
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 01 November 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.756100
This article is part of the Research Topic Rhizosphere Microbiology: Toward a Clean and Healthy Soil Environment View all 25 articles
 Chih-Wei Wang1,2,3,4†
Chih-Wei Wang1,2,3,4† Yu-Hsiang Yu5†
Yu-Hsiang Yu5† Chun-Yao Wu1
Chun-Yao Wu1 Ru-Ying Feng6
Ru-Ying Feng6 Kshitij Tandon2
Kshitij Tandon2 Ying-Lien Chen5,6*‡
Ying-Lien Chen5,6*‡ Sen-Lin Tang2,3,7*‡
Sen-Lin Tang2,3,7*‡Wilt disease of roselle (Hibiscus sabdariffa L.) is common in Taiwan; however, the causative agent remains unknown. The stems of wilted roselle are browned, slightly constricted, and covered by white aerial hyphae, suggesting that potential pathogens may originate from soil. To identify the potential pathogens, we conducted a rhizosphere microbiota survey in phenotypically healthy and diseased plants through fungal internal transcribed spacer (ITS) and bacterial 16S rRNA amplicon sequencing for uncovering the microbial compositions in the roselle rhizosphere. The fungal family Nectriaceae exhibited significantly higher abundance in diseased rhizospheres than in healthy rhizospheres, and this bacterial community was more specific to geography (i.e., plot-dependent) than to rhizosphere disease status. However, a few bacterial groups such as Bacilli were associated with the healthy rhizosphere. Fusarium species were the most dominant species of Nectriaceae in the survey and became the main target for potential pathogen isolation. We successfully isolated 119 strains from diseased plants in roselle fields. Koch’s postulates were used to evaluate the pathogenicity of these strains; our results indicated that Fusarium solani K1 (FsK1) can cause wilting and a rotted pith in roselles, which was consistent with observations in the fields. This is the first demonstration that F. solani can cause roselle wilt in Taiwan. Furthermore, these newly isolated strains are the most dominant operational taxonomic units detected in ITS amplicon sequencing in diseased rhizospheres, which serves as further evidence that F. solani is the main pathogen causing the roselle wilt disease. Administration of Bacillus velezensis SOI-3374, a strain isolated from a healthy roselle rhizosphere, caused considerable anti-FsK1 activity, and it can serve as a potential biocontrol agent against roselle wilt disease.
Roselle (Hibiscus sabdariffa L.) is a multipurpose crop that belongs to the Malvaceae family (Alshoosh, 1997; Islam et al., 2016). Roselle is native to West Africa and India and is currently grown in numerous tropical and subtropical regions. It is an annual or perennial, woody or herbaceous plant and can be divided into two common varieties that are cultivated for food (H. sabdariffa var. sabdariffa) and fiber (H. sabdariffa var. altissima Wester) (Alshoosh, 1997; Ansari et al., 2013). China and Thailand are the main growers of this crop in Asia; however, other regions including Mexico, Egypt, Senegal, Tanzania, Jamaica, and Mali are also major producers (Islam et al., 2016).
According to a national survey conducted by the Agricultural and Food Agency in Taiwan, as of 2019, the nationwide cultivation area of roselle was approximately 182.53 ha, with an annual yield of approximately 305,925 kg. Roselle grows at 18 to 35°C, optimally at 25°C, and is intolerant to low temperatures (Ansari et al., 2013). Several roselle cultivars are employed in Taiwan, among which Taitung No. 3 is the one with the highest yield, thickest calyx, and optimal processing quality (Chen and Chen, 2019). Various diseases occur in roselle plants in Taiwan, including gray mold (Botrytis cinerea), irregular leaf spot (Cercospora malayensis), anthracnose (Colletotrichum gloeosporioides), Sclerotinia rot (Sclerotinia sclerotiorum), Phytophthora infection (Phytophthora nicotianae), bacterial wilt (Ralstonia solanacearum), wrinkled leaves and phyllody disorder (16SrI phytoplasma), and diseases caused by nematodes and insect pests (Tzean, 2019).
Roselle wilt disease in Taiwan is prevalent from July to October, with a field incidence rate of up to 80%, leading to tremendous agricultural economic loss. Roselles are grown from seeds, and those that wilt do not flower, and they rot from the roots; the stems of wilted plants are browned, slightly constricted, and covered by white aerial hyphae. Rotted piths are also commonly found in the vertically dissected stem base. The causative agent of roselle wilt disease is yet to be identified in Taiwan, but it has been reported in several countries such as the United States, Malaysia, Mexico, Nigeria, and Egypt. For example, F. oxysporum causes vascular wilt on roselles in the United States and Malaysia (Ooi and Salleh, 1999; Ploetz et al., 2007); Phytophthora parasitica causes crown rot (black foot) along with necrosis at the bases of the stems as well as foliar wilt and death of roselles in Mexico (Estrada et al., 2001); F. oxysporum causes vascular wilt and stem blight on roselles in Nigeria (Amusa et al., 2005); F. solani and Macrophomina phaseolina cause roselle wilt; and F. oxysporum causes pre-emergence damping off on roselles in Egypt (Hassan et al., 2014).
The fungal genus Fusarium is a large pathogenic group in plants and animals. In this genus, F. solani is a common pathogen that causes root rot in many plants and causes aboveground symptoms such as wilt (Coleman, 2016). Sudden death syndrome in soybeans in North and South America (Westphal et al., 2008) and chili wilt disease in India (Sundaramoorthy et al., 2012) are well-known examples of plant diseases caused by F. solani infection. F. solani also causes complex diseases with other pathogens such as nematodes (Gomes et al., 2011).
Although numerous types of crops are infected and severely damaged by Fusarium species, effective disease control methods are limited. Fusarium species produce chlamydospores and thus survive in harsh environments (Hou et al., 2020); thus, infection with this species is challenging to control comprehensively. Along with the use of fungicides, the development of biological control (i.e., biocontrol) has been extensively studied in recent years. Biocontrol occurs naturally in soil environments, and its mechanisms can be categorized into three macro aspects: (1) competitive root colonization, (2) synthesis of allelochemicals, and (3) indirect plant growth promotion through induced systemic resistance (Borriss, 2020; Etesami and Adl, 2020). Biocontrol strains can be screened and isolated using diverse methods, with the microbiome survey having been employed in numerous microbiological and plant pathological studies (Benítez and Gardener, 2009; Poudel et al., 2016; O’Brien, 2017). Notably, the potential biocontrol strains and pathogens may inhabit the same space underground, and understanding the abundance and diversity of biocontrol microbes in healthy and diseased soils can provide mechanism-related insights regarding disease causes and opportunities for developing management strategies (Poudel et al., 2016). Numerous non-pathogenic bacteria and fungi have been used as biocontrol agents for disease management, including Bacillus spp., Pseudomonas spp., Streptomyces spp. and Trichoderma spp. (O’Brien, 2017). In Taiwan, 11 Bacillus strains (six B. amyloliquefaciens and three B. subtilis strains, one B. mycoides, and one B. velezensis) have been registered for plant disease control because Bacillus spp. form endospores that not only expand the shelf life of plant products but also exhibit strong control efficacy against various plant diseases (Borriss, 2020).
In this study, four main objectives were achieved: (1) fungal and bacterial communities were surveyed in the rhizospheres of healthy and diseased roselles; (2) we found that Fusarium species were overabundant in diseased roselle plants compared with their healthy counterparts; (3) the dominant fungal species of F. solani was successfully isolated and verified as being the most likely pathogen causing roselle wilt disease; (4) a bacterial strain, Bacillus velezensis SOI-3374, isolated from the healthy roselle rhizosphere exhibited a significant anti-F. solani effect in vitro, highlighting its potential as a biocontrol agent for this disease.
A schematic diagram of the hypothesis and methodology in this study was shown in Figure 1A. The experimental fields were located in Taitung City (Field_01; GPS: 22.745875°N; 121.150813°E), Zhiben (Field_02; GPS: 22.743759°N; 121.061361°E), and Taimali (Field_03; GPS: 22.581578°N; 120.993655°E) in southeastern Taiwan. There were no fertilizer used in these three fields. According to the information of Central Weather Bureau of Taiwan, the average temperature in July to October is 27.8°C (24.7°C for annual average); the average rainfall is 269.3 mm (144.8 mm for annual average), and the relative humidity is 75.3% (74.3% for annual average). From each field, rhizosphere soil samples of healthy (n = 10) and wilted (n = 10) roselle plants (H. sabdariffa) were collected, yielding 60 samples. Each selected plant was separated from others by over 2 m. The entire plant was dug out to collect the rhizosphere soil. After the loosely attached soil was removed, the soil tightly attached to roots was rapidly brushed out and collected in a 50-ml falcon tube. The rhizosphere soil samples were immediately placed on ice, transported to the laboratory, and stored at −80°C until DNA extraction.
