- College of Resources and Environment, Southwest University, Chongqing, China
Anaerobic digestion is used to treat diverse waste classes, and polycyclic aromatic hydrocarbons (PAHs) are a class of refractory compounds that common in wastes treated using anaerobic digestion. In this study, a microbial consortium with the ability to degrade phenanthrene under methanogenesis was enriched from paddy soil to investigate the cometabolic effect of glucose on methane (CH4) production and phenanthrene (a representative PAH) degradation under methanogenic conditions. The addition of glucose enhanced the CH4 production rate (from 0.37 to 2.25mg⋅L−1⋅d−1) but had no influence on the degradation rate of phenanthrene. Moreover, glucose addition significantly decreased the microbial α-diversity (from 2.59 to 1.30) of the enriched consortium but showed no significant effect on the microbial community (R2=0.39, p=0.10), archaeal community (R2=0.48, p=0.10), or functional profile (R2=0.48, p=0.10). The relative abundance of genes involved in the degradation of aromatic compounds showed a decreasing tendency with the addition of glucose, whereas that of genes related to CH4 synthesis was not affected. Additionally, the abundance of genes related to the acetate pathway was the highest among the four types of CH4 synthesis pathways detected in the enriched consortium, which averagely accounted for 48.24% of the total CH4 synthesis pathway, indicating that the acetate pathway is dominant in this phenanthrene-degrading system during methanogenesis. Our results reveal that achieving an ideal effect is diffcult via co-metabolism in a single-stage digestion system of PAH under methanogenesis; thus, other anaerobic systems with higher PAH removal efficiency should be combined with methanogenic digestion, assembling a multistage pattern to enhance the PAH removal rate and CH4 production in anaerobic digestion.
Introduction
Anaerobic digestion is a promising strategy for the disposal of diverse categories of waste materials, such as domestic wastewater, agricultural waste, food industry waste, and municipal solid waste, because it has the advantages of low energy consumption, less investment in land and management, nutrient reclamation, and biogas generation (Gallert and Winter, 1997; He et al., 2005; Stabnikova et al., 2008; Chan et al., 2009; Holm-Nielsen et al., 2009; Lytras et al., 2021). Methane (CH4) is the principal component of biogas produced by anaerobic digestion and can be used as clean, renewable bioenergy that can substitute fossil fuels (Holm-Nielsen et al., 2009; Haberl et al., 2012; Awe et al., 2017; Li et al., 2019). Additionally, many kinds of organic hazards, such as polycyclic aromatic hydrocarbons (PAHs), BTEX, chloramphenicol, polyacrylamide, and antibiotics, can be effectively removed from the initial substrates and converted into CH4 via anaerobic digestion (Kunapuli et al., 2007; Agarry and Owabor, 2011; Hu et al., 2018; Guo et al., 2019; Bianco et al., 2020). Therefore, how to efficiently harness the anaerobic digestion process for the improvement of CH4 production and removal the rate of organic pollutants has long been a global challenge.
Co-metabolism is a traditional approach adopted to enhance the degradation of refractory pollutants and CH4 production in the anaerobic digestion system, which can vigorously promote the growth and activity of indigenous microbes by supplying some popular carbon sources (e.g., glucose, fructose, and sucrose) for microbes to the target system (Li et al., 2016a,b; Xie et al., 2016; Khan et al., 2017; Hadibarata et al., 2018; Feng et al., 2019). Glucose is one of the most favorable carbon (energy) sources for heterotrophic microbes and has been frequently used as a co-metabolic substrate to improve CH4 production and accelerate the degradation of refractory pollutants in diverse environments (Ambrosoli et al., 2005; Jing et al., 2017; Khan et al., 2017; Shu et al., 2021; Zhou et al., 2021). For example, the addition of glucose was found to significantly enhance the degradation of fluorene, phenanthrene, and pyrene under an anaerobic denitrification system (Ambrosoli et al., 2005), and the addition of glucose effectively alleviated the inhibitory effect of gallic acid on CH4 production in an anaerobic digestion system (Mousa and Forster, 1999). Therefore, it seems that glucose can be used as a prioritized metabolic substrate to efficiently remove refractory pollutants in anaerobic digestion systems (Li et al., 2016b; Khan et al., 2017).
Polycyclic aromatic hydrocarbons are a group of persistent organic pollutants with intensive toxicity and wide distribution, posing a serious threat to environmental and human health (Xue and Warshawsky, 2005; Couling et al., 2010; Zhou et al., 2018). Previous investigations have reported that co-metabolism triggered by glucose could significantly accelerate the degradation of PAHs under both aerobic and anaerobic digestion environments (Ambrosoli et al., 2005; Ma et al., 2011; Qin et al., 2018; Tripathi et al., 2020; Al Farraj et al., 2021). For example, the addition of glucose was reported to significantly enhance the degradation of PAHs (benzopyrene, anthracene, and pyrene) when treated with some pure bacterial species, such as Cellulosimicrobium, Sphingomonas, and Bacillus (Khanna et al., 2012; Qin et al., 2018; Al Farraj et al., 2021). Additionally, PAH degradation under methanogenesis is thermodynamically feasible and the degradation of PAHs can be coupled with methanogenesis, suggesting that remediation of PAHs pollution via the anaerobic digestion system should be a potentially ideal method marked by the properties of energy conservation and resource reuse (Chang et al., 2005; Dolfing et al., 2009; Berdugo-Clavijo et al., 2012; Wan et al., 2012; Nzila, 2018; Ye et al., 2019; Wang et al., 2021b). Whether methanogens can directly participate in the degradation of PAHs remains unclear, but some potential keystone microbes in this process have been identified. In addition to methanogenic archaea Methanosaeta, Methanoculleus, and Methanosarcina, some syntrophic bacteria, such as Methylibium, Legionella, Clostridiaceae, Citrobacter, and Pseudomonas, have also been found essential for PAH degradation and CH4 generation (Berdugo-Clavijo et al., 2012; Zhang et al., 2012). Thus, in anaerobic digestion, methanogenesis is the pivotal step that not only can harness biogas (CH4) production but also can be coupled with PAH degradation. However, the co-metabolic effect induced by glucose on PAH degradation and CH4 production under methanogenesis has seldom been reported.
In this study, we enriched the phenanthrene-degrading microbial consortium under methanogenic conditions from a long-term rice paddy, which was used to investigate the co-metabolic effect of glucose on CH4 production and phenanthrene degradation during methanogenesis. The response of the microbial community and functional composition to the bio-stimulation of glucose in the enriched microbial consortium was further analyzed based on the results of metagenomic sequencing. This study aimed to examine whether the co-metabolic substrate (glucose) could enhance CH4 production and phenanthrene degradation under the methanogenesis system and explore the community and functional responses of the phenanthrene-degrading microbial consortium to the bio-stimulation of glucose, providing useful information for the enhancement of CH4 production and PAHs elimination during the anaerobic digestion of waste containing PAHs.
Materials and Methods
Enrichment of Methanogenesis Consortium of Phenanthrene Degradation
In this experiment, the methanogenesis consortium was enriched from a paddy soil sample collected from a long-term (approximately 30years) rice paddy field (N29°48', E106°48') in the Beibei District (Chongqing, China) in July 2020. A brief enrichment procedure is shown in Figure 1 and described as follows.
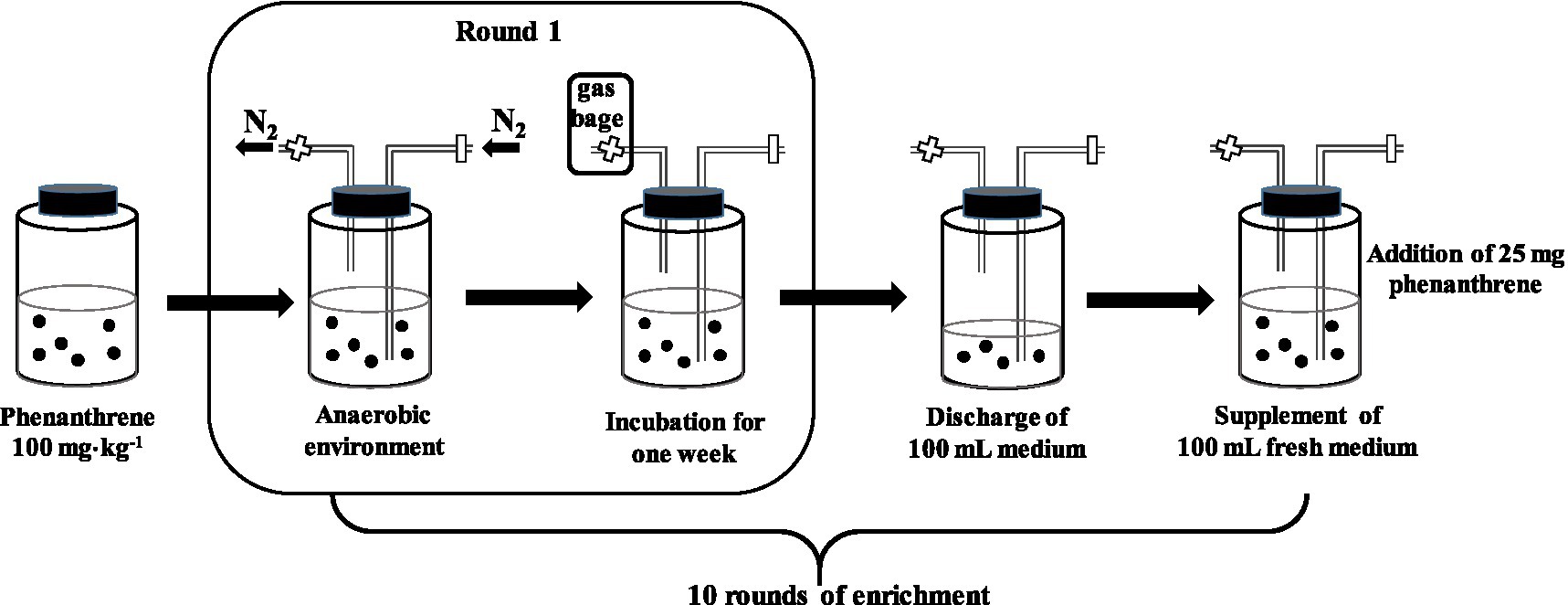
Figure 1. The diagrammatic sketch showing the enrichment procedure of the phenanthrene-degrading consortium under methanogenesis.
