- Institute of Aquaculture, Faculty of Natural Sciences, University of Stirling, Stirling, United Kingdom
Essential genes in bacterial pathogens are potential drug targets and vaccine candidates because disrupting their function is lethal. The development of new antibiotics, in addition to effective prevention measures such as vaccination, contributes to addressing the global problem of bacterial antibiotic resistance. The aim of this present study was to determine the essential genes of Vibrio anguillarum, a bacterial pathogen of aquatic animals, as a means to identify putative targets for novel drugs and to assist the prioritisation of potential vaccine candidates. Essential genes were characterised by a Tn-seq approach using the TnSC189 mariner transposon to construct a library of 52,662 insertion mutants. In total, 329 essential genes were identified, with 34.7% found within the core genome of this species; each of these genes represents a strong potential drug target. Seven essential gene products were predicted to reside in the cell membrane or be released extracellularly, thus serving as putative vaccine candidates. Comparison to essential gene data from five other studies of Vibrio species revealed 13 proteins to be conserved across the studies, while 25 genes were specific to V. anguillarum and not found to be essential in the other Vibrio spp. This study provides new information on the essential genes of Vibrio species and the methodology may be applied to other pathogens to guide the development of new drugs and vaccines, which will assist efforts to counter antibiotic resistance.
1. Introduction
Essential genes are those that when disrupted lead to the organism becoming non-viable because a critical biological process can no longer be accomplished (Jordan et al., 2002; Reznikoff and Winterberg, 2008), and this characteristic makes these genes and the products they encode important targets for the development of new antibiotics (Forsyth et al., 2002; Thanassi, 2002). The search for novel bacterial drug targets is now more urgent thanks to rising antibiotic resistance that renders many clinical classes of drugs potentially ineffective, with the situation particularly worrisome for Gram-negative pathogens (Aslam et al., 2018; Breijyeh et al., 2020; De Oliveira et al., 2020). Moreover, products of essential genes are strong candidates for vaccine development, particularly those encoding proteins that are released from the cell or expressed at the cell surface, as these may be recognised by the host to elicit long-term specific immune protection (García-Quintanilla et al., 2016b; Naz et al., 2019).
Essential genes are discovered through their inactivation, which may be achieved by targeted approaches such as transposon mutagenesis, CRISPR, and antisense RNA, though the most common approach is random transposon mutagenesis (van Opijnen and Camilli, 2013; Peters et al., 2016). Typically, this latter method requires conjugation of a suicide vector plasmid carrying a transposon from a donor bacterium into the recipient host, followed by transposon excision from the plasmid and insertion at a non-specific site in the host genome, thus disrupting the gene at that particular locus. Massively parallel sequencing approaches have greatly assisted the discovery of essential genes because the insertion sites of thousands of transposon-insertion mutants can be characterised in a single protocol (van Opijnen et al., 2009; Le Breton et al., 2015). The transposon-insertion sequencing (Tn-seq) method relies on a mariner-based transposon that inserts at TA dinucleotide sites, which occur approximately every 11–16 base pairs in bacterial genomes, meaning an excellent coverage of disrupted genes can be achieved (Judson and Mekalanos, 2000; Rubin et al., 2015).
Essential genes have been identified for human pathogens, such as Bacillus subtilis (Kobayashi et al., 2003; Peters et al., 2016), Burkholderia pseudomallei (Moule et al., 2014), Escherichia coli (Gerdes et al., 2003; Baba et al., 2006; Goodall et al., 2018; Martínez-Carranza et al., 2018), Pseudomonas aeruginosa (Gallagher et al., 2011; Turner et al., 2015; Poulsen et al., 2019), Salmonella enterica serovar Typhimurium (Barquist et al., 2013), Shigella flexneri (Freed et al., 2016), Staphylococcus aureus (Ji, 2001; Forsyth et al., 2002; Chaudhuri et al., 2009), and Vibrio cholerae (Cameron et al., 2008; Chao et al., 2013; Kamp et al., 2013); however, few studies concern bacteria species that infect fish, despite the problems caused by antibiotic-resistant pathogens in global aquaculture (Pham-Duc et al., 2019; Vincent et al., 2019; Reverter et al., 2020; FAO, 2021), which has led to a need for new therapies and vaccine candidates. Vaccination of fish has reduced antibiotic use considerably when farming certain species, such as Atlantic salmon (Sommerset et al., 2005), and this approach is suitable for other finfish. Still, most fish vaccines contain whole cells of inactivated pathogens and subunit vaccines, where only selected antigens are included in the preparation, could offer improved efficacy and safety profiles (Hansson et al., 2000; Adams, 2019; Ma et al., 2019).
Vibrio anguillarum is a fermentative, curved, Gram-negative bacterium responsible for vibriosis outbreaks in many species of fish, crustaceans and molluscs (Thakur et al., 2003; Toranzo et al., 2005; Frans et al., 2011; Marcos-López et al., 2013). Like other Vibrio spp., the V. anguillarum genome consists of two differently sized chromosomes, while plasmids may also be present such as the ca. 65-kDa pJM1 (and related plasmids) that encodes numerous virulence factors (Di Lorenzo et al., 2003; Okada et al., 2005; Naka et al., 2011). Vibriosis is a potentially lethal infection that is treated with antibiotics, but resistance can emerge and there are broader concerns for the impact of applying these agents in aquatic systems (Frans et al., 2011; Reverter et al., 2020; Desbois et al., 2021). Inactivated whole-cell vaccines are available to protect some species of fish against V. anguillarum (Colquhoun and Lillehaug, 2014), but these do not protect against every strain of the pathogen, and opportunities exist to develop more effective and safer subunit vaccines.
Essential genes in Vibrio spp. have been studied previously, and typically these have been consistent with work on many other free-living bacteria that show ca. 10–20% of genes to be essential for growth in vitro (Gerdes et al., 2003; Gil et al., 2004; Peters et al., 2016). Chao et al. (2013) identified 343 essential genes in V. cholerae, while Kamp et al. (2013) used a different isolate to identify 414 essential genes in this species. Cameron et al. (2008) determined 789 essential genes in V. cholerae but acknowledged that some of these were probably included by chance and Chao et al. (2013) provided some support to this suggestion. Chao et al. (2013) found most essential genes to be involved in “metabolism” and “translation,” but the functions of many other genes were uncharacterised. Hubbard et al. (2016) identified 565 essential genes in V. parahaemolyticus, and the functions of many of these genes were also undetermined. Still, there was a high degree of overlap in the essential gene sets in V. parahaemolyticus and the V. cholerae study of Chao et al. (2013), with 69% of the V. parahaemolyticus essential genes having homologs in the V. cholerae list (Hubbard et al., 2016). Guanhua et al. (2018) found 473 essential genes in V. anguillarum MVM425, many of which were hypothetical or of unknown function. Of note, these previous Vibrio spp. studies did not seek to identify gene products predicted to be released from the cell or expressed at the cell surface such that they could be developed as candidates for novel vaccines.
The aim of this present study was to determine the essential genes of V. anguillarum to identify putative targets for novel drugs and to assist the prioritisation of potential vaccine candidates for development and inclusion into a subunit vaccine. To achieve this, essential genes were identified by a Tn-seq approach, and then the subcellular locations of the products of each essential gene, their functions and metabolic pathways to which they contribute were determined. Finally, comparison was made to earlier studies of essential genes in V. cholerae, V. parahaemolyticus and V. anguillarum to shed light on the conservation of this set of fundamentally important genes across the genus and which guides the development of new antibacterial approaches against Vibrio spp.
