- 1Water Research Institute, IRSA-CNR, Rome, Italy
- 2Department of Chemistry, Sapienza University of Rome, Rome, Italy
Bioelectrochemical systems (BES) are attractive and versatile options for the bioremediation of organic or inorganic pollutants, including trichloroethylene (TCE) and Cr(VI), often found as co-contaminants in the environment. The elucidation of the microbial players’ role in the bioelectroremediation processes for treating multicontaminated groundwater is still a research need that attracts scientific interest. In this study, 16S rRNA gene amplicon sequencing and whole shotgun metagenomics revealed the leading microbial players and the primary metabolic interactions occurring in the biofilm growing at the biocathode where TCE reductive dechlorination (RD), hydrogenotrophic methanogenesis, and Cr(VI) reduction occurred. The presence of Cr(VI) did not negatively affect the TCE degradation, as evidenced by the RD rates estimated during the reactor operation with TCE (111±2 μeq/Ld) and TCE/Cr(VI) (146±2 μeq/Ld). Accordingly, Dehalococcoides mccartyi, the primary biomarker of the RD process, was found on the biocathode treating both TCE (7.82E+04±2.9E+04 16S rRNA gene copies g−1 graphite) and TCE/Cr(VI) (3.2E+07±2.37E+0716S rRNA gene copies g−1 graphite) contamination. The metagenomic analysis revealed a selected microbial consortium on the TCE/Cr(VI) biocathode. D. mccartyi was the sole dechlorinating microbe with H2 uptake as the only electron supply mechanism, suggesting that electroactivity is not a property of this microorganism. Methanobrevibacter arboriphilus and Methanobacterium formicicum also colonized the biocathode as H2 consumers for the CH4 production and cofactor suppliers for D. mccartyi cobalamin biosynthesis. Interestingly, M. formicicum also harbors gene complexes involved in the Cr(VI) reduction through extracellular and intracellular mechanisms.
Introduction
Trichloroethylene (TCE) is a toxic and persistent anthropogenic pollutant commonly found in groundwater and often detected in the aquifer with other co-contaminants, including heavy metals such as the carcinogenic Cr(VI) (Watts et al., 2015). Despite the persistence into the environment, TCE is a biodegradable compound, while Cr(VI) can be transformed into less toxic forms (i.e., Cr(III)).
The TCE transformation to cis-1,2-dichloroethene (cis-DCE) and vinyl chloride (VC) up to the harmless ethene occurs via the anaerobic reductive dechlorination (RD) in the presence of an electron donor and through the biological activity of specialized organohalide-respiring bacteria (OHRB; Strycharz et al., 2008; Löffler et al., 2013). Among them, Dehalococcoides mccartyi is the sole microorganism capable of performing RD to ethene. Thus, it is considered a robust biomarker for the RD monitoring and evaluation at laboratory and field scale (Taş et al., 2010; Saiyari et al., 2018). Dehalococcoides mccartyi strains are characterized by electron transport chain’s essential membrane-associated enzymes, known as reductive dehalogenases (RDases; Ahsanul Islam et al., 2010; Pérez-de-Mora et al., 2018; Hermon et al., 2019). Some RDases have been functionally characterized (i.e., TceA, VcrA, and BvcA), but most of them remain uncharacterized (RdhAses) among dechlorinating isolates or mixed cultures (Hug et al., 2013; Molenda et al., 2020). Dehalococcoides mccartyi is strictly anaerobic and sensitive to oxygen exposure, incapable of substrate fermentation as a source of electrons, and unable to synthesize corrinoids (i.e., cobalamin), fundamental cofactors for the functionality of RDases (Yan et al., 2016; Yang et al., 2017). These metabolic peculiarities make D. mccartyi growth and RD activity favored within mixed anaerobic consortia where non-dechlorinating microorganisms can supply exogenous cofactors.
In respect of Cr(VI) remediation, anaerobic microorganisms capable of Cr(VI) to Cr(III) reduction (i.e., Pseudomonas dechloromaticans, Enterobacter cloacae, Shewanella oneidensis, and Clostridium chromiireducens) and with Cr(VI) tolerance (i.e., Bacillus sp., Leucobacter sp., Exiguobacterium sp., Microcococcus sp., Rhodococcus sp., Arthrobacter sp., Achromobacter sp., and Ochrobactrum sp.) have been reported (Horitsu et al., 1987; Turick and Apel, 1997; Desai et al., 2008; Sarangi and Krishnan, 2008; Elangovan et al., 2010; Beretta et al., 2018; Kabir et al., 2018). Several studies also demonstrated that indirect Cr(VI) bioreduction is achievable through injections of biodegradable organic substrates that prompt anaerobic conditions and reductant production such as iron and sulfur species capable of mediating Cr(VI) reduction to Cr(III) (Kim et al., 2001; Somenahally et al., 2013; Beretta et al., 2018, 2019). Further, under anaerobic conditions, direct Cr(VI) reduction can be mediated by membrane-bound reductases encoded by mtrA, mtrB, mtrC genes, and soluble enzymes (e.g., soluble cytochrome c3; Belchik et al., 2011; Thatoi et al., 2014). Some studies also reported that Cr(VI) reduction occurs with H2 as electron donor and CO2 as a carbon source in anaerobic mixed cultures (Marsh and McInerney, 2001).
The simultaneous biodegradation of TCE and biotransformation of Cr(VI) to the less toxic form Cr(III) is a challenging remediation goal when co-contamination occurs. However, the available bioremediation technologies commonly treat TCE or Cr(VI) separately (Malaviya and Singh, 2011; Shukla et al., 2014; Jin et al., 2015; Sophia and Saikant, 2016; Wang et al., 2020). Among these, bioelectrochemical technologies emerged in the last years as the most attractive and versatile options being advantageous in terms of cost-effectiveness, side reactions control, and sustainability for in situ remediation of contaminated matrices (Fernando et al., 2019; Cecconet et al., 2020; Wang et al., 2020). Recently, there has been an ever-increasing interest in biocatalyzed reduction at the cathode (i.e., microbial electrolysis cells, MECs) of many pollutants, including TCE or Cr(VI) (Palma et al., 2018). These systems require an electrode acting as an electron donor (cathode), an electron sink (anode), and microorganisms driving reactions to exploit energy from toxic contaminants (Modin and Aulenta, 2017; Ivase et al., 2020). Several studies have demonstrated that MECs, under controlled potentials, may rapidly enhance RD by establishing dechlorinating consortia growing on the biocathode (Aulenta et al., 2007, 2011; Verdini et al., 2015; Zeppilli et al., 2019). Nevertheless, despite various aspects of MECs for TCE dechlorination have been investigated (i.e., the effect of cathode potential on TCE dechlorination rate, selectivity, electron transfer mechanisms; Aulenta et al., 2007, 2008, 2010, 2011; Chen et al., 2018; Wang et al., 2020), only a few reports documented the microbial composition of the TCE-reducing biocathode without the addition of external carbon substrates (Aulenta et al., 2010; Di Battista et al., 2012; Wang et al., 2015) and even less in the case of TCE/Cr(VI) co-contamination. Similarly, very little is known about the microbial players of the bioelectrochemical systems (BES) treating Cr(VI) contamination. A recent study conducted at the Cr(VI)-reducing biocathode revealed γ-Proteobacteria as the most abundant electrotrophic component and, among them, also the presence of known chromium reducing/resistant bacteria including Pseudomonas sp. and Ochrobactrum sp. (Romo et al., 2019). Pieces of evidence reported that the production of H2 at the cathode of a bioelectrochemical system could favor the autotrophic reduction of Cr(VI) by hydrogenotrophic bacteria.
Therefore, despite that the BES treating TCE or Cr(VI) have been previously described along with some biological data reporting the microbial players involved in the reductive processes, the feasibility of a bioelectrochemical system treating TCE in the presence of Cr(VI) as co-contaminants has been only recently investigated (Lai et al., 2021). Nonetheless, the microbial composition and interactions occurring at the biocathode treating TCE/Cr(VI) co-contamination still need to be analyzed. To this aim, metagenomics approaches provide reliable information about the key enzymes/genes involved in the degradation and detoxification of environmental pollutants to understand better the microbial community structure, functions, and interactions in contaminated matrices or engineered systems for the biological removal of pollutants, with implications for process optimization and bioremediation application at field scale (Bharagava et al., 2019; Lapidus and Korobeynikov, 2021). Metagenomic studies have been previously conducted on stable dechlorinating consortia to shed light on the metabolic features of the microbial components and their role in the RD processes under different conditions (Brisson et al., 2012; Maphosa et al., 2012; Dam et al., 2017; Wang et al., 2019; Kucharzyk et al., 2020). Similarly, some metagenomic studies have also been conducted on biological systems for Cr(VI) removal to explore the metabolic potential, including Cr(VI) remediation genes, primarily in wastewater treatment systems or microbial consortia (Miao et al., 2015; Sun et al., 2019; Pei et al., 2020; Rahman and Thomas, 2021). To the best of our knowledge, no metagenomic studies have been conducted on a bioelectrochemical system for simultaneous TCE/Cr(VI) reduction.
In this study, the biofilm growing on the biocathode of a bioelectrochemical system capable of complete TCE-to-VC/ethene reduction in the presence of Cr(VI) as a co-contaminant has been characterized for the first time. The RD process biomarkers, including D. mccartyi 16S rRNA and RDase genes, have been monitored. The metagenomic analysis has been performed to define the microbiome composition and metabolic features of the critical biological players of the processes occurring in the bioelectrochemical system.
Materials and Methods
Continuous Flow Reactor: Set Up and Operating Conditions
The continuous flow bioelectrochemical reactor scheme for the TCE and Cr(VI) removal used in this study is reported in Figure 1. The continuous flow bioelectrochemical reactor was composed of a 0.821L cathodic chamber (Figure 1A) filled with graphite granules (2 and 6mm; El Carb 100, Graphite sales, Inc., United States). The cathodic chamber adopted a graphite rod as a current collector and a 0.90L anodic chamber composed of an MMO Electrode (Magneto special Anodes, Netherland) inserted in a silica bed (Figure 1B).
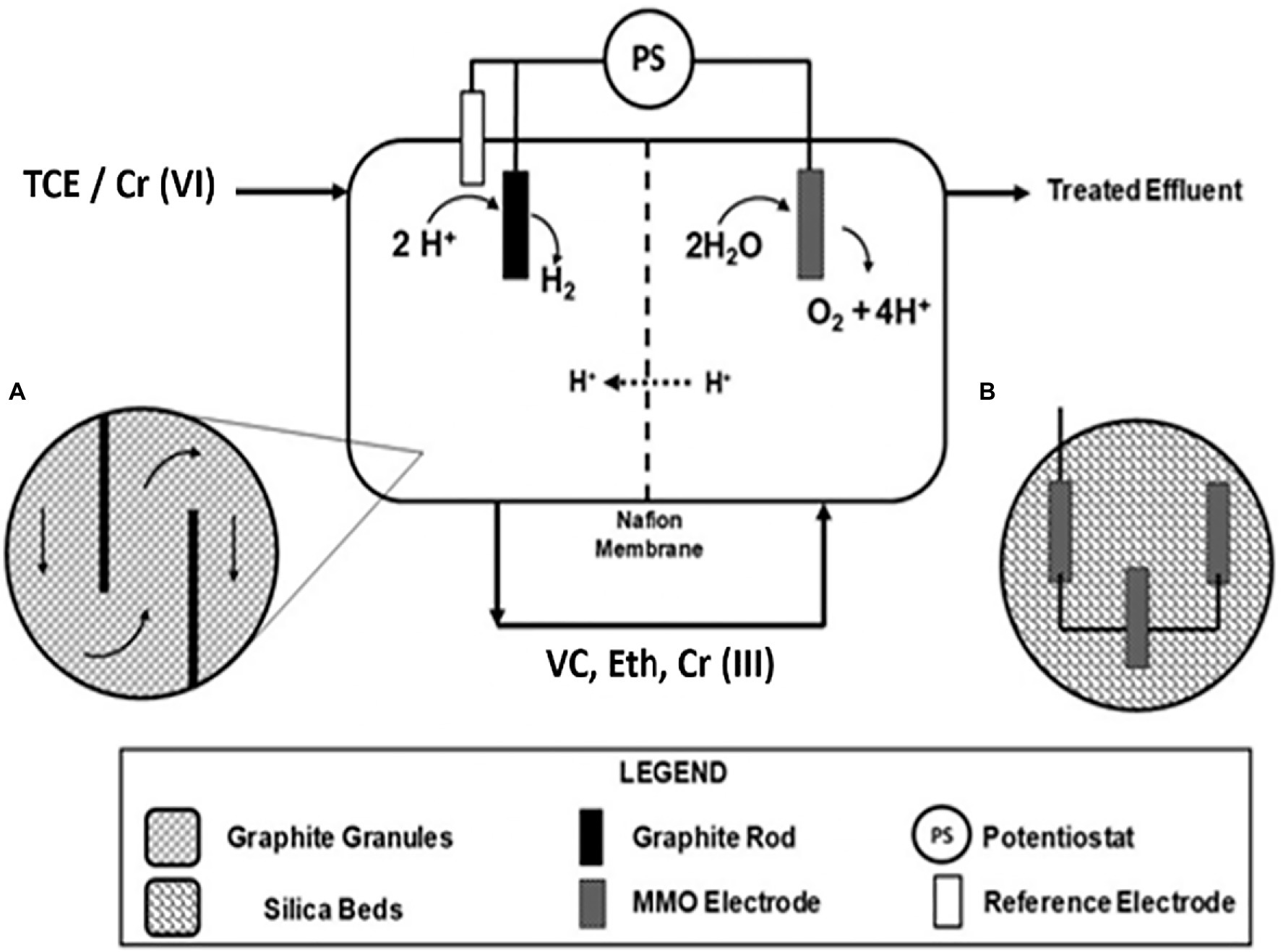
Figure 1. Scheme of the continuous flow bioelectrochemical reactor for the trichloroethylene (TCE) and Cr(VI) removal. Horizontal cross section detail of the cathodic chamber (A) and schematic representation of the MMO (Magneto special Anodes, Netherland) electrode inserted in the silica bed (B).
An anaerobic mineral medium (NH4Cl, 0.5; MgCl2x6H2O, 0.1; CaCl2x2H2O, 0.05; K2HPO4, 0.4gL−1) supplemented with a metal and vitamin solution was used as previously described (Lai et al., 2017). The cathode and anode chambers were physically separated by the Nafion® 117 proton exchange membrane, which allowed the proton migration to maintain electroneutrality. An Ag/AgCl reference electrode (+0.199 vs. standard hydrogen electrode, SHE; Amel, Milan, Italy) placed in the cathode chamber permitted the polarization of the reactor by a potentiostat (Amel Model 549, Milan) adopting a three-electrode configuration. Two Teflon® septa were inserted within the graphite granules in the cathodic chamber to obligate the flow direction along the total cathodic volume (Figure 1). The cathodic chamber was inoculated with a hydrogenophilic dechlorinating, while the anodic chamber was inoculated with an anaerobic dechlorinating mixed culture (Lai et al., 2021). The continuous flow reactor was operated with the TCE-contaminated mineral medium (TCE 50μM) and the TCE/Cr(VI)-contaminated mineral medium (Lai et al., 2021). The flow rate was 0.58l/d, corresponding to a cathodic hydraulic retention time (HRT) of 1.4day. The cathodic chamber of the bioelectrochemical reactor was polarized at −650mV vs. SHE. The average rates of RD (1) and Cr(VI) reduction (2) were calculated as follows:
where [cisDCE], [VC], [ETH], and [ETA] are the average liquid-phase compound concentration (μM); 2, 4, 6, and 8 are the number of moles of electrons required for the formation of RD intermediates from 1mol of TCE, while 3 is the equivalent moles required for the reduction of Cr(VI) to Cr(III); Q is the flow rate (L d−1); VC is the empty volume of the cathode chamber (L) (i.e., the volume without the electrode).