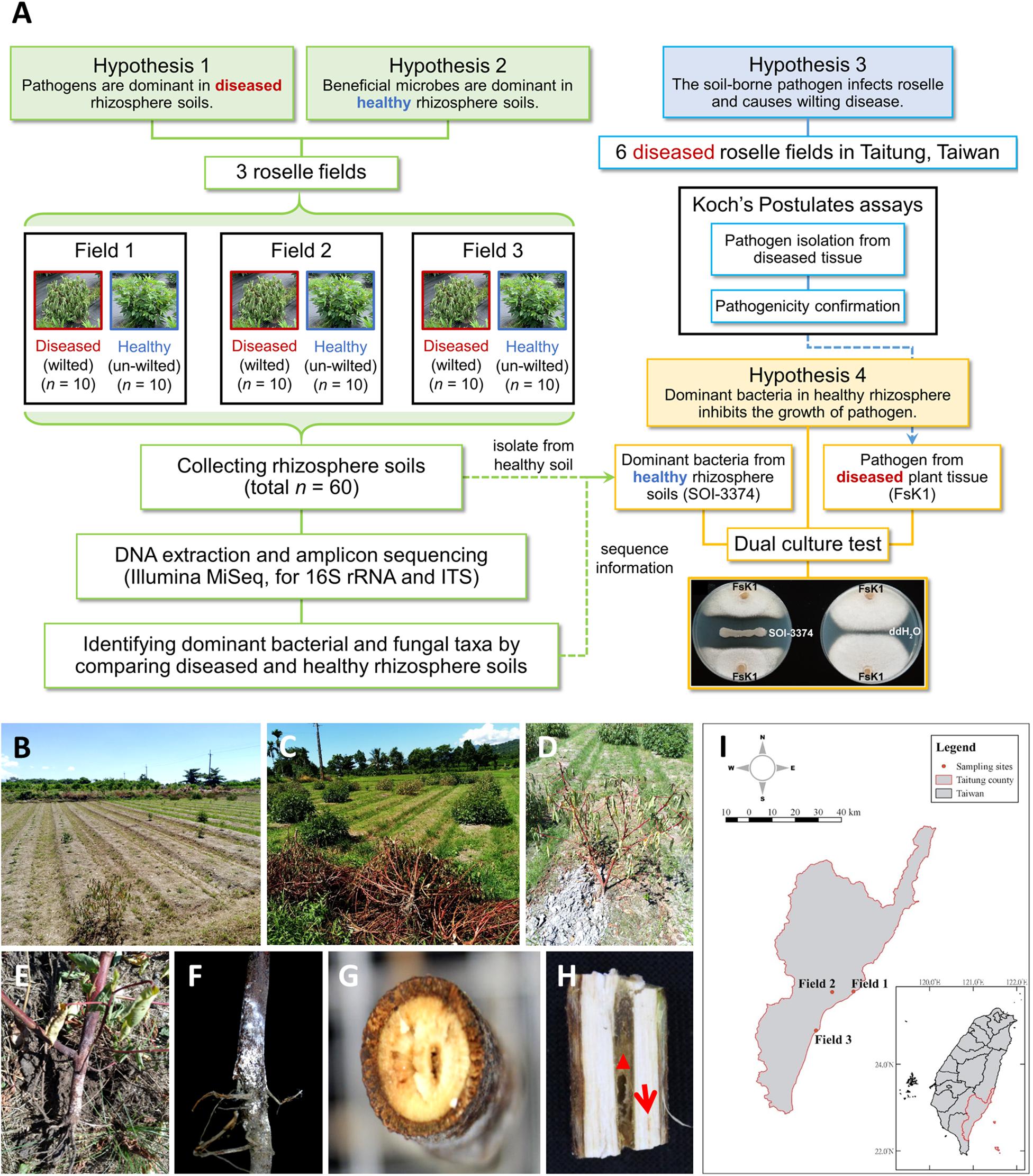
Figure 1. Schematic diagram of this study and the roselle wilt disease and symptoms found in Taitung, Taiwan. (A) A schematic diagram of the hypothesis and methodology. (B) and (C) Roselles were severely damaged by wilt disease. (D) Close-up shot of the wilting symptoms of roselle. (E) and (F) Stem base of wilted roselle colonized by fluffy, cotton-like aerial mycelia and tile mycelia with orange-white color. (G) Cross-section and (H) vertical dissection of diseased stem base, showing that the central pith (red triangle) was browning and rotten, with occasional lightly browning vascular bundles (red arrow). (I) Sampling sites of microbiome study in Taitung City (Field_01), Zhiben (Field_02), and Taimali (Field_03).
DNA extraction was performed using the DNeasy PowerSoil Kit (QIAGEN, MD, United States) according to the manufacturer’s protocol. For the bacterial composition survey, the V6 to V8 hypervariable region of 16S ribosomal RNA (rRNA) genes was amplified using polymerase chain reaction (PCR) with primers U968F and U1391R (Table 1), as previously described (Yang et al., 2016). All amplified and purified DNA were further added with specific barcodes according to the method of Yang et al. (2019). For the fungal composition survey, internal transcribed spacer (ITS) domain I of fungal rRNA genes was amplified using PCR with primers ITS1-F and ITS2 (Table 1), as previously described (Smith et al., 2018), with some modifications (Supplementary Material and Methods). Sixty bacterial and sixty fungal barcoded amplicon DNA samples were sent to Yourgene Bioscience (Taipei, Taiwan) and Tri-I Biotech (New Taipei City, Taiwan), respectively, for library construction and paired-end sequencing (2 × 300) on the Illumina MiSeq platform (Illumina, CA, United States). All of the bacterial and fungal community sequences were deposited in GeneBank (SRA accession PRJNA751843).
The raw reads were demultiplexed according to barcode into respective samples using Sabre1. On a per sample basis, R1 and R2 were first merged (–fastq_mergepairs), and then, the primers were removed (–search_pcr2) using USEARCH v11.0.667 (Edgar, 2010). The reads were further processed using MOTHUR v1.35.1 (Schloss et al., 2009) to retain high-quality reads that (1) had a length of 380–450 base pairs (bp), (2) contained homopolymers ≤ 8 bp, (3) did not have any ambiguous base, and (4) had an average Phred score of ≥ 20. Potential chimeras were identified (--uchime2_ref) and discarded (with options --mindiv 3 and --mode high_confidence) using USEARCH against the rdp_gold reference dataset2. To circumvent the memory limitation imposed by the 32-bit version of USEARCH, non-chimeric reads were clustered into operational taxonomic units (OTUs) by using VSEARCH v2.14.2 (Rognes et al., 2016) at a threshold of 97% identity, and OTU representative sequences were searched against the SILVA SSU Ref nr99 database v132 (Quast et al., 2013) using VSEARCH global alignment to identify the corresponding taxonomy of the best hit. Any OTU without a hit or with only a weak hit (i.e., an average percentage identity and percentage coverage < 93) was excluded. Finally, OTUs with the same affiliation were collapsed into the same OTU. Merging of OTUs that matched to the same reference sequence was conducted to avoid erroneous diversity inflation from sequences that were not well clustered.
The raw reads were demultiplexed based on barcodes into the corresponding samples. On a per sample basis, the read merging and quality control steps were similar to those for bacterial amplicons, except for the high-quality reads defined as those that (1) had a length of 210–450 bp, (2) contained homopolymers ≤ 15 bp, (3) did not have any ambiguous base, and (4) had an average Phred score of ≥ 20. Potential chimeras were identified (–uchime2_ref) and discarded (with options –mindiv 3 and –mode high_confidence) using USEARCH against the UNITE reference dataset v7.2. Non-chimeric reads were clustered into OTUs by using VSEARCH at a threshold of 99.9% identity, and OTU representative sequences were searched against the UNITE database v7.2 (Nilsson et al., 2019) using VSEARCH global alignment to identify the corresponding taxonomy of the most suitable hit. Any OTU without a hit or with only a weak hit was excluded. Finally, OTUs with the same affiliation were also collapsed into the same OTU to avoid the erroneous diversity inflation from sequences not well clustered. The taxonomy of three OTUs were manually annotated using BLASTn search against the NCBI database (Supplementary Table 2).
The roselle wilt disease samples were collected in July and October 2018 at six locations in Taitung, Taiwan: (1) Taitung District Agricultural Research and Extension Station (TDARES), (2) National Taitung University, (3) Beinan Township, (4) Donghe Township, (5) Dawu Township, and (6) Guanshan Township, and in October 2020 at TDARES. In total, 119 potential pathogens were isolated from infected tissues of roselles, including from the rotten root surface, constricted stem base, and browned pith. Tissues were cut and their surface sterilized for 1 min with 1% hypochlorous acid (repeated three times), washed with ddH2O, air dried, and placed on water agar (1% agar; BioShop, Burlington, ON, Canada). After incubation at 25°C for 3 d, the colonies of potential pathogens were purified and subcultured. Fungal strains were cultured on potato dextrose agar (PDA; 0.4% potato starch from infusion, 2% dextrose, and 1.5% agar; BioShop) at 28°C. Bacterial strains were cultured on nutrient agar (NA; 0.3% beef extract, 0.5% peptone, and 1.5% agar; BD Difco, Franklin Lakes, NJ, United States) at 30°C. All isolated fungal and bacterial strains (Supplementary Table 1) were stored at −80°C with 25% glycerol.
