
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 January 2022
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.756688
Whole genome analysis was performed on 501 isolates obtained from a previous survey which recovered 139 positive environmental sponge samples (i.e., up to 4 isolates per sample) from a total of 719 samples collected at 40 standardized sites in 3 commercial apple packinghouse facilities (i.e., P1, P2, and P3) over 3 successive seasons in a single production year. After excluding duplicated isolates, the data from 156 isolates revealed the clonal diversity of L. monocytogenes and allowed the detection of transient contamination, persistent contamination, and cross-area transmission events. Facility P2 with the poorest sanitary conditions had the least diversity (Shannon’s index of 0.38). P2 contained a Clonal Complex (CC) 554, serogroup IVb-v1 strain that persisted throughout the year and spread across the entire facility, a singleton Sequence Type (ST) 1003, lineage III strain that persisted through two seasons and spread across two areas of the facility, and 3 other clones from transient contaminations. P1 and P3, facilities with better sanitary conditions, had much higher diversity (i.e., 15 clones with a Shannon’s index of 2.49 and 10 clones with a Shannon’s index of 2.10, respectively) that were the result of transient contamination. Facilities P1 and P3 had the highest incidence (43.1%) of lineage III isolates, followed by lineage I (31.3%) and lineage II (25.5%) isolates. Only 1 isolate in the three facilities contained a premature stop codon in virulence gene inlA. Fourteen samples yielded 2–3 clones per sample, demonstrating the importance of choosing appropriate methodologies and selecting a sufficient number of isolates per sample for studying L. monocytogenes diversity. Only 1 isolate, belonging to CC5 and from facility P3, contained a known plasmid, and this was also the only isolate containing benzalkonium chloride tolerance genes. The persistent CC554 strain did not exhibit stronger sanitizer resistance than other isolates and did not contain any confirmed molecular determinants of L. monocytogenes stress resistance that were differentially present in other isolates, such as genes involved in sanitizer tolerance, heavy metal resistance, biofilm-forming, stress survival islet 1 (SSI-1), stress survival islet 2 (SSI-2) or Listeria genomic island (LGI2).
Listeria monocytogenes is estimated to cause over 1,600 illnesses and 250 deaths annually in the United States, making it one of the leading causes of death from foodborne illness (Silk et al., 2012). Older adults, persons with immunocompromising conditions, and pregnant women and their newborns are at higher risk of listeriosis than the general population (Silk et al., 2012). Most listeriosis outbreaks have historically been linked to ready-to-eat meats and dairy products (Silk et al., 2012). However, produce-associated listeriosis outbreaks are increasingly recognized (Chen et al., 2017c).
Traceback investigations in a caramel apple outbreak in 2014–2015 converged on a single apple grower, and samples from the apple packing facility and apples in the distribution chain yielded L. monocytogenes isolates highly related to patient isolates by whole genome sequencing (Angelo et al., 2017). The relatively large size of this outbreak (35 cases in 7 states) suggests persistence or repeated contamination of L. monocytogenes that allowed contamination of whole apples used to make caramel apples. Although fresh produce is now a well-established source of listeriosis, apples were an unexpected vehicle, given their high acidity that generally does not support L. monocytogenes growth. However, L. monocytogenes have been shown to survive for extended periods of time on experimentally contaminated Red Delicious, Fuji and Granny Smith apples and that the common practice of waxing could facilitate long-term survival of L. monocytogenes on apples (Macarisin et al., 2019).
Food safety research on tree fruits is not as extensive as that on leafy greens and vegetables, because these fruits are generally not in direct contact with soil, irrigation water, or organic fertilizers. The risks of apple contamination associated with current pre- and post-harvest practices, as well as environmental niches for L. monocytogenes in tree fruit production continuum, represents a large knowledge gap. A better understanding of the ecology, distribution, and persistence of L. monocytogenes in the tree fruit production environments is paramount to gaining a greater understanding of the sources and mechanisms of contamination.
We earlier had hypothesized that fruit packing/storage facilities can harbor transient and/or persistent L. monocytogenes strains leading to post-harvest contamination of fruits. We therefore conducted a 2-year environmental survey to obtain data on the incidence and prevalence of L. monocytogenes in 3 commercial apple packing and storage facilities (Simonetti et al., 2021). Samples were collected from fruit washing, waxing, sorting equipment and surroundings, cold storage rooms, walls, and floors. L. monocytogenes was most often found in the packing line areas, where moisture and fruit debris were commonly observed and less often in dry cold storage and packaging areas. Transient contamination was referred to contaminations that were detected in only one or two samplings and later eliminated. Persistent contamination was referred to those that occurred throughout entire sampling period. Persistent contamination was attributed to poor sanitation practices, including the inability of water drainage systems to prevent moisture accumulation on floors and equipment during peak production times and uncontrolled employee and equipment traffic throughout the facility (Simonetti et al., 2021). For the first year of that study, 139 positive samples from a total of 719 sponge samples were recovered yielding 501 isolates (i.e., up to 4 isolates per positive sponge sample). In the present study, our objective was to perform whole genome sequencing of these isolates to obtain highly resolved geo-spatial source distribution and genetic relationships among isolates.
Isolates analyzed in this study were obtained previously from 40 standardized non-food-contact surface locations in 3 packinghouses at two different times each within the fall depending on the facility (F1, September and F2, October or November), winter (W1, January or February and W2, February or March), and spring (S1, May and S2, June or July) seasons in the 2016–2017 production year (Table 1; Simonetti et al., 2021). Detailed descriptions of the conditions of each facility, sampling sites and dates, and L. monocytogenes detection and isolation methods were described in detail by Simonetti et al. (2021). Briefly, samples were enriched in Buffered Listeria Enrichment Broth (BLEB) (Oxoid Ltd., Basingstoke, Hamsphire, United Kingdom) for 4 h at 30°C. After the initial incubation, Listeria Selective Enrichment Supplement (SR0149A, Thermo Fisher Scientific, Lenexa, KS) was added to each sample and incubated for an additional 44 h at 30°C. The enriched samples were streaked (10 μl) onto Agar Listeria Ottaviani and Agosti (ALOA) and RAPID’ L. monocytogenes (RLM) (BioRad, Hercules, CA) media, then incubated for 24–48 h at 37°C. Presumptive L. monocytogenes isolates were confirmed with Qiagen Multiplex PCR kits (Qiagen Inc., Germantown, MD) using iap and lmo2234 primers specific for Listeria spp. and L. monocytogenes, respectively, as described by Chen and Knabel (2007).
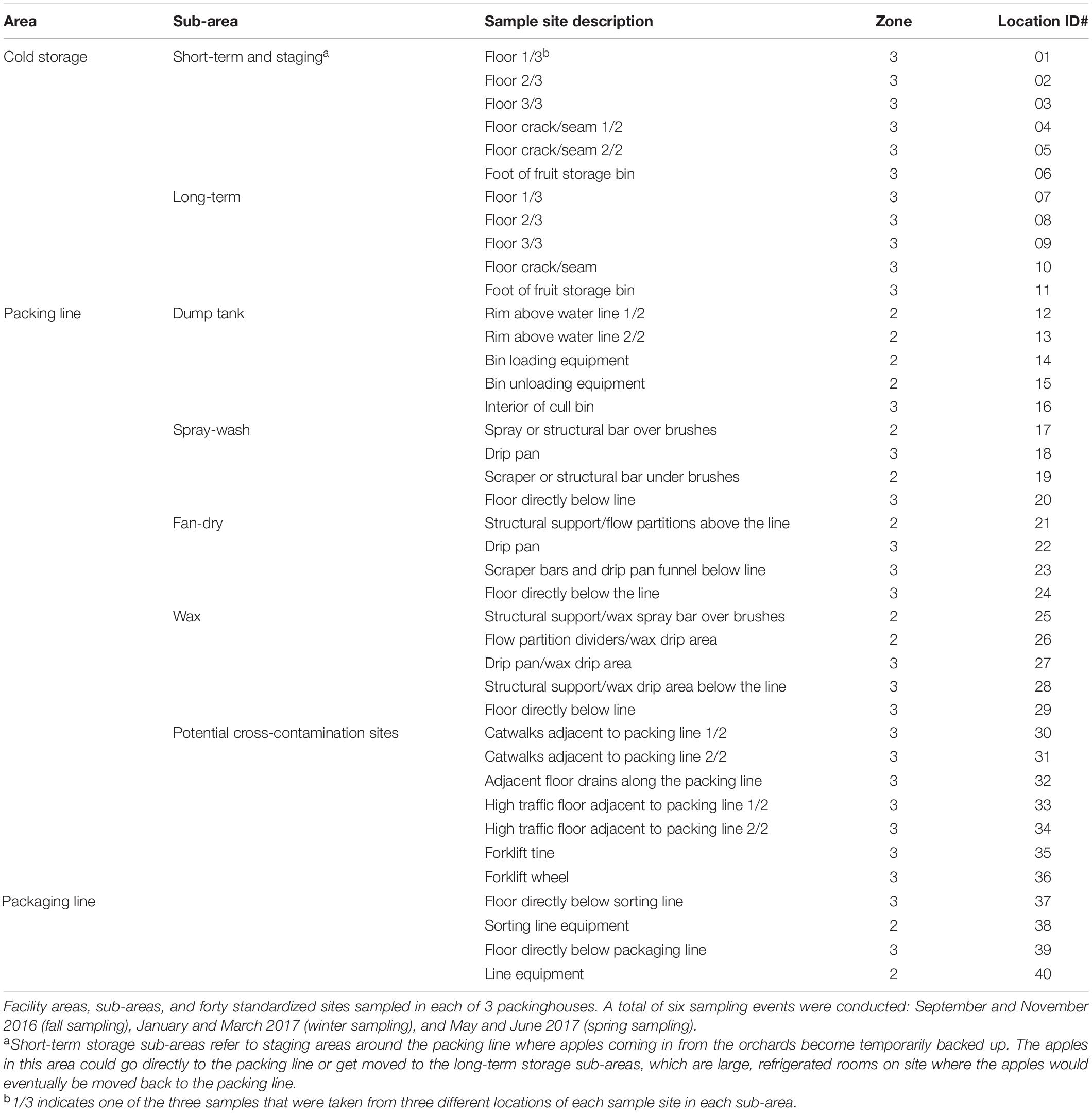
Table 1. Data from Simonetti et al. (2021).