The Coulombic efficiency (CERD) has been calculated as follows (3):
where ri is the average rate of the formation of RD compounds or reduction of Cr(VI) (μM d−1), F is the Faraday’s constant (96,485 C mol eq−1), and I is the electric current (μA; Lai et al., 2021). Methane production rate (4) and its relative contribution to the current consumption (i.e., Coulombic efficiency for methane generation) (4) were assessed as follows (Zeppilli et al., 2019, 2020):
where Q is the liquid flow rate, [CH4] represents the liquid methane concentration calculated according to the Henry’s law constant at room temperature, F is the Faraday constant (i.e., 96,485 C/mol e−), and s/d represents the seconds in a day (86,400s/day).
Sampling for Biomolecular Analysis and DNA Extraction
Samples for biomolecular analysis were collected in triplicate from the biocathode treating TCE only (sample hereafter cited as TCE) and TCE/Cr(VI) as co-contaminants (sample hereafter cited as TCE/Cr(VI)). One gram of biocathode (i.e., graphite granules) was collected from TCE and TCE/Cr(VI) samples. DNA extraction was performed with DNeasy PowerSoil Kit (Qiagen, Italy), following the manufacturer’s instructions. Purified DNA from each sample was eluted in 100μl sterile Milli-Q and stored at −20°C until further biomolecular analysis.
Real-Time PCR
Dehalococcoides mccartyi 16S rRNA gene and reductive dehalogenase genes tceA, bvcA, vcrA were quantified via absolute quantification qPCR assays. TaqMan® chemistry (6-carboxyfluorescein-FAM at the 5' end as reporter fluorophore; N, N, N, N,-tetramethyl-6-carboxyrhodamine-TAMRA at the 3' end as a quencher) was employed, and reactions were conducted in 20μl total volume including 3μl of template DNA, 300nM of each primer, 300nM TaqMan® probe and SsoAdvancedTM Universal Probes Supermix (Bio-Rad, Italy). Primers and probes used were previously reported (Ritalahti et al., 2006). Standard curves for the absolute quantification were constructed with the long PCR amplicons of the targeted genes (Matturro et al., 2013). Reactions were run in triplicate for each biological sample, and qPCR was performed with the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Italy). Quantitative data are reported as gene copy numbers g−1 of graphite granules.
Sequencing
The whole microbiome composition of the TCE and TCE/Cr(VI) biocathodes was analyzed by 16S rRNA gene amplicon sequencing. The biomolecular analysis was expanded through whole shotgun metagenomic sequencing and a genome-centric analysis. A de novo metagenome assembly was performed on quality-filtered reads, and individual genomes (genome bins) were subsequently extracted from the assembled metagenome using a customized bioinformatics approach. The individual bins were subjected to quality assessment, taxonomic classification, and gene annotation. In the following, the detailed methods for 16S rRNA gene and shotgun metagenomic sequencing are reported.
16S rRNA Gene Amplicon Sequencing and Bioinformatics
Four nanograms of DNA extracted from TCE and TCE/Cr(VI) biocathode was used for NGS analysis. DNA extraction 16S rRNA Amplicon Library Preparation targeting the V1–3 variable region was constructed as previously reported (Matturro et al., 2017). PCR reactions were performed in 25μl total volume containing Phusion Master Mix High Fidelity (Thermo Fisher Scientific, United States) and 0.5μM final concentration of the library adaptors with V1–V3 primers (Bacteria, 27F: 5'-AGAGTTTGATCCTGGCTCAG-3'; 534R: 5'-ATTACCGCGGCTGCTGG-3') and V3-V5 primers (Archaea, 340F: 5'-CCCTAHGGGGYGCASCA-3'; 915R: 5'-GWGCYCCCCCGYCAATTC-3'). All PCR reactions were run in duplicate and pooled afterward. Libraries were purified using the Agencourt® AMpure XP-beads protocol (Beckman Coulter, Italy), and the concentration was measured with Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Italy). Purified libraries were pooled in equimolar concentrations and diluted to 4nM. PhiX control (15%) was added at 10% in the pooled libraries to overcome issues often observed in 16S rRNA gene sequencing. Samples were paired-end sequenced (2×301bp) on a MiSeq (Illumina, United States) instrument using a MiSeq Reagent kit v3, 600cycles (Illumina, United States) following the standard guidelines. Raw data were processed and analyzed using QIIME2 software tools 2018.2 release (c). The reads were demultiplexed using demux plugin1, denoised, dereplicated, and chimera-filtered using the DADA2 algorithm (Callahan et al., 2016). The taxonomic analysis was based on a Naïve Bayes classifier trained on 16S rRNA gene sequences clustered at 99% similarities within the Silva 132–99 database (release December 2017),1 allowing to construct a data set of amplicon sequence variants (ASVs).
Metagenomic Sequencing and Bioinformatics
Sample DNA concentrations from TCE/Cr(VI) contaminated biocathode were measured using the Qubit dsDNA HS kit. The DNA quality and concentrations were evaluated using TapeStation with the Genomic ScreenTape (Agilent Technologies). The final concentration of 1.5ng/μl has been used for the library preparation. Sequencing libraries were prepared using the NEB Next Ultra II DNA library prep kit for Illumina (New England Biolabs, United States) following the manufacturer’s protocol. Library concentrations were measured in triplicate using the Qubit dsDNA HS kit and library size estimated using TapeStation with D1000 HS ScreenTape. The sequencing libraries were pooled in equimolar concentrations and diluted to 4nM. The samples were paired-end sequenced (2x301bp) on a MiSeq (Illumina, United States) using a MiSeq Reagent kit v3, 600cycles (Illumina, United States) following the standard guidelines for preparing and loading samples on the MiSeq. Raw Illumina reads were filtered for PhiX using Usearch11 (Edgar, 2010) subsequently trimmed using Cutadapt v. 2.10 (Martin, 2011). Forward and reverse reads were used to perform de novo assembly in megahit v. 1.2.9. The total assembly length of the metagenomes, the length of the longest contig, and the shortest contig length needed to cover 50% of the genome were calculated. Individual genomes (genome bins) were subsequently extracted from each sample metagenome in mmgenome2 v. 2.1.3. Bins were quality assessed with CheckM v. 1.1.3 (Parks et al., 2015). Classification of bacterial bins was conducted with the Genome Taxonomy Database toolkit (GTDB-TK) v. 1.3.0 (Chaumeil et al., 2019). Average nucleotide identities (ANI) were calculated using FastANI v. 1.32 (Goris et al., 2007; Jain et al., 2018). Genome annotations of bacterial and archaeal genomes were firstly conducted with Prokka v. 1.14.6. Further, NCBI Prokaryotic Genome Annotation Pipeline (PGAP) was also performed for the deepest annotation. The Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the multiple accession JADIIK000000000-JADIIN000000000. Raw data are deposited under the SRA accession SRR12879946 within the BioProject PRJNA670625.
Results
Performance of the Bioelectrochemical Reactor Operating With TCE and TCE/Cr(VI) Contamination
In the cathodic chamber under continuous flow, the complete TCE removal occurred also in the presence of Cr(VI) as co-contaminant. The dechlorination products observed in the cathodic chamber fed with TCE and TCE/Cr(VI) were VC, ethene, and ethane (Figure 2). RD rates obtained during TCE and TCE/Cr(VI) operating conditions were 111±2 and 146±2 μeq/Ld, respectively (Table 1).
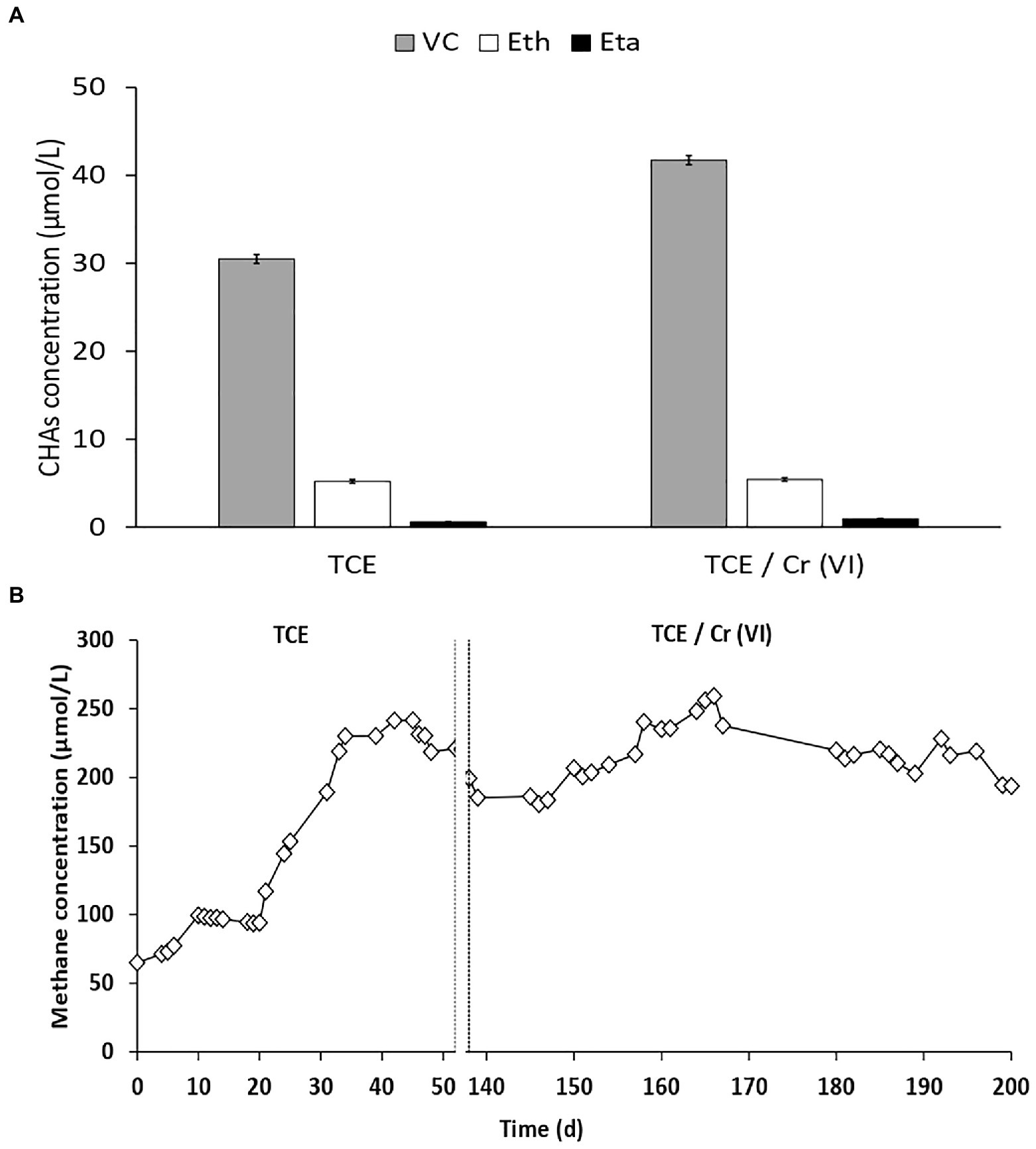
Figure 2. (A) Distribution of the reductive dechlorination (RD) products observed in the cathode outlet TCE and TCE/Cr(VI) operating conditions. (B) CH4 concentrations in the cathodic effluent of the bioelectrochemical reactor during all the system’s operation periods, including the runs with TCE and TCE/Cr(VI).
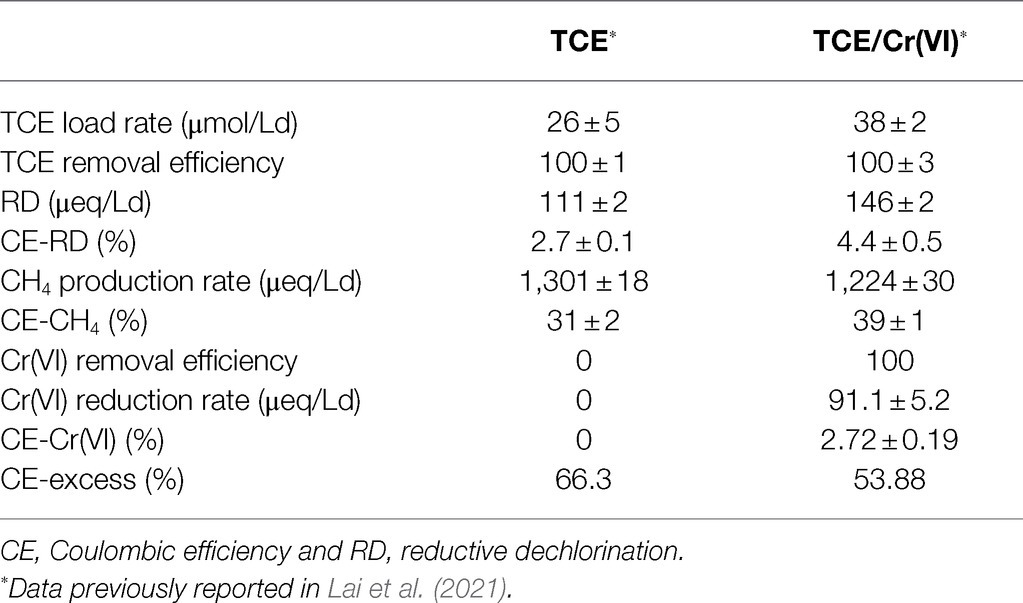
Table 1. Reductive dechlorination and methanogenesis performances during TCE and TCE/Cr(VI) operating conditions.
The Coulombic efficiency (i.e., the electrons utilized by the dehalorespiring bacteria to reduce the chlorinated compounds, CE-RD) allowed for the consumption of 2.7±0.1 and 4.4±0.5% of the electric current. Bioelectrochemical CO2 reduction into CH4 (i.e., bioelectromethanogenesis) was observed in the cathodic chamber during all the operating periods with a constant concentration of 231±1μmol/L (Figure 2B). In particular, during the run with TCE and with TCE/Cr(VI), similar CH4 production rates were observed with average values of 1,301±18 and 1,224±30 μeq/Ld, respectively (Table 1). These CH4 production rates corresponded to the consumption of 31 and 39% of the electrical current generated by the bioelectrochemical cell in the TCE and TCE/Cr(VI) runs, respectively. Moreover, Cr(VI) was completely removed with a reduction rate of 91.1±5.2 μeq/Ld and Coulombic efficiencies (CE-Cr(VI)) of 2.72±0.19%. Overall, the CE excess estimated in the TCE-fed and TCE/Cr(VI) fed bioreactor was 66.3 and 53.8%, respectively.
