
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 October 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.759208
This article is part of the Research TopicResistance to third-generation TetracyclinesView all 10 articles
 Yujiao Wang1†
Yujiao Wang1† Bo Zhu2†
Bo Zhu2† Min Liu3†
Min Liu3† Xiutao Dong1
Xiutao Dong1 Jianping Ma1
Jianping Ma1 Xiaofeng Li1
Xiaofeng Li1 Fang Cheng1
Fang Cheng1 Jianzhuang Guo1
Jianzhuang Guo1 Sumei Lu1
Sumei Lu1 Furong Wan1
Furong Wan1 Yingying Hao4
Yingying Hao4 Wanshan Ma1*
Wanshan Ma1* Mingju Hao1*
Mingju Hao1* Liang Chen5,6
Liang Chen5,6Tigecycline serves as one of the last-resort antibiotics to treat severe infections caused by carbapenem-resistant Enterobacterales. Recently, a novel plasmid-mediated resistance-nodulation-division (RND)-type efflux pump gene cluster, TmexCD1-ToprJ1, and its variants, TmexCD2-ToprJ2 and TmexCD3-ToprJ3, encoding tetracyclines and tigecycline resistance, were revealed. In this study, we reported three TmexCD2-ToprJ2-harboring Klebsiella species strains, collected from two teaching tertiary hospitals in China, including one K. quasipneumoniae, one K. variicola, and one K. michiganensis. The three strains were characterized by antimicrobial susceptibility testing (AST), conjugation assay, WGS, and bioinformatics analysis. AST showed that K. variicola and K. quasipneumoniae strains were resistant to tigecycline with MIC values of 4μg/ml, whereas the K. michiganensis was susceptible to tigecycline with an MIC value of 1μg/ml. The TmexCD2-ToprJ2 clusters were located on three similar IncHI1B plasmids, of which two co-harbored the metallo-β-lactamase gene blaNDM-1. Conjugation experiments showed that all three plasmids were capable of self-transfer via conjugation. Our results showed, for the first time, that this novel plasmid-mediated tigecycline resistance mechanism TmexCD2-ToprJ2 has spread into different Klebsiella species, and clinical susceptibility testing may fail to detect. The co-occurrence of blaNDM-1 and TmexCD2-ToprJ2 in the same plasmid is of particular public health concern as the convergence of “mosaic” plasmids can confer both tigecycline and carbapenem resistance. Its further spread into other clinical high-risk Klebsiella clones will likely exacerbate the antimicrobial resistance crisis. A close monitoring of the dissemination of TmexCD-ToprJ encoding resistance should be considered.
Tigecycline is one of the last-resort antibiotics used to treat severe infections caused by carbapenem-resistant Enterobacterales (Cheng et al., 2020). However, increasing studies reported the emergence of tigecycline resistance in clinical settings, which is frequently caused by the overexpression of non-specific active efflux pumps [tet(A) and tet(K)] or mutations within the drug-binding site in the ribosome [tet(M); Grossman, 2016; Linkevicius et al., 2016]. In Klebsiella pneumoniae, tigecycline resistance is also frequently associated with the overexpression of ramA, which can directly regulate multidrug resistance efflux pumps AcrAB and OqxAB (Ruzin et al., 2005, 2008). These tigecycline resistances are primarily mediated by chromosome-encoding mechanisms thus could not be easily transferred horizontally. The newly emerging mobile tigecycline resistance mechanism is of particular public health concern (He et al., 2019; Lv et al., 2020). The enzymatic modification gene variants [tet(X)] are highly transferable between species (Linkevicius et al., 2016; He et al., 2019; Sun et al., 2019).
Recently, a novel plasmid-mediated resistance-nodulation-division (RND)-type efflux pump gene cluster, TmexCD1-ToprJ1, has been identified in K. pneumoniae (Lv et al., 2020). This gene cluster was first identified in animal isolates but soon later was also found in clinical isolates (Sun et al., 2020). TmexCD1-ToprJ1 was initially reported in China, but it has now been found in clinical K. pneumoniae isolates outside of China (Hirabayashi et al., 2021), suggesting this resistance has started to spread into other global regions. Two homologous variants, TmexCD2-ToprJ2 (Wang et al., 2021a) and TmexCD3-ToprJ3 (Wang et al., 2021b), have also been identified in Raoultella ornithinolytica and Proteus mirabilis, displaying similar tigecycline resistance profiles. Worrisomely, the mobile tigecycline gene clusters have also been found in clinical carbapenem-resistant K. pneumoniae strains (Chiu et al., 2017). Here, we reported the identifications of three clinical TmexCD2-ToprJ2-encoding Klebsiella strains, including two carbapenem-resistant strains co-harboring blaVIM-8/NDM-5 or blaNDM-1.
Klebsiella variicola strain JNQH579 was recovered from a sputum sample of a 64-year male patient in intensive care units (ICUs) at a tertiary hospital in Jinan City, Shandong Province, in March 2021, who had been hospitalized for 17days due to severe pneumoniae and renal failure. The patient had received tigecycline treatment for 7days at a dosage of 100mg (IV) q12h before the isolation of the strain. Based on the antibiotic susceptibility testing results, antibiotic therapy was switched to aztreonam 0.5g (IV) q8h in combination with tigecycline. The respiratory symptoms improved after antibiotic treatment and the patient continued to be hospitalized for 3months due to cardiovascular and renal disease. Klebsiella quasipneumoniae strain JNQH473 was recovered from a urine sample of a 1-month-old infant with sepsis at a tertiary hospital in Xiamen City, Fujian Province, in August 2019, who was hospitalized at the department of pediatrics for 7days. During her hospitalization, the neonate received multiple antimicrobial treatments, including cefotaxime, vancomycin, and ceftazidime. The patient was fully recovered after antimicrobial and supportive treatment and was discharged on hospital day 13. Klebsiella michiganensis strain JNQH491 was recovered from a blood culture of a 48-year male patient at the department of oncology, who had received chemotherapy due to nasopharyngeal carcinoma in the same hospital as JNQH473. The patient was discharged home after a cycle of chemotherapy on hospital day 11. The overall strain features of the three strains are listed in Table 1. All the patients reported no recent travel abroad.