All isolates were sequenced on an Illumina MiSeq platform (250-bp, paired-end reads) (Illumina, Inc., San Diego, CA) using the Nextera XT Library Preparation Kit per the manufacturer’s instructions. The genomic sequence contigs for each isolate were de novo assembled using Qiagen CLC Genomics Workbench 11.1 (Aarhus, Denmark). We then analyzed these genomes by a previously developed cgMLST typing scheme implemented in Ridom SeqSphere + (Ridom© GmbH, Germany), targeting 1,827 core genes of L. monocytogenes (Chen et al., 2016b). A neighbor-joining tree was constructed using the pairwise allele differences. The combination of clustering in the neighbor-joining tree and number of allele differences was used to identify transmission and persistence events.
In silico MLST implemented in the SeqSphere + software was used to determine the sequence type of these isolates. Clonal complex (CC) and singleton were then assigned based on the definition by Ragon et al. (2008) and profiles curated in the Institute Pasteur MLST Listeria database.1 A CC is defined by the 7-gene multilocus sequence tying (MLST) scheme (Ragon et al., 2008) as a group of sequence types (STs) differing by no more than one allele from at least one other ST in the group, and a group having only one ST which differs from all other existing STs in the database by at least two alleles is defined as a singleton (Ragon et al., 2008). In silico serogroup identification was performed by determining the presence of genetic markers for the four major serogroups: IIa (1/2a or 3a); IIc (1/2c or 3c); IVb (4b, 4d or 4e), and IIb (1/2b or 3b) (Doumith et al., 2004). Although designation of those molecular serogroups (e.g., 1/2a, 3a), instead of serotypes (e.g., 1/2a) were achievable based on the WGS data, one serotype in each serogroup (e.g., 1/2a, 4b, 1/2b) is typically more prevalent. Lineage I or lineage II isolates were determined using the combination of serogroup information and cgMLST phylogeny. Lineage III and/or IV isolates were determined using cgMLST phylogeny.
BLAST was performed to determine the presence of select virulence and persistence genes among all the isolates. A threshold of ≥ 70% query coverage with ≥ 80% sequence identity of BLAST alignment indicated the presence of a gene or genomic island. The genes analyzed in this study included those in major Listeria pathogenicity islands and stress survival islets, those encoding internalins, including inlA, and those associated with sanitizer (i.e., quaternary ammonium compounds [QAC]) and heavy metal resistance, and tolerance to various adverse conditions (Moura et al., 2016; Maury et al., 2019). The inlA sequences were extracted from the whole genome sequences using CLC Genomics Workbench and aligned using MEGA 7.0; and premature stop codons (PMSCs) were determined manually. The contigs of each shotgun genome were compared with complete sequences of 60 Listeria plasmids deposited in the GenBank as of November 10, 2019 by BLASTn (Camacho et al., 2009) and the plasmid repA gene was compared with all shotgun genomes by BLASTn (Camacho et al., 2009). We considered a contig as a plasmid contig if it contained ≥ 60% of any plasmid sequences. We considered repA present in a shotgun genome if the BLAST had query coverage of ≥ 60% and sequence identity of ≥ 60%. Because 90% of the complete plasmids are ≥ 10 Kbp and the shortest plasmid length is 3.7 kb, we determined that a shotgun genome contained a plasmid if the total length of plasmid contigs in a shotgun genome exceeded 10 Kbp and repA was present in a plasmid contig. If the total length of plasmid contigs in a shotgun genome was less than 10 Kbp but exceeded 3.7 Kbp, we considered the result inconclusive.
For statistical analysis, we removed duplicate isolates from the same sample. During each sampling, 40 samples were taken and up to 4 typical colonies were picked from each sample for identification and WGS. If the 4 isolates from the same sample likely belonged to the same strain, we removed the duplication of 3 isolates from further analysis. If isolates from different samples likely belonged to the same strain, they were not considered duplicates. We calculated the percentage of isolates belonging to each lineage, serotype, and clones. We also calculated the Shannon’s index among isolates from different facilities using previously described methods; Shannon’s index measures the diversity of a strain collection (Shannon, 1948).
The resistance of a set of representative strains to low levels of Sani-T-10, a QAC-based sanitizer used by facility P2, was compared using a method adapted from Harrand et al. (2020), which suggested that the currently recognized QAC resistance genes of L. monocytogenes only confer resistance to low levels of QAC, not high levels. Sani-T-10 contains 5% of n-alkyl (60% C14, 30% C16, 5% C12, 5% C18) dimethyl benzyl ammonium chloride and 5% of n-alkyl (68% C12, 32% C14) dimethyl ethylbenzyl ammonium chloride. Final concentrations used for growth experiments included 1, 2, 3, 4, 5, 6, and 7 ppm. Sani-T-10 was prepared in BHI broth, and 100-μl overnight culture in BHI were then added to 7 ml BHI broth containing Sani-T-10 before incubation at 30°C for 24 h. The minimum inhibitory concentration (MIC) was defined as the lowest sanitizer concentration at which L. monocytogenes growth was inhibited. The growth was measured by OD600 with 0.010 as the threshold.
We performed whole genome sequence analysis of a total of 501 isolates obtained from the survey. After determining the genetic relatedness of all the isolate and removing duplicate isolates (i.e., isolates from the same swab sample that likely belonged to the same strain), we analyzed 156 isolates in detail. The NCBI Biosample ID, ST, CC, genetic lineage, molecular serogroup, facility ID, sampling date, location number and area designation listed in Supplementary Table 1 for the 156 isolates and in Supplementary Table 2 for the 501 isolates. Even though isolates belonging to epidemiologically-defined outbreaks could have varying degrees of genetic diversity (Chen et al., 2016b), our determination of duplicate isolates were not affected by the threshold of WGS diversity used to define a strain because when two or more isolates of the same sample in the current study belonged to the same clone, they always had ≤ 4 allele differences and thus were considered duplicates. When two or more isolates of the same sample differed by more than 4 alleles, they always belonged to different clones and thus were obviously different strains. In 125 of the 139 positive samples, all isolates of each sample differed by ≤ 4 alleles; in 11 positive samples, each sample yielded 2 strains; and in 3 positive samples, each sample yielded 3 strains. Overall, 345 isolates were removed as duplicates and the remaining 156 isolates were subjected to further analyses.
We determined the genetic relatedness among isolates from different samples to identify possible persistence and transmission events and this determination was not significantly affected by the threshold of WGS diversity. When isolates from different samples belonged to the same clone, the following scenarios were observed (Figure 1). In 12 clades, isolates differed by ≤ 3 alleles; in 1 clade, 3 ST1512 isolates differed by 0, 7, and 7 alleles in a pairwise comparison; we determined each clade to likely represent one strain. The 96 CC554 isolates in one clade differed by up to 8 alleles, and any neighboring isolates in a minimum-spanning tree (tree not shown) differed by 0–4 alleles. Considering that in previous studies isolates that were likely the same strain usually differed by less than 7 alleles (Chen et al., 2016b; Moura et al., 2016), we determined that this clade of 96 isolates likely represented one strain. All clades with diversity larger than the above-described clades contained isolates that differed by at least 21 alleles in pairwise comparisons. Specifically, the clade of 3 CC4 isolates differed from two other CC4 isolates by 21, 23, and 32 alleles in pairwise comparisons; the 2 ST1510 clades differed by ≥ 27 alleles; the clade of 2 CC369 isolates, exhibiting 2 allele differences, differed from another CC369 isolate by 29 alleles; the 2 ST368 isolates differed by 47 alleles; and the clade of 2 CC20 isolates, exhibiting 0 allele difference, differed by 49 alleles from another CC20 isolate. The isolates in these more diverse clades were relatively close genetically, however, we could not conclude whether they were the same strain and therefore did not use those isolates to infer transmission or persistence. All other more diverse clades contained isolates differing by ≥ 100 alleles, and we considered those isolates to represent different strains.
