al., 2012; Tiwari et al., 2019).
Aside from A/E lesions, another main pathogenic factor of EHEC O157:H7 causing severe diarrhea and HUS is the secretion of Shiga-like toxins (Stx1 and Stx2) (Leotta et al., 2008). The Stx molecule consists of one A subunit and five B subunits through covalent bonding via an AB5 structure, and its toxic effect on host cells has already been widely recognized (Laing et al., 2012; Page and Liles, 2013; Perera et al., 2015). Briefly, after adhesion and colonization in the colon, STEC, such as EHEC O157:H7, induced the delivery of Stx, which passes through the intestinal mucosa. Then toxins entered the bloodstream and traveled to target organs such as kidneys with toxin receptor Gb3 on the cell surface. Once bounded to Gb3, Stxs were internalized and delivered from early endosome to the trans-Golgi network, through the Golgi apparatus, to the endoplasmic reticulum (ER), leading to inhibition of protein synthesis, ER stress, ribotoxic stress, pro-inflammatory responses, and autophagy in host cells (Leyva-Illades et al., 2012; Lee et al., 2016, 2021). Therefore, Stx causes severe inflammation and extensive histological damage, resulting in bloody dysentery or bloody diarrhea and acute renal failure. In addition, Stx also causes programmed cell death or apoptosis (Jones et al., 2000; Suzuki et al., 2000).
Moreover, Stx maybe a virulence factor to affect intestinal tissue damage alone (Lee et al., 2021). Gigliucci et al. (2018) detected stx2a (encoding Stx2a) and eae (encoding intimin) in the feces of STEC-infected patients, which suggested that Stx not only works by migrating to the lamina propria but can also act in the intestinal lumen. Robinson et al. (2006) found that Stx2 was involved in increasing the colonization capacity of EHEC by increasing the expression of nucleolin in HEp-2 cells, which implied that Stxs affected the EHEC colonization of the intestine (Lee et al., 2021).
To determine the influence of the deletion of the ppk1 gene on the Stx virulence of EHEC O157:H7, real-time reverse-transcription PCR was performed. Our results showed that the ppk1 deletion significantly reduced the relative copy number of stx1A, stx1B, stx2A, and stx2B as compared to the WT and Cppk1 strains (Figure 5). This suggested that ppk1 affected the virulence of EHEC O157:H7 at a molecular level. Moreover, to identify the effect of ppk1 deletion on the bacterial invasion of cells, an invasion assay was performed via TEM and gentamicin protection experiments. Our results demonstrated that the invasion rate of Δppk1 was reduced to 0.39‰ as compared with WT (0.56‰) and Cppk1 (0.62‰) (Figures 3B,D).
Enterohemorrhagic Escherichia coli invasions is sometimes associated with recurrent infection and persistent diarrhea. It attained some degree of coexistence with the infected cell and survived in the intracellular milieu for a long time. That strategy seemed appropriate to assure the persistence of the micro-organism in the animal reservoir, as well as to sustain its continuous shedding to the environment (Cordeiro et al., 2013). On the other hand, host cell cytoskeletal rearrangement and phosphorylation-mediated signal transduction are also involved in cell invasion by pathogens, which leads to the intestinal injury (Cossart and Sansonetti, 2004). Based on the adhesion, A/E lesions and invasion results, we concluded that the Δppk1 was less adherent and invasive in LoVo cells. We further confirmed the effect of ppk1 deletion on intestinal injury through animal experiments.
Three commonly used species (pigs, rabbits, and mice) as animal models have been developed to facilitate study of EHEC pathogenesis in vivo. The piglet intestine is permissive for EHEC replication and shows typical signs of central nervous system (CNS) as human beings (Ritchie, 2014). But their breeding and maintenance require considerable veterinary skill, space, and financial support (Mohawk and O’Brien, 2011). Rabbits have been used both to study the toxic effects of Stx and the intestinal biology of the organism. Despite being sensitive to the intestinal manifestations of EHEC infection, suckling rabbits do not develop HUS or any other evidence of renal failure. Mouse is another small animal model systems are preferable for general use. Mice are naturally resistant to colonization by EHEC and fail to develop signs of intestinal disease following oral infection. Thus, we tried to use mitomycin C (MMC) (an alkylating agent regarded as an important chemotherapeutic drug and is widely used in the treatment of several malignant human tumors) as an agent to induce toxin expression and form animal model of EHEC O157:H7 infection (Fujii et al., 1994; Shimizu et al., 2003).