Four types of nutrition solutions, selective media for methanogens (solution A), inorganic salt solution (solution B), trace element solution (solution C), and vitamin solution (solution D) were prepared for the enrichment of the target microbial consortium, the ingredients of which are listed in Supplementary Table 1 (Ferguson and Mah, 1983; Brauer et al., 2006). First, 200g of fresh paddy and 200ml of sterilized and distilled water were added into a borosilicate glass bottle (1L) pre-added with 50mg phenanthrene, and then 100ml of mixed solution containing A, B, C, and D (A:B:C:D=20:1:0.2:0.2) was added into the bottle with a final phenanthrene concentration of 100mg⋅kg−1; the bottle was sealed using a butl rubber stopper that was pierced through by two glass tubes, one of which was inserted into the liquid and the other was above the liquid surface (Figure 1). The pure nitrogen gas (N2) was filled into the bottle via the gas inlet for 10min to expel the air via the outlet (Figure 1); after the inlet was closed and a gas collection bag was connected to the outlet, the bottle was shaken (100r⋅min−1) and incubated in the dark at 30°C for 1week for the first round of enrichment. Subsequently, 100ml of supernatant in the bottle was discharged and replaced by 100ml of fresh mixed solution (containing 25mg phenanthrene), and then the second round of enrichment was conducted via the same procedure as that for the first round. Using this method, six bottles of the target microbial consortium were obtained, each of which had undergone 10 rounds of enrichment (10weeks).
Experiment Design to Examine the Effect of Glucose on Methane Production and Phenanthrene Degradation
The experimental design is illustrated in Figure 2. The six bottles of consortium obtained were equally and randomly divided into two groups, denoted as PME (without glucose) and PMEG (with glucose), respectively. In a serum bottle (100ml) with 0.25mg phenanthrene, 20ml suspension from one bottle of the enriched consortium, 30ml mixed medium, and 6ml sterilized and distilled water was added for the PME treatment. For the PMEG treatment, sterilized and distilled water was substituted with glucose solution (0.1mol⋅L−1). After sealing with butyl rubber and an aluminum cap, the bottle was filled with N2 for 10min to generate an anaerobic environment, and then shaken (100 r⋅min−1) and incubated in the dark at 30°C for 21days. The CH4 concentration in the bottle was measured on days 3, 7, 14, and 21, and the phenanthrene concentration was detected on days 1 and 21. After incubation, genomic DNA was extracted from each microbial consortium for metagenomic sequencing.
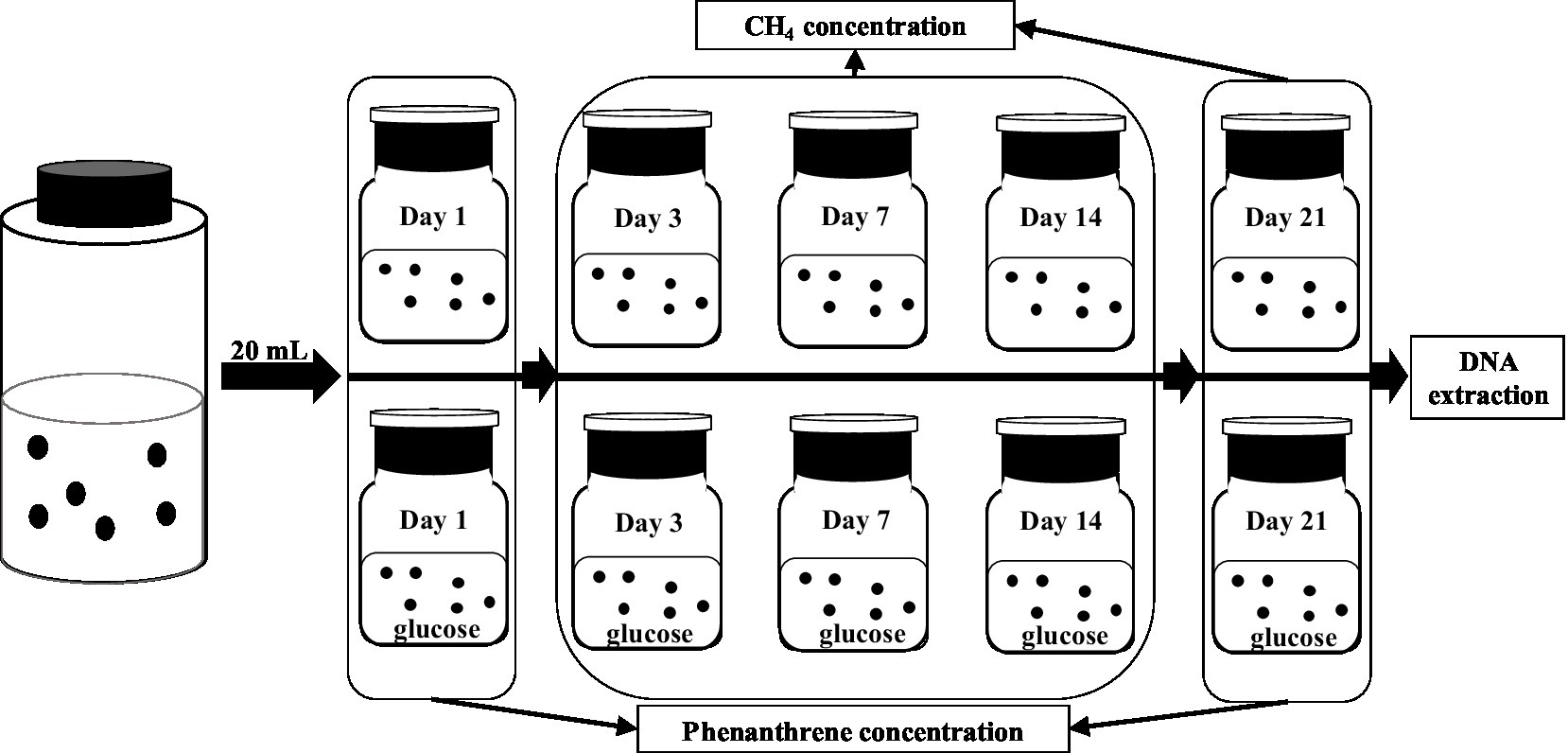
Figure 2. The diagram showing the experiment design to investigate the cometabolic effect of glucoe on the MPR and PDR of the enriched consortium.
Determinations of Methane Concentration and Phenanthrene Content
On days 3, 7, 14, and 21 of the incubation period, the CH4 concentration in the headspace of each serum bottle was sampled and then detected using a gas chromatograph (Agilent Technologies 7890A, United States) equipped with a flame ionization detector (FID) under the following conditions – carrier gas, N2; flow rate, 30cm3⋅min−1; oven temperature, 50°C; FID temperature, 200°C (standardized by Wang and Wang, 2003) – and the methane production rate (MPR, mg CH4⋅L−1⋅d−1) was calculated according to the method previously described by Zhang et al. (2011).
The phenanthrene concentration of each culture sample in the serum bottle was detected on days 1 and 21 during the incubation, and the brief detection procedure was as follows. The culture in each serum bottle was shaken well and then transferred into a separating funnel (250ml) with 1.5g NaCl. Subsequently, the bottle was rinsed with 50ml of n-hexane, also transferred into the funnel. After shaking for 5min, the funnel was statically placed to stratify the organic and aqueous (containing soil particles) phases. Next, the organic phase was retained, while the residual phase was re-extracted using n-hexane. After mixing and then clarifying using anhydrous sodium sulfate, the extracted organic phase was concentrated using a rotary evaporator. According to the method described by Zhou et al. (2017b), the solvent (n-hexane) was replaced with methanol, and then the phenanthrene content was detected using a gradient elution method (mobile phase: methanol and water; flow rate: 0.6ml⋅min−1; column temperatuer: 30°C; detection wavelength: 250nm) by an HPLC (Hitachi High-Technologies Corporation, Japan). Finally, the phenanthrene degradation rate (PDR, %) was determined using the following equation:
where C1 and C21 are the phenanthrene content (mg⋅L−1) of the culture in the serum bottle on days 1 and 21, respectively.
DNA Extraction and Metagenomic Sequencing of the PME and PMEG Samples
Total genomic DNA was extracted from 0.5ml culture in each serum bottle (thoroughly shaken for a sufficient homogeneity) using MoBio UltraClean™ soil DNA isolation kits (San Diego, CA, United States) according to the manufacturer’s instructions, and the integrity and purity of DNA were checked with 1% agarose gel electrophoresis and NanoDrop200 spectrophotometer (Thermo, United States), respectively. The extracted DNA was sent to Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) for metagenomic sequencing, and the brief procedure is described as follows.