The morphology of potential pathogens isolated from roselle samples was identified and observed using an inverted microscope (Olympus CKX53, Tokyo, Japan). A scanning electron microscope (SEM) was also used for sample observation (Supplementary Material and Methods).
First, for identifying fungi, the 119 isolated strains (Supplementary Table 1) were preliminarily identified using the ITS with primers ITS4 and ITS5 (JC1753 and JC976, Table 1) (White et al., 1990); 16S rDNA with primers JC889 and JC890 (Table 1) was used to identify bacteria (Singh et al., 2013). To determine the phylogenetic relationships among the 107 isolated fungal strains (Supplementary Table 1), sequences were first compared using BLAST + software (Camacho et al., 2009), and those with > 99% identity (> 98% identity for oomycete strains) were identified as having the same sequence. Finally, 23 representative sequences were retrieved, and multiple sequence alignment was performed using the MAFFT online service (Talavera and Castresana, 2007). The poorly aligned positions and divergent regions in the alignment were eliminated using Gblocks (Talavera and Castresana, 2007). The maximum likelihood (ML) phylogeny was computed using IQ-TREE (v1.6.12) with the TNe + G4 model and 1000 bootstraps. A consensus tree was visualized and edited in iTOL (v4) (Letunic and Bork, 2019). To compare potential fungal pathogens isolated from diseased tissues with fungal OTU amplicons detected in rhizosphere soils, 23 representative sequences were searched against all the fungal OTU sequences by using BLAST. Among the 23 representative strains, the OTU sequences with identity > 97% and a length over 217 bp were retrieved. The abundance of each OTU was calculated from the average abundance of 60 samples from the OTU table after the OTUs were rarefied to the smallest sample size and OTUs less than 3 reads were removed; the abundance of each OTU was presented in < 10% of the samples. For further molecular identification, 13 sequences of potentially pathogenic F solani strains were analyzed with 32 sequences of F. solani strains from a recent study (Sandoval-Denis et al., 2019). Translation elongation factor 1-α (TEF1α, JC1189, and JC1190) (O’Donnell et al., 1998) and RNA polymerase II (RPB2, JC2247, and JC2248) (O’Donnell et al., 2008) sequences were used for further identifying these strains (Table 1). Phylogenetic trees were constructed on the basis of the ITS, TEF1α, and RPB2 sequences, and the phylogenetic relationship among fungal species was inferred using the ML method described previously (Talavera and Castresana, 2007; Camacho et al., 2009; Katoh et al., 2019; Letunic and Bork, 2019) with the TIM2e + I + G4 model.
Fusarium solani and F. oxysporum were prepared as a conidia suspension. Four 6-mm mycelium disks were punched from the PDA culture, added into 150 ml potato dextrose broth, and incubated at 28°C under 180 rpm for 8 to 10 d. The conidia were subsequently filtered using a two-layered Miracloth (EMD Millipore Corp., MA, United States) and collected by centrifugation at 3,500 rpm for 10 min, and the conidia were resuspended in sterile dH2O. The concentration of the freshly prepared conidia solution was calculated with a hemocytometer, and the solution was diluted for further tests. Koch’s postulate tests were conducted to identify potential pathogens of roselle wilt disease in Taiwan. The Taitung No. 3 roselle cultivar was used in this study. Three-week-old roselle roots were cut 5 cm from root tip with a sterile blade and then immersed into 30 ml ddH2O or 5 × 106/ml of conidia suspension of F. solani K1 (GenBank accession number: MZ701961) for 30 min (Hou et al., 2020). After the inoculation, the roselles were planted in mixed soil (Silu Kudra peat moss: sandy loam [1:1]; Euler Humuswerk GmbH, Germany; Ming Sheng Industrial Co., Ltd., Pingtung, Taiwan), and 30 ml ddH2O or Fusarium was irrigated into the soil of each treatment, and roselle plants were grown in a growth chamber at 29°C. Five plants were used for each treatment, and a 16 h: 8 h, light: dark cycle was set for all treatments. During the period of pathogenicity tests, all plants were observed every day, and any symptom developed was recorded. The treatment temperature herein was set at 29°C, which is similar to the monthly mean temperature recorded in Taitung from July to August in 2018 according to the Taiwan Central Weather Bureau.
Five grams of roselle rhizosphere soil (Field_03, No. H3) that was stored at −80°C was resuspended in 50 ml of sterilized water and heated for 30 min in a 65°C water bath. In total, 200 μL of the soil suspension was spread on NA medium and incubated in a 30°C incubator until a single colony appeared. Each single colony was streaked out, purified three times using the streaking method, and stored at −80°C. A confrontation assay was performed against the F. solani K1 (FsK1) strain on PDA medium. Strain SOI-3374, which showed the best antagonist effect was identified as Bacillus velezensis through 16S rRNA gene sequencing.
In this study, roselle plants were severely damaged by wilt disease in fields (Figures 1B,C). The symptoms included water loss in all leaves; fluffy, cotton-like aerial mycelia; and tile mycelia colonization on the stem base. The central pith exhibited browning and rotting (Figures 1D–H). To investigate the fungal and bacterial variations between diseased and healthy rhizospheres, roselles from three fields were selected for sampling, with each field being at least 8 km from the others (Figure 1I).
In total, 53,352 bacterial and 6,725 fungal OTUs were identified from the 60 samples. Taxa not noted more than three times in at least 10% of the samples (n = 6) were excluded. After rarefaction, a total of 5,845 bacterial and 715 fungal OTUs were retained. The fungal Shannon diversity was significantly lower in diseased rhizosphere soils collected from Field_02 than in healthy samples (Figures 2A,B). Non-metric multidimensional scaling ordination indicated a marked difference between diseased and healthy rhizospheres in both fungal and bacterial communities (PERMANOVA, p < 0.05), although site-specific effects were noted (Figures 2C,D). Among the major fungal taxa, the fungal class Sordariomycetes had higher relative abundance in the diseased rhizosphere than in the healthy rhizosphere in all three fields (Figure 2E), and the fungal family Nectriaceae was dominant (Supplementary Figure 1); moreover, the OTU_JX371352 was the most abundant contributor to the differential abundance of Sordariomycetes between diseased and healthy rhizosphere soils (Figure 2F and Supplementary Table 2).
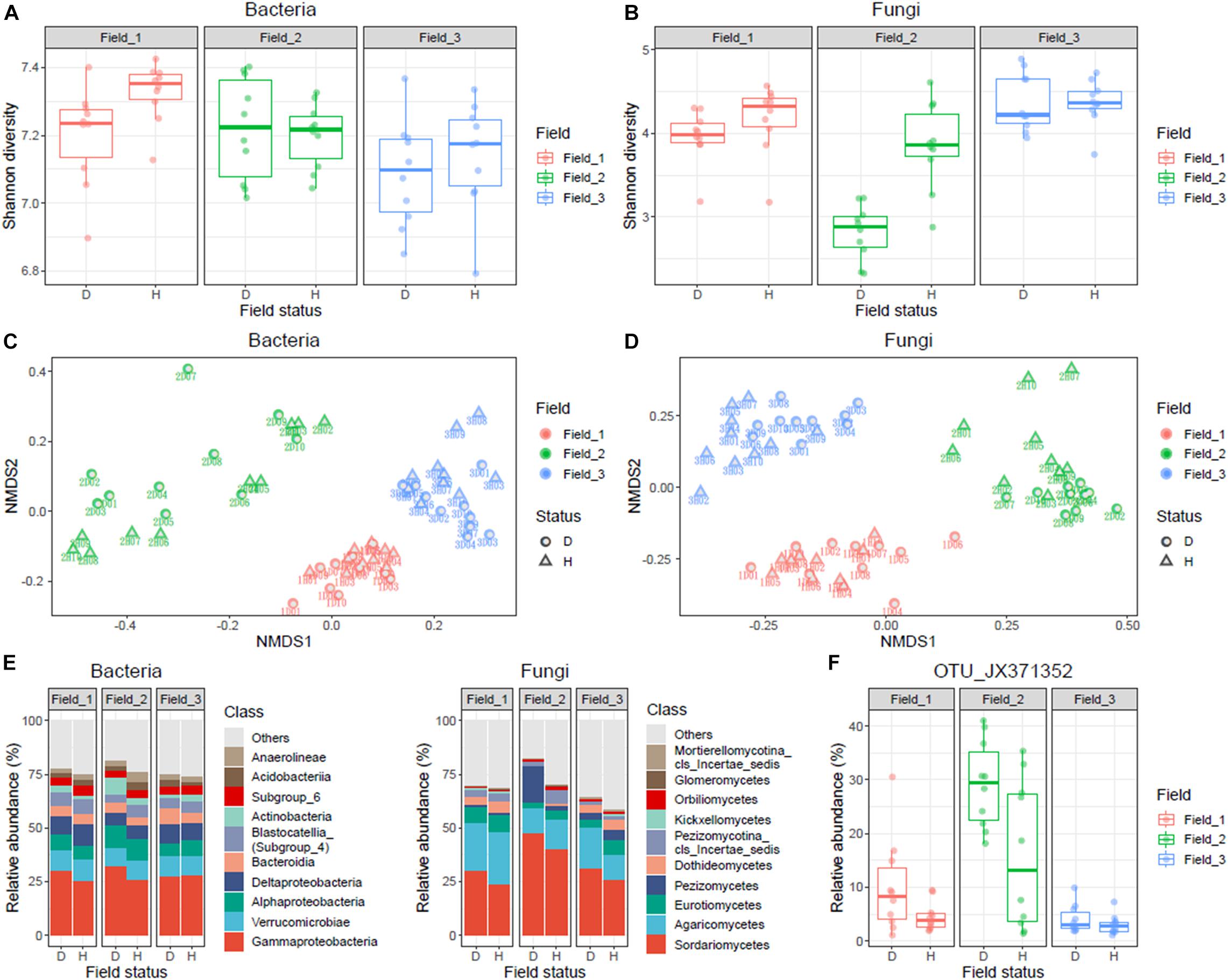
Figure 2. Statistical analysis and summary of bacterial and fungal communities in diseased and healthy roselle rhizosphere soils. Rhizosphere soil samples of healthy (n = 10) and wilted (n = 10) roselle plants were collected from three fields and a total of 60 samples were analyzed. (A) Shannon diversity of bacterial community. (B) Shannon diversity of fungal community. (C) nMDS analysis of bacterial OTUs. (D) nMDS analysis of fungal OTUs. (E) Bacterial and fungal community composition of the 10 most abundant classes on average. (F) The abundance distribution of fungal OTU_JX371352 across sampling fields. Field status, “D” for diseased and “H” for healthy.