Our experimental results showed that the diarrhea symptoms of WT and Cppk1 mice were obvious, but the diarrhea symptoms of Δppk1 were not, which could be seen on fecal formation (Figure 4A). Moreover, colonic HE staining and histologic damage scores showed that Δppk1 was less destructive to the colon, which was also an evidence of mild symptoms of enteritis, as shown in Figure 4B.
All these results indicated that PPK1 was a key factor in the pathogenesis of EHEC O157:H7. The ppk1-deficient strain exhibited defects in adhesion and invasion in human colonic epithelial cells, which was consistent with the results of others (Dong and Schellhorn, 2009; Peng et al., 2012; Srisanga et al., 2019).
In summary, we demonstrated that ppk1 was involved in the pathogenesis of EHEC O157:H7. PPK1 improved the stress response by regulating the expression of the stable phase regulatory gene rpoS, promoted A/E lesions, and increased the expression of virulence genes stx1 and stx2, which enhanced the pathogenicity of the bacteria. However, further research regarding the mechanism of PPK1-mediated regulation will improve our understanding of the pathogenesis and aid in the prevention of EHEC O157:H7 infection. PPK1 is not present in humans, and as such, the unique ATP-binding pocket in its structure is an important target for the development of PPK1 inhibitors, especially for new antibiotics. This study identified PPK1 as a potential target for the development of novel treatments for EHEC O157:H7 infection.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Southern Medical University Experimental Animal Ethics Committee.
Author Contributions
YD and XW did the conception and design of the work and drafted the article. YD, YH and KY collected the data. ZH, BZ, and WZ did the data analysis and interpretation. CW did the critical revision of the article. WZ and CW did the final approval of the version to be published. All authors contributed to the article and approved the submitted version.
Funding
This work were supported by the National Natural Science Foundation of China (No. 81902022), the Natural Science Foundation of Guangdong Province (Nos: 2018B030311063 and 2014A030313312), and the Shenzhen Polytechnic Fund (6021310006K).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Ruifu Yang (Institute of Microbiology and Epidemiology, Beijing, China) for providing pKD46 and pKD4. We also thank LetPub (www.letpub.com) for linguistic assistance and pre-submission expert review.
References
Abdallah, J., Caldas, T., Kthiri, F., Kern, R., and Richarme, G. (2007). YhbO protects cells against multiple stresses. J. Bacteriol. 189, 9140–9144. doi: 10.1128/JB.01208-07
Abramov, A. Y., Fraley, C., Diao, C. T., Winkfein, R., Colicos, M. A., Duchen, M. R., et al. (2007). Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. U.S.A. 104, 18091–18096. doi: 10.1073/pnas.0708959104
Aschar-Sobbi, R., Abramov, A. Y., Diao, C., Kargacin, M. E., Kargacin, G. J., French, R. J., et al. (2008). High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866. doi: 10.1007/s10895-008-0315-4
Ault-Riche, D., Fraley, C. D., Tzeng, C. M., and Kornberg, A. (1998). Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180, 1841–1847. doi: 10.1128/JB.180.7.1841-1847.1998
Campellone, K. G., Rankin, S., Pawson, T., Kirschner, M. W., Tipper, D. J., and Leong, J. M. (2004a). Clustering of Nck by a 12-residue Tir phosphopeptide is sufficient to trigger localized actin assembly. J. Cell Biol. 164, 407–416. doi: 10.1083/jcb.200306032
Campellone, K. G., Robbins, D., and Leong, J. M. (2004b). EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7, 217–228. doi: 10.1016/j.devcel.2004.07.004
Candon, H. L., Allan, B. J., Fraley, C. D., and Gaynor, E. C. (2007). Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189, 8099–8108. doi: 10.1128/JB.01037-07
Caprioli, A., Morabito, S., Brugere, H., and Oswald, E. (2005). Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36, 289–311. doi: 10.1051/vetres:2005002