2. Materials and Methods
2.1. Bacteria and Culture Media
Vibrio anguillarum NB10Sm, a spontaneous streptomycin [STR]-resistant strain of V. anguillarum NB10, a pathogenic isolate of the O1 serotype (Holm et al., 2015), was used as the recipient strain for transposon mutagenesis. The donor strain carrying the TnSC189 transposon on the pSC189 plasmid was E. coli SM10λpir (resistant to kanamycin [KAN]); pSC189 contains a resistance gene to ampicillin (AMP). TnSC189 had an MmeI restriction site introduced into the inverted repeat sequence at the 5′ end (van Opijnen and Camilli, 2013). Tryptone soy agar (TSA) and broth (TSB) and lysogeny agar (LBA) and broth (LBB) were prepared according to manufacturer instructions. To facilitate culture of V. anguillarum, the media were supplemented with 10–15 parts per thousand (ppt) sodium chloride (supplement in ppt given as superscript). Antibiotics were added to media as required (final concentration in mg/L given as superscript).
2.2. Transposon Mutagenesis and Extraction of Genomic DNA
Donor and recipient strains were incubated to early stationary phase (E. coli: LBB+AMP100, 37℃, 120 rpm, 4 h; V. anguillarum: TSA15 + STR200, 22℃, 150 rpm, 18 h), before combining at a CFU ratio of 1:1. Aliquots were pipetted onto 0.22-μm nitrocellulose filters on LBA10 plates and incubated (30℃, 24 h). Bacteria from the filters were collected in TSB15 and plated across TSA15+KAN250+STR100 and incubated (22℃, 72 h) to select for V. anguillarum containing the TnSC189 transposon. Colonies were collected, incubated (22℃, 150 rpm, 1 h), harvested by centrifugation and stored at −70℃ until required. This process was repeated to give three independent transposon insertion mutant libraries. Finally, duplicate 0.5-mL aliquots of each library were cultured in 5 mL TSB15 + KAN250 + STR100 (22℃, 150 rpm, 3 h) before genomic DNA from 3 mL of each of the six cultures was extracted according to Bartie et al. (2020).
2.3. Preparation of DNA for Sequencing
The genomic DNA was prepared for sequencing based on van Opijnen and Camilli (2013). The DNA was cleaved with MmeI that produces a staggered cut approximately 16-bp downstream of the transposon-located recognition site, thus within the genomic DNA of the bacterium, and which can be used to determine the insertion locus (van Opijnen and Camilli, 2013). The DNA was washed to remove protein, precipitated with sodium acetate, washed twice in 70% ethanol, and finally resuspended in water. Sequencing adaptors to allow amplification by PCR were ligated to the sticky ends resulting from digestion. DNA fragments (140 bp) across the junction of the transposon and genomic DNA were amplified by PCR following a modified version of the Illumina 16S Metagenomic DNA sequencing library preparation protocol (Illumina, 2016). Reaction products were purified by magnetic beads (Agencourt AMPure XP magnetic beads; Beckman Coulter UK Ltd, High Wycombe, UK). A second PCR attached the Illumina Nextera XT v2 index primers, with each sample amplified in duplicate and barcoded uniquely to distinguish them during the parallel sequencing of the libraries.
2.4. Sequencing of Libraries
Each of the 12 libraries were standardised to 20 nM in EB Buffer (Qiagen), pooled and adjusted to 10 nM. This master sample was diluted in HT1 buffer (Illumina) to 11 pM and PhiX (Illumina) was added to 20 pM to improve sequence read quality. The sample was sequenced on a MiSeq platform (Illumina) with a 50-cycle v2 reagent kit to generate up to 15 M single-end reads (Illumina).
2.5. Mapping of Transposon Insertion Locations
Reads from the sequenced libraries were filtered for quality (QC <25), length (50 nt) and complexity (entropy over 15) using fastp (Chen et al., 2018). Remaining reads were processed through cutadapt v2.10 (Martin, 2011) to remove the transposon sequences and Tn-seq primers/adaptors. Then, the genomic DNA sequences flanking the sites of transposon insertion were mapped to the V. anguillarum NB10 genome (Assembly GCF_000786425.1; NB10Sm has 107 fewer transposase genes than the published parent stain NB10) and plasmids using bowtie2 v2.3.5.1 (Langmead and Salzberg, 2012): Chromosomes I and II (NZ_LK021130.1 and NZ_LK021129.1, respectively) and 8 k (pL02, NC_009351.1), 6 k (pJV, NC_019325.1) and 67 k (p67vangNB10 virulence plasmid, NZ_LK021128.1) plasmids, allowing for no base mismatches. Reads mapping to multiple TA sites were distributed between the multiple possible sites (Hubbard et al., 2016). The number of unique insertion sites were counted, and their locations plotted with visual rendering generated by GView v1.7 (Petkau et al., 2010).
2.6. Identification of Essential Genes
A Python implementation of EL-ARTIST (Pritchard et al., 2014) was used to normalise the dataset for origin proximity and smooth the transposon insertion dataset using hidden Markov model analysis following sliding window training (50 bp; P-value > 0.005). Each gene was classified as “essential” (i.e., absence of an insertion in the gene sequence), “domain-essential” (i.e., insertions present only at the end of the sequence [continuously]) or “non-essential” (i.e., presence of insertions [dis-continuously or continuously in the totality of the sequence]). Sequence reads that failed to tally with a TA site in the V. anguillarum NB10 genome were kept only if coverage was higher than two reads, to account for divergence between the sequences of the reference and isolate used herein.
2.7. Predicted Functions and Subcellular Locations of Essential Genes
For genes identified as essential, work was performed to determine putative functions and subcellular locations in the bacterium. The annotated, hypothetical and pseudo-genes were functionally re-classified using InterProscan v5.44-79 (Jones et al., 2014; Mitchell et al., 2019) and KofamKOALA v95.0 (Aramaki et al., 2020). Protein subcellular locations were predicted using SignalP v5.0 (Almagro Armenteros et al., 2019), SecretomeP v2.0a (Bendtsen et al., 2005), and LipoP v1.0 (Juncker et al., 2003). The KEGG pathway enrichment analysis was done using bioconductor/DOSE v3.10 and R/clusterProfiler v3.14.3 (Yu et al., 2015) using the set of all V. anguillarum genes with KEGG annotation as reference. The Protein-protein interaction functional enrichment analysis was done using STRING v11.5 (Szklarczyk et al., 2021) and using the set of all V. anguillarum annotated genes as reference.
2.8. Comparison of Essential Gene Lists With Vibrio spp. Studies
Locus names/tags of essential genes from V. cholerae (Cameron et al., 2008; Chao et al., 2013; Kamp et al., 2013), V. parahaemolyticus (Hubbard et al., 2016) and V. anguillarum MVM425 (Guanhua et al., 2018) determined in previous studies were collected from the supplementary materials or by correspondence with the authors. These data were used to recover the nucleic acid sequence of each gene from the NCBI GenBank database. Sequence alignment using BlastN was performed to recover orthologous genes independent of the gene names because these can be misleading or incomplete. Gene sequences that had an alignment > 80% across > 80% of the gene length were considered to be orthologous. Subsequently, Venn diagrams to visualise the overlap in essential gene lists were generated by jVenn (Bardou et al., 2014).
2.9. Essential Genes in the Core Genome of V. anguillarum
A desirable drug or vaccine candidate would be conserved across the species, and so the essential genes identified in this present study were compared to the core genes (i.e., present in ≥ 95% of genomes) found in the 105 V. anguillarum genomes analysed by Coyle et al. (2020). PIRATE v1.0.4 (Bayliss et al., 2019) was used to build a comprehensive pangenome of V. anguillarum, and the essential genes identified using the Coyle et al. (2020) methodology. Analysis of the output was conducted using R v4.0.2 (R Core Team, 2020).
