
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 November 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.760873
This article is part of the Research Topic Bacterial Secretion Systems, Volume II View all 17 articles
Integral and membrane-anchored proteins are pivotal to survival and virulence of the dental pathogen, Streptococcus mutans. The bacterial chaperone/insertase, YidC, contributes to membrane protein translocation. Unlike Escherichia coli, most Gram-positive bacteria contain two YidC paralogs. Herein, we evaluated structural features that functionally delineate S. mutans YidC1 and YidC2. Bacterial YidCs contain five transmembrane domains (TMD), two cytoplasmic loops, and a cytoplasmic tail. Because S. mutans YidC1 (SmYidC1) and YidC2 (SmYidC2) cytoplasmic domains (CD) are less well conserved than are TMD, we engineered ectopic expression of the 14 possible YidC1-YidC2 CD domain swap combinations. Growth and stress tolerance of each was compared to control strains ectopically expressing unmodified yidC1 or yidC2. Acid and osmotic stress sensitivity are associated with yidC2 deletion. Sensitivity to excess zinc was further identified as a ΔyidC1 phenotype. Overall, YidC1 tolerated CD substitutions better than YidC2. Preferences toward particular CD combinations suggested potential intramolecular interactions. In silico analysis predicted salt-bridges between C1 and C2 loops of YidC1, and C1 loop and C-terminal tail of YidC2, respectively. Mutation of contributing residues recapitulated ΔyidC1- and ΔyidC2-associated phenotypes. Taken together, this work revealed the importance of cytoplasmic domains in distinct functional attributes of YidC1 and YidC2, and identified key residues involved in interdomain interactions.
The Gram-positive oral pathogen Streptococcus mutans is a leading cause of dental caries (Hamada and Slade, 1980), with certain strains also associated with bacterial endocarditis (Lockwood et al., 1974; McGhie et al., 1977; Moore et al., 1977; Smith et al., 1977; Vose et al., 1987; Nomura et al., 2006). This species’ ability to rapidly utilize available dietary sugars and produce large amounts of acids, coupled with high tolerance to numerous environmental stressors, gives it a competitive advantage among its co-colonizers in the oral cavity. In addition, an ability to form robust biofilms, a high level of genetic competence enabling robust adaptability, and production of anti-microbial compounds such as mutacins contribute to its pathogenic potential [reviewed in Lemos et al. (2019)]. Most known virulence attributes of S. mutans stem from secreted or integral membrane proteins. Thus, a more complete understanding of the components of S. mutans protein translocation and integral membrane insertion pathways is of significant interest.
Membrane biogenesis in bacteria is largely a co-translational process that involves the signal recognition particle (SRP) pathway working in conjunction with the SecYEG translocon and/or a membrane protein insertase called YidC [reviewed in Koch et al. (2003) and Steinberg et al. (2018)]. YidC is ubiquitously present in all three domains of life and belongs to the Oxa/Alb/YidC family of insertases found in mitochondria, chloroplasts, and bacteria, respectively. Most details related to membrane biogenesis in bacteria, including contributions by YidC, have come from research in the model Gram-negative bacterium Escherichia coli (Samuelson et al., 2000; Scotti et al., 2000; Urbanus et al., 2001; van Bloois et al., 2004; van der Laan et al., 2004; Yi et al., 2004; Price et al., 2010; Kumazaki et al., 2014b). However, Gram-positive bacteria differ from their Gram-negative counterparts both in terms of the composition of the translocation machinery and the essentiality of individual components for survival [reviewed in Lewis and Brady (2015)]. Three key distinctions that differentiate the protein translocation machinery of Gram-positive bacteria from Gram-negative bacteria include: 1) the non-essentiality of the SRP components, Ffh, scRNA, and/or the SRP receptor FtsY, 2) the presence of two or sometimes more paralogs of YidC in Gram-positive bacteria that can be eliminated individually without lethality, and 3) the presence of an accessory SRP protein, YlxM (Hasona et al., 2005; Williams et al., 2014). Phenotypic characterization of S. mutans mutants has revealed overlapping features of SRP- and YidC2-deficient strains, with both being growth impaired under environmental stress (acid/osmotic/oxidative) as well as non-stress conditions. In contrast, a ΔyidC1 mutant has previously shown no obvious growth or stress-sensitivity phenotype (Hasona et al., 2005; Palmer et al., 2012). Combinatorial deletion of ffh and yidC2, or yidC1 and yidC2, is lethal in S. mutans, while a Δffh/ΔyidC1 double deletion strain survives suggesting that the presence of YidC2 alone can confer viability in this organism (Mishra et al., 2019). Membrane proteomic analyses of Δffh, ΔyidC1, ΔyidC2, and Δffh/yidC1 strains suggested a high degree of functional redundancy between YidC1 and YidC2, with gain of function mutations within yidC1 in a ΔyidC2 background also identified (Mishra et al., 2019). A gain of function mutation has also been identified in Bacillus subtilis YidC1 (SpoIIIJ) enabling it to survive lethality caused by high σM levels that interfere with intrinsic resistance to cell wall targeting antibiotics (Zhao et al., 2019). Work to characterize S. mutans YidC1 and YidC2 (SmYidC1 and SmYidC2) interactomes via immunocapture and chemical crosslinking experiments revealed several differences between apparent YidC1 and YidC2 interaction partners, and suggested both overlapping and individual substrate preferences (Lara Vasquez et al., 2021). Taken together results of phenotypic, proteomic, and protein-protein interaction analyses of WT, ΔyidC1, and ΔyidC2 mutants have suggested significant unique features of SmYidC1 and SmYidC2 despite a high degree of functional overlap. Phenotypic distinctions between B. subtilis ΔyidC1(spoIIIJ) and ΔyidC2(yqjG) mutants have also been reported (Errington et al., 1992; Saller et al., 2011; Zhao et al., 2019). Therefore, it is of broad general interest to understand why Gram-positive bacteria generally contain two distinct paralogs of YidC, and to identify specific structural features that confer individual activities of YidC1 compared to YidC2.
Primary sequence comparison demonstrates ∼27% identity and ∼47% homology (Emboss-Needleman tool) between S. mutans YidC1 and YidC2. A key feature delineating YidC1 and YidC2 is the longer and more basic C-terminal tail of YidC2. Indeed, acid stress tolerance was partially restored in a ΔyidC2 background when the YidC1 tail was replaced with that of YidC2 (Palmer et al., 2012). Later, crystal structure characterization combined with site-directed mutagenesis of B. halodurans YidC2 showed that its C1 cytoplasmic loop is flexible, and that its deletion impairs membrane insertase activity (Kumazaki et al., 2014a). Primary sequence analyses and crystal structures illustrate that all mature YidCs consist of five transmembrane domains (TM2-TM6) that are interspersed with cytoplasmic loop 1 (C1), cytoplasmic loop 2 (C2), and a cytoplasmic C-terminal tail (T). In consideration of the fact that protein synthesis occurs in the cytoplasm, YidC cytoplasmic domains would be expected to participate not only in specific interactions with other co-translational translocation machinery components but also with substrates destined for insertion. In E. coli, incoming substrates have been hypothesized to interact initially with the YidC C1 loop followed by interaction with a conserved arginine residue within transmembrane domain TM2 for insertion (Kol et al., 2008; Kumazaki et al., 2014a,c). More recently, molecular dynamics simulation showed that the crystallographically unresolved cytoplasmic C2 loop of BhYidC2 plays an important role in stabilizing this insertase’s structure (Harkey et al., 2019).
In the current work, we engineered a panel of chimeric yidC1/2 constructs in which one or more YidC1 cytoplasmic domains were substituted for those of YidC2, and vice versa. The chimeric variants were integrated into the bacterial chromosome and ectopically expressed from the gtfA locus in a genetic background in which endogenous yidC1 and yidC2 had both been eliminated. Each strain was characterized in terms of growth and survival under non-stress and stress conditions including acid, osmotic, and metal excess. This revealed a novel phenotype for the ΔyidC1 mutant, that was not shared by the ΔyidC2 strain, namely sensitivity to excess Zn(II). Three dimensional (3D)-structure prediction tools were also utilized to evaluate structural constraints within each of the various YidC1/2 chimeric proteins compared to unmodified YidC1 and YidC2. Our experimental results suggest considerable plasticity of YidC1 function in that this paralog could accommodate any combination of cytoplasmic domain substitutions without significant impact on the phenotypes tested. In contrast, YidC2 structure appeared less malleable with a preference for specific combinations of cytoplasmic domains in order to support growth under non-stress as well as environmental stress conditions. Lastly, prediction tools identified putative interactions between cytoplasmic loops 1 and 2 of YidC1, and between cytoplasmic loop 1 and the C-terminal tail of YidC2, which appeared to stem from stabilizing salt-bridges within YidC1 and YidC2, respectively. Amino acids involved in these putative salt-bridges, K91 and E190 in YidC1, and E92 and K253 in YidC2, were substituted with alanine by site-directed mutagenesis and confirmed to contribute to paralog-specific functions.
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. All of the S. mutans mutants used in this study were derived from S. mutans strain UA159 (Ajdic et al., 2002). S. mutans strains were routinely grown in Todd-Hewitt broth (BBL, Becton Dickinson) supplemented with 0.3% yeast extract (THYE) at 37°C in 5% CO2/95% air atmosphere (v/v). Spectinomycin (1 mg ml–1), kanamycin (1 mg ml–1), or erythromycin (10 mg ml–1) were added, where appropriate, for the growth and selection of S. mutans strains. E. coli strain C2987 (NEB) was used for standard cloning procedures and routinely grown in Luria–Bertani medium (10 g l–1 tryptone, 5 g l–1 yeast extract and 5 g l–1 NaCl) at 37°C with vigorous shaking or on LB agar (1.5%) with appropriate selection. Ampicillin (100 μg ml–1), kanamycin (50 μg ml–1), or erythromycin (250 μg ml–1) were used for the growth and selection of E. coli transformants.
The Gibson assembly method was used to construct 14 different chimeric genes of yidC1 and yidC2 with DNA encoding one, two, or three cytoplasmic domains swapped with the corresponding regions from the other paralog. Details of the chimeric YidC1/2 proteins are provided in Supplementary Table 2. DNA fragments corresponding to the specific regions of the yidC1 and yidC2 within each chimera were PCR-amplified using primers listed in Supplementary Table 3. PCR products were assembled between NdeI/BamHI sites of pET15b using NEBuilder HiFi DNA Assembly kit (NEB) following the manufacturer’s instructions and transformed into C2987 competent E. coli (NEB) by heat shock. Chimeric yidC1/2 fragments subcloned in pET15b were PCR-amplified using end primers and subcloned between XbaI/BsrGI sites of the S. mutans integration vector, pBGE. These constructs were subsequently used for integration at the non-essential gtfA locus.
A standard PCR-based technique involving partially overlapping primers (Supplementary Table 3) was used for site-directed mutagenesis. pSM15 and pSM20 were used as templates for the mutagenesis of yidC1 and yidC2 genes, respectively (Zheng et al., 2004). For subsequent integration into S. mutans, genes containing the engineered point mutations were first subcloned into the integration vector, pBGE, and transformants were selected for resistance to erythromycin.
Construction of S. mutans strains expressing a single chimeric CD-substituted yidC1 or yidC2 backbone gene was done in three steps. First yidC1 or yidC2 was deleted from the UA159 genome. yidC1 was eliminated by allelic exchange with a spectinomycin resistant marker (aad9) using the pCR2.1-DyidC1Sp plasmid (Palmer et al., 2012). The resultant ΔyidC1 strain (SM2001) was next transformed with pBGE plasmids containing either the wild-type (WT) or a chimeric yidC1 gene and selected for resistance to spectinomycin (1 mg ml–1) or erythromycin (10 mg ml–1). Lastly yidC2 was deleted from the WT- or chimeric yidC1-expressing strain by allelic replacement with aphA-3 encoding kanamycin resistance. The ΔyidC2:aphA-3 cassette for yidC2 deletion was generated by allelic exchange with a kanamycin resistant marker using pAH342 plasmid (Hasona et al., 2005). Similarly, chimeric yidC2 expressing strains were constructed by initial deletion of yidC2 using the ΔyidC2:aphA-3 cassette followed by integration of chimeric yidC2 at the gtfA locus and subsequent deletion of yidC1 by allelic replacement with aad9. Expression of site-directed point mutants of yidC1 and yidC2 genes was carried out as described above for chimeric yidC1 and yidC2. Deletion of yidC1 and yidC2, and integration of point mutated or chimeric yidC1/2, were confirmed by PCR and sequencing of the relevant genomic loci. Production of the WT or chimeric YidC1/YidC2 polypeptide was confirmed in each genetically engineered strain by Western blot using monospecific anti-YidC1 and anti-YidC2 antibodies (Palmer et al., 2012; Supplementary Figure 1).