Antibiotic susceptibility testing (AST) was performed using the VITEK 2 (bioMérieux, Nürtingen, Germany) system. Minimum inhibitory concentrations (MICs) for tigecycline were performed by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. ATCC 25922 (Escherichia coli) and ATCC 27853 (Pseudomonas aeruginosa) were used as quality control strains for susceptibility testing. All the tests were performed in duplicate in different days. The breakpoints were interpreted according to CLSI guidelines except for tigecycline, of which the EUCAST epidemiological cutoff value >2μg/ml (for K. pneumoniae) was used.1
The combination Oxford Nanopore (MinION system) and Illumina sequencing (NovaSeq) were used to achieve a high-quality genome assembly. First, we derived fastq read sequences from MinION raw electric signal fast5 files using guppy 3.2.2 with the high accuracy flip-flop algorithm. Adapters were trimmed out with Porechop.2 Low-quality reads were filtered out using trimmomatic 0.38 (Bolger et al., 2014). The filtered nanopore reads were de novo assembled with Flye 2.8.3 (Kolmogorov et al., 2019). Then, the obtained assemblies were polished using Nanopore reads by Racon 1.4.3 (1–4 iterations; Vaser et al., 2017). Next, the polished sequences were additionally corrected using Illumina reads by Pilon 1.23 until no changes occur (Walker et al., 2014). The whole-genome sequences were annotated by Prokka (Seemann, 2014) and RAST (Brettin et al., 2015), followed by manually curations.
The in silico multilocus sequence typing (MLST) was carried out using MLST v. 2.19.0,3 and antibiotic resistance/plasmid replicon gene detections were carried out using ABRicate v. 0.9.94 using CARD (Jia et al., 2017) and PlasmidFinder (Carattoli et al., 2014) database, respectively. The intrinsic variants of oqxAB, chromosomal ampH and fosA, which confer resistance to quinolones, β-lactams and fosfomycin, respectively, in Enterobacterales rather than Klebsiella species, are therefore not reported (Henderson et al., 1997; Lam et al., 2021). Kleborate v. 1.0.0 (Lam et al., 2021) was used for Klebsiella K locus and O locus typing. The comparative analysis of TmexCD2-ToprJ2 harboring plasmids was done by BLASTn and illustrated using CGView Server (Grant and Stothard, 2008). Easyfig (Sullivan et al., 2011) was used to visualize the genetic context comparisons. Selected plasmids were compared with Mauve 2.3.1 (Darling et al., 2010), followed by visualization using genoPlotR (Guy et al., 2010). Plasmid distance trees were generated using Mashtree (Katz et al., 2019). Alignment between three TmexCD-ToprJ variants was done by Clustal Omega (Madeira et al., 2019). ISFinder5 was used to identify ISs. Tn number was identified using Tn Number Registry (Roberts et al., 2008). In order to examine the distribution and relationships of TmexCD2-ToprJ2 and its variants (TmexCD1-ToprJ1, TmexCD2-ToprJ2), plasmid sequences were downloaded from NCBI6 and compared.
Conjugation experiments were performed by plate mating using E. coli J53 (AziR) as the recipient as described in our previous study (Hao et al., 2021). Briefly, overnight cultures of the donor strains and the recipient strain E. coli J53 were mixed (1:1) and applied onto an LB agar plate, followed by overnight culture at 37°C. Transconjugants were selected on Mueller-Hinton (MH) agar containing sodium azide (200μg/ml) and tigecycline (0.5μg/ml). The presence of TmexCD2-ToprJ2 and IncHI1B replicons, as well as blaNDM-1 and sul1 resistance genes in transconjugants, was confirmed by PCR (Supplementary Table S1). Conjugation frequency was calculated by dividing the number of transconjugants by the number of recipient cells. AST of the E. coli J53 transconjugants was performed as described above.
The MICs for JNQH473, JNQH491, and JNQH579 were shown in Table 2. The MICs of tigecycline in JNQH473 and JNQH579 were both 4μg/ml. However, JNQH491 was susceptible to tigecycline with a MIC value of 1μg/ml. These strains were resistant to almost all tested β-lactam antibiotics except that JNQH491 and JNQH579 were susceptible to aztreonam. All the strains were susceptible to amikacin with an MIC value of less than 2μg/ml.
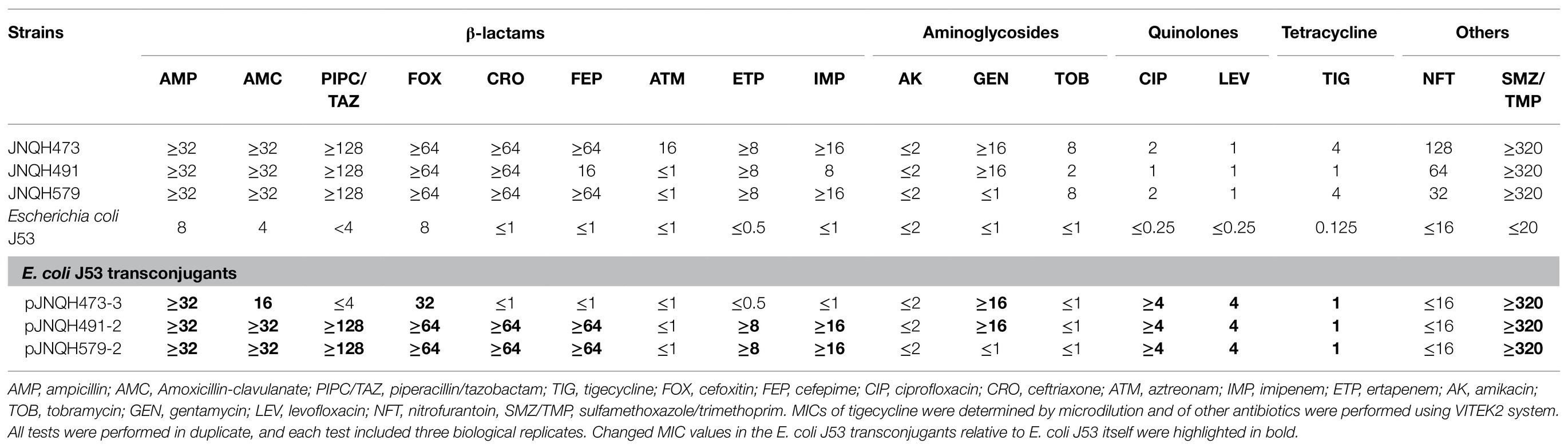
Table 2. Minimum inhibitory concentration (MIC) profiles of parental strains and trans-conjugants (μg/ml).