3. Results
3.1. Construction of Transposon Insertion Mutants
Three independent transposon insertion libraries were created in V. anguillarum NB10Sm using a mariner-based transposon that inserts at TA dinucleotide loci, and from these were collected an estimated 5,500, 9,500, and 15,300 mutant colonies, respectively; mean transformation efficiency was 1.10 × 10-7. The reads of three independent libraries that had been extracted and sequenced in duplicate were merged, giving a total of 5,802,645 reads. After filtering for sequence quality, length, and complexity, 4,727,608 reads (81.5%) were retained and aligned to the V. anguillarum NB10 genome and relevant plasmids (Supplementary Data 1). In total, 3,100,490 reads (65.6%) aligned exactly once to a complementary sequence in the reference genome, while 1,171,283 reads (24.8%) did not match any sequence; 455,835 reads (9.6%) aligned to more than one sequence and so were distributed evenly between the multiple possible sites (Figure 1). In total, 52,662 unique insertion locations were mapped from the three transposon insertion libraries, with the discrepancy in colonies vs. insertion locations most likely due to overlapping colonies not being counted separately.
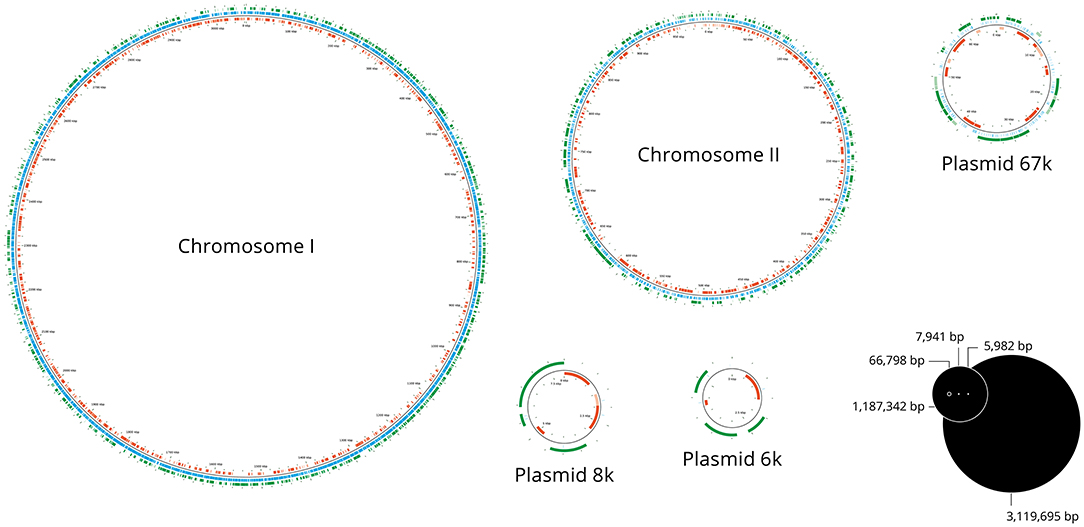
Figure 1. Map of transposon insertion sites. Representation of the two chromosomes and three plasmids of V. anguillarum NB10Sm showing the locations of the transposon insertions in the library of 52,662 mutants. The blue lines indicate the site of each transposon insertion, while green and red bars indicate the genes in forward and reverse orientations, respectively.
3.2. Distribution of Transposon Insertion Sites
Chromosomes I and II have similar median gene lengths and were found to contain approximately the same abundance of insertions per gene (Table 1) but, despite having a median gene length similar to the chromosomes, the 67-kb pJM1-like virulence plasmid contained approximately twice as many transposon insertions per gene as the chromosomes (Table 1). As expected, there was strong correlation between gene length and number of insertions (Figure 2), with the chromosomes showing similar linear regressions (slopes of 0.011 and 0.012, respectively, ANOVA P-value = 0.69) that differed significantly from the regression of the 67-kb plasmid (slope of 0.029, ANOVA P-value <10-15). The linear regressions for the two smaller plasmids were not significant, due to the very low number of genes these encoded (5 and 10 genes). Across the genome, 20.7% of the TA insertion sites were disrupted by the transposon.
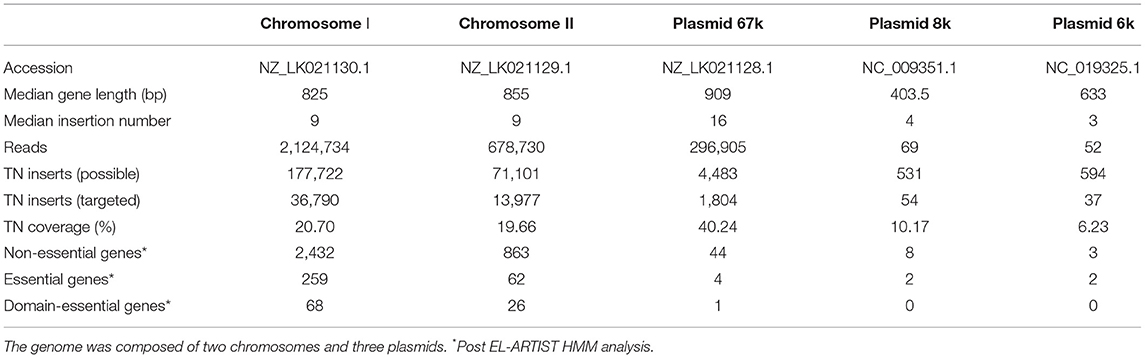
Table 1. Summary statistics of the V. anguillarum NB10Sm genome and transposon insertion mutant library.
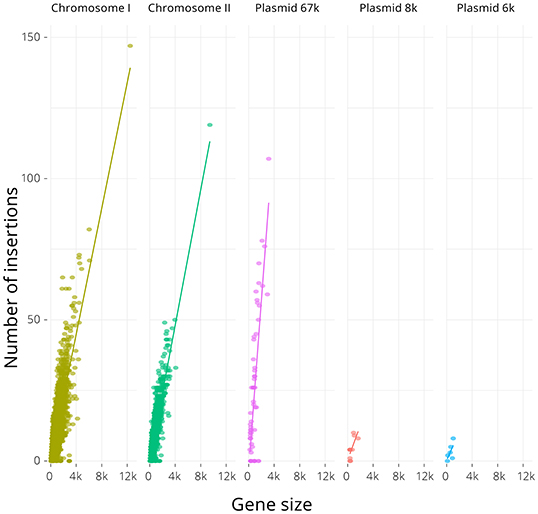
Figure 2. Relationship between gene sequence length (i.e., size) and number of transposon insertions for the two chromosomes and three plasmids in the V. anguillarum NB10Sm genome. There was significant correlation between gene sequence length and number of transposon insertions for the chromosomes and 67-kb plasmid and the R2 values were 0.61, 0.70, 0.60, 0.58, and 0.25 for Chromosome I, Chromosome II, Plasmid 67k, Plasmid 8k and Plasmid 6k, respectively. Slopes were calculated by regression where possible: 0.011, 0.012, and 0.029 for Chromosome I, Chromosome II, and 67-kb plasmid, respectively.
3.3. Essential Genes
Of the 3,774 genes on the two chromosomes and three plasmids annotated in the V. anguillarum NB10Sm genome, 329 (8.7%) genes were classified as essential, with 95 (2.5%) classified as domain-essential (Figure 3; Supplementary Data 2). The essential genes were grouped together based on whether they encoded rRNA, tRNA, or protein. Notably, all 25 rRNA genes were classified as essential whilst, of the 93 tRNAs, 89 were classified as essential, with the remaining four classified as domain-essential. The genes encoding essential proteins were functionally re-annotated using the KEGG and InterPro databases (Table 2), which shed further light on their known or potential functions, including the pathways that had been disrupted to lethal consequence. Interestingly, one of the largest groups of essential genes with an ascribed function was those identified as “transposases,” which accounted for 40 of the 212 proteins (18.9%) in total.