Growth of each S. mutans strain was monitored using a Bioscreen C instrument (Growth Curves United States, Piscataway, NJ, United States). Overnight (O/N) cultures were grown in THYE with appropriate antibiotic selection at 37°C in air with 5% CO2. For precultures, O/N cultures were diluted 1:50 (or 1:20 for very slow growing strains) into 2 ml THYE without selection and grown to OD600 of 0.2–0.4. Precultures were then diluted to OD600 = 0.005 in 1 mL of prewarmed THYE and 300 ml of each dilution was transferred in triplicate to wells of a Bioscreen micro-well plate. A sterile mineral oil overlay (50 μL) was added on top of cultures in each well to measure growth under anaerobic conditions. Cell density was measured spectrophotometrically (OD600) every 30 min for up to 48 h of growth at 37°C. To assess the effect of oxygen stress, the overlay of sterile mineral oil was omitted. To assess the effect of acid stress, cells were grown in THYE adjusted to pH 5.0. To assess osmotic stress, THYE was supplemented with 3% NaCl. Metal intoxication experiments involved addition of 5 mM FeSO4 (added from a stock of 1 M FeSO4 prepared in 0.1 N HCl), 5 mM MnCl2, or 2.5 mM ZnCl2 to THYE. Bioscreen plate wells were not overlayed with mineral oil when bacteria were grown under metal-excess conditions.
Strains were grown to mid-exponential phase (OD600 = 0.2–0.4) in THYE from an O/N culture in THYE with appropriate antibiotic selection. Cultures were then adjusted to OD600 = 0.2, and serial 10-fold dilutions were made in THYE. Four microliter aliquots of each dilution were spotted on THYE agar (pH 7.0, pH 5.0, 3% NaCl, 5 mM FeSO4, 5 mM MnCl2, or 2.5 mM ZnCl2). Plates were incubated at 37°C in air with 5% CO2 for 2 days before visual inspection and documentation.
YidC sequence logo was created by alignment of YidC1 and YidC2 sequences of ten Gram-positive bacterial species using the weblogo tool1 (Schneider and Stephens, 1990). The clustal omega alignment of YidC1 and YidC2 sequences was entered into the webtool to generate the sequence logo image. Physico-chemical properties of various chimeric YidC1 and YidC2 proteins were evaluated with the Protparam web tool2 using their primary amino acid sequences (Gasteiger et al., 2005). Three-dimensional structures of the wild-type proteins and chimeric YidC1 and YidC2 variants were predicted using I-TASSER (Roy et al., 2010). Protein structures were analyzed with Pymol (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC). Structure comparisons were done to determine the best alignment (lowest root mean square deviation over the largest number of amino acids) of two proteins using the cealign plugin in Pymol (Shindyalov and Bourne, 1998).
Functional differences in paralogous proteins often arise as a result of sequence-structure differences acquired through mutations (Conrad and Antonarakis, 2007), with other contributing factors including distinct gene neighborhood, operon organization, and genetic regulation (Zallot et al., 2016). A high resolution crystal structure of BhYidC2 lacking the N-terminal domain and C-terminal tail region is available (Kumazaki et al., 2014a); however, this information alone is not sufficient to explain distinct functional attributes of multiple YidC paralogs of Gram-positive bacteria.
To begin to explore the structural features that likely contribute to characteristic functional activities of SmYidC1 vs. SmYidC2, we first carried out in silico analysis and comparison of their structures. Pairwise comparison showed ∼27% identity and ∼47% similarity between SmYidC1 and SmYidC2 (EMBOSS Needle tool). Both paralogs are predicted to be lipoproteins with cleavable signal peptide sequences (PRED-LIPO and Signal P 4.0), and hydropathy analysis by various webtools indicates a post-cleavage structure containing five transmembrane domains (TMDs) connected by two cytoplasmic loops: C1 and C2, and a hydrophilic C-terminal tail for both proteins (Stoffel, 1993; Krogh et al., 2001; Bagos et al., 2008; Petersen et al., 2011; Figure 1A). Despite low sequence similarity, SmYidC1 and SmYidC2 are predicted to have similar topological architectures except that the cytoplasmic C-terminal tail region of YidC2 is significantly longer and more basic than that of YidC1 (63 compared to 38 amino acids). Overlaying topology predictions onto the aligned amino acid sequences illustrates that a greater degree of identity/homology (red boxes/texts in Figure 1B, respectively) is observed between the TMDs of SmYidC1 and SmYidC2 compared to their cytoplasmic loops and C-terminal tails (Figure 1B).
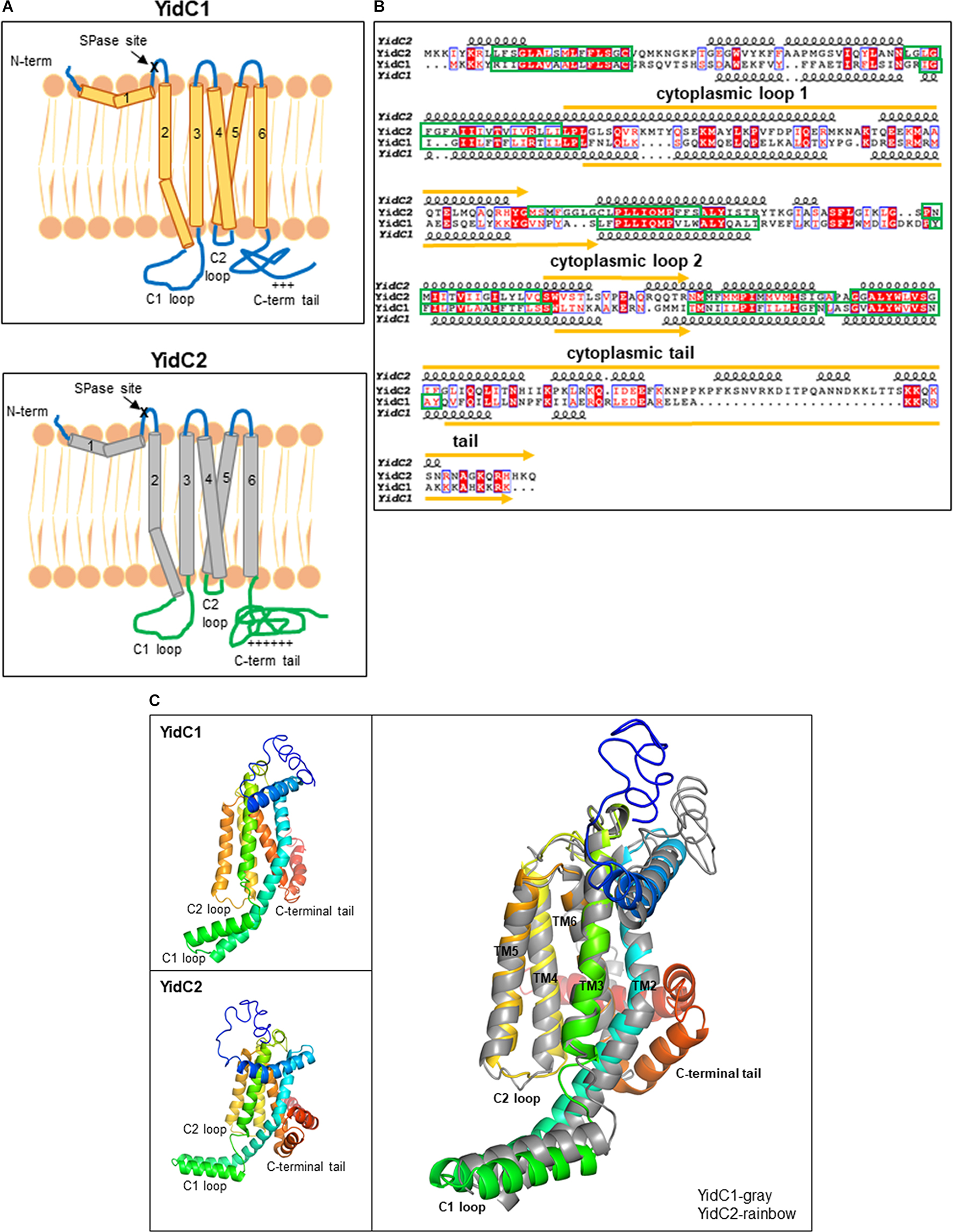
Figure 1. In silico comparison of S. mutans YidC1 and YidC2. (A) Predicted membrane topologies of YidC1 and YidC2 using TM-Pred [TMpred Server (vital-it.ch)]. The mature YidC1 and YidC2 proteins each contain five TM domains and three cytoplasmic regions: C1 loop, C2 loop, and C-terminal tail. “ + + + ” or “ + + + + + ” refers to the relative positivity of the charged C-terminal tails of Sm YidC1 and YidC2. (B) Alignment of primary and secondary structures of SmYidC1 and SmYidC2 predicted by I-TASSER [I-TASSER server for protein structure and function prediction (umich.edu)]. Amino acid sequences were aligned using the Clustal Omega Program (EMBOSS-Needle) and presented using the ESPript2.2 program (Gouet et al., 1999). Identical residues are highlighted by red boxes, while similar residues are highlighted by red text. Indicated secondary structural helices are based on iTasser-predicted 3D structures. Green boxes correspond to transmembrane domains and yellow lines represent cytoplasmic domains. (C) Cartoon representations of the superimposed I-TASSER-predicted 3D structures of YidC1 and YidC2 with YidC1 shown in gray and YidC2 shown as rainbow colored from N- to C-terminus. The YidC2 structure superimposes on YidC1 with a root-mean-square deviation of 4.11 Å for all the Cα atoms (for over 232 residues).
We also extended our pairwise comparison of SmYidC1 and SmYidC2 to the respective YidC orthologs from other Gram-positive bacteria to determine if the sequence diversity observed within the cytoplasmic regions is limited to S. mutans or extends to other species. The results of multiple YidC1/YidC2 sequence alignments from various Gram-positive bacteria are shown in Supplementary Figure 2. Conservation of residues was plotted using Weblogo (Supplementary Figure 3). Interestingly, in all cases of dual YidC paralogs, higher sequence conservation was observed within regions that align with the SmYidC1 and SmYidC2 TMDs, especially TM3. The highest degree of variation was observed among the YidC C-terminal tails, which did not align significantly enough to be included in the Weblogo representation (Supplementary Figures 2, 3).
Since crystallographic data is not available for SmYidC1/2, we used I-TASSER (Iterative Threading ASSEmbly Refinement) to generate their 3D-models (Roy et al., 2010; Figure 1C). Three dimensional models of SmYidC1 and SmYidC2 resemble the crystal structure of the BhYidC (PDB nos. 3WO6 and 3WO7) core region, which also contains TMDs (TM2-TM5) and three cytoplasmic domains: C1 loop, C2 loop, and C-terminal tail. When predicted 3D-models of SmYidC1 and SmYidC2 were compared to one another, the Cα atoms superimposed with a root mean square deviation (RMSD) of 4.11 Å over 232 residues (Figure 1C). The 3D structure comparison further showed that the C1 and C2 cytoplasmic loops of YidC1 and YidC2, as well as their C-terminal tails, did not superimpose as well as their TMDs. Taken together, sequence and structural analyses clearly indicate that the TMDs are more highly conserved than are the cytoplasmic domains of YidC1 and YidC2. We therefore hypothesized that cytoplasmic regions likely confer paralog-specific functional activities to the two S. mutans insertases.
Previous work from our laboratory revealed the significance of the C-terminal tails in conferring functional differences between SmYidC1 and SmYidC2. When a ΔyidC2 deletion strain was engineered to harbor tail-swapped versions of YidC1 or YidC2, stress-sensitivity of the mutant strain was partially restored by ectopic expression of chimeric YidC1 harboring the YidC2 tail, while the mutant phenotype was worsened by expression of chimeric YidC2 harboring the YidC1 tail (Palmer et al., 2012). Notably, these experiments were conducted in a ΔyidC2 mutant background, in which a functionally active native YidC1 remained in addition to the chimeric ectopic YidCs. In the current work, we extended our analysis by swapping all three cytoplasmic domains including loops C1, C2, and the C-terminal tails singly and in combination, and constitutively expressing each chimeric gene as the sole paralog from the ectopic gtfA locus in the genome so that the contribution of each individual domain toward the particular phenotype of interest could be specifically addressed. Combinatorial exchange of two and three different cytoplasmic domains were included in these experiments so that potential interactions among the YidC1 or YidC2 cytoplasmic domains could also be assessed. Schematic representations of YidC1, YidC2, and the 14 different domain swapped chimeric constructs are illustrated in Figure 2. Throughout the paper, the YidC1 backbone and cytoplasmic domains are indicated in regular type, while the YidC2 backbone and cytoplasmic domains are indicated in bold.
