- Key Laboratory of Microbial Resources Collection and Preservation, Ministry of Agriculture and Rural Affairs, Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
Pseudomonas fluorescens 2P24 is a plant growth-promoting rhizobacterium (PGPR) isolated from wheat take-all decline soil. Genomic analysis of strain 2P24 revealed the presence of a complete SPI-1 type III secretion system (T3SS) gene cluster on the chromosome with an organization and orientation similar to the SPI-1 T3SS gene clusters of Salmonella enterica and P. kilonensis F113. Phylogenetic analysis revealed that the SPI-1 T3SS gene cluster of strain 2P24 might be obtained from Salmonella and Shigella by horizontal gene transfer. Two transcriptional regulator homologs of HilA and InvF were found from the SPI-1 T3SS gene cluster of strain 2P24. HilA regulated the expression of the structural genes positively, such as invG, sipB, sipD, prgI, and prgK. Prediction of transcriptional binding sites and RNA-seq analysis revealed 14 genes were up-regulated by InvF in strain 2P24. Exploring potential roles of SPI-1 T3SS revealed that it was not associated with motility. However, 2P24ΔinvF reduced resistance against Fusarium graminearum significantly. 2P24ΔhilA enhanced formation of biofilm significantly at 48 h. All three mutants 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C reduced the chemotactic responses to glucose significantly. Finally, the determination of SPI-1 mutants to trigger innate immunity in Nicotiana benthamiana showed that 2P24ΔinvE-C reduced the ability to induce the production of reactive oxygen species compared with the wild type strain 2P24.
Introduction
Type III secretion system (T3SS) is a protein secretion apparatus that is widespread in animal and plant pathogenic bacteria. T3SS is a syringe-like complex composed of >20 proteins and consists of three parts: a ring component that spans the inner and outer membranes of the bacteria, an extracellular needle/pilus that extends from the outer membrane ring, and a transporter tip that is attached to the host cell membrane (Büttner, 2012; Deng et al., 2017). For adapting to diverse hosts and habitats, T3SS has evolved into seven types: Hrp1, Hrp2, SPI-1, SPI-2, Ysc, RhC, and ChL (Pallen et al., 2005; Troisfontaines and Cornelis, 2005). Among them, Ysc, SPI-1, and SPI-2 T3SS usually exist in animal pathogenic bacteria. Hrp1 and Hrp2 T3SS are mainly present in plant pathogenic bacteria. RhC T3SS is mostly found in plant symbiotic rhizobia. And ChL T3SS often appears in some bacteria that live in animals, insects, and protozoa (Nazir et al., 2017).
Animal and plant pathogens deploy T3SS to deliver type III effectors (T3Es) into host cells for pathogenesis. T3SS structural genes are usually conserved, but T3Es are diverse to adapt to different host environments. The distribution of T3SS and the mechanisms of T3Es in animal and plant pathogens have been studied well. However, the T3SS was also reported to be present in many plant growth-promoting rhizobacteria and especially in the Pseudomonas fluorescens group (Preston et al., 2001; Kimbrel et al., 2010; Marchi et al., 2013; Viollet et al., 2017; Stringlis et al., 2019). Preston et al. (2001) found that P. fluorescens SBW25 harbored a Hrp1 T3SS, and expression of the rscC gene, a hrcC homolog, was induced in the sugar beet rhizosphere. Mutations of the rsp regulators rspL and rspV in strain SBW25 resulted in a significant reduction in competitive colonization on the root tips of sugar beet seedlings (Jackson et al., 2005). Genomic analysis of P. kilonensis F113 revealed a complete Hrp1 T3SS gene cluster and four T3E homologs RopM, RopAA1, RopAA2, and RopB (Barret et al., 2013). Surprisingly, in addition to the Hrp1 T3SS, SPI-1 T3SS homologs were also found in P. kilonensis F113 and some other PGPR, such as P. fluorescens HK44 and P. fluorescens Q2-87 (Chauhan et al., 2011; Loper et al., 2012; Barret et al., 2013). One report showed that the SPI-1 T3SS of P. kilonensis F113 was involved in resistance to amoeboid grazing (Barret et al., 2013).
Pseudomonas fluorescens 2P24 is a PGPR isolated from wheat take-all decline soil in Shandong Province, China (Wei et al., 2004a). Strain 2P24 produces several secondary metabolites, such as 2, 4-diacetylphloroglucinol (2, 4-DAPG), hydrogen cyanide (HCN), and siderophore(s), and antagonizes a variety of plant pathogens, such as Ralstonia solanacearum, Rhizoctonia solani, and Fusarium oxysporum (Wei et al., 2004a). Genetic analysis revealed that the antibiotic 2, 4-DAPG is the key biocontrol factor in strain 2P24 (Wei et al., 2004b). In this study, a full length of SPI-1 T3SS gene cluster was identified from strain 2P24. We conducted phylogenetic analysis of the SPI-1 T3SS and transcriptomic analysis of two transcriptional regulators, HilA and InvF. The effects of SPI-I mutation on antagonism, motility, chemotaxis, biofilm formation, and plant immunity were also determined.
Materials and Methods
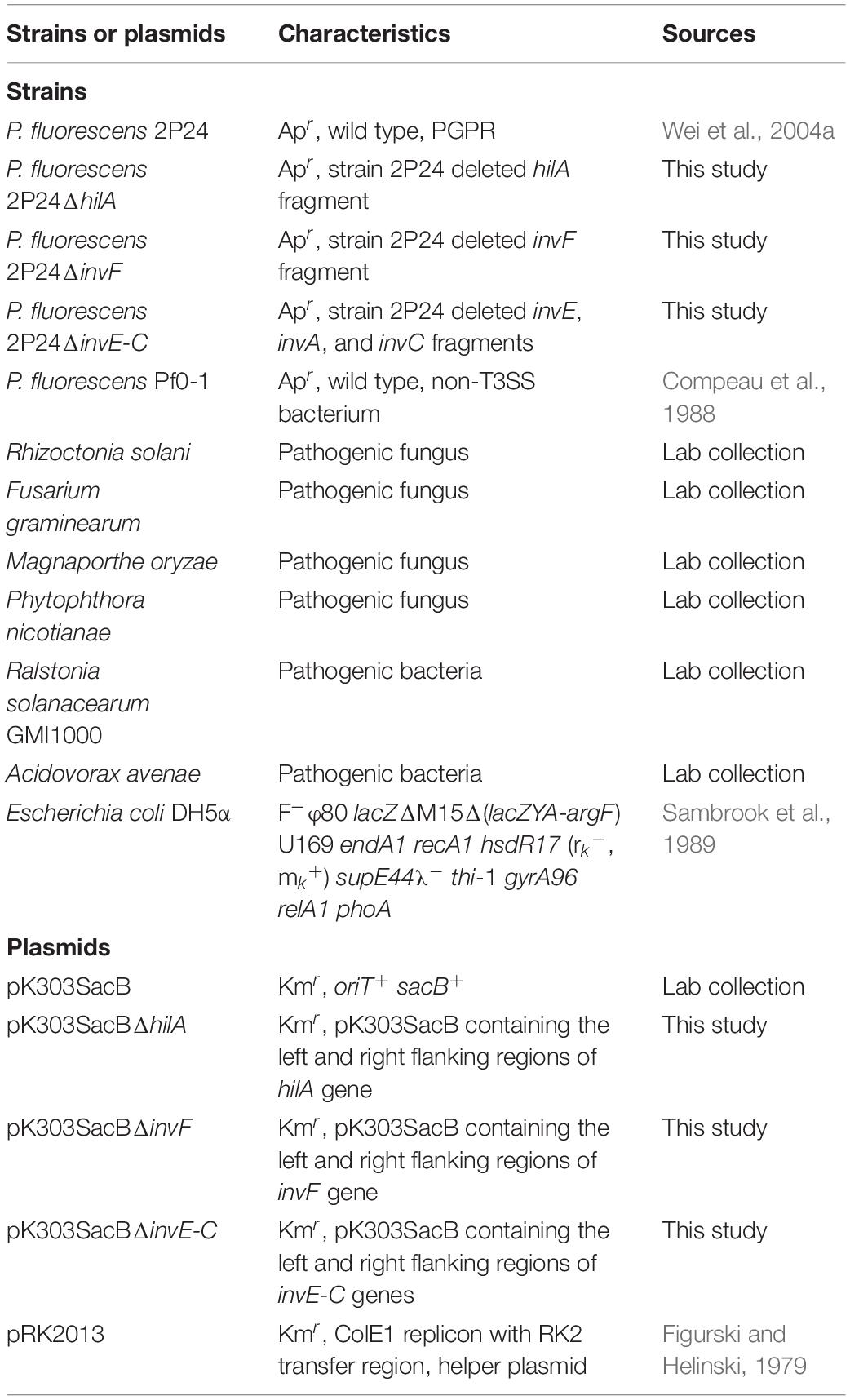
Strains, Plasmids, and Plants
Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C. P. fluorescens was grown in King’s medium B (KB) broth or mannitol-glutamate (MG) minimal medium at 28°C. Pathogenic fungi were grown on Potato Dextrose Agar (PDA) at 28°C, except Phytophthora, which was grown on V8 juice agar. Pathogenic bacteria Ralstonia solanacearum and Acidovorax avenae were grown in Nutrient Broth (NB) at 28°C (Table 1). The following concentrations of antibiotics were used: ampicillin at 50 μg/mL and kanamycin at 50 μg/mL. Nicotiana benthamiana was grown in a greenhouse with 16 h light/8 h dark, 65% humidity, and temperatures of 24°C during daylight and 22°C at night.
Bioinformatic Analyses
To locate the SPI-1 T3SS gene cluster, we performed a BlastP search in the complete genome of strain 2P24 (GenBank accession: CP025542.1), using protein sequences of T3SS conserved components that were identified previously in Salmonella enterica (InvA: QBG67165.1, SpaP: EBB2042894.1, and PrgK: NUD93968.1). DNAMAN Version 9 was used to analyze the identity of T3SS components between P. fluorescens 2P24, P. kilonensis F113, and S. enterica. To produce a phylogenetic analysis of SPI-1, we searched the National Center of Biotechnology Information (NCBI) database for InvA protein sequences and 16S rRNA sequences in five genera: Pseudomonas, Salmonella, Shigella, Yersinia, and Burkholderia. Phylogenetic trees were created with MEGA7 (Kumar et al., 2016) using the Maximum Likelihood method.
For scanning InvF binding sites from intergenic regions of strain 2P24, Geneious Prime® (Biomatters Ltd) was used to generate intergenic regions by retrieving the genome sequence of strain 2P24. Then, we input the sequence matrix of the InvF binding site ‘‘ANNGGNCNTTTTTTNAANGTT’’ and the intergenic region file to the Find Individual Motif Occurrences (FIMO)1 (Grant et al., 2011); we set the p value ≤ 10–5.