Chi, F., Wang, Y., Gallaher, T. K., Wu, C. H., Jong, A., and Huang, S. H. (2009). Identification of IbeR as a stationary-phase regulator in meningitic Escherichia coli K1 that carries a loss-of-function mutation in rpoS. J. Biomed. Biotechnol. 2009:520283. doi: 10.1155/2009/520283
Coico, R. (2005). Gram staining. Curr. Protoc. Microbiol. Appendix 3:Aendix3C. doi: 10.1002/9780471729259.mca03cs00
Cordeiro, F., da Silva, R. I. K., Vargas-Stampe, T. L. Z., Cerqueira, A. M. F., and Andrade, J. R. C. (2013). Cell invasion and survival of Shiga toxin-producing Escherichia coli within cultured human intestinal epithelial cells. Microbiology (Reading) 159(Pt 8), 1683–1694. doi: 10.1099/mic.0.064204-0
Cossart, P., and Sansonetti, P. J. (2004). Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248. doi: 10.1126/science.1090124
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Davis, T. K., Van De Kar, N., and Tarr, P. I. (2014). Shiga toxin/verocytotoxin-producing Escherichia coli Infections: practical clinical perspectives. Microbiol. Spectr. 2:EHEC–0025–2014. doi: 10.1128/microbiolspec.EHEC-0025-2014
Deibel, C., Kramer, S., Chakraborty, T., and Ebel, F. (1998). EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol. Microbiol. 28, 463–474. doi: 10.1046/j.1365-2958.1998.00798.x
Diaz, J. M., and Ingall, E. D. (2010). Fluorometric quantification of natural inorganic polyphosphate. Environ. Sci. Technol. 44, 4665–4671. doi: 10.1021/es100191h
Dong, T., and Schellhorn, H. E. (2009). Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10:349. doi: 10.1186/1471-2164-10-349
Elliott, S. J., Wainwright, L. A., McDaniel, T. K., Jarvis, K. G., Deng, Y. K., Lai, L. C., et al. (1998). The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28, 1–4. doi: 10.1046/j.1365-2958.1998.00783.x
Elsinghorst, E. A., and Kopecko, D. J. (1992). Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 60, 2409–2417. doi: 10.1128/iai.60.6.2409-2417.1992
Fujii, J., Kita, T., Yoshida, S., Takeda, T., Kobayashi, H., Tanaka, N., et al. (1994). Direct evidence of neuron impairment by oral infection with verotoxin-producing Escherichia coli O157:H- in mitomycin-treated mice. Infect. Immun. 62, 3447–3453. doi: 10.1128/iai.62.8.3447-3453.1994
Gigliucci, F., von Meijenfeldt, F. A. B., Knijn, A., Michelacci, V., Scavia, G., Minelli, F., et al. (2018). Metagenomic characterization of the human intestinal microbiota in fecal samples from STEC-infected patients. Front. Cell Infect. Microbiol. 8:25. doi: 10.3389/fcimb.2018.00025
Golan, L., Gonen, E., Yagel, S., Rosenshine, I., and Shpigel, N. Y. (2011). Enterohemorrhagic Escherichia coli induce attaching and effacing lesions and hemorrhagic colitis in human and bovine intestinal xenograft models. Dis. Model. Mech. 4, 86–94. doi: 10.1242/dmm.005777
Gomes, F. M., Ramos, I. B., Wendt, C., Girard-Dias, W., De Souza, W., Machado, E. A., et al. (2013). New insights into the in situ microscopic visualization and quantification of inorganic polyphosphate stores by 4’,6-diamidino-2-phenylindole (DAPI)-staining. Eur. J. Histochem. 57:e34. doi: 10.4081/ejh.2013.e34
Han, R., Xu, L., Wang, T., Liu, B., and Wang, L. (2017). A small regulatory RNA contributes to the preferential colonization of Escherichia coli O157:H7 in the large intestine in response to a low DNA concentration. Front. Microbiol. 8:274. doi: 10.3389/fmicb.2017.00274
Hengge-Aronis, R. (1993). Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72, 165–168. doi: 10.1016/0092-8674(93)90655-a
Hengge-Aronis, R. (2002). Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/MMBR.66.3.373-395.2002
Hoac, B., Kiffer-Moreira, T., Millan, J. L., and McKee, M. D. (2013). Polyphosphates inhibit extracellular matrix mineralization in MC3T3-E1 osteoblast cultures. Bone 53, 478–486. doi: 10.1016/j.bone.2013.01.020
Hua, Y., Wu, J., Fu, M., Liu, J., Li, X., Zhang, B., et al. (2020). Enterohemorrhagic Escherichia coli effector protein EspF interacts with host protein ANXA6 and triggers myosin light chain kinase (MLCK)-dependent tight junction dysregulation. Front. Cell Dev. Biol. 8:613061. doi: 10.3389/fcell.2020.613061