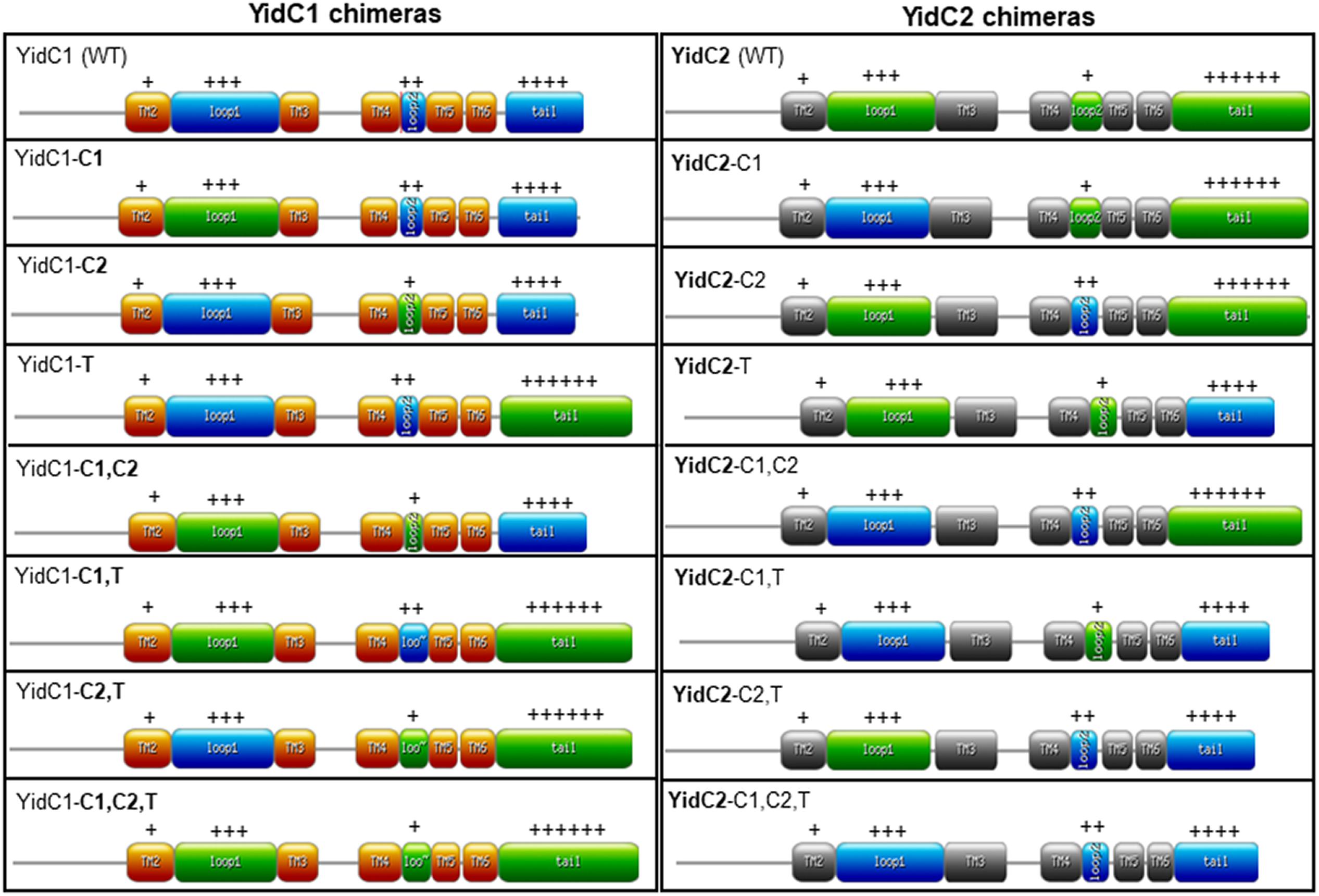
Figure 2. Schematic representation of unmodified and chimeric YidC1 and YidC2 polypeptides. YidC1 transmembrane (TM) domains are illustrated in orange and cytoplasmic domains (CD) in blue. YidC2 TM domains are illustrated in gray and CD in green. Chimeric constructs were designed using the web-based https://prosite.expasy.org/mydomains/tool. Regular text indicates YidC1-derived backbone or CD. Bold text indicates YidC2-derived backbone or CD. “ + + + ” or “ + + + + + ” refers to the relative positivity of the charged C-terminal tails of Sm YidC1 and YidC2.
In silico characterization of YidC1, YidC2, and their chimeric variants included determination of pI, number of positively and negatively charged amino acids, instability index (II), grand average hydropathy (GRAVY) using Exapsy ProtParam tools and structural comparison using predicted 3D models (Table 1 and Supplementary Figure 4). Theoretical pI calculations showed that both YidC1 and YidC2 are basic proteins (pI = 10 and 10.25, respectively), with the pI values of chimeric YidC1 and YidC2 proteins remaining close to those of the WT versions. Next, we determined the hydrophobicity of the proteins by calculating their GRAVY scores such that positive values represent hydrophobic proteins while negative values indicate hydrophilic proteins (Kyte and Doolittle, 1982). As expected for membrane proteins, GRAVY scores of YidC1 and YidC2 were both above 0; however, the C1 loop of YidC1 and C-terminal tail of YidC2 were more hydrophilic compared to their respective counterparts. Swapping these domains indeed lowered the predicted hydrophobicity of the chimeric proteins.
We were also interested to know the effect of individual and combinatorial domain substitutions on chimeric YidC1/2 structures (Supplementary Figure 4). We used I-TASSER to predict the 3D structures of each variant and compared them with the 3D structures of the WT proteins, evaluating similarity with each WT protein based on lowest RMSD values over the largest number of amino acids using the cealign plugin in Pymol (Shindyalov and Bourne, 1998; Table 1). Alignment data showed higher RMSD values for most of the YidC1 chimeras, which suggests that YidC1 structure is less rigid compared to that of YidC2. That is, most of the domain swaps impacted the 3D structure of YidC1. On the other hand, most of the chimeric YidC2 proteins, except YidC2-C1 and YidC2-C1,C2, aligned better with unmodified YidC2 than with unmodified YidC1. Therefore, the core domain structure of YidC2 appears more rigid with most of the domain swaps not substantially deviating from that predicted for unmodified YidC2. In agreement with physicochemical characterization of the chimeric YidC proteins, C1 loop substitutions in both YidC1 and YidC2 notably altered alignment with their respective unmodified proteins. Substitution of all three domains within YidC1 (YidC1-C1,C2,T) made this chimera dissimilar to both YidC1 and YidC2, while YidC2-C1,C2,T was similar to YidC2 but did not align well with YidC1.
We next evaluated S. mutans strains expressing the 14 different chimeric versions of yidC1 or yidC2. The chimeric yidC genes, or unmodified controls, were introduced into the non-essential gtfA locus for expression in a S. mutans genetic background in which native yidC1 and yidC2 had been both eliminated. Strain constructions were carried out in a step-wise manner to overcome the lethality caused by simultaneous elimination of genes encoding both YidCs. Thus, native yidC1 or yidC2 was deleted before integrating a chimeric or unmodified gene at the gtfA locus. This was followed by deletion of the residual native yidC2 or yidC1, and resulted in each engineered S. mutans strain ectopically expressing a single yidC variant under control of the gtfA promoter. Production of each WT or chimeric YidC1/2 polypetide was confirmed by Western blot with appropriate monospecific anti-YidC1 or YidC2 rabbit antisera generated against synthetic peptides corresponding to the cytoplasmic tail domains (Supplementary Figure 1).
After their construction, we first evaluated the growth of the S. mutans strains that ectopically expressed yidC1 or yidC2 in both single deletion and double ΔyidC1/ΔyidC2 deletion backgrounds compared to the UA159 parental strain under non-stress and environmental stress conditions (acid and osmotic) (Figure 3). As expected, growth under all three conditions was notably diminished only in the yidC2 single mutant. When growth was evaluated under aerated conditions in both broth culture and on agar plates, results were similar to those under non-stress conditions (Supplementary Figure 5). Under non-stress and osmotic stress conditions, the ΔyidC1/ΔyidC2-yidC1∗ ectopic expression strain grew at a rate comparable to that of the UA159 parent, while the ΔyidC1/ΔyidC2-yidC2∗ ectopic expression strain grew more slowly (Figures 3A,B,E,F and Supplementary Table 4). Neither ectopic expression of yidC1, nor yidC2, restored the WT level of growth under acid stress conditions (Figures 3C,D). Incomplete rescue when yidC1 or yidC2 were expressed ectopically rather than from their endogenous locations likely reflects the lack of native gene regulation when non-native promoters and loci are employed. Indeed, our prior membrane proteome characterization of WT, ΔyidC1, and ΔyidC2 strains showed that YidC2 levels were increased in a ΔyidC1 background, although a similar increase in YidC1 was not observed in the ΔyidC2 mutant (Mishra et al., 2019). Because the focus of the current work is a structure-function analysis of the YidC1 and YidC2 polypeptides themselves, and not an evaluation of gene regulation or expression, all subsequent functional assays were performed using the ectopically-expressed unmodified yidC1 or yidC2 genes in a ΔyidC1/ΔyidC2 background, rather than the UA159 parent strain, as the basis for of comparison of the chimeric yidC1/2 variants.
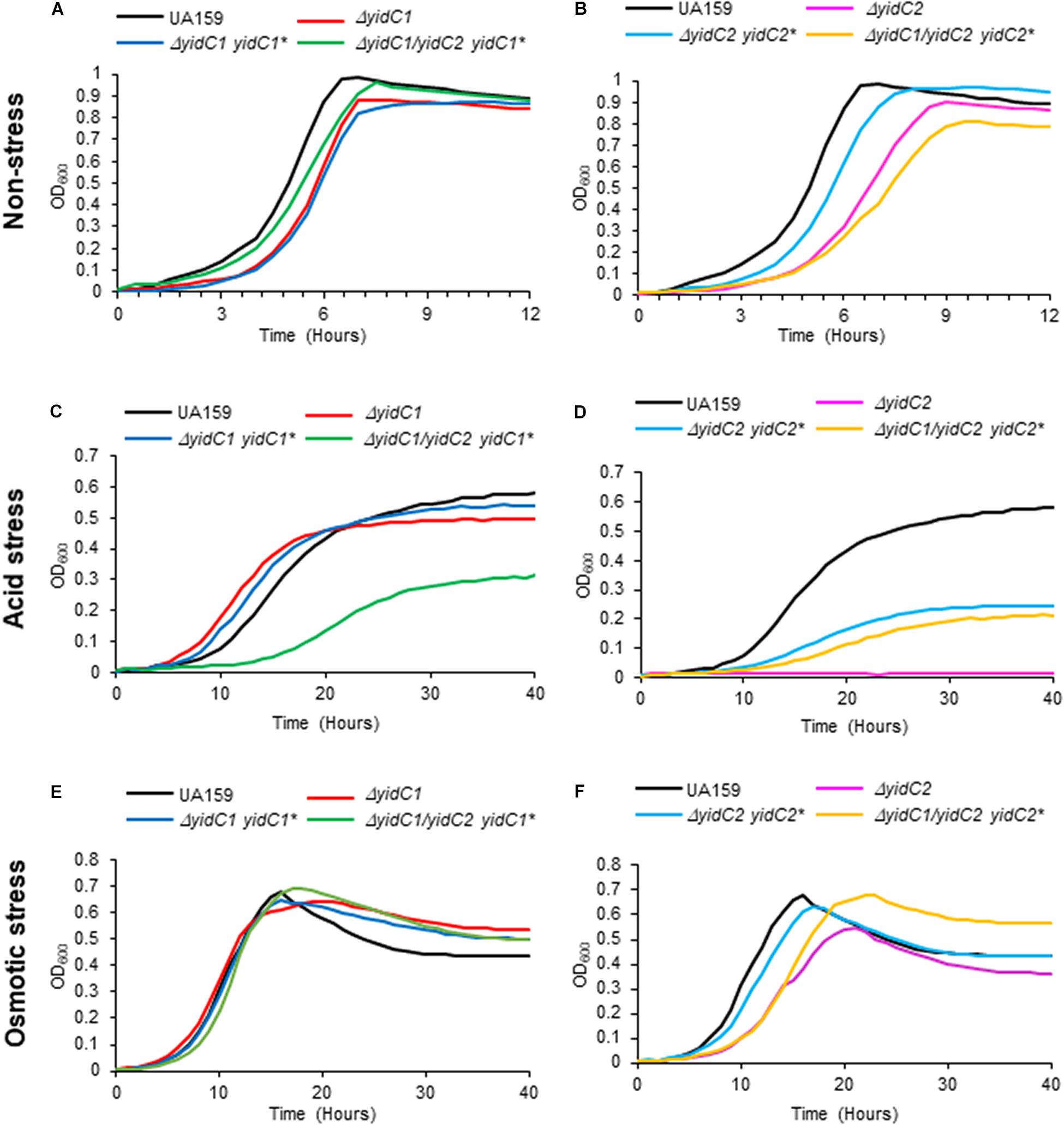
Figure 3. Evaluation of growth of S. mutans wild-type, DyidC1/2 mutants, and ectopic expression strains under non-stress, acid-stress, and salt-stress conditions. Growth of the wild-type UA159 parent strain compared to ΔyidC1 (A,C,E) or ΔyidC2 (B,D,F) deletion mutants, and strains complemented by ectopic expression (*) from the gtfA locus of yidC1 or yidC2 in single ΔyidC1/2 or double ΔyidC1/ΔyidC2 backgrounds. (A,B) Growth in THYE, pH 7). (C,D) Growth in THYE acidified to pH 5.0. (E,F) Growth in THYE supplemented with 3% NaCl. Growth measurements were carried out at least three times with each sample assayed in triplicate. Data shown are mean replicates of an independent experiment.
