
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 November 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.756519
 Li Zhao1,2†
Li Zhao1,2† Song Wang1,2†
Song Wang1,2† Jiahuan Dong1,2
Jiahuan Dong1,2 Jialu Shi1,2
Jialu Shi1,2 Jiaqi Guan1,2
Jiaqi Guan1,2 Deyu Liu1,2
Deyu Liu1,2 Fei Liu1,2
Fei Liu1,2 Bailiang Li1,2*
Bailiang Li1,2* Guicheng Huo1,2*
Guicheng Huo1,2*Increasing evidence has indicated that oxidative stress is associated with the health of infants. Bifidobacterium, especially B. longum subsp. longum strains, are abundant in the gut microbiota of infants, which may have the potential to ameliorate oxidative damage. Thus, this study aimed to isolate and screen B. longum subsp. longum strains with probiotic characters and antioxidant properties as infants’ dietary supplements. In this study, 24 B. longum subsp. longum strains were isolated from 15 healthy infants identified via 16S rRNA and heat shock protein 60 (hsp60) sequences. B. longum subsp. longum B13, F2, K4, K5, K10, K13, and K15 strains were selected based on high values obtained from autoaggregation, hydrophobicity, and adhesion assays to HT-29 cells. Among these seven strains, B. longum subsp. longum F2, K5, K10, and K15 were selected according to the high tolerance of gastrointestinal tract conditions compared to Bifidobacterium animalis subsp. lactis BB-12. Among these four strains, B. longum subsp. longum K5 was susceptible to common antibiotics and showed the highest intestinal epithelial cell proliferation of CCD 841 CoN. Additionally, B. longum subsp. longum K5 showed a strong antioxidant capacity, and its supernatant exhibited better activity of reducing power, hydroxyl radical scavenging, and DPPH radical scavenging than that of the intact cells with cell-free extracts. The findings indicated that B. longum subsp. longum K5 could be used as a probiotic candidate in infant nutrition.
Oxidative stress has been closely linked to the health of infants, especially preterm infants (Cong et al., 2016). Preterm infants are vulnerable to oxidative damage because of the absence of antioxidants at birth and the impairment of the synthesis ability of antioxidants (Hanson et al., 2016). The undeveloped antioxidant defense system of infants may increase the risk for complications of intestinal and systemic anomalies, such as retinopathy of prematurity, necrotizing enterocolitis, and bronchopulmonary dysplasia (Weber et al., 2014). Furthermore, it has been reported that the level of oxidative stress of preterm infants was higher than that of full-term infants (Cai et al., 2019). Furthermore, dietary antioxidants were related to reducing the risk of inflammation, diabetes, carcinogenesis, and other health problems associated with aging (Uttara et al., 2009). Thus, it is critical to find natural antioxidants which could apply in baby dietary supplements.
Bifidobacterium is one of the predominant genera of the infant gastrointestinal tract (Milani et al., 2015; Duranti et al., 2017; Nuriel-Ohayon et al., 2016; Ma et al., 2020). As probiotics, they inhibit pathogenic bacteria colonization and regulate immune response (Ruiz et al., 2017). Thus, Bifidobacteria were usually applied in dietary supplements as probiotics. Furthermore, it has been reported that Bifidobacteria possess excellent antioxidant properties (Mishra et al., 2015). However, most of the Bifidobacteria applied in baby dietary antioxidant supplements are not originated from infants. Before they reach the mature adult gut microbiota, the development proceedings of infant gut microbiota are influenced by many external environmental factors (e.g., antibiotics, food). The infant gut microbiota is inherent and less affected by the environment than the adult gut microbiota. Furthermore, the amount of Bifidobacterium strains decreases with age (Silvia et al., 2016), so the current research aims to isolate Bifidobacterium from infants. Additionally, evidence has been reported that the abundance and species of B. longum colonized in the intestine are closely associated with the health of the body (Akay et al., 2014; Zhang et al., 2019). Thus, it is still a challenge for us to discover potential Bifidobacteria, especially B. longum, with antioxidant properties derived from infants.
Being alive and providing beneficial effects in sufficient amounts were the requirements for the probiotic selection claimed by the World Health Organization (Morelli and Capurso, 2012; Nazhand et al., 2020). To be effective, the probiotic should possess the potential ability to adhesion to the intestinal epithelial cells and survive under the gastrointestinal tract (Vankerckhoven et al., 2008; Mandal et al., 2016). The adhesion ability may contribute to the bacteria against undesirable bacteria by competition for binding sites of host intestinal epithelial cells and increasing the chances to interrelate with the host (Celebioglu et al., 2017; Okochi et al., 2017; Monteagudo-Mera et al., 2019). Probiotics demonstrated that the antibiotic resistance genes they carried could be transferred to intestinal pathogens in the host gut (Salyers et al., 2004; Ammor et al., 2007; Sommer et al., 2009). Therefore, it is vital to evaluate the antibiotic resistance properties of bacteria. Furthermore, the development of the intestinal tract is significant in infants; immature intestines and slow intestinal immune development were associated with the high risk of postpartum growth restriction, lactose intolerance, allergy, necrotizing enterocolitis, and other intestinal related diseases (Zhuang et al., 2019; Osorio, 2020; Crovetto et al., 2021). Studies confirmed that probiotics were beneficial to intestinal development, intestinal epithelial renewal, and intestinal mucosa balance (Blumberg and Gensollen, 2017; Zhuang et al., 2019). Thus, it is vital to find effective probiotics to promote intestinal development with no side effects.
The objective of this study was to evaluate and screen B. longum subsp. longum isolated from infant feces and assess its adhesion ability, high tolerance during gastrointestinal digestion, antibiotic susceptibility, capacity for intestinal epithelial cell proliferation, and antioxidant, which could apply in infants’ dietary antioxidant supplements.
Isolates were anaerobically incubated in de Man, Rogosa and Sharpe broth supplemented with 0.05% L-cysteine hydrochloride (MRSL) (2% v/v) at 37°C for 24 h and subcultured twice before further experiments. Bifidobacterium animal subsp. Lactis BB-12 (BB-12) were obtained from the American Type Culture Collection (ATCC).
The normal human colonic epithelial cells CCD 841 CoN cells were kindly donated by Prof. Meng Xiangchen and stored in liquid nitrogen before use. The human colon cell line HT-29 cells were obtained from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM (HyClone, United States) supplemented with 10% fetal bovine serum (FBS, AlphaBio, South America) (DMEMF) at 37°C in a 5% CO2 atmosphere.
Fifteen fresh feces samples of healthy infants (aged 0–6 months) without taking antibiotics before the time of sampling were collected from Harbin, Heilongjiang Province of China. The samples were collected in sterile feces collectors and kept at 4°C until transported to the laboratory. All the operations were completed within 2 h for the isolation of Bifidobacteria.
The samples were diluted and spread on MRS agar with 0.05% L-cysteine (MRSL) and incubated at 37°C for 72 h under an anaerobic atmosphere (10% H2, 10% CO2, and 80% N2) (Bevilacqua et al., 2003). Gram-positive colonies were purified on MRSL and incubated under the same conditions. Purified colonies only grown under anaerobic conditions were selected for the assay of catalase activity. Catalase-negative colonies were inoculated in MRSL liquid medium at 37°C for 24 h.
The total DNA was extracted from a 1 mL culture medium of different isolated strains (Liu et al., 2015). The genomic DNA primer is shown in Table 1. The results were sequenced by KuMei Company, Harbin, China, and screened using the BLAST program. The sequence of strains was uploaded in the GenBank (Supplementary Table 1). A phylogenetic was constructed via the neighbor-joining method using the software of Mega 6.0.
All Bifidobacteria strains identified by 16S rRNA gene sequencing were discriminated via the hsp60 gene (Modesto et al., 2018). The genomic DNA is shown in Table 1. PCR amplification was set in 50 μL volume, including master mix (Yesen, Shanghai, China), 2 μM primer, 120 ng chromosomal DNA, and ddH2O. The cycling program was performed at 95°C for 5 min, 35 cycles of 94°C, 30 s, 59°C, 60 s, 72°C for 30 s, and the final extension was set at 72°C for 10 min. A phylogenetic was constructed as described above.
The SEM analysis was determined using a modified method as previously described by Guang-Yu et al. (2017). After incubation, the Bifidobacteria were centrifuged at 3,000 × g for 5 min. The pellets were washed three times using phosphate-buffered saline (PBS) solution, mixed with 2.5% glutaraldehyde (pH 6.8), and fixed for at least 1.5 h at 4°C. Samples were obtained by washing three times with PBS after centrifuged, then dehydrated with ethanol. Then the samples were freeze-dried and sputter-coated with 100 Å of gold after being replaced with tert-butanol. The results were obtained with Hitachi S-3400N (Hitachi, Japan) SEM.