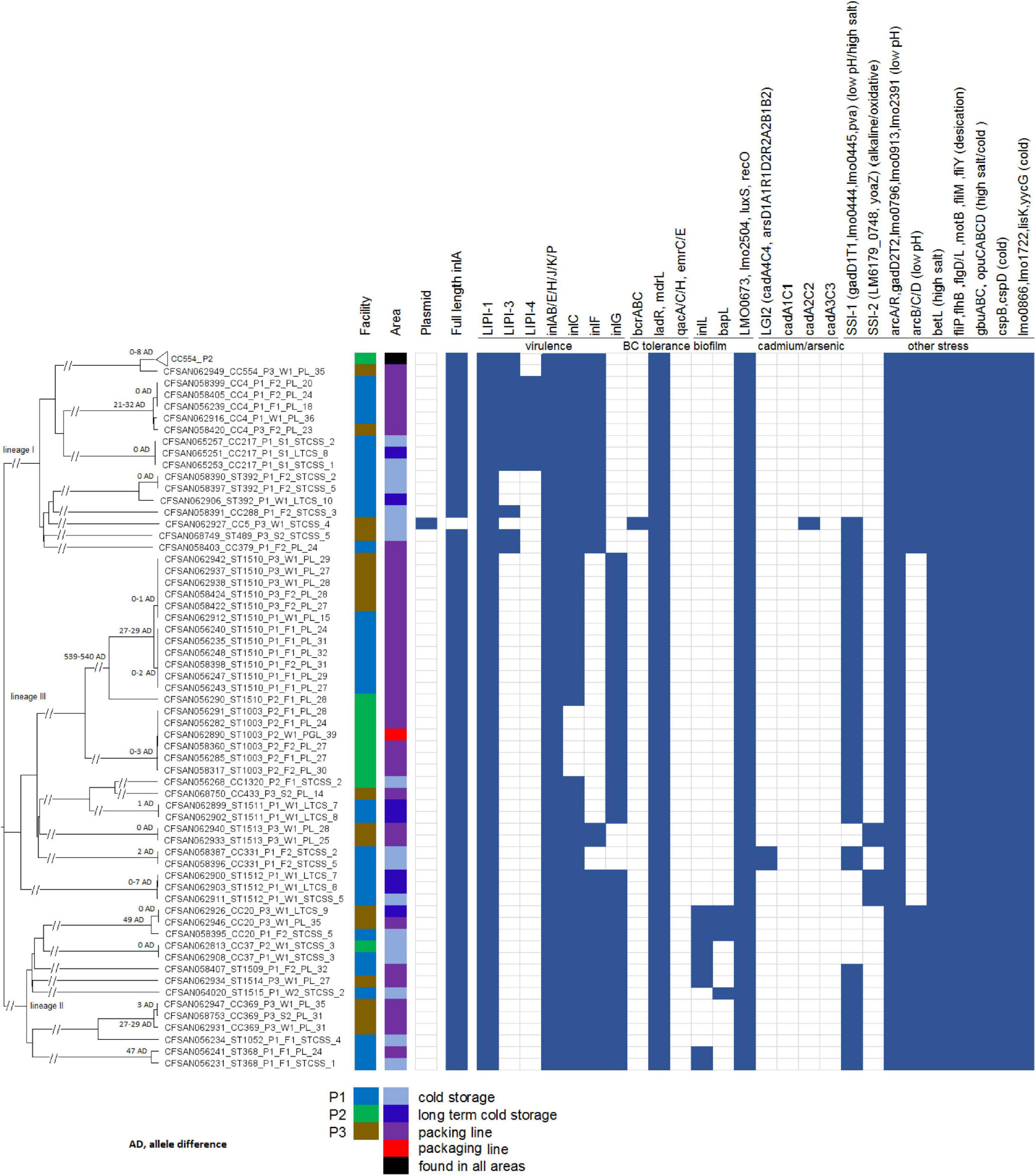
Figure 1. Neighbor-joining tree of the 156 deduplicated isolates collected in 3 apple packinghouses. The Biosample ID is followed by clonal complex or singleton designation, facility number, sampling, sub-area of the facility (i.e., STCSS, short-term cold storage and staging; LTCS, long-term cold storage; PL, packing line; PGL, packaging line) and the number corresponding to each subarea of the facility. The 96 CC554 isolates in facility P2 are collapsed into a triangle shown as the top taxa of the tree. The presence (solid blue color) and absence (empty box) of genes associated with virulence and stress resistance are shown. The pairwise allele differences (AD) of isolates in each clade are shown near the root of the clade.
In the description below, sequence type (ST) is followed by clonal designation [i.e., clonal complex (CC) or singleton] and molecular serogroup (e.g., IIa, IIb, IVb, IVb-v1). If an isolate belonged to a singleton, only ST is listed; if an isolate belonged to a CC, both ST and CC are listed. Serogroup IIb and IVb isolates belong to lineage I; serogroup IIa and IIc isolates belong to lineage II; and lineage III isolates could not be serotyped by molecular methods and were designated as LIII. During the sampling, samples were labeled as #1 through #40.
The sampling locations in each facility were standardized to allow easy comparison and description (Table 1). The details of these standardized sites were previously described (Simonetti et al., 2021). Each facility was divided to 3 areas, cold storage, packing line and packaging line. The packing line is the main processing area that includes washing, sorting, brushing, coating, drying, and grading. The packaging line is where the fruits are packaged into boxes or bags before storage (Simonetti et al., 2021). Each area was further divided to multiple sub-areas and multiple sampling sites were chosen in each sub-area; in some sampling sites, different locations were sampled, which yielded different samples (Table 1). Only deduplicated isolates are discussed below.
(1) In the F1 sampling, 8 positive samples yielded 9 isolates.
Two samples in the short-term storage and staging sub-area were positive. Sample #1 from the floor 1/3 near apple input yielded 1 isolate (ST368, IIa) and sample #4 from the floor crack/seam 1/2 yielded 1 isolate (ST1052, IIa). In this article floor 1/3 indicates the first of the 3 samples taken from 3 different locations of the floor sample site (Table 1); floor crack/seam 1/2 indicates that the first of the 2 samples taken from 2 different locations of the floor crack/seam sample site (Table 1).
Six samples from the packing line were positive. Sample #27 from a waxing drip pan, sample #29 from the floor below waxing, sample #31 from the catwalks adjacent to the packing line 2/2 (a potential cross-contamination site), and sample #32 from adjacent floor drains along the packing line (a potential cross-contamination site) yielded 4 isolates of ST1510 (LIII); sample #24 from the floor below fan-dry yielded 2 isolates (ST1510, LIII and ST368, IIa); and sample #18 from a spray-wash drip pan yielded 1 isolate (ST219/CC4, IVb). Among these isolates, all 5 ST1510 isolates likely belonged to the same strain, indicating cross-contamination between the waxing and fan-dry sub-areas, possibly due to the same strain present in the catwalks going through different sub-areas of the packing line. The drain common to the spray-wash, fan-dry and waxing sub-areas had a convergence of rinse water and product debris, which might explain that the drain was contaminated with the same strain as those packing line sub-areas.
(2) In the F2 sampling, 7 positive samples yielded 11 isolates.
Three samples in the short-term storage and staging sub-area were positive. Sample #2 from the floor 2/3 at the exit to packing yielded 2 isolates (ST331/CC331, LIII and ST392, IIb); sample #3 from the floor 3/3 at the room center yielded 1 isolate (ST323/CC288, IIb); and sample #5 from floor crack/seam 2/2 yielded 3 isolates (ST331/CC331, LIII; ST392, IIb and ST1508/CC20, IIa). Among these isolates, the CC331 isolates from samples #2 and #5 were likely the same strain; the ST392 isolates from samples #2 and #5 were also likely the same strain. The sample sites in the cold storage area were floors, floor cracks/seams and the feet of fruit storage bins, and therefore, cross-contamination due to foot or equipment traffic was highly possible.
Four samples in the packing line were positive. Sample #31 from the catwalks adjacent to the packing line 2/2 (a potential cross-contamination site) yielded 1 isolate (ST1510, LIII); sample #20 from the floor below the spray-wash sub-area yielded 1 isolate (ST219/CC4, IVb); sample #24 from the floor below the fan-dry sub-area yielded 2 isolates (ST379/CC379, IIb and ST219/CC4, IVb); and sample #32 from adjacent floor drains along the packing line yielded 1 isolate (ST1509, IIa). Among these isolates, the ST219/CC4 isolates from samples #20 and #24 were likely the same strain.
(3) Combining data from the two fall samplings, the ST1510 isolate from sample #31 from the F1 sampling likely belonged to the same strain as the ST1510 isolates from samples #24, #27, #29, #31 and #32 from the F2 sampling. This indicated that this strain spread across multiple sub-areas of the facility P1 packing line and persisted through F1 and F2 sampling. However, it is also possible that the same strain was reintroduced to the facility from external sources.
(1) In the W1 sampling, 7 positive samples yielded 9 isolates.
Five samples in the cold storage area were positive. Sample #10 from a floor crack/seam in long-term storage yielded 1 isolate (ST392, IIb); sample #7 from the floor 1/3 in the long-term storage sub-area yielded 2 isolates (ST1511, LIII and ST1512, LIII); sample #8 from the floor 2/3 in the long-term storage yielded 2 isolates (ST1511, LIII and ST1512, LIII); sample #3 from the floor 3/3 in the short-term staging sub-area yielded 1 isolate (ST37/CC37, IIa); and sample #5 from the floor crack/seam 2/2 in the short-term storage and staging sub-area yielded 1 isolate (ST1512, LIII). Among these isolates, the 2 ST1511 isolates from samples #7 and #8 were likely the same strain, and the 2 ST1522 isolates from samples #7 and #8 were likely the same strain. Among these isolates, the ST1512 isolates from samples #5, #7, and #8 likely belonged to the same strain.
Two samples in the packing line were positive. Sample #15 from a bin unloading equipment by dump tank yielded 1 isolate (ST1510, LIII). Sample #36 from a forklift wheel (a potential cross-contamination site) yielded 1 isolate (ST219/CC4, IVb).
(2) In the W2 sampling, only sample #2, from the floor 2/3 near exit of apple output in the short-term storage staging sub-area, was positive and yielded 1 isolate (ST1515, IIa).
(3) Combining data from the two samplings, no persistence or reintroduction events were observed.
(1) In the S1 sampling, 3 samples, from the cold storage area, were positive and yielded 3 isolates that likely belonged to the same strain (ST217/CC217, IVb). These samples were sample #1 from the floor 1/3 in the short-term storage and staging sub-area, sample #2 from the floor 2/3 in the short-term storage and staging and sample #8 from the floor 2/3 in long-term storage sub-area.