None of the individual or combinatorial substitutions of YidC2 cytoplasmic domains into YidC1 resulted in diminished growth of S. mutans compared to ectopic expression of unmodified yidC1 under non-stress conditions (Figure 4A and Supplementary Table 4). Similar results were obtained when each of the strains was grown on solid media (Supplementary Figure 6). These results suggest that any of the three cytoplasmic domains of YidC2 can successfully replace those of YidC1, individually or in combination, to support growth and survival of S. mutans in the absence of external environmental stressors. Next, we tested strains harboring chimeric YidC2 variants compared to the unmodified YidC2 polypeptide (Figure 4B). Most of the chimeric YidC2 variants, except for YidC2-C1,T, exhibited decreased growth compared to the unmodified YidC2 control strain. Similar results were observed when the strains were grown under aerated rather than anaerobic conditions except that the YidC2-C1,T variant more closely resembled that of the unmodified YidC2 control (Supplementary Figure 6 and Supplementary Table 4). Growth on solid media also followed a similar pattern to that observed in liquid media (Supplementary Figure 6). The strain harboring the YidC2-C2 chimera was noteworthy in that after an extended lag phase it achieved a substantially higher yield compared to the unmodified YidC2 control strain (Figure 4B). Subsequent subculture in THYE did not alter this strain’s growth kinetics suggesting that delayed growth and increased yield was not the result of a suppressor mutation, but rather physiological adaptation in broth culture. The negative impact of expression of most of the chimeric yidC2 variants on S. mutans growth illustrates that individual YidC1 and YidC2 cytoplasmic domains are not functionally equivalent. YidC2 structure appears less malleable and tolerant of substitutions than that of YidC1. YidC2 function was perturbed by all domain substitutions except for YidC2-C1,T. These results are consistent with the notion that putative interactions occur between cytoplasmic domains such that specific combinatorial substitutions are preferred. The C1 loops of YidC1 and YidC2 are of similar size suggesting that primary sequence rather than overall length plays an important role in C1 functional attributes. In contrast, the C2 loops and C-terminal tails of YidC1 and YidC2 vary in both size and sequence.
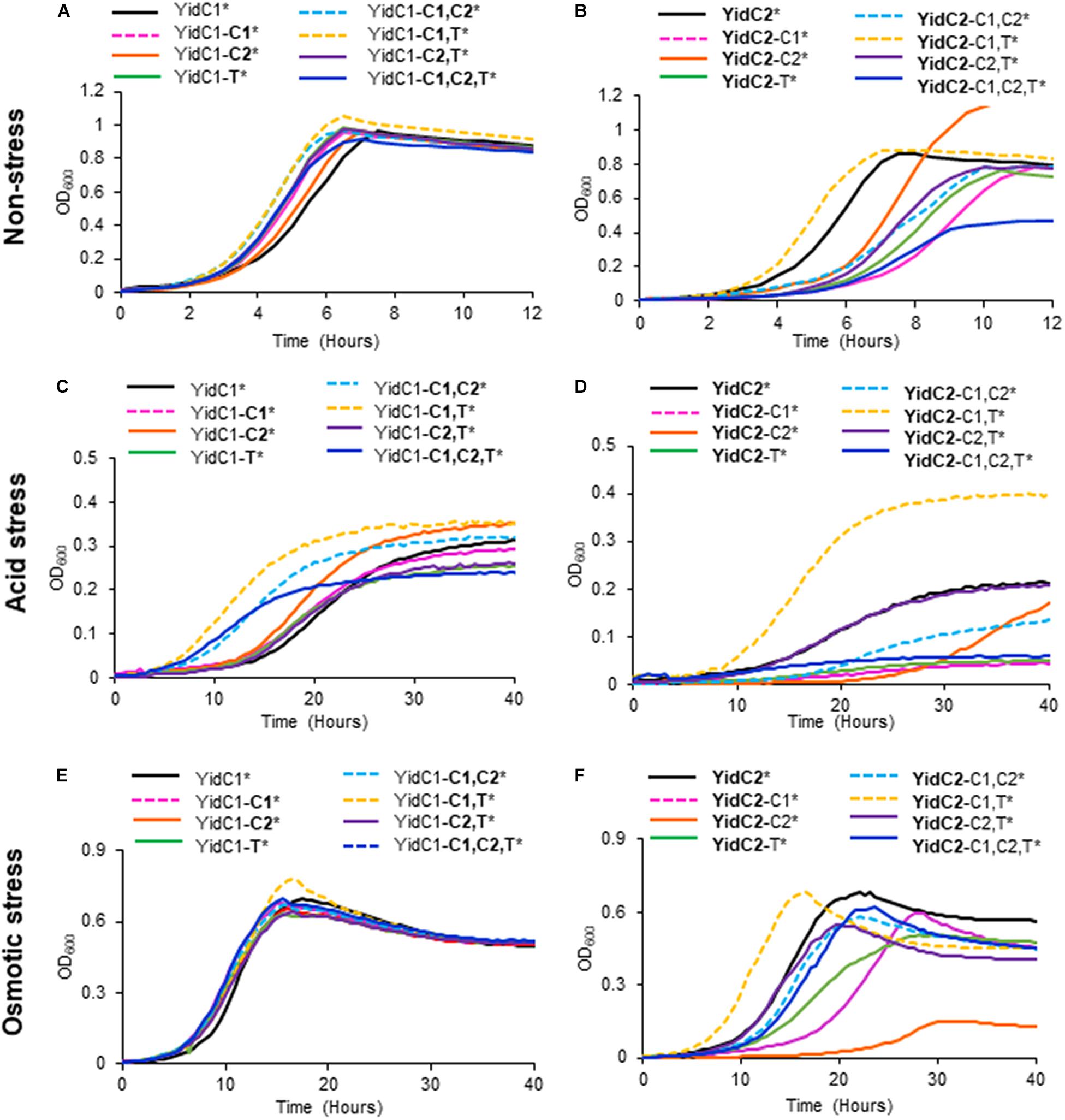
Figure 4. Effect of YidC1/2 cytoplasmic domain substitutions on S. mutans growth under non-stress, acid-stress, or salt-stress conditions. Growth of strains complemented by ectopic expression (*) from the gtfA locus of unmodified yidC1 (A,C,E) or yidC2 (B,D,F) compared to corresponding cytoplasmic domain (CD) swap variants in a double ΔyidC1/ΔyidC2 background. (A,B) Growth in THYE, pH 7). (C,D) Growth in THYE acidified to pH 5.0. (E,F) Growth in THYE supplemented with 3% NaCl. Growth measurements were carried out at least three times with each sample assayed in triplicate. Data shown are mean replicates of an independent experiment. Regular text indicates YidC1 backbone or CD. Bold text indicates YidC2 backbone or CD.
Growth in acidified medium (pH 5.0) of the strain with chimeric YidC1-T was negatively affected, whereas the strain harboring YidC1-C2 demonstrated improved growth, and that with YidC1-C1 appeared unaffected (Figure 4C and Supplementary Table 4). This suggests specific influences of the cytoplasmic tail and C2 loop on the structure and function of YidC1 compared to YidC2. Combinatorial replacement of YidC2 C1 and C2, or the YidC2 C1 and tail domains, into YidC1 revealed notably enhanced growth of the chimeric strains in acid conditions (Figure 4C and Supplementary Table 4). This suggests potential interactions between certain pairs of cytoplasmic domains that may not be replicated with heterologous partners. Surprisingly, growth yield under acid conditions was notably diminished in the YidC1-C1,C2,T triple chimeric strain. This pronounced negative affect implies a collective behavior of the YidC2 cytoplasmic domains in the context of the YidC1 backbone that cannot be attributed to a single domain substitution, or pairs of substitutions. Growth of the YidC1 chimeric strains on pH 5.0 agar plates was consistent with that in liquid media (Supplementary Figure 7), although differences among strains were not as pronounced, possibly because plates were grown in 5% v/v CO2 saturated air rather than anaerobically.
The impact of domain swapping on acid tolerance was even more apparent when YidC1 cytoplasmic domains were introduced into YidC2 (Figure 4D). With the exceptions of the simultaneous exchange of the YidC1 C1 loop and tail into YidC2 (YidC2-C1,T) or the YidC1 C2 loop and tail into YidC2 (YidC2-C2,T), all other combinations of domain substitutions resulted in worsened growth of the YidC2 chimeric strains. The negative impact of the YidC1 C1 loop was ameliorated, and growth of the chimeric YidC2 strain was notably enhanced, when the YidC1 tail was exchanged as well (YidC2-C1,T). Simultaneous substitution of YidC1’s C2 loop and tail with those of YidC2 (YidC2-C2,T) also alleviated the negative impact of single substitution of the C2 loop (YidC2-C2). However, simultaneous transfer of all three of YidC1 cytoplasmic domains into YidC2 (YidC2-C1,C2,T) again resulted in a seriously impaired chimeric insertase unable to contend with acid stress. Taken together, these results indicate specific functional attributes of YidC2 compared to YidC1 cytoplasmic domains that must be expressed in the proper combination and context to achieve optimal functional activity.
Exchange of any combination of YidC2 cytoplasmic domains into YidC1 had little to no effect on the ability of the YidC1 chimeric strains to grow in the presence of high salt in liquid culture (Figure 4E), or on agar plates (Supplementary Figure 7). In stark contrast, introducing YidC1 cytoplasmic domains into YidC2 had substantial negative effects on the ability of the YidC2 chimeric strains to grow during osmotic stress (Figure 4F and Supplementary Figure 7), the only exception being co-substitution of the YidC1 C1 loop and tail-domains into YidC2 (YidC2-C1,T) which modestly enhanced growth. The most detrimental impact on growth in high salt was observed when only the C2 loop of YidC1 was introduced into YidC2 (YidC2-C2). This effect was partially alleviated when the C2 loop was exchanged in combination with the YidC2 C1 loop or C-terminal tail (YidC2-C1,C2 or YidC2-C2,T). The chimeric strain harboring YidC2 with all three cytoplasmic domains exchanged for those of YidC1 (YidC2-C1,C2,T) was only minimally impacted during growth in the presence of high salt when compared to ectopic expression of unmodified yidC2. Thus, the results of domain swapping experiments vary considerably depending on the phenotype being evaluated, as well as on the backbone of the insertase under study. This information is expected to become even more enlightening as YidC1- compared to YidC2-specfic substrates responsible for predominant ΔyidC1 and ΔyidC2 mutant phenotypes are identified in future studies. Collectively, the results shown in Figure 4 suggest optimal function when the C1 loop and C-terminal tail are paired from the same paralog; however, that beneficial pairing can be obfuscated when all three domains are co-transferred.
