- 1Department of Medical Microbiology, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Selangor, Malaysia
- 2Borneo Medical and Health Research Center, Universiti Malaysia Sabah, Sabah, Malaysia
The zoonotic disease leptospirosis is caused by pathogenic species of the genus Leptospira. With the advancement of studies in leptospirosis, several new species are being reported. It has always been a query, whether Leptospira species, serovars, and strains isolated from different geographical locations contribute to the difference in the disease presentations and severity. In an epidemiological surveillance study performed in Malaysia, we isolated seven novel intermediate and saprophytic species (Leptospira semungkisensis, Leptospira fletcheri, Leptospira langatensis, Leptospira selangorensis, Leptospira jelokensis, Leptospira perdikensis, Leptospira congkakensis) from environments and three pathogenic species from rodents (Leptospira borgpetersenii strain HP364, Leptospira weilii strain SC295, Leptospira interrogans strain HP358) trapped in human leptospirosis outbreak premises. To evaluate the pathogenic potential of these isolates, we performed an in vivo and in silico virulence analysis. Environmental isolates and strain HP364 did not induce any clinical manifestations in hamsters. Strain SC295 caused inactivity and weight loss with histopathological changes in kidneys, however, all hamsters survived until the end of the experiment. Strain HP358 showed a high virulent phenotype as all infected hamsters died or were moribund within 7 days postinfection. Lungs, liver, and kidneys showed pathological changes with hemorrhage as the main presentation. In silico analysis elucidated the genome size of strain HP358 to be larger than strains HP364 and SC295 and containing virulence genes reported in Leptospira species and a high number of specific putative virulence factors. In conclusion, L. interrogans strain HP358 was highly pathogenic with fatal outcome. The constituent of Leptospira genomes may determine the level of disease severity and that needs further investigations.
Introduction
Leptospirosis is a zoonotic disease, with rodents being the main transmitting source. Humans get infected either by direct contact with the animals or indirectly from environments contaminated with animal urine. There are three classes of Leptospira: nonpathogenic, intermediate, and pathogenic. To date, there have been 66 Leptospira species (Masuzawa et al., 2018; Thibeaux et al., 2018; Vincent et al., 2019; Casanovas-Massana et al., 2020) and more than 300 serovars identified. The majority of the non-pathogens and intermediate species come from environments (soil and water), while the pathogens are excreted in the urine of reservoir animals (Levett, 2001; Putz and Nally, 2020). The pathogenic Leptospira species, serovars, and strains are known to cause leptospirosis and contribute to the broad spectrum of the disease presentations (Vinetz, 2001). The pathogenic potential of Leptospira species has been attributed to the presence of genes that encode the virulence phenotype (Fouts et al., 2016). There are unique genetic determinants that influence the pathogenic Leptospira to adhere, invade, disseminate, escape from host defense, and establish systemic infection. These genetic elements may be absent or less expressed in intermediate and nonpathogenic Leptospira. When compared to several other bacteria, pathogenic Leptospira species do not have any typical exotoxin-encoding genes (Ren et al., 2003; Nascimento et al., 2004). Similarly, the lipopolysaccharide in pathogenic Leptospira interrogans has been reported to have lower endotoxic activities than other bacteria (Isogai et al., 1986, 1989; Vinh et al., 1989; Bulach et al., 2000; Adler, 2015). Endostatin-like outer membrane proteins, such as LipL32/53, Lsa21/32/63, LenA, LigA, and LigB (Choy et al., 2007; Stevenson et al., 2007; Atzingen et al., 2008; Hauk et al., 2008; Oliveira et al., 2010; Vieira et al., 2010; Domingos et al., 2015), have been identified to participate in the adherence of pathogenic Leptospira to host cells by binding to fibronectin, laminin, and collagens of extracellular matrix, whereas collagenase (Kassegne et al., 2014), invA (Fraser and Brown, 2017), M16 type of metalloprotease/metallopeptidase (Ren et al., 2003), mammalian cell entry protein, and endoflagellum of pathogenic Leptospira have been reported to contribute to leptospiral invasiveness (Liao et al., 2009; Zhang et al., 2012). The hemolysins (sphingomyelinase-type hemolysins, sphH)—SphH pore-forming protein Sph1-3, HlpA, TlyA, and Loa22—are recognized as the strong inducer of interleukin 1β (IL-1β), IL-6, and tumor necrosis factor α production (Lee et al., 2002; Ristow et al., 2007; Wang et al., 2012; Hsu et al., 2021). The vWA proteins encoded by genes vwa-I and vwa-II have been shown to induce hemorrhage in L. interrogans (Fang et al., 2018). It is believed that there could be more virulent determinants that are yet to be discovered.
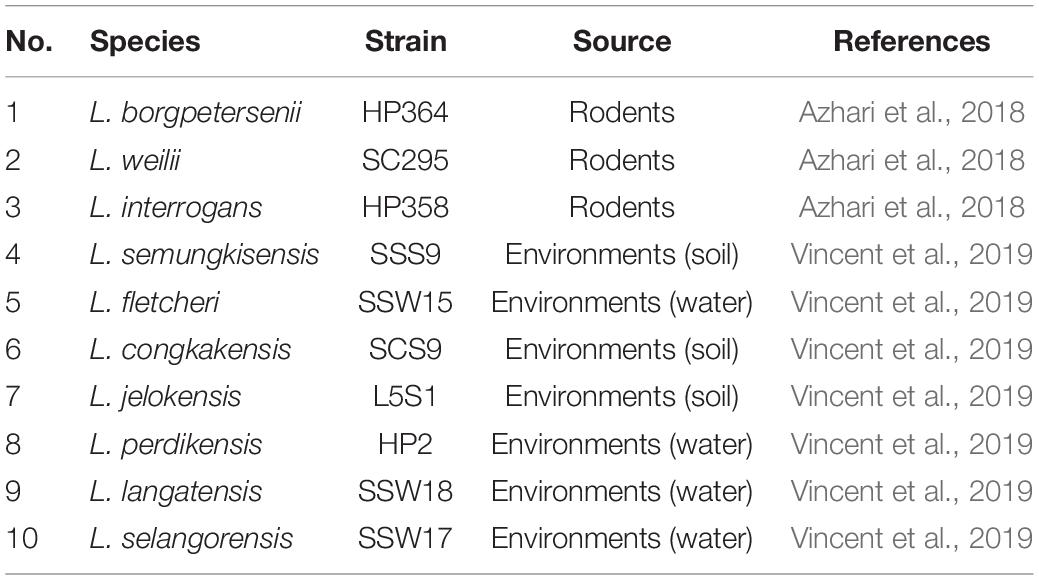
With the advancement in genomics, rise in leptospirosis cases, and studies from various geographical locations, more new species and serovars are being reported. It is vital to identify and characterize the circulating species and serovars in every country to understand the epidemiology and pathogenic potential to combat the illness. In an earlier study, we reported seven novel species classified as nonpathogenic isolated from soil and water samples from leptospirosis outbreak areas in Malaysia (Vincent et al., 2019). We also isolated a few pathogenic species such as L. interrogans, Leptospira kirschneri, Leptospira weilii, and Leptospira borgpetersenii from rodents captured in the same areas (Azhari et al., 2018). In a very recent study, we reported L. interrogans and L. kirschneri as the human-infecting Leptospira species in Malaysia (Philip et al., 2020). In the present study, we aimed to evaluate the in vivo virulence of the pathogenic (from rodents) and novel Leptospira species (from environments) in a hamster model and to perform a comparative genomics study to elucidate the virulence characteristics of Leptospira species and identify potential virulence factors.
Materials and Methods
Animal Experiments and Ethics Approval
Experiments were conducted following the guidelines of the Code of Practice for the Care and Use of Animals for Scientific Purposes, Universiti Putra Malaysia. Male golden Syrian hamsters purchased from Monash Universiti Malaysia, Bandar Sunway, Selangor, aged between 4 and 6 weeks, were housed individually in cages with sterile sawdust bedding. The animals were fed with their routine food, watered in sterile bottles throughout the study, and were acclimatized for 7 days before infection. All animal procedures carried out in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC), Universiti Putra Malaysia with Animal Use Protocol (AUP) number UPM/IACUC/AUP-R044/2018.
Infection, Monitoring, and Euthanasia of Animals
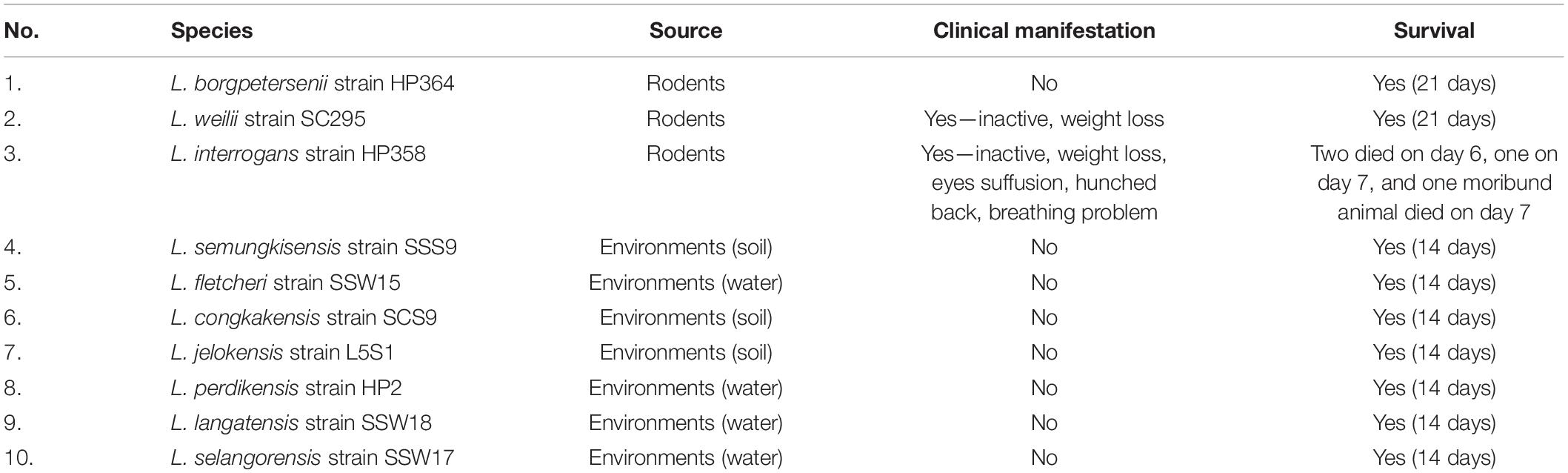
Upon completion of 7 days of acclimatization, the hamsters were intraperitoneally (IP) injected with 2 × 108 (Matsui et al., 2011) of second passages of leptospires cultures from environmental and animal isolates (Table 1) in 500 μL Ellinghausen, McCullough, Johnson and Harris (EMJH) medium. Control animals were injected (IP) with 500 μL sterile EMJH medium. Four animals were infected with leptospires in each Leptospira species and control. Hamsters injected with environmental isolates were monitored for 14 days, whereas for pathogenic isolates up to 21 days. Those animals that were moribund during the course of study characterized by significant weight loss ≥ 10%, lethargy, gait difficulty, dyspnea, and prostration were anesthetized with 100 mg/kg ketamine and 5 mg/kg xylazine IP injection and euthanized by atlanto-occipital dislocation and dissected after cardiac puncture. All animals that survived until the end of the study were euthanized on days 14 (environmental) and 21 (animal isolates) postinfection (p.i.). Prior to euthanization, blood was collected by cardiac puncture for direct culture in the EMJH medium and for leptospiral DNA detection in the EDTA tube. Approximately 25 mg of lungs, liver, and kidneys were collected in absolute ethanol for leptospiral DNA detection. The remaining part of the organs was preserved in neutral-buffered formalin for histopathology observation.