(2) In the S2 sampling, no sample yielded L. monocytogenes.
Overall, 33 isolates from 15 clones were recovered from P1, resulting in a Shannon’s diversity index of 2.49 (Figure 2). No isolate from different seasons belonged to the same strain, indicating no persistence across seasons. Therefore, the contamination was transient. Isolates from the cold storage area and those from the packing line did not belong to the same strain, indicating that the environmental isolates of L. monocytogenes did not spread between these two areas, suggesting proper traffic control of the facility. Samples from the packaging line did not yield any L. monocytogenes.
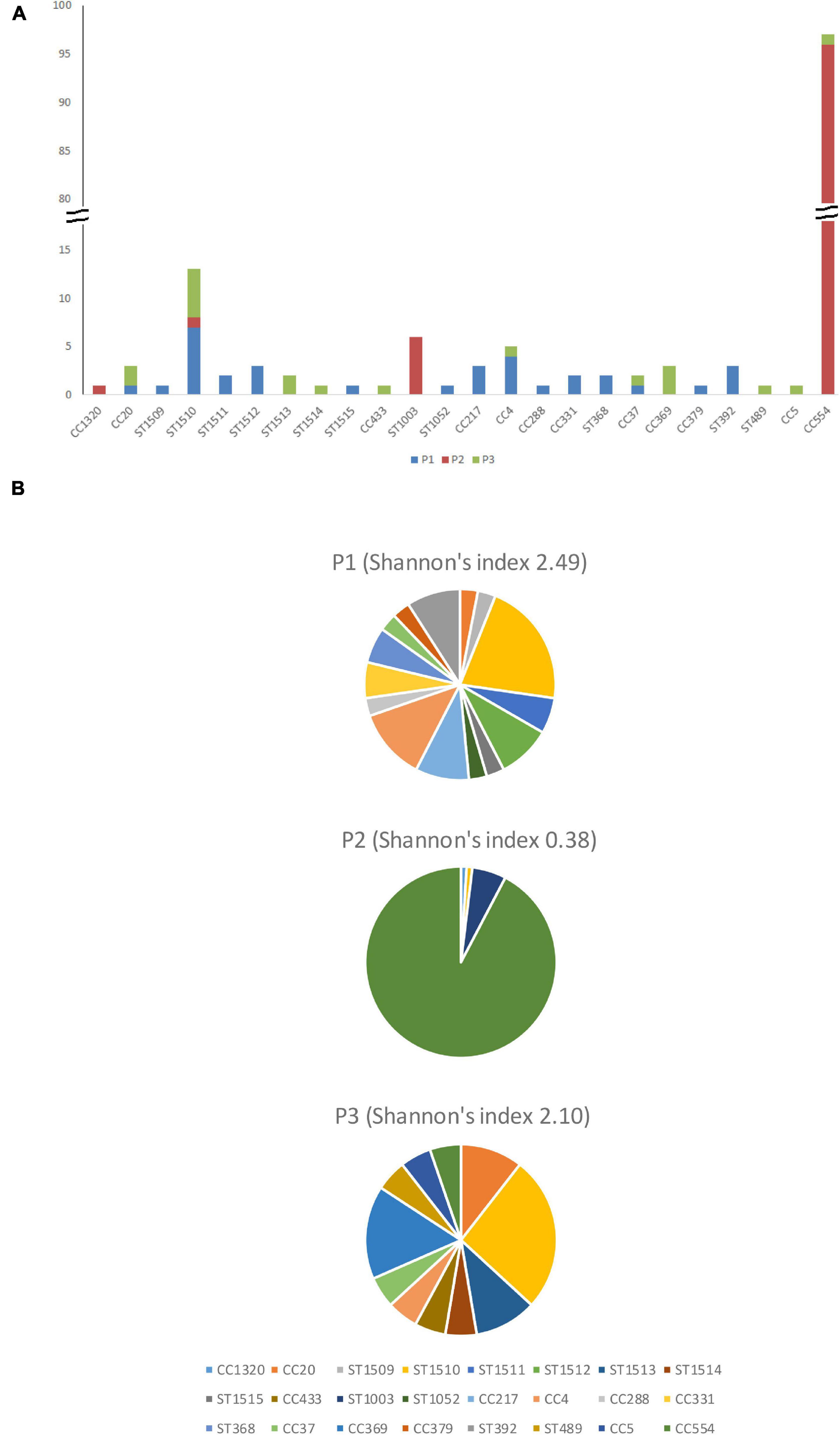
Figure 2. The number (A) and percentage (B) of isolates from different clonal complexes and singletons in facility P1, P2, and P3.
In this facility, many samples yielded ST554/CC554 (IVb-v1) isolates that were likely the same strain; many samples yielded ST1003 (LIII) isolates that were likely the same strain.
(1) In the F1 sampling, 8 positive samples yielded 12 isolates.
Two samples in the cold storage area were positive. Sample #2 from the floor 2/3 in the short-term storage and staging sub-area yielded 1 isolate (ST1507/CC1320, LIII); and sample #8 from the floor 2/3 in the long-term storage sub-area yielded 1 isolate (ST554/CC554).
Six samples in the packing line were positive. Sample #22 from the drip-pan of the fan-dry sub-area yielded 1 isolate (ST554/CC554); sample #23 from the scraper bar and drip pan funnel below the fan-dry sub-area yielded 1 isolate (ST554/CC554); sample #24 from the floor below the fan-dry sub-area yielded 2 isolates (ST554/CC554 and ST1003); sample #27 from the drip pan of wax dripping site yielded 2 isolates (ST554/CC554 and ST1003); sample #28 from the structural support of wax dripping site yielded 3 isolates (ST554/CC554, ST1003 and ST1510, LIII); and sample #29 from the floor below waxing yielded 1 isolate (ST554/CC554).
(2) In the F2 sampling, 17 positive samples yielded 18 isolates.
Sixteen samples in the packing line were positive. Among them, sample #30 from the catwalks adjacent to the packing line 1/2 (a potential cross-contamination site) yielded 1 isolate (ST1003); sample #27 from a drip pan/wax drip site yielded 2 isolates (ST554/CC554 and ST1003); and 14 samples (#17, #20, #21, #22, #23, #24, #25, #26, #28, #29, #31, #32, #33, and #34) from various sample sites of spray-wash, fan-dry, waxing, and potential cross-contamination sites each yielded 1 isolate, all of which were ST554/CC554.
One sample (#38) from the sorting line equipment in the packaging line was positive and yielded 1 isolate (ST554/CC554).
(3) In summary, the ST554/CC554 strain apparently spread to multiple sub-areas in the cold storage, packing line, and packaging line areas of the facility, and persisted through the two successive samplings that fall. The ST1003 strain spread to multiple sub-areas in the packing line and persisted through the two consecutive fall samplings.
(1) In the W1 sampling, 24 positive samples yielded 25 isolates.
Five samples in the cold storage area were positive. Sample #10 from a floor crack/seam in the long-term storage sub-area yielded 1 isolate (ST554/CC554). Others were in short-term storage and staging sub-area. Sample #1 from the floor 1/3 yielded 1 isolate (ST554/CC554); sample #2 from the floor 2/3 yielded 1 isolate (ST554/CC554); sample #3 from the floor 3/3 yielded 2 isolates (ST554/CC554 and ST37/CC37, IIa); and sample #5 from the floor crack/seam 2/2 yielded 1 isolate (ST554/CC554).
Sixteen samples in the packing line were positive, yielding 16 isolates, all of which were ST554/CC554. These samples were #16, #20, #21, #22, #23, #24, #25, #26, #27, #28, #29, #30, #31, #32, #33, and #34, from various sample sites in the dump tank, spray-wash, fan-dry, waxing and potential cross-contamination sites (Table 1).
Three samples from the packaging line were positive. Sample #37 from the floor below the sorting line yielded 1 isolate (ST554/CC554); sample #38 from a sorting line equipment yielded 1 isolate (ST554/CC554); and sample #39 from the floor below the packing line yielded 1 isolate (ST1003).
(2) In the W2 sampling, 24 positive samples yielded 24 isolates, all of which were ST554/CC554.
Six samples (#2, #5, #7, #8, #9, and #10) were from various sample sites in the long-term and short-term storage sub-areas.
Sixteen samples (#14, #20, #21, #22, #23, #24, #25, #26, #27, #28, #29, #30, #31, #32, #33, and #36) were from various sample sites of the dump tank, spray-wash, fan-dry, waxing and potential cross-contamination sites (Table 1).
Two samples (#39 and #40) were from the line equipment and the floor below the packaging line.
(3) In summary, the ST554/CC554 strain was widespread among multiple sub-areas in the cold storage, packing line, and packaging line of the facility, and persisted through the two consecutive winter samplings.
(1) In the S1 sampling, 14 positive samples yielded 14 isolates, all of which were ST554/CC554.
The three positive samples (#3, #7, and #10) were from various sample sites in the long-term and short-term storage sub-areas.
The eleven positive samples (#20, #21, #23, #24, #27, #28, #29, #30, #31, #32, and #33) were from various sample sites of the spray-wash, fan-dry, wax, and potential cross-contamination sites in the packing line (Table 1).
(2) In the S2 sampling, 12 positive samples yielded 12 isolates, all of which were ST554/CC554.
Eleven samples (#18, #20, #22, #23, #24, #26, #27, #28, #29, #30, and #32) were from various sample sites of the spray-wash, fan-dry, wax, and potential cross-contamination sites in the packing line.
One sample (#38) was from the sorting line equipment in the packaging line.