Our recent membrane proteomic analysis of ΔyidC1 and ΔyidC2 deletion mutants, as well as characterization of YidC1 and YidC2 interactomes, suggested a potential involvement of the YidC insertases in metal homeostasis (Supplementary Table 5; Mishra et al., 2019; Lara Vasquez et al., 2021). This is a logical supposition given the membrane localization of integral components of metal ion import and efflux systems. We therefore evaluated growth and survival of ΔyidC1 and ΔyidC2 mutants, and ectopic expression strains, in media supplemented with salts of transition metals including zinc, iron, and manganese (Figure 5 and Supplementary Table 4). These growth experiments were carried out under aerated conditions since transition metals are redox-active and their toxicity is therefore linked to oxygen (Storz and Imlay, 1999; Moore and Helmann, 2005). The most striking result of these experiments was a complete inability of the ΔyidC1 deletion strain to grow in the presence of excess Zn(II) (Figure 5A). Zn(II) is a highly abundant and tightly regulated transition metal that acts as a cofactor for multiple bacterial proteins [reviewed in Chandrangsu et al. (2017)]. Results were similar on agar plates containing 2.5 mM ZnCl2 (Supplementary Figure 8). Ectopic expression of yidC1 in the ΔyidC1 and ΔyidC1/ΔyidC2 backgrounds was largely able to complement the Zn toxicity phenotype. In contrast, deletion of yidC2 had far less impact on the ability of S. mutans to grow in the presence of excess Zn(II) (Figure 5B). Deletion of yidC1 had no obvious impact on S. mutans growth under conditions of Fe(II) or Mn(II) excess (Figures 5C,E), while growth of the ΔyidC2 was modestly diminished under these conditions (Figures 5D,F). Growth of the ΔyidC1/ΔyidC2-yidC1 ectopic expression strain was somewhat diminished compared to the UA159 parent strain in the presence of excess Fe(II). In contrast, the strain engineered to express ectopic yidC2, while viable in the presence of excess Zn(II), Fe(II), or Mn(II), was severely growth-impaired under these conditions (Figures 5B,D,F). The ability of this strain, which lacks endogenous or ectopic yidC1, to grow at all in the presence of excess Zn(II) was initially surprising, but can be explained if the YidC1 and YidC2 paralogs have differential effects on import of particular metals as opposed to efflux. For example, if YidC1 is responsible for insertion of a critical zinc exporter, then inefficient insertion of a metal importer by ectopic expression of yidC2 would render the cells less susceptible to normally toxic levels of Zn(II) in the extracellular environment. While identification of specific membrane-localized importer vs. exporter substrates is beyond the scope of the current study, evaluation of metal toxicity provides another useful phenotypic property to assess the impact of cytoplasmic domain swaps on YidC1 and YidC2 behavior compared to the unmodified insertases.
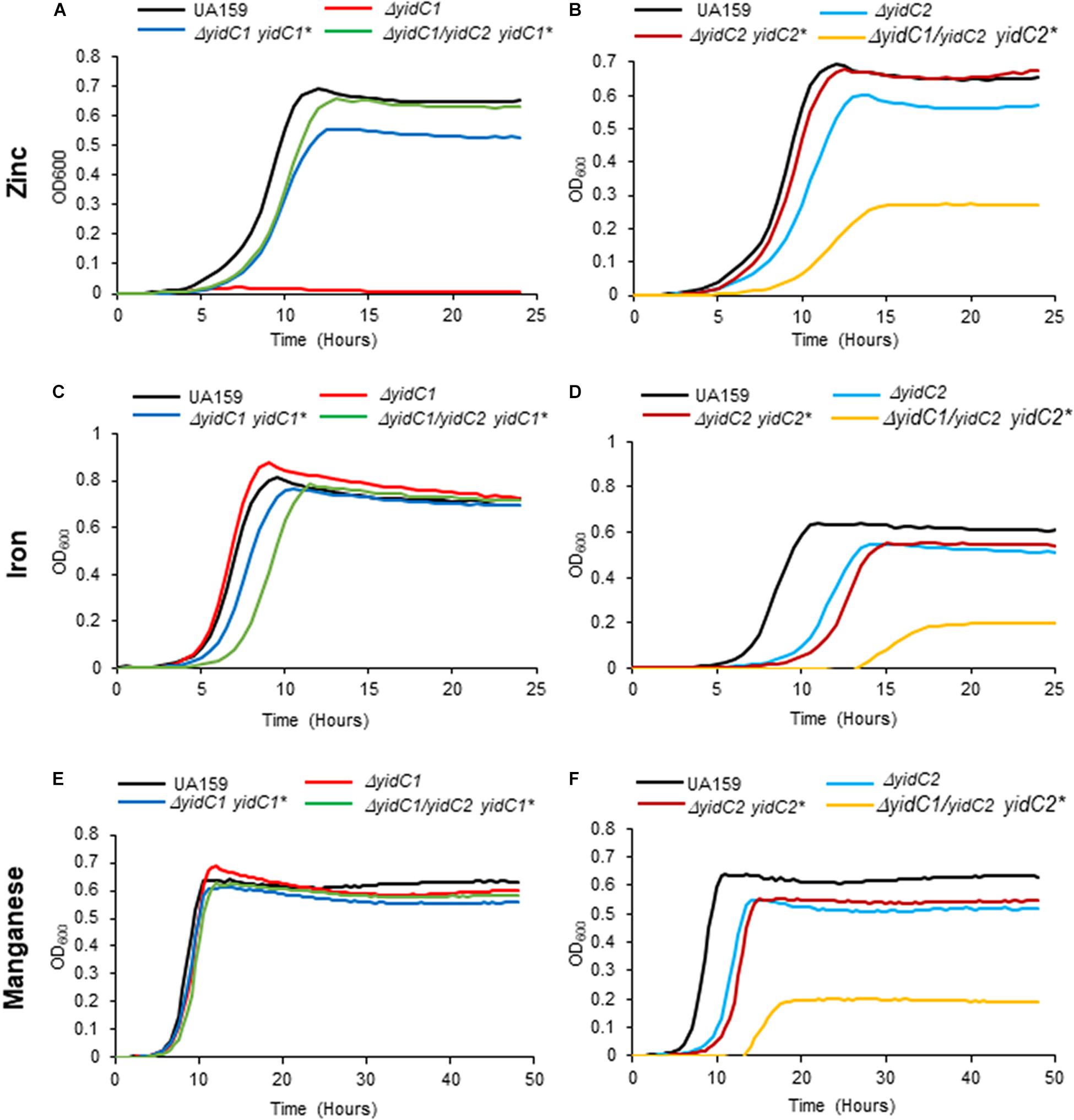
Figure 5. Evaluation of growth of S. mutans wild-type, ΔyidC1/2 mutants, and ectopic expression strains under excess transition metal conditions. Growth of the wild-type UA159 parent strain compared to ΔyidC1 (A,C,E) or ΔyidC2 (B,D,F) deletion mutants, and strains complemented by ectopic expression (*) from the gtfA locus of yidC1 or yidC2 in single ΔyidC1/2 or double ΔyidC1/DyidC2 backgrounds. (A,B) Growth in THYE, pH 7, supplemented with 2.5 mM ZnCl2 under aerating conditions. (C,D) Growth in THYE, pH 7, supplemented with 5 mM MnCl2 under aerating conditions. (E,F) Growth in THYE, pH 7, supplemented with 5 mM MnCl2 under aerating conditions. Growth measurements were carried out at least three times with each sample assayed in triplicate. Data shown are mean replicates of an independent experiment.
Substitution of the cytoplasmic domains of YidC2 into YidC1 had little impact on growth of the chimeric strains in Fe(II), Mn(II), or Zn(II) compared to the unmodified YidC1 control, except that the YidC1-C1,T variant grew slightly less well in the presence of excess Zn(II) and slightly better in the presence of excess Fe(II) than the other strains (Figures 6A,E). This strain also achieved a notably lower cell yield during growth in excess Mn(II), and is the first example thus far whereby a homologous pairing of C1 and C-terminal tail domains was apparently detrimental. Overall, these results demonstrate that YidC1 supports the growth of S. mutans in the presence of high levels of the metals tested, and that this paralog’s function in this regard is not severely impacted by exchange of its cytoplasmic domains with those of YidC2. The lower cell yield of the YidC1-C1,T variant during growth in Mn(II) could stem from increased import or decreased export, or both, of this particular metal. In the case of strains harboring chimeric YidC2 variants, all combinations of single, double, and triple domain swaps of YidC1 cytoplasmic domains into YidC2 improved growth in the presence of Mn(II), with the chimeric YidC2-C2 strain achieving a notably higher cell yield (Figure 6F). Results were similar in the presence of excess Fe(II) (Figure 6D), suggesting that overlapping import/efflux systems may contribute to Mn(II) and Fe(II) homeostasis, and that YidC1 cytoplasmic domains, particularly the C2 loop, are more effective than those of YidC2 in the insertion of relevant substrates that limit toxicity by these two metals. The impact of the chimeric YidC2 variants in the presence of excess Zn(II) was strikingly different than in the presence of excess Fe(II) or Mn(II) (Figure 6B). This suggests that S. mutans contends with Zn(II) in a different way than it does the other two metals. In this case, incorporation of either the YidC1 C1 loop (YidC2-C1), or C2 loop (YidC2-C2), into YidC2 severely diminished the ability of the variant insertases to contend with toxic levels of Zn(II). In contrast, all other combinations of domain exchanges substantially improved growth in excess Zn(II). Thus, when the YidC1 C1 and C2 loops were exchanged together, or in combination with the YidC1 C-terminal tail, the net effect on the YidC2 chimeric polypeptides was to enable the cells to better contend with a high level of environmental Zn(II)- again either due to diminished uptake or increased export of this particular metal. The amelioration of the detrimental impact of single substitution of the YidC1 C1 or C2 into YidC2 by co-exchange with other potentially interactive cytoplasmic domains highlights the likely importance of cooperative effects of the cytoplasmic domains of each insertase to accomplish appropriate membrane protein insertion to support specific cellular activities.
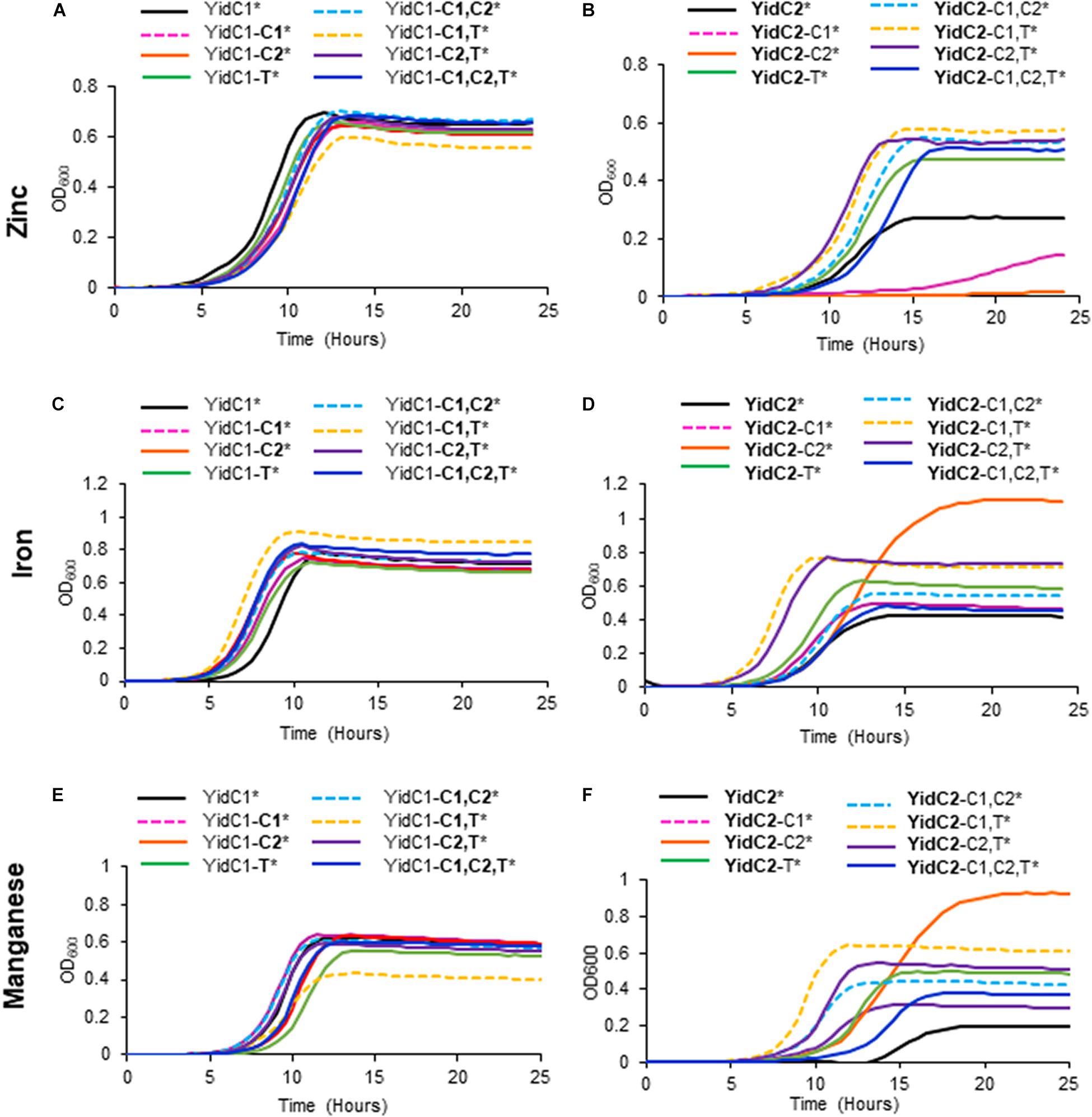
Figure 6. Effect of YidC1/2 cytoplasmic domain substitutions on S. mutans growth under excess transition metal conditions. Growth of strains complemented by ectopic expression (*) from the gtfA locus of unmodified yidC1 (A,C,E) or yidC2 (B,D,F) compared to corresponding cytoplasmic domain (CD) swap variants in a double ΔyidC1/ΔyidC2 background. (A,B) Growth in THYE, pH 7, supplemented with 2.5 mM ZnCl2 under aerating conditions. (C,D) Growth in THYE, pH 7, supplemented with 5 mM MnCl2 under aerating conditions. (E,F) Growth in THYE, pH 7, supplemented with 5 mM MnCl2 under aerating conditions. Growth measurements were carried out at least three times with each sample assayed in triplicate. Data shown are mean replicates of an independent experiment. Regular text indicates YidC1 backbone or CD. Bold text indicates YidC2 backbone or CD.