Microbiome Composition of the TCE and TCE/Cr(VI) Cathodic Biofilms
The microbiome composition of the TCE and TCE/Cr(VI) biocathode, including Bacteria and Archaea, was firstly characterized through 16S rRNA gene amplicon sequencing (Figure 3A). On the TCE biocathode, members of Chloroflexi phylum (27%), including mostly D. mccartyi (21.12%), and Deltaproteobacteria (28.9%) affiliated to Desulfovibrio (27%), were mainly found. In particular, D. mccartyi detected on the biocathode of the bioelectrochemical reactor running with TCE accounted for 7.82E+04±2.9E+04 16S rRNA gene copies g−1 graphite and were mostly tceA-carrying strains (Figure 4). Bacteroidetes phylum represented 14% of total ASVs and included Lentimicrobium species (12%). Gammaproteobacteria also colonized the biocathode (13%), including 7% of Rhodocyclaceae and 3% of Thiobacillus (Figure 3B).
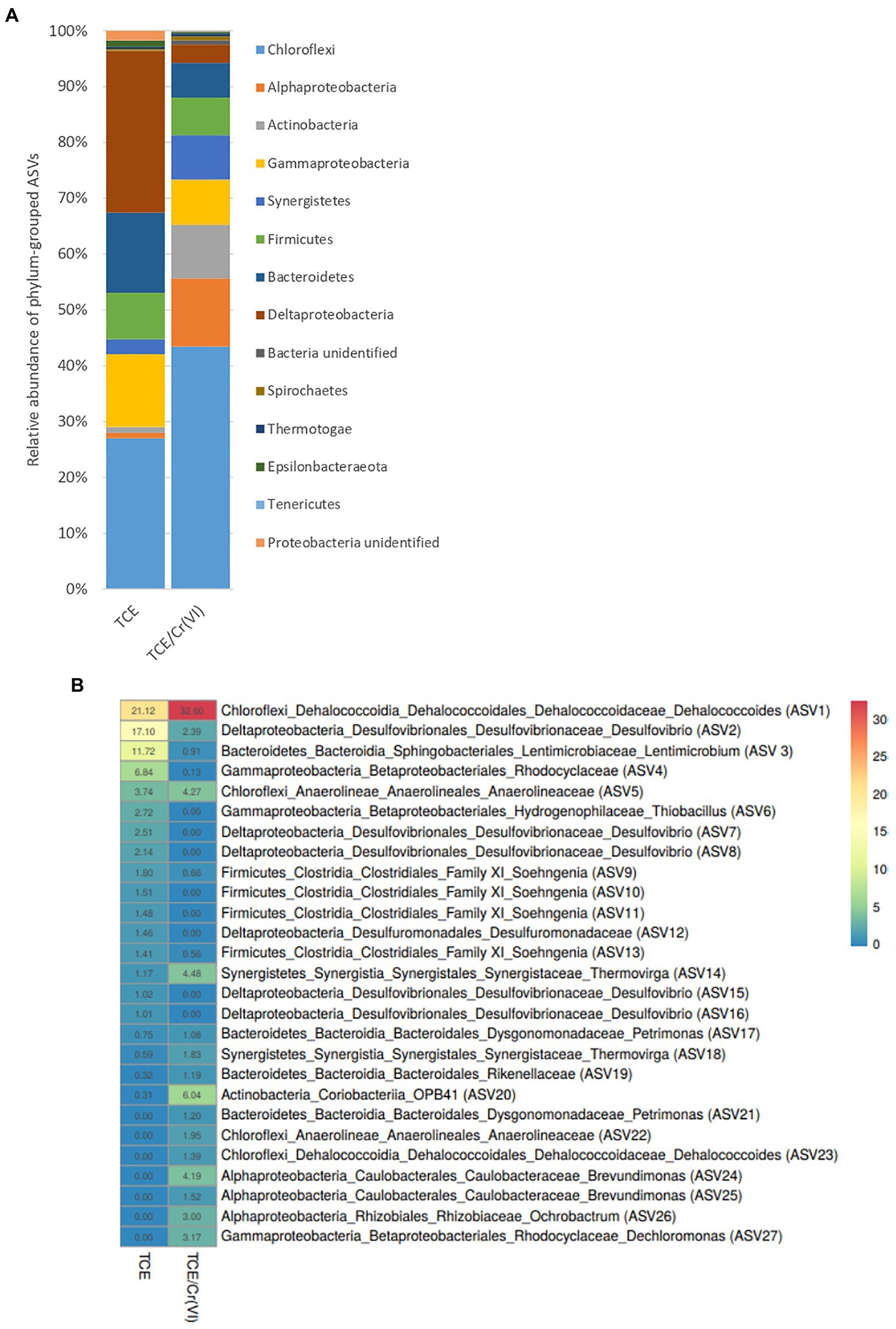
Figure 3. Microbiome composition of the cathodic biofilm at phylum level (A) and heatmap of the most abundant ASVs at genus level (B) found in the biofilm of the bioelectrochemical reactor treating TCE and TCE/Cr(VI).
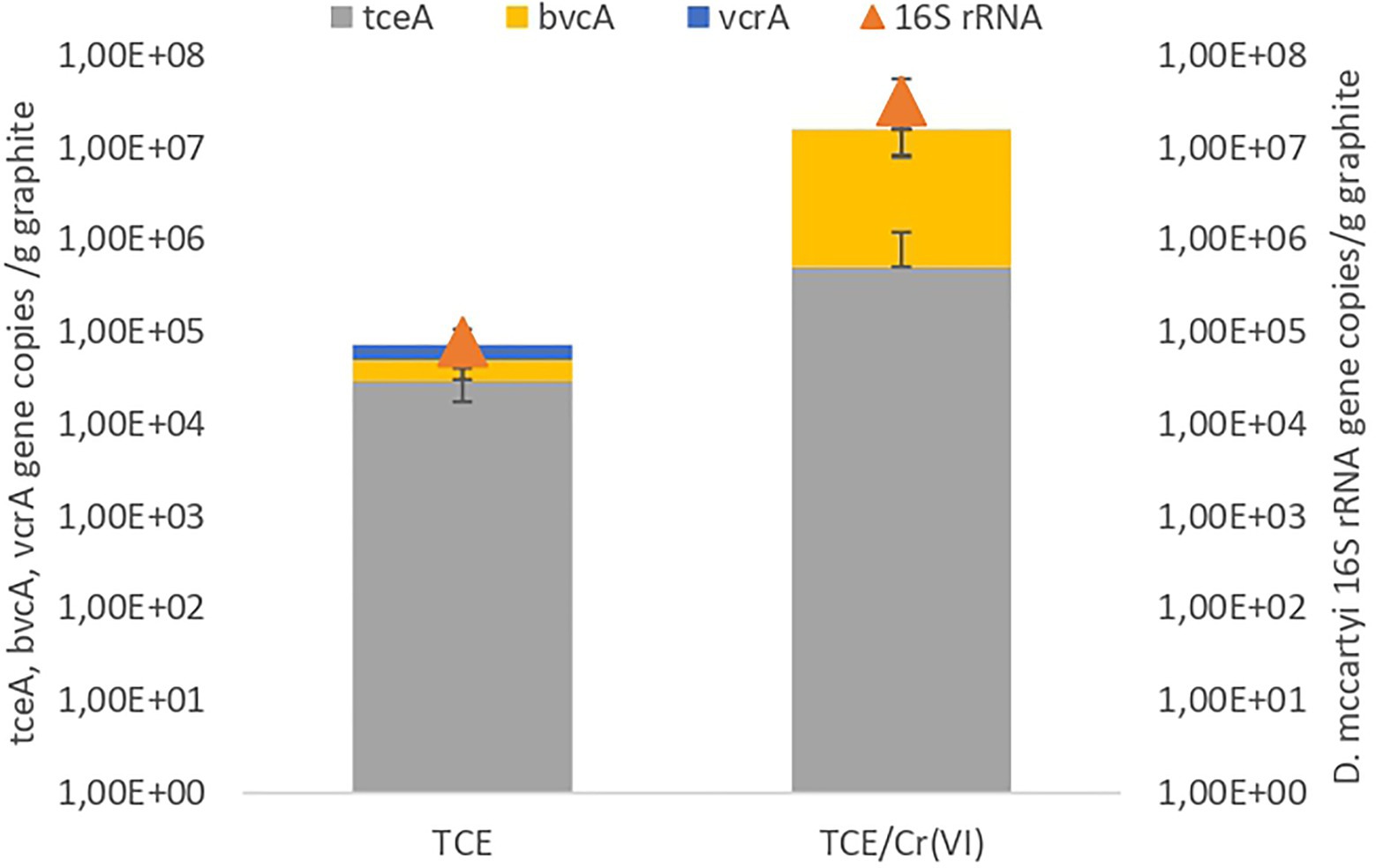
Figure 4. Quantification of 16S rRNA of Dehalococcoides mccartyi and strain-specific RDase genes conducted on the samples from the operating conditions with TCE and TCE/Cr(VI). Data are plotted in log scale (base 10).
Similarly, the TCE/Cr(VI) biofilm was mainly composed of Chloroflexi (43%), including 35.18% of D. mccartyi (Figure 3A), the latter accounted for 3.2E+07±2.37E+07 16S rRNA gene copies g−1 graphite, mostly tceA-carrying strains (Figure 4). Moreover, Alphaproteobacteria (12%) were detected and included Brevundimonas (6%) and Ochrobactrum species (4%), while Gammaproteobacteria (8%) included Thiobacillus (1%), Dechlomononas (3.2%), and unidentified Rhodocyclaceae (2%). At a lower extent, Thermovirga of the phylum Synergistetes (8%), Soehngenia (2.4%), and Acidaminobacter (1.8%) of the phylum Firmicutes (7%) and Desulfovibrio (2.6%) within Deltaproteobacteria (3%) were found (Figure 3A).
As for the most representative ASVs (Supplementary Table S1) at the genus level (Figure 3B), D. mccartyi (ASV1), Desulfovibrio (ASV2), Lentimicrobium (ASV3), and Thiobacillus (ASV6) species were mainly found in the TCE biocathode (Figure 3B). Unidentified species belonging to Rhodocyclaceae (ASV4) and Anaerolineaceae (ASV5) families were detected among the most abundant ASVs of the TCE biocathode (Figure 3B). The TCE/Cr(VI) biocathode showed a slightly modified microbial composition at the genus level than the reactor’s cathodic biofilm running only with TCE. Higher D. mccartyi relative abundance (ASV1) was observed (21.1% on the TCE biocathode; 32.6% on the TCE/Cr(VI) biocathode; Figure 3B). Additionally, Thermovirga (ASV14), Coriobacteria OPB41 (ASV20), Brevundimonas (ASV24, ASV25), Ochrobactrum (ASV26), and Dechloromonas (ASV27) species were also detected among the most abundant ASVs at genus level on the TCE/Cr(VI) biocathode (Figure 3B). Also, Desulfovibrio (ASV2) was found abundant in the system, even if with differences between TCE (17%) and TCE/Cr(VI) (2.4%) biocathode (Figure 3B). Interestingly, on the TCE/Cr(VI) biocathode, some known chromium-resistant bacteria, including Ochrobactrum (ASV26, 3%), Brevundimonas (ASV24 and ASV25, 5.71%), and Dechloromonas (ASV27, 3.17%), were found. Moreover, in line with CH4 formation performance (Table 1), the hydrogenotrophic methanogenic consortium with two specialized methanogens, Methanobrevibacter arboriphilus strain DH-1 and Methanobacterium formicicum strain MF, was identified both in TCE and TCE/Cr(VI) biocathodes by archaeal 16SrRNA gene amplicon sequencing (Figure 5).
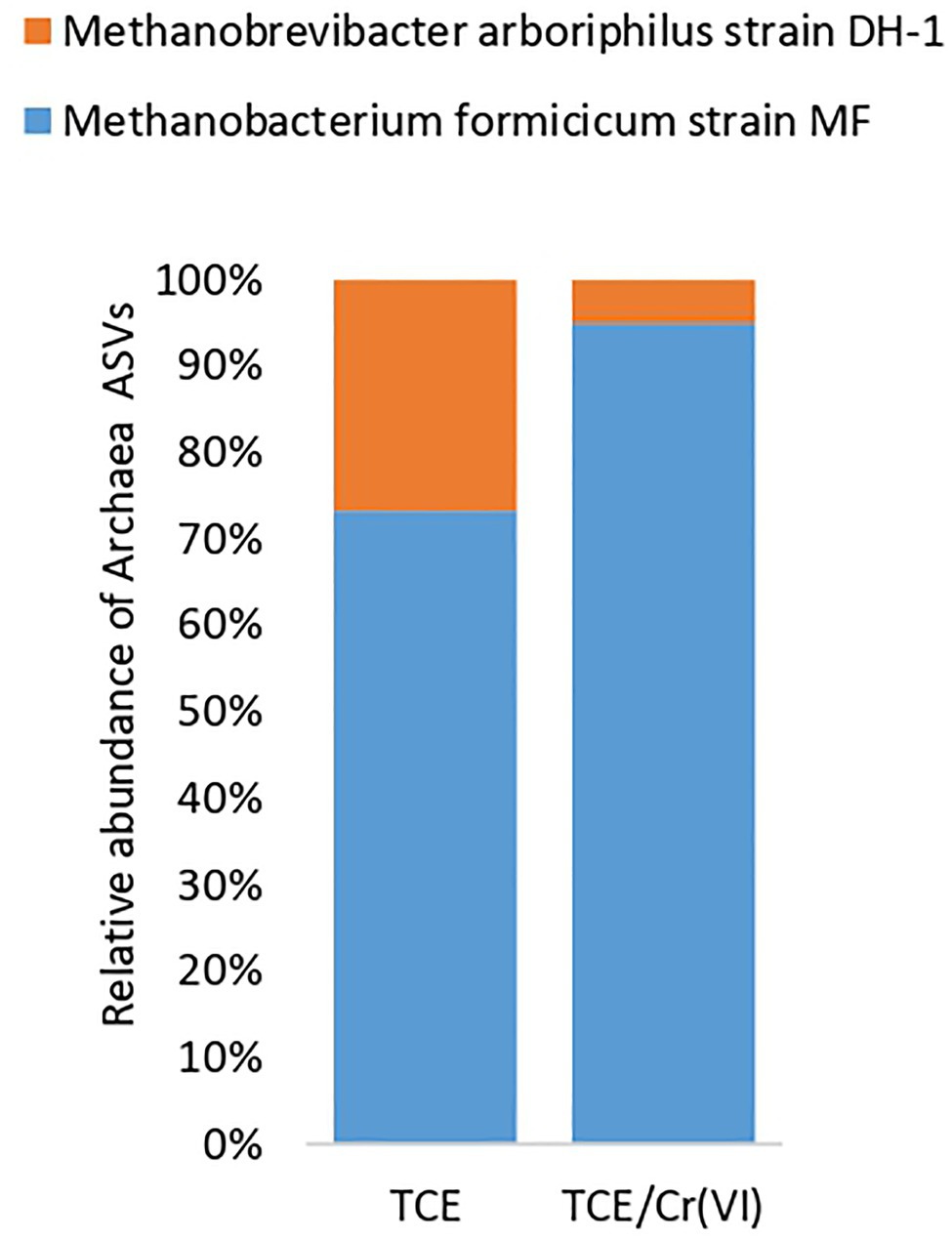
Figure 5. Archaea composition of the cathodic biofilm in the reactor operating with TCE and TCE/Cr(VI).