Strain JNQH473 was classified as sequence type 571 (ST571) based on the in silico MLST, and it belonged to KL64 capsule and O5 lipopolysaccharide serotypes (Table 1). It harbored a 5.39-Mb chromosome and four plasmids, designated pJNQH473-1 (229.2-Kb), pJNQH473-2 (70.9-Kb), pJNQH473-3 (297.9-Kb), and pJNQH473-4 (46.1-Kb), respectively (Table 3). Two metallo-β-lactamase (blaMBL) genes, blaIMP-8 and blaNDM-5, were located on incompatible FIB type plasmid pJNQH473-1 and IncX3 type plasmid pJNQH473-4, respectively. blaIMP-8 was located in class 1 integron In655 carrying the gene cassette of blaIMP-8-aacA4 (Jiang et al., 2017), and blaNDM-5 was located in an ΔTn125-like region containing bleMBL and IS26 genes downstream and IS3000, IS30, and IS5 family transposase genes upstream. The TmexCD2-ToprJ2 gene cluster was located on an IncHI1B type plasmid, pJNQH473-3. In addition to TmexCD2-ToprJ2 genes, nine antimicrobial resistance genes were found on the same plasmid, including two copies of sulfonamide resistance gene sul1 and single copy of aminoglycoside resistance gene aac(3)-IId and aadA16, quinolone-resistant gene qnrS1, macrolides resistance gene mphA, trimethoprim resistance gene dfrA27, the β-lactamase gene blaTEM-1D, and rifampicin resistance ribosyltransferase gene arr-3.
Strain JNQH491 belonged to ST109 type (Table 1). The K and O locus could not be classified according to the currently available K and O loci database. The genome contained a 6.04-Mb chromosome and two plasmids, pJNQH491-1 (205.5-Kb) and pJNQH491-2 (307.4-Kb; Table 3). The TmexCD2-ToprJ2 gene cluster was located on pJNQH491-2, which is an IncHI1B type plasmid, co-harboring multiple resistant genes, including the blaMBL and blaNDM-1 (Figure 1). The pJNQH491-1 plasmid had an IncFIB replicon and did not carry any known resistance genes.
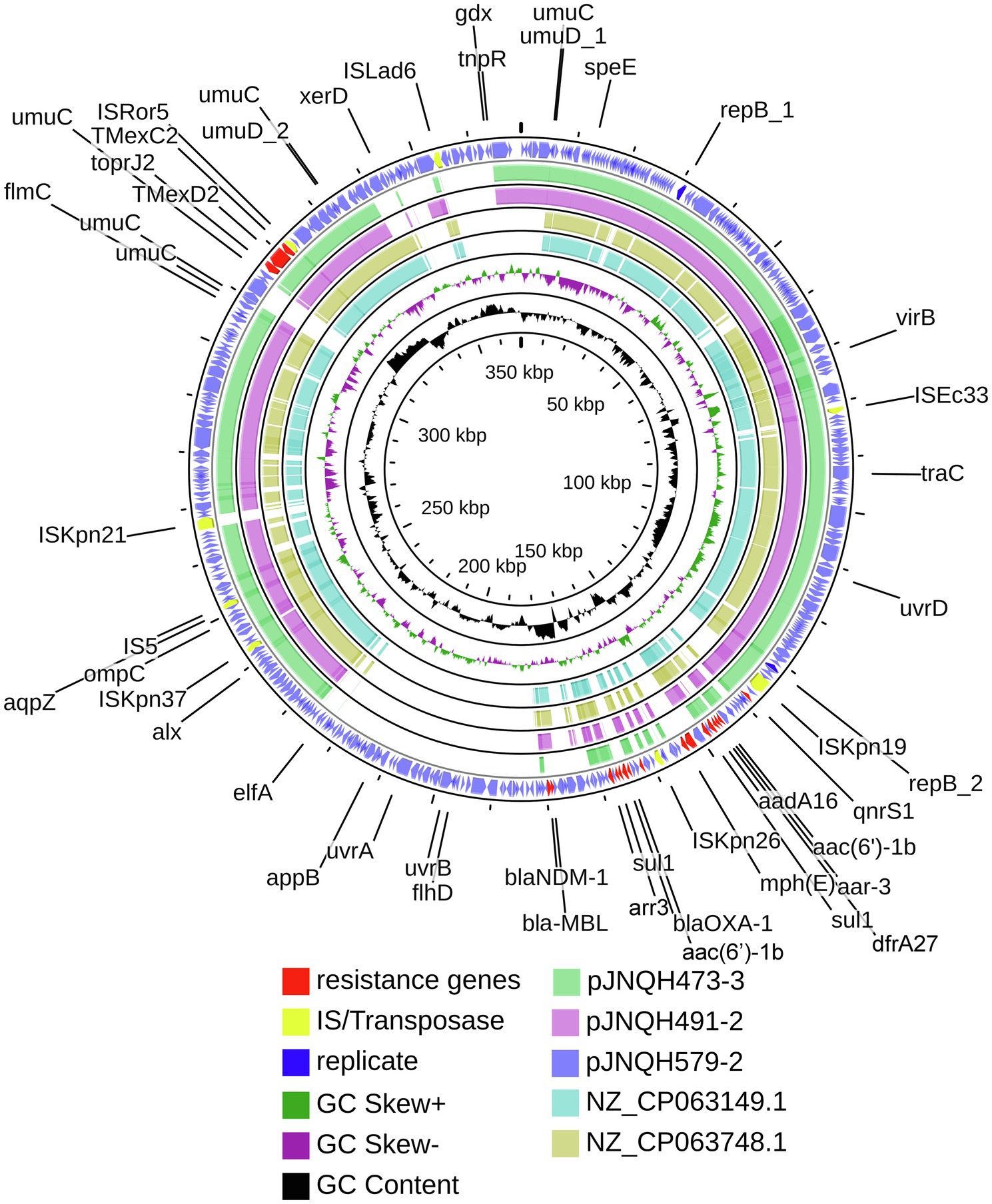
Figure 1. Comparative structural analysis of pJNQH473-3, pJNQH491-2, pJNQH579-2, and two TmexCD2-ToprJ2 harboring plasmids NZ_CP063149.1 and NZ_CP063748.1. Open reading frames (ORFs) of pJNQH579-2 are shown as the outermost ring, with plasmid replicons, insertion sequences (IS), and antimicrobial resistance genes highlighted.