Autoaggregation assay was performed according to the method of Krausova et al. (2019). with some modifications Bifidobacteria grown overnight were centrifuged (6,000 × g, 10 min) and adjusted to an optical density of 0.6 at 600 nm (OD 600, A0). 8 ml of each stain was incubated at 37°C for 1 h, 4 h, and 24 h (At) and then the upper suspension was determined at 600 nm. The autoaggregation was calculated with the following formula:
Hydrophobicity assay was performed with the modified method as previously described by Yan et al. (2020). CSH was determined with ethyl-acetate (basic polar solvent) and xylene (apolar solvent). Bifidobacteria were performed as described above. The cell pellets were suspended in PBS to a concentration of 0.8–1.0 at 600 nm (H0). Then, 1 mL ethyl-acetate or xylene was mixed with 3 mL cell suspension and shocking for 2 min. The aqueous phase was collected and measured at 600 nm (H1) after incubating at 37°C for 1 h. The value of hydrophobicity was calculated as follows:
HT-29 cells were incubated at 37°C in a 5% CO2 atmosphere using a pre-heated DMEMF medium. The adhesion assay was determined by the method of Li et al. (2014) with some modifications. HT-29 cells (3.0 × 105 cells/mL) were seeded in 12-well culture plates and changed the medium every 2 days until monolayers of HT-29 cells were confirmed. Then, 1 mL of strains with a 1.0 × 108 cfu/mL concentration was added per well and incubated at 37°C for 2 h. Each well was washed five times with PBS and added 0.25 mL trypsin-EDTA for 10 min to digest. The reaction was terminated via adding the same volume of FBS, and the samples of each well were collected and plated on MRSL agar. The results were assessed as follows:
Principal component analysis (PCA) reduces the dimensionality of the original dataset to a new set of non-correlated variables, called principal components (PCs), without losing much information. Principal component analysis was performed with SPSS 16.0 software on a data matrix containing autoaggregation at 1, 4, and 24 h, CSH in ethyl-acetate and xylene, and adhesion of Bifidobacteria to HT-29 cells of different B. longum subsp. longum strains. The total scores of strains were obtained according to the contribution of components and factor score and used to selected Bifidobacteria with good adhesion ability.
The survival rate of test strains in the simulated gastrointestinal tract was accessed by the modified method of the previous assay (Zhu et al., 2015). Strains were adjusted to 1.0 × 109 cfu/mL after incubated for 24 h. The cells were added into gastric juice for 2 h, 1 mL of the solution was taken into the intestinal fluid for 2 h incubation. The survival cells were valued by plate counting as the following formula:
Here, the survival strains were represented by N1, and N0 stands for the total strains before the treatment.
The antibiotic susceptibility of test strains was evaluated against antibiotics vancomycin, gentamicin, penicillin G, chloramphenicol, kanamycin, streptomycin, rifampicin, amoxicillin, tetracycline, erythromycin, roxithromycin, clindamycin, trimethoprim (MTP), ciprofloxacin, and ampicillin, using the disk diffusion method reported by Rajoka et al. (2017). After incubation for 48 h at 37°C, the inhibition zone diameter was measured and analyzed according to the commercial introduction provided by Hangzhou Binhe Microorganism Reagent Co., Ltd., China.
Cell proliferation of test strains was evaluated by the CCK-8 (Dalian Meilun Biotechnology Co., Ltd., China) method described by Liang et al. (2017). The CCD 841 CoN cells were seeded in 96-well plates and incubated for 24 h to ensure cell attachment. Then, Bifidobacteria strains (MOI 1:100) were added into the cells for 24 h incubation. The medium was discarded, and 100 μL 10% CCK-8 solution was added to each well. After 2 h incubation, each well’s optical density (OD) was detected at 450 nm using a microplate reader (Molecular Devices, United States). Cell proliferation of each sample was calculated with the following equation:
The test strains were incubated as above, and the cell-free supernatant (CFS) was collected by centrifugation (8,000 × g, 15 min) at 4°C. The intact cells (IC) were harvested by washing three times with PBS (pH 7.4) and then resuspended at a 1.0 × 109 cfu/mL concentration. The cell-free extracts (CFE) were acquired by ultrasonic in an ice bath for 15 min and centrifuged (8,000 × g, 15 min) at 4°C for the supernatant. The CFE and CFS were treated with 0.22 mm filter membranes for further experiments.
Reducing activity was determined by the method of Oyaizu (1988). A total of 0.5 mL of PBS (pH 6.6) and potassium ferricyanide (1%) were added into 0.5 mL of each sample. The mixture was cultured at 50°C for 20 min and then cooled to room temperature rapidly. Then, 0.5 mL of trichloroacetic acid (10%) was added, and 1 mL of the supernatant was added into 1.0 mL of ferric chloride (0.1%) after centrifuging (3,000 × g, 5 min). The absorbance was recorded at 700 nm after the mixture was placed at room temperature for 10 min. The standard expression of L-Cysteine assessed the value of reducing activity.
A previous study described by Makoto and Oyaizu (1988) analyzed the Hydroxyl radical scavenging assay. A total of 1.0 mL of each sample was added into the mixture contained 2.5 mM 1,10-phenanthroline (1.0 mL), 2.5 mM FeSO4 (1.0 mL), and PBS (1.0 mL, pH 7.4). The mixture was incubated in a 37°C water bath for 90 min after adding 20 mM H2O2 (1.0 mL). The absorbance was obtained at 517 nm. The Hydroxyl radical scavenging activity was calculated as follows:
Here, Asample was represented the absorbance of each test sample, the controls contained distilled water instead of H2O2 in the reaction system, and the blanks included distilled water instead of samples in the reaction system.
The DPPH radical scavenging activity was measured using the method with some modifications of Lin and Yen (1999). A total of 1.0 mL of each sample was mixed with 1.0 mL of 0.2 mM DPPH radical solution and placed for 30 min in the dark. Then, the mixture was centrifuged at 3,000 × g for 10 min. The absorbance was read at 517 nm. The scavenging ability of DPPH radical was assessed as follows:
Where Asample stands for the absorbance of each sample, the controls contain DPPH solution and distilled water, and the blanks include only the samples and ethanol.
According to a previous assay, the superoxide anion radical scavenging properties were assessed (Li et al., 2012). A total of 3.0 mL of Tris–HCl solution (pH 8.2) was mixed with 1.0 mL of samples and stood at room temperature for 20 min. The mixture was placed in the dark for 4 min after adding 0.4 mL of pyrogallol (25 mM). Then, 0.5 mL of HCL (8 mM) was used to end the reaction. The absorbance was recorded at 325 nm. The ability of superoxide anion radical scavenging was valued as follows:
Where Asample stands for the absorbance of each sample, and the blanks include distilled water, not samples in the reaction system.
All tests were executed in triplicate, and the data were expressed as the mean ± standard deviation. Analysis of variance (One-Way ANOVA) was realized with the method of Duncan’s multiple ranges (p < 0.05) via SPSS statistical software version 16.0 to value the statistical significance of differences between means.
The B. longum subsp. longum strains were generated circular, smooth surface, convex, and white colonies on MRSL (Figure 1A). Pleomorphism (dumbbell, V, and Y shaped) was found in the Gram staining of B. longum subsp. longum (Figure 1B). The morphology of B. longum subsp. longum was shown in Figure 1C via scanning electron microscopy.
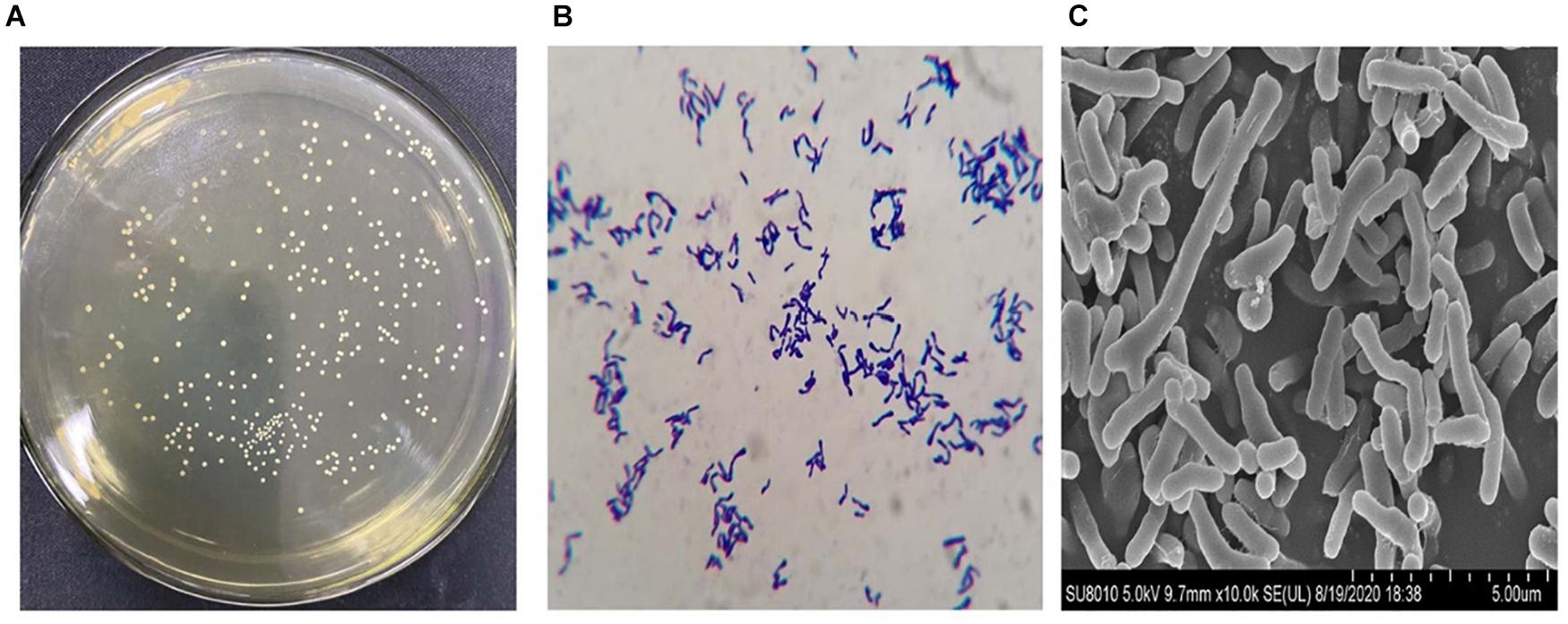
Figure 1. Representative morphologic characteristics of B. longum subsp. longum strains. (A) Colony morphology; (B) Gram staining morphology; (C) Scanning electron microscopy. Morphology of the B. longum subsp. longum strain was obtained using a scanning electron microscope ×10.0 K.