(3) In summary, the CC554 strain spread widely to multiple sub-areas in the cold storage, packing line, and packaging line of the facility, and persisted through the two consecutive spring samplings.
Overall, 105 isolates from 5 clones were collected in P2, resulting in a Shannon’s index of 0.38 (Figure 2). The CC554 strain spread widely to multiple sub-areas in cold storage, packing line, and packaging line of the facility, and persisted through all six samplings. The persistence of isolates in successive samplings suggested relatively poor sanitation conditions in that facility and is further discussed below.
(1) The F1 sampling did not yield any L. monocytogenes.
(2) In the F2 sampling, 3 samples, all in the packing line, yielded 3 isolates. Sample #23 from the scraper bars and the drip pan funnel below the fan-dry sub-area yielded 1 isolate (ST219/CC4, IVb), sample #27 from the drip pan/wax drip site yielded 1 isolate (ST1510, LIII) and sample #28 from the structural support/wax drip site below the waxing sub-area yielded 1 isolate (ST1510, LIII). Among these isolates, the ST1510 isolates from samples #27 and #28 were likely the same strain. Thus, different sampling sites in the waxing sub-area were likely contaminated by the same strain.
(1) In the W1 sampling, 8 samples yielded 12 isolates.
Two samples from the cold storage area were positive. Sample #9 from the floor 3/3 in the long-term storage sub-area yielded 1 isolate (ST1508/CC20, IIa) and sample #4 from the floor crack/seam 1/2 in the short-term storage and staging sub-area yielded 1 isolate (ST5/CC5, IIb).
Six samples from the packing line were positive. Sample #25 from the structural support/wax spray bar over waxing brushes yielded 1 isolate (ST1513, LIII); sample #27 from the drip pan/wax drip site yielded 2 isolates (ST1514, IIa, and ST1510, LIII); sample #28 from the structural support/wax drip site below the waxing line yielded 2 isolates (ST1513, LIII and ST1510, LIII); sample #29 from the floor below the waxing line yielded 1 isolate (ST1510, LIII); sample #31 from the catwalks adjacent to lines 2/2, a potential cross-contamination site, yielded 1 isolate (ST374/CC369, IIa); and sample #35 from a forklift tine, a potential cross-contamination site, yielded 3 isolates (ST1508/CC20, IIa; ST374/CC369, IIa and ST554/CC554, IVb-v1). Among these isolates, both ST1513 isolates from samples #25 and #28 were likely the same strain; the ST1510 isolates from samples #27, #28, and #29 were likely the same strain. Thus, multiple strains spread to different sample sites of the waxing sub-area.
Both ST1508 isolates from samples #9 and #35 were likely the same strain, indicating transmission between the long-term storage sub-area and the packing line.
(2) The W2 sampling did not yield any L. monocytogenes.
(1) The S1 sampling did not yield any L. monocytogenes.
(2) In the S2 sampling, 3 positive samples yielded 3 isolates.
One sample (#5) from the floor crack/seam 2/2 in the short-term storage and the staging sub-area yielded 1 isolate (ST489, IIb).
Two samples in the packing line were positive. Sample #14 from a bin loading equipment near the dump tank yielded 1 isolate (ST1516/CC443, LIII) and sample #31 from the catwalks adjacent to packing line 2/2, a potential cross-contamination site, yielded 1 isolate (ST374/CC369, IIa).
Overall, 18 isolates from 10 clones were collected in P3, resulting in a Shannon’s index of 2.10 (Figure 2). The ST1510 isolates collected in the F2 sampling and those collected in the W1 sampling likely belonged to the same strain, indicating persistence across two seasons. The CC369 isolates collected in the W1 sampling and those collected in the S2 sampling likely belonged to the same strain, indicating persistence across two seasons. Therefore, there were both transient contamination and short-term persistence in P3.
In order to study the transmission events, we divided each facility to 3 areas, the cold storage, the packing line, and the packaging line (Table 1). WGS data indicated no cross-area transmission or cross-season persistence events in P1, and only short-term persistence between two samplings in the same season. However, within each area, cross-contamination in different sub-areas was detected. In the cold storage area, we sampled sites such as floors, floor cracks and the foot of fruit storage bins; cross-contamination among these sites, possibly caused by employee or equipment traffic, was detected during multiple samplings. In the packing line, spray-wash, fan-dry and/or waxing sub-areas were contaminated by the same strain(s) during multiple samplings, and in one sampling, such cross-contamination was likely due to the same strain present in the catwalks along the packing line. The drain where rinse water and product debris from spray-wash, fan-dry and waxing sub-areas were collected was contaminated with the same strain. Even though facility P3 had the lowest incidence of L. monocytogenes among the 3 facilities, our data indicated a cross-area transmission event and two cross-season contamination events, which were not observed in facility P1. We only sampled Zone 2 and Zone 3, and thus, we could not determine if there was cross-area transmission in Zone 1. Nonetheless, it appears that sanitation practices in P1 and P3 effectively eliminated most of L. monocytogenes, however, constant reintroduction of different strains occurred. In contrast, there were persistence and wide spread of L. monocytogenes in P2. This difference could be explained by relatively poor sanitation controls in P2 compared to P1 and P3 (Tan et al., 2019). As a result, Shannon’s indexes of clone diversity for P1 and P3 was 2.49 and 2.10, respectively and those were significantly higher (p < 0.05) than that of P2 (Shannon’s index, 0.38). In our other study that analyzed the relationship between the prevalence of L. monocytogenes and the microbiome of the 3 facilities during the following year (i.e., Year 2), P2 had the highest quantity of L. monocytogenes (i.e., 1.5–3 log higher than those in P1 and P3) and lowest bacterial and fungal species richness (Tan et al., 2019). The authors hypothesized that lower diversity of microbial species could lead to less competition for nutrients and less inhibition from competitor flora, which might render P2 an environment that supports the persistence of L. monocytogenes (Tan et al., 2019). Furthermore, biofilm formers and indicators of unsanitary conditions, such as Pseudomonadaceae and Dipodascaceae, were predominant in P2, and the authors hypothesized that Pseudomonas might have formed biofilms and sheltered L. monocytogenes from cleaning and sanitizing treatments (Tan et al., 2019). Indeed, our targeted sequencing of isolates collected in the hotspots of P2 at the end of Year 2 confirmed that the CC554 strain was still present in P2 (our unpublished data). No quantification and microbiome analysis were conducted in Year 1 when isolates discussed in this study were collected; however, it is not unreasonable to speculate that the quantity of L. monocytogenes and species richness in these 3 facilities were not significantly different between Year 1 and Year 2. In addition, since our enrichment-based detection could introduce bias toward finding the more abundant strain(s) in the environment, we cannot exclude the possibility that higher than what we observed diversity of L. monocytogenes was present in P2.
The ST1510 isolate from the P1 packing line collected during the W1 sampling likely belonged to the same strain as the ST1510 isolates from the P3 packing line collected during the F2 and W1 sampling. The CC37 isolates from the P1 short-term storage sub-area during the W1 sampling belonged to the same strain as the CC37 isolates from the P2 short-term storage sub-area in the W1 sampling. Therefore, common sources of contamination of these facilities could exist.
The wide spread of a L. monocytogenes strain in a facility was also observed in several recent listeriosis outbreaks, in which WGS performed during traceback investigations revealed that multiple locations of the facility were contaminated by the outbreak strain (Chen et al., 2016a,2017a,2017b,2017c).
Facility P1 had 12 lineage I isolates (incidence: 36.3%), 7 lineage II isolates (21.2%) and 14 lineage III isolates (42.4%); the difference was not significant (p > 0.05) (Figure 3). Facility P2 had 96 lineage I isolates (91.4%), all of which belonged to CC554, 1 lineage II isolate (1%) and 8 lineage III isolates (7.6%), 6 of which were ST1003; The wide spread of a single CC554 strain and modest spread of an ST1003 strain across facility P2 contributed to much higher incidence of lineage I and much lower incidence of lineage II and III in this facility (p < 0.05). Facility P3 had 4 lineage I isolates (22.2%), 6 lineage II isolates (33.3%) and 8 lineage III isolates (44.4%); the difference was not significant (p > 0.05) (Figure 2). When combining data from only facilities P1 and P3, lineage III isolates had the highest incidence (43.1%), followed by lineage I (31.3%) and lineage II (25.5%) isolates, even though the incidence was not significantly different (p > 0.05) (Figure 3). In previous studies, lineage III isolates were predominantly isolated from animal sources, and rarely isolated from food processing environments (Orsi et al., 2011). Thus, our findings of high prevalence of lineage III isolates in P1 and P3 may provide new insights on L. monocytogenes contamination in tree fruit packing environments. Perhaps there were connections between the contamination sources of these packinghouses and wild animals since lineage III of L. monocytogenes was often found in animals (Orsi et al., 2011).