Construction of the hilA, invF, and invE-C Mutants
To make a hilA mutant of P. fluorescens 2P24, a 2.8 kb fragment that covered the full length of the hilA gene was amplified from strain 2P24 using primer pairs WHL140 (5′-TTCTAAGCTTGAGCAACAGCAGCG-3′) and WHL141 (5′-GGACACGCCACTTCTAGAAGTTGG-3′) and digested with HindIII and XbaI. The fragment was cloned into the same sites of pK303SacB. The resulting pK303SacB derivative was digested with NarI to eliminate a 0.9 kb fragment of the hilA gene from the insertion. The parental fragment was recycled and self-ligated. The final pK303SacB derivative pK303SacBΔhilA was constructed and transformed into strain 2P24. Colonies were selected from KB medium with 10% sucrose. Then kanamycin-sensitive mutants were selected and screened by PCR.
To make an invF mutant of strain 2P24, two 1.0 kb fragments that carried the left and right flanking regions of invF were amplified by PCR using primer pairs WHL92 (5′-CCAAAGCTTCAGGACTGGTCACGCC-3′) and WHL93 (5′-GGCATCGCTCATGTCGAAAAAAGC-3′), and primer pairs WHL94 (5′-CTCGCTGACCGATGTGGCACT-3′) and WHL95 (5′-TCGAGGATCCACAACGACAACTC-3′), respectively. The left and right regions were digested with HindIII and BamHI, respectively, and were cloned into the relevant sites of pK303SacB. The resulting pK303SacB derivative pK303SacBΔinvF was transformed into strain 2P24. The correct mutant was screened using the same method as hilA mutant, and the same protocol was used to make an invE-C mutant. The left and right flanking regions of invE-C were amplified by PCR using primer pairs WHL15 (5′- TGGCGAATTCATCCTCGGAACAAG-3′) and WHL16 (5′- CTGACAATTCGTCGCTGATCTGC-3′), and primer pairs WHL17 (5′-GCGTCTCGAGCAACTGCAGGTCT-3′) and WHL18 (5′-ATCAAGCTTGTATCGGCAGCGGT-3′), respectively. The left and right regions were digested with EcoRI and HindIII, respectively, and then cloned into the relevant sites of pK303SacB.
RNA Preparation
Strains 2P24, 2P24ΔinvF, and 2P24ΔhilA were grown on KB plates with 50 μg/mL ampicillin at 28°C. Fresh lawns were suspended in 5 mL of KB and incubated at 28°C overnight with 200 rpm shaking. Bacterial suspension was diluted in 50 mL of MG medium in flasks to a final OD600 of 0.1. The cultures were incubated at 28°C at 200 rpm on a rotary shaker for 6 h/12 h. The cell fractions were separated by centrifugation at 10, 000 rpm for 5 min at 4°C. Total RNA was isolated using a E.Z.N.A.® Bacterial RNA Kit (Omega Bio-tek, United States) as described by the manufacturer for RNA-seq and qRT-PCR.
RNA-Seq and qRT-PCR
A prokaryotic chain specific sequencing library was constructed using a NEBNext Ultra Directional RNA Library Prep Kit and then sequenced on an Illumina HiSeq platform. Gene expression was calculated using HTseq software (V 0.6.1) based on the FPKM (Expected number of Fragments Per Kilobase of transcript sequence Per Millions base pairs sequenced) method. Genetic differences were analyzed using DESEQ2 (V1.6.3) in the Bioconductor software package. For qRT-PCR, a FastKing RT Kit (With gDNase) (Tiangen, China) was used to synthesize cDNA from 50 ng-2 μg of total RNA. Luna Universal qPCR Master Mix (New England Biolabs, United States) was used for a real time PCR reaction to quantify the cDNA level of target genes in different samples. Reactions were run and data were collected using the ABI QuantStudio6 Flex real-time PCR system (Applied Biosystems, United States).
In vitro Tests of Pathogenic Antagonism
Antagonistic tests were performed as described by Gu et al. (2020). For antagonism of pathogenic fungi, a 1 cm diameter agar plug with mycelium was placed in the center of an agar plate, and 10 μL cultures of strains 2P24, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C were dropped on the plate at four directions, approximately 3 cm from the center. The plates were incubated at 28°C and checked for inhibition zones of mycelial growth. For antagonism of pathogenic bacteria, we incubated pathogenic bacteria in 5 mL of NB overnight and diluted in melting Nutrient Agar (NA) at a ratio of 1:100 for preparing plates. The cultures of strains 2P24, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C were inoculated and observed as above.
Motility Tests
We conducted motility tests as described by Rashid and Kornberg (2000), with slight modification. For swimming tests, we used KB medium that contained 0.2% agar, and for swarming tests, we used KB medium that contained 0.5% agar and 0.5% glucose. Freshly cultured strains were dipped using pipette tips and inoculated on the surface of the plates’ center. For twitching tests, we used KB medium that contained 1% agar. Freshly cultured strains were dipped using pipette tips and inoculated into the bottom of the plates’ center. Then, plates were placed stably in the incubator and cultured at 28°C.
The Chemotactic Detection of Glucose
Chemotactic responses of strain 2P24 and SPI-1 related mutants to glucose were performed according to a simplified capillary assay method (Mazumder et al., 1999). Bacteria grew to late log phase in KB liquid medium. Pellets were collected by centrifugation and washed twice with BHA (NH4NO3, 1 g/L; FeCl3, 0.05 g/L; KH2PO4, 1 g/L; K2HPO4, 1 g/L; MgSO4, 0.2 g/L; CaCl2, 0.02 g/L; pH 7.0) (Ugochukwu et al., 2013). Finally, bacteria were suspended into BHA (OD600 = 1.0). One hundred microliters of bacterial suspension were sucked into a 200 μL pipette tip, and 100 μL BHA that contained 0.1% glucose was drawn up through the needle into a 1 mL syringe, and 100 μL BHA without carbon source was drawn up through the needle into a 1 mL syringe as negative control. The needle-syringe capillary was inserted into the pipette tip that contained the bacterial suspension. After 45 min of incubation at room temperature, the contents from the needle-syringe were removed and diluted in 25 mM PBS buffer (pH 7.0) and plated onto LB medium. Accumulation in the capillary was calculated from the CFUs on the plates.
Biofilm Formation Test
The assay of biofilm formation was performed according to the classic approach (Christensen et al., 1982). One hundred microliters cultures of bacterial strains that were incubated overnight were added to 2 mL centrifuge tubes, and 900 μL KB was added to each tube. Bacterial strains were incubated at 28°C for 48 h statically. Then, the contents of each tube were emptied and washed three times with sterile water. The tubes were stained for 15 min with 200 μL of 1% crystal violet and then washed with sterile water. After the tubes were air dried, the dye bound to the adherent cells was resolubilized with 2 mL of 95% ethyl alcohol per tube. The OD of each tube was measured at 570 nm by using a L3S visible spectrophotometer (INESA, China).
Assay for Reactive Oxygen Species
For ROS assays, bacterial strains were injected into 5-wk-old N. benthamiana leaves at 5 × 108 cfu/mL. Leaf disks were excised 12 h later with a 0.5 cm diameter cork borer and placed into wells of 96-well plates to which 10 μL ddH2O had been pre-added. Then, 100 μL of 0.5 mM L-012 (Wako, Kyoto, Japan) in 10 mM morpholinepropanesulfonic acid-KOH buffer (pH 7.4) was added. Chemiluminescence was monitored immediately for 10 h using a Veritas luminometer (GENios Pro, TECAN, Switzerland).
Statistical Analyses
All tests in this study were performed at least triplicate, and standard deviations were calculated. One-way ANOVA was performed to determine significant changes.
Results
Characterization of the SPI-1 T3SS Components in Pseudomonas fluorescens 2P24
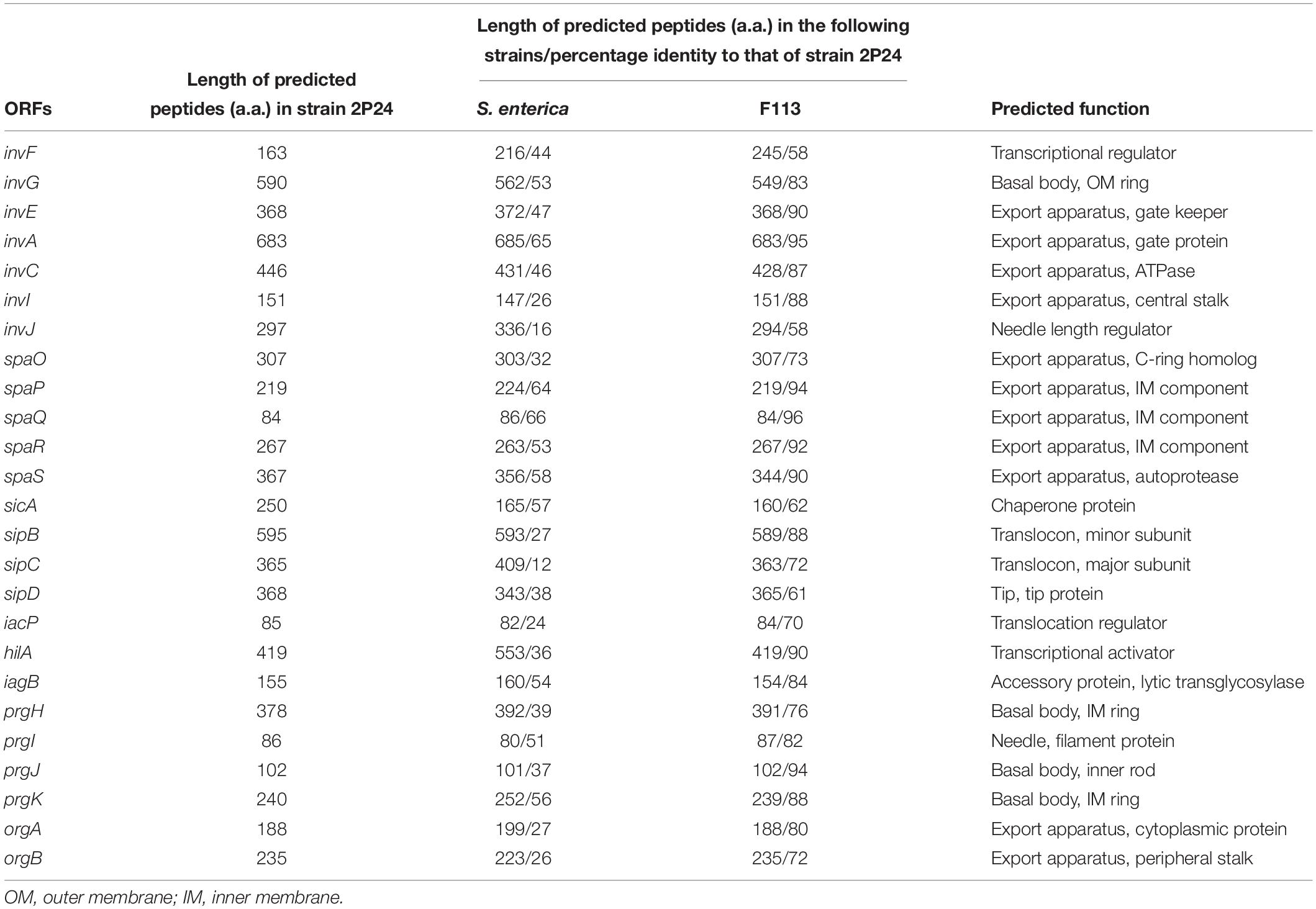
Through genome mining and comparison, a 23 kb SPI-1 T3SS gene cluster comprised of 25 predicted open reading frames (ORFs) was identified in P. fluorescens 2P24, and the gene names were assigned according to the SPI-1 T3SS of S. enterica. Comparing SPI-1 T3SS gene clusters of PGPR and mammalian pathogens indicated that the organization and orientation of SPI-1 T3SS in strain 2P24 were very close to those in S. enterica and P. kilonensis F113 (Figure 1). The SPI-1 T3SS of strain 2P24 retained all assembly proteins except InvH, which indicated that the structure of SPI-1 T3SS in strain 2P24 was conserved. Two transcriptional regulator homologs, HilA and InvF, were retained in strain 2P24. However, compared with the SPI-1 T3SS in S. enterica, effector proteins such as SipA, SptP, and OrgC were lost, and only the chaperone protein SicA was retained in strains 2P24 and F113. Amino acid sequence analysis of the SPI-1 T3SS components in strain 2P24 showed higher identity to strain F113 than to Salmonella (Table 2). The export apparatus (i.e., InvA, SpaP, SpaQ, SpaR, and SpaS), basal body proteins (InvG and PrgK), accessory protein (IagB), needle (PrgI), and chaperone protein (SicA) displayed >50% identity between S. enterica and strain 2P24, although the remaining accessory proteins (i.e., InvE, InvC, InvI, SpaO, PrgH, PrgJ, OrgA, and OrgB) showed less identity (between 25 and 50%). In addition, less conserved regulators and translocons suggested a changed function of SPI-1 T3SS in PGPR.