In total, 186 isolates were isolated from 15 feces samples of 0–6-months infants. Of these, 31 strains can only be grown under anaerobic conditions at 37°C. A phylogenetic tree was constructed according to the results of 16S rRNA sequences. The results demonstrated that all isolates belonged to Bifidobacterium, and 24 strains among them belonged to Bifidobacterium longum (Figure 2).

Figure 2. The phylogenetic tree based on 16S rRNA genes for 31 Bifidobacterium stains isolated from infant feces.
As shown in Figure 3, the phylogenetic tree was constructed according to the results of the PCR productions of hsp60 gene sequences using the neighboring method. The results supported the findings of 16S rRNA sequences.
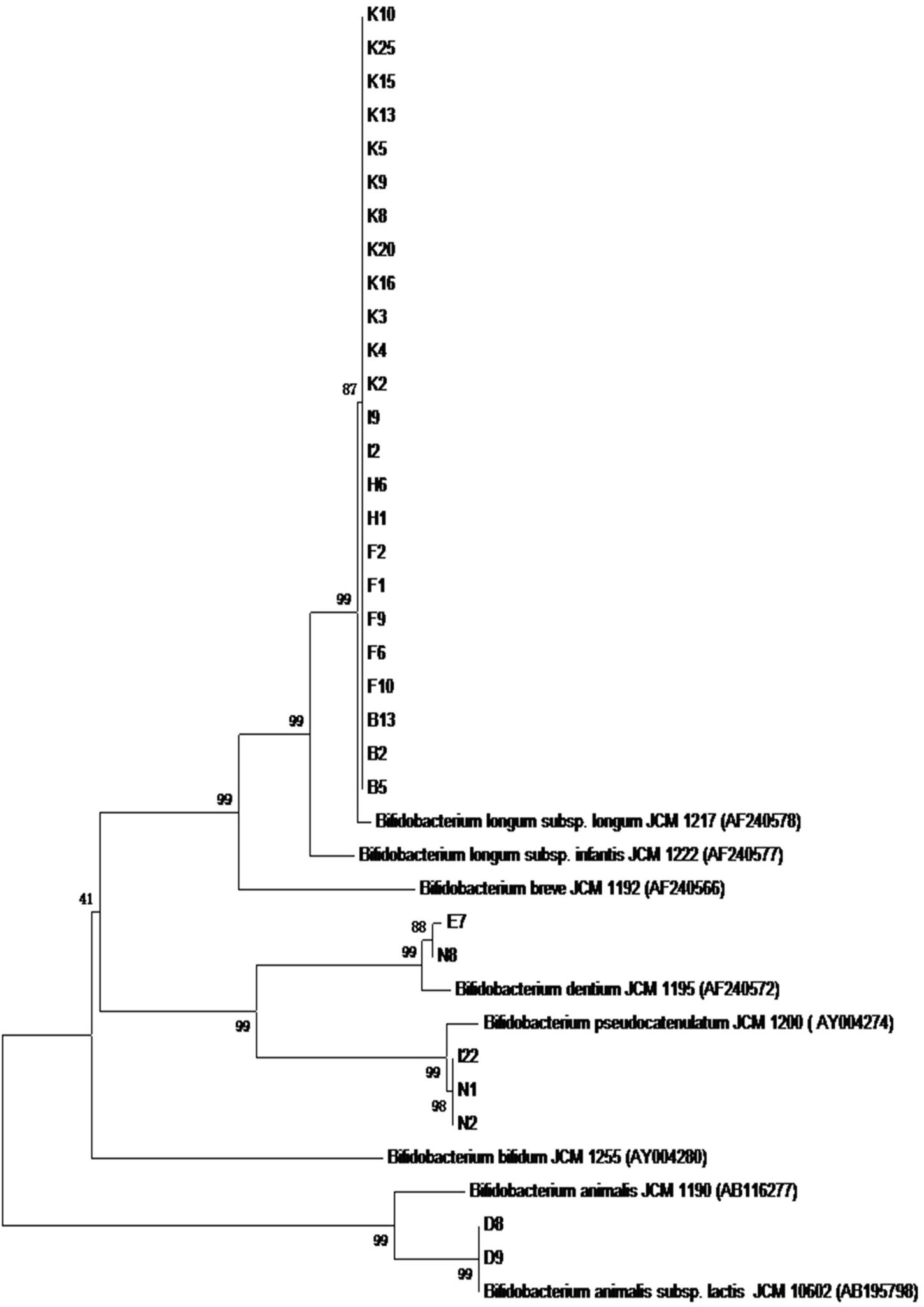
Figure 3. The phylogenetic tree based on the sequences of the PCR production of hsp60 gene for 31 Bifidobacterium strains.
In general, all B. longum subsp. longum strains showed a high autoaggregation after incubating for 24 h (Figure 4), the autoaggregation of all B. longum subsp. longum strains were higher than that of reference strain BB-12 at 4 h. Notably, B. longum subsp. longum K5 revealed the highest autoaggregation with 65.84% at 1 h, followed by B. longum subsp. longum K10 (61.26%).
As shown in Table 2, B. longum subsp. longum K5 presented the highest CSH level in xylene solution (69.67%), followed by K10 (64.86%) and K4 (61.45%), while B. longum subsp. longum K20 showed the lowest CSH level (21.70%). The CSH of B. longum subsp. longum isolates ranged from 21.05 to 72.18% in ethyl acetate solution. B. longum subsp. longum K5, K10, and K4 also showed significantly higher (p < 0.05) CSH than other strains, including the reference strain BB-12. These results indicated these 3 B. longum subsp. longum strains had good hydrophobicity ability.
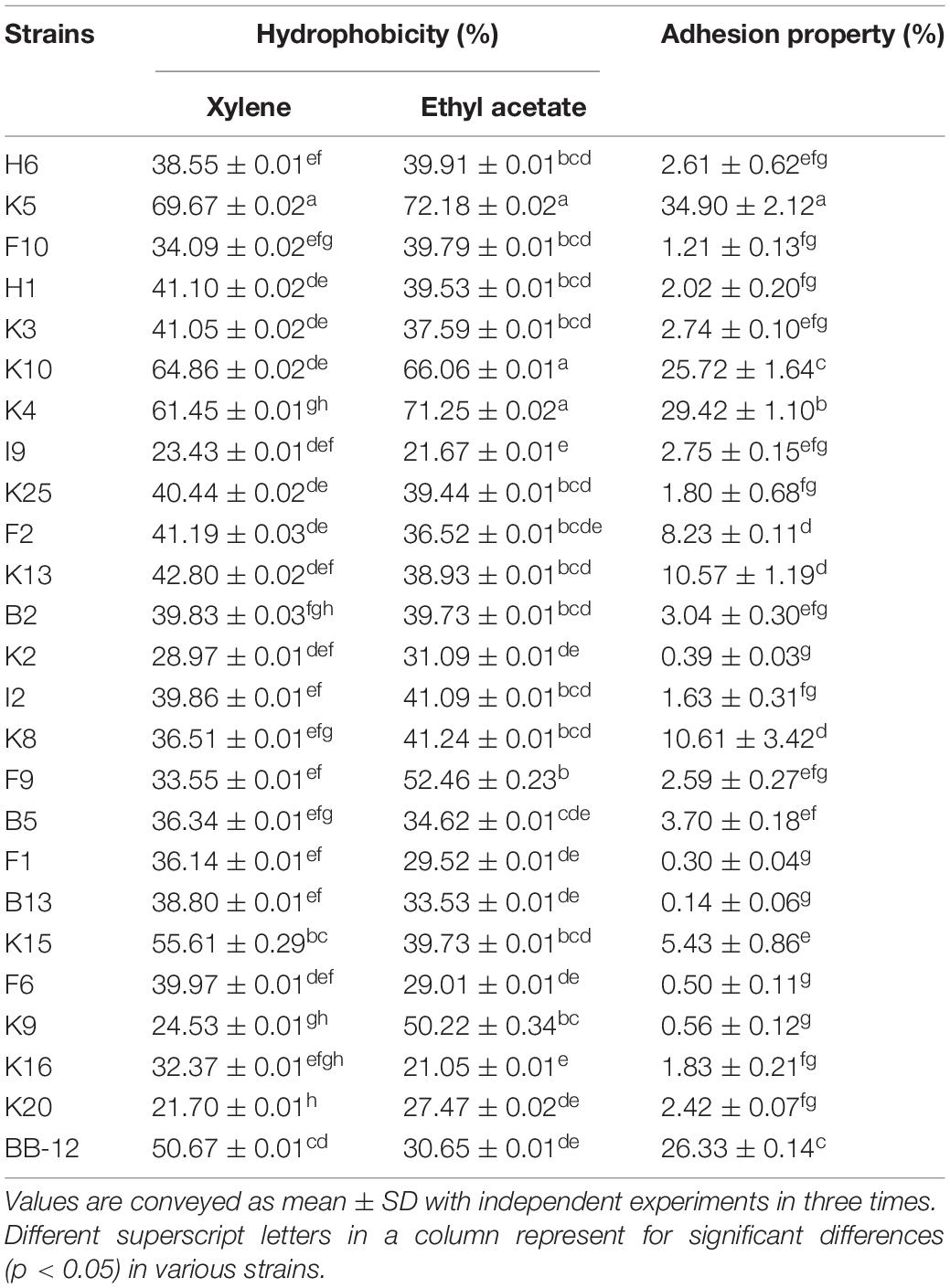
Table 2. Adhesion properties of B. longum subsp. longum strains: Cell surface hydrophobicity of in xylene solution and ethyl acetate solution, and adhesion property of B. longum subsp. longum strains to HT-29 cells.