Strain JNQH579 belonged to ST2013 and harbored an OL103 serotype (Table 1). The genome included a 5.58-Mb chromosome and two plasmids, designated pJNQH579-1 (197.4-Kb) and pJNQH579-2 (369.3-Kb), respectively (Table 3). The TmexCD2-ToprJ2 gene cluster was carried by a 34,827bp mobile region, which was located on pJNQH579-2 plasmid (Figure 2). pJNQH579-2 is also an IncHI1B type plasmid, co-harboring multiple resistance genes [aac(6′)-Ib, aadA16, qnrS1, mphE, msrE, catB3, catII.2, arr-3, sul1, dfrA27, blaOXA-1, and blaNDM-1; Table 3]. Notably, the blaNDM-1 carbapenem-resistant gene was co-existed with TmexCD2-ToprJ2 in pJNQH579-2 plasmid. blaNDM-1 was found within a truncated transposon Tn125 (Figure 3), with the structure of “ΔISAba125-blaNDM-1-bleMBL-tat-dvt-groESL- tnpAISCR21” (Poirel et al., 2012). The pJNQH579-1 plasmid had an IncFIB replicon but did not carry any known resistance genes.
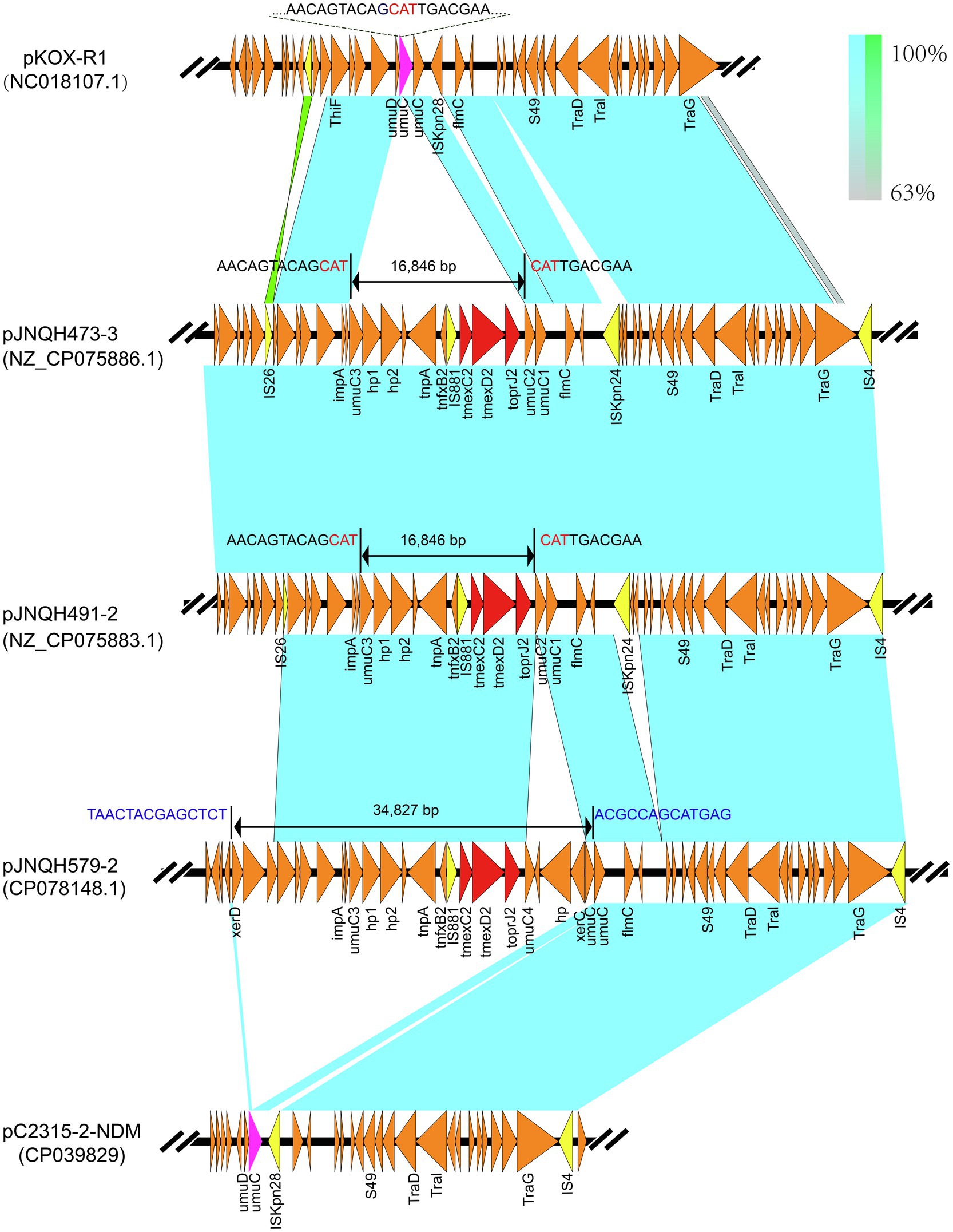
Figure 2. Linear comparisons of TmexCD2-ToprJ2-bearing genetic contexts in pJNQH473-3 and that in pJNQH579-2, pC2315-2-NDM, and plasmid pKOX_R1(NC018107.1). Light blue shading indicates shared regions of homology, while green shading indicates inversely displayed regions of homology. Colored arrows indicate ORFs. The red arrows indicate the antibiotic resistance genes. The yellow arrows indicate IS. The nucleotide sequence of the umuC gene (green arrow) representing the insertion site is shown above the gene. The 16,846bp putative “hp1-hp2-tnpA-tnfxB2-IS881-TmexCD2-ToprJ2” transposon located on pJNQH473-3 and 34,827bp larger putative transposon on pJNQH579-2 are marked with bilateral black arrows.