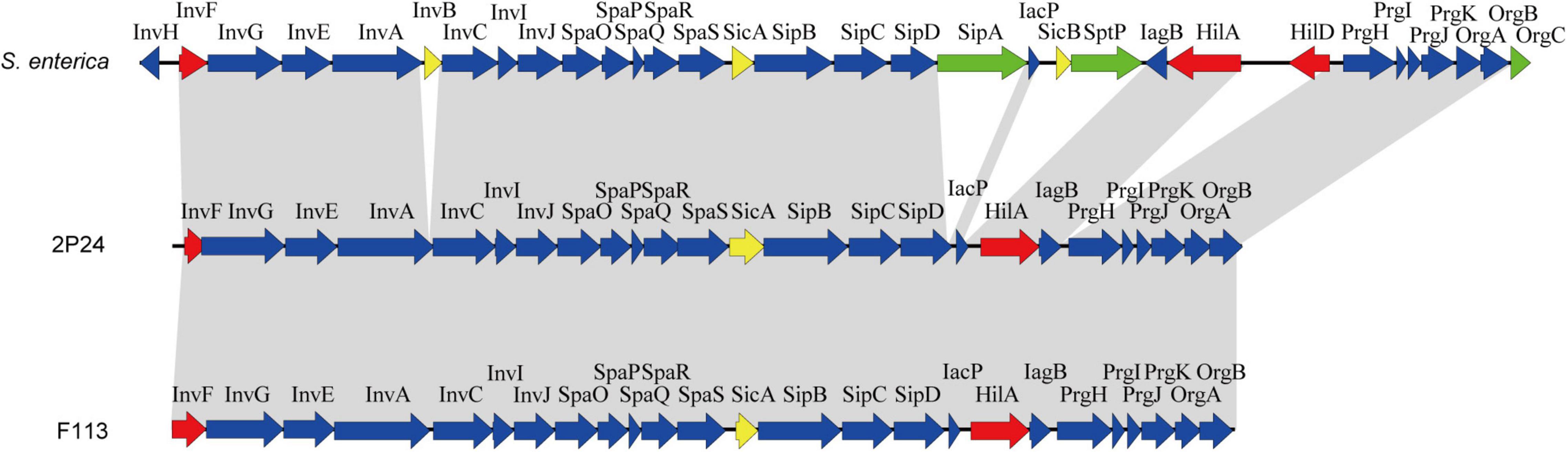
Figure 1. Genetic organization of the SPI-1 T3SS cluster of P. fluorescens 2P24 and comparison with the clusters of Salmonella enterica and P. kilonensis F113. Predicted open reading frames and their orientation are shown by large arrows. T3SS accessory protein encoding genes are shown as blue arrows, transcriptional regulator encoding genes are shown as red arrows, effector protein encoding genes are shown as green arrows, and chaperone protein encoding genes are shown as yellow arrows. Homologous genes are connected with gray shading.
Phylogenetic Analysis of SPI-1 T3SS Among Beneficial and Pathogenic Bacteria
A few reports showed that the SPI-1 T3SS was present in some plant beneficial bacteria, especially in fluorescent Pseudomonas (Chauhan et al., 2011; Loper et al., 2012). To investigate the relationship of SPI-1 T3SS among beneficial and pathogenic bacteria, the 16S rRNA sequences and InvA amino acid sequences were compared among Pseudomonas, Salmonella, Shigella, Yersinia, and Burkholderia spp. Based on alignment of the 16S rRNA, the maximum likelihood method was used to construct a phylogenetic tree that depicted evolutionary distance among these T3SS-containing bacteria (Figure 2A). There were two clades in the tree, and all of the fluorescent Pseudomonas strains were located on a separate subbranch of one clade, which indicated a distant evolutionary relationship with mammalian pathogens. Then, the SPI-1 T3SS phylogenetic tree was constructed based on InvA amino acid sequences using the same method (Figure 2B). The phylogenetic tree contained two clades, and fluorescent Pseudomonas strains were located in a subbranch together with Shigella spp. and Salmonella spp. The difference in phylogenetic relationship between 16S rRNA and InvA suggested a horizontal transfer of SPI-1 T3SS from mammalian pathogens, such as Salmonella and Shigella, to plant beneficial fluorescent Pseudomonas.
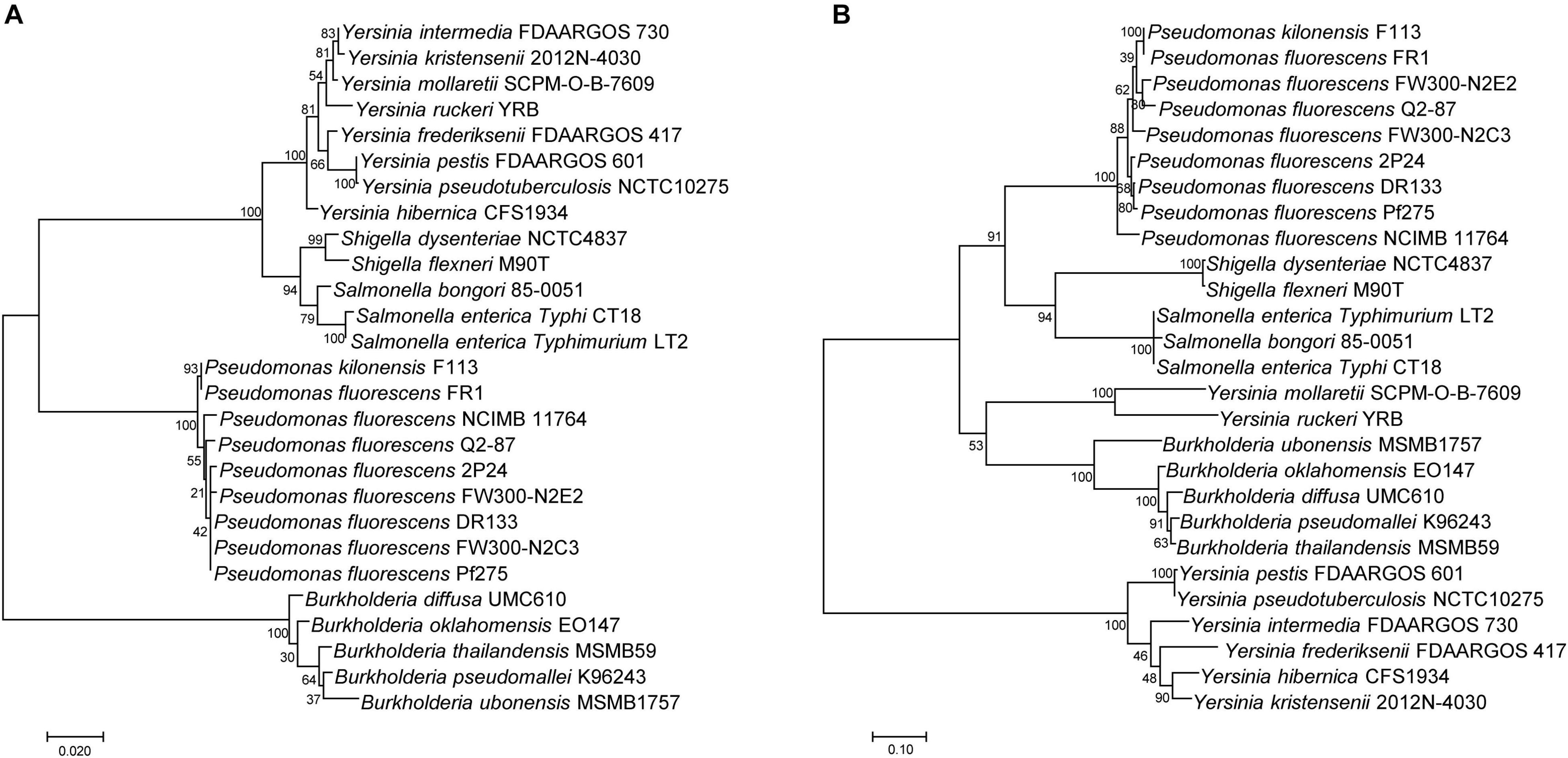
Figure 2. Phylogenetic analysis of SPI-1 T3SS gene clusters of the P. fluorescens group. (A) A distance tree was calculated from 16S rRNA homologs by using the Maximum Likelihood method based on the Tamura-Nei model. (B) A distance tree was calculated from InvA homologs by using the Maximum Likelihood method based on the JTT matrix-based model.
HilA Up-Regulates the Structural Genes of SPI-1 T3SS
HilA is a transcriptional activator, which regulates transcription of the SPI-1 T3SS apparatus genes directly (Ellermeier and Slauch, 2007). To verify whether HilA had a similar function in strain 2P24, the hilA deficient mutant 2P24ΔhilA was constructed, and the expression of five apparatus genes (invG, sipB, sipD, prgI, and prgK) of the SPI-1 T3SS complex in strains 2P24 and 2P24ΔhilA was detected by qRT-PCR. Compared to strain 2P24, the expression of these five apparatus genes was decreased significantly in 2P24ΔhilA (Figure 3). HilA of strain 2P24 had a transcriptional activation function similar to mammalian pathogens and regulated SPI-1 apparatus genes positively.