Three batches of wilted roselle samples were collected in July and October 2018 and in October 2020 in Taitung, Taiwan. A total of 119 isolates were obtained and initially verified using ITS (fungi) and 16S rDNA (bacteria) sequencing. These isolates included F. solani (52.1%, 62/119), Phytophthora nicotianae (11.8%, 14/119), F. equiseti (11.8%, 14/119), F. oxysporum (5.1%, 6/119), F. acuminatum (0.8%, 1/119), F. proliferatum (0.8%, 1/119), Phytopythium vexans (0.8%, 1/119), and other fungi or bacteria (16.8%, 20/119; Figure 3 and Supplementary Table 1).
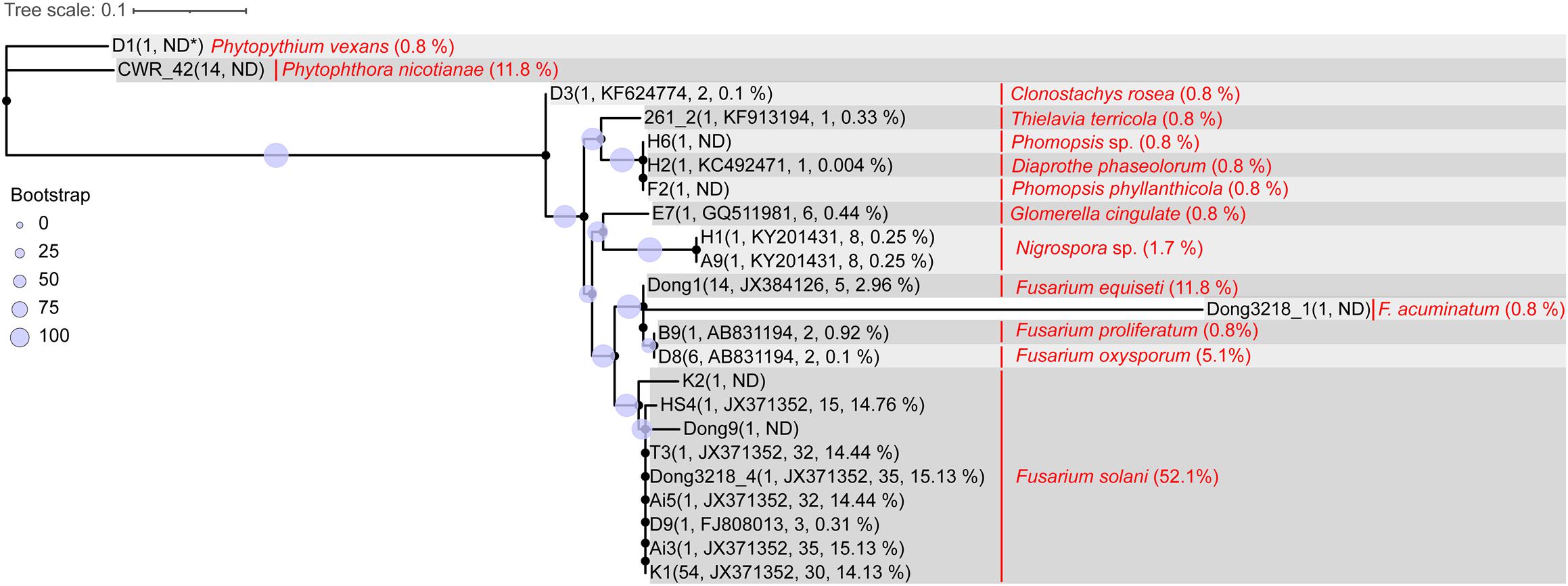
Figure 3. Sequence comparison of ITS1 region between isolated roselle wilt pathogens and rhizosphere amplicons. Parentheses following the strain ID indicate number of representative sequences, blasted most abundant OTU_ID, number of OTUs blasted, and sum of abundance (average percentage of the 60 samples). Words in red denote the species name of the isolate strain ID and parentheses indicate the percentage in 119 isolated strains. The maximum likelihood phylogeny was computed using IQ-TREE (v1.6.12) with the TNe + G4 model and 1000 bootstraps. A consensus tree was visualized and edited in iTOL (v4). (ND∗: the ITS sequences did not blast to any OTU amplicon sequence).
To determine whether the isolated potential pathogens were correlated with rhizosphere amplicons, the 23 fungal ITS representative sequences (clustered from 107 isolates) were searched against the fungal OTU representative amplicons using BLAST. The results indicated that 56% (60/107) of the isolates were nearly perfect hits to OTU_JX371352 (with identity > 99% on the aligned length of approximately 221 bp), which belongs to the fungal species F. solani, a major cause of roselle wilt disease (Figure 3).
The 10 most differentially abundant bacterial and fungal family taxa in diseased and healthy rhizosphere communities are listed in Table 2. In summary, the families Xanthomonadaceae, Microbacteriaceae, Enterobacteriaceae, Nectriaceae, and Ascobolaceae were more abundant in the diseased community, whereas Subgroup 6, Saccharimonadales, Anaerolineaceae, Bacillaceae, Chaetomiaceae, and Lasiosphaeriaceae were more abundant in healthy rhizosphere soils. In terms of genera, Microbacteria, Luteolibacter, Pseudoxanthomonas, and Fusarium were more abundant in diseased soils. Species belonging to Bacillus and Cladorrhinum were more abundant in healthy soils (Supplementary Table 3).