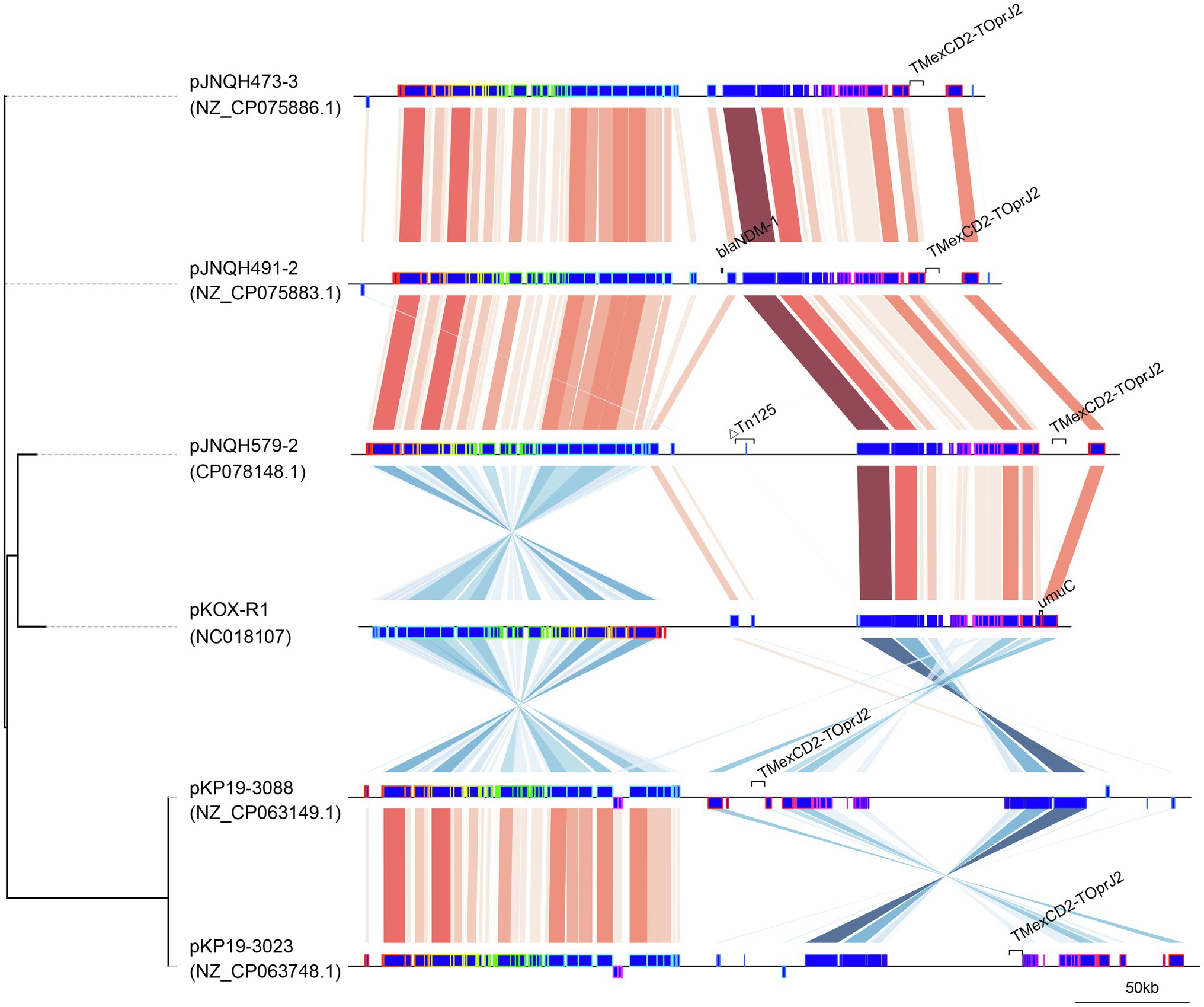
Figure 3. Comparison of linear plasmid maps of four TmexCD2-ToprJ2-bearing plasmids and the related plasmid pKOX-R1. The genomes were compared with Mauve and the elements were designated as Mauve blocks. Red shaded regions between plasmids indicates shared regions of homology, while blue shading indicates inversely displayed regions of homology. The tree was created using Mashtree.
The TmexCD2-ToprJ2 cluster in the three strains had almost 100% nucleotide identities to the cluster of the originally reported TmexCD2-ToprJ2 (Wang et al., 2021a). Plasmid sequence comparison demonstrated that pJNQH473-3 and pJNQH491-2 had highly conserved plasmid synteny and structure, with over 99.9% nucleotide identities (Figure 3). pJNQH491-2 had a blast query coverage of 74% and over 99% nucleotide identities with pJNQH579-2. Further plasmid BLAST query against GenBank database showed that pJNQH473-3 and pJNQH491-2 were closely related to plasmid pKOX-R1 (NC018107), isolated from Taiwan in K. michiganensis E718 (Figure 3). In comparison with pKOX-R1, a 16,846bp region, containing the TmexCD2-ToprJ2, was inserted into the umuC gene of pJNQH473-3, generated a 3bp (CAT) direct repeats (Figure 2). The three pJNQH plasmids contain the same TmexCD2-ToprJ2 structure of “hp1-hp2-tnpA-tnfxB2-IS881-TmexCD2-ToprJ2,” with an average GC content of 42.3%. pJNQH579-2 was found to be closely related to plasmid pC2315-2-NDM (CP039829). The putative “hp1-hp2-tnpA-tnfxB2-IS881-TmexCD2-ToprJ2” transposon was surrounded by XerC, XerD, and additional hypothetical genes. This region was also inserted into the umuC gene therefore constituted a putative larger transposon unit of 34,827bp (Figure 2).
TmexCD2-ToprJ2 cluster shares a high similarity to TmexCD1-ToprJ1, of which the variants of TmexC, TmexD, and ToprJ genes had 98.02, 96.75, and 99.93% nucleotide identities and 97.67, 97.61, and 99.79% amino acid identities between each other. Compared with TmexCD3-ToprJ3, the variants of TmexC, TmexD, and ToprJ genes shared 94, 96.72, and 99.86% nucleotide identities and 97.12, 98.08, and 99.79% amino acid identities. ToprJ gene shares the highest similarity at the amino acid level among the three variants, of which ToprJ2 differs from ToprJ1 and ToprJ3 by a single amino acid substitution (Ala47Thr; Supplementary Figure S1).