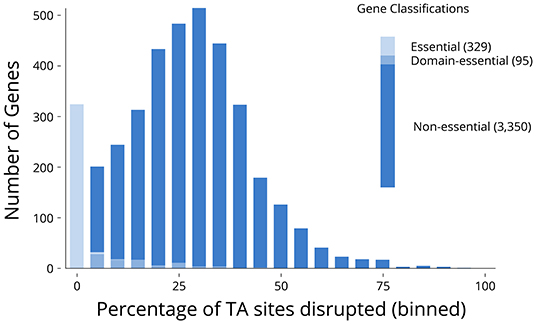
Figure 3. Classification of genes as essential, domain-essential, or non-essential. Distribution of percentage of TA sites disrupted in each gene (for those containing more than 10 TA sites in their sequence). Genes were categorised as essential, domain-essential, and non-essential within each bin (10-nt window) and in aggregate.
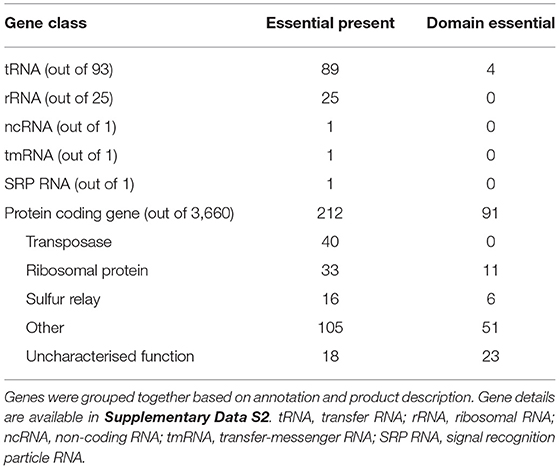
Table 2. Classifications of the 329 essential and 95 domain essential genes found in V. anguillarum NB10Sm.
3.4. Enrichment Analysis
Two KEGG pathways showed significant enrichment for essential genes (i.e., P-value <0.001). First is the “Ribosome” pathway (ko03010, Supplementary Figure 1A), where most of the 33 essential gene proteins contribute to the structure of the ribosome, in addition to the tRNA, rRNA, and the small number of elongation factors that are involved (adjusted P-value = 10-23); second is the “Sulfur relay system” pathway (ko04122, Supplementary Figure 1B), where 16 essential genes assist in the sulfur transfer steps of tRNA thiolation, folate biosynthesis, and thiamine and cysteine metabolism (adjusted P-value = 10-6). Similarly, STRING protein-protein interaction analysis revealed that “Ribosome and Protein biosynthesis,” where most of the 33 essential gene proteins contributed, was the most significant cluster (PPI enrichment P-value <10−16; expected interactions: 583; detected interactions: 2,869).
3.5. Core Genes
To assess the conservation of protein-encoding essential genes across the species V. anguillarum, the presence of the essential genes in the core genome of 105 sequenced isolates available at time of analysis was assessed according to the approach of Coyle et al. (2020). In total, 114 of the essential protein-encoding genes (53.8%) and 68 of the domain-essential genes (74.7%) were found within the core genome of V. anguillarum, and most of these core genes were annotated (110/114), with one transposase and only three genes of uncharacterised function (Figure 4). Fifty-four essential protein-encoding genes were within the accessory genome (i.e., found in at least 5% of genomes but <95% of genomes overall), which included ten uncharacterised proteins and eight transposase genes. Some of these essential genes determined to be part of the accessory genome may in fact be core genes, with the incompleteness of some published genomes meaning these genes are not found within a sufficient proportion of genomes to be classified as core. Of note, 44/212 (20.8%) of the essential protein-encoding genes were classified as unique (i.e., present in <5% of V. anguillarum genomes), whilst only eight essential genes with an annotation were unique.
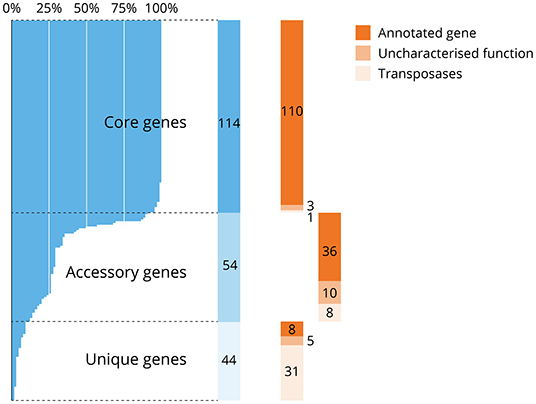
Figure 4. Distribution of protein-encoding essential genes in the pan-genome of V. anguillarum. Most essential protein-encoding genes (114/212, 53.8%) were found within the core genome of V. anguillarum, i.e., found in over 95% of the 105 genomes analysed, whilst 54/212 (25.5%) were in the accessory genome or detected uniquely in this isolate (44/212, 20.8%; in blue). Each essential gene was further characterised to be annotated, of or a transposase or a transposase (in orange). Most of the core genes were annotated (110/114, 96.5%) whilst most of the transposases were found uniquely in this isolate (31/44, 70.5%). Note that some core genes may have been incorrectly assigned to the accessory gene list due to available genomes being incomplete or of poor quality.
3.6. Comparison of Essential Gene Sets in Vibrio spp.
The essential gene list identified in this present study was compared with the lists determined in other Vibrio species and isolates (Figure 5; Supplementary Data 3). In total, 51 essential protein-encoding genes were shared between the two V. anguillarum isolates analysed (Figure 5A; Supplementary Data 3), and 36 (70.6%) of these genes were present within the core genome of this species. Comparison of the three studies to date on V. cholerae revealed 219 essential genes to be shared across these studies, with a further 171 genes essential in two of the three studies (Figure 5B; Supplementary Data 3). Taking together the three studies of V. cholerae, two studies of V. anguillarum and a single study of V. parahaemolyticus, there were 13 essential genes shared between the six studies, with nine of these genes encoding components of the 30S and 50S ribosomes and the other four genes described in the V. anguillarum NB10 genome to be a single-stranded DNA-binding protein; dihydropteroate synthase; co-chaperone HscB; and UDP-3-O-acyl-N-acetylglucosamine deacetylase (Figure 5C; Supplementary Data 3). Twenty-five essential genes were found exclusively in the V. anguillarum studies and were not present in the other Vibrio studies (Figure 5C; Supplementary Data 3). Finally, 118 protein-encoding genes in V. anguillarum NB10Sm were essential only in this isolate, though some of these genes are not reported to be present in the genomes of the isolates used in the other Vibrio studies (Figure 5C; Supplementary Data 3).
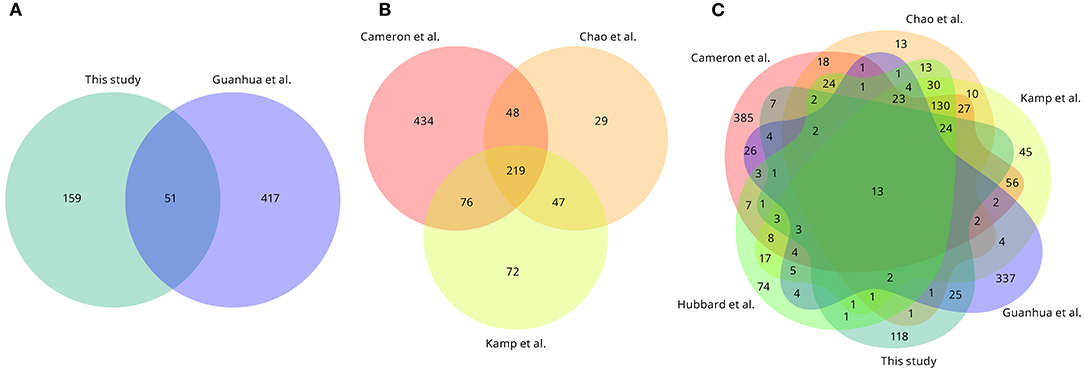
Figure 5. Conservation of essential protein-encoding genes across published Vibrio spp. studies. (A) Overlap of essential genes between V. anguillarum NB10Sm (this study) and V. anguillarum MVM425 (Guanhua et al., 2018). (B) Overlap of essential genes between three V. cholerae studies (Cameron et al., 2008; Chao et al., 2013; Kamp et al., 2013). (C) Overlap of essential genes between the six Vibrio studies. Note that for V. parahaemolyticus (Hubbard et al., 2016) only 402 genes out of 418 described were retrieved, as 16 loci were absent (e.g., VP0254). Duplicated genes are counted only once.