As detailed above, phenotypic characterization of strains producing chimeric YidC1 and YidC2 variants demonstrated preferences for combinatorial replacement of certain cytoplasmic domains over individual substitutions suggesting specific functional inter-domain interactions. Salt bridges (electrostatic interaction between charged residues) often play an important role in the conformational stability of proteins (Ban et al., 2019). All three cytoplasmic domains of YidC1, as well as those of YidC2, contain multiple charged residues that could potentially be involved in formation of salt bridges. We used an in silico approach to identify those amino acid residues within the YidC1 and YidC2 cytoplasmic domains most likely to form salt bridges, and measured the distance between the putative donor and acceptor atoms of the residues involved (Supplementary Table 6). Potential intra-domain interactions were identified within the C1 loop and C-terminal tail of YidC1, and within the C1 loop, C2 loop, and C-terminal tail of YidC2. In addition, putative inter-domain interactions between YidC1 amino acid residues K91 (C1 loop) and E190 (C2 loop) (Figure 7A), and YidC2 residues E92 (C1 loop) and K253 (C-terminal tail) (Figure 7B), were identified. The predicted distances between the side chains of both of these pairs of charged residues were <3Å, supporting the likelihood of their interaction. Multiple sequence alignment of YidC1s from various oral streptococci showed that both the K91 and E190 residues are highly conserved (Supplementary Figure 2). Alignment of YidC2 sequences from various oral streptococci showed YidC2 E92 and K253 also to be highly conserved (Supplementary Figure 2). Therefore, we hypothesized that these residues in particular are decisive contributors to the respective functions of YidC1 and YidC2.
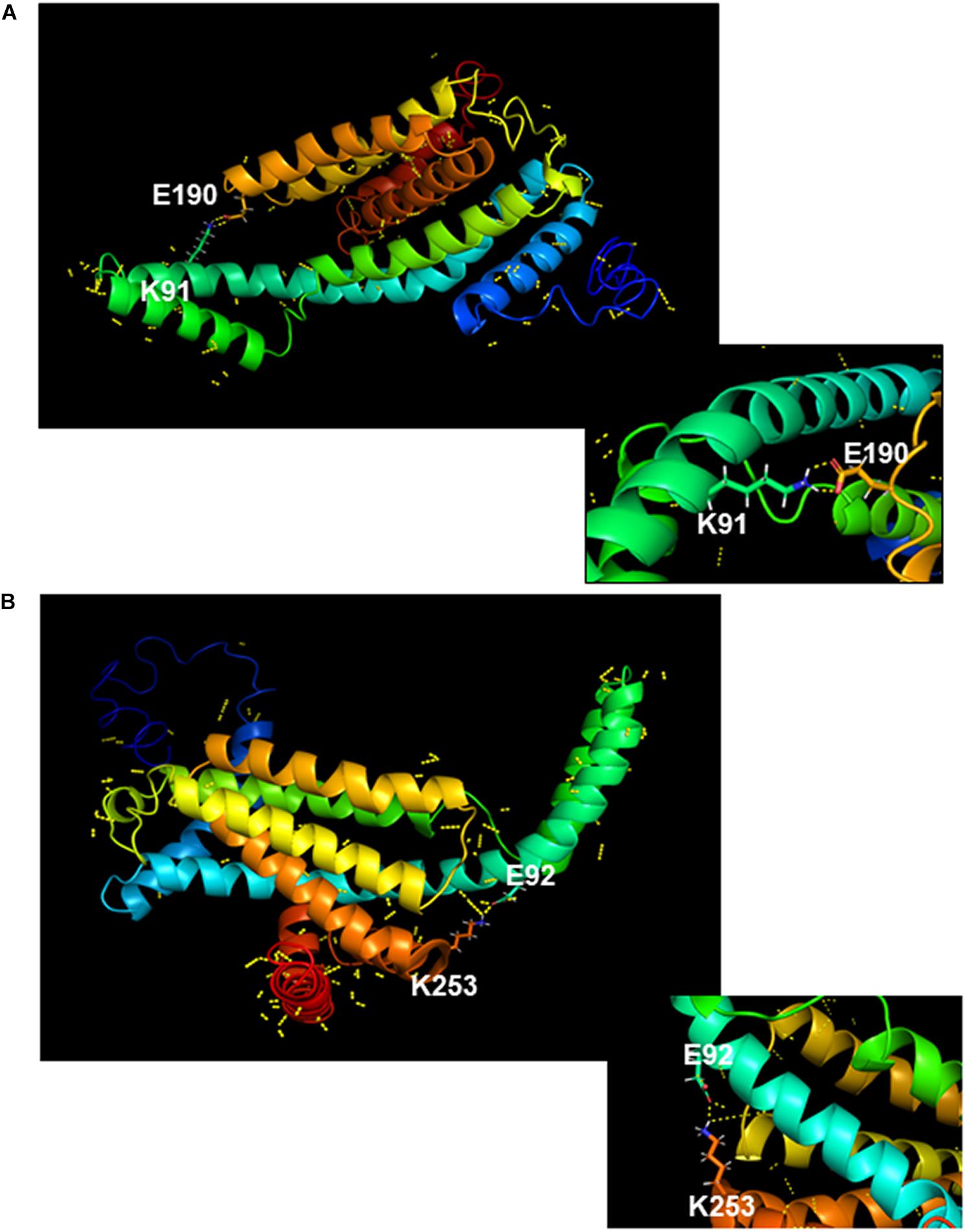
Figure 7. In silico modeling of salt bridge formation between cytoplasmic domains of YidC1 or YidC2. (A) Putative polar contacts on the side chains of YidC1 (yellow) were mapped and displayed using the Pymol Plugin. The salt bridge between residues K91 and E190 is shown enlarged in the box. (B) Putative polar contacts on the side chains of YidC2 (yellow) were mapped and displayed using the Pymol Plugin. The salt bridge between residues E92 and K253 is shown enlarged in the box.
To examine the functional relevance of key residues predicted by in silico analysis to form stabilizing interactions within the YidC1 or YidC2 structures, we used site-directed mutagenesis to change each of these charged residues to an alanine. The modified yidC1 or yidC2 genes were expressed similarly to the chimeric genes under control of the gtfA promoter in a ΔyidC1/ΔyidC2 background for phenotypic characterization. Measurement of growth kinetics under non-stress conditions showed that the strains engineered to produce YidC2E92A or YidC2K253A were clearly affected compared to the unmodified YidC2 control strain. The negative impact of the YidC2 E92 and K253 mutations on growth of the yidC2 ectopic expression strain was also observed by dilution plating of log-phase cultures. In contrast, strains producing YidC1K91A or YidC1E190A did not show any obvious difference in growth compared to ectopic expression of unmodified yidC1 (Figure 8A). Thus, the YidC1 point mutations did not appear to impact its general housekeeping functions. However, when the ability of the point mutant strains to tolerate environmental stress conditions reflective of previously identified YidC1- and YidC2-associated functions was assessed, the apparent importance of the putative salt bridges was striking. The ability of ectopically expressed yidC1 to support S. mutans growth in the presence of excess zinc was clearly abolished by the K91A and E190A point mutations (Figure 8B). Next, we measured the impact of point mutations within YidC2 on growth of S. mutans in THYE acidified to pH 5.0 (Figure 8C). Both the E92A and K253A point mutations destroyed the ability of YidC2 to contend with acid stress. Taken together, these results confirm that the residues predicted to form salt bridges within YidC1 and YidC2, respectively, do in fact contribute to the respective functional activities of each paralog under specific environmental conditions.
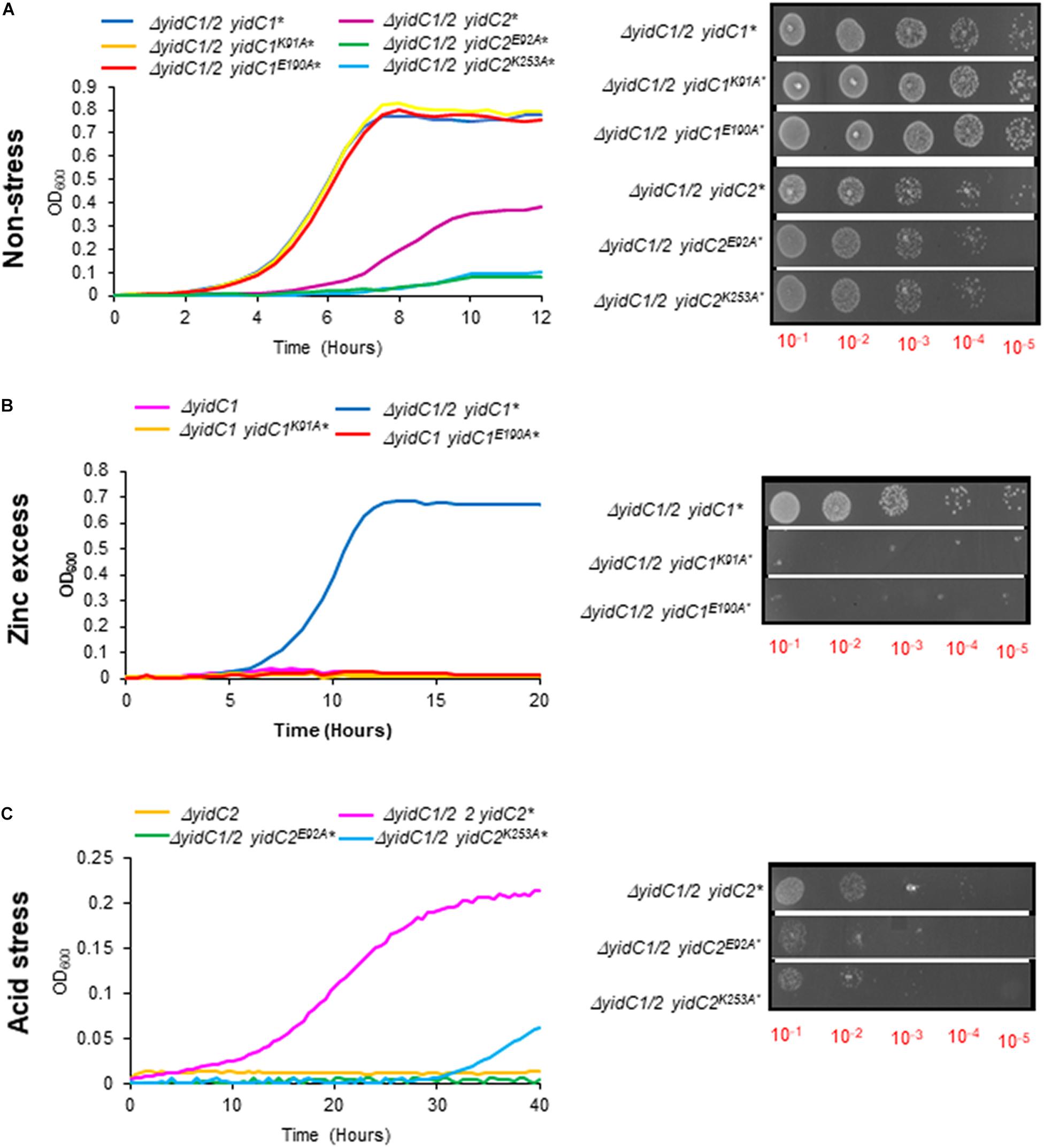
Figure 8. Growth kinetics and efficiency of plating assay of S. mutans strains expressing yidC1 or yidC2 containing point mutations of residues predicted to contribute to salt bridge formation. Growth of strains complemented by ectopic expression (*) from the gtfA locus of unmodified yidC1, yidC2, or yidC1 or yidC2 containing the indicated point mutations, in a double ΔyidC1/ΔyidC2 background under (A) non-stress (THYE, pH 7.0), (B) zinc excess (THYE containing 2.5 mM ZnCl2), or (C) acid stress (THYE, pH 5.0) conditions.