Detection of Leptospires in Blood and Organs
DNA from 200 μL of blood and 25 mg of organs was extracted using the DNeasy blood and Tissue kit (Qiagen, Germany) following the manufacturer’s instructions. Detection of lipL32 gene (Stoddard et al., 2009) was performed using the quantitative polymerase chain reaction (PCR) platform on Eppendorf MasterCycler® Realplex. Final reaction for the real-time PCR contained 12.5 μL Quantinova probe PCR kits (2 × concentration), 1 μL of 10 pmol of each primer, 0.5 μL of 10-pmol probe, 5 μL of RNase free water, and 5 μL of DNA extracted from blood or tissue samples in a final volume of 25 μL. The amplification protocol started with 8 min at 95°C, followed by 45 cycles of 95°C for 3 s and 58°C for 15 s, and finished with a cool cycle at 45°C for 90 s. Extracted DNA from a pure culture of L. interrogans strain HP358 and RNase free water were used as positive and negative controls.
Observation of Tissue Damages in the Infected Animals by Histopathology
For each organ, formalin-fixed paraffin-embedded tissue blocks were subjected to hematoxylin and eosin (H&E) stain. Formalin-fixed tissues were embedded in paraffin, cut into thin sections (∼4 mm), and stained with H&E by standard protocol. Tissue damages are recorded as per previously reported criteria (Marinho et al., 2009; Matsui et al., 2011; Villanueva et al., 2014).
Genome Sequencing, de novo Assembly, Phylogenetic Tree, and Annotation
The genomic DNA of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 was extracted from pure cultures using DNeasy Blood and Tissue kits (Qiagen) according to the manufacturer’s instruction. The DNA library for whole-genome sequencing was prepared using the IGP-NGSP Illumina Library and subsequently sequenced through the Miseq instruments platform. The quality of raw reads of the sequenced genomes was checked using FastQC and preprocessed using BBMap version 38.43 tool (Bushnell et al., 2017). The adapters and reads with less than 50 bp were trimmed based on the phred quality score (below Q30) using BBduk.sh. To avoid bias using reference mapping, de novo genome assembly was performed for all the three species using SPAdes version 3.11.1 (Bankevich et al., 2012).
Single-nucleotide polymorphism (SNP)–based phylogenetic tree was performed for the three pathogenic Leptospira strains described previously (L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358) and another 49 Leptospira strains (including the seven Leptospira species from environment isolates used in this study) (Supplementary File 1, Table 1). The whole-genome sequences of the 49 strains were extracted from NCBI GenBank. The core SNP was determined using the kSNP3 package (Gardner et al., 2015). All SNP matrices were aligned using CLUSTALW, and SNP-based phylogenetic tree was performed using the maximum likelihood method in MEGAX (Molecular Evolutionary Genetic Analysis) software (Kumar et al., 2018). The significance of the branching patterns was evaluated through bootstrap analysis of 1,000 replicates.
The generated contigs of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 were annotated using Rapid Annotation using Subsystem Technology (RAST) server version 2.0 (Aziz et al., 2008). Gene and putative protein-coding sequences of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 were predicted with the GeneMark program (Besemer et al., 2001).
Pan-Genome Analysis
A total of 30 genomes of Leptospira strains (Supplementary File 1, Table 2) were subjected to pan-genome analysis using Roary V3.11.2 (Page et al., 2015). The input files used were the GFF3 (General Feature Format version 3) format generated from annotated assembly (GeneMark) (L. borgpetersenii strain HP364, L. weilii strain SC295, L. interrogans strain HP358) and GenBank files obtained from the NCBI website (27 strains). For definition of core genes in all strains, the threshold of 99% was used, whereas for sequence comparison between strains, a user-defined percentage sequence identity (default 95%) was performed with BLASTP (Sitto and Battistuzzi, 2020). Using conserved gene neighborhood information, homologous groups containing paralogs were split into groups of true orthologs. The strains were clustered based on gene presence in the accessory genome, weighted by the total and shared genomes. A core-genome phylogenetic tree was constructed using the maximum likelihood method in MEGAX with a bootstrap analysis of 1,000 replicates. The list genes of the three pathogenic species (L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358) were extracted from the pan-genome data for comparison of core and specific gene.
Virulence Factor Analysis
A list of known virulence factors in Leptospira species was generated from literature reviews (Murray, 2015; Gomes-Solecki et al., 2017; Picardeau, 2017). The amino acid sequences of these virulence factors were retrieved from UniProt database and were subjected to a BLAST search against the genome of Leptospira species used in the in vivo study and 29 representatives Leptospira genomes using local blast (tblastn) in BlastSation software in the following link.1 Additional five virulence factors with nucleotide sequences were retrieved from NCBI GenBank database and blasted against the genomes of Leptospira using homology blastn method (Altschul et al., 1990). Besides the known virulence factors in Leptospira genome, potential virulence factors from other bacterial species were also investigated in L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans HP358 through ortholog gene identification present in full datasets of virulence factor database (VFDB) (Chen et al., 2016) and Victors database2 (Sayers et al., 2019) with criteria of more than 70% alignment coverage (bit score) and 40% identity (Rost, 1999) using Blastation software.
Results
Clinical Response to Infection
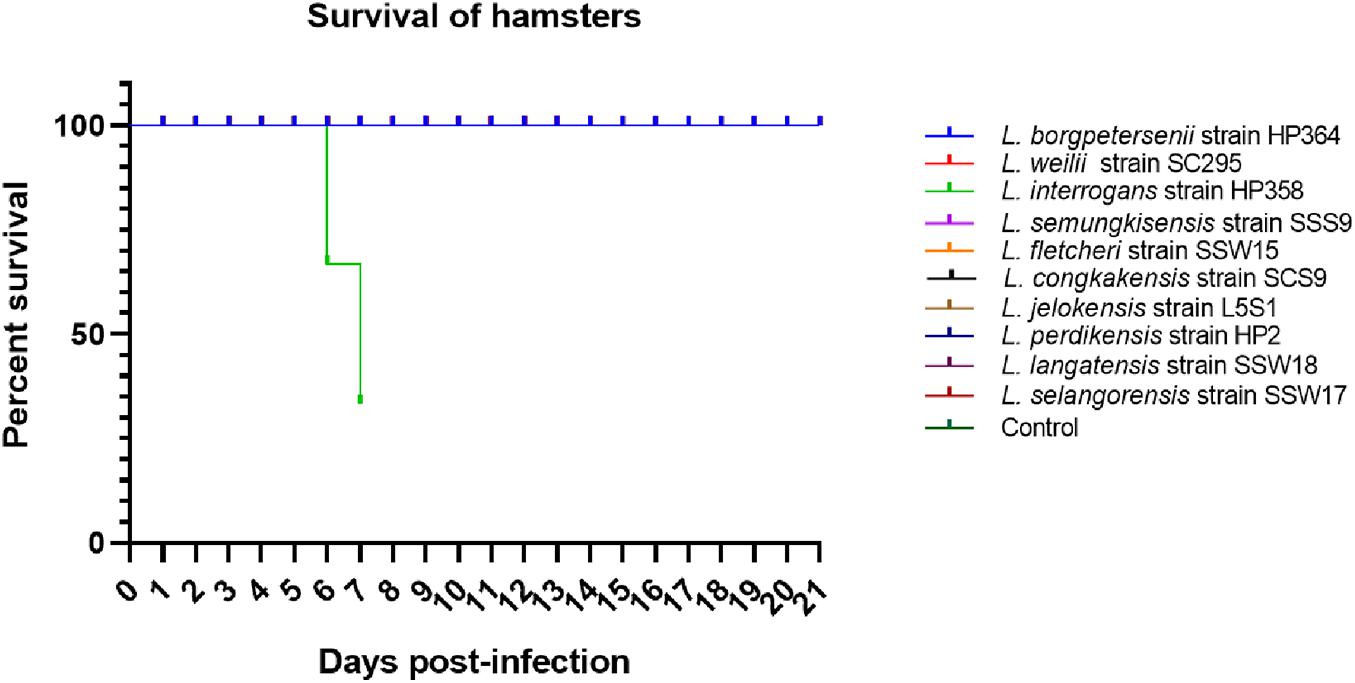
All hamsters infected with Leptospira isolates from environments survived, increased in body weight (Figure 1), and showed no clinical presentations until the animals were euthanized on day 14 (Table 2). Similar to the environment isolates, L. borgpetersenii strain HP364 isolated from rodents also did not show any clinical manifestations in hamsters, and all survived until they were euthanized (21st day). Although the hamsters infected with L. weilii strain SC295 survived until the end of the study, the animals showed mild weight loss and were less active between day 5 and 9 p.i. On the contrary, L. interrogans strain HP358 induced a fatal outcome in infected hamsters. The animals showed loss of appetite and presence of eyes suffusion, were less active, and had difficulty in breathing from day 5 p.i. onward. Two animals died on day 6 and one on day 7 p.i., whereas the remaining one animal was moribund, hence euthanized on day 7 (Figure 2: survival curve).