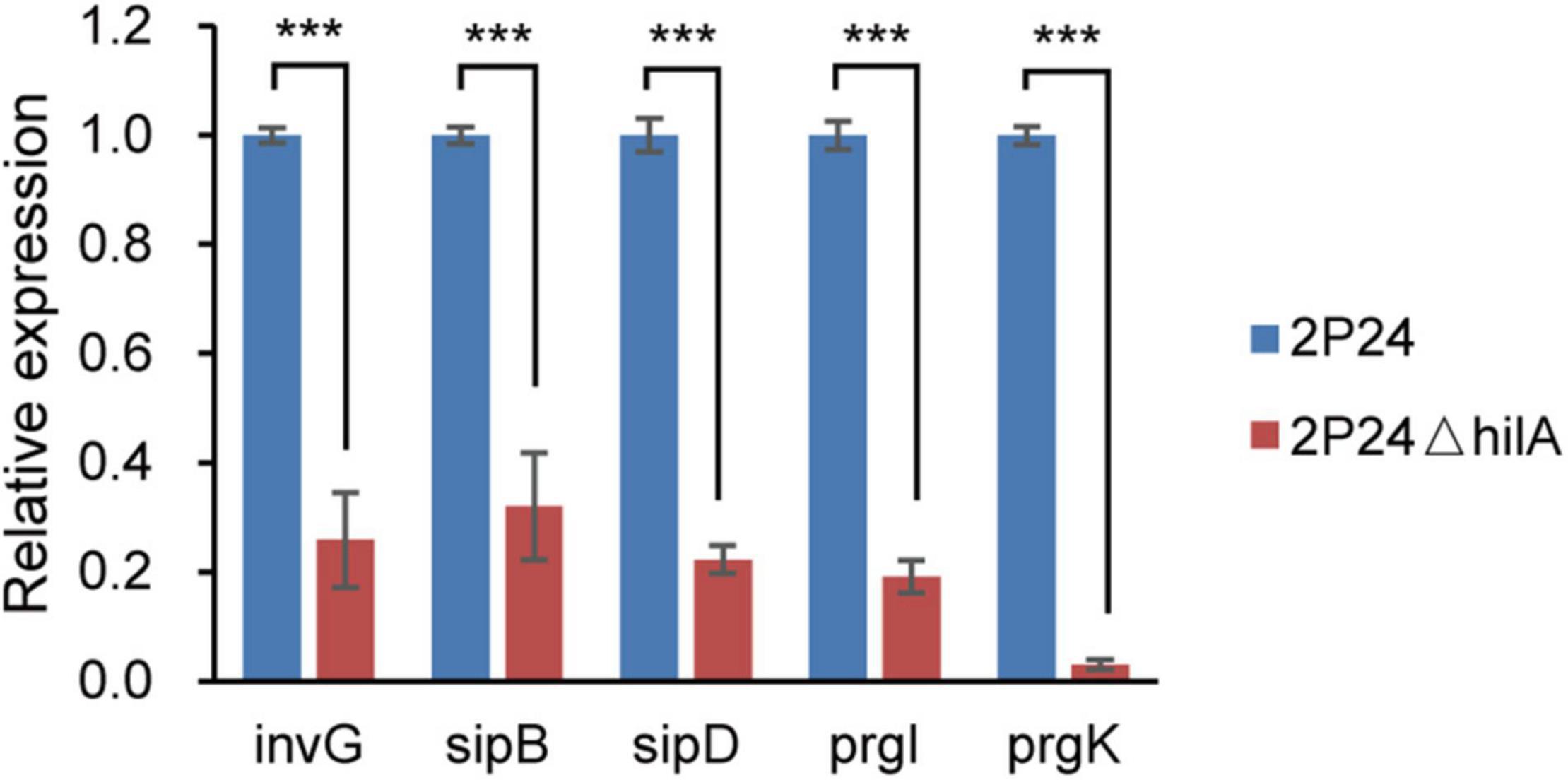
Figure 3. Expression profiles of invG, sipB, sipD, prgI, and prgK in strains 2P24 and 2P24ΔhilA by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Graph shows gene expression levels in strains 2P24 or 2P24ΔhilA at 12 h after being incubated in MG medium, as quantified by qRT-PCR. Gene expression was normalized to the expression level of reference gene recA using the 2–ΔΔCt method. Data are means of three replicates. Error bars represent SD. *** represents P < 0.001.
Identification of the Regulon of Transcriptional Regulator InvF
InvF, an AraC-type transcriptional activator, regulated the expression of genes that encoded the secreted effector molecules SipABCD, SigD, SptP, and SopE (Darwin and Miller, 1999; 2001). However, it seems that almost all effector homologs were lost in strain 2P24 (Figure 1). To identify the InvF regulon in strain 2P24, the invF deficient mutant 2P24ΔinvF was constructed, and we implemented transcriptome sequencing (GEO accession: GSE181272). A total of 127 significantly differentially expressed genes (DEGs) were obtained by comparing the transcriptional level of strain 2P24ΔinvF with 2P24 at 6 h and 12 h [Fold change ≥ 1.5; p value ≤ 0.05] (Figure 4A and Supplementary Table 1). KEGG analyses indicated that DEGs at 6 h and 12 h involved biosynthesis of secondary metabolites, biosynthesis of amino acids, and ABC transporters (Supplementary Figure 1). Fifty-two down-regulated genes were regarded as InvF regulon candidates. Among the InvF regulon candidates, 39 genes were selected for qRT-PCR verification and eight genes were confirmed to be regulated positively by InvF (Figure 4B).
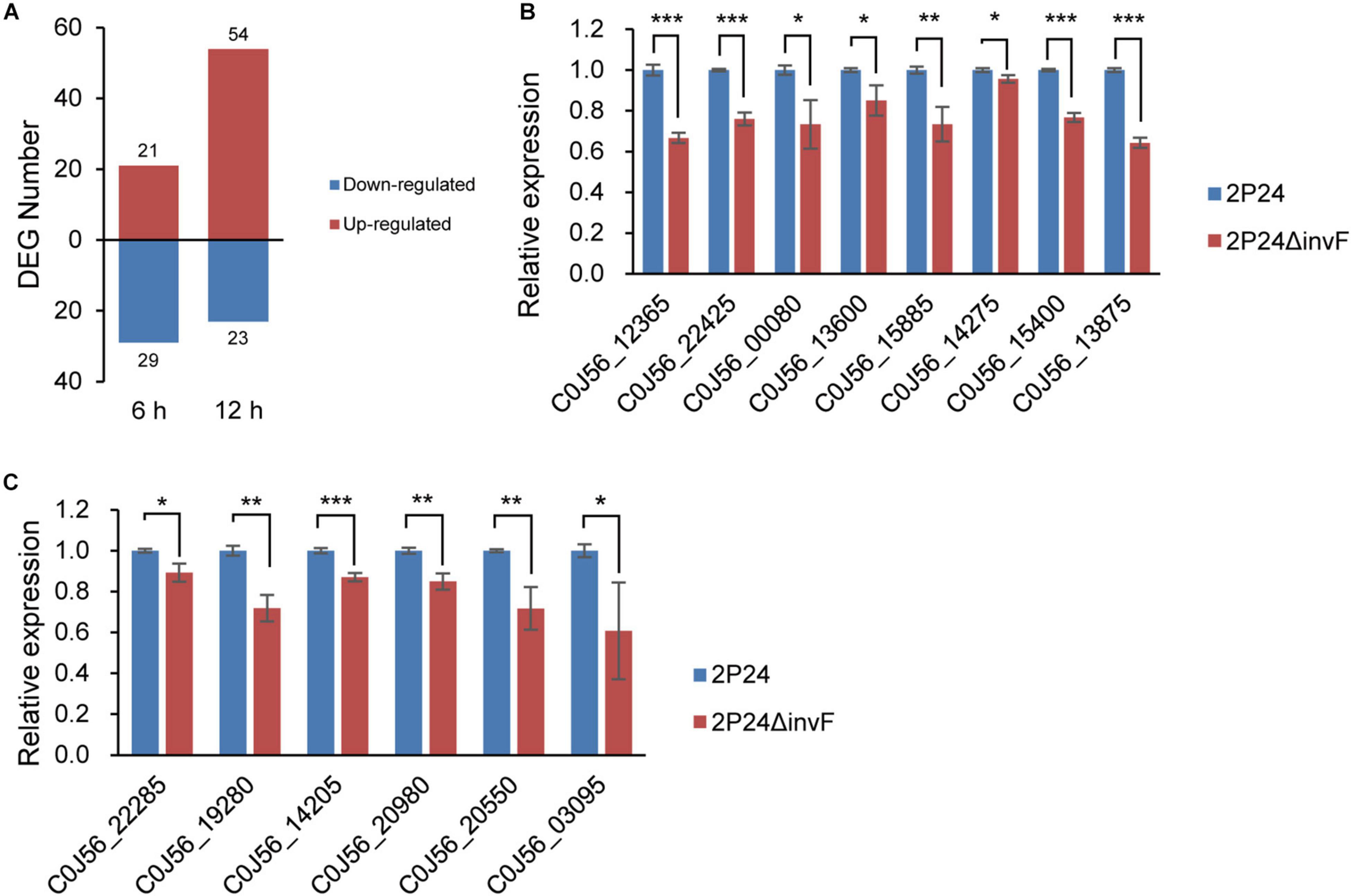
Figure 4. Identification of InvF regulons. (A) Number of up-regulated (red bars) and down-regulated (blue bars) differentially expressed genes (DEGs) from RNA-seq analysis of 2P24ΔinvF compared with strain 2P24 at 6 and 12 h after being incubated in MG medium [Fold change ≥ 1.5; p value ≤ 0.05]. (B) Expression profiles of InvF up-regulated genes selected from RNA-seq DEGs in strains 2P24 and 2P24ΔinvF by qRT-PCR. Graph shows gene expression levels in strains 2P24 or 2P24ΔinvF at 6 or 12 h after being incubated in MG medium, as quantified by qRT-PCR. (C) Expression profiles of InvF up-regulated genes selected from the adjacent downstream genes of predicted InvF binding sites in strains 2P24 and 2P24ΔinvF by qRT-PCR. Graph shows gene expression levels in strains 2P24 or 2P24ΔinvF at 6 h after being incubated in MG medium, as quantified by qRT-PCR. Gene expression was normalized to the expression level of reference gene recA using the 2–ΔΔCt method. Data are means of three replicates. Error bars represent SD. *, **, and *** represent P < 0.05, P < 0.01, and P < 0.001, respectively.