The extracted DNA was fragmented to an average length of approximately 400bp using Covaris M220 (Gene Company Limited, China) for the construction of paired-end (PE) library, and then NEXTflex™ Rapid DNA-Seq (Bioo Scientific, United States) was used to construct the PE library. After the ligation of specific DNA adapters with the blunt end of fragments, PE sequencing was performed on the Illumina NovaSeq platform (Illumina Inc., San Diego, CA, United States) using NovaSeq Reagent Kits according to the manufacturer’s standard procedure. Using fastp (version 0.20.0)1 on the free online cloud platform of Majorbio,2 the adaptor sequences and low-quality reads (length below 50bp, minimum quality lower than 20, and with N bases) in the raw reads of metagenomic sequencing were removed and trimmed. These pair-end and single-end reads with high quality were then assembled into contigs using MEGAHIF (version 1.1.2)3 based on succinct de Bruijn graphs. These assembled contigs being or longer than 300bp were preserved as the final assembly result. Open reading frames (ORFs) in these available contigs were identified using MetaGene (Noguchi et al., 2006), and the identified ORFs equal to or longer than 100bp were retrieved and translated into amino acid sequences using the NCBI translation table. The predicted contigs were clustered using CD-HIT (version 4.6.1)4 with 90% sequence identity and 90% coverage, and the longest gene sequence in each gene cluster was isolated as the representative for the construction of a non-redundant gene set. The high-quality reads in each sample were mapped to the non-redundant gene set under 95% identity using SOAPaligner (version 2.21),5 and the gene abundance in the sample was calculated. The functional annotation of the representative sequences in the non-redundant gene set was performed using Diamond (version 0.8.35)6 against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (version 94.2)7 with an e-value cutoff of 1e−5.
Data Analysis
The independent t-test in the SPSS 11.5 (SPSS, Chicago, United States) was used to check the difference of the parameters (MPR, PDR, and relative abundance of functional genes) between the PME and PMEG treatments, and p values below 0.05 were considered as significant. In R software with the Vegan package, the principal coordinate analysis (PCoA) and Adonis test (checking the difference in significance of the composition in the PME and PMEG treatments) were performed, and the Mental test was conducted with the command of “envit” to calculate the correlations between the composition (microbial and functional) and MPR or PDR. Variance partition analysis (VPART) was applied to quantify the explained variance of MPR and PDR for the microbial or functional composition, conducted using the Vegan package with the command of “rdavp.” All figures were generated using the Ggplot2 package in the R software.
Results
Effect of Glucose on MPR and PDR of the Phenanthrene-Degrading Consortium Under Methanogenesis
The MPR and PDR in the no-glucose (PME) and glucose-added (PMEG) treatments are shown in Figure 3. The MPR of the PME (0.37mg⋅L−1⋅d−1) treatment was significantly lower (p<0.05) than that of the PMEG (2.25mg⋅L−1⋅d−1) treatment (Figure 3A). Although the value of PDR of PME (48.69%) was lower than that of PMEG (62.10%), no significant difference was detected in the PDR between these two treatments (Figure 3B). This result indicated that the addition of glucose promoted the activity of methanogens in the enriched microbial consortium but did not affect the degradation of phenanthrene.
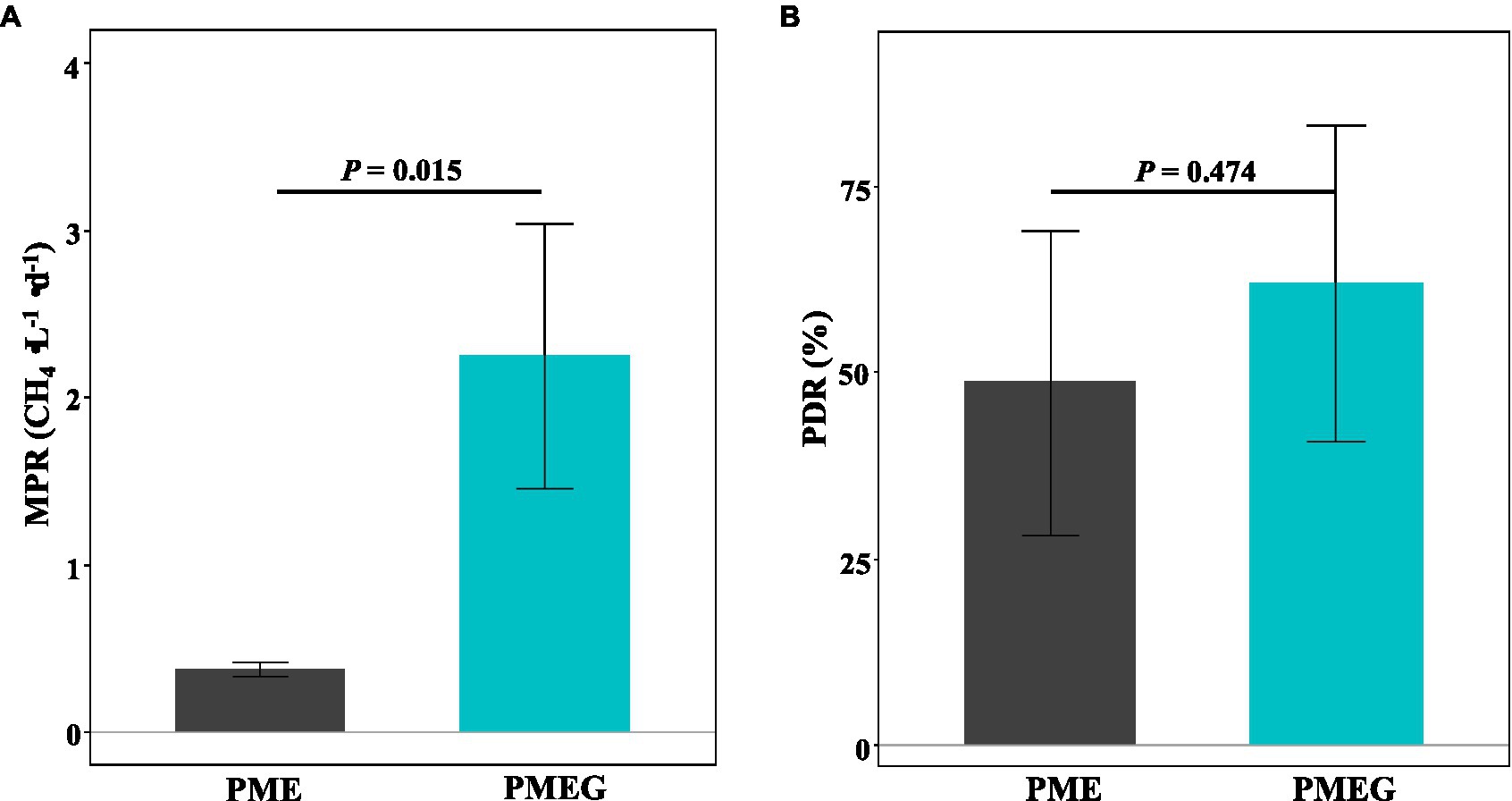
Figure 3. The MPR (A) and PDR (B) in the enriched consortium treatments of without (PME) and with (PMEG) additon of glucose. Error bars indicate SDs (n=3). The p values above the horizontal lines are the results of independent t-test.
Whole Microbial Community and Archaeal Community in the PME and PMEG Treatments
The result of microbial α-diversity (Supplementary Figure 1) indicates that the PME treatment had a significantly higher (p<0.05) α-diversity than the PMEG treatment. The PCoA profile (Figure 4A) constructed from the microbial species composition based on the metagenomic sequencing indicated that the points of the PME and PMEG treatments were in a narrow area (axis1, −0.27–0.43; axis2, −0.22–0.23). Moreover, the Adonis test revealed no significant difference (R2=0.39, p=0.10) between the microbial community structures of the PME and PMEG treatments (Figure 4A). However, the Mantel test indicated that MPR (r2=0.91, p=0.025) and PDR (r2=0.92, p=0.028) were significantly correlated with the microbial community structure in the enriched consortium (Table 1), which could explain 49 and 20% of the variance in the community structure, respectively (Figure 4B). The heatmap generated from the archaeal family composition in each treatment is shown in Figure 5 and indicates that all treatments had a similar composition of archaeal family with Methanoregulaceae and Methanosarcinaceae as the dominant classified families. Furthermore, our results showed that the ratio of archaea to total microbes in the PME was lower than that in the PMEG, but no significant difference was detected between them (Figure 4C). Additionally, no significant difference was detected in the archaeal community structure (based on the level of species) between the PME and PMEG treatments (Figure 4D), and only MPR had a significant relationship (r2=0.91, p=0.034) with the archaeal community structure of the enriched consortia (Table 1).