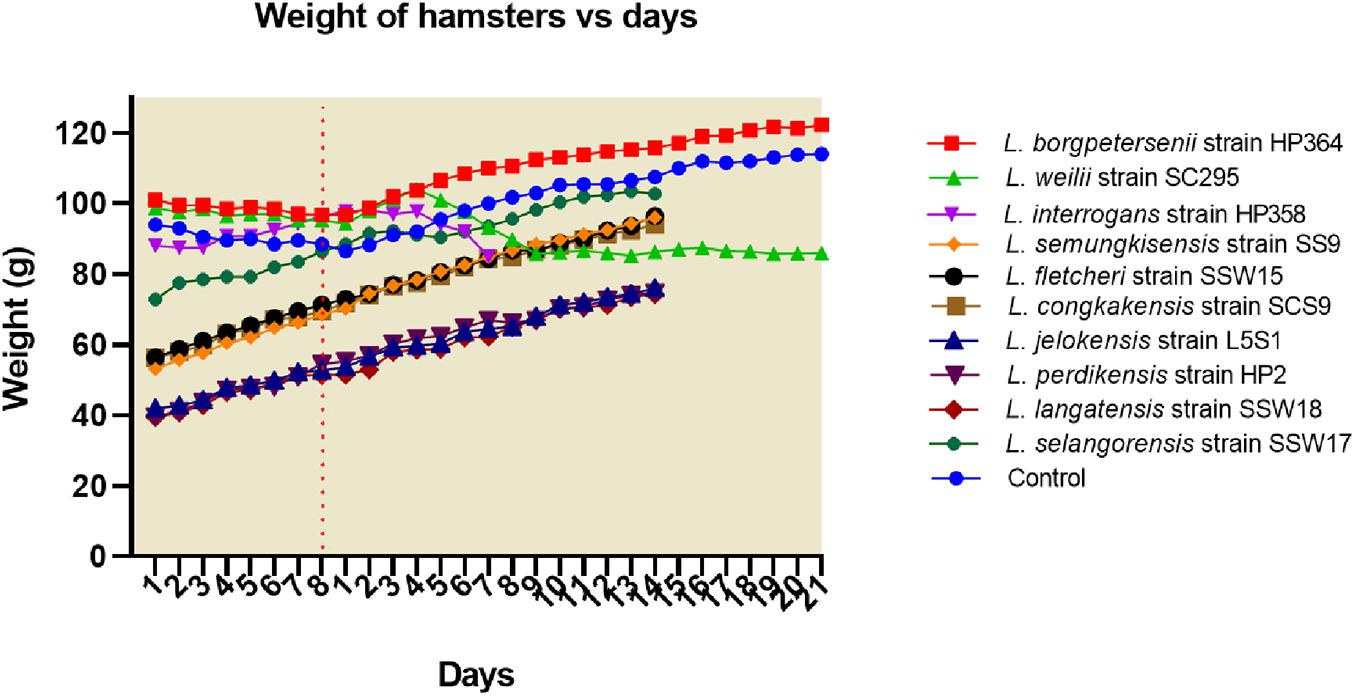
Figure 1. Weight of hamsters. Only hamsters infected with L. weilii strain SC295 and L. interrogans strain HP358 showed weight loss after infections. The dot line (day 8) represents the day in which the hamsters were infected with leptospires. Days 1–8 and 1–21 were the acclimatization and postinfection periods, respectively.
Macroscopic and Microscopic Observation of Lungs, Liver, and Kidneys Infected With Leptospira Species
Macroscopically, the lungs, liver, and kidneys of hamsters infected with environmental isolates and L. borgpetersenii strain HP364 showed normal morphology. Animals infected with L. weilii strain SC295 presented normal lungs and liver, whereas the kidneys appeared shrunken. L. interrogans strain HP358 induced changes in the lungs, liver, and kidneys of the infected hamsters. The changes included hemorrhage in the lungs, congestion in the liver, and paleness of kidneys (Figure 3).
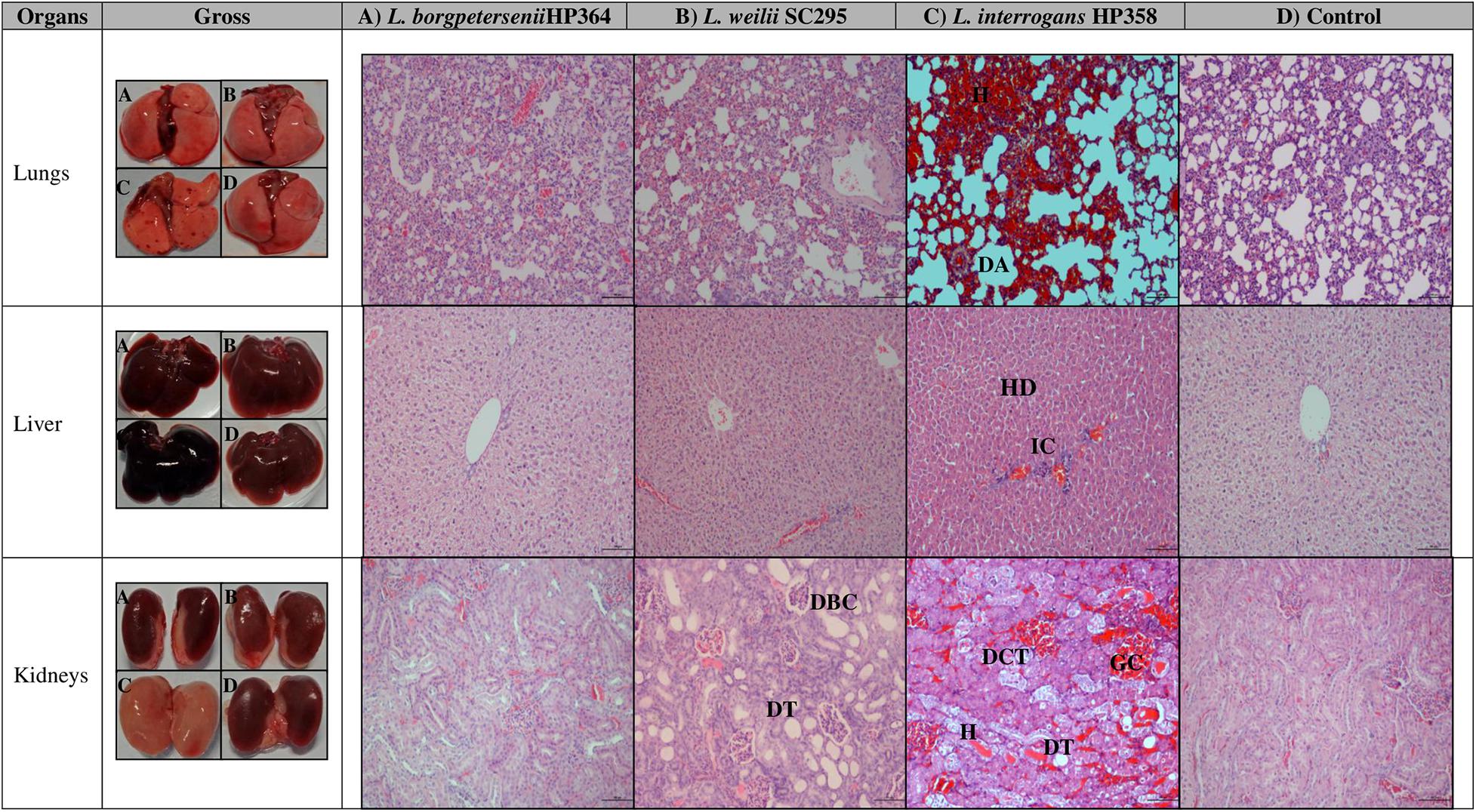
Figure 3. Gross and microscopic observation of organs in infected and control hamsters. H, hemorrhage; DA, dilated alveoli; HD, hepatic disarray; IC, infiltration on inflammatory cells; DBC, dilated Bowman capsule; DT, dilated tubule; DCT, disarrangement of cells in tubules; DT, dilated tubule; GC, glomerulus congestion. Magnification × 100, bar: 100 μm.
For microscopic analysis, organs (lungs, liver, and kidneys) from control and infected animals proceeded for H&E staining (Figure 3). Animals infected with L. borgpetersenii strain HP364 showed no pathological changes in lungs, liver, and kidneys. In L. weilii strain SC295–infected animals, lungs and liver appeared normal, but in kidneys, the tubules and Bowman capsule were dilated. All three organs in hamsters infected with L. interrogans strain HP358 showed severe damage. Lungs presented with congestion, hemorrhage, and mild dilation of alveoli, whereas in the liver, hepatic chord with ballooning of hepatocyte was observed. In kidneys, congestion and tubular hemorrhage, mild dilation of tubules, and derangement of epithelial cells were observed.
Recovery and Detection of Leptospires in Blood and Organs
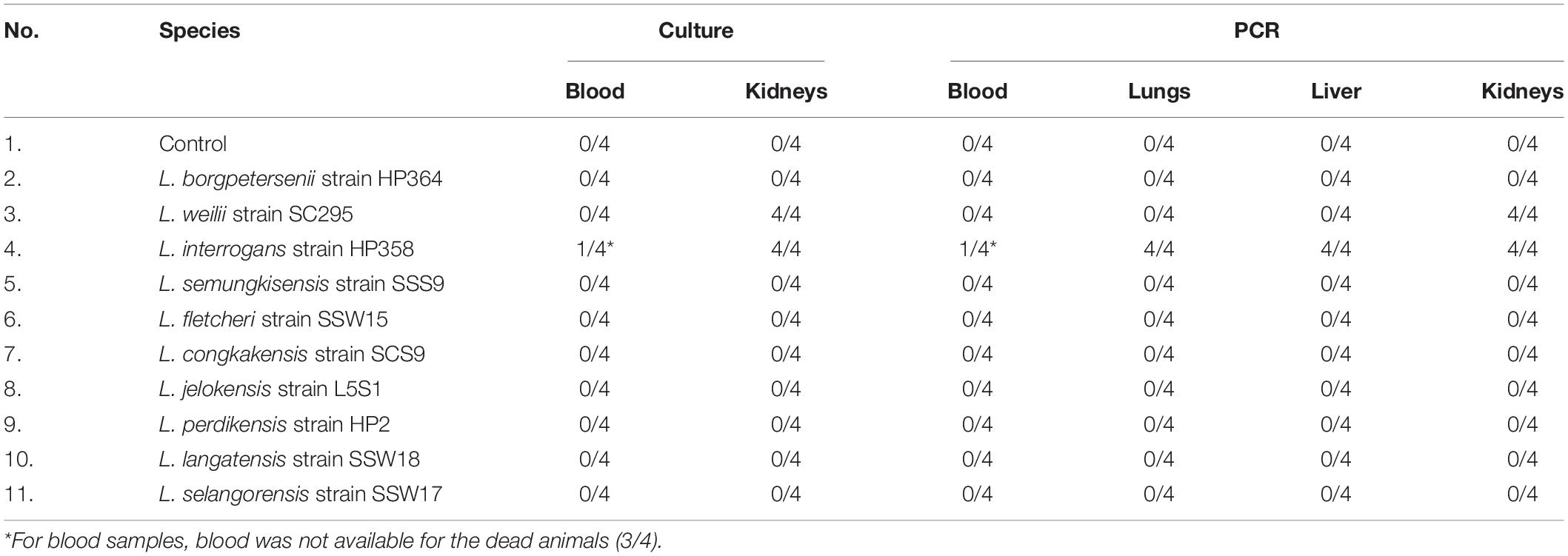
Leptospires were not detected in hamsters infected with environmental isolates and L. borgpetersenii strain HP364 (Table 3). For L. weilii strain SC295, leptospires were recovered and detected in kidneys and none from blood or other organs. In L. interrogans strain–infected animals, Leptospira was recovered and detected in blood, lungs, liver, and kidneys. The leptospiral load was high in all organs in hamsters infected with L. interrogans strain HP358 (Table 4).