Hua, Y., Yan, K., and Wan, C. (2018). Clever cooperation: interactions between EspF and Host proteins. Front. Microbiol. 9:2831. doi: 10.3389/fmicb.2018.02831
Jeong, J., Cho, N., Jung, D., and Bang, D. (2013). Genome-scale genetic engineering in Escherichia coli. Biotechnol. Adv. 31:804–810. doi: 10.1016/j.biotechadv.2013.04.003
Jones, N. L., Islur, A., Haq, R., Mascarenhas, M., Karmali, M. A., Perdue, M. H., et al. (2000). Escherichia coli Shiga toxins induce apoptosis in epithelial cells that is regulated by the Bcl-2 family. Am. J. Physiol. Gastrointest. Liver Physiol. 278, G811–G819. doi: 10.1152/ajpgi.2000.278.5.G811
Kenny, B., DeVinney, R., Stein, M., Reinscheid, D. J., Frey, E. A., and Finlay, B. B. (1997). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520. doi: 10.1016/s0092-8674(00)80437-7
Kim, K. S., Rao, N. N., Fraley, C. D., and Kornberg, A. (2002). Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U.S.A. 99, 7675–7680. doi: 10.1073/pnas.112210499
Kulakova, A. N., Hobbs, D., Smithen, M., Pavlov, E., Gilbert, J. A., Quinn, J. P., et al. (2011). Direct quantification of inorganic polyphosphate in microbial cells using 4’-6-diamidino-2-phenylindole (DAPI). Environ. Sci. Technol. 45, 7799–7803. doi: 10.1021/es201123r
Laing, C. R., Zhang, Y., Gilmour, M. W., Allen, V., Johnson, R., Thomas, J. E., et al. (2012). A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS One 7:e37362. doi: 10.1371/journal.pone.0037362
Lange, R., and Hengge-Aronis, R. (1991). Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 173, 4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991
Ledwaba, S. E., Costa, D. V. S., Bolick, D. T., Giallourou, N., Medeiros, P., Swann, J. R., et al. (2020). Enteropathogenic Escherichia coli infection induces diarrhea, intestinal damage, metabolic alterations, and increased intestinal permeability in a murine model. Front. Cell Infect. Microbiol. 10:595266. doi: 10.3389/fcimb.2020.595266
Lee, K. S., Jeong, Y. J., and Lee, M. S. (2021). Escherichia coli Shiga Toxins and gut microbiota interactions. Toxins (Basel) 13:416. doi: 10.3390/toxins13060416
Lee, M. S., Koo, S., Jeong, D. G., and Tesh, V. L. (2016). Shiga Toxins as multi-functional proteins: induction of host cellular stress responses, role in pathogenesis and therapeutic applications. Toxins (Basel) 8:77. doi: 10.3390/toxins8030077
Leotta, G. A., Miliwebsky, E. S., Chinen, I., Espinosa, E. M., Azzopardi, K., Tennant, S. M., et al. (2008). Characterisation of Shiga toxin-producing Escherichia coli O157 strains isolated from humans in Argentina, Australia and New Zealand. BMC Microbiol. 8:46. doi: 10.1186/1471-2180-8-46
Leyva-Illades, D., Cherla, R. P., Lee, M. S., and Tesh, V. L. (2012). Regulation of cytokine and chemokine expression by the ribotoxic stress response elicited by Shiga toxin type 1 in human macrophage-like THP-1 cells. Infect. Immun. 80, 2109–2120. doi: 10.1128/IAI.06025-11
Liu, Y., Han, R., Wang, J., Yang, P., Wang, F., and Yang, B. (2020). Magnesium sensing regulates Intestinal colonization of enterohemorrhagic Escherichia coli O157:H7. mBio 11, 1–17. doi: 10.1128/mBio.02470-20
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Z., Gong, S., Richard, H., Tucker, D. L., Conway, T., and Foster, J. W. (2003). GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49, 1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x
Magnusson, L. U., Farewell, A., and Nystrom, T. (2005). ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 13, 236–242. doi: 10.1016/j.tim.2005.03.008
Melton-Celsa, A. R. (2014). Shiga Toxin (Stx) classification, structure, and function. Microbiol. Spectr. 2:EHEC–0024–2013. doi: 10.1128/microbiolspec.EHEC-0024-2013
Menge, C. (2020). Molecular biology of Escherichia Coli Shiga Toxins’ effects on mammalian cells. Toxins (Basel) 12:345. doi: 10.3390/toxins12050345
Mohawk, K. L., and O’Brien, A. D. (2011). Mouse models of Escherichia coli O157:H7 infection and shiga toxin injection. J. Biomed. Biotechnol. 2011:258185. doi: 10.1155/2011/258185