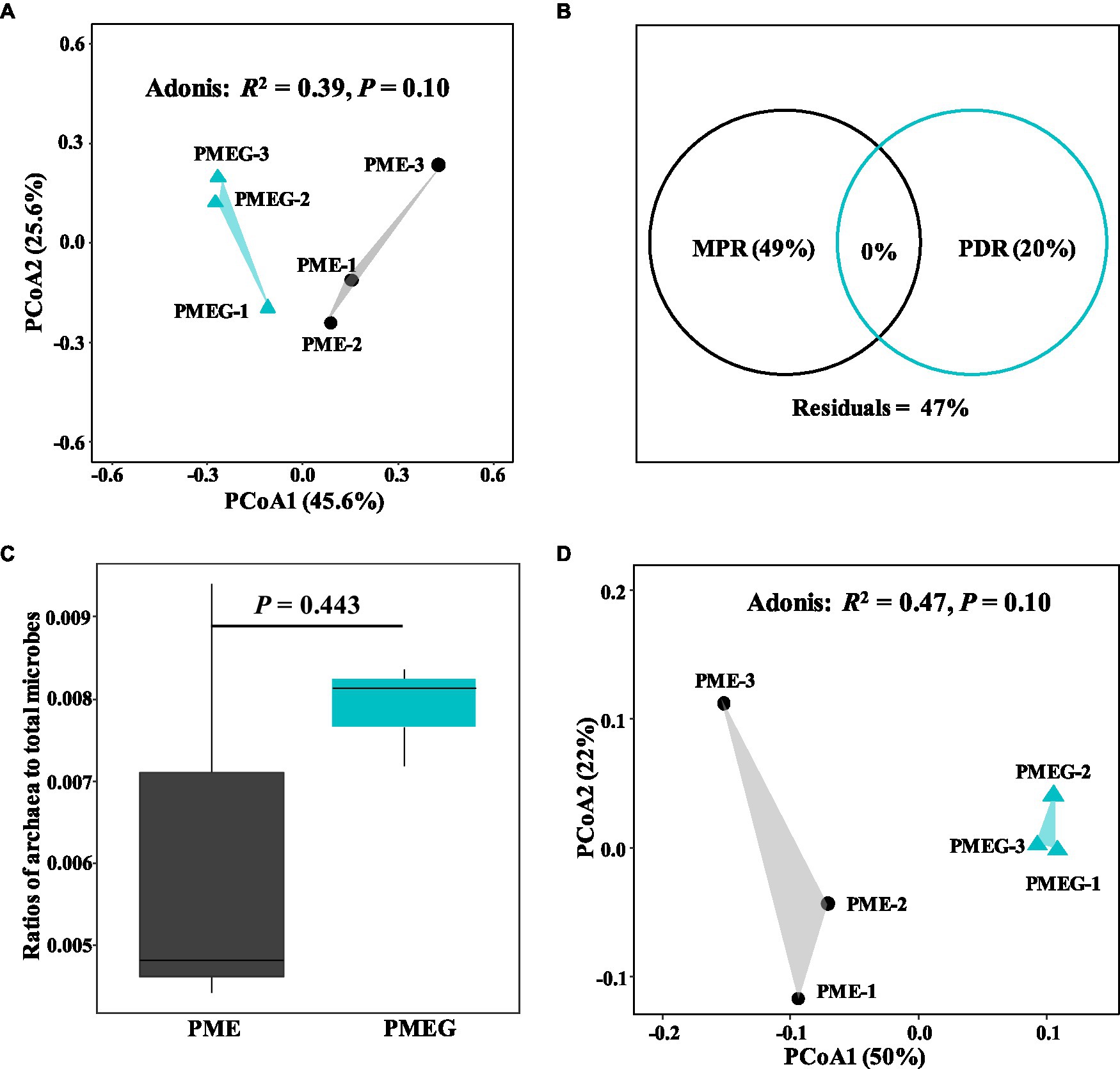
Figure 4. The principal coordinate analysis (PCoA) profile based on the microbial species composition in the PME and PMEG treatments (A) and variance partitioning analysis showing the portions explained by MPR and PDR for the microbial community structure (B); the ratios of archaea to total microbes in the PME and PMEG treatments (C) and the PCoA profile based on the archaeal species composition in the PME and PMEG treatments (D).
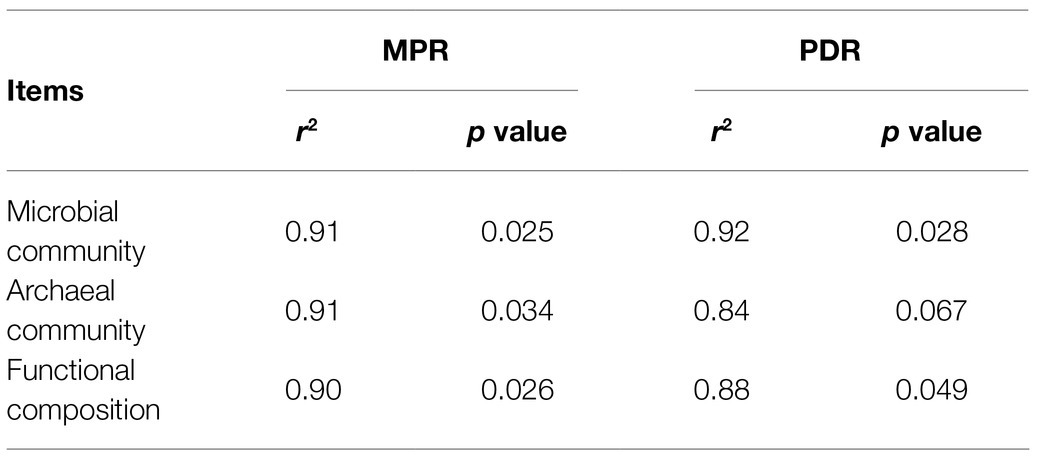
Table 1. Mantel correlations among microbial community, archaeal community, functioanl composition, methane production rate (MPR), and phenanthrene degradation rate (PDR) in the enriched consortium.
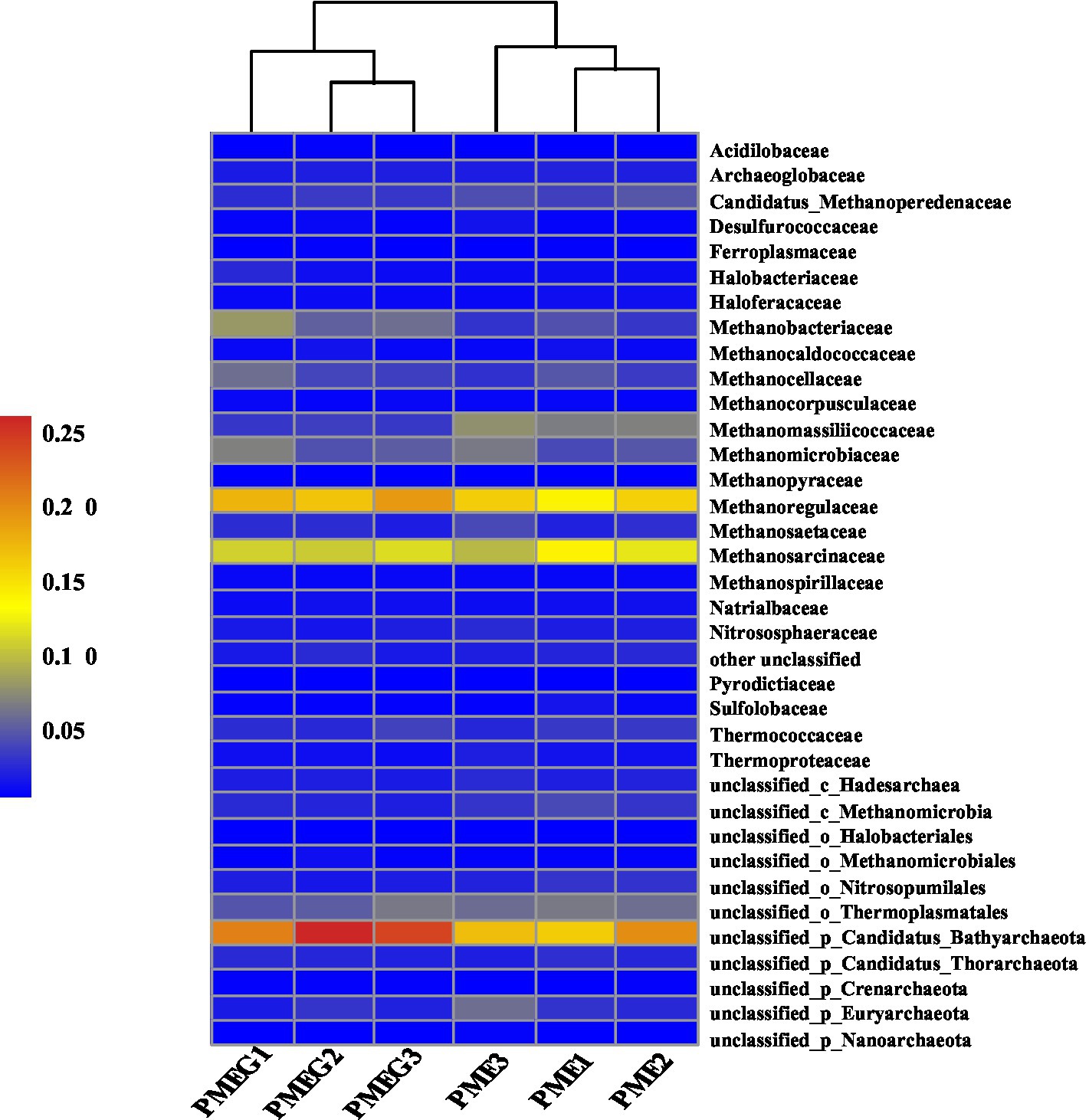
Figure 5. Heatmap showing the archaeal community composition based on the level of family in the PME and PMEG treatments.
PCoA Analysis of a Functional Profile Based on Metagenomic Sequencing in the PME and PMEG Treatments
No significant difference was detected in the macro-functional composition based on the KEGG annotation between the PME and PMEG treatments (Supplementary Figure 2; Supplementary Table 2). Thus, we further analyzed the composition of functional genes in these two treatments by using PCoA analysis; the results are shown in Figure 6A. Similar to the PCoA profile (Figure 4A) based on the microbial composition, the sample points of the PME and PMEG treatments were distributed in a narrow area (axis1, −0.14-0.99; axis2, −0.03-0.10), and the Adonis test showed no significant difference (R2=0.48, p=0.10) in the functional composition between these two treatments (Figure 6A). MPR and PDR were also significantly correlated with the functional components in the enriched microbial consortium (Table 1), explaining 39 and 10% of the variance in the functional composition, respectively (Figure 6B).