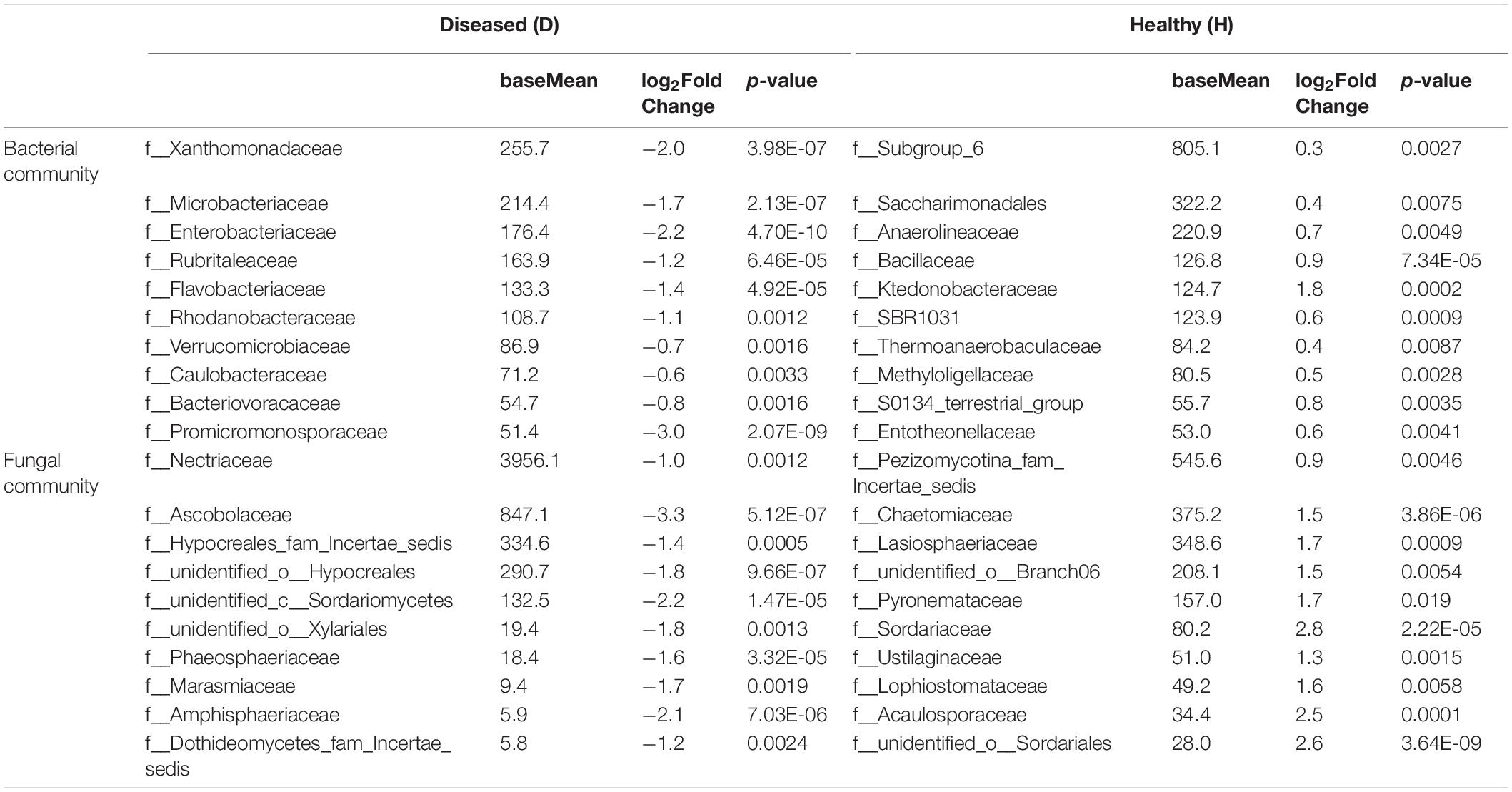
Table 2. Selected differential abundance of bacterial and fungal family taxa in healthy and diseased roselle rhizospheres.
Koch’s postulates were applied to verify the pathogenicity of our isolates to roselle wilt disease. Five to seven days after inoculation (dpi), conidia of FsK1 caused roselle wilt on 3-week-old roselles. The same symptoms were observed in both inoculation tests and fields (Figures 4, 5). The microbes reisolated from the FsK1-infected roselles were further identified as F. solani species complex by using ITS, TEF1α, and RPB2 sequence comparison. To the best of our knowledge, this is the first demonstration that F. solani species complex is the causative agent of roselle wilt disease in Taiwan. By contrast, isolated microbes other than F. solani (e.g., F. oxysporum) only caused leaf yellowing (Supplementary Figure 2). In virulence testing, another F. solani strain, K2, (isolated from the rotten pith of wilted roselle) in the field also caused roselle wilt in the inoculation test with conidia (Supplementary Figure 3).
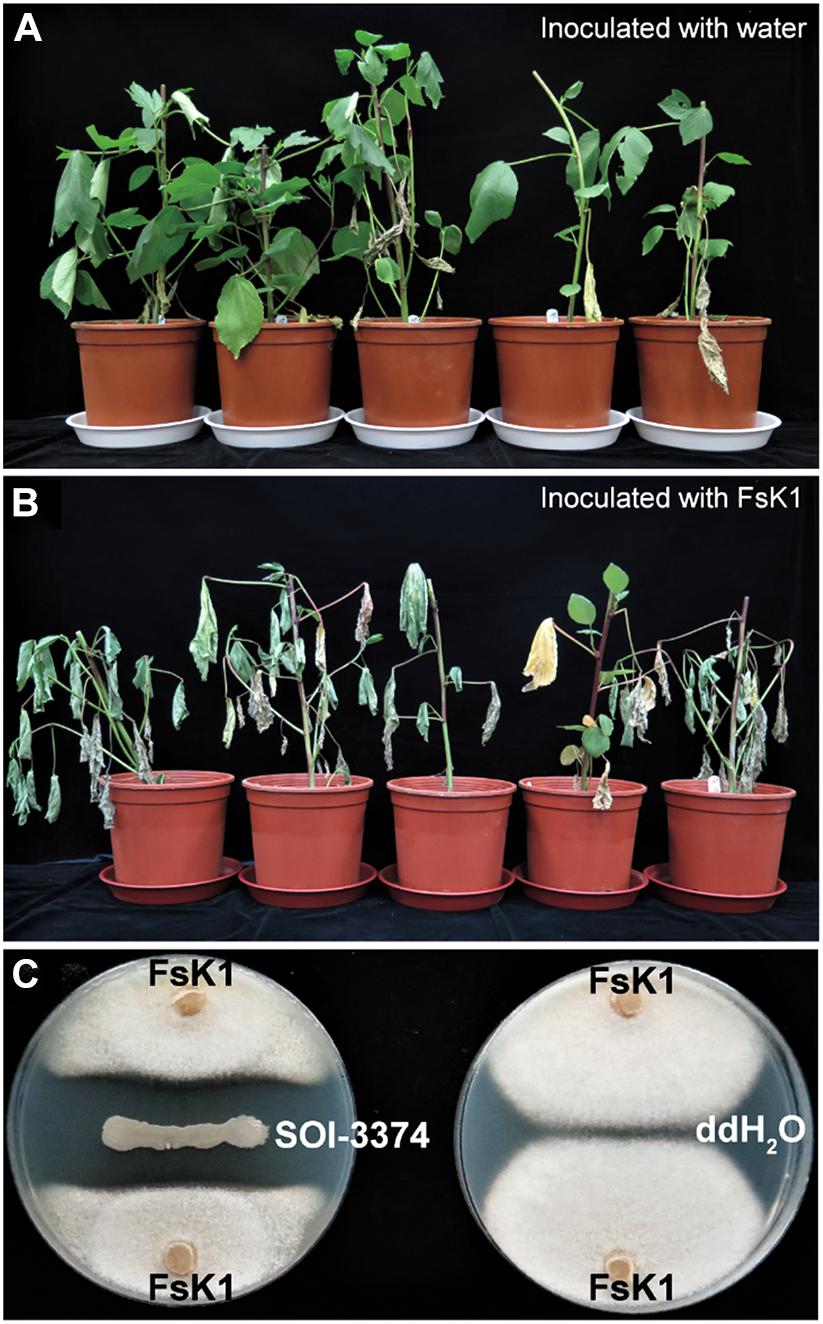
Figure 4. Fusarium solani K1 (FsK1) can cause roselle wilt disease. (A) Three-week-old roselles remained healthy after inoculation with ddH2O. (B) Those inoculated with the conidia of FsK1 exhibited wilting symptoms. (C) SOI-3374 showed antagonistic activity against FsK1 compared to ddH2O control.