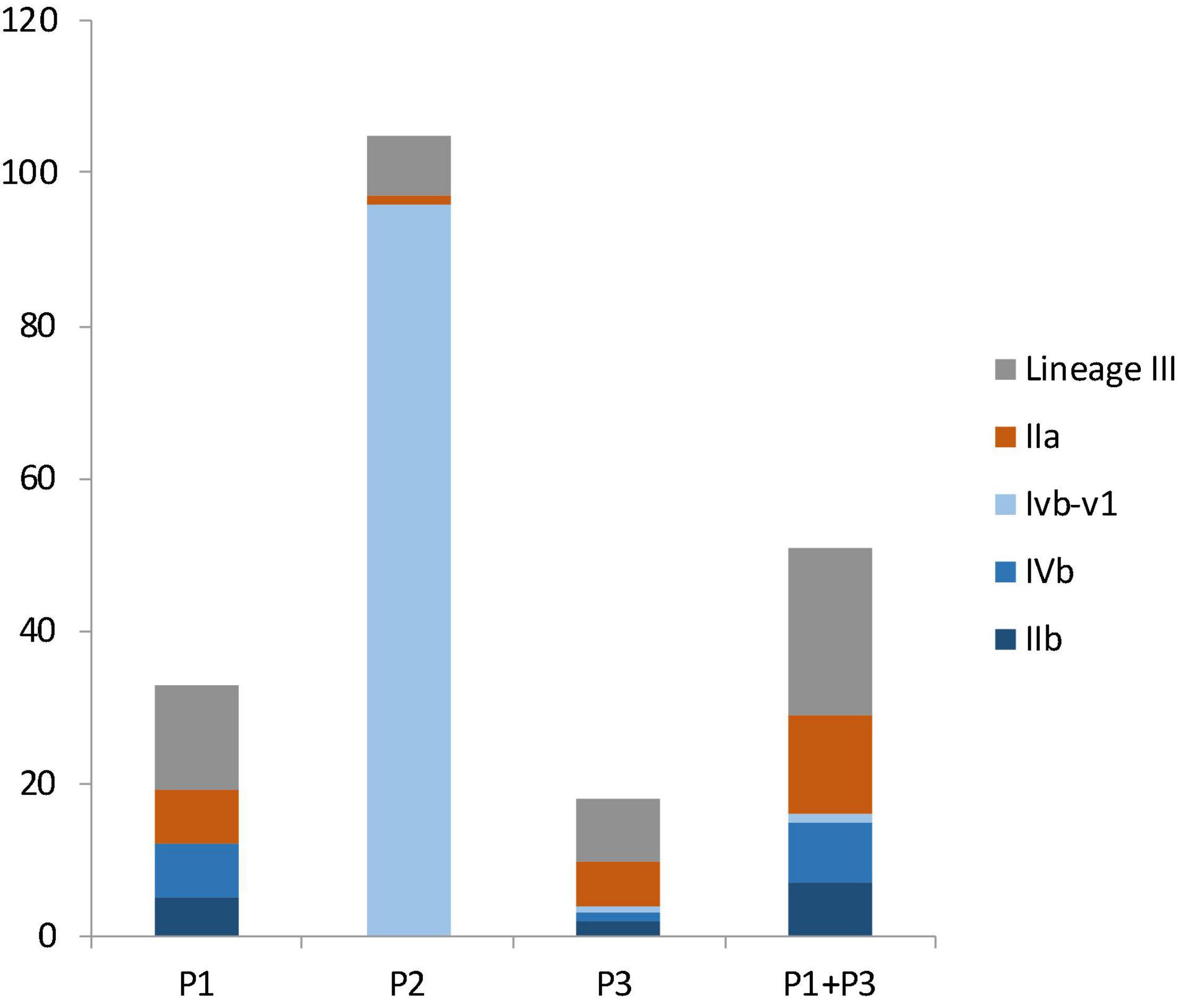
Figure 3. The number of isolates from different molecular serogroups in facility P1, P2, P3 and in both P1 and P3. Lineage III isolates, in gray color, were not assigned a serogroup. Serogroup IIa, in brown color, belongs to lineage II. Serogroups IIb, IVb, and IVb-v1, in three shades of blue colors, belong to lineage I.
Among lineage I isolates in facility P1 were 5 serogroup IIb isolates and 7 serogroup IVb isolates; among lineage I isolates in facility P3 were 2 serogroup IIb isolates, 1 serogroup IVb isolate, and 1 serogroup IVb-v1 isolate (Figure 3). When combing data of lineage I isolates from P1 and P3, there were 7 serogroup IIb isolates (43.8%), 8 serogroup IVb isolates (50%) and 1 serogroup IVb-v1 isolate (6.3%) (Figure 3); there was no significant difference between the occurrence of serogroup IVb and serogroup IIb (p > 0.05). All lineage II isolates in these facilities were serogroup IIa (Figure 3).
When combining data from P1 and P3, we did not observe any strong association between L. monocytogenes genetic lineages and facility areas (i.e., cold storage and packing line). No L. monocytogenes was isolated from packaging lines from P1 and P3.
In this study, in silico analysis of 7-gene MLST defined a total of 24 clones. Among them, 8 were novel clones that are first reported in this study, ST1507/CC1320, singletons ST1509, ST1510, ST1511, ST1512, ST1513, ST1514, and ST1515. In addition, two novel STs of existing clones were discovered, ST1516 of CC433 and ST1508 of CC20. Due to the varying degree of WGS diversity of a few MLST-defined clones, cgMLST diversity was proposed to redefine L. monocytogenes, and a new term, sublineage, was proposed (Moura et al., 2016). With the 1827-cgMLST scheme employed in the current study, isolates belonging to a typical MLST-defined clone differed by ≤ 167 alleles, and isolates belonging to a lineage I or II differed by ≤ ∼1,500 alleles (Chen et al., 2016b,2017c). In this study, isolates in all other clones except singleton ST1510 differed by ≤ 115 cgMLST alleles within each clone, thus, isolates in these clones would have been classified into the same clone if WGS diversity were used to define L. monocytogenes clones. In singleton ST1510, CFSAN056290 differed from the other 12 isolates by up to 540 alleles, and the other isolates differed by ≤ 29 alleles; thus, CFSAN056290 could belong to a different clone from the other 12 isolates if WGS diversity were used to define L. monocytogenes clones. Due to the relatively small number of isolates for each clone, we could not determine any strong association between clone and facility, except that CC554 dominated facility P2. The ST1512 isolates harbored ORF2819 in the molecular serogrouping scheme which indicated it was serogroup IIb, however, this singleton was determined to be lineage III according to the WGS phylogeny, therefore, we considered this multiplex PCR serogroup identification incorrect.
The clones associated with the 2014–2015 multistate listeriosis outbreak linked to caramel apples (i.e., CC1 and ST382) were not found in this study (Chen et al., 2017c). Among the clones identified in this study, CC4 was a hypervirulent clone previously associated with multiple listeriosis outbreaks outside United States (Chen et al., 2016b; Cabal et al., 2019); CC5 had been associated with multiple outbreaks associated with contaminated cheese, ice cream and cantaloupe (Chen et al., 2016b); CC554 had been associated with a 2014 multistate outbreak linked to contaminated mung bean sprouts (Centers for Disease Control and Prevention, 2015; Yang et al., 2020). The CC4, CC5 and CC554 isolates collected in this study did not match the strains of aforementioned outbreaks (our unpublished data). Several major hypervirulent clones, such as CC1, CC2, and CC6 (Maury et al., 2016), were not found in this study. This is consistent with observation in France that hypervirulent clones were not prevalent in foods and food processing environments.
Among isolates that were persistent in facility P2, we did not observe any significant temporal or spatial microevolution events of the CC554 isolates; isolates from different areas (i.e., cold storage, packing line and packaging line) did not form any specific clades, and isolates from different samplings did not form any specific clades. For most samples, multiple isolates from the same sample did not form a clade. In addition to the cgMLST targeting 1,827 genes, we also performed an analysis targeting the entire genome and reached the same conclusion (data not shown).
In this study, we used the 48 h enrichment cultures to analyze the environmental samples and picked up to 4 typical colonies from agar plates of each sample for identification and WGS. Fourteen out of 139 (10%) positive samples yielded more than one strain per sample. Different strains in each sample always belonged to different clones. Given the very large clonal diversity observed in P1 and P3, we speculate that more samples could be contaminated with more than one strain, and more clones might be present in these facilities, but the 48 h enrichment protocol we employed might confer disadvantage to strains that had less abundance. Perhaps, using shorter enrichment duration (e.g., 24 h) could lead to the detection of those less abundant strains, however, heavily stressed cells may not recover with shorter or no enrichment (Sheth et al., 2018). In addition, sampling more than 4 colonies per sample could also lead to the discovery of more strains. In our study, sampling of 40 locations per facility and picking of up to 4 isolates per sample could compensate the bias introduced by the 48 h enrichment, at least partially, since we expect that not every single location was co-contaminated by strains with differing abundance. Previous studies analyzing WGS data of isolates recovered using enrichment-based methods during outbreak traceback investigations also reported differing abundance of strains contaminating the food processing environments or foods. For example, the facility implicated in the 2014 stone fruit outbreak yielded a singleton ST382 strain from 3 environmental samples, a CC5 strain from 13 environmental samples and a singleton ST392 strain from only one sample, suggesting differing prevalence of these clones contaminating the facility (Chen et al., 2016a; Yang et al., 2020). Therefore, if the purpose of a study is to comprehensively survey all genotypes of L. monocytogenes in a food production environment, the detection methods need to be properly evaluated and sufficient number of locations need to be sampled. For another example, the CC5 outbreak strain was present predominantly in ice cream produced in a facility, but another non-outbreak CC5 strain was also isolated from a few ice cream samples produced in the same facility (Chen et al., 2017b). In that case, a simple 7-gene MLST-identified clone could not provide the precise picture of the genetic diversity of L. monocytogenes in that facility; rather, WGS was needed to provide the high resolution on L. monocytogenes genotypes.