Previous study found an InvF binding site in the promoter regions of sicA, sigD, and sopE that was recognized by InvF to activate downstream expression of genes (Darwin and Miller, 2001). To identify the InvF regulon more comprehensively, the intergenic regions of the genome of strain 2P24 were scanned for the InvF binding site using the FIMO website (see text footnote 1) (Grant et al., 2011). Eleven InvF binding sites were obtained at p value ≤ 1 × 10–5 (Supplementary Table 2). The adjacent downstream genes of these binding sites were searched, and their expression in strains 2P24 and 2P24ΔinvF was verified by qRT-PCR (Supplementary Table 2). Six genes among them were regulated positively by InvF (Figure 4C). Finally, 14 genes were identified as InvF regulons through transcriptional and promoter analysis (Table 3). However, all of the InvF regulons are not homologous to any effector proteins in pathogenic bacteria.
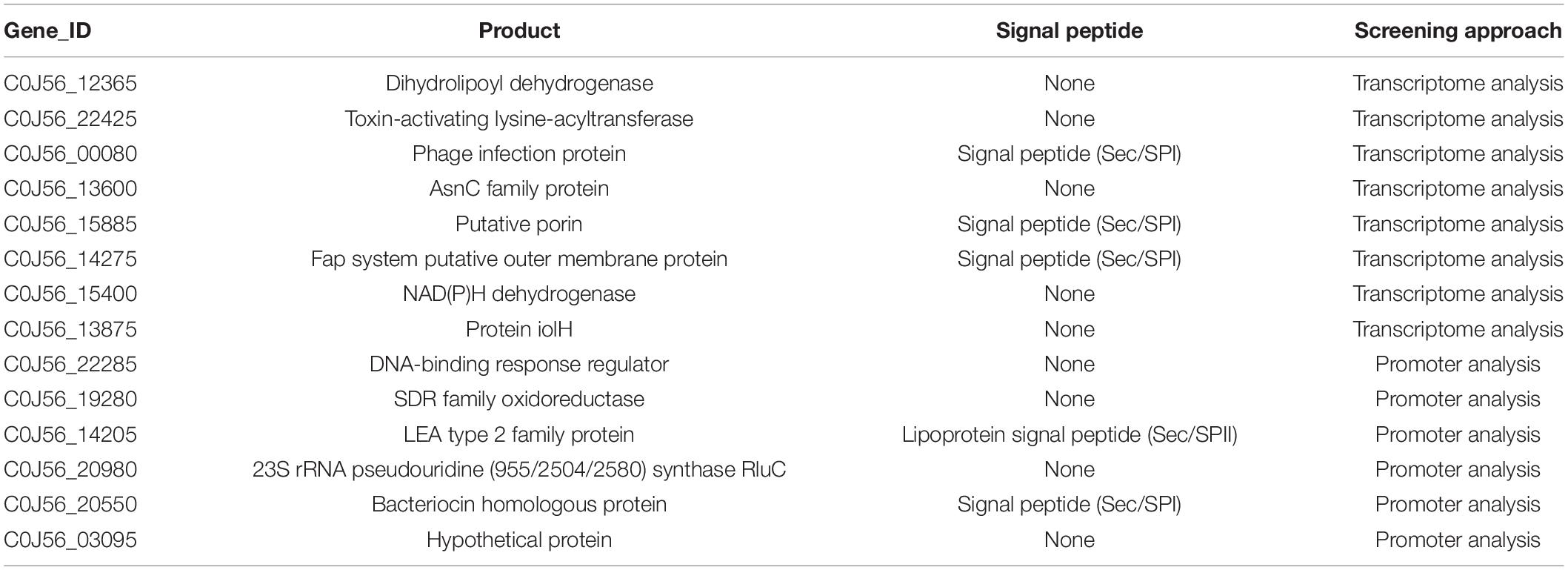
Table 3. InvF regulons up-regulated by InvF and the predicted information about signal peptide of each protein sequence.
InvF Partially Affects Antagonism of Strain 2P24 Against Fusarium graminearum
A previous study found that strain 2P24 antagonized multiple plant pathogens (Wei et al., 2004a). To investigate whether SPI-1 T3SS was involved in the antagonism, the T3SS deficient mutant 2P24ΔinvE-C was constructed. Then, the antagonistic ability of strains 2P24, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C against various plant pathogens was detected. 2P24ΔinvE-C and 2P24ΔhilA exhibited no significant difference with strain 2P24 in antagonism (Figure 5). 2P24ΔinvF also showed no significant difference with strain 2P24 in antagonism to a variety of pathogens, but it reduced antagonistic ability against F. graminearum significantly. We speculated that InvF might influence antagonism to F. graminearum indirectly by regulating the expression of some genes rather than the T3SS.
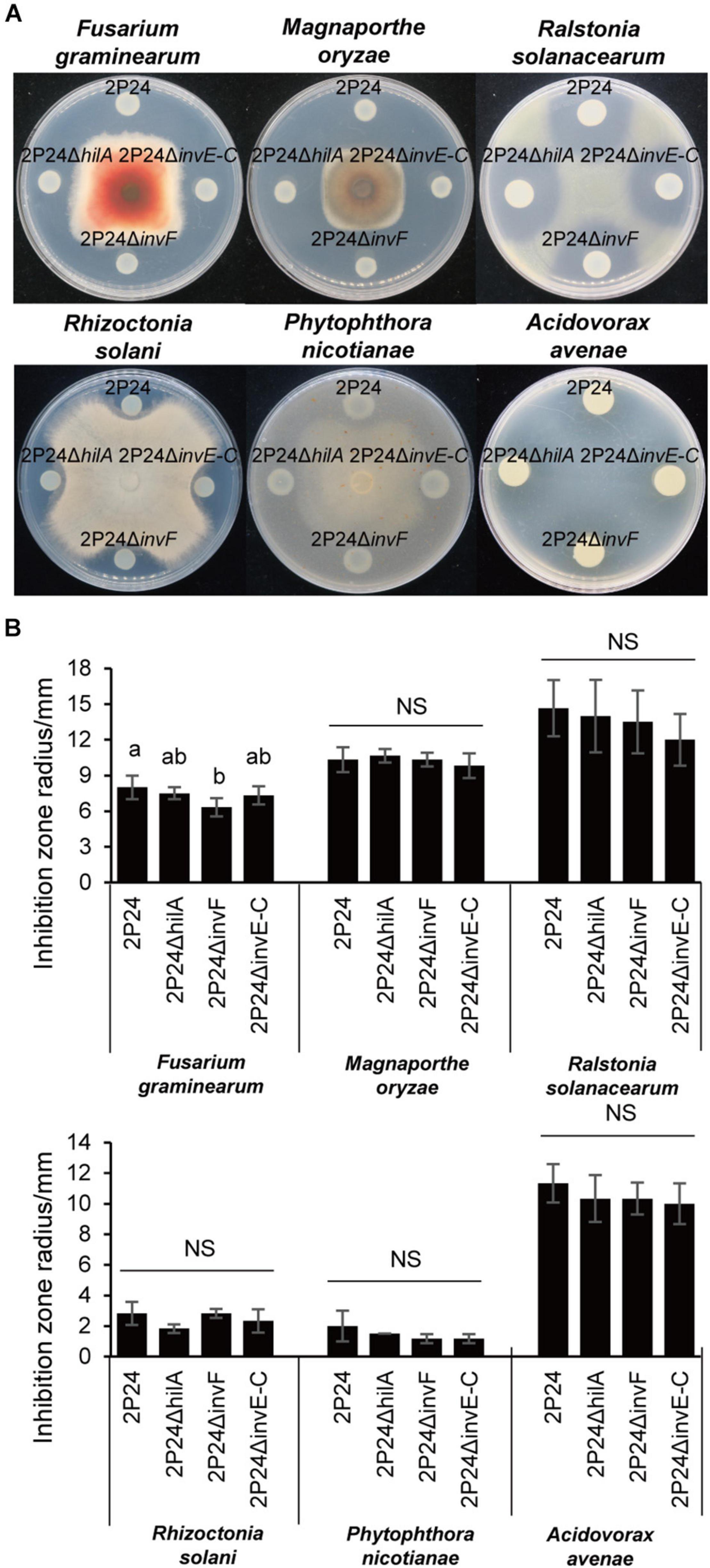
Figure 5. Antagonism of strain 2P24 and SPI-1 related mutants to plant pathogens. (A) Antagonistic effect on pathogenic fungi Fusarium graminearum, Magnaporthe oryzae, Rhizoctonia solani, and Phytophthora nicotianae and pathogenic bacteria Ralstonia solanacearum and Acidovorax avenae. (B) Quantitative analysis of antagonistic effect. Data are means of three replicates. Error bars represent SD. Means shown with the same letters are not different statistically at the 5% confidence level on the basis of Duncan’s multiple range test. NS means no significant difference.
SPI-1 T3SS Is Not Associated With Motility but Involved in Chemotaxis of Strain 2P24
Bacteria can carry out single bacterial swimming and multiple bacterial swarming by the rotational movement of flagella. In addition, the expansion and contraction of type IV secretion system (T4SS) can drive twitching (Henrichsen, 1972). To explore the influence of SPI-1 T3SS in the motility of strain 2P24, we detected swimming, swarming, and twitching of strain 2P24 and the SPI-1 mutants. Strains 2P24, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C exhibited similar swimming, swarming, and twitching motility under different concentrations of agar (Figure 6A). Statistical analysis showed that no significant difference was found between the strains (Figure 6B).