YidC belongs to the Oxa1/Alb3/YidC family of proteins present in all three domains of the life, and plays a pivotal role in the proper folding and insertion of membrane proteins in bacteria (Dalbey and Kuhn, 2015; Chen et al., 2019). When discovered in E. coli, YidC was demonstrated to be essential for viability (Samuelson et al., 2000). Later, discovery of two different paralogs in Gram-positive bacteria, including B. subtilis and S. mutans, showed that at least one YidC is required for growth and survival under various environmental conditions, although they are not equally effective (Errington et al., 1992; Tjalsma et al., 2003; Hasona et al., 2005; Saller et al., 2011; Palmer et al., 2012; Mishra et al., 2019; Zhao et al., 2019). Subsequent proteomic analysis of S. mutans membrane preparations from mutants lacking either yidC1 or yidC2, coupled with characterization of the YidC1 compared to YidC2 interactomes, further revealed paralog-specific activities as well as overlap in their functional capabilities (Mishra et al., 2019; Lara Vasquez et al., 2021). Thus, when one paralog is eliminated, the residual paralog must accomplish many of the basic housekeeping needs of the cell. However, each paralog cannot fully accomplish the specific functions mediated by the other, thereby manifesting in characteristic phenotypic consequences. As expected for paralogous bacterial proteins, divergent SmYidC1 and SmYidC2 amino acid sequences have retained low sequence identity (∼30%). Nevertheless, their similar membrane topologies and largely superimposable predicted 3D structures are consistent with their high degree of functional overlap. Other than disparate roles for the C-terminal tails (Palmer et al., 2012), specific structural features responsible for distinct YidC1- and YidC2-associated functional attributes have not been characterized. The absence of SmYidC1 and SmYidC2 crystallographic data, the numerous yet unidentified paralog-specific membrane-localized substrates, and the current lack of experimental methods to carry out membrane protein insertion assays using exclusively components of Gram-positive translation and translocation machineries have hindered our understanding of YidC1- vs. YidC2-specific structure-function features. In this study, we utilized insights obtained from in silico sequence-structure analyses to engineer all possible cytoplasmic domain-swapped versions of YidC1/2 and tested growth of the chimeric variants under those environmental conditions known (acid, high salt), or predicted (excess metal), to contribute to paralog-specific phenotypes.
An important breakthrough in the field of YidC structure-function occurred when crystal structures of BhYidC2 and E. coli YidC (EcYidC) were determined in 2014 (Kumazaki et al., 2014b,c). Both solved structures demonstrated the presence of a hydrophilic groove formed by the five core TMDs, which was proposed as a conserved structural feature of all YidCs. Structuralanalyses of both species’ YidCs further suggested their C1 loops as a primary point of contact for incoming substrates (Kumazaki et al., 2014a). Another unrelated study utilized EcYidC mutants containing segmental deletions of the C1 loop and demonstrated the significance of the second-half of this domain in rescuing a YidC-depletion strain; although this work proposed that the precise sequence of C1 is not essential for EcYidC function (Chen et al., 2014). More recently, Koch and coworkers used formaldehyde cross-linking of E. coli cells to demonstrate that the YidC C1 loop interacts with the signal recognition particle (SRP), the SRP receptor FtsY, SecY, SecE, and SecG, with a stronger interaction identified with SRP compared to SecYEG (Petriman et al., 2018). Unlike its C1 loop, the BhYidC2 C2 loop was not structurally resolved by X-ray crystallography (Kumazaki et al., 2014c). Molecular dynamic (MD) simulations, however, predicted a contribution of the C2 loop in stabilizing protein structure, especially in reducing flexibility of the C1 loop by formation of an inter-domain salt bridge between D205 (C2) and K109 (C1) (Harkey et al., 2019). Structural analysis of BhYidC2 also revealed that its C1 loop is assessable to the cytoplasm, while the smaller C2 loop appears to be embedded within the lipid bilayer and probably involved in an interaction with lipid heads.
Since the discovery of dual YidC paralogs in the overwhelming majority of Gram-positive bacteria, it has been of broad interest to understand how they are structurally and functionally distinguished. Distinctive lengths and charge distribution of their cytoplasmic tails is a striking feature. Previous work demonstrated the significance of the SmYidC2 tail with respect to its contribution to acid tolerance; however, the underlying mechanism for this result was not fully understood (Palmer et al., 2012). Driessen and coworkers used purified SmYidC1 and SmYidC2 to demonstrate that both of their tails appear to be involved in interactions with ribosomal proteins; although this work was carried out using E. coli ribosomes (Wu et al., 2013). Of note, the longer tails of marine Gram-negative bacteria such as Rhodopirellula baltica and Oceanicaulis alexandrii interacted with E. coli ribosomal proteins, whereas E. coli YidC with a shorter C-terminal tail failed to do so (Seitl et al., 2014). We analyzed the respective interactomes of YidC1 and YidC2, including assessment of the binding partners of their C-terminal tails by 2D-difference gel electrophoresis (DIGE) (Lara Vasquez et al., 2021). These data clearly suggested that the isolated C-terminal tails can interact with multiple ribosomal proteins but do not appear to play an important role in recognizing and binding substrates. Characterization of the YidC1/2 protein interactomes showed that of YidC1 to be larger than that of YidC2, and further identified YidC1-SecYEG and YidC2-SRP interactions in S. mutans (Lara Vasquez et al., 2021). Bacterial two-hybrid experiments revealed that YidC1 could interact with both SecY and Ffh, while YidC2 could only interact with the SRP protein Ffh (Lara Vasquez et al., 2021). This explained in part, why individual deletion of yidC2 or ffh results in similar growth defects and stress-sensitivity profiles, and co-deletion of yidC2 and ffh is lethal (Hasona et al., 2005; Mishra et al., 2019). Taken together with prior interactome data, and the in silico sequence-structure analyses performed in the current study, we hypothesized that their cytoplasmic domains play pivotal roles in conferring functional differences between S. mutans YidC1 and YidC2. The data we obtained support this hypothesis.
In silico analyses showed higher RMSD values, and therefore greater structural deviations from unmodified YidC1 for most of the chimeric YidC1 variants. Yet strains harboring chimeric YidC1 did not manifest significant growth phenotypes under numerous experimental conditions. This suggests that YidC1 can accommodate multiple structural perturbations without loss of function, or in other words, that its structural malleability is associated with functional plasticity. In contrast, in silico analyses of chimeric YidC2 variants showed less structural deviation from unmodified YidC2, with most domain substitutions resulting in impaired YidC2 function as manifested by an inability to support bacterial growth under the conditions tested. This suggests that the more rigid nature of YidC2 compared to YidC1 constrains its ability to mediate insertion of the membrane proteins necessary for bacterial growth and tolerance to environmental stress when its cytoplasmic domains are exchanged for those of YidC1.
An interesting outcome of the current work was the preference for certain combinations of cytoplasmic domains over others, which was especially notable among the chimeric YidC2 variants. The results of in silico analyses of chimeras compared with unmodified YidC1 and YidC2, as evidenced by small (<3 Å) RMSD values, further suggests that certain combinatorial substitutions result in a propensity for the tertiary structure of a given chimera to more closely align with YidC1 or YidC2. In light of in silico structure predictions, combined with insight gained from phenotypic assays, we hypothesized that specific interactions between cytoplasmic domains were responsible for particular structure-function attributes. In fact, Moradi and coworkers had proposed that formation of a salt-bridge between the C1 and C2 loops of B. halodurans YidC2 appears to stabilize the flexible C1 loop (Harkey et al., 2019). When the SmYidC1 and SmYidC2 cytoplasmic domains were evaluated, both paralogs contained multiple oppositely charged residues within close enough proximity (<3 Å) to form salt-bridges (Supplementary Table 6). Most were intra-domain interactions; however, we focused on the two inter-domain interactions identified within YidC1 [K91(C1)-E190(C2)] and YidC2 [E92(C1)-K253(tail)], respectively. Similar to deletion of yidC2, the YidC2K253A mutant had a severe growth phenotype. The other three point mutants also recapitulated environmental stress phenotypes associated with yidC1 or yidC2 deletion. Hence, the predicted inter-domain salt bridges appear to play important roles in stabilizing YidC1 and YidC2 structures that are necessary for paralog-specific activities.
An interesting outcome of the current work was the discovery of a novel yidC1 elimination phenotype. We previously identified pronounced sensitivity to acid, osmotic, or oxidative stress as a result of yidC2 elimination (Hasona et al., 2005). In contrast, elimination of yidC1 had far less obvious consequences apart from a modest impact on maturation of surface proteins and hyperadherence to salivary components (Palmer et al., 2012). However, until now a readily assayable clearcut phenotype associated primarily with yidC1 elimination had not been observed. Our proteomic characterization of S. mutans membrane preparations derived from WT and mutant strains, including ΔyidC1 and ΔyidC2, had suggested overlapping, as well as differential, impacts of elimination of these insertases on the levels of previously characterized, or predicted, membrane-localized metal transporters or regulators (Supplementary Table 5; Mishra et al., 2019). Thus, the survival of S. mutans ΔyidC1 and ΔyidC2 mutants, and the behavior of the ectopic-expression chimeric and control strains, in the presence of excess zinc, iron, and manganese were assessed as part of this work. S. mutans contains multiple transporters for Fe(II) including FeoABC, the SMU_995-998 system, and the Mn(II) transporters SloABC and MntH (Paik et al., 2003; Galvao et al., 2015; Ganguly et al., 2018). Deletion of yidC2 had a more apparent impact on S. mutans tolerance to Fe(II) and Mn(II) excess than deletion of yidC1, with most YidC2-backbone chimeric proteins being negatively impacted by substitution of YidC1 cytoplasmic domains. However, lack of complete complementation of iron and manganese tolerance by ectopic expression of yidC2 in a ΔyidC1/ΔyidC2 background suggests the existence of regulatory circuits that will require extensive future study.
While deletion of yidC2 had a slight impact on zinc tolerance, it did not approach that of yidC1 deletion. Ectopic expression of yidC1 in a ΔyidC1/ΔyidC2 background almost completely restored growth in excess Zn(II) to wild-type levels. Proteomic data revealed a pronounced decrease in levels of the putative metal transporter Smu_2057 associated with elimination of yidC1 compared to yidC2 (Supplementary Table 5). Blast search showed SMU_2057 to be 42% identical and 61% homologous to the B. subtilis metal transporting ATPase, PfeT. Mutation of pfeT led to improved growth in Zn(II) excess conditions (Gaballa and Helmann, 2002), but the molecular mechanism of the PfeT-dependent increase in growth remains elusive (Guan et al., 2015). It has been proposed to be mediated by mismetallation caused by Zn(II) under aerated conditions when PfeT-dependent iron-efflux is inhibited. The role of SMU_2057 in zinc homeostasis in S. mutans, and the potential role of YidC1 in its membrane insertion, will be of much interest in future studies.
In summary, this work successfully revealed distinct functional contributions of cytoplasmic domains to paralog-specific functions of S. mutans YidC1 and YidC2. The novel discovery of functionally significant residues within the S. mutans YidC1/2 cytoplasmic domains is important, especially since bacterial YidC TMDs have received more attention in prior structure-function studies given their membrane localization and direct involvement in substrate insertion. This newly acquired knowledge will provide additional insights as YidC1/2-specific substrates and relevant interactions with other components of the protein translocation machinery are identified in S. mutans and other Gram-positive bacteria.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
SM and LJB conceived the experiments, analyzed the data, and wrote the manuscript. SM performed the experiments and collected data. All authors contributed to the article and approved the submitted version.