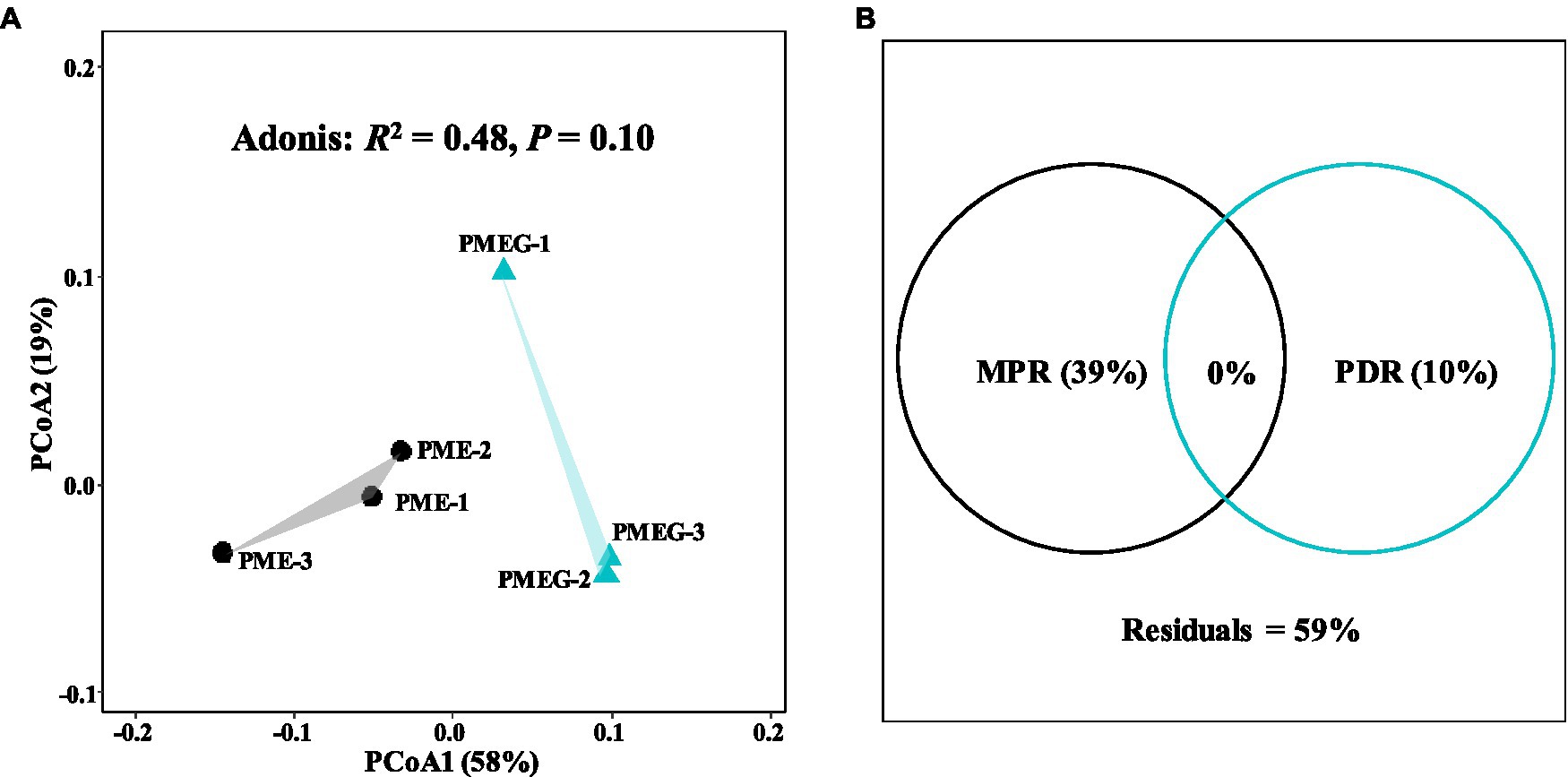
Figure 6. The PCoA profile based on the functional enzymes composition in the PME and PMEG treatments (A) and variance partitioning analysis showing the portions explained by MPR and PDR for functional enzymes composition (B).
Relative Abundance of Genes Related to Aromatic Compounds Degradation and CH4 Synthesis in the PME and PMEG Treatments
To investigate the potential mechanism by which glucose affects the MPR and PDR in the enriched microbial consortium, we further compared the relative abundance of genes related to the degradation of aromatic compounds and CH4 synthesis in the PME and PMEG treatments (Figure 7). Fourteen types of genes responsible for the degradation of aromatic compounds were detected based on the KEGG pond of metagenomic sequencing, 13 types of which showed higher values of relative abundance in the PME treatment than in the PMEG treatment (Figure 7A). The results of the independent t-test indicated that the relative abundance of genes associated with nitrotoluene (PME: 0.243%, PMEG: 0.235%), fluorobenzoate (PME: 0.039%, PMEG: 0.009%), xylene (PME: 0.226%, PMEG: 0.189%), aminobenzoate (PME: 0.206%, PMEG: 0.121%), and benzoate (PME: 0.809%, PMEG:0.537%) degradation in the PME treatment were significantly higher than those in the PMEG treatment (Figure 7A). For the genes involved in CH4 synthesis, genes related to 4 types of CH4 synthesis pathways – acetate (dominant in all treatments with an average proportion of 48.20%), CO2, methylamine, and methanol pathways – were detected based on the results of metagenomic sequencing, and no significant difference was observed between these two treatments (Figure 7B).
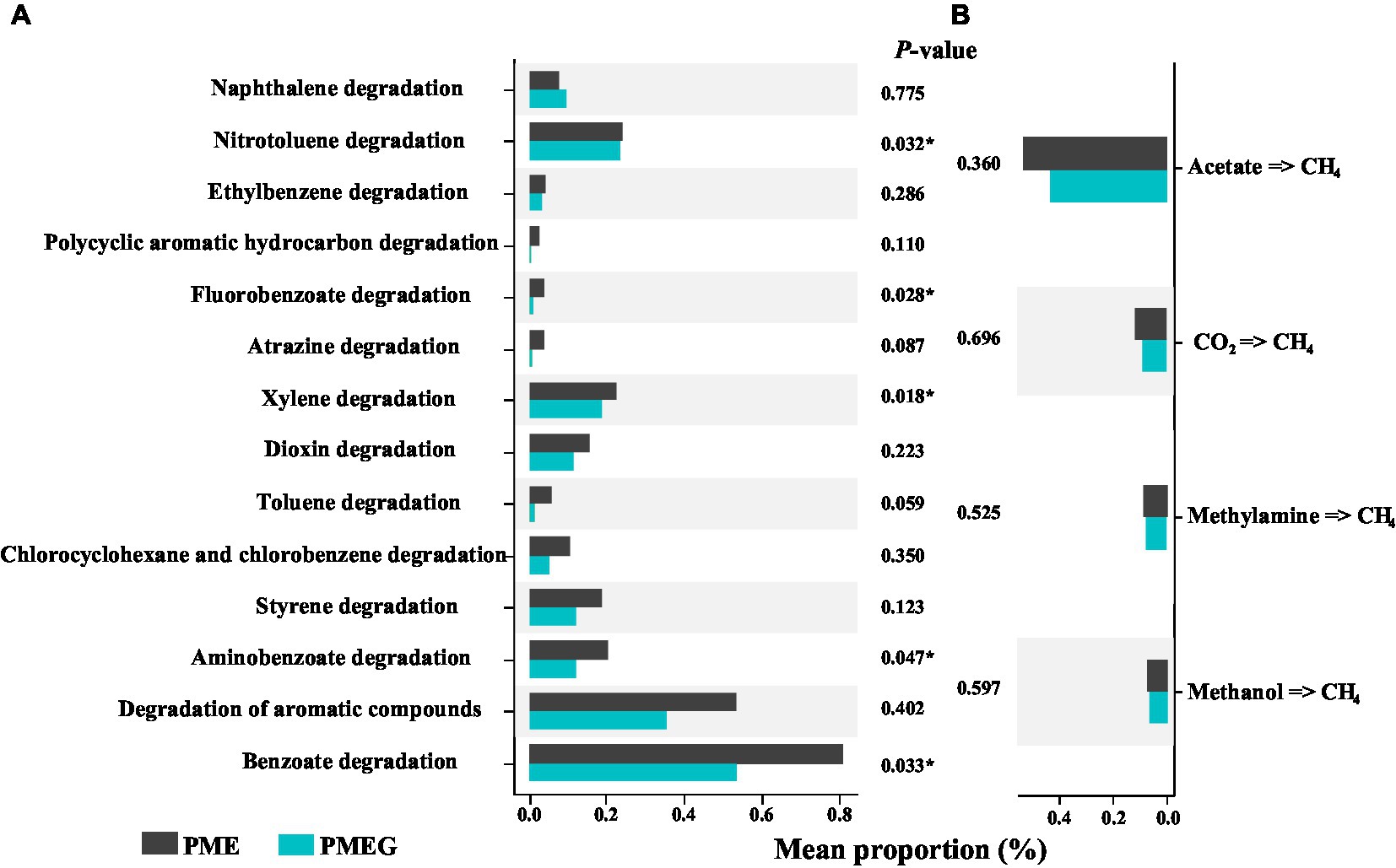
Figure 7. Comparision of the relative abundances of genes involved in the degradation of aromatic commpounds (A) and methane synthesis (B) between the PME and PMEG treatments.