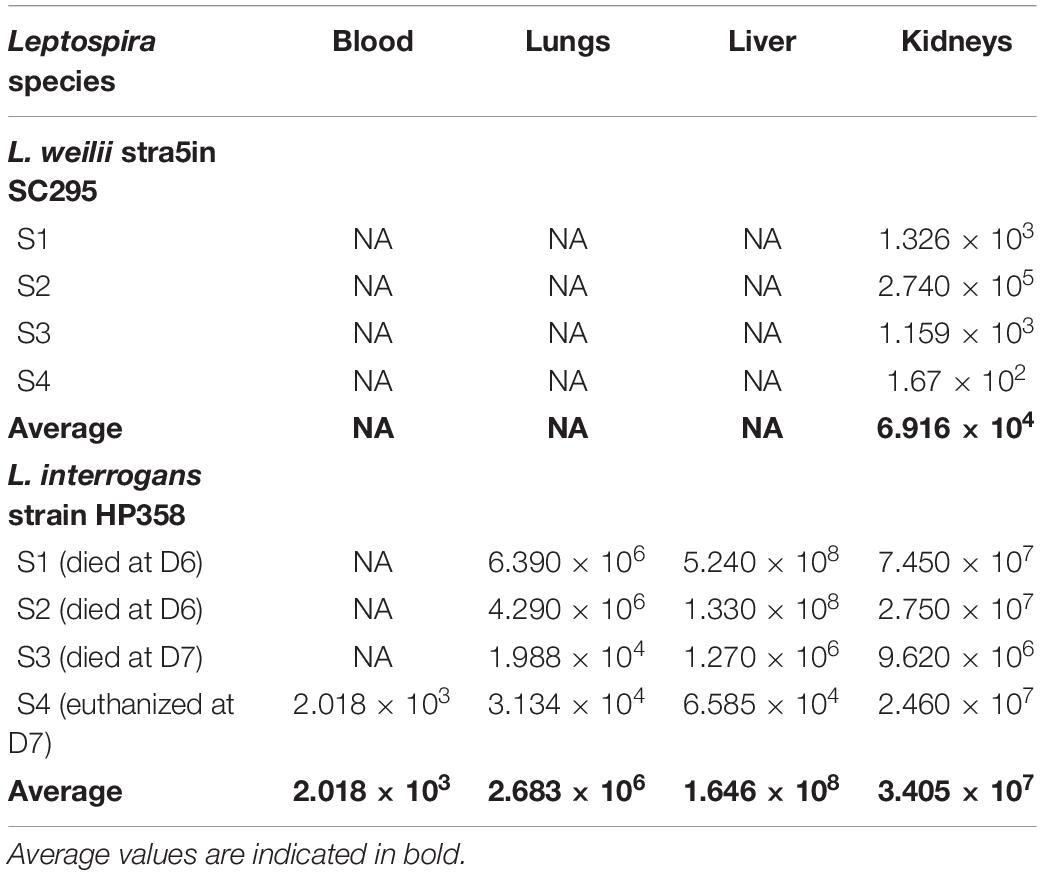
Table 4. The leptospiral load in organs infected with L. weilii strain SC295 and L. interrogans strain HP358.
General Genomic Descriptions
The whole-genome size of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 was determined to be 3,904,517; 4,111,826; and 4,808,724 bp with GC content (40.1, 40.2, and 35.0%) and predicted gene and protein-coding sequences (4,323; 4,691; and 4,733), respectively. The genetic variations based on SNP (Figure 4) confirm that L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 are pathogenic species and clustered among its respective species as established earlier (Azhari et al., 2018; Vincent et al., 2019). The low support value on the separation between the pathogenic and intermediate groups might be due to low samples size, and SNP is based on the genotype analysis. RAST categorized the functions of the coding sequences (CDSs) in strains HP364, SC295, and HP358 into 661 (16%), 648 (15%), and 661 (13%) subsystem coverage containing 946, 903, and 930 CDSs, respectively (Supplementary File 2). Most CDSs in strains HP364, SC295, and HP358 related to amino acids and derivatives (172/946, 161/903, 163/930), cofactors, vitamins, prosthetic groups, pigments (117/946, 99/903, 119/930), protein metabolism (121/946, 108/903, 100/930), motility and chemotaxis (72/946, 72/903, 73/930), and carbohydrates (76/946, 71/903, 64/930) (Figure 5).
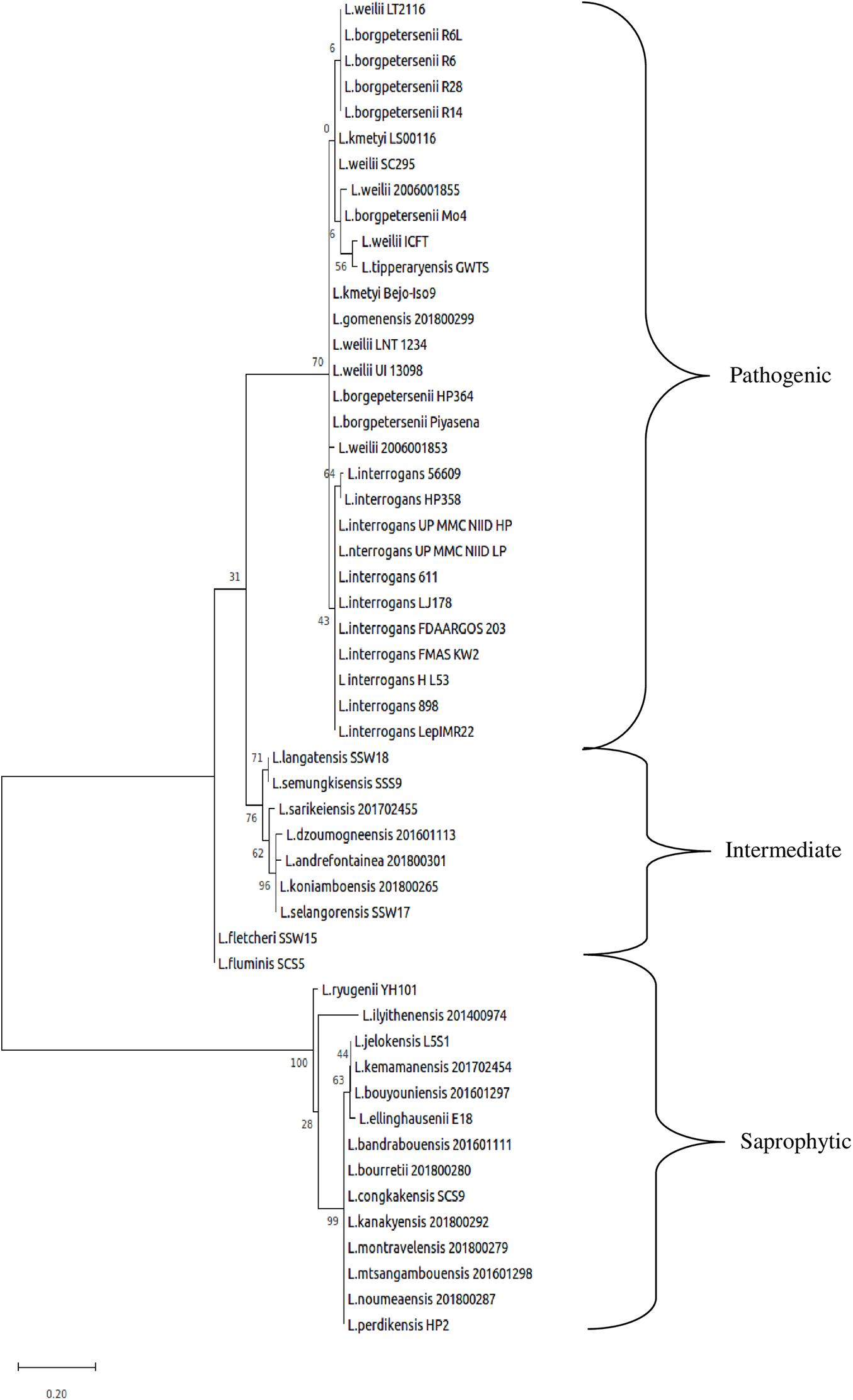
Figure 4. Whole-genome core single-nucleotide polymorphisms (core SNPs) phylogeny of Leptospira isolates based on the maximum likelihood method with 1,000 bootstrap replicates.
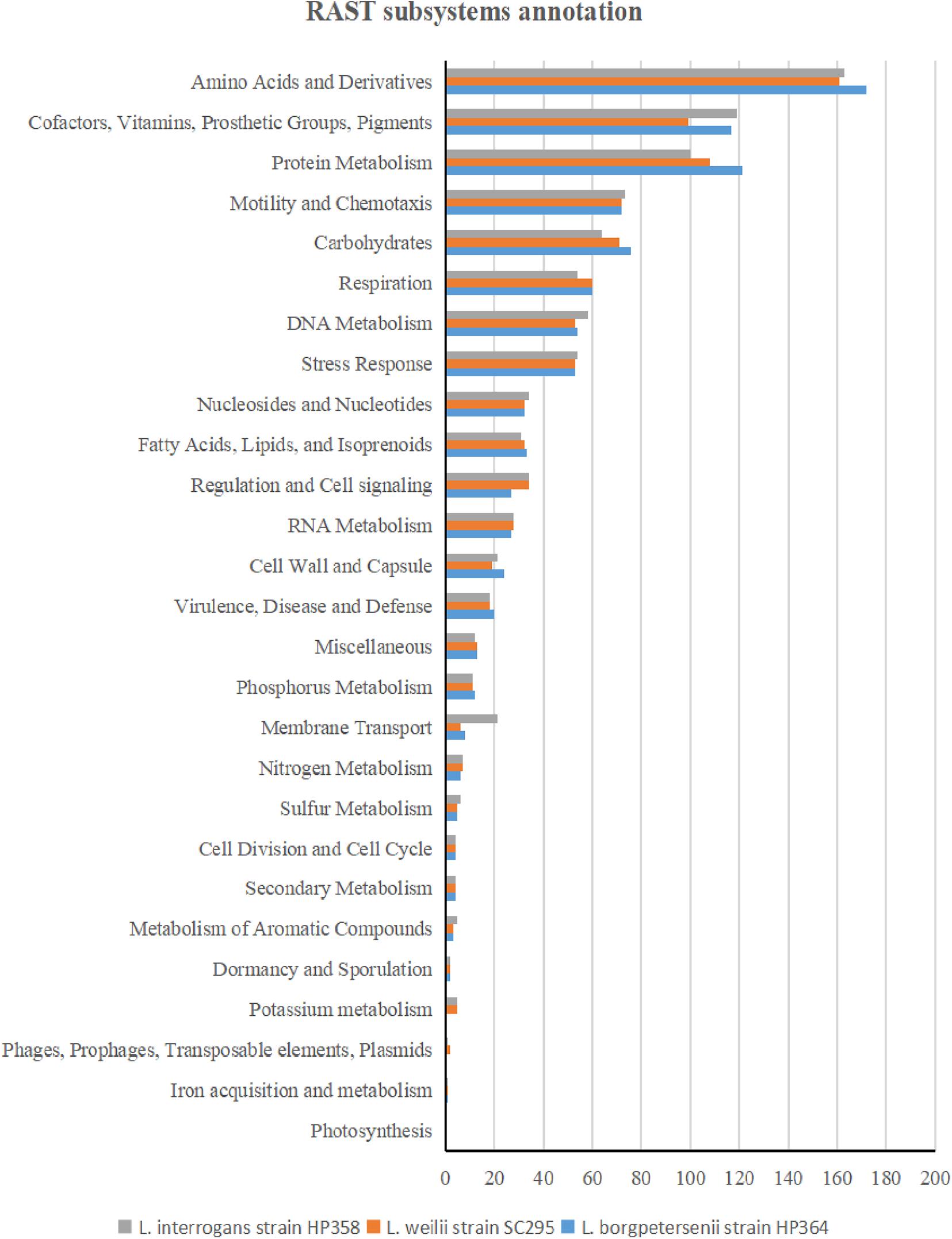
Figure 5. RAST subsystem annotation in L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358.