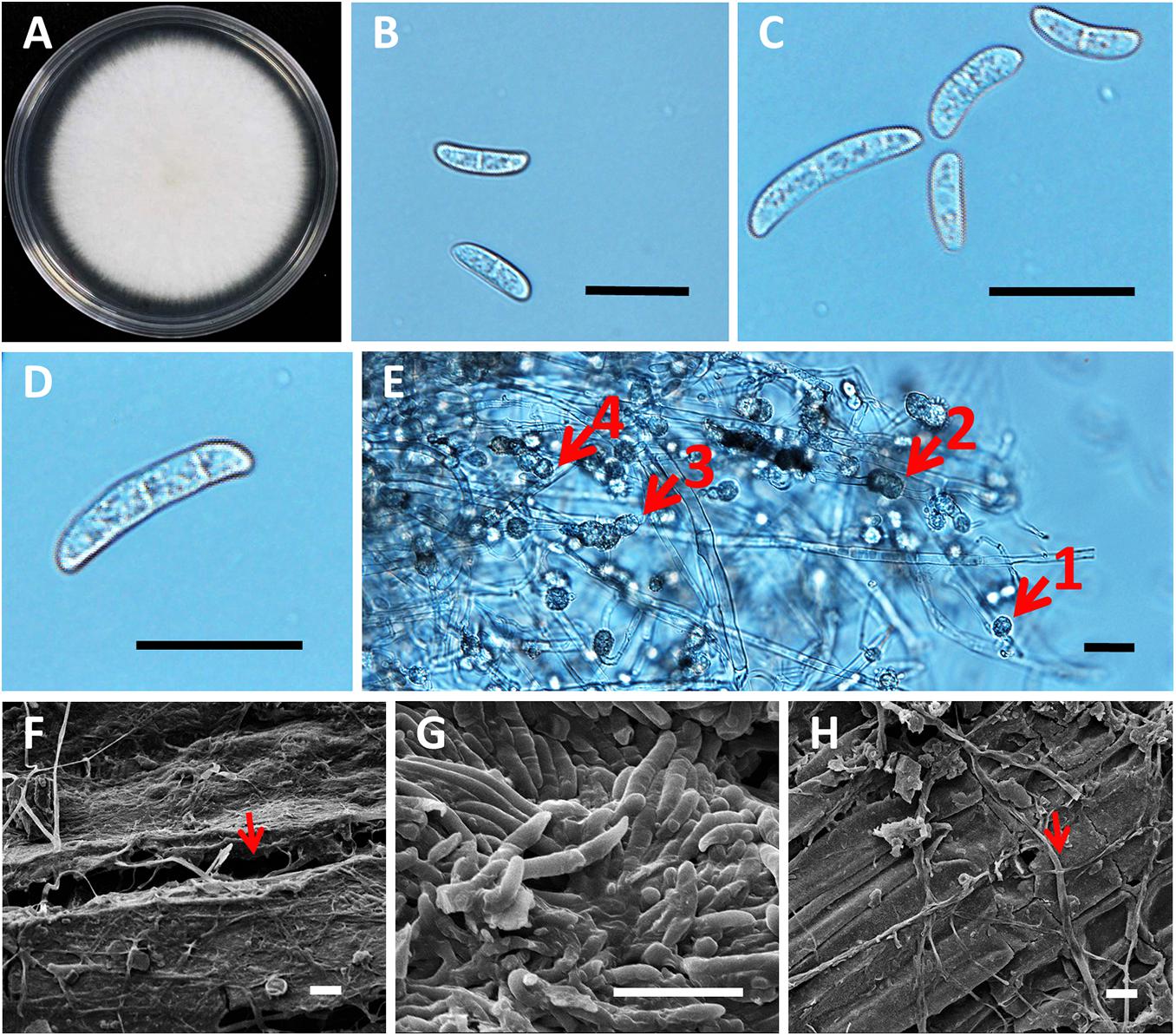
Figure 5. Morphology of roselle wilt fungus, FsK1. (A) Colony on PDA medium with white to light cream color, irregular margins, and sparse aerial mycelia. (B) Microconidia were 13.46 ± 3.52 μm × 4.46 ± 0.81 μm, hyaline, oval to ellipsoid, with zero to one septum. (C) Microconidia and macroconidia. (D) Macroconidia were 30.9 ± 7.12 μm × 5.95 ± 1.52 μm, hyaline, straight to falciform with three to four septa, whereas apical cells were blunt, and the foot shape of the basal cell was poorly developed. (E) Chlamydospores were 10.38 μm ± 2.94 μm, hyaline or dark brown, oval to globose, one to four cells growing intercalary or terminally on the hyphae, and usually with oil droplets. The numbered red arrows indicate 4 different clusters with 1 to 4 chlamydospores grown on a single chain of the hyphae. (F) Opening in the root surface with hyphae (red arrow). (G) Surface of the stem base was colonized with hyphae and conidia of Fusarium spp. (H) Hyphae (red arrow), but not conidia, were observed in the rotten pith. Scale bar: 20 μm.
The phylogenetic trees of FsK1 were constructed using ITS, TEF1α, and RPB2 sequences (Figure 6). Most of the 13 isolated strains were grouped together, including FsK1, which had the closest relationship with F. paranaense CBS 141593 and F. falciformis CBS 475.67, both of which belong to the F. solani species complex (Hypocreales, Nectriaceae) (Sandoval-Denis et al., 2019). The reference strain F. paranaense CBS 141593 was isolated from soybeans in Brazil, and the strain F. falciformis CBS 475.67 originated from human mycetoma in Puerto Rico (Sandoval-Denis et al., 2019). The morphology of FsK1 was observed under an optical microscope. The FsK1 colony on the PDA medium was white to cream color with irregular margins and sparse aerial mycelia (Figure 5A). The microconidia of FsK1 were 13.46 ± 3.52 μm × 4.46 ± 0.81 μm, hyaline, and oval to ellipsoid, with zero to one septum, and conidiogenous cells were monophalides with abundant growth and false heads (Figures 5B,C). The macroconidia of FsK1 were 30.9 ± 7.12 μm × 5.95 ± 1.52 μm, hyaline, and straight to falciform with 3-4 septa, whereas apical cells were blunt, and the foot shape of the basal cell was poorly developed (Figure 5C,D). The chlamydospores of FsK1 were 10.38 ± 2.94 μm and hyaline or dark brown; they contained one to four cells, were round to globose, and exhibited growth intercalary in or terminally on the hyphae, with a thick and smooth cell wall (Figure 5E). SEM observation revealed openings on the root surface with hyphae (red arrow, Figure 5F), and the surface of the stem base was colonized with hyphae and conidia of Fusarium spp. (Figure 5G). By contrast, hyphae (red arrow), but not conidia, were observed in the rotten pith (Figure 5H).

Figure 6. Phylogenetic analysis of Fusarium solani K1 (FsK1). A phylogenetic tree was constructed using ITS, TEF1α, and RPB2 sequences. FsK1 (K1 in the figure) exhibited the closest relationship with F. paranaense and F. falciformis, both belonging to the F. solani species complex. The phylogenetic relationship among fungal species was inferred using the maximum likelihood method with the TIM2e + I + G4 model and 1000 bootstraps. A consensus tree was visualized and edited in iTOL (v4).
Confrontation assay of bacteria isolated from healthy rhizosphere soil (Field_03) was conducted. Among hundreds of strains we isolated, strain SOI-3374 has the best antagonistic effects toward FsK1. The clear inhibition zone which FsK1 could not grow was formed between the two microorganisms on the plate (Figure 4C).
In this study, amplicon sequencing was used to explore the mycobiota and microbiota in healthy and diseased roselle rhizosphere soils; the results indicated that the abundant fungal OTU, OTU_JX371352 was considerably more common in diseased soil rhizospheres than in healthy soil rhizospheres and is thus a potential pathogen. Through pathogen isolation experiments, F. solani was isolated from the diseased roselle tissues with a high proportion of 52.1% (Figure 3 and Supplementary Table 1). Among the strains, FsK1 and F. solani K2 were pathogenic to roselles, as confirmed by Koch’s postulates. Moreover, according to the high Bacillus population in healthy soil samples, the B. velezensis SOI-3374 isolated from the healthy soil samples was verified to prevent FsK1 growth in vitro.
In our study, compared with healthy fields, all diseased fields had a higher abundance of pathogenic F. solani, suggesting that F. solani is the major causative agent of roselle wilt disease. In addition to F. solani (the Nectriaceae family), abundant populations of Xanthomonadaceae, Microbacteriaceae, Enterobacteriaceae, Flavobacteriaceae, Rubritaleaceae, and Ascobolaceae were detected in diseased soils. By contrast, large populations of Saccharimonadales, Subgroup 6, Bacillaceae, Anaerolineaceae, Chaetomiaceae, and Lasiosphaeriaceae were observed in healthy soils. In a related study investigating healthy or Fusarium-wilt-diseased soils, Yuan et al. found that F. oxysporum, Xanthomonadaceae, Bacillaceae, and Gibberella were more abundant in diseased soils, whereas more populations of Bradyrhizobiaceae, Comamonadaceae, Mortierella, Streptomyces mirabilis, and non-pathogenic Fusarium were observed in healthy soils (Yuan et al., 2020). In another study, Liu and Zhang compared healthy and Fusarium-diseased soils of cucumber fields and found that in the soil fungal community, Blastocladiomycota and Mycothermus were significantly more abundant in healthy soils and played an influential role in disease development (Liu and Zhang, 2021). In line with the findings of these studies, our results demonstrated that Blastocladiaceae (belonging to Blastocladiomycota) and Chaetomiaceae (containing the Mycothermus genus) were considerably more abundant in healthy roselle rhizosphere soils, whereas Xanthomonadaceae and pathogenic Fusarium were strongly enriched in diseased soils. Mycothermus has been reported as a potential pathogen-suppressive microorganism with an inverse proportion to the abundance of Fusarium (Huang et al., 2019), which may serve as a proxy to determine the risk probability of Fusarium diseases. Furthermore, the discovery of abundant Firmicutes in healthy soils (Liu and Zhang, 2021) agrees with our observation of abundant Bacillaceae (belonging to Firmicutes) in healthy soils. Within the family Bacillaceae, the Bacillus genus is a well-known bacterial group that has been applied in fields as biological control agents, and it is a taxon with a contrasting abundance between healthy and diseased rhizosphere soils (Beneduzi et al., 2012; Borriss, 2020).