Discussion
In this study, the CH4 production rate of the enriched microbial consortium was significantly improved by the addition of glucose (Figure 3A), which was consistent with findings in the literature that glucose could enhance CH4 production in anaerobic digestion systems (Bucha et al., 2018; Maletic et al., 2018; Vu and Min, 2019; Li et al., 2021). Compared with phenanthrene, glucose can be more quickly and easily utilized and subsequently converted into carbon dioxide (CO2), hydrogen (H2), and acetate by the microbes in the enriched consortium under anaerobic conditions, supplying feasible substrates for methanogens to synthesize CH4 (Khan et al., 2017; Zhang et al., 2019). However, our results indicated that the addition of glucose did not promote the degradation of phenanthrene (Figure 3B). Several studies have reported the enhancement of PAH degradation caused by glucose addition or other co-metabolic substrates under anaerobic conditions (Yuan and Chang, 2007; Ma et al., 2011; Li et al., 2016a). For example, Li et al. (2016a) reported that glucose addition enhanced the degradation rate of petroleum hydrocarbons by 200%, while the promotion of petroleum hydrocarbons degradation was mainly attributed to the stimulation of the biocurrent triggered by glucose. Maletic et al. (2018) found that glucose addition improved CH4 production but had no effect on the degradation of PAHs, which is consistent with our current results and should be explained by the following reasons. The utilization of phenanthrene was delayed by the addition of glucose, which should start after the complete consumption of glucose, and then the metabolism of glucose resulted in the improvement of CH4 production. However, the glucose addition might neither promote microbial activity to degrade phenanthrene nor induce the microbial consortium to produce phenanthrene-degrading enzymes during methanogenesis.
This study revealed that the addition of glucose decreased the α-diversity of the enriched consortium (Supplementary Figure 1), which is consistent with reports that glucose addition decreased the microbial α-diversity in some enriched consortia (Li et al., 2016a; Goldford et al., 2018; Zhang et al., 2019; Shu et al., 2021), indicating the selectivity of a single carbon source on the microbial community. Additionally, glucose addition had no significant influence on the microbial community of the enriched phenanthrene-degrading consortium (Figure 4A). The addition of co-metabolic substrates has usually been shown to trigger shifts in the microbial community structure in anaerobic digestion systems (Li et al., 2016a,b; Zhang et al., 2019). In these studies, the wastes treated in their systems contained inartificial microbes originating from soil or wastewater, which should be complex, unstable, and fictile, and then susceptible to external carbon sources such as glucose. By contrast, the microbial consortium in our study had undergone an enrichment period of 10 rounds supplied with the sole carbon source (phenanthrene) and nutrient components, which should be a stable ecosystem insensitive to the external addition of glucose. Sundh et al. (2003) also reported that glucose addition significantly enhanced CH4 yield, but only slightly affected the microbial community in an anaerobic digestion system fed with a synthetic substrate. Furthermore, our result showed that MPR and PDR were significantly correlated with the microbial community composition of the enriched consortium (Table 1). Generally accepted is that the microbial community structure determines its function (de Souza and Santos, 2018; Zhou et al., 2019). The close linkage between CH4 production and microbial communities in diverse ecosystems has been confirmed (Shrestha et al., 2011; Edwards et al., 2015; Wang et al., 2020; Watanabe et al., 2020). However, the strong selectivity of PAHs to environmental microbes has led to a strong relationship between PAH degradation and microbial community structure (Zhou et al., 2017a; Spini et al., 2018; Wang et al., 2021a). Moreover, our results revealed that MPR explained a greater percentage of the variance in the microbial community than PDR (Figure 4B), indicating that the former might be more dependent on the microbial community structure than the latter, which might be due to the different microbial mechanisms of CH4 synthesis and phenanthrene degradation. In detail, the CH4 synthesis process might involve a series of steps driven by diverse microbes, including the primary metabolism of carbon sources, the transformation of the intermediates, and the generation of substrates (CO2, H2, and acetate) for methanogens (Maletic et al., 2018), whereas the degradation rate of phenanthrene might be determined by limited microbes responsible for the cleavage of the benzene ring (Meckenstock and Mouttaki, 2011). Therefore, MPR should be more important in explaining the variance of the microbial community than PDR in the phenanthrene-degrading system during methanogenesis.
In this study, the PCoA profile based on the composition of functional genes indicated that the macro-function of the enriched consortium was not affected by the addition of glucose, which was consistent with the response of the microbial community structure to glucose (Figure 6A; Supplementary Figure 2). Additionally, the significant correlation between the functional composition and MPR or PDR (Table 1), and the result of VPART analysis based on the functional composition (Figure 6B), were similar to the results based on the microbial community structure, further confirming the uniformity between the microbial community and its functions. In this study, the genes involved in the degradation of some refractory compounds and CH4 biosynthesis were selected from the functional pool of KEGG for further analysis. The relative abundance of those degrading genes showed a decreasing tendency due to the addition of glucose (Figure 7A), indicating that the metabolic characteristic of the microbial community also followed the principle of “glucose repression.” Glucose (monosaccharide) will hamper the Escherichia coli from utilizing lactose (disaccharide), known as “glucose repression,” suggesting that an easy-utilized carbon source will be preferentially metabolized by microbes and thus hinder the utilization of the complex one (Rose et al., 1991). Nevertheless, the relative abundance values of degrading genes in the PME and PMEG treatments were of the same order of magnitude (Figure 7A), which the following reasons might explain. First, the DNA samples were collected at the end of the incubation (day 21) when the glucose had been exhausted; second, the metagenomic sequencing in this study was based on the DNA level, which could not reflect the actual transcription level of those degrading genes. The relative abundance of the genes related to CH4 synthesis pathways was not significantly different between the PME and PMEG treatments, and the pathway of CH4 synthesis via acetate was dominant in this enriched consortium (Figure 7B). The acetate pathway has been reported to be the key CH4 synthesis pathway in paddy soils (Conrad, 1999; Glissmann and Conrad, 2002; Masuda et al., 2018). Acetate is also an essential intermediate or terminal product in the degradation of crude oil (a major source of PAHs) or some PAHs (naphthalene, phenanthrene, anthracene, and pyrene) during methanogenesis (Dolfing et al., 2008, 2009; Jones et al., 2008). Therefore, CH4 synthesis via the acetate pathway should be dominant in the anaerobic phenanthrene digestion system during methanogenesis.
Conclusion
In this study, we examined the co-metabolic effect of glucose on CH4 production and phenanthrene degradation in a phenanthrene-degrading consortium under methanogenesis. We found that glucose addition significantly enhanced the CH4 production rate of the enriched consortium but had no influence on its ability to degrade phenanthrene. Glucose addition significantly decreased the α-diversity of the enriched microbial consortium but showed no significant effect on either the microbial community or the macro-function of the enriched consortium. Glucose addition only slightly decreased the relative abundance of the genes involved in the degradation of aromatic compounds, and the acetate pathway was dominant for CH4 synthesis in this anaerobic phenanthrene digestion system under methanogenesis. These results indicate that it is difficult to remarkably enhance PAH degradation in a single-stage anaerobic digestion system under methanogenesis via the addition of co-metabolic substrates. Therefore, we propose that a multistage and combined pattern comprising methanogenesis and other more efficient anaerobic digestion pathways of PAH (sulfate-reducing or denitrification) should be a promising strategy to enhance the CH4 production and PAH degradation rate in the anaerobic digestion of PAH-containing wastes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA744876.