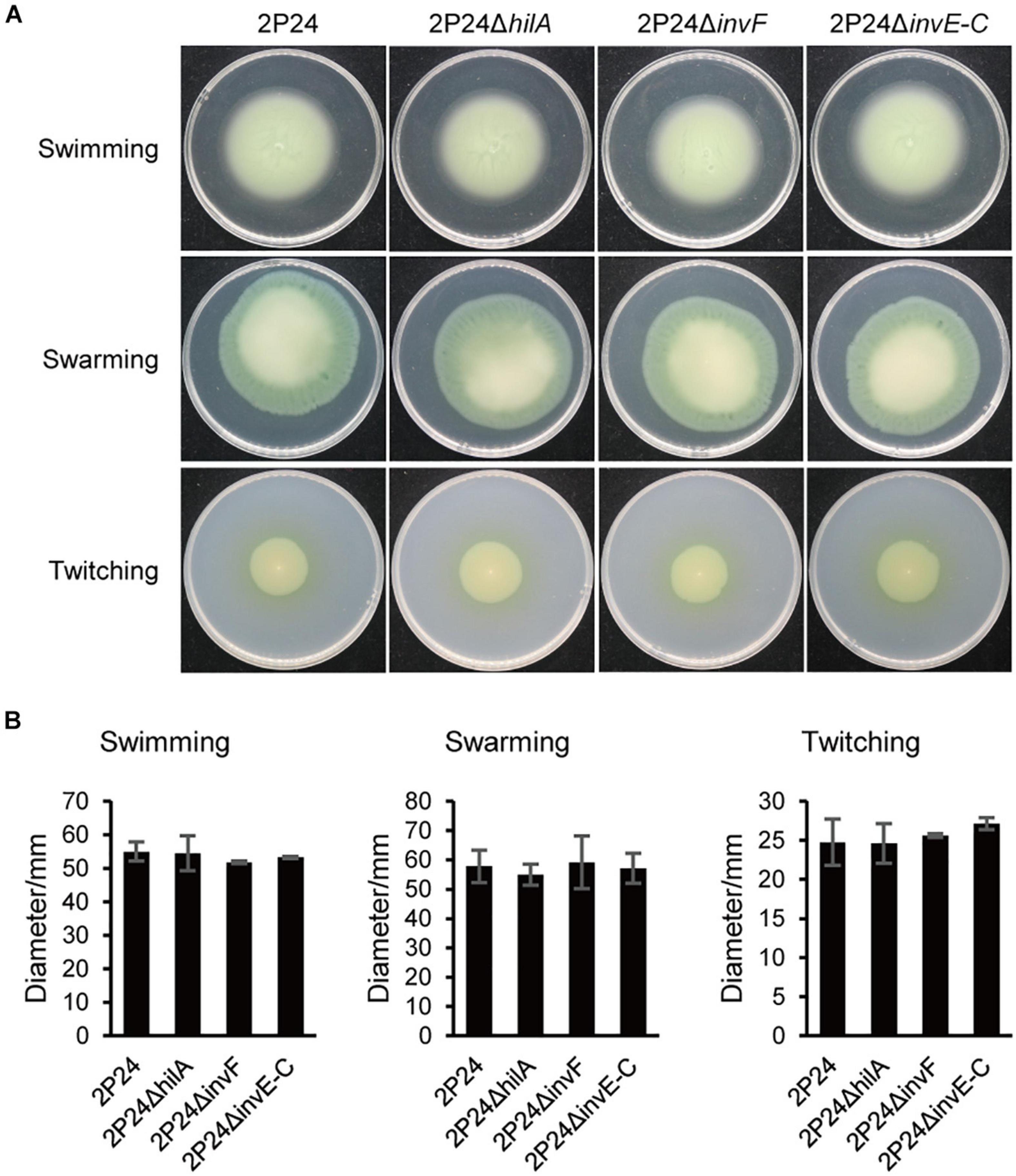
Figure 6. Motility of strain 2P24 and SPI-1 related mutants. (A) Swimming on the 0.2% agar-contained KB medium for 42 h, swarming on the 0.5% agar-contained KB medium for 48 h, and twitching in the 1% agar-contained KB medium for 96 h. (B) Quantitative analysis of swimming, swarming, and twitching of strain 2P24 and SPI-1 related mutants. Data are means of three replicates. Error bars represent SD.
Bacteria perceive the change of chemical concentration in the environment to produce an approach or a retreat response. This chemotactic behavior usually depends on the motility of flagella and can help bacteria survive better (Chet and Mitchell, 1976). In this study, we tested the influence of SPI-1 T3SS in perceiving exogenous glucose. Strain 2P24 had a strong chemotactic response to glucose. However, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C reduced the chemotactic response to glucose significantly (Figure 7). These results suggest that the SPI-1 T3SS of strain 2P24 is involved in chemotaxis independent on motility.
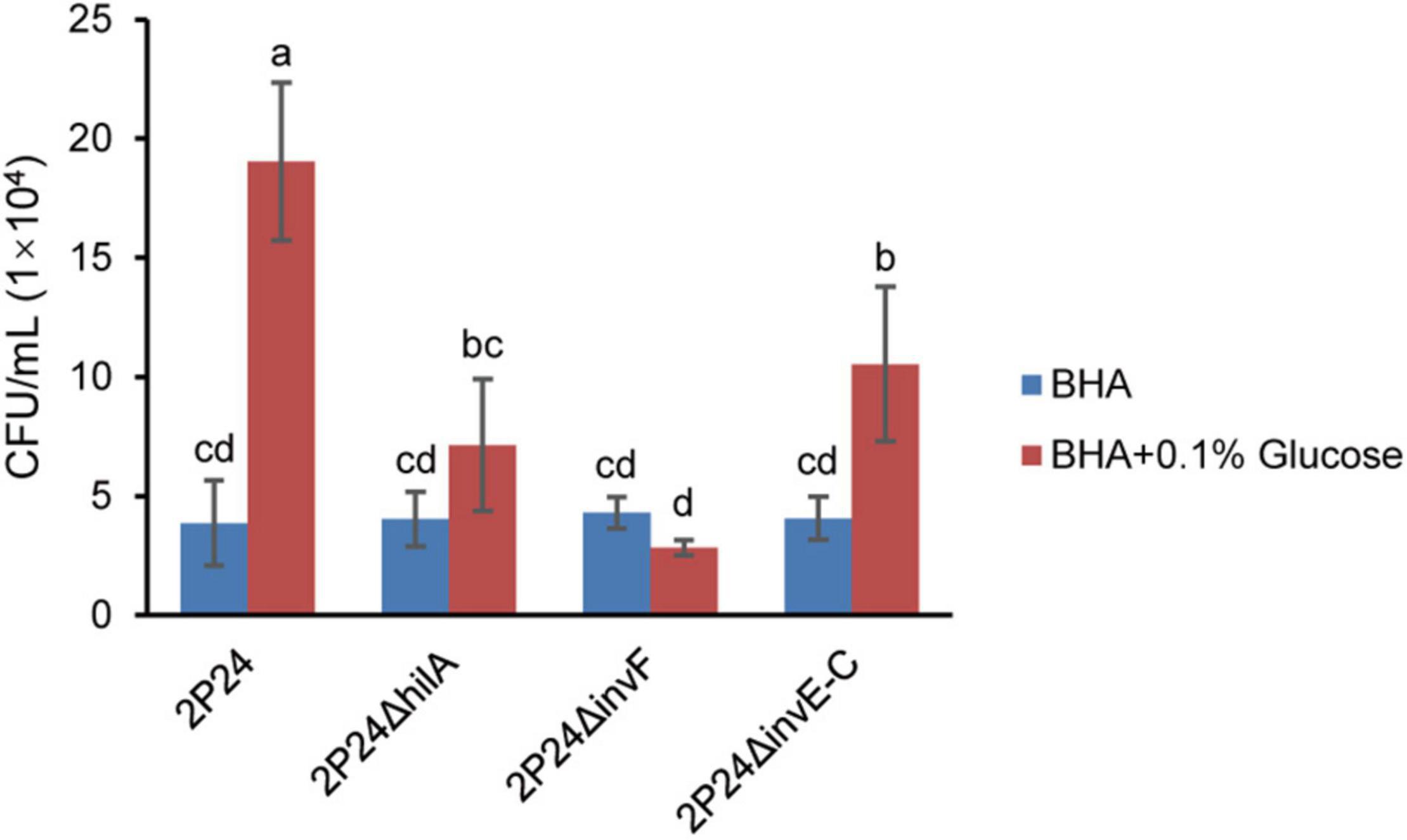
Figure 7. The chemotactic responses of strain 2P24 and SPI-1 related mutants to glucose. Data are means of three replicates. Error bars represent SD. Means shown with the same letters are not different statistically at the 5% confidence level on the basis of Duncan’s multiple range test.
2P24ΔhilA Enhances Biofilm Formation
Biofilm formation of bacteria is related to many factors. Ding and Wang (2009) reported that the T3SS mutant of Yersinia pseudotuberculosis reduced its ability to form biofilm significantly. To verify whether SPI-1 T3SS affected the ability of strain 2P24 to form biofilm, we detected the biofilm of strain 2P24 and related mutants of SPI-1 after static incubation at 28°C for 12, 24, and 48 h in 2 mL centrifuge tubes. Compared with negative control, strain 2P24 formed obvious biofilm at 48 h (Figure 8). 2P24ΔinvF and 2P24ΔinvE-C showed no significant difference with wild type, but 2P24ΔhilA enhanced biofilm formation significantly (Figure 8) which suggest that HilA plays important role in biofilm formation via a transcriptional regulation on some genes. However, the mechanism remains to be explored further.
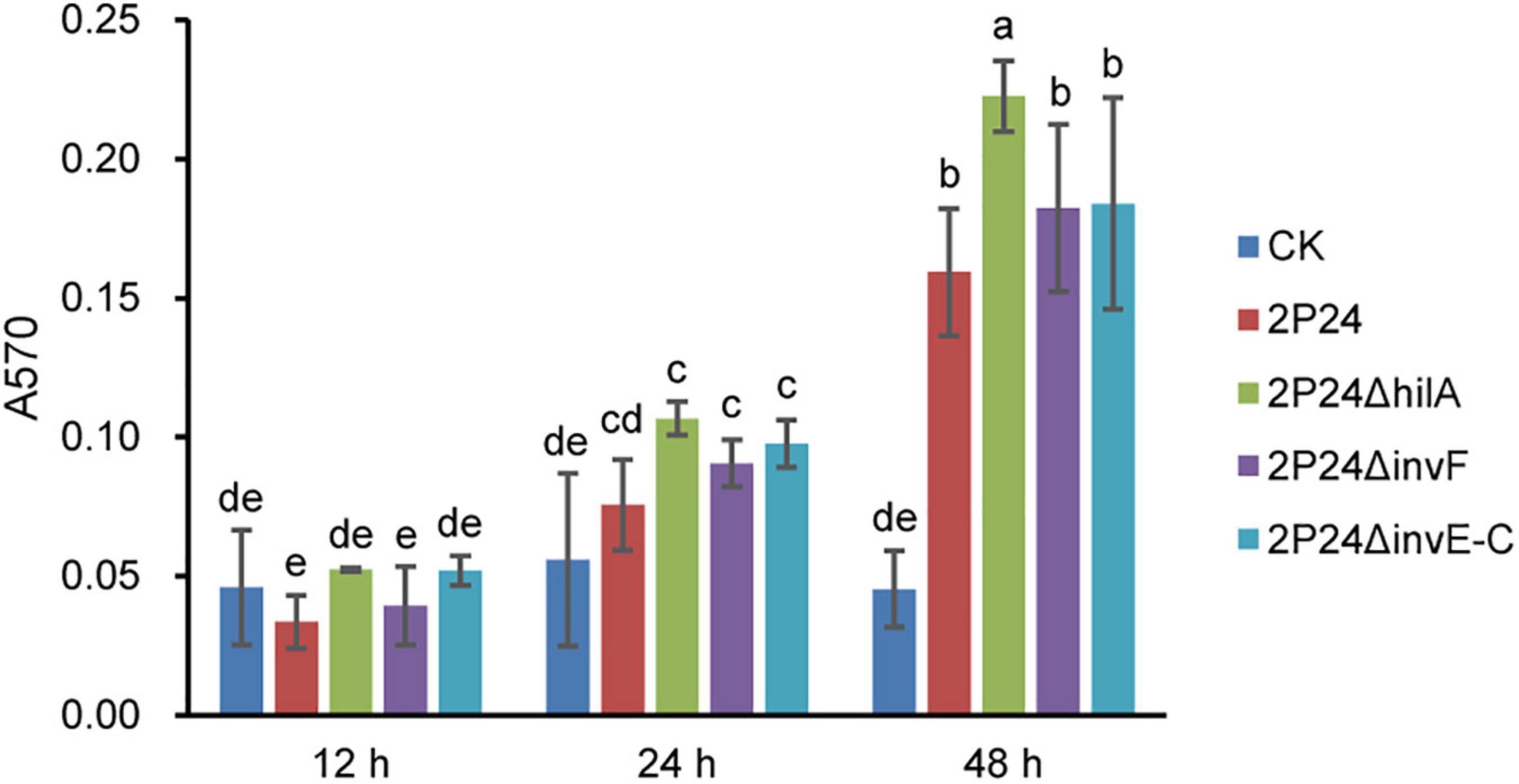
Figure 8. Biofilm formation of strain 2P24 and SPI-1 related mutants incubated in KB medium for 12, 24, and 48 h in 2 mL centrifuge tubes. Data are means of three replicates. Error bars represent SD. Means shown with the same letters are not different statistically at the 5% confidence level on the basis of Duncan’s multiple range test.