Although a plethora of evidence suggests a connection between microbial (including fungal and bacterial) communities and roselle wilt disease, technical limitations emerge when ITS1F and ITS2 primers are used to differentiate pathogenic or non-pathogenic Fusarium species unless accompanied with isolation and inoculation tests. The primer set is widely used in studies of Fusarium diseases, which have explored the resolution of universal primers; however, more evaluation is required in the future (Tedersoo et al., 2016; Liu and Zhang, 2021).
In this study, F. solani, and not F. oxysporum, was the main causative agent of roselle wilt disease (Figure 4). Different F. solani strains, K1 and K2, caused roselle wilt similar to that found in fields (Figure 4 and Supplementary Figure 3). In general, F. oxysporum exists in the vascular tissues and causes wilt, whereas F. solani exists in the crown or root and causes root rot (Westerlund et al., 1974; Trapero Casas and Jiménez Díaz, 1985; Ales and Lenka, 1997). Only five F. oxysporum strains were noted among our 119 isolates; thus, these five F. oxysporum strains are highly likely to be involved in secondary infection. Moreover, F. solani likely infects or colonizes the pith, vascular tissues, or both, which is supported by the following studies: (1) a histochemical study of yellow poplar revealed that F. solani infected the pith tissue and caused pith rot symptoms in plants (Arnett and Witcher, 1974); (2) in China, physic nut infected by F. solani caused root rot and pith rot symptoms (Wu et al., 2011); (3) yellowing and wilting symptoms of cannabis plants were observed in northern California, with F. solani causing pith tissue browning and plant death within 6 to 10 wk after inoculation (Punja et al., 2018); and (4) in passion fruits, F. solani caused the vascular bundle browning of plants (Cole et al., 1992). In a phylogenetic study, the reference strain F. paranaense CBS 141593 (F. solani species complex, close relationship to the K1 strain) was isolated from soybean, which is consistent with the finding of a previous study that F. solani causes symptoms such as sudden death syndrome in soybean (Westphal et al., 2008); we also suggest that the pathogenic strains isolated in the current study are closely related to the phytopathogenic F. solani species complex. Although the strain F. falciformis CBS 475.67 is closely related to the strains we isolated in this study, it originates from human mycetoma. The relationship between human pathogens and plant pathogens should be further studied to confirm whether human pathogens infect humans only or plants as well.
Regarding the development of control strategies for roselle wilt disease, Bacillus velezensis SOI-3374, isolated from the healthy roselle rhizosphere, can control roselle wilt, as demonstrated in confrontation assay. In general, chitin-degrading and other cell wall–degrading enzymes may act as crucial substances that enable Bacillus to inhibit Fusarium, whose cell wall is composed of α- and β-1, 3-glucans (Schoffelmeer et al., 1999; Khan et al., 2018). The biocontrol activity of Bacillus species against plant-pathogenic F. solani has been documented. For example, a Bacillus commercial formulation effectively controlled the crown and root rot of tomato caused by F. solani in field trials (Pastrana et al., 2016); B. subtilis reduced the damping off incidence of cucumber seedlings caused by F. solani (Al-Fadhal et al., 2019).
A recent study indicated that B. velezensis effectively prevented F. solani infection in passion fruits (Wang et al., 2021). B. velezensis strains with secondary-metabolite-secretion ability could inhibit F. solani and oomycetes, and these strains were reported to serve plant growth–promoting rhizobacteria (Cheffi et al., 2019). Furthermore, in a rhizosphere study, B. velezensis that inhabited the grass (Sporobolus airoides) rhizosphere was isolated and the whole genome sequenced; this B. velezensis strain exhibited strong antifungal activity against phytopathogens that cause root rot, including F. solani, F. oxysporum, Phytophthora capsici, and Rhizoctonia solani (Martínez-Raudales et al., 2017). Although B. velezensis holds promise for controlling plant pathogens, the mechanisms through which B. velezensis SOI-3374 controls roselle wilt remain to be explored.
Data were available within the article or its Supplementary Material. All of the bacterial and fungal community sequences were deposited in GeneBank (SRA accession PRJNA751843).
C-WW, Y-HY, Y-LC, and S-LT planned and designed the research. C-WW, Y-HY, C-YW, and R-YF performed experiments, conducted fieldwork, analyzed data etc. KT analyzed the sequence data. C-WW, Y-HY, Y-LC and S-LT wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by grants 109AS-1.1.5-ST-a5 and 109AS-7.7.3-ES-E1 from the Council of Agriculture, Executive Yuan, and 107-2320-B-002-061-MY3 from the Taiwan Ministry of Science and Technology and Biodiversity Research Center, Academia Sinica.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Miranda Loney and Ian Fritz for language editing help and to the Technology Commons, College of Life Science, National Taiwan University for scanning electron microscopy support. We thank Ching-Hung Tseng, and Germark Biotechnology, Co., Ltd., Taichung, Taiwan, for draft editing and sequence analysis. This manuscript was edited by Wallace Academic Editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.756100/full#supplementary-material
Ales, L., and Lenka, S. (1997). Variation in response of several wild Pisum spp. to Fusarium solani and Fusarium oxysporum. Cereal Res. Commun. 25, 845–846. doi: 10.1007/BF03543870
Al-Fadhal, F. A., Al-Abedy, A. N., and Alkhafije, D. A. (2019). Isolation and molecular identification of Rhizoctonia solani and Fusarium solani isolated from cucumber (Cucumis sativus L.) and their control feasibility by Pseudomonas fluorescens and Bacillus subtilis. Egypt. J. Biol. Pest Co. 29:47. doi: 10.1186/s41938-019-0145-5
Alshoosh, W. G. A. (1997). Chemical Composition of Some Roselle (Hibiscus sabdariffa) Genotypes, Sudan: University of Khartoum. [Master Thesis].
Amusa, N., Adegbite, A., and Oladapo, M. (2005). Vascular wilt of roselle (Hibiscus sabdariffa L. var. sabdariffa) in the humid forest region of south-western Nigeria. Plant Pathol. J. 4, 122–125. doi: 10.3923/ppj.2005.122.125
Ansari, M., Eslaminejad, T., Sarhadynejad, Z., and Eslaminejad, T. (2013). An overview of the roselle plant with particular reference to its cultivation, diseases and usages. Eur. J. Med. Plants 3, 135–145. doi: 10.9734/EJMP/2013/1889
Arnett, J., and Witcher, W. (1974). Histochemical studies of yellow poplar infected with Fusarium solani. Phytopathology 64, 414–418.
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/s1415-47572012000600020
Benítez, M. S., and Gardener, B. B. M. (2009). Linking sequence to function in soil bacteria: sequence-directed isolation of novel bacteria contributing to soilborne plant disease suppression. Appl. Environ. Microb. 75, 915–924. doi: 10.1128/AEM.01296-08
Borriss, R. (2020). “Phytostimulation and biocontrol by the plant-associated Bacillus amyloliquefaciens FZB42: an update,” in Phyto-Microbiome in Stress Regulation, eds M. Kumar, V. Kumar, and R. Prasad (Singapore: Springer Nature Singapore Pte Ltd), 1–20.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., and Bealer, K. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Cheffi, M., Chenari Bouket, A., Alenezi, F. N., Luptakova, L., Belka, M., Vallat, A., et al. (2019). Olea europaea L. root endophyte Bacillus velezensis OEE1 counteracts oomycete and fungal harmful pathogens and harbours a large repertoire of secreted and volatile metabolites and beneficial functional genes. Microorganisms 7:314. doi: 10.3390/microorganisms7090314
Chen, Y. F., and Chen, J. W. (2019). Effect of processing manufacture on the quility of roselle ‘Taitung No. 3’ products. Res. Bull. Tait. Distr. Agricult. Res. Extens. Stat. Taiwan 29, 25–34. doi: 10.6959/RBTDAIS
Cole, D., Hedges, T., and Ndowora, T. (1992). A wilt of passion fruit (Passiflora edulis f. edulis Sims) caused by Fusarium solani and Phytophthora nicotianae var. parasitica. Int. J. Pest Manage. 38, 362–366. doi: 10.1080/09670879209371728
Coleman, J. J. (2016). The Fusarium solani species complex: ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 17, 146–158. doi: 10.1111/mpp.12289
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Estrada, Y. I. E., Kawasoe, S. O., and Estrada, J. A. S. E. (2001). Variabilidad patogénica de Phytophthora parasitica Dastur en jamaica (Hibiscus sabdariffa L.). Rev. Mexicana Fitopatol. 19, 84–89.
Etesami, H., and Adl, S. M. (2020). “Plant growth-promoting rhizobacteria (PGPR) and their action mechanisms in availability of nutrients to plants,” in Phyto-Microbiome in Stress Regulation, eds M. Kumar, V. Kumar, and R. Prasad (Singapore: Springer Nature Singapore Pte Ltd), 147–203.