Author Contributions
ZiZ and YW conducted the experiment, analyzed the data, and wrote the original draft. MW arranged the investigation consortium and contributed to the phenanthrene and methane detection. ZhZ designed the experiment and contributed to the edit, data analysis, and review of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (41771501).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.749967/full#supplementary-material
Footnotes
1. ^https://github.com/OpenGene/fastp
3. ^https://github.com/voutcn/megahit
4. ^http://www.bioinformatics.org/cd-hit/
5. ^http://github.com/ShujiaHuang/SOAPaligner
References
Agarry, S. E., and Owabor, C. N. (2011). Anaerobic bioremediation of marine sediment artificially contaminated with anthracene and naphthalene. Environ. Technol. 32, 1357–1381. doi: 10.1080/09593330.2010.536788
Al Farraj, D. A., Alkufeidy, R. M., Alkubaisi, N. A., and Alshammari, M. K. (2021). Polynuclear aromatic anthracene biodegradation by psychrophilic Spingomonas sp., cultivated with tween-80. Chemosphere 263:128115. doi: 10.1016/j.chemosphere.2020.128115
Ambrosoli, R., Petruzzelli, L., Minati, J. L., and Marsan, F. A. (2005). Anaerobic PAH degradation in soil by a mixed bacterial consortium under denitrifying conditions. Chemosphere 60, 1231–1236. doi: 10.1016/j.chemosphere.2005.02.030
Awe, O. W., Zhao, Y. Q., Nzihou, A., Minh, D. P., and Lyczko, N. (2017). A review of biogas utilisation, purification and upgrading technologies. Waste Biomass Valori. 8, 267–283. doi: 10.1007/s12649-016-9826-4
Berdugo-Clavijo, C., Dong, X. L., Soh, J., Sensen, C. W., and Gieg, L. M. (2012). Methanogenic biodegradation of two-ringed polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 81, 124–133. doi: 10.1111/j.1574-6941.2012.01328.x
Bianco, F., Race, M., Papirio, S., and Esposito, G. (2020). Removal of polycyclic aromatic hydrocarbons during anaerobic biostimulation of marine sediments. Sci. Total. Environ. 709:136141. doi: 10.1016/j.scitotenv.2019.136141
Brauer, S. L., Yashiro, E., Ueno, N. G., Yavitt, J. B., and Zinder, S. H. (2006). Characterization of acid-tolerant H2/CO2-utilizing methanogenic enrichment cultures from an acidic peat bog in New York state. FEMS Microbiol. Ecol. 57, 206–216. doi: 10.1111/j.1574-6941.2006.00107.x
Bucha, M., Jedrysek, M. O., Kufka, D., Plesniak, L., Marynowski, L., Kubiak, K., et al. (2018). Methanogenic fermentation of lignite with carbon-bearing additives, inferred from stable carbon and hydrogen isotopes. Int. J. Coal. Geol. 186, 65–79. doi: 10.1016/j.coal.2017.11.020
Chan, Y. J., Chong, M. F., Law, C. L., and Hassell, D. G. (2009). A review on anaerobic–aerobic treatment of industrial and municipal wastewater. Chem. Eng. J. 155, 1–18. doi: 10.1016/j.cej.2009.06.041
Chang, W., Um, Y. S., Hoffman, B., and Holoman, T. R. P. (2005). Molecular characterization of polycyclic aromatic hydrocarbon (PAH)-degrading methanogenic communities. Biotechnol. Progr. 21, 682–688. doi: 10.1021/bp049579l
Conrad, R. (1999). Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28, 193–202. doi: 10.1111/j.1574-6941.1999.tb00575.x
Couling, N. R., Towell, M. G., and Semple, K. T. (2010). Biodegradation of PAHs in soil: influence of chemical structure, concentration and multiple amendment. Environ. Pollut. 158, 3411–3420. doi: 10.1016/j.envpol.2010.07.034
de Souza, T. A. F., and Santos, D. (2018). Effects of using different host plants and long-term fertilization systems on population sizes of infective arbuscular mycorrhizal fungi. Symbiosis 76, 139–149. doi: 10.1007/s13199-018-0546-3
Dolfing, J., Larter, S. R., and Head, I. M. (2008). Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2, 442–452. doi: 10.1038/ismej.2007.111
Dolfing, J., Xu, A., Gray, N. D., Larter, S. R., and Head, I. M. (2009). The thermodynamic landscape of methanogenic PAH degradation. Microb. Biotechnol. 2, 566–574. doi: 10.1111/j.1751-7915.2009.00096.x
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U. S. A. 112, 911–920. doi: 10.1073/pnas.1414592112
Feng, N. X., Yu, J., Xiang, L., Yu, L. Y., Zhao, H. M., Mo, C. H., et al. (2019). Co-metabolic degradation of the antibiotic ciprofloxacin by the enriched bacterial consortium XG and its bacterial community composition. Sci. Total. Environ. 665, 41–51. doi: 10.1016/j.scitotenv.2019.01.322
Ferguson, T. J., and Mah, R. A. (1983). Isolation and characterization of an H2-oxidizing thermophilic methanogen. Appl. Environ. Microbiol. 45, 265–274.
Gallert, C., and Winter, J. (1997). Mesophilic and thermophilic anaerobic digestion of source-sorted organic wastes: effect of ammonia on glucose degradation and methane production. Appl. Microbiol. Biotechnol. 48, 405–410. doi: 10.1007/s002530051071
Glissmann, K., and Conrad, R. (2002). Saccharolytic activity and its role as a limiting step in methane formation during the anaerobic degradation of rice straw in rice paddy soil. Biol. Fertil. Soils 35, 62–67. doi: 10.1007/s00374-002-0442-z
Goldford, J. E., Lu, N., Bajic, D., Estrela, S., Tikhonov, M., Sanchez-Gorostiaga, A., et al. (2018). Emergent simplicity in microbial community assembly. Science 361, 469–474. doi: 10.1126/science.aat1168
Guo, N., Ma, X. F., Ren, S. J., Wang, S. G., and Wang, Y. K. (2019). Mechanisms of metabolic performance enhancement during electrically assisted anaerobic treatment of chloramphenicol wastewater. Water Res. 156, 199–207. doi: 10.1016/j.watres.2019.03.032
Haberl, H., Sprinz, D., Bonazountas, M., Cocco, P., Desaubies, Y., Henze, M., et al. (2012). Correcting a fundamental error in greenhouse gas accounting related to bioenergy. Energ. Policy 45, 18–23. doi: 10.1016/j.enpol.2012.02.051
Hadibarata, T., Syafiuddin, A., Al-Dhabaan, F. A., Elshikh, M. S., and Rubiyatno, (2018). Biodegradation of mordant orange-1 using newly isolated strain Trichoderma harzianum RY44 and its metabolite appraisal. Bioproc. Biosyst. Eng. 41, 621–632. doi: 10.1007/s00449-018-1897-0
He, Y. L., Xu, P., Li, C. J., and Zhang, B. (2005). High-concentration food wastewater treatment by an anaerobic membrane bioreactor. Water Res. 39, 4110–4118. doi: 10.1016/j.watres.2005.07.030
Holm-Nielsen, J. B., Al Seadi, T., and Oleskowicz-Popiel, P. (2009). The future of anaerobic digestion and biogas utilization. Bioresour. Technol. 100, 5478–5484. doi: 10.1016/j.biortech.2008.12.046
Hu, H., Liu, J. F., Li, C. Y., Yang, S. Z., Gu, J. D., and Mu, B. Z. (2018). Anaerobic biodegradation of partially hydrolyzed polyacrylamide in long-term methanogenic enrichment cultures from production water of oil reservoirs. Biodegradation 29, 233–243. doi: 10.1007/s10532-018-9825-1
Jing, Y. P., Li, Y., Liu, Z. H., Zhang, Y. P., Liu, P., Sun, M., et al. (2017). Effects of bacterial-feeding nematodes and glucose on phenanthrene removal by Pseudomonas putida. Pedosphere 27, 165–171. doi: 10.1016/S1002-0160(15)60103-5
Jones, D. M., Head, I. M., Gray, N. D., Adams, J. J., Rowan, A. K., Aitken, C. M., et al. (2008). Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180. doi: 10.1038/nature06484
Khan, M. D., Khan, N., Nizami, A. S., Rehan, M., Sabir, S., and Khan, M. Z. (2017). Effect of cosubstrates on biogas production and anaerobic decomposition of pentachlorophenol. Bioresour. Technol. 238, 492–501. doi: 10.1016/j.biortech.2017.04.063
Khanna, P., Goyal, D., and Khanna, S. (2012). Characterization of pyrene utilizing Bacillus spp. from crude oil contaminated soil. Braz. J. Microbiol. 43, 606–617. doi: 10.1590/S1517-83822012000200024
Kunapuli, U., Lueders, T., and Meckenstock, R. U. (2007). The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J. 1, 643–653. doi: 10.1038/ismej.2007.73
Li, Y., Chen, Y. G., and Wu, J. (2019). Enhancement of methane production in anaerobic digestion process: a review. Appl. Energy 240, 120–137. doi: 10.1016/j.apenergy.2019.01.243
Li, Y. J., Wang, Q. S., Liu, L. Y., Tabassum, S., Sun, J., and Hong, Y. L. (2021). Enhanced phenols removal and methane production with the assistance of graphene under anaerobic co-digestion conditions. Sci. Total. Environ. 759:143523. doi: 10.1016/j.scitotenv.2020.143523
Li, X. J., Wang, X., Wan, L. L., Zhang, Y. Y., Li, N., Li, D. S., et al. (2016a). Enhanced biodegradation of aged petroleum hydrocarbons in soils by glucose addition in microbial fuel cells. J. Chem. Technol. Biotechnol. 91, 267–275. doi: 10.1002/jctb.4660
Li, Y. C., Zhou, J., Gong, B. Z., Wang, Y. M., and He, Q. (2016b). Cometabolic degradation of lincomycin in a sequencing batch biofilm reactor (SBBR) and its microbial community. Bioresour. Technol. 214, 589–595. doi: 10.1016/j.biortech.2016.04.085
Lytras, G., Lytras, C., Mathioudakis, D., Papadopoulou, K., and Lyberatos, G. (2021). Food waste valorization based on anaerobic digestion. Waste Biomass Valori. 12, 1677–1697. doi: 10.1007/s12649-020-01108-z
Ma, C., Wang, Y. Q., Zhuang, L., Huang, D. Y., Zhou, S. G., and Li, F. B. (2011). Anaerobic degradation of phenanthrene by a newly isolated humus-reducing bacterium, Pseudomonas aeruginosa strain PAH-1. J. Soil. Sediment. 11, 923–929. doi: 10.1007/s11368-011-0368-x