3.7. Subcellular Locations
Essential genes are potential drug targets, while extracellular products of essential genes and those expressed at the cell membrane would be strong candidates for immunogenicity assessment and consideration for possible inclusion into a vaccine. Hence, SecretomeP, SignalP, and LipoP were used to predict the subcellular locations of the essential gene products from V. anguillarum NB10Sm by identifying those that were non-classically secreted or contained a suitable signal peptide (Table 3). In total, seven essential gene products were predicted to be located to the inner cell membrane or the periplasm, including five genes found within the core genome of V. anguillarum (Table 3). Interestingly, two of these core genes (the SoxR reducing system protein gene rseC and the thiosulfate sulfurtransferase gene pspE), in addition to bcrA (benzoyl-CoA reductase subunit A gene) found in the accessory genome of V. anguillarum, were also essential in the other study performed on V. anguillarum MVM425.
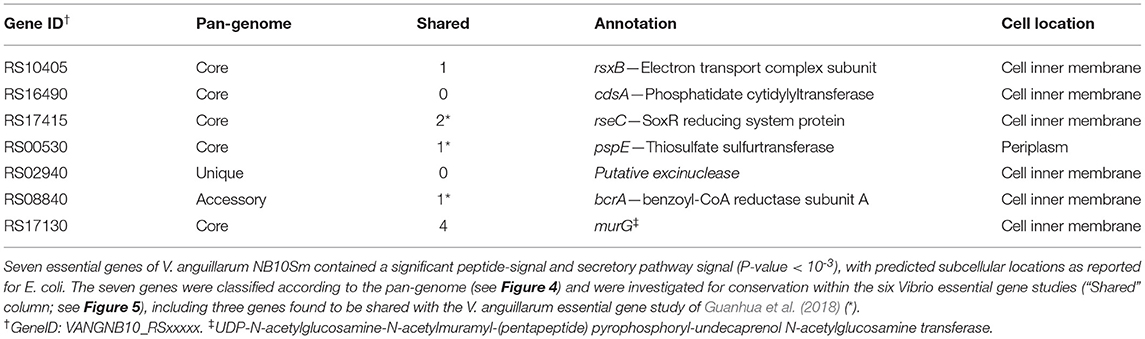
Table 3. Essential genes encoding products predicted to be released from the cell or expressed at the cell surface.
4. Discussion
In this present study, a Tn-seq approach to disrupting gene functions led to the determination of 329 essential and 95 domain-essential genes in the NB10Sm isolate of V. anguillarum, an aquatic pathogen that can infect many cultured aquatic species. Each of these essential genes is a potential target for the design of new chemotherapeutants, while subcellular location predictions in silico allowed for the generation of a shortlist of seven putative vaccine candidates containing proteins released from the cell or expressed at the cell surface. Comparison with studies of essential genes in another isolate of V. anguillarum and other Vibrio spp. allowed for the compilation of conserved lists of genes for prioritisation of the most promising antibacterial targets, whilst also advancing prospects for the development of genus- and species-specific antibacterial agents. Though essential genes have been identified in various human pathogens, including Vibrio spp., few studies have applied such an approach to fish pathogenic bacteria, and previous studies have not used these data to select possible Vibrio spp. vaccine candidates.
In total, 8.7% of the genes in the V. anguillarum NB10Sm genome were essential, with a further 2.5% classified as domain-essential, which is consistent with studies of other free-living bacteria that report typically 10–20% of total genes in a genome to be essential during culture in a rich medium in vitro (Gerdes et al., 2003; Gil et al., 2004; Peters et al., 2016). Other essential gene studies with Vibrio spp. employing various methods reported 343, 414, and 789 essential genes in V. cholerae, representing 9.9, 10.7, and 20.3%, respectively (Cameron et al., 2008; Chao et al., 2013; Kamp et al., 2013); 565 (12.7%) in V. parahaemolyticus (Hubbard et al., 2016); and 473 (12.5%) in V. anguillarum MVM425 (Guanhua et al., 2018).
Many of the essential protein-encoding genes in V. anguillarum NB10Sm were found within two pathways that contribute critically to protein synthesis, namely the “Ribosome” and “Sulfur relay system” pathways (ko03010 and ko04122, respectively), which is consistent with other studies that have found disruption of protein synthesis to be lethal in bacteria (Kobayashi et al., 2003; Chao et al., 2013; Hubbard et al., 2016). Indeed, several antibiotic groups disrupt ribosome function to lethal effect, including aminoglycosides and tetracyclines that target the 30S subunit (Fourmy et al., 1996; Chopra and Roberts, 2001) and the lincosamides and macrolides that bind to the 50S subunit (Spížek and Řezanka, 2017; Vázquez-Laslop and Mankin, 2018). The ribosome is the organelle where proteins are translated, meaning disruption of this key cellular machinery has a global impact on the cell's ability to synthesise proteins faithfully for normal cell functioning. All the tRNA genes were essential or domain-essential, which is to be expected given their vital role in protein synthesis, an observation with precedent (Reznikoff and Winterberg, 2008; Guanhua et al., 2018). Often tRNAs are excluded from essential gene lists due to their short sequence length (typically 76–90 bp in length), meaning there may not be sufficient confidence that the genes were not targeted by an insertion in Tn-seq libraries of lower saturation (Chao et al., 2016). However, in this present study, when the tRNA genes were considered as a concatenated sequence, the lack of transposon insertions was highly significant (P-value = 10−21), thus supporting their essentiality. Proteins in the sulfur relay system play crucial roles in tRNA thiolation, folate synthesis, and thiamine, cysteine and methionine metabolism, which are processes contributing to protein synthesis. Furthermore, rRNA composes the integral structure of the ribosome and all 25 rRNA genes in V. anguillarum NB10Sm were essential. This observation for the rRNA genes was made despite their redundancy, and this may be due to the mechanism of gene reversion that acts to maintain consistency of the genetic sequence at each locus (Liao, 2000; Santoya and Romero, 2005). As a result, the transposon-insertion sequence may have been either eliminated from each of the rRNA genes or propagated to each locus to likely lethal effect. Of course, the Tn-seq methodology only identifies the insertion locations in surviving bacteria, which would be those where the insertion had been eliminated, thus the genes would appear to lack insertions and be classified as essential. The high number of transposase or transposase-like genes classified as essential, particularly those belonging to the IS66 family, were mostly only found in the isolate used in this present study (31/40; 77.5%). The consistent lack of insertions into such genes may be due to the key roles they have in maintaining genome structure (Vigil-Stenman et al., 2017), but it may also be that these sequences possess mechanisms that protect against insertion. Finally, many essential genes were of uncharacterised function, which is familiar for studies of this nature (Hubbard et al., 2016), and it serves to draw attention to the shortcomings in our understanding of fundamental aspects of the biology of many genes in bacterial genomes.
Amongst the 13 essential genes conserved across each of the six studies of Vibrio spp. was the gene encoding UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, LpxC. LpxC is a key enzyme in the biosynthesis of Lipid A, a key structural component of the Gram-negative outer membrane, where it acts to anchor the lipopolysaccharide (Emiola et al., 2015; Joo, 2015). Compounds targeting this enzyme have been sought previously because it is conserved across Gram-negative species (Langklotz et al., 2011; Titecat et al., 2016) and its inactivation represents a novel means to combat bacterial pathogens (Williams et al., 2006; Tomaras et al., 2014; García-Quintanilla et al., 2016a). In addition to studies of various bacteria where LpxC is essential (Akerley et al., 2002; Barquist et al., 2013), the finding that this gene was essential in all six Vibrio spp. studies, confirms its value as a putative drug target and provides further support for the bioinformatics-driven approach to drug target prioritisation demonstrated herein.