Gomes, V. M., Souza, R. M., Mussi−Dias, V., Silveira, S. F. D., and Dolinski, C. (2011). Guava decline: a complex disease involving Meloidogyne mayaguensis and Fusarium solani. J. Phytopathol. 159, 45–50. doi: 10.1111/j.1439-0434.2010.01711.x
Hassan, N., Shimizu, M., and Hyakumachi, M. (2014). Occurrence of root rot and vascular wilt diseases in roselle (Hibiscus sabdariffa L.) in Upper Egypt. Mycobiology 42, 66–72. doi: 10.5941/MYCO.2014.42.1.66
Hou, Y. H., Hsu, L. H., Wang, H. F., Lai, Y. H., and Chen, Y. L. (2020). Calcineurin regulates conidiation, chlamydospore formation and virulence in Fusarium oxysporum f. sp. lycopersici. Front. Microbiol. 11:2629. doi: 10.3389/fmicb.2020.539702
Huang, X., Zhao, J., Zhou, X., Han, Y., Zhang, J., and Cai, Z. (2019). How green alternatives to chemical pesticides are environmentally friendly and more efficient. Eur. J. Soil Sci. 70, 518–529. doi: 10.1111/ejss.12755
Islam, A. K. M. A., Jamini, T. S., Islam, A. K. M. M., and Sabina, Y. (2016). Roselle: a functional food with high nutritional and medicinal values. Fund. Appl. Agric. 1, 44–49.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Khan, N., Martínez-Hidalgo, P., Ice, T. A., Maymon, M., Humm, E. A., Nejat, N., et al. (2018). Antifungal activity of Bacillus species against Fusarium and analysis of the potential mechanisms used in biocontrol. Front. Microbiol. 9:2363. doi: 10.3389/fmicb.2018.02363
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Liu, X., and Zhang, Y. (2021). Exploring the communities of bacteria, fungi and ammonia oxidizers in rhizosphere of Fusarium-diseased greenhouse cucumber. Appl. Soil Ecol. 161:103832. doi: 10.1016/j.apsoil.2020.103832
Martínez-Raudales, I., De La Cruz-Rodríguez, Y., Alvarado-Gutiérrez, A., Vega-Arreguín, J., Fraire-Mayorga, A., Alvarado-Rodríguez, M., et al. (2017). Draft genome sequence of Bacillus velezensis 2A-2B strain: a rhizospheric inhabitant of Sporobolus airoides (Torr.) Torr., with antifungal activity against root rot causing phytopathogens. Stand. Genomic Sci. 12, 1–10. doi: 10.1186/s40793-017-0289-4
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
O’Brien, P. A. (2017). Biological control of plant diseases. Australas Plant Path. 46, 293–304. doi: 10.1007/s13313-017-0481-4
O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. PNAS 95, 2044–2049. doi: 10.1073/pnas.95.5.2044
O’Donnell, K., Sutton, D. A., Fothergill, A., McCarthy, D., Rinaldi, M. G., Brandt, M. E., et al. (2008). Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46, 2477–2490.
Ooi, K., and Salleh, B. (1999). Vegetative compatibility groups of Fusarium oxysporum, the causal organism of vascular wilt on roselle in Malaysia. Biotropia 12, 31–41. doi: 10.11598/btb.1999.0.12.149
Pastrana, A. M., Basallote-Ureba, M. J., Aguado, A., Akdi, K., and Capote, N. (2016). Biological control of strawberry soil-borne pathogens Macrophomina phaseolina and Fusarium solani, using Trichoderma asperellum and Bacillus spp. Phytopathol. Mediterr. 55, 109–120. doi: 10.14601/Phytopathol_Mediterr-16363
Ploetz, R., Palmateer, A., Geiser, D., and Juba, J. (2007). First report of Fusarium wilt caused by Fusarium oxysporum on roselle in the United States. Plant Dis. 91:639. doi: 10.1094/PDIS-91-5-0639A
Poudel, R., Jumpponen, A., Schlatter, D. C., Paulitz, T., Gardener, B. M., Kinkel, L. L., et al. (2016). Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology 106, 1083–1096. doi: 10.1094/PHYTO-02-16-0058-FI
Punja, Z. K., Scott, C., and Chen, S. (2018). Root and crown rot pathogens causing wilt symptoms on field-grown marijuana (Cannabis sativa L.) plants. Can. J. Plant Pathol. 40, 528–541. doi: 10.1080/07060661.2018.1535470
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2584. doi: 10.7717/peerj.2584
Sandoval-Denis, M., Lombard, L., and Crous, P. W. (2019). Back to the roots: a reappraisal of Neocosmospora. Persoonia 43:90. doi: 10.3767/persoonia.2019.43.04
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schoffelmeer, E. A., Klis, F. M., Sietsma, J., and Cornelissen, B. J. (1999). The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27, 275–282. doi: 10.1006/fgbi.1999.1153
Singh, S., Moholkar, V. S., and Goyal, A. (2013). Isolation, identification, and characterization of a cellulolytic Bacillus amyloliquefaciens strain SS35 from rhinoceros dung. ISRN Microbiol. 2013, 1–7. doi: 10.1155/2013/728134
Smith, D. P., Peay, K. G., Ackermann, G., Apprill, A., Bauer, M., Berg-Lyons, D., et al. (2018). EMP ITS Illumina Amplicon Protocol. protocols. io. doi: 10.17504/protocols.io.pa7dihn
Sundaramoorthy, S., Raguchander, T., Ragupathi, N., and Samiyappan, R. (2012). Combinatorial effect of endophytic and plant growth promoting rhizobacteria against wilt disease of Capsicum annum L. caused by Fusarium solani. Biol. Control. 60, 59–67. doi: 10.1016/j.biocontrol.2011.10.002
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Tedersoo, L., Bahram, M., Cajthaml, T., Põlme, S., Hiiesalu, I., Anslan, S., et al. (2016). Tree diversity and species identity effects on soil fungi, protists and animals are context dependent. ISME J. 10, 346–362. doi: 10.1038/ismej.2015.116
Trapero Casas, A., and Jiménez Díaz, R. M. (1985). Fungal wilt and root rot diseases of chickpea in southern Spain. Phytopathology 75, 1146–1151. doi: 10.1094/phyto-75-1146
Tzean, S. S. (2019). List of Plant Diseases in Taiwan (fifth edition). Taiwan: Taiwan Phytopathological Society, 149–150.
Wang, C., Ye, X., Ng, T. B., and Zhang, W. (2021). Study on the biocontrol potential of antifungal peptides produced by Bacillus velezensis against Fusarium solani that infects the passion fruit Passiflora edulis. J. Agr. Food Chem. 69, 2051–2061. doi: 10.1021/acs.jafc.0c06106
Westerlund, F., Campbell, R., and Kimble, K. (1974). Fungal root rots and wilt of chickpea in California. Phytopathology 64, 432–436.
Westphal, A., Abney, T. S., Xing, L., and Shaner, G. (2008). Sudden death syndrome of soybean. Plant Health Instr. doi: 10.1094/PHI-I-2008-0102-01
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR prot. 18, 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wu, Y. K., Ou, G. T., and Yu, J. Y. (2011). First report of Nectria haematococca causing root rot disease of physic nut (Jatropha curcas) in China. Australas Plant Dis. Notes 6, 39–42. doi: 10.1007/s13314-011-0014-x
Yang, S. H., Chiang, P. W., Hsu, T. C., Kao, S. J., and Tang, S. L. (2016). Bacterial community associated with organs of shallow hydrothermal vent crab Xenograpsus testudinatus near Kuishan Island Taiwan. PLoS One. 11:e0150597. doi: 10.1371/journal.pone.0150597
Yang, S. H., Tandon, K., Lu, C. Y., Wada, N., Shih, C. J., Hsiao, S. S. Y., et al. (2019). Metagenomic, phylogenetic, and functional characterization of predominant endolithic green sulfur bacteria in the coral Isopora palifera. Microbiome 7, 1–13. doi: 10.1186/s40168-018-0616-z
Keywords: rhizosphere, microbiome, roselle wilt disease, Fusarium wilt, Fusarium solani, Bacillus velezensis
Citation: Wang C-W, Yu Y-H, Wu C-Y, Feng R-Y, Tandon K, Chen Y-L and Tang S-L (2021) Detection of Pathogenic and Beneficial Microbes for Roselle Wilt Disease. Front. Microbiol. 12:756100. doi: 10.3389/fmicb.2021.756100
Received: 10 August 2021; Accepted: 11 October 2021;
Published: 01 November 2021.
Edited by:
Wei Zhang, Michigan State University, United StatesReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandCopyright © 2021 Wang, Yu, Wu, Feng, Tandon, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Lien Chen, eWNoZW4yOEBudHUuZWR1LnR3; Sen-Lin Tang, c2x0YW5nQGdhdGUuc2luaWNhLmVkdS50dw==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.