Maletic, S., Murenji, S., Agbaba, J., Roncevic, S., Isakovski, M. K., Jazic, J. M., et al. (2018). Potential for anaerobic treatment of polluted sediment. J. Environ. Manage. 214, 9–16. doi: 10.1016/j.jenvman.2018.02.029
Masuda, Y., Itoh, H., Shiratori, Y., and Senoo, K. (2018). Metatranscriptomic insights into microbial consortia driving methane metabolism in paddy soils. Soil Sci. Plant. Nutr. 64, 455–464. doi: 10.1080/00380768.2018.1457409
Meckenstock, R. U., and Mouttaki, H. (2011). Anaerobic degradation of non-substituted aromatic hydrocarbons. Curr. Opin. Biotechnol. 22, 406–414. doi: 10.1016/j.copbio.2011.02.009
Mousa, L., and Forster, C. F. (1999). The use of glucose as a growth factor to counteract inhibition in anaerobic digestion. Process. Saf. Environ. Prot. 77, 193–198. doi: 10.1205/095758299530062
Noguchi, H., Park, J., and Takagi, T. (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630. doi: 10.1093/nar/gkl723
Nzila, A. (2018). Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons under anaerobic conditions: overview of studies, proposed pathways and future perspectives. Environ. Pollut. 239, 788–802. doi: 10.1016/j.envpol.2018.04.074
Qin, W., Fan, F. Q., Zhu, Y., Huang, X. L., Ding, A. Z., Liu, X., et al. (2018). Anaerobic biodegradation of benzo (a) pyrene by a novel Cellulosimicrobium cellulans CWS2 isolated from ploycyclic aromatic hydrocarbon-contaminated soil. Braz. J. Microbiol. 49, 258–268. doi: 10.1016/j.bjm.2017.04.014
Rose, M., Albig, W., and Entian, K. D. (1991). Glucose repression in Saccharomyces-cerevisiae is directly associated with hexose phosphorylation by hexokinase-PI and hexokinase-PII. Eur. J. Biochem. 199, 511–518. doi: 10.1111/j.1432-1033.1991.tb16149.x
Shrestha, M., Shrestha, P. M., and Conrad, R. (2011). Bacterial and archaeal communities involved in the in situ degradation of 13C-labelled straw in the rice rhizosphere. Environ. Microbiol. Rep. 3, 587–596. doi: 10.1111/j.1758-2229.2011.00267.x
Shu, W. H., Zhang, Y., Wen, D. H., Wu, Q. Y., Liu, H., Cui, M. H., et al. (2021). Anaerobic biodegradation of levofloxacin by enriched microbial consortia: effect of electron acceptors and carbon source. J. Hazard. Mater. 414:125520. doi: 10.1016/j.jhazmat.2021.125520
Spini, G., Spina, F., Poli, A., Blieux, A. L., Regnier, T., Gramellini, C., et al. (2018). Molecular and microbiological insights on the enrichment procedures for the isolation of petroleum degrading bacteria and fungi. Front. Microbiol. 9:2543. doi: 10.3389/fmicb.2018.02543
Stabnikova, O., Liu, X. Y., and Wang, J. Y. (2008). Anaerobic digestion of food waste in a hybrid anaerobic solid–liquid system with leachate recirculation in an acidogenic reactor. Biochem. Eng. J. 41, 198–201. doi: 10.1016/j.bej.2008.05.008
Sundh, I., Carlsson, H., Nordberg, A., Hansson, M., and Mathisen, B. (2003). Effects of glucose overloading on microbial community structure and biogas production in a laboratory-scale anaerobic digester. Bioresour. Technol. 89, 237–243. doi: 10.1016/S0960-8524(03)00075-0
Tripathi, V., Gaur, V. K., Dhiman, N., Gautam, K., and Manickam, N. (2020). Characterization and properties of the biosurfantant produced by PAH-degrading bacteria isolated from contaminated oily sludge environment. Environ. Sci. Pollut. Res. 27, 27268–27278. doi: 10.1007/s11356-019-05591-3
Vu, H. T., and Min, B. (2019). Integration of submersible microbial fuel cell in anaerobic digestion for enhanced production of methane and current at varying glucose levels. Int. J. Hydrogen. Energ. 44, 7574–7582. doi: 10.1016/j.ijhydene.2019.01.091
Wan, R., Zhang, S. Y., and Xie, S. G. (2012). Microbial community changes in aquifer sediment microcosm for anaerobic anthracene biodegradation under methanogenic condition. J. Environ. Sci. 24, 1498–1503. doi: 10.1016/S1001-0742(11)60959-5
Wang, B. B., Teng, Y., Yao, H. Y., and Christie, P. (2021a). Detection of functional microorganisms in benzene [a] pyrene-contaminated soils using DNA-SIP technology. J. Hazard. Mater. 407:124788. doi: 10.1016/j.jhazmat.2020.124788
Wang, Y. S., and Wang, Y. H. (2003). Quick measurement of CH4, CO2 and N2O emissions from a short-plant ecosystem. Adv. Atmos. Sci. 20, 842–844. doi: 10.1007/BF02915410
Wang, Y. Q., Wang, M. X., Chen, Y. Y., Li, C. M., and Zhou, Z. F. (2021b). Microbial community structure and co-occurrence are essential for methanogenesis and its contribution to phenanthrene degradation in paddy soil. J. Hazard. Mater. 417:126086. doi: 10.1016/j.jhazmat.2021.126086
Wang, Y. Q., Xiao, G. Q., Wang, M. X., Cheng, Y. Y., Sun, B. Y., and Zhou, Z. F. (2020). The linkage between methane production activity and prokaryotic community structure in the soil within a shale-gas field in China. Environ. Sci. Pollut. Res. 27, 7453–7462. doi: 10.1007/s11356-019-07454-3
Watanabe, T., Asakawa, S., and Hayano, K. (2020). Long-term submergence of non-methanogenic oxic upland field soils helps to develop the methanogenic archaeal community as revealed by pot and field experiments. Pedosphere 30, 62–72. doi: 10.1016/S1002-0160(19)60819-2
Xie, X. H., Liu, N., Yang, B., Yu, C. Z., Zhang, Q. Y., Zheng, X. L., et al. (2016). Comparison of microbial community in hydrolysis acidification reactor depending on different structure dyes by illumina miSeq sequencing. Int. Biodeter. Biodegr. 111, 14–21. doi: 10.1016/j.ibiod.2016.04.004
Xue, W. L., and Warshawsky, D. (2005). Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol. Appl. Pharmacol. 206, 73–93. doi: 10.1016/j.taap.2004.11.006
Ye, Q. H., Liang, C. Y., Chen, X. W., Fang, T. T., Wang, Y., and Wang, H. (2019). Molecular characterization of methanogenic microbial communities for degrading various types of polycyclic aromatic hydrocarbon. J. Environ. Sci. 86, 97–106. doi: 10.1016/j.jes.2019.04.027
Yuan, S. Y., and Chang, B. V. (2007). Anaerobic degradation of five polycyclic aromatic hydrocarbons from river sediment in Taiwan. J. Environ. Sci. Health B. 42, 63–69. doi: 10.1080/03601230601020860
Zhang, G. B., Ji, Y., Ma, J., Xu, H., and Cai, Z. C. (2011). Case study on effects of water management and rice straw incorporation in rice fields on production, oxidation, and emission of methane during fallow and following rice seasons. Soil Res. 49, 238–246. doi: 10.1071/SR10117
Zhang, S. Y., Wang, Q. F., and Xie, S. G. (2012). Molecular characterization of phenanthrene-degrading methanogenic communities in leachate-contaminated aquifer sediment. Int. J. Environ. Sci. Technol. 9, 705–712. doi: 10.1007/s13762-012-0098-7
Zhang, Q. Y., Xie, X. H., Liu, Y. B., Zheng, X. L., Wang, Y. Q., Cong, J. H., et al. (2019). Sugar sources as co-substrates promoting the degradation of refractory dye: a comparative study. Ecotoxicol. Environ. Saf. 184:109613. doi: 10.1016/j.ecoenv.2019.109613
Zhou, W. Z., Chen, X. G., Ismail, M., Wei, L., and Hu, B. L. (2021). Simulating the synergy of electron donors and different redox mediators on the anaerobic decolorization of azo dyes: can AQDS-chitosan globules replace the traditional redox mediators? Chemosphere 275:130025. doi: 10.1016/j.chemosphere.2021.130025
Zhou, Z. F., Wang, M. X., Zuo, X. H., and Yao, Y. H. (2017a). Comparative investigation of bacterial, fungal, and archaeal community structures in soils in a typical oilfield in Jianghan, China. Arch. Environ. Contam. Toxicol. 72, 65–77. doi: 10.1007/s00244-016-0333-1
Zhou, Z. F., Wei, W. L., Shi, X. J., Liu, Y. M., He, X. H., and Wang, M. X. (2019). Twenty-six years of chemical fertilization decreased soil RubisCO activity and changed the ecological characteristics of soil cbbL-carrying bacteria in an entisol. Appl. Soil. Ecol. 141, 1–9. doi: 10.1016/j.apsoil.2019.05.005
Zhou, Z. F., Yao, Y. H., Wang, M. X., and Zuo, X. H. (2017b). Co-effects of pyrene and nitrate on the activity and abundance of soil denitrifiers under anaerobic condition. Arch. Microbiol. 199, 1091–1101. doi: 10.1007/s00203-017-1380-3
Zhou, Z. F., Zhang, Z. Y., Wang, M. X., Liu, Y. M., and Dai, J. S. (2018). Effect of the nitrification inhibitor (3, 4-dimethylpyrazole phosphate) on the activities and abundances of ammonia-oxidizers and denitrifiers in a phenanthren polluted and waterlogged soil. Ecotoxicol. Environ. Saf. 161, 474–481. doi: 10.1016/j.ecoenv.2018.06.030
Keywords: anaerobic digestion, methanogenesis, phenanthrene degradation, methane production, co-metabolism
Citation: Zhou Z, Wang Y, Wang M and Zhou Z (2021) Co-metabolic Effect of Glucose on Methane Production and Phenanthrene Removal in an Enriched Phenanthrene-Degrading Consortium Under Methanogenesis. Front. Microbiol. 12:749967. doi: 10.3389/fmicb.2021.749967
Edited by:
Ruiyong Zhang, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Dhiraj Kumar Chaudhary, Korea University, South KoreaXiaojing Li, Agro-Environmental Protection Institute, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Zhou, Wang, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhifeng Zhou, emhvdXpoZkBzd3UuZWR1LmNu
 Ziyan Zhou
Ziyan Zhou Yanqin Wang
Yanqin Wang Mingxia Wang
Mingxia Wang Zhifeng Zhou
Zhifeng Zhou