2P24ΔinvE-C Reduces the Activation of ROS Burst in Nicotiana benthamiana
Previous study found that strain 2P24 triggered a ROS burst in N. benthamiana leaves (Liu et al., 2016). In this study, we detected whether SPI-1 T3SS was involved in triggering a ROS burst in N. benthamiana. After injection of N. benthamiana with wild type and SPI-1 mutants of strain 2P24 for 12 h, ROS was measured every 5 min for 10 h. Consistent with P. fluorescens Pf0-1, strain 2P24 triggered a strong ROS burst, and deficient mutants of hilA and invF did not influence this immunity (Figure 9). However, 2P24ΔinvE-C reduced the accumulation of ROS significantly (Figure 9). This indicated that the SPI-1 T3SS was involved in triggering a ROS burst in N. benthamiana.
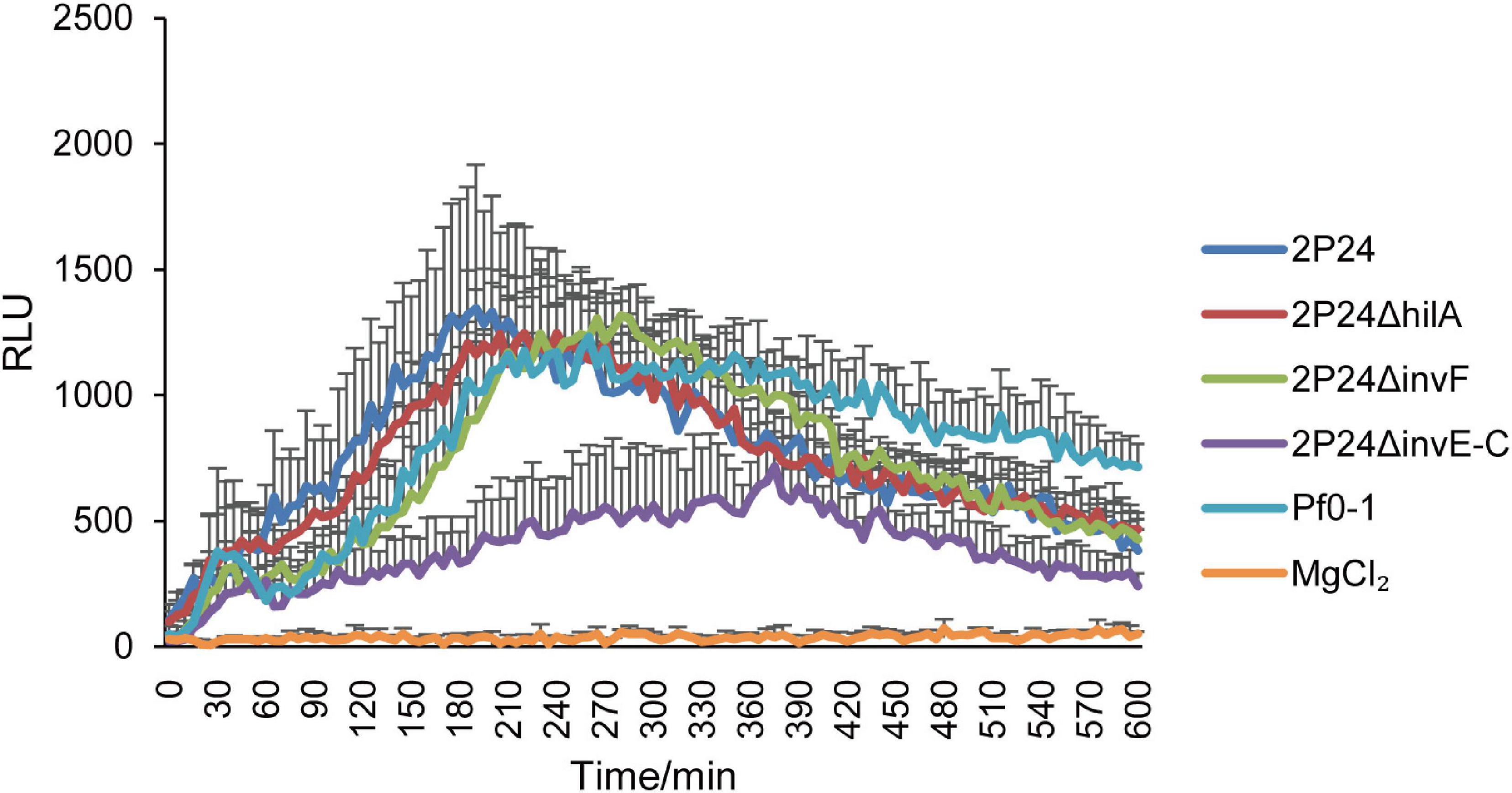
Figure 9. ROS assay of strain 2P24 and SPI-1 related mutants. The bacteria were infiltrated at 5 × 108 cfu/mL into N. benthamiana leaves and 12 h later assayed for ROS burst using L-012 chemiluminescence. P. fluorescens Pf0-1 was used as the positive control and 10 mM MgCl2 as the negative control. Data are means of six replicates. Error bars represent SD.
Discussion
The Salmonella Pathogenicity Island 1 (SPI-1) family T3SS mainly exists in mammalian pathogens as a pathogenic apparatus. Recently, SPI-1 T3SS was also found in plant-associated bacteria such as Xanthomonas (Alavi et al., 2008; Marguerettaz et al., 2011), Erwinia (Triplett et al., 2006), Pantoea (Correa et al., 2012; Kirzinger et al., 2015), and Pseudomonas species (Barret et al., 2013; Redondo-Nieto et al., 2013). However, the function of SPI-1 T3SS in these plant bacteria is unclear. In this study, SPI-1 T3SS of PGPR P. fluorescens 2P24 was characterized, and the function was determined.
A 23 kb SPI-1 T3SS gene cluster was found in the chromosome of strain 2P24, and its organization and orientation were similar to that in S. enterica (Figure 1). This cluster consisted of 25 ORFs encoding structural and regulatory proteins of T3SS, but it had lost almost all of the effector encoding genes (Figure 1 and Table 2). Previous studies have found that beneficial fluorescent Pseudomonas that contained Hrp1 T3SS also lost most of its effector homologs (Jackson et al., 2005; Kimbrel et al., 2010; Marchi et al., 2013; Stringlis et al., 2019). This phenomenon suggests that the function of T3SS in plant beneficial Pseudomonas may have changed compared to pathogens. PGPR strains live in the rhizosphere of plants, relying on the interaction with the surrounding environmental factors, and do not need to transport proteins into host cells. This might result in the loss of effector homologs in P. fluorescens 2P24.
To explore genetic evolution of SPI-1 T3SS further in fluorescent Pseudomonas, phylogenetic trees of 16S rRNA and InvA in Pseudomonas, Salmonella, Shigella, Yersinia, and Burkholderia spp. were constructed (Figure 2). The results suggested that SPI-1 T3SS in fluorescent Pseudomonas might be obtained from Salmonella and Shigella by horizontal gene transfer. A previous study also found that there was horizontal gene transfer of SPI-1 T3SS in Pantoea by analyzing its phylogenetic relationship and GC content (Kirzinger et al., 2015). The increase in recent genome sequences has shown that the homologs of SPI-1 T3SS are present in many other bacterial species, such as Sodalis, Erwinia, Pantoea, Pandoraea, Pectobacterium, Providencia, and Serratia (Egan et al., 2014). A more systematic evolutionary analysis of SPI-1 T3SS is needed.
The SPI-1 T3SS of strain 2P24 retained two transcription factor homologs, HilA and InvF. HilA performed a similar transcriptional activation function to SPI-1 T3SS structural genes as in mammalian pathogens. However, the verified 14 InvF regulons by qRT-PCR were not homologous to any effector proteins in pathogenic bacteria. Among the InvF regulons, five InvF regulons had N-terminal signal peptides (Table 3). Specifically, C0J56_14275 encodes an outer membrane protein of the Fap system which might be involved in the export of amyloid subunits for construction of a biofilm matrix (Rouse et al., 2017). C0J56_20550 is a homolog of bacteriocin which might be associated with antagonism against plant pathogens.
To determine the biological functions of SPI-1 T3SS, strains 2P24, 2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C were detected in antagonism, motility, chemotactic response to glucose, and biofilm formation. Only 2P24ΔinvF reduced resistance to F. graminearum significantly. In addition, SPI-1 T3SS was reported to be involved in the interaction of bacteria- protozoa. For instance, SPI-1 T3SS of P. kilonensis F113 was involved in the resistance to amoeboid grazing (Barret et al., 2013). Whether SPI-1 T3SS of strain 2P24 participates in the resistance to amoeba and nematodes in the soil is worthy of subsequent investigation.
2P24ΔhilA, 2P24ΔinvF, and 2P24ΔinvE-C reduced the chemotactic responses to glucose significantly. Production of bacterial chemotaxis relies on membrane surface chemical-recognition receptors, intracellular signaling systems, and the movement of flagella (Li and Mu, 2006). In this study, we found that SPI-1 T3SS was not associated with motility of strain 2P24. It is possible that the influence of SPI-1 mutants on the glucose chemotactic response may be due to the following reasons: (i) SPI-1 acted as a sensory receptor for glucose in the external environment, and the absence of the SPI-1 T3SS device reduced the sensitivity to glucose, which affected the chemotactic response to glucose; and (ii) transcriptional factors HilA and InvF mutants affected the expression of chemotactic sensing receptors or chemotactic signal-transduction related proteins.