The presence of major virulence genes and genes implicated in stress response and environmental persistence (Moura et al., 2016; Maury et al., 2019; Chen et al., 2020) were determined for all the isolates (Figure 1). Only the CC5 isolate, CFSAN062927, contained a premature stop codon (PMSC) in inlA. PMSC in inlA has been associated reduced virulence of some L. monocytogenes strains (Nightingale et al., 2008). A 65 amino acid (AA) region (606th AA to 671th AA), part of the B-repeat (Ebbes et al., 2011), of the ST1514 isolate (CFSAN062934) was not covered by shotgun sequencing, possibly due to sequencing artifacts. However, the examination of the sequencing-covered inlA region of this isolate did not reveal any premature stop codons. Previous studies showed that inlA PMSCs occurred more frequently in lineage II isolates than in lineage I isolates and that isolates with inlA PMSCs were often overrepresented in food and environmental samples, but underrepresented in clinical samples (Van Stelten et al., 2010, 2016). In the present study, only one isolate contained a PMSC. This is similar to studies performed by Gorski et al. (2016) who reported that only 2.7% of L. monocytogenes isolates from naturally contaminated watersheds contained inlA PMSCs, Wang et al. (2015) who reported 2.4% of L. monocytogenes isolates from retail deli environments containing inlA PMSCs, and (Kim et al., 2018) who reported 2.5% of isolates from milk, milk filters and milking equipment from United States dairies containing inlA PMSCs. Therefore, analyses of additional strain collections should be performed to determine the overall prevalence of inlA PMSCs in food, environmental and clinical isolates.
Listeria pathogenicity island-1 (LIPI-1), the first identified L. monocytogenes genomic island where virulence factors cluster together (Vazquez-Boland et al., 2001), was found in all isolates. LIPI-3, encoding listeriolysin S, was found in CC4, CC288, ST489 and CC379 isolates. LIPI-4, a newly identified pathogenicity island associated with neural and placental infections (Maury et al., 2016), was found in CC4 and CC217. Major internalins (Moura et al., 2016; Chen et al., 2020), which were surface proteins that are associated with attachment of L. monocytogenes host cells (Navarre and Schneewind, 1999), were found in the isolates except for 3 internalin genes. Specifically, inlC and inlF were not found in a portion of lineage III isolates and inlG was not found in lineage I isolates. Many genes involved in resistance to low pH, high salt concentrations, desiccation, alkaline stress, and oxidative stress were found in all the isolates. arcABC were not found in any lineage III isolates, while arcA and arcR were found in all isolates. arcABCDR was involved in resistance to low pH (Matereke and Okoh, 2020). Several plasmid-borne benzalkonium chloride (BC) tolerance genes have been confirmed to contribute to adaptation of L. monocytogenes to food processing environments (Moura et al., 2016; Maury et al., 2019). Among them, qacA, qacC, qacH, emrC, and emrE were not found in any isolates; the bcrABC cassette was found in the only CC5 isolate. Therefore, these BC tolerance determinants were not associated with the occurrence of L. monocytogenes in the 3 tree fruit packinghouses. Listeria stress survival islet 1 (SSI-1), which was involved in growth of Listeria under suboptimal conditions such as low pH and high salt concentrations (Ryan et al., 2010), was found in 3 lineage I isolates (18.8% when only counting P1 and P3), 9 lineage II isolates (64.3% of all lineage II isolates), and 25 lineage III isolates (83.3% of all lineage III isolates). Listeria stress survival islet 2 (SSI-2), associated with Listeria alkaline and oxidative stress responses (Harter et al., 2017), which may contribute to the wide spread of CC121 in food production environments in several countries of Europe (Schmitz-Esser et al., 2015; Rychli et al., 2017; Pasquali et al., 2018; Maury et al., 2019), was found only in isolates of ST1513 and ST1512 which both belonged to lineage III. Listeria genomic island 2 (LGI2), which was associated with arsenic and cadmium resistance and previously found in several hypervirulent clones of serotype 4b (Lee et al., 2017), was found only in isolates of CC331, lineage III. Thus, quite a few lineage III isolates contained genes associated with Listeria stress resistance. Another cadmium resistance cassette, cadA2C2, was found only in the CC5 isolate. Two genes involved in biofilm formations, inlL and bapL, were found only in isolates of lineage II. The only CC5 isolate, CFSAN062927 contained a plasmid which was nearly identical to the LM-F-131 plasmid (NCBI Accession, CM009923.1, 81.7Kbp), containing bcrABC and cadA2C2. Five other isolates contained plasmid contigs that partially matched the plasmid pLMIV (NCBI Accession, CM001470.1, 77.8 Kbp) of a lineage IV isolate (J1-208) and did not match any other plasmids deposited in GenBank as of November 10th, 2019. Specifically, the 3 CC369 (lineage II) isolates each contained plasmid contigs of ∼48 Kbp; the CC20 (Lineage II) isolate, CFSAN058395, contained plasmid contigs of ∼34 Kbp; and a ST1510 isolate, CFSAN056290, contained plasmid contigs of ∼61 Kbp. Plasmid pLMIV did not contain known heavy metal resistance genes or benzalkonium chloride resistance genes. This plasmid analysis corroborates our results showing that only CFSAN062927 contained known plasmid-borne stress resistance genes.
The widely spread CC554 strain in P2, which persisted for at least 2 years based on data in the present study and our unpublished data, did not contain the SSI-1, SSI-2, LGI2, any other heavy metal resistance genes, or any genes associated with BC tolerance.
The genes associated with biofilm formation found in the CC554 strain were also found in all other isolates. This indicated that the persistence of this CC554 strain might not be strongly associated with these identified stress resistance mechanisms; other factors, such as poor sanitary conditions, the protection from biofilm-forming background flora, and/or unidentified stress resistance mechanisms might have played major roles in the persistence of CC554.
The strains chosen to measure the resistance to Sani-T-10 were CFSAN062890 (ST1003), CFSAN062899 (ST1511), CFSAN062927 (CC5), CFSAN062933 (ST1513), CFSAN062942 (ST1510), CFSAN062946 (CC20), CFSAN062947 (CC369), and CFSAN071674 (CC554), and their MICs were 2, 2, 3, 3, 3, 2, 3, and 3 ppm, respectively. These strains were chosen to represent lineages I, II and III; and those strains that were repeatedly isolated in facility P2 (i.e., CFSAN062890 and CFSAN071674). In previous studies, many L. monocytogenes strains that contained BC tolerance genes and those that were persistent in food processing environments exhibited 2–4-fold increased resistance to low levels of BC-based sanitizers (Carpentier and Cerf, 2011; Muller et al., 2014; Ortiz et al., 2016; Moretro et al., 2017; Harrand et al., 2020). In our study, only one strain (CFSAN062927) contained BC tolerance genes and only two strains (CFSAN062890 and CFSAN071674) might have been persistent in the packing environment; and these three strains did not show increased resistance to low concentrations of Sani-T-10 compared to other isolates. Therefore, sanitizer resistance might not be a factor that contributed to the presence of these three strains in the packinghouses.
High resolution of WGS analysis of L. monocytogenes from 3 apple packinghouses collected in 1 year revealed the clonal diversity of L. monocytogenes and allowed the detection of transient contamination, persistent contamination, and cross-area transmission events. Different facilities exhibited distinct contamination patterns (i.e., persistent vs. transient, widespread vs. localized). The facility P2, with the poorest sanitary conditions, had the least diversity of L. monocytogenes clones; P2 contained a CC554, serogroup IVb-v1 strain persisting through the year and spreading across entire facility, an ST1003, lineage III strain persisting through two seasons and spreading across two areas of the facility, and several other strains due to transient contaminations. The facilities with better sanitary conditions, P1 and P3, had much higher diversity of L. monocytogenes clones, as a result of transient contamination, with little cross-area spread in the facilities.
The collected isolates exhibited unique genotypic characteristics. In facilities P1 and P3, lineage III isolates, which were predominantly isolated from animal sources previously, had the highest incidence (43.1%), followed by lineage I (31.3%) and lineage II (25.5%), indicating possible connection between animal sources and contamination sources of these two facilities. Only one isolate contained premature stop codons in inlA. The persistent strain CC554 did not contain major genes that are confirmed molecular determinants of L. monocytogenes stress resistance, and did not exhibit stronger sanitizer resistance than other isolates, demonstrating that persistence of L. monocytogenes in a packinghouse could be determined by multiple factors.
The datasets presented in this study can be found in online repositories are deposited in NCBI. The names of the repository/repositories and accession number(s)/biosample IDs can be found in the article/Supplementary Material.
YC: methodology, data analysis, and writing of the manuscript. TS: data collection. KP and LL: methodology, review, and editing. QJ: laboratory analysis, data collection, and analysis. EB: supervision of data collection and analysis. DM: conception and administration of the project, methodology, review, and editing. All authors contributed to the article and approved the submitted version.
This research was supported by the USDA National Institute of Food and Agriculture and Multistate/Regional Research Appropriations under Projects #PEN04666 and the United States Food and Drug Administration (FDA) under Contract # HHSF223201610124C. This research was also funded in part by an appointment to the Research Participation Program at the FDA Center for Food Safety and Applied Nutrition administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the United States Department of Energy and the FDA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the team of curators of the Institut Pasteur MLST system (Paris, France) for importing novel MLST profiles at http://www.pasteur.fr/mlst.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.756688/full#supplementary-material
Angelo, K. M., Conrad, A. R., Saupe, A., Dragoo, H., West, N., Sorenson, A., et al. (2017). Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol. Infect. 145, 848–856. doi: 10.1017/S0950268816003083
Cabal, A., Allerberger, F., Huhulescu, S., Kornschober, C., Springer, B., Schlagenhaufen, C., et al. (2019). Listeriosis outbreak likely due to contaminated liver pate consumed in a tavern, Austria, December 2018. Euro Surveill. 24:1900274. doi: 10.2807/1560-7917.ES.2019.24.39.1900274
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carpentier, B., and Cerf, O. (2011). Review–Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145, 1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005
Centers for Disease Control and Prevention (2015). Wholesome Soy Products, Inc. Sprouts and Investigation of Human Listeriosis Cases (Final Update). Atlanta, GA: Centers for Disease Control and Prevention.