In this study, all B. longum subsp. longum isolates showed the strain-specific property to HT-29 cells adhesion (Table 2). The adhesion property of B. longum subsp. longum strains to HT-29 cells ranged from 0.14 to 34.90%. B. longum subsp. longum K5 and K4 exhibited significantly higher (p < 0.05) adhesion ability than that of the reference strain BB-12 (26.33%), with the adhesion property of 34.9% and 29.42%.
In this study, PCA was applied for evaluating the adhesion properties of all B. longum subsp. longum strains. As shown in Figure 5, two principal components (PCs) were taken out independently. PC 1 represented 61.31% of the variance, including autoaggregation in 1, 4, and 24 h. PC 2 explained 16.76% of the variance, including CSH in ethyl-acetate and xylene and adhesion of Bifidobacteria to HT-29 cells. The total score was applied for ranking strains in this study (Supplementary Table 2). Among these strains, B. longum subsp. longum B13, F2, K4, K5, K10, K13, and K15 were extracted with a score greater than 0. The results indicated that these seven strains had good adhesion properties, and these strains were selected for further study.
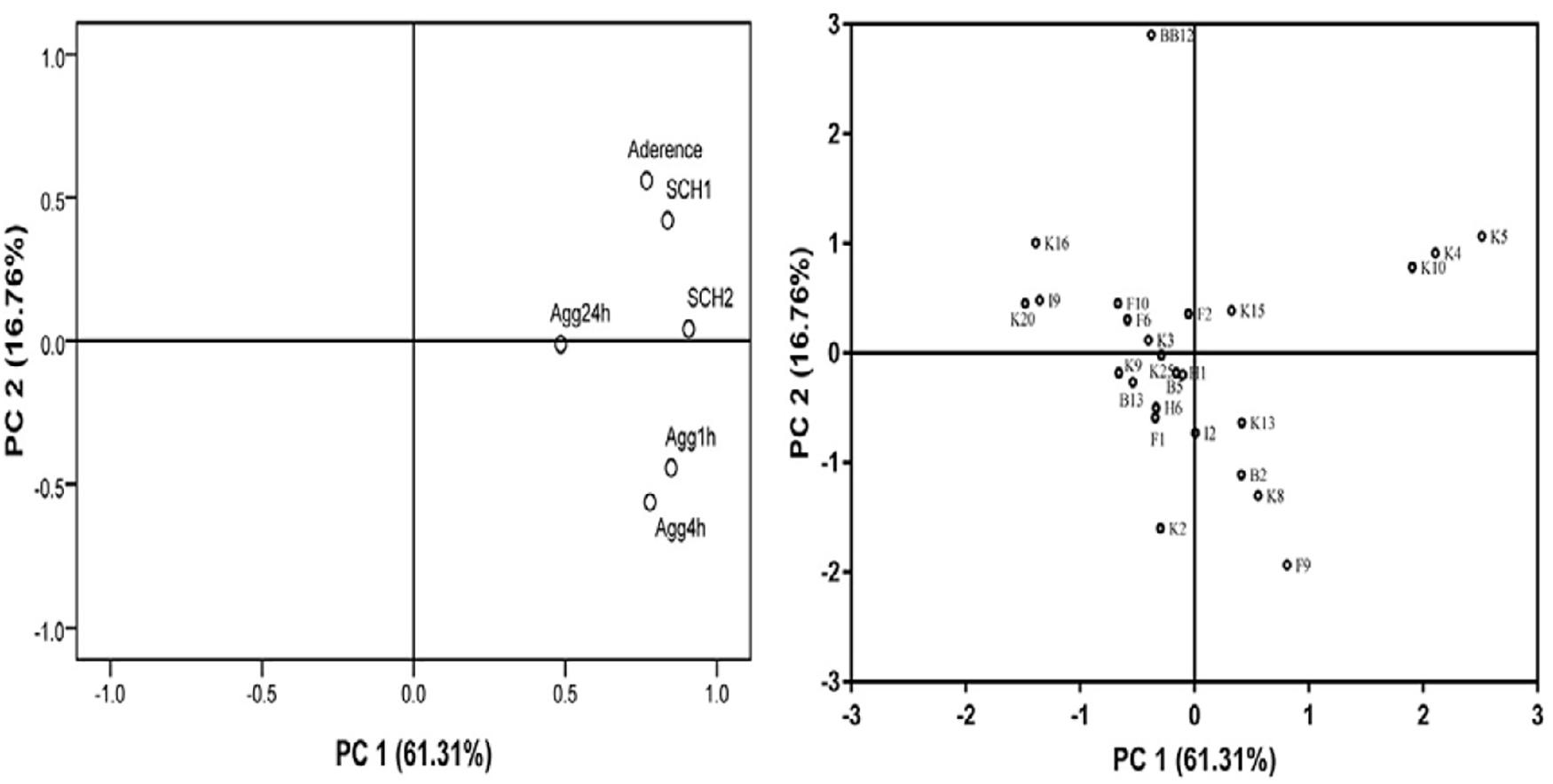
Figure 5. Loading plot of principal component analysis. The image in the left is loading plot represented for six independent assays of adhesion properties: autoaggregation in 1 h (Agg1 h), autoaggregation in 4 h (Agg4 h), autoaggregation in 24 h (Agg24 h), cell surface hydrophobicity level in xylene (CSH1) and ethyl acetate solution (CSH2), and adhesion property to HT-29 cells. The image in the right is score plot of the adhesion properties of B. longum subsp. longum strains.
Figure 6 showed a survival rate of 7 B. longum subsp. longum strains treated by artificial gastrointestinal digestion in vitro. B. longum subsp. longum K5 showed the highest tolerance under gastric fluid with a survival rate of 74.72%, followed by B. longum subsp. longum K7 (74.53%), F2 (74.15%), and K15 (72.53%), which exhibited no significant difference (p > 0.05) with the reference strain BB-12 (73.91%). B. longum subsp. longum B13, K4, and K13 showed negligible survival in artificial gastrointestinal digestion conditions, excluding them from further study. After treatment of the intestinal fluid, there was no significant difference (p > 0.05) in survival rate among B. longum subsp. longum F2, K5, K10, K15, and BB-12 strains. Thus, B. longum subsp. longum F2, K5, K10, and K15 were selected for further study.
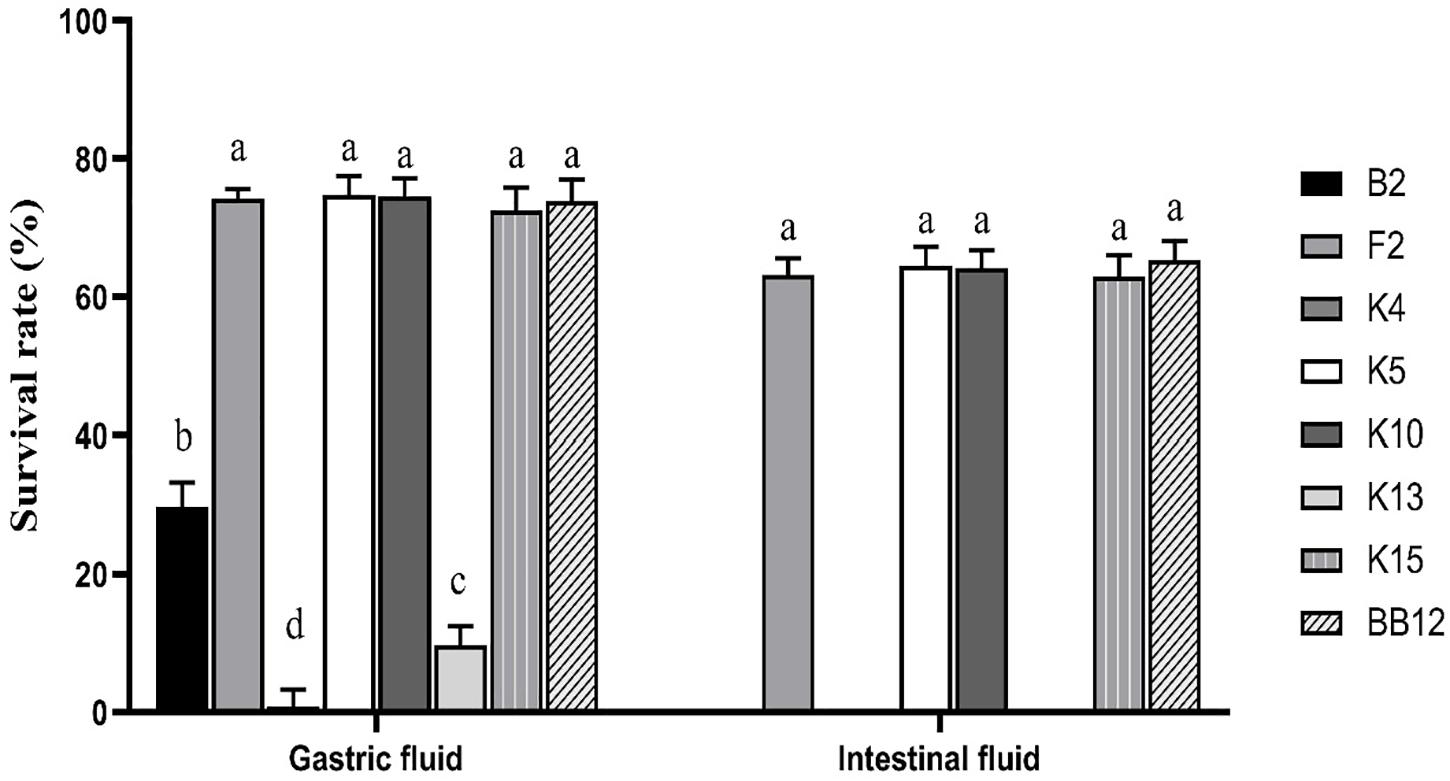
Figure 6. Survival rate of B. longum subsp. longum strains in the simulation of gastrointestinal digestion. P < 0.05 was considered to demonstrate statistically significant differences and recorded with different letters.