The analysis of plasmid database revealed that a total of 43 plasmids carried TMexCD1-ToprJ1 (n=23), TmexCD2-ToprJ2 (n=8), and TmexCD3-ToprJ3 (n=12) gene clusters (with 100% gene coverage and over 99.97% identity), which were distributed mainly in Enterobacteriaceae and P. aeruginosa (Figure 4). TmexCD1-ToprJ1 and TmexCD2-ToprJ2 were mostly identified in K. pneumoniae, whereas TmexCD3-ToprJ3 was more frequently found in P. aeruginosa. In addition to Klebsiella, the TmexCD2-ToprJ2 harboring plasmids were also found in R. ornithinolytica (n=1), Citrobacter freundii (n=1). Most TmexCD2-toprJ2 gene clusters were associated with IncFII (n=3), IncHI1B (n=3), and IncQ (n=2) plasmids. Of note, six TmexCD2-ToprJ2 harboring plasmids also co-harbored carbapenem-resistant genes, including four blaNDM-1 and two blaKPC-1 genes.
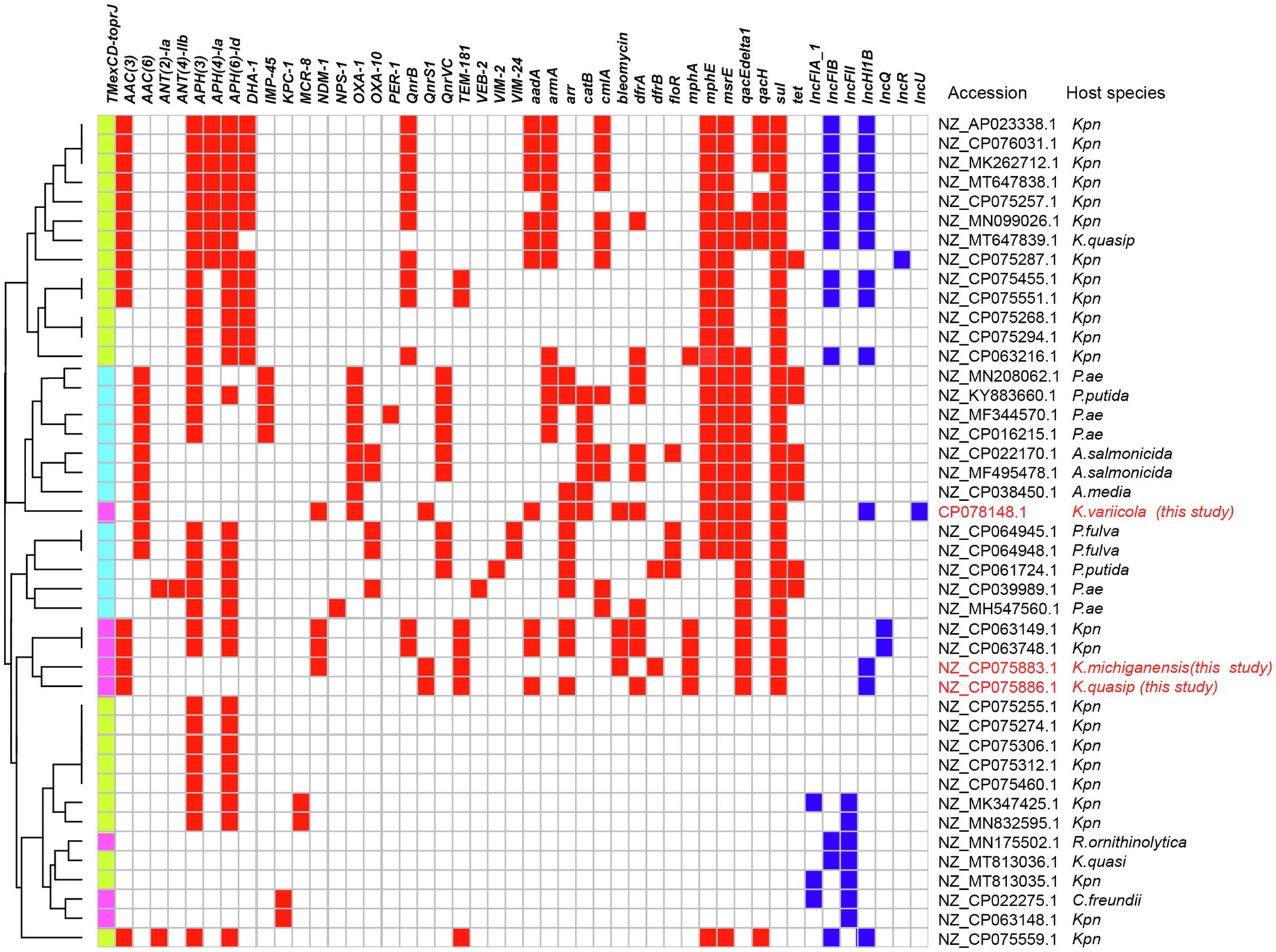
Figure 4. TMexCD-toprJ-harboring plasmids. The heatmap shows the distribution of plasmid replicons (dark blue boxes) and antibiotic resistance genes (red boxes) detected within 51 TMexCD-toprJ-harboring plasmids. The variants of TMexCD-toprJ resistance genes are indicated in green (TMexCD1-toprJ1), pink (TMexCD2-toprJ2), and light blue (TMexCD3-toprJ3) boxes. GenBank accession numbers and species are listed on the right-hand side. Kpn, Klebsiella pneumoniae; K. quasip, Klebsiella quasipneumoniae; Pae, Pseudomonas aeruginosa; P. putida, Pseudomonas putida; A. salmonicida, Aeromonas salmonicida; A. media, Aeromonas media; K. variicola, Klebsiella variicola; P. fulva, Pseudomonas fulva; P. sp., Pseudomonas species; R. ornithinolytica, Raoultella ornithinolytica; C. freundii, Citrobacter freundii.
The TmexCD2-ToprJ2 harboring IncHI1B plasmids were successfully transferred into E. coli J53 from three JNQH strains. Further PCRs confirmed that other plasmids in the donor strains were not co-transferred along with the TmexCD2-ToprJ2 harboring IncHI1B plasmids into the recipient E. coli J53 strain (Supplementary Figure S2). The conjugation frequency was 10−6, 10−6, and 10−5 for JNQH473, JNQH491, and JNQH579 per recipient cell, respectively. The MICs for the E.coli J53 transconjugants are shown in Table 2. The MICs of tigecycline against E. coli J53 transconjugants were 1μg/ml, which were 8-fold higher than that of the E. coli J53 itself. In addition, the transfer of blaNDM-1 together with the TmexCD2-ToprJ2 genes for JNQH491 and JNQH579 in the E. coli J53 transconjugants conferred resistance to all tested β-lactams (ampicillin, amoxicillin-clavulanate, piperacillin/tazobactam, cefoxitin, ceftriaxone, cefepime, imipenem, and ertapenem) except for aztreonam. All transconjugants exhibited resistance to quinolones (ciprofloxacin and levofloxacin, ≥4μg/ml), which was likely due to the combinational effects of both quinolone resistance determinant qnrS1 and the TmexCD2-ToprJ2. Further, E.coli J53 transconjugants for JNQH491 and JNQH579 exhibited resistance to gentamycin, while it was susceptible for JNQH473. Aac(3)-IId is likely the main source of the disparity on the basis that aac(3)-IId was co-harbored with TmexCD2-ToprJ2 in the same plasmids for JNQH473 and JNQH491 strains, while it was absent on the pJNQH579-2 plasmid.