The range of pan-genome size for all the 30 Leptospira strains used in the pan-genome analysis is 3,352–4,658 (Supplementary File 3, Tables 1, 2). The phylogenetic tree showing the clustering of the strains is shown in Figure 6. The pan-genome analysis performed on three pathogenic Leptospira species presented genome sizes of 3,352; 3,581; and 3,868 for L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358. From a total 9,144 genes, 346 were present in the core genome with 95% sequence identity among the species (Figure 7). A list of gene presence and absence from the pan-genome analysis in these three pathogenic species is shown in Supplementary File 3, Table 3.
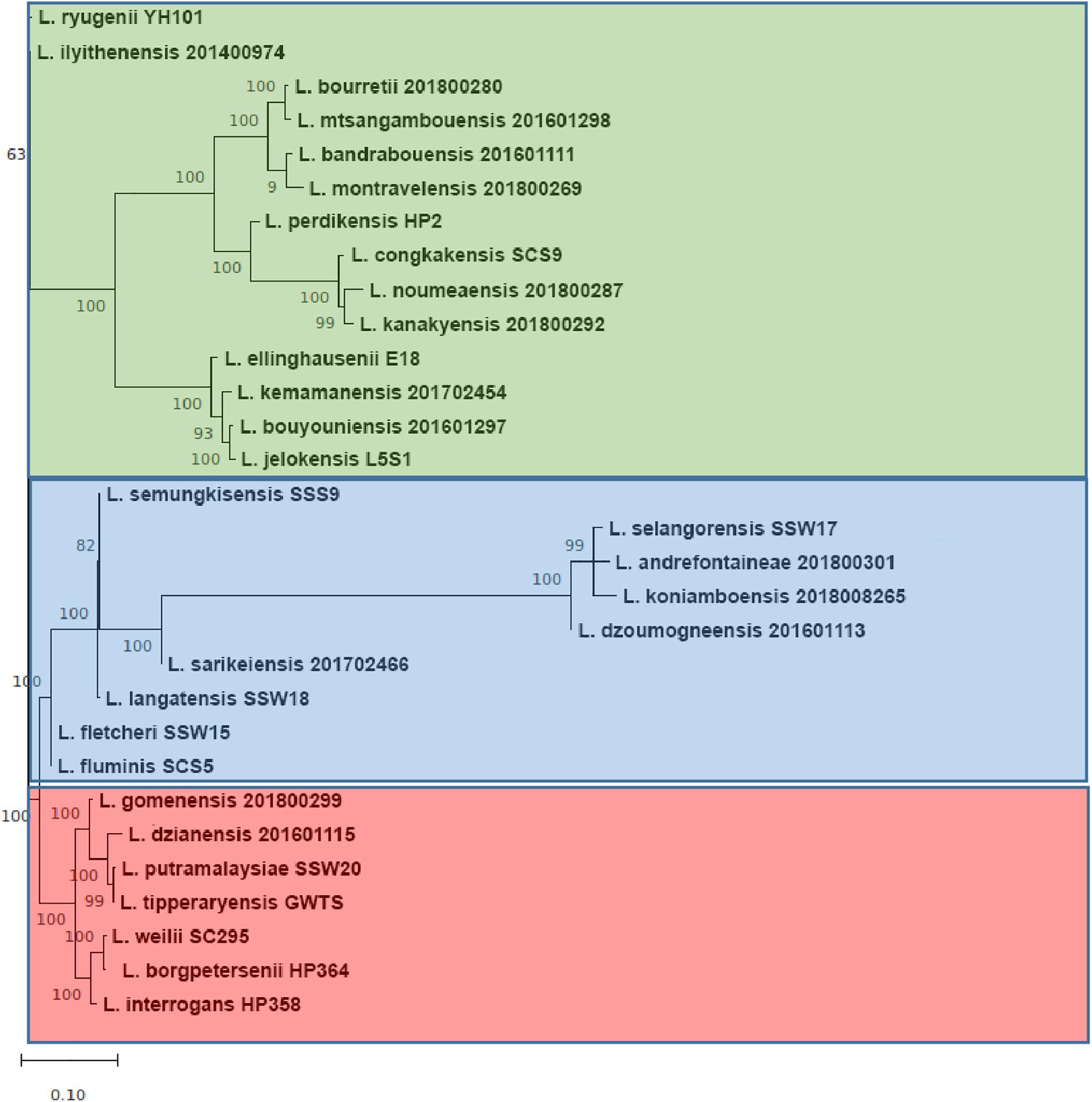
Figure 6. The core-genome phylogenetic tree. The group colored red, blue, and green corresponding to pathogenic, intermediate, and non-pathogenic clades, respectively. Each major branch of tree was supported with very high bootstrap 1,000.
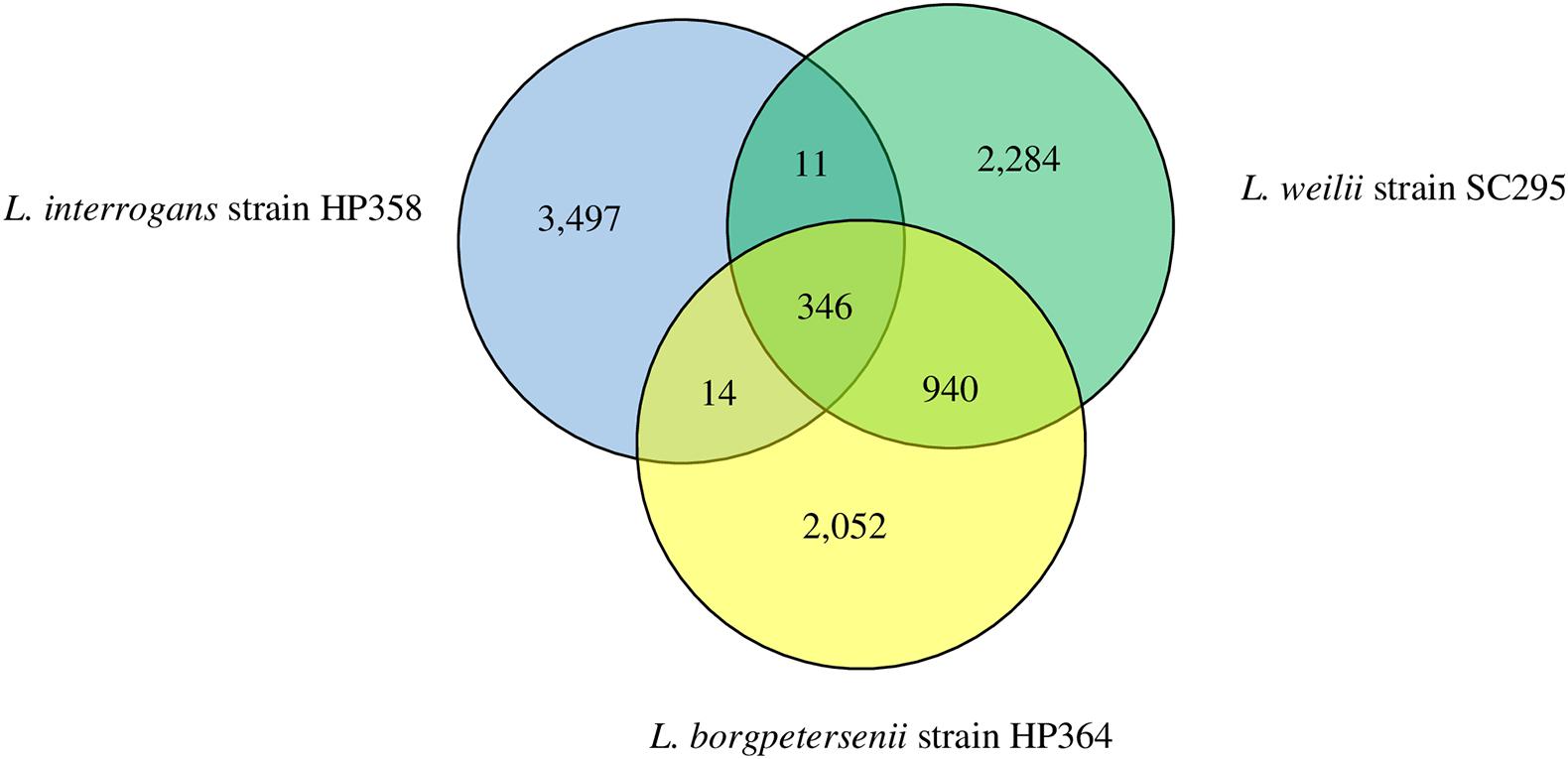
Figure 7. Pan-genome overview. The Venn diagram presents the core genome and the number of genes specific for L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358.
Presence of Known Virulence Factors
The basic genome characteristics of Leptospira species and strains used in known virulence analysis are summarized in Supplementary File 4. Among the 38 Leptospira-associated virulence factors, Loa22, FliN, ClpB, HtpG, LipL32, hemolysin B, and hemolysin C were found to be common among the pathogenic Leptospira species and strains with ≥ 90% identity (Supplementary File 5, Table 1). Two (leptospiral endostation-like protein D and thermolysin) and one (leptospiral endostation-like protein D) virulence factors in L. borgpetersenii strain HP364 and L. weilii strain SC295 were found to be less than 40% identity with the L. interrogans species and strains (Table 5). Gene Lsa21 was only detected among the L. interrogans strains. All virulence factors except for LigA-like protein and leptospiral endostatin-like protein F in L. interrogans strain HP358 revealed 97–100% similarity with the other L. interrogans strains. In intermediate and saprophytic Leptospira species, all virulence factors except hemolysin B have ≤ 80% identity and the alignment hit for vwa-I, vwa-II, Lsa21, and invA are not significant. Collagenase, putative lipoprotein (LB194), hypothetical protein (LA2786), LigA-like protein, LipL32, and sphingomyelinase C are not found in some of the intermediate and saprophytic species.