This work was supported by NIH NIDCR award DE008007 to LJB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.760873/full#supplementary-material
Ajdic, D., Mcshan, W. M., Mclaughlin, R. E., Savic, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99, 14434–14439. doi: 10.1073/pnas.172501299
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D., and Hamodrakas, S. J. (2008). Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J. Proteome Res. 7, 5082–5093. doi: 10.1021/pr800162c
Ban, X., Lahiri, P., Dhoble, A. S., Li, D., Gu, Z., Li, C., et al. (2019). Evolutionary stability of salt bridges hints its contribution to stability of proteins. Comput. Struct. Biotechnol. J. 17, 895–903. doi: 10.1016/j.csbj.2019.06.022
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Chen, Y., Shanmugam, S. K., and Dalbey, R. E. (2019). The principles of protein targeting and transport across cell membranes. Protein J. 38, 236–248. doi: 10.1007/s10930-019-09847-2
Chen, Y., Soman, R., Shanmugam, S. K., Kuhn, A., and Dalbey, R. E. (2014). The role of the strictly conserved positively charged residue differs among the Gram-positive, Gram-negative, and chloroplast YidC homologs. J. Biol. Chem. 289, 35656–35667. doi: 10.1074/jbc.M114.595082
Conrad, B., and Antonarakis, S. E. (2007). Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 8, 17–35. doi: 10.1146/annurev.genom.8.021307.110233
Dalbey, R. E., and Kuhn, A. (2015). Membrane insertases are present in all three domains of life. Structure 23, 1559–1560. doi: 10.1016/j.str.2015.08.002
Errington, J., Appleby, L., Daniel, R. A., Goodfellow, H., Partridge, S. R., and Yudkin, M. D. (1992). Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J. Gen. Microbiol. 138, 2609–2618. doi: 10.1099/00221287-138-12-2609
Gaballa, A., and Helmann, J. D. (2002). A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45, 997–1005. doi: 10.1046/j.1365-2958.2002.03068.x
Galvao, L. C., Miller, J. H., Kajfasz, J. K., Scott-Anne, K., Freires, I. A., Franco, G. C., et al. (2015). Transcriptional and phenotypic characterization of novel spx-regulated genes in Streptococcus mutans. PLoS One 10:e0124969. doi: 10.1371/journal.pone.0124969
Ganguly, T., Kajfasz, J. K., Miller, J. H., Rabinowitz, E., Galvao, L. C. C., Rosalen, P. L., et al. (2018). Disruption of a novel iron transport system reverses oxidative stress phenotypes of a dpr mutant strain of streptococcus mutans. J. Bacteriol. 2018:200. doi: 10.1128/JB.00062-18
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). The Proteomics Protocols Handbook. Totowa, N.J: Humana Press.
Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308. doi: 10.1093/bioinformatics/15.4.305
Guan, G., Pinochet-Barros, A., Gaballa, A., Patel, S. J., Arguello, J. M., and Helmann, J. D. (2015). PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol. Microbiol. 98, 787–803. doi: 10.1111/mmi.13158
Hamada, S., and Slade, H. D. (1980). Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44, 331–384. doi: 10.1128/mr.44.2.331-384.1980
Harkey, T., Govind Kumar, V., Hettige, J., Tabari, S. H., Immadisetty, K., and Moradi, M. (2019). The role of a crystallographically unresolved cytoplasmic loop in stabilizing the bacterial membrane insertase YidC2. Sci. Rep. 9:14451. doi: 10.1038/s41598-019-51052-9
Hasona, A., Crowley, P. J., Levesque, C. M., Mair, R. W., Cvitkovitch, D. G., Bleiweis, A. S., et al. (2005). Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. USA 102, 17466–17471. doi: 10.1073/pnas.0508778102
Koch, H. G., Moser, M., and Muller, M. (2003). Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146, 55–94. doi: 10.1007/s10254-002-0002-9
Kol, S., Nouwen, N., and Driessen, A. J. (2008). The charge distribution in the cytoplasmic loop of subunit C of the F1F0 ATPase is a determinant for YidC targeting. J. Biol. Chem. 283, 9871–9877. doi: 10.1074/jbc.M709408200
Krogh, A., Larsson, B., Vonheijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumazaki, K., Chiba, S., Takemoto, M., Furukawa, A., Nishiyama, K., Sugano, Y., et al. (2014a). Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520. doi: 10.1038/nature13167
Kumazaki, K., Kishimoto, T., Furukawa, A., Mori, H., Tanaka, Y., Dohmae, N., et al. (2014b). Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4:7299. doi: 10.1038/srep07299
Kumazaki, K., Tsukazaki, T., Nishizawa, T., Tanaka, Y., Kato, H. E., Nakada-Nakura, Y., et al. (2014c). Crystallization and preliminary X-ray diffraction analysis of YidC, a membrane-protein chaperone and insertase from Bacillus halodurans. Acta Crystallogr. F Struct. Biol. Commun. 70, 1056–1060. doi: 10.1107/S2053230X14012540
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Lara Vasquez, P., Mishra, S., Kuppuswamy, S. K., Crowley, P. J., and Brady, L. J. (2021). Protein Interactomes of Streptococcus mutans YidC1 and YidC2 Membrane Protein Insertases Suggest SRP pathway-independent- and -dependent functions, respectively. mSphere 6:e01308-20. doi: 10.1128/mSphere.01308-20
Lemos, J. A., Palmer, S. R., Zeng, L., Wen, Z. T., Kajfasz, J. K., Freires, I. A., et al. (2019). The Biology of Streptococcus mutans. Microbiol. Spectr. 7:512018. doi: 10.1128/9781683670131.ch27
Lewis, N. E., and Brady, L. J. (2015). Breaking the bacterial protein targeting and translocation model: oral organisms as a case in point. Mol. Oral Microbiol. 30, 186–197. doi: 10.1111/omi.12088
Lockwood, W. R., Lawson, L. A., Smith, D. L., Mcneil, K. M., and Morrison, F. S. (1974). Streptococcus mutans endocarditis. Report of a case. Ann. Intern. Med. 80, 369–370. doi: 10.7326/0003-4819-80-3-369
McGhie, D., Hutchison, J. G., Nye, F., and Ball, A. P. (1977). Infective endocarditis caused by Streptococcus mutans. Br. Heart J. 39, 456–458. doi: 10.1136/hrt.39.4.456
Mishra, S., Crowley, P. J., Wright, K. R., Palmer, S. R., Walker, A. R., Datta, S., et al. (2019). Membrane proteomic analysis reveals overlapping and independent functions of Streptococcus mutans Ffh, YidC1, and YidC2. Mol. Oral Microbiol. 34, 131–152. doi: 10.1111/omi.12261
Moore, C. M., and Helmann, J. D. (2005). Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8, 188–195. doi: 10.1016/j.mib.2005.02.007
Moore, J., Keane, C. T., and Tomkin, G. H. (1977). Streptococcus mutans endocarditis. Ir. J. Med. Sci. 146, 144–145. doi: 10.1007/BF03030949
Nomura, R., Nakano, K., Nemoto, H., Fujita, K., Inagaki, S., Takahashi, T., et al. (2006). Isolation and characterization of Streptococcus mutans in heart valve and dental plaque specimens from a patient with infective endocarditis. J. Med. Microbiol. 55, 1135–1140. doi: 10.1099/jmm.0.46609-0
Paik, S., Brown, A., Munro, C. L., Cornelissen, C. N., and Kitten, T. (2003). The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185, 5967–5975. doi: 10.1128/JB.185.20.5967-5975.2003
Palmer, S. R., Crowley, P. J., Oli, M. W., Ruelf, M. A., Michalek, S. M., and Brady, L. J. (2012). YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 158, 1702–1712. doi: 10.1099/mic.0.059139-0
Petersen, T. N., Brunak, S., Vonheijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Petriman, N. A., Jauss, B., Hufnagel, A., Franz, L., Sachelaru, I., Drepper, F., et al. (2018). The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci. Rep. 8:578. doi: 10.1038/s41598-017-19019-w
Price, C. E., Otto, A., Fusetti, F., Becher, D., Hecker, M., and Driessen, A. J. (2010). Differential effect of YidC depletion on the membrane proteome of Escherichia coli under aerobic and anaerobic growth conditions. Proteomics 10, 3235–3247. doi: 10.1002/pmic.201000284
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Saller, M. J., Otto, A., Berrelkamp-Lahpor, G. A., Becher, D., Hecker, M., and Driessen, A. J. (2011). Bacillus subtilis YqjG is required for genetic competence development. Proteomics 11, 270–282. doi: 10.1002/pmic.201000435
Samuelson, J. C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., et al. (2000). YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641. doi: 10.1038/35020586
Schneider, T. D., and Stephens, R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. doi: 10.1093/nar/18.20.6097
Scotti, P. A., Urbanus, M. L., Brunner, J., Degier, J. W., Von Heijne, G., Van Der Does, C., et al. (2000). YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549. doi: 10.1093/emboj/19.4.542
Seitl, I., Wickles, S., Beckmann, R., Kuhn, A., and Kiefer, D. (2014). The C-terminal regions of YidC from Rhodopirellula baltica and Oceanicaulis alexandrii bind to ribosomes and partially substitute for SRP receptor function in Escherichia coli. Mol. Microbiol. 91, 408–421. doi: 10.1111/mmi.12465
Shindyalov, I. N., and Bourne, P. E. (1998). Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11, 739–747. doi: 10.1093/protein/11.9.739
Smith, J. P., Marymont, J. H., and Schweers, J. H. (1977). Subacute bacterial endocarditis due to Streptococcus mutans. Am. J. Med. Technol. 43, 429–432.
Steinberg, R., Knupffer, L., Origi, A., Asti, R., and Koch, H. G. (2018). Co-translational protein targeting in bacteria. FEMS Microbiol. Lett. 365:95. doi: 10.1093/femsle/fny095
Stoffel, K. H. A. W. (1993). Tmbase-a database of membrane spanning protein segments. Biol. Chem. Hoppe. Seyler. 347:166.
Tjalsma, H., Bron, S., and Vandijl, J. M. (2003). Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278, 15622–15632. doi: 10.1074/jbc.M301205200
Urbanus, M. L., Scotti, P. A., Froderberg, L., Saaf, A., Degier, J. W., Brunner, J., et al. (2001). Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529. doi: 10.1093/embo-reports/kve108
van Bloois, E., Janhaan, G., Degier, J. W., Oudega, B., and Luirink, J. (2004). F(1)F(0) ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 576, 97–100. doi: 10.1016/j.febslet.2004.08.069
van der Laan, M., Nouwen, N., and Driessen, A. J. (2004). SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. J. Biol. Chem. 279, 1659–1664. doi: 10.1074/jbc.M306527200
Vose, J. M., Smith, P. W., Henry, M., and Colan, D. (1987). Recurrent Streptococcus mutans endocarditis. Am. J. Med. 82, 630–632. doi: 10.1016/0002-9343(87)90111-2
Williams, M. L., Crowley, P. J., Hasona, A., and Brady, L. J. (2014). YlxM is a newly identified accessory protein that influences the function of signal recognition particle pathway components in Streptococcus mutans. J. Bacteriol. 196, 2043–2052. doi: 10.1128/JB.01465-13
Wu, Z. C., De, K. E. Y. Z. E. R., Berrelkamp-Lahpor, G. A., and Driessen, A. J. (2013). Interaction of Streptococcus mutans YidC1 and YidC2 with translating and nontranslating ribosomes. J. Bacteriol. 195, 4545–4551. doi: 10.1128/JB.00792-13
Yi, L., Celebi, N., Chen, M., and Dalbey, R. E. (2004). Sec/SRP requirements and energetics of membrane insertion of subunits a, b, and c of the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 279, 39260–39267. doi: 10.1074/jbc.M405490200
Zallot, R., Harrison, K. J., Kolaczkowski, B., and Decrecylagard, V. (2016). Functional annotations of paralogs: a blessing and a curse. Life 6:39. doi: 10.3390/life6030039
Zhao, H., Sachla, A. J., and Helmann, J. D. (2019). Mutations of the Bacillus subtilis YidC1 (SpoIIIJ) insertase alleviate stress associated with sigmaM-dependent membrane protein overproduction. PLoS Genet 15:e1008263. doi: 10.1371/journal.pgen.1008263
Keywords: structure-function, YidC, Streptococcus mutans, insertase, paralogs, bacterial stress tolerance
Citation: Mishra S and Brady LJ (2021) The Cytoplasmic Domains of Streptococcus mutans Membrane Protein Insertases YidC1 and YidC2 Confer Unique Structural and Functional Attributes to Each Paralog. Front. Microbiol. 12:760873. doi: 10.3389/fmicb.2021.760873
Received: 18 August 2021; Accepted: 30 September 2021;
Published: 02 November 2021.
Edited by:
Ignacio Arechaga, University of Cantabria, SpainReviewed by:
Yoshihiro Yamaguchi, Osaka City University, JapanCopyright © 2021 Mishra and Brady. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: L. Jeannine Brady, amJyYWR5QGRlbnRhbC51ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.