TmexCD1-ToprJ1 is the first reported plasmid-encoded RND efflux pump, conferring resistance to multiple drugs including tigecycline (Lv et al., 2020). More recently, a variant of TmexCD2-ToprJ2 was identified in a clinical R. ornithinolytica strain NC189, which demonstrated similar tigecycline resistance as the TmexCD1-ToprJ1 (Wang et al., 2021a). In comparison with TmexCD1-ToprJ1, most of which were detected in K. pneumoniae (Figure 2), TmexCD2-ToprJ2 was identified among a variety of Enterobacteriaceae species, including R. ornithinolytica, C. freundii, Aeromonas hydrophila, and K. pneumoniae (Wang et al., 2021a). Our study demonstrated that the TmexCD2-ToprJ2 had spread into carbapenem-resistant K. pneumoniae species complex (KpSC), which is an emerging pathogenic species and frequently detected clinically (Wyres et al., 2020). The highly conserved synteny and structure of the TmexCD2-ToprJ2 harboring plasmids suggested the likelihood of horizontal transfer of a highly similar plasmid between different Klebsiella species. We also found the co-existence of TmexCD2-ToprJ2 and blaNDM-1 genes in pJNQH491-2 and pJNQH579-2, confirmed by the resistance profile of the transconjugants. The co-transfer of blaNDM-1 with TmexCD2-ToprJ2 for JNQH491 and JNQH579 in the E. coli J53 conjugants conferred resistance to tigecycline and all tested β-lactams except aztreonam. In addition, analysis of plasmids from the database from NCBI revealed frequent co-occurrence of TmexCD2-ToprJ2 and carbapenem-resistant genes in the same plasmid. Our finding is of particular public health concern as the convergence of “mosaic” plasmids can confer both tigecycline and carbapenem resistance, thus leading to a serious challenge to the treatment of bacterial infections.
It has been demonstrated in a previous study that TmexCD2-ToprJ2 functions as an efflux pump system by the efflux inhibition experiments (Wang et al., 2021a). TmexCD2-ToprJ2 exhibits a broad substrate spectrum toward tetracyclines, eravacycline, tigecycline (8-fold MIC increase), ciprofloxacin (4-fold MIC increase), and slightly decreased susceptibility (2-fold MIC increase) to cefotaxime and cefepime (Wang et al., 2021a). Our finding is consistent with previous study that TmexCD2-ToprJ2 gene cluster caused 8-fold increase in the tigecycline MICs in the E. coli transconjugants (Wang et al., 2021a). However, strain K. michiganensis JNQH491 was susceptible to tigecycline with a MIC value of 1μg/ml irrespective of the presence of TmexCD2-ToprJ2 gene. We speculated that the differences in plasmid copy numbers or transcription of promoter sequence in this K. michiganensis strain might contribute to the different susceptibility profiles observed in strains from this study (Shaheen et al., 2011). Further studies are needed to evaluate how much TmexCD2-ToprJ2 will affect the therapeutic effects of tigecycline in vivo, including clinical isolates with low MIC values.
An increasing number of studies have supported that the TmexCD-ToprJ-like efflux pump system may originate from the chromosome of Pseudomonas species, as their structures are closely related to the chromosomal MexCD-OprJ system in P. aeruginosa (Lv et al., 2020; Sun et al., 2020; Wang et al., 2021a,b). These findings suggested TmexCD-ToprJ-like clusters might be originated from chromosomal genes in Pseudomonas species, through horizontal transfer into Enterobacteriaceae species. The MexCD-OprJ family proteins act as efflux pumps, conferring intrinsic resistance to tetracycline, chloramphenicol, and norfloxacin in P. aeruginosa (Poole et al., 1993). The rapid expansion of the TmexCD-ToprJ cluster has been attributed to various mobile genetic elements, such as ICEs, transposons (e.g., Tn5393), or IS element (e.g., IS26; Sun et al., 2020; Wan et al., 2020; Wang et al., 2021b; Yang et al., 2021). In our study, a genetic structure (hp1-hp2-tnpA-tnfxB2-IS881-TmexCD2-ToprJ2) constitutes a putative transposon system. Further, a larger putative transposon comprised of 34,827bp harboring TmexCD2-ToprJ2 was also inserted into the umuC-like gene of pJNQH579-2. As such, the umuC gene appears to be a “hotspot” for TmexCD2-ToprJ2 integration in IncHIB plasmids, while the molecular mechanism underlying the site-specific integration deserves further studies.
Overall, we report the first time of three IncHI1B type plasmids encoding efflux pump TmexCD2-ToprJ2 in carbapenem-resistant K. variicola, K. michiganensis, and K. quasipneumoniae species. The sequence analysis identified a putative transposon element for TmexCD2-ToprJ2 transmission, with the genetic structure of “hp1-hp2-tnpA-tnfxB2-IS881-TmexCD2-ToprJ2.” Of note, the co-existence of blaNDM-1 and TmexCD2-ToprJ2 is of particular public health concern. The emergence of such Klebsiella strains underscores the importance of clinical awareness of this pathotype and the need for continued monitoring of TmexCD-ToprJ family resistance genes in China and around the world.
Complete sequences of the chromosomes and plasmids of strain JNQH473, JNQH491, and JNQH579 have been deposited in the GenBank databases under accession numbers NZ_CP075884.1-NZ_CP075888.1, NZ_CP075881.1-NZ_CP075883.1, and NZ_CP078146.1-NZ_CP078148.1, respectively.