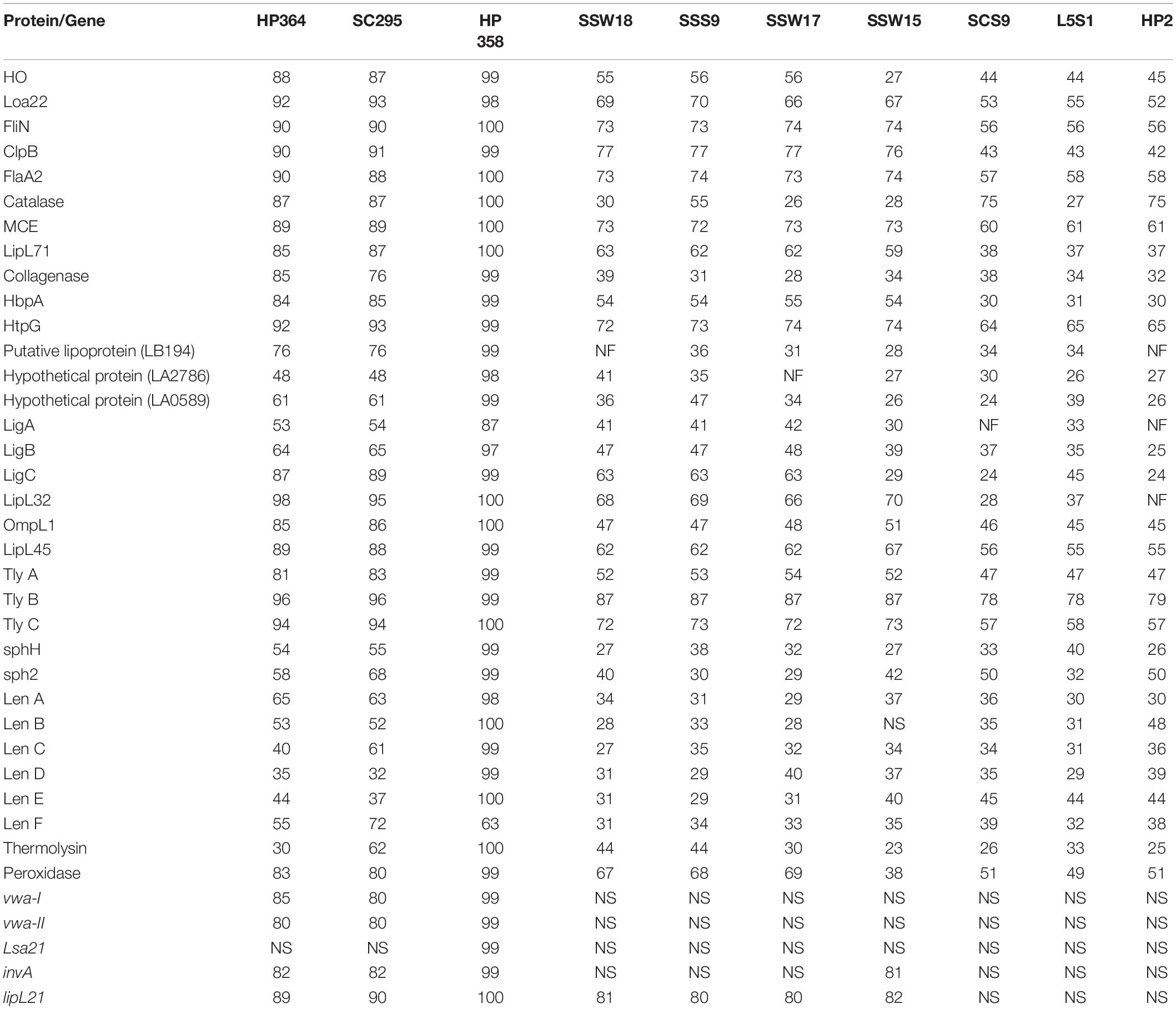
Table 5. The presence/absence and percentage of homology of the known virulence factors in each tested strain.
Potential Putative Virulence Factors
From VFDB database, the genomes of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 were shown to encode 109, 109, and 110 putative virulence factors, respectively (Supplementary File 5, Tables 2–4). Victors database analysis provided a higher number of predictive virulence factors, with 165, 166, and 161 in L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358, respectively (Supplementary File 5, Tables 5–7). Among all the predicted virulence factors, 65 (VFDB) and 128 (Victors) are common in the three Leptospira species (Supplementary File 5, Tables 8–9). The predicted virulence factors in VFDB databases are involved in adherence, metabolic adaptation, endotoxin, invasion, motility, stress protein, immune evasion, antiphagocytosis, secretion system, enzyme, regulation, amino acid, and purine metabolism, colonization, biofilm formation, phase variation, intracellular survival and entry, immunomodulator, structural mimicry, serum resistance, and toxin. The common predictive virulence factors shared between L. borgpetersenii strain HP364 and L. weilii strain SC295 (VFDB: 20, Victors: 18) are higher than in L. borgpetersenii strain HP364/L. interrogans strain HP358 (VFDB and Victors: 8) and L. weilii strain SC295/L. interrogans strain HP358 (VFDB: 8, Victors: 10) (Supplementary File 5, Tables 10–12). Both VFDB and Victors databases revealed the highest number of specific putative virulence factors in L. interrogans strain HP358 genome (VFDB: 29, Victors: 15) (Supplementary File 5, Table 13). In addition to the aforementioned roles/functions, one specific putative virulence factor, sphingomyelinase C (SMase) in L. interrogans HP358, is involved in exoenzyme or membrane damaging. In L. borgpetersenii strain HP364 and L. weilii strain SC295, the numbers of specific putative virulence factors are 16, 16 (VFDB) and 11, 10 (Victors) (Supplementary File 5, Tables 14, 15), respectively. From the pan-genome analysis, a list of genes that were characterized as virulence factors in other bacterial species was identified in L. interrogans HP358 genome. These genes included Lon protease, E3 ubiquitin–protein ligase sspH1, colicin I receptor (cirA), and gluP endotoxin (Rhomboid protease GluP) (Supplementary File 3, Table 3).
Discussion
The diversity of Leptospira species, serogroups, and serovars contributes to the wide variety of clinical presentations of leptospirosis. In this study, we investigated the in vivo (hamster model) virulence potential of Leptospira species isolated from the environments and rodents captured from the leptospirosis outbreak areas in Malaysia. None of the Leptospira species isolated from environments induced illness in hamsters, confirming the molecular characterization of these species as non-pathogens. The three pathogenic Leptospira species showed distinct virulence phenotype characteristics. Although L. borgpetersenii belongs to the pathogenic group of Leptospira, strain HP364 did not induce any clinical signs in hamsters. Reports from studies elsewhere elucidated mixed pathogenic potentials. L. borgpetersenii strain 2014TM FMNH228 isolated from bat (Triaenops menamena) in Madagascar did not induce any clinical signs in hamsters (Cordonin et al., 2019). Two strains of L. borgpetersenii serovar Hardjo, JB197 and 203 isolated from steers in the United States, were able to successfully infect hamsters but differed widely in clinical outcomes; the strain JB197 developed rapidly debilitating disease with a fatal outcome, whereas with the strain 203, although it established chronic renal infections, the hamsters remained asymptomatic (Miller et al., 1991; Zuerner et al., 2012). In other studies, L. borgpetersenii serovar Javanica strain K6 (isolated from rats in the Philippines) and Ballum strain 4E (isolated from Mus musculus in Brazil) induced clinical presentations typical of acute leptospirosis (Silva et al., 2010; Diniz et al., 2011; Villanueva et al., 2014). For hamsters infected with L. weilii strain SC295, animals developed mild symptoms, although damages in the kidney were observed. Similar findings were also reported from China, where L. weilii serovar Heyan (isolated from a patient) exhibited low virulence in guinea pig animal model (Xu et al., 2017).
L. interrogans strain HP358 induced severe illness in hamsters, causing fatality as early as day 6 p.i. This severe presentation was characterized as sudden death in some animals after the onset of clinical presentations. In previous studies performed in hamsters or guinea pig infected with different serovars/strains of L. interrogans, the strain 2013RR GLM983 isolated from Rattus rattus in Western Indian Ocean (Cordonin et al., 2019), serovar Manilae strain K64, Losbanos strain K37 and Ratnapura strain K5 isolated from rats in the Philippines (Villanueva et al., 2014), serovar Icterohaemorrhagiae strain Verdun (Reference Collection of the Institut Pasteur in Paris, France) (Matsui et al., 2011), serovar Icterohaemorrhagiae strain no 1143 (FIOCRUZ laboratory, Brazil) (Marinho et al., 2009), serovar Copenhageni strains RJ15958 and RJ1644 isolated from patients with severe pulmonary form of leptospirosis in Brazil (Nally et al., 2004), serovar Manilae strain UPMMC isolated from human patients with severe leptospirosis in the Philippines (Koizumi and Watanabe, 2003), and serotype Icterohaemorrhagiae strain NADL-H14 (van den Ingh and Hartman, 1986) developed acute leptospirosis and resulted in fatal outcomes. In general, hemorrhages associated with fatality were the main clinical presentations in the hosts infected with L. interrogans in most of the studies.
It was observed from this study that the presence of leptospires in organs contributes to the clinical consequences in host cells. As shown in Table 4, there were differences in the leptospiral colonization and number in animals infected with L. weilii strain SC295 and L. interrogans strain HP358. Leptospires were detected only in the kidney infected with L. weilii strain SC295, which was in accordance with the pathological changes observed in the kidney and none in the lungs and liver. In hamsters infected with L. interrogans strain HP358, leptospires were detected in all organs (lungs, liver, and kidneys), in accordance with the pathological changes in these organs. There were no leptospires detected in animals infected with L. borgpetersenii strain HP364, and thus, no pathological changes were observed in the lungs, liver, and kidneys. Hence, it can be concluded that besides the exacerbated cytokines responses (Cagliero et al., 2018; Senavirathna et al., 2020), the leptospiral dynamics also contribute to the clinical consequences in the host cells as reported in previous studies (Miyahara et al., 2014; Xu et al., 2020).
The differences in the virulence level among the species could be attributed to the wide variety of genes contained in the genome of each Leptospira isolates. The identification and characterization of virulence factors are important for understanding leptospiral pathogenesis, interaction with the host, and developing diagnostics, vaccines, and new drugs. Through RAST analysis, the genomes of the L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 were found to have approximately similar numbers of CDs and subsystems, which include amino acids and derivatives, cofactors, vitamins, prosthetic groups, pigments, protein metabolism, motility and chemotaxis, and carbohydrates. However, as subsystems coverage in RAST is only between 13 and 16% from the whole genomes and as revealed from the pan-genome analysis that these three species share only 346 core genes, there are many more genes that need to be explored and elucidated to understand the determinants of virulence characteristics. Indeed, from the known virulence factor analysis, the nucleotide sequence identity in the genome of L. interrogans strain HP358 differs from L. borgpetersenii strain HP364 and L. weilii strain SC295. Leptospiral endostation-like protein D and thermolysin, which are involved in adherence and invasion of Leptospira, showed less than 40% identity in L. borgpetersenii strain HP364 and L. weilii strain SC295. L. interrogans strain HP358 encodes the Lsa21 gene (absent in L. borgpetersenii strain HP364 and L. weilii strain SC295) and had been reported to induce strong production of proinflammatory cytokines via TLR2 and TLR4 signaling in mouse macrophage (Faisal et al., 2016), and this might also explain the high virulence capability of L. interrogans strain HP358. As expected, most of the known virulence factors showed less than 80% identity in the genomes of intermediate and saprophytic species. Overall, the absence or low similarity of virulence factors among the pathogenic Leptospira species may explain the difference in their capability to cause disease in infected hosts.