The results of antibiotic susceptibility of B. longum subsp. longum strains were accessed by using common antibiotics, as shown in Table 3. All of the B. longum subsp. longum strains were resistant to gentamicin, kanamycin, streptomycin, and ciprofloxacin. Additionally, B. longum subsp. longum K10 also exhibited resistance to clindamycin and trimethoprim (MTP). The results indicated that B. longum subsp. longum K10 were antibiotic-resistant compared to the other strains.
As exhibited in Figure 7, the cell proliferation of CCD 841 CoN cells was evaluated by CCK-8. The cell proliferation of B. longum subsp. longum K5 (64.62%) was significantly higher (p < 0.05) than that of other strains, including the reference strain BB-12. There was no significant difference (p > 0.05) in cell proliferation among B. longum subsp. longum F2, K15, and BB-12. The results indicated that B. longum subsp. longum K5 possessed the excellent cell proliferation of CCD 841 CoN cells.
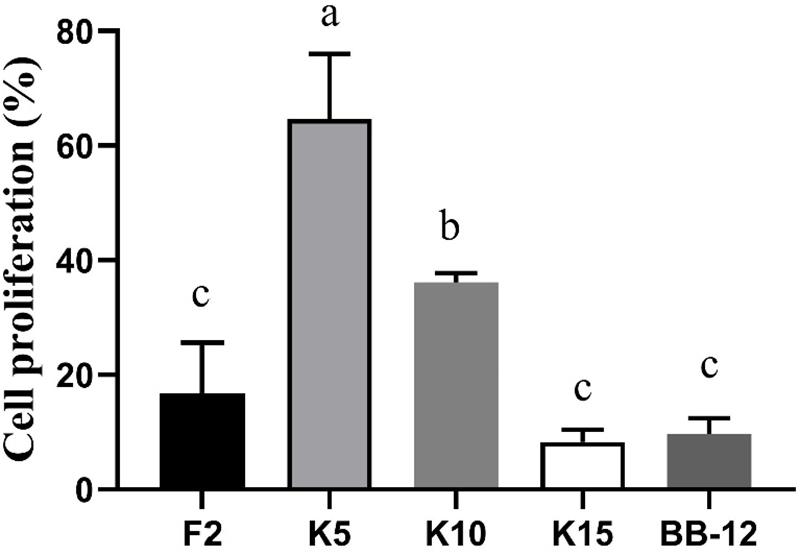
Figure 7. CCD 841 CoN cells proliferation of the selected B. longum subsp. longum strains. P < 0.05 was considered to demonstrate statistically significant differences and recorded with different letters.
All the test strains exhibited varying degrees of reducing power (Figure 8A). The IC of B. longum subsp. longum K10 showed the highest reducing power among these five strains, followed by B. longum subsp. longum K5. The IC of B. longum subsp. longum K5 exhibited no significant difference (p > 0.05) with BB-12. The CFE and CFS of B. longum subsp. longum K5 possessed significantly higher (p < 0.05) reducing power than that of reference strain BB-12. The CFS of all test strains showed higher reducing power than IC and CFE of strains.
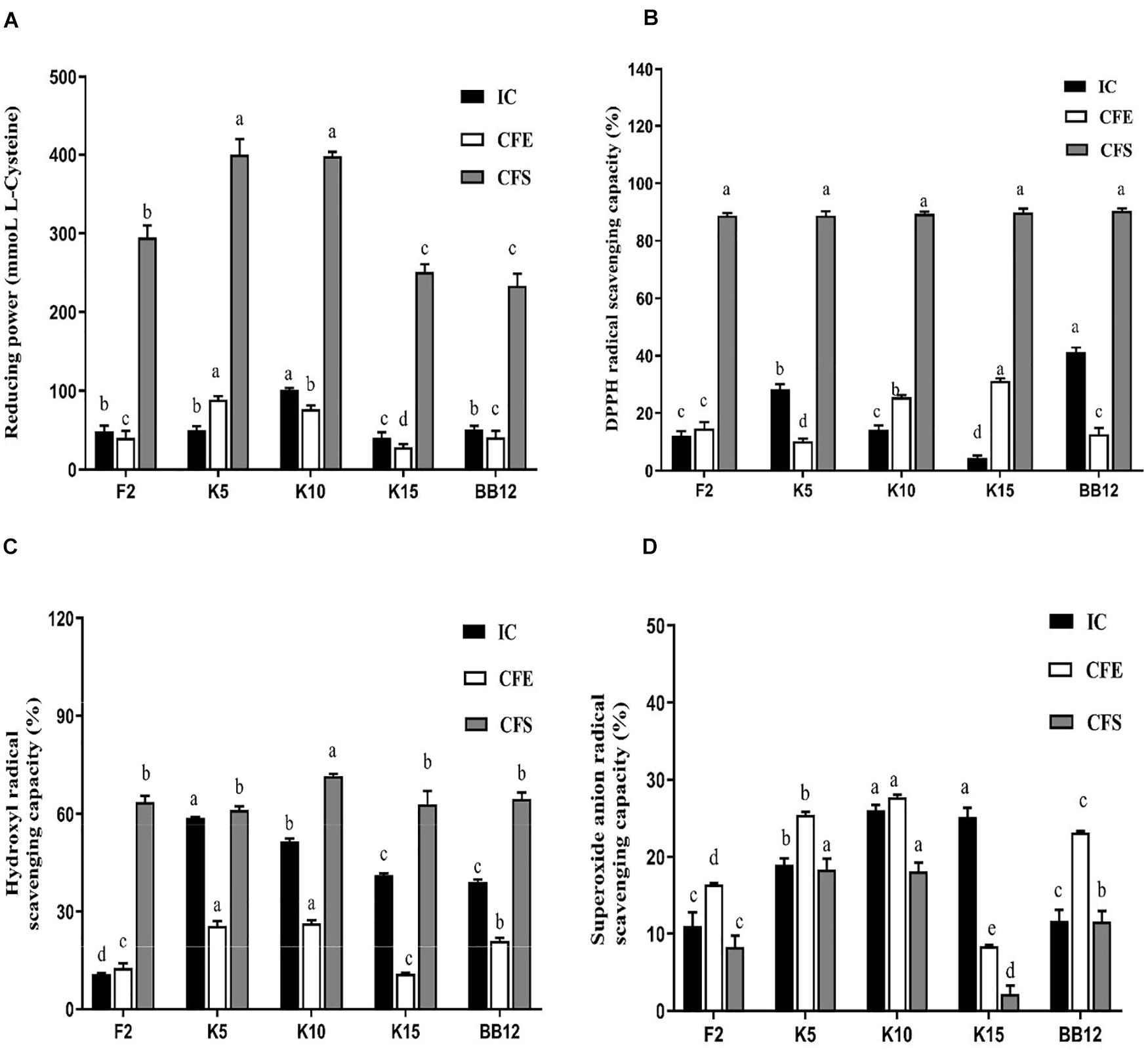
Figure 8. Antioxidant properties of B. longum subsp. longum strains, including reducing power (A), hydroxyl radical scavenging capacity (B), DPPH radical scavenging capacity (C), and superoxide anion radical scavenging capacity (D). IC, intact cells; CFS, cell-free supernatant; CFE, cell-free extracts. P < 0.05 was considered to demonstrate statistically significant differences and recorded with different letters.
As shown in Figure 8B, the IC of BB-12 showed the highest capacity in DPPH radical scavenging, followed by B. longum subsp. longum K5. The CFE of B. longum subsp. longum K15 exhibited significantly higher (p < 0.05) scavenging capacity of DPPH radical than that of reference strain BB-12. The CFS of 4 B. longum subsp. longum strains displayed an excellent capacity of DPPH radical scavenging with no significant difference (p > 0.05). The CFS of all test strains showed higher DPPH radical scavenging capacity than IC and CFE of strains.
In this study, the IC and CFE of B. longum subsp. longum K5 had the highest hydroxyl radical scavenging capacity (58.76 and 25.57%), which were significantly higher (p < 0.05) than those of BB-12 (39.00 and 21.02%). The CFS of B. longum subsp. longum K10 expressed significantly higher (p < 0.05) scavenging capacity of hydroxyl radical than that of other strains (Figure 8C). The CFS of B. longum subsp. longum K5 showed no significant difference (p > 0.05) with BB-12. The CFS of all test strains showed higher hydroxyl radical scavenging capacity than IC and CFE of strains.