Twenty-five essential genes were found exclusively in the V. anguillarum studies and were not present in the other studies of Vibrio spp., which could be exploited to develop more specific-acting antibacterials. This may be desirable because many non-pathogenic Vibrio spp. are important to the normal growth, development and health of the farmed aquatic animals, and they are often found within the animals' microbiota and in the culture water (Gajardo et al., 2016; Bone et al., 2021; Lorgen-Ritchie et al., 2021). Essential genes within the core genome of a bacterial species will be of greater interest for drug and vaccine development, particularly if they are essential across multiple isolates, because this allows for the effective targeting of most strains of a pathogen. A shift away from application of broad-spectrum antibiotics to agents with a narrow spectrum that target only the intended bacterium is one approach to reducing the problem of bacterial antibiotic resistance, as this avoids applying selection pressure for resistance on non-target species (Melander et al., 2018).
This present study demonstrated a new approach to identifying and prioritising potential candidates for incorporation into subunit vaccines for Vibrio spp. by applying bioinformatics tools to identify proteins of essential genes predicted to be located in the cell membrane or released extracellularly. All seven of the proteins identified by this approach in V. anguillarum NB10Sm, including three identified to be essential in V. anguillarum MVM425 (Guanhua et al., 2018), were predicted to be located at the inner cell membrane or in the periplasm, perhaps suggesting they may not be as detectable by host immune cells where the adaptive response is ideally desired. None of these seven genes were found amongst the 13 essential genes conserved across the six Vibrio spp. studies (Figure 5). Although, follow-up work would need to assess the immunogenicity of the surface-expressed or extracellular proteins, this present study does demonstrate the potential usefulness of such an approach.
Importantly, the differences between the essential gene lists derived from two phylogenetically closely related isolates, i.e., V. anguillarum NB10Sm and MVM425 (Coyle et al., 2020), and determined by a similar approach, demonstrates the value of characterising essential genes in multiple isolates of the same species when seeking conserved targets (Martínez-Carranza et al., 2018). An unexplained distinction between the V. anguillarum studies was the presence of 91% of the essential genes in MVM425 to be located on Chromosome I (Guanhua et al., 2018) despite comprising 72% of the genome, which compared to 79% of essential genes on Chromosome I in this present study where this chromosome accounted for 73% of the genome. Collectively, inconsistencies between similar studies exposes our still primitive understanding of essential genes, including their roles in the host and “rules” underpinning essentiality, further justifies their continued investigation.
The approach described in this present study is limited by the application of different approaches to identifying and classifying essential genes between studies. Even so, such distinctions between experiments allow for further rationalisation of conserved gene lists and provide stronger support for targeting the genes ultimately identified. Unquestionably, the use of pharmacological agents with specific action against the products of the essential genes, or the generation of targeted mutants, would have strengthened the case for essentiality and such approaches will be necessary to confirm this trait for each candidate essential gene (Falconer et al., 2011; Fields et al., 2017). Likewise, the essentiality of the genes has been determined in vitro only and investigation of their importance in more natural conditions, such as in seawater or during an infection, are likely to uncover slightly different gene lists, as these depend entirely on the conditions in which they are generated (Freed et al., 2016; Fields et al., 2017). Nevertheless, the combination of transposon-based gene disruption and high-throughput sequencing technologies allowing for rapid mapping of insertion locations across entire genomes is revolutionising our understanding of the essentiality of genes in bacterial genomes, whilst assisting to uncover gene functions through experiments under selective conditions (van Opijnen and Camilli, 2013; Chao et al., 2016). This present study adds to the increasing number of reports on essential genes in bacteria, which provide fundamental insights into genetic and metabolic networks that can also inform the creation of synthetic microorganisms with minimised genomes, thereby helping to unlock their exciting potential (Gil et al., 2004; Hutchison et al., 2016).
In conclusion, the approach demonstrated here provides a means to find new vaccine candidates and bacterial targets for the development of novel antibiotics, including agents of varying specificity. The methodology can be applied to different pathogens to guide the discovery of new measures to combat infectious diseases. Multiple approaches, including the discovery of new antibacterial agents and effective vaccination, are needed to address the global issue of antibiotic resistance, a classical One Health problem with human, animal, and environmental components.
Data Availability Statement
The Tn-seq data were submitted to the EBI ENA database under project number PRJEB39186. The scripts and pipeline used to process the Tn-seq reads are available at https://github.com/pseudogene/vibrio-tnseq.
Ethics Statement
This study was performed in accordance with the ethical review procedures of the University of Stirling.
Author Contributions
AD and SM conceived and designed research and conducted the experiments. MB contributed new analytical tools. NG, MB, and AD analysed the data. MB and AD wrote the manuscript. All authors read and approved the manuscript.
Funding
SM was funded in part by a University of Stirling Collaborative Research Studentship with the Centre for Environment, Fisheries and Aquaculture Science.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Sincere thanks to Prof. Matthew Waldor (Harvard Medical School, USA) and Prof. David Nelson (University of Rhode Island, USA) for kindly gifting E. coli SM10λpir (pSC189) and V. anguillarum NB10.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.755801/full#supplementary-material
References
Adams, A. (2019). Progress, challenges and opportunities in fish vaccine development. Fish Shell. Immunol. 90, 210–214. doi: 10.1016/j.fsi.2019.04.066
Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N., and Mekalanos, J. J. (2002). A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 99, 966–971. doi: 10.1073/pnas.012602299
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2020). KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. doi: 10.1038/msb4100050
Bardou, P., Mariette, J., Escudié, F., Djemiel, C., and Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. doi: 10.1186/1471-2105-15-293
Barquist, L., Langridge, G. C., Turner, D. J., Phan, M.-D., Turner, A. K., Bateman, A., et al. (2013). A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res. 41, 4549–4564. doi: 10.1093/nar/gkt148
Bartie, K. L., Taslima, K., Bekaert, M., Wehner, S., Syaifudin, M., Taggart, J. B., et al. (2020). Species composition in the Molobicus hybrid tilapia strain. Aquaculture 526:735433. doi: 10.1016/j.aquaculture.2020.735433
Bayliss, S. C., Thorpe, H. A., Coyle, N. M., Sheppard, S. K., and Feil, E. J. (2019). PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience 8:giz119. doi: 10.1093/gigascience/giz119
Bendtsen, J. D., Kiemer, L., Fausbøll, A., and Brunak, S. (2005). Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. doi: 10.1186/1471-2180-5-58
Bone, A., Bekaert, M., Papadopoulou, A., McMillan, S., Adams, A., Davie, A., et al. (2021). Bacterial communities of ballan wrasse (Labrus bergylta) eggs at a commercial marine hatchery. Curr. Microbiol. 78, 114–124. doi: 10.1007/s00284-020-02286-8
Breijyeh, Z., Jubeh, B., and Karaman, R. (2020). Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 25:1340. doi: 10.3390/molecules25061340
Cameron, D. E., Urbach, J. M., and Mekalanos, J. J. (2008). A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 105, 8736–8741. doi: 10.1073/pnas.0803281105
Chao, M. C., Abel, S., Davis, B. M., and Waldor, M. K. (2016). The design and analysis of transposon insertion sequencing experiments. Nat. Rev. Microbiol. 14, 119–128. doi: 10.1038/nrmicro.2015.7
Chao, M. C., Pritchard, J. R., Zhang, Y. J., Rubin, E. J., Livny, J., Davis, B. M., et al. (2013). High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 41, 9033–9048. doi: 10.1093/nar/gkt654
Chaudhuri, R. R., Allen, A. G., Owen, P. J., Shalom, G., Stone, K., Harrison, M., et al. (2009). Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. doi: 10.1186/1471-2164-10-291
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). FASTP: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Colquhoun, D. J., and Lillehaug, A. (2014). “Chapter 15: Vaccination against Vibriosis,” in Fish Vaccination, eds R. Gudding, A. Lillehaug, and Ø. Evensen (Chichester: John Wiley & Sons, Ltd.), 172–184. doi: 10.1002/9781118806913.ch15
Coyle, N. M., Bartie, K. L., Bayliss, S. C., Bekaert, M., Adams, A., McMillan, S., et al. (2020). A hopeful sea-monster: a very large homologous recombination event impacting the core genome of the marine pathogen Vibrio anguillarum. Front. Microbiol. 11:1430. doi: 10.3389/fmicb.2020.01430
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33:e00181–19. doi: 10.1128/CMR.00181-19
Desbois, A. P., Garza, M., Eltholth, M., Hegazy, Y. M., Mateus, A., Adams, A., et al. (2021). Systems-thinking approach to identify and assess feasibility of potential interventions to reduce antibiotic use in tilapia farming in Egypt. Aquaculture 540:736735. doi: 10.1016/j.aquaculture.2021.736735
Di Lorenzo, M., Stork, M., Tolmasky, M. E., Actis, L. A., Farrell, D., Welch, T. J., et al. (2003). Complete sequence of virulence plasmid pjm1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 185, 5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003
Emiola, A., George, J., and Andrews, S. S. (2015). A complete pathway model for lipid a biosynthesis in Escherichia coli. PLoS ONE 10:e0121216. doi: 10.1371/journal.pone.0121216
Falconer, S. B., Czarny, T. L., and Brown, E. D. (2011). Antibiotics as probes of biological complexity. Nat. Chem. Biol. 7, 415–423. doi: 10.1038/nchembio.590
FAO (2021). Antimicrobial Resistance. Available online at: http://www.fao.org/antimicrobial-resistance/en/ (accessed May 25, 2021).