The studies involving human participants were reviewed and approved by the First Affiliated Hospital of Shandong First Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
MH and WM conceived the study and designed the experimental procedures. BZ and XD collected the strains. YW, BZ, ML, JM, XL, and FC performed the experiments. MH, JG, and SL analyzed the data. WM, YH, and YW contributed to reagents and materials. MH and LC wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.759208/full#supplementary-material
Supplementary Figure S1 | Multiple sequence alignment of amino acids of TmexCD1-ToprJ1, TmexCD2-ToprJ2, and TmexCD3-ToprJ3. Conserved residues were displayed in dark blue background and mutants were highlighted in white background.
Supplementary Figure S2 | PCR amplification of plasmid replicons and resistance genes of JNQH473, 491, 579, and their E. coli J53 transconjugants.
1. ^https://eucast.org/clinical_breakpoints/
2. ^https://github.com/rrwick/Porechop
3. ^https://github.com/tseemann/mlst
4. ^https://github.com/tseemann/abricate
5. ^https://isfinder.biotoul.fr/
6. ^https://ftp.ncbi.nlm.nih.gov/refseq/release/plasmid, latest update on July 15, 2021.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cheng, Y., Chen, Y., Liu, Y., Guo, Y., Zhou, Y., Xiao, T., et al. (2020). Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg. Microbes Infect. 9, 1843–1852. doi: 10.1080/22221751.2020.1803769
Chiu, S. K., Chan, M. C., Huang, L. Y., Lin, Y. T., Lin, J. C., Lu, P. L., et al. (2017). Tigecycline resistance among carbapenem-resistant Klebsiella pneumoniae: clinical characteristics and expression levels of efflux pump genes. PLoS One 12:e0175140. doi: 10.1371/journal.pone.0175140
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Guy, L., Kultima, J. R., and Andersson, S. G. (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335. doi: 10.1093/bioinformatics/btq413
Hao, M., Schuyler, J., Zhang, H., Shashkina, E., Du, H., Fouts, D. E., et al. (2021). Apramycin resistance in epidemic carbapenem-resistant Klebsiella pneumoniae ST258 strains. J. Antimicrob. Chemother. 76, 2017–2023. doi: 10.1093/jac/dkab131
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Henderson, T. A., Young, K. D., Denome, S. A., and Elf, P. K. (1997). AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179, 6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997
Hirabayashi, A., Ha, V. T. T., Nguyen, A. V., Nguyen, S. T., Shibayama, K., and Suzuki, M. (2021). Emergence of a plasmid-borne tigecycline resistance in Klebsiella pneumoniae in Vietnam. J. Med. Microbiol. 70:001320. doi: 10.1099/jmm.0.001320
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, X., Yin, Z., Yin, X., Fang, H., Sun, Q., Tong, Y., et al. (2017). Sequencing of blaIMP-carrying IncN2 plasmids, and comparative genomics of IncN2 plasmids harboring class 1 integrons. Front. Cell. Infect. Microbiol. 7:102. doi: 10.3389/fcimb.2017.00102
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2016). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/AAC.02465-15
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., Zhang, S., Den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4:1762. doi: 10.21105/joss.01762
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. MBio 11:e02930-19. doi: 10.1128/mBio.02930-19
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Poole, K., Krebes, K., Mcnally, C., and Neshat, S. (1993). Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175, 7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Ruzin, A., Immermann, F. W., and Bradford, P. A. (2008). Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52, 3430–3432. doi: 10.1128/AAC.00591-08
Ruzin, A., Visalli, M. A., Keeney, D., and Bradford, P. A. (2005). Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49, 1017–1022. doi: 10.1128/AAC.49.3.1017-1022.2005
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shaheen, B. W., Nayak, R., Foley, S. L., Kweon, O., Deck, J., Park, M., et al. (2011). Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob. Agents Chemother. 55, 5666–5675. doi: 10.1128/AAC.00656-11
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Sun, S., Gao, H., Liu, Y., Jin, L., Wang, R., Wang, X., et al. (2020). Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg. Microbes Infect. 9, 1102–1113. doi: 10.1080/22221751.2020.1768805
Vaser, R., Sovic, I., Nagarajan, N., and Sikic, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wan, M., Gao, X., Lv, L., Cai, Z., and Liu, J. H. (2020). IS26 mediate the acquisition of tigecycline resistance gene cluster tmexCD1-toprJ1 by IncHI1B-FIB plasmids in Klebsiella pneumoniae and Klebsie lla quasipneumoniae from food market sewage. Antimicrob. Agents Chemother. 65:e02178-20. doi: 10.1128/AAC.02178-20
Wang, C. Z., Gao, X., Yang, Q. W., Lv, L. C., Wan, M., Yang, J., et al. (2021a). Novel transferable resistance-nodulation-division pump gene cluster tmexCD2-toprJ2 that confers tigecycline resistance in Raoultella ornithinolytica. Antimicrob. Agents Chemother. 65:e02229-20. doi: 10.1128/AAC.02229-20
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021b). Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to tigecycline in Proteus mirabilis, and its associated integrative conjugative element. Antimicrob. Agents Chemother. 65:e0271220. doi: 10.1128/AAC.02712-20
Wyres, K. L., Lam, M. M. C., and Holt, K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359. doi: 10.1038/s41579-019-0315-1
Keywords: TmexCD2-ToprJ2, carbapenem resistance, blaNDM-1, blaNDM-5, Klebsiella variicola, Klebsiella quasipneumoniae, Klebsiella michiganensis, tigecycline resistance
Citation: Wang Y, Zhu B, Liu M, Dong X, Ma J, Li X, Cheng F, Guo J, Lu S, Wan F, Hao Y, Ma W, Hao M and Chen L (2021) Characterization of IncHI1B Plasmids Encoding Efflux Pump TmexCD2-ToprJ2 in Carbapenem-Resistant Klebsiella variicola, Klebsiella quasipneumoniae, and Klebsiella michiganensis Strains. Front. Microbiol. 12:759208. doi: 10.3389/fmicb.2021.759208
Received: 16 August 2021; Accepted: 10 September 2021;
Published: 06 October 2021.
Edited by:
Seok Hoon Jeong, Yonsei University, South KoreaReviewed by:
Martijn Zwama, Osaka University, JapanCopyright © 2021 Wang, Zhu, Liu, Dong, Ma, Li, Cheng, Guo, Lu, Wan, Hao, Ma, Hao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingju Hao, aGFvbWluZ2p1QDE2My5jb20=; Wanshan Ma, d3NfbWFAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.