Chen, Y., Gonzalez-Escalona, N., Hammack, T. S., Allard, M. W., Strain, E. A., and Brown, E. W. (2016b). Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl. Environ. Microbiol. 82, 6258–6272. doi: 10.1128/AEM.01532-16
Chen, Y., Burall, L. S., Luo, Y., Timme, R., Melka, D., Muruvanda, T., et al. (2016a). Listeria monocytogenes in stone fruits linked to a multistate outbreak: enumeration of cells and whole-genome sequencing. Appl. Environ. Microbiol. 82, 7030–7040. doi: 10.1128/AEM.01486-16
Chen, Y., Chen, Y., Pouillot, R., Dennis, S., Xian, Z., Luchansky, J. B., et al. (2020). Genetic diversity and profiles of genes associated with virulence and stress resistance among isolates from the 2010-2013 interagency Listeria monocytogenes market basket survey. PLoS One 15:e0231393. doi: 10.1371/journal.pone.0231393
Chen, Y., and Knabel, S. J. (2007). Multiplex PCR for simultaneous detection of bacteria of the genus Listeria, Listeria monocytogenes, and major serotypes and epidemic clones of L. monocytogenes. Appl. Environ. Microbiol. 15:e0231393. doi: 10.1371/journal.pone.0231393
Chen, Y., Luo, Y., Pettengill, J., Timme, R., Melka, D., Doyle, M., et al. (2017c). Singleton sequence type 382, an emerging clonal group of Listeria monocytogenes associated with three multistate outbreaks linked to contaminated stone fruit, caramel apples, and leafy green salad. J. Clin. Microbiol. 55, 931–941. doi: 10.1128/JCM.02140-16
Chen, Y., Luo, Y., Carleton, H., Timme, R., Melka, D., Muruvanda, T., et al. (2017a). Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, United States, 2013. Appl. Environ. Microbiol. 83:e00633-17. doi: 10.1128/AEM.00633-17
Chen, Y., Luo, Y., Curry, P., Timme, R., Melka, D., Doyle, M., et al. (2017b). Assessing the genome level diversity of Listeria monocytogenes from contaminated ice cream and environmental samples linked to a listeriosis outbreak in the United States. PLoS One 12:e0171389. doi: 10.1371/journal.pone.0171389
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
Ebbes, M., Bleymuller, W. M., Cernescu, M., Nolker, R., Brutschy, B., and Niemann, H. H. (2011). Fold and function of the InlB B-repeat. J. Biol. Chem. 286, 15496–15506. doi: 10.1074/jbc.M110.189951
Gorski, L., Parker, C. T., Liang, A. S., Walker, S., and Romanolo, K. F. (2016). The majority of genotypes of the virulence gene inlA are intact among natural watershed isolates of Listeria monocytogenes from the central California Coast. PLoS One 11:e0167566. doi: 10.1371/journal.pone.0167566
Harrand, A. S., Jagadeesan, B., Baert, L., Wiedmann, M., and Orsi, R. H. (2020). Evolution of Listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 86:e02493-19. doi: 10.1128/AEM.02493-19
Harter, E., Wagner, E. M., Zaiser, A., Halecker, S., Wagner, M., and Rychli, K. (2017). Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 83:e00827-17. doi: 10.1128/AEM.00827-17
Kim, S. W., Haendiges, J., Keller, E. N., Myers, R., Kim, A., Lombard, J. E., et al. (2018). Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS One 13:e0197053. doi: 10.1371/journal.pone.0197053
Lee, S., Ward, T. J., Jima, D. D., Parsons, C., and Kathariou, S. (2017). The arsenic resistance-associated listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 83.
Macarisin, D., Sheth, I., Hur, M., Wooten, A., Kwon, H. J., Gao, Z., et al. (2019). Survival of outbreak, food, and environmental strains of Listeria monocytogenes on whole apples as affected by cultivar and wax coating. Sci. Rep. 9:12170. doi: 10.1038/s41598-019-48597-0
Matereke, L. T., and Okoh, A. I. (2020). Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: a review. Pathogens 9:528. doi: 10.3390/pathogens9070528
Maury, M. M., Bracq-Dieye, H., Huang, L., Vales, G., Lavina, M., Thouvenot, P., et al. (2019). Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 10:2488. doi: 10.1038/s41467-019-10380-0
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313.
Moretro, T., Schirmer, B. C. T., Heir, E., Fagerlund, A., Hjemli, P., and Langsrud, S. (2017). Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 241, 215–224. doi: 10.1016/j.ijfoodmicro.2016.10.025
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Muller, A., Rychli, K., Zaiser, A., Wieser, C., Wagner, M., and Schmitz-Esser, S. (2014). The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 361, 166–173. doi: 10.1111/1574-6968.12626
Navarre, W. W., and Schneewind, O. (1999). Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63, 174–229. doi: 10.1128/MMBR.63.1.174-229.1999
Nightingale, K. K., Ivy, R. A., Ho, A. J., Fortes, E. D., Njaa, B. L., Peters, R. M., et al. (2008). inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74, 6570–6583. doi: 10.1128/AEM.00997-08
Orsi, R. H., Den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Ortiz, S., Lopez-Alonso, V., Rodriguez, P., and Martinez-Suarez, J. V. (2016). The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: evidence from comparative genome analysis. Appl. Environ. Microbiol. 82, 308–317. doi: 10.1128/AEM.02824-15
Pasquali, F., Palma, F., Guillier, L., Lucchi, A., De Cesare, A., and Manfreda, G. (2018). Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: persistence and ecophysiology. Front. Microbiol. 9:596. doi: 10.3389/fmicb.2018.00596
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Ryan, S., Begley, M., Hill, C., and Gahan, C. G. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi: 10.1111/j.1365-2672.2010.04726.x
Rychli, K., Wagner, E. M., Ciolacu, L., Zaiser, A., Tasara, T., Wagner, M., et al. (2017). Comparative genomics of human and non-human Listeria monocytogenes sequence type 121 strains. PLoS One 12:e0176857. doi: 10.1371/journal.pone.0176857
Schmitz-Esser, S., Muller, A., Stessl, B., and Wagner, M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6:380. doi: 10.3389/fmicb.2015.00380
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 and 623–656.
Sheth, I., Li, F., Hur, M., Laasri, A., Jesus, A. J. D., Kwon, H. J., et al. (2018). Comparison of three enrichment schemes for the detection of low levels of desiccation-stressed Listeria spp. from select environmental surfaces. Food Control 84, 493–498.
Silk, B. J., Date, K. A., Jackson, K. A., Pouillot, R., Holt, K. G., Graves, L. M., et al. (2012). Invasive Listeriosis in the foodborne diseases active surveillance network (FoodNet), 2004-2009: further targeted prevention needed for higher-risk groups. Clin. Infect. Dis. 54, (Suppl. 5) S396–S404. doi: 10.1093/cid/cis268
Simonetti, T., Peter, K., Chen, Y., Jin, Q., Zhang, G., Laborde, L. F., et al. (2021). Prevalence and distribution of Listeria monocytogenes in three commercial tree fruit packinghouses. Front. Microbiol. 12:652708. doi: 10.3389/fmicb.2021.652708
Tan, X., Chung, T., Chen, Y., Macarisin, D., Laborde, L., and Kovac, J. (2019). The occurrence of Listeria monocytogenes is associated with built environment microbiota in three tree fruit processing facilities. Microbiome 7:115. doi: 10.1186/s40168-019-0726-2
Van Stelten, A., Roberts, A. R., Manuel, C. S., and Nightingale, K. K. (2016). Listeria monocytogenes isolates carrying virulence-attenuating mutations in Internalin a are commonly isolated from ready-to-eat food processing plant and retail environments. J. Food Prot. 79, 1733–1740. doi: 10.4315/0362-028X.JFP-16-145
Van Stelten, A., Simpson, J. M., Ward, T. J., and Nightingale, K. K. (2010). Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76, 2783–2790. doi: 10.1128/AEM.02651-09
Vazquez-Boland, J. A., Dominguez-Bernal, G., Gonzalez-Zorn, B., Kreft, J., and Goebel, W. (2001). Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3, 571–584. doi: 10.1016/s1286-4579(01)01413-7
Wang, J., Ray, A. J., Hammons, S. R., and Oliver, H. F. (2015). Persistent and transient Listeria monocytogenes strains from retail deli environments vary in their ability to adhere and form biofilms and rarely have inlA premature stop codons. Foodborne Pathog. Dis. 12, 151–158. doi: 10.1089/fpd.2014.1837
Yang, H., Hoffmann, M., Allard, M. W., Brown, E. W., and Chen, Y. (2020). Microevolution and gain or loss of mobile genetic elements of outbreak-related Listeria monocytogenes in food processing environments identified by whole genome sequencing analysis. Front. Microbiol. 11:866. doi: 10.3389/fmicb.2020.00866
Keywords: Genomics, diversity, Listeria, packinghouse, apple
Citation: Chen Y, Simonetti T, Peter K, Jin Q, Brown E, LaBorde LF and Macarisin D (2022) Genetic Diversity of Listeria monocytogenes Isolated From Three Commercial Tree Fruit Packinghouses and Evidence of Persistent and Transient Contamination. Front. Microbiol. 12:756688. doi: 10.3389/fmicb.2021.756688
Received: 10 August 2021; Accepted: 13 December 2021;
Published: 10 January 2022.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
Lina Sheng, University of California, Davis, United StatesCopyright © 2022 Chen, Simonetti, Peter, Jin, Brown, LaBorde and Macarisin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Chen, eWkuY2hlbkBmZGEuaGhzLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.