Fields, F. R., Lee, S. W., and McConnell, M. J. (2017). Using bacterial genomes and essential genes for the development of new antibiotics. Biochem. Pharmacol. 134, 74–86. doi: 10.1016/j.bcp.2016.12.002
Forsyth, R. A., Haselbeck, R. J., Ohlsen, K. L., Yamamoto, R. T., Xu, H., Trawick, J. D., et al. (2002). A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x
Fourmy, D., Recht, M. I., Blanchard, S. C., and Puglisi, J. D. (1996). Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274, 1367–1371. doi: 10.1126/science.274.5291.1367
Frans, I., Michiels, C. W., Bossier, P., Willems, K. A., Lievens, B., and Rediers, H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34, 643–661. doi: 10.1111/j.1365-2761.2011.01279.x
Freed, N. E., Bumann, D., and Silander, O. K. (2016). Combining Shigella Tn-seq data with gold-standard E. coli gene deletion data suggests rare transitions between essential and non-essential gene functionality. BMC Microbiol. 16:203. doi: 10.1186/s12866-016-0818-0
Gajardo, K., Rodiles, A., Kortner, T. M., Krogdahl, Å., Bakke, A. M., Merrifield, D. L., et al. (2016). A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci. Rep. 6:30893. doi: 10.1038/srep30893
Gallagher, L. A., Shendure, J., and Manoil, C. (2011). Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10
García-Quintanilla, M., Caro-Vega, J. M., Pulido, M. R., Moreno-Martínez, P., Pachón, J., and McConnell, M. J. (2016a). Inhibition of LpxC increases antibiotic susceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 5076–5079. doi: 10.1128/AAC.00407-16
García-Quintanilla, M., Pulido, M. R., Carretero-Ledesma, M., and McConnell, M. J. (2016b). Vaccines for antibiotic-resistant bacteria: possibility or pipe dream? Trends Pharmacol. Sci. 37, 143–152. doi: 10.1016/j.tips.2015.10.003
Gerdes, S. Y., Scholle, M. D., Campbell, J. W., Balázsi, G., Ravasz, E., Daugherty, M. D., et al. (2003). Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185, 5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003
Gil, R., Silva, F. J., Peretó, J., and Moya, A. (2004). Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68, 518–537. doi: 10.1128/MMBR.68.3.518-537.2004
Goodall, E. C. A., Robinson, A., Johnston, I. G., Jabbari, S., Turner, K. A., Cunningham, A. F., et al. (2018). The essential genome of Escherichia coli K-12. mBio 9:e02096–17. doi: 10.1128/mBio.02096-17
Guanhua, Y., Wang, C., Wang, X., Ma, R., Zheng, H., Liu, Q., et al. (2018). Complete genome sequence of the marine fish pathogen Vibrio anguillarum and genome-wide transposon mutagenesis analysis of genes essential for in vivo infection. Microbiol. Res. 216, 97–107. doi: 10.1016/j.micres.2018.08.011
Hansson, M., Nygren, P.-Å., and Ståhl, S. (2000). Design and production of recombinant subunit vaccines. Biotechnol. Appl. Biochem. 32:95. doi: 10.1042/BA20000034
Holm, K. O., Nilsson, K., Hjerde, E., Willassen, N.-P., and Milton, D. L. (2015). Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia. Stand. Genom. Sci. 10:60. doi: 10.1186/s40793-015-0060-7
Hubbard, T. P., Chao, M. C., Abel, S., Blondel, C. J., Abel zur Wiesch, P., Zhou, X., et al. (2016). Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc. Natl. Acad. Sci. U.S.A. 113, 6283–6288. doi: 10.1073/pnas.1601718113
Hutchison, C. A., Chuang, R.-Y., Noskov, V. N., Assad-Garcia, N., Deerinck, T. J., Ellisman, M. H., et al. (2016). Design and synthesis of a minimal bacterial genome. Science 351:aad6253. doi: 10.1126/science.aad6253
Illumina (2016). 16S Metagenomic Sequencing Library Preparation 15044223 B. Available online at: https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (Accessed November 27, 2018).