Type III secretion system was recognized redundant with flagella in mammalian and plant pathogens due to their similarities in the secretion systems that serve flagellar biogenesis and type III effector delivery (Miao et al., 2006; Wei et al., 2013). SPI-1 T3SS of Salmonella secreted and translocated flagellin into host cytoplasm, which activated a potent host-defense pathway (Miao et al., 2006). Our results showed that mutation of the SPI-1 T3SS in strain 2P24 reduced the flagellin-triggered ROS burst in planta. This result suggests that the flagellin might be secreted through the SPI-1 T3SS in strain 2P24. Loss of SPI-1 T3SS reduced flagellin output, which led to a decline in ROS burst.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
H-LW designed the research. JW, YL, and YG performed the research. JW and H-LW analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2019YFD1002000), the Program of Science and Technology of Beijing, China (Z191100004019025), the National Natural Science Foundation of China (31871924), and the Central Public-interest Scientific Institution Basal Research Fund (Y2019XK07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.749037/full#supplementary-material
Supplementary Figure 1 | KEGG pathway analyses of DEGs of 2P24ΔinvF that compared with strain 2P24 at 6 h (A) and 12 h (B) after being incubated in MG medium.
Footnotes
References
Alavi, S. M., Sanjari, S., Durand, F., Brin, C., Manceau, C., and Poussier, S. (2008). Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl. Environ. Microbiol. 74, 3295–3301. doi: 10.1128/AEM.02507-07
Barret, M., Egan, F., Moynihan, J., Morrissey, J. P., Lesouhaitier, O., and O’Gara, F. (2013). Characterization of the SPI-1 and RSP type three secretion systems in Pseudomonas fluorescens F113. Environ. Microbiol. Rep. 5, 377–386. doi: 10.1111/1758-2229.12039
Büttner, D. (2012). Protein export according to schedule: architecture, assembly and regulation of type III secretion systems from plant and animal pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310.
Chauhan, A., Layton, A. C., Williams, D. E., Smartt, A. E., Ripp, S., Karpinets, T. V., et al. (2011). Draft genome sequence of the polycyclic aromatic hydrocarbon-degrading, genetically engineered bioluminescent bioreporter Pseudomonas fluorescens HK44. J. Bacteriol. 193, 5009–5010. doi: 10.1128/JB.05530-11
Chet, I., and Mitchell, R. (1976). Ecological aspects of microbial chemotactic behavior. Ann. Rev. Microbiol. 30, 221–239. doi: 10.1146/annurev.mi.30.100176.001253
Christensen, D. G., Simpson, W. A., Bisno, A. L., and Beachey, E. H. (1982). Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37, 318–326.
Compeau, G., Al-Achi, B. J., Platsouka, E., and Levy, S.B. (1988). Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl. Environ. Microbiol. 54, 2432–2438. doi: 10.1128/AEM.54.10.2432-2438.1988
Correa, V. R., Majerczak, D. R., Ammar el, D., Merighi, M., Pratt, R. C., Hogenhout, S. A., et al. (2012). The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl. Environ. Microbiol. 78, 6327–6336. doi: 10.1128/AEM.00892-12
Darwin, K. H., and Miller, V. L. (1999). InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181, 4949–4954.
Darwin, K. H., and Miller, V. L. (2001). Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20, 1850–1862. doi: 10.1093/emboj/20.8.1850
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Ding, L., and Wang, Y. (2009). Relationship between flagella-dependent motility and biofilm in bacteria. Acta Microbiol. Sin. 49, 417–422. doi: 10.3321/j.issn:0001-6209.2009.04.002
Egan, F., Barret, M., and O’Gara, F. (2014). The SPI-1-like Type III secretion system: more roles than you think. Front. Plant Sci. 5:34. doi: 10.3389/fpls.2014.00034
Ellermeier, J. R., and Slauch, J. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in salmonella enterica serovar typhimurium. Curr. Opin. Microbiol. 10, 24–29. doi: 10.1016/j.mib.2006.12.002
Figurski, D., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). Fimo: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Gu, Y., Wang, J., Xia, Z., and Wei, H. L. (2020). Characterization of a versatile plant growth-promoting rhizobacterium Pseudomonas mediterranea strain S58. Microorganisms 8:334. doi: 10.3390/microorganisms8030334
Henrichsen, J. (1972). Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36, 478–503. doi: 10.1128/MMBR.36.4.478-503.1972
Jackson, R. W., Preston, G. M., and Rainey, P. B. (2005). Genetic characterization of Pseudomonas fluorescens SBW25 rsp gene expression in the phytosphere and in vitro. J. Bacteriol. 187, 8477–8488. doi: 10.1128/JB.187.24.8477-8488.2005
Kimbrel, J. A., Givan, S. A., Halgren, A. B., Creason, A. L., Mills, D. I., Banowetz, G. M., et al. (2010). An improved, high-quality draft genome sequence of the germination-arrest factor-producing Pseudomonas fluorescens WH6. BMC Genomics 11:522. doi: 10.1186/1471-2164-11-522
Kirzinger, M., Butz, C. J., and Stavrinides, J. (2015). Inheritance of pantoea type III secretion systems through both vertical and horizontal transfer. Mol. Genet. Genomics 290, 2075–2088. doi: 10.1007/s00438-015-1062-2
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 7:1870. doi: 10.1093/molbev/msw054
Li, Y., and Mu, B. (2006). Progress in chemotaxis of bacteria. Chin. J. Appl. Environ. Biol. 12, 135–139. doi: 10.3321/j.issn:1006-687X.2006.01.033
Liu, P., Wei, Z., Zhang, L. Q., Liu, X., and Wei, H. L. (2016). Supramolecular structure and functional analysis of the type III secretion system in Pseudomonas fluorescens 2P24. Front. Plant Sci. 6:1190. doi: 10.3389/fpls.2015.01190
Loper, J. E., Hassan, K. A., Mavrodi, D. V., Davis, E. W., Lim, C. K., Shaffer, B. T., et al. (2012). Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8:e1002784. doi: 10.1371/journal.pgen.1002784
Marchi, M., Boutin, M., Gazengel, K., Rispe, C., Gauthier, J. P., Guillerm-Erckelboudt, A. Y., et al. (2013). Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots. Environ. Microbiol. Rep. 5, 393–403. doi: 10.1111/1758-2229.12048
Marguerettaz, M., Pieretti, I., Gayral, P., Puig, J., Brin, C., Cociancich, S., et al. (2011). Genomic and evolutionary features of the SPI-1 type III secretion system that is present in Xanthomonas albilineans but is not essential for xylem colonization and symptom development of sugarcane leaf scald. Mol. Plant Microbe Interact. 24, 246–259. doi: 10.1094/MPMI-08-10-0188
Mazumder, R., Phelps, T. J., Krieg, N. R., and Benoit, R. E. (1999). Determining chemotactic responses by two subsurface microaerophiles using a simplified capillary assay method. J. Microbiol. Methods 37, 255–263. doi: 10.1016/j.scriptamat.2010.02.026
Miao, E. A., Alpuche-Aranda, C. M., Dors, M., Clark, A. E., Bader, M. W., Miller, S. I., et al. (2006). Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575. doi: 10.1038/ni1344
Nazir, R., Mazurier, S., Yang, P., Lemanceau, P., and Van Elsas, J. D. (2017). The ecological role of type three secretion systems in the interaction of bacteria with fungi in soil and related habitats is diverse and context-dependent. Front. Microbiol. 8:38. doi: 10.3389/fmicb.2017.00038
Pallen, M. J., Beatson, S. A., and Bailey, C. M. (2005). Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a darwinian perspective. FEMS Microbiol. Rev. 29, 201–229. doi: 10.1016/j.femsre.2005.01.001
Preston, G. M., Bertrand, N., and Rainey, P. B. (2001). Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol. Microbiol. 41, 999–1014. doi: 10.1046/j.1365-2958.2001.02560.x
Rashid, M. H., and Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 4885–4890. doi: 10.1073/pnas.060030097
Redondo-Nieto, M., Barret, M., Morrissey, J., Germaine, K., Martinez-Granero, F., Barahona, E., et al. (2013). Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics 14:54. doi: 10.1186/1471-2164-14-54
Rouse, S. L., Hawthorne, W. J., Berry, J. L., Chorev, D. S., Ionescu, S. A., Lambert, S., et al. (2017). A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis. Nat. Commun. 8:263. doi: 10.1038/s41467-017-00361-6
Sambrook, E., Fritsch, F., and Maniatis, T. (1989). Molecular cloning. Cold spring harbor press, cold spring harbor. Am. J. Hum. Genet. 73, 1162–1169.
Stringlis, I. A., Zamioudis, C., Berendsen, R. L., Bakker, P. A. H. M., and Pieterse, C. M. J. (2019). Type III secretion system of beneficial rhizobacteria Pseudomonas simiae WCS417 and Pseudomonas defensor WCS374. Front. Microbiol. 10:1631. doi: 10.3389/fmicb.2019.01631
Triplett, L. R., Zhao, Y., and Sundin, G. W. (2006). Genetic differences between blight-causing Erwinia species with differing host specificities, identified by suppression subtractive hybridization. Appl. Environ. Microbiol. 72, 7359–7364. doi: 10.1128/AEM.01159-06
Troisfontaines, P., and Cornelis, G. R. (2005). Type III secretion: more systems than you think. Physiology 20, 326–339. doi: 10.1152/physiol.00011.2005
Ugochukwu, U. C., Jones, M. D., Head, I. M., Manning, D. A., and Fialips, C. I. (2013). Compositional changes of crude oil SARA fractions due to biodegradation and adsorption on colloidal support such as clays using Iatroscan. Environ. Sci. Pollut. Res. 20, 6445–6454. doi: 10.1007/s11356-013-1635-8
Viollet, A., Pivato, B., Mougel, C., Cleyet-Marel, J. C., Gubry-Rangin, C., Lemanceau, P., et al. (2017). Pseudomonas fluorescens C7R12 type III secretion system impacts mycorrhization of Medicago truncatula and associated microbial communities. Mycorrhiza 27, 23–33. doi: 10.1007/s00572-016-0730-3
Wei, H. L., Chakravarthy, S., Worley, J. N., and Collmer, A. (2013). Consequences of flagellin export through the type III secretion system of Pseudomonas syringae reveal a major difference in the innate immune systems of mammals and the model plant Nicotiana benthamiana. Cell. Microbiol. 15, 601–618. doi: 10.1111/cmi.12059
Wei, H. L., Wang, Y., Zhang, L. Q., and Tang, W. H. (2004a). Identification and characterization of biocontrol bacterial strain 2P24 and CPF-10. Acta Phytopathol. Sin. 34, 80–85.
Keywords: Pseudomonas fluorescens, PGPR, type III secretion system, transcriptional regulator, ROS
Citation: Wang J, Luo Y, Gu Y and Wei H-L (2021) Characterization of the SPI-1 Type III Secretion System in Pseudomonas fluorescens 2P24. Front. Microbiol. 12:749037. doi: 10.3389/fmicb.2021.749037
Received: 28 July 2021; Accepted: 31 August 2021;
Published: 21 September 2021.
Edited by:
Xiuling Yang, Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Qian Guoliang, Nanjing Agricultural University, ChinaBo Zhu, Shanghai Jiao Tong University, China
Copyright © 2021 Wang, Luo, Gu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Lei Wei, d2VpaGFpbGVpQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
 Jing Wang
Jing Wang Yuan Luo
Yuan Luo Yilin Gu
Yilin Gu Hai-Lei Wei
Hai-Lei Wei