Metagenome Classification, Sequencing, and Assembly Statistics of the TCE/Cr(VI) Biocathode
The metagenomic analysis has been conducted to gain insights into the metabolic features of the leading microbial players occurring at the TCE/Cr(VI) biocathode. A total of 2,110,164 raw DNA sequences were obtained. After the base quality and PhiX-filtered, a total of 2,071,082 trimmed reads before de novo metagenome assembly were generated (BioProject: PRJNA670625). The total assembly length of the TCE/Cr(VI) biocathode metagenome was 44.3Mb. The TCR/Cr(VI) metagenome was well assembled and contained few taxa (Supplementary Figure S1A). Four genome bins were extracted based on differential abundance using a kmer-based tSNE approach (t-distributed stochastic neighbor embedding; Supplementary Figure S1B). The extracted genome bins included D. mccartyi (Bin 1), M. formicicum (Bin 2), Aeromicrobium sp. (Bin 3), and M. arboriphilus (Bin 4). General characteristics of the four genome bins extracted from the TCE/Cr(VI) biocathode are reported in Supplementary Tables S2 and S3. The classification against the Global Taxonomy Database (GTDB) was subsequently used to infer taxonomy for the extracted genome bins, which were nearly complete (95–100%) and contained low contamination levels (0–1.8%). In the annotated genomes, the identified cluster of orthologous groups of proteins (COGs) mostly fell within functional categories linked to metabolism, information storage and processing, cellular process, and signaling (Supplementary Table S4; Supplementary Figure S2). Within the annotated coding sequences (CDSs), enzymes related to transferases, hydrolases, and oxidoreductases were the most abundant (Supplementary Figure S3).
Discussion
Overview of the Microbial Player Involved in the Bioprocesses Occurring in the Bioelectrochemical System
The bioelectrochemical reactor investigated in this study allowed a complete TCE removal via RD. Kinetic data demonstrated that the addition of Cr(VI) in the system did not negatively affect the performance of the bioelectrochemical reactor. Dehalococcoides mccartyi was found on the biocathode of the system operating both with TCE and TCE/Cr(VI), further demonstrating that the addition of Cr(VI) as a co-contaminant did not negatively affect the RD rate nor the growth of D. mccartyi. As reported in Figure 2, the abundance of D. mccartyi estimated in the run with TCE/Cr(VI) was higher compared to the initial run with only TCE. This finding was likely due to the establishment of the optimum growth conditions and kinetic performance after about 200days of operating conditions of the bioelectrochemical system (Figure 2).
The Coulombic efficiencies estimated in the reactor evidenced electron consuming mechanisms other than RD, including Cr(VI) to Cr(III) reduction and bioelectromethanogenesis for CO2 reduction into CH4. The microbiome characterization, including the metagenomic analysis, revealed that a specialized microbial community led the processes occurring in the bioelectrochemical system.
Dehalococcoides mccartyi was the most abundant microorganism found in the microbiome of the biocathode of the reactor operating with TCE and TCE/Cr(VI). Additionally, Desulfovibrio species were found. They are metabolically versatile microorganisms capable of anaerobic sulfate reduction and RD (Men et al., 2013; Mao et al., 2017; Wen et al., 2017; Lim et al., 2018), already detected in TCE-consortia enriched on a variety of electron donors (Kotik et al., 2013). Some Desulfovibrio species (i.e., D. vulgaris and D. desulfuricans) were previously described as capable of heavy metal reduction in anaerobic environments (Franco et al., 2018). Nevertheless, Desulfovibrio ASVs found in our study, including ASV2, ASV7, ASV8, ASV15, and ASV16 (Supplementary Table S1), are phylogenetically affiliated to Desulfovibrio sulfodismutans strain ThAcO1 (GenBank: NR_026480.1; Blast alignment: ≥95% similarity), a heterotrophic microorganism capable of growth via sulfur disproportionation. It can also grow via sulfate reduction coupled to the oxidation of small organic compounds, although slower than disproportionation (Ward et al., 2020). Previous studies highlighted the involvement of Desulfovibrio spp. in H2 production by direct electron uptake from the biocathode (Carepo et al., 2002; Perona-Vico et al., 2020), suggesting that a similar mechanism may occur in the bioelectrochemical system here described. The latter would be in line with the CE-excess percentage estimated in the system (Table 1).
In addition, on the TCE/Cr(VI) biocathode, some sequences affiliated with known chromium-resistant bacteria were found, corroborating the hypothesis that Cr(VI) reduction in the reactor was probably the result of the combination of both the applied reducing potential and microbial activity. In particular, Ochrobactrum, Brevundimonas, and Dechloromonas species were found. Among them, ASV26 (Supplementary Table S1) was affiliated with Ochrobactrum anthropic, a chromium-resistant microorganism isolated from Cr(VI) contaminated environments with Cr(VI) reduction potential being a carrier of chromate resistance genes. Brevundimonas ASV24 and ASV25 showed ≥99% similarity to Brevundimonas diminuta, a microorganism isolated from a magnetite mine drainage sample with solid tolerance to Cr(VI) (Lu et al., 2011). Additionally, the Dechloromonas genus is known for its ability to remove multiple inorganic contaminants, including perchlorate, nitrate, and selenium (Upadhyaya et al., 2019), and was already found as a dominant microorganism in anaerobic Cr(VI)-removing reactors (Chung et al., 2006). Although the analysis showed microorganisms with tolerance and resistance capabilities to Cr(VI), known bacteria directly involved in Cr(VI) bioreduction processes were not detected.
Further, M. arboriphilus strain DH-1 and M. formicicum strain MF colonized the biocathode. Methanobrevibacter arboriphilus strain DH1 (order Methanobacteriales) is an autotrophic methanogen isolated from the wetwood of methane-emitting trees (Enzmann et al., 2018), known to grow hydrogenotrophically, utilizing CO2 and H2 for CH4 production (Zeikus and Henning, 1975). Similarly, M. formicicum is a hydrogenotrophic methanogen (Bryant and Boone, 1987), and it has been previously reported with M. arboriphilus in a bioelectrochemical methanogenic reactor (Sasaki et al., 2013). Interestingly, previous studies evidenced that together with the reduction of CO2 to CH4 by H2 oxidation, Cr(VI) reduction might also be led by methanogens in the presence of specialized electron transfer complex, especially when H2 (or direct electrons) are not limiting in the environment (Viti et al., 2014).
Overall, gene amplicon sequencing provided an overview of the microbial player involved in the bioprocesses (i.e., RD, hydrogenotrophic methanogenesis, and Cr(VI) reduction) occurring in the bioelectrochemical system here reported. D. mccartyi led TCE-dechlorination. In contrast, Desulfovibrio sp. may be involved in bioelectrochemical H2 production. Further, M. arboriphilus and M. formicicum are primarily engaged in hydrogenotrophic CH4 production and might play a role in Cr(VI) reduction.
Metabolic Features of D. mccartyi and Methanogens in the TCE/Cr(VI) Biocathode
Metagenomic analysis conducted on the biofilm growing on the TCE/Cr(VI) biocathode reported organochloride respiration and hydrogenotrophic methanogenesis as the driving processes occurring in the bioelectrochemical system. A selected microbial consortium established on the biocathode was highlighted and included the most relevant dechlorinating microorganism D. mccartyi and the hydrogenotrophic methanogens M. arboriphilus and M. formicicum. In line with the 16S rRNA gene amplicon sequencing data, metagenomic investigations did not provide evidence for the presence of the known chromium reducing bacteria directly involved in the Cr(VI) to Cr(III) reduction, strengthening the hypothesis that hydrogenotrophic methanogens may be responsible for Cr(VI) reduction at the biocathode. Several studies conducted on the metabolic pathways of interest (i.e., cobalamin synthesis, methionine synthesis, oxygen scavenging, hydrogen production, and electron donor metabolism) of stable dechlorinating mixed cultures (i.e., dechlorinating cultures: KB1, ANAS, DonnaII) demonstrated that the presence of non-dechlorinating microorganisms, including methanogens, is essential to sustain anaerobic RD processes and D. mccartyi growth (Men et al., 2013). In particular, methanogens are crucial for D. mccartyi functionality to provide essential metabolites, including corrinoids, and often act as oxygen scavengers. Indeed, despite that methanogens have been often considered hydrogen competitors, several pieces of evidence suggested that this assumption may not be applicable in particular when electrons are exceeded (Hug et al., 2012). This is the case of the bioelectrochemical reactor here reported where unrecovered reducing power accounted for 53.9% in the TCE/Cr(VI) system (Table 1). It is conceivable that interactions instead of competitions were established between D. mccartyi and hydrogenotrophic methanogens based on kinetic and metagenomic data. We screened the annotated genomes extracted from TCE/Cr(VI) biocathode, mainly focusing on the primary metabolic features, including RDases as fundamental for the RD process, corrinoid compounds as an essential cofactor for the corresponding RDases, and on the hydrogenases as crucial enzymes for H2 metabolism in D. mccartyi. The analysis included also the annotated genomes of M. formicicum and M. arboriphilus as H2 consumers for CH4 production, cofactor suppliers to D. mccartyi for cobalamin biosynthesis, and Cr(VI) reducers.
RDases of D. mccartyi From TCE/Cr(VI) Biocathode
Among the 1,471 CDSs with proteins of the TCE/Cr(VI) biocathode D. mccartyi extracted genome, a total of 29 dehalogenase genes have been annotated (Table 2), including 13 reductive dehalogenase genes (rdhA) predicted to encode the catalytically active enzyme (RdhA), one rdhB gene encoding the membrane anchor protein, additional 14 genes coding for dehalogenase hypothetical proteins, and one gene encoding a haloacid dehalogenase (GenPept: MBF4482892).
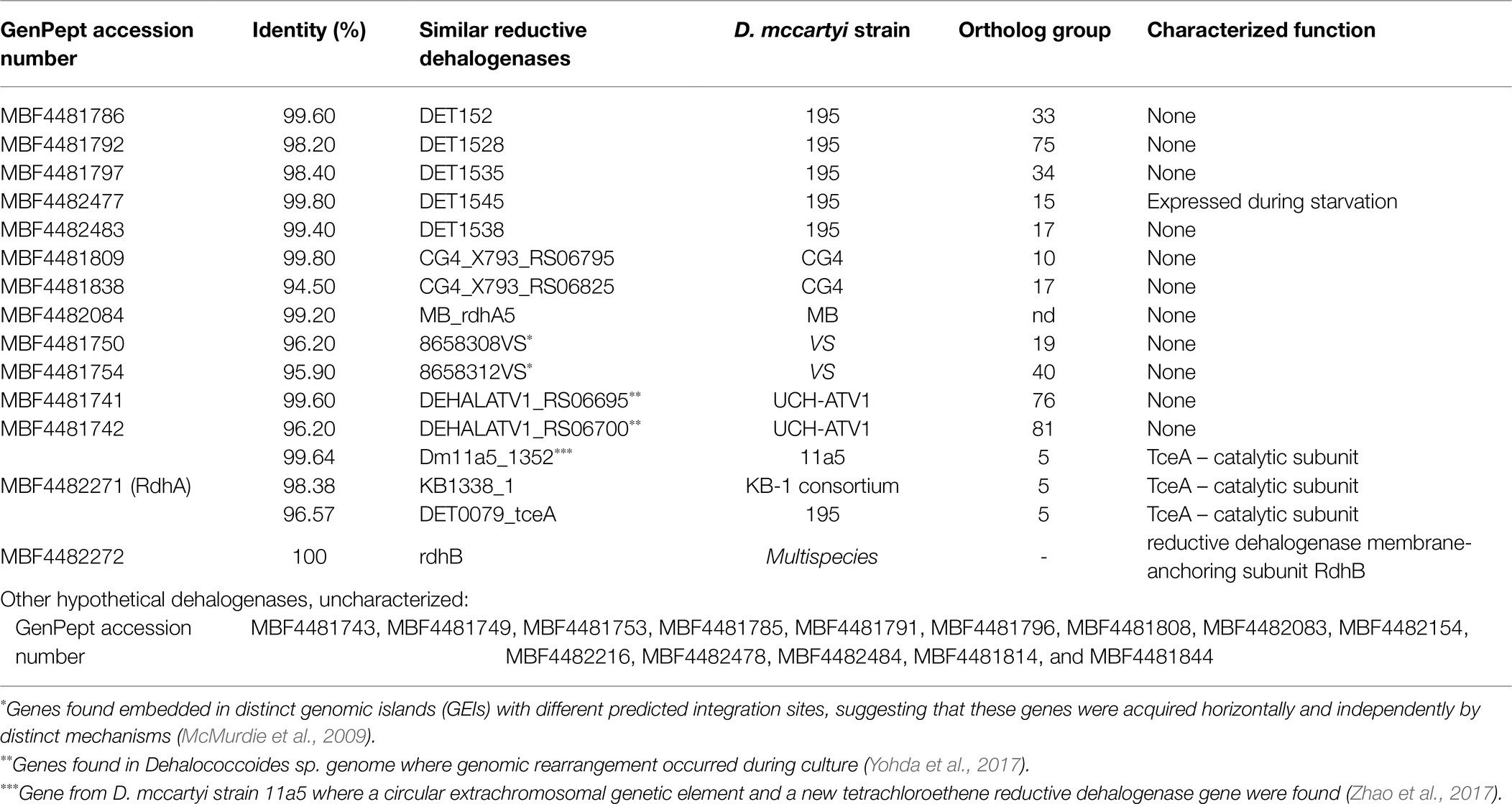
Table 2. Reductive dehalogenase genes annotated in the genome of D. mccartyi extracted from the TCE/Cr(VI) biocathode.