From the VFDB and Victors databases, approximately 109–110 and 161–166 genes in the genome of L. borgpetersenii strain HP364, L. weilii strain SC295, and L. interrogans strain HP358 were predicted to share orthologs with the virulence factors in other bacteria species. L. interrogans strain HP358 revealed a higher number of specific putative virulence factors compared to the other two Leptospira species. Besides VFDB and Victors analyses, the absence and presence of genes in pan-genome analysis also revealed that the genome of L. interrogans strain HP358 encodes several genes that are known to be virulence factors in other bacterial species such as Lon protease. The Lon mutants Salmonella, Campylobacter, and Pseudomonas aeruginosa showed less severe infection in various animal models (Takaya et al., 2003; Boddicker and Jones, 2004; Brazas et al., 2007; Cohn et al., 2007; Marr et al., 2007; Breidenstein et al., 2008). Another potential virulence factor is cirA, a gene that encodes the colicin I receptor. CirA mutant Salmonella enteritidis strain C50336 showed a sharp decrease in biofilm formation and impaired antibiotic resistance (Zhang et al., 2020). The genome of L. interrogans strain HP358 also contains sspH1 gene that encodes the E3 ubiquitin–protein ligase SspH1. This protein (also known as effector protein) has been reported to alter the host cell physiology and promote bacterial survival by interfering with the host’s ubiquitination pathway and target the host proteins for proteasomal degradation (Pisano et al., 2018). L. interrogans strain HP358 also encodes the gene gluP, which has been reported as an endotoxin in Helicobacter (Saeb et al., 2014). However, to understand the expression and function of these known virulence genes of other organisms in Leptospira, mutant studies, functional analysis, and expression studies are vital.
L. interrogans strain HP358 induced hemorrhage in hamster model, typical infection by most L. interrogans serovars/strain. Indeed, patients with severe leptospirosis reported in Malaysia had hemorrhagic manifestations. Leptospirosis with pulmonary hemorrhage was observed in patients and returned travelers from Malaysia (Wagenaar et al., 2004; Leung et al., 2011; Nor et al., 2016; Lim et al., 2018). Although the infecting Leptospira species or serovars were not known except for one case caused by serovar Lai: Langkawi (Wagenaar et al., 2004), it showed that the leptospires circulating in Malaysia have the capability to cause severe disease with hemorrhagic manifestations. The known virulence factors in the genome of L. interrogans strain HP358 also share a similar identity with the other two L. interrogans isolates in Malaysia (L. interrogans serovar Batavia strain LepIMR 22 and L. interrogans serovar Icterohaemorrhagiae strain 898). Therefore, it is imperative to perform a detailed study on L. interrogans strains isolated in Malaysia to understand the extent of damages to different organs and the inflammatory response to effectively understand the prognosis of the illness and efficient management.
Conclusion
In conclusion, Leptospira species plays an important role in determining the wide clinical presentation of leptospirosis. The new L. interrogans strain HP358 isolate showed a high level of virulence. This strain not only harbors the known virulence genes of Leptospira but also encodes genes known to be virulence factors in other bacteria. Further studies are recommended to confirm the role of these potential virulence factors in the virulence of this strain and in other Leptospira species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC), Universiti Putra Malaysia with Animal Use Protocol (AUP) number: UPM/IACUC/AUP-R044/2018.
Author Contributions
NP performed the study, interpreted the data, and drafted the manuscript. JJ performed the bioinformatic analysis and wrote that part of the manuscript. NA collected the Leptospira isolates. ZS helped with study design. VKN designed and supervised the study and reviewed and finalized the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the Ministry of Education, Malaysia through the Fundamental Research Grant Scheme FRGS/1/2019/WAB13/UPM/02/2 with project code 07-01-19-2134FR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Mohd Hafidz Mohd Izhar and Prof. Dato Dr. Tengku Azmi Tengku Ibrahim for their guidance in monitoring the animals and in all the procedures performed in this study. We also thank the staff in Animal Research Facility—Low level infection at Faculty of Veterinary, UPM for their help in providing the materials needed for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.753328/full#supplementary-material
Footnotes
- ^ https://www.blaststation.com/intl/en/blaststation2.php
- ^ http://www.phidias.us/victors/download.php
References
Adler, B. (2015). Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology. Heidelberg: Springer.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Atzingen, M. V., Barbosa, A. S., De Brito, T., Vasconcellos, S. A., de Morais, Z. M., Lima, D. M., et al. (2008). Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 8:70. doi: 10.1186/1471-2180-8-70
Azhari, N. N., Ramli, S. N. A., Joseph, N., Philip, N., Mustapha, N. F., Ishak, S. N., et al. (2018). Molecular characterization of pathogenic Leptospira sp. in small mammals captured from the human leptospirosis suspected areas of Selangor state, Malaysia. Acta Trop. 188, 68–77. doi: 10.1016/j.actatropica.2018.08.020
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Boddicker, J. D., and Jones, B. D. (2004). Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 72, 2002–2013. doi: 10.1128/IAI.72.4.2002-2013.2004
Brazas, M. D., Breidenstein, E. B. M., Overhage, J., and Hancock, R. E. W. (2007). Role of lon, an ATP-dependent protease homolog, in resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 51, 4276–4283. doi: 10.1128/AAC.00830-07
Breidenstein, E. B. M., Khaira, B. K., Wiegand, I., Overhage, J., and Hancock, R. E. W. (2008). Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52, 4486–4491. doi: 10.1128/AAC.00222-08
Bulach, D. M., Kalambaheti, T., de la Pena-Moctezuma, A., and Adler, B. (2000). Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2, 375–380.
Bushnell, B., Rood, J., and Singer, E. (2017). BBMerge-Accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/journal.pone.0185056
Cagliero, J., Villanueva, S. Y. A. M., and Matsui, M. (2018). Leptospirosis pathophysiology: into the storm of cytokines. Front. Cell. Infect. Microbiol. 8:204. doi: 10.3389/fcimb.2018.00204
Casanovas-Massana, A., Hamond, C., Santos, L. A., de Oliveira, D., Hacker, K. P., Balassiano, I., et al. (2020). Leptospira yasudae sp. nov. and Leptospira stimsonii sp. nov., two new species of the pathogenic group isolated from environmental sources. Int. J. Syst. Evol. Microbiol. 70, 1450–1456. doi: 10.1099/ijsem.0.003480
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Choy, H. A., Kelley, M. M., Chen, T. L., Moller, A. K., Matsunaga, J., and Haake, D. A. (2007). Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75, 2441–2450. doi: 10.1128/IAI.01635-06
Cohn, M. T., Ingmer, H., Mulholland, F., Jørgensen, K., Wells, J. M., and Brøndsted, L. (2007). Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 73, 7803–7813. doi: 10.1128/AEM.00698-07
Cordonin, C., Turpin, M., Bascands, J. L., Dellagi, K., Mavingui, P., Tortosa, P., et al. (2019). Three Leptospira strains from Western Indian Ocean wildlife show highly distinct virulence phenotypes through hamster experimental infection. Front. Microbiol. 10:382. doi: 10.3389/fmicb.2019.00382
Diniz, J. A., Félix, S. R., Bonel-Raposo, J., Neto, A. C. P. S., Vasconcellos, F. A., Grassmann, A. A., et al. (2011). Highly virulent Leptospira borgpetersenii strain characterized in the hamster model. Am. J. Trop. Med. Hyg. 85, 271–274. doi: 10.4269/ajtmh.2011.11-0013
Domingos, R. F., Fernandes, L. G., Romero, E. C., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2015). Novel Leptospira interrogans protein Lsa32 is expressed during infection and binds laminin and plasminogen. Microbiology 161, 851–864. doi: 10.1099/mic.0.000041
Faisal, S. M., Varma, V. P., Subathra, M., Azam, S., Sunkara, A. K., Akif, M., et al. (2016). Leptospira surface adhesin (Lsa21) induces Toll like receptor 2 and 4 mediated inflammatory responses in macrophages. Sci. Rep. 6:39530. doi: 10.1038/srep39530
Fang, J. Q., Imran, M., Hu, W. L., Ojcius, D. M., Li, Y., Ge, Y. M., et al. (2018). vWA proteins of Leptospira interrogans induce hemorrhage in leptospirosis by competitive inhibition of vWF/GPIb-mediated platelet aggregation. EBioMedicine 37, 428–441. doi: 10.1016/j.ebiom.2018.10.033
Fouts, D. E., Matthias, M. A., Adhikarla, H., Adler, B., Amorim-Santos, L., Berg, D. E., et al. (2016). What makes a bacterial species pathogenic?: comparative genomic analysis of the genus Leptospira. PLoS Negl. Trop. Dis. 10:e0004403. doi: 10.1371/journal.pntd.0004403
Fraser, T., and Brown, P. D. (2017). Temperature and oxidative stress as triggers for virulence gene expression in pathogenic Leptospira spp. Front. Microbiol. 8:783. doi: 10.3389/fmicb.2017.00783
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Gomes-Solecki, M., Santecchia, I., and Werts, C. (2017). Animal models of leptospirosis: of mice and hamsters. Front. Immunol. 8:58. doi: 10.3389/fimmu.2017.00058
Hauk, P., Macedo, F., Romero, E. C., Vasconcellos, S. A., de Morais, Z. M., Barbosa, A. S., et al. (2008). In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect. Immun. 76, 2642–2650. doi: 10.1128/IAI.01639-07
Hsu, S. H., Chang, M. Y., Lin, S. M., Ko, Y. C., Chou, L. F., Tian, Y. C., et al. (2021). Peptidoglycan mediates Leptospira outer membrane protein Loa22 to toll-like receptor 2 for inflammatory interaction: a novel innate immune recognition. Sci. Rep. 11:1064. doi: 10.1038/s41598-020-79662-8
Isogai, E., Isogai, H., Kurebayashi, Y., and Ito, N. (1986). Biological activities of leptospiral lipopolysaccharide. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261, 53–64. doi: 10.1016/s0176-6724(86)80062-1
Isogai, E., Kitagawa, H., Isogai, H., Matsuzawa, T., Shimizu, T., Yanagihara, K., et al. (1989). Effects of leptospiral lipopolysaccharide on rabbit platelets. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 271, 186–196. doi: 10.1016/s0934-8840(89)80072-6
Kassegne, K., Hu, W. L., Ojcius, D. M., Sun, D., Ge, Y. M., Zhao, J. F., et al. (2014). Identification of collagenase as a critical virulence factor for invasiveness and transmission of pathogenic Leptospira species. J. Infect. Dis. 209, 1105–1115. doi: 10.1093/infdis/jit659
Koizumi, N., and Watanabe, H. (2003). Identification of a novel antigen of pathogenic Leptospira spp. that reacted with convalescent mice sera. J. Med. Microbiol. 52, 585–589. doi: 10.1099/jmm.0.05148-0
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, S. H., Kim, S., Park, S. C., and Kim, M. J. (2002). Cytotoxic activities of Leptospira interrogans hemolysin SphH as a pore-forming protein on mammalian cells. Infect. Immun. 70, 315–322. doi: 10.1128/IAI.70.1.315-322.2002
Leung, V., Luong, M. L., and Libman, M. (2011). Leptospirosis: pulmonary hemorrhage in a returned traveller. CMAJ 183, E423–E427. doi: 10.1503/cmaj.092203
Levett, P. N. (2001). Leptospirosis. Clin. Microbiol. 14, 296–326. doi: 10.1128/CMR.14.2.296-326.2001
Liao, S. M., Sun, A. H., Ojcius, D. M., Wu, S. L., Zhao, J. F., and Yan, J. (2009). Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9:253. doi: 10.1186/1471-2180-9-253
Lim, T. S. C., Jun, T. Y., and Fah, T. R. (2018). Pulmonary hemorrhage associated with severe leptospirosis – the role of low dose intravenous methylprednisolone. Mal. J. Med. Health Sci. 14, 69–72.