As shown in Figure 8D, the superoxide anion radical scavenging capacity of IC and CFE of B. longum subsp. longum K10 (26.01 and 27.71%) and B. longum subsp. longum K5 (18.98 and 25.44%) was significantly higher than BB-12 (11.62 and 23.15%). The CFS of B. longum subsp. longum K5 and K10 showed significantly better (p < 0.05) superoxide anion radical scavenging capacity than other strains, including the reference strain BB-12.
Bifidobacterium is rich in breast-fed infants, especially infants aged from the first week to the sixth month (Hill C. J. et al., 2017). Bifidobacterium was confirmed to be beneficial to the host throughout the whole lifespan (Turroni et al., 2018). Thus, we try to isolate Bifidobacterium strains from infants. However, Bifidobacterium strains could not be isolated from all the feces samples (Tannock et al., 2016); 18% of the samples were found absent from Bifidobacteria reported by Eshaghi et al. (2017). In this study, we isolated 31 Bifidobacterium strains from 7 infant samples. Moreover, it was reported that the Bifidobacterium genus contained various species, which were rough to distinguish using 16S rRNA gene sequencing, especially B. longum, while the hsp60 gene was more accurate than the 16S rRNA gene to discriminate diversified species of Bifidobacterium (Baffoni et al., 2013; Stenico et al., 2014). Thus, these two methods were both used in our study to identify the strains isolated from infant feces. Twenty-four strains belonged to B. longum subsp. longum in this study. We found that most of the Bifidobacterium strains (77.42%) belonged to B. longum subsp. longum. This finding is consistent with that of Yan et al., who isolated 93 B. longum subsp. longum strains in 30 samples contained 15 feces of adults and 15 feces of infants (Yan et al., 2017). As previously reported, B. longum, especially B. longum subsp. longum, have a high abundance in the gut microbiota composition of infants (Arrieta et al., 2014). Furthermore, B. longum subsp. longum verified that it could assist non-HMO metabolizing Bifidobacterium strains to grow moderately via metabolizing HMO and then benefited the development of infant gut microbiota, even the long-term health of infants (Lawson et al., 2020). Therefore, B. longum subsp. longum strains were selected for further study.
Adhesion to intestinal mucosal surfaces was considered the prerequisite for Bifidobacterium to exert beneficial effects on the host (Monteagudo-Mera et al., 2019). The mechanism of Bifidobacterium was varied from strain to strain. Thus, to evaluate the adhesion ability of Bifidobacterium strains, a case-by-case assay was needed. The strong autoaggregation was viewed as the first step of bacteria in the process of adhesion to cause the barrier formed by the bacteria to prevent the adhesion of pathogenic bacteria (Saito et al., 2019). In this study, most strains exhibited excellent autoaggregation, as shown in Figure 4. Moreover, cell surface hydrophobicity was considered the driver to promote the adhesion ability, and microbial adhesion to hydrocarbon was the classical method to value the level of cell surface hydrophobicity (Haddaji et al., 2015). B. longum subsp. longum B13, F2, K4, K5, K10, K13, and K15 showed higher cell surface hydrophobicity among the 24 strains in this study. Furthermore, the adhesion ability of Bifidobacterium strains was commonly assessed by epithelial cells such as Caco-2 and HT-29 in vitro to mimic adhesion to intestinal epithelial cells of humans (Tuo et al., 2013; Zuo et al., 2016; Sun et al., 2020). In this study, B13, F2, K4, K5, K10, K13, and K15 showed excellent adhesion to HT-29 cells, as exhibited in Table 2. In order to access the adhesion ability of B. longum subsp. longum strains, PCA were used to analyze the autoaggregation, cell surface hydrophobicity, and adhesion ability to HT-29 cells. As a result, B. longum subsp. longum B13, F2, K4, K5, K10, K13, and K15 possessed excellent adhesion capacity were selected for further study.
Survival in the human digestive tract (GIT) is essential for exerting probiotic function (Rabah et al., 2017; Gut et al., 2018). The reference strain BB-12 is a commercial probiotic with excellent gastric intestinal fluid tolerance, as we all know (Liu et al., 2015). Shang et al. (2020) have also assessed the ability of gastric intestinal fluid tolerance of Bifidobacterium strains according to the reference strain BB-12. In this study, B. longum subsp. longum F2, K5, K10, and K15 showed high tolerance of gastrointestinal digestion fluid with no significant difference with BB-12. B. longum subsp. longum B13, K4, and K13 could hardly survive in the same condition (excluded for further study).
It is essential to evaluate the antibiotics susceptibility of probiotics before being used as a dietary supplement (Borriello et al., 2003). In this study, all of the B. longum subsp. longum strains were resistant to gentamicin, kanamycin, and streptomycin only. The result was consistent with the finding of Min et al., who also evaluated the antibiotics susceptibility of B. bifidum BGN4 and found it was resistant to gentamicin, kanamycin, and streptomycin (Min et al., 2018). It has been reported that Bifidobacterium strains were generally resistant to gentamicin, kanamycin, and streptomycin and found that their resistances were intrinsic (Mtt et al., 2007). The results indicated that B. longum subsp. longum F2, K5, and K15 were susceptible to antibiotics.
The immature intestines may cause several intestinal-related diseases such as necrotizing enterocolitis and postpartum growth restriction (Hill D. R. et al., 2017; Meng et al., 2020). Thus, it is vital to promote the development of the intestine of infants. Several studies have been proved to promote the maturation of the intestines (Hill D. R. et al., 2017; Westrm et al., 2020). At the same time, the strains were not derived from the infants. This study assessed the cell proliferation of CCD 841 CoN cells used as human fetal colon epithelial cells. B. longum subsp. longum K5 possessed the excellent cell proliferation of CCD 841 CoN cells, which indicated that B. longum subsp. longum K5 could be used for drives of intestinal epithelial cell growth.
Excessive oxidative stress was harmful to humans, even infants (Hong et al., 2012; Weber et al., 2014). Excessive ROS may cause oxidative injury and cell damage (Xu et al., 2011). Thus, probiotics with the ability to reduce ROS were considered an excellent potential antioxidant. DPPH radical, reducing power (Xie et al., 2010), hydroxyl radical, and superoxide radical (Zhong et al., 2013) were used to evaluate the antioxidant capacity because of its high sensitivity and rapidity. In this study, B. longum subsp. longum K5 and K10 exhibited excellent reducing power, DPPH radical scavenging capacity, hydroxyl radical scavenging capacity, and superoxide anion radical scavenging capacity (Figure 5). It is worth noting that the CFS of test strains showed a higher antioxidant ability than that of the IC and CFE. The findings were consistent with the results of previous studies; the CFS of B. animalis 01 showed higher antioxidant ability than that of the IC (Shen et al., 2011). It may be caused by kinds of metabolic products, such as soluble protein (Zhang et al., 2012) and exopolysaccharides (Xu et al., 2011). It has been reported that peptides produced by lactic acid bacteria after fermentation of whey have antioxidant properties (Embiriekah et al., 2018). Liu et al. stated that exopolysaccharides possessed the antioxidant activity (Liu et al., 2011), monosaccharide composition and glycosidic linkage were proved related with the free radical scavenging activity (Torino et al., 2010).