The annotated dehalogenases have been run against the Reductive Dehalogenase Database2 recently released and based on the orthologous sequence similarity (i.e., Ortholog Groups, OGs; Hug et al., 2013; Molenda et al., 2020). Remarkably, none of the putative reductive dehalogenases encoding proteins shared 100% identity with those identified in the database (Table 2). The rdhB gene encoding the membrane anchor protein shared 100% similarity to the TCE reductive dehalogenase membrane-anchoring subunit RdhB (NCBI Reference Sequence: WP_010935885.1), already found in several D. mccartyi species (Table 2). Diversely, most of the rdhA genes found encoding the catalytically active enzyme showed uncharacterized functions, except for the MBF4482477 RdhA with 99.8% of similarity to DET1545 – highly expressed during starvation in D. mccartyi 195 – and the MBF4482271 RdhA with similarity ≥98% to the genes KB1338_1 (from KB-1 consortium) and Dm11a5_1352 (D. mccartyi strain 11a5) that corresponded to the TceA catalytic subunit (Table 2). Interestingly, this protein showed only 96.57% of similarity to the gene DET0079_tceA of D. mccartyi 195. Among the RdhA found, five shared high similarities (from 98.2 to 99.6%) to known enzymes already detected in D. mccartyi 195, including DET152, DET1528, DET1535, DET1545, and DET1538 (Table 2). We also found RdhAses (GenPept: MBF4481741, MBF4481742, MBF4481741, MBF4481742, MBF4482271) similar to those already found in D. mccartyi strains VS, UCH-ATV1 and 11a5 (8658308VS, 658312VS, DEHALATV1_RS06695, DEHALATV1_RS06700, Dm11a5_1352). The rdhA genes of these strains were found embedded in distinct genomic islands with different predicted integration sites (i.e., 8658308VS, 8658312VS; McMurdie et al., 2009), or reported in genomes where genomic rearrangement occurred during culturing (i.e., DEHALATV1_RS06695, DEHALATV1_RS06700; Yohda et al., 2017) or found in D. mccartyi strain (11a5) with circular extrachromosomal genetic elements allowing gene mobilization/horizontal transfer (Zhao et al., 2017). Overall, in these strains, gene mobilization occurs. The reductive dehalogenase genes mobilization may occur by distinct mechanisms, and recent studies showed that CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats – CRISPR associated) system might play a role in D. mccartyi genes transfer (Molenda et al., 2019). Interestingly, D. mccartyi from TCE/Cr(VI) biocathode is a CRISPR-containing genome (Supplementary Table S3) harboring one CRISPR array with genes coding for the CRISPR-Cas system (Table 3). CRISPR is an adaptive immune system constituted by one or more CRISPR arrays (i.e., AT-rich leader sequence followed by short repeats of 21–48bp that are separated by unique spacers of 26–76bp, homologous to sequences in mobile genetic elements) and several CRISPR-associated (cas) genes: When “invading” genetic elements are identified in the genome, CRISPR-Cas system interferes by adding these elements to the CRISPR array.
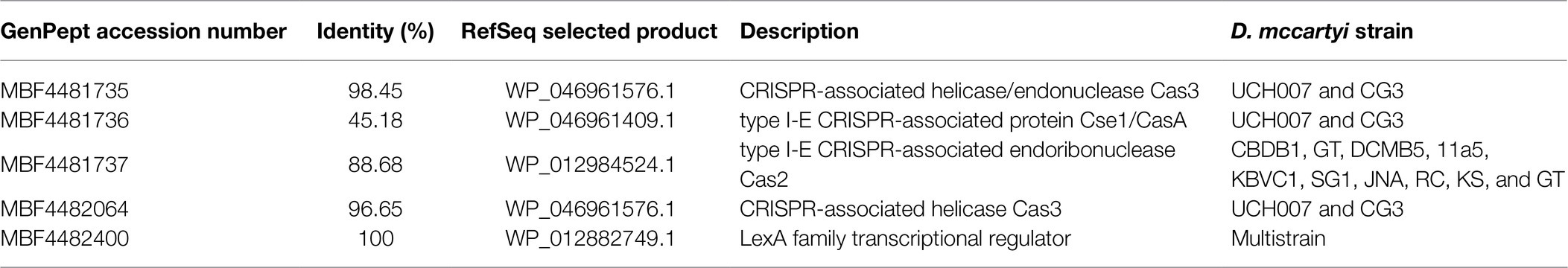
Till now, only five out of 24 total D. mccartyi genomes sequenced so far have been reported to contain CRISPR-Cas genes (strains GT, CBDB1, DCMB5, KBVC1, and KBDCA3), mostly coding for the Class I type I-E system (Huo et al., 2014; Makarova et al., 2015), involved in facilitating or blocking the lateral transfer of rdhAB genes among D. mccartyi strains. To date, only Molenda et al. (2019) investigated D. mccartyi CRISPR-Cas systems, demonstrating that phages and integrative or mobile elements (IMEs) are the most common targets through a site-specific integration in the DNA and that transcriptional regulator (i.e., XRE, LexA, or Cro/IC family) and translocases (i.e., Ftsk/SpoIIIE domain) are involved in this DNA modification system. The analysis of the CRISPR-Cas systems across a growing set of Dehalococcoides genomes enabled the discovery of different types of actively replicating extrachromosomal elements targeted by CRISPR-Cas and associated with the mobilization of rdh genes that are often located in genomic islands (GIs) and appear to have been acquired horizontally (i.e., vcrAB operon, bvcA gene; Molenda et al., 2019). These findings explain the common occurrence of a higher rdhA/16S rRNA gene copies ratio, mostly found in DNA samples from environmental samples (Molenda et al., 2019). In the annotated D. mccartyi genome from the TCE/Cr(VI) biocathode, genes linked to CRISPR-associated helicase/endonuclease (GenPept: MBF4481735), type I-E CRISPR-associated protein Cse1/CasA (GenPept: MBF4481736), type I-E CRISPR-associated endoribonuclease Cas2 (GenPept: MBF4481737) and to CRISPR-associated helicase Cas3 (GenPept: MBF4482064) were found (Table 3). They shared only low similarity (from 45.18 to 98.45%) to CRISPR-associated proteins previously characterized in other D. mccartyi strains. The only regulator gene of the CRISPR-Cas system found in D. mccartyi from the TCE/Cr(VI) biocathode is the LexA family transcriptional regulator (GenPept: MBF4482400), already present in the majority of D. mccartyi strains. These findings, together with the occurrence of rdhA genes with high similarity to those already found in strains where rdhA mobilization and/or horizontal transfers were reported, suggest that also the genome of D. mccartyi extracted from the TCE/Cr(VI) biocathode may harbor metabolic features linked to gene mobilization. However, additional genome walking, including the searching for extrachromosomal DNA, and comparative genomic studies are necessary to gain more insights into this important metabolic feature of the D. mccartyi strain extracted from the biocathode here described.
Corrinoid Synthesis in D. mccartyi and Cofactors Supply From Methanobacterium formicicum and Methanobrevibacter arboriphilus
Cobalamins, including vitamin B12, are corrinoid-based essential cofactors for the activity of RDases (Yan et al., 2016). Indeed, RDases are iron–sulfur clusters and cobalamin-containing membrane-bound components of the electron transfer chain that catalyze the H2-dependent dechlorination of the substrates (Seshadri et al., 2005). Nevertheless, it is well known that D. mccartyi is incapable of de novo cobalamin biosynthesis, and all strains harbor only genes involved in the downstream corrinoid biosynthesis pathway, from cobyrinate a, c diamide to vitamin B12 coenzyme (KEGG pathway: M00122). Thus, D. mccartyi is a corrinoid auxotroph microorganism that needs cobamide-producing microbes to supply the required corrinoid cofactors: This implies that the co-presence of non-dechlorinating members (mostly fermentative and/or acetogenic bacteria and/or methanogens) is essential for D. mccartyi growth (Yan et al., 2016; Yang et al., 2017). This feature makes it capable of scavenging metabolites (i.e., cofactors) from other microorganisms present in the same environment, such as methanogenic Archaea, and transport them into the cell. Metagenomics and kinetic performances suggest that in the TCE/Cr(VI) biocathode, RD activity driven by D. mccartyi was also sustained by corrinoid-producing hydrogenotrophic methanogens M. arboriphilus and M. formicicum. They can perform the upstream corrin ring biosynthesis from uroporphyrinogen III to sirohydrochlorin and finally to cobyrinate a,c-diamide (KEGG pathway: M00924), a precursor of the vitamin B12 coenzyme. The simplified pathway for the upstream (KEGG: M00924) and downstream (KEGG: M00122) corrinoid biosynthesis pathways of vitamin B12 coenzyme is reported in Figure 6.
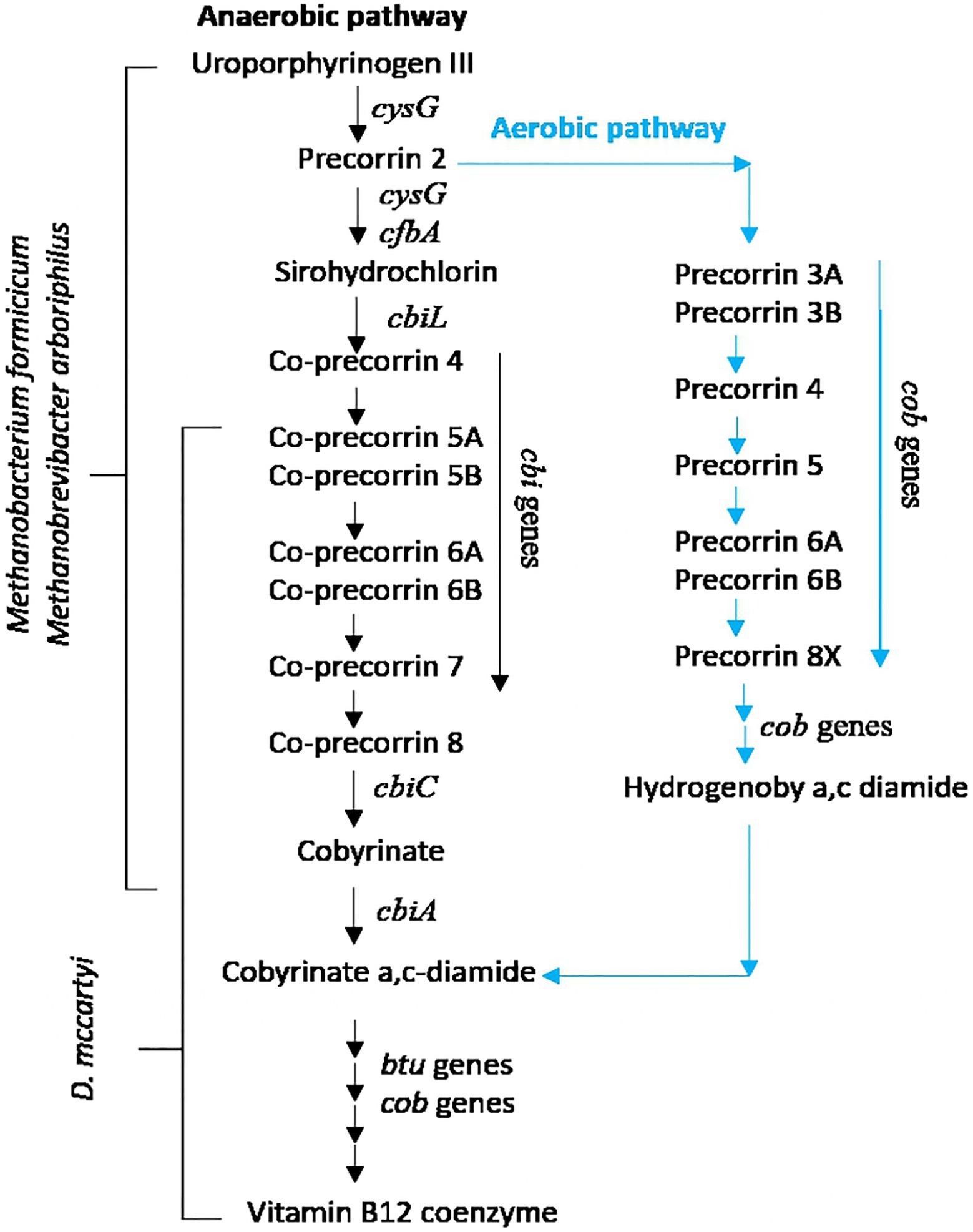
Figure 6. Upstream (KEGG: M00924) and downstream (KEGG: M00122) corrinoid biosynthesis pathways of vitamin B12 coenzyme.
Methanobrevibacter arboriphilus and M. formicicum genomes extracted from the biocathode harbor all the anaerobic cobalamin’s genes biosynthesis pathway from uroporphyrinogen III to sirohydrochlorin and finally to cobyrinate a,c-diamide (KEGG pathway: M00924), as a precursor of the vitamin B12 coenzyme. The first gene involved in this pathway is cysG, followed by genes cbi (cbiC, cbiD, cbiE, cbiG, cbiL, cbiM, cbiQ, and cbiT), cfbA, and cob (cobI, cobM, cobJ, cobK, cobH, and cobB). Diversely, in the TCE/Cr(VI) biocathode D. mccartyi genome, cob genes (cobC, cobD, cobU, cobS, cobT, and cobN genes, Table 4) were found.
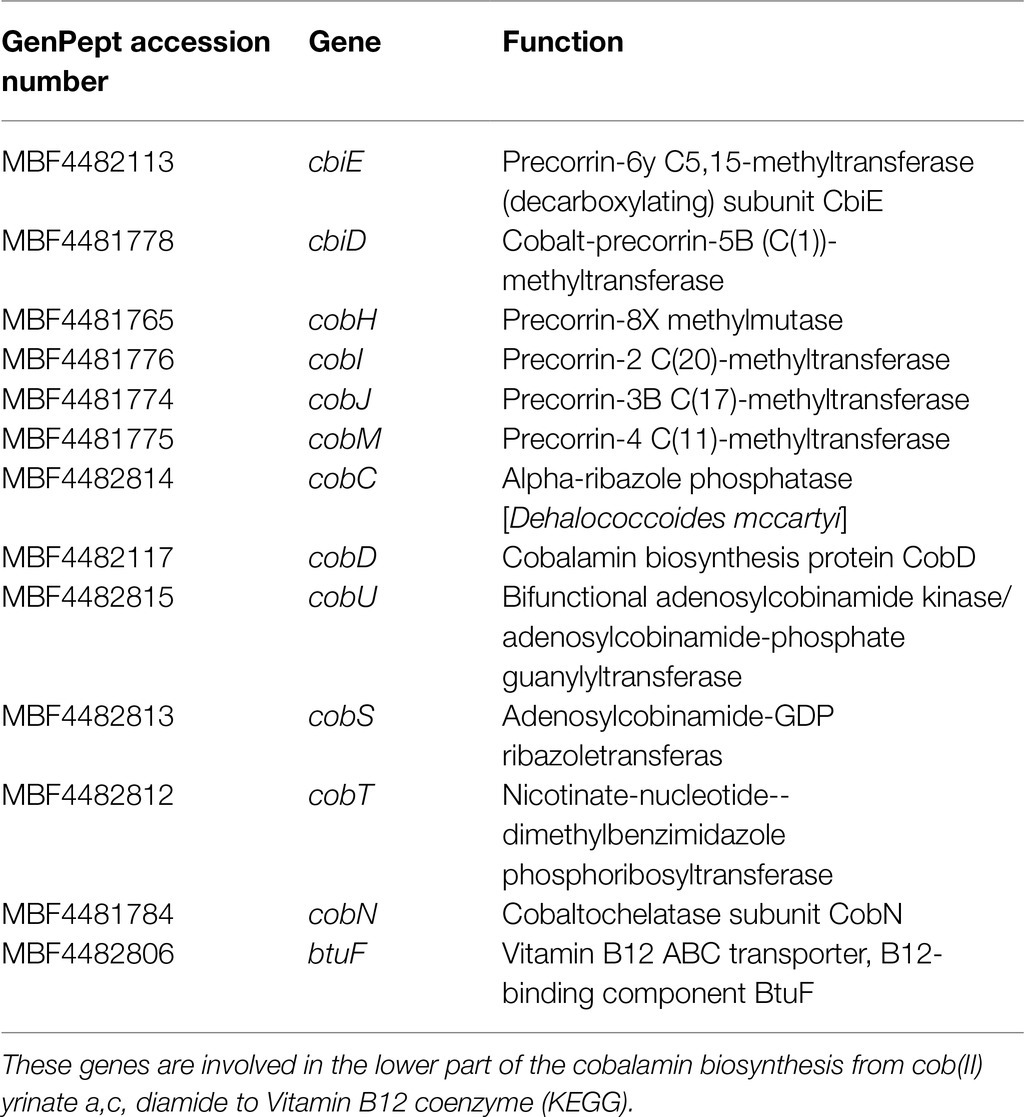
Table 4. Genes involved in the anaerobic cobalamin synthesis pathway in the genome of D. mccartyi from TCE/Cr(VI) biocathode.