Marinho, M., Oliveira-Júnior, I. S., Monteiro, C. M., Perri, S. H., and Salomão, R. (2009). Pulmonary disease in hamsters infected with Leptospira interrogans: histopathologic findings and cytokine mRNA expressions. Am. J. Trop. Med. Hyg. 80, 832–836.
Marr, A. K., Overhage, J., Bains, M., and Hancock, R. E. W. (2007). The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153(Pt 2), 474–482. doi: 10.1099/mic.0.2006/002519-0
Masuzawa, T., Sakakibara, K., Saito, M., Hidaka, Y., Villanueva, S. Y. A. M., Yanagihara, Y., et al. (2018). Characterization of Leptospira species isolated from soil collected in Japan. Microbiol. Immunol. 62, 55–59. doi: 10.1111/1348-0421.12551
Matsui, M., Rouleau, V., Bruyere-Ostells, L., and Goarant, C. (2011). Gene expression profiles of immune mediators and histopathological findings in animal models of leptospirosis: comparison between susceptible hamsters and resistant mice. Infect. Immun. 79, 4480–4492. doi: 10.1128/IAI.05727-11
Miller, D. A., Wilson, M. A., and Beran, G. W. (1991). Survey to estimate prevalence of Leptospira interrogans infection in mature cattle in the United States. Am. J. Vet. Res. 52, 1761–1765.
Miyahara, S., Saito, M., Kanemaru, T., Villanueva, S. Y. A. M., Gloriani, N. G., and Yoshida, S. (2014). Destruction of the hepatocyte junction by intercellular invasion of Leptospira causes jaundice in a hamster model of Weil’s disease. Int. J. Exp. Path. 95, 271–281. doi: 10.1111/iep.12085
Murray, G. L. (2015). The molecular basis of leptospiral pathogenesis. Curr. Top. Microbiol. Immunol. 387, 139–186. doi: 10.1007/978-3-662-45059-8_7
Nally, J. E., Chantranuwat, C., Wu, X. Y., Fishbein, M. C., Pereira, M. M., Da Silva, J. J., et al. (2004). Alveolar septal deposition of immunoglobulin and complement parallels pulmonary hemorrhage in a guinea pig model of severe pulmonary leptospirosis. Am. J. Pathol. 164, 1115–1127. doi: 10.1016/S0002-9440(10)63198-7
Nascimento, A. L., Ko, A. I., Martins, E. A., Monteiro-Vitorello, C. B., Ho, P. L., Haake, D. A., et al. (2004). Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186, 2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004
Nor, M. B. M., Ralib, A. M., Ibrahim, N. A., and Abdul-Ghani, M. R. (2016). High frequency oscillatory ventilation in leptospirosis pulmonary hemorrhage syndrome: a case series study. Indian J. Crit. Care Med. 20, 342–348. doi: 10.4103/0972-5229.183906
Oliveira, T. R., Longhi, M. T., Goncales, A. P., de Morais, Z. M., Vasconcellos, S. A., and Nascimento, A. L. (2010). LipL53, a temperature regulated protein from Leptospira interrogans that binds to extracellular matrix molecules. Microb. Infect. 12, 207–217. doi: 10.1016/j.micinf.2009.12.004
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Philip, N., Bahtiar Affendy, N., Ramli, S. N. A., Arif, M., Raja, P., Nagandran, E., et al. (2020). Leptospira interrogans and Leptospira kirschneri are the dominant Leptospira species causing human leptospirosis in Central Malaysia. PLoS Negl. Trop. Dis. 14:e0008197. doi: 10.1371/journal.pntd.0008197
Picardeau, M. (2017). Virulence of the zoonotic agent of leptospirosis: still terra incognita? Nat. Rev. Microbiol. 15, 297–307. doi: 10.1038/nrmicro.2017.5
Pisano, A., Albano, F., Vecchio, E., Renna, M., Scala, G., Quinto, I., et al. (2018). Revisiting bacterial ubiquitin ligase effectors: weapons for host exploration. Int. J. Mol. Sci. 19:3576. doi: 10.3390/ijms19113576
Putz, E. J., and Nally, J. E. (2020). Investigating the immunological and biological equilibrium of reservoir hosts and pathogenic Leptospira: balancing the solution to an acute problem? Front. Microbiol. 11:2005. doi: 10.3389/fmicb.2020.02005
Ren, S. X., Fu, G., Jiang, X. G., Zeng, R., Miao, Y. G., Xu, H., et al. (2003). Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422, 888–893. doi: 10.1038/nature01597
Ristow, P., Bourhy, P., da Cruz McBride, F. W., Figueira, C. P., Huerre, M., Ave, P., et al. (2007). The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3:e97. doi: 10.1371/journal.ppat.0030097
Rost, B. (1999). Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94. doi: 10.1093/protein/12.2.85
Saeb, A. T. M., David, S. K., and Al-Brahim, H. (2014). In silico detection of virulence gene homologues in the human pathogen Sphingomonas Spp. Evol. Bioinform. Online 10, 229–238. doi: 10.4137/EBO.S20710
Sayers, S., Li, L., Ong, E., Deng, S., Fu, G., Lin, Y., et al. (2019). Victors: a web based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 47, D693–D700. doi: 10.1093/nar/gky999
Senavirathna, I., Rathish, D., and Agampodi, S. (2020). Cytokine response in human leptospirosis with different clinical outcomes: a systematic review. BMC Infect. Dis. 20:268. doi: 10.1186/s12879-020-04986-9
Silva, ÉF., Felix, S. R., Cerqueira, G. M., Fagundes, M. Q., Neto, A. C. P. S., Grassmann, A. A., et al. (2010). Preliminary characterization of Mus musculus-derived pathogenic strains of Leptospira borgpetersenii serogroup Ballum in a hamster model. Am. J. Trop. Med. Hyg. 83, 336–337. doi: 10.4269/ajtmh.2010.10-0120
Sitto, F., and Battistuzzi, F. U. (2020). Estimating pangenomes with roary. Mol. Biol. Evol. 37, 933–939. doi: 10.1093/molbev/msz284
Stevenson, B., Choy, H. A., Pinne, M., Rotondi, M. L., Miller, M. C., Demoll, E., et al. (2007). Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2:e1188. doi: 10.1371/journal.pone.0001188
Stoddard, R. A., Gee, J. E., Wilkins, P. P., McCaustland, K., and Hoffmaster, A. R. (2009). Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the lipL32 gene. Diagn. Microbiol. Infect. Dis. 64, 247–255. doi: 10.1016/j.diagmicrobio.2009.03.014
Takaya, A., Suzuki, M., Matsui, H., Tomoyasu, T., Sashinami, H., Nakane, A., et al. (2003). Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect. Immun. 71, 690–696. doi: 10.1128/IAI.71.2.690-696.2003
Thibeaux, R., Iraola, G., Ferres, I., Bierque, E., Girault, D., Soupe-Gilbert, M. E., et al. (2018). Deciphering the unepxlored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb. Genom. 4:e000144.
van den Ingh, T. S., and Hartman, E. G. (1986). Pathology of acute Leptospira interrogans serotype icterohaemorrhagiae infection in the Syrian hamster. Vet. Microbiol. 12, 367–376. doi: 10.1016/0378-1135(86)90086-6
Vieira, M. L., de Morais, Z. M., Goncales, A. P., Romero, E. C., Vasconcellos, S. A., and Nascimento, A. L. (2010). Lsa63, a newly identified surface protein of Leptospira interrogans binds laminin and collagen IV. J. Infect. 60, 52–64. doi: 10.1016/j.jinf.2009.10.047
Villanueva, S. Y., Saito, M., Tsutsumi, Y., Segawa, T., Baterna, R. A., Chakraborty, A., et al. (2014). High virulence in hamsters of four dominant Leptospira serovars isolated from rats in the Philippines. Microbiology 160, 418–428. doi: 10.1099/mic.0.072439-0
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, E., Thibeaux, R., et al. (2019). Revisiting the taxanomy and evolution of pethogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 13:e0007270. doi: 10.1371/journal.pntd.0007270
Vinetz, J. M. (2001). Leptospirosis. Curr. Opin. Infect. Dis. 14, 527–538. doi: 10.1097/00001432-200110000-00005
Vinh, T. U., Shi, M. H., Alder, B., and Faine, S. (1989). Characterization and taxonomic significance of lipopolysaccharide of Leptospira interrogans serovar hardjo. J. Gen. Microbiol. 135, 2663–2673. doi: 10.1099/00221287-135-10-2663
Wagenaar, J. F. P., de Vries, P. J., and Hartskeerl, R. A. (2004). Leptospirosis with pulmonary hemorrhage, caused by a new strain of serovar Lai: Langkawi. J. Travel Med. 11, 379–382. doi: 10.2310/7060.2004.19208
Wang, H., Wu, Y. F., Ojcius, D. M., Yang, X. F., Zhang, C., Ding, S., et al. (2012). Leptospiral hemolysins induce proinflammatory cytokines through Toll-like receptor 2- and 4-mediated JNK and NF-kB signaling pathways. PLoS One 7:e42266. doi: 10.1371/journal.pone.0042266
Xu, J., Koizumi, N., and Nakamura, S. (2020). Crawling motility on the host tissue surfaces is associated with the pathogenicity of the zoonotic spirochete Leptospira. Front. Microbiol. 11:1886. doi: 10.3389/fmicb.2020.01886
Xu, Y., Zheng, H., Zhang, Y., Wang, Y., Zhang, J., Li, Z., et al. (2017). Genomic analysis of a new serovar of Leptospira weilii serogroup Manhao. Front. Microbiol. 8:149. doi: 10.3389/fmicb.2017.00149
Zhang, L., Zhang, C. L., Ojcius, D. M., Sun, D., Zhao, J. F., Lin, X. A., et al. (2012). The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Mol. Microbiol. 83, 1006–1023. doi: 10.1111/j.1365-2958.2012.07985.x
Zhang, Z., Du, W., Wang, M., Li, Y., Su, S., Wu, T., et al. (2020). Contribution of the colicin receptor CirA to biofilm formation, antibiotic resistance, and pathogenicity of Salmonella Enteriditis. J. Basic Microbiol. 60, 72–81. doi: 10.1002/jobm.201900418
Keywords: Leptospira, virulence, in vivo, in silico, hamster, genome
Citation: Philip N, Jani J, Azhari NN, Sekawi Z and Neela VK (2021) In vivo and in silico Virulence Analysis of Leptospira Species Isolated From Environments and Rodents in Leptospirosis Outbreak Areas in Malaysia. Front. Microbiol. 12:753328. doi: 10.3389/fmicb.2021.753328
Received: 04 August 2021; Accepted: 08 October 2021;
Published: 05 November 2021.
Edited by:
Xiaohui Zhou, Fudan University, ChinaReviewed by:
Mônica Larucci Vieira, Federal University of Minas Gerais, BrazilShuichi Nakamura, Tohoku University, Japan
Lingbing Zeng, Shanghai Jiao Tong University School of Medicine, China
Eneas Carvalho, Butantan Institute, Brazil
Copyright © 2021 Philip, Jani, Azhari, Sekawi and Neela. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vasantha Kumari Neela, dmFzYW50aGFAdXBtLmVkdS5teQ==
 Noraini Philip1
Noraini Philip1 Jaeyres Jani
Jaeyres Jani Vasantha Kumari Neela
Vasantha Kumari Neela