In summary, 24 B. longum subsp. longum strains were isolated from healthy infant feces. Among them, B. longum subsp. longum K5 had high adhesion ability, tolerance of artificial digestive tract, antibiotics susceptibility, intestinal cell proliferation, and antioxidant capacity. Overall, our study indicated B. longum subsp. longum K5 has excellent potential probiotic properties and antioxidant capacity and can be used as the candidate for infants’ dietary antioxidant supplements. However, most in vivo and clinical investigations are required before B. longum subsp. longum K5 is used for dietary supplements.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Northeast Agricultural University Hospital Medical Science Ethics Committee. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
LZ, BL, and GH conceived the study and designed the project. SW helped to collect fecal samples of infants and investigate infant information. LZ, JD, JS, and DL performed the experiment. LZ, JG, and FL analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Grants from the “Hundred, Thousand and Ten Thousand” Science and Technology Major Special Project of Heilongjiang Province: Dairy Products and Meat Processing (No. 2020ZX07B01), National Natural Science Youth Foundation of China (No. 31801518), “Young Talents” Project of Northeast Agricultural University (18QC52), National Natural Science Foundation of China (32072190), and Academic Backbone Plan of Northeast Agricultural University (No. 19YJXG10) funded this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the mothers for providing their infants’ fecal samples.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.756519/full#supplementary-material
Akay, H. K., Tokman, H. B., Hatipoglu, N., Hatipoglu, H., Siraneci, R., Demirci, M., et al. (2014). The relationship between Bifidobacteria and allergic asthma and/or allergic dermatitis: a prospective study of 0-3 years-old children in Turkey. Anaerobe 28, 98–103. doi: 10.1016/j.anaerobe.2014.05.006
Ammor, M. S., Florez, A. B., and Mayo, B. (2007). Antibiotic resistance in non-enterococcal lactic acid bacteria and Bifidobacteria. Food Microbiol. 24, 559–570. doi: 10.1016/j.fm.2006.11.001
Arrieta, M.-C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M., and Finlay, B. (2014). The intestinal microbiome in early life: health and disease. Front. Immunol. 5:427. doi: 10.3389/fimmu.2014.00427
Baffoni, L., Stenico, V., Strahsburger, E., Gaggìa, F., Di Gioia, D., Modesto, M., et al. (2013). Identification of species belonging to the Bifidobacterium genus by PCR-RFLP analysis of a hsp60 gene fragment. BMC Microbiol. 13:149. doi: 10.1186/1471-2180-13-149
Bevilacqua, L., Ovidi, M., Di Mattia, E., Trovatelli, L. D., and Canganella, F. (2003). Screening of Bifidobacterium strains isolated from human faeces for antagonistic activities against potentially bacterial pathogens. Microbiol. Res. 158, 179–185. doi: 10.1078/0944-5013-00192
Blumberg, R. S., and Gensollen, T. (2017). Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 139, 1084–1091. doi: 10.1016/j.jaci.2017.02.011
Borriello, S., Hammes, W., Holzapfel, W., Marteau, P., Schrezenmeir, J., Vaara, M., et al. (2003). Safety of probiotics that contain lactobacilli or Bifidobacteria. Clin. Infect. Dis. 36, 775–780. doi: 10.1086/368080
Cai, C., Zhang, Z., Morales, M., Wang, Y., Khafipour, E., and Friel, J. (2019). Feeding practice influences gut microbiome composition in very low birth weight preterm infants and the association with oxidative stress: a prospective cohort study. Free Radic. Biol. Med. 142, 146–154. doi: 10.1016/j.freeradbiomed.2019.02.032
Celebioglu, H. U., Olesen, S. V., Prehn, K., Lahtinen, S. J., Brix, S., Abou Hachem, M., et al. (2017). Mucin-and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J. Proteom. 163, 102–110. doi: 10.1016/j.jprot.2017.05.015
Cong, X., Xu, W., Janton, S., Henderson, W. A., Matson, A., McGrath, J. M., et al. (2016). Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One 11:e0152751. doi: 10.1371/journal.pone.0152751
Crovetto, F., Crispi, F., Borras, R., Paules, C., Casas, R., Martín-Asuero, A., et al. (2021). Mediterranean diet, mindfulness-based stress reduction and usual care during pregnancy for reducing fetal growth restriction and adverse perinatal outcomes: IMPACT BCN (Improving Mothers for a better PrenAtal Care Trial BarCeloNa): a study protocol for a randomized controlled trial. Trials 22:362. doi: 10.1186/s13063-021-05309-2
Duranti, S., Lugli, G. A., Mancabelli, L., Armanini, F., Turroni, F., James, K., et al. (2017). Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5:66. doi: 10.1186/s40168-017-0282-6
Embiriekah, S., Bulatovic, M., Boric, M., Zaric, D., and Rakin, M. (2018). Antioxidant activity, functional properties and bioaccessibility of whey protein hydrolysates. Int. J. Dairy Technol. 71, 243–252.
Eshaghi, M., Bibalan, M. H., Rohani, M., Esghaei, M., Douraghi, M., Talebi, M., et al. (2017). Bifidobacterium obtained from mother’s milk and their infant stool; A comparative genotyping and antibacterial analysis. Microb. Pathog. 111, 94–98. doi: 10.1016/j.micpath.2017.08.014
Guang-Yu, W., Ming, L., Fang, M., Hu-Hu, W., Xing-Lian, X., and Guang-Hong, Z. (2017). Physicochemical properties of Pseudomonas fragi isolates response to modified atmosphere packaging. FEMS Microbiol. Lett. 364:fnx106. doi: 10.1093/femsle/fnx106
Gut, A. M., Vasiljevic, T., Yeager, T., and Donkor, O. N. (2018). Salmonella infection-prevention and treatment by antibiotics and probiotic yeasts: a review. Microbiology 164, 1327–1344. doi: 10.1099/mic.0.000709
Haddaji, N., Mahdhi, A. K., Krifi, B., Ismail, M. B., and Bakhrouf, A. (2015). Change in cell surface properties of Lactobacillus casei under heat shock treatment. FEMS Microbiol. Lett. 362:fnv047. doi: 10.1093/femsle/fnv047
Hanson, C., Lyden, E., Furtado, J., Van Ormer, M., and Anderson-Berry, A. (2016). A comparison of nutritional antioxidant content in breast milk, donor milk, and infant formulas. Nutrients 8:681. doi: 10.3390/nu8110681
Hill, C. J., Lynch, D. B., Murphy, K., Ulaszewska, M., Jeffery, I. B., O’Shea, C. A., et al. (2017). Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET cohort. Microbiome 5:4. doi: 10.1186/s40168-016-0213-y
Hill, D. R., Sha, H., Nagy, M. S., Yadagiri, V. K., Courtney, F., Dishari, M., et al. (2017). Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. ELife 6:e29132. doi: 10.7554/eLife.29132.031
Hong, Z., Yu, C. H., Jiang, Y. P., Cheng, P., He, K., Tang, J. Y., et al. (2012). Protective effects of polydatin from polygonum cuspidatum against carbon tetrachloride-induced liver injury in mice. PLoS One 7:e46574. doi: 10.1371/journal.pone.0046574
Krausova, G., Hyrslova, I., and Hynstova, I. (2019). In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 5:100. doi: 10.3390/fermentation5040100
Lawson, M. A., O’Neill, I. J., Kujawska, M., Javvadi, S. G., Wijeyesekera, A., Flegg, Z., et al. (2020). Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 14, 635–648. doi: 10.1038/s41396-019-0553-2
Li, L., Jiang, Y. J., Yang, X. Y., Liu, Y., Wang, J. Y., and Man, C. X. (2014). Immunoregulatory effects on caco-2 cells and mice of exopolysaccharides isolated from Lactobacillus acidophilus NCFM. Food Funct. 5, 3261–3268. doi: 10.1039/C4FO00565A
Li, S., Zhao, Y., Zhang, L., Zhang, X., Huang, L., Li, D., et al. (2012). Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 135, 1914–1919. doi: 10.1016/j.foodchem.2012.06.048
Liang, T., Guan, R., Wang, Z., Shen, H., Xia, Q., and Liu, M. (2017). Comparison of anticancer activity and antioxidant activity between cyanidin-3-O-glucoside liposomes and cyanidin-3-O-glucoside in Caco-2 cells in vitro. RSC Adv. 7, 37359–37368. doi: 10.1039/C7RA06387C
Lin, M.-Y., and Yen, C.-L. (1999). Antioxidative ability of lactic acid bacteria. J. Agric. Food Chem. 47, 1460–1466. doi: 10.1021/jf981149l
Liu, C., Tseng, K. C., Chiang, S. S., Lee, B. H., Hsu, W. H., and Pan, T. M. (2011). Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J. Sci. Food Agric. 91, 2284–2291. doi: 10.1002/jsfa.4456
Liu, G., Ren, L., Song, Z., Wang, C., and Sun, B. (2015). Purification and characteristics of bifidocin A, a novel bacteriocin produced by Bifidobacterium animals BB04 from centenarians’ intestine. Food Control 50, 889–895. doi: 10.1016/j.foodcont.2014.10.049
Ma, J., Li, Z., Zhang, W., Zhang, C., Zhang, Y., Mei, H., et al. (2020). Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci. Rep. 10:15792. doi: 10.1038/s41598-020-72635-x
Mandal, H., Jariwala, R., and Bagchi, T. (2016). Isolation and characterization of lactobacilli from human faeces and indigenous fermented foods for their potential application as probiotics. Can. J. Microbiol. 62, 349–359. doi: 10.1139/cjm-2015-0576
Meng, D., Sommella, E., Salviati, E., Campiglia, P., Ganguli, K., Djebali, K., et al. (2020). Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr. Res. 88, 209–217. doi: 10.1038/s41390-019-0740-x
Milani, C., Mancabelli, L., Lugli, G. A., Duranti, S., Turroni, F., Ferrario, C., et al. (2015). Exploring vertical transmission of Bifidobacteria from mother to child. Appl. Environ. Microbiol. 81, 7078–7087. doi: 10.1128/AEM.02037-15