They typically encode proteins involved in the anaerobic cobalamin synthesis (KEGG pathway: M00122; Yan et al., 2016). cbiD and cbiE genes were also found: They are involved in the KEGG pathway M00924, from co-precorrin 5B to co-precorrin-6A and from co-precorrin-7 to co-precorrin-8 (Figure 6), and are present in some D. mccartyi strains (IBARAKI, DCMB5, CBDB, MB, FL2, 11a5, and CG1/4/5) and ANAS consortium (Brisson et al., 2012). Remarkably, we also found cobH, cobI, cobJ, and cobM genes (Table 4). These genes are typical of the upstream aerobic corrin ring synthesis pathway (KEGG pathway: M00925) from precorrin-2 to cobyrinate a,c-diamide. Nevertheless, a recent study showed that cbi genes in the anaerobic pathway are orthologous to cob genes in the aerobic pathway and are not exclusive to genomes with the aerobic or anaerobic pathways (Shelton et al., 2019). Therefore, the authors suggested that aerobic or anaerobic corrin ring biosynthesis pathways cannot be distinguished based on their annotated gene content, presumably because portions of the two pathways share orthologous genes. Additionally, B12-binding component BtuF of the vitamin B12 transporter has been found in the D. mccartyi genome from the TCE/Cr(VI) biocathode. Our findings suggest that in the TCE/Cr(VI) biocathode, M. formicicum and M. arboriphilus play a role in D. mccartyi growth by sustaining the complete cobalamin synthesis pathway (Figure 6).
H2 Metabolism
Dehalococcoides mccartyi can exclusively use H2 as an electron donor for the anaerobic dechlorination making H2 metabolism fundamental for its physiology. Dehalococcoides mccartyi commonly harbors genes encoding five different hydrogenase complexes such as cytoplasmic (Vhu) and four membrane-bound (Hup, Hyc, Ech, and Hym) hydrogenases (Kube et al., 2005; Seshadri et al., 2005; Morris et al., 2007; Schipp et al., 2013). Most of them are [NiFe]-hydrogenases, while the H2-uptake (Hup) hydrogenase is a [Fe]-hydrogenase, the main enzyme involved in H2-driven organohalide respiration (Seshadri et al., 2005). The Hup enzyme involves a direct transfer of the electrons derived from H2 oxidation via protein–protein interactions, including ferredoxin-like proteins, which resemble electron-transferring subunits of oxidoreductases (Kublik et al., 2016; Hartwig et al., 2017; Seidel et al., 2018). In line with the general metabolic features of D. mccartyi, in the genome extracted from the TCE/Cr(VI) biocathode, hydrogenase complexes Hyp, Hyc, Hym, Vhu, and Ech were found (Table 5).
In detail, genes encoding protein subunits linked to membrane-bound [Ni-Fe]-hydrogenase included hyc genes (hycC/hybG) encoding the formate hydrogenlyase subunits of the membrane-bound hydrogenase, and hyp genes (hypD, hypE, hypB, and hypD) encoding the carbamoyl dehydratase (HypE), carbamoyltransferase (HypF) subunits and the hydrogenase maturation factors (HypB and HypD). Additionally, hyb genes (hybB, hybD, and hybF) encoding the putative [Ni-Fe]-hydrogenase 2 b-type cytochrome subunit (HybB), the hydrogenase 2 maturation protease (HybD), and the hydrogenase maturation factor (HybF) were detected. hydB gene encoding the periplasmic [Ni-Fe]-hydrogenase large subunit and hyf genes (hyfB and hyfG) encoding the hydrogenase-4 components were also present. Vhu and Ech enzymes were found, the latter probably involved in generating the low-potential electrons for biosynthesis rather than being involved with organohalide respiration (Morris et al., 2007). Moreover, the gene cdhC encoding the CO dehydrogenase/CO-methylating acetyl-CoA synthase complex (GenPept: MBF4482820) and the genes frhB/fhdB encoding the coenzyme F420 hydrogenase/dehydrogenase complex (GenPept: MBF4482137) were also present in the TCE/Cr(VI) biocathode D. mccartyi genome (Table 5). Additionally, in line with the bioelectromethanogenesis occurring in the system, all the genes involved in CH4 formation from H2 and CO2 (KEGG pathway: M00567) were found in the annotated genomes of M. arboriphilus and M. formicicum (Table 6).
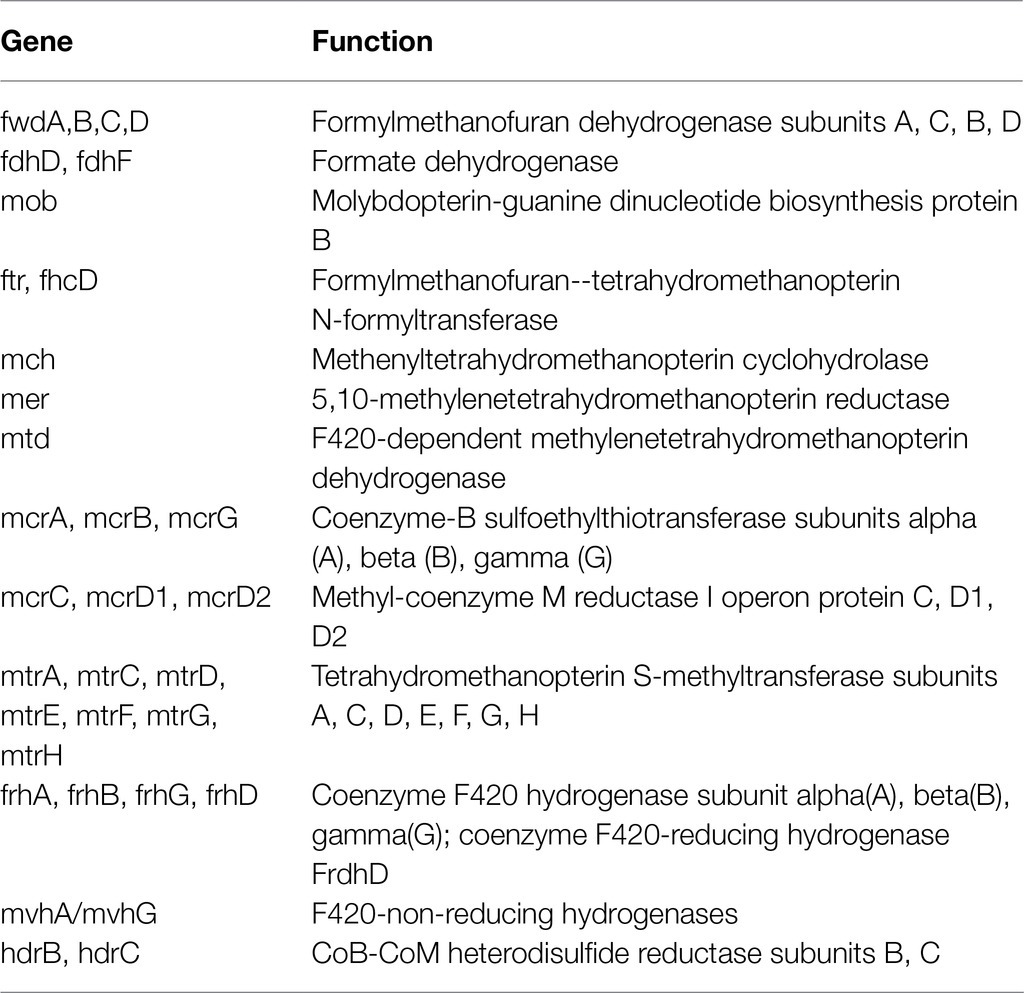
Table 6. Genes involved in the CH4 formation pathway from H2 and CO2 (KEGG: M00567) found in the annotated genome of Methanobacterium formicicum (GenBank: JADIIL000000000) and Methanobrevibacter arboriphilus (GenBank: JADIIN000000000) from the TCE/Cr(VI) biocathode and the corresponding functions.
Cr(VI)-to-Cr(III) Bioreduction
Cr(VI) to Cr(III) reduction occurring in the bioelectrochemical system resulted from the combination of both the applied reducing potential and microbial activity. Nevertheless, amplicon sequencing showed only the presence of AVS26 (3%) affiliated with the known Cr(VI) resistant Ochrobactrum, and bacteria linked to direct Cr(VI) reduction were not detected. Interestingly, previous studies reported that extracellular Cr(VI) reduction might occur with H2 as an electron donor in the presence of hydrogenases and through the action of c-type cytochromes and membrane reductase, such as the mtrCAB complex, essential components for extracellular electron transfer (Marsili et al., 2008; von Canstein et al., 2008). Previous studies demonstrated that this Cr(VI) reduction mechanism, together with the reduction of CO2 to methane by H2 oxidation, might be led by methanogens in the presence of the electron transfer c-cytochrome/mtrCAB complex, especially when H2 (or direct electrons) are not limiting in the environment (Viti et al., 2014; Singh et al., 2015). In line with these previous reports, we found that M. formicicum genome of the TCE/Cr(VI) biocathode harbors genes encoding c-cytochrome (GenPept: MBF4475800) and mtrCAB complex (GenPept: MBF4473862, MBF4474248, MBF4473860; Table 6). Diversely, M. arboriphilus from the TCE/Cr(VI) biocathode harbors genes encoding mtrCAB complex, but no cytochromes have been found in the annotated genome. Additionally, intracellular Cr(VI) reduction mechanisms have been reported in methanogens in the presence of H2 as electron donor and with available transport systems for inorganic solutes formed by the ABC family of the ATP-dependent transporters. This system allows Cr(VI) transport into the cells, mainly if moderate Cr(VI) concentrations are present in the medium to avoid the cytotoxic effect. Thus, Cr(III) reduction occurs within the cell by proteins similar to NADPH-dependent FMN reductase in Methanobacterium sp. (NCBI accession number: YP_004518865; Smith et al., 1997). Interestingly, in the TCE/Cr(VI) biocathode M. formicicum genome, we found 16 genes encoding ABC-transporter ATP-dependent, 17 genes encoding ABC-transporter permeases, and one metal ABC transporter permease in addition to the gene npdG encoding the NADPH-dependent F420 reductase (GenPept: MBF4475652). Diversely, in the M. arboriphilus genome of the TCE/Cr(VI) biocathode, 11 genes encoding ABC-transporter ATP-dependent and 10 genes encode the ABC-transporter permeases were found (Table 6). At the same time, no NADPH-dependent F420 reductase was present. These findings suggested that M. formicicum on the TCE/Cr(VI) biocathode may be responsible for the extracellular and/or intracellular hydrogenotrophic Cr(VI) reduction, together with hydrogenotrophic CH4 production.
Conclusion
The present study reported the microbial and (meta)genomic characterization of a bioelectrochemical system treating TCE- and TCE/Cr(VI)-contaminated water, allowing to elucidate the microbial interactions occurring at the biofilm growing on the cathode (Figure 7). The main processes occurring in the system were RD, hydrogenotrophic methanogenesis, and Cr(VI) reduction. The genomic and metagenomic analysis of the microbial community growing at the biocathode allowed the identification of the key-microbial players: D. mccartyi as the unique OHRB, Desulfovibrio as H2 producer, M. formicicum, and M. arboriphilus as H2 consumers for CH4 production, cofactor suppliers for cobalamin biosynthesis and Cr(VI) reducers. D. mccartyi genome extracted from the TCE/Cr(VI) biocathode showed high similarity with D. mccartyi 195 even if it harbors several uncharacterized RdhAse, including some similar to those found in D. mccartyi strains with IMEs (VS, UCH-ATV1, and 11a5), and genes involved in the CRISPR-Cas system. These findings suggest that the genome of D. mccartyi extracted from the TCE/Cr(VI) biocathode may harbor metabolic features linked to rdhA gene mobilization. Further genomic and/or transcriptomic evaluation is necessary to gain more insights into the gene-transfer capabilities. Concerning the H2 uptake, D. mccartyi genome analysis confirmed this as the only electron supply mechanism. As the genome lacks systems for the direct electron transfer into the cell (i.e., cytochromes), most likely electroactivity is not a property of D. mccartyi. Pure culture studies are necessary to fulfil this hypothesis.
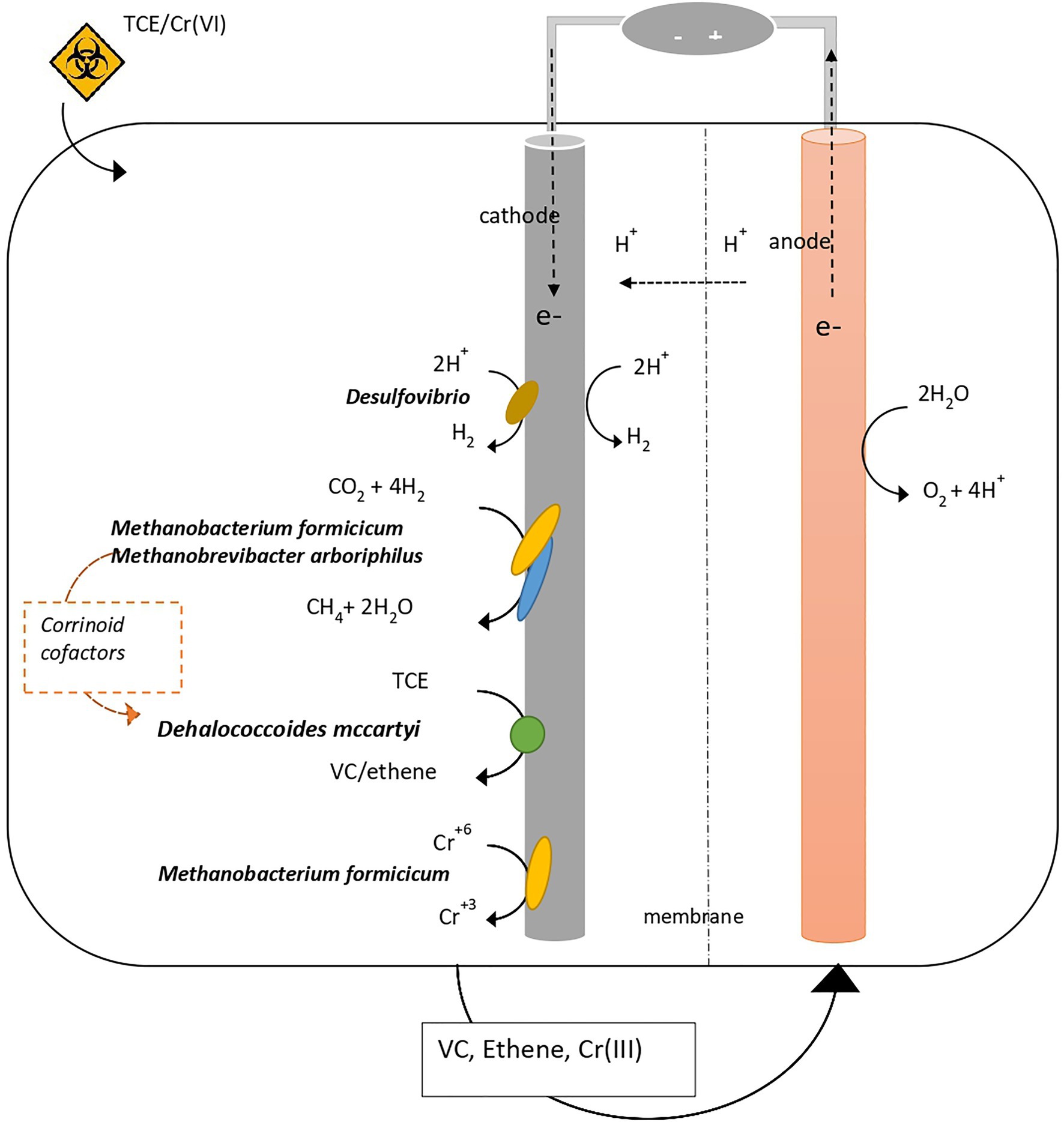
Figure 7. Microbial interactions revealed at the biocathode of the bioelectrochemical system where RD, hydrogenotrophic methanogenesis, and Cr(VI) reduction occur.