Muller, D., Benz, I., Liebchen, A., Gallitz, I., Karch, H., and Schmidt, M. A. (2009). Comparative analysis of the locus of enterocyte effacement and its flanking regions. Infect. Immun. 77, 3501–3513. doi: 10.1128/IAI.00090-09
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11, 142–201. doi: 10.1128/CMR.11.1.142
Nguyen, Y., and Sperandio, V. (2012). Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell Infect. Microbiol. 2:90. doi: 10.3389/fcimb.2012.00090
Pacheco, A. R., Lazarus, J. E., Sit, B., Schmieder, S., Lencer, W. I., Blondel, C. J., et al. (2018). CRISPR screen reveals that EHEC’s T3SS and Shiga toxin rely on shared host factors for infection. mBio 9:e01003–18. doi: 10.1128/mBio.01003-18
Page, A. V., and Liles, W. C. (2013). Enterohemorrhagic Escherichia coli infections and the hemolytic-uremic syndrome. Med. Clin. North Am. 97, 681–695. doi: 10.1016/j.mcna.2013.04.001
Peng, L., Luo, W. Y., Zhao, T., Wan, C. S., Jiang, Y., Chi, F., et al. (2012). Polyphosphate kinase 1 is required for the pathogenesis process of meningitic Escherichia coli K1 (RS218). Future Microbiol. 7, 411–423. doi: 10.2217/fmb.12.3
Perera, A., Clarke, C. M., Dykes, G. A., and Fegan, N. (2015). Characterization of Shiga Toxigenic Escherichia coli O157 and Non-O157 isolates from ruminant feces in Malaysia. Biomed. Res. Int. 2015:382403. doi: 10.1155/2015/382403
Perna, N. T., Plunkett, G. III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533. doi: 10.1038/35054089
Rangel, J. M., Sparling, P. H., Crowe, C., Griffin, P. M., and Swerdlow, D. L. (2005). Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11, 603–609. doi: 10.3201/eid1104.040739
Rao, N. N., and Kornberg, A. (1996). Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178, 1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996
Rao, N. N., Gomez-Garcia, M. R., and Kornberg, A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647.
Rao, N. N., Liu, S., and Kornberg, A. (1998). Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180, 2186–2193. doi: 10.1128/JB.180.8.2186-2193.1998
Riley, L. W., Remis, R. S., Helgerson, S. D., McGee, H. B., Wells, J. G., Davis, B. R., et al. (1983). Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308, 681–685. doi: 10.1056/NEJM198303243081203
Ritchie, J. M. (2014). Animal models of enterohemorrhagic Escherichia coli infection. Microbiol. Spectr. 2:EHEC–0022–2013. doi: 10.1128/microbiolspec.EHEC-0022-2013
Ritchie, J. M., Thorpe, C. M., Rogers, A. B., and Waldor, M. K. (2003). Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 71, 7129–7139. doi: 10.1128/IAI.71.12.7129-7139.2003
Robinson, C. M., Sinclair, J. F., Smith, M. J., and O’Brien, A. D. (2006). Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. U.S.A. 103, 9667–9672. doi: 10.1073/pnas.0602359103
Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y., Kameda, A., Tanaka, K., et al. (1997). Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. U.S.A. 94, 11210–11215. doi: 10.1073/pnas.94.21.11210
Shimizu, K., Asahara, T., Nomoto, K., Tanaka, R., Hamabata, T., Ozawa, A., et al. (2003). Development of a lethal Shiga toxin-producing Escherichia coli-infection mouse model using multiple mitomycin C treatment. Microb. Pathog. 35, 1–9. doi: 10.1016/s0882-4010(03)00065-2
Siegele, D. A., and Kolter, R. (1992). Life after log. J. Bacteriol. 174, 345–348. doi: 10.1128/jb.174.2.345-348.1992
Srisanga, K., Suthapot, P., Permsirivisarn, P., Govitrapong, P., Tungpradabkul, S., and Wongtrakoongate, P. (2019). Polyphosphate kinase 1 of Burkholderia pseudomallei controls quorum sensing, RpoS and host cell invasion. J. Proteomics 194, 14–24. doi: 10.1016/j.jprot.2018.12.024
Stevens, M. P., and Frankel, G. M. (2014). The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol. Spectr. 2:EHEC–0007–2013. doi: 10.1128/microbiolspec.EHEC-0007-2013
Suzuki, A., Doi, H., Matsuzawa, F., Aikawa, S., Takiguchi, K., Kawano, H., et al. (2000). Bcl-2 antiapoptotic protein mediates verotoxin II-induced cell death: possible association between bcl-2 and tissue failure by E. coli O157:H7. Genes Dev. 14, 1734–1740.