Min, K., Seockmo, K., Sun, K., Hyun, L., Jin, H., Kang, S., et al. (2018). Safety evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI. Int. J. Mol. Sci. 19:1422. doi: 10.3390/ijms19051422
Mishra, V., Shah, C., Mokashe, N., Chavan, R., Yadav, H., and Prajapati, J. (2015). Probiotics as potential antioxidants: a systematic review. J. Agric. Food Chem. 63, 3615–3626. doi: 10.1021/jf506326t
Modesto, M., Michelini, S., Sansosti, M. C., Filippo, C. D., Cavalieri, D., Qviris, L., et al. (2018). Bifidobacterium callitrichidarum sp. nov. from the faeces of the emperor tamarin (Saguinus imperator). Int. J. Syst. Evol. Microbiol. 68, 141–148. doi: 10.1099/ijsem.0.002472
Monteagudo-Mera, A., Rastall, R. A., Gibson, G. R., Charalampopoulos, D., and Chatzifragkou, A. (2019). Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 103, 6463–6472. doi: 10.1007/s00253-019-09978-7
Morelli, L., and Capurso, L. (2012). FAO/WHO guidelines on probiotics: 10 years later. J. Clin. Gastroenterol. 46, S1–S2. doi: 10.1097/MCG.0b013e318269fdd5
Mtt, J., Hoek, A., Domig, K. J., Saarela, M., Floréz, A., Brockmann, E., et al. (2007). Susceptibility of human and probiotic Bifidobacterium spp. to selected antibiotics as determined by the Etest method. Int. Dairy J. 17, 1123–1131. doi: 10.1016/j.idairyj.2007.01.008
Nazhand, A., Souto, E. B., Lucarini, M., Souto, S. B., Durazzo, A., and Santini, A. (2020). Ready to use therapeutical beverages: focus on functional beverages containing probiotics, prebiotics and synbiotics. Beverages 6:26. doi: 10.3390/beverages6020026
Nuriel-Ohayon, M., Neuman, H., and Koren, O. (2016). Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 7:1031. doi: 10.3389/fmicb.2016.01031
Okochi, M., Sugita, T., Asai, Y., Tanaka, M., and Honda, H. (2017). Screening of peptides associated with adhesion and aggregation of Lactobacillus rhamnosus GG in vitro. Biochem. Eng. J. 128, 178–185. doi: 10.1016/j.bej.2017.10.004
Osorio, J. S. (2020). Gut health, stress, and immunity in neonatal dairy calves: the host side of host-pathogen interactions. J. Anim. Sci. Biotechnol. 11:105. doi: 10.1186/s40104-020-00509-3
Oyaizu, M. (1988). Antioxidative activities of browning products of glucosamine fractionated by organic solvent and thin-layer chromatography. Nippon Shokuhin Kogyo Gakkaishi 35, 771–775. doi: 10.3136/nskkk1962.35.11_771
Rabah, H., Rosa do Carmo, F. L., and Jan, G. (2017). Dairy propionibacteria: versatile probiotics. Microorganisms 5:24. doi: 10.3390/microorganisms5020024
Rajoka, M. R., Mehwish, H. M., Siddiq, M., Haobin, Z., Zhu, J., Yan, L., et al. (2017). Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 84, 271–280. doi: 10.1016/j.lwt.2017.05.055
Ruiz, L., Delgado, S., Ruas-Madiedo, P., Sánchez, B., and Margolles, A. (2017). Bifidobacteria and their molecular communication with the immune system. Front. Microbiol. 8:2345. doi: 10.3389/fmicb.2017.02345
Saito, K., Tomita, S., and Nakamura, T. (2019). Aggregation of Lactobacillus brevis associated with decrease in pH by glucose fermentation. Biosci. Biotechnol. Biochem. 83, 1523–1529. doi: 10.1080/09168451.2019.1584522
Salyers, A. A., Gupta, A., and Wang, Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Shang, J., Wan, F., Zhao, L., Meng, X., and Li, B. (2020). Potential immunomodulatory activity of a selected strain Bifidobacterium bifidum H3-R2 as evidenced in vitro and in immunosuppressed mice. Front. Microbiol. 11:2089. doi: 10.3389/fmicb.2020.02089
Shen, Q., Shang, N., and Li, P. (2011). In vitro and in vivo antioxidant activity of Bifidobacterium animalis 01 isolated from centenarians. Curr. Microbiol. 62, 1097–1103. doi: 10.1007/s00284-010-9827-7
Silvia, A., Claire, W., Catherine, S., and Paul, R. R. (2016). Gut bifidobacteria populations in human health and aging. Front. Microbiol. 7:e10667. doi: 10.3389/fmicb.2016.01204
Sommer, M., Dantas, G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. doi: 10.1126/science.1176950
Stenico, V., Michelini, S., Modesto, M., Baffoni, L., Mattarelli, P., and Biavati, B. (2014). Identification of Bifidobacterium spp. using hsp60 PCR-RFLP analysis: an update. Anaerobe 26, 36–40. doi: 10.1016/j.anaerobe.2013.12.004
Sun, L., Tian, W., Guo, X., Zhang, Y., Liu, X., Li, X., et al. (2020). Lactobacillus gasseri JM1 with potential probiotic characteristics alleviates inflammatory response by activating the PI3K/Akt signaling pathway in vitro. J. Dairy Sci. 103, 7851–7864. doi: 10.3168/jds.2020-18187
Tannock, G. W., Lee, P. S., Wong, K. H., and Lawley, B. (2016). Why don’t all infants have Bifidobacteria in their stool? Front. Microbiol. 7:834. doi: 10.3389/fmicb.2016.00834
Torino, M. I., Taranto, M. P., Sesma, F., and Valdez, G. (2010). Heterofermentative pattern and exopolysaccharide production by Lactobacillus helveticus ATCC 15807 in response to environmental pH. J. Appl. Microbiol. 91, 846–852. doi: 10.1046/j.1365-2672.2001.01450.x
Tuo, Y., Yu, H., Ai, L., Wu, Z., Guo, B., and Chen, W. (2013). Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 96, 4252–4257. doi: 10.3168/jds.2013-6547
Turroni, F., Milani, C., Duranti, S., Ferrario, C., Lugli, G. A., Mancabelli, L., et al. (2018). Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell. Mol. Life Sci. 75, 103–118. doi: 10.1007/s00018-017-2672-0
Uttara, B., Singh, A. V., Zamboni, P., and Mahajan, R. T. (2009). Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 7, 65–74. doi: 10.2174/157015909787602823
Vankerckhoven, V., Huys, G., Vancanneyt, M., Vael, C., Klare, I., Romond, M.-B., et al. (2008). Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trend Food Sci. Technol. 19, 102–114. doi: 10.1016/j.tifs.2007.07.013
Weber, D., Stuetz, W., Bernhard, W., Franz, A., Raith, M., Grune, T., et al. (2014). Oxidative stress markers and micronutrients in maternal and cord blood in relation to neonatal outcome. Eur. J. Clin. Nutr. 68, 215–222. doi: 10.1038/ejcn.2013.263
Westrm, B., Sureda, E. A., Pierzynowska, K., Pierzynowski, S. G., and Pérez-Cano, F. (2020). The immature gut barrier and its importance in establishing immunity in newborn mammals. Front. Immunol. 11:1153. doi: 10.3389/fimmu.2020.01153
Xie, J. H., Xie, M. Y., Nie, S. P., Shen, M. Y., Wang, Y. X., and Li, C. (2010). Isolation, chemical composition and antioxidant activities of a water-soluble polysaccharide from Cyclocarya paliurus (Batal.) Iljinskaja. Food Chem. 119, 1626–1632. doi: 10.1016/j.foodchem.2009.09.055
Xu, R., Shang, N., and Li, P. (2011). Invitro and invivo antioxidant activity of exopolysaccharide fractions from Bifidobacterium animalis RH. Anaerobe 17, 226–231. doi: 10.1016/j.anaerobe.2011.07.010
Yan, F., Li, N., Yue, Y., Wang, C., Zhao, L., Evivie, S. E., et al. (2020). Screening for potential novel probiotics with dipeptidyl peptidase IV- inhibiting activity for type 2 diabetes attenuation in vitro and in vivo. Front. Microbiol. 10:2855. doi: 10.3389/fmicb.2019.02855
Yan, S., Zhao, G., Liu, X., Zhao, J., Zhang, H., and Chen, W. (2017). Production of exopolysaccharide by Bifidobacterium longum isolated from elderly and infant feces and analysis of priming glycosyltransferase genes. RSC Adv. 7, 31736–31744. doi: 10.1039/C7RA03925E
Zhang, C., Yu, Z., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2019). Colonization and probiotic function of Bifidobacterium longum. J. Funct. Foods 53, 157–165. doi: 10.1016/j.jff.2018.12.022
Zhang, J., Wang, D., Zhang, J., and Wang, Y. (2012). Extraction and antioxidant activity of the soluble protein from Lactobacillus rhamnosus broth. Food Sci. Technol. 37, 235–239.
Zhong, W., Liu, N., Xie, Y., Zhao, Y., Song, X., and Zhong, W. (2013). Antioxidant and anti-aging activities of mycelial polysaccharides from Lepista sordida. Int. J. Biol. Macromol. 60, 355–359. doi: 10.1016/j.ijbiomac.2013.06.018
Zhu, D.-Q., Liu, F., Sun, Y., Yang, L.-M., Xin, L., and Meng, X.-C. (2015). Genome-wide identification of small RNAs in Bifidobacterium animalis subsp. lactis KLDS 2.0603 and their regulation role in the adaption to gastrointestinal environment. PLoS One 10:e0117373. doi: 10.1371/journal.pone.0117373
Zhuang, L., Chen, H., Zhang, S., Zhuang, J., Li, Q., and Feng, Z. (2019). Intestinal microbiota in early life and its implications on childhood health. Genomics Proteomics Bioinformatics 17, 13–25. doi: 10.1016/j.gpb.2018.10.002
Keywords: Bifidobacterium longum subsp. longum strain, infant, potential probiotics, antibiotics susceptibility, cell proliferation, antioxidant
Citation: Zhao L, Wang S, Dong J, Shi J, Guan J, Liu D, Liu F, Li B and Huo G (2021) Identification, Characterization, and Antioxidant Potential of Bifidobacterium longum subsp. longum Strains Isolated From Feces of Healthy Infants. Front. Microbiol. 12:756519. doi: 10.3389/fmicb.2021.756519
Received: 10 August 2021; Accepted: 27 September 2021;
Published: 02 November 2021.
Edited by:
Marciane Magnani, Federal University of Paraíba, BrazilReviewed by:
Sabrina Neves Casarotti, Federal University of Mato Grosso, BrazilCopyright © 2021 Zhao, Wang, Dong, Shi, Guan, Liu, Liu, Li and Huo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bailiang Li, MTU4NDYwOTIzNjJAMTYzLmNvbQ==; Guicheng Huo, Z3VpY2hlbmdodW9AMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.