The role of hydrogenotrophic methanogens M. formicicum and M. arboriphilus in supporting the growth of D. mccartyi has been here highlighted. Indeed, the presence of a complete cobalamin synthesis pathway represents an essential supply of corrinoid-based crucial cofactors for the activity of RDases in D. mccartyi. Further, evidence of Cr(VI) to Cr(III) reduction mediated by M. formicicum in the TCE/Cr(VI) biocathode has been reported. Indeed, optimal conditions (i.e., H2 availability and moderate Cr(VI) concentration) and the presence of c-cytochrome/mtrCAB complex in the annotated genome suggested that M. formicicum harbors the metabolic feature for extracellular Cr(VI) reduction. Additionally, the metabolic potential for intracellular Cr(VI) reduction in M. formicicum is also sustained by the presence in the genome of genes encoding transport systems for inorganic solutes, and NADPH-dependent reductases, whose involvement in Cr(VI) reduction has been already reported in Cr(VI)-reducing bacteria (i.e., Shewanella sp.).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, JADIIK000000000-JADIIN000000000.
Author Contributions
BM: conceptualization, methodology, data curation, and writing – original draft preparation. MZ: conceptualization, investigation, methodology, and writing – original draft preparation. AL: conceptualization, investigation, and methodology. MM and SR: conceptualization, writing – review and editing, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the European Union’s Horizon 2020 Project ELECTRA (www.electra.site) under grant agreement no. 826244.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.747670/full#supplementary-material
Footnotes
References
Ahsanul Islam, M., Edwards, E. A., and Mahadevan, R. (2010). Characterizing the metabolism of dehalococcoides with a constraint-based model. PLoS Comput. Biol. 6:e1000887. doi: 10.1371/journal.pcbi.1000887
Aulenta, F., Canosa, A., Majone, M., Panero, S., Reale, P., and Rossetti, S. (2008). Trichloroethene dechlorination and H2 evolution are alternative biological pathways of electric charge utilization by a dechlorinating culture in a bioelectrochemical system. Environ. Sci. Technol. 42, 6185–6190. doi: 10.1021/es800265b
Aulenta, F., Catervi, A., Majone, M., Panero, S., Reale, P., and Rossetti, S. (2007). Electron transfer from a solid-state electrode assisted by methyl viologen sustains efficient microbial reductive dechlorination of TCE. Environ. Sci. Technol. 41, 2554–2559. doi: 10.1021/es0624321
Aulenta, F., Reale, P., Canosa, A., Rossetti, S., Panero, S., and Majone, M. (2010). Characterization of an electro-active biocathode capable of dechlorinating trichloroethene and cis-dichloroethene to ethene. Biosens. Bioelectron. 25, 1796–1802. doi: 10.1016/j.bios.2009.12.033
Aulenta, F., Tocca, L., Verdini, R., Reale, P., and Majone, M. (2011). Dechlorination of trichloroethene in a continuous-flow bioelectrochemical reactor: effect of cathode potential on rate, selectivity, and electron transfer mechanisms. Environ. Sci. Technol. 45, 8444–8451. doi: 10.1021/es202262y
Belchik, S. M., Kennedy, D. W., Dohnalkova, A. C., Wang, Y., Sevinc, P. C., Wu, H., et al. (2011). Extracellular reduction of hexavalent chromium by cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 77, 4035–4041. doi: 10.1128/AEM.02463-10
Beretta, G., Daghio, M., Espinoza Tofalos, A., Franzetti, A., Mastorgio, A. F., Saponaro, S., et al. (2019). Progress towards bioelectrochemical remediation of hexavalent chromium. WaterSA 11:2336. doi: 10.3390/w11112336
Beretta, G., Mastorgio, A. F., Pedrali, L., Saponaro, S., and Sezenna, E. (2018). Support tool for identifying in situ remediation technology for sites contaminated by hexavalent chromium. WaterSA 10:1344. doi: 10.3390/w10101344
Bharagava, R. N., Purchase, D., Saxena, G., and Mulla, S. I. (2019). “Chapter 26: applications of metagenomics in microbial bioremediation of pollutants: from genomics to environmental cleanup,” in Microbial Diversity in the Genomic Era. eds. S. Das and H. R. Dash (Oxford, UK: Academic Press), 459–477.
Brisson, V. L., West, K. A., Lee, P. K., Tringe, S. G., Brodie, E. L., and Alvarez-Cohen, L. (2012). Metagenomic analysis of a stable trichloroethene-degrading microbial community. ISME J. 6, 1702–1714. doi: 10.1038/ismej.2012.15
Bryant, M. P., and Boone, D. R. (1987). Isolation and characterization of Methanobacterium formicicum. Int. J. Syst. Evol. Microbiol. 37:171. doi: 10.1099/00207713-37-2-171
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carepo, M., Baptista, J. F., Pamplona, A., Fauque, G., Moura, J. J., and Reis, M. A. (2002). Hydrogen metabolism in Desulfovibrio desulfuricans strain New Jersey (NCIMB 8313)--comparative study with D. vulgaris and D. gigas species. Anaerobe 8, 325–332. doi: 10.1016/S1075-9964(03)00007-6
Cecconet, D., Sabba, F., Devecseri, M., Callegari, A., and Capodaglio, A. G. (2020). In situ groundwater remediation with bioelectrochemical systems: a critical review and future perspectives. Environ. Int. 137:105550. doi: 10.1016/j.envint.2020.105550
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chen, F., Li, Z.-L., Yang, J.-Q., Liang, B., Lin, X.-Q., Nan, J., et al. (2018). Effects of different carbon substrates on performance, microbiome community structure and function for bioelectrochemical-stimulated dechlorination of tetrachloroethylene. Chem. Eng. J. 352, 730–736. doi: 10.1016/j.cej.2018.07.082
Chung, J., Nerenberg, R., and Rittmann, B. E. (2006). Bio-reduction of soluble chromate using a hydrogen-based membrane biofilm reactor. Water Res. 40, 1634–1642. doi: 10.1016/j.watres.2006.01.049
Dam, H. T., Vollmers, J., Kaster, A. K., and Häggblom, M. M. (2017). Reconstructed genomes of novel Dehalococcoides mccartyi strains from 1,2,3,4-tetrachlorodibenzo-p-dioxin-dechlorinating enrichment cultures reveal divergent reductive dehalogenase gene profiles. FEMS Microbiol. Ecol. 93:fix151. doi: 10.1093/femsec/fix151
Desai, C., Jain, K., and Madamwar, D. (2008). Hexavalent chromate reductase activity in cytosolic of pseudomonas sp. G1DM21 isolated from Cr (VI) contaminated industrial landfill. Proceedings in Biochemistry 43, 713–721. doi: 10.1016/j.procbio.2008.02.015
Di Battista, A., Verdini, R., Rossetti, S., Pietrangeli, B., Majone, M., and Aulenta, F. (2012). CARD-FISH analysis of a TCE-dechlorinating biocathode operated at different set potentials. New Biotechnol. 30, 33–38. doi: 10.1016/j.nbt.2012.06.002
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Elangovan, R., Philip, L., and Chandraraj, K. (2010). Hexavalent chromium reduction by free and immobilized cell-free extract of Arthrobacter rhombi-RE. Appl. Biochem. Biotechnol. 160, 81–97. doi: 10.1007/s12010-008-8515-6
Enzmann, F., Mayer, F., Rother, M., and Holtmann, D. (2018). Methanogens: biochemical background and biotechnological applications. AMB Express 8:1. doi: 10.1186/s13568-017-0531-x
Fernando, E. Y., Keshavarz, T., and Kyazze, G. (2019). The use of bioelectrochemical systems in environmental remediation of xenobiotics: a review. Chem. Technol. and Biotechnol. 94, 2070–2080. doi: 10.1002/jctb.5848
Franco, L. C., Steinbeisser, S., Zane, G. M., Wall, J. D., and Fields, M. W. (2018). Cr(VI) reduction and physiological toxicity are impacted by resource ratio in Desulfovibrio vulgaris. Appl. Microbiol. Biotechnol. 102, 2839–2850. doi: 10.1007/s00253-017-8724-4
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Hartwig, S., Dragomirova, N., Kublik, A., Türkowsky, D., von Bergen, M., Lechner, U., et al. (2017). A H2-oxidizing, 1,2,3-trichlorobenzene-reducing multienzyme complex isolated from the obligately organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1. Environ. Microbiol. Rep. 9, 618–625. doi: 10.1111/1758-2229.12560
Hermon, L., Hellal, J., Denonfoux, J., Vuilleumier, S., Imfeld, G., Urien, C., et al. (2019). Functional genes and bacterial communities during organohalide respiration of chloroethenes in microcosms of multi-contaminated groundwater. Front. Microbiol. 10:89. doi: 10.3389/fmicb.2019.00089
Horitsu, H., Futo, S., Miyazawa, Y., Ogai, S., and Kawai, K. (1987). Enzymatic reduction of hexavalent chromium by hexavalent chromium tolerant Pseudomonas ambigua G-1. Agric. Biol. Chem. 51, 2417–2420. doi: 10.1080/00021369.1987.10868422
Hug, L. A., Beiko, R. G., Rowe, A. R., Richardson, R. E., and Edwards, E. A. (2012). Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics 13:327. doi: 10.1186/1471-2164-13-327
Hug, L. A., Maphosa, F., Leys, D., Löffler, F. E., Smidt, H., Edwards, E. A., et al. (2013). Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 368:20120322. doi: 10.1098/rstb.2012.0322
Huo, Y., Nam, K. H., Ding, F., Lee, H., Wu, L., Xiao, Y., et al. (2014). Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 21, 771–777. doi: 10.1038/nsmb.2875
Ivase, T. J. P., Nyakuma, B. B., Oladokun, O., Abu, P. T., and Hassan, M. N. (2020). Review of the principal mechanisms, prospects, and challenges of bioelectrochemical systems. Environ. Prog. Sustain. Energy 39:e13298. doi: 10.1002/ep.13298
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jin, W., Du, H., Zheng, S., and Zhang, Y. (2015). Electrochemical processes for the environmental remediation of toxic Cr(VI): a review. Electrochim. Acta 191, 1044–1055. doi: 10.1016/j.electacta.2016.01.130
Kabir, M. M., Fakhruddin, A., Chowdhury, M., Pramanik, M. K., and Fardous, Z. (2018). Isolation and characterization of chromium(VI)-reducing bacteria from tannery effluents and solid wastes. World J. Microbiol. Biotechnol. 34:126. doi: 10.1007/s11274-018-2510-z
Kim, C., Zhou, Q., Deng, B., Thornton, E. C., and Xu, H. (2001). Chromium(VI) reduction by hydrogen sulfide in aqueous media: stoichiometry and kinetics. Environ. Sci. Technol. 35, 2219–2225. doi: 10.1021/es0017007
Kotik, M., Davidová, A., Voříšková, J., and Baldrian, P. (2013). Bacterial communities in tetrachloroethene-polluted groundwaters: a case study. Sci. Total Environ. 454-455, 517–527. doi: 10.1016/j.scitotenv.2013.02.082
Kube, M., Beck, A., Zinder, S. H., Kuhl, H., Reinhardt, R., and Adrian, L. (2005). Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23, 1269–1273. doi: 10.1038/nbt1131
Kublik, A., Deobald, D., Hartwig, S., Schiffmann, C. L., Andrades, A., von Bergen, M., et al. (2016). Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement. Environ. Microbiol. 18, 3044–3056. doi: 10.1111/1462-2920.13200
Kucharzyk, K. H., Meisel, J. E., Kara-Murdoch, F., Murdoch, R. W., Higgins, S. A., Vainberg, S., et al. (2020). Metagenome-guided proteomic quantification of reductive dehalogenases in the dehalococcoides mccartyi-containing consortium SDC-9. J. Proteome Res. 19, 1812–1823. doi: 10.1021/acs.jproteome.0c00072
Lai, A., Astolfi, M. L., Bertelli, V., Agostinelli, V. G., Zeppilli, M., and Majone, M. (2021). Chromate fate and effect in bioelectrochemical systems for remediation of chlorinated solvents. New Biotechnol. 60, 27–35. doi: 10.1016/j.nbt.2020.06.006
Lai, A., Aulenta, F., Mingazzini, M., Palumbo, M. T., Papini, M. P., Verdini, R., et al. (2017). Bioelectrochemical approach for reductive and oxidative dechlorination of chlorinated aliphatic hydrocarbons (CAHs). Chemosphere 169, 351–360. doi: 10.1016/j.chemosphere.2016.11.072
Lapidus, A. L., and Korobeynikov, A. I. (2021). Metagenomic data assembly: the way of decoding unknown microorganisms. Front. Microbiol. 12:613791. doi: 10.3389/fmicb.2021.613791
Lim, S. S., Kim, B. H., Li, D., Feng, Y., Daud, W., Scott, K., et al. (2018). Effects of applied potential and reactants to hydrogen-producing biocathode in a microbial electrolysis cell. Front. Chem. 6:318. doi: 10.3389/fchem.2018.00318
Löffler, F. E., Yan, J., Ritalahti, K. M., Adrian, L., Edwards, E. A., Konstantinidis, K. T., et al. (2013). Dehalococcoides mccartyi gen. Nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales Ord. Nov. and family Dehalococcoidaceae fam. Nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635. doi: 10.1099/ijs.0.034926-0
Lu, Z., An, X., and Zhang, W. (2011). Isolation and phylogenetic analysis of chromium(VI) reducing bacteria of a magnetite mine drainage from Hebei China. Math. Models Methods Appl. Sci. 5:113. doi: 10.5539/mas.v5n2p113
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Malaviya, P., and Singh, A. (2011). Physicochemical technologies for remediation of chromium-containing waters and wastewaters. Crit. Rev. Environ. Sci. Technol. 41, 1111–1172. doi: 10.1080/10643380903392817
Mao, X., Polasko, A., and Alvarez-Cohen, L. (2017). Effects of sulfate reduction on trichloroethene dechlorination by dehalococcoides-containing microbial communities. Appl. Environ. Microbiol. 83, e03384–e03416. doi: 10.1128/AEM.03384-16
Maphosa, F., van Passel, M. W., de Vos, W. M., and Smidt, H. (2012). Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ. Microbiol. Rep. 4, 604–616. doi: 10.1111/j.1758-2229.2012.00376.x