Ji, Y. (2001). Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293, 2266–2269. doi: 10.1126/science.1063566
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Joo, S. H. (2015). Lipid A as a drug target and therapeutic molecule. Biomol. Therap. 23, 510–516. doi: 10.4062/biomolther.2015.117
Jordan, I. K., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12, 962–968. doi: 10.1101/gr.87702
Judson, N., and Mekalanos, J. J. (2000). Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8, 521–526. doi: 10.1016/S0966-842X(00)01865-5
Juncker, A. S., Willenbrock, H., von Heijne, G., Brunak, S., Nielsen, H., and Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662. doi: 10.1110/ps.0303703
Kamp, H. D., Patimalla-Dipali, B., Lazinski, D. W., Wallace-Gadsden, F., and Camilli, A. (2013). Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9:e1003800. doi: 10.1371/journal.ppat.1003800
Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., et al. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683. doi: 10.1073/pnas.0730515100
Langklotz, S., Scha kermann, M., and Narberhaus, F. (2011). Control of lipopolysaccharide biosynthesis by FtsH-mediated proteolysis of LpxC is conserved in enterobacteria but not in all Gram-negative bacteria. J. Bacteriol. 193, 1090–1097. doi: 10.1128/JB.01043-10
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Le Breton, Y., Belew, A. T., Valdes, K. M., Islam, E., Curry, P., Tettelin, H., et al. (2015). Essential genes in the core genome of the human pathogen Streptococcus pyogenes. Sci. Rep. 5:9838. doi: 10.1038/srep09838
Liao, D. (2000). Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J. Mol. Evol. 51, 305–317. doi: 10.1007/s002390010093
Lorgen-Ritchie, M., Clarkson, M., Chalmers, L., Taylor, J. F., Migaud, H., and Martin, S. A. M. (2021). A temporally dynamic gut microbiome in Atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci. 8:711797. doi: 10.3389/fmars.2021.711797
Ma, J., Bruce, T. J., Jones, E. M., and Cain, K. D. (2019). A review of fish vaccine development strategies: conventional methods and modern biotechnological approaches. Microorganisms 7:569. doi: 10.3390/microorganisms7110569
Marcos-López, M., Donald, K., Stagg, H., and McCarthy, Ú. (2013). Clinical Vibrio anguillarum infection in lumpsucker Cyclopterus lumpus in Scotland. Vet. Rec. 173:319. doi: 10.1136/vr.101763
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Martínez-Carranza, E., Barajas, H., Alcaraz, L.-D., Servín-González, L., Ponce-Soto, G.-Y., and Soberón-Chávez, G. (2018). Variability of bacterial essential genes among closely related bacteria: the case of Escherichia coli. Front. Microbiol. 9:1059. doi: 10.3389/fmicb.2018.02330
Melander, R. J., Zurawski, D. V., and Melander, C. (2018). Narrow-spectrum antibacterial agents. MedChemComm 9, 12–21. doi: 10.1039/C7MD00528H
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Moule, M. G., Hemsley, C. M., Seet, Q., Guerra-Assunção, J. A., Lim, J., Sarkar-Tyson, M., et al. (2014). Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5:e00926-13. doi: 10.1128/mBio.00926-13
Naka, H., Dias, G. M., Thompson, C. C., Dubay, C., Thompson, F. L., and Crosa, J. H. (2011). Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect. Immun. 79, 2889–900. doi: 10.1128/IAI.05138-11
Naz, K., Naz, A., Ashraf, S. T., Rizwan, M., Ahmad, J., Baumbach, J., et al. (2019). PanRV: Pangenome-reverse vaccinology approach for identifications of potential vaccine candidates in microbial pangenome. BMC Bioinformatics 20:123. doi: 10.1186/s12859-019-2713-9
Okada, K., Iida, T., Kita-Tsukamoto, K., and Honda, T. (2005). Vibrios commonly possess two chromosomes. J. Bacteriol. 187, 752–757. doi: 10.1128/JB.187.2.752-757.2005
Peters, J. M., Colavin, A., Shi, H., Czarny, T. L., Larson, M. H., Wong, S., et al. (2016). A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506. doi: 10.1016/j.cell.2016.05.003
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Pham-Duc, P., Cook, M. A., Cong-Hong, H., Nguyen-Thuy, H., Padungtod, P., Nguyen-Thi, H., et al. (2019). Knowledge, attitudes and practices of livestock and aquaculture producers regarding antimicrobial use and resistance in Vietnam. PLoS ONE 14:e0223115. doi: 10.1371/journal.pone.0223115
Poulsen, B. E., Yang, R., Clatworthy, A. E., White, T., Osmulski, S. J., Li, L., et al. (2019). Defining the core essential genome of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 116, 10072–10080. doi: 10.1073/pnas.1900570116
Pritchard, J. R., Chao, M. C., Abel, S., Davis, B. M., Baranowski, C., Zhang, Y. J., et al. (2014). ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 10:e1004782. doi: 10.1371/journal.pgen.1004782
R Core Team (2020). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online at: https://www.r-project.org/
Reverter, M., Sarter, S., Caruso, D., Avarre, J.-C., Combe, M., Pepey, E., et al. (2020). Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 11:1870. doi: 10.1038/s41467-020-15735-6
Reznikoff, W. S., and Winterberg, K. M. (2008). “Transposon-based strategies for the identification of essential bacterial genes,” in Microbial Gene Essentiality: Protocols and Bioinformatics, Vol. 416 of Methods in Molecular Biology, eds A. L. Osterman and S. Y. Gerdes (Totowa, NJ: Humana Press), 13–26. doi: 10.1007/978-1-59745-321-9_2
Rubin, B. E., Wetmore, K. M., Price, M. N., Diamond, S., Shultzaberger, R. K., Lowe, L. C., et al. (2015). The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. U.S.A. 112, E6634–E6643. doi: 10.1073/pnas.1519220112
Santoya, G., and Romero, D. (2005). Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29, 169–183. doi: 10.1016/j.femsre.2004.10.004
Sommerset, I., Krossøy, B., Biering, E., and Frost, P. (2005). Vaccines for fish in aquaculture. Expert Rev. Vacc. 4, 89–101. doi: 10.1586/14760584.4.1.89
Spížek, J., and Řezanka, T. (2017). Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 133, 20–28. doi: 10.1016/j.bcp.2016.12.001
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi: 10.1093/nar/gkaa1074
Thakur, A. B., Vaidya, R. B., and Suryawanshi, S. A. (2003). Pathogenicity and antibiotic susceptibility of Vibrio species isolated from moribund shrimps. Indian J. Mar. Sci. 32, 71–75.
Thanassi, J. A. (2002). Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30, 3152–3162. doi: 10.1093/nar/gkf418
Titecat, M., Liang, X., Lee, C.-J., Charlet, A., Hocquet, D., Lambert, T., et al. (2016). High susceptibility of MDR and XDR gram-negative pathogens to biphenyl-diacetylene-based difluoromethyl- all -threonyl-hydroxamate LpxC inhibitors. J. Antimicrob. Chemother. 71, 2874–2882. doi: 10.1093/jac/dkw210
Tomaras, A. P., McPherson, C. J., Kuhn, M., Carifa, A., Mullins, L., George, D., et al. (2014). LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. mBio 5:e01551-14. doi: 10.1128/mBio.01551-14
Toranzo, A. E., Magari nos, B., and Romalde, J. L. (2005). A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246, 37–61. doi: 10.1016/j.aquaculture.2005.01.002
Turner, K. H., Wessel, A. K., Palmer, G. C., Murray, J. L., and Whiteley, M. (2015). Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115. doi: 10.1073/pnas.1419677112
van Opijnen, T., Bodi, K. L., and Camilli, A. (2009). TN-SEQ: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772. doi: 10.1038/nmeth.1377
van Opijnen, T., and Camilli, A. (2013). Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442. doi: 10.1038/nrmicro3033
Vázquez-Laslop, N., and Mankin, A. S. (2018). How macrolide antibiotics work. Trends Biochem. Sci. 43, 668–684. doi: 10.1016/j.tibs.2018.06.011
Vigil-Stenman, T., Ininbergs, K., Bergman, B., and Ekman, M. (2017). High abundance and expression of transposases in bacteria from the Baltic sea. ISME J. 11, 2611–2623. doi: 10.1038/ismej.2017.114
Vincent, A. T., Gauthier, J., Derome, N., and Charette, S. J. (2019). “Chapter 1: The rise and fall of antibiotics in aquaculture,” in Microbial Communities in Aquaculture Ecosystems, ed N. Derome (Cham: Springer International Publishing), 1–19. doi: 10.1007/978-3-030-16190-3_1
Williams, A. H., Immormino, R. M., Gewirth, D. T., and Raetz, C. R. H. (2006). Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide. Proc. Natl. Acad. Sci. U.S.A. 103, 10877–10882. doi: 10.1073/pnas.0604465103
Keywords: antimicrobial resistance (AMR), aquaculture, transposon-insertion sequencing (Tn-seq), pathogen evolution, reverse vaccinology, Vibrio cholerae, Vibrio parahaemolyticus
Citation: Bekaert M, Goffin N, McMillan S and Desbois AP (2021) Essential Genes of Vibrio anguillarum and Other Vibrio spp. Guide the Development of New Drugs and Vaccines. Front. Microbiol. 12:755801. doi: 10.3389/fmicb.2021.755801
Received: 09 August 2021; Accepted: 24 September 2021;
Published: 20 October 2021.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Rabindra Kumar Mandal, University of Louisville, United StatesLi Sun, Anhui Medical University, China
Copyright © 2021 Bekaert, Goffin, McMillan and Desbois. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew P. Desbois, YW5kcmV3LmRlc2JvaXNAc3Rpci5hYy51aw==
 Michaël Bekaert
Michaël Bekaert Nikki Goffin
Nikki Goffin Stuart McMillan
Stuart McMillan Andrew P. Desbois
Andrew P. Desbois