Tiwari, P., Gosain, T. P., Singh, M., Sankhe, G. D., Arora, G., Kidwai, S., et al. (2019). Inorganic polyphosphate accumulation suppresses the dormancy response and virulence in Mycobacterium tuberculosis. J. Biol. Chem. 294, 10819–10832. doi: 10.1074/jbc.RA119.008370
Tzeng, C. M., and Kornberg, A. (1998). Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol. Microbiol. 29, 381–382. doi: 10.1046/j.1365-2958.1998.00887.x
Varas, M., Valdivieso, C., Mauriaca, C., Ortiz-Severin, J., Paradela, A., Poblete-Castro, I., et al. (2017). Multi-level evaluation of Escherichia coli polyphosphate related mutants using global transcriptomic, proteomic and phenomic analyses. Biochim. Biophys. Acta Gen. Subj. 1861, 871–883. doi: 10.1016/j.bbagen.2017.01.007
Vingadassalom, D., Campellone, K. G., Brady, M. J., Skehan, B., Battle, S. E., Robbins, D., et al. (2010). Enterohemorrhagic E. coli requires N-WASP for efficient type III translocation but not for EspFU-mediated actin pedestal formation. PLoS Pathog. 6:e1001056. doi: 10.1371/journal.ppat.1001056
Wang, X., Du, Y., Hua, Y., Fu, M., Niu, C., Zhang, B., et al. (2017). The EspF N-terminal of enterohemorrhagic Escherichia coli O157:H7 EDL933w imparts stronger toxicity effects on HT-29 cells than the C-Terminal. Front. Cell Infect. Microbiol. 7:410. doi: 10.3389/fcimb.2017.00410
Wang, Y., and Kim, K. S. (2000). Effect of rpoS mutations on stress-resistance and invasion of brain microvascular endothelial cells in Escherichia coli K1. FEMS Microbiol. Lett. 182, 241–247. doi: 10.1111/j.1574-6968.2000.tb08902.x
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F., and Hengge, R. (2005). Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005
Weiss, S. M., Ladwein, M., Schmidt, D., Ehinger, J., Lommel, S., Stading, K., et al. (2009). IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe 5, 244–258. doi: 10.1016/j.chom.2009.02.003
Yi, Y., Ma, Y., Gao, F., Mao, X., Peng, H., Feng, Y., et al. (2010). Crystal structure of EHEC intimin: insights into the complementarity between EPEC and EHEC. PLoS One 5:e15285. doi: 10.1371/journal.pone.0015285
Keywords: polyphosphate kinase 1 (PPK1), polyphosphate (polyP), enterohemorrhagic Escherichia coli O157:H7, pathogenesis, RpoS
Citation: Du Y, Wang X, Han Z, Hua Y, Yan K, Zhang B, Zhao W and Wan C (2021) Polyphosphate Kinase 1 Is a Pathogenesis Determinant in Enterohemorrhagic Escherichia coli O157:H7. Front. Microbiol. 12:762171. doi: 10.3389/fmicb.2021.762171
Received: 21 August 2021; Accepted: 08 October 2021;
Published: 27 October 2021.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Runhua Han, University of Texas at Austin, United StatesJincai Ma, Jilin Unversity, China
Ming chun Li, Nankai University, China
James L. Bono, United States Department of Agriculture, United States
Copyright © 2021 Du, Wang, Han, Hua, Yan, Zhang, Zhao and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengsong Wan, Z3p3Y3NAc211LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Yanli Du
Yanli Du Xiangyu Wang
Xiangyu Wang Zongli Han4
Zongli Han4 Chengsong Wan
Chengsong Wan