Marsh, T. L., and McInerney, M. J. (2001). Relationship of hydrogen bioavailability to chromate reduction in aquifer sediments. Appl. Environ. Microbiol. 67, 1517–1521. doi: 10.1128/AEM.67.4.1517-1521.2001
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Matturro, B., Frascadore, E., and Rossetti, S. (2017). High-throughput sequencing revealed novel Dehalococcoidia in dechlorinating microbial enrichments from PCB-contaminated marine sediments. FEMS Microbiol. Ecol. 93:fix134. doi: 10.1093/femsec/fix134
Matturro, B., Heavner, G. L., Richardson, R. E., and Rossetti, S. (2013). Quantitative estimation of Dehalococcoides mccartyi at laboratory and field scale: comparative study between CARD-FISH and real time PCR. J. Microbiol. Methods 93, 127–133. doi: 10.1016/j.mimet.2013.02.011
McMurdie, P. J., Behrens, S. F., Müller, J. A., Göke, J., Ritalahti, K. M., Wagner, R., et al. (2009). Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. doi: 10.1371/journal.pgen.1000714
Men, Y., Lee, P. K., Harding, K. C., and Alvarez-Cohen, L. (2013). Characterization of four TCE-dechlorinating microbial enrichments grown with different cobalamin stress and methanogenic conditions. Appl. Microbiol. Biotechnol. 97, 6439–6450. doi: 10.1007/s00253-013-4896-8
Miao, Y., Liao, R., Zhang, X. X., Wang, Y., Wang, Z., Shi, P., et al. (2015). Metagenomic insights into Cr(VI) effect on microbial communities and functional genes of an expanded granular sludge bed reactor treating high-nitrate wastewater. Water Res. 76, 43–52. doi: 10.1016/j.watres.2015.02.042
Modin, O., and Aulenta, F. (2017). Three promising applications of microbial electrochemistry for the water sector. Environ. Sci. Water Res. Technol. 3, 391–402. doi: 10.1039/C6EW00325G
Molenda, O., Puentes Jácome, L. A., Cao, X., Nesbø, C. L., Tang, S., Morson, N., et al. (2020). Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification. Environ. Sci. Process. Impacts 22, 663–678. doi: 10.1039/C9EM00605B
Molenda, O., Tang, S., Lomheim, L., Gautam, V. K., Lemak, S., Yakunin, A. F., et al. (2019). Extrachromosomal circular elements targeted by CRISPR-Cas in Dehalococcoides mccartyi are linked to mobilization of reductive dehalogenase genes. ISME J. 13, 24–38. doi: 10.1038/s41396-018-0254-2
Morris, R. M., Fung, J. M., Rahm, B. G., Zhang, S., Freedman, D. L., Zinder, S. H., et al. (2007). Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73, 320–326. doi: 10.1128/AEM.02129-06
Palma, E., Daghio, M., Franzetti, A., Petrangeli Papini, M., and Aulenta, F. (2018). The bioelectric well: a novel approach for in situ treatment of hydrocarbon-contaminated groundwater. Microb. Biotechnol. 11, 112–118. doi: 10.1111/1751-7915.12760
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pei, Y., Tao, C., Ling, Z., Yu, Z., Ji, J., Khan, A., et al. (2020). Exploring novel Cr(VI) remediation genes for Cr(VI)-contaminated industrial wastewater treatment by comparative metatranscriptomics and metagenomics. Sci. Total Environ. 742:140435. doi: 10.1016/j.scitotenv.2020.140435
Pérez-de-Mora, A., Lacourt, A., McMaster, M. L., Liang, X., Dworatzek, S. M., and Edwards, E. A. (2018). Chlorinated electron acceptor abundance drives selection of Dehalococcoides mccartyi (D. mccartyi) strains in dechlorinating enrichment cultures and groundwater environments. Front. Microbiol. 9:812. doi: 10.3389/fmicb.2018.00812
Perona-Vico, E., Feliu-Paradeda, L., Puig, S., and Bañeras, L. (2020). Bacteria coated cathodes as an in-situ hydrogen evolving platform for microbial electrosynthesis. Sci. Rep. 10:19852. doi: 10.1038/s41598-020-76694-y
Rahman, Z., and Thomas, L. (2021). Chemical-assisted microbially mediated chromium (Cr) (VI) reduction under the influence of various electron donors, redox mediators, and other additives: an outlook on enhanced Cr(VI) removal. Front. Microbiol. 11:619766. doi: 10.3389/fmicb.2020.619766
Romo, D. M. R., Gutiérrez, N. H. H., Pazos, J. O. R., Figueroa, L. V. P., and Ordóñez, L. A. O. (2019). Bacterial diversity in the Cr(VI) reducing biocathode of a microbial fuel cell with salt bridge. Rev. Argent. Microbiol. 51, 110–118. doi: 10.1016/j.ram.2018.04.005
Ritalahti, K. M., Amos, B. K., Sung, Y., Wu, Q., Koenigsberg, S. S., and Löffler, F. E. (2006). Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72, 2765–2774. doi: 10.1128/AEM.72.4.2765-2774.2006
Saiyari, D. M., Chuang, H. P., Senoro, D. B., Lin, T. F., Whang, L. M., Chiu, Y. T., et al. (2018). A review in the current developments of genus Dehalococcoides, its consortia and kinetics for bioremediation options of contaminated groundwater. Sustain. Environ. Res. 28, 149–157. doi: 10.1016/j.serj.2018.01.006
Sarangi, A., and Krishnan, C. (2008). Comparison of in vitro Cr(VI) reduction by CFEs of chromate resistant bacteria isolated from chromate contaminated soil. Bioresour. Technol. 99, 4130–4137. doi: 10.1016/j.biortech.2007.08.059
Sasaki, D., Sasaki, K., Watanabe, A., Morita, M., Igarashi, Y., and Ohmura, N. (2013). Efficient production of methane from artificial garbage waste by a cylindrical bioelectrochemical reactor containing carbon fiber textiles. AMB Express 3:17. doi: 10.1186/2191-0855-3-17
Schipp, C. J., Marco-Urrea, E., Kublik, A., Seifert, J., and Adrian, L. (2013). Organic cofactors in the metabolism of Dehalococcoides mccartyi strains. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 368:20120321. doi: 10.1098/rstb.2012.0321
Seidel, K., Kühnert, J., and Adrian, L. (2018). The complexome of Dehalococcoides mccartyi reveals its organohalide respiration-complex is modular. Front. Microbiol. 9:1130. doi: 10.3389/fmicb.2018.01130
Seshadri, R., Adrian, L., Fouts, D. E., Eisen, J. A., Phillippy, A. M., Methe, B. A., et al. (2005). Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108. doi: 10.1126/science.1102226
Shelton, A. N., Seth, E. C., Mok, K. C., Han, A. W., Jackson, S. N., Haft, D. R., et al. (2019). Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 13, 789–804. doi: 10.1038/s41396-018-0304-9
Shukla, A. K., Upadhyay, S. N., and Dubey, S. K. (2014). Current trends in trichloroethylene biodegradation: a review. Crit. Rev. Biotechnol. 34, 101–114. doi: 10.3109/07388551.2012.727080
Singh, R., Dong, H., Liu, D., Zhao, L., Marts, A. R., Farquhar, E., et al. (2015). Reduction of hexavalent chromium by the thermophilic methanogen Methanothermobacter thermautotrophicus. Geochim. Cosmochim. Acta 148, 442–456. doi: 10.1016/j.gca.2014.10.012
Smith, D. R., Doucette-Stamm, L. A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., et al. (1997). Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179, 7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997
Somenahally, A. C., Mosher, J. J., Yuan, T., Podar, M., Phelps, T. J., Brown, S. D., et al. (2013). Hexavalent chromium reduction under fermentative conditions with lactate stimulated native microbial communities. PLoS One 8:e83909. doi: 10.1371/journal.pone.0083909
Sophia, A. C., and Saikant, S. (2016). Reduction of chromium(VI) with energy recovery using microbial fuel cell technology. J. Water Process. Eng. 11, 39–45. doi: 10.1016/j.jwpe.2016.03.006
Strycharz, S. M., Woodard, T. L., Johnson, J. P., Nevin, K. P., Sanford, R. A., Löffler, F. E., et al. (2008). Graphite electrode as a sole electron donor for reductive dechlorination of tetrachlorethene by Geobacter lovleyi. Appl. Environ. Microbiol. 74, 5943–5947. doi: 10.1128/AEM.00961-08
Sun, F. L., Fan, L. L., Wang, Y. S., and Huang, L. Y. (2019). Metagenomic analysis of the inhibitory effect of chromium on microbial communities and removal efficiency in A2O sludge. J. Hazard. Mater. 368, 523–529. doi: 10.1016/j.jhazmat.2019.01.076
Taş, N., van Eekert, M. H., de Vos, W. M., and Smidt, H. (2010). The little bacteria that can - diversity, genomics and ecophysiology of 'Dehalococcoides' spp. in contaminated environments. Microb. Biotechnol. 3, 389–402. doi: 10.1111/j.1751-7915.2009.00147.x
Thatoi, H., Das, S., Mishra, J., Rath, B. P., and Das, N. (2014). Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. J. Environ. Manag. 146, 383–399. doi: 10.1016/j.jenvman.2014.07.014
Turick, C., and Apel, W. (1997). A bioprocessing strategy that allows for the selection of Cr(VI)-reducing bacteria from soils. J. Ind. Microbiol. Biotechnol. 18, 247–250. doi: 10.1038/sj.jim.2900373
Upadhyaya, G., Dunagan, B., Mattingly, C., and Brown, J. (2019). Resilient biological hexavalent chromium removal with a two-stage, fixed-bed biotreatment system. AWWA Water Sci. 1:e1151. doi: 10.1002/aws2.1151
Verdini, R., Aulenta, F., de Tora, F., Lai, A., and Majone, M. (2015). Relative contribution of set cathode potential and external mass transport on TCE dechlorination in a continuous-flow bioelectrochemical reactor. Chemosphere 136, 72–78. doi: 10.1016/j.chemosphere.2015.03.092
Viti, C., Marchi, E., Decorosi, F., and Giovannetti, L. (2014). Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol. Rev. 38, 633–659. doi: 10.1111/1574-6976.12051
von Canstein, H., Ogawa, J., Shimizu, S., and Lloyd, J. R. (2008). Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74, 615–623. doi: 10.1128/AEM.01387-07
Wang, X., Aulenta, F., Puig, S., Esteve-Núñez, A., He, Y., Mu, Y., et al. (2020). Microbial electrochemistry for bioremediation. Environ. Sci. Ecotechnol. 1:100013. doi: 10.1016/j.ese.2020.100013
Wang, S., Chen, C., Zhao, S., and He, J. (2019). Microbial synergistic interactions for reductive dechlorination of polychlorinated biphenyls. Sci. Total Environ. 666, 368–376. doi: 10.1016/j.scitotenv.2019.02.283
Wang, H., Luo, H., Fallgren, P. H., Jin, S., and Ren, Z. J. (2015). Bioelectrochemical system platform for sustainable environmental remediation and energy generation. Biotechnol. Adv. 33, 317–334. doi: 10.1016/j.biotechadv.2015.04.003
Ward, L. M., Bertran, E., and Johnston, D. T. (2020). Draft genome sequence of desulfovibrio sulfodismutans ThAc01, a heterotrophic sulfur-disproportionating member of the desulfobacterota. Microbiol. Resour. Announc. 9, e00202–e00220. doi: 10.1128/MRA.00202-20
Watts, M. P., Coker, V. S., Parry, S. A., Thomas, R. A., Kalin, R., and Lloyd, J. R. (2015). Effective treatment of alkaline Cr(VI) contaminated leachate using a novel Pd-bionanocatalyst: impact of electron donor and aqueous geochemistry. Appl. Catal. B: Environ. 170-171, 162–172. doi: 10.1016/j.apcatb.2015.01.017
Wen, L. L., Chen, J. X., Fang, J. Y., Li, A., and Zhao, H. P. (2017). Effects of 1,1,1-trichloroethane and triclocarban on reductive dechlorination of trichloroethene in a TCE-reducing culture. Front. Microbiol. 8:1439. doi: 10.3389/fmicb.2017.01439
Yan, J., Şimşir, B., Farmer, A. T., Bi, M., Yang, Y., Campagna, S. R., et al. (2016). The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J. 10, 1092–1101. doi: 10.1038/ismej.2015.197
Yang, Y., Higgins, S. A., Yan, J., Şimşir, B., Chourey, K., Iyer, R., et al. (2017). Grape pomace compost harbors organohalide-respiring Dehalogenimonas species with novel reductive dehalogenase genes. ISME J. 11, 2767–2780. doi: 10.1038/ismej.2017.127
Yohda, M., Ikegami, K., Aita, Y., Kitajima, M., Takechi, A., Iwamoto, M., et al. (2017). Isolation and genomic characterization of a Dehalococcoides strain suggests genomic rearrangement during culture. Sci. Rep. 7:2230. doi: 10.1038/s41598-017-02381-0
Zeikus, J. G., and Henning, D. L. (1975). Methanobacterium arbophilicum sp. nov. an obligate anaerobe isolated from wetwood of living trees. Antonie Van Leeuwenhoek 41, 543–552. doi: 10.1007/BF02565096
Zeppilli, M., Dell’Armi, E., Cristiani, L., Petrangeli Papini, M., and Majone, M. (2019). Reductive/oxidative sequential bioelectrochemical process for perchloroethylene removal. Water 11:2579. doi: 10.3390/w11122579
Zeppilli, M., Matturro, B., Dell’Armi, E., Cristiani, L., Petrangeli Papini, M., Rossetti, S., et al. (2020). Reductive/oxidative sequential bioelectrochemical process for perchloroethylene (PCE) removal: effect of the applied reductive potential and microbial community characterization. J. Environ. Chem. Eng. 1:104657. doi: 10.1016/j.jece.2020.104657
Keywords: reductive dechlorination, Cr(VI) reduction, bioelectrochemical remediation, Dehalococcoides mccartyi, Methanobacterium formicicum, Methanobrevibacter arboriphilus
Citation: Matturro B, Zeppilli M, Lai A, Majone M and Rossetti S (2021) Metagenomic Analysis Reveals Microbial Interactions at the Biocathode of a Bioelectrochemical System Capable of Simultaneous Trichloroethylene and Cr(VI) Reduction. Front. Microbiol. 12:747670. doi: 10.3389/fmicb.2021.747670
Edited by:
Shanquan Wang, Sun Yat-sen University, ChinaReviewed by:
Ali Raza, Shanghai Jiao Tong University, ChinaAng Li, Harbin Institute of Technology, China
Copyright © 2021 Matturro, Zeppilli, Lai, Majone and Rossetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruna Matturro, YnJ1bmEubWF0dHVycm9AaXJzYS5jbnIuaXQ=
 Bruna Matturro
Bruna Matturro Marco Zeppilli
Marco Zeppilli Agnese Lai2
Agnese Lai2